Graphical abstract
Keywords: COVID-19, Vaccine, Spike glycoprotein, Epitope engineering, Structural vaccinology, EvoDesign
Abstract
The development of effective and safe vaccines is the ultimate way to efficiently stop the ongoing COVID-19 pandemic, which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Built on the fact that SARS-CoV-2 utilizes the association of its Spike (S) protein with the human angiotensin-converting enzyme 2 (ACE2) receptor to invade host cells, we computationally redesigned the S protein sequence to improve its immunogenicity and antigenicity. Toward this purpose, we extended an evolutionary protein design algorithm, EvoDesign, to create thousands of stable S protein variants that perturb the core protein sequence but keep the surface conformation and B cell epitopes. The T cell epitope content and similarity scores of the perturbed sequences were calculated and evaluated. Out of 22,914 designs with favorable stability energy, 301 candidates contained at least two pre-existing immunity-related epitopes and had promising immunogenic potential. The benchmark tests showed that, although the epitope restraints were not included in the scoring function of EvoDesign, the top S protein design successfully recovered 31 out of the 32 major histocompatibility complex (MHC)-II T cell promiscuous epitopes in the native S protein, where two epitopes were present in all seven human coronaviruses. Moreover, the newly designed S protein introduced nine new MHC-II T cell promiscuous epitopes that do not exist in the wildtype SARS-CoV-2. These results demonstrated a new and effective avenue to enhance a target protein’s immunogenicity using rational protein design, which could be applied for new vaccine design against COVID-19 and other pathogens.
1. Introduction
The current Coronavirus Disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in over 77 million confirmed cases and over 1.7 million deaths globally as of December 24, 2020, according to the World Health Organization [1]. Tremendous efforts have been made to develop effective and safe vaccines against this viral infection. The Pfizer-BioNTech BNT162b2 mRNA vaccine showed 95% effectiveness in preventing COVID-19 [2], and the Moderna mRNA-1273 induced strong immune responses among recipients between the age of 18 and 55 during phase III clinical trials [3]. The FDA issued an Emergency Use Authorization for the mRNA-1273 and BNT162b2 mRNA vaccines in record time. On the other hand, the Inovio INO-4800 DNA vaccine not only showed protection from the viral infection in rhesus macaques but was also reported to induce long-lasting memory [4]. In addition to these two vaccines, there are over a hundred COVID-19 vaccines currently in clinical trials, including other types of vaccines such as the Oxford-AstraZeneca adenovirus-vectored vaccine (ChAdOx1 nCoV-19) [5], CanSino’s adenovirus type-5 (Ad5)-vectored COVID-19 vaccine [6], and Sinovac’s absorbed COVID-19 (inactivated) vaccine (ClinicalTrials.gov Identifier: NCT04456595). Among all the vaccines, a vast majority of them select the spike glycoprotein as their primary target.
The SARS-CoV-2 Spike (S) protein is a promising vaccine target, and many clinical studies reported anti-S protein neutralizing antibodies in COVID-19 recovered patients [7]. After the SARS outbreak in 2003 [8], clinical studies reported neutralizing antibodies targeting the SARS-CoV S protein [9], [10], which was selected as the target of vaccine development [11], [12]. Since SARS-CoV-2 shares a high sequence identity with SARS-CoV [13], and both viruses utilize the attachment of the S protein to the human angiotensin-converting enzyme 2 (ACE2) receptor to invade host cells, neutralization of the SARS-CoV-2 S protein could induce protection in COVID-19 vaccine development [14]. Many computational studies utilizing reverse vaccinology and immuno-informatics reported the S protein to be a promising vaccine antigen [15], [16], [17], and clinical studies identified anti-S protein neutralizing antibodies in patients that have recovered from COVID-19 [18], [19], [20]. The cryo-EM structures of the S protein [21] and the neutralizing antibodies that bind to the S protein [22], [23] were determined. Besides neutralizing antibodies, studies have also shown the importance of the CD4 T cell response in the control of SARS-CoV-2 infection and possible pre-existing immunity in healthy individuals without exposure to SARS-CoV-2 [7], [24], [25]. Kalita et al. has proposed a multi-peptide subunit-based epitope vaccine that is comprised of B cell, helper T cell, and cytotoxic T cell epitopes [26]. Overall, successful vaccination is likely linked to a robust and long-term humoral response to the SARS-CoV-2 S protein, which could be further enhanced by the rational structural design of the protein.
Structural vaccinology has shown success in improving vaccine candidates’ immunogenicity through protein structural modification. The first proof-of-concept was achieved by fixing the conformation-dependent neutralization-sensitive epitopes on the fusion glycoprotein of the respiratory syncytial virus [27]. A similar strategy has been applied to SARS-CoV-2 to conformationally control the S protein’s receptor-binding domain (RBD) domain between the “up” and “down” configurations [28] and stabilizing the protein in its prefusion form [79] to induce immunogenicity. Besides the structure-based rational design approaches, directed evolution, which is classified as an irrational method, has also been widely used in diverse fields, such as enzyme engineering [29], protein-RNA interaction [30], and COVID-19 therapeutic strategies [31]. The directed evolution usually first applies random mutagenesis to generate a large pool of variants, followed by screening for candidates with the preferred properties using high-throughput strategies. The advantage of directed evolution is that it works well without structural information. However, once a high-quality protein structure can be obtained either from the experimental determination or computational prediction, the structure-based approach is more suitable as it can efficiently explore a much larger sequence/conformational space using computer programs, yielding a few potential candidates for further screening and validation, which is more time-, money-, and labor-saving compared to a typical directed evolution process.
In this study, we extended structural vaccinology to a structure-based computational design of the SARS-CoV-2 S protein. Briefly, we used a protein design approach, EvoDesign [32], to generate multiple stable S protein variants without perturbing the surface amino acids to maintain the same B cell epitope profile. Meanwhile, we introduced mutations to the residues buried inside the S protein so that more major histocompatibility complex (MHC)-II T cell epitopes would be added into the newly designed S protein to potentially induce a stronger immune response. Finally, we evaluated the computationally designed protein candidates and compared them to the native S protein.
