Abstract
High throughput cDNA sequencing technologies have advanced our understanding of transcriptome complexity and regulation. However, these methods lose information contained in biological RNA because the copied reads are often short and because modifications are not retained. We address these limitations using a native poly(A) RNA sequencing strategy developed by Oxford Nanopore Technologies (ONT). Our study generated 9.9 million aligned sequence reads for the human cell line GM12878, using thirty MinION flow cells at six institutions. These native RNA reads had a median length of 771 bases, and a maximum aligned length of over 21,000 bases. Mitochondrial poly(A) reads provided an internal measure of read length quality. We combined these long nanopore reads with higher accuracy short-reads and annotated GM12878 promoter regions, to identify 33,984 plausible RNA isoforms. We describe strategies for assessing 3′ poly(A) tail length, base modifications, and transcript haplotypes.
INTRODUCTION
Sequencing by synthesis (SBS) strategies have dominated RNA sequencing since the early 1990s1. They involve generation of cDNA templates by reverse transcription (RT)2,3 coupled with PCR amplification4. Nanopore RNA strand sequencing has emerged as an alternative single molecule strategy5,6,7. It differs from SBS-based platforms in that native RNA nucleotides, rather than copied DNA nucleotides, are identified as they thread through and touch a nanoscale sensor. Nanopore RNA strand sequencing shares the core features of nanopore DNA sequencing, i.e. a processive helicase motor regulates movement of a bound polynucleotide driven through a protein pore by an applied voltage. As the polynucleotide advances through the nanopore in single nucleotide steps, ionic current impedance reports on the structure and dynamics of nucleotides in or proximal to the channel as a function of time. This continuous ionic current series is converted into nucleotide sequence using an ONT neural network algorithm trained with known RNA molecules.
Here we describe sequencing and analysis of a human poly(A) transcriptome from the GM12878 cell line using the Oxford Nanopore (ONT) platform. We demonstrate that long native RNA reads allow for discovery and characterization of polyA RNA molecules that are difficult to observe using short read cDNA methods8,9. Data and resources are posted online at: (https://github.com/nanopore-wgs-consortium/NA12878/blob/master/RNA.md).
RESULTS
RNA preparation, nanopore sequencing, and computational pipeline
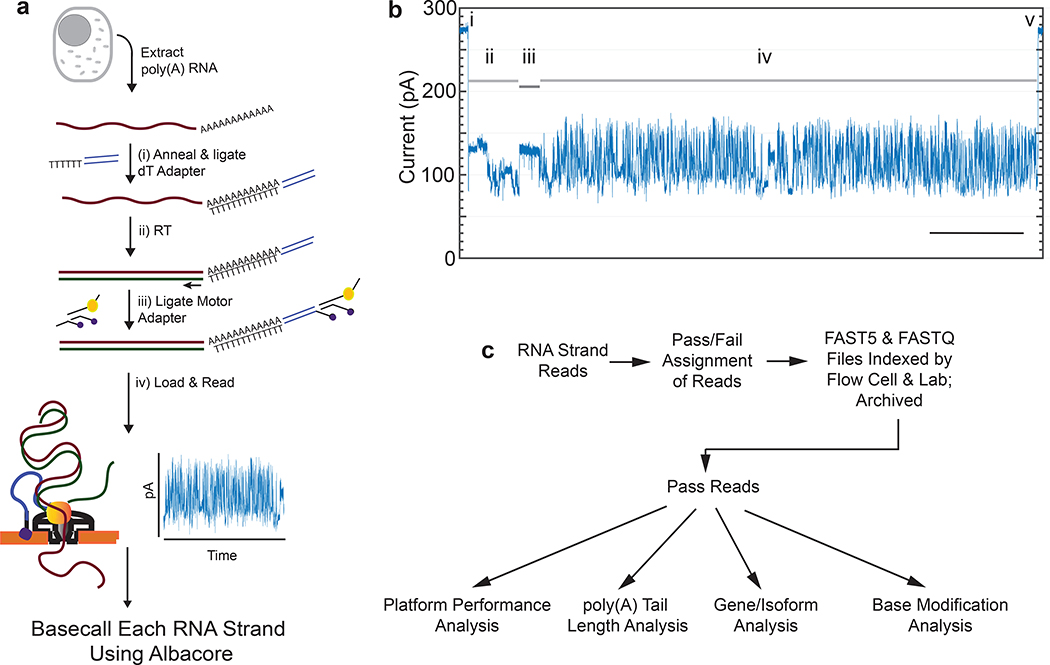
The protocol we used to isolate and sequence native poly(A) RNA from a human B-lymphocyte cell line (GM12878) is summarized in Figure 1a and detailed in Online Methods. A typical ionic current trace during TP53 mRNA translocation through a nanopore is shown in Figure 1b. The ionic current readout for each poly(A) RNA strand was basecalled using Albacore version 2.1.0 (ONT).
Figure 1.
Nanopore native poly(A) RNA sequencing pipeline. (a) RNA is isolated from cells followed by poly(A) selection using poly(dT) beads. Poly(A) RNA is then prepared for nanopore sequencing using the following steps: (i) A duplex adapter bearing a poly(dT) overhang is annealed to the RNA poly(A) tail, followed by ligation of the strand abutting the poly(A) tail; ii) the poly(dT) complement is extended by reverse transcription. This step improves throughput, but it is not necessary, and the cDNA strands are not read; iii) a proprietary ONT adapter bearing a motor enzyme is ligated to the first adapter; and (iv) the product is loaded onto the ONT flow cell for reading by ionic current impedance. The ionic current trace for each poly(A) RNA strand is base called using a proprietary ONT algorithm (Albacore). (b) A representative ionic current trace for a 2.3 kb TP3 transcript. Ionic current components: (i) Strand capture; ii) ONT adapter translocation; iii) poly(A) RNA tail translocation; iv) mRNA translocation; and (v) exit of the strand into the trans compartment. Bar is 5 seconds. (c) Processing of the RNA strand reads in silico, followed by data analysis.
We also performed nanopore cDNA sequencing using the identical GM12878 RNA sample and analysis pipeline, but with modified parameters appropriate for cDNA sequencing (Online Methods). Both the RNA and cDNA data were archived and used for downstream analyses (Figure 1c).
Native poly(A) RNA sequencing statistics
Six laboratories each performed five nanopore sequencing runs (Supplementary Table 1). These thirty runs produced 13.0 million poly(A) RNA strand reads, of which 10.3 million passed quality filters (PHRED>7). Throughput varied between 50K and 831K pass reads per flow cell, with an N50 length of 1,334 bases, and a median of 771 bases. Of these, 9.9 million aligned using minimap210 to the GRCh38 human genome reference. The 360,000 unaligned pass reads had a median read length of 211 bases.
We next aligned the RNA pass reads to the GENCODE v27 transcriptome reference using minimap210. The aligned reads ranged in length from 85 nt (a fragment of an mRNA encoding Ribosomal Protein RPL39), to 21kb (an mRNA encoding spectrin repeat containing nuclear envelope protein 2 (SYNE2)). A comprehensive list of the genes and isoforms can be found on GitHub and in Supplementary Tables 2 and 3 respectively.
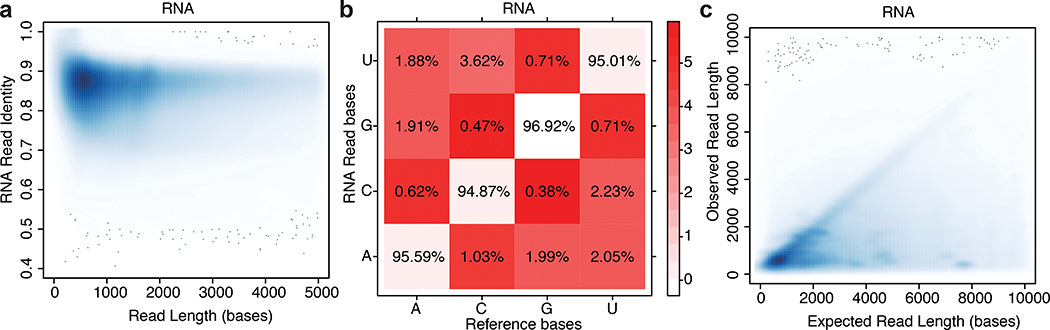
MarginStats (version 0.1)11 was employed to calculate percent identity and the number of matches, mismatches, and indels per aligned read in this population (Supplementary Table 4). Median identity was 86 +/− 0.86% (Figure 2a) and error profiles are given in Figure 2b. We compared the observed read length vs expected transcript length as defined by GENCODE v27, and found general agreement (Figure 2c). The discrete clusters below the diagonal represent incorrect assignments to GENCODE isoforms, and the diffuse shading represents fragmented RNA (see text below concerning RNA truncation).
