The first olfactory processing center in the larval Drosophila brain uses information about feeding state to shape behavior.
Abstract
Animals exhibit different behavioral responses to the same sensory cue depending on their internal state at a given moment. How and where in the brain are sensory inputs combined with state information to select an appropriate behavior? Here, we investigate how food deprivation affects olfactory behavior in Drosophila larvae. We find that certain odors repel well-fed animals but attract food-deprived animals and that feeding state flexibly alters neural processing in the first olfactory center, the antennal lobe. Hunger differentially modulates two output pathways required for opposing behavioral responses. Upon food deprivation, attraction-mediating uniglomerular projection neurons show elevated odor-evoked activity, whereas an aversion-mediating multiglomerular projection neuron receives odor-evoked inhibition. The switch between these two pathways is regulated by the lone serotonergic neuron in the antennal lobe, CSD. Our findings demonstrate how flexible behaviors can arise from state-dependent circuit dynamics in an early sensory processing center.
INTRODUCTION
Hunger influences decisions about food-related sensory cues in many animal species. Whereas well-fed individuals can afford to be selective, individuals facing starvation must consider any available source of nutrition (1–4). Because odors are commonly used to locate and identify food, olfactory responses can likewise vary with feeding state. In adult Drosophila, food deprivation has been shown to modulate the presynaptic excitability of certain olfactory receptor neurons (ORNs) (5, 6) and also the activity of dopaminergic input neurons in a higher brain region (7). In contrast, the fixed connectivity of the antennal lobe, the first olfactory processing center in the Drosophila brain, is thought to provide generic formatting of sensory inputs for use by various downstream circuits (8).
We asked whether the Drosophila larva responds differently to odors after food deprivation, and how its reduced nervous system might integrate internal state information to produce flexible innate behaviors. The larva has only 21 ORNs per hemisphere, each expressing a unique receptor type and innervating a distinct glomerulus in the larval antennal lobe (lAL). The complete wiring diagram of the lAL has been mapped by serial-section electron microscopy, providing a framework for understanding the algorithmic basis of larval olfaction (9). In addition to GABAergic broad local interneurons, well studied in the adult (10), the lAL connectome also contains glutamatergic picky local interneurons (pLNs), whose function is as yet unknown. Another notable but poorly understood feature of the lAL is the existence of two types of projection neurons targeting different brain areas. Uniglomerular projection neurons (uPNs) relay signals from individual ORNs to the mushroom body calyx for associative learning and to the lateral horn for innate olfactory processing (11). Multiglomerular projection neurons (mPNs) receive input from different subsets of ORNs and innervate a wide variety of brain regions (9). Similar mPNs also exist in the adult fly, but most of them project to the lateral horn (12, 13). The lAL also contains a single, prominent serotonergic neuron (CSD) that is common to the larvae and adults of many insect species (14, 15). This unexpected diversity of cell types hints at additional unknown computational functions.
We found that the larva’s behavioral response to an aversive odor depends on its feeding state. For example, geranyl acetate (GA), which is innately aversive to fed larvae, becomes attractive to food-deprived larvae. This drastic change in behavioral response arises within the lAL circuitry. We observed no significant modulation in the amplitude of odor-evoked ORN responses, in contrast to what is seen in the adult. However, the two projection neuron output pathways show opposite state-dependent changes in odor-evoked activity and promote opposite innate behavioral responses. We also found that food deprivation leads to an increase in odor-evoked CSD activity. Serotonergic signals from CSD directly excite the uPN pathway, which promotes odor attraction. Serotonin from CSD also recruits local glutamatergic inhibition and thereby indirectly suppresses the mPN pathway responsible for odor avoidance in the fed state. In summary, we reveal how state-dependent neuromodulation reconfigures early odor processing by shifting the excitatory-inhibitory balance between two separate output pathways, thus enabling opposite behavioral responses to exactly the same sensory input.
RESULTS
Feeding state determines the response to an odor
Food deprivation can alter food choice behaviors. A starving animal might approach food cues that a fed animal would avoid or ignore. To test this possibility in Drosophila larvae, we investigated olfactory behavior under different feeding states. Most monomolecular odorants are innately attractive to fed larvae (16, 17). We screened a panel of 21 odorants and found two, GA and menthol, that repel fed larvae but attract food-deprived larvae (Fig. 1, A and B, and fig. S1, A and B). Both GA and menthol occur naturally as antiherbivore compounds in green leaves (18, 19), which are not a preferred food source for fly larvae. We also tested the behavioral response to ethyl acetate (EA), a fruit odorant that is innately attractive to fed larvae (16). While we did not observe a change in the sign of the behavioral response, EA becomes significantly more attractive after food deprivation (fig. S1C).
Fig. 1. Feeding state determines odor responses by opposite modulation of lAL outputs.
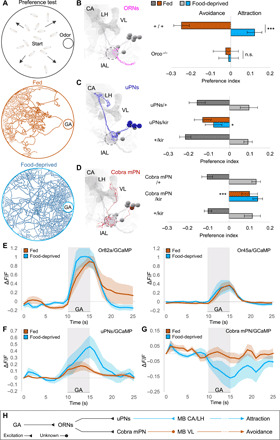
(A) For preference assay, 15 larvae are tracked in an odor gradient for 15 min. Trajectories demonstrate avoidance/attraction to GA by fed/food-deprived larvae. (B to D) Electron microscopy (EM) reconstructions (left) and behavioral phenotypes (right) for ORNs, uPNs, and cobra mPN [mushroom body calyx (CA), vertical lobe (VL), larval antennal lobe (lAL; circled), lateral horn (LH)]. (B) GA elicits avoidance in fed state (one-sample t test, P < 0.001) and attraction after food deprivation (P < 0.01). Behavior diverges between states (two-sample t test, ***P < 0.001). Anosmic larvae (Orco−/−) ignore GA [one-sample/two-sample t tests, not significant (n.s.); n = 12 to 18]. (C) Silencing uPNs (GH146-GAL4) specifically abolishes food-deprived attraction [one-way analysis of variance (ANOVA), *P < 0.05; n = 8 to 10]. (D) Silencing cobra mPN (GMR32E03-GAL4) specifically abolishes fed avoidance (one-way ANOVA, ***P < 0.001; n = 6 to 10). (E to G) Calcium responses to GA (10−6). (E) ORN-Or82a and ORN-Or45a exhibit similar responses in both states (n = 5 to 6). (F and G) After food deprivation, uPNs exhibit increased responses (n = 7 to 9), whereas cobra mPN exhibits more negative responses (n = 8 to 9). (H) Parallel lAL output pathways (uPNs and cobra mPN) support opposite behavioral responses and receive opposite state-dependent modulation.
