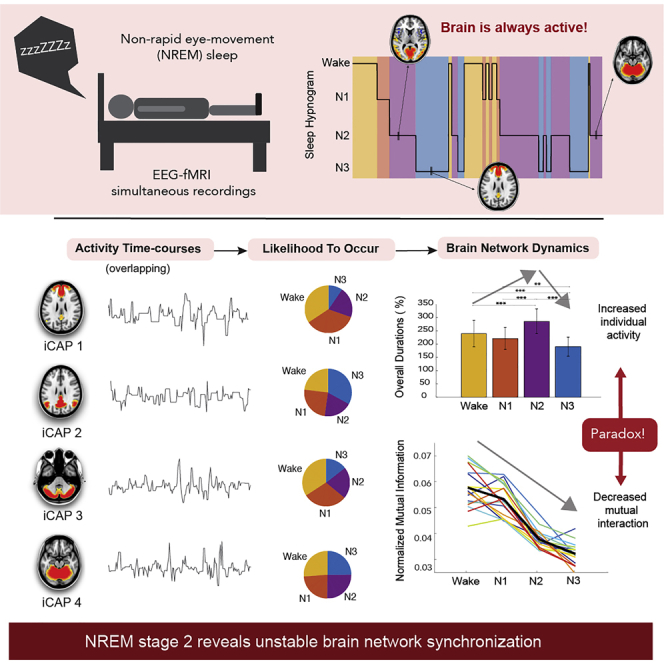
Summary
Functional dissociations in the brain observed during non-rapid eye movement (NREM) sleep have been associated with reduced information integration and impaired consciousness that accompany increasing sleep depth. Here, we explored the dynamical properties of large-scale functional brain networks derived from transient brain activity using functional magnetic resonance imaging. Spatial brain maps generally display significant modifications in terms of their tendency to occur across wakefulness and NREM sleep. Unexpectedly, almost all networks predominated in activity during NREM stage 2 before an abrupt loss of activity is observed in NREM stage 3. Yet, functional connectivity and mutual dependencies between these networks progressively broke down with increasing sleep depth. Thus, the efficiency of information transfer during NREM stage 2 is low despite the high attempt to communicate. Critically, our approach provides relevant data for evaluating functional brain network integrity and our findings robustly support a significant advance in our neural models of human sleep and consciousness.
Subject Areas: Medical Imaging, Systems Neuroscience, Cognitive Neuroscience, Techniques in Neuroscience
Graphical Abstract
Highlights
-
•
We examined the dynamics of large-scale functional brain networks during NREM sleep
-
•
Brain network activity peaks during NREM stage 2 and decreases during NREM stage 3
-
•
Interactions between brain networks break down with increasing sleep depth
-
•
NREM stage 2 reveals unstable functional network synchronization
Medical Imaging; Systems Neuroscience; Cognitive Neuroscience; Techniques in Neuroscience
Introduction
Spontaneous brain activity, as assessed by resting-state functional magnetic resonance imaging (fMRI), has provided key insights into the functional architecture of the brain. Resting-state networks (RSNs) identify sets of brain regions that exhibit synchronized fluctuations of activity over the whole duration of a resting-state session (typically 10-20 min of continuous scanning). The main hypothesis underlying most functional connectivity (FC) studies is that different RSNs reflect distinct ongoing cognitive/affective processes/states. For instance, the default mode network (DMN) typically shows reduced activity when subjects perform an externally oriented task (Greicius et al., 2003) and, contrastingly, the DMN becomes more engaged when self-referential processes or internal mentation predominate (Andrews-Hanna, 2012). According to this interpretation of RSNs, we expect that if conscious awareness dissipates as the brain transitions from wakefulness to deep sleep, we should observe a parallel and net decrease in activity and/or FC across regions of the brain involved in higher-order cognitive operations, such as reasoning, monitoring, or metacognition. Yet, some early evidence also suggested that several RSNs, including those encompassing association networks, persisted or even increased their connectivity during the descent from wakefulness to light sleep (Larson-Prior et al., 2009) or during anesthesia and coma, when conscious awareness is presumed to be completely abolished (Boly et al., 2008). Therefore, RSNs may reflect intrinsic dynamical properties of the brain's functional organization that are maintained across distinct levels of consciousness.
From a behavioral point of view, the brain in sleep undergoes marked and well-characterized physiological changes. Based on polysomnography, which is a combined use of EEG, electro-oculography, and electromyography, natural sleep can be broadly divided into rapid eye movement (REM) and non-rapid eye movement (NREM) periods. The latter is further subdivided into different sleep stages characterized by relaxed wakefulness (N1) to light sleep (N2), up to slow-wave sleep (SWS) or deep sleep (N3). It is therefore not surprising that, although RSNs may be detected across different sleep stages, their connectivity patterns (Horovitz et al., 2009; Sämann et al., 2011) and FC strengths undergo significant modifications (Tagliazucchi et al., 2013). For instance, upon reaching N3, the DMN has been found to dissociate into subcomponents, with a decrease in the connectivity between the medial prefrontal cortex (MPFC) and the posterior cingulate cortex (PCC) (Horovitz et al., 2009). Furthermore, several studies looked at changes in FC for regions associated with the reticular activation system that regulate the physiological state of arousal during sleep (e.g. thalamus, hypothalamus) and reported decreased connectivity between these regions and the rest of the cortex during light and deep sleep (Hale et al., 2016; Picchioni et al., 2014; Tagliazucchi and Laufs, 2014). These changes in brain network integrity, particularly its marked reduction from wakefulness to deep sleep, have been associated to diminished level of information integration (Tononi, 2004). That is, when the brain switches to more local cortical processing, this would lead to a global loss of information integration and a concomitant reduction in consciousness (Tononi and Koch, 2015).
Methodological tools allowing the detection of prevalent brain spatial patterns, such as FC analyses, are particularly useful when assessing imaging data collected outside of a predefined experimental paradigm (or in the absence of any temporally defined independent variable), namely when regression techniques cannot be applied. This is typically the case for continuous data collected during varying levels of arousal or states of consciousness (e.g. resting-state, sleep, anesthesia, and drugs). The simplest approach to investigating changes in FC is to use a sliding-window technique, where time courses from sets of brain regions (e.g. from atlas-based parcellation) are segmented into successive temporal windows so that various assessments of FC (e.g. bivariate Pearson correlations) can be applied to obtain time-evolving connectivity matrices. Another approach is to derive analogous information on resting-state FC based on time points where the regional BOLD signal exceeds a particular threshold (Tagliazucchi et al., 2012). Temporal clustering can also be applied to activity patterns occurring at these active fMRI time points to obtain patterns of co-activity among regions, also known as co-activation patterns (CAPs) (Liu and Duyn, 2013).
Furthermore, to account for the fundamental dynamic nature of the changes in neural FC, the latest developments on non-stationary FC approaches have started to successfully incorporate methods of temporal modeling. Using a dynamic Bayesian approach (i.e., Hidden Markov Model; HMM), Stevner et al. (2019) could recently demonstrate that some specific whole-brain functional connections are associated to each of the different stages of NREM. They found that networks with high specificity to occur in stages N2 and N3 generally expressed longer mean lifetimes, with each HMM state lasting from a few seconds to tens of seconds. They also examined the transition probabilities between the networks by extracting modules of HMM states that transitioned more often between each other than to other states (Vidaurre et al., 2017), allowing them to identify key trajectories of network activity from wake to NREM sleep.
Despite these major methodological advancements, it remains unsettling that coordinated network activity is assumed to be strictly temporally segregated, and that whole-brain states are not overlapping in time (i.e., only one RSN can occur per time instance). To overcome this limitation, we recently proposed to consider innovation-driven co-activation patterns (iCAPS), which capture transient brain activity, i.e., physiologically significant moments of regional activation and deactivation (Karahanoğlu and Van De Ville, 2015; Karahanoğlu et al., 2013), instead of the actual activity time points. This framework allows for the recovery of RSNs that are both spatially and temporally overlapping, providing a more plausible and thus putatively more accurate description of functional brain organization. Furthermore, unlike standard CAPs and other data-driven approaches (e.g., independent component analysis; Smith et al., 2012), the iCAP approach explicitly accounts for temporal blurring by the hemodynamic response function (HRF) by incorporating a deconvolution step in the preprocessing of the fMRI signal.