Figure 2.
Performance statistics for nanopore native RNA sequencing. (a) Alignment identity vs. read length for native RNA reads. b) Substitution matrix for native RNA reads. The x-axis is the known base identity for the GENCODE v27 transcriptome at positions that aligned to nanopore reads. The y-axis is base identity at the same position for nanopore reads. The values within boxes are the percentage of times nanopore basecalls corresponded to correct (diagonal) or incorrect (red shaded) calls according to the reference. The color intensity in the boxes represents the negative natural log probability of basecall matches or mismatches (see color key at right). (c) Observed vs. expected read length for ~9.7 million native RNA reads. The discrete clusters below the diagonal represent incorrect assignments to GENCODE isoforms, and the diffuse shading represents fragmented RNA.
For nanopore cDNA data, we observed a median identity of 85% which is comparable to recent published nanopore DNA results12. The substitution error patterns for cDNA data were similar to those for native RNA data (not shown).
Kmer coverage
Previous Kmer analyses indicated that some nucleotide sequences are over- or under-represented in nanopore-based DNA sequence reads11,12. We assessed nanopore RNA and cDNA 5-mer coverage using reads aligned to GENCODE v27 isoforms. Only reads that covered 90% or more of a given reference sequence were chosen; this selected 2.9 million of the total 10.3 million RNA reads. Of the 15.1 million pass cDNA reads, 3.9 million pass cDNA reads were selected. These reads included all 1024 possible 5-mers (see Supplementary Figures 1a and 1b for normalized native RNA and cDNA counts respectively).
5-mers that were under-represented in native RNA and over-represented in cDNA are shown in Supplementary Tables 5 and 6 respectively. Similar to previous studies11,12, the largest deviation from expectation occurred for homopolymer-rich kmers.
Nanopore sequencing performance assessed using mitochondrially-encoded RNA
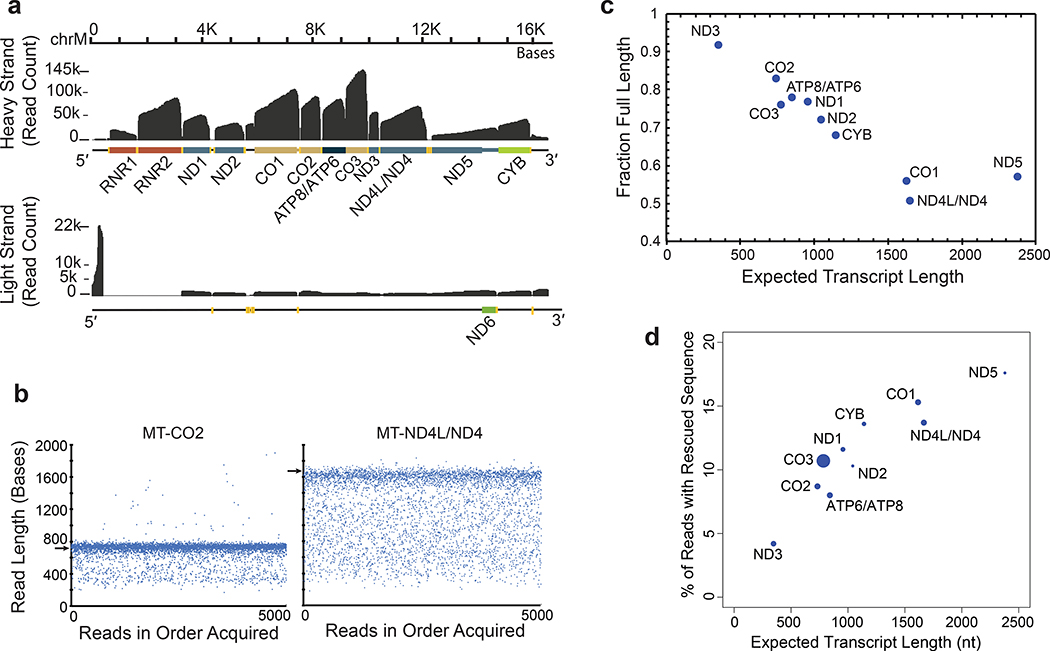
We reasoned that mitochondrial poly(A) transcripts could be used to benchmark nanopore sequencing performance because they are abundant in all human cells, are single exon, and vary substantially in length (349–2,379 nt). Approximately 10% (950,879) of reads aligned to the mitochondrial genome (Figure 3a and public UCSC track https://goo.gl/erWFyu). As expected, most of these poly(A) transcripts corresponded to mitochondrial ribosomal RNA or to mitochondrial mRNA. Overall, the nanopore RNA reads recapitulated known features of the human MT-transcriptome (Supplementary Figures 2–3). We also observed poly(A) RNA strands that are difficult to observe by conventional means (Supplementary Figures 4–5).
Figure 3.
Mitochondrially-encoded poly(A) RNA transcripts. (a) Read coverage of the H strand (top) and the L strand (bottom). Dark grey is base coverage along the MT genome. Labeled colored bars represent protein coding genes including known UTRs, or ribosomal RNA (RNR1, RNR2). Text denotes specific genes without the MT prefix. Yellow bars represent tRNA genes. (b) Distribution of nanopore read lengths for MT-CO2 and MT-ND4L/ND4 transcripts. Each point represents one of approximately 5000 reads in the order acquired from a single Lab 1 MinION experiment. Horizontal arrows are expected transcript read lengths. (c) Relationship between expected transcript read length and fraction of nanopore poly(A) RNA reads that were full length. Each point is for a protein coding transcript on the H strand. Labels are for mitochondrial genes without the MT prefix. See Online Methods for definition of ‘Full Length’. (d) Percent of artificially truncated strand reads where sequence was recovered from the ionic current signal. Points are for protein coding transcripts as in panel c.
MT-RNA read length analysis was revealing. Figure 3b shows 5,000 reads that aligned to MT-CO2 or to MT-ND4L/ND4 genes. In each panel, a dominant band corresponded closely to the expected transcript length (732 nt and 1,673 nt for MT-CO2 and MT-ND4L/ND4 respectively). However, for each of these, a population of truncated reads was randomly distributed between the dominant band and about 300 nt in length. When we quantified the fraction of truncated reads as a function of nominal transcript length for ten MT-mRNA of the heavy strand (Online Methods), we found a strong linear anti-correlation in most cases (Figure 3c). The single outlier was MT-ND5 which is the mitochondrial transcript with a 568 nt 3′ UTR.
These MT-poly(A) RNA truncations could occur at any of several non-biological steps during the sequencing process, or they could arise from regulated enzymatic degradation in the mitochondrion13. Here we considered three possible non-biological causes that were specific to the nanopore platform.
One systematic cause of read truncations occurred because the enzyme that controls translocation through the pore is 10–15 nt from the nanopore sensor. When the enzyme releases the last base at the 5′ end, the strand is rapidly driven through the pore which prevents reading the terminal 10–15 nt. This phenomenon was evident by close inspection of read coverage at the 5′ end of mitochondrial mRNA transcripts (https://goo.gl/erWFyu), and is expected for all direct RNA reads in the present ONT protocol.
Another possible cause was ionic current signal artifacts associated with enzyme stalls during RNA translocation, or with extraneous voltage spikes (Supplementary Figure 6a). Similar artifacts have been shown to disrupt strand reads during MinION sequencing of DNA 14. Systematic analysis of 2,729 MT-CO1 reads within bulk FAST5 files from Lab 1 identified 527 reads which started or ended abnormally (Online Methods). By including ionic current segments that were identified before or after many of these truncations, we reconstructed 300 reads with longer alignments to MT-CO1 (Supplementary Figure 6, Supplementary Table 7). This phenomenon was length dependent (Figure 3d), ranging from 4.2% of reads with rescued segments for ND3 (346 nt nominal length) to 17.6% for ND5 (2379 nt nominal length).
A third possible cause was strand breaks during nanopore sequencing runs. We analyzed MT-CO1 read-length distribution for each of the six laboratories as a function of time on ONT flow cells. We found that the read frequency at all lengths declined steadily over 36 hours as expected, however the full-length fraction declined by only 5% (Supplementary Figure 7). This analysis also revealed that RNA from Lab 6 had degraded prior to the sequencing run. Therefore, isoform-level analyses (see below) focused on 8.17 million aligned poly(A) RNA reads from Labs 1–5.
