Abstract
Mutations in the core RNA splicing factor SF3B1 are prevalent in leukemias and uveal melanoma, but hotspot SF3B1 mutations are also seen in epithelial malignancies such as breast cancer. Although hotspot mutations in SF3B1 alter hematopoietic differentiation, whether SF3B1 mutations contribute to epithelial cancer development and progression is unknown. Here, we identify that SF3B1 mutations in mammary epithelial and breast cancer cells induce a recurrent pattern of aberrant splicing leading to activation of AKT and NF-κB, enhanced cell migration, and accelerated tumorigenesis. Transcriptomic analysis of human cancer specimens, MMTV-cre Sf3b1K700E/WT mice, and isogenic mutant cell lines identified hundreds of aberrant 3′ splice sites (3′ss) induced by mutant SF3B1. Consistently between mouse and human tumors, mutant SF3B1 promoted aberrant splicing (dependent on aberrant branchpoints as well as pyrimidines downstream of the cryptic 3′ss) and consequent suppression of PPP2R5A and MAP3K7, critical negative regulators of AKT and NF-κB. Coordinate activation of NF-κB and AKT signaling was observed in the knockin models, leading to accelerated cell migration and tumor development in combination with mutant PIK3CA but also hypersensitizing cells to AKT kinase inhibitors. These data identify hotspot mutations in SF3B1 as an important contributor to breast tumorigenesis and reveal unique vulnerabilities in cancers harboring them.
Keywords: Oncology
Keywords: Breast cancer, RNA processing
Introduction
Comprehensive genomic analyses of cancer have identified change-of-function mutations in RNA splicing factors to be recurrent across a variety of disparate cancer types (1). Akin to driver oncoproteins, these mutations occur as heterozygous mutations recurrently affecting the same nucleotide positions (so-called hotspot mutations). Hotspot mutations occur in both the protein (2–5) as well as RNA components of the spliceosome (6, 7) and globally alter pre-mRNA recognition by the spliceosome in a manner distinct from loss of function (8–10). Malignancies of the hematopoietic system, including myeloid neoplasms and chronic lymphocytic leukemia (CLL), harbor a high frequency of these alterations and have been subject to detailed mechanistic investigations into the consequences of mutation on splicing, transcription, and cancer phenotypes (11–14).
SF3B1 is the most commonly mutated RNA splicing factor, and studies of hotspot mutations in SF3B1 have demonstrated that these mutations promote usage of aberrant branchpoint nucleotides (8, 15). Recent functional investigations of aberrant splicing driven by mutant SF3B1 in the context of myelodysplastic syndromes (MDS), CLL, and uveal melanoma (UVM) have begun to elucidate specific aberrant splicing events required for the maintenance of SF3B1 mutant cancers (11). In addition, a number of studies in the context of myeloid leukemias have identified that SF3B1 mutations confer therapeutic vulnerabilities to further modulation of splicing (16) as well as specific metabolic perturbations (17). However, to date, the biological consequences of expression of the same hotspot mutations in SF3B1 in epithelial-derived malignancies are largely unknown and make for an intriguing counterpoint. While kinase oncoproteins like BRAF or NTRK function as targetable drivers in different tissue types (18–21), it is unknown whether large-scale modification of RNA splicing in different cell types is similarly oncogenic and uses the same pathways within distinct tissues to derive tumor phenotypes.
In this study, we investigated the consequences of SF3B1 mutations in breast cancer, where across a cohort of more than 5000 patients, SF3B1 alterations are observed in approximately 3% of unselected cases. The effect of mutation upon global splicing, RNA expression, tumorigenesis, and tumor phenotypes highlights how aberrant splicing patterns are conserved but lead to lineage-specific effectors and phenotypes as well as novel therapeutic opportunities. Our data identify that mutations in SF3B1 promote breast cancer development and progression via aberrant splicing and expression of intermediary signaling proteins that normally negatively regulate AKT and NF-κB signaling in mammary epithelial cells.
Results
SF3B1 mutations are enriched in estrogen receptor–positive (ER+) breast cancer and associate with poor outcomes.
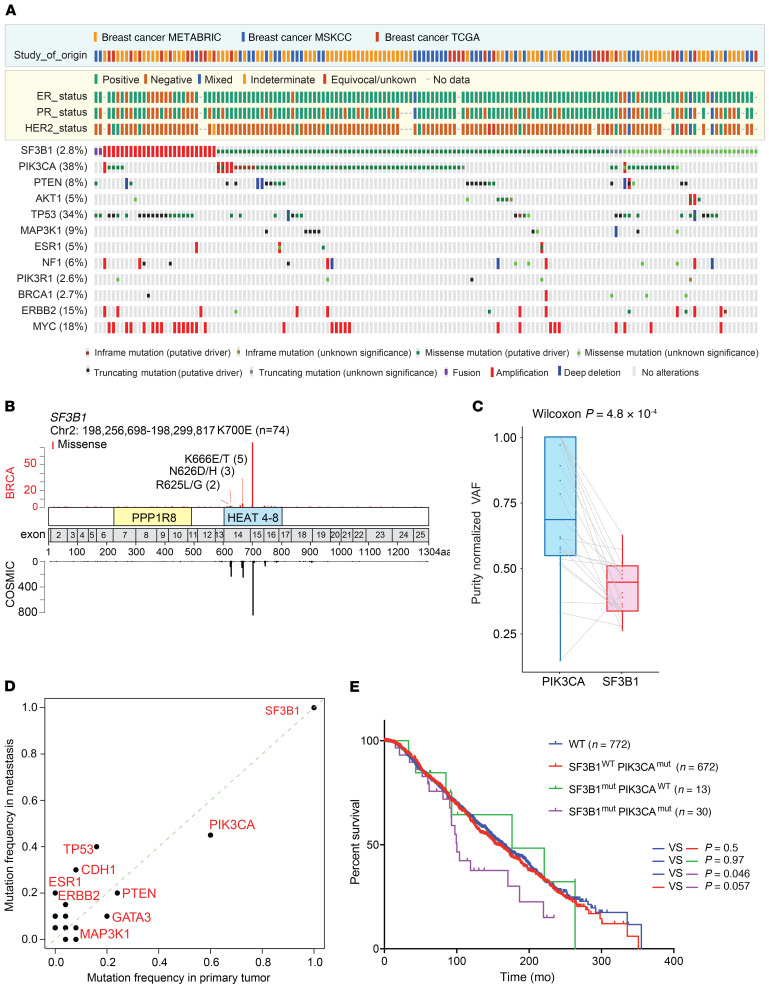
To systematically establish the prevalence and significance of SF3B1 mutations in breast cancer, we performed a large-scale analysis of genomic/exomic sequencing data from 5366 patients with breast cancer, including prior data from the METABRIC, TCGA, and MSK-IMPACT databases (22–24) (Figure 1A and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI138315DS1). Genetic alterations in SF3B1, including mutations, gene fusions, and copy number amplification, were identified in 152 patients (2.83%) (Figure 1A), among which 90 patients (1.68%) carried hotspot mutations previously identified as genomic hotspots (25). The K700E (n = 74) substitution in SF3B1 was the dominant SF3B1 mutation in patients with breast cancer, followed by hotspot mutations at K666 (n = 5), N626 (n = 3), and R625 (n = 2) residues (Figure 1B). Among the patients with hotspot SF3B1 mutations, ER status was available for 89 patients, only 2 of which were ER– (Figure 1A and Supplemental Table 2). These 2 patients both had hormone receptor positive primary cancer and later developed metastatic ER– tumors. Within the METABRIC and TCGA cohorts where Pam50 and claudin low subtyping is annotated, we found 84% (45/53) of SF3B1 mutations occurred in luminal A or B subtypes, and 60% (32/53) of the cases were significantly enriched in luminal A breast cancer (P = 0.002) (Supplemental Figure 1). In terms of other genomic alterations, SF3B1 hotspot mutations significantly co-occurred with PIK3CA mutations (n = 55; 2.76% in patients with PIK3CA mutations; log2 odds ratio = 1.382; q < 0.001) (Supplemental Figure 1). Interestingly, most SF3B1 mutant samples that did not carry PIK3CA mutations harbored mutations in PTEN or AKT1, each of which converges on activating PI3K signaling.
Figure 1. SF3B1 hotspot mutations are recurrent in breast cancer and are significantly associated with mutations activating PI3K signaling and shortened survival.
(A) Oncoprint of somatic alterations in SF3B1 and other breast cancer drivers across 5366 patients from the METABRIC (23, 65), MSK-IMPACT (24), and TCGA (22) breast cancer cohorts. ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2. (B) SF3B1 mutation maps showing the counts, amino acid change, position, and evidence of mutational hotspots, based on COSMIC database information. The y axis counts at the bottom of the maps reflect the number of identified mutations in the COSMIC database. (C) Purity normalized variant allele frequency (VAF) of PIK3CA and SF3B1 mutations among 51 double-mutated samples in the MSK-IMPACT cohort. (D) Frequency of somatic mutations in patients from the MSK-IMPACT cohort (n = 94) harboring SF3B1 hotspot mutations. Mutation frequency was calculated for each reported gene in 57 primary samples (x axis) and 45 metastasis samples (y axis). (E) Kaplan-Meier curve of disease-free survival in SF3B1 hotspot mutant (n = 13), SF3B1 WT PIK3CA mutant (n = 672), and SF3B1/PIK3CA double-mutant (n = 30) versus WT (n = 772) ER+ breast cancer patients from METABRIC. P values were derived from log-rank test. See also Supplemental Figure 1.
Given recent identification that ER+ breast cancers frequently harbor several subclonal mutations that arise under the selection of endocrine-targeted therapy (24, 26, 27), we next evaluated the clonality of SF3B1 mutations in our MSK-IMPACT cohort. Across tumors, mutations in SF3B1 were clonal with a median allele frequency of about 50%, without manifesting allelic imbalances characteristic of mutant PIK3CA (Figure 1C). This latter finding is consistent with recent work from our group and others identifying essentiality of the WT allele in cells bearing hotspot change-of-function mutations in SF3B1 and other RNA splicing factors (28–30). In considering the progression of tumors from primary to metastatic disease, genetic analysis of primary tumors and metastases from a series of patients with the SF3B1 mutant identified frequent co-occurrence of SF3B1 and PIK3CA mutations across primary tumors and metastases cases (Figure 1D).
We further analyzed the clinical outcomes of patients with SF3B1 hotspot mutation and PIK3CA mutations (Figure 1E). The median survival time of patients with ER+ breast cancer with no somatic alteration in either SF3B1 or PIK3CA was 164 months, whereas that of patients with ER+ disease harboring both SF3B1 and PIK3CA mutations was 100.1 months (hazard ratio = 1.787, P = 0.046). Neither SF3B1 nor PIK3CA mutation alone was associated with significantly shorter survival. These findings point to SF3B1 mutation as a recurrent alteration in ER+ and PIK3CA mutant breast cancer that appears to be an early event and linked to poor prognosis.
