Abstract
Gene specific recruitment of bromodomain-containing proteins to chromatin is affected by post-translational acetylation of lysine on histones. Whereas interactions of the bromodomain with acetylation patterns of native histones (H2A, H2B, H3, and H4) have been well characterized, the motif for recognition for histone variants H2A.Z I and H2A.Z II by bromodomains has yet to be fully investigated. Elucidating these molecular mechanisms is crucial for understanding transcriptional regulation in cellular processes involved in both development and disease. Here, we have used protein-observed fluorine NMR to fully characterize the affinities of H2A.Z I and II acetylation patterns for BPTF’s bromodomain and found the diacetylated mark of lysine 7 and 13 on H2A.Z II to have the strongest interaction with K7ac preferentially engaging the binding site. We further examined the selectivity of H2A.Z histones against a variety of bromodomains, revealing that the bromodomain of CECR2 binds with the highest affinity and specificity for acetylated H2A.Z I over isoform II. These results support a possible role for different H2A.Z transcriptional activation mechanisms that involve recruitment of chromatin remodeling complexes.
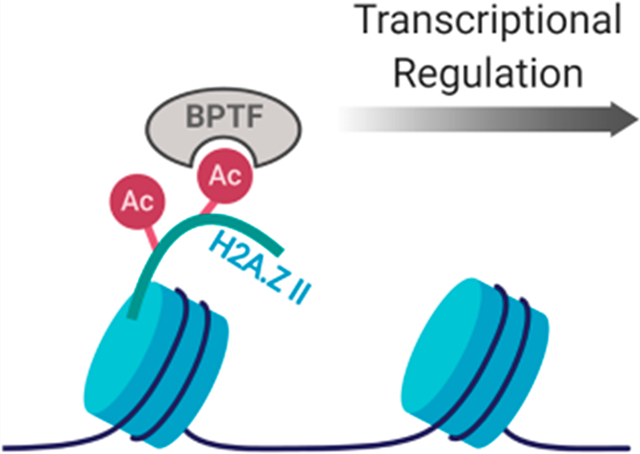
Graphical Abstract
Eukaryotic DNA is packaged within the nucleus of the cell in a condensed chromatin structure containing histone proteins within repeating nucleosomal subunits.1 Each nucleosome consists of ∼147 bp of DNA wrapped left-handedly around an octameric bundle of two copies of each conserved histone (H2A, H2B, H3, and H4).2 Various patterns and types of chemical modifications to the protruding N- and C-termini of histones directly affect the compaction of this chromatin assembly into euchromatic (transcriptionally active) and heterochromatic (transcriptionally repressive) states. This histone code plays a key role in the expression or silencing of genes not only by regulating genome accessibility but also by recruiting various transcriptional effector proteins in a gene specific manner.3
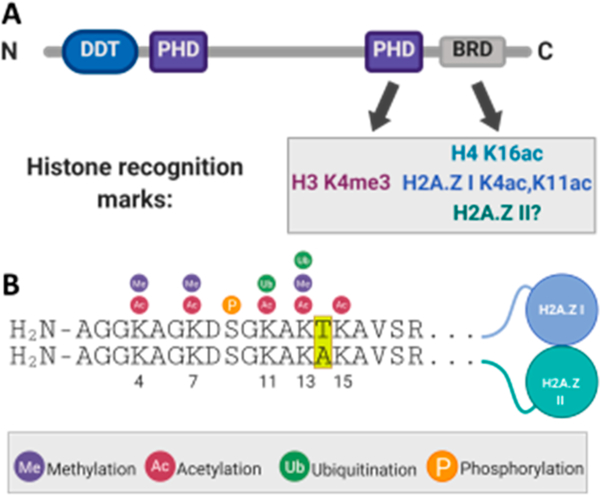
Bromodomains are structural motifs within epigenetic regulatory proteins that function through recognition of the N-ε-acetylation of lysine on histones, playing an integral role in the transcription of growth-promoting and cell cycle-regulating genes.4 Aberrant levels of bromodomain-containing proteins and dysregulation of their interactions have been implicated in many types of cancers,5–11 inflammation,12 and neurological diseases.13 Therefore, there is a need to map histone modification motifs that bromodomains recognize to better understand gene transcription during these cellular processes. One such bromodomain-containing protein is the bromodomain- and PHD finger-containing transcription factor (BPTF). BPTF is the largest protein subunit of the nucleosome remodeling factor (NURF) and contains an N-terminal DNA binding domain, two copies of a plant homeodomain (PHD) finger, and a bromodomain near its C-terminus (Figure 1A).14 The bromodomain of BPTF has been characterized as interacting with H4 K16ac,15–17 and its C-terminal PHD domain recognizes H3 K4me3.18,19 The association of BPTF with chromatin is likely bivalent, through engagement of both of these domains with doubly modified nucleosomes.15
Figure 1.
BPTF engages chromatin through multiple domains that recognize specific potential histone PTMs, including those on H2A.Z. (A) Domain diagram of BPTF showing its DNA binding and differential transcription factor domain (DDT), two PHD domains, and the bromodomain (BRD). Shown below are the histone modifications that have been characterized to interact with the C-terminal PHD domain and BRD. (B) Sequence alignment of the N-terminal tails of H2A.Z isoforms with their divergent amino acids highlighted in yellow. Shown above the sequence are their known post-translational modifications.
Adding to the complexity of the chromatin landscape are histone variants, which are encoded by genes that are separate from their canonical counterparts and differ in primary sequence. Their deposition into nucleosomes can dramatically alter higher-order chromatin structure and, in turn, gene transcription.20,21 H2A.Z is a H2A variant that is abundant in the promoter regions of genes.22–24 It is expressed as two isoforms [I and II (Figure 1B and Figure S1)] that have been shown to have unique, yet poorly understood, cellular roles.25–28 For example, overexpression of H2A.Z I has been associated with liver cancer,29 while H2A.Z II has been shown to drive malignant melanoma.30 In the context of chromatin, H2A.Z II nucleosomes are enriched with H3 K4me3 versus H2A.Z I-containing nucleosomes.25 Alternatively, bromodomain-containing protein BRD2 selectively immunoprecipitates H2A.Z I-containing nucleosomes.31 These results support differential mechanisms based on the primary sequence of the H2A.Z isoforms and subsequent protein–protein interactions.
Both bromodomain-containing proteins and H2A.Z are related to oncogenic function, and hyperacetylation of H2A.Z is localized at transcription start sites and enhancer regions.32 Therefore, transcriptional activation by the recruitment of chromatin remodelers or transcription factors through a direct bromodomain–H2A.Z interaction is feasible. Both BPTF and H2A.Z are found at gene promoters, supporting this transcriptional activation mechanism.33,34 Additionally, BPTF was shown to co-immunoprecipitate with H2A.Z-containing nucleosomes in bladder cancer, while the activation of H2A.Z target genes was largely erased with the suppression of BPTF expression in a shRNA-based knockdown.35 Whereas interactions of the bromodomain with acetylation patterns of native histones have been well characterized, the recognition motifs for H2A.Z I and II have yet to be fully investigated. The first direct interaction of the bromodomain of BPTF and H2A.Z was characterized using acetylated H2A.Z I peptides, where diacetylation at lysine 4 and lysine 11 had the greatest affinity for BPTF (Kd = 780 μM).36 The sequence of H2A.Z I differs from that of H2A.Z II by three amino acid residues (T14 vs A14, S38 vs T38, and V127 vs A127), and only one of these residues lies in the N-terminal tail region. Such a difference may affect how BPTF engages this tail region on the two isoforms.
