Abstract
The tree of life shows the relationship between all organisms based on their common ancestry. Until 1977, it comprised two major branches: prokaryotes and eukaryotes. Work by Carl Woese and other microbiologists led to the recategorization of prokaryotes and the proposal of three primary domains: Eukarya, Bacteria and Archaea. Microbiological, genetic and biochemical techniques were then needed to study the third domain of life. Haloferax volcanii, a halophilic species belonging to the phylum Euryarchaeota, has provided many useful tools to study Archaea, including easy culturing methods, genetic manipulation and phenotypic screening. This review will focus on DNA replication and DNA repair pathways in H. volcanii, how this work has advanced our knowledge of archaeal cellular biology, and how it may deepen our understanding of bacterial and eukaryotic processes.
Keywords: Archaea, Haloferax volcanii, DNA replication, DNA repair, homologous recombination
1. Haloferax volcanii
Pioneering work in the 1970s by Carl Woese and other microbiologists led to a profound reorganization of the tree of life. Woese's discovery of Archaea was initially based on small-subunit ribosomal RNA sequences [1], but was soon consolidated by work from Wolfram Zillig on RNA polymerase [2] and Otto Kandler on cell membranes [3]. Eventually, archaea took their place as members of a bona fide domain, alongside Eukarya and Bacteria [4]. Archaea share morphological features with bacteria—both are prokaryotic cells—but they show dramatic differences at the enzymatic level. The information processing machinery found in archaea, which includes the enzymes involved in DNA replication, is strikingly similar to that of eukaryotes. In the decades since their discovery, archaea have been shown to be neither ‘exotic bacteria’ nor ‘simplified eukaryotes’; instead, they display a mosaic of eukaryotic, bacterial and uniquely archaeal features. Furthermore, the recent discovery of Asgard archaea has provided support for a two-domain tree of life, where eukaryotes emerge from within the archaeal clade [5–7]. Thus, further study of archaea is needed to deepen our understanding of fundamental processes such as DNA replication and repair, and to shed light on our evolutionary history.
One of the model archaeal species is Haloferax volcanii, which is a member of the phylum Euryarchaeota. It is a halophile with disc-shaped cells and grows optimally at 45°C in 1.7–2.5 M NaCl, similar to the conditions found in the Dead Sea where it was first isolated in 1975 [8]. Haloferax volcanii cells do not possess a rigid cell wall but are instead surrounded by a glycoprotein surface (S-) layer, which can be a target for glycosylation [9]. Haloferax volcanii use a ‘salt-in’ mechanism to deal with the highly halophilic environment; this mechanism ensures that the internal salt concentration is maintained at the same molarity as the external environment [10,11]. The genome of H. volcanii is highly polyploid, with a copy number of approximately 20 copies per cell, as well as being relatively GC rich (approx. 65%) [12,13].
In the 1980s and 1990s, ground-breaking work from the groups of W. Ford Doolittle, Moshe Mevarech and Mike Dyall-Smith developed techniques for the transformation and genetic manipulation of H. volcanii, enabling researchers to use this organism to study halophilic archaea [9,14,15]. Since then, a variety of genetic, molecular and biochemical tools have been developed, making H. volcanii one of the key model organisms within the Archaea [16].
1.2. Genetics, molecular biology and biochemistry tools for H. volcanii
-
—
Ability to grow in complex and defined media, in both broth and agar, in a wide range of salinities;
-
—
antibiotic selection including novobiocin resistance and mevinolin resistance [15,17];
-
—
auxotrophic selection including selectable markers for uracil, leucine, tryptophan and thymidine biosynthesis [18–20];
-
—
efficient markerless gene deletion methods based on selection for uracil biosynthesis and counter-selection of resistance to 5-fluoroorotic acid [18,19];
-
—
reporter genes including ß-galactosidase [21], GFP and related fluorescent proteins [22,23], and luciferase [24];
-
—
shuttle vectors based on different H. volcanii replication origins [17,19,25,26];
-
—
inducible gene expression based on a tryptophan-inducible promoter [27], and constitutive gene expression using a strong synthetic promoter [28,29];
-
—
random genome insertion mutagenesis library [30];
-
—
utilization of own CRISPR system as a method of gene interference (CRISPRi) [31,32];
-
—
natural gene transfer system (cell mating), which can be used for combining mutations [14,33,34];
-
—
genome sequence with manually curated annotation [35];
-
—
protein overexpression and purification, and other biotechnology applications [29,36];
-
—
proteomic methods using metabolic labelling (SILAC) along with pulse-chase lipid analysis [16,37];
-
—
mapping of post-translational modifications [38];
-
—
pioneer species in the Archaeal Proteome Project (ArcPP) [39].
The ease with which H. volcanii can be cultured in broth and on solid media, and the extensive range of genetic, molecular and biochemical tools that have been developed, have made this organism ideal to compare and contrast fundamental cellular processes with other halophiles, other archaea and other domains of life. Here, we focus on DNA replication and repair pathways in archaea, and in particular in H. volcanii. The knowledge gained on mechanisms of DNA replication and repair in H. volcanii has highlighted both similarities and differences to bacteria and eukaryotes, and has contributed to an appreciation of the diversity (and grandeur) in this view of life.
2. DNA replication
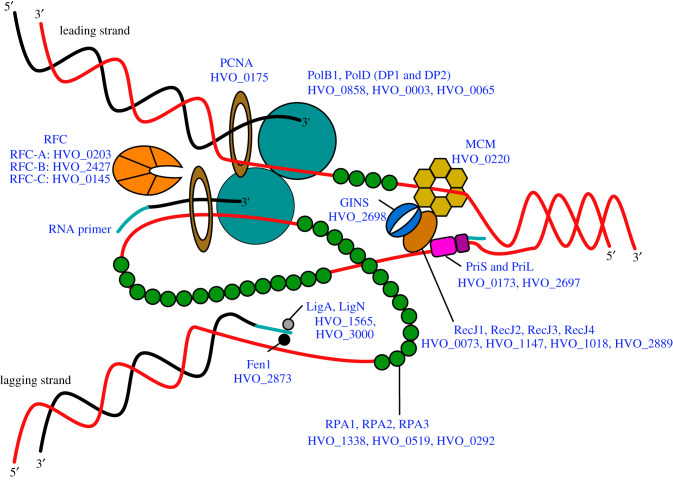
DNA replication is a fundamental cellular process and can be divided into three stages: initiation, elongation and termination. The initiation of DNA replication occurs at specific chromosomal sites termed origins and relies on the binding of initiator proteins at these sites [40]. Origins contain AT-rich sequences named duplex unwinding elements (DUEs), where weaker hydrogen bonding facilitates DNA strand opening. Binding of initiator proteins triggers the recruitment of a helicase that, when active, further unwinds the DNA double helix, exposing single-stranded DNA (ssDNA; outlined in figure 1 and table 1). The ssDNA is protected by single-stranded DNA-binding proteins (SSBs) that have an additional role in the downstream recruitment of replication factors, including primases and DNA polymerases. The formation of a replisome complex initiates bidirectional DNA synthesis in opposing directions away from the origin. During elongation, primases generate short RNA primers from which DNA polymerases prime synthesis of the leading strand continuously in a 5′–3′ direction, while replication of the lagging strand occurs discontinuously via the formation of Okazaki fragments. Additional components of the replisome include clamp loader proteins, which act to load sliding clamp proteins that act both as a molecular toolbelt and processivity factor for DNA polymerases. Termination of DNA replication occurs when replication forks meet and resolve, allowing for correct chromosome segregation upon completion of DNA synthesis.
Figure 1.
Structural components of the replisome. The CMG replicative helicase complex (RecJ:MCM:GINS in H. volcanii) unwinds DNA to expose single-stranded DNA (ssDNA). It remains unknown which of the four RecJ proteins in H. volcanii forms part of the CMG complex. The ssDNA is protected from damage by binding protein RPA and is used as a template for the synthesis of RNA primers by the primase activities of PriS and PriL. Replicative DNA polymerases (PolB1 and PolD) extend the RNA primer to initiate DNA replication. Clamp loader RFC removes primases from the replication fork and the open DNA structure is held in place by the sliding clamp PCNA. H. volcanii gene loci (HVO_#) for each component of the replisome are indicated.
Table 1.
DNA replication and repair enzymes and gene loci in H. volcanii.
| process | function | enzyme | HVO_ gene locus | notes |
|---|---|---|---|---|
| replication initiation | origin binding | Orc1 | 0001 | oriC1 |
| Orc2 | 0634 | oriC3 | ||
| Orc3 | A0001 | ori-pHV4 | ||
| Orc5 | 1725 | oriC2 | ||
| Orc6 | B0001 | ori-pHV3 | ||
| Orc10 | C0001 | ori-pHV1 | ||
| replisome formation | CMG replicative helicase complex | MCM | 0220 | |
| GINS | 2698 | |||
| RecJ1 | 0073 | alternative GAN proteins, Cdc45 orthologue not yet determined | ||
| RecJ2 | 1147 | |||
| RecJ3 | 1018 | |||
| RecJ4 | 2889 | |||
| primer generation (primase) | DnaG | 2321 | ‘bacterial’ primase, unlikely to act in replication | |
| PriS | 2697 | ‘eukaryotic’ primase | ||
| PriL | 0173 | |||
| clamp loader | RFC-A | 0203 | ||
| RFC-B | 2427 | |||
| RFC-C | 0145 | |||
| clamp protein | PCNA | 0175 | ||
| single-stranded DNA-binding protein | RPA1 | 1338 | only RPA2 essential, RPA1/3 unlikely to play major role in replication | |
| RPA2 | 0519 | |||
| RPA3 | 0292 | |||
| DNA ligase | LigA | 1565 | alternative and redundant ligases | |
| LigN | 3000 | |||
| DNA synthesis | replicative DNA polymerase | PolB1 | 0858 | |
| PolD1 (DP1) | 0003 | small exonuclease subunit of PolD | ||
| PolD2 (DP2) | 0065 | large subunit of PolD | ||
| termination of DNA replication | dimer and superhelical torsion resolution | XerC-like | 1422 | involvement in termination of replication yet to be shown |
| 2259 | ||||
| 2273 | ||||
| 2290 | ||||
| TopoIA | 0681 | ‘bacterial’ topoisomerase | ||
| TopoVI-A | 1570 | ‘archaeal’ topoisomerases | ||
| TopoVI-B | 1571 | |||
| GyrA | 1572 | ‘bacterial’ topoisomerases | ||
| GyrB | 1573 | |||
| removal of RNA primers from replicated DNA/DNA:RNA hybrids | RNaseH-A | 2438 | Type I RNase H | |
| RNaseH-B | 1978 | Type II RNase H | ||
| RNaseH-C | A0463 | Type I RNase H | ||
| RNaseH-D | A0277 | |||
| RNaseH-E | 0732 | Type I RNase H | ||
| flap endonuclease | Fen1 | 2873 | also acts in various repair pathways | |
| direct DNA repair | photolyase | Phr1 | 2911 | |
| Phr2 | 2843 | |||
| Phr3 | 1234 | as yet uncharacterized | ||
| type IV restriction enzyme | Mrr | 0682 | ||
| methyltransferase | A0006 | targets cytosine at Cm4TAG motifs | ||
| Zim | 0794 | |||
| A0237 | ||||
| rmeRMS | 2269–2271 | targets adenine at GCAm6BN6VTGC motifs | ||
| base excision repair | DNA glycosylase | Udg1 | 0231 | uracil DNA glycosylase |
| Udg2 | 2792 | |||
| Udg3 | 0444 | |||
| Udg4 | 1038 | |||
| OGG | 1681 | DNA N-glycosylase | ||
| AlkA | 2814 | DNA-3-methyladenine glycosylase | ||
| MutY1 | 2896 | A/G-specific adenine glycosylase | ||
| MutY2 | 2834 | |||
| AP endonuclease | Apn1 | 0573 | endonuclease IV | |
| NthA | 0848 | endonuclease III | ||
| NthB | 0878 | |||
| EndIV | 2708 | |||
| EndV | 0726 | |||
| EndVb | 0443 | endonuclease V homologue | ||
| nucleotide excision repair | damaged DNA recognition | UvrA | 0393 | |
| helicase | UvrB | 0029 | ||
| endonuclease | UvrC | 3006 | ||
| helicase | UvrD | 0415 | redundant function with other helicases | |
| mismatch repair | predicted ATPase | MutLa | 1939 | active in mismatch repair |
| MutLb | 0551 | |||
| mismatch repair ATPase | MutS1a | 1940 | ||
| MutS1b | 0552 | |||
| MutS5a | 0191 | not involved in mismatch repair | ||
| MutS5b | 1354 | |||
| branched structure endonuclease | NucS | 0486 | also called EndoMS | |
| translesion synthesis | translesion polymerase | PolY | 1302 | |
| microhomology-mediated end joining (end resection) | ATPase | Rad50 | 0854 | work together in Mre11-Rad50 complex |
| exonuclease | Mre11 | 0853 | ||
| homologous recombination | recombinase | RadA | 0104 | |
| recombinase mediator | RadB | 2383 | ||
| strand displacement | Hel308 | 0014 | ||
| Hef | 3010 | |||
| Holliday junction resolvase | Hjc | 0170 | alternative and redundant resolvases | |
| Hef | 3010 |
2.1. Initiation of DNA replication
Bacteria generally have a single circular chromosome with a single origin of replication, oriC. Initiation of replication begins when initiator protein DnaA binds oriC at sequence-specific sites called DnaA boxes. The cooperative binding of DnaA forces open the duplex at the DUE, forming a ssDNA bubble [41], while bacterial SSB binds to the exposed ssDNA. The helix opening at oriC allows access to the helicase loader DnaC, which acts as a chaperone to recruit replicative helicase DnaB onto the lagging strand. Activation of the helicase is dictated by DnaC; when DnaC is bound by ATP, DnaB is inactive, but when DnaC is bound by ADP DnaB helicase is activated [42,43]. Active DnaB unwinds double-stranded DNA (dsDNA), increasing the size of the replication bubble and allowing downstream recruitment of the remainder of the replication components including primase, DNA polymerase and clamp protein ß. Only a single hexamer of DnaB is loaded per replication fork [44].
DNA replication initiation in eukaryotes is inherently more complex than in bacteria; multiple origins are present along the length of multiple linear chromosomes, with initiation being triggered by the binding of a multimer of initiation proteins known as the origin recognition complex (ORC). The ORC complex consists of six origin recognition proteins (termed Orc1-Orc6) [45]; Orc1–5 proteins contain a winged-helix (WH) domain that facilitates their binding at the origin [46]. Prior to S-phase, the ORC complex, together with the regulator cell division cycle 6 protein (Cdc6) and the licensing factor Cdc10-dependent transcript 1 protein (Cdt1), load the replicative helicase mini-chromosome maintenance (MCM2–7; consisting of 6 paralogous proteins) to form the pre-replicative complex (pre-RC) [47,48]. The ATPase AAA+ domains of Orc1–5 initiator proteins interact with the C-terminal WH domain of MCM in an ATP-dependent reaction. Any exposed ssDNA is coated with eukaryotic SSB protein, named replication protein A (RPA), for protection. Upon recruitment to the pre-RC, MCM helicase is inactive; activation must occur for elongation to begin. Activation of the replicative complex occurs in S phase, whereupon ORC, Cdc6 and Cdt1 are no longer required and will dissociate. MCM helicase is loaded onto the leading strand and, unlike the situation in bacteria, multiple MCM molecules can associate with a single replication fork [49,50].
Archaea have circular chromosomes and can use single or multiple origins to initiate DNA replication [51]. Despite the similarity in genome organization between archaea and bacteria, DNA replication mechanisms used by archaea differ widely from those used in bacteria; archaeal cells possess eukaryotic-like replication mechanisms employing multiple origins and Orc1/Cdc6-like proteins (referred to onwards as Orc).
Similar to bacterial and eukaryotic origins, archaeal replication origins are AT-rich regions (DUE), but in archaea are surrounded by origin recognition box (ORB) sequences, of which pairs are often inverted around the DUE [52]. ORBs direct the binding of Orc proteins onto DNA, with one Orc monomer binding a single ORB sequence with a defined polarity. ORBs are found on the minor groove of DNA and contain a signature string of guanine nucleotides known as the G-string [53]. Strand opening enables stable binding of the N-terminal AAA+ domain of the Orc protein(s) to the G-string, while the C-terminal WH DNA-binding domain of the Orc protein determines the binding affinity to the origin through binding to the ORB more generally [54,55].
Orc binding at the origin does not cause further melting of the dsDNA; instead, it facilitates the recruitment of MCM helicase (a single polypeptide in archaea, homologous to eukaryotic MCM2–7) to mark the origin for replisome loading [51,52]. Only the ATP-bound Orc is capable of MCM recruitment, while ADP-bound Orc is unable to sustain interactions with MCM [52,56]. Archaeal MCM forms a homohexameric ring, which is loaded onto the leading strand of DNA as a double hexamer, the pair of inverted Orc proteins surrounding the DUE each load a single hexamer of MCM.
An initial study on archaeal DNA replication mechanisms mapped the single replication origin of Pyrococcus abyssi using nucleotide skew analysis [57]. Since then, more advanced techniques, including two-dimensional gels, whole-genome microarrays and marker frequency analysis (MFA), have enabled the identification and mapping of origins in over 20 archaeal species (reviewed in [58]). It is now clear that the number of origins and Orc proteins varies considerably throughout the archaea [40], but one of the striking consistencies is that the gene encoding the Orc protein is (nearly) always found directly adjacent to its cognate origin. The association of origin and Orc in close proximity allows independent control of each origin and reduces competition between origins and initiators [56].
The origins of replication for H. volcanii were first mapped in 2007 [59]. The genomic architecture of H. volcanii consists of one main circular chromosome (2.8 Mb) and three circular mini-chromosomes; pHV4 (636 kb), pHV3 (438 kb) and pHV1 (85 kb) [35]; the 6 kb plasmid pHV2 has been cured from the laboratory strain and its derivatives [15]. The main chromosome features three active origins, each associated with their own corresponding Orc initiator protein (oriC1 is associated with Orc1, oriC2 with Orc5 and oriC3 with Orc2) [35,59]. As with the main chromosome, each mini-chromosome of H. volcanii has its own origin and corresponding Orc protein [40]. Origins vary in activity; oriC1 the most active origin in H. volcanii [60] and deletion of oriC1 and its corresponding Orc protein Orc1 results in a reduced ploidy, suggesting an additional regulatory role for this origin [61,62]. Subsequent studies have confirmed these initial findings and the replication profile of the main chromosome of H. volcanii is now mapped in detail [60,63].
The laboratory strain of H. volcanii contains a fourth replication origin on the main chromosome, ori-pHV4 (and its corresponding Orc protein, Orc3). A fusion event between insertion sequence (IS) elements on pHV4 and the main chromosome led to the stable integration of pHV4 and thus a newly acquired main chromosomal origin [60]. Additional stable genomic rearrangements in H. volcanii have been observed, where a recombination event between two near-identical sod1 and sod2 genes led to the creation of a stable mini-chromosome [64].
Alongside the three origin-associated Orc proteins in H. volcanii, there are 12 additional Orc proteins whose genes are not linked to origins. Their function is currently unknown, but they are most likely to be dormant Orc proteins that have been orphaned due to the integration of foreign genetic elements. For example, the genes encoding Orc11 and Orc14 are both located within a 50 kb prophage region [35]. Such integration events, coupled with the fluctuating genome configurations of archaeal species, hints at evolutionary mechanisms that have facilitated a multi-origin replication system, including horizontal gene transfer (HGT) and gene duplication events [61,65]. For example, the replicon takeover hypothesis postulates that the host chromosome becomes dependent on extra-chromosomal elements for its propagation [66]. The apparent fluidity of the H. volcanii genome architecture provides a tool for study of how the loss or gain of an origin and/or Orc can lead to the multi-origin chromosomes seen in several archaeal species.
2.2. Elongation
For DNA replication to proceed away from the origin, a full replisome must be established. All domains of life share basic mechanisms of DNA synthesis, but differ primarily in the proteins used [67,68]. In eukaryotes, the pre-replication complex (pre-RC) must be activated prior to elongation; this activation provides a further level of regulation above that of bacteria.
Formation of a replisome in bacteria is relatively simple; following activation of DnaB helicase, the remaining components are recruited in a stepwise manner. Primase DnaG, a DNA-dependent RNA polymerase, acts to synthesize short (approx. 8–12 nucleotide) primers used by DNA polymerase to prime synthesis [69]. Pol-III is the main replicative polymerase in bacteria, while Pol-I is involved in Okazaki fragment maturation (discussed in more detail later). Bacterial clamp protein ß ensures Pol-III remains associated with the template and increases the processivity of the polymerase during synthesis.
In eukaryotes, the activation of MCM2–7, and therefore activation of the pre-RC, is dependent upon two events: phosphorylation by kinases cyclin-dependent kinase (CDK) and Dbf4-dependent kinase (DDK) [70], and the formation of the CMG replicative helicase complex (consisting of Cdc45, MCM and GINS proteins). Assembly of the CMG complex in eukaryotes causes switching of MCM binding from dsDNA to ssDNA, activating the helicase for helix unwinding [71,72]. At this point, further replication components can be loaded, such as replicative DNA polymerases (Pol-α, Pol-δ and Pol-ε), primases (PriS/L) and clamp protein proliferating cell nuclear antigen (PCNA). Primase acts in complex with Pol-α (as complex PrimPol) to synthesize an approximately 30 bp RNA-DNA primer for extension by Pol-ε to synthesize the leading strand, while Pol-δ performs synthesis of the lagging strand.
Elongation in archaea is akin to the more complex system of eukaryotes: MCM associates with Cdc45-like and GINS-like proteins to form an archaeal CMG complex, following which the remainder of replication components are loaded and replication proceeds bidirectionally away from the origin. Archaeal replication components will be discussed in further detail below.
2.2.1. The CMG replicative helicase complex
2.2.1.1. Mini-chromosome maintenance helicase
Despite the ubiquitous function of MCM helicases, there is significant genetic and structural diversity within this family of proteins. Most archaeal species encode a single MCM homologue thought to act as the replicative helicase [73]. Where species encode more than one mcm homologue, such as Thermococcus kodakarensis and Methanococcus maripaludis, which possess three and four MCM homologues, respectively, only one will be essential for viability [74–76]. The essential MCM protein in species with multiple paralogues shares structural and sequence similarity with the single MCM proteins in other species [75,76].
As a hexamer, archaeal MCM possesses 3′–5′ helicase activity that opens the DNA duplex while translocating along the leading strand. Archaeal MCM proteins are members of the AAA+ ATPase superfamily and are made up of a non-catalytic N-terminal domain, a central catalytic AAA+ domain and a C-terminal winged-helix-turn-helix (wHTH) domain [77]. However, structural variations of archaeal MCM have been found with some homologues lacking an N-terminal domain or helicase activity, while others consist only of a partial C-terminal domain [73,78]. The N-terminal portion is important for hexamer formation, enzyme regulation and DNA binding [73]. The catalytic region contains residues associated with other AAA+ ATPases, with Walker A and Walker B motifs being required for ATP binding and hydrolysis, respectively. The presence of an arginine finger motif within the catalytic domain is characteristic of MCM as a protein of this superfamily, with the string of positively charged residues instigating a strong interaction with negatively charged DNA [77]. At the interface of N-terminal and catalytic domains is the allosteric control loop (ACL); the ACL consists of a β7–β8 β-hairpin loop and acts to regulate interactions between the N- and C-terminus of MCM [79,80]. The C-terminal wHTH domain is implicated in the regulation of MCM but is yet to be fully characterized [73,81].
Unlike eukaryotic MCM, archaeal MCM has basal activity without the requirement for interactors Cdc45 and GINS [82–84]. Archaeal MCM proteins can form a range of structures in solution, but only hexameric MCM has been shown to possess helicase activity [85]. Double hexameric MCM has been shown to be more active than the monomeric form, suggesting double hexameric MCM acts during canonical replication [86]. The crystal structure for Sulfolobus solfataricus MCM has revealed a feature specific to archaea; each monomer of MCM encodes four β-hairpins, three positioned within the main channel and one externally [79]. Mutational analysis has since revealed these structures play a key role in both DNA-binding and helicase activity [78].
H. volcanii encodes a single essential MCM homologue (HVO_0220), which forms a homohexameric structure akin to other archaeal MCM complexes [77,87]. Structurally, H. volcanii MCM is made up of a zinc-binding N-terminal domain, an AAA+ catalytic core and a C-terminal wHTH domain. It uses a zinc-cofactor to break the hydrogen bonding of the DNA double helix [77]. Mutagenesis studies have shown short β7-β8 β-hairpin loop deletions and large β9-β10 β-hairpin loop deletions within the N-terminal domain are intolerable to the cells; it is speculated that these loops are crucial for the coordination of zinc binding [77]. Furthermore, a G187A mutation and alanine substitutions of conserved zinc-binding cysteines show these residues play a critical role in MCM function. Like other archaeal MCM homologues, it is essential for cell viability; deletion of the full-length gene is not possible (T.A. 2020, unpublished) and specific β7–β8 loop and zinc-binding domain mutants of H. volcanii MCM could not be generated [77].
2.2.1.2. GINS complex
GINS complex (named after Japanese numbers 5-1-2-3, go-ichi-ni-san, representing subunits of the eukaryotic complex Sld5, Psf1, Psf2 and Psf3, respectively) is known to play an essential role in eukaryotic replication [88]. The four subunits of eukaryotic GINS are predicted to be paralogous [89], but can be clustered into two groupings based on protein structures; A-domains contain high amounts of α-helices, while B-domains are smaller and rich in β-strands. This structural grouping places Sld5 and Psf1 together with an AB domain organization, and Psf2 and Psf3 together with a BA domain organization [90]. Archaeal GINS complex, as with MCM, is a simplified version of the eukaryotic counterpart. Structurally, archaeal and eukaryotic GINS are comparable, but archaeal GINS is encoded by only one (gins51) or two (gins51 and gins23) genes, depending on species [89,91,92]. Archaeal GINS51 protein shares structural similarity with Sld5 and Psf1 (AB-type), while GINS23 shares similarity with Psf2 and Psf3 (BA-type). Species can either form a dimer of dimers where GINS51 and GINS23 are encoded, or a homotetramer of GINS51 alone in the absence of GINS23.
The first archaeal GINS homologue to be identified and characterized was from S. solfataricus. Its GINS complex forms a tetrameric structure made up of GINS51 and GINS23 dimers. In S. solfataricus, these genes are found in the same operon as both MCM and primase. A clear interaction has since been demonstrated between GINS23, MCM and primase, providing evidence for a functional and eukaryotic-like CMG complex in S. solfataricus [84,91]. Eukaryotic-like GINS has also been identified and characterized in T. kodakarensis, where the crystal structure of GINS is directly comparable to that of human GINS complex [93]. As in S. solfataricus, T. kodakarensis GINS forms a dimer of dimers of GINS51 and GINS23 proteins [93].
Interactions between archaeal MCM and GINS are well documented, with GINS interaction boosting the ATPase and helicase activities of MCM. In species encoding both GINS51 and GINS23 subunits, interactions with MCM are mediated by GINS23 [91,94]. Thermoplasma acidophilum encodes only GINS51 subunits, forming a homotetramer, but has been shown to carry out MCM : GINS interactions in a GINS51-dependent manner [83,95]. A GINS51-MCM interaction has yet to be observed in species carrying both GINS51 and GINS23 subunits.
A single homologue of GINS has been identified in H. volcanii. The gene HVO_2968 encodes a GINS51-type protein and is located within an operon with primase gene priS (HVO_2697) [87]. Structurally, H. volcanii GINS is larger than the eukaryotic counterparts, due to the presence of a large sequence insertion between the conserved A and B domains. Such an insertion is seen in numerous halophilic species; however, the function of such an insertion remains unknown and warrants further study [92]. As yet, interactions between H. volcanii GINS and other components of replication have not been described; like MCM, GINS is essential for cell viability (T.A. and Stuart MacNeill 2020, unpublished).
2.2.1.3. Cdc45/recJ/GAN
While archaeal MCM and GINS homologues are easy to identify based on similarity to their eukaryotic counterparts, identification of a Cdc45-like protein in archaea has been less straightforward. Bioinformatic analysis of Cdc45 revealed it is the eukaryotic orthologue of bacterial and archaeal RecJ phosphodiesterase/nuclease family proteins [96,97]. Specifically, the N-terminus of eukaryotic Cdc45 shows similarity to the DHH domain associated with RecJ family nucleases [98]. However, unlike well-characterized RecJ proteins, Cdc45 is known to lack catalytic activity; this can be explained by the loss of key catalytic residues within the DHH domain [96,99]. Instead, eukaryotic Cdc45 plays an essential structural role within the CMG complex. Akin to RecJ nucleases, eukaryotic Cdc45 has maintained the ability to bind ssDNA [100], which may account for its role at the replication fork.
