Abstract
The gut microbiota and the host are intimately connected. The host physiology dictates the intestinal environment through regulation of pH, ion concentration, mucus production, etc., all of which exerts a selective pressure on the gut microbiota. Since different regions of the gastrointestinal tract are characterized by their own physicochemical conditions, distinct microbial communities are present in these locations. While it is widely accepted that the intestinal microbiome influences the host (tight junctions, cytokine/immune responses, diarrhea, etc.), the reciprocal interaction of the host on the microbiome is under-explored. This review aims to address these gaps in knowledge by focusing on how the host intestinal ion transport influences the luminal environment and thereby modulates the gut microbiota composition.
Abbreviations: GI, gastrointestinal; NHE3, sodium-hydrogen exchanger isoform 3; NHE2, sodium-hydrogen exchanger isoform 2; CFTR, cystic fibrosis transmembrane regulator; ClC, chloride channel; DRA, down-regulated in adenoma; GLUT2, glucose transporter 2; SGLT1, sodium glucose co-transporter 1; NKCC1, Na+-K+-2Cl− co-transporter; ENaC, epithelial Na+ channel; OTUs, operational taxonomic units
Keywords: Gastrointestinal, Microbiota, Microbiome, Ion transport, NHE3, NHE2, CFTR, DRA, GLUT2
1. Intestinal architecture and function
The gastrointestinal tract is essentially a series of hollow interconnected organs lined by epithelial cells which performs multiple functions including digestion, absorption of water, uptake of nutrients, immune tolerance/homeostasis, regulation of the intestinal environment, and maintenance of the gut microbiota [2]. The architecture of the gastrointestinal tract facilitates these numerous functions. The intestine is divided length wise based on location (Fig. 1). In mammals, the small intestine consists of the duodenum, jejunum and ileum, while the large intestine consists of the colon. In humans the small intestine is approximately (~) 6 m long. In mice the small intestine is approximately ~350 mm [3]. In terms of structure, the small intestine consists of a single layer of columnar epithelium with crypts and villi. The crypts of Lieberkuhn harbor the proliferating stem cells, and finger-like villi contain the majority of differentiated absorptive cells [4], [5]. The adult small intestinal epithelium is composed of different cell lineages: absorptive enterocytes (~90% of cells), mucus-secreting goblet cells (~5%), antimicrobial secreting Paneth cells (~3%), hormone secreting enteroendocrine cells (~1%), chemo-sensing tuft cells (<1%) and proliferative stem cells (~1%) [5], [6], [7], [8], [9], [10]. Enterocytes, goblet cells and enteroendocrine cells are located in the intestinal villi, while Paneth and stem cells are located in the crypts [6], [7], [11], [12]. Proliferation of the stems cells results in renewal of the epithelium every 3–6 days in humans [9] and ~2–3 days in mice [13].
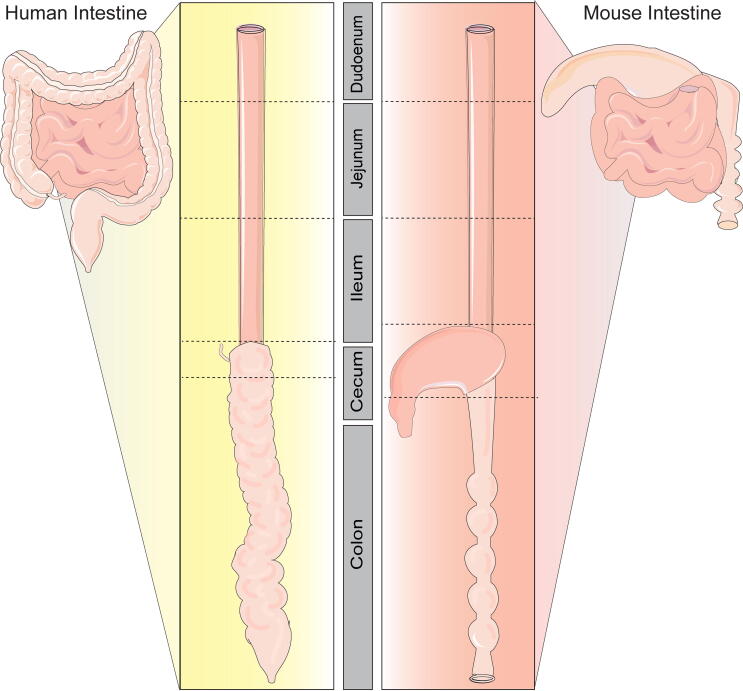
Fig. 1.
Graphical representation of the intestinal tract of humans and mice. Both human and mouse intestine can be separated into small and large intestine (also known as colon). Humans and mice have 3 segments of the small intestine: duodenum, jejunum and ileum. The cecum of humans is a small pouch at the beginning of the colon and is connected to the appendix. The cecum of mice is a large fermentation region that is distinct from both small and large intestine. In both humans and mice, the colon is segmented into different regions with distinct structures and function.
The different regions of the intestine have unique functions. The duodenum begins at the base of the stomach’s pyloric sphincter. A major function of the duodenum region is to neutralize the acidic chyme, or partially processed food material, from the stomach and enzymatically breakdown food. Enzymes from the pancreas and duodenum aid in digesting proteins and starches as well as emulsifying fats. The duodenum is also the site of amino acid, fatty acid, monoglycerides, phosphorus, and mono and/or disaccharides, iron, calcium, vitamin A, vitamin B12, and water absorption [5]. The jejunum is adjacent to the duodenum and is a site for the absorption of amino acids, fatty acids, oligosaccharides, minerals, electrolytes, vitamins, and water [5]. The final section of the small intestine is the ileum, which is responsible for absorption of bile salts and fats, as well as vitamin B12 and water.
The large intestine, also known as the colon, is largely responsible for reclaiming electrolytes and water. The colon in humans is ~1.5 m in length [5], while the colon in mice is ~110 mm [3]. Similar to the small intestine, the colon can be further subdivided based on anatomic divisions. In humans, the colon is divided into the ascending colon, transverse colon, descending colon, sigmoid colon, rectum, and anus [5]. In mice, the colon is commonly divided into proximal, mid and distal colon [14]. In contrast to the small intestine, the large intestine consists of crypts without villi [5], [9]. These crypts are rapidly renewed by stem cells at the crypt base [15]. Colonic epithelial cells include absorptive enterocytes at the top of the crypts and mucus producing goblet cells that line the colonic crypts. Mucus is critical in the colon as it provides lubrication for the feces and protects the underlying epithelium and immune cells from interacting with luminal antigens.
Although considerable anatomical and physiological features of the intestinal tract are shared between humans and mice, a key distinction is the cecum. In humans, the cecum is 6 cm and of minor importance for intestinal homeostasis [16]. In mice, the cecum is large, being 3–4 cm in length, and functions as a microbial fermentation vessel. It has been speculated that in mice, the cecum functions to restore the colonic microbiome after insult. In terms of size, the human cecum per kg of body weight roughly calculates to 0.09 cm per kg, while in mice, the cecum is 175 cm per kg body weight [16]. These calculations illustrate that, relative to body weight, the cecum is a much larger organ in mice than in humans. Since the cecum serves very different functions in mice versus humans, acting as a bioreactor for microbes in mice and serving as a reservoir for luminal contents passing from the small to large intestine in humans, this review will only focus on similar anatomical and functional segments (small and large intestine) in relationship to the microbiome in mice and humans.
2. Intestinal ion transport
The combination of digestion, nutrient absorption, water absorption, ion absorption and/or secretion determines the pH and ion composition of intestinal fluid and sets the environmental conditions for the growth of the intestinal microbiota [14], [17], [18], [19], [20], [21]. Overall, intestinal ion transport is characterized by net absorption of NaCl, nutrients and water and net secretion of bicarbonate (HCO3−) and KCl [22], [23]. The opposing functions of absorption and secretion are accomplished by transporters located in cells in both villus and crypt regions (Fig. 2). In general, cells in the villus facilitate absorption, while cells in the crypts promote secretion [22], [23], [24], [25], [26]. Transporters are found at both the apical (facing the lumen) and basal (facing the blood) membranes, allowing for transport in both directions [22].
