Graphical abstract
Keywords: DEAD/H box, Helicase, Lung cancer, Biomarker, Clinical significance, Bioinformatics
Abstract
DEAD/H box helicases are implicated in lung cancer but have not been systematically investigated for their clinical significance and function. In this study, we aimed to evaluate the potential of DEAD/H box helicases as prognostic biomarkers and therapeutic targets in lung cancer by integrated bioinformatic analysis of multivariate large-scale databases. Survival and differential expression analysis of these helicases enabled us to identify four biomarkers with the most significant alterations. These were found to be the negative prognostic factors DDX11, DDX55 and DDX56, and positive prognostic factor DDX5. Pathway enrichment analysis indicates that MYC signalling is negatively associated with expression levels of the DDX5 gene while positively associated with that of DDX11, DDX55 and DDX56. High expression levels of the DDX5 gene is associated with low mutation levels of TP53 and MUC16, the two most frequently mutated genes in lung cancer. In contrast, high expression levels of DDX11, DDX55 and DDX56 genes are associated with high levels of TP53 and MUC16 mutation. The tumour-infiltrated CD8 + T and B cells positively correlate with levels of DDX5 gene expression, while negatively correlate with that of the other three DEAD box helicases, respectively. Moreover, the DDX5-associated miRNA profile is distinguished from the miRNA profiles of DDX11, DDX55 and DDX56, although each DDX has a different miRNA signature. The identification of these four DDX helicases as biomarkers will be valuable for prognostic prediction and targeted therapeutic development in lung cancer.
1. Introduction
Lung cancer is the most common cancer and the leading cause of cancer-related death. It accounts for 11.6% of all cancers worldwide, with approximately 2.1 million new cases in 2018 alone [1]. In addition to its prevalence, most lung cancer patients have a poor prognosis, with a 5-year predicted survival of around 17.8% [2]. There is a disproportionate number of lung cancer-related deaths when compared with other cancers of a similar incidence, such as breast cancer. In 2018 lung cancer was responsible for 1.75 million deaths, almost 20% of all cancer-related deaths in that year, whereas breast cancer, despite having an almost equal number of diagnoses, led to only 0.6 million deaths [1]. This discrepancy may potentially be due to late diagnosis, leading to the delayed onset of treatment and allowing more time for cancer to develop in both stage and grade. As a result, a large number of lung cancer patients present late with metastasis, and miss the optimal time window for localised surgical intervention. Besides, a larger number of lung cancer patients that have metastasis will miss the optimal time window for surgical treatment when they are diagnosed. The majority of these patients have to undergo numerous cycles of chemotherapy and thus risk the severe adverse effects that accompany it [3]. This demonstrates the need to develop new treatments capable of limiting lung cancer progression, to improve the efficacy of chemotherapy and the survival of these patients.
Lung cancer is a heterogeneous disease comprising two subtypes. Small cell lung cancer (SCLC) is believed to arise in the neuroendocrine cells of the lung, while non-small cell lung cancer (NSCLC) accounts for 85% of cases and is further divided into 3 subtypes: squamous-cell carcinoma (LUSC), adenocarcinoma (LUAD), and large-cell carcinoma (LCC) [2], [4]. The most common forms of NSCLC are LUAD and LUSC [2]. LUAD begins in type II alveolar cells of the lung, responsible for maintaining alveolar surface tension, and the stem cells that accumulate in the bronchioalveolar duct junction [5]. LUSC arises from the squamous epithelial cells of the bronchus [5]. Despite their shared classification of NSCLC, the two subtypes possess distinct mechanisms of tumorigenesis that determines their heterogeneous histology and tumour progression [6]. Despite recent advances in the treatments of lung cancer, mortality rates are still substantially high in these patients; therefore improved therapeutic targeting of lung cancer cells may be vital to improving lung cancer survival [7].
DEAD/H box RNA helicases are a superfamily of molecules which possess crucial roles in all stages of RNA metabolism, from transcription to translation or degradation [8], [9]. All DEAD/H box proteins possess the same basic structure consisting of a structurally conserved helicase core which consists of two RecA-like domains [9], [10]. Conserved amino acid motifs, which confer the ATP and RNA binding ability of the helicases, are spread out across these regions and act together in a complex to confer the helicase function. The first member of the family to be discovered was eIF4A, a member of the translation initiation complex, the structure of which is restricted to a helicase core and RecA-like domains, possessing only a minimal N terminus chain [11], [12], [13]. It was observed that eIF4A shared several common motifs with other proteins possessing a similar function; this discovery led to the foundation of the DEAD/H box RNA helicase family [13]. A search of GenBank revealed that 57 members of the helicase family have been identified in humans, all possessing the conserved motifs held within the helicase core and RecA-like domains [14]. One of these shared motifs is the DEAD, or DEAH, amino acid sequences [13]. The RecA domains generate a cleft around the helicase core where both ATP and RNA can bind [9]. The RNA binding sites bind the sugar-phosphate backbone of the RNA strand and hold it in place opposite to the ATP binding site. Without the need for complementary base pairs to associate with a target sequence, the helicases are not selective for specific RNA [15]. As helicases, the DEAD/H box proteins are capable of binding and unwinding RNA in an ATP-dependent manner, however, in DEAD/H box helicases this process is limited to short RNA duplexes [16]. These RNA duplexes are capable of forming complex secondary structures, which arise through interactions between the bases of the RNA sequence, and these are capable of acting as regulators of RNA processing [17]. Since these structures are very stable, the RNA duplexes will remain in their conformation unless acted on by external factors, such as a helicase capable of remodelling the duplex to facilitate proper RNA processing. To achieve its association with ATP and RNA the binding cleft must be in a highly defined conformation and subtle changes to these motifs can impact the ability of the protein to perform its function [10]. Because of this, the variation in the DEAD/H box protein function is determined by the presence of variable C and N terminal auxiliary domains. These domains can possess their own function, such as nuclease activity, or be responsible for the recruitment of other proteins to a dynamic complex around the RNA, for example the spliceosome or translation complexes.
This motif variability allows the helicases to be involved in all aspects of RNA processing with each member of the family playing a distinct role. In RNA processing, each of the steps requires DEAD/H box RNA helicases to mediate progression and modification, either directly or through the recruitment of complexes [9]. As gene expression is a complex process involving several heavily regulated stages, the DEAD/H box RNA helicases must be correctly targeted and regulated to ensure appropriate RNA processing. RNA unwinding is an ATP-dependent process and a primary function of the helicases involving the conserved RecA-like domains and helicase core; many helicases mediate this action by loading onto a single-stranded region of an RNA duplex and displacing the complementary strand during ATP dependent unidirectional translocation [20]. However, an alternative mode of action has been suggested for some members of the DEAD/H box family. In this mechanism the helicase associates with a single-stranded region distinct from the target duplex but in close spatial proximity, this single-stranded DNA or RNA mediates helicase loading onto the duplex and subsequent duplex unwinding by local strand separation [20], [21]. Despite possessing ATP dependent helicase activity, some members of the DEAD/H box family are capable of mediating their actions through ATP independent mechanisms [9].
Among the DEAD/H box helicases, 16 are DEAH-box members (DHXs) while the others are DEAD-box members (DDXs). Although sharing the two evolutionarily-conserved RecA-like core domains, DHXs and DDXs unwind RNA structures through distinct mechanisms. DDXs bind and unwind RNA duplexes or ribonucleoproteins on their surfaces directly. In contrast, some DHXs require translocation inside the RNA structures along the 3′–>5′ direction before resolving nucleic acid, and some DHXs can unwind both RNA and DNA with a G-quadruplex structure [18], [19].
Notably, the DEAD/H box helicases also have other multiple biochemical or biological functions which are often independent of their conserved helicase core domains [22]. Individual helicase proteins can interact with proteins and participate in posttranslational modification through their N-terminal and/or C-terminal domains specifically [23]. For instance, the N-terminal domain of DDX3 plays a role in nuclear export by interacting with exportin 1 [24]. DDX3 is considered to play dual roles in cancer development and progression [25]. The N-terminal domain of DDX5 can regulate some transcriptional coactivators such as PIAS1, Fibrillarin, c-ABL kinase and RNA polymerase II by protein interaction [26]. The C-terminal of DHX9 contains a nuclear localisation and export signalling domain [27]. The N-terminal domain of DHX36 is essential for cell viability, required for the nuclear localization, and specific interaction with the spliceosome [28]. Instead, the C-terminal domain of DHX36 is involved in the interaction with single-stranded DNA at the 3′-end with a G4 structure [29]. The C-terminal domain of DDX20 can interact with p53 and NF-κB, and play a role in abnormal cell signal transduction [30].
Dysregulation of DEAD/H box helicases disrupts tightly controlled homeostasis of biological processes involving RNA, such as pre-mRNA splicing, RNA export, transcription and translation. The production of aberrant RNA and subsequent dysregulation of downstream transcription, can have profound consequences on uncontrolled fate, phenotype transition, migration and invasion of cancer cells during tumour development and metastasis [31], [32]. In addition, as a consequence of altering RNA metabolism or interacting with specific proteins, some DEAD/H box helicases may mediate genome stability and stress-induced DNA damage repair thus be linked with mutation occurrence or frequency [33], [34]. There is also evidence that a helicase can modulate methylation of mRNAs and microRNAs (miRNAs) [35].
These enzymes have been implicated in the development, progression and metastasis of cancer as well as in the response to chemotherapeutic agents [36], [37]. They can be both tumour suppressive, as demonstrated by the role of DDX3 in mediating p21 expression, and tumorigenic, as DHX32 was shown to upregulate VEGFA expression and contributed to the increased growth and vascularisation of colorectal cancer [38], [39]. Therefore, whilst they present promising targets to alter the expression of oncogenes and tumour suppressors, the role of the DEAD/H box helicases is very dependent on the context in which they are found. There is a particular interest in the potential of these enzymes for diagnostic and prognostic biomarkers or as novel pharmacological drug targets in cancer [40], [41]. There are several DEAD/H box helicase therapeutics currently in development in preclinical and clinical trials. Small molecules, capable of modulating RNA binding and ATP-dependent helicase activity, or disrupting certain protein–protein interactions implicated in cancer progression, have been demonstrated to possess anticancer activity [42], [43]. However, there has been no systematic investigation of the clinical relevance and biomarker potential of the DEAD/H box helicase family members. This means that the associations that exist in one cancer may not exist in another, so a systematic investigation is necessary to determine which helicases present potential targets for investigation.
