SUMMARY
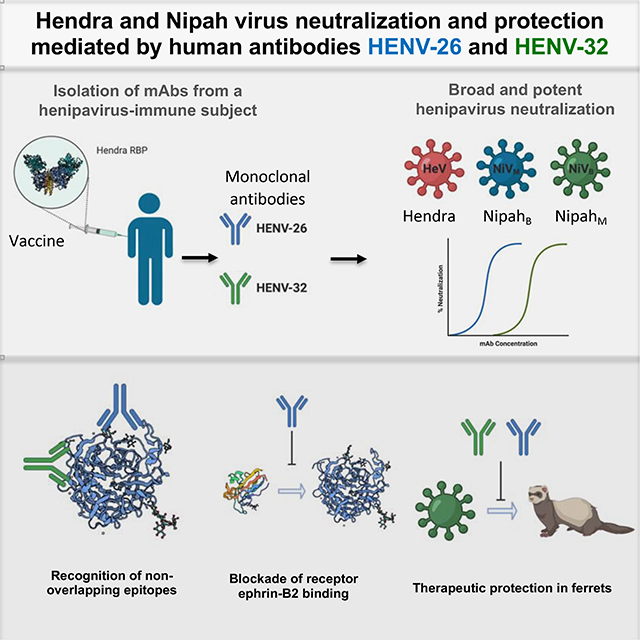
Hendra (HeV) and Nipah (NiV) viruses are emerging zoonotic pathogens in the Henipavirus genus causing outbreaks of disease with very high case fatality rates. Here, we report the first naturally occurring human monoclonal antibodies (mAbs) against HeV receptor binding protein (RBP). All isolated mAbs neutralized HeV, and some also neutralized NiV. Epitope binning experiments identified five major antigenic sites on HeV-RBP. Animal studies demonstrated that the most potent cross-reactive neutralizing mAbs, HENV-26 and HENV-32, protected ferrets in lethal models of infection with NiV Bangladesh 3 days after exposure. We solved the crystal structures of mAb HENV-26 in complex with both HeV-RBP and NiV-RBP and of mAb HENV-32 in complex with HeV-RBP. The studies reveal diverse sites of vulnerability on RBP recognized by potent human mAbs that inhibit virus by multiple mechanisms. These studies identify promising prophylactic antibodies and define protective epitopes that can be used in rational vaccine design.
Keywords: Hendra Virus, Nipah Virus, Henipavirus Infections, Antibodies, Viral, B-Lymphocytes, Antigen-Antibody Reactions, Antibodies, Monoclonal, Pre-Exposure Prophylaxis, Therapy: Epitopes, B-Lymphocyte, Receptor Binding Protein, Molecular Structure
eTOC blurb
Antibodies isolated from a healthy donor with exposure to Hendra G protein provide post-exposure protection against NipahBangladesh in experimental animals. Representative antibodies recognize diverse conserved sites on the henipavirus G protein and neutralize by diverse molecular mechanisms.
Graphical Abstract
INTRODUCTION
Hendra virus (HeV) and Nipah virus (NiV), belonging to the Henipavirus genus in the Paramyxoviridae family, are zoonotic pathogens that cause severe viral disease in humans characterized by serious respiratory illness and encephalitis with high mortality (Weatherman et al., 2018). Fruit bats of the Pteropus genus are natural reservoirs of both HeV and NiV, and the viruses are understood to have co-evolved with these bats (Halpin et al., 2011; Vidgen et al., 2015). Transmission of HeV to humans can occur indirectly from fruit bats following direct human contact with infected horses (Field, 2016; Murray et al., 1995; Selvey et al., 1995). Transmission of NiV to humans may occur directly from fruit bats, infected pigs, or from infected humans (Clayton et al., 2012; Weatherman et al., 2018). There are two distinct major strains of NiV, designated NiV Malaysia (NiVM) and NiV Bangladesh (NiVB) (Lo et al., 2012). NiVB may be more pathogenic than NiVM, as suggested by differences in mortality rates and transmission patterns (Gurley et al., 2007; Homaira et al., 2010; Mire et al., 2016). Recently, 20 new species of viruses in the Henipavirus genus, including Ghana virus (GH-M74a henipavirus) and Cedar virus, were identified in bats in Africa (Drexler et al., 2012) or Australia (Marsh et al., 2012). In 2014, a novel henipavirus-like virus, designated Mòjiāng virus, whose genes have high nucleotide sequence identities to those of the known henipaviruses, was found in yellow-breasted rats (Rattus flavipectus) in China after miners in the region succumbed to irregular pneumonia with unknown etiology (Wu et al., 2014). These viruses have high potential to cause significant human epidemics following their spillover from wildlife reservoirs to humans and domestic animals due to their wide host tropism and high pathogenicity (Smith and Wang, 2013). Henipavirus spillovers are appreciated to be increasing in frequency and distribution due to changes in wild reservoir species distribution and food sources (such as due to changing climate and human related habitat losses) resulting in increased contact with human populations and agriculture (Kessler et al., 2018; Martin et al., 2018; Plowright et al., 2015; Walsh et al., 2017). Furthermore, human-to-human transmission of NiV in India and Bangladesh shows that a large human outbreak is possible (Chadha et al., 2006; Gurley et al., 2007). There is also concern about the potential to weaponize henipaviruses (Luby, 2013). There are no licensed human vaccines or antiviral treatments for HeV or NiV infections (Broder et al., 2013).
The two henipavirus surface proteins, RBP and F, mediate viral entry by viral attachment to cells and fusion between the viral envelope and host cell membrane (Aguilar and Iorio, 2012). HeV or NiV first attach to host cells by binding to the receptors ephrinB2 or ephrinB3 using the viral receptor binding proteins HeV-RBP or NiV-RBP (Bonaparte et al., 2005; Negrete et al., 2005; Negrete et al., 2006). Like other members in the Orthoparamyxovirinae subfamily, henipavirus attachment proteins are required to enable fusion proteins to function in fusion, and conformational changes of the attachment proteins caused by receptor binding activate F proteins to undergo the transition from the pre-fusion to the post-fusion form in order to complete the fusion process (Bossart et al., 2013; Jardetzky and Lamb, 2014; Wong et al., 2017).
HeV-RBP and NiV-RBP proteins consist of an N-terminal cytoplasmic tail, a single transmembrane helix, a stalk region, and a globular C-terminal receptor binding domain (RBD) with a 6-bladed propeller fold. The two RBPs have about 80% amino acid sequence identity to each other, but < 30% amino acid sequence identity to the other henipaviruses. In paramyxoviruses, the ectodomains of the RBPs typically assemble into homotetramers (Bose et al., 2015). The RBP stalk regions form a parallel four helix bundle, while the head domains are organized into a tetramer of two separate dimers (Bose et al., 2011; Welch et al., 2013; Yuan et al., 2008; Yuan et al., 2011; Yuan et al., 2005). In contrast, although isolated head domains of paramyxovirus RBPs can be expressed as monomers in solution, these proteins can dimerize in crystalline phase with the same or similar dimeric interfaces as seen in the naturally occurring ectodomains (Crennell et al., 2000; Lawrence et al., 2004; Santiago et al., 2010). Similarly, the RBP ectodomain or the full-length HeV-RBP and NiV-RBP form tetramers in solution by forming disulfide bonds in the stem region, and head domains are monomers in solution (Bowden et al., 2010; Maar et al., 2012). A negative-stain EM study of NiV-RBP ectodomain showed that the protein assembles into an asymmetric tetramer, with a dimer of head domains at its apex and two monomeric head domains on sides of a central stalk (Wong et al., 2017). In the crystalline state, isolated HeV-RBP head domains can form dimers with a conserved dimeric interface, as occurs with other paramyxovirus RBPs (Bowden et al., 2010). In summary, henipaviruses possess quaternary structures of RBPs in which the stem regions form a major homo-tetrameric interface, while the head domains can associate as dimers or tetramers with a very dynamic quaternary arrangement.
To date, naturally occurring human monoclonal antibodies (mAbs) for HeV or NiV isolated from immune individuals have not been described. MAbs binding to HeV/NiV have been isolated by phage display from a henipavirus-naïve human Fab library (Zhu et al., 2006). An affinity-matured variant of one of those clones, designated m102.4, was converted to a recombinant IgG1 form in which it exhibited neutralization of HeV and NiV (Zhu et al., 2008) and protected animals after lethal NiV Malaysia challenge (Bossart et al., 2011; Bossart et al., 2009; Rockx et al., 2010). Crystal structures of the HeV-RBP head domain in complex with a derivative of the Fab m102.4 (designated m102.3) revealed that binding of the mAb heavy chain complementarity determining region 3 (CDRH3) loop binds to the receptor binding site on RBP for ephrinB2/ephrinB3 (Xu et al., 2013). m102.4 was well tolerated and exhibited linear pharmacokinetics in a recent Phase 1 human trail and has been used on compassionate grounds in the postexposure therapy of 14 humans following high-risk HeV exposures since 2010, highlighting the benefit and practicality of mAb postexposure therapy (Playford et al., 2020).
The demonstrated activity of the m102.4 antibody represents an important conceptual advance. However, the treatment failure of m102.4 in a day 5 and 7 treatment regimen of NiVB in African green monkeys (Mire et al., 2016) suggests a more potent antibody regimen may be of benefit. Second, cocktails of human mAbs that recognize diverse sites on viral glycoproteins and neutralize by differing mechanisms may be desirable for therapeutic development for RNA viruses that easily escape virus neutralization. Third, it is preferable in human therapeutic antibody development to use naturally occurring human mAbs from immune donors that possess naturally paired heavy and light chains and naturally occurring somatic mutations.
Here, we report the identification and characterization of naturally occurring human mAbs against HeV and NiV isolated directly from an immune human individual. We determined the crystal structures of the HeV-RBP or NiV-RBP head domain in complex with the two most potent neutralizing cross-reactive human mAbs, HENV-26 and HENV-32. The structures suggest that the two mAbs neutralize these viruses by very different molecular mechanisms. These antibodies could be developed as promising mAb prophylactic or therapeutic molecules, and the protective epitopes defined by recognition of these human antibodies informs rational vaccine development and testing for these lethal viruses.
RESULTS
Isolation of human mAbs.
To generate human cell lines secreting human mAbs to HeV, we obtained peripheral blood mononuclear cells from an individual in Australia with occupation-related exposure to the equine HeV-RBP subunit vaccine (Equivac®). At the time of study, the individual had a serum 50% virus neutralization titer of 1:40, 1:16 or ≤1:4 for HeV, NiVM or NiVB, respectively. We transformed B cells in the blood sample with Epstein-Barr virus, as described in the Experimental Procedures section. We screened supernatants from EBV-transformed B cell lines for binding to HeV-RBP and NiV-RBP head domain proteins and fused the resulting B cell lines to make hybridomas secreting fully human naturally occurring mAbs. A total of 32 transformed cell lines secreting RBP-reactive antibodies were fused, and 12 independent hybridoma cell lines were recovered after selection that were still secreting RBP-reactive antibodies. After flow cytometric sorting at a single-cell level for biological cloning, clones for 11 of those 12 lines were recovered. One of the clones recovered (designated HENV-13) produced IgG poorly and was not studied further. Thus, 10 cloned hybridomas secreting RBP-reactive mAbs were carried forward; after all functional studies were completed, antibody variable gene sequencing revealed the independently-derived hybridoma clones HENV-18 and HENV-19 shared identical antibody variable gene sequences (Table S1).
Binding activity of human mAbs to HeV-RBP head domain in ELISA.
In order to determine the breadth of mAb binding, we screened the mAbs in ELISA for binding to recombinant RBP head domain proteins from multiple henipaviruses: HeV, NiVM [strain Malaysia], or NiVB [strain Bangladesh] (Bowden et al., 2008). Determination of half maximal effective concentration (EC50) for binding of each mAb against the autologous HeV-RBP or heterologous NiV-RBP head domain proteins revealed that the clones bound at low concentrations; all bound HeV RBP at ≤0.86 μg/mL and 6 of 10 bound at ≤0.25 μg/mL (Table 1 and Figure S1A). Four of the HeV-reactive clones also cross-reacted with NiVB-RBP head domain.
Table 1.
Binding and neutralizing activity of human monoclonal antibodies
| Competitionbinding groupa | Clone | Binding to RBP head domain from indicated virus in ELISA EC50 valuesb in μg/mL (Area under the curve) |
Neutralization of indicated virus | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IC50 valuesd in μg/mL (Area under the curve) | IC80 valuesd in μg/mL (Max % Inhibition) | |||||||||
| HeV | NiVB | NiVM | HeV | NiVB | NiVM | HeV | NiVB | NiVM | ||
| A | HENV-1 | 0.19 (3.27) | >c (0.61) | > (0.67) | 0.63 (187) | > (7.98) | > (22.6) | 1.70 (100) | > (32.7) | > (34.9) |
| B | HENV-32 | 0.36 (3.71) | 0.96 (6.6) | 0.42 (8.0) | 0.27 (198) | 0.38 (194) | 0.31 (201) | 1.26 (100) | 1.14 (100) | 2.15 (100) |
| HENV-21 | 0.42 (3.30) | 2.44 (5.5) | 0.87 (7.2) | 1.94 (133) | 1.91 (140) | 2.01 (129) | 5.54 (100) | 4.09 (100) | 8.44 (92.4) | |
| HENV-10 | 0.21 (5.49) | > (0.63) | > (0.76) | 0.62 (181) | 1.07 (140) | 0.22 (152) | 1.71 (100) | 9.42 (96.3) | 1.44 (84.2) | |
| B/C | HENV-2 | 0.13 (5.81) | 0.51 (6.4) | 0.28 (9.0) | 0.78 (166) | 1.39 (143) | 0.75 (186) | 2.57 (100) | 4.88 (95.8) | 2.67 (100) |
| C | HENV-9 | 0.60 (3.19) | > (0.57) | 0.75 (4.2) | 0.37 (195) | > (11.9) | 8.93 (69.2) | 1.88 (100) | > (35.2) | > (61.5) |
| HENV-43 | 0.86 (6.87) | > (0.59) | 1.49 (1.4) | 0.61 (178) | > (9.2) | > (45.2) | 2.39 (100) | > (40.0) | > (51.9) | |
| D | HENV-18 e | 0.21 (5.80) | > (0.56) | > (0.60) | 0.38 (211) | > (5.7) | > (51.1) | 1.16 (100) | > (31.4) | > (50.8) |
| HENV-19 e | 0.25 (3.69) | > (0.52) | > (0.61) | 0.35 (209) | > (7.2) | 8.08 (43.1) | 0.89 (100) | > (33.9) | > (49.2) | |
| E | HENV-26 | 0.14 (5.39) | 0.09 (10.5) | 0.07 (11.0) | 0.07 (281) | 0.03 (289) | 0.040 (293) | 0.19 (100) | 0.12 (100) | 0.11 (100) |
Experiments were conducted with 2 or 3 biological replicates, each with 2 technical replicates, with consistent results. Binding data from one representative experiment are shown, with neutralization data combined from 3 independent experiments.
Competition-binding group, as determined by data in Figure 1.
50% maximal effective concentration.
The “>” symbol indicates half maximal binding or neutralization is not achieved below the highest concentration tested: 20 μg/mL for HeV RBP binding, 50 μg/mL for NiVM or NiVB binding, or 50 μg/mL for neutralization of each of the three viruses.
50% maximal inhibitory concentration.
After all functional studies were completed, antibody variable gene sequencing later revealed the independently-derived clones HENV-18 and -19 shared identical antibody variable gene sequences (Table S1).
Neutralizing activity of human mAbs.
To evaluate the inhibitory activity of the isolated mAbs, we tested the mAbs in an in vitro neutralization assay using HeV. All 10 of the HeV-RBP-reactive mAbs neutralized HeV, with half-maximal inhibitory concentration (IC50) values for 9 of 10 mAbs < 0.78 μg/mL (Table 1 and Figure S1B). The ELISA binding results discussed above suggested that cross-reactive mAbs in our panel might possess neutralizing activity to multiple henipaviruses. To test this hypothesis, we screened the mAbs in NiVB and NiVM neutralization assays and found that 5 of the 10 mAbs also neutralized the heterologous NiVB strain, while 7 of 10 neutralized the NiVM strain (Table 1). Five of the 6 remaining mAbs neutralized HeV well but neutralized NiV only incompletely, and one mAb (HENV-43) did not neutralize NiV (Table 1). In parallel, we tested the m102.4 antibody for comparative purposes and found the IC50 values to be 0.26 (HeV), 0.02 ng/mL (NiVM) or 0.049 μg/mL (NiVB) (Figure S1).
Binding activity of human mAbs to HeV, NiVM, or NiVB RBPs on the surface of mammalian cells.
The antibody discovery experiments and the ELISAs above were conducted with recombinant soluble forms of henipavirus RBP head domains. We next sought to determine the binding capacity of the most potent mAbs, HENV-26 and HENV-32, to full-length RBPs expressed on the surface of mammalian cells. We transfected 3 × 107 cells with cDNAs encoding the full-length HeV, NiVM, or NiVB RBP, allowed the cells to express the proteins, and then incubated the transfected cells with mAbs and tested for cell surface binding by flow cytometric detection. The results showed that these antibodies bound to the authentic full-length RBPs at low concentrations (Figure S2). The EC50 values for binding of HENV-26 or HENV-32 ranged from 325 to 343 ng/mL or 680 to 836 ng/mL respectively for binding to HeV, NiVM, or NiVB. We tested kinetics of binding of these antibodies to RBPs on a biosensor to determine affinity (Figure S2B). The KD for HENV-26 was 2.9, 2.2 or 1.0 nM and for HENV-32 was 2.1, 2.2, or 2.8 nM for HeV, NiVM, or NiVB, respectively.
Major antigenic sites recognized by human mAbs.
