Summary
Soluble ligand-bound Mn(III) can support anaerobic microbial respiration in diverse aquatic environments. Thus far, Mn(III) reduction has only been associated with certain Gammaproteobacteria. Here, we characterized microbial communities enriched from Mn-replete sediments of Lake Matano, Indonesia. Our results provide the first evidence for the biological reduction of soluble Mn(III) outside the Gammaproteobacteria. Metagenome assembly and binning revealed a novel betaproteobacterium, which we designate ‘Candidatus Dechloromonas occultata.’ This organism dominated the enrichment and expressed a porin-cytochrome c complex typically associated with iron-oxidizing Betaproteobacteria and a novel cytochrome c-rich protein cluster (Occ), including an undecaheme putatively involved in extracellular electron transfer. This occ gene cluster was also detected in diverse aquatic bacteria, including uncultivated Betaproteobacteria from the deep subsurface. These observations provide new insight into the taxonomic and functional diversity of microbially driven Mn(III) reduction in natural environments.
Introduction
Manganese(III) is a strong oxidant with a reduction potential close to molecular oxygen (Kostka et al., 1995). Mn(III) is short-lived and unstable, but its stability is greatly increased when bound to ligands (Luther III et al., 2015). Ligand-bound Mn(III) is often the most abundant dissolved Mn species in sediment pore waters (Madison et al., 2013; Oldham et al., 2019) and soils (Heintze and Mann, 1947), with the potential to facilitate one-electron redox reactions in a variety of biogeochemical cycles (Luther III et al., 2015). Microbes accelerate the oxidation and reduction of Mn by orders of magnitude compared with abiotic mechanisms (Hem, 1963; Diem and Stumm, 1984; Morgan, 2005; Tebo et al., 2005; Learman et al., 2011; Luther et al., 2018; Jung et al., 2020; Yu and Leadbetter 2020). Yet, despite clear evidence for the environmental importance of Mn(III), knowledge about microbial Mn(III) cycling pathways remains fragmented.
To date, only Shewanella spp. (Gammaproteobacteria) have been confirmed to respire soluble Mn(III) (Kostka et al., 1995; Szeinbaum et al., 2014). Shewanella respire Mn(III) using the Mtr pathway (Szeinbaum et al., 2017), a porin-cytochrome (PCC) conduit that transports electrons across the periplasm for extracellular respiration of Mn(III/IV), Fe(III) and other metals (Richardson et al., 2012; Shi et al., 2016). Many Fe(II)-oxidizing Betaproteobacteria also contain PCCs (MtoAB, generally lacking the C subunit), which are proposed to oxidize Fe(II) to Fe(III) by running the PCC in reverse (Emerson et al., 2013; Kato et al., 2015; He et al., 2017). In some metal-reducing Gammaproteobacteria and Deltaproteobacteria, extracellular undecaheme (11-heme) UndA is thought to play a key functional role in soluble Fe(III) reduction (Fredrickson et al., 2008; Shi et al., 2011; Smith et al., 2013; Yang et al., 2013). UndA’s crystal structure shows a surface-exposed heme surrounded by positive charges, which may bind negatively charged soluble iron chelates (Edwards et al., 2012). Environmental omics suggest that metal reduction by Betaproteobacteria may be widespread in the deep subsurface (Anantharaman et al., 2016; Hernsdorf et al., 2017). However, only a few Fe(III)-reducing Betaproteobacteria isolates have been characterized to date (Cummings et al., 1999; Finneran et al., 2003), and little is known about metal reduction pathways in Betaproteobacteria.
Manganese reduction coupled to methane (CH4) oxidation is a novel metabolism only recently discovered in cultures enriched in Archaea (Ettwig et al., 2016; Leu et al., 2020). Biological and geochemical evidence suggest that this metabolism may be found in a variety of environments (Beal et al., 2009; Crowe et al., 2011; Riedinger et al., 2014), including Fe-rich Lake Matano, Indonesia. In an attempt to explore whether CH4 can fuel microbial Mn(III) reduction in enrichments inoculated with sediments from Lake Matano, Indonesia, which has active and pronounced microbial Mn and CH4 cycles (Jones et al., 2011), we uncovered a novel betaproteobacterium as the most dominant and active member of our Mn(III)-reducing enrichment culture. Our results provide the first evidence for the biological reduction of soluble Mn(III) outside Gammaproteobacteria and provide evidence for a new biochemical pathway involved in extracellular electron transfer.
Results and discussion
Enrichment of Mn(III)-reducing populations
Lake Matano, Indonesia, is a permanently stratified ultra-oligotrophic lake (Crowe et al., 2008). Below its oxic surface waters, Lake Matano’s permanently anoxic and stratified waters are highly enriched in iron and manganese, and support the activity of Mn cycling organisms with organic carbon and CH4 as potential sources of electrons (Crowe et al., 2011; Jones et al., 2011; Kuntz et al., 2015; Sturm et al., 2019). We designed an enrichment strategy to select for microbes capable of anaerobic CH4 oxidation coupled to soluble Mn(III) reduction by incubating anoxic Lake Matano sediment communities with soluble Mn(III)-pyrophosphate as the electron acceptor (with 2% O2 in a subset of bottles), and CH4 as the sole electron donor and carbon source after pre-incubation to deplete endogenous organic carbon (see Supporting Information for enrichment details). Enrichment cultures were transferred into fresh media after Mn(III) was completely reduced to Mn(II), for a total of five transfers over 395 days. By the fourth transfer, cultures with CH4 headspace (with or without 2% O2) reduced ~80% of soluble Mn(III) compared with ~30% with N2 headspace (Fig. 1). 16S rRNA gene sequences were dominated by Betaproteobacteria (Rhodocyclales; 8%–35%) and Deltaproteobacteria (Desulfuromonadales; 13%–26%; Fig. S1). 13CH4 oxidation to 13CO2 was undetectable (Fig. S2).
Fig. 1.
