Abstract
Avian reproductive behavior is regulated through the neuroendocrine system. The transition from laying to brooding is strictly controlled by the hypothalamus-pituitary-gonadal (HPG) axis. Cross talk on the HPG axis relies on the circulatory system, where the dynamics of serum proteins can be observed during different reproductive phases. Some canonical hormones, such as prolactin and luteinizing hormone, play important roles in the transition through reproductive phases. However, little is known at the whole-proteome level. To discover novel serum proteins, we employed isobaric tags for relative and absolute quantification to assay the serum proteome during different reproductive phases in chicken. We identified a total of 1,235 proteins from chicken serum; 239 of these proteins showed differential expression between the laying and brooding stages, including a low concentration of steroid metabolism-related proteins and a high concentration of calcium signaling-related proteins (fold change ≥1.5 or ≤0.66; P < 0.05). Pathway analysis and protein–protein interaction networks predicated the difference in follicle development between the brooding stage and laying stages and were related to the 14-3-3 protein family, which is associated with oocyte meiosis and maturation. Together, these results provided a proteomics foundation for investigating the dynamic changes taking place in the circulatory system during reproductive phase transition, and also uncovered new insights regarding follicle development that underlie the avian reproductive cycle.
Key words: reproductive cycle, iTRAQ analysis, chicken serum proteome, incubation behavior
Introduction
Incubation is a specific reproductive behavior in birds that differs from other vertebrates, and is essential for breeding success. However, natural incubation behavior leads to the cessation of egg production and consequent significant economic losses for the poultry industry. Understanding the genetic mechanisms underlying incubation behavior will help in improving chicken reproduction and has scientific significance for understanding the regulation of complex behaviors.
Reproductive activity in chickens is driven by photostimulation and neuroendocrine regulation; the coordinated activity of hormones in the hypothalamus-pituitary-gonadal (HPG) axis initiates and maintains ovarian follicle development (Mauro et al., 1989; EI Halawani et al., 1993; Dunn et al., 2004; Johnson et al., 2015). The transition from laying to brooding is also strictly controlled by the HPG axis and involves the inhibition of gonadotropin secretion and the stimulation of prolactin (PRL) secretion (Youngren et al., 1991; Sharp et al., 1998; Huang et al., 2008). Hormone levels change markedly from the laying phase to the brooding phase in birds, and high levels of PRL and low concentrations of luteinizing hormone (LH) and ovarian steroids are known to be hallmarks of incubation behavior (Donham., 1979; Ramsey et al., 1985). It is also known that incubation behavior can be induced via the combined action of estradiol, progesterone, and PRL in ovariectomized female avians (El-Halawani et al., 1986). Thus, we pondered on how the circulatory system responds to the HPG axis hormonal signaling during the reproductive cycle. In this study, we aimed to investigate novel serum proteins that are different between the brooding and laying stages.
Female birds can rapidly shift from the laying phase to the brooding phase, and this shift is accompanied by significant morphological changes in the ovaries, including follicle degeneration and atresia (Kovasc et al., 1992). Broody chickens share a consistent pattern of greatly inhibited steroidogenic enzyme genes in the pituitary gland (Shen et al., 2016). This decline in steroid biosynthesis leads to a loss of progesterone and estradiol secretion that underlies the brooding state. In response to these changes, follicle development becomes quiescent in the ovaries of broody chickens (Liu et al., 2018). The circulatory system plays a critical role in transmitting hormonal signals from the HPG axis to the target tissues. Therefore, understanding serum protein dynamics during the transition between the reproductive phases will assist in understanding the mechanisms that trigger and maintain the drastic change of function in follicular development.
However, the interactions taking place among endocrine signals in the circulatory system are still unclear. Because the regulation of the reproductive cycle in chicken depends on dynamic interactions in the neuroendocrine system, examining the global protein profile in the serum would be an effective strategy for understanding the polygenic networks associated with chicken reproductive traits and the protein-level responses to reproductive phase transition. Protein dynamics in serum reflect the molecular and phenotypic changes of target tissues. Isobaric tags for relative and absolute quantification (iTRAQ) is a protein assay method based on high-precision mass spectrometry that provides accurate quantitation for detecting differential serum proteins (DSP) related to biological questions (Ouyang et al., 2017; Ning et al., 2019). We measured serum protein profiles using iTRAQ during different reproductive stages in chickens and investigated the major triggers of stage transitions by these serum protein levels, for providing candidate markers for monitoring reproductive phase transition.
Materials and methods
Ethics Statement
This study was carried out in strict accordance with the Experimental Animal Management protocols of Zhongkai University of Agriculture and Engineering. The protocol was approved by the Committee on the Ethics and Welfare of Animal Experiments of Zhongkai University of Agriculture and Engineering. All efforts were made to minimize animal suffering.
Animals and Sample Collection
Chicken of the Chinese native breed, Huiyang Bearded chicken, were obtained from the Institute of Animal Science, Guangdong Academy of Agricultural Sciences. We used 6 hens in the present study; they were randomly selected at an age of 280 d. The laying group had been continuously laying for 18 to 20 d prior to sampling. Hens in the broody group had been continuously brooding for 3 d prior to sampling. Two hens were housed per cage and maintained under a daily light period of 16 h. Blood samples were collected via venipuncture and incubated for blood coagulation and then centrifuged at 4°C. Serum was separated and then stored at −80°C for further analysis.
iTRAQ Assays and Data Analysis
The 6 serum samples were used for iTRAQ assays (Applied Biosystems, Foster City, CA). Highly abundant proteins were removed for serum proteomic analysis and subsequently quantified using Bradford protein assays (Bio-Rad, Hercules, CA) and detected using SDS-PAGE.
