Abstract
Transfer tRNAs (tRNAs) are small non-coding RNAs that are highly conserved in all kingdoms of life. Originally discovered as the molecules that deliver amino acids to the growing polypeptide chain during protein synthesis, tRNAs have been believed for a long time to play exclusive role in translation. However, recent studies have identified key roles for tRNAs and tRNA-derived small RNAs in multiple other processes, including regulation of transcription and translation, posttranslational modifications, stress response, and disease. These emerging roles suggest that tRNAs may be central players in the complex machinery of biological regulatory pathways. Here we overview these non-canonical roles of tRNA in normal physiology and disease, focusing largely on eukaryotic and mammalian systems.
Keywords: tRNA, tsRNA, arginylation, posttranslational modifications, RNA-mediated signaling
Introduction
Transfer RNAs (tRNAs) are adaptor molecules that translate genetic information into protein sequence by delivering amino acids to the protein synthesis machinery during translation. Mature tRNAs, formed from pre-tRNA through specialized steps of cleavage and posttranscriptional modifications (Figure 1A), are 73–93 nucleotides (nt) in length. Their secondary structure resembles a cloverleaf shape with four stem-loops: the acceptor stem where the amino acid is attached, the anticodon loop containing the anticodon triplet that recognizes the specific complementary codons on mRNA during protein synthesis, the deoxyuridine stem-loop (D-arm), the TΨC stem-loop (T-arm), and the variable loop (V-loop) (Giege, 2008; Figure 1B). The tRNA cloverleaf further folds into an L-shaped tertiary structure through base pairing of nucleotides in the D-loop and T-loop (Figure 1C).
FIGURE 1.
tRNA maturation and structure. (A) Maturation of tRNA involves 5′ and 3′ processing, intron splicing, modification of nucleotides, and addition of 3′ CCA end. Mature tRNAs are charged with cognate amino acids by aminoacyl-tRNA synthetases (aaRSs). This reaction involves conjugation of cognate amino acid to the 3′ terminal adenosine of tRNA by an ester bond (Berg and Offengand, 1958). (B) The generic secondary structure of tRNA with its constituent domains marked in different colors: acceptor stem (red); dihydrouridine (D-) stem and loop (blue); anticodon stem and loop (green); variable (V-) loop (gray); and TΨC (T-) stem and loop (yellow). Anticodon sequence is depicted in darker blue-green within the anticodon loop and is numbered (nt 34–36). Discriminator base (N73) aiding in aaRS recognition is located upstream of the 3′CCA tail (Giege, 2008). (C) Schematic representation of the L-shaped tertiary structure of yeast tRNAPhe. Loops are color-coded similarly to those in panel (B). The structure was derived from reported crystal structure (Protein Data Bank code 1EHZ) (Shi and Moore, 2000).
Transfer RNAs are synthesized as precursors by RNA polymerase III (PolIII) and undergo a multistep maturation process, involving removal of the 5′ leader, trimming of the 3′ trailer, splicing of introns, modifications of nucleotides, and addition of the 3′ terminal CCA tail that serves as a site for amino acid charging (Figure 1A). After processing of pre-tRNAs in the nucleoplasm, mature tRNAs are transported to the cytoplasm through the nuclear pore complexes, which serve as quality control to ensure that only correctly processed tRNAs can pass into the cytoplasm (Chatterjee et al., 2018). In the cytoplasm, mature tRNAs are aminoacylated by their cognate aminoacyl-tRNA synthetases (aaRSs) (Berg and Offengand, 1958), which enables them to deliver the attached amino acids to the ribosome for incorporation into the growing polypeptide chain. This reaction involves conjugation of cognate amino acid to 2′ or 3′ OH of the invariant 3′ terminal adenosine in the tRNA by an ester bond. Each aaRS recognizes specific identity elements within the cognate tRNA, e.g., the anticodon sequence, the discriminator base (N73) located upstream of the 3′ CCA end, and structural elements unique to each particular tRNA (Pang et al., 2014). Following amino acid conjugation, aminoacyl-tRNA complexed with elongation factor (eEF1α in eukaryotes) reaches the ribosome where the peptide bonds are formed.
Each of the 20 amino acids is usually encoded by several codons, which are referred to as synonymous codons. The frequency of synonymous codons is not uniform in the genome. Abundant/preferred codons are represented more than others, while rare/unpreferred codons are less frequent. This phenomenon is known as “codon usage bias.” In prokaryotes and unicellular eukaryotes, the frequency of a codon correlates with the corresponding tRNA gene copy number and the abundance of the cognate tRNA (Ikemura, 1985; Dong et al., 1996; Percudani et al., 1997). This co-adaptation is especially prominent in faster-growing bacteria (Wei et al., 2019), even though the relationship between codon usage and tRNA abundance in higher eukaryotes is more complex. It is believed that codon usage and cellular tRNA pool coevolved for an accurate and efficient protein production (Bulmer, 1987; Shields, 1990; Duret, 2000). In support, highly expressed genes, e.g., those encoding ribosomal proteins, generally contain preferred codons which are recognized by abundant tRNA species. However, finding direct correlations between codon usage and tRNA abundance in multicellular eukaryotes is complicated due to heterologous structure of the genome (i.e., isochoric structure), variations in individual gene structure (the length of the coding sequence and introns), and higher tRNA gene redundancy (Hurst and Williams, 2000; Kanaya et al., 2001; Comeron, 2004; dos Reis et al., 2004; Semon et al., 2006).
Development of high-throughput tools, including tRNA-based microarrays (Dittmar et al., 2004; Polte et al., 2019), and more recently, optimized methods for high-throughput tRNA sequencing (Zheng et al., 2015; Gogakos et al., 2017; Shigematsu et al., 2017; Erber et al., 2020; Pinkard et al., 2020) have provided more efficient means for detection of individual tRNA-level fluctuations and enabled the discovery of key trends that regulate cell homeostasis in a tRNA-dependent way. These studies revealed that tissue-specific tRNA expression generally reflects codon preference of highly expressed genes in human, indicating the importance of translation regulation via tRNA abundance in different tissues (Dittmar et al., 2006). This is consistent with the idea that codon-mediated translation control can regulate tissue-specific protein expression (Plotkin et al., 2004). Some of the more complex trends have also been revealed, suggesting that abundance and expression of specific tRNAs can drive reprogramming of different cell states, such as proliferation, differentiation, and metastatic potential (Gingold et al., 2014; Goodarzi et al., 2016; Zhang et al., 2018b). Opposing tRNA-level signatures have been reported in proliferating and differentiating cells, and it has been found that changes in individual tRNA abundance correlate with, and potentially coordinate, selective gene expression profiles operating different states of the cell (Gingold et al., 2014). Elevated levels of individual tRNAs promote cells’ metastatic progression by enhancing stability and translation of metastasis-driven genes enriched for their cognate codons, thereby shifting the cellular program toward disease-promoting state (Goodarzi et al., 2016). Thus, changes in translation efficiencies driven by the combination of codon usage and tRNA abundance constitute a powerful mechanism of protein regulation.
For the last 50 years, tRNA has been studied exclusively as an adaptor molecule between mRNA nucleotide sequence and amino acids in protein synthesis. New studies provide evidence that tRNAs perform a number of previously unanticipated regulatory roles in various metabolic pathways, from modulation of global gene expression to regulation of cell death. In prokaryotes, tRNAs are known to be used in cell wall biosynthesis, antibiotic biogenesis, and transcription attenuation, i.e., via T-box mechanism [see (Katz et al., 2016) for a recent review of non-canonical roles of tRNAs in prokaryotes].
Here we discuss the roles tRNAs play in critical regulatory pathways, in addition to their conventional roles in translation, focusing largely on eukaryotic and mammalian systems.
tRNAs Modulate Global Gene Expression
All cells are surrounded by a changing environment, which modulates cellular homeostasis and serves as a source of diverse forms of signals, including stress. Cells develop various mechanisms to respond to stress conditions, including reprogramming of gene expression that enables cells to adapt and maintain their normal physiology. Some of this reprogramming links directly to the protein synthesis machinery and tRNA.
Different types of stress trigger phosphorylation of serine 51 in the eukaryotic translation initiation factor 2α (eIF2α). This phosphorylation reduces the formation of the ternary complex (eIF2-GTP-Met-tRNAiMet) that transfers Met-tRNAiMet to the ribosome, therefore leading to a decrease in global protein synthesis (Merrick and Pavitt, 2018). Four eIF2α kinases are regulated by different stimuli: protein kinase R-like ER kinase (PERK) by unfolded proteins in the ER, protein kinase R (PKR) by double-stranded RNA in virus-infected cells, heme-regulated inhibitor (HRI) by heme deficiency in erythroid cells, and general control non-repressible kinase 2 (GCN2) by serum starvation (Taniuchi et al., 2016; Wek, 2018).
Cytoplasmic tRNAs rapidly translocate into the nucleus under a number of stress conditions, including nutrient starvation (Shaheen and Hopper, 2005; Takano et al., 2005; Hurto et al., 2007; Shaheen et al., 2007; Whitney et al., 2007; Dhakal et al., 2019), heat shock (Miyagawa et al., 2012; Watanabe et al., 2013), viral infection (Zaitseva et al., 2006), and oxidative stress (Schwenzer et al., 2019). This translocation not only directly depletes tRNAs from protein synthesis but also modulates stress response by activation of the GCN2 kinase (Castilho et al., 2014). GCN2 specifically binds deacylated tRNA via its histidyl-tRNA synthetase-like domain, which discriminates against aminoacyl-tRNAs (Dong et al., 2000; Zaborske et al., 2010; Figure 2). This binding leads to a conformational change of GCN2 and activates the kinase, which in turn phosphorylates eIF2α (Dever et al., 1992; Anda et al., 2017). Phosphorylated eIF2α is a competitive inhibitor of the guanine nucleotide exchange factor eIF2B (Adomavicius et al., 2019), and thus phosphorylation inhibits the recycling of eIF2 between the cycles of translation, resulting in downregulation of the global protein synthesis. Simultaneously, this pathway triggers increased translation of a select number of mRNAs containing short upstream open reading frames (uORF). Two of those mRNAs encode activating transcription factor 4 (ATF4) in mammals and general control protein (GCN4) in yeast (Hinnebusch et al., 2016; Young and Wek, 2016), which are required for selective expression of amino acid biosynthesis genes upon stress. Under normal conditions, translation of uORFs leads to repression of main ORFs (mORF) of ATF4 and GCN4. During nutrient deficiency, phosphorylated eIF2α reduces the ternary complex concentration, which leads the 40S ribosomal subunit to bypass the uORF start codon, instead initiating translation at ATF4 and GCN4 mORFs (Somers et al., 2013; Hinnebusch et al., 2016).
