Hematopoietic tumors develop through the acquisition of multiple molecular cytogenetic lesions, which confer growth advantage to the neoplastic cells, favoring the emergence of clones resistant to treatment. Not surprisingly, a high number of chromosome aberrations represents an unfavorable prognostic factor in several hematologic neoplasias, including acute myeloid leukemia,1 myelodysplastic syndrome,2 myelofibrosis3 and chronic lymphocytic leukemia (CLL).4
Conventional banding analysis (CBA) is a well-established method allowing for a complete overview of the cell genome, with high specificity and low sensitivity. The presence of three or more chromosome lesions in the same clone, usually referred to as complex karyotype, was associated with a shorter overall survival in CLL as early as 1990.5 Since then, reports have documented the negative prognostic significance of a complex karyotype in patients treated with chemoimmunotherapy and with novel mechanism-based therapy.6-10
While prognostic factors associate with outcome independently of the treatment, predictive factors are able to identify patients who are likely to obtain improved survival with a specific therapy and can only be identified in comparative trials.11 Because different types of treatment can be offered to CLL patients, biological factors predicting outcome with a specific drug are very useful in clinical practice and CBA was proposed as a desirable procedure in the diagnostic workup within prospective clinical studies.12 Interestingly, high efficacy of venetoclax plus obinutuzumab in patients with a complex karyotype and CLL was recently documented in the CLL14 trial,13 pointing to a possible role of complex karyotype as a predictive marker.
Despite the introduction of effective mitogens, CBA is quite a cumbersome procedure requiring fresh dividing cells. For this reason, molecular methods such as array comparative genomic hybridization (CGH), single nucleotide insertional polymorphisms and next-generation sequencing have been applied in CLL, consistently showing that a high number of genetic lesions is associated with an inferior outcome.14-16
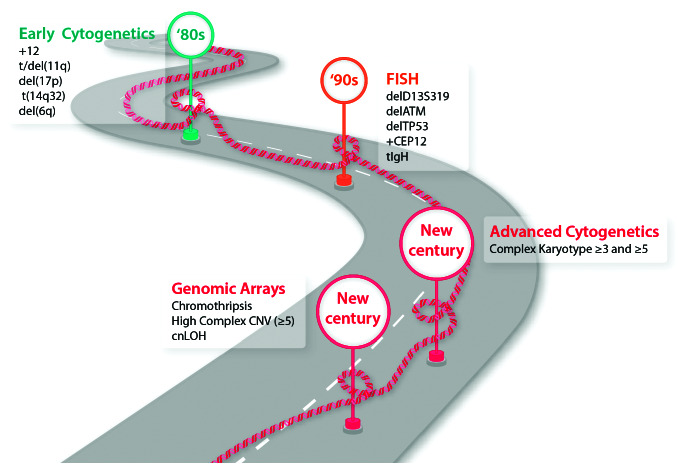
Figure 1.
The long road of genetic advances with a clinical impact on chronic lymphocytic leukemia. CNV: copy number variation; cnLOH: copy neutral loss of heterozygosity; ≥3 and ≥5: three or more, or five or more aberrations in the same clone, respectively.
In this issue of Haematologica, Leeksma and co-workers17 report the results of high-resolution genome-wide detection of copy-number alterations (CNA) in a large multicenter cohort of CLL patients, showing that genomic arrays are more sensitive than molecular cytogenetic methods and that high genomic complexity (i.e., 5 or more CNA) represents an independent adverse prognosticator.
In their analysis, 13 laboratories connected to the European Research Initiative in CLL (ERIC) group analyzed 2,293 patients by array-CGH and performed concurrent fluorescence in situ hybridization (FISH) in 260 patients using the classical four-probe panel detecting 17p-, 11q-, +12 and 13q- and concurrent CBA in 122 patients. Nine-hundred seventy-two patients, known to be untreated at the time of sampling, were included in the overall survival analyses.
Overall, CNA were detected in 78.9% of the cases and, as expected, previously treated patients and patients with high-risk CLL (i.e., in advanced stage or with unfavorable genetic features) had a significantly higher number of CNA.
In one-third of the cases genomic lesions detected by array-CGH were not detected by FISH. However, despite a high concordance between the two approaches, subclonal chromosome lesions detected by FISH, including 11q- and 17p-, were not detected by array-CGH.
In previous analyses, similar results were observed when comparing FISH and CBA.18,19 Interestingly, Leeksma and co-workers suggest that CGH may be more sensitive than CBA, given that a direct comparison of results from 122 patients showed chromosomal abnormalities in 86.1% of patients by genomic arrays versus 73.8% by CBA (P=0.015).
Among 972 untreated patients, 57 (5.8%) had high genomic complexity (i.e., ≥5 CNA). This parameter was an independent prognostic factor associated with shorter overall survival (hazard ratio = 2.19; 95% confidence interval: 1.35-3.54) and shorter time to first treatment (hazard ratio = 2.00; 95% confidence interval: 1.28-3.14), thus confirming the results of a recent international report on CBA.8 It is worth noting that, as the authors wisely pointed out, this series included patients treated with different chemoimmunotherapy regimens and that the role of genomic complexity in patients treated with novel agents is still unknown.
Besides the classical biomarkers and molecular genetic lesions associated with inferior prognosis, array-CGH identified gains of 8q, and losses of 9p and 18p as candidate markers possibly associated with a shorter overall survival in univariable Cox regression analysis. Interestingly, a minority of patients (n=32) showed very complex rearrangements, consisting of ≥10 oscillating copy number alterations involving two or three copy number states on one chromosome, a pattern suggestive of catastrophic mitotic events, usually referred to as chromothripsis. 20 Putative chromothripsis was associated with a dismal outcome in this series.