2. Materials and methods
2.1. Computational redesign of SARS-CoV-2 S protein
Fig. 1 illustrates the workflow for redesigning the SARS-CoV-2 S protein to improve its immunogenic potential for vaccine design. The full-length structure model (1,273 amino acids for the S monomer) of SARS-CoV-2 S assembled by C-I-TASSER [33] was used as the template for fixed-backbone protein sequence design using EvoDesign [32]. Although the cryo-EM structure for SARS-CoV-2 S is available (PDB ID: 6VSB) [21], it contains a large number of missing residues, and therefore, the full-length C-I-TASSER model was used for S protein design instead. The C-I-TASSER model used the cryo-EM density map to assemble the individual domain models and to refine the structure. The model showed a high similarity to the cryo-EM structure with a TM-score [34] of 0.87 and root-mean-square deviation (RMSD) of 3.4 Å in the commonly aligned regions, indicating a good model quality. The residues in the S protein were categorized into three groups: core, surface, and intermediate [35], according to their solvent accessible surface area ratio (SASAr). Specifically, SASAr is defined as the ratio of the absolute SASA of a residue in the structure to the maximum area of the residue in the GXG state [36], where X is the residue of interest and G is a glycine residue. The most extended GXG conformation measures the maximum exposure degree of the residue X in the solvated environment taking into account the local protein backbone. The SASAr ratios were calculated using the ASA web-server (http://cib.cf.ocha.ac.jp/bitool/ASA/), where the maximum area of each of the 20 canonical amino acid residues is provided. The core and surface residues were defined by us as those with SASAr < 5% and > 25%, respectively, while the other residues were regarded as intermediate. Since the surface residues may be involved in the interactions with other proteins (e.g., the formation of the S homotrimer, S-ACE2 complex, and S-antibody interaction) and may partially constitute the B cell epitopes, these residues were excluded from design, and more rigorously, their side-chain conformations were kept constant as well. Additionally, the residues that may form B cell epitopes reported by Grifoni et al. [16] were also fixed. The remaining core residues were subjected to design, allowing amino acid substitution, whereas the intermediate residues were repacked with conformation substitution. Specifically, 243, 275, and 755 residues were designed, repacked, and fixed, respectively; a list of these residue positions is shown in Supplementary Table S1. The 243 designable core residues were also compared to the global S protein mutations (global frequencies > 0.001) recorded in the GISAID database (as of December 7, 2020) [37], [38]. These residues were also evaluated for their intrinsic disorder predisposition based on the reported disorder regions in the DisProt database [39]. The corresponding Jensen-Shannon Divergence (JSD) scores (higher scores indicate greater conservation) of these core residues residing within the disordered regions were reported [40]. During protein design, the evolution term in EvoDesign was turned off as this term would introduce evolutionary constraints on the sequence simulation search, which were not needed for this design [41]; therefore, only the physical energy function, EvoEF2 [35], was used for design scoring to broaden sequence diversity and help to identify more candidates with increased immunogenicity. In previous studies, EvoEF2 has been appropriately utilized to model the binding interactions between the SARS-CoV-2 S-RBD and a large number of ACE2 orthologs to identify the zoonotic origin of this novel coronavirus [42] and to design multiple anti-SARS-CoV-2 peptide therapeutics [41]. We performed 20 independent design simulations and collected all the simulated sequence decoys. A total of 5,963,235 sequences were obtained, and the best-scoring sequence had stability energy of −4100.97 EvoEF2 energy unit (EEU). A set of 22,914 non-redundant sequences that were within a 100 EEU window of the lowest energy and had > 5% of the design residues mutated were retained for further analysis (Fig. 1). We also utilized another popular protein design software, Rosetta [43], to generate 1,000 low-energy S variants using the “fixbb” protocol due to lower computational efficiency. The same surface-intermediate-core criterion was applied to the Rosetta protein design process. The EvoDesign and Rosetta designs were then analyzed and compared to examine the advantages/limitations of EvoDesign designs.
Fig. 1.
The workflow for designing and screening immunogenicity-enhanced SARS-CoV-2 S proteins. The procedure started by defining the full-length SARS-CoV-2 native S protein into surface, intermediate, and core residues. This information was then fed into EvoDesign to generate structurally stable designs that introduce mutations to the core residues while keeping the surface conformation unchanged. The output design candidates from EvoDesign were then evaluated based on their immunogenic potential. The top ten candidates were also compared and evaluated in comparison to the native S protein.
2.2. MHC-II T cell epitope prediction and epitope content score calculation
The full-length S protein sequence was divided into 15-mers with ten amino-acid overlaps. For each 15-mer, the T cell MHC-II promiscuous epitopes were predicted using NetMHCIIpan v3.2 [44]. In brief, the percentile ranks of an epitope binding to each of the seven MHC-II alleles (i.e., HLA-DRB1*03:01, HLA-DRB1*07:01, HLA-DRB1*15:01, HLA-DRB3*01:01, HLA-DRB3*02:02, HLA-DRB4*01:01, and HLA-DRB5*01:01) were calculated, where the percentile rank was generated by comparing the 15-mer predicted binding affinity to the MHC-II molecule against that of a large set of similarly sized peptides randomly selected from the SWISS-PROT database [45]. An epitope was considered a promiscuous epitope if the median percentile rank was ≤ 20% by binding the 15-mer to any of the seven MHC-II alleles [46]. The selection of these seven MHC-II alleles aimed to predict the dominant MHC-II T cell epitopes across different ethnicities and HLA polymorphisms [47]. The MHC-II promiscuous epitopes of the native SARS-CoV-2 S protein (QHD43416) predicted using this method were also validated and compared to the dominant T cell epitopes mapped by Grifoni et al. [16]. In brief, Grifoni et al. mapped the experimentally verified SARS-CoV T cell epitopes reported in the Immune Epitope Database (IEDB) database, which includes experimentally verified T cell MHC-II epitope data, to the SARS-CoV-2 S protein based on sequence homology and reported as the dominant T cell epitopes [46]. The epitope content score (ECS) for a full-length S protein was calculated as the average value of the median percentile ranks for all the 15-mers spanning the whole sequence.
2.3. Human epitope similarity and human similarity score calculation
The human proteome included 20,353 reviewed (Swiss-Prot) human proteins downloaded from Uniprot (as of July 1, 2020) [48]. A total of 261,908 human MHC-II T cell promiscuous epitopes were predicted, as described above. The human epitope similarity between a peptide of interest (e.g., a peptide of the S protein) and a human epitope was then calculated using a normalized peptide similarity metric proposed by Frankild et al. [49]. In brief, the un-normalized peptide similarity score, , was first determined by the BLOSUM35 matrix [50] for all the positions between a target peptide (y) and a human epitope (x), which was subsequently normalized using the minimum and maximum similarity scores for the human epitope (Eq. (1)). The maximum and minimum similarity scores were determined from a range of similarity scores between a human epitope and all of its possible amino acid substitutions. Finally, the normalized similarity score of a 15-mer peptide was calculated by comparing it to all the predicted human MHC-II T cell promiscuous epitopes. The human similarity score (HSS) of the full-length S protein was calculated by averaging the human epitope similarity of all the 15-mers.
| (1) |
2.4. Pre-existing immunity evaluation of the designed proteins
The pre-existing immunity of the designed proteins was evaluated and compared to that of the native S protein of seven human coronaviruses (HCoVs) (i.e., SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1). The sequences of the seven HCoV S proteins were downloaded from Uniprot [48] (Table S2), and the MHC-II T cell epitopes were predicted as described above. The conserved epitopes were determined by the IEDB epitope clustering tool [45] and aligned using SEAVIEW [51].
2.5. Foldability assessment of the designed proteins
Since EvoDesign only produces a panel of mutated sequences, it is important to examine if the designed sequences can fold into the desired structure that the native S protein adopts. To examine their foldability, we used C-I-TASSER to model the structure of the designed sequences, where the structural similarity between the native and designed S proteins was assessed by TM-score [52]. Here, C-I-TASSER is a recently developed protein structure prediction program, which constructs full-length structure folds by assembling fragments threaded from the PDB, under the guidance of deep neural-network learning-based contact maps [53], [54]. The ectodomain of the S homotrimers and the functional domains including the N-terminal domain (NTD), receptor-binding domain (RBD), fusion peptide (FP), heptapeptide repeat sequence 1 (HR1), and connector domain (CD) [28], [55] were visualized via PyMOL [56]. Sequence logo plots for the top ten and worst ten S protein designs were also generated [57]. The multiple sequence alignment of the top four EvoDesign S protein candidates with balanced ECS and HSS were aligned to the native S protein using SEAVIEW [51].
