Abstract
Legionella spp. are widespread bacteria in aquatic environments with a growing impact on human health. Between the 61 species, Legionella pneumophila is the most prevalent in human diseases; on the contrary, Legionella non-pneumophila species are less detected in clinical diagnosis or during environmental surveillance due to their slow growth in culture and the absence of specific and rapid diagnostic/analytical tools. Reliable and rapid isolate identification is essential to estimate the source of infection, to undertake containment measures, and to determine clinical treatment. Matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI–TOF MS), since its introduction into the routine diagnostics of laboratories, represents a widely accepted method for the identification of different bacteria species, described in a few studies on the Legionella clinical and environmental surveillance. The focus of this study was the improvement of MALDI–TOF MS on Legionella non-pneumophila species collected during Legionella nosocomial and community surveillance. Comparative analysis with cultural and mip-gene sequencing results was performed. Moreover, a phylogenetic analysis was carried out to estimate the correlations amongst isolates. MALDI–TOF MS achieved correct species-level identification for 45.0% of the isolates belonging to the Legionella anisa, Legionella rubrilucens, Legionella feeleii, and Legionella jordanis species, displaying a high concordance with the mip-gene sequencing results. In contrast, less reliable identification was found for the remaining 55.0% of the isolates, corresponding to the samples belonging to species not yet included in the database. The phylogenetic analysis showed relevant differences inside the species, regruped in three main clades; among the Legionella anisa clade, a subclade with a divergence of 3.3% from the main clade was observed. Moreover, one isolate, identified as Legionella quinlivanii, displayed a divergence of 3.8% from the corresponding reference strain. However, these findings require supplementary investigation. The results encourage the implementation of MALDI–TOF MS in routine diagnostics and environmental Legionella surveillance, as it displays a reliable and faster identification at the species level, as well as the potential to identify species that are not yet included in the database. Moreover, phylogenetic analysis is a relevant approach to correlate the isolates and to track their spread, especially in unconventional reservoirs, where Legionella prevention is still underestimated.
Keywords: Legionella non-pneumophila species, Legionella identification, MALDI-TOF MS, MALDI Biotyper system, agglutination test, mip-gene sequencing
Introduction
Legionella spp. are Gram-negative bacteria ubiquitous in natural fresh water such as rivers, lakes, and thermal springs. They can also be found in moist soil and mud in which the association with amoebae, protozoa, and biofilms plays a key role in the life cycle of bacteria (Rowbotham, 1980; Fields, 1996; Diederen, 2008).
However, all cases, clusters, or outbreaks are linked to artificial freshwater environments, which represent a main reservoir of Legionella and a significant health risk from an epidemiological point of view since the main vehicle of microorganism diffusion and exposure to humans is the aerosol produced by some devices, such as condensers, showers, faucets, humidifiers, whirlpool baths, and medication nebulizer devices (Hines et al., 2014; Mercante and Winchell, 2015; Carlson et al., 2020).
Legionella have been found in drinking water distribution systems, cooling towers, and water supply systems, where the water temperature is higher than the environment temperature, sustaining its growth (Diederen, 2008). In fact, Legionella grow and replicate between temperatures of 25 and 45°C, with optimal growth between 32 and 42°C (Arvand et al., 2011). In addition, the presence of biofilm in water distribution systems increases the risk of infection, due to the ability of Legionella spp. to obtain a high level of nutrients inside biofilm and protection from environmental stresses (Abdel-Nour et al., 2013; Di Pippo et al., 2018). The ability of Legionella to replicate inside protozoa cells allows bacteria to then also infect human cells, such as alveolar macrophages, causing the disease in humans known as Legionellosis—an aggressive form of pneumonia—and the Pontiac fever, a febrile and generally benign non-pulmonary disease form (Newton et al., 2010; Mercante and Winchell, 2015; Cunha et al., 2016).
Legionella pneumophila (L. pneumophila) is the most common and the most studied Legionella pathogenic species in humans. This species is divided into 16 serogroups, and the majority of infection cases, confirmed by the use of specific diagnostic criteria (i.e., positive urinary antigen, antibody title movement, and isolation of strain), are attributable to the L. pneumophila serogroup 1 (SG1), as confirmed by epidemiological data (Fields et al., 2002; Yu et al., 2002; Beauté, 2017; European Centre for Disease Prevention and Control, 2019; Rota et al., 2019).
However, in addition to L. pneumophila, there are more than 60 known species (LPSN Bacterio.net, 2018), some of which are associated with human diseases. Therefore, the risk of infection related to Legionella non-pneumophila species (Muder and Yu, 2002; Potts et al., 2013; Amemura-maekawa et al., 2018), represents a serious problem in clinical settings (e.g., healthcare facilities, and hospitals), in addition to water for human consumption (Muder and Yu, 2002; World Health Organization, 2007; Decker and Palmore, 2014) and it should not be underestimated.
Among them, Legionella anisa (L. anisa) is frequently isolated together with L. pneumophila in the hospital plumbing systems and can be used as a probable indicator of epidemic risk, beyond being associated with some cases of human infections (Fallon and Stack, 1990; Fenstersheib et al., 1990; Van Der Mee-Marquet et al., 2006; Vaccaro et al., 2016). Moreover, cases of endocarditis due to L. anisa and co-infection in HIV-associated pneumonia have been reported (Compain et al., 2015; Head et al., 2019).
In Australia and New Zealand, L. longbeachae isolated from potting soil mixes, represents the main source of human infection with 30.4% of community-acquired Legionellosis (Steele et al., 1990; Yu et al., 2002; Whiley and Bentham, 2011).
Due to its abundance in the environment and the level of pathogenicity, different techniques are applied for the detection and identification of Legionella spp., each of which present both advantages and disadvantages.
According to the Italian Guidelines for the prevention and control of Legionellosis (Italian Health Ministry, 2015), the culture technique represents the “gold standard” method for the isolation of Legionella strains, as well as for the typing, which is routinely performed by serological and molecular techniques such as the agglutination test, the direct fluorescent antibody (DFA) test, indirect immunofluorescent assay (IFA), sequence-based typing (SBT), and amplification of the macrophage infectivity potentiator (mip) gene (Luck, 2002).
In detail during environmental investigations, the isolates from water samples using the culture technique are mainly identified by biochemical and serological tests such as the agglutination test. The agglutination test is routinely used to type Legionella isolates, but it is also associated with negative or ambiguous results, leading to inaccurate evaluations of Legionella (Orsini et al., 2011). Nevertheless, only an identification at the genus level can be achieved, and in most cases, the quality of the results depends on the experience of the laboratory staff.
Since standard methods are lab-intensive, time-consuming, and prone to delivering false-negative results, several molecular techniques have been developed to detect Legionella (Templeton et al., 2003; Blyth et al., 2009).
TheEuropean Working Group for Legionella Infection (EWGLI) developed the SBT approach for clinical and environmental L. pneumophila strain typing, currently, this tecnique is recognized as the gold standard for the genotyping of L. pneumophila strains (Gaia et al., 2005; Ratzow et al., 2007).
Regarding the typing of Legionella non-pneumophila species, Ratcliff et al. (1998) presented a classification scheme for the Legionella genus based on the mip gene, encoding for a membrane protein referred to as the “macrophage infectivity potentiator,” that for its genetic stability and specificity is the most reliable and recommended method for identification at the species level (Ratcliff et al., 1997; Fry et al., 2007; Haroon et al., 2012).