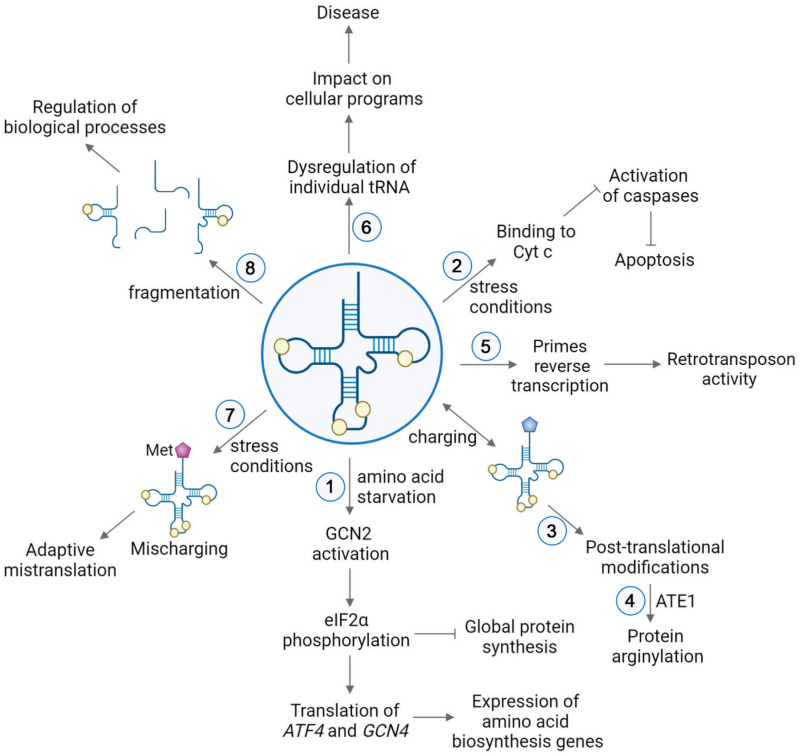
FIGURE 2.
tRNA functions in various pathways in cell physiology. (1) Upon nutrient deficiency, binding of deacylated tRNAs to GCN2 leads to conformational changes of the kinase GCN2, resulting in phosphorylation of eIF2α. This causes repression of global protein synthesis, along with translation of activating transcription factor 4 (ATF4) in mammals and general control protein (GCN4) in yeast. These proteins induce selective expression of amino acid biosynthesis genes upon stress (Hinnebusch et al., 2016; Young and Wek, 2016). (2) Stress conditions trigger the release of cytochrome c (Cyt c) from mitochondria to the cytoplasm. In the cytoplasm, binding of tRNAs to Cyt c prevents association of Cyt c with Apaf-1. Thereby, the caspase cascade required for apoptosis is precluded (Mei et al., 2010; Suryanarayana et al., 2012; Gorla and Sepuri, 2014; Liu et al., 2016). (3) Aminoacyl-tRNAs serve as a donors in posttranslational modifications. (4) In protein arginylation, ATE1 transfers Arg from arginyl-tRNAArg onto a protein substrate. (5) tRNA is used as a primer in the reverse transcription of LTR retrotransposons (Martinez, 2017). (6) Dysregulation of individual tRNA contributes to the onset and severity of different diseases by impacting cellular programs. (7) Mismethylation of tRNAs is induced in response to stress conditions. Extra incorporation of Met in newly synthesized proteins serves an evolutionary strategy for expanding the genetic information in proteins (Netzer et al., 2009; Wiltrout et al., 2012; Lee et al., 2014; Wang and Pan, 2015; Schwartz et al., 2016). (8) Generation of tRNA-derived small RNAs leads to global regulatory responses (Keam and Hutvagner, 2015; Xie et al., 2020).
Similar to mammalian cells, nutrient deprivation decreases amino acid availability and causes deacylated tRNA accumulation in prokaryotes (Potrykus and Cashel, 2008; Steinchen and Bange, 2016). The binding of deacylated tRNAs to the A-site of the ribosome induces the production of alarmones, referred to as (p)ppGpp, through the ribosome-associated protein RelA (Brown et al., 2016). In E. coli, (p)ppGpp directly binds to RNA polymerase and changes its affinity for sigma factors, leading to modulation of global gene expression (Ross et al., 2013, 2016). The binding of (p)ppGpp to RNA polymerase limits transcription of rRNA genes required for rapid growth while promoting the expression of stress response and amino acid biosynthesis genes. (p)ppGpp also binds to various proteins, leading to inhibition of cellular processes, such as translation and nucleotide biosynthesis (Kanjee et al., 2012; Wang et al., 2019). This in turn reprograms bacterial metabolism to slow down growth and reallocate resources during nutrient deficiency.
In addition to this generalized stress response, individual tRNAs also play regulatory roles in gene expression in humans. Misfolded tRNAAspGUC isodecoder, referred to as tRNAAsp7, binds to Alu RNA element present in the 3′ UTR of the AspRS mRNA sequence (Rudinger-Thirion et al., 2011). This interaction alters 3′ UTR folding, thereby reshaping the accessibility of the two alternative polyadenylation sites and leading to induction of expression of AspRS (Rudinger-Thirion et al., 2011). Thus, a single tRNA isodecoder regulates translation of an essential enzyme in the cell.
tRNAs Control Cell Death
Apoptosis is a cellular process of elimination of damaged or unwanted cells, characterized by distinct morphological changes, including membrane blebbing and nuclear fragmentation. Apoptosis plays major roles in normal physiology and disease and is considered a vital component of organismal homeostasis [see, e.g., (Tang et al., 2019) for a recent review]. This pathway can be initiated through one of the two routes: an extrinsic pathway, in which extracellular ligands bind to cell-surface death receptors, or an intrinsic pathway, which originates within the cell and is mediated by mitochondria (Pistritto et al., 2016). Both types of apoptosis are primarily executed by caspases (cysteinyl, aspartate-specific proteases) that induce global proteolysis and thus trigger cell death. There are two types of caspases: initiator caspases, e.g., caspase-8 and -9, which are activated by auto-processing, and effector caspases, e.g., caspase-3, -6, and -7, which become active upon proteolytic cleavage by initiator caspases (McIlwain et al., 2013).
The intrinsic apoptotic pathway is mediated by intracellular stimuli that converge at the mitochondrial level under various stress conditions, e.g., irradiation, viral infection, and oncogene activation. This pathway causes the loss of mitochondrial membrane potential, which leads to the release of cytochrome c (Cyt c) into the cytoplasm (Tait and Green, 2010; Figure 3). Cyt c is an essential mitochondrial protein which builds an electrochemical gradient driving ATP synthesis. In the cytoplasm, Cyt c binds to the death adaptor apoptotic protease-activating factor-1 (Apaf-1) and ATP or dATP, which triggers assembly of the oligomeric apoptosome complex (Acehan et al., 2002; Figure 3). This complex recruits procaspase-9, leading to its auto-proteolytic activation (Rodriguez and Lazebnik, 1999; Zou et al., 1999; Li et al., 2017; Figure 3). Then, caspase-9 activates effector caspases, e.g., caspase-3/7, and eventually degrades many cellular proteins and results in cell death (Figure 3).
FIGURE 3.
tRNAs play a protective role in the intrinsic apoptosis pathway. An internal stimulus, e.g., DNA damage or viral infection, causes the loss of mitochondrial membrane potential and leads to a release of cytochrome c (Cyt c) to the cytoplasm. Cytosolic Cyt c binds to the death adaptor apoptotic protease-activating factor-1 (Apaf-1) and ATP, triggering assembly of the oligomeric apoptosome complex, which recruits procaspase-9, leading to its activation. Caspase-9 activates effector caspases, e.g., caspase-3/7, and eventually leads to cell death. In this pathway, tRNAs directly bind to Cyt c and prevent Cyt c association with Apaf-1 and apoptosis.
Transfer RNAs have been shown to modulate apoptosis (Mei et al., 2010; Suryanarayana et al., 2012; Gorla and Sepuri, 2014; Liu et al., 2016; Figures 2, 3). Addition of RNase to cell extracts enhances Cyt c-induced caspase-9 activation, while exogenous cellular RNA supplementation reduces auto-activation of caspase-9 in a dose-dependent manner (Mei et al., 2010). Systematic analysis showed that tRNAs directly bind to Cyt c, leading to prevention of Cyt c association with Apaf-1 (Mei et al., 2010). Microinjection of tRNA into living cells inhibits apoptosis, while tRNA degradation enhances caspase activation (Mei et al., 2010). Taken together, these results show the critical role of tRNA in programmed cell death (Figures 2, 3).
Transfer RNAs bind to Cyt c through its heme domain, which defines the redox state of the protein (Suryanarayana et al., 2012; Gorla and Sepuri, 2014). It was proposed that tRNA interaction prevents the positively charged residues of Cyt c from being exposed to the Apaf-1 complex (Gorla and Sepuri, 2014). There are conflicting studies on how tRNA binding affects the redox state of Cyt c. Gorla and colleagues found that oxidized Cyt c is not able to interact with tRNA (Gorla and Sepuri, 2014). However, another study revealed that oxidized Cyt c can bind to tRNA with a 2-fold weaker affinity compared to the reduced form and, furthermore, that this binding promotes Cyt c reduction (Liu et al., 2016). Moreover, tRNA interaction hinders the peroxidase activity of Cyt c, which plays a role in the activation of the caspase cascade and Cyt c release from the mitochondria (Liu et al., 2016). Hence, this study concludes that tRNA is capable of regulating apoptosis by switching off the active form of Cyt c.
Recently, it has been found that Cyt c recognizes the L-shaped tertiary structure of tRNAs, particularly the core region (Liu et al., 2016). The binding affinity of Cyt c to tRNA is at a similar range to other tRNA-binding proteins [Kd 1–3.5 μM for Cyt c (Liu et al., 2016), compared to 1–3 μM for aaRS (Zhang et al., 2006; Liu et al., 2007, 2011) and 0.8–3.3 μM for tRNA nucleotidyltransferase (Kim et al., 2009)], therefore it is unknown how Cyt c competes with other tRNA-binding proteins in vivo.
Altogether, these results unveil an important role of tRNA in controlling cell death by preventing Cyt c from Apaf-1 activation. Notably, both tRNA and Cyt c are highly conserved molecules. Cyt c is a crucial mitochondrial protein that carries electrons from Cyt c reductase to Cyt c oxidase as part of the electron transport chain. This function has been evolutionarily conserved in eukaryotes over 1.5 billion years (Ow et al., 2008). Likewise, tRNA exists in all forms of life and its structure is identical in all organisms. Therefore, tRNA:Cyt c interaction has been proposed to be ancient (Hou and Yang, 2013; Raina and Ibba, 2014) and might have been established early in evolution as an indicator of the balance between protein synthesis, energy production, and cell survival.
tRNAs Are Utilized for Protein Modifications
Posttranslational protein modifications are fundamental processes that significantly amplify the repertoire of the proteome. Covalent addition of various chemical groups to proteins plays essential roles in physiological and pathological processes. In addition to a number of groups that are different from any elements of protein structure, proteins can be extensively modified by posttranslational addition of amino acids (Kaji et al., 1963; Kaji, 1968; Fung and Fahlman, 2015; Gadadhar et al., 2017; He et al., 2018; Kimura et al., 2018; Chen and Kashina, 2019). Some of these modifications involve direct amino acid transfer; however, a specific class of these modifications, including arginylation in eukaryotes (Kaji et al., 1963; Kaji, 1968; Chen and Kashina, 2019), L/F transfer in prokaryotes (Fung and Fahlman, 2015), and the recently demonstrated amino acid addition to the primary amines of the Lys side chains (He et al., 2018), involve transfer of amino acids from the charged tRNA. Here we overview arginylation, a posttranslational modification of emerging importance that regulates fundamental mechanisms in eukaryotes and mammalian cells (Figure 2).
Arginylation was discovered more than 50 years ago, when researchers observed addition of radioactively labeled amino acids to proteins in tRNA-dependent but ribosome-independent manner (Kaji et al., 1963; Kaji, 1968). Arginylation is mediated by arginyltransferase 1 (ATE1) that transfers arginine from aminoacyl-tRNAs onto proteins. Initial studies revealed that ATE1 targets N-terminally exposed acidic residues (Glu and Asp) of proteins (Soffer, 1971). Cys was also found to be arginylated in mammalian cells (Gonda et al., 1989). More recently, it has been shown that Cys oxidization greatly facilitates its arginylation, and this mechanism has been proposed to mediate intracellular oxygen sensing in mammalian cells (Hu et al., 2005) and plants (Gibbs et al., 2011). Lately, it was shown that arginylation is not limited to N-terminal residues; mid-chain Glu and Asp residues were found to be arginylated on their side chains in intact proteins (Wang et al., 2014).
Arginylation is essential for regulation of physiological functions of key proteins in vivo. Ate1 deletion in mice leads to embryonic lethality and severe defects in cardiovascular development and angiogenesis (Kwon et al., 2002). N-terminal arginylation of β-actin facilitates cell motility and was proposed as a mechanism that contributes to cell migration in vivo (Karakozova et al., 2006; Kurosaka et al., 2010). Arginylated calreticulin regulates stress granules scaffolding and enhances apoptotic response (Carpio et al., 2013; Comba et al., 2019). Moreover, lack of arginylation leads to neurodegeneration (Wang et al., 2017). Altogether, arginylation acts as a global regulator to control various biological conditions in cells (Saha and Kashina, 2011; Rassier and Kashina, 2019).