Bacterial RecJ is relatively well characterized; it has been implicated in a number of DNA repair pathways, including mismatch repair (MMR), homologous recombination (HR) and base excision repair (BER), along with a role in the restart of stalled replication forks [101–104]. Bacterial RecJ is composed of an N-terminally located catalytic core, made up of DHH and DHHA1 domains, and a C-terminal oligonucleotide-binding (OB)-fold domain responsible for binding ssDNA. DHHA1 is a subfamily of DHH superfamily proteins, found in both bacteria and archaea but absent from eukaryotes; this subfamily domain is involved in substrate specificity [105,106]. The C-terminal positioning of the OB-fold is specific to bacterial RecJ and is not found within Cdc45 or archaeal RecJ proteins.
The DHH superfamily has undergone an expansion event within the archaea (specifically within the phylum Euryarchaea), where multiple species now encode several RecJ-like proteins. To date, every archaeal species studied encodes at least one RecJ protein [107] and while some homologues retain previous identities, others evolved quickly and developed novel roles [96]. RecJ proteins in archaea have now been implicated in DNA repair and replication, some have lost all activity, while the roles of many remain unknown [96].
RecJ proteins were first implicated in archaeal DNA replication following the identification of in vivo interactors with a range of replication components in T. kodakarensis [108]. Two RecJ-like proteins were identified in T. kodakarensis, termed GAN (GINS-associated nuclease) and HAN (Hef-associated nuclease) [109,110]. GAN was primarily identified as an in vivo interactor with GINS, interacting specifically with the GINS51 subunit, while HAN interacted with stalled fork repair protein Hef, favouring GAN as the Cdc45-like protein in T. kodakarensis [110–112]. Since its discovery in T. kodakarensis, GAN homologues have been identified in various archaeal species, reflecting the complexity and diversity of archaeal DNA replication factors [96]. RecJ-like/GAN proteins are thought to be the Cdc45-like protein within the CMG complex of archaea (also called GMG for GAN : MCM : GINS). For all characterized archaeal CMG complex interactions mapped to date, GINS is essential to bridge the interaction between MCM and Cdc45 [82–84].
The GAN : GINS complex acts to boost the helicase activity of MCM, akin to the role of Cdc45 in eukaryotic CMG complexes [82]. The crystal structure of GAN has revealed similarities to eukaryotic Cdc45; the DHH domain contains a CID (CMG-interacting domain), as in Cdc45 [109,113]. However, unlike its eukaryotic counterpart, GAN remains catalytically active as a processive 5′–3′ ssDNA exonuclease [112]. GAN has been shown to form a complex with GINS and MCM in vitro, and the interaction with GINS51 stimulates the exonuclease activity of GAN [82]. By contrast, the alternative RecJ protein HAN does not interact with GINS [110], suggesting an alternative role for this RecJ homologue. It has recently been shown that the T. kodakarensis replicative DNA polymerase subunit PolD2 interacts with GAN via the GINS complex, placing GAN at the heart of the replication complex as in eukaryotes [108,109,112,114].
Similar to T. kodakarensis, Pyrococcus furiosus encodes two RecJ-like proteins, one sharing sequence and structural similarity with GAN (PF2055; here called PfuRecJ) and the other sharing characteristics with HAN (PF0399). The DHH domain of PfuRecJ is intact, facilitating 5′–3′ DNA exonuclease activity, 3′–5′ RNA exonuclease activity, and interactions with GINS51, implicating PfuRecJ in DNA replication as a member of the CMG complex. Interaction of PfuRecJ and the SSB protein RPA results in 3′–5′ exonuclease activity on both ssRNA and dsRNA/DNA hybrids; such an activity could be used at the replication fork to deal with Okazaki fragments [115,116]. The crystal structure of PfuRecJ has been solved and is comparable to that of T. kodakarensis GAN [116]; PfuRecJ is therefore is a strong candidate for the Cdc45-like protein in P. furiosus.
However, this pattern is not observed in all Euryarchaea. Two RecJ homologues, TaRecJ1 and TaRecJ2, have been identified in Thermoplasma acidophilum, both bearing resemblance to T. kodakarensis GAN [117]. TaRecJ1 possesses 5′–3′ ssDNA-specific exonuclease activity, while TaRecJ2 possesses 3′–5′ exonuclease activity specific for RNA. Interactions between TaRecJ2 and GINS51 occur in a stable fashion and it has been shown that TaRecJ2, not TaRecJ1, in combination with GINS, stimulates the helicase activity of T. acidophilum MCM [117]. A CMG-like complex comprising TaRecJ2 : MCM : GINS was recapitulated in vitro and also observed in vivo, making TaRecJ2 the true ‘GAN’ of this species [117]. The 5′-3′ directionality of TaRecJ1 is akin to that of bacterial RecJ, and it is possible that TaRecJ1 has gained a role in DNA repair akin to bacterial RecJ; this is yet to be confirmed.
Unlike the full-length RecJ proteins acting as Cdc45 in the aforementioned examples, the crenarchaeon S. solfataricus instead uses a truncated form of RecJ [91]. Primarily identified through its interaction with GINS51, RecJdbh (RecJ DNA-binding homologue) or ‘Cdc45’ shares only the DNA-binding portion of bacterial RecJ. These ‘inactive’ RecJ proteins have been shown to form CMG-like complexes and boost the helicase activity of MCM, making them bona fide Cdc45-like proteins [84,91]. RecJdbh shares very little homology to characterized GAN proteins, carrying a degenerate DHH domain and lacking any exonucleolytic activity. This suggests that there is more than one type of RecJ protein able to act in the manner of Cdc45 in archaea and warrants further study.
The identity of the Cdc45 homologue in H. volcanii is also an open question. Preliminary work has identified four RecJ homologues: RecJ1 (HVO_0073), RecJ2 (HVO_1147), RecJ3 (HVO_1018) and RecJ4 (HVO_2889) [107]. Based on similarity to Cdc45, GAN and RecJ proteins in general, it has been proposed that one (or more) of these RecJ homologues function as the Cdc45 homologue in H. volcanii [71]. Analysis of catalytic residues within RecJ1 and RecJ3 suggest they have retained nuclease activity, but in vitro and in vivo studies are needed to determine whether these nucleases are active, and whether they act in DNA repair or replication. By contrast, RecJ2 and RecJ4 are both predicted to have lost key catalytic residues and therefore nuclear activity. Comparison of H. volcanii RecJ proteins by arCOG grouping (archaeal clusters of orthologous genes) suggests that RecJ1 is the best candidate for GAN, while RecJ3 and RecJ4 are HAN candidates; RecJ2 has diverged away from the other H. volcanii RecJ proteins and its function remains unknown. RecJ1, RecJ3 and RecJ4 are all non-essential (even in combination) but the cellular requirement for RecJ2 is an open question (T.A. and Stuart MacNeill 2020, unpublished). Further work is needed to decipher which RecJ(s) play the role of Cdc45 and whether there is any redundancy between the four RecJ proteins in H. volcanii.
2.2.2. Other replisome components
2.2.2.1. Primases
Bacteria use a single subunit primase protein, DnaG, while eukaryotes encode heterodimers consisting of catalytic (PriS/p48) and regulatory (PriL/p58) subunits that work in tandem to synthesize short primers [69,118].
Archaea encode both bacteria-like and eukaryotic-like primases, depending on the species. The eukaryotic-like replicative primase found in archaea is a two-subunit complex consisting of a small catalytic subunit (PriS/p41) and a large regulatory subunit (PriL/p46); fusion events of PriS and PriL have been seen within nanoarchaeal genomes [61,68]. Unlike bacterial and eukaryotic primases that are only capable of synthesizing ribonucleotides, archaeal primases have been shown capable of synthesizing both RNA and DNA [119]. DNA synthesis can reach lengths of several kilobases, meaning archaeal primases in some cases can be defined as non-canonical DNA polymerases [119,120]. PriS/L-like primases in archaea have also been implicated in functions outside of replication, including gap-filling and strand displacement activities [121]. Bacterial-like DnaG proteins in archaea do not appear to act in DNA replication. For example in S. solfataricus, DnaG has been strongly implicated in RNA degradation and has only limited primer synthesis activity [122,123]; instead, S. solfataricus uses PriS/L to carry out primer synthesis during DNA replication [123,124].
H. volcanii also encodes homologues of both bacterial and eukaryotic primases. Bacterial DnaG primase (HVO_2321) can be deleted from H. volcanii without any effect on cell viability [121], suggesting that this protein has no role in DNA replication. As in S. solfataricus, DnaG may have gained an alternative role in RNA degradation but this requires further testing. By contrast, eukaryotic-like PriS and PriL genes (HVO_2697 and HVO_0173 respectively) are essential for cell viability [121], most likely priming DNA replication at the replisome. Significant work on the activities of PriS/L is still needed, including the length of RNA/DNA primers synthesized, polymerase specificity and any additional roles in the cell.
2.2.2.2. Clamp loader replication factor C
Sliding clamp proteins are required to boost the otherwise low processivity of replicative DNA polymerases. Sliding clamps are stable ring proteins and thus cannot self-assemble onto DNA; instead, they are assembled onto DNA by a clamp loader protein [125]. Clamp loader proteins facilitate the opening and loading of the clamp protein (ß-clamp protein in bacteria, PCNA in eukaryotes and archaea) onto a primer-template junction in an ATP-dependent manner [126]. Bacteria use clamp loader γ-complex while eukaryotes and archaea rely on replication factor C (RFC). The ability of clamp loader proteins to distinguish ssDNA : dsDNA junctions allows loading of clamp proteins specifically at primer-template junctions [126]. The primase, at the time of clamp recruitment, remains associated with the primer. Both primases and clamp loaders interact with SSB and this facilitates the handoff from primase to clamp loader protein binding the primer-template junction [127]. Due to the discontinuous priming of the lagging strand, clamp proteins are continuously recruited, meaning there is a constant requirement for clamp loaders during processive replication [128].
Eukaryotic RFC is pentameric and is composed of one large subunit (Rfc1) and four small subunits (Rfc2–5). Most archaea encode two homologues of RFC: one corresponding to the small eukaryotic RFC subunit (RFCS) and the other corresponding to the large subunit (RFCL) [44,125]. Akin to eukaryotic RFC, archaeal RFC forms a pentamer consisting of four RFCS subunits and one RFCL subunit [129]. Stimulation of PCNA by RFC has been characterized in Pyrococcus horikoshii, whereby RFC enables PCNA to recruit and activate both replicative DNA polymerases [130,131].
H. volcanii possesses three homologues of RFC (RFC-A, HVO_0203; RFC-B, HVO_2427; RFC-C, HVO_0145), all of which are essential for growth [132]. All three homologues possess AAA+ domains that enable ATP-dependent DNA binding. The larger of the three, RFC-B, carries an additional C-terminal PIP box that is absent from the smaller RFC subunits. A PIP box (PCNA-interacting protein peptide box, discussed in more detail later) facilitates interaction with PCNA, suggesting that RFC-B acts to stimulate PCNA for polymerase recruitment. Further work is required to decipher the roles of the RFC subunits in H. volcanii.
2.2.2.3. Proliferating cell nuclear antigen
The clamp protein proliferating cell nuclear antigen (PCNA) acts as a platform for the recruitment of DNA polymerases and other replicative proteins in eukaryotes and archaea. The protein binds dsDNA in a sequence-independent manner where it can move bidirectionally. PCNA acts as a clamp at the replication fork to tether replication factors onto DNA via the opening and closing of its ring structure around dsDNA (aided by clamp loader protein). Bacteria have a differing clamp protein, named β, which forms a homodimer, while both eukaryotes and archaea use trimeric protein PCNA [125].
Regarding clamp proteins in archaea, there appears to be a division along phylogenetic lines: in most euryarchaea, there is a single PCNA homologue that forms a homotrimer. Only one euryarchaeal species, T. kodakarensis, carries two PCNA homologues; however, one is believed to have been acquired relatively recently by lateral gene transfer (LGT) [133]. On the other hand, crenarchaea commonly encode multiple PCNA homologues and have been shown to form both homo- and hetero-trimeric structures [134].
H. volcanii, as a euryarchaeon, encodes a single homologue of PCNA (HVO_0175). PCNA is essential for viability in H. volcanii and forms a homotrimer in solution, with monomers interacting in a head-to-tail manner [135–137]. H. volcanii PCNA has been predicted to interact with numerous replication components, including replicative DNA polymerases PolB1 and PolD, clamp loader protein RFC-B, endonuclease Fen1 and ribonuclease RNase H2 [137]. All of these proteins contain a PIP box, a defined region of the protein made up of bulky aromatic groups containing conserved residues QxxLxxFx (where x represents any amino acid) [137]. Interactions of PCNA with proteins via PIP boxes is well-characterized throughout archaeal and eukaryotic species [68,138] and underlines a key role for PCNA in DNA replication. Alongside its role in replication, PCNA has also been linked to proteins associated with DNA repair via the identification of PIP boxes; these links are discussed in detail later. The ability of PCNA to interact with multiple proteins simultaneously has given rise to the ‘molecular toolbelt’ model, where PCNA acts to bring together replication and repair proteins at the site they will be required.
Structural studies of PCNA in H. volcanii have advanced our knowledge of protein adaptation to high intracellular salt concentrations [135,136]. Bacterial and eukaryotic PCNA homologues feature positively charged residues (commonly lysine and arginine) in the two α-helices that make up the inner channel of the ring structure. This facilitates strong interactions between PCNA and negatively charged DNA. Due to the high internal salt concentration of H. volcanii cells, proteins have adapted by increasing their surface acidity (specifically by increasing the percentage of aspartate and alanine residues), along with increasing the number of bound cations and intermolecular ion pairs. The crystal structure of H. volcanii PCNA shows a notable increase in surface acidic residues to alter the overall electrostatic charge distribution of the protein. This enables the protein to function with only two basic residues per monomer in the channel. H. volcanii PCNA also has increased cation binding to potentially facilitate a reduction in positively charged atoms at the pore region with three Na+ ions over two sites in each monomer. These adaptation mechanisms enable PCNA be stable at a wide range of salt concentrations while still facilitating the critical interaction of PCNA with DNA.
Interestingly, the deletion of proteasome-activating nucleotidase A (PanA; HVO_0850) increases the half-life of PCNA, demonstrating that H. volcanii PCNA is a target of proteasomal degradation [139,140]. This study, using pulse-chase labelling, immobilized metal affinity chromatography (IMAC) and immunoprecipitation, is one of the first to demonstrate any post-translational regulatory mechanisms during DNA replication in H. volcanii. It is suggested that post-translational phosphorylation events may also target H. volcanii PCNA as these same techniques purify phosphopeptides. Significant work needs to be carried to understand the intricacy of post-translational events occurring in H. volcanii.
2.2.2.4. DNA polymerases
Replicative DNA polymerases (DNAPs) function in a 5′ to 3′ manner, extending RNA primers to replicate the genome. Due to their directionality, synthesis of the leading strand is a continuous process, requiring only a single priming event, while the lagging strand must be synthesized discontinuously as Okazaki fragments.
Based on amino acid sequence, DNAPs were assigned to six main families: A, B, C, D and Y [141]. More recently, reverse transcriptase (RT) enzymes have also been defined as DNA polymerases of a separate novel family [142]. Replicative DNAPs used in each of the three domains differ, spreading across families A, B, C and D [143]. The identity and role of bacterial and eukaryotic replicative polymerases are relatively well defined. Although the families of DNAPs used by archaea have been identified, the definition of which replicative polymerase acts on which strand still remains a matter of contention.
Genome replication in bacteria is reliant on Pol-III (Family C polymerase) [141]. Two copies of Pol-III replicate both the leading and lagging strands simultaneously. The Pol-III core is tightly associated with the replisome via interactions with both clamp loader γ and clamp protein ß. Alongside the catalytic subunit, Pol-III also encodes subunits possessing 3′–5′ exonuclease proofreading activity. Gram-negative bacteria with a low GC content use two distinct copies of Pol-III, named PolC and DnaE, for leading and lagging strand synthesis, respectively [144,145]. Bacteria also encode family A polymerases, such as Pol-I. These function primarily in the processing and maturation of Okazaki fragments and removal of RNA primers [146].
Eukaryotes can encode up to 15 family B DNA polymerases. The main eukaryotic replicative DNAPs fall within this family, named Pol-α, Pol-ε and Pol-δ. These are all multi-subunit enzymes containing a catalytic core identifiable as a family B polymerase, alongside various accessory domains depending on the polymerase [147]. PrimPol generates a short RNA primer, which is then extended for approximately 40 nucleotides by low-fidelity Pol-α [68,148]. The bulk of synthesis is completed by high-fidelity replicative polymerases Pol-ε and Pol-δ [67,149,150].
Interestingly, there is a phylogenetic divide in the distribution of DNAP families in the archaea. While all archaea possess family B polymerases, archaeal-specific family D DNAPs are absent from crenarchaeal species. Work is beginning to elucidate the roles and functions of these polymerases but the question of which polymerase(s) acts at the leading and/or lagging strand remains under dispute.
Archaeal family B polymerases share homology with the catalytic subunit of family B replicative polymerases in eukaryotes [151,152]. Archaeal family B polymerases have been isolated, with some now being routinely used for PCR applications [153]. Three groups of archaeal PolB polymerases exist, historically termed PolB1, PolB2 and PolB3 [154]. PolB1 and PolB3 are active polymerases, while PolB2 proteins generally carry disrupted catalytic and exonuclease domains which can result in either an active or inactive PolB2 protein [154–156]. A single species can encode single or multiple copies of PolB, with all archaea encoding at least one PolB polymerase. It is usually present as a single protein, with one polypeptide encoding both the catalytic and proofreading activities; the exception is Methanothermobacter thermautotrophicus, where PolB is encoded by two polypeptides [154,157]. The distribution of specific PolB proteins changes throughout the archaeal domain; PolB1 is missing in Euryarchaeota and PolB3 is missing in Thaumarchaeota, while members of the PolB2 group are scattered across archaea [154]. Several groups of archaea carry multiple inteins within PolB3 genes, sometimes up to three per gene [158]. Inteins are selfish genetic elements that insert themselves into a coding sequence and self-splice once translated; they typically encode an endonuclease that propagates further intein insertions [159,160]. Across species carrying PolB3 proteins, intein insertion sites are generally conserved; however, some are lineage-specific [87,158].
Since Crenarchaeota species possess only family B polymerases, it is hypothesized that PolB alone must be capable of both leading and lagging strand synthesis [154,161]. However, crenarchaea typically possess multiple family B polymerases; it is possible that the multiple PolB polymerases within a strain have gained specialized functions and act on different strands. This is known to be the case for S. solfataricus: PolB1 (Dpo1) has been implicated in leading strand synthesis, while PolB3 (Dpo3) has been for lagging strand synthesis.
Archaeal family B DNAPs generally feature a polymerase core (made up of three domains; palm, fingers and thumb), an N-terminal 3′–5′ exonuclease domain and an uracil-recognition domain specific to archaea [154,162,163]. The uracil-recognition domain provides archaea with a unique damage sensing mechanism whereby the polymerase scans ahead of the catalytic site, pausing at misincorporated uracil or hypoxanthine moieties that have escaped canonical repair by uracil-N-glycosylase [164,165]. PolB is capable of extending DNA-primed templates efficiently; however, it struggles to extend RNA primers [166]. This suggests that the inherent DNA polymerase activity of archaeal primases or, in non-crenarchaeal species, family D polymerases are used to extent RNA primers with a short DNA template, prior to handover to PolB.
Recent studies have revealed that PolB is not essential for viability in all archaea, but can be deleted in some euryarchaeal species (which also encode PolD). In Thermococcus barophilus, T. kodakarensis and M. maripaludis, it has been shown that PolB is dispensable and PolD alone is essential [167–169]. The ability to delete PolB but not PolD in these species suggests PolD has the ability to carry out both leading and lagging strand replicative DNA synthesis, while PolB may carry out DNA synthesis as part of DNA repair. Cells of T. kodakarensis deleted for PolB have been shown to be sensitive to gamma irradiation, consistent with the suggestion that PolB carries out DNA synthesis during homologous recombination (which would be used to repair DNA double-strand breaks) [170]. Recent in vitro reconstitution studies in P. furiosus have shown that both PolB and PolD are capable of extending RadA recombinase-primed recombination intermediates [171], but that PolB was more efficient than PolD. This activity, of extending a D-loop recombination intermediate, is consistent with the role of PolB as a DNA repair polymerase.
However, the ability to replicate in the absence of PolB is not true of all euryarchaea. In the halophile Halobacterium sp. NRC-1, both PolB and PolD are essential for cell viability [172] and similar findings have been made in H. volcanii (T.A. 2020, unpublished). It is possible that in some species PolB has gained a novel role, or that the high ploidy associated with halophiles increases demand on replication proteins in general. Further work is required to explain the differing requirements for DNA polymerases (specifically PolB) within the euryarchaea.
The family D DNAPs were initially discovered in P. furiosus by the Ishino laboratory, with the discovery changing the classification system for DNA polymerases [161,173,174]. Family D polymerases form heterotetramers, encoded by subunits DP1 and DP2. DP1 is a small subunit with 3′–5′ proofreading activity and is structurally similar to the exonuclease domain of eukaryotic family B polymerases, while DP2 is the catalytic subunit [175,176]. It has been shown that interaction between DP1 and DP2 is required for PolD to achieve the maximum polymerase and exonuclease activities [173,177]. DP1 is made up of a ssDNA-binding OB fold and 3′–5′ exonuclease domain (from the metallophosphatase MPP family), which functions in the proofreading and removal of erroneously incorporated nucleotides during DNA synthesis [105,178]. The catalytic fold of this calcineurin-like phosphodiesterase family subunit has recently been shown to be specific to family D polymerases [179,180].
Structurally, family D polymerases display a close resemblance to RNA polymerases (RNAPs) [181,182]. The recent publication of a crystal structure of PolD elucidated this link in further detail [175]. While DP1 shows similarity to non-catalytic subunits of eukaryotic family B polymerases, DP2 shows homology to the two-DPBB (double-psi beta barrels) ‘two-barrel’ superfamily of polymerases [175]. Members of the two-barrel superfamily include both DNA- and RNA-dependent transcriptases, along with RNA silencing RNAPs and atypical viral RNAPs [179,181–183]. PolD is the first DNAP to be placed within this superfamily, extending the repertoire of known catalytic folds able to perform DNA synthesis [151,184]. The evolutionary history of replication posits that RNA was used as a genetic material prior to DNA [143], leading to the suggestion that PolD may be the ancestral replicative DNA polymerase of the last universal common ancestor (LUCA) [184].
Early studies in Pyrococcus showed that archaea-specific family D polymerase PolD can efficiently extend both RNA and DNA primers [173]. More recently, it has been shown that PolD can extend RNA primers with a greater efficiency than PolB [166]. Given this information, it has been theorized that in species encoding both PolB and PolD, PolD carries out preliminary synthesis from the RNA : DNA primer before handing over to PolB for the bulk of synthesis, akin to the mechanism seen in bacteria with Pol-I and Pol-III. However, questions remain regarding this mechanism. If PolD is the lagging strand polymerase, strand displacement activity would be required to remove the primers associated with Okazaki fragments on the lagging strand. Currently, this has only been shown in P. abyssi [185]. PolB has been shown to have strand displacement activity, implicating it in Okazaki fragment processing [166].
The RNA extension activity of PolD, and its processivity, requires stimulation from PCNA [175,185]. Interaction of PolD and PCNA occurs at multiple sites throughout both DP1 and DP2 subunits, including a conserved PIP motif encoded at the C-terminus of DP2. Studies on Thermococcus species have implicated a role for PolD at the replication fork; the DP1 exonuclease subunit associates with the GINS-GAN complex via interaction with GINS51. However, this interaction inhibits the exonuclease activity of PolD. This exonuclease activity may have a function elsewhere or may be used in the removal of replication components [114]. Recently, the three-dimensional structure of the PolD-PCNA-DNA complex in Thermococcus kodakarensis was determined using single-particle cryo-electron microscopy (EM). It was shown that a glutamate residue at position 171 of PCNA mediates the interaction with the DP1 and DP2 subunits, locking the PolD structure into a conformation that is competent for enzymatic activity [186].
As a euryarchaeon, H. volcanii contains homologues of both family B and family D DNA polymerases. Two PolB homologues are found; one is an active polymerase and one is predicted to be inactive. The active PolB, PolB1 (HVO_0858), is a member of the PolB3 family of polymerases common to euryarchaea, while the predicted-inactive PolB2 (HVO_A0065) is a member of the PolB2 group associated with often inactivated polymerases [87,156]. Little work has been carried out on H. volcanii PolB1 thus far; a structural analysis of the role of intein present in the C-terminus of PolB1 and its associated homing endonuclease (HEN) showed the loss of the intein sequence from the polB1 gene resulted in no growth defects, indicating this sequence has no active role in H. volcanii [187]. An association between a RadA recombinase-like gene and PolB2 has been observed in Sulfolobales [47]. The polB2 gene of H. volcanii is located near a radA-like gene, indicating a possible link between PolB2 group polymerases and DNA repair; however, the deletion of the polB2 gene (but not the polB1 gene) is possible in H. volcanii (T.A. 2020, unpublished).
H. volcanii encodes a family D polymerase PolD, consisting of subunits DP1 and DP2. The gene encoding the small exonuclease subunit DP1 (HVO_0003) is located in close proximity to oriC1 and the gene encoding Orc1, while the gene for the large catalytic subunit DP2 (HVO_0065) is located distal to the origin. Both DP1 and DP2 are essential for PolD activity; the two subunits stimulate the activity of one another, as seen for Pyrococcus species [177]. Sequence and domain analyses show that DP1 and DP2 are similar to other euryarchaeal family D polymerase subunits, and DP2 contains a C-terminal PIP domain for interaction with PCNA [87]. Thus far, little work has been carried out into the function of PolD in H. volcanii, apart from establishing that PolD is essential for cell viability (T.A. 2020, unpublished).
2.2.2.5. Single-stranded DNA-binding proteins
ssDNA-binding proteins (SSBs) play a central role in DNA replication, recombination and repair across all domains of life but share limited sequence conservation [188–193]. They function to coat ssDNA exposed during DNA replication, protecting it from degradation or chemical modification. In addition, SSBs can assist in homologous recombination by inhibiting secondary structure formation on ssDNA [194,195] or by interacting with RadA recombinase to promote strand exchange, as has been observed in the euryarchaeon P. furiosus [196]. Consistent with their role in DNA replication, SSBs are generally essential for viability in bacteria [197,198], eukaryotes [199,200] and archaea [201,202].
Bacteria use homotetrameric SSB, which forms nucleoprotein filaments along ssDNA. Eukaryotes bind ssDNA with hetero-trimeric replication protein A (RPA), which is structurally and functionally analogous to bacterial SSB [203]. Depending on species, the ssDNA-binding protein in archaea can be bacterial-like (SSB) or eukaryotic-like (RPA). For example, S. solfataricus uses a protein structurally akin to SSB [204], while P. abyssi encodes a heterotrimer showing homology to eukaryotic RPA [196]. A group of 10 species of Crenarchaea, belonging to the clade Thermoproteales, lack a canonical SSB; instead they encode a protein termed ThermoDBP that supplies the essential ssDNA-binding activity in the absence of SSB [205].
H. volcanii encodes three homologues of a eukaryotic-like SSB: RPA1 (HVO_1338), RPA2 (HVO_0519) and RPA3 (HVO_0292) [201]. All three RPA homologues contain OB folds that facilitate DNA binding, with each OB-fold consisting of five ß-sheet strands folded into a barrel-like structure. The binding of this barrel around ssDNA stabilizes the DNA and prevents attack by nucleases. Although structurally similar, each RPA protein has a unique function and they do not form a hetero-trimeric complex as seen in P. abyssi [196,206]. RPA2 is the only homologue essential for cellular survival, while RPA1 and RPA3 are both non-essential [201]. Formation of RPA2 foci has been seen in cells treated with aphidicolin, an inhibitor for PolB, indicating an essential role for RPA2 in overcoming replication stress [207,208]. RPA2 foci formation has also been observed in cells treated with ultraviolet (UV) light, suggesting an additional role for RPA2 in DNA repair.
Deletion of the gene encoding RPA1 results in no increase in sensitivity of cells to DNA damaging agents, indicating no major role in DNA repair [201]. RPA1 has been genetically linked with RPAP1 (RPA-associated protein 1; HVO_1337), an OB-fold protein predicted to assist RPA and facilitate ssDNA binding. RPA1 and RPAP1 are both located in the same operon with co-purification studies indicating an in vivo association between the proteins [209]. By contrast, cells deleted for RPA3 are sensitive to DNA damaging agents including UV radiation, phleomycin and methyl methane sulfonate (MMS). This indicates a role for RPA3 in DNA repair, in particular double-strand break (DSB) repair, as the aforementioned agents promote DSB formation [201]. Akin to RPA1, RPA3 is also encoded within an operon alongside an RPA-associated OB-fold protein, RPAP3 (HVO_0291). A similar increased sensitivity to multiple DNA damaging agents was seen upon the deletion of RPAP3 [209]. Whether this role in DNA repair also extrapolates to DNA replication is yet to be investigated; given that RPA3 is not essential, any role in DNA replication is likely to be minor.
