The spread of carbapenem- and polymyxin-resistant Enterobacteriaceae poses a significant threat to public health, challenging clinicians worldwide with limited therapeutic options. This review describes the current coding and noncoding genetic and transcriptional mechanisms mediating carbapenem and polymyxin resistance, respectively.
KEYWORDS: carbapenem resistance, polymyxin resistance, Enterobacteriaceae, transcription factors, resistance mechanisms, Enterobacteriales, antimicrobial resistance mechanisms, colistin, carbapenems
ABSTRACT
The spread of carbapenem- and polymyxin-resistant Enterobacteriaceae poses a significant threat to public health, challenging clinicians worldwide with limited therapeutic options. This review describes the current coding and noncoding genetic and transcriptional mechanisms mediating carbapenem and polymyxin resistance, respectively. A systematic review of all studies published in PubMed database between 2015 to October 2020 was performed. Journal articles evaluating carbapenem and polymyxin resistance mechanisms, respectively, were included. The search identified 171 journal articles for inclusion. Different New Delhi metallo-β-lactamase (NDM) carbapenemase variants had different transcriptional and affinity responses to different carbapenems. Mutations within the Klebsiella pneumoniae carbapenemase (KPC) mobile transposon, Tn4401, affect its promoter activity and expression levels, increasing carbapenem resistance. Insertion of IS26 in ardK increased imipenemase expression 53-fold. ompCF porin downregulation (mediated by envZ and ompR mutations), micCF small RNA hyperexpression, efflux upregulation (mediated by acrA, acrR, araC, marA, soxS, ramA, etc.), and mutations in acrAB-tolC mediated clinical carbapenem resistance when coupled with β-lactamase activity in a species-specific manner but not when acting without β-lactamases. Mutations in pmrAB, phoPQ, crrAB, and mgrB affect phosphorylation of lipid A of the lipopolysaccharide through the pmrHFIJKLM (arnBCDATEF or pbgP) cluster, leading to polymyxin resistance; mgrB inactivation also affected capsule structure. Mobile and induced mcr, efflux hyperexpression and porin downregulation, and Ecr transmembrane protein also conferred polymyxin resistance and heteroresistance. Carbapenem and polymyxin resistance is thus mediated by a diverse range of genetic and transcriptional mechanisms that are easily activated in an inducing environment. The molecular understanding of these emerging mechanisms can aid in developing new therapeutics for multidrug-resistant Enterobacteriaceae isolates.
INTRODUCTION
Antibiotic resistance is a significant problem worldwide, and its spread is a threat to public health and veterinary medicine due to the resultant restriction or depletion of therapeutic options, increased health care costs, unlimited transmission, and alarming mortality rates (1–3). Bacteria belonging to the Enterobacteriaceae family are of clinical concern owing to their association with carbapenem and polymyxin resistance worldwide (4). Specifically, carbapenem-resistant Enterobacteriaceae are designated by the World Health Organization (WHO) as critical priority pathogens due to their multidrug resistance (MDR) phenotypes and associated morbidities and mortalities (5). This is understandable since carbapenems are the treatment of choice and last-resort agents used against severe infections caused by MDR Enterobacteriaceae, which are usually resistant to clinically available antibiotics, including β-lactams, fluoroquinolones, and aminoglycosides (4, 6, 7). Although not categorized by the WHO yet, polymyxin-resistant Gram-negative bacteria (Enterobacteriaceae) also present a grave clinical challenge due to the importance of polymyxin as a reserved agent in treating carbapenem-resistant bacterial infections (8). The most frequently prescribed carbapenems include ertapenem, imipenem, and meropenem. Unlike other β-lactam antibiotics, carbapenems have the broadest antibacterial spectrum (2).
The use of carbapenems for MDR bacterial infections created a selection pressure within the clinical setting, resulting in the emergence of carbapenem resistance (2). Carbapenem resistance is primarily mediated by carbapenemase genes found on mobile genetic elements such as plasmids, integrons, insertion sequences, and transposons, allowing for easier horizontal transfer of genes across and within different bacterial species (9–12). Carbapenemases are a group of β-lactamases that hydrolyze the β-lactam ring of antibiotics, rendering them inactive (11). Other carbapenem resistance mechanisms include porin alteration, target modification, overproduction of extended-spectrum β-lactamases (ESBLs), and overexpression of efflux pumps (Fig. 1) (13).
FIG 1.
Summary of the carbapenem resistance mechanisms seen in Gram-negative bacteria (Enterobacteriaceae). (A) High-level resistance to carbapenems can be mediated by the alteration of membrane permeability due to porin mutations, restricting the entry of antibiotics into the periplasmic space. (B and C) Further, the hydrolysis of carbapenems (green balls) by highly expressed (increased concentrations of) AmpCs, and extended-spectrum β-lactamases (ESBLs) (B), as well as carbapenemases (C), respectively, also confer resistance to carbapenems. (D) Increased efflux pump activity also decreases the concentration of antibiotics (carbapenems) in the periplasmic space, reducing susceptibility to the antibiotic. (E) Finally, mobile genetic elements (MGEs), such as plasmids (with high copy numbers), transposons, and insertion sequences (upstream or within promoter sequences), can increase the expression levels of carbapenemases, AmpCs, and ESBLs, leading to higher levels of resistance to carbapenems. The figure was constructed using chemix.org and Paint 3D.
The emergence of carbapenem resistance in Enterobacteriaceae led to the reintroduction of polymyxin as a therapeutic option (14). Polymyxins, which are made up of polymyxin E and B (also known as colistin), were discovered in 1947 and used for the treatment of Gram-negative bacterial infections. Polymyxin acts by binding to the lipid A of the outer membrane of Gram-negative bacteria (15, 16). This interaction results in the disruption of the bacterial membrane, leading to cell death (13, 15). The use of polymyxin, however, was diminished in the 1970s due to its neurotoxicity and nephrotoxicity (16) but was widely used in veterinary medicine for the treatment of diarrhea in food-producing animals and as a growth promoter (17, 18). Its use as a growth promoter resulted in increased reports on polymyxin-resistant bacteria. Thereafter, a worldwide dissemination of the plasmid-mediated mcr-1 gene through the food-chain proliferated (19). Hence, several countries banned or restricted the use of polymyxin as a feed additive in veterinary medicine (20). Despite polymyxin’s limitations in human medicine, the increasing incidence of carbapenem-resistant Enterobacteriaceae led to the revival of polymyxin as a last-line treatment (21, 22).
The use of polymyxin in both human and veterinary medicine has led to the emergence of polymyxin-resistant Enterobacteriaceae (8). Polymyxin resistance is primarily mediated through covalent modification of the lipid A moiety of the bacterial lipopolysaccharide (LPS), through the addition of 4-amino-4-deoxy-arabinose or phosphoethanolamine residues (8, 23, 24). Specific chromosomal mutations within the two-component systems pmrA/pmrB and phoP/phoQ cause these modifications and within the genes that regulate these systems (23, 25). Recently, plasmid-mediated mcr-type genes encoding a phosphoethanolamine transferase enzyme were discovered and found to be responsible for the horizontal transfer of polymyxin resistance (8, 25). These modifications reduce the negative net charge of the LPS, reducing the affinity of the polycationic polymyxin peptide to the outer membrane of bacteria and thus decreasing the bacterial susceptibility to polymyxin (8, 15). Other resistance mechanisms include the use of efflux pumps, the formation of capsules, and the decrease in outer membrane proteins (8).
The occurrence of carbapenem and polymyxin resistance within Enterobacteriaceae reduces the therapeutic options for the treatment of MDR bacterial infections and increases the incidence of infections and mortality rates. This systematic review aims to describe the current resistance mechanisms that are known and explained in literature and to identify gaps within the field.
EVIDENCE BEFORE THIS REVIEW
In terms of polymyxin resistance mechanisms, numerous reviews have evaluated both the different resistance mechanisms and the epidemiology of mcr-type genes and their role in mediating polymyxin resistance (26, 27). Carbapenem resistance mechanisms have also been described in several reviews for both carbapenemase-producing and -nonproducing Enterobacteriaceae (28). This review, however, provides an in-depth characterization of emerging coding and noncoding genomic and transcriptional mechanisms that mediate resistance in polymyxin- and carbapenem-resistant Enterobacteriaceae. Particularly, other genome-based but noncoding elements mediating polymyxin and carbapenem resistance are also highlighted since their roles in polymyxin and carbapenem resistance have been less reviewed. Each resistance mechanism is supplemented with evidence from the literature to emphasize its role in mediating resistance.
LITERATURE SEARCH STRATEGY
A comprehensive literature search was carried out using the PubMed database. English journal articles published within the last 5 years (January 2015 to October 2020) were retrieved and screened with the following keywords: “carbapenem” and “colistin” in permutation and combination with “imipenem OR ertapenem OR meropenem OR doripenem OR polymyxin” and “resistance AND Enterobacteriaceae” in a factorial fashion. The search was focused on journal articles that evaluated the role of specific genes, noncoding elements, and transcriptional factors mediating resistance in Enterobacteriaceae using molecular intervention (mutagenesis, gene editing, etc.) assays. Therefore, studies that involved reviews, diagnostics, case reports, case studies, risk factors, epidemiology, and surveillance were excluded. Studies that performed antibiotic sensitivity testing and identified the presence of carbapenemase and mcr-type genes in isolates without evaluating the role of genes in mediating resistances were regarded as epidemiological studies and were excluded. Nevertheless, epidemiological studies that identified these resistance genes and further investigated whether the transfer of genes conferred resistance were included. The following data were extracted from the included articles: Enterobacteriaceae species, sample sources, molecular techniques used, and resistance genes (Table 1). The inclusion and exclusion protocols used in this review are given in Fig. S1 in the supplemental material.
TABLE 1.
Emerging genomic and transcriptional mechanisms mediating carbapenem resistance mechanisms in Enterobacteriaceae
| Resistance mechanism | Species | Resistance determinant(s) | Reference(s) |
|---|---|---|---|
| Narrow- and extended-spectrum β-lactamases | Enterobacter cloacae complex | AmpC | 100 |
| Escherichia coli | AmpC | 101 | |
| ESBL | 1, 54, 58 | ||
| Klebsiella pneumoniae | AmpC, ESBL | 101 | |
| Carbapenemase | Citrobacter freundii | LMB-1 | 39 |
| NDM, KPC | 11 | ||
| OXA | 102, 103 | ||
| Escherichia coli | IMP | 47 | |
| KPC-variants | 3, 37, 46 | ||
| NDM | 3, 44, 45, 51, 103–109 | ||
| NDM, KPC, IMP | 110 | ||
| OXA | 41, 111, 112 | ||
| Enterobacter cloacae complex | FRI-3 | 113, 114 | |
| FLC-1 | 115 | ||
| GES | 116 | ||
| IMP | 115 | ||
| KPC | 46 | ||
| LMB | 117 | ||
| MIR-17 | 118 | ||
| NDM | 119 | ||
| OXA | 102, 120 | ||
| Klebsiella pneumoniae | KPC | 9, 32, 121–126 | |
| IMP | 127 | ||
| NDM-4 | 43, 44, 128–132 | ||
| OXA | 42, 102, 111, 130, 133–136 | ||
| VIM | 125, 137 | ||
| Klebsiella aerogenes | NDM | 138 | |
| Klebsiella quasipneumoniae | KHM-1 | 139 | |
| E. coli, E. cloacae, and K. pneumoniae | NDM | 12 | |
| Efflux pumps | Escherichia coli | AcrAB-TolC | 3, 55, 59, 63, 65, 67, 140 |
| Klebsiella pneumoniae | AcrAB-TolC | 3, 65, 67, 125 | |
| AcrAB, RamA | 141 | ||
| Porin deficiency | Escherichia coli | OmpK35 and/or OmpK36 | 49, 51, 52, 54, 58, 67, 101, 125, 142, 143 |
| PhoE | 144 | ||
| Enterobacter cloacae complex | OmpK35 and/or OmpK36 | 1, 9, 64, 100, 145 | |
| MicC and MicF | 53 | ||
| Klebsiella pneumoniae | OmpK35 and/or OmpK36 | 32, 33, 67, 111, 125, 136, 145–147 | |
| Raoultella orithinolytica | OmpK35 and/or OmpK36 | 6 | |
| Target modifications | Escherichia coli | MdrA | 148, 149 |
Flow chart showing the literature search strategy, inclusion and exclusion criteria, and the final number of manuscripts used for the review. Download FIG S1, PDF file, 0.1MB (83.9KB, pdf) .
Copyright © 2020 Mmatli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CARBAPENEM RESISTANCE MECHANISMS
Carbapenem resistance in carbapenem-resistant Enterobacteriaceae (CRE) is mediated by a variation and synchrony of different resistance mechanisms: the loss of major porin proteins, increased activity of efflux pumps, and the production of β-lactamases, i.e., carbapenemases, ESBLs, and cephalosporinases (AmpCs) (Fig. 1). The β-lactamases include ESBLs (TEM, PER, VEB, SHV, LEN, and CTX-M), carbapenemases (such as KPC, GES-5, IMI, VCC, OXA-48, IMP, VIM, and NDM), and AmpC-type β-lactamases (i.e., CMY, FOX, MOX, ACT, MIR, DHA, etc.) (29–31). These enzymes are frequently plasmid-borne genes, allowing for the dissemination of genes within and between Gram-negative bacteria species. This results in resistance toward penicillin, cephalosporins, carbapenems, and/or monobactams (29, 202, 203). Carbapenem resistance in Enterobacteriaceae is mediated mainly by the production of carbapenemases (11). However, elevated carbapenem resistance in carbapenemase-producing Enterobacteriaceae is usually through the overproduction of carbapenemases and/or alteration of membrane permeability (Table 1) (32).
CARBAPENEMASE PRODUCTION
Carbapenemases are broad-spectrum β-lactamases that hydrolyze the β-lactam ring of carbapenems and other β-lactam antibiotics (11, 33). There are three groups of carbapenemases: Ambler class A, class B that is made up of metallo-β-lactamases (MBLs), and class D β-lactamases (34–36). Class A has a broad spectrum of activity and utilizes a serine residue in its active site during cleavage of the β-lactam ring of penicillin, cephalosporins, classic β-lactamase inhibitors (sulbactam and tazobactam), aztreonam, and carbapenems (37, 38). Class B depends on zinc as a cofactor in its active site but has a similar spectrum of activity as class A, sparing aztreonam (39, 40). Class D, similar to class A, utilizes a serine residue in its active site but has a unique spectrum of activity, i.e., reduced carbapenem susceptibility, high resistance to penicillin, and intermediate resistance to cephalosporins; it is inactive against aztreonam (41, 42). Klebsiella pneumoniae carbapenemase (KPC) of class A, New Delhi MBL (NDM) carbapenemase of class B, and oxacillin-hydrolyzing carbapenemase (OXA-48/-181) from class D are responsible for most carbapenem resistance in CRE (35).
