Abstract
Background
Height and body mass index (BMI) have both been positively associated with melanoma risk, although findings for BMI have been less consistent than height. It remains unclear, however, whether these associations reflect causality or are due to residual confounding by environmental and lifestyle risk factors. We re-evaluated these associations using a two-sample Mendelian randomization (MR) approach.
Methods
We identified single nucleotide polymorphisms (SNPs) for BMI and height from separate genome-wide association study (GWAS) meta-analyses. We obtained melanoma SNPs from the most recent melanoma GWAS meta-analysis comprising 12 874 cases and 23 203 controls. We used the inverse variance-weighted estimator to derive separate causal risk estimates across all SNP instruments for BMI and height.
Results
Based on the combined estimate derived from 730 SNPs for BMI, we found no evidence of an association between genetically predicted BMI and melanoma [odds ratio (OR) per one standard deviation (1 SD) (4.6 kg/m2) increase in BMI 1.00, 95% confidence interval (CI): 0.91–1.11]. In contrast, we observed a positive association between genetically-predicted height (derived from a pooled estimate of 3290 SNPs) and melanoma risk [OR 1.08, 95% CI: 1.02–1.13, per 1 SD (9.27 cm) increase in height]. Sensitivity analyses using two alternative MR methods yielded similar results.
Conclusions
These findings provide no evidence for a causal association between higher BMI and melanoma, but support the notion that height is causally associated with melanoma risk. Mechanisms through which height influences melanoma risk remain unclear, and it remains possible that the effect could be mediated through diverse pathways including growth factors and even socioeconomic status.
Keywords: Body mass index, height, body size, skin cancer, melanoma, causality, Mendelian randomization
Key Messages
Observational studies examining the association between body mass index and height and melanoma risk have yielded inconsistent findings.
To resolve this inconsistency, we conducted Mendelian randomization analyses using large genome-wide association study datasets.
We found no evidence to suggest that the association between higher body mass index and melanoma risk is causal, but height was found to be associated with melanoma.
Mechanisms through which height influences melanoma risk remain unclear; numerous pathways have been proposed.
Introduction
Exposure to ultraviolet (UV) radiation and having a sun-sensitive phenotype are established risk factors for cutaneous melanoma (hereafter referred to as melanoma) among susceptible people.1,2 The associations with other factors are less clear, although some studies suggest a possible link between anthropometric factors and melanoma risk.3–5 Previous observational studies have reported positive associations with body mass index (BMI) and height, but findings varied across studies. Positive associations between high BMI and melanoma have been reported in some4,6 but not all7–9 cohort studies. With regard to height, most previous observational studies of melanoma have reported an increased risk among taller people.4,5,10–13
It remains unclear whether the reported associations represent true causal relationships or are explained by bias or confounding by other factors simultaneously associated with BMI, height and melanoma risk. For example, some studies have speculated that obesity and height might be causally associated with melanoma through increased body surface area and larger number of target cells at risk14,15 or, conversely, that melanoma risk might be decreased among obese people through limited outdoor recreational activities and difference in sun-seeking behaviours compared with their non-obese counterparts. Obesity may also be associated with other unknown or unmeasured lifestyle factors, and the possibility of residual confounding by such factors remains a limitation of all observational studies. Finally, information regarding childhood illness and nutrition status, which are potential modifiable factors of height, have not been assessed in previous studies.
One approach to circumvent some of the threats to validity and limitations found in conventional observational studies is to conduct instrumental variable analyses using genetic variants as proxy markers for risk factors, a technique known as Mendelian randomization (MR).16 MR uses genetic variants associated with an exposure (or a biological intermediate) to estimate its effects on the outcome.17 Because genetic variants associated with adult BMI and height are randomly assigned from parents to their offspring at conception, MR studies are closer to the random assignment of exposure in a randomized controlled trial, in which known and unknown genetic confounders are randomly distributed across different treatment arms, assuming various MR assumptions are met. We conducted MR analyses of BMI and height in relation to the risk of melanoma using the very large international genome-wide association datasets from the Genetic Investigation of ANthropometric Traits (GIANT) consortium18 and consortium data from the melanoma GWAS meta-analysis.19
Methods
We applied a two-sample Mendelian randomization approach to evaluate whether genetically predicted BMI and height are risk factors for melanoma, using publicly available summary data from the meta-analyses of genome-wide association studies (GWAS).
Instrumental variables
Single nucleotide polymorphisms (SNPs) were identified from the largest 2018 GWAS meta-analysis of measured BMI and height in adulthood, from the GIANT consortium.18 This meta-analysis included data from a total of ∼700 000 participants of European descent, comprising ∼250 000 participants from the earlier GWAS meta-analyses (conducted in 2014 and 2015)20,21 and new GWAS data from ∼450 000 participants in the UK Biobank. In total, 754 and 3290 independent SNPs known to be associated at P <5 x 10–8 with BMI and height, respectively, were used as instruments for these analyses. Detailed information regarding sample and SNP selection, summary statistics, quality control and meta-analyses have been reported previously.18 We extracted data on major and minor alleles for each SNP together with the allele frequencies, beta coefficient, standard error (SE) of the beta coefficient and P-value for the relevant association.