Mutant larvae (Orco−/−) lacking functional ORNs showed no response to GA, menthol, or EA when either fed or food-deprived (Fig. 1B and fig. S1, B and C). The state-dependent change in the behavioral response to these odorants requires the olfactory system. As every odorant activates different subsets of ORNs (GA, 82a/45a; menthol, 49a/63a/1a; EA, 42b/42a) and other cells downstream (20), we focused our study on a single odorant, GA, to facilitate precise dissection of the mechanisms underlying the observed behavioral switch. The transition from GA avoidance to attraction after 5 to 7 hours of food deprivation is highly reproducible across different odorant concentrations and larval stages (fig. S2, A to C).
Feeding state modulates two lAL output pathways in opposite ways
The larval ORNs most sensitive to GA are those that express Or82a and Or45a (20). What are the downstream targets of these sensory neurons? Each ORN connects to one uPN that innervates the mushroom body calyx and lateral horn (Fig. 1C). The GH146-GAL4 line (11) drives expression in 19 of 21 uPNs (fig. S2E), including all uPNs activated by GA (20). From the wiring diagram of the lAL, we also identified an mPN called “cobra” that receives direct synaptic input from several ORNs including those activated by GA (Fig. 1D). Cobra mPN projects to the vertical lobe of the mushroom bodies (9). We also identified a driver line (GMR32E02-GAL4) specific for cobra mPN expression (fig. S2F).
To probe the involvement of uPNs and cobra mPN in the innate behavioral response to GA, we inactivated each pathway by expression of Kir2.1, an inwardly rectifying potassium channel (21). Inactivation of uPNs made food-deprived larvae unable to switch their behavioral responses from avoidance to attraction (Fig. 1C). In contrast, inactivation of cobra mPN caused fed larvae to be attracted to GA without affecting attraction in food-deprived larvae (Fig. 1D).
To determine how feeding state affects odor-evoked activity of GA-sensing ORNs, uPNs, and the cobra mPN, we next recorded calcium dynamics by imaging GCaMP, a genetically encoded calcium indicator, in intact, immobilized larvae (20). First, we quantified odor-evoked activity in axon terminals of ORN-Or82a or ORN-Or45a or in all ORNs (Fig. 1E; fig. S2D for statistics). We found no statistically significant change in the amplitude of odor-evoked ORN responses after food deprivation. In contrast, we found that food deprivation both increased odor-evoked uPN responses and inhibited cobra mPN (Fig. 1, F and G; fig. S2, E and F, for statistics).
These observations suggest that this state-dependent change in olfactory behavior might be computed within the antennal lobe circuit by changing how the same ORN input leads to different responses of the uPN and mPN output pathways (Fig. 1H). Higher uPN and lower mPN activity correlate with GA attraction. Lower uPN and higher mPN activity correlate with GA aversion.
The mPN pathway receives glutamatergic inhibition in food-deprived larvae
We sought synaptic mechanisms within the lAL that might up-regulate the uPN pathway and down-regulate the cobra mPN pathway. The glutamatergic pLNs are well positioned to down-regulate the mPN pathway because they preferentially synapse onto mPNs (9). Moreover, GluClα, a glutamate-gated chloride channel that makes glutamate inhibitory, is widely expressed in the adult antennal lobe (22).
When we reduced GluClα expression specifically in cobra mPN, both fed and food-deprived animals exhibited GA avoidance (Fig. 2A). Food deprivation decreased odor-evoked mPN responses in control larvae by a similar amount to what we observed in previous imaging experiments (Fig. 2B; compare Fig. 1G), up to slight differences in the baseline response most likely due to nonolfactory inputs not in our direct control. However, after GluClα knockdown, we found no significant change in the peak amplitude of GA-evoked calcium responses between fed and food-deprived animals (Fig. 2, B and C; fig. S3A for statistics). Removing glutamatergic inhibition of cobra mPN appears to prevent inactivation of the odor avoidance pathway after food deprivation, and odor response does not shift to attraction.
Fig. 2. Upon food deprivation, the mPN pathway receives glutamatergic inhibition from pLNs.
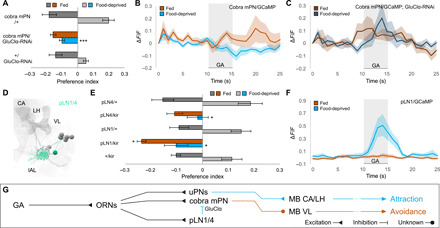
(A) RNA interference (RNAi) knockdown of GluClα receptor in cobra mPN specifically abolishes attraction in food-deprived state (Kruskal-Wallis test, ***P < 0.001; n = 4 to 8) without affecting avoidance in fed state (P > 0.05). (B and C) Calcium responses of cobra mPN to GA (10−6) in wild-type (light) and GluClα-RNAi larvae (dark). GluClα knockdown abolishes state-dependent inhibition of cobra mPN seen in wild-type larvae (n = 8). (D) EM reconstruction of picky local interneurons pLN1/4 [mushroom body calyx (CA), vertical lobe (VL), larval antennal lobe (lAL; circled), lateral horn (LH)]. (E) Silencing either pLN1 or pLN4 with UAS-Kir2.1 abolishes attraction in the food-deprived state (Kruskal-Wallis test, *P < 0.05; n = 8 to 10). Silencing pLN1 (but not pLN4) modestly increases avoidance in fed state (*P < 0.05). (F) pLN1 exhibits enhanced calcium responses to GA (10−6) in food-deprived state (n = 7). (G) pLNs provide glutamatergic inhibition onto cobra mPN via the GluClα receptor.
There are four glutamatergic pLNs that receive input from GA-sensitive ORNs and send output to the cobra mPN: pLN0, pLN1, pLN2, and pLN4 (9). The wiring diagram only contains one other pLN (pLN3), which shares synapses with neither the GA-sensitive ORNs nor cobra mPN. We identified three cell-specific split GAL4 lines for pLN1, pLN3, and pLN4 (Fig. 2D and fig. S3, B to G). Targeted inactivation of pLN1, pLN3, or pLN4 had no effect on GA avoidance in fed larvae, but inactivating either pLN1 or pLN4 eliminated GA attraction in food-deprived larvae (Fig. 2E and fig. S3H). With imaging, we found that GA-evoked calcium responses in pLN1 were elevated in the food-deprived state and undetectable in the fed state (Fig. 2F; fig. S3I for statistics). We conclude that food deprivation may down-regulate the cobra mPN pathway by increasing glutamatergic inhibition from pLN1 and pLN4 (Fig. 2G).
