Abstract
Starting shortly after parturition, and continuing throughout our lifetime, the gut microbiota coevolves with our metabolic and neurological programming. This symbiosis is regulated by a complex interplay between the host and environmental factors, including diet and lifestyle. Not surprisingly, the development of this microbial community is of critical importance to health and wellness. In this targeted review, we examine the gut microbiome from birth to two years of age to characterize the role human milk oligosaccharides play in early formation of microbial flora.
Keywords: human milk oligosaccharide, prebiotic, antimicrobial, microbiome
I. Introduction
For every ten human cells, there are thirteen microbial cells living in and on the body (1). Of the eight major microbial communities that comprise the human microbiome (nose, mouth, lungs, stomach, small intestine, colon, urogenital, and skin), the gastrointestinal tract features the densest microbial community. Not surprisingly, the intestinal flora executes functions vital to neonatal health and development.
Breastfeeding is a major factor guiding the establishment of the gut flora in early life. Human milk contains essential nutrients such as carbohydrates, fatty acids, and proteins, as well as a collection of bioactive small molecules and biologics critical to the protection and development of an infant (3). Breast milk itself contains a variety of bacterial species, most predominantly Staphylococcus and Streptococcus species (4–6). However, research indicates that other breast milk components may guide microbiome development more than the direct bacterial components within. Recent studies have shown signficant differences in the microbiota population between breast-fed infants and formula-fed infants (Figure 1) (2, 7–9). Specifically, breast-fed infants see an increased colonization of beneficial Bacteroides species juxtaposed with a decrease in colonization by the Streptococcus and Clostridia species (2) correlated with diarrhea and other adverse gastrointestinal issues. A closer look at the macromolecular content of breast milk identifies non-digestible sugars, known as human milk oligosaccharides (HMOs), as key modulators of the infant gut microbiome (10). In terms of chemical microbiology, HMOs serve multiple functions (Figure 2). First, they are prebiotics for a variety of commensal microbes. Secondly, they possess antimicrobial activity against a number of bacterial, viral, parasitic, and fungal pathogens. These dual functions indicate that HMOs play a key regulatory role in the maintenance of a symbiotic microbiome during early childhood.
Figure 1:
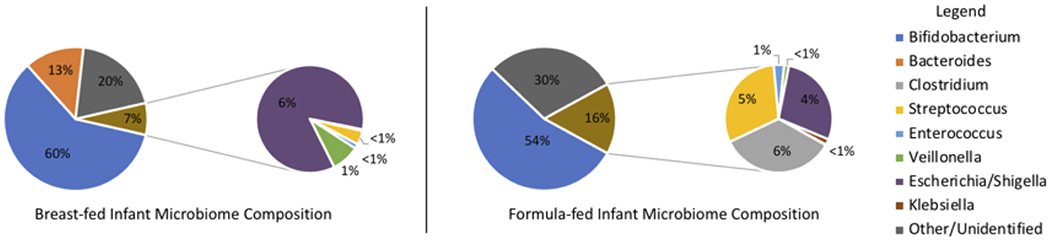
Comparison of infant gut microbiome population in breast-fed and formula-fed infants (2).
Figure 2:

Mechanisms by which HMOs impact microbiome symbiosis
Herein, we review the established interactions between HMOs and members of the gut microbiome, specific microbial colonization processes that are impacted, and the methodologies used to illuminate this complex host-microbe dialogue.
II. HMO Structure and Biosynthesis
HMOs are a class of structurally diverse oligosaccharides comprising approximately 8% of the macromolecules in human breast milk (Figure 2A). Currently, the structures of over 200 unique HMOs have been elucidated using a combination of biosynthetic studies and analysis by mass spectrometry and nuclear magnetic resonance (11–13). The concentration and structural diversity present in any particular sample of breast milk is dependent on the mother’s genetics (Lewis blood group and secretor status) and the stage of lactation. It has been shown that concentration of HMOs is highest at the initial time of birth and decreases in total concentration overtime (14). Moreover, the relative abundance of specific oligosaccharides can change over the course of breastfeeding. For example, the presence of fucosylated HMOs are significantly reduced in mature milk (15). While our understanding of the enzymatic machinery involved in HMO biosynthesis is continually evolving (16, 17), it is well-established that molecular composition of breast milk varies as infant maturation proceeds, making breastmilk a personalized medicine.
Structurally, HMOs consist of five pyranose monosaccharide residues (11, 18–20); β-D-galactose (Gal), β-D-glucose (Glc), N-acetyl-β-D-glucosamine (GlcNAc), α-L-fucose (Fuc), and the sialic acid N-acetyl-α-D-neuraminic acid (NeuNAc or Sia). HMO biosynthesis begins in the Golgi apparatus of the mammary gland, where lactose is first synthesized via β-(4) connection of Gal-Glc. Next, lactose is functionalized using N-acetyllactosamine as an elongation residue or lacto-N-biose (LNB) as a terminating residue to produce linear (iso-) or branched (para-) HMOs (Figure 2B). From this core oligosaccharide, subsequent fucosylation or sialylation is genetically guided based on secretor status and Lewis (Le) blood group (16). Secretor status is determined by the presence of a gene encoding for the α−2-fucosyltransferase (FUT2), while Le blood group encodes for the α3/4-fucosyltransferase (FUT3). These enzymes, among others that are poorly described in the literature, are responsible for the installation of fucose onto HMO core oligosaccharides. Similarly, sialic acid decoration occurs after initial HMO elongation (19, 21), although less is understood about the enzymes that govern the sialylation process. Several previously published reviews contain a complete analysis of known HMOs, and their chemical structures (11, 17). Overall, of the HMOs present in human breast milk, approximately 62% are fucosylated, 25% are non-fucosylated, 12% are sialylated, and <1% are both sialylated and fucosylated (Figure 2A) (2).
III -. HMOs as Prebiotics
The human gut does not possess the enzymatic machinery needed to metabolize HMOs (22). Instead, commensals metabolize HMOs to gain a growth advantage over pathogens. Indeed, adherence to the intestinal epithelium is critical to temporal development of the gut community, as it prevents the elimination of bacteria via peristalsis (23). Adhesion also promotes the modulation of the immune system (24) and prevents pathogens from mucosal attachment (25). Several microbes adhere to the mucosa and metabolize HMOs, including Bifidobacteria, Bacteroides, Lactobacillus, and a variety of other firmicutes (Table 1) (26).
