Abstract
Hepatitis C virus (HCV) proliferates by hijacking the host lipid machinery. In vitro replication systems revealed many aspects of the virus life cycle; in particular, viral utilization of host lipid metabolism during HCV proliferation. HCV interacts with lipid droplets (LDs) before starting the process of virus capsid formation at the lipid-rich endoplasmic reticulum (ER) membrane compartment. HCV buds into the ER via lipoprotein assembly and secretion. Exchangeable apolipoproteins, represented by apolipoprotein E (apoE), play pivotal roles in enhancing HCV-specific infectivity. HCV virions are likely to interact with other lipoproteins circulating in blood vessels and incorporate apolipoproteins as well as lipids. This review focuses on virus assembly and egress by briefly describing the recent advances in this area.
The causative agents of chronic non-A and non-B hepatitis are present in patients at much lower titers than hepatitis viruses such as hepatitis A virus (HAV) and hepatitis B virus (HBV); therefore, isolation of genomic information was difficult until the advent of the bacteriophage expression vector λgt11 (Choo et al. 1989). Analysis of the cloned hepatitis C virus (HCV) genome revealed high sequence similarity with flaviviruses and pestiviruses. HCV is now classified into the family Flaviviridae, genus Hepacivirus. Compared with other members of this family, HCV shows peculiar physicochemical characteristics, most notably differences in virus density in circulating blood (Carrick et al. 1992; Thomssen et al. 1992; Hijikata et al. 1993b; Prince et al. 1996; André et al. 2002, 2005; Pumeechockchai et al. 2002; Nielsen et al. 2006, 2008). This varies from low (∼1.05 g/mL) to high (∼1.25 g/mL). The low-density HCV fraction associates with very low-density lipoproteins (VLDLs) and contains apolipoprotein A (apoA), apoA1, apoB, apoC, apoE, and phospholipids, such as phosphatidylcholine (PC) and sphingomyelin (SM), along with free and esterified cholesterol (Chang et al. 2007; Dreux et al. 2007; Meunier et al. 2008; Merz et al. 2011; Catanese et al. 2013; Sun et al. 2013). Thus, circulating HCV is referred to as a lipoviroparticle (LVP) (André et al. 2002).
Development of in vitro culture systems (sub- and full genome replication systems adapted to cultured cell lines) (Lohmann et al. 2001; Lindenbach et al. 2005; Wakita et al. 2005; Zhong et al. 2005) and pseudotype HCV proliferation systems (Bartosch et al. 2003) has revealed the importance of lipids to the HCV life cycle: (1) there is a functional association between lipid droplets (LDs) and virus replication; (2) the virus uses host lipid secretion machinery to facilitate secretion; (3) there is increased association between apoE and HCV both inside and outside infected cells; and (4) removal of lipid components from HCV decreases its infectivity, suggesting a strong association between HCV and lipid components. However, the detailed mechanism(s) underlying the association between lipids and HCV proliferation is unclear.
In this review, I discuss the mechanisms underlying virus egress by dissecting the process into three parts: (1) LD to virus envelopment; (2) envelopment to secretion of virus into the endoplasmic reticulum (ER) lumen; and (3) postegress events. I also refer the reader to several other review articles (Bartenschlager et al. 2011; Lindenbach and Rice 2013; Aizawa et al. 2015; Grassi et al. 2016).
HETEROGENEOUS DENSITY OF HCV PARTICLES
Measurement of HCV RNA in the plasma of infected individuals has shown that virus density varies from 1.03 to 1.25 g/mL. HCV particles at low density are highly infectious (Bradley et al. 1991; Hijikata et al. 1993b). The density of HCV particles varies according to host cell type. Infectious viruses from cultured cells show characteristics similar to those from HCV-infected patients (Bradley et al. 1991; Cai et al. 2005; Lindenbach et al. 2005; Wakita et al. 2005; Zhong et al. 2005; Gastaminza et al. 2006; Chang et al. 2007).
Intracellular virions produced by in vitro systems are characterized by high density and low infectivity (Gastaminza et al. 2006), suggesting that increased infectivity is attributable to association with host lipoproteins before egress. HCV produced by chimpanzees or mice harboring human liver grafts infected with in vitro–cultured HCV shows higher infectivity and lower average buoyant density than cultured HCV (Lindenbach et al. 2006; Calattini et al. 2015).
THE MEMBRANOUS NICHE IN WHICH VIRUS RNA SYNTHESIS OCCURS
HCV induces remodeling of the primary ER-derived membrane, termed the membranous web (Gosert et al. 2003). A similar altered membrane structure is observed in the liver of HCV-infected chimpanzees and humans (Pfeifer et al. 1980; De Vos et al. 2002; Egger et al. 2002; Falcón et al. 2003). HCV structural proteins are not essential for establishing this web structure in which HCV RNA synthesis takes place. The altered membranous structures are observed in cells that express autonomously self-replicating HCV subgenomes that lack coding sequences for virus structural proteins (HCV core proteins [core] and HCV envelope proteins [E1 and E2]). Newly synthesized HCV RNA, detected by metabolic labeling with 5-bromouridine, colocalizes with the membranous web, suggesting that the web structure contains an active site for HCV replication (Gosert et al. 2003). Details of the membranous structures induced by HCV-infected cells have been revealed by electron microscopy (EM) (Romero-Brey et al. 2012; Ferraris et al. 2013). Several host factors contribute to membranous web formation (Dreux et al. 2009; Ferraris et al. 2010; Neufeldt et al. 2013; Paul et al. 2013).
Digitonin is a detergent that permeabilizes the plasma membrane without affecting the inner cellular membrane structure. Cells bearing HCV subgenomic replicons do not show altered HCV RNA synthesis before or after the treatment with digitonin (Miyanari et al. 2007). Most HCV proteins in permeabilized cells are retained inside the cell membrane, indicating that the proteins are associated with membrane components. Incubation of permeabilized cells with a protease reduces the amount of HCV proteins, including nonstructural (NS) proteins NS3, NS4A, NS5A, and NS5B, to <10% of the amount present before treatment. However, importantly, the cells retain the ability to synthesize the subgenomic HCV RNA and replicate almost as efficiently as before protease treatment (Miyanari et al. 2007). Thus, only a small proportion of HCV proteins maintain HCV RNA synthesis. However, treatment of permeabilized cells with detergents (e.g., 1% NP40) followed by protease treatment results in almost complete clearance of HCV RNA and protein, suggesting that small amounts of NS HCV proteins and cellular membranous components constitute an active “replication niche” for HCV RNA synthesis. The majority of remaining HCV proteins is localized on the cytoplasmic side of the cell membrane. Because a very small percentage of NS HCV proteins is responsible for genome replication, membranous webs observed by EM might reflect the vast majority of the HCV proteins that are not directly involved in active “replication niches.” It is possible that the “replication niche” is in close proximity to other HCV protein complexes and that the majority of HCV proteins that are not involved in the “replication niche” play other important roles that maintain virus proliferation, for example, during the pre- and postreplication stages. Indeed, accumulating evidence shows that HCV proteins not involved in virus components do contribute to virus assembly and egress. Also, it is possible that both the niche and excess NS proteins outside the niches are required to protect viral RNA from host defenses (Horner 2015; Neufeldt et al. 2016).
Although this study suggests that HCV RNA synthesis is sensitive to detergent, Shi et al. (2003) reported detergent resistance. However, the RNA synthesis was reduced to a level less than one-third of that of untreated virus (Shi et al. 2003), indicating that the RNA synthesis is sensitive to detergent. It is likely that the effect of detergent depends on the amounts of lipid components within the membranes in the sample analyzed.
ASSOCIATION OF HCV CORE PROTEIN WITH CYTOPLASMIC LDs
HCV core protein forms the viral capsid and is associated with the ER membrane and the surface of cytoplasmic LDs in mammalian cells (Moradpour et al. 1996; Barba et al. 1997; Yasui et al. 1998). Based on hydrophobicity and clustering of basic amino acids within the protein (Hope and McLauchlan 2000), the HCV core comprises two regions: the amino-terminal region (D1; amino acid [aa] residues 1–118) and a central domain (D2; aa residues 119–177). D1 harbors basic aa residues that interact with viral RNA (Boulant et al. 2005; Ivanyi-Nagy et al. 2006). D2 is hydrophobic and is important for the interaction with LDs (Hope and McLauchlan 2000; Boulant et al. 2006).
