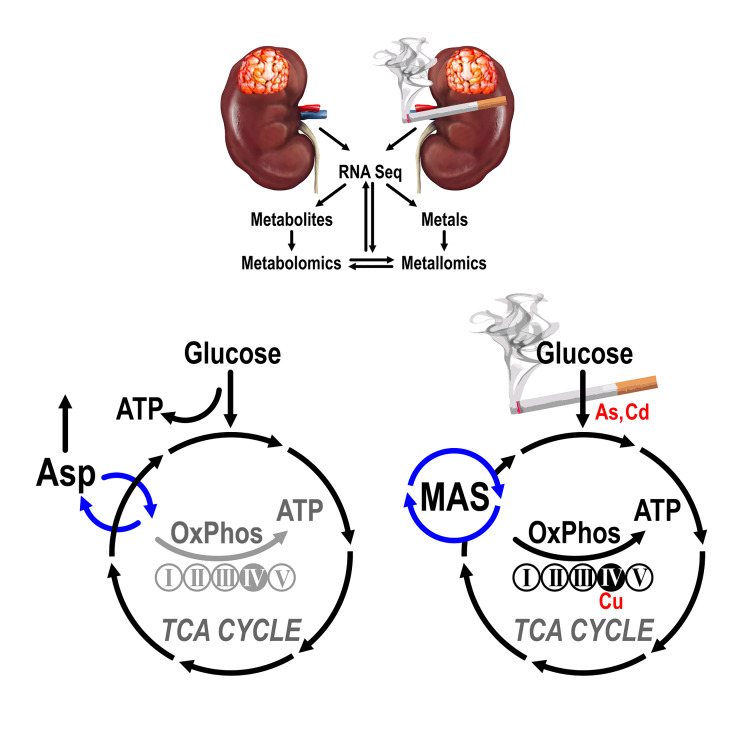
Abstract
BACKGROUND
Clear cell renal cell carcinoma (ccRCC) is the most common histologically defined renal cancer. However, it is not a uniform disease and includes several genetic subtypes with different prognoses. ccRCC is also characterized by distinctive metabolic reprogramming. Tobacco smoking (TS) is an established risk factor for ccRCC, with unknown effects on tumor pathobiology.
METHODS
We investigated the landscape of ccRCCs and paired normal kidney tissues using integrated transcriptomic, metabolomic, and metallomic approaches in a cohort of white males who were long-term current smokers (LTS) or were never smokers (NS).
RESULTS
All 3 Omics domains consistently identified a distinct metabolic subtype of ccRCCs in LTS, characterized by activation of oxidative phosphorylation (OXPHOS) coupled with reprogramming of the malate-aspartate shuttle and metabolism of aspartate, glutamate, glutamine, and histidine. Cadmium, copper, and inorganic arsenic accumulated in LTS tumors, showing redistribution among intracellular pools, including relocation of copper into the cytochrome c oxidase complex. A gene expression signature based on the LTS metabolic subtype provided prognostic stratification of The Cancer Genome Atlas ccRCC tumors that was independent of genomic alterations.
CONCLUSION
The work identified the TS-related metabolic subtype of ccRCC with vulnerabilities that can be exploited for precision medicine approaches targeting metabolic pathways. The results provided rationale for the development of metabolic biomarkers with diagnostic and prognostic applications using evaluation of OXPHOS status. The metallomic analysis revealed the role of disrupted metal homeostasis in ccRCC, highlighting the importance of studying effects of metals from e-cigarettes and environmental exposures.
FUNDING
Department of Defense, Veteran Administration, NIH, ACS, and University of Cincinnati Cancer Institute.
Keywords: Metabolism, Oncology
Keywords: Cancer
Introduction
Clear cell renal cell carcinoma (ccRCC) is characterized by loss of tumor suppressors localized to the short arm of chromosome 3, including the von Hippel Lindau gene (VHL), which serves in the E3 ubiquitin ligase complex targeting HIFαs for proteasomal degradation, and 3 genes with chromatin remodeling functions, PBRM1, BAP1, and SED2 (1–3). Recent studies identified several molecular subtypes of ccRCC exhibiting different patterns of mutations and clinical outcomes (3). A cardinal feature of ccRCC is metabolic reprogramming, characterized by induction of glycolysis, nucleotides, and lipid biosynthetic pathways (1, 4, 5), and downregulation of multiple other metabolic genes (6).
Tobacco smoking (TS) is a dose-dependent and reversible risk factor for the development of ccRCC, particularly in white males (7–9). TS contains at least 70 recognized carcinogens and cocarcinogens, including metals and metalloids (10). The molecular effects of TS in ccRCC differ from the carcinogenic effects of TS in lung or head and neck oncogenesis because the kidneys are exposed to different sets of carcinogens compared with tissues in direct contact with TS. This is supported by a global analysis of TS-induced mutation patterns in renal cancers showing enrichment for mutation signature 5 independent of TS, rather than signature 4 that characterizes tumors derived from epithelia directly exposed to TS, suggesting the absence of major, direct TS-induced genomic effects (11). Considering that TS carcinogens act in the context of kidney proximal tubule function, it is possible that toxic elements in TS such as cadmium and arsenic, which have their final repository or excretory route in the kidneys and are considered risk factors, contribute to ccRCC pathobiology (12–15). However, little is known about the relation of TS with the molecular subtypes and metabolic profiles of ccRCC. None of the existing databases, including The Cancer Genome Atlas (TCGA), has sufficient information about smoking status, creating a need for well-designed cohorts to study the role of TS in ccRCC.
Here we collected a well-matched cohort of white males who were long-term smokers (LTS) or who were never smokers (NS) at the time of diagnosis and performed integrated landscape analysis using transcriptomics, metabolomics, and metallomics of the primary tumors and paired normal kidney tissues (NKTs).
Results
LTS were defined as consuming at least 1 pack of cigarettes a day for 15 years at the time of surgery, whereas NS were described as smoking less than 100 cigarettes in their lifetime. The cohorts of NS and LTS white males did not differ significantly in terms of age, BMI, tumor grade distributions, or frequency of altered VHL (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI140522DS1). The cohorts also matched well in terms of age and ccRCC tumor grade with the TCGA cohort of 309 white males (TCGA-KIRC, Firehose Legacy). When analyzed together, the NS and LTS cohorts will be referred to as the Cincinnati Tobacco Smoking Cohort (CTSC).
RNA expression profiles show induction of OXPHOS genes in ccRCCs from LTS.
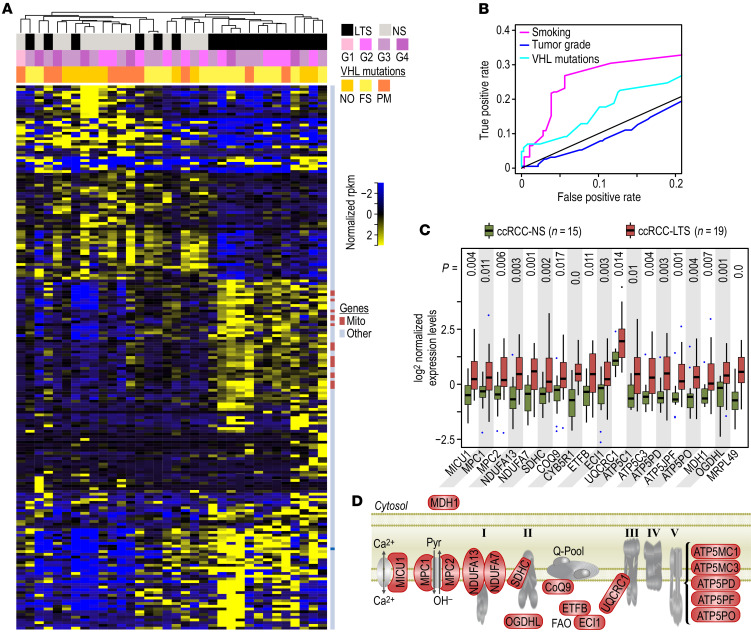
ccRCCs and NKTs from LTS show respectively a 63% induction (125 out of 199 genes) and 84% repression (234 out of 280 genes) of differentially expressed genes compared with NS, with little overlap (Supplemental Figure 1, A and B; Supplemental Table 2A; and Supplemental Table 3A). Unsupervised clustering using differentially expressed genes (FDR < 0.1) and a Pearson correlation–based distance measure stratified the majority of ccRCCs and NKTs by smoking status (Figure 1A and Supplemental Figure 1C). Importantly, in ccRCCs, gene expression patterns correlated with smoking status more clearly than with either tumor grade or VHL status, suggesting a dominant effect of TS exposure in the etiology or functional status of ccRCC. The accuracy for the classification of the samples into groups defined by smoking status was supported by the receiver operating characteristic (ROC) curves that indicated a good performance by a large AUC (Figure 1B). Importantly, 38 out of 125 (30%) of upregulated genes regulated metabolism, whereas 18 genes were associated with the mitochondrial respiratory chain, as shown by the results of gene set enrichment analysis (GSEA) (Supplemental Figure 1D and Supplemental Table 2, B and C). The significant induction of respiratory chain genes in the ccRCCs from LTS was confirmed by nonparametric analysis (Figure 1C and Supplemental Figure 1E). These included genes for mitochondrial calcium and pyruvate transporters; subunits of complex I, II, III, V; and malate dehydrogenase 1 (MDH1), a cytosolic enzyme necessary for the activity of the malate-aspartate shuttle (MAS) (Figure 1D and Supplemental Table 2B). ChIP enrichment analysis (ChEA) and ENCODE analysis of transcription factors regulating 18 mitochondrial genes upregulated by TS in ccRCC, performed using Enrichr, showed significant (P = 0.01974) enrichment for transcription factor YY1, a multifunctional transcription factor member of the polycomb group protein family regulating mitochondrial oxidative function (16).
Figure 1. Identification of genes differentially regulated by TS in ccRCCs.
(A) Heatmap of all differentially regulated genes shows stratification with TS (marked by black in the top bar). Pink bar represents tumor grades, and yellow bar represents mutations in VHL (WT: wild type, PM: point mutations, FS: frameshift mutations). Dark blue marks in the bar on the right of the heatmap indicate mitochondrial genes induced in the ccRCCs from LTS. (B) ROC curves for the classification of the samples into classes defined by TS (magenta), tumor grade (blue), or VHL mutation status (cyan). Random classifier is drawn as a diagonal gray line, and classification accuracy is represented by the AUC indicating that observed clustering provides the best classification for TS status. (C) Box-whisker plots for expression of mitochondrial genes in ccRCCs from NS and LTS. The boxes represent lower and upper quartiles separated by the median (thick horizontal line) and the whiskers extend to the minimum and maximum values, excluding points that are outside the 1.5 IQR from the box (marked as circles). P values from Mann-Whitney-Wilcoxon test are provided at the top. (D) Model showing localization of the 18 induced genes (marked in red) in the context of mitochondrial electron transport chain. ccRCCs from 15 NS and 19 LTS were analyzed.
