Abstract
In eukaryotic translation, termination and ribosome recycling phases are linked to subsequent initiation of a new round of translation by persistence of several factors at ribosomal sub‐complexes. These comprise/include the large eIF3 complex, eIF3j (Hcr1 in yeast) and the ATP‐binding cassette protein ABCE1 (Rli1 in yeast). The ATPase is mainly active as a recycling factor, but it can remain bound to the dissociated 40S subunit until formation of the next 43S pre‐initiation complexes. However, its functional role and native architectural context remains largely enigmatic. Here, we present an architectural inventory of native yeast and human ABCE1‐containing pre‐initiation complexes by cryo‐EM. We found that ABCE1 was mostly associated with early 43S, but also with later 48S phases of initiation. It adopted a novel hybrid conformation of its nucleotide‐binding domains, while interacting with the N‐terminus of eIF3j. Further, eIF3j occupied the mRNA entry channel via its ultimate C‐terminus providing a structural explanation for its antagonistic role with respect to mRNA binding. Overall, the native human samples provide a near‐complete molecular picture of the architecture and sophisticated interaction network of the 43S‐bound eIF3 complex and the eIF2 ternary complex containing the initiator tRNA.
Keywords: ABCE1, eIF3, cryo‐EM, translation initiation, ribosome recycling
Subject Categories: Structural Biology, Translation & Protein Quality
Cryo‐EM structures of native human and yeast translation initiation complexes reveal a novel hybrid conformation of the recycling factor ATPase ABCE1, which is stabilized by a dimer of translation initiation factor eIF3j.
Introduction
Translation of an mRNA into a polypeptide sequence is a central cellular process, which is highly regulated and linked to other cellular processes like ribosome biogenesis, mRNA turnover, and ribosome quality control. Most decisive for translational efficiency and regulation is the initiation phase; however, in eukaryotes the individual phases of translation were found to be coupled, especially termination with ribosome recycling and a new round of initiation. Two prominent examples are the conserved multisubunit complex eIF3, which has been described as a factor functioning across the translation cycle (Valasek et al, 2017), as well as the ATP‐binding cassette (ABC) ATPase ABCE1 (Rli1 in Saccharomyces cerevisiae), which was shown to enhance termination activity of the eRF1 release factor and which represents the key enzyme for ATP‐dependent ribosome recycling (Pisarev et al, 2010; Shoemaker & Green, 2011). Moreover, ABCE1 was found associated with initiation factors (Chen et al, 2006; Dong et al, 2004) and as a part of eIF3‐containing 43S or 48S pre‐initiation complexes (Andersen & Leevers, 2007; Preis et al, 2014; Mancera‐Martinez et al, 2017).
The ABCE1 ATPase consists of two nucleotide‐binding domains (NBDs) that are forming two nucleotide‐binding sites (NBSs) at their interface, as well as an essential iron–sulfur cluster domain (FeSD) at its N‐terminus (Barthelme et al, 2007; Hopfner, 2016). ABCE1 binds the 80S ribosome during canonical stop codon‐dependent termination or during rescue of stalled ribosomes and splits the 80S ribosomes into 40S and 60S small (SSU) and large (LSU) subunits, respectively. This recycling reaction requires an A site factor in the ribosome, either release factor eRF1 (after termination) or its homologue Pelota (Dom34 in S.c.; for ribosome rescue), in order to form part of the interaction network for ABCE1 (Becker et al, 2012; Brown et al, 2015; Preis et al, 2014). ABCE1 binds these pre‐splitting complexes in a semi‐open state with respect to its NBSs. Splitting requires binding of ATP and site‐occlusion to both NBS (Barthelme et al, 2011; Gouridis et al, 2019; Nurenberg‐Goloub et al, 2018). According to current models, the conformational change occurring during site‐occlusion would be transmitted via the FeSD of ABCE1 to the bound A site factor (eRF1 or Dom34), whereby the FeSD exerts a force on the A site factor which ultimately leads to ribosome splitting (Becker et al, 2012; Heuer et al, 2017; Nürenberg‐Goloub et al, 2020). The splitting reaction can be recapitulated in vitro (Becker et al, 2012; Nurenberg‐Goloub & Tampe, 2019; Pisareva et al, 2011; Shao et al, 2015; Shoemaker & Green, 2011), where ABCE1 was observed to remain bound to the 40S small subunit to form a post‐splitting complex (PSC), in which the two NBDs are present in a closed, nucleotide‐occluding state (Heuer et al, 2017; Kiosze‐Becker et al, 2016; Nürenberg‐Goloub et al, 2020). Therefore, it was assumed that in vivo as well, ABCE1 may remain bound to the 40S for a defined time span (Gerovac & Tampe, 2019) to prevent re‐association of the LSU (Heuer et al, 2017) or to coordinate assembly of initiation factors on the 40S subunit. However, a direct physical involvement of ABCE1 in the translation initiation process has not been shown to date.
