Significance
The CLC-2 ion channel facilitates selective passage of Cl– ions across cell membranes. In the central nervous system, CLC-2 is expressed in both neurons and glia and is proposed to regulate electrical excitability and ion homeostasis. CLC-2 has been implicated in various central nervous system disorders, including certain types of epilepsy and leukodystrophy. Establishing a causative role for CLC-2 in neuropathologies, however, has been limited by the absence of selective reagents that enable acute and specific channel modulation. Our studies have resulted in the identification of a highly potent, small-molecule inhibitor that enables specific block of CLC-2 Cl– currents in hippocampal brain slices. This precise molecular tool should enable future efforts to identify and treat CLC-2–related disease.
Keywords: chloride channel, CLC-2, inhibitor
Abstract
CLC-2 is a voltage-gated chloride channel that is widely expressed in mammalian tissues. In the central nervous system, CLC-2 appears in neurons and glia. Studies to define how this channel contributes to normal and pathophysiological function in the central nervous system raise questions that remain unresolved, in part due to the absence of precise pharmacological tools for modulating CLC-2 activity. Herein, we describe the development and optimization of AK-42, a specific small-molecule inhibitor of CLC-2 with nanomolar potency (IC50 = 17 ± 1 nM). AK-42 displays unprecedented selectivity (>1,000-fold) over CLC-1, the closest CLC-2 homolog, and exhibits no off-target engagement against a panel of 61 common channels, receptors, and transporters expressed in brain tissue. Computational docking, validated by mutagenesis and kinetic studies, indicates that AK-42 binds to an extracellular vestibule above the channel pore. In electrophysiological recordings of mouse CA1 hippocampal pyramidal neurons, AK-42 acutely and reversibly inhibits CLC-2 currents; no effect on current is observed on brain slices taken from CLC-2 knockout mice. These results establish AK-42 as a powerful tool for investigating CLC-2 neurophysiology.
The CLC-2 ion channel is one of nine mammalian chloride channel (CLC) homologs that comprise a family of Cl–-selective channels and transporters (1). CLC-2 is expressed in nearly every organ, including brain, heart, intestines, and lungs (2), and is critical for electrogenesis, homeostatic control of cell volume, and maintenance of ion gradients. Malfunction and dysregulation of CLC-2 underlie disparate human pathologies (1, 3–9). In renal tissue, CLC-2 mutations are linked to primary aldosteronism, the most common cause of secondary arterial hypertension in humans (10–13). In the central nervous system (CNS), CLC-2 mutations are found in various rare forms of leukodystrophy, characterized by leukoencephalopathy, male infertility, and secondary paroxysmal kinesigenic dyskinesia (an episodic, involuntary movement disorder) (14–20). A phenotype reminiscent of the human disease is observed in CLC-2 genetic knockout mice, which exhibit severe retinal degeneration/blindness, infertility, and vacuolization of myelin in brain tissue (21–23). Additionally, CLC-2 activity influences neuronal excitability, as indicated by in vitro electrophysiology recordings of neurons from wild-type and Clcn2−/− knockout animals (24–30). The influence of CLC-2 on electrical excitability in inhibitory signaling pathways may play a role in idiopathic generalized epilepsies (31–37); however, a direct causal link between CLC-2 mutations and epilepsy has not been definitively established (38). These findings motivate subsequent research efforts to gain a deeper understanding of CLC-2 function in the CNS.
Unraveling the details of CLC-2 physiology is challenged, in part, by the absence of selective chemical reagents for modulating channel activity. While mouse knockout models have provided important insights into CLC-2 function, compensatory changes in gene expression and developmental abnormalities that accompany from-birth genetic deletion of CLC-2 complicate phenotypic analysis. A potent, selective chemical reagent targeting CLC-2 could inhibit channel function rapidly and reversibly, a decided advantage over genetic (irreversible) methods for investigating channel physiology. Available Cl– channel inhibitors (39), however, suffer from low potency, with mid-micromolar to millimolar concentrations required to inhibit Cl– current, and lack specificity against different Cl– channel subtypes (1, 40–42). As such, the usefulness of these tool compounds for cellular and organismal studies is severely limited. For CLCs, the most potent and selective small-molecule inhibitors (IC50 ∼0.5 µM) are those targeting CLC-Ka, one of two CLC homologs expressed in the kidney (43–46). For CLC-2, a peptide toxin, GaTx2, has been identified as a selective inhibitor of CLC-2 at concentrations of ∼20 pM (47); however, reduction in peak current saturates at ∼50% with GaTx2 application, thus restricting use of this toxin as a pharmacological probe. All other reported CLC-2 inhibitors are not selective and require application of high concentrations (∼1 mM) (1, 2, 48–51) to inhibit channel function.
The dearth of high-affinity, high-precision tool compounds for investigating CLC-2 physiology inspires the studies described herein. Through a systematic structure–activity analysis, we have developed the most potent small-molecule inhibitor known to date for any member of the CLC protein family. This compound, AK-42, has an IC50 of 17 ± 1 nM against human CLC-2 and displays ∼10,000× greater potency toward CLC-2 compared to the most closely related CLC homolog, CLC-1. In addition, AK-42 is specific for CLC-2 over a diverse panel of 61 CNS receptors, channels, and transporters. We have validated the suitability of AK-42 for physiological studies of the CNS by performing brain slice recordings on both wild-type and CLC-2 knockout mice. Computational and mutagenesis studies identify an AK-42 binding site on the extracellular side of the channel pore, laying the groundwork for understanding the mechanistic underpinnings of channel inhibition and for further development of related tools. The availability of AK-42 as a potent, highly selective reagent for pharmacological knockout of CLC-2 should greatly accelerate physiological studies of this channel.
Results
Screening for New CLC-2 Inhibitors.
Using the IonWorks Barracuda (IWB) automated patch-clamp platform, we performed a focused screen of Food and Drug Administration-approved drug molecules against human CLC-2 stably expressed in Chinese hamster ovary (CHO) cells and identified meclofenamate (MCFA) as a “hit” compound (Fig. 1, SI Appendix, Fig. S1 and Table S1, and Dataset S1). MCFA was selected over other hits due to the ease of synthesis and modification of the diarylamine core. Examination of other nonsteroidal antiinflammatory drugs within our initial screen (SI Appendix, Table S1) revealed that the interaction of MCFA with CLC-2 is complex and does not simply derive from nonspecific Coulombic attraction of the anionic carboxylate group to the electropositive ion conduction pore. Most notably, mefenamic acid and diclofenac, which are structurally analogous to MCFA, have negligible inhibitory activity at 30 µM (SI Appendix, Table S1). A small collection of angiotensin (AT1) receptor antagonists called sartans, which are anionic at physiological pH (due to the presence of a tetrazole, carboxylic acid, or both), also failed to inhibit Cl– currents to any appreciable extent at 30 µM (SI Appendix, Table S1). Other known small-molecule modulators of CLC homologs (e.g., BIM1, BIM4, DPC, and NFA) (SI Appendix, Fig. S1) displayed no efficacy against CLC-2 (SI Appendix, Table S1). Additionally, lubiprostone, a reported CLC-2 activator (52, 53), had no effect on CLC-2 currents at concentrations up to 120 μM, in agreement with prior reports (54–56) (SI Appendix, Table S1).
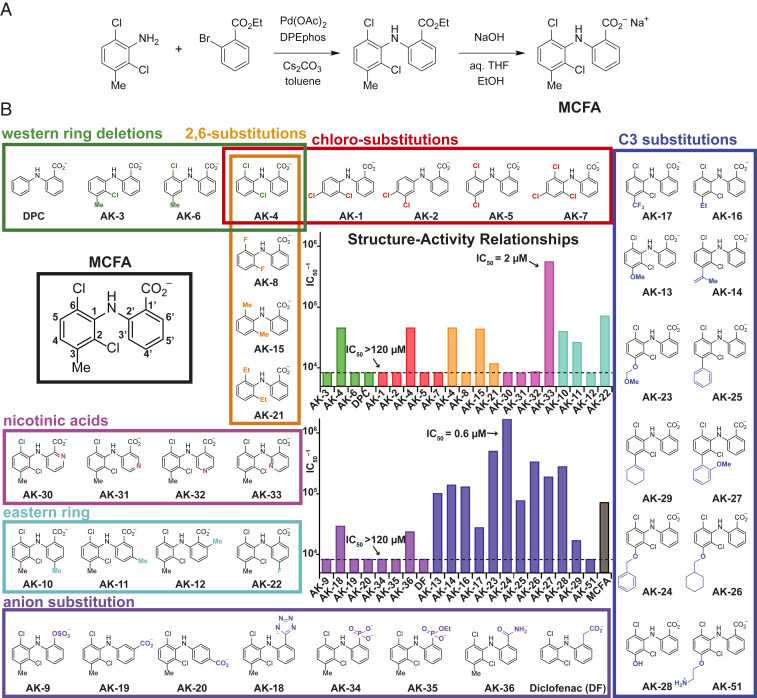
Fig. 1.
SAR studies to develop a selective CLC-2 inhibitor based on the MCFA hit compound. (A) General synthetic route for the preparation of MCFA and analogs. (B) SAR of MCFA derivatives. The structure of MCFA is shown left (black box) with carbon numbers of the eastern and western rings labeled. The bar graphs show the inverse IC50 (IC50–1) for each compound against CLC-2, estimated from inhibition at four concentrations (n = 3 to 4 for each concentration). Compounds are grouped by substitution position, as indicated by bar colors that match the box outlines around each group of structures. Compounds that inhibited weakly at the highest concentration tested (usually 120 µM) are indicated with the dotted line (IC50 > 120 µM). Results are tabulated in Table 1.