Genes downregulated in ccRCCs from LTS (Supplemental Table 2) did not show enrichment for any pathways identified by GSEA or Enrichr. One downregulated gene was AKR1B10, an aldo-keto reductase that utilizes NADH/NADPH as cofactors, and which has been reported to be upregulated by TS in the airway epithelium (17). Interestingly, 36% of genes upregulated in tumors from LTS (45 out of 125) were favorable predictors, while 35% of genes downregulated in ccRCCs from LTS (26 out of 74) were unfavorable predictors in renal cancer according to the Human Protein Atlas (Supplemental Table 2A).
Genes upregulated in NKTs from LTS were identified by GSEA as genes associated with response to arsenic toxicity (Supplemental Figure 1, F–H, and Supplemental Table 3). Consistently, analysis of all genes upregulated in NKTs from LTS using GO Biological Process 2018 revealed pathways related to response to copper, cadmium, and zinc. Two induced genes, AKR1C3 and EPHX1, participated in metabolism of benzo(a)pyrene, a group 1 carcinogen present in TS.
Most genes were downregulated in NKTs from LTS (Supplemental Figure 1F). The GSEA revealed genes regulating extracellular matrix and genes controlled by polycomb repressor complex 2 (PRC2) (Supplemental Figure 1F and Supplemental Table 3). These data indicate that kidney tissue from LTS represented a different macroenvironment for tumor growth.
Metabolomic profiling of ccRCCs and NKTs from LTS and NS.
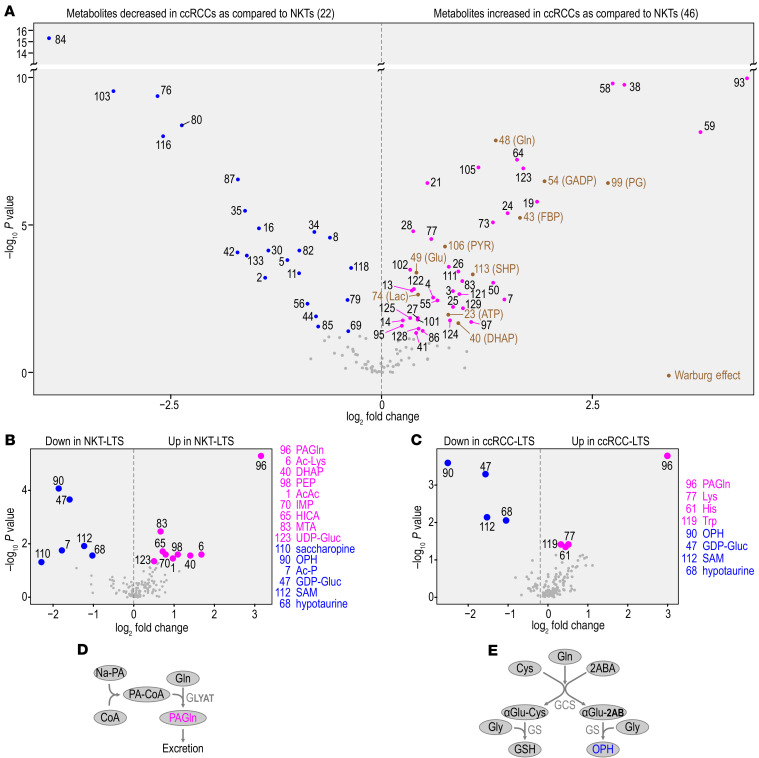
Considering the major effect of TS exposure in transcriptional classification indicating metabolic reprogramming of ccRCCs, we analyzed metabolic profiles according to TS exposure in ccRCCs and NKTs using high resolution LC-MS metabolomics. Samples were standardized by addition of equal volumes of a balanced mixture of heavy labeled metabolite extracts obtained from cells cultured in 13C heavy labeled media (18). Unsupervised clustering of 133 endogenous metabolites matched with 13C labeled exogenous metabolites (FDR < 0.05, Supplemental Table 4 and Supplemental Table 5A) revealed differential abundances of 68 metabolites between NKTs and tumors (Figure 2A and Supplemental Figure 2A), of which 46 showed higher abundance and 22 showed lower abundance in ccRCCs compared with NKTs (Figure 2A and Supplemental Table 5B). Metabolic pathway enrichment analysis (MetaboAnalyst) using all metabolites with increased abundance in ccRCCs revealed the Warburg effect at the top of the list (Figure 2A and Supplemental Figure 2B), consistent with the well-established role of this pathway in ccRCC; however, there was no difference in abundances of glycolytic intermediates in ccRCCs from NS and LTS. These data validate our metabolomic analysis in reference to previously reported data (1, 5).
Figure 2. Metabolic profiling of ccRCCs and NKTs from LTS and NS.
(A) Volcano plot shows metabolites with differential abundance (P < 0.05, fold change, Wilcoxon nonparametric test) between all NKTs (n = 36) and all ccRCCs (n = 37). Differentially abundant metabolites are labeled by numbers (see Supplemental Table 5B for the identification of the indicated metabolites). Intermediates of the Warburg effect labeled in brown. (B) Volcano plots show metabolites with differential abundance in NKTs from LTS compared with NS. (C) Volcano plots show metabolites with differential abundance in ccRCCs from LTS compared with NS. (D) Pathway for metabolism of phenylacetylglutamine (PAGln). (E) Pathway of OPH and GSH biosynthesis.
Analysis of the steady-state levels of metabolites from NKTs and tumors by TS exposure at FDR less than 0.05 showed 15 metabolites changed in kidney tissues (Figure 2B and Supplemental Table 5C), and 8 metabolites changed in tumors (Figure 2C and Supplemental Table 5C). The metabolite with the highest abundance in both tissues from LTS was phenylacetylglutamine (PAGln), a metabolite alternative to urea in nitrogen excretion. This likely represents an adaptive response to ammonia present in TS (19). The enzyme synthesizing PAGln, glycine-N-acyltransferase, is also involved in detoxification of xenobiotics, including those in TS. Synthesis of PAGln consumes large amounts of glutamine (Figure 2D). Interestingly, 3 amino acids, lysine, tryptophan, and histidine, showed higher abundance in ccRCCs from LTS. This may be related to the augmented gene expression for amino acid transporters determined in RNA-Seq (Supplemental Table 2B).
Four metabolites with lower abundance in both NKTs and ccRCCs from LTS are ophthalmic acid (OPH), S-adenosyl methionine (SAM), hypotaurine, and GDP-glucose. OPH is γ-glutamyl-L-2-aminobutyryl-glycine tripeptide, synthesized in parallel to glutathione (GSH) (ref. 20 and Figure 2E) and considered a biomarker of oxidative stress reversely correlated with GSH consumption (21). The decrease in the levels of SAM, a donor of methyl groups, implies major changes in the utilization of SAM in the cellular processes requiring methylation, such as DNA and histone methylation or arsenic methylation.
Alterations in metabolites’ intercorrelations indicate major TS-induced metabolic reprogramming.
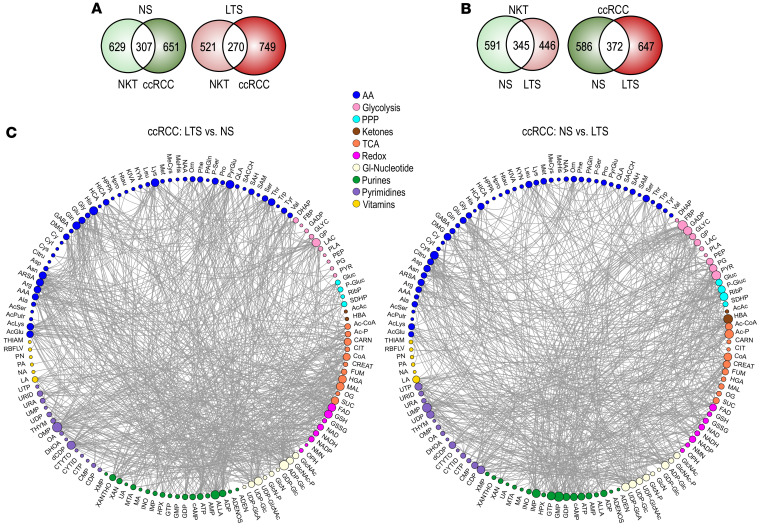
Analysis of metabolites’ steady-state levels provides only a limited view of the metabolic landscape. Isotope tracing can reveal metabolic flux in primary tumors from patients (22), but these approaches are limited by the adequate uptake of the labeled metabolites during presurgery infusion and their distribution within the tumor. However, analysis of the patterns of correlations among metabolites contributes insights into the metabolic state that can be more mechanistically interpreted (23, 24). We used Spearman’s correlation analysis to identify overall shifts in the distributions of squared pairwise correlations (variance explained) for all pairs of 133 metabolites in each tissue and to map strong and highly significant correlations, defined as those with Spearman’s correlation coefficient values greater than or equal to 0.5 (Supplemental Table 6). Analysis revealed predominantly positive correlations among the metabolites in all tissues. They were visualized using Circos, where edges show connections between individual metabolites and the size of the nodes is proportional to the number of connections for each metabolite (Figure 3).
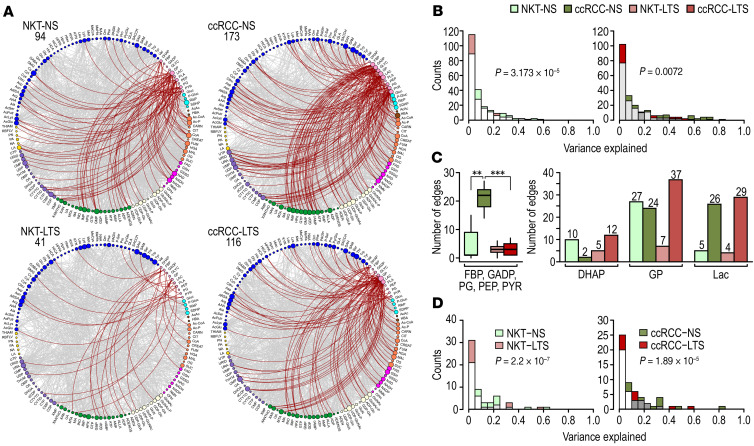
Figure 3. Correlational analysis of metabolic connections demonstrates TS-induced metabolic reprogramming.
(A) Venn diagrams show unique versus common edges for NKTs and ccRCCs in NS and LTS. We used 18 tissue pairs from NS and 19 NKTs and 18 ccRCCs from LTS. (B) Venn diagrams show unique versus common edges for NKTs and ccRCCs from NS versus LTS, respectively. (C) Circos visualization of unique correlations was used among all 133 metabolites in ccRCCs from NS and LTS. Colors of nodes represent categories of metabolites. (Acronyms explained in Supplemental Table 5D).
We identified a significant number of unique edges, i.e., pairs of metabolites uniquely coupled in NKTs and in ccRCCs, irrespective of smoking status (Figure 3A and Supplemental Figure 3A), consistent with tumor metabolic reprogramming. Importantly, we established that TS exposure induced a large number of unique edges in ccRCCs and NKTs (Figure 3, B and C, and Supplemental Figure 3B), an indication of substantial metabolic alterations. Thus, correlational analysis of the metabolites’ connections uncovered major and global TS-dependent metabolic reprogramming that was not revealed by analysis of the metabolites’ mean abundances.
TS shifts cellular metabolism away from glycolysis.