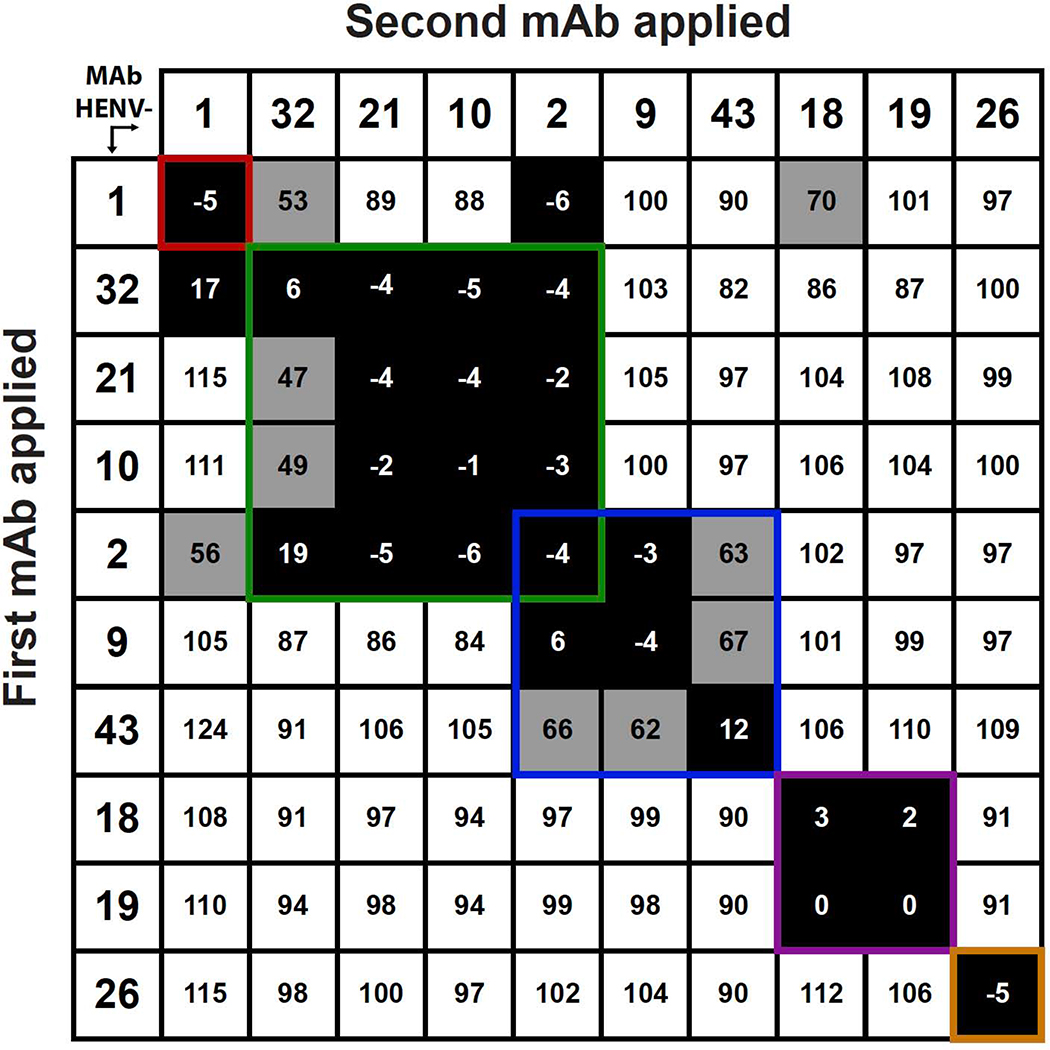
To determine whether Abs from distinct binding groups targeted different antigenic regions on the HeV-RBP surface, we performed a quantitative competition-binding assay using a real-time biosensor. We tested all mAbs in a tandem blocking assay in which HeV-RBP was attached to the biosensor. The data suggest that mAbs in this panel form at least 5 major competition-binding groups, consistent with recognition of 5 different antigenic regions on the HeV-RBP head domain (Figure 1). The potently neutralizing mAbs HENV-26 and HENV-32 segregated into different competition-binding groups. Interestingly, mAb HENV-2 competed with 5 other mAbs, suggesting that it may bind to overlapping regions from 2 or 3 antigenic sites.
Figure 1. Competition-binding assay data enabling binning into groups of mAbs recognizing common antigenic sites.
Ten human IgG mAbs were competed for binding to HeV RBP head domain using biolayer interferometry. Numbers in boxes are the percentage binding signal of the second mAb applied after binding of the first mAb, compared with binding signal of the second mAb alone. The antibodies were defined as competing antibodies if the first antibody reduced binding of the second antibody by more than 70 percent. The antibodies were defined as non-competing antibodies if the first antibody reduced binding of the second antibody by less than 40 percent. Binding signals 40 to 70% were considered intermediate competition (grey boxes with black numbers). Inferred competition-binding groups A to E (designated in order left to right and top to bottom) are indicated with colored boxes, A) red, B) green, C) blue, D) purple, E) yellow.
Competition-binding studies with the ephrinB2 receptor.
Henipaviruses use the human ephrinB2 protein as a receptor for attachment and entry (Bonaparte et al., 2005; Negrete et al., 2005). We sought to determine if anti-HeV-RBP mAbs neutralized virus by blocking virus attachment to ephrinB2 by studying three mAbs (HENV-19, −26 and −32) representing each of three main competition-binding groups. We competed recombinant ephrinB2 protein with mAbs or buffer for binding to biosensor tips coated with HeV-RBP head domain. MAb HENV-26 reciprocally competed with ephrinB2 for binding to HeV-RBP, while mAbs HENV-19 and HENV-32 did not (Figure S3A), suggesting that HENV-26 neutralizes by binding to the receptor binding site on HeV-RBP. MAb HENV-26 also competed with ephrinB2 for binding to full-length HeV-RBP expressed on the surface of 293F cells (Figure S3B).
Crystal structures of HENV-26 in complex with HeV-RBP or NiV-RBP proteins.
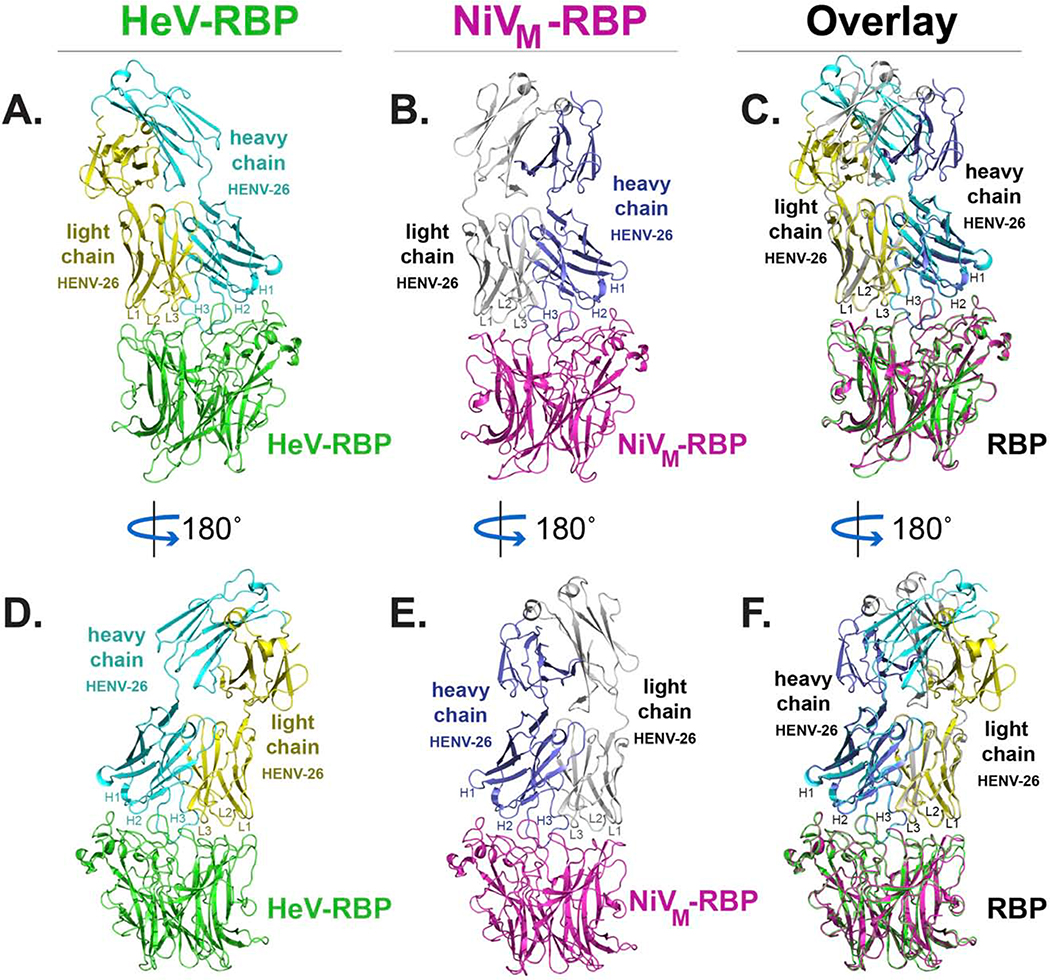
Next, we determined the structure of antigen-antibody complexes for two mAbs using crystallography. We selected the two most potent and cross-reactive mAbs from the panel, HENV-26 and HENV-32, for crystallographic studies and first determined their heavy and light chain variable gene sequences (Table S1). HENV-26 Fab complexes with HeV-RBP or NiVM-RBP head domains were crystallized in spacegroups P22121 or P322, and the crystal structures were solved at 2.60 Å or 3.40 Å, respectively (Table S2). Electron density for both structures was well defined except for several loop regions, e.g., S128 – T131 of the HENV-26 heavy chain in the HENV-26/HeV-RBP complex, S127 – A136 of the HENV-26 heavy chain in the HENV-26/HeV-RBP complex, P126 – A137 of HENV −26 heavy chain, K156 – V159 of HENV −26, A157 – G158 of HENV-26 light chain, and I237 – V244 of NiV-RBP in the HENV-26/NiV-RBP complex. The buried surface areas for the HENV-26/HeV-RBP complex or the HENV-26/NiV-RBP complex are 1,282.2 Å2 or 1,323.1 Å2, respectively. The overall structures of the two complexes are superimposable for the RBPs and the antibody variable domains, with an RMSD of 0.97 Å for 643 Cα atoms (Figure 2). The relative orientations of Fab constant domains differed significantly because of the flexibility of the antibody elbow region and differing crystal contacts in the two crystals. HENV-26 has a relatively long CDRH3 loop (19 residues) and it adopts a spoon-shaped conformation that targets the central cavity of the HeV-RBP or NiV-RBP proteins (Figure 2). In this engagement, HENV-26 shares some structural features with the antibody m102.3. (Figure 2). In the m102.3/HeV-RBP complex (PDB 6CMG; PDB 6CMI), the paratope residues interacting with HeV-RBP are located mostly in the CDRH3 with an additional 3 interacting residues in CDRH2, 1 in CDRH1, and 1 in CDRL1 (Xu et al., 2013). In contrast, all CDRs of HENV-26 (except CDRL2 in the HENV-26/NiV-RBP complex), the light chain DE loop, and the heavy chain β strand C′′ of HENV-26 participate in antigen binding. The CDRH3 contributes more to antigen binding than the other structural elements (Figures 2 and 3). The HENV-26 CDRH3 and CDRH2/CDRH1/CDRL3 form a saddle-shaped conformation, straddling the RBP rim region formed by the β4S4-β5S1 loop and the β5S2-β5S3 loop, with CDRL1 and/or CDRL2 interacting with the β5S4-β6S1 loop, β6S2-β6S3 loop, and β1S2-β1S3 loops. Although the CDRH3s of both HENV-26 and m102.3 target the receptor-binding site of the RBPs, the two antibodies exhibit significantly different binding modes. This difference in binding modes between m102.3 and HENV-26 may be due to different conformations of their CDRH3s, since m102.3 has a relatively long, protruding CDRH3 with a β-hairpin conformation (Xu et al., 2013). The binding mode difference between the naturally occurring HENV-26 and the phage library-derived m102.3 is consistent with previous studies showing that naturally occurring human mAbs typically recognize their antigens using multiple CDRs to form an integrated interface, while phage library-derived mAbs often rely dominantly or even exclusively on their CDRH3 loops (Burkovitz and Ofran, 2016).
Figure 2. Crystal structures of Fab HENV-26 in complex with HeV-RBP or NiV-RBP head domains.
Heavy chain CDRs are labeled as H1, H2, and H3, and light chain CDRs labeled as L1, L2, and L3. A) The structure of HENV-26 in complex with HeV-RBP head domain. HeV-RBP is colored in green, the HENV-26 heavy chain in cyan, and the light chain in yellow. B) The structure of HENV-26 in complex with NiV-RBP head domain. NiV-RBP is colored in magenta, the HENV-26 heavy chain in light blue, and the light chain in grey. C) The superimposition of the two structures. D), E), or F) are 180° rotation views along the designated axis of panel A, B, or C, respectively.
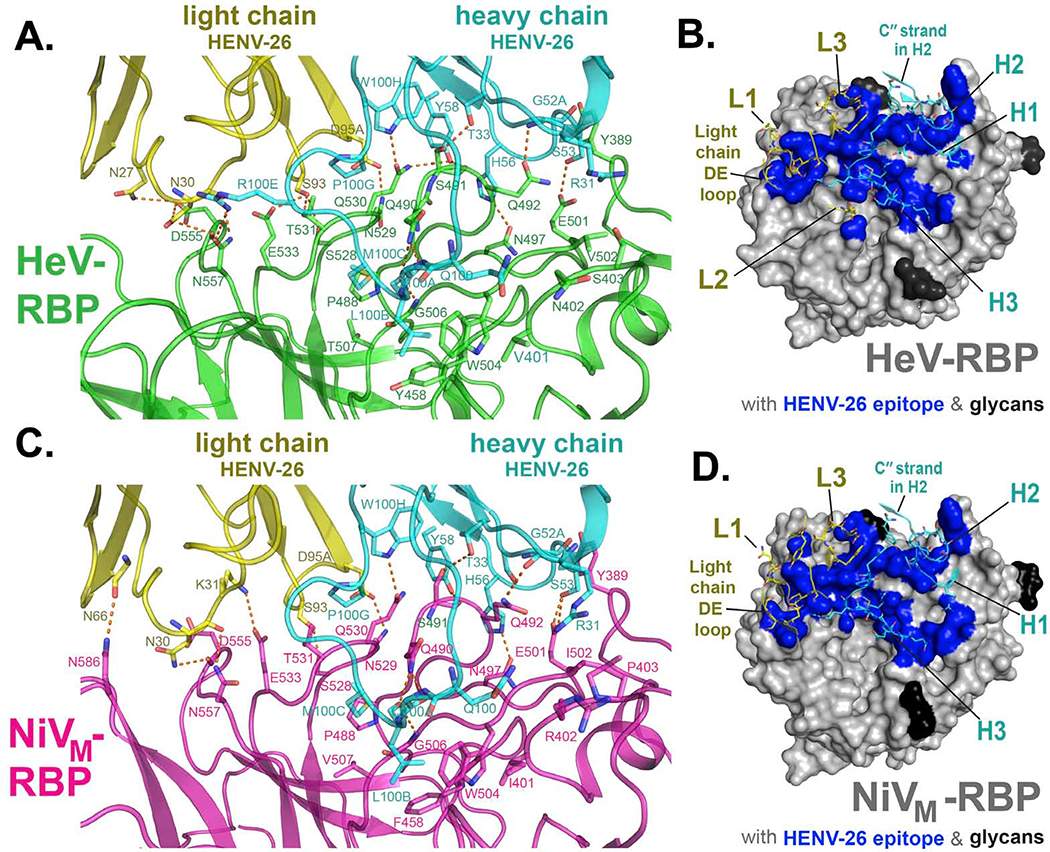
Figure 3. Interface of mAb HENV-26 in complex with HeV-RBP or NiV-RBP.
Residues in the crystal structures of protein complexes with an interatomic distance between antigen (RBP) and antibody of less than 5 Å were designated as participating in the interface.
A) Side view of the interface between mAb HENV-26 and the HeV-RBP head domain. HeV-RBP head domain is colored green, HENV-26 heavy chain in cyan, and light chain in yellow. The interface residues are shown in stick representation. Polar interactions between HeV-RBP and HENV-26 are represented as broken orange lines. The interface residues are labeled in green, cyan, or yellow respectively for HeV-RBP, the heavy chain, or the light chain.
B) Top view of the interface between HENV-26 and HeV-RBP head domain. HeV-RBP head domain is shown in grey as surface representation, glycans on HeV-RBP are colored in black, and interface atoms of HeV-RBP are colored in blue. Interface residues and neighboring residues of HENV-26 are shown as cartoon representation, and interface residues as stick representation. CDRs, the light chain DE loop, and heavy chain C′′ strand of the mAb are labeled.
C) Side view of the interface between HENV-26 and NiV-RBP head domain. NiV-RBP head domain is colored pink, HENV-26 heavy chain in cyan, and light chain in yellow. The interface residues are shown in stick representation. Polar interactions between NiV-RBP and HENV-26 are represented as broken orange lines. The interface residues are labeled in pink, cyan, or yellow for NiV-RBP, the heavy chain, or the light chain, respectively.
D) Top view of the interface between HENV-26 and NiV-RBP head domain. NiV-RBP head domain is shown in grey as surface representation, glycans on NiV-RBP are colored in black, and interface atoms of HeV-RBP are colored in blue. Interface residues and neighboring residues of HENV-26 are shown as cartoon representation, and interface residues as stick representation. CDRs, the light chain DE loop, and heavy chain C′′ strand of the mAb are labeled.
Overlays of the epitopes on the surface of HeV-RBP or NiV-RBP recognized by HENV-26 with that of the ephrinB2 receptor binding sites show that the antibody epitopes overlap greatly with the receptor binding sites (Figure S3C), consistent with the competition-binding experiments (Figures S3A, S3B). Therefore, HENV-26 neutralizes HeV or NiV by competitive inhibition of viral attachment to the viral receptor.
