Abstract
Tumor-infiltrating immune cells as integral component of the tumor microenvironment are associated with tumor progress, prognosis and responses to immunotherapy. Genetic variants have been demonstrated to impact tumor-infiltrating, underscoring the heritable character of immune landscape. Therefore, identification of immunity quantitative trait loci (immunQTLs), which evaluate the effect of genetic variants on immune cells infiltration, might present a critical step toward fully understanding the contribution of genetic variants in tumor development. Although emerging studies have demonstrated the determinants of germline variants on immune infiltration, no database has yet been developed to systematically analyze immunQTLs across multiple cancer types. Using genotype data from TCGA database and immune cell fractions estimated by CIBERSORT, we developed a computational pipeline to identify immunQTLs in 33 cancer types. A total of 913 immunQTLs across different cancer types were identified. Among them, 5 immunQTLs are associated with patient overall survival. Furthermore, by integrating immunQTLs with GWAS data, we identified 527 immunQTLs overlapping with known GWAS linkage disequilibrium regions. Finally, we constructed a user-friendly database, CancerImmunityQTL (http://www.cancerimmunityqtl-hust.com/) for users to browse, search and download data of interest. This database provides an informative resource to understand the germline determinants of immune infiltration in human cancer and benefit from personalized cancer immunotherapy.
INTRODUCTION
Tumor microenvironment (TME) comprises a complex milieu of non-malignant cells including vascular vessels, fibroblasts, extracellular matrix and immune infiltrates, which can interact closely with tumor cells and affect tumor growth and metastasis (1,2). As the integral component of tumor microenvironment, the immune infiltrates play a critical role in tumor progress, clinical outcome and responses to immunotherapy. Correlations between the levels of immune cell infiltration in tumors and clinical outcome have been investigated in many cancers, identifying several cell types that could be regarded as prognosis markers (3). The immune infiltrate is often a heterogeneous mixture of distinct cell types which have different effects on tumor control. The density and location of these immune cells could vary according to cancer type and are very diverse from patient to patient, resulting in only a subset of patients benefit from immunotherapy (3,4). To explore the role of this underappreciated determinant of heterogeneity among patients with cancer, researchers tried to link germline genetic variants to cancer risk by altering immune cell contents in the corresponding cancer target tissue (5,6).
Individual genetic characteristics play an essential part in the development of tumor, which also are crucial factors in determining tumor susceptibility. The contribution of inherited genetic factors to the causation of cancer is about 27–42% (7). Apart from the rare high penetrance mutation, single nucleotide polymorphisms (SNPs) as the most frequent genetic variants in humans could be responsible for the development of malignant diseases (8,9). Large-scale genome-wide association studies (GWAS) have identified great amount of trait/disease-associated variants, while these variants explain only a small fraction of the heritability of complex traits, and the identification of causal variants, genes, and disease mechanisms still needs to be further explored (10–12). Quantitative trait locus (QTL) analysis is considered as a useful approach to evaluate the effects of genetic variants on intermediate molecular phenotypes and discern the causal variant within GWAS loci (13–15). Inherited genetic variants have been demonstrated to impact baseline and induced host immune responses, underscoring the heritable character of immune landscape (16,17). Moreover, results from GWAS have identified risk-associated variants that could affect anti-tumor immune response. For example, the first GWAS of cervical cancer identified a locus in MICA which encodes a membrane-bound protein acting as a ligand for NKG2D to activate natural killer (NK) and T cells (18). Inflammation-related genetic variations could influence cancer immune response thereby affecting prognosis (19). The evaluation of common genetic variation affecting immune infiltration which refers to leading different fractions of infiltrating immune cells between individuals, is defined as immunity quantitative trait loci (immunQTLs) analysis. Thus, identification of immunQTLs might provide a resource linking immunology with genomics in human cancer and contribute to understanding the importance of genetic variants in tumor development.
Considering the significance of immunQTLs, emerging studies about the influence of genetic variation on immune infiltration have been conducted both in solid tumors and hematological malignancies (6,20–22). These studies revealed widespread impact of genetic variants on immune landscape in various cancer type, which could serve as potential prognostic and treatment markers. However, no database has been developed to systematically analyze the immunQTLs across multiple cancer types. To bridge this gap, we developed a computational pipeline to systematically perform immunQTL analysis by integrating genotype data and immune cell proportions profiled by RNA-seq data from The Cancer Genome Atlas (TCGA). We further associated the identified immunQTLs with patient overall survival time and loci in GWAS linkage disequilibrium (LD) regions. We identified hundreds of immunQTLs reached significant level across different cancer types, and constructed a user-friendly database, CancerImmunityQTL (http://www.cancerimmunityqtl-hust.com/), for users to conveniently browse, search and download data of interest. This comprehensive immunQTL resource helps to effectively evaluate the impact of germline SNPs on cancer-immune phenotypes, and support a path toward personalized cancer immunotherapy.
