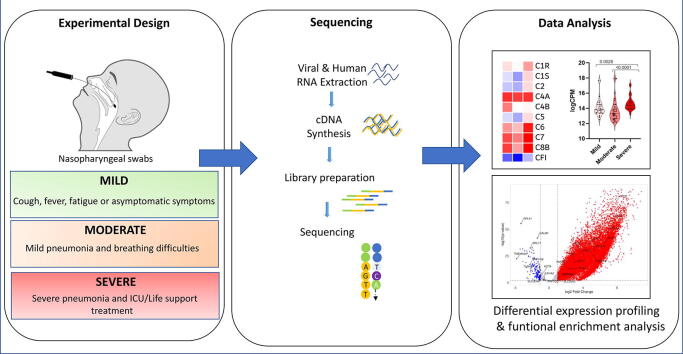
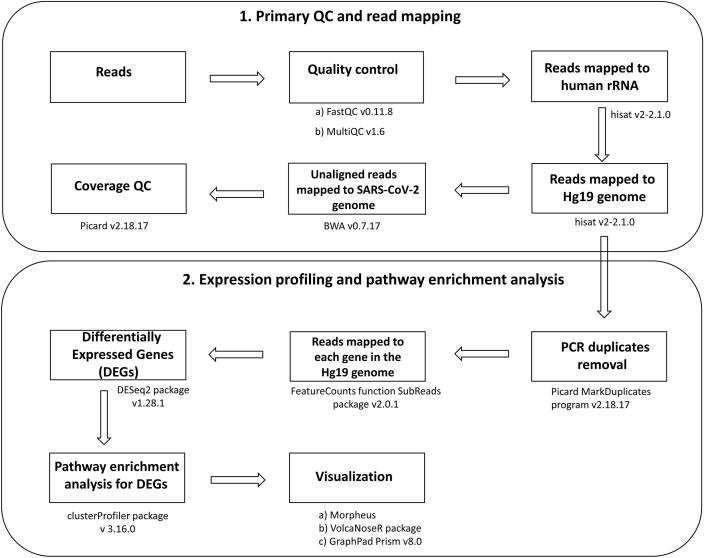
Graphical abstract
Keywords: Transcriptome sequencing, COVID-19, Disease severity, Expression signature, Nasopharyngeal swabs
Abstract
Characterizing key molecular and cellular pathways involved in COVID-19 is essential for disease prognosis and management. We perform shotgun transcriptome sequencing of human RNA obtained from nasopharyngeal swabs of patients with COVID-19, and identify a molecular signature associated with disease severity. Specifically, we identify globally dysregulated immune related pathways, such as cytokine-cytokine receptor signaling, complement and coagulation cascades, JAK-STAT, and TGF- β signaling pathways in all, though to a higher extent in patients with severe symptoms. The excessive release of cytokines and chemokines such as CCL2, CCL22, CXCL9 and CXCL12 and certain interferons and interleukins related genes like IFIH1, IFI44, IFIT1 and IL10 were significantly higher in patients with severe clinical presentation compared to mild and moderate presentations. Differential gene expression analysis identified a small set of regulatory genes that might act as strong predictors of patient outcome. Our data suggest that rapid transcriptome analysis of nasopharyngeal swabs can be a powerful approach to quantify host molecular response and may provide valuable insights into COVID-19 pathophysiology.
1. Introduction
Since the first case reported in December 2019 in Wuhan, SARS-CoV-2 has spread very quickly across 188 different countries leading to over 31,887,485 COVID-19 cases and 976,789 associated deaths worldwide as of 24th September 2020 [1]. The common signs and symptoms of SARS-CoV-2 include fever, muscle pain, cough, fatigue, shortness of breath, and chest CT abnormalities [2], [3]. Some patients with COVID-19 quickly develop severe pulmonary symptoms including acute respiratory distress syndrome (ARDS), pulmonary edema, intense kidney injury and multiple organ failure [4] whereas other patients present with no symptoms or with only mild disease [5]. The development of ARDS and episodes of thromboembolism leads to disseminated intravascular coagulation (DIC) representing primary causes of lethality during COVID-19 infection [6]. Patients with severe progression of COVID-19 also show signs of hyperinflammatory syndrome i.e. secondary haemophagocytic lymphohistiocytosis (HLH), a potentially fatal cytokine storm with multiorgan failure [7]. Similarly lymphopenia, increased serum ferritin, D-dimer, C-reactive protein (CRP), and lactic-dehydrogenase (LDH), levels are also considered to be the predictors of poor outcome of COVID infection [8]. Pathogen load and prevalence of infections are higher in males than females demonstrating that sex physiology plays a role in infectious disease pathogenesis [9]. Meng and colleagues [10] showed that men who died of COVID-19 had elevated levels of systemic inflammatory markers such as neutrophil-to-lymphocyte ratio and C-reactive protein. Nevertheless, the mechanism behind increased mortality among older adults and males with COVID-19 remains speculative.
Despite the worldwide spreading, the host immune response against SARS-CoV-2 infection remains poorly characterized. Dysregulation of host immune response and activation of inflammatory cytokines, known as the “cytokine storm”, is associated with disease severity and poor prognosis [11], [12], [13]. A recent host transcriptome study on patients with COVID-19 revealed distinct host inflammatory cytokine profiles and highlighted the association between SARS-CoV-2 pathogenesis and excessive release of cytokine such as CCL2/MCP-1, CXCL10/IP-10, CCL3/MIP-1A, and CCL4/MIP1B [14]. Additionally, Lieberman et al. showed upregulation of antiviral factors such as OAS1-3 and IFIT1-3, and Th1 chemokines CXCL9/10/11 upon SARS-CoV2 induced antiviral response. This study also showed immune responses may underlie disparities between males and females, and among the elderly compared to younger age groups [15].
