Abstract
We present Peryton (https://dianalab.e-ce.uth.gr/peryton/), a database of experimentally supported microbe-disease associations. Its first version constitutes a novel resource hosting more than 7900 entries linking 43 diseases with 1396 microorganisms. Peryton's content is exclusively sustained by manual curation of biomedical articles. Diseases and microorganisms are provided in a systematic, standardized manner using reference resources to create database dictionaries. Information about the experimental design, study cohorts and the applied high- or low-throughput techniques is meticulously annotated and catered to users. Several functionalities are provided to enhance user experience and enable ingenious use of Peryton. One or more microorganisms and/or diseases can be queried at the same time. Advanced filtering options and direct text-based filtering of results enable refinement of returned information and the conducting of tailored queries suitable to different research questions. Peryton also provides interactive visualizations to effectively capture different aspects of its content and results can be directly downloaded for local storage and downstream analyses. Peryton will serve as a valuable source, enabling scientists of microbe-related disease fields to form novel hypotheses but, equally importantly, to assist in cross-validation of findings.
INTRODUCTION
Over the past decade, microbiome research has experienced an exponential growth (1). The latest refined calculations suggest the human body harbors ∼3.8 × 1013 microbial cells (2), with the majority of them residing in the gastrointestinal tract and spanning over 1000 species. A rough estimate of 2000 genes per species yields a total of 2 million non-redundant genes (3), a number that is ∼100 times higher than the total number of human genes. Bacteroidetes, Firmicutes and Actinobacteria are three of the most abundant phyla of the human gut representing more than 90% of the total bacterial population (4). This complex and dynamic system is to a degree personal (5) and is affected by a variety of environmental factors that can introduce imbalances to the microbial composition of the host (6,7). Consortium-level efforts and the development of cutting-edge high-throughput experimental and insilico methodologies have expanded our view of the implication of microorganisms to a wide range of human pathologies (3,8–9). Numerous studies identify associative relationships between the bacterial abundance and the existence, progress and outcomes of several diseases starting from disorders of the gastrointestinal tract (10), but surprisingly extending to other systems, including malignancies (11,12), neurodegenerative disorders (13) and cardiovascular diseases (14). A number of studies blaze a trail in microbiota research, going beyond the qualitative model and focusing on the causative effects of the microbiome. For instance, Fusobacterium nucleatum has been found to selectively stimulate the growth of colorectal tumor cells in a colorectal cancer progression model comprising cell lines from human colonic adenoma (15). The stimulation is achieved mainly using the Adhesin A protein expressed by FadA gene. Campylobacter jejuni has been shown to promote colorectal tumorigenesis by utilizing a cytolethal distending toxin (16) and F. nucleatum with a number of co-occurring bacteria have been found present in both primary tumors and distant metastasis sites (17). Finally, in a breakthrough publication in 2016, Sampson et al. (13) have provided a functional link between short-chain fatty acids produced by gut bacteria and Parkinson disease, in a mouse model. Besides disease occurrence and progression, bacteria have been shown to promote chemoresistance and to affect other types of treatment too (18). Contrarily, microbiome-based diagnostic and therapeutic strategies for treatment of gastrointestinal disorders, neurodegenerative diseases and other types of pathologies are being actively pursued (19).
The systematic cataloging of the rapidly expanding volume of microbe-disease associations is indispensable to basic and applied microbiome research and is partly addressed by the online repositories Disbiome (20) and gutMDisorder (21). We present here Peryton, a novel resource that, for the first time, (i) supports advanced queries in a user-friendly manner, (ii) hosts advanced exploratory visualizations and (iii) incorporates associations that extend far beyond the disease-healthy phenotype scheme. Peryton is devoted specifically to the manual collection of significant associations of microbes with neurodegenerative, gastrointestinal, cardiovascular and neoplasmic diseases.
METHODS AND RESULTS
Data collection and curation process
Peryton is a database comprising experimentally supported microbe-disease associations. From the thousands of studies investigating this topic and the overabundance of proposed associated microbes, we selected for manual curation those covering the wide spectrum of neurodegenerative diseases, gastrointestinal disorders, cardiovascular diseases and cancer. In order to only include high-quality associations, all entries were manually curated and extracted from available primary research articles (i.e. not from reviews). The PubMed database was queried using a collection of relevant keywords, such as ‘microbiome’, ‘disease’, ‘bacteria’, ‘cancer’, ‘microbiota’, ‘neurodegenerative disease’, ‘microbe’ and ‘gut’, providing a list of more than 2000 publications. This list was refined by reading publication abstracts and results, to keep ∼350 publications comprising associations in one of the four aforementioned disease categories. Through meticulously manual curation the information from selected studies was collected into an Excel sheet with pre-defined fields. The minimum criteria required to record an entry were the following: (i) a valid NCBI Taxonomy ID should exist for every microorganism participating in an entry, (ii) the association had to refer to one of the four disease categories, (iii) associations had to be statistically significant and (iv) detailed information on the experimental design and the sample extraction/type should be provided by the authors. From the ∼350 publications, 314 were found to contain associations fulfilling those criteria.