Isoform detection and analysis
Long nanopore reads could improve resolution of RNA exon-exon connectivity, allowing for discovery of unannotated RNA isoforms. However, these reads averaged 14% per-read basecall errors, confounding precise determination of splice sites. Also, biological RNA processing and in vitro 5′-end truncations (see above) can make it difficult to define transcription start sites (TSS).
To overcome these limitations we employed FLAIR23 (Full-Length Alternative Isoform Analysis of RNA, Online Methods). We first replaced any nanopore-based splice sites bearing apparent sequencing errors with splice sites supported by GENCODE v27 annotations or by Illumina GM12878 cDNA data (Supplementary Figure 8)15,16. Second, to overcome TSS uncertainty caused by truncated RNA reads, we considered only reads with 5′ ends proximal to promoter regions (defined by ENCODE promoter chromatin states for GM1287817–19). Third, we used FLAIR to group reads into isoforms according to chains of splice junctions.
We compiled two FLAIR isoform sets (Supplementary Table 8) using different supporting read criteria (see Online Methods, Supplementary Figure 9):
A FLAIR-sensitive set that included isoforms with three or more uniquely mapped reads (see GitHub link). This large set could be useful for isoform discovery, at the risk of false positives.
A FLAIR-stringent set that was compiled by filtering set (i) for isoforms having three or more supporting reads that spanned ≥80% of the isoform with ≥25nt coverage into the first and last exon.
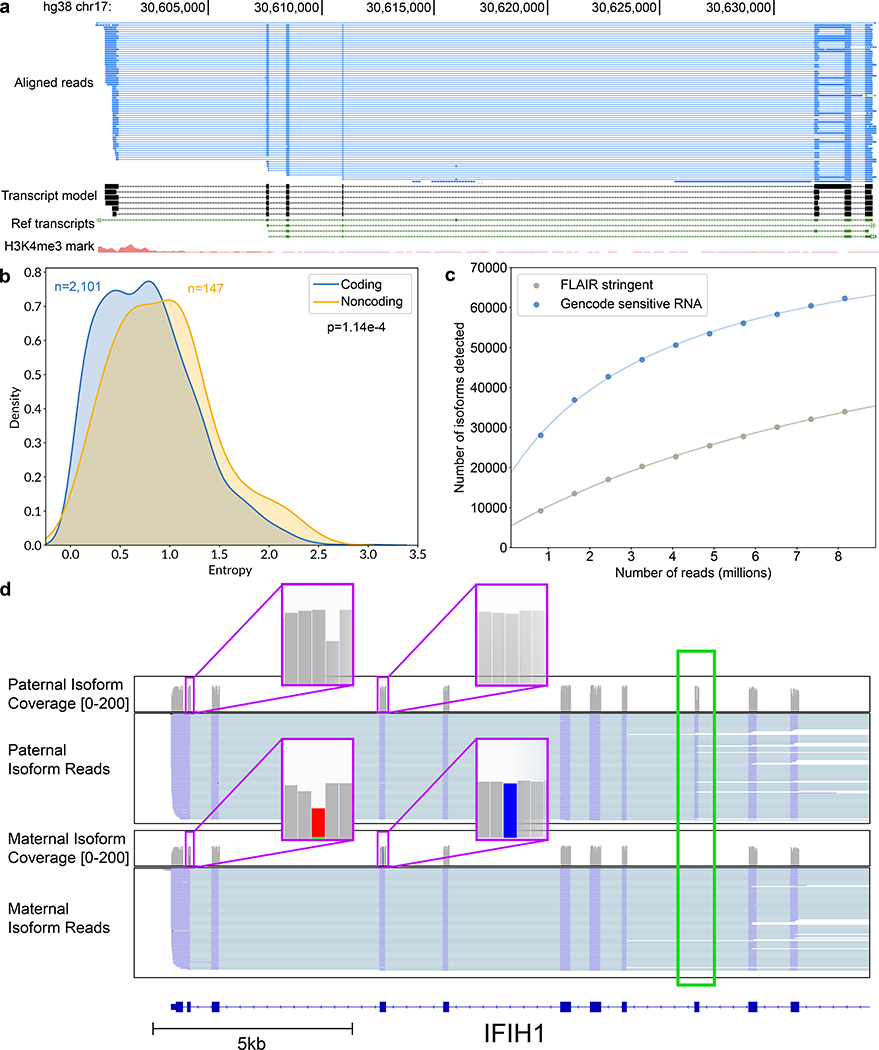
We screened for unannotated isoforms within the FLAIR-stringent dataset. Of the 33,984 isoforms representing 10,793 genes (Supplementary Table 9), 52.6% had a splice junction chain that was unannotated in GENCODE (13.0% of total assigned reads). Figure 4a shows an example set of lncRNA isoforms arising from an unannotated transcription start site with multiple splice variants. We observed that non-coding genes had more complex splicing patterns per gene than did coding genes (Figure 4b), in agreement with prior studies demonstrating increased alternative splicing in non-coding exons20,21.
Figure 4.
Isoform-level analysis of GM12878 native poly(A) RNA sequence reads. (a) Genome browser view of unannotated isoforms that aligned to SMURF2P1-LRRC37BP1. The tracks are: a subset of the aligned native RNA reads (blue); the FLAIR-defined isoforms (black); SMURF2P1-LRRC37BP1 annotated isoforms from GENCODE v27 comprehensive set (green); transcription regulatory histone methylation marks (red). (b) Shannon entropy of isoform expression for coding versus noncoding genes detected by FLAIR. Only genes with at least 50 reads and more than two isoforms were used. The p-value was calculated from a Mann-Whitney U test. (c) Saturation plot showing the number of isoforms discovered (y-axis) versus the number of native RNA reads (x-axis). (d) IGV view of allele-specific isoforms for IFIH1. Purple boxes (insets) indicate the location of SNPs used to assign allele specificity (gray reference; red and blue SNPs). The alternatively spliced exon is indicated by a green box.
As a conservative alternative to FLAIR, we compiled two GENCODE-based isoform sets (Supplementary Table 8):
A GENCODE-sensitive set that included isoforms with one or more reads that mapped uniquely to GENCODE v27. We implemented a lower coverage threshold than we did for FLAIR because GENCODE is curated.
A GENCODE-stringent set that was compiled by filtering set (iii) for isoforms having one or more supporting reads that spanned ≥80% of the isoform with ≥25nt coverage into the first and last exon.
To estimate the sequencing depth required to completely characterize the GM12878 transcriptome, we plotted the number of isoforms detected in the GENCODE-sensitive and FLAIR-stringent isoform sets versus the number of subsampled reads in 10% increments. We then fitted a hyperbolic function to the data (Figure 4c, Supplementary Figure 10, Supplementary Table 10). It is evident that the curves did not saturate and that additional reads would be required to capture a complete GM12878 transcriptome.
Assignment of transcripts to parental alleles
Allele-specific expression (ASE) is the preferential transcription of RNA from the paternal or maternal copy of a gene. Although the importance of this phenomenon has been characterized22, the consequences are not fully understood. This is partly due to technical limitations of haplotype identification using short read sequencing technologies.
We reasoned that the long nanopore RNA reads would be easier to assign to the parental allele of origin due to the greater chance of encountering a heterozygous SNP. Reads with at least two heterozygous SNPs were assigned to the parental allele of origin using HapCUT223. To discover the most possible genes, we used the FLAIR-sensitive data-set. In it, we found 3,751 genes with at least 10 haplotype informative reads. 3,707 of these genes were from autosomal chromosomes and 44 were from the X-chromosome (Supplementary Table 11). Among autosomal genes, 228 (6.1%) showed significant ASE (binomial test, p<0.001), and among X-chromosome genes, 23 (95.7%) showed significant ASE (binomial test, p<0.001). X-chromosome expression was biased, with 22/23 allele-specific X-linked genes originating from the maternal allele, consistent with previous results for this cell line24. The sole paternally expressed X-linked locus encoded the lncRNA XIST (Supplementary Figure 11), which is transcribed from the inactive X-chromosome and recruits epigenetic silencing machinery for X-inactivation in females25. The remaining genes were expressed equally from both parental alleles.
We combined these allele-specific reads with isoforms from the FLAIR-sensitive set to mine for allele-specificity (Online Methods). We identified 5 genes with one isoform expressed from one allele and another isoform expressed from the other allele (binomial test, P<0.001, Supplementary Table 12). One of these genes, IFIH1, had a paternal isoform with exon 8 retained, while the maternal isoform did not retain exon 8 (Figure 4d, Supplementary Figure 12). We note that the closest SNV used in allele-assignment was 886 nt away from the alternative splicing event in this transcript. This would be undetectable using short read sequencing.