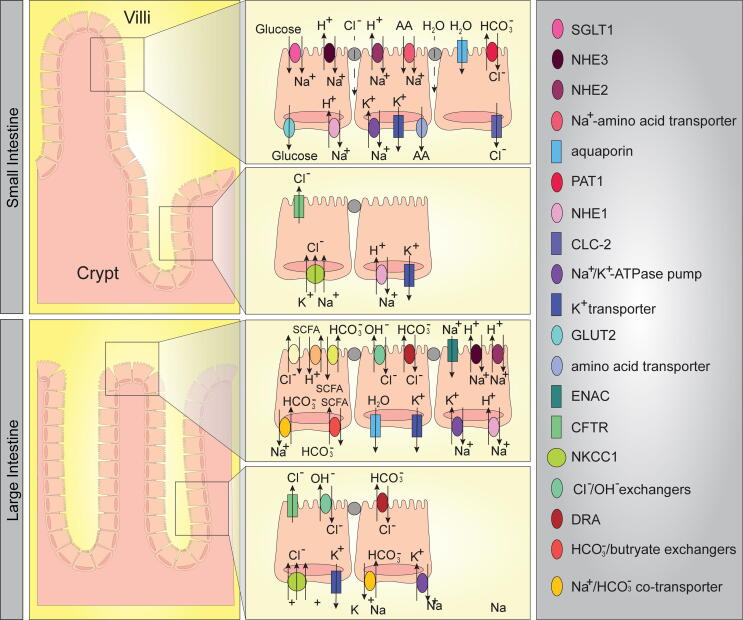
Fig. 2.
Graphical representation of intestinal ion transport. Transport mechanisms in the small intestine and colon are depicted. As shown, ion transporters are differentially distributed, creating unique microenvironments. These transporters are present in mouse and human.
Small intestinal fluid absorption predominantly occurs via the net movement of Na+. Na+ absorption is accomplished by either direct absorption of Na+ by Na+/H+ Exchangers (NHEs) or through coordination of Na+ absorption with other components such as glucose, transported by Na+-glucose co-transporter 1 (SGLT1) [17], [18], [19], [23]. In the intestine, there are three major NHEs: NHE1, NHE2 and NHE3. Apical NHE3 plays a major role in Na+ absorption, pH and cell volume regulation [23], [27], [28], while NHE2 appears to contribute to pH regulation and only minimally in Na+ absorption [17], [19], [21], [29]. Basolateral NHE1 also participates in volume and pH regulation. In addition to Na+ and water, nutrients such as glucose and amino acids are primarily absorbed in the small intestine. Glucose enters the enterocyte via apical SGLT1 and exits enterocytes via the basolateral glucose transporter 2 (GLUT-2) [23]. Amino acids enter the enterocyte via apical Na+-coupled and/or H+/dipeptide co-transporters and exit via various basal amino acid transporters.
In contrast to absorption, secretion in the small intestine involves the basal Na+/K+ pump, which drives the uphill entry of Cl− by the basal Na+-K+-2Cl− co-transporter (NKCC1) [30]. Secretion is also supported by basal K+ channels, which repolarize the cell and maintain the driving force for Cl− exit. At the apical surface, Cl− can exit the enterocyte by Cystic Fibrosis Transmembrane Regulator (CFTR, family Cl− channels (ClC)), and/or the Ca2+-activated Cl− channels (ClCA) [22], [31], [32], [33]. In addition to Cl− secretion, enterocytes also actively secrete bicarbonate, which plays a critical role in defining the intestinal pH. In the duodenum in particular, bicarbonate secretion is crucial for luminal alkalization. Bicarbonate secretion is accomplished by the anion exchanger down-regulated in adenoma protein (DRA) [34] and the putative anion transporter PAT1 [35]. Bicarbonate secretion via DRA and Cl− secretion via CFTR are also essential for mucus excretion from goblet cells [36], [37].
Compared to the small intestine, there are far fewer nutrient transporters in the colon (Fig. 2). In contrast to the small intestine where Na+ and glucose drive water absorption, in the colon water absorption is only driven by Na+ absorption [23]. The proximal segments of the colon expresses NHE2 and NHE3, which contribute to the absorption of residual water and maintenance of colonic pH. The more distal segments of colon can also express NHE2, NHE3, but their expression is lower than the proximal colon. Additionally, the distal colon expresses the epithelial Na+ channel (ENaC) [22], [33]. The colon also harbors secretory transporters. The bicarbonate transporter DRA is predominantly expressed in the distal colon [14], [23]. In mice, NHE3 is highly expressed in the proximal colon with decreasing expression in the distal colon, while DRA is highly expressed at the most distal end of the colon with decreasing expression toward the proximal colon [14]. The specificity of these ion transporters likely reflects specific intestinal environment regulation and dictates potential environmental changes. The colon also secretes and absorbs K+ by apical K+ transporters and the H+/K+-ATPase [23]. Collectively, the balance of absorption and secretion work in concert for the proper absorption of nutrients and water, as well as maintaining the proper intestinal ion composition and pH. This absorption/secretion balance produces feces low in salt, nutrients and water content (<2%). When this absorption/secretion balance is disrupted, diarrhea ensues [23]. Together these ion transporters regulate the intestinal environment of the small and large intestine.
3. Mouse and human gut microbiomes
In both humans and mice, the gut microbiota is dominated by two major phyla: Firmicutes and Bacteroidetes [38], [39], [40], [41], [42]. Although mice and humans appear to harbor similar bacterial communities at the high-taxonomic levels (phyla, class, order), they differ at the lower-taxonomic levels (genera, species, subspecies) [43], [44], [45], [46], [47]. A comparison of fecal 16S rDNA data from four public datasets from healthy adults [48], [49] and five murine studies [50], [51], [52], [53], [54] by Nguyen et al. revealed that mice and humans harbor 79 shared genera [55]. Clostridium (Firmicutes), Bacteroides (Bacteroidetes) and Blautia (Firmicutes) were found in both humans and mice at similar relative abundance. However, in human samples, Prevotella (Bacteroidetes), Faecalibacterium (Firmicutes) and Ruminococcus (Firmicutes) were found in high abundance, while Lactobacillus (Firmicutes), Alistipes (Bacteroidetes) and Turicibacter (Firmicutes) were more abundant in mice [55]. Other studies have identified differences between Mucispirillum schaedleri (Deferribacteres) [56] and segmented filamentous bacteria (Firmicutes) [57], [58], [59], [60], [61], which both appear higher in mice than humans. Despite lower-taxonomic differences, the microbiota of both humans and mice share similar metagenomic core functions [44], [45], [48], [62], [63], [64]. In mouse and human gut microbiome cores, 25 genera have been identified as shared [63]. Moreover, almost 80% of annotated functions were found in common between the mouse and human microbiome, indicating significant functional overlap in microbiome function.
