Summary
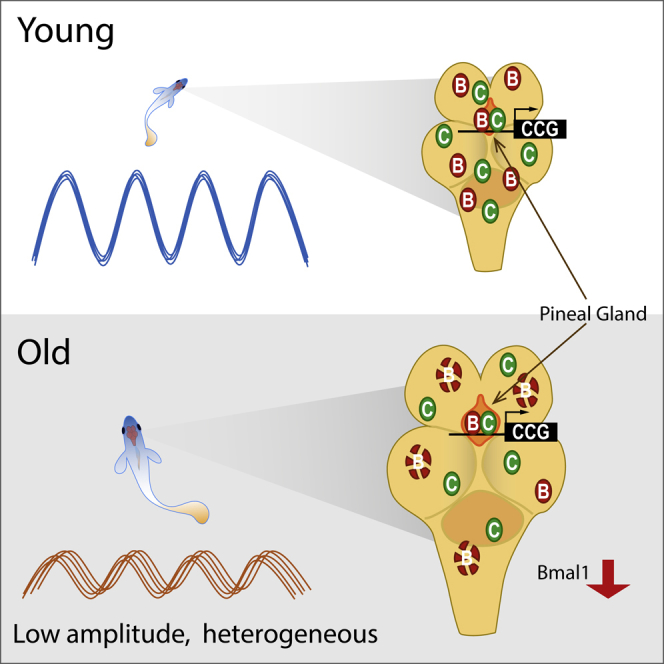
Circadian rhythm is altered during aging, although the underlying molecular mechanisms remain largely unknown. Here, we used the turquoise killifish as a short-lived vertebrate model to examine the effects of aging on the major circadian network comprising the four mammalian clock protein homologs, Bmal1, Clockb, Cry1b, and Per3, which are highly conserved in the killifish with 50%–85% amino acid sequence identity to their human counterparts. The amplitude of circadian rhythm was smaller in old fish (14 weeks) than in young fish (6 weeks). In old fish brain, the Bmal1 protein level was significantly downregulated. However, the Bmal1 interaction with Clockb and chromatin binding of Bmal1 to its downstream target promoters were retained. Furthermore, Bmal1 was relatively well maintained in the pineal gland compared with other regions of the old fish brain. The results suggest that the circadian clock system in the killifish becomes spatially confined to the pineal gland upon aging.
Subject areas: Biological Sciences, Physiology, Molecular Biology, Chronobiology
Graphical Abstract
Highlights
-
•
The amplitude of free-running circadian rhythms decreases during aging in killifish
-
•
Core clock genes are highly conserved in the turquoise killifish genome
-
•
Bmal1 protein expression decreases in whole brain tissue with aging
-
•
Bmal1 expression is relatively well maintained in the pineal gland with aging
Biological Sciences; Physiology; Molecular Biology; Chronobiology
Introduction
Most organisms have a circadian system characterized by an endogenous biological rhythm with a period of approximately 24 h, which is critical for physiological, metabolic, and developmental adaptation to daily environmental changes. This rhythm evolved in response to regular repetitive changes in environmental factors such as light and temperature resulting from the Earth's rotation, but it can also be maintained under a constant environment as an internal clock (Panda et al., 2002b). The circadian clock controls a wide range of biological responses, including transcriptional and translational regulation, hormone secretion, metabolic cycling, and the sleep-wake cycle (Farhud and Aryan, 2018). In mammals, approximately 20% of transcripts undergo daily oscillations in expression (Du et al., 2014; Koike et al., 2012; Menet et al., 2012; Mure et al., 2018; Panda et al., 2002a; Ueda et al., 2002; Zhang et al., 2014), implying that a significant portion of physiological processes is under the control of the circadian clock. The circadian clock system consists of three main components: the input pathway that transfers the external environmental changes into the core system, the core oscillator that generates the 24 h rhythm, and the output pathway that leads to rhythmic activities (Lowrey and Takahashi, 2004; Quintero et al., 2003). In animals, the core oscillator consists of transcription/translation feedback loops formed by multiple components that modulate the abundance, interactions, and localization of various proteins during the entire day (Bell-Pedersen et al., 2005; Hardin, 2006; Panda et al., 2002b; Partch et al., 2014).
Four groups of genes encode the proteins that form the core oscillatory feedback loop in mammalian circadian systems: brain and muscle Aryl Hydrocarbon Receptor Nuclear Translocator-like 1/Brain and Muscle ARNT-Like 1 (ARNTL/BMAL1), circadian locomotor output cycles kaput (CLOCK), cryptochromes (CRYs), and periods (PERs). In the morning, BMAL1 and CLOCK form heterodimers that bind to the E-boxes, which are abundant in the promoters of PERs and CRYs, and lead to a gradual increase in the expression of PERs and CRYs during the day (Bunger et al., 2000; Gekakis et al., 1998; King et al., 1997; Kume et al., 1999; Yoo et al., 2005; Zheng et al., 2001). This accumulation is attenuated by casein-kinase-dependent protein phosphorylation and SCF ubiquitin ligase-dependent degradation (Camacho et al., 2001; Eide et al., 2005; Shirogane et al., 2005; Vanselow et al., 2006). As night falls, PER and CRY form a complex that is protected from further degradation and transported into the nucleus. The nuclear PER-CRY complex then negatively regulates the gene expression of PERs and CRYs by disrupting the BMAL1/CLOCK dimer (Griffin et al., 1999; Kume et al., 1999; Lee et al., 2001; Sato et al., 2006).
In the mammalian circadian system, the suprachiasmatic nucleus (SCN) of the brain is the center of the master clock oscillator and is the main site that controls circadian physiology and behavior (Moore and Eichler, 1972; Stephan and Zucker, 1972; Yamazaki et al., 2000). The core circadian oscillator in SCN generates approximately 24-h cycling rhythms by receiving light input from eye photoreceptors. The circadian signals from the SCN are delivered to other tissues, such as the pineal gland and liver. Unlike mammals, teleost fish tissues and cells can generate their own rhythms by directly sensing external light (Whitmore et al., 2000); therefore, defining a site of the master circadian clock in teleost fish has been difficult.
As an organism ages, the circadian clock system and the rhythmic physiological cycles undergo changes (Banks et al., 2015; Dijk et al., 1999; Espiritu, 2008; Farajnia et al., 2012; Pandi-Perumal et al., 2005; Roenneberg et al., 2007; Van Cauter et al., 1998). However, the mechanisms underlying age-dependent changes of circadian clock system and physiological rhythms have yet to be clearly defined. For example, It is still unclear which accessory and/or core clock components undergo age-dependent changes in function and which are responsible for the age-dependent changes in physiological rhythms (Banks et al., 2016; Bonaconsa et al., 2014; Weinert et al., 2001). Conversely, it is known that defects in circadian components can affect longevity and age-related phenotypes (Kondratov et al., 2006, 2009; Sun et al., 2006). Knockout of the key clock components, CLOCK and/or BMAL1, reduces the lifespan of mice (Dubrovsky et al., 2010; Kondratov et al., 2006, 2009; Sun et al., 2006). Knockout of BMAL1 in mice causes neurodegeneration, sarcopenia, and early mortality (Kondratov et al., 2006). Thus, aging and the circadian clock system greatly affect each other, although the molecular mechanisms underlying their interactions remain unknown. Here, we investigated age-dependent alterations of the circadian clock system in Nothobranchius furzeri (the turquoise killifish), a short-lived vertebrate model organism, and suggest a possible molecular mechanism that explains age-associated alterations in circadian physiology.