The core protein is produced as a precursor peptide (191 aa residues) (Hijikata et al. 1991), which is cleaved to yield the mature form, p21 (Yasui et al. 1998). Retention of the hydrophobic sequence at the carboxyl terminus (aa 178–191) anchors the protein to the phospholipid bilayer of the membrane. p21 can traverse the phospholipid monolayer membrane of the LD or the cytoplasmic leaflet of the ER in the double layer membrane structure (McLauchlan et al. 2002; Targett-Adams et al. 2008).
LDs are thought to be important for production of infectious virus (Boulant et al. 2007; Miyanari et al. 2007; Shavinskaya et al. 2007). The replication-competent HCV replicon, HCVJFH1, increases the number of large LDs in a core-dependent manner (Miyanari et al. 2007). LD size varies depending on the core sequence in the HCV replicon. LD growth occurs through fusion of smaller LDs, transfer of lipid esters between adjoining LDs, and via activity of biosynthetic enzymes (Wilfling et al. 2013; Ohsaki et al. 2014). HCV cell culture (HCVcc) variants derived from cell culture may show differences in the regulation of the function that determines LD morphogenesis (Perlemuter et al. 2002; Tsutsumi et al. 2002; Dharancy et al. 2005; Blackham et al. 2010; Mankouri et al. 2010; Bose et al. 2014; Kim et al. 2014). The D2 core mutant does not associate with LDs, thereby inhibiting virus production (Boulant et al. 2007; Miyanari et al. 2007; Shavinskaya et al. 2007). Inhibiting the signal peptidase responsible for cleaving the carboxy-terminal region of the HCV core also reduces virus production (Targett-Adams et al. 2008). These observations indicate that productive infection requires association of the HCV core with LDs or the ER membrane.
More importantly, core-associating LDs are closely associated with membrane structures rich in other HCV proteins such as E1/E2 and NS proteins such as NS5A and NS4A/B (Miyanari et al. 2007). Because membrane structures do not accumulate near LDs in cells not expressing HCV core, or in cells expressing an HCV core mutant lacking association with LDs, association between core and LDs is important for recruitment of membrane components to the area in which core-associating LDs reside. It is possible that the biochemically prepared LD fractions in this study might have been copurified with ER membrane fractions harboring HCV RNA synthesizing activity (Miyanari et al. 2007). This is likely because HCV proteins such as E2, NS3, NS4B, and NS5B (which do not associate directly with LDs but do associate with ER membranes) were present in the prepared LD fraction (Miyanari et al. 2007). This result strongly suggests close proximity of the core-bound LDs with the replication niche and the site of assembly on the ER membrane.
NS5A associates with LD and interacts with HCV core. NS5A may play roles, at least in part, in assembling the membranous structure around core-associating LDs because NS5A not only interacts with core-associating LDs but also with the ER membrane (Shi et al. 2002). NS5A interacts with the amino-terminal basic region of HCV core through domain I, rather than domain III, of NS5A (Gawlik et al. 2014).
Several host factors are involved in association of HCV core with LDs. Diacylglycerol acyltransferase-1 (DGAT1) interacts with HCV core and NS5A and is required for trafficking of HCV core and NS5A to LDs (Herker et al. 2010). Inhibition of DGAT1 activity or RNA interference (RNAi)-mediated knockdown of DGAT1 severely impairs production of infectious virions (Camus et al. 2013). Small interfering RNA (siRNA)-mediated knockdown of heat shock cognate 70 (HSC70) reduces the volume of LDs and inhibits virus release (Parent et al. 2009). The mitogen-activated protein kinase (MAPK)-regulated cytosolic phospholipase A2, group IVA (PLA2G4A) plays as a role in recruiting HCV core to LDs and in specific cleavage of lipids containing arachidonic acid, which is essential for production of highly infectious viral particles (Menzel et al. 2012). Lipolysis by PLA2G4A also reduces the amount of HCV core protein on LDs (Menzel et al. 2012).
The genetic determinants of association of HCV core with LDs depends on the HCV core variant (Etienne et al. 2015). Replacing ten amino acid residues in the core protein of HCVJFHI that differ from those residues that are conserved in other HCV genotypes alters core localization and HCV infectivity. In cells expressing this mutant HCV genome, termed C10M-JFH1, most core proteins localize to the ER membrane rather than LDs. NS5A also colocalizes with this core protein at the ER membrane. Replication of C10M-JFH-1 is higher than that of JFH-1, indicating an inverse correlation between extracellular viral titer and association of HCV core proteins with LDs (Etienne et al. 2015). This may explain why the processes of encapsidation and virus envelopment are rate limiting in the case of HCVJFH1. Different genotypes show varying levels of core association with the LD (Galli et al. 2013; Kim et al. 2014). Colocalization of core and NS5A at the membrane is observed irrespective of genotype (Galli et al. 2013). Core and NS5A proteins are highly associated with LDs at 12 hours postinfection, but are associated mostly with the ER at later times (Galli et al. 2013). These results suggest that core-bound LD brings the replication niche and assembly platform into close proximity. Once this structure is established, the number of core molecules on the LD decreases as they accumulate at the ER membrane and the virus is assembled.
These data suggest that the direct involvement of core-bound LD in virus assembly (i.e., interaction between LD-associated NS5A and core proteins with HCV RNA) needs to be reevaluated using different HCV genotypes.
STAGES INVOLVED IN VIRUS ASSEMBLY AND EGRESS
From LD to Envelopment of Virus Particles
Studies of HCV assembly show that almost all virus proteins are involved in assembly, together with the coordinated actions of cellular proteins. The core and E1/E2 proteins are the minimum integral protein components of an HCV particle. Among the other virus proteins, the roles of NS5A, p7, and NS2 in virus assembly have been relatively well studied.
Trafficking to and from LDs to the ER is an important part of lipid metabolism. Several host factor(s) are involved in functional association between LDs and the ER (Ohsaki et al. 2014; Salo et al. 2016). Lipid mobilization from cytoplasmic LDs favors the morphogenesis and secretion of HCV particles (Vieyres et al. 2016; Beilstein et al. 2017). On HCV infection, hepatocyte cell lines and primary human hepatocytes decrease production of lysophosphatidylcholine acyltransferase 1 (LPCAT1) transcript and protein; this results in altered lipid metabolism, characterized by LD remodeling, increased triacylglycerol storage, and increased secretion of VLDL. In infected cells, LPCAT1 depletion increases production of viral particles with the lowest density and highest infectivity (Beilstein et al. 2017). α/β hydrolase domain-containing protein 5 (ABHD5) associates with LDs, thereby triggering their hydrolysis. ABHD5 mobilizes lipids for HCV virus production (Vieyres et al. 2016). These data suggest that an active functional interaction between LDs and the ER facilitates production of both VLDL and HCV.
Virion assembly requires recruitment of HCV core to an appropriate site on the ER membrane. Adaptor-related protein complex 2 subunit mu 1 (AP2M1) traffics HCV core from LDs to the site of virus budding at the ER (Neveu et al. 2012). HCV NS2, in complex with other HCV proteins, acts as a scaffold that bridges structural and NS proteins during HCV assembly (Ma et al. 2008; Jirasko et al. 2010; Popescu et al. 2011; Stapleford and Lindenbach 2011). NS2 is also required for dynamic HCV core movement and for functional association with NS5A (Coller et al. 2012).
The p7-NS2-E2 complex functions at an early stage of virion morphogenesis, before envelopment of infectious virus (Jones et al. 2007). In vitro studies show that the ion channel activity of p7 and the protease domain of NS2 (but not its catalytic active site) within the complexes are required for the function (Jones et al. 2007). Over time, NS2 accumulates in ER-derived dotted structures and colocalizes with HCV E1, E2, and components of the replication complex in close proximity to the HCV core protein (Popescu et al. 2011). Genetic and coimmunoprecipitation data suggest that NS2 interacts with HCV E2 (Selby et al. 1994; Steinmann et al. 2007; Phan et al. 2009; Yi et al. 2009). In addition, NS4B associates with NS3 during HCV assembly (Jones et al. 2009; Yan et al. 2017); thus, both NS5A and NS2 play pivotal roles in virus assembly.
Interaction between NS5A and HCV core is crucial for productive infection (Masaki et al. 2008; Gawlik et al. 2014). A core mutant lacking interaction with NS5A abolishes production of infectious virus, despite maintaining synthesis of HCV RNA (Gawlik et al. 2014). In addition, NS5A associates with HCV RNA (Huang et al. 2005). The amino terminus of NS5A comprises an amphipathic α-helix embedded into the cytosolic leaflet of the ER membrane (Penin et al. 2004). This specific structure shows high affinity for lipid-rich components (Shi et al. 2002).