In eukaryotes, the start of translation initiation requires the assembly of the 43S pre‐initiation complex (PIC). It consists of the 40S subunit, eIF3, eIF1, eIF1A, eIF5, and the ternary complex (TC) formed by the trimeric eIF2, initiator methionyl tRNA (tRNAi), and GTP. After 43S PIC assembly, the mRNA—in collaboration with the eIF4F complex (the cap‐binding protein eIF4E, the helicase eIF4A, and the scaffolding protein eIF4G)—can be recruited to the 43S PIC, forming the 48S initiation complex (IC). This event is coordinated by interactions between eIF3 and eIF4F as well as eIF4B, a single‐stranded RNA‐binding protein that attaches to the 40S subunit (Walker et al, 2013) and stimulates the helicase activity of eIF4A. The 48S complex then scans the mRNA for the first cognate AUG codon. After start‐codon recognition, inorganic phosphate (Pi) is released from the eIF2 complex, which is stimulated by eIF5 acting as a GTPase‐activating protein, likely via an arginine‐finger mechanism (Algire et al, 2005; Das et al, 2001; Paulin et al, 2001). Subsequently, initiation factors apart from eIF1A and eIF3 dissociate (Mohammad et al, 2017; Sha et al, 2009) and subunit joining with the 60S LSU is then mediated by the GTPase eIF5B (Acker et al, 2006; Acker et al, 2009; Lee et al, 2002; Pestova et al, 2000).
An important regulatory and scaffolding role in these processes is taken on by the multisubunit complex eIF3 (Cate, 2017; Hinnebusch, 2006), which can be structurally divided into the so‐called PCI‐MPN core and the peripheral subunits. In yeast, the PCI‐MPN core consists of the two subunits eIF3a (Rpg1/Tif32) and eIF3c (Nip1), whereas in mammals, it is formed by an octamer of eIFs 3a, 3c, 3e, 3f, 3h, 3i, 3k, and 3l (Valasek et al, 2017). The peripheral subunits consist of the so‐called yeast‐like core (YLC) module, containing eIF3b (Prt1), eIF3g (Tif35), and eIF3i (Tif34), as well as the C‐terminus of eIF3a, the N‐terminal domain of eIF3c that interacts with eIF1 and eIF5 (Valasek et al, 2003; Valasek et al, 2004; Yamamoto et al, 2005; Zeman et al, 2019), and in mammals eIF3d. In addition, eIF3j is associated with eIF3 but does not belong to its core, and plays a special role (Block et al, 1998; Valasek et al, 1999). It was shown that eIF3j participates during termination by recycling eRF3 (Beznoskova et al, 2013) and during ribosome recycling by assisting ABCE1 in subunit splitting (Young & Guydosh, 2019). Furthermore, it is involved in dissociation of mRNA from the 40S subunit (Pisarev et al, 2007; Pisarev et al, 2010). In the context of initiation, eIF3j is believed to participate in the recruitment of eIF3 to the 40S (Elantak et al, 2010; Fraser et al, 2004; Nielsen et al, 2006), to antagonize premature mRNA recruitment (Fraser et al, 2007), and to regulate start‐site selection (Elantak et al, 2010).
For a better mechanistic understanding of this complicated interplay, a number of cryo‐EM structures of 43S PICs and partial 48S ICs gave first insights into the architectural variety of initiation complexes (Aylett et al, 2015; des Georges et al, 2015; Eliseev et al, 2018; Erzberger et al, 2014; Hashem et al, 2013; Hussain et al, 2014; Llacer et al, 2015; Llacer et al, 2018; Mancera‐Martinez et al, 2017). During 43S assembly, the 40S subunit gets prepared to thread the mRNA into the mRNA‐binding channel between the 40S body and the head. The main constriction for mRNA is at the so‐called “latch”, a structural element formed between ribosomal RNA (rRNA) helix h18 and ribosomal protein (r‐protein) uS12 on the 40S body, and h34 and uS3 on the head (Schluenzen et al, 2000). Empty or only ABCE1‐bound 40S usually does not adopt a defined head conformation, and the latch is rather closed (Heuer et al, 2017; Passmore et al, 2007). Binding of eIF1 and especially eIF1A, which bridges the body with the head, seems to prime and confine the 40S by inducing a small rotation of the 40S head (Llacer et al, 2015; Passmore et al, 2007), but the latch still remains in a closed position (Llacer et al, 2015). Latch opening was only observed in in vitro reconstituted partial 48S ICs containing mRNA and both eIF3 and the eIF2 TC in addition to eIF1 and eIF1A (Llacer et al, 2015; Llacer et al, 2018). Here, two conformations of the 48S IC can be distinguished: the open POUT and the closed PIN conformation, which differ in the orientation of the 40S head and the TC. Compared to the empty and eIF1/1A‐bound structures, the head is moved upwards away from the body in the POUT conformation. This leads to widening of the latch and the P site tRNAi in the TC is only bound via the anticodon loop (AL) to the 40S head but not the body. In the PIN conformation, the AL moves down and engages in stable codon–anticodon interactions with the cognate start codon in the P site, accompanied by a downward movement of the 40S head.
