SUMMARY
Interleukin-27 (IL-27) is an immunoregulatory cytokine that suppresses inflammation through multiple mechanisms, including induction of IL-10, but the transcriptional network mediating its diverse functions remains unclear. Combining temporal RNA profiling with computational algorithms, we predict 79 transcription factors induced by IL-27 in T cells. We validate 11 known and discover 5 positive (Cebpb, Fosl2, Tbx21, Hlx, and Atf3) and 2 negative (Irf9 and Irf8) Il10 regulators, generating an experimentally refined regulatory network for Il10. We report two central regulators, Prdm1 and Maf, that cooperatively drive the expression of signature genes induced by IL-27 in type 1 regulatory T cells, mediate IL-10 expression in all T helper cells, and determine the regulatory phenotype of colonic Foxp3+ regulatory T cells. Prdm1/Maf double-knockout mice develop spontaneous colitis, phenocopying ll10-deficient mice. Our work provides insights into IL-27-driven transcriptional networks and identifies two shared Il10 regulators that orchestrate immunoregulatory programs across T helper cell subsets.
Graphical Abstract
In Brief
Zhang et al. construct a transcriptional network for IL-27-mediated Il10 production in CD4 T cells, characterize the function of 16 Il10 regulators, and uncover the role of two transcription factors, Prdm1 and Maf, in driving Il10 production in all T helper cells and in maintaining immune homeostasis in the colon.
INTRODUCTION
Interleukin-27 (IL-27) is an immunoregulatory cytokine that regulates immune responses by multiple mechanisms, including inhibition of differentiation of effector T cell subsets (Artis et al., 2004; Stumhofer et al., 2006; Villarino et al., 2006; Yoshida and Hunter, 2015), induction of a “co-inhibitory” gene module to promote T cell exhaustion (Chihara et al., 2018; DeLong et al., 2019), and polarization of Foxp3+ regulatory T (Treg) cells to a T-bet+ subset that specializes in controlling Th1 immunity (Hall et al., 2012). In addition, we and others have described that IL-27 can differentiate naive T cells into type 1 regulatory T (Tr1) cells (Awasthi et al., 2007; Fitzgerald et al., 2007; Stumhofer et al., 2007; Wang et al., 2011), a Foxp3− IL-10-producing regulatory cell population identified in mouse and human, that suppresses tissue inflammation, autoimmune reactions, and graft versus host disease (GVHD) largely via IL-10 (Roncarolo et al., 1988). IL-27 has the unique capability to induce IL-10 production from a wide range of cell types, including Th1, Th2, Th17, and Treg cells (Awasthi et al., 2007; Fitzgerald et al., 2007; Hall et al., 2012; Stumhofer et al., 2006, 2007). Consistent with these observations, Il27ra−/− T cells have defects in producing IL-10 in vitro and in vivo (Batten et al., 2008). Il27ra−/− mice suffer from lethal immunopathology in parasitic diseases, which is reminiscent of Il10−/− mice (Villarino et al., 2003), and they are more susceptible to experimental autoimmune encephalomyelitis (Batten et al., 2006; Fitzgerald et al., 2007). The molecular mechanisms by which IL-27 induces these diverse regulatory functions in T cells are not fully understood.
The anti-inflammatory cytokine IL-10 has an indispensable role in maintaining immune tolerance and limiting immunopathology during homeostasis, inflammation, infection, and autoimmune diseases (Iyer and Cheng, 2012; Ouyang et al., 2011). Mutations in IL-10 or IL-10R lead to early-onset inflammatory bowel disease (IBD) in humans (Shim, 2019), and mice deficient in IL-10 or IL-10R develop spontaneous colitis (Kühn et al., 1993; Spencer et al., 1998). Importantly, all T helper cell subsets, including Th1, Th2, and Th17 cells, can produce IL-10 to mitigate hyperactive immune responses (Gabryšová et al., 2014). Several transcription factors (TFs) have been shown to regulate IL-10 (Gabryšová et al., 2014; Zhang and Kuchroo, 2019). However, a comprehensive model that systemically examines the dynamic transcriptional network that regulates Il10 in a temporal context of induction and maintenance is lacking.
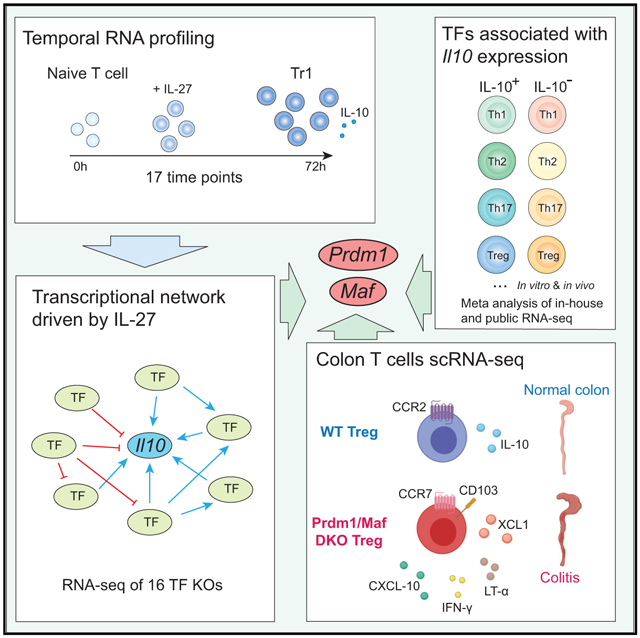
Here we combined computational algorithms with high-resolution temporal transcriptional profiling to predict the TF network driven by IL-27 during Tr1 differentiation. Network analysis systematically identified regulators for Il10 and highlighted Prdm1 and Maf as two central nodes of the Il10 regulatory circuits that cooperatively promoted IL-10 production not only in Tr1 cells but also in Th1, Th2, Th17, and Treg cells. Genetic deletion of Prdm1 and Maf in T cells (Prdm1/Maf DKO), but not either alone, led to spontaneous colitis in mice that exhibits features of human IBD, underscoring the importance of Prdm1 and Maf crosstalk in regulating immune homeostasis in vivo. Single-cell RNA sequencing (scRNA-seq) of colonic CD4+ T cells in DKO mice identified a unique cluster of Treg cells that lost Il10 expression and acquired proinflammatory signatures.
RESULTS
Building a Predictive Model for the IL-27-Driven Transcriptional Program in CD4 T Cells by High-Resolution Temporal Transcriptional Profiling
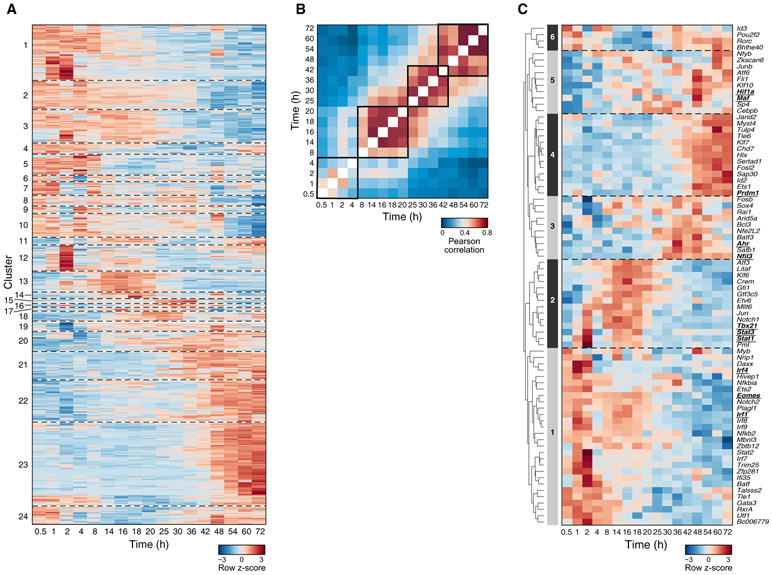
To understand the gene expression program induced by IL-27 in CD4 T cells, we activated naive CD4 T cells in vitro in the presence (Tr1) or absence (Th0) of IL-27 for 72 h and performed whole-genome microarrays at 17 time points. 790 genes were differentially expressed in Tr1 cells compared with Th0 cells, which partitioned into 24 co-expression clusters with distinct temporal profiles (Figure 1A). After activation with IL-27, the cells underwent several transcriptional waves before acquiring a stable phenotype (Figure 1B): an early phase from 0–4 h, when the global transcriptional profile changes dynamically; a stable early phase from 8–20 h; an intermediate phase from 25–42 h; and a late phase from 48–72 h. The dynamic transcriptional changes during the first 4 h are a distinct feature of Tr1 cell differentiation compared with Th17 cells, which manifest a relatively stable profile during the early phase from 0–2 h (Yosef et al., 2013).
Figure 1. Building a Predicative Model of the IL-27-Driven Transcriptional Program in CD4 T Cells by High-Resolution Temporal Transcriptional Profiling.
Gene expression profiles during IL-27-driven in-vitro Tr1 differentiation were measured by microarray at 17 time points with the Th0 condition as a control.
(A) Relative expression (log2(Tr1/Th0)) of 790 differentially expressed genes (rows).
(B) Pearson correlation matrix of the transcriptome at every pair of time points.
(C) Relative expression (log2(Tr1/Th0)) of 79 TFs predicted to regulate gene clusters. Underlined are TFs known to regulate Tr1 differentiation.
To identify TFs that drive the distinct transcriptional waves, we hypothesized that genes co-expressed in a cluster (Figure 1A) are likely to share regulators that are active at the relevant time point. We predicted regulator-target associations in the IL-27-driven transcriptional programs based on significant overlap between genes in a specific cluster and a regulator’s putative targets in a regulator-target association database (Yosef et al., 2013). This generated a predictive network containing 79 TFs that were putative regulators of the gene clusters induced by IL-27 (Figure 1C). The 79 TFs fall into six major expression patterns, each containing both known and previously uncharacterized regulators, that are (1) highly expressed during the dynamic early phase (0–4 h), including Irf1 and Batf, which are the pioneer TFs of Tr1 cell differentiation (Karwacz et al., 2017), as well as Eomes (Zhang et al., 2017); (2) increased during the stable early phase (8–20 h), including Stat1 and Stat3, which mediate signaling downstream of the IL-27 receptor (Stumhofer et al., 2007) and IL-21 receptor (Leonard and Wan, 2016; Pot et al., 2009); (3) increased during the intermediate phase (25–42 h), including Ahr (Apetoh et al., 2010); (4) increased during the late phase (48–72 h), such as Prdm1 (Montes de Oca et al., 2016); (5) increased gradually over time, such as Maf (Pot et al., 2009) and Hif1a (Mascanfroni et al., 2015); and (6) decreased specifically during the stable early phase and may act as gate-keepers for the IL-27-induced gene program, which, interestingly, include a potent IL-10 inhibitor, Bhlhe40 (Huynh et al., 2018; Lin et al., 2014; Yu et al., 2018). Our computational analysis identified almost all TFs known to be required for Tr1 cells, indicating a good predictive power, and predicted 69 TFs that were not implicated in Tr1 differentiation.
Experimental Validation of the IL-27 Predicative Network Identifies TFs that Regulate IL-10 In Vitro
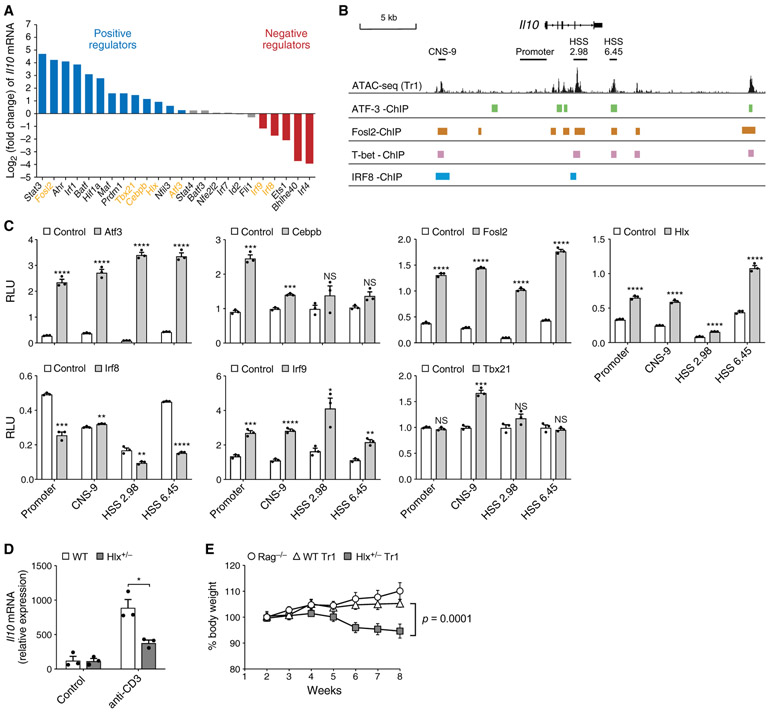
Because IL-10 production is the most representative feature of IL-27-induced Tr1 cells, we used IL-10 expression as a readout to validate the predicative IL-27 network. We gathered 24 available knockout mice, differentiated their naive CD4 T cells into Tr1 cells with IL-27, and compared their Il10 mRNA levels with the respective controls (Figure 2A). We validated 11 known Il10 regulators in T cells: 8 positive regulators (Prdm1, Stat3, Ahr, Maf, Irf1, Batf, Hif1a, and Nfil3) and 3 negative regulators (Irf4, Bhlhe40, and Ets1) (Huynh et al., 2018; Karwacz et al., 2017; Lee et al., 2012; Lin et al., 2014; Yu et al., 2018). We found that Cebpb, a TF that induces ll10 in M2 macrophages (Liu et al., 2003), was also required for Il10 production in Tr1 cells. Importantly, among the 12 tested factors that were not known to regulate Il10, we discovered 4 positive (Atf3, Fosl2, Tbx21, and Hlx) and 2 negative (Irf9 and Irf8) Il10 regulators. These TFs also regulated IL-10 at the protein level (Figure S1A). Of the 24 genetically perturbed TFs, we successfully validated 18 (75%) of our computational predictions.