Structure–Activity Relationship Studies.
To enable structure–activity relationship (SAR) studies of MCFA, a general synthesis of substituted diarylamines was developed (Fig. 1A). Using this chemistry, a total of 51 MCFA derivatives were prepared. The results from evaluation of the first 37 derivatives tested (Fig. 1B) guided the design of an additional 14 compounds (Fig. 2).
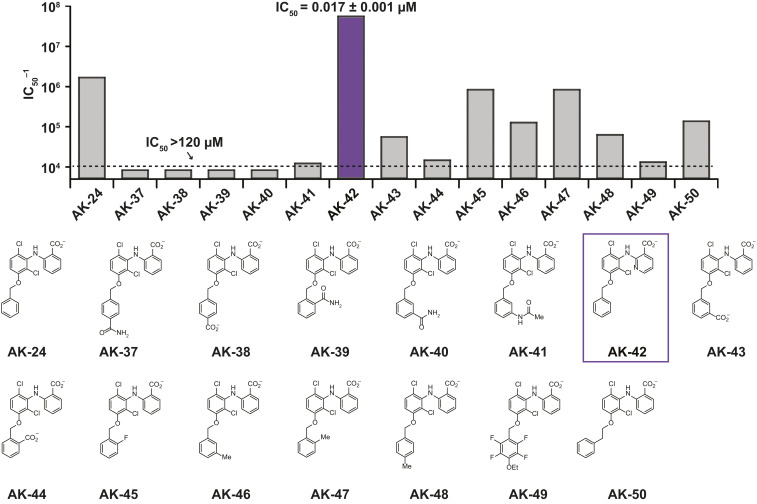
Fig. 2.
SAR studies around the AK-24 scaffold. The bar graph shows the inverse IC50 estimated from inhibition at four concentrations (n = 3 to 4 for each concentration), as in Fig. 1. Substitutions around the C3-OBn ring are destabilizing. Combining the substitutions of AK-24 and AK-33 has a synergistic effect on inhibitor potency (AK-42, purple). Data are given in Table 2.
An initial systematic examination of the western aryl ring of the MCFA scaffold was performed to assess the effects of -Cl and -Me substitution (Fig. 1, Table 1, green, orange, and red boxes). Removal of either the C2- or C6-Cl group (AK-3, AK-6) resulted in an ∼50% decrease in potency relative to MCFA. In contrast, the potency of AK-4, which lacks the C3-Me group, was relatively unchanged. Other di- and trichlorinated analogs—including AK-1, AK-2, AK-5, and AK-7—were ineffective at blocking CLC-2 (IC50 ≥ 120 µM). C2,C6-Substitution with Cl or Me groups (AK-4 and AK-15, respectively) proved imperative; introduction of sterically smaller (e.g., -F, AK-8) or larger (e.g., -Et, AK-21) substituents significantly reduced inhibitor potency. These data suggest that a twisted geometry of the diarylamine scaffold is necessary for inhibition.
Table 1.
SAR studies with MCFA analogs: IC50 values of fenamate derivatives against CLC-2
| Compound | IC50 (µM) | Compound | IC50 (µM) |
| Western Ring Deletions | 2,6-Substitutions | ||
| AK-3 | >120 (43%) | AK-4 | 22 |
| AK-4 | 22 | AK-8 | >120 (41%) |
| AK-6 | >120 (48%) | AK-15 | 23 |
| DPC | >312 (27%) | AK-21 | 86 |
| Chloro Substitutions | Nicotinic Acids | ||
| AK-1 | >120 (13%) | AK-30 | >222 (12%) |
| AK-2 | >120 (12%) | AK-31 | >42 (16%) |
| AK-4 | 22 | AK-32 | 115 |
| AK-5 | >120 (23%) | AK-33 | 2 |
| AK-7 | >120 (22%) | Eastern Ring | |
| C3-Substitutions | AK-10 | 25 | |
| AK-13 | 10 | AK-11 | 38 |
| AK-14 | 7 | AK-12 | >102 (37%) |
| AK-16 | 8 | AK-22 | 14 |
| AK-17 | 36 | Anion Substitutions | |
| AK-23 | 2 | AK-9 | >120 (28%) |
| AK-24 | 0.6 | AK-18 | 33 |
| AK-25 | 13 | AK-19 | >141 (36%) |
| AK-26 | 3 | AK-20 | >144 (8%) |
| AK-27 | 5 | AK-34 | >195 (6%) |
| AK-28 | 4 | AK-35 | >354 (41%) |
| AK-29 | 49 | AK-36 | 42 |
| AK-51 | >120 (7%) | DF | >363 (5%) |
| MCFA | 14 | ||
Approximate IC50 values of fenamate derivatives against CLC-2, determined from four concentrations using the IWB platform. Compounds are listed according to position of modification, as shown in Fig. 1B, and groups are color-coded accordingly. If the IC50 was greater than the highest concentration tested, this concentration is listed along with the corresponding % inhibition (in parenthesis).
The small reduction in potency of AK-4 vs. MCFA (IC50 = 22 µM vs. 14 µM) led us to query the effect of replacing the C3-Me group in MCFA with alternative substituents (Fig. 1, Table 1, blue box). Initially, a series of four compounds was prepared in which a -OMe, isopropenyl, -Et, or -CF3 group was introduced at C3 (AK-13, AK-14, AK-16, and AK-17, respectively). With the exception of AK-17, these ligands were nearly twofold more potent than MCFA. In a second round of SAR, incorporation of sterically larger groups at C3, which included -OBn and -OCH2cHx (AK-24 and AK-26, respectively), resulted in an additional boost in potency. Among these derivatives, AK-24 was identified as the most effective inhibitor with an IC50 of 0.6 µM.
We next evaluated the influence of the MCFA carboxylate group on ligand binding (Fig. 1, Table 1, purple). This study included moving the position of the carboxylate from C1′ to C5′ and C6′ on the eastern ring (AK-20 and AK-19, respectively) and replacing the carboxylate unit with other charged substituents, including -OSO3–, -PO32–, -PO2(OEt)–, -tetrazolate, and -CH2CO2– (AK-9, AK-34, AK-35, AK-18, and diclofenac, respectively). A neutral analog of MCFA, AK-36, in which the carboxylate group was converted to a primary carboxamide, was also prepared. All eight compounds displayed reduced potency relative to MCFA. From these data, the efficacy of carboxamide AK-36 is perhaps most surprising, as this derivative is only approximately threefold less potent than MCFA despite lacking the anionic charge. This result indicates that ligand–protein binding is not purely dictated by charge–charge interactions.
Additional ligand SAR data were obtained by varying substituent groups on the eastern ring of MCFA with the western ring held fixed (Fig. 1, Table 1, magenta and cyan, and Table 2). A Me-scan at positions C4′, C5′, and C6′ (AK-10, AK-11, and AK-12, respectively) showed diminished potency for all three isomers, with the most significant loss resulting from C6′ substitution. A C4′-F derivative (AK-22) also proved less potent than MCFA. Collectively, these findings intimate that the eastern ring of MCFA is sequestered in a restricted binding pocket. Given these findings, we prepared four isomeric nicotinic acid derivatives (AK-30, AK-31, AK-32, and AK-33), reasoning that introduction of a N-atom in this region of the inhibitor would alter steric size and potentially enable a stabilizing hydrogen bonding interaction with the protein. While three of these compounds displayed substantially reduced potency, one derivative, AK-33, was improved sevenfold over MCFA (IC50 = 2 µM). Because the four isomers—AK-30, AK-31, AK-32, and AK-33—have similar cLogP values and polar surface areas, this finding suggests that the N-atom at the 3′ position engages in a specific interaction between AK-33 and CLC-2.
Table 2.
SAR studies with MCFA analogs: IC50 data summary for derivatives of AK-24
| Compound | IC50 (µM) | Compound | IC50 (µM) |
| AK-24 | 0.6 | AK-44 | 68 |
| AK-37 | >120 (15%) | AK-45 | 1 |
| AK-38 | >120 (4%) | AK-46 | 8 |
| AK-39 | >120 (0%) | AK-47 | 1 |
| AK-40 | >120 (5%) | AK-48 | 16 |
| AK-41 | 82 | AK-49 | 76 |
| AK-42 | <0.12 (90%) | AK-50 | 7 |
| AK-43 | 18 |
IC50 data summary for derivatives of AK-24, determined from four concentrations using the IWB platform. If the IC50 was greater than the highest concentration tested, this concentration is listed along with the corresponding % inhibition (in parenthesis).
In light of our results with AK-24 and AK-33, we synthesized AK-42, a derivative that combines the C3-OBn substituent on the western ring with the C3′-nicotinic acid on the eastern half (Fig. 2, Table 2, and Fig. 3A). AK-42 is almost three orders-of-magnitude more potent than MCFA (Fig. 2, Table 2, Fig. 3B). Manual patch-clamp experiments on CHO cells transiently transfected with rat CLC-2 indicate comparable AK-42 potency (IC50 = 14 ± 1 nM for rat CLC-2) (SI Appendix, Fig. S2) to that determined for human CLC-2 using the IWB (17 ± 1 nM). To our knowledge, AK-42 is the most potent small-molecule inhibitor against any CLC homolog.