Database content and statistics
The curation of 314 publications yielded more than 7900 microbe-disease associations. Summarized statistics are listed in Table 1. From the total of 7977 associations, 3777 entries (47.35%) emerged by comparing a disease group against healthy individuals and 4200 (52.65%) by comparing different disease states. Peryton incorporates 43 diseases and 1396 microorganisms. Diseases comprise 23 cancer types, 10 gastrointestinal disorders, 7 cardiovascular diseases and 3 neurodegenerative disorders. From a taxonomic perspective, Peryton's entries span over all known ranks, with the genus-related associations having the highest frequency (3680, 46%). Importantly, 1718 entries (21.54%) provide information at species level or below. For each database entry, information including bacterial abundance, the groups under study (including group sizes, mean age and sex ratios), experimental design, study cohorts, sample type, the applied high- or low-throughput techniques, Next-Generation Sequencing (NGS) sample accession numbers and article metadata are meticulously annotated and catered to users. Interestingly, in more than 50% of the associations, the sample size (i.e. the number of individuals participated in the study) is >50. Finally, diseases and microorganisms are provided in a systematic, standardized manner by using the vocabularies provided by MeSH and the NCBI Taxonomy database (22), respectively.
Table 1.
Summary of Peryton's content and brief database statistics
| Peryton's content | Associations | Percentage (%) | |
|---|---|---|---|
| Metrics | Sample size > 50 | 4611 | 57.80 |
| Sample size < 50 | 3366 | 42.20 | |
| Disease group against control group | 3777 | 47.35 | |
| Contrasts of disease states | 4200 | 52.65 | |
| Species level or below | 1718 | 21.54 | |
| Total | 7977 | 100.00 | |
| Diseases per type | Entries per type | ||
| Disease types | Cancers | 23 | 4065 |
| Gastrointestinal disorders | 10 | 2918 | |
| Cardiovascular diseases | 7 | 125 | |
| Neurodegenerative disorders | 3 | 869 | |
| Total | 43 | 7977 | |
| Number | |||
| General | Microorganisms | 1396 | |
| Taxonomic ranks | 8 | ||
| Sample types | 73 | ||
| Experimental methods | 43 | ||
| Curated articles | 314 | ||
Interface and modules
We designed a user-friendly interface (Figure 1) that provides a number of functionalities to enhance user experience and enable ingenious use of Peryton. One or more microorganisms and/or diseases can be queried at the same time. Advanced filtering options to refine search results can be applied for taxonomic rank, experimental methodology, disease type, sample type, sample size, publication year and relative abundance type. Users can also choose to only return associations in comparison to healthy individuals. Direct text-based filtering of results enables refinement of returned information and the conducting of tailored queries suitable to different research questions. Peryton supports the interconnection with NCBI Taxonomy database (22), PubMed database and MeSH terms, via entry-specific hyperlinks, enabling the direct access to relevant information on microorganisms, publications and diseases, respectively. Additionally, we compiled a list of common contaminants following Eisenhofer et al. (23) and integrated it in the database. Therefore, users can see whether or not a microorganism participating in an association has been deemed a common contaminant, requiring extra caution in result interpretation and handling.
Figure 1.
Peryton’s main user interface. (A) Querying and filtering options. Users may search for one or more of the available taxon names (1) or diseases (2) and helpful drop-down menus emerge. Extensive filtering options are provided and enable even query-free searches (3). Filters include taxonomic rank(s), applied method(s), disease type(s), sample origin(s), studied relationship(s) among groups, cohort size, publication year and even the option to keep only associations contrasting healthy versus disease (4). Users are given the choice to highlight known potential contaminants in the resulting entries, if they require so (5). Upon defining search criteria, they may perform a search, or clear them to conduct a different query (6). (B) Table of results. Standardized disease (7) and microorganism (8) names are provided in the results. Bacterial abundance (9) relative to group 2 is indicated, among other main details (10), including compared groups, experimental group, studied species and study PubMed ID link. When available, dataset accession numbers from popular repositories are provided for entries derived from high-throughput experiments (11). Upon selecting a specific entry, supplementary meta-information including disease MeSH term links (12), taxonomy details and links (13) and additional cohort information (14) are catered. Using a word-based on-the-fly filter on top (15) users may narrow down the results list, while all results can be instantly stored locally as tab-separated files (16).