Figure 2. Experimental Validation of IL-27 Predicative Network Identifies TFs that Regulate Il10 In Vitro and In Vivo.
(A) Log2 fold change of Il10 mRNA levels in WT versus KO Tr1 cells differentiated in vitro with IL-27 for 72 h, quantified by qPCR. Blue, positive regulator; red, negative regulator; gray, not statistically significant. Data are displayed as mean of 2–3 replicates.
(B) Statistically significant ChIP-seq binding sites of ATF-3 ATF-3, Fosl2, T-bet, and IRF8 in the Il10 locus.
(C) Luciferase activity in 293T cells transfected with luciferase reporters for the indicated cis-regulatory elements of Il10 and plasmids encoding the depicted TFs. Firefly luciferase activity is normalized to constitutive Renilla luciferase activity.
(D) WT and Hlx+/− mice were injected intraperitoneally (i.p.) with anti-CD3. Il10 mRNA in CD4+ T cells MACS purified from mesenteric lymph nodes was measured by qPCR.
(E) 5 x 105 in vitro differentiated WT (diamonds) and Hlx+/−(squares) Tr1 cells were transferred i.p. into Rag1−/− recipients. Rag1−/− (circles) did not receive any cells. Changes in body weight were monitored weekly. n = 5.
We examined binding of the aforementioned Il10 regulators to the Il10 locus in public chromatin immunoprecipitation sequencing (ChIP-seq) data. ATF-3 (Garber et al., 2012), T-bet (Nakayamada et al., 2011), Fosl2 (Ciofani et al., 2012), and IRF8 (Xu et al., 2015) have significant binding in the Il10 locus, some of which lies in chromatin-accessible regions in Tr1 cells (Figure 2B), indicating that they may directly regulate Il10 transcription in Tr1 cells. To investigate whether these TFs can trans-activate or inhibit Il10, we performed luciferase reporter assays in 293T cells using reporters for the proximal Il10 promoter and the CNS-9, HSS+2.98, and HSS+6.45 enhancers (Hedrich and Bream, 2010; Figure 2C). Atf3, Fosl2, and Hlx transactivated the Il10 promoter and the three enhancers. Transactivation by Cebpb was more restricted to the promoter and CNS-9 region, and T-bet only transactivated the CNS-9 region. Irf8 inhibited the baseline activity of the Il10 promoter, HSS+2.98, HSS+6.45, and, to a lesser extent, CNS-9. We found that transactivation of Il10 by the pioneer factor Irf1 (Karwacz et al., 2017) was completely blocked by Irf8 co-expression at the three enhancers but not the proximal promoter (Figure S1B). In contrast, the putative negative regulator Irf9 transactivated Il10 at all four cis-regulatory sites, indicating that inhibition of Il10 by Irf9 in Tr1 cells may be mediated by indirect mechanisms (Figure 2C). In summary, we validated 11 known and discovered 7 direct and indirect regulators of Il10 during IL-27-driven Tr1 differentiation.
Hlx Regulates Il10 Expression and Tr1 Function In Vivo
It has been shown previously that Hlx cooperates with T-bet to promote interferon (IFN)-γexpression in Th1 cells in vitro (Mullen et al., 2002); however, whether it regulates T cell function in vivo and whether it has an immunoregulatory role has not been investigated. To address these questions, we first tested the role of Hlx in a model of self-limiting inflammation induced by intraperitoneal injection of an anti-CD3 antibody, which spontaneously resolves in a IL-10-dependent manner (Huber et al., 2011; Kamanaka et al., 2006). Because Hlx deficiency is embryonically lethal (Hentsch et al., 1996), we compared Il10 expression in CD4 T cells from Hlx+/− and WT mice following anti-CD3 injection and observed less Il10 in Hlx+/− T cells (Figure 2D). We next investigated how Hlx regulates the immunoregulatory function of Tr1 cells in a T cell transfer colitis model. Rag−/− mice receiving wild-type (WT) Tr1 cells were able to maintain their body weight because Tr1 cells normally do not induce colitis. However, the recipients of Hlx+/− Tr1 cells lost weight over time, indicating that Tr1 cells haplodeficient for Hlx might lose the regulatory phenotype and become proinflammatory (Figure 2E).
A Comprehensive Transcriptional Network Focused on Regulation of IL-10 by IL-27
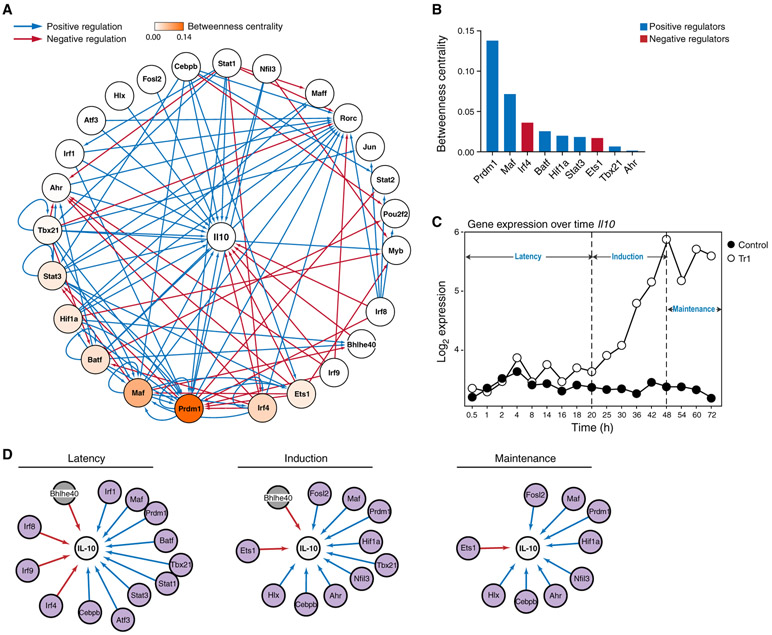
We identified causal genetic targets of the Il10-regulating TFs in the IL-27 network by performing RNA-seq on Tr1 cells genetically deficient in each of them, generating a comprehensive network showing the effect of the TFs on Il10 as well as on the expression of each other (Figure 3A). The network showed dense inter-connectedness between multiple negative and positive Il10 regulators, with substantial cross-regulation between them, explaining how indirect regulators affect Il10 expression through direct regulation of other regulators (Figure 3A). Besides direct inhibition of Il10 by Irf8, the negative regulators, including Irf8, Irf4, Ets1, and Irf9, may regulate Il10 by inhibiting the expression of positive regulators such as Prdm1, Maf, Ahr, and Batf. Except for inhibition of Irf4 by Stat1 and Bhlhe40 by Maf, very few positive regulators inhibited expression of the negative regulators; rather, they reinforced the expression of each other. Quantification of TF connectivity within the network by betweenness centrality score revealed Prdm1 and Maf as the most central positive regulators and Irf4 as the most central negative regulator, identifying these TFs as central hubs for regulation of Il10 in Tr1 cells (Figure 3B).
Figure 3. A Comprehensive Transcriptional Network Focused on Regulation of IL-10 by IL-27.
(A) General network of Il10 regulation by TFs in Tr1 cells, visualized using Cytoscape. Edges indicate causal regulatory targets identified using genetic perturbation by RNA-seq or qPCR. Blue and red edges indicate positive and negative regulations, respectively. Nodes are colored by betweenness centrality score.
(B) Betweenness centrality scores of the regulators in (A). Blue, positive regulator; red, negative regulator.
(C) Temporal expression of Il10 in Tr1 versus Th0 cells measured by microarray.
(D) Temporal regulation of Il10 in Tr1 cells, divided into 3 main phases: latency (0–20 h), induction (25–48 h), and maintenance (54–72 h). Purple nodes, increased by IL-27; gray nodes, decreased by IL-27.
We observed three distinct phases of Il10 expression during Tr1 differentiation from the temporal microarray data (Figure 3C): a latency phase (0–20 h) with no detectable Il10, an induction phase (20–48 h), and a maintenance phase (48–72 h). To further understand the temporal dynamics of Il10 regulation, we divided the global network into three phase-specific networks based on the regulator’s temporal expression pattern (Figure 3D). The IRFs and Atf3 were mainly increased during the latency phase; Hlx during the induction phase; Tbx21, which has been shown to cooperate with Hlx (Mullen et al., 2002), at the latency and induction phase; and Fosl2, during the late induction phase and the maintenance phase. Notably, Prdm1, Maf, and Cebpb were increased at all three phases. Multiple regulators that suppress Il10 were expressed at the latency phase and decreased at the induction and maintenance phases. This may be one of the reasons why Il10 induction is relatively late during Tr1 differentiation.
Prdm1 and Maf Have Complementary but Indispensable Roles in Regulating Tr1 Identity at the Transcriptional and Chromatin Level
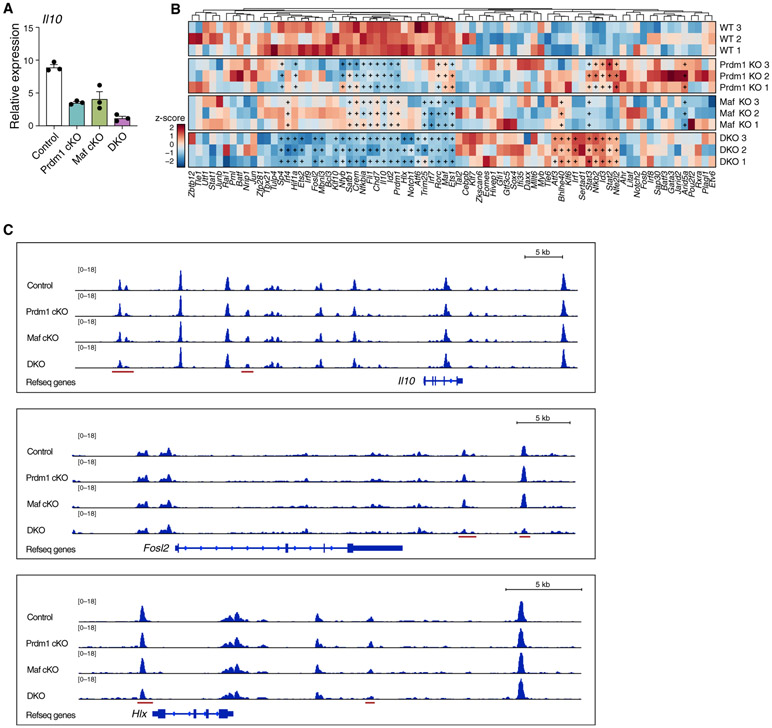
Despite being the most central nodes in the Il10 regulatory network, Il10 expression in Prdm1 or Maf single-knockout (cKO) Tr1 cells is only partially reduced, suggesting a complementary relationship between the two TFs. We therefore generated mice that lack both Prdm1 and Maf (DKO) in T cells, using conditional deletion driven by Cd4-Cre. Prdm1/Maf double deficiency led to almost complete loss of Il10 in Tr1 cells (Figure 4A).
Figure 4. Prdm1 and Maf Have Complementary but Indispensable Roles in Regulating Tr1 Identity at the Transcriptional and Chromatin Level.
(A) Naive CD4 T cells from the indicated mice were differentiated in vitro into Tr1 cells with IL-27. Il10 expression was measured by qPCR on day 3.
(B and C) Control, Prdm1 cKO, Maf cKO, and Prdm1/Maf DKO Tr1 cells generated as described in (A) were analyzed by RNA-seq (B) and ATAC-seq (C).
(B) Heatmap showing expression of 79 predicted regulators in the Tr1 network. “+” indicates statistically significant differential expression.
(C) Chromatin accessibility in the Il10, Fosl2, and Hlx loci in Tr1 cells of the indicated genotype. Red bars represent regions with differential chromatin accessibility in DKO cells.
To investigate how loss of Prdm1 and Maf influences the Il10 regulatory network (Figure 3A), we performed RNA-seq on single- and double-KO Tr1 cells at 72 h (Figure 4B). Deficiency in Prdm1 and Maf led to a collapse in expression of several TFs important for Il10 expression, including Fosl2, Hif1a, Hlx, and Notch1 (Rutz et al., 2008), which was not observed in Prdm1 or Maf single KO. In addition, a number of other transcriptional regulators that are induced by IL-27 were also specifically decreased in DKO cells, such as Sp1, Ets2, Mbnl3, Klf10, Nfyb, Satb1, Crem, Nfkbia, Chd7, Trim25, Klf6, and Nfkb2, although their role in regulating Il10 remains to be investigated. Furthermore, Prdm1 and Maf transcriptionally regulated each other’s expression (Figure 4B). Bhlhe40, a potent Il10 inhibitor, was increased dramatically in DKO mice, indicating that Prdm1 and Maf are critical not only for driving Il10 expression but also for antagonizing the expression of TFs that repress Il10. Although some positive regulators, such as Irf1 and Atf3, were increased in DKO cells, these early-stage Il10 inducers could not rescue the loss of Il10 in the absence of Prdm1 and Maf, perhaps because of their inability to overcome inhibition (Figure S1B).
To assess whether Prdm1 or Maf regulated the chromatin landscape of Tr1 cells, we profiled chromatin accessibility in single- and double-KO Tr1 cells using Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq). Although the chromatin landscape in the Il10 locus remains largely unchanged in Prdm1 or Maf single-KO cells, we detected a reduction in accessibility at specific enhancer regions in the Il10 locus in DKO cells (Figure 4C). In addition, we found that Fosl2 and Hlx, two other positive regulators of Il10, also became less accessible in DKO but not either single-KO cells (Figure 4C), which is consistent with their gene expression profile. Moreover, DKO Tr1 cells showed a unique reduction in chromatin accessibility in co-inhibitory receptor gene loci such as Ctla4, Pdcd1 (PD-1), Tigit, Havcr2 (Tim-3) (Figure S2), another hallmark of Tr1 cells that is transcriptionally regulated by Prdm1 and Maf (Chihara et al., 2018). In summary, these data suggest that Prdm1 and Maf have complementary but indispensable roles in regulating the hallmark genes for Tr1 identity at the transcriptional and chromatin levels.