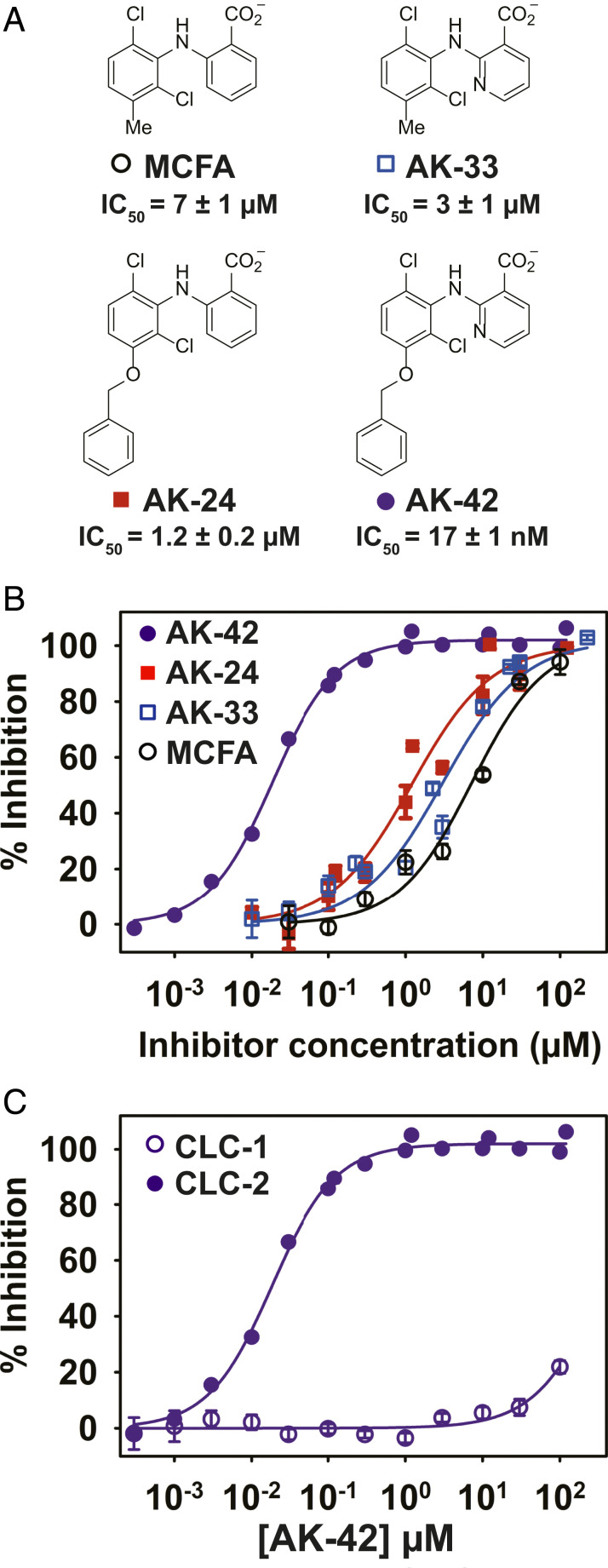
Fig. 3.
Combining the most potent compounds identified in the SAR studies results in a highly potent and selective CLC-2 inhibitor, AK-42. (A) Structures, including the initial hit compound MCFA, are shown. IC50 values were determined from data in B; these IC50 values differ slightly from those obtained in the initial screen (Fig. 1), which used only four concentrations of each compound. (B) Percent inhibition at −120 mV was determined using the IWB, n = 3 to 4 for each concentration ± SEM. Fits to the Hill equation yield IC50 values of 7 ± 1 µM, 1.2 ± 0.2 µM, 3 ± 1 µM, and 0.017 ± 0.001 µM for MCFA, AK-24, AK-33, and AK-42, respectively (SI Appendix, Table S2). (C) Inhibition curves for AK-42 against CLC-1 and CLC-2 determined using the IWB, illustrating that AK-42 is ∼10,000 times more potent against CLC-2 compared to CLC-1, the most closely related CLC homolog (data are tabulated in SI Appendix, Table S3).
To further characterize AK-42 for use in physiological studies, we measured the time course of CLC-2 inhibition and washout in CHO cells. At 30 nM, onset of inhibition by AK-42 occurs in <10 s (SI Appendix, Fig. S3) and is completely reversible within ∼10 min upon washout with extracellular buffer (SI Appendix, Fig. S2). The potency of AK-42, together with rapid inhibition kinetics and reversibility, are particularly desirable properties of this reagent for acute pharmacological knockout and rescue of channel function.
AK-42 Selectivity and Specificity.
To assess the utility of AK-42 as a tool compound for studies of CLC-2 physiology in the CNS, we evaluated the specificity of this ligand for CLC-2 over other protein targets. Electrophysiological recordings of CLC-1, the most closely related homlog to CLC-2 (∼55% sequence identical), revealed greater than four orders-of-magnitude reduced sensitivity to AK-42 (Fig. 3C and SI Appendix, Fig. S4). By comparison, the original hit compound, MCFA, exhibits only modest selectivity (<10-fold) between CLC-1 and CLC-2 (SI Appendix, Table S2). Additionally, MCFA inhibition of the kidney channel, CLC-Ka, requires mid-micromolar concentrations (IC50 ∼40 µM) (57). Collectively, these results support AK-42 as a uniquely selective agent for targeting CLC-2 over other CLCs.
In certain cell types, namely astrocytes and oligodendrocytes, CLC-2 is associated with an auxiliary subunit known as GlialCAM, which increases Cl– current amplitudes and changes channel voltage-dependence (58). Accordingly, we investigated whether AK-42 would block CLC-2 when coexpressed with GlialCAM. Electrophysiology recordings were performed with 30 nM AK-42 against CHO cells coexpressing both CLC-2 and GlialCAM (SI Appendix, Fig. S2 C and D). Comparing these data versus equivalent results obtained with cells expressing only the pore-forming CLC-2 subunit revealed that the magnitude of current inhibition was similar for both sets of experiments. Thus, it follows that AK-42 should block both neuronal and glial CLC-2 currents in the CNS with comparable efficacy.
The value of AK-42 as a tool compound for studying channel function in the CNS was further assessed by measuring ligand potency against a diverse panel of potential off-target CNS receptors, channels, and transporters. To obtain these data, we employed a comprehensive screen developed and conducted by the National Institutes of Health Psychoactive Drug Screening Program (NIH PDSP) (59). Depending on the identity of the receptor, primary binding assays or functional assays were performed with each target at 10 µM AK-42, a concentration well above the IC50 of this compound for CLC-2 and at least four times higher than the inhibitor concentrations used in our primary cell experiments (see Validation of AK-42 in Brain Slice Recordings). In these assays, no notable off-target protein binding effects were noted; for the eight targets exhibiting a response of >50% at 10 µM (table 1 in Dataset S2), secondary binding assays confirmed that the effects were weak or negligible (<50% mean response at 10 µM) (figure 1 in Dataset S2). In addition to the proteins tested in the PDSP screen, AK-42 was evaluated against other anion channels. Application of 10 µM AK-42 to the volume-regulated anion channel (VRAC), cystic fibrosis transmembrane conductance regulator (CFTR), and TMEM16A resulted in < 30% inhibition of protein activity (figures 2–4 in Dataset S2). Thus, at the low-nanomolar concentrations of AK-42 required for physiological experiments to block CLC-2 function, complications resulting from off-target engagement of these channels are not expected.
Computational Modeling of AK-42•CLC-2 Binding.
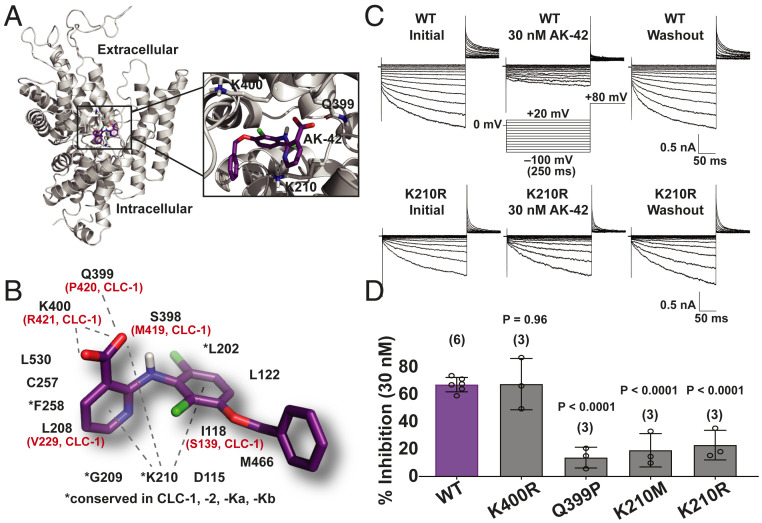
Computational docking was performed to generate binding site predictions for AK-42 and to gain insight into the molecular basis for CLC homolog selectivity. Absent a high-resolution structure of CLC-2, we capitalized on a CLC-2 structural homology model that we developed and refined with 600 µs of molecular dynamics simulations (60). Guided by prior mutagenesis and computational modeling studies of fenamate inhibitors (in the same structural class as AK-42) against CLC-Ka and CLC-Kb, AK-42 was docked to an extracellular site on the channel. This places AK-42 near the outer vestibule of the ion permeation pathway, in a similar location to that targeted by anionic inhibitors of CLC-Ka (45, 46). Notably, docking studies of AK-42 to the analogous extracellular site on CLC-1 [6COY (61), structure unrefined by molecular dynamics] did not produce any docking poses, in line with our functional studies on selectivity (Fig. 3C). While the top docking poses in our CLC-2 model vary in the specific interactions predicted, all place AK-42 in the same general binding site. A representative docking pose to the putative CLC-2 binding site, along with a schematic of the 14 residues in the vicinity of AK-42 (compiled from multiple high-scoring docking models), is shown in Fig. 4 A and B.