Although sensitive and specific, molecular methods are expensive and require specialized laboratories and well-trained staff. Moreover, molecular techniques are affected by their inability to quantify the real risk to humans, as they do not allow for discrimination between dead and alive bacteria, and this leads to a lack of correlation between genomic units and bacterial loads [expressed in colony formant units (CFUs)]. Therefore, the culture-based approach is considered as a reference method in Legionella environmental surveillance (Lee et al., 2011; Italian Health Ministry, 2015), although the need to support the culture technique with rapid and cost-effective methods to improve diagnosis and the adoption of the appropriate antibiotic treatment.
In recent years, matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI–TOF MS) has emerged as an innovative, rapid, and inexpensive technique for species-level microbial identification through the analysis of ribosomal protein patterns. This technique has improved the routine practice of clinical microbiology laboratories, replacing most traditional biochemical or molecular techniques (Maier et al., 2006; Croxatto et al., 2012; Singhal et al., 2015).
Although the use of MALDI–TOF MS is widespread across the world, few data have been published regarding its application for the identification of Legionella spp. in clinical and environmental samples (Moliner et al., 2010; Fujinami et al., 2011; Gaia et al., 2011; He et al., 2011; Svarrer and Uldum, 2012; Dilger et al., 2016; Trnková et al., 2018). Moreover, in Italy, to the best of our knowledge, MALDI–TOF MS is not widely used in the clinical or environmental laboratories where Legionella surveillance is carried out.
The aim of this study is to evaluate the MALDI Biotyper system for the identification of Legionella isolates in the environment. We focused our attention on Legionella non-pneumophila species isolated from different facilities (i.e., hospitals, healthcare settings, companies, and community areas). Moreover, some of these environments (e.g., homes, companies, and hotels) are supplied by water distribution systems that do not have a water safety plan that includes Legionella control measures (i.e., temperature controls, disinfection treatment, and maintenance water system programs), representing a risk of subsequent Legionella infections.
The MALDI Biotyper system was compared to other identification methods, such as the agglutination test and mip-gene sequencing, to evaluate its performance and its potential use in clinical and environmental Legionella surveillance.
Materials and Methods
The isolates evaluated in this study were collected during the Legionella environmental surveillance programs of several municipal water distribution systems, supplying different environments usually associated with risk of Legionella infections such as hospitals, healthcare facilities, companies, and community areas (e.g., spas, private apartments, and hotels). The dataset used in the study is shown in Table 1.
TABLE 1.
Distribution of isolates between man-made environments.
| Number of isolates (n = 202) | Man-made environments |
| 121 | Hospitals |
| 35 | Private apartments |
| 24 | Hotels |
| 16 | Companies |
| 3 | Fitness centers |
| 2 | Wellness centers |
| 1 | Bathhouses |
Legionella Culture and Isolate Selection
Water sampling was performed according to UNI EN International Standard Organization (ISO) 19458:2006 (EN ISO 19458:2006, 2006, 2006 Water quality—Sampling for microbiological analysis) and Italian guidelines (Italian Health Ministry, 2015). Two liters of a hot water sample were collected in sterile polytetrafluoroethylene (PTFE) bottles containing sodium thiosulphate solution (20 mg/L) and then stored at 4°C and processed within 24 h of collection.
The isolation of Legionella was performed by the culture technique according to ISO 11731:2017 (ISO 11731:2017, 2017 Water quality—Enumeration of Legionella). Briefly, 2 L of the sample was concentrated by filtration on a 0.22-μm polyethersulfone membrane (Sartorius, Bedford, MA, United States). Different aliquots (from 0.2 to 0.1 mL) of the untreated sample or the filtered, heated, and acid-treated sample were seeded on plates of the selective medium glycine–vancomycin–polymyxin B–cycloheximide (GVPC) (Thermo Fisher Diagnostic, Basingstoke, United Kingdom), and incubated at 35 ± 2°C with 2.5% CO2 for a maximum of 15 days. During the incubation period, the growth of Legionella was evaluated every 2 days, examining the plates for the presence of colonies with specific characteristics ascribable to Legionella spp.
Suspected colonies were sub-cultured on buffered charcoal yeast extract (BCYE) agar supplemented with L-cysteine (cys +) and without L-cysteine (cys-) supplementation (Thermo Fisher Diagnostics, Basingstoke, United Kingdom). Positive Legionella colonies grow on Legionella BCYE cys + agar, but fail to grow on Legionella BCYE cys- agar. Furthermore, to provide more evidence that the suspect colonies do not belong to the genus Legionella, they were isolated on a blood agar plate (Tryptone Soya Agar + 5% sheep blood) (Thermo Fisher Diagnostics, Basingstoke, United Kingdom): The colonies that grew on blood agar were considered as cysteine non-dependent and were reported as non-Legionella spp.
Serological and Biochemical Typing
A total of 202 isolates of Legionella detected in the hot water sample growth on BCYE cys +, without growth on blood agar and with or without a positive reaction to the latex agglutination test, were considered eligible for the study. All colonies grown on BCYE cys +, identified presumably as Legionella non-pneumophila species, underwent serological typing by the latex agglutination test (Legionella latex test kit, Thermo Fisher Diagnostic, Basingstoke, United Kingdom). The test allowed to identify L. pneumophila SG1, L. pneumophila SG 2–14, and Legionella non-pneumophila species. Regarding the Legionella non-pneumophila group, the pool of antibody provided by the manufacturer recognized only a few species most commonly associated with clinical cases, such as L. anisa, L. bozemanii 1 and 2, L. gormanii, L. longbeachae 1 and 2, L. dumoffii, and L. jordanis.
Three clinical isolates of L. pneumophila previously typed as SBT as sequence type 1 (ST1) were included as a positive control. Moreover, five isolates grown on GVPC medium, but which failed to grow on BCYE cys +, were inserted as the negative control. These five isolates were sub-cultured on tryptic soy agar (TSA) (Biolife, Milan, Italy) and the colonies were successively biochemically typed by the Remel RapID NF Plus system (Thermo Fisher Diagnostic) as Brevundimonas diminuita (n = 1), Acinetobacter junii (n = 2), and Flavobacterium lindanitolerans (n = 1), and by Rapid ANA II as Streptococcus sanguinis (n = 1), according to the manufacturer’s instructions. To confirm the results obtained, the isolates were also analyzed by the MALDI Biotyper system.
Identification by MALDI–TOF MS
All 202 isolates grown on BCYE cys + with positive, negative, or ambiguous results of the Legionella agglutination test were analyzed by the MALDI Biotyper system (Bruker Daltonik GmbH, Bremen, Germany). Spectra acquisition and processing were performed using the Microflex LT mass spectrometer (2,000–20,000 Da, linear positive mode) and the MALDI Biotyper Compass 4.1 software, whose library (version BDAL 7854) included the spectra of 39 Legionella strains, as shown in Table 2.
TABLE 2.