Other clinically relevant carbapenemases include MBLs that belong to subclass B1: Verona Integron-encoded MBLs (VIM) and imipenemase (IMP) (34, 43). As carbapenemases spread within Enterobacteriaceae, amino acid substitutions occur, producing different variants of the carbapenemase. This results in changes in the carbapenemase activity and its affinity to carbapenems (43). Paul et al. (44) showed that NDM variants (NDM-1 and NDM-5) had different transcriptional responses to different carbapenems, where NDM-5 had a 10-fold increase in expression when exposed to ertapenem, and NDM-1 had only a 2-fold increase. Paul et al. (44) further speculated that new variants of NDM are evolving inducibility in the presence of carbapenem drugs, resulting in elevated NDM production. A similar study revealed how molecular differences between NDM-17 and NDM-5 carbapenemases affected their carbapenemase activity (45). NDM-17 had an E170K (glutamic acid to lysine) amino acid substitution that was responsible for higher affinity and an increased carbapenemase activity compared to NDM-5, resulting in elevated ertapenem and meropenem resistance (45). In KPC, mutations within its mobile transposon affects the promoter activity of blaKPC and, subsequently, carbapenem resistance (Table 1).
The blaKPC gene is usually located within a 10-kb mobile transposon, Tn4401, allowing for its dissemination within the Enterobacteriaceae family and other Gram-negative bacteria such as Pseudomonas and Acinetobacter species (46). Cheruvanky et al. studied the different Tn4401 isoforms within Enterobacteriaceae and identified three isoforms—Tn4401b, Tn4401a, and Tn4401h—which were mostly found in Klebsiella (48%), Enterobacter (37%), and Citrobacter (12%) spp. Tn4401a and Tn4401h were mutational variations of Tn4401b. Genomic comparison analysis found that Tn4401a and Tn4401h had 99- and 188-bp deletions, respectively, between the P1 and P2 regions of the putative promoter sequences (46). These mutations increased the promoter activity of these isoforms that resulted in a 23- and 4-fold increase in KPC expression in Tn4401a and Tn4401h, respectively, compared to the Tn4401b isoform (46).
In electrocompetent E. coli Genehog cells, the three different isoforms were introduced. This resulted in meropenem MIC values of 1, 16, and 4 μg/ml for Tn4401b, Tn4401a, and Tn4401h, respectively. Tn4401a had the highest KPC production, which conferred the highest meropenem resistance (46). Huang et al. performed a similar study characterizing three Tn3-Tn4401 chimera isoforms: CTA, CTB, and CTC. The chimeras had different combinations of P1, PY, and PX promoters, and the study evaluated how it affected the expression of blaKPC and carbapenem susceptibility in KPC-producing isolates. Huang et al. and Cheruvanky et al. both showed that mutations within the putative promoter sequence of Tn4401 affect the expression of the blaKPC gene and carbapenem susceptibility in isolates.
The overproduction of carbapenemase in an IMP-harboring E. coli isolates was achieved through the insertion of an insertion element (IS26) within the ardK gene of the IncN plasmid during meropenem selection (47). ardK encodes a putative transcription factor that negatively modulates the transcription of the blaIMP-6 gene (47). The disruption of this gene with an IS26 element resulted in 53- and 256-fold increases in IMP production and meropenem resistance, respectively (47). Although the parental E. coli strain harbored IMP and was carbapenem susceptible, it plays a potential role in the dissemination of IMP-6-harboring plasmids that can, under selection, mediate a high-level carbapenem resistance (47). Wu et al. (11) identified a clinical Citrobacter freundii ST88 isolate that harbored two carbapenemase-harboring plasmids encoding blaKPC-2 and blaNDM-1, respectively. The transformation of E. coli J53 with both plasmids conferred 2- or 4-fold increases in imipenem and meropenem MICs compared to J53 isolate alone or with either plasmid. Thus, the coexistence of blaKPC-2 and blaNDM-1 resulted in a synergistic effect, conferring high-level carbapenem resistance, resulting in MIC values of 1,024 and 512 μg/ml for imipenem and meropenem, respectively, in the C. freundii isolate (Table 1).
Unlike the other resistance mechanisms, carbapenemase production is adequate to confer clinical resistance for carbapenems. This was shown by Choudhury et al., who transformed E. coli J53 competent cells with an NDM-4-harboring plasmid. The donor isolate, E. coli ST448, and the transformant both had MICs above the imipenem, meropenem, and ertapenem breakpoints (48).
β-LACTAMASE PRODUCTION
In non-carbapenemase-producing carbapenem-resistant Enterobacteriaceae, carbapenem nonsusceptibility is observed in ESBL- and/or AmpC β-lactamase-producing Enterobacteriaceae. When ESBL production is coupled with the loss of the two major outer membrane porin groups, including OmpC and OmpF, clinical carbapenem resistance is observed (49). OmpC and OmpF are responsible for the nonspecific transport of solutes across the outer membrane into the cytoplasm (4). OmpC and OmpF are homologues of OmpK36 and OmpK35, respectively (6), and will be used interchangeably in this review.
The production of extended-spectrum and AmpC β-lactamases alone without membrane impermeability is insufficient to confer clinical carbapenem resistance. van Boxtel et al. (1) showed that the transformation of E. coli isolates with a plasmid encoding blaCMY-2 gene resulted in reduced meropenem susceptibility of E. coli but did not confer clinical meropenem resistance (Table 1).
AmpC and ESBLs have previously been shown to hydrolyze carbapenems weakly, and van Boxtel et al. further showed that CMY-2 hydrolysis of meropenem was below the detection limit during β-lactamase activity evaluation assays. Clinical meropenem resistance was observed when a blaCMY-2- harboring plasmid transformed a porin-deficient Escherichia coli isolate (1). Guiana ESBL (GES) belonging to Amber class A acquires carbapenemase activity due to the glycerin substitution at position 170 with either a serine or an asparagine (13). Streling et al. (13) reported that GES-16, which had the Gly170Ser amino acid substitution, had a broad-spectrum hydrolysis profile, hydrolyzing penicillin, cephamycin, cephalosporins, and carbapenems. Although GES-16 showed some carbapenemase activity, it conferred low-level resistance to carbapenems. Moreover, it remains to be seen if GES-16 shall be classified as a carbapenemase like GES-5 (50) since they had similar kinetic parameters toward carbapenems (13).
ALTERATION OF MEMBRANE PERMEABILITY
The loss of major outer membrane proteins, OmpK36 and OmpK35, is frequently observed in CRE, resulting in reduced permeability of the outer membrane due to structural changes in porin channels restricting the uptake of charged molecules through the bacterial cell wall (Fig. 1) (3, 49, 51). These structural changes were due to mutations that either reduced the channel size of porins or modified its electrostatics (52). This phenotype is frequently observed in resistance-induced mutants through serial passage assays of non-carbapenemase-producing isolates (53). Hao et al. (53) revealed that clinical Enterobacteriaceae isolates acquired carbapenem resistance through mutations within both OmpK36 and OmpK35 proteins. Isolates with mutations in both OmpK36 and OmpK35 had elevated resistance (8 to 32 μg/ml) to ertapenem, imipenem, and meropenem compared to isolates with only OmpK36 mutations (0.25 to 2 μg/ml). This correlates with the findings of Hamzaoui et al. (2) in K. pneumoniae isolates, which showed that the loss of both major porins or mutations within genes seen to regulate the porin system resulted in elevated carbapenem resistance. These genes include the two-component transduction regulatory system envZ-ompR, micF, and micC genes (4, 53). These mutations include single-base deletion, insertion, or substitution in the coding sequence resulting in the inactivation of proteins (53).
EnvZ is an inner membrane sensor kinase that is encoded by the EnvZ-OmpR regulatory system that regulates the expression of the two major porin groups, OmpC and OmpF (54). This is accomplished through phosphorylation or dephosphorylation of OmpR, the transcriptional factor responsible for porin gene activation (54). The phosphorylation of OmpR results in a structural change in the protein, increasing its binding affinity to the major porin transcriptional factor binding site (51). Kong et al. (51) revealed that the Gly63Ser amino acid substitution within the N-terminal phosphorylation domain of OmpR affects the phosphorylation of OmpR by EnvZ. The OmpR mutant, after that, failed to initiate porin transcription, resulting in a change in membrane permeability and, subsequently, carbapenem resistance (51). The study further went on to show the synergistic effect of OmpR mutants and carbapenemase activity, with the transformation of OmpR mutants with a NDM-harboring plasmid resulting in a 100-fold increase in the carbapenem MIC (Table 1) (51).
SDS-PAGE analysis of the outer membrane porins of OmpR mutants revealed a sharp decrease in the expression of both major porin groups (49). Adler et al. (55) reported that mutations within both envZ and ompR are early genetic events that mediate carbapenem resistance during serial passage. OmpR mutants had decreased expression of both ompC and ompF, whereas envZ mutations led to downregulation of ompF and upregulation of ompC. In ompCF-deleted porin-deficient isolates, envZ mutations were still observed and resulted in a 6-fold increase in carbapenem resistance, illustrating that envZ mutations are critical for carbapenem resistance (55). The role of envZ in mediating resistance in porin-deficient isolates is still unknown (54).
The loss of the major porin groups can also be achieved through the change in expression of micC and micF genes. These genes are part of the outer membrane genes and encode small antisense RNA that negatively regulates OmpC and OmpF genes (4, 53). Serial passage of Enterobacter aerogenes (currently Klebsiella aerogenes) performed by Hao et al. (53) produced two carbapenem-resistant isolates with the loss of both major porin groups. SDS-PAGE analysis of the outer membrane proteins revealed the loss of both porins; however, no mutations within ompCF were observed compared to E. aerogenes strain NCTC10336, which led to the investigation of micC and micF gene expression. Transcriptional analysis revealed that the overexpression of both micC and micF in isolates results in the significant downregulation of OmpK36 and OmpK35, respectively (53). Similar results were observed in E. coli, where the upregulation of micF resulted in the downregulation of ompF genes (4). Interestingly, the upregulation of micC leads to the downregulation of ompC but to an increase in ompF to compensate for the loss of OmpC (4).
PORIN DEFICIENCY AND β-LACTAMASE PRODUCTION
The loss of both major outer membrane proteins, OmpK35 and OmpK36, in E. coli and K. aerogenes was seen in some clinical isolates to mediate high-level carbapenem resistance (51–53). However, in K. pneumoniae, Salmonella enterica serotype Typhimurium, and other Enterobacteriaceae species, including Enterobacter cloacae complex (E. asburiae and E. cloacae) and Raoultella ornithinolytica, porin deficiency reduces the susceptibility of isolates to carbapenem but does not confer clinical resistance, and β-lactamase activity is required (56, 57). The transformation of porin-deficient isolates with an NDM-harboring plasmid revealed a synergistic effect mediating high-level carbapenem resistance (51). Porin deficiency reduces the uptake of antibiotics into the periplasm, which reduces the concentrations of antibiotics in the periplasmic space and cytosol, amplifying the β-lactamase effect (due to reduced intracellular antibiotic concentrations) and resulting in a synergistic effect (Fig. 1) (49). van Boxtel et al. (1) demonstrated that for clinical meropenem resistance in CMY-2-harboring E. coli isolate, mutations that disrupt porin expression and increase CMY-2 expression were required. Individually, the loss of porin and the upregulation of CMY-2 expression reduced meropenem susceptibility in isolate but did not confer clinical resistance. Clinical resistance was achieved when both mechanisms were found in the isolate, resulting in a meropenem MIC of >32 μg/ml (1) (Table 1 and Fig. 1).
OVERPRODUCTION OF EFFLUX PUMPS
In ESBL-producing Enterobacteriaceae, carbapenem resistance is also achieved through the overexpression of efflux pumps (58, 59). The overexpression of the efflux pump phenotype is usually observed when there is a significant increase in carbapenem susceptibility when an isolate is incubated with a carbapenem and the appropriate efflux pump inhibitor. There are different types of efflux pump inhibitors (EPIs) based on their mechanisms of action (60). Carbonyl cyanide m-chlorophenylhydrazine (CCCP) is an example of a protonophore that indirectly affects the activity of proton pumps by disrupting the proton motive force, reducing ATP production and resulting in an increased membrane permeability (60, 61). The disruption of the proton motive force across the membrane leads to membrane depolarization, eradicating the electrochemical concentration gradient across the membrane (60).
Osei Sekyere and Amoako (60) hypothesized that the cytoplasmic ion imbalance caused by the depolarized membrane created by CCCP disrupts the optimal activity of carbapenemase, which requires energy (ATP) and zinc to function. This was observed when some carbapenemase-producing Enterobacteriaceae species resulted in a 2-fold reduction in meropenem resistance in the presence of CCCP. At the same time, CCCP did not affect carbapenem susceptibility in non-carbapenemase-producing Enterobacteriaceae isolates (60). More research evaluating the effects of a depolarized membrane on carbapenemase activity is required to support this hypothesis.
Another mechanism of action of EPIs is the direct binding of an EPI to the functional efflux pump, reducing the ability of antibiotics to be pumped out of the cell by efflux pumps. This is the mechanism of action of phenylalanine-arginine β-naphthylamide (PAβN) (61). Lee and coworkers identified an E. cloacae ST74 clinical isolate whose imipenem susceptibility increased from 64 mg/ml to 0.5 mg/ml in the presence of PAβN, revealing the active role of efflux pumps in mediating carbapenem resistance and the synergistic effect of PAβN and carbapenemases in increasing carbapenem susceptibility in E. coli (Fig. 1) (60, 62).
AcrAB-TolC is a well-known multidrug efflux pump system that confers resistances toward a wide variety of agents, including β-lactams and is responsible for the MDR phenotype in E. coli (59, 63). It belongs to the resistance nodulation division (RND) superfamily and has been shown to synergistically work with other mechanisms to confer high-level resistance (3, 59). AcrAB-TolC is a tripartite efflux pump system that is made up of acrA, acrB, and tolC genes that encode a periplasmic membrane fusion protein, an inner membrane transporter, and an outer membrane protein, respectively (Fig. 1) (3, 59).
Saw et al. (3) evaluated the role of the AcrAB-TolC efflux pump system in Enterobacteriaceae species, viz., E. coli, K. pneumoniae, and Salmonella enterica serotype Typhimurium. In all three isolates, mutations within acrAB and tolC had no significant effect on carbapenem susceptibility. The transformation of acrAB and tolC mutants of E. coli and K. pneumoniae with KPC-harboring plasmid resulted in 4- and 8-fold increases in ertapenem and meropenem resistance, respectively. In KPC-producing E. coli and K. pneumoniae, the introduction of acrAB mutations resulted in a 4-fold increase in ertapenem MIC in E. coli and a 2- to 8-fold increase in carbapenem resistance in K. pneumoniae. acrAB mutations in carbapenemase-harboring K. pneumoniae and E. coli isolates created a synergistic effect with the β-lactamase activity, causing high-level carbapenem resistance (3).
The introduction of tolC mutations and KPC- and NDM-harboring plasmids into S. Typhimurium isolates resulted in 2-, 250-, and 1,000-fold increases in the ertapenem MICs, respectively. The introduction of carbapenemases in Salmonella enterica serotype Typhimurium isolates, therefore, results in elevated carbapenem resistance (3). The introduction of acrAB mutations in KPC and NDM-producing isolates did not affect nor contribute to carbapenem resistance in S. Typhimurium isolates. In comparison, the introduction of tolC mutations resulted in 2- and 4-fold increases in the ertapenem MICs in S. Typhimurium isolates (3).