A confounding factor in studying the microbiome of mice and humans is the techniques employed by each individual study [65], [66]. Common techniques include 16S rRNA sequencing and shot-gun metagenomic approaches. 16S rRNA gene sequencing employs PCR to target and amplify portions of the hypervariable regions (V1-V9) of the bacterial 16S rRNA gene. Amplicons are then given molecular barcodes, pooled together, and sequenced. Raw sequencing data undergoes trimming, error correction, and comparison to a 16S reference database which assigns phylogenetic rank to reads. In contrast with 16S sequencing, which only targets 16S rRNA genes, shotgun metagenomic sequencing sequences all the genomic DNA. The workflow for library preparation is similar to regular whole genome sequencing. Similar to 16S sequencing, shotgun metagenomic sequencing also includes quality trimming and comparison to a reference database comprising whole genomes or marker genes to generate a taxonomy profile. Since shotgun metagenomic sequencing covers all genetic information, the data can be used for additional analyses, e.g. metabolic function, antibiotic resistance, metabolite prediction, etc. A major difference between 16S and shotgun metagenomics is the taxonomic resolution: 16S is limited to the genus level, while shotgun can obtain high resolution at the species and strain level. For both 16S sequencing and shotgun metagenomic sequencing, multiple reference databases exist, as well as different versions of each database, and these databases may generate different taxonomy assignment; leading to potential differences in microbiome output. Another consideration is sequencing depth. Jovel et al. randomly sampled libraries at depths of 500, 1000, 5000, 10,000, 50,000, and 100,000 to investigate the minimal sequencing depth sufficient for accurately profiling bacterial community composition in stool samples [66]. The authors found that sequencing depths of 1000 and 50,000 were remarkably consistent, but the assignments of some bacteria required increasing sequencing depth to augment artifacts. A recent study comparing 16S rRNA gene-based analyses to shotgun metagenomic sequencing identified that many aspects of bacterial community characterization were consistent across methods [67]. In this study, Rausch et al. found that single-step amplification of the V3- V4 region yield more comparable results to shotgun metagenomics than multi-step amplification and use of the V1-V2 region of the 16S rRNA gene [67]. Based on these studies, it is evident that the microbiome composition of both mouse and human studies rely in part on the sequencing methodologies and thus information should be interpreted with caution. Despite these variables, it has been speculated that mouse microbiome studies can yield valuable information on the potential function of the microbiome in the setting of disease.
4. The gut microbiome composition along the length of the intestine
The gastrointestinal tract harbors diverse and dynamic microbial communities [68], [69], [70], [71]. In general, the gut microbiota is dominated by the phyla Firmicutes and Bacteroidetes, with Proteobacteria, Actinobacteria, Fusobacteria and Verruomicrobia present in lower abundance [39], [44], [72], [73], [74], [75], [76]. The composition of these microbial communities differs based on intestinal locations (duodenum, jejunum, ileum, colon) as well as proximity to the host (luminal vs mucosa-associated) [14], [19], [21], [45], [77], [78], [79], [80], [81], [82], [83], [84]. The differences in microbiota composition reflect the differences in the local environments: the microbiota is exposed to varying pH conditions (acidic stomach and alkaline intestine), various ion concentrations, intestinal motility, redox potential, nutrient supplies, and host secretions (e.g. hydrochloric acid, digestive enzymes, bile juice, pancreatic secretion and mucus) [1], [85], [86], [87].
The microbiota increases in number and diversity from the stomach to the colon, with the colon being the most densely populated region [86], [88]. The Human Microbiome Project (HMP) and the Metagenomics of the Human Intestinal Tract (MetaHIT) initiatives have helped define the healthy human microbiome and identify region specific microbial ecosystems [89], [90]. Based on these initiatives and current literature, we know that the small intestine commonly harbors Streptococcus (Firmicutes), Lactobacillus (Firmicutes), Clostridium (Firmicutes), and Prevotella (Bacteroidetes) [91], [92], [93]. Analysis from luminal contents along the length of the small intestine in two separate studies has revealed a dominance of Streptococcaceae (Firmicutes, ~55% of sequences) in the duodenum and jejunum followed by Veilonellacaea (Firmicutes, ~35%), and Lactobacillaceae (Firmicutes, ~5%), with lower abundance of Lachnospiraceae (Firmicutes), Clostridiaceae (Firmicutes), Erysipelotrichaceae (Firmicutes), Pasteurellaceae (Proteobacteria) and Prevotella (Bacteroidetes) [94], [95] (see graphical representation in Fig. 3, Table 1). In contrast, the feces of these healthy individuals were dominated by Ruminococcaceae (Firmicutes), Blautia (Firmicutes), Bifidobacteria (Actinobacteria), and Lachnospiraceae (Firmicutes)(~35%). Analysis of ileal luminal contents from a separate study has shown that the ileum harbored high levels of Bacilli (which include Streptoccoccus and Lactobacilli, Firmicutes, ~60%), followed by Bacteroides (Bacteroidetes, ~10%) and Clostridium (Firmicutes, ~25%) [96]. These findings are consistent with ileal swabs which also confirmed the dominance of Streptococcus (Firmicutes, ~60%), followed by Lactobacillus (Firmicutes) and other microbes [97]. In one study, the pH of the luminal contents was correlated with microbial operational taxonomic units (OTUs) [95]. In this study, Seekatz et al. demonstrated that 15 OTUs significantly correlated with pH changes. Six OTUs classified as Bacteroidetes, mainly Prevotella, and two OTUs were classified as Pasteurellaceae (Proteobacteria) were found to negatively correlate with pH (with decreased abundance at higher pHs). The other OTUs, classified as Firmicutes, mainly Streptococcus and Lactobacobacillaleas, as well as Actinomyces (Actinobacteria) were found to positively correlate with pH [95]. These findings highlight the link between intestinal pH and microbe composition.
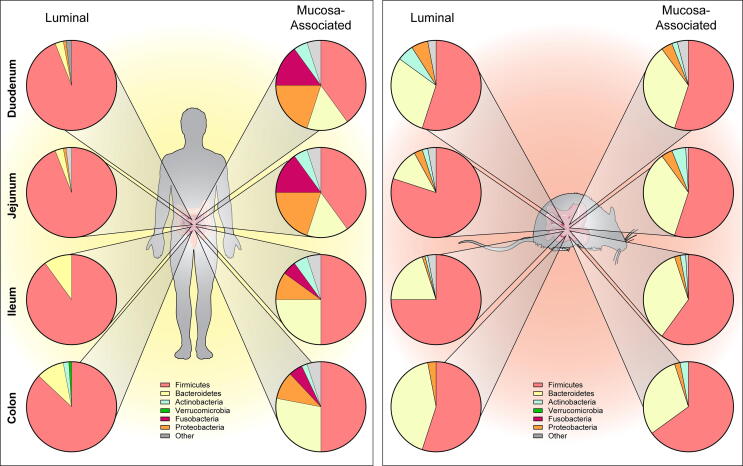
Fig. 3.
Graphical representation of the microbiome composition along the length of the intestine. Pie graphs depict the relative abundance of the major phyla (Firmicutes (pink), Bacteroidetes (yellow), Actinobacteria (blue), Verrucomicrobia (green), Fusobacteria (fusia), Proteobacteria (orange) and other (grey)). Composition is noted for both the luminal population as well as the mucosa-associated microbiome for human and mouse. Composition is approximated from published literature. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Bacteria that are commonly identified in human luminal and mucosa-associated microbiota from various intestinal segments.