Number of Legionella strains spectra included in matrix-assisted laser desorption ionization (MALDI) Biotyper software library.
| L. anisa (n = 9) | L. feeleii (n = 8) | L. longbeachae (n = 9) | L. rubrilucens (n = 1) |
| L. beliardensis (n = 1) | L. geestiana (n = 1) | L. maceachernii (n = 3) | L. sainthelensi (n = 3) |
| L. birminghamensis (n = 6) | L. gormanii (n = 2) | L. micdadei (n = 5) | L. santicrucis (n = 3) |
| L. bozemanii (n = 10) | L. gratiana (n = 5) | L. moravica (n = 1) | Legionella sp. (n = 1) |
| L. brunensis (n = 2) | L. hackeliae (n = 1) | L. oakridgensis (n = 2) | L. tucsonensis (n = 2) |
| L. cherrii (n = 4) | L. impletisoli (n = 1) | L. parisiensis (n = 1) | L. wadsworthii (n = 1) |
| L. cincinnatiensis (n = 2) | L. israelensis (n = 1) | L. pneumophila (n = 3) | L. waltersii (n = 1) |
| L. dresdenensis (n = 1) | L. jamestowniensis (n = 1) | L. pneumophila ssp fraseri (n = 4) | L. worsleiensis (n = 1) |
| L. dumoffii (n = 7) | L. jordanis (n = 2) | L. pneumophila ssp pascullei (n = 2) | L. yabuuchiae (n = 1) |
| L. erythra (n = 1) | L. lansingensis (n = 1) | L. pneumophila ssp pneumophila (n = 6) |
During the first part of the experiments a comparison between direct smear and full extraction methods was performed for a subset of strains (20/202, 10%), following manufacturer’s instructions. No significant differences were found in term of identification and quality of spectra. Therefore, considering that Legionella is a Gram-negative bacterium, and the full extraction method is recommended for species with thick cell wall, such as Gram-positives, Actinomycetales, and fungi, the direct smear method was applied on the remain Legionella isolates.
Briefly, a small amount of bacterial biomass, picked from a fresh plate culture (24–48 h of incubation) was spotted in duplicate onto a MALDI Biotyper target, overlaid with 1 μl of MALDI Biotyper matrix solution, and allowed to air dry before the measurement. The identification result was considered reliable when the log (score) was ≥ 2.0 (“high confidence level”) or between 1.7 and 1.99 (“low confidence level”). In addition, in case of log (score) results between 1.4 and 1.59, identification was considered reliable when the first four proposed results were identical (Christner et al., 2010; Schubert et al., 2011). Identification was considered reliable also in those cases in which the difference in the log (score) values between the first- and second-best matches was ≥ 0.3 (Martiny et al., 2012). In contrast, the presence of score of 0.00–1.69 indicated a non-reliable identification.
Determination of the sensitivity, specificity, and confidence interval (CI) at the 95% level of significance was performed using GraphPad Prism software version 8.0.1 for Windows (GraphPad Software, San Diego, California, United States).
Identification of Legionella spp. by mip-Gene Sequencing
The DNA extraction was carried out using the InstaGene Purification Matrix (Bio-Rad, Hercules, CA), and DNA concentrations were determined using the Qubit fluorometer (Thermo Fisher Scientific, Paisley, United Kingdom). The PCR for all Legionella non-pneumophila species isolates was performed using the protocols for the mip gene suggested by EWGLI, as described by Ratcliff et al. (1998). The mip-gene amplification was carried out using degenerate primers and modified by M13 tailing to avoid noise in the DNA sequence (Mentasti et al., 2012). The mip-gene amplification was carried out in a 50-μL reaction containing DreamTaq Green PCR Master Mix 2 × (Thermo Fisher Diagnostic Basingstoke, United Kingdom) and 40 picomoles of each primer; 100 ng of the DNA extracted from the presumptive colonies was added as template.
Specifically, the mip amplicons were sequenced using tailed M13 forward and reverse primers (mip-595R-M13R caggaaacagctatgaccCATATGCAAGACCTGAGGGAAC and mip-74F-M13F tgtaaaacgacggccagtGCTGCAACCGATGCCAC) to obtain a complete coverage of the sequenced region of interest (Mentasti et al., 2012). Amplification was performed in a thermocycler under the following conditions: Pre-denaturation for 3 min at 96°C, then 35 cycles consisting of 1 min at 94°C for denaturation, 2 min at 58°C for annealing, and 2 min at 72°C for extension, followed by a final extension at 72°C for 5 min. The reaction mixtures were then held at 4°C.
The PCR products were visualized by electrophoresis on 2% agarose gel and stained with ethidium bromide. Following purification, DNA was sequenced using BigDye Chemistry and analyzed on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA).
Raw sequencing data were assembled using CLC Main Workbench 7.6.4 software. The sequences were compared to sequences deposited in the Legionella mip-gene sequence database using a similarity analysis tool. EWGLI have established an accessible web database1 that contains sequence data from described species and allows the identification of Legionella non-pneumophila species. The species-level identification as done on the basis of ≥ 98% similarity to a sequence in the database (Fry et al., 2007). For one strain with a sequence generically identified in the mip-gene sequence database as Legionella species (L. spp.), the sequence was also queried in the GenBank database using the basic local alignment search tool (BLAST), and it showed the best match with Legionella species H (L. species H), (Ratcliff et al., 1998). The 205 nucleotide mip sequences generated for this study were submitted to GenBank. The provided accession numbers were as follows: MW021138, MW052863-MW053066.
Phylogenetic and Allelic Diversity Analysis
To estimate the relationship among the Legionella species found, a multiple sequence alignment (MSA) and a phylogenetic tree were performed on the 202 mip-gene sequences and the three positive controls. For each taxon, identified as previously described, the reference mip sequence of the correspondent American Type Culture Collection (ATCC) and Institute of Medical and Veterinary Science (IMVS) strain was retrieved and added to the analysis. When required, a manual editing was performed on the sequences, trimming them to the same length as the reference sequence. The nucleotide sequences were aligned by the multiple sequence comparison by log-expectation (MUSCLE) program (Edgar, 2004), executed in Geneious Prime 2020.1.22, retaining the default settings. The phylogenetic tree was built by the Geneious Tree Builder, using Tamura–Nei (Tamura and Nei, 1993) as a genetic distant model and neighbor-joining (Saitou and Nei, 1987) as a tree building method, and then bootstrapped using 100 replicates.
Results
Legionella Identification by the Agglutination Test
Overall, 34/202 (16.8%) isolates resulted positive and 158/202 (78.2%) negative in the agglutination test, while 10/202 (5.0%) isolates provided an ambiguous result. All of the strains that were included as controls delivered the expected results. Examples of the agglutination results are shown in Figure 1.
FIGURE 1.
Positive (A), negative (B), and ambiguous (C) Legionella non-pneumophila species results of the agglutination test.
MALDI Biotyper System Results
Applying the cut-offs recommended by the manufacturer, the MALDI Biotyper system identified at the genus level 90/202 (44.5%) isolates. Among them, 59/90 (65.5%) were identified at the species level with a high confidence level (n = 50 L. anisa; n = 1 L. feeleii; and n = 8 L. rubrilucens), and 31/90 (34.4%) were identified at the genus level with a low confidence level (L. anisa, L. feeleii, L. jordanis, and L. rubrilucens; Table 3); 112/202 isolates (56.0%) remain without identification. The application of the additional interpretation criteria for identification results with a log (score) ≤ 1.69 enabled a further 40 strains to be identified from 112 un-identified isolates, achieving a total of 130/202 (64.4%) successful results. In contrast, 72 isolates remain unidentified. The summary of the MALDI Biotyper system results is shown in Table 3.