Here, we adopt an iCAP approach to recover functional brain networks from fMRI data recorded during wakefulness and NREM sleep stages. We obtain temporally overlapping activity time courses of each network for each participant and evaluate their temporal properties to capture important features of cortical organization as the brain goes from a fully awake state to deep sleep. We observe that RSNs such as control, salience, visual and sensory networks (Shirer et al., 2012) persist until the deep sleep, and that some of these networks dissociate into subcomponents (e.g., DMN). Moreover, since the activity time courses are not constrained to be temporally segregated, many networks tend to overlap in time, and that these numbers are altered across the different levels of sleep. To test these hypotheses, temporal measures, such as their overall accumulated durations and mean lifetimes, were computed, as well as their co-occurrences and temporal overlaps. Using a more accurate model to extract network-level representations of brain organization, we put the focal point into detecting the temporal alterations of large-scale brain activity across the distinct vigilance states.
Results
Data distribution across study 1 and study 2
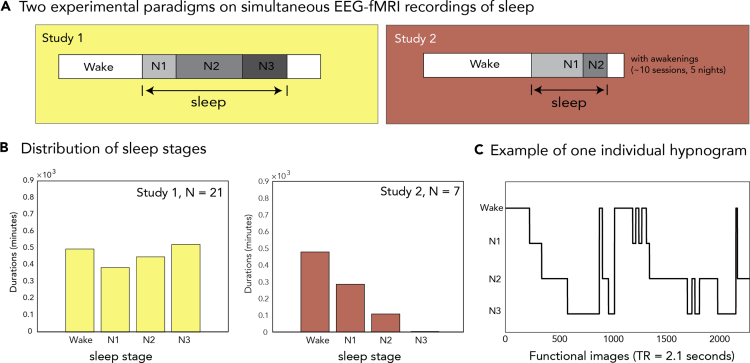
Figure 1 illustrates the two experimental paradigms on simultaneously acquired EEG and fMRI recordings of sleep. We used a total of 21 subjects in study 1 (13 reached N3), and a total of 7 subjects in the study 2 (all of them reached N1 sleep). The distribution of accumulated data from each sleep stage for each dataset is also shown in Figure 1, with Study 1 generating a substantial amount of N3 sleep (more than 8 hr in total), while study 2 was predominated by wake and N1 sleep (about 8h and 4h of data, respectively).
Figure 1.
Experimental paradigms
(A) The experiment for study 1 lasted between 51 min and 2 hr 40 min while study 2 lasted between 5 min up to 20 min.
(B) The distribution of accumulated data from each NREM sleep stage (in minutes of acquisition) shows that study 1 covered up to N3, while study 2 covered mostly wake and N1.
(C) Example of sleep scoring (or hypnogram) for one participant in study 1.
Spatial patterns in sleep and waking state
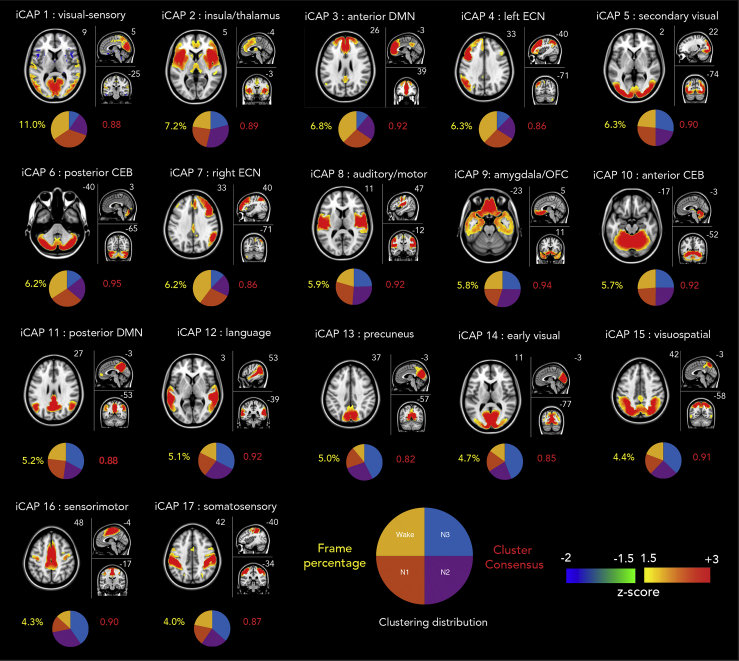
We applied a deconvolution process called total activation (TA) to all functional volumes of study 1 and study 2. Transient frames (i.e., moments of activity changes) are extracted through a derivative step that is incorporated in the TA framework. These frames correspond to time points when a change is observed in the amplitude of the fMRI BOLD signal. Significant transient frames are then selected using a two-step thresholding process (see Transparent Methods or (Karahanoğlu and Van De Ville, 2015)). Frames that survived are concatenated together and underwent a clustering procedure known as the iCAPs framework to obtain the most prevalent brain spatial patterns. We observed 17 large-scale brain networks displayed in Figure 2, representing the different functional maps that dominate brain activity from wakefulness to deep sleep. The iCAPs are ordered in descending order according to the number of times that they appeared in the significant transient frames. We then looked at the wake/sleep stages where these significant transient points occurred, and identified which iCAP they corresponded to. Figure 2 also shows the clustering distribution for each iCAP in pie charts, revealing the proportion of transient frames that the clustered iCAP occurred in each wake/sleep stage. Spatial similarities between iCAPs generated separately using study 1 and study 2 are shown in Figure S4.
Figure 2.
Spatial patterns of the 17 innovation-driven co-activation patterns (iCAPs) derived from all recordings across both studies
The iCAPs are numbered according to the percentage of significant transients that contributed to the recovery of that network (descending order), which are shown below the functional maps in yellow font. The cluster consensus of each iCAP is written in red. Pie charts indicate the distribution of each iCAP across sleep stage. MNI coordinates of each brain slice are indicated in white font. The names of the iCAPs are derived according to their correspondence with Greicius networks (Shirer et al., 2012) which are presented in the Supplemental Information (Table S1). CEB, cerebellum, DMN, default mode network, ECN, executive control network, OFC, orbitofrontal cortex.
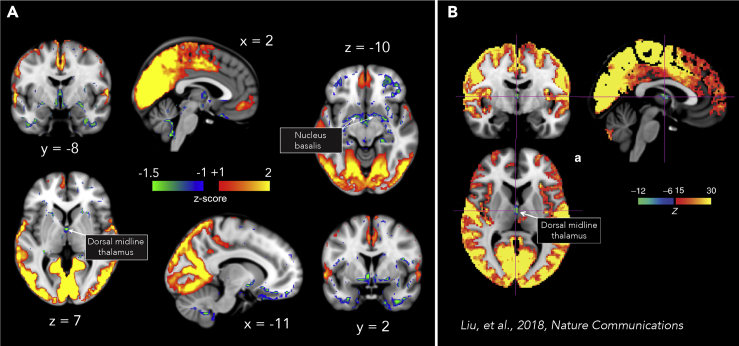
The first iCAP (iCAP 1) featured both visual and somatosensory regions, and resembles a previously observed functional pattern that distinguishes sleep from waking conditions. These regions were found to be more prevalent in N1 and N2 sleep here, like in previous work (Tagliazucchi and Laufs, 2014; Tagliazucchi et al., 2013). Unlike any other iCAP extracted, this iCAP also reveals a negative activation in subcortical regions, very much similar to what was observed by Liu and colleagues (Liu et al., 2018), as is shown side-by-side in Figure 3.
Figure 3.
Visual-sensory iCAP reveals negative activations in subcortical regions
(A) Visual-sensory iCAP also shows negative patterns in subcortical regions.
(B) Arousal-related network showing the deactivation in dorsal midline thalamus (figure adapted from previous study, Liu et al., 2018).