The uPN pathway receives serotonergic excitation through the 5-HT7 receptor
How might the activity of the uPN pathway be up-regulated in food-deprived animals? Serotonin can be a prominent neuromodulator, and the excitatory 5-HT7 is the name of the receptor (no expanded form) receptor is expressed in the uPN pathway (Fig. 3A and fig. S4A) (23, 24). We found that inactivating 5-HT7–expressing neurons did not impair GA avoidance in fed animals, consistent with previous observations (Fig. 3B) (23). However, inactivating 5-HT7–expressing neurons caused food-deprived larvae to avoid GA, similar to the phenotype obtained by inactivating all uPNs. We also specifically removed 5-HT7 from the uPN pathway using a CRISPR-Cas9–based cell type–specific gene knockout system (25, 26). Without 5-HT7 in the uPNs, food-deprived larvae also avoided GA (Fig. 3C). With imaging, we found that odor-evoked calcium activity in the uPNs lacking 5-HT7 was weak in both food-deprived and fed animals and did not increase after food deprivation (Fig. 3, D and E; fig. S4B for statistics). We conclude that serotonin is required to elevate odor-evoked uPN activity and to switch the behavioral response to attraction in food-deprived animals.
Fig. 3. Upon food deprivation, CSD activates uPNs via the excitatory 5-HT7 receptor.
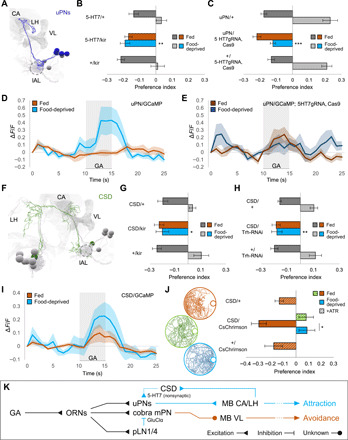
(A) EM reconstruction of uPNs [mushroom body calyx (CA), vertical lobe (VL), larval antennal lobe (lAL; circled), lateral horn (LH)]. (B) Silencing 5-HT7–expressing neurons specifically abolishes food-deprived attraction (one-way ANOVA, **P < 0.01; n = 8). (C) CRISPR knockout of 5-HT7 in uPNs specifically abolishes food-deprived attraction (one-way ANOVA, ***P < 0.001; n = 8 to 14). (D and E) uPN calcium responses to GA (10−6) in wild-type (light) and uPN-specific 5-HT7 knockout larvae (dark). 5-HT7 knockout abolishes state-dependent enhancement of uPN responses seen in wild-type (n = 6 to 7). (F) EM reconstruction of CSD neuron [neuropil annotations as in (A)]. (G) Silencing CSD (R60F02-GAL4) specifically abolishes food-deprived attraction (Kruskal-Wallis test, *P < 0.05; n = 6 to 8). (H) Disrupting 5-HT synthesis in CSD via Trh knockdown specifically abolishes food-deprived attraction (Kruskal-Wallis test, **P < 0.01; n = 8). (I) CSD exhibits increased calcium responses to GA (10−8) following food deprivation (n = 8 to 10). (J) Optogenetic activation of CSD in fed larvae reproduces the state-dependent behavioral switch (one-way ANOVA, *P < 0.05; n = 12 to 20). (K) Upon food deprivation, elevated 5-HT from CSD excites uPNs via 5-HT7 receptor, promoting behavioral attraction.
Serotonin from CSD induces a state-dependent switch in odor response
A single serotonergic neuron in each hemisphere called CSD widely innervates higher brain areas as well as the antennal lobes in both the larva and adult fly (Fig. 3F and fig. S4C) (15, 27–29). We found that cell-specific inactivation of CSD using GMR60F02-GAL4 (but not inactivation of other serotonergic neurons; fig. S4D) causes GA avoidance in both fed and food-deprived larvae (Fig. 3G). This phenotype was also obtained by cell-specific knockdown of serotonin synthesis in CSD and exhibited by serotonin-synthesis mutants (Fig. 3H and fig. S4E).
CSD receives only a few synapses from ORNs but substantial indirect olfactory input from neurons in the lateral horn that integrate signals from uPNs (9). We measured GA-evoked calcium dynamics in CSD and found an increase in food-deprived animals compared to fed animals (Fig. 3I; fig. S4F for statistics). To determine whether elevated CSD activity is linked to a change in behavior, we used optogenetics. We expressed the red-shifted optogenetic effector CsChrimson in CSD and tested behavior. Fed larvae that normally show GA avoidance will tend to exhibit attraction when CSD is artificially activated by continuous illumination with red light (Fig. 3J and fig. S5A). We note that CSD activation does not affect basal locomotion patterns (e.g., lengths of crawling movements) in the same way as food deprivation (fig. S5B). CSD activation changes olfactory responses, but not the ability to crawl toward an olfactory cue.
Inhibition of the serotonin transporter SerT, which localizes in presynaptic membranes and recycles released neurotransmitter, is a method for increasing serotonergic neurotransmission (30). RNA interference (RNAi) knockdown of SerT in CSD reduced GA avoidance in fed larvae (fig. S4G). In contrast, overexpression of SerT (which presumably lowers serotonergic transmission) causes GA avoidance in both fed and food-deprived larvae (fig. S4G). These phenotypes are consistent with optogenetic activation of CSD and constitutive inactivation of CSD, respectively. These phenotypes also argue against the possibility that state-dependent changes in receptor expression occur in the projection neurons themselves.
Notably, the wiring diagram reveals no direct synapses from CSD to the uPNs (Fig. 4, A and B). One possibility is that 5-HT7 receptors in the uPNs might be activated by extrasynaptic release or synaptic spillover after food deprivation (Fig. 3K) (31, 32).
Fig. 4. A simple dynamical model recapitulates state-dependent changes in odor valence.
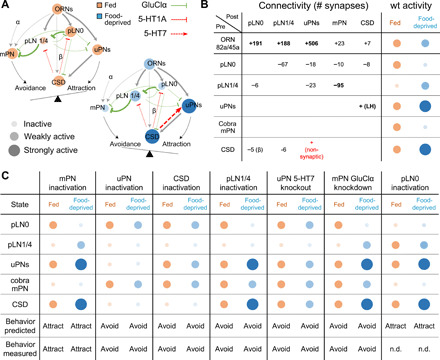
(A) Connectivity and state-dependent dynamics of lAL model. Each node represents a cell type; node size and intensity represent the predicted activity in fed (orange) and food-deprived (blue) states. Edges terminating in arrowheads and bars represent excitatory and inhibitory connections, respectively; edge thickness indicates connection strength. (B) EM-derived synapse counts between all modeled cell types and model predictions for neuronal activity in the fed (orange) and food-deprived (blue) states. Assignment of excitatory (+) or inhibitory (−) sign to each connection is based on experimental data. The positive connection from CSD to uPNs models the effect of putatively nonsynaptic 5-HT release. We also assume excitatory feedback from uPNs to CSD via the lateral horn (LH). Connections with ≥25 synapses are assigned weight 1, and connections with <25 synapses are assigned weight w < 1. The weak connection from CSD to pLN0 is assigned weight β. wt, wild type. (C) Model predictions for activity of each cell type and overall behavioral output in the fed (orange) and food-deprived (blue) states under simulated perturbations. All model predictions are corroborated by experimental results except for the case of pLN0 inactivation (n.d., no data).