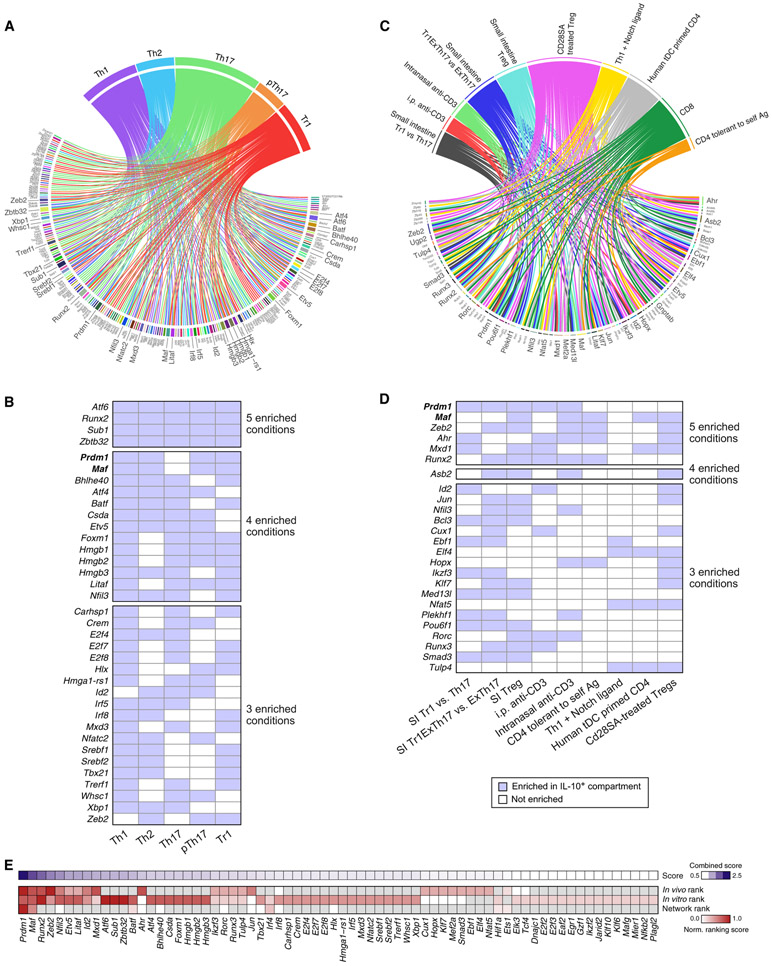
IL-10 Regulators Are Induced in Diverse IL-10-Producing T Helper Cells
All T helper cells can produce IL-10, but the regulation of IL-10 expression in these contexts is unclear. We therefore examined whether the regulators we identified in the IL-27 network were also utilized for IL-10 regulation in other T helper cells. We differentiated naive CD4 T cells from IL-10Thy1.1 reporter mice (10BiT) (Maynard et al., 2007) into Th1, Th2, non-pathogenic Th17 (Th17), pathogenic Th17 (pTh17), and Tr1 cells; sorted out the IL-10+ and IL-10− compartments; and performed RNA-seq. We also analyzed the RNA profiles of IL-10+ versus IL10− T cells purified from several other in vivo and in vitro conditions (Boks et al., 2016; Burton et al., 2014; Gagliani et al., 2015; Langenhorst et al., 2012; Neumann et al., 2014; Trandem et al., 2011; Table S1). We identified TFs whose expression was associated with IL-10 in each T cell subset or condition (Figures 5A and 5C) and ranked them based on the number of conditions where their expression is enriched in the IL-10-producing compartment (Figures 5B and 5D). Many of the regulators identified in our IL-27 network were also identified by this analysis, including Prdm1, Maf, Hlx, Tbx21, Batf, Nfil3, Ahr, Bhlhe40, and Irf8, indicating that they might also regulate IL-10 in other contexts (Figures 5A and 5C). Prdm1 and Maf, the two positive regulators with highest centrality in the IL-27 network (Figure 3B), were enriched in the IL-10-producing compartment of all T helper cell subsets (Figure 5B) as well as under many other in vivo and in vitro conditions (Figure 5D), whereas the other TFs were more restricted to certain conditions. A combined ranking scheme of IL-10 regulators evaluating the centrality score in the IL-27 network and generalizability in other IL-10-producing T cell subsets derived from in vitro and in vitro contexts revealed Prdm1 and Maf as the two top TFs that regulate IL-10 production across T helper cells (Figure 5E).
Figure 5. TFs Associated with IL-10 Production in Different T Helper Cells.
(A and C) TFs enriched in the IL-10+ compartments compared with their IL-10− compartments in (A) in-vitro-generated T helper cell subsets, (C) 10 in vivo/ex vivo conditions where a direct comparison between the transcriptome of IL-10+ and IL-10− cells was made in public data (Table S1). TFs that are enriched under at least 3 conditions were magnified.
(B and D) A different display of same data in (A) and (C), respectively, showing TFs that are enriched under at least 3 conditions and the conditions where their expression is enriched in the IL-10+ compartment. SI, small intestine.
(E) A ranking scheme (STAR Methods) for all potential regulators of IL-10, taking into account network centrality (Figure 3B), enrichment in in vitro conditions (Figure 5A), and enrichment in public datasets (Figure 5C).
Role of Prdm1 and Maf in Regulating IL-10 in Different T Helper Cells
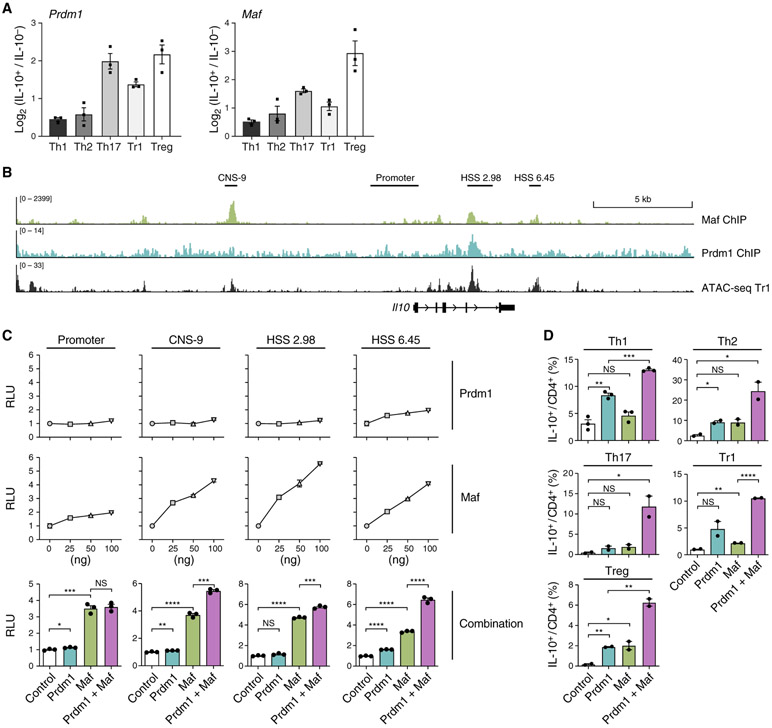
We validated the association of Prdm1 and Maf expression with Il10 in various contexts by qPCR. The IL-10+ compartment of Th1, Th2, Th17, Tr1, and Treg cells expressed higher levels of Prdm1 and Maf than their IL-10− counterparts (Figure 6A). Moreover, analysis of public ChIP-seq datasets confirmed binding of Prdm1 and Maf at accessible chromatin regions in the Il10 locus (Figure 6B). These findings further support the hypothesis that Prdm1 and Maf may be critical regulators of IL-10 in multiple settings.
Figure 6. Prdm1 and Maf Synergistically Regulate IL-10 in All T Helper Cells.
(A) Enrichment of Prdm1 and Maf mRNA in in-vitro-generated T helper cells validated by qPCR.
(B) ChIP-seq of Maf in Th17 cells and Prdm1 in tissue-resident memory T cells aligned with ATAC-seq data of Tr1 cells differentiated in vitro at 72 h.
(C) Luciferase activity in 293T cells transfected with Il10 luciferase reporters along with constructs encoding Prdm1, Maf, or both. n = 3.
(D) T helper cells differentiated in vitro were transduced with two retroviruses expressing Prdm1 and Maf, respectively. IL-10 expression in control cells, Prdm1-overexpressing cells, Maf-overexpressing cells, and cells overexpressing Prdm1 and Maf was measured by flow cytometry 48 h after transduction.
We tested the interaction between Prdm1 or Maf in regulating Il10 using luciferase assays. Although Prdm1 alone had very limited capability to transactivate Il10, it significantly enhanced transactivation of Il10 by Maf (Figure 6C), suggesting that cooperation between Prdm1 and Maf is required for optimal IL-10 production. Of note, the synergistic effect between Prdm1 and Maf is specific to enhancer regions (tested by the interaction term in the linear regression model; CNS-9, p = 0.00255; HSS+6.45, p = 0.000154) but not the promoter. We further studied the synergy between Prdm1 and Maf in regulating Il10 in primary CD4 T cells using a gain-of-function approach by transducing Th1, Th2, Th17, Tr1, and Treg cells with retroviruses encoding Prdm1 (MSCV-IRES-Thy1.1) and Maf (MSCV-IRES-GFP). IL-10 production was enhanced dramatically when Prdm1 and Maf were co-expressed (Figure 6D). Importantly, although Prdm1 and Maf cooperatively promoted IL-10 production across all T helper cells, they did not inhibit production of signature cytokines of the T helper cell subsets (Figure S3). Thus, Prdm1 and Maf enabled expression of a gene module that induced IL-10 in all T helper cell subsets without disrupting their cell differentiation program.
Genetic Deficiency of Prdm1 and Maf, but Not Either Alone, in T Cells Leads to Human IBD-like Spontaneous Colitis Driven by a Unique Cluster of Treg Cells
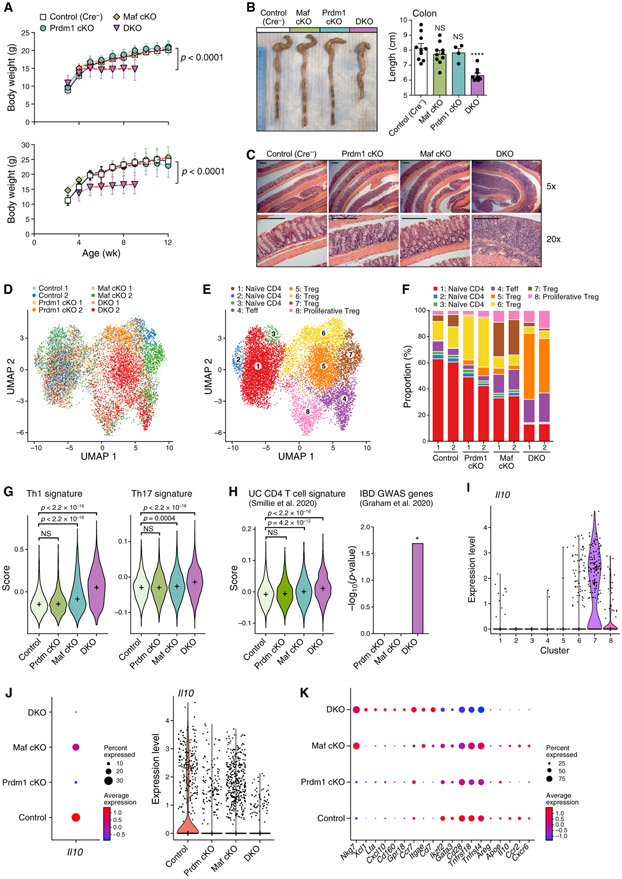
IL-10 has a critical role in maintaining intestinal homeostasis. Mutations in IL-10 or IL-10R are associated with human ulcerative colitis presenting in early childhood (Zhu et al., 2017). We therefore monitored Prdm1/Maf DKO mice for spontaneous development of colitis. Strikingly, loss of Prdm1 and Maf in T cells led to spontaneous weight loss over time (Figure 7A), similar to that observed in IL-10-deficient mice (Kühn et al., 1993). The presence of one copy of the Maf allele protected the mice from weight loss until 16 weeks of age, and one copy of Prdm1 protected the mice until at least 24 weeks of age (Figure S4A). DKO mice, but not single-KO mice, had shorter colons (Figure 7B), and histological analysis of the entire intestine confirmed the presence of active colitis in DKO mice (Figure 7C; Figure S4B) with features reminiscent of human ulcerative colitis. Severe chronic active colitis with cryptitis, crypt abscess, and crypt loss with mucosal ulcers reminiscent of ulcerative colitis was the most prevalent pathology in DKO mice. DKO mice occasionally showed flask-shaped aphthous erosions that are common in human Crohn’s disease, but other defining features of Crohn’s disease, such as transmural lymphoid aggregates, were absent in all mice (Figure 7C; Figure S4C). Although Cd4-Cre; Prdm1fl/fl mice have been reported to develop spontaneous colitis (Martins et al., 2006), we observed spontaneous intestinal disease very rarely only in male but not in any female mice. Moreover, the pathology in these mice was mainly located in the proximal end of the small intestine rather than the colon (Figures S4D and S4E).
Figure 7. Genetic Deficiency of Prdm1 and Maf, but Not Either Alone, in T Cells Leads to Human IBD-like Spontaneous Colitis Driven by a Unique Cluster of Treg Cells.
(A) Weekly monitored body weights. Top: female mice. Bottom: male mice. n ≥ 8. Data are represented as mean ± SD.
(B) Colon length of the indicated mice, presented as seen by gross anatomy and measurement.