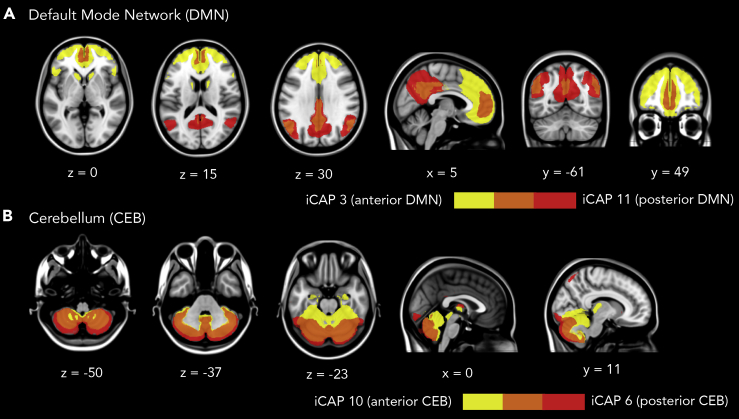
ICAP 2 featured regions of the salience network and presented a strong and selective activation of the insula, as well as portions of the thalamus. This network showed the highest likelihood to occur during N2 sleep. ICAP 3 predominantly occurred during wakefulness, displaying the anterior portion of the DMN, and in particular the anterior cingulate. By contrast, iCAP 11 (posterior DMN) and iCAP 13 (precuneus, ventral DMN) were predominantly active during deep sleep (N3), and displayed stronger activation in the posterior regions (e.g., PCC, precuneus, IPC). Notice that the anterior and posterior DMN were separately extracted during the clustering procedure despite the close similarity of their spatial patterns as shown in Figure 4A. This reflects a strong dissociation between the DMN sub-networks in terms of their temporal dynamics across the different sleep stages.
Figure 4.
Dissociation of DMN and cerebellum into posterior and anterior parts
(A) Overlay of iCAPs corresponding to anterior DMN (yellow) and posterior DMN (red).
(B) Overlay of anterior cerebellum (yellow) and posterior cerebellum (red). We also see some co-activation in the thalamus in the anterior cerebellum.
Meanwhile, attention-related networks such as the iCAPs 4 and 7 (left and right executive control networks [ECNs]), and iCAP 15 (visuospatial) were also observed. Interestingly, both ECN networks mostly resulted from transients occurring during wakefulness, whereas the visuospatial network predominated during N3 sleep. ICAP 9 contained the amygdala and the orbitofrontal cortex, with limbic-emotional iCAPs predominating during N2. We also found various networks corresponding to sensory areas corresponding to transients from the deep sleep (N3). These included iCAP 8 (auditory/motor), iCAP 14 (early visual), iCAP 16 (sensorimotor), and iCAP 17 (somatosensory).
Finally, similar to the DMN, we also observed a dissociation of the cerebellum in the form of iCAPs 6 and 10, both featuring the anterior and posterior regions of the cerebellum, respectively. These iCAPs are overlaid in Figure 4B. A detailed description of the regions in each iCAP using AAL regions (Tzourio-Mazoyer et al., 2002) and their similarity to Greicius networks (Shirer et al., 2012) are displayed in the Supplemental Information (Table S1).
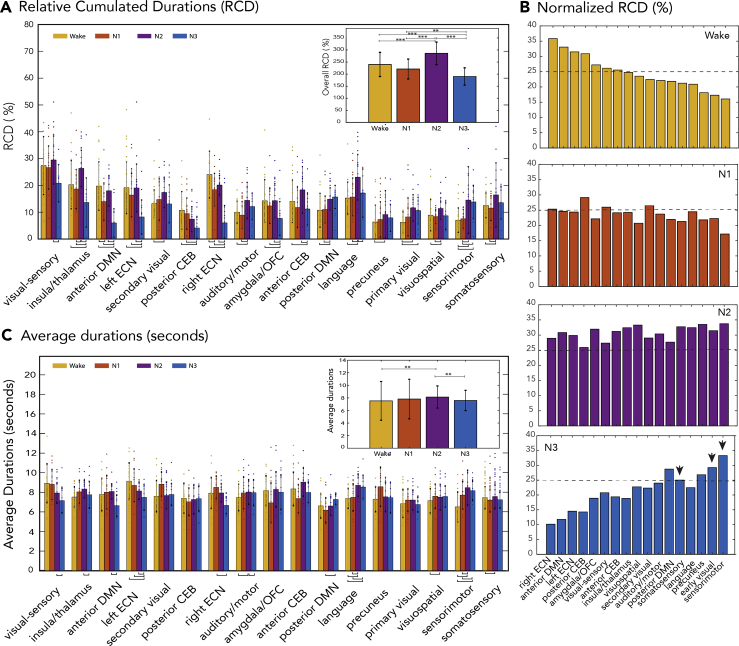
iCAP relative cumulated durations are consistent with the cluster distribution of significant transients, and both reveal stage-dependent network activity
The iCAPs were obtained by clustering the significant transients from the data (i.e., instances when functional maps markedly increased or decreased activity). The pie charts in Figure 2 display how the significant transients contributing to each iCAP were distributed across wake/sleep stages. In order to obtain the activity time courses of iCAPs per participant in fMRI TR resolution (i.e., frame-wise), and be able to compute their durations (in contrast to only transients or activity changes), we performed a spatiotemporal transient-informed back-projection of the iCAPs onto the HRF-deconvolved frames.
In Figure 5A, we display the relative cumulated durations (RCDs) or the likelihood of each iCAP to appear in a specific sleep stage. This is a measure describing the cumulated time that a particular iCAP was active divided by the total time that the participant spent in a particular sleep stage. This normalization thus ensured that the reported persistence of an iCAP within one sleep stage was independent of the duration of that stage. Unsurprisingly, the relative cumulated iCAP durations appeared mostly consistent with the clustering distribution observed in Figure 2. Attention-related iCAPs such as the left and right ECN showed predominant sustained activity in wakefulness. Conversely, sensory-related iCAPs (e.g., early visual, somatosensory, sensorimotor) were more persistent in N2 and N3 sleep compared to wakefulness and N1. The insula/thalamus iCAP also displayed predominant activity in N2 sleep. In addition, we found that the RCDs of iCAPs related to anterior DMN is higher during wakefulness, while the posterior DMN iCAP showed higher likelihood to appear in N3. These findings are well in line with previous observations of DMN dissociating into posterior and anterior parts upon reaching deep sleep (Larson-Prior et al., 2009; Sämann et al., 2011). Interestingly, we also observed the posterior (but not the anterior) cerebellum to be preferentially activated during wakefulness compared to N3. The corresponding test statistics (e.g., p values, t-statistic, and effect sizes) for individual networks are reported in Table S2.
Figure 5.
Duration of iCAPs in different sleep stages
(A) Relative cumulated durations (RCD, in %) of iCAPs depicting each network's likelihood to occur in different sleep stages. Data are represented as mean ± SEM. The inset shows the overall trend of the iCAP durations in wakefulness/sleep stages, which are all above 100%, reflecting the tendency of iCAPs to overlap in time.
(B) RCD divided by the number of time points that an iCAP is active in the whole time course. The bars are ordered according to their descending representation in wakefulness. The broken horizontal line in each subplot indicates 25% likelihood, while the arrows in N3 subplot indicate visual and sensory-related iCAPs that are above the 25% line.
(C) Average durations (in seconds) of iCAPs, reflecting the length of continuous activity of iCAPs. The inset corresponds to the overall trend of iCAP average durations. Data in the insets are represented as mean ± SD. The horizontal lines at the bottom of the bar plot correspond to significant differences evaluated through paired t-tests and permutation testing. Horizontal lines with 3 stars and 2 stars in the insets represent significant differences with p values less than 0.001 and 0.01, respectively (corrected using Tukey's range test).