Food deprivation modulates inhibitory interactions in the pLN circuit
Our results suggest that a pLN circuit regulates mPN activity by providing glutamatergic inhibition in the food-deprived state. But what inactivates pLN1/4 in the fed state? The wiring diagram suggests that pLNs inhibit one another in a hierarchical manner: pLN1/4 receive strong glutamatergic inhibition from pLN0 (Fig. 4, A and B) (9). We were not able to study pLN0 due to the lack of a cell-specific driver. However, we found that reducing GluClα expression in pLN1 and pLN4 eliminates GA avoidance in the fed state (fig. S6, A and B). Thus, pLN1/4 receive glutamatergic inhibition, presumably from pLN0, only when larvae are fed.
In addition, the local interneurons of the lAL express an inhibitory serotonergic receptor, 5-HT1A (fig. S6C) (23). CRISPR-mediated knockout of the 5-HT1A receptor from pLN1/4 similarly abolished GA avoidance in the fed state (fig. S6D). According to the wiring diagram, pLN1/4 receive direct synaptic inputs from CSD that seem to provide enough serotonin in the fed state to achieve this inhibition (Fig. 4B). Thus, in the fed state, pLN1/4 receive joint inhibition from glutamatergic and serotonergic signals, thereby lowering their ability to inhibit the mPN pathway for odor avoidance (fig. S6E).
In the food-deprived state, knockdown of GluClα in pLN1/4 does not affect odor attraction. This suggests that they receive less glutamatergic inhibition (presumably from pLN0) and are thus able to down-regulate the mPN pathway (fig. S6, A and B). Both glutamatergic and serotonergic inputs seem to be required for inhibition of pLN1/4 and the resulting disinhibition of the mPN pathway, as increased serotonergic inhibition from CSD in the food-deprived state appears to be insufficient to inactivate pLN1/4 (fig. S6D). In contrast, inhibition of pLN0 is likely independent of glutamate, as it lies exclusively upstream of the rest of the pLN network. Because all five pLNs originate from the same neuroblast lineage (9, 33), pLN0 is also likely to express the inhibitory 5-HT1A receptor and therefore be subject to inhibition from CSD under food deprivation.
A computational model of the state-dependent switch in olfactory response
To integrate our findings about synaptic properties and state-dependent neuromodulatory dynamics with the known circuit connectivity, we built a computational model encompassing all major cell types of the lAL network (Fig. 4, A and B). Having uncovered the sign of the relevant synaptic and nonsynaptic interactions, we set out to test whether the resulting dynamics would give rise to decision-making, namely, the appropriate changes in activity in the uPN and mPN output pathways. In our model, the weight of every synaptic connection is calculated from the number of synapses between cell types in the connectome (Fig. 4B) (9). The only plausible cellular input to pLN1/4 that provides glutamatergic inhibition is pLN0, which, like pLN1/4, is modeled as receiving direct serotonergic inhibition from CSD.
Our model, based on the established connectivity and empirically determined synaptic properties, recapitulates the observed changes in neuronal activity. Activation of CSD in the food-deprived state up-regulates the uPN pathway directly through serotonergic excitation and inhibits the mPN pathway indirectly through recruitment of glutamatergic pLNs (Fig. 4, A and B). In contrast, weak CSD activity in the fed state shifts activity toward the mPN pathway.
Our model also allows us to test the consequences of all the molecular and cellular perturbations that we performed to uncover circuit properties. We simulated the effects of removing individual neurons from the circuit (akin to chronic inactivation by Kir2.1) as well as individual synapses (akin to receptor knockdown or knockout) and found that, in every case, we could predict whether the circuit output would be shifted towards avoidance or attraction in either fed or food-deprived animals (Fig. 4C). In addition, our model predicts that silencing pLN0 should cause an attractive response to GA in both states (Fig. 4C). Although a suitable driver line is currently unavailable to directly test this prediction, our model parsimoniously reproduces all state-dependent shifts in lAL circuit dynamics that we have observed experimentally.
DISCUSSION
The insect antennal lobe, like the mammalian olfactory bulb, is typically regarded as a preprocessing stage whose basic function is to format inputs in a generic way for use by various downstream circuits (8). In contrast, we have demonstrated a crucial role for the lAL in actively implementing a state-dependent behavioral switch between avoidance and attraction to certain odors. In particular, we show that food deprivation reverses the larva’s innate aversion to GA. We also reproduce our key observations about GA using menthol, the only other odorant that we found to elicit consistent avoidance behavior in well-fed larvae (fig. S7, A to G). Both odorants are monoterpenoids, a class of volatile organic compounds that plants secrete to defend against herbivory (34). Nevertheless, it may be adaptive for larvae to suppress their default aversion to these potential toxins when faced with starvation.
We find that this behavioral choice is implemented by a state-dependent shift in the relative activity of two differentially projecting lAL output pathways. In the adult fly, both the uPNs and most mPNs project to the lateral horn, where their activity is integrated to generate innate behaviors (35, 36). In the larva, uPNs project to the mushroom body calyx and lateral horn and promote attractive behavior, while the previously uncharacterized cobra mPN projects to the mushroom body vertical lobe and promotes aversive behavior. By regulating the state-dependent switch between these pathways, the lAL contributes to the assignment of innate odor valence. Understanding how the uPNs and mPN organize locomotory behavior toward or away from odors will require mapping downstream pathways to the motor circuit (37, 38). However, we find that the key neuromodulatory switch between these output pathways is implemented within the lAL, which must therefore be regarded as a bona fide decision-making circuit.
In the adult fly, feeding state directly adjusts the sensitivity of ORNs via presynaptic modulation (5, 6). This is accomplished through regulation of the neuropeptide receptors sNPFR1 and DTKR in specific subsets of ORNs following a starvation-dependent decrease in insulin signaling. Other studies have also implicated monoamine neuromodulators such as octopamine and serotonin and neuropeptides such as NPF or SIFamide in hunger-gated modulation of various brain regions in adult flies [reviewed in (39)]. However, the fact that larvae and adults often exhibit completely different behavioral responses to the same odors suggests that the mechanisms that give rise to state-dependent behavior may differ between these two life stages.