We tested the validity of the BMI and height instrumental variables in an independent dataset of 17 965 participants in the QSkin cohort. The QSkin Sun and Health Study is a population-based cohort consisting of 43 794 men and women aged between 40–69 years, who were randomly sampled from the Queensland population.22 The study was approved by the Human Research Ethics Committee of the QIMR Berghofer Medical Research Institute, and each participant provided written informed consent. Genome-wide polygenic risk scores (PRS) for BMI and height were calculated for eligible cohort participants who provided a DNA sample (n = 17 222). We generated the PRS using summary statistics from the same set of SNPs as used for MR analyses. PRS were generated using the --score function of plink V1.90b6.6.23
Association of BMI and height genetic variants with cutaneous melanoma
We obtained GWAS summary statistics on melanoma from the largest meta-analysis of GWAS on cutaneous melanoma to date, which included 12 874 histologically confirmed cases and 23 203 controls from 11 independent GWAS in people of European ancestry in UK, Australia, USA, Germany, France and Greece.19 Details regarding GWAS quality control and study samples have been published previously.19 For each identified BMI or height SNP instrument, we extracted the per allele log odds ratio (OR) for melanoma together with its SE and allele frequencies from the melanoma GWAS meta-analysis. Since the two-sample MR involves combining data from two independently generated datasets, we harmonized the data by comparing allele frequencies between the BMI, height and melanoma datasets, thereby ensuring that reference alleles for each locus were concordant across the datasets. Palindromic strands with minor allele frequency threshold for alignment above 0.3 are non-inferable and hence were excluded from the analyses.
Two-sample Mendelian randomization methods
In contrast to traditional two-stage least squares MR, whereby individual-level genotype and phenotype data are obtained from the same sample, we used a two-sample MR strategy in which the SNP exposure and SNP outcome associations are estimated using summary statistics from independent samples.24 The two-sample method typically offers greater statistical power than the one-sample method, because one can use large, independent datasets to, first, derive the instruments, and then to test the associations. The association between genetically predicted BMI or height and melanoma risk per SNP was evaluated using a Wald-type ratio estimator.25 The 95% confidence intervals (95% CIs) were calculated from the SE of each Wald ratio. We combined individual Wald ratio estimates for all SNPs for each trait (i.e. BMI or height) using the inverse variance-weighted method (IVW) to obtain a weighted average of the effect estimates.25 We tested for heterogeneity in Wald ratios using Cochran's Q statistic.26
Sensitivity analyses
The IVW method assumes that there is no horizontal pleiotropy for all SNPs (that is, that the effect of genetic variants on the outcome operates entirely via the exposure of interest), and that all SNPs are valid instruments. However, because testing the validity of these assumptions is difficult in practice, two additional MR analyses, namely MR-Egger regression27 and weighted median estimator,28 were conducted to check for robustness of the estimates from IVW.27 MR-Egger regression is similar to IVW except that the intercept is not constrained to pass through the origin, with a non-zero intercept suggesting possibility of directional pleiotropy. The weighted median estimator method has the advantage that it is possible to estimate the effect as long as at least 50% of the variants in the analysis come from SNPs that satisfy the MR assumptions. These techniques address potential concern on the causal estimate due to weak violation of MR assumptions, but they require large sample sizes to detect effects.
Because deriving BMI and height instruments from datasets that include patients with melanoma could conceivably induce bias in odds ratios, we performed an additional sensitivity analysis restricted to 390 628 cancer-free White British participants in the UK Biobank only. For this analysis we used 520 and 2059 independent SNPs that were associated with BMI or height, respectively, at genome-wide significance as previously described.29,30 Cancer-free participants were defined as any individual without cancer, or benign or in situ tumours, recorded in the cancer registry using the International Classification of Disease, 10th edition (ICD-10). Prevalent and incident cancer cases in the UK Biobank were identified through linkage to the national cancer registries and hospital inpatient data. To assess whether our estimates were sensitive to the choice of instrument, we also conducted a separate analysis restricted to 97 BMI-associated SNPs and 697 height-associated SNPs identified in earlier published studies in the GIANT consortium only.20,21 We used R software (TwoSampleMR package) for Mendelian randomization analyses.31
Results
Mendelian randomization analyses of the association between BMI and melanoma risk
The primary analyses included 754 independent genetic variants obtained from the UK Biobank and the GIANT consortia as instruments for BMI. Five variants that were not available in the melanoma GWAS dataset and 19 that were palindromic strands were excluded, leaving 730 variants for analysis (Table 1). These variants explained approximately 8% of the variance in BMI in the UK Biobank and GIANT (Table 1). In an independent cohort (QSkin), a PRS derived from these variants explained ∼5% of the variance in BMI (Supplementary Figures 1 and 2, available as Supplementary data at IJE online).
Table 1.
The characteristics and source of genetic instruments used in Mendelian randomisation analyses
| Instruments from | Body mass index |
Height |
||||||
|---|---|---|---|---|---|---|---|---|
| Sample size | Number of SNPs obtained | Number of SNPs used | Total variance explained(r2) a | Sample size | Number of SNPs obtained | Number of SNPs used | Total variance explained (r2) a | |
| UK Biobank + GIANT | 681 275 | 754 | 730 | 7.8% | 693 529 | 3290 | 3163 | 19% |
| GIANT only | 339 224 | 97 | 79 | 2.7% | 253 288 | 697 | 360 | 13% |
| UK Biobank only | 390 628 | 520 | 495 | 7% | 310 793 | 2059 | 1810 | 17% |
SNP, single-nucleotide polymorphism; GIANT, Genetic Investigation of ANthropometric Traits.