Here, we more fully characterize the affinities of acetylation patterns of N-terminal tail peptides of both isoforms of H2A.Z for BPTF. Using protein-observed fluorine NMR (PrOF NMR) as a primary assay to quantify these weak interactions, we found diacetylated H2A.Z II K7ac,K13ac to have the strongest interaction (Kd = 310 ± 30 μM) with BPTF. This affinity was validated using surface plasmon resonance (SPR) giving a Kd of 195 ± 30 μM and is comparable to the affinity of the physiologically relevant canonical histone target of BPTF, H4 K16ac (Kd = 99–210 μM).15 Structural analyses using 1H Carr–Purcell–Meiboom–Gill (CPMG) NMR and X-ray crystallography confirmed the BPTF binding site preferentially engages a single acetylated lysine residue in the peptide sequence, K7ac. We further assessed the selectivity of H2A.Z histone peptides against a panel of bromodomains, revealing that the bromodomain of CECR2, a member of the CERF nucleosomal remodeling complex, binds with the highest affinity (Kd = 130 μM) and specificity for acetylated H2A.Z I over H2A.Z II. These results support a H2A.Z transcriptional activation mechanism that involves recruitment of chromatin remodeling complexes.
MATERIALS AND METHODS
Peptide Synthesis.
Peptides were synthesized using standard N-9-fluorenylmethoxycarbonyl (Fmoc) solid phase synthesis methods on NovaSyn TGR resin (Novabiochem, 0.25 mmol/g) using a Liberty Blue automated microwave synthesizer and N,N′-diisopropylcarbodiimide (DIC) and Oxyma for amino acid activation. All peptides were cleaved from the solid support in a 95/2.5/2.5 trifluoroacetic acid (TFA)/triisopropylsilane/water mixture for 2–5 h followed by evaporation of solvent under a nitrogen stream. The crude peptides were precipitated into cold diethyl ether and purified by reverse phase high-performance liquid chromatography (HPLC) on a C-18 column using 0.1% TFA water and CH3CN as solvents (4 to 24% CH3CN gradient over 30 min). The peptide molecular weight was confirmed using an Ab-Sciex 5800 matrix-assisted laser desorption ionization (MALDI) time-of-flight mass spectrometer (representative spectra shown in Figure S2). Peptide theoretical and observed masses are listed in Table S1, and HPLC traces can be found in the Supporting Information (pp S22–S32).
Unlabeled and Fluorinated Protein Expression.
The pNIC28-Bsa4 plasmid containing the bromodomain of BPTF was a kind gift from S. Knapp (Nuffield Department of Medicine, University of Oxford, Oxford, U.K.). The pNIC28-Bsa4 plasmids containing the bromodomain of CECR2 (Addgene plasmid 39066; http://n2t.net/addgene:39066; RRID, Addgene_39066), the first bromodomains of BRD4 (Addgene plasmid 38943; http://n2t.net/addgene:38943; RRID, Addgene_38943) and BRDT (Addgene plasmid 38898; http://n2t.net/addgene:38898; RRID, Addgene_38898), and the second bromodomain of BRD2 (Addgene plasmid 39074; http://n2t.net/addgene:39074; RRID, Addgene_39074) were a kind gift from N. Burgess-Brown. The pET28a(+) plasmid containing the first bromodomain of BRD2 (residues 71–194), the second bromodomain of BRD4 (residues 333–460), and the PCAF bromodomain was purchased from GenScript. The procedure for fluorinated protein expression of Gee et al. was followed.37 Escherichia coli strain BL21 (DE3) containing the pRARE plasmid was transformed with the plasmid containing the desired protein gene and plated on agar plates containing kanamycin and chloramphenicol. The plate was incubated overnight at 37 °C. A 5 mL LB culture containing antibiotics was inoculated using a single colony from this plate and grown overnight at 25 °C while being shaken at 220 rpm. The primary culture was used to inoculate 1 L of LB medium containing chloramphenicol (35 mg/L) and kanamycin (100 mg/L). This secondary culture was grown at 37 °C and 220 rpm until the optical density at 600 nm had reached 0.6–0.8. At this point, for unlabeled protein expression, an equilibration time of 60 min at 20 °C and 220 rpm was followed by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) to induce protein expression. For 5FW labeling, the cells were pelleted by centrifugation and resuspended in 1 L of defined medium and 5-fluoroindole (80 mg) dissolved in dimethyl sulfoxide (DMSO, 200 μL) was added. After a recovery time of 60 min at 37 °C and 250 rpm, followed by a 30 min cooling to the induction temperature of 20 °C, the culture was induced with 1 mM IPTG and allowed to shake for 16–20 h. Cells were pelleted by centrifugation at 8000g and stored at −20 °C until they were purified (method included in the Supporting Information). Quadrupole time-of-flight (Q-TOF) LC/MS was used to confirm protein identity and to determine percent fluorine incorporation (Figure S3 and Table S2).
Protein-Observed Fluorine NMR (PrOF NMR).
All experiments were performed on a Bruker Avance III HD 500 instrument with a 5 mm Prodigy TCI inverse cryoprobe (19F S:N 2000:1). 5FW-labeled bromodomains were diluted in 50 mM Tris, 100 mM NaCl, pH 7.4 buffer by the addition of D2O and 0.1% TFA to final concentrations of 5% and 0.05%, respectively. Two one-dimensional 19F NMR spectra of the control protein sample were recorded at an O1P of −75 ppm, NS = 16, D1 = 1, and AQ = 0.5 (TFA reference set to −75.25 ppm) and an O1P of −125 ppm, NS = 750–1000, D1 = 0.7, and AQ = 0.05s (protein resonances). Peptide stock solutions of 15–35 mM were prepared in Milli-Q water and titrated into bromodomain protein solutions (40–50 μM). The change in the chemical shift of the protein resonance (Δδobs) was plotted as a function of ligand concentration to generate binding isotherms using eq 1
| (1) |
where Δδmax is the maximum change in the fluorine chemical shift, [L] is the ligand concentration, [P] is the protein concentration, and [PL] is the bound complex concentration. The PrOF NMR titration of 5FW-BRD2(1) with bromosporine (BSP) to assign the WPF shelf 5FW resonance is shown in Figure S4. All peptide titrations were performed in a single replicate unless otherwise specified. Raw NMR data and binding isotherms for each titration are included in the Supporting Information.
1H CPMG NMR.