Therefore, this study aims to unveil whether DEAD/H box RNA helicases have a clinical significance in lung cancer and whether they possess the potential to become a therapeutic target or prognostic biomarker. With the advances of the Bioconductor, R and other statistical computing tools, we undertook integrated bioinformatic analysis by integrating publicly-available large-scale databases, including KM survival, gene expression, mutation, miRNA expression, immune cell correlation and methylation profiles, for this purpose,
2. Results
2.1. Association of the DEAD/H box helicase genes with the survival of lung cancer.
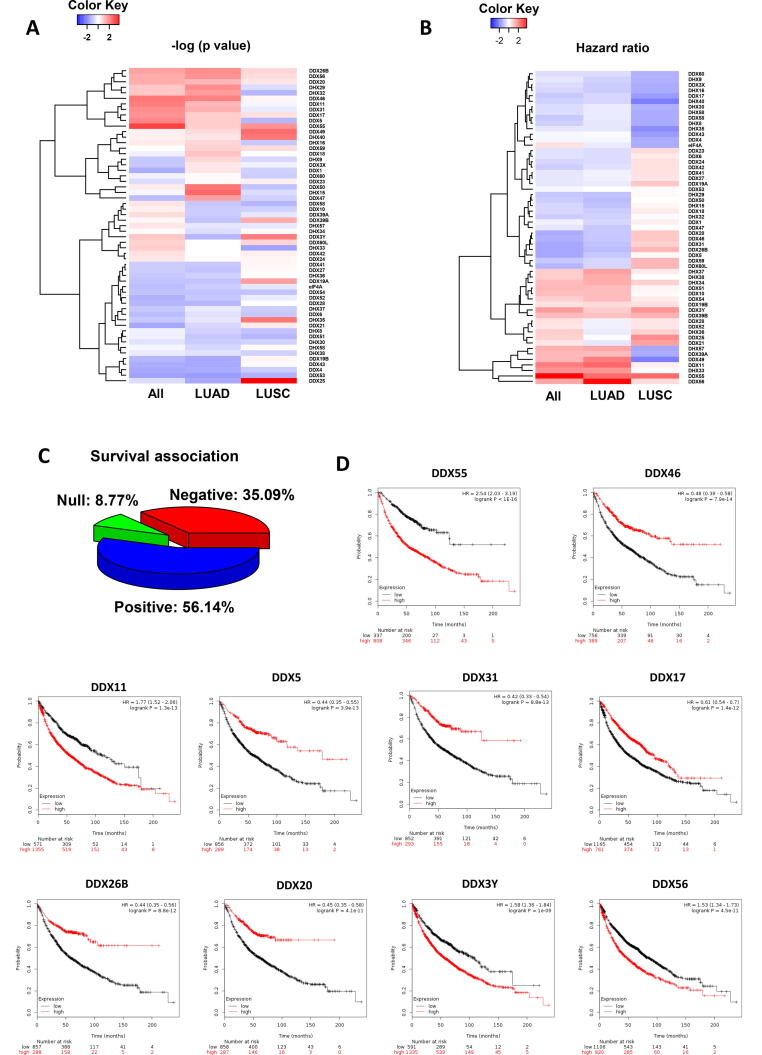
In order to understand the prognostic values of the 57 DEAD/H box helicases in lung cancer, we performed the KM survival analysis of their gene expression respectively. In all lung cancer patients, 92.44% of the helicases were significantly associated with overall survival (OS) (52/57) (p < 0.01). In LUAD, the significant helicases numbered 92.88% (53/57). In contrast, the percentage of significant helicases was only 10.53% (6/57) in LUSC. The heat map of the association of the individual helicase genes with the OS is shown in Fig. 1A after the p-values were converted to negative log10 (p-values). In general, LUAD is more similar in all lung cancer cases, while LUSA is quite different. In Fig. 1B, the hazard ratio from survival analysis is shown for each DEAD/H box helicase gene. In all lung cancer cases, 56.14% of the helicases are positively associated with the survival rate, while 35.09% possess a negative association (Fig. 1C). The KM plots of the first 10 ranked DEAD/H box genes in ascending order of log-rank p-values are shown in Fig. 1D-1L. The database of the p-values and HR of all the DEAD/H box genes are provided in Supplement Table 1.
Fig. 1.
Survival analysis of the DEAD/H box helicases in lung cancer. (A) Heatmap of log-rank test p-values (negative log10 scale) showing the association with overall survival (OS). (B) Heatmap of hazard ratio (HR) showing the direction of the risk association (HR > 1, positive; HR < 1, negative; HR = 1, null). (C) Pie chart summarising the percentage of the helicases associated with OS. (D) The OS plot of the first 10 most significant genes in lung cancer.
2.2. The association of DEAD/H box members with the clinical parameters of lung cancer
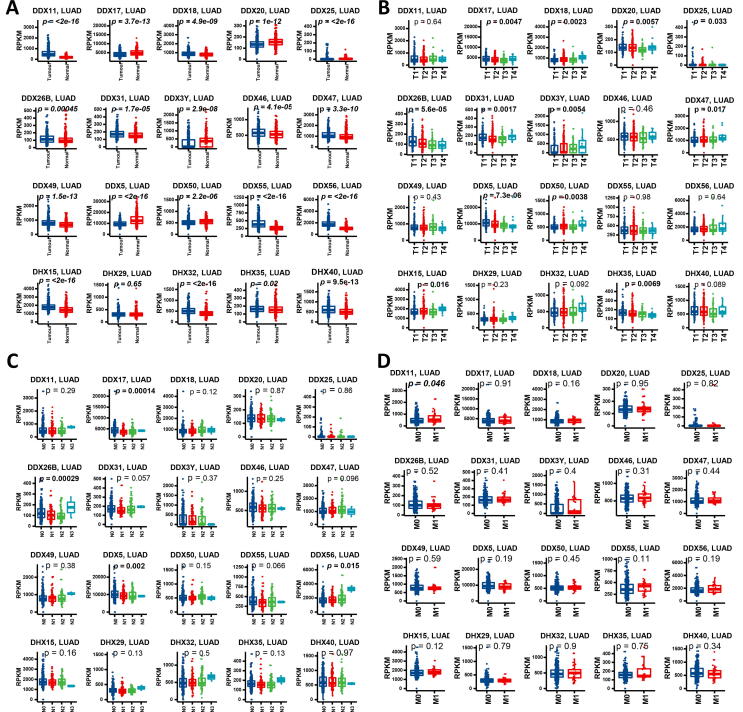
We then selected the top 20 DEAD/H box helicase genes according to the p-values of KM analysis. The selection criteria were based on the p-values after KM analysis: all the DEAD/H box helicase members in LUSC with a p < 0.01 (n = 6 in total) and the 14 with the smallest p-values in LUAD.
We investigated the association of the top 20 DEAD/H box genes with clinical parameters of lung cancer. In LUAD, as shown in Fig. 2, 19 of the 20 genes showed differential expression when tumour and normal tissues were compared. The top 5 genes with higher levels of expression in tumour included DDX56, DDX11, DDX55, DDX15 and DDX32 (p < 2e-16), while DDX5 is one of the genes which has lower expression in tumours (p < 2e-16). Among the different T stages in the TNM classification, 12 of the selected DDX members showed significance. Among them, DDX5 was the most significant which was downregulated in the advanced T stages (p < 7.3e-06) followed by DDX26B which also showed downregulation with T stage progression (p < 5.6e-06). Among the different N stages, only four of the selected genes showed significance, which were DDX17 (p = 0.00014), DDX26B (p = 0.00029), DDX5 (p = 0.002) and DDX56 (p = 0.015). Among the different M stages, only DDX11 showed statistical significance (p < 0.05). We also performed gene expression analysis of the overall stages, and found that DDX17 (p = 0.0005), DDX26B (p = 3.6E-05), DDX47 (p = 0.044) and DDX5 (p = 0.0014) were significantly different. Among the different ages, only DDX5 and DDX56 showed differences. Between the genders, DDX3Y (p < 0.0001), DDX47(p = 0.016), DHX40 (p = 0.016), DDX26B (p = 0.020) and DHX15 (p = 0.028) showed difference of gene expression (Supplement Fig. 1).
Fig. 2.
Gene expression profiles of the DEAD/H box helicase genes among tissue types and TNM stages in LUAD. (A) Gene expression between tumour and normal tissues. (B) Gene expression at different primary tumour (T) stages in the TNM system. (C) Gene expression at different regional lymph nodes (N) stages in the TNM system. (D) Gene expression at different distant metastasis (M) stages in the TNM system. The p-values<0.05 were highlighted in bold.
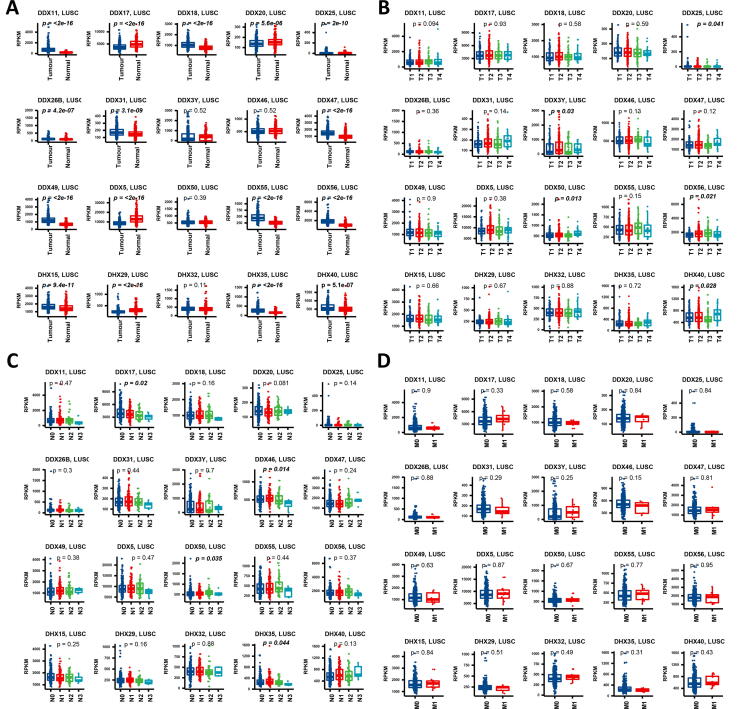
In LUSC, 16 of the 20 genes showed differential expression when tumour and normal tissues were compared. The top five genes with higher levels of expression in tumour were DDX11, DDX56, DDX49, DDX55 and DDX47 (p < 0.0001), while DDX5 was also the gene which had the most significant low expression in tumour (p < 0.0001) followed by DDX29 (p < 0.0001) and DDX17 (p < 0.0001) (Fig. 3). Among the different T stages in LUSC, DDX50 (p = 0.013), DDX56 (p = 0.021), DHX40 (p = 0.028), DDX3Y (p = 0.03) and DDX25 (p = 0.041) showed significance. Among the N stages in LUSA, DDX46 (p = 0.014), DDX17 (p = 0.020), DDX50 (p = 0.035) and DDX35 (p = 0.044) showed differential expression. Among the M-stages, no significance was observed in terms of the expression of the DEAD/H box helicase genes.
Fig. 3.
Gene expression profiles of the DEAD/H box helicase genes at different types and TNM stages in LUSC. (A) Gene expression between tumour and normal tissues. (B) Gene expression at different primary tumour (T) stages in the TNM system. (C) Gene expression at different regional lymph nodes (N) stages in the TNM system. (D) Gene expression at different distant metastasis (M) stages in the TNM system. The p-values <0.05 were highlighted in bold.
We also performed gene expression analysis of the overall stages, and found that DDX20 (p = 0.002), DDX50 (p = 0.017), DDX17 (p = 0.026) and DDX46 (p = 0.030) were significantly different. Among the different ages, only DDX31 (p < 0.007) showed differential expression. Between the genders, only DDX3Y (p < 0.0001) and DDX47 (p = 0.015) showed a difference in gene expression (Supplement Fig. 2).
2.3. Correlation of the DEAD/H box helicase gene expression
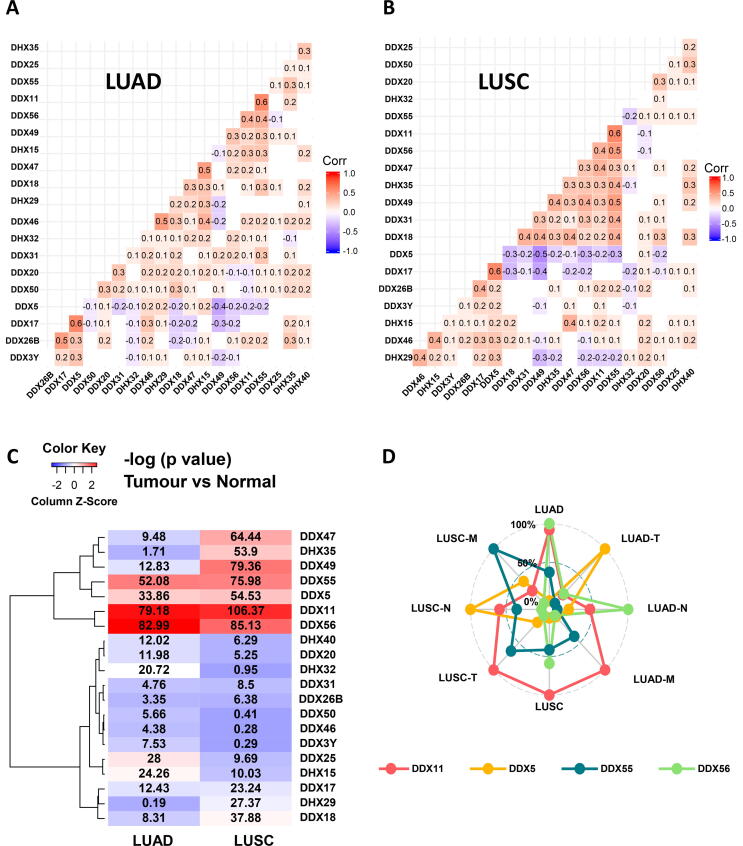
We then performed a matrix correlation analysis using the Pearson method. As shown in Fig. 4A, in LUAD, DDX55 had a more positive correlation with other DDX members such as DDX11 (correlation coefficient (r = 0.6) and DDX56 (r = 0.4), while DDX5 had the most negative correlation with other DDX members such as DDX49 (r = -0.4). Similarly, in LUSC, DDX55 also had the most frequent positive correlation with other DDX members such as DDX11 (r = 0.6), DDX56 (r = 0.5) and DDX49 (r = 0.5), while DDX5 also had the most frequent negative correlation with other DDX members such as DDX49 (r = −0.5) (Fig. 4B).