Total variance explained by instrument is computed based on the genetic influence of the body mass index or height SNPs instruments on measured body mass index or height from the White-British participants from the UK Biobank cohort.
We estimated the overall odds ratio of developing melanoma per one SD increase in BMI (1 SD = 4.6 kg/m2). Based on the combined estimate derived from 730 genetic variants for BMI, we found no evidence of an association between higher genetically predicted BMI and melanoma (OR per 1 SD increase in BMI 1.00, 95% CI: 0.91–1.11) (Table 2 and Figure 1).
Table 2.
Association between increased body mass index and height and risk of melanoma using two-sample Mendelian randomisationa
| Instruments from | MR method | Body mass index |
Height |
||||
|---|---|---|---|---|---|---|---|
| Odds ratio | 95% CI | P-value | Odds ratio | 95% CI | P-value | ||
| UK Biobank + GIANTb | IVW | 1.00 | 0.91–1.11 | 0.99 | 1.08 | 1.02–1.13 | 0.004 |
| MR-Eggere | 0.99 | 0.74–1.33 | 0.97 | 1.05 | 0.96–1.14 | 0.14 | |
| Weighted median | 1.01 | 0.86–1.17 | 0.92 | 1.11 | 1.02–1.20 | 0.02 | |
| Simple mode | 0.85 | 0.51–1.43 | 0.55 | 1.07 | 0.80–1.45 | 0.61 | |
| Weighed mode | 0.98 | 0.73–1.32 | 0.89 | 1.13 | 0.95–1.36 | 0.15 | |
| GIANT onlyc | IVW | 0.96 | 0.80–1.15 | 0.66 | 1.09 | 1.02–1.18 | 0.01 |
| MR-Eggere | 0.99 | 0.56–1.73 | 0.25 | 1.02 | 0.84–1.25 | 0.8 | |
| Weighted median | 0.86 | 0.66–1.11 | 0.82 | 1.14 | 1.02–1.27 | 0.02 | |
| Simple mode | 0.78 | 0.45–1.34 | 0.36 | 1.14 | 0.81–1.60 | 0.44 | |
| Weighted mode | 0.99 | 0.72–1.38 | 0.98 | 1.14 | 0.87–1.49 | 0.34 | |
| UK Biobank onlyd | IVW | 1.01 | 0.98–1.03 | 0.76 | 1.07 | 1.01–1.13 | 0.01 |
| MR-Eggere | 1.03 | 0.96–1.11 | 0.31 | 1.04 | 0.87–1.25 | 0.25 | |
| Weighted median | 1.00 | 0.97–1.04 | 0.81 | 1.07 | 0.99–1.16 | 0.08 | |
| Simple mode | 0.95 | 0.86–1.07 | 0.45 | 1.03 | 0.97–1.07 | 0.33 | |
| Weighted mode | 1.01 | 0.93–1.08 | 0.83 | 1.02 | 0.98–1.04 | 0.26 | |
SNP, single nucleotide polymorphism; CI, confidence interval; GIANT, Genetic Investigation of ANthropometric Traits; IVW, inverse variance-weighted.
The estimates are given per one SD increase in body mass index (1 SD = 4.6 kg/m2) and per one SD increase in height (1 SD = 9.27 cm).
UK Biobank + GIANT: genetic variants obtained from the 2018 body mass index and height GWAS meta-analysis in the GIANT consortium.
GIANT only: genetic variants obtained from the 2015 body mass index genome-wide association study meta-analysis and from the 2014 height genome-wide association study meta-analysis in the GIANT consortium.
UK Biobank only: body mass index and height genetic variants obtained from the UK Biobank only.
For MR-Egger analyses, the standard error (SE) for each exposure-outcome estimate was obtained by bootstrapping the distributions of the SNP effect estimates for both exposure and outcome 1000 times.
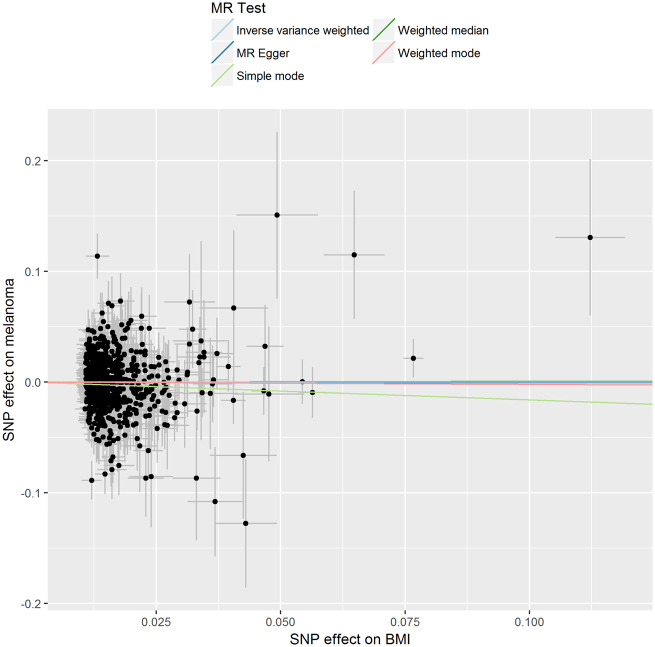
Figure 1.