AcrAB-TolC efflux pump systems in the Enterobacteriaceae family are regulated by the local regulators AcrR and the global regulators MarA, SoxS, and RamA (59, 64). Mutations within ramA and ramB have been shown via quantitative-PCR to upregulate RamA and AcrA transcripts, increasing efflux pump activity and decreasing ompCF expression (64). A novel AraC-type regulator called regulator of antibiotic resistance A, RarA, regulates the efflux pump system conferring the MDR phenotype in Enterobacteriaceae (63). Chetri et al. (63) evaluated its transcriptional response with the increase of carbapenem concentration. The expression of RarA was directly proportional to the concentration of ertapenem, resulting in the upregulation of AcrAB expression, reducing carbapenem susceptibility in E. coli clinical isolates (Table 1).
The study showed that RarA acts as a positive regulator of AcrAB, independent of the global regulators MarA, SoxS, and RamA (63). The transformation of E. coli DH5α with a plasmid encoding rarA resulted in MIC values of >32 μg/ml for ertapenem, meropenem, and imipenem (65). Pavez et al. evaluated the AcrAB efflux pump expression under imipenem stress and found that MarA and SdeR were responsible for the increased expression of AcrAB efflux pump in E. coli, E. cloacae, and K. pneumoniae (65). The global regulatory pathways are interconnected and function to downregulate porin expression and upregulate efflux systems (64). The overexpression of acrAB decrease the expression of porin genes, ompC and ompF (59). Pal et al. (66) reported an active role of acrB expression in augmenting ompC reduction in E. coli and K. pneumoniae isolates. The mechanisms mediating this role is unknown; however, the global regulatory system is assumed to play a role (64). Mutations within AcrD, a transporter of the RND superfamily, have been shown to compensate for the loss of AcrB, increasing the export of carbapenems out of the periplasm and mediating carbapenem resistance (Fig. 1) (55). This was seen in E. coli isolates with acrD and acrB double mutants (55). Mutations in the local AcrAB-TolC regulator, AcrR, increases AcrB expression, increasing carbapenem nonsusceptibility in E. coli isolates (55). An increase in efflux pump activity, mediated by mutations in the regulators stated above, only confers clinical carbapenem resistance in Enterobacteriaceae, and aids in mediating high-level carbapenem resistance when coupled with β-lactamase/carbapenemase production (Fig. 1) (3, 67).
POLYMYXIN RESISTANCE MECHANISMS
Polymyxin resistance in Enterobacteriaceae includes modification of the LPS in the outer membrane layer of the bacterium, neutralizing the negative charge of the outer membrane (23). This results in a weak interaction or binding affinity between the positively charged polymyxin and LPS molecules, viz., lipid A (21, 22). These modifications include the transfer of 4-amino-4-deoxy-l-arabinose (Ara4N) and phosphoethanolamine (PEtN) to the 4-phosphate and the 1-phosphate groups of lipid A, respectively, through the pbgP operon and PmrC or mcr-type gene products (Fig. 2) (22).
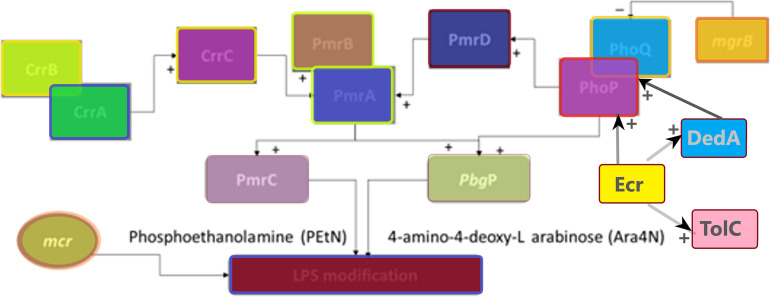
FIG 2.
Representation of the various mechanisms and determinants interacting to mediate polymyxin resistance in Gram-negative bacteria. The LPS of the outer membrane of Gram-negative bacteria is modified through PmrC, the pbgP operon, and mcr-type genes. The PmrC and PbgP genes are regulated by three two-component systems—PhoPQ, PmrAB, and CrrAB—that are interconnected by CrrC and PmrD proteins. The newly discovered DedA and Ecr proteins also activate PbgP through PhoPQ, whereas Ecr also activates DedA and TolC. Heteroresistance in Enterobacter sp. is thought to be mediated by Ecr membrane proteins. A “+” indicates activation toward upregulation; a “–” indicates repression/inhibition toward downregulation. The diagram was constructed using Paint 3D with a structure based on one by Cheng et al. (75).
The pbgP operon encodes the endogenous LPS modification system that is regulated by PhoPQ and PmrAB two-component regulatory systems (68, 69). The pbgP operon encodes enzymes that synthesize Ara4N from UDP glucuronic acid and mediates the addition of Ara4N to the 1-phosphate group of lipid A (70). These regulatory systems are responsible for the biosynthesis and transfer of Ara4N to lipid A; chromosomal mutations within the phoP, phoQ, and pmrB genes upregulate these systems, mediating polymyxin resistance (Fig. 2) (23, 71).
PmrAB TWO-COMPONENT REGULATORY SYSTEM
The PmrAB system is made up of a pmrABC operon that encodes three proteins: a cytoplasmic membrane-bound sensor kinase, PmrB; a regulatory protein, PmrA; and a PEtN transferase, PmrC (21). PmrB activates PmrA protein through phosphorylation, which then binds to the pbgP operon for Ara4N modification (21). The cytoplasmic membrane-bound kinase is further activated by extracellular stimulants, such as a high concentration of iron (Fe3+) and aluminum (Al3+), as well as an acidic pH (<5.5) (69). Mutations within pmrB increase PmrB kinase activity, resulting in autophosphorylation of PmrA and leading to an increased expression of the pbgP operon (21). Cannatelli et al. (204) showed that pmrB mutation leads to the constitutive activation of pmrA that increases the expression of pmrK (of pbgP operon), resulting in colistin resistance in the E. coli ST59 isolate.
In Salmonella enterica subsp. enterica serovar Newport ST45, colistin resistance was achieved through a 12-nucleotide deletion within pmrB, conferring colistin resistance (16 mg/liter) (72). Phan et al. (21) showed that pmrB mutations have a feedback loop onto pmrC and pmrA genes located upstream of the pmrB gene. PmrC (also known as eptA) is responsible for the biosynthesis of PEtN transferase that is regulated by the response regulator PmrA (21). Hence, the upregulation of PmrA by PmrB mutants activates both pmrC and pbgP, resulting in both PEtN and Ara4N modification of the LPS (Fig. 2) (16, 73). The deletion of pmrAB decreases the expression of pmrC and pbgP operon and colistin susceptibility (Table 2) (22).
TABLE 2.
Emerging genomic and transcriptional mechanisms mediating polymyxin resistance mechanisms in Enterobacteriaceae
| Resistance mechanism | Species | Resistance determinant(s) | Reference(s) |
|---|---|---|---|
| Efflux pumps | Escherichia coli | AcrAB | 150 |
| MarA, AcrAB | 151 | ||
| Enterobacter cloacae complex | TolC, SoxRS | 152 | |
| Klebsiella pneumoniae | RamA, SoxS | 80 | |
| Salmonella enterica serovar Typhimurium | AcrB, CpxR | 153 | |
| mcr-type genes | Citrobacter braakii | MCR-1 | 154 |
| Citrobacter freundii | MCR-1 | 155 | |
| Escherichia coli | MCR-1 | 19, 82, 85, 93, 155–175 | |
| MCR-2 | 176 | ||
| MCR-3 | 162, 169, 177, 178 | ||
| MCR-5 | 95, 96 | ||
| MCR-9 | 98 | ||
| Enterobacteria cloacae complex | MCR-4.3 | 97 | |
| Klebsiella pneumoniae | MCR-1 | 25, 174, 179 | |
| MCR-7.1 | 94 | ||
| MCR-8 | 180 | ||
| Salmonella enterica | MCR-1 | 181 | |
| MCR-2 | 181 | ||
| MCR-9 | 92 | ||
| Salmonella enterica serovar Paratyphi B | MCR-5 | 99 | |
| Salmonella enterica serovar Typhimurium | MCR-9 | 91 | |
| Shigella sonnei | MCR-1 | 182 | |
| Shigella flexneri | MCR-1 | 183 | |
| TCS PEtN modification | Citrobacter | PhoPQ-PmrAB | 184 |
| Escherichia coli | PhoPQ-PmrAB | 18, 21, 23, 24, 68, 185 | |
| QseBC-PmrAB | 98, 186, 187 | ||
| Enterobacter | PhoPQ-PmrAB | 76, 184 | |
| Klebsiella pneumoniae | PhoPQ-PmrAB | 69, 71, 81, 188–190 | |
| CrrAB | 16, 22, 73–75, 77 | ||
| Salmonella enterica serovar Typhimurium | PhoPQ-PmrAB | 72, 191 | |
| Yersinia pestis | PhoPQ | 192 | |
| PEtN modification | Escherichia coli | MgrB | 68, 82, 83, 86 |
| Klebsiella pneumoniae | MgrB | 7, 25, 133, 193–197 | |
| Escherichia coli, Klebsiella pneumoniae, and Salmonella enterica | EptA | 181, 185 | |
| Escherichia coli | EptA, EptB, EptC | 198 | |
| Salmonella enterica serovar Typhimurium | MgrB, SroC, EptB | 199 | |
| Membrane permeability | Salmonella enterica serovar Typhimurium | Mig-14 | 200 |
| Unknown mechanisms | Salmonella enteritidis | Not applicable | 201 |
CrrAB TWO-COMPONENT REGULATORY SYSTEM
The PmrAB regulatory system is regulated by the CrrAB two-component regulatory system, which encodes a sensor kinase (crrB), a regulatory protein (crrA), and a modulator (crrC) that regulates the pbgP operon (Fig. 2) (16, 22). An increase in crrC expression, mediated by crrB mutations, increases PEtN and Ara4N modifications through the pbgP operon and the pmrC gene, respectively (22, 74). Jayol et al. (74) identified four crrB mutations—an F84S mutation within the HAMP domain and N141Y, P151L, and G183V mutations within the histidine kinase A domain, respectively—in four K. pneumoniae isolates, which conferred high-level colistin resistance (74). Each isolate had a colistin MIC value of >128 μg/ml and, when transformed with a plasmid with an intact crrB gene, colistin susceptibility was restored (74).
CrrAB regulates pmrAB operon through crrC expression, and this allows for the activation of both pmrC genes and pbgP operon through pmrA (22, 75). CrrC acts as a connection protein between CrrAB and PmrAB and is regulated by crrA (75). Cheng et al. showed that crrB mutations also result in an increased expression of H239_3064, a putative efflux pump, resulting in a reduced polymyxin susceptibility (Table 2) (75).
PhoPQ TWO-COMPONENT REGULATORY SYSTEM
The PhoPQ system encodes a regulatory protein (PhoP) and a membrane-bound sensor kinase (PhoQ); LPS modification is mediated through PhoQ activation of PhoP via phosphorylation (71). Activated PhoP either binds directly to pbgP operon or indirectly by binding to PmrD, a connector protein of PhoPQ and the PmrAB system, which protects PmrA from dephosphorylation by PmrB kinase (Fig. 2) (21, 74). The PmrD protein is, however, not found in all Enterobacteriaceae species, being mainly found in E. coli, S. enterica, and K. pneumoniae (76). In E. coli, however, PmrD does not connect the two regulatory systems (77).
Similar to PmrB, the cytoplasmic membrane-bound kinase, PhoQ, is activated by environmental stimulants such as a low concentration of magnesium (Mg2+) and calcium (Ca2+) (69). Jayol et al. showed that an phoP mutation, Asp191Tyr, which caused significant modification to the secondary structure of the protein interrupting the α-helix, resulted in elevated colistin resistance (MIC of 12 μg/ml). A phoP mutation upregulated phoP, phoQ, pmrD, and pmrK (69). Cain et al. demonstrated that a single mutation (K46Q) in the phoQ phosphate domain resulted in the loss-of-function of PhoQ, resulting in colistin resistance during serial passage of K. pneumoniae (16). Site-directed mutagenesis in phoQ (Leu26Pro) performed by Cheng et al. in K. pneumoniae resulted in elevated colistin resistance with a 32-fold increase in the MIC (Table 2) (68).
Huang et al. (78) investigated the polymyxin resistance mechanisms of heterogeneously resistant E. cloacae using Tn5 mutagenesis. These authors found that mutations within the DedA protein may mediate heteroresistance to polymyxin through the PhoPQ system (78). The DedA protein is part of a superfamily of membrane proteins and is proposed to be a substrate of the protein-motive-force-dependent drug efflux (78, 79). Though its role in the PhoPQ system was not investigated, Huang et al. found that Tn5 insertion mutations within the dedA gene resulted in E. cloacae susceptibility to polymyxin (MIC of 1 mg/liter). The complementation of dedAEcl mutants with plasmids carrying phoP-phoQ or wild-type dedAEcl with its natural promoters restored the heteroresistance phenotype (MIC of 256 mg/liter) (78).
The polymyxin heteroresistance seen in E. cloacae was, however, mediated by a new small transmembrane protein-encoding gene, ecr, which was hypothesized to activate the pbgP operon via the PhoPQ system (78). The transformation of Enterobacter mori strain A6008 with ecr on a pCR-BluntII-TOPO vector conferred high-level resistance to colistin (MIC 256 mg/liter) and resulted in significant changes in the expression profile of the pbgP operon and PhoPQ system (78). Ecr is suspected to act on the PhoPQ system, activating the pbgP operon and increasing LPS modification. The introduction of ecr into A6008 further resulted in the upregulation of tolC and dedA expression. Ecr was found to be widely spread in the Enterobacter genus, and thus its role in mediating resistance should further be investigated (78).
The mgrB gene encodes a small transmembrane lipoprotein that is responsible for the negative regulation of the PhoPQ system by inhibiting the kinase activity of PhoQ (73, 80). Multiple studies have shown that the deletion or inactivation of mgrB leads to the upregulation of the phoPQ operon, resulting in enhanced LPS modification in K. pneumoniae (Fig. 2) (68, 81, 82). The mgrB gene has been reported to be inactivated through various mutations such as deletion, nonsense, missense, and insertional mutations (83–85). Formosa et al. (86) investigated the difference in the surface properties of the extracellular capsule using atomic force microscopy in K. pneumoniae isolates with or without polymyxin. The capsule of the K. pneumoniae isolates with an inactivated mgrB gene was tightly bound to the bacterial cell wall and, when exposed to polymyxin, the capsule became harder with increasing concentration. In contrast, polymyxin was able to remove the capsule from K. pneumoniae isolates with an intact mgrB gene, resulting in lysis (86). This study demonstrated that inactivation of the mgrB gene directly affects the organization of capsules during polymyxin exposure (86).