|
Luminal Microbiota |
Mucosa-associated Microbiota |
|||||
|---|---|---|---|---|---|---|
| Small Intestine | Feces | Small Intestine | Colon | |||
| Dudoenum | Jejunum | Ileum | Colon | Dudoenum | Ileum | Colon |
| Streptococcaceae | Streptococcaceae | Streptoccoccus | Ruminococcaceae | Bacillales | Lachnospiraceae | Lachnospiraceae |
| Veilonellacaea | Veilonellacaea | Lactobacilli | Blautia | Streptococcaceae | Bacteroidaceae | Bacteroidaceae |
| Lactobacillaceae | Lactobacillaceae | Bacteroide | Bifidobacteria | Veillonellaceae | Ruminoccocaceae | Ruminoccocaceae |
| Lachnospiraceae | Lachnospiraceae | Clostridium | Lachnospiraceae | Pseudomonadaceae | Enterobacteriaceae | Enterobacteriaceae |
| Clostridiaceae | Clostridiaceae | Fusobacteriaceae | Fusobacteriaceae | Fusobacteriaceae | ||
| Erysipelotrichaceae | Erysipelotrichaceae | Akkermansia | ||||
| Pasteurellaceae | Pasteurellaceae | |||||
| Prevotella | Prevotella |
Analysis of biopsy specimens from healthy humans has revealed that the mucosa-associated microbiota differs significantly from the luminal microbiota. In a study examining mucosal biopsies along the length of the gastrointestinal tract, the authors found that the duodenum mucosa-associated microbiome contained Bacillales (Firmicutes, ~25%), Streptococcaceae (Firmicutes, ~20%), Veillonellaceae (Firmicutes, ~10%), Pseudomonadaceae (Proteobacteria, ~10%), and Fusobacteriaceae (Fusobacteria, ~5%), with lower levels of other microbes [98]. Interestingly, the ileum mucosa-associated microbiome was found to harbor Lachnospiraceae (Firmicutes, ~35%), Bacteroidaceae (Bacteroidetes, ~30%) Ruminoccocaceae (Firmicutes ~5%), Enterobacteriaceae (Proeobacteria ~5%) and Fusobacteriaceae (Fusobacteria ~3%). This composition greatly differs from the documented members of the luminal microbiome. Similarities were found in the mucosa-associated bacterial populations of the ascending and descending colon mucosa-associated microbiomes to that of the ileum, which also harbored Lachnospiraceae (~40%), Bacteroidaceae (~30%) Ruminoccocaceae (~5%), Enterobacteriaceae (~5%) and Fusobacteriaceae (~2%). These human studies establish relative community structures of the healthy microbiome along the gastrointestinal tract.
Similar to the human microbiome, the mouse microbiome has also been shown to vary along the length of the intestine (Fig. 3, Table 2). Whole segment microbiome analysis has shown that the small intestine (duodenum, jejunum and ileum) contained higher levels of Lactobacillaceae (Firmicutes ~30%), Bacteroidales (Bacteroidetes ~30%) followed by Lachnospiraceae (Firmicutes, ~10%), while the colon contained Bacteroidales (Bacteroidetes, ~40%), Clostridia (Firmicutes, ~30%), Lachnospiraceae (Firmicutes, ~20%), and Ruminococcus (~4%) in mice [99]. Analysis of luminal contents has revealed a dominance of Lactobacilli (Firmicutes, ~20%), Clostridium (Firmicutes, ~20%) and other Firmicutes members (~20%), followed by Bacteroides (Bacteroidetes, 5%) and other microbes in the ileum of mice [19], [21], [100]. In the mouse colon, the lumen was dominated by Clostridium (Firmicutes, ~35%) and Bacteroides (Bacteroidetes, ~20%), followed by Prevotella (Bacteroidetes, ~5%) and other microbes. In contrast, the mucosa-associated population was found to contain higher levels of Bacteroides, Prevotella and Mouse Intestinal Bacteroides (MIB) in the ileum and higher levels of Clostridium and MIB in the mucosa of the mouse colon than the luminal population [19], [21], [100]. These studies set the stage for examining how intestinal ion transport shapes the microbiome.
Table 2.
Bacteria that are commonly identified in mouse luminal and mucosa-associated microbiota from various intestinal segments.
|
Luminal Microbiota |
Mucosa-associated Microbiota |
||
|---|---|---|---|
| Small Intestine | Feces | Small Intestine | Feces |
| Ileum | Colon | Ileum | Colon |
| Clostridium | Clostridium | Bacteroides | Bacteroides |
| Lactobacilli | Bacteroides | Prevotella | Prevotella |
| Bacteroides | Prevotella | Clostridium | Clostridium |
| Prevotella | Lactobacillus | Lactobacillus | Lactobacillus |
| Enteroccocus | Lachnospiraceae | Akkermansia | |
5. Connecting ion transport with the gut microbiome
The interconnection between the intestinal environment and the microbiome has been elegantly demonstrated using various animal knockout models. Among the best characterized ion transport knockout mice are the NHE3, NHE2, CFTR, DRA and GLUT2 deficient mice (Fig. 4).
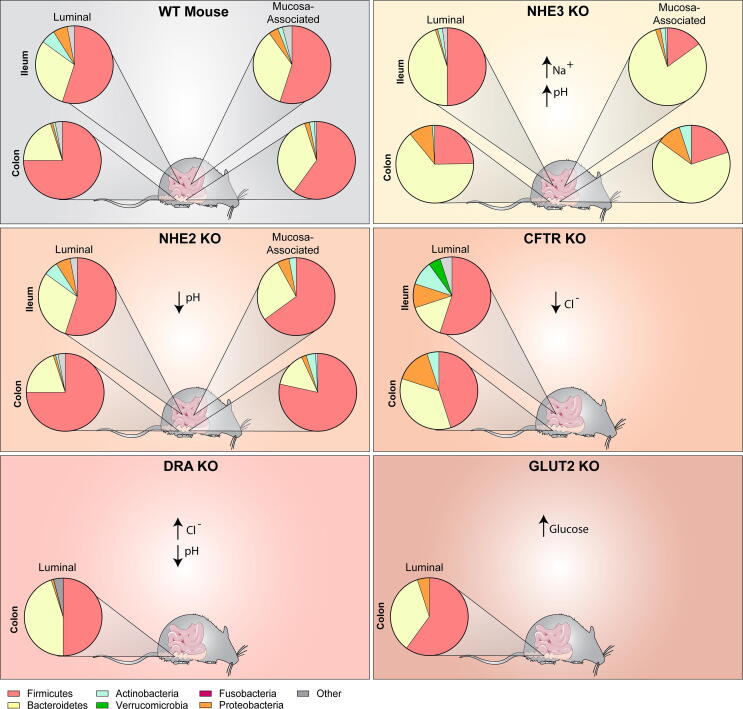
Fig. 4.
Graphical representation of the microbiome of WT type mice highlighting the communities in the ileum and colon, for both luminal and mucosa-associated communities. Pie graphs depict the relative abundance of the major phyla (Firmicutes (pink), Bacteroidetes (yellow), Actinobacteria (blue), Verrucomicrobia (green), Fusobacteria (fusia), Proteobacteria (orange) and other (grey)). In contrast to WT mice, NHE3, NHE2, CFTR, DRA and GLUT2 knockout (KO) mice harbor an altered intestinal environment and bacterial populations. Composition is approximated from published literature. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
5.1. NHE3 and NHE2 knockout mouse microbiome
Given the importance of NHE3 in sodium and water absorption, NHE3−/− mice exhibit chronic diarrhea with an alkaline intestinal fluid high in sodium compared with wild type (WT) littermates [17], [18], [19], [21]. Two different groups have reported on the microbiome of NHE3−/− mice [19], [101], [102]. NHE3−/− mice housed at one university exhibited an ileal and colonic luminal microbiome higher in Bacteroidetes compared to WT littermates [19]. Interestingly, the mucosa-associated microbiome exhibited even higher levels of Bacteroidetes in both the ileal and colonic microbial populations in the NHE3−/− mice compared to WT littermates and compared to the luminal contents. At another institution, NHE3−/− mice were likewise found to have increased Bacteroidetes in both the luminal and mucosa-associated colonic microbiomes, as well as expanded Proteobacteria [102]. These microbiome findings are consistent with previous studies that indicate that Bacteroidetes have improved growth at slightly higher pHs [103]. Select Bacteroides, like B. thetaiotaomicron, which were increased in NHE3−/− mice, were found to have optimal growth in conditions that resemble the NHE3−/− ileal environment in vitro [19]. Mice in the second facility exhibited decreased colonic mucus and developed spontaneous colitis, exhibited increased sensitivity to dextran-sodium-sulfate (DSS)-induced colitis, and when crossed with Rag2−/− mice for T-cell transfer experiments, NHE3−/−Rag2−/− mice experienced dramatically accelerated and exacerbated disease in a microbiome dependent manner [101], [102], [104]. These studies highlight the role of NHE3 in dictating the microbiome, which in turn can promote inflammation in the right setting.