Table 1:
HMOs metabolized by bacterial species
| Genus | Species | Metabolized HMOs[a] | Reference |
|---|---|---|---|
| Bifidobacterium | bifidum | 2’-FL, 3-FL, 3’-SL, 6’SL, LNT-II, LNT, LNnT | (30) |
| longum | LNB, 2’-FL, 3-FL, 3’-SL, 6’SL, LNT, LNnT, LDFT, LNFP-I | (30–32) | |
| breve | LNB, 3-FL, 6’-SL, LNT-II, LNT, LNnT, LSTb, LSTc, | (30, 32, 33) | |
| infantis | 2’-FL, 3-FL, 3’-SL, 6’-SL, LNT-II, LNT, LNnT, LDFT, LNFP-III | (30, 31) | |
| kashiwanohense | 2’-FL | (32) | |
| Bacteroides | thetaiotaomicron | 2’-FL, 3-FL, 3’-SL, 6’-SL | (31) |
| fragilis | 2’-FL, 3-FL, 6’-SL, LDFT | (31) | |
| vulgatus | 2’-FL, 3-FL, 3’-SL, 6’-SL, LDFT | (31) | |
| Lactobacillus | acidophilus | 2’-FL, 3-FL, 6’-SL, LNT | (34) |
| plantarum | 2’-FL, 3-FL, 6’-SL, LNT | (34) | |
| delbrueckii | 2’-FL, 3-FL, 6’-SL | (31) | |
| Enterococcus | faecalis | 2’FL, 3-FL | (31) |
| Staphylococcus | thermophilus | 2’-FL, 3-FL | (31) |
HMO Abbreviations: lacto-N-biose (LNB), 2’-fucosyllactose (2’-FL), 3-fucosyllactose (3-FL), 3’-sialyllactose (3’-SL), 6’-sialyllactose (6’-SL), lacto-N-triose II (LNT-II), lacto-N-tetraose (LNT), lacto-N-neotetraose (LNnT), lactodifucotetraose (LDFT), lacto-N-fucopentaose I (LNFP-I), lacto-N-fucopentaose III (LNFP-III), LS-tetrasaccharide b (LSTb), LS-tetrasaccharide c (LSTc).
While our knowledge of the species that metabolize HMOs is increasing, their origins in the infant microbiome are far more uncertain. The first microbes to colonize an infant are vertically transmitted from mother during labor and delivery. Children that are vaginally delivered are initially colonized by aerobic bacteria which deoxygenate the gut, setting the stage for latter colonization by beneficial anaerobes (27). The next wave of microbes arrive during breastfeeding, as the average mothers milk contains 400-800 species of bacteria, alongside other microbes (28). Additionally, the baby is inoculated during skin contact with community members (6, 29).
A. Bifidobacteria
As one of the earliest colonizers of the infant gut microbiome (35) Bifidobacteria, a gram-positive anaerobe (32), thrives in the infant microbiome due to its ability to metabolize HMOs (36). After colonization, metabolism of HMOs occurs via one of two mechanisms; (i) extracellular hydrolysis and subsequent trafficking of monosaccharides into the cell or (ii) internalization of intact HMOs and subsequent hydrolysis. From the generated monosaccharide residues, further digestion leads to the generation of short chain fatty acids (31, 37). Figure 3 illustrates how Bifidobacteria metabolize monosaccharide residues into short chain fatty acids (38, 39).
Figure 3:
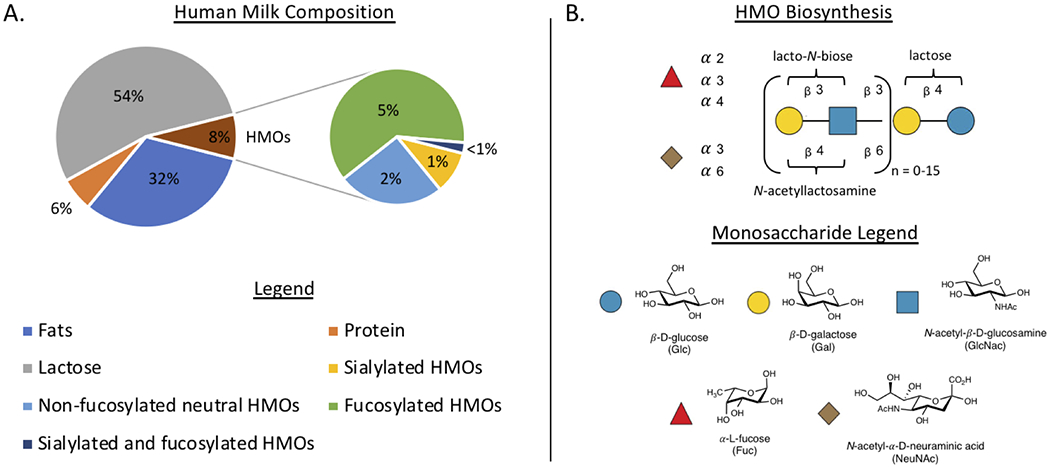
Macromolecular composition of human milk and the HMO biosynthetic pathway (A.) Average composition of human breast milk and percent abundances of key HMO residues (B.) Pictogram representation of HMO biosynthesis and chemical structures of monosaccharides used in HMO biosynthesis
Initial reports revealed that HMOs are substrates for a variety of Bifidobacteria sup-species, such as Bifidobacterium bifidum, Bifidobacterium longum subsp. infantis and Bifidobacterium breve (30, 32, 35, 40). Among these microbes, fucosylated HMOs are recognized and metabolized at a faster rate than other neutral or negatively charged HMOs. B. bifidium has been shown to utilize LNnT and sialylated variants via extracellular hydrolysis of fucose and sialic acid residues (41, 42). Bifidobacterium longum subsp. infantis (43) and Bifidobacterium breve (44) traffic fully intact HMOs such as LNT, LNnT and LNB intracellularly before further degradation. Recently, a newly studied species, Bifidobacterium kashiwanohense, was identified as being able to survive solely on 2’-FL metabolism (32). 2’-FL is one of the most abundant HMOs in the breast milk of women of European descent (11, 12, 45).
The unique mechanisms of HMO degradation and substrate specificity amongst Bifidobacteria species highlight the symbiotic nature of Bifidobacteria co-colonization. To that end, a recent report from Katayama illustrated the cross-feeding capability of Bifidobacteria species and specifically identified B. bifidium as a critical strain in the promotion of growth for many other bifidobacterial co-colonizers (46). B. bifidium was demonstrated to have a robust set of enzymatic machinery that digests HMOs and creates increased concentrations of available disaccharides and monosaccharides, such as lactose, lacto-N-biose, fucose, and glucose. Stemming from initial HMO degradation by B. bifidium, other Bifidobacteria saw promoted growth upon the increased availability of smaller carbohydrates. This symbiotic development is key to the diversity of Bifidobacteria in the infant microbiome and demonstrates why Bifidobacteria are such prolific colonizers. Representing more than 50% of gut microbes, Bifidobacteria effectively use HMOs as carbohydrate sources and modern formula contains additives to help mimic HMO-mediated growth of Bifidobacteria (7, 47–49). It has been shown that formula additives such as galactooligosaccharides (GOS), oligofructose, and long-chain inulin (fructooligosaccharides, FOS) can act as HMO surrogates to promote Bifidobacteria colonization via metabolism to short chain fatty acids (50). Advances such as this, continue to improve formula and its benefits for infants.