Sf3b1K700E promotes accelerated mammary tumor progression.
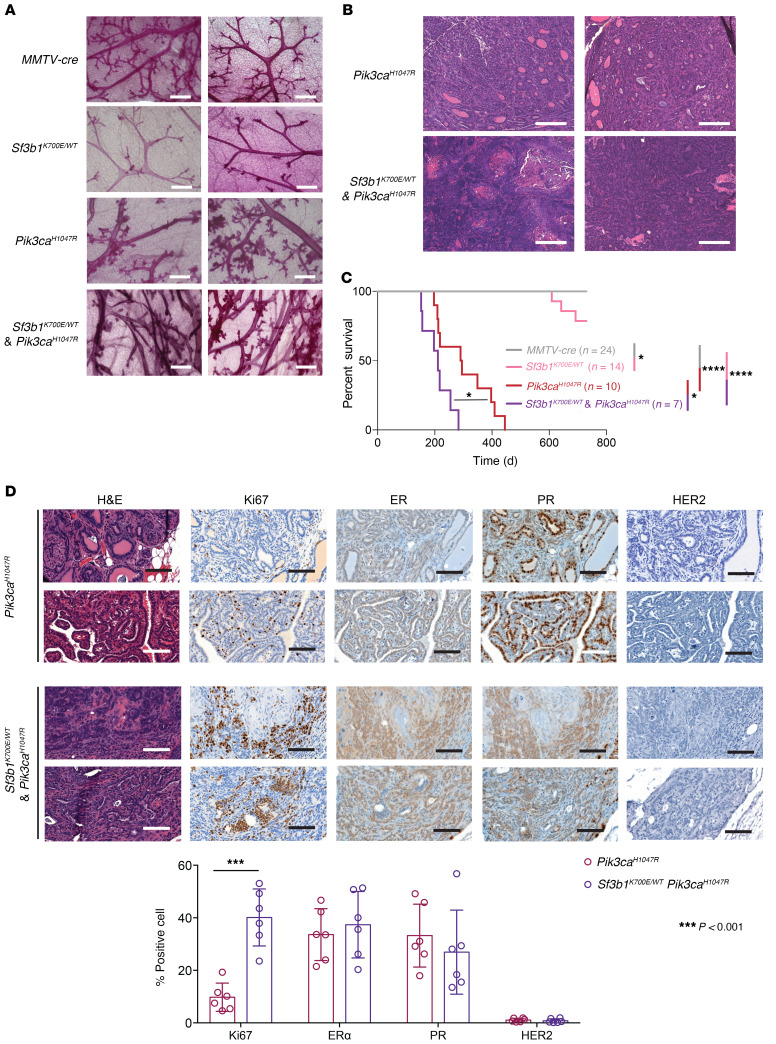
To determine the functional role of SF3B1 hotspot mutations in breast tumorigenesis and progression, we generated a conditional knockin mouse model to express the most common SF3B1 mutation found in patients with breast cancer (the K700E mutant) from the endogenous Sf3b1 locus in the mouse mammary gland (31). Heterozygous MMTV-cre Sf3b1K700E/WT mice expressed mutant Sf3b1 transcripts in mammary epithelial cells (sorted live, CD45– CD24+ cells) at a 50% allelic frequency in RNA (similar to allelic frequency observed in human breast cancers; Figure 1C and Supplemental Figure 2, A–C). Due to the significant co-occurrence of SF3B1 mutation and alterations activating the PI3K pathway, particularly PIK3CA mutations in patients with breast cancer (Supplemental Figure 1), we further crossed MMTV-cre Sf3b1K700E/WT mice with mice bearing a transgenic kinase domain mutation in Pik3ca from the Rosa26 locus (Rosa26 lox-stop-lox Pik3caH1047R (R26-LSL-Pik3caH1047R)) (32) to evaluate the combinatorial effects of these mutations on mammary tumor development and progression.
Whole-mount examination of mammary glands from 4-month-old nulliparous mice of each genotype revealed ductal hyperplasia in mice expressing Pik3caH1047R alone (MMTV-cre R26-LSL-Pik3caH1047R) and in combination with mutant Sf3b1 (Figure 2A and Supplemental Figure 2D). Interestingly, reduced branching of mammary ducts was observed in MMTV-cre Sf3b1K700E/WT mice (Supplemental Figure 2D). With further age, PIK3CA single-mutant and Sf3b1K700E/WT/Pik3caH1047R double-mutant mice (MMTV-cre Sf3b1K700E/WT R26-LSL-Pik3caH1047R) went on to develop mammary gland tumors (Figure 2, B and C), while MMTV-cre Sf3b1K700E/WT mice did not exhibit any clear protumorigenic phenotype, which is consistent with previous studies with hematopoietic stem cells (13, 29) and B cells (33, 34) where Sf3b1 K700E mutation alone was not sufficient to mediate tumorigenesis. Strikingly, the life span of double-mutant mice (median = 211 days) was significantly shorter than Pik3ca single-mutant mice (median = 293 days, P = 0.0125, Figure 2C). Akin to the tumors MMTV-cre R26-LSL-Pik3caH1047R mice developed, MMTV-cre Sf3b1K700E/WT R26-LSL-Pik3caH1047R mice develop lesions within the spectrum of tumors reported in Pik3ca conditional mouse models (35), including adenomyoepitheliomas and adenosquamous carcinomas (Supplemental Figure 3A). The adenomyoepitheliomas were composed of a proliferation of gland-like structures lined by a dual cell population of luminal epithelial (CK8/18 and Gata3 staining in Supplemental Figure 3A) and myoepithelial cells (Sma staining in Supplemental Figure 3A), whereas the adenosquamous carcinomas were composed of a complex admixture of gland-like structures, sheets of squamous cells (CK14 staining in Supplemental Figure 3A) and occasional squamous pearls; areas of necrosis were observed. No other histologic abnormalities were detected in the liver, lungs, kidneys, adrenals, spleen, heart, and central nervous system of these mice. Given that MMTV-cre has been reported to be expressed in tissues besides mammary epithelial cells (including expression in salivary gland and lymphocytes) (36), we examined both the salivary gland and performed a comprehensive analysis of hematopoietic tissues but did not identify overt malignancies in these tissues at the time of breast tumor formation (Supplemental Figure 3B and Supplemental Figure 4). To characterize the mammary gland tumors that developed in these mice, we probed ER, progesterone receptor (PR), HER2, and Ki67 in representative tumor sections. While all tumors collected were ER+, PR+, and HER2–, Ki67 staining was significantly higher in tumors from MMTV-cre Sf3b1K700E/WT Pik3caH1047R mice than in those from Pik3caH1047R single-mutant mice (Figure 2D). We next sought to understand the mechanistic basis by which SF3B1 mutation could contribute to accelerated progression that was observed both in the mouse and human data.
Figure 2. Sf3b1K700E mutation promotes mammary tumorigenesis in conjunction with mutant Pik3ca.
(A) Whole-mount hematoxylin staining of mammary glands from MMTV-cre control, MMTV-cre Sf3b1K700E/WT, MMTV-cre R26-LSL-Pik3caH1047R, and MMTV-cre Sf3b1K700E/WT R26-LSL-Pik3caH1047R at the age of 4 months. Two representative examples are shown. Scale bars: 500 μM. (B) Representative sections of H&E-stained mammary glands from the mice shown in A at time of death. Two representative examples are shown. Scale bars: 250 μM. (C) Kaplan-Meier survival curve of the mice shown in A. The endpoint criterion was presence of tumors at least 1 cm3. P values calculated by log-rank (Mantel-Cox) test in Prism 7 software *P <0.05, ****P < 0.0001. (D) H&E and immunohistochemical staining of breast tumors developing in MMTV-cre R26-LSL-Pik3caH1047R transgenic and compound Sf3b1K700E/WT Pik3caH1047R transgenic mice. ER, estrogen receptor; PR, progesterone receptor. Scale bars: 100 μM. Quantitation of percentage of positively stained cells (n = 6 tumors/group) is shown below. ***P < 0.001 (Student’s t test). See also Supplemental Figures 2–4.
Pan-cancer and breast cancer–specific aberrant splicing events induced by mutant SF3B1.
Prior work in hematopoietic-derived cells and UVM has revealed that cancer-associated mutations in SF3B1 result in splicing events that use aberrant branchpoint nucleotides, manifesting in increased use of aberrant (or so-called cryptic) intron-proximal 3′ splice sites (3′ss) (15, 37). To understand the impact of SF3B1 mutations on splicing in breast cancers compared with other malignancies, we performed a pan-cancer transcriptomic analysis to identify and quantify cryptic 3′ss usage across SF3B1 WT versus mutant cancers (Figure 3). For breast cancer, RNA-seq data from 20 tumors harboring SF3B1 mutations and 20 randomly selected, ER+ WT tumors were collected from the TCGA breast cancer (BRCA) database (Supplemental Table 3). In this analysis, we also included an isogenic pair of parental and SF3B1 K700E knockin MCF10A breast epithelial cells, as well as MCF7 and T47D breast cancer cells ectopically expressing SF3B1 WT or SF3B1 K700E mutant via a doxycycline-inducible expression system (Supplemental Figure 5). The expression of the SF3B1 K700E mutant did not significantly affect cell proliferation and dependence on ER of these breast cancer cells under in vitro culture conditions (Supplemental Figure 5). For the comparison of SF3B1 mutant breast cancer to other cancer types bearing SF3B1 mutations, RNA-seq data of 15 patients with CLL and 16 patients with UVM together with same number of randomly chosen WT samples of the same histologic types of cancer were also included (Supplemental Table 3). Principal component analysis and unsupervised hierarchical clustering of cryptic 3′ss usage using the percent spliced in (PSI) matrix (Figure 3A, Supplemental Figure 6 and Supplemental Methods) robustly separated samples with SF3B1 hotspot mutations from WT cases, with the exception of one breast cancer sample with an SF3B1 K666N mutation (a result likely explained by the low allelic frequency [7%] of the mutation in the RNA-seq data from this sample). Samples with non–hotspot SF3B1 mutations (e.g., S455V, E802Q) clustered together with SF3B1 WT samples (Supplemental Figure 6 and Supplemental Table 3), suggesting that these mutations do not cause aberrant 3′ss usage. Within the SF3B1 mutant group, samples clustered separately into subgroups by their cancer types. Breast cancer patient and cell line samples (triangles in Figure 3A) showed a very similar pattern of aberrant splicing, compared with CLL (circles in Figure 3A) or UVM samples (squares in Figure 3A), suggesting the aberrant 3′ss usage induced by SF3B1 mutations in distinct cancers may be due to intrinsic gene expression patterns unique to cell lineage.
Figure 3. Differential aberrant 3′ splicing events across SF3B1 mutant cancers.