Results
Changes in free-running circadian rhythms in old fish under LL and DD conditions
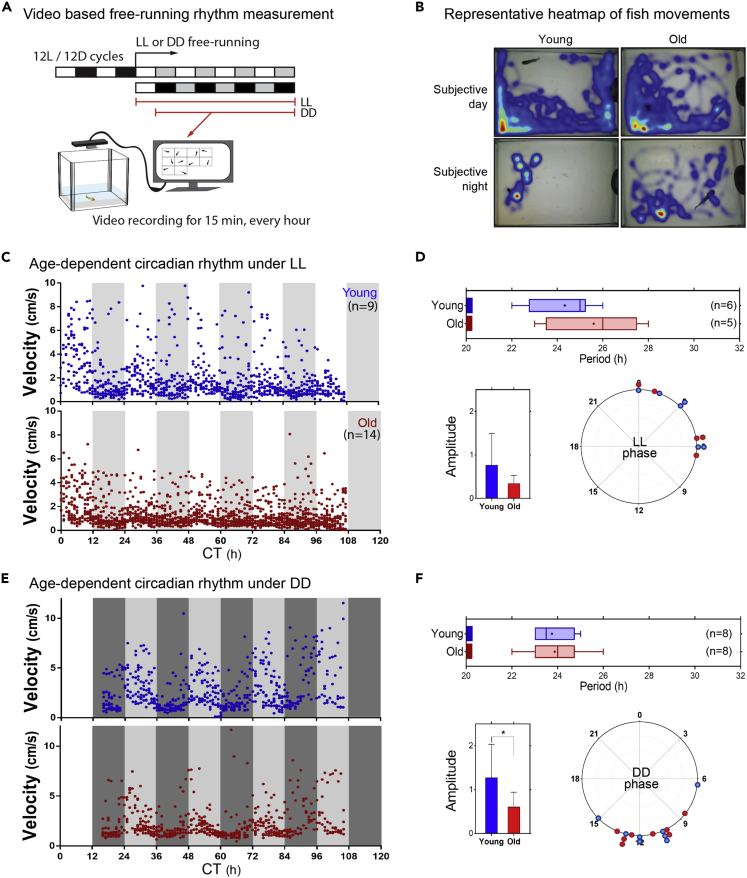
To monitor the behavior of turquoise killifish under free-running conditions, we first developed a circadian rhythm measurement system using a culture facility equipped with continuous flow-through water circulation. Monitoring locomotor activity is a typical non-invasive assay for measuring circadian rhythms. As the fish move three-dimensionally in water, monitoring the locomotor activity of fish is conventionally performed using laser sensors installed at the bottom of the tank and/or near the water surface in separate behavior rooms or facilities, and the circadian rhythm is assessed by measuring the number of times the fish touch the sensors over time (Hurd et al., 1998; Lucas-Sanchez et al., 2011). However, this assay, despite its convenience, generates a square-shaped output, so it is intuitively difficult to find the key parameters (period, phase, and amplitude) of the circadian rhythm. To improve the non-invasive characterization of circadian rhythmicity in a conventional locomotor activity measurement system, we developed a cost-effective video-based LL (continuous light) and DD (continuous darkness) free-running rhythm monitoring system with a continuous water flow in a culture facility (Figure 1A, see Transparent methods for details).
Figure 1.
Changes in free-running circadian rhythms in old fish under LL and DD conditions
(A) Experimental setup of a video-based circadian rhythm measurement system under free-running conditions.
(B) Representative images of fish movement during the subjective day and night. Cumulative fish movements are presented as a heatmap.
(C–F) (C and E) The mean velocity of young (6 weeks old) and old (14 weeks old) fish, respectively, under LL (C) and DD (E) free-running conditions. Blue and red dots represent individual young and old fish, respectively. The gray (C) and the dark gray backgrounds (E) denote subjective night. CT denotes “Circadian time.” (D and F) Characterization of circadian rhythmicity of young and old fish under LL (D) and DD (F). The rhythmic behavior of individual fish was analyzed with JTK_CYCLE (Adj. p < 0.1, see Table S1). For amplitude, data are expressed as the mean ± SD, ∗p < 0.05.
Using the custom-built video-based free-running monitoring system, we examined for potential changes in circadian rhythms during fish aging. GRZ-AD, the shortest-lived strain used in this study, had a median lifespan of approximately 16 weeks under our culture conditions. Thus, we measured endogenous circadian locomotor activity of young (6 weeks old, after sexual maturation) and old (14 weeks old, approximate median lifespan) fish under free-running conditions. Fish activity was first measured under LL by determining how many times the fish entered a specific sector during 15 minutes every hour, which was reported previously to represent locomotor activity (Lucas-Sanchez et al., 2011). We also analyzed movement duration, velocity, and total distance covered by the moving fish (Figure S1).
The raw video and heatmap of movement revealed differences in the general movement of young fish between day and night compared with that of older fish (Figure 1B and Video S1). The overall movement throughout the free-running cycles was higher in young fish than in old fish. Similar results were obtained by measuring mean velocity, cumulative movement duration, and total distance of movement, so mean velocity was chosen for further analysis (Figure S1).
The mean velocity of each fish was calculated to analyze endogenous rhythmicity under LL and DD free-running conditions. In the LL condition, we first noticed that the proportion of fish with rhythmic circadian activity was far smaller among old fish than among young fish; six of nine young fish and five of fourteen old fish showed significant rhythmic behavior under the LL condition (Table S1A). The circadian periods of the young and old fish with rhythmic circadian activity (adjusted p < 0.1) were 24.3 and 25.6 h under the LL condition, respectively, but the difference was not statistically significant (Figure 1C and Table S1A). In addition, old fish with rhythmic circadian activity showed more variation in circadian periods than young fish (Figure 1D). Under the DD condition, circadian rhythmicity was measurable in all young and old individuals, but neither their circadian periods (23.75 and 23.88 h for young and old fish, respectively) nor their phases differed significantly (Figure 1E and Table S1B). However, the amplitude of circadian rhythm was significantly smaller in old fish than in young fish under DD conditions (Figure 1F). These results suggested that free-running circadian rhythmicity was altered in old fish.