In contrast to NHE3−/−, NHE2−/− mice do not exhibit diarrhea, but have an acidic intestinal fluid throughout the gastrointestinal tract [17], [18], [19], [21]. Interestingly, no differences were observed in the microbial communities of the luminal contents in the NHE2−/− mice compared to WT littermates [21]. However, significant differences were observed in mucosa-associated ileal and colonic microbiomes. In the ileal mucosa-associated microbiome, there were dramatic increases in Actinobacteria and decreases in Bacteroides (Bacteroidetes), MIB (Bacteroidetes), and other Firmicutes in NHE2−/− mice compared to WT controls. In the colon, significant increases were observed in Clostridia and Lactobacillus (Firmicutes) in NHE2−/− mice. Shifts in Lactobacillus and Clostridium/Ruminococcus correlates with changes in host mucus oligosaccharide composition [21]. These studies were among the first to identify a direct role of intestinal ion transport in shaping the intestinal environment and the microbiome composition.
5.2. CFTR knockout mouse microbiome
The microbiome is also altered in chloride transporter cystic fibrosis transmembrane conductance regulator (CFTR) deficient mice [105], [106]. CFTR−/− mice exhibit decreased luminal Cl− and increased mucus secretion with no change in pH [105]. In the terminal ileum of CFTR−/− mice, total bacteria were increased with enrichment of Enterobacteriaceae (Proteobacteria), Mycobacteria (Actinobacteria) and Bacteroides (Bacteroidetes), with an associated reduction in Lactobacilliales (Firmicutes) and Acinetobacter lwoffii (Proteobacteria) [105]. In a separate study, analysis of small intestinal luminal contents found increased Firmicutes and decreased Verrucomicrobia in CFTR−/− mice compared to WT mice [107]. OTU classification revealed increased abundance of Lactobacillus (Firmicutes) and Porphyromonadaceae (Bacteroidetes), with decreased abundance of Akkermansia (Verrucomicrobia) in CFTR−/− mice. Interestingly, another study examining small intestinal luminal contents found the reverse. In CFTR−/− mice they observed increased Akkermanisa (Verrucomicrobia) and Erysipelotrichaceae (Proteobacteria), and decreased Firmicutes (Lactobacillus) [108]. These studies highlight the need to identify region specific differences and emphasizes the microbiome variations that can be observed in different animal housing facilities. Fecal analysis of CFTR−/− mice from another study indicated a significant increase in E. coli (Proteobacteria) compared to WT controls [109]. To the best our knowledge, there are no mucosa-associated microbiome studies in CFTR−/− mice. Collectively, these studies confirm the importance of ion transport and particularly the role of Cl− in modulating the microbiome composition and depict the need for more studies on the CFTR microbiome.
5.3. DRA knockout mouse microbiome
Recently, two groups have established the microbiome of DRA−/− mice [110], [111]. Mutations in mouse DRA resembles that of congenital chloride-losing diarrhea in humans. Mice exhibit diarrhea with a high chloride, volume depletion, and growth defects [112]. DRA−/− mice also have an acidic colonic pH‐microclimate, similar to NHE2−/− mice [111]. At an institution in the United States, DRA−/− mice exhibited an expansion in Bacteroidacaea (Bacteriodetes) with significant increases in Parabacteroides and B. ovatus in the feces [110]. DRA−/− mice also had an expansion in Erysipelotrichaceae (Firmicutes) and Porphromonadaceae (Bacteroidetes) and a retraction in Actinobacteria and Bacteroidales family S24-7 (Bacteroidetes). In an institution in Germany, DRA−/− mice also exhibited expansion of Bacteroidetes and retraction of Firmicutes in the proximal and distal colon [111]. These DRA−/− knockout mice also had decreased levels of Actinobacteria. DRA−/− mice from both institutions exhibited decreased colonic mucus and inflammation [110], [111]. These two studies highlight the importance of DRA in regulating the gut microbiota and intestinal homeostasis.
5.4. GLUT2 knockout mouse microbiome
The transporter GLUT2 facilitates the passage of dietary sugars, glucose, fructose, and galactose in the intestine [113]. Reduced GLUT2 leads to higher sugar content in the distal intestine. Animals deficient in GLUT2 exhibit decreased microvillus length in the jejunum compared to control mice and reduction in absorptive epithelial cells [114]. Sequencing demonstrates that GLUT2−/− mice have increased fecal levels of Clostridium cluster IV (Firmicutes) and Enterococcus (Firmicutes) compared to control mice. These findings indicate that glucose persistence in the intestinal lumen can impact gut bacterial composition.
6. Conclusions
Although multiple aspects of host physiology can influence the microbiome, including oxygen content [115], [116], [117], [118], bile acids [119], [120], [121], [122], antibiotic use [123], [124], [125], diet [126], [127], [128], [129], mucus [130], [131], [132], [133], [134], etc., in this review we have focused on the unique link between intestinal ion transport and the gut microbiota. The studies presented herein demonstrate that endogenous ion transport (luminal Na+, K+, Cl−, pH) alters the intestinal microenvironment making it inhabitable for particular bacterial groups. Since multiple factors can influence the gut microbiome, at present we lack a strong understanding of exactly how transported ions impact the gut microbiota. In the future, studies using bioreactors with human and mice feces might shed more insight into how ion composition and pH can directly modulate gut microbes (independent of the immune system, mucus secretion and subsequent host responses, etc.). These types of studies would provide valuable information of microbial tolerance and niche development and may provide insights into how to shift an altered microbiome back towards a healthy composition.
The highlighted studies emphasize that knowledge of the stool microbiota does not necessarily fully reflect intestinal changes upstream (small intestine) or fully reflect the mucosa-associated bacterial populations, which dramatically change in the setting of altered host ion transport. Since it can be challenging to obtain human small intestinal samples, mouse models provide a valuable scientific tool for these types of analyses. Analyzing the microbiota regionally along the length of the intestine and by population (luminal or mucosa-associated) could be used in the future to determine mechanistic interactions. Since mucosa-associated bacteria live in closer proximity to the intestinal epithelium, it is likely they execute different functions within the GI ecosystem compared with luminal microbiota [1].
Although mouse models can be useful, mice differ from humans in some key aspects. Mouse diet, fur, and behavior (e.g., nocturnal behavior, grooming practices, and coprophagia) clearly differ from humans [44], [135]. These differences likely influence the gut microbiota composition and confound analysis. Furthermore the mouse immune system differs from the human immune system [135], which affects the way the host responds to the gut microbiota. As a result, mouse disease does not always reflect human disease. Despite these differences mouse models can still be a useful tool for unraveling mechanisms of host–microbiota interactions. Kostic et al. eloquently stated “acknowledging this complexity and the potential pitfalls is not meant to suggest that using mice for host–microbiota studies is a flawed approach; rather, the point is to highlight that studying host–microbiota interactions in mice requires careful experimental design” [44].
Mouse genetic background has been shown to impact the composition, diversity, and richness of the gut microbiota in both WT and knockout mice [136], [137]. In a study by Campbell et al. eight core inbred strains were examined by 16S rRNA. Effects were shown to exist in the gut microbiota based on litter, co-housing and in some mouse strains, gender [138]. Effects of mouse background can provide an advantage for selecting specific traits and determining how they influence the gut microbiota. However, it can also confound data analysis, and as such mouse background should be considered in experimental design. Animal age is also an important consideration as the microbiome of both rodents and humans shifts overtime.
Despite the potential impact of microbial communities on human health and disease, our understanding of how microbial communities are maintained in the gut remains incomplete. Although much is known about basic host physiology and stool gut microbiota composition, more studies are needed for a better understanding of the interplay between the gut microbiota and host environment set by ion transport. Current work in the field supports the notion that ion transport shapes the microbiota composition and ultimately the microbe-host interactions. Knowledge of how the intestinal environment affects specific bacteria will likely aid in the development of future therapies for diseases with abnormal bacterial composition.
Funding
This study was supported by the NIH K01DK121869 (ACE) and K01DK123195 (MAE).