Association of individual single nucleotide polymorphisms with body mass index and melanoma risk. Instrumental variable estimates were derived from 730 body mass index single nucleotide polymorphisms instruments identified in the body mass index genome-wide association study meta-analysis using samples from Genetic Investigation of ANthropometric Traits and the UK Biobank cohort. Error bars represent 95% confidence intervals. The gradients of regression lines colours correspond to the instrumental variable estimates of the effect of body mass index on melanoma risk with different Mendelian randomization (MR) methods compared.
We performed sensitivity analyses to check whether the null association might have arisen through violations of the MR assumptions. We found no evidence that our risk estimates were influenced by directional pleiotropy, as the average pleiotropic effect of the MR-Egger regression intercept was close to null (MR-Egger intercept: 0.0001, P-value = 0.9 (Table 3). Graphical assessment of bias in the MR funnel plot suggested that the dispersion of individual estimates was symmetrical (Supplementary Figure 3, available as Supplementary data at IJE online), indicating that our estimates were not driven by individual outliers. Taken together, the sensitivity analyses suggest that the null findings were very unlikely to be due to violating the assumption imposed by the exclusion restriction criterion. Finally, we checked whether our inferences were influenced by the choice of instruments, by repeating the analyses using 79 of the 97 BMI-associated variants identified by the GIANT consortium. We also repeated the same analysis using the 495 of the 520 BMI variants identified from the UK Biobank. The results were essentially the same regardless of the source of instrument (Table 2).
Table 3.
Estimates of Egger intercept to evaluate evidence for directional pleiotropy in Mendelian randomisation association
| Instruments from | Body mass index |
Height |
||||
|---|---|---|---|---|---|---|
| Egger intercept | SE of Egger intercept | P-value | Egger intercept | SE of Egger intercept | P-value | |
| UK Biobank + GIANT | 0.0001 | 0.002 | 0.9 | 0.001 | 0.0008 | 0.6 |
| UK Biobank only | −0.002 | 0.02 | 0.3 | 0.003 | 0.001 | 0.5 |
| GIANT only | −0.005 | 0.005 | 0.4 | 0.004 | 0.003 | 0.2 |
GIANT, Genetic Investigation of ANthropometric Traits.
Here, P-value refers to the P-value of the estimated Egger intercept being null. A significant P-value (P <0.05) would present evidence that the MR causal estimates derived via the inverse variance-weighted model were biased by directional pleiotropy.
Mendelian randomization analyses of the association between height and melanoma risk
We obtained data for a total of 3290 height-related genetic variants from UK Biobank and the GIANT consortium as potential instrumental variables. However, 117 of these 3290 variants were not available in the melanoma GWAS dataset and 10 variants were palindromic, so were excluded from the analysis. The remaining 3163 variants explained ∼19% of variance in height in UK Biobank and GIANT (Table 1). A PRS, derived from these height variants, explained ∼12% of variance in height in the QSkin cohort (Supplementary Figures 4 and 5, available as Supplementary data at IJE online). Using the IVW method to pool estimates from individual variants, genetically predicted height was statistically significantly associated with increased melanoma risk [OR 1.08, 95% CI: 1.02–1.13, per 1 SD (9.27 cm) increase in height] (Table 2 and (Figure 2).
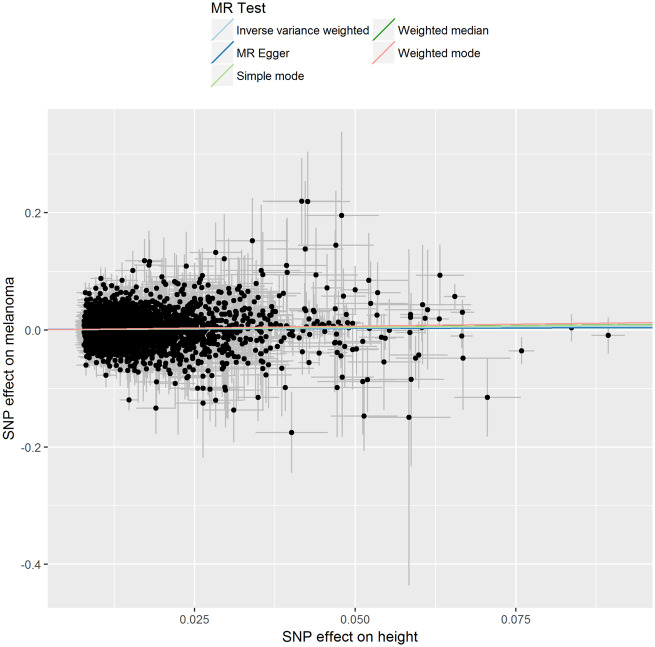
Figure 2.