(A) Principal component analysis of differential 3′ splice sites (3′ss) in SF3B1 mutant and WT samples. The PSI input matrix contains 618 cryptic 3′ss events identified from the pan-cancer data set are shown. BRCA, breast cancer; CLL, chronic lymphocytic leukemia; UVM, uveal melanoma. (B) Histogram of statistically significant (q < 0.05) cryptic 3′ss identified in BRCA samples: WT (black) versus mutant (red) for SF3B1 and their distance in nucleotides (nt) from the associated canonical 3′ss. (C) Consensus 3′ss motif near the associated cryptic 3′ AG dinucleotide (top) and canonical 3′ AG dinucleotide (bottom) for 398 aberrant 3′ss events more than 10 nt from the canonical 3′ss as well as 90 aberrant 3′ss events more than 50 nt from the canonical 3′ss. See also Supplemental Figures 5 and 6.
Consistent with previous studies in other cancer types (15, 37), most aberrant 3′ss in SF3B1 mutant breast tumors were located 10 to 30 nucleotides upstream of the canonical 3′ss (Figure 3B). For most unaffected 3′ss, a stretch of pyrimidine nucleotides (known as a polypyrimidine tract) is present within the intron, near the canonical 3′ss. To understand the mechanism by which SF3B1 mutations affect 3′ss recognition, we analyzed the sequences around cryptic 3′ss promoted in SF3B1 mutant cells (see Methods; Figure 3C). As previously reported, this revealed that polypyrimidine tracts at cryptic 3′ss were interrupted by adenines, indicating a shorter distance between the cryptic 3′ss and branchpoint nucleotides (8, 15). At the same time, we also identified an enrichment of pyrimidines downstream of the aberrant 3′ss. Since most cryptic 3′ss were within 30 nucleotides upstream of the canonical 3′ss, this enrichment could be due to a bias from the polypyrimidine track associated with the canonical 3′ss. To eliminate this bias, we performed a sequence analysis exclusively on cryptic sites 50 nucleotides away from the canonical 3′ss. This analysis continued to reveal significant enrichment of pyrimidines within these 90 aberrant 3′ss (P < 0.0001, Fisher’s exact test, Figure 3C). These data indicate that recognition of cryptic 3′ss by mutant SF3B1 may require downstream pyrimidine-rich motifs, a finding which was evident across SF3B1 mutant cancer types.
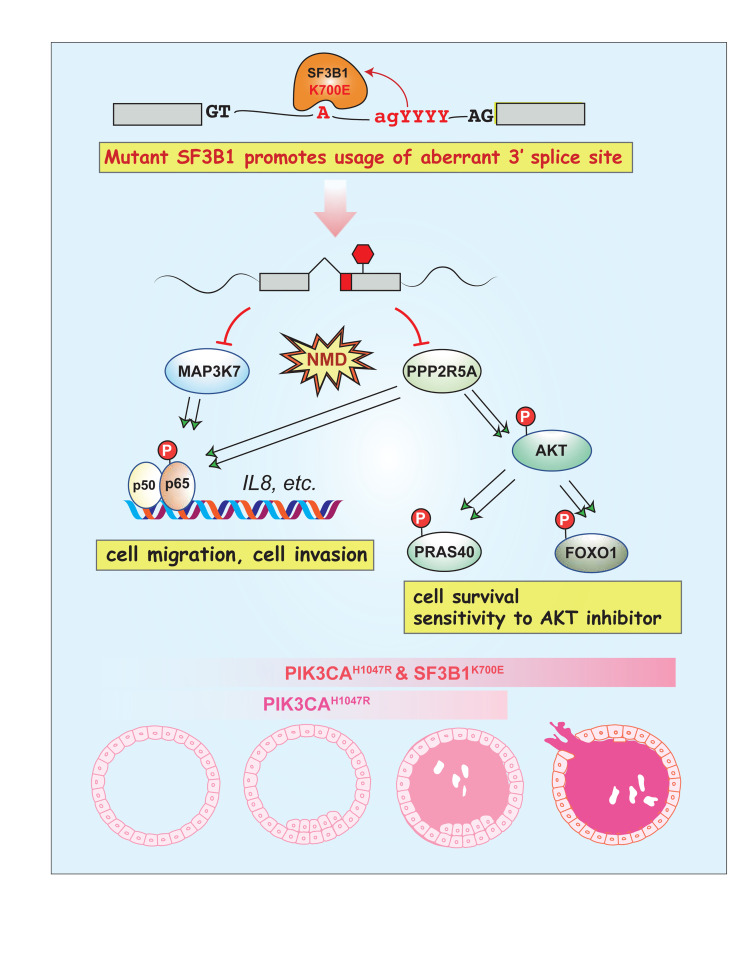
Aberrant splicing mediated suppression of PPP2R5A and MAP3K7 in SF3B1-mutant breast cancers.
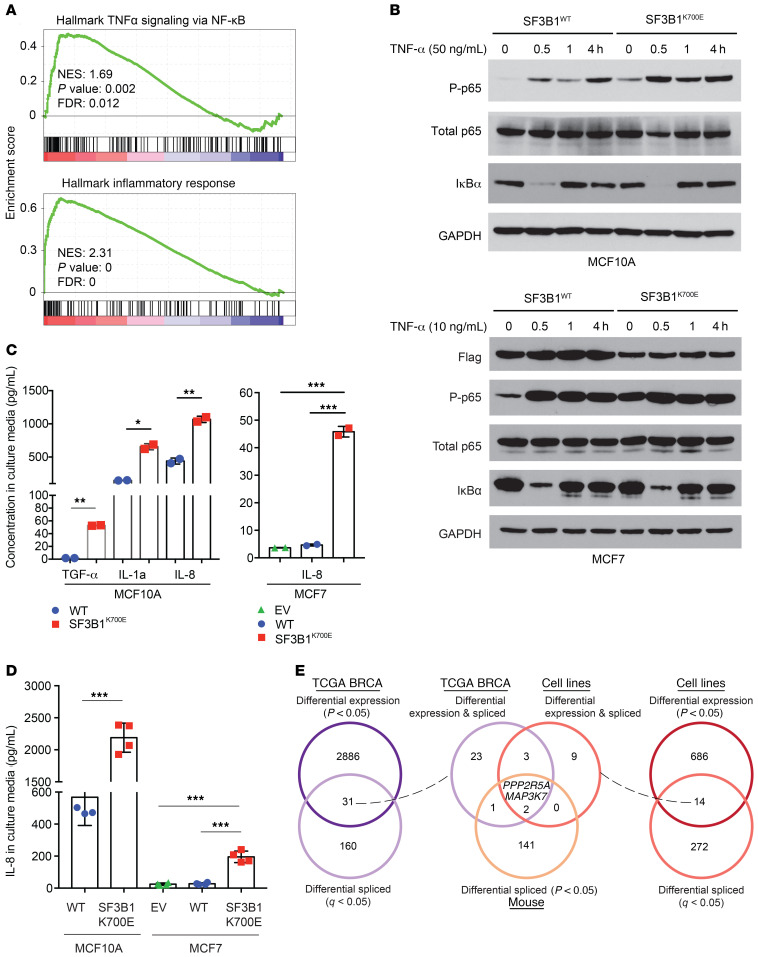
To identify the key cellular processes that mutant SF3B1 alters in accelerating breast cancer progression, we performed a gene set enrichment analysis (GSEA) on the RNAs derived from WT and SF3B1-mutant breast cancer cell lines. This revealed that inflammatory response pathways (P = 0), and NF-κB signaling (P = 0.002) in particular, were significantly altered in SF3B1 K700E-expressing breast cancer cell lines (Figure 4A). Moreover, GSEA analysis on the TCGA BRCA gene expression data also revealed upregulation of these same pathways in the patient samples bearing hotspot SF3B1 mutations (Supplemental Figure 7A). To verify activation of NF-κB signaling as a result of mutant SF3B1 expression, we performed Western blot analysis to assess phosphorylation of p65 (RelA), a core subunit of NF-κB complex, in isogenic SF3B1 K700E knockin MCF10A cells and MCF7 cells expressing mutant SF3B1 (Figure 4B and Supplemental Figure 7B). Phosphorylation of p65 was upregulated by 3- to 5-fold in mutant SF3B1-expressing cells, and the levels of phosphorylation were further enhanced following stimulation by TNF-α (Figure 4B and Supplemental Figure 7B). To further assess the significance of this activation of NF-κB, we examined the effect of SF3B1 mutation on the expression of inflammatory cytokines known to be regulated by NF-κB. We measured the quantity of secreted cytokines in the culture media of MCF10A and MCF7 WT versus SF3B1 K700E-expressing cells. IL-1a, IL-8, and TGF-α were all increased in the media derived from SF3B1 K700E MCF10A cells (Figure 4C), and mutant SF3B1-expressing MCF7 cells produced 10-fold more IL-8 than SF3B1 WT counterpart isogenic cells (Figure 4C), consistent with the observed upregulation of p65 phosphorylation in these cells. Upregulated IL-8 secretion in both MCF10A and MCF7 cells expressing mutant SF3B1 was also confirmed by ELISA (Figure 4D). These data together confirm the induction of NF-κB signaling observed in the GSEA analysis of SF3B1 mutant tumors.
Figure 4. Increased inflammation and activation of NF-κB signaling in SF3B1 mutant mouse and human breast tumors.
(A) GSEA identifies enrichment of hallmark gene sets of TNF-α signaling via NF-κB (top) and inflammatory response (bottom) in SF3B1 mutant versus WT human breast cell lines. FDR, false discovery rate; NES, normalized enrichment score. (B) Activation of phospho-p65 (P-p65) upon TNF-α stimulation of MCF10A SF3B1K700E/WT mutant knockin cells (top) and MCF7 with transgenic expression of SF3B1K700E versus SF3B1WT cDNA (bottom). Quantitation of 3 independent experiments is shown in Supplemental Figure 7B. (C) Levels of TGF-α, IL-1a, and IL-8 in culture media from MCF10A cells shown in B or IL-8 in culture media from MCF7 cells with expression of an empty vector (EV), SF3B1 WT, or SF3B1 K700E cDNAs measured by Milliplex cytokine/chemokine multiplex assay. (D) Levels of IL-8 in culture media shown in C measured by ELISA. Mean levels with standard deviation shown. (*P < 0.05, **P < 0.01, ***P < 0.0001; Student’s t test for MCF10A and Dunnett’s multiple comparisons test for MCF7). (E) Venn diagrams of genes significantly differentially expressed (P < 0.05; DESeq2) and spliced (q < 0.05; t test) across SF3B1 mutant versus WT human breast cancers from TCGA (22) (left), isogenic human breast cell lines (right), and overlapping across human tumors, human cell lines, and mouse mammary epithelial cells mutant for Sf3b1 (middle). See also Supplemental Figure 7.