3′ poly(A) analysis
Transcript poly(A) tails are thought to play a role in post-transcriptional regulation, including mRNA stability and translational efficiency26. However, these homopolymers can be several hundred nucleotides long making them difficult to measure using short-read SBS data27,28.
We measured poly(A) tail lengths directly using a low variance ionic current signal associated with the 3′ end of each poly(A) strand (Figure 1b, iii). We developed a computational method (‘nanopolish-polya’, https://github.com/jts/nanopolish) to segment this signal and estimate how many ionic current samples were drawn from the poly(A) tail region. By correcting for the rate at which the RNA molecule passes through the pore, nanopolish-polya estimates the length of the poly(A) tail. Algorithmic details can be found in Supplementary Note 1.
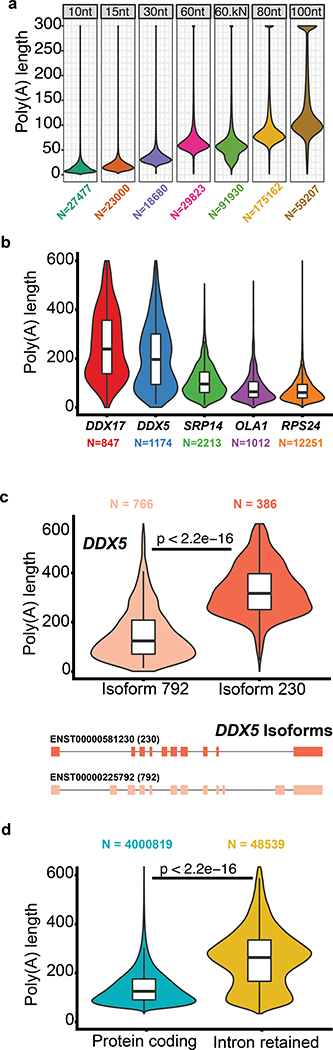
To test this method we obtained six MinION-derived poly(A) RNA control datasets generated by ONT (ENA accession PRJEB28423). These datasets consisted of ionic current traces for synthetic S. cerevisiae enolase transcripts appended with 3′ poly(A) tails of 10, 15, 30, 60, 80 or 100 nucleotides. A second version of the 60nt poly(A) tailed construct (60nt-kN) contained a 10nt randomer between the enolase sequence and the 3′ poly(A). Poly(A) tail length estimates for these synthetic controls are shown in Figure 5a (see Supplementary Table 13 for statistics). For algorithmic details and discussion on the poly(A) estimator, see Supplementary Note 1.
Figure 5.
Testing and implementation of the poly(A) tail length estimator nanopolish-polya. (a) Estimate of poly(A) lengths for a synthetic enolase control transcript bearing 3′ poly(A) tails of 10, 15, 30, 60, 80 or 100 nucleotides. ‘60kN’ contained all possible combinations of a 10nt random sequence inserted between the enolase sequence and the 3′ poly(A) 60mer. (b) Violin/box plots showing poly(A) tail length distributions for genes with the longest (DDX5, DDX17), median (SRP14), and shortest (RPS24, OLA1) values from a ranked list of 1043 genes. (c) Distribution of poly(A) tail lengths and gene models for two isoforms of DDX5. (d) Distribution of poly(A) tail lengths for representative intron-retaining and intron-free transcripts identified using the GENCODE-Sensitive isoform set. Kruskal-Wallis p-value are denoted.
We applied this poly(A) length estimator to the complete GM12878 native poly(A) RNA sequence dataset. Overall, the poly(A) length distribution centered at ~50nt, with mitochondrial transcripts averaging at 52nt and almost no poly(A) tail lengths greater than 100nt. This is consistent with results for mitochondrial poly(A) RNA from other human cell lines29. Conversely, nuclear transcripts showed a broader length distribution, with a peak at 58nt, a mean of 112nt, and a large number of poly (A) tails greater than 200nt.
Next, we measured poly(A) tail length differences between genes with at least 500 reads (Supplementary Table 14). Figure 5b shows poly(A) tail length distributions for representatives from a list of 1043 genes ranked by median values. For some genes, e.g. the RNA-binding protein DDX5, multiple peaks were observed (Figure 5b), suggesting the presence of isoform-specific poly(A) tail-length sub-populations. To explore this, we analyzed genes in the GENCODE-sensitive dataset, and found 215 genes that had isoforms with significantly different poly(A) lengths (Supplementary Figure 13).
When we compared two GENCODE isoforms of DDX5, we noted that an intron-retaining isoform (ENST00000581230, ‘230’) had a median poly(A) tail length of 327nt, compared with the protein-coding isoform (ENST00000225792,’792’), which had a median poly(A) tail length of 125nt. (Figure 5c). This difference motivated us to explore the relationship between poly(A) tail length and RNA intron-retention. We classified each isoform in GENCODE-sensitive as either protein-coding or intron-retaining. The subset of transcripts with retained introns tended to have longer poly(A) tails (median 232nt) than did transcripts without introns (median 91nt) (t-test p-value < 2.2e-16, Figure 5d).
Modification detection
Nanopore sequencing has been used to identify base modifications in DNA30,31 and RNA5,7. N6-methyladenine (m6A) is the most common internal modification on mRNA32, and has been implicated in many facets of RNA metabolism33. m6A dysregulation has been linked to human diseases, including obesity and cancer34. Because m6A modifications are enriched in 3′ UTRs, with two-thirds of these containing miRNA sites35, the impact of this modification appears to be largely regulatory, as opposed to altering protein coding sequence.
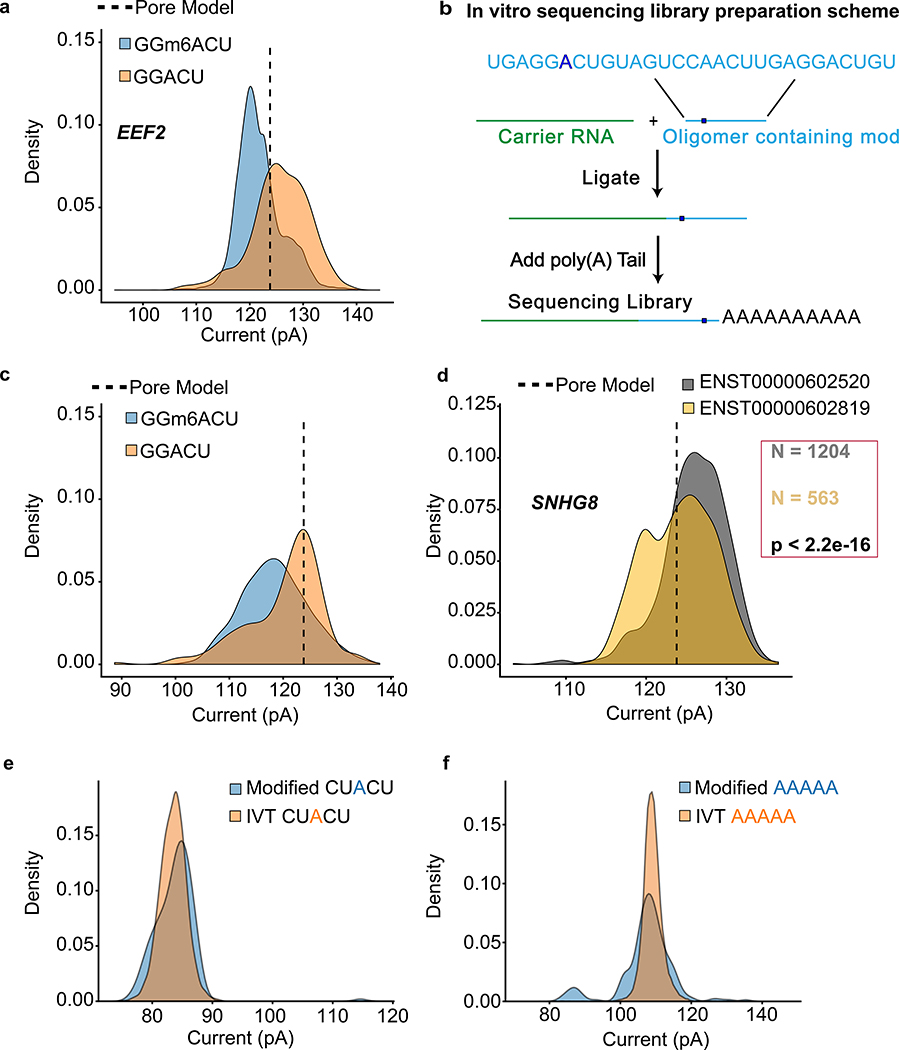
We focused our studies on the GGACU binding motif of METTL3, a subunit of the m6A methyltransferase complex36. As an example, we compared the raw current signal at a putative m6A site (chr19:3976327) in eukaryotic elongation factor 2 (EEF2) versus the signal for an in vitro transcribed copy (Online Methods). This comparison revealed an ionic current change attributable to m6A (Figure 6a). To validate this result, we used synthetic oligomers that were identical except for the presence or absence of m6A within the GGACU motif (Figure 6b). This revealed a clear current difference (Figure 6c) consistent with the EEF2 result.