One of the most striking effects of TS was the decrease in the number of connections between glycolytic intermediates and all other metabolites in tissues from LTS compared with NS, although the number of correlations in ccRCCs from NS and LTS was higher than in the respective NKTs (Figure 4A). In particular, the significant concordance of glycolytic metabolites with purines found in ccRCCs and NKTs from NS was lost in LTS (Figure 4B and Supplemental Table 7, A and B).
Figure 4. Inhibition of the glycolytic pathway in ccRCCs from LTS.
(A) Circos visualization of all correlations for all glycolytic metabolites in the indicated tissues. Numbers indicate total number of correlations (red edges) for all glycolytic metabolites. Size of the nodes is proportional to the number of correlations for each metabolite. (B) Histograms showing distribution of variance explained for the concordance for all glycolytic metabolites and all purines in ccRCCs and NKTs from NS and LTS. P values were calculated by χ2 analysis. (C) Box-whisker plot shows quantification of the connections for the indicated glycolytic intermediates in the indicated tissues (left). P values from 2-tailed t test. Numbers above bars indicate total number of correlations for each metabolite (right). (D) Histograms showing distribution of variance explained for the concordance among all glycolytic metabolites in ccRCCs and NKTs from NS and LTS. P values were calculated by χ2 analysis.
TS also induced qualitative differences in the association of specific glycolytic intermediates (Supplemental Table 7C). Fructose bisphosphate (FBP), glyceraldehyde phosphate (GADP), phosphoglyceric acid (PG), phosphoenolpyruvate (PEP), and pyruvate (PYR) had a significantly lower number of connections in tumors from LTS (Figure 4C and Supplemental Figure 4A). Notably, in ccRCCs from NS, we found significant concordance among glycolytic intermediates (Figure 4D and Supplemental Table 7, A and C), which supports the rapid glycolytic equilibrium reaction (23, 24) consistently measured in metabolic flux experiments. Except for one, these glycolytic intercorrelations were absent in tumors from LTS (Figure 4, C and D).
In contrast, the number of strong connections for dihydroxyacetone phosphate (DHAP) and glycerol phosphate (GP) in particular was higher in tumors from LTS, whereas there was no difference in the number of connections for lactate (Lac) (Figure 4C). GP participates in glycerol phosphate shuttle and can be derived from glycerol generated during fatty acid oxidation by glycerol kinase.
Analysis of all metabolites correlated with FBP, GADP, PGA, and PEP using MetaboAnalyst revealed a significant enrichment (FDR values from 2.52 × 10–9 to 2.18 × 10–2) for 53 metabolic pathways, including the Warburg effect, Pentose Phosphate Pathway (PPP), and purine and pyrimidine metabolism, in ccRCCs from NS (Supplemental Figure 4B), further supporting the role of glycolysis as a central metabolic hub essential for the function of multiple pathways. In contrast, analogous analysis in the case of ccRCCs from LTS showed only 8 metabolic pathways with FDR from 5.14 × 10–4 to 0.05, including the urea cycle, ammonia recycling, glutamate and aspartate metabolism, and MAS (Supplemental Figure 4C).
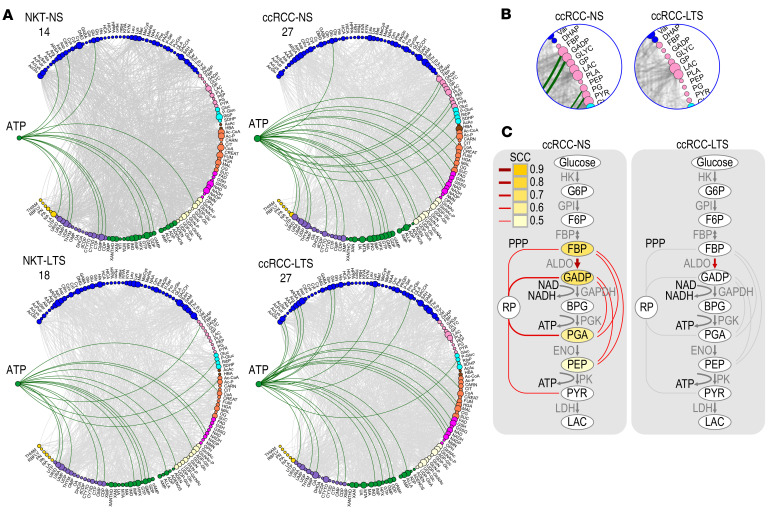
The main glycolytic energy product, ATP, showed increased steady-state levels (Figures 2A and Supplemental Figure 5A) and a higher number of correlations (Figure 5A) in ccRCCs compared with NKTs. However, the correlations of ATP differed in ccRCCs from NS and LTS: ATP correlated with 4 glycolytic intermediates, FBP, GADP, PG, and PEP, only in ccRCCs from NS, whereas these connections were absent in ccRCCs from LTS (Figure 5, B and C, and Supplemental Table 7B).
Figure 5. Disconnection of ATP from glycolysis in ccRCCs from LTS.
(A) Circos visualization of all metabolic correlations for ATP in the indicated tissues. Numbers indicate total number of correlations (green edges) in the indicated tissues. (B) Inserts from Circos shown in panel 4E to visualize connections between ATP and 4 glycolytic intermediates in tumors from NS but not LTS. (C) Schematic presentation of glycolytic pathway and correlations of intermediates with ATP (yellow). Red lines indicate correlations among intermediates and with ribose phosphate (RP), intermediate of PPP. SCC: Spearman’s correlation coefficient.
Importantly, TS changed correlations of the first fully formed purine nucleotide, IMP (Supplemental Figure 5B and Supplemental Table 7B). In ccRCCs from NS, IMP correlated with FBP, GADP, PG, and PEP as well as with ribose phosphate and aspartate, an indication of de novo purine synthesis. These correlations were not present in tumors from LTS.
TS induces a metabolic shift supporting activity of oxidative phosphorylation and malate.
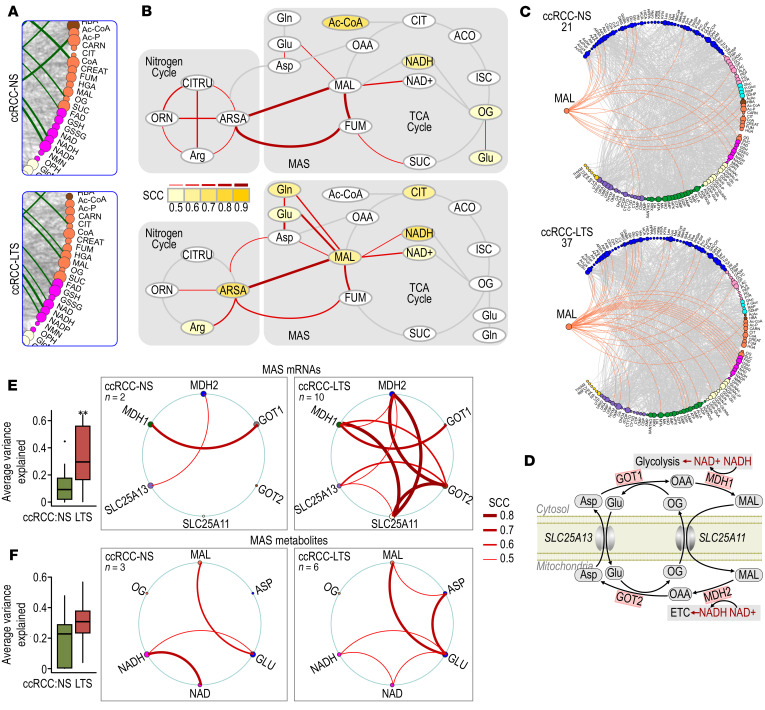
In view of the lost connection between glycolytic intermediates and ATP, we analyzed correlations of ATP to metabolites contributing to oxidative phosphorylation (OXPHOS). We found an increased number of correlations of ATP with TCA cycle intermediates in ccRCCs from LTS compared with NS. In tumors from LTS, ATP correlated with malate, citrate, NAD, and NADH, while it correlated only with acetyl-CoA, oxoglutarate, and NAD in ccRCCs from NS (Figure 6, A and B, and Supplemental Table 7B). Correlation of ATP with malate and NAD/NADH in tumors from LTS implies a role for malate dehydrogenase 2 (MDH2) as a source of NADH that enters respiratory complex I leading to the production of ATP by OXPHOS.
Figure 6. TS-induced reprogramming of TCA cycle and MAS.
(A) Inserts derived from the Circos visualization in Figure 5A show correlations of ATP with TCA cycle metabolites in ccRCCs from NS and LTS. (B) Schematic representation of the ATP correlations (yellow) with metabolites of TCA and urea cycles in ccRCCs from LTS. Red bars indicate correlations among indicated metabolites. Scales: values of Spearman’s correlation coefficient (SCC). (C) Circos visualization show correlations of malate (orange edges) in ccRCCs from NS and LTS. Numbers indicate total number of malate connections. (D) Schematic representation of the MAS. (E) Box-whisker plot shows average-variance-explained analysis (mean ± SEM), and Circos visualization of individual connections for 6 MAS RNAs (MDH1, MDH2, GOT1, GOT2, SLC25A11, SLC25A13) in ccRCCs from NS and LTS. n = numbers of connections among MAS genes. Thickness of the connecting lines is representative of SCC. **P < 0.01 (Wilcoxon test). (F) Box-whisker plot shows average-variance-explained analysis (mean ± SEM), and Circos visualizations of individual connections for 6 MAS metabolites (MAL, OG, Glu, Asp, NAD, NADH) in ccRCCs from NS and LTS. Thickness of the connecting lines is representative of SCC. n = number of connections among MAS metabolites.
Consistent with the predicted activity of MDH2 in tumors from LTS, the number of correlations for malate increased from 21 to 37 in ccRCCs from LTS compared with NS (Figure 6C and Supplemental Table 7C). Importantly, malate gained connections to aspartate, glutamate, NAD, and NADH in tumors form LTS that were absent in NS (Figure 6, B and C). These data point to the effects of TS on the activity of MAS.
The MAS transfers reducing equivalents between cytosol and mitochondria (Figure 6D). On the cytosolic site, MDH1 catalyzes reduction of oxaloacetate (OAA) to malate, regenerating NAD, necessary for the key glycolytic enzyme GAPDH. Malate is then transported into the mitochondria in exchange for 2-oxoglutarate by the SLC25A11 carrier, and is oxidized by the TCA cycle enzyme, MDH2, to OAA. This reaction utilizes NAD and generates NADH, which is available for OXPHOS by respiratory complexes producing ATP. OAA undergoes transamination to aspartate by GOT2, and aspartate is transported out of mitochondria in exchange for glutamate by the SLC25A13 carrier. In the cytoplasm it undergoes transamination to OAA by GOT1.
The effect of TS on MAS is also supported by the induction of gene expression for MDH1 measured in RNA-Seq analysis (Figure 1C). Importantly, we measured significantly higher average variance explained and an increased number of correlations in expression of all 6 MAS mRNAs (SLC25A11, SLC25A13, MDH1, MDH2, GOT1, and GOT2) in LTS (Figure 6E). MAS metabolites were linked to twice as many metabolites in tumors from LTS compared with tumors from NS, although the significance in average variance explained was not reached (Figure 6F).