Biochemical analyses identified domain I, domain II, and domain III in NS5A (Tellinghuisen et al. 2004). Domain I has RNA-binding ability and is essential for RNA replication (Huang et al. 2005). A large portion of domain II and all of domain III are not essential for RNA replication (Tellinghuisen et al. 2008b). However, domain III is necessary for virus assembly (Appel et al. 2008; Masaki et al. 2008; Tellinghuisen et al. 2008a). Domain III is rich in serine and threonine and is a target for phosphorylation (Tanji et al. 1995). Phosphorylation of domain III affects the interaction with LD and core, as well as the association with NS2 because the phosphorylated form is more stable (Hughes et al. 2009; Popescu et al. 2011).
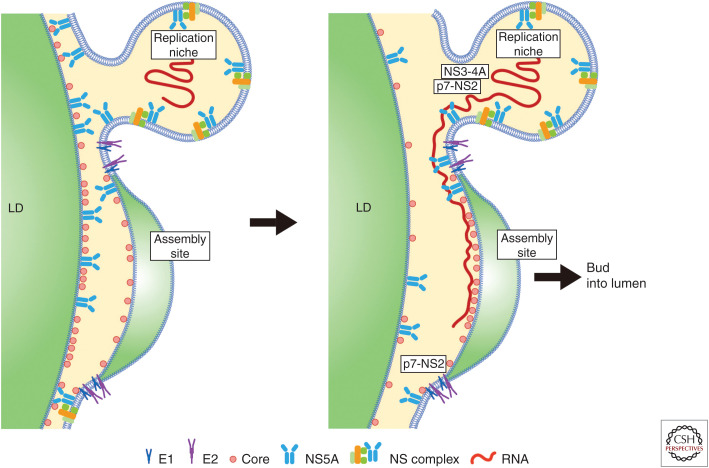
Among the many host factors involved in virus assembly, annexin A2 (ANXA2), present in the membrane fraction of HCV replication complexes (Backes et al. 2010), colocalizes with NS5A and plays a role in HCV assembly. TIP47 interacts with NS5A and recruits NS5A-associated viral RNA to the site of HCV assembly site on the surface of LDs (Ploen et al. 2013; Vogt et al. 2013). Heterogeneous nuclear ribonucleoprotein K (HNRNPK) interacts specifically with HCV RNA. This protein is recruited to sites in close proximity to LDs and colocalizes with HCV core protein and HCV plus-strand RNA (Poenisch et al. 2015). A concise model of early HCV assembly before encapsidation virus capsid formation is shown in Figure 1.
Figure 1.
Model of early hepatitis C virus (HCV) assembly before encapsidation. (Left) Association of HCV core protein with lipid droplets (LDs) brings the HCV replication niche and the assembly platform into close proximity. (From data in Miyanari et al. 2007, with permission, from the authors.) (Right) RNA synthesis occurs in the niche in which nonstructural (NS) proteins NS3–NS5B play a role. HCV RNA is transferred from the RNA replication niche to the site of assembly at the ER membrane. NS5A and NS3-4A are involved in transferring RNA from the niche to the site of assembly. p7-NS2 plays a bridging function to associate structural and NS proteins at the assembly site (see main text for details; also see Bartenschlager et al. 2011; Lindenbach and Rice 2013; Aizawa et al. 2015; Grassi et al. 2016). LDs may not be required once coordination between the replication niche and assembly platforms is established (Galli et al. 2013; Etienne et al. 2015).
From Envelopment of Particles to Egress into ER Lumen
Mainly, in vitro studies of the HCV life cycle are performed using Huh7 cells or its derivative cell lines infected with HCVcc. The mechanism of virus assembly described here is mainly based on experiments conducted using these systems. Several host factors participate in budding of viruses into the ER lumen. These factors are often involved in VLDL synthesis. Taking advantage of the fact that most HCV proteins associate with cell membranes (Hijikata et al. 1993a; Moradpour et al. 2004), HCV proteins complexes were coimmunoprecipitated together with ER membrane fractions using tagged NS5A as a probe. The obtained vesicles, which are rich in proteins required for VLDL assembly (i.e., apoB, apoE, and microsomal triglyceride transfer protein [MTP]), are used to analyze the effect of MTP and apoB on HCV secretion (Huang et al. 2007). Blocking VLDL assembly using an inhibitor of MTP and an siRNA specific for apoB (Huang et al. 2007) inhibits HCV production. Analysis of HCV production by Huh7 or its derivatives reveal functional association between HCV and the VLDL secretion machinery (Gastaminza et al. 2008). Furthermore, intracellular (premature) HCV that is poorly lipidated during assembly is degraded quickly (Gastaminza et al. 2008). Also, other studies reported a possible functional association between HCV secretion and VLDL (Nahmias et al. 2008; Yao and Ye 2008).
However, another study indicates that production of HCV with high infectivity is more dependent on apoE than on apoB (Chang et al. 2007). The density of HCVcc produced in vitro varies from 1.03 to 1.23 g/mL (Chang et al. 2007). Particles in the low-density fractions are more infectious than those in higher density fractions. Strikingly, the fractions containing most infectious virus contains the highest amounts of apoE and no correlation between the amount of apoB and specific infectivity of HCV. Knockdown of apoE using siRNA suppresses formation of HCV particles. In contrast, apoB-specific antibodies and siRNAs had no significant effect on HCV infectivity and production, respectively (Jiang and Luo 2009). Thus, it seems that apoB does not play an essential role in the HCV life cycle (Jiang and Luo 2009). Furthermore, inhibitors of MTP more severely affect secretion of HCV than that of VLDL (Jiang and Luo 2009). This suggests that HCV assembly does not rely completely on the mechanism underlying VLDL secretion in cells; rather, assembly occurs via some modification of the machinery.
Alternatively, HCV assembly may occur via mechanisms different from those underlying VLDL secretion. Imaging analysis of HCV core trafficking in live cells revealed colocalization of HCV core with apoE, but not with apoB (Coller et al. 2012). HCV-mediated modification of VLDL secretion has been reported (Perlemuter et al. 2002). HCV core inhibits MTP activity (Perlemuter et al. 2002; Mancone et al. 2012). NS5A dysregulates VLDL secretion by inducing ferritin heavy chain, which inhibits production of apoB (Mancone et al. 2012). It is possible that HCV suppresses VLDL secretion such that HCV can use excess lipid to generate lipid-rich viruses. Thus, HCV assembly may not be fully dependent on the VLDL secretion system. Instead, HCV may modify the system for its own benefit.
Huh7 cells produce poorly lipidated VLDL; thus, they produce dense apoB-containing particles (Icard et al. 2009; Meex et al. 2011). It is possible that the mechanisms underlying involvement of lipoproteins during HCV assembly in this cell line (or its derivatives) may only reflect one side of the virus assembly process, which may not be sufficient in in vivo systems. Investigating HCV assembly in hepatocytes that have normal lipoprotein assembly systems will reveal the precise role of lipoprotein synthesis in HCV assembly.
Production of infectious HCVs independent of VLDL secretion is observed in nonhepatic cells. Exogenous expression of defined host factors in human nonhepatic HeLa and 293 cells reconstitutes the entire HCV life cycle, thereby allowing robust HCV entry and RNA replication but not production of infectious virus (Da Costa et al. 2012; Hueging et al. 2014). However, ectopic expression of apoE, followed by infection with HCV, allows production of infectious viruses (Da Costa et al. 2012). Because these cell lines do not produce VLDL, infectious virus production does not require apoB- and MTP-related VLDL production.
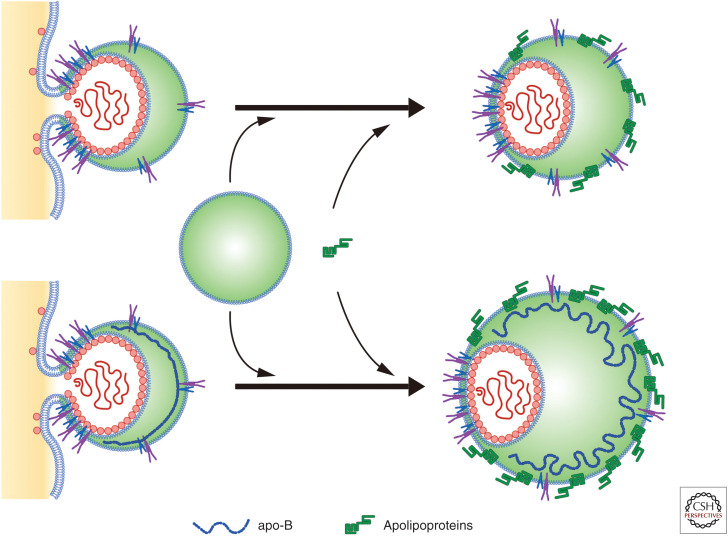
Studies have used siRNA to suppress apoB (Huang et al. 2007; Jiang and Luo 2009); however, it is unclear whether apoB is dispensable or required (even in small amounts) for HCV biogenesis. Knockout of apoB or apoE in Huh7 cells reduced HCV production to <1/10 of control levels, respectively. However, double knockout of apoB and apoE inhibited secretion of infectious viruses by 100-fold. Ectopic expression of apoE to apoB/apoE double knockout cells restored production of infectious virus. Ectopic expression of apoE or MTP in apoB/MTP double knockout cells restored production of infectious virus to high or moderate levels, respectively (Fukuhara et al. 2014). In sum, these studies have led to the proposal of apoB-dependent and -independent virion assembly pathways (Fig. 2).