At present, it is not known if the host gene expression profile varies among patients with mild, moderate, or severe clinical outcomes. Identification of such transcriptomic differences can be useful for predicting COVID-19 outcomes and for better management and earlier interventions especially for patient groups with severe outcomes. In this study, we performed RNA sequencing on nasopharyngeal tissue of patients with mild, moderate, and severe disease to characterize transcriptomic regulation and immune response differences among these patients, and to identify markers for early identification of patients with possible severe disease outcomes.
2. Methods
2.1. Patient cohort and ethics statement
This study was approved by the Dubai Scientific Research Ethics Committee - Dubai Health Authority (approval number #DSREC-04/2020_02). The Ethics committee waived the requirement for informed consent since this study was part of a public health surveillance and outbreak investigation in the UAE. The electronic medical records of patients with laboratory confirmed SARS-CoV-2 were reviewed and important clinical data were extracted using the World Health Organization (WHO) case report form.
Our patient cohort consisted of fifty patients with COVID-19 (36% female) who tested positive for SARS-CoV-2 by RT-qPCR with a mean age of 40.9 years (ranging from 3 to 70 years) at diagnosis. Cases were categorized into mild, moderate, and severe as previously described [16]. Briefly, mild cases were asymptomatic or had mild non-life-threatening symptoms, while moderate cases presented with symptoms (such as persistent fever) requiring medical attention and/or hospitalization. Severe cases presented with advanced disease and pneumonia requiring admission to intensive care units and life-support treatment (such as mechanical ventilation). In total, 37 patients (21 males and 16 females; mean age 36.4 years) had mild disease, 10 patients (9 males and 1 female; mean age 49.3 years) had moderate presentations, while 3 patients (2 males and 1 female; mean age 68.0 years) had severe/critical disease. All patients were non-smokers. All 3 patients with severe disease had chronic health issues (hypertension, chronic kidney disease and/or diabetes mellitus) (Table 1).
Table 1.
Clinical characteristics of COVID-19 patients in this study.
| Characteristic | All patients (N = 50) | Stratified by disease severity |
||
|---|---|---|---|---|
| Asymptomatic/mild (N = 37) | Moderate (N = 10) | Severe/critical (N = 3) | ||
| Age Mean (SD), yr | 40.92 (16.03) | 36.4 (14.04) | 49.28 (14.35) | 68 (3.56) |
| Female Sex – No./total no. (%) | 18/50 (36%) | 16/37 (43.24%) | 1/10 (10%) | 1/3 (33.33%) |
| Body Mass (kg) – Mean (SD) | 66.2 (30.84) | 63.4 (28.65) | 73.07 (41.83) | 77.93 (8.29) |
| Current Smoker – no./total no. (%) | 0/50 (0%) | 0/37 (0%) | 0/10 (0%) | 0/3 (0%) |
| Coexisting Disorder - no./total no. (%) | ||||
| Chronic Cardiac Disease (not Hypertension) | 3/50 (6%) | 1/37 (2.7%) | 2/10 (20%) | 0/3 (0%) |
| Hypertension | 9/50 (18%) | 5/37 (13.51%) | 3/10 (30%) | 1/3 (33.33%) |
| Chronic Pulmonary Disease | 0/50 (0%) | 0/37 (0%) | 0/10 (0%) | 0/3 (0%) |
| Asthma | 0/50 (0%) | 0/37 (0%) | 0/10 (0%) | 0/3 (0%) |
| Chronic kidney disease | 1/50 (2%) | 0/37 (0%) | 0/10 (0%) | 1/3 (33.33%) |
| Chronic liver disease | 0/50 (0%) | 0/37 (0%) | 0/10 (0%) | 0/3 (0%) |
| Chronic Neurological Disorder | 0/50 (0%) | 0/37 (0%) | 0/10 (0%) | 0/3 (0%) |
| Diabetes Mellitus | 8/50 (16%) | 3/37 (8.1%) | 3/10 (30%) | 2/3 (66.66%) |
| Malignant Neoplasms | 0/50 (0%) | 0/37 (0%) | 0/10 (0%) | 0/3 (0%) |
2.2. Sample preparation, RNA isolation, library construction and sequencing
RNA was extracted from nasopharyngeal swabs using the QIAamp Viral RNA Mini or the 213 EZ1 DSP Virus Kits (Qiagen, Hilden, Germany). All patients in this study tested positive for SARS-CoV-2 by RT-qPCR performed at Dubai Health Authority Hospitals. RNA libraries were then prepared for shotgun transcriptome sequencing using the TruSeq Stranded Total RNA Library kit from Illumina (San Diego, CA, USA) as previously described [17], [18]. Libraries were then sequenced using the NovaSeq SP Reagent kit (2 X 150 cycles) from Illumina (San Diego, CA, USA) to generate a minimum of 15 M reads per sample (Supplementary Table 1).