To understand the molecular mechanisms for NF-κB activation in SF3B1 mutant cells, we examined the intersection of differentially expressed and differentially misspliced genes in human tumors, cell lines, and our mouse model (Figure 4E and Supplemental Figure 7C). From this intersection, only 2 genes were identified: MAP3K7 and PPP2R5A, both of which were significant reduced in expression in SF3B1 mutants (Figure 4E and Supplemental Tables 4–6). Of note, these missplicing events in MAP3K7 and PPP2R5A have been reported in hematopoietic malignancies with SF3B1 mutations recently by Lee et al. (29) and Liu et al. (34).
Mutant SF3B1 causes aberrant splicing of MAP3K7, stimulating NF-κB signaling.
MAP3K7 has known roles in regulating NF-κB signaling (38–40) and so we initially sought to verify its missplicing and biologic contribution to mutant SF3B1–mediated pathogenesis. To confirm a consistent effect of SF3B1 mutation on MAP3K7 splicing and expression, we examined the RNA-seq data of the isogenic cell lines, mouse mammary tissue, and human tumors at the candidate intron where missplicing was implicated (Figure 5, A–C). Across these samples, mutant SF3B1 led to aberrant splicing of MAP3K7 via use of a cryptic 3′ss that was conserved in human breast cell lines and tumors and mouse mammary gland expressing Sf3b1K700E (Figure 5, A and B). As expected, NMD caused by missplicing led to reduced MAP3K7 transcript and protein levels in mutant SF3B1-expressing cells and mouse mammary gland tumors (Figure 5, C–E, and Supplemental Figure 8A).
Figure 5. Aberrant splicing and downregulation of MAP3K7 by mutant SF3B1 promotes NF-κB activation in breast cancer.
Representative RNA-seq coverage plots showing cryptic 3′ss usage in MAP3K7 in (A) human isogenic breast cancer cell lines (MCF10A, MCF7, T47D) expressing WT or SF3B1 K700E (K700E) and human breast tumor samples from TCGA (22) as well as (B) mouse mammary epithelial cells from MMTV-cre control, MMTV-cre Sf3b1K700E/WT, MMTV-cre R26-LSL-Pik3caH1047R transgenic, and compound MMTV-cre Sf3b1K700E/WT R26-LSL-Pik3caH1047R mice. PSI value is shown on the right of each coverage plot. (C) MAP3K7 mRNA expression (normalized log2RPKM values) in SF3B1 mutant (MUT) versus WT human breast tumors from TCGA and cell lines shown in A. Box indicates upper and lower quartiles; thick bar indicates the median value. Whiskers indicate the largest/smallest value no further than 1.5 times the interquartile range. (D) RT-PCR of MAP3K7 splicing event (top) and MAP3K7 protein levels in isogenic MCF10A cells with or without SF3B1K700E knockin. The 3’ss used is shown in the schematic. (E) MAP3K7 protein levels in MCF7 cells expressing doxycycline-inducible FLAG-SF3B1WT or FLAG-SF3B1K700E vectors. RT-PCR of MAP3K7 splicing is shown on bottom. (F) RT-PCR of MAP3K7 splicing event derived from MAP3K7 minigene with mutations at aberrant branchpoint or polypyrimidine region downstream of the aberrant 3’ss. RT-PCR of MAP3K7 splicing event derived from endogenous MAP3K7 is shown on bottom. A schematic of sequences around the normal and aberrant branchpoint and splice site is shown on top. (G) MAP3K7 and phospho- and total p65 and p38 levels in MCF7 and MCF10A cells transduced with anti-MAP3K7 shRNA. (H) MAP3K7 and phospho- and total p65 and p38 levels in MCF7 and MCF10A cells expressing SF3B1 K700E mutant from D and E transduced with or without MAP3K7-GFP cDNA. Quantitation of 3 independent experiments is shown in Supplemental Figure 8. See also Supplemental Figure 9.
To understand this aberrant splicing event in MAP3K7 in greater molecular detail, we mapped the branchpoints in the aberrantly spliced intron by lariat RT-PCR. Sequencing of the PCR products identified the branchpoints used for the canonical as well as cryptic 3′ss (Supplemental Figure 9A). The branchpoint for the canonical 3′ss was an adenine 22 nucleotides upstream of the intron-exon boundary, whereas the branchpoint for the cryptic 3′ss mapped to an adenine 14 nucleotides upstream of the aberrant intron-exon boundary (Figure 5F). To identify cis nucleotides required for mutant SF3B1–specific aberrant splicing of MAP3K7, we generated a minigene construct that contains sequences from the misspliced intron and flanking exons. As with the endogenous MAP3K7 transcript, mutant SF3B1 also enhanced usage of a cryptic 3′ss in the minigene-derived MAP3K7 RNA (Figure 5F). Substitution of adenines at the branchpoint with guanines blocked the usage of the cryptic 3′ss (Figure 5F). A pyrimidine-rich motif was located in the sequences downstream of the cryptic 3′ss. We mutated thymine nucleotides within this pyrimidine-rich motif to test if these residues were necessary for mutant SF3B1–specific splicing. Substitution of either 2 or 3 thymines to adenines completely abolished the usage of the cryptic 3′ss and aberrant inclusion of the 20-nucleotide intronic sequences (Figure 5F). These data identify the requirement of the aberrant branchpoint as well as pyrimidines downstream of the aberrant branchpoint in missplicing MAP3K7.
We next sought to establish the biologic role for MAP3K7 in NF-κB signaling in breast cancer cells as its regulation of NF-κB signaling, which has been previously shown to be cell-type specific (38–40). To characterize the impact of MAP3K7 on NF-κB activation in breast cells specifically, we manipulated MAP3K7 activity in MCF7 and MCF10A cells. Knockdown of MAP3K7 greatly increased phosphorylation of p65 and reduced p38 phosphorylation (Figure 5G and Supplemental Figure 8B), which was also observed with pharmacological inhibition of MAP3K7 using 5Z-7-oxozeaenol (41) (Supplemental Figure 8C). Consistent with this, restored expression of MAP3K7 (via expression of MAP3K7 cDNA) in SF3B1 K700E isogenic MCF10A cells or MCF7 expressing mutant SF3B1 greatly reduced NF-κB activation and promoted p38 phosphorylation (Figure 5H and Supplemental Figure 8D). Taken together, these data establish mutant SF3B1-directed missplicing of MAP3K7 to be a key mediator of enhanced NF-κB signaling in breast cancer.
Mutant SF3B1 causes aberrant splicing of PPP2R5A, enhancing AKT activation and inflammatory signaling.
In addition to missplicing of MAP3K7, SF3B1 K700E mutation also induced usage of a cryptic 3′ss 13 nucleotides upstream the normal 3′ss of intron 4 of PPP2R5A in human and mouse cells (Figure 6, A and B). PPP2R5A encodes the alpha isoform of the B56 regulatory subunit of PP2A (the serine/threonine-protein phosphatase 2A). PP2A is 1 of 4 major serine/threonine phosphatases, and it is a heterotrimeric protein consisting of a catalytic C subunit, 1 structural A subunit, and a regulatory B subunit. Thirteen distinct genes encoding for the different regulatory B subunits and the specific B subunit expressed regulates substrate selectivity and catalytic activity of PP2A.
Figure 6. Aberrant splicing and downregulation of PPP2R5A by mutant SF3B1 promotes AKT activation and inflammatory signaling in breast cancer.
Representative RNA-seq coverage plots showing cryptic 3′ss usage in PPP2R5A in (A) human isogenic breast cancer cell lines (MCF10A, MCF7, T47D) expressing WT or SF3B1 K700E (K700E) and human breast tumor samples from TCGA (22) as well as (B) mouse mammary epithelial cells from MMTV-cre control, MMTV-cre Sf3b1K700E/WT, MMTV-cre R26-LSL-Pik3caH1047R transgenic, and compound MMTV-cre Sf3b1K700E/WT R26-LSL-Pik3caH1047R mice. PSI value of each event is shown on the right of each coverage plot. (C) PPP2R5A mRNA expression (normalized log2RPKM values) in SF3B1 mutant (MUT) versus WT primary human breast tumors from TCGA and cell lines shown in A. Box indicates upper and lower quartiles; thick bar indicates the median value. Whiskers indicate the largest/smallest value no further than 1.5 times the interquartile range. (D) PPP2R5A, phospho- and total AKT, and p65 levels in MCF7 and MCF10A with and without mutant SF3B1 expression (EV, empty vector). Quantitation of 3 independent experiments is shown in Supplemental Figure 10A. (E) RT-PCR of PPP2R5A splicing event derived from PPP2R5A minigene with mutations at aberrant branchpoint or polypyrimidine region downstream of the aberrant 3′ss. RT-PCR of PPP2R5A splicing event derived from endogenous PPP2R5A is shown on the bottom. A schematic of sequences around the normal and aberrant branchpoint and splice site is shown on top. (F) PPP2R5A, phospho- and total AKT, and p65 levels in MCF7 and MCF10A with and without PPP2R5A shRNA knockdown. Quantitation of 3 independent experiments is shown in Supplemental Figure 10B. (G) Relative mRNA expression of IL1A, CXCL1, IL8, CDKN1A, and PPP2R5A in the cells from D and F. Mean ± standard deviation shown; asterisks represent P values of Dunnett’s test compared with shRenilla control (shRE). See also Supplemental Figures 9 and 10.
Mutant SF3B1 promoted the inclusion of 13 intronic nucleotides in exon 5 by using an upstream cryptic 3′ss, resulting in a reading frame shift (Figure 6, A and B). This is predicted to cause NMD of the aberrantly spliced PPP2R5A transcript and, consistent with this, SF3B1 mutant patient breast tumors had significantly reduced PPP2R5RA mRNA and protein expression compared with WT (Figure 6, C and D, and Supplemental Figure 10A). As with MAP3K7, missplicing of PPP2R5A by mutant SF3B1K700E could be replicated within a PPP2R5A minigene and this missplicing was abolished by disruption of branchpoint (Figure 6E and Supplemental Figure 9B) or interruption of the poly-pyrimidine tract with non-pyridimines (Figure 6E).
PP2A has been reported to regulate phosphorylation of a variety of signaling proteins, including AKT, MEK1/2, and MYC, the downstream consequence of which could be NF-κB signaling. We therefore probed known PP2A substrates in the context of breast epithelial cells by Western blot and found increased phosphorylation of AKT at residue T308 in breast cells expressing mutant SF3B1 (Figure 6D and Supplemental Figure 10A). Knockdown of PPP2R5A in MCF10A and MCF7 cells by shRNA also upregulated phosphorylation of p65 and AKT (at T308) (Figure 6F and Supplemental Figure 10B). Furthermore, depletion of PPP2R5A in cells also induced expression of inflammatory response genes, including IL-8, CDKN1A, IL-1A, and CXCL1 (Figure 6G). Restoration of PPP2R5A expression in MCF7 cells expressing mutant SF3B1 or isogenic SF3B1K700E/WT MCF10A cells showed consistently reduced phosphorylation of p65 and AKT (Supplemental Figure 10C). Given the known roles of AKT in regulating IKK and NF-κB activation, our data point to PPP2R5A downregulation as a second mechanism whereby mutant SF3B1 promotes NF-κB signaling in breast cancer.