Fig. 4.
Correlation and expression analysis of the first twenty most significant DEAD/H box helicase genes. (A) LUAD. (B) LUSC. (C) Heatmap of the differential expression of the first 20 most significant DDXs in both LUAD and LUSC types. (D) Radar chart showing the association of four DDX genes with TNM-stage parameters of the LUAD and LUSC subtypes.
2.4. Identification of the lead DEAD/H box helicase gene candidates
We then plotted the levels of the differential expression (–log 10 (p-value)) to generate a heat map of all the 20 DDX genes in the two subtypes. Based on this analysis, we identified that DDX11, DDX56, DDX55 and DDX5 showed the most significant differential expression profiles in both subtypes of the NSCLC (Fig. 4C). Using a radar chart (Fig. 4D), we were able to illustrate the multivariate association of these four DDX genes with the whole aspect and three individual TNM-staging parameters of the LUAD and LUSC subtypes. It showed that DDX11 covered more significant association aspects with both subtypes, the M-stage of LUAD and the T-stage of LUSC. DDX55 showed a more significant association with the M-stage of LUSC. DDX5 showed a more significant association with the N-stage of LUSC and the T-stage of LUAD. DDX56 showed a more significant association with the whole LUAD subtype and the N-stage of LUAD.
2.5. Function signatures indicated by GSEA
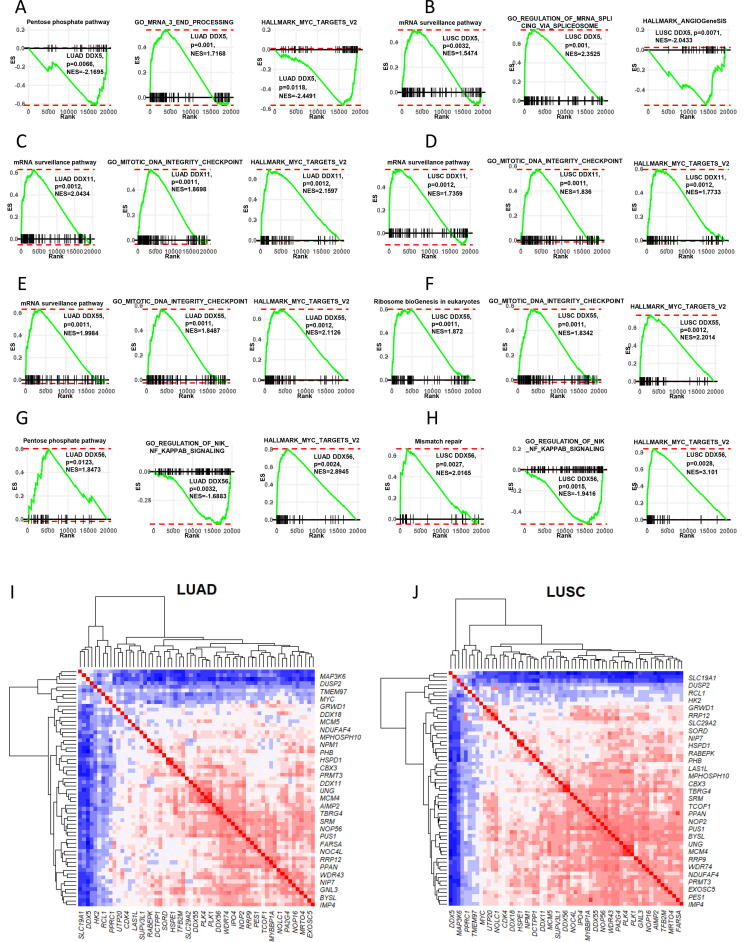
By GSEA analysis, we were able to identify the most significant signalling pathways as the functional signatures of the DDX molecules. In both subtypes of the NSCLC, DDX5-associated genes were found to be enriched in the positive regulation of the mRNA surveillance pathway (LUAD: p = 0.0234; LUSC: p = 0.0011) and mRNA 3′ end processing (LUAD: p = 0.0010; LUSC: p = 0.0011). On the other hand, DDX5-associated genes were enriched in the negative regulation of the pentose phosphate pathway (LUAD: p = 0.0057; LUSC: p = 0.0058) and protein secretion (LUAD: p = 0.0227; LUSC: p = 0.0200). DDX5-associated genes in LUAD were also enriched in the negative regulation of MYC targets (p = 0.0119) (Fig. 5A–B).
Fig. 5.
GSEA of the four DDX candidates. Three significant functional annotations of each DDX were presented. (A) DDX5 in LUAD. (B) DDX5 in LUSC. (C) DDX11 in LUAD. (D) DDX11 in LUSC. (E) DDX55 in LUAD. (F) DDX56 in LUSC. (G) DDX56 in LUAD. (H) DDX56 in LUSC. (I) Heatmap showing the correlation matrix of the MYC target genes and four DDXs in LUAD. (J) Heatmap showing the correlation matrix of the MYC target genes and four DDXs in LUSC.
DDX11-associated genes were found to be enriched in the positive regulation of a cluster of signalling pathways including mRNA surveillance pathway (LUAD: p = 0.0012; LUSC: p = 0.0012), mitotic DNA integrity checkpoint (LUAD: p = 0.0011; LUSC: p = 0.0011) and MYC targets (LUAD: p = 0.0012; LUSC: p = 0.0012). In contrast, DDX11-associated genes were enriched in the negative regulation of protein secretion (LUAD: p = 0.0080; LUSC: p = 0.0080) and IL6-JAK-STAT3 signalling (LUAD: p = 0.0072; LUSC: p = 0.0069) (Fig. 5C–D).
DDX55-associated genes were found to be enriched in the positive regulation of a cluster of signalling pathways including mRNA surveillance pathway (LUAD: p = 0.0011; LUSC: p = 0.0011), Ribosome biogenesis in eukaryotes (LUAD: p = 0.0011; LUSC: p = 0.0012), mitotic DNA integrity checkpoint (LUAD: p = 0.0011; LUSC: p = 0.0011) and MYC targets (LUAD: p = 0.0012; LUSC: p = 0.0012). In contrast, DDX55-associated genes were enriched in the negative regulation of IL6-JAK-STAT3 signalling (LUAD: p = 0.0097; LUSC: p = 0.0092) and protein secretion (LUAD: p = 0.0102; LUSC: p = 0.0095) (Fig. 5E–F).
In both subtypes, DDX56-associated genes were found to be enriched in the positive regulation of a cluster of signalling pathways including homologous recombination (LUAD: p = 0.0022; LUSC: p = 0.0026), proteasome (LUAD: p = 0.00; LUSC: p = 0.00) and MYC targets (LUAD: p = 0.00; LUSC: p = 0.00). In contrast, DDX56-associated genes were enriched in the negative regulation of nik/NFκb signalling (LUAD: p = 0.00; LUSC: p = 0.00), protein secretion (LUAD: p = 0.00; LUSC: p = 0.00), TGF-β signalling (LUAD: p = 0.00; LUSC: p = 0.00), IL6_JAK_STAT3 signalling (LUAD: p = 0.00; LUSC: p = 0.00) and HEDGEHOG signalling (LUAD: p = 0.00; LUSC: p = 0.00) (Fig. 5G–H).
We further performed multiple correlation analysis on the four DDXs and 58 MYC target genes. As shown by the correlation heat map plots in Fig. 5I and J, both in LUAD and LUSC, DDX5 negatively correlated with the majority of these gene targets. In contrast, the other three DDXs correlated positively with the majority of the MYC targets. This further confirmed the result of the GSEA. The correlation coefficient and p-values of each MYC target pair are provided in Supplement Table 2.
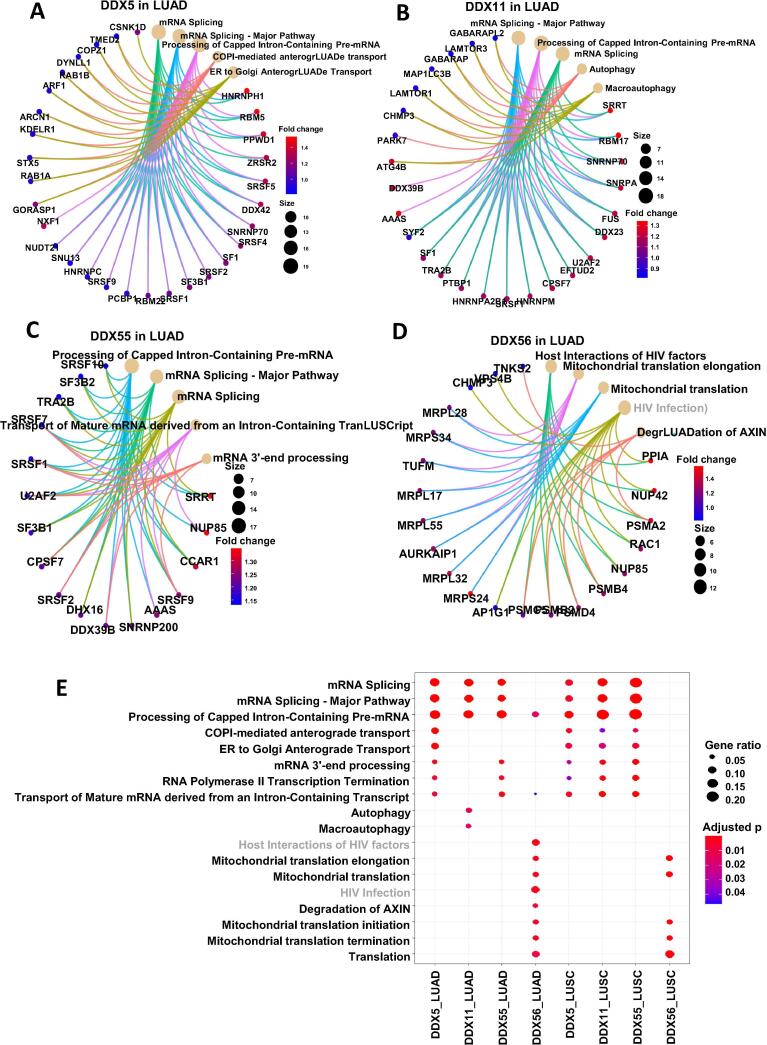
The pathway viewing analysis indicated that DDX5, DDX11 and DDX55 played their roles in mediating RNA splicing and process through regulating their associated genes in lung cancer (Fig. 6). Interestingly, DDX56-associated genes participated in a role in regulating multiple steps of mitochondrial translation.
Fig. 6.