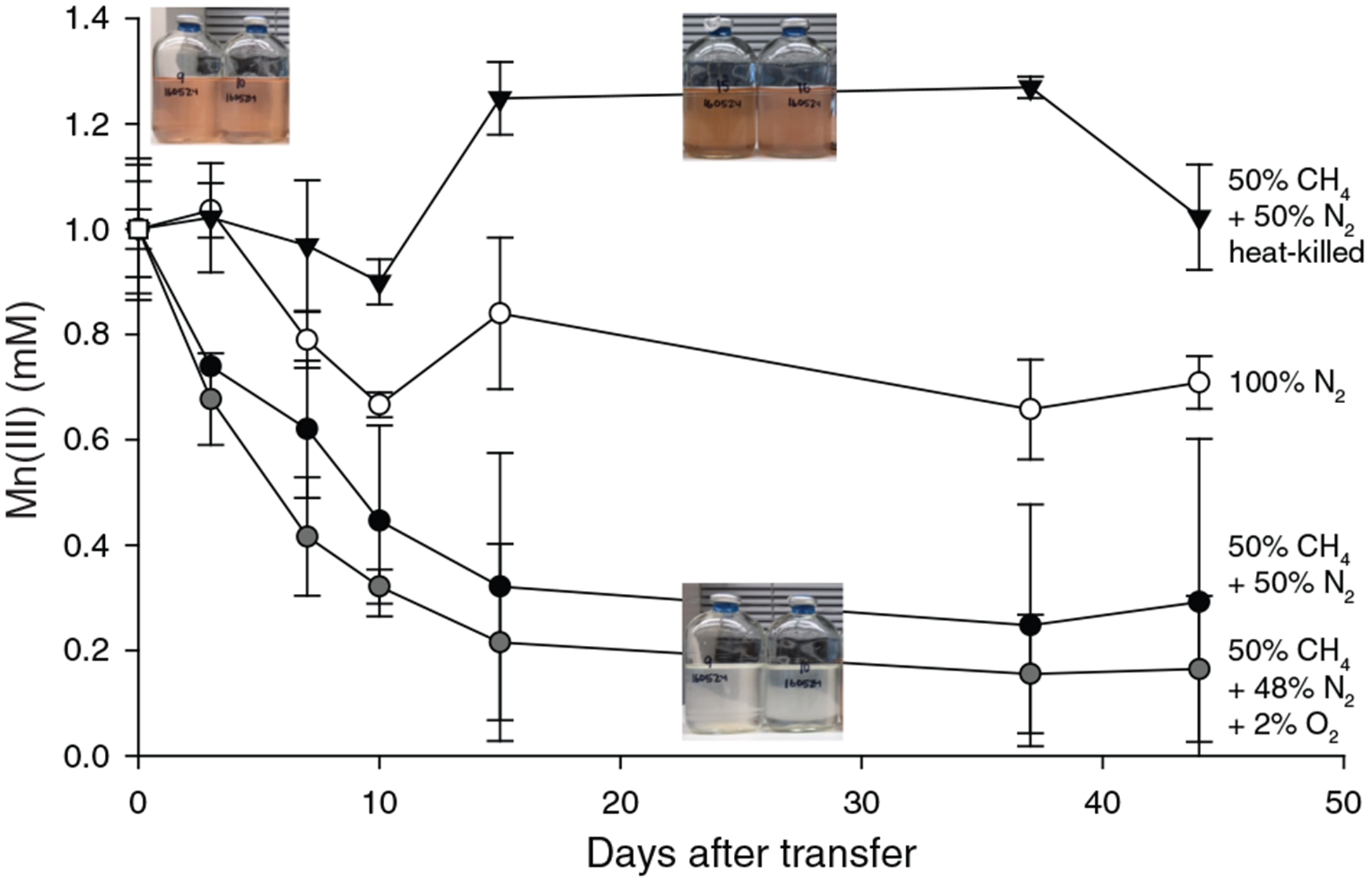
Consumption of Mn(III) in Lake Matano enrichments in the presence and absence of methane. Sediment-free cultures (transfer 4), from 335 days after the initial enrichment, were incubated for 45 days with 1 mM Mn(III) pyrophosphate as the sole electron acceptor. One set was incubated with Mn(III) and 2% O2. Initial bottle headspace contained 50% CH4 + 50% N2 (black circles), 50% CH4 + 48% N2 + 2% O2 (grey circles), 100% N2 (white circles) and 50% CH4 + 50% N2 heat-killed controls (black triangles). Error bars are standard deviations from duplicate experiments. Colour change from red to clear indicates Mn(III) reduction.
Samples for metagenomic and metaproteomic analysis were harvested from the fifth transfer (Fig. S1). Out of 2952 proteins identified in the proteome, 90% were assigned to Betaproteobacteria; of those, 72% mapped to a 99.5% complete metagenome-assembled genome (MAG; Rhodocyclales bacterium GT-UBC; NCBI accession QXPY01000000) with 81%–82% average nucleotide identity and phylogenetic affiliation to Dechloromonas spp. (Table S1; Fig. S3). This MAG is named here ‘Candidatus Dechloromonas occultata’ sp. nov.; etymology: occultata; (L. fem. adj. ‘hidden’). The remaining 10% of proteins mapped to Deltaproteobacteria; of those, 70% mapped to a nearly complete MAG (Desulfuromonadales bacterium GT-UBC; NCBI accession RHLS01000000) with 80% ANI to Geobacter sulfurreducens. This MAG is named here ‘Candidatus Geobacter occultata’.
Cytochrome expression during Mn(III) reduction
Cytochromes containing multiple c-type hemes are key for electron transport during microbial metal transformations, and therefore also expected to play a role in Mn(III) reduction. Numerous mono-, di-, and multi (>3)-heme cytochromes (MHCs) were expressed by ‘Ca. D. occultata’ in Mn(III)-reducing cultures. Nine out of 15 MHCs encoded by the ‘Ca. D. occultata’ MAG were expressed, including two decahemes similar to MtoA in Fe(II)-oxidizing Betaproteobacteria (Tables 1, Tables S2, S3; Fig. 2A, Fig. S4). Several highly expressed MHCs were encoded on a previously unreported 19-gene cluster with 10 cytochrome-c proteins, hereafter occA-S (Table 1; Fig. 2B, Figs S5 and S6). OccP was predicted to be an extracellular undecaheme protein of ~100 kDa (922 amino acids). ‘Ca. Dechloromonas occultata’ may reduce Mn(III) using the novel extracellular undecaheme OccP as the terminal Mn(III) reductase. Experimental verification of the function of the putative Occ complex is currently limited by the scarcity of genetically tractable Betaproteobacteria.
Table 1.
Expression levels for select ‘Ca. Dechloromonas occultata’ and ‘Ca. Geobacter occultata’ proteins in the presence of CH4 and N2.
| Protein sequence predictions | Normalized peptide abundance | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme complex/category | Function | Proteins | NCBI ID | Motifs | By treatment | Differential peptide | ||||||||
| SP | TMH | CxxCH | P-sort | CH4 | SD | N2 | SD | Avg | SD | p-value | ||||
| Ca. Dechloromonas occultata | ||||||||||||||
| Mto-1 | Outer membrane porin-cytochrome c electron conduit | MtoX-1 (cyt-b) | RIX49676 | N | 5 | 0 | IM | |||||||
| MtoY-1 (MCP) | RIX49677 | N | 2 | 1 | IM | 2.7 | 0.5 | 3.6 | 0.2 | 0.8 | 0.2 | 0.2 | ||
| MtoB-1 (porin) | RIX49678 | Y | 0 | 0 | OM | 10 | 2 | 15 | 2 | 0.6 | 0.1 | 0.004 | ||
| MtoA-1 | RIX49874 | Y | 1 | 10 | P | 5 | 1 | 2.5 | 0.1 | 1.9 | 0.4 | 0.1 | ||
| MtoD-1 | RIX49875 | N | 0 | 1 | P | |||||||||
| Mto-2 | Outer membrane porin-cytochrome c electron conduit | MtoX-2 (cyt-b) | RIX48942 | N | 4 | 0 | IM | |||||||
| MtoB-2 (porin) | RIX48943 | Y | 0 | 0 | OM | 8 | 1 | 16 | 0.2 | 0.5 | 0.1 | 0.04 | ||
| MtoA-2 | RIX48944 | Y | 1 | 10 | P | 7.3 | 0.8 | 4 | 2 | 2.1 | 1.3 | 0.2 | ||
| MtoD-2 | RIX48945 | Y | 1 | 1 | U | 2.6 | 0.3 | 0.7 | 0.3 | 4.0 | 1.4 | 0.003 | ||