B. Bacteroides
While early microbiome research has focused primarily on the consumption of HMOs by Bifidobacteria, recent studies have demonstrated that Bacteroides colonization is more variable between breast-fed and formula-fed infants (2, 51). Analysis has demonstrated Bacteroides colonization to be as little as <1% in formula-fed infants, whereas colonization can increase upwards of 10% in breast-fed infants. This large shift in colonization between the two feeding methods has elicited interest in characterizing the mechanisms by which Bacteroides metabolize HMOs.
Bacteroides are gram-negative obligate anaerobes. Sub-species Bacteroides thetaiotaomicron, Bacteroides fragilis, and Bacteroides vulgatus efficiently metabolize both fucosylated and sialylated HMOs (26, 31, 51). Bacteroides species have a preference for larger, mucin like oligosaccharides due to their ability to upregulate mucin glycan degradation pathways and polysaccharide utilization loci (40, 51, 52). Clearly, Bacteroides species represent another highly efficient metabolizer of HMOs in the infant gut, but data indicates current infant formulas poorly mimic HMO-promoted Bacteroides growth. As a result, new research toward appropriate formula additives for Bacteroides promotion are needed.
C. Lactobacillus and Other Species
Additional species digest HMOs, albeit to a lesser degree than Bacteroides and Bifidobacteria (30, 34). Lactobacillus acidophilus and Lactobacillus plantarum digest both fucosylated and sialylated HMOs, but do so with preference for tri- and tetra-saccharides over larger oligosaccharides (34). These gram-positive facultative anaerobes convert sugars into lactic acid and have long been used as probiotics (53, 54), although Lactobacillus themselves are less prevalent colonizers of the infant gut.
HMOs can also be digested and promote the growth of Clostridium, Enterococcus, and Staphylococcus strains (26). Nevertheless, it is widely believed that in the microbiome HMOs promote the growth of Bifidobacteria and Bacteroides species to a far greater extent, ultimately helping to decrease overall colonization of these firmicutes.
IV -. HMOs as Antimicrobial Agents
While the prebiotic activity of HMOs is established, their antimicrobial properties are an emerging area of research. HMOs have well documented antibacterial properties, in addition to antibiofilm effects in infectious pathogens. HMOs have also been implicated in decreases in viral adhesion and parasite infectivity. Both viruses and parasites are common causes of diarrhea and infant gastroenteritis (55). These antimicrobial effects are often attributed to HMO interactions with carbohydrate-recognizing proteins that impact microbial adhesion and pathogenesis. These multifaceted roles identify HMOs as key compounds in globally structuring a healthy infant microbiome.
A. Antibacterial effects
HMOs possess antibacterial effects on a variety of microfloral pathogens. HMOs directly decrease bacterial growth of Group B Streptococcus (GBS) across multiple strains and serotypes (56–60). GBS passively colonizes between 20-50% of pregnant mothers (61, 62), but can be transmitted vertically to infants (63). Once transmitted, it can cause neonatal sepsis and meningitis, and has a high correlation to adverse pregnancy outcomes. Metabolomic analysis has identified the antimicrobial effects of HMOs on GBS are due to global increase in cell permeability and disruption in cell membrane affiliated metabolites (64). Work has also revealed that HMOs have direct antibacterial effects on Acinetobacter baumanii (57). Additionally, HMOs act as antibiotic adjuvants (65, 66), increasing the activity of clinically used antibiotics in the treatment of infectious disease.
While some antibacterial effects are linked to direct HMO interactions with bacterial cells and stunting of bacterial growth, HMOs also mediate antibacterial responses through other mechanisms in the microbiome. For example, HMOs have been shown to decrease pathogenesis of Campylobacter jejuni (11, 67, 68). α−2-fucosylated HMOs are recognized by C. jejuni and prevent its ability to bind to epithelial glycans, causing net reduction in adhesion of C. jejuni in the gastrointestinal tract.
B. Antibiofilm effects
While there are a significant number of planktonic bacteria found in the luminal region of the gut, research suggests that microbial populations lining the epithelial surface live predominantly in the biofilm state (10, 69, 70). Bacterial biofilms form when planktonic bacteria irreversibly attach to a surface and produce protective exopolysaccharide (EPS) matrix. This biofilm impacts bacterial virulence and confers antibiotic resistance (71). These denser biofilm organizations are also thought to play a larger role in immunological modulation, due to their close proximity to epithelial cell surfaces (72–74).
Research has demonstrated that HMOs have significant antibiofilm effects in GBS (60) and Staphylococcus aureus (57) models. Pooled HMOs (from multiple donors) consistently demonstrate antibiofilm effects within GBS, while single donor HMO samples can range in antibiofilm effects. This has been demonstrated by scanning electron microscopy (SEM), where clear morphological changes in GBS biofilm architecture occur upon treatment with HMOs from some individual donors and not others (Figure 5) (60). GBS conventionally grows in long linear strands of diplococci (Figure 5A), however upon treatment with certain HMO isolates, the biofilm architecture can become truncated and globular in structure (Figure 5B). These SEM images help visualize the differences in organization of streptococcal cells upon interaction with HMOs, and how perturbations to surface interactions impede further biofilm maturation. This ability to impact biofilm production amongst pathogenic bacteria, identifies HMOs as candidate compounds for the treatment of GBS and S. aureus infections. However, since not all donor HMO samples induce such a morphological change in biofilm architecture, research is ongoing to identify specific oligosaccharides necessary for the desired antibiofilm effects. Even less is known about HMO-mediated impacts on commensal bacterial biofilm formation.