Figure 2.
Envelopment to egress into the endoplasmic reticulum (ER) lumen. At least two mechanisms may be involved in hepatitis C virus (HCV) secretion into the lumen: apolipoprotein B (apoB)-dependent and -independent. One model shows the production of a fused form of HCV with very low-density lipoproteins (VLDLs). Another model is budding of HCV without apoB. All nascent virus particles may be further lipidated by luminal lipoproteins and may incorporate exchangeable apolipoproteins. E1/E2 dimers may distribute both in the ER membrane–derived phospholipid bilayers and in the phospholipid monolayer membrane of lipid rich HCV (Icard et al. 2009). (Based on data in Bartenschlager et al. 2011.)
Importantly, ectopic expression of exchangeable apolipoproteins, such as apoA1, A2, C1, C2, and C3, in apoB/apoE double knockout cells restored production of infectious viruses to the same level as that observed after expression of apoE. This effect was observed by expressing the peptides of amphipathic α-helices containing the amino-terminal domain of apoE (Fukuhara et al. 2014; Li et al. 2017). This suggests that production of infectious virus particles is facilitated by factors involved in lipoprotein synthesis and secretion. It may be that production of infectious virus is regulated redundantly by exchangeable apolipoproteins expressed in the liver. Generation of apoB-independent large lipoproteins in the ER lumen occurs under conditions of ER stress (Sołtysik et al. 2019). Similarly, HCV may induce ER stress to generate large lipoproteins that facilitate its own assembly.
ApoE and other exchangeable apolipoproteins participate in secondary lipidation of VLDL precursors by fusing with luminal LDs (Sundaram and Yao 2012). ApoE may associate with HCV either during or after the envelopment of viruses by interacting with HCV E2 (Boyer et al. 2014; Hueging et al. 2014; Lee et al. 2014). Association of apoE with HCV E2 is detected in the ER (Boyer et al. 2014), suggesting association of apoE with HCV at the early stage of assembly. In this scenario, copresentation of E1/E2 and apoE on HCV particles during virion envelopment seems to occur. In contrast, apoE may be incorporated at a late stage of assembly, such as particle maturation, because apoE depletion does not affect formation of the nucleocapsid or virus envelopment (Lee et al. 2014). There may be no strict spatiotemporal limitation with respect to apoE interaction with HCV. As described below, association with apoE also occurs extracellularly. The effects of apoE on virus assembly and specific infectivity vary, dependent on the HCVcc isolate (Weller et al. 2017).
Secretion of HCV from the ER Lumen
HCV release depends on components of the endosomal sorting complex required for transport (Corless et al. 2010; Ariumi et al. 2011). Trans-Golgi network (TGN)-associating adaptors play a pivotal role during HCV assembly and release (Coller et al. 2012; Mankouri et al. 2016). HCV RNA is enriched in highly buoyant COPII vesicle fractions, and is cofractionated with apoB, apoE, and the HCV core and E1/E2 (Syed et al. 2017). Ultrastructural analysis reveals the presence of HCV structural proteins, RNA, and apolipoproteins in the Golgi stacks, supporting the hypothesis that HCV assembles with apolipoproteins in the ER and is then transported to the Golgi compartment in COPII vesicles, where it enters the Golgi secretory route (Syed et al. 2017). VLDL secretion is unaffected by silencing Rabs and TGN-endosome adaptors that block HCV egress, whereas inhibiting apoE secretion using monensin, a polyether antibiotic, does not impair HCV release suggesting they use distinct secretion pathways (Mankouri et al. 2016). ABHD5 colocalizes with apoE and may play a role in HCV secretion (Vieyres et al. 2016). Mature HCV is sorted into secretory vesicles (e.g., multivesicular bodies) and recycling endosomes before transport to the plasma membrane. Early-endosome proteins such as Rab5 are necessary for HCV genome replication, suggesting involvement of the early-endosome at multiple stages of HCV proliferation (Stone et al. 2007; Manna et al. 2010). Taken together, the results suggest that release of infectious HCV occurs via a TGN-endosomal secretion pathway that is distinct from that of VLDL.
AFTER EGRESS
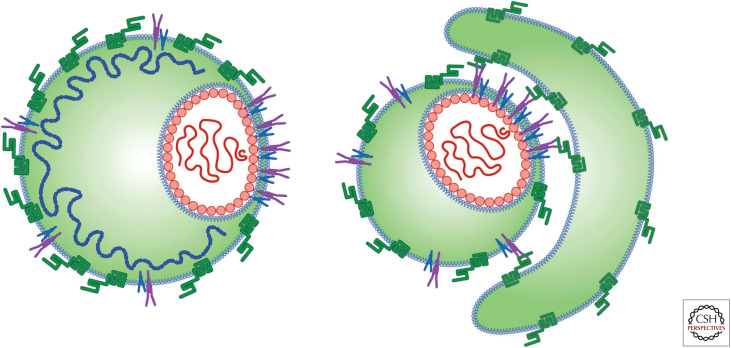
The average buoyant density of HCV generated in chimpanzees or mice harboring human liver grafts infected with HCV in vitro cultured from Huh7-derived cells is lower than that of cultured HCV (Lindenbach et al. 2006). Assuming that Huh7 cells produce high-density HCV caused by aberrant VLDL secretion mechanisms (Icard et al. 2009; Meex et al. 2011), we would expect conversion to low-density viruses as the animals are likely to have normal VLDL production in liver tissues. Also, it is possible that HCV gains lipids from serum lipoproteins (Fig. 3).
Figure 3.
Different architectures of lipid-rich hepatitis C virus (HCV) particles. Fusion of HCV with serum lipoprotein may result in lipidated HCVs with different architectures. The association of serum lipoprotein may not be required for fusion to HCV particles; an interaction not involving fusion may be sufficient for efficient HCV infection. However, the role played by different particles in the virus life cycle still remains unknown.
The blood of HCV-infected patients contains HCV particles harboring apoB100 or apoB48. This indicates that a significant fraction of plasma HCV is associated with apoB48-containing lipoproteins (Diaz et al. 2006). ApoB48 is a truncated form of apoB100 and is major protein in lipoprotein particles secreted by intestinal enterocytes. It is unlikely that intestinal enterocytes produce HCV particles; rather, HCV is likely to associate with apoB48-containing lipoproteins when circulating in blood. Interaction with serum apoB48-containing lipoproteins is likely to be both transient and reversible (Diaz et al. 2006; Felmlee et al. 2010), but may facilitate HCV infectivity.
Accumulating evidence suggests the importance of extracellular association between apoE and HCV. This interaction enhances specific infectivity and may aid the virus in evading neutralizing antibodies (Yang et al. 2016; Bankwitz et al. 2017; Li et al. 2017). Addition of secreted apoE-associated lipoprotein increases the infectivity of HCV produced by apoE-deficient cells. Exchange of apoE between HCV and lipoproteins maintains adequate apoE levels on HCV, facilitating efficient attachment to cell surfaces (Yang et al. 2016). Secreted HA-tagged apoE produced by plasmid-transduced 293T cells bind to HCV produced by primary human hepatocytes (Li et al. 2017). Soluble apoE genetic isoforms affected HCV infectivity on association with HCV (Li et al. 2017), as shown by a study examining the effect of apoE genetic variants on production of HCV by infected cells (Hishiki et al. 2010). Serum-derived HCV particles harbor higher amounts of apoE than cell culture–derived HCV particles (Bankwitz et al. 2017), suggesting that permissiveness of cells for HCV infection is determined mainly by exogenous apoE-associated lipoproteins, which are enriched in HCV-infected patients. Infectivity of HCV particles from primary human hepatocytes is enhanced by addition of exogenous secreted apoE.