MOBILIZED POLYMYXIN RESISTANCE (mcr) GENES
The second type of LPS modification seen to achieve polymyxin resistance is the transfer of phosphoethanolamine (PEtN) mediated by PmrC and mobilized colistin resistance (mcr-type) genes (21, 82). mcr-type genes are plasmid-mediated (some have also been detected on chromosomes) genes that encode enzymes that modify lipid A through the addition of PEtN (23, 87). This phenotype was observed in mcr-positive E. coli isolates using mass spectrometry with a PEtN-modified lipid A peak at m/z 1,919 instead of m/z 1,796 in mcr-negative E. coli isolates (87–89). These genes are responsible for the horizontal transfer of polymyxin resistance in Enterobacteriaceae through mobile genetic elements (MGEs) (23, 90). To date, there are 10 mcr genes that have been identified, with mcr-1 genes being the most prevalent and predominantly found in E. coli (91, 92). The transformation of E. coli ST7314 isolates with an mcr-1-bearing plasmid resulted in a 32-fold increase in colistin MIC (85, 93). The acquisition of mcr-type genes, therefore, has a significant clinical impact by conferring high-level resistance to colistin (87). Sato et al. (18) showed that colistin-resistant E. coli had elevated eptA and arnT expression levels compared to colistin-susceptible E. coli ST131 isolates and exhibited PEtN modifications.
The decrease in polymyxin susceptibility is seen across most mcr genes, including mcr-7.1 isolated from K. pneumoniae in chickens (94), mcr-5.1 isolated from E. coli in retail chicken rice (95), and mcr-5 isolated from pigs in E. coli (96). mcr-4.3, identified in a clinical E. cloacae isolate in China, was found to not confer polymyxin resistance (97). Chavda et al. (97) compared mcr-4.3 to mcr-4 and identified two amino acid substitutions in mcr-4.3 that significantly altered the function of mcr-4, resulting in no modifications to lipid A. mcr-9, however, remains the most identified variant after mcr-1 and is common in several Enterobacteriales species, although it is particularly common in Enterobacter hormaechei and other Enterobacter sp. (205).
mcr-type genes may sometimes be polymyxin induced; Kieffer et al. (98) showed that mcr-9 mRNA expression was induced by colistin, where an increase in colistin concentration increased the number of mcr-9 transcripts (98). This feature, however, was only seen with mcr-9 genes and not with mcr-1 and was reported to be regulated by the two-component system located downstream of the mcr-9 gene (98). In Salmonella Paratyphi B, Borowiak et al. (99) showed that an increase in plasmid copy number resulted in a higher degree of colistin resistance. Sun et al. (93) showed that one plasmid copy number results in PEtN modification and, subsequently, a reduced polymyxin susceptibility in E. coli isolates; moreover, Zhang et al. (206) demonstrated that the plasmid types hosting the mcr gene also affects mcr expression and polymyxin resistance.
Kieffer et al. (98) and Cha et al. (92) explored the genomic context of mcr-9 genes in an E. coli 68A strain and a Salmonella enterica isolate and found that the inducible expression and transferability of mcr-9 genes was due to the QseC-QseB two-component system. In the E. coli isolate the mcr-9 gene was located on a IncH2 plasmid, and in S. enterica it was located on an IncX1 plasmid. On both plasmids, the mcr-9 gene was located between two insertion sequences, and qseC and qseB genes were located downstream of the mcr-9 gene (92, 98).
Though a high level of resistance is observed in most mcr-positive isolates, Zhang et al. (25) reported that it does not confer the same level of resistance to polymyxin as an inactivated mgrB gene. Zhang et al. (25) further reported that the transformation of inactivated mgrB K. pneumoniae isolates with an mcr-harboring plasmid does not result in a synergistic activity i.e., no change in polymyxin MIC value. Sato et al. (18) and Kieffer et al. (98) both showed that mcr-type genes do not affect PmrAB genes; thus, only PEtN modifications are observed in mcr-positive isolates (Fig. 2).
POLYMYXIN RESISTANCE INDUCTION
In polymyxin inducing environments, mcr-negative isolates acquire resistance through mutations that increase the expression of PhoPQ and PmrAB regulatory systems (71). Mutations within the phoQ and insertions in mgrB have been identified in K. pneumoniae isolates, resulting in the upregulation of phoQ, pmrD, and pbgP (71). A K. pneumoniae isolate was identified with crrB mutations resulting in increased pmrB expression (71). A serial passage performed by Cain et al. (16) reported a change in membrane permeability, contributing to polymyxin resistance. The expression of several efflux pumps—BN373_11321 (RND-family), BN373_15271 (MacA), and BN373_26531 and BN373_36071 (RND-family)—was increased under polymyxin selection (16). These efflux pumps have been previously reported to act as multidrug transporters (16). The outer membrane porins OmpA and OmpC that allow for active antibiotic uptake were significantly decreased in K. pneumoniae isolates (16). Although these mechanisms were not actively investigated, the change in membrane permeability may result in the reduction of polymyxin susceptibility (Table 2).
CONCLUSION
The misuse of antibiotics creates a selection pressure, resulting in mutations or transmission of resistance genes mediating antibiotic resistance. This is mainly seen in β-lactamase- and mcr-negative Enterobacteriaceae isolates that mutate and acquire high-level resistance in antibiotic-inducing environments through target modifications, overexpression of efflux pumps, and loss of major porin groups. This emphasizes the need for the correct usage of carbapenem and polymyxin antibiotics during therapy to ensure therapeutic success instead of the production of resistant clinical isolates. This review provides an in-depth molecular characterization of current and emerging resistance mechanisms that mediate carbapenem and polymyxin resistance. The mechanisms, however, of RamA, an efflux pump regulator, and micC and micF, which are small RNAs, in downregulating the major porin groups is unknown. Thus, further research into factors influencing this phenotype through these negative regulators is required. The role of micC and micF also shows the importance of sRNA and siRNA in gene regulation and antimicrobial resistance. Hence, studies investigating the global role of these small regulatory RNAs in microbial resistance is needed.
Within Enterobacterales, members of the tribe Proteeae and Serratia sp. are known to be intrinsically resistant to polymyxin. Mechanisms mediating resistance to these species were not discussed here since they were not found in the included articles. Future studies might interrogate transcriptional, coding, and noncoding genetic elements mediating intrinsic resistance in these clinically important species. Finally, studies demonstrating the transcriptional effects of noncoding and coding genetic elements in OXA-48 carbapenemase-mediated carbapenem resistance were not included in this review because they did not meet the inclusion criteria. Hence, further transcriptional and mutagenesis studies are required to confirm the effects of noncoding and coding genetic elements on the transcriptional levels and subsequent phenotypic resistance levels of OXA-48-type carbapenemases in Enterobacterales.
REFERENCES
- 1.van Boxtel R, Wattel AA, Arenas J, Goessens WHF, Tommassen J. 2017. Acquisition of carbapenem resistance by plasmid-encoded-AmpC-expressing Escherichia coli. Antimicrob Agents Chemother 61:01413-16. doi: 10.1128/AAC.01413-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamzaoui Z, Ocampo-Sosa A, Maamar E, Fernandez Martinez M, Ferjani S, Hammami S, Harbaoui S, Genel N, Arlet G, Saidani M, Slim A, Boutiba-Ben Boubaker I, Martinez-Martinez L. 2018. An outbreak of NDM-1-producing Klebsiella pneumoniae, associated with OmpK35 and OmpK36 porin loss in Tunisia. Microb Drug Resist 24:1137–1147. doi: 10.1089/mdr.2017.0165. [DOI] [PubMed] [Google Scholar]
- 3.Saw HTH, Webber MA, Mushtaq S, Woodford N, Piddock LJV. 2016. Inactivation or inhibition of AcrAB-TolC increases resistance of carbapenemase-producing Enterobacteriaceae to carbapenems. J Antimicrob Chemother 71:1510–1519. doi: 10.1093/jac/dkw028. [DOI] [PubMed] [Google Scholar]
- 4.Chetri S, Singha M, Bhowmik D, Nath K, Chanda DD, Chakravarty A, Bhattacharjee A. 2019. Transcriptional response of OmpC and OmpF in Escherichia coli against differential gradient of carbapenem stress. BMC Res Notes 12:138. doi: 10.1186/s13104-019-4177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 6.Iovleva A, Mettus RT, McElheny CL, Griffith MP, Mustapha MM, Pasculle AW, Shields RK, Cooper VS, Doi Y. 2019. High-level carbapenem resistance in OXA-232-producing Raoultella ornithinolytica triggered by ertapenem therapy. Antimicrob Agents Chemother 64:01335-19. doi: 10.1128/AAC.01335-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaidane N, Bonnin RA, Mansour W, Girlich D, Creton E, Cotellon G, Chaouch C, Boujaafar N, Bouallegue O, Naas T. 2017. Genomic insights into colistin-resistant Klebsiella pneumoniae from a Tunisian teaching hospital. Antimicrob Agents Chemother 62:e01601-17. doi: 10.1128/AAC.01601-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou K, Luo Q, Wang Q, Huang C, Lu H, Rossen JWA, Xiao Y, Li L. 2018. Silent transmission of an IS1294b-deactivated mcr-1 gene with inducible colistin resistance. Int J Antimicrob Agents 51:822–828. doi: 10.1016/j.ijantimicag.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Ye Y, Xu L, Han Y, Chen Z, Liu C, Ming L. 2018. Mechanism for carbapenem resistance of clinical Enterobacteriaceae isolates. Exp Ther Med 15:1143–1149. doi: 10.3892/etm.2017.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potter RF, Wallace MA, McMullen AR, Prusa J, Stallings CL, Burnham CAD, Dantas G. 2018. blaIMP-27 on transferable plasmids in Proteus mirabilis and Providencia rettgeri. Clin Microbiol Infect 24:1019.e5–1019.e8. doi: 10.1016/j.cmi.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu W, Espedido B, Feng Y, Zong Z. 2016. Citrobacter freundii carrying blaKPC-2 and blaNDM-1: characterization by whole-genome sequencing. Sci Rep 6:30670. doi: 10.1038/srep30670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta S, Mitra S, Chattopadhyay P, Som T, Mukherjee S, Basu S. 2017. Spread and exchange of blaNDM-1 in hospitalized neonates: role of mobilizable genetic elements. Eur J Clin Microbiol Infect Dis 36:255–265. doi: 10.1007/s10096-016-2794-6. [DOI] [PubMed] [Google Scholar]
- 13.Streling AP, Barbosa PP, Marcondes MF, Nicoletti AG, Picao RC, Pinto EC, Marques EA, Oliveira V, Gales AC. 2018. Genetic and biochemical characterization of GES-16, a new GES-type beta-lactamase with carbapenemase activity in Serratia marcescens. Diagn Microbiol Infect Dis 92:147–151. doi: 10.1016/j.diagmicrobio.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Osei Sekyere J, Govinden U, Bester LA, Essack SY. 2016. Colistin and tigecycline resistance in carbapenemase-producing Gram-negative bacteria: emerging resistance mechanisms and detection methods. J Appl Microbiol 121:601–617. doi: 10.1111/jam.13169. [DOI] [PubMed] [Google Scholar]
- 15.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. 2017. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother 72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 16.Cain AK, Boinett CJ, Barquist L, Dordel J, Fookes M, Mayho M, Ellington MJ, Goulding D, Pickard D, Wick RR, Holt KE, Parkhill J, Thomson NR. 2018. Morphological, genomic, and transcriptomic responses of Klebsiella pneumoniae to the last-line antibiotic colistin. Sci Rep 8:9868–9868. doi: 10.1038/s41598-018-28199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zurfluh K, Nuesch-Inderbinen M, Klumpp J, Poirel L, Nordmann P, Stephan R. 2017. Key features of mcr-1-bearing plasmids from Escherichia coli isolated from humans and food. Antimicrob Resist Infect Control 6:91. doi: 10.1186/s13756-017-0250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato T, Shiraishi T, Hiyama Y, Honda H, Shinagawa M, Usui M, Kuronuma K, Masumori N, Takahashi S, Tamura Y, Yokota SI. 2018. Contribution of novel amino acid alterations in PmrA or PmrB to colistin resistance in mcr-negative Escherichia coli clinical isolates, including major multidrug-resistant lineages O25b:H4-ST131-H30Rx and Non-x. Antimicrob Agents Chemother 62:e00864-18. doi: 10.1128/AAC.00864-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu F, Zhang R, Yang Y, Li H, Wang J, Lan J, Li P, Zhu Y, Xie Z, Jiang S. 2020. Occurrence and molecular characteristics of mcr-1-positive Escherichia coli from healthy meat ducks in Shandong Province of China. Animals (Basel) 10:1299. doi: 10.3390/ani10081299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li R, Yu H, Xie M, Chen K, Dong N, Lin D, Chan EW-C, Chen S. 2018. Genetic basis of chromosomally-encoded mcr-1 gene. Int J Antimicrob Agents 51:578–585. doi: 10.1016/j.ijantimicag.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 21.Phan M-D, Nhu NTK, Achard MES, Forde BM, Hong KW, Chong TM, Yin W-F, Chan K-G, West NP, Walker MJ, Paterson DL, Beatson SA, Schembri MA. 2017. Modifications in the pmrB gene are the primary mechanism for the development of chromosomally encoded resistance to polymyxins in uropathogenic Escherichia coli. J Antimicrob Chemother 72:2729–2736. doi: 10.1093/jac/dkx204. [DOI] [PubMed] [Google Scholar]