In the HENV-26/HeV-RBP complex, 30 residues of HENV-26 (including 9 residues from CDRH3) and 31 residues plus the N-acetylglucosamine of the glycan at N529 of HeV-RBP from the Ab-Ag interface. The interaction contains a total of 19 hydrogen bonds (H-bonds) and 1 ionic interaction between Ab and Ag, and the H-bonds distribute relatively evenly among the CDRs, i.e., 5 H-bonds for CDRH3, 3 for CDRH2, 1 for CDRH1, 4 for CDRL3, 4 for CDRL1, and 2 for the heavy chain framework C′′ strand (Figure 3A). Hydrophobic effects drive a portion of the Ab-Ag binding, as seen mainly between the tip of CDRH3 and the HeV-RBP cavity. CDRH3 residue M100C of HENV-26 is surrounded by residues P488, G489, T507, A532, residue L100B is surrounded by Y458, W504, and G506 mainchain atoms, and the Cβ and Cγ residues of Q100 stack on hydrophobic residues V401 and W504 (Figures 3A, 3B). In the ephrinB2/HeV-RBP complex, residues F111, P119, L121, and W122 of the ephrinB2 G-H loop occupy 4 hydrophobic pockets of the central cavity of HeV-RBP (here we designated them as pocket F111, P119, L121, or W122, respectively), while L105, P107, and P109 on the m102.3 CDRH3 interact with the first three pockets (Bowden et al., 2008; Xu et al., 2013). HENV-26 CDRH3 residues M100C, L100B, or Q100 occupy similar positions to that of the ephrinB2 residues P119, L121, or W122, respectively, mimicking their interactions with HeV-RBP. In addition, atom Sδ of the M100C sidechain has relatively weak S···O polar interactions with the mainchain oxygen atoms of P488 and G489 on HeV-RBP, further strengthening the interaction between the HENV-26 CDRH3 and HeV-RBP. Importantly, the mainchain O atoms of L100B and Q100A make three H-bonds with the HeV-RBP Q490 Nε2 and G506 mainchain N atoms (Figure 3A). It is most likely that the HENV-26 CDRH3 tip is a critical region for the Ab-Ag interaction. There are extensive van der Waals interactions between CDRH3/CDRH2/CDRL3/CDRL1 and HeV-RBP because of the high shape complementarity between these CDRs and the antigen (Figures 3A, 3B). Interestingly, 14 water molecules with well-defined electron density can be found at the Ab-Ag interface, forming H-bonds with antibody CDRs and HeV-RBP loops, and they distribute evenly at the interface (Figure S4A). Given the high affinity of binding between HENV-26 and HeV-RBP, the H-bond network mediated by these water molecules must contribute positively to the binding free energy, i.e., the enthalpy gain from these H-bonds overcomes the entropy loss due to the loss of translational freedom of the water molecules.
As discussed above, the HENV-26/NiV-RBP complex shows very similar structure features to those of the HENV-26/HeV-RBP complex. The HeV and NiV RBPs have very high sequence identity (>80%). Mapping the epitopes on HeV-RBP or NiV-RBP recognized by HENV-26 onto their amino sequences at points of contact explains why HENV-26 cross-reacts with both RBPs (Figure S4E). Most Ab-Ag interface interactions in the HENV-26/HeV-RBP complex described above are conserved in the HENV-26/NiVHeV-RBP complex due to sequence conservation of RBPs at these regions. This observation is noted especially for the heavy chain CDRs and /CDRL3, and 3 regions of the RBPs (P488 – E501, W504 – T/V507 with F/Y459, and S528 – E533), indicating that these CDRs (except heavy chain R31) likely contribute most to the binding energy (Figure 3, Figure S4E). In contrast, for CDRL1 and RBP region D555 – Q559, there are significant rearrangements of polar interactions between the two structures, which most likely relate to conformational differences at the β6S2-β6S3 and β6S4-β1S1 loops between HeV-RBP and NiV-RBP, suggesting a lesser contribution to the binding. Other minor interactions, such as a salt bridge formed between light chain D53 and HeV-RBP R242 in the HENV-26/HeV-RBP complex and an H-bond between light chain N66 mainchain oxygen and NiV-RBP N586 Nδ2 in the HENV-26/NiV-RBP complex, are not conserved, but are located at the periphery of the interface, suggesting that they do not contribute significantly to the binding energy. N402 in HeV-RBP and R402 in NiV-RBP make non-specific van der Waals interactions with CDRH3 Q100, and sequence conservation at this site should affect binding affinity only minimally.
Crystal structures of HENV-32 in complex with HeV-RBP protein.
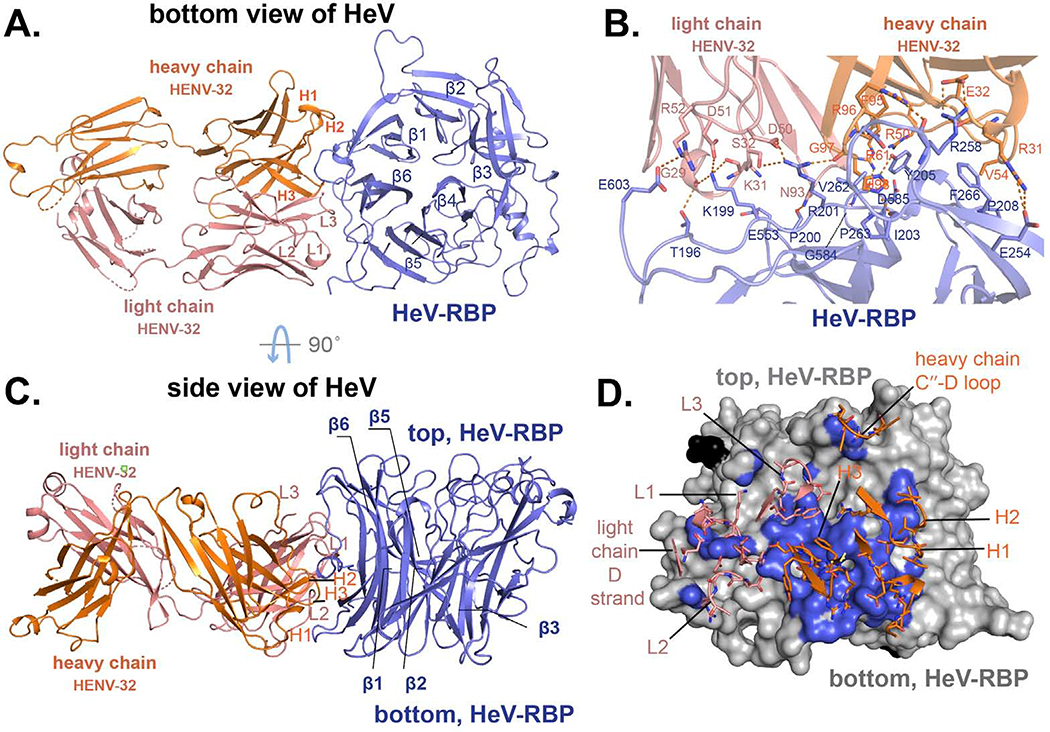
It was apparent from the competition-binding studies with HENV-26 or with ephrinB2 shown above that the potent neutralizing mAb HENV-32 bound to an antigenic site distinct from the receptor-binding domain epitope recognized by HENV-26. Therefore, we next determined the structure of antigen-antibody complexes for HENV-32 with HeV-RBP using crystallography. The structure revealed the molecular details of HENV-32 binding to an epitope distinct from that of HENV-26. HENV-32 in complex with HeV-RBP head domain crystallized in spacegroup C2 with a resolution of 2.0 Å (Table S2). There are two copies of HENV-32/HeV-RBP complex in one asymmetric unit (ASU). The structures of the two copies of the complex are very similar, with an RMSD of 1.05 Å for 640 Cα atoms of HeV-RBP and variable domains. The relatively high RMSD for the two copies in an ASU is likely due to motion of the HeV-RBP head domain and/or the difference of crystal contacts of the two copies. When we overlaid antibody CDRs and interface regions of the HeV-RBP head domain, the RMSD for 264 mainchain atoms was only 0.36 Å. The constant domains adopt very different relative orientation to the variable domains in the two copies due to the high flexibility of the antibody elbow region and the crystal lattice packing. The buried surface areas for the two copies of the complex are 1,157 or 1,092 Å2, respectively. HENV-32 mainly interacts with the highly flexible N-terminal segment T196 – I209 and the β1S3/β1S4 β-turn on the bottom side of the head domain (Figures 4, S4B, and S5A). All CDRs of HENV-32 contribute to antigen binding, with CDRH3 contributing the most among them. The N-terminal segment consists of the β6S4 strand and part of β6S4/β1S4 loop of HeV-RBP. The epitope overlaps the putative dimeric interface of HeV-RBP head domain (Bowden et al., 2010), especially at the N-terminal segment (Figure S5B). HeV-RBP or NiV-RBP head domain alone exists as a monomeric form in solution but can self-associate as dimers in the crystalline state (Bowden et al., 2010), suggesting that the capacity of HeV-RBP or NiV-RBP head domain proteins to self-associate is weak.
Figure 4. Crystal structure of HENV-32 in complex with HeV-RBP head domain.
A) and C) show cartoon representations of the crystal structures. HeV-RBP head domain is colored in light blue, HENV-32 heavy chain in orange, and the light chain in salmon. Individual CDRs are labeled. The six blades of HeV-RBP head domain are labeled (β1 to β6). Panel A) is the bottom view of HeV-RBP head domain, and panel C) is side view. B) and D) show the interface of the complex (residues with an interatomic distance between antigen and mAb of less than 5 Å). In panel B, interface residues are shown as cartoon representation. The residues of HeV-RBP are labeled in blue, those of the heavy chain in orange, and those of the light chain in salmon. The polar interactions are shown as broken orange lines. In panel D, HeV-RBP head domain is shown as surface representation and colored in grey; the interface atoms from HeV-RBP are colored in light blue. Again, the paratope residues are shown as sticks. CDRs, loop between the heavy chain C′′ strands and D loop, and light chain D strand of the mAb are labeled.
The interface between Ab and Ag in the HENV-32/HeV-RBP complex comprises 29 or 30 residues from HENV-32 and 27 residues from the HeV-RBP head domain (depending on the copy in the ASU). There is a minor difference in the interface between the two copies in one ASU, for example, the heavy chain residues R61 and R64 form a polar interaction and salt bridge with G584 mainchain O atom and D585 side chain respectively in one copy, while the interactions are missing in the other copy. The interactions are highly exposed to solvent, and they do not contribute significantly to the binding free energy. 17 or 13 electrostatic interactions (including H-bonds, salt bridges, interactions between positively charged sidechain and mainchain O atoms) can be found between Ab and Ag, with 13 interactions shared by the two copies. Among these interactions, 12 of them most likely are important for binding. There are three focal interface locations for electrostatic interactions. The HENV-32 CDRH3 residue H98 sidechain forms two H-bonds with the mainchain O atoms of Y205 and P263 of the Ag, and the G97 mainchain N interacts with the HeV-RBP D260 sidechain via an H-bond (Figure 4B). The positive charge on residue K199 of HeV-RBP is neutralized by the mainchain O atoms of residues G29 and K31 and the D51 sidechain of the light chain, and R201 is neutralized by the light chain D50 and mainchain O atom from the heavy chain CDRH3 G97. The R201 side chain also makes an H-bond with the light chain S32 Oγ atom. In addition to these three focal sites of contact, there are three more electrostatic interaction pairs: salt bridges between CDRH1 residue E32 and Ag residue R258, CDRH1 residue R31 and Ag residue E254, and H-bond between CDRL3 residue N93 sidechain and Ag residue P200 main chain O atom. Other important interactions include π-π stacking between CDRH3 F95 and Ag Y205, cation-π interactions between heavy chain residue R50 and HeV-RBP Y205, and CDRH1 R31 and Ag F266 (Figure 4B). A hydrophobic effect can be seen between the CDRH2 I54 and Ag F266 residues, CDRH2 V55 and Ag P208 residues, CDRH3 F95 and Ag Y205 residues.
As mentioned above, Nδ1, and Nε2 of HENV-32 CDRH3 residue H98 form 2 H-bonds with the Y205 hydroxyl group (D-A distance = 2.6Å) and P263 mainchain oxygen (D-A distance = 2.7Å), and its sidechain also interacts extensively with surrounding residues from HeV-RBP via van der Waals force, e.g., a small pocket formed by I203, Y205, T206, V262, P263, and S264 (Figures 4B, 4D, and 5B). Thus, CDRH3 residue H98 is the most important residue for the Ab-Ag interaction. Consistent with the highly hydrophilic interface of the HENV-32/HeV-RBP complex, there is a total of 34 visible water molecules, 15 of which are screened from bulk water, at the interface forming an H-bond network and mediating the Ab-Ag interaction (Figure S4B). Given the tight binding of HENV-32 to HeV-RBP (Table 1 and Figures S1, S2) and relatively small buried area of the complex, the number of interface water molecules of the complex is unusual because there is, on average, about one interface water per 100 Å2 of interface area (Lo Conte et al., 1999). Taken together, water-mediated Ab-Ag interactions likely are a major factor in the high binding affinity of HENV-32 to RBPs. In sum, the binding of HENV-32 to HeV-RBP appears to be highly driven by enthalpy in a process involving extensive charge-charge interactions and water-mediated hydrogen bonding.
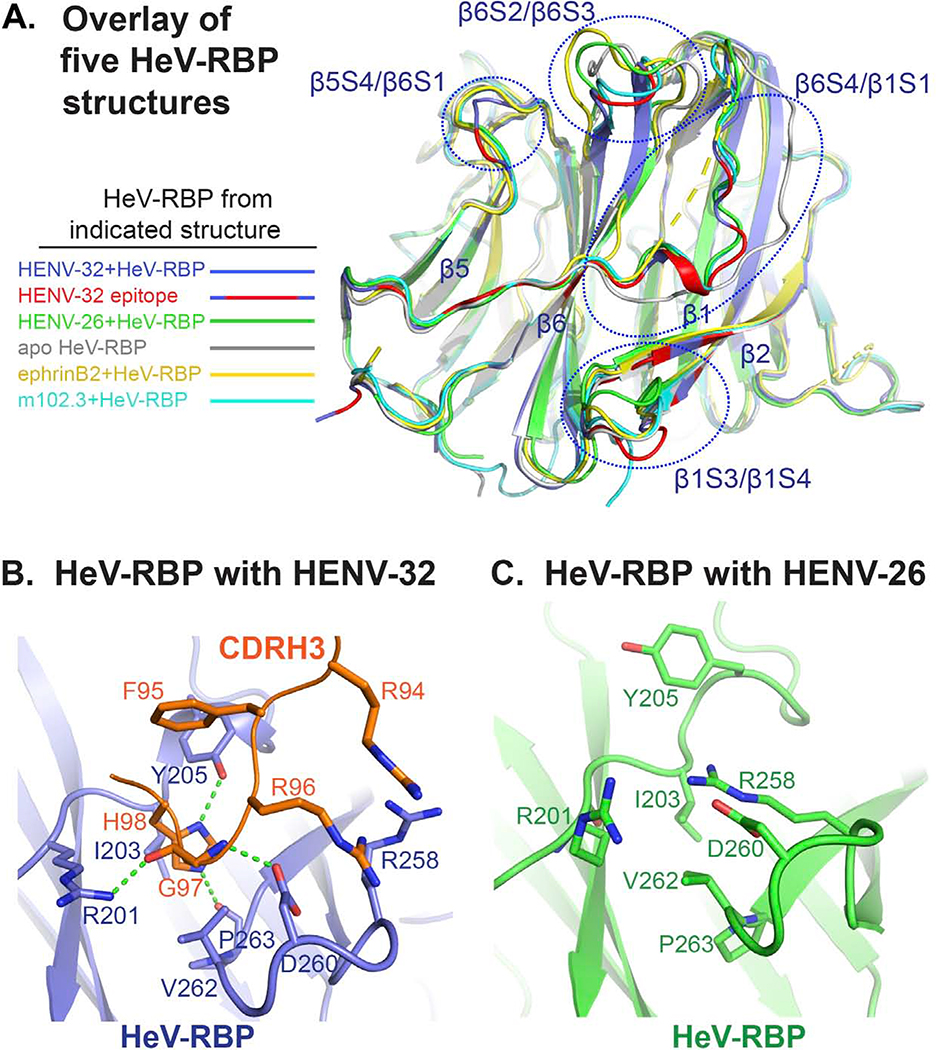
Figure 5. Functional significance of conformational changes of HeV-RBP head domain upon HENV-32 binding.
A) Overlay of five HeV-RBP head domain crystal structures. All structures are shown as cartoon representation. Light blue: HeV-RBP in the HENV-32/HeV-RBP complex, and the epitope recognized by HENV-32 is highlighted in red; white: HeV-RBP apo form; yellow: HeV-RBP in the ephrinB2/HeV-RBP complex; green: HeV-RBP in the ENV-26/HeV-RBP complex; cyan: HeV-RBP in the mAb m102.3/HeV-RBP complex.
B) Structural details of the binding interface of HENV-32 CDRH3 and HeV-RBP head domain. Interacting residues are shown as stick representation, and the hydrogen bonds between CDRH3 and HeV-RBP are shown as broken green lines.
C) Structural details of HENV-32 CDRH3 binding site in the crystal structure of the HeV-RBP/HENV-26 complex are shown for comparison. Residues interacting with the HENV-32 CDRH3 are shown as sticks. Comparison of the two structures of HeV-RBP head domain reveals extensive conformational changes at the site caused by CDRH3 binding. In the crystal structure of HENV-32/HeV-RBP complex, HENV-32 CDRH3 residue H98 is surrounded by a pocket on HeV-RBP formed by residues I203, Y205, V262, and P263, while in the crystal structure of HENV-26/HeV-RBP complex, there is no pocket at this site. The strong ionic bonding between HeV-RBP residues R258 and D260 forms a lid covering the site. HENV-32 HCDR3 remodels the site by inducing an outward flipping of the main chain of the β1/S3-S4 loop, residues L256 – S264, and corresponding side chain rearrangements (especially the outward flipping of R258 side chain). The major driving force for this induced fit might be the interactions between HENV-32 R94/R96 and HeV-RBP R258/D260, and interaction of HENV-32 H98 with the surrounding cavity residues of HeV-RBP.