The correlations of aspartate and glutamate were affected by TS in a manner supporting MAS activity. The number of connections for aspartate diminished from 16 to 9, disrupting connections representative of biosynthetic and storage functions, such as IMP, N-acetylaspartate, asparagine, glutamine, and pyroglutamate, but inducing correlations that indicated activity of MAS such as with malate and glutamate (Supplemental Figure 6, A and B, and Supplemental Table 7D).
The correlations of glutamate showed major rewiring of connectivity toward TCA cycle metabolites and amino acids, away from nucleotide synthesis (Supplemental Figure 6, C and D, and Supplemental Table 7D). Stronger connections of glutamate to aspartate, malate, fumarate, and NAD+/NADH in ccRCCs from LTS implicates glutamate anaplerosis through MAS.
Strong correlations of glutamate with histidine, lysine, and glutamine in ccRCCs from LTS (Supplemental Figure 6, E–H) suggest metabolism of these amino acids to glutamate. Importantly, the abundance of histidine was augmented in LTS ccRCCs (Figure 2C), corresponding with a dramatic increase in its connectivity throughout the metabolome, from none in ccRCCs from NS to 27 strongly correlated metabolites in ccRCCs from LTS (Supplemental Figure 6E and Supplemental Table 7D). Likewise, the number of metabolites correlating with lysine was increased in LTS ccRCCs (Supplemental Figure 6F and Supplemental Table 7D). The data suggest that histidine- and lysine-derived glutamate may contribute to MAS activity in LTS. This is also supported by correlations between histidine and lysine and malate (Figure 6C). The potential utilization of histidine and lysine as a source for glutamate may be related to the decreased expression of SLC1A7, a glutamate transporter in ccRCCs from LTS (Supplemental Table 2B). Glutamine correlations with glutamate, malate, NADH, and ATP in tumors from LTS support its connection to MAS activity (Supplemental Figure 6, G–H). The contribution of the amino acid anaplerosis to ATP production is also implicated by strong correlations of ATP with several amino acids in ccRCCs from LTS (Supplemental Figure 6I).
Metallomic analysis reveals increased formation of Cu-cytochrome c oxidase complex in ccRCCs from LTS and presence of free inorganic arsenic.
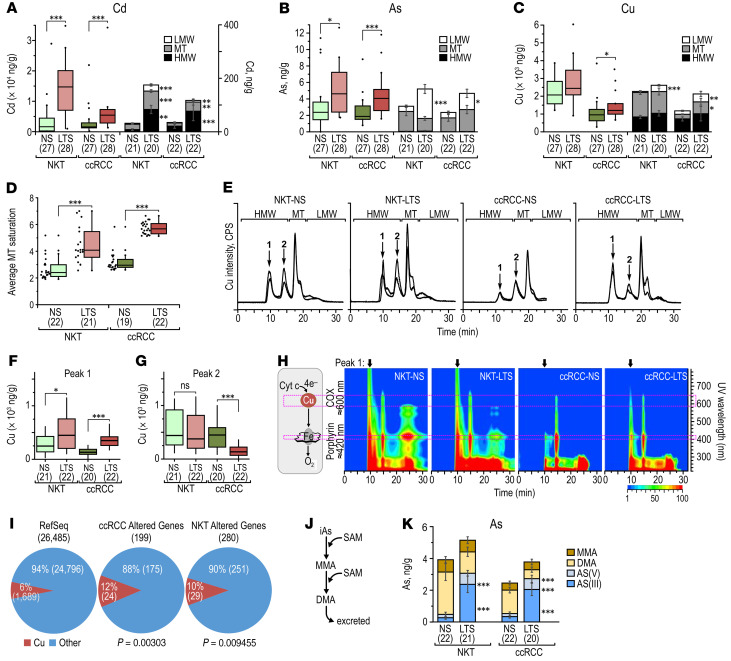
The identification of arsenic-related pathways in the transcriptomic signature upregulated by TS in NKTs prompted us to determine the concentrations of 15 metals and metalloids (Al, As, Cd, Co, Cr, Cu, Fe, Mn, Ni, Pb, Se, Sb, U, V, Zn) in NKTs and ccRCCs from NS and LTS using inductively coupled plasma mass spectrometry (ICP-MS) (Supplemental Table 8). There was a clear separation of all metal concentrations between ccRCCs and NKTs (Supplemental Figure 7A), likely related to the decreased abundance of metallothioneins (MTs), small cysteine-rich proteins binding free metals, in tumors (Supplemental Figure 7B). We found significantly higher accumulation of Cd and As in NKTs and ccRCCs from LTS (Figure 7, A and B) and increased accumulation of Cu in ccRCCs from LTS (Figure 7C).
Figure 7. Metallomic analysis of TS effects in NKTs and ccRCCs from NS and LTS.
(A) Box-whisker plots for the total Cd content and stacked bar graphs for Cd distribution for NKTs and ccRCCs from NS and LTS. (B) Box-whisker plots for the total As content and stacked bar graphs for As distribution for NKTs and ccRCCs from NS and LTS. (C) Box-whisker plots for the total Cu content and stacked bar graphs for Cu distribution for NKTs and ccRCCs from NS and LTS. (D) Saturation of MTs in the indicated tissues. (E) SEC-ICP-MS chromatograms from indicated tissues. Y-axis on the left represents the intensity of signal for the metals. HMW: high molecular weight fraction, LMW: low molecular weight fraction, MT: metallothioneins. Peak 1 corresponds to Cu bound to cytochrome oxidase C, and peak 2 to Cu bound to other proteins. (F) Quantification of Cu in the fraction corresponding to cytochrome C oxidase peak 1 in the indicated tissues. (G) Quantification of Cu in the fraction corresponding to peak 2 in the indicated tissues. (H) UV-Vis isoabsorbance plots. The specific absorption bands at 420 and 600–700 nm are respectively specific for porphyrin rings and CuA-red and CuB-Ox clusters. Scale:1–100 mAU log10. (I) Pie charts show proportion of genes encoding proteins using Cu among the protein-coding genes in RefSeq database and among differentially expressed genes in NKTs and ccRCCs from NS and LTS. P values were determined by 2-sample test for equality of proportion with continuity correction. (J) Schematic representation of As methylation pathway. (K) Stacked bar graphs showing speciation of As in the indicated tissues. MMA: monomethylated As; DMA: demethylated As. P values calculated by Mann-Whitney-Wilcoxon test. Numbers in parentheses indicate number of samples used in each experiment.
Next, we analyzed metal distributions using SEC-UV-Vis-ICP-MS in 3 fractions: high molecular weight proteins (> 10 kDa); low molecular weight proteins containing MTs (10–5 kDa); and the low molecular weight fraction (< 5 kDa), which includes metals that are free or bound to small metabolites (Figure 7E and Supplemental Figure 7C). The total increase in the accumulation of Cd in ccRCCs from LTS was accompanied by its augmented distribution across all 3 fractions (Figure 7A). Increased levels of As in ccRCCs from LTS were primarily due to the accumulation of free arsenic, with no changes in its fraction bound to MT (Figure 7B), likely resulting from saturation of MTs with Cd (Figure 7D). There was significantly increased distribution of Cu to the low molecular weight fraction and to a lesser degree to MTs (Figure 7C).
Importantly, while the total allocation of Cu to high molecular weight did not differ between ccRCCs from NS and LTS, there was a significant enrichment for Cu in a high molecular weight peak corresponding to the cytochrome C oxidase (COX) complex (Figure 7, E and F). In contrast, there was a decrease in the second high molecular weight Cu peak where Cu was bound to other proteins (Figure 7G). The identity of the COX complex-Cu peak (peak 1) was confirmed by UV-Vis absorbance at 420 nm that corresponds to the porphyrin ring and at 600 to 700 nm that corresponds to copper A and copper B clusters (ref. 25 and Figure 7H). The mitochondrial COX complex is the terminal metallo-enzyme in the electron transport chain that transfers electrons to molecular O2. There was also a significant enrichment for genes encoding proteins related to Cu among genes differentially regulated in ccRCCs from LTS compared with the overall percentage of such genes in the RefSeq database (Figure 7I).
Activities of As depend on its oxidoreduction state and methylation as well as its intracellular distribution and molecular association (13). TS contains mainly inorganic arsenate (iAsV) (26). As undergoes several steps of oxidoreductive methylations by arsenic 3-methyl transferase using SAM as a source of methyl groups to generate mono- and dimethyl derivatives (13) (Figure 7J). Dimethyl arsenic (DMA) is the primary derivative excreted by kidneys into urine, and methylation is considered a process of detoxification; however, methylated forms also have oncogenic effects (13). Both NKTs and ccRCCs from LTS showed increased levels of inorganic As, and tumors also showed a decrease in DMA (Figure 7K), which may have been related to the decreased level of SAM in both tissues from LTS (Figure 2, B and C).
Metabolic signature is a prognostic factor stratifying ccRCCs in TCGA.
We selected a set of 154 relevant metabolic genes (MG-154), including genes upregulated in RNA-Seq analysis of ccRCCs from LTS (Figure 1), genes encoding subunits of the mitochondrial respiratory complexes, and enzymes of the TCA cycle and glycolysis (Supplemental Table 9A), in order to further interrogate whether such a defined gene set represents a signature of distinct metabolic subtypes with potential clinical implications. A substantial number of genes in MG-154 (89 out of 154) had been determined to be favorable prognostic factors in the Human Protein Atlas (Supplemental Table 9B). An increased average gene expression for MG-154 relative to the average expression of all genes was measured in the CTSC discovery cohort (Supplemental Figure 8A) and in the TCGA KIRC cohort of 309 white males (Supplemental Figure 8B). Unsupervised clustering analysis using standard hierarchical clustering and differential coexpression analysis were performed with the goal of identifying distinct subtypes associated with patterns of metabolic gene expression in each cohort.
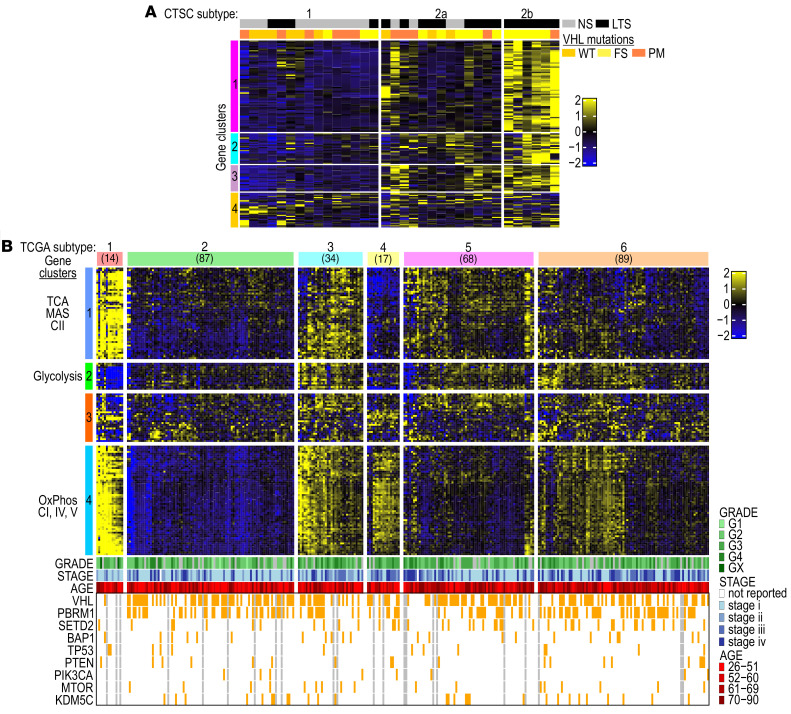
Using the CTSC cohort, we found 4 clusters of genes, of which cluster 1 included genes of OXPHOS, TCA cycle, and MAS; the other clusters were more heterogenous (Supplemental Table 9C). The MG-154 signature stratified CTSC tumors into 3 subtypes and the coexpression pattern of OXPHOS/TCA cycle genes correlated with smoking (Figure 8A). Subtype 1 with the lowest expression of these genes had 27% of LTS, intermediate subtype 2a had 70% of LTS, and subtype 2b with the highest levels of gene expression had 100% of LTS (Figure 8A). The average expression for each gene in MG-154 was significantly higher in subtype 2b as compared with subtypes 1 and 2a (Supplemental Figure 8C).