Association of individual single nucleotide polymorphisms with height and melanoma risk. Instrumental variable estimates were derived from 3290 height single nucleotide polymorphisms instruments identified in the height genome-wide association study meta-analysis using samples from Genetic Investigation of ANthropometric Traits and the UK Biobank cohort. Error bars represent 95% confidence intervals. The gradients of regression lines colours correspond to the instrumental variable estimates of the effect of height on melanoma risk with different Mendelian randomization (MR) methods compared.
After excluding non-inferable palindromic SNPs and SNPs that could not be obtained from the melanoma GWAS dataset, we performed sensitivity analyses initially using 1810 height-associated SNPs from the UK Biobank only, and secondly using a restricted list of 360 height-associated SNPs from the earlier GIANT consortium analysis.21 These analyses made little difference to the main findings (Table 2). Additional sensitivity analysis using MR-Egger regression to assess whether the causal estimates could have been affected by directional pleiotropy showed no such evidence (intercept = −0.001, P-value = 0.6) (Table 3). Visual assessment of directional pleiotropy using a funnel plot showed that variants were symmetrically distributed (Supplementary Figure 6, available as Supplementary data at IJE online).
We observed evidence of heterogeneity across SNP estimates [Q = 937, P <0.0001 for BMI; Q = 2757, P <0.0001 for height (Supplementary Table 1, available as Supplementary data at IJE online)]. SNPs that showed strong evidence of heterogeneity (Q >3.84) were removed and the analyses were repeated. The adjusted MR estimates showed no evidence of heterogeneity. However, the effect estimates were generally unchanged.
Discussion
We conducted two-sample MR analyses using summary statistics from the largest GWAS meta-analyses of BMI, height and melanoma. Overall, we found no evidence that genetically predicted BMI was associated with increased risk of melanoma, but found evidence to suggest that genetically predicted height conferred increased risk of melanoma.
Investigating a possible causal association between obesity and melanoma is relevant, given the heterogeneity observed across previous observational studies, the substantial increase in obesity prevalence worldwide32 and the rapid increases in melanoma incidence observed in many populations.33 To our knowledge, this is the first analysis to use MR techniques. While observational studies can identify associations, they cannot always establish whether relationships are causal, notably in instances where confounding is believed to be present but not fully controlled. For example, many epidemiological studies rely on self-reported weight and height measurements8,12,34 which are subject to misclassification.35,36 In addition, few studies adjusted for the confounding effect of sun exposure,6,37 the major risk factor for melanoma.
Our null findings suggest that the increased risk of melanoma among overweight and obese people, reported in previous epidemiological studies,3,4,38,39 may be due to other, non-causal explanations such as ascertainment biases, misclassification or residual confounding inherent in observational studies. Earlier experimental studies had suggested that an association between BMI and melanoma might be plausible. For example, there were reports that obese mice exposed to UVB radiation had higher levels of pro-inflammatory cytokines in their skin than their non-obese counterparts.40 Those laboratory findings gave some credence to a possible link between obesity and skin cancer, on the basis that inflammatory responses in the skin might increase the risk of neoplasia.41,42 Our data indicate that even if obesity modifies cutaneous responses to sunlight in some animal models, the effects do not necessarily translate into measurable changes in melanoma risk in humans.
In contrast to the lack of association between BMI and melanoma, the MR findings in relation to a positive association between height and melanoma are broadly consistent with the observational literature, albeit with a smaller effect size estimate from MR. For instance, the UK Million Women Study, with ∼3500 melanoma cases, reported a 30% increased melanoma risk per 10-cm increase in height.5 Subsequently, in a large population-based cohort in Norway including more than 3000 melanoma cases among ∼300 000 men and women, a 50% increased melanoma risk was observed for men and women in the fifth versus first quintile of height.4 Increased risk of melanoma with greater height was also reported in a study conducted in a high ambient sunlight setting (Queensland, Australia), although the estimates lack precision due to low sample size.9 Finally, the evidence from a pooled analysis including eight case-control studies provides additional support for an association between height and melanoma in women.11
The MR findings described in this study in relation to the two-sample MR generally agree with previously reported findings from one-sample MR analyses conducted in the UK Biobank. In those analyses, no association between genetically determined higher BMI and melanoma was found (OR: 0.98, 95% CI: 0.85–1.14),30 but a 12% increase in melanoma risk was reported per 10-cm increase in genetically predicted height (OR: 1.12, 95% CI: 1.001–1.26).29 Height has also been reported as a risk factor for other types of cancer in other MR studies, although the effect of height on melanoma reported in our study was relatively lower for some cancer types, but similar for others. For example, previous research has reported that for every 10-cm increase in genetically predicted height, the risk ratio was 1.22 (95% CI: 1.13–1.32) for breast cancer,43 1.07 (95% CI: 1.01–1.14) for colorectal cancer,44 1.23 (95% CI 1.06–1.42) for endometrial cancer29 and 1.12 (95% CI: 1.02–1.23) for ovarian cancer.45
Adult height is determined by various growth mechanisms, childhood environment and possibly epigenetic factors, any of which may influence melanoma risk. Previous investigators have speculated that height is a proxy for the total number of cells (including stem cells) in the body, and that this presumably increases the probability of mutations and hence malignancy.14,15 However, this hypothesis does not accord with the higher rates of malignancy observed across species (for example, between mice and humans which differ in volume by orders of magnitude).46 The lack of relationship between body size and cancer risk across species, known as Peto’s Paradox,47 suggests that larger organisms might have evolved cancer-suppressing mechanisms in order to live longer.48 For example, a recent study has shown that the genome of an elephant has 20 copies of the P53 tumour suppressor gene, whereas humans have only one copy.49 Thus, body size is an imperfect predictor of cancer risk in an organism across species, but body size and cancer risk are positively correlated within members of the same species7–10 BMI is a reliable measure of body size, but mature adipocytes do not undergo mitoses as they are a fully differentiated cell type, and thus provide fewer targets for oncogenic mutations.