RPA homologues present in the closely related species Halobacterium have also been implicated in DNA repair and deletion mutants show increased sensitivity to various DNA damaging agents [202]. Halobacterium salinarium possesses 5 SSB homologues (2 eukaryotic-like, 2 bacterial and one euryarchaea-specific). Upon deletion of these homologues, cells display increased sensitivity to infrared (IR) and UV radiation, and mitomycin C (MMC) treatment, with the strain deleted for the euryarchaeal-specific RPA homologue being most sensitive [202]. Despite the high degree of homology between these two halophiles, there are functional differences regarding DNA replication and repair mechanisms; H. volcanii RPA proteins are implicated only in DSB repair, while Halobacterium homologues appear to be playing a role in multiple DNA repair pathways [201,202,209].
2.2.2.6. Other replisome components
Lagging strand maturation requires the removal of RNA primers on Okazaki fragments; the resulting gap is filled, and nicks are ligated to give a continuous DNA strand. RNase H proteins act to remove RNA primers associated with Okazaki fragments, flap endonucleases remove any flap structures generated in displacing primers, and DNA ligase seals any remaining nicks to give a complete product. In eukaryotes, gap filling is an early event, occurring prior to removal of the CMG complex from dsDNA [210].
The RNase H family of proteins acts to remove RNA primers from fully replicated Okazaki fragments; they also degrade R-loops (RNA-DNA hybrids) in a sequence-independent manner. RNase H enzymes are evolutionarily conserved and although not essential for cell survival, their deletion leads to strong sensitivity to DNA damaging agents in eukaryotes [211,212]. They can be categorized into three groups: RNase H1 proteins are present in bacteria, archaea and eukaryotes, and in reverse transcriptases from retroviruses and retroelements; RNase H2 proteins are present in all domains of life, usually together with a RNase H1 [213]; RNase H3 proteins are found in some bacteria and archaea, and show structural similarities to RNase H2 [213]. RNase H proteins generally share low sequence similarity, but both RNase H1 and RNase H2 group proteins use a highly conserved two metal ion catalytic mechanism [214,215]. All archaea encode an RNase H2 similar to the eukaryotic enzyme [213], together with an RNase H1 or RNase H3 [216].
H. volcanii possesses encodes five RNase H homologues, three of type 1 (RNase H-E, HVO_0732; RNase H-A, HVO_2438; RNase H-C, HVO_A0463) and a single type 2 protein (RNase H-B HVO_1978). RNase H-D (HVO_A0277) does not fit clearly into either group and its function remains unknown. Type 2 RNase H-B is non-essential in H. volcanii. It encodes a C-terminal PIP domain, implicating this RNase H at the replication fork. In vitro reconstitution of RNA primer removal in P. abyssi has implicated RNase H in cutting the RNA : DNA hybrid at Okazaki fragments as an early step, allowing subsequent strand displacement by PolB/PolD [217]. The roles of the three type 1 RNase H genes in H. volcanii remain unknown and warrant further study.
Fen1 (Flap endonuclease 1) is a structure-specific endonuclease that acts to remove 5′ overhangs generated during Okazaki fragment maturation as a result of strand displacement. Replicative DNA polymerases will then act on the newly generated 3′ end to fill the gap and DNA ligase will seal the nick. The eukaryotic polymerase responsible for final synthesis (gap filling) still remains a controversial topic. In both Caenorhabditis elegans and Xenopus laevis, Pol-ε, but not Pol-δ, has been shown to interact with the post-replication CMG complex [218]. DNA incorporation studies in P. abyssi have shown a reduced incorporation of nucleotides in the absence of Fen1, indicating a possible role for Fen1 in archaeal DNA replication [137].
H. volcanii encodes a single Fen1 homologue (HVO_2873), with deletion of this nuclease being viable [219]. This is in contrast with Halobacterium sp. NRC-1 where its single Fen1 homologue, rad2, is essential [172]. Rad2 has been implicated as a key player in UV damage repair in Halobacterium [220]; similarly, H. volcanii strains lacking Fen1 display increased sensitivity to UV and DNA cross-linking agents [219]. The fact that fen1 can be deleted from H. volcanii suggests redundant systems are in place to deal with DNA damage in this species.
2.3. Termination of replication
Termination of DNA replication involves the convergence of two replication forks, either at random or at a defined location(s) depending on the organism, followed by dissociation of the replisome and decatenation of the chromosomes to allow correct segregation into daughter cells [221].
2.3.1. Sites of termination
In bacteria, specific regions on the chromosome called termination (Ter) sites dictate where replication is halted, they are generally located at the furthest point from the origin. Ter sites act as a polar block to the DNA replication machinery, causing the replication fork(s) to stall within the defined termination region. Up to 10 ter, sites (named TerA-J) are bound by the DNA replication terminus-binding protein Tus in a specific orientation [222]. The replication fork is able to bypass 5 Tus-bound Ter sites with the ter sites terminating the replisome. Unlike bacteria, eukaryotes do not have sequence-defined termination sites. Termination events occur midway between two origins, with more active origins displaying more defined regions of termination [223–225]. Some studies have indicated the convergence of CMG complexes is a key step in the initiation of termination in eukaryotes [226].
While archaea share a circular genome architecture with bacteria, their chromosomes lack defined termination sites [60,227]. Instead, the termination of replication appears to occur in ‘zones’ where replication forks meet randomly, as in eukaryotes. This is visible on replication profiles (MFA plots), where termination zones map as broad valleys; this contrasts with the sharp ‘canyons’ seen for bacteria, which represent defined termination sites. Work carried out on Sulfolobus species has shown replication to be asynchronous, suggesting both number of origins and rate of initiation may affect where termination occurs [228].
Little is known about the details of termination of DNA replication in H. volcanii. The broad termination zones seen equidistant to origins of replication on MFA plots suggest that H. volcanii does not encode defined ter sites, and the relocation of such termination zones upon deletion of origins confirms there is no sequence specificity to termination in this species [60].
2.3.2. Dissociation of the replisome
Prior to completion of replication, components of the replisome must be removed to prevent over-replication and to allow segregation of the newly synthesized DNA. During DNA synthesis in eukaryotes, the CMG replicative helicase complex encircles ssDNA, opening the helix to allow processive elongation. When converging with another fork, the CMG complex will bypass the CMG complex of the oncoming replisome and switch from binding ssDNA to binding dsDNA; the location of the CMG complex on the leading strand of both replisomes ensures there is no steric clash, and no decrease in synthesis rate is observed at termination sites in Xenopus [210]. The switching of binding of the CMG complex from ssDNA to dsDNA acts as a marker for downstream events. Polyubiquitylation of MCM subunit MCM7 by specific E3 ligases leads to unloading of the CMG complex by the activity of ATPase Cdc48/p97 [229,230]. Dissociation of MCM from the heart of the replisome is hypothesized to cause dissociation of the entire replisome. However, some predictions have been made that it is the unloading of PCNA, and its numerous associated proteins, which results in an unloading of further replicative factors. Such a model for coordinating termination of replication has been proposed in archaea for S. solfataricus, whereby PCNA coordinates the termination activity of PolB1, flap endonuclease 1 (Fen1) and DNA ligases, due to the presence of PIP domains on these proteins [231].
Re-replication events are not seen in eukaryotes, indicating the presence of strict regulatory mechanisms in termination; the use of ubiquitylation adds a layer of complexity to eukaryotic termination, which is not seen for bacteria [232]. Little is understood regarding the removal of replication components in archaeal species, including H. volcanii, but preliminary evidence suggests a system more complex than that of bacteria. It remains to be elucidated if post-translational modifications play a role in archaeal termination and replisome unloading, but given that homologues of Cdc48/p97 and ubiquitin-like proteins are both found in H. volcanii [233], this remains a distinct possibility.
2.3.3. Decatenation and resolution
Unwinding of DNA during replication will lead to overwinding of the duplex ahead of the replication fork, forming supercoils. If left unresolved during replication, this increased torsional stress would prevent the replication fork from proceeding along with the duplex and at termination would prevent equal segregation of DNA to daughter cells.
In bacteria, topoisomerases act to control the level of torsion in DNA during replication [234]. Type II topoisomerases are important in termination: DNA gyrase acts to relieve positive supercoils formed as a product of DNA unwinding while TopoIV resolves pre-catenanes, allowing fork convergence to occur and to be resolved successfully [235,236]. Following the resolution of torsional stress, RecG translocase and RecBCD helicase-nuclease resolve overlapping sequences at the terminus, giving a product suitable for dissolution and segregation [237,238]. Should an odd number of crossover events occur, chromosome dimers can be created [239]. Such structures must be separated prior to segregation to ensure each daughter cell receives a full genome complement. In bacteria, Xer site-specific recombinases act at specific loci named Dif (differential induced filamentation) sites, which are in close proximity to ter sites, and resolve chromosome dimers into monomers [240].
Argonaute family proteins (AGO) are found across all domains of life. In eukaryotes, short RNA guides act to target AGO proteins against transposons and viruses while in bacteria, AGO proteins have been shown to defend against transformation by DNA plasmids. Recent work has implicated Argonaute protein in termination and decatenation of DNA replication in the bacterium T. thermophilus [241]. When DNA gyrase is inhibited, Argonaute is capable of completing DNA synthesis and ensuring correct decatenation of the chromosome [241]. In the absence of both DNA gyrase and Argonaute activities, chromosome resolution does not occur [241]. Such a critical function for AGO proteins in DNA replication has not previously been observed, and further work is warranted to see if AGO proteins act in chromosome resolution in archaea.
For eukaryotes, the completion of DNA replication will lead to daughter molecules that are catenated to one another. Any pre-catenanes present would also be converted to full catenanes for processing [242]. The specific details of resolution in eukaryotes remain unknown; however it is believed that topoisomerase II (TopoII; type II topoisomerase) is essential for the process [243,244]—inactivation of TopoII leads to stalling in G2 phase, resulting in a build-up of catenanes and failure to complete replication [245].
Although no Ter sites or Tus homologues have been identified in archaea, homologues of Dif and Xer have been identified [68,227,246]. Some archaeal species (e.g. Thermococcus) possess Dif sequences at zones of termination, suggesting coordination of chromosome monomerization and replication termination [246]. However, in Sulfolobales, Dif sites are situated far away from termination zones, suggesting that these two processes may be less tightly coupled [227].
H. volcanii possesses multiple XerC/D-like homologues, suggesting the possibility of Dif sites. Of the 12 xerC/D genes, four have been deleted without impacting viability (HVO_1422, deleted by Uri Gophna, HVO_2259, HVO_2273 and HVO_2290 deleted by T.A.; T.A. 2020, unpublished). Whether these XerC/D-like enzymes have a role in the termination of replication remains to be determined. The presence of broad zones of termination coupled with the presence of Dif sites hints at archaea carrying both bacterial- and eukaryotic-like mechanisms of chromosome resolution and termination.
Topological stress in H. volcanii can be imagined to be a large problem. There are approximately 20 genome copies within each cell, which are replicated asynchronously due to the lack of a defined cell cycle. Relieving superhelical torsion is carried out by the action of three topoisomerases: DNA topoisomerase IA (TopoIA; HVO_0681), DNA topoisomerase VI comprising subunit A (HVO_1570) and B (HVO_1571), and DNA gyrase comprising subunit A (HVO_1572) and B (HVO_1573). The laboratory strain of H. volcanii displays sensitivity towards novobiocin, an inhibitor of DNA gyrase, indicating that DNA gyrase is essential for viability [247,248]. Both subunits of DNA TopoVI have also been shown to be essential (T.A. 2020, unpublished). Further work is needed to assess the interplay of the different topoisomerases in H. volcanii and how they act together to resolve chromosome structures for segregation.
Following decatenation, RNA primers on Okazaki fragments are removed; the resulting gap is filled by replicative DNA polymerases, and nicks are ligated to give a continuous DNA strand. RNase H proteins act to remove RNA primers associated with Okazaki fragments; flap endonucleases remove any flap structures generated in displacing primers, and ATP-dependent (and in some species, NAD-dependent) DNA ligases (to be discussed in detail later) seals any remaining nicks to give a complete product. In eukaryotes, gap filling is an early event, occurring prior to removal of the CMG complex from dsDNA [210].
2.4. DNA repair
Environmental and endogenous factors threaten the genome integrity of all living organisms. Damage to DNA can lead to mutagenesis, genome instability, senescence and cell death [249]. The majority of DNA damage lesions arise from endogenous sources during normal cellular metabolic processes, generating oxidation, hydrolysis and alkylation damage, along with the insertion of mismatched DNA bases. Environmental agents such as UV light, ionizing radiation and various chemical agents generate base lesions including the deamination of cytosines, adenines and guanines, depurination of bases, oxidative damage, as well as DNA double-strand breaks (DSBs) [249–253].
While evolution is driven by rare advantageous mutations, efficient DNA repair is a requirement of all forms of life as large amounts of unrepaired damage cannot be tolerated. This is especially true for many archaeal species, which inhabit demanding environments. Extremes of salinity, temperature and pH can increase the load of DNA damage faced by these organisms and thus they require robust methods of repair. A recent study has estimated the genome-wide mutation rate and spectrum in H. volcanii; the base substitution rate of 3.15 × 10−10 per site per generation is similar to that seen in mesophilic species [254].
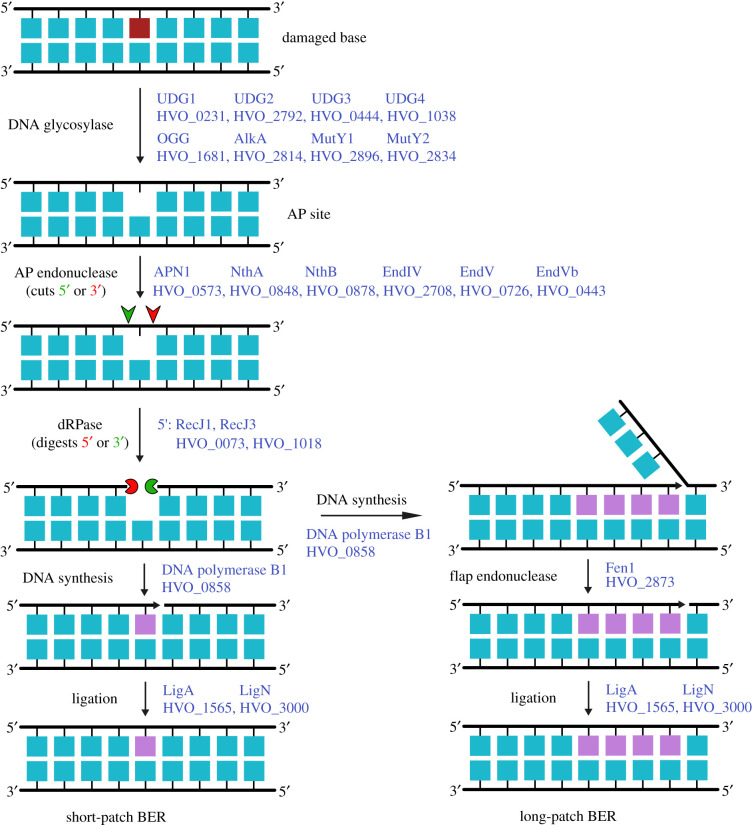
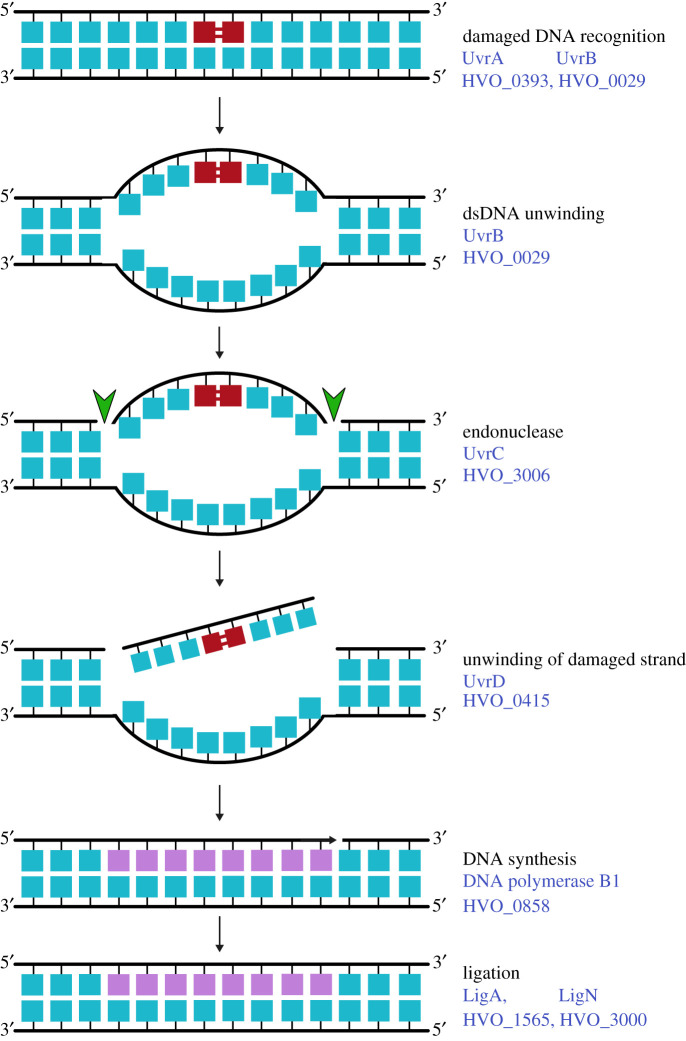
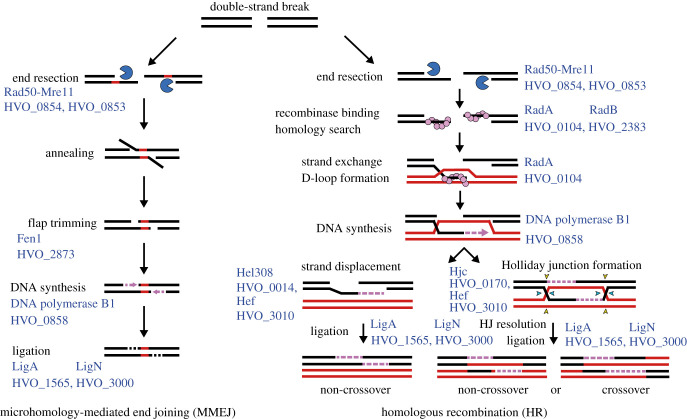
Cells have developed a plethora of DNA repair mechanisms, and generally, these repair mechanisms differ depending on the type of DNA damage incurred [255]. While a small number of specific chemical modifications can be repaired by a single protein without a requirement for cutting of the DNA backbone, a process known as direct repair, mismatched and damaged bases are more commonly repaired by one of four excision pathways: MMR, BER, nucleotide excision repair (NER) and ribonucleotide excision repair (RER) (table 1).
One of the most harmful DNA lesions is the DSB, since it affects both strands of the duplex. If unrepaired, DSBs can give rise to large-scale genome rearrangements, chromosomal translocations, significant mutagenesis and cell death. Due to the potential danger of DSBs, the most complex DNA repair mechanisms are responsible for repairing this type of DNA damage. The major DSB repair pathways are HR, non-homologous end joining (NHEJ) and microhomology-mediated end joining (MMEJ). HR is a high-fidelity mechanism that generates error-free products, while NHEJ and MMEJ pathways are quicker but error-prone processes, which can result in deletions, insertions and translocations [256–260].
Generally, DNA repair processes are highly conserved throughout evolution. Archaea share many components of their DNA repair machinery with eukaryotes [253,261], but halophilic archaea also carry numerous enzymes that have been acquired by LGT from bacteria. Furthermore, halophilic archaea, which inhabit UV-intense hypersaline environments, have gained further strategies to prevent damage. Polyploidy provides an evolutionary strategy for DNA damage resistance [65,262], an option that is not usually available in organisms like Saccharomyces cerevisiae [263], and many halophiles use photoprotective membrane-associated pigments such as carotenoids [264].
2.5. Direct repair
2.5.1. Photolyases
Halophilic archaea experience a significant dose of UV light in their natural environments and need efficient mechanisms to repair UV-induced DNA damage. Photoreactivation is a light-dependent direct repair mechanism catalysed by photolyase. In a single step, photolyase is able to reverse cyclobutene pyrimidine dimer (CPD) and 6–4 pyrimidine-pyrimidine photoproduct (6–4PP) lesions formed as a consequence of solar UV radiation [265,266]. Photoreactivation is the most important repair mechanism for surviving high doses of UV light in nature [267].
Photolyases are present in several archaea and share homology with those present in bacteria and eukaryotes, suggesting that this mechanism arose early during evolution [255,268,269]. Few archaeal species encode more than one photolyase homologue; Halobacterium species are known to encode two as a result of a gene duplication event, encoded by genes phr1 and phr2 [267]. H. volcanii also encodes two photolyase homologues, phr1 and phr2 (HVO_2911 and HVO_2843 respectively), alongside an uncharacterized photolyase-related gene phr3 (HVO_1234). The requirement for more than one photolyase in these halophiles may be a consequence of the high amounts of UV damage experienced in their environment. However, genetic studies have shown not all phr homologues within a species are active, suggesting some redundancy [270].
2.5.2. Methyltransferases
Restriction-modification (RM) systems have evolved to protect cells from invading DNA; RM systems comprise restriction endonucleases (RE) and DNA methyltransferases (MTases). RM systems are present in bacteria and archaea and allow the cell to differentiate between its own methylated DNA and foreign unmethylated DNA, which can be recognized and digested by the RE [271,272]. H. volcanii encodes a putative type IV Mrr RE (HVO_0682), which cleaves DNA that is methylated at GAm6TC sites; this includes Dam-methylated plasmid DNA extracted from Escherichia coli, hindering transformation protocols. Gene deletions of Mrr RE have been carried out to resolve such limitations [36].
MTases encoded in the absence of a cognate RE, known as orphans MTases, play essential roles in cellular processes such as DNA replication, DNA repair and gene expression [273]. These defence mechanisms have been extensively characterized in bacteria but are only poorly defined in archaea. The use of deletion mutants of genes predicted to be methyltransferases in combination with single-molecule real-time (SMRT) sequencing has allowed the detection and mapping of methylated bases throughout the genome [274]. Two methylated motifs were detected in the H. volcanii genome: Cm4TAG and GCAm6BN6VTGC (where B stands for C, G or T, V stands for A, C or G, and N stands for any base). Genes responsible for DNA methylation in H. volcanii include HVO_A0006, HVO_0794 and HVO_A0237 that methylate cytosine at Cm4TAG, and the type I RM operon rmeRMS (HVO_2269–2271) that methylate adenine at GCAm6BN6VTGC motifs [275].
2.6. Excision repair
2.6.1. Base excision repair
Base excision repair is conserved across all domains of life (figure 2 and table 1). The canonical pathway involves the action of a DNA glycosylase specific for a damaged base, which cleaves the N-glycosyl bond between the damaged base and the sugar, generating an apurinic or apyrimidinic (AP) site. Most DNA glycosylases are mono-functional but some are bi-functional with a coupled β-lyase activity that cleaves 3′ of the AP site by β-elimination. Alternatively, the AP site is cleaved on the 5′ side by an AP endonuclease that acts by hydrolysis. The AP product is then degraded by a 3′ or 5′ phosphodiesterase, respectively, leaving a single-nucleotide gap with a 3′ hydroxyl and either a 5′ deoxyribose phosphate (5′dRP) or a 5′ phosphate. The generation of the 5′ end allows a family X DNA repair polymerase (Pol-β in eukaryotes) to begin synthesis, filling the gap. While 5′ phosphate can only be repaired by short-patch repair, 5′dRP can be repaired by both short- and long-patch repair [276]. Short-patch repair is where the family X polymerase, Pol-β, synthesizes the single nucleotide, removing the 5′dRP with its inherent lyase activity. Long-patch repair is more complex and involves the insertion of a further 4–6 nucleotides to generate a flap structure that displaces oncoming DNA, which is subsequently removed by flap endonuclease (FEN-1 in eukaryotes) [277,278]. While long-patch repair still uses Pol-β, family B DNA polymerases Pol-δ and Pol-ε have also been implicated in this mechanism of BER.
Figure 2.
Base excision repair. The damaged base (red) is recognized and removed by DNA glycosylases, which cleave the N-glycosyl bond between the damaged base and the sugar to generate an apurinic or apyrimidinic (AP) site. AP endonucleases cleave 5′ of the abasic site or ß lyase cleaves 3′ of the site, and the backbone is removed by phosphodiesterases. Short-patch BER uses DNA polymerases (PolB1; HVO_0858 in H. volcanii) to insert the missing nucleotide (purple) with DNA ligases (LigA; HVO_1565 or LigN; HVO_3000) linking the newly synthesized nucleotide to the sugar backbone. In long-patch BER, DNA polymerases insert 2–6 nucleotides at the gap to generate a flap structure. Flap endonuclease Fen1 (HVO_2873) cleaves the displaced strand and DNA ligases seal the DNA backbone.
In some archaea, evidence supports the family B polymerase PolB as the candidate for BER synthesis [276]. Among replicative DNA polymerases, PolB has been described to be involved in DNA repair, but not the archaea-specific family D polymerase PolD [167,170]. In contrast with eukaryotes, few family X DNA polymerases have been identified in archaea thus far, suggesting an alternative method of BER [279]. In archaea, Fen1 is implicated in the removal of RNA primers from Okazaki fragments during lagging strand DNA replication and Fen1 orthologues have been described in both Euryarchaeota and Crenarchaeota [280–284]. To date, only Fen1 from M. thermautrophicus has been shown to be actively involved in BER [285]. The Rad2-family flap endonuclease is essential in Halobacterium sp. NRC-1 [172], potentially offering an alternate flap endonuclease used by archaea for resolution of long-patch repair.
Both long- and short-patch BER result in a nick that is targeted for ligation by a DNA ligase. Bacterial and eukaryotic ligases show no homology. While bacterial ligases are NAD dependent, eukaryotic ligases are ATP dependent. Most archaea possess eukaryotic-like ATP-dependent DNA ligases [286] but haloarchaeal species can encode more than one type of DNA ligase, including bacterial-like NAD-dependent ligases.
In H. volcanii, in addition to the ATP-dependent ligase LigA (HVO_1565), an NAD-dependent ligase LigN (HVO_3000) is present. Halophilic species are known to undergo large amounts of gene transfer, thus it is likely this NAD-dependent ligase has been acquired by LGT from bacteria [287,288]. Neither DNA ligase alone is essential, but a double mutant is inviable, indicating that the two ligases are redundant for their essential function; most likely the ligation of Okazaki fragments during lagging strand DNA synthesis. H. volcanii strains lacking eukaryotic-like ligase ligA show higher UV sensitivity compared with wild type, indicating that this enzyme could play a role in DNA damage resolution in BER and/or NER [288,289].
An alternative excision repair (AER) pathway has been described which is initiated by an endonuclease rather than a glycosylase [290]. Two endonucleases able to nick damage-containing DNA have been characterized: Endonuclease V (EndoV) and Endonuclease Q (EndoQ). EndoQ cuts 5′ of uracil, hypoxanthine and abasic sites [291,292], while EndoV shows a preference for cutting 3′ of lesions [293]. EndoV is widely conserved in bacteria, eukarya and archaea [294], while EndoQ has a much narrower distribution throughout the three domains [292]. Within archaea, EndoQ is found within a subset of the phylum Euryarchaeota, including Thermococcales and numerous methanogenic orders, and is often found in combination with EndoV. This is in contrast with the majority of crenarchaeal and halophilic species, including H. volcanii, which encode only EndoV (HVO_0726). EndoQ from T. kodakarensis and P. furiosus has been shown to be stimulated by interaction with PCNA in vitro [295], which may coordinate its action at the replication fork.
2.6.2. Nucleotide excision repair
Nucleotide excision repair is a DNA repair mechanism that recognizes and removes a large number of different helix-distorting lesions [296,297]. Examples of NER substrates are CPDs and 6–4PPs generated by UV radiation, reactive oxygen species (ROS)-induced base modifications or base adducts created by exogenous chemical agents. NER is the primary mechanism to repair UV-induced DNA lesions in the absence of photoreactivation; because NER is light-independent, it is often referred to as ‘dark repair’ [298]. The basic steps of the process are conserved in all domains of life, but bacterial and eukaryotic NER proteins show very little homology.
The NER pathway in bacteria is catalysed by the UvrABC excision repair machinery: UvrA is responsible for damage recognition, UvrB helicase separates the two DNA strands and UvrC nuclease cuts at both sides of the lesion. Primarily, UvrA : UvrB will scan DNA, with the UvrA subunit recognizing bulky lesions in the template. Upon recognition, UvrA dissociates and UvrC binds UvrB (giving UvrBC). UvrBC will act to cleave up- and downstream of the lesion. UvrD is a superfamily I helicase member and moves in a 3′–5′ direction [296], acting to peel away the excised segment containing the damaged DNA. This permits access to family A polymerase DNA Pol I and DNA ligase, which act to fill and seal the gap, respectively. A small number of archaeal species carry homologues of the bacterial NER proteins; they are primarily found in mesophilic methanogens and halophiles [219,299].
