Significance
Anorexia or decreased appetite is a common feature of infection. The effects of acute nutritional stress on effector T cells, which are required for pathogen-specific responses and protective immunological memory, are not fully understood. We show that reduced food intake during infection decreases the numbers of effector T cells in a manner dependent on the Farnesoid X Receptor (FXR). FXR was previously implicated in controlling hepatic responses to fasting. Our findings suggest that FXR functions in effector T cells to promote coherent physiological responses to decreased feeding, allowing organisms to scale their immune responses according to food availability.
Keywords: anorexia, infection, T cells, FXR
Abstract
Reduced nutrient intake is a widely conserved manifestation of sickness behavior with poorly characterized effects on adaptive immune responses. During infectious challenges, naive T cells encountering their cognate antigen become activated and differentiate into highly proliferative effector T cells. Despite their evident metabolic shift upon activation, it remains unclear how effector T cells respond to changes in nutrient availability in vivo. Here, we show that spontaneous or imposed feeding reduction during infection decreases the numbers of splenic lymphocytes. Effector T cells showed cell-intrinsic responses dependent on the nuclear receptor Farnesoid X Receptor (FXR). Deletion of FXR in T cells prevented starvation-induced loss of lymphocytes and increased effector T cell fitness in nutrient-limiting conditions, but imparted greater weight loss to the host. FXR deficiency increased the contribution of glutamine and fatty acids toward respiration and enhanced cell survival under low-glucose conditions. Provision of glucose during anorexia of infection rescued effector T cells, suggesting that this sugar is a limiting nutrient for activated lymphocytes and that alternative fuel usage may affect cell survival in starved animals. Altogether, we identified a mechanism by which the host scales immune responses according to food intake, featuring FXR as a T cell-intrinsic sensor.
Acute infection is often accompanied by physiological and behavioral alterations including changes in body temperature, anhedonia, and anorexia. Reduced feeding is a widely conserved manifestation of sickness behavior that affects host fitness in a context-dependent manner (1–3). While the effects of infection-induced anorexia on tissue function are well documented (2–5), its impact on immune responses remains poorly characterized. Despite enhancing bacterial clearance early during infection (6), starvation can reduce host resistance to subsequent challenges (7), suggesting that nutrient deprivation may limit adaptive immune responses.
During an infectious challenge, adaptive lymphocytes quickly exit quiescence and undergo vigorous proliferation, with populations of antigen-specific cells expanding over 20,000-fold numerically (8). To support their heightened metabolic demands, activated lymphocytes greatly increase their glycolytic capacity and the ability to acquire nutrients from the environment (9). Although the connection between cell-intrinsic metabolism and the effector function of lymphocytes has been extensively investigated, the responses of immune cells to changes in the metabolic state of the host, particularly in the context of anorexia of infection, are not fully understood.
We hypothesized that animals scale their immune responses according to nutrient availability and that lymphocytes display cell-intrinsic responses to the metabolic state of the host. We report that decreased feeding during infection causes a reduction in lymphocyte numbers that can be restored by provision of glucose. We further show that the nuclear receptor Farnesoid X Receptor (FXR), which regulates hepatic responses to fasting and refeeding (10, 11), is up-regulated upon T cell activation and plays a cell-intrinsic role in mediating starvation-induced loss of lymphocytes and in their fuel utilization. Impairing FXR-dependent T cell contraction in response to diminished nutrient availability resulted in greater weight loss and lower glycemia, indicating that the ability to scale the immune compartment according to food availability may affect organismal energy homeostasis during infection. Our results suggest that induction of FXR expression in activated T cells renders the effector T cell compartment sensitive to changes in host metabolism and that specific fuel utilization programs may support lymphocyte survival under nutrient-limiting conditions.
Results
Reduced Feeding during Infection Decreases Splenic Lymphocyte Numbers.
Since lymphocytes undergo massive clonal expansion during infectious challenges, we hypothesized that changes in nutrient intake may affect this bioenergetically costly process. Infection with lymphocytic choriomeningitis virus (LCMV) Armstrong causes no weight loss and induces limited anorexic responses that are negligible after 96 h (Fig. 1A), making this model amenable to perturbation-style experiments to dissect the effects of reduced food intake on adaptive lymphocytes. We thus assessed the impact of nutritional stress on emerging adaptive immune responses by comparing hosts fed ad libitum or subjected to 24 h of fasting on day 5 to day 6 (D5 to D6) post infection followed by a short period of refeeding (see Fig. 1B for experimental setup). Limiting nutrient intake led to lower splenic weights and decreased total leukocyte counts at the peak of adaptive immune responses on D7 (Fig. 1 C and D), affecting the numbers of T and B cells (Fig. 1E). Decreased cell counts were not yet detectable at D6 (SI Appendix, Fig. S1 A–C). However, prolonging fasting from D5 until the moment of analysis on D7 reduced cell counts similarly to transient food deprivation (SI Appendix, Fig. S1 D and E). These results imply that refeeding is dispensable to the numerical decrease in splenocytes and suggest that the observed changes on D7 are a delayed manifestation of responses to reduced food intake.
Fig. 1.
Reduced feeding during infection decreases splenic lymphocyte numbers. (A–G) Six- to 10-wk-old C57Bl6/N mice were infected with LCMV Armstrong. (A) Food consumption and body weight changes during the course of infection. (B) Schematics of food deprivation experiments. Mice were allowed to feed ad libitum or subjected to 24 h of fasting on D5 post infection beginning at 6 PM. Analyses of splenocytes were carried out on D7 after 18 h of refeeding. (C) Splenic masses. (D) Total counts of CD45+ cells. (E) Lymphocyte counts. (F) Cell counts for naive (Tn, CD44−CD62L+), central memory (Tcm, CD44+CD62L+), and effector (Teff, CD44+CD62L−) CD8+ T cells. (G) Representative FACS plots showing the percentage of H-2Kb/GP33 tetramer+ cells. Gated on effector CD8+ T cells. Data are plotted as mean ± SD. Data in A (n = 10) are representative of three independent experiments. Data in C–G (n = 10) are pooled from two independent experiments. Statistical significance determined by a two-tailed t test (C and D) or two-way ANOVA followed by Sidak’s correction for multiple comparisons (E and F). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
We then assessed the effects of transient food deprivation on the T cell compartment. While the counts for naive and central memory T cells were not altered by fasting and refeeding, effector T cell numbers on D7 were decreased compared to ad-libitum–fed controls (Fig. 1F and SI Appendix, Fig. S1F). The frequencies of Foxp3+ CD4+ regulatory T cells remained unchanged (SI Appendix, Fig. S1G). Likewise, the percentage of H2-Kb/GP33 tetramer+ cells was not altered by food deprivation (Fig. 1G), indicating that virus-specific cells decreased proportionally with the effector CD8+ T cell population.