CPMG data were acquired on a Bruker 700-MHz Avance instrument with a CryoProbe 5 mm TXI and excitation sculpting water suppression. For these experiments the peptides were dissolved in water, unlabeled BPTF was buffer exchanged into CPMG buffer [50 mM phosphate and 100 mM NaCl (pH 7.4) in D2O], and all dilutions were performed using the CPMG buffer. A series of three experiments were conducted: (1) peptide only (50 μM), (2) peptide (50 μM) with BPTF or CECR2 (10–30 μM), and (3) peptide (50 μM) with BPTF or CECR2 (10–30 μM) and TP-238 (25 μM). The following parameters for 1H CPMG were used: τ = 0.004 s, L = 120, number of scans = 128, and acquisition time = 2 s.
Surface Plasmon Resonance (SPR).
For analysis of histone peptides, the anti-glutathione S-transferase (GST) antibody was immobilized on a CM5 chip using the GST capture kit (GE Healthcare) according to the manufacturer’s specifications. GST-BPTF was immobilized by injecting 10 μg/mL protein for 180 s. Peptide–protein interactions were analyzed with a 60 s association time followed by a 90 s dissociation. The surface was regenerated with a 10 mM glycine-HCl (pH 2.1) solution for 120 s, followed by reimmobilization before the beginning of each eight-point titration. Data were analyzed using Biaevaluation software.
AlphaScreen.
The AlphaScreen assay procedure for BPTF was adapted from the manufacturer’s protocol (PerkinElmer). Nickel chelate (Ni-NTA) acceptor beads and streptavidin donor beads were purchased from PerkinElmer. The biotinylated histone H4 K5ac,8ac,12ac,16ac peptide was purchased from EpiCypher, with the sequence Ac-SGRGK-(Ac)GGK(Ac)GLGK(Ac)GGAK(Ac)RHRKVLR-Peg(Biot).
All reagents were diluted in the assay buffer [50 mM HEPES-Na+ (ChemImpex), 100 mM NaCl (SigmaAldrich), 0.05% CHAPS (RPI), and 0.1% BSA (SigmaAldrich) (pH 7.4)]. Final assay concentrations (after the addition of all assay components) of 30 nM for the His9-tagged BPTF bromodomain and 50 nM for the biotinylated peptide were used. Three-fold serial dilutions were prepared with varying concentrations of the test peptide and a fixed protein concentration. Five microliters of these solutions was added to a 384-well plate (ProxiPlate-384, PerkinElmer). The plate was sealed and kept at room temperature for 30 min, followed by the addition of 5 μL of the biotinylated peptide. Five microliters of nickel chelate acceptor beads was added to each well under low-light conditions (<100 lx), to a final concentration of 20 μg/mL, and the plate was incubated at room temperature in the dark for 30 min. This was followed by the addition of 5 μL (final concentration of 20 μg/mL) of streptavidin donor beads under low-light conditions. After incubation for 30 min in the dark, the plate was read in AlphaScreen mode using a PerkinElmer EnSpire plate reader. The test peptides were run in duplicate, and IC50 values were calculated in GraphPad Prism 5.
RESULTS
Evaluation of Interactions of the Acetylated H2A.Z I and II Peptides with 5FW-BPTF Using PrOF NMR.
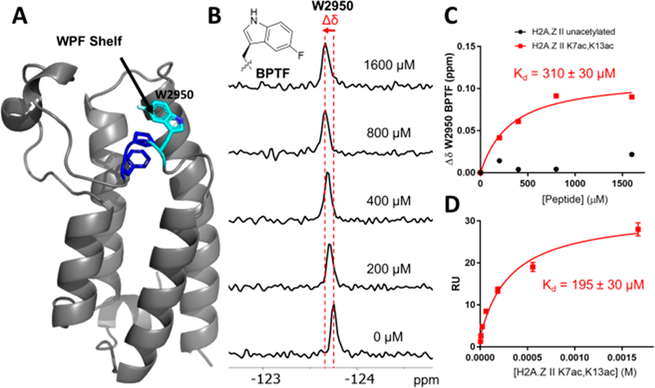
Interactions of bromodomains with acetylated histone substrates are relatively weak, with affinities in the low micromolar to millimolar Kd range.38 PrOF NMR is an attractive tool for measuring these interactions because fluorine chemical shifts are highly sensitive to changes in the environment.39 Fluorine was incorporated into the BPTF bromodomain biosynthetically using a fluorinated precursor of tryptophan, 5-fluoroindole. This resulted in the sequence selective labeling of W2950 as 5-fluorotryptophan (5FW), which lies in a three-residue hydrophobic motif in BPTF common to several bromodomains near the acetyl lysine binding site that is known as the WPF shelf (Figure 2A).40 Fluorination has been shown not to have a significant effect on protein structure or its binding interactions, making the 5FW a useful reporter for readily characterizing peptide binding events.41 This construct, 5FW-BPTF, was used to evaluate the affinities of the acetylated H2A.Z peptides.
Figure 2.
Binding experiment with H2A.Z II binding to 5FW-BPTF. (A) X-ray crystal structure of 5FW-BPTF with WPF residues colored blue and 5FW of 2950 colored cyan.42 (B) Stacked 19F NMR spectra with 44 μM 5FW-BPTF and increasing concentrations of the H2A.Z II K7ac,K13ac peptide showing the shift in the 5-fluorotryptophan resonance of W2950. (C) Binding isotherms for titrations of 5FW-BPTF with unacetylated and K7ac,K13ac H2A.Z II peptides. (D) Binding isotherm from the SPR sensorgram of H2A.Z II K7ac,K13ac with GST-BPTF.
Peptides were synthesized representing the 19 N-terminal amino acids of H2A.Z with an additional N-terminal tyrosine residue acting as a chromophore for accurate concentration determination. In this region, there are five lysine residues (K4, K7, K11, K13, and K15) with the potential to be acetylated. The two isoforms of H2A.Z differ by a single amino acid residue at position 14 in this region: a threonine in H2A.Z I and an alanine in H2A.Z II. To determine the preferred acetylation state of H2A.Z that is “read” by BPTF, mono-, di-, and two triacetylated patterns of H2A.Z II peptides were synthesized. The H2A.Z I peptides with equivalent acetylation patterns not previously reported36 were also synthesized for comparison. Measuring the chemical shift response of the BPTF 5FW resonance in the 19F NMR spectrum over a ligand titration series allowed each peptide’s affinity to be quantified (Figure 2B,C).