In all eIF3‐containing structures, the PCI‐MPN core was located on the back of the 40S subunit, from where peripheral subunits stretch out. In 43S PICs, the YLC was found close to the mRNA entry site of the 40S (Aylett et al, 2015; des Georges et al, 2015; Eliseev et al, 2018; Erzberger et al, 2014), however only at low resolution. Moreover, the YLC module has been shown to relocate to the intersubunit space (ISS), as observed in in vitro reconstituted partial 48S complexes (Llacer et al, 2015), thereby occupying the position of ABCE1. The other peripheral subunits eIF3d and the eIF3c N‐terminal domain have been localized near the mRNA exit site (eIF3d: Eliseev et al, 2018) and in the ISS (eIF3c‐NTD: Llacer et al, 2015; Obayashi et al, 2017). Interestingly, two structures of partial native 43S/48S complexes exist in which ABCE1 could be visualized in substantial quantities (Simonetti et al, 2016, re‐interpreted in Mancera‐Martinez et al, 2017; Heuer et al, 2017). Notably, both samples were obtained after adding non‐hydrolyzable AMP‐PNP and/or GMP‐PNP to either yeast (Heuer et al, 2017) or rabbit reticulocyte (Simonetti et al, 2016) lysates and subsequent isolation of the 43S peak from a sucrose gradient. This may have led to non‐physiological locking of ABCE1 on the 40S subunit, thereby limiting any conclusions about a putative role of ABCE1 during the phase connecting recycling with initiation. Furthermore, apart from a low‐resolution cryo‐EM map (Aylett et al, 2015) no structural data exist on eIF3j in the context of the native 43S PIC. Therefore, the native structural landscape enabling the transition from translation termination via recycling to initiation is not yet well‐understood.
Results
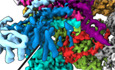
In this work, we set out to provide a structural inventory of ABCE1‐containing 43S or 48S initiation complexes from native small ribosomal subunits (SSU). We first asked if substantial amounts of ABCE1 are associated with initiation factor‐bound 40S under native conditions. To that end, lysates from a yeast strain (S.c.) containing TAP‐tagged ABCE1 (Rli1) were subjected to density gradient centrifugation followed by Western blotting of fractions (Fig EV1A). In agreement with previous studies (Andersen & Leevers, 2007; Pisarev et al, 2010; Pisareva et al, 2011), we observed that ABCE1 was especially enriched on 40S and 80S ribosomes. We further performed affinity purification from the lysates under varying buffer conditions but without any stabilizing non‐hydrolyzable ATP or GTP analogs, and analyzed the elution fractions by quantitative mass spectrometry (LC‐MS/MS) (Figs 1A and EV1B and C). We found that the expected SSU proteins but also eIF3 core components and especially eIF3j (Hcr1) were enriched by ABCE1 affinity purification, indicating that both proteins were indeed integral components of native pre‐initiation complexes. Because of this finding and since eIF3j was implicated in ABCE1‐dependent ribosome splitting in vivo (Young & Guydosh, 2019), we tested if eIF3j together with ABCE1 had a direct impact on ribosome splitting in a reconstituted system. To this end, we performed in vitro splitting assays in yeast and tested if eIF3j can play a stimulatory role. Purified 80S ribosomes were incubated with the purified splitting factors Dom34, Hbs1, Rli1 (ABCE1), eIF6 to prevent re‐association of ribosomal subunits, ATP and GTP as well as different amounts of eIF3j. Splitting efficiency was assessed from sucrose density gradient UV profiles by monitoring 80S versus ribosomal subunit amounts (Figs 1B and C, and EV1D). Indeed, we observed that an addition of eIF3j in molar excess increased the ratio of split subunits to 80S when compared to a reaction containing the splitting factors only (Fig 1C). Increasing amounts of eIF3j resulted in higher splitting activity. However, eIF3j alone did not exhibit any activity (Fig EV1E). In addition, we found that eIF3j and substoichiometric amounts of ABCE1 remained bound to the 40S after splitting (Fig EV1F). To further confirm that eIF3j can still be associated with the 40S‐ABCE1 complex after splitting, we employed the “facilitated splitting” assay as described before (Heuer et al, 2017). In this assay, ribosomes are allowed to dissociate under splitting‐promoting conditions (low Mg2+ and high salt) and in the presence of putative subunit‐binding factors (see Materials and Methods). Indeed, in this assay we observed that eIF3j remained on the 40S SSU together with ABCE1, confirming that the two factors remain together on the 40S for downstream events such as initiation after collaborating during splitting (Fig EV1G and H).