2.3. Data analysis
The sequencing read quality was checked using FastQC v0.11.8 [19] and MultiQC v1.6 [20]. High quality reads (Q ≥ 30) were first mapped to rRNA sequences to remove potential rRNA reads using hisat v2-2.1.0 [21] with the default parameter. The remaining reads were then mapped to the GRCh37 (hg19). The unmapped reads were then mapped to the SARS-CoV-2 genome (GenBank Accession number: NC_045512.2) using BWA v0.7.17. PCR duplicates were removed with Picard MarkDuplicates program v2.18.17 [22]. Samples with more than 6 M reads aligned to the GRCh37 (hg19) were considered for further analysis. RNA sequencing data obtained from nasopharyngeal swabs of samples confirmed to be COVID-19 negative by RT-qPCR, hereafter referred to as controls (n = 32), were downloaded from GSE152075 [15].
The number of reads mapped to each gene in the genome (GRCh37) was calculated using the FeatureCounts program in the SubReads package v2.0.1 [23]. DESeq2 package v1.28.1 [24] was applied to perform batch effects and normalization. In brief, DESeq2 uses the median of ratios method to normalize data and estimate size factors (which control for differences in the library size of the sequencing experiment). We have also incorporated multi-factor design to control additional variation arising due to different library sizes. Distribution of Control and our cohorts was plotted using Counts per million (CPM) (Supplementary Fig. 1). CPM values are calculated by normalizing the read counts for a given gene by the total counts per sample. Further, gene expression change was calculated with respect to control for each groups using Wald test and p-values and log2 fold change was extracted. The resulting genes p-value was adjusted using the Benjamini and Hochberg method. These adjusted p-value (adj p-value) are calculated as: gene p-value × (m/i)) where m is the total number of genes, i is the gene p-value rank. Here we have considered a fraction of 5% false positives acceptable hence the genes with adj p-value < 0.05 were called as significant.
Pathway enrichment analysis was performed using the clusterProfiler package v 3.16.0 [25] to identify shared pathways among DEGs. Pathways with adj p-value < 0.05 were reported as significant. Heatmaps were generated using Morpheus [26] and volcano plots were generated using VolcaNoseR package [27]. CPM values were used to compare expression between certain genes and violin plots were generated using GraphPad Prism v8.0 [28]. Two tailed Mann–Whitney U test P-values are reported. A schematic illustration of data analysis is represented in Fig. 1.
Fig. 1.
Schematic workflow for Transcriptomic analysis. Sequencing data underwent pre-processing which includes primary QC and read mapping, followed by differentially expressed genes (DEG) analysis and downstream pathway enrichment analysis and visualization.
2.4. Data availability
The processed data from this study have been deposited in the Gene Expression Omnibus (GEO), under the accession number: GSE162835.
3. Results
3.1. Global transcriptomic changes in patients with COVID-19
To understand the host mechanisms of SARS-CoV-2 infections, we explored transcriptomic profiling of patients with COVID-19 using RNA extracted from nasopharyngeal samples (see Methods). We first mapped high quality reads to the human genome and the SARS-CoV-2 genome (GenBank accession number: NC_045512.2) and summarized sequencing statistics in Supplementary Table 1. On average, ~2% (ranging from 0.1% to 13.6%) of the reads mapped to the SARS-CoV-2 genome confirming the presence of the virus, albeit coverage over the viral genome varied across patients most likely due to differences in viral loads. On the other hand, ~65% (ranges from 24.3% to 91.0%) of the RNA reads mapped to the human genome, enabling host transcriptomic analysis.
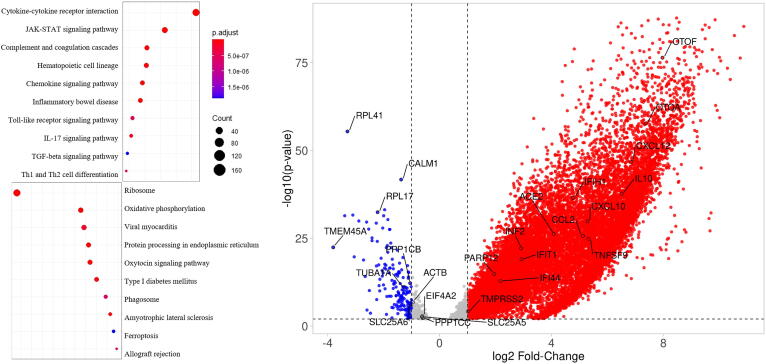
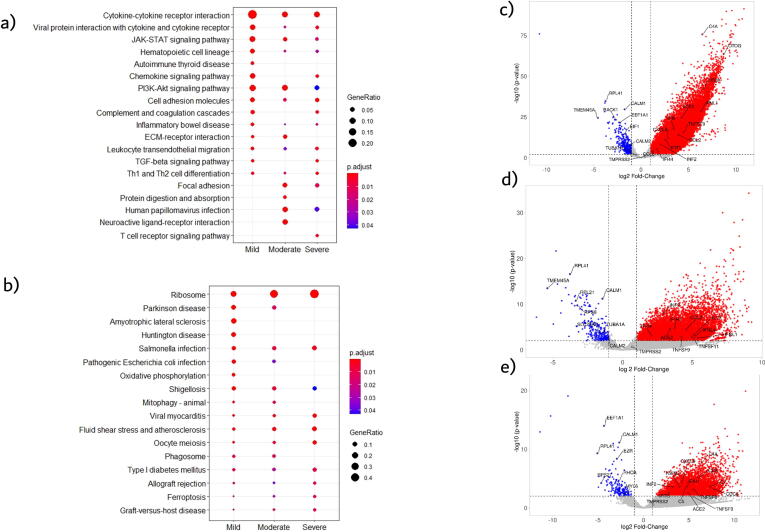
To detect global signature genes in patients with COVID-19 we combined transcriptomic data from patients with mild, moderate, and severe disease and made a comprehensive cohort called “COVID” to compare against controls for DEG analysis (see Methods). We detected 3547 upregulated genes and 410 downregulated genes with adj p-value < 0.05 and fold-change cutoff >2 (Supplementary File). Several interferon, cytokine and immune-related genes, such as CXCL5, CXCL12, CCL2, CCL4, CXCL10, IFIH1, IFI44, IFIT1 and IL6, IL10 were upregulated, whereas metabolic pathways or housekeeping genes, like RPL41, RPL17, SLC25A6, CALM1 and TUBA1A, were downregulated (Fig. 2).