We then looked at the general trend of network activity across the different sleep stages (inset of Figure 5A), and found that the RCD of iCAPs significantly increased during N2 with respect to wakefulness and N1, followed by a steep decrease in N3. Please note that values above 100% are due to the temporal overlapping nature of iCAPs, with two or more iCAPs occurring at the same time. We then computed a network-based normalization of RCD, displayed in Figure 5B. This is equivalent to normalizing the number of active time points in each sleep stage by successive division to two factors: (1) overall number of time instances that an iCAP is present over the whole duration of the recording and (2) the total time that the participant spent in a particular sleep stage. The bar plots in Figure 5B show that almost all iCAPs displayed proportions above 25% in N2 sleep (broken horizontal line). We observed a marked disparity in iCAP proportions during wakefulness and N3, in contrast to the more uniform distribution in N1 and N2 sleep. Moreover, iCAPs that were more represented in wakefulness were less represented in N3, which is consistent with the RCD measures of each iCAP across wake/sleep stages in Figure 5A (all p < 0.05). Specifically, iCAPs that reached more than 25% activity during specific sleep/wake stages are: (1) wakefulness—visual-sensory, left and right ECN and posterior CEB, (2) N1—posterior CEB, visual-sensory, secondary visual, and (3) N3—posterior DMN, somatosensory, precuneus, early visual, sensorimotor.
Contrasting with the RCD above, the average duration or mean lifetimes of an iCAP represents the average bouts (in seconds) of continuous activity. Overall, we observed iCAPs to be active between 5 and 10 s (7.3 1.7 s; Figure 5C). In general, we also found iCAP activity to have longer durations in N2 compared to N3 (p < 0.01), and compared to wakefulness (p < 0.01). For some of the networks, the average durations were proportional to their relative propensity to occur in each sleep stage. For instance, the left ECN displayed the least likelihood to occur in N3, and also exhibited the shortest average duration during that stage.
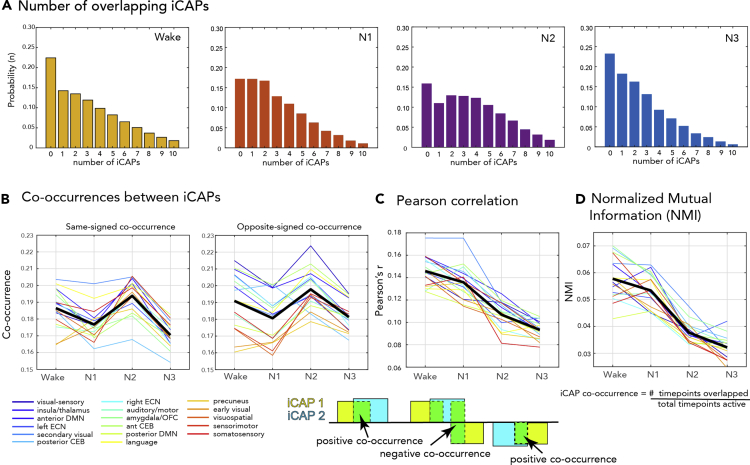
Alterations in network co-occurrences in different sleep stages
To go beyond the iCAPs' individual temporal properties, we looked at the probability of having either none, one, two, or more iCAPs occurring at one time point. Figure 6A shows the likelihood for different numbers of overlapping iCAPs to occur in wakefulness and in different sleep stages. We observed the overlaps to be maximally N = 10, and thus we limit the histogram for the range {N = 0,1,2, …, 10}. Among all vigilance states, N2 displayed a relatively flatter distribution especially compared to wakefulness and N3 (both p < 0.05), with a higher likelihood of having 2 to 5 overlapping iCAPs. For a table of test-statistics, see Table S3. Given that individual iCAPs had higher likelihood to occur in N2 compared to other sleep stages, it is not surprising that iCAPs are also more likely to overlap in time during N2.
Figure 6.
Interactions between iCAPs across different sleep stages
(A) The probability of different numbers of iCAPs to overlap is displayed for wakefulness and different sleep stages.
(B) The iCAP co-occurrence pertains to the number of time points during which a pair of iCAPs were both active, divided by the total number of time points that at least one of them was active. This was computed separately for similar signed activations (same-signed co-occurrences) or opposite-signed activations (opposite-signed co-occurrences). Co-occurrence was computed for each network (i.e., pairwise co-occurrence of one network with all the other networks).
(C) Mean of classical FC metric (Pearson's r) applied to iCAP time courses.
(D) Mean of normalized mutual information (NMI) of pairwise iCAP time courses. The computation for the co-occurrence between pairs of iCAP is illustrated on the bottom of the figure.
We then evaluated the pairwise co-occurrence between each iCAP and all other networks (Figure 6B). The co-occurrence is a normalized measure of the number of time points at which an overlap occurs between two pairs of iCAPs, divided by the total number of time points that at least either one is active. This is different from the temporal overlap measure observed in Figure 6A because we focus on pairwise co-occurrence between two iCAPs. This can also be referred to as the Jaccard similarity score of two iCAP time courses. We also take into account the signs of the activations, see illustrations in Figure 6. In general, all iCAPs showed a high mean same-signed co-occurrence and opposite-signed co-occurrence in wakefulness and N2, and a decrease in N1 and N3 (p < 0.001, see Table S4). Next, we compared these observations using classical FC analysis (i.e., Pearson correlation) and normalized mutual information (NMI) applied to the iCAP time courses. We observed that the pairwise network FC across iCAPs decreased with increasing sleep depth (with no increase in N2). The same trend can be observed in Figure 6D which shows the overall NMI of iCAP time courses, suggesting that the mutual dependence between iCAPs decreases from wakefulness to deep sleep. These observations are consistent with the general notion that connectivity breaks down with increasing sleep depth (Tagliazucchi and Laufs, 2014). Both FC and the NMI also has the lowest variance (Figure S5) during N2 and N3 sleep.
Robustness of results across datasets and across choice of parameters
In order to see whether the resulting iCAPs are robust with regards to the data set from which the contributing frames belong to, we perform an averaging of significant transient frames corresponding to iCAP indices within a similar dataset (i.e., among those belonging study 1 and among those belonging to study 2). Study 1 contributes 58,373 frames while the study 2 contributes 39,235, giving a total of 97,608 significant frames. The number of contributing frames coming from each data set is shown in Figure S5 and is written in red and yellow fonts for study 1 and study 2, respectively. The distribution of clustering assignments is particularly telling, especially on the contribution of each dataset reflecting the tendency of the iCAP to appear in either study 1 (reaching deep sleep) or in study 2 (wake to N1). We observe the visual-sensory, anterior DMN, posterior cerebellum, and the left and right executive control networks to be mostly contributed by frames belonging to the study 2 data set. On the other hand, the study 1 contributed more on the recovery of the remaining networks. We computed the spatial similarity of the resulting iCAPs. Figure S4 provides an overlay of the iCAPs computed using frames belonging to study 1 versus study 2. Visually, all iCAPs display similar spatial patterns in both the study 1 and study 2, which is also well supported by their Dice coefficients.
Moreover, we also evaluated the robustness of the results in terms of the observed temporal characteristics using the data from study 2. We computed the occurrences and co-occurrences of iCAPs using only subjects from study 2. These are displayed in Figures S6 and S7, respectively. In these figures, there are no values for N3 since subjects from study 2 only reach NREM stage 2. Consistent with the findings in study 1, we find a surge in iCAP activity during NREM stage 2. We also observe a higher likelihood for 3 or more iCAPs to overlap in this stage of sleep. Meanwhile, the iCAPs overlap the least during wakefulness.
Finally, we evaluated the robustness of the results with respect to the choice of the number of clusters, K. The choice of the number of clusters was motivated by a quantitative approach for optimal class discovery (Monti et al., 2003). To assess whether the choice of K affects the final outcome of the analysis, we also repeated the clustering procedure to the whole dataset using K = 20. Figure S8 displays the side-by-side analysis done using K = 17 and K = 20. Here, we find that the general spatial characteristics are the same for both analyses, and that the distribution (pie charts) show similar proportions across NREM sleep stages. Moreover, we also find that increasing the number of K, beyond the optimally observed K = 17, results in the recovery of similar repeating spatial patterns that are already present for K = 17.
