Abstract
This study was aimed at the genome-wide identification, a comprehensive in silico characterization of NHX genes from soybean (Glycine max L.) and their tissue-specific expression under varied levels (0-200 mM NaCl) of salinity stress. A total of nine putative NHX genes were identified from soybean. The phylogenetic analysis confirmed a total of five sub-groups and GmNHXs were distributed in three of them. Bioinformatics analyses confirmed all GmNHXs as ion transporters in nature, and all were localized on the vacuolar membrane. Several cis-acting regulatory elements involved in hormonal signal-responsiveness and abiotic stress including salinity responses were identified in the promoter regions of GmNHXs. Amiloride, which is a known Na+/H+ exchanger activity inhibitor, binding motifs were observed in all the GmNHXs. Furthermore, the identified GmNHXs were predicted-targets of 75 different miRNA candidates. To gain an insight into the functional divergence of GmNHX transporters, qRT-PCR based gene expression analysis was done in control and salt-treated root, stem and leaf tissues of two contrasting Indian soybean varieties MAUS-47 (tolerant) and Gujosoya-2 (sensitive). The gene up-regulation was tissue-specific and varied amongst the soybean varieties, with higher induction in tolerant variety. Maximum induction was observed in GmNHX2 in root tissues of MAUS-47 at 200 mM NaCl stress. Overall, identified GmNHXs may be explored further as potential gene candidates for soybean improvement.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02555-0) contains supplementary material, which is available to authorized users.
Keywords: Gene expression, Na+/H+ antiporter (NHX), Na+ compartmentalization, Na+ transport, Salt tolerance, Soybean
Introduction
Soil salinity is one of the major abiotic stresses and a serious global threat to agricultural crop production (Wani et al. 2020). It has, therefore, become imperative to understand and explore molecular mechanisms underlying salinity stress responses so as to minimize the negative impacts, projected to cause 50% arable land loss by 2050. Salinity-driven cellular ionic imbalances and/or toxicities are considered primary cause for hampered growth and development of plants growing in hyper-saline conditions (Wu et al. 2019). Plants respond to this ionic or nutrient imbalance in a coordinated manner mainly through maintenance of ionic homeostasis through regulating the influx/efflux of the excessive ions, and accumulation and compartmentalization of the toxic ions (Khare et al. 2015). Sodium (Na+) is a dominant cation of saline soils (Munns and Tester 2008) and is often attributed for the toxic impacts exerted by the salinity stress, as reported in several crops, barley (Tavakkoli et al. 2011), faba bean (Tavakkoli et al. 2010), rice (Kumar and Khare 2016) and soybean (Shelke et al. 2019). The Na+/H+ antiporters (NHXs), located on plasma membranes and tonoplast, play key role in maintaining Na+ homeostasis via transporting it from the cytoplasm to extracellular spaces or vacuoles, and are, therefore, attaining greater attention amongst the sodium transporters (Pehlivan et al. 2016).
The Na+ influx in plant system is mediated via non-selective cation channels which are permeable to many monovalent cations. These non-selective cation channels are classified into voltage-dependent/independent cation channels, and the voltage-independent channels are mainly responsible for the movement of Na+ across the plant tissues (Wani et al. 2020). Long distance movement of the Na+ from roots to shoots is modulated by the NHX transporters which belong to the cation proton antiporter family 1 (CPA1) (Bassil and Blumwald 2014). The evolutionary analysis suggests that the CPA1 family is evolved from the prokaryotic ancestral sodium-proton antiporter (NhaP) (Rodríguez-Rosales et al. 2009) and is involved in cytoplasmic pH regulation and extrusion of H+ produced during cellular metabolism in exchange of Na+ in cytoplasm/vacuole (Leidi et al. 2010). Several reports have confirmed the involvement of NHX proteins in cell expansion, pH regulation, K+ homeostasis and cellular vesicle trafficking and in salinity stress responses (Apse et al. 2003; Ohnishi et al. 2005; Jiang et al. 2010; Qiu 2012; Huertas et al. 2013; Wu et al. 2019). Owing to these vital roles, NHXs have gained attention as potent candidates for engineering crop plants for improved salt tolerance (Bao et al. 2016; Li et al. 2017). There are many evidences of utilization of NHXs from halophytes for production of salt tolerant crops using genetic engineering approaches (Wani et al. 2020 and the relevant references therein).
Soybean (Glycine max L.) is one of the premium agricultural oilseed crops, widely grown around the world as a main source of protein and oil, and is extensively used for food and livestock feed (Yasuta and Kokubun 2014). However, salinity stress is a major limiting factor for soybean growth and crop yield (Sun et al. 2019). Considering the availability of G. max genome sequence, it is possible to identify the NHX genes at whole genome level, but the information and functional characterization of NHX gene families of G. max still remains largely missing. Furthermore, there are not many attempts on using G. max as a source of NHX genes to raise transgenic plants with improved salinity tolerance, except a few instances like GmNHX1-overexpressing transgenic lines of a model plant Arabidopsis (Sun et al. 2019) and Lemna turionifera (Yang et al. 2017). Therefore, the identification and characterization of endogenous genes governing the salt tolerance levels in soybean would be considerably advantageous for the aim of engineering salt tolerant plants.
Considering the importance of NHX genes in regulating salt tolerance, a genome-wide identification and characterization of NHXs was attempted in G. max. Though genomic and functional investigations have been carried out in a number of plant species, soybean NHX family is yet to be described and/or explored fully. The current study involves orthology-based identification and characterization of putative NHXs from G. max genome, analysis of different motifs and domains present therein and analyzing their promoter regions. The study also investigated the protein–protein interactions and miRNA interaction networks involving the identified NHXs. Finally, qRT-PCR-based expression patterns of identified NHXs were investigated under saline conditions in the soybean cultivars to infer the genotype based differential behavior of these transporters in different plant parts. Our findings hold significance in shedding light on the molecular properties and regulatory roles of G. max NHXs in salinity tolerance of soybean and may provide candidate gene resources for further exploration in developing salt tolerant soybean varieties.
Materials and methods
Plant material, growth conditions and treatments
Two Glycine max L. cultivars with contrasting salt tolerance, Gujosoya-2 (sensitive) and MAUS-47 (tolerant) were used in this study. The seeds were first washed with the water containing 0.05% Tween-20 followed by the surface sterilization with 0.1% mercuric chloride for 3 min. The seeds were then sown in the soil (red clay, sieved through 2 mm mesh, pH 6.03 ± 0.5; EC 1.21 dS m−1), allowed to germinate and grow at 25 ± 2 °C, 75% relative humidity, 16/8 photoperiod till the trifoliate stage was achieved (~ 21 days). Hoagland medium was used as basic growth medium for all the experiments. After this, the plants were randomly categorized into three groups followed by the application of salt treatments [0 mM (control), 100 mM and 200 mM NaCl]. Plants were harvested 24 h after the treatment and thoroughly washed with distilled water. Leaf, stem and root tissues were separated, flash-freezed in the liquid nitrogen and successively used for total RNA extraction.
Identification and phylogeny of GmNHXs
The putative G. max NHX orthologs were retrieved using the known NHX proteins from ten different plant genus namely Arabidopsis, Brassica, Cucumus, Lupinus, Medicago, Phaseolus, Solanum, Trifolium, Vigna and Vitis from Gramene database (http://www.gramene.org/). Orthologous sequences from G. max showing similarity score ≥ 70% were considered for further analysis. Obtained orthologs were then checked for the presence of Na+/H+ exchanger activity using HMMER web server (http://hmmer.org/) (Finn et al. 2011) and EggNOG mapper (http://eggnog5.embl.de/#/app/home) (Huerta-Cepas et al. 2019). The CDS, peptide, genomic sequences of the finalized genes were retrieved from the Gramene database for further use (renamed as GmNHXs). To investigate the phylogenetic relationship between the identified GmNHXs and other reference NHXs, the peptide sequences of GmNHXs were aligned with reference peptides using Clustal Omega tools (https://www.ebi.ac.uk/Tools/msa/clustalo/). Phylogenetic tree was constructed by MEGA-X (https://www.megasoftware.net) using the neighbor-joining (NJ) method, with 1000 bootstrap replicates (Kumar et al. 2018) and visualized using FigTree.