SF3B1 mutations promote invasion and migration of breast cancer cells.
NF-κB signaling has been implicated in regulating cell migration of breast cancer cells (42–44). To test if SF3B1 mutation facilitates cell migration, we initially performed wound-healing assays. MCF7 cells expressing mutant SF3B1 traversed wounds significantly faster than the cells expressing WT SF3B1 or empty vector (Figure 7A) irrespective of stimulation by TNF-α. Comparable differences were also seen with MCF10A isogenic pairs (Figure 7B).
Figure 7. SF3B1 mutations promote migration and invasion of breast cancer cells.
(A) Representative photos (top) at 0, 24, and 48 hours following wound creation in cell culture of MCF7 cells expressing an empty vector (EV), SF3B1WT, or SF3B1K700E cDNA. Scale bars: 200 μM. Relative scratch width over time shown below (n = 3; mean value ± standard deviation shown below; P values by ANOVA). (B) Quantification and representative photos (left) of wound at 0, 8, and 24 hours following wound creation and exposure to 0, 2, and 10 ng/mL TNF-α in cell culture of MCF10A with or without knockin of SF3B1K700E. Right, mean levels with standard deviation shown (n = 3; P value by Student’s test). Scale bars: 500 μM. (C) Same as A but for parental MCF7 cells exposed to medium from cells in A. Scale bars: 200 μM. (D) Number of invaded MCF10A cells with or without knockin of SF3B1K700E migrating toward EGF in a Transwell invasion assay (left). Mean ± standard deviation (n = 3, P value by Student’s t test) shown. Right, photos of hematoxylin staining of the migrated cells (photos taken using a microscope with a 10× objective (total magnification ×100); scale bars: 200 μM). (E) Growth of MCF7 tumors expressing either WT or mutant SF3B1 in NSG mice. Size of individual tumor (n = 10 in each group) and the mean are shown (P value by ANOVA). See also Supplemental Figures 11 and 12.
Since SF3B1 mutant cells secrete more inflammatory cytokines (including IL-8), we hypothesized that the higher migration might be mediated by cytokines within the culture media. To address this possibility, we harvested media from cells expressing WT or mutant SF3B1 and studied the effect of media addition upon migration of parental MCF7 cells in wound-healing assays. WT MCF7 cells exposed to media from mutant SF3B1-expressing cells healed wounds faster than controls (Figure 7C). In addition to cellular motility, NF-κB signaling has been linked to breast cancer invasiveness and we thus investigated the effects of SF3B1 mutation in Transwell Matrigel invasion assays. MCF10A breast epithelial cells hardly invaded Matrigel, whereas significantly more SF3B1 K700E knockin MCF10A cells migrated through the Matrigel layer (Figure 7D). Moreover, we assessed for enhanced invasiveness using a xenograft assay (Figure 7E), in which limiting quantities of WT and mutant SF3B1 expressing MCF7 cells were implanted into NSG mice, and observed that the SF3B1 mutant tumors grew to significantly larger sizes despite equivalent growth rates for the corresponding cell line models in 2D culture conditions.
As missplicing of MAP3K7 and PPP2R5A induced by mutant SF3B1 mediates dysregulation of NF-κB, we further tested the role of these proteins on cell migration. Knockdown of either MAP3K7 or PPP2R5A enhanced cell migration (Supplemental Figure 11, A and B and Supplemental Figure 12, A and B), and restoring these proteins in SF3B1 mutant cells restrain cell migration as assessed in wound-healing assays (Supplemental Figure 11, C and D, and Supplemental Figure 12, C and D). These data indicate that mutant SF3B1-mediated dysregulation of NF-κB signaling enhances breast cancer migration and invasion.
SF3B1 K700E mutations sensitize breast cancer cells to AKT inhibitors.
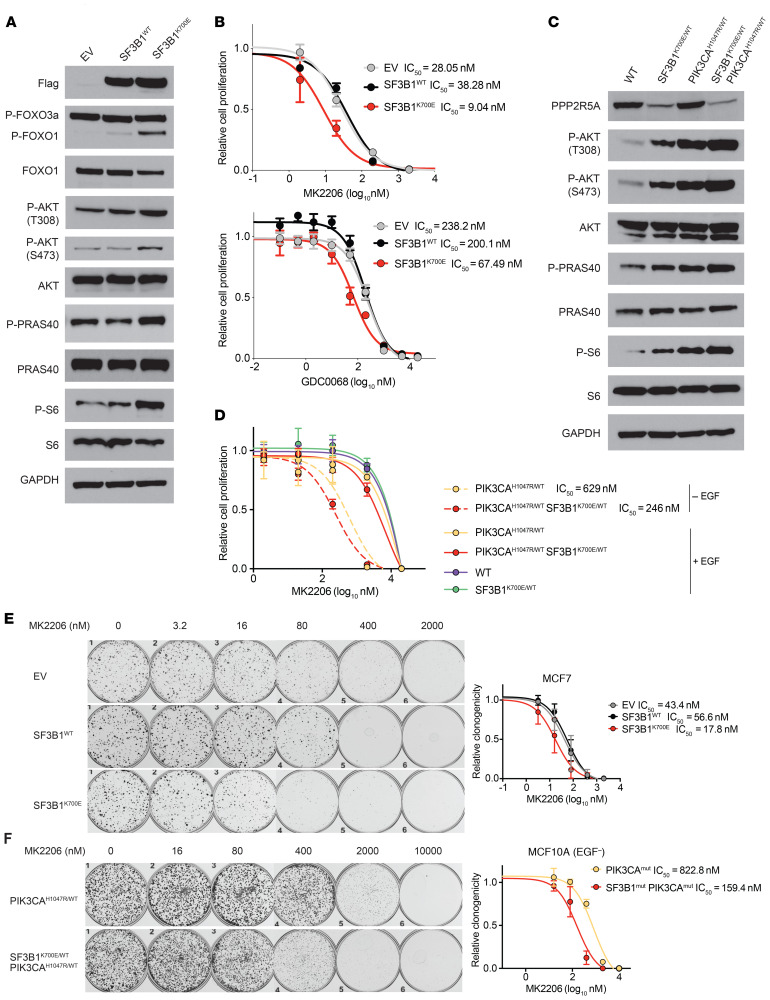
While the independent splicing events identified above converge upon promoting NF-κB signaling and associated phenotypes, therapeutic approaches targeting NF-κB have proven challenging. By contrast, PI3K/AKT inhibition has shown some efficacy in breast cancer and so we sought to determine whether suppression of PPP2R5A might increase tumor dependence on AKT for growth and viability. Given the induction of AKT T308 phosphorylation in SF3B1 K700E knockin cells, we evaluated the effects of SF3B1 mutation upon a set of established downstream substrates of activated AKT in breast cancer cells. In isogenic MCF7 breast cancer cells, expression of SF3B1 K700E increased phosphorylation of FOXO1, PRAS40, and ribosomal protein S6 (Figure 8A and Supplemental Figure 13A), further confirming AKT activation. Recent studies have suggested that increased signaling output through PI3K/AKT can enforce heightened dependence and drug sensitivity. We therefore evaluated the effect of expression of mutant SF3B1K700E on selective inhibitors of AKT. We examined the effect of both an allosteric pan-AKT inhibitor, MK2206, as well as an ATP-competitive pan-AKT inhibitor, GDC0068, in MCF7 cells, which are PIK3CA mutant and sensitive to AKT inhibitors, and observed a 2.5 times to 4 times further reduction in the IC50 as a result of SF3B1 mutation (Figure 8B). Finally, to examine the sensitivity of MCF10A cells to AKT inhibitors, we created MCF10A cells with heterozygous knockin of PIK3CAH1047R in the endogenous locus of PIKC3A with or without combined SF3B1K700E/WT knockin (Supplemental Figure 13B). These cells exhibited comparable splicing alterations of MAP3K7 and PPP2R5A, as well as activation of NF-κB signaling (Supplemental Figure 14A). Mutant PIK3CAH1047R induced expression of inflammatory response genes and promoted cell migration, which was further enhanced in the cells with both SF3B1K700E and PIK3CAH1047R mutations (Supplemental Figure 14). Knockin of PIK3CAH1047R relieved the dependency of MCF10A cells on epidermal growth factor (EGF) supplemented to culture media and activated the PI3K-AKT signaling pathway (Figure 8C and Supplemental Figure 13C). Again, in these cells, expression of mutant SF3B1 enhanced the sensitivity of PIK3CA mutant cells to inhibition of AKT (Figure 8D). This enhanced sensitivity of PIK3CA mutant cells induced by SF3B1 mutation was also confirmed by clonogenic survival assays (Figure 8, E and F), in which 2.5 times to 5 times reduction in IC50 was associated with expression of the SF3B1 mutation. Together, these data suggest that SF3B1 mutant tumors are a unique subset of breast cancers wherein coordinate mutational activation of signaling together with suppression of key negative regulator PPP2R5A leads to enhanced sensitivity of AKT inhibitors, which are under current investigation in the clinic.
Figure 8. SF3B1K700E mutation sensitizes breast cancer cells to AKT inhibitors.
(A) Western blot of MCF7 cells expressing an empty vector (EV), SF3B1 WT, or SF3B1 K700E cDNA. (B) IC50 of MCF7 cells shown in A following 5 days of exposure to MK2206 (left) or GDC0068 (right). Data are represented as mean ± SD; n = 6. (C) Western blot of MCF10A WT cells as well as SF3B1K700E/WT knockin cells, PIK3CAH1047R/WT knockin cells, and SF3B1K700E/WT PIK3CAH1047R/WT double knockin cells cultured in the absence of EGF. (D) IC50 to MK2206 of MCF10A cells shown in C in the presence or absence of EGF. Data are represented as mean ± SD; n = 6. (E) Clonogenic survival assay of MCF7 cells shown in A following 21 days of exposure to MK2206. IC50 curve is shown at right. (F) Clonogenic survival assay of MCF710A cells shown in C following 14 days of exposure to MK2206 in absence of EGF. IC50 curve is shown at right. See also Supplemental Figures 13 and 14.
Discussion
Identifying and targeting oncogenic pathways stimulated by gain-of-function genomic alterations has resulted in major clinical advances in outcomes for patients with breast cancer (45–49). Recent comprehensive genomic analyses of thousands of patients with breast cancer have identified a number of subgroups of patients with recurrent genomic alterations ranging from 1%–10% and for which deeper understanding of their contribution to tumor growth is lacking (22, 50, 51). As a result, functional investigation of the genetic alterations enriched in these specific subsets of patients with breast cancer has become paramount. Successful examples include the use of PD1 blockade in mismatch repair-deficient tumors (52), PARP inhibitors in germline BRCA1/2-mutant breast cancer (53), or anti-androgen therapy in androgen receptor–positive triple-negative breast cancer (54, 55). In this work, we identified what we believe is a novel class of recurrent genetic alterations that appear to promote breast tumorigenesis through modifying the function of the core RNA splicing factor SF3B1. Our data revealed SF3B1 mutant breast cancers to be a unique transcriptomic subtype of ER+ breast cancers with strong coenrichment with PI3K/AKT alterations, augmenting AKT and NF-κB signaling.