The H2A.Z II monoacetylated peptides (K4, K11, K13, and K15) all showed weak affinities for BPTF in the range of 1.3–1.7 mM, whereas K7ac and the unacetylated peptide showed nonsaturating and nonbinding behavior, respectively (Table 1). Diacetylation enhanced the affinity of the peptide for BPTF. Of the diacetylated patterns, the H2A.Z II peptide with K7ac and K13ac had the highest affinity with a Kd of 310 ± 30 μM (Figure 2C). To validate the affinity of the H2A.Z K7ac,K13ac peptide for BPTF, we characterized the Kd in a direct binding SPR assay using immobilized glutathione S-transferase (GST)-tagged BPTF that was not labeled with fluorine. This gave a closely agreeing Kd of 195 ± 30 μM (Figure 2D and Figure S5), suggesting 5FW labeling minimally perturbs BPTF binding and supports PrOF NMR as a reliable assay for characterizing BPTF–histone interactions. This affinity determined by SPR for the H2A.Z II K7ac,K13ac peptide is very similar to the reported binding affinity of the H4 K16ac peptide for BPTF (Kd = 99–210 μM), supporting a physiologically relevant affinity for the interaction of H2A.Z II with BPTF.15
Table 1.
Dissociation Constants for H2A.Z I and II Acetylated Peptides for 5FW-BPTF Determined by PrOF NMR Binding Experiments
|
Kd (μM)a |
||
|---|---|---|
| acetylation pattern | H2A.Z I | H2A.Z II |
| unacetylated | NB | NB |
| monoacetylated | ||
| K4ac | 1300d | 1290 |
| K7ac | 1140d | NS |
| K11ac | 1300d | 1360 |
| K13ac | 1810d | 1730 |
| K15ac | NSd | 1700 |
| diacetylated | ||
| K4ac,K7ac | 990d | 660 |
| K4ac,K11ac | 780d | 760 |
| K4ac,K13ac | 780 | 790 |
| K4ac,K15ac | 630 | 430b |
| K7ac,K11ac | 1200d | 780 |
| K7ac,K13ac | 520 | 310c |
| K4me,K7ac,K13ac | ND | 340 |
| K7ac,K15ac | 750 | 370 |
| K11ac,K13ac | 1440 | 790 |
| K11ac,K15ac | 950 | 880 |
| K13ac,K15ac | 830 | 910 |
| triacetylated | ||
| K4ac,K7ac,K11ac | 1510d | 810 |
| K7ac,K13ac,K15ac | 650 | 960 |
NB = nonbinding. NS = nonsaturating. ND = not determined.
Average of two experimental replicates.
Average of three experimental replicates (SD ± 30 μM).
Affinities previously reported by Perell et al.36
On the basis of the increased affinity of BPTF for diacetylated peptides over monoacetylated peptides, we next investigated higher acetylation states of H2A.Z and an additional PTM. Interestingly, a triacetylated H2A.Z II peptide including K7ac and K13ac (H2A.Z II K7ac,K13ac,K15ac) did not enhance BPTF binding. Rather, a loss of affinity was observed, up to 3-fold, in comparison with those of most diacetylated motifs. This suggests the greater affinity of diacetylated peptides over their monoacetylated counterparts is not simply due to an avidity effect and supports secondary interactions at the protein–peptide interface that contribute to binding. Similarly, incorporating another known PTM of H2A.Z,43 monomethylated lysine at position 4, into the sequence in combination with K7ac and K13ac did not lead to enhanced affinity, which has been reported for certain bromodomain–histone recognition events in other contexts.38
Next, we compared H2A.Z II sequences to H2A.Z I sequences with an identical acetylation pattern. This builds on the previously reported affinities of 5FW-BPTF for acetylated H2A.Z I peptides,36 with the untested diacetylated motifs synthesized and tested via PrOF NMR in this report (Table 1). Given an identical modification pattern, strong selectivity for one isoform of H2A.Z versus the other was not observed. However, it is worth noting that the peptide affinities of H2A.Z II, as a whole, are equivalent to or slightly stronger than those of H2A.Z I. The previously reported highest-affinity isoform I peptide, H2A.Z I K4ac,K11ac (Kd = 780 μM), shows an affinity similar to that of the equivalent isoform II peptide (Kd = 760 μM). Interestingly, the acetylation pattern of H2A.Z II we found to produce the highest affinity for BPTF was not evaluated in the original report, but we find that diacetylation at K7ac and K13ac is also the highest-affinity acetylation pattern of isoform I (Kd = 520 μM). Given the similarities in affinity, we cannot reach a conclusion about a significant rationale for a difference in isoform function based solely on the recruitment of the bromodomain of BPTF, despite the amino acid difference.
AlphaScreen and 1H CPMG NMR Competition Assays.
We next sought to test bromodomain binding site engagement by H2A.Z II versus a nonfunctional binding mode using an AlphaScreen assay. H2A.Z II K7ac,K13ac competed with a tetraacetylated H4 peptide and gave an average IC50 value of 830 μM, confirming a specific interaction (Figure S6). Given that our results support direct engagement with the bromodomain binding site, we sought to obtain higher-resolution structural information to evaluate the importance of the second acetylation modification. Whereas BET bromodomains tend to engage doubly acetylated histones,38 interactions of BPTF with canonical histones typically engage singly modified histones.15 We obtained the co-crystal structures of both H2A.Z I K4ac,K11ac (Figure S7A and Table S3) and H2A.Z II K7ac,K13ac (Figure S7B and Table S3) with BPTF, revealing a single Kac residue lodged in the hydrophobic binding pocket. Unfortunately, the electron density in the region of the peptide backbone was not sufficiently resolved despite the high resolution of these co-crystal structures [1.22 and 1.51 Å (Table S3)]. Therefore, assignment of the bound acetylated peptides was not possible and suggests either of the acetylated lysines is potentially engaged by the histone binding site. The results indicate that the regions flanking the acetylated lysines lack sufficient binding potential with BPTF residues beyond the Kac site, explaining the relatively weak binding affinity of these peptides.
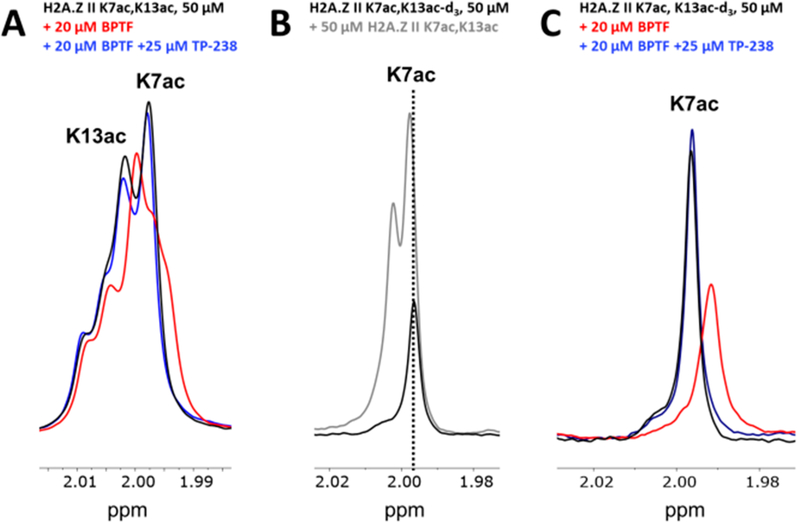
To further test bromodomain binding site engagement by H2A.Z II and to identify which acetyllysine is engaged by BPTF, we turned to a ligand-observed 1H CPMG NMR competition experiment. This ligand-observed NMR experiment takes advantage of the transfer of a protein’s short transverse relaxation time (T2) to bound ligand, detecting ligand binding interactions as a function of the decrease in signal intensity of specific ligand resonances (H2A.Z peptides in this case).44 Recovery of the peptide resonance intensities can be induced upon the addition of a competitor, which displaces the ligand from the binding site.45 1H NMR spectra of H2A.Z II K7ac,13ac show two singlet resonances near 1.9 ppm corresponding to the protons of the two acetyl methyl groups. When BPTF was added, decreases in the resonance intensity of both methyl resonances were observed but to varying degrees. The upfield resonance decreased more than the downfield resonance in a protein-dependent manner (Figure 3A and Figure S8), indicating that while each Kac on the peptide engages BPTF’s binding site to some extent, a single Kac residue is preferred. Addition of BPTF bromodomain chemical probe TP-23846 restores the resonance intensity, supporting a binding site specific interaction (Figure 3A).