Alongside global genomic NER (GG-NER), transcription-coupled NER (also known as transcription-coupled repair, TCR) forms a sub-pathway of NER. TCR functions to remove RNA polymerase-arresting DNA lesions from the template of actively transcribed genes [300]. Usually, repair by TCR is quicker than that of canonical NER, thus favouring correction of lesions within the transcribed strand of DNA [301]. TCR is initiated when RNA polymerase stalls at a lesion in the transcribed strand of DNA. In bacteria, RNA polymerase is displaced by NER proteins, which are recruited to the site of damage by a transcription-repair coupling factor (TRCF), otherwise known as Mfd protein [300,302].
Eukaryotes use a more complex pathway encoded by 9 proteins (named XPA to XPG). While differing in complexity, eukaryotic NER follows the same general principle as bacterial NER. Akin to bacteria, eukaryotic NER can be split into two sub-pathways: GG-NER and TCR. GG-NER can occur throughout the genome, while TCR is responsible for timely repair of lesions in the transcribed strand of active genes. In eukaryotic GG-NER, damage recognition is carried out by XPC-Rad23B (or by DDB1/2 in heterochromatin) and the DNA is opened by transcription factor IIH (TFIIH), a multi-protein complex containing helicase subunits XPB and XPD [303]. Following opening, binding of XPA and SSB protein RPA results in the recruitment of nucleases ERCC1-XPF and XPG to cleave either side of the lesion (dual incision). Canonical family B replicative DNA polymerases Pol-δ and Pol-ε, along with error-prone family Y translesion synthesis polymerase Pol-κ, have been implicated in gap filling [304], with requirements having been shown to change depending on cell cycle stage [305]. The same applies to the type of DNA ligase used for sealing the nick; DNA ligase IIIα and DNA ligase I have both been linked to NER in eukaryotes [305,306]. In eukaryotic TCR, the stalled RNA polymerase itself acts as the signal for recruitment of NER proteins; the stalled polymerase works in the place of XPC-Rad23B, leading to recruitment of downstream components as previously described.
H. volcanii carries homologues of UvrABC proteins, encoded by genes uvrA (HVO_0393), uvrB (HVO_0029) and uvrC (HVO_3006) (figure 3 and table 1) [35]. The bacterial NER genes are non-essential in H. volcanii and deletion mutants in uvrABC show enhanced sensitivity to UV damage in the absence of photo-reactivating light [219], implicating UvrA, UvrB and UvrC in ‘dark repair’ of UV lesions in H. volcanii. Cells deficient in UvrD (HVO_0415) show no such sensitivity phenotype and it has been proposed that a redundant helicase may substitute for UvrD [219]. Furthermore, uvrABC mutants, but not uvrD, exhibit increased sensitivity to the DNA inter-strand cross-linking agent MMC [307]. Similar results have been shown for other haloarchaeal species, including Halobacterium sp. NRC-1 [265] where deletion of uvrABC genes results in UV sensitivity, indicating that the bacterial Uvr system is required for the repair of UV-induced DNA damage. Intriguingly, Halobacterium also encodes homologues of eukaryotic-like NER proteins XPB and XPD (helicases), as well as XPF (endonuclease).
Figure 3.
Nucleotide excision repair. In H. volcanii, bulky DNA adducts (red) are recognized by UvrA (HVO_0393). UvrB (HVO_0029) initiates DNA unwinding around the damage site through its helicase activity. Incisions 5′ and 3′ to the damaged bases are carried out by the endonuclease UvrC (HVO_3006). Unwinding of the damaged strand is carried out by a helicase such as UvrD (HVO_0415). The remaining gap is filled (purple) by replicative DNA polymerase PolB1 (HVO_0858) and the newly synthesized DNA is attached to the backbone by the activity of DNA ligases (LigA; HVO_1565 or LigN; HVO_3000).
Genetic experiments in H. volcanii have revealed a link between the bacterial NER protein UvrA and NreA (HVO_0734), a member of the archaea-specific Nre family of proteins [307]. Most archaea encode at least one Nre homologue with a C-terminal PIP motif, while some species encode a second protein, NreB, with a less well-defined PIP motif [308]. H. volcanii encodes only NreA, which is not essential but cells lacking NreA are sensitive to MMC treatment. Double deletion mutants of nreA and uvrA are no more sensitive than single mutants, suggesting NreA and UvrABC act in the same pathway in H. volcanii [307]. NreA has been proposed to be involved in the repair of MMC-induced DSBs, acting in combination with the UvrABC NER system, and the interaction of NreA and PCNA has been shown to be essential for this role [307].
The majority of archaeal species, including hyperthermophiles, carry homologues of eukaryotic-like NER proteins, including endonucleases XPF/XPG and helicases XPB/XPD [253,289,309–313]. S. islandicus and T. kodakarensis mutants lacking XPB and XPD helicases are only slightly sensitive to UV radiation, MMC and MMS treatments, indicating that these enzymes do not play an essential role in NER [111,314]. Some archaea carry both bacterial UvrABC-like proteins and an incomplete eukaryotic XP system, for example Methanosarcina mazei [289] and various halophilic species [255]. As previously discussed, H. volcanii encodes homologues of the bacterial Uvr system. However, it also encodes homologues of endonucleases XPF (Hef, HVO_3010) and XPG (Fen1, HVO_2873) [35]. Unlike XPF in eukaryotes, Hef in H. volcanii has been shown to play no role in NER, instead being implicated in the restart of stalled replication forks [219].
TCR, while observed in some archaeal species, does not seem to be universally conserved throughout the domain. The crenarchaeon S. solfataricus does not favour repair of transcribed strands suggesting that the system for TCR in this species (if present) is no faster than that of GG-NER [315,316]. However, the RNAP of euryarchaeon T. kodakarensis has been shown to pause at a variety of DNA lesions suggesting damage recognition by the RNAP itself, akin to mechanisms of TCR in other domains of life [317]. To date, no homologues of bacterial TRCF have been identified in archaea [255]. While TCR is known to occur in halophilic species, the specifics of the mechanism remain unknown. In H. volcanii, TCR has been shown to occur in the absence of UvrA, unlike Halobacterium sp. NRC-1, which is unable to efficiently repair UV damage without UvrA [318]. In the latter species, UvrA protein seems essential for the initial recognition of the DNA damage in both GG-NER and TCR. By contrast, the initial recognition event for TCR in H. volcanii is UvrA independent; this could be performed by the RNA polymerase itself (as seen in eukaryotes and T. kodakarensis) or by an as yet unidentified coupling factor [318,319].
2.6.3. Mismatch repair
The MMR machinery is conserved across bacteria and eukaryotes, but most archaea lack key components of the canonical pathway [255]. In bacteria and eukaryotes, the canonical MutS-MutL MMR pathway is able to detect and repair mismatched nucleotides that arise as a consequence of misincorporation by DNA polymerases [320]. MutS or MutL homologues are absent from many archaeal species [313,321], but are found in halophiles, methanogens and a few other euryarchaea; these species are all subject to LGT and their mutSL genes are thought to have been acquired from bacteria [322–325]. Archaea lacking MutSL proteins show a rate of spontaneous mutation similar to organisms that possess MutSL [321,326], indicating the presence of an alternative MMR mechanism. Furthermore, the deletion of mutS and mutL genes in Halobacterium sp. NRC-1 does not increase the mutation rate [327], suggesting that some archaea may carry more than one active MMR system.
EndoMS (also named NucS) is an endonuclease first characterized in vitro in P. abyssi, where it acts on branched DNA structures containing flapped and splayed DNA [328–331]. EndoMS is present in bacteria of the phylum Actinobacteria and in most members of archaea that lack functional MutSL homologues. EndoMS was initially proposed as a potential NER endonuclease due to its ability to cleave flapped and splayed structures, akin to XPF [312]. Both S. solfataricus XPF and P. abyssi and T. kodakarensis EndoMS/NucS are able to interact with PCNA, and this interaction is required for their specificity and endonuclease activity [329,330,332,333]. PCNA may also assist EndoMS and XPF to access the site of damage and facilitate the following steps of the repair by enabling interactions with other repair proteins.
Ishino et al. [333] have demonstrated that T. kodakarensis EndoMS has the ability to specifically cleave dsDNA containing a base pair mismatch in vitro, indicating a role for EndoMS in a novel MMR pathway. Other structural studies have supported the hypothesis of EndoMS specifically recognizing and binding mismatched bases by a unique dual base flipping mechanism [334,335]. In T. kodakarensis, the endoMS gene is in an operon with the radA recombinase gene [329], but this genetic link is only conserved within the genus Thermococcus. Nevertheless, a functional link between EndoMS and HR has been observed in Sulfolobus acidocaldarius, where EndoMS has been shown to act in HR-mediated stalled fork repair to remove helix-distorting DNA lesions, overlapping with the role of XPF and suggesting a potential role for EndoMS in NER [336]. A new function for NucS was recently described in T. gammatolerans, where NucS is capable of cleaving uracil- and hypoxanthine-containing dsDNA, indicating an alternative to BER for the repair of deaminated bases in this species [337].
Interestingly, some haloarchaeal species encode both NucS and MutSL homologues, including H. volcanii; it encodes four MutS (mutS1a HVO_1940; mutS1b HVO_0552; mutS5a HVO_0191; mutS5b HVO_1354), two MutL (mutLa HVO_1939; mutLb HVO_0551) and one NucS protein (HVO_0486). NucS is not essential and no phenotype is observed in cells lacking nucS [330]. Two of the four mutS genes (namely mutS1a and mutS1b) are located in operons with a mutL partner and are predicted to function in MMR [35]; deletion of these canonical mutSL genes leads to an increase in the spontaneous mutation rate of H. volcanii (T.A. 2020, unpublished). The other two MutS proteins belong to a subfamily that does not seem to be involved in DNA repair in other organisms [255,338]. The possible interplay between MutS-MutL and NucS pathways in H. volcanii remains to be elucidated.
2.6.4. Ribonucleotide excision repair
Ribonucleotides (rNMPs) that are misincorporated into genomic DNA are recognized and removed by the RER pathway. Archaeal D family DNA polymerases have been shown to incorporate 1 rNTP every approximately 1000 bases, and archaeal B family DNA polymerases every approximately 2500 bases [339,340]. RNase H2 creates a nick 5′ to the misincorporated rNMP. The 3′ end of this gap is displaced by DNA polymerases and cleaved by Fen1. The remaining gap is then sealed by the activity of DNA ligases, mirroring the mechanism for Okazaki fragment maturation. In T. kodakarensis, this pathway has been identified using computational methodology [340]. The activity of RNase H2 proteins, Fen1 and DNA polymerases in RER in H. volcanii remains to be demonstrated.
2.7. Translesion synthesis
Family Y DNA polymerases have a specialized function, whereby they are able to bypass various forms of DNA damage that block DNA synthesis by canonical replicative polymerases; this process is known as translesion synthesis (TLS) [341]. Family Y DNA polymerases are conserved across the three domains of life [342]. They have a larger and more accommodating active site with smaller thumb and fingers domains, which make little or no close or specific contacts with the nascent base pair [343]. The closed conformation of the fingers domain suggests that the canonical ‘induced-fit’ mechanism to ensure correct Watson–Crick base pairing does not take place [344]. Moreover, TLS polymerases do not have an exonuclease domain or any proofreading activity. Instead, these error-prone enzymes carry out low-fidelity DNA synthesis, with the aim of bypassing the lesion without halting the replication fork. On undamaged DNA, TLS polymerases incorporate an incorrect nucleotide once every 10−1–10−3 bases [341,345,346]. Some translesion polymerases are better than others at incorporating the correct base opposite particular DNA lesions, suggesting an element of specificity depending on the species [347]. Thus, the product of TLS can be error-free or error-prone, depending of the type of lesion encountered.
Most eukaryotes encode four-family Y DNA polymerases, each with specificity for different types of the lesion [348,349]. Meanwhile, bacteria and archaea generally only encode one or two TLS polymerases, each with a broader lesion specificity than their eukaryotic counterpart. Due to the error-prone nature of TLS, this pathway is a potential source of genome instability and thus requires tight regulation. In eukaryotes, this is controlled by post-translational modification of both PCNA and family Y DNA polymerases [350]. All eukaryotic family Y DNA polymerases contain PIP motifs, which facilitate the interaction with PCNA [349].
The properties, roles and functions of archaeal family Y DNAPs are not well understood, since most have been tested in vitro under non-physiological temperatures. The TLS polymerases found in archaea belong to the DinB subfamily of family Y DNAPs. This subfamily is the most diverse and is found throughout all domains of life. DinB polymerases are prone to making single-nucleotide deletions in vivo and in vitro, caused by template slippage where repetitive sequences are present [343,351,352].
Hyperthermophilic archaea encode only one family Y DNA polymerase. Depending on strain, S. solfataricus encodes either Dpo4 (polymerase IV) or Dbh (DinB homologue) [343,344,353–355]. All archaeal DinB-like polymerases studied to date are capable of replicating past abasic sites, and in the case of Dbh, incorporating dATP [344,356]. Dpo4 can bypass a UV-induced cis-syn cyclobutene thymine dimer and 8-oxo guanines with relative efficiency, compared with other family Y DNA polymerases, since it can accommodate two adjacent template bases into its active site [355,357]; similar observations have been made in yeast [358] and humans [359]. This feature is not observed for other DinB-like polymerases [360–363]. Similar to human family Y DNA polymerases, Dpo4 shows limited activity on G4 structures [364,365].
Until now, only family Y polymerases were believed to act in TLS, but recent work in S. islandicus has implicated a family B enzyme, Dpo2, in TLS repair of helix-distorting lesions [366]. The potential TLS activity of Dpo2 may compensate for the lack of a canonical NER pathway in this crenarchaeal lineage [253]. In S. solfataricus, a complex has been described where both family Y and family B polymerases are bound to PCNA and DNA [367]. This is consistent with the ‘tool-belt’ model proposed for bacteria [368] and eukaryotes [369]. Tethering to PCNA allows for the coordination of DNA synthesis and TLS activities at the replication fork, with rapid switching from the canonical polymerase to TLS enzyme at the site of damage, and vice versa upon bypass.
H. volcanii encodes one family Y polymerase, PolY (HVO_1302), which shares homology with bacterial DinB. As in eukaryotic family Y polymerases, H. volcanii PolY encodes a C-terminal PIP motif, but it has diverged somewhat from the canonical sequence [136]. Degeneracy of the PIP motif has been observed in other family Y polymerases, including eukaryotic proteins [370]. Deletion of polY is possible in H. volcanii (T.A. 2020, unpublished), suggesting that other pathways are available for the repair or bypass of bulky lesions that block DNA synthesis by canonical polymerases.
2.8. Double-strand break repair
DSBs are considered to be one of the most critical forms of DNA damage that cells can suffer. A break in both strands of the DNA double helix can lead to inhibition of key processes including DNA replication, transcription and cell division, alongside major genome rearrangements. Therefore, the major pathways of double-strand break repair (DSBR) are crucial for maintaining genome stability. The most accurate form of DSBR is homologous recombination (HR), but this is a complex and energetically costly process, and therefore less-demanding pathways of DSBR operate alongside HR (figure 4 and table 1).
Figure 4.
Double-strand break repair. Double-strand break repair in H. volcanii occurs by either microhomology-mediated end joining (MMEJ) or homologous recombination (HR). MMEJ begins with end resection at the break site which is carried out by Rad50 (HVO_0854) and Mre11 (HVO_0853). Homologous DNA strands around the break (red) is annealed and any displaced DNA is trimmed by Fen1 (HVO_2873). Gaps in both strands are filled (purple) by replicative DNA polymerase PolB1 (HVO_0858) and ligated by DNA ligases; in H. volcanii either LigA (HVO_1565) or LigN (HVO_3000). HR also involves initial end resection at the break site by the combined activities of Rad50 and Mre11. The single-stranded DNA ends are bound by the recombinase RadA (HVO_0104), aided by the recombination mediator RadB (HVO_2383), which then promotes the search for homologous DNA duplex (red). Strand exchange with the homologous duplex generates a D-loop (displacement loop) with a 3′ invading end, from which DNA is synthesized (purple) by the action of replicative polymerase PolB1 (HVO_0858). At this stage, either a non-crossover event occurs due to displacement of the invading strand by helicases such as Hel308 (HVO_0014) or Hef (HVO_3010); the free strand anneals with and is ligated to the other end of the DNA break using either LigA (HVO_1565) or LigN (HVO_3000). Alternatively, a Holliday junction is formed that is processed by resolvases Hjc (HVO_0170) or Hef (HVO_3010) to cut the four-way DNA junction, leading to either a crossover or non-crossover event between recombining chromosomes. The remaining nicks in DNA are resolved by either LigA or LigN.
2.8.1. Non-homologous end joining
Classical non-homologous end joining (NHEJ) repairs DSBs by ligating DNA ends using little or no complementary base pairing. The first step is recognition of the DSB by Ku protein, comprising a Ku homodimer in bacteria and a Ku70/Ku80 heterodimer in eukaryotes. Ku acts as a scaffold protein to recruit other proteins involved in DSB repair, including nucleases to resect the damaged DNA, family B and family X DNAPs to fill the gap, and DNA ligase to ligate the DNA strands [251]. NHEJ is common in eukaryotes; the lack of requirement for a homologous partner means this is an effective method of DSB repair during the G1 phase of the cell cycle, when only a single genome copy is present.
Ku proteins are conserved in bacteria, yeast and higher eukaryotes, but to date only a few archaeal species have been shown to encode Ku proteins, which have most likely been acquired from bacteria by LGT [258,371]. To date, a complete NHEJ system has only been identified in a single archaeon, Methanocella paludicola [371]. Homologues of Ku are not found in H. volcanii [372,373]; instead an alternative mechanism of microhomology-mediated end joining (MMEJ) operates to repair DSBs [374,375].
2.8.2. Microhomology-mediated end joining
While NHEJ does not require any homology, microhomology-mediated end joining (MMEJ) involves the annealing of short homologous sequences at broken DNA ends. The products of MMEJ are always associated with gene deletions and contribute to chromosome translocations and genome rearrangements. The basic mechanism for MMEJ involves resection of DNA ends, annealing of the micro-homologous region, the removal of the DNA flaps by the structure-specific nuclease Fen1, filling of the gap by DNA polymerases and ligation by DNA ligase [376–378] (figure 4a and table 1).
MMEJ has been observed directly in a small number of archaeal species, including H. volcanii and S. islandicus [374,375,379]. However, the detailed enzymology of MMEJ in archaea remains unknown. Due to the lack of signature proteins dedicated to MMEJ (in contrast with the requirement for Ku protein in NHEJ), the prevalence of MMEJ is unknown but likely to be widespread. H. volcanii uses MMEJ to repair DSBs; this process is stimulated (directly or indirectly) by the Rad50-Mre11 complex, which restrains HR and instead promotes MMEJ as the immediate pathway of DSB repair [374]. HR is later used to restore the repaired allele.
2.8.3. Homologous recombination
The mechanism of HR is conserved throughout the three domains of life. HR is an essential process in archaea that provides a high-fidelity DSB repair mechanism, compared with other error-prone end-joining processes. This high fidelity is due to the use of homologous DNA duplex as the template for repair. Regulation of HR is essential to maintain genome integrity and avoid DNA rearrangements [380,381]. When a DSB occurs, it must be processed to generate ssDNA tails (pre-synapsis). This ssDNA tail then invades nearby duplex DNA with sequence homology, creating a D-loop (displacement loop) (synapsis). The final step of HR is post-synapsis, in which resolution of HR structures occurs, generating a crossover (where genetic exchange takes place) or a non-crossover (no genetic exchange) product (figure 4b and table 1).
HR is the best-studied DSB pathway in archaea [382]. Alongside its role in DSB repair, HR has been implicated in recombination-dependent replication (RDR), the restart of stalled DNA replication forks and in increasing genetic diversity [60,383,384]. The H. volcanii genome is highly polyploid with a genome copy number of approximately 20 copies per cell [12]; having multiple copies of DNA can be advantageous for efficient repair by HR as this increases the chances of having a non-damaged homologous template for DNA repair.
2.8.4. Pre-synapsis
2.8.4.1. Rad50-Mre11 complex
The Rad50-Mre11 complex is present in all domains of life [385,386]. In eukaryotes, Rad50-Mre11 is involved in the early steps of DSB repair, being one of the primary protein complexes recruited to the site of damage. Mre11 is an ATP-independent dsDNA exonuclease and ssDNA endonuclease, while Rad50 has ATP-dependent DNA-binding activity [387–391].
Rad50-Mre11 processes DSB ends by resecting the 5′ strand, generating a short single-stranded 3′ overhang. This overhang is then coated by a recombinase protein (RadA in archaea) to facilitate strand invasion [392,393]. In thermophilic archaea (e.g. S. islandicus and T. kodakarensis), Rad50 and Mre11 are essential for cell viability [111]. By contrast, in the halophile Halobacterium sp. NRC-1, Mre11 is essential while Rad50 is dispensable, indicating a Rad50-independent function of Mre11 [394]. In H. volcanii, deletions of rad50 (HVO_0854) and mre11 (HVO_0853) are viable and mutants show increased resistance to various types of DNA damage, including UV, ionizing radiation and MMS. However, these mutants recover slowly and exhibit higher rates of HR at DSBs than the wild type [374]. In a polyploid organism like H. volcanii, the action of Rad50-Mre11 in temporarily restraining HR may help prevent DNA ends from engaging with multiple homologous partners; after the number of available DNA ends has been reduced by MMEJ, DSBs are ultimately repaired by HR [374].
In addition, the H. volcanii Rad50-Mre11 complex has been shown to be involved in nucleoid compaction following genomic stress. Such nucleoid compaction is another mechanism to ensure DSB ends remain within close proximity [395], which may aid the search for intact DNA partners during HR and faster recruitment of DNA repair proteins to the sites of damage; similar mechanisms are seen in eukaryotes [391,396,397]. This compaction process is independent of the recombinase protein RadA (HVO_0104) [395], unlike in bacteria [398,399].
2.8.4.2. HerA-NurA
In thermophilic archaea, herA and nurA are encoded in the same operon as rad50 and mre11 [400]. HerA and NurA have been shown to interact with each other both in vitro and in vivo [401]. The HerA-NurA ATP-dependent helicase-nuclease complex cooperates with Rad50-Mre11 to coordinate the repair of DSBs, although the mechanism has not yet been described in detail [400,402–404]. The ATPase activity of HerA, the nuclease activity of NurA and their interaction are essential for Sulfolobus viability [405]. HerA and NurA proteins are not present in H. volcanii. Bacterial homologues of HerA and NurA have been identified and were initially thought to be essential for cell viability [111]. Genetic studies have shown this is not the case in bacterial species Deinococcus radiodurans and Thermus thermophilus [406]; in the latter, deletion of nurA and herA has no effect on cell growth. Unexpectedly, T. thermophilus cells lacking nurA and herA show increased resistance to UV irradiation and MMC treatment [401].
2.8.5. Synapsis
2.8.5.1. Rada
The 3′ ssDNA tail generated during pre-synapsis acts to invade a homologous duplex; this is facilitated by coating of the ssDNA with a recombinase protein. The strand exchange protein, or recombinase, promotes homology search and catalyses strand invasion, giving rise to a D-loop (displacement loop) [381,407]. Archaea contain homologues of eukaryotic HR proteins, including the evolutionarily conserved RecA-family strand exchange protein, called RadA in archaea, RecA in bacteria and Rad51 in eukaryotes.
Archaeal RadA is more similar to eukaryotic Rad51 than to bacterial RecA [408]. Like bacterial cells lacking RecA, RadA-deficient H. volcanii cells show growth defects, a lack of homologous recombination, and increased sensitivity to DNA damage agents [409–412], indicating an important role for HR in archaea. In thermophilic archaea, RadA is essential [111], suggesting these organisms are critically reliant on HR for repair and/or replication.
2.8.5.2. RadB
Paralogues of RecA-family proteins are present in many organisms where they generally function as recombination mediators [413–418]. Eukaryotic recombination mediators, such as BRCA2 in humans, and Rad52 and Rad55–Rad57 in yeast, act to displace RPA and facilitate the loading of recombinase Rad51 onto DNA [419,420]. Rad55–Rad57 are paralogues of Rad51 and have been shown to stabilize Rad51 filament formation [421]; BRCA2 and Rad52, while recombination mediators, are not paralogues of Rad51.
In archaea, RadB is a paralogue of RadA that is present only in Euryarchaea [422]. In H. volcanii, RadB (HVO_2383) acts as a recombination mediator that interacts with RadA and promotes its polymerization on ssDNA [412]. Strains in which radB has been deleted show a phenotype similar to cells lacking radA; defects include growth retardation, low levels of recombination and DNA damage sensitivity [411,412]. Two suppressor mutations that alleviate the ΔradB phenotype have been identified in H. volcanii RadA, S101P and A196 V, which suggest that RadB induces a conformational change in RadA, promoting its polymerization on ssDNA [412].
2.8.6. Post-synapsis
2.8.6.1. Hjc
During HR, four-way DNA intermediates (Holliday junctions; HJs) are formed by strand exchange between homologous DNA molecules. Resolvases, a group of highly specialized structure-specific metal-dependent endonucleases catalyse the cleavage of HJs into two DNA duplexes [423]. Resolvases are ubiquitous and are found in bacteria, eukaryotes, archaea and even in some viruses, although they are not directly related [424,425].
In E. coli, HJs are resolved by the RuvABC complex. The initial RuvAB complex, comprising a RuvA tetramer and two RuvB hexamers, specifically binds the HJ and promotes branch migration, following which RuvC cleaves the junction [426]. Eukaryotic HJ resolution is more complex than the bacterial counterpart and multiple endonucleases have been implicated in the resolution of HJs, including Yen1 (GEN1), Mus81-Mms4 (Eme1), Slx1-Slx4 and XPF-ERCC1 [427,428].
In archaea, the Hjc resolvase has been implicated in the cleavage of HJs. Hjc is present in all archaeal species and has been shown to interact with various proteins involved in DNA replication and repair, including the helicase Hel308 [429], clamp protein PCNA [430] and ATPase HerA [431]. Some Sulfolobus species contain an additional resolvase Hje, which shows higher DNA cleavage activity than Hjc [431,432]. H. volcanii Hjc (HVO_0170) is non-essential, has been shown to act in the same pathway as RadA and is necessary for efficient growth in the presence of cross-linking agent MMC [219].
In S. islandicus, Hjc is regulated by phosphorylation, which acts to inhibit its catalytic activity [433]. Interestingly, Hjc phosphomimetic mutants of S. islandicus exhibit increased resistance to DNA damaging agents, suggesting that phosphorylation acts to redirect repair to avoid HR that is dependent on Hjc [433]. While the serine residues targeted for phosphorylation in S. islandicus Hjc are not conserved in other archaeal species, modification of Hjc and other HR proteins by phosphorylation in S. acidocaldarius is well documented [434]. This regulatory mechanism could have evolved early during evolution, as the activity of several HJ resolvases is also regulated by phosphorylation in eukaryotes [435–437]. However, it remains unknown whether such post-translational modifications are used to regulate Hjc in H. volcanii.
2.8.6.2. Hef
Hef (helicase-associated endonuclease fork-structure DNA) is a member of the XPF family of structure-specific endonucleases known to act on branched, flapped and forked DNA structures [438]. Hef features two distinct domains: an N-terminal helicase domain and C-terminal endonuclease domain [438–441]. All archaea encode an XPF family endonuclease, but Hef is specific to euryarchaeal species.
In H. volcanii, Hef (HVO_3010) is not involved in NER and instead a bacterial Uvr system is used [219]. However, Hef has been implicated in NER in other species [111]. Both H. volcanii and T. kodakarensis mutants deleted for hef show sensitivity to MMC [111,307]. In H. volcanii, Hef is non-essential but cannot be deleted from cells lacking Hjc, suggesting these two proteins participate in alternative mechanisms for the resolution of recombination intermediates [219]. Redundancy of proteins involved in HJ resolution has also been described for other species of archaea; for example, HJ resolvases Hje and Hjc in S. islandicus are redundant [431]. In H. volcanii, Hef is required for cell viability in the absence of HR and is recruited to sites of DNA replication fork arrest [219,442]. This indicates a key role for Hef in the restart of stalled replication forks by RDR. Accordingly, Hef has been shown to interact with PCNA in T. kodakarensis [443], P. abysii [444] and T. acidophilum [445]. This interaction likely ensures availability of Hef at the replication fork, facilitating its role in RDR. While an interaction with PCNA has not yet been shown in H. volcanii, Hef in this species features a PIP box, indicating that Hef : PCNA interactions are common to most euryarchaea.
Alongside Hef, archaea from the phylum Euryarchaea encode a conserved protein HAN (Hef-associated nuclease), a RecJ-like protein displaying 3′–5′ exonuclease activity [443]. Hef and HAN interact with PCNA in T. kodakarensis, although Hef cannot bind both PCNA and HAN simultaneously [443]. Deletion of han in H. volcanii (HVO_1018; also called recJ3) results in increased sensitivity to DNA damage agent MMS, but not to other genotoxic agents such as H2O2, 4NQO or MMC [446]. It has been proposed that HAN acts to coordinate the helicase and nuclease activities of Hef during the processing of stalled replication forks [110,442].