Figure EV1. Enrichment of ABCE1 and eIF3j on 40S complexes and assessment of their role in splitting of 80S ribosomes.
- Total cellular extracts from yeast cells expressing ABCE1‐TAP were separated on a sucrose gradient (10–50%) by ultracentrifugation. Proteins of each fraction were analyzed by Western blot using a PAP antibody for the detection of the ABCE1‐TAP fusion protein and anti‐Nog1 antibody to mark the 60S fraction. AU–absorption units at 260 nm.
- Silver‐stained NuPAGE gel showing elution from affinity purification using ABCE1‐TAP performed in Tris or HEPES buffer (see Methods for details).
- Volcano plot showing the fold enrichment of proteins in the elution fraction from the ABCE1‐TAP purification in Tris buffer followed by mass spectrometry analysis (LC‐MS/MS). The enrichment was calculated relative to an “input” corresponding to an aliquot of the ABCE1‐TAP cell lysate used for the affinity purification. It is represented, on the x‐axis, as log2(LFQ ABCE1‐TAP/LFQ input) where LFQ stands for label‐free quantification. The y‐axis represents the P‐value distribution (‐log10 ‐p‐value) calculated using Student’s t‐test for all identified proteins represented by a circle. Proteins above the curved lines show a statistically significant enrichment according to the t‐test value. The assay was performed in triplicates.
- UV profiles from in vitro splitting reaction triplicates with and without splitting factors (SF; ABCE1, Dom34, Hbs1, and eIF6) and eIF3j. Samples were separated on a sucrose gradient (10–50%) by ultracentrifugation. (+) = 4‐fold molar excess of eIF3j; (++) = 20‐fold molar excess of eIF3j.
- Relative abundance of 80S and subunits in each experiment, as calculated from triplicates shown in (D).
- SDS–PAGE of the 40S, 60S, and 80S peaks obtained from the in vitro splitting experiment (D) containing SFs and high amounts of eIF3j.
- UV profiles from in vitro “facilitated” splitting reactions. Samples were separated on a sucrose gradient (10–50%) by ultracentrifugation.
- SDS–PAGE of the input factors (eIF3j and ABCE1) as well as 40S and 80S peaks from the “facilitated” splitting experiment. ABCE1 and eIF3j are marked by arrows, respectively.
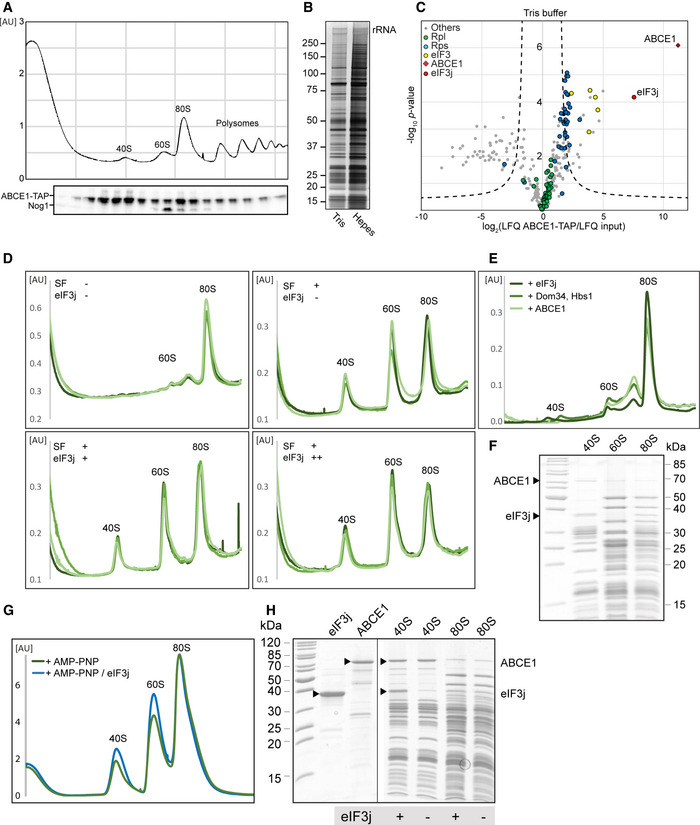
Figure 1. Biochemical analysis and cryo‐EM structures of yeast ABCE1‐containing initiation complexes.