- 22.Cheng YH, Lin TL, Lin YT, Wang JT. 2016. Amino acid substitutions of CrrB responsible for resistance to colistin through CrrC in Klebsiella pneumoniae. Antimicrob Agents Chemother 60:3709–3716. doi: 10.1128/AAC.00009-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cannatelli A, Giani T, Aiezza N, Di Pilato V, Principe L, Luzzaro F, Galeotti CL, Rossolini GM. 2017. An allelic variant of the PmrB sensor kinase responsible for colistin resistance in an Escherichia coli strain of clinical origin. Sci Rep 7:5071. doi: 10.1038/s41598-017-05167-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jayol A, Nordmann P, Andre C, Dubois V, Poirel L. 2017. Increased colistin resistance upon acquisition of the plasmid-mediated mcr-1 gene in Escherichia coli isolates with chromosomally encoded reduced susceptibility to polymyxins. Int J Antimicrob Agents 50:503–504. doi: 10.1016/j.ijantimicag.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Zhao D, Shi Q, Quan J, Li X, Yu Y. 2018. mcr-1 gene has no effect on colistin resistance when it coexists with inactivated mgrB gene in Klebsiella pneumoniae. Microb Drug Resist 24:1117–1120. doi: 10.1089/mdr.2017.0291. [DOI] [PubMed] [Google Scholar]
- 26.Gharaibeh MH, Shatnawi SQ. 2019. An overview of colistin resistance, mobilized colistin resistance genes dissemination, global responses, and the alternatives to colistin: a review. Vet World 12:1735–1746. doi: 10.14202/vetworld.2019.1735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quiroga C, Nastro M, Di Conza J. 2019. Current scenario of plasmid-mediated colistin resistance in Latin America. Rev Argent Microbiol 51:93–100. doi: 10.1016/j.ram.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Suay-García B, Pérez-Gracia MT. 2019. Present and future of carbapenem-resistant Enterobacteriaceae (CRE) infections. Antibiotics 8:122. doi: 10.3390/antibiotics8030122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goering R, Dockrell H, Zuckerman M, Chiodini P. 2018. Mims’ medical microbiology and immunology, 6th ed. Elsevier, New York, NY. [Google Scholar]
- 30.Jacoby GA. 2009. AmpC β-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangat CS, Boyd D, Janecko N, Martz S-L, Desruisseau A, Carpenter M, Reid-Smith RJ, Mulvey MR. 2016. Characterization of VCC-1, a novel Ambler class A carbapenemase from Vibrio cholerae isolated from imported retail shrimp sold in Canada. Antimicrob Agents Chemother 60:1819–1825. doi: 10.1128/AAC.02812-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalmolin TV, Bianchini BV, Rossi GG, Ramos AC, Gales AC, Trindade PA, de Campos MMA. 2017. Detection and analysis of different interactions between resistance mechanisms and carbapenems in clinical isolates of Klebsiella pneumoniae. Braz J Microbiol 48:493–498. doi: 10.1016/j.bjm.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams-Sapper S, Gayoso A, Riley LW. 2018. Stress-adaptive responses associated with high-level carbapenem resistance in KPC-producing Klebsiella pneumoniae. J Pathog 2018:3028290. doi: 10.1155/2018/3028290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Somboro AM, Osei Sekyere J, Amoako DG, Essack SY, Bester LA. 2018. Diversity and proliferation of metallo-β-lactamases: a clarion call for clinically effective metallo-β-lactamase inhibitors. Appl Environ Microbiol 84:e00698-18. doi: 10.1128/AEM.00698-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kopotsa K, Osei Sekyere J, Mbelle NM. 2019. Plasmid evolution in carbapenemase‐producing Enterobacteriaceae: a review. Ann N Y Acad Sci 1457:61–91. doi: 10.1111/nyas.14223. [DOI] [PubMed] [Google Scholar]
- 36.Osei Sekyere J, Govinden U, Essack S. 2016. The molecular epidemiology and genetic environment of carbapenemases detected in Africa. Microb Drug Resist 22:59–68. doi: 10.1089/mdr.2015.0053. [DOI] [PubMed] [Google Scholar]
- 37.Oueslati S, Iorga BI, Tlili L, Exilie C, Zavala A, Dortet L, Jousset AB, Bernabeu S, Bonnin RA, Naas T. 2019. Unravelling ceftazidime/avibactam resistance of KPC-28, a KPC-2 variant lacking carbapenemase activity. J Antimicrob Chemother 74:2239–2246. doi: 10.1093/jac/dkz209. [DOI] [PubMed] [Google Scholar]
- 38.Huang J, Ding H, Shi Y, Zhao Y, Hu X, Ren J, Huang G, Wu R, Zhao Z. 2018. Further spread of a blaKPC-harboring untypeable plasmid in Enterobacteriaceae in China. Front Microbiol 9:1938. doi: 10.3389/fmicb.2018.01938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dabos L, Rodriguez CH, Nastro M, Dortet L, Bonnin RA, Famiglietti A, Iorga BI, Vay C, Naas T. 2020. LMB-1-producing Citrobacter freundii from Argentina, a novel player in the field of MBLs. Int J Antimicrob Agents 55:105857. doi: 10.1016/j.ijantimicag.2019.11.014. [DOI] [PubMed] [Google Scholar]
- 40.Lisa M-N, Palacios AR, Aitha M, González MM, Moreno DM, Crowder MW, Bonomo RA, Spencer J, Tierney DL, Llarrull LI, Vila AJ. 2017. A general reaction mechanism for carbapenem hydrolysis by mononuclear and binuclear metallo-β-lactamases. Nat Commun 8:538–538. doi: 10.1038/s41467-017-00601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oueslati S, Retailleau P, Marchini L, Berthault C, Dortet L, Bonnin RA, Iorga BI, Naas T. 2020. Role of the arginine 214 in the substrate specificity of OXA-48. Antimicrob Agents Chemother 64. doi: 10.1128/AAC.02329-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iovleva A, Mettus RT, McElheny CL, Mustapha MM, Van Tyne D, Shields RK, Pasculle AW, Cooper VS, Doi Y. 2019. Reduced ceftazidime and ertapenem susceptibility due to production of OXA-2 in Klebsiella pneumoniae ST258. J Antimicrob Chemother 74:2203–2208. doi: 10.1093/jac/dkz183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar G, Issa B, Kar D, Biswal S, Ghosh AS. 2017. E152A substitution drastically affects NDM-5 activity. FEMS Microbiol Lett 364. doi: 10.1093/femsle/fnx008. [DOI] [PubMed] [Google Scholar]
- 44.Paul D, Garg A, Bhattacharjee A. 2017. Occurrence of blaNDM-1 and blaNDM-5 in a tertiary referral hospital of North India. Microb Drug Resist 23:815–821. doi: 10.1089/mdr.2016.0124. [DOI] [PubMed] [Google Scholar]
- 45.Liu Z, Wang Y, Walsh TR, Liu D, Shen Z, Zhang R, Yin W, Yao H, Li J, Shen J. 2017. Plasmid-mediated novel blaNDM-17 gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli strain. Antimicrob Agents Chemother 61:e02233-16. doi: 10.1128/AAC.02233-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheruvanky A, Stoesser N, Sheppard AE, Crook DW, Hoffman PS, Weddle E, Carroll J, Sifri CD, Chai W, Barry K, Ramakrishnan G, Mathers AJ. 2017. Enhanced Klebsiella pneumoniae carbapenemase expression from a novel Tn4401 deletion. Antimicrob Agents Chemother 61:e00025-17. doi: 10.1128/AAC.00025-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Segawa T, Sekizuka T, Suzuki S, Shibayama K, Matsui M, Kuroda M. 2018. The plasmid-encoded transcription factor ArdK contributes to the repression of the IMP-6 metallo-β-lactamase gene blaIMP-6, leading to a carbapenem-susceptible phenotype in the blaIMP-6-positive Escherichia coli strain A56-1S. PLoS One 13:e0208976. doi: 10.1371/journal.pone.0208976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choudhury NA, Paul D, Chakravarty A, Bhattacharjee A, Dhar Chanda D. 2018. IncX3 plasmid mediated occurrence of blaNDM-4 within Escherichia coli ST448 from India. J Infect Public Health 11:111–114. doi: 10.1016/j.jiph.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 49.Dupont H, Choinier P, Roche D, Adiba S, Sookdeb M, Branger C, Denamur E, Mammeri H. 2017. Structural alteration of OmpR as a source of ertapenem resistance in a CTX-M-15-producing Escherichia coli O25b:H4 sequence type 131 clinical isolate. Antimicrob Agents Chemother 61:e00014-17. doi: 10.1128/AAC.00014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pedersen T, Osei Sekyere J, Govinden U, Moodley K, Sivertsen A, Samuelsen O, Essack SY, Sundsfjord A. 2018. Spread of plasmid-encoded NDM-1 and GES-5 carbapenemases among extensively drug-resistant and pandrug-resistant clinical Enterobacteriaceae in Durban. Antimicrob Agents Chemother 62:e02178-17. doi: 10.1128/AAC.02178-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kong H-K, Pan Q, Lo W-U, Liu X, Law COK, Chan T-F, Ho P-L, Lau TC-K. 2018. Fine-tuning carbapenem resistance by reducing porin permeability of bacteria activated in the selection process of conjugation. Sci Rep 8:15248. doi: 10.1038/s41598-018-33568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bajaj H, Scorciapino MA, Moynie L, Page MGP, Naismith JH, Ceccarelli M, Winterhalter M. 2016. Molecular basis of filtering carbapenems by porins from beta-lactam-resistant clinical strains of Escherichia coli. J Biol Chem 291:2837–2847. doi: 10.1074/jbc.M115.690156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hao M, Ye M, Shen Z, Hu F, Yang Y, Wu S, Xu X, Zhu S, Qin X, Wang M. 2018. Porin deficiency in carbapenem-resistant Enterobacter aerogenes strains. Microb Drug Resist 24:1277–1283. doi: 10.1089/mdr.2017.0379. [DOI] [PubMed] [Google Scholar]
- 54.Gandra S, Choi J, McElvania E, Green SJ, Harazin M, Thomson RB, Dantas G, Singh KS, Das S. 2020. Faropenem resistance causes in vitro cross-resistance to carbapenems in ESBL-producing Escherichia coli. Int J Antimicrob Agents 55:105902. doi: 10.1016/j.ijantimicag.2020.105902. [DOI] [PubMed] [Google Scholar]
- 55.Adler M, Anjum M, Andersson DI, Sandegren L. 2016. Combinations of mutations in envZ, ftsI, mrdA, acrB, and acrR can cause high-level carbapenem resistance in Escherichia coli. J Antimicrob Chemother 71:1188–1198. doi: 10.1093/jac/dkv475. [DOI] [PubMed] [Google Scholar]
- 56.Senchyna F, Gaur RL, Sandlund J, Truong C, Tremintin G, Kültz D, Gomez CA, Tamburini FB, Andermann T, Bhatt A, Tickler I, Watz N, Budvytiene I, Shi G, Tenover FC, Banaei N. 2019. Diversity of resistance mechanisms in carbapenem-resistant Enterobacteriaceae at a health care system in Northern California, from 2013 to 2016. Diagn Microbiol Infect Dis 93:250–257. doi: 10.1016/j.diagmicrobio.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chung H-S, Yong D, Lee M. 2016. Mechanisms of ertapenem resistance in Enterobacteriaceae isolates in a tertiary university hospital. J Invest Med 64:1042–1049. doi: 10.1136/jim-2016-000117. [DOI] [PubMed] [Google Scholar]
- 58.Nuramrum S, Chanawong A, Lunha K, Lulitanond A, Sangka A, Wilailuckana C, Angkititrakul S, Charoensri N, Wonglakorn L, Chaimanee P, Chetchotisakd P. 2017. Molecular characterization of carbapenemase-nonproducing clinical isolates of Escherichia coli (from a Thai University hospital) with reduced carbapenem susceptibility. Jpn J Infect Dis 70:628–634. doi: 10.7883/yoken.JJID.2017.156. [DOI] [PubMed] [Google Scholar]
- 59.Chetri S, Dolley A, Bhowmik D, Chanda DD, Chakravarty A, Bhattacharjee A. 2018. Transcriptional response of AcrEF-TolC against fluoroquinolone and carbapenem in Escherichia coli of clinical origin. Indian J Med Microbiol 36:537–540. doi: 10.4103/ijmm.IJMM_18_308. [DOI] [PubMed] [Google Scholar]
- 60.Osei Sekyere J, Amoako DG. 2017. Carbonyl cyanide m-chlorophenylhydrazine (CCCP) reverses resistance to colistin, but not to carbapenems and tigecycline in multidrug-resistant Enterobacteriaceae. Front Microbiol 8:228–228. doi: 10.3389/fmicb.2017.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharma A, Gupta VK, Pathania R. 2019. Efflux pump inhibitors for bacterial pathogens: from bench to bedside. Indian J Med Res 149:129–145. doi: 10.4103/ijmr.IJMR_2079_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee JH, Bae IK, Lee CH, Jeong S. 2017. Molecular characteristics of first IMP-4-producing Enterobacter cloacae sequence type 74 and 194 in Korea. Front Microbiol 8:2343. doi: 10.3389/fmicb.2017.02343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chetri S, Singha K, Bhowmik D, Chanda DD, Chakravarty A, Bhattacharjee A. 2018. Subinhibitory concentration of ertapenem induces overexpression of regulator of antibiotic resistance A in Escherichia coli. Indian J Med Microbiol 36:569–571. doi: 10.4103/ijmm.IJMM_18_436. [DOI] [PubMed] [Google Scholar]
- 64.Philippe N, Maigre L, Santini S, Pinet E, Claverie J-M, Davin-Regli A-V, Pages J-M, Masi M. 2015. In vivo evolution of bacterial resistance in two cases of Enterobacter aerogenes infections during treatment with imipenem. PLoS One 10:e0138828. doi: 10.1371/journal.pone.0138828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pavez M, Vieira C, de Araujo MR, Cerda A, de Almeida LM, Lincopan N, Mamizuka EM. 2016. Molecular mechanisms of membrane impermeability in clinical isolates of Enterobacteriaceae exposed to imipenem selective pressure. Int J Antimicrob Agents 48:78–85. doi: 10.1016/j.ijantimicag.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 66.Pal T, Ghazawi A, Darwish D, Villa L, Carattoli A, Hashmey R, Aldeesi Z, Jamal W, Rotimi V, Al-Jardani A, Al-Abri SS, Sonnevend A. 2017. Characterization of NDM-7 carbapenemase-producing Escherichia coli isolates in the Arabian Peninsula. Microb Drug Resist 23:871–878. doi: 10.1089/mdr.2016.0216. [DOI] [PubMed] [Google Scholar]
- 67.Pal A, Dhara L, Tripathi A. 2019. Contribution of acrB upregulation and OmpC/OmpK36 loss over the presence of blaNDM towards carbapenem resistance development among pathogenic Escherichia coli and Klebsiella spp. Indian J Med Res 149:528–538. doi: 10.4103/ijmr.IJMR_716_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng Y-H, Lin T-L, Pan Y-J, Wang Y-P, Lin Y-T, Wang J-T. 2015. Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob Agents Chemother 59:2909–2913. doi: 10.1128/AAC.04763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jayol A, Nordmann P, Brink A, Poirel L. 2015. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother 59:2780–2784. doi: 10.1128/AAC.05055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aghapour Z, Gholizadeh P, Ganbarov K, Bialvaei AZ, Mahmood SS, Tanomand A, Yousefi M, Asgharzadeh M, Yousefi B, Kafil HS. 2019. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect Drug Resist 12:965–975. doi: 10.2147/IDR.S199844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choi M-J, Kim S, Ko KS. 2016. Pathways regulating the pbgP operon and colistin resistance in Klebsiella pneumoniae strains. J Microbiol Biotechnol 26:1620–1628. doi: 10.4014/jmb.1604.04016. [DOI] [PubMed] [Google Scholar]