DATA COLLECTION AND PROCESSING
Estimate of immune cell fractions and processing
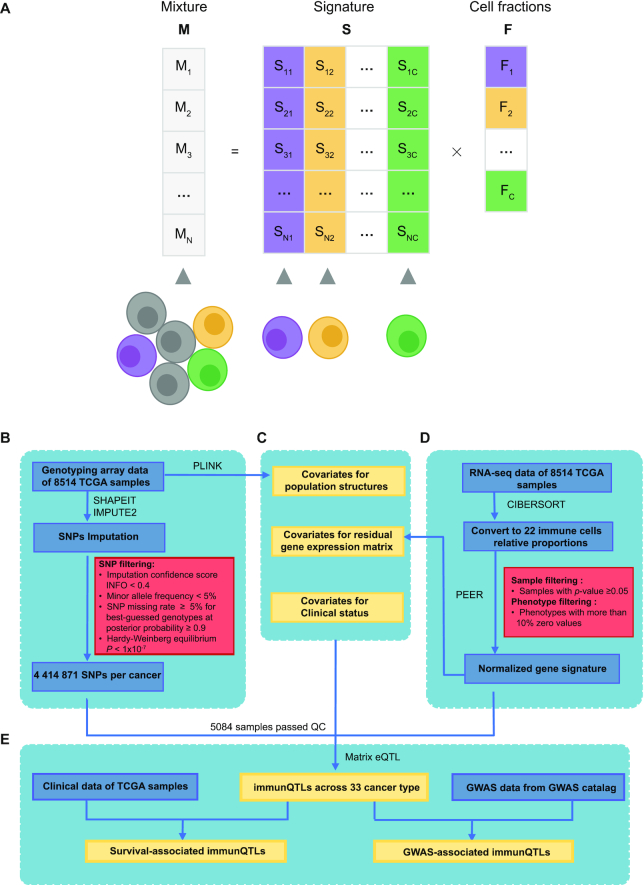
Using bioinformatics approaches based on immune-specific marker genes or expression signature, tumor-infiltrating immune cells can be quantified from microarray or RNA sequencing (RNA-seq) data of human tumors. Deconvolution algorithms consider gene expression profiles of a heterogeneous sample as the convolution of the gene expression levels of the different cells, and estimate the unknown cell fractions leveraging on a signature matrix describing the cell-type-specific expression profiles (23). Through this approach, the relative fractions of interested cell types can be estimated quantitatively (Figure 1A). CIBERSORT is one of the methods based on deconvolution algorithms to characterize leukocyte composition with LM22 (24), which is a signature matrix file consisting of 547 genes that accurately distinguish 22 mature human hematopoietic populations (Table 1). With the application of CIBERSORT algorithm, we quantified relative percent of 22 immune cell subsets based on RNA-seq data downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/), and an empirically defined global P-value for the deconvolution was also calculated for each sample. For each cancer type, tumor samples were filtered using the following criterion: samples with P-value ≥ 0.05. Besides, immune cell subsets were filtered by the following criterion: phenotypes with more than 10% zero values which represent negative estimates. Finally, 17 immune cell subsets were included for analysis. To minimize the effects of outliers on the regression scores (25,26), the relative percent for each immune cell subset across samples per cancer type were transformed into a standard normal distribution based on rank (Figure 1D).
Figure 1.
Identification of immunQTLs in the CancerImmunityQTL database. (A) Deconvolution method to estimate the relative fractions of cell types. In the mixture M, the expression of a gene is considered as a linear combination of the expression of that gene in different cell types weighted by the relative fractions F of the cell types in mixture. Signature matrix S represents a summary of average expression profiles. (B) The procedure for collecting and processing genotype data. (C) Covariates included in immunQTL mapping. (D) The procedure of estimate on immune cell fractions and processing. (E) Identification of immunQTLs, survival-associated immunQTLs and GWAS-related immunQTLs.
Table 1.
Overview of 22 immune cell types
| LM22 cells | Cell type description |
|---|---|
| B cells | B cells naive |
| B cells memory | |
| PCs | Plasma cells |
| CD8 T cells | T cells CD8 |
| CD4 T cells | T cells CD4 naive |
| T cells CD4 memory resting | |
| T cells CD4 memory activated | |
| T cells follicular helper | |
| T cells regulatory (Tregs) | |
| Gamma delta T cells | T cells gamma delta |
| NK cells | NK cells resting |
| NK cells activated | |
| Monocytes and macrophages | Monocytes |
| Macrophages M0 | |
| Macrophages M1 | |
| Macrophages M2 | |
| Dendritic cells | Dendritic cells resting |
| Dendritic cells activated | |
| Mast cells | Mast cells resting |
| Mast cells activated | |
| Eos | Eosinophils |
| PMNs | Neutrophils |
Genotype data collection, imputation and processing
Genotype data (level 2) of 10 944 samples across 33 cancer types were obtained from the TCGA data portal (https://portal.gdc.cancer.gov/), which detected the genotypes using Affymetrix SNP Array 6.0 containing 898 620 SNPs. To increase the power for immunQTL discovery, we conducted imputation for autosomal variants in all samples across all cancer type, with 1000 Genomes Phase 3 as the reference panel as described in our previous study (27). To improve computation efficiency, we adopted the two-step procedure: first to produce haplotype estimates with SHAPEIT, and then used IMPUTE2 to impute untyped genotypes (28). Following criteria were used to exclude SNPs: (i) imputation confidence score, INFO < 0.4, (ii) minor allele frequency (MAF) < 5%, (iii) SNP missing rate ≥ 5% for best-guessed genotypes at posterior probability ≥ 0.9 and (iv) Hardy–Weinberg Equilibrium P-value < 1 × 10−7 estimated by Hardy–Weinberg R package (Figure 1B). After imputation and quality filtering, an average of 4 414 871 SNPs per cancer type were remained in immunQTL analysis.