Chromosomal location and sequence analysis of GmNHXs
The chromosomal coordinates of the identified GmNHXs were retrieved from the Glycine max genome at the Gramene database (Glycine_max_v2.1) (http://www.gramene.org/) and the physical map of the chromosomal locations of the genes was constructed using MapGene2Chromosoem v2 (http://mg2c.iask.in/mg2c_v2.0/). The molecular weight [MW], isoelectric point [pI], instability index, aliphatic index and the grand average of hydropathicity [GRAVY] was computed using the ProtParam tool by ExPASy (https://web.expasy.org/protparam/) (Gasteiger et al. 2005). Sub-cellular localization of the proteins was predicted using Plant-mPLoc server (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/). To identify the disordered regions in the proteins, the prediction of the intrinsically unstructured regions was achieved using IUPred2A web interphase (https://iupred2a.elte.hu/) (Mészáros et al. 2018). The phosphorylation sites in the proteins were predicted using the kinase specific predictions by NetPhos 3.1 Server (http://www.cbs.dtu.dk/services/NetPhos/) (Blom et al. 1999). The secondary structural elements were predicted using the PSIPRED Workbench (http://bioinf.cs.ucl.ac.uk/psipred/). The transmembrane helices in the proteins were predicted using the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) (Möller et al. 2001). Gene annotations of the identified GmNHXs were extracted from the Gene Ontology Resource (http://geneontology.org/) (Carbon et al. 2019) and visualized using the Web Gene Ontology Annotation Plot (WEGO 2.0, http://wego.genomics.org.cn/) (Ye et al. 2018).
Gene structure, conserved motifs and domain architecture analysis of GmNHXs
The gene structure (number of introns/exons and up-/down-stream regions) was visualized using the Gene Structure Display Server (GSDS 2.0, http://gsds.cbi.pku.edu.cn/index.php) (Hu et al. 2015). The MEME program (http://meme-suite.org/) and Pfam tool (https://pfam.xfam.org/) were used to search for motifs and conserved domain in the GmNHX sequences, respectively (Bailey et al. 2009; El-Gebali et al. 2019); subsequently, the domain and motif diagrams were drawn using the TBtools software. Multiple sequence alignment of all the GmNHX peptides was obtained using MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle/) and was visualized using ESPript 3.0 tool (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) (Robert and Gouet 2014) to trace the motif sequences.
The Cis-element analysis in the promoter region of GmNHXs
The promoter region, 2000 bp upstream of the CDS was retrieved for each GmNHX and the cis-acting regulatory elements were identified using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot 2002).
In silico prediction of protein specific SSRs and miRNA networks
Gene specific molecular markers were predicted using the genomic transcripts of identified NHX using BatchPrimer3 v1.0 server (https://wheat.pw.usda.gov/demos/BatchPrimer3/) (You et al. 2008). Furthermore, putative miRNAs targeting the GmNHXs were isolated genes using psRNATarget server (http://plantgrn.noble.org/psRNATarget/) (Dai et al. 2018) with default parameters. The interaction network between the identified miRNAs and their targets was build using Cytoscape software (http://www.cytoscape.org) (Otasek et al. 2019).
Three-dimensional structural predictions of GmNHXs
Three-dimensional (3-D) structures of the identified GmNHXs were predicted using I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) (Yang and Zhang 2015) and Phyre2 (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index) tools (Kelley et al. 2015). The structural modeling was achieved using the same (single) template as the best hit for all the GmNHXs. The structures with best accuracy were selected on the basis of C-score values.
Prediction of protein–protein interactions
The protein–protein interaction network was constructed using the STRING database v11.0 (https://string-db.org/) (Szklarczyk et al. 2019) by utilizing the peptide sequences of all the GmNHXs.
Quantitative real-time (qRT)-PCR analysis of the GmNHX genes
Total RNA was isolated from the samples using HiPurA™ total RNA purification kit (Himedia, India) using the spin column based approach by following the manufacturer’s instructions. Furthermore, the cDNA was synthesized for each sample using Hi-cDNA synthesis kit (Himedia, India). The cDNA was then subjected to qRT-PCR for all the GmNHXs in the study using gene specific primers (Supplementary Table S1) and Hi-SYBr master mix (Himedia, India). Tubulin was used as house-keeping gene. The qRT-PCR conditions were: initial denaturation of 95 °C for 2 min, 40 cycles of 94 °C denaturation for 30 s, 60 °C annealing for 45 s and 72 °C extension for 30 s. The analysis was conducted with three biological replicates. The relative expression levels (RQ) of the GmNHXs were calculated as relative changes by the 2−ΔΔCt method. The numeric outcomes of assays were represented as mean ± standard error, and means were compared using Duncan’s multiple range test (DMRT) at p ≤ 0.05 using MSTAT-C package. The hierarchical clustering was performed using the normalized mean values and heatmap was generated using MeV software (http://mev.tm4.org/).
Results
Identification and phylogeny of GmNHXs
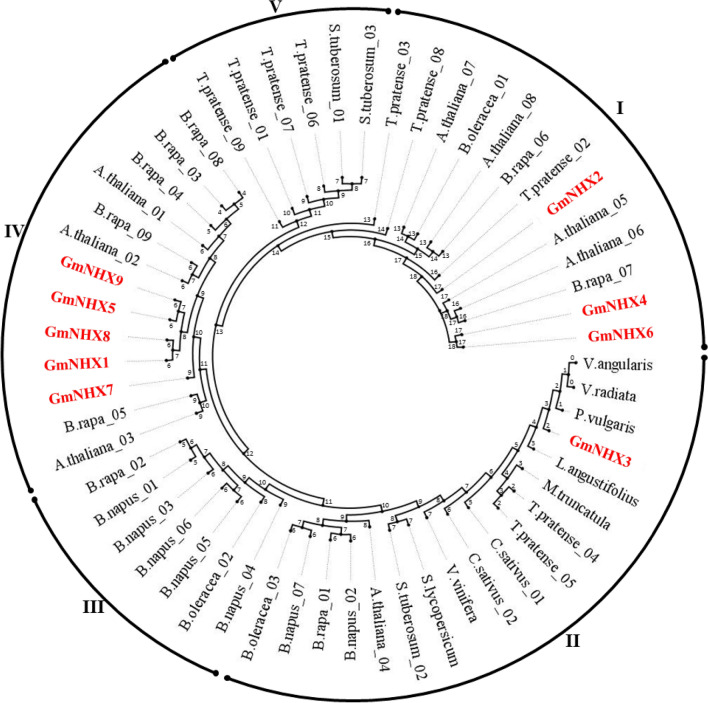
A total of 14 NHX genes, orthologous to the reference NHXs, were identified in G. max on the basis of percent identity (≥ 70%). Identified orthologs were subjected to the protein domain identification using HMMER web server and EggNOG mapper. Based on the presence of sodium/hydrogen exchanger domain in the orthologs (Supplementary Tables S2 and S3), nine putative NHXs were shortlisted in G. max. The identified orthologs were named as GmNHX1-9 and analyzed for their phylogenetic positions amongst the NHXs from the reference plants using the peptide sequences. The phylogenetic tree revealed the formation of five different groups, as illustrated in Fig. 1. Group IV in the phylogenetic tress displayed presence of 5 GmNHXs (GmNHX1, GmNHX5, GmNHX7, GmNHX8, and GmNHX9), whereas 3 other GmNHXs were placed in Group I (GmNHX2, GmNHX4, and GmNHX6). Only GmNHX3 was positioned in Group III. The positioning of the GmNHXs in the tree reveals their closeness towards the NHXs from Brassica and Arabidopsis (Group I and IV). Besides, GmNHX1, GmNHX5, GmNHX7, GmNHX8, and GmNHX9 were closely related to each other than other identified genes. The GmNHX3, which was positioned in group III, displayed the closeness towards the NHXs from Fabaceae family members (Vigna and Phaseolus).
Fig. 1.
Phylogenetic relationship between the NHXs of G. max and the reference plants (Arabidopsis thaliana, Brassica rapa, Brassica oleracea, Brassica napus, Cucumus sativus, Lupinus angustifolius, Medicago trancatula, Phaseolus, Solanum tuberosum, Solanum lycopersicum, Trifolium pratense, Vigna radiata, Vigna angularis and Vitis vinifera) (Table S8). The peptide sequences of GmNHXs were aligned with reference peptides using Clustal Omega tools and phylogenetic tree was constructed by MEGA-X using the neighbor-joining (NJ) method, with 1000 bootstrap replicates
Chromosomal location and sequence analysis of GmNHXs
The physical chromosomal locations of the identified GmNHXs were retrieved from the Gramene database (Glycine_max_v2.1), and they found to be distributed in nine different chromosomes of G. max (Supplementary Fig. S1). All GmNHXs displayed the negative orientation (strand) except the GmNHX9 on chromosome 20 which displayed positive orientation. GmNHX2 was the smallest peptide with 455 amino acids, while GmNHX1 and GmNHX8 were largest sequences (547 amino acids). All other peptides varied with amino acid numbers ranging from 527 to 546 (Table 1). Molecular weight of the GmNHXs ranged from 50.05 to 60.79 kD, while pI values were observed in between 5.38 and 9.06. The protein instability indices confirmed the stable nature of proteins in the test-tube, as most of the proteins had the instability index below 40, except GmNHX2 (40.34) and GmNHX7 (40.62). The aliphatic indices for all the proteins were > 100, with highest and lowest values displayed by GmNHX3 and GmNHX4 (118.37 and 102.64, respectively). The grand average of hydropathicity (GRAVY) for all the proteins were positive (ranging from 0.460 to 0.734) suggesting all proteins being hydrophobic in nature (Table 1).