Fig. 4.
Analysis of AK-42 selectivity and the inhibitory binding site. (A) One subunit of the CLC-2 homodimer is shown. AK-42 docks near the extracellular entrance to the chloride permeation pathway, which is tilted toward the viewer. The expanded view highlights three of the residues in the predicted binding pocket. (B) Illustration of one of the top poses adopted by AK-42 in molecular docking to CLC-2, together with a schematic of the 14 residues proximal to AK-42 in all docking poses. Residues differing between CLC-1 and CLC-2 are indicated in red. (C) Representative CLC-2 current traces from manual patch-clamp experiments. (Upper) WT CLC-2 currents, illustrating reversible CLC-2 inhibition by 30 nM of AK-42, consistent with an extracellular binding site. (Lower) K210R CLC-2 currents, illustrating a diminution in AK-42 potency upon mutating K210 to R, supporting the docking model in B. (D) Testing other residues predicted to interact with AK-42 in the CLC-2 binding site. Mutations of K210 and Q399 produced statistically significant (by unpaired t test) effects on inhibitor binding at 30 nM, whereas the K400R mutation did not. Number of data points per mutant is shown in parenthesis. Summary data are tabulated in SI Appendix, Table S4. These data support that AK-42 binds in the pocket predicted by docking studies.
To test the binding site predictions of this model, we first examined mutations of the two charged residues located within this proposed pocket, K210 and K400, reasoning that at least one of these could contribute to electrostatic stabilization of the anionic inhibitor molecule. Mutations at K210, a highly conserved residue, resulted in significantly diminished sensitivity to AK-42 (Fig. 4 C and D). The effectiveness of AK-42 is reduced even against the charge-conserved mutant, K210R. Accordingly, K210 may play a critical role in fixing the inhibitor geometry within the binding pocket, as highlighted in Fig. 4B. In contrast, no change in inhibitor sensitivity is measured against the K400R mutant as compared to wild-type. These results reveal that K400 does not contribute to inhibitor selectivity for CLC-2 over CLC-1, the latter of which bears an arginine at this position. Finally, introduction of proline at Q399, the residue present at the equivalent position in CLC-1, reduces inhibition by AK-42 (Fig. 4D). While further studies are needed to rule out the possible allosteric influence of these residues on ligand binding, our mutagenesis data support a working model for AK-42 binding to an extracellular site on CLC-2. In addition, we note that the kinetics of inhibition by AK-42 (SI Appendix, Fig. S3) are rapid and reversible and fit well to a 1:1 AK-42•CLC-2 stoichiometry, consistent with our working model.
Validation of AK-42 in Brain-Slice Recordings.
To validate the effectiveness of AK-42 for neurophysiological studies, we measured CLC-2 currents in CA1 pyramidal cells of acute brain slices using whole-cell patch-clamp electrophysiology. The presence of a voltage-gated chloride conductance in CA1 pyramidal neurons was first reported by Madison et al. (62). Subsequent in situ hybridization studies (63) and immunohistochemistry (64) identified CLC-2 mRNA and protein expression in CA1 pyramidal cells. Further evidence for presumed CLC-2 currents in these cells came from Rinke et al. (28), who demonstrated that hyperpolarization-induced chloride currents in CA1 pyramidal cells were selective for Cl– over I– (a hallmark of CLCs in contrast to other chloride channel families) and were sensitive to low extracellular pH (characteristic of CLC-2); these currents were abolished in CLC-2 knockout mice. Other physiological studies by Földy et al. (27) corroborated these results, reporting a Cl– current in mouse CA1 pyramidal cells that is consistent with the findings of Madison et al. (62) and that is absent in CLC-2 deficient neurons.
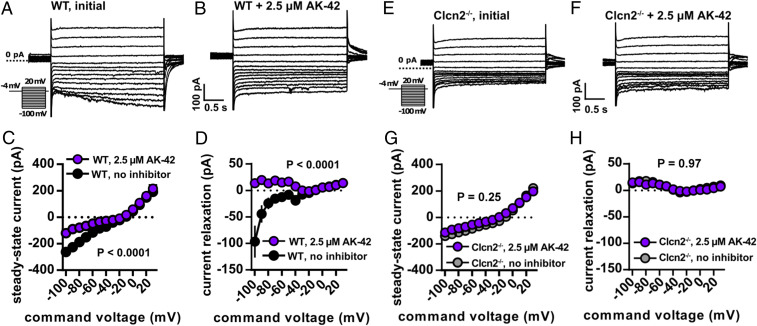
CLC-2 currents were isolated by blocking synaptic transmission, currents mediated by voltage-gated sodium channels, and hyperpolarization-activated cyclic nucleotide-gated (HCN) channels (Materials and Methods). To evoke CLC-2 currents, cells were stepped from −4 mV, a membrane potential at which CLC-2 channels have low open probability, to test potentials ranging from −100 to +30 mV. During the 3-s test pulse, CLC-2 current is observed as a slow increase in inward current at the most negative potentials (Fig. 5A). These data are as expected based on the previous studies described above (24, 27, 28, 62, 63, 65). Relative to other neuronal ion channels, the CLC-2 channel activates slowly (seconds) following hyperpolarization, thereby facilitating current isolation (62). We measured CLC-2 channel activity in terms of the final steady-state current, as well as change in current over the 4-s test step (relaxation current). AK-42 attenuated steady-state currents and eliminated relaxation currents in recorded neurons (Fig. 5 A–D); in contrast, AK-42 had no effect on either measurement in neurons taken from homozygous CLC-2 knockout (Clcn2−/−) mice, thereby demonstrating target specificity (Fig. 5 E–H). An overlay of recordings from untreated Clcn2−/− versus wild-type animals treated with AK-42 shows no discernable difference (SI Appendix, Fig. S5 A and B), confirming that inhibition of CLC-2 current by this compound in wild-type hippocampal CA1 neurons is complete and highly selective.
Fig. 5.
AK-42 inhibits hyperpolarization-activated current in hippocampal cells. (A and B) Whole-cell voltage-clamp recordings (protocol in Inset) of WT mouse CA1 pyramidal neurons with and without AK-42. Capacitive transients are clipped for display purposes. (C) Whole-cell I-V relationship at steady-state both with and without AK-42 (n = 5 cells, 6 slices, and 2 animals per group. P < 0.0001 via two-way repeated-measures ANOVA). Error bars represent ± SEM throughout. (D) Current relaxation measured as the difference between the beginning and end of a 4-s pulse, showing that development of inward current was blocked by AK-42 (n = 6 cells, 6 slices, and 2 animals per group. P < 0.0001 via two-way repeated-measures ANOVA). (E and F) Whole-cell currents from voltage-clamp recordings of Clcn2−/− CA1 pyramidal neurons before and after AK-42 treatment. Unlike WT responses, steady-state responses show no differences after AK-42 treatment at negative membrane potentials. Steady-state currents (G) and current relaxation (H) in Clcn2−/− cells show no difference after AK-42 treatment (n = 6 cells, 6 slices, and 2 animals per group. P = 0.25 and 0.97 for steady-state and current relaxation measurements, respectively, via two-way repeated-measures ANOVA).
In the pilot studies described above, a high concentration of AK-42 (2.5 µM) was used to ensure efficient penetration of the inhibitor into the tissue slice. To confirm potency, we repeated these experiments using a lower, but still saturating, concentration of AK-42 (100 nM). Under these conditions, AK-42 also attenuates relaxation current and peak steady-state current, providing additional validation of our initial results (SI Appendix, Fig. S5 C–E). Current-clamp recordings with 100 nM AK-42 show no differences in firing rate, resting membrane potential, input resistance, or membrane time constant, indicating that this compound does not influence neuronal excitability through off-target effects (SI Appendix, Fig. S5 F–I).
Using the same tissue preparation, the time course for inhibition by AK-42 (100 nM) was measured (SI Appendix, Fig. S5C). Steady-state block of CLC-2 current recorded at −80 mV in hippocampal neurons was achieved within 5 to 10 min of compound application to brain slices. The inhibitor had no effect on currents recorded at −20 mV, a voltage at which little to no CLC-2 current is expected, thus providing additional evidence of target specificity. The 5- to 10-min time course for onset of inhibition is slower than the rate measured in CHO cells (SI Appendix, Figs. S2 and S3), but such differences are expected for a compound that must penetrate tissue to reach the intended target. Following steady-state block, we evaluated the reversibility of CLC-2 inhibition by washing out the compound (perfusion with buffer solution) over several minutes (SI Appendix, Fig. S5C). Partial recovery of hyperpolarization-induced current (66%) was achieved over 25 min of washing. Together, these data establish that application of AK-42 can provide acute, selective, and reversible block of CLC-2 in primary neurons.
Discussion
The numerous outstanding questions surrounding the complex physiology of CLC-2 in the CNS inspired our development of AK-42 as a high-precision molecular tool. Systematic modification of an initial hit compound, MCFA, yielded a small library of 51 derivatives from which the critical molecular features responsible for inhibitor potency were identified. This work culminated in the design and synthesis of AK-42.