HENV-32 binding causes conformational changes of the HeV-RBP head domain.
When we superimposed all five available HeV-RBP crystal structures (Figure 5A), we observed extensive conformational variations at the epitope recognized by HENV-32 among these structures (note: residues Y205 – R212 of β6S4/β1S1 loop are missing in the crystal structure of the ephrinB2/HeV-RBP complex). RMSD values (Å) between Cα atoms of HeV-RBP structures in the different binding states are shown in Table S3. There are 4 major regions of the epitope showing large structural variations: the β5S4/β6S1 loop, β6S4/β1S1 loop, β6S2/β6S3 loop, and β1S3/β1S4 β-turn. From the discussion above, we know that the β5S4/β6S1 loop might not contribute significantly to binding energy. In addition, in the second copy of the HENV-32/HeV-RBP complex, the conformation of the β5S4/β6S1 loop is similar to that in the HENV-26/HeV-RBP complex, suggesting that the structural variations at this loop do not affect the binding or function of HENV-32 and only reflect the innate structural flexibility of the loop and/or represent a crystallization artifact. Conformational changes in the β6S2/β6S3 loop can be seen in both copies of the ASU in the HENV-32/HeV-RBP complex. However, as above, this region also may not be critical for binding either. Therefore, these conformational changes may be secondary conformational adjustments induced by the conformational changes of the adjacent β6S4/β1S1 loop, which is one major region of the interface of the complex. From the structure overlay, it is clear that β6S4/β1S1 loop is the most flexible region of HeV-RBP (Figure 5A). HENV-32 binding stabilizes this region in certain conformations (there are still subtle conformational differences between the two copies in one ASU of the crystal structure). The conformation of this loop in the HENV-32 bound form is similar to, but different from, those in the HENV-26-bound or m102.3-bound forms (Figures 5A and S5A), suggesting that the conformational changes result from a combination of conformation selection and induced fit. The β1S3/β1S4 β-turn also shows large conformational variations among the HeV-RBP structures (Figures 5A, 5B, 5C). The β1S3/β1S4 β-turn and β6S2/β6S3 loop, especially residues R201 – N210, constitute the major epitope region interacting with mainly with the heavy chain CDRs. Their conformational changes are essential in order for HENV-32 to bind. The small pocket of HeV-RBP between the β1S3/β1S4 β-turn and the β6S2/β6S3 loop is recognized by HENV-32 CDRH3 residue H98 (Figures 4B S4B, and 5B), as discussed above. In all other HeV-RBP crystal structures, this pocket is covered by a salt bridge pair from the β1S3/β1S4 loop, R258 and D260 (Figures 5B, S6), although the mainchain conformation varies among these structures (Figure 5A). When HENV-32 binds, the sidechain of residue R258 moves away from the pocket, accompanying opening movement of the mainchain of the loop. Meanwhile, residue R96 of the HENV-32 CDRH3, replacing HeV-RBP residue R258, forms a new salt bridge with residue D260 of the loop (Figure 5B). The positively-charged residues R94 and R96 of CDRH3 may provide a repulsive force for the opening movement of β1S3/β1S4 loop and stabilize the position of D260. The CDRH3 residue H98 probably also participates in remodeling the pocket by interacting with HeV-RBP residue P263 and I203 (by H-bonding and van der Waals force). These features support the case for an induced fit mechanism of protein-protein interaction. On the other hand, a conformation selection mechanism also may contribute to the conformational changes, due to the flexibility of the β1S3/β1S4 β-turn seen in the structure overlay (Figure 5A). A mixed mechanism could be used to explain the conformational changes of HeV-RBP when HENV-32 binds. We suggest that HENV-32 may bind to highly flexible regions of RBP head domains via a “mixed mechanism”, suggesting that its heavy chain CDRs should be rigid to minimize entropy cost to the binding reaction. We tested whether or not HENV-32 could disrupt soluble HeV-RBP oligomers, but it did not (Figure S4D).
As above, HENV-32 cross-reacts with both HeV-RBP and NiV-RBP (Table 1). Mapping the HeV-RBP epitope recognized by HENV-32 onto the amino acid sequences of HeV-RBP and NiV-RBP (Figure S4E) shows that 26 of the 30 interface residues are conserved in NiV-RBP and HeV-RBP. There is a H-bond between residue T197 side chain in HeV-RBP and HENV-32 light chain G29 carbonyl oxygen in one copy of HENV-32/HeV-RBP complex, but it is missing in the other. Thus, this H-bond is not critical for binding. The substitution of the position with Q197 in NiV-RBP could make van der Waals contacts with HENV-32 CDRL1 as T197 does, causing minimal effect on binding. Residue R201 of HeV-RBP is neutralized by D50 of HENV-32 CDRL2, and forms 2 H-bonds with S32 of CDRL1 and the G97 mainchain oxygen of CDRH3. The substitution with K201 in NiV-RBP would preserve the salt bridge and most probably one H-bond with G97 of CDRH3 or S32 of CDRL1. HeV-RBP residue N210 makes van der Waals contacts with HENV-32 residue V56. Sequence variation from asparagine to valine at this position in NiV-RBP would maintain the van der Waals contacts in addition to improving the hydrophobic effect between NiV-RBP V210 and HENV-32 V54/V56. Residues E553 and E602 of HeV-RBP form ionic interactions with HENV-32 CDRL1 K31 and CDRL2 R52, respectively, in one copy in an ASU of the complex, but the interactions are missing in the other, suggesting that they likely are dispensable for Ab-Ag binding. To summarize, HENV-32 can recognize both HeV-RBP and NiV-RBP (including both Bangladesh and Malaysia strains) because of the conservation of the sequences and structures of the epitope in these strains. The epitope recognized by HENV-32 is located in the putative dimeric interface of RBP head domains, especially the site bound by the heavy chain CDRs. If RBP dimerization is functionally important for henipaviruses, it may be difficult for HeV or NiV to generate mutant viruses that escape neutralization from HENV-32 mAb.
Lack of cross-reactivity with RBPs from Cedar virus or Ghanaian bat henipavirus.
We also examined whether or not HENV-26 of HENV-32 could recognize more distantly related henipaviruses including Cedar virus (CedV) and Ghanaian bat henipavirus (GhV). The RBP of the more distantly related henipavirus Mòjiāng virus was not tested as it is more divergent in sequence, is antigenically distinct, and lacks an ephrinB2/B3 binding domain (Rissanen et al., 2017). The HENV mAbs did not bind to recombinant forms of RBPs from CedV or GhV in ELISA, whereas CedV- or GhV- -specific control antibodies did bind (Figure S4C). There are numerous differences in the epitopes when RBP sequence alignments between HeV/NiV and CedV or GhV are compared that suggest why the antibodies do not recognize CedV or GhV (Figure S4E), and these differences can be understood in the context of the RBP structures. Superimposition of the CedV-RBP or GhV-RBP crystal structure onto that of HeV-RBP in the HENV-26/HeV-RBP complex explains the absence of binding of HENV-26 to CedV-RBP or GhV-RBP (Figure S4F). Sequence variations of CedV-RBP and GhV-RBP at several key positions from those of HeV-RBP or NiV-RBP cause loss of several interactions including H-bonds. The side chains of residues N497, S491, T531, and N529 in HeV-RBP form H-bonds with residues of HENV-26, while the residue variations at the structurally corresponding positions in CedV-RBP (residues H518, G512, L552, and D550) would abolish these H-bonds. Furthermore, L552 in CedV-RBP would make steric clashes with residue S93 of HENV-26 light chain in the superimposed structures, disrupting potential H-bond between S93 of HENV-26 light chain and residue S549 of CedV-RBP. Similarly, residue variations at V541, G500, E539, and T511 in GhV-RBP (corresponding to residues T531, Q490, N529, and E501 respectively in HeV-RBP) disrupt 5 potential H-bonds between GhV-RBP and HENV-26. Additionally, residue V468 in GhV-RBP (Y458 in HeV-RBP ) could significantly weaken hydrophobic/van der Waals interaction between this position and residue L100B of CDRH3 of HENV-26. Due to significant sequence differences of CedV-RBP (residues P219 – Q233) or GhV-RBP (residues P212 – T226) from HeV-RBP (residues T196 – N210) or NiV-RBP (residues Q196 – V210) in one major epitope region recognized by HENV-32, the backbone conformations of CedV-RBP and GhV-RBP deviate significantly from those of HeV-RBP and NiV-RBP at this region. This finding indicates that the energy barrier is prohibitively high for CedV-RBP or GhV-RBP to adopt a similar backbone conformation to that of HeV-RBP in the Ab-Ag complex at the region, explaining the inability of binding of CedV-RBP and GhV-RBP by HENV-32.
Post-exposure efficacy of human mAbs in a ferret model of henipavirus infection.
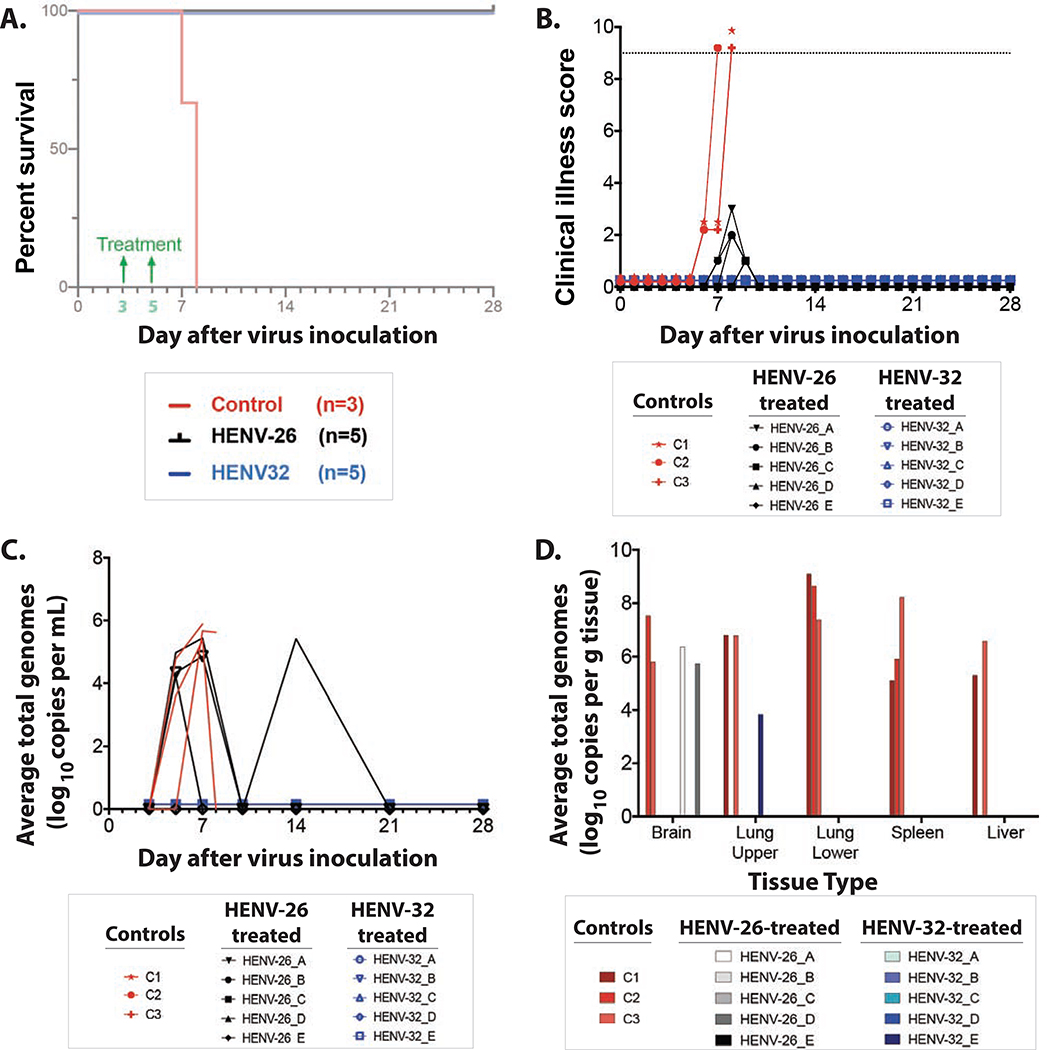
To determine the therapeutic activity of these cross-neutralizing Abs, we tested two antibodies in ferrets. We focused on potent cross-reactive antibodies for challenge with NiVB. We selected the two mAbs HENV-26 and HENV-32, because they bound non-overlapping antigenic regions in the competition-binding experiments and structural studies. Female ferrets (~3–5 months old) received 15 mg/kg of antibody by the intraperitoneal route on days 3 and 5 (for a total of 30 mg/kg cumulative dose) after intranasal inoculation with 5,000 PFU of NiVB. The serum 50% virus neutralizing titers for NiVB for ferrets treated with HENV-26 were 1:369 (day 5) and 1:765 (day 7), while the titers for ferrets treated with HENV-32 were 1: 135 (day 5) and 1:132 (day 7). HENV-26 and HENV-32 each reduced disease and protected ferrets from death when delivered 3 and 5 days after virus challenge (Figure 6A,B). All untreated control animals exhibited a clinical course and pathology consistent with previous reports of henipavirus infection in ferrets including: pulmonary complications, lymphopenia, neutrophilia, thrombocytopenia, and hypoalbuminemia (Mire et al., 2013). Circulating viral genomes were detected beginning on day 5, with a mean value of 5.64 (+/− 0.26 SD) log10 genomes/mL (Figure 6C). Viral genomes were detected in all tissues tested (Figure 6D), and infectious virus was detected at low levels in spleen, kidney, adrenal glands, lung (data not shown). All control animals displayed gross and histologic lesions consistent with NiV infection (Figure S7). Significant lesions included necro-hemorrhagic hepatitis, splenitis, and pneumonia. Positive immunolabeling for specific anti-NiV N protein was noted in the hepatic sinusoidal lining cells, endothelium of small caliber vessels and rarely mononuclear cells and/or hepatocytes, endothelium and mononuclear cells of the spleen, endothelium of small caliber vessels within the neural parenchyma, and the endothelium of the pulmonary alveolar septa, endothelium of small caliber vessels, mononuclear cells within alveolar septa, alveolar macrophages and rarely the lower airway epithelium. None of the HENV-32 treated subjects exhibited overt signs of clinical disease, although evidence of infection was detected through the presence of transient hematological changes. In HENV-26 treated subjects, 4 of 5 exhibited observable clinical signs that included depression and mild respiratory signs (Table S4). Circulating viral genomes were not detected from any animal from the HENV32 treated group; however, in the HENV-26 treated group between 4 and 5 log10 of viral genome/mL was detected on day 5 (in 3 of 5 subjects) and on day 14 (in 1 of 5 subjects) (Figure 6C). The significance of this observation of late viremia is uncertain. It is possible that viral RNA had been cleared temporarily from circulation but had otherwise already become established in the tissues to some degree, whereby on day 14 it was then detected, likely harbored by scavenging antigen presenting cells in circulation. Another factor that cannot be ruled out is rapid clearance of the therapeutic human antibody allowing for a secondary phase of viremia, which may have been brought under control by the emerging host immune responses.
Figure 6. Ferret protection studies.
A) Kaplan-Meier survival curve of ferrets infected with NiV-Bangladesh.
B) Clinical scores of ferrets infected with NiV-Bangladesh. Dotted line represents the threshold for euthanasia criteria.
C) Circulating viral genomes from ferrets infected with NiV-Bangladesh.
D) Viral genomes present in select tissues at study endpoints.
All ten treated ferrets in the study were euthanized at the study end point on day 28 after challenge. Gross inspection at necropsy revealed that all ten animals failed to display significant lesions associated with NiV infection. Two ferrets from the group treated with HENV-26 (designated HENV-26_B & HENV-26_E) and one ferret from the group treated with HENV-32 (designated HENV-32_B) had minimal lymphocytic aggregates within the liver and in at least one lung lobe (data not shown); however, no other significant lesions or significant immunolabeling were noted. One of the HENV-26 treated animals (HENV-26A) had minimal lymphocytic aggregates within the liver, in at least one lung lobe, and in the brainstem; however, no other significant lesions or significant immunolabeling were present in this animal (data not shown). A different ferret from the same treatment group (HENV-26_D) had minimal lymphocytic aggregates within the liver, in at least one lung lobe, and in the brainstem and had significant immunolabeling of neurons in the brainstem (Figure S7). Every animal was sampled for the presence of virus once from brain, spleen, and liver. Lung lobes from the left upper and lower lobes were also sampled, so each lung was sampled in two different regions of the lung. Sampled areas from any tissue were largely representative of the entire organ in terms of severity of gross pathology since the pathology revealed appeared to be uniform throughout the organs. Viral genomes were detected in the brain of two HENV-26-treated subjects (HENV-26_A & HENV-26_D) and in the upper quadrant of the lung of one HENV-32 -treated subject (HENV-32_E) (Figure 6D). We tested both the spleen and liver tissue of each of the 10 animals in the two treatment groups (five animals per group), and infectious virus was not detected in any organ sampled from any animal.
DISCUSSION
We obtained the first panel of naturally occurring human mAbs from a human individual immune to HeV and found mAbs that were potently neutralizing, including 4 that exhibited breadth of recognition for the major strains of NiV. The two most potent, cross-reactive mAbs, HENV-26 and HENV-32, afforded post-exposure protection against the notably more pathogenic Bangladesh strain of NiV in an animal model (Clayton et al., 2012; Lo et al., 2012; Mire et al., 2016). There is no FDA-approved HeV or NiV vaccine or effective treatment for these viruses, and NiV can be transmitted person to person. These two mAbs could be considered lead candidates for prophylaxis or therapy of HeV or NiV infections. HENV-26 directly competes with ephrinB2 for RBP binding, while HENV-32 does not, and crystal structures revealed very distinct antigenic sites. The two mAbs do not compete with each other for HeV/NiV binding, and they neutralize the viruses by very different mechanisms. Therefore, a combination prevention or treatment formulation combining these two mAbs could be considered. MAb combinations may be desirable for treatment of RNA virus infections to prevent virus escape and may produce cooperative effects.