Table 1.
List of identified NHXs with their accession IDs (from Gramene database), Chromosomal location and strand, size (aa: amino acid), Iso-electric point (pI), molecular weight (MW) (kDa), instabilty index, aliphatic Index, GRAVY, sub-cellular localization (V: Vacuole) and transmembrane helixes (predicted no., number of aa in TMH)
| Gene | Accession ID | Chromosomal location and strand (±) |
Size (aa) | pI | MW (kDa) | Instability Index | Aliphatic Index | GRAVY | Sub-cellular Localization | Predicted TMHs | aas in TMHs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GmNHX1 | GLYMA_03G243000 | Chr3:44,079,352-44,085,682 (-) | 547 | 8.77 | 60.79 | 34.44 | 108.14 | 0.466 | V | 12 | 251.54 |
| GmNHX2 | GLYMA_07G270400 | Chr7:44,306,021-44,315,600(-) | 455 | 6.20 | 50.05 | 40.34 | 112.26 | 0.734 | V | 09 | 229.41 |
| GmNHX3 | GLYMA_08G324200 | Chr8:44,233,968-44,242,320(-) | 527 | 7.69 | 58.72 | 37.41 | 118.37 | 0.645 | V | 10 | 234.59 |
| GmNHX4 | GLYMA_09G018200 | Chr9:1,420,668-1,431,498(-) | 534 | 5.38 | 58.45 | 39.20 | 102.64 | 0.460 | V | 10 | 239.39 |
| GmNHX5 | GLYMA_10G158700 | Chr10:39,253,048-39,262,228(-) | 546 | 6.78 | 60.12 | 36.47 | 113.52 | 0.558 | V | 10 | 232.14 |
| GmNHX6 | GLYMA_15G124100 | Chr15:9,849,463-9,859,551(-) | 534 | 5.53 | 58.39 | 40.62 | 102.66 | 0.475 | V | 10 | 240.26 |
| GmNHX7 | GLYMA_17G022500 | Chr17:1,643,783-1,649,939(-) | 536 | 9.06 | 59.64 | 37.88 | 116.92 | 0.626 | V | 11 | 234.36 |
| GmNHX8 | GLYMA_19G240500 | Chr19:48,830,897-48,836,752(-) | 547 | 8.99 | 60.74 | 36.10 | 110.46 | 0.474 | V | 12 | 253.82 |
| GmNHX9 | GLYMA_20G229900 | Chr20:46,409,048-46,417,710(+) | 546 | 6.78 | 60.10 | 36.66 | 112.45 | 0.531 | V | 10 | 233.17 |
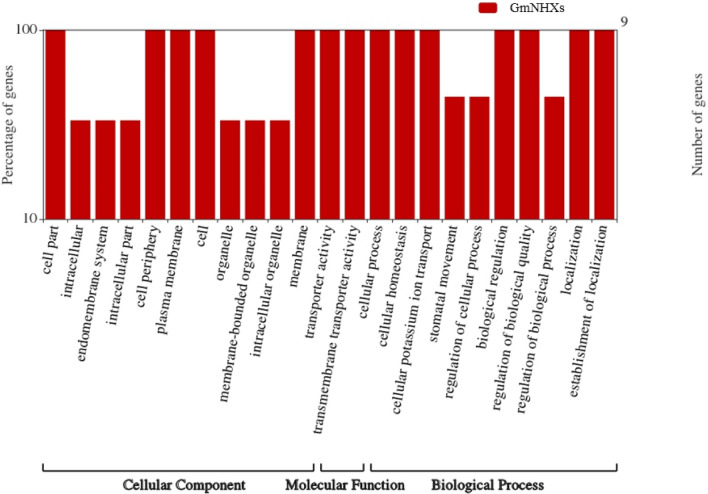
Sub-cellular locations predicted using Plant-mPLoc server indicated that all the proteins were present on the vacuolar membrane. The identification of the disordered protein regions was carried out via IUPred2A analysis, and it revealed that the region harboring Na+/H+ exchanger domain was without any disorder or disordered binding regions (Supplementary Fig. S2). The phosphorylation sites were predicted with the help of NetPhos 3.1 server which showed that all the GmNHXs contained high number of PKCs followed by PKAs, CKs and CDCs (Supplementary Fig. S3). Secondary structure elements in the GmNHXs predicted using PSIPRED confirmed that the helices and coils were predominantly present in all the proteins (Supplementary Fig. S4). TMHMM Server v. 2.0 showed the presence of at least 9 to 12 transmembrane helices in GmNHXs suggesting the compactly arranged membrane bound nature of the identified GmNHXs (Supplementary Fig. S5, Table 1). The MEMSATSVM service of PSIPRED Workbench was used to get the topology of the transmembrane helices in GmNHXs (Supplementary Fig. S6). The amino acid position indicated that five GmNHXs (GmNHX1, GmNHX5, GmNHX7, GmNHX8, and GmNHX9) had the N-terminus end of peptide in the extracellular region and C-terminal end in the cytoplasmic region. GmNHX2, GmNHX4 and GmNHX6 showed both the ends in the cytoplasmic region, while GmNHX3 had C-terminus end in extracellular region and N-terminus end in the cytoplasmic region. Each of the GmNHXs contains 229 to 253 amino acids in their transmembrane helices (Table 1). The WEGO 2.0 tool further confirmed the membrane-bound nature of all nine GmNHXs, as shown in Fig. 2. This figure also indicated the transmembrane transporter activity as a molecular function of all the identified GmNHXs, and this validated the fact that all the identified GmNHXs were indeed transporters in nature.
Fig. 2.
Gene annotations obtained for every GmNHXs, represented using the WEGO tool (http://wego.genomics.org.cn/). Gene annotations of the identified GmNHXs were extracted from the Gene Ontology (GO) Resource. Both the Y axis quantitates the genes (exact number and percentage). The X axis represents the allotted gene annotation and the annotations are categorized as cellular component, molecular function and biological process
Gene structure, conserved motifs and domain architecture analysis of GmNHXs
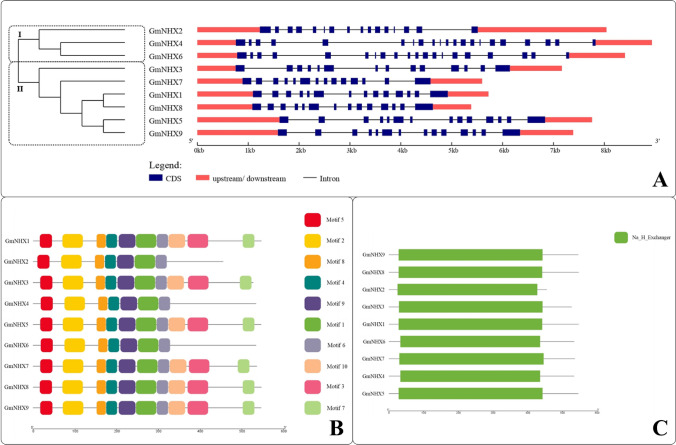
To examine the structural characteristics of GmNHX genes, the intron/exon organizations of these genes were analyzed and compared using nucleotide sequences. Based on the results obtained by Gene Structure Display Server, we categorized GmNHXs in two groups (Fig. 3a). The length and number of introns as well as intron phases were comparatively conserved in group II genes, where all the genes possessed 13 introns, unlike the group I, where unequal number of introns were seen. The analysis further revealed that all the genes have upstream and downstream regions.
Fig. 3.
a Phylogenetic relation between the identified GmNHXs and their genetic structure (CDS, intron/exon regions) constructed using the gene structure display server (GSDS 2.0, http://gsds.cbi.pku.edu.cn/index.php). The phylogenetic tree is divided in two groups representing two different branches. CDS: coding sequence, kb: kilobases; b Identification and distribution of the conserved motifs in GmNHXs. The identification, distribution and positioning of ten different motifs was obtained using online MEME server (http://meme-suite.org/). The diagram was constructed using TBtools software; c Distribution of the conserved Na+/H+ exchanger domain (Na_H_Exchanger) in GmNHX proteins. The domain analysis was done using the HMMER and the diagram was constructed using TBtools software
To gain a better understanding about structural diversity of GmNHX proteins, the motif distributions in proteins were investigated using the MEME program, which identified a total of 10 putative motifs (Fig. 3b), and out of which only 4 conserved motifs were designated to encode for the function of Na+/H+ exchanger domain, as revealed by the annotations from Pfam (Table S4). Motifs 1, 2, 3 and 4 belonged to the Na+/H+ exchanger family (CL0064, Pfam). The multiple sequence alignment of the GmNHX peptides characterized the positioning of the four conserved motifs (Supplementary Fig. S7). The results confirmed that the motif 1, 2 and 4 were represented by all 9 GmNHXs, whereas motif 3 was present in GmMHX1, GmMHX3, GmMHX5, GmMHX7, GmMHX8, and GmMHX9. Apart from these principal motifs, 5, 6, 8 and 9 motifs were also present in the GmNHXs (Fig. 3b). A conserved domain analysis revealed that all the GmNHX proteins had Na+/H+ exchanger domain (Fig. 3c), thus qualifying them for the basic characteristic feature of an NHX transporter.