An earlier study also reported a positive association between the number of naevi and height, but not weight.50 The study argued that both naevus count and melanoma are associated with longer telomeres. Telomere length has been reported as a genetic marker of reduced senescence and increased growth,51 and is believed to play important roles in carcinogenesis.52 Various hormones implicated in childhood growth, such as insulin-like growth factor (IGF)-I,53 also regulate cell turnover, apoptosis and tumour progression,54 and thus could be implicated in cancer development. These mechanisms are presumably largely under genetic control, and the genetic associations between height and melanoma might be mediated through these pathways.
While biological mechanisms to explain the association between height and melanoma have intuitive appeal, it is also possible that height might mediate its effects on melanoma risk through other pathways, at least in some populations. Recently, a Mendelian randomization study conducted within the UK Biobank (using instruments very similar to those that we used) reported that genetically predicted height was significantly associated with four different measures of socioeconomic status (SES), with strong positive associations observed for job class and annual household income.55 Previous observational studies, particularly those conducted in settings of low ambient sunlight (such as northern Europe), have reported significantly higher melanoma incidence among people in high SES categories compared with those in low SES categories. In those settings, it has been postulated that greater affluence has given greater access to holidays in sunny locations and sunburns, thereby conferring an increased risk of melanoma.56–58 Occupational studies from the UK59 and Sweden60 corroborate this hypothesis, showing that indoor workers have significantly higher risks of melanoma than outdoor workers in those countries. Thus, at a population-level, genetically determined height is possibly causally associated with melanoma through a pathway of social class and sun exposure.
A limitation common to all MR analyses relates to potential pleiotropy, whereby a genetic variant is independently associated with the outcome, but not through the exposure of interest. We assessed potential pleiotropy using the MR-Egger method and observed no evidence that the exclusion restriction criteria assumption was violated. While it is also possible that some of the variants used in the analysis might be associated with confounders of the height and melanoma association, such an effect would likely be small because our genetic instrument was generated from more than 3000 variants explaining ∼19% of variance in height, which further reduces the likelihood of bias from violating MR assumptions.61 Our analyses also intrinsically assume a linear relationship between BMI/height and the log(OR) on melanoma. Here, the MR estimates capture a population-averaged effect across different strata of exposure, which might differ from the association of BMI/height on melanoma for individuals at extreme ends of the distribution. We argue that this is unlikely a major concern for our analyses, given the overall null finding for BMI (except for extreme circumstances where the opposing direction of effect across two ends of a stratum directly cancel out), however a larger sample size will be required to comprehensively evaluate any non-linear relationships between height and melanoma.
In conclusion, these large-scale Mendelian randomization analyses found little evidence to suggest that the association between higher BMI and melanoma risk is causal but, in accord with earlier observational studies, height was found to be associated with melanoma. Mechanisms through which greater height might lead to increased risk of melanoma remain unclear, and it is possible that the effect is mediated through various pathways, ranging from direct hormonal effects through to social class and sun exposure. While the most effective way to reduce cancer risks involves elimination of the causal risk factor, it is not feasible to modify adult height. However, the specific mechanisms (which might be modifiable) underlying this association may provide valuable insights into carcinogenesis. It is also possible that height may contribute to future risk stratification algorithms which could be used to target people for early detection activities.
Funding
This work was supported by a programme grant from the National Health and Medical Research Council (NHMRC) of Australia (grant numbers APP1073898, APP1063061 and APP1123248). This work was conducted using the UK Biobank Resource (application number 25331). D.C.W. and R.E.N. are supported by NHMRC Research Fellowships. S.M. is supported by the Australian Research Council Future Fellowship. The funders had no role in study design, data collection and analysis or decision to publish.
Supplementary Material
Acknowledgements
The full acknowledgements for the melanoma meta-analysis can be found in the Supplementary Note of the 2015 melanoma GWAS meta-analysis.19
Conflict of interest
None declared.