Functional pathway analysis of the differential genes of the four DDX candidates. (A) DDX5 in LUAD. (B) DDX11 in LUAD. (C) DDX55 in LUAD. (D) DDX56 in LUAD. (E) Comparison of the gene function profiles associated with the four DDXs in both LUAD and LUSC types.
2.6. Differential miRNA profiles associated with the DDXs
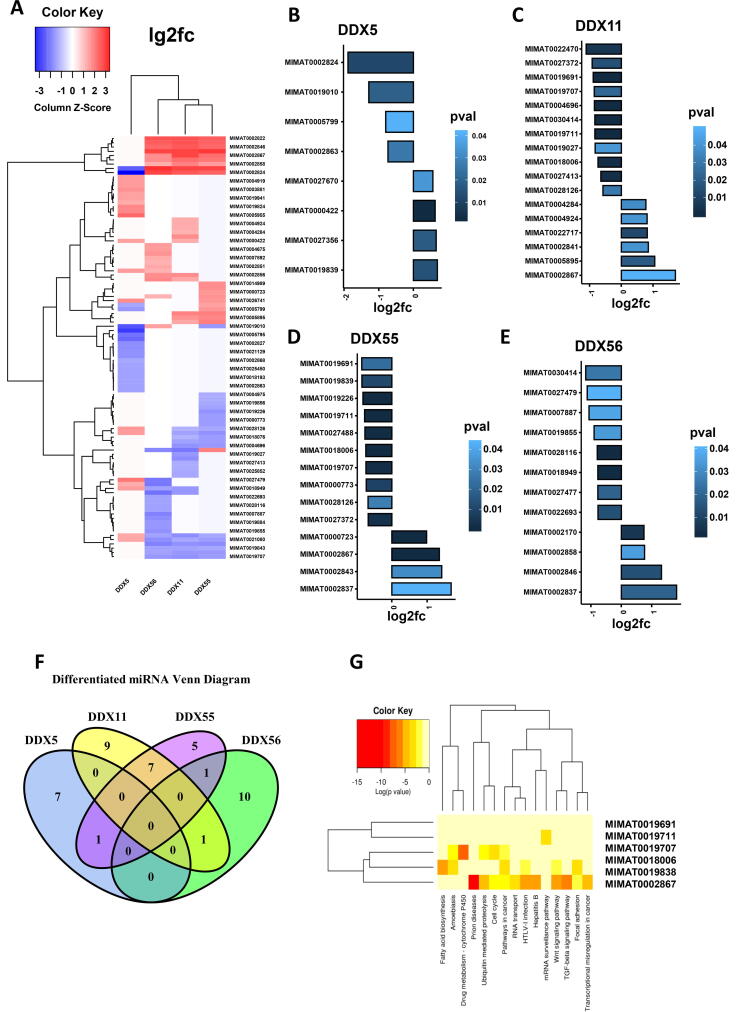
We investigated the correlation of the four DDXs with the miRNA expression by database integration. There were 466 samples filtered after the match of the gene expression database and the miRNA expression database. By differential data analysis, the miRNAs with significant alteration were identified (p < 0.05) in response to the gene expression levels of the four individual DDXs, respectively (i.e. high vs low). As shown in Fig. 7A, the pooled heat map analysis of the lg2FC values of these miRNAs indicated that DDX5 correlated with a distinctive miRNA profile compared to DDX11, DDX55 and DDX56, despite the fact that each DDX showed different miRNA signatures. The miRNAs which were significantly associated with the four individual DDXs are shown in Fig. 7B–E, respectively. The logical relationships of the distinguished miRNAs from different DDX groups are shown using a Venn diagram (Fig. 7F). DDX5 only shared one miRNA with DDX55 which was MIMAT0019839 (hsa-miR-4723-3p). However, DDX5 correlated with MIMAT0019839 positively (lg2fc = 0.69, p < 0.05) while the correlation of DDX55 with this miRNA was negative (lg2fc = −0.85, p < 0.05). DDX5 did not share any differentiated miRNAs with DDX11 and DDX56. MIMAT0030414 (hsa-miR-4433b-3p) negatively correlated with both DDX11 (lg2fc = −0.84, p < 0.05) and DDX56 (lg2fc = −1.17, p < 0.05). MIMAT0002837 (hsa-miR-519b-3p) correlated positively with both DDX55 (lg2fc = 1.68, p < 0.05) and DDX56 (lg2fc = 1.82.84, p < 0.05). DDX11 and DDX55 shared seven differentiated miRNAs which were MIMAT0019691 (hsa-miR-4634), MIMAT0019711 (hsa-miR-4649-5p), MIMAT0018006 (hsa-miR-3622b-3p), MIMAT0027372 (hsa-miR-6735-3p), MIMAT0019707 (hsa-miR-4646-5p), MIMAT0028126 (hsa-miR-7114-3p) and MIMAT0002867 (hsa-miR-520h).
Fig. 7.
Analysis of miRNA profilings which are associated with the four DDX candidates in LUAD. (A) Heatmap of differential miRNAs between the expression levels of the DDXs. (B) Significantly differentiated miRNAs (p < 0.05) between the expression level of DDX5. (C) Significantly differentiated miRNAs between the expression level of DDX11. (D) Significantly differentiated miRNAs between the expression level of DDX55. (E) Significantly differentiated miRNAs between the expression level of DDX56. (F) Venn diagram showing the logical relations of the four DDXs through their-associated miRNAs. (G) Function enrichment analysis of the miRNA targets.
We further investigated the functional enrichment of these differentiated miRNAs using the mirPATH in Diana Tools. As shown in Fig. 7G, these miRNAs are involved in several cancer-cell related signalling pathways including Wnt signalling (MIMAT0002867, MIMAT0009838), focal adhesion (MIMAT0002867, MIMAT0009838), cell cycle (MIMAT0002867, MIMAT0008006) and ubiquitin-mediated proteolysis (MIMAT0002867, MIMAT0008006).
2.7. Mutation profiling associated with DDX
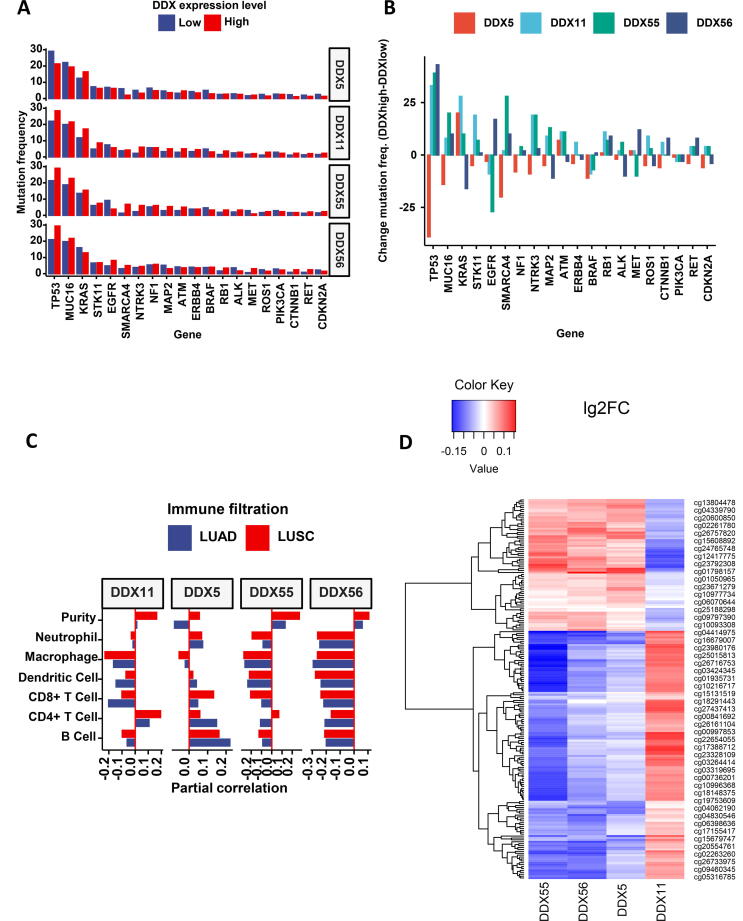
As dysregulated DDXs are implicated in the process of RNA regulation and transcription in cancer, we investigated whether DDX is associated with the mutation of cancer-related genes. As shown in Fig. 8A and 8B, in lung cancer, there were three genes with high frequencies of mutation, TP53, MUC16 and KRAS. High expression of DDX5 is associated with a low level of TP53 mutation, while high expression levels of DDX11, DDX55 and DDX56 are associated with high levels of TP53 mutation. A similar pattern was also observed between the association of MUC16 mutation with the expression levels of these four DDX genes. For KRAS, its mutation frequency is associated negatively with DDX56 expression, while it is associated positively with DDX11, DDX55 and DDX56. Individually, DDX5 expression also is associated negatively with the mutation frequency of SMARCA4 (BRG1). DDX55 however is associated positively with the mutation of SMARCA4, but associated negatively with the mutation of EGFR.
Fig. 8.
The association of the DDX candidates with gene mutation, tumour-infiltrating immune cells and DNA methylation. (A) Mutation frequency of the top 20 genes with the most frequent mutation. (B) Comparison of the absolute mutated number. (C) Tumour-infiltrating immune cell profile. (D) Heatmap of the top 50 most frequent DNA methylation sites.
2.8. Association of DDXs with immune filtration in lung cancer
To investigate whether the interaction of cancer cells with the tumour-infiltrating immune cells may influence the production of DDX molecules, we investigated the association of DDX expression levels with the filtrated immune cells in lung cancer tissues. As shown in Fig. 8C, DDX5 expression positively correlated with CD8 + T cells and B cells. In contrast, DDX11, DDX55 and DDX56 negatively correlated with infiltrated CD8 + T and B cells. The correlations of DDX5 with macrophages and dendritic cells were marginal, while DDX11, DDX55 and DDX56 negatively correlated with infiltrated macrophages and dendritic cells. Overall, this indicated that the immune cell profiles associated with DDX5 were distinct to those associated with DDX11, DDX55 and DDX56. The data of partial correlation coefficient and p-value of the tumour-infiltrating immune cells and DDXs are summarised in Supplement Table 3.
2.9. Association of DNA methylation with DDX expression
We investigated the correlation of DDX gene expression with DNA methylation by dataset combination. As shown in Fig. 8D, the DNA methylation pattern associated with DDX11 was distinctive from those associated with DDX5, DDX55 and DDX56. The top 50 DNA methylation sites associated with these four DDXs are summarised in Supplement Table 4.