Figure 6.
Nanopore detection of m6A and inosine base modifications. (a) Comparing current signal from m6A-modified and unmodified GGACU motifs in the native RNA dataset for EEF2 and in vitro transcribed dataset. Pore model (indicated by a dashed line) is defined as the mean current amplitude (pA) for the canonical GGACU 5-mer in the ONT model. (b) Schematic for the oligomer-ligation. A synthetic RNA oligomer (Trilink Biotechnologies) containing canonical and modified m6A bearing GGACU 5-mer was ligated to a carrier RNA. This was followed by in vitro polyadenylation. (c) Comparison of ionic current signals for m6A-modified and canonical GGACU motifs. The data were acquired using the assay described in (b). (d) Ionic current distributions for GGACU motifs within SNHG8 gene isoforms (see gene models in Supplemental Figure 7). (e) Ionic current distributions for putative inosine-bearing CUACU 5-mer in the 3′-UTR region of the AHR gene. Blue is native RNA and orange is IVT RNA. (f) Ionic current distributions for putative inosine-bearing AAAAA 5-mer in the 3′-UTR region of the AHR gene. Blue is native RNA and orange is IVT RNA.
To determine if m6A modifications differed between isoforms of the same gene, we screened GENCODE-sensitive isoforms for ionic current changes at the GGACU motif. We found 86 genes (198 isoforms) where the median current levels at a single GGACU were significantly different between gene isoforms (Kruskal-Wallis, Student’s t-test, and Kolmogorov-Smirnov statistical testing with Bonferroni multiple testing correction). An example is illustrated for the SNHG8 gene (Figure 6d, isoform models in Supplementary Figure 14).
Another post-transcriptional modification, A-to-I RNA editing37, plays a role in splicing and regulating innate immunity38,39. NGS detects A-to-I editing as an A-to-G nucleotide variant in cDNA sequences.
Previous nanopore experiments documented the presence of systematic base miscalls in regions of E. coli 16S rRNA bearing modified RNA bases7. We found systematic base miscalls at putative inosine bearing positions in the GM12878 aryl hydrocarbon receptor (AHR) data (Supplementary Figure 15). To cross-validate, we compared our cDNA sequence data relative to the GM12878 reference and found that putative inosines were detected as an A-to-G base change as expected (i.e. a single inosine for the CUACU 5-mer, and multiple inosines for the AAAAA 5-mer).
The ionic current distribution for the putative single inosine 5-mer (CUACU) was modestly different from the canonical 5-mer (Figure 6e). The ionic current distribution for the inosine containing AAAAA 5-mer was more complex, possibly reflecting the presence of multiple inosines (Figure 6f).
DISCUSSION
Nanopore RNA sequencing has two useful features: 1) The sequence composition of each strand is read as it existed in the cell. This permits direct detection of post-transcriptional modifications including nucleotide alterations and polyadenylation; 2) reads can be continuous over many thousands of nucleotides providing splice-variant and haplotype phasing. Although each of these features is useful in itself, the combination is unique and likely to provide new insights into RNA biology. The two principal drawbacks of the present ONT nanopore RNA sequencing platform is the relatively high error rate (compared to Illumina cDNA sequencing), and uncertainty about the 5’ end of the transcript.
We were concerned about read fragmentation due to RNA degradation during sequencing. However, we found minimal (~5%) reduction in the full-length fraction of a 1.6 kb mRNA (MT-CO1) over 36 hours. Preliminary analysis indicated that read truncations were more often caused by electronic signal noise due to current spikes of unknown origin. We showed that meaningful biological signals can be recovered from bulk Fast5 files around these truncations, suggesting that future improvements to the MinKNOW read segmentation pipeline are needed.
When combined with more accurate short Illumina reads, long nanopore reads allowed for end-to-end documentation of RNA transcripts bearing numerous splice junctions, which would not be possible using either platform alone. We documented a high proportion (52.6%) of unannotated isoforms, similar to other long-read transcriptome sequencing studies (e.g., 35.6% and 49%)40,41. While many of these unannotated isoforms are low abundance and their protein coding potential unknown, it is important to catalog them because subtle splicing changes can impact function42,43. We also note that the number of detected isoforms did not saturate using the nanopore poly(A) RNA dataset, indicating that greater sequence depth will be necessary to give a comprehensive picture of the GM12878 poly(A) transcriptome.
A variety of techniques have been used to examine allele-specific expression (ASE)15,24. However, identification of ASE is limited using short read platforms because heterozygous variants are rare within any given window of a few hundred nucleotides. Nanopore sequencing has the advantage of long reads, albeit limited by errors. We attempted to mitigate the effects of these errors by requiring multiple heterozygous variants and a stringent false-discovery rate (FDR) during ASE analysis. Therefore, the number of genes that we report as demonstrating ASE (167) is likely an underestimation. We report nearly exclusive use of the maternal X-chromosome, with the only paternal transcripts originating from the XIST locus, consistent with previous findings24. We have shown that nanopore sequencing enables allele-specific isoform studies, especially in cases where the splicing variation does not have a heterozygous variant within range of conventional short-read sequencing.
Polyadenylation of RNA 3′ ends regulates RNA stability and translation efficiency by modulating RNA-protein binding and RNA structure26. However, transcriptome-wide poly(A) analysis has been difficult due to basecalling and dephasing errors28. Recently implemented modifications to the Illumina strategy address these limitations28,27; but can not resolve distal relationships, such as between splicing and poly(A) length. Nanopore poly(A) tail length estimation using nanopolish-polya offers the advantages of both direct length assessment and maintenance of information about isoform and modification status per transcript. Our preliminary studies revealed differences in poly(A) length distribution between mitochondrial and nuclear genes, between different nuclear genes, and between different isoforms of the same gene. We note in particular an increase in poly(A) tail length for some intron-retaining isoforms. This is consistent with previous work showing that hyper-adenylation targets intron-retaining nuclear transcripts for degradation through recognition by a poly(A)-binding protein (PABPN1)44. Additionally, deadenylation of cytoplasmic transcripts is a core part of the RNA degradation pathway45, suggesting that time course experiments investigating RNA decay kinetics46 could be possible with this technology.
We have demonstrated detection of N6-methyladenosine and inosine modifications in human poly(A) RNA. This validates prior work which showed modification-dependent ionic current shifts associated with m6A (S. cerevisiae)5. Differences in m6A modification level proved to be discernible at the isoform level for human SNGH8 mRNA (Figure 6d), documenting splicing variation and modification changes simultaneously.
Although other methods exist for high throughput analysis of RNA modifications47, they often require enrichment which limits quantification, and they are usually short-read based. The latter precludes analysis of long-distance interactions between modifications, and between modifications and other RNA features such as splicing and poly(A) tail length. The capacity to detect these long-range interactions is likely to be important given recent work suggesting links between RNA modifications, splicing regulation, and RNA transport and lifetime48,49. We argue that nanopore native RNA sequencing could deliver this long-range information for entire transcriptomes. However, this will require algorithms trained on large, cross-validated datasets as has been accomplished for cytosine and adenine methylation in genomic DNA30,31.
ONLINE METHODS
Unless otherwise noted, kit based protocols described below followed the manufacturer’s instructions.