The cross-reactivity of the antibodies we isolated for recognition and neutralization of HeV, NiVM and NiVB, and protection against those viruses, is desirable since it is plausible that a single regimen of monotherapy or a cocktail of these antibodies could prevent or treat each of the three viruses. We tested for breadth of binding for other more distantly related henipaviruses but did not detect cross-reactivity to other viruses. This finding was not surprising, since the RBPs of NiV and HeV have been reported to elicit only a limited cross-reactive antibody response, and cross-protection between Mòjiāng virus or Ghana virus, and the highly pathogenic henipaviruses was not detected (Li et al., 2020).
To explore structural mechanisms of binding and neutralization of the two mAbs, we solved the crystal structures of the mAbs in complex with HeV-RBP and/or NiV-RBP head domains. HENV-26 targets the central cavity and top loops of mainly blade 4, 5, and 6 of the propeller-fold of HeV-RBP and NiV-RBP head domains, overlapping the ephrinB2/B3 binding sites, thus directly competing with ephrinB2/B3 for RBP binding. Therefore, HENV-26 neutralizes HeV or NiV by blocking the receptor binding site of the viruses, thus protecting animals against viral infection by inhibition of viral entry. The major interacting residues of RBPs are conserved between HeV-RBP and NiV-RBP (Figure S4E), making the mAb cross reactive to both viruses. All of the HENV-26 CDRs participate in the formation of the Ab-Ag interface, in contrast to the interaction mode of the previously described phage display library derived antibody m102.3 (Xu et al., 2013).
In contrast, HENV-32 binding causes conformational changes at the β5S4/β6S1 loop and β6S4/β1S1 loops. If HENV-32 bound HeV-RBP is superimposed onto ephrinB2-bound HeV-RBP, the conformational changes at these two loops result in steric clashes between the ephrinB2 G-H loop and residues in these loops of HeV-RBP (Figure S5C). We considered whether HENV-32 could compete with the binding of ephrinB2 via an allosteric effect. However, we did not observe competition between HENV-32 and ephrinB2 for HeV-RBP binding in a BLI assay (Figure S3A). Therefore, the binding of HENV-32 to the putative dimeric interface of RBP head domains likely neutralizes HeV or NiV by altering dynamic features of the surface protein on virions. There is extensive literature defining the dimeric architecture of the RBP (Bowden et al., 2010), which forms a functional tetrameric unit when two disulfide-linked dimers associate (Bossart et al., 2005; Bowden et al., 2008; Negrete et al., 2007), and a disulfide bond in the stalk stabilizes the tetramer (Maar et al., 2012). The henipavirus RBP interacts with host cellular B class ephrins, triggering conformational alterations in RBP that lead to the activation of the F glycoprotein, which facilitates the membrane fusion process (Bradel-Tretheway et al., 2019; Navaratnarajah et al., 2020; Steffen et al., 2012).
Possibly, HENV-32 binding causes rearrangement of the quaternary structure of the RBPs in the head domains in such a way that the orientation of the receptor-binding sites of the RBPs are no longer suitable for receptor binding, preventing viral attachment to cells. However, we found that a soluble recombinant form of ephrinB2 can bind in the presence of HENV-32. Another possibility is that HENV-32 interferes with the activation of HeV/NiV fusion proteins by RBPs, in a mechanism suggest by previous studies of a rabbit antibody with inhibitory activity that likely binds near the epitope recognized by the human mAb HENV-32 (Aguilar et al., 2009). The rearrangement of quaternary structure caused by HENV-32 binding also might make the activation residues in the stem regions of the RBPs inaccessible to the fusion proteins, thus inhibiting viral entry to cells. Further studies are needed to clarify these possibilities.
The solved crystal structures of these Ab-Ag complexes also inform opportunities for future rational antibody engineering efforts to improve binding affinities of the mAbs. HENV-26 CDRH1 residue R31 makes only loose van der Waals interactions with HeV-RBP V502 or NiV-RBP residues I502 and P403. Insertions and mutations at R31 position might enhance binding. As mentioned above, there are significant rearrangements of polar interactions between the HENV-26/HeV-RBP and HENV-26/NiV-RBP complexes at the interface between CDRL1 and RBP region D555 – Q559. This finding suggests that CDRL1 may be a malleable region for improvement of binding. HENV-32 CDRL3 residue N93 interacts with a hydrophobic patch on HeV-RBP (comprising residues P200, L202, and F593) and with the mainchain carbonyl oxygen atom of residue P200 via an H-bond. It could be interesting to examine the effects of mutations of CDRL3 residue N93 to aliphatic residues (valine, leucine, or isoleucine) on binding affinity, because the mutation might improve the hydrophobic effect between N93 and the hydrophobic patch but lose a H-bond between Ab and Ag.
Vaccine development for NiV is a high priority for many recommending bodies. The epitopes recognized by these broad and potent antibodies could be used in structure-based reverse vaccinology design programs to design new vaccine candidates. Thus, the studies provide important new conceptual data on henipavirus immunity, but there are significant limitations of these studies. First, the mAbs in this study were isolated from a single human immune individual, and it is uncertain how generalizable these findings would be in a population. Second, the studies focus on antibodies to RBP, since the individual studied had exposure to a vaccine RBP and we screened for antibodies reacting to the RBP head domain; from these studies we cannot determine the role for antibodies to the RBP stem, potential complex quaternary epitopes at the head/stem interface, or fusion protein in immunity to henipaviruses and more antigenic sites are possible. Third, there may be a role for non-neutralizing antibodies to henipaviruses, but we did not explore that mode of immunity here. These limitations point to the need for additional in-depth studies of this type for immunity to henipaviruses.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, James E. Crowe, Jr. (james.crowe@vumc.org).
Materials Availability
Materials described in this paper are available for distribution for nonprofit use using templated documents from Association of University Technology Managers “Toolkit MTAs”, available at: https://autm.net/surveys-and-tools/agreements/material-transferagreements/mta-toolkit.
Data and Code Availability
Accession numbers -
The crystal structures are deposited at the Protein Data Bank (PDB). Atomic coordinates and structure factors for the crystal structures of HENV-26 or HENV-32 in complex with HeV-RBP or NiV-RBP head domain have been deposited in the Protein Data Bank with the accession codes 6VY6 (HENV-26 in complex with HeV-RBP head domain), 6VY5 (HENV-26 in complex with NiV-RBP head domain), and 6VY4 (HENV-32 in complex with HeV-RBP head domain).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Source of Human B Cells
The study was approved by the Vanderbilt University Medical Center Institutional Review Board. Peripheral blood was collected at Vanderbilt after written informed consent from a healthy male donor with prior history of inadvertent inoculation with recombinant HeV-G in an equine HeV vaccine.
Ferret Model
The animal studies were performed at the Galveston National Laboratory, University of Texas Medical Branch at Galveston (UTMB) and were approved by the UTMB Institutional Animal Care and Use Committee (IACUC). This facility is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
METHOD DETAILS
Expression and purification of HeV and NiV attachment glycoproteins.
The DNA segments correspondent to the head domain of HeV-RBP (residues 185 – 604), head domain of NiVM-RBP (residues 183 – 602) (Bowden et al., 2008), and head domain of NiVB-RBP (residues 185 – 602) were sequenced optimized for expression, synthesized, and cloned into the pcDNA3.1 (+) (HeV and NiVM) or pcDNA3.1 (+)-C-6His (NiVB) expression DNA plasmid downstream of the signal peptide from the pHLsec vector (MGILPSPGMPALLSLVSLLSVLLMGCVA) or osteonectin (MRAWIFFLLCLAGRALA) (GenScript). A TEV protease cleavage site and a His-tag also were incorporated at the C-terminus of HeV and NiVM constructs to facilitate protein purification. Expi293F cells were transfected transiently with plasmids encoding HeV-RBP, NiVM-RBP, or NiVB-RBP head domains, and culture supernatants were harvested after 6 to 7 days. The head domains were purified from the supernatants by nickel affinity chromatography with HisTrap Excel columns (GE Healthcare Life Sciences). For protein production used in crystallization trials, 5 μM kifunensine was included in the culture medium to produce the head domains with high mannose glycans. The high mannose glycoproteins subsequently were treated with endoglycosidase F1 (Millipore) to obtain homogeneously deglycosylated HeV-RBP or NiVM-RBP head domains (Bowden et al., 2008).
PBMC isolation and hybridoma generation.
Peripheral blood was collected at Vanderbilt after written informed consent from a healthy donor with prior history of inadvertent inoculation with recombinant HeV-RBP in an equine HeV vaccine. PBMCs from the donor were isolated by density gradient separation on Ficoll, cryopreserved and stored in the vapor phase of liquid nitrogen until use. Generation of human hybridoma cell lines secreting human mAbs was performed as described previously (Smith et al., 2012). Briefly, human B cells in the PBMC suspension were immortalized by transformation with EBV in the presence of CpG10103, cyclosporin A, and a Chk2 inhibitor and plated in 384-well culture plates. On day 7 to 10 after EBV transformation, the supernatants from transformed B cells were used to screen for the presence of antibodies binding to recombinant HeV-RBP head domain in ELISA. Cells from the wells containing B cells secreting HeV-RBP-reactive antibodies were fused with HMMA2.5 myeloma cells using a BTX ECM 2001 electro cell manipulator by an electrofusion method (Yu et al., 2008). After fusion, human hybridomas were selected in medium with HAT solution containing ouabain. The hybridomas were cloned by flow cytometric sorting of single cells into 384-well plates and then expanded in culture. Particular clones for downstream studies were selected by choosing the clone for each independently derived hybridoma line that exhibited the highest level of IgG secretion.
Production of IgG for mAbs from hybridoma cells.
The selected cloned cell lines secreting mAbs were grown initially in hybridoma growth medium (ClonaCell-HY medium E from STEMCELL Technologies, 03805) and then switched to serum-free medium (GIBCO Hybridoma-SFM, Invitrogen, 12045084) for antibody expression and purification. Cloned hybridoma cells were expanded sequentially to 225 cm2 flasks for mAb production. The supernatants from hybridoma cultures were filtered with 0.45 μm pore diameter filter flasks, and then the IgG from the hybridoma cell line supernatants was purified by affinity chromatography using protein G columns (GE Life Sciences, Protein G HP Columns). Purified IgG generated from hybridomas was used for all EC50 and IC50 studies, competition-binding studies, HDX-MS studies, and animal studies. To generate the corresponding fragment antigen-binding (Fab) fragments for crystallization trials, papain digestion of purified mAb IgG was performed using the Pierce Fab Preparation Kit (Thermo Fisher Scientific). The resulting Fabs were purified from the digestions by affinity chromatography by coupling a protein G affinity column and an anti-human CH1 column (GE Healthcare Life Sciences).
Characterization of antibody isotype, subclass, and variable genes.
The isotype and subclass of secreted antibodies were determined by ELISA. Antibody heavy and light chain variable region genes were sequenced from antigen-specific hybridoma lines that had been cloned biologically using flow cytometric single cell sorting. Briefly, total RNA was extracted using the RNeasy Mini kit (Qiagen, 74106) and reverse-transcriptase PCR (RT-PCR) amplification of the antibody gene cDNAs was performed using the PrimeScript One Step RT-PCR kit (Clontech, RR055A) according to the manufacturer’s protocols with gene-specific primers as shown in Supplemental Table 3 of a previous report (Thornburg et al., 2016). PCR products were purified using Agencourt AMPure XP magnetic beads (Beckman Coulter) and sequenced directly using an ABI3700 automated DNA sequencer without cloning. The identities of gene segments and mutations from germlines were determined by alignment using ImMunoGeneTics database (Brochet et al., 2008; Giudicelli and Lefranc, 2011).
Determination of half maximal effective concentration (EC50) for binding.
To determine EC50 concentrations for binding, we performed ELISA using 384-well plates that were coated overnight at 4°C with 2 μg/mL of a recombinant form of soluble head domain of HeV-RBP or NiV-RBP protein. The plates were blocked for 1 hour with 2% non-fat dry milk and 2% goat serum in PBS-T. After washing the plates 4 times with PBS-T, primary mAbs or hybridoma cell culture supernatants were applied to wells, and the plates were incubated at room temperature for 1 hour. Alkaline phosphatase-conjugated secondary antibodies (goat anti-human IgG Fc, Meridian Life Science), with a dilution of 1:4,000 in blocking solution, were placed into each well following plate wash with PBS-T. After 1-hour incubation, the plates were washed 4 times with PBS-T, and substrate solution (1 mg/mL pNPP disodium salt hexahydrate, Sigma) was added to each well. The plates were incubated at room temperature for approximately 30 min before reading the optical density at 405 nm with a Biotek plate reader. To obtain half maximal effective concentration (EC50) values of human mAbs binding to HeV-RBP or NiV-RBP, ELISA experiments were performed with purified antibodies in three-fold serial dilutions, starting at 20 μg/mL for HeV, and 50 μg/mL for NiVM-RBP or NiVB-RBP, and EC50 values were estimated by a sigmoidal dose-response nonlinear curve fitting procedure with Prism software (GraphPad). Each dilution was performed in quadruplicate, and the experiment was conducted twice independently.
KD determination by bio-layer interferometry (BLI).
Kinetic assays with BLI were performed on an Octet RED biosensor instrument (Pall FortéBio, Menlo Park). Recombinant histidine-tagged RBP (head domain) was immobilized to HIS1K biosensor tips (FortéBio) at 10 μg/mL in proprietary kinetics buffer (FortéBio). After a brief baseline step, serial dilutions of HENV-26 or HENV-32 Fab starting at 200 nM then were associated to coated biosensor tips for 300 seconds, followed by a 900 second dissociation step in 10x kinetics buffer. Data Analysis HT 11.0.2 software was used for curve-fitting to extrapolate equilibrium dissociation constant values. Association and dissociation steps were aligned to reference wells to account for dissociation of antigen from the biosensor tip. Global fitting using a 1:1 model with Savitzky-Golay filtering was used to fit curves.
Biolayer interferometry (BLI) to determine competition-binding groups.
Competition-binding experiments were performed on the Octet RED biosensor, as described previously (Flyak et al., 2015). In brief, HeV-RBP or NiV-RBP with a C-terminal His-tag at 20 μg/mL was loaded onto Ni-NTA coated biosensor tips for 2 min. After 1 min wash in a kinetic buffer (1% BSA, 0.002% Tween 20 in PBS), the biosensor tips were dipped into the first antibody solution at a concentration of 50 μg/mL for 5 min, and then biosensors were switched into a second antibody solution at a concentration of 50 μg/mL for 5 min. The ratio of the maximal signal from the seconding antibody after the first antibody binding to the maximal signal of the second antibody tested alone was calculated and expressed as a percentage.
Biolayer interferometry to test for mAb blocking of HeV-RBP protein binding to the host receptor ephrinB2.
The human antibodies also were used in competition binding with a recombinant form of the host receptor ephrinB2 to determine if the mechanism of neutralization was blockade of receptor binding. The studies were performed using BLI on an Octet RED instrument. Streptavidin (SA) sensor tips were coated in 5 μg/mL biotinylated, recombinant HeV-RBP head domain protein diluted in proprietary Kinetics Buffer 10X (Pall FortéBio) for 30 seconds. Following a brief baseline step, 25 μg/mL HENV-26, HENV-32, or soluble ephrinB2 in buffer was associated to the coated sensor tips for 100 seconds. Tips then were dipped into wells containing a second antibody or ephrinB2. The data were analyzed using FortéBio software, with percentage binding determined by comparing the maximal binding signal of the second protein associated to that of the same protein associated alone.
Cell-surface display flow cytometric assay to test for mAb blocking of HeV-RBP protein binding to the host receptor ephrinB2.
A suspension of 293F cells was transfected with cDNA encoding the full length HeV-RBP protein using PEI for 72 hours. Transfected cells were harvested and plated in V-bottom 96-well plates at 50,000 cells/well. After a wash step, cells were incubated with 50 μg/mL soluble ephrinB2 protein or FACS buffer for 30 minutes. Without washing, 2 μg/mL HENV-26 labeled with AlexaFluor-647 (Invitrogen) was added to cells and incubated for 30 minutes. Cells then were washed and analyzed using an Intellicyt iQue flow cytometry instrument. Binding of HENV-26 in the presence or absence of ephrinB2 was expressed as mean fluorescence intensity (MFI).
Crystallization and structural determination of antibody-antigen complexes.
Purified Fabs were mixed with deglycosylated HeV-RBP or NiV-RBP head domain in a molar ratio of 1:1, and the mixtures were purified further by size-exclusion chromatography with a Superdex-200 HiLoad column (GE Healthcare Life Sciences) to obtain antibody-antigen complexes. The complexes were concentrated to about 10 mg/mL and subjected to crystallization trials. HeV-RBP head domain in complex with the Fab HENV-26 was crystallized in 30% MPD, 0.1 imidazole pH 6.5, 0.2 M ammonium sulfate, and 10% PEG 3350, and NiV-RBP head domain in complex with the Fab HENV-26 was in 1.0 M sodium malonate pH 7.0, 0.1 M Bis-Tris propane pH 7.0. Protein crystals were flash-frozen in liquid nitrogen after a quick soaking in the corresponding cryo-protection solutions (same as the crystallization solution for HeV-RBP/HENV26 complex, the solution of 25% sodium malonate pH 7.0 and 0.1 M Bis-Tris propane pH 7.0 for NiV-RBP/HENV-26 complex). Diffraction data were collected at the beamline 21-ID-G at the Advanced Photon Source. The diffraction data were processed with XDS (Kabsch, 2010) and CCP4 suite (Winn et al., 2011). The crystal structures were solved by molecular replacement using the structure of the head domain of HeV-RBP or NiV-RBP in human ephrinB2-HeV-RBP or ephrinB2-NiV-RBP complex (PDB ID 2VSK and 2VSM) and Fab structure of MR78 (PDB ID 5JRP) with the program Phaser (McCoy et al., 2007).The structure was refined and rebuilt manually with Phenix (Adams et al., 2010) and Coot (Emsley and Cowtan, 2004), respectively. The models have been deposited into the Protein Data Bank. PyMOL software (Schrodinger, 2015) was used to make all of the structural figures.