Notably, we observe no statistically significant change in odor-evoked calcium responses at larval ORN axons following food deprivation. Instead, we find that feeding state modifies olfactory preferences by shifting the equilibrium between the uPN and mPN output pathways. Thus, in the Drosophila larva, food deprivation acts downstream of the ORN layer and by a substantially different circuit mechanism than what has been observed in the adult. Because of the broad similarities in the antennal lobe circuitry of the larva and adult, we speculate that the adult antennal lobe may also be a locus of state-dependent modulation. Recent findings in the adult fly suggest that neuromodulatory feedback also conveys internal-state information from the higher brain to early visual processing layers (40–42). Thus, the mechanism that we have uncovered may reflect a general principle in the organization of diverse sensory processing systems.
Serotonergic neuromodulation specifically has been implicated in the regulation of numerous state-dependent behaviors in both vertebrates and invertebrates (43–45). For instance, activation of a small set of serotonergic neurons in the central brain of a sated adult fly causes the animal to behave as though starved (46). We find that CSD, a prominent serotonergic neuron whose processes span the Drosophila olfactory system, controls the switch between attraction and aversion to GA and menthol in fly larvae. CSD exhibits greater odor-evoked activity in the food-deprived state than in the fed state. When an animal is food-deprived, high levels of serotonin released from CSD both activate the uPN pathway (via the excitatory 5-HT7 receptor) and increase glutamatergic inhibition onto the mPN pathway (via modulation of inhibitory glutamatergic local interneurons). As there are no direct synapses from CSD onto uPNs, serotonergic transmission likely involves either synaptic overspill or nonsynaptic neurotransmitter release (31). Serotonin from CSD has also been shown to excite uPNs in the adult fly (47). However, the adult CSD neuron exhibits more elaborate patterns of innervation (48) and compartmentalized odor-evoked responses. Moreover, each cell type in the adult antennal lobe expresses multiple types of serotonin receptors, suggesting the possibility for cross-talk (24). By exploiting the numerical simplicity of the lAL, we have elucidated a neuromodulatory mechanism that generates flexible behavior from a fixed wiring diagram.
The neural circuit for early olfactory processing in mammals, the olfactory bulb, is notably similar in its molecular and circuit architecture to the insect antennal lobe (49). Here, we have shown that CSD activity encodes information about feeding state that the lAL uses to select an appropriate behavioral response. In the mouse, serotonergic projection neurons from the raphe nucleus innervate the olfactory bulb and also modulate distinct local interneurons and output pathways (50, 51). The circuit logic and behavioral role of serotonergic signaling in mammalian olfaction is not well understood. However, homologous 5-HT receptors are known to regulate appetite and seeking/craving behaviors, suggesting a conserved function for serotonergic regulation of behaviors that depend on feeding state (45, 52, 53). By systematically analyzing the circuit-level effects of state-dependent neuromodulation in the lAL, our study suggests a potentially general mechanism by which internal state can modify early sensory processing to determine behavior.
MATERIALS AND METHODS
Fly husbandry
Flies were reared at 22°C under a 12-hour light/12-hour dark cycle and 60% relative humidity in vials containing standard cornmeal agar–based medium. For larval experiments, adult flies were transferred to larvae collection cages (Genesee Scientific) containing grape juice agar plates and 180 mg of fresh yeast paste per cage. Flies were allowed to lay eggs on the agar plate for 1 to 2 days before the plate was removed for collection of larvae in the different developmental stages. Behavioral experiments were performed with L2 larvae [3 to 4 days after egg laying (AEL)], unless otherwise stated (fig. S2A). Calcium imaging experiments and anatomical studies were performed in L1 larvae (2 days AEL). Most transgenic stocks were obtained from Bloomington Drosophila Stock Center (BDSC; see table S1).
Cell-specific CRISPR-Cas9–mediated knockout
We generated UAS-gRNA lines targeting the 5-HT7 and 5-HT1A receptors, as described previously (54). In short, we digested the pCFD6 vector (a gift from S. Bullock, Addgene #73915) with Bbs I [New England Biolabs (NEB)] and used a Gibson Assembly (NEB) to incorporate polymerase chain reaction (PCR) products. We generated two PCR fragments each harboring three guide sequences with homology to the CDS of either 5-HT7 or 5-HT1A (see table S2). The resulting colonies were sequenced, and the correct constructs were inserted into the attP1 landing site on chromosome II (BDSC #8621) by ΦC31-mediated recombination (Rainbow Transgenic Flies, Camarillo, CA, USA). Transgenic flies were backcrossed to w1118 and balanced using BDSC #3703 (25, 26).
Behavioral assays
Pure odorants were diluted in deionized (DI) water and stored for no more than 1 week. Our initial screen of behavioral responses in fed larvae (fig. S1A) involved an odorant panel of natural constituents of ripe fruit from B. Smith (55). All diluted odorants and odor solutions were stored in the dark in separate glass bottles to avoid photodegradation and contamination. For behavior experiments, the following odorants were obtained from Sigma-Aldrich and used at the indicated dilutions (v/v): GA, 10−4 (except see fig. S2B; CAS #105-87-3); menthol, 10−3 (CAS #89-78-1); and EA, 10−6 (CAS #141-78-6). The larval olfactory choice assay is illustrated in Fig. 1A. For food deprivation, larvae were picked from grape juice plates 5 to 7 hours before testing (except as otherwise stated; fig. S2C), washed in DI water, and placed in a small petri dish (VWR, #60872-306, 60 mm by 15 mm) filled with 2% agarose and covered by a thin layer of tap water. Both fed larvae (picked directly from grape juice plates) and food-deprived larvae were washed in DI water immediately before testing. At the start of the test, 15 larvae were placed in the center of a 10-cm petri dish filled with 2% agarose and equipped with a 12-mm plastic cup at one edge (VWR, #25384-318). Before each experiment, the cup was loaded with 200 μl of odorant solution. Larvae were free to explore the arena for 15 min; the lid was placed over the dish for the duration of testing. We tracked the number of larvae in each quadrant of the arena over time and computed a preference index
| (1) |
where Q+ denotes the number of larvae in the quadrant containing the odor cup, Q− is the number of larvae in the quadrant opposite the odor cup, and N is the total number of larvae in the arena.
Each odor preference test used freshly picked and previously untested larvae. Different larvae were tested under fed and food-deprived conditions. Experiments were performed at room temperature under uniform illumination with a point light source (desk lamp) on a clean bench, except for the optogenetic experiments shown in Fig. 3J (see below). In all experiments, we alternated the position of the odor cup in the petri dish between the left or right sides to avoid spatial bias.