TABLE 3.
Isolates identification by MALDI Biotyper system according to the manufacturer’s threshold and the revised sub-criteria.
| Identification | Isolates | Manufacturer thresholds and number (n) of isolates | Total manufacturer identification (n) | * Sub-Criteriafor score ≤ 1.69 | Total(n) | ||
| 2.00–3.00 | 1.70–1.99 | 0.00–1.69 * | |||||
| Legionella | L. anisa | 50 | 2 | 52 | 52 | ||
| L. feeleii | 1 | 8 | 9 | 2* | 11 | ||
| L. jordanis | 1 | 1 | 1 | ||||
| L. rubrilucens | 8 | 20 | 28 | 38* | 66 | ||
| Total | 59 | 31 | 90 | 40* | 130 | ||
| Not Identified* | 112 * | 112 | 72 | 72 | |||
| Total | 202 | 202 | |||||
The isolates used as controls were identified with a high confidence level as L. pneumophila and other bacteria (i.e., Acinetobacter junii, Brevundimonas dimunita, Flavobacterium lindanitolerans, and Streptococcus sanguinis).
Comparison Between the Agglutination Test and MALDI Biotyper System Results
The 34 isolates with positive agglutination results were identified by the MALDI Biotyper system as L. anisa. The main isolates involved in the study (n = 158) displayed negative agglutination results, of which the MALDI Biotyper identified 89/158 (56.3%) of the isolates as Legionella genus, while the other 69/158 (43.7%) were not identified.
Regarding the isolates with ambiguous agglutination results (n = 10), the MALDI Biotyper system correctly identified Legionella in seven cases (n = 4 L anisa with a high score, and n = 3 L. rubrilucens with a revised low score); in contrast, 3/10 isolates were not identified.
Full concordance between the two techniques was found for the control strains. However the three positive controls identified by the agglutination reaction as L. pneumophila SG1, 3, and 6 were not distinguished at the serogroup level by the MALDI Biotyper system.
mip-Gene Sequencing
The results obtained by mip-gene sequencing with the respective range of matches to the reference strains (i.e., ATCC, IMVS-911, and IMVS-3376) are shown in Table 4.
TABLE 4.
Results of the mip-gene sequencing and the number of isolates identified.
| mip-gene sequencing results | Isolates identified n (%) | % of match with reference strains |
| L. anisa | 50 (24.8%) | 100% |
| 2 (1.0%) | 96.7% | |
| L. feeleii | 1 (0.5%) | 99.4% |
| 1 (0.5%) | 98.4% | |
| 9 (4.5%) | 98.2% | |
| L. jordanis | 1 (0.5%) | 100% |
| L. londiniensis | 7 (3.5%) | 100% |
| L. nautarum | 15 (7.4%) | 100% |
| L. quateirensis | 4 (1.9%) | 98.2% |
| L. quinvilanii | 1 (0.5%) | 96.2% |
| L. rubrilucens | 30 (14.9%) | 100% |
| L. species H | 1 (0.5%) | 100% |
| L. steelei | 1 (0.5%) | 99.8% |
| L. taurinensis | 79 (39.1%) | 100% |
| TOTAL | 202 (100%) |
The most common species detected by the sequencing analysis were L. taurinensis (39.1%), followed by L. anisa and L. rubrilucens (Table 4). Among the L. anisa subset, two isolates, identical to each other, were found to match the reference sequence strain at 96.7%, showing 20 mismatches with the main L. anisa group.
Only one isolates generically identified as L. spp. In the mip-gene database, were matched in GenBank and identified as L. species H (Ratcliff et al., 1998). The positive control was confirmed to belong to L. pneumophila; in contrast, the negative controls were not included in the mip-gene sequences analysis.
Comparison Between the MALDI Biotyper System and mip-Gene Sequencing Results
Among the species included in the MALDI Biotyper system database, the concordance between this system and the sequencing was 91/94 (96.9%), as follows: L. anisa (n = 52/52), L. rubrilucens (n = 27/30), L. feeleii (n = 11/11), and L. jordanis (n = 1/1). Therefore, among the species included in the MALDI Biotyper system database, the overall sensitivity of this system was 96.8% (95% CI: 0.91–0.99).
In the remaining 108 isolates identified by mip-gene sequencing as L. londiniensis, L. nautarum, L. quateirensis, L. quinlivanii, L. species H, L. steelei, and L. taurinensis, whose spectra are not included in the MALDI Biotyper system database, the MALDI Biotyper system did not identify 69 isolates and led to a misidentification in 39 cases (L. rubrilucens instead of L. taurinensis). Therefore, the specificity of the MALDI Biotyper system was 69/108 = 63.9% (95% CI: 0.55–0.72).
A comparison between results delivered by the three techniques evaluated in this study is shown in Table 5.
TABLE 5.
Comparison of the results of the three techniques.
| Mip-gene sequencingresults | Latex agglutination test results | MALDI Biotyper system results | ||||
| Isolate identification (n) | positive (+), negative (–), and ambiguous (±) results (n) | |||||
| + | – | ± | 2.00–3.00(n) | 1.70–1.99(n) | 0.00–1.69 *(n) | |
| L. anisa (52) | 34 | 14 | 4 | L. anisa(50) | L. anisa(2) | |
| L. feeleii (11) | 11 | L. feeleii(1) | L. feeleii(8) | L. feeleii(2) | ||
| L. jordanis (1) | 1 | L. jordanis(1) | ||||
| L. londiniensis (7) | 5 | 2 | Not Identified (7) | |||
| L. nautarum (15) | 14 | 1 | Not Identified (15) | |||
| L. quateirensis (4) | 4 | Not Identified (4) | ||||
| L. quinvilanii (1) | 1 | Not Identified (1) | ||||
| L. rubrilucens (30) | 30 | L. rubrilucens(8) | L. rubrilucens(17) |
L. rubrilucens(2) Not Identified (3) |
||
| L. species H (1) | 1 | Not Identified (1) | ||||
| L. steelei (1) | 1 | Not Identified (1) | ||||
| L. taurinensis (79) | 76 | 3 | L. rubrilucens (3) |
L. rubrilucens (36) Not Identified (40) |
||
* Manufacturer’s criteria plus sub-criteria (Christner et al., 2010; Schubert et al., 2011; Martiny et al., 2012). In bold type the concordance between techniques.