References
- 1.International Agency for Research on Cancer. IARC Monograph on the Evaluation of Carcinogenic Risks to Humans; Ultraviolet Radiation. Lyon, France: International Agency for Research on Cancer, 1992. [Google Scholar]
- 2. Gandini S, Sera F, Cattaruzza MS et al. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur J Cancer 2005;41:2040–059. [DOI] [PubMed] [Google Scholar]
- 3. Sergentanis TN, Antoniadis AG, Gogas HJ et al. Obesity and risk of malignant melanoma: a meta-analysis of cohort and case-control studies. Eur J Cancer 2013;49:642–57. [DOI] [PubMed] [Google Scholar]
- 4. Stenehjem JS, Veierod MB, Nilsen LT et al. Anthropometric factors and cutaneous melanoma: prospective data from the population-based Janus Cohort. Int J Cancer 2018;142:681–90. [DOI] [PubMed] [Google Scholar]
- 5. Green J, Cairns BJ, Casabonne D, Wright FL, Reeves G, Beral V. Height and cancer incidence in the Million Women Study: prospective cohort, and meta-analysis of prospective studies of height and total cancer risk. Lancet Oncol 2011;12:785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lukanova A, Bjor O, Kaaks R et al. Body mass index and cancer: results from the Northern Sweden Health and Disease Cohort. Int J Cancer 2006;118:458–66. [DOI] [PubMed] [Google Scholar]
- 7. Freedman DM, Sigurdson A, Doody MM, Rao RS, Linet MS. Risk of melanoma in relation to smoking, alcohol intake, and other factors in a large occupational cohort. Cancer Causes Control 2003;14:847–57. [DOI] [PubMed] [Google Scholar]
- 8. Pothiawala S, Qureshi AA, Li Y, Han J. Obesity and the incidence of skin cancer in US Caucasians. Cancer Causes Control 2012;23:717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lahmann PH, Hughes MC, Williams GM, Green AC. A prospective study of measured body size and height and risk of keratinocyte cancers and melanoma. Cancer Epidemiol 2016;40:119–25. [DOI] [PubMed] [Google Scholar]
- 10. Shors AR, Solomon C, McTiernan A, White E. Melanoma risk in relation to height, weight, and exercise (United States). Cancer Causes Control 2001;12:599–606. [DOI] [PubMed] [Google Scholar]
- 11. Olsen CM, Green AC, Zens MS et al. Anthropometric factors and risk of melanoma in women: a pooled analysis. Int J Cancer 2007;122:1100–08. [DOI] [PubMed] [Google Scholar]
- 12. Kvaskoff M, Bijon A, Mesrine S, Vilier A, Clavel-Chapelon F, Boutron-Ruault MC. Anthropometric features and cutaneous melanoma risk: a prospective cohort study in French women. Cancer Epidemiol 2014;38:357–63. [DOI] [PubMed] [Google Scholar]
- 13. Benyi E, Linder M, Adami J, Kieler H, Palme M, Sävendahl L. Adult height is associated with risk of cancer and mortality in 5.5 million Swedish women and men. J Epidemiol Community Health 2019;73:730–36. jech-2018-211040. [DOI] [PubMed] [Google Scholar]
- 14. Nunney L. Size matters: height, cell number and a person's risk of cancer. Proc Biol Sci 2018;285:1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Albanes D, Winick M. Are cell number and cell proliferation risk factors for cancer? J Natl Cancer Inst 1988;80:772–74. [DOI] [PubMed] [Google Scholar]
- 16. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133–63. [DOI] [PubMed] [Google Scholar]
- 17. Evans DM, Davey Smith G. Mendelian randomization: new applications in the coming age of hypothesis-free causality. Annu Rev Genom Hum Genet 2015;16:327–50. [DOI] [PubMed] [Google Scholar]
- 18. Yengo L, Sidorenko J, Kemper KE et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet 2018;27:3641–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Law MH, Bishop DT, Lee JE et al. Genome-wide meta-analysis identifies five new susceptibility loci for cutaneous malignant melanoma. Nat Genet 2015;47:987–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Locke AE, Kahali B, Berndt SI et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wood AR, Esko T, Yang J et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 2014;46:1173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Olsen CM, Green AC, Neale RE et al. Cohort Profile: The QSkin Sun and Health Study. Int J Epidemiol 2012;41:929. [DOI] [PubMed] [Google Scholar]
- 23. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigasci 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pierce BL, Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol 2013;178:1177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 2013;37:658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burgess S, Bowden J. Integrating summarized data from multiple genetic variants in Mendelian randomization: bias and coverage properties of inverse-variance weighted methods. arXiv 2015;preprint arXiv:151204486.