Core circadian clock components are highly conserved in the turquoise killifish
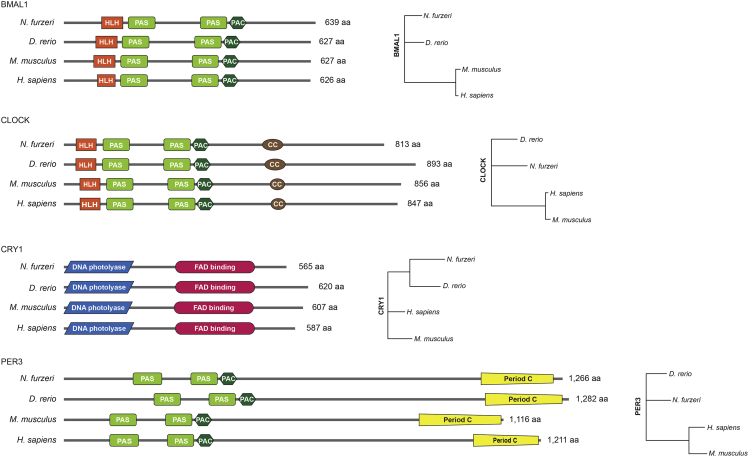
Having confirmed that the turquoise killifish exhibits typical circadian behavior with a free running rhythm in swimming behavior, we searched for homologs of the core circadian clock components, BMAL1, CLOCK, CRY1, and PER3, in the turquoise killifish genome. The turquoise killifish Bmal1, Clockb, Cry1b, and Per3 showed 85%, 65%, 81%, and 50% overall amino acid sequence identity with the corresponding human proteins, respectively, and they showed the highest amino acid sequence homology with human counterparts (Figures 2 and S2 and Table S1C); the amino acid identity is notably higher for Bmal1 and Cry1b than for Clockb and Per3. Multiple ancient paralogs of the circadian clock genes have been discovered based on amino acid comparisons. In the turquoise killifish genome there are three bmal genes (bmal1, bmal2a, and bmal2b), two clock genes (clocka and clockb), five cry genes (cry1a, cry1b, cry2a, cry2b, and cry5), and four per genes (per1b, per2a, per2b, and per3) (Table S1C). The core clock proteins with the highest homology to their human counterparts were further compared with those of mice and zebrafish, and their relationships were visualized as a phylogenetic tree (Figures 2 and S2). The turquoise killifish homologs of the four core clock proteins contained all the conserved functional domains of core clock proteins (Figure 2). Together, these results indicated overall high conservation of core clock proteins in the turquoise killifish.
Figure 2.
Core circadian clock components are highly conserved in the turquoise killifish
The functional domains of the core clock proteins in four species, Nothobranchius furzeri (turquoise killifish), Danio rerio (zebrafish), Mus musculus (mouse), and Homo sapiens (human) are shown on the left. Translated amino acid sequences of turquoise killifish, zebrafish, mice, and human BMAL1 (XM_015965280 [bmal1], NM_131577 [bmal1a], NM_007489, and NM_001030272, respectively), CLOCK (XM_015971720 [clockb], BC163244 [clock1], AF000998, and AF011568, respectively), CRY1 (XM_015964931 [cry1b], NM_001077297 [cry1aa], NM_007771, and NM_004075, respectively) and PER3 (XM_015959552 [per3], AF254792, NM_011065, and NM_001289862, respectively) were used for the analysis. The phylogenetic trees (right) were constructed using neighbor-joining clustering.
See also Figure S2.
The mRNA expression of core circadian clock genes changes slightly during aging
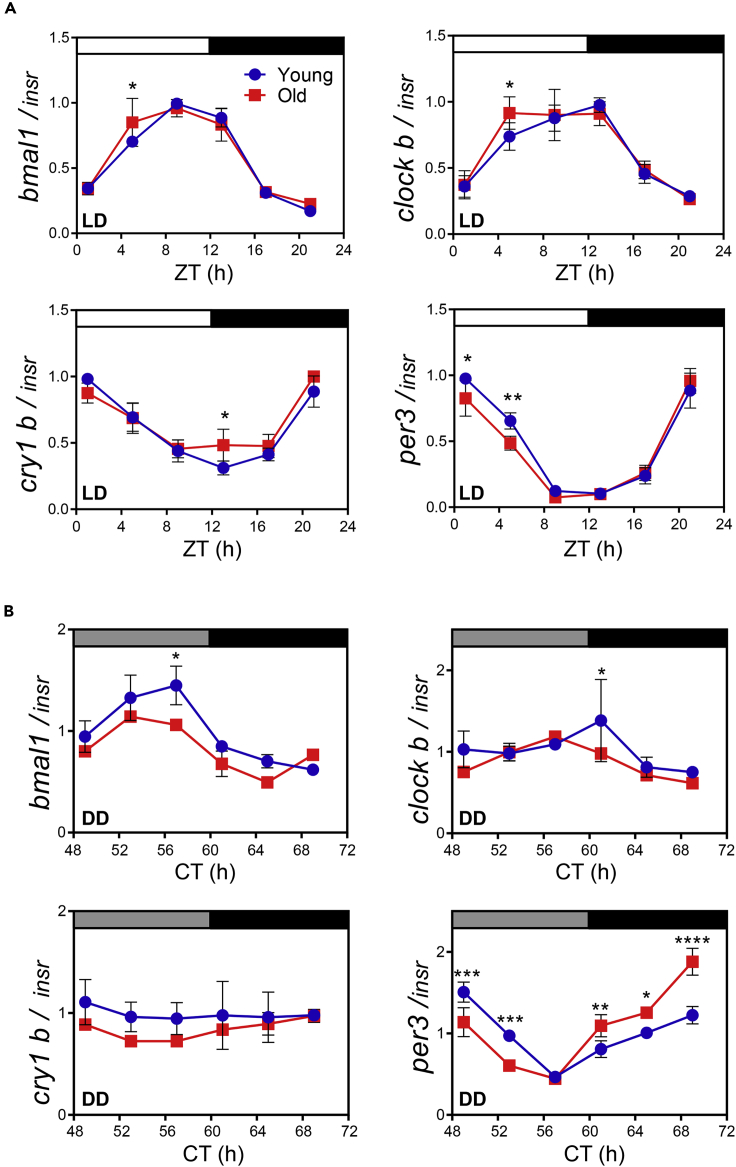
To determine whether the four core clock genes show rhythmic expression in the fish brain and, if so, whether their rhythmic expression is altered in old fish, their mRNA expression in brains of young and old fish was measured throughout the day. Brain aging was first observed by staining brain sections with Fluoro-Jade B, and the level of neurodegeneration was greater in old fish than in young fish (Figure S3). In the LD (light-dark cycles) condition, expression of bmal1 and clockb showed peaks around 10 h after light on (zeitgeber time [ZT] 10 h), whereas that of cry1b and per3 exhibited a relatively higher peak around ZT 22 h (Figures 3A and S4A and Table S1D), as expected for turquoise killifish, a diurnal animal, whose expression of these genes is anti-phasic with respect to the corresponding genes in nocturnal animals such as mice. The circadian control of turquoise killifish clock gene expression was robust under the LD condition, as indicated by the >3-fold difference between peak and trough expression of all genes tested. bmal1/clockb and cry1b/per3 were controlled by antagonistic cycles, suggesting that they can form a transcriptional/translational feedback loop similar to that in other model organisms (Figure S4A). The circadian amplitude in the expression of the four clock genes was not significantly different between young and old fish brains. These results indicated that aging has a minor effect on the rhythmic mRNA expression of the core clock genes under the LD condition.
Figure 3.
mRNA expression of core circadian clock genes changes slightly during aging
(A and B) Expression of the four core clock genes, bmal1, clockb, cry1b, and per3, in the whole brain of young (6-week-old) and old (14-week-old) killifish under LD (A) and DD (B). The expression of each gene was normalized to the expression of insr, a gene that shows constant expression during aging. Data are expressed as the mean ± SD of five fish at each time point. ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.001 and ∗∗∗∗p<0.0001 after two-way ANOVA followed by the Sidak's multiple comparison test.
See also Figures S3 and S4 and Table S1.