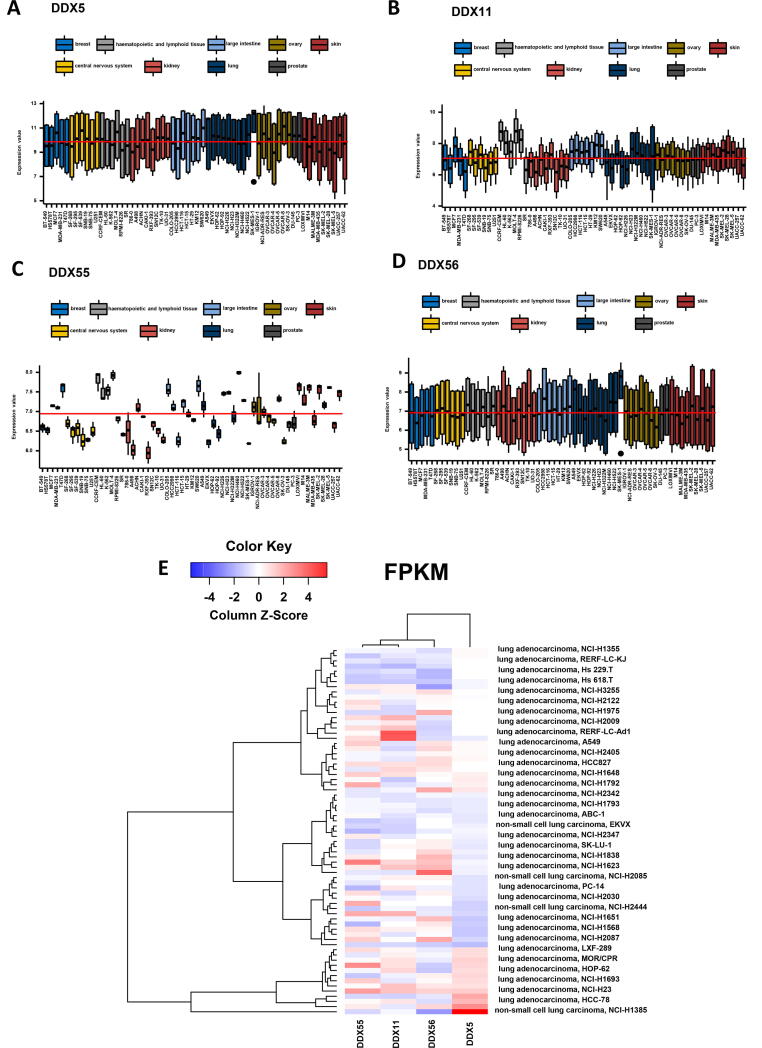
2.10. DDX expression in cancer cell lines
We investigated how these four DDX genes are differentially expressed in the lung cancer cell lines compared to other cancer cell lines. Despite the large variation of the pooled data, the expression levels of DDX5 were similar to the cell lines from other cancer types, although one lung cancer cell line, SK-MES-1 showed high expression (Fig. 9A). The expression levels of DDX11 and DDX55 were varied in lung cancer cell lines, although they are in the range of other cancer cell lines (Fig. 9B and C). Interestingly the variation of the DDX55 expression data is relatively narrow. The expression levels of DDX56 were similar to the cell lines from other cancer types, although SK-MES-1 showed high expression (Fig. 9D). We further examined the expression patterns of the four DDX genes only in lung cancer cell lines. The clustered analysis indicated that in the 71 lung cancer cell lines, the gene expression profile of DDX5 is distinguished from those of DDX11, DDX55 and DDX56 (Fig. 9E). The databases of the gene repression of the four DDXs in pan-cancer and lung cancer cell lines are provided in Supplement Tables 5 and 6, respectively.
Fig. 9.
Expression levels of the DDX genes in cancer cell lines. (A) Gene expression level of DDX5 in pan-cancer cell lines. (B) Gene expression level of DDX11 in pan-cancer cell lines. (C) Gene expression level of DDX55 in pan-cancer cell lines. (D) Gene expression level of DDX56 in pan-cancer cell lines. (E) Heatmap of the gene expression levels of the four DDXs in 71 lung cancer cell lines using the data from Expression Atlas.
3. Discussion
In this study, we present a whole image of the prognosis comparison of all the DEAD/H box helicases, considering their possible differences between LUAD and LUSC as well as all types in lung cancer. The prognostic values of a biomolecule can be potentially cancer-type dependent or sometimes the opposite, in different (sub)types or even in different stages of one cancer [44], [45]. There are various mechanisms which could count for this discrepancy. A protein biomarker may have varied spliced isoforms with diverse functions, be localised to different cellular compartments (e.g. nuclear/cytoplasm), be signalled in a complex to modulate different downstream signalling pathways, may interact to a loss/gain function (e.g. mutation) of its effector/partner in different subtypes, be regulated/interacted by other cells in a different tumour microenvironment, or mediates a substrate with multiple functions (e.g. INOS) [46]. For example, in a breast cancer subtype with wild-type p53, in response to DNA damage stress, DDX3 associates with p53, leading to the accumulation of p53 in the nucleus, and the activation of its downstream target p21 expression, to modulate DNA damage-induced apoptosis. However, in a cancer subtype with non-functional or mutant p53, DDX3 otherwise inhibits the cellular apoptosis process [25]. In non-small cell lung cancer, the subtypes of LUAD and LUSC differ in their pathological types, molecular features and treatment responses [47]. As a transcript signature, one gene cluster (e.g. MUC5B, HABP2, MUC21, and KCNK5) can be up-regulated in LUAD but down-regulated in LUSC whilst another cluster (e.g. CSTA, P2RY1, and ANXA8) can be vice-versa [48]. Further, some gene mutations have been particularly linked with the prognostic outcomes of LUSC [49]. Therefore, to identify biomarker candidates with prognostic significance, it is vital to take into account the subtypes of cancer, in this case, LUAD and LUSC in NSCLC.
Among the lead DDX candidates we identified by integrative bioinformatic analysis, DDX5 is associated positively with prognosis in NSCLC. Lower expression of DDX5 genes predicted a worse survival including OS, first progression (FP) and post-progression survival (PPS). One previous study reported a significantly higher DDX5 expression in tumour samples than in paired adjacent normal tissue, indicated by Western blots and immunohistochemistry of limited samples and without follow-up information [50]. This contrasted with the observations through our bioinformatic analysis. Lung cancer is a heterogeneous disease in which differences between and within the subtypes are driven by genetic and environmental factors, therefore there can be a large amount of variability between patients [51], [52]. Limited numbers of samples involved in a study may not be robust enough to resist being skewed by extreme data points, so the results could be vulnerable to the introduction of bias [53]. In contrast, our use of large-scale datasets such as TCGA and GEO allowed a substantial number of normalised samples to be grouped for analysis; with more samples, the data should provide a more representative view of the population which, in theory, would increase the reliability and relevance of the data. Whilst it is still possible for the associations to have occurred by chance, the large amount of data implemented should decrease the probability of this event occurring. As a drawback, the inclusion of tumour and paired adjacent normal tissues from each patient is not available with the Kaplan-Meier plotter and the TCGA dataset. We instead ran a paired analysis using a GEO dataset, GSE18842, with 44 paired lung cancer tissue samples. This analysis again indicated that there is lower DDX5 gene expression in tumour tissues than in paired adjacent normal tissue, which supports our observations in this study using the TCGA dataset and suggests this is unlikely to be biased by genetic or environmental factors not related to cancer.
DDX5 dysregulation has been found in a range of cancers, including hepatocellular carcinoma where low DDX5 expression correlates with a worse prognosis [54]. As DDX5 plays an important role in the initial stages of miRNA processing in the Drosha complex, its downregulation may limit the ability of the cell to perform post-transcriptional inhibition resulting in the upregulation of oncogenes [55]. It is important to note that, due to its ability to perform a wide range of functions, the role of DDX5 is very dependent on the context in which it is found. Therefore, although miRNA is involved in the mechanism by which DDX5 promotes hepatocellular carcinoma, it is unclear as to whether this would be the same in lung cancer. However, as this is the only other current example of DDX5 downregulation, it may be appropriate to determine whether similar associations exist in lung cancer. There is already evidence for a negative association between Dicer, another gene involved in miRNA synthesis downstream of DDX5, expression and survival in lung cancer [56]. The previous study on Dicer however did not show a correlation between Drosha expression and survival, and did not measure the expression of the proteins which complex with Drosha and are necessary for its function, such as DDX5. The downregulation of DDX5 shown in the bioinformatic data highlights the potential for a similar correlation in lung cancer as in hepatocellular carcinoma. In breast cancer, however, DDX5 is shown as an oncogenic coactivator of transcription factor Fra-1 [57]. DDX5 regulates DNA replication prior to proliferation and regulates miRNAs involved in mediating cytoskeletal reorganization in breast cancer cells [58]. In prostate cancer, DDX5 is overexpressed and considered as a transcriptional coactivator of androgen receptor, probably through interactions with β-Catenin and RNAP II [59]. Therefore, the expression of DDX5 helicase is cancer dependent.
The other three DDX candidates (i.e. DDX11, DDX55 and DDX56) are observed as unfavourable prognostic indicators which are unlike DDX5. DDX11 has been reported to be negatively associated with the survival of patients with lung cancer [60]. DDX11 is also a survival indicator in other solid cancers such as melanomas and is essential for melanoma metastasis, by participating in chromosome segregation, telomere and cell apoptosis [61]. Functionally, the DDX11 helicase is involved in the regulation of the G2/M transition and is required for sister chromatid cohesion. The depletion of DDX11 results in centromeric loss and arm cohesion defects, which is JunB dependent in ALK-positive anaplastic large cell lymphoma [62]. DDX55 and DDX56 have not yet been linked with lung cancer in the literature. However, DDX55 has been considered as one of the dysregulated genes to predict poor early recurrence-free survival of hepatocellular carcinoma after a microscopically margin-negative (R0) resection [63]. DDX56 is a negative prognostic factor in colorectal cancer and osteosarcoma [64], [65]. The expression of DDX56 is associated with lymphatic invasion and distant metastasis. There is a report that DDX56 may play a role in ribosome assembly and maturation [66].
It is believed that miRNAs play a vital role in the initiation, progression and metastasis of cancer by translationally repressing or silencing oncogenic or tumour-suppressing mRNA targets. The biosynthesis of miRNAs contains multiple steps which include the cleavage of pri-miRNAs to pre-miRNAs, the nucleic exit of pre-miRNA, cleavage of pre-miRNA to miRNA/miRNA duplex and the formation of mature miRNAs. DDX5 protein can bind directly with the nuclear microprocessor complex Drosha/DGCR8 (Pasha) to facilitate the processes of pri-miRNA cleavage [67], [68]. DDX5 functions as a stress sensor to tune the cleavage activity of Drosha/DGCR8 in pathological conditions such as cancer. Stress-induced factors, including TP53 and TGFβ, can trigger the mediation of DDX5 and lead to the production of particular clusters of pre-miRNAs, respectively [69], [70]. Among the eight DDX5-correlated miRNAs, miR-124 suppresses tumorigenesis and enhances radiation-induced apoptosis in lung cancer [71], [72] while miR-128 enhances mutant p53 gain of function and promotes chemoresistance-associated metastasis in lung cancer [73], [74]. hsa-miR-124-3p is predicted to target DDX5 mRNA at the 3′ UTR (position 1295-1301) as well. Among the DDX11-associated miRNAs, miR-675 appears to suppress NSCLC by targeting a pro-carcinogenic putative cannabinoid receptor GPR55 [75] with hsa-miR-4646-3p predicted to target the DDX11 3′ UTR at position 2906-2913. Among the miRNAs correlated with DDX55, there is no evidence that any are implicated in lung cancer. Among the miRNAs correlated with DDX56, has-miR-204 is positively associated with the survival of lung cancer including the LUAD and LUSC subtypes. We have identified that the correlated miRNA profile of DDX5 is distinctive from those which correlate with DDX11, DDX55 and DDX56. However, how these differences happen requires further investigation in the future.
It is known that tumour-infiltrating immune cells have a profound impact on tumour progression and the efficacy of anti-tumour therapies. Therefore, there have been various approaches developed to track their dynamics [76]. By examining the association of the DDXs with the infiltrated immune cells, we found that DDX55 and DDX56 are quite similar, while DDX5 is different from the other three. In particular, DDX5 positively correlates with B cells and CD8 + T cells, unlike the others. It would be interesting to ask these questions regarding the mechanism: Is it possible that DDX5 produced by cancer cells attracts the infiltration of these immune cells? Also, can different types of infiltrated immune cells produce different levels of DDX5 and other DDXs in response to the tumour microenvironment?
The combination of distinctive protein domains determines the function of a protein. All four DDXs share one protein domain called P-loop_NTPase. The P-loop_NTPase domain catalyses the hydrolysis of both GTP and ATP. DDX5 has only one different domain, RNA helicase p68 repeat (p68_rpt), compared with the other three DDXs. This p68_rpt domain plays a role in the regulation of the alternative splicing of pre-mRNA such as tau [77]. It is difficult to understand how a difference of only one domain could contribute to the distinctive association of DDX5 with prognosis, gene mutation and tumour immune cell profiling. It might be that there are other mechanisms underlining the DDX5 function which have not yet been recognised. DDX55 and DDX56 share the same pattern of seven protein domains. DDX11 shares only the common P-loop_NTPase with others but has considerable extra domains, which cannot be seen in DDX5, DDX55 and DDX56. The specific domains of DDX11 such as CHL1-like DNA helicase and Rad3 may confer its important role in DNA repair in cancer [78].