GM12878 cell tissue culture
GM12878 cells (passage 4) were received from the Coriell Institute and cultured in RPMI media (Invitrogen cat# 21870076) supplemented with 15% non heat-inactivated FBS (Lifetech cat# 12483020) and 2mM L-Glutamax (Lifetech cat# 35050061). Cells were grown to a density of 1 × 106 / ml before subsequent dilution of ⅓ every ~3 days and expanded to 9 x T75 flasks (45 ml of media in each). Cells were centrifuged for 10 min at 100 x g (4°C), washed in 1/10th volume of PBS (pH 7.4) and combined for homogeneity. The cells were then evenly split between 8 × 15ml tubes and pelleted at 100g for 10 mins at 4°C. The cell pellets were then snap frozen in liquid nitrogen and immediately stored at −80°C before shipping on dry ice. Two tubes of 5 × 107 frozen GM12878 cell pellets from passage 10 from a single passage, cultured at UBC, were distributed and used at UBC, OICR, JHU, and UCSC. Two tubes of cells from passage 11 were distributed to UoN from UBC, and an independently cultured passage of GM12878 was used at UoB. (University of British Columbia (UBC), University of Birmingham (UoB), Ontario Institute of Cancer Research (OICR), Johns Hopkins University (JHU), University of Nottingham (UoN), and University of California Santa Cruz (UCSC))
Total RNA Isolation
The following protocol was used by each of the six institutions. Four ml of TRI-Reagent (Invitrogen AM9738) was added to a frozen pellet of 5 × 107 GM12878 cells and vortexed immediately. This sample was incubated at room temperature for 5 minutes. Four hundred μl BCP (1-Bromo-3-chloro-propane) or 200 μl CHCl3 (Chloroform) was added per ml of sample, vortexed, incubated at room temperature for 5 minutes, vortexed again, and centrifuged for 10 minutes at 12,000g (4°C). The aqueous phase was pooled in a LoBind Eppendorf tube and combined with an equal volume of isopropanol. The tube was mixed, incubated at room temperature for 15 minutes, and centrifuged for 15 minutes at 12,000g (4°C). The supernatant was removed, the RNA pellet was washed with 750 μl 80% ethanol and then centrifuged for 5 minutes at 12,000g (4°C). The supernatant was removed. The pellet was air-dried for 10 minutes, resuspended in nuclease free water (100 μl final volume), quantified, and either stored at −80°C or processed further by poly(A) purification.
Poly(A) RNA isolation
One hundred μg aliquots of total RNA were diluted in 100 μl of nuclease free water and poly(A) selected using NEXTflex Poly(A) Beads (BIOO Scientific Cat#NOVA-512980). Resulting poly(A) RNA was eluted in nuclease free water and stored at −80°C.
MinION native RNA sequencing of GM12878 poly(A) RNA
Biological poly(A) RNA (500–775 ng) and a synthetic control (Lexogen SIRV Set 3, 5 ng) were prepared for nanopore direct RNA sequencing generally following the ONT SQK-RNA001 kit protocol, including the optional reverse transcription step recommended by ONT. One difference from the standard ONT protocol was in the use of Superscript IV (Thermo Fisher) for reverse transcription. RNA sequencing on the MinION and GridION platforms was performed using ONT R9.4 flow cells and the standard MinKNOW (version 1.7.14) protocol script (NC_48Hr_sequencing_FLO-MIN106_SQK-RNA001) recommended by ONT, with one exception, i.e. we restarted the sequencing runs at several time points to improve active pore counts and throughput during the first 24hrs.
cDNA synthesis
First strand cDNA synthesis was performed using Superscript IV (Thermo Fisher) and 100 ng of poly(A) purified RNA. Reverse transcription and strand-switching primers were provided by ONT in the SQK-PCS108 kit. After reverse transcription, PCR was performed using LongAmp Taq Master Mix (NEB) under the following conditions: 95°C for 30 seconds, 11–15 cycles (95°C for 15 seconds, 62°C for 15 seconds, 65°C for 15 minutes), 65°C for 15 minutes, hold at 4°C. The 15 cycle PCR was performed when using the SQK-PCS108 kit and 11 cycle PCR was performed when using the SQK-LSK308 kit. PCR products were purified using 0.8X AMPure XP beads.
MinION sequencing of GM12878 cDNA
cDNA sequencing libraries were prepared using 1 μg of cDNA following the standard ONT protocol for SQK-PCS108 (1D sequencing) or SQK-LSK308 (1D^2 sequencing) with one exception. That is, we used 0.8X aAMPure XP beads for cleanup. We used standard ONT MinKNOW scripts for MinION sequencing with one exception. That is, we restarted the sequencing runs at several time points to improve active pore counts and throughput during the first 24 hours.
Acquiring continuous data for nanopore sequencing runs and resegmenting reads
For a subset of runs, “bulk FAST5 files” containing continuous raw current traces and read decisions made by MinKNOW were recorded for more detailed analysis. This can be enabled in MinKNOW by looking at “Additional options” under “Output” when configuring a run to start in MinKNOW. Options were set to capture raw signal data and the read table. Events were not captured to reduce file size14. Bulk FAST5 files were investigated using BulkVis14 and scripts available on GitHub (https://github.com/nanopore-wgs-consortium/NA12878/tree/master/nanopore-human-transcriptome/scripts/bulk_signal_read_correction). To identify reads with abnormal start or ends the read classifications made by MinKNOW in the 2 seconds before and after each read start or end respectively. Read starts should include ‘pore’, ‘good_single’, ‘inrange’ or ‘unblocking’ classifications14. Read ends should also end with these categories. Reads which did not start or end with these classifications were considered as potentially abnormal. Additional signal before and after the read was extracted from the bulk FAST5 file and a new synthetic read created for base calling (using Albacore version 2.1.3). For abnormal read starts, signal up to the start of the previous read was prepended. For abnormal read ends, signal up to the start of the following read was appended. Base calling is disrupted by signal incorrectly classified as open pore. Therefore these incorrect signal chunks were replaced with signal matching the mean for each read to generate a corrected read. These reads were recalled and mapped against the candidate targets using minimap2 with standard ONT parameters. This method can result in incorrectly concatenated reads and so mapping to the target was used to filter out such sequences. The difference in target coverage for each read was used to indicate recovery of sequence data as summarised in Supplementary Figure 7 and Supplementary Table 7. All corrected read files, basecalls, mapping files and scripts used to generate them are available on GitHub (link cited above).
Length analysis of mitochondrial protein-coding transcripts
In this analysis, we limited the test population for each gene to reads that aligned to a 50 nt sequence at the 3′ prime end of its ORF, except for MT-ND5 where alignment was to a 50 nt sequence at the end of its 568 nt 3′ UTR. Full length was defined as extending to at least within 25 nt of the genes expected 5′ terminus. This limit was chosen because the processive enzyme that regulates RNA translocation is distal from the CsgG nanopore limiting aperture and necessarily falls off before the 5′ end is read. The sharpest coverage drop-off is typically at 10 nt from the 5′ transcript end; we chose the 25 nt limit to ensure that all likely full length reads were captured in the count.
In vitro transcription
cDNA synthesis was performed according to ONT instructions (SQK-PCS108 kit) by combining Superscript IV (Thermo Fisher), RT and ONT strand switching primers, and 100 ng of poly(A) purified RNA. Next, an 11 cycle PCR reaction was performed using the ONT SQK-LSK308 kit but with a modified version of the primer that included a T7 promoter as recommended by NEB (Catalog number E2040S). The PCR reaction was run under the following conditions: 95°C for 30 seconds, 11 cycles (95°C for 15 seconds, 62°C for 15 seconds, 65°C for 15 minutes), 65°C for 15 minutes, hold at 4°C.
PCR products were purified using 0.8X AMPure XP beads. Next, in vitro transcription was performed using the NEB HiScribe T7 High Yield RNA Synthesis Kit following NEB instructions. The IVT product was poly(A) tailed using the same kit. The resulting IVT RNA was purified using LiCl precipitation and then adapted for RNA sequencing on the MinION the using SQK-RNA001 kit.
Oligomer Ligation
The oligomer containing the N6-methyladenosine modification was obtained as a lyophilized pellet from Trilink BioTechnologies and resuspended to 20 μM using TE buffer (Quality Biological Cat#351–011-721). The firefly luciferase (FLuc) transcript used as the carrier molecule was produced by in vitro transcription using the HiScribe™ ARCA mRNA Kit (with tailing) (NEB Cat#E2060) and supplied protocol with the following exception: after DNase treatment, the reaction was terminated and the RNA purified using 1X Agencourt RNAClean XP beads (Beckman Coulter A63987). The oligomer was then treated with T4 polynucleotide kinase (PNK) (NEB Cat#M0201) to phosphorylate the 5′ end for ligation. After phosphorylation, the oligomer was purified using the Oligo Clean & Concentrator kit (Zymo Research Cat#D4060). The phosphorylated oligomer and FLuc transcript were quantified, combined in equimolar amounts, and ligated using T4 RNA Ligase 1 (NEB Cat#M0204). The reaction mixture was incubated at 16°C overnight. After incubation, the RNA was purified using RNAClean XP beads. The ligated product was poly(A) tailed using E. coli Poly(A) Polymerase (NEB HiScribe™ ARCA mRNA Kit) according to the supplier’s instructions. After A-tailing, the RNA was purified using RNAClean XP beads. The isolated RNA was poly(A) selected using NEXTflex Poly(A) Beads. The resulting poly(A) RNA was eluted in nuclease free water and immediately prepared for sequencing using Oxford Nanopore’s direct RNA sequencing kit (SQK-RNA001) and protocol.