Fig. 2.
Left, Dotplot visualization of enriched Pathway terms in all COVID-19 patients. The color of the dots represents adj p-value for each enriched pathway, and size represents the percentage of genes enriched in the total gene set. Right, Volcano plot representing upregulated and downregulated genes. X-axis represents log2 fold change of genes and Y-axis represents –log10P-value in differentially expressed gene (DEG) analysis.
Pathway enrichment analysis of the DEGs was performed to interrogate signaling programs induced by SARS-CoV-2. Up-regulated genes were related to cytokine-cytokine receptor interaction, JAK-STAT signaling pathway, complement and coagulation cascades and other inflammatory pathways. Whereas, several metabolic pathways were negatively enriched, including ribosome and ER protein processing, which suggests a global reduction in the production of proteins related to cellular energy production (Fig. 2). Oxidative phosphorylation pathway was mainly downregulated in patients with mild disease. This could be due to immune cells, such as macrophages/monocytes and immune sentinels, triggering intracellular cascades which in turn alter mitochondrial metabolism and impede mitochondrial oxidative phosphorylation for ATP production in favor of cytosolic aerobic glycolysis [29].
3.2. Transcriptome analysis of patients with mild, moderate and severe COVID-19
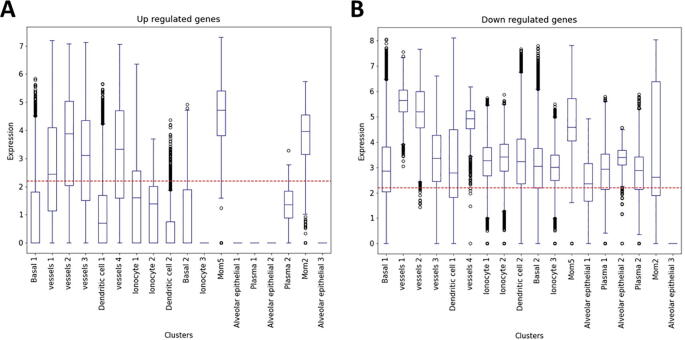
Next, we compared the transcriptomic profiles of patients with mild, moderate, and severe disease each separately with controls to determine if the above global signature, and the identified cytokine storm, hold similarly or varies by disease severity. Overall, we detected 2315 upregulated and 418 downregulated genes in mildly, 3413 upregulated and 302 downregulated genes in moderately, and 2691 upregulated and 215 downregulated genes in severely ill patient samples. Most of the immune response genes were significantly upregulated in all patient groups (Supplementary Fig. 2). Interestingly, single cell transcriptomic analysis from bronchoalveolar fluid (BALF) of patients with severe COVID-19 (from another study) showed that dysregulated genes identified here overlap with different cell types, including monocyte derived alveolar macrophages (MoMs) (Supplementary Fig. 3).
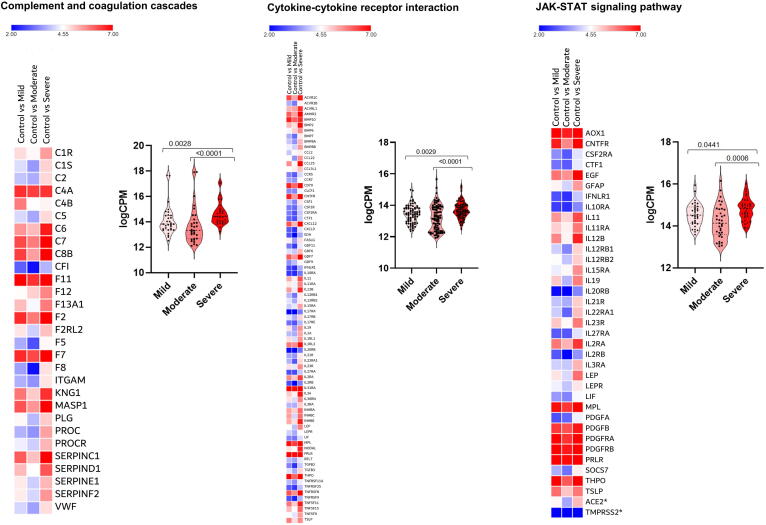
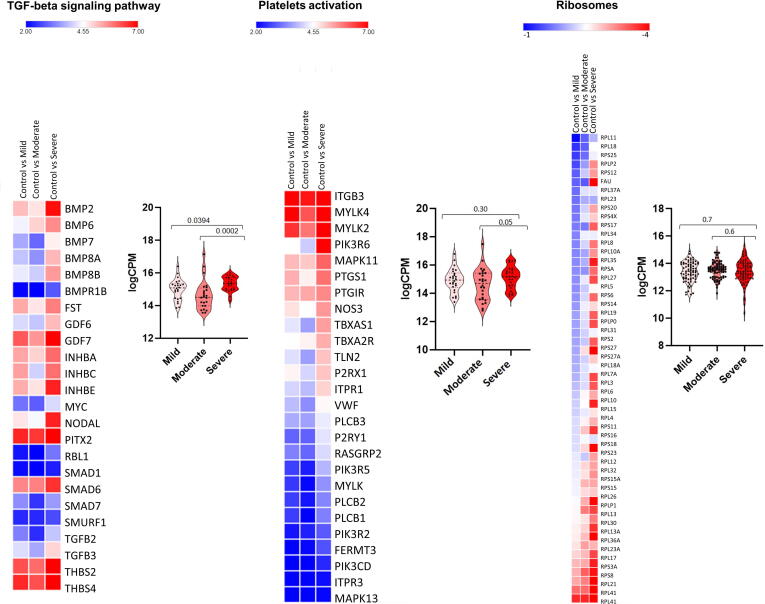
Furthermore, pathway enrichment analysis showed modulation, to varying extents (Fig. 3), of several overlapping pathways among mildly, moderately, and severely affected patients (Supplementary Fig. 2). In addition to upregulation of immune-related response genes, there was consistent disruption of the ribosome pathway. The SARS-CoV-2 receptor ACE2 is an interferon-regulated receptor and is upregulated in response to SARS-CoV-2 infection [30]. We also found that ACE2 expression was highest in patients with severe disease compared to patients with mild and moderate disease (Fig. 3).