(C) Hematoxylin and eosin staining of colon Swiss rolls. Pictures are representative of 10 control, 4 Prdm1 cKO, 6 Maf cKO, and 7 DKO mice. Scale bars represent 250 μm.
(D–K) CD4 T cells from colonic lamina propria were profiled by scRNA-seq.
(D and E) Uniform manifold approximation and projection (UMAP) plots show 13,535 cells (dots) colored by genotype (D) or cluster (E).
(F) Distribution of cells with different genotypes in clusters.
(G and H) Left: distribution of gene signature scores of Tconv cells (clusters 1–4) by genotype. “+” indicates median. (H) right: enrichment of IBD-associated GWAS genes that are involved in adaptive immunity in differentially expressed genes of Prdm1 cKO, Maf cKO, and DKO Tconv cells, respectively, compared with the control. Significance of enrichment was tested by hypergeometric test.
(I-K) Gene expression level represented as log(TP10K+1).
(I) Il10 expression by control (WT) cells across clusters.
(J) Il10 expression by Treg cells (clusters 5–8) across genotypes.
(K) Representative differentially expressed genes of DKO Treg cells compared with all other genotypes. Dot size represents the fraction of cells in the cluster that express the gene; color indicates mean expression in expressing cells relative to other genotypes.
To characterize the transcriptional changes in T cells that lead to spontaneous colitis in DKO mice, we performed scRNA-seq on CD4 T cells from the colonic lamina propria at 3 weeks of age before disease onset. We included two biological replicates for each genotype (Figure 7D) and collected a total of 13,535 high-quality single-cell profiles that were partitioned into eight distinct clusters (Figure 7E). Cluster identity was designated based on bulk RNA-seq-derived gene signatures (Immgen) and confirmed by expression of key marker genes (Figures S5A and S5B). Cells of the four genotypes distributed evenly within the naive T cell clusters (clusters 1–3), indicating negligible batch effects between samples and similar phenotypes of naive T cells in the absence of Prdm1 or Maf. However, Maf cKO and DKO cells formed distinct sub-clusters within the effector T cell cluster (cluster 4), and each of the Treg cell clusters (clusters 5–8) was dominated by a different genotype (Figures 7D-7F; Figure S5C).
The proportion of effector T cells was increased in both Maf cKO and DKO (Figure 7F), but these cells were qualitatively different (Figures 7G and 7H) in that DKO effector cells had dramatically increased expression of the Th1 gene signature, which is a major pathogenic cell population implicated in IBD (Ito and Fathman, 1997; Neurath et al., 2002). The Th17 gene signature was also increased significantly in DKO but to a lesser extent (Figure 7G). A subset of effector cells in DKO mice acquired a signature that resembles CD4 T cells from inflamed intestinal lesions of humans with ulcerative colitis (Smillie et al., 2019). In addition, the differentially expressed genes in DKO effector T cells (compared with control cells) showed unique enrichment for IBD-associated genome-wide association study (GWAS) genes that are involved in adaptive immunity (Graham and Xavier, 2020; Figure 7H). These data indicate that loss of Prdm1 and Maf leads to spontaneous colitis that resembles human IBD in terms of not only pathological features but also molecular signatures.
Consistent with previous reports (Maynard et al., 2007), Treg cells (clusters 7 and 8) were a major source of IL-10 in the colon (Figure 7I). We observed that the average expression level of Il10 was reduced in colonic Treg cells in the absence of Maf; the average expression level and percentage of Il10-positive cells were reduced in the absence of Prdm1, and Il10 expression was barely detectable in the absence of both (Figure 7J). Therefore, Prdm1 and Maf were also required for Il10 expression in Treg cells in vivo.
Besides downregulation of Il10, DKO Treg cells, compared with single-KO or control Treg cells, exhibited a unique gene expression profile (cluster 5; Figures 7D-7F). DKO Treg cells lost immunoregulatory phenotypes, including expression of TFs critical for Treg cell stability and function (e.g., Ikzf2 and Gata3), co-stimulatory receptors (e.g., Cd28, Tnfrsf18, and Tnfrsf4), and soluble immunosuppressive molecules (e.g., Areg and Apoe). On the other hand, DKO Treg cells acquired Th1- associated (e.g., Nkg7, Xcl1, Lta, and Cxcl10) and Cytotoxic T Lymphocytes (CTL)-associated (e.g., Cd160 and Gpr18) genes that are known to actively promote inflammation. Moreover, DKO Treg cells showed profound changes in their chemotaxis profile, turning off Ccr2 and Cxcr6 while dramatically upregulating Ccr7 (Figure 7K). These data indicated that Prdm1 and Maf cooperatively regulate the identity and function of Treg cells and are indispensable for immune tolerance in vivo.
DISCUSSION
Computational inference of gene regulation from temporal profiling of gene expression has shown great potential to delineate the dynamic transcriptional circuits that regulate T cell differentiation. We and others have successfully built models of regulatory networks for Th17 cells (Wu et al., 2013; Yosef et al., 2013; Ciofani et al., 2012) and Th2 cells (Henriksson et al., 2019), discovering key regulators and revealing general principles governing T cell differentiation. Here we computed a transcriptional network induced by IL-27 in CD4 T cells that showed strong predicative power for identification of Il10 regulators: 18 (75%) of the 24 predicted TFs we validated experimentally were confirmed to be regulating Il10 expression. In addition to IL-10, IL-27-induced Tr1 cells feature expression of IFN-γ (Awasthi et al., 2007; Pot et al., 2009) and co-inhibitory receptors (Chihara et al., 2018; DeLong et al., 2019). Therefore, the predicted TFs in the IL-27 network presented here could also be utilized for defining the transcriptional regulation of these molecules in a manner similar to what is presented here for Il10.
By genetically perturbing the IL-27 network, we identified critical regulators of Il10, which may shed light on previously unappreciated roles of IL-10 in physiological processes and deepen our understanding of IL-10-related immune disorders such as IBD (Zhu et al., 2017). For example, Atf3 is a TF that is induced by endoplasmic reticulum stress (Schmitz et al., 2018) with anti-inflammatory properties (De Nardo et al., 2014; Gilchrist et al., 2006); induction of IL-10 by Atf3 may provide a negative feedback loop to dampen endoplasmic reticulum (ER) stress (Hasnain et al., 2013; Shkoda et al., 2007) and suppress inflammation. Further, Fosl2 is a member of the AP1 family, which contains several members that were implicated in Il10 regulation (Hu et al., 2006; Kremer et al., 2007). Fosl2 has been shown previously to regulate the pathogenicity of Th17 cells (Ciofani et al., 2012), which play an important role in the pathogenesis of IBD. A single-nucleotide polymorphism (SNP) in Fosl2 (rs925255) has been linked genetically to IBD in a GWAS (Jostins et al., 2012). Our study raises the possibility that the SNP in Fosl2 may further influence susceptibility to IBD by regulating IL-10 expression. Last, we identified Hlx as a regulator of IL-10 and showed that its haplodeficiency is sufficient to convert Tr1 cells to proinflammatory cells that exacerbate T cell transfer colitis. Interestingly, the Hlx locus has been shown to be hypermethylated in epithelial cells in humans with IBD, and these data suggest that Hlx might have a role in regulating immune responses beyond T cells.
The lineage-defining TFs for Th2 (GATA-3) and Th17 (ROR-γt) have been shown to contribute to IL-10 expression (Shoemaker et al., 2006; Wang et al., 2015). However, the role of T-bet, the master TF for Th1 cells, in Il10 regulation has been controversial. One study has reported that IL-10 production is increased in CD4 T cells in the absence of T-bet (Shin et al., 2014), which could be due to the indirect effect of a decrease in IFN-γ (Hu et al., 2006). Other studies have reported that T-bet can induce IL-10 production but under conditions where T-bet or other TFs are overexpressed (Rutz et al., 2008; Zhu et al., 2015). Here we show that genetic deficiency in T-bet leads to impaired IL-10 production in Tr1 cells. Additionally, we show, by ChIP-seq and luciferase assays, that T-bet can directly bind and transactivate Il10. Thus, the master TFs for all T helper cell subsets can induce immunoregulatory genes to mitigate overexuberant responses. Eomes, another T-box TF that is highly homologous to T-bet in its DNA binding domain (Pearce et al., 2003), has been reported to regulate IL-10 in a GVHD model (Zhang et al., 2017). Our study suggests that the relative contribution of these two TFs to IL-10 regulation may be highly context dependent (Zhang et al., 2017).
Master TFs have been identified for other T cell subsets but not for Tr1 cells. Our network analysis in Tr1 cells highlighted Prdm1 and Maf as central hubs in regulating Il10. Not only are they heavily regulated, but, more importantly, they orchestrate a regulatory circuit composed of multiple other transcriptional modulators. Prdm1/Maf DKO Tr1 cells, but not either single KO Tr1 cells, exhibited complete loss of Il10 and collapse of the Il10 regulatory circuit, both accompanied by reduced chromatin accessibility, and a notable upregulation of the Il10 repressor Bhlhe40. Expression of co-inhibitory receptors, another hallmark of Tr1 cells (Brockmann et al., 2018; Chihara et al., 2018; DeLong et al., 2019), are induced by IL-27 and controlled by Prdm1 and Maf transcriptionally and at the chromatin level. These data suggest that the key signature of Tr1 cells might be established through collaboration between two TFs with complementary roles.
We and others have shown that c-Maf is a universal regulator of IL-10 in Th1, Th2, Th17, Tr1, as well as Treg cells (Gabryšová et al., 2018). Further, we discovered that Maf needs to cooperate with other TFs, such as Ahr, to achieve robust Il10 transcription (Apetoh et al., 2010). In this study, we identified Prdm1 as a critical partner of Maf and that together they synergistically transactivate Il10 in all CD4 T cell subsets, including IL-10-producing Tr1 cells.
Commitment of T helper cells to specific subsets requires induction of master TFs that not only induce specific transcriptional programs that push T cell subsets in one direction but also initiate repressive programs that antagonize other fates (Sungnak et al., 2019). Interestingly, we observed that, although Prdm1 and Maf synergistically promote IL-10 production, they do not inhibit production of signature cytokines of the different T helper cell subsets, enabling them to co-produce IL-10 while maintaining their original transcriptional program.
We found that Prdm1/Maf DKO mice, but not single KO mice, phenocopy Il10-deficient mice and develop spontaneous colitis that presents pathological and molecular features of human IBD. Prdm1 is a well-recognized GWAS gene associated with IBD (Ellinghaus et al., 2013). It would therefore be interesting to investigate whether SNPs related to Maf can further enhance IBD susceptibility. With scRNA-seq analysis, we discovered a unique cluster of colonic Treg cells in Prdm1/Maf DKO mice that was not observed when Prdm1 (Cretney et al., 2011; Garg et al., 2019; Ogawa et al., 2018) or Maf (Neumann et al., 2019; Xu et al., 2018) was perturbed individually. These DKO Treg cells lose immunoregulatory phenotypes, including production of IL-10, and acquire strong Th1- and CTL-associated gene signatures, indicating potential to actively exacerbate inflammation. In addition, this DKO Treg cell cluster shows a profound shift in the use of chemokine receptors from those that drive T cells to tissue with active inflammation (e.g., Ccr2 and Cxc6) (Hamano et al., 2014; Loyher et al., 2016; Mondini et al., 2019; Zhang et al., 2009) to Ccr7, which, together with two other markers highly expressed by DKO Treg cells, Lta (Upadhyay and Fu, 2013) and Itgae (Leithäuser et al., 2006), are associated with development of lymphoid-like structures. Therefore, it would be interesting to further study how Prdm1 and Maf regulate the migration and location of Treg cells.
We have shown that Prdm1 and Maf co-operatively induce the co-inhibitory receptor gene module on exhausted CD8 T cells (Chihara et al., 2018), which not only have a dysfunctional effector program but also co-produce IL-10 to actively suppress the immune responses in chronic viral infections and cancer. Further, Maf was also implicated in IL-10 expression in B cells (Liu et al., 2018) and macrophages (Cao et al., 2005), and Prdm1 has a regulatory role in dendritic cells (Kim et al., 2011; Watchmaker et al., 2014). These data, together with our current study, emphasize the importance of cooperativity between Prdm1 and Maf in regulating immunoregulatory gene programs across multiple immune cell types.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Vijay Kuchroo (vkuchroo@evergrande.hms.harvard.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Data generated in this paper has been deposited in the Gene Expression Omnibus (GEO) under accession number GEO: GSE159208.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice and Ethics Statement
C57BL/6, BALB/cJ, dLckCre, Hlx+/− (Hentsch et al., 1996), Tbx21−/− (Finotto et al., 2002), Irf8fl/fl (Feng et al., 2011), Prdm1fl/fl (Shapiro-Shelef et al., 2003), Nfe2l2−/− (Chan et al., 1996), Hif1afl/fl (Ryan et al., 2000), Ahr−/− (Schmidt et al., 1996), Batf3−/− (Hildner et al., 2008), Stat3fl/fl (Moh et al., 2007), Stat4−/− (Kaplan et al., 1996), Irf1−/− (Matsuyama et al., 1993), Batf−/− (Schraml et al., 2009), Irf4fl/fl (Klein et al., 2006), Bhlhe40−/− (Jiang et al., 2008) and 10BiT (Maynard et al., 2007) mice were purchased from Jackson Laboratory. Cd4Cre (Lee et al., 2001) mouse was purchased from Taconic. Maffl/fl (Wende et al., 2012), Nfil3fl/fl (Gascoyne et al., 2009), Id2fl/fl (Seillet et al., 2013), Ets1−/− (Muthusamy et al., 1995) and Foxp3-GFP mouse (Bettelli et al., 2006) has been previously described. Other previously described mutant strains were kindly provided by the following researchers: Fosl2fl/fl (Karreth et al., 2004), D. Littman; Atf3fl/fl Actb-Cre(Taketani et al., 2012), H. Weiner; Cebpbfl/fl Lck-Cre+ (Sterneck et al., 2006), M. Rincon. In addition, spleens from Irf7−/−, Fli1+/−, Irf9−/− mice were obtained from Ian R. Rifkin, Maria Trojanowska, and Paul J. Utz, respectively. In vitro experiments were performed using 6-10 weeks old female and male mice.