CRediT authorship contribution statement
Amy C. Engevik: Conceptualization, Writing - original draft, Writing - review & editing, Funding acquisition. Melinda A. Engevik: Conceptualization, Writing - review & editing, Visualization, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Gerritsen J. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 2011;6(3):209–240. doi: 10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huffnagle G., Noverr M.C. GI microbiota and regulation of the immune system. Preface Adv Exp Med Biol. 2008;635:v–vi. [PubMed] [Google Scholar]
- 3.Wolczuk K. Morphometric characteristics of the small and large intestines of Mus musculus during postnatal development. Folia Morphol (Warsz) 2011;70(4):252–259. [PubMed] [Google Scholar]
- 4.Montgomery R.K., Mulberg A.E., Grand R.J. Development of the human gastrointestinal tract: twenty years of progress. Gastroenterology. 1999;116(3):702–731. doi: 10.1016/s0016-5085(99)70193-9. [DOI] [PubMed] [Google Scholar]
- 5.Rao, J.N. and J.Y. Wang, Regulation of Gastrointestinal Mucosal Growth, in Regulation of Gastrointestinal Mucosal Growth. 2010: San Rafael (CA).
- 6.Pinto D. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17(14):1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clavel T., Haller D. Molecular interactions between bacteria, the epithelium, and the mucosal immune system in the intestinal tract: implications for chronic inflammation. Curr Issues Intest Microbiol. 2007;8(2):25–43. [PubMed] [Google Scholar]
- 8.Salzman N.H., Underwood M.A., Bevins C.L. Paneth cells, defensins, and the commensal microbiota: a hypothesis on intimate interplay at the intestinal mucosa. Semin Immunol. 2007;19(2):70–83. doi: 10.1016/j.smim.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Wells J.M. Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4607–4614. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vieira P.L. IL-10-secreting regulatory T cells do not express Foxp3 but have comparable regulatory function to naturally occurring CD4+CD25+ regulatory T cells. J Immunol. 2004;172(10):5986–5993. doi: 10.4049/jimmunol.172.10.5986. [DOI] [PubMed] [Google Scholar]
- 11.Potten C.S. Stem cells in gastrointestinal epithelium: numbers, characteristics and death. Philos Trans R Soc Lond B Biol Sci. 1998;353(1370):821–830. doi: 10.1098/rstb.1998.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potten C.S., Loeffler M. Stem cells: attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development. 1990;110(4):1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- 13.Creamer B., Shorter R.G., Bamforth J. The turnover and shedding of epithelial cells. I. The turnover in the gastro-intestinal tract. Gut. 1961;2:110–118. doi: 10.1136/gut.2.2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talbot C., Lytle C. Segregation of Na/H exchanger-3 and Cl/HCO3 exchanger SLC26A3 (DRA) in rodent cecum and colon. Am J Physiol Gastrointest Liver Physiol. 2010;299(2):G358–G367. doi: 10.1152/ajpgi.00151.2010. [DOI] [PubMed] [Google Scholar]
- 15.Sato T. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 16.Hugenholtz F., de Vos W.M. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci. 2018;75(1):149–160. doi: 10.1007/s00018-017-2693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gawenis L.R. Intestinal NaCl transport in NHE2 and NHE3 knockout mice. Am J Physiol Gastrointest Liver Physiol. 2002;282(5):G776–G784. doi: 10.1152/ajpgi.00297.2001. [DOI] [PubMed] [Google Scholar]
- 18.Schultheis L.M., Baldwin B.G. Molecular phylogenetics of Fouquieriaceae: evidence from nuclear rDNA ITS studies. Am J Bot. 1999;86(4):578–589. [PubMed] [Google Scholar]
- 19.Engevik M.A. Loss of NHE3 alters gut microbiota composition and influences Bacteroides thetaiotaomicron growth. Am J Physiol Gastrointest Liver Physiol. 2013;305(10):G697–G711. doi: 10.1152/ajpgi.00184.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engevik M.A. Prebiotic Properties of Galursan HF 7K on Mouse Gut Microbiota. Cell Physiol Biochem. 2013;32(7):96–110. doi: 10.1159/000356631. [DOI] [PubMed] [Google Scholar]
- 21.Engevik M.A. Acidic Conditions in the NHE2 Mouse Intestine Result in an Altered Mucosa-Associated Bacterial Population with Changes in Mucus Oligosaccharides. Cell Physiol Biochem. 2013;32(7):111–128. doi: 10.1159/000356632. [DOI] [PubMed] [Google Scholar]
- 22.Kunzelmann K., Mall M. Electrolyte transport in the mammalian colon: mechanisms and implications for disease. Physiol Rev. 2002;82(1):245–289. doi: 10.1152/physrev.00026.2001. [DOI] [PubMed] [Google Scholar]
- 23.Rao M.C. Oral rehydration therapy: new explanations for an old remedy. Annu Rev Physiol. 2004;66:385–417. doi: 10.1146/annurev.physiol.66.032902.134726. [DOI] [PubMed] [Google Scholar]
- 24.Field M. Intestinal ion transport and the pathophysiology of diarrhea. J Clin Invest. 2003;111(7):931–943. doi: 10.1172/JCI18326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sangan P. Cloning and expression of a chloride-dependent Na+-H+ exchanger. J Biol Chem. 2002;277(12):9668–9675. doi: 10.1074/jbc.M110852200. [DOI] [PubMed] [Google Scholar]
- 26.Chu J., Chu S., Montrose M.H. Apical Na+/H+ exchange near the base of mouse colonic crypts. Am J Physiol Cell Physiol. 2002;283(1):C358–C372. doi: 10.1152/ajpcell.01380.2000. [DOI] [PubMed] [Google Scholar]
- 27.Counillon L., Pouyssegur J. The expanding family of eucaryotic Na(+)/H(+) exchangers. J Biol Chem. 2000;275(1):1–4. doi: 10.1074/jbc.275.1.1. [DOI] [PubMed] [Google Scholar]
- 28.Wakabayashi S., Shigekawa M., Pouyssegur J. Molecular physiology of vertebrate Na+/H+ exchangers. Physiol Rev. 1997;77(1):51–74. doi: 10.1152/physrev.1997.77.1.51. [DOI] [PubMed] [Google Scholar]
- 29.Guan Y. NHE2 is the main apical NHE in mouse colonic crypts but an alternative Na+-dependent acid extrusion mechanism is upregulated in NHE2-null mice. Am J Physiol Gastrointest Liver Physiol. 2006;291(4):G689–G699. doi: 10.1152/ajpgi.00342.2005. [DOI] [PubMed] [Google Scholar]
- 30.Haas M., Forbush B., 3rd The Na-K-Cl cotransporter of secretory epithelia. Annu Rev Physiol. 2000;62:515–534. doi: 10.1146/annurev.physiol.62.1.515. [DOI] [PubMed] [Google Scholar]
- 31.Fuller C.M., Benos D.J. Ca(2+)-Activated Cl(-) Channels: A Newly Emerging Anion Transport Family. News Physiol Sci. 2000;15:165–171. [PubMed] [Google Scholar]
- 32.Frizzell R.A. Ten years with CFTR. Physiol Rev. 1999;79(1 Suppl):S1–S2. doi: 10.1152/physrev.1999.79.1.S1. [DOI] [PubMed] [Google Scholar]
- 33.Jentsch T.J. Molecular structure and physiological function of chloride channels. Physiol Rev. 2002;82(2):503–568. doi: 10.1152/physrev.00029.2001. [DOI] [PubMed] [Google Scholar]
- 34.Jacob P. Down-regulated in adenoma mediates apical Cl-/HCO3- exchange in rabbit, rat, and human duodenum. Gastroenterology. 2002;122(3):709–724. doi: 10.1053/gast.2002.31875. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z. Identification of an apical Cl(-)/HCO3(-) exchanger in the small intestine. Am J Physiol Gastrointest Liver Physiol. 2002;282(3):G573–G579. doi: 10.1152/ajpgi.00338.2001. [DOI] [PubMed] [Google Scholar]
- 36.De Lisle R.C. Pass the bicarb: the importance of HCO3- for mucin release. J Clin Invest. 2009;119(9):2535–2537. doi: 10.1172/JCI40598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia M.A., Yang N., Quinton P.M. Normal mouse intestinal mucus release requires cystic fibrosis transmembrane regulator-dependent bicarbonate secretion. J Clin Invest. 2009;119(9):2613–2622. doi: 10.1172/JCI38662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen T.L. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8(1):1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eckburg P.B. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ley R.E. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ley R.E. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 42.Engevik M.A., Versalovic J. Biochemical Features of Beneficial Microbes: Foundations for Therapeutic Microbiology. Microbiol Spectr. 2017;5(5) doi: 10.1128/microbiolspec.bad-0012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ley R.E. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kostic A.D., Howitt M.R., Garrett W.S. Exploring host-microbiota interactions in animal models and humans. Genes Dev. 2013;27(7):701–718. doi: 10.1101/gad.212522.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho I., Blaser M.J. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huse S.M. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4(11) doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krych L. Quantitatively different, yet qualitatively alike: a meta-analysis of the mouse core gut microbiome with a view towards the human gut microbiome. PLoS ONE. 2013;8(5) doi: 10.1371/journal.pone.0062578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yatsunenko T. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagy-Szakal D. Maternal micronutrients can modify colonic mucosal microbiota maturation in murine offspring. Gut Microbes. 2012;3(5):426–433. doi: 10.4161/gmic.20697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riboulet-Bisson E. Effect of Lactobacillus salivarius bacteriocin Abp118 on the mouse and pig intestinal microbiota. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0031113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ubeda C. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun. 2013;81(3):965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ward N.L. Intestinal aganglionosis is associated with early and sustained disruption of the colonic microbiome. Neurogastroenterol Motil. 2012;24(9):874–e400. doi: 10.1111/j.1365-2982.2012.01937.x. [DOI] [PubMed] [Google Scholar]
- 54.Zenewicz L.A. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol. 2013;190(10):5306–5312. doi: 10.4049/jimmunol.1300016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aagaard K. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 2013;27(3):1012–1022. doi: 10.1096/fj.12-220806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robertson B.R. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol. 2005;55(Pt 3):1199–1204. doi: 10.1099/ijs.0.63472-0. [DOI] [PubMed] [Google Scholar]
- 57.Gaboriau-Routhiau V. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 58.Snel J. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of “Candidatus Arthromitus”. Int J Syst Bacteriol. 1995;45(4):780–782. doi: 10.1099/00207713-45-4-780. [DOI] [PubMed] [Google Scholar]
- 59.Ivanov I.I. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y. Induction of Intestinal Th17 Cells by Flagellins From Segmented Filamentous Bacteria. Front Immunol. 2019;10:2750. doi: 10.3389/fimmu.2019.02750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yin Y. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J. 2013;7(3):615–621. doi: 10.1038/ismej.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muegge B.D. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332(6032):970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qin J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turnbaugh P.J., Gordon J.I. The core gut microbiome, energy balance and obesity. The Journal of physiology. 2009;587(Pt 17):4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laudadio I. Next-Generation Metagenomics: Methodological Challenges and Opportunities. OMICS. 2019;23(7):327–333. doi: 10.1089/omi.2019.0073. [DOI] [PubMed] [Google Scholar]
- 66.Jovel J. Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front Microbiol. 2016;7:459. doi: 10.3389/fmicb.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rausch P. Comparative analysis of amplicon and metagenomic sequencing methods reveals key features in the evolution of animal metaorganisms. Microbiome. 2019;7(1):133. doi: 10.1186/s40168-019-0743-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sender R., Fuchs S., Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016;14(8) doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bull M.J., Plummer N.T. Part 1: The Human Gut Microbiome in Health and Disease. Integr Med (Encinitas) 2014;13(6):17–22. [PMC free article] [PubMed] [Google Scholar]
- 70.Rath C.M., Dorrestein P.C. The bacterial chemical repertoire mediates metabolic exchange within gut microbiomes. Curr Opin Microbiol. 2012;15(2):147–154. doi: 10.1016/j.mib.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Flint H.J. The impact of nutrition on the human microbiome. Nutr Rev. 2012;70(Suppl 1):S10–S13. doi: 10.1111/j.1753-4887.2012.00499.x. [DOI] [PubMed] [Google Scholar]
- 72.Mariat D. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guarner F., Malagelada J.R. Gut flora in health and disease. Lancet. 2003;361(9356):512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 74.Suau A. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65(11):4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu J., Gordon J.I. Honor thy symbionts. Proc Natl Acad Sci U S A. 2003;100(18):10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Backhed F. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 77.Berkes J. Intestinal epithelial responses to enteric pathogens: effects on the tight junction barrier, ion transport, and inflammation. Gut. 2003;52(3):439–451. doi: 10.1136/gut.52.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Salzman N.H. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11(1):76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berg R.D. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4(11):430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 80.Mackie R.I., Sghir A., Gaskins H.R. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69(5):1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 81.Hooper L.V., Falk P.G., Gordon J.I. Analyzing the molecular foundations of commensalism in the mouse intestine. Curr Opin Microbiol. 2000;3(1):79–85. doi: 10.1016/s1369-5274(99)00055-7. [DOI] [PubMed] [Google Scholar]
- 82.Macfarlane G.T., Cummings J.H. Probiotics and prebiotics: can regulating the activities of intestinal bacteria benefit health? West J Med. 1999;171(3):187–191. [PMC free article] [PubMed] [Google Scholar]
- 83.Macfarlane S., Hopkins M.J., Macfarlane G.T. Toxin synthesis and mucin breakdown are related to swarming phenomenon in Clostridium septicum. Infect Immun. 2001;69(2):1120–1126. doi: 10.1128/IAI.69.2.1120-1126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zoetendal E.G. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68(7):3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Falk P.G. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev. 1998;62(4):1157–1170. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dunne C. Adaptation of bacteria to the intestinal niche: probiotics and gut disorder. Inflamm Bowel Dis. 2001;7(2):136–145. doi: 10.1097/00054725-200105000-00010. [DOI] [PubMed] [Google Scholar]
- 87.Booijink C.C. Microbial communities in the human small intestine: coupling diversity to metagenomics. Future Microbiol. 2007;2(3):285–295. doi: 10.2217/17460913.2.3.285. [DOI] [PubMed] [Google Scholar]
- 88.Greenblum S., Turnbaugh P.J., Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A. 2012;109(2):594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Turnbaugh P.J. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ehrlich, S.D., MetaHIT: The European Union Project on Metagenomics of the Human Intestinal Tract, in Metagenomics of the Human Body, K.E. Nelson, Editor . New York, NY; New York: 2011. Springer; pp. 307–316. [Google Scholar]
- 91.Wang M. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005;54(2):219–231. doi: 10.1016/j.femsec.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 92.Ahmed S. Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol. 2007;73(22):7435–7442. doi: 10.1128/AEM.01143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hayashi H. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J Med Microbiol. 2005;54(Pt 11):1093–1101. doi: 10.1099/jmm.0.45935-0. [DOI] [PubMed] [Google Scholar]
- 94.Leite G.G.S. Mapping the Segmental Microbiomes in the Human Small Bowel in Comparison with Stool: A REIMAGINE Study. Dig Dis Sci. 2020;65(9):2595–2604. doi: 10.1007/s10620-020-06173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Seekatz A.M. Spatial and Temporal Analysis of the Stomach and Small-Intestinal Microbiota in Fasted Healthy Humans. mSphere. 2019;4(2) doi: 10.1128/mSphere.00126-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stolaki M. Microbial communities in a dynamic in vitro model for the human ileum resemble the human ileal microbiota. FEMS Microbiol Ecol. 2019;95(8) doi: 10.1093/femsec/fiz096. [DOI] [PubMed] [Google Scholar]