Fig. 3.
Heat map and Violin plots of pathways in patients with mild, moderate or severe disease. Pathways are: a) Complement and coagulation cascades, b) Cytokine-cytokine receptor interaction, c) JAK-STAT signaling pathway, d) TGF-beta signalling, e) Platelets activation, and f) Ribosome. Heat map depicts the log2 fold change of differentially expressed gene (DEGs) of COVID-19 patients compared with controls. Genes included have a log2 fold change of more than 1 and a p-adjusted value of <0.05. For every gene in a given pathway, the average counts per million (CPM) was calculated in cases with mild (n = 37), moderate (n = 10) and severe (n = 3) disease, and these CPM values (dark dots) were plotted (Violin plots) indicating median and quartiles as well as minima and maxima bounds. *Expression of ACE2 and TMPRSS2.
3.3. Comparison of functional enrichment among patients with mild, moderate, and severe COVID-19
To further delineate the differences among these groups, we compared the expression levels of genes and associated pathways (complement and coagulation cascades, cytokine-cytokine receptor interaction, JAK STAT signaling, TGF beta signaling pathway, platelets activation and ribosome pathway) between patients with mild, moderate, and severe disease. Gene expression in most regulated pathways was significantly higher (adj p-value < 0.05) in patients with severe disease compared to those with mild or moderate disease (Fig. 3). For example, expression of certain interleukins (such as IL11, IL12, IL19, IL34), interleukin receptors (like IL10RA, IL21R and IL11RA), chemokines and tumour necrosis factor genes (such as CXCL12, CXCL9, CCL25, CCL2, CCR5, CCR7, TNFSF9, TNFSF15, TNFRSF25 and TNFRSF9) were significantly higher in patients with severe disease compared to patients with mild or moderate disease (adj p-value < 0.05; Fig. 3). Furthermore, complement and coagulation cascades (e.g. C2, C5, C6, F12 and F8) were activated to a higher extent in patients with severe disease. Finally, patients with severe disease had higher expression of STAT4, STAT5A and STAT5B which are important components of JAK-STAT pathway and play a critical role in the fate of T helper cells [31].
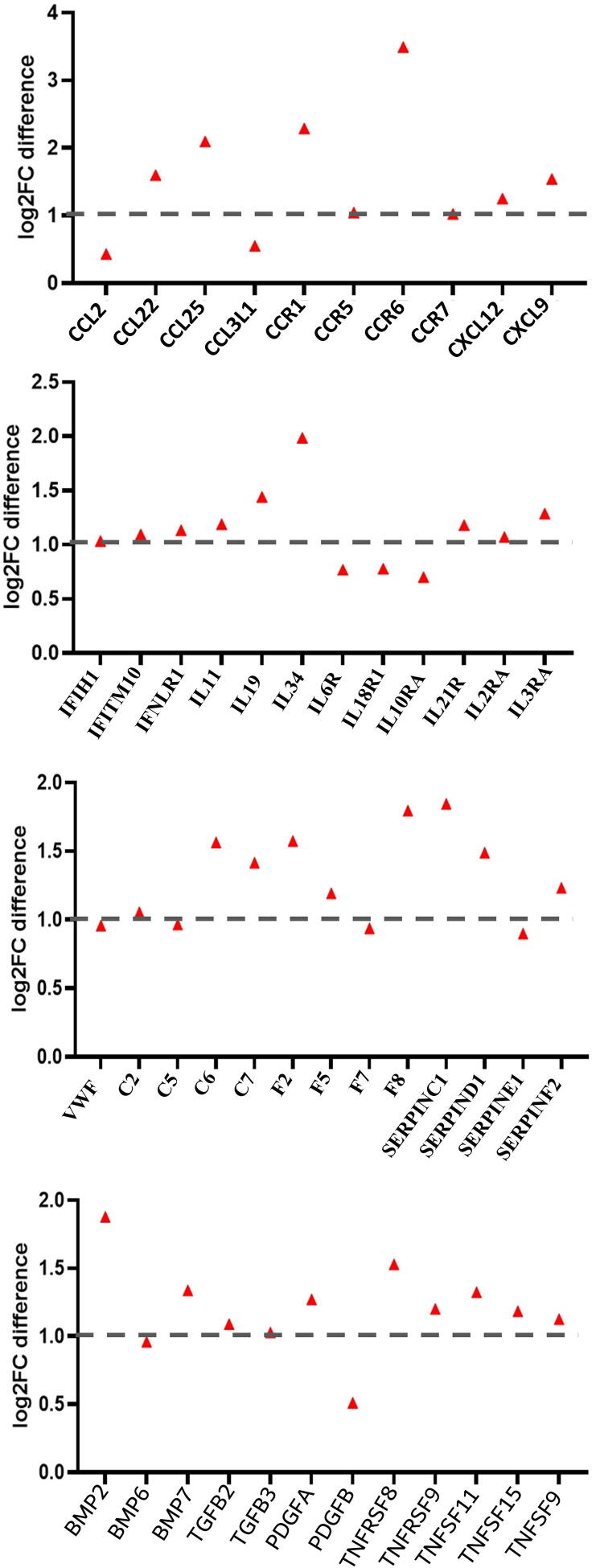