All animals were housed and maintained in conventional pathogen-free facilities at the Harvard Institute of Medicine and Hale Building for Transformative Medicine in Boston (IUCAC protocols: 2016N000444 (V.K.K.)). All experiments were performed in accordance to guidelines outlined by Harvard Medical Area Standing Committee on Animals and the Brigham and Women’s Hospital Institutional Animal Care and Use Committee.
METHOD DETAILS
Experimental methods
T cell sorting and in-vitro T helper cell differentiation
For the generation of time-course microarray data of Tr1 cells and validation of Il10 regulators in Tr1 cells in vitro, CD4+CD44−CD62L+CD25− naive cells were sorted from WT B6 or indicated KO and their corresponding control mice with BD FACSAria sorter, and then activated with plate-bound anti-CD3 and anti-CD28 (both at 1ug/ml) in the presence of 25ng/ml rmIL-27 (R & D systems). 10ug/ml anti-TGFβ (Bioxcell, Clone# 1D11.16.8) was also added for the microarray experiment.
For RNA-seq and qPCR analysis of IL-10 producing and non-producing T helper cells, naive CD4+CD44−CD62L+GFP− cells were sorted from Foxp3-GFP; Il10-Thy1.1 double reporter mice using BD FACSAria sorter and were activated with irradiated splenocytes depleted of CD4 T cells (at the T: APC ratio of 1:6) and 2.5ug/ml soluble anti-CD3 in the presence of polarizing cytokines. Concentration of cytokines are as follows: 20ng/ml rmIL-12 (R & D systems) for Th1; 20ng/ml rmIL-4 (Miltenyi Biotec) for Th2; 2ng/ml of rhTGFb1 and 25ng/ml rmIL-6(both from Miltenyi Biotec) for non-pathogenic Th17; 20ng/ml rmIL-1β(Miltenyi Biotec), 25ng/ml rmIL-6 (Miltenyi Biotec), and 20ng/ml rmIL-23 (R&D systems) for pathogenic Th17; 25ng/ml of rmIL-27 (R & D systems) for Tr1. IL-10 positive (Thy1.1+) and negative (Thy1.1+) 7-AAD−TCRβ+CD4+GFP− cells were re-sorted at 72 hours.
Isolation of lymphocytes from colonic lamina propria
To remove epithelial cells, colons were first washed for 20min in RPMI medium (GIBCO) with 3% FBS (Sigma-Aldrich), 5mM EDTA (Invitrogen) and 1mM DTT (Sigma) in a shaking incubator at 400rpm at 37°C, followed by three other washes each for 30 s by vibrant vortexing in RPMI with 2mM EDTA. The tissue was then cut into little pieces and digested for 30min in RPMI with 100ug/mL Liberase TL (Sigma) and 500ug/mL DNase I (Sigma) Digestion in a shaking incubator at 400rpm at 37°C. Digestion was terminated by addition of ice-cold RPMI with 3% FCS. Cells were washed twice in RPMI with 3% FCS, passed through a 40μm cell strainer and resuspended in ice-cold RPMI with 3% FCS and 1mM EDTA for sorting.
RNA profiling by microarrays, population RNA-seq and single cell RNA-seq
The temporal gene expression profiling of Tr1 cells at 17 time points during in vitro differentiation by IL-27 were measured by Affymetrix GeneChip Mouse Genome 430 2.0 Arrays. For genetic validation of the 24 TFs, naive CD4+ T cells were isolated from spleen of knockout mice and matched controls and differentiated in vitro in Tr1 polarizing conditions for 72 hours. Cells were collected and processed using an adaptation of the SMART-Seq 2 protocol (Tirosh et al., 2016), using 5uL of lysate from bulk CD4+ T cells as the input for each sample during RNA cleanup via SPRI beads (~2,000 cells lysed on average in RLT). Libraries were prepared using the Nextera XT DNA Sample Prep Kit (Illumina), quantified, pooled, and then sequenced on the HiSeq 2500 (Illumnia) to an average depth of 20M reads.
For scRNA-seq profiling of colonic CD4 T cells in control, Prdm1 cKO, Maf cKO, and DKO mice, live (7-AAD−) TCRβ+CD4+ cells were sorted from colonic lamina propria of each mouse with two biological replicates for each genotype. Cells were processed using Chromium Single Cell 3′ Reagent Kits v2 according to manufacturer’s protocol (10X Genomics). For each biological replicate, an input of 7,000 single cells was added to an individual channel with a recovery rate of approximately 1,300~2100 cells. The generated scRNA-seq libraries were sequenced on HiSeq X Ten.
ATAC-seq
Control, Prdm1 cKO, Maf cKO, and DKO Tr1 cells were cultured as described above for 72h with IL-27. Three to five replicates were included for each genotype. Subsequently, 6,000 viable Tr1 cells were sorted and frozen in BambankerTM cell freezing media (LYMPHOTEC Inc.) at 80°C. For ATAC-seq library preparation, cells were thawed at 37°C, washed once with PBS, and lysed and tagmented in 1X TD Buffer, 0.2ml TDE1 (provided in Nextera® DNA Sample Preparation Kit from Illumina), 0.01% digitonin, and 0.3X PBS in 40ml reaction volume following the protocol described by Corces et al. (2016). The DNA was purified immediately with the MinElute PCR purification kit (QIAGEN), and then PCR amplified and quantified as we previously described (Wallrapp et al., 2019). The library was sequenced on an Illumina NextSeq 550 system with paired-end reads of 37 base pairs in length.
Quantitative RT-PCR
RNA was extracted using RNeasy Plus Mini Kit (QIAGEN), cDNA was prepared using iScript Reverse Transcription Supermix (Bio-rad) and used as template for real-time qPCR run with TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific) on the ViiA 7 Real-Time PCR System (Applied Biosystems). Expression was normalized to Actb. The following probes used for qPCR were purchased from Applied Biosystems: Il10 (Mm01288386_m1), Maf (Mm02581355_s1), Prdm1 (Mm00476128_m1), Actb (Mm00607939_s1).
Flow Cytometry
Single cell suspensions were stained with antibodies against surface molecules. Fixable viability dye eF506 or 7-AAD was used to exclude dead cells. For intra-cytoplasmic cytokine staining, cells were stimulated with 12-myristate 13-acetate (PMA) (50ng/ml, Sigma), ionomycin (1 μg/ml, Sigma) in the presence of Brefeldin A (Golgiplug, BD Biosciences) and Monensin (Golgistop, BD Biosciences) for 4-5 hours prior to staining with antibodies against surface proteins followed by fixation, permeabilization with Fixation/Permeabilization Solution Kit (BD Biosciences) and staining with antibodies against intracellular cytokines. Data was analyzed with Flowjo.
Luciferase assays
5x104 293T cells were seeded in 96 well plate one day before transfection and then transfected with Firefly luciferase reporter constructs for Il10, Renilla luciferase reporter (internal control) and plasmids expressing specific transcription factors using PolyJet In Vitro DNA Transfection Reagent (SignaGen Laboratories). Cells were analyzed 48h later with Dual-Luciferase Reporter Assay System (Promega). To construct reporters for Il10 enhancers, previously described enhancer regions, including the CNS-9, HSS+2.98, and HSS+6.45 (Lee et al., 2009), were cloned upstream of the Il10 minimal promoter. Fragments containing the proximal Il10 promoter (−1.5 kb including the HSS-0.12 site) or the aforementioned enhancers were cloned into pGL4.10 Luciferase reporter plasmid (Promega).
In-vivo treatment of anti-CD3
Mice were treated with 20 μg anti-CD3 monoclonal antibody (clone 145-2C11, Bio X Cell) or an isotype control (Bio X Cell) intraperitoneally every 3 days for a total of three times. Mice were sacrificed 4h after the last treatment. CD4 T cells were purified from mesenteric lymph node by MACS® cell separation and Il10 expression was measured by qPCR.
T cell transfer colitis
CD4+CD62L+ T cells were sorted from WT and Hlx+/− mice and cultured with plate-bound anti-CD3 and anti-CD28 antibody in the presence of 25ng IL-27. 72 h later cells were detached from the plate and rested for 48h before transferred into Rag1−/− recipients. 5 × 105 WT or Hlx heterozygous Tr1 cells were transferred intraperitoneally into Rag1−/− animals and changes in body weight were monitored weekly.
Retroviral infection
T cells activated with plate-bound anti-CD3 and anti-CD28 antibody in the presence of polarizing cytokines were transduced with MSCV expressing Prdm1 (marked by Thy1.1) and Maf (marked by GFP) at 24h after activation. IL-10 expression in control (Thy1.1−GFP−), Prdm1-overexpressing (Thy1.1+GFP−), Maf-overexpressing (Thy1.1−GFP+) and cells overexpressing both (Thy1.1+GFP+) was analyzed by flow cytometry. For preparation of retroviruses, Plat-E cells were transfected with MSCV vectors with PolyJet. Supernatant containing virus was harvested 48hr after transfection of Plat-E cells and then used for spin transduction of T cells with polybrene (8 μg/ml) at 2000rpm, 32°C for 1hr.
Computational Methods
Microarray data pre-processing and analysis
Individual .CEL files were RMA normalized and merged to an expression matrix using the ExpressionFileCreator of GenePattern with default parameters (Reich et al., 2006). Gene-specific intensities were then computed by taking for each gene j and sample i the maximal probe value observed for that gene. Samples were then transferred to log-space by taking log2(intensity).
Differentially expressed genes (comparing to the Th0 control) were found using a method we previously described (Yosef et al., 2013). Briefly, genes that were detected in two of the four methods used were defined as differentially expressed: (1) Fold change. Requiring a 2-fold change (up or down) during at least two time points. (2) Polynomial fit. We used the EDGE software (Leek et al., 2006; Storey et al., 2005), designed to identify differential expression in time course data, with a threshold of q-value ≤ 0.01. (3) Sigmoidal fit. We used an algorithm similar to EDGE while replacing the polynomials with a sigmoid function, which is often more adequate for modeling time course gene expression data (Chechik and Koller, 2009). We used a threshold of q-value ≤ 0.01. (4) ANOVA. Gene expression was modeled by time (using only time points for which we have more than one replicate) and treatment. The model takes into account each variable independently, as well as their interaction. We report cases in which the P value assigned with the treatment parameter or the interaction parameter passed an FDR threshold of 0.01.
To associate the regulation activity of a differentially expressed transcription factor with the three phases of IL-10 expression (latency, induction and maintenance) we segmented our time course dataset into three corresponding time windows: 0-20h, 25-48h and 54-72h. TFs were assigned to specific phases if they were differential expressed (> 1.8 Fold change) anytime during this time window.
Prediction of TFs regulating the IL-27 network
Using approaches as we previously described (Yosef et al., 2013), we identified potential regulators of Tr1 differentiation by computing overlaps between their putative targets and sets of differentially expressed genes grouped by k-means clustering. For every TF in our database, we computed the statistical significance of the overlap between its putative targets and each of the groups defined above using Fisher’s exact test. We included cases where p < 5 × 10−5 and the fold enrichment > 1.5.
Population RNA-seq data pre-processing and analysis
RNA-seq reads were aligned using Tophat (Trapnell et al., 2009) and RSEM-based quantification (Li and Dewey, 2011) using known transcripts (mm9), followed by further processing using the Bioconductor package DESeq2 in R (Anders and Huber, 2010). The data was normalized using TMM normalization. The TMM method estimates scale factors between samples that can be incorporated into currently used statistical methods for DE analysis. Post-processing and statistical analysis was carried out in R (Li and Dewey, 2011).
For the analysis of the effect of different regulator KOs, differentially expressed genes were defined as genes with abs (logFC between control and KO) > 1.
For comparison between IL-10+ and IL-10− cells, differentially expressed genes were defined based on the raw counts with a single call to the function DESeq2 (Love et al., 2014) (FDR-adjusted P value < 0.05). Heatmap figures were generated using pheatmap package (https://cran.r-project.org/web/packages/pheatmap/index.html).
ATAC-seq analysis
Generation and analysis of ATAC-seq data for in-vitro differentiated Tr1 cells at 24 h and 72 h were performed in our previously published study (Karwacz et al., 2017). A publicly available ATAC-seq pipeline (Lee et al., 2016) was used for the processing of ATAC-seq on Prdm1 cKO, Maf cKO and DKO Tr1 cells. Briefly, reads were aligned to the mm10 genome using Bowtie2 and filtered to remove duplicates and mitochondrial reads. Biological replicate for each group were merged peaking-calling using MACS2 (Zhang et al., 2008). Integrative Genomics Viewer (IGV) was used for visualization of ATAC-seq peaks.
Single cell RNA-seq analysis
Data preprocessing.
De-multiplexing, alignment to the mm10 mouse transcriptome and UMI-collapsing were performed using the Cellranger toolkit (version 2.1.0, 10X Genomics). Subsequent analysis was performed with R package Seurat v3 (Butler et al., 2018). For downstream processing we filtered out low quality cells that had (1) a low number (< 500) of unique detected genes, and (2) a high mitochondrial content (15%) determined by the ratio of reads mapping to the mitochondria. A small proportion of cells were identified as contamination by macrophages, innate lymphoid cells, intraepithelial lymphocytes and fibroblasts, and were excluded from downstream analysis. To account for differences in sequencing depth across cells, UMI counts were normalized by the total number of UMIs per cell and converted to transcripts-per-10,000 before being log transformed (henceforth “log(TP10K+1)”).