Viral clearance during acute LCMV infection is critically dependent upon CD8+ T cell responses (12, 13). Since nutrient deprivation reduced the effector CD8+ T cell compartment, we sought to compare viral loads in mice fed ad libitum (AL) versus subjected to fasting. Quantification of splenocytes expressing the LCMV nuclear protein (NP) showed no differences between control and fasted mice, with comparable mean fluorescence intensity for NP staining across groups (SI Appendix, Fig. S1 H and I). Moreover, determination of viral RNA copies in spleen homogenates by qPCR revealed that fed and fasted mice had similar viral loads (SI Appendix, Fig. S1 J and K), indicating that reduced feeding is not associated with poor control of acute infection. In line with these results, while we did not find consistent changes in the fraction of T cells producing cytokines following ex vivo restimulation (SI Appendix, Fig. S1 L–N), fasting increased IFNγ levels on a per-cell basis (SI Appendix, Fig. S1O). These results suggest that enhanced cell-intrinsic responses may compensate for reduced lymphocyte numbers during nutrient deprivation or, nonexclusively, that the metabolic state of the host during fasting may directly interfere with viral replication.
FXR Renders Effector T Cells Sensitive to Reduced Feeding.
Next, we sought to determine the mechanisms involved in the population-level responses of lymphocytes to reduced food intake. Caloric restriction at the steady state was shown to increase systemic glucocorticoid (GC) levels, which promoted peripheral CD8+ T lymphopenia and the relocation of cells to the bone marrow in a GC-receptor–dependent manner (14). Interestingly, we found that, while infection increased serum corticosterone concentration, food deprivation failed to further enhance GC levels (SI Appendix, Fig. S2A). Serum corticosterone levels still displayed day-to-night fluctuation in infected mice (SI Appendix, Fig. S2A), indicating that the hypothalamic–pituitary–adrenal axis remained subject to circadian regulation but could no longer relay information regarding nutritional stress during an infectious challenge. Consistent with these results, we found no changes in the cellular counts or composition of the bone marrow (SI Appendix, Fig. S2 B and C). These findings suggest that a distinct mechanism regulates the responses of effector lymphocytes to nutritional stress during infection.
The nutrient-sensing nuclear receptors Peroxisome-Proliferating Activating Receptor alpha and FXR are key regulators of transcriptional responses to fasting and refeeding (15–17). FXR acts as a fed-state sensor in the liver and represses catabolic processes in nutrient-replete conditions (10, 11). We postulated that a similar mechanism might be operational in effector T cells and thus affect their responses to reduced food intake during infection. In support of this idea, we found that FXR expression, while undetectable in naive CD8+ T cells, can be induced upon T cell activation in vitro (SI Appendix, Fig. S3 A and B). The molecular weights of FXR protein varied across ileum, liver, and T cell lysates (SI Appendix, Fig. S3A), likely reflecting cell-type–specific expression of FXR isoforms (18). To assess a cell-intrinsic role for FXR in effector T cells, we induced T cell-specific ablation of this nuclear receptor in CD4-CreNr1h4fl/fl (T∆FXR) mice. T∆FXR mice were outwardly normal and had no observable alterations in immune cell populations at the steady state (SI Appendix, Fig. S3 C–F). Upon challenge with LCMV Armstrong, T∆FXR and wild type (WT) littermate control mice displayed similar frequencies of effector T cells and antiviral CD8+ T cells, with no differences in cytokine production following ex vivo restimulation (Fig. 2 A–E). However, while WT mice displayed the expected reduction in T cell numbers upon fasting, T∆FXR mice failed to do so (Fig. 2F). Of note, uninfected WT and T∆FXR mice showed similar decreases in T cell numbers after food deprivation (SI Appendix, Fig. S3G), which is consistent with FXR expression being induced upon T cell activation (SI Appendix, Fig. S3A).
Fig. 2.
FXR expression renders effector T cells sensitive to reduced feeding. Six- to 10-wk-old wild type (TWT) and TΔFXR mice were infected with LCMV Armstrong. Mice were fed ad libitum (A–F) or fasted for 24 h on D5 post infection (F–L). Splenocytes were analyzed by FACS on D7. (A and D) Frequencies of naive (Tn), central memory (Tcm), and effector (Teff) cells among indicated T cell types. (B and E) Percentage of IFNγ-producing effector T cells upon ex vivo restimulation with PMA and Ionomycin in the presence of brefeldin A and monensin. (C) Representative FACS plots showing the percentage of H-2Kb/GP33 tetramer+ cells. Gated on effector CD8+ T cells. (F) Total splenic T cell counts on D7. Fasted mice were refed for 18 h before euthanization. (G) Weight loss at the end of fasting. (H–J) Organ-to-body-weight ratios for the eWAT, liver, and tibialis anterior. (K and L) Ex vivo oxygen consumption measurements for the liver and tibialis anterior. Data are plotted as mean ± SD. Data in A–E and G–L (n = 8 to 13) are pooled from at least two independent experiments. Data in F (n = 4 to 5) are representative of three independent experiments. Statistical significance was determined by a two-tailed t test (B, E, G–L) or a two-way ANOVA followed by Sidak’s correction for multiple comparisons (A, D, and F). *P < 0.05; ns, not significant.
We conjectured that lymphocyte-intrinsic responsiveness to food intake may be important for metabolic homeostasis, and that FXR expression in effector T cells may allow for a coherent organismal response to reduced feeding during infection. Indeed, we observed that LCMV-infected T∆FXR mice had increased weight loss after fasting compared to WT littermates (Fig. 2G). Consistent with their similar decreases in the T cell compartment after starvation (SI Appendix, Fig. S3G), uninfected WT and T∆FXR mice showed comparable weight loss upon fasting (SI Appendix, Fig. S3H). To further characterize the consequences of impaired lymphocyte responses to reduced feeding on host metabolism during infection, we assessed the weights and oxygen consumption rates of key metabolic tissues. WT and T∆FXR mice showed similar organ-to-body-mass ratios for the liver, epididymal white adipose tissue (eWAT), and the tibialis anterior (Fig. 2 H–J). Liver and skeletal muscle respiration were also unchanged (Fig. 2 K and L), suggesting that the increased weight loss of T∆FXR mice is unlikely driven by higher metabolic activity of these tissues. Overall, these results suggest that FXR expression renders effector T cells sensitive to reduced feeding and that the inability to scale T cell numbers according to food intake affects organismal energy homeostasis without imposing widespread changes in physiology.
Fasting-Induced Transcriptional Changes in Effector T Cells Suggest Utilization of Alternative Fuels.