- 72.Olaitan AO, Dia NM, Gautret P, Benkouiten S, Belhouchat K, Drali T, Parola P, Brouqui P, Memish Z, Raoult D, Rolain J-M. 2015. Acquisition of extended-spectrum cephalosporin-and colistin-resistant Salmonella enterica subsp. enterica serotype Newport by pilgrims during Hajj. Int J Antimicrob Agents 45:600–604. doi: 10.1016/j.ijantimicag.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 73.Yang T-Y, Wang S-F, Lin J-E, Griffith BTS, Lian S-H, Hong Z-D, Lin L, Lu P-L, Tseng S-P. 2020. Contributions of insertion sequences conferring colistin resistance in Klebsiella pneumoniae. Int J Antimicrob Agents 55:105894. doi: 10.1016/j.ijantimicag.2020.105894. [DOI] [PubMed] [Google Scholar]
- 74.Jayol A, Nordmann P, Brink A, Villegas M-V, Dubois V, Poirel L. 2017. High-level resistance to colistin mediated by various mutations in the crrB gene among carbapenemase-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 61:e01423-17. doi: 10.1128/AAC.01423-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng Y-H, Lin T-L, Lin Y-T, Wang J-T. 2018. A putative RND-type efflux pump, H239_3064, contributes to colistin resistance through CrrB in Klebsiella pneumoniae. J Antimicrob Chemother 73:1509–1516. doi: 10.1093/jac/dky054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guérin F, Isnard C, Sinel C, Morand P, Dhalluin A, Cattoir V, Giard J-C. 2016. Cluster-dependent colistin hetero-resistance in Enterobacter cloacae complex. J Antimicrob Chemother 71:3058–3061. doi: 10.1093/jac/dkw260. [DOI] [PubMed] [Google Scholar]
- 77.Kim SJ, Ko KS. 2018. Diverse genetic alterations responsible for post-exposure colistin resistance in populations of the same strain of Klebsiella pneumoniae. Int J Antimicrob Agents 52:425–429. doi: 10.1016/j.ijantimicag.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 78.Huang L, Feng Y, Zong Z. 2019. Heterogeneous resistance to colistin in Enterobacter cloacae complex due to a new small transmembrane protein. J Antimicrob Chemother 74:2551–2558. doi: 10.1093/jac/dkz236. [DOI] [PubMed] [Google Scholar]
- 79.Kumar S, Doerrler WT. 2014. Members of the conserved DedA family are likely membrane transporters and are required for drug resistance in Escherichia coli. Antimicrob Agents Chemother 58:923–930. doi: 10.1128/AAC.02238-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Naha S, Sands K, Mukherjee S, Roy C, Rameez MJ, Saha B, Dutta S, Walsh TR, Basu S. 2020. KPC-2-producing Klebsiella pneumoniae ST147 in a neonatal unit: clonal isolates with differences in colistin susceptibility attributed to AcrAB-TolC pump. Int J Antimicrob Agents 55:105903. doi: 10.1016/j.ijantimicag.2020.105903. [DOI] [PubMed] [Google Scholar]
- 81.Jasim R, Baker MA, Zhu Y, Han M, Schneider-Futschik EK, Hussein M, Hoyer D, Li J, Velkov T. 2018. A comparative study of outer membrane proteome between paired colistin-susceptible and extremely colistin-resistant Klebsiella pneumoniae strains. ACS Infect Dis 4:1692–1704. doi: 10.1021/acsinfecdis.8b00174. [DOI] [PubMed] [Google Scholar]
- 82.Hadjadj L, Riziki T, Zhu Y, Li J, Diene SM, Rolain J-M. 2017. Study of mcr-1 gene-mediated colistin resistance in Enterobacteriaceae isolated from humans and animals in different countries. Genes 8:394. doi: 10.3390/genes8120394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singh SK, Mishra M, Sahoo M, Patole S, Mohapatra H. 2017. Efflux-mediated colistin resistance in diverse clones of Klebsiella pneumoniae from aquatic environment. Microb Pathog 102:109–112. doi: 10.1016/j.micpath.2016.11.024. [DOI] [PubMed] [Google Scholar]
- 84.Haeili M, Javani A, Moradi J, Jafari Z, Feizabadi MM, Babaei E. 2017. MgrB alterations mediate colistin resistance in Klebsiella pneumoniae isolates from Iran. Front Microbiol 8:2470–2470. doi: 10.3389/fmicb.2017.02470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang Y-Q, Li Y-X, Song T, Yang Y-X, Jiang W, Zhang A-Y, Guo X-Y, Liu B-H, Wang Y-X, Lei C-W, Xiang R, Wang H-N. 2017. Colistin resistance gene mcr-1 and its variant in Escherichia coli isolates from chickens in China. Antimicrob Agents Chemother 61:e01204-16. doi: 10.1128/AAC.01204-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Formosa C, Herold M, Vidaillac C, Duval RE, Dague E. 2015. Unravelling of a mechanism of resistance to colistin in Klebsiella pneumoniae using atomic force microscopy. J Antimicrob Chemother 70:2261–2270. doi: 10.1093/jac/dkv118. [DOI] [PubMed] [Google Scholar]
- 87.Liu Y-Y, Chandler CE, Leung LM, McElheny CL, Mettus RT, Shanks RMQ, Liu J-H, Goodlett DR, Ernst RK, Doi Y. 2017. Structural modification of lipopolysaccharide conferred by mcr-1 in Gram-negative ESKAPE pathogens. Antimicrob Agents Chemother 61:e00580-17. doi: 10.1128/AAC.00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu B-T, Song F-J, Zou M, Zhang Q-D, Shan H. 2017. High incidence of Escherichia coli strains coharboring mcr-1 and blaNDM from chickens. Antimicrob Agents Chemother 61:e02347-16. doi: 10.1128/AAC.02347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu L, Feng Y, Zhang X, McNally A, Zong Z. 2017. New variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM-5. Antimicrob Agents Chemother 61:e01757-17. doi: 10.1128/AAC.01757-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim ES, Chong YP, Park S-J, Kim M-N, Kim S-H, Lee S-O, Choi S-H, Woo JH, Jeong J-Y, Kim YS. 2017. Detection and genetic features of MCR-1-producing plasmid in human Escherichia coli infection in South Korea. Diagn Microbiol Infect Dis 89:158–160. doi: 10.1016/j.diagmicrobio.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 91.Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. 2019. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. mBio 10:e00853-19. doi: 10.1128/mBio.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cha MH, Woo GJ, Lee W, Kim SH, Woo JH, Kim J, Ryu JG, Kwak HS, Chi YM. 2020. Emergence of transferable mcr-9 gene-carrying colistin-resistant Salmonella enterica Dessau ST14 isolated from retail chicken meat in Korea. Foodborne Pathog Dis 17:720–727. doi: 10.1089/fpd.2020.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun J, Li X-P, Fang L-X, Sun R-Y, He Y-Z, Lin J, Liao X-P, Feng Y, Liu Y-H. 2018. Co-occurrence of mcr-1 in the chromosome and on an IncHI2 plasmid: persistence of colistin resistance in Escherichia coli. Int J Antimicrob Agents 51:842–847. doi: 10.1016/j.ijantimicag.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 94.Yang Y-Q, Li Y-X, Lei C-W, Zhang A-Y, Wang H-N. 2018. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother 73:1791–1795. doi: 10.1093/jac/dky111. [DOI] [PubMed] [Google Scholar]
- 95.Guo S, Tay MYF, Thu AK, Seow KLG, Zhong Y, Ng LC, Schlundt J. 2019. Conjugative IncX1 plasmid harboring colistin resistance gene mcr-5.1 in Escherichia coli isolated from chicken rice retailed in Singapore. Antimicrob Agents Chemother 63:e01043-19. doi: 10.1128/AAC.01043-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hammerl JA, Borowiak M, Schmoger S, Shamoun D, Grobbel M, Malorny B, Tenhagen B-A, Kasbohrer A. 2018. mcr-5 and a novel mcr-5.2 variant in Escherichia coli isolates from food and food-producing animals, Germany, 2010 to 2017. J Antimicrob Chemother 73:1433–1435. doi: 10.1093/jac/dky020. [DOI] [PubMed] [Google Scholar]
- 97.Chavda B, Lv J, Hou M, Chavda KD, Kreiswirth BN, Feng Y, Chen L, Yu F. 2018. Coidentification of mcr-4.3 and blaNDM-1 in a clinical Enterobacter cloacae isolate from China. Antimicrob Agents Chemother 62:e00649-18. doi: 10.1128/AAC.00649-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kieffer N, Royer G, Decousser JW, Bourrel AS, Palmieri M, Ortiz De La Rosa JM, Jacquier H, Denamur E, Nordmann P, Poirel L. 2019. mcr-9, an inducible gene encoding an acquired phosphoethanolamine transferase in Escherichia coli, and its origin. Antimicrob Agents Chemother 63:e00965-19. doi: 10.1128/AAC.00965-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Borowiak M, Hammerl JA, Fischer J, Szabo I, Malorny B. 2017. Complete genome sequence of Salmonella enterica subsp. enterica serovar Paratyphi B sequence type 28 harboring mcr-1. Genome Announc 5:00991-17. doi: 10.1128/genomeA.00991-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Flury BB, Ellington MJ, Hopkins JL, Turton JF, Doumith M, Woodford N. 2016. The differential importance of mutations within AmpD in cephalosporin resistance of Enterobacter aerogenes and Enterobacter cloacae. Int J Antimicrobial Agents 48:555–558. doi: 10.1016/j.ijantimicag.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 101.Hamzaoui Z, Ocampo-Sosa A, Fernandez Martinez M, Landolsi S, Ferjani S, Maamar E, Saidani M, Slim A, Martinez-Martinez L, Boutiba-Ben Boubaker I. 2018. Role of association of OmpK35 and OmpK36 alteration and blaESBL and/or blaAmpC genes in conferring carbapenem resistance among non-carbapenemase-producing Klebsiella pneumoniae. Int J Antimicrob Agents 52:898–905. doi: 10.1016/j.ijantimicag.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 102.Samuelsen Ø, Hansen F, Aasnæs B, Hasman H, Lund BA, Leiros H-KS, Lilje B, Janice J, Jakobsen L, Littauer P, Søes LM, Holzknecht BJ, Andersen LP, Stegger M, Andersen PS, Hammerum AM. 2018. Dissemination and characteristics of a novel plasmid-encoded carbapenem-hydrolyzing class D β-lactamase, OXA-436, found in isolates from four patients at six different hospitals in Denmark. Antimicrob Agents Chemother 62:e01260-17. doi: 10.1128/AAC.01260-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhu B, Ying C, Xu H, Ying J. 2018. Coexistence of NDM-1-producing Escherichia coli and Citrobacter freundii in the same patient. J Glob Antimicrob Resist 15:79–81. doi: 10.1016/j.jgar.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 104.Liu Z, Li J, Wang X, Liu D, Ke Y, Wang Y, Shen J. 2018. Novel variant of New Delhi metallo-beta-lactamase, NDM-20, in Escherichia coli. Front Microbiol 9:248. doi: 10.3389/fmicb.2018.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rahman M, Mukhopadhyay C, Rai RP, Singh S, Gupta S, Singh A, Pathak A, Prasad KN. 2018. Novel variant NDM-11 and other NDM-1 variants in multidrug-resistant Escherichia coli from South India. J Glob Antimicrob Resist 14:154–157. doi: 10.1016/j.jgar.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 106.Novović K, Trudić A, Brkić S, Vasiljević Z, Kojić M, Medić D, Ćirković I, Jovčić B. 2017. Molecular epidemiology of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae in Serbia from 2013 to 2016. Antimicrob Agents Chemother 61:e02550-16. doi: 10.1128/AAC.02550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mancini S, Keller PM, Greiner M, Bruderer V, Imkamp F. 2019. Detection of NDM-19, a novel variant of the New Delhi metallo-β-lactamase with increased carbapenemase activity under zinc-limited conditions, in Switzerland. Diagn Microbiol Infect Dis 95:114851–114851. doi: 10.1016/j.diagmicrobio.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 108.Peng Z, Li X, Hu Z, Li Z, Lv Y, Lei M, Wu B, Chen H, Wang X. 2019. Characteristics of carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-1 and MCR-1 from pig farms in China. Microorganisms 7:482. doi: 10.3390/microorganisms7110482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Choudhury NA, Paul D, Das BJ, Dhar Chanda D, Bhattacharjee A. 2019. Adaptation of blaNDM through IncP plasmid within broad host range. Indian J Med Microbiol 37:527–530. doi: 10.4103/ijmm.IJMM_20_48. [DOI] [PubMed] [Google Scholar]
- 110.Sidjabat HE, Gien J, Kvaskoff D, Ashman K, Vaswani K, Reed S, McGeary RP, Paterson DL, Bordin A, Schenk G. 2018. The use of SWATH to analyze the dynamic changes of bacterial proteome of carbapenemase-producing Escherichia coli under antibiotic pressure. Sci Rep 8:3871. doi: 10.1038/s41598-018-21984-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lunha K, Chanawong A, Lulitanond A, Wilailuckana C, Charoensri N, Wonglakorn L, Saenjamla P, Chaimanee P, Angkititrakul S, Chetchotisakd P. 2016. High-level carbapenem-resistant OXA-48-producing Klebsiella pneumoniae with a novel OmpK36 variant and low-level, carbapenem-resistant, non-porin-deficient, OXA-181-producing Escherichia coli from Thailand. Diagn Microbiol Infect Dis 85:221–226. doi: 10.1016/j.diagmicrobio.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 112.Paul D, Ingti B, Bhattacharjee D, Maurya AP, Dhar D, Chakravarty A, Bhattacharjee A. 2017. An unusual occurrence of plasmid-mediated blaOXA-23 carbapenemase in clinical isolates of Escherichia coli from India. Int J Antimicrob Agents 49:642–645. doi: 10.1016/j.ijantimicag.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 113.Schauer J, Gatermann SG, Marschal M, Pfennigwerth N. 2019. Genetic and biochemical characterization of FRI-3, a novel variant of the Ambler class A carbapenemase FRI-1. J Antimicrob Chemother 74:2891–2894. doi: 10.1093/jac/dkz295. [DOI] [PubMed] [Google Scholar]
- 114.Kubota H, Uwamino Y, Matsui M, Sekizuka T, Suzuki Y, Okuno R, Uchitani Y, Ariyoshi T, Aoki W, Suzuki S, Kuroda M, Shinkai T, Yokoyama K, Sadamasu K, Funakoshi T, Murata M, Hasegawa N, Iwata S. 2018. FRI-4 carbapenemase-producing Enterobacter cloacae complex isolated in Tokyo, Japan. J Antimicrob Chemother 73:2969–2972. doi: 10.1093/jac/dky291. [DOI] [PubMed] [Google Scholar]
- 115.Brouwer MSM, Tehrani KHME, Rapallini M, Geurts Y, Kant A, Harders F, Mashayekhi V, Martin NI, Bossers A, Mevius DJ, Wit B, Veldman KT. 2019. Novel carbapenemases FLC-1 and IMI-2 encoded by an Enterobacter cloacae complex isolated from food products. Antimicrob Agents Chemother 63:e02338-18. doi: 10.1128/AAC.02338-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cuzon G, Bogaerts P, Bauraing C, Huang T-D, Bonnin RA, Glupczynski Y, Naas T. 2016. Spread of plasmids carrying multiple GES variants. Antimicrob Agents Chemother 60:5040–5043. doi: 10.1128/AAC.00360-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lange F, Pfennigwerth N, Hartl R, Kerschner H, Achleitner D, Gatermann SG, Kaase M. 2018. LMB-1, a novel family of class B3 MBLs from an isolate of Enterobacter cloacae. J Antimicrob Chemother 73:2331–2335. doi: 10.1093/jac/dky215. [DOI] [PubMed] [Google Scholar]