2.6. Molecular dynamics (MD) simulation
The extracellular domain (amino acids 1-1146) of the trimeric wildtype S and the top design (i.e., Design-10705, see Results) was subjected to MD simulation using GROMACS [58] with the CHARMM36 force field [59]. The initial full-length (amino acids 1-1273) trimers were built by C-I-TASSER and residues 1147-1273 were deleted; glycosylation was not considered during structure modeling and the simulation. In each simulation case, a dodecahedron box was constructed with a distance of 10 Å from the solute, and TIP3P [60] water molecules were filled into the box. The system was then neutralized by the addition of an appropriate number of Na+ or Cl− ions. After the system was assembled, energy minimization was carried out using steepest descent minimization with a maximum force of 10 kJ/mol. The system was then equilibrated at 300 K using 100 ps NVT simulations and 100 ps NPT simulations with position restraints (1000 kJ/mol) on the heavy atoms of the protein. After the two equilibration phases, the system was well-equilibrated at the desired temperature and pressure. Unconstrained production MD was then carried out at 300 K for 50 ns as suggested in similar MD simulation studies [61], [62]. The LINCS [63] algorithm was used to restrain the hydrogen bonds. Non-bonded interactions were truncated at 12 Å, and the Particle Mesh Ewald [64] method was utilized for long-range electrostatic interactions. The velocity-rescaling thermostat [65] and Parrinello-Rahman barostat [66] were used to couple the temperature and pressure, respectively. About 5,000 snapshots were saved with a time interval of 10 ps and utilized for further analysis using the built-in GROMACS command-line tools.
3. Results
The epitope content score (ECS) and human similarity score (HSS) of the S proteins from seven HCoV strains (severe HCoV: SARS-CoV-2, SARS-CoV, and MERS-CoV; mild HCoV: HCoV-229, HCoV-HKU1, HCoV-NL63, and HCoV-OC43) were computed. The ECS for the severe HCoV S proteins (mean = 49.3, standard deviation (SD) = 24.7) was significantly different (p = 0.0016, Mann-Whitney) from that of the mild ones (mean = 45.8, SD = 24.5). In terms of HSS, the severe HCoV S proteins (mean = 0.640, SD = 0.03) tended to be less self-like (p = 0.097, Mann-Whitney) than the mild ones (mean = 0.642, SD = 0.03). Overall, it was shown that both ECS and HSS might be used as indicators of the immunogenic potential of the designed S proteins.
On the other hand, previous studies suggested the potential role of pre-existing immunity in fighting COVID-19 [7], [24], [25]. Therefore, the predicted MHC-II T cell promiscuous epitopes of the SARS-CoV-2 S protein were compared to those from the other six HCoVs. There were two SARS-CoV-2 predicted MHC-II T cell promiscuous epitopes, which were also present on all of the seven HCoV S proteins (Fig. 2) and could be potentially linked to pre-existing immunity. Therefore, the designs were subsequently filtered based on the availability of these pre-existing immunity-related epitopes (Fig. 1). In particular, the SARS-CoV-2 promiscuous epitope S816-D830 overlapped with the dominant B cell epitope F802-E819 reported by Grifoni et al. [16].
Fig. 2.
The two pre-existing immunity-related SARS-CoV-2 MHC-II T cell promiscuous epitopes. The first SARS-CoV-2 promiscuous epitope is located within residues 816–830 (indexed by SARS-CoV-2).
EvoDesign generated a total of 22,914 low-energy S protein designs, in which 243 core residues were subjected to substitution (see Methods for details). As SARS-CoV-2 has been reported to have substantial mutations in its genome [67], it is important to compare the EvoDesign mutations to the natural mutations (global mutation frequency of > 0.001) reported by GISAID (Table S3). There were two EvoDesign core residues (D80 and S98) that also had natural mutations, and these two core residues had different mutation rates in EvoDesign in comparison to the natural infection. Specifically, for the D80 core residue, the natural mutation frequency (D80Y) was 0.005, but the EvoDesign mutations, D80N, D80A, and D80S, had frequencies of 0.149, 0.106, and 0.003, respectively. For S98, the natural mutation (S98F) frequency was 0.007, but EvoDesign mutations S98T and S98A had frequencies of 0.837 and 0.008, respectively. To further investigate whether the EvoDesign candidates’ mutations were related to the intrinsic disorder predisposition, the 243 core residues were aligned to the DisProt database [39] and the aligned residues’ sequence conservations were evaluated. Twenty core residues were annotated as intrinsically disordered in DisProt, but these residues showed relatively high levels of conservation, with JSD scores ranging from 0.76 to 0.85 (Table S4), in the top 10 designs.
Among the 22,914 designs with relatively low (favorable) stability energy, 19,063 candidates that contained the two pre-existing immunity-related epitopes were ranked based on ECS and HSS (Fig. 3A). Using the ECS and HSS of the native SARS-CoV-2 S as the cutoff, we obtained 301 candidates with better immunogenic potential (i.e., lower ECS and HSS) (Fig. 3B). Ten candidates with balanced ECS and HSS were selected and evaluated (Table 1, full-length sequences in Table S5). The EvoDesign energy and sequence identity of all designs were plotted, and the top 10 designs were highlighted (Fig S1). The S protein variants generated by EvoDesign had consistently better ECS in comparison to the Rosetta designs, although the latter had a better HSS score (Fig S2). All 1,000 Rosetta designs had higher ECS (thus worse immunogenic potential) than the native S, whereas EvoDesign was able to produce a few designs with both lower ECS and HSS, affirming EvoDesign’s ability to design vaccine candidates with better T cell promiscuous epitopes.
Fig 3.
The epitope content score (ECS) and human similarity score (HSS) for designed S proteins. (A) All 22,914 designs. Each design is shown as a blue dot, whereas the native SARS-CoV-2 S was plotted as a black diamond marker. The dashed-line box defines the 301 candidates with both lower ECS and HSS scores than the native. (B) The shaded area contains the top ten candidates (highlighted by red circles) with balanced ECS and HSS scores. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Summary of the features for the top 10 designs. The table is ranked based on the EvoDesign energy score (from low to high) except for the native S protein.
| Design ID | PEC | RECa | ECS | HSS | EE (EEU) | RMSD (Å)b | TM-scoreb | SI (%) |
|---|---|---|---|---|---|---|---|---|
| 10,705 | 40 | 31 | 48.78 | 0.6394 | −4051.21 | 3.45 | 0.931 | 94.9 |
| 10,763 | 40 | 31 | 48.80 | 0.6394 | −4051.04 | 3.06 | 0.944 | 95.0 |
| 12,865 | 40 | 31 | 48.76 | 0.6396 | −4044.99 | 3.14 | 0.939 | 95.0 |
| 19,356 | 41 | 30 | 48.44 | 0.6399 | −4020.14 | 3.12 | 0.929 | 94.7 |
| 20,348 | 38 | 30 | 48.99 | 0.6390 | −4014.74 | 3.33 | 0.929 | 95.4 |
| 20,467 | 38 | 30 | 48.97 | 0.6391 | −4014.10 | 4.32 | 0.901 | 95.4 |
| 20,671 | 37 | 28 | 48.83 | 0.6395 | −4013.03 | 3.36 | 0.940 | 94.7 |
| 22,676 | 36 | 28 | 48.37 | 0.6399 | −4001.70 | 3.35 | 0.939 | 95.0 |
| 22,769 | 38 | 28 | 48.51 | 0.6398 | −4001.11 | 3.27 | 0.937 | 95.0 |
| 22,869 | 38 | 28 | 48.55 | 0.6398 | −4000.23 | 3.24 | 0.919 | 94.7 |
| Native | 32 | – | 49.61 | 0.6401 | – | – | – | – |
PEC: Promiscuous Epitope Count; REC: Recovered Epitope Count; ECS: Epitope Content Score; HSS: Human Similarity Score; EE: EvoDesign Energy (in EEU, EvoEF2 energy unit); RMSD: Root-Mean-Square Deviation; TM: TM-score; SI: Sequence identity.