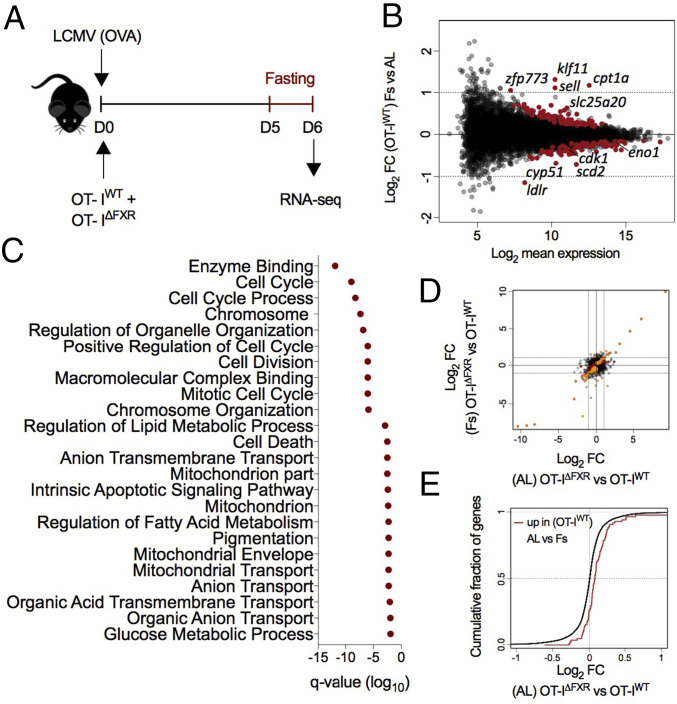
To further investigate the effects of reduced feeding on effector T cells and the role of FXR in these responses, we generated T∆FXR mice expressing the transgenic TCR OT-I, which recognizes the SIINFEKL peptide from ovalbumin (OVA) in the context of H-2Kb. Lymphoreplete hosts received congenically marked OT-IWT and OT-I∆FXR CD8+ T cells and were challenged with LCMV (OVA). On D5 post infection, recipients were fasted for 24 h (FS) or fed AL as previously described. Since in LCMV-infected animals the decrease in lymphocyte numbers does not manifest immediately after fasting (SI Appendix, Fig. S1 B and C), we used fluorescence-activated cell sorting (FACS) to purify effector OT-I cells for RNA-sequencing on D6 to ensure the analysis of comparable populations (for experimental scheme, see Fig. 3A). Overall, the magnitude of the changes induced by nutrient deprivation was small (Fig. 3B), indicating that transcriptional responses to reduced feeding in lymphocytes are subtle and may be complemented by posttranscriptional regulatory mechanisms promoting adaptation to environmental cues (9, 19, 20).
Fig. 3.
Fasting-induced transcriptional changes in effector CD8+ T cells. (A) Experimental scheme for data shown in B–E. Congenically marked naive OT-IWT and OT-IΔFXR cells were cotransferred into lymphoreplete hosts. Recipient mice were challenged with LCMV (OVA) and allowed to feed AL or FS for 24 h on D5. At the end of the fasting, adoptively transferred cells were FACS-purified for gene expression analysis by RNA sequencing. (B) Transcriptional profiling of OT-IWT cells exposed to fasting showing the mean expression level (x axis) and log2 fold-change (FC) between OT-IWT FS and OT-IWT AL (y axis). Significantly altered transcripts (adjusted P value <0.05) are highlighted in red, and two-fold change thresholds are indicated by dotted lines. (C) GSEA was performed on genes differentially expressed by OT-IWT cells in response to fasting. KEGG pathways significantly affected by reduced food intake are shown. (D) FC vs. FC plot showing genes differentially expressed between OT-IΔFXR and OT-IWT in AL (x axis, highlighted in red) or FS (y axis, highlighted in orange) state. (E) Cumulative distribution function (CDF) plot comparing starvation-induced genes (up-regulated in OT-IWT FS vs. OT-IWT AL, red line) to the overall gene expression changes between OT-IΔFXR and OT-IWT in AL hosts (black line). Statistical significance was determined by a one-sided Kolmogorov–Smirnov test (P = 2.11e−7). Data are from one experiment (n = 3).
To gain insight into the cellular processes affected by food deprivation, we performed gene-set enrichment analysis (GSEA). We identified cell cycle and apoptosis as among the pathways differentially regulated in OT-IWT cells (Fig. 3C). These findings further indicate that the fasting-induced decrease in lymphocyte numbers during infection is likely a delayed manifestation of changes in the rates of proliferation and cell death after the host experiences reduced feeding.
In addition to the processes above, reduced feeding also altered glucose and lipid-related pathways (Fig. 3C). Genes induced upon nutrient deprivation included Klf11, a transcriptional regulator modulated by fasting in the liver (21, 22); Acadvl, Acaa2, Cpt1a and Slc25a20, involved in the degradation and mitochondrial oxidation of fatty acids; and the negative regulators of pyruvate dehydrogenase, Pdk1 and Pdk2. These changes suggest that effector CD8+ T cells may oxidize alternative substrates in fasted animals.
Next, we assessed the effects of FXR deficiency on the transcriptome of effector CD8+ T cells. Analyses of OT-I∆FXR and OT-IWT cells revealed that most FXR-dependent changes in gene expression were observed regardless of the absorptive state of the host (Fig. 3D). Furthermore, we detected a significant enrichment for genes induced upon fasting (up in OT-IWT FS versus AL) among the genes up-regulated in OT-I∆FXR relative to OT-IWT AL (Fig. 3E). Altogether, these data indicate that reduced feeding affects essential cellular processes including proliferation, apoptosis, and metabolism and suggest that FXR may repress the expression of starvation-induced genes in effector T cells.
FXR Affects the Metabolism of Effector T Cells.
To directly assess the effects of FXR expression on CD8+ T cell metabolism, we performed ex vivo extracellular flux measurement assays. Bulk effector CD8+ T cells sorted from T∆FXR mice challenged with LCMV showed increased basal respiration compared to their WT counterparts, regardless of the absorptive state of the animal (Fig. 4A). To interrogate whether FXR expression affects the use of distinct energetic substrates, we measured changes in oxygen consumption rates (OCR) in response to inhibitors of the mitochondrial pyruvate carrier (UK5099), glutaminase (BPTES), and carnitine palmitoyl transferase (Etomoxir; for schematic of inhibitors and pathways, see Fig. 4). While WT effector T cells showed a decrease in OCR upon UK5099 injection, FXR-deficient cells displayed minimal changes in response to this inhibitor (Fig. 4B), implying compensation via utilization of alternative substrates. Indeed, administration of BPTES + Etomoxir following the injection of UK5099 reduced the respiration of FXR-deficient, but not -sufficient, cells (Fig. 4B). These results suggest that FXR expression limits the metabolic flexibility of effector T cells by restricting the utilization of glutamine and fatty acids to fuel respiration.
Fig. 4.