Covariates
To increase the sensitivity in QTL analyses, covariates are often included to correct for known and unknown confounders (29). We performed principal components analysis (PCA) by PLINK (30) and considered the top five principal components as covariates to control for ethnicity differences. Furthermore, to remove confounding factors such as batch effects from expression data, we included first 15 PEER factors that captured the global variance in gene expression using PEER software (31). Since the PEER factors were highly correlated with estimated cell fractions, which would decrease detection sensitivity in immunQTL analysis. Therefore, we regressed estimated cell fractions from all gene within RNA-seq data for all tissues, and the resulting residual gene expression matrix could be used to generate a new group of 15 PEER factors. Besides, other common confounders such as age, sex and tumor stage were also included as covariates to remove the potential effects of clinical status (26,32,33) (Figure 1C).
Identification of immunQTLs
For each cancer type, the effects of genetic variation on immune infiltration were evaluated by linear regression using MatrixEQTL (34) with the adjustment of covariates mentioned above (Figure 1E). SNPs with false discovery rate (FDR) < 0.1 calculated by MatrixEQTL were defined as immunQTLs.
Identification of survival-associated immunQTLs
Since the levels of infiltrating immune cells are associated with patient outcome (35), immunQTLs may influence the prognosis by affecting infiltrating immune cells. To prioritize promising immunQTLs, we further explored the association between immunQTLs and patient survival times. For each immunQTL, samples were classified into three groups: homozygous genotype AA, heterozygous genotype Aa and homozygous genotype aa (A and a respectively represent major and minor allele of one SNP). We compared the differences of survival time between three groups by using log-rank test, and Kaplan–Meier (KM) curves were constructed to display the results. immunQTL with FDR < 0.05 based on Benjamini & Hochberg method were defined as survival-associated immunQTLs (36).
Identification of GWAS-associated immunQTLs
Although GWAS have identified an unprecedented number of genetic variants, these approaches do not necessarily pinpoint the causal variants (11). Thus, we integrated the immunQTLs with existing GWAS risk loci to facilitate interpretation of GWAS results. Risk tag SNPs identified in GWAS studies were downloaded from the National Human Genome Research Institute (NHGRI) GWAS catalog (http://www.ebi.ac.uk/gwas/, accessed by June 2020) (37). Then we obtained GWAS linkage disequilibrium (LD) regions of these risk tag SNPs from SNAP database (https://personal.broadinstitute.org/plin/snap/ldsearch.php) (38) with parameters (SNP data set: 1000 Genomes; r2 (the square of the Pearson correlation coefficient of linkage disequilibrium) threshold: 0.5; population panel: CEU (Utah Residents with Northern and Western European Ancestry), and distance limit: 100 kb). immunQTLs that overlapped with GWAS tag SNPs and LD SNPs (r2 ≥ 0.5) were defined as GWAS-related immunQTLs.
Variants annotation
All the genetic variants identified in immunQTL analysis have been annotated using SnpEff software, which enables rapid analyses of whole-genome sequencing data (39). The process of annotation is based on variants genomic locations, the annotated genomic locations include intronic, untranslated region, upstream, downstream, splice site, or intergenic regions. In addition to the effects of variants in genome sequences, their located or nearby genes were also available.
Enrichment analysis
To assess the enrichment of immunQTLs in GWAS linkage disequilibrium regions (r2 ≥ 0.8), we generated a control data set of non-immunQTL SNPs with minor allele frequency (MAF) matched to immunQTL SNPs among each cancer type (40). Two-tailed Pearson's χ2 test or Fisher's exact test were applied to perform enrichment analysis for the following 2 × 2 tables: columns; immunQTL SNPs and non-immunQTL SNPs, rows; SNPs within and not within GWAS loci.