Optogenetics
For optogenetic experiments with CsChrimson, experimental larvae additionally received food supplemented with 0.1 mM all-trans-retinal (Sigma-Aldrich, CAS #116-31-4). Larval collection cages and grape juices plates were wrapped in foil and kept in the dark during egg laying and larval development (56). Experiments were performed in a petri dish filled with 2% agarose and equipped with an odor cup (as above), placed inside a lightproof box. Olfactory response behavior was recorded at 4 Hz using a Mightex charge-coupled device (CCD) camera equipped with a long-pass infrared filter (cutoff, 740 nm). Larvae received 500-Hz pulsed stimulation with spatially uniform red light (625 nm; pulse width, 20 μs; 0.62 W/m2 ± 0.5%) to activate the optogenetic effector throughout the entire 15-min test period. Larval trajectories were analyzed with a custom tracking script (57). For each frame, the number of larvae in the two halves of the petri dish was enumerated automatically. The preference index was calculated as
| (2) |
where S+ denotes the number of larvae in the half of the plate containing the odor cup, S− is the number of larvae in the half opposite the odor cup, and N is the total number of larvae in the arena.
Functional imaging and microfluidics
We used a previously described method for microfluidic delivery of odorants in aqueous form with simultaneous imaging of calcium activity in intact larvae (20). All experiments used an eight-channel microfluidic chip equipped with a vacuum port to stabilize the animal’s head. The same odorants were used as in the behavioral experiments, but at lower concentrations (GA, 10−8 and 10−6; menthol, 10−4). Stimuli consisted of 5-s odor pulses interleaved with 15-s water washout periods. An L1 larva was washed with DI water and loaded into the microfluidic device using a 1-ml syringe filled with Triton X-100 [0.1% (v/v)] solution. The animal was pushed to the end of the loading channel, with its dorsal side facing the objective. GCaMP signal was recorded using an inverted Nikon Ti-E spinning disc confocal microscope and a 60× water immersion objective [numerical aperture (NA), 1.2]. A CCD microscope camera (Andor iXon EMCCD) captured frames at 30 Hz. The CSD neuron and projection neurons were recorded by scanning the entire volume (step size, 1.5 μm) of the brain, ranging from the antennal lobe to the processes in the higher brain. Orco::RFP (red fluorescent protein) was used to label the lAL. Recordings from at least five to nine larvae were collected for each genotype and condition. All samples were used for analysis unless dendritic varicosities developed in the ORNs over the course of the recording, indicating neuronal damage due to mechanical stress.
Anatomical studies
Green fluorescent protein (GFP) expression patterns of GAL4 and split-GAL4 lines were imaged in intact larvae using an inverted Nikon Ti-E spinning disc confocal microscope with a 60× water-immersion objective (NA, 1.2) or 20× air objective. Larvae were immobilized between two glass slides using a micromanipulator. Orco::RFP was used to label the lAL.
Computational circuit model
To better understand how the neural circuitry in the lAL dictates the state-dependent shift of odor valence, we describe a dynamical model based on connectomic data and the observed activity of larval neurons. We assume that the neural activities for pLNs and cobra mPN are binary (0 and 1), while uPNs and CSD have three states (0, 1, and 2). We denote the state of neuron i at discrete time t as si(t) ∈ {0,1,2}, where i = 1..5. For simplicity, we have treated pLN1 and pLN4 as a single unit (pLN1/4). The connection weights are binned across two levels: “strong” (with absolute strength 1) and “weak” (with absolute strength w), as shown in table S3. Experimental data indicate strong feedback from CSD to uPNs after food deprivation. Hence, we modeled this nonsynaptic interaction by setting the weight from CSD to uPNs to be 2, i.e., W5,3 = 2 under food deprivation. CSD inhibits pLN0 through serotonin, modeled by parameter β. Cobra mPN is assigned a basal (nonolfactory) input α. As shown in table S3, a neuron receives several inputs: from ORN, pLN, CSD, or baseline input. In vector notation, we have
| (3) |
where xin is the input vector from ORNs, x0 is the basal (nonolfactory) input vector, and W is the interaction matrix between pLNs, PNs, and CSD. The state of neuron i changes in response to its total input hi according to the following update rule
| (4) |
for i = 1, 2, and 4 (corresponding to pLN0, pLN1/4, and cobra mPN, respectively), or
| (5) |
for i = 3 and 5 (corresponding to uPNs and CSD, respectively).
To fit our model parameters, we used the steady-state activity from experiments in fed and food-deprived wild-type larvae, shown in table S4. These neural activity patterns are stable and thus impose a constraint on the model parameters
| (6a) |
| (6b) |
| (6c) |
Data from perturbation experiments additionally imply β < 1 − w. To simulate the circuit (Fig. 4C), we choose w = 0.25, α = 0.5, and β = 0.5. To relate neural activity to behavioral preference, we assume a binary readout
| (7) |
The exact value of the coefficient of smPN does not matter, but it must be greater than 2 to generate the observed avoidance behavior. This simple model predicts all behavioral results from perturbation experiments.
Statistical analysis
Imaging data were analyzed with custom MATLAB code, available from GitHub. Data from behavioral experiments were preprocessed in LabVIEW and analyzed with MATLAB and Microsoft Excel. Data were tested for normality (Shapiro-Wilk test) and analyzed by parametric or nonparametric statistics as appropriate: one-way analysis of variance (ANOVA) or the Kruskal-Wallis test. For post hoc pairwise comparisons, two-tailed one- or two-sample t tests or Mann-Whitney U tests were performed as appropriate. The significance level of hypothesis tests was set to α = 0.05. Only the most conservative statistical result of multiple pairwise comparisons is indicated. No statistical methods were used to determine sample sizes in advance, but sample sizes are similar to those reported in other studies in the field. Sample sizes (n), P values, and other relevant summary statistics are shown in the appropriate figure legends. Solid lines in raw calcium imaging data indicate mean normalized fluorescence change (ΔF/F), shaded regions: ±1 SEM. Bar graphs represent pooled data from 5 to 15 min during testing (mean ± SEM). Box and whisker plots show single data points, median (50th percentile), quartiles (25th/75th percentile), and range of data.