Figure 5:

Scanning electron micrographs of GBS10/84 biofilm formation after 24 hrs when grown in Todd-Hewitt broth + 1% glucose. (A.) Untreated GBS10/84 biofilm formation (B.) GBS10/84 biofilm formation when dosed with pooled HMOs at 5 mg mL−1
C. Antiviral effects
Recent work has indicated that HMOs possess strong antiviral activity, specifically in the context of microbiome-based interactions with norovirus, rotavirus, human immunodeficiency virus (HIV), and influenza (75, 76). For example, norovirus pathogenesis, a leading cause of gastroenteritis (77), has been shown to be impacted upon treatment with HMOs. Specifically, norovirus pathogenesis begins with viral binding to histo-blood group antigens (HBGAs) on the surface of epithelial cells. Studies indicate that several HMOs, especially 2’-FL, decrease norovirus binding in epithelial models due to competitive binding with HBGAs (76). Structural analysis reveals that carbohydrate-mimicry of these surface glycans, allows HMOs to act as decoys and stunt norovirus adhesion. This decoy mechanism has also been demonstrated in the case of rotavirus, another leading cause of infant gastroenteritis (78, 79). Similarly, HIV is known to be passed from mother to infant via mucosal barriers and interactions of viral glycoproteins and dendritic integrins (11, 75). These integrins have been shown to also have an affinity for Le blood group antigens, 2’-FL, and 3-FL (80). As a result, it is believed that HMO introduction can block HIV binding sites in the epithelium and prevent HIV transmission. Indeed, it has been shown that 80-90% of breast-fed infants are not infected with HIV (81, 82), despite exposure to the virus through breast milk. Additionally, in influenza viruses, the hemagglutinin glycoproteins responsible for infectivity are known to bind sialylated HMOs, such as 6’-SL and 3’-SL (83). This suggests that HMOs could possess antiviral effects in the upper-respiratory microbiome as well. The receptor decoy mechanisms of HMOs ultimately illicit a variety of antiviral effects in the microbiome.
D. Antifungal and antiparasitic effects
Other organisms that contribute to microbiome symbiosis include fungi and parasites (84, 85). While not as widely researched, HMOs have been shown to impact both fungi and parasite mediated infections in a handful of cases. One such fungal pathogen, Candida albicans, is a frequent fungal colonizer of the neonatal gut and is correlated to intestinal disorders (86). Studies indicate that treatment of C. albicans with HMOs stunts the growth of the fungal hyphal necessary for invasion, thereby hindering C. albicans pathogenesis (87). Additionally, Bode and coworkers have studied the effects of HMOs on the attachment of the parasite Entamoeba histolytica (88). This work revealed that HMOs could promote detachment of E. histolytica trophozoites in a dose-dependent manner, as well as decrease cytotoxicity. These effects were speculated to be due to competitive binding of HMOs at surface Gal/GalNAc lectins. Research such as this helps to explain why breast-fed infants are less likely to face certain parasitic and fungal infections, and offer clues to broader regulatory capabilities of HMOs in the microbiome.
V -. HMOs and the Infant Immune System
HMOs directly influence multiple aspects of the infant immune system. Non-charged HMOs participate in active transport over intestinal epithelial monolayers to reach systemic circulation (89). Indeed, HMOs have been detected in the urine, feces, and blood (albeit at lower levels) of infants (90). This transport leads to HMO-mediated immunomodulatory effects throughout the human body, most of which occur through HMO interactions with lectins (carbohydrate binding proteins).
Lectins, such as galectins, siglecs, and selectins, have a wide range of functions and ligand specificities and it has been shown that HMOs bind each of these receptors on human immune cells. Galectins are expressed on T-cells, intestinal epithelial cells, antigen presenting cells, and granulocytes. After binding an appropriate ligand, galectins participate in cell-signaling. Galectins are also secreted and bind to glycoproteins or receptors at other cell surfaces. Structurally, galectins bind N-acetyllactosamine and lactose containing sugars (91–93). Galectins also bind sialylated and fucosylated galactose residues. Indeed, the Klassen and Cummings labs have independently demonstrated that galectins bind several HMOs featuring these substructures.
Another family of lectins involved in HMO binding are siglecs, which are sialic acid binding immunoglobulin-like lectins. Siglecs are known to bind sialylated HMOs (94). Siglecs 1–16 are expressed on a number of blood cells, including dendritic cells, monocytes, macrophages, neutrophils, and NK cells (95, 96) and, in contrast to galectins, siglecs are not expressed by intestinal epithelial cells.
In addition to galectins and siglecs, HMOs engage the selectins (97, 98), a family of lectins involved in a wide variety of immune system functions, such as modulating cellular adhesion. Selectins bind to oligosaccharides featuring sialylated Le blood group epitopes (i.e. fucosylated and sialylated lacto-N-biose or N-acetyllactosamine). From an immunological perspective, selectins mediate the early stages of leukocyte trafficking (99, 100). During inflammation, leukocytes migrate from the blood to the endothelium, where selectin expression is induced by pro-inflammatory cytokines. Selectins then bind glycoconjugates on the surface of leukocytes, slowing their movement and directing these immune cells towards the desired location. As sialylated HMOs are capable of competitively binding to siglecs, they can impact leukocyte recruitment (101) and subsequent inflammatory responses.
In addition to lectin binding, several reports indicate that HMOs engage Toll-like receptor (TLR) family members. These receptors typically engage pathogen-related antigens. For example, TLR-4 dependent effects of two HMOs, 3′-SL (102) and LNFP-III (103), have been described wherein in vivo evaluation of the HMOs required the expression of TLR-4 for their effects. Since TLRs modulate downstream immune cell activation and cytokine production, it is believed that select HMOs also impact these processes.
VI -. Summary
Human milk oligosaccharides (HMOs) are the third largest macromolecule present in breast milk after fat and lactose – yet they possess no nutritive activity for the growing infant. Instead, HMOs are responsible for directly defending the neonate by promoting the growth of commensal bacteria, strengthening gut barrier function, and preventing the adhesion and growth of pathogens. While research in human milk science has long revealed the complexity of breast milk and the benefits it gives for the baby, frontier studies focusing on HMOs reveals that these macromolecules play a key role in infant health and wellness.
Figure 4:

Representative metabolism of the non-glucose residues of an HMO into short chain fatty acids by Bifidobacteria. Abbreviations: acetate kinase (AckA), aldehyde-alcohol dehydrogenase 2 (Adh2), enolase (Eno), galactokinase (GalK), galactose mutarotase (GalM), glyceraldehyde-3-phosphate dehydrogenase C (GAPDH), glucose 6-phosphate isomerase (Gpi), phosphoglycerate mutase (Gpm), fructose-6-phosphoketolase (F6PPK), L-fucose isomerase (FucI), L-fuculose kinase (FucK), L-fuculose-1P aldolase (FucA), lactate dehydrogenase (Ldh2), lacto-N-biose phosphorylase (LNBP), phosphate (P), phosphoglyceric kinase (Pgk), phosphoglucomutase (Pgm), formate acetyltransferase (Pfl), pyruvate kinase (Pyk), transaldolase (Tal), triosephosphate isomerase (TpiA), UTP-glucose-1-phosphate uridylyltransferase (UgpA)
Perspectives.