There is a positive mutual correlation between DDX11, DDX55 and DDX56, implying that they may be co-expressed in lung cancer. The abundance of viral nucleic acid in lung cancer may shift the expression levels of DDXs. There is evidence that some DEAD/H box RNA helicases play a role as anti-pathogen immune sensors. In response to certain pathogens, they can transport and participate in the formation of a protein complex which interacts with viral RNA/DNAs and facilitate their nuclear export [79], [80]. A previous study has suggested that most LUSC and LUAD contain either retroviral or HPV DNA [81]. However, as there is no large-scale gene expression dataset that contains the information of viral infection of samples, this could not be investigated thoroughly.
By function enrichment analysis, we found that DDX5 is involved in the negative regulation of MYC, while DDX11, DDX55 and DDX56 are all involved in the positive regulation of MYC. One study demonstrated that DDX5 may unwind the G4 structure of the MYC promoters which leads to the activation of MYC transcription [82]. However, there is no significant correlation of the gene expression of DDX5 and MYC in lung cancer. As a well-characterised proto-oncogenic transcript factor, MYC can be dysregulated at transcriptional and post-transcriptional levels. MYC transcription can be enhanced by growth factors or cytokines such as NOTCH, WNT, IGF1, TGFα, EGF and IL-6. In another way, the TGF-β and Type I interferon (IFN-alpha/-beta) signalling pathways may play a suppressing role in MYC expression. DDX5 has been reported as a negative regulator of the Wnt/β-catenin pathway in cancer [83], [84], which may partially explain the negative relationship of DDX5 and MYC. There is no report on the relationship of MYC and DDX11/DDX55/DDX56 genes. At the protein level, however, there is evidence that MYC protein can bind DDX11 directly, or regulate the function of DDX11 by activating cohesion regulator noncoding RNA (CONCR).
Our data indicate that DDX5 is negatively associated with the mutation frequency of TP53 and MUC16. The gain-of-function mutation of MUC16, frequently observed in lung cancer, confers a protecting role in cancer epithelial cells from chemotherapeutic drugs [85]. It has also been reported that MUC16 mutation may enhance the infiltrated cytotoxic T cells in the tumour microenvironment [86]. The tumour-infiltrated cytotoxic T cells might contribute to the alteration of DDX expression although further investigation is needed. The loss-of-function mutation of p53 may be caused by exposure to carcinogenic substances or a genetic defect. As a risk factor of cancer development, p53 mutants drive cancer cells to proliferate and spread beyond control. As DDXs play an essential role in RNA-associated genomic instability and DNA repair [87], they may mediate the DNA mutation frequency as a consequence of stress-induced DNA damage. Aberrant promoter methylations of tumour suppressor genes have been frequently observed in lung cancer. There is evidence that miRNA clusters can control the DNA methylation dynamics [88], [89], [90]. We hypothesise that DDXs may alter the DNA methylation patterns by participating in miRNA processing, which would be interesting to investigate.
4. Conclusion
To date, there is no integrated bioinformatic analysis of large-scale datasets available to investigate whether DEAD/H box helicases can be identified as biomarkers, to predict the prognosis of lung cancer. In this study, we conducted and identified four DDX box helicases (namely, DDX5, DDX11, DDX55 and DDX56) with the most clinical significance by carrying out an integral bioinformatic analysis of multivariate lung cancer databases. We demonstrated that these four DDX biomarkers have diverse functional links with the MYC-signalling pathway and are implicated in the mutation levels of p53 and MUC16, the top two most frequently mutated genes in lung cancer. We also elucidated the distinctive association of these four biomarkers with tumour-infiltrated immune cells, miRNA expression patterns and DNA methylation profile.
We propose a new avenue, following the identification of four DDX biomarker candidates, in which our understanding remains limited. It would be worthy to further pinpoint the cellular and molecular functional roles of these DDX candidates in lung cancer, in order to enhance our understanding of their prognostic and therapeutic potential. Furthermore, given the size of the gene family and the seemingly diverse roles in different cancer types, it would be necessary to explore the wider context of the connection both with other family members and in other tumour types.
5. Methods
5.1. Survival analysis
The analysis of the survival was performed using the datasets (Version on 25 May 2020) from the Kaplan Meier plotter (https://kmplot.com) [91]. The pooled database repository (n = 1925) contained the data from Gene Expression Omnibus (GEO) (n = 1288), caArray (n = 504) and the Cancer Genome Atlas (TCGA) (n = 133). The summary of data sources is provided in Supplement Table 7. Patients were stratified using the option of Auto select best cut off for KM plotting.
5.2. Correlation of the DEAD/H box helicase genes
The RNASeq datasets, including TCGA normal and tumour datasets of lung cancer and GTEX dataset of the normal lung, were obtained from Github (http://github.com/mskcc/RNAseqDB). The datasets were values of FPKM (fragments per kilobase of exon model per million reads mapped) with quantile normalisation and correction for batch effects [92]. The clinical data of the TCGA samples were obtained from UCSC Xena (https://tcga.xenahubs.net) using the UCSCXenaTools [93]. They were then merged according to sample names and sample types. The correlation of the DEAD/H box helicase genes was performed using the “ggcorrplot” package in R [94].
5.3. Associations with clinicopathological parameters
These clinical-related associations were evaluated using those DEAD/H box helicase genes with huge significance in survival analysis. We investigated the clinical features including type, TNM stage, grade, pathological stage, age, gender and smoking intensity.
5.4. Gene set enrichment analysis (GSEA)
To prepare the data set for a GSEA analysis, the TCGA RNASeq expression data were first divided into two levels (high & low) by the median expression values of a DEAD box helicase (DDX) gene. The log2 of the fold change (log2fc) of each gene in the RNASeq database was then calculated with this equation: log2fc = log2(high)-log2(low). The p-value and adjusted p-value (padj) of each gene were calculated using the “limma” package in R [95]. The rank of each gene was calculated using this equation: rank = −log10 (padj)/log2fc. The gene expression profiles associated with each DDX are provided in Supplement Table 8. The annotated gene sets for GSEA analysis were obtained from the Molecular Signatures Database (MSigDB) (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp). We then conducted the GSEA analysis using a “fgsea” package [96]. The gene set collections which we applied were obtained from Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO) and Hallmark. We then sought to highlight the top 10 signalling pathways from each gene set, according to their normalised enrichment scores (NES), if there was significance (p < 0.05). Maximally, five signalling pathways were enriched from NES > 0 while another five were from NES < 0, depending on the ascending order of their p-values. The gene function profiles associated with each DDX were also compared using a “clusterProfiler” package in R and the Reactome pathway database [97], [98].
5.5. miRNA profiling
The dataset of miRNAs was obtained from UCSC Xena which included 495 samples and 2228 miRNA targets. This dataset was then merged with a sub-dataset from TCGA RNAseq which contained the expression levels of the DDX genes. The log2fc values of miRNAs were then calculated according to the expression levels of a DDX gene. The miRNAs with the top 20 and bottom 20 of the log2fc values associated with each DDX were selected for the calculation of the p-values. The data of differential miRNAs for all DDXs were then pooled for the heatmap plotting analysis (Supplement Table 9). The miRNAs with significance (p < 0.05) for each DDX were plotted with a bidirectional bar chart. A Venn diagram was used to evaluate the logical relations of the DDXs through their-associated miRNAs. The predicted miRNA targets and their functional analysis was performed using DIANA-miRPath (v3.0) (http://diana.imis.athena-innovation.gr/DianaTools) [99].
5.6. Mutation
The dataset of mutation was obtained from UCSC Xena which included 514 samples and 40,543 gene targets. This dataset was then merged with the expression levels of the DDX genes. The top 20 genes with the most frequent mutation were compared for their difference between the levels of individual DDX genes (Supplement Table 10).
5.7. Methylation
The dataset of DNA methylation was obtained from UCSC Xena which included 493 samples and 485,577 methylation sites. This dataset was then merged with the expression levels of the DDX genes according to the matched sample names (n = 477). The difference of the DDX methylation frequencies associated with each DDX gene was analysed using the limma Bioconductor package.
5.8. Tumour filtrating immune cell profiling
The correlation of tumour-infiltrating immune cell profile and gene correlation was obtained from TIMER (https://cistrome.shinyapps.io/timer) [100].
5.9. Gene expression of lung cancer cell lines
The data of DDX gene expression in pan-cancer cell lines were obtained from CellExpress (http://cellexpress.cgm.ntu.edu.tw). The obtained database was originally from three datasets including CCLE (GSE36133) [101], NCI-60 (GSE32474) [102] and Sanger Cell Line Project (GSE68950) [103]. There were 60 cell lines from 9 primary cancer sites. The boxplots were made using the “ggpubr” package in R. For an extended investigation of lung cancer cell lines, the gene expression data including 71 lung cancer cell lines were obtained from Expression Atlas (https://www.ebi.ac.uk/gxa).
5.10. Correlation of DDX genes with the MYC target genes
The list of MYC target genes was obtained from GSEA (https://www.gsea-msigdb.org/gsea). The correlation of these with DDX genes was conducted using the “ggcorrplot” package in R.
5.11. Domain predication
The domains of the DEAD/H box helicase proteins were predicted using InterPro (https://www.ebi.ac.uk/interpro).
5.12. Data analysis
The data were processed and analysed on the platform of Rstudio (Version 1.3.959) [104] equipped with R (Version 3.6.3) [105]. The R scripts and package version information are provided in Supplement Table 11.
Authors' contributions
Conception and design: Y. Cui; Development of methodology: Y. Cui; Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Y. Cui; Writing, review, and/or revision of the manuscript: Y Cui, A Hunt, E. Birkin, Z. Li, J. Lane, F. Ruge, W.G. Jiang; Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Y. Cui. Study supervision: Y. Cui and W.G. Jiang.
Funding
This work was financially supported by the Realcan Fellowship.
Ethical Approval and Consent to participate
Not applicable.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank Dr Hywel Williams for his valuable comments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.12.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Gene expression profiles of the DEAD/H box helicase genes at different gender, age, overall stage and smoking intensity in LUAD.
Gene expression profiles of the DEAD/H box helicase genes among different genders, ages, overall stages and smoking intensity in LUSC.
Data of the p-values and HR of all the DEAD/H box genes following KM-plotting analysis.
Correlation coefficient and p-values of the individual MYC targets.
Data of the correlation between tumour-infiltrating immune cells and the four DDXs.
Data of the top 50 methylation sites with the most significant difference between the expression levels of each DDX.
Database of the gene repression of the four DDXs in pan-cancer cell lines.
Database of the gene repression of the four DDXs in lung cancer cell lines.
Clinicopathological features of the lung cancer samples.
Database of the gene expression profiles associated with DDX candidates.
Data of differential miRNAs which are associated with the DDXs for the heatmap plotting analysis.
Data of the top 20 genes with the most frequent mutation and their association with the expression levels of the four DDX genes.