: The number of predicted promiscuous epitopes in designs that overlap with those in the native S protein.
: The RMSD and TM-score compared to the C-I-TASSER model of the native S protein.
The multiple sequence alignment of the top four candidates showed that 88 of the 243 core residues were mutated at least once (Fig. 4). There were 32 core residues substituted to the same amino acids (R34T, V62I, I100M, R102Q, C136T, V143T, Y145S, E191V, T250A, Y279F, R328K, V341I, V350S, W353A, D420A, Y423M, C432V, S438V, V512I, T523N, T599L, S673T, N777T, S875A, T881I, L916Y, C1043A, F1052H, S1055A, C1241G, S1242G, and C1248T) in all top four designs. Additionally, the top ten and worst ten designs were also plotted to infer functionally important mutations to enhance immunogenicity (Fig S3). Specifically, there were 12 core residues in both the top-scoring and worst-scoring designs that were substituted to the same amino acids in comparison to the native S protein (V62I, C136T, V143T, Y145S, E191V, R328K, V341I, D420A, C432V, T599L, S1055A, and C1241G). In particular, two remained unmutated in the top-scoring designs but were mutated in the worst-scoring designs (Y265W and V267T), suggesting mutations of these two residues might result in reduced immunogenicity.
Fig. 4.
The multiple sequence alignment of the top four designed S proteins in comparison to the native SARS-CoV-2 S protein. The four EvoDesign S proteins (Design-10705, 10763, 12865, and 19356) were selected based on their high structural similarity to the native S protein and promising immunogenic potential (in terms of promiscuous epitope count, ECS, and HSS scores). The solid red boxes highlight the core residues that were subjected to mutations by EvoDesign. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Design-10705 was overall the best candidate with high structural similarity to the native S protein and good immunogenic potential (in terms of promiscuous epitope count, ECS, and HSS scores) amongst the top ten candidates. The candidate Design-10705 had a 93.9% sequence identity to the native S protein with a TM-score of 0.931 and an RMSD of 3.45 Å to the C-I-TASSER model of the native S protein. The homo-trimer 3D structure of Design-10705 was visualized and compared to the S protein C-I-TASSER and cryo-EM structural models (Fig. 5). In terms of immunogenicity, it had the second-highest number of promiscuous epitopes. Table 2 shows the complete MHC-II T cell epitope profile of Design-10705. There were 32 predicted promiscuous epitopes in the native S protein (Table S6), and 31 of them were recovered in Design-10705. The two pre-existing immunity-related epitopes, V991-Q1005 and S816-D830, were both recovered in the new design. Besides these two epitopes, there were 19 epitopes identical to the native S protein epitopes, while ten epitopes had at least one mutation in Design-10705. Compared with the native S protein, the only missing MHC-II epitope in Design-10705 was V911-N926, which was predicted to have reduced binding affinity to HLA-DRB1*03:01 and HLA-DRB4*01:01. Critically, this design introduced nine new MHC-II T cell promiscuous epitopes, which could potentially induce a stronger immune response with minimal perturbation compared to the native S protein.
Fig. 5.
The 3D structures of C-I-TASSER S protein trimer (A), cryo-EM trimer (B), Design-10705 trimer (C), and Design-10705 monomer (D). The ectodomain of Design-10705 was modeled using C-I-TASSER. Both the homo-trimer and monomer of Design-10705 are rendered. The NTD, RBD, HR1, FP, and CD domains are also highlighted in the Design-10705 monomer. The mutations introduced in Design-10705 are shown in red spheres. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2.
The predicted promiscuous MHC-II T cell epitopes of Design-17050.
| Epitope | Start | End | Median Percentile Rank | Comment |
|---|---|---|---|---|
| VQLDRLITGRLQSLQ | 991 | 1005 | 17 | Pre-existing immunity-related epitopes |
| SFIEDLLFNKVTLAD | 816 | 830 | 16 | |
| VYYPDKVFRSSVLHS | 36 | 50 | 11 | Identical epitopes to native S protein |
| KVFRSSVLHSTQDLF | 41 | 55 | 17 | |
| SLLIVNNATNVVIKV | 116 | 130 | 6.5 | |
| EFRVYSSANNCTFEY | 156 | 170 | 18 | |
| FKIYSKHTPINLVRD | 201 | 215 | 14 | |
| SVLYNSASFSTFKCY | 366 | 380 | 18 | |
| YLYRLFRKSNLKPFE | 451 | 465 | 5.7 | |
| SIIAYTMSLGAENSV | 691 | 705 | 4.7 | |
| YGSFCTQLNRALTGI | 756 | 770 | 19 | |
| LLFNKVTLADAGFIK | 821 | 835 | 17 | |
| CAQKFNGLTVLPPLL | 851 | 865 | 19 | |
| GAALQIPFAMQMAYR | 891 | 905 | 18 | |
| IPFAMQMAYRFNGIG | 896 | 910 | 3.7 | |
| QMAYRFNGIGVTQNV | 901 | 915 | 19 | |
| TLVKQLSSNFGAISS | 961 | 975 | 14 | |
| TYVTQQLIRAAEIRA | 1006 | 1020 | 20 | |
| QLIRAAEIRASANLA | 1011 | 1025 | 12 | |
| AEIRASANLAATKMS | 1016 | 1030 | 7.9 | |
| REGVFVSNGTHWFVT | 1091 | 1105 | 9.4 | |
| LPFFSNITWFHAIHV | 56 | 70 | 7.1 | Mutated epitopes |
| VFVYKNIDGYFKIYS | 191 | 205 | 13 | |
| IGINITRFMTIRASS | 231 | 245 | 6.2 | |
| TRFMTIRASSRSYLA | 236 | 250 | 1.2 | |
| YVGYLQPRTFLLKFN | 266 | 280 | 12 | |
| SNFRVQPTETIVKFP | 316 | 330 | 14 | |
| IFNATRFASSYAANR | 341 | 355 | 13 | |
| RFASSYAANRKRISN | 346 | 360 | 17 | |
| VILSFELLHAPANVC | 511 | 525 | 14 | |
| KLIANQFNSAIGKLQ | 921 | 935 | 17 | |
| NITWFHAIHVSGTNG | 61 | 75 | 20 | New epitopes |
| FNDGVYFAATLKTNM | 86 | 100 | 14 | |
| GKQGNFKNLRVFVYK | 181 | 195 | 13 | |
| LVDLPIGINITRFMT | 226 | 240 | 20 | |
| GVVIAWNVNNLDAKV | 431 | 445 | 11 | |
| TDEMIAQYTAALLAG | 866 | 880 | 19 | |
| VVNQLAQALNTLVKQ | 951 | 965 | 19 | |
| GAISSVMNDILSRLD | 971 | 985 | 20 | |
| VFLHVNLVPAQEKNF | 1061 | 1075 | 16 | |
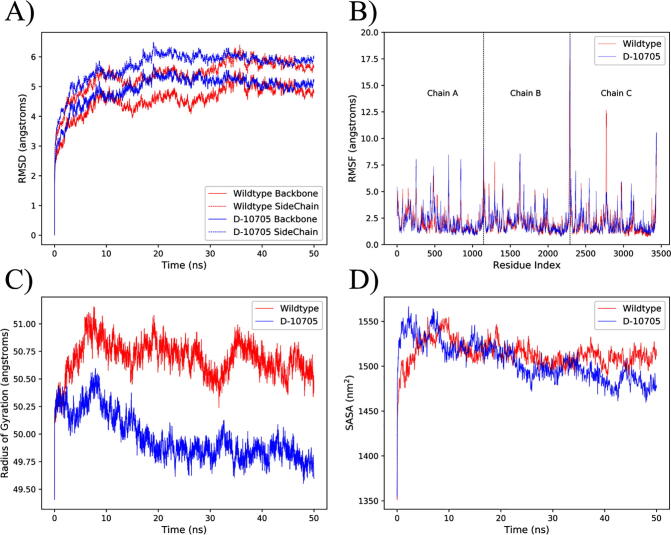