- 118.Nepal S, Bonn F, Grasso S, Stobernack T, de Jong A, Zhou K, Wedema R, Rosema S, Becher D, Otto A, Rossen JW, van Dijl JM, Bathoorn E. 2018. An ancient family of mobile genomic islands introducing cephalosporinase and carbapenemase genes in Enterobacteriaceae. Virulence 9:1377–1389. doi: 10.1080/21505594.2018.1509666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shi Z, Zhao H, Li G, Jia W. 2017. Molecular characteristics of carbapenem-resistant Enterobacter cloacae in Ningxia Province, China. Front Microbiol 8:94. doi: 10.3389/fmicb.2017.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kotsakis SD, Flach C-F, Razavi M, Larsson DGJ. 2018. Characterization of the first OXA-10 natural variant with increased carbapenemase activity. Antimicrob Agents Chemother 63:e01817-18. doi: 10.1128/AAC.01817-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wong JLC, Romano M, Kerry LE, Kwong H-S, Low W-W, Brett SJ, Clements A, Beis K, Frankel G. 2019. OmpK36-mediated carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo. Nat Commun 10:3957. doi: 10.1038/s41467-019-11756-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Barría-Loaiza C, Pincheira A, Quezada M, Vera A, Valenzuela P, Domínguez M, Lima CA, Araya I, Araya P, Prat S, Aguayo C, Fernández J, Hormazábal JC, Bello-Toledo H, González-Rocha G. 2016. Molecular typing and genetic environment of the blaKPC gene in Chilean isolates of Klebsiella pneumoniae. J Glob Antimicrob Resist 4:28–34. doi: 10.1016/j.jgar.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 123.Papagiannitsis CC, Di Pilato V, Giani T, Giakkoupi P, Riccobono E, Landini G, Miriagou V, Vatopoulos AC, Rossolini GM. 2016. Characterization of KPC-encoding plasmids from two endemic settings, Greece and Italy. J Antimicrob Chemother 71:2824–2830. doi: 10.1093/jac/dkw227. [DOI] [PubMed] [Google Scholar]
- 124.Chen Y, Marimuthu K, Teo J, Venkatachalam I, Cherng BPZ, De Wang L, Prakki SRS, Xu W, Tan YH, Nguyen LC, Koh TH, Ng OT, Gan Y-H. 2020. Acquisition of plasmid with carbapenem-resistance gene blaKPC2 in hypervirulent Klebsiella pneumoniae, Singapore. Emerg Infect Dis 26:549–559. doi: 10.3201/eid2603.191230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Castanheira M, Deshpande LM, Mills JC, Jones RN, Soave R, Jenkins SG, Schuetz AN. 2016. Klebsiella pneumoniae isolate from a New York City hospital belonging to sequence type 258 and carrying blaKPC-2 and blaVIM-4. Antimicrob Agents Chemother 60:1924–1927. doi: 10.1128/AAC.01844-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang J, Hu X, Zhao Y, Shi Y, Ding H, Xv J, Ren J, Wu R, Zhao Z. 2019. Genetic factors associated with enhanced blaKPC expression in Tn3/Tn4401 chimeras. Antimicrob Agents Chemother 64:e01836-19. doi: 10.1128/AAC.01836-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kubota H, Suzuki Y, Okuno R, Uchitani Y, Ariyoshi T, Takemura N, Mihara F, Mezaki K, Ohmagari N, Matsui M, Suzuki S, Sekizuka T, Kuroda M, Yokoyama K, Sadamasu K. 2019. IMP-68, a novel IMP-type metallo-β-lactamase in imipenem-susceptible Klebsiella pneumoniae. mSphere 4:e00736-19. doi: 10.1128/mSphere.00736-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sharma R, Patel S, Abboud C, Diep J, Ly NS, Pogue JM, Kaye KS, Li J, Rao GG. 2017. Polymyxin B in combination with meropenem against carbapenemase-producing Klebsiella pneumoniae: pharmacodynamics and morphological changes. Int J Antimicrob Agents 49:224–232. doi: 10.1016/j.ijantimicag.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Low YM, Chong CW, Yap IKS, Chai LC, Clarke SC, Ponnampalavanar S, Abdul Jabar K, Md Yusof MY, Teh CSJ. 2018. Elucidating the survival and response of carbapenem-resistant Klebsiella pneumoniae after exposure to imipenem at sublethal concentrations. Pathog Glob Health 112:378–386. doi: 10.1080/20477724.2018.1538281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee H, Shin J, Chung Y-J, Park M, Kang KJ, Baek JY, Shin D, Chung DR, Peck KR, Song J-H, Ko KS. 2020. Co-introduction of plasmids harboring the carbapenemase genes, blaNDM-1 and blaOXA-232, increases fitness and virulence of bacterial host. J Biomed Sci 27:8–8. doi: 10.1186/s12929-019-0603-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu Y, Zhang H, Zhang X, Jiang N, Zhang Z, Zhang J, Zhu B, Wang G, Zhao K, Zhou Y. 2019. Characterization of an NDM-19-producing Klebsiella pneumoniae strain harboring 2 resistance plasmids from China. Diagn Microbiol Infect Dis 93:355–361. doi: 10.1016/j.diagmicrobio.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 132.Duzgun AO. 2018. Effect of amino acid substitution in New Delhi metallo-beta-lactamase on carbapenem susceptibility. Acta Microbiol Immunol Hung 65:1–333. doi: 10.1556/030.65.2018.022. [DOI] [PubMed] [Google Scholar]
- 133.Sonnevend Á, Ghazawi A, Hashmey R, Haidermota A, Girgis S, Alfaresi M, Omar M, Paterson DL, Zowawi HM, Pál T. 2017. Multihospital occurrence of pan-resistant Klebsiella pneumoniae sequence type 147 with an ISEcp1-directed blaOXA-181 insertion in the mgrB gene in the United Arab Emirates. Antimicrob Agents Chemother 61:e00418-17. doi: 10.1128/AAC.00418-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dabos L, Bogaerts P, Bonnin RA, Zavala A, Sacré P, Iorga BI, Huang DT, Glupczynski Y, Naas T. 2018. Genetic and biochemical characterization of OXA-519, a novel OXA-48-like β-lactamase. Antimicrob Agents Chemother 62:e00469-18. doi: 10.1128/AAC.00469-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mancini S, Poirel L, Tritten M-L, Lienhard R, Bassi C, Nordmann P. 2018. Emergence of an MDR Klebsiella pneumoniae ST231 producing OXA-232 and RmtF in Switzerland. J Antimicrob Chemother 73:821–823. [DOI] [PubMed] [Google Scholar]
- 136.Ma P, Laibinis HH, Ernst CM, Hung DT. 2018. Carbapenem resistance caused by high-level expression of OXA-663 beta-lactamase in an OmpK36-deficient Klebsiella pneumoniae clinical isolate. Antimicrob Agents Chemother 62:e01281-18. doi: 10.1128/AAC.01281-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Esposito EP, Gaiarsa S, Del Franco M, Crivaro V, Bernardo M, Cuccurullo S, Pennino F, Triassi M, Marone P, Sassera D, Zarrilli R. 2017. A novel IncA/C1 group conjugative plasmid, encoding VIM-1 metallo-beta-lactamase, mediates the acquisition of carbapenem resistance in ST104 Klebsiella pneumoniae isolates from neonates in the intensive care unit of V. Monaldi Hospital in Naples. Front Microbiol 8:2135. doi: 10.3389/fmicb.2017.02135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shen X, Liu L, Yu J, Cao X, Zhan Q, Guo Y, Wang L, Yu F. 2019. Coexistence of blaNDM-1 and rmtC on a transferrable plasmid of a novel ST192 Klebsiella aerogenes clinical isolate. Infect Drug Resist 12:3883–3891. doi: 10.2147/IDR.S228130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Suzuki Y, Ida M, Kubota H, Ariyoshi T, Murakami K, Kobayashi M, Kato R, Hirai A, Suzuki J, Sadamasu K. 2019. Multiple β-lactam resistance gene-carrying plasmid harbored by Klebsiella quasipneumoniae isolated from urban sewage in Japan. mSphere 4:e00391-19. doi: 10.1128/mSphere.00391-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chetri S, Bhowmik D, Paul D, Pandey P, Chanda DD, Chakravarty A, Bora D, Bhattacharjee A. 2019. AcrAB-TolC efflux pump system plays a role in carbapenem non-susceptibility in Escherichia coli. BMC Microbiol 19:210–210. doi: 10.1186/s12866-019-1589-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wan Nur Ismah WAK, Takebayashi Y, Findlay J, Heesom KJ, Avison MB. 2018. Impact of OqxR loss of function on the envelope proteome of Klebsiella pneumoniae and susceptibility to antimicrobials. J Antimicrob Chemother 73:2990–2996. [DOI] [PubMed] [Google Scholar]
- 142.Pantel A, Dunyach-Remy C, Ngba Essebe C, Mesureur J, Sotto A, Pagès J-M, Nicolas-Chanoine M-H, Lavigne J-P. 2016. Modulation of membrane influx and efflux in Escherichia coli sequence type 131 has an impact on bacterial motility, biofilm formation, and virulence in a Caenorhabditis elegans model. Antimicrob Agents Chemother 60:2901–2911. doi: 10.1128/AAC.02872-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Scorciapino MA, D’Agostino T, Acosta-Gutierrez S, Malloci G, Bodrenko I, Ceccarelli M. 2016. Exploiting the porin pathway for polar compound delivery into Gram-negative bacteria. Future Med Chem 8:1047–1062. doi: 10.4155/fmc-2016-0038. [DOI] [PubMed] [Google Scholar]
- 144.Bialek-Davenet S, Mayer N, Vergalli J, Duprilot M, Brisse S, Pagès J-M, Nicolas-Chanoine M-H. 2017. In vivo loss of carbapenem resistance by extensively drug-resistant Klebsiella pneumoniae during treatment via porin expression modification. Sci Rep 7:6722–6722. doi: 10.1038/s41598-017-06503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kádár B, Kocsis B, Tóth Á, Kristóf K, Felső P, Kocsis B, Böddi K, Szabó D. 2017. Colistin resistance associated with outer membrane protein change in Klebsiella pneumoniae and Enterobacter asburiae. Acta Microbiol Immunol Hung 64:217–227. doi: 10.1556/030.64.2017.017. [DOI] [PubMed] [Google Scholar]
- 146.Vera-Leiva A, Carrasco-Anabalón S, Lima CA, Villagra N, Domínguez M, Bello-Toledo H, González-Rocha G. 2018. The efflux pump inhibitor phenylalanine-arginine β-naphthylamide (PAβN) increases resistance to carbapenems in Chilean clinical isolates of KPC-producing Klebsiella pneumoniae. J Glob Antimicrob Resist 12:73–76. doi: 10.1016/j.jgar.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 147.Lev AI, Astashkin EI, Shaikhutdinova RZ, Platonov ME, Kartsev NN, Volozhantsev NV, Ershova ON, Svetoch EA, Fursova NK. 2017. Identification of IS1R and IS10R elements inserted into ompk36 porin gene of two multidrug-resistant Klebsiella pneumoniae hospital strains. FEMS Microbiol Lett 364. doi: 10.1093/femsle/fnx072. [DOI] [PubMed] [Google Scholar]
- 148.Lange F, Pfennigwerth N, Höfken L-M, Gatermann SG, Kaase M. 2019. Characterization of mutations in Escherichia coli PBP2 leading to increased carbapenem MICs. J Antimicrob Chemother 74:571–576. doi: 10.1093/jac/dky476. [DOI] [PubMed] [Google Scholar]
- 149.Ranjitkar S, Reck F, Ke X, Zhu Q, McEnroe G, Lopez SL, Dean CR. 2019. Identification of mutations in the mrdA gene encoding PBP2 that reduce carbapenem and diazabicyclooctane susceptibility of Escherichia coli clinical isolates with mutations in ftsI (PBP3) and which carry blaNDM-1. mSphere 4:e00074-19. doi: 10.1128/mSphere.00074-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ramos PIP, Custódio MGF, Quispe Saji GDR, Cardoso T, da Silva GL, Braun G, Martins WMBS, Girardello R, de Vasconcelos ATR, Fernández E, Gales AC, Nicolás MF. 2016. The polymyxin B-induced transcriptomic response of a clinical, multidrug-resistant Klebsiella pneumoniae involves multiple regulatory elements and intracellular targets. BMC Genomics 17:737–737. doi: 10.1186/s12864-016-3070-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu J, Huang Z, Ruan B, Wang H, Chen M, Rehman S, Wu P. 2020. Quantitative proteomic analysis reveals the mechanisms of polymyxin B toxicity to Escherichia coli. Chemosphere 259:127449–127449. doi: 10.1016/j.chemosphere.2020.127449. [DOI] [PubMed] [Google Scholar]
- 152.Telke AA, Olaitan AO, Morand S, Rolain J-M. 2017. soxRS induces colistin hetero-resistance in Enterobacter asburiae and Enterobacter cloacae by regulating the acrAB-tolC efflux pump. J Antimicrob Chemother 72:2715–2721. doi: 10.1093/jac/dkx215. [DOI] [PubMed] [Google Scholar]
- 153.Zhai Y-J, Huang H, Liu J, Sun H-R, He D, Pan Y-S, Hu G. 2018. CpxR overexpression increases the susceptibility of acrB and cpxR double-deleted Salmonella enterica serovar Typhimurium to colistin. J Antimicrob Chemother 73:3016–3024. doi: 10.1093/jac/dky320. [DOI] [PubMed] [Google Scholar]
- 154.Liu J, Yang Y, Li Y, Liu D, Tuo H, Wang H, Call DR, Davis M, Zhang A. 2018. Isolation of an IncP-1 plasmid harboring mcr-1 from a chicken isolate of Citrobacter braakii in China. Int J Antimicrob Agents 51:936–940. doi: 10.1016/j.ijantimicag.2017.12.030. [DOI] [PubMed] [Google Scholar]
- 155.Hu Y-Y, Wang Y-L, Sun Q-L, Huang Z-X, Wang H-Y, Zhang R, Chen G-X. 2017. Colistin resistance gene mcr-1 in gut flora of children. Int J Antimicrob Agents 50:593–597. doi: 10.1016/j.ijantimicag.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 156.Wang X, Lu Q, Qi J, Chai Y, Wang Y, Gao GF. 2018. Structural and functional insights into MCR-2 mediated colistin resistance. Sci China Life Sci 61:1432–1436. [DOI] [PubMed] [Google Scholar]
- 157.Bai P-Y, Qin S-S, Chu W-C, Yang Y, Cui D-Y, Hua Y-G, Yang Q-Q, Zhang E. 2018. Synthesis and antibacterial bioactivities of cationic deacetyl linezolid amphiphiles. Eur J Med Chem 155:925–945. doi: 10.1016/j.ejmech.2018.06.054. [DOI] [PubMed] [Google Scholar]
- 158.Khalifa HO, Ahmed AM, Oreiby AF, Eid AM, Shimamoto TT, Shimamoto TT. 2016. Characterization of the plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli isolated from animals in Egypt. Int J Antimicrob Agents 47:413–414. [DOI] [PubMed] [Google Scholar]