Figure 8. Identification of ccRCC subtypes based on stratification using metabolic set of genes.
(A) Heatmap shows MG-154 stratifying CTSC cohort of ccRCCs into 3 subtypes according to the smoking status. (B) Heatmap shows MG-154 stratifying the TCGA cohort of ccRCCs into 6 subtypes with a distinct pattern of metabolic gene coexpression.
Next, we applied the same approach to interrogate the TCGA cohort of ccRCCs (Figure 8B). We identified 4 gene clusters (Supplemental Table 9D). Cluster 1 included genes encoding TCA cycle enzymes, subunits of respiratory complex II and MAS; clusters 2 and 3 had several glycolytic genes; and cluster 4 consisted mostly of genes encoding subunits of respiratory complex I, IV, and V. Importantly, the MG-154 gene set stratified the TCGA cohort into 6 subtypes (Figure 8B). Each of these subtypes had different patterns of coexpression in gene clusters. Subtype 1 showed a pattern of gene expression very similar to that in NKTs. Subtype 2 revealed coordinated low gene expression in clusters 1 and 4, whereas subtype 3 presented high expression in all gene clusters. In contrast, subtype 4 demonstrated decoupling of gene expression between clusters 1 and 4, with high expression of genes in cluster 4 but low expression of genes in cluster 1. Subtypes 5 and 6 showed less coordinated expression across the clusters. Mapping of the 3 CTSC subtypes into the 6 TCGA subtypes using the nearest centroid approach revealed a high degree of similarity between CTSC subtype 1 and TCGA subtype 2 (80%) and between CTSC subtype 2b and TCGA subtype 3 (100%). CTSC subtype 2a showed the highest similarity with TCGA subtype 6 and some overlap with subtype 5 (Supplemental Figure 8D). The average expression for each gene in MG-154 was significantly higher in subtypes 1 and 3 compared with the other subtypes (Supplemental Figure 8, E and F).
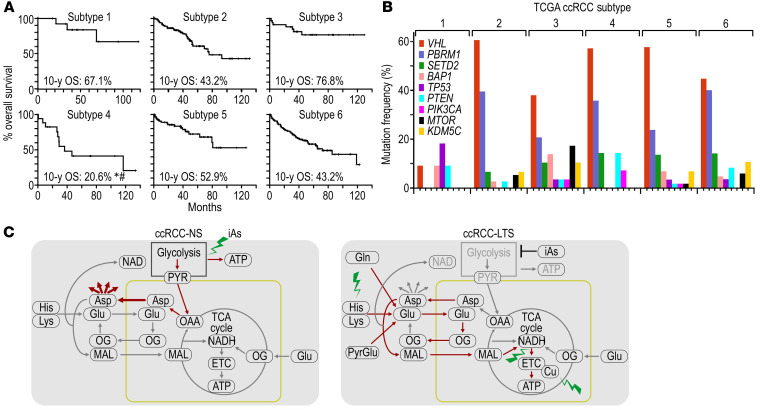
The 6 TCGA subtypes were associated with different 10-year overall survival (OS) as measured by log-rank test (Figure 9A). Subtype 4 had an OS of 20.6%, significantly lower compared with subtype 1 (OS 67.1%, P = 0.0487) and subtype 3 (OS 76.8%, P = 0.0131) (Figure 9A). Subtype 4 was not significantly different from subtypes 1, 3, 5, and 6 in terms of the frequency of tumor grades, i.e., G1 + G2 versus G3 + G4 (Supplemental Figure 9A), and from subtypes 3, 5, and 6 in terms of frequency of tumor stage, i.e. I + II versus III + IV (Supplemental Figure 9B) or in the frequency of metastatic disease (Supplemental Figure 9C).
Figure 9. Metabolic subtypes have different survival rates and are independent from molecular subtypes.
(A) Kaplan-Meier curves and 10-year overall survival (OS) for each of the 6 TCGA subtypes. OS for subtype 4 was significantly different from OS in subtype 1 (#P = 0.049) and subtype 3 (*P = 0.013). P value determined by log-rank test. (B) Percentage of tumors with mutation in the indicated ccRCC tumor–suppressing genes in the TCGA cohort subtypes. (C) Model representing reprogramming of the central carbon and ATP production in tumors from LTS as compared with NS. Red lines indicate activated pathways. Green lightning symbols mark subtype-specific vulnerabilities.
Except for subtype 1 with overall few mutations, all subtypes showed the highest number of mutations in VHL, followed by PBRM1 and SETD2 (Figure 9B). There were no significant differences in average expression of MG-154 between tumors without or with mutations in VHL, PBRM1, SETD2, or BAP1 (Supplemental Figure 9D), further supporting independence of metabolic and molecular subtypes, at least at this stage of analysis.
Discussion
We performed a comprehensive analysis of primary ccRCCs from LTS versus NS using correlational analyses of metabolomic and transcriptomic datasets (23, 24). The suitability of this approach was validated by independent confirmation of the well-established activation of glycolysis and its contributions to cancer metabolism in ccRCCs from NS. Most importantly, correlational analysis revealed TS-induced metabolic activation of OXPHOS and MAS activity, which was not apparent from first-order analysis of steady-state metabolite levels. Independent validation from transcriptomics data showed induction of OXPHOS and MAS genes, which corresponded with an increase in Cu-COX complex formation.
We hypothesize that MAS is at the center of this reprogramming as a mechanism maintaining balance between mitochondrial/cytosolic pools of NAD+/NADH, aspartate, glutamate, malate, and oxoglutarate and connecting the utilization of these metabolites in the biosynthetic pathways with mitochondrial generation of ATP. This agrees with previous observations of a critical role for aspartate in cancer cell line proliferation (27–29).
We propose that metals contribute to the TS-dependent metabolic reprogramming. High levels of Cd saturate cellular metal buffers, MTs, promoting the accumulation of labile As and Cu. High levels of bioavailable Cu can stimulate OXPHOS and affect tumor growth and can be therapeutically controlled (30, 31). The activities of As, interpreted in the context of As speciation, methylation, and excretion by the kidneys, likely evolve during the course of long-term exposures depending on the use and availability of SAM. Monomethyl As derivative selectively inhibits respiratory complex II and IV (32) and leads to induction of glycolysis (33–37). The significant decrease in SAM in tissue from LTS and loss of As methylation contribute to increased levels of inorganic As in low molecular weight fraction. Inorganic As cannot be excreted, but binds to and inhibits the activity of many metabolic enzymes, including glycolytic enzymes (38). Inorganic As also undergoes oxidative arsenylation, i.e., a futile cycle that creates ADP-arsenate, which quickly hydrolyzes to ADP with concomitant reduction of arsenate to arsenite, a process that requires active OXPHOS (39, 40). Thus, induction of OXPHOS by TS may represent a mechanism of As detoxification once SAM is unavailable for methylation. Both effects of As, inhibition of glycolysis and activation of OXPHOS, would have tumor-suppressing effects in ccRCC.
This work demonstrates potential vulnerabilities in ccRCCs from smokers compared with nonsmokers (Figure 9D). There is a growing interest in the use of inhibitors of OXPHOS in treatment of cancers (41). This includes IACS-010759 and antidiabetic biguanides metformin and phenformin, which are respiratory complex I inhibitors being evaluated in clinical trials (42, 43). In addition, Cu chelation to suppress Cu-dependent respiratory complexes may serve a similar therapeutic purpose (31). Another example of targetable metabolic vulnerability in ccRCC associated with TS is the remarkably increased utilization of histidine. Degradation of histidine uses tetrahydrofolate, and therefore sensitizes leukemia cells to treatment with methotrexate, which inhibits dihydrofolate reductase (44). Thus, extensive histidine metabolism in ccRCCs from LTS may create sensitivity to the inhibitors of this pathway.
This work identifies a number of therapeutic opportunities specific to ccRCC metabolic subtypes. Specifically, the tumors that rely on OXPHOS, and therefore molecular oxygen, may respond better to antiangiogenic therapies, which inhibit access to oxygen. The ccRCC subtype 4 characterized by desynchronization between TCA enzymes and OXPHOS genes implicates activity of respiratory complexes in the absence of TCA cycle input of electrons. Electrons for OXPHOS may be supplied by dihydroorotate dehydrogenase (DHODH), a mitochondrial enzyme in de novo pyrimidine biosynthesis, which donates electrons to CoQ (45). This would link OXPHOS to the biosynthetic pathway essential for cancer cell proliferation. Importantly, small molecules targeting DHODH such as brequinar (46) are being evaluated for treatment of cancer (47).
A notable feature of the multiomics analysis is the consistency among the datasets, despite inclusion of specimens from 3 independent centers. We attribute this to the uniformity in the patient population that was analyzed. The analysis reemphasizes the need for further in-depth clinical stratifications in ccRCC studies supporting precision medicine approaches, with better classification of more defined subtypes taking into consideration not only genetics, but also metabolomic profiles, environmental exposures, and risk factors, as well as gender and ethnicity.
Our study has some limitations. At this stage, we did not provide genomic analysis of tumors from smokers and nonsmokers. However, we did not determine any correlations between the mutations in major ccRCC tumor suppressors and metabolic subtypes in TCGA. Thus, it possible that metabolic subtypes are coupled to epigenetic rather than genetic aberrations, and this can also be true for the tumors from smokers. Diminished levels of SAM in LTS and lack of any major metabolic correlations for this epigenetically essential metabolite may indicate its utilization in epigenetic events. The overall small number of metabolic correlations for oxoglutarate may also support extensive utilization of this metabolite by epigenetic enzymes. It is important to note that TS-induced risk of ccRCC is reversible (48), and a potential explanation of this reversibility would be epigenetic rather than genetic driving mechanisms.