Analysis of Cis-elements in GmNHXs promoters
The promoter region of each GmNHXs was analyzed for the identification and enumeration of the cis-acting elements with an aim to further explore the regulatory roles of GmNHX genes. As described in Table 2, cis-acting elements involved in hormonal responses (ABRE, ERE), stress responses (ARE, MYC, DRE, MYB, TC-rich, W Box) and other cellular functioning (A-box, O2-SITE, F-BOX, G-BOX, GT1-MOTIF, CIRCADIAN) were identified in the promoter regions of GmNHXs. Phytohormone responsive element ABRE was present in promoter region of all the GmNHXs, whereas ERE was present in all except the GmNHX4 promoter. Stress responsive elements, namely ARE, MYC, MYB, W Box were present in promoter regions of all the GmNHXs. The stress responsive DRE element was observed only in five GmNHX promoters (GmNHX2, GmNHX3, GmNHX4, GmNHX5 and GmNHX6). Amongst the other cis-acting elements, light responsive (G-BOX, GT1-MOTIF) elements were again present in all the promoters. The O2 site, which is involved in zein metabolism, was present in the promoters of all GMNHXs except GmNHX7. The element involved in circadian control was present in the promoter region of GmNHX1, GmNHX2, GmNHX3, GmNHX6, GmNHX7, GmNHX8, and GmNHX9. The presence of diverse kinds of cis-regulatory elements in the promoter region of GmNHXs implies the complicated control of the expression levels of GmNHXs at transcriptional level. This indicate the potential roles of GmNHXs in phytohormonal signal-responsiveness and stress adaptations of plants.
Table 2.
Cis-acting elements analyzed from the promoter region of the identified GmNHX
| Motif | Description | GmNHX1 | GmNHX2 | GmNHX3 | GmNHX4 | GmNHX5 | GmNHX6 | GmNHX7 | GmNHX8 | GmNHX9 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phytohormone response | ABRE | Involved in responses to abscisic acid | 15 | 18 | 8 | 6 | 9 | 12 | 10 | 12 | 27 |
| ERE | Ethylene responsive element | 3 | 5 | 1 | – | 8 | 3 | 2 | 9 | 4 | |
| Stress response | ARE | Element essential for anaerobic induction | 9 | 8 | 9 | 9 | 8 | 8 | 5 | 9 | 5 |
| MYC | Stress response regulators | 12 | 19 | 17 | 21 | 21 | 22 | 15 | 15 | 13 | |
| DRE | Dehydration responsive | – | 1 | 1 | 1 | 1 | 1 | – | – | – | |
| MYB | Stress response regulators | 17 | 18 | 13 | 19 | 19 | 15 | 13 | 12 | 22 | |
| TC-RICH | Defense and responses to stress | 1 | 3 | 4 | 2 | 2 | 3 | 1 | – | 3 | |
| W-BOX | Stress responsive element | 2 | 4 | 1 | 5 | 2 | 6 | 2 | 2 | 1 | |
| Other | A-box | Elicitor of light response | 1 | 1 | 0 | 3 | – | – | – | – | 1 |
| O2-SITE | Involved in zein metabolism regulation | 1 | 2 | 2 | 6 | 4 | 5 | – | 1 | 3 | |
| F-BOX | Regulation of cell cycle | 1 | 1 | – | 1 | 1 | – | 1 | 1 | – | |
| G-BOX | Involved with light responses | 10 | 11 | 6 | 6 | 5 | 6 | 8 | 7 | 16 | |
| GT1-MOTIF | Light responsive element | 6 | 6 | 6 | 10 | 12 | 7 | 3 | 4 | 4 | |
| CIRCADIAN | Involved in circadian control | 1 | 3 | 2 | – | – | 1 | 2 | 1 | 1 |
In silico prediction of protein specific SSRs and miRNA networks
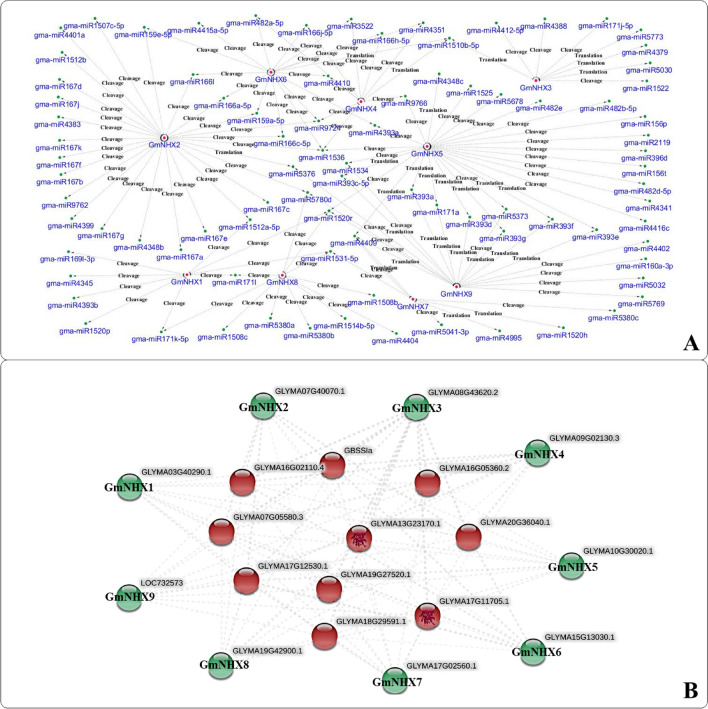
We detected a total of 30 SSRs from the genomic transcripts of identified GmNHXs (Supplementary Table S5, Supplementary Fig. S8). Majority (40%) of the detected SSRs belonged to the dinucleotide category (12/30), followed by the tetranucleotide (37%). Trinucleotide SSRs shared 20% (6/30) of the total SSRs, whereas only one pentanucletide SSR was identified. A total of 75 different miRNA candidates were predicted that can potentially target the identified 9 GmNHXs. The number of miRNAs predicted to target GmNHX1-9 were 9, 7, 8, 10, 19, 16, 9, 11 and 17, respectively (Supplementary Table S6). The miRNA network analysis revealed the complex interactions between the putative miRNAs targeting the GmNHXs along with their mode of action (cleavage or translation based inhibition) (Fig. 4a). The GmNHX5 was targeted by highest (19) number of miRNA candidates, which includes two miRNA families (miR393 and miR482). The GmNHX8 could potentially be a target of 11 miRNAs from 3 miRNA families- miR171, miR1508 and miR5380. Similarly, GmNHX9 is a target of 17 different miRNAs including miR393 family, while 9 different miRNAs were identified to target GmNHX1 and GmNHX8. GmNHX2 came out to be a target of least number of miRNAs. Interestingly, miRNA interaction network analysis also revealed some common miRNAs targeting multiple GmNHXs (Fig. 4a), for instance, miR393 family targeted GmNHX5 and GmNHX9. Similarly, GmNHX4 and GmNHX6 were the targets of miR166 family members, and miRNA candidates belonging to miR171 family targeted GmNHX1 and GmNHX8. Apart from these, there were several other miRNAs having common GmNHX targets such as miR1510b-5p (targeting GmNHX3, GmNHX4 and GmNHX6), miR1536 (targeting GmNHX2, GmNHX4, GmNHX5, and GmNHX6), miR1520r (targeting GmNHX1, GmNHX5, GmNHX8, GmNHX9) and miR9724 (targeting GmNHX2, GmNHX4, and GmNHX6).