Supplementary Material
Acknowledgments
We thank members of the Samuel and de Bivort laboratories for helpful discussions; we also thank Y. Fisher, H. Yang, R. Wilson, M. Herre, V. Murthy, A. Bahl, D. Galili, B. Kottler, and K. Blum for their comments on the manuscript. Funding: K.V. was supported by a DFG research fellowship (project no. 345729665); S.Q. and C.P. were supported by a grant from the NIH (U01NS111697-01); A.D.T.S. was supported by grants from the NIH (R01 GM130842-01) and NSF (IOS-1555914). Author contributions: K.V. performed behavioral experiments, functional imaging, anatomical studies, and optogenetics; D.M.Z. performed functional imaging; M.S. and M.R. provided genetic reagents; K.M. performed behavioral experiments; L.H.-N. analyzed data from optogenetic experiments; S.Q. and C.P. designed the computational model; A.C. provided access to connectomic data; K.V. conceived the project; K.V. and A.D.T.S. designed the study, interpreted the results, and wrote the manuscript, with input from all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/1/eabd6900/DC1
REFERENCES AND NOTES
- 1.DiBattista D., Bedard M., Effects of food deprivation on hunger motivation in golden hamsters (Mesocricetus auratus). J. Comp. Psychol. 101, 183–189 (1987). [Google Scholar]
- 2.Wu Q., Zhao Z., Shen P., Regulation of aversion to noxious food by Drosophila neuropeptide Y– and insulin-like systems. Nat. Neurosci. 8, 1350–1355 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Inagaki H. K., Panse K. M., Anderson D. J., Independent, reciprocal neuromodulatory control of sweet and bitter taste sensitivity during starvation in Drosophila. Neuron 84, 806–820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crossley M., Staras K., Kemenes G., A central control circuit for encoding perceived food value. Sci. Adv. 4, eaau9180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Root C. M., Ko K. I., Jafari A., Wang J. W., Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell 145, 133–144 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ko K. I., Root C. M., Lindsay S. A., Zaninovich O. A., Shepherd A. K., Wasserman S. A., Kim S. M., Wang J. W., Starvation promotes concerted modulation of appetitive olfactory behavior via parallel neuromodulatory circuits. eLife 4, e08298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsao C.-H., Chen C.-C., Lin C.-H., Yang H.-Y., Lin S., Drosophila mushroom bodies integrate hunger and satiety signals to control innate food-seeking behavior. eLife 7, e35264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson R. I., Early olfactory processing in Drosophila: Mechanisms and principles. Annu. Rev. Neurosci. 36, 217–241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berck M. E., Khandelwal A., Claus L., Hernandez-Nunez L., Si G., Tabone C. J., Li F., Truman J. W., Fetter R. D., Louis M., Samuel A. D., Cardona A., The wiring diagram of a glomerular olfactory system. eLife 5, e14859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen S. R., Wilson R. I., Lateral presynaptic inhibition mediates gain control in an olfactory circuit. Nature 452, 956–960 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masuda-Nakagawa L. M., Gendre N., O’Kane C. J., Stocker R. F., Localized olfactory representation in mushroom bodies of Drosophila larvae. Proc. Natl. Acad. Sci. U.S.A. 106, 10314–10319 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka N. K., Endo K., Ito K., Organization of antennal lobe-associated neurons in adult Drosophila melanogaster brain. J. Comp. Neurol. 520, 4067–4130 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Bates A. S., Schlegel P., Roberts R. J. V., Drummond N., Tamimi I. F. M., Turnbull R., Zhao X., Marin E. C., Popovici P. D., Dhawan S., Jamasb A., Javier A., Li F., Rubin G. M., Waddell S., Bock D. D., Costa M., Jefferis G. S. X. E., Complete connectomic reconstruction of olfactory projection neurons in the fly brain. Curr. Biol. 30, 3183–3199.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Python F., Stocker R. F., Immunoreactivity against choline acetyltransferase, γ-aminobutyric acid, histamine, octopamine, and serotonin in the larval chemosensory system of Drosophila melanogaster. J. Comp. Neurol. 453, 157–167 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Dacks A. M., Christensen T. A., Hildebrand J. G., Phylogeny of a serotonin-immunoreactive neuron in the primary olfactory center of the insect brain. J. Comp. Neurol. 498, 727–746 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Fishilevich E., Domingos A. I., Asahina K., Naef F., Vosshall L. B., Louis M., Chemotaxis behavior mediated by single larval olfactory neurons in Drosophila. Curr. Biol. 15, 2086–2096 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Kreher S. A., Mathew D., Kim J., Carlson J. R., Translation of sensory input into behavioral output via an olfactory system. Neuron 59, 110–124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernasconi M. L., Turlings T. C. J., Ambrosetti L., Bassetti P., Dorn S., Herbivore-induced emissions of maize volatiles repel the corn leaf aphid, Rhopalosiphum maidis. Entomol. Exp. Appl. 87, 133–142 (1998). [Google Scholar]
- 19.Lin Y., Qasim M., Hussain M., Akutse K. S., Avery P. B., Dash C. K., Wang L., The herbivore-induced plant volatiles methyl salicylate and menthol positively affect growth and pathogenicity of entomopathogenic fungi. Sci. Rep. 7, 40494 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Si G., Kanwal J. K., Hu Y., Tabone C. J., Baron J., Berck M., Vignoud G., Samuel A. D. T., Structured odorant response patterns across a complete olfactory receptor neuron population. Neuron 101, 950–962 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baines R. A., Uhler J. P., Thompson A., Sweeney S. T., Bate M., Altered electrical properties in Drosophila neurons developing without synaptic transmission. J. Neurosci. 21, 1523–1531 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W. W., Wilson R. I., Glutamate is an inhibitory neurotransmitter in the Drosophila olfactory system. Proc. Natl. Acad. Sci. U.S.A. 110, 10294–10299 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huser A., Eschment M., Güllü N., Collins K. A. N., Böpple K., Pankevych L., Rolsing E., Thum A. S., Anatomy and behavioral function of serotonin receptors in Drosophila melanogaster larvae. PLOS ONE 12, e0181865 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sizemore T. R., Dacks A. M., Serotonergic modulation differentially targets distinct network elements within the antennal lobe of Drosophila melanogaster. Sci. Rep. 6, 37119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlichting M., Díaz M. M., Xin J., Rosbash M., Neuron-specific knockouts indicate the importance of network communication to Drosophila rhythmicity. eLife 8, e48301 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delventhal R., O’Connor R. M., Pantalia M. M., Ulgherait M., Kim H. X., Basturk M. K., Canman J. C., Shirasu-Hiza M., Dissection of central clock function in Drosophila through cell-specific CRISPR-mediated clock gene disruption. eLife 8, e48308 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy B., Singh A. P., Shetty C., Chaudhary V., North A., Landgraf M., VijayRaghavan K., Rodrigues V., Metamorphosis of an identified serotonergic neuron in the Drosophila olfactory system. Neural Dev. 2, 20 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X., Gaudry Q., Functional integration of a serotonergic neuron in the Drosophila antennal lobe. eLife 5, e16836 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huser A., Rohwedder A., Apostolopoulou A. A., Widmann A., Pfitzenmaier J. E., Maiolo E. M., Selcho M., Pauls D., von Essen A., Gupta T., Sprecher S. G., Birman S., Riemensperger T., Stocker R. F., Thum A. S., The serotonergic central nervous system of the Drosophila larva: Anatomy and behavioral function. PLOS ONE 7, e47518 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giang T., Rauchfuss S., Ogueta M., Scholz H., The serotonin transporter expression in Drosophila melanogaster. J. Neurogenet. 25, 17–26 (2011). [DOI] [PubMed] [Google Scholar]