All the samples underwent trypsin digestion to obtain peptides and were then labeled individually with iTRAQ Reagent 8Plex One Assay Kit (SCIEX, Foster City, CA). Liquid phase separation of the labeled peptides was performed using a liquid phase system (LC-20AB, Shimadzu, Kyoto, Japan). Subsequently, the peptide was desalted using 5-μm 4.6 × 250 mm Gemini C18 solid phase extraction and submitted for electrospray ionization tandem mass spectrometry (TripleTOF 5600, SCIEX, Framingham, MA). Data were collected under the following conditions: a spray voltage of 2,300V, a nebulizer gas of 30 pounds/inch2, and spray interface temperature maintained at 150°C.
The tandem mass spectrometry data were analyzed with iQuant software (Wenbo, Beijing Genomics Institute, Shenzhen, China) (Wen et al., 2014) and searched in the UniProt database (Gallus gallus). A target-decoy-based strategy was applied to control peptide-level false discovery rates with a threshold of 1%. Only unique peptides were used for protein quantification, and normalization to protein medians was used to correct experimental bias. The minimum number was set to 200. For protein quantitation, the ratios of each sample were weighted and normalized by making a comparison with the control group as the denominator. For quantitative changes, we set a ≥1.5- or ≤0.66-fold change and a P-value (t test) < 0.05 as the cutoff for DSP.
Multiple Reaction Monitoring (MRM) Validation and Data Analysis
The Multiple Reaction Monitoring (MRM) method was used to validate the 6 different serum proteins. Serum samples, from which the highly abundant proteins had been removed, were quantified by using Bradford protein assays (Bio-Rad); subsequently, proteins were digested with Trypsin Gold (Promega, Madison, WI) at 37°C for 16 h. After trypsin digestion, the peptides were dried by vacuum centrifugation and then reconstituted in 0.5 M triethylammonium bicarbonate buffer. For liquid chromatography-MRM mass spectrometry, the samples were digested as described previously and spiked with 50 fmol of β-galactosidase for data normalization. MRM analyses were performed on a QTRAP 5500 mass spectrometer (SCIEX, Framingham, MA) equipped with an LC-20AD Nano HPLC system (Shimadzu). The peptides were separated on a C18 column (0.075 × 150 mm column, 3.6 μm) at 300 nL/min and eluted with a gradient of 5 to 30% solvent B for 38 min, 30 to 80% solvent B for 4 min, and maintenance at 80% for 8 min. For the QTRAP 5500 mass spectrometer, a spray voltage of 2,400 V, a nebulizer gas of 23 pounds/inch2, and a dwell time of 10 ms were used. Multiple MRM transitions were monitored using unit resolution in both the Q1 and Q3 quadrupoles to maximize specificity.
We used Skyline software (Brendan MacLean, University of Washington, Seattle, WA) to integrate the raw file generated by QTRAP 5500. An indexed retention time strategy was used to define the chromatography of a given peptide against a spectral library. All transitions for each peptide were used for quantitation unless interference from the matrix was observed. A spike of β-galactosidase was used for label-free data normalization. Mass spectrometry statistics with the linear mixed-effects model was used for analysis. The P-values were adjusted to control the false discovery rate. All proteins with a P-value <0.05 and a fold change >1.5 were considered significant.
Bioinformatic Analysis
Principal component analysis was performed to evaluate the relationship among the 6 samples by using factoextra package in R program (R Foundation, Vienna, Austria) based on protein abundance values. Gene ontology (GO) analysis was performed using the Blast2GO software (https://www.blast2go.com/). We used DAVID 6.7 (online software: https://david.ncifcrf.gov/) (Huang et al., 2009) for Kyoto Encyclopedia of Genes and Genomes pathway analysis by uploading the DSP list. Graphics were plotted by the ggplot2 package in R program. The protein–protein interaction (PPI) network was obtained based on the online STRING database (http://string-db.org). Subsequently, Cytoscape software (v.3.7.2, http://cytoscape.org/) was used to analyze and visualize the PPI networks acquired from STRING.