Although arginylation was discovered almost 60 years ago, its molecular mechanism is not fully elucidated. Our recent study shows that ATE1-mediated arginylation is highly specific to tRNAArg conjugated with Arg (Avcilar-Kucukgoze et al., 2020). Mouse tRNAArg species can all participate in arginylation, albeit with somewhat different efficiencies, suggesting potential in vivo preferences of ATE1 enzyme for specific tRNAArg (Avcilar-Kucukgoze et al., 2020). However, it is difficult to estimate whether these preferences amount to different efficiencies of arginylation by different tRNAs in vivo, since tRNA repertoire is highly dynamic (Torrent et al., 2018). On the other hand, abundance of arginyl-tRNAArg also depends on availability of intracellular arginine, which, in addition to its role in protein synthesis, also serves as a precursor for the synthesis of important molecules such as nitric oxide, urea, polyamines, proline, glutamate, creatine, and agmatine (Szefel et al., 2019). Thus, it is possible that intracellular level of Arg plays a key role in balancing all these processes. In support, numerous cancers are defective in arginine biosynthesis due to reduced expression of arginine biosynthesis enzymes, argininosuccinate synthetase, and argininosuccinate lyase (Karatsai et al., 2020). Arg deprivation has been directly linked to impairments in actin arginylation and cell migration in invasive glioblastomas (Pavlyk et al., 2015). Ate1 deletion results in carcinogenic transformation of cultured fibroblasts (Rai et al., 2016). Hence, deficiency of intracellular Arg may affect fundamental processes, potentially due to altered arginyl-tRNAArg availability.
tRNA Prime Reverse Transcription of Retroviruses and Retrotransposons
Retrotransposons are mobile genetic elements abundantly found in eukaryotic genomes. Their transposition mechanism involves an RNA intermediate. Based on this mechanism, retrotransposons are divided into two groups: (1) Long terminal repeat (LTR) retrotransposons and (2) non-LTR retrotransposons (Mita and Boeke, 2016).
Long terminal repeat retrotransposons are characterized by the repeats of a few hundred base pairs on both ends. They replicate via reverse transcription using a cytoplasmic tRNA, with a sequence complementary to the primer binding site (PBS) at the 5′ end, acting as a primer. After reverse transcription, RNA template is partially degraded and full-length dsDNA is integrated into the chromosomal DNA [see (Martinez, 2017) for the recent review of tRNA role in retrotransposition]. LTR retrotransposons are structurally and evolutionary related to retroviruses, and the mechanism of LTR transposon replication via tRNA-primed reverse transcription and genome integration resembles that used by the retroviruses. The difference between retroviruses and retrotransposons is the PBS size. While PBS size is 18 nts in retroviruses, it varies between 8 and 18 nts in retrotransposons (Wilhelm and Wilhelm, 2001).
Specific tRNAs utilized as primers for different retrotransposons and retroviruses are determined by their PBS. For example, human immunodeficiency virus type 1 (HIV-1) uses human tRNALys,3 (Wain-Hobson et al., 1985), HTLV-1 uses tRNAPro (Seiki et al., 1983) while retrotransposons of the copia group use mostly tRNAMet as a primer (Kikuchi et al., 1986). In the case of HIV-1, human tRNALys,3 is selectively packaged into the virus particles and the level of tRNALys,3 in the virion correlates with the level of viral infectivity (Gabor et al., 2002). Lysyl-tRNA synthetase (LysRS) has been also found in HIV-1 virions, and it has been suggested that its interaction with the viral structural proteins Gag and Gag-Pol is required for tRNALys,3 encapsidation. It has been suggested that phosphorylation of LysRS on Ser207 promotes its release from the multisynthetase complex and its association with Gag (Duchon et al., 2017). Another study found that only mitochondrial LysRS (mLysRS) is present in HIV-1 extracts (Kaminska et al., 2007). Recently, the molecular details of the interaction between mLysRS and Gag-Pol have been revealed (Phongsavanh et al., 2020). The C-terminal integrase subunit of Gag-Pol is shown to stabilize the tRNALys,3:mLysRS interaction (Khoder-Agha et al., 2018; Phongsavanh et al., 2020).
tRNAs Play Emerging Roles in Disease
Above and beyond its role in viral infection, recent studies have proved that the fluctuation of tRNA repertoire plays a crucial role in shaping the cellular proteomic landscape. Dysregulation of individual tRNAs contributes to the onset and severity of various diseases, including metastatic and non-metastatic cancers, Huntington disease, cystic fibrosis, seizures, multiple myeloma, and neurodegeneration (Pavon-Eternod et al., 2009, 2013; Zhou et al., 2009; Girstmair et al., 2013; Ishimura et al., 2014; Birch et al., 2016; Goodarzi et al., 2016; Kirchner et al., 2017; Zhang et al., 2018b; Kapur et al., 2020; Figure 2).
The activity of PolIII is known to be upregulated in cancer, providing sufficient tRNA abundance to support protein synthesis in rapidly proliferating cells (Kantidakis et al., 2010; Arimbasseri and Maraia, 2016; Sriskanthadevan-Pirahas et al., 2018). tRNA levels are elevated up to 10 fold in breast cancer and multiple myeloma cell lines (Pavon-Eternod et al., 2009; Zhou et al., 2009). Interestingly, the increase of tRNAs is not uniform. tRNAArgCCU, tRNAArgUCU, tRNAThrCGU, tRNALeuUAA, tRNATyrGUA, and tRNASerGCU are more strongly overexpressed than others (Pavon-Eternod et al., 2009). tRNAGluUUC and tRNAArgCCG have been found to promote breast cancer metastasis (Goodarzi et al., 2016). The upregulation of these tRNAs increases translation efficiency of disease-promoting genes, which are enriched in the corresponding codons (Goodarzi et al., 2016). tRNAArg was reported to be pervasively upregulated in eight cancer types, and tRNAAsn was upregulated in five cancer types (Zhang et al., 2018b). Moreover, elevated level of tRNAiMet was reported to boost metabolic activity and proliferation in various cancer cells by reshaping the entire tRNA landscape (Pavon-Eternod et al., 2013; Gingold et al., 2014; Birch et al., 2016). Interestingly, the tRNAiMet level is regulated posttranscriptionally by miR-34a, which suppresses breast carcinogenesis and inhibits the proliferation of breast cancer cells (Wang et al., 2018). Thus, tRNAiMet has been proposed to be an oncogene by itself (Wang et al., 2018). Collectively, these studies suggest that individual tRNA expression is involved in tumor progression [see (Santos et al., 2019) for additional examples of tRNA deregulation in cancer].
Accumulating evidence supports the importance of tRNA expression in shaping various cellular programs. tRNA repertoire influences the onset and severity of diseases in a tissue-specific manner by modulating translation of specific genes. Kirchner and coworkers studied the impact of a specific synonymous single-nucleotide polymorphism (SNP) in the cystic fibrosis transmembrane conductance regulator (CFTR), which substitutes Thr ACT codon to ACG codon (Kirchner et al., 2017). This synonymous SNP alters protein stability and function. Researchers revealed that ACG codon is encoded by low-abundance tRNAThrCGU, particularly in human bronchial epithelia, which leads to slower local translation speed at Thr854 codon (Kirchner et al., 2017). The elevated level of tRNAThrCGU rescues the effects of synonymous SNP and restores the CFTR protein conformation and function (Kirchner et al., 2017). tRNAGlnCUG is implicated in Huntington’s disease (HD) (Girstmair et al., 2013), caused by extensive repeat of Gln CAG codon in the protein of huntingtin. Expression of an expanded CAG stretch depletes tRNAGlnCUG, leading to translational frameshifting and marked changes in the aggregation of huntingtin protein in vivo (Girstmair et al., 2013). Thus, depletion of a single tRNA may change translation dynamics and aggravate a major disease phenotype.
Mutations in tRNA genes cause many diseases. Two examples of best-characterized syndromes linked to mt-tRNA mutations are myoclonic epilepsy and ragged-red fiber (MERRF), and mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS) (Kirino and Suzuki, 2005). Majority of MELAS and MERRF cases are caused by single-point mutations [A3243G or T3271C for MELAS (Goto et al., 1990, 1991), A8344G for MERRF (Shoffner et al., 1990)] of mt-tRNALeuUAA and mt-tRNALysUUU, respectively. These pathogenic mutations hinder the taurine modification (5-taurinomethyluridine, i.e., τm5U, for mt-tRNALeuUAA and 5-taurinomethyl-2-thiouridine, i.e., τm5s2U, for mt-tRNALysUUU) in the wobble position (U34). While wild-type mt-tRNALeuUAA decode both UUA and UUG codons, the wobble modification deficiency in MELAS disrupts the decoding of UUG codon due to destabilization of U:G wobble base pairing (Yasukawa et al., 2000). This decoding bias specifically reduces the translation of the UUG-enriched NADH dehydrogenase 6 gene (MT-ND6), which is the component of the mitochondrial complex I. Similarly, in the case of MERRF, a 50–60% decrease in mt-tRNALysUUU aminoacylation capacity results in severe protein synthesis impairment due to premature termination of translation at Lys codons (AAA and AAG) (Enriquez et al., 1995). The pathological mutations are found at different locations within the tRNA molecule outside the anticodon, and it appears that they disturb the recognition of the RNA-modifying enzymes, which are responsible for producing the 5-taurinomethyl and 2-thio groups of the wobble bases of the mt-tRNAs (Koga et al., 2012). Since uridine modifications at the wobble position are crucial for precise and efficient codon recognition (Novoa et al., 2012; Agris et al., 2018), mutant mt-tRNAs induce translational defects of cognate codons and cause considerable decoding disorders. Considering that tRNA functions require multiple posttranscriptional modifications (Pan, 2018), it is expected that tRNA modification disorders develop widely in human diseases. Chronic ophthalmoplegia, cardiomyopathy, and hypertension are some examples of diseases caused by mutant mt-tRNAs. A complete list in MITOMAP (Brandon et al., 2005) and an extensive list of such diseases are included in Boczonadi and Horvath (2014), Kirchner and Ignatova (2015), Tahmasebi et al. (2018), Schaffer et al. (2019).
In mice, a point mutation (C50T in T-stem loop) of the central nervous system (CNS)-specific nuclear-coded tRNAArgUCU isodecoder (n-Tr20) results in a significant reduction of aminoacylation level and ribosome stalling at the AGA codons, although mouse genome contains four other tRNAArgUCU genes (Ishimura et al., 2014). tRNAArg C50T variants, including in the UCU isoacceptor, have also been observed in humans (Lant et al., 2019); however, no evidence has been found that these variants cause human disease. It is elusive how the expression of a single tRNA isodecoder is regulated in a tissue-specific manner, because the promoter sequences used by PolIII are identical among the five members of the tRNAArgUCU family. Interestingly, loss of wild-type (wt) n-Tr20 alters signaling pathways regulating transcription and translation, leading to changes in synaptic transmission and reduced seizure susceptibility (Kapur et al., 2020). Deletion of wt n-Tr20 regulates the transcription of 236 genes and splicing of 377 genes, demonstrating the role of a single tRNA isodecoder at different stages of RNA metabolism (Kapur et al., 2020).
Regulated tRNA-Dependent Mistranslation Serves as Adaptation Mechanism Upon Stress
For many decades since the original discovery of translation, it has generally been assumed that translational machinery is optimized to maintain translation errors at a minimum to sustain the cellular fitness. However, regulated mistranslation through tRNA misacylation may serve a beneficial mechanism to adapt the stress conditions (Figure 2). While technically translation-dependent, this role of tRNAs is unconventional enough to merit inclusion into this review.
Stress conditions induce tRNA misacylation with Met in mammalian cells (Netzer et al., 2009; Lee et al., 2014; Wang and Pan, 2015), yeast (Wiltrout et al., 2012), and bacteria (Jones et al., 2011; Schwartz et al., 2016; Schwartz and Pan, 2017). In mammalian cells, mismethionylated tRNAs constitute ∼1% compared to methyl-tRNAsMet under normal conditions; however, Met-misacylation increases up to 10 fold upon oxidative stress (Netzer et al., 2009). Mammalian tRNA mismethionylation is regulated through phosphorylation of two Ser residues of MetRS by the extracellular signal-regulated kinase (ERK) (Lee et al., 2014). The incorporation of Met at non-Met codons has been detected in various proteins (Netzer et al., 2009; Wiltrout et al., 2012; Lee et al., 2014; Wang and Pan, 2015), indicating that mismethionylated tRNAs are directly utilized in translation. Extra-genetic incorporation of Met residues protects enzyme active sites against reactive oxygen species (ROS)-mediated damage through their reactive sulfur group (Levine et al., 1996; Luo and Levine, 2009). Upon Ca+2 stress, extra incorporation of Met has been found in Ca+2/calmodulin-dependent protein kinase II (CaMKII), a multifunctional protein required for cellular Ca+2 homeostasis (Wang and Pan, 2015). Met-mistranslated CaMKII shows elevated activity especially when the cells are treated with Ca+2 (Wang and Pan, 2015). Met-mutant CaMKII proteins have distinct expression levels and subcellular localizations compared to the wild type protein under Ca+2 stress (Wang and Pan, 2015). Moreover, human AlaRS mischarges tRNACys through recognition of G4:U69 base pair in the acceptor stem, resulting in Cys-to-Ala substitution in the overexpressed reporter protein in HEK293T cells (Sun et al., 2016). The biological meaning of this AlaRS mischarging mechanism is unknown (Sun et al., 2016), but studies suggest that such regulated tRNA misacylation represents an evolutionary strategy for expanding the genetic information in proteins, especially under stress conditions.