Figure 3.
Ligand-observed 1H NMR CPMG competition experiment evaluating H2A.Z II K7,13ac for BPTF binding site engagement. (A) Experimental spectra of the H2A.Z II K7,13ac peptide alone (black) or with BPTF (red) or competitor TP-238 (blue) are overlaid. (B) Overlaid spectra of the H2A.Z II K7ac,13ac-d3 peptide alone (black) or with the H2A.Z II K7,13 peptide (gray). The dashed line shows the alignment of the K7ac resonance. (C) Experimental spectra for the H2A.Z II K7ac,13ac-d3 peptide alone (black) or with BPTF (red) or the BPTF bromodomain competitor TP-238 (blue) are overlaid.
To identify which acetyllysine is engaging BPTF in the binding site, we synthesized the H2A.Z II peptide with a deuterated acetyllysine at position 13 (K13ac-d3) and K7ac. The 1H NMR spectrum of this peptide shows a single resonance corresponding to the K7 acetyl methyl group (Figure 3B). Spiking in the nondeuterated H2A.Z K7ac,13ac peptide shows the increased intensity of the upfield resonance, allowing us to assign the downfield resonance to K13ac and the upfield resonance to K7ac. A CPMG competition experiment using the deuterated peptide reveals a decrease in the K7ac resonance upon addition of BPTF with restoration by TP-238 (Figure 3C). The complementary CPMG competition experiment using the H2A.Z II K7ac-d3,13ac deuterated peptide reveals an insignificant decrease in the K13ac resonance upon addition of BPTF without restoration by TP-238 (Figure S9). These results show that BPTF preferentially engages K7ac on H2A.Z II. This is supported by proteomics studies of H2A.Z acetylation levels.25 In this case, acetylation near the N-terminus appears to be more prevalent, with the most abundant marks being K4ac, K7ac, and K11ac. It is possible that K13ac results in increased affinity due to the elimination of an unfavorable interaction at the protein–peptide interface through the neutralization of positive charge or additional noncovalent interactions outside the binding site.
Determining H2A.Z Selectivity against a Panel of Bromodomains.
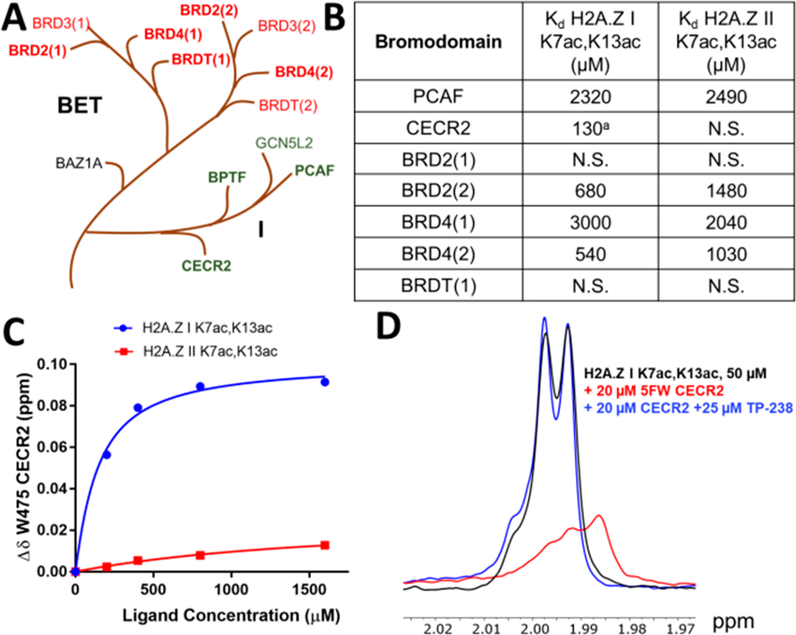
To address whether an H2A.Z interaction was specific to the bromodomain of BPTF, we selected seven additional bromodomains to characterize interactions using our PrOF NMR method. There are 61 human bromodomains across 46 different proteins separated into families based on similarity of sequence and structure.38 For our panel, we chose two bromodomains in the same family as BPTF: PCAF and CECR2. Similar to BPTF, CECR2 is a component of an imitation switch (ISWI) nucleosome remodeling complex, the CECR2-containing remodeling factor (CERF).47 We also chose five bromodomains in the bromodomain and extra terminal (BET) family, BRD2(1), BRD2(2), BRD4(1), BRD4(2), and BRDT(1) (Figure 4A). These bromodomains each contain a WPF shelf and were expressed as 5FW-labeled constructs to be used in PrOF NMR experiments. Each bromodomain’s affinity for both H2A.Z I K7ac,K13ac and H2A.Z II K7ac,K13ac peptides was determined to evaluate a possible isoform specific binding interaction (Figure 4).
Figure 4.
Evaluation of H2A.Z selectivity against a panel of bromodomains. (A) Branches of the human BRD phylogenetic tree displaying family I, to which BPTF belongs, and the closely related BET family. Bromodomains for which H2A.Z affinity was determined using PrOF NMR are represented in bold. (B) Dissociation constants for H2A.Z K7ac,K13ac peptides determined by PrOF NMR for various 5FW-labeled bromodomain constructs (N.S. = nonsaturating). (C) Binding isotherms for 5FW-CECR2 and H2A.Z I (blue) and II (red) K7ac,13ac peptides. (D) Ligand-observed 1H NMR CPMG competition experiment evaluating H2A.Z I K7ac,K13ac for CECR2 binding site engagement. Experimental spectra for the peptide alone (black) or with 5FW-CECR2 (red) or BPTF bromodomain competitor TP-238 (blue) are overlaid. aAverage of two technical replicates.