The potency and specificity of AK-42 for CLC-2 stands in contrast to all other known Cl– channel small-molecule modulators. Application of this compound to cells expressing CLC-2 enables acute channel inhibition, thus avoiding the interpretive challenges of genetic knockout or gene-silencing methods. Electrophysiology slice recordings on hippocampal CA1 neurons provide compelling evidence in support of AK-42 specificity; current vs. voltage plots from data collected on Clcn2−/− neurons in the absence and presence of AK-42 mirror those obtained from application of AK-42 to wild-type CA1 neurons. Furthermore, primary binding and functional assays using 10 µM AK-42 (≥100× the concentration required to elicit complete CLC-2 current block in hippocampal slices) show no substantive off-target engagement against a panel of 61 common membrane proteins, including the VRAC, CFTR, and TMEM16A anion channels.
The availability of AK-42 enables subsequent experiments to interrogate CLC-2 structure and channel biophysics. While all CLCs share a homodimeric architecture with two independent Cl– permeation pathways, channel-gating properties differ between CLC homologs. The disclosures of two mammalian CLC structures have revealed only subtle structural differences between channels (61, 66), and the detailed biophysical machinery driving functional diversity within the CLC family remains largely enigmatic. Small molecules, such as AK-42, are indispensable tools to advance such investigations.
A desire to understand the basis for AK-42 selectivity and specificity prompted our efforts to identify the inhibitor binding site. Fast and reversible inhibition by AK-42 in electrophysiological recordings of heterologously expressed CLC-2 suggest that AK-42 acts extracellularly. This conclusion is supported by computational docking models of AK-42 to a CLC-2 homology model, as well as by mutagenesis experiments with CLC-2 at positions K210 and Q399 (which markedly reduce AK-42 potency at 30 nM). Even conservative mutation of K210 to arginine (K210R) perturbs the ability of AK-42 to block Cl– current. In contrast, arginine mutation of K400 (K400R) has no influence on AK-42 affinity. From our modeling studies, the putative AK-42 binding pocket occupies an extracellular vestibule, located in the same region as the benzimidazole sulfonate, BIM1, receptor site in CLC-Ka (46). Our results, however, indicate that BIM1 does not inhibit CLC-2 at concentrations exceeding 100 µM, suggesting that subtle differences between channels in this locus can be exploited to target individual CLC homologs. Additional investigations are required to determine exact details of AK-42 binding to CLC-2, as our ligand-docking model comprises multiple high-scoring poses within the extracellular pocket. Future studies that leverage available SAR data, along with our ability to generate additional AK-42 derivatives and CLC-2 mutants, should reveal the precise molecular interactions between AK-42 and the channel.
The development of AK-42, together with cell-type–specific genetic manipulation of CLC-2, will facilitate future investigations into CLC-2 function in both neurons and glia, including studies to determine the effects of this channel on network excitability, cell volume, and ion homeostasis. The widespread expression of CLC-2 in different cell types found in the CNS (21, 24, 28, 58, 62–64, 67) intimates multiple functional roles for this channel in proper brain function. One hypothesis is that the strong inward rectification of CLC-2 at hyperpolarized potentials maintains low intracellular [Cl–] in hippocampal neurons to facilitate GABAergic inhibition (24, 25, 27). Alternatively, it has been argued that CLC-2 regulates neuronal excitability by mediating Cl– influx under physiological conditions (26). Other studies have suggested that fast-spiking parvalbumin-basket cells (PV-BCs) specifically associate with CLC-2-containing GABAergic synapses, where the channel is thought to act as a safety valve for Cl– efflux during intense synchronized network oscillations mediated by PV-BC interneurons (27, 29). In glia, CLC-2 is expressed in oligodendrocytes and astrocytes and is associated with an adhesion molecule known as GlialCAM (58, 64, 68). Glial CLC-2 is postulated to regulate ion and fluid homeostasis, and may indirectly influence neuronal excitability by facilitating uptake of extracellular K+ following intense neuronal firing (17, 21, 69–71). While compelling, these mechanisms have been difficult to parse in the absence of acute and specific methods for modulating CLC-2 function. With the advent of AK-42, we expect that new insights to these questions will be revealed.
Materials and Methods
IWB Electrophysiology Recording Conditions.
Data acquisition and analyses were performed using the IWB systems (Molecular Devices, operation software v2.5.3.359) in the Population Patch Clamp (PPC) Mode. Test article (TA) and positive control concentrations were prepared fresh daily by diluting stock solutions into an extracellular buffer solution, and the TA formulations were loaded in a 384-well compound plate. Currents were recorded from groups of 64 cells. HEPES-buffered intracellular solution (50 mM CsCl, 90 mM CsF, 5 mM MgCl2, 5 mM EGTA, 10 mM HEPES, adjusted to pH = 7.4 with CsOH) for whole-cell recordings was loaded into the intracellular compartment of the PPC planar electrode. Extracellular buffer (HB-PS; 137 mM NaCl, 4 mM KCl, 3.8 mM CaCl2, 1 mM MgCl2, 10 mM Hepes, 10 mM glucose, adjusted to pH = 7.3 with NaOH) was loaded into PPC planar electrode plate wells (11 µL per well). The cell suspension was pipetted into the wells of the PPC planar electrode (9 µL per well). After establishment of a whole-cell configuration (the perforated patch; 12 µM escin, 7 min), membrane currents were recorded using a patch-clamp amplifier in the IWB system. The current recordings were performed one time before TA application to the cells (baseline) and one time after application of TA or reference compound. Seal resistance prior to compound application was ≥200 MΩ. Reference compound concentrations were applied to naïve cells (n = 4, where n = replicates/concentration). Each application consisted of a 20 μL addition of a 2× concentrated test article solution to the total 40-μL final volume of the extracellular well of the PPC electrode. The addition was followed by a single mixing of the PPC well content. Duration of exposure to each compound concentration was 5 min.
Prior to initial screening, we validated our assay by using Cd2+ and NPPB as positive controls for CLC-2 inhibition. Eight dose–response curves were generated in each experiment on 2 different days (n = 3 to 4 per concentration). All Z′ values (a standard metric for evaluating assay quality) exceeded 0.5, indicating acceptable assay variability (72). The Z′ factor was calculated as: Z′ = 1–(3 × SDVC + 3 × SDPC)/ABS (MeanVC – MeanPC), where MeanVC and SDVC are the mean and SD values for a vehicle control, MeanPC and SDPC are the mean and SD values for a positive control (300 µM NPPB or 1 mM Cd2+). Initial screening for hit compounds was performed using the ENZO Life Sciences SCREEN-WELL Food and Drug Administration-approved drug library, v2.0 (Product # BML-2843-0100). All compounds in this library are reported in Dataset S1. Vehicle controls of 0.3 or 1.0% DMSO (without compound) in recording solution were applied in replicate as additional controls in each experiment.
Cell Line Generation.
All CLC-2 recordings using the IWB platform were performed with a stably expressing CHO cell line. CHO cells were transfected with human CLC-2 cDNAs (CLCN2, GenBank accession no. NM_004366.3). Stable transfectants were selected by expression with the antibiotic-resistance gene (Geneticin/G418) incorporated into the expression plasmid [pcDNA3.1(+)]. Selection pressure was maintained by including selection antibiotics in the culture medium. Cells were cultured in Ham’s F-12 media (Life Technologies) supplemented with 10% fetal bovine serum (HyCloneTM Fetal Bovine Serum), 100 U/mL penicillin G sodium, 100 µg/mL streptomycin sulfate, and 500 µg/mL G418. Before testing, cells in culture dishes were washed twice with HBSS (Life Technologies), treated with Accutase (Innovative Cell Technologies) solution for 20 min at room temperature, and resuspended in HBSS (4–6 × 106 cells in 10 mL). Immediately before use, the cells were washed twice in HBSS to remove the Accutase and resuspended in 5 mL of HBSS. CLC-1 recordings were performed using a stably expressing CHO cell line that was generated according to the same methods described for CLC-2.
IWB Voltage Protocol and Data Analysis.
From a holding potential of −30 mV, CLC-2 currents were elicited by applying 2-s test pulses from −120 to +20 mV in 20-mV increments. Following each test pulse, a 500-ms tail pulse to 0 mV was applied. We required initial currents at the 0-mV tail pulse to be ≥0.2 nA to be included in analysis. All IC50 data reported in the manuscript are given according to measurements at the maximum current at −120 mV, the potential at which the maximum number of CLC-2 channels are open. The decrease in current amplitude after compound application (TA) was used to calculate the percent block relative to the positive control, according to the equation: % Inhibition = (1 – (ITA / IBaseline)) × 100%, where IBaseline and ITA are the currents measured before and after addition of compound, respectively. The data were then corrected for run-down according to the equation: %Block′ = 100% – ((%Block – %PC) × (100%/(%VC–%PC))), where %VC and %PC are the mean values of the current inhibition with the vehicle control and the highest concentration of a reference compound (300 µM Cd2+). The inhibition curves were fit to the equation: % Inhibition = 100/[1 + ([IC50]/[TA])n], where [TA] is the concentration of test article, IC50 is the concentration of TA producing half-maximal inhibition, n is the Hill coefficient, and % block is the percentage of current inhibited at each concentration of a TA. Nonlinear least-squares fits were solved with the XLfit add-in for Excel (Microsoft) (initial screening data) or using the Sigmaplot 13.0 (Systat Software) regression analysis (final analyses).