Generation of tRNA Fragments Serves Regulatory Roles in vivo
Earlier studies reported the existence of stable tRNA-derived small RNAs (tsRNA), presumably generated by cleavage of mature tRNA (Borek et al., 1977; Speer et al., 1979). Initially, these tsRNAs have been assumed to be by-products of random degradation of tRNAs, but recent advances in high-throughput sequencing methods revealed the existence of numerous stable tsRNAs in all three domains of life. A growing body of evidence unveils diverse biological roles of tsRNAs in cellular physiology and suggests that these molecules constitute a class of small non-coding RNAs involved in global cellular regulation (Keam and Hutvagner, 2015; Xie et al., 2020; Figure 2).
Based on their length and the tRNA cleavage sites involved in their generation, tsRNAs are mainly divided into two groups. One is stress-induced tRNA fragments (tiRNAs), produced by a specific cleavage at or near the anticodon loop, generating two fragments of 31–40 nts in length (Xie et al., 2020; Figure 4). In mammals, this cleavage is mediated by angiogenin (ANG) [a member of the ribonuclease (RNase) A superfamily], generating 5′- and 3′-tRNA halves (Fu et al., 2009; Yamasaki et al., 2009; Elkordy et al., 2018; Su et al., 2019; Figure 4). In yeast, tRNA halves are generated by Rny1p (a member of RNase T2 family) (Thompson and Parker, 2009). tiRNA accumulation is generally induced by various stress conditions, e.g., nutrition deficiency, UV radiation, heat shock, and oxidative stress (Thompson et al., 2008; Fu et al., 2009; Yamasaki et al., 2009; Elkordy et al., 2018).
FIGURE 4.
Biogenesis and classification tsRNAs. tsRNAs are divided into two main types, tiRNA (red) and tRF (blue). tiRNAs are produced by a specific cleavage at the anticodon loop of mature tRNA, generating two fragments, 5′-tiRNA and 3′-tiRNA. This cleavage is mediated by angiogenin (ANG), mostly under stress conditions. 5- and 3-tRF are produced by a specific cleavage at or near D- and T-loops of mature tRNAs. “a,” “b,” and “c” for 5-tRFs and 3-tRFs to define fragments of different length (∼15–18, 22, and 31 nt, respectively). The enzymes mediating this cleavage are not fully characterized but are believed to involve endonuclease Dicer and/or RNase T2. 2-tRF is derived from the internal region and generated by the unknown cleavage method. 1-tRF contains 3′-trailer, produced by RNase Z/ELAC2. tsRNA, tRNA-derived small RNA, tiRNA, stress induced tRNA fragment, tRF, tRNA-derived fragment.
The second group is tRNA-derived fragments (tRFs), produced by a specific cleavage at or near D- and T-loops, yielding fragments of 14–30 nt in length, corresponding to 5′-derived tRFs (5-tRF) and 3′-derived tRFs (3-tRF) (Figure 4), as well as the internal piece corresponding to the tRNA sequence in between (2-tRF, also referred to as tRF-i) (Telonis et al., 2015) and a 3′-trailer (1-tRF). 5-tRF and 3-tRF come at different lengths, and thereby these fragments are further classified into 5a-tRFs (∼15 nt), 5b-tRFs (∼22 nts), 5c-tRFs (∼31 nts), 3a-tRFs (∼18 nt), and 3b tRFs (∼22 nt) (Kumar et al., 2015). tRNA cleavage generating these fragments is believed to be mediated by endonuclease Dicer, a member of the RNase III family (Babiarz et al., 2008; Cole et al., 2009; Yeung et al., 2009; Haussecker et al., 2010; Maute et al., 2013); however, other studies show that Dicer deficiency does not abolish tRF generation, suggesting that other RNase(s) are also involved (Li et al., 2012; Kumar et al., 2014; Kuscu et al., 2018). In plants, it has been proposed that RNase T2, rather than Dicer, is the main enzyme generating these regulatory tRFs (Megel et al., 2019), and it is possible that RNAse T2 also acts in this pathway in other organisms. Overall, the questions of potential additional enzymes involved in this process, and their hierarchy in vivo, require further investigation. It appears likely that different RNases can mediate tRF generation under different conditions to modulate tRF-regulated processes in the cell.
Transfer RNA modifications have an impact on the tsRNAs biogenesis. NSun2 and Dnmt2 mediate posttranscriptional methylation of tRNA at cytosine-5 (m5C) (Bohnsack et al., 2019). Loss of mammalian Dnmt2 and NSun2 resulted in hypomethylated tRNAs and accumulation of tsRNAs (Tuorto et al., 2012; Blanco et al., 2016; Zhang et al., 2018a). It was found that lack of m5C at C38 position significantly changes the RNA secondary structure (Zhang et al., 2018a), which might facilitate tRNA cleavage. Highly expressed PUS7 catalyzes the modification of uridine (U) to pseudouridine and its deletion in human embryonic stem cells affects specific tRFs, especially those derived from tRNA containing a 5′ terminal oligoguanine (TOG) (Guzzi et al., 2018). These results show that RNA modifications contribute to the generation of tsRNA. tsRNAs levels can vary in different cells and tissues, as well as in different diseases; therefore, several databases listing the entire repertoire of cellular tsRNAs have been generated (Kumar et al., 2015; Selitsky and Sethupathy, 2015; Zheng et al., 2016; Pliatsika et al., 2018; Li et al., 2020; Yao et al., 2020; Zuo et al., 2020). Potentially, modulation of tsRNA levels can be involved in disease-related cellular reprogramming and constitute conceptually novel targets for therapeutics.
Stress-induced tRNA halves, especially 5′-tiRNAs, are known to reduce global protein synthesis by ∼20% (Yamasaki et al., 2009). This reduction cannot be a direct consequence of depletion of mature cytoplasmic tRNA pool, because only a small fraction (<5%) of total tRNA is fragmented upon stress (Fu et al., 2009; Yamasaki et al., 2009; Saikia et al., 2012). Evidence suggests that tiRNAs, upon cleavage, cause translational repression. It was found that 5′-tiRNAs, but not 3′-tiRNAs, derived from tRNAAla and tRNACys, inhibit translation initiation (Ivanov et al., 2011). 5′-tiRNAs interact with the cold shock domain of the translation repressor Y-box-binding protein 1 (YB-1), which leads to displacement of cap-binding complex eIF4F from the cap structures (m7G) (Ivanov et al., 2011; Figure 5). tiRNA-induced translation inhibition also induces the assembly of stress granules, which constitute translationally stalled mRNAs, associated pre-initiation factors, and signaling proteins (Emara et al., 2010; Ivanov et al., 2011). 5′-tiRNAAla/Cys possess a unique oligoguanine motif at their 5′ terminus, which forms G-quadruplex (G4) structures required for translation inhibition (Ivanov et al., 2014). On the other hand, tsRNAs can directly interact with the ribosomes and inhibit global protein synthesis in various organisms (Gebetsberger et al., 2012; Sobala and Hutvagner, 2013; Gonskikh et al., 2020). A recent study suggests that 5′-tiRNAPro, produced in a stress-independent manner, represses global protein synthesis through binding to the ribosomes in mammalian cells (Gonskikh et al., 2020; Figure 5). Interestingly, addition of 5′-tiRNAPro leads to accumulation of peptidyl-tRNA inside the arrested ribosomes (Gonskikh et al., 2020; Figure 5). It is unknown whether the peptidyl-tRNA product possesses a biological role by itself or if it is a by-product of stalled ribosomes (Gonskikh et al., 2020). Moreover, some tsRNAs have functions similar to known miRNAs and piRNAs, repressing expression of a target gene by binding to complementary sites in 3′ UTR.
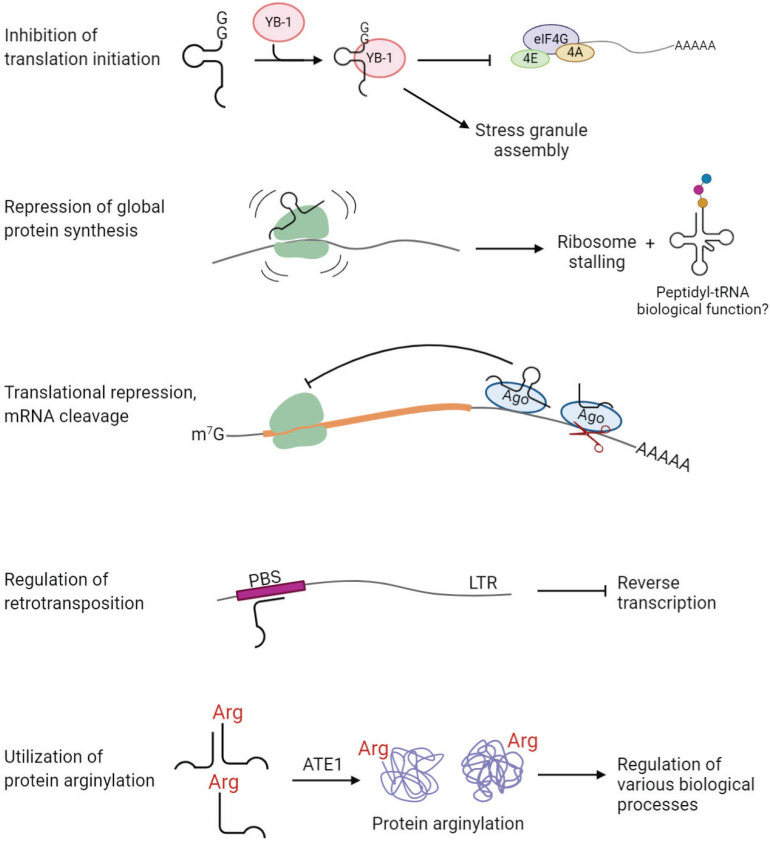
FIGURE 5.
tsRNAs play regulatory functions in cell physiology. Examples of tsRNA functions include inhibition of translation initiation, repression of global protein synthesis, translational repression and mRNA cleavage, regulation of retrotransposition, and utilization of protein arginylation.
Several studies revealed an association of tsRNAs with Argonaute (Ago) proteins, the key players in RNA silencing (Cole et al., 2009; Haussecker et al., 2010; Li et al., 2012; Kumar et al., 2014; Kuscu et al., 2018; Guan et al., 2020) [reviewed in Shigematsu and Kirino (2015)] (Figure 5). In HeLa cells, Dicer-dependent 5′-tRFGlnCUG is associated with Ago1 and Ago2, even though the binding is weak (Cole et al., 2009). Another study showed that 1-tRFs and 3-tRFs preferentially bind to Ago3/4 over Ago1/2 in HEK293 cells (Haussecker et al., 2010). 3-tRFHisGTG and 3-tRFLeuCAG were detected in Ago2-IP fractions in MEFs (Li et al., 2012). Analysis of PAR-CLIP data revealed that both 5-tRFs and 3-tRFs bind to Ago1, 3, and 4, but not Ago2, in HEK293 (Kumar et al., 2014). Moreover, 3-tRFs derived from tRNALeuAAG, tRNACysGCA and tRNALeuTAA downregulate expression of luciferase reporters in Ago-dependent manner (Kuscu et al., 2018). Importantly, the elevated level of 3-tRFLeuTAA significantly decreases the expression of target genes in vivo in HEK293T cells (Kuscu et al., 2018). A recent study found that 3-tRFAlaAGC interacts with Ago2 to modulate the expression of tumor suppressor FBZO47 by binding to 3′ UTR, resulting in enhanced cell proliferation, migration and invasion in gastric cancer (Zhang et al., 2020). Altogether, accumulating evidence suggests that tsRNAs are capable of repressing target genes posttranscriptionally through binding to Ago proteins, similar to miRNAs.
Some, recent studies classify the roles of tsRNAs in translational regulation into two global pathways: Ago-dependent, which targets specific mRNAs to induce translation inhibition, and Ago-independent, which acts by exerting structural effects on mRNA and rRNA independently of the Ago proteins. These aspects of tsRNA-dependent translation regulation have been recently reviewed in Shi et al. (2019).