Importance: HMOs are the next frontier in neonatal nutrition, health, and wellness as they are a major factor in the protection conferred by breast milk. HMOs are prebiotics, promoting the growth of commensal bacteria in the baby’s digestive tract where 70% of immune system functions originate. While breast feeding is the unquestioned gold standard in infant nutrition, HMOs are an interesting tool that can be used to narrow the health gap between breast milk and formula.
-
Current understanding and challenges: While there have been significant advances in our knowledge of HMO chemistry and biochemistry, there are a number of key roadblocks preventing our ability to leverage these molecules for human health and wellness. First, characterization of the therapeutic efficacy of HMOs has been hindered due to limited access to individual structures in appropriate quantity and purity for exhaustive biological evaluation. Frontier methods for characterizing HMO biochemistry depend on probing heterogeneous mixtures of HMOs and isolating individual structures from pooled milk using chromatography. Neither strategy provides a facile platform for characterizing individual HMOs. Accordingly, target identification, structure-activity relationships, and pre-clinical development have been stifled.
Second, while there is momentum to include HMOs in commercially available infant formulas, basic science researchers do not play a large enough role in determining which HMOs should be studied, their dosage, and the duration of administration. Specific clinical benchmarks, with anticipated results, should be met to justify supplementation as it is currently not clear that all babies benefit from all HMOs.
Future directions: As small clinical trials have shown that HMO supplementation is an attractive alternative for newborns who cannot be breast-fed, a number of groups are focused on developing robust synthetic and chemoenzymatic strategies to access well-defined HMOs, and associated tool compounds, for biological evaluation (104–110). Future directions in the area of human milk science, and more globally glycoscience, is primed to make large advances in the production and biological evaluation of HMOs.
Acknowledgements
This work was supported by the National Institutes of Health under Grant No. 1R35GM133602 to S.D.T. S.A.C. was supported by the Vanderbilt Chemical Biology Interface (CBI) training program (T32 GM065086)
References
- 1.Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016;14(8):e1002533–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang M, Li M, Wu S, Lebrilla CB, Chapkin RS, Ivanov I, et al. Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J Pediatr Gastroenterol Nutr. 2015;60(6):825–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballard O, Morrow AL. Human milk composition: nutrients and bioactive factors. Pediatr Clin North Am. 2013;60(1):49–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunt KM, Foster JA, Forney LJ, Schütte UME, Beck DL, Abdo Z, et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PloS one. 2011;6(6):e21313–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collado MC, Delgado S, Maldonado A, Rodríguez JM. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Letters in Applied Microbiology. 2009;48(5):523–8. [DOI] [PubMed] [Google Scholar]
- 6.Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, et al. Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome. JAMA Pediatrics. 2017;171(7):647–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho NT, Li F, Lee-Sarwar KA, Tun HM, Brown BP, Pannaraj PS, et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nature communications. 2018;9(1):4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunz C Historical aspects of human milk oligosaccharides. Adv Nutr. 2012;3(3):430S–9S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timmerman HM, Rutten NBMM, Boekhorst J, Saulnier DM, Kortman GAM, Contractor N, et al. Intestinal colonisation patterns in breastfed and formula-fed infants during the first 12 weeks of life reveal sequential microbiota signatures. Scientific reports. 2017;7(1):8327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiology and Molecular Biology Reviews. 2017;81(4):e00036–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bode L Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22(9):1147–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobata A. Structures and application of oligosaccharides in human milk. Proc Jpn Acad Ser B Phys B Sci. 2010;86(7):731–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Craft KM, Townsend SD. Mother Knows Best: Deciphering the Antibacterial Properties of Human Milk Oligosaccharides. Acc Chem Res. 2019;52(3):760–8. [DOI] [PubMed] [Google Scholar]
- 14.Coppa GV, Gabrielli O, Pierani P, Catassi C, Carlucci A, Giorgi PL. Changes in Carbohydrate Composition in Human Milk Over 4 Months of Lactation. Pediatrics. 1993;91(3):637. [PubMed] [Google Scholar]
- 15.Bai Y, Tao J, Zhou J, Fan Q, Liu M, Hu Y, et al. Fucosylated Human Milk Oligosaccharides and N-Glycans in the Milk of Chinese Mothers Regulate the Gut Microbiome of Their Breast-Fed Infants during Different Lactation Stages. mSystems. 2018;3(6):e00206–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blank D, Dotz V, Geyer R, Kunz C. Human Milk Oligosaccharides and Lewis Blood Group: Individual High-Throughput Sample Profiling to Enhance Conclusions From Functional Studies. Advances in nutrition (Bethesda, Md). 2012;3:440S–9S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X Chapter Four - Human Milk Oligosaccharides (HMOS): Structure, Function, and Enzyme-Catalyzed Synthesis. In: Baker DC, Horton D, editors. Advances in Carbohydrate Chemistry and Biochemistry. 72: Academic Press; 2015. p. 113–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bode L, Jantscher-Krenn E. Structure-function relationships of human milk oligosaccharides. Adv Nutr. 2012;3(3):383S–91S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lis-Kuberka J, Orczyk-Pawiłowicz M. Sialylated Oligosaccharides and Glycoconjugates of Human Milk. The Impact on Infant and Newborn Protection, Development and Well-Being. Nutrients. 2019;11(2):306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeuner B, Teze D, Muschiol J, Meyer AS. Synthesis of Human Milk Oligosaccharides: Protein Engineering Strategies for Improved Enzymatic Transglycosylation. Molecules. 2019;24(11):2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan J, Ding J, Jin G, Yu D, Yu L, Long Z, et al. Profiling of Sialylated Oligosaccharides in Mammalian Milk Using Online Solid Phase Extraction-Hydrophilic Interaction Chromatography Coupled with Negative-Ion Electrospray Mass Spectrometry. Analytical Chemistry. 2018;90(5):3174–82. [DOI] [PubMed] [Google Scholar]