Fig. 4.
a Interacting network of the miRNAs targeting the GmNHXs. The putative miRNAs targeting the GmNHXs were isolated using psRNATarget server (http://plantgrn.noble.org/psRNATarget/) and the interaction network between the identified miRNAs and their targets was constructes using Cytoscape software (http://www.cytoscape.org). Pink dot represents the GmNHXs, green dot represents the corresponding miRNAs. The mode of miRNA mediated regulation (cleavage/translation) is mentioned for every connection; b protein–protein interaction network constructed between the GmNHXs and the respective interacting proteins. The protein–protein interaction network was constructed using the STRING database v11.0 (https://string-db.org/) by utilizing the peptide sequences of all the GmNHXs. The GmNHXs are represented at the periphery of the network (green)
Analysis of GmNHX protein structure
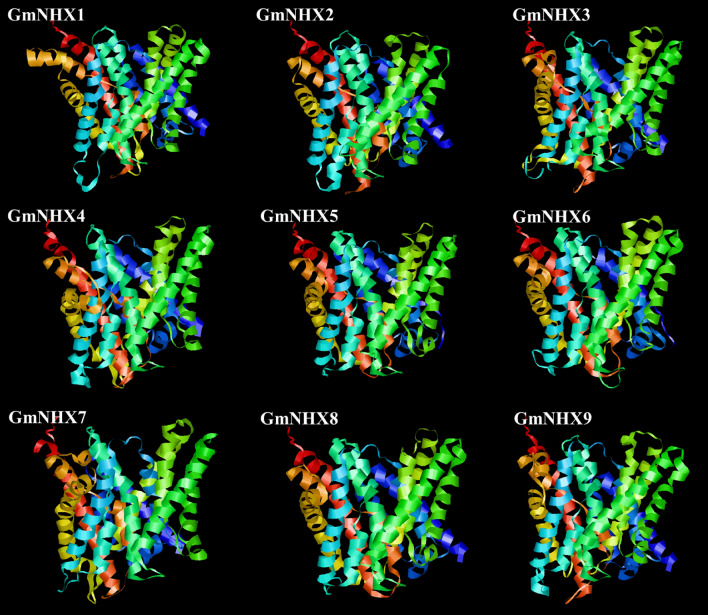
To understand the putative structural and functional characters of GmNHX proteins in soybean, the 3-D structures were modeled using Phyre2 web portal and I-TASSER server. Three-dimensional models were constructed through selection of the best structural template and utilization of their crystal structures from the protein data bank (PDB) (Fig. 5). All the structures were constructed using the same template (PDB Hit: 4cz8A). The best model was selected on the basis of the C-score (from − 5 to 2) with the higher C-score representing better confidence level for the model. All the modeled GmNHX structures were well within the stipulated C-score range (− 1.74 to − 0.33) (Supplementary Table S7), confirming the high accuracy of predicted GmNHX structures.
Fig. 5.
Structural analysis (modeling of the three-dimensional structure) of GmNHXs. Three-dimensional (3-D) structures of the identified GmNHXs were predicted using I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). The best protein data bank analog used as template for all the transporters: 4cz8A (PDB ID)
Prediction of protein–protein interactions
Protein–protein interaction network was constructed using STRING database to further understand the potential functions of GmNHXs during their interactions with other cellular proteins. Amongst the GmNHXs, no immediate interaction was observed (Fig. 4b); however, most of the proteins were sharing the putatively interacted proteins. Similar interactions were observed in the interactive (Fig. 4b) as well as the individual protein networks (Supplementary Fig. S9). Such proteins include NADH-cytochrome b5 reductase (GLYMA17G12530.1, GLYMA05G08480.1; belongs to the flavoprotein pyridine nucleotide cytochrome reductase family), starch synthase (GLYMA16G02110.4, GLYMA20G36040.1, and GLYMA07G05580.3; chloroplastic/amyloplastic-granule-bound protein from glycosyltransferase-1 family), bifunctional dihydrofolate reductase–thymidylate synthase (GLYMA12G04480.2; bifunctional enzyme involved in de novo dTMP biosynthesis during folate metabolism), proteins with H_HPPase domain (GLYMA17G11705.1, GLYMA13G23170.1; pyrophosphatase involved in hydrolysis of inorganic pyrophosphate) and proteins from potassium transporter family (GLYMA07G15590.2, GLYMA12G16040.1, and GLYMA17G31011.1).
Expression pattern of the GmNHX genes under NaCl stress
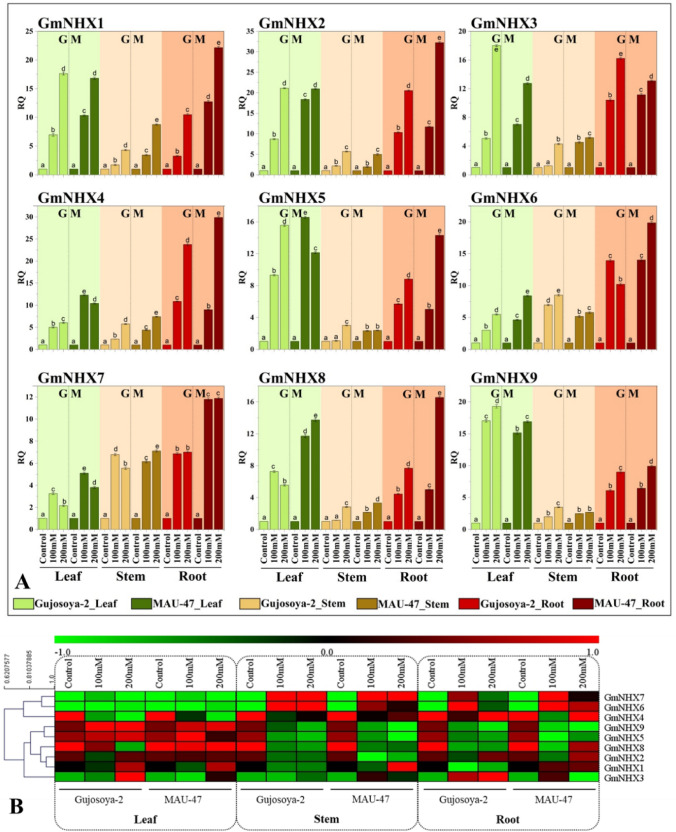
In an attempt to gain insights into the possible roles GmNHXs play in soybean responses to salinity stress, we compared their expression patterns quantitatively under control and salt-stress conditions using qRT-PCR analysis. Results confirmed that salinity induced all the GmNHXs genes in leaf, stem and root tissues of both salt tolerant (MAUS-47) and sensitive (Gujosoya-2) soybean cultivars in a concentration-dependent manner (Fig. 6a). Amongst the tissues, roots showed highest induction followed by leaves and stems. Salinity-mediated upregulation of GmNHX genes was higher in MAUS-47 than Gujosoya-2.
Fig. 6.
a Effect of NaCl (0, 100, 200 mM NaCl) on expression levels of GmNHXs in leaf (green), stem (brown) and root (dark brown) of soybean cultivars with contrasting salt tolerance nature G: Gujosoya-2 (sensitive), M: MAUS-47 (tolerant). Values are represented as mean ± standard error (n = 3). The DMRT at p ≤ 0.05 was applied. Treatments followed by the same lower-case letter for a particular genotype do not differ statistically. RQ: relative quantitative expression; b Heat map of the expression analysis of the identified GmNHXs in leaf, stem, and roots of the Gujosoya-2 (salt sensitive) and MAU-47 (salt tolerant) under saline conditions (0, 100 and 200 mM NaCl)
Out of nine GmNHX genes, six genes GmNHX1, GmNHX2, GmNHX4, GmNHX6, GmNHX7 and GmNHX8 showed highest induction, with 22-, 32-, 30-, 20-, 12- and 17-fold increase in expression levels over their respective controls in roots of MAUS-47 under 200 mM NaCl. On the other hand, expression levels of GmNHX3, GmNHX5 and GmNHX9 were highest in leaves of MAUS-47 treated with 200 mM NaCl with 18-, 16- and 19-fold increase, respectively. Expression levels of GmNHX1 in MAUS-47 roots treated with 100 and 200 mM NaCl treatment showed 13- and 22-fold rise against 3- and tenfold increment in Gujosoya-2 roots under similar NaCl levels. Similar trends were shown by GmNHX2, GmNHX6, GmNHX7, GmNHX8 and GmNHX9 in roots. Whereas, GmNHX4, GmNHX6, GmNHX7 and GmNHX8 expression levels were higher in leaves of MAUS-47 (10-, 8-, 4- and 14-times, respectively) than Gujosoya-2 (6-, 5-, 2-, 6-times, respectively) at 200 mM NaCl treatment. Salinity stress clearly upregulated GmNHXs in leaves of both the cultivars, albeit with a higher magnitude in salt tolerant cultivar. In stem tissues, GmNHX genes especially GmNHX1, GmNHX3, GmNHX4, GmNHX7 and GmNHX8 showed higher expression levels in salt tolerant cultivar. Overall, the gene expression levels of each of the GmNHXs induced in leaf, stem and root tissues in response to salinity stress, and the results were in concurrence with the salt tolerance capabilities of two soybean varieties.
The expression levels of all the genes in all three tissues from both the cultivars were used for hierarchical clustering of genes showing the similar expression patterns (Fig. 6b). The heat-map and the hierarchical clustering analysis revealed that the genes can be clustered into two groups. Cluster 1 represents GmNHX4, GmNHX6 and GmNHX7, whereas the remaining genes (GmNHX1, GmNHX2, GmNHX3, GmNHX5, GmNHX8, and GmNHX9) represented the cluster 2. GmNHX5 and GmNHX9 were closely placed in cluster 2. Similarly, GmNHX1, GmNHX2 and GmNHX8 were placed close to each other too. On the other hand, GmNHX3 was placed distinctly from the other members of cluster 2 genes. In case of cluster 1, GmNHX6 and GmNHX7 were placed in the same branch, whereas GmNHX4 was represented by the separate branch.