For the kinetics experiments (SI Appendix, Fig. S3), current was elicited with a 2,000-ms voltage step to a potential of −120 mV, followed by a 500-ms voltage step to a potential of 0 mV; the holding potential was −30 mV. The test pulses were repeated with a frequency of 0.1 Hz. Six stimulations were applied before application of test articles (1-min baseline) and 30 stimulations were performed after application of test articles (5 min). In the time course for AK-42 application, the channel occupies both the open and closed states; thus, the results reflect a weighted average of AK-42 binding to the different channel conformations. More detailed experiments are necessary to address whether AK-42 inhibition is state-dependent. Findings from our experiments establish AK-42 binding to an extracellular (efficiently reversible) site with a 1:1 AK-42/channel subunit stoichiometry. Current amplitudes were measured as the minimal inward current at test potential −120 mV and the maximal outward current at test potential 0 mV.
The vehicle control data (0.3% DMSO; n = 32) were averaged for each time point. All data were normalized to the time-matching averaged vehicle control. Then, data were normalized to the baseline current recorded before application of test articles.
Cell Culture and Recording Protocols (Manual Patch-Clamp).
CHO K1 cells (ATCC CCL-61) were cultured at 37 °C in F12K media (ATCC, Catalog no. 30-2004) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Gibco). At 70 to 90% confluency, cells were transfected with full-length rat CLC-2 cDNA (449 ng) using Lipofectamine LTX, opti-MEM, and PLUS reagent (Invitrogen). The rat CLC-2 gene was contained in a custom plasmid vector called pFROG (ampicillin resistance), provided by Michael Pusch, Istituto di Biofisica, CNR, Genoa, Italy. Cotransfection with GFP (177 ng, 2.5:1 CLC-2/GFP) was used to estimate transfection efficiency. Cells were transfected 18 h prior to recording. For CLC-2 with GlialCAM experiments, rat CLC-2 DNA was cotransfected with human GlialCAM (530 ng, 1:1 CLC-2/GlialCAM) in CHO cells. Chloride currents from CLC-2 with GlialCAM were recorded 18 to 22 h following transfection. The human GlialCAM gene was contained in a pcDNA3 construct (ampicillin resistance), provided by Raúl Estévez, Department of Physiological Sciences, University of Barcelona, Barcelona, Spain. Currents were elicited from a holding potential of 0 mV for 20 ms, followed by steps from −100 mV to + 40 mV in 20-mV intervals with a tail pulse of +40 mV for 50 ms. Borosilicate glass micropipettes (Sutter Instruments) were pulled and fire-polished to a tip diameter with a resistance of 1.5 to 3.0 MΩ. For whole-cell patch-clamp recordings, the external solution was composed of 148 mM CsCl, 2 mM CaCl2, 10 mM HEPES, and 100 mM d-mannitol (adjusted to pH 7.4 with 2 M aqueous CsOH). The internal solution was composed of 146 mM CsCl, 5 mM EGTA, 10 mM HEPES, 5 mM NaF, and 60 mM d-mannitol (adjusted to pH 7.4 with 2 M aqueous CsOH). If necessary, the osmolarity was adjusted with d-mannitol such that it was slightly higher outside than inside in order to prevent cell swelling, which can make it difficult to establish a high-resistance seal. CLC-2 currents were measured in whole-cell patch-clamp mode using an Axopatch-200B amplifier (Axon Instruments), an InstruTECH ITC-16 interface (HEKA Instruments), and a Mac Mini computer (Apple).
Igor Pro (WaveMetrics) software was used for stimulation and data collection. The data were sampled at 5 kHz and filtered at 1 kHz. From a holding potential of 0 mV, currents were elicited with 250-ms voltage steps of −100 to +20 mV in 10-mV increments, followed by a 100-ms tail pulse to +80 mV (Fig. 4C). Note, we used modest hyperpolarizing potentials, compared to those used to study the full voltage-dependence of CLC-2 (73, 74), to ensure stability of the voltage-clamp seals over the course of AK-42 application and washout. All experiments were carried out at room temperature (22–25 °C). Following initial measurement of currents (“initial”), AK-42 was flowed into the recording chamber, and current was measured again after 5 to 10 min. At least two voltage-step families were measured in order to ensure that steady-state inhibition had been reached (“inhibited”). Currents were measured again following ∼10 min of washout (“washout”), again ensuring steady-state had been attained. For fitting dose–response curves (SI Appendix, Fig. S2), we excluded experiments in which there was poor reversibility of inhibition. Poor reversibility was defined as when reversal current differed by >20% from initial current and inhibition calculated using washout current differed by >10% from inhibition calculated using initial current. An IC50 value and a Hill coefficient (n) were obtained with Sigmaplot 13.0 (Systat Software) by fitting dose–response curves to the following equation: I (%) = 1/{1 + (IC50/[D])n}, in which I (%) is the percentage current inhibition {I (%) = [1 − Iinhibited/Iinitial] × 100} at the −100-mV test potential and [D] represents various concentrations of inhibitor. Results were each expressed as a mean ± SEM. ANOVA was used for statistical analysis.
Mutagenesis Strategy.
CLC-2 mutants were generated by site-directed mutagenesis using a QuikChange XL Site-Directed Mutagenesis Kit (Agilent Technologies). Table 3 lists primers used to generate CLC-2 mutants in this study.
Table 3.
Primers used to generate mutants in pFROG-CLC-2
| CLC-2 Mutant | Primer | Sequence |
| K400R | Forward | GACAGCTCTCACAGCGGGAGACCC |
| Reverse | GGGTCTCCCGTGTGAGAGCTGTCC | |
| Q399P | Forward | CAGCTCTCACCCAAAGAGACCCTGGTCAC |
| Reverse | GGTCTCTTTGGGTGAGAGCTGTCCGGCC | |
| K210M | Forward | CCTTGGAATGGAGGGACCCTTTGTGCAC |
| Reverse | GTCCCTCCATTCCAAGGGGCATCCC | |
| K210R | Forward | CCTTGGAAGAGAGGGACCCTTTGTGCAC |
| Reverse | GTCCCTCTCTTCCAAGGGGCATCCC |
PDSP Screening Results and Off-Target Assay of VRAC, CFTR, and TMEM16A Anion Channels.
AK-42 was screened against a panel of receptors, transporters, and ion channels to examine specificity for CLC-2 in CNS tissue. A comprehensive primary binding assay screen was performed through the NIH PDSP at the University of North Carolina at Chapel Hill. Secondary or functional assays were performed for compounds showing >50% activity in the primary assay. All results are shown in Dataset S2. The activities of VRAC (LRRC8A), CFTR, and TMEM16A channels were measured by manual and automated patch-clamp methods using 10 µM AK-42. All methods for data collection and results are described in Dataset S2.
CLC-2 KO Mice.
Animal procedures and care were performed according to protocols approved by the Stanford Institutional Animal Care and Use Committee. Heterozygous CLC-2 KO mice (Clcn2+/−) (23) were bred and litters genotyped to identify littermate Clcn2−/− and Clcn2+/+ mice, which were paired in each experiment. On any experimental day, brain slices from a pair of young mice (postnatal days 30–45) were prepared, and then slices separated by group with the identity blinded to the investigator.
Electrophysiological Recordings in Acute Hippocampal Slices.
Coronal slices (250-µM thick) were prepared from 30 to 35-d-old wild-type C57/B6J or KO (Clcn2−/−) mice, as described previously (75). After recovery for 1 h, slices were transferred to a recording chamber at 23.5 °C containing oxygenated artificial cerebrospinal fluid (ACSF): 2.5 mM KCl, 10 mM glucose, 10 mM CsCl, 116 mM NaCl, 1.25 mM sodium phosphate hydrate, 1.0 mM MgSO4, 2.0 mM CaCl2, 26 mM NaHCO3. Whole-cell voltage-clamp recordings were made under constant perfusion at room temperature from CA1 hippocampal pyramidal cells under conditions that isolate and promote stable recordings of hyperpolarization-activated CLC-2 currents. These include Cs+ as the main internal monovalent cation to block K+ currents, high internal [Cl–] to make ECl = 0 mV and facilitate recordings at a physiological membrane potential of −80 mV, 10 mM external Cs+ to block HCN channels, and 50 µM picrotoxin/1.5 mM kynurenic acid to block synaptic responses. Internal solution for voltage-clamp contained the following: 10 mM HEPES 130 mM CsCl, 10 mM EGTA, 3 mM MgATP, 0.3 mM Na2GTP hydrate, 5 mM QX-314. For long recordings in SI Appendix, Fig. S5, internal solution was modified to include an ATP regenerating system and was comprised of: 10 mM HEPES, 125 mM CsCl, 10 mM EGTA, 4 mM MgATP, 0.3 mM Na2GTP, 5 mM QX-314, 15 mM phosphocreatine disodium hydrate, 50 U/mL creatine phosphokinase. Internal solution for current-clamp included: 105 mM K-gluconate, 10 mM HEPES, 10 mM NaCl, 20 mM KCl, 0.6 mM EGTA, 5 mM MgATP, 0.3 mM Na2GTP. ACSF for current-clamp omitted picrotoxin and cesium. Recordings were performed using an Axon Instruments Multiclamp 700A amplifier and Digidata 1550B digitizer (Molecular Devices); 10-KHz bessel filtering was applied during the recording.