| Occ | Membrane-spanning electron transport cytochromes | OccA | RIX49688 | Y | 1 | 3 | P | 4 | 0.5 | 0.7 | 0.6 | 7.8 | 5.7 | 0.01 |
| OccB | RIX49689 | Y | 0 | 3 | U | 41 | 4 | 19 | 2 | 2.2 | 0.0 | 0.03 | ||
| OccC | RIX49877 | N | 0 | 1 | U | |||||||||
| OccD | RIX49878 | N | 0 | 3 | U | |||||||||
| OccE (6-NHL) | RIX49690 | N | 1 | 0 | U | 22 | 2.1 | 20.5 | 0.2 | 1.1 | 0.1 | 0.2 | ||
| OccF | RIX49691 | Y | 2 | 4 | E | 13 | 0.7 | 10.1 | 0.1 | 1.3 | 0.1 | 0.06 | ||
| OccG (PPIase) | RIX49692 | N | 0 | 0 | U | 14 | 1 | 3.3 | 0.5 | 4.2 | 0.3 | 0.01 | ||
| OccH | RIX49693 | N | 0 | 0 | OM/E | 6.0 | 0.2 | 7.7 | 0.6 | 0.8 | 0.1 | 0.10 | ||
| Occl | RIX49694 | N | 1 | 3 | U | 7 | 2.5 | 2.3 | 0.0 | 2.9 | 1.1 | 0.1 | ||
| OccJ | RIX49879 | Y | 0 | 4 | U | 44 | 0.2 | 19 | 3 | 2.4 | 0.4 | 0.03 | ||
| OccK | RIX49880 | N | 0 | 0 | C | 39 | 6 | 13 | 1 | 3.0 | 0.2 | 0.04 | ||
| OccL | RIX49695 | N | 1 | 3 | U | |||||||||
| OccM | RIX49881 | N | 0 | 3 | U | |||||||||
| OccN (6 NHL) | RIX49696 | N | 2 | 0 | U | 5.7 | 0.3 | 6 | 1 | 0.9 | 0.1 | 0.2 | ||
| OccO (6 NHL) | RIX49882 | N | 0 | 0 | U | 1.2 | 0.8 | 4.2 | 0.4 | 0.3 | 0.2 | 0.03 | ||
| OccP | RIX49697 | N | 0 | 11 | E | 14 | 2 | 12 | 3 | 1.2 | 0.5 | 0.4 | ||
| OccQ | RIX49698 | Y | 4 | 0 | IM | |||||||||
| OccR | RIX49883 | N | 8 | 0 | IM | |||||||||
| OccS | RIX49699 | N | 12 | 0 | IM | |||||||||
| Cyt c | Mono- and di-heme c-type cytochromes involved in electron transfer | Cyt c5 | RIX47670 | N | 1 | 1 | U | 27 | 2 | 9 | 3 | 3.2 | 0.8 | 0.01 |
| Cyt c5 | RIX40984 | Y | 1 | 2 | P | 19 | 2 | 6 | 1 | 3.3 | 1.0 | 0.06 | ||
| Cyt c’/C_2 | RIX44710 | Y | 1 | 1 | P | 17 | 5 | 3.6 | 0.8 | 4.8 | 2.3 | 0.09 | ||
| Cyt c’/C_2 | RIX49630 | Y | 1 | 1 | P | 7 | 1 | 1.2 | 0.9 | 8.2 | 6.6 | 0.07 | ||
| Cyt C551/C552 | RIX49087 | Y | 0 | 1 | P | 13 | 3 | 2.8 | 0.0 | 4.8 | 1.1 | 0.06 | ||
| Cyt c4 | RIX48804 | Y | 0 | 2 | P | 16 | 0.8 | 9.8 | 0.8 | 1.6 | 0.2 | 0.06 | ||
| Cyt c4 | RIX44782 | Y | 0 | 2 | P | 4 | 2 | 1.7 | 0.7 | 2.6 | 0.1 | 0.08 | ||
| Cyt c4 | RIX45018 | Y | 0 | 2 | P | 7 | 0.6 | 2.2 | 0.2 | 3.0 | 0.0 | 0.02 | ||
| Nap | Periplasmic nitrate reductase | NapA | RIX41011 | Y | 0 | 0 | P | 76 | 2 | 67 | 3 | 1.1 | 0.1 | 0.1 |
| NapB | RIX41010 | Y | 1 | 2 | P | 15 | 1 | 5 | 2 | 3.2 | 0.9 | 0.02 | ||
| NapC | RIX41009 | N | 1 | 4 | IM | 12 | 3 | 13 | 1 | 1.0 | 0.2 | 0.1 | ||
| Nir | Nitrite reductase | NirS | RIX44719 | Y | 0 | 1 | P | 58 | 2 | 44 | 4 | 1.3 | 0.2 | 0.1 |
| NirB | RIX44720 | Y | 1 | 2 | P | 14 | 3 | 10 | 2 | 1.5 | 0.6 | 0.2 | ||
| NirC | RIX44788 | N | 0 | 1 | P | |||||||||
| NirF | RIX44721 | Y | 1 | 0 | P or C | 2 | 1 | 7 | 1 | 0.3 | 0.1 | 0.02 | ||
| Nor | Nitric oxide reductase | NorC | RIX45182 | N | 1 | 1 | IM | 3.5 | 0.7 | 3.2 | 0.7 | 1.1 | 0.0 | 0.1 |
| NorB | RIX45183 | N | 12 | 1 | IM | |||||||||
| cNos | Type II nitrous oxide reductase | cNosZ | RIX42539 | Y | 0 | 0 | P | 77 | 17 | 66 | 8 | 1.2 | 0.3 | 0.2 |
| cNosCI | RIX42538 | Y | 1 | 1 | P | 16 | 2 | 4 | 2 | 4.9 | 3.3 | 0.08 | ||
| cNosC2 | RIX42537 | Y | 1 | 2 | P | 10 | 0.1 | 3.9 | 0.3 | 2.6 | 0.1 | 0.02 | ||
| cNosB | RIX42536 | N | 6 | 0 | IM | |||||||||
| cNosD | RIX42535 | N | 0 | 0 | P | |||||||||
| cNosG | RIX42534 | N | 1 | 0 | C | |||||||||
| cNosH | RIX42533 | N | 4 | 0 | IM | |||||||||
| Qcr | Menaquinolcytochrome c reductase complex | QcrA | RIX41976 | N | 9 | 0 | CM | |||||||
| QcrB | RIX41977 | N | 9 | 0 | CM | |||||||||
| QcrC | RIX41978 | N | 1 | 0 | CM | |||||||||
| Proteases | Serine protease | RIX49468 | N | 0 | 0 | P | 27 | 2 | 1.0 | 0.3 | 29.0 | 9.9 | 0.02 | |
| Carboxyl-terminal protease (S41) | RIX48818 | N | 1 | 0 | CM | 18.5 | 0.8 | 8.0 | 0.9 | 2.3 | 0.1 | 0.0002 | ||
| Membrane/ Extracellular | DUF4214 protein | RIX44180 | N | 0 | 0 | OM/E | 146 | 25 | 43 | 0.6 | 3.4 | 0.5 | 0.05 | |
| S-layer protein | RIX44181 | N | 0 | 0 | U | 8 | 0.5 | 10 | 0.6 | 0.8 | 0.1 | 0.14 | ||