Overall, BET bromodomains showed weaker affinity for these H2A.Z diacetylated peptides than BPTF (Figure 4B). The second bromodomains of BRD2 and BRD4 showed tighter binding than their first bromodomains and also had ∼2-fold higher affinity for H2A.Z I. The BRD2 results are supported by immunoprecipitation studies of H2A.Z-containing nucleosomes, resulting in co-purification of BRD2.31 PCAF showed a weaker affinity for H2A.Z than BPTF (Kd = 2.3–2.5 mM). Surprisingly, CECR2 showed the strongest binding to H2A.Z I (Kd = 130 μM), whereas its affinity for H2A.Z II remained weak (Figure 4B,C). A direct interaction of H2A.Z I with unlabeled CECR2 was furthered supported by the competitive CPMG experiment described above (Figure 4D and Figure S10).
DISCUSSION
In this study, we find that H2A.Z II K7ac,13ac attains moderate affinity (Kd = 195–310 μM by SPR and PrOF NMR) for BPTF and approaches that of previously characterized H4 K16ac (Kd = 99–210 μM by ITC and fluorescence polarization15). This affinity is not likely enough for BPTF enrichment of modified H2A.Z-containing nucleosomes and selectivity over other bromodomains, which is more likely the result of a multivalent interaction. The deposition of H2A.Z into chromatin positively correlates with H3 K4me3,48 a modification that facilitates a higher-affinity interaction with the PHD domain of BPTF (Kd = 2.7 μM).49 This implicates a bivalent recognition event of BPTF’s bromodomain with acetylated H2A.Z and its PHD domain with methylated H3 may be possible. Ruthenburg et al. uncovered a similar multivalent interaction using modified histones in synthetic nucleosome constructs.15 The incorporation of H3 K4me3 along with acetylated H4 histones into nucleosomes led to enhanced affinity and specificity of the BPTF bromodomain for H4 K16ac over other H4 acetylation patterns. Moving beyond the scope of using isolated synthetic peptides, future studies placing H2A.Z histones in a nucleosome context may help elucidate affinity differences and isoform specific recognition interaction of H2A.Z with BPTF.50
Comparing our H2A.Z II peptide affinities to those previously reported for H2A.Z I, we found BPTF does not show strong selectivity for either H2A.Z I or II sequences given an identical acetylation pattern. Although the N-terminal tail region of isoform II differs from that of isoform I only by the amino acid at position 14 (T14 vs A14), it is possible that this single change could affect BPTF binding. Wysocka et al. found that the single amino acid change between isoforms I and II in the core histone sequence (S38 vs T38) altered the protein conformation and therefore increased the instability of H2A.Z II nucleosomes.51 Isoform II of H2A.Z is more enriched in H3 K4me3-containing nucleosomes,25 which suggests an isoform specific modification pattern recognized by BPTF could be possible in a multivalent chromatin binding event. Further investigation of these interactions in a more native context is necessary to probe BPTF’s selectivity for H2A.Z isoform modification patterns.
Seven other bromodomains were also evaluated for their affinity for H2A.Z peptides with K7,13 acetylation patterns. Of these constructs, BET bromodomains had weaker affinities for H2A.Z than BPTF, with most interactions in the millimolar Kd range. The second bromodomain of BRD2 showed modest affinity for H2A.Z I (Kd = 680 μM), representing a 2-fold selectivity over H2A.Z II. This isoform selectivity is supported by previous studies of association of BRD2 with H2A.Z-containing nucleosomes.31 Although it is believed that this association occurs primarily through interactions between the bromodomains of BRD2 and acetylated H4 sites, experiments revealed that more BRD2 co-purified with H2A.Z I than H2A.Z II when both nucleosomes had similar levels of H4 acetylation. The weak binding and poor selectivity observed may indicate that members of the BET family of bromodomain-containing proteins interact promiscuously with acetylated H2A.Z to facilitate chromatin signaling events.
An unexpected finding was the bromodomain of CECR2 showing high selectivity for H2A.Z I over isoform II, with a Kd of 130 μM. Although a direct interaction between CECR2 and H2A.Z has not been reported, our findings are supported by immunoprecipitation studies showing H2A.Z co-purifies significant levels of SNF2L,31 the common ATP-dependent chromatin remodeling subunit of NURF and CERF complexes that contain BPTF52 and CECR2,47 respectively. Additionally, H2A.Z incorporation has been shown to stimulate in vitro nucleosome remodeling activity of the ISWI class of mammalian remodelers, to which both NURF and CERF belong.53
CONCLUSION
By studying the patterns of N-terminal acetylation of H2A.Z histone variants that directly interact with bromodomains, we identified a diacetylated motif that is responsible for BPTF binding. Because both bromodomains and H2A.Z have been linked to transcriptional activity in various cancers, these results support a direct interaction between the BPTF bromodomain and acetylated H2A.Z as a possible mechanism for regulating disease progression. Additional investigations of multivalent interactions of BPTF with H2A.Z-containing nucleosomes may be necessary to determine if an isoform specific recognition event can occur under more physiologically relevant conditions. Similarly, the relative abundance of BPTF and CECR2 in specific tissues may be relevant in dictating divergent transcriptional activation mechanisms involving H2A.Z isoforms.
Supplementary Material
ACKNOWLEDGMENTS
The table of contents figure and Figure 1 were created with BioRender. We thank the Moffitt Chemical Biology Core for use of the protein crystallography facility (National Cancer Institute grant P30-CA076292).
Funding
Research reported in this publication was supported by National Institute of General Medical Sciences Grant R01GM121414. J.A.J. and N.M.O. were supported by National Institutes of Health Biotechnology Training Grants 5T32GM008347-27 and 5T32GM008347-29, respectively.
ABBREVIATIONS
- BPTF
bromodomain- and PHD finger-containing transcription factor
- CMPG
Carr–Purcell–Meiboom–Gill
- BET
bromodomain and extraterminal
- BRD
bromodomain
- DDT
DNA binding homeobox and different transcription factor
- PHD
plant homeodomain
- NURF
nucleosome remodeling factor
- CERF
CECR2-containing remodeling factor
- CECR2
cat eye syndrome chromosome region candidate 2
- PrOF NMR
protein-observed fluorine nuclear magnetic resonance
- PCAF
p300/CBP-associated factor
- SPR
surface plasmon resonance
- ITC
isothermal titration calorimetry
- 5FW
5-fluorotryptophan
- PTM
post-translational modification
- SNF2L
SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 1
- ISWI
imitation switch
Footnotes
Notes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.0c00159.
Protein binding experiments, peptide characterization, and crystallography methods (PDF)
Contributor Information
Noelle M. Olson, Department of Chemistry, University of Minnesota, Minneapolis, Minnesota 55455, United States
Samantha Kroc, Department of Chemistry, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Jorden A. Johnson, Department of Chemistry, University of Minnesota, Minneapolis, Minnesota 55455, United States
Huda Zahid, Department of Chemistry, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Peter D. Ycas, Department of Chemistry, University of Minnesota, Minneapolis, Minnesota 55455, United States
Alice Chan, Drug Discovery Department, H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida 33612, United States.
Jennifer R. Kimbrough, Department of Chemistry, University of Minnesota, Minneapolis, Minnesota 55455, United States
Prakriti Kalra, Department of Chemistry, University of Minnesota, Minneapolis, Minnesota 55455, United States.