2.8.6.3. Hel308
Hel308 is a Ski2 family 3′–5′ helicase found in archaea and metazoans (where it is named HelQ/PolQ) [447–450]; however, Hel308 is absent from fungi and bacteria. Hel308 is able to unwind various dsDNA structures in vitro, showing the preference for forked DNA and D-loops, and can remove bound proteins during translocation; these activities suggest a role in HR and the restart of stalled replication forks [411]. Hel308 has been shown to interact with other proteins involved in HR, including the BCDX2-Rad51 paralogue complex in humans [450,451], Hjc resolvase in S. tokodaii [429] and RPA in M. thermautotrophicus [452]. Hel308 has been proposed to act at blocked DNA replication forks, whereby it unwinds the parental strands and facilitates the loading of other factors required for the restart of the stalled replication fork.
Hel308 is not essential in the majority of archaeal species that encode it, with the exception of S. tokodaii and S. islandicus [429,453]. Mutants of Hel308 in H. volcanii (HVO_0014) are viable but slow-growing and show sensitivity to DNA cross-linking agents such as MMC (T.A. 2020, unpublished data); a similar phenotype has been seen in human ΔhelQ mutants [450]. This implicates H. volcanii Hel308 in the processing and repair of inter-strand cross-linked DNA lesions.
2.9. Recombination-dependent DNA replication
Replication origins were originally thought to be indispensable for cellular life, and deletion of origins was found to lead to impaired growth or cell death [454]. Work in bacterial model species E. coli showed that in the absence of origins, DNA replication can be primed from D-loops or R-loops (RNA displacement loops); these structures undergo remodelling to form a canonical replication fork [455]. For utilization of an R-loop, the invading RNA strand must remain intact and since RNase H proteins usually act to degrade RNA : DNA hybrids, RNase H gene(s) must be inactivated to allow R-loop-mediated replication to occur [454,455]. Both DNA- and RNA-dependent mechanisms of originless replication have been shown to be reliant on bacterial recombinase, RecA, indicating that the process uses HR; hence, it is known asRDR [456].
Surprisingly, it is possible to delete all chromosomal origins of replication in H. volcanii without affecting viability; in fact, strains deleted for origins grow 7.5% faster than wild type [60]. As in E. coli, cells deleted for replication origins become dependent upon the recombinase protein (here RadA), indicating that originless strains use RDR and hence HR becomes essential [60]. Marker frequency analysis (MFA) in originless H. volcanii shows a flat replication profile, indicating a lack of specific initiation and termination sites [60]. Similarly, the euryarchaeon T. kodakarensis has been shown to be capable of DNA replication in the absence of origins, again using a mechanism dependent on HR [457]. As in H. volcanii, MFA analysis of T. kodakarensis shows a flat replication profile in originless strains. But unlike H. volcanii, a flat replication profile is also seen in wild-type T. kodakarensis, suggesting that the origin, while present, is not used under laboratory conditions [457].
Both H. volcanii and T. kodakarensis are highly polyploid species. Having a large number of genome copies increases the chance of finding a homologous DNA duplex to invade and carry out HR; this may explain why RDR can occur in these organisms with relative ease. Similar findings have been made with polyploid species of cyanobacteria, which are able to carry out efficient DNA replication in the absence of the initiator protein DnaA [458]. Such a link between high ploidy and a capacity for RDR may explain why organisms with low ploidy, including E. coli and higher eukaryotes, are reliant upon origins to replicate their DNA; the lack of homologous DNA sequences would restrict the potential for RDR. Within archaea, members of the crenarchaea have been shown to contain only 1–2 genome copies and, unlike polyploid euryarchaea, have a defined cell cycle [459,460]. When ploidy is reduced to a single copy at the G1 stage of the cell cycle, RDR is no longer possible. This has been shown for S. islandicus, where one of its three replication origins must be maintained for cell viability [56].
However, not all polyploid Euryarchaea are capable of originless replication, including two close relatives of H. volcanii: Haloarcula hispanica and Haloferax mediterranei. The two replication origins of H. hispanica cannot be deleted at the same time, indicating that at least one origin is essential for viability [461]. In H. mediterranei, deletion of all three chromosomal origins is possible but this leads to the activation of a dormant origin [462]; the dormant origin appears to have been acquired by LGT and is not found in H. volcanii. The variety of responses to origin deletion in archaea hints at complex differences between species in their capacity for RDR, and further work is needed to elucidate the specific mechanisms involved.
3. Conclusion
Over the past three decades, the development and application of genetic and biochemical tools for H. volcanii have accelerated. These tools have, in turn, increased our understanding of the mechanisms of DNA replication and repair, both in this organism, in halophiles and in archaea generally. The increasing number of model archaeal species has provided additional insights into diversity across the domain, where species within a phylum may use radically different mechanisms for basic processes such as DNA replication.
The identification and study of archaeal DNA repair and replication enzymes has provided invaluable information on their eukaryotic counterparts. On the other hand, the development of tools specific for archaea has shed light on adaptations and proteins specific to this domain. Such a two-pronged approach—to determine what is common with eukaryotes and what is unique to archaea—is needed to uncover the evolutionary history of DNA replication and repair.
The ease of genetics and availability of DNA damaging agents have allowed the identification of key DNA repair enzymes in H. volcanii. The pathways used are counterparts of either entire eukaryotic or bacterial systems, with limited substitution within a pathway. Such preservation of pathway integrity is explained by the complexity hypothesis [463], which states that genes encoding proteins that function in complexes (e.g. DNA repair) are less frequently transferred via LGT and then only as entire operons. By contrast, the genes encoding proteins that do not form complexes (e.g. those that act in central metabolism) can be transferred successfully by LGT. In the case of H. volcanii, LGT has led to wholesale displacements (e.g. bacterial UvrABC has displaced the archaeal NER pathway), alternative activities that are used interchangeably (e.g. archaeal LigA and bacterial LigN are both active as DNA ligases) or acquisitions that are not used (e.g. bacterial DnaG is not used as primase, instead archaeal PriS/L plays that role). These three scenarios illustrate how H. volcanii is not just a good model organism for the study of DNA replication and repair mechanisms, but is also invaluable for the study of LGT and evolution.
While our understanding of DNA replication and repair in H. volcanii has undoubtedly increased, numerous questions remain. The ever-increased repertoire of genetic and biochemical tools for H. volcanii cements its place as a bona fide archaeal model organism. These tools will prove invaluable to answer open questions and thereby increase our understanding of DNA replication and repair in archaea.
Acknowledgements
We are grateful to all present and past members of the T.A. group for their work on H. volcanii.
Data accessibility
This article has no additional data.
Authors' contributions
P.P.-A., A.D. and V.S. contributed equally to the manuscript.
Competing interests
The authors declare that there are no conflicts of interest.
Funding
Work in the T.A. laboratory has been supported by the Biotechnology and Biological Sciences Research Council (BBSRC) (grant numbers BB/N016491/1 and BB/R007543/1 to P.P.-A. and A.D., respectively). The funders had no role in study design, data collection and interpretation, nor the decision to submit the work for publication.
References
- 1.Woese CR, Fox GE. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc.Natl Acad. Sci. USA 74, 5088–5090. ( 10.1073/pnas.74.11.5088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huet J, Schnabel R, Sentenac A, Zillig W. 1983. Archaebacteria and eukaryotes possess DNA-dependent RNA polymerases of a common type. EMBO J. 2, 1291–1294. ( 10.1002/j.1460-2075.1983.tb01583.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kandler O, König H. 1993. Cell envelopes of archaea: structure and chemistry. In The biochemistry of archaea (archaebacteria) (eds Kates M, Kushner DJ, Matheson AT), pp. 223–259. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 4.Woese CR, Kandler O, Wheelis ML. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. USA 87, 4576–4579. ( 10.1073/pnas.87.12.4576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imachi H, et al. 2020. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 577, 519– 525 ( 10.1038/s41586-019-1916-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spang A, et al. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179. ( 10.1038/nature14447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams TA, Cox CJ, Foster PG, Szollosi GJ, Embley TM. 2020. Phylogenomics provides robust support for a two-domains tree of life. Nat. Ecol. Evol. 4, 138–147. ( 10.1038/s41559-019-1040-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mullakhanbhai MF, Larsen H. 1975. Halobacterium volcanii spec. nov., a Dead Sea halobacterium with a moderate salt requirement. Arch. Microbiol. 104, 207–214. ( 10.1007/BF00447326) [DOI] [PubMed] [Google Scholar]
- 9.Cline SW, Lam WL, Charlebois RL, Schalkwyk LC, Doolittle WF. 1989. Transformation methods for halophilic archaebacteria. Can. J. Microbiol. 35, 148–152. ( 10.1139/m89-022) [DOI] [PubMed] [Google Scholar]
- 10.Christian JH, Waltho JA. 1962. Solute concentrations within cells of halophilic and non-halophilic bacteria. Biochim. Biophys. Acta. 65, 506–508. ( 10.1016/0006-3002(62)90453-5) [DOI] [PubMed] [Google Scholar]
- 11.Siglioccolo A, Paiardini A, Piscitelli M, Pascarella S. 2011. Structural adaptation of extreme halophilic proteins through decrease of conserved hydrophobic contact surface. BMC Struct. Biol. 11, 50 ( 10.1186/1472-6807-11-50) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breuert S, Allers T, Spohn G, Soppa J. 2006. Regulated polyploidy in halophilic archaea. PLoS ONE 1, e92 ( 10.1371/journal.pone.0000092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludt K, Soppa J. 2019. Polyploidy in halophilic archaea: regulation, evolutionary advantages, and gene conversion. Biochem. Soc. Trans. 47, 933–944. ( 10.1042/BST20190256) [DOI] [PubMed] [Google Scholar]
- 14.Mevarech M, Werczberger R. 1985. Genetic transfer in Halobacterium volcanii. J. Bacteriol. 162, 461–462. ( 10.1128/JB.162.1.461-462.1985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wendoloski D, Ferrer C, Dyall-Smith ML. 2001. A new simvastatin (mevinolin)-resistance marker from Haloarcula hispanica and a new Haloferax volcanii strain cured of plasmid pHV2. Microbiology. 147, 959–964. ( 10.1099/00221287-147-4-959) [DOI] [PubMed] [Google Scholar]
- 16.Pohlschroder M, Schulze S. 2019. Microbe of the month Haloferax volcanii. Trends Microbiol. 27, 86–87. ( 10.1016/j.tim.2018.10.004) [DOI] [PubMed] [Google Scholar]
- 17.Holmes ML, Dyall-Smith ML. 1990. A plasmid vector with a selectable marker for halophilic archaebacteria. J. Bacteriol. 172, 756–761. ( 10.1128/JB.172.2.756-761.1990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bitan-Banin G, Ortenberg R, Mevarech M. 2003. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185, 772–778. ( 10.1128/JB.185.3.772-778.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allers T, Ngo HP, Mevarech M, Lloyd RG. 2004. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70, 943–953. ( 10.1128/AEM.70.2.943-953.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ortenberg R, Rozenblatt-Rosen O, Mevarech M. 2000. The extremely halophilic archaeon Haloferax volcanii has two very different dihydrofolate reductases. Mol. Microbiol. 35, 1493–1505. ( 10.1046/j.1365-2958.2000.01815.x) [DOI] [PubMed] [Google Scholar]
- 21.Holmes ML, Dyall-Smith ML. 2000. Sequence and expression of a halobacterial beta-galactosidase gene. Mol. Microbiol. 36, 114–122. ( 10.1046/j.1365-2958.2000.01832.x) [DOI] [PubMed] [Google Scholar]
- 22.Reuter CJ, Maupin-Furlow J. 2004. Analysis of proteasome-dependent proteolysis in Haloferax volcanii cells, using short-lived green fluorescent proteins. Appl. Environ. Microbiol. 70, 7530–7538. ( 10.1128/AEM.70.12.7530-7538.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Born J, Pfeifer F. 2019. Improved GFP variants to study gene expression in haloarchaea. Front. Microbiol. 10, 1200 ( 10.3389/fmicb.2019.01200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis CR, Johnson CH, Robertson JB. 2020. A bioluminescent reporter for the halophilic archaeon Haloferax volcanii. Extremophilesm 24, 773–785. ( 10.1007/s00792-020-01193-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes M, Pfeifer F, Dyall-Smith M. 1994. Improved shuttle vectors for Haloferax volcanii including a dual-resistance plasmid. Gene. 146, 117–121. ( 10.1016/0378-1119(94)90844-3) [DOI] [PubMed] [Google Scholar]
- 26.Lam WL, Doolittle WF. 1989. Shuttle vectors for the archaebacterium Halobacterium volcanii. Proc. Natl Acad. Sci. USA 86, 5478–5482. ( 10.1073/pnas.86.14.5478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Large A, Stamme C, Lange C, Duan Z, Allers T, Soppa J, Lund PA. 2007. Characterization of a tightly controlled promoter of the halophilic archaeon Haloferax volcanii and its use in the analysis of the essential cct1 gene. Mol. Microbiol. 66, 1092–1106. ( 10.1111/j.1365-2958.2007.05980.x) [DOI] [PubMed] [Google Scholar]
- 28.Haque RU, Paradisi F, Allers T. 2019. Haloferax volcanii as immobilised whole cell biocatalyst: new applications for halophilic systems. Appl. Microbiol. Biotechnol. 103, 3807–3817. ( 10.1007/s00253-019-09725-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haque RU, Paradisi F, Allers T. 2020. Haloferax volcanii for biotechnology applications: challenges, current state and perspectives. Appl. Microbiol. Biotechnol. 104, 1371–1382. ( 10.1007/s00253-019-10314-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiljunen S, Pajunen MI, Dilks K, Storf S, Pohlschroder M, Savilahti H. 2014. Generation of comprehensive transposon insertion mutant library for the model archaeon, Haloferax volcanii, and its use for gene discovery. BMC Biol. 12, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maier LK, Dyall-Smith M, Marchfelder A. 2015. The adaptive immune system of Haloferax volcanii. Life (Basel) 5, 521–537. ( 10.3390/life5010521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stachler AE, Marchfelder A. 2016. Gene repression in Haloarchaea Using the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas I-B system. J. Biol. Chem. 291, 15 226–15 242. ( 10.1074/jbc.M116.724062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenshine I, Tchelet R, Mevarech M. 1989. The mechanism of DNA transfer in the mating system of an archaebacterium. Science 245, 1387–1389. ( 10.1126/science.2818746) [DOI] [PubMed] [Google Scholar]
- 34.Shalev Y, Turgeman-Grott I, Tamir A, Eichler J, Gophna U. 2017. Cell surface glycosylation is required for efficient mating of Haloferax volcanii. Front. Microbiol. 8, 1253 ( 10.3389/fmicb.2017.01253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartman AL, et al. 2010. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS ONE 5, e9605 ( 10.1371/journal.pone.0009605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allers T, Barak S, Liddell S, Wardell K, Mevarech M. 2010. Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl. Environ. Microbiol. 76, 1759–1769. ( 10.1128/AEM.02670-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMillan LJ, Hwang SM, Farah RE, Koh J, Chen SX, Maupin-Furlow JA. 2018. Multiplex quantitative SILAC for analysis of archaeal proteomes: a case study of oxidative stress responses. Environ. Microbiol. 20, 385–401. ( 10.1111/1462-2920.14014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eichler J, Maupin-Furlow J. 2013. Post-translation modification in Archaea: lessons from Haloferax volcanii and other haloarchaea. Fems Microbiol. Rev. 37, 583–606. ( 10.1111/1574-6976.12012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schulze S, et al. 2020. The Archaeal Proteome Project advances knowledge about archaeal cell biology through comprehensive proteomics. Nature Communications 11, 1–4. ( 10.1038/s41467-020-16784-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ausiannikava D, Allers T. 2017. Diversity of DNA replication in the Archaea. Genes (Basel) 8, 56 ( 10.3390/genes8020056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaper S, Messer W. 1995. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 270, 17 622–17 626. ( 10.1074/jbc.270.29.17622) [DOI] [PubMed] [Google Scholar]
- 42.Davey MJ, Fang LH, McInerney P, Georgescu RE, O'Donnell M. 2002. The DnaC helicase loader is a dual ATP/ADP switch protein. Embo J. 21, 3148–3159. ( 10.1093/emboj/cdf308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mott ML, Erzberger JP, Coons MM, Berger JM. 2008. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 135, 623–634. ( 10.1016/j.cell.2008.09.058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barry ER, Bell SD. 2006. DNA replication in the archaea. Microbiol. Mol. Biol. Rev. 70, 876–887. ( 10.1128/MMBR.00029-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Stillman B. 2012. The origin recognition complex: a biochemical and structural view. Subcell Biochem. 62, 37–58. ( 10.1007/978-94-007-4572-8_3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bleichert F, Botchan MR, Berger JM. 2015. Crystal structure of the eukaryotic origin recognition complex. Nature 519, 321–326. ( 10.1038/nature14239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Makarova KS, Koonin EV. 2013. Archaeology of eukaryotic DNA replication. Cold Spring Harb. perspect. Biol. 5, a012963 ( 10.1101/cshperspect.a012963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bell SP, Dutta A. 2002. DNA replication in eukaryotic cells. Ann. Rev. Biochem. 71, 333–374. ( 10.1146/annurev.biochem.71.110601.135425) [DOI] [PubMed] [Google Scholar]
- 49.Li H, O'Donnell ME. 2018. The eukaryotic CMG helicase at the replication fork: emerging architecture reveals an unexpected mechanism. Bioessays 40, 1700208 ( 10.1002/bies.201700208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kelman LM, O'Dell WB, Kelman Z. 2020. Unwinding 20 years of the archaeal minichromosome maintenance helicase. J. Bacteriol. 202, e00729-19 ( 10.1128/JB.00729-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Z, Liu J, Yang H, Xiang H. 2014. DNA replication origins in archaea. Front microbiol. 5, 179 ( 10.3389/fmicb.2014.00179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samson RY, Abeyrathne PD, Bell SD. 2016. Mechanism of archaeal MCM helicase recruitment to DNA replication origins. Mol. Cell. 61, 287–296. ( 10.1016/j.molcel.2015.12.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu ZF, Liu HL, Liu JF, Liu XQ, Xiang H. 2012. Diversity and evolution of multiple orc/cdc6-adjacent replication origins in haloarchaea. BMC genomics. 13, 16 ( 10.1186/1471-2164-13-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. 2007. Structural basis of DNA replication origin recognition by an ORC protein. Science 317, 1213–1216. ( 10.1126/science.1143664) [DOI] [PubMed] [Google Scholar]
- 55.Kawakami H, Ohashi E, Kanamoto S, Tsurimoto T, Katayama T. 2015. Specific binding of eukaryotic ORC to DNA replication origins depends on highly conserved basic residues. Sci. Rep. 5, 14 ( 10.1038/srep14929) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Samson RY, et al. 2013. Specificity and function of archaeal DNA replication initiator proteins. Cell Rep. 3, 485–496. ( 10.1016/j.celrep.2013.01.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Myllykallio H, Lopez P, Lopez-Garcia P, Heilig R, Saurin W, Zivanovic Y, Philippe H, Forterre P. 2000. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science. 288, 2212–2215. ( 10.1126/science.288.5474.2212) [DOI] [PubMed] [Google Scholar]
- 58.Zatopek KM, Gardner AF, Kelman Z. 2018. Archaeal DNA replication and repair: new genetic, biophysical and molecular tools for discovering and characterizing enzymes, pathways and mechanisms. Fems Microbiol. Rev. 42, 477–488. ( 10.1093/femsre/fuy017) [DOI] [PubMed] [Google Scholar]
- 59.Norais C, Hawkins M, Hartman AL, Eisen JA, Myllykallio H, Allers T. 2007. Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet. 3, e77 ( 10.1371/journal.pgen.0030077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hawkins M, Malla S, Blythe MJ, Nieduszynski CA, Allers T. 2013. Accelerated growth in the absence of DNA replication origins. Nature 503, 544–547. ( 10.1038/nature12650) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raymann K, Forterre P, Brochier-Armanet C, Gribaldo S. 2014. Global phylogenomic analysis disentangles the complex evolutionary history of DNA replication in Archaea. Genome Biol. Evol. 6, 192–212. ( 10.1093/gbe/evu004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maurer S, Ludt K, Soppa J. 2018. Characterization of copy number control of two Haloferax volcanii replication origins using deletion mutants and haloarchaeal artificial chromosomes. J. Bacteriol. 200, 15 ( 10.1128/JB.00517-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pelve EA, Martens-Habbena W, Stahl DA, Bernander R. 2013. Mapping of active replication origins in vivo in thaum- and euryarchaeal replicons. Mol. Microbiol. 90, 538–550. ( 10.1111/mmi.12382) [DOI] [PubMed] [Google Scholar]
- 64.Ausiannikava D, Mitchell L, Marriott H, Smith V, Hawkins M, Makarova KS, Koonin EV, Nieduszynski CA, O'connell M. 2018. Evolution of genome architecture in Archaea: spontaneous generation of a new chromosome in Haloferax volcanii. Mol. Biol. Evol. 35, 1855–1868. ( 10.1093/molbev/msy075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ludt K, Soppa J. 2018. Influence of origin recognition complex proteins on the copy numbers of three chromosomes in Haloferax volcanii. J. Bacteriol. 200, 13 ( 10.1128/JB.00161-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McGeoch AT, Bell SD. 2008. Extra-chromosomal elements and the evolution of cellular DNA replication machineries. Nat. Rev. Mol. Cell Biol. 9, 569–574. ( 10.1038/nrm2426) [DOI] [PubMed] [Google Scholar]
- 67.Burgers PMJ, Kunkel TA. 2017. Eukaryotic DNA replication fork. Annu. Rev. Biochem. 86, 417–438. ( 10.1146/annurev-biochem-061516-044709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kelman LM, Kelman Z. 2014. Archaeal DNA replication. Annu. Rev. Genet. 48, 71–97. ( 10.1146/annurev-genet-120213-092148) [DOI] [PubMed] [Google Scholar]
- 69.Rowen L, Kornberg A. 1978. Primase, DnaG protein of Escherichia coli—enzyme which starts DNA chains. J. Biol. Chem. 253, 758–764. [PubMed] [Google Scholar]
- 70.Tanaka S, Araki H. 2013. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb. perspect. biol. 5, a010371 ( 10.1101/cshperspect.a010371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Onesti S, MacNeill SA. 2013. Structure and evolutionary origins of the CMG complex. Chromosoma. 122, 47–53. ( 10.1007/s00412-013-0397-x) [DOI] [PubMed] [Google Scholar]
- 72.Evrin C, Clarke P, Zech J, Lurz R, Sun JC, Uhle S, Li H, Stillman B, Speck C. 2009. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl Acad. Sci. USA 106, 20 240–20 245. ( 10.1073/pnas.0911500106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sakakibara N, Kelman LM, Kelman Z. 2009. Unwinding the structure and function of the archaeal MCM helicase. Mol. Microbiol. 72, 286–296. ( 10.1111/j.1365-2958.2009.06663.x) [DOI] [PubMed] [Google Scholar]
- 74.Walters AD, Chong JPJ. 2017. Non-essential MCM-related proteins mediate a response to DNA damage in the archaeon Methanococcus maripaludis. Microbiology (UK) 163, 745–753. ( 10.1099/mic.0.000460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ishino S, Fujino S, Tomita H, Ogino H, Takao K, Daiyasu H, Kanai T, Atomi H, Ishino Y. 2011. Biochemical and genetical analyses of the three mcm genes from the hyperthermophilic archaeon, Thermococcus kodakarensis. Genes to Cells. 16, 1176–1189. ( 10.1111/j.1365-2443.2011.01562.x) [DOI] [PubMed] [Google Scholar]
- 76.Pan M, Santangelo TJ, Li Z, Reeve JN, Kelman Z. 2011. Thermococcus kodakarensis encodes three MCM homologs but only one is essential. Nucleic Acids Res. 39, 9671–9680. ( 10.1093/nar/gkr624) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kristensen TP, Cherian RM, Gray FC, MacNeill SA. 2014. The haloarchaeal MCM proteins: bioinformatic analysis and targeted mutagenesis of the beta 7-beta 8 and beta 9-beta 10 hairpin loops and conserved zinc binding domain cysteines. Front. Microbiol. 5, 13 ( 10.3389/fmicb.2014.00123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brewster AS, Chen XJS. 2010. Insights into the MCM functional mechanism: lessons learned from the archaeal MCM complex. Critical Rev. Biochem. Mol. Biol. 45, 243–256. ( 10.3109/10409238.2010.484836) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brewster AS, Wang GG, Yu X, Greenleaf WB, Carazo JM, Tjajadia M, Klein MG, Chen XS. 2008. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA plus hexameric helicase. Proc. Natl Acad. Sci. USA 105, 20 191–20 196. ( 10.1073/pnas.0808037105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sakakibara N, Kasiviswanathan R, Melamud E, Han M, Schwarz FP, Kelman Z. 2008. Coupling of DNA binding and helicase activity is mediated by a conserved loop in the MCM protein. Nucleic Acids Res. 36, 1309–1320. ( 10.1093/nar/gkm1160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barry ER, McGeoch AT, Kelman Z, Bell SD. 2007. Archaeal MCM has separable processivity, substrate choice and helicase domains. Nucleic Acids Res. 35, 988–998. ( 10.1093/nar/gkl1117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nagata M, Ishino S, Yamagami T, Ogino H, Simons J-R, Kanai T, Atomi H, Ishino Y. 2017. The Cdc45/RecJ-like protein forms a complex with GINS and MCM, and is important for DNA replication in Thermococcus kodakarensis. Nucleic Acids Res. 45, 10 693–10 705. ( 10.1093/nar/gkx740) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ogino H, Ishino S, Mayanagi K, Haugland GT, Birkeland NK, Yamagishi A, Ishino Y. 2011. The GINS complex from the thermophilic archaeon, Thermoplasma acidophilum may function as a homotetramer in DNA replication. Extremophiles. 15, 529–539. ( 10.1007/s00792-011-0383-2) [DOI] [PubMed] [Google Scholar]
- 84.Xu Y, Gristwood T, Hodgson B, Trinidad JC, Albers SV, Bell SD. 2016. Archaeal orthologs of Cdc45 and GINS form a stable complex that stimulates the helicase activity of MCM. Proc. Natl Acad. Sci. USA. 113, 13 390–13 395. ( 10.1073/pnas.1613825113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shin JH, Heo GY, Kelman Z. 2009. The methanothermobacter thermautotrophicus MCM helicase is active as a hexameric ring. J. Biol. Chem. 284, 540–546. ( 10.1074/jbc.M806803200) [DOI] [PubMed] [Google Scholar]
- 86.Fletcher RJ, Shen J, Gómez-Llorente Y, Martín CS, Carazo JM, Chen XS. 2005. Double hexamer disruption and biochemical activities of methanobacterium thermoautotrophicum MCM. J. Biol. Chem. 280, 42 405–42 410. ( 10.1074/jbc.M509773200) [DOI] [PubMed] [Google Scholar]
- 87.MacNeill SA. 2009. The haloarchaeal chromosome replication machinery. Biochem. Soc. Trans. 37, 108–113. ( 10.1042/BST0370108) [DOI] [PubMed] [Google Scholar]
- 88.Kamada K. 2012. The GINS complex: structure and function. Subcellular Biochem. 62, 135–156. ( 10.1007/978-94-007-4572-8_8) [DOI] [PubMed] [Google Scholar]