PCA and clustering
Highly variable genes were selected using the ‘mean.var.plot’ method in FindVariableFeatures function with default settings, resulting in 341 genes which are then used for PCA analysis by RunPCA function. We used the first 40 PCs for subsequent analyses as they capture the majority of signal in an elbow plot, but we also confirmed that the resulting analyses were not particularly sensitive to the above-mentioned choice of parameters. The cells were clustered via Seurat’s FindClusters function, which optimizes modularity on a K-nearest-neighbor (KNN) graph computed from the top eigenvectors using Louvain algorithm, with nn.eps at 0.5, resolution at 0.4, and n.start at 10. These parameters resulted in clusters that captured major genotype- related separations, known T cell subgroups, and statistically validated transcriptional distinct sections of interest while avoiding subdivisions of relatively uniform parts of the data. To visualize the data, UMAP plots were generated using Seurat’s RunUMAP function with min.dist at 0.75.
Cell type assignment
To identify which T cell subtype each cluster represents, we identified markers of each cluster using Seurat’s FindAllMarkers function with min.pct at 0.25. The top 200 genes of each cluster were then used as input for My Geneset module of Immgen (immgen.org). We assigned identity to each cluster based on the cell population in the Immgen database that display highest expression of its marker genes and confirmed the designation by expression of known marker genes (Figure S5A).
Gene signatures
Scoring gene signature was performed using AddModuleScore function of Seurat based on strategies described by Tirosh et al. (2016). Markers of cell cycles including G2/M phase and S phase were provided by Tirosh et al. (2016). Th1 signature was manually curated based on literature. Th17 signature was generated by comparing microarrays of in vitro cultured Th17 cells to other T helper cells, including naive, Th1, Th2, iTreg and nTreg cells (Wei et al., 2009; Xiao et al., 2014). CD4 T cells signature from ulcerative colitis patients contains genes upregulated in CD4 T cells from biopsies of inflamed intestinal tissue in patients compared to those from healthy tissue in healthy controls profiled by scRNA-seq (Smillie et al., 2019). IBD associated GWAS genes were compiled from literature (Graham and Xavier, 2020).
Differential expression analysis
Differentially expressed genes were tested using MAST (Finak et al., 2015) by calling FindMarkers function in Seurat. To find unique markers for Prdm1/Maf DKO Tregs, DKO Tregs were compared against Treg cells in both single KO and control groups.
QUANTIFICATION AND THE STATISTICAL ANALYSIS
Unless otherwise specified, all statistical analyses were performed using the two-tail Student’s t test using GraphPad Prism software. P value less than 0.05 is considered significant (p < 0.05 = *; p < 0.01 = **; p < 0.001 = ***, p < 0.0001 = ****. Data were represented as mean ± s.e.m. unless otherwise specified. For certain types of numeric computations for transcriptomic data, the smallest P value that R can report is < 2.2 × 10−16.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| InVivoMab anti-mouse CD3ε | Bio X Cell | Cat# BE0001-1 |
| InVivomAb anti-mouse CD28 | Bio X Cell | Cat# BE0015-5 |
| InVivomAb anti-mouse TGF-β | Bio X Cell | Cat# BE0057 |
| InVivomAb polyclonal Armenian hamster IgG | Bio X Cell | Cat# BE0091 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant Mouse IL-27 (NS0-expressed) Protein | R&D SYSTEMS | Cat# 2799-ML-010 |
| Recombinant Mouse IL-12 Protein | R&D SYSTEMS | Cat# 419-ML-050 |
| Mouse IL-4, research grade | Miltenyi Biotec | Cat# 130-097-757 |
| Recombinant Mouse IL-1 beta/IL-1F2 Protein | R&D SYSTEMS | Cat# 419-ML-010 |
| Recombinant Mouse IL-6 Protein | R&D SYSTEMS | Cat# 406-ML-025 |
| Recombinant Mouse IL-23 Protein | R&D SYSTEMS | Cat# 1887-ML-010 |
| Human TGF-β1, premium grade | Miltenyi Biotec | Cat# 130-095-067 |
| Liberase TL Research Grade | Sigma | Cat# 5401020001 |
| DNase I | Sigma | Cat# 10104159001 |
| Fixable viability dye eFluor506 | eBioscience | Cat# 65-0866-14 |
| 7AAD | BD Biosciences | Cat# 559925 |
| Digitonin | Promega | Cat# G9441 |
| Critical Commercial Assays | ||
| RNeasy Plus Mini Kit | QIAGEN | Cat# 74134 |
| iScript Reverse Transcription Supermix | Bio-Rad | Cat# 1708841 |
| TaqMan Fast Advanced Master Mix | Thermo Fisher Scientific | Cat# 4444557 |
| PolyJet In Vitro DNA Transfection Reagent | SignaGen Laboratories | Cat# SL100688 |
| Dual-Luciferase® Reporter Assay System | Promega | Cat# E1960 |
| GeneChip Mouse Genome 430 2.0 Array | Affymetrix | Cat# 900497 |
| Nextera® DNA Sample Preparation Kit | Illumina | Cat# FC-121-1030 |
| MinElute Reaction Cleanup kit | QIAgen | Cat# 28204 |
| Chromium Single Cell 3′ Library & Gel Bead Kit v2 | 10x Genomics | Cat# PN-120237 |
| Chromium Single Cell A Chip Kit | 10x Genomics | Cat# PN-1000009 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE159208 |
| Tr1 ATAC-seq (related to Figures 2B and 6B) | Karwacz et al., 2017 | GEO: GSE92993 |
| Atf3 ChIP-seq (related to Figure 2B) | Garber et al., 2012 | GEO: GSE36104 |
| Fosl2 ChIP-seq (related to Figure 2B) | Ciofani et al., 2012 | GEO: GSE40918 |
| Tbx21 ChIP-seq (related to Figure 2B) | Nakayamada et al., 2011 | GEO: GSE33802 |
| Irf8 ChIP-seq (related to Figure 2B) | Xu et al., 2015 | GEO: GSE70712 |
| Experimental Models: Cell Lines | ||
| 293T cells | GenHunter | Cat# Q401 |
| Platinum-E (Plat-E) Retroviral Packaging Cell Line | Cell Biolabs | Cat# RV-101 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664 |
| BALB/cJ | The Jackson Laboratory | JAX: 000651 |
| Mouse: Foxp3-GFP | Bettelli et al., 2006 | N/A |
| Mouse: 10BiT | Maynard et al., 2007 | N/A |
| Mouse: Prdm1f/f | Shapiro-Shelef et al., 2003 | JAX: 008100 |
| Mouse: Maff/f | Wende et al., 2012 | N/A |
| Mouse: Ahr KO | Schmidt et al., 1996 | JAX: 002831 |
| Mouse: Atf3f/f | Taketani et al., 2012 | N/A |
| Mouse: Batf KO | Schraml et al., 2009 | JAX: 013758 |
| Mouse: Cebpbf/f | Sterneck et al., 2006 | N/A |
| Mouse: Ets1 KO | Muthusamy et al., 1995 | N/A |
| Mouse: Fosl2f/f | Karreth et al., 2004 | N/A |
| Mouse: Hif1af/f | Ryan et al., 2000 | JAX:007561 |
| Mouse: Irf1 KO | Matsuyama et al., 1993 | JAX: 002762 |
| Mouse: Irf4 KO | Klein et al., 2006 | JAX: 009380 |
| Mouse: Irf8f/f | Feng et al., 2011 | JAX:014175 |
| Mouse: Irf9 KO | Gift from Paul J. Utz | RIKEN: RBRC00915 |
| Mouse: Nfil3f/f | Gascoyne et al., 2009 | N/A |
| Mouse: Stat3f/f | Moh et al., 2007 | JAX: 016923 |
| Mouse: Tbx21 KO | Finotto et al., 2002 | JAX: 004648 |
| Mouse: Bhlhe40 KO | Jiang et al., 2008 | JAX: 029732 |
| Mouse: Hlx+/− | Hentsch et al., 1996 | JAX: 008313 |
| Mouse: Stat4 | Kaplan et al., 1996 | JAX: 002826 |
| Mouse: Batf3 KO | Hildner et al., 2008 | JAX: 013755 |
| Mouse: Nfe2l2 KO | Chan et al., 1996 | JAX: 017009 |
| Mouse: Irf7 KO | Gift from Ian Rifkin | N/A |
| Mouse: Id2f/f | Seillet et al., 2013 | N/A |
| Mouse: Fli1+/− | Gift from Maria Trojanowska | N/A |
| Mouse: Cd4-Cre | Lee et al., 2001 | Taconic: 4196 |
| Mouse: Actin-Cre | Lewandoski et al., 1997 | JAX: 033984 |
| Mouse: Lck-Cre | Hennet et al., 1995 | JAX: 003802 |
| Mouse: dLck-Cre | Wang et al., 2001 | JAX: 012837 |
| Recombinant DNA | ||
| pGL4.10[luc2] Vector | Promega | Cat# E665A |
| pGL4.10-Il10 proximal promoter-luc2 | This paper | N/A |
| pGL4.10-Il10 CNS-9-luc2 | This paper | N/A |
| pGL4.10-Il10 HSS+2.98-luc2 | This paper | N/A |
| pGL4.10-Il10 HSS+6.45-luc2 | This paper | N/A |
| MSCV-IRES-GFP | Gift from Tannishtha Reya | Addgene #20672 |
| MSCV-Maf-IRES-GFP | This paper | N/A |
| MSCV-IRES-Thy1.1 | Gift from Philippa Marrack | N/A |
| MSCV-Prdm1-IRES-Thy1.1 | This paper | N/A |
| Software and Algorithms | ||
| GenePattern | Reich et al., 2006 | https://www.genepattern.org/ |
| EDGE | Leek et al., 2006 | https://www.bioconductor.org/ |
| Tophat | Trapnell et al., 2009 | https://github.com/infphilo/tophat |
| RSEM | Li and Dewey, 2011 | http://deweylab.github.io/RSEM/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| RStudio | RStudio | https://www.rstudio.com/ |
| ATAC-seq pipeline | Lee et al., 2016 | https://zenodo.org/record/211733 |
| Integrative Genomics Viewer | Robinson et al., 2017 | http://software.broadinstitute.org/software/igv/ |
| R package Seurat v3 | Butler et al., 2018 | https://satijalab.org/seurat/ |
| FlowJo | FlowJo | https://www.flowjo.com |
| Prism | GraphPad | https://www.graphpad.com |
Highlights.
IL-27-driven transcriptional network in CD4 T cells unravels key Il10 regulators
Systematic characterization of the function of 16 Il10 regulators by RNA-seq
Identification of transcription factors associated with Il10 in multiple T cell subsets
Prdm1 and Maf are critical for Il10 production and intestinal immune homeostasis
ACKNOWLEDGMENTS
The authors thank Christophe Benoist, Arlene Sharpe, Mikael Pittet, Mary Collins, and Karen Dixon for constructive criticism and discussions and Deneen Kozoriz, Rajesh K. Krishnan, Leslie Gaffney, Sarah Zaghouani, Haoxin Li, Ruihan Tang, Qianxia Zhang, Yu Hou, Jingwen Shi, Danyang He, Sheng Xiao, and Ido Amit for technical assistance. This work was supported by NIH grants R01NS30843, R01AI144166, P01AI073748, P01AI039671, P01AI056299, and P01AI129880 (to V.K.K.) and the Klarman Cell Observatory (to A.R.). A.R. is an Investigator of the Howard Hughes Medical Institute. A.C.A. is a recipient of the Brigham and Women’s President’s Scholar Award and is supported by NIH grant CA229400. A.M. was supported by the Alon Fellowship for Outstanding Young Scientists, Israel Council for Higher Education. L.A. was supported by the European Research Council (grant agreement 677251). A.A. was supported by DST-SERB grant CRG/2018/002653 from the Department of Science and Technology, Government of India.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108433.
DECLARATION OF INTERESTS
A.R. is a founder and equity holder of Celsius Therapeutics, an equity holder in Immunitas Therapeutics, and, until August 31, 2020, an SAB member of Syros Pharmaceuticals, Neogene Therapeutics, Asimov, and Thermo Fisher Scientific. From August 1, 2020, A.R. is an employee of Genentech, a member of the Roche Group. V.K.K. has an ownership interest in Tizona Therapeutics, Celsius Therapeutics, and Bicara Therapeutics. V.K.K. has financial interests in Biocon Biologic, BioLegend, Elpiscience Biopharmaceutical Ltd., Equilium Inc., and Syngene Intl. V.K.K. is a member of SABs for Elpiscience Biopharmaceutical Ltd., GSK, Kintai Therapeutics, Repertoir Immune Medicines, Rubius Therapeutics, and Tizona Therapeutics. A.C.A. is a member of SAB for Tizona Therapeutics, Compass Therapeutics, Zumutor Biologics, ImmuneOncia, and Astellas Global Pharma Development Inc. A.C.A.’s and V.K.K.’s interests were reviewed and managed by the Brigham and Women’s Hospital and Partners Healthcare and A.R.’s interests by the Broad Institute and HHMI in accordance with their conflict of interest policies.