Phylogenetic Analysis
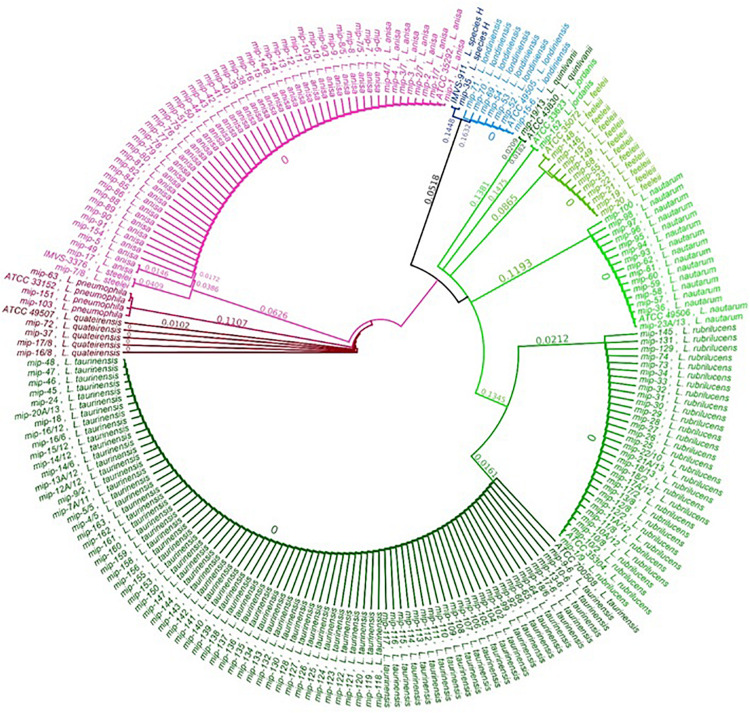
The relationship between the 202 Legionella isolates, positive controls, and reference strains was studied by a phylogenetic tree of the sequenced mip gene in order to display a graphical representation of the inter- and intra-specific genetic variability among the taxa previously identified (Figure 2). The phylogenetic tree shows that the clusters corresponding to the different Legionella species are clearly separate and distinct for each taxon, with every identified sequence falling on the same branch of the correspondent ATCC/IMVS reference mip gene. Therefore, our dataset can be separated into three major clades, in concordance with previous studies (Ratcliff et al., 1998; Burstein et al., 2016): One clade that includes L. quinlivanii, L. jordanis, L. feeleii, L. nautarum, L. rubrilucens, and L. taurinensis; one clade represented by L. anisa, L. steelei, L. pneumophila (positive control), and L. quateirensis; and one that includes L. londiniensis and the so-called L. species H. Interestingly, the tree shows how L. rubrilucens and L. taurinensis form a monophyletic group, revealing a high genetic similarity (96.3%).
FIGURE 2.
Phylogenetic tree obtained by the mip sequence analyses of isolates and strain types.
Concerning the intra-species analysis, the tree reveals the presence of a subclade inside the L. anisa principal clade, formed by two isolates sequences (i.e., mip-17 and mip-49), which share only 96.7% pairwise identity with the L. anisa reference mip sequence (i.e., ATCC 35392). The mip gene of the L. quinlivanii isolate shares only 96.2% sequence similarity with its correspondent reference strain (i.e., ATCC 43830).
Discussion
The role of Legionella in human respiratory infections emphasizes the importance of its surveillance in conventional and unconventional artificial environments that could represent a reservoir of infections (Szewzyk et al., 2000).
One of the main concerns during environmental and clinical Legionella surveillance is the need of a rapid and sensitive technique that could improve therapeutic and epidemiological choices.
First, it could allow the timely adoption of an appropriate antibiotic treatment, as well as the rapid identification of the source of infection by comparison between strains isolated from environmental and clinical samples, thus enabling the adoption of the correct prevention measures to control the infection. The long incubation time, the poor sensitivity of serological agglutination methods, and the long and laborious molecular techniques have long been the object of scientific discussion, suggesting the need to enhance Legionella identification with faster, cheaper, and more sensitive methods. MALDI–TOF MS is nowadays the universal method for microbial identification at the species level in routine microbial identification laboratories. In this study, a comparison between three different techniques (i.e., the agglutination test, mip-gene sequencing, and MALDI–TOF MS) was performed in order to assess whether MALDI–TOF MS technology could be a useful and valid tool to identify environmental Legionella species strains, reducing analytical times and costs.
The strains included in this study were collected from different artificial water reservoirs, chosen considering the presence of a routine surveillance program (e.g., hospitals and healthcare facilities), and environments where a water safety plan is not required or is missed (e.g., homes, companies, and recreational communities). Starting from the cultural and diagnostic techniques commonly used on L. pneumophila, we evaluated the possibility of carrying out species-level identification using MALDI–TOF MS, including Legionella non-pneumophila isolates. Although some species are linked to the epidemiology of Legionella diseases, a standardized diagnostic approach for environmental and clinical samples has not yet been defined. These species are abundant in the environment, but are clinically less associated with human cases due to a lack of culture, a low growth rate, and the poor sensitivity of diagnostic techniques (Fields et al., 2002; Svarrer et al., 2012; Mercante and Winchell, 2015).
All of these limits with regard to Legionella non-pneumophila species research lead to a misidentification of strains and to an underestimation of the real risk represented by environments in which they can survive and proliferate.
In our study, we reproduced routine laboratory workflows for Legionella surveillance, i.e., sampling, standardized culture, and biochemical–serological testing to elaborate an analytical report. Our culture results showed that most of the isolates grew after more than 10 days of incubation. Despite the reference technical guidelines provided by standard institutions, which suggest an incubation time of a minimum of 10 days, in routine laboratory practice, the time of incubation is typically no longer than 10 days, with a loss of isolate growth after this time (Italian Health Ministry, 2015; ISO 11731:2017 Water quality—Enumeration of Legionella, 2017).
The growth of colonies on BCYE cys + is the only discriminating element for routine investigations: Isolates with positive growth are subjected to the latex agglutination test and, when agglutination fails, the results are reported in terms of counts of colonies (CFU/L) followed by a generic identification of “Legionella non-pneumophila species detection.” The misidentification of isolates leads to an underestimation of circulating species in the environment and, consequently, to inappropriate preventive measures to contain the risk.
In the current research, the agglutination test returned positive results for only 34/202 (16.8%) isolates, failing to detect most of the Legionella non-pneumophila species isolates, i.e., 158/202 (78.2%). This result is related to limited equipment that does not recognize all of the Legionella non-pneumophila species; therefore, this assay is not suitable for discrimination within the Legionella genus, according to Orsini et al. (2011), and highlights the need to establish, in routine analysis, a further technique able to accurately identify these Legionella species.
The MALDI Biotyper system, following the manufacturer’s criteria, overall identified 90/202 (44.5%) Legionella isolates at the species level, corresponding to all of the species for which reference spectra are present in the database. A further significant number of isolates (n = 40) could have been identified by applying only the additional criteria described by Christner et al. (2010); Schubert et al. (2011), and Martiny et al. (2012).
The failed identification of the remaining 72 isolates is easily explained by the absence of the species in the MALDI Biotyper database (Moliner et al., 2010; Fujinami et al., 2011; Dilger et al., 2016).
The version of manufacturer’s database is closed and can be improved only by the manufacturer and it is covered by intellectual property rights. Therefore, the Legionella spectra identification produced in the study, could be represent our first “in-house database” for developing Legionella MALDI Biotyper application. These isolates were identified successively only by mip-gene sequencing as L. taurinensis, L. londiniensis, L. nautarum, L. species H, L. quateirensis, L. quinlivanii, and L. steleei.