Core clock gene expression was also assessed 3 days after transferring the fish to a DD free-running condition (Figure 3B). bmal1, clockb, and per3 cycled throughout the day, similar to the pattern observed under the LD condition; however, cyclic cry1b expression was almost completely abolished in both young and old fish under the DD condition. The phase of bmal1 and per3 expression was more advanced in old fish that in young fish under the DD condition (Figures 3B and S4B and Table S1E). However, the expression patterns of the four core genes under the DD condition did not show a marked change in the brain of old fish. These results show that the rhythmic expression of cry1b is more critically affected by the DD condition than by the LL condition, but the amplitude and period of expression of the four core clock genes in the brain of old fish were similar between the DD condition and LD condition.
The Bmal1 protein level decreases dramatically in the brain of old turquoise killifish
Next, we examined potential post-transcriptional changes associated with organismal age (Figure 4). For this purpose, we used human BMAL1 and CLOCK antibodies to detect killifish Bmal1 and Clock proteins, because antibodies against killifish Bmal1 and Clockb were not available and Bmal1 and Clockb proteins show high amino acid identities to their human homologs (85% and 65%, respectively). First, we tested the specificity of human BMAL1 and CLOCK antibodies for the detection of synthetic Bmal1 and Clockb proteins of the turquoise killifish. Both anti-BMAL1 and anti-CLOCK antibodies detected ectopic Bmal1 and Clock proteins of the turquoise killifish and had sufficient specificity to detect endogenous Bmal1 and Clockb in the killifish brain (Figure S5).
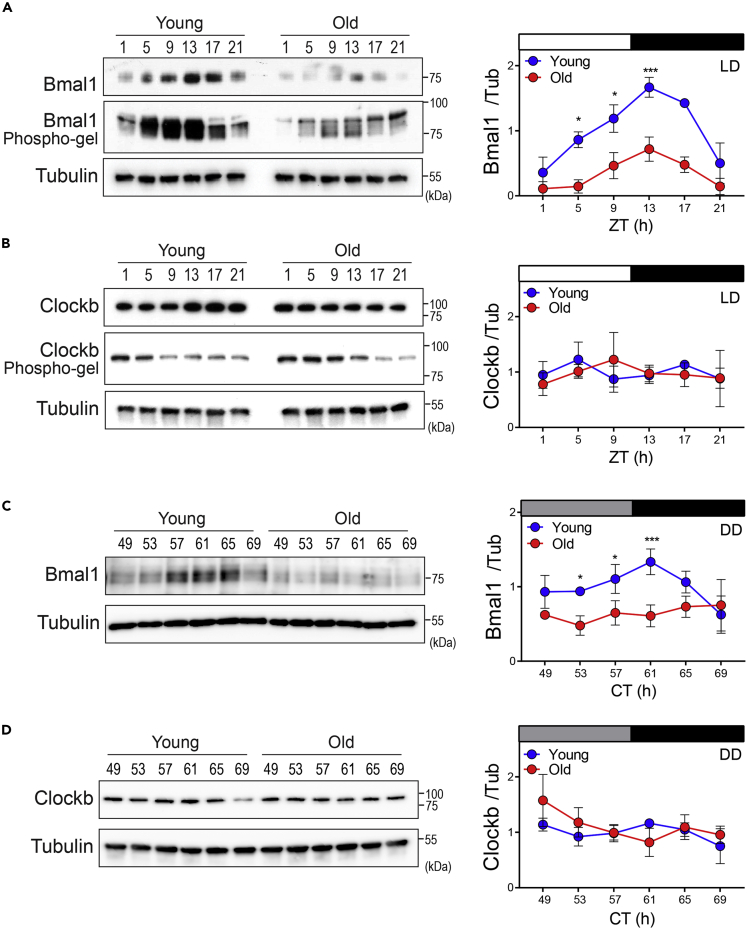
Figure 4.
The Bmal1 protein level decreases dramatically in the brain of old turquoise killifish
(A and B) Protein levels, phosphorylation patterns (left), and quantified protein levels (right) of Bmal1 (A) and Clockb (B) under LD.
(C and D) (C) Bmal1 and Clockb protein levels (left) and their quantification (right) under DD. The protein levels were normalized to that of tubulin. Data are presented as the mean ± SD from three biological replicates. ∗p < 0.05 and ∗∗∗p < 0.001 after two-way ANOVA followed by the Sidak's multiple comparison test.
See also Figures S5 and S6 and Table S1.
Assessment of Bmal1 and Clockb protein expression in killifish brain tissue under LD and DD conditions showed that Bmal1 protein expression cycled during the day in both young and old brains. The expression of the Bmal1 protein peaked at approximately ZT13 under the LD condition, and a 3-fold difference in the abundance of Bmal1 protein was observed between its peak and the trough levels. In old fish brain, the abundance and amplitude of Bmal1 protein expression were lower than in young brain, whereas the phase and period were not different under the LD condition (Figures 4A and S6A and Table S1F). Clockb protein was expressed at a constant level during the day, and its abundance was not significantly different between old and young brains (Figure 4B). Similar patterns of Bmal1 and Clockb protein expression were observed under the DD condition (Figure 4C). Bmal1 protein abundance and its amplitude were significantly lower in old fish brain than in young fish brain, but there were no changes in the phase and period of its expression. Clockb protein expression did not differ significantly between young and old fish under the DD condition (Figures 4C and S6B and Table S1G).
We also examined the phosphorylation of Bmal1 and Clockb in young and old fish brains by measuring in-gel differences in the mobility of phosphorylated and non-phosphorylated forms of these proteins (See Transparent methods for details). Bmal1 from young fish brain was detected as four separate bands, and the relative intensities of the four bands varied throughout the day, indicating that Bmal1 underwent diurnal changes in phosphorylation. The abundance of phosphorylated Bmal1 was lower in old fish brain (Figure 4A), and the relative intensities of the four bands were different from those of young fish brain during the day, indicating that Bmal1 in old fish brain was subject to different types of phosphorylation throughout the day. Clockb protein from young and old turquoise killifish brains migrated as a single band suggesting that it might be uniformly phosphorylated or not phosphorylated (Figure 4B).
The amount and function of the Bmal1/Clockb heterodimer are retained in the old killifish brain
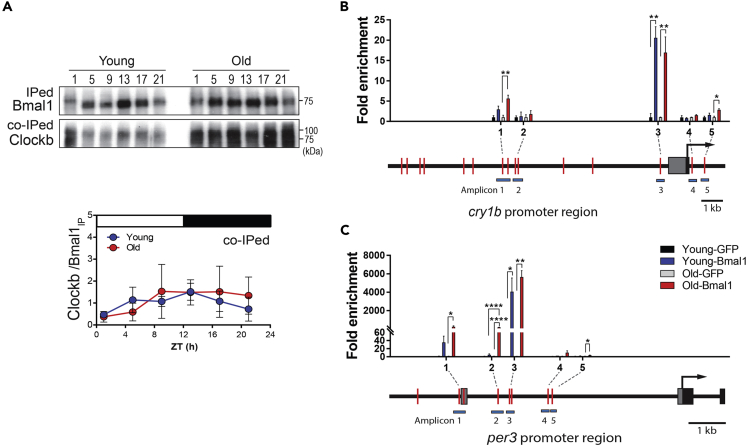
In circadian clock system, Bmal1 and Clockb form a complex to control downstream genes including the evening clock genes, cry1b and per3. As fish age, the expression of Bmal1 dramatically decreased; thus we tested if this age-dependent change in Bmal1 expression affects the Bmal1/Clockb complex pathway. We first confirmed that Bmal1 and Clockb proteins interact in the killifish brain by performing bidirectional immunoprecipitation using anti-BMAL1 or anti-CLOCK antibodies (Figure S7). The Bmal1/Clockb interaction was monitored throughout the day in young and old fish brains (Figure 5A). After normalizing the Bmal1/Clockb interaction using immunoprecipitated Bmal1, we noticed that the relative Bmal1/Clockb interaction was not markedly different between old fish brain and young fish brain (Figure 5A). Although the Bmal1 protein abundance decreased during aging, the Bmal1/Clockb interaction was maintained or protected in the aging brain.