DATABASE CONTENT
Samples in CancerImmunityQTL
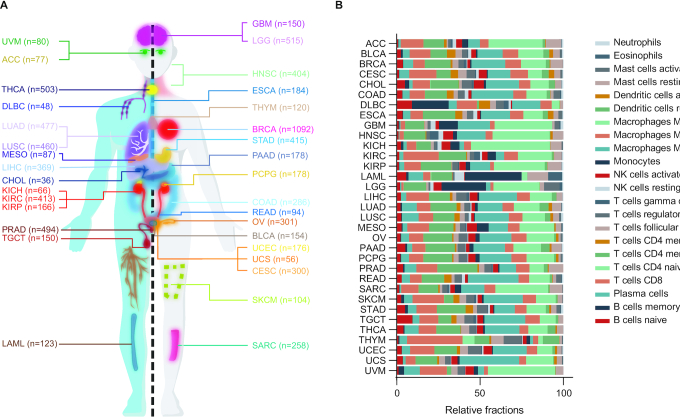
Before sample filter, 8514 tumor samples with both genotype data and phenotype data available for 33 cancer types were included. After quality control for samples based on P-value calculated in CIBERSORT, a total of 5084 tumor samples with both genotype data and credible estimated immune cell fractions were used for immunQTL analysis (Figure 2A). The sample size for each cancer type ranged from 10 in adrenocortical carcinoma (ACC) or pheochromocytoma and paraganglioma (PCPG) to 823 in invasive breast carcinoma (BRCA) (Supplementary Table S1). After genotype imputation and quality control, an average of 4 414 871 SNPs per cancer type were used for analyses, ranging from 2 944 254 for ACC to 5 120 270 for acute myeloid leukemia (LAML). After removing estimated cell fractions with more than 10% zero values, 17 types of immune cells were used for analyses.
Figure 2.
immunQTLs statistics across 33 cancer types. (A) The included cancer types in our study and sample size for each cancer type. (B) Bar plot indicates relative percent of 22 immune cells estimated by CIBERSORT, the sum up of all cell types fractions is equal to one.
immunQTLs in CancerImmunityQTL
The CancerImmunityQTL mainly consists of three datasets which are immunQTLs, survival-immunQTLs and GWAS-immunQTLs. We have listed SNPs with P-value <1 × 10−3 on the website and SNPs with P-value < 0.05 could be acquired in download page, the summary of these parts could be found in Supplementary Tables S1 and S2, respectively. In the immunQTL analysis, a total of 913 immunQTL-immune cell pairs across 11 cancer types were identified at the level of FDR < 0.1, with a median of 83 immunQTLs per cancer type, ranging from 1 in BRCA or liver hepatocellular carcinoma (LIHC) to 253 in stomach adenocarcinoma (STAD) (Table 2). The detected number hierarchy of immunQTLs was consistent with previous study (6). Since the limited number of immunQTLs, the association between number of immunQTLs and number of samples did not attain statistical significance, but presented a tendency of positively correlated (Spearman correlation Rs = 0.032, P-value = 0.621, Supplementary Figure S1). Macrophages M2 are the most frequent infiltrating immune cells across all cancer types, which is widely considered to favor tumor growth and spreading (41). Macrophages M0, T cells CD4 memory resting and T cells CD8 are also the common immune cell types in tumor microenvironment (Figure 2B). The germline variants derived from genotype imputation accounted for an average of 78.14% of immunQTLs in 11 cancer types (Supplementary Table S3).
Table 2.
Summary of immunQTLs at FDR < 0.1 for each cancer type in CancerImmunityQTL
| Cancer type | Full name | No. of sample | No. of sample (after QC) | No. of SNPs | immunQTL-immune cell pairs | immunQTLs | Immune cells | Survival-immunQTLs | GWAS-immunQTLs |
|---|---|---|---|---|---|---|---|---|---|
| BRCA | Breast invasive carcinoma | 1092 | 823 | 4115366 | 1 | 1 | 1 | 0 | 0 |
| COAD | Colon adenocarcinoma | 286 | 131 | 4491421 | 126 | 126 | 6 | 5 | 32 |
| KICH | Kidney Chromophobe | 66 | 11 | 3771773 | 50 | 50 | 1 | 0 | 6 |
| KIRP | Kidney renal papillary cell carcinoma | 166 | 163 | 4894174 | 8 | 8 | 2 | 0 | 7 |
| LIHC | Liver hepatocellular carcinoma | 369 | 58 | 4156507 | 1 | 1 | 1 | 0 | 1 |
| PRAD | Prostate adenocarcinoma | 494 | 44 | 4823458 | 243 | 243 | 7 | 0 | 205 |
| SARC | Sarcoma | 258 | 167 | 4081096 | 8 | 8 | 2 | 0 | 0 |
| STAD | Stomach adenocarcinoma | 415 | 257 | 4306085 | 253 | 253 | 6 | 0 | 210 |
| THCA | Thyroid carcinoma | 503 | 171 | 4870332 | 31 | 31 | 2 | 0 | 0 |
| THYM | Thymoma | 120 | 112 | 4892278 | 187 | 187 | 6 | 0 | 65 |
| UCEC | Uterine Corpus Endometrial Carcinoma | 176 | 94 | 4941208 | 5 | 5 | 2 | 0 | 1 |
Abbreviation: QC, quality control; SNPs, single nucleotide polymorphisms.
The quality control for samples were based on empirically defined global P-value calculated by CIBERSORT, samples with P-value ≥ 0.05 were filtered.