Given prior work identifying how SF3B1 mutations alter splice site sequence preference with attendant effects of select genes (11, 29, 37), we sought to profile aberrant splicing across breast cancers and compare these results with other cancers where SF3B1 mutations are found. Although the results demonstrated aberrant 3′ss usage shared across SF3B1 mutant tumors, only a handful of events were shared between different breast cancer samples, isogenic ER+ and ER– breast cancer cell lines, and our mouse breast cancer models. Specifically, our consensus overlap identified only 2 transcripts shared between cell lines and tumors with convergent alterations in 3′ss usage and mRNA downregulation — PPP2R5A and MAP3K7. Importantly, these 2 events are also observed in other cancer types, such as MDS and CLL, which identifies them as potentially critical mediators of the effects of mutant SF3B1 across diverse cancer lineages. For both of these mRNAs, mutant SF3B1 promoted usage of an aberrant branchpoint adenosine nucleotide that was required for missplicing, and the presence of a polypyrimidine tract downstream of the aberrant branchpoint was also necessary for mutant SF3B1 missplicing.
Our data demonstrated that SF3B1 mutation can downregulate expression of both PPP2R5A and MAP3K7 simultaneously and thereby promote AKT and NF-κB signaling output with resulting increases in breast cancer migration/invasion. Akin to SF3B1 mutation in hematopoietic stem cells (13, 29) as well as B cells (33, 34), SF3B1 mutation alone does not appear sufficient to mediate tumorigenesis in the mammary gland. However, when mutations in SF3B1 and PIK3CA were paired, major acceleration in tumor phenotypes was observed, with the double mutants significantly accelerating tumor formation and lethality. Coinciding with this observation from the mice was the finding that patients with mutations in both SF3B1 and PIK3CA had an inferior prognosis compared with WT patients.
We speculate that addition of the SF3B1 mutation to a PIK3CA mutant model can serve to reinforce signals initially elicited by PI3K activation. It is well established that PI3K activation promotes feedback suppression of the pathway through a variety of mechanisms such as suppression of RTK signaling (56) as well as inhibition of parallel pathways such as the NF-κB pathway. Given the widespread mechanisms of feedback suppression of signaling as a result of oncogenic PI3K signaling, there may be stronger selection for genomic alterations that coordinately remodel multiple signaling pathways akin to changes in the epigenome or transcription factors.
Finally, our functional interrogation of the misspliced events in SF3B1 mutant breast cancers nominated specific pathways for therapy of these tumors that might not emerge as obvious from the genotypes alone. We noted that the hyperactivation of AKT signaling that was induced as a result of combined PIK3CA and SF3B1 mutation might engender further therapeutic significance. Recent work has demonstrated that multiple mutations in PIK3CA can lead to increases in AKT output that ultimately increases the sensitivity of these tumors to PI3K/AKT inhibition (57). The alteration in PPP2R5A results in changes that serve to further activate AKT signaling and, in like manner, appeared to increase the sensitivity of these tumors to AKT kinase inhibitors. AKT inhibitors are currently under study in ER+ breast cancers harboring AKT E17K mutations, and our data suggest that SF3B1 mutation could provide an additional patient subset that may be sensitized to these therapies.
Methods
Human subjects.
A total of 1918 breast tumor specimens from 1756 patients with breast cancer who underwent prospective clinical genomic profiling were included for study. The demographic and clinical characteristics of the cohort are presented in Supplemental Table 2.
Mutation oncoprint analysis.
The mutual mutational pattern of SF3B1, PIK3CA, PTEN, AKT1, TP53, MAP3K1, ESR1, NF1, PIK3R1, BRCA1, ERBB3, and MYC was investigated on 3 breast cancer cohorts: METABRIC, MSK-IMPACT, and TCGA (Figure 1A and Supplemental Figure 1). The analysis was performed using the online tool from the cBioportal website (https://www.cbioportal.org/oncoprinter). The raw input files are provided in Supplemental Table 1.
Mutation and allele frequency analysis.
We compared the allele frequency of SF3B1 and PIK3CA mutations in 23 double-mutated samples from the MSK-IMPACT breast cancer cohort. Variant allele frequency (VAF) was corrected by tumor purity estimated by immunohistochemistry. Wilcoxon rank sum test was applied to give statistics between the VAF of 2 mutations. We analyzed the frequency of somatic mutations in patients from the MSK-IMPACT cohort (n = 41) harboring SF3B1 hotspot mutations. Mutation frequency was calculated for each reported gene in 25 primary samples and 20 metastasis samples.
Animals.
All mice were housed at Memorial Sloan Kettering Cancer Center (MSKCC). Generation and genotyping of the Sf3b1K700E/WT and R26-LSL-Pik3caH1047R as well as the MMTV-cre transgenic mice have been previously described (13, 32). The mice were monitored closely for signs of disease or morbidity daily and were killed when they showed a volume of the visible tumor formation above 1 cm3, failed to thrive, experienced weight loss greater than 10% total body weight, or showed open skin lesions, bleeding, or any signs of infection.
Whole mount staining of mouse mammary gland.
The fourth pair of mammary glands were excised from 4-month-old female mice and stained as previously described (58). Briefly, mammary glands were placed flattened on glass slides and fixed with Carnoy’s fixative (ethanol/chloroform/acetic acid, 6:3:1) at room temperature for 4 hours. Then the glands were gradually rehydrated with 70%, 35%, 17.5% ethanol, and distilled water. The mammary glands were stained in carmine alum solution (0.2% carmine natural red [MilliporeSigma], 0.5% aluminum potassium sulfate [MilliporeSigma] in water) overnight at room temperature, and then dehydrated progressively in 70%, 95%, 100% ethanol. The mammary fat pads were then cleared in xylene overnight. The slides were mounted with Permount (Thermo Fisher Scientific) and scanned by Pannoramic Scanner (3DHISTECH). The mammary epithelial area (MEA) and the number of branching points (center) were analyzed using FIJI software as previously described (59). The number of terminal buds were counted manually in 10 random 2 mm × 2 mm squares on each slide with QuPath software (60) and presented as number of terminal bud per square millimeter.
Immunohistochemistry.
Mouse tissue was collected and fixed in 10% neutral buffered formalin for 48 hours. Samples were then processed, embedded in paraffin, sectioned at 4 μm thickness, and stained with hematoxylin and eosin by the Laboratory of Comparative Pathology at MSKCC. Immunohistochemical staining was performed on a Ventana Stains System with primary antibodies (Supplemental Table 7) and OmniMap DAB anti-Rb Detection Kit or OmniMap DAB anti-Ms Detection Kit (Ventana). The slides were scanned by Pannoramic Scanner (3DHISTECH) and positively stained cell number was counted with QuPath software (60).
Cell lines.
Cell lines were maintained in 37°C incubator with 5% CO2 in a humidified atmosphere. MCF7 and MCF10A cells were obtained from ATCC. HEK293T cells were cultured in DMEM with 10% FBS (Gibco). MCF7 cells were grown in DMEM/F12 supplemented with 10% FBS (Gibco), 100 μg/mL penicillin, 100 mg/mL streptomycin (Corning), and 4 mM glutamine, and MCF10A cells were grown in DMEM/F12 supplemented with 5% horse serum (Gibco), 20 ng/mL EGF (MilliporeSigma), 10 μg/mL insulin (MilliporeSigma), 0.5 μg/mL hydrocortisone (MilliporeSigma), 0.1 μg/mL cholera toxin (MilliporeSigma), 100 μg/mL penicillin, and 100 mg/mL streptomycin (Corning). T47D cells were cultured in RPMI1640 with 10% FBS (Gibco), 100 μg/mL penicillin, and 100 mg/mL streptomycin (Corning). For cell proliferation assay, 1000 cells were seeded in each well of 96-well plates, and proliferation was measured with resazurin reagent (Thermo Fisher Scientific).
MCF10A SF3B1K700E/WT knockin cell lines were generated by Horizon Discovery. MCF10A PIK3CAH1047R single and PIK3CAH1047R SF3B1K700E/WT double knockin cells were generated by CRISPR and homology-directed repair (HDR) as previously described (11). Briefly, guide RNA sequence was cloned into px458-GFP vector (Addgene 48138) and nucleofected into MCF10A WT or SF3B1K700E/WT cells together with a single-stranded donor oligonucleotide (ssODN) as a template for HDR using the Lonza Nucleofector V kit and program T-024 on the nucleofector device. The ssODN sequence was CGAAAGACCCTAGCCTTAGATAAAACTGAGCAAGAGGCTTTGGAGTATTTCATGAAACAAATGAACGACGCGCGTCACGGTGGCTGGACAACAAAAATGGATTGGATCTTCCACACAATTAAACAGCATGCATTGAACTGA. Additional silent mutations were introduced with H1047R to avoid Cas9 recognition on the knockin allele. GFP-positive nucleofected cells were single-cell sorted 48 hours after nucleofection. Clones were screened for knockin of PIK3CAH1047R by PCR with primers GTTTCAGGAGATGTGTTACAAG and GTGTGGAATCCAGAGTGAGC and subsequent Sanger sequencing.
Plasmids.
The SF3B1-expressing lentiviral vector was generated by cloning cDNA of WT SF3B1 or SF3B1 K700E with flag tagged at N-terminal end into pInducer20 (Addgene 44012). Lentivirus was prepared in HEK293T cells with pInducer20 empty vector or the plasmid with SF3B1 (WT or mutant) cDNA, and then used to transduce breast cell lines. Transduced cells were selected in medium supplemented with 1 mg/mL Geneticin (Invivogen) for 2 weeks before being used in the experiments.
shRNAs against PPP2R5A or renilla luciferase were cloned into MLS-E-GFP vector. The shRNA sequences were PPP2R5A-1: TTAGTTGAAACATACTCAACCA; PPP2R5A-2: TTTAATTATATTATACTGATGA; Renilla: CAGGAATTATAATGCTTATCTA. Retrovirus was prepared in HEK293T cells and then used to transduce MCF7 or MCF10A cells. GFP+ cells stably expressing shRNAs were selected by FACS 3 days after transduction.
shRNAs against MAP3K7 or renilla luciferase were cloned into LT3GEPIR vector. The shRNA sequences were MAP3K7-1: TTAGGTAAATTTTTTATCAGTG and MAP3K7-2: TTTTCAACAATTTTGATTCTAA. Lentivirus was prepared in HEK293T cells and then used to transduce MCF7 and MCF10A cells. Transduced cells were selected in medium supplemented with 1 μg/mL puromycin (Thermo Fisher Scientific) for 1 week before being used in the experiments.