| PEP-CTERM sorting protein | RIX45463 | Y | 1 | 0 | E | 68 | 6 | 33 | 10 | 2.1 | 0.5 | 0.03 | ||
| Tol-Pal system protein | TolB | RIX44015 | Y | 0 | 0 | P | 20 | 2 | 12 | 1 | 1.7 | 0.0 | 0.03 | |
| Peptidoglycan-associated lipoprotein | Pal | RIX44016 | N | 0 | 0 | OM | 27.3 | 0.2 | 10 | 3 | 2.7 | 0.7 | 0.04 | |
| Tol-Pal system protein | YbgF | RIX44017 | Y | 0 | 0 | U | 10.8 | 0.4 | 4 | 2 | 3.7 | 2.2 | 0.06 | |
| Pilus assembly protein | RIX46961 | N | 0 | 0 | U | 54 | 5 | 30 | 5 | 1.8 | 0.1 | 0.001 | ||
| Other | Ethanol/methanol dehydrogenase | RIX45050 | Y | 0 | 0 | P | 37 | 4 | 17 | 1 | 2.2 | 0.1 | 0.03 | |
| Alcohol dehydrogenase | RIX45053 | Y | 0 | 0 | P | 12.4 | 1.4 | 14.2 | 1.7 | 0.9 | 0.0 | 0.04 | ||
| Aldehyde dehydrogenase | RIX45061 | Y | 0 | 0 | P | 125 | 31 | 221 | 75 | 0.6 | 0.1 | 0.10 | ||
| Phasin family granuleassociated protein | RIX40682 | N | 0 | 0 | U | 49 | 2 | 22 | 1 | 2.2 | 0.2 | 0.03 | ||
| Phasin family granuleassociated protein | RIX40683 | Y | 0 | 0 | U | 34 | 4 | 16 | 1 | 2.1 | 0.0 | 0.03 | ||
| High potential ironsulfur protein | RIX49681 | Y | 0 | 0 | U | 10.79 | 0.01 | 6.5 | 0.4 | 1.7 | 0.1 | 0.02 | ||
| Electron transfer flavoprotein | FixA | RIX43544 | N | 0 | 0 | C | 16 | 3 | 10 | 2 | 1.7 | 0.0 | 0.04 | |
| NAD-reducing hydrogenase | HoxH | RIX46736 | N | 0 | 0 | C | 22.9 | 0.7 | 37 | 3 | 0.6 | 0.1 | 0.07 | |
| Ca. Geobacter occultata | ||||||||||||||
| Hydrogenase | [Ni/Fe] hydrogenase, group 1, small subunit | HyaA | RNC64339 | Y | 0 | 0 | P | 11.1 | 0.4 | 3 | 1 | 5 | 2 | 0.06 |
| [Ni/Fe] hydrogenase, group 1, large subunit | HyaB | RNC64340 | N | 0 | 0 | P | 32 | 0 | 11 | 5 | 3 | 1 | 0.01 | |
| E-pilus | Type IV pilin | PilA | RNC67631 | N | 1 | 0 | E | 93 | 3 | 18 | 3 | 5.6 | 0.6 | 0.02 |
Grey boxes indicate membrane proteins. SP: signal peptide (Y: present/N: absent); TMH: numbers of transmembrane helices; CxxCH: number of heme-binding motifs; P-sort: predicted cellular location based on Psortb v.3.0. MCP: methyl-accepting chemotaxis protein; PPIase: Peptidyl-proline isomerase; P: periplasm, C: cytoplasm; OM: outer membrane; IM: inner membrane, E: extracellular; U: unknown. MtoX and MtoY were predicted to be an inner membrane cytochrome-b protein and a methyl-accepting chemotaxis protein respectively. Membrane proteins may be under-represented by mass spectrometry-based metaproteomic analyses, which inherently favour soluble over insoluble membrane-bound or hydrophobic proteins. Bold proteins indicate proteins that were significantly more expressed with CH4 than N2 (CH4/N2 > 1; p < 0.05). p values indicate significance of abundance difference between CH4 and N2 treatments.
Fig. 2.
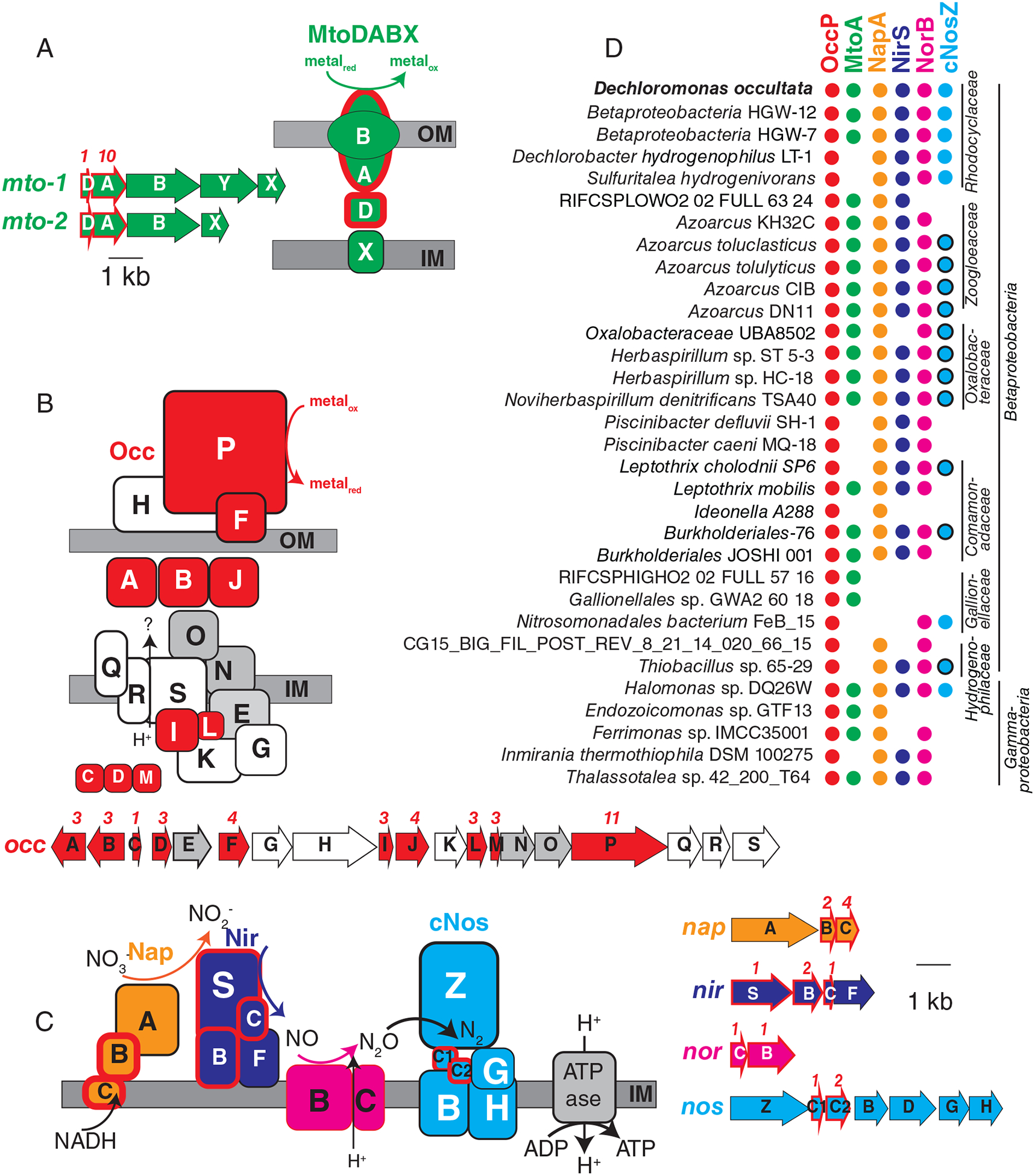
Gene arrangement, predicted protein location and taxonomic distribution of major expressed respiratory complexes in ‘Ca. D. occultata’. A: MtoDAB(Y)X porin-cytochrome c electron conduit; B: OccA-S; C: denitrification complexes (Nap, Nir, Nor and cNos); D: Occurrence of key marker genes in Betaproteobacteria and Gammaproteobacteria with >95% complete genomes that encode OccP. Protein sequences from ‘Ca. D. occultata’ were used as a query against a genome database and searched using PSI BLAST. Matches with identities >40%, query coverage >80% and E values <10−5 were considered positive. Red fill around genes and proteins indicate cytochrome-c proteins. Black outlines around blue circles in D indicate type I nitrous oxide reductase to distinguish from blue dots (type II/cytochrome-nitrous oxide reductase). Grey-shaded genes on the occ gene cluster indicate 6-NHL repeat proteins. Protein locations shown are based on P-sort predictions. Numbers above genes indicate the number of CxxCH motifs predicted to bind cytochrome c. IM: inner membrane; OM: outer membrane. For more details, see Table 1 and Table S3.