Results
Quantification of Serum Proteomes in Laying and Brooding Chickens
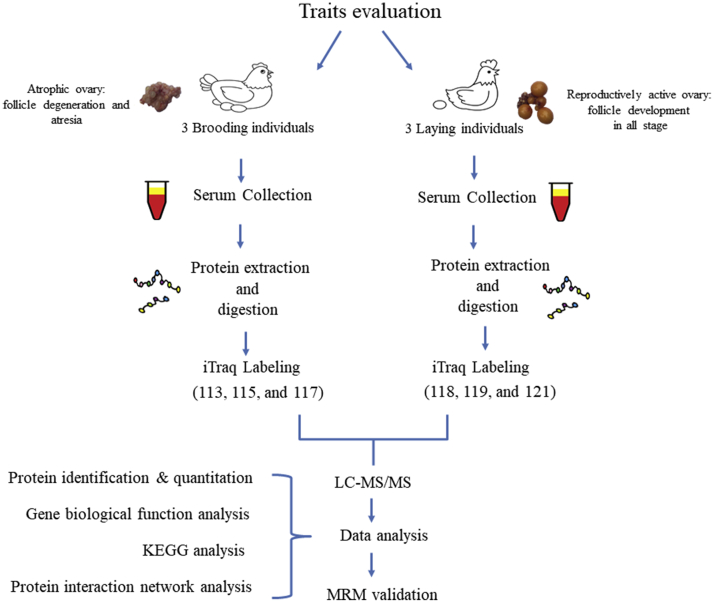
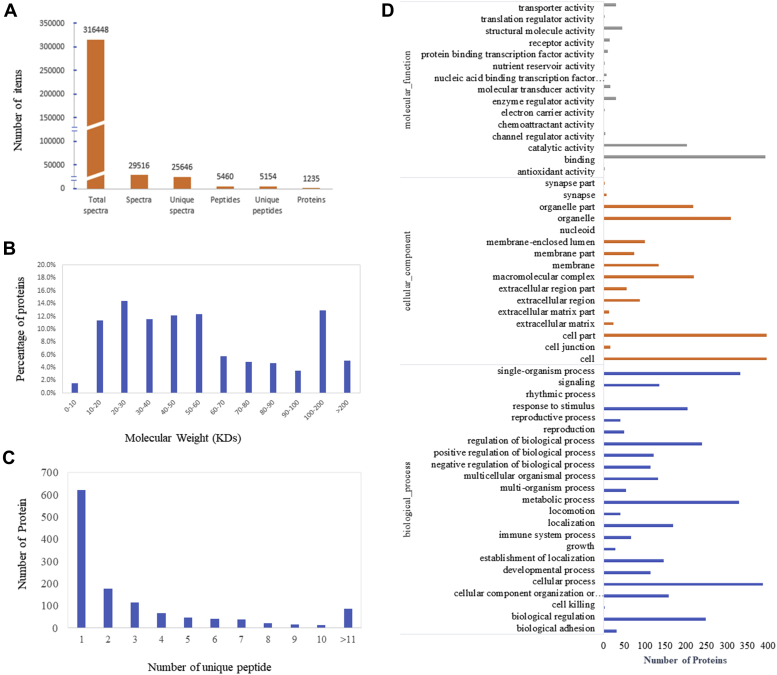
Three biological replicates were collected for each stage and serum was separated for protein assays by iTRAQ (Figure 1). We detected 316,448 spectra in total, which corresponded to 5,154 unique peptides and 5,235 proteins, in both the brooding and laying stages (Figure 2A). The proteins identified by iTRAQ had molecular weights ranging from 5 to 620 kD (Figure 2B), lengths ranging from 45 to 5,432 amino acids, with approximately 43.7% of these proteins covered by no less than 3 peptides (Figure 2C, Supplementary Table 1). Principal component analysis showed that the detected proteins were clearly distinguishable between the broody group and laying group (Supplementary Figure 1). To understand the function of serum proteins, we performed GO term analysis and annotated them according to molecular function, biological process, and cellular component (Figure 2D). We found that the serum proteins showed statistically significant enrichment in molecular function of binding and catalytic activity (P < 0.05, Supplementary Table 2).
Figure 1.
Workflow for proteomic assay in chicken serum by iTRAQ. Abbreviations: iTRAQ, isobaric tags for relative and absolute quantification; LC, liquid chromatography; MRM, multiple reaction monitoring; MS/MS, tandem mass spectrometry.
Figure 2.
Protein identification and GO annotation. (A) Basic information of the iTRAQ assay. (B) The coverage of the identified proteins. (C) Molecular weight spectrum of proteins identified by iTRAQ. (D) GO annotation for the identified proteins in chicken serum (P < 0.05). Abbreviations: iTRAQ, isobaric tags for relative and absolute quantification; GO, gene ontology.
Identification of DSP
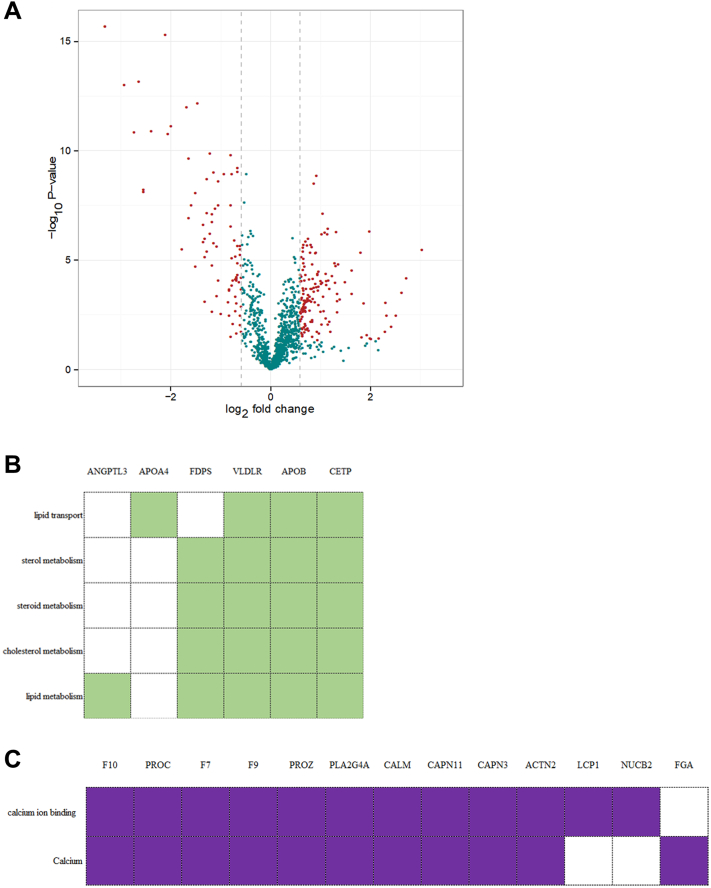
To assess protein dynamics during the reproductive cycle, we analyzed differences in protein abundance between the laying and brooding stages and identified 239 significantly different serum proteins (fold change ≥1.5 and P < 0.05; Figure 3A). In the brooding stage, 151 of these DSP were upregulated, while 88 were downregulated. Six DSP were randomly selected for validation via MRM assay and their difference in expression was consistent with the results from the iTRAQ assay (Table 1). A complete list of these DSP is shown in Supplementary Table 3.