Figure 5.
The amount and function of the Bmal1/Clockb heterodimer are retained in old killifish brain
(A) Bmal1/Clockb heterodimer formation throughout the day in the brain of young and old fish. Co-immunoprecipitated (co-IPed) Clockb was normalized against IPed-Bmal1 for each trial. Data are presented as the mean ± SD from three replicates.
(B and C) Binding affinity of Bmal1 to the promoter regions of cry1b (B) and per3 (C) in the brains of young and old fish. Schematic diagrams of the promoter regions of cry1b and per3 are shown along with the amplicon positions. E-box, red bar; gray box, 5′-UTR; black box, protein coding sequence; solid arrow, start codon; pale blue bar, amplicon position. Data are presented as the mean ± SE from three replicates. Asterisks indicate ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001 after two-way ANOVA followed by the Tukey's multiple comparison test.
See also Figure S7.
Next, we examined the chromatin binding of Bmal1 to the E-box regions of circadian clock-regulated gene promoters. The region upstream of the start codon in the cry1b and per3 genes was searched to see if it contained canonical E-box (CACGTG) and non-canonical E-box (CACGTT and CAGCTT) sequences (Nakahata et al., 2008; Salero et al., 2003; Zhang et al., 2012). To analyze for age-dependent differences in Bmal1-chromatin association in the killifish brain, five E-box-containing regions in each of the cry1b and per3 promoter regions were targeted. Among the five amplicons tested, three cry1b and four per3 E-box-containing genomic regions were identified as Bmal1-binding sites, but not in the GFP control. Only one target sequence in the per3 promoter (amplicon 2 in Figure 5C, −4,720 to −4,917 before the start codon) showed a significant age-dependent increase in Bmal1 chromatin association. These results indicate that despite the large downregulation of Bmal1 in old fish brain, Bmal1/Clockb heterodimer formation and Bmal1 chromatin binding to E-boxes were relatively well maintained in old fish brain. This was consistent with the subtle changes in the age-dependent expression of cry1b and per3 mRNA expression.
Bmal1 is relatively well maintained in the pineal gland of old turquoise killifish brain
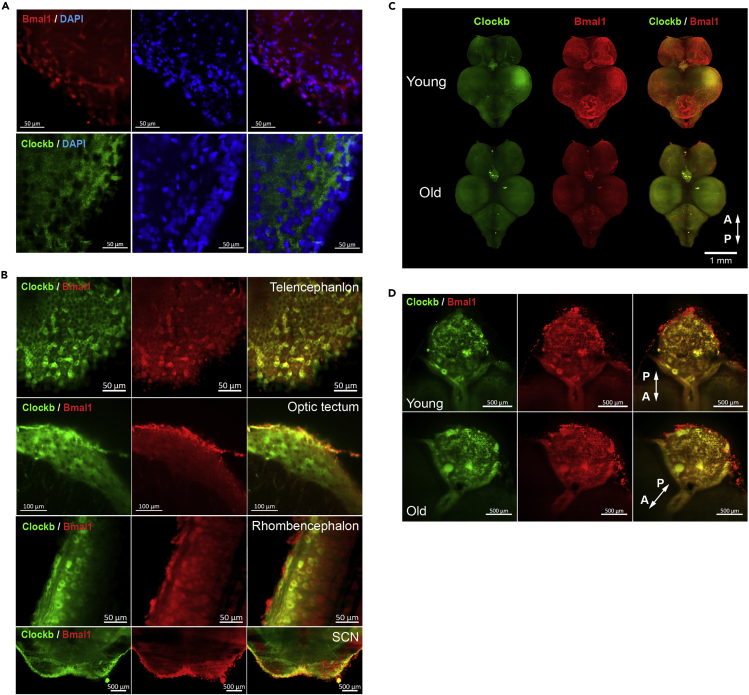
It was curious that the Bmal1/Clockb interaction and Bmal1-chromatin association were retained in old killifish brain despite the significant decrease in the Bmal1 protein level. One possibility was that Bmal1/Clockb heterodimer formation occurs in a specific location in the brain where the local expression of Bmal1 for Bmal1/Clockb heterodimer formation is relatively well sustained during aging. To test this idea, we used whole brain and detected Bmal1 and Clockb proteins in the killifish brain after making the brain transparent.
To get an insight into the function of Bmal1 and Clockb, we first examined their intracellular and tissue localizations in young fish brain. Bmal1 protein was localized to both the nucleus and cytoplasm, whereas Clockb protein was preferentially localized to the cytoplasm (Figure 6A). The two proteins were detected throughout the brain but their spatial distribution differed slightly (Figure 6B and Videos S2, S3, and S4). For example, both Bmal1 and Clockb were detected on the surface of the brain, but Clockb protein was detected in a deeper layer than Bmal1 in the optic tectum (Figure 6B and Video S3).
Figure 6.
Bmal1 is relatively well maintained in the pineal gland of old turquoise killifish brain
(A) Subcellular localization of Bmal1 and Clockb in the turquoise killifish brain. Green, red, and blue colors represent Clockb, Bmal1, and DAPI (nuclear marker) staining, respectively.
(B) Co-localization of Bmal1 and Clockb in the telencephalon, optic tectum, rhombencephalon, and SCN of the turquoise killifish brain.
(C) Whole brain immunostaining of Bmal1 and Clockb in young and old turquoise killifish brains.
(D) Bmal1 and Clockb localization in the pineal gland of young and old fish brains. Abbreviations “A” and “P” in the figure represent directions toward the anterior and posterior sides of the brain, respectively.
We then analyzed the effect of aging on the whole brain localization of these two proteins. Fluorescence detection of Bmal1 revealed that its level decreased more drastically than that of Clockb in old fish brain (Figures 6C and S8), which was consistent with the analysis of their expression at the protein level (Figure 4). However, Bmal1 fluorescence was better maintained in the pineal gland than in other parts of the brain. Bmal1 and Clockb co-localized in an internal area of the pineal gland, whereas Bmal1 alone was detected in the epidermal region of the pineal gland (Figures 6C and 6D). These data indicate that Bmal1/Clockb complex formation might be specifically maintained in the pineal gland of old killifish brain.
Discussion
The circadian rhythm is weakened in old killifish
A custom-built non-invasive and cost-effective endogenous circadian locomotor monitoring system was used to obtain a robust and clear picture of the locomotor circadian rhythms of the short-lived turquoise killifish Nothobranchius furzeri, strain GRZ-AD, which includes mean velocity, cumulative movement duration, and total distance of movement under LL and DD conditions. Continuous monitoring of the circadian activity using this system revealed a typical wave-like circadian oscillatory pattern.