Proteins with 40%–60% identity to the expressed ‘Ca. D. occultata’ OccP protein were widely distributed in Betaproteobacteria from diverse freshwaters and deep subsurface groundwaters, as well as in several Gammaproteobacteria and one alphaproteobacterium (Fig. 2D; Table S3). Most occP-containing bacteria also possessed mtoA and denitrification genes (Fig. 2D; Fig. S7). These results widen the phylogenetic diversity of candidate extracellular MHCs that may be involved in microbial Mn(III) reduction.
Heme-copper oxidases in ‘Ca. D. occultata’
‘Ca. D. occultata’ expressed high-affinity cbb3-type cytochrome c oxidase (CcoNOQP) associated with micro-aerobic respiration (Table S4). Features of the ‘Ca. D. occultata’ occS gene product, including conserved histidine residues (H-94, H-411 and H-413) that bind hemes a and a3, as well as the H-276 residue that binds CuB (Fig. S6), suggest that OccS may function similarly to CcoN, the terminal heme-copper oxidase proton pump in aerobic respiration. All identified OccS amino acid sequences lack CuB ligands Y-280 and H-403, and most lack CuB ligands H-325 and H-326. OccS sequences also lack polar and ionizable amino acids that comprise the well-studied D and K channels involved in proton translocation in characterized cytochrome c oxidases (Blomberg and Siegbahn, 2014), but contain conserved H, C, E, D and Y residues that may serve in alternate proton translocation pathways, similar to those recently discovered in qNOR (Gonska et al., 2018). OccS homologues were also found in Azoarcus spp. and deep subsurface Betaproteobacteria (Fig. S6).
Expression of denitrification proteins and possible sources of oxidized nitrogen species
Periplasmic nitrate reductase (NapA), cytochrome nitrite reductase (NirS) and type II atypical nitrous oxide reductase (cNosZ; Fig. S7) were highly expressed by ‘Ca. D. occultata’ (Table 1). Expression of the denitrification pathway was not expected because oxidized nitrogen species were not added to the medium, to which the only nitrogen supplied was 0.2 mM NH4Cl (along with headspace N2). Oxidized nitrogen species could result from the oxidation of NH4Cl, but we did not find any of the canonical genes for aerobic nor anaerobic ammonia oxidation, nor did we measure any ammonium oxidation in experimental bottles from the transfer used to make Fig. 1.
The expression of denitrification genes is controlled by a diverse array of transcriptional regulators that depend on different signals including low levels of oxygen, even in the absence of nitrate (Spiro, 2012; Lin et al., 2018). The close redox potential of Mn(III)-pyrophosphate (~0.8 V; Yamaguchi and Sawyer, 1985) to oxidized nitrogen species (0.35–0.75 V) at circumneutral pH and the lack of oxygen in the media could have induced the expression of denitrification genes simultaneously with Mn(III)-reduction genes. Gammaproteobacteria, for example, reduce Mn(III) even in the presence of nitrate (Kostka et al., 1995), and there is precedent for microbial use of multiple electron acceptors, e.g. ‘co-respiration’ of oxygen and nitrate during aerobic denitrification (Chen and Strous, 2013; Ji et al., 2015).
Because solid-phase Mn(III) is known to chemically oxidize NH4+ (Aigle et al., 2017; Boumaiza et al., 2018), we tested for abiotic NH4+ oxidation by soluble Mn(III) (1 mM). Ammonium concentrations remained unchanged, and no N2O or NOx− production was observed (Fig. S8), likely because our experiments lacked solid surfaces to mediate electron transfer. Similarly, N2O levels in the headspace of our experimental bottles with Mn(III)-reducing cultures were near or below the detection limit (data not shown). These findings are consistent with the lack of detectable ammonium oxidation by Mn(III) pyrophosphate in estuarine sediments (Crowe et al., 2012).
Electron donors
Methane was the only electron donor added intentionally to the enrichment cultures, to select for organisms that oxidize methane anaerobically. Yet, we did not detect 13CO2 after the addition of 13CH4 (Fig. S2). One explanation is that 13CO2 was produced, but was subsequently assimilated by other members of the microbial community such as abundant Deltaproteobacteria (Fig. S1), as observed in previous studies (Wegener et al., 2008). A filtration step included in our protocol to measure 13CO2 would have excluded 13C-enriched biomass from our analyses. Alternatively, we considered other electron donors that might have been unintentionally present in trace amounts, but sufficiently abundant to drive the observed ~300–600 μM Mn(III) reduction (Fig. 1). The ethanol catabolism pathway (PQQ-dependent methanol/ethanol dehydrogenase (RIX45050), quinoprotein alcohol dehydrogenase (RIX45053) and an NAD+-dependent aldehyde dehydrogenase-II (RIX45061)) were all highly expressed in ‘Ca. D. occultata’ (Table 1). Ethanol could have been introduced to the bottles during culture preparation during sterilization of bottle stoppers. Based on the stoichiometry of ethanol oxidation coupled to Mn(III) reduction:
One-hundred and fifty micromolar ethanol would be required to reduce 600 μM of Mn(III), which equates to ~1 μl of 70% ethanol (12 M) into 100 ml culture medium. We conclude that trace contamination of ethanol was likely the major electron donor to our cultures.
It is also possible that other substrates, such as H2 from fermentation by other microbes in the enrichment or from impurities in the headspace gas, could have supplied another source of electrons. Indeed, an NAD-reducing hydrogenase (RIX44099–100) was expressed by ‘Ca. D. occultata’ (Table 1). Based on the stoichiometry of H2 oxidation coupled to Mn(III) reduction:
Six hundred micromolar H2 would be required to reduce 600 μM of Mn(III). Thus, H2 may have contributed electrons to Mn(III) reduction but is not likely sole electron donor. A combination of ethanol, H2 and other trace contaminants would likely have been necessary to provide enough electrons for the additional reduction of Mn(III) observed in the 13CH4-amended cultures compared with the controls lacking 13CH4. There is precedent for other metal-reducers simultaneously using H2 and an organic electron donor (Brown et al., 2005).
Another trace source of organics to our cultures could have been leaching from the rubber stoppers, which were black bromobutyl and pre-boiled in 0.1 N NaOH. A previous study reported that organics leaked an array of nalkanes (C16–C34) and unidentified organic contaminants in black bromobutyl stoppers (Niemann et al., 2015). It is also conceivable that trace organic was introduced as impurities in solid Mn(III) oxide powder (99% purity) used to synthesize Mn(III)-pyrophosphate.