Discussion
It is well-documented that NHX gene families-encoded Na+/H+ antiporters play crucial roles in ion homeostasis and membrane trafficking especially in salt-stressed plant cells (Pehlivan et al. 2016). In this study, we attempted to identify NHX genes from the soybean genome and further characterize them via deciphering their phylogenetic relationship, chromosomal localization, identifying conserved motifs, their protein structure prediction, protein–protein interactions and their expression profiling under variable salinity stress.
We obtained a total of 68 NHX orthologs in Soybean (G. max) in primary homology search, and finally, nine NHX members were confirmed. Though previously Chen et al. (2015) reported 10 GmNHXs in soybean genome; however, in the present investigation, we used the cut off value of 70% of the Target %id (i.e., percentage of the orthologous sequence matching the reference plant sequence) to predict the putative GmNHXs with high accuracy. Besides, unlike Chen et al. (2015) we also considered the presence of Na+/H+ exchange activity (using HMMER web server and EggNOG mapper) as an essential factor to confirm the orthologs as putative NHXs in soybean.
The presence of Na+/H+ exchanger domain in all nine GmNHXs (Fig. 3c) confirmed their involvement in Na+ transportation in soybean. The transmembrane-transporter nature of identified GmNHXs was confirmed by the annotation results visualized via WEGO plot (Fig. 2). Phylogenetic analysis of the GmNHXs indicated the closeness between identified and the reference NHXs. Five GmNHXs belonged to group IV of the NHX peptides in the phylogeny, which also contained representatives from Arabidopsis thaliana and Brassica rapa (Fig. 1). All the peptides from group IV were localized on the vacuolar membrane. Similarly, three GmNHXs from group I were also localized on the vacuolar membrane along with their close relatives from Arabidopsis and Brassica. The GmNHX3 from group II peptides also localized on vacuolar membrane followed the same pattern, where peptide represented the closeness towards the representatives from the genus Vigna and Phaseolus. Previous investigations on similar line have demonstrated that the NHX genes are occasionally clustered in three different clades depending upon their localization, mainly the endosomal-, vacuolar-, or plasma membrane-positioning of the protein (Kumari et al. 2018; Wu et al. 2019). All the nine GmNHXs in present investigation were localized on vacuolar membrane. The molecular weight and the pI values for all the GmNHXs were equivalent, indicating the possible similarities in their composition (Table 1). The phosphorylation sites namely PKA and PKC are associated with the membrane transporters (Vialaret et al. 2014), and phosphorylation site analysis also revealed the presence of considerable PKA and PKC phosphorylation sites along with many other sites in the current study (Supplementary Fig. S3). Most of the NHX proteins were encompassing 10 – 12 transmembrane helices, as reported previously (Yamaguchi et al. 2003).
The gene structural variability in terms of number of introns/exons provides the additional evidence regarding the phylogenetic relationship amongst the GmNHXs and the current results were in agreement with the previous such reports of NHXs in soybean (Chen et al. 2015), poplar (Tian et al. 2017), sugar beet (Wu et al. 2019) and grapevine (Ayadi et al. 2020). Ten conserved motifs were identified in the GmNHXs, out of which four were categorized as a part of Na+/H+ exchanger family (Pfam clan:CL0064; Supplementary Table S4) indicating the specific and similar functions for all the putative GmNHXs. The characteristic features of membrane-bound NHX-transporters in plants include highly conserved membrane-spanning pore and cation-binding domain comprising the nonapeptide “FFIYLLPPI” (amiloride-binding site) (Tian et al. 2017; Wu et al. 2019). This site is known for inhibition of cation/H+ exchange in presence of amiloride. Noticeably, we found this sequence to be conserved in the second motif of all the GmNHX peptides suggesting the functional as well as sequence-based resemblance amongst the putative GmNHXs (Supplementary Table S4, Supplementary Fig. S7). Similar kinds of “FFIYLLPPI” based characters of NHXs were observed in other plants including Arabidopsis (Aharon et al. 2003), Ipomoea batatas (Wang et al. 2016) and Zygophyllum xanthoxylum (Wu et al. 2011). During putative 3-D structure prediction, all the GmNHXs exhibited the same protein template (4cz9A) (Supplementary Table S7). All the putative protein models were constructed using same template with highest level of confidence and accuracy. Collectively, the analyses of motifs, domains and 3-D structures revealed a high level of similarity amongst putative GmNHXs. The analyses also reveal the link between motifs/domains/3-D structures with their probable function, as all the motifs/domains/3-D structures demonstrated the sodium exchange as their prime function.
The cis-acting elements are key molecular switches for transcriptional regulation of gene, which govern the gene patterns to regulate biological processes (hormonal fluctuations, environmental stress responsiveness, developmental processes etc.) (Ding et al. 2018; Verma et al. 2019). The phytohormones including ethylene and abscisic acid are crucial for plant growth, development and stress responses (Wang and Huang 2019). The presence of ABREs and EREs in promoter region of GmNHXs hence suggest that they are apparently involved in abscisic acid and ethylene signaling during developmental phases or stress responses in G. max (Table 2). Likewise, many stress responsive elements were also identified in the promoter regions of GmNHXs in present study, similar to previous reports from Poplar trichocarpa (Tian et al. 2017) and Sorghum bicolor (Kumari et al. 2018). Besides, cis-acting elements involved in light responses, circadian control, cell cycle regulation and zein metabolism were also identified in the promoter region of GmNHXs. This suggest possible involvement of the GmNHXs in crucial plant regulatory events such as phytohormonal alterations, stress responses and cellular growth/metabolism. Besides, the occurrence of different cis-elements also indicate cross-talk among the diverse signalling and that based on a specific element, specific signalling pathway could be induced into action for better cellular homeostasis under stress conditions (Agarwal et al. 2018).
Currently there are 684 miRNA precursors and 756 mature miRNA sequences of soybean present in the miRBase (release 22.1, Kozomara et al. 2019). The extensive miRNA network constructed using the GmNHXs and their respective targeting-miRNAs showed 75 different miRNAs interconnected with nine different GmNHXs (Fig. 4a). These miRNAs belonged to different miRNA families like miR167, miR171, miR393, miR482, miR1508, miR1512, and miR5380. All these miRNAs from G. max have been characterized as stress-responsive (Ramesh et al. 2019). The miR167 family is known to directly regulate certain members of auxin responsive factors (ARFs) (Han et al. 2014). In crops like maize, miR167 was identified as negative regulator of phosholopase C and displayed inhibition under drought as well as abscisic acid (Wei et al. 2009). The nodule specific miR171 family is known to target the GRAS family transcription factors, and is reported to be involved in low temperature stress responses (Ramesh et al. 2019). Furthermore, miR393 is also a nodule specific family which has been demonstrated to play vital roles during the nodule development. MiR393 plays crucial role of auxin signaling via mediating the expression levels of auxin receptors TIR1/AFB3 (Cai et al. 2017). The miR482 targets primarily resistance (R) gene receptor kinases (majorly the defense-related kinases), whereas the miR1512 targets the genes encoding protein Copine I-like calmodulin-binding proteins. Both the miR482 and miR1512 play important roles in nodule development (Li et al. 2010). The miR1508 members are known to regulate the PPR genes (Pentatricopeptide Repeats) which modulates cold and drought mediated stress responses in soybean (Sun et al. 2020). The miRNA network in current investigation demonstrated the involvement of all the above-mentioned miRNA families with the putative GmNHXs as their targets. Apart from these miRNA families, the miRNAs with common GmNHX target (miR1510b-5p, miR1536, and miR1520r) have also been proved as stress-responsive (Ramesh et al. 2019). Collectively, these miRNA families are proved to play the role of the crucial regulatory factors during the stress responses (drought, cold, abscisic acid etc.) and development (nodules). This in turns highlights the importance of predicted GmNHXs during the stress responses and plant development. The differential expression patterns of the GmNHXs in the current investigation hence can be linked with the stress responsive miRNAs targeting the GmNHXs.
Protein–protein interaction network analysis confirms the interaction of GmNHXs with other proteins including NADH-cytochrome b5 reductase, starch synthase, proteins containing H_HPPase domain and proteins from potassium transporter family (Fig. 4b). The NADH-cytochrome b5 reductase is known to be an electron donor during important biochemical processes such as fatty acid desaturation and increasing the unsaturated fatty acid levels. This in turn stimulates the membrane H+-ATPase (Wayne et al. 2013; Oh et al. 2016). This function of the NADH-cytochrome b5 reductase indicates its link with the NHXs, which is also shown to be functionalized via electrochemical proton gradient across the membrane generated by two H+-pumps; H+-ATPase and H+-PPase (Bao et al. 2009; Wu et al. 2011). The H+-PPase related proteins are potassium dependent membrane bound pyrophosphatases (proton transporters) which are important during the stress responses and plant growth and development (Zhang et al. 2020). Current study also displayed the presence of starch synthase; a prime player during the process of starch synthesis, which is activated by the potassium (Parveen et al. 2020).