FXR regulates the metabolism of effector CD8+ T cells. (A–F) Bioenergetic profiling of wild type (TWT) and TΔFXR effector CD8+ T cells. Extracellular flux measurements were carried out in the presence of glucose (11 mM) and L-glutamine (2 mM). (A and B) TWT and TΔFXR mice were infected with LCMV Armstrong. Mice were fed ad libitum or fasted for 24 h on D5 post infection as described in Fig. 1B. CD8+ effector T cells were FACS-purified at the end of the fasting. (A) Baseline OCR. (B–E) Mitochondrial fuel utilization showing changes in OCR over time in response to the mitochondrial pyruvate carrier inhibitor UK5099 (2 μM), the CPT1 inhibitor etomoxir (Eto, 4 μM), or the glutaminase inhibitor BPTES (3 μM). (D–F) Naive CD8+ T cells were activated in vitro with CD3/CD28 beads in the presence of IL-2 for 3 d to generate effector cells (D and E). Contribution of indicated fuels to mitochondrial respiration. Data in E are plotted as the percentage of basal OCR after injection of inhibitor. (F) In-vitro–generated effector cells were exposed to various concentrations of glucose in the presence or absence of glutamine for 24 h. Cell survival was determined by FACS. Data are plotted as mean ± SD (n = 3 to 11), representative of at least two independent experiments. Statistical significance determined by one- or two-way ANOVA followed by a Dunnet (A), Sidak (E), or Tukey (F) correction for multiple comparisons. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To minimize potential confounding effects of cell-extrinsic signals present in vivo, we also performed extracellular flux measurements on in-vitro–generated effector T cells. In line with our ex vivo assessment, T∆FXR cells showed a modest but highly reproducible increase in their basal OCR compared to WT (SI Appendix, Fig. S4A). T∆FXR and WT effector T cells displayed similar basal extracellular acidification rates (SI Appendix, Fig. S4B), indicating that anaerobic glycolysis was not affected. By interrogating the use of individual substrates to support respiration, we confirmed that FXR-deficient effector T cells were less sensitive to inhibition of the mitochondrial pyruvate carrier with UK5099 compared to their WT counterparts (Fig. 4D). Furthermore, we found that FXR deficiency increased the contribution of glutamine and long-chain fatty acids to the oxidative metabolism of T cells even when pyruvate usage was not impeded (Fig. 4D).
We then carried out carbon-tracing experiments to gain further insights into the metabolic alterations imposed by FXR deficiency. Consistent with their increased oxidative metabolism, T∆FXR effector T cells showed greater labeling of citric acid cycle intermediates from C13-glucose and C13-glutamine compared to WT (SI Appendix, Fig. S4 C and D). In addition to glutamine, other amino acids including serine and aspartate can be used to fuel the TCA cycle. Despite presenting a slight increase in the synthesis of serine and aspartate from glucose and glutamine (SI Appendix, Fig. S4 C and D), FXR-deficient T cells showed substantially decreased levels of these amino acids (SI Appendix, Fig. S4E), possibly indicating higher catabolism of these molecules.
Our results suggested that FXR deficiency increased the contribution of fuels other than glucose to respiration. To assess whether this greater metabolic flexibility could impact cell survival, we cultured in-vitro–generated effector cells in various amounts of glucose. FXR-deficient effector CD8+ T cells retained higher viability in low sugar compared to WT cells (Fig. 4F). Of note, the survival advantage of these cells was contingent on the presence of glutamine (Fig. 4F), suggesting that the oxidation of alternative fuels may enable their survival in glucose-limiting conditions. Altogether, our results support a role for FXR in restricting the metabolic flexibility of effector CD8+ T cells, which may reduce their ability to survive when glucose is limiting.
Lymphocytes Respond to Changes in Circulating Macronutrients during Anorexia of Infection.
The above results led us to hypothesize that changes in the levels of energetic substrates may relate to the decrease in cell numbers in vivo. To address this question, we first profiled the levels of macronutrients in LCMV-infected mice at the end of the 24-h fasting period. Starved animals presented higher serum triacylglycerides (SI Appendix, Fig. S5A), which was in agreement with previously reported increases in circulating lipids during infection-related anorexia (23). As expected, reduced feeding led to a significant decrease in glycemia and to an increase in the abundance of ketones (SI Appendix, Fig. S5A). Blood urea nitrogen levels were elevated in fasted animals, suggesting increased amino acid catabolism (SI Appendix, Fig. S5A).
Considering the correlation between glycemia and spleen size (Fig. 5A) and the prominent role of glycolytic metabolism in effector T cells (24–30), we set out to test whether glucose supplementation was sufficient to prevent the loss of these cells upon food deprivation. For this purpose, LCMV-infected animals were subjected to fasting and refeeding as previously described and supplied with glucose (10%) in the drinking water during food withdrawal (Fig. 5B). Provision of glucose reverted metabolic changes caused by fasting and supported effector T cell numbers in the spleen (Fig. 5C and SI Appendix, Fig. S5B), suggesting that sugar, either by serving as fuel or preventing metabolic changes associated with reduced food intake, is sufficient to sustain T cells during infection.
Fig. 5.
Lymphocytes respond to changes in circulating macronutrients during anorexia of infection. (A–C) Six- to 10-wk-old C57Bl6/N mice were infected with LCMV Armstrong and fed ad libitum or fasted for 24 h on D5 post infection. (A) Correlation between blood glucose levels on D6 (y axis) and spleen size on D7 (x axis). (B) Schematics of glucose rescue experiments shown in C. Glucose (10%) was provided in the drinking water for the duration of the fasting. (C) Splenic effector CD8+ T cell counts on D7. (D–G) Six- to 10-wk-old C57Bl6/N mice were infected i.v. with L. mono (OVA). (D) Food consumption and body-weight changes during the course of infection. (E) Blood glucose levels of L. mono (OVA)-challenged mice on D6 post infection and age-matched, sex-matched, uninfected control animals. (F) Animals infected with L. mono (OVA) received injections of glucose (1 mg/mouse) or vehicle i.p. twice daily on D5 to D6 (four injections total), showing counts for effector CD8+ T cells in the spleen on D7. (G) Congenically marked OT-IWT and OT-IΔFXR naive CD8+ T cells were cotransferred into lymphoreplete hosts. Recipient mice were challenged with L. mono (OVA) showing total counts of transferred cells in the spleen on D7. Data are plotted as mean +/− SD. Data in A and C (n = 10 to 24) are pooled from three experiments. Data in E–G (n = 5 to 10) are representative of at least two independent experiments. Statistical significance determined by Pearson correlation analysis (A: P = 0.0018, R2 = 0.3633), one-way ANOVA followed by a Dunnett’s correction for multiple comparisons (C), or a two-tailed t test (E–G). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
Next, we measured the glycemia of LCMV-infected WT and T∆FXR animals to assess whether potential differences could explain the higher numbers of effector T cells in the latter. Unexpectedly, we found that blood glucose levels were significantly lower in T∆FXR compared to WT following food deprivation (SI Appendix, Fig. S5C). These results reinforce the notion that the inability to scale the T cell compartment size according to food intake negatively affects organismal metabolism and suggest that FXR-deficient effector T cells can persist in lower glucose levels in vivo.
Bile acids (BA) act as fed-state signals, repressing catabolic processes in an FXR-dependent manner (10, 11). Food intake induces BA secretion from the gall bladder into the intestinal lumen and increases serum BA levels via enterohepatic circulation. To test whether BA levels affect T cell numbers, we fed LCMV-infected mice a diet containing cholestyramine, which is clinically used to prevent intestinal BA reabsorption and enhance fecal cholesterol excretion. Cholestyramine failed to change splenic T cell counts (SI Appendix, Fig. S5D). These results suggest that reduced abundance of FXR ligands is not sufficient to alter T cell numbers in the absence of other physiological alterations imposed by fasting.