Basecalling, alignments, and percent identity calculations
We used the ONT Albacore workflow (version 2.1.0) for basecalling direct RNA and cDNA data. A strand read with an average sequence quality of 7 or higher (Q7) was classified as pass (default setting for Albacore (version 2.1.0)). We used minimap2 version 2.110 (recommended parameters i.e. -ax splice -uf -k14 for alignments to the human genome and -ax map-ont for alignments to the human transcriptome) to align the nanopore RNA and cDNA reads to the GRCh38 human genome reference50 and to the GENCODE v27 transcriptome reference51. This algorithm was chosen because it aligns nanopore reads to exons while spanning across introns52. We used marginStats (version 0.1)53 to calculate alignment identities and errors for pass RNA strand reads and pass 1D cDNA strand reads. Substitutions were calculated using custom scripts available within marginAlign (version 0.1)11.
Kmer analysis
We assessed nanopore RNA and cDNA 5-mer coverage using GENCODE isoforms. The read sequences were filtered by length and only reads covering 90% or more of the respective reference sequence were chosen. We calculated expected 5-mer counts from the set of reference sequences and observed 5-mer counts from the set of read sequences. For plotting purposes, we normalized the read and reference counts to coverage per megabase. The scripts are available within marginAlign11.
Isoform detection and characterization
To define isoforms from the sets of native RNA and cDNA reads, we used FLAIR v1.4, a version of FLAIR52 with additional considerations for native RNA nanopore data. For our analysis, we first removed reads generated by lab 6, because a disproportionate number of those molecules appeared to be truncated prior to addition to the nanopore flow cell. We also removed 71,276 aligned reads with deletions greater than 100 bases caused by minimap2 version 2.1. We then selected reads that had TSSs within promoter regions that were computationally derived from ENCODE ChIP-Seq data18,19. Using FLAIR-correct, we corrected primary genomic alignments for pass reads based on splice junction evidence from GENCODE v27 annotations and Illumina short-read sequencing of GM12878. This step also removes reads containing non-canonical splice junctions not present in the annotation or short-read data. The filtered and corrected reads were then processed by FLAIR-collapse which generates a first-pass isoform set by grouping reads on their splice junctions chains. Next, pass reads were realigned to the first-pass isoform set, retaining alignments with MAPQ>0. Isoforms with fewer than 3 supporting reads or those which were subsets of a longer isoform were filtered out to compile the FLAIR-sensitive isoform set. A FLAIR-stringent isoform set was also compiled by filtering the FLAIR-sensitive set for isoforms which had 3 supporting reads that spanned ≥80% of the isoform and a minimum of 25nt into the first and last exons. Unannotated isoforms were defined as those with a unique splice junction chain not found in GENCODE v27. Isoforms were considered intron-retaining if they contained an exon which completely spanned another isoform’s splice junction. Isoforms with unannotated exons were defined as those with at least one exon that did not overlap any existing annotated exons in GENCODE v27. Genes that did not contain an annotated start codon were considered non-coding genes.
Defining promoter regions in GM12878 for isoform filtering
Promoter chromatin states for GM12878 were downloaded from the UCSC Genome Browser in BED format from the hg18 genome reference. Chromatin states were derived from an HMM based on ENCODE ChIP-Seq data of nine factors18,19. The liftover tool54 was used to convert hg18 coordinates to hg38. The active, weak, and poised promoter states were used.
Haplotype Assignment and Allele-Specific Analysis
We obtained genotype information for GM12878 from existing phased Illumina platinum genome data generated by deep sequencing of the cell donors’ familial trio55. The bcftools package was used to filter for only variants that are heterozygous in GM12878. Starting with aligned reads, we used the extractHAIRS utility of the haplotype-sensitive assembler HapCUT223 to identify reads with allele-informative variants. For allelic assignment, we required a read to contain at least two variants, and required that greater than 75% of identified variants agreed on the parental allele of origin -- this stringent threshold was selected to reduce the chances of incorrect assignment from nanopore sequencing errors. Through this approach, each read was annotated as maternal, paternal or unassigned. To identify genes that demonstrated a very strong bias for a single allele, we performed a binomial test of all reads assigned to a parental allele, with an FDR of 0.001. We also visually inspected numerous genes displaying genes demonstrating allele-specificity using IGV, to increase our confidence in proper mapping of the reads and evaluate the presence of variants.
We further integrated this haplotype-specific analysis with our isoform pipeline to explore for the presence of allele-specific isoforms. If reads for a specific isoform originated from a single parental allele (binomial test, FDR 0.001), the isoform was assigned as allele specific. We then filtered for any genes which contained both maternal and paternal allele-specific isoforms, and visually inspected these isoforms using IGV to compare location of variants and splicing events.
Poly(A) tail length analysis
Supplemental Note 1 describes use of nanopolish-polya version 0.10.2 (https://github.com/jts/nanopolish) to estimate polyadenylated tail lengths of nanopore native RNA sequence reads. We used the Kruskal-Wallis test as implemented in Python to determine statistically significant changes between isoforms; code is available at [ https://github.com/nanopore-wgs-consortium/NA12878/tree/master/nanopore-human-transcriptome/scripts ]
Modification detection and analysis
We focused our initial efforts on m6A modification in genes previously identified as enriched in modifications from m6A immunoprecipitation sequencing data on human cell lines36,56. We aligned native RNA reads and IVT RNA reads to candidate genes and then extracted ionic current information (mean current and standard deviation in pA) for specific 5-mers using nanopolish eventalign (version 0.10.2). We compared ionic current kernel density estimates (KDE) for GGACU within the 3′ UTR of the EEF2 gene in native RNA with the KDE for its canonical IVT RNA counterpart. The extent and directionality of current shifts observed by m6A modification within the GGACU motif were orthogonally investigated using an in-vitro oligomer ligation assay, as described above. We compared KDEs for the modified and unmodified GGACU motifs within the synthetic oligomer. Statistical testing (Kruskal-Wallis, Student’s t-test, Kolmogorov-Smirnov and Bonferroni correction) was implemented in Python with code available at [https://github.com/nanopore-wgs-consortium/NA12878/tree/master/nanopore-human-transcriptome/scripts].
For detecting A-to-I editing, we focused on the 3′-UTR region of the human aryl hydrocarbon receptor (AHR) gene. Using the UCSC Genome Browser, we identified systematic G base variant calls in AHR cDNA data (probable inosine substitutions in RNA). We then tested for systematic base miscalls at the corresponding positions in native RNA data. Next, we used nanopolish eventalign (version 0.10.2) to extract ionic current information for two putative inosine-containing 5-mers (CUACU and AAAAA), and for their respective IVT-derived canonical 5-mers from chromosome 7. Ionic current distributions for CUACU and AAAAA 5-mers between the biological and IVT data were compared using kernel density estimates.
DATA AVAILABILITY
Sequence data including raw signal files (FAST5), event-level data (FAST5), base-calls (FASTQ) and alignments (BAM) are available as an Amazon Web Services Open Data set for download from https://github.com/nanopore-wgs-consortium/NA12878. The scripts used for various analyses are also available from the same GitHub under nanopore-human-transcriptome/scripts.
CODE AVAILABILITY
General scripts available at: https://github.com/nanopore-wgs-consortium/NA12878/tree/master/nanopore-human-transcriptome/scripts. Poly(A) caller (‘nanopolish-polya’, https://github.com/jts/nanopolish) and isoform analysis code for FLAIR (https://github.com/BrooksLabUCSC/flair).
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful for support from the following individuals. Libby Snell, Botond Sipos and Dan Turner (ONT) provided materials and advice relevant to the 3′ poly(A) standards used to test nanopolish-polya. Daniel Garalde (ONT) provided early advice on use of the MinION for RNA sequencing. Nicholas Conrad gave insight into the correlation of intron retention and poly(A) tail length. Mark Diekhans reviewed the isoform analysis. Zofia M. Chrzanowska-Lightowlers, Tom Suzuki, and Shunpei Okada commented on early drafts of the manuscript. Andrew Beggs, Louise Tee and Tom Nieto (University of Birmingham, UK) provided cell cultures used in the Birmingham sequencing runs. The project was supported by the following grants: NIH HG010053 (ANB, BP, & MA), NIH 5T32HG008345 (ADT), NIH HG010538 (WT), NIH U54HG007990 (BP), U01 HL137183-02 (BP), Oxford Nanopore Research Grant SC20130149 (MA), National Institutes of Health Research Surgical Reconstruction and Microbiology Research Centre (JQ), Medical Research Council CLIMB Fellowship (NL), Wellcome Trust 204843/Z/16/Z (ML), BBSRC BB/N017099/1 and BB/M020061/1 (ML), the Canada Research Chair in Biotechnology and Genomics-Neurobiology (TPS), the Canadian Institutes of Health Research (#10677; TPS), the Canadian Epigenetics, Environment and Health Research Consortium (TPS), the Koerner Foundation (TPS), the Ontario Institute for Cancer Research through funds provided by the Government of Ontario (JTS).