Figure 3.
Differential serum proteins between the brooding and laying stages. (A)Volcano plot showing the DSP detected in the serum between the laying stage and brooding stage. The X-axis refers to the log2 fold change of the proteins in the brooding stage compared to the laying stage, while the Y-axis shows the −log10 (adjusted P-value) of that comparison. DSP (fold change ≥ 1.5, P-value < 0.001, FDR < 0.05) are in red and the other in gray. (B) Function analysis of downregulated proteins in the broody stage. (C) Function analysis of upregulated proteins in the broody stage. Proteins in column and function terms in row. Abbreviations: DSP, differential serum proteins; FDR, false discovery rate.
Table 1.
Verification of DSP between broody stage and laying stage by MRM.
| Protein | Gene symbol | Fold change1 |
|---|---|---|
| ENSGALP00000004753 | SERPINF1 | 0.65 |
| ENSGALP00000007185 | GPX3 | 1.24 |
| ENSGALP00000010888 | YWHAH | 2.57 |
| ENSGALP00000011922 | SERPING1 | 1.83 |
| ENSGALP00000016512 | VLDLR | 0.08 |
| ENSGALP00000027989 | YWHAG | 2.02 |
Abbreviations: DSP, differential serum proteins; MRM, multiple reaction monitoring.
The ratio of broody stage vs. laying stage.
Annotation and Functional Enrichment of DSP
To understand the function of these DSP, we annotated these DSP using DAVID. The downregulated serum proteins in the broody stage were mainly involved in lipid metabolism, such as cholesterol metabolism, terpenoid metabolism, lipid concentration, steroid transport, and response to progesterone functions (Figure 3B). Meanwhile, the upregulated serum proteins were related to the regulation of calcium binding and metal binding functions (Figure 3C).
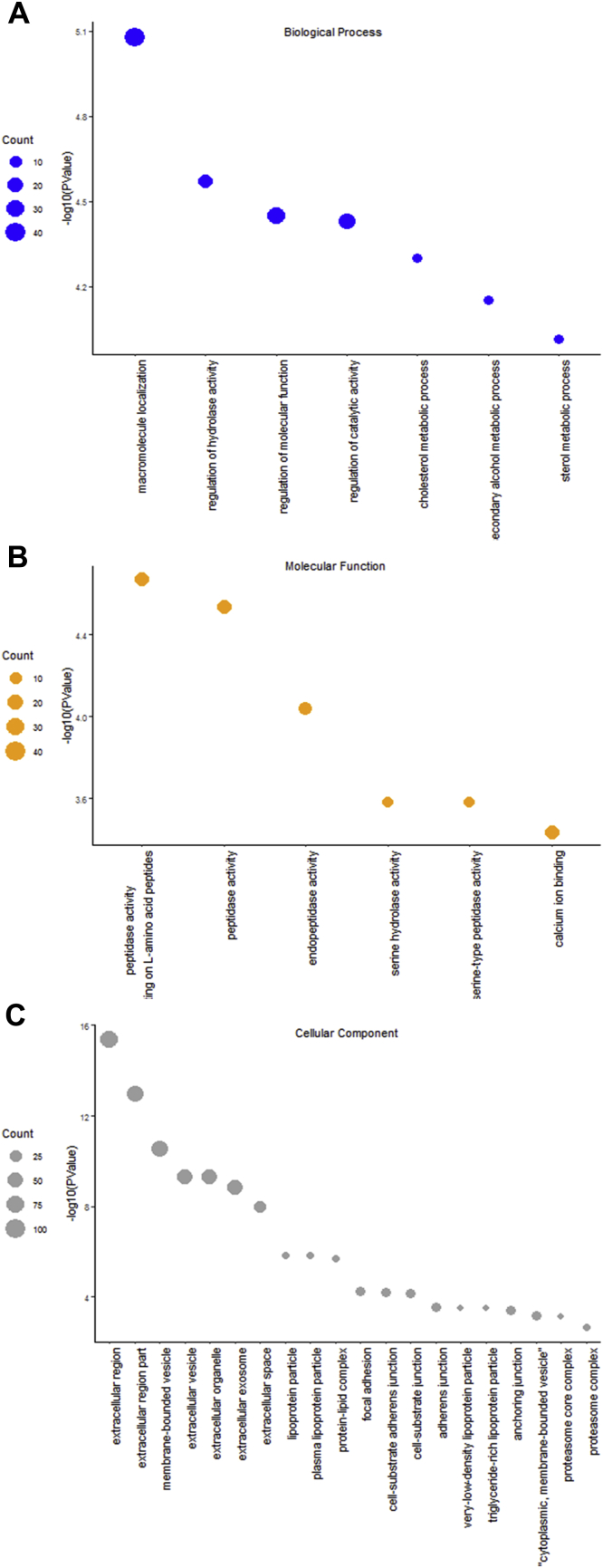
We found the most significantly enriched GO terms belonged to metabolism in the biology process category, such as regulation of catalytic activity, cholesterol metabolic process, secondary alcohol metabolic process, and sterol metabolic process (Figure 4A). For cellular components, the DSP were mainly enriched in the extracellular region and lipoprotein, such as extracellular vesicle, extracellular organelle and extracellular exosomes, lipoprotein particle, plasma lipoprotein particle, protein-lipid complex, and very low-density lipoprotein particle (Figure 4B). For molecular function, the significantly enriched terms were peptidase activity, such as endopeptidase activity, serine hydrolase activity, serine-type peptidase activity, and calcium ion binding (Figure 4C).
Figure 4.