It is worth noting that, despite heterogeneity of the patients and of the methods employed in the different laboratories (i.e., total blood or purified CD19-positive lymphocytes, different array-CGH platforms and protocols) the study by Leeksma et al. has the following merits: (i) rigorous criteria were applied for the detection of CNA, with reference to the 5 Mb cut-off that is close to the chromosome band size; (ii) virtually all the results corresponded to established cytogenetic imbalances expected to occur in CLL; and (iii) the highly complex karyotype (i.e., ≥5 changes) was detected, with results in patients being superimposable to those with concurrent CBA.
On the other hand, an intrinsic limit of genomic arrays, like that of other genomic techniques, is the lack of identification of subclones and translocations; furthermore, the major specific contributions generated in this analysis, i.e., chromothripsis and copy neutral loss of heterozygosity, could not be analyzed in detail.
Although not yet validated in prospective studies, genomic arrays could become an accurate tool for CLL risk stratification, as they may represent a widely applicable method for the detection of DNA gains and losses. Indeed, while FISH remains the gold standard for the detection of 11q- and 17p-, the introduction of array- CGH in clinical trials may be advantageous as it may allow for investigations on the role of genomic complexity as a prognostic/predictive factor in clinical trials, assisting clinicians in the choice of the most appropriate treatment for their patients.
Supplementary Material
References
- 1.Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenberg PL, Tuechler H, Schanz J, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012; 120(12):2454-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gangat N, Caramazza D, Vaidya R, et al. DIPSS plus: a refined Dynamic International Prognostic Scoring System for primary myelofibrosis that incorporates prognostic information from karyotype, platelet count, and transfusion status. J Clin Oncol. 2011;29(4):392-397. [DOI] [PubMed] [Google Scholar]
- 4.Rigolin GM, del Giudice I, Formigaro L, et al. Chromosome aberrations detected by conventional karyotyping using novel mitogens in chronic lymphocytic leukemia: clinical and biologic correlations. Genes Chromosomes Cancer. 2015;54(12):818-826. [DOI] [PubMed] [Google Scholar]
- 5.Juliusson G, Oscier DG, Fitchett M, et al. Prognostic subgroups in Bcell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med. 1990;323(11):720-724. [DOI] [PubMed] [Google Scholar]
- 6.Cavallari M, Cavazzini F, Bardi A, et al. Biological significance and prognostic/predictive impact of complex karyotype in chronic lymphocytic leukemia. Oncotarget. 2018;9(76):34398-34412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rigolin GM, Saccenti E, Guardalben E, et al. In chronic lymphocytic leukaemia with complex karyotype, major structural abnormalities identify a subset of patients with inferior outcome and distinct biological characteristics. Br J Haematol. 2018;181(2):229-233. [DOI] [PubMed] [Google Scholar]
- 8.Baliakas P, Jeromin S, Iskas M, et al. Cytogenetic complexity in chronic lymphocytic leukemia: definitions, associations, and clinical impact. Blood. 2019;133(11):1205-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson PA, O'Brien SM, Wierda WG, et al. Complex karyotype is a stronger predictor than del(17p) for an inferior outcome in relapsed or refractory chronic lymphocytic leukemia patients treated with ibrutinib-based regimens. Cancer. 2015;121(20):3612-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson MA, Tam C, Lew TE, et al. Clinicopathological features and outcomes of progression of CLL on the BCL2 inhibitor venetoclax. Blood. 2017;129(25):3362-3370. [DOI] [PubMed] [Google Scholar]
- 11.Ballman KV. Biomarker: predictive or prognostic? J Clin Oncol. 2015;33(33):3968-3971. [DOI] [PubMed] [Google Scholar]
- 12.Hallek M, Cheson BD, Catovsky D, et al. IwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood. 2018;131(25):2745-2760. [DOI] [PubMed] [Google Scholar]
- 13.Al-Sawaf O, Lilienweiss E, Bahlo J, et al. High efficacy of venetoclax plus obinutuzumab in patients with complex karyotype and chronic lymphocytic leukemia. Blood. 2020;135(11):866-870. [DOI] [PubMed] [Google Scholar]
- 14.Delgado J, Salaverria I, Baumann T, et al. Genomic complexity and IGHV mutational status are key predictors of outcome of chronic lymphocytic leukemia patients with TP53 disruption. Haematologica. 2014;99(11): e231-e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunnarsson R, Isaksson A, Mansouri M, et al. Large but not small copy-number alterations correlate to high-risk genomic aberrations and survival in chronic lymphocytic leukemia: a high-resolution genomic screening of newly diagnosed patients. Leukemia. 2010;24(1):211-215. [DOI] [PubMed] [Google Scholar]
- 16.Rigolin GM, Saccenti E, Bassi C, et al. Extensive next-generation sequencing analysis in chronic lymphocytic leukemia at diagnosis: clinical and biological correlations. J Hematol Oncol. 2016;9(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leeksma AC, Baliakas P, Moysiadis T, et al. Genomic arrays identify high-risk chronic lymphocytic leukemia with genomic complexity: a multicenter study. Haematologica. 2021;106(1):87-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haferlach C, Dicker F, Schnittger S, et al. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgVH status and immunophenotyping. Leukemia. 2007; 21(12):2442-2451. [DOI] [PubMed] [Google Scholar]
- 19.Rigolin GM, Cibien F, Martinelli S, et al. Chromosome aberrations detected by conventional karyotyping using novel mitogens in chronic lymphocytic leukemia with “normal” FISH: correlations with clinicobiologic parameters. Blood. 2012; 119(10):2310-2313. [DOI] [PubMed] [Google Scholar]
- 20.Stephens PJ, Greenman CD, Fu B, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144(1):27-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.