The protective role of tRNAs against apoptosis, described earlier in this review, also involves tRNA-derived fragments. A recent study showed that the released Cyt c preferentially binds ANG-induced tiRNAs, but not full-length tRNAs, in response to hyperosmotic stress in mouse embryonic fibroblasts, leading to attenuation of Apaf-1 oligomerization and apoptosome activity (Saikia et al., 2014). This report points that tiRNAs also play a critical role in the activation of the apoptosis pathway under stress.
An interesting study published in 2017 has unveiled a “safeguard” role of tsRNAs in genome integrity in mammalian cells (Schorn et al., 2017; Figure 5). LTR retrotransposons integrated into the genome are tightly regulated to prevent mutations, especially during early embryonic development, when epigenetic suppression of transposon elements is inconceivable. This study found that abundant endogenous 3-tRFs can specifically bind tRNA primer binding site (PBS) at the 5′ UTR of LTR retrotransposons, where tRNA normally binds during LTR retrotransposon replication (Schorn et al., 2017). These 3-tRFs repress retrotranspositions of the two most active mouse transposons by inhibiting reverse transcription in preimplantation stem cells (Schorn et al., 2017). It is still unknown how the cell arranges silencing of transposable elements by generating tRFs in certain conditions.
The role of tsRNA in intercellular communication is only beginning to be explored. tsRNAs are enriched in extracellular vehicles (EVs) more than any other class of RNA (Chiou et al., 2018). For T cells, 45% of tRFs are at least 1.5-fold enriched in EVs; some are enriched up to 10-fold (Chiou et al., 2018). Interestingly, silencing of EV-enriched tRFs promotes T cell activation, suggesting that these tRFs are actively released by secretion to prevent repression of immune activation (Chiou et al., 2018). tRNA halves can be delivered to the sperm during its maturation in epididymis, potentially via EVs (Sharma et al., 2016, 2018). These tRNA halves regulate embryonic gene expression of metabolic pathways depending on parental diet (Chen et al., 2016; Sharma et al., 2016). Stable 5′ tRNA halves are found to circulate in mouse blood, most likely as part of a nucleoprotein complex (Dhahbi et al., 2013). A recent study unraveled the generation of tRNA halves in the extracellular environment by RNase 1, a highly active secreted nuclease, after the release of full-length tRNAs into the extracellular space (Nechooshtan et al., 2020). However, their cellular targets are not yet known (see (Tosar and Cayota, 2020) for a recent review of extracellular tRNA-derived fragments).
In principle, amino acid-charged tRNAs can be cleaved by tsRNA-generating machinery to produce aminoacyl-tsRNAs. Such aminoacyl-tsRNAs would then be incapable of participating in translation, due to the lack of anticodon loop, but can potentially still serve as amino acid donors for other reactions utilizing tRNA-bound amino acids. Given that tRNA secondary and tertiary structure is generally very stable, it appears possible that after tRNA cleavage, 5-tRF and 3-tRFs stick together due to the complementary base pairing. While this has never been directly demonstrated, some examples of amino acid-charged tsRNAs have been identified. A previous study found abundant aminoacylated tiRNAs derived from tRNAAspGUC, referred to as SHOT-RNAs, in breast and prostate cancers (Honda et al., 2015). Our recent study showed that arginyl-tRFArg can be generated in vitro from Arg-tRNAArg using RNase T2 and that such Arg-tRFArg can mediate arginylation (Avcilar-Kucukgoze et al., 2020; Figure 5). As translation and arginylation compete for the same substrate, arginyl-tRNAArg, such aminoacyl-tRF generation in this case, if it happens in vivo, may serve as a switch between these two processes (Avcilar-Kucukgoze et al., 2020). In support, lack of Ate1 significantly alters the ratio of tRNAArg:tRFArg in MEFs, suggesting a functional link between tRFArg and arginylation in vivo (Avcilar-Kucukgoze et al., 2020). We propose that arginyl-tRFArg generation is a mechanism that maintains the balance between translation and arginylation and can potentially shift this balance in favor of one or the other process in response to physiological stimuli. This study illustrates that aminoacyl-tRFs may serve as amino acid donors in pathways other than translation, and opens up a possibility of other pathways that depend on the regulated balance between translation and ribosome-independent utilization of tRNA-conjugated amino acids.
Conclusion
Emerging data from multiple studies suggest that, in addition to the tRNAs’ role in translation, these small non-coding RNAs play a wealth of other functions that regulate and fine-tune multiple processes in normal physiology and disease. These studies so far have just scratched the surface of the complexity involved in the biological functions of tRNAs. In this review, we described the usage of tRNAs in various mechanisms and pathways, which are critical for the fate of the cell. Some of these processes involve uncharged tRNA as a regulator; however, some of them consume aminoacyl-tRNA which is also a substrate for translational machinery. It is an important question how ribosome and other aminoacyl-tRNA-dependent processes compete for the common substrate, i.e., aminoacyl-tRNAs. For example, L/F transferase has a strong preference for the most abundant tRNALeuCAG (Fung et al., 2014). However, under starvation, the tRNALeuCAG aminoacylation level drops to 5% (Dittmar et al., 2005). How does L/F transferase compete with the ribosome, especially when the leucyl-tRNALeu gets scarce? In fact, it is a general question for all of the processes which rely on the use of charged tRNAs. Although it remains largely elusive, tRNA fragmentation may serve a mechanism that potentially shifts the balance depending on the demand of the cell for either translation or arginylation (Avcilar-Kucukgoze et al., 2020). On the other hand, tRNAs with specific posttranscriptional modifications may be dedicated to particular non-translational processes. There is no doubt that development of new tools will provide insights into the competition for the aminoacyl-tRNAs in the future.
Author Contributions
Both authors conceptualized, planned, and wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by NIH R35GM122505 and R01NS102435 to AK.
References
- Acehan D., Jiang X., Morgan D. G., Heuser J. E., Wang X., Akey C. W. (2002). Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol. Cell 9 423–432. [DOI] [PubMed] [Google Scholar]
- Adomavicius T., Guaita M., Zhou Y., Jennings M. D., Latif Z., Roseman A. M., et al. (2019). The structural basis of translational control by eIF2 phosphorylation. Nat. Commun. 10:2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris P. F., Eruysal E. R., Narendran A., Vare V. Y. P., Vangaveti S., Ranganathan S. V. (2018). Celebrating wobble decoding: half a century and still much is new. RNA Biol. 15 537–553. 10.1080/15476286.2017.1356562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anda S., Zach R., Grallert B. (2017). Activation of Gcn2 in response to different stresses. PLoS One 12:e0182143. 10.1371/journal.pone.0182143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri A. G., Maraia R. J. (2016). RNA polymerase III advances: structural and tRNA functional views. Trends Biochem. Sci. 41 546–559. 10.1016/j.tibs.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avcilar-Kucukgoze I., Gamper H., Polte C., Ignatova Z., Kraetzner R., Shtutman M., et al. (2020). tRNA(Arg)-derived fragments can serve as arginine donors for protein arginylation. Cell Chem. Biol. 27 839-849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz J. E., Ruby J. G., Wang Y., Bartel D. P., Blelloch R. (2008). Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, dicer-dependent small RNAs. Genes Dev. 22 2773–2785. 10.1101/gad.1705308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg P., Offengand E. J. (1958). An enzymatic mechanism for linking amino acids to RNA. Proc. Natl. Acad. Sci. U.S.A. 44 78–86. 10.1073/pnas.44.2.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch J., Clarke C. J., Campbell A. D., Campbell K., Mitchell L., Liko D., et al. (2016). The initiator methionine tRNA drives cell migration and invasion leading to increased metastatic potential in melanoma. Biol. Open 5 1371–1379. 10.1242/bio.019075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S., Bandiera R., Popis M., Hussain S., Lombard P., Aleksic J., et al. (2016). Stem cell function and stress response are controlled by protein synthesis. Nature 534 335–340. 10.1038/nature18282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczonadi V., Horvath R. (2014). Mitochondria: impaired mitochondrial translation in human disease. Int. J. Biochem. Cell Biol. 48 77–84. 10.1016/j.biocel.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack K. E., Hobartner C., Bohnsack M. T. (2019). Eukaryotic 5-methylcytosine (m(5)C) RNA Methyltransferases: mechanisms, Cellular Functions, and Links to Disease. Genes 10:102. 10.3390/genes10020102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borek E., Baliga B. S., Gehrke C. W., Kuo C. W., Belman S., Troll W., et al. (1977). High turnover rate of transfer RNA in tumor tissue. Cancer Res. 37 3362–3366. [PubMed] [Google Scholar]
- Brandon M. C., Lott M. T., Nguyen K. C., Spolim S., Navathe S. B., Baldi P., et al. (2005). MITOMAP: a human mitochondrial genome database–2004 update. Nucl. Acids Res. 33 D611-D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A., Fernandez I. S., Gordiyenko Y., Ramakrishnan V. (2016). Ribosome-dependent activation of stringent control. Nature 534 277–280. 10.1038/nature17675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M. (1987). Coevolution of codon usage and transfer RNA abundance. Nature 325 728–730. 10.1038/325728a0 [DOI] [PubMed] [Google Scholar]
- Carpio M. A., Decca M. B., Lopez Sambrooks C., Durand E. S., Montich G. G., Hallak M. E. (2013). Calreticulin-dimerization induced by post-translational arginylation is critical for stress granules scaffolding. Int. J. Biochem. Cell Biol. 45 1223–1235. 10.1016/j.biocel.2013.03.017 [DOI] [PubMed] [Google Scholar]
- Castilho B. A., Shanmugam R., Silva R. C., Ramesh R., Himme B. M., Sattlegger E. (2014). Keeping the eIF2 alpha kinase Gcn2 in check. Biochim. Biophys. Acta 1843 1948–1968. 10.1016/j.bbamcr.2014.04.006 [DOI] [PubMed] [Google Scholar]
- Chatterjee K., Nostramo R. T., Wan Y., Hopper A. K. (2018). tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: location, location, location. Biochim. Biophys. Acta Gene Regulat. Mech. 1861 373–386. 10.1016/j.bbagrm.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Kashina A. (2019). Quantification of intracellular N-terminal beta-actin arginylation. Sci. Rep. 9:16669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J., et al. (2016). Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351 397–400. 10.1126/science.aad7977 [DOI] [PubMed] [Google Scholar]
- Chiou N. T., Kageyama R., Ansel K. M. (2018). Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep. 25 3356.e4-3370.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C., Sobala A., Lu C., Thatcher S. R., Bowman A., Brown J. W., et al. (2009). Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15 2147–2160. 10.1261/rna.1738409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comba A., Bonnet L. V., Goitea V. E., Hallak M. E., Galiano M. R. (2019). Arginylated calreticulin increases apoptotic response induced by bortezomib in glioma cells. Mol. Neurobiol. 56 1653–1664. 10.1007/s12035-018-1182-x [DOI] [PubMed] [Google Scholar]
- Comeron J. M. (2004). Selective and mutational patterns associated with gene expression in humans: influences on synonymous composition and intron presence. Genetics 167 1293–1304. 10.1534/genetics.104.026351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Feng L., Wek R. C., Cigan A. M., Donahue T. F., Hinnebusch A. G. (1992). Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68 585–596. 10.1016/0092-8674(92)90193-g [DOI] [PubMed] [Google Scholar]
- Dhahbi J. M., Spindler S. R., Atamna H., Yamakawa A., Boffelli D., Mote P., et al. (2013). 5’ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genom. 14:298. 10.1186/1471-2164-14-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakal R., Tong C., Anderson S., Kashina A. S., Cooperman B., Bau H. H. (2019). Dynamics of intracellular stress-induced tRNA trafficking. Nucl. Acids Res. 47 2002–2010. 10.1093/nar/gky1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar K. A., Goodenbour J. M., Pan T. (2006). Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2:e221. 10.1371/journal.pgen.0020221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar K. A., Mobley E. M., Radek A. J., Pan T. (2004). Exploring the regulation of tRNA distribution on the genomic scale. J. Mol. Biol. 337 31–47. 10.1016/j.jmb.2004.01.024 [DOI] [PubMed] [Google Scholar]