R scripts and Session Information used in this study.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Zappa C., Mousa S.A. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res. 2016;5(3):288–300. doi: 10.21037/tlcr.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis P.M., Vandermeer R. Delays in the diagnosis of lung cancer. J Thoracic Dis. 2011;3:183. doi: 10.3978/j.issn.2072-1439.2011.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karachaliou N., Pilotto S., Lazzari C., Bria E., de Marinis F., Rosell R. Cellular and molecular biology of small cell lung cancer: an overview. Transl Lung Cancer Res. 2016;5:2–15. doi: 10.3978/j.issn.2218-6751.2016.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutherland K.D., Berns A. Cell of origin of lung cancer. Mol Oncol. 2010;4:397–403. doi: 10.1016/j.molonc.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Relli V., Trerotola M., Guerra E., Alberti S. Distinct lung cancer subtypes associate to distinct drivers of tumor progression. Oncotarget. 2018;9(85):35528–35540. doi: 10.18632/oncotarget.26217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones G.S., Baldwin D.R. Recent advances in the management of lung cancer. Clin Med. 2018;18(Suppl 2):s41–s46. doi: 10.7861/clinmedicine.18-2-s41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linder P. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucl Acids Res 2006, 34, 4168-4180. [DOI] [PMC free article] [PubMed]
- 9.Linder P., Jankowsky E. From unwinding to clamping — the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12(8):505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 10.Fairman-Williams M.E., Guenther U.-P., Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20(3):313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers G.W., Jr., Komar A.A., Merrick W.C. eIF4A: the godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- 12.Andreou A.Z., Klostermeier D. The DEAD-box helicase eIF4A: paradigm or the odd one out? RNA Biol. 2013;10(1):19–32. doi: 10.4161/rna.21966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linder P., Lasko P.F., Ashburner M., Leroy P., Nielsen P.J., Nishi K., Schnier J., Slonimski P.P. Birth of the D-E-A-D box. Nature. 1989;337(6203):121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 14.Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucl Acids Res. 2011;39(Database):D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sengoku T., Nureki O., Nakamura A., Kobayashi S., Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 16.Jarmoskaite I., Russell R. DEAD-box proteins as RNA helicases and chaperones: DEAD-box proteins. WIREs RNA. 2011;2(1):135–152. doi: 10.1002/wrna.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bevilacqua P.C., Ritchey L.E., Su Z., Assmann S.M. Genome-wide analysis of RNA secondary structure. Annu Rev Genet. 2016;50:235–266. doi: 10.1146/annurev-genet-120215-035034. [DOI] [PubMed] [Google Scholar]
- 18.Gilman B., Tijerina P., Russell R. Distinct RNA-unwinding mechanisms of DEAD-box and DEAH-box RNA helicase proteins in remodeling structured RNAs and RNPs. Biochem Soc Trans. 2017;45:1313–1321. doi: 10.1042/bst20170095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamann F., Enders M., Ficner R. Structural basis for RNA translocation by DEAH-box ATPases. Nucleic Acids Res. 2019;47:4349–4362. doi: 10.1093/nar/gkz150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Q., Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13(11):981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 21.Yang Q., Del Campo M., Lambowitz A.M., Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28(2):253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 22.Fuller-Pace F.V. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paine I., Posey J.E., Grochowski C.M., Jhangiani S.N., Rosenheck S., Kleyner R., Marmorale T., Yoon M., Wang K., Robison R., Cappuccio G., Pinelli M., Magli A., Coban Akdemir Z., Hui J., Yeung W.L., Wong B.K.Y., Ortega L., Bekheirnia M.R., Bierhals T., Hempel M., Johannsen J., Santer R., Aktas D., Alikasifoglu M., Bozdogan S., Aydin H., Karaca E., Bayram Y., Ityel H., Dorschner M., White J.J., Wilichowski E., Wortmann S.B., Casella E.B., Kitajima J.P., Kok F., Monteiro F., Muzny D.M., Bamshad M., Gibbs R.A., Sutton V.R., Van Esch H., Brunetti-Pierri N., Hildebrandt F., Brautbar A., Van den Veyver I.B., Glass I., Lessel D., Lyon G.J., Lupski J.R. Paralog studies augment gene discovery: DDX and DHX genes. Am J Human Genet. 2019;105(2):302–316. doi: 10.1016/j.ajhg.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fröhlich A., Rojas-Araya B., Pereira-Montecinos C., Dellarossa A., Toro-Ascuy D., Prades-Pérez Y., García-de-Gracia F., Garcés-Alday A., Rubilar P.S., Valiente-Echeverría F., Ohlmann T., Soto-Rifo R. DEAD-box RNA helicase DDX3 connects CRM1-dependent nuclear export and translation of the HIV-1 unspliced mRNA through its N-terminal domain. Biochimica et Biophysica Acta (BBA) – Gene Regulatory Mechanisms. 2016;1859(5):719–730. doi: 10.1016/j.bbagrm.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Zhao L., Mao Y., Zhou J., Zhao Y., Cao Y., Chen X. Multifunctional DDX3: dual roles in various cancer development and its related signaling pathways. Am. J. Cancer Res. 2016;6:387–402. [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson B.J., Bates G.J., Nicol S.M., Gregory D.J., Perkins N.D., Fuller-Pace F.V. The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol Biol. 2004;5:11. doi: 10.1186/1471-2199-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang H., McDonald D., Middlesworth T., Hope T.J., Wong-Staal F. The carboxyl terminus of RNA helicase A contains a bidirectional nuclear transport domain. Mol Cell Biol. 1999;19(5):3540–3550. doi: 10.1128/MCB.19.5.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Guthrie C. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA. 1998;4:1216–1229. [PMC free article] [PubMed] [Google Scholar]
- 29.Giri B., Smaldino P.J., Thys R.G., Creacy S.D., Routh E.D., Hantgan R.R., Lattmann S., Nagamine Y., Akman S.A., Vaughn J.P. G4 resolvase 1 tightly binds and unwinds unimolecular G4-DNA. Nucleic Acids Res. 2011;39:7161–7178. doi: 10.1093/nar/gkr234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curmi F., Cauchi R.J. The multiple lives of DEAD-box RNA helicase DP103/DDX20/Gemin3. Biochem. Soc. Trans. 2018;46:329–341. doi: 10.1042/bst20180016. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D., Weinberg R.A. The Hallmarks of Cancer. Cell. 2000;100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 32.Sadikovic B., Al-Romaih K., Squire J.A., Zielenska M. Cause and consequences of genetic and epigenetic alterations in human cancer. Curr Genomics. 2008;9:394–408. doi: 10.2174/138920208785699580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newman M., Sfaxi R., Saha A., Monchaud D., Teulade-Fichou M.-P., Vagner S. The G-Quadruplex-Specific RNA Helicase DHX36 Regulates p53 Pre-mRNA 3′-End Processing Following UV-Induced DNA Damage. J Mol Biol. 2017;429(21):3121–3131. doi: 10.1016/j.jmb.2016.11.033. [DOI] [PubMed] [Google Scholar]
- 34.Song C., Hotz-Wagenblatt A., Voit R., Grummt I. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 2017;31(13):1370–1381. doi: 10.1101/gad.300624.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah A., Rashid F., Awan H.M., Hu S., Wang X., Chen L., Shan G.e. The DEAD-Box RNA Helicase DDX3 Interacts with m 6 A RNA Demethylase ALKBH5. Stem Cells Int. 2017;2017:1–11. doi: 10.1155/2017/8596135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuller-Pace F.V. DEAD box RNA helicase functions in cancer. RNA Biol. 2013;10(1):121–132. doi: 10.4161/rna.23312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicol S.M., Bray S.E., Black H.D., Lorimore S.A., Wright E.G., Lane D.P. The RNA helicase p68 (DDX5) is selectively required for the induction of p53-dependent p21 expression and cell-cycle arrest after DNA damage. Oncogene. 2013;32:3461–3469. doi: 10.1038/onc.2012.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chao C.-H., Chen C.-M., Cheng P.-L., Shih J.-W., Tsou A.-P., Lee Y.-H.-W. DDX3, a DEAD box RNA helicase with tumor growth–suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res. 2006;66:6579–6588. doi: 10.1158/0008-5472.CAN-05-2415. [DOI] [PubMed] [Google Scholar]
- 39.Lin H., Liu W., Fang Z., Liang X., Li J., Bai Y., Lin L., You H., Pei Y., Wang F., Zhang Z.-Y. Overexpression of DHX32 contributes to the growth and metastasis of colorectal cancer. Sci Rep. 2015;5(1) doi: 10.1038/srep09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taniguchi T., Iizumi Y., Watanabe M., Masuda M., Morita M., Aono Y., Toriyama S., Oishi M., Goi W., Sakai T. Resveratrol directly targets DDX5 resulting in suppression of the mTORC1 pathway in prostate cancer. Cell Death Disease. 2016;7 doi: 10.1038/cddis.2016.114. e2211–e2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tajirika T., Tokumaru Y., Taniguchi K., Sugito N., Matsuhashi N., Futamura M., Yanagihara K., Akao Y., Yoshida K. DEAD-box protein RNA-helicase DDX6 regulates the expression of HER2 and FGFR2 at the post-transcriptional step in gastric cancer cells. Int J Mol Sci. 2005;2018:19. doi: 10.3390/ijms19072005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Voss M.R.H., van Diest P.J., Raman V. Targeting RNA helicases in cancer: The translation trap. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2017;1868:510–520. doi: 10.1016/j.bbcan.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barkovich K.J., Moore M.K., Hu Q., Shokat K.M. Chemical genetic inhibition of DEAD-box proteins using covalent complementarity. Nucleic Acids Res. 2018;46:8689–8699. doi: 10.1093/nar/gky706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y., Wang W., Chen Y., Huang Y., Zhang J., He S., Tan Y., Qiang F., Li A., Røe O.D., Wang S., Zhou Y., Zhou J. The opposite prognostic significance of nuclear and cytoplasmic p21 expression in resectable gastric cancer patients. J Gastroenterol. 2014;49(11):1441–1452. doi: 10.1007/s00535-013-0900-4. [DOI] [PubMed] [Google Scholar]
- 45.Mzoughi S., Tan Y.X., Low D., Guccione E. The role of PRDMs in cancer: one family, two sides. Curr Opin Genet Dev. 2016;36:83–91. doi: 10.1016/j.gde.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Vannini F., Kashfi K., Nath N. The dual role of iNOS in cancer. Redox Biol. 2015;6:334–343. doi: 10.1016/j.redox.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shea M., Costa D.B., Rangachari D. Management of advanced non-small cell lung cancers with known mutations or rearrangements: latest evidence and treatment approaches. Therapeutic Adv Respiratory. 2016;10(2):113–129. doi: 10.1177/1753465815617871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lucchetta M., da Piedade I., Mounir M., Vabistsevits M., Terkelsen T., Papaleo E. Distinct signatures of lung cancer types: aberrant mucin O-glycosylation and compromised immune response. BMC Cancer. 2019;19(1) doi: 10.1186/s12885-019-5965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012, 489, 519-525, doi:10.1038/nature11404 [DOI] [PMC free article] [PubMed]
- 50.Wang Z., Luo Z., Zhou L., Li X., Jiang T., Fu E. DDX 5 promotes proliferation and tumorigenesis of non-small-cell lung cancer cells by activating β-catenin signaling pathway. Cancer Sci. 2015;106:1303–1312. doi: 10.1111/cas.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Z., Fillmore C.M., Hammerman P.S., Kim C.F., Wong K.-K. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marino F.Z., Bianco R., Accardo M., Ronchi A., Cozzolino I., Morgillo F., Rossi G., Franco R. Molecular heterogeneity in lung cancer: from mechanisms of origin to clinical implications. Int. J. Med. Sci. 2019;16(7):981–989. doi: 10.7150/ijms.34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pannucci C.J., Wilkins E.G. Identifying and avoiding bias in research. Plast Reconstr Surg. 2010;126(2):619–625. doi: 10.1097/PRS.0b013e3181de24bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kitagawa N., Ojima H., Shirakihara T., Shimizu H., Kokubu A., Urushidate T. Downregulation of the micro RNA biogenesis components and its association with poor prognosis in hepatocellular carcinoma. Cancer Sci. 2013;104:543–551. doi: 10.1111/cas.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen J., Hung M.-C. Signaling-mediated regulation of MicroRNA processing. Cancer Res. 2015;75(5):783–791. doi: 10.1158/0008-5472.CAN-14-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karube Y., Tanaka H., Osada H., Tomida S., Tatematsu Y., Yanagisawa K. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He H., Song D., Sinha I., Hessling B., Li X., Haldosen L.-A., Zhao C. Endogenous interaction profiling identifies DDX5 as an oncogenic coactivator of transcription factor Fra-1. Oncogene. 2019;38(28):5725–5738. doi: 10.1038/s41388-019-0824-4. [DOI] [PubMed] [Google Scholar]