The turquoise killifish were active during the day and calm at late night, as well expected for a diurnal animal. In the free-running LL condition, the proportion of fish with significant rhythmic behavior decreased, and the circadian period tended to be longer in old fish. An age-dependent decline in circadian rhythmicity has been reported for other model organisms, including teleost fishes such as Danio rerio (zebrafish) and Nothobranchius korthausae (Lucas-Sanchez et al., 2011; Zhdanova et al., 2008). However, the lifespans of these organisms are relatively long; the median lifespans of zebrafish and N. korthausae are 42 months and 72 weeks (Gerhard et al., 2002; Lucas-Sanchez et al., 2011), respectively. Investigation of age-associated changes in circadian activity using these model organisms requires 4 years for zebrafish and 72 weeks for N. korthausae (Lucas-Sanchez et al., 2011; Zhdanova et al., 2008). The turquoise killifish strain used in this study, GRZ-AD, is short-lived vertebrate with a median lifespan of only 16 weeks under laboratory conditions, and a decrease in circadian locomotor activity was observed even in 14-week-old fish, demonstrating that the killifish model is a reliable and useful system for the study of age-dependent changes in circadian rhythm and physiology. It should also be noted that the killifish is a diurnal animal, like human but unlike mouse, which makes it additionally advantageous as a vertebrate circadian model system.
Free-running circadian locomotor activity was different between LL and DD conditions. Under the DD condition, all young and old fish showed significant circadian behavior unlike under the LL condition. Interestingly, a phase difference between circadian rhythms under LL and DD conditions was also observed and the circadian phase was advanced in LL and delayed in DD (Figures 1D and 1F). The circadian period in DD was not significantly different in old fish; however, we speculate that, with advancing age, the circadian period will get longer in DD as well. As individual variability in circadian locomotor behavior was lower under DD, we propose that the DD condition allows a better estimation of the internal circadian rhythm in turquoise killifish.
The turquoise killifish has highly conserved circadian components
The core circadian system in animals is highly conserved across species, including flies, zebrafish, and mammals (Hardin, 2011; Lowrey and Takahashi, 2004; Vatine et al., 2011). The core circadian oscillatory component complexes in these animals, BMAL1/CLOCK and CRY1/PER3, form an interlocking feedback loop and are well conserved in the turquoise killifish genome. This suggests that the basic molecular mechanisms controlling the circadian clock are comparable between the turquoise killifish and other vertebrate models. There are multiple paralogs of these circadian core genes in the turquoise killifish genome, reflecting their early emergence during evolution and speciation from other teleost fishes (Table S1C). For example, eight ancient paralogs of bmal1 have been identified in the killifish genome and one of them is clockb. All the bmal1 paralogs showed <50% sequence identity with bmal1, and they evolved from bilateral animals, namely, arthropods and nematodes (Ecdysozoa). The clockb gene has one paralog (clocka) and seven ancient paralogs in the killifish genome that are also present in bilateral animals. The clocka gene has the highest amino acid sequence identity (62.26%) with Clockb and is duplicated in teleost fish (Clupeocephala). cry1b has one paralog (cry1a), which is a duplicate from Osteoglossocephalai, and five ancient paralogs that are similar to those found in Saccharomyces cerevisiae. per3 has three ancient paralogs that are similar to those found in arthropods and nematodes (Ecdysozoa). It would be interesting to investigate the key features of the turquoise killifish circadian clock network that distinguish it from the clock regulatory pathways of other vertebrate and non-vertebrate organisms.
Organismal age has little impact on the Bmal1 and Clockb pathway in the brain of the turquoise killifish
The turquoise killifish core clock components show the following conserved properties across species: (1) mRNA cycling of bmal1, clockb, cry1b, and per3 with an opposite phase between bmal1/clockb and cry1b/per3; (2) cycling of the Bmal1 protein; (3) phosphorylation of Bmal1 protein; (4) Bmal1 and Clockb heterodimer formation; and (5) chromatin binding of Bmal1 to the E-Box sequences of promoters of circadian clock-controlled genes. The turquoise killifish Bmal1 and Clockb proteins form a heterodimer that binds to cry1b and per3 promoters, inducing the expression of these genes. This indicates that the negative-positive feedback loop in animal core clocks system is also operational in turquoise killifish.
The age-dependent changes of circadian core gene expression in mammals and flies still remain unclear (Asai et al., 2001; Rakshit et al., 2012). In the other fish model, zebrafish, bmal1a and per1a expression is lower in 4-year-old fish than in 1-year-old fish, whereas clocka expression does not change with age (Zhdanova et al., 2008). In our study using turquoise killifish, bmal1, clockb, cry1b, and per3 showed only minor differences in their rhythmic expression between young (6 weeks old, after sexual maturation) and old (14 weeks old, approximate median lifespan) fish brain (Figure 3). However, overall Bmal1 protein abundance was dramatically lower in old fish brain than in young fish brain throughout the day (Figure 4). This observation provides a clue to understanding the age-dependent change in the fish circadian clock; the post-transcriptional regulation of Bmal1, a core clock protein, occurs before a change occurs in its transcript level. Another important finding is that aging, despite its negative effect on Bmal1 protein abundance, had a minimal effect on Bmal1/Clockb heterodimer formation and E-box binding affinity of Bmal1 (Figure 5). These results suggest that the circadian core oscillator system involving Bmal1 remained robust in old fish, especially in the pineal gland of the brain as explained below.
Bmal1 is preferentially maintained in the pineal gland of old killifish brain
The master circadian clock oscillator is located in the SCN in the mammalian brain to control circadian physiology (Yamazaki et al., 2000; Yoo et al., 2004). However, in teleost fishes including zebrafish, the location of the master clock is unclear, because the light sensing capacity of every cell can generate endogenous rhythms (Whitmore et al., 2000). In young turquoise killifish brain, Bmal1 and Clockb proteins were widely expressed, whereas their cellular localizations were distinct. Analysis of whole brain localization of these proteins in the turquoise killifish showed that Bmal1 and Clockb localized to distinct regions of the brain (Figure 6B). Whole brain imaging of Bmal1 and Clockb suggested that the circadian network associated with Bmal1 or Clockb can form throughout the brain but can also be restricted to specific regions of the brain through the spatiotemporal co-localization of Bmal1 and Clockb.
Whole brain immunostaining provided information on the age-dependent localization of Bmal1 and Clockb in the killifish brain. The fluorescence intensity of Bmal1 was decreased in whole brain images of older fish, in agreement with the decreased overall protein level. However, the fluorescence intensity in one small area of the dorsal brain, the pineal gland, remained constant relative to that in other parts of the brain, and Bmal1 and Clockb co-localized in cells of the pineal gland (Figure 6C). This result suggests that Bmal1 acts as a critical modulator of the circadian network during aging, and that the pineal gland is a major sub-organ involved in the maintenance of circadian activity, most likely via the Bmal1/Clockb complex, in old killifish.
In this study, we identified age-dependent changes in the highly conserved core circadian clock components of the turquoise killifish. We showed that downregulation of the core clock protein Bmal1 occurs mainly at the translational level, but that its expression is maintained in the pineal gland of old killifish brain, probably to maintain circadian gene expression. The pineal gland, which secretes melatonin, is an important region for neuro-endocrine activity (Wurtman et al., 1963) and also acts as a central circadian oscillator in many non-mammalian vertebrates (Fukada and Okano, 2002). The present results suggest that the master clock of the old turquoise killifish might be located in the pineal gland. Our findings thus provide an important insight into the change that occurs in the spatiotemporal regulation of circadian clock system in the brain of aging vertebrates.