- 31.De-Miguel F. F., Trueta C., Synaptic and extrasynaptic secretion of serotonin. Cell. Mol. Neurobiol. 25, 297–312 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Trueta C., De-Miguel F. F., Extrasynaptic exocytosis and its mechanisms: A source of molecules mediating volume transmission in the nervous system. Front. Physiol. 3, 319 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das A., Gupta T., Davla S., Prieto-Godino L. L., Diegelmann S., Reddy O. V., Raghavan K. V., Reichert H., Lovick J., Hartenstein V., Neuroblast lineage-specific origin of the neurons of the Drosophila larval olfactory system. Dev. Biol. 373, 322–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.P. J. Rice, J. R. Coats, Structural requirements for monoterpenoid activity against insects, in Bioregulators for Crop Protection and Pest Control, P. A. Hedin, Ed. (American Chemical Society, 1994), vol. 557, chap. 8, pp. 92–108. [Google Scholar]
- 35.Strutz A., Soelter J., Baschwitz A., Farhan A., Grabe V., Rybak J., Knaden M., Schmuker M., Hansson B. S., Sachse S., Decoding odor quality and intensity in the Drosophila brain. eLife 3, e04147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K., Gong J., Wang Q., Li H., Cheng Q., Liu Y., Zeng S., Wang Z., Parallel pathways convey olfactory information with opposite polarities in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 111, 3164–3169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tastekin I., Khandelwal A., Tadres D., Fessner N. D., Truman J. W., Zlatic M., Cardona A., Louis M., Sensorimotor pathway controlling stopping behavior during chemotaxis in the Drosophila melanogaster larva. eLife 7, e38740 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tastekin I., Riedl J., Schilling-Kurz V., Gomez-Marin A., Truman J. W., Louis M., Role of the subesophageal zone in sensorimotor control of orientation in Drosophila larva. Curr. Biol. 25, 1448–1460 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Lin S., Senapati B., Tsao C.-H., Neural basis of hunger-driven behaviour in Drosophila. Open Biol. 9, 180259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng K. Y., Frye M. A., Neuromodulation of insect motion vision. J. Comp. Physiol. A 206, 125–137 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Sampson M. M., Meyers Gschweng K. M., Hardcastle B. J., Bonanno S. L., Sizemore T. R., Arnold R. C., Gao F., Dacks A. M., Frye M. A., Krantz D. E., Serotonergic modulation of visual neurons in Drosophila melanogaster. PLOS Genet. 16, e1009003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strother J. A., Wu S.-T., Rogers E. M., Eliason J. L. M., Wong A. M., Nern A., Reiser M. B., Behavioral state modulates the ON visual motion pathway of Drosophila. Proc. Natl. Acad. Sci. U.S.A. 115, E102–E111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dayan P., Huys Q. J. M., Serotonin in affective control. Annu. Rev. Neurosci. 32, 95–126 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Kim S. M., Su C.-Y., Wang J. W., Neuromodulation of innate behaviors in Drosophila. Annu. Rev. Neurosci. 40, 327–348 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Bacqué-Cazenave J., Bharatiya R., Barrière G., Delbecque J.-P., Bouguiyoud N., Di Giovanni G., Cattaert D., De Deurwaerdère P., Serotonin in animal cognition and behavior. Int. J. Mol. Sci. 21, 1649 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albin S. D., Kaun K. R., Knapp J.-M., Chung P., Heberlein U., Simpson J. H., A subset of serotonergic neurons evokes hunger in adult Drosophila. Curr. Biol. 25, 2435–2440 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Dacks A. M., Green D. S., Root C. M., Nighorn A. J., Wang J. W., Serotonin modulates olfactory processing in the antennal lobe of Drosophila. J. Neurogenet. 23, 366–377 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coates K. E., Majot A. T., Zhang X., Michael C. T., Spitzer S. L., Gaudry Q., Dacks A. M., Identified serotonergic modulatory neurons have heterogeneous synaptic connectivity within the olfactory system of Drosophila. J. Neurosci. 37, 7318–7331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaudry Q., Serotonergic modulation of olfaction in rodents and insects. Yale J. Biol. Med. 91, 23–32 (2018). [PMC free article] [PubMed] [Google Scholar]
- 50.Kapoor V., Provost A. C., Agarwal P., Murthy V. N., Activation of raphe nuclei triggers rapid and distinct effects on parallel olfactory bulb output channels. Nat. Neurosci. 19, 271–282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brunert D., Tsuno Y., Rothermel M., Shipley M. T., Wachowiak M., Cell-type-specific modulation of sensory responses in olfactory bulb circuits by serotonergic projections from the raphe nuclei. J. Neurosci. 36, 6820–6835 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hauser S. R., Hedlund P. B., Roberts A. J., Sari Y., Bell R. L., Engleman E. A., The 5-HT7 receptor as a potential target for treating drug and alcohol abuse. Front. Neurosci. 8, 448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu L., He J., Kaiser A., Gräber N., Schläger L., Ritze Y., Scholz H., A single pair of serotonergic neurons counteracts serotonergic inhibition of ethanol attraction in Drosophila. PLOS ONE 11, e0167518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Port F., Bullock S. L., Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat. Methods 13, 852–854 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keesey I. W., Knaden M., Hansson B. S., Olfactory specialization in Drosophila suzukii supports an ecological shift in host preference from rotten to fresh fruit. J. Chem. Ecol. 41, 121–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hernandez-Nunez L., Belina J., Klein M., Si G., Claus L., Carlson J. R., Samuel A. D. T., Reverse-correlation analysis of navigation dynamics in Drosophila larva using optogenetics. eLife 4, e06225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gershow M., Berck M., Mathew D., Luo L., Kane E. A., Carlson J. R., Samuel A. D. T., Controlling airborne cues to study small animal navigation. Nat. Methods 9, 290–296 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Becnel J., Johnson O., Luo J., Nässel D. R., Nichols C. D., The serotonin 5-HT7Dro receptor is expressed in the brain of Drosophila, and is essential for normal courtship and mating. PLOS ONE 6, e20800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo J., Becnel J., Nichols C. D., Nässel D. R., Insulin-producing cells in the brain of adult Drosophila are regulated by the serotonin 5-HT1A receptor. Cell. Mol. Life Sci. 69, 471–484 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/1/eabd6900/DC1