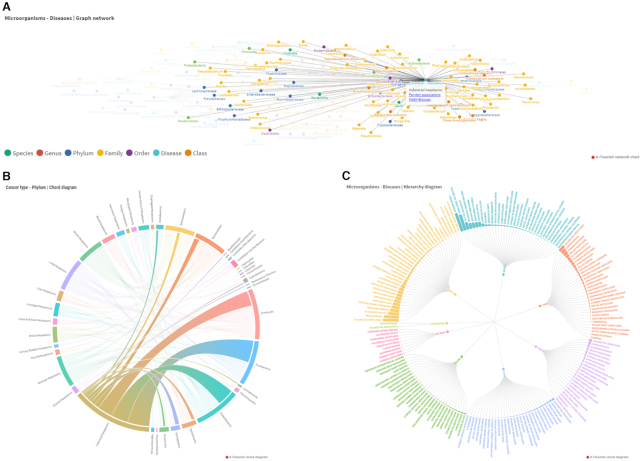
Peryton also provides state-of-the-art interactive visualizations to effectively capture different aspects of its content. Via Network graphs, Chord diagrams and Hierarchy diagrams, users can browse into the available content and perform observations about microbe-disease relationships using information from the latest relevant literature. The Graph network (Figure 2A) offers an interactive graph visualization of the strongest microbe-disease associations (i.e. associations reproduced independently in different studies). Color-coded nodes represent diseases and microorganisms grouped under their taxonomic rank. Taxonomic ranks may be filtered-out or -in, depending on the users’ choice. Each node can be selected and moved and their neighboring nodes will move at different speed, according to their link status (i.e. ‘hub’ nodes will be heavier, while less linked nodes will move more freely). By clicking on a node, a pop-up window appears, allowing users to directly navigate to the node-relevant associations in a new tab, or move to the respective entry on external resources (i.e. NCBI taxonomy DB for microbe-nodes or the MeSH browser for disease-nodes). The Chord diagram (Figure 2B), allows the interactive observation and comparison of available microbe-disease associations. Each string's width is relative to the number of supporting entries, which appears by hovering. Relationships of interest can be highlighted temporarily by hovering or permanently by clicking on specific arcs and/or strings. The Chord diagram comprises associations between all available phyla and cancer types. The Hierarchy diagram (Figure 2C) is a multi-layered exploratory tool containing information on all entries. Diseases and number of entries are provided in the circle's arcs for each taxonomic rank. Upon selecting a specific rank (e.g. species), a zoomed-in depiction of the database content for this rank is provided. Users may select among available diseases from this view to zoom-in more, or use the upward-facing arrow to zoom-out one level. Users may also directly move to the deepest available layer by clicking on a bar of interest (e.g. ‘Species | Colorectal Cancer’). The deepest layer contains information on a single disease, the microorganisms linked to this disease and the number of times each association is included in Peryton. Notably, the deepest layer is also clickable, facilitating direct transition from the ‘Visualizations’ page to specific query results in the main ‘Associations’ page.
Figure 2.
Visualization options offered in Peryton. (A) In Graph network, available associations are depicted in an interactive network. Users can explore the graph, highlight nodes of interest and filter-in and -out taxonomic ranks, according to taste. Importantly, by moving a selected node, connected nodes will move as well, with velocity depending on their connectivity status, allowing fast identification of hub or unitary nodes. Via pop-up boxes, each node in Graph network directs to its corresponding query results on the main ‘Associations’ page and NCBI Taxonomy or MeSH Browser, accordingly. (B) Chord diagram provides an interactive view of available cancer-related associations. Diseases and microorganisms are deployed along the circle's arcs, and chords of width relative to the number of existing associations depict connections. Users may select on or more components (i.e. arcs and/or chords) to highlight them permanently, or hover over them to highlight them temporarily. (C) Users can utilize Hierarchy diagram to browse Peryton's content in a hierarchically structured manner. For each taxonomic rank, numbers of available associations are provided as bars surrounding the circle. By selecting on bars and/or taxonomic ranks, a zoomed-in depiction of the relevant content is offered, enabling focused examination on associations of interest. Deepest layers in the Hierarchy diagram are also inter-connected with the Associations page via hyperlinks in microbe-disease-specific pop-up boxes.