We further linked immunQTLs to patient survival data and known GWAS loci to prioritize promising immunQTLs. A total of five immunQTLs associated with patient overall survival at FDR < 0.05. Based on the GWAS studies gathered in NHGRI GWAS Catalog, we identified 527 immunQTLs that overlapped with GWAS linkage disequilibrium (LD) regions of one or multiple traits at an LD threshold of 0.5. The number of GWAS-immunQTLs ranged from one in LIHC or uterine corpus endometrial carcinoma (UCEC) to 210 in STAD. Enrichment analysis also showed that immunQTL are significantly enriched in GWAS loci compared to non-immunQTL (Supplementary Table S4).
DATABASE CONSTRUCTION AND WEB INTERFACE
CancerImmunityQTL was built based on the NodeJS 8.10.0 (https://nodejs.org/en/) framework, all results mentioned above were stored in MongoDB 3.6.5 (https://www.mongodb.com/) database. As a database with user-friendly web interface for browsing, searching and downloading, CancerImmunityQTL runs on a Linux-based Nginx Web server, combined with ReactJS (https://reactjs.org/) as the JavaScript library. We have tested it on various web browsers, including Google Chrome (preferred), Firefox, Internet Explorer and Safari of macOS. The CancerImmunityQTL website is available online (http://www.CancerImmunityQTL-hust.com/) and requires no registration.
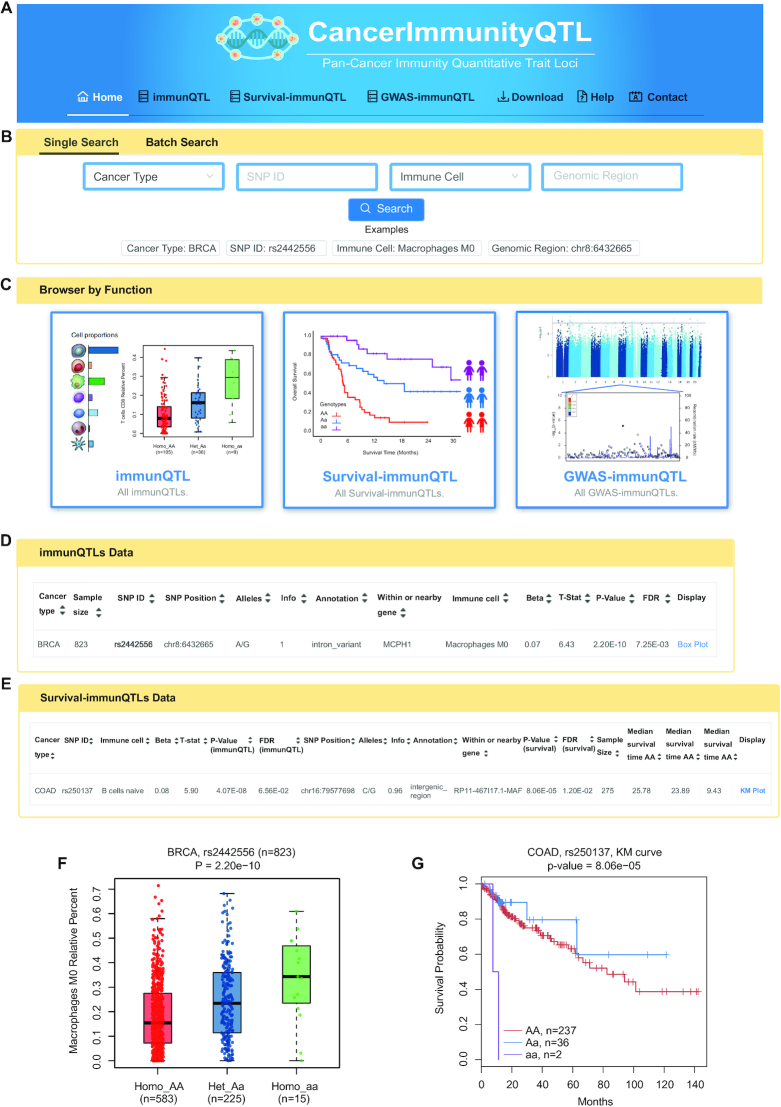
CancerImmunityQTL provides data browsing and querying of three modules. On the ‘home’ page, by clicking on the corresponding button in the browser bar (Figure 3A) or the corresponding images in the ‘Modules’ section at the bottom of this page (Figure 3C), users can enter the ‘immunQTL/survival-immunQTL/GWAS-immunQTL’ pages. Both ‘Single Search’ and ‘Batch Search’ are available for comprehensive queries across all three datasets (Figure 3B). In the ‘Single Search’ section, users can select a specific cancer type or immune cell type and input an SNP ID or genomic region to search immunQTLs across all datasets. After inputting the search terms, the results would be presented as three dynamic tables containing that three datasets. If users do not select specific cancer type, it will return results for all cancer types. The ‘Batch Search’ section allows users to input multiple cancer types, SNPs, immune cells or genomic regions of interest. We also displayed a summary of the sample size, immunQTL pairs number and relative percent of immune cell on the ‘home’ page. Putting the cursor over a cancer name on the-hand side of human anatomy diagram, the matched results will be displayed on the right figures. If users click certain cancer name, the web interface will turn to the search result of this cancer type. All three datasets for each cancer type can be downloaded from the ‘Download’ page. A detailed tutorial showing how the data were collected and processed is available on the ‘Help’ page. CancerImmunityQTL welcomes any feedback by email to the address provided in the ‘Contact’ page.