- 89.Makarova KS, Koonin EV. 2005. Evolutionary and functional genomics of the Archaea. Curr. Opin. Microbiol. 8, 586–594. ( 10.1016/j.mib.2005.08.003) [DOI] [PubMed] [Google Scholar]
- 90.Kamada K, Kubota Y, Arata T, Shindo Y, Hanaoka F. 2007. Structure of the human GINS complex and its assembly and functional interface in replication initiation. Nat. Struct. Mol. Biol. 14, 388–396. ( 10.1038/nsmb1231) [DOI] [PubMed] [Google Scholar]
- 91.Marinsek N, Barry ER, Makarova KS, Dionne I, Koonin EV, Bell SD. 2006. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep. 7, 539–545. ( 10.1038/sj.embor.7400649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.MacNeill SA. 2010. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem. J. 425, 489–500. ( 10.1042/BJ20091531) [DOI] [PubMed] [Google Scholar]
- 93.Oyama T, et al. 2011. Architectures of archaeal GINS complexes, essential DNA replication initiation factors. BMC Biol. 9, 12 ( 10.1186/1741-7007-9-28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yoshimochi T, Fujikane R, Kawanami M, Matsunaga F, Ishino Y. 2008. The GINS complex from Pyrococcus furiosus stimulates the MCM helicase activity. J. Biol. Chem. 283, 1601–1609. ( 10.1074/jbc.M707654200) [DOI] [PubMed] [Google Scholar]
- 95.Ogino H, Ishino S, Haugland GT, Birkeland NK, Kohda D, Ishino Y. 2014. Activation of the MCM helicase from the thermophilic archaeon, Thermoplasma acidophilum by interactions with GINS and Cdc6-2. Extremophiles 18, 915–924. ( 10.1007/s00792-014-0673-6) [DOI] [PubMed] [Google Scholar]
- 96.Makarova KS, Koonin EV, Kelman Z. 2012. The CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes. Biol. direct. 7, 7 ( 10.1186/1745-6150-7-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pellegrini L. 2017. Structural insights into Cdc45 function: was there a nuclease at the heart of the ancestral replisome? Biophys. Chem. 225, 10–14. ( 10.1016/j.bpc.2016.11.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sanchez-Pulido L, Ponting CP. 2011. Cdc45: the missing RecJ ortholog in eukaryotes? Bioinformatics. 27, 1885–1888. ( 10.1093/bioinformatics/btr332) [DOI] [PubMed] [Google Scholar]
- 99.Krastanova I, Sannino V, Amenitsch H, Gileadi O, Pisani FM, Onesti S. 2012. Structural and functional insights into the DNA replication factor Cdc45 reveal an evolutionary relationship to the DHH family of phosphoesterases. J. Biol. Chem. 287, 4121–4128. ( 10.1074/jbc.M111.285395) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Szambowska A, Tessmer I, Kursula P, Usskilat C, Prus P, Pospiech H, Grosse F. 2014. DNA binding properties of human Cdc45 suggest a function as molecular wedge for DNA unwinding. Nucleic Acids Res. 42, 2308–2319. ( 10.1093/nar/gkt1217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Burdett V, Baitinger C, Viswanathan M, Lovett ST, Modrich P. 2001. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl Acad. Sci. USA 98, 6765–6770. ( 10.1073/pnas.121183298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dianov G, Sedgwick B, Daly G, Olsson M, Lovett S, Lindahl T. 1994. Release of 5′-terminal deoxyribose-phosphate residues from incised abasic sites in DNA by the Escherichia coli RecJ protein. Nucleic Acids Res. 22, 993–998. ( 10.1093/nar/22.6.993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chow MH, Courcelle J. 2007. RecBCD and RecJ/RecQ initiate DNA degradation on distinct substrates in UV-irradiated Escherichia coli. Radiation Res. 168, 499–506. ( 10.1667/RR1033.1) [DOI] [PubMed] [Google Scholar]
- 104.Courcelle J, Hanawalt PC. 1999. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. General Genet. 262, 543–551. ( 10.1007/s004380051116) [DOI] [PubMed] [Google Scholar]
- 105.Aravind L, Koonin EV. 1998. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 26, 3746–3752. ( 10.1093/nar/26.16.3746) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cheng KY, Xu H, Chen XY, Wang LY, Tian B, Zhao Y, Hua Y. 2016. Structural basis for DNA 5′-end resection by RecJ. Elife 5, e14294 ( 10.7554/elife.14294) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.MacNeill SA. 2018. The archaeal RecJ-like proteins: nucleases and ex-nucleases with diverse roles in replication and repair. Emerging Topics Life Sci. 2, 493–501. ( 10.1042/etls20180017) [DOI] [PubMed] [Google Scholar]
- 108.Li Z, Santangelo TJ, Cubonova L, Reeve JN, Kelman Z. 2010. Affinity purification of an archaeal DNA replication protein network. MBio. 1, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oyama T, Ishino S, Shirai T, Yamagami T, Nagata M, Ogino H, Kusunoki M, Ishino Y. 2016. Atomic structure of an archaeal GAN suggests its dual roles as an exonuclease in DNA repair and a CMG component in DNA replication. Nucleic Acids Res. 44, 9505–9517. ( 10.1093/nar/gkw789) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nagata M, Ishino S, Yamagami T, Simons JR, Kanai T, Atomi H, Ishino Y. 2017. Possible function of the second RecJ-like protein in stalled replication fork repair by interacting with Hef. Sci. Rep. 7, 16949 ( 10.1038/s41598-017-17306-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fujikane R, Ishino S, Ishino Y, Forterre P. 2010. Genetic analysis of DNA repair in the hyperthermophilic archaeon, Thermococcus kodakaraensis. Genes Genet Syst. 85, 243–257. ( 10.1266/ggs.85.243) [DOI] [PubMed] [Google Scholar]
- 112.Li Z, Pan M, Santangelo TJ, Chemnitz W, Yuan W, Edwards JL, Hurwitz J, Reeve JN, Kelman Z. 2011. A novel DNA nuclease is stimulated by association with the GINS complex. Nucleic Acids Res. 39, 6114–6123. ( 10.1093/nar/gkr181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Simon AC, Sannino V, Costanzo V, Pellegrini L. 2016. Structure of human Cdc45 and implications for CMG helicase function. Nat. commun. 7, 11638 ( 10.1038/ncomms11638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lu SH, Zhang XS, Chen KY, Chen ZM, Li YX, Qi ZQ, Shen Y, Li Z. 2019. The small subunit of DNA polymerase D (DP1) associates with GINS-GAN complex of the thermophilic archaea in Thermococcus sp. 4557. Microbiologyopen 8, 15 ( 10.1002/mbo3.848) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yuan H, Liu XP, Han Z, Allers T, Hou JL, Liu JH. 2013. RecJ-like protein from Pyrococcus furiosus has 3′-5′ exonuclease activity on RNA: implications for proofreading of 3′-mismatched RNA primers in DNA replication. Nucleic Acids Res. 41, 5817–5826. ( 10.1093/nar/gkt275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li MJ, et al. 2017. The crystal structure of Pyrococcus furiosus RecJ implicates it as an ancestor of eukaryotic Cdc45. Nucleic Acids Res. 45, 12 551–12 564. ( 10.1093/nar/gkx887) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ogino H, Ishino S, Kohda D, Ishino Y. 2017. The RecJ2 protein in the thermophilic archaeon Thermoplasma acidophilum is a 3′-5′ exonuclease that associates with a DNA replication complex. J. Biol. Chem. 292, 7921–7931. ( 10.1074/jbc.M116.767921) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arezi B, Kuchta RD. 2000. Eukaryotic DNA primase. Trends in Biochem. Sci. 25, 572–576. ( 10.1016/S0968-0004(00)01680-7) [DOI] [PubMed] [Google Scholar]
- 119.Lao-Sirieix SH, Bell SD. 2004. The heterodimeric primase of the hyperthermophilic archaeon Sulfolobus solfataricus possesses DNA and RNA primase, polymerase and 3′-terminal nucleotidyl transferase activities. J. Mol. Biol. 344, 1251–1263. ( 10.1016/j.jmb.2004.10.018) [DOI] [PubMed] [Google Scholar]
- 120.Galal WC, Pan M, Kelman Z, Hurwitz J. 2012. Characterization of DNA primase complex isolated from the archaeon, Thermococcus kodakaraensis. J. Biol. Chem. 287, 16 209–16 219. ( 10.1074/jbc.M111.338145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Le Breton M, Henneke G, Norais C, Flament D, Myllykallio H, Querellou J, Raffin J-P. 2007. The heterodimeric primase from the euryarchaeon Pyrococcus abyssi: a multifunctional enzyme for initiation and repair? J. Mol. Biol. 374, 1172–1185. ( 10.1016/j.jmb.2007.10.015) [DOI] [PubMed] [Google Scholar]
- 122.Evguenieva-Hackenberg E, Walter P, Hochleitner E, Lottspeich F, Klug G. 2003. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep. 4, 889–893. ( 10.1038/sj.embor.embor929) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Walter P, Klein F, Lorentzen E, Ilchmann A, Klug G, Evguenieva-Hackenberg E. 2006. Characterization of native and reconstituted exosome complexes from the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 62, 1076–1089. ( 10.1111/j.1365-2958.2006.05393.x) [DOI] [PubMed] [Google Scholar]
- 124.Zuo Z, Rodgers CJ, Mikheikin AL, Trakselis MA. 2010. Characterization of a functional DnaG-type primase in archaea: implications for a dual-primase system. J. Mol. Biol. 397, 664–676. ( 10.1016/j.jmb.2010.01.057) [DOI] [PubMed] [Google Scholar]
- 125.Yao NY, O'Donnell M. 2012. The RFC clamp loader: structure and function. Subcell. Biochem. 62, 259–279. ( 10.1007/978-94-007-4572-8_14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.O'Donnell M, Kuriyan J. 2006. Clamp loaders and replication initiation. Curr. Opin. Struct. Biol. 16, 35–41. ( 10.1016/j.sbi.2005.12.004) [DOI] [PubMed] [Google Scholar]
- 127.Yuzhakov A, Kelman Z, O'Donnell M. 1999. Trading places on DNA-a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell 96, 153–163. ( 10.1016/S0092-8674(00)80968-X) [DOI] [PubMed] [Google Scholar]
- 128.Stukenberg PT, Turner J, Odonnell M. 1994. An explanation for lagging strand replication: polymerase hopping among DNA sliding clamps. Cell 78, 877–887. ( 10.1016/S0092-8674(94)90662-9) [DOI] [PubMed] [Google Scholar]
- 129.Indiani C, O'Donnell M. 2006. The replication clamp-loading machine at work in the three domains of life. Nat. Rev. Mol. Cell Biol. 7, 751–761. ( 10.1038/nrm2022) [DOI] [PubMed] [Google Scholar]
- 130.Oyama T, Ishino Y, Cann IKO, Ishino S, Morikawa K. 2001. Atomic structure of the clamp loader small subunit from Pyrococcus furiosus. Mol. Cell. 8, 455–463. ( 10.1016/S1097-2765(01)00328-8) [DOI] [PubMed] [Google Scholar]
- 131.Cann IKO, Ishino S, Yuasa M, Daiyasu H, Toh H, Ishino Y. 2001. Biochemical analysis of replication factor C from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 183, 2614–2623. ( 10.1128/JB.183.8.2614-2623.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Giroux X, MacNeill SA. 2015. Molecular genetic methods to study DNA replication protein function in Haloferax volcanii, a model archaeal organism. Methods Mol Biol. 1300, 187–218. ( 10.1007/978-1-4939-2596-4_13) [DOI] [PubMed] [Google Scholar]
- 133.Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15, 352–363. ( 10.1101/gr.3003105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Daimon K, Kawarabayasi Y, Kikuchi H, Sako Y, Ishino Y. 2002. Three proliferating cell nuclear antigen-like proteins found in the hyperthermophilic archaeon Aeropyrum pernix: interactions with the two DNA polymerases. J. Bacteriol. 184, 687–694. ( 10.1128/JB.184.3.687-694.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Morgunova E, Gray FC, Macneill SA, Ladenstein R. 2009. Structural insights into the adaptation of proliferating cell nuclear antigen (PCNA) from Haloferax volcanii to a high-salt environment. Acta Crystallogr. D Biol. Crystallogr. 65, 1081–1088. ( 10.1107/S0907444909029321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Winter JA, Christofi P, Morroll S, Bunting KA. 2009. The crystal structure of Haloferax volcanii proliferating cell nuclear antigen reveals unique surface charge characteristics due to halophilic adaptation. BMC Struct. Biol. 9, 55 ( 10.1186/1472-6807-9-55) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Meslet-Cladiere L, Norais C, Kuhn J, Briffotaux J, Sloostra JW, Ferrari E, Hübscher U, Flament D, Myllykallio H. 2007. A novel proteomic approach identifies new interaction partners for proliferating cell nuclear antigen. J. Mol. Biol. 372, 1137–1148. ( 10.1016/j.jmb.2007.06.056) [DOI] [PubMed] [Google Scholar]
- 138.Prestel A, et al. 2019. The PCNA interaction motifs revisited: thinking outside the PIP-box. Cell. Mol. Life Sci. 76, 4923–4943. ( 10.1007/s00018-019-03150-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kirkland PA, Maupin-Furlow JA. 2009. Stabilization of an archaeal DNA-sliding clamp protein, PCNA, by proteasome-activating nucleotidase gene knockout in Haloferax volcanii. Fems Microbiol. Lett. 294, 32–36. ( 10.1111/j.1574-6968.2009.01547.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kirkland PA, Gil MA, Karadzic IM, Maupin-Furlow JA. 2008. Genetic and proteomic analyses of a proteasome-activating nucleotidase A mutant of the haloarchaeon Haloferax volcanii. J. Bacteriol. 190, 193–205. ( 10.1128/JB.01196-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ito J, Braithwaite DK. 1991. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 19, 4045–4057. ( 10.1093/nar/19.15.4045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nakamura TM, Cech TR. 1998. Reversing time: origin of telomerase. Cell 92, 587–590. ( 10.1016/S0092-8674(00)81123-X) [DOI] [PubMed] [Google Scholar]
- 143.Leipe DD, Aravind L, Koonin EV. 1999. Did DNA replication evolve twice independently? Nucleic Acids Res. 27, 3389–3401. ( 10.1093/nar/27.17.3389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Inoue R, Kaito C, Tanabe M, Kamura K, Akimitsu N, Sekimizu K. 2001. Genetic identification of two distinct DNA polymerases, DnaE and PolC, that are essential for chromosomal DNA replication in Staphylococcus aureus. Mol. Genet. Genomics. 266, 564–571. ( 10.1007/s004380100564) [DOI] [PubMed] [Google Scholar]
- 145.Zhao X-Q, Hu J-F, Yu J. 2006. Comparative analysis of eubacterial DNA polymerase III alpha subunits. Genomics Proteomics Bioinformatics. 4, 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Allen JM, et al. 2011. Roles of DNA polymerase I in leading and lagging-strand replication defined by a high-resolution mutation footprint of ColE1 plasmid replication. Nucleic Acids Res. 39, 7020–7033. ( 10.1093/nar/gkr157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hubscher U, Maga G, Spadari S. 2002. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 71, 133–163. ( 10.1146/annurev.biochem.71.090501.150041) [DOI] [PubMed] [Google Scholar]
- 148.Frick DN, Richardson CC. 2001. DNA primases. Annu. Rev. Biochem. 70, 39–80. ( 10.1146/annurev.biochem.70.1.39) [DOI] [PubMed] [Google Scholar]
- 149.Miyabe I, Kunkel TA, Carr AM. 2011. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 7, e1002407 ( 10.1371/journal.pgen.1002407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. 2007. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science 317, 127–130. ( 10.1126/science.1144067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Raia P, Delarue M, Sauguet L. 2019. An updated structural classification of replicative DNA polymerases. Biochem. Soc. Trans. 47, 239–249. ( 10.1042/BST20180579) [DOI] [PubMed] [Google Scholar]
- 152.Kazlauskas D, Krupovic M, Guglielmini J, Forterre P, Venclovas C. 2020. Diversity and evolution of B-family DNA polymerases. Nucleic Acids Res. 48, 10 142–10 156. ( 10.1093/nar/gkaa760) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ishino S, Ishino Y. 2014. DNA polymerases as useful reagents for biotechnology: the history of developmental research in the field. Front. Microbiol. 5, 465 ( 10.3389/fmicb.2014.00465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Makarova KS, Krupovic M, Koonin EV. 2014. Evolution of replicative DNA polymerases in Archaea and their contributions to the eukaryotic replication machinery. Front. Microbiol. 5, 10 ( 10.3389/fmicb.2014.00354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Guy L, Ettema TJ. 2011. The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 19, 580–587. ( 10.1016/j.tim.2011.09.002) [DOI] [PubMed] [Google Scholar]
- 156.Rogozin IB, Makarova KS, Pavlov YI, Koonin EV. 2008. A highly conserved family of inactivated archaeal B family DNA polymerases. Biol. Direct. 3, 5 ( 10.1186/1745-6150-3-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kelman Z, Pietrokovski S, Hurwitz J. 1999. Isolation and characterization of a split B-type DNA polymerase from the archaeon Methanobacterium thermoautotrophicum deltaH. J. Biol. Chem. 274, 28 751–28 761. ( 10.1074/jbc.274.40.28751) [DOI] [PubMed] [Google Scholar]
- 158.Perler FB. 2002. InBase: the intein database. Nucleic Acids Res. 30, 383–384. ( 10.1093/nar/30.1.383) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gogarten JP, Doolittle WF, Lawrence JG. 2002. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 19, 2226–2238. ( 10.1093/oxfordjournals.molbev.a004046) [DOI] [PubMed] [Google Scholar]
- 160.Liu J, He B, Qing H, Kow YW. 2000. A deoxyinosine specific endonuclease from hyperthermophile, Archaeoglobus fulgidus: a homolog of Escherichia coli endonuclease V. Mutat Res. 461, 169–177. ( 10.1016/S0921-8777(00)00054-9) [DOI] [PubMed] [Google Scholar]
- 161.Ishino Y, Komori K, Cann IK, Koga Y. 1998. A novel DNA polymerase family found in Archaea. J. Bacteriol. 180, 2232–2236. ( 10.1128/JB.180.8.2232-2236.1998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gawel D, Pham PT, Fijalkowska IJ, Jonczyk P, Schaaper RM. 2008. Role of accessory DNA polymerases in DNA replication in Escherichia coli: analysis of the dnaX36 mutator mutant. J. Bacteriol. 190, 1730–1742. ( 10.1128/JB.01463-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wardle J, et al. 2008. Uracil recognition by replicative DNA polymerases is limited to the archaea, not occurring with bacteria and eukarya. Nucleic Acids Res. 36, 705–711. ( 10.1093/nar/gkm1023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Greagg MA, Fogg MJ, Panayotou G, Evans SJ, Connolly BA, Pearl LH. 1999. A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proc. Natl Acad. Sci. USA 96, 9045–9050. ( 10.1073/pnas.96.16.9045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Connolly BA. 2009. Recognition of deaminated bases by archaeal family-B DNA polymerases. Biochem. Soc. Trans. 37, 65–68. ( 10.1042/BST0370065) [DOI] [PubMed] [Google Scholar]
- 166.Greenough L, Kelman Z, Gardner AF. 2015. The roles of family B and D DNA polymerases in Thermococcus species 9 degrees N Okazaki fragment maturation. J. Biol. Chem. 290, 12 514–12 522. ( 10.1074/jbc.M115.638130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cubonova L, Richardson T, Burkhart BW, Kelman Z, Connolly BA, Reeve JN, Santangelo TJ. 2013. Archaeal DNA polymerase D but not DNA polymerase B is required for genome replication in Thermococcus kodakarensis. J. Bacteriol. 195, 2322–2328. ( 10.1128/JB.02037-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sarmiento F, Mrazek J, Whitman WB. 2013. Genome-scale analysis of gene function in the hydrogenotrophic methanogenic archaeon Methanococcus maripaludis. Proc. Natl Acad. Sci. USA 110, 4726–4731. ( 10.1073/pnas.1220225110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Birien T, Thiel A, Henneke G, Flament D, Moalic Y, Jebbar M. 2018. Development of an effective 6-methylpurine counterselection marker for genetic manipulation in Thermococcus barophilus. Genes (Basel) 9, 77 ( 10.3390/genes9020077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kushida T, Narumi I, Ishino S, Ishino Y, Fujiwara S, Imanaka T, Higashibata H. 2019. Pol B, a Family B DNA polymerase, in Thermococcus kodakarensis is important for DNA repair, but not DNA replication. Microbes Environ. 34, 316–326. ( 10.1264/jsme2.ME19075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hogrel G, Lu Y, Alexandre N, Bosse A, Dulermo R, Ishino S, Ishino Y, Flament D. 2020. Role of RadA and DNA polymerases in recombination-associated DNA synthesis in hyperthermophilic Archaea. Biomolecules 10, 1045 ( 10.3390/biom10071045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Berquist BR, DasSarma P, DasSarma S. 2007. Essential and non-essential DNA replication genes in the model halophilic archaeon, Halobacterium sp. NRC-1. BMC Genet. 8, 31 ( 10.1186/1471-2156-8-31) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Uemori T, Sato Y, Kato I, Doi H, Ishino Y. 1997. A novel DNA polymerase in the hyperthermophilic archaeon, Pyrococcus furiosus: gene cloning, expression, and characterization. Genes Cells. 2, 499–512. ( 10.1046/j.1365-2443.1997.1380336.x) [DOI] [PubMed] [Google Scholar]
- 174.Imamura M, Uemori T, Kato I, Ishino Y. 1995. A non-alpha-like DNA polymerase from the hyperthermophilic archaeon Pyrococcus furiosus. Biol. Pharm. Bull. 18, 1647–1652. ( 10.1248/bpb.18.1647) [DOI] [PubMed] [Google Scholar]
- 175.Raia P, et al. 2019. Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases. PLoS Biol. 17, e3000122 ( 10.1371/journal.pbio.3000122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Takashima N, Ishino S, Oki K, Takafuji M, Yamagami T, Matsuo R, Mayanagi K, Ishino Y. 2019. Elucidating functions of DP1 and DP2 subunits from the Thermococcus kodakarensis family D DNA polymerase. Extremophiles 23, 161–172. ( 10.1007/s00792-018-1070-3) [DOI] [PubMed] [Google Scholar]
- 177.Shen YL, Tang XF, Yokoyama H, Matsui E, Matsui I. 2004. A 21-amino acid peptide from the cysteine cluster II of the family D DNA polymerase from Pyrococcus horikoshii stimulates its nuclease activity which is Mre11-like and prefers manganese ion as the cofactor. Nucleic Acids Res. 32, 158–168. ( 10.1093/nar/gkh153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jokela M, Eskelinen A, Pospiech H, Rouvinen J, Syvaoja JE. 2004. Characterization of the 3′ exonuclease subunit DP1 of Methanococcus jannaschii replicative DNA polymerase D. Nucleic Acids Res. 32, 2430–2440. ( 10.1093/nar/gkh558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sauguet L. 2019. The extended ‘two-barrel’ polymerases superfamily: structure, function and evolution. J.Mol. Biol. 431, 4167–4183. ( 10.1016/j.jmb.2019.05.017) [DOI] [PubMed] [Google Scholar]
- 180.Sauguet L, Raia P, Henneke G, Delarue M. 2016. Shared active site architecture between archaeal PolD and multi-subunit RNA polymerases revealed by X-ray crystallography. Nat. commun. 7, 12227 ( 10.1038/ncomms12227) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Iyer LM, Koonin EV, Aravind L. 2003. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct Biol. 3, 1–23. ( 10.1186/1472-6807-3-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ruprich-Robert G, Thuriaux P. 2010. Non-canonical DNA transcription enzymes and the conservation of two-barrel RNA polymerases. Nucleic Acids Res. 38, 4559–4569. ( 10.1093/nar/gkq201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fouqueau T, Blombach F, Werner F. 2017. Evolutionary origins of two-barrel RNA polymerases and site-specific transcription initiation. Annu. Rev. Microbiol. 71, 331–348. ( 10.1146/annurev-micro-091014-104145) [DOI] [PubMed] [Google Scholar]
- 184.Koonin EV, Krupovic M, Ishino S, Ishino Y. 2020. The replication machinery of LUCA: common origin of DNA replication and transcription. BMC Biol. 18, 61 ( 10.1186/s12915-020-00800-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Henneke G, Flament D, Hubscher U, Querellou J, Raffin JP. 2005. The hyperthermophilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J. Mol. Biol. 350, 53–64. ( 10.1016/j.jmb.2005.04.042) [DOI] [PubMed] [Google Scholar]
- 186.Mayanagi K, et al. 2020. Two conformations of DNA polymerase D-PCNA-DNA, an archaeal replisome complex, revealed by cryo-electron microscopy. BMC Biol. 18, 152 ( 10.1186/s12915-020-00889-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Naor A, Lazary R, Barzel A, Papke RT, Gophna U. 2011. In Vivo characterization of the homing endonuclease within the polB gene in the halophilic archaeon Haloferax volcanii. PLoS ONE 6, 7 ( 10.1371/journal.pone.0015833) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kunkel TA, Meyer RR, Loeb LA. 1979. Single-strand binding protein enhances fidelity of DNA synthesis in vitro. Proc. Natl Acad. Sci. USA 76, 6331–6335. ( 10.1073/pnas.76.12.6331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lohman TM, Ferrari ME. 1994. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 63, 527–570. ( 10.1146/annurev.bi.63.070194.002523) [DOI] [PubMed] [Google Scholar]
- 190.Iftode C, Daniely Y, Borowiec JA. 1999. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34, 141–180. ( 10.1080/10409239991209255) [DOI] [PubMed] [Google Scholar]
- 191.Wold MS. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92. ( 10.1146/annurev.biochem.66.1.61) [DOI] [PubMed] [Google Scholar]
- 192.Chedin F, Seitz EM, Kowalczykowski SC. 1998. Novel homologs of replication protein A in archaea: implications for the evolution of ssDNA-binding proteins. Trends biochem. sci. 23, 273–277. ( 10.1016/S0968-0004(98)01243-2) [DOI] [PubMed] [Google Scholar]
- 193.Kelly TJ, Simancek P, Brush GS. 1998. Identification and characterization of a single-stranded DNA-binding protein from the archaeon Methanococcus jannaschii. Proc. Natl Acad. Sci. USA 95, 14 634–14 639. ( 10.1073/pnas.95.25.14634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sung P, Krejci L, Van Komen S, Sehorn MG. 2003. Rad51 recombinase and recombination mediators. J.Biol. Chem. 278, 42 729–42 732. ( 10.1074/jbc.R300027200) [DOI] [PubMed] [Google Scholar]
- 195.Rolfsmeier ML, Haseltine CA. 2010. The single-stranded DNA binding protein of Sulfolobus solfataricus acts in the presynaptic step of homologous recombination. J. Mol. Biol. 397, 31–45. ( 10.1016/j.jmb.2010.01.004) [DOI] [PubMed] [Google Scholar]
- 196.Komori K, Ishino Y. 2001. Replication protein A in Pyrococcus furiosus is involved in homologous DNA recombination. J. Biol. Chem. 276, 25 654–25 660. ( 10.1074/jbc.M102423200) [DOI] [PubMed] [Google Scholar]
- 197.Meyer RR, Glassberg J, Kornberg A. 1979. An Escherichia coli mutant defective in single-strand binding protein is defective in DNA replication. Proc. Natl Acad. Sci. USA. 76, 1702–1705. ( 10.1073/pnas.76.4.1702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kobayashi K, et al. 2003. Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA. 100, 4678–4683. ( 10.1073/pnas.0730515100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Brill SJ, Stillman B. 1991. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 5, 1589–1600. ( 10.1101/gad.5.9.1589) [DOI] [PubMed] [Google Scholar]
- 200.Heyer WD, Rao MR, Erdile LF, Kelly TJ, Kolodner RD. 1990. An essential Saccharomyces cerevisiae single-stranded DNA binding protein is homologous to the large subunit of human RP-A. EMBO J. 9, 2321–2329. ( 10.1002/j.1460-2075.1990.tb07404.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Skowyra A, MacNeill SA. 2012. Identification of essential and non-essential single-stranded DNA-binding proteins in a model archaeal organism. Nucleic Acids Res. 40, 1077–1090. ( 10.1093/nar/gkr838) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Evans JJ, Gygli PE, McCaskill J, DeVeaux LC. 2018. Divergent roles of RPA homologs of the model archaeon Halobacterium salinarum in survival of DNA damage. Genes. 9, 15 ( 10.3390/genes9040223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.O'Donnell M, Langston L, Stillman B. 2013. Principles and concepts of DNA replication in Bacteria, Archaea, and Eukarya. Cold Spring Harb. perspect. biol. 5, a010108. ( 10.1101/cshperspect.a010108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wadsworth RI, White MF. 2001. Identification and properties of the crenarchaeal single-stranded DNA binding protein from Sulfolobus solfataricus. Nucleic Acids Res. 29, 914–920. ( 10.1093/nar/29.4.914) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Paytubi S, McMahon SA, Graham S, Liu H, Botting CH, Makarova KS, Koonin EV, Naismith JH, White MF. 2012. Displacement of the canonical single-stranded DNA-binding protein in the Thermoproteales. Proc. Natl Acad. Sci. USA 109, E398–E405. ( 10.1073/pnas.1113277108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lin Y, Lin LJ, Sriratana P, Coleman K, Ha T, Spies M, Cann IKO. 2008. Engineering of functional replication protein a homologs based on insights into the evolution of oligonucleotide/oligosaccharide-binding folds. J. Bacteriol. 190, 5766–5780. ( 10.1128/JB.01930-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Forterre P, Elie C, Kohiyama M. 1984. Aphidicolin inhibits growth and DNA synthesis in halophilic archaebacteria. J. Bacteriol. 159, 800–802. ( 10.1128/JB.159.2.800-802.1984) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Delpech F, Collien Y, Mahou P, Beaurepaire E, Myllykallio H, Lestini R. 2018. Snapshots of archaeal DNA replication and repair in living cells using super-resolution imaging. Nucleic Acids Res. 46, 10 757–10 770. ( 10.1093/nar/gky829) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Stroud A, Liddell S, Allers T. 2012. Genetic and biochemical identification of a novel single-stranded DNA-binding complex in Haloferax volcanii. Front. Microbiol. 3, 1–14. ( 10.3389/fmicb.2012.00224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Dewar JM, Budzowska M, Walter JC. 2015. The mechanism of DNA replication termination in vertebrates. Nature. 525, 345–350. ( 10.1038/nature14887) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Arudchandran A, Cerritelli S, Narimatsu S, Itaya M, Shin DY, Shimada Y, Crouch R. 2000. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells. 5, 789–802. ( 10.1046/j.1365-2443.2000.00373.x) [DOI] [PubMed] [Google Scholar]