Results can be directly downloaded as tab-separated files with additional complementary metadata (e.g. disease and publication information, additional comments, etc.) for local storage and downstream analyses. Basic statistics of the database (e.g. top 15 diseases and top 15 microorganisms) are presented through simple bar plots in a dedicated page. A help page with rich material (screenshots and text-based walkthroughs) is available to guide users and ensure a smooth experience for first-time visitors. Finally, we also provide a dedicated page were users can submit their own associations using one of two options: an advanced submission form comprising multiple association fields, or a simple form were only publication's DOI, PubMed ID and a submitter comment are needed. New user-submitted associations will be taken into consideration by Peryton's curation team in each major update of the database content.
Comparison with existing resources
Disbiome (20) and gutMDisorder (21) are two online resources hosting content similar to that of Peryton. A major advantage of Peryton over both Disbiome and gutMDisorder is that it incorporates associations that extend far beyond the disease-healthy phenotype scheme. Specifically, Peryton hosts associations between different cancer grades, between metastatic versus non-metastatic cancer groups, benign and malignant tumor samples, symptomatic versus asymptomatic disease phenotypes etc. These entries push the envelope in the microbe-disease association field, making available, for the first time, a systematic set that can be used to investigate the potential involvement of these microorganisms in mechanisms of cancer progression, metastasis, onset of symptoms and many more. Disbiome and gutMDisorder focus on associations in contrast with healthy phenotypes. Disbiome hosts a number of entries in contrast with non-healthy states (8.85% of total associations), but this field is not used to construct queries, filter, or even annotate returned results.
Apart from its primary content, Peryton delivers a number of interface novelties that enhance user experience. Its advanced visualization capacity allows users to perform observations about microbe-disease relationships in an exploratory and interactive manner. Peryton supports advanced queries using one or more diseases and/or microorganism names, one or multiple taxonomic ranks, as well as the application of smart filtering options, including ‘disease’, ‘experimental method’, ‘sample origin’, ‘sample size’ and a check box for including only associations derived using healthy controls. gutMDisorder only permits querying and refining returned associations using species name, disorder/intervention and microorganism name. On the other hand, disbiome allows for complex queries but its free-text search engine is not designed to be field-aware, frequently returning irrelevant entries too. Peryton's user-interface is minimal, easy to follow and addresses most query combinations a user may need.
Finally, some fine annotation and curation choices further differentiate Peryton from existing resources: (i) Information and awareness on the existence of common contaminants is highlighted in query results, (ii) disease names and microorganisms follow a systematic, uniform nomenclature and (iii) the largest number of cancer-related associations is achieved (i.e. more than 2-fold increase from the second largest resource).
Database architecture and implementation
Peryton is a relational database built using the MVC architecture and hosted on Apache HTTP server 2.4. The back-end consists of PostgreSQL server 11.8 (https://www.postgresql.org/) and the PHP framework Laravel 7.14 (https://laravel.com/) (PHP 7.2), while the front-end is designed using Angular 9.1 (https://angular.io/) and the Angular Material UI library (https://material.angular.io/). Peryton’s data are stored in multiple tables connected relationally inside the PostgreSQL database and Laravel handles the connection to them for storing and/or retrieval. Finally, on the presentation layer, the database statistics are presented using Chart JS (https://www.chartjs.org/), while Flourish (https://flourish.studio/) is utilized for the more complex visualizations.
DISCUSSION AND FUTURE WORK
Investigation of the human microbiome and its importance in physiological and pathological states is a research hotspot (24). Currently, numerous research projects worldwide are examining microbiome applications in diagnostics, therapeutic interventions and the mechanisms by which the microbiome affects the promotion and progression of diseases, or even outcomes of therapeutic strategies (19,25–26). Reference databases indexing such information are invaluable for scientists working on similar fields. Peryton is the first version of an effort to systematically catalogue and annotate high quality statistically significant associations between microbes and diseases, with an emphasis in cancer. The database will be updated every year, expanding its content into new diseases and microorganisms, but also adding more associations to the already existing diseases. Future versions will be supported by text-mining strategies (27,28) and broader taxonomy schemes (29), as well as fully user-customizable visualization options. We believe that Peryton will serve as a valuable source, enabling scientists of microbe-related human disease fields to form novel hypotheses, refine and enhance existing ones and cross-validate their experimental findings.