After achieving whole-cell configuration, cells were allowed to stabilize for 2 min before beginning each recording. Recordings in SI Appendix, Fig. S5 were obtained (two per slice), one before AK-42 was added and one after washing the slice for 10 min with AK-42. Cell membrane parameters were monitored before and after each voltage-clamp protocol to ensure the stability of each recording. Recordings were excluded if the series resistance was greater than 20 MΩ or changed by more than 25%, or if the holding current at −60 mV exceeded −200 pA. Recordings in Fig. 4 were obtained one per slice in ACSF containing either 2.5 µM AK-42 or no inhibitor. Access resistance was measured every 4 min. Voltage-step protocols were 2 s at −4 mV, followed by a 3-s step at the test voltage ranging between −100 mV and +20 mV in 10-mV increments before stepping back to −4 mV for 0.5 s. Continuous monitoring of whole-cell currents in SI Appendix, Fig. S5 C–E used a modified voltage protocol utilizing a 4-s conditioning step at −4 mV and a 4-s test step to either −20 or −80 mV. Protocols were run every 30 s along with a membrane test.
Data were measured using Clampfit 10.6 (Molecular Devices). Steady-state currents were measured as the RMS current in the last 200 ms of the test voltage step. Current relaxation was measured to be the RMS current between 200 and 400 ms into the test voltage step minus the steady-state current. Two-way repeated measures ANOVA was run in GraphPad Prism 6.01 (GraphPad Software) to compare treatment groups.
Computational Docking.
Initial docking studies involved preparation of the receptor and ligand using the dock prep tool in the University of California, San Francisco (UCSF) Chimera package (76). The receptor surface and ligand were parameterized using the AMBER parm99 force field (77). A spacing of 0.3 Å was used for the receptor grid. Initial grids included the entire extracellular surface of the protein. Once prepared, docking calculations were performed using Dock6 (78). These calculations employed flexible docking and yielded 1,000 ligand orientations. From this dataset, the highest scoring conformations were analyzed. The results revealed clustering of ligands in a single extracellular pocket above the ion permeation pathway. Following identification of a general binding site, additional docking poses were generated for the most potent lead compounds using the Schrödinger Maestro software suite (v2019-2, Schrödinger 2019). Prior to docking, ligands were optimized using the OPLS3e (79) force field in LigPrep (Schrödinger 2019). Possible ligand tautomers and protonation states at pH 7.0 ± 2.0 were sampled using Epik (80) (Schrödinger 2019). Ligands were docked to grids centered on the general binding site (residue K210) for each of the conformational states developed from the CLC-2 homology model described in McKiernan et al. (60) as well as CLC-2 homology models generated from the 5TQQ and 5TR1 cryoelectron microscopy structures of bovine CLC-K (61). Alignment of the CLC-2 sequence with these structures for generation of the homology models was accomplished with the UCSF Chimera sequence alignment tool (76). The CLC-2 sequence was then mapped onto the template structure using the Modeler package (81). Unresolved regions of the template structure were also refined using Modeler. Docking to the CLC-1 cryoelectron microscopy structure (6COY) (66) was performed using a grid centered on the equivalent lysine residue for this homolog (K231). Docking grids were generated using the default Maestro settings for receptor rigidity (Van der Waals radius scaling factor of 1.0 with a partial charge cutoff of 0.25), temperature, and pH (protonation states). Prepared ligands were docked to these grids using the default settings for Schrödinger Glide (82) (Schrödinger 2019, Van der Waals radius scaling factor of 0.80 with a partial charge cutoff of 0.15). Standard precision docking was performed with flexible ligand sampling (sampling of nitrogen inversion and ring conformations), and postdocking minimization was performed to generate a maximum of 10 poses per ligand. A representative docking pose is illustrated in Fig. 4.
Chemical Synthesis.
For protocols for synthesizing AK-42 and related compounds, see Dataset S3.
Supplementary Material
Acknowledgments
Receptor-binding profiles and agonist/antagonist functional data were generously provided by the National Institute of Mental Health’s Psychoactive Drug Screening Program (NIMH PDSP) Contract HHSN-271-2018-00023-C. The NIMH PDSP is directed by Bryan L. Roth at the University of North Carolina at Chapel Hill and Project Officer Jamie Driscoll at National Institute of Mental Health. Clcn2−/− mice were generously provided by J. Melvin (University of Rochester). The CLC-2 and GlialCAM constructs used for manual voltage-clamp recordings were kindly provided by M. Pusch (Istituto di Biofisica, Genoa, Italy) and R. Estevez (University of Barcelona, Barcelona, Spain), respectively, with permission from Thomas Jentsch. This work was supported by a Stanford Bio-X Seed grant (to J.D.B. and M.M.) and NIH Grant R01NS113611 (to J.D.B., M.M., and J.R.H.). A.K.K. and K.A.M. were supported by the Stanford Center for Molecular Analysis and Design. A.K.K. was also supported by a Stanford Interdisciplinary Graduate Fellowship through the Stanford ChEM-H Institute.
Footnotes
Competing interest statement: A.K.K., J.D.B., and M.M. have filed for a patent “Compositions and Methods to Modulate Chloride Ion Channel Activity,” USSN 16/449,021 from the U.S. Patent & Trademark Office, January 16, 2020.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009977117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Jentsch T. J., Pusch M., CLC chloride channels and transporters: Structure, function, physiology, and disease. Physiol. Rev. 98, 1493–1590 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Thiemann A., Gründer S., Pusch M., Jentsch T. J., A chloride channel widely expressed in epithelial and non-epithelial cells. Nature 356, 57–60 (1992). [DOI] [PubMed] [Google Scholar]
- 3.Walz W., Chloride/anion channels in glial cell membranes. Glia 40, 1–10 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Wilson C. S., Mongin A. A., The signaling role for chloride in the bidirectional communication between neurons and astrocytes. Neurosci. Lett. 689, 33–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stauber T., Jentsch T. J., Chloride in vesicular trafficking and function. Annu. Rev. Physiol. 75, 453–477 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Rahmati N., Hoebeek F. E., Peter S., De Zeeuw C. I., Chloride homeostasis in neurons with special emphasis on the olivocerebellar system: Differential roles for transporters and channels. Front. Cell. Neurosci. 12, 101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe M., Fukuda A., Development and regulation of chloride homeostasis in the central nervous system. Front. Cell. Neurosci. 9, 371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poroca D. R., Pelis R. M., Chappe V. M., ClC channels and transporters: Structure, physiological functions, and implications in human chloride channelopathies. Front. Pharmacol. 8, 151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi M. M., et al. , Chloride channelopathies of ClC-2. Int. J. Mol. Sci. 15, 218–249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandes-Rosa F. L., et al. , A gain-of-function mutation in the CLCN2 chloride channel gene causes primary aldosteronism. Nat. Genet. 50, 355–361 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Scholl U. I., et al. , CLCN2 chloride channel mutations in familial hyperaldosteronism type II. Nat. Genet. 50, 349–354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schewe J., et al. , Elevated aldosterone and blood pressure in a mouse model of familial hyperaldosteronism with ClC-2 mutation. Nat. Commun. 10, 5155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Göppner C., et al. , Pathogenesis of hypertension in a mouse model for human CLCN2 related hyperaldosteronism. Nat. Commun. 10, 4678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Depienne C., et al. , Brain white matter oedema due to ClC-2 chloride channel deficiency: An observational analytical study. Lancet Neurol. 12, 659–668 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Di Bella D., et al. , Subclinical leukodystrophy and infertility in a man with a novel homozygous CLCN2 mutation. Neurology 83, 1217–1218 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Hanagasi H. A., et al. , Secondary paroxysmal kinesigenic dyskinesia associated with CLCN2 gene mutation. Parkinsonism Relat. Disord. 21, 544–546 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Gaitán-Peñas H., et al. , Leukoencephalopathy-causing CLCN2 mutations are associated with impaired Cl- channel function and trafficking. J. Physiol. 595, 6993–7008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeydan B., et al. , Identification of 3 novel patients with CLCN2-related leukoencephalopathy due to CLCN2 mutations. Eur. Neurol. 78, 125–127 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Giorgio E., et al. , A novel homozygous change of CLCN2 (p.His590Pro) is associated with a subclinical form of leukoencephalopathy with ataxia (LKPAT). J. Neurol. Neurosurg. Psychiatry 88, 894–896 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Guo Z., et al. , CLCN2-related leukoencephalopathy: A case report and review of the literature. BMC Neurol. 19, 156 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanz J., et al. , Leukoencephalopathy upon disruption of the chloride channel ClC-2. J. Neurosci. 27, 6581–6589 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bösl M. R., et al. , Male germ cells and photoreceptors, both dependent on close cell-cell interactions, degenerate upon ClC-2 Cl– channel disruption. EMBO J. 20, 1289–1299 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nehrke K., et al. , Loss of hyperpolarization-activated Cl– current in salivary acinar cells from Clcn2 knockout mice. J. Biol. Chem. 277, 23604–23611 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Staley K., The role of an inwardly rectifying chloride conductance in postsynaptic inhibition. J. Neurophysiol. 72, 273–284 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Staley K., Smith R., Schaack J., Wilcox C., Jentsch T. J., Alteration of GABAA receptor function following gene transfer of the CLC-2 chloride channel. Neuron 17, 543–551 (1996). [DOI] [PubMed] [Google Scholar]
- 26.Ratté S., Prescott S. A., ClC-2 channels regulate neuronal excitability, not intracellular chloride levels. J. Neurosci. 31, 15838–15843 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Földy C., Lee S. H., Morgan R. J., Soltesz I., Regulation of fast-spiking basket cell synapses by the chloride channel ClC-2. Nat. Neurosci. 13, 1047–1049 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rinke I., Artmann J., Stein V., ClC-2 voltage-gated channels constitute part of the background conductance and assist chloride extrusion. J. Neurosci. 30, 4776–4786 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armstrong C., Soltesz I., Basket cell dichotomy in microcircuit function. J. Physiol. 590, 683–694 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smart T. G., Handling accumulated internal Cl− at inhibitory synapses. Nat. Neurosci. 13, 1043–1044 (2010). [DOI] [PubMed] [Google Scholar]
- 31.D’Agostino D., et al. , Mutations and polymorphisms of the CLCN2 gene in idiopathic epilepsy. Neurology 63, 1500–1502 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Saint-Martin C., et al. , Two novel CLCN2 mutations accelerating chloride channel deactivation are associated with idiopathic generalized epilepsy. Hum. Mutat. 30, 397–405 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Kleefuss-Lie A., et al. , CLCN2 variants in idiopathic generalized epilepsy. Nat. Genet. 41, 954–955 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Everett K., et al. , Linkage and mutational analysis of CLCN2 in childhood absence epilepsy. Epilepsy Res. 75, 145–153 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Stogmann E., et al. , Mutations in the CLCN2 gene are a rare cause of idiopathic generalized epilepsy syndromes. Neurogenetics 7, 265–268 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Bertelli M., et al. , Quantification of chloride channel 2 (CLCN2) gene isoforms in normal versus lesion- and epilepsy-associated brain tissue. Biochim. Biophys. Acta 1772, 15–20 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Combi R., et al. , Clinical and genetic familial study of a large cohort of Italian children with idiopathic epilepsy. Brain Res. Bull. 79, 89–96 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Niemeyer M. I., et al. , No evidence for a role of CLCN2 variants in idiopathic generalized epilepsy. Nat. Genet. 42, 3 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Sigma-Aldrich, The Handbook of Receptor Classification and Signal Transduction (2019), https://www.sigmaaldrich.com/technical-documents/articles/biology/rbi-handbook/ion-channels/chloride-channels.html. Accessed 18 November 2020.