Finally, we sought to extend our findings to a spontaneous model of anorexia of infection. Intravenous injection of Listeria monocytogenes (L. mono) expressing OVA as a model antigen induces a long-lasting anorexic response that is present when adaptive immune responses arise on D5 post infection (Fig. 5D). In addition to weight loss, L. mono (OVA)-challenged mice showed reduced glycemia compared to noninfected controls (Fig. 5E). Corroborating our observations in LCMV-infected mice subjected to experimental anorexia, administration of glucose on D5 to D6 boosted the number of effector CD8+ T cells (Fig. 5F), indicating that sugar is also a limiting nutrient for lymphocytes during naturally occurring anorexia of infection.
To assess the role of cell-intrinsic FXR expression in CD8+ T cells during anorexia of infection, we cotransferred congenically marked OT-I∆FXR and OT-IWT cells immediately before challenge with L. mono (OVA) and analyzed recipients 7 d later. As expected, OT-I∆FXR cells showed a competitive advantage over their WT counterparts (Fig. 5G), confirming our finding that FXR limits the effector CD8+ T cell pool in nutrient-deprived hosts. Overall, these experiments confirm that effector T cells are sensitive to the nutritional status of the host during infection and that FXR, whose expression is up-regulated upon T cell activation, is a key regulator of this biological process.
Discussion
The survival and reproductive success of living organisms depend on the optimal allocation of finite resources into growth, reproduction, and strategies to thrive in specific niches (31). Mounting an immune response often impairs growth and reproduction in plants and animals (32, 33). Although the incompatibility between these energy-consuming processes has long been acknowledged, the mechanisms deployed to balance the costs of immunity and indispensable physiological functions remain poorly understood. Conditions where the energetic budget of an organism is put under stress have revealed the existence of metabolic trade-offs to support physiological functions of higher priority. Infected mammals exposed to cold temperatures present with increased pathogen burden (23), implying that homeothermy is maintained at the expense of immunological defenses. These trade-offs corroborate the general paradigm in life history theory that unfavorable environmental conditions promote reallocation of resources from anabolic processes into catabolic maintenance mechanisms and suggest that distinct organismal functions may be prioritized according to energy availability.
In addition to increases in expenditure, the energetic budget of an organism may become compromised by decreased food intake. Intriguingly, infection and the ensuing immune responses are frequently accompanied by anorexia, imposing a double tax on energy reserves. The wide conservation of anorexia of infection from insects to mammals suggests that these responses evolved prior to emergence of adaptive immunity. Therefore, the persistence of such stereotypical manifestations in higher vertebrates was likely driven by mechanisms that enhance host fitness independently of antigen-specific responses, such as modulation of innate immunity or preservation of essential tissue functions. Clonal expansion of B and T cells coincides with the beginning of recovery from infection-induced anorexia. Thus, this energetically costly process segregates temporally, yet only partially, from reduced food intake. In addition to this not fully synchronous timing, we propose that mammals may scale their adaptive immune cell compartment in response to feeding, potentially to ensure proper resource allocation during infection.
In this regard, our study identified FXR as an important modulator of effector T cell responses to reduced food intake. FXR expression is induced upon T cell activation, suggesting that a cell-intrinsic “failsafe” mechanism is built into the differentiation program of effector T cells to limit their population size and potentially ensure effective organismal recovery after anorexia. While other cell-intrinsic and -extrinsic mechanisms have been implicated in the responses of naive and central memory T cell subsets to feeding restriction (14, 34), their role in effector T cells remains unexplored. It is possible that those mechanisms have a nonredundant role in effector T cell responses to starvation or that the same pathways are “repurposed” in this subset to integrate additional context-specific cues (35).
Our findings revealed that FXR reduced the capacity of effector T cells to engage in oxidative metabolism and restrained the use of glutamine and fatty acids as substrates for mitochondrial respiration. Since reduced feeding changes the profile of circulating macronutrients, the heightened survival of FXR-deficient T cells in starved animals may be related to their ability to use fuels other than glucose when this substrate becomes depleted. In support of this possibility, we found that FXR-deficient T cells showed modest, but significantly increased, survival in glucose-limiting conditions, but only when glutamine was present. We further demonstrated that exogenous provision of glucose is sufficient to support lymphocytes in nutrient-deprived hosts, highlighting the key role of sugar in dictating the magnitude of adaptive immune responses. Although nutritional supplementation early during infection was shown to have context-dependent effects on tissue function and disease outcome (1–3), we envision that timed provision of glucose could be employed therapeutically and with potentially few deleterious effects on host physiology in instances where heightened adaptive immune responses are desired.
The seemingly narrow gene expression changes caused by nutrient deprivation may be indicative of how these responses are regulated in vivo or reflect a difficulty in capturing differences between the fed and fasted states, which could arise from animals displaying discordant feeding behavior before fasting or in the hours preceding the analysis. Despite these confounders, we were still able to detect significant changes in the levels of a few key metabolic genes. Given that our analyses were performed on bulk effector T cell populations, it is possible that these changes are present only in a subset of cells that may successfully adapt and survive nutrient deprivation. It remains to be determined whether feeding restriction and other environmental constraints limit the transcriptional heterogeneity of effector T cell populations or merely induce a temporary convergence toward metabolic programs associated with cell survival.
In conclusion, our work demonstrated that T cells display cell-intrinsic responses to the nutritional status of the host during infection and that FXR is a key mediator of this process, limiting the metabolic flexibility of effector CD8+ T cells.
Methods
Mice.
C57Bl6/N mice were purchased from Charles River Laboratories. Genetically modified strains were bred in-house. The generation of Nr1h4fl/fl mice was described by Sinal et al. (36). Nr1h4fl/fl animals were used as WT controls for CD4CreNr1h4fl/fl mice. Six- to 10-wk-old male or female mice were used in all experiments. In each independent experiment, mice were all of the same sex, and the age of animals within and across experimental groups did not vary by more than 2 wk. Animals were maintained in standard husbandry conditions with 12 h/12 h light:dark cycles (6 AM/6 PM) and housed in cages with sani-chip bedding. Mice were fed a standard rodent diet (5053, PicoLab). Cholestyramine (Sigma, 2% wt/wt) and control Teklad diets were manufactured by Envigo. For fasting and refeeding experiments, at the end of the photoperiod mice were moved into a fresh cage without food, but with free access to water. After 24 h of fasting, food was added back to the cage. Food consumption and body weight measurements were performed at 6 PM. Animal husbandry and experiments were carried out in accordance with guidelines determined by the Institutional Animal Care and Use Committee at Memorial Sloan Kettering Cancer Center (MSKCC).
Infection.
For viral challenges, mice were infected with 2 × 105 plaque-forming units (PFU) of LCMV Armstrong intraperitoneally (i.p. ) or 2 × 105 PFU of LCMV (OVA) intravenously. For L. monocytogenes infection, bacteria were grown overnight in Brain-Heart-Infusion broth (BD) with agitation (250 × g) at 37 °C and then split 1:10 and grown for an additional 2 to 3 h until OD600 = 0.3 to 0.5. Each mouse received 5 × 104 colony-forming units via retro-orbital injection. All infections were performed at 6 PM. For glucose supplementation experiments, sugar was provided in the drinking water (10%) ad libitum or injected intraperitoneally (10 mg/mL, 100 μL per mouse) twice daily at 10 AM and 10 PM.
Quantification of Viral Loads.