GO enrichment for DSPs. Significant GO terms (P < 0.01 and FDR < 0.05) are shown in the bubble plot and ordered by P-values. Size of the circle represents the number of genes. (A) Molecular function; (B) cellular component; (C) biology process. Abbreviations: DSP, differential serum proteins; GO, gene ontology.
Pathway Analysis
To further examine protein functions, pathway analysis was performed. We found that DSP were significantly enriched in the meiosis pathway and cell cycle pathway (Table 2). DSP annotated to oocyte meiosis and the cell cycle pathway were upregulated in the brooding stage compared to the laying stage. The 2 pathways shared 5 YWHA (14-3-3) proteins, which have been reported to be associated with the control of ovarian follicular development in vertebrate ovaries (De et al., 2012). The 5 DSP belonging to the 14-3-3 protein family were significantly upregulated in the brooding stage (fold change >1.5 and P < 0.01, Table 3).
Table 2.
DSP involved in oocyte meiosis and cell cycle pathway.
| Pathway | Protein ID | Gene symbol | P-value | FDR (Benjamini) | Regulation |
|---|---|---|---|---|---|
| ENSGALP00000010447 | SKP1 | 4.20E-06 | 2.90E-04 | + | |
| ENSGALP00000064090 | CALM2 | ||||
| ENSGALP00000015963 | PPP2R5A | ||||
| ENSGALP00000019298 | PPP2R5E | ||||
| Oocyte Meiosis | ENSGALP00000045068 | RPS6KA3 | |||
| ENSGALP00000039133 | YWHAE | ||||
| ENSGALP00000043305 | YWHAB | ||||
| ENSGALP00000010888 | YWHAH | ||||
| ENSGALP00000027989 | YWHAG | ||||
| ENSGALP00000036122 | YWHAQ | ||||
| ENSGALP00000005503 | CCD3 | 9.70E-04 | 3.30E-02 | + | |
| ENSGALP00000039133 | YWHAE | ||||
| ENSGALP00000036122 | YWHAQ | ||||
| ENSGALP00000032274 | MCM6 | ||||
| Cell cycle | ENSGALP00000027989 | YWHAG | |||
| ENSGALP00000010447 | SKP1 | ||||
| ENSGALP00000043305 | YWHAB | ||||
| ENSGALP00000010888 | YWHAH |
Pathway analysis using DAVID (https://david.ncifcrf.gov/); significantly enriched pathway (P < 0.01 and FDR< 0.05) was ordered by P-value. “+” represents the upregulated proteins in broody chicken serum.
Abbreviations: DSP, differential serum proteins; MRM, multiple reaction monitoring.
Table 3.
14-3-3 proteins upregulated in the broody stage.
| Gene symbol | Mass1 | Number2 | Fold change (brooding vs. laying) | P-value |
|---|---|---|---|---|
| YWHAE | 29308.46984 | 10 | 1.81 | 3.23E-09 |
| YWHAQ | 28031.81995 | 4 | 1.57 | 2.07E-06 |
| YWHAG | 28437.96658 | 3 | 1.73 | 2.08E-06 |
| YWHAB | 27985.80754 | 3 | 1.61 | 1.44E-06 |
| YWHAH | 28477.11581 | 2 | 1.57 | 2.07E-03 |
Molecular weight of protein (KDs).
Indicates the number of unique peptides covering the protein.
PPI
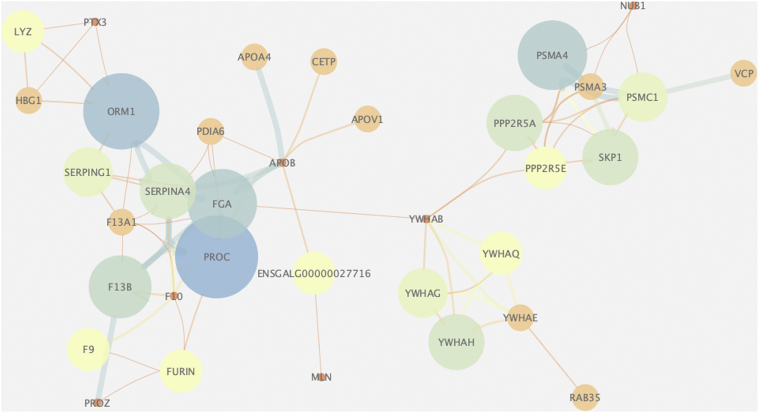
In endocrine systems, hormones interact with each other and affect change or maintenance of biological functions such as reproduction and growth. To further explore the interaction networks of DSP, we analyzed protein interaction networks using Cytoscape. One hundred and eighty two DSP were provided to STRING to obtain PPI information, and the PPI network was visualized using Cytoscape. Two separated networks with high confidence interactions (CI >0.9) are shown in Figure 5. Proteins related to oocyte meiosis (SKP1, PPP2R5A, PPP2R5E, YWHAB, YWHAQ, YWHAH, YWHAE, and YWHAG) had a pivotal role in the DSP network. The other set of proteins (APOA4, APOB, APOV1, and CETP) were related to lipid and steroid metabolism. It is suggested that oocyte meiosis and steroid metabolism play an important role in the modulation of protein networks in the serum level.
Figure 5.
Protein–protein interaction network of DSP. Protein interconnectivity shows important proteins involved in oocyte meiosis (SKP1, PPP2R5A, PPP2R5E, YWHAB, YWHAQ, YWHAH, YWHAE, YWHAG). Abbreviation: DSP, differential serum proteins.