Current findings postulate the inter-connected network of the H+-ATPase related cytochrome b5 reductase, H+-PPase related potassium dependent transporters, starch synthase as well as proteins from potassium transporters family. The maintenance of the Na+/K+ is one of the essential event during the stress (salinity and drought) mediated adaptations in plants (Khare et al. 2015). Ultimately, balanced Na+/K+ ratio is essentially required for the optimal stomatal function, protein synthesis, cell osmoregulation, photosynthesis and turgor maintenance (Parveen et al. 2020). Therefore, involvement of the proteins related to the potassium transport in the protein interaction networks along with GmNHXs in current investigation is justifiable.
Further to establish the relationship between the salinity stress and the corresponding expression levels of GmNHXs in the two G. max cultivars with contrasting salt tolerance abilities, quantitative real-time (qRT) PCR analysis was performed using the transcripts from different plant organs including roots, stem with or without varying levels of salinity stress. Membrane-bound transporters like NHX are known to be involved in Na+ exclusion from the cytoplasm and are, therefore, crucial in efflux, compartmentalization, and homeostasis of Na+ across the cell (Blumwald 2000; Zhu 2003; Kumari et al. 2018). Since a probable association between stress responsive cis-acting element and the stress-responsive miRNAs with GmNHXs was already proved, the variability in the expression patterns of GmNHXs was predictable. In current study, irrespective of the level of salinity imposed and salt tolerance abilities of the cultivars, GmNHX genes were up-regulated in leaf, stem and root tissues of both the cultivars as a response to salinity stress (Fig. 6a). Roots, being the first organ/tissue exposed to the salinity have shown considerably higher level of induction in GmNHX genes. Leaves also exhibited the induction of GmNHXs; however, the extent was lesser than the roots, while lowest salinity-driven gene(s) upregulation was observed in stems. The higher expression patterns of these sodium transporters may be responsible for the exclusion as well as better compartmentalization of the Na+; which will directly help to maintain the Na+ homeostasis across the cells. In summary, all GmNHXs were upregulated by salinity in leaf, stem and root tissues and the results were in concurrence with the salt tolerance capabilities of two soybean varieties. The behavioral pattern of these GmNHXs under saline conditions can thus be considered as an important character defining their salt sensitivity or tolerance capabilities. Salinity mediated differential expression patterns in the NHXs were previously reported in five NHXs from Beta vulgaris (Wu et al. 2019), eight NHXs from Populus trichocarpa (Tian et al. 2017) and six NHXs from Sorghum bicolor (Kumari et al. 2018). The hierarchical clustering (Fig. 6b) revealed that two clusters separated as per the activities of GmNHXs in all the tissues under saline conditions from both the cultivars.
Conclusion
The present study on genome-wide analysis and screening of G. max signifies the presence of nine GmNHX genes in the soybean genome. Bioinformatics analysis confirmed that all the identified GmNHXs were sodium transporters positioned on vacuolar membrane and involved in sodium transport across the cells. The amiloride binding site in NHXs implied their apparent involvement in ABA signalling pathway. The cis-elements present in NHX promoter regions revealed their roles plant’s responsiveness for salinity stress and guarding the cells. The identified GmNHXs were predicted to be potential targets of 75 different miRNAs. Real-time quantitative PCR analysis indicated that all GmNHX genes in different plant tissues were significantly up-regulated by salinity stress, more so in salt-tolerant cultivar, thus confirming the association of these genes with salinity stress responses of soybean. These findings hold significance in providing theoretical basis and functional validation of candidate gene resources in relation to salinity stress tolerance in crop species.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was financially supported by the Department of Atomic Energy, Board of Research in Nuclear Sciences (DAE-BRNS), Government of India [37(1)/14/30/2018-BRNS]. Authors also acknowledge the use of facilities created under DST-FIST program and DBT-STAR STATUS schemes of Departments of Science and Technology and Biotechnology, respectively.
Author contributions
VK conceived the idea. SJ, KK and TK conducted the experiment, AKS, PS and VK supervised the work. Each author contributed substantially to the work. All the authors have seen and approved the manuscript and its contents.
Complaince with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Agarwal PK, Dave A, Agarwal P. Transcriptional regulation of salinity stress: role and spatio-temporal expressions of ion-transporter gene promoters. Biol Plant. 2018;62:641–646. doi: 10.1007/s10535-018-0815-2. [DOI] [Google Scholar]
- Aharon GS, Apse MP, Duan S, et al. Characterization of a family of vacuolar Na+/H+ antiporters in Arabidopsis thaliana. Plant Soil. 2003;253:245–256. doi: 10.1023/a:1024577205697. [DOI] [Google Scholar]
- Apse MP, Sottosanto JB, Blumwald E. Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 2003;36:229–239. doi: 10.1046/j.1365-313x.2003.01871.x. [DOI] [PubMed] [Google Scholar]
- Ayadi M, Martins V, Ben Ayed R, et al. Genome wide identification, molecular characterization, and gene expression analyses of grapevine NHX antiporters suggest their involvement in growth, ripening, seed dormancy, and stress response. Biochem Genet. 2020;58:102–128. doi: 10.1007/s10528-019-09930-4. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao AK, Wang SM, Wu GQ, et al. Overexpression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.) Plant Sci. 2009;176:232–240. doi: 10.1016/j.plantsci.2008.10.009. [DOI] [Google Scholar]
- Bao A-K, Du B-Q, Touil L, et al. Co-expression of tonoplast Cation/H + antiporter and H + -pyrophosphatase from xerophyte Zygophyllum xanthoxylum improves alfalfa plant growth under salinity, drought and field conditions. Plant Biotechnol J. 2016;14:964–975. doi: 10.1111/pbi.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil E, Blumwald E. The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr Opin Plant Biol. 2014;22:1–6. doi: 10.1016/j.pbi.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Blumwald E. Sodium transport and salt tolerance in plants. Curr Opin Cell Biol. 2000;12:431–434. doi: 10.1016/S0955-0674(00)00112-5. [DOI] [PubMed] [Google Scholar]
- Cai Z, Wang Y, Zhu L, Tian Y, Chen L, Sun Z, Ullah I, Li X. GmTIR1/GmAFB3-based auxin perception regulated by miR393 modulates soybean nodulation. New Phytol. 2017;215:672–686. doi: 10.1111/nph.14632. [DOI] [PubMed] [Google Scholar]
- Carbon S, Douglass E, Dunn N, et al. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HT, Chen X, Wu BY, et al. Whole-genome identification and expression analysis of K+ efflux antiporter (KEA) and Na+/H+ antiporter (NHX) families under abiotic stress in soybean. J Integr Agric. 2015;14:1171–1183. doi: 10.1016/s2095-3119(14)60918-7. [DOI] [Google Scholar]
- Dai X, Zhuang Z, Zhao PX. PsRNATarget: a plant small RNA target analysis server (2017 release) Nucleic Acids Res. 2018;46:W49–W54. doi: 10.1093/nar/gky316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Li J, Pan Y, et al. Genome-wide identification and expression analysis of the UGlcAE gene family in tomato. Int J Mol Sci. 2018;19:1583. doi: 10.3390/ijms19061583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, et al. (2005) Protein identification and analysis tools on the ExPASy Server. In: The Proteomics Protocols Handbook. Humana Press, pp 571–607
- Han YQ, Hu Z, Zheng DF, Gao YM. Analysis of promoters of microRNAs from a Glycine max degradome library. J Zhejiang Univ Sci B. 2014;15(2):125–132. doi: 10.1631/jzus.b1300179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Jin J, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Heller D, et al. EggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas R, Rubio L, Cagnac O, et al. The K+/H+ antiporter LeNHX2 increases salt tolerance by improving K+ homeostasis in transgenic tomato. Plant, Cell Environ. 2013;36:2135–2149. doi: 10.1111/pce.12109. [DOI] [PubMed] [Google Scholar]
- Jiang X, Leidi EO, Pardo JM. How do vacuolar NHX exchangers function in plant salt tolerance? Plant Signal Behav. 2010;5:792–795. doi: 10.4161/psb.5.7.11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare T, Kumar V, Kishor PBK. Na+ and Cl− ions show additive effects under NaCl stress on induction of oxidative stress and the responsive antioxidative defense in rice. Protoplasma. 2015;252:1149–1165. doi: 10.1007/s00709-014-0749-2. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Birgaoanu M, Griffiths-Jones S. MiRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Khare T. Differential growth and yield responses of salt-tolerant and susceptible rice cultivars to individual (Na+ and Cl−) and additive stress effects of NaCl. Acta Physiol Plant. 2016;38:1–9. doi: 10.1007/s11738-016-2191-x. [DOI] [Google Scholar]