REFERENCES
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D, Burns EJ, Sherr DH, Weiner HL, and Kuchroo VK (2010). The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol 11, 854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D, Villarino A, Silverman M, He W, Thornton EM, Mu S, Summer S, Covey TM, Huang E, Yoshida H,et al. (2004). The IL-27 receptor(WSX-1) is an inhibitor of innate and adaptive elements of type 2 immunity. J. Immunol 173, 5626–5634. [DOI] [PubMed] [Google Scholar]
- Awasthi A, Carrier Y, Peron JPS, Bettelli E, Kamanaka M, Flavell RA, Kuchroo VK, Oukka M, and Weiner HL (2007). A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nat. Immunol 8, 1380–1389. [DOI] [PubMed] [Google Scholar]
- Batten M, Li J, Yi S, Kljavin NM, Danilenko DM, Lucas S, Lee J, de Sauvage FJ, and Ghilardi N (2006). Interleukin 27 limits autoimmune encephalomyelitis by suppressing the development of interleukin 17-producing T cells. Nat. Immunol 7, 929–936. [DOI] [PubMed] [Google Scholar]
- Batten M, Kljavin NM, Li J, Walter MJ, de Sauvage FJ, and Ghilardi N (2008). Cutting edge: IL-27 is a potent inducer of IL-10 but not FoxP3 in murine T cells. J. Immunol 180, 2752–2756. [DOI] [PubMed] [Google Scholar]
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, and Kuchroo VK (2006). Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441,235–238. [DOI] [PubMed] [Google Scholar]
- Boks MA, Kager-Groenland JR, van Ham SM, and ten Brinke A (2016). IL-10/IFNγ co-expressing CD4(+) T cells induced by IL-10 DC display a regulatory gene profile and downmodulate T cell responses. Clin. Immunol 162, 91–99. [DOI] [PubMed] [Google Scholar]
- Brockmann L, Soukou S, Steglich B, Czarnewski P, Zhao L, Wende S, Bedke T, Ergen C, Manthey C, Agalioti T, et al. (2018). Molecular and functional heterogeneity of IL-10-producing CD4+ T cells. Nat. Commun 9, 5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton BR, Britton GJ, Fang H, Verhagen J, Smithers B, Sabatos-Peyton CA, Carney LJ, Gough J, Strobel S, and Wraith DC (2014). Sequential transcriptional changes dictate safe and effective antigen-specific immunotherapy. Nat. Commun 5, 4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Liu J, Song L, and Ma X (2005). The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages. J. Immunol 174, 3484–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Lu R, Chang JC, and Kan YW (1996). NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. USA 93, 13943–13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chechik G, and Koller D (2009). Timing of gene expression responses to environmental changes. J. Comput. Biol 16, 279–290. [DOI] [PubMed] [Google Scholar]
- Chihara N, Madi A, Kondo T, Zhang H, Acharya N, Singer M, Nyman J, Marjanovic ND, Kowalczyk MS, Wang C, et al. (2018). Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature 558, 454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkhurst CN, Muratet M, et al. (2012). A validated regulatory network for Th17 cell specification. Cell 151, 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. (2016). Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet 48, 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL, and Kallies A (2011). The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol 12, 304–311. [DOI] [PubMed] [Google Scholar]
- De Nardo D, Labzin LI, Kono H, Seki R, Schmidt SV, Beyer M, Xu D, Zimmer S, Lahrmann C, Schildberg FA, et al. (2014). High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nat. Immunol 15, 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong JH, O’Hara Hall A, Rausch M, Moodley D, Perry J, Park J, Phan AT, Beiting DP, Kedl RM, Hill JA, et al. (2019). IL-27 and TCR Stimulation Promote T Cell Expression of Multiple Inhibitory Receptors. Immunohorizons 3, 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D, Zhang H, Zeissig S, Lipinski S, Till A, Jiang T, Stade B, Bromberg Y, Ellinghaus E, Keller A, et al. (2013). Association between variants of PRDM1 and NDP52 and Crohn’s disease, based on exome sequencing and functional studies. Gastroenterology 145, 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Wang H, Shin D-M, Masiuk M, Qi C-F, and Morse HC 3rd. (2011). IFN regulatory factor 8 restricts the size of the marginal zone and follicular B cell pools. J. Immunol 186, 1458–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, and Linsley PS (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finotto S, Neurath MF, Glickman JN, Qin S, Lehr HA, Green FHY, Ackerman K, Haley K, Galle PR, Szabo SJ, et al. (2002). Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science 295, 336–338. [DOI] [PubMed] [Google Scholar]
- Fitzgerald DC, Zhang G-X, El-Behi M, Fonseca-Kelly Z, Li H, Yu S, Saris CJM, Gran B, Ciric B, and Rostami A (2007). Suppression of autoimmune inflammation of the central nervous system by interleukin 10 secreted by interleukin 27-stimulated T cells. Nat. Immunol 8, 1372–1379. [DOI] [PubMed] [Google Scholar]
- Gabryšová L, Howes A, Saraiva M, and O’Garra A (2014). The regulation of IL-10 expression. Curr. Top. Microbiol. Immunol 380, 157–190. [DOI] [PubMed] [Google Scholar]
- Gabryšová L, Alvarez-Martinez M, Luisier R, Cox LS, Sodenkamp J, Hosking C, Pérez-Mazliah D, Whicher C, Kannan Y, Potempa K, et al. (2018). c-Maf controls immune responses by regulating disease-specific gene networks and repressing IL-2 in CD4+ T cells. Nat. Immunol 19, 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliani N, Amezcua Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW, de Zoete MR, Licona-Limón P, Paiva RS, Ching T, et al. (2015). Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 523, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, Robinson J, Minie B, Chevrier N, Itzhaki Z, et al. (2012).A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol. Cell 47, 810–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg G, Muschaweckh A, Moreno H, Vasanthakumar A, Floess S, Lepennetier G, Oellinger R, Zhan Y, Regen T, Hiltensperger M, et al. (2019). Blimp1 prevents methylation of foxp3 and loss of regulatory T cell identity at sites of inflammation. Cell Rep. 26, 1854–1868.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, and Brady HJM (2009). The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol 10, 1118–1124. [DOI] [PubMed] [Google Scholar]
- Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, Kennedy K, Hai T, Bolouri H, and Aderem A (2006). Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 441, 173–178. [DOI] [PubMed] [Google Scholar]
- Graham DB, and Xavier RJ (2020). Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 578, 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AO, Beiting DP, Tato C, John B, Oldenhove G, Lombana CG, Pritchard GH, Silver JS, Bouladoux N, Stumhofer JS, et al. (2012). The cytokines interleukin 27 and interferon-γ promote distinct Treg cell populations required to limit infection-induced pathology. Immunity 37, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamano R, Baba T, Sasaki S, Tomaru U, Ishizu A, Kawano M, Yamagishi M, and Mukaida N (2014). Ag and IL-2 immune complexes efficiently expand Ag-specific Treg cells that migrate in response to chemokines and reduce localized immune responses. Eur. J. Immunol 44, 1005–1015. [DOI] [PubMed] [Google Scholar]
- Hasnain SZ, Tauro S, Das I, Tong H, Chen AC-H, Jeffery PL, McDonald V, Florin TH, and McGuckin MA (2013). IL-10 promotes production of intestinal mucus by suppressing protein misfolding and endoplasmic reticulum stress in goblet cells. Gastroenterology 144, 357–368.e9. [DOI] [PubMed] [Google Scholar]
- Hedrich CM, and Bream JH (2010). Cell type-specific regulation of IL-10 expression in inflammation and disease. Immunol. Res 47, 185–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennet T, Hagen FK, Tabak LA, and Marth JD (1995). T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc. Natl. Acad. Sci. USA 92, 12070–12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson J, Chen X, Gomes T, Ullah U, Meyer KB, Miragaia R, Duddy G, Pramanik J, Yusa K, Lahesmaa R, and Teichmann SA (2019). Genome-wide CRISPR Screens in T Helper Cells Reveal Pervasive Crosstalk between Activation and Differentiation. Cell 176, 882–896.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentsch B, Lyons I, Li R, Hartley L, Lints TJ, Adams JM, and Harvey RP (1996). Hlx homeo box gene is essential for an inductive tissue interaction that drives expansion of embryonic liver and gut. Genes Dev. 10, 70–79. [DOI] [PubMed] [Google Scholar]
- Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, et al. (2008). Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Paik PK, Chen J, Yarilina A, Kockeritz L, Lu TT, Woodgett JR, and Ivashkiv LB (2006). IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity 24, 563–574. [DOI] [PubMed] [Google Scholar]
- Huber S, Gagliani N, Esplugues E, O’Connor W Jr., Huber FJ, Chaudhry A, Kamanaka M, Kobayashi Y, Booth CJ, Rudensky AY, et al. (2011). Th17 cells express interleukin-10 receptor and are controlled by Foxp3− and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity 34, 554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh JP, Lin C-C, Kimmey JM, Jarjour NN, Schwarzkopf EA, Bradstreet TR, Shchukina I, Shpynov O, Weaver CT, Taneja R, et al. (2018). Bhlhe40 is an essential repressor of IL-10 during Mycobacterium tuberculosis infection. J. Exp. Med 215, 1823–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, and Fathman CG (1997). CD45RBhigh CD4+ T cells from IFN-gamma knockout mice do not induce wasting disease. J. Autoimmun 10, 455–459. [DOI] [PubMed] [Google Scholar]
- Iyer SS, and Cheng G (2012). Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit. Rev. Immunol 32, 23–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Tian F, Du Y, Copeland NG, Jenkins NA, Tessarollo L, Wu X, Pan H, Hu X-Z, Xu K, et al. (2008). BHLHB2 controls Bdnf promoter 4 activity and neuronal excitability. J. Neurosci 28, 1118–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. ; International IBD Genetics Consortium (IIBDGC) (2012). Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491,119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamanaka M, Kim ST, Wan YY, Sutterwala FS, Lara-Tejero M, Galán JE, Harhaj E, and Flavell RA (2006). Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity 25, 941–952. [DOI] [PubMed] [Google Scholar]
- Kaplan MH, Sun YL, Hoey T, and Grusby MJ (1996). Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature 382, 174–177. [DOI] [PubMed] [Google Scholar]
- Karreth F, Hoebertz A, Scheuch H, Eferl R, and Wagner EF (2004). The AP1 transcription factor Fra2 is required for efficient cartilage development. Development 131, 5717–5725. [DOI] [PubMed] [Google Scholar]
- Karwacz K, Miraldi ER, Pokrovskii M, Madi A, Yosef N, Wortman I, Chen X, Watters A, Carriero N, Awasthi A, et al. (2017). Critical role of IRF1 and BATF in forming chromatin landscape during type 1 regulatory cell differentiation. Nat. Immunol 18, 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Zou YR, Goldstein J, Reizis B, and Diamond B (2011). Tolerogenic function of Blimp-1 in dendritic cells. J. Exp. Med 208, 2193–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, and Dalla-Favera R (2006). Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol 7, 773–782. [DOI] [PubMed] [Google Scholar]
- Kremer KN, Kumar A, and Hedin KE (2007). Haplotype-independent costimulation of IL-10 secretion by SDF-1/CXCL12 proceeds via AP-1 binding to the human IL-10 promoter. J. Immunol 178, 1581–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn R, Löhler J, Rennick D, Rajewsky K, and Mülller W (1993). Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274. [DOI] [PubMed] [Google Scholar]
- Langenhorst D, Gogishvili T, Ribechini E, Kneitz S, McPherson K, Lutz MB, and Hünig T (2012). Sequential induction of effector function, tissue migration and cell death during polyclonal activation of mouse regulatory T-cells. PLoS ONE 7, e50080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Christoforo G, Foo CS, Probert C, Kundaje A, Boley N, kohpangwei, Dacre M, and Kim D (2016). kundajelab/atac_ dnase_pipelines: 0.3.3. 10.5281/zenodo.211733. [DOI] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Pérez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. (2001). A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774. [DOI] [PubMed] [Google Scholar]
- Lee C-G, Kang K-H, So J-S, Kwon H-K, Son J-S, Song M-K, Sahoo A, Yi H-J, Hwang K-C, Matsuyama T, et al. (2009). A distal cis-regulatory element, CNS-9, controls NFAT1 and IRF4-mediated IL-10 gene activation in T helper cells. Mol. Immunol 46, 613–621. [DOI] [PubMed] [Google Scholar]
- Lee C-G, Kwon H-K, Sahoo A, Hwang W, So J-S, Hwang J-S, Chae C-S, Kim G-C, Kim J-E, So H-S, et al. (2012). Interaction of Ets-1 with HDAC1 represses IL-10 expression in Th1 cells. J. Immunol 188, 2244–2253. [DOI] [PubMed] [Google Scholar]
- Leek JT, Monsen E, Dabney AR, and Storey JD (2006). EDGE: extraction and analysis of differential gene expression. Bioinformatics 22, 507–508. [DOI] [PubMed] [Google Scholar]
- Leithäuser F, Meinhardt-Krajina T, Fink K, Wotschke B, Möller P, and Reimann J (2006). Foxp3-expressing CD103+ regulatory T cells accumulate in dendritic cell aggregates of the colonic mucosa in murine transfer colitis. Am. J. Pathol 168, 1898–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard WJ, and Wan C-K (2016). IL-21 Signaling in Immunity. F1000Res. 5, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M, Meyers EN, and Martin GR (1997). Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol 62, 159–168. [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C-C, Bradstreet TR, Schwarzkopf EA, Sim J, Carrero JA, Chou C, Cook LE, Egawa T, Taneja R, Murphy TL, et al. (2014). Bhlhe40 controls cytokine production by T cells and is essential for pathogenicity in autoimmune neuroinflammation. Nat. Commun 5, 3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-W, Tseng H-P, Chen L-C, Chen B-K, and Chang W-C (2003). Functional cooperation of simian virus 40 promoter factor 1 and CCAAT/enhancer-binding protein beta and delta in lipopolysaccharide-induced gene activation of IL-10 in mouse macrophages. J. Immunol 171, 821–828. [DOI] [PubMed] [Google Scholar]