Limitations of the study
The pineal gland-specific mechanisms of Bmal1/Clockb complex regulation of circadian rhythms require further investigation. The physiological roles of age-specific and pineal gland-specific maintenance of Bmal1 with cell-type specific markers of pineal gland should help improve our understanding of circadian physiology during aging. Comparison of the spatiotemporal patterns of Bmal1 expression across species could provide further insights into the evolution of the circadian system and the role of Bmal1 in the circadian system of aging vertebrates.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yumi Kim (yumikim@ibs.re.kr).
Materials availability
All unique reagents generated in this study are available from the Lead Contact without restriction.
Data and code availability
All data are available in the main text or Supplemental information. Raw data images can be found Mendeley Data: https://data.mendeley.com/datasets/gy57ymzh67/draft?a=b36c53c2-250b-4b92-bbe3-d1990378e123.
This study did not contain any newly generated datasets or codes.
Methods
All methods can be found in the accompanying Transparent methods supplemental file.
Acknowledgments
We thank Vu Minh Hung for initial setup of the circadian rhythm measurement system. This work was supported by the Institute for Basic Science (IBS-R013-D1).
Author contributions
Y.K. conceived the project. S.L. and Y.K. designed and performed experiments. Y.K., S.L., and H.G.N. wrote the manuscript.
Declaration of interests
The authors have no competing interests to declare.
Published: January 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2020.101905.
Contributor Information
Hong Gil Nam, Email: nam@dgist.ac.kr.
Yumi Kim, Email: yumikim@ibs.re.kr.
Supplemental information
(A) JTK output of LL free-running rhythm.
(B) JTK output of DD free-running rhythm.
(C) Paralogs of circadian clock genes in the turquoise killifish genome.
(D) JTK output of circadian core gene expression under LD.
(E) JTK output of circadian core gene expression under DD.
(F) JTK output of Bmal1 and Clock protein expression under LD.
(G) JTK output of Bmal1 and Clock protein expression under DD.
References
- Asai M., Yoshinobu Y., Kaneko S., Mori A., Nikaido T., Moriya T., Akiyama M., Shibata S. Circadian profile of Per gene mRNA expression in the suprachiasmatic nucleus, paraventricular nucleus, and pineal body of aged rats. J. Neurosci. Res. 2001;66:1133–1139. doi: 10.1002/jnr.10010. [DOI] [PubMed] [Google Scholar]
- Banks G., Heise I., Starbuck B., Osborne T., Wisby L., Potter P., Jackson I.J., Foster R.G., Peirson S.N., Nolan P.M. Genetic background influences age-related decline in visual and nonvisual retinal responses, circadian rhythms, and sleep. Neurobiol. Aging. 2015;36:380–393. doi: 10.1016/j.neurobiolaging.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks G., Nolan P.M., Peirson S.N. Reciprocal interactions between circadian clocks and aging. Mamm. Genome. 2016;27:332–340. doi: 10.1007/s00335-016-9639-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D., Cassone V.M., Earnest D.J., Golden S.S., Hardin P.E., Thomas T.L., Zoran M.J. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaconsa M., Malpeli G., Montaruli A., Carandente F., Grassi-Zucconi G., Bentivoglio M. Differential modulation of clock gene expression in the suprachiasmatic nucleus, liver and heart of aged mice. Exp. Gerontol. 2014;55:70–79. doi: 10.1016/j.exger.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Bunger M.K., Wilsbacher L.D., Moran S.M., Clendenin C., Radcliffe L.A., Hogenesch J.B., Simon M.C., Takahashi J.S., Bradfield C.A. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho F., Cilio M., Guo Y., Virshup D.M., Patel K., Khorkova O., Styren S., Morse B., Yao Z., Keesler G.A. Human casein kinase Idelta phosphorylation of human circadian clock proteins period 1 and 2. FEBS Lett. 2001;489:159–165. doi: 10.1016/s0014-5793(00)02434-0. [DOI] [PubMed] [Google Scholar]
- Dijk D.J., Duffy J.F., Riel E., Shanahan T.L., Czeisler C.A. Ageing and the circadian and homeostatic regulation of human sleep during forced desynchrony of rest, melatonin and temperature rhythms. J. Physiol. 1999;516(Pt 2):611–627. doi: 10.1111/j.1469-7793.1999.0611v.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du N.H., Arpat A.B., De Matos M., Gatfield D. MicroRNAs shape circadian hepatic gene expression on a transcriptome-wide scale. Elife. 2014;3:e02510. doi: 10.7554/eLife.02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky Y.V., Samsa W.E., Kondratov R.V. Deficiency of circadian protein CLOCK reduces lifespan and increases age-related cataract development in mice. Aging (Albany NY) 2010;2:936–944. doi: 10.18632/aging.100241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide E.J., Woolf M.F., Kang H., Woolf P., Hurst W., Camacho F., Vielhaber E.L., Giovanni A., Virshup D.M. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espiritu J.R. Aging-related sleep changes. Clin. Geriatr. Med. 2008;24:1–14. doi: 10.1016/j.cger.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Farajnia S., Michel S., Deboer T., vanderLeest H.T., Houben T., Rohling J.H.T., Ramkisoensing A., Yasenkov R., Meijer J.H. Evidence for neuronal desynchrony in the aged suprachiasmatic nucleus clock. J. Neurosci. 2012;32:5891–5899. doi: 10.1523/JNEUROSCI.0469-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhud D., Aryan Z. Circadian rhythm, lifestyle and health: a narrative review. Iran. J. Public Health. 2018;47:1068–1076. [PMC free article] [PubMed] [Google Scholar]
- Fukada Y., Okano T. Circadian clock system in the pineal gland. Mol. Neurobiol. 2002;25:19–30. doi: 10.1385/MN:25:1:019. [DOI] [PubMed] [Google Scholar]
- Gekakis N., Staknis D., Nguyen H.B., Davis F.C., Wilsbacher L.D., King D.P., Takahashi J.S., Weitz C.J. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Gerhard G.S., Kauffman E.J., Wang X., Stewart R., Moore J.L., Kasales C.J., Demidenko E., Cheng K.C. Life spans and senescent phenotypes in two strains of Zebrafish (Danio rerio) Exp. Gerontol. 2002;37:1055–1068. doi: 10.1016/s0531-5565(02)00088-8. [DOI] [PubMed] [Google Scholar]
- Griffin E.A., Jr., Staknis D., Weitz C.J. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Hardin P.E. Essential and expendable features of the circadian timekeeping mechanism. Curr. Opin. Neurobiol. 2006;16:686–692. doi: 10.1016/j.conb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Hardin P.E. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv. Genet. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd M.W., Debruyne J., Straume M., Cahill G.M. Circadian rhythms of locomotor activity in zebrafish. Physiol. Behav. 1998;65:465–472. doi: 10.1016/s0031-9384(98)00183-8. [DOI] [PubMed] [Google Scholar]