Interesting results were observed among L. tauriniensis, as 39/79 isolates were misidentified (with a low confidence level) as L. rubrilucens, likely due to the close relatedness of these two species (Lo Presti et al., 1997; Baladron et al., 2006). In the phylogenetic tree, the two species are part of two distinct clades, although the differences between them only account for 3.7% (24 mismatches), regrouping them in the same monophyletic group. All L.tauriniensis isolates showed negative or ambiguous agglutination results and their identification was achieved only via the genotyping approach. The misclassification of L. taurinensis with other Legionella non-pneumophila species (i.e., L. rubrilucens, L. erythra, and L. spiritiensis) occurred already in the first characterization of the species, performed on the 16srRNA gene, that led to considering them as belonging to the same species (Lo Presti et al., 1997; Baladron et al., 2006). The subsequent characterization by the mip gene was able to distinguish between four different species. The MALDI–TOF MS approach, which establishes bacteria identification based on proteomic profiles, is likely unable to detect low differences at the genomic level, and thus returned no results. Different studies in the fields of bacteria identification suggest to combine MALDI Biotyper system analysis with Fourier-transfrom infrared spectroscopy (FTIR), which can be used to analyze carbohydrate and glycoproteins compounds, is able to provide a unique “fingerprint” spectrum for each species of bacteria exceeding the limit previously described (Maity et al., 2013; Feng et al., 2020). The three positive controls, belonging to different serogroups (i.e., 1, 3, and 6), were identified only at the species level without serogroup identification, confirming that the MALDI Biotyper system is unable to discriminate between L. pneumophila among serogroups.
A concordance between the three methodologies included in this study was observed only for L. anisa. Interestingly, only two out of the 52 L. anisa isolates (i.e., mip-49 and mip-17) identified by the MALDI Biotyper system with a low confidence level corresponded to the sub-clade exhibiting a difference of 3.3% (20 mismatches) to the principal clade. We could speculate that these genetic differences can cause differences at the ribosomal protein level that could be underrepresented in the MALDI Biotyper system database, which includes nine L. anisa reference spectra. However, the differences found for the two sub-clades of L. anisa need further investigation.
The comparison of the mip-gene sequencing of one isolate of L. quinlivanii showed a similarity of 95.91% to the reference strain ATCC 43830, with only 3.8% of differences linked to 23 mismatches.
Considering the criteria adopted by Ratcliff to develop the mip-gene database (Ratcliff et al., 1998), we found some discrepancies that could be improved by further investigation.
Finally, we found a different isolate from all of the characterized species, matched to the L. species H described by Ratcliff et al. (1998), close to the L. londiniensis clade, with differences of 34.7%. These findings will be improved by the whole-genome sequencing (WGS) approach in order to obtain more information regarding the isolates’ characteristics and their evolutionary adaptations in the environment.
These findings show that the MALDI Biotyper system is a powerful tool for the identification of Legionella non-pneumophila species other than L. pneumophila, as demonstrated by its high sensitivity (96.8%) for the species included in the database and its low specifity (63.9%) that could be improved by amplifying the database with species missing or less represented (e.g., only one spectrum for L. rubrilucens). The limit of this study is linked to the need to improve the manufacturer’s database and re-evaluation of the identification score criteria that could support the introduction of MALDI-TOF MS into routine clinical and environmental Legionella surveillance. The rievaluation of our “in-house database” with the new one from manufacturer represent the areas for future research.
Moreover, the approach used in this study could develop the knowledge regarding the relationship between strains in water distribution systems, in addition to supporting the rapid identification of the source of infection, the match to clinical strains, and the adoption of corrective actions to limit the spread of bacteria and to control nosocomial and community infection.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found below: NCBI GenBank (accession numbers: MW021138 and MW052863–MW053066).
Author Contributions
SC and MP conceived and designed the experiments and wrote the manuscript. MM and LG performed the samples collection and the experiments. SS performed the phylogenetic analysis. AG and MLS performed mip-gene sequencing. MC, PDM, and FB performed MALDI-TOF MS analysis and data interpretation. MBS and MV supply communities Legionella isolates. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Graziella Ciani and Federica Marino for their support with the microbiological analyses.
Funding. This work was partially supported by the University of Bologna, Italy.
References
- Abdel-Nour M., Duncan C., Low D. E., Guyard C. (2013). Biofilms: the stronghold of Legionella pneumophila. Int. J. Mol. Sci. 14 21660–21675. 10.3390/ijms141121660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemura-maekawa J., Kura F., Chida K., Ohya H., Kanatani J., Isobe J., et al. (2018). Legionella pneumophila and other legionella species isolated from legionellosis patients in Japan between 2008 and 2016. Appl. Environ. Microbiol. 84 1–9. 10.1007/978-0-387-70896-6_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvand M., Jungkind K., Hack A. (2011). Contamination of the cold water distribution system of health care facilities by Legionella pneumophila: do we know the true dimension? Eurosurveillance 16 1–6. 10.2807/ese.16.16.19844-en [DOI] [PubMed] [Google Scholar]
- Baladron B., Gil V., Pelaz C. (2006). “Serological versus sequence-based methods for Legionella identification,” in Legionella: State of the Art 30 Years After Its Recognition, eds Cianciotto S. M. S., Kwaik Y. A., Edelstein P. H., Fields B. S., Geary D. F., Harrison T. G., et al. (Washington DC: American Society for Microbiology; ), 58–62. 10.1128/9781555815660.ch16 [DOI] [Google Scholar]
- Beauté J. (2017). Legionnaires’ disease in Europe, 2011 to 2015. Eurosurveillance 22 1–8. 10.2807/1560-7917.ES.2017.22.27.30566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyth C. C., Adams D. N., Chen S. C. A. (2009). Diagnostic and typing methods for investigating Legionella infection. N. S. W. Public Health Bull. 20 157–161. 10.1071/nb08062 [DOI] [PubMed] [Google Scholar]