One concern is that the top design, Design-10705, might lose the desired structure and protein function due to reduced stability caused by redesigning the core regions. To examine this concern, 50-ns MD simulations were carried out to compare the stability and flexibility of Design-10705 and the native S. As shown in Fig. 6A, Design-10705 and the wildtype showed similar RMSD shifts for both the backbones and side-chains after convergence at about 30 ns. The root-mean-square fluctuation (RMSF) measurement showed that the two proteins exhibited similar fluctuating profiles and thus comparable flexibility (Fig. 6B). Moreover, the radius of gyration and solvent-accessible surface area (SASA) as a function of simulation time were also analyzed for the two proteins. Design-10705 showed a slightly smaller radius of gyration (Fig. 6C) and a smaller SASA than the wildtype S (Fig. 6D), indicating that Design-10705 had a slightly more compact structure. Taken together, Design-10705 is expected to be sufficiently stable with the desired biological function (e.g., increased immunogenicity).
Fig. 6.
Analysis of molecular dynamics simulation results for the wildtype S and Design-10705. Design-10705 is denoted as D-10705 in the plot. (A) Root-mean-square deviation (RMSD) for the wildtype S and D-10705 backbone and side-chains. (B) Root-mean-square fluctuation (RMSF) for all the residues in the trimeric protein. The three chains are separated by black dashed lines (Chain A: amino acids 1–1146; Chain B: 1147–2292; Chain C: 2293–3438). (C) Radius of gyration. (D) Solvent-accessible surface area (SASA).
4. Discussion
The subunit, DNA, and mRNA vaccines are typically considered to be safer but often induce weaker immune responses than the live-attenuated and inactivated vaccines. Although the addition of adjuvant or better vaccination strategies can compensate for the immunogenicity, the addition of new epitopes to the antigen provides an alternative way to induce stronger immune responses [68], [69]. During the protein design process, we applied design constraints so that the surface conformation, and in particular, B cell epitopes of the designed S protein variants were unchanged. For the designed S proteins with at least 5% of the core residues mutated, the immunogenicity potential of these candidates was evaluated and was structurally compared to the native S protein. The top candidate (Design-10705) recovered 31 out of 32 MHC-II promiscuous epitopes, and the two pre-existing immunity-related epitopes (V991-Q1005 and S816-D830) were present in the design. In addition to the 31 recovered epitopes, Design-10705 also introduced nine new MHC-II promiscuous epitopes with the potential to induce stronger CD4 T cell response. MD analysis of Design-10705 and the native S protein showed that the two proteins shared similar stability and flexibility (Fig. 6). Overall, the newly designed S protein should preserve the native S protein’s structure and function with enhanced immunogenicity.
The concept of manipulating epitopes to decrease the immunogenicity has been applied to therapeutic proteins. King at el. disrupted the MHC-II T cell epitopes in GFP and Pseudomonas exotoxin A using the Rosetta protein design protocol [70], [71]. The EpiSweep program was also applied to structurally redesign bacteriolytic enzyme lysostaphin as an anti-staphylococcal agent with reduced immunogenicity to the host [72], [73]. In this study, a similar strategy, but to improve immunogenicity, was applied to redesign the SARS-CoV-2 S protein as an enhanced vaccine candidate; specifically, we aimed to increase immunogenicity by introducing more MHC-II T cell promiscuous epitopes to the protein without reducing the number of B cell epitopes.
The addition of epitopes to induce stronger immune responses has been previously applied to develop H7N9 vaccines. The H7N9 hemagglutinin (HA) vaccine-elicited non-neutralizing antibody responses in clinical trials [74], [75]. Rudenko et al. reported that there were fewer CD4 T cell epitopes found in H7N9 HA in comparison to the seasonal H1 and H3 HA proteins [76]. Based on this finding, Wada et al. improved the H7N9 vaccine by introducing a known H3 immunogenic epitope to the H7 HA protein without perturbing its conformation, which resulted in an over 4-fold increase in the HA-binding antibody response [68]. However, the number of epitopes is not the only factor that influences protective immunity. Studies have reported that CD8 T cell epitopes might induce regulatory T cell responses [49], [77], and pathogens adapted to include CD4 and CD8 epitopes with high similarity to human peptides as a means to suppress host immunity for its survival [78]. Therefore, we examined the significance of ECS and HSS in the context of mild versus severe forms of HCoV infection and then utilized these two scores to evaluate the designed S protein candidates.
The computational design of the SARS-CoV-2 S protein could be coupled with some other structural modifications for a more rational structure-based vaccine design. The present study aims to introduce new epitopes to the S protein while keeping the surface residues unchanged to minimize the structural change of the designed proteins, and according to the protein structure prediction results, the designed candidates were predicted to be structurally similar to the native S protein (Table 1 & Fig. 5). The structural modifications performed on the native S protein, such as stabilizing the protein in its prefusion form [79], or fixing the RBD in the “up” or “down” state [28], could still be applied to the final candidate in this study. The combination of these structural vaccinology techniques into the current pipeline could further enhance the immunogenicity of the S protein as a vaccine target. However, a major limitation of the present study is the wet-lab experimental validation of the designed proteins. First, the newly designed protein sequences need to be folded properly with a structure comparable to that of the native S protein. Second, the capability of the newly added epitopes for binding MHC-II molecules and subsequently inducing immune responses needs to be validated. Finally, these candidates should be tested for their protectiveness and safety in animal models.