After recording total organ weight, samples of ∼10 mg were taken from spleens, placed in 2 mL of TRIzol, and homogenized with a tissue homogenizer (Ultra-Turrax homogenizer, IKA). RNA extraction was performed using phase-lock tubes according to the TRIzol manufacturer’s protocol followed by a cleanup step with RNEasy mini kit (Qiagen) to eliminate excess salt. The complementary DNA (cDNA) was synthesized with qScript Supermix (95048-025, Quanta Bio). Viral RNA copy numbers were determined by RT-qPCR. SYBR Green PCR Master Mix (4309155, ThermoFisher) was used in a final volume of 10 μL per reaction. A standard curve of plasmid DNA containing a single copy of the LCMV envelope glycoprotein (GP) gene (37) was used to calculate absolute numbers of viral particles. Primer sequences were as follows: LCMV GP Fw—5′ CATTCACCTGGACTTTGTCAGACT 3′; LCMV GP Rv—5′ GCAACTGCTGTGTTCCCGAAAC 3′.
Blood and Serum Chemistry.
Blood glucose and β-hydroxybutyrate levels were measured with handheld monitors (Contour and NovaMax plus, respectively). At the time of measurement, animals were put on top of the cage without restraining to minimize distress, and a small cut at the tip of the tail was made for blood collection. For serum chemistry, animals were euthanized, and blood was collected via cardiac puncture into serum separator tubes (BD). After clotting for at least 1 h at room temperature (RT), tubes were centrifuged at 13,000 x g for 3 min, and serum was transferred into Eppendorf tubes and stored at −80 °C until analysis. Serum chemistry analyses were performed by the Center for Comparative Medicine and Pathology at MSKCC.
Continuous Oxygen Measurement with Clark-type Electrode.
Tissue oxygen consumption was measured with a Clark-type oxygen polarographic electrode. Immediately after euthanization of animals, tissues were dissected and weighed. After mincing into 0.2- to 0.5-mm diameter fragments with sterile surgical tools, tissue aliquots were transferred into an oxygenated PBS solution at RT. Tissue suspensions were put into a respirometer microcell linked to an oxygen meter (Strathkelvin Instruments MS200A) and continuously stirred during measurements. Oxygen consumption detection was made with a platinum cathode silver anode electrode connected by a saturated potassium chloride solution (Strathkelvin Instruments 1302) and shielded with a polypropylene membrane jacket (Strathkelvin Instruments SI020). Oxygen consumption rates were recorded for ∼1 min after sealing the microcell with a plunger. For each tissue, oxygen consumption rates were calculated for the same time interval, starting at 20 to 30 s after the chamber was sealed. Rates were normalized to tissue weight.
RNA Sequencing.
RNA extraction and sequencing were performed by the Integrative Genomics Operation Core at MSKCC. Samples were sequenced at a depth of 30 to 40 million reads per sample. Reads were aligned to the mouse genome assembly mm10.GRCm38. Gene annotations from GENCODE vM17 were used for all analyses. Analysis was done using R v3.4.0 (2017-04-21) and custom scripts. Reads were aligned to the mouse genome using HISAT2 v2.1.0 (38) with default parameters including splice sites obtained from gene annotations. Uniquely aligned reads were extracted using grep with the parameters “-v 'NH:i:[2-9]'” and SAMtools v1.3.1 (39) with parameters “view -h -F 4 -q 20 -b” and sorted and indexed using SAMtools. Reads in genes were counted using Rsubread v1.28.1 (40). Differential gene expression between any pair of samples was assessed with DESeq2 v1.18.1 (41), using default false discovery rate adjustment of P values for multiple hypothesis testing. Pathway analyses were carried out with GSEA (https://www.gsea-msigdb.org/gsea/index.jsp).
In Vitro Generation of Effector Cells.
Naive (CD44−CD62L+) CD8+ T cells were FACS-purified from spleen and peripheral (pooled inguinal, brachial, axial, and submandibular) lymph nodes after a CD8-enrichment step (Dynabeads11447D, Invitrogen) performed as per manufacturer’s instructions. Cells were activated with Mouse T activator CD3/CD28 Dynabeads (Gibco) at 1:1 bead-to-cell ratio in the presence of 100 U/mL of IL-2 (Biological Resources Branch, National Cancer Institute [NCI]) in complete RPMI (RPMI 1640 with 10 mM Hepes buffer [ThermoFisher], 1% penicillin/streptomycin [ThermoFisher], 1% L-glutamine [ThermoFisher], 55 μM BME [Gibco], and 10% fetal bovine serum [FBS] [ThermoFisher, 35010CV]) for 3 d. Beads were removed by magnetic separation before cells were used in in vitro assays.
Extracellular Flux Measurements.
Assays were performed using a Seahorse XP96 analyzer (Agilent). To coat assay plates, Cell Tak (Corning) was diluted in sterile 0.1 M bicarbonate buffer, pH 8.0, at 20 μg/mL. For every part of Cell Tak reagent used, an equal volume of sterile 1 M NaOH was added to the coating solution immediately before use as per manufacturer’s recommendation. A total of 25 μL of Cell Tak coating solution was dispensed into each well, and plates were incubated at RT for 20 min. After aspiration, coated plates were washed twice with 200 μL sterile water per well and allowed to air-dry inside a biosafety cabinet before use or storage according to manufacturer’s instructions. Each plate well received a total of 2 × 105 cells in 30 μL of Assay Medium (Seahorse XF Base medium [Agilent] freshly supplemented with 1 mM pyruvate [ThermoFisher], 10 mM glucose [Gibco] and 2 mM L-glutamine [ThermoFisher]). Corner wells were loaded with medium only to serve as references. After centrifugation at 450 x g for 5 min, 150 μL of previously warmed assay medium was gently added to each well in order to preserve the cell monolayer. Cells were placed in a non-CO2 incubator at 37 °C for 30 to 45 min for adhesion. During this incubation, a hydrated Seahorse cartridge was loaded with 10x stocks of the relevant mitochondrial interrogation compounds and put into the Seahorse XP96 analyzer to equilibrate. The run was set up as follows: Calibrate; Equilibrate; Base line readings (Loop 3 times): Mix—3 min; Wait—2 min; Measure—3 min; End loop; Inject port X (Loop 3 times): Mix—3 min; Wait—2 min; Measure—3 min; End loop. For Mito Stress and Mito Fuel Flex (103015-100 and 103260–100, Agilent) assays, powdered stocks were freshly reconstituted from the Seahorse XF Cell Mito Stress Test or Seahorse XF Mito Fuel Flex Test kits (103015-100 and 103260-100, Agilent) in assay medium according to the manufacturer’s instructions. Final well concentrations of compounds were the following: 1 μM Oligomycin; 1.5 μM carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP); 100 nM Rotenone; 1 μM Antimycin A; 3 μM BPTES; 4 μM Etomoxir, and 2 μM UK5099.
Stable Isotopologue Labeling and Analysis.