- 159.Peng Z, Hu Z, Li Z, Li X, Jia C, Zhang X, Wu B, Chen H, Wang X. 2019. Characteristics of a colistin-resistant Escherichia coli ST695 harboring the chromosomally-encoded mcr-1 gene. Microorganisms 7:558. doi: 10.3390/microorganisms7110558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li H, Wang YY, Meng Q, Wang YY, Xia G, Xia X, Shen J. 2019. Comprehensive proteomic and metabolomic profiling of mcr-1-mediated colistin resistance in Escherichia coli. Int J Antimicrob Agents 53:795–804. doi: 10.1016/j.ijantimicag.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 161.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 162.Sun J, Xu Y, Gao R, Lin J, Wei W, Srinivas S, Li D, Yang R-S, Li X-P, Liao X-P, Liu Y-H, Feng Y. 2017. Deciphering MCR-2 colistin resistance. mBio 8:e00625-17. doi: 10.1128/mBio.00625-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Li B, Ke B, Zhao X, Guo Y, Wang W, Wang X, Zhu H. 2018. Antimicrobial resistance profile of mcr-1-positive clinical isolates of Escherichia coli in China from 2013 to 2016. Front Microbiol 9:2514. doi: 10.3389/fmicb.2018.02514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. 2016. Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob Agents Chemother 60:4394–4397. doi: 10.1128/AAC.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yassin AK, Zhang J, Wang J, Chen L, Kelly P, Butaye P, Lu G, Gong J, Li M, Wei L, Wang Y, Qi K, Han X, Price S, Hathcock T, Wang C. 2017. Identification and characterization of mcr-mediated colistin resistance in extraintestinal Escherichia coli from poultry and livestock in China. FEMS Microbiol Lett 364. doi: 10.1093/femsle/fnx242. [DOI] [PubMed] [Google Scholar]
- 166.Lee J-Y, Lim S-K, Choi Y, Moon D-C, Shin J, Ko KS. 2018. Whole sequences and characteristics of mcr-1-harboring plasmids of Escherichia coli strains isolated from livestock in South Korea. Microb Drug Resist 24:489–492. doi: 10.1089/mdr.2017.0369. [DOI] [PubMed] [Google Scholar]
- 167.Wu L, Chen J, Wang L, Wu Z. 2018. Whole-genome sequence of an MCR-1-carrying, extended-spectrum β-lactamase (ESBL)-producing Escherichia coli ST746 isolate recovered from a community-acquired urinary tract infection. J Glob Antimicrob Resist 13:171–173. doi: 10.1016/j.jgar.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 168.Moosavian M, Emam N, Pletzer D, Savari M. 2020. Rough-type and loss of the LPS due to lpx genes deletions are associated with colistin resistance in multidrug-resistant clinical Escherichia coli isolates not harboring mcr genes. PLoS One 15:e0233518. doi: 10.1371/journal.pone.0233518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li H, Yang L, Liu Z, Yin W, Liu D, Shen Y, Walsh T, Shao B, Wang Y. 2018. Molecular insights into functional differences between mcr-3- and mcr-1-mediated colistin resistance. Antimicrob Agents Chemother 62:e00366-18. doi: 10.1128/AAC.00366-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yu H, Qu F, Shan B, Huang B, Jia W, Chen C, Li A, Miao M, Zhang X, Bao C, Xu Y, Chavda KD, Tang Y-W, Kreiswirth BN, Du H, Chen L. 2016. Detection of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae from different hospitals in China. Antimicrob Agents Chemother 60:5033–5035. doi: 10.1128/AAC.00440-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zheng B, Huang C, Xu H, Guo L, Zhang J, Wang X, Jiang X, Yu X, Jin L, Li X, Feng Y, Xiao Y, Li L. 2017. Occurrence and genomic characterization of ESBL-producing, mcr-1-harboring Escherichia coli in farming soil. Front Microbiol 8:2510. doi: 10.3389/fmicb.2017.02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhu L, Zhou Z, Liu Y, Lin Z, Shuai X, Xu L, Chen H. 2020. Comprehensive understanding of the plasmid-mediated colistin resistance gene mcr-1 in aquatic environments. Environ Sci Technol 54:1603–1613. doi: 10.1021/acs.est.9b05919. [DOI] [PubMed] [Google Scholar]
- 173.Zurfluh K, Kieffer N, Poirel L, Nordmann P, Stephan R. 2016. Features of the mcr-1 cassette related to colistin resistance. Antimicrob Agents Chemother 60:6438–6439. doi: 10.1128/AAC.01519-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Huang B, He Y, Ma X, Cai R, Zeng J, Lu Y, Zhang W, Lan K, E S, Tang Y-W, Kreiswirth BN, Chen C, Chen L. 2018. Promoter variation and gene expression of mcr-1-harboring plasmids in clinical isolates of Escherichia coli and Klebsiella pneumoniae from a Chinese hospital. Antimicrob Agents Chemother 62:e00018-18. doi: 10.1128/AAC.00018-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhou Y, Wang T, Guo Y, Liu S, Wang J, Shen Y, Tang S, Wang Y, Deng X. 2018. In vitro/vivo activity of potential mcr-1 inhibitor in combination with colistin against mcr-1-positive Klebsiella pneumoniae. Front Microbiol 9:1615. doi: 10.3389/fmicb.2018.01615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang X, Wang Y, Wang Y, Zhang S, Shen Z, Wang S. 2018. Emergence of the colistin resistance gene mcr-1 and its variant in several uncommon species of Enterobacteriaceae from commercial poultry farm surrounding environments. Vet Microbiol 219:161–164. doi: 10.1016/j.vetmic.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 177.Yang QE, MacLean C, Papkou A, Pritchard M, Powell L, Thomas D, Andrey DO, Li M, Spiller B, Yang W, Walsh TR. 2020. Compensatory mutations modulate the competitiveness and dynamics of plasmid-mediated colistin resistance in Escherichia coli clones. ISME J 14:861–865. doi: 10.1038/s41396-019-0578-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang H, Zhao D, Quan J, Hua X, Yu Y. 2019. mcr-1 facilitated selection of high-level colistin-resistant mutants in Escherichia coli. Clin Microbiol Infect Dis 25:517.e1–517–e4.. doi: 10.1016/j.cmi.2018.12.014. [DOI] [PubMed] [Google Scholar]
- 179.Wang R, Liu Y, Zhang Q, Jin L, Wang Q, Zhang Y, Wang X, Hu M, Li L, Qi J, Luo Y, Wang H. 2018. The prevalence of colistin resistance in Escherichia coli and Klebsiella pneumoniae isolated from food animals in China: coexistence of mcr-1 and blaNDM with low fitness cost. Int J Antimicrob Agents 51:739–744. doi: 10.1016/j.ijantimicag.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 180.Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, Zhang S, Shen J, Shen Z, Wang Y. 2018. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect 7:122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Xu Y, Wei W, Lei S, Lin J, Srinivas S, Feng Y. 2018. An evolutionarily conserved mechanism for intrinsic and transferable polymyxin resistance. mBio 9:e02317-17. doi: 10.1128/mBio.02317-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Thanh DP, Tuyen HT, Nguyen TNT, The HC, Wick RR, Thwaites GE, Baker S, Holt KE. 2016. Inducible colistin resistance via a disrupted plasmid-borne mcr-1 gene in a 2008 Vietnamese Shigella sonnei isolate. J Antimicrob Chemother 71:2314–2317. doi: 10.1093/jac/dkw173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Liang B, Roberts AP, Xu X, Yang C, Yang X, Wang J, Yi S, Li Y, Ma Q, Wu F, Qiu S, Song H. 2018. Transferable plasmid-borne mcr-1 in a colistin-resistant Shigella flexneri isolate. Appl Environ Microbiol 84:e02655-17. doi: 10.1128/AEM.02655-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wand ME, Sutton JM. 2020. Mutations in the two component regulator systems PmrAB and PhoPQ give rise to increased colistin resistance in Citrobacter and Enterobacter spp. J Med Microbiol 69:521–529. doi: 10.1099/jmm.0.001173. [DOI] [PubMed] [Google Scholar]
- 185.Hjort K, Nicoloff H, Andersson DI. 2016. Unstable tandem gene amplification generates heteroresistance (variation in resistance within a population) to colistin in Salmonella enterica. Mol Microbiol 102:274–289. doi: 10.1111/mmi.13459. [DOI] [PubMed] [Google Scholar]
- 186.Breland EJ, Zhang EW, Bermudez T, Martinez CR III, Hadjifrangiskou M. 2017. The histidine residue of QseC is required for canonical signaling between QseB and PmrB in uropathogenic Escherichia coli. J Bacteriol 199:e00060-17. doi: 10.1128/JB.00060-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Guckes KR, Breland EJ, Zhang EW, Hanks SC, Gill NK, Algood HMS, Schmitz JE, Stratton CW, Hadjifrangiskou M. 2017. Signaling by two-component system noncognate partners promotes intrinsic tolerance to polymyxin B in uropathogenic Escherichia coli. Sci Signal 10:eaag1775. doi: 10.1126/scisignal.aag1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Borsa BA, Demirci M, Gungordu-Dalar Z, Karabiyik G, Aygun G, Kucukbasmaci O. 2019. Molecular mechanisms of colistin resistance among Klebsiella pneumoniae strains. Clin Lab 65. doi: 10.7754/Clin.Lab.2019.180705. [DOI] [PubMed] [Google Scholar]
- 189.Wand ME, Bock LJ, Sutton JM. 2017. Retention of virulence following colistin adaptation in Klebsiella pneumoniae is strain-dependent rather than associated with specific mutations. J Med Microbiol 66:959–964. doi: 10.1099/jmm.0.000530. [DOI] [PubMed] [Google Scholar]
- 190.Kidd TJ, Mills G, Sá-Pessoa J, Dumigan A, Frank CG, Insua JL, Ingram R, Hobley L, Bengoechea JA. 2017. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol Med 9:430–447. doi: 10.15252/emmm.201607336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hong S-K, Wu P-K, Park J-I. 2018. Gene expression kinetics governs stimulus-specific decoration of the Salmonella outer membrane. Sci Signal 42:11–20. doi: 10.1126/scisignal.aar7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fukuto HS, Vadyvaloo V, McPhee JB, Poinar HN, Holmes EC, Bliska JB. 2018. A single amino acid change in the response regulator PhoP, acquired during Yersinia pestis evolution, affects PhoP target gene transcription and polymyxin B susceptibility. J Bacteriol 200:e00050-18. doi: 10.1128/JB.00050-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Jayol A, Nordmann P, Desroches M, Decousser J-W, Poirel L. 2016. Acquisition of broad-spectrum cephalosporin resistance leading to colistin resistance in Klebsiella pneumoniae. Antimicrob Agents Chemother 60:3199–3201. doi: 10.1128/AAC.00237-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sun Q-L, Gu D, Wang Q, Hu Y, Shu L, Hu J, Zhang R, Chen G-X. 2019. Dynamic colonization of Klebsiella pneumoniae isolates in gastrointestinal tracts of intensive care patients. Front Microbiol 10:230. doi: 10.3389/fmicb.2019.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Silva MM, Fernandes MR, Sellera FP, Cerdeira L, Medeiros LKG, Garino F, Azevedo SS, Lincopan N. 2018. Multidrug-resistant CTX-M-15-producing Klebsiella pneumoniae ST231 associated with infection and persistent colonization of dog. Diagn Microbiol Infect Dis 92:259–261. doi: 10.1016/j.diagmicrobio.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 196.Matheeussen V, Xavier BB, Mermans I, De Weerdt A, Lammens C, Goossens H, Jansens H, Malhotra-Kumar S. 2019. Emergence of colistin resistance during treatment of recurrent pneumonia caused by carbapenemase producing Klebsiella pneumoniae. Diagn Microbiol Infect Dis 94:407–409. doi: 10.1016/j.diagmicrobio.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 197.Pishnian Z, Haeili M, Feizi A. 2019. Prevalence and molecular determinants of colistin resistance among commensal Enterobacteriaceae isolated from poultry in northwest of Iran. Gut Pathog 11:2–2. doi: 10.1186/s13099-019-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Salazar J, Alarcón M, Huerta J, Navarro B, Aguayo D. 2017. Phosphoethanolamine addition to the heptose I of the lipopolysaccharide modifies the inner core structure and has an impact on the binding of polymyxin B to the Escherichia coli outer membrane. Arch Biochem Biophys 620:28–34. doi: 10.1016/j.abb.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 199.Acuña LG, Barros MJ, Peñaloza D, Rodas PI, Paredes-Sabja D, Fuentes JA, Gil F, Calderón IL. 2016. A feed-forward loop between SroC and MgrR small RNAs modulates the expression of eptB and the susceptibility to polymyxin B in Salmonella Typhimurium. Microbiology (Reading) 162:1996–2004. doi: 10.1099/mic.0.000365. [DOI] [PubMed] [Google Scholar]
- 200.Sheng X, Wang W, Chen L, Zhang H, Zhang Y, Xu S, Xu H, Huang X. 2019. Mig-14 may contribute to Salmonella enterica serovar Typhi resistance to polymyxin B by decreasing the permeability of the outer-membrane and promoting the formation of biofilm. Int J Med Microbiol 309:143–150. doi: 10.1016/j.ijmm.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 201.Rule R, Mbelle N, Osei Sekyere J, Kock M, Hoosen A, Said M. 2019. A rare case of colistin-resistant Salmonella enteritidis meningitis in an HIV-seropositive patient. BMC Infect Dis 19:806–806. doi: 10.1186/s12879-019-4391-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kopotsa K, Mbelle NM, Osei Sekyere J. 10 November 2020. Epigenomics, genomics, resistome, mobilome, virulome and evolutionary phylogenomics of carbapenem-resistant Klebsiella pneumoniae clinical strains. Microb Genomics doi: 10.1099/mgen.0.000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Osei Sekyere J, Reta MA. 2020. Genomic and resistance epidemiology of Gram-negative bacteria in Africa: a systematic review and phylogenomic analyses from a One Health perspective. mSystems 5:e00897-20. doi: 10.1128/mSystems.00897-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Cannatelli A, Giani T, Aiezza N, Di Pilato V, Principe L, Luzzaro F, Galeotti CL, Rossolini GM. 2017. An allelic variant of the PmrB sensor kinase responsible for colistin resistance in an Escherichia coli strain of clinical origin. Sci Rep 7:5071. doi: 10.1038/s41598-017-05167-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Osei Sekyere J, Maningi NE, Modipane L, Mbelle NM. 2020. Emergence of mcr-9.1 in extended-spectrum-β-lactamase-producing clinical Enterobacteriaceae in Pretoria, South Africa: global evolutionary phylogenomics, resistome, and mobilome. mSystems 5:e00148-20. doi: 10.1128/mSystems.00148-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhang H, Miao M, Yan J, Wang M, Tang Y, Kreiswirth BN, Zhang X, Chen L, Du H. 2017. Expression characteristics of the plasmid-borne mcr-1 colistin resistance gene. Oncotarget 8:107596–107602. doi: 10.18632/oncotarget.22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow chart showing the literature search strategy, inclusion and exclusion criteria, and the final number of manuscripts used for the review. Download FIG S1, PDF file, 0.1MB (83.9KB, pdf) .
Copyright © 2020 Mmatli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.