ApoE is more abundant than VLDL in the HCV fraction from HCV patients (Nielsen et al. 2006). What is the reason for this? Assuming that HCV E2 has high affinity for apoE (Boyer et al. 2014; Lee et al. 2014), interaction between the E2 and apo-E in blood vessels may lead to accumulation of apoE, rather than VLDL, on HCV.
CONCLUDING REMARKS
Development of in vitro HCV culture systems has progressed tremendously and has revealed many aspects of the virus life cycle (Lohmann et al. 1999; Lindenbach et al. 2005; Wakita et al. 2005; Zhong et al. 2005). However, a consensus in understanding the HCV life cycle is still needed to facilitate future work, because current understanding relies heavily on a limited number of HCV variants and cell lines, and the results obtained in current studies sometimes yield conflicting results. Difficulties in HCV particle purification also hinder the study of the precise mechanisms of virus proliferation.
Considering the rapid progress that has been made in HCV replication systems over the last decade, an understanding of the precise mechanisms of virus replication of serum-derived HCV in primary hepatocytes may soon be within our grasp.
ACKNOWLEDGMENTS
This work was supported by the Research Program on Hepatitis from the Japan Agency for Medical Research and Development (AMED).
Footnotes
Editors: Arash Grakoui, Jean-Michel Pawlotsky, and Glenn Randall
Additional Perspectives on Hepatitis C Viruses: The Story of a Scientific and Therapeutic Revolution available at www.perspectivesinmedicine.org
REFERENCES
- Aizawa Y, Seki N, Nagano T, Abe H. 2015. Chronic hepatitis C virus infection and lipoprotein metabolism. World J Gastroenterol 21: 10299–10313. 10.3748/wjg.v21.i36.10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- André P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Brechot C, Paranhos-Baccala G, Lotteau V. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol 76: 6919–6928. 10.1128/JVI.76.14.6919-6928.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- André P, Perlemuter G, Budkowska A, Bréchot C, Lotteau V. 2005. Hepatitis C virus particles and lipoprotein metabolism. Semin Liver Dis 25: 93–104. 10.1055/s-2005-864785 [DOI] [PubMed] [Google Scholar]
- Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog 4: e1000035 10.1371/journal.ppat.1000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y, Kuroki M, Maki M, Ikeda M, Dansako H, Wakita T, Kato N. 2011. The ESCRT system is required for hepatitis C virus production. PLoS ONE 6: e14517 10.1371/journal.pone.0014517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backes P, Quinkert D, Reiss S, Binder M, Zayas M, Rescher U, Gerke V, Bartenschlager R, Lohmann V. 2010. Role of annexin A2 in the production of infectious hepatitis C virus particles. J Virol 84: 5775–5789. 10.1128/JVI.02343-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankwitz D, Doepke M, Hueging K, Weller R, Bruening J, Behrendt P, Lee JY, Vondran FWR, Manns MP, Bartenschlager R, et al. 2017. Maturation of secreted HCV particles by incorporation of secreted ApoE protects from antibodies by enhancing infectivity. J Hepatol 67: 480–489. 10.1016/j.jhep.2017.04.010 [DOI] [PubMed] [Google Scholar]
- Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman MJ, Miyamura T, et al. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci 94: 1200–1205. 10.1073/pnas.94.4.1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R, Penin F, Lohmann V, André P. 2011. Assembly of infectious hepatitis C virus particles. Trends Microbiol 19: 95–103. 10.1016/j.tim.2010.11.005 [DOI] [PubMed] [Google Scholar]
- Bartosch B, Dubuisson J, Cosset FL. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1–E2 envelope protein complexes. J Exp Med 197: 633–642. 10.1084/jem.20021756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilstein F, Lemasson M, Pène V, Rainteau D, Demignot S, Rosenberg AR. 2017. Lysophosphatidylcholine acyltransferase 1 is downregulated by hepatitis C virus: impact on production of lipo-viro-particles. Gut 66: 2160–2169. 10.1136/gutjnl-2016-311508 [DOI] [PubMed] [Google Scholar]
- Blackham S, Baillie A, Al-Hababi F, Remlinger K, You S, Hamatake R, McGarvey MJ. 2010. Gene expression profiling indicates the roles of host oxidative stress, apoptosis, lipid metabolism, and intracellular transport genes in the replication of hepatitis C virus. J Virol 84: 5404–5414. 10.1128/JVI.02529-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose SK, Kim H, Meyer K, Wolins N, Davidson NO, Ray R. 2014. Forkhead box transcription factor regulation and lipid accumulation by hepatitis C virus. J Virol 88: 4195–4203. 10.1128/JVI.03327-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant S, Vanbelle C, Ebel C, Penin F, Lavergne JP. 2005. Hepatitis C virus core protein is a dimeric α-helical protein exhibiting membrane protein features. J Virol 79: 11353–11365. 10.1128/JVI.79.17.11353-11365.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant S, Montserret R, Hope RG, Ratinier M, Targett-Adams P, Lavergne JP, Penin F, McLauchlan J. 2006. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J Biol Chem 281: 22236–22247. 10.1074/jbc.M601031200 [DOI] [PubMed] [Google Scholar]
- Boulant S, Targett-Adams P, McLauchlan J. 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J Gen Virol 88: 2204–2213. 10.1099/vir.0.82898-0 [DOI] [PubMed] [Google Scholar]
- Boyer A, Dumans A, Beaumont E, Etienne L, Roingeard P, Meunier JC. 2014. The association of hepatitis C virus glycoproteins with apolipoproteins E and B early in assembly is conserved in lipoviral particles. J Biol Chem 289: 18904–18913. 10.1074/jbc.M113.538256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D, McCaustland K, Krawczynski K, Spelbring J, Humphey C, Cook EH. 1991. Hepatitis C virus: buoyant density of the factor VIII-derived isolate in sucrose. J Med Virol 34: 206–208. 10.1002/jmv.1890340315 [DOI] [PubMed] [Google Scholar]
- Cai Z, Zhang C, Chang KS, Jiang J, Ahn BC, Wakita T, Liang TJ, Luo G. 2005. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J Virol 79: 13963–13973. 10.1128/JVI.79.22.13963-13973.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calattini S, Fusil F, Mancip J, Dao Thi VL, Granier C, Gadot N, Scoazec JY, Zeisel MB, Baumert TF, Lavillette D, et al. 2015. Functional and biochemical characterization of hepatitis C Virus (HCV) particles produced in a humanized liver mouse model. J Biol Chem 290: 23173–23187. 10.1074/jbc.M115.662999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus G, Herker E, Modi AA, Haas JT, Ramage HR, Farese RV Jr, Ott M. 2013. Diacylglycerol acyltransferase-1 localizes hepatitis C virus NS5A protein to lipid droplets and enhances NS5A interaction with the viral capsid core. J Biol Chem 288: 9915–9923. 10.1074/jbc.M112.434910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrick RJ, Schlauder GG, Peterson DA, Mushahwar IK. 1992. Examination of the buoyant density of hepatitis C virus by the polymerase chain reaction. J Virol Methods 39: 279–289. 10.1016/0166-0934(92)90101-I [DOI] [PubMed] [Google Scholar]
- Catanese MT, Uryu K, Kopp M, Edwards TJ, Andrus L, Rice WJ, Silvestry M, Kuhn RJ, Rice CM. 2013. Ultrastructural analysis of hepatitis C virus particles. Proc Natl Acad Sci 110: 9505–9510. 10.1073/pnas.1307527110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KS, Jiang J, Cai Z, Luo G. 2007. Human apolipoprotein E is required for infectivity and production of hepatitis C virus in cell culture. J Virol 81: 13783–13793. 10.1128/JVI.01091-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244: 359–362. 10.1126/science.2523562 [DOI] [PubMed] [Google Scholar]
- Coller KE, Heaton NS, Berger KL, Cooper JD, Saunders JL, Randall G. 2012. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog 8: e1002466 10.1371/journal.ppat.1002466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corless L, Crump CM, Griffin SD, Harris M. 2010. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J Gen Virol 91: 362–372. 10.1099/vir.0.017285-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa D, Turek M, Felmlee DJ, Girardi E, Pfeffer S, Long G, Bartenschlager R, Zeisel MB, Baumert TF. 2012. Reconstitution of the entire hepatitis C virus life cycle in nonhepatic cells. J Virol 86: 11919–11925. 10.1128/JVI.01066-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos R, Verslype C, Depla E, Fevery J, Van Damme B, Desmet V, Roskams T. 2002. Ultrastructural visualization of hepatitis C virus components in human and primate liver biopsies. J Hepatol 37: 370–379. 10.1016/S0168-8278(02)00236-2 [DOI] [PubMed] [Google Scholar]