The highest fold change (>2) in patients with severe disease was among the pro-inflammatory cytokines and chemokine receptors CCR1 (CXCL8/CXCL6 receptor), CCR6, CCR22, CCR25, IL3RA, IL11, IL19, and IL21RA, the TGF-β signaling genes BMP2, BMP7, PDGFA, and TNFSF11, and the complement and coagulation cascade genes C6, C7, F2, F5, SERPINC1, and SERPIND1 (Fig. 4). This distinction is likely due to viral infection resulting in release of inflammatory responses to a higher extent in patients with severe disease as compared to patients with mild or moderate disease.
Fig. 4.
Scatter plot of fold change gene expression differences between patients with mild/moderate and severe outcomes. Fold change of mild/moderate was averaged and subtracted from severe fold change when compared to controls.
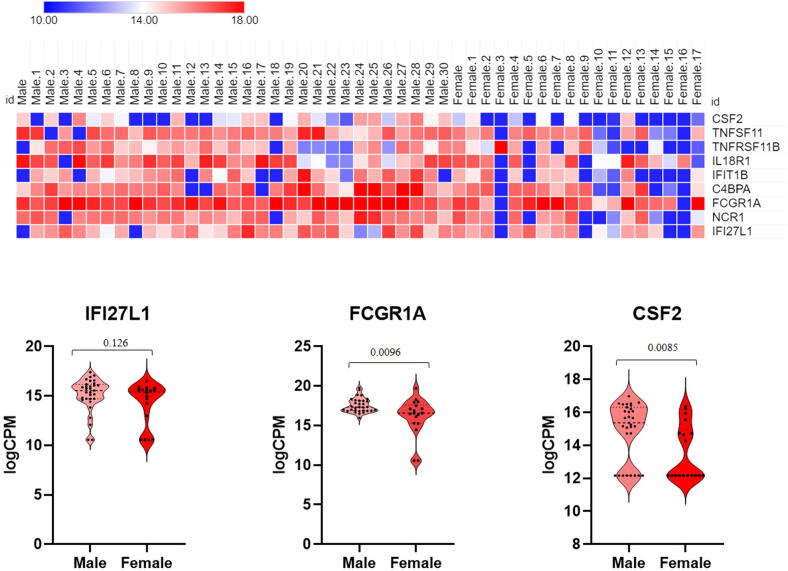
Finally, we compared transcriptomic changes in male and female COVID-19 patients with mild and moderate outcomes. Several immune related genes (CSF2, TNFSF11, TNFRSF11B, IL18R1, IFIT1B and C4BPA) were upregulated in male patients compared to females (Fig. 5).
Fig. 5.
Heat map (top) and Violin plots (bottom) of differentially expressed genes in males and females. Heat map depicts the log2 fold change of differentially expressed gene (DEGs) of COVID-19 patients compared with controls. Genes included have a log2 (fold change) of more than 1 and adj p-value < 0.05. Violin plots represent each gene as CPM (counts per million) for each male of female patients. Shown are the median and 75th quartiles as well as minima and maxima bounds of CPM (counts per million) of highlighted differentially regulated genes.
4. Discussion
Transcriptomic analysis of nasopharyngeal tissues from patients with COVID-19 reveals a robust induction of cytokine and immune-related profiles by SARS-CoV-2 infection. Those results were highly consistent with those from Lieberman et al. study from which controls were used in our analysis. Our findings are also similar to those recently observed by Butler et al. [32] in New York (United States) and Qin et al. [33] in Wuhan (China). The latter work reported that an increase in ‘serum’ cytokine and chemokine levels, as well as in neutrophil–lymphocyte-ratio (NLR) in SARS-CoV-2 infected patients is correlated with the severity of the disease and adverse outcomes, suggesting a role for hyper-inflammatory responses in COVID-19 pathogenesis. In addition to immune-related pathways, elevated levels of complement and coagulation cascades were also observed in our study. Inflammation-induced coagulation pathways, which can themselves be regulated by the complement system, are pivotal in controlling pathogenesis associated with infections and have recently been shown to correlate with adverse outcomes [34].