Figure 3.
Overview of the CancerImmunityQTL database. (A) Browser bar in CancerImmunityQTL. (B) The single and batch search boxes in CancerImmunityQTL. (C) Three modules in CancerImmunityQTL, including immunQTLs, survival-associated immunQTLs, and GWAS-related immunQTLs. (D, F) An example of immunQTL results on the ‘immunQTL’ page and the corresponding boxplot. (E, G) An example of survival-immunQTL results in ‘survival-immunQTL’ page and the corresponding Kaplan–Meier plot.
On the ‘immunQTLs’ page, users can search by selecting a specific cancer type, immune cell type from a pull-down menu or by entering a SNP ID. We also set sample size, imputation score of SNP and P-value or FDR of immunQTL as search criteria for users to filter. After completing filter and click the ‘Search’ button, the query results will be displayed in a table containing sample size, SNP ID, SNP genomic position, SNP alleles, SNP INFO, SNP annotation, located or nearby gene symbol, immune cell type, beta value (effect size of SNP on immune cell fraction), P-value and FDR of immunQTL (Figure 3D). Besides, a vector diagram of a boxplot was embedded to display the association between SNP genotypes and immune cell fraction for each record. This boxplot could not only be browsed by clicking the hyperlink ‘Box Plot’ on the right side of table, but also available for downloading. For example, our analysis showed that individuals carrying the homozygote rs2442556 aa has significantly higher percent of macrophages M0 than that of individuals carrying the homozygote rs2442556 AA and heterozygous rs2442556 Aa in BRCA (P-value = 2.20 × 10−10, Figure 3F).
On the ‘survival-immunQTLs’ page, search boxes are designed to retrieve specific cancer type, SNP ID, sample size, imputation score of SNP, P-value or FDR of immunQTL. A table with SNP ID, immune cell type, beta value (effect size of SNP on immune cell fraction), P-value and FDR of immunQTL, SNP genomic position, SNP alleles, SNP INFO, SNP annotation, located or nearby gene symbol, P-value and FDR for log-rank test and median survival time for each genotype group will be displayed after submitting search terms (Figure 3E). For each record, a vector diagram of KM Plot will reveal the association between SNP genotypes and overall survival times. For example, our analysis showed that colon adenocarcinoma (COAD) patients with rs250137 AA allele have a better prognosis than patients with rs250137 aa allele (P-value = 8.06 × 10−5, Figure 3G).
On the ‘GWAS-immunQTLs’ page, in addition to search by cancer type, phenotype or SNP ID, various LD thresholds, sample size, imputation score of SNP, P-value or FDR of immunQTL are designed in the dropdown box to restrict SNPs. The information of SNP, sample size, located or nearby gene, regulated immune cell and related GWAS-traits are listed on the search result. For example, rs78239497, as a tag SNP of metabolite levels, was found to be significantly associated with relative fraction of B cells naive in STAD.
SUMMARY AND FUTURE DIRECTIONS
We comprehensively evaluated the effects of genetic variants on immune infiltration in large cancer samples across 33 cancer type, and provided a user-friendly database, CancerImmunityQTL, for users to query, browse, and download immunQTLs. We also presented massive vector diagrams of immunQTL box plots and KM plots for scientific usage. The immunQTLs we identified were further analyzed their association with patient survival times or linkage disequilibrium with known GWAS loci, which will facilitate the interpretation of identified genetic variants. Biologists can download entire datasets for further integrative studies.
With the rapid development of immunogenomic in cancer, we expect the number of cancer samples with genotype and infiltrating immune cell profiles to increase dramatically. We also anticipate the development of single-cell studies characterizing simultaneously transcriptional, genomic and epigenetic states, which will enable the elucidation of immune cell-cancer crosstalk. In the future, we will continue to update CancerImmunityQTL to include more genetic and immune landscape in cancer samples and maintain it as a useful resource for the research community. We believe that CancerImmunityQTL will provide important resource for understanding the germline determinants of immune infiltration in human cancer, which could enable more precise development of immune intervention approaches.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge Weilin Nie for helping design website and build database. We are also grateful to members of the Miao lab for valuable suggestions.