- Burstein D., Amaro F., Zusman T., Lifshitz Z., Cohen O., Gilbert J. A., et al. (2016). Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat. Genet. 48, 167–175. 10.1038/ng.3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson K. M., Boczek L. A., Chae S., Ryu H. (2020). Legionellosis and recent advances in technologies for Legionella control in premise plumbing systems: a review. Water 12 1–22. 10.3390/w12030676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christner M., Rohde H., Wolters M., Sobottka I., Wegscheider K., Aepfelbacher M. (2010). Rapid identification of bacteria from positive blood culture bottles by use of matrix-assisted laser desorption-ionization time of flight mass spectrometry fingerprinting. J. Clin. Microbiol. 48 1584–1591. 10.1128/jcm.01831-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compain F., Bruneval P., Jarraud S., Perrot S., Aubert S., Napoly V., et al. (2015). Chronic endocarditis due to Legionella anisa: a first case difficult to diagnose. New Microbes New Infect. 8 113–115. 10.1016/j.nmni.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxatto A., Prod’hom G., Greub G. (2012). Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev. 36 380–407. 10.1111/j.1574-6976.2011.00298.x [DOI] [PubMed] [Google Scholar]
- Cunha B. A., Burillo A., Bouza E. (2016). Legionnaires’ disease. Lancet 387 376–385. [DOI] [PubMed] [Google Scholar]
- Decker B. K., Palmore T. N. (2014). Hospital water and opportunities for infection prevention. Curr. Infect. Dis. Rep. 16 1–13. 10.1007/s11908-014-0432-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pippo F., Di Gregorio L., Congestri R., Tandoi V., Rossetti S. (2018). Biofilm growth and control in cooling water industrial systems. FEMS Microbiol. Ecol. 94:fiy044. 10.1093/femsec/fiy044/4935158 [DOI] [PubMed] [Google Scholar]
- Diederen B. M. W. (2008). Legionella spp. and Legionnaires’ disease. J. Infect. 56 1–12. 10.1016/j.jinf.2007.09.010 [DOI] [PubMed] [Google Scholar]
- Dilger T., Melzl H., Gessner A. (2016). Rapid and reliable identification of waterborne Legionella species by MALDI-TOF mass spectrometry. J. Microbiol. Methods 127 154–159. 10.1016/j.mimet.2016.05.028 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EN ISO 19458:2006 (2006). Water Quality – Sampling for Microbiological Analysis. Available online at: http://store.uni.com/catalogo/en-iso-19458-2006 (accessed May 10, 2020) [Google Scholar]
- European Centre for Disease Prevention and Control (2019). “Legionnaires’ disease,” in ECDC. Annual Epidemiological Report for 2017, (Solna Municipality: European Centre for Disease Prevention and Control; ). [Google Scholar]
- Fallon R. J., Stack B. H. R. (1990). Legionnaires’ disease due to Legionella anisa. J. Infect. 20 227–229. 10.1016/0163-4453(90)91144-3 [DOI] [PubMed] [Google Scholar]
- Feng B., Shi H., Xu F., Hu F., He J., Yang H., et al. (2020). FTIR-assisted MALDI-TOF MS for the identification and typing of bacteria. Anal. Chim. Acta 1111 75–82. 10.1016/j.aca.2020.03.037 [DOI] [PubMed] [Google Scholar]
- Fenstersheib M. D., Miller M., Diggins C., Liska S., Detwiler L., Werner S. B., et al. (1990). Outbreak of pontiac fever due to Legionella anisa. Lancet 336 35–37. 10.1016/0140-6736(90)91532-F [DOI] [PubMed] [Google Scholar]
- Fields B., Benson R., Besser R. E. (2002). Legionella and Legionnaires’ disease: 25 years of investigation. Clin. Microbiol. Rev. 15 506–526. 10.1128/CMR.15.3.506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B. S. (1996). The molecular ecology of legionellae. Trends Microbiol. 4 286–290. 10.1016/0966-842X(96)10041-X [DOI] [PubMed] [Google Scholar]
- Fry N. K., Afshar B., Bellamy W., Underwood A. P., Ratcliff R. M., Harrison T. G., et al. (2007). Identification of Legionella spp. by 19 European reference laboratories: results of the European working group for Legionella infections external quality assessment scheme using DNA sequencing of the macrophage infectivity potentiator gene a. Clin. Microbiol. Infect. 13 1119–1124. 10.1111/j.1469-0691.2007.01808.x [DOI] [PubMed] [Google Scholar]
- Fujinami Y., Kikkawa H. S., Kurosaki Y., Sakurada K., Yoshino M., Yasuda J. (2011). Rapid discrimination of Legionella by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Microbiol. Res. 166 77–86. 10.1016/j.micres.2010.02.005 [DOI] [PubMed] [Google Scholar]
- Gaia V., Casati S., Tonolla M. (2011). Rapid identification of Legionella spp. by MALDI-TOF MS based protein mass fingerprinting. Syst. Appl. Microbiol. 34 40–44. 10.1016/j.syapm.2010.11.007 [DOI] [PubMed] [Google Scholar]
- Gaia V., Fry N. K., Afshar B., Lück P., Meugnier H., Etienne J., et al. (2005). Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J. Clin. Microbiol. 43 2047–2052. 10.1128/JCM.39.11.4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon A., Koide M., Higa F., Tateyama M., Fujita J. (2012). Identification of Legionella pneumophila serogroups and other Legionella species by mip gene sequencing. J. Infect. Chemother. 18 276–281. 10.1007/s10156-011-0324-0 [DOI] [PubMed] [Google Scholar]
- He Y., Chang T. C., Li H., Shi G., Tang Y. W. (2011). Matrix-assisted laser desorption ionization timeof-flight mass spectrometry and database for identification of legionella species. Can. J. Microbiol. 57 533–538. 10.1139/w11-039 [DOI] [PubMed] [Google Scholar]
- Head B. M., Trajtman A., Bernard K., Burdz T., Vélez L., Herrera M., et al. (2019). Legionella co-infection in HIV-associated pneumonia. Diagn. Microbiol. Infect. Dis. 95 71–76. 10.1016/j.diagmicrobio.2019.03.005 [DOI] [PubMed] [Google Scholar]
- Hines S. A., Chappie D. J., Lordo R. A., Miller B. D., Janke R. J., Lindquist H. A., et al. (2014). Assessment of relative potential for Legionella species or surrogates inhalation exposure from common water uses. Water Res. 56 203–213. 10.1016/j.watres.2014.02.013 [DOI] [PubMed] [Google Scholar]
- ISO 11731:2017 (2017). Water Quality — Enumeration of Legionella. Available online at: https://www.iso.org/standard/61782.html (accessed May 12, 2020). [Google Scholar]
- Italian Health Ministry (2015). Guidelines for Prevention and Control of Legionellosis. Approvate in Conferenza Stato-Regioni Seduta del 7 maggio 2015. Rome: Italian Health Ministry. [Google Scholar]
- Lee J. V., Lai S., Exner M., Lenz J., Gaia V., Casati S., et al. (2011). An international trial of quantitative PCR for monitoring Legionella in artificial water systems. J. Appl. Microbiol. 110 1032–1044. 10.1111/j.1365-2672.2011.04957.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti F., Riffard S., Meugnier H., Reyrolle M., Lasne Y., Grirnontf P. A. D., et al. (1997). Legionella taurinensis sp. nov., a new species antigenically similar to Legionella spiritensis. Int. J. Syst. Bacteriol. 49 397–403. [DOI] [PubMed] [Google Scholar]
- LPSN Bacterio.net (2018). LPSN – List of Prokaryotic Names With Standing in Nomenclature Legionella. Available online at: https://www.bacterio.net/genus/legionella (accessed May 20, 2020). [Google Scholar]
- Luck P. C., Helbig J. H., Schuppler M. (2002). Epidemiology and laboratory diagnosis of Legionella infections. Infektiologie 26 174–182. 10.1515/LabMed.2002.023 [DOI] [Google Scholar]
- Maier T., Klepel S., Renner U., Kostrzewa M. (2006). Fast and reliable MALDI-TOF MS-based microorganism identification. Nat. Methods 3 i–ii. 10.1038/nmeth870 [DOI] [Google Scholar]
- Maity J. P., Kar S., Lin C. M., Chen C. Y., Chang Y. F., Jean J. S., et al. (2013). Identification and discrimination of bacteria using Fourier transform infrared spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 116 478–484. 10.1016/j.saa.2013.07.062 [DOI] [PubMed] [Google Scholar]