- Dittmar K. A., Sorensen M. A., Elf J., Ehrenberg M., Pan T. (2005). Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 6 151–157. 10.1038/sj.embor.7400341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Nilsson L., Kurland C. G. (1996). Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260 649–663. 10.1006/jmbi.1996.0428 [DOI] [PubMed] [Google Scholar]
- Dong J., Qiu H., Garcia-Barrio M., Anderson J., Hinnebusch A. G. (2000). Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol. Cell 6 269–279. 10.1016/s1097-2765(00)00028-9 [DOI] [PubMed] [Google Scholar]
- dos Reis M., Savva R., Wernisch L. (2004). Solving the riddle of codon usage preferences: a test for translational selection. Nucl. Acids Res. 32 5036–5044. 10.1093/nar/gkh834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchon A. A., St Gelais C., Titkemeier N., Hatterschide J., Wu L., Musier-Forsyth K. (2017). HIV-1 Exploits a Dynamic Multi-aminoacyl-tRNA synthetase complex to enhance viral replication. J. Virol. 91:e01240-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L. (2000). tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends genet. 16 287–289. 10.1016/s0168-9525(00)02041-2 [DOI] [PubMed] [Google Scholar]
- Elkordy A., Mishima E., Niizuma K., Akiyama Y., Fujimura M., Tominaga T., et al. (2018). Stress-induced tRNA cleavage and tiRNA generation in rat neuronal PC12 cells. J. Neurochem. 146 560–569. 10.1111/jnc.14321 [DOI] [PubMed] [Google Scholar]
- Emara M. M., Ivanov P., Hickman T., Dawra N., Tisdale S., Kedersha N., et al. (2010). Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 285 10959–10968. 10.1074/jbc.m109.077560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez J. A., Chomyn A., Attardi G. (1995). MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNA(Lys) and premature translation termination. Nat. Genet. 10 47–55. 10.1038/ng0595-47 [DOI] [PubMed] [Google Scholar]
- Erber L., Hoffmann A., Fallmann J., Betat H., Stadler P. F., Morl M. (2020). LOTTE-seq (Long hairpin oligonucleotide based tRNA high-throughput sequencing): specific selection of tRNAs with 3’-CCA end for high-throughput sequencing. RNA Biol. 17 23–32. 10.1080/15476286.2019.1664250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Feng J., Liu Q., Sun F., Tie Y., Zhu J., et al. (2009). Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 583 437–442. 10.1016/j.febslet.2008.12.043 [DOI] [PubMed] [Google Scholar]
- Fung A. W., Fahlman R. P. (2015). The molecular basis for the post-translational addition of amino acids by L/F transferase in the N-end rule pathway. Curr. Prot. Peptide Sci. 16 163–180. 10.2174/1389203716666150112095726 [DOI] [PubMed] [Google Scholar]
- Fung A. W., Leung C. C., Fahlman R. P. (2014). The determination of tRNALeu recognition nucleotides for Escherichia coli L/F transferase. RNA 20 1210–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabor J., Cen S., Javanbakht H., Niu M., Kleiman L. (2002). Effect of altering the tRNA(Lys)(3) concentration in human immunodeficiency virus type 1 upon its annealing to viral RNA, GagPol incorporation, and viral infectivity. J. Virol. 76 9096–9102. 10.1128/jvi.76.18.9096-9102.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadadhar S., Dadi H., Bodakuntla S., Schnitzler A., Bieche I., Rusconi F., et al. (2017). Tubulin glycylation controls primary cilia length. J. Cell Biol. 216 2701–2713. 10.1083/jcb.201612050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebetsberger J., Zywicki M., Kunzi A., Polacek N. (2012). tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012:260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D. J., Lee S. C., Isa N. M., Gramuglia S., Fukao T., Bassel G. W., et al. (2011). Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479 415–418. 10.1038/nature10534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giege R. (2008). Toward a more complete view of tRNA biology. Nat. Struct. Mol. Biol. 15 1007–1014. 10.1038/nsmb.1498 [DOI] [PubMed] [Google Scholar]
- Gingold H., Tehler D., Christoffersen N. R., Nielsen M. M., Asmar F., Kooistra S. M., et al. (2014). A dual program for translation regulation in cellular proliferation and differentiation. Cell 158 1281–1292. 10.1016/j.cell.2014.08.011 [DOI] [PubMed] [Google Scholar]
- Girstmair H., Saffert P., Rode S., Czech A., Holland G., Bannert N., et al. (2013). Depletion of cognate charged transfer RNA causes translational frameshifting within the expanded CAG stretch in huntingtin. Cell Rep. 3 148–159. 10.1016/j.celrep.2012.12.019 [DOI] [PubMed] [Google Scholar]
- Gogakos T., Brown M., Garzia A., Meyer C., Hafner M., Tuschl T. (2017). Characterizing expression and processing of precursor and mature human tRNAs by Hydro-tRNAseq and PAR-CLIP. Cell Rep. 20 1463–1475. 10.1016/j.celrep.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda D. K., Bachmair A., Wunning I., Tobias J. W., Lane W. S., Varshavsky A. (1989). Universality and structure of the N-end rule. J. Biol. Chem. 264 16700–16712. [PubMed] [Google Scholar]
- Gonskikh Y., Gerstl M., Kos M., Borth N., Schosserer M., Grillari J., et al. (2020). Modulation of mammalian translation by a ribosome-associated tRNA half. RNA Biol. 17:1125-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H., Nguyen H. C. B., Zhang S., Dill B. D., Molina H., Tavazoie S. F. (2016). Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell 165 1416–1427. 10.1016/j.cell.2016.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorla M., Sepuri N. B. (2014). Perturbation of apoptosis upon binding of tRNA to the heme domain of cytochrome c. Apoptosis 19 259–268. 10.1007/s10495-013-0915-6 [DOI] [PubMed] [Google Scholar]
- Goto Y., Nonaka I., Horai S. (1990). A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348 651–653. 10.1038/348651a0 [DOI] [PubMed] [Google Scholar]
- Goto Y., Nonaka I., Horai S. (1991). A new mtDNA mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS). Biochim. Biophys. Acta 1097 238–240. 10.1016/0925-4439(91)90042-8 [DOI] [PubMed] [Google Scholar]
- Guan L., Karaiskos S., Grigoriev A. (2020). Inferring targeting modes of Argonaute-loaded tRNA fragments. RNA Biol. 17 1070–1080. 10.1080/15476286.2019.1676633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi N., Ciesla M., Ngoc P. C. T., Lang S., Arora S., Dimitriou M., et al. (2018). Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173 1204-1216 e1226. [DOI] [PubMed] [Google Scholar]
- Haussecker D., Huang Y., Lau A., Parameswaran P., Fire A. Z., Kay M. A. (2010). Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16 673–695. 10.1261/rna.2000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X. D., Gong W., Zhang J. N., Nie J., Yao C. F., Guo F. S., et al. (2018). Sensing and transmitting intracellular amino acid signals through reversible lysine aminoacylations. Cell Metab. 27 151.e6-166.e6. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Ivanov I. P., Sonenberg N. (2016). Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science 352 1413–1416. 10.1126/science.aad9868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S., Loher P., Shigematsu M., Palazzo J. P., Suzuki R., Imoto I., et al. (2015). Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc. Natl. Acad. Sci. U.S.A. 112 E3816-E3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. M., Yang X. (2013). Regulation of cell death by transfer RNA. Antiox. Redox Signal. 19 583–594. 10.1089/ars.2012.5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R. -G., Sheng J., Qi X., Xu Z., Takahashi T. T., Varshavsky A. (2005). The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437 981–986. 10.1038/nature04027 [DOI] [PubMed] [Google Scholar]
- Hurst L. D., Williams E. J. (2000). Covariation of GC content and the silent site substitution rate in rodents: implications for methodology and for the evolution of isochores. Gene 261 107–114. 10.1016/s0378-1119(00)00489-3 [DOI] [PubMed] [Google Scholar]
- Hurto R. L., Tong A. H., Boone C., Hopper A. K. (2007). Inorganic phosphate deprivation causes tRNA nuclear accumulation via retrograde transport in Saccharomyces cerevisiae. Genetics 176 841–852. 10.1534/genetics.106.069732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. (1985). Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2 13–34. [DOI] [PubMed] [Google Scholar]
- Ishimura R., Nagy G., Dotu I., Zhou H., Yang X. L., Schimmel P., et al. (2014). RNA function, Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345 455–459. 10.1126/science.1249749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P., Emara M. M., Villen J., Gygi S. P., Anderson P. (2011). Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 43 613–623. 10.1016/j.molcel.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P., O’Day E., Emara M. M., Wagner G., Lieberman J., Anderson P. (2014). G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. U.S.A. 111 18201–18206. 10.1073/pnas.1407361111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. E., Alexander R. W., Pan T. (2011). Misacylation of specific nonmethionyl tRNAs by a bacterial methionyl-tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 108 6933–6938. 10.1073/pnas.1019033108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji H. (1968). Further studies on the soluble amino acid incorporating system from rat liver. Biochemistry 7 3844–3850. 10.1021/bi00851a009 [DOI] [PubMed] [Google Scholar]
- Kaji H., Novelli G. D., Kaji A. (1963). A soluble amino acid-incorporating system from rat liver. Biochim. Biophys. Acta 76 474–477. 10.1016/0926-6550(63)90070-7 [DOI] [PubMed] [Google Scholar]
- Kaminska M., Shalak V., Francin M., Mirande M. (2007). Viral hijacking of mitochondrial lysyl-tRNA synthetase. J. virol. 81 68–73. 10.1128/jvi.01267-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya S., Yamada Y., Kinouchi M., Kudo Y., Ikemura T. (2001). Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J. Mol. Evol. 53 290–298. 10.1007/s002390010219 [DOI] [PubMed] [Google Scholar]
- Kanjee U., Ogata K., Houry W. A. (2012). Direct binding targets of the stringent response alarmone (p)ppGpp. Mol. Microbiol. 85 1029–1043. 10.1111/j.1365-2958.2012.08177.x [DOI] [PubMed] [Google Scholar]
- Kantidakis T., Ramsbottom B. A., Birch J. L., Dowding S. N., White R. J. (2010). mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc. Natl. Acad. Sci. U.S.A. 107 11823–11828. 10.1073/pnas.1005188107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur M., Ganguly A., Nagy G., Adamson S. I., Chuang J. H., Frankel W. N., et al. (2020). Expression of the neuronal tRNA n-Tr20 regulates synaptic transmission and seizure susceptibility. Neuron 108 193.e9-208.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakozova M., Kozak M., Wong C. C., Bailey A. O., Yates J. R., 3rd, Mogilner A., et al. (2006). Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science 313 192–196. 10.1126/science.1129344 [DOI] [PubMed] [Google Scholar]
- Karatsai O., Stasyk O., Redowicz M. J. (2020). Effects of arginine and its deprivation on human glioblastoma physiology and signaling. Adv. Exp. Med. Biol. 1202 243–258. 10.1007/978-3-030-30651-9_12 [DOI] [PubMed] [Google Scholar]
- Katz A., Elgamal S., Rajkovic A., Ibba M. (2016). Non-canonical roles of tRNAs and tRNA mimics in bacterial cell biology. Molecular microbiology 101 545–558. 10.1111/mmi.13419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keam S. P., Hutvagner G. (2015). tRNA-Derived Fragments (tRFs): emerging new roles for an ancient RNA in the regulation of gene expression. Life 5 1638–1651. 10.3390/life5041638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoder-Agha F., Dias J. M., Comisso M., Mirande M. (2018). Characterization of association of human mitochondrial lysyl-tRNA synthetase with HIV-1 Pol and tRNA3(Lys). BMC Biochem. 19:2. 10.1186/s12858-018-0092-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Ando Y., Shiba T. (1986). Unusual priming mechanism of RNA-directed DNA synthesis in copia retrovirus-like particles of Drosophila. Nature 323 824–826. 10.1038/323824a0 [DOI] [PubMed] [Google Scholar]
- Kim S., Liu C., Halkidis K., Gamper H. B., Hou Y. M. (2009). Distinct kinetic determinants for the stepwise CCA addition to tRNA. RNA 15 1827–1836. 10.1261/rna.1669109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y., Tsutsumi K., Konno A., Ikegami K., Hameed S., Kaneko T., et al. (2018). Environmental responsiveness of tubulin glutamylation in sensory cilia is regulated by the p38 MAPK pathway. Sci. Rep. 8:8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner S., Cai Z., Rauscher R., Kastelic N., Anding M., Czech A., et al. (2017). Alteration of protein function by a silent polymorphism linked to tRNA abundance. PLoS Biol. 15:e2000779. 10.1371/journal.pbio.2000779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner S., Ignatova Z. (2015). Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 16 98–112. 10.1038/nrg3861 [DOI] [PubMed] [Google Scholar]
- Kirino Y., Suzuki T. (2005). Human mitochondrial diseases associated with tRNA wobble modification deficiency. RNA Biol. 2 41–44. 10.4161/rna.2.2.1610 [DOI] [PubMed] [Google Scholar]