CedV-RBP and GhV-RBP ELISA.
Constructs for CedV-RBP head domain and GhV-RBP full ectodomain were transfected transiently into Expi293F cells using ExpiFectamine transfection reagents (Thermo Fisher). Cell supernatants were harvested 7 days post-transfection. CedV-RBP head domain was purified using HisTrap affinity chromatography as described above for HeV-RBP and NiV-RBP head domains (SigmaAldrich). Full-length ectodomain GhV-RBP containing a GCN tetramerization domain was purified using S-protein agarose (EMD Millipore). To test HENV-26 and HENV-32 for binding to CedV-RBP and GhV-RBP, 384-well plates were coated with 5 μg/mL CedV-RBP head domain or cell supernatant from GhV-RBP transfected cells and incubated overnight at 4°C. The following day, plates were blocked with DPBS-T containing 2% milk and 1% goat serum at room temperature for 1 hour. After a wash step, 3-fold serial dilutions of HENV-26, HENV-32, or control mAbs for CedV (14F3) or GhV (10D5) kindly provided by Christopher Broder were added to plates and incubated for 1 hour at room temperature. Secondary antibody (goat anti-human IgG-HRP for HENV-26 and HENV-32, goat anti-mouse human adsorbed Ig-HRP for controls) diluted 1:1,000 in DBPS-T containing 1% milk and 1% goat serum were added to plates. TMB substrate was used to develop plates, and the reaction was quenched using 1N HCl 10–15 minutes later. Absorbance at 450 nm was read using a Biotek plate reader, and binding curves were generated using non-linear regression analysis in GraphPad Prism software.
HeV and NiV viruses.
Nipah virus number 1999011924 was obtained from a patient from the 1999 outbreak in Malaysia. The passage 3 (P3) virus stock of NiVM we used for used for neutralization assays is known to have an N277K polymorphism in the RBP (Mire et al., 2016). The isolate of NiVB was 200401066 and was obtained from a fatal human case during the outbreak in Rajbari, Bangladesh in 2004 and passaged on Vero E6 cell monolayer cultures twice, making this a passage 2 virus. Hendra virus was obtained from a patient from the 1994 outbreak in Australia. All viruses were kindly provided by Dr. Thomas Ksiazek, UTMB. Each virus was propagated on Vero E6 cells in Eagle’s minimal essential medium supplemented with 10% fetal calf serum. The NiVM, NiVB and HeV challenge virus stocks were assessed for the presence of endotoxin using The Endosafe-Portable Test System (PTS) (Charles River Laboratories, Wilmington, MA). Each virus preparation was diluted 1:10 in Limulus Amebocyte Lysate (LAL) Reagent Water per the manufacturer’s instructions, and endotoxin levels were tested in LAL Endosafe-PTS cartridges as directed by the manufacturer. Each preparation was found to be below detectable limits, whereas positive controls showed that the tests were valid. All experiments involving infectious henipaviruses were carried out at the UTMB Galveston National Laboratory under biosafety level 4 conditions.
Neutralization assays.
The virus neutralizing activity concentrations were determined for NiVM, NiVB, and HeV using a plaque reduction assay. Briefly, antibodies were diluted serially two-fold from 50 μg/mL to extinction and incubated with a target of ~100 plaque-forming units (pfu) of NiVM, NiVB, or HeV for 45 min at 37 °C. Virus and antibody mixtures then were added to individual wells of six-well plates of Vero76 cells. Plates were stained with neutral red two days after infection, and plaques were counted 24 h after staining. Neutralization potency was calculated based on pfu for each virus in the well without antibody. The neutralization experiments were performed in triplicate, with independent virus preparations and duplicate readings for each replicate. Mean half-maximal inhibitory concentrations were calculated from the plaque counts using GraphPad Prism software following the step-by-step protocol outlined explicitly in a previous report (Ferrara and Temperton, 2018).
Protection study in ferrets.
Thirteen healthy immunocompetent female ferrets without previous enrollment in studies weighing 0.75–1 kg were housed socially and placed into cohorts for treatment or no treatment (Table S4). For virus challenge and procedures, animals were anesthetized by isoflurane inhalation. Animals were inoculated intranasally (i.n.) with ~5 × 103 plaque-forming units (pfu) of NiVB in 0.5 mL Dulbecco’s minimal essential medium (Sigma-Aldrich, St. Louis, MO) on day 0. After challenge, ferrets in the treated cohorts were given mAb HENV-26 or HENV-32 by intraperitoneal (i.p.) injection on day 3 and 5 after challenge at a 15 mg/kg dose, a dosage lower than that used in prior studies in ferrets and nonhuman primates with an antiviral mAb (Bossart et al., 2011; Bossart et al., 2009; Geisbert et al., 2014; Mire et al., 2019; Mire et al., 2016). Animals were anesthetized for clinical examination including body weight, temperature, respiration quality, and blood collection on days 0, 3, 5, 7, 10, 14, and 28 after challenge. Before and after challenge, animals were assessed daily for clinical score on a scale of 0 of 12 for clinical observations based on coat appearance, body weight loss, social behavior, and provoked behavior; animals scoring 9 or greater were euthanized per the established UTMB IACUC protocol. The remaining subjects were euthanized at the study endpoint on day 28 after challenge.
Specimen collection and processing in NiV- and HeV-infected ferrets.
On sampling days, blood was collected and placed in MiniCollect EDTA tubes (Greiner Bio-One, Monroe, NC) for virus load and hematology analysis or MiniCollect serum tubes (Greiner Bio-One) for clinical chemistry analysis. Necropsy was performed on all ferrets, and tissues sampled included lungs, liver, spleen, kidney, adrenal gland, pancreas, and brain (frontal cortex). Ten percent tissue homogenates of liver, spleen, kidney, adrenal gland, and brain were used for virus load analysis.
Measurement of infectious virus load in ferret tissues.
Virus titration was performed by plaque assay with Vero cells from all tissue homogenates (10% w/v). In brief, increasing 10-fold dilutions of the samples were adsorbed to Vero cell monolayers in duplicate wells (200 μL); the limit of detection was 25 pfu/mL for whole blood and 250 pfu/gram for tissue.
RNA isolation from ferret tissues.
Immediately following sampling, 100 μL of blood was added to 600 μL of AVL viral lysis buffer (Qiagen) for RNA extraction. For tissues, approximately 100 mg was stored in 1 ml RNAlater (Qiagen) for 7 days to stabilize RNA. RNAlater was completely removed, and tissues were homogenized in 600 μL RLT buffer (Qiagen) in a 2-mL cryovial using a tissue lyser (Qiagen) and ceramic beads. The tissues sampled included cerebral spinal cord, brain stem, brain (frontal cortex), lung (left upper and left lower), spleen, and liver. All blood samples were inactivated in AVL viral lysis buffer, and tissue samples were homogenized and inactivated in RLT buffer prior to removal from the BSL-4 laboratory. Subsequently, RNA was isolated from blood using the QIAamp viral RNA kit (Qiagen), and from tissues using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions supplied with each kit.
Detection of viral genomes in ferret samples.
RNA was isolated from blood or tissues and analyzed using primers/probe targeting the nucleoprotein (N) gene and intergenic region between N and phosphoprotein (P) of NiV for quantitative real-time PCR (qRT-PCR), with the probe used here being 6-carboxyfluorescein (6FAM)-5′ CGT CAC ACA TCA GCT CTG ACA A 3′−6 carboxytetramethylrhodamine (TAMRA) (Life Technologies, Carlsbad, CA). NiV RNA was detected using the CFX96 detection system (Bio-Rad) in One-step probe qRT-PCR kits (Qiagen) with the following cycle conditions: 50°C for 10 minutes, 95°C for 10 seconds, and 40 cycles of 95°C for 10 seconds and 57°C for 30 seconds. Threshold cycle (CT) values representing NiV genomes were analyzed with CFX Manager Software, and data are shown as genome equivalents (GEq). To create the GEq standard, RNA from NiV challenge stocks was extracted and the number of NiV genomes was calculated using Avogadro’s number and the molecular weight of the NiV genome.
Hematology and serum biochemistry.
Blood and sera were collected via the anterior vena cava from all 11 ferrets on days 0, 3, 5, 7, 10, 14, 21, and 28 after challenge or at euthanasia. Complete blood counts of total white blood cells, white blood cell differentials, red blood cells, platelets, hematocrit values, total hemoglobin concentrations, mean cell volumes, mean corpuscular volumes, and mean corpuscular hemoglobin concentrations were analyzed from blood collected in EDTA tubes (Greiner Bio One) using a VetScan HM5 hematology instrument per the manufacturer’s instructions (Abaxis). Serum analysis of blood chemistries was completed using a Piccolo point-of-care analyzer and Biochemistry Panel Plus analyzer discs (Abaxis), which measured concentrations of albumin, amylase, alanine aminotransferase (ALT) aspartate aminotransferase (AST), alkaline phosphatase (ALP), gamma-glutamyltransferase (GGT), glucose, cholesterol, total protein, total bilirubin (TBIL), blood urea nitrogen (BUN), creatine (CRE), and C-reactive protein (CRP). All blood and serum samples were processed and analyzed immediately after collection.
Histopathology and immunohistochemistry.
Necropsy was performed on all subjects. Tissue samples of all major organs were collected for histopathologic and immunohistochemical examination and were immersion-fixed in 10% neutral buffered formalin for at least 21 days in BSL-4. Subsequently, formalin was changed; specimens were removed from BSL-4, processed in BSL-2 by conventional methods and embedded in paraffin and sectioned at 5 μm thickness. For immunohistochemistry, specific anti-NiV immunoreactivity was detected using an anti-NiV N protein rabbit primary antibody (kindly provided by Dr. Christopher Broder) at a 1:5,000 dilution for 30 minutes. The tissue sections were processed for immunohistochemistry using the Dako Autostainer (Dako, Carpinteria, CA). The secondary antibody used was biotinylated goat anti-rabbit IgG (Vector Laboratories, Burlingame, CA) at 1:200 for 30 minutes followed by Dako LSAB2 streptavidin-HRP (Dako) for 15 minutes. Slides were developed with Dako DAB chromagen (Dako) for 5 minutes and counterstained with hematoxylin for one minute. Non-immune rabbit IgG was used as a negative staining control.
QUANTIFICATION AND STATISTICAL ANALYSIS
In ELISA experiments, binding curves were generated using non-linear regression analysis in GraphPad Prism software. In the ferret studies, the constraints of high-containment work using animal studies in biosafety level 4 restrict the number of animal subjects and the volume of biological samples, which affects the ability to repeat assays independently and thus limit statistical analysis. Data are presented as the mean calculated from replicate samples, not from replicate assays, and error bars represent the standard deviation (s.d.) between replicates.
Supplementary Material
Figure S1, Related to Table 1. Full dilution curve data for neutralization or binding of 10 human monoclonal antibodies to HeV, NiVM, or NiVB. A. Neutralization was tested with live viruses; percent neutralization is shown over a varying antibody concentration range. m102.4 is shown for comparative purposes. B. Binding in ELISA to recombinant RBPs. Optical density at 405 nm is shown over a varying antibody concentration range.
Figure S2, Related to Table 1.
A. Binding assays of HENV-26 or HENV-32 to full-length HeV-RBP or NiV-RBP proteins detected by flow cytometric analysis. Binding to cells transfected with HeV-RBP (red), NiVM-RBP (blue), NiVB-RBP (green), or un-transfected (black) cells was analyzed using an Intellicyt iQue instrument. Values are expressed as mean fluorescence intensity (MFI) of PE-conjugated secondary antibody signal and were plotted in GraphPad to interpolate EC50 values by non-linear regression analysis. Serial dilutions of HENV-26 or HENV-32 were performed in triplicate, with data representative of three independent assays shown. Error bars indicate SEM.
B. Kinetics of binding of HENV-26 or HENV-32 to full-length HeV-RBP or NiV-RBP proteins in biolayer interferometry to determine affinity of binding. Binding kinetics of Fab fragments corresponding to HENV-26 and HENV-32 were performed on an Octet RED instrument (FortéBio). Recombinant histidine-tagged RBP head domain was immobilized to HIS1K biosensor tips (FortéBio) at 10 μg/mL in kinetics buffer. After a brief baseline step, serial dilutions of HENV-26 or HENV-32 Fab starting at 200 nM then were associated to coated biosensor tips, followed by a dissociation step in 1x kinetics buffer. Curve-fitting was performed to extrapolate equilibrium dissociation constant values.
Figure S3, Related to Figures 2 and 3. Comparison of the eprhrinB2 binding site on HeV-RBP with the epitope for binding of HENV-26
A. Competition-binding assay of three human mAbs with ephrinB2 for binding to HeV-RBP head domain using biolayer interferometry. Numbers in boxes are the percentage binding signal of the second protein applied after binding of the first protein, compared with binding signal of the second protein alone. The proteins were defined as competing if the first protein reduced binding of the second protein by more than 70 percent. The proteins were defined as non-competing if the first protein reduced binding of the second protein by less than 40 percent.
B. Cell surface display flow cytometric assay to test for mAb blocking of HeV-RBP protein binding to the host receptor ephrinB2. 293F cells were transfected to display the full-length HeV-RBP protein on the cell surface. Cells were incubated with soluble ephrinB2 protein or FACS buffer, then HENV-26 labeled with AlexaFluor-647 was added to cells and incubated. Cells then were washed and analyzed using an Intellicyt iQue flow cytometry instrument. Binding of HENV-26 in the presence or absence of ephrinB2 was expressed as mean fluorescence intensity (MFI).
C-E. Binding site mapping of ephrinB2 and m102.3 onto the HeV-RBP or NiV-RBP head domain surface. The surfaces of HeV-RBP or NiV-RBP head domains are colored in grey, with glycans colored in black.
C. The ephrinB2 binding site is mapped and colored in green onto HeV-RBP head domain surface. The surface envelope (shown as blue mesh) of the epitope of HeV-RBP head domain recognized by HENV-26 is overlaid onto the HeV-RBP head domain.
D. The mAb m102.3 binding site is mapped and colored in yellow onto the HeV-RBP head domain surface. The surface envelope (shown as blue mesh) of the epitope on HeV-RBP head domain recognized by HENV-26 is overlaid onto the HeV-RBP head domain.
E. The ephrinB2 binding site is mapped and colored in pink onto the NiV-RBP head domain surface. The surface envelope (shown as blue mesh) of the epitope on NiV-RBP head domain recognized by HENV-26 is overlaid on to the NiV-RBP head domain.
Figure S4, Related to Figures 3 and 4. Molecular recognition of HeV-RBP head domain by mAbs.
A) Water-mediated interactions between HeV-RBP head domain and mAb HENV-26. HeV-RBP head domain is colored in green, HENV-26 heavy chain in cyan, and the light chain in yellow. Water molecules are represented as magenta spheres, and involved residues are shown as sticks and labeled. Hydrogen bonds are represented as broken orange lines.
B) Water-mediated interactions between HeV-RBP head domain and mAb HENV-32. HeV-RBP head domain is colored in green, HENV-32 heavy chain in orange, and the light chain in salmon. Water molecules are represented as magenta spheres, and involved residues are shown as sticks, and labeled. Hydrogen bonds are represented as broken orange lines.
C) CedV-RBP or GhV-RBP ELISA. Binding of HENV-26 or HENV-32 was tested in ELISA using recombinant CedV-RBP head domain or GhV-RBP full ectodomain. MAbs to CedV-RBP (14F3) or GhV-RBP (10D5) were used as controls.
D) Lack of dimer disruption mediated by HENV-26 or HENV-32. HeV-RBP full ectodomain was expressed in Expi293F cells and purified sequentially by HisTrap affinity chromatography followed by size exclusion chromatography (SEC). Size exclusion peaks corresponding to dimeric (SEC peak #2) or tetrameric (SEC peak #1) species were each incubated with HENV-26 or HENV-32 Fab for 1 hour at room temperature. Each preparation was then analyzed by 4–12% Bis-Tris SDS-PAGE using SimplyBlue SafeStain Coomassie G-250 stain.
E) Epitope mapping of RBP head domains of HeV, NiVM, and NiVB. The RBP amino acid sequences of the three viruses were aligned with MUSCLE (1), and the figure was made with sequence alignment editor, ALINE (2). For comparison, the sequences of Cedar virus RBP (CedV-RBP) and Ghana virus RBP (GhV-RBP) also are shown.
Upper panel: The epitope residues recognized by HENV-26 (in complex with HeV-RBP [PDB ID 6VY6] or NiV-RBP Malaysia [NiV-RBPm; PDB ID 6VY5] are highlighted with red boxes, the epitope residues recognized by HENV-32 (in complex with HeV-RBP [PDB ID 6VY4]) with orange boxes, and epitope resides recognized by m102.3 [PDB ID 6CMG] are indicated with purple lines over the alignment.
Lower panel: The ephrinB2 binding residues from complexes with HeV-RBP [PDB ID 6PDL] or NiV-RBPm [PDB ID 2VSM] are highlighted with red boxes, and the ephrinB3 binding residues from a complex with NiV-RBPm [PDB ID 3D12] with yellow boxes.
F) Superimposition of crystal structures for comparative purposes
Upper panel: The crystal structure of CedV-RBP (PDB ID: 6P72) is superimposed onto that of the HeV-RBP/HENV-26 complex. CedV-RBP structure is shown in dark grey; HeV-RBP in green; HENV-26 heavy chain in cyan, and HENV-26 light chain in yellow. Sequence variations at several positions in CedV-RBP (e.g., G512, H518, D550, L552) result in loss of H-bonds seen in the corresponding positions in the HeV-RBP complex.