The MAP3K7 and PPP2R5A expression vector was generated by cloning MAP3K7 or PPP2R5A cDNA into pMSCV-GFP retroviral vector to express MAP3K7 or PPP2R5A protein. Retrovirus was prepared in HEK293T cells and then used to transduce MCF7 or MCF10A cells. GFP+ cells stably expressing MAP3K7 or PPP2R5A were selected by FACS 3 days after transduction.
Antibodies.
The antibodies used are listed in Supplemental Table 7.
Western blotting.
Protein samples were extracted from cultured cells with RIPA buffer (Thermo Fisher Scientific) and quantified by BCA assay. Protein fractionated on NuPAGE 4%–12% Bis-Tris gels (Life Technologies) was transferred onto nitrocellulose membranes. All primary antibodies (Supplemental Table 7) were used with 1:1000 dilution, except for GAPDH antibody, which was used at 1:5000 dilution in TBST containing 5% BSA and 0.02% sodium azide. Peroxidase-conjugated goat anti-rabbit IgG secondary antibody (MilliporeSigma) was used at 1:10000 dilution in TBST containing 2.5% skim milk. The blot was then visualized by chemiluminescence. Developed films were scanned and quantitated with FIJI software.
RT-PCR and quantitative RT-PCR.
Total RNA was extracted with the QIAGEN RNeasy Mini Kit, and cDNA was generated with qScript cDNA SuperMix (Quantabio) according to the manufacturers’ instructions. For RT-PCR, the amplification was performed with DreamTaq Hot Start Green PCT Master Mix (Thermo Fisher Scientific) and specific primers. PCR products were fractionated on 10% TBE gel (Life Technologies) and visualized with ethidium bromide. For quantitative RT-PCR, 0.5 μL cDNA was added to 10 μ PCR reactions prepared with TaqMan Universal PCR Master Mix (Thermo Fisher Scientific) and gene-specific TaqMan probes. All quantitative RT-PCR analysis was performed on an Applied Biosystems ViiA 7 Real-Time PCR System. Relative gene expression levels were calculated using the comparative CT method. Primers and TaqMan probes used in this study are listed in Supplemental Table 8.
Cell migration assay.
About 1 × 105 cells were seeded into each well of 12-well plates and cultured until 100% confluence. The “scratch” was created with a p200 pipet tip on the cell monolayer through the center of the well. The debris were removed by washing the well with 1 mL culture medium and then 1 mL medium (with 2% serum) specific for the cell migration assay was added into each well. The plate was incubated at 37°C, and images of the scratch were captured at various time points with a Zeiss Axiovert Microscope. The width of scratch was measured with FIJI software.
Cell invasion assay.
MCF10A cells were cultured to about 70% confluence and serum starved overnight. Harvested by trypsinization, 5 × 104 cells resuspended in serum-free medium (without EGF) were seeded to the Matrigel invasion chamber (8 μm pore size, Corning) in a 24-well plate. Complete culture medium for MCF10A was added to the bottom chambers of each well. After 24-hour incubation, the cells at the bottom side of the chamber were fixed and stained with hematoxylin. The membrane was then cut off from the chamber and the number of migrating cells were quantified by counting cells under microscope.
Cytokine multiplex assay.
Forty-one cytokines and chemokines were analyzed simultaneously with bead-based multiplex assay using Milliplex Map Human Cytokine/Chemokine Magnetic Bead Panel (HCYTMAG-60K-PX41, Millipore) on the Luminex FlexMap 3D instrument according to the manufacturer’s instruction. Briefly, cells were seeded in a T25 flask and media were refreshed when the cells approached 70% confluence. Cell culture supernatant was harvested 48 hours after media change and multiplex cytokine assay was performed on the Luminex FlexMap 3D instrument. The results were normalized to cell numbers in each flask.
IL-8 ELISA.
IL-8 concentration in the culture supernatant was quantified by human IL-8 ELISA kit (RayBiotech) according to the manufacturer’s instruction. Briefly, cells were seeded in a T25 flask and culture medium was changed when the cells approached 70% confluence. Cell culture supernatant was harvested 48 hours after media change and IL-8 ELISA was performed on SpectraMax M5 microplate reader (Molecular Devices). The results were normalized to cell numbers in each flask.
Cell proliferation.
Cell proliferation was measured as previously described (61). Briefly, cells (1000/well) were seeded into 96-well plates and treated with different concentrations of indicated drugs. Media and drug were changed every week. On days 0, 3, 5, and 7, 25 μL Resazurin (R&D Systems) was added to each well and incubated for 4 hours. Fluorescence in the plate was measured using a SpectraMax M5 microplate reader (Molecular Devices). Cell growth data from day 5 were then plotted as percentage inhibition against the log concentration of indicated drugs. IC50 was determined using a sigmoidal regression model using GraphPad Prism 7.0.
Minigene assay.
The PPP2R5A minigene construct was generated by inserting the DNA fragment containing the PPP2R5A genomic sequence from exon 4 to exon 5 in between the KpnI and XhoI restriction sites of pcDNA3.1 vector. The MAP3K7 minigene construct was generated by inserting the DNA fragment containing the MAP3K7 genomic sequence from exon 4 to exon 5 at HindIII restriction site of pcDNA3.1 vector. The sequences of the inserted fragments were verified by Sanger sequencing. Mutagenesis of minigene constructs was performed with the Agilent QuikChange II site-directed mutagenesis kit with specific primers according to the manufacturer’s directions. For transient transfection experiments, cells were seeded into a 24-well plate with culture medium containing doxycycline 48 hours before transfection of minigene constructs in the presence of X-tremeGENE HP DNA transfection reagent (Roche) according to the manufacturer’s directions. Forty-eight hours after transfection, cells were collected and RNA was extracted using the QIAGEN RNeasy Mini Kit. Minigene-derived and endogenous MAP3K7 and PPP2R5A transcripts were analyzed by RT-PCR using specific primers (Supplemental Table 8).
Clonogenic survival assay.
Clonogenic survival assay was performed on a 6-well plate. Five hundred cells in single cell suspension were seeded into each well of a 6-well plate and treated with various doses of MK2206 on the next day, following which the plate was incubated at 37°C with 5% CO2 in a humidified atmosphere for 14 days (for MCF10A cells) or 21 days (for MCF7 cells). After incubation, the colonies were fixed with formalin and stained with 0.01% crystal violet. The plate was then photographed and analyzed with the Oxford GelCount colony counter.
Xenograft assay.
NOD-SCID gamma (NSG) female mice ages 6–8 weeks were obtained from Jackson Laboratory and maintained in pressurized ventilated caging. MCF7 xenograft tumors expressing WT or mutant SF3B1 were established in NSG mice by subcutaneously implanting 0.72 mg sustained release 17β-estradiol pellets with a 10 g trocar into one flank followed by injecting 1 × 106 cells suspended 1:1 (vol/vol) with reconstituted basement membrane (Matrigel, Collaborative Research) on the opposite side 3 days later. On the same day of cell inoculation, the mice were fed with filter water containing 2 mg/mL doxycycline and 2% sucrose, which was refreshed twice a week. Tumor dimensions were measured with vernier calipers starting from 14 days after cell implantation and tumor volumes were calculated as (π/6 × larger diameter × (smaller diameter)2). The experiment was closed 7 weeks after implantation. Based on our previous work measuring the variability in size and growth of MCF7 xenografts, we estimated 10 mice per group would allow us to detect tumor size differences of greater than 200 mm3.
RNA-seq of breast cancer cell lines and mouse tissues.
Total RNA was extracted with the Direct-Zol RNA miniprep kit (Zymo). RNA-seq libraries were prepared and sequenced by Genewiz with 150-bp pair-end reads with 100 million read depth.
Heatmap and hierarchical clustering.
To explore potential tumor/hotspot specificity or similarity of 3′ss alteration, principal component analysis and unsupervised hierarchical clustering were performed using 618 significantly (q value < 0.001) differential splicing junctions between all SF3B1 mutant and all WT samples (see Supplemental Methods for novel cryptic 3′ss identification). Command prcomp from R package stats was used for the principal component analysis. Command heatmap.2 from R package gplots was adopted for the clustering analysis using (1-Pearson correlation coefficient)/2 as distance. This analysis was performed on TCGA BRCA, MSK breast cancer cell lines (MCF7, T47D and MCF10A), ICGC CLL, dbGaP CLL, and TCGA UVM samples.
Sequence motif analysis.
We performed motif analysis at aberrant 3′ss as well as the associated canonical 3′ss. We first identified sets of cryptic 3′ss significantly differentially used between SF3B1 mutant and WT samples by combining TCGA breast cancer samples and MSK cell line data. Next, motif logo was plotted based on the intron nucleotide composition up to 50 nt upstream and 20 nt downstream from the AG dinucleotide of cryptic 3′ss as well as the associated canonical 3′ AG site, using R package motifStack. Fisher’s exact test was performed to compare the enrichment of pyrimidines in the 10 nt downstream from the AG of cryptic 3′ss to the associated canonical 3′ AG site.
GSEA analysis.
We used GSEA software (62) with defined hallmark genesets (MSigDB, v6.1) to investigate molecular profiles enriched in SF3B1 mutant breast cancer samples. Parameters used were as follows: collapse data: false; permutation type: gene set, 1000 permutations; chip platform: gene symbol. The input gene expression matrix contains 10,165 genes, which are significantly differentially expressed (DE) between SF3B1 mutations and WT on cell line data (combining the DE lists from MCF7, T47D, and MCF10A).
Expression data set and differential expression.
To generate mRNA expression matrix for transcriptome analysis, we used featureCounts (63) from package Subread to call read counts from STAR realigned bam files. Genes with low read depths across the cohort were removed. Then, read counts were transformed into RPKM values followed by log2 transformation, and quantile normalized on the sample level. We also performed DESeq2 analysis (64) (using the raw read counts) to call DE genes between SF3B1 mutant and WT TCGA breast cancer samples and MSK cell line data.
Data availability.
The RNA sequencing data were deposited in the NCBI Sequence Read Archive (SRA) under accession number GSE145471.
Statistics.
Statistical significance was determined by (a) unpaired or paired 2-sided Student’s t test after testing for normal distribution, (b) 1-way or 2-way ANOVA followed by Dunnett’s or Tukey’s multiple comparisons tests, or (c) Kruskal-Wallis tests with Dunn’s test corrected for multiple comparisons. Data were plotted using GraphPad Prism 7 software as individual points with mean values and error bars representing standard deviation.
Study approval.
This study was approved by the Memorial Sloan Kettering Cancer Center Institutional Review Board (IRB) and all patients provided written informed consent for tumor sequencing and review of patient medical records for detailed demographic, pathologic, and treatment information (NCT01775072). All mouse procedures and experiments were completed in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees at MSKCC (MSKCC#12-10-016).