Contributor Information
Jianbo Tian, Department of Epidemiology and Biostatistics, Key Laboratory of Environmental Health of Ministry of Education, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
Yimin Cai, Department of Epidemiology and Biostatistics, Key Laboratory of Environmental Health of Ministry of Education, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
Yue Li, Department of Epidemiology and Biostatistics, Key Laboratory of Environmental Health of Ministry of Education, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
Zequn Lu, Department of Epidemiology and Biostatistics, Key Laboratory of Environmental Health of Ministry of Education, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
Jinyu Huang, Department of Epidemiology and Biostatistics, Key Laboratory of Environmental Health of Ministry of Education, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
Yao Deng, Department of Epidemiology and Biostatistics, Key Laboratory of Environmental Health of Ministry of Education, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
Nan Yang, Department of Epidemiology and Biostatistics, Key Laboratory of Environmental Health of Ministry of Education, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
Xiaoyang Wang, Department of Epidemiology and Biostatistics, Key Laboratory of Environmental Health of Ministry of Education, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
Pingting Ying, Department of Epidemiology and Biostatistics, Key Laboratory of Environmental Health of Ministry of Education, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
Shanshan Zhang, Department of Epidemiology and Biostatistics, Key Laboratory of Environmental Health of Ministry of Education, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
Ying Zhu, Department of Epidemiology and Biostatistics, Key Laboratory of Environmental Health of Ministry of Education, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
Huilan Zhang, Department of Respiratory and Critical Care Medicine, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
Rong Zhong, Department of Epidemiology and Biostatistics, Key Laboratory of Environmental Health of Ministry of Education, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
Jiang Chang, Department of Epidemiology and Biostatistics, Key Laboratory of Environmental Health of Ministry of Education, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
Xiaoping Miao, Department of Epidemiology and Biostatistics, Key Laboratory of Environmental Health of Ministry of Education, School of Public Health, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Fund for Distinguished Young Scholars of China [NSFC-81925032]; National Natural Science Foundation of China [81974456]. Funding for open access charge: National Science Fund for Distinguished Young Scholars of China [NSFC-81925032].
Conflict of interest statement. None declared.
REFERENCES
- 1. Meurette O., Mehlen P.. Notch signaling in the tumor microenvironment. Cancer Cell. 2018; 34:536–548. [DOI] [PubMed] [Google Scholar]
- 2. Turley S.J., Cremasco V., Astarita J.L.. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat. Rev. Immunol. 2015; 15:669–682. [DOI] [PubMed] [Google Scholar]
- 3. Fridman W.H., Pages F., Sautes-Fridman C., Galon J.. The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer. 2012; 12:298–306. [DOI] [PubMed] [Google Scholar]
- 4. Gentles A.J., Newman A.M., Liu C.L., Bratman S.V., Feng W., Kim D., Nair V.S., Xu Y., Khuong A., Hoang C.D. et al.. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015; 21:938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Palomero L., Galvan-Femenia I., de Cid R., Espin R., Barnes D.R., Cimba, Blommaert E., Gil-Gil M., Falo C., Stradella A. et al.. Immune cell associations with cancer risk. iScience. 2020; 23:101296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lim Y.W., Chen-Harris H., Mayba O., Lianoglou S., Wuster A., Bhangale T., Khan Z., Mariathasan S., Daemen A., Reeder J. et al.. Germline genetic polymorphisms influence tumor gene expression and immune cell infiltration. Proc. Natl Acad. Sci. U.S.A. 2018; 115:E11701–E11710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lichtenstein P., Holm N.V., Verkasalo P.K., Iliadou A., Kaprio J., Koskenvuo M., Pukkala E., Skytthe A., Hemminki K.. Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000; 343:78–85. [DOI] [PubMed] [Google Scholar]
- 8. Ostendorf B.N., Bilanovic J., Adaku N., Tafreshian K.N., Tavora B., Vaughan R.D., Tavazoie S.F.. Common germline variants of the human APOE gene modulate melanoma progression and survival. Nat. Med. 2020; 26:1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tian J., Lou J., Cai Y., Rao M., Lu Z., Zhu Y., Zou D., Peng X., Wang H., Zhang M. et al.. Risk SNP-Mediated Enhancer-Promoter interaction drives colorectal cancer through both FADS2 and AP002754.2. Cancer Res. 2020; 80:1804–1818. [DOI] [PubMed] [Google Scholar]
- 10. Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A.. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tam V., Patel N., Turcotte M., Bosse Y., Pare G., Meyre D.. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019; 20:467–484. [DOI] [PubMed] [Google Scholar]
- 12. Tian J., Chang J., Gong J., Lou J., Fu M., Li J., Ke J., Zhu Y., Gong Y., Yang Y. et al.. Systematic functional interrogation of genes in GWAS loci identified ATF1 as a key driver in colorectal cancer modulated by a Promoter-Enhancer interaction. Am. J. Hum. Genet. 2019; 105:29–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M. et al.. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016; 48:481–487. [DOI] [PubMed] [Google Scholar]
- 14. Vandiedonck C. Genetic association of molecular traits: A help to identify causative variants in complex diseases. Clin. Genet. 2018; 93:520–532. [DOI] [PubMed] [Google Scholar]