- Koga Y., Povalko N., Nishioka J., Katayama K., Yatsuga S., Matsuishi T. (2012). Molecular pathology of MELAS and L-arginine effects. Biochim. Biophys. Acta 1820 608–614. 10.1016/j.bbagen.2011.09.005 [DOI] [PubMed] [Google Scholar]
- Kumar P., Anaya J., Mudunuri S. B., Dutta A. (2014). Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 12:78. 10.1186/s12915-014-0078-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Mudunuri S. B., Anaya J., Dutta A. (2015). tRFdb: a database for transfer RNA fragments. Nucl. Acids Res. 43 D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaka S., Leu N. A., Zhang F., Bunte R., Saha S., Wang J., et al. (2010). Arginylation-dependent neural crest cell migration is essential for mouse development. PLoS Genet. 6:e1000878. 10.1371/journal.pgen.1000878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C., Kumar P., Kiran M., Su Z., Malik A., Dutta A. (2018). tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24 1093–1105. 10.1261/rna.066126.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y. T., Kashina A. S., Davydov I. V., Hu R. G., An J. Y., Seo J. W., et al. (2002). An essential role of N-terminal arginylation in cardiovascular development. Science 297 96–99. 10.1126/science.1069531 [DOI] [PubMed] [Google Scholar]
- Lant J. T., Berg M. D., Heinemann I. U., Brandl C. J., O’Donoghue P. (2019). Pathways to disease from natural variations in human cytoplasmic tRNAs. J. Biol. Chem. 294 5294–5308. 10.1074/jbc.rev118.002982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Kim D. G., Kim B. G., Yang W. S., Hong J., Kang T., et al. (2014). Promiscuous methionyl-tRNA synthetase mediates adaptive mistranslation to protect cells against oxidative stress. J. Cell Sci. 127 4234–4245. 10.1242/jcs.152470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine R. L., Mosoni L., Berlett B. S., Stadtman E. R. (1996). Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. U.S.A. 93 15036–15040. 10.1073/pnas.93.26.15036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Shan N., Lu L., Wang Z. (2020). tRFtarget: a database for transfer RNA-derived fragment targets. Nucl. Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhou M., Hu Q., Bai X. C., Huang W., Scheres S. H., et al. (2017). Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme. Proc. Natl. Acad. Sci. U.S.A. 114 1542–1547. 10.1073/pnas.1620626114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Ender C., Meister G., Moore P. S., Chang Y., John B. (2012). Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucl. Acids Res. 40 6787–6799. 10.1093/nar/gks307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Gamper H., Liu H., Cooperman B. S., Hou Y. M. (2011). Potential for interdependent development of tRNA determinants for aminoacylation and ribosome decoding. Nat. Commun. 2:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Gamper H., Shtivelband S., Hauenstein S., Perona J. J., Hou Y. M. (2007). Kinetic quality control of anticodon recognition by a eukaryotic aminoacyl-tRNA synthetase. J. Mol. Biol. 367 1063–1078. 10.1016/j.jmb.2007.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Stonestrom A. J., Christian T., Yong J., Takase R., Hou Y. M., et al. (2016). Molecular Basis and Consequences of the Cytochrome c-tRNA Interaction. J. Biol. Chem. 291 10426–10436. 10.1074/jbc.m115.697789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Levine R. L. (2009). Methionine in proteins defends against oxidative stress. FASEB J 23 464–472. 10.1096/fj.08-118414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G. (2017). tRNAs as primers and inhibitors of retrotransposons. Mobile Genet. Elem. 7 1–6. 10.1080/2159256x.2017.1393490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maute R. L., Schneider C., Sumazin P., Holmes A., Califano A., Basso K., et al. (2013). tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 110 1404–1409. 10.1073/pnas.1206761110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain D. R., Berger T., Mak T. W. (2013). Caspase functions in cell death and disease. Cold Spring Harbor Perspect. Biol. 5:a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megel C., Hummel G., Lalande S., Ubrig E., Cognat V., Morelle G., et al. (2019). Plant RNases T2, but not Dicer-like proteins, are major players of tRNA-derived fragments biogenesis. Nucl. Acids Res. 47 941–952. 10.1093/nar/gky1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y., Yong J., Liu H., Shi Y., Meinkoth J., Dreyfuss G., et al. (2010). tRNA binds to cytochrome c and inhibits caspase activation. Mol. Cell 37 668–678. 10.1016/j.molcel.2010.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick W. C., Pavitt G. D. (2018). Protein synthesis initiation in eukaryotic cells. Cold Spring Harbor Perspect. Biol. 10:a033092. 10.1101/cshperspect.a033092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita P., Boeke J. D. (2016). How retrotransposons shape genome regulation. Curr. Opin. Genet. Dev. 37 90–100. 10.1016/j.gde.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa R., Mizuno R., Watanabe K., Ijiri K. (2012). Formation of tRNA granules in the nucleus of heat-induced human cells. Biochem. Biophys. Res. Commun. 418 149–155. 10.1016/j.bbrc.2011.12.150 [DOI] [PubMed] [Google Scholar]
- Nechooshtan G., Yunusov D., Chang K., Gingeras T. R. (2020). Processing by RNase 1 forms tRNA halves and distinct Y RNA fragments in the extracellular environment. Nucl. Acids Res. 48 8035-8049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer N., Goodenbour J. M., David A., Dittmar K. A., Jones R. B., Schneider J. R., et al. (2009). Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462 522–526. 10.1038/nature08576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa E. M., Pavon-Eternod M., Pan T., Ribas de Pouplana L. (2012). A role for tRNA modifications in genome structure and codon usage. Cell 149 202–213. 10.1016/j.cell.2012.01.050 [DOI] [PubMed] [Google Scholar]
- Ow Y. P., Green D. R., Hao Z., Mak T. W. (2008). Cytochrome c: functions beyond respiration. Nat. Rev. Mol. Cell Biol. 9 532–542. 10.1038/nrm2434 [DOI] [PubMed] [Google Scholar]
- Pan T. (2018). Modifications and functional genomics of human transfer RNA. Cell Res. 28 395–404. 10.1038/s41422-018-0013-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y. L., Poruri K., Martinis S. A. (2014). tRNA synthetase: tRNA aminoacylation and beyond. Wiley Interdiscipl. Rev. RNA 5 461–480. 10.1002/wrna.1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlyk I., Rzhepetskyy Y., Jagielski A. K., Drozak J., Wasik A., Pereverzieva G., et al. (2015). Arginine deprivation affects glioblastoma cell adhesion, invasiveness and actin cytoskeleton organization by impairment of beta-actin arginylation. Amino Acids 47 199–212. 10.1007/s00726-014-1857-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavon-Eternod M., Gomes S., Geslain R., Dai Q., Rosner M. R., Pan T. (2009). tRNA over-expression in breast cancer and functional consequences. Nucl. Acids Res. 37 7268–7280. 10.1093/nar/gkp787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavon-Eternod M., Gomes S., Rosner M. R., Pan T. (2013). Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA 19 461–466. 10.1261/rna.037507.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani R., Pavesi A., Ottonello S. (1997). Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 268 322–330. 10.1006/jmbi.1997.0942 [DOI] [PubMed] [Google Scholar]
- Phongsavanh X., Al-Qatabi N., Shaban M. S., Khoder-Agha F., El Asri M., Comisso M., et al. (2020). How HIV-1 Integrase Associates with Human Mitochondrial Lysyl-tRNA Synthetase. Viruses 12:1202. 10.3390/v12101202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkard O., McFarland S., Sweet T., Coller J. (2020). Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nat. Commun. 11:4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistritto G., Trisciuoglio D., Ceci C., Garufi A., D’Orazi G. (2016). Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging 8 603–619. 10.18632/aging.100934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliatsika V., Loher P., Magee R., Telonis A. G., Londin E., Shigematsu M., et al. (2018). MINTbase v2.0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all the cancer genome Atlas projects. Nucl. Acids Res. 46 D152-D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin J. B., Robins H., Levine A. J. (2004). Tissue-specific codon usage and the expression of human genes. Proc. Natl. Acad. Sci. U.S.A. 101 12588–12591. 10.1073/pnas.0404957101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polte C., Wedemeyer D., Oliver K. E., Wagner J., Bijvelds M. J. C., Mahoney J., et al. (2019). Assessing cell-specific effects of genetic variations using tRNA microarrays. BMC Genom. 20:549. 10.1186/s12864-019-5864-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus K., Cashel M. (2008). (p)ppGpp: still magical? Annu. Rev. Microbiol. 62 35–51. 10.1146/annurev.micro.62.081307.162903 [DOI] [PubMed] [Google Scholar]
- Rai R., Zhang F., Colavita K., Leu N. A., Kurosaka S., Kumar A., et al. (2016). Arginyltransferase suppresses cell tumorigenic potential and inversely correlates with metastases in human cancers. Oncogene 35 4058–4068. 10.1038/onc.2015.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina M., Ibba M. (2014). tRNAs as regulators of biological processes. Front. Genet. 5:171. 10.3389/fgene.2014.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassier D. E., Kashina A. (2019). Protein arginylation of cytoskeletal proteins in the muscle: modifications modifying function. Am. J. Physiol. Cell Physiol. 316 C668-C677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J., Lazebnik Y. (1999). Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 13 3179–3184. 10.1101/gad.13.24.3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Sanchez-Vazquez P., Chen A. Y., Lee J. H., Burgos H. L., Gourse R. L. (2016). ppGpp Binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol. Cell 62 811–823. 10.1016/j.molcel.2016.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Vrentas C. E., Sanchez-Vazquez P., Gaal T., Gourse R. L. (2013). The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 50 420–429. 10.1016/j.molcel.2013.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudinger-Thirion J., Lescure A., Paulus C., Frugier M. (2011). Misfolded human tRNA isodecoder binds and neutralizes a 3’ UTR-embedded Alu element. Proc. Natl. Acad. Sci. U.S.A. 108 E794-E802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Kashina A. (2011). Posttranslational arginylation as a global biological regulator. Dev. Biol. 358 1–8. 10.1016/j.ydbio.2011.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M., Jobava R., Parisien M., Putnam A., Krokowski D., Gao X. H., et al. (2014). Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol. Cell. Biol. 34 2450–2463. 10.1128/mcb.00136-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M., Krokowski D., Guan B. J., Ivanov P., Parisien M., Hu G. F., et al. (2012). Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J. Biol. Chem. 287 42708–42725. 10.1074/jbc.m112.371799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M., Fidalgo A., Varanda A. S., Oliveira C., Santos M. A. S. (2019). tRNA deregulation and its consequences in cancer. Trends Mol. Med. 25 853–865. 10.1016/j.molmed.2019.05.011 [DOI] [PubMed] [Google Scholar]
- Schaffer A. E., Pinkard O., Coller J. M. (2019). tRNA metabolism and neurodevelopmental disorders. Annu. Rev. Genom. Hum. Genet. 20 359–387. 10.1146/annurev-genom-083118-015334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn A. J., Gutbrod M. J., LeBlanc C., Martienssen R. (2017). LTR-retrotransposon control by tRNA-derived small RNAs. Cell 170 61.e11-71.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. H., Pan T. (2017). tRNA misacylation with methionine in the mouse gut microbiome in situ. Microb. Ecol. 74 10–14. 10.1007/s00248-016-0928-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. H., Waldbauer J. R., Zhang L., Pan T. (2016). Global tRNA misacylation induced by anaerobiosis and antibiotic exposure broadly increases stress resistance in Escherichia coli. Nucl. Acids Res. 44 10292–10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenzer H., Juhling F., Chu A., Pallett L. J., Baumert T. F., Maini M., et al. (2019). Oxidative stress triggers selective tRNA retrograde transport in human cells during the integrated stress response. Cell Rep. 26 3416.e5-3428.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiki M., Hattori S., Hirayama Y., Yoshida M. (1983). Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc. Natl. Acad. Sci. U.S.A. 80 3618–3622. 10.1073/pnas.80.12.3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selitsky S. R., Sethupathy P. (2015). tDRmapper: challenges and solutions to mapping, naming, and quantifying tRNA-derived RNAs from human small RNA-sequencing data. BMC Bioinform. 16:354. 10.1186/s12859-015-0800-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semon M., Lobry J. R., Duret L. (2006). No evidence for tissue-specific adaptation of synonymous codon usage in humans. Mol. Biol. Evol. 23 523–529. 10.1093/molbev/msj053 [DOI] [PubMed] [Google Scholar]