Our study does not have direct information about the survival of NS and LTS patients. The TS-associated signature indicating OXPHOS is a favorable prognostic marker in TCGA for the white males points to TS as a positive prognostic factor. This is consistent with the increased expression of TCA cycle genes as an indicator of better ccRCC prognosis (1). Yet, the role of TS as a favorable prognostic factor is counterintuitive because studies showed that TS promotes tumor progression in metastatic disease (49–52). One explanation is that the TS-altered macro- and microenvironment in normal kidney tissue may play a role in promoting tumor progression, even if the tumor is more indolent. The critique of the previous epidemiological studies is that these are retrospective studies analyzing large cohorts of patients, where well-controlled stratifications and normalizations were not consistently applied. Moreover, the rigorous interpretation of the association of ccRCC survival with TS requires information about smoking status after tumor removal and during metastatic disease. Clearly, this information is limited in the existing studies. The role of a risk factor as a favorable prognostic factor is not unprecedented. Obesity, an established risk factor for ccRCCs, is paradoxically a positive prognostic factor related to the decreased levels of fatty acid synthase (53, 54). Similarly, in the case of head and neck squamous cell carcinoma (HNSCC), although HPV is a risk factor, HNSCCs (especially those involving the oropharynx) related to HPV have a better prognosis (55). Moreover, As implicated in TS effects is recognized for its oncogenic and chemotherapeutic activities (56).
Overall, this study identified previously unrecognized extensive metabolic reprogramming in primary ccRCCs from LTS that creates opportunities for treatments that would not be predicted based on genetic subtypes. This work highlights the importance of considering metabolites and metal content as well as more in-depth analysis of metabolic signatures in identifying clinical subtypes to establish best practices for precision medicine.
Methods
Tissues and reagents.
Specimens were obtained from tumor banks at the University of Cincinnati and VA Medical Center (Cincinnati, Ohio, USA), Urology Oncology Branch at the National Cancer Institute (Bethesda, Maryland, USA), and University of North Carolina at Chapel Hill (Chapel Hill, North Carolina, USA). Fresh-frozen and OCT embedded tissues were used. All samples were reviewed by genitourinary pathologists and derived from regions with more than 80% cancer cells. Supplemental Table 1 provides information about the patient cohorts. Supplemental Table 10 describes the use of each specimen in the individual analyses. Supplemental Table 11 lists all used reagents. DNA was isolated using DNAzol (MRC) or QIAamp DNA micro kit (Qiagen). VHL was sequenced as described before (57).
RNA-Seq.
RNA was extracted using RNAlater ICE (Ambion) and miRNA isolation kit (Ambion). The quality of RNA was checked using Bioanalyzer RNA 6000 Nano kit (Agilent). PolyA RNA was extracted using NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB) and used as input for RNA-Seq. RNA-Seq libraries were prepared using NEBNext Ultra II Directional RNA Library Prep Kit (NEB). After library quality control analysis using Bioanalyzer High Sensitivity DNA kit (Agilent) and library quantification using NEBNext Library Quant kit (NEB), the sequencing was performed under the setting of single read 1 × 51 bp to generate approximately 30 million reads per sample on HiSeq 1000 sequencer (Illumina).
Data analysis.
Reads were first aligned to the human reference genome (hg19) and current gene definitions using TopHat2 aligner (58). Reads aligning to each known transcript were counted and the follow-up analyses performed using the Bioconductor package for next-generation sequencing data analysis (59). The quality of raw, aligned, and counts data was assessed using fastQC (60) and RNA-SeQC (61) tools, and by examining correlation patterns of normalized data between different samples. Two samples that showed poor correlation with the rest of the samples and unusually high levels of read duplication were excluded from the analysis. The differential gene expression analysis was based on the negative-binomial statistical model of read counts as implemented in the edgeR Bioconductor package (62) for each separate comparison. Differential expressions with FDR-adjusted P values (63) less than 0.1 were considered to be statistically significant. For cluster analysis, gene expression levels were normalized by calculating log2 reads per kilobase million (RPKM) levels (64). Expression profiles of differentially expressed genes in the heatmap were clustered using the Bayesian infinite mixture model (65), and the samples were clustered using average linkage hierarchical clustering based on pairwise Pearson’s correlations as the measure of similarity. The enrichment analysis of up- and downregulated genes in gene sets defined by MSigDB (66) and Gene Ontologies (67) was performed using logistic regression–based LRPath methodology (68) as implemented in the CLEAN package (69). The enrichment analysis of transcription factor targets was performed by submitting lists of differentially expressed genes to Enrichr (70). The genes associated with Cu were identified by searching GeneCards (71). The statistical significance of enrichment of genes related to copper was assessed using Fisher’s exact test (72). Enrichr has been used to perform enrichment analysis using ChEA and ENCODE libraries of curated sets of transcription factor targets identified by ChIP-Seq. All raw RNA-Seq and demographic data from patients who granted informed consent to share such data are made available at the Database of Genotypes and Phenotypes (dbGaP) under accession number phs2083.v1.
Differential coexpression analysis of mRNA abundance.
Unsupervised gene differential coexpression analysis was used (73) to capture context-specific patterns of gene coexpression that revealed patient strata, i.e., groups of samples with coordinated expression. GimmR package with default options was used for differential coexpression analysis; clustering and heatmaps were generated using R package ComplexHeatmap. Strata identified in the CTSC discovery cohort by using the MG-154 metabolic signature, in conjunction with differential coexpression analysis, were subsequently characterized in terms of mutational profiles and association with smoking.
Histogram analysis.
Variance-explained data for each of the 4 tissues were binned into 20 segments ranging from 0 to 1. Comparisons were then performed for NKT-NS versus NKT-LTS and ccRCC-NS versus ccRCC-LTS. Overlaid histograms were generated for these comparisons using R’s base statistics and graphics functions (hist, plot). For each comparison, statistical significance was obtained via the chisq.test function in R base statistics with a significance threshold at 0.05. The difference in variance explained for the 2 comparisons was computed for each metabolite pair, and the resulting values were sorted to find metabolite pairs that contributed the maximum degree of difference between the 2 tissues under consideration. The histogram distances were computed using the HistogramTools package in R and included Manhattan, Euclidean, Intersect distance, and Kullback-Leibler divergence.
TCGA subtype analysis.
The TCGA-KIRC cohort of 309 white males was used to determine the pattern of stratification by MG-154 signature. cBioportal (https://www.cbioportal.org/study/summary?id=kirc_tcga, Firehose Legacy) was used to extract sample-specific metadata, including overall survival and mutational profiles, whereas normalized RNA-Seq expression profiles were obtained using iLINCS (http://www.ilincs.org/ilincs/dataset/TCGA_KIRC_RNA-seqV2). The strata observed using the MG-154 signature and differential coexpression analysis were characterized in terms of stage, grade, survival, and mutational status. The R package Survival and cBioportal were used to generate Kaplan-Meier curves and assess statistical significance of differences in overall survival using a log-rank test. A χ2 test was used to determine differences in frequencies of stage, grade, and metastatic status of the subtypes. Subtypes observed in CTSC were mapped into TCGA subtypes using the nearest centroid analysis with Euclidean distance, where centroids for each TCGA subtype were defined as arithmetic averages of gene expression within each subtype, and each sample in the CTSC discovery cohort was mapped to the TCGA subtype centroid with the smallest distance.
Preparation of tissues for metabolomic analyses.
Tissues (15 mg) were homogenized in a solvent mixture of MeOH:ACN:H2O (50:30:20) in a TissueLyser II (Qiagen) at 4°C, followed by flash freezing in liquid nitrogen, and then centrifuged and passed through a Nanosep 3K molecular weight cutoff filter (Life Sciences Technologies). Tissue lysates were mixed 1:1 with 13C labeled internal standard mix balanced for the metabolites of interest. The balanced internal standard was generated by combining IROA yeast extract (IROA Technologies) with 13C-labeled lysates from several human cell lines grown in the presence of 5.5 mM U-13C glucose (Cambridge Isotopes) for 3 passages.
Chromatographic separation and MS analyses.
Chromatographic separation was accomplished by hydrophobic interaction liquid chromatography using a Luna NH2 3 μm, 2 mm × 100 mm column, (Phenomenex) on a Vanquish Flex Quaternary UHPLC system (Thermo Fisher Scientific). Mobile phase A consisted of 10 mM NH4HCO3, 2% NH4OH (Thermo Fisher Scientific). Mobile phase B consisted of acetonitrile (Honeywell Burdick & Jackson).
MS analyses were performed on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) interfaced with an H-ESI electrospray source (Thermo Fisher Scientific). General instrumental conditions were RF 30%; sheath gas, auxiliary gas, and sweep gas of 35, 11, and 0 arbitrary units, respectively; ion transfer tube temperature of 300°C; vaporizer temperature of 250°C; and spray voltage of 2500 V for positive mode and 3500 V for negative mode. Data were collected for each sample in negative mode, using 2 different mass ranges (70–700 m/z and 220–900 m/z) to enhance sensitivity for larger, less-abundant compounds, and in positive mode (70–900 m/z) (74, 75).
Metabolite identification.
A custom library of retention times and m/z ratios was compiled using unlabeled and 13C-labeled reference compounds and ms/ms fragmentation patterns. These data were referenced to the MZCloud (mzcloud.org) and Mass Bank (massbank.eu) databases. All data were converted to MZXML format using MassMatrix. Peak areas, including isotopically enriched metabolites, were obtained using Maven (76, 77) for targeted analysis, and untargeted analysis was performed using both IROA ClusterFinder and Compound Discoverer. Differentially abundant metabolites were identified by computing fold change and statistical significance using Wilcoxon nonparametric test. Volcano plots were generated for each pairwise comparison of NKTs and ccRCCs to illustrate the effect size and statistical significance for each metabolite.
Analysis and visualization of metabolites’ correlation patterns.
The correlations between abundance levels of 133 metabolites was determined using Spearman’s correlation coefficient for each pair of metabolites within each tissue. The overall distributions of correlations squared, i.e. variance explained, were plotted for relevant pathways in each tissue type. Strong correlations defined as Spearman’s correlation coefficient of more than 0.5 were illustrated using Circos generated by Pinet Server (78) where nodes denote metabolites, edges represent correlations (with width proportional to the strength of correlation), and the size of a node represents the number of edges originating from that node, i.e. its degree.
Multielemental analysis.
Samples (1–5 mg) were digested with nitric acid, diluted with ultra-pure water, and loaded into the ICP-MS-MS (triple quad Agilent 8800x ICP-MS-MS) to be run in helium and no-gas mode (79). Integration time was adjusted according to the concentration range for each element. The external calibration method was used from 0.01 ng mL–1 to 2500 ng mL–1 with a mixture of scandium, yttrium, indium, and bismuth as internal standards at 5 ng mL–1. Internal mass index elemental tags were used in the form of phosphorus and sulfur instead of the sample mass. Data were analyzed using Agilent MassHunter software. Quality control samples included NIST toxic elements in frozen human urine standard reference material (SRM 2668) and NIST bovine muscle powder (SRM 8414).
SEC-ICP-MS analysis.