Our RNA work, using nasopharyngeal tissue, corroborates and extends those findings to show that those pathways are more significantly altered in patients with severe disease outcomes. Specifically, SARS-CoV-2 seems to induce expression of certain genes (Fig. 4) to higher extents in patients with severe, compared to those with mild or moderate disease. This finding has direct prognostic and therapeutic implications. Identifying molecules targeting the pathways where those genes are involved might help ameliorate or avoid adverse clinical outcomes due to SARS-CoV-2. Moreover, this expression signature can be used to predict the clinical course of disease, so that early interventions can be implemented. However, a cautionary note should be made since the differentially expressed genes presented in our study have been obtained from small cohort of severe patients (n = 3). Hence the data may be biased, due to the high degree of inter-individual variability that characterize SARS-CoV-2 infection. In addition, since 2 out of the 3 patients in our cohort suffered from diabetes mellitus (Table 1), it is possible that our differential gene expression is affected by this comorbidity. Larger datasets are needed to validate this expression signature and to show that it can reliably predict disease outcomes in the general population.
While preparing this manuscript, several groups published additional data consistent with our findings regarding the elevated cytokine expression profile in COVID-19 patients, and specifically those with severe outcomes. Single-cell RNA sequencing (scRNA-seq) of nasopharyngeal and bronchial samples suggested that chemokine and chemokine receptor expression of the different cell populations increased markedly in the critical compared to the moderate cases [35]. In addition, another study identified distinct expression and cellular immunopathological patterns in the lungs of fatal COVID-19 [36]. Cavalli et al. identified humoral immune expression profiles in branchoalveolar lavage fluid samples from COVID-19 patients [37].
Certain molecular pathways require specific attention. In the majority of patients with COVID-19, respiratory failure is the primary cause of death and is often associated with uncontrolled inflammatory responses, edema, and lung fibrosis [38]. In our analysis, the TGF-β pathway was significantly upregulated in patients with severe disease suggesting they are more prone to respiratory failure [39]. Another main manifestation of COVID-19 is arterial or venous thrombosis, our data shows that complement and coagulation factors and of inhibitors of the fibrinolytic system, in particular SERPINE1 and SERPINF2, are up-regulated. Those genes may be responsible for the important development of thrombosis in COVID-19 patients [40].
The PI3K/Akt and JAK/STAT signaling pathways are two other noteworthy pathways given that several cellular and immune responses, including host cell immune response to counteract viral infection [41], activation of interferon-stimulated genes (ISGs) [42], and activation of several regulatory and pro-inflammatory cytokines [43] converge on those two pathways.
Our gender-specific transcriptomic analysis revealed a few genes which were upregulated in male patients compared to females (Fig. 5). It is highly likely that many other key factors, which are involved in sex-specific responses to SARS-CoV-2, were not detected in our study. It is possible that our cohort was not large enough to perform appropriate comparisons on a large number of patients adequately stratified for age, sex, and disease severity.
Taken together, our study highlights key molecular and functional pathways involved in COVID-19 pathogenesis and characterizes a specific Supplementary Figs. 1–3 expression signature associated with severe disease outcomes due to SARS-CoV-2.
5. Authors Statement
AAT was responsible for conception and design of the study. DH generated the data; RJ, SR, MU, and AAT analysed the data. RJ and AAT drafted the manuscript. AAT edited the manuscript. MU, TL, NN, HA, RV, ZD, AA, HK, and AA provided overall feedback and edited the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We would like to thank all members at Al Jalila Genomics Center and Dubai Health Authority Microbiology laboratory for facilitating this work.
Financial support and competing interests Statement
This work was supported by internal funds from the College of Medicine, Mohammed Bin Rashid University of Medicine and Health Sciences. Authors have no conflicts of interest to disclose.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.12.016.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
Supplementary figure 2.
Supplementary figure 3.
References
- 1.Johns Hopkins Center for Systems Sciences and Engineering. COVID-19 Dashboard. https://www.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6, 2020.
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciaffi J., Meliconi R., Ruscitti P., Berardicurti O., Giacomelli R., Ursini F. Rheumatic manifestations of COVID-19: a systematic review and meta-analysis. BMC Rheumatol. 2020;4:65. doi: 10.1186/s41927-020-00165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uddin M., Mustafa F., Rizvi T.A., Loney T., Al Suwaidi H., Al-Marzouqi A.H.H. SARS-CoV-2/COVID-19: viral genomics, epidemiology, vaccines, and therapeutic interventions. Viruses. 2020;12:1–18. doi: 10.3390/v12050526. https://www.mdpi.com/1999-4915/12/5/526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavalli E., Bramanti A., Ciurleo R., Tchorbanov A.I., Giordano A., Fagone P. Entangling COVID-19 associated thrombosis into a secondary antiphospholipid antibody syndrome: diagnostic and therapeutic perspectives (Review) Int J Mol Med. 2020;46:903–912. doi: 10.3892/ijmm.2020.4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramos-Casals M., Brito-Zerón P., López-Guillermo A., Khamashta M.A., Bosch X. Adult haemophagocytic syndrome. Lancet. 2014;383(9927):1503–1516. doi: 10.1016/s0140-6736(13)61048-x. [DOI] [PubMed] [Google Scholar]