- Martiny D., Dediste A., Vandenberg O., Vandenberg O. (2012). Comparison of an in-house method and the commercial SepsityperTM kit for bacterial identification directly from positive blood culture broths by matrix-assisted laser desorption-ionisation time-of-flight mass spectrometry. Eur. J. Clin. Microbiol. Infect. Dis. 31 2269–2281. 10.1007/s10096-012-1566-1 [DOI] [PubMed] [Google Scholar]
- Mentasti M., Fry N. K., Afshar B., Palepou-Foxley C., Naik F. C., Harrison T. G. (2012). Application of Legionella pneumophila-specific quantitative real-time PCR combined with direct amplification and sequence-based typing in the diagnosis and epidemiological investigation of Legionnaires’ disease. Eur. J. Clin. Microbiol. Infect. Dis. 31 2017–2028. 10.1007/s10096-011-1535-0 [DOI] [PubMed] [Google Scholar]
- Mercante J. W., Winchell J. M. (2015). Current and emerging Legionella diagnostics for laboratory and outbreak investigations. Clin. Microbiol. Rev. 28 95–133. 10.1128/cmr.00029-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moliner C., Ginevra C., Jarraud S., Flaudrops C., Bedotto M., Couderc C., et al. (2010). Rapid identification of Legionella species by mass spectrometry. J. Med. Microbiol. 59 273–284. 10.1099/jmm.0.014100-0 [DOI] [PubMed] [Google Scholar]
- Muder R. R., Yu V. L. (2002). Infection due to Legionella species other than L. pneumophila. Clin. Infect. Dis. 35 990–998. 10.1086/342884 [DOI] [PubMed] [Google Scholar]
- Newton H. J., Ang D. K. Y., Van Driel I. R., Hartland E. L. (2010). Molecular pathogenesis of infections caused by Legionella pneumophila. Clin. Microbiol. Rev. 23 274–298. 10.1128/cmr.00052-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini M., Cristino S., Grottola A., Romano-Spica V. (2011). Bacteria misagglutination in Legionella surveillance programmes. J. Hosp. Infect. 79 179–180. 10.1016/j.jhin.2011.05.021 [DOI] [PubMed] [Google Scholar]
- Potts A., Donaghy M., Marley M., Othieno R., Stevenson J., Hyland J., et al. (2013). Cluster of Legionnaires’ disease cases caused by Legionella longbeachae serogroup 1, Scotland, August to September 2013. Eurosurveillance 18 1–5. 10.2807/1560-7917.ES2013.18.50.20656 [DOI] [PubMed] [Google Scholar]
- Ratcliff R. M., Donnellan S. C., Lanser J. A., Manning P. A., Heuzenroeder M. W. (1997). Interspecies sequence differences in the Mip protein from the genus Legionella: implications for function and evolutionary relatedness. Mol. Microbiol. 25 1149–1158. 10.1046/j.1365-2958.1997.5471908.x [DOI] [PubMed] [Google Scholar]
- Ratcliff R. M., Lanser J. A., Manning P. A., Heuzenroeder M. W. (1998). Sequence-based classification scheme for the genus Legionella targeting the mip gene. J. Clin. Microbiol. 36 1560–1567. 10.1128/jcm.36.6.1560-1567.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzow S., Gaia V., Helbig J. H., Fry N. K., Lück P. C. (2007). Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J. Clin. Microbiol. 45 1965–1968. 10.1128/jcm.00261-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota M. C., Caporali M. G., Bella A., Scaturro M., Giannitelli S., Ricci M. L. (2019). Annual report of Legionellosis in Italy – 2018. Not. dell’Iastituto Super. di Sanità 32. Rome, Italy: National legionella Laboratory, ISS. [Google Scholar]
- Rowbotham T. J. (1980). Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33 1179–1183. 10.1136/jcp.33.12.1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- Schubert S., Weinert K., Wagner C., Gunzl B., Wieser A., Maier T., et al. (2011). Novel, improved sample preparation for rapid, direct identification from positive blood cultures using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. J. Mol. Diagnostics 13 701–706. 10.1016/j.jmoldx.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N., Kumar M., Kanaujia P. K., Virdi J. S. (2015). MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front. Microbiol. 6:791. 10.3389/fmicb.2015.00791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele T. W., Lanser J., Sangster N. (1990). Isolation of Legionella longbeachae serogroup 1 from potting mixes. Appl. Environ. Microbiol. 56 49–53. 10.1128/aem.56.1.49-53.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svarrer C. W., Lück C., Elverdal P. L., Uldum S. A. (2012). Immunochromatic kits Xpect Legionella and BinaxNOW Legionella for detection of Legionella pneumophila urinary antigen have low sensitivities for the diagnosis of Legionnaires’ disease. J. Med. Microbiol. 61 213–217. 10.1099/jmm.0.035014-0 [DOI] [PubMed] [Google Scholar]
- Svarrer C. W., Uldum S. A. (2012). The occurrence of Legionella species other than Legionella pneumophila in clinical and environmental samples in Denmark identified by mip gene sequencing and matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 18 1004–1009. 10.1111/j.1469-0691.2011.03698.x [DOI] [PubMed] [Google Scholar]
- Szewzyk U., Szewyk R., Manz W., Schleifer K. H. (2000). Microbiological safety of drinking water. Annu. Rev. Microbiol. 54 81–127. 10.1201/9781315159126-6 [DOI] [PubMed] [Google Scholar]
- Tamura K., Nei M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10 512–526. 10.1093/oxfordjournals.molbev.a040023 [DOI] [PubMed] [Google Scholar]
- Templeton K. E., Scheltinga S. A., Sillekens P., Crielaard J. W., Van Dam A. P., Goossens H., et al. (2003). Development and clinical evaluation of an internally controlled, single-tube multiplex real-time PCR assay for detection of Legionella pneumophila and other Legionella species. J. Clin. Microbiol. 41 4016–4021. 10.1128/JCM.41.9.4016-4021.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trnková K., Kotrbancová M., Špaleková M., Fulová M., Boledovièová J., Vesteg M. (2018). MALDI-TOF MS analysis as a useful tool for an identification of Legionella pneumophila, a facultatively pathogenic bacterium interacting with free-living amoebae: a case study from water supply system of hospitals in Bratislava (Slovakia). Exp. Parasitol. 184 97–102. 10.1016/j.exppara.2017.12.002 [DOI] [PubMed] [Google Scholar]
- Vaccaro L., Izquierdo F., Magnet A., Hurtado C., Salinas M. A., Gomes T. S., et al. (2016). First case of legionnaire’s disease caused by Legionella anisa in Spain and the limitations on the diagnosis of Legionella non-pneumophila infections. PLoS One 11:e0159726. 10.1371/journal.pone.0159726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Mee-Marquet N., Domelier A. S., Arnault L., Bloc D., Laudat P., Hartemann P., et al. (2006). Legionella anisa, a possible indicator of water contamination by Legionella pneumophila. J. Clin. Microbiol. 44 56–59. 10.1128/JCM.44.1.56-59.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley H., Bentham R. (2011). Legionella longbeachae and legionellosis. Emerg. Infect. Dis. 17 579–583. 10.3201/eid1704.100446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2007). Legionella and the Prevention of Legionellosis. Geneva: WHO. [Google Scholar]
- Yu V. L., Plouffe F. J., Pastoris M. C., Stout J. E., Schousboe M., Widmer A., et al. (2002). Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired Legionellosis: an International collaborative survey. Infect. Dis. Clin. Pract. 11:258 10.1097/00019048-200205000-00039 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found below: NCBI GenBank (accession numbers: MW021138 and MW052863–MW053066).