Tissues were homogenized and sonicated in protein extracting solution (50 mM SDS, 10 mM NaCl, 50 mM Tris-HCL, pH 7.4), filtered and injected to a 1200 HPLC system composed of a thermostated autosampler set to 4°C, a vacuum membrane degasser, a binary pump, a column oven compartment, and a UV-Vis diode array detector (Agilent). The size exclusion chromatography (SEC) column was a TSK gel 3000SW 7.8 × 30 mm, 10 μm particle size with 50 mM NH4Ac buffer in 0.1% MeOH at pH 7.4 at the mobile phase at 0.5 mL min–1 (Tosoh Global). The outlet of the HPLC system was connected to the ICP-MS-MS nebulizer by a 65 cm PEEK capillary of 0.17 mm of internal diameter. The ICP-MS was operated in time-resolved analysis with an integration time of 0.15 seconds per isotope. A 5-point calibration was generated for 75As, 63Cu, 111Cd, 56Fe, 60Ni, 78Se, 207Pb, 34S, 31P, 55Mn, 59Co and 66Zn with a spiked FBS sample in order to avoid column contamination by free metals and to replicate the protein matrix of the real samples. FBS was spiked with 0.2 ppm of Pb, Cd, Se, Co, Mn, and P with inorganic salts and incubated for 1 hour at 37°C. The unbound metals were filtered out and a total metal analysis was carried out in the remaining FBS. An aliquot of metal-enriched protein sample was filtered and diluted 1:1 with the SEC mobile phase and injected in increasing volumes to generate a calibration curve.
SEC-ICP-MS data analysis.
The integration of the SEC-ICP-MS chromatograms was performed in the Origin X software package (OriginLab) after exporting them from the Agilent Mass Hunter ICP-MS software. The areas obtained were used to quantify the metal content of the SEC fraction and normalized to the starting tissue mass (80). The retention times for the molecular mass fraction were 10 to 19 minutes (> 500–10 kDa), 19.1–22 minutes (10–5 kDa), and 22.1–27 minutes (< 5 kDa).
Total MT quantification and saturation by SEC-ICP-MS.
Standards for MTs were prepared from rabbit liver (Sigma Aldrich). The total concentration of MTs was determined by the sulfur content in the 20-minute peak. An average of 20 cysteine residues per molecule of MT was considered for a 1:20 molar ratio of MT:S. The kidney and tumor samples were analyzed in the same batch. The major Cd signal coeluted with the MT standard confirmed the identity of the peak in the analyzed tissues. The saturation rate was defined as the molar ratio of MT:Cd+Zn+Cu+Ni+Mn in the 20-minute peak obtained from the tissue analysis by SEC-ICP-MS.
Arsenic speciation analysis.
The homogenized tissues were extracted for As compounds with 10 mM (NH4)2HPO4, and 30 mmol L–1 HNO3, centrifuged, filtered, and transferred to 1200 HPLC vessels (Agilent). An anion exchange separation column PRP-X100, 250 × 4.1 mm, 10 μm (Hamilton) and a guard column PRP-X100 20 × 2 mm, 10 μm (Hamilton) were used. The elution was performed isocratically at a flow rate of 1.0 mL min–1 (81). Calibration curves were obtained using stock solutions for As(III), As(V), DMA, and monomethylated As (MMA) in the mobile phase.
Principal component analysis.
The total concentration of all metals was used to generate a descriptive model based on principal component analysis using Unscrambler X (CAMO Software) software package. The model was constructed with down weight of 1/y for all variables. The explained variance of the model was 99.1% for PC1 + PC2 + PC3. The grouping was done in the scores plot by tissue type.
Statistics.
Sigma plot 12.0 was used to evaluate the significance of the comparisons between results using the Mann-Whitney rank-sum test, with a P value of less than 0.05 considered significant.
Study approval.
In all cases, tissues were obtained after acquiring written informed consent and the approval of the respective IRBs from the University of Cincinnati and VA Medical Center, the Urology Oncology Branch at the National Cancer Institute, and the University of North Carolina at Chapel Hill, and samples were deidentified.
Author contributions
MFCK, JM, and JLF designed the study, supervised the work, and wrote the manuscript. JR and JB performed bioinformatics analysis of the data and wrote part of the manuscript. DS performed metallomic experiments and analysis. CW performed metabolomic experiments. BS performed visualization of the metabolomic data and generated Circos illustrations. JC, YZ, NT, MEB, and YZ performed DNA and RNA extractions, VHL sequencing, and sample preparation. CT, KT, HA, AS, SR, LSS, and WML provided surgical samples and edited the manuscript. HA provided pathological analysis. XZ performed RNA sequencing. MM supervised bioinformatics analysis of RNA-Seq and coexpression analyses and wrote part of the manuscript. JTC and DRP contributed to the development of metabolic analyses, writing of the manuscript, and conceptual development of the project. Order of the co–first authors was based on the contribution to (a) the bioinformatic integration of the multiomics data; (b) generation and analysis of individual omics data; (c) time spent on the project.
Supplementary Material
Acknowledgments
The work was supported by the following grants: DoD Idea Award W81XWH-14-1-0347, R01CA122346, R01GM128216, 2I01BX001110 BLR&D VA Merit, VA Spark, and UCCI funds to MFCK. JM was supported by UC P30-ES006096 CEG, R01MH107487, and R01DK091566. DRP was supported by R01CA168815. CW was supported by 5T32CA117846 and ACS PF-17-199. BS was supported by 1T32CA236764. JTC was supported by R01CA230904 and R35GM133561. YZ and YZ were partially supported by an overseas study stipend from Shandong University, Jinan, China. LSS was supported in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN26120080001E. We thank R. Ross for help with metabolomic technology; Shuchi Gulati and Chris Beecher for reading the manuscript; Mathew LaMacchia for initial preparation of DNA samples; Maddie Worman for preparation of the tables; B. Peace for professional editing; and G. Doerman for preparing the figures.
Version 1. 09/24/2020
In-Press Preview
Version 2. 01/04/2021
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(1):e140522.https://doi.org/10.1172/JCI140522.
Contributor Information
James Reigle, Email: reiglej@mail.uc.edu.
Dina Secic, Email: secicda@mail.uc.edu.
Jacek Biesiada, Email: biesiajk@uc.edu.
Collin Wetzel, Email: wetzelcollin@gmail.com.
Behrouz Shamsaei, Email: shamsabz@ucmail.uc.edu.
Johnson Chu, Email: chuj@apsistewch.com.
Yuanwei Zang, Email: zangyuanwei@126.com.
Xiang Zhang, Email: zhanx5@uc.edu.
Nicholas J. Talbot, Email: ntalbot14@yahoo.com.
Megan E. Bischoff, Email: bischomg@ucmail.uc.edu.
Yongzhen Zhang, Email: zyzlover87@163.com.
Charuhas V. Thakar, Email: thakarcv@UCMAIL.UC.EDU.
Krishnanath Gaitonde, Email: gaitonk@ucmail.uc.edu.
Abhinav Sidana, Email: sidanaav@ucmail.uc.edu.
Hai Bui, Email: hai.bui@va.gov.
John T. Cunningham, Email: cunnijn@ucmail.uc.edu.
Qing Zhang, Email: qing_zhang@med.unc.edu.
Laura S. Schmidt, Email: schmidtl@ncifcrf.gov.
W. Marston Linehan, Email: wml@nih.gov.
Mario Medvedovic, Email: medvedm@UCMAIL.UC.EDU.
David R. Plas, Email: plasd@uc.edu.
Julio A. Landero Figueroa, Email: landerjo@ucmail.uc.edu.
Jarek Meller, Email: mellerj@ucmail.uc.edu.
Maria F. Czyzyk-Krzeska, Email: Maria.Czyzykkrzeska@uc.edu.
References
- 1.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell TJ, et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx renal. Cell. 2018;173(3):611–623.e17. doi: 10.1016/j.cell.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turajlic S, et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell. 2018;173(3):595–610.e11. doi: 10.1016/j.cell.2018.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenza GL. HIF-1 mediates the Warburg effect in clear cell renal carcinoma. J Bioenerg Biomembr. 2007;39(3):231–234. doi: 10.1007/s10863-007-9081-2. [DOI] [PubMed] [Google Scholar]
- 5.Hakimi AA, et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29(1):104–116. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gatto F, Nookaew I, Nielsen J. Chromosome 3p loss of heterozygosity is associated with a unique metabolic network in clear cell renal carcinoma. Proc Natl Acad Sci U S A. 2014;111(9):E866–E875. doi: 10.1073/pnas.1319196111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt JD, van der Hel OL, McMillan GP, Boffetta P, Brennan P. Renal cell carcinoma in relation to cigarette smoking: meta-analysis of 24 studies. Int J Cancer. 2005;114(1):101–108. doi: 10.1002/ijc.20618. [DOI] [PubMed] [Google Scholar]
- 8.Cote ML, et al. Cigarette smoking and renal cell carcinoma risk among black and white Americans: effect modification by hypertension and obesity. Cancer Epidemiol Biomarkers Prev. 2012;21(5):770–779. doi: 10.1158/1055-9965.EPI-11-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, et al. Dose-response relationships between cigarette smoking and kidney cancer: A systematic review and meta-analysis. Crit Rev Oncol Hematol. 2019;142:86–93. doi: 10.1016/j.critrevonc.2019.07.019. [DOI] [PubMed] [Google Scholar]
- 10.Talhout R, Schulz T, Florek E, van Benthem J, Wester P, Opperhuizen A. Hazardous compounds in tobacco smoke. Int J Environ Res Public Health. 2011;8(2):613–628. doi: 10.3390/ijerph8020613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexandrov LB, et al. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;354(6312):618–622. doi: 10.1126/science.aag0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moulis JM. Cellular mechanisms of cadmium toxicity related to the homeostasis of essential metals. Biometals. 2010;23(5):877–896. doi: 10.1007/s10534-010-9336-y. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe T, Hirano S. Metabolism of arsenic and its toxicological relevance. Arch Toxicol. 2013;87(6):969–979. doi: 10.1007/s00204-012-0904-5. [DOI] [PubMed] [Google Scholar]
- 14.Song Jk, et al. Association between cadmium exposure and renal cancer risk: a meta-analysis of observational studies. Sci Rep. 2015;5:17976. doi: 10.1038/srep17976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreccio C, et al. Case-control study of arsenic in drinking water and kidney cancer in uniquely exposed Northern Chile. Am J Epidemiol. 2013;178(5):813–818. doi: 10.1093/aje/kwt059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1α transcriptional complex. Nature. 2007;450(7170):736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 17.Wang R, et al. Smoking-induced upregulation of AKR1B10 expression in the airway epithelium of healthy individuals. Chest. 2010;138(6):1402–1410. doi: 10.1378/chest.09-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jong FA, Beecher C. Addressing the current bottlenecks of metabolomics: isotopic ratio outlier analysis, an isotopic-labeling technique for accurate biochemical profiling. Bioanalysis. 2012;4(18):2303–2314. doi: 10.4155/bio.12.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callicutt CH, et al. The role of ammonia in the transfer of nicotine from tobacco to mainstream smoke. Regul Toxicol Pharmacol. 2006;46(1):1–17. doi: 10.1016/j.yrtph.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Servillo L, et al. Ophthalmic acid is a marker of oxidative stress in plants as in animals. Biochim Biophys Acta Gen Subj. 2018;1862(4):991–998. doi: 10.1016/j.bbagen.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Dello SA, et al. Systematic review of ophthalmate as a novel biomarker of hepatic glutathione depletion. Clin Nutr. 2013;32(3):325–330. doi: 10.1016/j.clnu.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Courtney KD, et al. Isotope tracing of human clear cell renal cell carcinomas demonstrates suppressed glucose oxidation in vivo. Cell Metab. 2018;28(5):793–800.e2. doi: 10.1016/j.cmet.2018.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steuer R. Review: on the analysis and interpretation of correlations in metabolomic data. Brief Bioinformatics. 2006;7(2):151–158. doi: 10.1093/bib/bbl009. [DOI] [PubMed] [Google Scholar]