- 22.Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. The American Journal of Clinical Nutrition. 2000;71(6):1589–96. [DOI] [PubMed] [Google Scholar]
- 23.Tuomola E, Crittenden R, Playne M, Isolauri E, Salminen S. Quality assurance criteria for probiotic bacteria. The American Journal of Clinical Nutrition. 2001;73(2):393s–8s. [DOI] [PubMed] [Google Scholar]
- 24.He F, Ouwehand AC, Isolauri E, Hashimoto H, Benno Y, Salminen S. Comparison of mucosal adhesion and species identification of bifidobacteria isolated from healthy and allergic infants. FEMS Immunology & Medical Microbiology. 2001;30(1):43–7. [DOI] [PubMed] [Google Scholar]
- 25.Resta-Lenert S, Barrett KE. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive <em>Escherichia coli</em> (EIEC). Gut. 2003;52(7):988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, et al. Consumption of Human Milk Oligosaccharides by Gut-Related Microbes. J Agric Food Chem. 2010;58(9):5334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore RE, Townsend SD. Temporal development of the infant gut microbiome. Open Biology.9(9):190128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwab C, Voney E, Ramirez Garcia A, Vischer M, Lacroix C. Characterization of the Cultivable Microbiota in Fresh and Stored Mature Human Breast Milk. Frontiers in Microbiology. 2019;10(2666). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis EC, Wang M, Donovan SM. The role of early life nutrition in the establishment of gastrointestinal microbial composition and function. Gut Microbes. 2017;8(2):143–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zúñiga M, Monedero V, Yebra MJ. Utilization of Host-Derived Glycans by Intestinal Lactobacillus and Bifidobacterium Species. Frontiers in Microbiology. 2018;9(1917). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Z- T, Chen C, Newburg DS. Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology. 2013;23(11):1281–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James K, Bottacini F, Contreras JIS, Vigoureux M, Egan M, Motherway MOc, et al. Metabolism of the predominant human milk oligosaccharide fucosyllactose by an infant gut commensal. Scientific Reports. 2019;9(1):15427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Underwood MA, Davis JCC, Kalanetra KM, Gehlot S, Patole S, Tancredi DJ, et al. Digestion of Human Milk Oligosaccharides by Bifidobacterium breve in the Premature Infant. J Pediatr Gastroenterol Nutr. 2017;65(4):449–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwab C, Gänzle M. Lactic acid bacteria fermentation of human milk oligosaccharide components, human milk oligosaccharides and galactooligosaccharides. FEMS Microbiology Letters. 2011;315(2):141–8. [DOI] [PubMed] [Google Scholar]
- 35.Duranti S, Lugli GA, Milani C, James K, Mancabelli L, Turroni F, et al. Bifidobacterium bifidum and the infant gut microbiota: an intriguing case of microbe-host co-evolution. Environmental Microbiology. 2019;21(10):3683–95. [DOI] [PubMed] [Google Scholar]
- 36.German JB, Freeman SL, Lebrilla CB, Mills DA. Human milk oligosaccharides: evolution, structures and bioselectivity as substrates for intestinal bacteria. Nestle Nutr Workshop Ser Pediatr Program. 2008;62:205–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parada Venegas D, De la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, et al. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol. 2019;10(277). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Callaghan A, van Sinderen D. Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Frontiers in Microbiology. 2016;7(925). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pokusaeva K, Fitzgerald GF, van Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes & Nutrition. 2011;6(3):285–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, et al. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. The Journal of biological chemistry. 2011;286(40):34583–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, et al. Two distinct α-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology. 2009;19(9):1010–7. [DOI] [PubMed] [Google Scholar]
- 42.Kiyohara M, Tanigawa K, Chaiwangsri T, Katayama T, Ashida H, Yamamoto K. An exo-α-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology. 2010;21(4):437–47. [DOI] [PubMed] [Google Scholar]
- 43.Xiao J-z, Takahashi S, Nishimoto M, Odamaki T, Yaeshima T, Iwatsuki K, et al. Distribution of in vitro fermentation ability of lacto-N-biose I, a major building block of human milk oligosaccharides, in bifidobacterial strains. Applied and environmental microbiology. 2010;76(1):54–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.James K, Motherway MOC, Bottacini F, van Sinderen D. Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways. Scientific Reports. 2016;6(1):38560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Craft KM, Townsend SD. Mother Knows Best: Deciphering the Antibacterial Properties of Human Milk Oligosaccharides. Acc Chem Res. 2019;ASAP. [DOI] [PubMed] [Google Scholar]
- 46.Gotoh A, Katoh T, Sakanaka M, Ling Y, Yamada C, Asakuma S, et al. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Scientific Reports. 2018;8(1):13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veereman-Wauters G, Staelens S, Broek H, Plaskie K, Wesling F, Roger L, et al. Physiological and Bifidogenic Effects of Prebiotic Supplements in Infant Formulae. J Pediatr Gastroenterol Nutr. 2011;52:763–71. [DOI] [PubMed] [Google Scholar]
- 48.Oozeer R, van Limpt K, Ludwig T, Ben Amor K, Martin R, Wind RD, et al. Intestinal microbiology in early life: specific prebiotics can have similar functionalities as human-milk oligosaccharides. The American Journal of Clinical Nutrition. 2013;98(2):561S–71S. [DOI] [PubMed] [Google Scholar]
- 49.Madan JC, Hoen AG, Lundgren SN, Farzan SF, Cottingham KL, Morrison HG, et al. Association of Cesarean Delivery and Formula Supplementation With the Intestinal Microbiome of 6-Week-Old Infants. JAMA pediatrics. 2016;170(3):212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knol J, Scholtens P, Kafka C, Steenbakkers J, Gro S, Helm K, et al. Colon Microflora in Infants Fed Formula with Galacto- and Fructo-Oligosaccharides: More Like Breast-Fed Infants. J Pediatr Gastroenterol Nutr. 2005;40(1). [DOI] [PubMed] [Google Scholar]
- 51.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10(5):507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Comstock LE. Importance of Glycans to the Host-Bacteroides Mutualism in the Mammalian Intestine. Cell Host Microbe. 2009;5(6):522–6. [DOI] [PubMed] [Google Scholar]