- Kumar S, Stecher G, Li M, et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari PH, Kumar SA, Ramesh K, et al. Genome-wide identification and analysis of Arabidopsis sodium proton antiporter (NHX) and human sodium proton exchanger (NHE) homologs in Sorghum bicolor. Genes (Basel) 2018;9:236. doi: 10.3390/genes9050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidi EO, Barragán V, Rubio L, et al. The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 2010;61:495–506. doi: 10.1111/j.1365-313x.2009.04073.x. [DOI] [PubMed] [Google Scholar]
- Lescot M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Deng Y, Wu T, Subramanian S, Yu O. Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol. 2010;153(4):1759–1770. doi: 10.1104/pp.110.156950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Wang X, Ma B, et al. Expression of a Na+/H+ antiporter RtNHX1 from a recretohalophyte Reaumuria trigyna improved salt tolerance of transgenic Arabidopsis thaliana. J Plant Physiol. 2017;218:109–120. doi: 10.1016/j.jplph.2017.07.015. [DOI] [PubMed] [Google Scholar]
- Mészáros B, Erdös G, Dosztányi Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018;46:W329–W337. doi: 10.1093/nar/gky384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Oh YJ, Kim H, Seo SH, et al. Cytochrome b5 reductase 1 triggers serial reactions that lead to iron uptake in plants. Mol Plant. 2016;9:501–513. doi: 10.1016/j.molp.2015.12.010. [DOI] [PubMed] [Google Scholar]
- Ohnishi M, Fukada-Tanaka S, Hoshino A, et al. Characterization of a novel Na+/H+ antiporter gene InNHX2 and comparison of InNHX2 with InNHX1, which is responsible for blue flower coloration by increasing the vacuolar pH in the Japanese morning glory. Plant Cell Physiol. 2005;46:259–267. doi: 10.1093/pcp/pci028. [DOI] [PubMed] [Google Scholar]
- Otasek D, Morris JH, Bouças J, et al. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019;20:185. doi: 10.1186/s13059-019-1758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parveen Anwar-Ul-Haq M, Aziz T, Aziz O, Maqsood L. Potassium induces carbohydrates accumulation by enhancing morpho-physiological and biochemical attributes in soybean under salinity. Arch Agron Soil Sci. 2020 doi: 10.1080/03650340.2020.1769075. [DOI] [Google Scholar]
- Pehlivan N, Sun L, Jarrett P, et al. Co-overexpressing a plasma membrane and a vacuolar membrane sodium/proton antiporter significantly improves salt tolerance in transgenic Arabidopsis plants. Plant Cell Physiol. 2016;57:1069–1084. doi: 10.1093/pcp/pcw055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS. Plant and yeast NHX antiporters: roles in membrane trafficking. J Integr Plant Biol. 2012;54:66–72. doi: 10.1111/j.1744-7909.2012.01097.x. [DOI] [PubMed] [Google Scholar]
- Ramesh SV, Govindasamy V, Rajesh MK, Sabana AA, Praveen S. Stress-responsive miRNAome of Glycine max (L.) Merrill: molecular insights and way forward. Planta. 2019;249:1267–1284. doi: 10.1007/s00425-019-03114-5. [DOI] [PubMed] [Google Scholar]
- Ramesh SV, Govindasamy V, Rajesh MK, et al. Stress-responsive miRNAome of Glycine max (L.) Merrill: molecular insights and way forward. Planta. 2019;249:1267–1284. doi: 10.1007/s00425-019-03114-5. [DOI] [PubMed] [Google Scholar]
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rosales MP, Gálvez FJ, Huertas R, et al. Plant NHX cation/proton antiporters. Plant Signal Behav. 2009;4:265–276. doi: 10.4161/psb.4.4.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelke DB, Nikalje GC, Chambhare MR, et al. Na + and Cl − induce differential physiological, biochemical responses and metabolite modulations in vitro in contrasting salt-tolerant soybean genotypes. 3 Biotech. 2019;9:1–15. doi: 10.1007/s13205-019-1599-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TJ, Fan L, Yang J, et al. A Glycine max sodium/hydrogen exchanger enhances salt tolerance through maintaining higher Na + efflux rate and K+/Na+ ratio in Arabidopsis. BMC Plant Biol. 2019;19:469. doi: 10.1186/s12870-019-2084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Jing Y, Wang X, Zhang Y, Zhang Y, Ai J, Li J, Jin L, Li W, Li Y. Gma-miR1508a confers dwarfing, cold tolerance, and drought sensitivity in soybean. Mol Breed. 2020;40:44. doi: 10.1007/s11032-020-01116-w. [DOI] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakkoli E, Rengasamy P, McDonald GK. High concentrations of Na+ and Cl− ions in soil solution have simultaneous detrimental effects on growth of faba bean under salinity stress. J Exp Bot. 2010;61:4449–4459. doi: 10.1093/jxb/erq251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakkoli E, Fatehi F, Coventry S, et al. Additive effects of Na+ and Cl− ions on barley growth under salinity stress. J Exp Bot. 2011;62:2189–2203. doi: 10.1093/jxb/erq422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Chang E, Li Y, et al. Expression and integrated network analyses revealed functional divergence of NHX-type Na+/H+ exchanger genes in poplar. Sci Rep. 2017;7:1–17. doi: 10.1038/s41598-017-02894-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Lakhanpal N, Singh K. Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa. BMC Genomics. 2019;20:227. doi: 10.1186/s12864-019-5593-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialaret J, Di Pietro M, Hem S, et al. Phosphorylation dynamics of membrane proteins from Arabidopsis roots submitted to salt stress. Proteomics. 2014;14:1058–1070. doi: 10.1002/pmic.201300443. [DOI] [PubMed] [Google Scholar]
- Wang J, Huang R. Modulation of ethylene and ascorbic acid on reactive oxygen species scavenging in plant salt response. Front Plant Sci. 2019;10:319. doi: 10.3389/fpls.2019.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhai H, He S, et al. A vacuolar Na+/H+ antiporter gene, IbNHX2, enhances salt and drought tolerance in transgenic sweetpotato. Sci Hortic. 2016;201:153–166. doi: 10.1016/j.scienta.2016.01.027. [DOI] [Google Scholar]
- Wani SH, Kumar V, Khare T, et al. Engineering salinity tolerance in plants: progress and prospects. Planta. 2020;251:76. doi: 10.1007/s00425-020-03366-6. [DOI] [PubMed] [Google Scholar]
- Wayne LL, Wallis JG, Kumar R, et al. Cytochrome b5 reductase encoded by CBR1 is essential for a functional male gametophyte in Arabidopsis. Plant Cell. 2013;25:3052–3066. doi: 10.1105/tpc.113.113324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Zhang D, Xiang F, Zhang Z. Differentially expressed miRNAs potentially involved in the regulation of defense mechanism to drought stress in maize seedlings. Int J Plant Sci. 2009;170:979–989. doi: 10.1086/605122. [DOI] [Google Scholar]
- Wu GQ, Wang Q, Bao AK, Wang SM. Amiloride reduces sodium transport and accumulation in the succulent xerophyte Zygophyllum xanthoxylum under salt conditions. Biol Trace Elem Res. 2011;139:356–367. doi: 10.1007/s12011-010-8662-9. [DOI] [PubMed] [Google Scholar]
- Wu G-Q, Wang J-L, Li S-J. Genome-Wide identification of Na+/H+ antiporter (NHX) genes in sugar beet (Beta vulgaris L.) and their regulated expression under salt stress. Genes. 2019;10:401. doi: 10.3390/genes10050401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Apse MP, Shi H, Blumwald E. Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc Natl Acad Sci USA. 2003;100:12510–12515. doi: 10.1073/pnas.2034966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43:W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Han Y, Wu D, et al. Salt and cadmium stress tolerance caused by overexpression of the Glycine Max Na+/H+ Antiporter (GmNHX1) gene in duckweed (Lemna turionifera 5511) Aquat Toxicol. 2017;192:127–135. doi: 10.1016/j.aquatox.2017.08.010. [DOI] [PubMed] [Google Scholar]
- Yasuta Y, Kokubun M. Salinity tolerance of super-nodulating soybean genotype En-b0-1. Plant Prod Sci. 2014;17:32–40. doi: 10.1626/pps.17.32. [DOI] [Google Scholar]
- Ye J, Zhang Y, Cui H, et al. WEGO 2.0: a web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018;46:W71–W75. doi: 10.1093/nar/gky400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You FM, Huo N, Gu YQ, et al. BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics. 2008;9:253. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng X, Wang L, et al. The structure, functional evolution, and evolutionary trajectories of the H+-PPase gene family in plants. BMC Genomics. 2020;21:195. doi: 10.1186/s12864-020-6604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003;6:441–445. doi: 10.1016/S1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.