Discussion
Incubation behavior is induced by the combined actions of neurological and endocrine regulation. The transition is under the control of hormones, especially PRL and LH. It is known that broody chickens have high levels of plasma PRL (Sharp, 1979, 1988), and that PRL can successfully induce incubation behavior in pigeons and chickens (Youngren et al., 1991). The plasma concentrations of LH and steroid hormones decrease during broodiness, resulting in regression of the ovaries, an inhibitory effect caused by increasing PRL. To uncover the broad proteomics rather than the high concentration sets in serum, high concentration proteins were removed before performing iTRAQ. This explains why PRL was not observed in our DSP, in contrast to the traditional radioimmunoassay of hormones. The iTRAQ method provided a proteomics view of the protein dynamics and detected other DSP involved in the response to the reproductive phase transition.
Ovarian morphology changes significantly in the brooding stage, as autophagy levels of follicles are enhanced (Yu et al., 2016) and primary to secondary follicle transition is inhibited (Liu et al., 2018). We observed that downregulated DSP in the broody stage mainly involved lipid metabolism, steroid metabolism, and steroid transport. It has been suggested that steroidogenesis weakened in brooding chickens, coinciding with the decline of gonadal steroid hormones in plasma (Romanov, 2001; Sharp, 2009).
In contrast to the downregulated serum proteins, a cluster of calcium signaling-related proteins exhibited significantly high levels in the broody stage. A Ca2+ pool is required for reproductive activity and pituitary gonadotropin release ( Batesand Conn, 1984), and intracellular Ca2+ concentration is essential for neurotransmitters and pituitary hormone release (Martin, 2003).
Among DSP, we found that 5 members of the14-3-3 protein family (YWHAQ, YWHAH, YWHAE, and YWHAG) were significantly enriched in both the cell cycle and the oocyte meiosis pathway. The 14-3-3 protein family has 7 members (Aitken, 2006), and these 14-3-3 proteins are known to be key regulators of cell fate decisions, playing an inhibitory effect on apoptosis and cell cycle control (van Hemert et al., 2001; Pennington et al., 2018). Furthermore, high expression levels of 14-3-3 proteins can result in the inhibition of transition from G2 to M phase (Hermeking et al., 1997). Moreover, the 14-3-3 protein family was predicated to associate with various signal transduction pathways (Wakui et al., 1997; Zilliacus et al., 2001; Quayle and Sadar, 2007).
In almost all species, oocytes are arrested at prophase I of meiosis (Sagata, 1996). It is known that 14-3-3 proteins play an essential role in oocyte meiosis and oocyte maturation (Meng et al., 2013). In murine oogenesis, 14-3-3 proteins play a role in maintaining meiotic arrest, and oogenesis and oocyte maturation (Eisa et al., 2019), with higher concentrations appearing in immature oocytes than in mature oocytes (De et al., 2012). 14-3-3 proteins also play an important role in maintaining the bipolar spindle during meiosis in Drosophila oocytes (Robin et al., 2017).
In chickens, progesterone is secreted from the granulosa cells of mature follicles and impacts follicular maturation (Pierce and Parson, 1981); however, low concentrations of progesterone are observed in the broody stage (Cogger et al., 1979). Regarding mRNA level, we detected that progesterone receptor was downregulated in the broody stage in pituitary tissues (Shen et al., 2016); moreover, low levels of progesterone influence follicular maturation in the ovary (Hammond et al., 1981). The relationship between progesterone and 14-3-3 protein is still unknown in avians. However, the 14-3-3 protein family was found to be involved in progesterone signaling (Wang et al., 2012). In human uterine cells, 14-3-3 protein is a progesterone-responsive gene and modulates progesterone signaling (Ito et al., 2012). Moreover, 14-3-3 proteins play a key role in the transcriptional activation of estrogen receptors (Kim et al., 2011).
It has been suggested that 14-3-3 proteins in serum may act on the ovary and result in meiotic arrest during follicle development. All these evidences suggest that the upregulation of 14-3-3 family proteins in the broody stage may affect oocyte maturation in chicken ovaries; this is a promising candidate for further investigation. Our results indicated that five 14-3-3 DSP may be used as biomarkers for monitoring ovarian development in chickens.
In summary, our data offer a valuable resource regarding the global profile and dynamics of serum proteins during the reproductive phase transition in chickens. These results provide a new insight into the circulatory response to reproductive phase transition. This information may ultimately assist in the improvement of egg production in the poultry industry.
Acknowledgments
We sincerely thank Professor Jin Xu of Sun Yat-Sen University for valuable and useful comments on this manuscript. We would like to express our gratitude to Aimei Dai, Yushuai Wang, and Zhongqi Liufu for helpful discussions and their help with data analysis. Many thanks to the Institute of Animal Science, Guangdong Academy of Agricultural Sciences for providing Huiyang Bearded chicken. This work was supported by the National Natural Science Foundation of China (31101718 and 31301010) and a grant from the National Key Technologies R & D Program of China (2016YFD0500510).
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.09.058.
Disclosures
The authors declare that they have no competing interests.