- Liu M, Zhao X, Ma Y, Zhou Y, Deng M, and Ma Y (2018). Transcription factor c-Maf is essential for IL-10 gene expression in B cells. Scand. J. Immunol 88, e12701. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyher P-L, Rochefort J, Baudesson de Chanville C, Hamon P, Lescaille G, Bertolus C, Guillot-Delost M, Krummel MF, Lemoine FM, Combadière C, and Boissonnas A (2016). CCR2 influences T regulatory cell migration to tumors and serves as a biomarker of cyclophosphamide sensitivity. Cancer Res. 76, 6483–6494. [DOI] [PubMed] [Google Scholar]
- Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, Magnusdottir E, and Calame K (2006). Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat. Immunol 7, 457–465. [DOI] [PubMed] [Google Scholar]
- Mascanfroni ID, Takenaka MC, Yeste A, Patel B, Wu Y, Kenison JE, Siddiqui S, Basso AS, Otterbein LE, Pardoll DM, et al. (2015). Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-α. Nat. Med 21, 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kundig TM, Amakawa R, Kishihara K, Wakeham A, et al. (1993). Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell 75, 83–97. [PubMed] [Google Scholar]
- Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, Rudensky AY, and Weaver CT (2007). Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3-precursor cells in the absence of interleukin 10. Nat. Immunol 8, 931–941. [DOI] [PubMed] [Google Scholar]
- Moh A, Iwamoto Y, Chai G-X, Zhang SS-M, Kano A, Yang DD, Zhang W, Wang J, Jacoby JJ, Gao B, et al. (2007). Role of STAT3 in liver regeneration: survival, DNA synthesis, inflammatory reaction and liver mass recovery. Lab. Invest 87, 1018–1028. [DOI] [PubMed] [Google Scholar]
- Mondini M, Loyher P-L, Hamon P, Gerbé de Thoré M, Laviron M, Berthelot K, Clémenson C, Salomon BL, Combadière C, Deutsch E, and Boissonnas A (2019). CCR2-Dependent Recruitment of Tregs and Monocytes Following Radiotherapy Is Associated with TNFα-Mediated Resistance. Cancer Immunol. Res 7, 376–387. [DOI] [PubMed] [Google Scholar]
- Montes de Oca M, Kumar R, de Labastida Rivera F, Amante FH, Sheel M, Faleiro RJ, Bunn PT, Best SE, Beattie L, Ng SS, et al. (2016). Blimp-1-Dependent IL-10 Production by Tr1 Cells Regulates TNF-Mediated Tissue Pathology. PLoS Pathog. 12, e1005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Hutchins AS, High FA, Lee HW, Sykes KJ, Chodosh LA, and Reiner SL (2002). Hlx is induced by and genetically interacts with T-bet to promote heritable T(H)1 gene induction. Nat. Immunol 3, 652–658. [DOI] [PubMed] [Google Scholar]
- Muthusamy N, Barton K, and Leiden JM (1995). Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature 377, 639–642. [DOI] [PubMed] [Google Scholar]
- Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, et al. (2011). Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity 35,919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann C, Heinrich F, Neumann K, Junghans V, Mashreghi M-F, Ahlers J, Janke M, Rudolph C, Mockel-Tenbrinck N, Kühl AA, et al. (2014). Role of Blimp-1 in programing Th effector cells into IL-10 producers. J. Exp. Med 211, 1807–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann C, Blume J, Roy U, Teh PP, Vasanthakumar A, Beller A, Liao Y, Heinrich F, Arenzana TL, Hackney JA, et al. (2019). c-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host-microbiota homeostasis. Nat. Immunol 20, 471–481. [DOI] [PubMed] [Google Scholar]
- Neurath MF, Weigmann B, Finotto S, Glickman J, Nieuwenhuis E, Iijima H, Mizoguchi A, Mizoguchi E, Mudter J, Galle PR, et al. (2002). The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn’s disease. J. Exp. Med 195, 1129–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa C, Bankoti R, Nguyen T, Hassanzadeh-Kiabi N, Nadeau S, Porritt RA, Couse M, Fan X, Dhall D, Eberl G, et al. (2018). Blimp-1 Functions as a Molecular Switch to Prevent Inflammatory Activity in Foxp3+ RORγt+ Regulatory T Cells. Cell Rep. 25, 19–28.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Rutz S, Crellin NK, Valdez PA, and Hymowitz SG (2011). Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol 29, 71–109. [DOI] [PubMed] [Google Scholar]
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao C-A, et al. (2003). Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 302, 1041–1043. [DOI] [PubMed] [Google Scholar]
- Pot C, Jin H, Awasthi A, Liu SM, Lai C-Y, Madan R, Sharpe AH, Karp CL, Miaw S-C, Ho I-C, and Kuchroo VK (2009). Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J. Immunol 183, 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, and Mesirov JP (2006). GenePattern 2.0. Nat. Genet 38, 500–501. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Wenger AM, Zehir A, and Mesirov JP (2017). Variant Review with the Integrative Genomics Viewer. Cancer Res. 77, e31–e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncarolo MG, Yssel H, Touraine JL, Betuel H, De Vries JE, and Spits H (1988). Autoreactive T cell clones specific for class I and class II HLA antigens isolated from a human chimera. J. Exp. Med 167, 1523–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz S, Janke M, Kassner N, Hohnstein T, Krueger M, and Scheffold A (2008). Notch regulates IL-10 production by T helper 1 cells. Proc. Natl. Acad. Sci. USA 105, 3497–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan HE, Poloni M, McNulty W, Elson D, Gassmann M, Arbeit JM, and Johnson RS (2000). Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 60, 4010–4015. [PubMed] [Google Scholar]
- Schmidt JV, Su GH, Reddy JK, Simon MC, and Bradfield CA (1996). Characterization of a murine Ahr null allele: involvement of the Ah receptor in hepatic growth and development. Proc. Natl. Acad. Sci. USA 93, 6731–6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz ML, Shaban MS, Albert BV, Gökçen A, and Kracht M (2018). The Crosstalk of Endoplasmic Reticulum (ER) Stress Pathways with NF-κB: Complex Mechanisms Relevant for Cancer, Inflammation and Infection. Biomedicines 6, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraml BU, Hildner K, Ise W, Lee W-L, Smith WA-E, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, et al. (2009). The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 460, 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C, Jackson JT, Markey KA, Brady HJM, Hill GR, Macdonald KPA, Nutt SL, and Belz GT (2013). CD8α+ DCs can be induced in the absence of transcription factors Id2, Nfil3, and Batf3. Blood 121, 1574–1583. [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Lin K-I, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, and Calame K (2003). Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity 19, 607–620. [DOI] [PubMed] [Google Scholar]
- Shim JO (2019). Recent advance in very early onset inflammatory bowel disease. Pediatr. Gastroenterol. Hepatol. Nutr 22, 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin B, Poholek C, Yeh WI, and Harrington L (2014). T-bet controls intestinal chronic inflammation via regulation of IL-10 production by CD4 T cells (MUC8P.806). J. Immunol 192, 198.7. [Google Scholar]
- Shkoda A, Ruiz PA, Daniel H, Kim SC, Rogler G, Sartor RB, and Haller D (2007). Interleukin-10 blocked endoplasmic reticulum stress in intestinal epithelial cells: impact on chronic inflammation. Gastroenterology 132, 190–207. [DOI] [PubMed] [Google Scholar]
- Shoemaker J, Saraiva M, and O’Garra A (2006). GATA-3 directly remodels the IL-10 locus independently of IL-4 in CD4+ T cells. J. Immunol 176, 3470–3479. [DOI] [PubMed] [Google Scholar]
- Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, et al. (2019). Intra-and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 178, 714–730.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SD, Di Marco F, Hooley J, Pitts-Meek S, Bauer M, Ryan AM, Sordat B, Gibbs VC, and Aguet M (1998). The orphan receptor CRF2-4 is an essential subunit of the interleukin 10 receptor. J. Exp. Med 187, 571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterneck E, Zhu S, Ramirez A, Jorcano JL, and Smart RC (2006). Conditional ablation of C/EBP beta demonstrates its keratinocyte-specific requirement for cell survival and mouse skin tumorigenesis. Oncogene 25, 1272–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Xiao W, Leek JT, Tompkins RG, and Davis RW (2005). Significance analysis of time course microarray experiments. Proc. Natl. Acad. Sci. USA 102, 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumhofer JS, Laurence A, Wilson EH, Huang E, Tato CM, Johnson LM, Villarino AV, Huang Q, Yoshimura A, Sehy D, et al. (2006). Interleukin 27 negatively regulates the development of interleukin 17-producing T helper cells during chronic inflammation of the central nervous system. Nat. Immunol 7, 937–945. [DOI] [PubMed] [Google Scholar]
- Stumhofer JS, Silver JS, Laurence A, Porrett PM, Harris TH, Turka LA, Ernst M, Saris CJM, O’Shea JJ, and Hunter CA (2007). Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat. Immunol 8, 1363–1371. [DOI] [PubMed] [Google Scholar]
- Sungnak W, Wang C, and Kuchroo VK (2019). Multilayer regulation of CD4 T cell subset differentiation in the era of single cell genomics. Adv. Immunol 141, 1–31. [DOI] [PubMed] [Google Scholar]
- Taketani K, Kawauchi J, Tanaka-Okamoto M, Ishizaki H, Tanaka Y, Sakai T, Miyoshi J, Maehara Y, and Kitajima S (2012). Key role of ATF3 in p53-dependent DR5 induction upon DNA damage of human colon cancer cells. Oncogene 31, 2210–2221. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trandem K, Zhao J, Fleming E, and Perlman S (2011). Highly activated cytotoxic CD8 T cells express protective IL-10 at the peak of coronavirus-induced encephalitis. J. Immunol 186, 3642–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay V, and Fu Y-X (2013). Lymphotoxin signalling in immune homeostasis and the control of microorganisms. Nat. Rev. Immunol 13, 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino A, Hibbert L, Lieberman L, Wilson E, Mak T, Yoshida H, Kastelein RA, Saris C, and Hunter CA (2003). The IL-27R (WSX-1) is required to suppress T cell hyperactivity during infection. Immunity 19, 645–655. [DOI] [PubMed] [Google Scholar]
- Villarino AV, Stumhofer JS, Saris CJM, Kastelein RA, de Sauvage FJ, and Hunter CA (2006). IL-27 limits IL-2 production during Th1 differentiation. J. Immunol 176, 237–247. [DOI] [PubMed] [Google Scholar]
- Wallrapp A, Burkett PR, Riesenfeld SJ, Kim S-J, Christian E, Abdulnour R-EE, Thakore PI, Schnell A, Lambden C, Herbst RH, et al. (2019). Calcitonin Gene-Related Peptide Negatively Regulates Alarmin-Driven Type 2 Innate Lymphoid Cell Responses. Immunity 51, 709–723.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Strong J, and Killeen N (2001). Homeostatic competition among T cells revealed by conditional inactivation of the mouse Cd4 gene. J. Exp. Med 194, 1721–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Meng R, Li Z, Yang B, Liu Y, Huang F, Zhang J, Chen H, and Wu C (2011). IL-27 induces the differentiation of Tr1-like cells from human naive CD4+ T cells via the phosphorylation of STAT1 and STAT3. Immunol. Lett 136, 21–28. [DOI] [PubMed] [Google Scholar]
- Wang C, Yosef N, Gaublomme J, Wu C, Lee Y, Clish CB, Kaminski J, Xiao S, Meyer Zu Horste G, Pawlak M, et al. (2015). CD5L/AIM regulates lipid biosynthesis and restrains th17 cell pathogenicity. Cell 163, 1413–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watchmaker PB, Lahl K, Lee M, Baumjohann D, Morton J, Kim SJ, Zeng R, Dent A, Ansel KM, Diamond B, et al. (2014). Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nat. Immunol 15, 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh T-Y, Watford WT, et al. (2009). Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wende H, Lechner SG, Cheret C, Bourane S, Kolanczyk ME, Pattyn A, Reuter K, Munier FL, Carroll P, Lewin GR, and Birchmeier C (2012). The transcription factor c-Maf controls touch receptor development and function. Science 335, 1373–1376. [DOI] [PubMed] [Google Scholar]
- Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y, Regev A, and Kuchroo VK (2013). Induction of pathogenic TH17 cells by inducible saltsensing kinase SGK1. Nature 496, 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Yosef N, Yang J, Wang Y, Zhou L, Zhu C, Wu C, Baloglu E, Schmidt D, Ramesh R, et al. (2014). Small-molecule RORγt antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity 40, 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Chaudhri VK, Wu Z, Biliouris K, Dienger-Stambaugh K, Rochman Y, and Singh H (2015). Regulation of bifurcating B cell trajectories by mutual antagonism between transcription factors IRF4 and IRF8. Nat. Immunol 76, 1274–1281. [DOI] [PubMed] [Google Scholar]
- Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, Galan C, Belkaid Y, Bonneau R, and Littman DR (2018). c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, et al. (2013). Dynamic regulatory network controlling TH17 cell differentiation. Nature 496, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, and Hunter CA (2015). The immunobiology of interleukin-27. Annu. Rev. Immunol 33, 417–443. [DOI] [PubMed] [Google Scholar]
- Yu F, Sharma S, Jankovic D, Gurram RK, Su P, Hu G, Li R, Rieder S, Zhao K, Sun B, and Zhu J (2018). The transcription factor Bhlhe40 is a switch of inflammatory versus antiinflammatory Th1 cell fate determination. J. Exp. Med 215, 1813–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, and Kuchroo V (2019). Epigenetic and transcriptional mechanisms for the regulation of IL-10. Semin. Immunol 44, 101324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Schröppel B, Lal G, Jakubzick C, Mao X, Chen D, Yin N, Jessberger R, Ochando JC, Ding Y, and Bromberg JS (2009). Regulatory T cells sequentially migrate from inflamed tissues to draining lymph nodes to suppress the alloimmune response. Immunity 30, 458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Lee JS, Gartlan KH, Schuster IS, Comerford I, Varelias A, Ullah MA, Vuckovic S, Koyama M, Kuns RD, et al. (2017). Eomesodermin promotes the development of type 1 regulatory T (TR1) cells. Sci. Immunol 2, eaah7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Sakuishi K, Xiao S, Sun Z, Zaghouani S, Gu G, Wang C, Tan DJ, Wu C, Rangachari M, et al. (2015). An IL-27/NFIL3 signalling axis drives Tim-3 and IL-10 expression and T-cell dysfunction. Nat. Commun 6, 6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Shi T, Zhong C, Wang Y, Chang M, and Liu X (2017). IL-10 and IL-10 Receptor Mutations in Very Early Onset Inflammatory Bowel Disease. Gastroenterol. Res 10, 65–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated in this paper has been deposited in the Gene Expression Omnibus (GEO) under accession number GEO: GSE159208.