- 8.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46(5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.vom Steeg L.G., Klein S.L. SeXX matters in infectious disease pathogenesis. PLoS Pathog. 2016;12:1–6. doi: 10.1371/journal.ppat.1005374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng Y., Wu P., Lu W., Liu K., Ma K., Huang L. Sex-specific clinical characteristics and prognosis of coronavirus disease-19 infection in Wuhan, China: a retrospective study of 168 severe patients. PLoS Pathog. 2020;16:1–13. doi: 10.1371/journal.ppat.1008520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fehr A.R., Channappanavar R., Perlman S. Middle east respiratory syndrome: emergence of a pathogenic human coronavirus. Annu Rev Med. 2017;68:387–399. doi: 10.1146/annurev-med-051215-031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newton A.H., Cardani A., Braciale T.J. The host immune response in respiratory virus infection: balancing virus clearance and immunopathology. Semin Immunopathol. 2016;38:471–482. doi: 10.1007/s00281-016-0558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Wit E., Van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lieberman NAP, Peddu V, Xie H, Shrestha L, Huang ML, Mears MC, et al. In vivo antiviral host response to SARS-CoV-2 by viral load, sex, and age. bioRxiv 2020. https://doi.org/10.1101/2020.06.22.165225. [DOI] [PMC free article] [PubMed]
- 16.Tayoun A.A., Loney T., Khansaheb H., Ramaswamy S., Harilal D., Deesi Z.O. Multiple early introductions of SARS-CoV-2 into a global travel hub in the Middle East. Sci Rep. 2020;10:17720. doi: 10.1038/s41598-020-74666-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harilal D., Ramaswamy S., Loney T., Al Suwaidi H., Khansaheb H., Alkhaja A. SARS-CoV-2 whole genome amplification and sequencing for effective population-based surveillance and control of viral transmission. Clin Chem. 2020;9:1–9. doi: 10.1093/clinchem/hvaa187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alm E., Broberg E.K., Connor T., Hodcroft E.B., Komissarov A.B., Maurer-Stroh S. Geographical and temporal distribution of SARS-CoV-2 clades in the WHO European Region, January to June 2020. Eurosurveillance. 2020;25:1–8. doi: 10.2807/1560-7917.ES.2020.25.32.2001410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010; Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 20.Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.http://broadinstitute.github.io/picard/.
- 23.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2013;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 24.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu G., Wang L.G., Han Y., He Q.Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.https://software.broadinstitute.org/morpheus/.
- 27.Goedhart J, Luijsterburg MS. VolcaNoseR – a web app for creating, exploring and sharing volcano plots. bioRxiv 2020. https://doi.org/10.1101/2020.05.07.082263. [DOI] [PMC free article] [PubMed]
- 28.GraphPad Prism version 8.00 for Windows, GraphPad Software, La Jolla California USA, www.graphpad.com.
- 29.Reiter R.J., Sharma R., Ma Q., Dominquez-Rodriguez A., Marik P.E. Melatonin inhibits COVID-19-induced cytokine storm by reversing aerobic glycolysis in immune cells: a mechanistic analysis. Med Drug Discov. 2020;6 doi: 10.1016/j.medidd.2020.100044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(1016–1035) doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seif F., Khoshmirsafa M., Aazami H., Mohsenzadegan M., Sedighi G., Bahar M. The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun Signal. 2017;15:1–13. doi: 10.1186/s12964-017-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butler DJ, Mozsary C, Meydan C, Danko D, Foox J, Rosiene J, et al. Shotgun Transcriptome and Isothermal Profiling of SARS-CoV-2 Infection Reveals Unique Host Responses, Viral Diversification, and Drug Interactions. bioRxiv 2020. https://doi.org/10.1101/2020.04.20.048066. [DOI] [PMC free article] [PubMed]
- 33.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y. Dysregulation of Immune Response in Patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramlall V., Thangaraj P.M., Meydan C., Foox J., Butler D., Kim J. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat Med. 2020 doi: 10.1038/s41591-020-1021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol. 2020;38(8):970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 36.Nienhold R., Ciani Y., Koelzer V.H., Tzankov A., Haslbauer J.D., Menter T. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat Commun. 2020;11(1):5086. doi: 10.1038/s41467-020-18854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavalli E., Petralia M.C., Basile M.S., Bramanti A., Bramanti P., Nicoletti F. Transcriptomic analysis of COVID-19 lungs and bronchoalveolar lavage fluid samples reveals predominant B cell activation responses to infection. Int J Mol Med. 2020;46:1266–1273. doi: 10.3892/ijmm.2020.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respirat Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen W. A potential treatment of COVID-19 with TGF-β blockade. Int J Biol Sci. 2020 Apr 21;16(11):1954–1955. doi: 10.7150/ijbs.46891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazzaroni M.G., Piantoni S., Masneri S., Garrafa E., Martini G., Tincani A. Coagulation dysfunction in COVID-19: The interplay between inflammation, viral infection and the coagulation system. Blood Rev. 2020;100745 doi: 10.1016/j.blre.2020.100745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diehl N., Schaal H. Make yourself at home: viral hijacking of the PI3K/Akt signaling pathway. Viruses. 2013;5:3192–3212. doi: 10.3390/v5123192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ezell S.A., Polytarchou C., Hatziapostolou M., Guo A., Sanidas I., Bihani T. The protein kinase Akt1 regulates the interferon response through phosphorylation of the transcriptional repressor EMSY. Proc Natl Acad Sci USA. 2012;109:613–621. doi: 10.1073/pnas.1115029109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villarino A.V., Kanno Y., Ferdinand J.R., O'Shea J.J. Mechanisms of Jak/STAT signaling in immunity and disease. J Immunol. 2015;194:21–27. doi: 10.4049/jimmunol.1401867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed data from this study have been deposited in the Gene Expression Omnibus (GEO), under the accession number: GSE162835.