Discussion
General findings
Here we extracted large-scale brain networks or iCAPs by clustering moments of significantly changing brain activity from two studies using simultaneous EEG and fMRI recordings during sleep. The use of transient fMRI activity allowed us to obtain temporally overlapping spatial patterns together with subject-specific time courses at a time-scale of seconds. Altogether, our findings help establish the following: (1) that coordinated activation of regional brain areas during wakefulness was largely preserved in N1, N2, and N3 sleep, yet with specific relative distributions and dynamic modulations across NREM stages, (2) that functional brain networks sustain continuous bouts of activity on an average of about 7.31.7 seconds, and for some networks, their mean lifetimes changed accordingly to their likelihood to occur in each sleep stages, (3) that these networks exhibit maximal activity in N2 sleep and the least in N3, and (4) that the pairwise network co-occurrences generally show marked increase in N2 sleep followed by a decrease in N3, even though (5) FC and mutual information between these networks progressively broke down from wakefulness to N3.
The first observation is largely consistent with previous work reporting the persistence of large-scale brain networks during sleep (Horovitz et al., 2009; Sämann et al., 2011), with various modifications when reaching deeper sleep stages, such as decreased connection strengths in RSNs (Larson-Prior et al., 2009), changes of hierarchical organization into smaller independent modules (Boly et al., 2008), or reduced long-range temporal dependencies in the BOLD signal (Tagliazucchi et al., 2013). For instance, the well-known attention-related networks, such as the bilateral ECN, occurred preferentially during wakefulness and N1, whereas networks associated with primary sensory systems were most present during N2 and N3, consistent with some earlier observations (Horovitz et al., 2008; Larson-Prior et al., 2009).
In the next subsections, we discuss the most important findings of the work and give interpretations to the observed dynamical characteristics of cross-network interactions.
Visual-sensory iCAP confirms the emergence of arousal-related network
The first iCAP (iCAP 1, Figure 2), corresponding to the network of regions contributing to the highest number of significant transients in our dataset, was the visual-sensory iCAP, which predominated during the transition to sleep (N1). It included a large portion of the visual cortex, as well as some sensory and motor regions. This network also displayed a unique characteristic of having negative activations in subcortical areas. Using combined electrophysiological and fMRI signals, Liu et al. (2018) described a very similar spatial pattern, with negative activation in basal forebrain and thalamus paralleled by positive activations in sensory cortices, which arose during momentary drops in arousal, indexed by a spectral shift in local field potentials toward low frequencies. In Figure 3, we provide a side-by-side comparison between our visual-sensory iCAP 1 and the arousal-related network observed by Liu et al. While both networks displayed positive activations in visual, sensory, and motor regions, they also exhibited negative activation in the dorsal midline thalamus, as well as the nucleus basalis. Because the thalamus holds an important role in regulating the physiological state of arousal during sleep (Gent et al., 2018; Saper et al., 2010) and is responsible for relaying motor and sensory signals to the cerebral cortex, we can suggest that reduced thalamic activity favors sleep onset while limiting incoming external sensory stimulation. This interpretation is also supported by the fact that most of the frames that contributed to this iCAP came from wake and N1, suggesting that this iCAP might play a key role in the onset of sleep. Interestingly, however, we found that this network exhibited a persistent likelihood in terms of the cumulated duration to occur in N2, before decreasing in N3. Fittingly, it has also been found in previous studies that there is an increase in BOLD signal variance in sensory and motor cortices during N1 and N2 sleep, which has been used as diagnostic for detecting sleep in typical resting-state recordings. On the other hand, high activities in frontal, parietal, and temporal cortices are typically associated with wake conditions (Tagliazucchi and Laufs, 2014; Tagliazucchi et al., 2013). These two brain spatial patterns bear resemblance to the visual-sensory iCAP and the bilateral ECN, respectively, whose temporal properties show consistent behavioral stage-dependence. In addition, Stevner et al. (2019) reported the same visual-sensory network to be implicated also in N1.
DMN and cerebellum dissociates into posterior and anterior regions
The DMN has been well studied particularly in light of its connectivity changes when subjects transition from wake to light sleep, and deep sleep. The initial hypothesis was that the DMN supports a range of self-related mental processes, such as unconstrained self-referential thought and recollection. However, previous studies have found the DMN to persist during light sleep, as well as in deep sleep with reduced connectivity between the MPFC and the rest of the network (Horovitz et al., 2009; Larson-Prior et al., 2009). In the present work, we observed a dissociation of the DMN subcomponents into its posterior and anterior parts (Figure 4A). The pie-chart distributions of cluster frame indices displayed in Figure 2 and the cumulated durations in Figure 5A reveal that the anterior region of the DMN mostly occurred during wake and N1 sleep, whereas the posterior DMN predominated in N2 and N3 sleep. We found a similar antero-posterior dissociation in the cerebellum (overlap shown in Figure 4B). Studies that investigated FC in the posterior and the anterior regions of the cerebellum have remained inconclusive regarding their potential roles in sleep. The cerebellum has been found to be related to motor control and motor memory formation (Gao et al., 2012). It has also been observed to show sleep stage-dependent activity, whose impairments disrupt the sleep-wake cycle which then leads to sleep disorders (DelRosso and Hoque, 2014). As a general observation, signals in the cerebellum appear to be lower during N1 as compared to wakefulness (Hiroki et al., 2005; Kaufmann et al., 2006). However, a map summarizing major cerebellar research in sleep using fMRI and PET revealed a localized change in cerebellar activity during SWS surrounding the cerebellum's larger lobules (parts IV, V, VI, and VII), as well as marked correlations with slow spindles in N2 (Canto et al., 2017). These lobules make up the anterior region of the cerebellum (Dang-Vu et al., 2008). Moreover, it has been previously shown that the sensorimotor domain is related to the anterior cerebellar lobe, while the cognitive domain corresponds to the posterior cerebellar region (Stoodley and Schmahmann, 2010). This particular finding is consistent with the observed co-activation of the anterior cerebellum with regions corresponding to the sensorimotor network represented by iCAP 10 (see also Shirer et al., 2012) and Table S1) Meanwhile, the inferior posterior lobe has been associated with the frontal gray matter, a well-known core area of executive function (Jung et al., 2019; Tiemeier et al., 2010). Altogether, these studies support our current findings on the preferential tendency of posterior and anterior cerebellum to persist in wakefulness and N2, respectively.
Alterations in network temporal characteristics and interactions during sleep reveal paradoxical trend
Previous connectivity analyses of brain regions found a general decrease of FC with increasing sleep depth (Haimovici et al., 2017; Spoormaker et al., 2010; Tagliazucchi et al., 2016). Classical FC approaches capture statistical interdependencies between activity from two brain regions (Friston, 2011). Typically, if two regional time courses exhibit simultaneous positive (or negative) values over some time-windows, and exactly opposite values on some other time-windows, the final correlation (and derived FC) between both regions would be close to zero, despite the synchronized deviations from the baseline, albeit the incoherent signs. Therefore, it follows that a decrease in FC does not mean a decrease in global network activity. This is also true in measures of mutual information.
In this work, we demonstrated that network activations increased when participants went from wakefulness to N2, while they decreased when reaching N3. This important observation was captured because we were able to extract network temporal activity at the frame-wise level for each individual iCAP. In contrast, classical dynamic FC analyses only capture the statistical relationship among ROIs comprising the networks, and not the actual activity of the networks themselves. Complementary, NMI measures the general relationship and detects both linear and non-linear mutual dependencies between these networks. Furthermore, because the iCAP approach allows networks to overlap in time, we were able to observe that functional networks show a higher likelihood to overlap (e.g., 2 to 4 iCAPs active at the same time) in N2 compared to other stages. Congruently, we also found high co-occurrences in N2. By contrast, N1 and N3 generally showed much lower co-occurrences. What is remarkable is the simultaneous increase in both the same-signed and opposite-signed activations in N2 (Figure 6B). Meanwhile, classical Pearson correlation measure applied to iCAP time courses displayed a decreasing trend from wakefulness to deep sleep, consistent with FC findings based on regional fMRI time courses (Haimovici et al., 2017; Kung et al., 2019; Spoormaker et al., 2010; Tagliazucchi et al., 2016). This loss of FC is supported by the observed decline of iCAP mutual dependence based on NMI measures in Figure 6D further strengthening the observed paradox between network activity and network interactions from wakefulness to deep sleep.