Middle panel: The crystal structure of GhV-RBP (PDB ID: 4UF7) is superimposed onto that of the HeV-RBP/HENV-26 complex. GhV-RBP structure is shown in light grey; HeV-RBP in green; HENV-26 heavy chain in cyan, and HENV-26 light chain in yellow. Sequence variations at several positions in GhV-RBP (e.g., G500, T511, E539, V541) result in loss of H-bonds seen in the corresponding positions in the HeV-RBP complex.
Lower panel: The CedV-RBP and GhV-RBP structures are superimposed on that of the HeV-RBP/HENV-32 structure. The major epitope of HeV-RBP (shown in blue, residues T196 – N210) and corresponding residues of CedV-RBP (in dark grey, residues P219 – Q233) or GhV-RBP (in light grey, residues P212 – T226) are shown in cartoon to represent backbone conformations. Sequence variations at this region among the different RBPs result in significant conformational variation and may create a prohibitively high energy barrier to prevent CedV-RBP or GhV-RBP from adopting backbone conformations similar to that of HeV-RBP or NiV-RBP.
Figure S5, Related to Figures 2, 4 and 5. Overlays of HENV-26 or 32 for comparative purposes.
A) Superimposition of the crystal structures of HeV-RBP head domain in complex with HENV-26 or HENV-32. HeV-RBP head domain in the HeV-RBP/HENV-26 complex is colored in green, and that in the HeV-RBP/HENV-32 in light blue. The HENV-26 heavy chain is colored in cyan, HENV-26 light chain in yellow, HENV-32 heavy chain in orange, and HENV-32 light chain in salmon. The CDRs of both mAbs and the individual blades of the HeV-RBP head domain are labeled. Regions in the HeV-RBP head domain with large structural differences between structures are indicated within broken red lines.
B) Superimposition of the crystal structures of the dimeric HeV-RBP head domain and HeV-RBP head domain in complex with HENV-32. HeV-RBP head domain in the HeV-RBP/HENV-32 is shown in light blue, HENV-32 heavy chain in orange, and HENV-32 light chain in salmon. One protomer of the HeV-RBP head domain is colored in magenta and the other in cyan. Individual blades of the superimposed HeV-RBP head domains are labeled (β1 to β6).
C) Superimposition of HeV-RBP structures in the HENV-32/HeV-RBP complex and ephrinB2/HeV-RBP complex. The HeV-RBP structure in the HENV-32/HeV-RBP complex is shown in light blue, and HeV-RBP in the ephrinB2/HeV-RBP complex in yellow. EphrinB2 in the complex is shown in grey. The overlay suggests that the conformation of HeV-RBP β6/S2-S3 and β5/S4- β6/S1 loops in the HENV-32/HeV-RBP complex causes potential steric clashes between the loops and the ephrinB2 G-H loop. The potential steric clashes between the HENV-32 bound HeV-RBP structure and ephrinB2 G-H loop are represented with broken red lines, and the residues with potential steric clashes are shown in stick, and corresponding residues of the ephrinB2 bound HeV-RBP structure are shown as line representation.
Figure S6, Related to Figure 5. Structural details of the HENV-32 CDRH3 binding site in the crystal structures of HeV-RBP-apo (dimeric form) (panel A) or the HeV-RBP/m102.3 complex (panel B). Residues interacting with HENV-32 CDRH3 are shown as sticks. Consistent with the structure in the HeV-RBP/HENV-26 complex, the HENV-32 CDRH3 binding pocket is hidden underneath salt bridges between residue R258 and D260 in both structures.
(panels C - F) Surface representation of the HENV-32 CDRH3 binding site in four different crystal structure forms of HeV-RBP. The residues interacting with the HENV-32 CDRH3 are indicated; R201 is colored in red, L202 in yellow, I203 in green, Y205 in americium, R258 in white on blue, G259 in cyan, D260 in wheat, V262 in pink, P263 in olive, and S264 in magenta.
C) HENV-32/HeV-RBP complex; the HENV-32 CDRH3 paratope is shown in orange as both ribbon and stick representations and labeled in cyan.
D) HENV-26/HeV-RBP complex.
E) m102.3/HeV-RBP complex.
F) HeV-RBP apo (dimeric form). Residue P263 in the HeV-RBP/HENV-26 complex and HeV-RBP/m102.3 complex is not solvent accessible.
Figure S7, Related to Figure 6. Representative histopathology of brain and liver tissues from ferrets infected with NiVB and treated with recombinant anti-HENV mAbs, compared with untreated control animals.
A to D) Tissues from tissue from subject C1:
A) H&E staining of brain tissue did not show significant histological lesions;
B) immunolabeling of brain tissue showed the endothelium of small caliber vessels within the neuronal parenchyma multifocally had diffuse cytoplasmic immunolabeling.
C) H&E staining of lung tissue showed mild interstitial pneumonia with multifocal nodular formation composed of necrotic debris, hemorrhage and mixed inflammatory infiltrates of neutrophils and macrophages;
D) immunolabeling of lung tissue showed diffuse cytoplasmic immunolabeling of segmental regions of the alveolar septa largely centered on the necrotic nodules, mononuclear cells within alveolar septa, mononuclear cells free within alveolar spaces (alveolar macrophages), endothelium of small to medium caliber vessels, and rarely lower airway epithelium (D).
E to H) Tissues from subject HENV-32_A did not show significant histological lesions or immunolabeling:
E) H&E staining of brain;
F) Immunolabeling of brain;
G) H&E staining of lung;
H) Immunolabeling of lung.
I to L) Tissues from subject HENV-26_D:
I) H&E staining of brain showed moderate cuffing of small caliber vessels with lymphocytes in the brainstem and in vessels surrounding the ventricular system;
J) Immunolabeling of brain showed diffuse cytoplasmic immunolabeling of neurons in association with the lymphocytic infiltrates (inset and arrow);
K) H&E staining of lung showed rare cuffing of small caliber vessels of the lung with lymphocytes and plasma cells present;
L) Immunolabeling of lung was not detected. (I).
Highlights.
Antibodies isolated from a healthy donor with exposure to Hendra G protein
Antibodies protect against Hendra and Nipah infections in animals
Antibodies recognize diverse conserved sites on the henipavirus G protein
Antibodies to differing sites neutralize by diverse molecular mechanisms
ACKNOWLEDGEMENTS
We thank Rachel Nargi from Vanderbilt in the Crowe laboratory for technical support with antibody production and purification, Robert Carnahan for intellectual contributions to antibody production and sequencing, and Christopher Broder for helpful scientific conversations. We also thank Mathew Hyde and the UTMB Animal Resource Center for their assistance with animal procedures. This work was supported by a grant from the National Institutes of Health (NIH) U19 AI142764 and departmental funds from UTMB to Thomas Geisbert. Was supported by NIH fellowship grant F31 AI152332. The project described was supported by CTSA award No. UL1 TR002243 from the National Center for Advancing Translational Sciences (NCATS). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIAID or NIH. X-ray diffraction data were collected at Beamline 21-ID-G at the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). Support for crystallography was provided from the Vanderbilt Center for Structural Biology. The graphical abstract was made in part with BioRender.com.
Footnotes
Declaration of interests. J.E.C. has served as a consultant for Takeda Vaccines, Sanofi Pasteur, Pfizer, and Novavax, is on the Scientific Advisory Boards of CompuVax, GigaGen, Meissa Vaccines, and is Founder of IDBiologics, Inc. Vanderbilt University has applied for a patent related to antibodies described in this paper. All other authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information including supplemental experimental procedures, figures, and tables can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar HC, Ataman ZA, Aspericueta V, Fang AQ, Stroud M, Negrete OA, Kammerer RA, and Lee B (2009). A novel receptor-induced activation site in the Nipah virus attachment glycoprotein (G) involved in triggering the fusion glycoprotein (F). J Biol Chem 284, 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar HC, and Iorio RM (2012). Henipavirus membrane fusion and viral entry. Curr Top Microbiol Immunol 359, 79–94. [DOI] [PubMed] [Google Scholar]
- Bonaparte MI, Dimitrov AS, Bossart KN, Crameri G, Mungall BA, Bishop KA, Choudhry V, Dimitrov DS, Wang LF, Eaton BT, et al. (2005). Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci U S A 102, 10652–10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S, Jardetzky TS, and Lamb RA (2015). Timing is everything: Fine-tuned molecular machines orchestrate paramyxovirus entry. Virology 479–480, 518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S, Welch BD, Kors CA, Yuan P, Jardetzky TS, and Lamb RA (2011). Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J Virol 85, 12855–12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart KN, Crameri G, Dimitrov AS, Mungall BA, Feng YR, Patch JR, Choudhary A, Wang LF, Eaton BT, and Broder CC (2005). Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J Virol 79, 6690–6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart KN, Fusco DL, and Broder CC (2013). Paramyxovirus entry. Adv Exp Med Biol 790, 95–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart KN, Geisbert TW, Feldmann H, Zhu Z, Feldmann F, Geisbert JB, Yan L, Feng YR, Brining D, Scott D, et al. (2011). A neutralizing human monoclonal antibody protects african green monkeys from hendra virus challenge. Sci Transl Med 3, 105ra103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart KN, Zhu Z, Middleton D, Klippel J, Crameri G, Bingham J, McEachern JA, Green D, Hancock TJ, Chan YP, et al. (2009). A neutralizing human monoclonal antibody protects against lethal disease in a new ferret model of acute nipah virus infection. PLoS Pathog 5, e1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden TA, Aricescu AR, Gilbert RJ, Grimes JM, Jones EY, and Stuart DI (2008). Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat Struct Mol Biol 15, 567–572. [DOI] [PubMed] [Google Scholar]
- Bowden TA, Crispin M, Harvey DJ, Jones EY, and Stuart DI (2010). Dimeric architecture of the Hendra virus attachment glycoprotein: evidence for a conserved mode of assembly. J Virol 84, 6208–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradel-Tretheway BG, Zamora JLR, Stone JA, Liu Q, Li J, and Aguilar HC (2019). Nipah and Hendra virus glycoproteins induce comparable homologous but distinct heterologous fusion phenotypes. J Virol 93, e00577–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochet X, Lefranc MP, and Giudicelli V (2008). IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36, W503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder CC, Xu K, Nikolov DB, Zhu Z, Dimitrov DS, Middleton D, Pallister J, Geisbert TW, Bossart KN, and Wang LF (2013). A treatment for and vaccine against the deadly Hendra and Nipah viruses. Antiviral Res 100, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkovitz A, and Ofran Y (2016). Understanding differences between synthetic and natural antibodies can help improve antibody engineering. MAbs 8, 278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadha MS, Comer JA, Lowe L, Rota PA, Rollin PE, Bellini WJ, Ksiazek TG, and Mishra A (2006). Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis 12, 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton BA, Middleton D, Bergfeld J, Haining J, Arkinstall R, Wang L, and Marsh GA (2012). Transmission routes for Nipah virus from Malaysia and Bangladesh. Emerg Infect Dis 18, 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crennell S, Takimoto T, Portner A, and Taylor G (2000). Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol 7, 1068–1074. [DOI] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Muller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Cottontail VM, Rasche A, Yordanov S, et al. (2012). Bats host major mammalian paramyxoviruses. Nat Commun 3, 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Ferrara F, and Temperton N (2018). Pseudotype neutralization assays: From laboratory bench to data analysis. Methods Protoc 1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field HE (2016). Hendra virus ecology and transmission. Curr Opin Virol 16, 120–125. [DOI] [PubMed] [Google Scholar]
- Flyak Andrew I., Ilinykh Philipp A., Murin Charles D., Garron T, Shen X, Fusco Marnie L., Hashiguchi T, Bornholdt Zachary A., Slaughter James C., Sapparapu G, et al. (2015). Mechanism of human antibody-mediated neutralization of Marburg virus. Cell 160, 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Mire CE, Geisbert JB, Chan YP, Agans KN, Feldmann F, Fenton KA, Zhu Z, Dimitrov DS, Scott DP, et al. (2014). Therapeutic treatment of Nipah virus infection in nonhuman primates with a neutralizing human monoclonal antibody. Sci Transl Med 6, 242ra282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli V, and Lefranc MP (2011). IMGT/junctionanalysis: IMGT standardized analysis of the V-J and V-D-J junctions of the rearranged immunoglobulins (IG) and T cell receptors (TR). Cold Spring Harb Protoc 2011, 716–725. [DOI] [PubMed] [Google Scholar]
- Gurley ES, Montgomery JM, Hossain MJ, Bell M, Azad AK, Islam MR, Molla MA, Carroll DS, Ksiazek TG, Rota PA, et al. (2007). Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis 13, 1031–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin K, Hyatt AD, Fogarty R, Middleton D, Bingham J, Epstein JH, Rahman SA, Hughes T, Smith C, Field HE, et al. (2011). Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am J Trop Med Hyg 85, 946–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homaira N, Rahman M, Hossain MJ, Epstein JH, Sultana R, Khan MS, Podder G, Nahar K, Ahmed B, Gurley ES, et al. (2010). Nipah virus outbreak with person-to-person transmission in a district of Bangladesh, 2007. Epidemiol Infect 138, 1630–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardetzky TS, and Lamb RA (2014). Activation of paramyxovirus membrane fusion and virus entry. Curr Opin Virol 5, 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler MK, Becker DJ, Peel AJ, Justice NV, Lunn T, Crowley DE, Jones DN, Eby P, Sanchez CA, and Plowright RK (2018). Changing resource landscapes and spillover of henipaviruses. Ann N Y Acad Sci 1429, 78–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MC, Borg NA, Streltsov VA, Pilling PA, Epa VC, Varghese JN, McKimm-Breschkin JL, and Colman PM (2004). Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J Mol Biol 335, 1343–1357. [DOI] [PubMed] [Google Scholar]
- Li Y, Li R, Wang M, Liu Y, Yin Y, Zai X, Song X, Chen Y, Xu J, and Chen W (2020). Fc-based recombinant henipavirus vaccines elicit broad neutralizing antibody responses in mice. Viruses 12, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, and Janin J (1999). The atomic structure of protein-protein recognition sites. J Mol Biol 285, 2177–2198. [DOI] [PubMed] [Google Scholar]
- Lo MK, Lowe L, Hummel KB, Sazzad HM, Gurley ES, Hossain MJ, Luby SP, Miller DM, Comer JA, Rollin PE, et al. (2012). Characterization of Nipah virus from outbreaks in Bangladesh, 2008–2010. Emerg Infect Dis 18, 248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby SP (2013). The pandemic potential of Nipah virus. Antiviral Res 100, 38–43. [DOI] [PubMed] [Google Scholar]
- Maar D, Harmon B, Chu D, Schulz B, Aguilar HC, Lee B, and Negrete OA (2012). Cysteines in the stalk of the nipah virus G glycoprotein are located in a distinct subdomain critical for fusion activation. J Virol 86, 6632–6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh GA, de Jong C, Barr JA, Tachedjian M, Smith C, Middleton D, Yu M, Todd S, Foord AJ, Haring V, et al. (2012). Cedar virus: a novel Henipavirus isolated from Australian bats. PLoS Pathog 8, e1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Yanez-Arenas C, Chen C, Plowright RK, Webb RJ, and Skerratt LF (2018). Climate change could increase the geographic extent of hendra virus spillover risk. Ecohealth 15, 509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J Appl Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mire CE, Chan YP, Borisevich V, Cross RW, Yan L, Agans KN, Dang HV, Veesler D, Fenton KA, Geisbert TW, et al. (2019). A cross-reactive humanized monoclonal antibody targeting fusion glycoprotein function protects ferrets against lethal Nipah virus and Hendra virus infection. J Infect Dis 221,(Supplement 4):S471–S479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mire CE, Satterfield BA, Geisbert JB, Agans KN, Borisevich V, Yan L, Chan YP, Cross RW, Fenton KA, Broder CC, et al. (2016). Pathogenic differences between Nipah virus Bangladesh and Malaysia strains in primates: Implications for antibody therapy. Sci Rep 6, 30916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mire CE, Versteeg KM, Cross RW, Agans KN, Fenton KA, Whitt MA, and Geisbert TW (2013). Single injection recombinant vesicular stomatitis virus vaccines protect ferrets against lethal Nipah virus disease. Virol J 10, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K, Selleck P, Hooper P, Hyatt A, Gould A, Gleeson L, Westbury H, Hiley L, Selvey L, Rodwell B, et al. (1995). A morbillivirus that caused fatal disease in horses and humans. Science 268, 94–97. [DOI] [PubMed] [Google Scholar]
- Navaratnarajah CK, Generous AR, Yousaf I, and Cattaneo R (2020). Receptor-mediated cell entry of paramyxoviruses: Mechanisms, and consequences for tropism and pathogenesis. J Biol Chem 295, 2771–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrete OA, Chu D, Aguilar HC, and Lee B (2007). Single amino acid changes in the Nipah and Hendra virus attachment glycoproteins distinguish ephrinB2 from ephrinB3 usage. J Virol 81, 10804–10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrete OA, Levroney EL, Aguilar HC, Bertolotti-Ciarlet A, Nazarian R, Tajyar S, and Lee B (2005). EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436, 401–405. [DOI] [PubMed] [Google Scholar]
- Negrete OA, Wolf MC, Aguilar HC, Enterlein S, Wang W, Muhlberger E, Su SV, Bertolotti-Ciarlet A, Flick R, and Lee B (2006). Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog 2, e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Playford EG, Munro T, Mahler SM, Elliott S, Gerometta M, Hoger KL, Jones ML, Griffin P, Lynch KD, Carroll H, et al. (2020). Safety, tolerability, pharmacokinetics, and immunogenicity of a human monoclonal antibody targeting the G glycoprotein of henipaviruses in healthy adults: a first-in-human, randomised, controlled, phase 1 study. Lancet Infect Dis 20, 445–454. [DOI] [PubMed] [Google Scholar]
- Plowright RK, Eby P, Hudson PJ, Smith IL, Westcott D, Bryden WL, Middleton D, Reid PA, McFarlane RA, Martin G, et al. (2015). Ecological dynamics of emerging bat virus spillover. Proc Biol Sci 282, 20142124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissanen I, Ahmed AA, Azarm K, Beaty S, Hong P, Nambulli S, Duprex WP, Lee B, and Bowden TA (2017). Idiosyncratic Mojiang virus attachment glycoprotein directs a host-cell entry pathway distinct from genetically related henipaviruses. Nat Commun 8, 16060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B, Bossart KN, Feldmann F, Geisbert JB, Hickey AC, Brining D, Callison J, Safronetz D, Marzi A, Kercher L, et al. (2010). A novel model of lethal Hendra virus infection in African green monkeys and the effectiveness of ribavirin treatment. J Virol 84, 9831–9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago C, Celma ML, Stehle T, and Casasnovas JM (2010). Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol 17, 124–129. [DOI] [PubMed] [Google Scholar]
- Schrodinger LLC (2015). The PyMOL Molecular Graphics System, Version 1.8. [Google Scholar]
- Selvey LA, Wells RM, McCormack JG, Ansford AJ, Murray K, Rogers RJ, Lavercombe PS, Selleck P, and Sheridan JW (1995). Infection of humans and horses by a newly described morbillivirus. Med J Aust 162, 642–645. [DOI] [PubMed] [Google Scholar]
- Smith I, and Wang LF (2013). Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr Opin Virol 3, 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Zhou Y, Olivarez NP, Broadwater AH, de Silva AM, and Crowe JE Jr. (2012). Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J Virol 86, 2665–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen DL, Xu K, Nikolov DB, and Broder CC (2012). Henipavirus mediated membrane fusion, virus entry and targeted therapeutics. Viruses 4, 280–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg NJ, Zhang H, Bangaru S, Sapparapu G, Kose N, Lampley RM, Bombardi RG, Yu Y, Graham S, Branchizio A, et al. (2016). H7N9 influenza virus neutralizing antibodies that possess few somatic mutations. J Clin Invest 126, 11482–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidgen ME, de Jong C, Rose K, Hall J, Field HE, and Smith CS (2015). Novel paramyxoviruses in Australian flying-fox populations support host-virus co-evolution. J Gen Virol 96, 1619–1625. [DOI] [PubMed] [Google Scholar]
- Walsh MG, Wiethoelter A, and Haseeb MA (2017). The impact of human population pressure on flying fox niches and the potential consequences for Hendra virus spillover. Sci Rep 7, 8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherman S, Feldmann H, and de Wit E (2018). Transmission of henipaviruses. Curr Opin Virol 28, 7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch BD, Yuan P, Bose S, Kors CA, Lamb RA, and Jardetzky TS (2013). Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain. PLoS Pathog 9, e1003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JJW, Young TA, Zhang J, Liu S, Leser GP, Komives EA, Lamb RA, Zhou ZH, Salafsky J, and Jardetzky TS (2017). Monomeric ephrinB2 binding induces allosteric changes in Nipah virus G that precede its full activation. Nat Commun 8, 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang L, Yang F, Ren X, Jiang J, Dong J, Sun L, Zhu Y, Zhou H, and Jin Q (2014). Novel Henipa-like virus, Mojiang Paramyxovirus, in rats, China, 2012. Emerg Infect Dis 20, 1064–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Rockx B, Xie Y, DeBuysscher BL, Fusco DL, Zhu Z, Chan YP, Xu Y, Luu T, Cer RZ, et al. (2013). Crystal structure of the Hendra virus attachment G glycoprotein bound to a potent cross-reactive neutralizing human monoclonal antibody. PLoS Pathog 9, e1003684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, McGraw PA, House FS, and Crowe JE (2008). An optimized electrofusion-based protocol for generating virus-specific human monoclonal antibodies. J Immunol Methods 336, 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Leser GP, Demeler B, Lamb RA, and Jardetzky TS (2008). Domain architecture and oligomerization properties of the paramyxovirus PIV 5 hemagglutinin-neuraminidase (HN) protein. Virology 378, 282–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Swanson KA, Leser GP, Paterson RG, Lamb RA, and Jardetzky TS (2011). Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc Natl Acad Sci U S A 108, 14920–14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Thompson TB, Wurzburg BA, Paterson RG, Lamb RA, and Jardetzky TS (2005). Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13, 803–815. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Bossart KN, Bishop KA, Crameri G, Dimitrov AS, McEachern JA, Feng Y, Middleton D, Wang LF, Broder CC, et al. (2008). Exceptionally potent cross-reactive neutralization of Nipah and Hendra viruses by a human monoclonal antibody. J Infect Dis 197, 846–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Dimitrov AS, Bossart KN, Crameri G, Bishop KA, Choudhry V, Mungall BA, Feng YR, Choudhary A, Zhang MY, et al. (2006). Potent neutralization of Hendra and Nipah viruses by human monoclonal antibodies. J Virol 80, 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, Related to Table 1. Full dilution curve data for neutralization or binding of 10 human monoclonal antibodies to HeV, NiVM, or NiVB. A. Neutralization was tested with live viruses; percent neutralization is shown over a varying antibody concentration range. m102.4 is shown for comparative purposes. B. Binding in ELISA to recombinant RBPs. Optical density at 405 nm is shown over a varying antibody concentration range.