Finally, we considered the possibility that 0.2 mM NH4+, added to the cultures as a nitrogen source, could have provided the electron donor, via an unknown pathway. Based on the stoichiometry of NH4+ oxidation coupled to Mn(III) reduction:
About 0.2 mM of NH4+ would supply 1.6 mM electron equivalents, which is more than enough to account for the observed reduction of 600 μM of Mn(III). This process could operate cryptically if the oxidized products were reduced to N2 via denitrification enzymes, such as nitrous oxide reductase (cNosZ), which was one of the most abundant proteins expressed in Mn(III)-reducing cultures (Fig. 2c, Table 1).
Carbon metabolism
‘Ca. D. occultata’ appeared to be growing mixotrophically. ‘Ca. D. occultata’ encoded several central metabolic pathways, including a complete TCA cycle with a glyoxylate bypass, an incomplete (acetate-dependent) 3-hydroxypropionate bicycle, a modified Calvin-Benson-Bassham (CBB) pathway and a pathway for synthesis of polyhydroxybutyrate (Fig. S9). In addition, ‘Ca. D. occultata’ encoded genes for organic carbon transport, and lactate, acetate, and propionate utilization (Fig. S10). Like D. agitata and D. denitrificans, the CBB pathway of ‘Ca. D. occultata’ did not encode RuBisCO and sedoheptulose-1,7-bisphosphatase (SHbisPase; Fig. S10); SHbisPase may be replaced by 6-phosphofructokinase and an energy-generating pyrophosphatase (RIX41248; Kleiner et al., 2012; Zorz et al., 2018). The presence of incomplete carbon fixation pathways and organic carbon utilization pathways suggests that ‘Ca. D. occultata’ relies on organic carbon to fix inorganic carbon mixotrophically. The source of this organic carbon could have been ethanol, which is converted to acetate via the pathway discussed in the previous section.
Effect of methane
Although we did not measure appreciable 13CH4 oxidation to 13CO2, CH4 stimulated Mn(III) reduction and cytochrome expression in ‘Ca. D. occultata’ enrichment cultures. While the specific role of CH4 in Mn(III) reduction remains unknown, the addition of CH4 appeared to significantly stimulate expression of many cytochrome c proteins, including OccABGJK, MtoD-2 and cytochrome-c4 and -c5 proteins associated with anaerobic respiration (p < 0.05; Table 1-). Expression of several ‘Ca. D. occultata’ proteins involved in outer membrane structure and composition—including an extracellular DUF4214 protein located next to an S-layer protein similar to those involved in manganese binding and deposition (Wang et al., 2009), a serine protease possibly involved in Fe(III) particle attachment (Burns et al., 2009), an extracellular PEP-CTERM sorting protein for protein export (Haft et al., 2006) and a Tol-Pal system for outer membrane integrity—was higher in the presence of CH4 (Table 1).
Transporters and sensors
Numerous transporters were present in the ‘Ca. D. occultata’ genome, including 26 TonB-dependent side-rophore transporters, 13 TRAP transporters for dicarboxylate transport, as well as ABC transporters for branched-chained amino acids and dipeptides and polypeptides (Table S4). ‘Ca. D. occultata’ also contained a large number of environmental sensing genes: 52 bacterial haemoglobins with PAS-PAC sensors, eight TonB-dependent receptors and eight NO responsive regulators (Dnr: Crp/fr family; Table S4). Uniquely in ‘Ca. D. occultata’, PAC-PAS sensors flanked accessory genes nosFLY on the c-nosZ operon (Fig. S7). Comparison of these flanking PAC-PAS sensors in ‘Ca. D. occultata’ with O2-binding sensors revealed that an arginine ~20 aa upstream from the conserved histidine as the distal pocket ligand for O2-binding is not present in either sensor (Fig. S11), suggesting that the sensor may bind a different ligand, possibly NO, consistent with the placement of these genes next to cNosZ (Shimizu et al., 2015).
Nutrient storage
Active synthesis of storage polymers suggested that ‘Ca. D. occultata’ was experiencing electron acceptor starvation at the time of harvesting, consistent with Mn(III) depletion in the bottles (Liu et al., 2015; Guanghuan et al., 2018). Polyphosphate-related proteins, including phosphate transporters, polyphosphate kinase, polyphosphatase and poly-3-hydroxybutyrate synthesis machinery were detected in the proteome (Table S4). Polyphosphate-accumulating organisms store poly-phosphates with energy generated from organic carbon oxidation during aerobic respiration or denitrification. These stored compounds are later hydrolyzed when respiratory electron acceptors for ATP production are limiting. Cyanophycin was actively synthesized for nitrogen storage.
Geobacter
‘Ca. Geobacter occultata’ expressed proteins in the TCA cycle at moderate abundance. ‘Ca. G. occultata’ contained 17 multiheme c-type cytochromes, none of which were detected in the proteome. The lack of expression of electron transport and metal-reducing pathways makes it unlikely that ‘Ca. G. occultata’ was solely responsible for Mn(III) reduction observed in the incubations. A periplasmic group I Ni-Fe hydrogenase (RNC64340; 91% identity to a protein (RLB64899) from Geobacter MAG from terrestrial hot spring sediment) and a type IV pilin (RNC67631; 10% aromatics, 87% identity to Geobacter pickeringii (Holmes et al., 2016)) were significantly more expressed in the presence of CH4 than N2 in the ‘Ca. G. occultata’ proteome (p < 0.05; Table 1). It is possible that ‘Ca. G. occultata’ transferred electrons to ‘Ca. D. occultata’ via e-pilins (e.g. direct interspecies electron transfer), contributing to the higher rates of Mn(III) reduction in the presence of CH4 vs. N2. The possible involvement of Geobacter e-pilins in Mn(III) reduction remains an open question, due to the lack of studies examining the possibility of Mn(III) reduction in Deltaproteobacteria.
Conclusions
To our knowledge, this study provides the first evidence for the biological reduction of soluble Mn(III) by a bacterium outside of the Gammaproteobacteria class. The dominant bacterium in Mn(III)-reducing enrichment cultures was ‘Ca. D. occultata’, a member of the Rhodocyclales order of Betaproteobacteria. ‘Ca. D. occultata’ expressed decahemes similar to the Mto pathway, and occ genes, including a novel extracellular undecaheme (OccP), which are predicted to encode a new respiratory electron transport pathway. The novel occ operon was found to be widespread in Betaproteobacteria from the deep subsurface, where metal cycling can fuel microbial metabolism. We also found highly expressed peptides from various central metabolic cycles and organic substrate utilization pathways, suggesting that ‘Ca. D. occultata’ may have been using multiple pathways simultaneously for energy generation and carbon assimilation during Mn(III) reduction.
Puzzles remain about whether ‘Ca. D. occultata’ can transform two potent greenhouse gases: methane and nitrous oxide. Although ‘Ca. D. occultata’ was enriched with CH4 as the sole electron donor and cultures reduced Mn(III) more rapidly in the presence of CH4, no CH4 oxidation activity was measured in Mn(III)-reducing cultures, and proteomic data suggested that ‘Ca. D. occultata’ was growing mixotrophically rather than assimilating CH4. Furthermore, although we did not add oxidized nitrogen compounds to our media, and Mn(III) did not chemically oxidize NH4+ under our culture conditions, type II nitrous oxide reductase (cNosZ) was one of the most abundant proteins expressed in Mn(III)-reducing cultures. The role of cNosZ and other denitrification enzymes in ‘Ca. D. occultata’ metabolism, and their possible connection to Mn(III) reduction, remain to be investigated.