ACKNOWLEDGEMENTS
We would like to thank Maria Zioga (University of Crete, School of Medicine) for designing the image displayed on Peryton's home page as well as for her critical comments regarding visualizations.
Contributor Information
Giorgos Skoufos, Department of Electrical & Computer Engineering, Univ. of Thessaly, Volos 38221, Greece; Hellenic Pasteur Institute, Athens 11521, Greece.
Filippos S Kardaras, Hellenic Pasteur Institute, Athens 11521, Greece; DIANA-Lab, Department of Computer Science and Biomedical Informatics, Univ. of Thessaly, Lamia 351 31, Greece.
Athanasios Alexiou, Hellenic Pasteur Institute, Athens 11521, Greece; DIANA-Lab, Department of Computer Science and Biomedical Informatics, Univ. of Thessaly, Lamia 351 31, Greece.
Ioannis Kavakiotis, Department of Electrical & Computer Engineering, Univ. of Thessaly, Volos 38221, Greece; Hellenic Pasteur Institute, Athens 11521, Greece.
Anastasia Lambropoulou, Hellenic Pasteur Institute, Athens 11521, Greece.
Vasiliki Kotsira, Hellenic Pasteur Institute, Athens 11521, Greece.
Spyros Tastsoglou, Department of Electrical & Computer Engineering, Univ. of Thessaly, Volos 38221, Greece; Hellenic Pasteur Institute, Athens 11521, Greece.
Artemis G Hatzigeorgiou, Department of Electrical & Computer Engineering, Univ. of Thessaly, Volos 38221, Greece; Hellenic Pasteur Institute, Athens 11521, Greece; DIANA-Lab, Department of Computer Science and Biomedical Informatics, Univ. of Thessaly, Lamia 351 31, Greece.
FUNDING
This work was supported by ‘ELIXIR-GR: The Greek Research Infrastructure for Data Management and Analysis in Life Sciences’ [MIS-5002780] which is implemented under the Action ‘Reinforcement of the Research and Innovation Infrastructure’, funded by the Operational Programme ‘Competitiveness, Entrepreneurship and Innovation’ [NSRF 2014 –2020] and co-financed by Greece and the European Union (European Regional Development Fund). It was also co-financed by a donation from the Stavros Niarchos Foundation, by a postdoctoral scholarship from University of Thessaly funded by “Stavros Niarchos Foundation" and (Greece and European Social Fund-ESF) through the Operational Programme ‘Human Resources Development, Education and Lifelong Learning’ in the context of the project ‘Strengthening Human Resources Research Potential via Doctorate Research’ [MIS-5000432], implemented by the State Scholarships Foundation (IKY), in the form of two PhD Scholarships. Funding for open access charge: ELIXIR-GR: The Greek Research Infrastructure for Data Management and Analysis in Life Sciences [MIS-5002780].
Conflict of interest statement. None declared.
REFERENCES
- 1. Proctor L., LoTempio J., Marquitz A., Daschner P., Xi D., Flores R., Brown L., Ranallo R., Maruvada P., Regan K. et al.. A review of 10 years of human microbiome research activities at the US National Institutes of Health, fiscal years 2007–2016. Microbiome. 2019; 7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sender R., Fuchs S., Milo R.. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016; 14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I.. The human microbiome project. Nature. 2007; 449:804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tap J., Mondot S., Levenez F., Pelletier E., Caron C., Furet J.P., Ugarte E., Muñoz-Tamayo R., Paslier D.L., Nalin R.. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 2009; 11:2574–2584. [DOI] [PubMed] [Google Scholar]
- 5. Franzosa E.A., Huang K., Meadow J.F., Gevers D., Lemon K.P., Bohannan B.J.M., Huttenhower C.. Identifying personal microbiomes using metagenomic codes. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E2930–E2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hasan N., Yang H.. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ. 2019; 7:e7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gilbert J.A., Blaser M.J., Caporaso J.G., Jansson J.K., Lynch S.V., Knight R.. Current understanding of the human microbiome. Nat. Med. 2018; 24:392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Almeida A., Nayfach S., Boland M., Strozzi F., Beracochea M., Shi Z.J., Pollard K.S., Sakharova E., Parks D.H., Hugenholtz P. et al.. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol. 2020; doi:10.1038/s41587-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Proctor L.M., Creasy H.H., Fettweis J.M., Lloyd-Price J., Mahurkar A., Zhou W., Buck G.A., Snyder M.P., Strauss J.F., Weinstock G.M. et al.. The integrative human microbiome project. Nature. 2019; 569:641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ohkusa T., Sato N., Ogihara T., Morita K., Ogawa M., Okayasu I.. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J. Gastroenterol. Hepatol. 2002; 17:849–853. [DOI] [PubMed] [Google Scholar]