- Shaheen H. H., Hopper A. K. (2005). Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S. A. 102 11290–11295. 10.1073/pnas.0503836102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen H. H., Horetsky R. L., Kimball S. R., Murthi A., Jefferson L. S., Hopper A. K. (2007). Retrograde nuclear accumulation of cytoplasmic tRNA in rat hepatoma cells in response to amino acid deprivation. Proc. Natl. Acad. Sci. U.S.A. 104 8845–8850. 10.1073/pnas.0700765104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U., Conine C. C., Shea J. M., Boskovic A., Derr A. G., Bing X. Y., et al. (2016). Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351 391–396. 10.1126/science.aad6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U., Sun F., Conine C. C., Reichholf B., Kukreja S., Herzog V. A., et al. (2018). Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev. Cell 46 481-494 e486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Moore P.B. (2000). The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA 6 1091-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zhang Y., Zhou T., Chen Q. (2019). tsRNAs: the swiss army knife for translational regulation. Trends Biochem. Sci. 44 185–189. 10.1016/j.tibs.2018.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields D. C. (1990). Switches in species-specific codon preferences: the influence of mutation biases. J. Mol. Evol. 31 71–80. 10.1007/bf02109476 [DOI] [PubMed] [Google Scholar]
- Shigematsu M., Honda S., Loher P., Telonis A. G., Rigoutsos I., Kirino Y. (2017). YAMAT-seq: an efficient method for high-throughput sequencing of mature transfer RNAs. Nucl. Acids Res. 45:e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu M., Kirino Y. (2015). tRNA-Derived Short Non-coding RNA as Interacting Partners of Argonaute Proteins. Gene Regul. Syst. Biol. 9 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoffner J. M., Lott M. T., Lezza A. M., Seibel P., Ballinger S. W., Wallace D. C. (1990). Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell 61 931–937. 10.1016/0092-8674(90)90059-n [DOI] [PubMed] [Google Scholar]
- Sobala A., Hutvagner G. (2013). Small RNAs derived from the 5’ end of tRNA can inhibit protein translation in human cells. RNA Biol. 10 553–563. 10.4161/rna.24285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffer R. L. (1971). Enzymatic modification of proteins. V. Protein acceptor specificity in the arginine-transfer reaction. J Biol. Chem. 246 1602–1606. [PubMed] [Google Scholar]
- Somers J., Poyry T., Willis A. E. (2013). A perspective on mammalian upstream open reading frame function. Int. J. Biochem. Cell Biol. 45 1690–1700. 10.1016/j.biocel.2013.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer J., Gehrke C. W., Kuo K. C., Waalkes T. P., Borek E. (1979). tRNA breakdown products as markers for cancer. Cancer 44 2120–2123. [DOI] [PubMed] [Google Scholar]
- Sriskanthadevan-Pirahas S., Deshpande R., Lee B., Grewal S. S. (2018). Ras/ERK-signalling promotes tRNA synthesis and growth via the RNA polymerase III repressor Maf1 in Drosophila. PLoS Genet. 14:e1007202. 10.1371/journal.pgen.1007202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinchen W., Bange G. (2016). The magic dance of the alarmones (p)ppGpp. Mol. Microbiol. 101 531–544. 10.1111/mmi.13412 [DOI] [PubMed] [Google Scholar]
- Su Z., Kuscu C., Malik A., Shibata E., Dutta A. (2019). Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3-mediated gene silencing. J. Biol. Chem. 294 16930–16941. 10.1074/jbc.ra119.009272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Gomes A. C., He W., Zhou H., Wang X., Pan D. W., et al. (2016). Evolutionary gain of alanine mischarging to noncognate tRNAs with a G4:U69 base pair. J. Am. Chem. Soc. 138 12948–12955. 10.1021/jacs.6b07121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryanarayana T., Uppala J. K., Garapati U. K. (2012). Interaction of cytochrome c with tRNA and other polynucleotides. Mol. Biol. Rep. 39 9187–9191. 10.1007/s11033-012-1791-9 [DOI] [PubMed] [Google Scholar]
- Szefel J., Danielak A., Kruszewski W. J. (2019). Metabolic pathways of L-arginine and therapeutic consequences in tumors. Advances in medical sciences 64 104–110. 10.1016/j.advms.2018.08.018 [DOI] [PubMed] [Google Scholar]
- Tahmasebi S., Khoutorsky A., Mathews M. B., Sonenberg N. (2018). Translation deregulation in human disease. Nat. Rev. Mol. Cell Biol. 19 791–807. 10.1038/s41580-018-0034-x [DOI] [PubMed] [Google Scholar]
- Tait S. W., Green D. R. (2010). Mitochondria and cell death: outer membrane permeabilization and beyond. Nature reviews Mol. Cell Biol. 11 621–632. 10.1038/nrm2952 [DOI] [PubMed] [Google Scholar]
- Takano A., Endo T., Yoshihisa T. (2005). tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309 140–142. 10.1126/science.1113346 [DOI] [PubMed] [Google Scholar]
- Tang D., Kang R., Berghe T. V., Vandenabeele P., Kroemer G. (2019). The molecular machinery of regulated cell death. Cell Res. 29 347–364. 10.1038/s41422-019-0164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi S., Miyake M., Tsugawa K., Oyadomari M., Oyadomari S. (2016). Integrated stress response of vertebrates is regulated by four eIF2alpha kinases. Sci. Rep. 6:32886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telonis A. G., Loher P., Honda S., Jing Y., Palazzo J., Kirino Y., et al. (2015). Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget 6 24797–24822. 10.18632/oncotarget.4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D. M., Lu C., Green P. J., Parker R. (2008). tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14 2095–2103. 10.1261/rna.1232808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D. M., Parker R. (2009). The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 185 43–50. 10.1083/jcb.200811119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrent M., Chalancon G., de Groot N. S., Wuster A., Madan Babu M. (2018). Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci. Signal. 11: eaat6409. 10.1126/scisignal.aat6409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosar J. P., Cayota A. (2020). Extracellular tRNAs and tRNA-derived fragments. RNA biology 17 1149–1167. 10.1080/15476286.2020.1729584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuorto F., Liebers R., Musch T., Schaefer M., Hofmann S., Kellner S., et al. (2012). RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 19 900–905. 10.1038/nsmb.2357 [DOI] [PubMed] [Google Scholar]
- Wain-Hobson S., Sonigo P., Danos O., Cole S., Alizon M. (1985). Nucleotide sequence of the AIDS virus, LAV. Cell 40 9–17. 10.1016/0092-8674(85)90303-4 [DOI] [PubMed] [Google Scholar]
- Wang B., Dai P., Ding D., Del Rosario A., Grant R. A., Pentelute B. L., et al. (2019). Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 15 141–150. 10.1038/s41589-018-0183-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Li D., Kovalchuk I., Apel I. J., Chinnaiyan A. M., Woycicki R. K., et al. (2018). miR-34a directly targets tRNAi(Met) precursors and affects cellular proliferation, cell cycle, and apoptosis. Proc. Natl. Acad. Sci. U.S. A. 115 7392–7397. 10.1073/pnas.1703029115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Han X., Leu N. A., Sterling S., Kurosaka S., Fina M., et al. (2017). Protein arginylation targets alpha synuclein, facilitates normal brain health, and prevents neurodegeneration. Sci. Rep. 7 11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Han X., Wong C. C., Cheng H., Aslanian A., Xu T., et al. (2014). Arginyltransferase ATE1 catalyzes midchain arginylation of proteins at side chain carboxylates in vivo. Chem. Biol. 21 331–337. 10.1016/j.chembiol.2013.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Pan T. (2015). Methionine mistranslation bypasses the restraint of the genetic code to generate mutant proteins with distinct activities. PLoS Genet. 11:e1005745. 10.1371/journal.pgen.1005745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Miyagawa R., Tomikawa C., Mizuno R., Takahashi A., Hori H., et al. (2013). Degradation of initiator tRNAMet by Xrn1/2 via its accumulation in the nucleus of heat-treated HeLa cells. Nucl. Acids Res. 41 4671–4685. 10.1093/nar/gkt153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Silke J. R., Xia X. (2019). An improved estimation of tRNA expression to better elucidate the coevolution between tRNA abundance and codon usage in bacteria. Sci. Rep. 9:3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C. (2018). Role of eIF2alpha kinases in translational control and adaptation to cellular stress. Cold Spring Harbor Perspect. Biol. 10:a032870. 10.1101/cshperspect.a032870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney M. L., Hurto R. L., Shaheen H. H., Hopper A. K. (2007). Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol. Biol. Cell 18 2678–2686. 10.1091/mbc.e07-01-0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M., Wilhelm F. X. (2001). Reverse transcription of retroviruses and LTR retrotransposons. Cell. Mol. Life Sci. 58 1246–1262. 10.1007/pl00000937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltrout E., Goodenbour J. M., Frechin M., Pan T. (2012). Misacylation of tRNA with methionine in Saccharomyces cerevisiae. Nucl. Acids Res. 40 10494–10506. 10.1093/nar/gks805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Yao L., Yu X., Ruan Y., Li Z., Guo J. (2020). Action mechanisms and research methods of tRNA-derived small RNAs. Signal Trans. Target. Ther. 5:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S., Ivanov P., Hu G. F., Anderson P. (2009). Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 185 35–42. 10.1083/jcb.200811106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao D., Sun X., Zhou L., Amanullah M., Pan X., Liu Y., et al. (2020). OncotRF: an online resource for exploration of tRNA-derived fragments in human cancers. RNA Biol. 17 1081–1091. 10.1080/15476286.2020.1776506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T., Suzuki T., Suzuki T., Ueda T., Ohta S., Watanabe K. (2000). Modification defect at anticodon wobble nucleotide of mitochondrial tRNAsLeu(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J. Biol. Chem. 275 4251–4257. 10.1074/jbc.275.6.4251 [DOI] [PubMed] [Google Scholar]
- Yeung M. L., Bennasser Y., Watashi K., Le S. Y., Houzet L., Jeang K. T. (2009). Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucl. Acids Res. 37 6575–6586. 10.1093/nar/gkp707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young S. K., Wek R. C. (2016). Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J. Biol. Chem. 291 16927–16935. 10.1074/jbc.r116.733899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborske J. M., Wu X., Wek R. C., Pan T. (2010). Selective control of amino acid metabolism by the GCN2 eIF2 kinase pathway in Saccharomyces cerevisiae. BMC Biochem. 11:29. 10.1186/1471-2091-11-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitseva L., Myers R., Fassati A. (2006). tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol. 4:e332. 10.1371/journal.pbio.0040332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. M., Perona J. J., Ryu K., Francklyn C., Hou Y. M. (2006). Distinct kinetic mechanisms of the two classes of Aminoacyl-tRNA synthetases. J. Mol. Biol. 361 300–311. 10.1016/j.jmb.2006.06.015 [DOI] [PubMed] [Google Scholar]
- Zhang F., Shi J., Wu Z., Gao P., Zhang W., Qu B., et al. (2020). A 3’-tRNA-derived fragment enhances cell proliferation, migration and invasion in gastric cancer by targeting FBXO47. Arch. Biochem. Biophys. 690:108467 10.1016/j.abb.2020.108467 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang X., Shi J., Tuorto F., Li X., Liu Y., et al. (2018a). Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat. Cell Biol. 20 535–540. 10.1038/s41556-018-0087-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Ye Y., Gong J., Ruan H., Liu C. J., Xiang Y., et al. (2018b). Global analysis of tRNA and translation factor expression reveals a dynamic landscape of translational regulation in human cancers. Commun. Biol. 1:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Qin Y., Clark W. C., Dai Q., Yi C., He C., et al. (2015). Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12 835–837. 10.1038/nmeth.3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L. L., Xu W. L., Liu S., Sun W. J., Li J. H., Wu J., et al. (2016). tRF2Cancer: a web server to detect tRNA-derived small RNA fragments (tRFs) and their expression in multiple cancers. Nucl. Acids Res. 44 W185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Goodenbour J. M., Godley L. A., Wickrema A., Pan T. (2009). High levels of tRNA abundance and alteration of tRNA charging by bortezomib in multiple myeloma. Biochem. Biophys. Res. Commun. 385 160–164. 10.1016/j.bbrc.2009.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Li Y., Liu X., Wang X. (1999). An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 274 11549–11556. 10.1074/jbc.274.17.11549 [DOI] [PubMed] [Google Scholar]
- Zuo Y., Zhu L., Guo Z., Liu W., Zhang J., Zeng Z., et al. (2020). tsRBase: a comprehensive database for expression and function of tsRNAs in multiple species. Nucl. Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]