The remarkable increase in network activity during N2 is particularly telling and perhaps not coincidental, as it is to be noted that N2 sleep acts as an intermediate epoch between drowsiness and deep sleep. This is also the period when various paroxysmal events occur, i.e., spindles and K complexes (Jahnke et al., 2012). Functionally, the role of K complexes is under debate whether it acts to promote deeper sleep, or whether it does the opposite and correlates with cortical arousal (Bastien et al., 2000). As such, it is believed to often emerge at times of brain instability. Thus, it cannot be discounted that the general increase of network activity and co-occurrences in N2 may be due to a common cause or a global drive that occurs at this sleep stage. Meanwhile, the overall decrease in network activity and co-occurrences during N3 fit previous findings on global increase in functional segregation during deep sleep (Boly et al., 2012; Horovitz et al., 2009; Spoormaker et al., 2012). Deep sleep is first and foremost characterized by high-amplitude, low-frequency brain waves. It is manifested by unresponsiveness, and the decline of the ability to react to external stimuli (Cirelli and Tononi, 2008). Fittingly, similar findings have been shown for other unconscious brain states, such as in propofol anesthesia (Monti et al., 2013).
We therefore summarize three important findings regarding network temporal characteristics of sleep stage N2: (1) FC and NMI between networks decreased with sleep depth, (2) there is a higher likelihood for stereotyped networks or iCAPs to emerge in N2 compared to wakefulness and other sleep stages, and (3) both same-signed and opposite-signed co-occurrences between iCAPs increased in N2. In line with previous works, we speculate that the decreased in FC and more strongly the reduced NMI, both reflect a global reduced efficiency of information transfer between networks. These observations are possible manifestations of reduced brain network integrity (Nofzinger, 2006) that is commonly interpreted to reflect reduced consciousness during sleep (Cirelli and Tononi, 2008). This interpretation is consistent with the general consensus among previous studies that view consciousness not as a persistence of functional brain networks to occur, but rather the degree of interactions among them (Horovitz et al., 2009; Nofzinger, 2006; Sämann et al., 2011; Spoormaker et al., 2010). Intriguingly, despite the decrease in mutual dependence between networks in N2, a general increase in network activity is observed, possibly reflecting the unstable dynamics among iCAPs. In other words, observation (1) likely corresponds to a decrease in global network integration, while observations (2) and (3) can be presumed to express network attempts to communicate specifically during N2 resulting in an increased inter-network co-occurrence with nonetheless unstable synchronizations. This latter finding is in agreement with a very recent observation by Kung et al. (2019), who reported a very high variance in the dynamic FC evaluated across sliding windows during N2, compared to during wakefulness, N1, and N3. High dynamic FC variance in N2 was interpreted in their study as elevated instability of information transfer between networks, whereas low mean dynamic FC was deduced to reflect decreased intra-network consistency.
Concerning N3, we observed (1) an overall decrease of global network occurrence, (2) a decrease in network mean lifetimes, and (3) a decrease in functional association characterized by lowest cross-network FC and mutual dependence through NMI. Altogether, our results are consistent with the interpretation of a more stable brain state (Jobst et al., 2017) yet more localized signal integration in SWS (Deco et al., 2017). Spatially, the huge drop in anterior DMNs likelihood to occur in deep sleep compared to other stages, together with the high persistence of unimodal primary sensory networks, provide further support to the notion of a breakdown of long-distance functional connections (Tagliazucchi et al., 2013) in favor of short-distance associations (Boly et al., 2012). The global decrease of network activity, mean lifetimes in N3, and mutual dependence between networks are, altogether, potential indicators of a more stable state of the system, but also a general loss of effective information integration.
In summary, we investigated the dynamic properties of large-scale functional networks during wakefulness and NREM sleep. We used the TA and iCAPs framework to capture precise moments of transient brain activity, giving us a quantitative view of how the brain dynamically evolves across the different stages of NREM sleep. We found new networks that are largely related to regions that support the physiological organization of sleep and arousal. We also uncovered whole-brain spatial patterns that resemble currently known RSNs whose temporal profiles are consistent with previous findings. The temporal dynamics these networks exhibited alterations between different sleep stages. In particular, we observed a global dissociation/decrease of brain activity both in spatial and temporal domains, from wakefulness to deep sleep. Unexpectedly, we found an increase in network activity in N2 and a global increase in simultaneous positive and negative network co-occurrence, signaling instability of network synchronization and ineffective brain integration. Altogether, these findings support the general consensus of relating cortical integration to consciousness dissipation during sleep and provide new evidence for the presence of unstable yet distributed global inter-regional co-activation in N2 sleep.
Limitations of the study
The iCAPs framework has already been applied to resting-state fMRI data of healthy (Karahanoğlu and Van De Ville, 2015), clinical populations (Zöller et al., 2019), and spinal cord fMRI (Kinany et al., 2020). The recovered iCAPs in the present study were highly similar to the ones observed in the first two studies, while additional spatial patterns unique in the sleep were also found (e.g., visual-sensory network with deactivations in subcortical regions, cerebellum, and insula). While dynamic FC analyses relying on direct statistical interdependencies of brain regions are effective in capturing coherent fluctuations between brain areas, it does not capture individual brain activity at a single time point. The framework is unique in its ability to detect transients which allows for the extraction of spatially and temporally overlapping functional networks at each time point, which is a particular advantage that is beyond the classical methodologies applied in sleep studies. Using this advantage, we were able to show individual network activity at precise fMRI temporal resolution across sleep stages and found a distinct stage-dependent activity for each network.
Although the method used is advantageous particularly in our goal to characterize functional brain dynamics during sleep, it also comes with some limitations. Unlike classical FC studies where one can evaluate the integrity of connectivity within a network (i.e., correlate one ROI to another ROI within the same network), the iCAP approach extracts the spatial patterns as a whole. Thus, we are not able to evaluate the strength of connectivity within a network. Nevertheless, the clustering step itself reveals data-driven dissociations of well-known spatial patterns, such as the DMN and the cerebellum, which essentially reflects the spatial modifications that the brain undergoes as it transitions to SWS. Furthermore, while our approach does remove hemodynamic effects resulting to a much cleaner representation of the BOLD activity, the model used to deconvolve the HRF is assumed to be the same across all brain voxels, following the general practice in the fMRI field. Nevertheless, for the research questions that we aim to answer in this work, these effects can be reasonably ignored for our main findings.
Lastly, while the extracted temporal overlaps and co-occurrences between iCAPs give useful insights on the overall global brain dynamics, these measures do not reflect causal influence between these networks and therefore cannot be used to directly assess different levels of consciousness. In contrast, FC, and more strongly, NMI, capture linear and non-linear interdependencies between iCAPs. The agreeable outcome between these two measures provides further evidence on reduced information integration that accompany increasing sleep depth, and give stronger confidence on the observed paradox between the activity of large-scale brain networks and their mutual dependencies. Nevertheless, in order to more accurately quantify the level of consciousness across NREM sleep, additional analysis is suggested using previously prescribed measures of integrated information (Krohn and Ostwald, 2017; Tononi, 2008; Tononi et al., 2016).
Resource availability
Lead contact
Further information and requests for data and code should be directed to and will be fulfilled by the lead contact Anjali Tarun (anjali.tarun@epfl.ch).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The datasets supporting the current study have not been deposited in a public repository but are available from the corresponding author on request. The code of the TA and iCAPs pipeline are available in a public repository (https://c4science.ch/source/iCAPs/).
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
A.T. is supported by the Swiss National Science Foundation under the Project Grant 205321-163376. D.W.A is supported by the National Institute for the Humanities and Social Sciences (NIHSS) under the Project Grant SDS14/1117, the German Academic Exchange Service (DAAD) Grant 91570813, the National Research Foundation of South Africa (NRF) Grant 83341, and the Harry Oppenheimer Memorial Trust, South Africa Grant OMT 20027. This work was supported by the National Center of Competence in Research (NCCR) Affective Sciences financed by the Swiss National Science Foundation (grant number: 51NF40-104897) and hosted by the University of Geneva, and by individual grants from the Swiss National Science Foundation (to S.S., grant numbers: 320030-159862 and 320030-135653), and by the Mercier Foundation. Study 1 was conducted on the imaging platform at the Brain and Behavior Lab (BBL) and benefited from support of the BBL technical staff.