- 27. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ong J-S, An J, Law MH et al. Height and overall cancer risk and mortality: evidence from a Mendelian randomisation study on 310,000 UK Biobank participants. Br J Cancer 2018;118:1262–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gharahkhani P, Ong JS, An J et al. Effect of increased body mass index on risk of diagnosis or death from cancer. Br J Cancer 2019;120:565–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hemani G, Zheng J, Elsworth B et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ng M, Fleming T, Robinson M et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014;384:766–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Whiteman DC, Green AC, Olsen CM. The growing burden of invasive melanoma: projections of incidence rates and numbers of new cases in six susceptible populations through 2031. J Invest Dermatol 2016;136:1161–71. [DOI] [PubMed] [Google Scholar]
- 34. Tang JY, Henderson MT, Hernandez-Boussard T et al. Lower skin cancer risk in women with higher body mass index: the women's health initiative observational study. Cancer Epidemiol Biomarkers Prev 2013;22:2412–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shields M, Connor Gorber S, Tremblay MS. Estimates of obesity based on self-report versus direct measures. Health Rep 2008;19:61–76. [PubMed] [Google Scholar]
- 36. Connor Gorber S, Tremblay M, Moher D, Gorber B. A comparison of direct vs. self-report measures for assessing height, weight and body mass index: a systematic review. Obes Rev 2007;8:307–26. [DOI] [PubMed] [Google Scholar]
- 37. Bhaskaran K, Douglas I, Forbes H, dos-Santos-Silva I, Leon DA, Smeeth L. Body-mass index and risk of 22 specific cancers: a population-based cohort study of 5.24 million UK adults. Lancet 2014;384:755–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet 2008;371:569–78. [DOI] [PubMed] [Google Scholar]
- 39. Dobbins M, Decorby K, Choi BC. The association between obesity and cancer risk: a meta-analysis of observational studies from 1985 to 2011. ISRN Prev Med 2013;2013:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sharma SD, Katiyar SK. Leptin deficiency-induced obesity exacerbates ultraviolet B radiation-induced cyclooxygenase-2 expression and cell survival signals in ultraviolet B-irradiated mouse skin. Toxicol Appl Pharmacol 2010;244:328–35. [DOI] [PubMed] [Google Scholar]
- 41. Clement E, Lazar I, Muller C, Nieto L. Obesity and melanoma: could fat be fueling malignancy? Pigment Cell Melanoma Res 2017;30:294–306. [DOI] [PubMed] [Google Scholar]
- 42. Brandon EL, Gu JW, Cantwell L, He Z, Wallace G, Hall JE. Obesity promotes melanoma tumor growth: role of leptin. Cancer Biol Ther 2009;8:1871–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang B, Shu XO, Delahanty RJ et al. Height and breast cancer risk: evidence from prospective studies and Mendelian randomization. J Natl Cancer Inst 2015;107:djv219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thrift AP, Gong J, Peters U et al. Mendelian randomization study of height and risk of colorectal cancer. Int J Epidemiol 2015;44:662–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dixon-Suen SC, Nagle CM, Thrift AP et al. Adult height is associated with increased risk of ovarian cancer: a Mendelian randomisation study. Br J Cancer 2018;118:1123–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nagy JD, Victor EM, Cropper JH. Why don't all whales have cancer? A novel hypothesis resolving Peto's paradox. Integr Comp Biol 2007;47:317–28. [DOI] [PubMed] [Google Scholar]
- 47. Peto R. Epidemiology, multistage models, and short-term mutagenicity tests 1. Int J Epidemiol 2016;45:621–37. [DOI] [PubMed] [Google Scholar]
- 48. Caulin AF, Maley CC. Peto's Paradox: evolution's prescription for cancer prevention. Trends Ecol Evol 2011;26:175–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Abegglen LM, Caulin AF, Chan A et al. Potential mechanisms for cancer resistance in elephants and comparative cellular response to DNA damage in humans. JAMA 2015;314:1850–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ribero S, Glass D, Aviv A, Spector TD, Bataille V. Height and bone mineral density are associated with naevus count supporting the importance of growth in melanoma susceptibility. PLoS One 2015;10:e0116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bernadotte A, Mikhelson VM, Spivak IM. Markers of cellular senescence. Telomere shortening as a marker of cellular senescence. Aging 2016;8:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Han J, Qureshi AA, Prescott J et al. A prospective study of telomere length and the risk of skin cancer. J Invest Dermatol 2009;129:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Clayton PE, Banerjee I, Murray PG, Renehan AG. Growth hormone, the insulin-like growth factor axis, insulin and cancer risk. Nat Rev Endocrinol 2011;7:11–24. [DOI] [PubMed] [Google Scholar]
- 54. Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer 2008;8:915–28. [DOI] [PubMed] [Google Scholar]
- 55. Tyrrell J, Jones SE, Beaumont R et al. Height, body mass index, and socioeconomic status: Mendelian randomisation study in UK Biobank. BMJ 2016;352:i582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pérez-Gómez B, Aragonés N, Gustavsson P, Lope V, López-Abente G, Pollán M. Socio-economic class, rurality and risk of cutaneous melanoma by site and gender in Sweden. BMC Public Health 2008;8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Elwood JM, Whitehead SM, Davison J, Stewart M, Galt M. Malignant melanoma in England: risks associated with naevi, freckles, social class, hair colour, and sunburn. Int J Epidemiol 1990;19:801–10. [DOI] [PubMed] [Google Scholar]
- 58. Petersen B, Triguero-Mas M, Maier B et al. Sun behaviour and personal UVR exposure among Europeans on short term holidays. J Photochem Photobiol B 2015;151:264–69. [DOI] [PubMed] [Google Scholar]
- 59. Beral V, Robinson N. The relationship of malignant melanoma, basal and squamous skin cancers to indoor and outdoor work. Br J Cancer 1981;44:886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vagero D, Persson G. Occurrence of cancer in socioeconomic groups in Sweden. An analysis based on the Swedish Cancer Environment Registry. Scand J Soc Med 1986;14:151–60. [DOI] [PubMed] [Google Scholar]
- 61. VanderWeele TJ, Tchetgen Tchetgen EJ, Cornelis M, Kraft P. Methodological challenges in mendelian randomization. Epidemiology 2014;25:427–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.