- 53.Walter J Ecological Role of Lactobacilli in the Gastrointestinal Tract: Implications for Fundamental and Biomedical Research. Applied and Environmental Microbiology. 2008;74(16):4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Azad MAK, Sarker M, Li T, Yin J. Probiotic Species in the Modulation of Gut Microbiota: An Overview. Biomed Res Int. 2018;2018:9478630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elliott EJ. Acute gastroenteritis in children. BMJ. 2007;334(7583):35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin AE, Autran CA, Szyszka A, Escajadillo T, Huang M, Godula K, et al. Human milk oligosaccharides inhibit growth of group B Streptococcus. J Biol Chem. 2017;292(27):11243–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ackerman DL, Craft KM, Doster RS, Weitkamp JH, Aronoff DM, Gaddy JA, et al. Antimicrobial and Antibiofilm Activity of Human Milk Oligosaccharides against Streptococcus agalactiae, Staphylococcus aureus, and Acinetobacter baumannii. ACS Infect Dis. 2018;4(3):315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Craft KM, Thomas HC, Townsend SD. Sialylated variants of lacto-N-tetraose exhibit antimicrobial activity against Group B Streptococcus. Org Biomol Chem. 2018;17(7):1893–900. [DOI] [PubMed] [Google Scholar]
- 59.Craft KM, Thomas HC, Townsend SD. Interrogation of Human Milk Oligosaccharide Fucosylation Patterns for Antimicrobial and Antibiofilm Trends in Group B Streptococcus. ACS Infect Dis. 2018;4:1755–65. [DOI] [PubMed] [Google Scholar]
- 60.Ackerman DL, Doster RS, Weitkamp JH, Aronoff DM, Gaddy JA, Townsend SD. Human Milk Oligosaccharides Exhibit Antimicrobial and Antibiofilm Properties against Group B Streptococcus. ACS Infect Dis. 2017;3(8):595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jones N, Oliver K, Jones Y, Haines A, Crook D. Carriage of group B streptococcus in pregnant women from Oxford, UK. J Clin Pathol. 2006;59(4):363–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kwatra G, Adrian PV, Shiri T, Buchmann EJ, Cutland CL, Madhi SA. Serotype-Specific Acquisition and Loss of Group B Streptococcus Recto-Vaginal Colonization in Late Pregnancy. PLOS ONE. 2014;9(6):e98778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raabe VN, Shane AL. Group B Streptococcus (Streptococcus agalactiae). Microbiol Spectr. 2019;7(2): 10.1128/microbiolspec.GPP3-0007-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chambers SA, Moore RE, Craft KM, Thomas HC, Das R, Manning SD, et al. A Solution to Antifolate Resistance in Group B Streptococcus: Untargeted Metabolomics Identifies Human Milk Oligosaccharide-Induced Perturbations That Result in Potentiation of Trimethoprim. mBio. 2020;11(2):e00076–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Craft KM, Gaddy JA, Townsend SD. Human Milk Oligosaccharides (HMOs) Sensitize Group B Streptococcus to Clindamycin, Erythromycin, Gentamicin, and Minocycline on a Strain Specific Basis. ACS Chem Biol. 2018;13(8):2020–6. [DOI] [PubMed] [Google Scholar]
- 66.Chambers SA, Moore RE, Craft KM, Thomas HC, Das R, Gaddy JA and Townsend SD. A Solution to Antifolate Resistance in Group B Streptococcus: Un-targeted Metabolomics Identifies Human Milk Oligosaccharide-Induced Perturbations that Result in Potentiation of Trimethoprim. mBio. 2020;ASAP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morrow AL, Ruiz-Palacios GM, Altaye M, Jiang X, Lourdes Guerrero M, Meinzen-Derr JK, et al. Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J Pediatr. 2004;145(3):297–303. [DOI] [PubMed] [Google Scholar]
- 68.Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni Binds Intestinal H(O) Antigen (Fucα1, 2Galβ1, 4GlcNAc), and Fucosyloligosaccharides of Human Milk Inhibit Its Binding and Infection. Journal of Biological Chemistry. 2003;278(16):14112–20. [DOI] [PubMed] [Google Scholar]
- 69.Macfarlane S, Bahrami B, Macfarlane GT. Chapter 4 - Mucosal Biofilm Communities in the Human Intestinal Tract. In: Laskin AI, Sariaslani S, Gadd GM, editors. Advances in Applied Microbiology. 75: Academic Press; 2011. p. 111–43. [DOI] [PubMed] [Google Scholar]
- 70.de Vos WM. Microbial biofilms and the human intestinal microbiome. npj Biofilms and Microbiomes. 2015;1(1):15005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Craft KM, Nguyen JM, Berg LJ, Townsend SD. Methicillin-resistant Staphylococcus aureus (MRSA): antibiotic-resistance and the biofilm phenotype. MedChemComm. 2019;10(8):1231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Triantis V, Bode L, van Neerven RJJ. Immunological Effects of Human Milk Oligosaccharides. Front Pediatr. 2018;6:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nolan LS, Parks OB, Good M. A Review of the Immunomodulating Components of Maternal Breast Milk and Protection Against Necrotizing Enterocolitis. Nutrients. 2019;12(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ayechu-Muruzabal V, van Stigt AH, Mank M, Willemsen LEM, Stahl B, Garssen J, et al. Diversity of Human Milk Oligosaccharides and Effects on Early Life Immune Development. Front Pediatr. 2018;6(239). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morozov V, Hansman G, Hanisch F- G, Schroten H, Kunz C. Human Milk Oligosaccharides as Promising Antivirals. Molecular Nutrition & Food Research. 2018;62(6):1700679. [DOI] [PubMed] [Google Scholar]
- 76.Weichert S, Koromyslova A, Singh BK, Hansman S, Jennewein S, Schroten H, et al. Structural Basis for Norovirus Inhibition by Human Milk Oligosaccharides. J Virol. 2016;90(9):4843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. The New England journal of medicine. 2009;361(18):1776–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crawford SE, Ramani S, Tate JE, Parashar UD, Svensson L, Hagbom M, et al. Rotavirus infection. Nature Reviews Disease Primers. 2017;3(1):17083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramani S, Stewart CJ, Laucirica DR, Ajami NJ, Robertson B, Autran CA, et al. Human milk oligosaccharides, milk microbiome and infant gut microbiome modulate neonatal rotavirus infection. Nature Communications. 2018;9(1):5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Noll Alexander J, Yu Y, Lasanajak Y, Duska-McEwen G, Buck Rachael H, Smith David F, et al. Human DC-SIGN binds specific human milk glycans. Biochemical Journal. 2016;473(10):1343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Read DJS, Breastfeeding T, Groupa HITS. Late Postnatal Transmission of HIV-1 in Breast-Fed Children: An Individual Patient Data Meta-Analysis. The Journal of Infectious Diseases. 2004;189(12):2154–66. [DOI] [PubMed] [Google Scholar]