Ernst Schönbrunn, Drug Discovery Department, H. Lee Moffitt Cancer Center and Research Institute, Tampa, Florida 33612, United States.
William C. K. Pomerantz, Department of Chemistry, University of Minnesota, Minneapolis, Minnesota 55455, United States.
REFERENCES
- (1).Kornberg RD (1974) Chromatin Structure: A Repeating Unit of Histones and DNA Published by: American Association for the Advancement of Science Linked references are available on JSTOR for this article: Chromatin Structure: A Repeating Unit of Histones and DNA Chromatin. Science 184, 868–871. [DOI] [PubMed] [Google Scholar]
- (2).Luger K, Mäder AW, Richmond RK, Sargent DF, and Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- (3).Strahl BD, and Allis CD (2000) The language of covalent histone modifications. Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- (4).Muller S, Filippakopoulos P, and Knapp S (2011) Bromodomains as therapeutic targets. Expert Rev. Mol. Med 13, No. e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Roussy M, Bilodeau M, Jouan L, Tibout P, Laramée L, Lemyre E, Léveillé F, Tihy F, Cardin S, Sauvageau C, Couture F, Louis I, Choblet A, Patey N, Gendron P, Duval M, Teira P, Hébert J, Wilhelm BT, Choi JK, Gruber TA, Bittencourt H, and Cellot S (2018) NUP98-BPTF gene fusion identified in primary refractory acute megakaryoblastic leukemia of infancy. Genes, Chromosomes Cancer 57, 311–319. [DOI] [PubMed] [Google Scholar]
- (6).Jordan-Sciutto KL, Dragich JM, Caltagarone J, Hall DJ, and Bowser R (2000) Fetal Alz-50 clone 1 (FAC1) protein interacts with the myc-associated zinc finger protein (ZF87/MAZ) and alters its transcriptional activity. Biochemistry 39, 3206–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Dar AA, Nosrati M, Bezrookove V, De Semir D, Majid S, Thummala S, Sun V, Tong S, Leong SPL, Minor D, Billings PR, Soroceanu L, Debs R, Miller JR, Sagebiel RW, and Kashani-Sabet M (2015) The role of BPTF in melanoma progression and in response to BRAF-targeted therapy. J. Natl. Cancer Inst 107, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Xiao S, Liu L, Fang M, Zhou X, Peng X, Long J, and Lu X (2015) BPTF Associated with EMT Indicates Negative Prognosis in Patients with Hepatocellular Carcinoma. Dig. Dis. Sci 60, 910–918. [DOI] [PubMed] [Google Scholar]
- (9).Sun H, Liu J, Zhang J, Shen W, Huang H, Xu C, Dai H, Wu J, and Shi Y (2007) Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4. Biochem. Biophys. Res. Commun 358, 435–441. [DOI] [PubMed] [Google Scholar]
- (10).Dai M, Lu J-J, Guo W, Yu W, Wang Q, Tang R, Tang Z, Xiao Y, Li Z, Sun W, Sun X, Qin Y, Huang W, Deng W, and Wu T (2015) BPTF promotes tumor growth and predicts poor prognosis in lung adenocarcinomas. Oncotarget 6, 33878–33892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Buganim Y, Goldstein I, Lipson D, Milyavsky M, Polak-Charcon S, Mardoukh C, Solomon H, Kalo E, Madar S, Brosh R, Perelman M, Navon R, Goldfinger N, Barshack I, Yakhini Z, and Rotter V (2010) A novel translocation breakpoint within the BPTF gene is associated with a pre-malignant phenotype. PLoS One 5, No. e9657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Filippakopoulos P, and Knapp S (2014) Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat. Rev. Drug Discovery 13, 337–356. [DOI] [PubMed] [Google Scholar]
- (13).Stankiewicz P, Khan TN, Szafranski P, Slattery L, Streff H, Vetrini F, Bernstein JA, Brown CW, Rosenfeld JA, Rednam S, Scollon S, Bergstrom KL, Parsons DW, Plon SE, Vieira MW, Quaio CRDC, Baratela WAR, Acosta Guio JC, Armstrong R, Mehta SG, Rump P, Pfundt R, Lewandowski R, Fernandes EM, Shinde DN, Tang S, Hoyer J, Zweier C, Reis A, Bacino CA, Xiao R, Breman AM, Smith JL, Katsanis N, Bostwick B, Popp B, Davis EE, and Yang Y (2017) Haploinsufficiency of the Chromatin Remodeler BPTF Causes Syndromic Developmental and Speech Delay, Postnatal Microcephaly, and Dysmorphic Features. Am. J. Hum. Genet 101, 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Jones MH, Hamana N, and Shimane M (2000) Identification and characterization of BPTF, a novel bromodomain transcription factor. Genomics 63, 35–39. [DOI] [PubMed] [Google Scholar]
- (15).Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, and Allis CD (2011) Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145, 692–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Philpott M, Yang J, Tumber T, Fedorov O, Uttarkar S, Filippakopoulos P, Picaud S, Keates T, Felletar I, Ciulli A, Knapp S, and Heightman TD (2011) Bromodomain-peptide displacement assays for interactome mapping and inhibitor discovery. Mol. BioSyst 7, 2899–2908. [DOI] [PubMed] [Google Scholar]
- (17).Sudhamalla B, Dey D, Breski M, Nguyen T, and Islam K (2017) Site-specific azide-acetyllysine photochemistry on epigenetic readers for interactome profiling. Chem. Sci 8, 4250–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, and Allis CD (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86–90. [DOI] [PubMed] [Google Scholar]
- (19).Chen P, Guo Z, Chen C, Tian S, Bai X, Zhai G, Ma Z, Wu H, and Zhang K (2020) Identification of dual histone modification-binding protein interaction by combining mass spectrometry and isothermal titration calorimetric analysis. J. Adv. Res 22, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Maze I, Noh K, Soshnev AA, and Allis CD (2014) Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat. Rev. Genet 15, 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Talbert PB, and Henikoff S (2017) Histone variants on the move: Substrates for chromatin dynamics. Nat. Rev. Mol. Cell Biol 18, 115–126. [DOI] [PubMed] [Google Scholar]
- (22).Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, and Madhani HD (2005) Histone variant H2A.Z Marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123, 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, and Felsenfeld G (2009) H3.3/H2A.Z double variant-containing nucleosomes mark “nucleosome-free regions” of active promoters and other regulatory regions. Nat. Genet 41, 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Guillemette B, Bataille AR, Gévry N, Adam M, Blanchette M, Robert F, and Gaudreau L (2005) Variant histone H2A.z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3, e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Dryhurst D, Ishibashi T, Rose KL, Eirín-López JM, McDonald D, Silva-Moreno B, Veldhoen N, Helbing CC, Hendzel MJ, Shabanowitz J, Hunt DF, and Ausió J (2009) Characterization of the histone H2A.Z-1 and H2A.Z-2 isoforms in vertebrates. BMC Biol. 7, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Eirín-López JM, González-Romero R, Dryhurst D, Ishibashi T, and Ausió J (2009) The evolutionary differentiation of two histone H2A.Z variants in chordates (H2A.Z-1 and H2A.Z-2) is mediated by a stepwise mutation process that affects three amino acid residues. BMC Evol. Biol 9, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JRE, Taylor H, Matthaei K, Rathjen PD, Tremethick DJ, and Lyons I (2001) Histone variant H2A.Z is required for early mammalian development. Curr. Biol 11, 1183–1187. [DOI] [PubMed] [Google Scholar]