We would also like to acknowledge all participants in the two experimental studies included in this work, and Carina Zoellner and Annika Rips for help with data acquisition in Study 2.
Author contributions
Conceptualization, A.T. and D.V.D.; Methodology, A.T.; Validation, A.T.; Formal Analysis, A.T.; Investigation, D.W.A. and V.S.; Resources, D.W.A., V.S., L.P., L.B. and M.S.; Data curation, A.T., D.W.A., V.S., L.P., and L.B.; Writing – Original Draft, A.T.; Writing – Review & Editing, A.T., D.W.A., L.P., N.A., S.O.S. and D.V.D.; Visualization, A.T.; Supervision; S.O.S. and D.V.D.; Funding Acquisition, M.S., S.O.S., and D.V.D.
Declaration of interests
The authors declare no competing interests.
Published: January 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2020.101923.
Supplemental information
Percentiles indicate the fraction of voxels belonging to a network or region that has a z-score > 1.5. Only networks and regions that have at least 20% surviving percentile are included in the list. iCAP 6 is not included due to the excluded cerebellum in Greicius atlas. Specific regions that are not part of the cerebellum but are co-activated with iCAP 10 (anterior cerebellum) are included.
References
- Andrews-Hanna J.R. The brain’s default network and its adaptive role in internal mentation. Neuroscience. 2012;18:251–270. doi: 10.1177/1073858411403316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien C.H., Ladouceur C., Campbell K.B. EEG characteristics prior to and following the evoked K-Complex. Can. J. Exp. Psychol. 2000;54:255–265. doi: 10.1037/h0087345. [DOI] [PubMed] [Google Scholar]
- Boly M., Phillips C., Tshibanda L., Vanhaudenhuyse A., Schabus M., Dang-Vu T.T., Moonen G., Hustinx R., Maquet P., Laureys S. Intrinsic brain activity in altered states of consciousness. Ann. N. Y. Acad. Sci. 2008;1129:119–129. doi: 10.1196/annals.1417.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boly M., Perlbarg V., Marrelec G., Schabus M., Laureys S., Doyon J., Pelegrini-Issac M., Maquet P., Benali H. Hierarchical clustering of brain activity during human nonrapid eye movement sleep. Proc. Natl. Acad. Sci. U S A. 2012;109:5856–5861. doi: 10.1073/pnas.1111133109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C.B., Onuki Y., Bruinsma B., van der Werf Y.D., De Zeeuw C.I. The sleeping cerebellum. Trends Neurosci. 2017;40:309–323. doi: 10.1016/j.tins.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Cirelli C., Tononi G. Is sleep essential? Plos Biol. 2008;6:e216. doi: 10.1371/journal.pbio.0060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang-Vu T.T., Schabus M., Desseilles M., Albouy G., Boly M., Darsaud A., Gais S., Rauchs G., Sterpenich V., Vandewalle G. Spontaneous neural activity during human slow wave sleep. Proc. Natl. Acad. Sci. U S A. 2008;105:15160–15165. doi: 10.1073/pnas.0801819105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G., Tagliazucchi E., Laufs H., Sanjuán A., Kringelbach M.L. Novel intrinsic ignition method measuring local-global integration characterizes wakefulness and deep sleep. Eneuro. 2017;4 doi: 10.1523/ENEURO.0106-17.2017. ENEURO.0106-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelRosso L.M., Hoque R. The cerebellum and sleep. Neurol. Clin. 2014;32:893–900. doi: 10.1016/j.ncl.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Friston K.J. Functional and effective connectivity: a Review. Brain Connect. 2011;1:13–36. doi: 10.1089/brain.2011.0008. [DOI] [PubMed] [Google Scholar]
- Gao Z., van Beugen B.J., De Zeeuw C.I. Distributed synergistic plasticity and cerebellar learning. Nat. Rev. Neurosci. 2012;13:619–635. doi: 10.1038/nrn3312. [DOI] [PubMed] [Google Scholar]
- Gent T.C., Bandarabadi M., Herrera C.G., Adamantidis A.R. Thalamic dual control of sleep and wakefulness. Nat. Neurosci. 2018;21:974–984. doi: 10.1038/s41593-018-0164-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius M.D., Krasnow B., Reiss A.L., Menon V. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc. Natl. Acad. Sci. U S A. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovici A., Tagliazucchi E., Balenzuela P., Laufs H. On wakefulness fluctuations as a source of BOLD functional connectivity dynamics. Sci. Rep. 2017;7:5908. doi: 10.1038/s41598-017-06389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale J.R., White T.P., Mayhew S.D., Wilson R.S., Rollings D.T., Khalsa S., Arvanitis T.N., Bagshaw A.P. Altered thalamocortical and intra-thalamic functional connectivity during light sleep compared with wake. Neuroimage. 2016;125:657–667. doi: 10.1016/j.neuroimage.2015.10.041. [DOI] [PubMed] [Google Scholar]
- Hiroki M., Uema T., Kajimura N., Ogawa K., Nishikawa M., Kato M., Watanabe T., Nakajima T., Takano H., Imabayashi E. Cerebral white matter blood flow is constant during human non-rapid eye movement sleep: a positron emission tomographic study. J. Appl. Physiol. 2005;98:1846–1854. doi: 10.1152/japplphysiol.00653.2004. [DOI] [PubMed] [Google Scholar]
- Horovitz S.G., Fukunaga M., de Zwart J.A., van Gelderen P., Fulton S.C., Balkin T.J., Duyn J.H. Low frequency BOLD fluctuations during resting wakefulness and light sleep: a simultaneous EEG-fMRI study. Hum. Brain Mapp. 2008;29:671–682. doi: 10.1002/hbm.20428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz S.G., Braun A.R., Carr W.S., Picchioni D., Balkin T.J., Fukunaga M., Duyn J.H. Decoupling of the brain’s default mode network during deep sleep. Proc. Natl. Acad. Sci. U S A. 2009;106:11376–11381. doi: 10.1073/pnas.0901435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnke K., von Wegner F., Morzelewski A., Borisov S., Maischein M., Steinmetz H., Laufs H. To wake or not to wake? The two-sided nature of the human K-complex. Neuroimage. 2012;59:1631–1638. doi: 10.1016/j.neuroimage.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Jobst B.M., Hindriks R., Laufs H., Tagliazucchi E., Hahn G., Ponce-Alvarez A., Stevner A.B.A., Kringelbach M.L., Deco G. Increased stability and breakdown of brain effective connectivity during slow-wave sleep: mechanistic insights from whole-brain computational modelling. Sci. Rep. 2017;7:1–16. doi: 10.1038/s41598-017-04522-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K.-I., Park M.-H., Park B., Kim S.-Y., Kim Y.O., Kim B.-N., Park S., Song C.-H. Cerebellar gray matter volume, executive function, and insomnia: gender differences in adolescents. Sci. Rep. 2019;9:855. doi: 10.1038/s41598-018-37154-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahanoğlu F.I., Van De Ville D. Transient brain activity disentangles fMRI resting-state dynamics in terms of spatially and temporally overlapping networks. Nat. Commun. 2015;6:7751. doi: 10.1038/ncomms8751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahanoğlu F.I., Caballero-Gaudes C., Lazeyras F., Van De Ville D. Total activation: fMRI deconvolution through spatio-temporal regularization. Neuroimage. 2013;73:121–134. doi: 10.1016/j.neuroimage.2013.01.067. [DOI] [PubMed] [Google Scholar]
- Kaufmann C., Wehrle R., Wetter T.C., Holsboer F., Auer D.P., Pollmächer T., Czisch M. Brain activation and hypothalamic functional connectivity during human non-rapid eye movement sleep: an EEG/fMRI study. Brain. 2006;129:655–667. doi: 10.1093/brain/awh686. [DOI] [PubMed] [Google Scholar]
- Kinany N., Pirondini E., Micera S., Van De Ville D. Dynamic functional connectivity of resting-state spinal cord fMRI reveals fine-grained intrinsic architecture. Neuron. 2020;108:424–435.e4. doi: 10.1016/j.neuron.2020.07.024. [DOI] [PubMed] [Google Scholar]