- 97.Villmones H.C. Species Level Description of the Human Ileal Bacterial Microbiota. Sci Rep. 2018;8(1):4736. doi: 10.1038/s41598-018-23198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vasapolli R. Analysis of Transcriptionally Active Bacteria Throughout the Gastrointestinal Tract of Healthy Individuals. Gastroenterology. 2019 doi: 10.1053/j.gastro.2019.05.068. [DOI] [PubMed] [Google Scholar]
- 99.Gu S. Bacterial community mapping of the mouse gastrointestinal tract. PLoS ONE. 2013;8(10) doi: 10.1371/journal.pone.0074957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jakobsson H.E. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015;16(2):164–177. doi: 10.15252/embr.201439263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Laubitz D. Reduced Epithelial Na+/H+ Exchange Drives Gut Microbial Dysbiosis and Promotes Inflammatory Response in T Cell-Mediated Murine Colitis. PLoS ONE. 2016;11(4) doi: 10.1371/journal.pone.0152044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Larmonier C.B. Reduced colonic microbial diversity is associated with colitis in NHE3-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2013;305(10):G667–G677. doi: 10.1152/ajpgi.00189.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ilhan Z.E. pH-Mediated Microbial and Metabolic Interactions in. Fecal Enrichment Cultures. mSphere. 2017;2(3) doi: 10.1128/mSphere.00047-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kiela P.R. Changes in mucosal homeostasis predispose NHE3 knockout mice to increased susceptibility to DSS-induced epithelial injury. Gastroenterology. 2009;137(3) doi: 10.1053/j.gastro.2009.05.043. p. 965-75, 975 e1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thomsson K.A. Intestinal mucins from cystic fibrosis mice show increased fucosylation due to an induced Fucalpha1-2 glycosyltransferase. Biochem J. 2002;367(Pt 3):609–616. doi: 10.1042/BJ20020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Norkina O., Burnett T.G., De Lisle R.C. Bacterial overgrowth in the cystic fibrosis transmembrane conductance regulator null mouse small intestine. Infect Immun. 2004;72(10):6040–6049. doi: 10.1128/IAI.72.10.6040-6049.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bazett M., Bergeron M.E., Haston C.K. Streptomycin treatment alters the intestinal microbiome, pulmonary T cell profile and airway hyperresponsiveness in a cystic fibrosis mouse model. Sci Rep. 2016;6:19189. doi: 10.1038/srep19189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bazett M. Cystic fibrosis mouse model-dependent intestinal structure and gut microbiome. Mamm Genome. 2015;26(5–6):222–234. doi: 10.1007/s00335-015-9560-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Debray D. Diet-Induced Dysbiosis and Genetic Background Synergize With Cystic Fibrosis Transmembrane Conductance Regulator Deficiency to Promote Cholangiopathy in Mice. Hepatol Commun. 2018;2(12):1533–1549. doi: 10.1002/hep4.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kumar A. A novel role of SLC26A3 in maintenance of intestinal epithelial barrier integrity. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kini A. Slc26a3 deletion alters pH-microclimate, mucin biosynthesis, microbiome composition and increases the TNFalpha expression in murine colon. Acta Physiol (Oxf) 2020;230(2) doi: 10.1111/apha.13498. [DOI] [PubMed] [Google Scholar]
- 112.Schweinfest C.W. slc26a3 (dra)-deficient mice display chloride-losing diarrhea, enhanced colonic proliferation, and distinct up-regulation of ion transporters in the colon. J Biol Chem. 2006;281(49):37962–37971. doi: 10.1074/jbc.M607527200. [DOI] [PubMed] [Google Scholar]
- 113.Thorens B. GLUT2, glucose sensing and glucose homeostasis. Diabetologia. 2015;58(2):221–232. doi: 10.1007/s00125-014-3451-1. [DOI] [PubMed] [Google Scholar]
- 114.Schmitt C.C. Intestinal invalidation of the glucose transporter GLUT2 delays tissue distribution of glucose and reveals an unexpected role in gut homeostasis. Mol Metab. 2017;6(1):61–72. doi: 10.1016/j.molmet.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Albenberg L. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147(5):1055–63 e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Litvak Y., Byndloss M.X., Baumler A.J. Colonocyte metabolism shapes the gut microbiota. Science. 2018;362(6418) doi: 10.1126/science.aat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yasuda K. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe. 2015;17(3):385–391. doi: 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Scheithauer T.P. Causality of small and large intestinal microbiota in weight regulation and insulin resistance. Mol Metab. 2016;5(9):759–770. doi: 10.1016/j.molmet.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hylemon P.B., Harris S.C., Ridlon J.M. Metabolism of hydrogen gases and bile acids in the gut microbiome. FEBS Lett. 2018;592(12):2070–2082. doi: 10.1002/1873-3468.13064. [DOI] [PubMed] [Google Scholar]
- 120.Ridlon J.M. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30(3):332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sagar N.M. The interplay of the gut microbiome, bile acids, and volatile organic compounds. Gastroenterol Res Pract. 2015;2015 doi: 10.1155/2015/398585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Singh J. Review on Bile Acids: Effects of the Gut Microbiome, Interactions with Dietary Fiber, and Alterations in the Bioaccessibility of Bioactive Compounds. J Agric Food Chem. 2019;67(33):9124–9138. doi: 10.1021/acs.jafc.8b07306. [DOI] [PubMed] [Google Scholar]
- 123.Iizumi T. Gut Microbiome and Antibiotics. Arch Med Res. 2017;48(8):727–734. doi: 10.1016/j.arcmed.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 124.Fujisaka S. Antibiotic effects on gut microbiota and metabolism are host dependent. J Clin Invest. 2016;126(12):4430–4443. doi: 10.1172/JCI86674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yassour M. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016;8(343):p. 343ra81. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Leeming E.R. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients. 2019;11(12) doi: 10.3390/nu11122862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Singh R.K. Influence of diet on the gut microbiome and implications for human health. Journal of translational medicine. 2017;15(1) doi: 10.1186/s12967-017-1175-y. 73–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Flint H.J. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9(10):577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 129.Valdes A.M. Role of the gut microbiota in nutrition and health. BMJ. 2018;361 doi: 10.1136/bmj.k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schroeder B.O. Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep (Oxf) 2019;7(1):3–12. doi: 10.1093/gastro/goy052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Corfield A.P. The Interaction of the Gut Microbiota with the Mucus Barrier in Health and Disease in Human. Microorganisms. 2018;6(3) doi: 10.3390/microorganisms6030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Derrien M. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut microbes. 2010;1(4):254–268. doi: 10.4161/gmic.1.4.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kim Y.S., Ho S.B. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep. 2010;12(5):319–330. doi: 10.1007/s11894-010-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sicard J.F. Interactions of Intestinal Bacteria with Components of the Intestinal Mucus. Front Cell Infect Microbiol. 2017;7:387. doi: 10.3389/fcimb.2017.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rongvaux A. Human hemato-lymphoid system mice: current use and future potential for medicine. Annu Rev Immunol. 2013;31:635–674. doi: 10.1146/annurev-immunol-032712-095921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Esworthy R.S., Smith D.D., Chu F.F. A Strong Impact of Genetic Background on Gut Microflora in Mice. Int J Inflam. 2010;2010(2010) doi: 10.4061/2010/986046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Buchler G. Strain-specific colitis susceptibility in IL10-deficient mice depends on complex gut microbiota-host interactions. Inflamm Bowel Dis. 2012;18(5):943–954. doi: 10.1002/ibd.21895. [DOI] [PubMed] [Google Scholar]
- 138.Campbell J.H. Host genetic and environmental effects on mouse intestinal microbiota. ISME J. 2012;6(11):2033–2044. doi: 10.1038/ismej.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]