- (28).Vardabasso C, Gaspar-Maia A, Hasson D, Pünzeler, Valle-Garcia, Straub T, Keilhauer EC, Strub T, Dong J, Panda T, Chung CY, Yao JL, Singh R, Segura MF, Fontanals-Cirera B, Verma A, Mann M, Hernando E, Hake SB, and Bernstein E (2015) Histone Variant H2A.Z.2 Mediates Proliferation and Drug Sensitivity of Malignant Melanoma. Mol. Cell 59, 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Yang HD, Kim P-J, Eun JW, Shen Q, Kim HS, Shin WC, Ahn YM, Park WS, Lee JY, and Nam SW (2016) Oncogenic potential of histone-variant H2A.Z.1 and its regulatory role in cell cycle and epithelial-mesenchymal transition in liver cancer. Oncotarget 7, 11412–11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Vardabasso C, Gaspar-Maia A, Hasson D, Punzeler S, Valle-Garcia D, Straub T, Keilhauer EC, Strub T, Dong J, Panda T, Chung C-Y, Yao JL, Singh R, Segura MF, Fontanals-Cirera B, Verma A, Mann M, Hernando E, Hake SB, and Bernstein E (2015) Histone Variant H2A.Z.2 Mediates Proliferation and Drug Sensitivity of Malignant Melanoma. Mol. Cell 59, 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Draker R, Ng MK, Sarcinella E, Ignatchenko V, Kislinger T, and Cheung P (2012) A Combination of H2A.Z and H4 Acetylation Recruits Brd2 to Chromatin during Transcriptional Activation. PLoS Genet. 8, No. e1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Valdés-Mora F, Song JZ, Statham AL, Strbenac D, Robinson MD, Nair SS, Patterson KI, Tremethick DJ, Stirzaker C, and Clark SJ (2012) Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome Res. 22, 307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Landry J, Sharov AA, Piao Y, Sharova LV, Xiao H, Southon E, Matta J, Tessarollo L, Zhang YE, Ko MSH, Kuehn MR, Yamaguchi TP, and Wu C (2008) Essential role of chromatin remodeling protein bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 4, No. e1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Landry JW, Banerjee S, Taylor B, Aplan PD, Singer A, and Wu C (2011) Chromatin remodeling complex NURF regulates thymocyte maturation. Genes Dev. 25, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kim K, Punj V, Choi J, Heo K, Kim JM, Laird PW, and An W (2013) Gene dysregulation by histone variant H2A.Z in bladder cancer. Epigenet. Chromatin 6, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Perell GT, Mishra NK, Sudhamalla B, Ycas PD, Islam K, and Pomerantz WCK (2017) Specific Acetylation Patterns of H2A.Z Form Transient Interactions with the BPTF Bromodomain. Biochemistry 56, 4607–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Gee C, Arntson K, Urick AK, Mishra NK, Hawk LML, Wisniewski A, and Pomerantz WCK (2016) Protein Observed 19F-NMR for fragment screening, affinity quantification and druggability assessment. Nat. Protoc 11, 1414–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Müller S, Pawson T, Gingras AC, Arrowsmith CH, and Knapp S (2012) Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Gerig JT Fluorine NMR. 2001, 1–35. [Google Scholar]
- (40).Romero FA, Taylor AM, Crawford TD, Tsui V, Côté A, and Magnuson S (2016) Disrupting Acetyl-Lysine Recognition: Progress in the Development of Bromodomain Inhibitors. J. Med. Chem. 59, 1271–1298. [DOI] [PubMed] [Google Scholar]
- (41).Mishra NK, Urick AK, Ember SWJ, Schonbrunn E, and Pomerantz WC (2014) Fluorinated aromatic amino acids are sensitive 19f NMR probes for bromodomain-ligand interactions. ACS Chem. Biol 9, 2755–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Kirberger SE, Ycas PD, Johnson JA, Chen C, Ciccone MF, Woo RWL, Urick AK, Zahid H, Shi K, Aihara H, Mcallister SD, Kashani-Sabet M, Shi J, Dickson A, dos Santos CO, and Pomerantz WCK (2019) Selectivity, ligand deconstruction, and cellular activity analysis of a BPTF bromodomain inhibitor. Org. Biomol. Chem 17, 2020–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Giaimo BD, Ferrante F, Herchenröther A, Hake SB, and Borggrefe T (2019) The histone variant H2A.Z in gene regulation. Epigenet. Chromatin 12, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Meiboom S, and Gill D (1958) Modified spin-echo method for measuring nuclear relaxation times. Rev. Sci. Instrum 29, 688–691. [Google Scholar]
- (45).Urick AK, Calle LP, Espinosa JF, Hu H, and Pomerantz WCK (2016) Protein-observed fluorine NMR is a complementary ligand discovery method to1H CPMG ligand-observed NMR. ACS Chem. Biol 11, 3154–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).TP-238, a chemical probe for CECR2/BPTF bromodomains.
- (47).Banting GS, Barak O, Ames TM, Burnham AC, Kardel MD, Cooch NS, Davidson CE, Godbout R, Mcdermid HE, and Shiekhattar R (2005) CECR2, a protein involved in neurulation, forms a novel chromatin remodeling complex with SNF2L. Hum. Mol. Genet 14, 513–524. [DOI] [PubMed] [Google Scholar]
- (48).Ku M, Jaffe JD, Koche RP, Rheinbay E, Endoh M, Koseki H, Carr SA, and Bernstein BE (2012) H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biol. 13, R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, and Patel DJ (2006) Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Morrison EA, Bowerman S, Sylvers KL, Wereszczynski J, and Musselman CA (2018) The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome. eLife 7, e31481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Greenberg RS, Long HK, Swigut T, and Wysocka J (2019) Single Amino Acid Change Underlies Distinct Roles of H2A. Z Subtypes in Human Syndrome. Cell 178, 1421–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Tsukiyama T, Daniel C, Tamkun J, and Wu C (1995) ISWI, a Member of the SliW2KNF2 ATPase Family, Encodes the 140 kDa Subunit of the Nucleosome Remodeling Factor. Cell 83, 1021–1026. [DOI] [PubMed] [Google Scholar]
- (53).Goldman JA, Garlick JD, and Kingston RE (2010) Chromatin Remodeling by Imitation Switch (ISWI) Class ATP-dependent Remodelers Is Stimulated by Histone Variant H2A.Z. J. Biol. Chem 285, 4645–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.