- Dharancy S, Malapel M, Perlemuter G, Roskams T, Cheng Y, Dubuquoy L, Podevin P, Conti F, Canva V, Philippe D, et al. 2005. Impaired expression of the peroxisome proliferator-activated receptor α during hepatitis C virus infection. Gastroenterology 128: 334–342. 10.1053/j.gastro.2004.11.016 [DOI] [PubMed] [Google Scholar]
- Diaz O, Delers F, Maynard M, Demignot S, Zoulim F, Chambaz J, Trepo C, Lotteau V, André P. 2006. Preferential association of hepatitis C virus with apolipoprotein B48-containing lipoproteins. J Gen Virol 87: 2983–2991. 10.1099/vir.0.82033-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M, Boson B, Ricard-Blum S, Molle J, Lavillette D, Bartosch B, Pécheur EI, Cosset FL. 2007. The exchangeable apolipoprotein ApoC-I promotes membrane fusion of hepatitis C virus. J Biol Chem 282: 32357–32369. 10.1074/jbc.M705358200 [DOI] [PubMed] [Google Scholar]
- Dreux M, Gastaminza P, Wieland SF, Chisari FV. 2009. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci 106: 14046–14051. 10.1073/pnas.0907344106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol 76: 5974–5984. 10.1128/JVI.76.12.5974-5984.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne L, Blanchard E, Boyer A, Desvignes V, Gaillard J, Meunier JC, Roingeard P, Hourioux C. 2015. The replacement of 10 non-conserved residues in the core protein of JFH-1 hepatitis C virus improves its assembly and secretion. PLoS ONE 10: e0137182 10.1371/journal.pone.0137182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcón V, Acosta-Rivero N, Chinea G, Gavilondo J, de la Rosa MC, Menéndez I, Dueñas-Carrera S, Viña A, Garcı´a W, Gra B, et al. 2003. Ultrastructural evidences of HCV infection in hepatocytes of chronically HCV-infected patients. Biochem Biophys Res Commun 305: 1085–1090. 10.1016/S0006-291X(03)00884-2 [DOI] [PubMed] [Google Scholar]
- Felmlee DJ, Sheridan DA, Bridge SH, Nielsen SU, Milne RW, Packard CJ, Caslake MJ, McLauchlan J, Toms GL, Neely RD, et al. 2010. Intravascular transfer contributes to postprandial increase in numbers of very-low-density hepatitis C virus particles. Gastroenterology 139: 1774–1783.e6. 10.1053/j.gastro.2010.07.047 [DOI] [PubMed] [Google Scholar]
- Ferraris P, Blanchard E, Roingeard P. 2010. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J Gen Virol 91: 2230–2237. 10.1099/vir.0.022186-0 [DOI] [PubMed] [Google Scholar]
- Ferraris P, Beaumont E, Uzbekov R, Brand D, Gaillard J, Blanchard E, Roingeard P. 2013. Sequential biogenesis of host cell membrane rearrangements induced by hepatitis C virus infection. Cell Mol Life Sci 70: 1297–1306. 10.1007/s00018-012-1213-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara T, Wada M, Nakamura S, Ono C, Shiokawa M, Yamamoto S, Motomura T, Okamoto T, Okuzaki D, Yamamoto M, et al. 2014. Amphipathic α-helices in apolipoproteins are crucial to the formation of infectious hepatitis C virus particles. PLoS Pathog 10: e1004534 10.1371/journal.ppat.1004534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli A, Scheel TK, Prentoe JC, Mikkelsen LS, Gottwein JM, Bukh J. 2013. Analysis of hepatitis C virus core/NS5A protein co-localization using novel cell culture systems expressing core-NS2 and NS5A of genotypes 1-7. J Gen Virol 94: 2221–2235. 10.1099/vir.0.053868-0 [DOI] [PubMed] [Google Scholar]
- Gastaminza P, Kapadia SB, Chisari FV. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J Virol 80: 11074–11081. 10.1128/JVI.01150-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. 2008. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol 82: 2120–2129. 10.1128/JVI.02053-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawlik K, Baugh J, Chatterji U, Lim PJ, Bobardt MD, Gallay PA. 2014. HCV core residues critical for infectivity are also involved in core-NS5A complex formation. PLoS ONE 9: e88866 10.1371/journal.pone.0088866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol 77: 5487–5492. 10.1128/JVI.77.9.5487-5492.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassi G, Di Caprio G, Fimia GM, Ippolito G, Tripodi M, Alonzi T. 2016. Hepatitis C virus relies on lipoproteins for its life cycle. World J Gastroenterol 22: 1953–1965. 10.3748/wjg.v22.i6.1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV Jr, Ott M. 2010. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med 16: 1295–1298. 10.1038/nm.2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci 88: 5547–5551. 10.1073/pnas.88.13.5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M, Mizushima H, Tanji Y, Komoda Y, Hirowatari Y, Akagi T, Kato N, Kimura K, Shimotohno K. 1993a. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc Natl Acad Sci 90: 10773–10777. 10.1073/pnas.90.22.10773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M, Shimizu YK, Kato H, Iwamoto A, Shih JW, Alter HJ, Purcell RH, Yoshikura H. 1993b. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J Virol 67: 1953–1958. 10.1128/JVI.67.4.1953-1958.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishiki T, Shimizu Y, Tobita R, Sugiyama K, Ogawa K, Funami K, Ohsaki Y, Fujimoto T, Takaku H, Wakita T, et al. 2010. Infectivity of hepatitis C virus is influenced by association with apolipoprotein E isoforms. J Virol 84: 12048–12057. 10.1128/JVI.01063-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope RG, McLauchlan J. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J Gen Virol 81: 1913–1925. 10.1099/0022-1317-81-8-1913 [DOI] [PubMed] [Google Scholar]
- Horner SM. 2015. Insights into antiviral innate immunity revealed by studying hepatitis C virus. Cytokine 74: 190–197. 10.1016/j.cyto.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Hwang J, Sharma SD, Hargittai MR, Chen Y, Arnold JJ, Raney KD, Cameron CE. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J Biol Chem 280: 36417–36428. 10.1074/jbc.M508175200 [DOI] [PubMed] [Google Scholar]
- Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M Jr, Ye J. 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci 104: 5848–5853. 10.1073/pnas.0700760104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueging K, Doepke M, Vieyres G, Bankwitz D, Frentzen A, Doerrbecker J, Gumz F, Haid S, Wolk B, Kaderali L, et al. 2014. Apolipoprotein E codetermines tissue tropism of hepatitis C virus and is crucial for viral cell-to-cell transmission by contributing to a postenvelopment step of assembly. J Virol 88: 1433–1446. 10.1128/JVI.01815-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M, Griffin S, Harris M. 2009. Domain III of NS5A contributes to both RNA replication and assembly of hepatitis C virus particles. J Gen Virol 90: 1329–1334. 10.1099/vir.0.009332-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icard V, Diaz O, Scholtes C, Perrin-Cocon L, Ramière C, Bartenschlager R, Penin F, Lotteau V, André P. 2009. Secretion of hepatitis C virus envelope glycoproteins depends on assembly of apolipoprotein B positive lipoproteins. PLoS ONE 4: e4233 10.1371/journal.pone.0004233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R, Kanevsky I, Gabus C, Lavergne J, Ficheux D, Penin F, Fossé P, Darlix J. 2006. Analysis of hepatitis C virus RNA dimerization and core-RNA interactions. Nucleic Acids Res 34: 2618–2633. 10.1093/nar/gkl240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Luo G. 2009. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J Virol 83: 12680–12691. 10.1128/JVI.01476-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirasko V, Montserret R, Lee JY, Gouttenoire J, Moradpour D, Penin F, Bartenschlager R. 2010. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Pathog 6: e1001233 10.1371/journal.ppat.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol 81: 8374–8383. 10.1128/JVI.00690-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DM, Patel AH, Targett-Adams P, McLauchlan J. 2009. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J Virol 83: 2163–2177. 10.1128/JVI.01885-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Date T, Yokokawa H, Kono T, Aizaki H, Maurel P, Gondeau C, Wakita T. 2014. Development of hepatitis C virus genotype 3a cell culture system. Hepatology 60: 1838–1850. 10.1002/hep.27197 [DOI] [PubMed] [Google Scholar]