- 40.Suh K. S., Yuspa S. H., Intracellular chloride channels: Critical mediators of cell viability and potential targets for cancer therapy. Curr. Pharm. Des. 11, 2753–2764 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Pusch M., et al. , Mechanisms of block of muscle type CLC chloride channels (Review). Mol. Membr. Biol. 19, 285–292 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Cabantchik Z. I., Greger R., Chemical probes for anion transporters of mammalian cell membranes. Am. J. Physiol. 262, C803–C827 (1992). [DOI] [PubMed] [Google Scholar]
- 43.Liantonio A., et al. , Molecular switch for CLC-K Cl– channel block/activation: Optimal pharmacophoric requirements towards high-affinity ligands. Proc. Natl. Acad. Sci. U.S.A. 105, 1369–1373 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gradogna A., Pusch M., Molecular pharmacology of kidney and inner ear CLC-K chloride channels. Front. Pharmacol. 1, 130 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liantonio A., et al. , Kidney CLC-K chloride channels inhibitors: Structure-based studies and efficacy in hypertension and associated CLC-K polymorphisms. J. Hypertens. 34, 981–992 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Koster A. K., et al. , A selective class of inhibitors for the CLC-Ka chloride ion channel. Proc. Natl. Acad. Sci. U.S.A. 115, E4900–E4909 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson C. H., Freeman C. S., French R. J., McCarty N. A., Pharmacological charcterization of GaTx2, a peptide inhibitor of ClC-2 chloride channels. Biophys. J. 96, 470a (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clark S., Jordt S. E., Jentsch T. J., Mathie A., Characterization of the hyperpolarization-activated chloride current in dissociated rat sympathetic neurons. J. Physiol. 506, 665–678 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furukawa T., Ogura T., Katayama Y., Hiraoka M., Characteristics of rabbit ClC-2 current expressed in Xenopus oocytes and its contribution to volume regulation. Am. J. Physiol. 274, C500–C512 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Jordt S. E., Jentsch T. J., Molecular dissection of gating in the ClC-2 chloride channel. EMBO J. 16, 1582–1592 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zúñiga L., et al. , The voltage-dependent ClC-2 chloride channel has a dual gating mechanism. J. Physiol. 555, 671–682 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moeser A. J., Nighot P. K., Engelke K. J., Ueno R., Blikslager A. T., Recovery of mucosal barrier function in ischemic porcine ileum and colon is stimulated by a novel agonist of the ClC-2 chloride channel, lubiprostone. Am. J. Physiol. Gastrointest. Liver Physiol. 292, G647–G656 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Cuppoletti J., et al. , SPI-0211 activates T84 cell chloride transport and recombinant human ClC-2 chloride currents. Am. J. Physiol. Cell Physiol. 287, C1173–C1183 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Flores C. A., ClC-2 and intestinal chloride secretion. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G775 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Chan W. W., Mashimo H., Lubiprostone increases small intestinal smooth muscle contractions through a prostaglandin E receptor 1 (EP1)-mediated pathway. J. Neurogastroenterol. Motil. 19, 312–318 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Norimatsu Y., Moran A. R., MacDonald K. D., Lubiprostone activates CFTR, but not ClC-2, via the prostaglandin receptor (EP(4)). Biochem. Biophys. Res. Commun. 426, 374–379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liantonio A., et al. , Activation and inhibition of kidney CLC-K chloride channels by fenamates. Mol. Pharmacol. 69, 165–173 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Jeworutzki E., et al. , GlialCAM, a protein defective in a leukodystrophy, serves as a ClC-2 Cl– channel auxiliary subunit. Neuron 73, 951–961 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Besnard J., et al. , Automated design of ligands to polypharmacological profiles. Nature 492, 215–220 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McKiernan K. A., Koster A. K., Maduke M., Pande V. S., Dynamical model of the CLC-2 ion channel reveals conformational changes associated with selectivity-filter gating. PLOS Comput. Biol. 16, e1007530 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park E., Campbell E. B., MacKinnon R., Structure of a CLC chloride ion channel by cryo-electron microscopy. Nature 541, 500–505 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Madison D. V., Malenka R. C., Nicoll R. A., Phorbol esters block a voltage-sensitive chloride current in hippocampal pyramidal cells. Nature 321, 695–697 (1986). [DOI] [PubMed] [Google Scholar]
- 63.Smith R. L., Clayton G. H., Wilcox C. L., Escudero K. W., Staley K. J., Differential expression of an inwardly rectifying chloride conductance in rat brain neurons: A potential mechanism for cell-specific modulation of postsynaptic inhibition. J. Neurosci. 15, 4057–4067 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sík A., Smith R. L., Freund T. F., Distribution of chloride channel-2-immunoreactive neuronal and astrocytic processes in the hippocampus. Neuroscience 101, 51–65 (2000). [DOI] [PubMed] [Google Scholar]
- 65.Mager R., Ferroni S., Schubert P., Adenosine modulates a voltage-dependent chloride conductance in cultured hippocampal neurons. Brain Res. 532, 58–62 (1990). [DOI] [PubMed] [Google Scholar]
- 66.Park E., MacKinnon R., Structure of the CLC-1 chloride channel from Homo sapiens. eLife 7, e36629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clayton G. H., Staley K. J., Wilcox C. L., Owens G. C., Smith R. L., Developmental expression of CLC-2 in the rat nervous system. Brain Res. Dev. Brain Res. 108, 307–318 (1998). [DOI] [PubMed] [Google Scholar]
- 68.Jeworutzki E., et al. , GlialCAM, a CLC-2 Cl– channel subunit, activates the slow gate of CLC chloride channels. Biophys. J. 107, 1105–1116 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maduke M. C., Reimer R. J., Biochemistry to the rescue: A ClC-2 auxiliary subunit provides a tangible link to leukodystrophy. Neuron 73, 855–857 (2012). [DOI] [PubMed] [Google Scholar]
- 70.Hoegg-Beiler M. B., et al. , Disrupting MLC1 and GlialCAM and ClC-2 interactions in leukodystrophy entails glial chloride channel dysfunction. Nat. Commun. 5, 3475 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Sirisi S., et al. , Depolarization causes the formation of a ternary complex between GlialCAM, MLC1 and ClC-2 in astrocytes: Implications in megalencephalic leukoencephalopathy. Hum. Mol. Genet. 26, 2436–2450 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Zhang J. H., Chung T. D., Oldenburg K. R., A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 (1999). [DOI] [PubMed] [Google Scholar]
- 73.Niemeyer M. I., Cid L. P., Yusef Y. R., Briones R., Sepúlveda F. V., Voltage-dependent and -independent titration of specific residues accounts for complex gating of a ClC chloride channel by extracellular protons. J. Physiol. 587, 1387–1400 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Jesús-Pérez J. J., et al. , Gating the glutamate gate of CLC-2 chloride channel by pore occupancy. J. Gen. Physiol. 147, 25–37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huguenard J. R., Prince D. A., Intrathalamic rhythmicity studied in vitro: Nominal T-current modulation causes robust antioscillatory effects. J. Neurosci. 14, 5485–5502 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pettersen E. F., et al. , UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 77.Case D. A., et al. , The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Allen W. J., et al. , DOCK 6: Impact of new features and current docking performance. J. Comput. Chem. 36, 1132–1156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harder E., et al. , OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 12, 281–296 (2016). [DOI] [PubMed] [Google Scholar]
- 80.Greenwood J. R., Calkins D., Sullivan A. P., Shelley J. C., Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J. Comput. Aided Mol. Des. 24, 591–604 (2010). [DOI] [PubMed] [Google Scholar]
- 81.Sali A., Blundell T. L., Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993). [DOI] [PubMed] [Google Scholar]
- 82.Friesner R. A., et al. , Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 49, 6177–6196 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.