Supplementary Material
Acknowledgements
This research was funded by NASA Exobiology grant NNX14AJ87G. Support was also provided by a Center for Dark Energy Biosphere Investigations (NSF-CDEBI OCE-0939564) small research grant and supported by the NASA Astrobiology Institute (NNA15BB03A) and a NASA Astrobiology Postdoctoral Fellowship to N.S. S.A.C. was supported through NSERC CRC, CFI, and Discovery grants. We thank Marcus Bray, Andrew Burns, Caleb Easterly, Ellery Ingall, Pratik Jagtap, Cory Padilla, Angela Peña, Johnny Striepen, Yael Toporek and Rowan Wolschleger for technical assistance. We thank Karen Lloyd, Nagissa Mahmoudi and Emily Weinert for helpful discussions.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site:
Appendix S1: Supporting Information
Table S3: Supporting information
Table S4: Supporting information
References
- Aigle A, Bonin P, Iobbi-Nivol C, Mejean V, and Michotey V (2017) Physiological and transcriptional approaches reveal connection between nitrogen and manganese cycles in Shewanella algae C6G3. Sci Rep 7: 44725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, et al. (2016) Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun 7: 13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal EJ, House CH, and Orphan VJ (2009) Manganese- and iron-dependent marine methane oxidation. Science 325: 184–187. [DOI] [PubMed] [Google Scholar]
- Blomberg MR, and Siegbahn PE (2014) Proton pumping in cytochrome c oxidase: energetic requirements and the role of two proton channels. Biochim Biophys Acta 1837: 1165–1177. [DOI] [PubMed] [Google Scholar]
- Boumaiza H, Coustel R, Despas C, Ruby C, and Bergaoui L (2018) Interaction of ammonium with birnessite: evidence of a chemical and structural transformation in alkaline aqueous medium. J Solid State Chem 258: 543–550. [Google Scholar]
- Brown DG, Komlos J, and Jaffé PR (2005) Simultaneous utilization of acetate and hydrogen by Geobacter sulfurreducens and implications for use of hydrogen as an indicator of redox conditions. Environ Sci Technol 39: 3069–3076. [DOI] [PubMed] [Google Scholar]
- Burns JL, Ginn BR, Bates DJ, Dublin SN, Taylor JV, Apkarian RP, et al. (2009) Outer membrane-associated serine protease involved in adhesion of Shewanella oneidensis to Fe(III) oxides. Environ Sci Technol 44: 68–73. [DOI] [PubMed] [Google Scholar]
- Chen J, and Strous M (2013) Denitrification and aerobic respiration, hybrid electron transport chains and co-evolution. Biochimica et Biophysica Acta (BBA)-Bioenergetics 1827: 136–144. [DOI] [PubMed] [Google Scholar]
- Crowe SA, Canfield DE, Mucci A, Sundby B, and Maranger R (2012) Anammox, denitrification and fixed-nitrogen removal in sediments from the lower St. Lawrence Estuary. Biogeosciences 9: 4309–4321. [Google Scholar]
- Crowe SA, Katsev S, Leslie K, Sturm A, Magen C, Nomosatryo S, et al. (2011) The methane cycle in ferruginous Lake Matano. Geobiology 9: 61–78. [DOI] [PubMed] [Google Scholar]
- Crowe SA, O’Neill AH, Katsev S, Hehanussa P, Haffner GD, Sundby B, et al. (2008) The biogeochemistry of tropical lakes: a case study from Lake Matano, Indonesia. Limnol Oceanogr 53: 319–331. [Google Scholar]
- Cummings DE, Caccavo F, Spring S, and Rosenzweig RF (1999) Ferribacterium limneticum, gen. nov., sp. nov., an Fe(III)-reducing microorganism isolated from mining-impacted freshwater lake sediments. Arch Microbiol 171: 183–188. [Google Scholar]
- Diem D, and Stumm W (1984) Is dissolved Mn2+ being oxidized by O2 in absence of Mn-bacteria or surface catalysts? Geochim Cosmochim Acta 48: 1571–1573. [Google Scholar]
- Edwards MJ, Hall A, Shi L, Fredrickson JK, Zachara JM, Butt JN, et al. (2012) The crystal structure of the extracellular 11-heme cytochrome UndA reveals a conserved 10-heme motif and defined binding site for soluble iron chelates. Structure 20: 1275–1284. [DOI] [PubMed] [Google Scholar]
- Emerson D, Field EK, Chertkov O, Davenport KW, Goodwin L, Munk C, et al. (2013) Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front Microbiol 4: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig KF, Zhu B, Speth D, Keltjens JT, Jetten MSM, and Kartal B (2016) Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc Natl Acad Sci U S A 113: 12792–12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finneran KT, Johnsen CV, and Lovley DR (2003) Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe(III). Int J Syst Evol Microbiol 53: 669–673. [DOI] [PubMed] [Google Scholar]
- Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, et al. (2008) Towards environmental systems biology of Shewanella. Nat Rev Microbiol 6: 592–603. [DOI] [PubMed] [Google Scholar]
- Gonska N, Young D, Yuki R, Okamoto T, Hisano T, Antonyuk S, et al. (2018) Characterization of the quinol-dependent nitric oxide reductase from the pathogen Neisseria meningitidis, an electrogenic enzyme. Sci Rep 8: 3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guanghuan G, Jianqiang Z, Aixia C, Bo H, Ying C, Kun G, et al. (2018) Nitrogen removal and nitrous oxide emission in an anaerobic/oxic/anoxic sequencing biofilm batch reactor. Environ Eng Sci 35: 19–26. [Google Scholar]
- Haft DH, Paulsen IT, Ward N, and Selengut JD (2006) Exopolysaccharide-associated protein sorting in environmental organisms: the PEP-CTERM/EpsH system. Application of a novel phylogenetic profiling heuristic. BMC Biol 4: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Barco RA, Emerson D, and Roden EE (2017) Comparative genomic analysis of neutrophilic iron(II) oxidizer genomes for candidate genes in extracellular electron transfer. Front Microbiol 8: 1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintze S, and Mann P (1947) Soluble complexes of manganic manganese. J Agric Sci 37: 23–26. [Google Scholar]
- Hem JD (1963) Chemical equilibria and rates of manganese oxidation, Washington DC: US Government Printing Office. [Google Scholar]
- Hernsdorf AW, Amano Y, Miyakawa K, Ise K, Suzuki Y, Anantharaman K, et al. (2017) Potential for microbial H2 and metal transformations associated with novel bacteria and archaea in deep terrestrial subsurface sediments. ISME J 11: 1915–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DE, Dang Y, Walker DJ, and Lovley DR (2016) The electrically conductive pili of Geobacter species are a recently evolved feature for extracellular electron transfer. Microb Genom 2: e000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji B, Yang K, Zhu L, Jiang Y, Wang H, Zhou J, and Zhang H (2015) Aerobic denitrification: a review of important advances of the last 30 years. Biotechnology and Bioprocess Engineering 20: 643–651. [Google Scholar]
- Jones C, Crowe SA, Sturm A, Leslie KL, MacLean LCW, Katsev S, et al. (2011) Biogeochemistry of manganese in ferruginous Lake Matano, Indonesia. Biogeosciences 8: 2977–2991. [Google Scholar]