- Lee JY, Acosta EG, Stoeck IK, Long G, Hiet MS, Mueller B, Fackler OT, Kallis S, Bartenschlager R. 2014. Apolipoprotein E likely contributes to a maturation step of infectious hepatitis C virus particles and interacts with viral envelope glycoproteins. J Virol 88: 12422–12437. 10.1128/JVI.01660-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Li Y, Bi Y, Zhang H, Yao Y, Li Q, Cun W, Dong S. 2017. Extracellular interactions between hepatitis C virus and secreted apolipoprotein E. J Virol 91: e02227-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. 2013. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol 11: 688–700. 10.1038/nrmicro3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309: 623–626. 10.1126/science.1114016 [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, Lanford RE, Feinstone SM, Major ME, Leroux-Roels G, et al. 2006. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci 103: 3805–3809. 10.1073/pnas.0511218103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285: 110–113. 10.1126/science.285.5424.110 [DOI] [PubMed] [Google Scholar]
- Lohmann V, Korner F, Dobierzewska A, Bartenschlager R. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J Virol 75: 1437–1449. 10.1128/JVI.75.3.1437-1449.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Yates J, Liang Y, Lemon SM, Yi M. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J Virol 82: 7624–7639. 10.1128/JVI.00724-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancone C, Montaldo C, Santangelo L, Di Giacomo C, Costa V, Amicone L, Ippolito G, Pucillo LP, Alonzi T, Tripodi M. 2012. Ferritin heavy chain is the host factor responsible for HCV-induced inhibition of apoB-100 production and is required for efficient viral infection. J Proteome Res 11: 2786–2797. 10.1021/pr201128s [DOI] [PubMed] [Google Scholar]
- Mankouri J, Tedbury PR, Gretton S, Hughes ME, Griffin SD, Dallas ML, Green KA, Hardie DG, Peers C, Harris M. 2010. Enhanced hepatitis C virus genome replication and lipid accumulation mediated by inhibition of AMP-activated protein kinase. Proc Natl Acad Sci 107: 11549–11554. 10.1073/pnas.0912426107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri J, Walter C, Stewart H, Bentham M, Park WS, Heo WD, Fukuda M, Griffin S, Harris M. 2016. Release of infectious hepatitis C virus from Huh7 cells occurs via a trans-Golgi network-to-endosome pathway independent of very-low-density lipoprotein secretion. J Virol 90: 7159–7170. 10.1128/JVI.00826-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna D, Aligo J, Xu C, Park WS, Koc H, Do Heo W, Konan KV. 2010. Endocytic Rab proteins are required for hepatitis C virus replication complex formation. Virology 398: 21–37. 10.1016/j.virol.2009.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, Date T, Matsuura Y, Miyamura T, Wakita T, et al. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J Virol 82: 7964–7976. 10.1128/JVI.00826-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J, Lemberg MK, Hope G, Martoglio B. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J 21: 3980–3988. 10.1093/emboj/cdf414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meex SJ, Andreo U, Sparks JD, Fisher EA. 2011. Huh-7 or HepG2 cells: which is the better model for studying human apolipoprotein-B100 assembly and secretion? J Lipid Res 52: 152–158. 10.1194/jlr.D008888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel N, Fischl W, Hueging K, Bankwitz D, Frentzen A, Haid S, Gentzsch J, Kaderali L, Bartenschlager R, Pietschmann T. 2012. MAP-kinase regulated cytosolic phospholipase A2 activity is essential for production of infectious hepatitis C virus particles. PLoS Pathog 8: e1002829 10.1371/journal.ppat.1002829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz A, Long G, Hiet MS, Brügger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. 2011. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem 286: 3018–3032. 10.1074/jbc.M110.175018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier JC, Russell RS, Engle RE, Faulk KN, Purcell RH, Emerson SU. 2008. Apolipoprotein c1 association with hepatitis C virus. J Virol 82: 9647–9656. 10.1128/JVI.00914-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9: 1089–1097. 10.1038/ncb1631 [DOI] [PubMed] [Google Scholar]
- Moradpour D, Englert C, Wakita T, Wands JR. 1996. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology 222: 51–63. 10.1006/viro.1996.0397 [DOI] [PubMed] [Google Scholar]
- Moradpour D, Brass V, Bieck E, Friebe P, Gosert R, Blum HE, Bartenschlager R, Penin F, Lohmann V. 2004. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J Virol 78: 13278–13284. 10.1128/JVI.78.23.13278-13284.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahmias Y, Goldwasser J, Casali M, van Poll D, Wakita T, Chung RT, Yarmush ML. 2008. Apolipoprotein B-dependent hepatitis C virus secretion is inhibited by the grapefruit flavonoid naringenin. Hepatology 47: 1437–1445. 10.1002/hep.22197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeldt CJ, Joyce MA, Levin A, Steenbergen RH, Pang D, Shields J, Tyrrell DL, Wozniak RW. 2013. Hepatitis C virus-induced cytoplasmic organelles use the nuclear transport machinery to establish an environment conducive to virus replication. PLoS Pathog 9: e1003744 10.1371/journal.ppat.1003744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeldt CJ, Joyce MA, Van Buuren N, Levin A, Kirkegaard K, Gale M Jr, Tyrrell DL, Wozniak RW. 2016. The hepatitis C virus-induced membranous web and associated nuclear transport machinery limit access of pattern recognition receptors to viral replication sites. PLoS Pathog 12: e1005428 10.1371/journal.ppat.1005428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu G, Barouch-Bentov R, Ziv-Av A, Gerber D, Jacob Y, Einav S. 2012. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLoS Pathog 8: e1002845 10.1371/journal.ppat.1002845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SU, Bassendine MF, Burt AD, Martin C, Pumeechockchai W, Toms GL. 2006. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J Virol 80: 2418–2428. 10.1128/JVI.80.5.2418-2428.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SU, Bassendine MF, Martin C, Lowther D, Purcell PJ, King BJ, Neely D, Toms GL. 2008. Characterization of hepatitis C RNA-containing particles from human liver by density and size. J Gen Virol 89: 2507–2517. 10.1099/vir.0.2008/000083-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y, Suzuki M, Fujimoto T. 2014. Open questions in lipid droplet biology. Chem Biol 21: 86–96. 10.1016/j.chembiol.2013.08.009 [DOI] [PubMed] [Google Scholar]
- Parent R, Qu X, Petit MA, Beretta L. 2009. The heat shock cognate protein 70 is associated with hepatitis C virus particles and modulates virus infectivity. Hepatology 49: 1798–1809. 10.1002/hep.22852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. 2013. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J Virol 87: 10612–10627. 10.1128/JVI.01370-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penin F, Brass V, Appel N, Ramboarina S, Montserret R, Ficheux D, Blum HE, Bartenschlager R, Moradpour D. 2004. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J Biol Chem 279: 40835–40843. 10.1074/jbc.M404761200 [DOI] [PubMed] [Google Scholar]
- Perlemuter G, Sabile A, Letteron P, Vona G, Topilco A, Chrétien Y, Koike K, Pessayre D, Chapman J, Barba G, et al. 2002. Hepatitis C virus core protein inhibits microsomal triglyceride transfer protein activity and very low density lipoprotein secretion: a model of viral-related steatosis. FASEB J 16: 185–194. 10.1096/fj.01-0396com [DOI] [PubMed] [Google Scholar]
- Pfeifer U, Thomssen R, Legler K, Böttcher U, Gerlich W, Weinmann E, Klinge O. 1980. Experimental non-A, non-B hepatitis: four types of cytoplasmic alteration in hepatocytes of infected chimpanzees. Virchows Arch B Cell Pathol Incl Mol Pathol 33: 233–243. 10.1007/BF02899184 [DOI] [PubMed] [Google Scholar]
- Phan T, Beran RK, Peters C, Lorenz IC, Lindenbach BD. 2009. Hepatitis C virus NS2 protein contributes to virus particle assembly via opposing epistatic interactions with the E1–E2 glycoprotein and NS3–NS4A enzyme complexes. J Virol 83: 8379–8395. 10.1128/JVI.00891-09 [DOI] [Google Scholar]
- Ploen D, Hafirassou ML, Himmelsbach K, Sauter D, Biniossek ML, Weiss TS, Baumert TF, Schuster C, Hildt E. 2013. TIP47 plays a crucial role in the life cycle of hepatitis C virus. J Hepatol 58: 1081–1088. 10.1016/j.jhep.2013.01.022 [DOI] [PubMed] [Google Scholar]