In-vitro–generated effector cells were washed in tracing media (RPMI 1640 without glucose without glutamine with 10% dialyzed FBS [Gemini] with 10 mM Hepes [ThermoFisher], with 1% penicillin/streptomycin [ThermoFisher] with 55 μM 2-mercaptoethanol [Gibco]) and resuspended at 20 × 106 cells/mL. Tracing media was supplemented with either 12C-glucose (Sigma) and 12C-glutamine (Gibco) or the 13C versions of each metabolite, [U-13C] glucose or [U-13C] glutamine (Cambridge Isotope Laboratories) to produce 13C-tracing media with a final concentration of 11 mM (glucose) and 2 mM (glutamine). A total of 2 × 106 cells in 3 mL of 13C tracing media were added to each well of six-well plates and incubated for 6 h at 37 °C, 5% CO2. At the end of the incubation period, cells were pelleted at 300 × g for 2 min at 4°C and resuspended in 1 mL of ice-cold 80% methanol containing 2 μM deuterated 2-hydroxyglutarate (D-2-hydroxyglutaric-2,3,3,4,4-d5 acid, d5-2HG) as an internal standard. After overnight incubation at −80 °C, samples were vortexed and centrifuged at 20,000 × g for 20 min to remove protein. Supernatants were dried in a vacuum evaporator (Genevac EZ-2 Elite) for 2 h. Dried samples were incubated in 50 μL of 40 mg/mL methoxyamine hydrochloride in pyridine at 30 °C for 2 h and derivatized with addition of 80 μL of N-Methyl-N-trimethylsilyltrifluoroacetamide + 1% 2,2,2-Trifluoro-N-methyl-N-(trimethylsilyl)-acetamide, Chlorotrimethylsilane (MSTFA + TCMS [Thermo Scientific]) and 70 μL ethyl acetate (Sigma) by shaking at 37 °C for 30 min. Samples were analyzed using an Agilent 7890 GC coupled to an Agilent 5977 mass selective detector. The GC was operated in splitless mode with constant helium carrier gas flow of 1 mL/min and with a HP-5MS column (Agilent Technologies). The injection volume was 1 μL, and the GC oven temperature was ramped from 60 °C to 290 °C over 25 min. Peaks representing compounds of interest were extracted and integrated using MassHunter vB.08.00 (Agilent Technologies) and then normalized to both the internal standard (deuterated-2HG) peak area and biomass (cell number x cell volume). Ions used for quantification of metabolite levels were as follows: d5-2HG m/z 354, 2HG m/z 349, alpha-ketoglutarate m/z 304, aspartate m/z 334, citrate m/z 465, fumarate m/z 245, glutamate m/z 363, malate m/z 335, succinate m/z 247, and serine m/z 306. All peaks were manually inspected and verified relative to known spectra for each metabolite. Enrichment of 13C was assessed by quantifying the abundance of the following ions: alpha-ketoglutarate m/z 304 to 313, aspartate m/z 334 to 343, citrate m/z 465 to 477, fumarate m/z 245 to 255, glutamate m/z 363 to 375, malate m/z 335 to 346, and serine m/z 306 to 314. Correction for natural isotope abundance was performed using IsoCor software (42).
Survival in Low Glucose.
A total of 1 × 105 in-vitro–generated effector cells were plated onto flat-bottom 96-well plates (USA Scientific) in tracing media. Concentrated stocks (4x) of glucose (Gibco) and L-glutamine (ThermoFisher) freshly prepared in tracing media were added into each well to a final volume of 200 μL. Cells were incubated at 37 °C, 5% CO2, for 24 h. Viability was analyzed by flow cytometry.
Flow Cytometry.
Foxp3 staining was performed using the eBioscience transcription factor staining buffer set (eBioscience). For cytokine production analyses, cells were incubated for 3 h at 37 °C, 5% CO2, in restimulation media (complete RPMI 1640 with 5% FBS, 50 ng/mL phorbol-12-myristate-13-acetate [Sigma]), 500 ng/mL ionomycin [Sigma], 1 μg/mL brefeldin A [Sigma], and 2 μM monensin [Sigma]). Extracellular antigens were stained for 15 min at 4 °C with an antibody staining mix containing Ghost Dye Red 780 viability dye diluted in PBS. Cells were fixed and permeabilized with BD Cytofix/Cytoperm for 20 min at 4 °C. Antibodies against intracellular antigens were diluted in 1x BD Perm/Wash buffer, and cells were stained for 30 min at 4 °C. Biotinylated H-2Kb/GP33 monomers (NIH tetramer core) were tetramerized/conjugated to phycoerythrin in house and used to label virus-specific cells before the extracellular staining step by incubation at RT for 30 min. The 123count eBeads (Invitrogen) were added at 5,000 beads per sample to quantify absolute cell numbers. Cytometry data were acquired on a LSRII (Becton Dickinson) and analyzed on FlowJo.
Western Blot.
Nuclear extracts were obtained with the Nuclear Complex Co-IP kit (Active Motif). For tissue extraction, pieces of liver or small intestine (terminal ileum) were placed in ice-cold hypotonic lysis buffer and homogenized with a tissue homogenizer (Ultra-Turrax homogenizer, IKA) until fragments were no longer visible. Naive and activated CD8+ T cells were washed three times in cold PBS before processing. Nuclear extract protein content was determined with the Pierce BCA Protein Assay Kit (Thermo Fisher). Protein (40 to 60 µg) was separated on a 12% sodium dodecyl sulfate/polyacrylamide gel electrophoresis gel (Thermo Scientific) and transferred onto a polyvinylidene difluoride membrane (Immobilon-P, 0.45 µm, Millipore). Membranes were blocked with 5% nonfat milk powder in Tris-buffered saline (TBS) + 0.1% Tween20 (blocking buffer [BB]) for 1 h at RT before an overnight incubation with primary antibodies at 4 °C. After four washes with TBS + 0.1% Tween20 (TBST), membranes were incubated with secondary antibodies for 1 h at RT, followed by four more washes with TBST before addition of chemiluminescent substrates. Detection of FXR was done with a monoclonal antibody (D-3, SCBT) diluted at 1:500, followed by incubation with an HRP-conjugated donkey anti-mouse at 1:5,000. Lamin B was used as a loading control and detected with a polyclonal antibody (1:500, sc6217, Santa Cruz Biotechnology) combined with a horseradish peroxidase (HRP)-conjugated mouse anti-goat (1:5,000). Antibodies were diluted in BB, and all incubations were done under gentle shaking at RT unless stated otherwise. Luminata Forte and Luminata Classico ECL reagents (Millipore) were used as HRP substrates for FXR and Lamin B blots, respectively.
RT-qPCR.
For tissue samples, pieces of liver or small intestine (terminal ileum) were placed in TRIzol and homogenized with a tissue homogenizer (Ultra-Turrax homogenizer, IKA) until fragments were no longer visible. RNA extraction was performed using phase-lock tubes according to the TRIzol manufacturer’s protocol, followed by a cleanup step with RNEasy mini kit (Qiagen) to eliminate excess salt. cDNA was synthesized with qScript Supermix (95048-025, Quanta Bio). SYBR Green PCR Master Mix (4309155, ThermoFisher) was used in a final volume of 10 μL per reaction. FXR expression was normalized to beta-actin. Primer sequences were as follows: FXR Fw—5′ CACAGCGATCGTCATCCTCTCT 3′; FXR Rv—5′ TCTCAGGCTGGTACATCTTGC 3′; β-actin Fw—5′ CGCAGCCACTGTCGAGTC 3′; β-actin Rv—5′ GTCATCCATGGCGAACTGGT 3′.