- 212.Lazzaro F, et al. 2012. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol. Cell. 45, 99–110. ( 10.1016/j.molcel.2011.12.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hyjek M, Figiel M, Nowotny M. 2019. RNases H: structure and mechanism. DNA Repair (Amst) 84, 102672 ( 10.1016/j.dnarep.2019.102672) [DOI] [PubMed] [Google Scholar]
- 214.Chapados BR, Chai Q, Hosfield DJ, Qiu J, Shen B, Tainer JA. 2001. Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication. J. Mol. Biol. 307, 541–556. ( 10.1006/jmbi.2001.4494) [DOI] [PubMed] [Google Scholar]
- 215.Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. 2007. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol. Cell. 28, 264–276. ( 10.1016/j.molcel.2007.08.015) [DOI] [PubMed] [Google Scholar]
- 216.Kochiwa H, Tomita M, Kanai A. 2007. Evolution of ribonuclease H genes in prokaryotes to avoid inheritance of redundant genes. BMC Evol. Biol. 7, 128 ( 10.1186/1471-2148-7-128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Henneke G. 2012. In vitro reconstitution of RNA primer removal in Archaea reveals the existence of two pathways. Biochem. J. 447, 271–280. ( 10.1042/BJ20120959) [DOI] [PubMed] [Google Scholar]
- 218.Sonneville R, Moreno SP, Knebel A, Johnson C, Hastie CJ, Gartner A, Gambus A, Labib K. 2017. CUL-2(LRR-1) and UBXN-3 drive replisome disassembly during DNA replication termination and mitosis. Nat. cell biol. 19, 468 ( 10.1038/ncb3500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lestini R, Duan Z, Allers T. 2010. The archaeal Xpf/Mus81/FANCM homolog Hef and the Holliday junction resolvase Hjc define alternative pathways that are essential for cell viability in Haloferax volcanii. DNA repair. 9, 994–1002. ( 10.1016/j.dnarep.2010.06.012) [DOI] [PubMed] [Google Scholar]
- 220.Vafadarnejad E, Amoozgar MA, Khansha J, Fallahzade R. 2015. The rad2 gene of haloarchaeum Halobacterium salinarum is functional in the repair of ultraviolet light induced DNA photoproducts. Microbiol. Res. 173, 44–49. ( 10.1016/j.micres.2015.01.012) [DOI] [PubMed] [Google Scholar]
- 221.Dewar JM, Low E, Mann M, Raschle M, Walter JC. 2017. CRL2(Lrr1) promotes unloading of the vertebrate replisome from chromatin during replication termination. Genes Dev. 31, 275–290. ( 10.1101/gad.291799.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Berghuis BA, et al. 2015. Strand separation establishes a sustained lock at the Tus-Ter replication fork barrier. Nat. Chem. Biol. 11, 579–U75. ( 10.1038/nchembio.1857) [DOI] [PubMed] [Google Scholar]
- 223.Berezney R, Dubey DD, Huberman JA. 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 108, 471–484. ( 10.1007/s004120050399) [DOI] [PubMed] [Google Scholar]
- 224.McGuffee SR, Smith DJ, Whitehouse I. 2013. Quantitative, genome-wide analysis of eukaryotic replication initiation and termination. Mol. Cell. 50, 123–135. ( 10.1016/j.molcel.2013.03.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Greenfeder SA, Newlon CS. 1992. Replication forks pause at yeast centromeres. Mol. Cell Biol. 12, 4056–4066. ( 10.1128/MCB.12.9.4056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Fu YV, et al. 2011. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA Helicase. Cell 146, 931–941. ( 10.1016/j.cell.2011.07.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Duggin IG, Dubarry N, Bell SD. 2011. Replication termination and chromosome dimer resolution in the archaeon Sulfolobus solfataricus. EMBO J. 30, 145–153. ( 10.1038/emboj.2010.301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Lundgren M, Andersson A, Chen L, Nilsson P, Bernander R. 2004. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc. Natl Acad. Sci. USA 101, 7046–7051. ( 10.1073/pnas.0400656101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Maric M, Maculins T, De Piccoli G, Labib K. 2014. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science 346, 1253596 ( 10.1126/science.1253596) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Moreno SP, Bailey R, Campion N, Herron S, Gambus A. 2014. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science 346, 477–481. ( 10.1126/science.1253585) [DOI] [PubMed] [Google Scholar]
- 231.Beattie TR, Bell SD. 2012. Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment maturation. Embo J. 31, 1556–1567. ( 10.1038/emboj.2012.12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Merlet J, Burger J, Tavernier N, Richaudeau B, Gomes JE, Pintard L. 2010. The CRL2(LRR-1) ubiquitin ligase regulates cell cycle progression during C. elegans development. Development 137, 3857–3866. ( 10.1242/dev.054866) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Humbard MA, Miranda HV, Lim JM, Krause DJ, Pritz JR, Zhou G, Chen S, Wells L, Maupin-Furlow JA. 2010. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature 463, 54–60. ( 10.1038/nature08659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wang JC. 1996. DNA topoisomerases. Annu. Rev. Biochem. 65, 635–692. ( 10.1146/annurev.bi.65.070196.003223) [DOI] [PubMed] [Google Scholar]
- 235.Espeli O, Lee C, Marians KJ. 2003. A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J. Biol. Chem. 278, 44 639–44 644. ( 10.1074/jbc.M308926200) [DOI] [PubMed] [Google Scholar]
- 236.Hiasa H, Marians KJ. 1994. Topoisomerase III, but not Topoisomerase I, can support nascent chain elongation during Theta-type DNA replication. J. Biol. Chem. 269, 32 655–32 659. [PubMed] [Google Scholar]
- 237.Beattie TR, Reyes-Lamothe R. 2015. A replisome's journey through the bacterial chromosome. Front. microbiol. 6, 562 ( 10.3389/fmicb.2015.00562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Rudolph CJ, Upton AL, Stockum A, Nieduszynski CA, Lloyd RG. 2013. Avoiding chromosome pathology when replication forks collide. Nature. 500, 608–611. ( 10.1038/nature12312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lesterlin C, Barre FX, Cornet F. 2004. Genetic recombination and the cell cycle: what we have learned from chromosome dimers. Mol. Microbiol. 54, 1151–1160. ( 10.1111/j.1365-2958.2004.04356.x) [DOI] [PubMed] [Google Scholar]
- 240.Reyes-Lamothe R, Nicolas E, Sherratt DJ. 2012. Chromosome replication and segregation in Bacteria. Annu. Rev. Genet. 46, 121–143. ( 10.1146/annurev-genet-110711-155421) [DOI] [PubMed] [Google Scholar]
- 241.Jolly SM, et al. 2020. Thermus thermophilus Argonaute functions in the completion of DNA replication. Cell 182, 1545–1559; e18. ( 10.1016/j.cell.2020.07.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ullsperger CJ, Vologodskii AV, Cozzarelli NR. 1995. Unlinking of DNA by topoisomerases during DNA replication. Berlin, Germany: Springer. [Google Scholar]
- 243.Bailey R, Priego Moreno S, Gambus A. 2015. Termination of DNA replication forks: ‘breaking up is hard to do’. Nucleus 6, 187–196. ( 10.1080/19491034.2015.1035843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Goto T, Wang JC. 1984. Yeast DNA Topoisomerase II is encoded by a single-copy, essential gene. Cell 36, 1073–1080. ( 10.1016/0092-8674(84)90057-6) [DOI] [PubMed] [Google Scholar]
- 245.Baxter J, Diffley JFX. 2008. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol. Cell. 30, 790–802. ( 10.1016/j.molcel.2008.04.019) [DOI] [PubMed] [Google Scholar]
- 246.Cortez D, Quevillon-Cheruel S, Gribaldo S, Desnoues N, Sezonov G, Forterre P, Serre M-C. 2010. Evidence for a Xer/dif system for chromosome resolution in archaea. PLoS Genetics. 6, 11 ( 10.1371/journal.pgen.1001166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Sioud M, Possot O, Elie C, Sibold L, Forterre P. 1988. Coumarin and quinolone action in archaebacteria: evidence for the presence of a DNA gyrase-like enzyme. J. Bacteriol. 170, 946–953. ( 10.1128/JB.170.2.946-953.1988) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Atomi H, Imanaka T, Fukui T. 2012. Overview of the genetic tools in the Archaea. Front. Microbiol. 3, 13 ( 10.3389/fmicb.2012.00337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Lindahl T, Wood RD. 1999. Quality control by DNA repair. Science 286, 1897–1905. ( 10.1126/science.286.5446.1897) [DOI] [PubMed] [Google Scholar]
- 250.Lindahl T. 1993. Instability and decay of the primary structure of DNA. Nature 362, 709–715. ( 10.1038/362709a0) [DOI] [PubMed] [Google Scholar]
- 251.Lieber MR. 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211. ( 10.1146/annurev.biochem.052308.093131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Tubbs A, Nussenzweig A. 2017. Endogenous DNA damage as a source of genomic instability in cancer. Cell 168, 644–656. ( 10.1016/j.cell.2017.01.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.White MF, Allers T. 2018. DNA repair in the Archaea: an emerging picture. FEMS Microbiol. Rev. 42, 514–526. ( 10.1093/femsre/fuy020) [DOI] [PubMed] [Google Scholar]
- 254.Kucukyildirim S, Behringer M, Williams EM, Doak TG, Lynch M. 2020. Estimation of the genome-wide mutation rate and spectrum in the archaeal species Haloferax volcanii. Genetics 215, 1107–1116. ( 10.1534/genetics.120.303299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Eisen JA, Hanawalt PC. 1999. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 435, 171–213. ( 10.1016/S0921-8777(99)00050-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Niu H, et al. 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467, 108–111. ( 10.1038/nature09318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Longhese MP, Bonetti D, Manfrini N, Clerici M. 2010. Mechanisms and regulation of DNA end resection. EMBO J. 29, 2864–2874. ( 10.1038/emboj.2010.165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Blackwood JK, Rzechorzek NJ, Bray SM, Maman JD, Pellegrini L, Robinson NP. 2013. End-resection at DNA double-strand breaks in the three domains of life. Biochem. Soc. Trans. 41, 314–320. ( 10.1042/BST20120307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Symington LS. 2014. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb. perspect. biol. 6, a016436 ( 10.1101/cshperspect.a016436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Mimitou EP, Symington LS. 2009. DNA end resection: many nucleases make light work. DNA Repair (Amst) 8, 983–995. ( 10.1016/j.dnarep.2009.04.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Allers T, Mevarech M. 2005. Archaeal genetics: the third way. Nat. Rev. Genet. 6, 58–73. ( 10.1038/nrg1504) [DOI] [PubMed] [Google Scholar]
- 262.Hildenbrand C, Stock T, Lange C, Rother M, Soppa J. 2011. Genome copy numbers and gene conversion in methanogenic archaea. J. Bacteriol. 193, 734–743. ( 10.1128/JB.01016-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Mable BK, Otto SP. 2001. Masking and purging mutations following EMS treatment in haploid, diploid and tetraploid yeast (Saccharomyces cerevisiae). Genet. res. 77, 9–26. ( 10.1017/S0016672300004821) [DOI] [PubMed] [Google Scholar]
- 264.Schmid AK, Allers T, DiRuggiero J. 2020. SnapShot: microbial extremophiles. Cell 180, 818–8e1. ( 10.1016/j.cell.2020.01.018) [DOI] [PubMed] [Google Scholar]
- 265.Crowley DJ, Boubriak I, Berquist BR, Clark M, Richard E, Sullivan L, Dassarma S, Mccready S. 2006. The uvrA, uvrB and uvrC genes are required for repair of ultraviolet light induced DNA photoproducts in Halobacterium sp. NRC-1. Saline Syst. 2, 11 ( 10.1186/1746-1448-2-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.McCready S. 1996. The repair of ultraviolet light-induced DNA damage in the halophilic archaebacteria, Halobacterium cutirubrum, Halobacterium halobium and Haloferax volcanii. Mutat. Res. 364, 25–32. ( 10.1016/0921-8777(96)00018-3) [DOI] [PubMed] [Google Scholar]
- 267.McCready S, Marcello L. 2003. Repair of UV damage in Halobacterium salinarum. Biochem. Soc. Trans. 31(Pt 3), 694–698. ( 10.1042/bst0310694) [DOI] [PubMed] [Google Scholar]
- 268.Jones DL, Baxter BK. 2017. DNA repair and photoprotection: mechanisms of overcoming environmental ultraviolet radiation exposure in halophilic Archaea. Front. Microbiol. 8, 1882 ( 10.3389/fmicb.2017.01882) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Sancar GB. 2000. Enzymatic photoreactivation: 50 years and counting. Mutat. Res. 451, 25–37. ( 10.1016/S0027-5107(00)00038-5) [DOI] [PubMed] [Google Scholar]
- 270.Baliga NS, Bjork SJ, Bonneau R, Pan M, Iloanusi C, Kottemann MC, Hood L, DiRuggiero J. 2004. Systems level insights into the stress response to UV radiation in the halophilic archaeon Halobacterium NRC-1. Genome Res. 14, 1025–1035. ( 10.1101/gr.1993504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Bickle TA, Kruger DH. 1993. Biology of DNA restriction. Microbiol. Rev. 57, 434–450. ( 10.1128/MMBR.57.2.434-450.1993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Tock MR, Dryden DT. 2005. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 8, 466–472. ( 10.1016/j.mib.2005.06.003) [DOI] [PubMed] [Google Scholar]
- 273.Adhikari S, Curtis PD. 2016. DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol. Rev. 40, 575–591. ( 10.1093/femsre/fuw023) [DOI] [PubMed] [Google Scholar]
- 274.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. 2010. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. methods. 7, 461–465. ( 10.1038/nmeth.1459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Ouellette M, Gogarten JP, Lajoie J, Makkay AM, Papke RT. 2018. Characterizing the DNA methyltransferases of Haloferax volcanii via bioinformatics. Gene Deletion, and SMRT Sequencing. Genes (Basel) 9, 129 ( 10.3390/genes9030129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Grasso S, Tell G. 2014. Base excision repair in Archaea: back to the future in DNA repair. DNA Repair (Amst) 21, 148–157. ( 10.1016/j.dnarep.2014.05.006) [DOI] [PubMed] [Google Scholar]
- 277.Hitomi K, Iwai S, Tainer JA. 2007. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 6, 410–428. ( 10.1016/j.dnarep.2006.10.004) [DOI] [PubMed] [Google Scholar]
- 278.Wallace SS, Murphy DL, Sweasy JB. 2012. Base excision repair and cancer. Cancer letters. 327, 73–89. ( 10.1016/j.canlet.2011.12.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Moen MN, Knaevelsrud I, Haugland GT, Grosvik K, Birkeland NK, Klungland A, Bjelland S. 2011. Uracil-DNA glycosylase of Thermoplasma acidophilum directs long-patch base excision repair, which is promoted by deoxynucleoside triphosphates and ATP/ADP, into short-patch repair. J. Bacteriol. 193, 4495–4508. ( 10.1128/JB.00233-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Mase T, Kubota K, Miyazono K, Kawarabayasi Y, Tanokura M. 2011. Structure of flap endonuclease 1 from the hyperthermophilic archaeon Desulfurococcus amylolyticus. Acta crystallographica. 67, 209–213. ( 10.1107/S1744309110053030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Matsui E, Musti KV, Abe J, Yamasaki K, Matsui I, Harata K. 2002. Molecular structure and novel DNA binding sites located in loops of flap endonuclease-1 from Pyrococcus horikoshii. J. Biol. Chem. 277, 37 840–37 847. ( 10.1074/jbc.M205235200) [DOI] [PubMed] [Google Scholar]
- 282.Hosfield DJ, Mol CD, Shen B, Tainer JA. 1998. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell 95, 135–146. ( 10.1016/S0092-8674(00)81789-4) [DOI] [PubMed] [Google Scholar]
- 283.Hwang KY, Baek K, Kim HY, Cho Y. 1998. The crystal structure of flap endonuclease-1 from Methanococcus jannaschii. Nat. Struct. Biol. 5, 707–713. ( 10.1038/1406) [DOI] [PubMed] [Google Scholar]
- 284.Chapados BR, Hosfield DJ, Han S, Qiu J, Yelent B, Shen B, Tainer JA. 2004. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell 116, 39–50. ( 10.1016/S0092-8674(03)01036-5) [DOI] [PubMed] [Google Scholar]
- 285.Schomacher L, Chong JP, McDermott P, Kramer W, Fritz HJ. 2009. DNA uracil repair initiated by the archaeal ExoIII homologue Mth212 via direct strand incision. Nucleic Acids Res. 37, 2283–2293. ( 10.1093/nar/gkp102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Martin IV, MacNeill SA. 2002. ATP-dependent DNA ligases. Genome biol. 3, REVIEWS3005 ( 10.1186/gb-2002-3-4-reviews3005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Poidevin L, MacNeill SA. 2006. Biochemical characterisation of LigN, an NAD+-dependent DNA ligase from the halophilic euryarchaeon Haloferax volcanii that displays maximal in vitro activity at high salt concentrations. BMC Mol. Biol. 7, 44 ( 10.1186/1471-2199-7-44) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Zhao A, Gray FC, Macneill SA. 2006. ATP- and NAD-dependent DNA ligases share an essential function in the halophilic archaeon Haloferax volcanii. Mol. Microbiol. 59, 743–752. ( 10.1111/j.1365-2958.2005.04975.x) [DOI] [PubMed] [Google Scholar]
- 289.White MF. 2003. Archaeal DNA repair: paradigms and puzzles. Biochem. Soc. Trans. 31, 690–693. ( 10.1042/bst0310690) [DOI] [PubMed] [Google Scholar]
- 290.Yasui A. 2013. Alternative excision repair pathways. Cold Spring Harb. perspect. Biol. 5, a012617 ( 10.1101/cshperspect.a012617) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Ishino S, Makita N, Shiraishi M, Yamagami T, Ishino Y. 2015. EndoQ and EndoV work individually for damaged DNA base repair in Pyrococcus furiosus. Biochimie 118, 264–269. ( 10.1016/j.biochi.2015.06.015) [DOI] [PubMed] [Google Scholar]
- 292.Shiraishi M, Ishino S, Yamagami T, Egashira Y, Kiyonari S, Ishino Y. 2015. A novel endonuclease that may be responsible for damaged DNA base repair in Pyrococcus furiosus. Nucleic Acids Res. 43, 2853–2863. ( 10.1093/nar/gkv121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Kiyonari S, Egashira Y, Ishino S, Ishino Y. 2014. Biochemical characterization of endonuclease V from the hyperthermophilic archaeon, Pyrococcus furiosus. J. Biochem. 155, 325–333. ( 10.1093/jb/mvu010) [DOI] [PubMed] [Google Scholar]
- 294.Cao W. 2013. Endonuclease V: an unusual enzyme for repair of DNA deamination. Cell Mol. Life Sci. 70, 3145–3156. ( 10.1007/s00018-012-1222-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Shiraishi M, Ishino S, Yoshida K, Yamagami T, Cann I, Ishino Y. 2016. PCNA is involved in the EndoQ-mediated DNA repair process in Thermococcales. Sci. Rep. 6, 25532 ( 10.1038/srep25532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kisker C, Kuper J, Van Houten B. 2013. Prokaryotic nucleotide excision repair. Cold Spring Harb. perspect. Biol. 5, a012591 ( 10.1101/cshperspect.a012591) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Van Houten B, Croteau DL, DellaVecchia MJ, Wang H, Kisker C. 2005. 'Close-fitting sleeves’: DNA damage recognition by the UvrABC nuclease system. Mutat. Res. 577, 92–117. ( 10.1016/j.mrfmmm.2005.03.013) [DOI] [PubMed] [Google Scholar]
- 298.Friedberg E, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. 2006. DNA repair and mutagenesis, 2nd edn Washington, DC: ASM Press. [Google Scholar]
- 299.Costa RM, Chigancas V, Galhardo Rda S, Carvalho H, Menck CF. 2003. The eukaryotic nucleotide excision repair pathway. Biochimie 85, 1083–1099. ( 10.1016/j.biochi.2003.10.017) [DOI] [PubMed] [Google Scholar]
- 300.Savery NJ. 2007. The molecular mechanism of transcription-coupled DNA repair. Trends Microbiol. 15, 326–333. ( 10.1016/j.tim.2007.05.005) [DOI] [PubMed] [Google Scholar]
- 301.Mellon I, Hanawalt PC. 1989. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature 342, 95–98. ( 10.1038/342095a0) [DOI] [PubMed] [Google Scholar]
- 302.Ganesan A, Spivak G, Hanawalt PC. 2012. Transcription-coupled DNA repair in prokaryotes. Prog. Mol. Biol. Transl. Sci. 110, 25–40. ( 10.1016/B978-0-12-387665-2.00002-X) [DOI] [PubMed] [Google Scholar]
- 303.Fuss JO, Tainer JA. 2011. XPB and XPD helicases in TFIIH orchestrate DNA duplex opening and damage verification to coordinate repair with transcription and cell cycle via CAK kinase. DNA Repair (Amst) 10, 697–713. ( 10.1016/j.dnarep.2011.04.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Scharer OD. 2013. Nucleotide excision repair in eukaryotes. Cold Spring Harb. perspect. Biol. 5, a012609 ( 10.1101/cshperspect.a012609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Moser J, Kool H, Giakzidis I, Caldecott K, Mullenders LHF, Fousteri MI. 2007. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol. Cell. 27, 311–323. ( 10.1016/j.molcel.2007.06.014) [DOI] [PubMed] [Google Scholar]
- 306.Iyama T, Wilson DM III, 2013. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst) 12, 620–636. ( 10.1016/j.dnarep.2013.04.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Giroux X, MacNeill SA. 2016. A novel archaeal DNA repair factor that acts with the UvrABC system to repair mitomycin C-induced DNA damage in a PCNA-dependent manner. Mol. Microbiol. 99, 1–14. ( 10.1111/mmi.13210) [DOI] [PubMed] [Google Scholar]
- 308.MacNeill SA. 2016. PCNA-binding proteins in the archaea: novel functionality beyond the conserved core. Curr. Genet. 62, 527–532. ( 10.1007/s00294-016-0577-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Galagan JE, et al. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12, 532–542. ( 10.1101/gr.223902) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Ogrunc M, Becker DF, Ragsdale SW, Sancar A. 1998. Nucleotide excision repair in the third kingdom. J. Bacteriol. 180, 5796–5798. ( 10.1128/JB.180.21.5796-5798.1998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Grogan DW. 2000. The question of DNA repair in hyperthermophilic archaea. Trends Microbiol. 8, 180–185. ( 10.1016/S0966-842X(00)01729-7) [DOI] [PubMed] [Google Scholar]
- 312.Rouillon C, White MF. 2011. The evolution and mechanisms of nucleotide excision repair proteins. Res. microbiol. 162, 19–26. ( 10.1016/j.resmic.2010.09.003) [DOI] [PubMed] [Google Scholar]
- 313.Grogan DW. 2015. Understanding DNA repair in hyperthermophilic Archaea: persistent gaps and other reasons to focus on the fork. Archaea. 2015, 942605 ( 10.1155/2015/942605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.She Q, Zhang C, Deng L, Peng N, Chen Z, Liang YX. 2009. Genetic analyses in the hyperthermophilic archaeon Sulfolobus islandicus. Biochem. Soc. Trans. 37, 92–96. ( 10.1042/BST0370092) [DOI] [PubMed] [Google Scholar]
- 315.Dorazi R, Gotz D, Munro S, Bernander R, White MF. 2007. Equal rates of repair of DNA photoproducts in transcribed and non-transcribed strands in Sulfolobus solfataricus. Mol. Microbiol. 63, 521–529. ( 10.1111/j.1365-2958.2006.05516.x) [DOI] [PubMed] [Google Scholar]
- 316.Romano V, Napoli A, Salerno V, Valenti A, Rossi M, Ciaramella M. 2007. Lack of strand-specific repair of UV-induced DNA lesions in three genes of the archaeon Sulfolobus solfataricus. J. Mol. Biol. 365, 921–929. ( 10.1016/j.jmb.2006.10.045) [DOI] [PubMed] [Google Scholar]
- 317.Gehring AM, Santangelo TJ. 2017. Archaeal RNA polymerase arrests transcription at DNA lesions. Transcription. 8, 288–296. ( 10.1080/21541264.2017.1324941) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Stantial N, Dumpe J, Pietrosimone K, Baltazar F, Crowley DJ. 2016. Transcription-coupled repair of UV damage in the halophilic archaea. DNA Repair (Amst) 41, 63–68. ( 10.1016/j.dnarep.2016.03.007) [DOI] [PubMed] [Google Scholar]
- 319.Naor A, Altman-Price N, Soucy SM, Green AG, Mitiagin Y, Turgeman-Grott I, Davidovich N, Gogarten JP, Gophna U. 2016. Impact of a homing intein on recombination frequency and organismal fitness. Proc. Natl Acad. Sci. USA 113, E4654–E4661. ( 10.1073/pnas.1606416113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Iyer RR, Pluciennik A, Burdett V, Modrich PL. 2006. DNA mismatch repair: functions and mechanisms. Chem. Rev. 106, 302–323. ( 10.1021/cr0404794) [DOI] [PubMed] [Google Scholar]
- 321.Grogan DW. 2004. Stability and repair of DNA in hyperthermophilic Archaea. Curr. Issues Mol. Biol. 6, 137–144. [PubMed] [Google Scholar]
- 322.Sachadyn P. 2010. Conservation and diversity of MutS proteins. Mutat. Res. 694, 20–30. ( 10.1016/j.mrfmmm.2010.08.009) [DOI] [PubMed] [Google Scholar]
- 323.Lin Z, Nei M, Ma H. 2007. The origins and early evolution of DNA mismatch repair genes multiple horizontal gene transfers and co-evolution. Nucleic Acids Res. 35, 7591–7603. ( 10.1093/nar/gkm921) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Mizrahi V, Andersen SJ. 1998. DNA repair in Mycobacterium tuberculosis: what have we learnt from the genome sequence? Mol. Microbiol. 29, 1331–1339. ( 10.1046/j.1365-2958.1998.01038.x) [DOI] [PubMed] [Google Scholar]
- 325.Banasik M, Sachadyn P. 2014. Conserved motifs of MutL proteins. Mutat. Res. 769, 69–79. ( 10.1016/j.mrfmmm.2014.07.006) [DOI] [PubMed] [Google Scholar]
- 326.Grogan DW, Carver GT, Drake JW. 2001. Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc. Natl Acad. Sci. USA. 98, 7928–7933. ( 10.1073/pnas.141113098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Busch CR, DiRuggiero J. 2010. MutS and MutL are dispensable for maintenance of the genomic mutation rate in the halophilic archaeon Halobacterium salinarum NRC-1. PLoS ONE 5, e9045 ( 10.1371/journal.pone.0009045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Zhang LK, Jiang DH, Wu M, Yang ZH, Oger PM. In press. New insights into DNA repair revealed by NucS endonucleases from hyperthermophilic archaea. Front. Microbiol. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Ren B, Kuhn J, Meslet-Cladiere L, Briffotaux J, Norais C, Lavigne R, Flament D, Ladenstein R, Myllykallio H. 2009. Structure and function of a novel endonuclease acting on branched DNA substrates. Embo J. 28, 2479–2489. ( 10.1038/emboj.2009.192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Creze C, et al. 2012. Modulation of the Pyrococcus abyssi NucS endonuclease activity by replication clamp at functional and structural levels. J. Biol. Chem. 287, 15 648–15 660. ( 10.1074/jbc.M112.346361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Creze C, Lestini R, Kuhn J, Ligabue A, Becker HF, Czjzek M, Flament D, Myllykallio H. 2011. Structure and function of a novel endonuclease acting on branched DNA substrates. Biochem. Soc. Trans. 39, 145–149. ( 10.1042/BST0390145) [DOI] [PubMed] [Google Scholar]
- 332.Roberts JA, Bell SD, White MF. 2003. An archaeal XPF repair endonuclease dependent on a heterotrimeric PCNA. Mol. Microbiol. 48, 361–371. ( 10.1046/j.1365-2958.2003.03444.x) [DOI] [PubMed] [Google Scholar]
- 333.Ishino S, Nishi Y, Oda S, Uemori T, Sagara T, Takatsu N, Yamagami T, Shirai T, Ishino Y. 2016. Identification of a mismatch-specific endonuclease in hyperthermophilic Archaea. Nucleic Acids Res. 44, 2977–2986. ( 10.1093/nar/gkw153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Ariyoshi M, Morikawa K. 2016. A dual base flipping mechanism for archaeal mismatch repair. Structure 24, 1859–1861. ( 10.1016/j.str.2016.10.004) [DOI] [PubMed] [Google Scholar]
- 335.Nakae S, Hijikata A, Tsuji T, Yonezawa K, Kouyama KI, Mayanagi K, Ishino S, Ishino Y, Shirai T. 2016. Structure of the EndoMS-DNA complex as mismatch restriction endonuclease. Structure 24, 1960–1971. ( 10.1016/j.str.2016.09.005) [DOI] [PubMed] [Google Scholar]
- 336.Suzuki S, Kurosawa N. 2019. Endonucleases responsible for DNA repair of helix-distorting DNA lesions in the thermophilic crenarchaeon Sulfolobus acidocaldarius in vivo. Extremophiles 23, 613–624. ( 10.1007/s00792-019-01120-9) [DOI] [PubMed] [Google Scholar]