- 82.Bode L, Kuhn L, Kim H-Y, Hsiao L, Nissan C, Sinkala M, et al. Human milk oligosaccharide concentration and risk of postnatal transmission of HIV through breastfeeding. The American journal of clinical nutrition. 2012;96(4):831–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duska-McEwen G, Senft A, Ruetschilling T, Barrett E, Buck R. Human Milk Oligosaccharides Enhance Innate Immunity to Respiratory Syncytial Virus and Influenza in Vitro. Food and Nutrition Sciences. 2014;5:1383–95. [Google Scholar]
- 84.Leung JM, Graham AL, Knowles SCL. Parasite-Microbiota Interactions With the Vertebrate Gut: Synthesis Through an Ecological Lens. Frontiers in microbiology. 2018;9:843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kapitan M, Niemiec MJ, Steimle A, Frick JS, Jacobsen ID. Fungi as Part of the Microbiota and Interactions with Intestinal Bacteria. In: Rodrigues ML, editor. Fungal Physiology and Immunopathogenesis. Cham: Springer International Publishing; 2019. p. 265–301. [DOI] [PubMed] [Google Scholar]
- 86.Saiman L, Ludington E, Dawson JD, Patterson JE, Rangel-Frausto S, Wiblin RT, et al. Risk factors for Candida species colonization of neonatal intensive care unit patients. The Pediatric Infectious Disease Journal. 2001;20(12):1119–24. [DOI] [PubMed] [Google Scholar]
- 87.Gonia S, Tuepker M, Heisel T, Autran C, Bode L, Gale CA. Human Milk Oligosaccharides Inhibit Candida albicans Invasion of Human Premature Intestinal Epithelial Cells. The Journal of Nutrition. 2015;145(9):1992–8. [DOI] [PubMed] [Google Scholar]
- 88.Jantscher-Krenn E, Lauwaet T, Bliss LA, Reed SL, Gillin FD, Bode L. Human milk oligosaccharides reduce Entamoeba histolytica attachment and cytotoxicity in vitro. British Journal of Nutrition. 2012;108(10):1839–46. [DOI] [PubMed] [Google Scholar]
- 89.Gnoth MJ, Rudloff S, Kunz C, Kinne RKH. Investigations of the in Vitro Transport of Human Milk Oligosaccharides by a Caco-2 Monolayer Using a Novel High Performance Liquid Chromatography-Mass Spectrometry Technique. Journal of Biological Chemistry. 2001;276(37):34363–70. [DOI] [PubMed] [Google Scholar]
- 90.Dotz V, Rudloff S, Meyer C, Lochnit G, Kunz C. Metabolic fate of neutral human milk oligosaccharides in exclusively breast-fed infants. Molecular Nutrition & Food Research. 2015;59(2):355–64. [DOI] [PubMed] [Google Scholar]
- 91.Rapoport EM, Kurmyshkina OV, Bovin NV. Mammalian galectins: Structure, carbohydrate specificity, and functions. Biochemistry (Moscow). 2008;73(4):393–405. [DOI] [PubMed] [Google Scholar]
- 92.Shams-Ud-Doha K, Kitova EN, Kitov PI, St-Pierre Y, Klassen JS. Human Milk Oligosaccharide Specificities of Human Galectins. Comparison of Electrospray Ionization Mass Spectrometry and Glycan Microarray Screening Results. Analytical Chemistry. 2017;89(9):4914–21. [DOI] [PubMed] [Google Scholar]
- 93.El-Hawiet A, Chen Y, Shams-Ud-Doha K, Kitova EN, St-Pierre Y, Klassen JS. High-Throughput Label- and Immobilization-Free Screening of Human Milk Oligosaccharides Against Lectins. Analytical Chemistry. 2017;89(17):8713–22. [DOI] [PubMed] [Google Scholar]
- 94.Koliwer-Brandl H, Siegert N, Umnus K, Kelm A, Tolkach A, Kulozik U, et al. Lectin inhibition assays for the analysis of bioactive milk sialoglycoconjugates. International dairy journal. 2011;2011 v.21 no.6(no. 6):pp. 413–20. [Google Scholar]
- 95.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nature Reviews Immunology. 2007;7(4):255–66. [DOI] [PubMed] [Google Scholar]
- 96.O’Reilly MK, Paulson JC. Siglecs as targets for therapy in immune-cell-mediated disease. Trends in pharmacological sciences. 2009;30(5):240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chichlowski M, German JB, Lebrilla CB, Mills DA. The Influence of Milk Oligosaccharides on Microbiota of Infants: Opportunities for Formulas. Annual Review of Food Science and Technology. 2011;2(1):331–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rudloff S, Stefan C, Pohlentz G, Kunz C. Detection of ligands for selectins in the oligosaccharide fraction of human milk. European Journal of Nutrition. 2002;41(2):85–92. [DOI] [PubMed] [Google Scholar]
- 99.Osborn L Leukocyte adhesion to endothelium in inflammation. Cell. 1990;62(1):3–6. [DOI] [PubMed] [Google Scholar]
- 100.Springer TA. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 1994;76(2):301–14. [DOI] [PubMed] [Google Scholar]
- 101.Bode L, Kunz C, Muhly-Reinholz M, Mayer K, Seeger W, Rudloff S. Inhibition of monocyte, lymphocyte, and neutrophil adhesion to endothelial cells by human milk oligosaccharides. Thrombosis and haemostasis. 2004;92(6):1402–10. [DOI] [PubMed] [Google Scholar]
- 102.Kurakevich E, Hennet T, Hausmann M, Rogler G, Borsig L. Milk oligosaccharide sialyl(α2,3)lactose activates intestinal CD11c<sup>+</sup> cells through TLR4. Proceedings of the National Academy of Sciences. 2013;110(43):17444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thomas PG, Carter MR, Atochina O, Da’Dara AA, Piskorska D, McGuire E, et al. Maturation of Dendritic Cell 2 Phenotype by a Helminth Glycan Uses a Toll-Like Receptor 4-Dependent Mechanism. The Journal of Immunology. 2003;171(11):5837. [DOI] [PubMed] [Google Scholar]
- 104.McArthur JB, Yu H, Chen X. A Bacterial β1–3-Galactosyltransferase Enables Multigram-Scale Synthesis of Human Milk Lacto-N-tetraose (LNT) and Its Fucosides. ACS Catalysis. 2019;9(12):10721–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Craft KM, Townsend SD. Synthesis of lacto-N-tetraose. Carb Res. 2017;440–441: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bandara MD, Stine KJ, Demchenko AV. Chemical synthesis of human milk oligosaccharides: lacto-N-neohexaose (Galβ1 → 4GlcNAcβ1→)2 3,6Galβ1 → 4Glc. Organic & Biomolecular Chemistry. 2020;18(9):1747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bandara MD, Stine KJ, Demchenko AV. Chemical Synthesis of Human Milk Oligosaccharides: Lacto-N-hexaose Galβ1→3GlcNAcβ1→3 [Galβ1→4GlcNAcβ1→6] Galβ1→4Glc. The Journal of Organic Chemistry. 2019;84(24):16192–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bandara MD, Stine KJ, Demchenko AV. The chemical synthesis of human milk oligosaccharides: Lacto-N-neotetraose (Galβ1→4GlcNAcβ1→3Galβ1→4Glc). Carbohydrate Research. 2019;483:107743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bandara MD, Stine KJ, Demchenko AV. The chemical synthesis of human milk oligosaccharides: Lacto-N-tetraose (Galβ1→3GlcNAcβ1→3Galβ1→4Glc). Carbohydrate Research. 2019;486:107824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chambers SA, Townsend SD. Bioorthogonal human milk oligosaccharide probes for antimicrobial target identification within Streptococcus agalactiae. Carb Res. 2020;488:107895. [DOI] [PubMed] [Google Scholar]