- Krohn S., Ostwald D. Computing Integrated Information. Neurosci. Conscious. 2017;2017:nix017. doi: 10.1093/nc/nix017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung Y., Li C., Chen S., Chen S.C., Lo C.Z., Lane T.J., Biswal B., Wu C.W., Lin C. Instability of brain connectivity during nonrapid eye movement sleep reflects altered properties of information integration. Hum. Brain Mapp. 2019;40:3192–3202. doi: 10.1002/hbm.24590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson-Prior L.J., Zempel J.M., Nolan T.S., Prior F.W., Snyder A.Z., Raichle M.E. Cortical network functional connectivity in the descent to sleep. Proc. Natl. Acad. Sci. U S A. 2009;106:4489–4494. doi: 10.1073/pnas.0900924106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Duyn J.H. Time-varying functional network information extracted from brief instances of spontaneous brain activity. Proc. Natl. Acad. Sci. U S A. 2013;110:4392–4397. doi: 10.1073/pnas.1216856110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., De Zwart J.A., Schölvinck M.L., Chang C., Ye F.Q., Leopold D.A., Duyn J.H. Subcortical evidence for a contribution of arousal to fMRI studies of brain activity. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-017-02815-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti M.M., Lutkenhoff E.S., Rubinov M., Boveroux P., Vanhaudenhuyse A., Gosseries O., Bruno M.A., Noirhomme Q., Boly M., Laureys S. Dynamic change of global and local information processing in propofol-induced loss and recovery of consciousness. PLoS Comput. Biol. 2013;9:e1003271. doi: 10.1371/journal.pcbi.1003271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti S., Tamayo P., Mesirov J., Golub T. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Mach. Learn. 2003;52:91–118. [Google Scholar]
- Nofzinger E.A. Neuroimaging of sleep and sleep disorders. Curr. Neurol. Neurosci. Rep. 2006;6:149–155. doi: 10.1007/s11910-996-0038-3. [DOI] [PubMed] [Google Scholar]
- Picchioni D., Pixa M.L., Fukunaga M., Carr W.S., Horovitz S.G., Braun A.R., Duyn J.H. Decreased connectivity between the thalamus and the neocortex during human nonrapid eye movement sleep. Sleep. 2014;37:387–397. doi: 10.5665/sleep.3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sämann P.G., Wehrle R., Hoehn D., Spoormaker V.I., Peters H., Tully C., Holsboer F., Czisch M. Development of the brain’s default mode network from wakefulness to slow wave sleep. Cereb. Cortex. 2011;21:2082–2093. doi: 10.1093/cercor/bhq295. [DOI] [PubMed] [Google Scholar]
- Saper C.B., Fuller P.M., Pedersen N.P., Lu J., Scammell T.E. Sleep state switching. Neuron. 2010;68:1023–1042. doi: 10.1016/j.neuron.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirer W.R., Ryali S., Rykhlevskaia E., Menon V., Greicius M.D. Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cereb. Cortex. 2012;22:158–165. doi: 10.1093/cercor/bhr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Miller K.L., Moeller S., Xu J., Auerbach E.J., Woolrich M.W., Beckmann C.F., Jenkinson M., Andersson J., Glasser M.F. Temporally-independent functional modes of spontaneous brain activity. Proc. Natl. Acad. Sci. U S A. 2012;109:3131–3136. doi: 10.1073/pnas.1121329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoormaker V.I., Schroter M.S., Gleiser P.M., Andrade K.C., Dresler M., Wehrle R., Samann P.G., Czisch M. Development of a large-scale functional brain network during human non-rapid eye movement sleep. J. Neurosci. 2010;30:11379–11387. doi: 10.1523/JNEUROSCI.2015-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoormaker V.I., Gleiser P.M., Czisch M. Frontoparietal connectivity and hierarchical structure of the brain’s functional network during sleep. Front. Neurol. 2012;3:1–10. doi: 10.3389/fneur.2012.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevner A.B.A., Vidaurre D., Cabral J., Rapuano K., Nielsen S.F.V., Tagliazucchi E., Laufs H., Vuust P., Deco G., Woolrich M.W. Discovery of key whole-brain transitions and dynamics during human wakefulness and non-REM sleep. Nat. Commun. 2019;10:1035. doi: 10.1038/s41467-019-08934-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoodley C.J., Schmahmann J.D. Evidence for topographic organization in the cerebellum of motor control versus cognitive and affective processing. Cortex. 2010;46:831–844. doi: 10.1016/j.cortex.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E., Laufs H. Decoding wakefulness levels from typical fMRI resting-state data reveals reliable drifts between wakefulness and sleep. Neuron. 2014;82:695–708. doi: 10.1016/j.neuron.2014.03.020. [DOI] [PubMed] [Google Scholar]
- Tagliazucchi E., von Wegner F., Morzelewski A., Brodbeck V., Laufs H. Dynamic BOLD functional connectivity in humans and its electrophysiological correlates. Front. Hum. Neurosci. 2012;6:1–22. doi: 10.3389/fnhum.2012.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E., von Wegner F., Morzelewski A., Brodbeck V., Jahnke K., Laufs H. Breakdown of long-range temporal dependence in default mode and attention networks during deep sleep. Proc. Natl. Acad. Sci. U S A. 2013;110:15419–15424. doi: 10.1073/pnas.1312848110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E., Crossley N., Bullmore E.T., Laufs H. Deep sleep divides the cortex into opposite modes of anatomical–functional coupling. Brain Struct. Funct. 2016;221:4221–4234. doi: 10.1007/s00429-015-1162-0. [DOI] [PubMed] [Google Scholar]
- Tiemeier H., Lenroot R.K., Greenstein D.K., Tran L., Pierson R., Giedd J.N. Cerebellum development during childhood and adolescence: a longitudinal morphometric MRI study. Neuroimage. 2010;49:63–70. doi: 10.1016/j.neuroimage.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G. An information integration theory of consciousness an information integration theory of consciousness. BMC Neurosci. 2004;5:42–64. doi: 10.1186/1471-2202-5-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G. Consciousness as integrated information: a provisional manifesto. Biol. Bull. 2008;215:216–242. doi: 10.2307/25470707. [DOI] [PubMed] [Google Scholar]
- Tononi G., Koch C. Consciousness: here, there and everywhere? Philos. Trans. R. Soc. B Biol. Sci. 2015;370:20140167. doi: 10.1098/rstb.2014.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G., Boly M., Massimini M., Koch C. Integrated information theory: from consciousness to its physical substrate. Nat. Rev. Neurosci. 2016;17:450–461. doi: 10.1038/nrn.2016.44. [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N., Mazoyer B., Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Vidaurre D., Smith S.M., Woolrich M.W. Brain network dynamics are hierarchically organized in time. Proc. Natl. Acad. Sci. U S A. 2017;114:12827–12832. doi: 10.1073/pnas.1705120114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöller D., Sandini C., Karahanoğlu F.I., Padula M.C., Schaer M., Eliez S., Van De Ville D. Large-scale brain network dynamics provide a measure of psychosis and anxiety in 22q11.2 deletion syndrome. Biol. Psychiatry Cogn. Neurosci. Neuroimaging. 2019;4:881–892. doi: 10.1016/j.bpsc.2019.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Percentiles indicate the fraction of voxels belonging to a network or region that has a z-score > 1.5. Only networks and regions that have at least 20% surviving percentile are included in the list. iCAP 6 is not included due to the excluded cerebellum in Greicius atlas. Specific regions that are not part of the cerebellum but are co-activated with iCAP 10 (anterior cerebellum) are included.
Data Availability Statement
The datasets supporting the current study have not been deposited in a public repository but are available from the corresponding author on request. The code of the TA and iCAPs pipeline are available in a public repository (https://c4science.ch/source/iCAPs/).