- Jung H, Taillefert M, Sun J, Wang Q, Borkiewicz OJ, Liu P, et al. (2020) Redox cycling driven transformation of layered manganese oxides to tunnel structures. J Am Chem Soc 142: 2506–2513. [DOI] [PubMed] [Google Scholar]
- Kato S, Ohkuma M, Powell DH, Krepski ST, Oshima K, Hattori M, et al. (2015) Comparative genomic insights into ecophysiology of neutrophilic, micro-aerophilic iron oxidizing bacteria. Front Microbiol 6: 1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner M, Wentrup C, Lott C, Teeling H, Wetzel S, Young J, et al. (2012) Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use. Proc Natl Acad Sci U S A 109: 1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostka JE, Luther GW, and Nealson KH (1995) Chemical and biological reduction of Mn(III)-pyrophosphate complexes - potential importance of dissolved Mn(III) as an environmental oxidant. Geochim Cosmochim Acta 59: 885–894. [Google Scholar]
- Kuntz LB, Laakso TA, Schrag DP, and Crowe SA (2015) Modeling the carbon cycle in Lake Matano. Geobiology 13: 454–461. [DOI] [PubMed] [Google Scholar]
- Learman D, Wankel S, Webb S, Martinez N, Madden A, and Hansel C (2011) Coupled biotic–abiotic Mn (II) oxidation pathway mediates the formation and structural evolution of biogenic Mn oxides. Geochem Cosmochim Acta 75: 6048–6063. [Google Scholar]
- Leu AO, Cai C, McIlroy SJ, Southam G, Orphan VJ, Yuan Z, et al. (2020) Anaerobic methane oxidation coupled to manganese reduction by members of the Methanoperedenaceae. ISME J 14: 1030–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y-C, Sekedat MD, Cornell WC, Silva GM, Okegbe C, Price-Whelan A, et al. (2018) Phenazines regulate nap-dependent denitrification in Pseudomonas aeruginosa biofilms. J Bacteriol 200: e00031–e00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Peng L, Guo J, Chen X, Yuan Z, and Ni B-J (2015) Evaluating the role of microbial internal storage turnover on nitrous oxide accumulation during denitrification. Sci Rep 5: 15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther GW, Thibault de Chanvalon A, Oldham VE, Estes ER, Tebo BM, and Madison AS (2018) Reduction of manganese oxides: thermodynamic, kinetic and mechanistic considerations for one- versus two-electron transfer steps. Aquatic Geochem 24: 257–277. [Google Scholar]
- Luther GW III, Madison AS, Mucci A, Sundby B, and Oldham VE (2015) A kinetic approach to assess the strengths of ligands bound to soluble Mn (III). Mar Chem 173: 93–99. [Google Scholar]
- Madison AS, Tebo BM, Mucci A, Sundby B, and Luther GW 3rd. (2013) Abundant porewater Mn(III) is a major component of the sedimentary redox system. Science 341: 875–878. [DOI] [PubMed] [Google Scholar]
- Morgan JJ (2005) Kinetics of reaction between O2 and Mn (II) species in aqueous solutions. Geochim Cosmochim Acta 69: 35–48. [Google Scholar]
- Niemann H, Steinle L, Blees J, Bussmann I, Treude T, Krause S, et al. (2015) Toxic effects of lab-grade butyl rubber stoppers on aerobic methane oxidation. Limnol Oceangr: Methods 13: 40–52. [Google Scholar]
- Oldham VE, Siebecker MG, Jones MR, Mucci A, Tebo BM, and Luther GW (2019) The speciation and mobility of Mn and Fe in estuarine sediments. Aquat Geochem 25: 3–26. [Google Scholar]
- Richardson DJ, Butt JN, Fredrickson JK, Zachara JM, Shi L, Edwards MJ, et al. (2012) The ‘porin-cytochrome’ model for microbe-to-mineral electron transfer. Mol Microbiol 85: 201–212. [DOI] [PubMed] [Google Scholar]
- Riedinger N, Formolo MJ, Lyons TW, Henkel S, Beck A, and Kasten S (2014) An inorganic geochemical argument for coupled anaerobic oxidation of methane and iron reduction in marine sediments. Geobiology 12: 172–181. [DOI] [PubMed] [Google Scholar]
- Shi L, Belchik SM, Wang Z, Kennedy DW, Dohnalkova AC, Marshall MJ, et al. (2011) Identification and characterization of UndAHRCR-6, an outer membrane endecaheme c-type cytochrome of Shewanella sp. strain HRCR-6. Appl Environ Microbiol 77: 5521–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Dong H, Reguera G, Beyenal H, Lu A, Liu J, et al. (2016) Extracellular electron transfer mechanisms between microorganisms and minerals. Nat Rev Microbiol 14: 651. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Huang D, Yan F, Stranava M, Bartosova M, Fojtíková V, and Martínková M.t. (2015) Gaseous O2, NO, and CO in signal transduction: structure and function relationships of heme-based gas sensors and heme-redox sensors. Chem Rev 115: 6491–6533. [DOI] [PubMed] [Google Scholar]
- Smith JA, Lovley DR, and Tremblay PL (2013) Outer cell surface components essential for Fe(III) oxide reduction by Geobacter metallireducens. Appl Environ Microbiol 79: 901–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro S (2012) Nitrous oxide production and consumption: regulation of gene expression by gas-sensitive transcription factors. Philos Trans R Soc Lond B Biol Sci 367: 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Fowle DA, Jones C, Leslie K, Nomosatryo S, Henny C, et al. (2019) Rates and pathways of CH4 oxidation in ferruginous Lake Matano, Indonesia. Geobiology 17: 294–307. [DOI] [PubMed] [Google Scholar]
- Szeinbaum N, Burns JL, and DiChristina TJ (2014) Electron transport and protein secretion pathways involved in Mn(III) reduction by Shewanella oneidensis. Environ Microbiol Rep 6: 490–500. [DOI] [PubMed] [Google Scholar]
- Szeinbaum N, Lin H, Brandes JA, Taillefert M, Glass JB, and DiChristina TJ (2017) Microbial manganese(III) reduction fuelled by anaerobic acetate oxidation. Environ Microbiol 19: 3475–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebo BM, Johnson HA, McCarthy JK, and Templeton AS (2005) Geomicrobiology of manganese(II) oxidation. Trends Microbiol 13: 421–428. [DOI] [PubMed] [Google Scholar]
- Wang X, Schröder HC, Schloßmacher U, and Müller WE (2009) Organized bacterial assemblies in manganese nodules: evidence for a role of S-layers in metal deposition. Geo-Mar Lett 29: 85–91. [Google Scholar]
- Wegener G, Niemann H, Elvert M, Hinrichs KU, and Boetius A (2008) Assimilation of methane and inorganic carbon by microbial communities mediating the anaerobic oxidation of methane. Environ Microbiol 10: 2287–2298. [DOI] [PubMed] [Google Scholar]
- Yamaguchi KS, and Sawyer DT (1985) The redox chemistry of manganese(III) and manganese(IV) complexes. Isr J Chem 25: 164–176. [Google Scholar]
- Yang Y, Chen J, Qiu D, and Zhou J (2013) Roles of UndA and MtrC of Shewanella putrefaciens W3-18-1 in iron reduction. BMC Microbiol 13: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, and Leadbetter JR (2020). Bacterial chemolithoautotrophy via manganese oxidation. Nature, 583, (7816), 453–458. 10.1038/s41586-020-2468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorz JK, Kozlowski JA, Stein LY, Strous M, and Kleiner M (2018) Comparative proteomics of three species of ammonia-oxidizing bacteria. Front Microbiol 9: 938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