- King D.P., Zhao Y., Sangoram A.M., Wilsbacher L.D., Tanaka M., Antoch M.P., Steeves T.D., Vitaterna M.H., Kornhauser J.M., Lowrey P.L. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N., Yoo S.H., Huang H.C., Kumar V., Lee C., Kim T.K., Takahashi J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov R.V., Kondratova A.A., Gorbacheva V.Y., Vykhovanets O.V., Antoch M.P. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov R.V., Vykhovanets O., Kondratova A.A., Antoch M.P. Antioxidant N-acetyl-L-cysteine ameliorates symptoms of premature aging associated with the deficiency of the circadian protein BMAL1. Aging. 2009;1:979–987. doi: 10.18632/aging.100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K., Zylka M.J., Sriram S., Shearman L.P., Weaver D.R., Jin X., Maywood E.S., Hastings M.H., Reppert S.M. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Lee C., Etchegaray J.P., Cagampang F.R., Loudon A.S., Reppert S.M. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lowrey P.L., Takahashi J.S. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu. Rev. genomics Hum. Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas-Sanchez A., Almaida-Pagan P.F., Madrid J.A., de Costa J., Mendiola P. Age-related changes in fatty acid profile and locomotor activity rhythms in Nothobranchius korthausae. Exp. Gerontol. 2011;46:970–978. doi: 10.1016/j.exger.2011.08.009. [DOI] [PubMed] [Google Scholar]
- Menet J.S., Rodriguez J., Abruzzi K.C., Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R.Y., Eichler V.B. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972;42:201–206. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- Mure L.S., Le H.D., Benegiamo G., Chang M.W., Rios L., Jillani N., Ngotho M., Kariuki T., Dkhissi-Benyahya O., Cooper H.M. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science. 2018;359:1232. doi: 10.1126/science.aao0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y., Yoshida M., Takano A., Soma H., Yamamoto T., Yasuda A., Nakatsu T., Takumi T. A direct repeat of E-box-like elements is required for cell-autonomous circadian rhythm of clock genes. BMC Mol. Biol. 2008;9:1. doi: 10.1186/1471-2199-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S., Antoch M.P., Miller B.H., Su A.I., Schook A.B., Straume M., Schultz P.G., Kay S.A., Takahashi J.S., Hogenesch J.B. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Panda S., Hogenesch J.B., Kay S.A. Circadian rhythms from flies to human. Nature. 2002;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Pandi-Perumal S.R., Zisapel N., Srinivasan V., Cardinali D.P. Melatonin and sleep in aging population. Exp. Gerontol. 2005;40:911–925. doi: 10.1016/j.exger.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Partch C.L., Green C.B., Takahashi J.S. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero J.E., Kuhlman S.J., McMahon D.G. The biological clock nucleus: a multiphasic oscillator network regulated by light. J. Neurosci. 2003;23:8070–8076. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakshit K., Krishnan N., Guzik E.M., Pyza E., Giebultowicz J.M. Effects of aging on the molecular circadian oscillations in Drosophila. Chronobiol. Int. 2012;29:5–14. doi: 10.3109/07420528.2011.635237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roenneberg T., Kuehnle T., Juda M., Kantermann T., Allebrandt K., Gordijn M., Merrow M. Epidemiology of the human circadian clock. Sleep Med. Rev. 2007;11:429–438. doi: 10.1016/j.smrv.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Salero E., Gimenez C., Zafra F. Identification of a non-canonical E-box motif as a regulatory element in the proximal promoter region of the apolipoprotein E gene. Biochem. J. 2003;370:979–986. doi: 10.1042/BJ20021142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T.K., Yamada R.G., Ukai H., Baggs J.E., Miraglia L.J., Kobayashi T.J., Welsh D.K., Kay S.A., Ueda H.R., Hogenesch J.B. Feedback repression is required for mammalian circadian clock function. Nat. Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirogane T., Jin J., Ang X.L., Harper J.W. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- Stephan F.K., Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc. Natl. Acad. Sci. U S A. 1972;69:1583–1586. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Yang Z., Niu Z., Wang W., Peng J., Li Q., Ma M.Y., Zhao Y. The mortality of MOP3 deficient mice with a systemic functional failure. J. Biomed. Sci. 2006;13:845–851. doi: 10.1007/s11373-006-9108-4. [DOI] [PubMed] [Google Scholar]
- Ueda H.R., Chen W.B., Adachi A., Wakamatsu H., Hayashi S., Takasugi T., Nagano M., Nakahama K., Suzuki Y., Sugano S. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- Van Cauter E., Plat L., Leproult R., Copinschi G. Alterations of circadian rhythmicity and sleep in aging: endocrine consequences. Horm. Res. 1998;49:147–152. doi: 10.1159/000023162. [DOI] [PubMed] [Google Scholar]
- Vanselow K., Vanselow J.T., Westermark P.O., Reischl S., Maier B., Korte T., Herrmann A., Herzel H., Schlosser A., Kramer A. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatine G., Vallone D., Gothilf Y., Foulkes N.S. It's time to swim! Zebrafish and the circadian clock. FEBS Lett. 2011;585:1485–1494. doi: 10.1016/j.febslet.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Weinert H., Weinert D., Schurov I., Maywood E.S., Hastings M.H. Impaired expression of the mPer2 circadian clock gene in the suprachiasmatic nuclei of aging mice. Chronobiology Int. 2001;18:559–565. doi: 10.1081/cbi-100103976. [DOI] [PubMed] [Google Scholar]
- Whitmore D., Foulkes N.S., Sassone-Corsi P. Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature. 2000;404:87–91. doi: 10.1038/35003589. [DOI] [PubMed] [Google Scholar]
- Wurtman R.J., Axelrod J., Phillips L.S. Melatonin synthesis in the pineal gland: control by light. Science. 1963;142:1071–1073. doi: 10.1126/science.142.3595.1071. [DOI] [PubMed] [Google Scholar]
- Yamazaki S., Numano R., Abe M., Hida A., Takahashi R., Ueda M., Block G.D., Sakaki Y., Menaker M., Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- Yoo S.H., Ko C.H., Lowrey P.L., Buhr E.D., Song E.J., Chang S.W., Yoo O.J., Yamazaki S., Lee C., Takahashi J.S. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl. Acad. Sci. U S A. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.H., Yamazaki S., Lowrey P.L., Shimomura K., Ko C.H., Buhr E.D., Siepka S.M., Hong H.K., Oh W.J., Yoo O.J. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Lahens N.F., Ballance H.I., Hughes M.E., Hogenesch J.B. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. U S A. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.P., Patel S.P., McCarthy J.J., Rabchevsky A.G., Goldhamer D.J., Esser K.A. A non-canonical E-box within the MyoD core enhancer is necessary for circadian expression in skeletal muscle. Nucleic Acids Res. 2012;40:3419–3430. doi: 10.1093/nar/gkr1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdanova I.V., Yu L., Lopez-Patino M., Shang E., Kishi S., Gueling E. Aging of the circadian system in zebrafish and the effects of melatonin on sleep and cognitive performance. Brain Res. Bull. 2008;75:433–441. doi: 10.1016/j.brainresbull.2007.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B.H., Albrecht U., Kaasik K., Sage M., Lu W.Q., Vaishnav S., Li Q., Sun Z.S., Eichele G., Bradley A. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) JTK output of LL free-running rhythm.
(B) JTK output of DD free-running rhythm.
(C) Paralogs of circadian clock genes in the turquoise killifish genome.
(D) JTK output of circadian core gene expression under LD.
(E) JTK output of circadian core gene expression under DD.
(F) JTK output of Bmal1 and Clock protein expression under LD.
(G) JTK output of Bmal1 and Clock protein expression under DD.
Data Availability Statement
All data are available in the main text or Supplemental information. Raw data images can be found Mendeley Data: https://data.mendeley.com/datasets/gy57ymzh67/draft?a=b36c53c2-250b-4b92-bbe3-d1990378e123.
This study did not contain any newly generated datasets or codes.