Figure S2, Related to Table 1.
A. Binding assays of HENV-26 or HENV-32 to full-length HeV-RBP or NiV-RBP proteins detected by flow cytometric analysis. Binding to cells transfected with HeV-RBP (red), NiVM-RBP (blue), NiVB-RBP (green), or un-transfected (black) cells was analyzed using an Intellicyt iQue instrument. Values are expressed as mean fluorescence intensity (MFI) of PE-conjugated secondary antibody signal and were plotted in GraphPad to interpolate EC50 values by non-linear regression analysis. Serial dilutions of HENV-26 or HENV-32 were performed in triplicate, with data representative of three independent assays shown. Error bars indicate SEM.
B. Kinetics of binding of HENV-26 or HENV-32 to full-length HeV-RBP or NiV-RBP proteins in biolayer interferometry to determine affinity of binding. Binding kinetics of Fab fragments corresponding to HENV-26 and HENV-32 were performed on an Octet RED instrument (FortéBio). Recombinant histidine-tagged RBP head domain was immobilized to HIS1K biosensor tips (FortéBio) at 10 μg/mL in kinetics buffer. After a brief baseline step, serial dilutions of HENV-26 or HENV-32 Fab starting at 200 nM then were associated to coated biosensor tips, followed by a dissociation step in 1x kinetics buffer. Curve-fitting was performed to extrapolate equilibrium dissociation constant values.
Figure S3, Related to Figures 2 and 3. Comparison of the eprhrinB2 binding site on HeV-RBP with the epitope for binding of HENV-26
A. Competition-binding assay of three human mAbs with ephrinB2 for binding to HeV-RBP head domain using biolayer interferometry. Numbers in boxes are the percentage binding signal of the second protein applied after binding of the first protein, compared with binding signal of the second protein alone. The proteins were defined as competing if the first protein reduced binding of the second protein by more than 70 percent. The proteins were defined as non-competing if the first protein reduced binding of the second protein by less than 40 percent.
B. Cell surface display flow cytometric assay to test for mAb blocking of HeV-RBP protein binding to the host receptor ephrinB2. 293F cells were transfected to display the full-length HeV-RBP protein on the cell surface. Cells were incubated with soluble ephrinB2 protein or FACS buffer, then HENV-26 labeled with AlexaFluor-647 was added to cells and incubated. Cells then were washed and analyzed using an Intellicyt iQue flow cytometry instrument. Binding of HENV-26 in the presence or absence of ephrinB2 was expressed as mean fluorescence intensity (MFI).
C-E. Binding site mapping of ephrinB2 and m102.3 onto the HeV-RBP or NiV-RBP head domain surface. The surfaces of HeV-RBP or NiV-RBP head domains are colored in grey, with glycans colored in black.
C. The ephrinB2 binding site is mapped and colored in green onto HeV-RBP head domain surface. The surface envelope (shown as blue mesh) of the epitope of HeV-RBP head domain recognized by HENV-26 is overlaid onto the HeV-RBP head domain.
D. The mAb m102.3 binding site is mapped and colored in yellow onto the HeV-RBP head domain surface. The surface envelope (shown as blue mesh) of the epitope on HeV-RBP head domain recognized by HENV-26 is overlaid onto the HeV-RBP head domain.
E. The ephrinB2 binding site is mapped and colored in pink onto the NiV-RBP head domain surface. The surface envelope (shown as blue mesh) of the epitope on NiV-RBP head domain recognized by HENV-26 is overlaid on to the NiV-RBP head domain.
Figure S4, Related to Figures 3 and 4. Molecular recognition of HeV-RBP head domain by mAbs.
A) Water-mediated interactions between HeV-RBP head domain and mAb HENV-26. HeV-RBP head domain is colored in green, HENV-26 heavy chain in cyan, and the light chain in yellow. Water molecules are represented as magenta spheres, and involved residues are shown as sticks and labeled. Hydrogen bonds are represented as broken orange lines.
B) Water-mediated interactions between HeV-RBP head domain and mAb HENV-32. HeV-RBP head domain is colored in green, HENV-32 heavy chain in orange, and the light chain in salmon. Water molecules are represented as magenta spheres, and involved residues are shown as sticks, and labeled. Hydrogen bonds are represented as broken orange lines.
C) CedV-RBP or GhV-RBP ELISA. Binding of HENV-26 or HENV-32 was tested in ELISA using recombinant CedV-RBP head domain or GhV-RBP full ectodomain. MAbs to CedV-RBP (14F3) or GhV-RBP (10D5) were used as controls.
D) Lack of dimer disruption mediated by HENV-26 or HENV-32. HeV-RBP full ectodomain was expressed in Expi293F cells and purified sequentially by HisTrap affinity chromatography followed by size exclusion chromatography (SEC). Size exclusion peaks corresponding to dimeric (SEC peak #2) or tetrameric (SEC peak #1) species were each incubated with HENV-26 or HENV-32 Fab for 1 hour at room temperature. Each preparation was then analyzed by 4–12% Bis-Tris SDS-PAGE using SimplyBlue SafeStain Coomassie G-250 stain.
E) Epitope mapping of RBP head domains of HeV, NiVM, and NiVB. The RBP amino acid sequences of the three viruses were aligned with MUSCLE (1), and the figure was made with sequence alignment editor, ALINE (2). For comparison, the sequences of Cedar virus RBP (CedV-RBP) and Ghana virus RBP (GhV-RBP) also are shown.
Upper panel: The epitope residues recognized by HENV-26 (in complex with HeV-RBP [PDB ID 6VY6] or NiV-RBP Malaysia [NiV-RBPm; PDB ID 6VY5] are highlighted with red boxes, the epitope residues recognized by HENV-32 (in complex with HeV-RBP [PDB ID 6VY4]) with orange boxes, and epitope resides recognized by m102.3 [PDB ID 6CMG] are indicated with purple lines over the alignment.
Lower panel: The ephrinB2 binding residues from complexes with HeV-RBP [PDB ID 6PDL] or NiV-RBPm [PDB ID 2VSM] are highlighted with red boxes, and the ephrinB3 binding residues from a complex with NiV-RBPm [PDB ID 3D12] with yellow boxes.
F) Superimposition of crystal structures for comparative purposes
Upper panel: The crystal structure of CedV-RBP (PDB ID: 6P72) is superimposed onto that of the HeV-RBP/HENV-26 complex. CedV-RBP structure is shown in dark grey; HeV-RBP in green; HENV-26 heavy chain in cyan, and HENV-26 light chain in yellow. Sequence variations at several positions in CedV-RBP (e.g., G512, H518, D550, L552) result in loss of H-bonds seen in the corresponding positions in the HeV-RBP complex.
Middle panel: The crystal structure of GhV-RBP (PDB ID: 4UF7) is superimposed onto that of the HeV-RBP/HENV-26 complex. GhV-RBP structure is shown in light grey; HeV-RBP in green; HENV-26 heavy chain in cyan, and HENV-26 light chain in yellow. Sequence variations at several positions in GhV-RBP (e.g., G500, T511, E539, V541) result in loss of H-bonds seen in the corresponding positions in the HeV-RBP complex.
Lower panel: The CedV-RBP and GhV-RBP structures are superimposed on that of the HeV-RBP/HENV-32 structure. The major epitope of HeV-RBP (shown in blue, residues T196 – N210) and corresponding residues of CedV-RBP (in dark grey, residues P219 – Q233) or GhV-RBP (in light grey, residues P212 – T226) are shown in cartoon to represent backbone conformations. Sequence variations at this region among the different RBPs result in significant conformational variation and may create a prohibitively high energy barrier to prevent CedV-RBP or GhV-RBP from adopting backbone conformations similar to that of HeV-RBP or NiV-RBP.
Figure S5, Related to Figures 2, 4 and 5. Overlays of HENV-26 or 32 for comparative purposes.
A) Superimposition of the crystal structures of HeV-RBP head domain in complex with HENV-26 or HENV-32. HeV-RBP head domain in the HeV-RBP/HENV-26 complex is colored in green, and that in the HeV-RBP/HENV-32 in light blue. The HENV-26 heavy chain is colored in cyan, HENV-26 light chain in yellow, HENV-32 heavy chain in orange, and HENV-32 light chain in salmon. The CDRs of both mAbs and the individual blades of the HeV-RBP head domain are labeled. Regions in the HeV-RBP head domain with large structural differences between structures are indicated within broken red lines.
B) Superimposition of the crystal structures of the dimeric HeV-RBP head domain and HeV-RBP head domain in complex with HENV-32. HeV-RBP head domain in the HeV-RBP/HENV-32 is shown in light blue, HENV-32 heavy chain in orange, and HENV-32 light chain in salmon. One protomer of the HeV-RBP head domain is colored in magenta and the other in cyan. Individual blades of the superimposed HeV-RBP head domains are labeled (β1 to β6).
C) Superimposition of HeV-RBP structures in the HENV-32/HeV-RBP complex and ephrinB2/HeV-RBP complex. The HeV-RBP structure in the HENV-32/HeV-RBP complex is shown in light blue, and HeV-RBP in the ephrinB2/HeV-RBP complex in yellow. EphrinB2 in the complex is shown in grey. The overlay suggests that the conformation of HeV-RBP β6/S2-S3 and β5/S4- β6/S1 loops in the HENV-32/HeV-RBP complex causes potential steric clashes between the loops and the ephrinB2 G-H loop. The potential steric clashes between the HENV-32 bound HeV-RBP structure and ephrinB2 G-H loop are represented with broken red lines, and the residues with potential steric clashes are shown in stick, and corresponding residues of the ephrinB2 bound HeV-RBP structure are shown as line representation.
Figure S6, Related to Figure 5. Structural details of the HENV-32 CDRH3 binding site in the crystal structures of HeV-RBP-apo (dimeric form) (panel A) or the HeV-RBP/m102.3 complex (panel B). Residues interacting with HENV-32 CDRH3 are shown as sticks. Consistent with the structure in the HeV-RBP/HENV-26 complex, the HENV-32 CDRH3 binding pocket is hidden underneath salt bridges between residue R258 and D260 in both structures.
(panels C - F) Surface representation of the HENV-32 CDRH3 binding site in four different crystal structure forms of HeV-RBP. The residues interacting with the HENV-32 CDRH3 are indicated; R201 is colored in red, L202 in yellow, I203 in green, Y205 in americium, R258 in white on blue, G259 in cyan, D260 in wheat, V262 in pink, P263 in olive, and S264 in magenta.
C) HENV-32/HeV-RBP complex; the HENV-32 CDRH3 paratope is shown in orange as both ribbon and stick representations and labeled in cyan.
D) HENV-26/HeV-RBP complex.
E) m102.3/HeV-RBP complex.
F) HeV-RBP apo (dimeric form). Residue P263 in the HeV-RBP/HENV-26 complex and HeV-RBP/m102.3 complex is not solvent accessible.
Figure S7, Related to Figure 6. Representative histopathology of brain and liver tissues from ferrets infected with NiVB and treated with recombinant anti-HENV mAbs, compared with untreated control animals.
A to D) Tissues from tissue from subject C1:
A) H&E staining of brain tissue did not show significant histological lesions;
B) immunolabeling of brain tissue showed the endothelium of small caliber vessels within the neuronal parenchyma multifocally had diffuse cytoplasmic immunolabeling.
C) H&E staining of lung tissue showed mild interstitial pneumonia with multifocal nodular formation composed of necrotic debris, hemorrhage and mixed inflammatory infiltrates of neutrophils and macrophages;
D) immunolabeling of lung tissue showed diffuse cytoplasmic immunolabeling of segmental regions of the alveolar septa largely centered on the necrotic nodules, mononuclear cells within alveolar septa, mononuclear cells free within alveolar spaces (alveolar macrophages), endothelium of small to medium caliber vessels, and rarely lower airway epithelium (D).
E to H) Tissues from subject HENV-32_A did not show significant histological lesions or immunolabeling:
E) H&E staining of brain;
F) Immunolabeling of brain;
G) H&E staining of lung;
H) Immunolabeling of lung.
I to L) Tissues from subject HENV-26_D:
I) H&E staining of brain showed moderate cuffing of small caliber vessels with lymphocytes in the brainstem and in vessels surrounding the ventricular system;
J) Immunolabeling of brain showed diffuse cytoplasmic immunolabeling of neurons in association with the lymphocytic infiltrates (inset and arrow);
K) H&E staining of lung showed rare cuffing of small caliber vessels of the lung with lymphocytes and plasma cells present;
L) Immunolabeling of lung was not detected. (I).
Data Availability Statement
Accession numbers -
The crystal structures are deposited at the Protein Data Bank (PDB). Atomic coordinates and structure factors for the crystal structures of HENV-26 or HENV-32 in complex with HeV-RBP or NiV-RBP head domain have been deposited in the Protein Data Bank with the accession codes 6VY6 (HENV-26 in complex with HeV-RBP head domain), 6VY5 (HENV-26 in complex with NiV-RBP head domain), and 6VY4 (HENV-32 in complex with HeV-RBP head domain).