Overall, this study presents a strategy to improve the immunogenicity and antigenicity of a vaccine candidate by manipulating the MHC-II T cell epitopes through computational protein design. In the current settings, the immunogenicity evaluation was carried out after the standard protein design simulations with EvoDesign. In the future, the assessment of the immunogenic potential could be incorporated into the protein design process so that the sequence decoy generated at each step will be guided by balancing both the protein stability and immunogenicity. Moreover, with proper prior knowledge of known epitopes (e.g., both MHC-I and MHC-II from the pathogen proteome), it is also possible to create a chimeric protein, which integrates epitopes from antigens other than the target protein.
Financial Disclosure Statement
YH is supported by the National Institute of Allergy and Infectious Diseases (AI081062). YZ is supported by the National Institute of Allergy and Infectious Diseases (AI134678), the National Institute of General Medical Sciences (GM136422, S10OD026825), and the National Science Foundation (IIS1901191, DBI2030790). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Author contributions
YZ and YH conceived and designed the project. EO and XH performed studies on protein design and analyses. RP participated in the discussion. EO drafted the manuscript. All authors performed result interpretation, edited, and approved the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.12.039.
Contributor Information
Yang Zhang, Email: zhng@umich.edu.
Yongqun He, Email: yongqunh@med.umich.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. 2020 [cited 24 Dec 2020]. Available: https://covid19.who.int/.
- 2.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A.B., Flach B., Lin B.C., Doria-Rose N.A., O’Dell S., Schmidt S.D., Corbett K.S., Swanson P.A., Padilla M., Neuzil K.M., Bennett H., Leav B., Makowski M., Albert J., Cross K., Edara V.V., Floyd K., Suthar M.S., Martinez D.R., Baric R., Buchanan W., Luke C.J., Phadke V.K., Rostad C.A., Ledgerwood J.E., Graham B.S., Beigel J.H. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N Engl J Med. 2020;383(25):2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel A, Walters J, Reuschel EL, Schultheis K, Parzych E, Gary EN, et al. Intradermal-delivered DNA vaccine provides anamnestic protection in a rhesus macaque SARS-CoV-2 challenge model. bioRxiv [Preprint] 2020. Available: https://www.biorxiv.org/content/10.1101/2020.07.28.225649v1. [DOI] [PMC free article] [PubMed]
- 5.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., Dold C., Faust S.N., Finn A., Flaxman A.L., Hallis B., Heath P., Jenkin D., Lazarus R., Makinson R., Minassian A.M., Pollock K.M., Ramasamy M., Robinson H., Snape M., Tarrant R., Voysey M., Green C., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Pollard A.J., Aboagye J., Adams K., Ali A., Allen E., Allison J.L., Anslow R., Arbe-Barnes E.H., Babbage G., Baillie K., Baker M., Baker N., Baker P., Baleanu I., Ballaminut J., Barnes E., Barrett J., Bates L., Batten A., Beadon K., Beckley R., Berrie E., Berry L., Beveridge A., Bewley K.R., Bijker E.M., Bingham T., Blackwell L., Blundell C.L., Bolam E., Boland E., Borthwick N., Bower T., Boyd A., Brenner T., Bright P.D., Brown-O'Sullivan C., Brunt E., Burbage J., Burge S., Buttigieg K.R., Byard N., Cabera Puig I., Calvert A., Camara S., Cao M., Cappuccini F., Carr M., Carroll M.W., Carter V., Cathie K., Challis R.J., Charlton S., Chelysheva I., Cho J.-S., Cicconi P., Cifuentes L., Clark H., Clark E., Cole T., Colin-Jones R., Conlon C.P., Cook A., Coombes N.S., Cooper R., Cosgrove C.A., Coy K., Crocker W.E.M., Cunningham C.J., Damratoski B.E., Dando L., Datoo M.S., Davies H., De Graaf H., Demissie T., Di Maso C., Dietrich I., Dong T., Donnellan F.R., Douglas N., Downing C., Drake J., Drake-Brockman R., Drury R.E., Dunachie S.J., Edwards N.J., Edwards F.D.L., Edwards C.J., Elias S.C., Elmore M.J., Emary K.R.W., English M.R., Fagerbrink S., Felle S., Feng S., Field S., Fixmer C., Fletcher C., Ford K.J., Fowler J., Fox P., Francis E., Frater J., Furze J., Fuskova M., Galiza E., Gbesemete D., Gilbride C., Godwin K., Gorini G., Goulston L., Grabau C., Gracie L., Gray Z., Guthrie L.B., Hackett M., Halwe S., Hamilton E., Hamlyn J., Hanumunthadu B., Harding I., Harris S.A., Harris A., Harrison D., Harrison C., Hart T.C., Haskell L., Hawkins S., Head I., Henry J.A., Hill J., Hodgson S.H.C., Hou M.M., Howe E., Howell N., Hutlin C., Ikram S., Isitt C., Iveson P., Jackson S., Jackson F., James S.W., Jenkins M., Jones E., Jones K., Jones C.E., Jones B., Kailath R., Karampatsas K., Keen J., Kelly S., Kelly D., Kerr D., Kerridge S., Khan L., Khan U., Killen A., Kinch J., King T.B., King L., King J., Kingham-Page L., Klenerman P., Knapper F., Knight J.C., Knott D., Koleva S., Kupke A., Larkworthy C.W., Larwood J.P.J., Laskey A., Lawrie A.M., Lee A., Ngan Lee K.Y., Lees E.A., Legge H., Lelliott A., Lemm N.-M., Lias A.M., Linder A., Lipworth S., Liu X., Liu S., Lopez Ramon R., Lwin M., Mabesa F., Madhavan M., Mallett G., Mansatta K., Marcal I., Marinou S., Marlow E., Marshall J.L., Martin J., McEwan J., McInroy L., Meddaugh G., Mentzer A.J., Mirtorabi N., Moore M., Moran E., Morey E., Morgan V., Morris S.J., Morrison H., Morshead G., Morter R., Mujadidi Y.F., Muller J., Munera-Huertas T., Munro C., Munro A., Murphy S., Munster V.J., Mweu P., Noé A., Nugent F.L., Nuthall E., O'Brien K., O'Connor D., Oguti B., Oliver J.L., Oliveira C., O'Reilly P.J., Osborn M., Osborne P., Owen C., Owens D., Owino N., Pacurar M., Parker K., Parracho H., Patrick-Smith M., Payne V., Pearce J., Peng Y., Peralta Alvarez M.P., Perring J., Pfafferott K., Pipini D., Plested E., Pluess-Hall H., Pollock K., Poulton I., Presland L., Provstgaard-Morys S., Pulido D., Radia K., Ramos Lopez F., Rand J., Ratcliffe H., Rawlinson T., Rhead S., Riddell A., Ritchie A.J., Roberts H., Robson J., Roche S., Rohde C., Rollier C.S., Romani R., Rudiansyah I., Saich S., Sajjad S., Salvador S., Sanchez Riera L., Sanders H., Sanders K., Sapaun S., Sayce C., Schofield E., Screaton G., Selby B., Semple C., Sharpe H.R., Shaik I., Shea A., Shelton H., Silk S., Silva-Reyes L., Skelly D.T., Smee H., Smith C.C., Smith D.J., Song R., Spencer A.J., Stafford E., Steele A., Stefanova E., Stockdale L., Szigeti A., Tahiri-Alaoui A., Tait M., Talbot H., Tanner R., Taylor I.J., Taylor V., Te Water Naude R., Thakur N., Themistocleous Y., Themistocleous A., Thomas M., Thomas T.M., Thompson A., Thomson-Hill S., Tomlins J., Tonks S., Towner J., Tran N., Tree J.A., Truby A., Turkentine K., Turner C., Turner N., Turner S., Tuthill T., Ulaszewska M., Varughese R., Van Doremalen N., Veighey K., Verheul M.K., Vichos I., Vitale E., Walker L., Watson M.E.E., Welham B., Wheat J., White C., White R., Worth A.T., Wright D., Wright S., Yao X.L., Yau Y. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396(10249):467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu F.-C., Li Y.-H., Guan X.-H., Hou L.-H., Wang W.-J., Li J.-X., Wu S.-P., Wang B.-S., Wang Z., Wang L., Jia S.-Y., Jiang H.-D., Wang L., Jiang T., Hu Y.i., Gou J.-B., Xu S.-B., Xu J.-J., Wang X.-W., Wang W., Chen W. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395(10240):1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu R., Zhao X., Li J., Niu P., Yang B.o., Wu H., Wang W., Song H., Huang B., Zhu N.a., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L.i., Chen J., Meng Y., Wang J.i., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Temperton N.J., Chan P.K., Simmons G., Zambon M.C., Tedder R.S., Takeuchi Y., Weiss R.A. Longitudinally profiling neutralizing antibody response to SARS coronavirus with pseudotypes. Emerg Infect Dis. 2005;11(3):411–416. doi: 10.3201/eid1103.040906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan J.F.W., Lau S.K.P., To K.K.W., Cheng V.C.C., Woo P.C.Y., Yuen K.-Y. Middle East Respiratory syndrome coronavirus: Another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28(2):465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shim B.-S., Park S.-M., Quan J.-S., Jere D., Chu H., Song M. Intranasal immunization with plasmid DNA encoding spike protein of SARS-coronavirus/polyethylenimine nanoparticles elicits antigen-specific humoral and cellular immune responses. BMC Immunol. 2010;11(1):65. doi: 10.1186/1471-2172-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z.-Y., Kong W.-P., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428(6982):561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X.i., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ong E., Wong M.U., Huffman A., He Y. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. Front Immunol. 2020;11:1581. doi: 10.3389/fimmu.2020.01581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27(671–680) doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enayatkhani M., Hasaniazad M., Faezi S., Guklani H., Davoodian P., Ahmadi N. Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in silico study. J Biomol Struct Dyn. 2020;1–16 doi: 10.1080/07391102.2020.1756411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu F, Wang A, Liu M, Wang Q, Chen J, Xia S. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv [Preprint] 2020 doi: 10.2139/ssrn.3566211. [DOI] [Google Scholar]