- Poenisch M, Metz P, Blankenburg H, Ruggieri A, Lee JY, Rupp D, Rebhan I, Diederich K, Kaderali L, Domingues FS, et al. 2015. Identification of HNRNPK as regulator of hepatitis C virus particle production. PLoS Pathog 11: e1004573 10.1371/journal.ppat.1004573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu CI, Callens N, Trinel D, Roingeard P, Moradpour D, Descamps V, Duverlie G, Penin F, Héliot L, Rouillé Y, et al. 2011. NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PLoS Pathog 7: e1001278 10.1371/journal.ppat.1001278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince AM, Huima-Byron T, Parker TS, Levine DM. 1996. Visualization of hepatitis C virions and putative defective interfering particles isolated from low-density lipoproteins. J Viral Hepat 3: 11–17. 10.1111/j.1365-2893.1996.tb00075.x [DOI] [PubMed] [Google Scholar]
- Pumeechockchai W, Bevitt D, Agarwal K, Petropoulou T, Langer BC, Belohradsky B, Bassendine MF, Toms GL. 2002. Hepatitis C virus particles of different density in the blood of chronically infected immunocompetent and immunodeficient patients: implications for virus clearance by antibody. J Med Virol 68: 335–342. 10.1002/jmv.10208 [DOI] [PubMed] [Google Scholar]
- Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, et al. 2012. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog 8: e1003056 10.1371/journal.ppat.1003056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo VT, Belevich I, Li S, Karhinen L, Vihinen H, Vigouroux C, Magré J, Thiele C, Hölttä-Vuori M, Jokitalo E, et al. 2016. Seipin regulates ER–lipid droplet contacts and cargo delivery. EMBO J 35: 2699–2716. 10.15252/embj.201695170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby MJ, Glazer E, Masiarz F, Houghton M. 1994. Complex processing and protein:protein interactions in the E2:NS2 region of HCV. Virology 204: 114–122. 10.1006/viro.1994.1515 [DOI] [PubMed] [Google Scholar]
- Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J Biol Chem 282: 37158–37169. 10.1074/jbc.M707329200 [DOI] [PubMed] [Google Scholar]
- Shi ST, Polyak SJ, Tu H, Taylor DR, Gretch DR, Lai MM. 2002. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology 292: 198–210. 10.1006/viro.2001.1225 [DOI] [PubMed] [Google Scholar]
- Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J Virol 77: 4160–4168. 10.1128/JVI.77.7.4160-4168.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sołtysik K, Ohsaki Y, Tatematsu T, Cheng J, Fujimoto T. 2019. Nuclear lipid droplets derive from a lipoprotein precursor and regulate phosphatidylcholine synthesis. Nat Commun 10: 473 10.1038/s41467-019-08411-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleford KA, Lindenbach BD. 2011. Hepatitis C virus NS2 coordinates virus particle assembly through physical interactions with the E1–E2 glycoprotein and NS3–NS4A enzyme complexes. J Virol 85: 1706–1717. 10.1128/JVI.02268-10 [DOI] [Google Scholar]
- Steinmann E, Penin F, Kallis S, Patel AH, Bartenschlager R, Pietschmann T. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog 3: e103 10.1371/journal.ppat.0030103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone M, Jia S, Heo WD, Meyer T, Konan KV. 2007. Participation of rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J Virol 81: 4551–4563. 10.1128/JVI.01366-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HY, Lin CC, Lee JC, Wang SW, Cheng PN, Wu IC, Chang TT, Lai MD, Shieh DB, Young KC. 2013. Very low-density lipoprotein/lipo-viro particles reverse lipoprotein lipase-mediated inhibition of hepatitis C virus infection via apolipoprotein C-III. Gut 62: 1193–1203. 10.1136/gutjnl-2011-301798 [DOI] [PubMed] [Google Scholar]
- Sundaram M, Yao Z. 2012. Intrahepatic role of exchangeable apolipoproteins in lipoprotein assembly and secretion. Arterioscler Thromb Vasc Biol 32: 1073–1078. 10.1161/ATVBAHA.111.241455 [DOI] [PubMed] [Google Scholar]
- Syed GH, Khan M, Yang S, Siddiqui A. 2017. Hepatitis C virus lipoviroparticles assemble in the endoplasmic reticulum (ER) and bud off from the ER to the Golgi compartment in COPII vesicles. J Virol 91: e00499-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji Y, Kaneko T, Satoh S, Shimotohno K. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol 69: 3980–3986. 10.1128/JVI.69.7.3980-3986.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targett-Adams P, Hope G, Boulant S, McLauchlan J. 2008. Maturation of hepatitis C virus core protein by signal peptide peptidase is required for virus production. J Biol Chem 283: 16850–16859. 10.1074/jbc.M802273200 [DOI] [PubMed] [Google Scholar]
- Tellinghuisen TL, Marcotrigiano J, Gorbalenya AE, Rice CM. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J Biol Chem 279: 48576–48587. 10.1074/jbc.M407787200 [DOI] [PubMed] [Google Scholar]
- Tellinghuisen TL, Foss KL, Treadaway J. 2008a. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog 4: e1000032 10.1371/journal.ppat.1000032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. 2008b. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J Virol 82: 1073–1083. 10.1128/JVI.00328-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomssen R, Bonk S, Propfe C, Heermann KH, Köchel HG, Uy A. 1992. Association of hepatitis C virus in human sera with β-lipoprotein. Med Microbiol Immunol 181: 293–300. 10.1007/BF00198849 [DOI] [PubMed] [Google Scholar]
- Tsutsumi T, Suzuki T, Shimoike T, Suzuki R, Moriya K, Shintani Y, Fujie H, Matsuura Y, Koike K, Miyamura T. 2002. Interaction of hepatitis C virus core protein with retinoid X receptor α modulates its transcriptional activity. Hepatology 35: 937–946. 10.1053/jhep.2002.32470 [DOI] [PubMed] [Google Scholar]
- Vieyres G, Welsch K, Gerold G, Gentzsch J, Kahl S, Vondran FW, Kaderali L, Pietschmann T. 2016. ABHD5/CGI-58, the Chanarin–Dorfman syndrome protein, mobilises lipid stores for hepatitis C Virus production. PLoS Pathog 12: e1005568 10.1371/journal.ppat.1005568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt DA, Camus G, Herker E, Webster BR, Tsou CL, Greene WC, Yen TS, Ott M. 2013. Lipid droplet-binding protein TIP47 regulates hepatitis C Virus RNA replication through interaction with the viral NS5A protein. PLoS Pathog 9: e1003302 10.1371/journal.ppat.1003302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M, et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11: 791–796. 10.1038/nm1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller R, Hueging K, Brown RJP, Todt D, Joecks S, Vondran FWR, Pietschmann T. 2017. Hepatitis C virus strain-dependent usage of apolipoprotein E modulates assembly efficiency and specific infectivity of secreted virions. J Virol 91: e00422-17 10.1128/JVI.00422-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Fröhlich F, et al. 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell 24: 384–399. 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, He Y, Boson B, Wang X, Cosset FL, Zhong J. 2017. A point mutation in the N-terminal amphipathic helix α0 in NS3 promotes hepatitis C virus assembly by altering core localization to the endoplasmic reticulum and facilitating virus budding. J Virol 91: e02399-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wang X, Chi X, Zhao F, Guo J, Ma P, Zhong J, Niu J, Pan X, Long G. 2016. Neglected but important role of apolipoprotein E exchange in hepatitis C virus infection. J Virol 90: 9632–9643. 10.1128/JVI.01353-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Ye J. 2008. Long chain acyl-CoA synthetase 3-mediated phosphatidylcholine synthesis is required for assembly of very low density lipoproteins in human hepatoma Huh7 cells. J Biol Chem 283: 849–854. 10.1074/jbc.M706160200 [DOI] [PubMed] [Google Scholar]
- Yasui K, Wakita T, Tsukiyama-Kohara K, Funahashi SI, Ichikawa M, Kajita T, Moradpour D, Wands JR, Kohara M. 1998. The native form and maturation process of hepatitis C virus core protein. J Virol 72: 6048–6055. 10.1128/JVI.72.7.6048-6055.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Ma Y, Yates J, Lemon SM. 2009. Trans-complementation of an NS2 defect in a late step in hepatitis C virus (HCV) particle assembly and maturation. PLoS Pathog 5: e1000403 10.1371/journal.ppat.1000403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci 102: 9294–9299. 10.1073/pnas.0503596102 [DOI] [PMC free article] [PubMed] [Google Scholar]