- 337.Zhang L, Li Y, Shi H, Zhang D, Yang Z, Oger P, Zheng J. 2019. Biochemical characterization and mutational studies of the 8-oxoguanine DNA glycosylase from the hyperthermophilic and radioresistant archaeon Thermococcus gammatolerans. Appl. Microbiol. Biotechnol. 103, 8021–8033. ( 10.1007/s00253-019-10031-w) [DOI] [PubMed] [Google Scholar]
- 338.Eisen JA. 1998. A phylogenomic study of the MutS family of proteins. Nucleic Acids Res. 26, 4291–4300. ( 10.1093/nar/26.18.4291) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Gardner AF, Jack WE. 1999. Determinants of nucleotide sugar recognition in an archaeon DNA polymerase. Nucleic Acids Res. 27, 2545–2553. ( 10.1093/nar/27.12.2545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Heider MR, Burkhart BW, Santangelo TJ, Gardner AF. 2017. Defining the RNaseH2 enzyme-initiated ribonucleotide excision repair pathway in Archaea. J. Biol. Chem. 292, 8835–8845. ( 10.1074/jbc.M117.783472) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Friedberg EC, Wagner R, Radman M. 2002. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296, 1627–1630. ( 10.1126/science.1070236) [DOI] [PubMed] [Google Scholar]
- 342.Woodgate R. 1999. A plethora of lesion-replicating DNA polymerases. Genes Dev. 13, 2191–2195. ( 10.1101/gad.13.17.2191) [DOI] [PubMed] [Google Scholar]
- 343.Ling H, Boudsocq F, Woodgate R, Yang W. 2001. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107, 91–102. ( 10.1016/S0092-8674(01)00515-3) [DOI] [PubMed] [Google Scholar]
- 344.Zhou BL, Pata JD, Steitz TA. 2001. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell. 8, 427–437. ( 10.1016/S1097-2765(01)00310-0) [DOI] [PubMed] [Google Scholar]
- 345.Goodman MF, Tippin B. 2000. The expanding polymerase universe. Nat. Rev. Mol. Cell Biol. 1, 101–109. ( 10.1038/35040051) [DOI] [PubMed] [Google Scholar]
- 346.Ohmori H, et al. 2001. The Y-family of DNA polymerases. Mol. Cell. 8, 7–8. ( 10.1016/S1097-2765(01)00278-7) [DOI] [PubMed] [Google Scholar]
- 347.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. 2006. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 4, e11 ( 10.1371/journal.pbio.0040011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Prakash S, Johnson RE, Prakash L. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74, 317–353. ( 10.1146/annurev.biochem.74.082803.133250) [DOI] [PubMed] [Google Scholar]
- 349.Sale JE, Lehmann AR, Woodgate R. 2012. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 13, 141–152. ( 10.1038/nrm3289) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.McIntyre J, Woodgate R. 2015. Regulation of translesion DNA synthesis: posttranslational modification of lysine residues in key proteins. DNA Repair (Amst) 29, 166–179. ( 10.1016/j.dnarep.2015.02.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Kim J, Dordick JS. 1997. Unusual salt and solvent dependence of a protease from an extreme halophile. Biotechnol. Bioeng. 55, 471–479. () [DOI] [PubMed] [Google Scholar]
- 352.DeLucia AM, Grindley ND, Joyce CM. 2007. Conformational changes during normal and error-prone incorporation of nucleotides by a Y-family DNA polymerase detected by 2-aminopurine fluorescence. Biochemistry 46, 10 790–10 803. ( 10.1021/bi7006756) [DOI] [PubMed] [Google Scholar]
- 353.Kulaeva OI, Koonin EV, McDonald JP, Randall SK, Rabinovich N, Connaughton JF, Levine AS, Woodgate R. 1996. Identification of a DinB/UmuC homolog in the archeon Sulfolobus solfataricus. Mutat. Res. 357, 245–253. ( 10.1016/0027-5107(96)00164-9) [DOI] [PubMed] [Google Scholar]
- 354.Silvian LF, Toth EA, Pham P, Goodman MF, Ellenberger T. 2001. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nat. Struct. Biol. 8, 984–989. ( 10.1038/nsb1101-984) [DOI] [PubMed] [Google Scholar]
- 355.Boudsocq F, Iwai S, Hanaoka F, Woodgate R. 2001. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic poleta. Nucleic Acids Res. 29, 4607–4616. ( 10.1093/nar/29.22.4607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Potapova O, Grindley ND, Joyce CM. 2002. The mutational specificity of the Dbh lesion bypass polymerase and its implications. J. Biol. Chem. 277, 28 157–28 166. ( 10.1074/jbc.M202607200) [DOI] [PubMed] [Google Scholar]
- 357.Cranford MT, Kaszubowski JD, Trakselis MA. 2020. A hand-off of DNA between archaeal polymerases allows high-fidelity replication to resume at a discrete intermediate three bases past 8-oxoguanine. Nucleic Acids Res. 48, 10 986–10 997. ( 10.1093/nar/gkaa803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Johnson RE, Prakash S, Prakash L. 1999. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science 283, 1001–1004. ( 10.1126/science.283.5404.1001) [DOI] [PubMed] [Google Scholar]
- 359.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. 1999. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 18, 3491–3501. ( 10.1093/emboj/18.12.3491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Tang M, Pham P, Shen X, Taylor JS, O'Donnell M, Woodgate R, Goodman MF. 2000. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404, 1014–1018. ( 10.1038/35010020) [DOI] [PubMed] [Google Scholar]
- 361.Johnson RE, Prakash S, Prakash L. 2000. The human DINB1 gene encodes the DNA polymerase Poltheta. Proc. Natl Acad. Sci. USA. 97, 3838–3843. ( 10.1073/pnas.97.8.3838) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. 2000. Error-prone bypass of certain DNA lesions by the human DNA polymerase kappa. Genes Dev. 14, 1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 363.Zhang Y, Yuan F, Wu X, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. 2000. Error-prone lesion bypass by human DNA polymerase eta. Nucleic Acids Res. 28, 4717–4724. ( 10.1093/nar/28.23.4717) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Edwards DN, Machwe A, Wang Z, Orren DK. 2014. Intramolecular telomeric G-quadruplexes dramatically inhibit DNA synthesis by replicative and translesion polymerases, revealing their potential to lead to genetic change. PLoS ONE 9, e80664 ( 10.1371/journal.pone.0080664) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Berroyer A, Alvarado G, Larson ED. 2019. Response of Sulfolobus solfataricus Dpo4 polymerase in vitro to a DNA G-quadruplex. Mutagenesis. 34, 289–297. ( 10.1093/mutage/gez010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Feng X, Liu X, Xu R, Zhao R, Feng W, Liao J, Han W, She Q. 2020. A unique B-family DNA polymerase facilitating error-prone DNA damage tolerance in Crenarchaeota. Front. microbiol. 11, 1585 ( 10.3389/fmicb.2020.01585) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Cranford MT, Chu AM, Baguley JK, Bauer RJ, Trakselis MA. 2017. Characterization of a coupled DNA replication and translesion synthesis polymerase supraholoenzyme from Archaea. Nucleic Acids Res. 45, 8329–8340. ( 10.1093/nar/gkx539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Indiani C, McInerney P, Georgescu R, Goodman MF, O'Donnell M. 2005. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol. Cell. 19, 805–815. ( 10.1016/j.molcel.2005.08.011) [DOI] [PubMed] [Google Scholar]
- 369.Boehm EM, Spies M, Washington MT. 2016. PCNA tool belts and polymerase bridges form during translesion synthesis. Nucleic Acids Res. 44, 8250–8260. ( 10.1093/nar/gkw563) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Bunting KA, Roe SM, Pearl LH. 2003. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. EMBO J. 22, 5883–5892. ( 10.1093/emboj/cdg568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Brissett NC, Martin MJ, Bartlett EJ, Bianchi J, Blanco L, Doherty AJ. 2013. Molecular basis for DNA double-strand break annealing and primer extension by an NHEJ DNA polymerase. Cell Reports 5, 1108–1120. ( 10.1016/j.celrep.2013.10.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Aravind L, Koonin EV. 2001. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 11, 1365–1374. ( 10.1101/gr.181001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Bowater R, Doherty AJ. 2006. Making ends meet: repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genet. 2, e8 ( 10.1371/journal.pgen.0020008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Delmas S, Shunburne L, Ngo HP, Allers T. 2009. Mre11-Rad50 promotes rapid repair of DNA damage in the polyploid archaeon Haloferax volcanii by restraining homologous recombination. PLoS Genet. 5, e1000552 ( 10.1371/journal.pgen.1000552) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Stachler AE, Turgeman-Grott I, Shtifman-Segal E, Allers T, Marchfelder A, Gophna U. 2017. High tolerance to self-targeting of the genome by the endogenous CRISPR-Cas system in an archaeon. Nucleic Acids Res. 45, 5208–5216. ( 10.1093/nar/gkx150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Wang H, Xu X. 2017. Microhomology-mediated end joining: new players join the team. Cell Biosci. 7, 1–6. ( 10.1186/s13578-017-0136-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Sinha S, Villarreal D, Shim EY, Lee SE. 2016. Risky business: microhomology-mediated end joining. Mutat. Res. 788, 17–24. ( 10.1016/j.mrfmmm.2015.12.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Seol JH, Shim EY, Lee SE. 2018. Microhomology-mediated end joining: Good, bad and ugly. Mutat. Res. 809, 81–87. ( 10.1016/j.mrfmmm.2017.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Zhang C, Whitaker RJ. 2018. Microhomology-mediated high-throughput gene inactivation strategy for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 84, e02167-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Heyer WD, Ehmsen KT, Liu J. 2010. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139. ( 10.1146/annurev-genet-051710-150955) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Krejci L, Altmannova V, Spirek M, Zhao X. 2012. Homologous recombination and its regulation. Nucleic Acids Res. 40, 5795–5818. ( 10.1093/nar/gks270) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.White MF. 2011. Homologous recombination in the Archaea: the means justify the ends. Biochem. Soc. Trans. 39, 15–19. ( 10.1042/BST0390015) [DOI] [PubMed] [Google Scholar]
- 383.Naor A, Lapierre P, Mevarech M, Papke RT, Gophna U. 2012. Low species barriers in halophilic archaea and the formation of recombinant hybrids. Curr. Biol. 22, 1444–1448. ( 10.1016/j.cub.2012.05.056) [DOI] [PubMed] [Google Scholar]
- 384.van Wolferen M, Ajon M, Driessen AJ, Albers SV. 2013. Molecular analysis of the UV-inducible pili operon from Sulfolobus acidocaldarius. Microbiologyopen. 2, 928–937. ( 10.1002/mbo3.128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Oh J, Symington LS. 2018. Role of the Mre11 complex in preserving genome integrity. Genes 9, 25 ( 10.3390/genes9010025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Paull TT. 2018. 20 Years of Mre11 biology: no end in sight. Mol. Cell. 71, 419–427. ( 10.1016/j.molcel.2018.06.033) [DOI] [PubMed] [Google Scholar]
- 387.D'Amours D, Jackson SP. 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3, 317–327. ( 10.1038/nrm805) [DOI] [PubMed] [Google Scholar]
- 388.de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C. 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell. 8, 1129–1135. ( 10.1016/S1097-2765(01)00381-1) [DOI] [PubMed] [Google Scholar]
- 389.Lisby M, Rothstein R. 2004. DNA repair: keeping it together. Curr. Biol. 14, R994–R996. ( 10.1016/j.cub.2004.11.020) [DOI] [PubMed] [Google Scholar]
- 390.Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. 2005. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature 437, 440–443. ( 10.1038/nature03927) [DOI] [PubMed] [Google Scholar]
- 391.Williams RS, Williams JS, Tainer JA. 2007. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. cell biol.=Biochimie et biologie cellulaire. 85, 509–520. ( 10.1139/O07-069) [DOI] [PubMed] [Google Scholar]
- 392.Hopfner KP, Karcher A, Shin D, Fairley C, Tainer JA, Carney JP. 2000. Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J. Bacteriol. 182, 6036–6041. ( 10.1128/JB.182.21.6036-6041.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Hopkins BB, Paull TT. 2008. The P. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell 135, 250–260. ( 10.1016/j.cell.2008.09.054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Kish A, DiRuggiero J. 2008. Rad50 is not essential for the Mre11-dependent repair of DNA double-strand breaks in Halobacterium sp. strain NRC-1. J. Bacteriol. 190, 5210–5216. ( 10.1128/JB.00292-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Delmas S, Duggin IG, Allers T. 2013. DNA damage induces nucleoid compaction via the Mre11-Rad50 complex in the archaeon Haloferax volcanii. Mol. Microbiol. 87, 168–179. ( 10.1111/mmi.12091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM. 2012. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat. Cell Biol. 14, 502–509. ( 10.1038/ncb2465) [DOI] [PubMed] [Google Scholar]
- 397.Mine-Hattab J, Rothstein R. 2012. Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 14, 510–517. ( 10.1038/ncb2472) [DOI] [PubMed] [Google Scholar]
- 398.Levin-Zaidman S, Frenkiel-Krispin D, Shimoni E, Sabanay I, Wolf SG, Minsky A. 2000. Ordered intracellular RecA-DNA assemblies: a potential site of in vivo RecA-mediated activities. Proc. Natl Acad. Sci. USA. 97, 6791–6796. ( 10.1073/pnas.090532397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Smith BT, Grossman AD, Walker GC. 2002. Localization of UvrA and effect of DNA damage on the chromosome of Bacillus subtilis. J. Bacteriol. 184, 488–493. ( 10.1128/JB.184.2.488-493.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Constantinesco F, Forterre P, Elie C. 2002. NurA, a novel 5′-3′ nuclease gene linked to rad50 and mre11 homologs of thermophilic Archaea. EMBO Rep. 3, 537–542. ( 10.1093/embo-reports/kvf112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Fujii Y, Inoue M, Fukui K, Kuramitsu S, Masui R. 2018. Resistance to UV irradiation caused by inactivation of nurA and herA genes in Thermus thermophilus. J. Bacteriol. 200, e00201-18 ( 10.1128/JB.00201-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Constantinesco F, Forterre P, Koonin EV, Aravind L, Elie C. 2004. A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic Archaea. Nucleic Acids Res. 32, 1439–1447. ( 10.1093/nar/gkh283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Blackwood JK, Rzechorzek NJ, Abrams AS, Maman JD, Pellegrini L, Robinson NP. 2012. Structural and functional insights into DNA-end processing by the archaeal HerA helicase-NurA nuclease complex. Nucleic Acids Res. 40, 3183–3196. ( 10.1093/nar/gkr1157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Ahdash Z, et al. 2017. Mechanistic insight into the assembly of the HerA-NurA helicase-nuclease DNA end resection complex. Nucleic Acids Res. 45, 12 025–12 038. ( 10.1093/nar/gkx890) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Huang Q, Liu L, Liu J, Ni J, She Q, Shen Y. 2015. Efficient 5′-3′ DNA end resection by HerA and NurA is essential for cell viability in the crenarchaeon Sulfolobus islandicus. BMC Mol. Biol. 16, 2 ( 10.1186/s12867-015-0030-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Cheng K, Chen X, Xu G, Wang L, Xu H, Yang S, Zhao Y, Hua Y. 2015. Biochemical and functional characterization of the NurA-HerA complex from Deinococcus radiodurans. J. Bacteriol. 197, 2048–2061. ( 10.1128/JB.00018-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Lin Z, Kong H, Nei M, Ma H. 2006. Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc. Natl Acad. Sci. USA 103, 10 328–10 333. ( 10.1073/pnas.0604232103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Shin DS, et al. 2003. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 22, 4566–4576. ( 10.1093/emboj/cdg429) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Clark AJ, Margulies AD. 1965. Isolation and characterization of recombination-deficient mutants of Escherichia coli K-12. Proc. Natl Acad. Sci. USA 53, 451–459. ( 10.1073/pnas.53.2.451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Woods WG, Dyall-Smith ML. 1997. Construction and analysis of a recombination-deficient (radA) mutant of Haloferax volcanii. Mol. Microbiol. 23, 791–797. ( 10.1046/j.1365-2958.1997.2651626.x) [DOI] [PubMed] [Google Scholar]
- 411.Guy CP, Haldenby S, Brindley A, Walsh DA, Briggs GS, Warren MJ, Allers T, Bolt EL. 2006. Interactions of RadB, a DNA repair protein in archaea, with DNA and ATP. J. Mol. Biol. 358, 46–56. ( 10.1016/j.jmb.2006.02.010) [DOI] [PubMed] [Google Scholar]
- 412.Wardell K, Haldenby S, Jones N, Liddell S, Ngo GHP, Allers T. 2017. RadB acts in homologous recombination in the archaeon Haloferax volcanii, consistent with a role as recombination mediator. DNA Repair (Amst) 55, 7–16. ( 10.1016/j.dnarep.2017.04.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Lovett ST. 2006. Replication arrest-stimulated recombination: dependence on the RecA paralog, RadA/Sms and translesion polymerase. DinB. DNA Repair (Amst 5, 1421–1427. ( 10.1016/j.dnarep.2006.06.008) [DOI] [PubMed] [Google Scholar]
- 414.Sung P. 1997. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11, 1111–1121. ( 10.1101/gad.11.9.1111) [DOI] [PubMed] [Google Scholar]
- 415.Takata M, et al. 2000. The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell Biol. 20, 6476–6482. ( 10.1128/MCB.20.17.6476-6482.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. 2001. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell Biol. 21, 2858–2866. ( 10.1128/MCB.21.8.2858-2866.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Rashid N, Morikawa M, Imanaka T. 1996. A RecA/RAD51 homologue from a hyperthermophilic archaeon retains the major RecA domain only. Mol. Gen. Genet. 253, 397–400. ( 10.1007/s004380050337) [DOI] [PubMed] [Google Scholar]
- 418.Komori K, Miyata T, DiRuggiero J, Holley-Shanks R, Hayashi I, Cann IK, Mayanagi K, Shinagawa H, Ishino Y. 2000. Both RadA and RadB are involved in homologous recombination in Pyrococcus furiosus. J. Biol. Chem. 275, 33 782–33 790. ( 10.1074/jbc.M004557200) [DOI] [PubMed] [Google Scholar]
- 419.Sugiyama T, Kowalczykowski SC. 2002. Rad52 protein associates with replication protein a (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 277, 31 663–31 672. ( 10.1074/jbc.M203494200) [DOI] [PubMed] [Google Scholar]
- 420.Liu J, Doty T, Gibson B, Heyer WD. 2010. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1260–1262. ( 10.1038/nsmb.1904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer WD. 2011. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature 479, 245–248. ( 10.1038/nature10522) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Haldenby S, White MF, Allers T. 2009. RecA family proteins in archaea: RadA and its cousins. Biochem. Soc. Trans. 37, 102–107. ( 10.1042/BST0370102) [DOI] [PubMed] [Google Scholar]
- 423.Wyatt HD, West SC. 2014. Holliday junction resolvases. Cold Spring Harb. perspect. biol. 6, a023192 ( 10.1101/cshperspect.a023192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Lilley DM, White MF. 2000. Resolving the relationships of resolving enzymes. Proc. Natl Acad. Sci. USA. 97, 9351–9353. ( 10.1073/pnas.97.17.9351) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Lilley DM, White MF. 2001. The junction-resolving enzymes. Nat. Rev. Mol. Cell Biol. 2, 433–443. ( 10.1038/35073057x) [DOI] [PubMed] [Google Scholar]
- 426.West SC. 1997. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 31, 213–244. ( 10.1146/annurev.genet.31.1.213) [DOI] [PubMed] [Google Scholar]
- 427.Lilley DMJ. 2017. Holliday junction-resolving enzymes-structures and mechanisms. FEBS Lett. 591, 1073–1082. ( 10.1002/1873-3468.12529) [DOI] [PubMed] [Google Scholar]
- 428.West SC, Chan YW. 2017. Genome instability as a consequence of defects in the resolution of recombination intermediates. Cold Spring Harb. Symp. Quant. Biol. 82, 207–212. ( 10.1101/sqb.2017.82.034256) [DOI] [PubMed] [Google Scholar]
- 429.Hong Y, Chu M, Li Y, Ni J, Sheng D, Hou G, She Q, Shen Y. 2012. Dissection of the functional domains of an archaeal Holliday junction helicase. DNA Repair (Amst) 11, 102–111. ( 10.1016/j.dnarep.2011.10.009) [DOI] [PubMed] [Google Scholar]
- 430.Dorazi R, Parker JL, White MF. 2006. PCNA activates the Holliday junction endonuclease Hjc. J. Mol. Biol. 364, 243–247. ( 10.1016/j.jmb.2006.09.011) [DOI] [PubMed] [Google Scholar]
- 431.Huang Q, Li Y, Zeng C, Song T, Yan Z, Ni J, She Q, Shen Y. 2015. Genetic analysis of the Holliday junction resolvases Hje and Hjc in Sulfolobus islandicus. Extremophiles. 19, 505–514. ( 10.1007/s00792-015-0734-5) [DOI] [PubMed] [Google Scholar]
- 432.Kvaratskhelia M, White MF. 2000. An archaeal Holliday junction resolving enzyme from Sulfolobus solfataricus exhibits unique properties. J. Mol. Biol. 295, 193–202. ( 10.1006/jmbi.1999.3363) [DOI] [PubMed] [Google Scholar]
- 433.Huang Q, Mayaka JB, Zhong Q, Zhang C, Hou G, Ni J, Shen Y. 2019. Phosphorylation of the archaeal holliday junction resolvase Hjc inhibits its catalytic activity and facilitates DNA repair in Sulfolobus islandicus REY15A. Front. microbiol. 10, 1214 ( 10.3389/fmicb.2019.01214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Reimann J, et al. 2013. Archaeal signal transduction: impact of protein phosphatase deletions on cell size, motility, and energy metabolism in Sulfolobus acidocaldarius. Mol. Cell Proteomics. 12, 3908–3923. ( 10.1074/mcp.M113.027375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Dehe PM, et al. 2013. Regulation of Mus81-Eme1 Holliday junction resolvase in response to DNA damage. Nat. Struct. Mol. Biol. 20, 598–603. ( 10.1038/nsmb.2550) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Blanco MG, Matos J, West SC. 2014. Dual control of Yen1 nuclease activity and cellular localization by Cdk and Cdc14 prevents genome instability. Mol. Cell. 54, 94–106. ( 10.1016/j.molcel.2014.02.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Chan YW, West SC. 2014. Spatial control of the GEN1 Holliday junction resolvase ensures genome stability. Nat. commun. 5, 4844 ( 10.1038/ncomms5844) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Nishino T, Komori K, Tsuchiya D, Ishino Y, Morikawa K. 2005. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure 13, 143–153. ( 10.1016/j.str.2004.11.008) [DOI] [PubMed] [Google Scholar]
- 439.Nishino T, Komori K, Ishino Y, Morikawa K. 2003. X-ray and biochemical anatomy of an archaeal XPF/Rad1/Mus81 family nuclease: similarity between its endonuclease domain and restriction enzymes. Structure 11, 445–457. ( 10.1016/S0969-2126(03)00046-7) [DOI] [PubMed] [Google Scholar]
- 440.Nishino T, Komori K, Ishino Y, Morikawa K. 2005. Structural and functional analyses of an archaeal XPF/Rad1/Mus81 nuclease: asymmetric DNA binding and cleavage mechanisms. Structure 13, 1183–1192. ( 10.1016/j.str.2005.04.024) [DOI] [PubMed] [Google Scholar]
- 441.Komori K, Hidaka M, Horiuchi T, Fujikane R, Shinagawa H, Ishino Y. 2004. Cooperation of the N-terminal helicase and C-terminal endonuclease activities of archaeal Hef protein in processing stalled replication forks. J. Biol. Chem. 279, 53 175–53 185. ( 10.1074/jbc.M409243200) [DOI] [PubMed] [Google Scholar]
- 442.Lestini R, Laptenok SP, Kuhn J, Hink MA, Schanne-Klein MC, Liebl U, Myllykallio H. 2013. Intracellular dynamics of archaeal FANCM homologue Hef in response to halted DNA replication. Nucleic Acids Res. 41, 10 358–10 370. ( 10.1093/nar/gkt816) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Ishino S, et al. 2014. Multiple interactions of the intrinsically disordered region between the helicase and nuclease domains of the archaeal Hef protein. J. Biol. Chem. 289, 21 627–21 639. ( 10.1074/jbc.M114.554998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Pluchon PF, et al. 2013. An extended network of genomic maintenance in the archaeon Pyrococcus abyssi highlights unexpected associations between eucaryotic homologs. PLoS ONE 8, e79707 ( 10.1371/journal.pone.0079707) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Rohleder F, Huang J, Xue Y, Kuper J, Round A, Seidman M, Wang W, Kisker C. 2016. FANCM interacts with PCNA to promote replication traverse of DNA interstrand crosslinks. Nucleic Acids Res. 44, 3219–3232. ( 10.1093/nar/gkw037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Feng L, et al. 2018. The trimeric Hef-associated nuclease HAN is a 3–5′ exonuclease and is probably involved in DNA repair. Nucleic Acids Res. 46, 9027–9043. ( 10.1093/nar/gky707) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Marini F, Wood RD. 2002. A human DNA helicase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 277, 8716–8723. ( 10.1074/jbc.M110271200) [DOI] [PubMed] [Google Scholar]
- 448.Guy CP, Bolt EL. 2005. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res. 33, 3678–3690. ( 10.1093/nar/gki685) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Fujikane R, Komori K, Shinagawa H, Ishino Y. 2005. Identification of a novel helicase activity unwinding branched DNAs from the hyperthermophilic archaeon, Pyrococcus furiosus. J. Biol. Chem. 280, 12 351–12 358. ( 10.1074/jbc.M413417200) [DOI] [PubMed] [Google Scholar]
- 450.Takata K, Reh S, Tomida J, Person MD, Wood RD. 2013. Human DNA helicase HELQ participates in DNA interstrand crosslink tolerance with ATR and RAD51 paralogs. Nat. commun. 4, 2338 ( 10.1038/ncomms3338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Adelman CA, et al. 2013. HELQ promotes RAD51 paralogue-dependent repair to avert germ cell loss and tumorigenesis. Nature 502, 381–384. ( 10.1038/nature12565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Woodman IL, Brammer K, Bolt EL. 2011. Physical interaction between archaeal DNA repair helicase Hel308 and Replication Protein A (RPA). DNA Repair (Amst) 10, 306–313. ( 10.1016/j.dnarep.2010.12.001) [DOI] [PubMed] [Google Scholar]
- 453.Zhang C, Tian B, Li S, Ao X, Dalgaard K, Gokce S, Liang Y, She Q. 2013. Genetic manipulation in Sulfolobus islandicus and functional analysis of DNA repair genes. Biochem. Soc. Trans. 41, 405–410. ( 10.1042/BST20120285) [DOI] [PubMed] [Google Scholar]
- 454.Ogawa T, Pickett GG, Kogoma T, Kornberg A. 1984. RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc. Natl Acad. Sci. USA. 81, 1040–1044. ( 10.1073/pnas.81.4.1040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Kogoma T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61, 212–238. ( 10.1128/.61.2.212-238.1997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Masai H, Arai K. 1996. Mechanisms of primer RNA synthesis and D-loop/R-loop-dependent DNA replication in Escherichia coli. Biochimie 78, 1109–1117. ( 10.1016/S0300-9084(97)86737-5) [DOI] [PubMed] [Google Scholar]
- 457.Gehring AM, Astling DP, Matsumi R, Burkhart BW, Kelman Z, Reeve JN, Jones KL, Santangelo TJ. 2017. Genome replication in Thermococcus kodakarensis independent of Cdc6 and an origin of replication. Front. microbiol. 8, 2084 ( 10.3389/fmicb.2017.02084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Ohbayashi R, Hirooka S, Onuma R, Kanesaki Y, Hirose Y, Kobayashi Y, Fujiwara T, Furusawa C, Miyagishima S. 2020. Evolutionary changes in DnaA-dependent chromosomal replication in Cyanobacteria. Front. microbiol. 11, 786 ( 10.3389/fmicb.2020.00786) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Poplawski A, Bernander R. 1997. Nucleoid structure and distribution in thermophilic Archaea. J. Bacteriol. 179, 7625–7630. ( 10.1128/JB.179.24.7625-7630.1997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Bernander R, Poplawski A. 1997. Cell cycle characteristics of thermophilic Archaea. J. Bacteriol. 179, 4963–4969. ( 10.1128/JB.179.16.4963-4969.1997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Wu Z, Yang H, Liu J, Wang L, Xiang H. 2014. Association between the dynamics of multiple replication origins and the evolution of multireplicon genome architecture in haloarchaea. Genome Biol. Evol. 6, 2799–2810. ( 10.1093/gbe/evu219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Yang H, Wu Z, Liu J, Liu X, Wang L, Cai S, Xiang H. 2015. Activation of a dormant replication origin is essential for Haloferax mediterranei lacking the primary origins. Nat. commun. 6, 8321 ( 10.1038/ncomms9321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Jain R, Rivera MC, Lake JA. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl Acad. Sci. USA 96, 3801–3806. ( 10.1073/pnas.96.7.3801) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.