- 19.Ni L., Ye F., Cheng M.-L., Feng Y., Deng Y.-Q., Zhao H. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barnes C.O., West A.P., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182(4):828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Hoffmann M., Pöhlmann S., Graham B.S., Callewaert N., Schepens B., Saelens X., McLellan J.S. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181(5):1004–1015.e15. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 25.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587(7833):270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 26.Kalita P., Padhi A.K., Zhang K.Y.J., Tripathi T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microb Pathog. 2020;145:104236. doi: 10.1016/j.micpath.2020.104236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLellan J.S., Chen M., Joyce M.G., Sastry M., Stewart-Jones G.B.E., Yang Y., Zhang B., Chen L., Srivatsan S., Zheng A., Zhou T., Graepel K.W., Kumar A., Moin S., Boyington J.C., Chuang G.-Y., Soto C., Baxa U., Bakker A.Q., Spits H., Beaumont T., Zheng Z., Xia N., Ko S.-Y., Todd J.-P., Rao S., Graham B.S., Kwong P.D. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342(6158):592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Gobeil S.M.C. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat Struct Mol Biol. 2020;27(10):925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall B.G. Experimental evolution of a new enzymatic function. II. Evolution of multiple functions for EBG enzyme in E. coli. Genetics. 1978 doi: 10.1093/genetics/89.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morozova N., Allers J., Myers J., Shamoo Y. Protein-RNA interactions: Exploring binding patterns with a three-dimensional superposition analysis of high resolution structures. Bioinformatics. 2006;22(22):2746–2752. doi: 10.1093/bioinformatics/btl470. [DOI] [PubMed] [Google Scholar]
- 31.Padhi AK, Kalita P, Zhang KYJ, Tripathi T. High Throughput Designing and Mutational Mapping of RBD-ACE2 Interface Guide Non-Conventional Therapeutic Strategies for COVID-19. bioRxiv [Preprint] 2020. Available: 10.1101/2020.05.19.104042. [DOI]
- 32.Pearce R., Huang X., Setiawan D., Zhang Y. EvoDesign: designing protein-protein binding interactions using evolutionary interface profiles in conjunction with an optimized physical energy function. J Mol Biol. 2019;431(13):2467–2476. doi: 10.1016/j.jmb.2019.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C., Zheng W., Huang X., Bell E.W., Zhou X., Zhang Y. Protein structure and sequence reanalysis of 2019-nCoV genome refutes snakes as its intermediate host and the unique similarity between its spike protein insertions and HIV-1. J Proteome Res. 2020;19(4):1351–1360. doi: 10.1021/acs.jproteome.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X, Pearce R, Zhang Y. EvoEF2: Accurate and fast energy function for computational protein design. Bioinformatics. 2020;36: 1135–1142. [DOI] [PMC free article] [PubMed]
- 36.Tian Y., Huang X., Zhu Y. Computational design of enzyme–ligand binding using a combined energy function and deterministic sequence optimization algorithm. J Mol Model. 2015;21:191. doi: 10.1007/s00894-015-2742-x. [DOI] [PubMed] [Google Scholar]
- 37.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob Challenges. 2017;1(1):33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatos A, Hajdu-Soltész B, Monzon AM, Palopoli N, Álvarez L, Aykac-Fas B. Intrinsic protein disorder annotation in 2020. Nucleic Acids Res. 2020;48:D269–D276. doi: 10.1093/nar/gkz975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Capra J.A., Singh M. Predicting functionally important residues from sequence conservation. Bioinformatics. 2007;23:1875–1882. doi: 10.1093/bioinformatics/btm270. [DOI] [PubMed] [Google Scholar]
- 41.Huang X., Pearce R., Zhang Y. De novo design of protein peptides to block association of the SARS-CoV-2 spike protein with human ACE2. Aging. 2020;12(12):11263–11276. doi: 10.18632/aging.103416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang X., Zhang C., Pearce R., Omenn G.S., Zhang Y. Identifying the zoonotic origin of SARS-CoV-2 by modeling the binding affinity between the spike receptor-binding domain and host ACE2. J Proteome Res. 2020;19(12):4844–4856. doi: 10.1021/acs.jproteome.0c00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leaver-Fay A., Tyka M., Lewis S.M., Lange O.F., Thompson J., Jacak R. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jensen K.K., Andreatta M., Marcatili P., Buus S., Greenbaum J.A., Yan Z., Sette A., Peters B., Nielsen M. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology. 2018;154(3):394–406. doi: 10.1111/imm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhanda SK, Mahajan S, Paul S, Yan Z, Kim H, Jespersen MC, et al. IEDB-AR: immune epitope database—analysis resource in 2019. Nucleic Acids Res. 2019;47: W502–W506. [DOI] [PMC free article] [PubMed]
- 46.Fleri W., Paul S., Dhanda S.K., Mahajan S., Xu X., Fleri W. The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front Immunol. 2017;8:1–16. doi: 10.3389/fimmu.2017.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paul S., Lindestam Arlehamn C.S., Scriba T.J., Dillon M.B.C., Oseroff C., Hinz D., McKinney D.M., Carrasco Pro S., Sidney J., Peters B., Sette A. Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J Immunol Methods. 2015;422:28–34. doi: 10.1016/j.jim.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.The UniProt Consortium The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D193–197. doi: 10.1093/nar/gkl929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frankild S., de Boer R.J., Lund O., Nielsen M., Kesmir C., Zhang L. Amino acid similarity accounts for T cell cross-reactivity and for “holes” in the T cell repertoire. PLoS ONE. 2008;3(3):e1831. doi: 10.1371/journal.pone.0001831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henikoff S., Henikoff J.G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gouy M., Guindon S., Gascuel O. Sea view version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27(2):221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y., Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins Struct Funct Genet. 2004;57(4):702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 53.Li Y., Zhang C., Bell E.W., Yu D.-J., Zhang Y. Ensembling multiple raw coevolutionary features with deep residual neural networks for contact-map prediction in CASP13. Proteins Struct Funct Bioinform. 2019;87(12):1082–1091. doi: 10.1002/prot.25798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Hu J, Zhang C, Yu DJ, Zhang Y. ResPRE: High-accuracy protein contact prediction by coupling precision matrix with deep residual neural networks. Bioinformatics. 2019;35: 4647–4655. [DOI] [PMC free article] [PubMed]
- 55.Huang Y., Yang C., Xu X.-F., Xu W., Liu S.-W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schrödinger L. The PyMol Molecular Graphics System, Version~1.8. 2015 [cited 15 May 2020]. Available: https://pymol.org.