- 15. Tian J., Wang Z., Mei S., Yang N., Yang Y., Ke J., Zhu Y., Gong Y., Zou D., Peng X. et al.. CancerSplicingQTL: a database for genome-wide identification of splicing QTLs in human cancer. Nucleic Acids Res. 2019; 47:D909–D916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Orru V., Steri M., Sole G., Sidore C., Virdis F., Dei M., Lai S., Zoledziewska M., Busonero F., Mulas A. et al.. Genetic variants regulating immune cell levels in health and disease. Cell. 2013; 155:242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patin E., Hasan M., Bergstedt J., Rouilly V., Libri V., Urrutia A., Alanio C., Scepanovic P., Hammer C., Jonsson F. et al.. Natural variation in the parameters of innate immune cells is preferentially driven by genetic factors. Nat. Immunol. 2018; 19:302–314. [DOI] [PubMed] [Google Scholar]
- 18. Chen D., Juko-Pecirep I., Hammer J., Ivansson E., Enroth S., Gustavsson I., Feuk L., Magnusson P.K., McKay J.D., Wilander E. et al.. Genome-wide association study of susceptibility loci for cervical cancer. J. Natl. Cancer Inst. 2013; 105:624–633. [DOI] [PubMed] [Google Scholar]
- 19. Pu X., Hildebrandt M.A., Lu C., Roth J.A., Stewart D.J., Zhao Y., Heist R.S., Ye Y., Chang D.W., Su L. et al.. Inflammation-related genetic variations and survival in patients with advanced non-small cell lung cancer receiving first-line chemotherapy. Clin. Pharmacol. Ther. 2014; 96:360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dufva O., Polonen P., Bruck O., Keranen M.A.I., Klievink J., Mehtonen J., Huuhtanen J., Kumar A., Malani D., Siitonen S. et al.. Immunogenomic landscape of hematological malignancies. Cancer Cell. 2020; 38:380–399. [DOI] [PubMed] [Google Scholar]
- 21. Shahamatdar S., He M.X., Reyna M.A., Gusev A., AlDubayan S.H., Van Allen E.M., Ramachandran S.. Germline features associated with immune infiltration in solid tumors. Cell Rep. 2020; 30:2900–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Q., Gu J., Wang L., Chang D.W., Ye Y., Huang M., Roth J.A., Wu X.. Genetic associations of T cell cancer immune response-related genes with T cell phenotypes and clinical outcomes of early-stage lung cancer. J. Immunother. Cancer. 2020; 8:e000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Finotello F., Trajanoski Z.. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol. Immunother. 2018; 67:1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A.. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015; 12:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Consortium G.T. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015; 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gaunt T.R., Shihab H.A., Hemani G., Min J.L., Woodward G., Lyttleton O., Zheng J., Duggirala A., McArdle W.L., Ho K. et al.. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016; 17:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gong J., Mei S., Liu C., Xiang Y., Ye Y., Zhang Z., Feng J., Liu R., Diao L., Guo A.Y. et al.. PancanQTL: systematic identification of cis-eQTLs and trans-eQTLs in 33 cancer types. Nucleic Acids Res. 2018; 46:D971–D976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Howie B.N., Donnelly P., Marchini J.. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009; 5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Consortium G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J.. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015; 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stegle O., Parts L., Piipari M., Winn J., Durbin R.. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 2012; 7:500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ongen H., Andersen C.L., Bramsen J.B., Oster B., Rasmussen M.H., Ferreira P.G., Sandoval J., Vidal E., Whiffin N., Planchon A. et al.. Putative cis-regulatory drivers in colorectal cancer. Nature. 2014; 512:87–90. [DOI] [PubMed] [Google Scholar]
- 33. Schulz H., Ruppert A.K., Herms S., Wolf C., Mirza-Schreiber N., Stegle O., Czamara D., Forstner A.J., Sivalingam S., Schoch S. et al.. Genome-wide mapping of genetic determinants influencing DNA methylation and gene expression in human hippocampus. Nat. Commun. 2017; 8:1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shabalin A.A. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012; 28:1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Coussens L.M., Zitvogel L., Palucka A.K.. Neutralizing tumor-promoting chronic inflammation: a magic bullet. Science. 2013; 339:286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Benjamini Y., Hochberg Y.. Controlling the false Discovery Rate: A practical and powerful approach to multiple testing. J. Roy. Statist. Soc.: Ser. B (Methodological). 1995; 57:289–300. [Google Scholar]
- 37. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. et al.. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017; 45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Johnson A.D., Handsaker R.E., Pulit S.L., Nizzari M.M., O’Donnell C.J., de Bakker P.I.. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008; 24:2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M.. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012; 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song M., Yang X., Ren X., Maliskova L., Li B., Jones I.R., Wang C., Jacob F., Wu K., Traglia M. et al.. Mapping cis-regulatory chromatin contacts in neural cells links neuropsychiatric disorder risk variants to target genes. Nat. Genet. 2019; 51:1252–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mantovani A., Allavena P., Sica A., Balkwill F.. Cancer-related inflammation. Nature. 2008; 454:436–444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.