MA holds options in Oxford Nanopore Technologies (ONT). MA is a paid consultant to ONT. REW, WT, TG, JRT, JQ, NJL, JTS, NS, ANB, MA, HEO, MJ, and ML received reimbursement for travel, accommodation and conference fees to speak at events organised by ONT. NL has received an honorarium to speak at an ONT company meeting. WT has two patents (8,748,091 and 8,394,584) licensed to Oxford Nanopore. MA is an inventor on 11 UC patents licensed to ONT (6,267,872, 6,465,193, 6,746,594, 6,936,433, 7,060,50, 8,500,982, 8,679,747, 9,481,908, 9,797,013, 10,059,988, and 10,081,835). JTS, ML, and MA received research funding from ONT.
Footnotes
COMPETING FINANCIAL INTERESTS
REFERENCES
- 1.Complementary DNA sequencing: expressed sequenced tags and human genome project M.D. Adams et al. Science 252, 1651–1656. Trends Genet. 7, 281 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Temin HM & Mizutani S RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 226, 1211–1213 (1970). [DOI] [PubMed] [Google Scholar]
- 3.Baltimore D Viral RNA-dependent DNA Polymerase: RNA-dependent DNA Polymerase in Virions of RNA Tumour Viruses. Nature 226, 1209 (1970). [DOI] [PubMed] [Google Scholar]
- 4.Saiki RK et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239, 487–491 (1988). [DOI] [PubMed] [Google Scholar]
- 5.Garalde DR et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods (2018). doi: 10.1038/nmeth.4577 [DOI] [PubMed] [Google Scholar]
- 6.Jenjaroenpun P et al. Complete genomic and transcriptional landscape analysis using third-generation sequencing: a case study of Saccharomyces cerevisiae CEN.PK113–7D. Nucleic Acids Res. (2018). doi: 10.1093/nar/gky014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith AM, Jain M, Mulroney L, Garalde DR & Akeson M Reading canonical and modified nucleobases in 16S ribosomal RNA using nanopore native RNA sequencing. PLOS ONE 14, e0216709 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steijger T et al. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 10, 1177–1184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venturini L, Caim S, Kaithakottil GG, Mapleson DL & Swarbreck D Leveraging multiple transcriptome assembly methods for improved gene structure annotation. Gigascience 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics (2018). doi: 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain M et al. Improved data analysis for the MinION nanopore sequencer. Nat. Methods 12, 351–356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jain M et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szczesny RJ et al. RNA degradation in yeast and human mitochondria. Biochim. Biophys. Acta 1819, 1027–1034 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Payne A, Holmes N, Rakyan V & Loose M BulkVis: a graphical viewer for Oxford nanopore bulk FAST5 files. Bioinformatics (2018). doi: 10.1093/bioinformatics/bty841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tilgner H, Grubert F, Sharon D & Snyder MP Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc. Natl. Acad. Sci. U. S. A. 111, 9869–9874 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho H et al. High-resolution transcriptome analysis with long-read RNA sequencing. PLoS One 9, e108095 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein BE et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120, 169–181 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Ernst J & Kellis M Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 28, 817–825 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst J et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deveson IW et al. Universal Alternative Splicing of Noncoding Exons. Cell Syst 6, 245–255.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Gonzàlez-Porta M, Frankish A, Rung J, Harrow J & Brazma A Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene. Genome Biol. 14, R70 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baralle FE & Giudice J Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edge P, Bafna V & Bansal V HapCUT2: robust and accurate haplotype assembly for diverse sequencing technologies. Genome Res. 27, 801–812 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rozowsky J et al. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol. Syst. Biol. 7, 522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown CJ et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38 (1991). [DOI] [PubMed] [Google Scholar]
- 26.Eckmann CR, Rammelt C & Wahle E Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA 2, 348–361 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Subtelny AO, Eichhorn SW, Chen GR, Sive H & Bartel DP Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang H, Lim J, Ha M & Kim VN TAIL-seq: genome-wide determination of poly(A) tail length and 3’ end modifications. Mol. Cell 53, 1044–1052 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Temperley RJ, Wydro M, Lightowlers RN & Chrzanowska-Lightowlers ZM Human mitochondrial mRNAs—like members of all families, similar but different. Biochimica et Biophysica Acta (BBA) - Bioenergetics 1797, 1081–1085 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson JT et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Rand AC et al. Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Methods 14, 411–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu N & Pan T N6-methyladenosine–encoded epitranscriptomics. Nat. Struct. Mol. Biol. 23, 98–102 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Dai D, Wang H, Zhu L, Jin H & Wang X N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 9, 124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sibbritt T, Patel HR & Preiss T Mapping and significance of the mRNA methylome. Wiley Interdiscip. Rev. RNA 4, 397–422 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Meyer KD et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149, 1635–1646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roost C et al. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc. 137, 2107–2115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Licht K, Kapoor U, Mayrhofer E & Jantsch MF Adenosine to Inosine editing frequency controlled by splicing efficiency. Nucleic Acids Res. 44, 6398–6408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishikura K Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tajaddod M, Jantsch MF & Licht K The dynamic epitranscriptome: A to I editing modulates genetic information. Chromosoma 125, 51–63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tardaguila M et al. SQANTI: extensive characterization of long-read transcript sequences for quality control in full-length transcriptome identification and quantification. Genome Res. (2018). doi: 10.1101/gr.222976.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anvar SY et al. Full-length mRNA sequencing uncovers a widespread coupling between transcription initiation and mRNA processing. Genome Biol. 19, 46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L et al. Transcriptomic Characterization of SF3B1 Mutation Reveals Its Pleiotropic Effects in Chronic Lymphocytic Leukemia. Cancer Cell 30, 750–763 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bradley RK, Merkin J, Lambert NJ & Burge CB Alternative splicing of RNA triplets is often regulated and accelerates proteome evolution. PLoS Biol. 10, e1001229 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bresson SM, Hunter OV, Hunter AC & Conrad NK Canonical Poly(A) Polymerase Activity Promotes the Decay of a Wide Variety of Mammalian Nuclear RNAs. PLoS Genet. 11, e1005610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi H et al. PABP Cooperates with the CCR4-NOT Complex to Promote mRNA Deadenylation and Block Precocious Decay. Mol. Cell 70, 1081–1088.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Parker R & Song H The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11, 121–127 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Li X, Xiong X & Yi C Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods 14, 23–31 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Roundtree IA, Evans ME, Pan T & He C Dynamic RNA Modifications in Gene Expression Regulation. Cell 169, 1187–1200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee M, Kim B & Kim VN Emerging roles of RNA modification: m(6)A and U-tail. Cell 158, 980–987 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Index of /1000genomes/ftp/technical/reference/GRCh38_reference_genome. Available at: https://ftp-trace.ncbi.nih.gov/1000genomes/ftp/technical/reference/GRCh38_reference_genome/ (Accessed: 20th February 2018)
- 51.gencode. GENCODE - GENCODE Release Files. Available at: https://www.gencodegenes.org/releases/current.html (Accessed: 20th February 2018)
- 52.Tang AD et al. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. bioRxiv 410183 (2018). doi: 10.1101/410183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain M et al. Improved data analysis for the MinION nanopore sequencer. Nat. Methods 12, 351–356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hinrichs AS et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 34, D590–8 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eberle MA et al. A reference data set of 5.4 million phased human variants validated by genetic inheritance from sequencing a three-generation 17-member pedigree. Genome Res. (2016). doi: 10.1101/gr.210500.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Molinie B et al. m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat. Methods 13, 692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data including raw signal files (FAST5), event-level data (FAST5), base-calls (FASTQ) and alignments (BAM) are available as an Amazon Web Services Open Data set for download from https://github.com/nanopore-wgs-consortium/NA12878. The scripts used for various analyses are also available from the same GitHub under nanopore-human-transcriptome/scripts.