Author contributions
BL, ZL, RR, OAW, and S. Chandarlapaty designed the study. BL, S. Chen, MK, CE, QC, and EDS performed the animal experiments. BHD and JSRF assisted with pathological analysis. BL, ZL, YS, and RR performed RNA-seq analyses. BL, OAW, and S. Chandarlapaty performed in vitro cell line experiments. BL, ZL, OAW, and S. Chandarlapaty wrote the manuscript.
Supplementary Material
Acknowledgments
OAW and S. Chandarlapaty are supported by the Department of Defense Breast Cancer Research Program (W81XWH-18-1-0383), the Functional Genomics Initiative of Memorial Sloan Kettering Cancer Center, and the NCI Cancer Center Support Grant (CCSG, P30 CA08748). S. Chandarlapaty acknowledges support by the Breast Cancer Research Foundation for this work.
Version 1. 10/08/2020
In-Press Preview
Version 2. 01/04/2021
Print issue publication
Footnotes
Conflict of interest: OAW has served as a consultant for H3B Biomedicine, Foundation Medicine Inc., Merck, and Janssen, and is on the scientific advisory board of Envisagenics Inc. and AIChemy. He has received prior research funding from H3B Biomedicine and LOXO Oncology. JSRF has received personal/consultancy fees from Goldman Sachs, Paige AI, and REPARE Therapeutics. He is a member of the scientific advisory boards of VolitionRx, REPARE Therapeutics, and Paige AI; the Board of Directors of Grupo Oncoclinicas; and he is an ad hoc member of the scientific advisory boards of Roche Tissue Diagnostics, Ventana Medical Systems, Novartis, Genentech, and InVicro. RR is founder of GenoTwin and is a member of the scientific advisory board of AimedBio. SC has served as a consultant for Eli Lilly, Novartis, Context Therapeutics, Sermonix, Paige AI, and Bristol Myers Squibb. He has received prior research funding from Daiichi-Sankyo.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(1):e138315.https://doi.org/10.1172/JCI138315.
Contributor Information
Bo Liu, Email: liub@mskcc.org.
Zhaoqi Liu, Email: liuzq@big.ac.cn.
Sisi Chen, Email: chens7@mskcc.org.
Michelle Ki, Email: mck96@cornell.edu.
Caroline Erickson, Email: ericksc1@mskcc.org.
Jorge S. Reis-Filho, Email: reisfilj@mskcc.org.
Benjamin H. Durham, Email: durhamb@mskcc.org.
Qing Chang, Email: changq@mskcc.org.
Elisa de Stanchina, Email: destance@mskcc.org.
Yiwei Sun, Email: ys3235@cumc.columbia.edu.
Raul Rabadan, Email: rr2579@cumc.columbia.edu.
Omar Abdel-Wahab, Email: abdelwao@mskcc.org.
Sarat Chandarlapaty, Email: ChandarS@MSKCC.ORG.
References
- 1.Seiler M, et al. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 2018;23(1):282–296.e4. doi: 10.1016/j.celrep.2018.01.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 3.Wang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365(26):2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papaemmanuil E, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365(15):1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graubert TA, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2011;44(1):53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki H, et al. Recurrent noncoding U1 snRNA mutations drive cryptic splicing in SHH medulloblastoma. Nature. 2019;574(7780):707–711. doi: 10.1038/s41586-019-1650-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shuai S, et al. The U1 spliceosomal RNA is recurrently mutated in multiple cancers. Nature. 2019;574(7780):712–716. doi: 10.1038/s41586-019-1651-z. [DOI] [PubMed] [Google Scholar]
- 8.Darman RB, et al. Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep. 2015;13(5):1033–10345. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 9.Kim E, et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27(5):617–630. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ilagan J, et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Research. 2015;25(1):14–26. doi: 10.1101/gr.181016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue D, et al. Spliceosomal disruption of the non-canonical BAF complex in cancer. Nature. 2019;574(7778):432–436. doi: 10.1038/s41586-019-1646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshimi A, et al. Coordinated alterations in RNA splicing epigenetic regulation drive leukaemogenesis. Nature. 2019;574(7777):273–277. doi: 10.1038/s41586-019-1618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Obeng EA, et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell. 2016;30(3):404–417. doi: 10.1016/j.ccell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, et al. Transcriptomic characterization of SF3B1 mutation reveals its pleiotropic effects in chronic lymphocytic leukemia. Cancer Cell. 2016;30(5):750–763. doi: 10.1016/j.ccell.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeBoever C, et al. Transcriptome sequencing reveals potential mechanism of cryptic 3’ splice site selection in SF3B1-mutated cancers. PLoS Comput Biol. 2015;11(3):e1004105. doi: 10.1371/journal.pcbi.1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seiler M, et al. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nature Med. 2018;24(4):497–504. doi: 10.1038/nm.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalton WB, et al. Hotspot SF3B1 mutations induce metabolic reprogramming and vulnerability to serine deprivation. J Clin Invest. 2019;129(11):4708–4723. doi: 10.1172/JCI125022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drilon A, et al. Efficacy of arotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378(8):731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pratilas CA, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009;106(11):4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyman DM, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med. 2015;373(8):726–736. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diamond EL, et al. Efficacy of MEK inhibition in patients with histiocytic neoplasms. Nature. 2019;567(7749):521–524. doi: 10.1038/s41586-019-1012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Razavi P, et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34(3):427–438.e6. doi: 10.1016/j.ccell.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang MT, et al. Accelerating discovery of functional mutant alleles in cancer. Cancer Discov. 2018;8(2):174–183. doi: 10.1158/2159-8290.CD-17-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toy W, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet. 2013;45(12):1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertucci F, et al. Genomic characterization of metastatic breast cancers. Nature. 2019;569(7757):560–564. doi: 10.1038/s41586-019-1056-z. [DOI] [PubMed] [Google Scholar]
- 28.Bielski CM, et al. Widespread selection for oncogenic mutant allele imbalance in cancer. Cancer Cell. 2018;34(5):852–862.e4. doi: 10.1016/j.ccell.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SC, et al. Synthetic lethal and convergent biological effects of cancer-associated spliceosomal gene mutations. Cancer Cell. 2018;34(2):225–241.e8. doi: 10.1016/j.ccell.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Q, et al. A chemical genetics approach for the functional assessment of novel cancer genes. Cancer Res. 2015;75(10):1949–1958. doi: 10.1158/0008-5472.CAN-14-2930. [DOI] [PubMed] [Google Scholar]
- 31.Obeng EA, et al. Mutant splicing factor 3b subunit 1 (SF3B1) causes dysregulated erythropoiesis and a stem cell disadvantage. Blood. 2014;124(21):828 [Google Scholar]
- 32.Adams JR, et al. Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res. 2011;71(7):2706–2717. doi: 10.1158/0008-5472.CAN-10-0738. [DOI] [PubMed] [Google Scholar]
- 33.Yin S, et al. A murine model of chronic lymphocytic leukemia based on B cell-restricted expression of sf3b1 mutation and atm deletion. Cancer Cell. 2019;35(2):283–296.e5. doi: 10.1016/j.ccell.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, et al. Mutations in the RNA splicing factor SF3B1 promote tumorigenesis through MYC stabilization. Cancer Discov. 2020;10(6):806–821. doi: 10.1158/2159-8290.CD-19-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koren S, et al. PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature. 2015;525(7567):114–118. doi: 10.1038/nature14669. [DOI] [PubMed] [Google Scholar]
- 36.Wagner KU, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25(21):4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darman RB, et al. Cancer-associated SF3B1 hotspot mutations induce cryptic 3’ splice site selection through use of a different branch point. Cell Rep. 2015;13(5):1033–1034. doi: 10.1016/j.celrep.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 38.Sato S, et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol. 2005;6(11):1087–1095. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- 39.Singh A, et al. TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell. 2012;148(4):639–650. doi: 10.1016/j.cell.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ajibade AA, et al. TAK1 negatively regulates NF-κB and p38 MAP kinase activation in Gr-1+CD11b+ neutrophils. Immunity. 2012;36(1):43–54. doi: 10.1016/j.immuni.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu J, et al. Mechanism and in vitro pharmacology of TAK1 inhibition by (5Z)-7-oxozeaenol. ACS Chem Biol. 2013;8(3):643–650. doi: 10.1021/cb3005897. [DOI] [PubMed] [Google Scholar]
- 42.Helbig G, et al. NF-kappaB promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J Biol Chem. 2003;278(24):21631–21638. doi: 10.1074/jbc.M300609200. [DOI] [PubMed] [Google Scholar]
- 43.Sovak MA, et al. Aberrant nuclear factor-kappaB/Rel expression and the pathogenesis of breast cancer. J Clin Invest. 1997;100(12):2952–2960. doi: 10.1172/JCI119848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS. Selective activation of NF-kappa B subunits in human breast cancer: potential roles for NF-kappa B2/p52 and for Bcl-3. Oncogene. 2000;19(9):1123–1131. doi: 10.1038/sj.onc.1203412. [DOI] [PubMed] [Google Scholar]
- 45.Slamon DJ, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 46.Baselga J, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2012;366(2):109–119. doi: 10.1056/NEJMoa1113216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.André F, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019;380(20):1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 48.Baselga J, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366(6):520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finn RS, et al. Palbociclib and letrozole in advanced breast cancer. N Engl J Med. 2016;375(20):1925–1936. doi: 10.1056/NEJMoa1607303. [DOI] [PubMed] [Google Scholar]
- 50.Ciriello G, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163(2):506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang MT, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34(2):155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le DT, et al. PD-1 Blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robson M, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377(6):523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 54.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gucalp A, et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic Breast Cancer. Clin Cancer Res. 2013;19(19):5505–5512. doi: 10.1158/1078-0432.CCR-12-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chandarlapaty S. Negative feedback and adaptive resistance to the targeted therapy of cancer. Cancer Discov. 2012;2(4):311–319. doi: 10.1158/2159-8290.CD-12-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vasan N, et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors. Science. 2019;366(6466):714–723. doi: 10.1126/science.aaw9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plante I, Stewart MK, Laird DW. Evaluation of mammary gland development and function in mouse models. J Vis Exp. 2011;(53):2828. doi: 10.3791/2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stanko JP, Fenton SE. Quantifying branching density in rat mammary gland whole-mounts using the sholl analysis method. J Vis Exp. 2017;(125):55789. doi: 10.3791/55789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bankhead P, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Z, et al. Loss of the FAT1 tumor suppressor promotes resistance to CDK4/6 inhibitors via the Hippo pathway. Cancer Cell. 2018;34(6):893–905.e8. doi: 10.1016/j.ccell.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 64.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pereira B, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data were deposited in the NCBI Sequence Read Archive (SRA) under accession number GSE145471.