Supplementary Material
Acknowledgments
We thank members of the Donald and Catherine C. Marron Cancer Metabolism Center for their assistance with extracellular flux measurement assays; Dr. Frank Gonzalez (NCI) for providing mice carrying the FXR floxed allele; Dr. Daniel Pinschewer (University of Basel) for providing LCMV (OVA); Dr. Shane Crotty (La Jolla Institute for Immunology) for providing plasmids containing a single copy of the LCMV-GP protein used for determination of viral loads; Daniel Konstantinovsky for providing general help with preliminary experiments that were not included in the manuscript; and all members of the A.Y.R. laboratory for helpful discussion and technical assistance. This work was supported by NCI Cancer Center Support Grants P30 CA008748 and NCI U54 CA20997; NIH Grant R01 AI034206; the Hilton-Ludwig Cancer Prevention Initiative (Conrad N. Hilton Foundation and Ludwig Cancer Research); and the Ludwig Center at the Memorial Sloan Kettering Cancer Center. A.Y.R. is an investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020619117/-/DCSupplemental.
Data Availability.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE157439 (43). All study data are included in the article and supporting information.
References
- 1.Murray M. J., Murray A. B., Anorexia of infection as a mechanism of host defense. Am. J. Clin. Nutr. 32, 593–596 (1979). [DOI] [PubMed] [Google Scholar]
- 2.Ayres J. S., Schneider D. S., The role of anorexia in resistance and tolerance to infections in Drosophila. PLoS Biol. 7, e1000150 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang A., et al. , Opposing effects of fasting metabolism on tissue tolerance in bacterial and viral inflammation. Cell 166, 1512–1525.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao S., et al. , Pathogen-mediated inhibition of anorexia promotes host survival and transmission. Cell 168, 503–516.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang A., et al. , Glucose metabolism mediates disease tolerance in cerebral malaria. Proc. Natl. Acad. Sci. U.S.A. 115, 11042–11047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wing E. J., Young J. B., Acute starvation protects mice against Listeria monocytogenes. Infect. Immun. 28, 771–776 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wing E. J., Magee D. M., Barczynski L. K., Acute starvation in mice reduces the number of T cells and suppresses the development of T-cell-mediated immunity. Immunology 63, 677–682 (1988). [PMC free article] [PubMed] [Google Scholar]
- 8.Badovinac V. P., Haring J. S., Harty J. T., Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity 26, 827–841 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hukelmann J. L., et al. , The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat. Immunol. 17, 104–112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J. M., et al. , Nutrient-sensing nuclear receptors coordinate autophagy. Nature 516, 112–115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seok S., et al. , Transcriptional regulation of autophagy by an FXR-CREB axis. Nature 516, 108–111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moskophidis D., Cobbold S. P., Waldmann H., Lehmann-Grube F., Mechanism of recovery from acute virus infection: Treatment of lymphocytic choriomeningitis virus-infected mice with monoclonal antibodies reveals that Lyt-2+ T lymphocytes mediate clearance of virus and regulate the antiviral antibody response. J. Virol. 61, 1867–1874 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kägi D., et al. , Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature 369, 31–37 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Collins N., et al. , The bone marrow protects and optimizes immunological memory during dietary restriction. Cell 178, 1088–1101.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cariou B., et al. , Transient impairment of the adaptive response to fasting in FXR-deficient mice. FEBS Lett. 579, 4076–4080 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto T., et al. , Defect in peroxisome proliferator-activated receptor alpha-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J. Biol. Chem. 275, 28918–28928 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Kim K. H., Moore D. D., Regulation of liver energy balance by the nuclear receptors farnesoid X receptor and peroxisome proliferator activated receptor α. Dig. Dis. 35, 203–209 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Huber R. M., et al. , Generation of multiple farnesoid-X-receptor isoforms through the use of alternative promoters. Gene 290, 35–43 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Araki K., et al. , Translation is actively regulated during the differentiation of CD8+ effector T cells. Nat. Immunol. 18, 1046–1057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rollings C. M., Sinclair L. V., Brady H. J. M., Cantrell D. A., Ross S. H., Interleukin-2 shapes the cytotoxic T cell proteome and immune environment-sensing programs. Sci. Signal. 11, eaap8112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renga B., Mencarelli A., Vavassori P., Brancaleone V., Fiorucci S., The bile acid sensor FXR regulates insulin transcription and secretion. Biochim. Biophys. Acta 1802, 363–372 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Zhang H., et al. , Mouse KLF11 regulates hepatic lipid metabolism. J. Hepatol. 58, 763–770 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Ganeshan K., et al. , Energetic trade-offs and hypometabolic states promote disease tolerance. Cell 177, 399–413.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cham C. M., Gajewski T. F., Glucose availability regulates IFN-gamma production and p70S6 kinase activation in CD8+ effector T cells. J. Immunol. 174, 4670–4677 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Chang C. H., et al. , Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gubser P. M., et al. , Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat. Immunol. 14, 1064–1072 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Ho P. C., et al. , Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 162, 1217–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michalek R. D., et al. , Cutting edge: Distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186, 3299–3303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phan A. T., et al. , Constitutive glycolytic metabolism supports CD8+ T cell effector memory differentiation during viral infection. Immunity 45, 1024–1037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang R., et al. , The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stearns S. C., The Evolution of Life Histories (Oxford University Press, 1992). [Google Scholar]
- 32.Karasov T. L., Chae E., Herman J. J., Bergelson J., Mechanisms to mitigate the trade-off between growth and defense. Plant Cell 29, 666–680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lochmiller R. L., Deerenberg C., Trade-offs in evolutionary immunology: Just what is the cost of immunity? Oikos 88, 87–98 (2000). [Google Scholar]
- 34.Saucillo D. C., Gerriets V. A., Sheng J., Rathmell J. C., Maciver N. J., Leptin metabolically licenses T cells for activation to link nutrition and immunity. J. Immunol. 192, 136–144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong J. Y., et al. , Long-term programming of CD8 T cell immunity by perinatal exposure to glucocorticoids. Cell 180, 847–861.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinal C. J., et al. , Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102, 731–744 (2000). [DOI] [PubMed] [Google Scholar]
- 37.McCausland M. M., Crotty S., Quantitative PCR technique for detecting lymphocytic choriomeningitis virus in vivo. J. Virol. Methods 147, 167–176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L., Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H. et al.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao Y., Smyth G. K., Shi W., The subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Millard P., Letisse F., Sokol S., Portais J. C., IsoCor: Correcting MS data in isotope labeling experiments. Bioinformatics 28, 1294–1296 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Pritykin Y., et al. , Data from "FXR mediates T cell-intrinsic responses to reduced feeding during infection." Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157439. Deposited 3 September 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE157439 (43). All study data are included in the article and supporting information.