- 57.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- 59.Best R.B., Zhu X., Shim J., Lopes P.E.M., Mittal J., Feig M. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 Dihedral Angles. J Chem Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 61.Li Q., Huang X., Zhu Y. Evaluation of active designs of cephalosporin C acylase by molecular dynamics simulation and molecular docking. J Mol Model. 2014;20:2314. doi: 10.1007/s00894-014-2314-5. [DOI] [PubMed] [Google Scholar]
- 62.Xue J., Huang X., Zhu Y. Using molecular dynamics simulations to evaluate active designs of cephradine hydrolase by molecular mechanics/Poisson-Boltzmann surface area and molecular mechanics/generalized Born surface area methods. RSC Adv. 2019;9(24):13868–13877. doi: 10.1039/c9ra02406a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–1472. [Google Scholar]
- 64.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
- 65.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126(1):014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 66.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52(12):7182–7190. [Google Scholar]
- 67.Padhi A.K., Tripathi T. Can SARS-CoV-2 accumulate mutations in the S-protein to increase pathogenicity? ACS Pharmacol Transl Sci. 2020;3(5):1023–1026. doi: 10.1021/acsptsci.0c00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wada Y., Nithichanon A., Nobusawa E., Moise L., Martin W.D., Yamamoto N. A humanized mouse model identifies key amino acids for low immunogenicity of H7N9 vaccines. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-01372-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hewitt J.S., Karuppannan A.K., Tan S., Gauger P., Halbur P.G., Gerber P.F., De Groot A.S., Moise L., Opriessnig T. A prime-boost concept using a T-cell epitope-driven DNA vaccine followed by a whole virus vaccine effectively protected pigs in the pandemic H1N1 pig challenge model. Vaccine. 2019;37(31):4302–4309. doi: 10.1016/j.vaccine.2019.06.044. [DOI] [PubMed] [Google Scholar]
- 70.King C., Garza E.N., Mazor R., Linehan J.L., Pastan I., Pepper M., Baker D. Removing T-cell epitopes with computational protein design. Proc Natl Acad Sci. 2014;111(23):8577–8582. doi: 10.1073/pnas.1321126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fleishman S.J., Leaver-Fay A., Corn J.E., Strauch E.-M., Khare S.D., Koga N., Ashworth J., Murphy P., Richter F., Lemmon G., Meiler J., Baker D., Uversky V.N. Rosettascripts: a scripting language interface to the rosetta macromolecular modeling suite. PLoS ONE. 2011;6(6):e20161. doi: 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blazanovic K., Zhao H., Choi Y., Li W., Salvat R.S., Osipovitch D.C. Structure-based redesign of lysostaphin yields potent antistaphylococcal enzymes that evade immune cell surveillance. Mol Ther – Methods Clin Dev. 2015;2:15021. doi: 10.1038/mtm.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choi Y., Verma D., Griswold K.E., Bailey-Kellogg C. EpiSweep: computationally driven reengineering of therapeutic proteins to reduce immunogenicity while maintaining function. In: Samish I., editor. Computational Protein Design. New York, NY; Springer, New York: 2017. pp. 375–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mulligan M.J., Bernstein D.I., Winokur P., Rupp R., Anderson E., Rouphael N. Serological responses to an avian influenza A/H7N9 vaccine mixed at the point-of-use with MF59 adjuvant a randomized clinical trial. JAMA – J Am Med Assoc. 2014;312(14):1409. doi: 10.1001/jama.2014.12854. [DOI] [PubMed] [Google Scholar]
- 75.Guo L.i., Zhang X.i., Ren L., Yu X., Chen L., Zhou H., Gao X., Teng Z., Li J., Hu J., Wu C., Xiao X., Zhu Y., Wang Q., Pang X., Jin Q.i., Wu F., Wang J. Human antibody responses to avian influenza A(H7N9) virus, 2013. Emerg Infect Dis. 2014;20(2):192–200. doi: 10.3201/eid2002.131094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rudenko L., Isakova-Sivak I., Naykhin A., Kiseleva I., Stukova M., Erofeeva M., Korenkov D., Matyushenko V., Sparrow E., Kieny M.-P. H7N9 live attenuated influenza vaccine in healthy adults: a randomised, double-blind, placebo-controlled, phase 1 trial. Lancet Infect Dis. 2016;16(3):303–310. doi: 10.1016/S1473-3099(15)00378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Calis J.J.A., de Boer R.J., Keşmir C., Peters B. Degenerate T-cell recognition of peptides on MHC molecules creates large holes in the T-cell repertoire. PLoS Comput Biol. 2012;8(3):e1002412. doi: 10.1371/journal.pcbi.1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moise L., Gutierrez A.H., Bailey-Kellogg C., Terry F., Leng Q., Abdel Hady K.M., VerBerkmoes N.C., Sztein M.B., Losikoff P.T., Martin W.D., Rothman A.L., De Groot A.S. The two-faced T cell epitope: examining the host-microbe interface with JanusMatrix. Hum Vaccin Immunother. 2013;9(7):1577–1586. doi: 10.4161/hv.24615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bos R., Rutten L., van der Lubbe J.E.M., Bakkers M.J.G., Hardenberg G., Wegmann F. Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 Spike immunogen induces potent humoral and cellular immune responses. NPJ Vaccines. 2020;5(1) doi: 10.1038/s41541-020-00243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.