Supplementary data
References
- Aitken A. 14-3-3 proteins: a historic overview. Semin. Cancer Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Bates M.D., Conn P.M. Calcium mobilization in the pituitary gonadotrope: relative roles of intra- and extracellular sources. Endocrinology. 1984;115:1380–1385. doi: 10.1210/endo-115-4-1380. [DOI] [PubMed] [Google Scholar]
- Cogger E.A., Burke W.H., Ogren L.A. Serum luteinizing hormone, progesterone, and estradiol levels in relation to broodiness in the turkey (Meleagris gallapavo) Poult. Sci. 1979;58:1355–1360. doi: 10.3382/ps.0581355. [DOI] [PubMed] [Google Scholar]
- De S., Marcinkiewicz J.L., Vijayaraghavan S., Kline D. Expression of 14- 3-3 protein isoforms in mouse oocytes, eggs and ovarian follicular development. BMC Res. Notes. 2012;5:57. doi: 10.1186/1756-0500-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donham R.S. Annual cycle of plasma luteinizing hormone and sex hormones in male and female mallards (Anas platyrhynchos) Biol. Reprod. 1979;21:1273–1285. doi: 10.1095/biolreprod21.5.1273. [DOI] [PubMed] [Google Scholar]
- Dunn I.C., Miao Y.W., Morris A., Romanov M.N., Wilson P.W., Waddington D. A study of association between genetic markers in candidate genes and reproductive traits in one generation of a commercial broiler breeder hen population. Heredity. 2004;92:128–134. doi: 10.1038/sj.hdy.6800396. [DOI] [PubMed] [Google Scholar]
- Eisa A.A., De S., Detwiler A., Gilker E., Ignatious A.C., Vijayaraghavan S., Kline D. YWHA (14-3-3) protein isoforms and their interactions with CDC25B phosphatase in mouse oogenesis and oocyte maturation. BMC Dev. Biol. 2019;19:20. doi: 10.1186/s12861-019-0200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Halawani M.E., Rozenboim I. The ontogeny and control of incubation behavior in turkeys. Poult. Sci. 1993;72:906–911. [Google Scholar]
- El Halawani M.E., Silsby J.L., Behnke E.J., Fehrer S.C. Hormonal induction of incubation behavior in ovariectomized female turkeys (Meleagris gallopavo) Biol. Reprod. 1986;35:59–67. doi: 10.1095/biolreprod35.1.59. [DOI] [PubMed] [Google Scholar]
- Hammond R.W., Burke W.H., Hertelendy F. Influence of follicular maturation on progesterone release in chicken granulosa cells in response to turkey and ovine gonadotropins. Biol. Reprod. 1981;24:1048–1055. doi: 10.1095/biolreprod24.5.1048. [DOI] [PubMed] [Google Scholar]
- Hermeking H., Lengauer C., Polyak K., He T.C., Zhang L., Thiagalingam S., Kinzler K.W., Vogelstein B. 14-3-3 sigma is a p53-Regulated inhibitor of G2/M Progression. Mol. Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang Y.M., Shi Z.D., Liu Z., Liu Y., Li X.W. Endocrine regulations of reproductive seasonality, follicular development and incubation in Magang geese. Anim. Reprod. Sci. 2008;104:344–358. doi: 10.1016/j.anireprosci.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Ito M., Urano T., Hiroi H., Momoeda M., Saito M., Hosokawa Y., Tsutsumi R., Zenri F., Koizumi M., Nakae H., Horie-Inoue K., Fujii T., Yano T., Kozuma S., Inoue S., Taketani Y. The progesterone-responsive gene 14-3-3τ enhances the transcriptional activity of progesterone receptor in uterine cells. J. Mol. Endocrinol. 2012;49:193–202. doi: 10.1530/JME-12-0112. [DOI] [PubMed] [Google Scholar]
- Johnson A.L. Ovarian follicle selection and granulosa cell differentiation. Poult. Sci. 2015;94:781–785. doi: 10.3382/ps/peu008. [DOI] [PubMed] [Google Scholar]
- Kim Y., Kim H., Jang S.W., Ko J. The role of 14-3-3β in transcriptional activation of estrogen receptor α and its involvement in proliferation of breast cancer cells, Biochem. Biophys. Res. Commun. 2011;414:199–204. doi: 10.1016/j.bbrc.2011.09.056. [DOI] [PubMed] [Google Scholar]
- Kovas c.J., Forgó V., Péczely P. The fine structure of the follicular cells in growing and atretic ovarian follicles of the domestic goose. Cell Tissue Res. 1992;267:561–569. doi: 10.1007/BF00319379. [DOI] [PubMed] [Google Scholar]
- Liu L.B., Xiao Q.H., Gilbert E.R., Cui Z.F., Zhao X.L., Wang Y., Yin H.D., Li D.Y., Zhang H.H., Zhu Q. Whole transcriptome analysis of atrophic ovaries in broody chickens reveals regulatory pathways associated with proliferation and apoptosis. Sci. Rep. 2018;8:7231. doi: 10.1038/s41598-018-25103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T.F. Tuning exocytosis for speed: fast and slow modes, Biochim. Biophys. Acta. 2003;1641:157–165. doi: 10.1016/s0167-4889(03)00093-4. [DOI] [PubMed] [Google Scholar]
- Mauro L.J., Elde R.P., Youngren O.M., Phillips R.E., El Halawani M.E. Alterations in hypothalamic vasoactive intestinal peptide-like immunoreactivity are associated with reproduction and prolactin release in the female Turkey. Endocrinology. 1989;125:1795–1804. doi: 10.1210/endo-125-4-1795. [DOI] [PubMed] [Google Scholar]