- 24.Rosato A, Tenori L, Cascante M, De Atauri Carulla PR, Martins Dos Santos VAP, Saccenti E. From correlation to causation: analysis of metabolomics data using systems biology approaches. Metabolomics. 2018;14(4):37. doi: 10.1007/s11306-018-1335-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karu TI. Multiple roles of cytochrome c oxidase in mammalian cells under action of red and IR-A radiation. IUBMB Life. 2010;62(8):607–610. doi: 10.1002/iub.359. [DOI] [PubMed] [Google Scholar]
- 26.Iwai T, Chiba K, Narukawa T. Arsenic speciation and cadmium determination in tobacco leaves, ash and smoke. Anal Sci. 2016;32(9):957–962. doi: 10.2116/analsci.32.957. [DOI] [PubMed] [Google Scholar]
- 27.Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell. 2015;162(3):540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gui DY, et al. Environment dictates dependence on mitochondrial complex I for NAD+ and aspartate production and determines cancer cell sensitivity to metformin. Cell Metab. 2016;24(5):716–727. doi: 10.1016/j.cmet.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell. 2015;162(3):552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishida S, Andreux P, Poitry-Yamate C, Auwerx J, Hanahan D. Bioavailable copper modulates oxidative phosphorylation and growth of tumors. Proc Natl Acad Sci U S A. 2013;110(48):19507–19512. doi: 10.1073/pnas.1318431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denoyer D, Masaldan S, La Fontaine S, Cater MA. Targeting copper in cancer therapy: ‘Copper That Cancer’. Metallomics. 2015;7(11):1459–1476. doi: 10.1039/C5MT00149H. [DOI] [PubMed] [Google Scholar]
- 32.Naranmandura H, et al. Mitochondria are the main target organelle for trivalent monomethylarsonous acid (MMA(III))-induced cytotoxicity. Chem Res Toxicol. 2011;24(7):1094–1103. doi: 10.1021/tx200156k. [DOI] [PubMed] [Google Scholar]
- 33.WARBURG O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–270. [PubMed] [Google Scholar]
- 34.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 35.Zhao F, Severson P, Pacheco S, Futscher BW, Klimecki WT. Arsenic exposure induces the Warburg effect in cultured human cells. Toxicol Appl Pharmacol. 2013;271(1):72–77. doi: 10.1016/j.taap.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luz AL, Godebo TR, Smith LL, Leuthner TC, Maurer LL, Meyer JN. Deficiencies in mitochondrial dynamics sensitize Caenorhabditis elegans to arsenite and other mitochondrial toxicants by reducing mitochondrial adaptability. Toxicology. 2017;387:81–94. doi: 10.1016/j.tox.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luz AT, et al. Arsenite uncouples mitochondrial respiration and induces a Warburg-like effect in Caenorhabditis elegans. Toxicol Sci. 2016;154(1):195. doi: 10.1093/toxsci/kfw185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang HN, et al. Systematic identification of arsenic-binding proteins reveals that hexokinase-2 is inhibited by arsenic. Proc Natl Acad Sci U S A. 2015;112(49):15084–15089. doi: 10.1073/pnas.1521316112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Németi B, Regonesi ME, Tortora P, Gregus Z. Polynucleotide phosphorylase and mitochondrial ATP synthase mediate reduction of arsenate to the more toxic arsenite by forming arsenylated analogues of ADP and ATP. Toxicol Sci. 2010;117(2):270–281. doi: 10.1093/toxsci/kfq141. [DOI] [PubMed] [Google Scholar]
- 40.Németi B, Gregus Z. Mitochondria work as reactors in reducing arsenate to arsenite. Toxicol Appl Pharmacol. 2002;182(3):208–218. doi: 10.1006/taap.2002.9443. [DOI] [PubMed] [Google Scholar]
- 41.Ashton TM, McKenna WG, Kunz-Schughart LA, Higgins GS. Oxidative phosphorylation as an emerging target in cancer therapy. Clin Cancer Res. 2018;24(11):2482–2490. doi: 10.1158/1078-0432.CCR-17-3070. [DOI] [PubMed] [Google Scholar]
- 42.Chae YK, et al. Repurposing metformin for cancer treatment: current clinical studies. Oncotarget. 2016;7(26):40767–40780. doi: 10.18632/oncotarget.8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molina JR, et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med. 2018;24(7):1036–1046. doi: 10.1038/s41591-018-0052-4. [DOI] [PubMed] [Google Scholar]
- 44.Kanarek N, et al. Histidine catabolism is a major determinant of methotrexate sensitivity. Nature. 2018;559(7715):632–636. doi: 10.1038/s41586-018-0316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loffler M, Carrey EA, Knecht W. The pathway to pyrimidines: the essential focus on dihydroorotate dehydrogenase, the mitochondrial enzyme coupled to the respiratory chain. Nucleosides Nucleotides Nucleic Acids. 2020:1–25. doi: 10.1080/15257770.2020.1723625. [DOI] [PubMed] [Google Scholar]
- 46.Hurt DE, Sutton AE, Clardy J. Brequinar derivatives and species-specific drug design for dihydroorotate dehydrogenase. Bioorg Med Chem Lett. 2006;16(6):1610–1615. doi: 10.1016/j.bmcl.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 47.Madak JT, Bankhead A, Cuthbertson CR, Showalter HD, Neamati N. Revisiting the role of dihydroorotate dehydrogenase as a therapeutic target for cancer. Pharmacol Ther. 2019;195:111–131. doi: 10.1016/j.pharmthera.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 48.Rini BI, Campbell SC, Rathmell WK. Renal cell carcinoma. Curr Opin Oncol. 2006;18(3):289–296. doi: 10.1097/01.cco.0000219260.60714.c4. [DOI] [PubMed] [Google Scholar]
- 49.Parker A, Lohse C, Cheville J, Leibovich B, Igel T, Blute M. Evaluation of the association of current cigarette smoking and outcome for patients with clear cell renal cell carcinoma. Int J Urol. 2008;15(4):304–308. doi: 10.1111/j.1442-2042.2008.01994.x. [DOI] [PubMed] [Google Scholar]
- 50.Kroeger N, et al. Smoking negatively impacts renal cell carcinoma overall and cancer-specific survival. Cancer. 2012;118(7):1795–1802. doi: 10.1002/cncr.26453. [DOI] [PubMed] [Google Scholar]
- 51.Ehdaie B, Furberg H, Zabor EC, Hakimi AA, Russo P. Comprehensive assessment of the impact of cigarette smoking on survival of clear cell kidney cancer. J Urol. 2014;191(3):597–602. doi: 10.1016/j.juro.2013.08.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fajkovic H, et al. Impact of smoking status on survival after cytoreductive nephrectomy for metastatic renal cell carcinoma. World J Urol. 2016;34(10):1411–1419. doi: 10.1007/s00345-016-1767-9. [DOI] [PubMed] [Google Scholar]
- 53.Hakimi AA, et al. An epidemiologic and genomic investigation into the obesity paradox in renal cell carcinoma. J Natl Cancer Inst. 2013;105(24):1862–1870. doi: 10.1093/jnci/djt310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanchez A, et al. Transcriptomic signatures related to the obesity paradox in patients with clear cell renal cell carcinoma: a cohort study. Lancet Oncol. 2020;21(2):283–293. doi: 10.1016/S1470-2045(19)30797-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kobayashi K, Hisamatsu K, Suzui N, Hara A, Tomita H, Miyazaki T. A review of HPV-related head and neck cancer. J Clin Med. 2018;7(9):E241. doi: 10.3390/jcm7090241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rehman K, Naranmandura H. Double-edged effects of arsenic compounds: anticancer and carcinogenic effects. Curr Drug Metab. 2013;14(10):1029–1041. doi: 10.2174/13892002113146660116. [DOI] [PubMed] [Google Scholar]
- 57.Yi Y, et al. von Hippel-Lindau-dependent patterns of RNA polymerase II hydroxylation in human renal clear cell carcinomas. Clin Cancer Res. 2010;16(21):5142–5152. doi: 10.1158/1078-0432.CCR-09-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huber W, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12(2):115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Andrews S. Fast QC: A quality control tool for high throughput sequence data. Babraham Institute website. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ Updated August 1, 2019. Accessed September 21, 2020.
- 61.DeLuca DS, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28(11):1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 64.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 65.Freudenberg JM, Sivaganesan S, Wagner M, Medvedovic M. A semi-parametric Bayesian model for unsupervised differential co-expression analysis. BMC Bioinformatics. 2010;11:234. doi: 10.1186/1471-2105-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sartor MA, Leikauf GD, Medvedovic M. LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics. 2009;25(2):211–217. doi: 10.1093/bioinformatics/btn592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Freudenberg JM, Joshi VK, Hu Z, Medvedovic M. CLEAN: CLustering Enrichment ANalysis. BMC Bioinformatics. 2009;10:234. doi: 10.1186/1471-2105-10-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Safran M, et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010;2010:baq020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. J Royal Stat Soc. 1922;85(1):87–94. doi: 10.2307/2340521. [DOI] [Google Scholar]
- 73.Freudenberg JM, Sivaganesan S, Wagner M, Medvedovic M. A semi-parametric Bayesian model for unsupervised differential co-expression analysis. BMC Bioinformatics. 2010;11:234. doi: 10.1186/1471-2105-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A. 2006;1125(1):76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 75.Liu Z, Rochfort S. A fast liquid chromatography-mass spectrometry (LC-MS) method for quantification of major polar metabolites in plants. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;912:8–15. doi: 10.1016/j.jchromb.2012.10.040. [DOI] [PubMed] [Google Scholar]
- 76.Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem. 2010;82(23):9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clasquin MF, Melamud E, Rabinowitz JD. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr Protoc Bioinformatics. 2012;Chapter 14:Unit14.11. doi: 10.1002/0471250953.bi1411s37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shamsaei B, et al. piNET: a versatile web platform for downstream analysis and visualization of proteomics data. Nucleic Acids Res. 2020;48(W1):W85–W93. doi: 10.1093/nar/gkaa436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Figueroa JA, Stiner CA, Radzyukevich TL, Heiny JA. Metal ion transport quantified by ICP-MS in intact cells. Sci Rep. 2016;6:20551. doi: 10.1038/srep20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Subramanian Vignesh K, Landero Figueroa JA, Porollo A, Caruso JA, Deepe GS. Granulocyte macrophage-colony stimulating factor induced Zn sequestration enhances macrophage superoxide and limits intracellular pathogen survival. Immunity. 2013;39(4):697–710. doi: 10.1016/j.immuni.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmidt L, et al. A feasible method for As speciation in several types of seafood by LC-ICP-MS/MS. Food Chem. 2018;255:340–347. doi: 10.1016/j.foodchem.2018.02.079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.