- 11. Luu T.H., Michel C., Bard J.M., Dravet F., Nazih H., Bobin-Dubigeon C.. Intestinal proportion of blautia sp. is associated with clinical stage and histoprognostic grade in patients with early-stage breast cancer. Nutr. Cancer. 2017; 69:267–275. [DOI] [PubMed] [Google Scholar]
- 12. Ohigashi S., Sudo K., Kobayashi D., Takahashi O., Takahashi T., Asahara T., Nomoto K., Onodera H.. Changes of the intestinal microbiota, short chain fatty acids, and fecal pH in patients with colorectal cancer. Dig. Dis. Sci. 2013; 58:1717–1726. [DOI] [PubMed] [Google Scholar]
- 13. Sampson T.R., Debelius J.W., Thron T., Janssen S., Shastri G.G., Ilhan Z.E., Challis C., Schretter C.E., Rocha S., Gradinaru V.. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell. 2016; 167:1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toya T., Corban M.T., Marrietta E., Horwath I.E., Lerman L.O., Murray J.A., Lerman A.. Coronary artery disease is associated with an altered gut microbiome composition. PLoS One. 2020; 15:e0227147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rubinstein M.R., Baik J.E., Lagana S.M., Han R.P., Raab W.J., Sahoo D., Dalerba P., Wang T.C., Han Y.W.. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β-catenin modulator Annexin A1. EMBO Rep. 2019; 20:e47638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He Z., Gharaibeh R.Z., Newsome R.C., Pope J.L., Dougherty M.W., Tomkovich S., Pons B.. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut. 2019; 68:289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bullman S., Pedamallu C.S., Sicinska E., Clancy T.E., Zhang X., Cai D., Neuberg D., Huang K., Guevara F., Nelson T. et al.. Analysis of <em>Fusobacterium</em>persistence and antibiotic response in colorectal cancer. Science. 2017; 358:1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu T., Guo F., Yu Y., Sun T., Ma D., Han J., Qian Y., Kryczek I., Sun D., Nagarsheth N.. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017; 170:548–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wong A.C., Levy M.. New approaches to microbiome-based therapies. mSystems. 2019; 4:e00122-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Janssens Y., Nielandt J., Bronselaer A., Debunne N., Verbeke F., Wynendaele E., Van Immerseel F., Vandewynckel Y.-P., De Tré G., De Spiegeleer B.. Disbiome database: linking the microbiome to disease. BMC Microbiol. 2018; 18:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheng L., Qi C., Zhuang H., Fu T., Zhang X.. gutMDisorder: a comprehensive database for dysbiosis of the gut microbiota in disorders and interventions. Nucleic Acids Res. 2019; 48:D554–D560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Federhen S. The NCBI taxonomy database. Nucleic Acids Res. 2012; 40:D136–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eisenhofer R., Minich J.J., Marotz C., Cooper A., Knight R., Weyrich L.S.. Contamination in low microbial biomass microbiome Studies: Issues and recommendations. Trends Microbiol. 2019; 27:105–117. [DOI] [PubMed] [Google Scholar]
- 24. Shreiner A.B., Kao J.Y., Young V.B.. The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 2015; 31:69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schlaberg R. Microbiome diagnostics. Clin. Chem. 2019; 66:68–76. [DOI] [PubMed] [Google Scholar]
- 26. Durack J., Lynch S.V.. The gut microbiome: relationships with disease and opportunities for therapy. J. Exp. Med. 2019; 216:20–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Badal V.D., Wright D., Katsis Y., Kim H.-C., Swafford A.D., Knight R., Hsu C.-N.. Challenges in the construction of knowledge bases for human microbiome-disease associations. Microbiome. 2019; 7:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ren J., Li G., Ross K., Arighi C., McGarvey P., Rao S., Cowart J., Madhavan S., Vijay-Shanker K., Wu C.H.. iTextMine: integrated text-mining system for large-scale knowledge extraction from the literature. Database. 2018; 2018:bay128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Balvočiūtė M., Huson D.H.. SILVA, RDP, Greengenes, NCBI and OTT—how do these taxonomies compare. BMC Genomics. 2017; 18:114–114. [DOI] [PMC free article] [PubMed] [Google Scholar]