- Meng J., Cui C., Liu Y.C., Jin M.L., Wu D.D., Chao L., Wang E.H., Yu B.Z. The role of 14-3-3ε Interaction with phosphorylated cdc25B at its ser321 in the release of the mouse oocyte from prophase I arrest. PLoS One. 2013;8:e53633. doi: 10.1371/journal.pone.0053633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning H.M., Cui Y.L., Song X.C., Chen L.L., Yin Z.H., Hua L.S., Ren F., Suo Y., Wang X.R., Zhang H.L., Hu D.F., Ge Y.M. iTRAQ-based proteomic analysis reveals key proteins affecting cardiac function in broilers that died of sudden death syndrome. Poult. Sci. 2019;98:6472–6482. doi: 10.3382/ps/pez532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang H.J., Wang Z.J., Chen X.L., Yu J., Li Z.H., Nie Q.H. Proteomic analysis of chicken skeletal muscle during embryonic development. Front. Physiol. 2017;8:281. doi: 10.3389/fphys.2017.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce J.G., Parson T.S. Glycoprotein hormones: structure and function, Annu. Rev. Biochem. 1981;50:465–495. doi: 10.1146/annurev.bi.50.070181.002341. [DOI] [PubMed] [Google Scholar]
- Pennington K.L., Chan T.Y., Torres M.P., L Andersen J. The dynamic and stress-adaptive signaling hub of 14-3-3: emerging mechanisms of regulation and context-dependent protein-protein interactions. Oncogene. 2018;37:5587–5604. doi: 10.1038/s41388-018-0348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quayle S.N., Sadar M.D. 14-3-3 sigma increases the transcriptional activity of the androgen receptor in the absence of androgens. Cancer Lett. 2007;254:137–145. doi: 10.1016/j.canlet.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey S.M., Goldsmith A.R., Silver R. Stimulus requirements for prolactin and LH secretion in incubating ring doves. Gen. Comp. Endocrinol. 1985;59:246–256. doi: 10.1016/0016-6480(85)90376-4. [DOI] [PubMed] [Google Scholar]
- Robin B., Bastos R.N., Spanos C., Romé P., Cullen C.F., Rappsilber J., Giet R., Goshima G., Ohkura H. 14-3-3 regulation of Ncd reveals a new mechanism for targeting proteins to the spindle in oocytes. J. Cell Biol. 2017;216:3029–3039. doi: 10.1083/jcb.201704120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov M.N. Genetics of broodiness in poultry - a review. Asian-Australas. J. Anim. Sci. 2001;14:1647–1654. [Google Scholar]
- Sagata N. Meiotic metaphase arrest in animal oocytes: its mechanisms and biological significance. Trends Cell Biol. 1996;6:22–28. doi: 10.1016/0962-8924(96)81034-8. [DOI] [PubMed] [Google Scholar]
- Sharp P.J., Macnamee M.C., Sterling R.J., Lea R.W., Pedersen H.C. Relationships between prolactin, LH and broody behavior in Bantam hens. J. Endocrinol. 1988;18:279–286. doi: 10.1677/joe.0.1180279. [DOI] [PubMed] [Google Scholar]
- Sharp P.J., Scanes C.G., Williams J.B., Harvey S., Chadwick A. Variations in concentrations of prolactin, luteinizing hormone, growth hormone and in the plasma of broody bantams (Gallus domesticus) J. Endocrinol. 1979;80:51–57. doi: 10.1677/joe.0.0800051. [DOI] [PubMed] [Google Scholar]
- Sharp P.J., Dawson A., Lea R.W. Control of luteinizing hormone and prolactin secretion in birds. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 1998;119:275–282. doi: 10.1016/s0742-8413(98)00016-4. [DOI] [PubMed] [Google Scholar]
- Sharp P.J. CABI; Wallingford, UK: 2009. Broodiness and Broody Control; pp. 181–205. [Google Scholar]
- Shen X., Bai X., Xu J., Zhou M., Xu H.P., Nie Q.H., Lu X.M., Zhang X.Q. Transcriptome sequencing reveals genetic mechanisms underlying the transition between the laying and brooding phases and gene expression changes associated with divergent reproductive phenotypes in chickens. Mol. Biol. Rep. 2016;43:977–989. doi: 10.1007/s11033-016-4033-8. [DOI] [PubMed] [Google Scholar]
- van Hemert M.J., Steensma H.Y., van Heusden G.P. 14-3-3 proteins: key regulators of cell division, signaling and apoptosis. Bioessays. 2001;23:936–946. doi: 10.1002/bies.1134. [DOI] [PubMed] [Google Scholar]
- Wakui H., Wright A.P., Gustafsson J., Zilliacus J. Interaction of the ligand- activated glucocorticoid receptor with the 14-3-3η protein. J. Biol. Chem. 1997;272:8153–8156. doi: 10.1074/jbc.272.13.8153. [DOI] [PubMed] [Google Scholar]
- Wang L.Q., Huang H., Liu D. Evaluation of 14-3-3 protein family levels and associated receptor expression of estrogen and progesterone in human uterine leiomyomas. Gynecol. Endocrinol. 2012;28:665–668. doi: 10.3109/09513590.2012.650768. [DOI] [PubMed] [Google Scholar]
- Wen B., Zhou R., Feng Q., Wang Q., Wang J., Liu S. IQuant: an automated pipeline for quantitative proteomics based upon isobaric tags. Proteomics. 2014;4:2280–2285. doi: 10.1002/pmic.201300361. [DOI] [PubMed] [Google Scholar]
- Youngren O.M., el Halawani M.E., Silsby J.L., Phillips R.E. Intracranial prolactin perfusion induce incubation behavior in Turkey. Biol. Reprod. 1991;44:425–431. doi: 10.1095/biolreprod44.3.425. [DOI] [PubMed] [Google Scholar]
- Yu Jin g., Lou Y.P., Zhao A.Y. Transcriptome analysis of follicles reveals the importance of autophagy and hormones in regulating broodiness of Zhedong white goose. Sci. Rep. 2016;6:36877. doi: 10.1038/srep36877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilliacus J., Holter E., Wakui H., Tazawa H., Treuter E., Gustafsson J.A. Regulation of glucocorticoid receptor activity by 14–3-3-dependent intracellular relocalization of the corepressor RIP140, Mol. Endocrinol. 2001;15:501–511. doi: 10.1210/mend.15.4.0624. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.