- 58.Wang D., Huang J., Hu Z. RNA helicase DDX5 regulates microRNA expression and contributes to cytoskeletal reorganization in basal breast cancer cells. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.011932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clark E.L., Hadjimichael C., Temperley R., Barnard A., Fuller-Pace F.V., Robson C.N. p68/DdX5 supports β-catenin & RNAP II during androgen receptor mediated transcription in prostate cancer. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0054150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li J., Liu L., Liu X., Xu P., Hu Q., Yu Y. The role of upregulated DDX11 as A potential prognostic and diagnostic biomarker in lung adenocarcinoma. J. Cancer. 2019;10(18):4208–4216. doi: 10.7150/jca.33457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhattacharya C., Wang X., Becker D. The DEAD/DEAH box helicase, DDX11, is essential for the survival of advanced melanomas. Mol Cancer. 2012;11(1):82. doi: 10.1186/1476-4598-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pérez-Benavente B., García J.L., Rodríguez M.S., Pineda-Lucena A., Piechaczyk M., Font de Mora J., Farràs R. GSK3-SCFFBXW7 targets JunB for degradation in G2 to preserve chromatid cohesion before anaphase. Oncogene. 2013;32(17):2189–2199. doi: 10.1038/onc.2012.235. [DOI] [PubMed] [Google Scholar]
- 63.Yu B., Liang H., Ye Q., Wang Y. Establishment of a Genomic-Clinicopathologic Nomogram for Predicting Early Recurrence of Hepatocellular Carcinoma After R0 Resection. J Gastrointestinal Surgery. 2020 doi: 10.1007/s11605-020-04554-1. [DOI] [PubMed] [Google Scholar]
- 64.Kouyama Y., Masuda T., Fujii A., Ogawa Y., Sato K., Tobo T. Oncogenic splicing abnormalities induced by DEAD-Box Helicase 56 amplification in colorectal cancer. Cancer Sci. 2019;110:3132–3144. doi: 10.1111/cas.14163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu C., Zhang X., Kourkoumelis N., Shen Y., Huang W. Integrated analysis of DEAD-box helicase 56: a potential oncogene in osteosarcoma. Front Bioeng Biotechnol. 2020;8:588. doi: 10.3389/fbioe.2020.00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J., Liu J., Ye M., Liu F., Wu S.u., Huang J., Shi G. Ddx56 maintains proliferation of mouse embryonic stem cells via ribosome assembly and interaction with the Oct4/Sox2 complex. Stem Cell Res Ther. 2020;11(1) doi: 10.1186/s13287-020-01800-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Kouwenhove M., Kedde M., Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 68.Fuller-Pace F.V., Moore H.C. RNA helicases p68 and p72: multifunctional proteins with important implications for cancer development. Future Oncol. 2011;7:239–251. doi: 10.2217/fon.11.1. [DOI] [PubMed] [Google Scholar]
- 69.Suzuki H.I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 70.Davis B.N., Hilyard A.C., Lagna G., Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin H., Li Q., Cao F., Wang S.-N., Wang R.-T., Wang Y., Tan Q.-Y., Li C.-R., Zou H., Wang D., Xu C.-X. miR-124 Inhibits Lung Tumorigenesis Induced by K-ras Mutation and NNK. Mol Ther Nucleic Acids. 2017;9:145–154. doi: 10.1016/j.omtn.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang M., Meng B., Liu Y., Yu J., Chen Q., Liu Y. MiR-124 Inhibits growth and enhances radiation-induced apoptosis in non-small cell lung cancer by inhibiting STAT3. Cell Physiol Biochem. 2017;44:2017–2028. doi: 10.1159/000485907. [DOI] [PubMed] [Google Scholar]
- 73.Cai J., Fang L., Huang Y., Li R., Xu X., Hu Z. Publisher Correction: Simultaneous overactivation of Wnt/β-catenin and TGFβ signalling by miR-128-3p confers chemoresistance-associated metastasis in NSCLC. Nat Commun. 2018;9:16196. doi: 10.1038/ncomms16196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Donzelli S., Fontemaggi G., Fazi F., Di Agostino S., Padula F., Biagioni F., Muti P., Strano S., Blandino G. MicroRNA-128-2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function. Cell Death Differ. 2012;19(6):1038–1048. doi: 10.1038/cdd.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.He D., Wang J., Zhang C., Shan B., Deng X., Li B., Zhou Y., Chen W., Hong J., Gao Y., Chen Z., Duan C. Down-regulation of miR-675-5p contributes to tumor progression and development by targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol Cancer. 2015;14(1) doi: 10.1186/s12943-015-0342-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finotello F., Trajanoski Z. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol Immunother. 2018;67(7):1031–1040. doi: 10.1007/s00262-018-2150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kar A., Fushimi K., Zhou X., Ray P., Shi C., Chen X. RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5′ splice site. Mol Cell Biol. 2011;31:1812–1821. doi: 10.1128/MCB.01149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hirota Y., Lahti J.M. Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucleic Acids Res. 2000;28:917–924. doi: 10.1093/nar/28.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Diot C., Fournier G., Dos Santos M., Magnus J., Komarova A., van der Werf S., Munier S., Naffakh N. Influenza A virus polymerase recruits the RNA helicase DDX19 to promote the nuclear export of viral mRNAs. Sci Rep. 2016;6(1) doi: 10.1038/srep33763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reid C.R., Hobman T.C. The nucleolar helicase DDX56 redistributes to West Nile virus assembly sites. Virology. 2017;500:169–177. doi: 10.1016/j.virol.2016.10.025. [DOI] [PubMed] [Google Scholar]
- 81.Robinson L.A., Jaing C.J., Pierce Campbell C., Magliocco A., Xiong Y., Magliocco G., Thissen J.B., Antonia S. Molecular evidence of viral DNA in non-small cell lung cancer and non-neoplastic lung. Br J Cancer. 2016;115(4):497–504. doi: 10.1038/bjc.2016.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu G., Xing Z., Tran E.J., Yang D. DDX5 helicase resolves G-quadruplex and is involved in MYC gene transcriptional activation. Proc Natl Acad Sci. 2019;116:20453–20461. doi: 10.1073/pnas.1909047116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma Z., Feng J., Guo Y., Kong R., Ma Y., Sun L., Yang X., Zhou B., Li S., Zhang W., Jiang J., Zhang J., Qiao Z., Cheng Y., Zha D., Liu S. Knockdown of DDX5 Inhibits the Proliferation and Tumorigenesis in Esophageal Cancer. Oncol Res. 2017;25(6):887–895. doi: 10.3727/096504016X14817158982636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang M., Weng W., Zhang Q., Wu Y., Ni S., Tan C. The lncRNA NEAT1 activates Wnt/β-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J Hematol Oncol. 2018;11:1–13. doi: 10.1186/s13045-018-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lakshmanan I., Salfity S., Seshacharyulu P., Rachagani S., Thomas A., Das S., Majhi P.D., Nimmakayala R.K., Vengoji R., Lele S.M., Ponnusamy M.P., Batra S.K., Ganti A.K. MUC16 regulates TSPYL5 for lung cancer cell growth and chemoresistance by suppressing p53. Clin Cancer Res. 2017;23(14):3906–3917. doi: 10.1158/1078-0432.CCR-16-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu J., Sun J. MUC16 mutations improve patients’ prognosis by enhancing the infiltration and antitumor immunity of cytotoxic T lymphocytes in the endometrial cancer microenvironment. OncoImmunology. 2018;7(10):e1487914. doi: 10.1080/2162402X.2018.1487914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abe T., Ooka M., Kawasumi R., Miyata K., Takata M., Hirota K., Branzei D. Warsaw breakage syndrome DDX11 helicase acts jointly with RAD17 in the repair of bulky lesions and replication through abasic sites. PNAS. 2018;115(33):8412–8417. doi: 10.1073/pnas.1803110115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Benetti R., Gonzalo S., Jaco I., Muñoz P., Gonzalez S., Schoeftner S., Murchison E., Andl T., Chen T., Klatt P., Li E.n., Serrano M., Millar S., Hannon G., Blasco M.A. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15(3):268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ortiz I.M.D.P., Barros-Filho M.C., dos Reis M.B., Beltrami C.M., Marchi F.A., Kuasne H., do Canto L.M., de Mello J.B.H., Abildgaard C., Pinto C.A.L., Kowalski L.P., Rogatto S.R. Loss of DNA methylation is related to increased expression of miR-21 and miR-146b in papillary thyroid carcinoma. Clin Epigenet. 2018;10(1) doi: 10.1186/s13148-018-0579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sinkkonen L., Hugenschmidt T., Berninger P., Gaidatzis D., Mohn F., Artus-Revel C.G., Zavolan M., Svoboda P., Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15(3):259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 91.Gyorffy B., Surowiak P., Budczies J., Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS one. 2013;8 doi: 10.1371/journal.pone.0082241. e82241–e82241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Q., Armenia J., Zhang C., Penson A.V., Reznik E.d., Zhang L., Minet T., Ochoa A., Gross B.E., Iacobuzio-Donahue C.A., Betel D., Taylor B.S., Gao J., Schultz N. Unifying cancer and normal RNA sequencing data from different sources. Sci Data. 2018;5(1) doi: 10.1038/sdata.2018.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang S., Liu X. The UCSCXenaTools R package: a toolkit for accessing genomics data from UCSC xena platform, from Cancer multi-omics to single-cell RNA-seq. J Open Source Software. 2019;4:1627. [Google Scholar]
- 94.Kassambara A, Kassambara MA. Package ‘ggcorrplot’. R package version 0.1 2019, 3
- 95.Wickham H., Francois R., Henry L., Müller K. dplyr: A grammar of data manipulation. R package version. 2015;(4):3. [Google Scholar]
- 96.Sergushichev A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. BioRxiv 2016, 060012
- 97.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Matthews L., Gopinath G., Gillespie M., Caudy M., Croft D., de Bono B., Garapati P., Hemish J., Hermjakob H., Jassal B., Kanapin A., Lewis S., Mahajan S., May B., Schmidt E., Vastrik I., Wu G., Birney E., Stein L., D'Eustachio P. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37(Database):D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vlachos I.S., Zagganas K., Paraskevopoulou M.D., Georgakilas G., Karagkouni D., Vergoulis T., Dalamagas T., Hatzigeorgiou A.G. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S., Li B.o., Liu X.S. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shoemaker R.H. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6(10):813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 103.Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Team, R. RStudio: integrated development for R. RStudio. Inc., Boston, MA 2015, 700.
- 105.Team, R.C. R development core team. RA Lang Environ Stat Comput 2013, 55, 275-286
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression profiles of the DEAD/H box helicase genes at different gender, age, overall stage and smoking intensity in LUAD.
Gene expression profiles of the DEAD/H box helicase genes among different genders, ages, overall stages and smoking intensity in LUSC.
Data of the p-values and HR of all the DEAD/H box genes following KM-plotting analysis.
Correlation coefficient and p-values of the individual MYC targets.
Data of the correlation between tumour-infiltrating immune cells and the four DDXs.
Data of the top 50 methylation sites with the most significant difference between the expression levels of each DDX.
Database of the gene repression of the four DDXs in pan-cancer cell lines.
Database of the gene repression of the four DDXs in lung cancer cell lines.
Clinicopathological features of the lung cancer samples.
Database of the gene expression profiles associated with DDX candidates.
Data of differential miRNAs which are associated with the DDXs for the heatmap plotting analysis.
Data of the top 20 genes with the most frequent mutation and their association with the expression levels of the four DDX genes.
R scripts and Session Information used in this study.