-
AVolcano plot representing the statistical analysis of the fold enrichment of proteins after affinity purification in HEPES buffer of ABCE1‐TAP followed by label‐free quantification (LFQ) using liquid chromatography–tandem mass spectrometry (LC‐MS/MS). Proteins above the curved lines show a statistically significant enrichment according to the t‐test value.
-
B, CSucrose density gradient UV profile after in vitro splitting assays (B) and relative abundance of 80S and subunits as calculated from triplicates and displayed as mean ± SD. (C); SF = splitting factors including Dom34, Hbs1, ABCE1, and eIF6; (+) = 4‐fold molar excess of eIF3j; (++) = 20‐fold molar excess of eIF3j.
-
D, ECryo‐EM maps low‐pass filtered at 6 Å and models of the yeast subclasses representing an ABCE1‐ and eIF3j‐containing 43S PIC (D) and an ABCE1‐ and eIF5‐containing partial 48S IC (E).
Source data are available online for this figure.
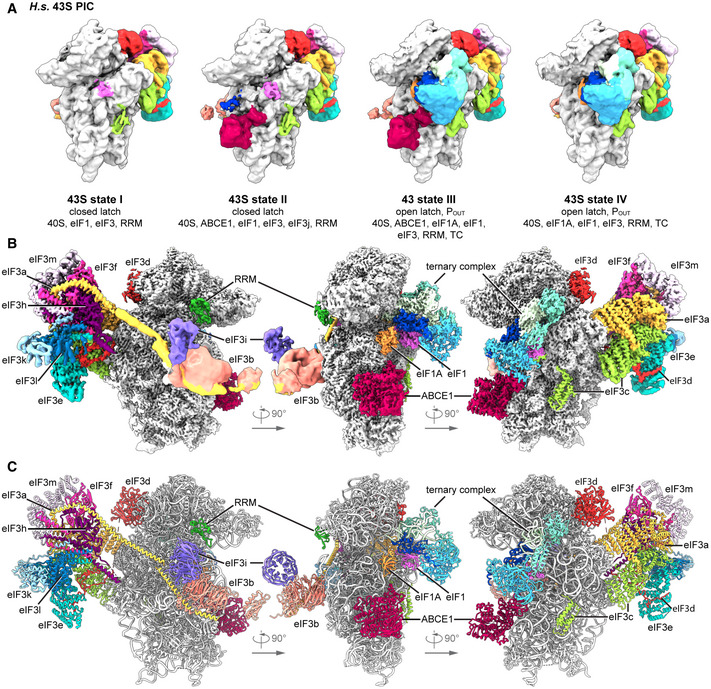
To gain further insights into the composition of native small subunits in yeast and human cells, we adopted a shotgun cryo‐EM approach. Yeast SSU complexes were obtained after harvesting the crude 43S/48S peak from a preparative sucrose density gradient of yeast cell lysate that was not further treated or stabilized with a non‐hydrolyzable nucleotide analog. Similarly, human native 40S was obtained from untreated lysates of HEK Flp‐In 293 T‐Rex cells after serendipitous non‐specific enrichment on sepharose material during unrelated affinity pullouts (see Materials and Methods). Of these samples, large enough cryo‐EM data sets were collected in order to analyze their complex composition by extensive 3D classification (Appendix Figs S1 and S2).
In the yeast data set, as expected, the selected particles contained pre‐initiation complexes, which could be further classified into defined states varying in composition and conformation of eIF‐associated 40S subunits. The majority of these complexes (62%) contained ABCE1, and the most interesting classes consisted of 43S particles containing ABCE1, eIF3, eIF1, eIF1A, and eIF3j on the 40S (Aylett et al, 2015; Heuer et al, 2017). The mRNA path (latch) was in the closed conformation (Passmore et al, 2007), and at the mRNA entry, we found a density for a typical RNA recognition motif (RRM) (see below). Importantly, in these classes we observed an interaction between the FeSD of ABCE1 and eIF3j (Fig 1D). Moreover, we found one class of particles with mRNA bound, apparently representing a partial 48S IC complex. It contained eIF3, eIF1, tRNAi in the PIN conformation, as well as the N‐terminal domain (NTD) of eIF5 as observed before (Llacer et al, 2018), and, to our surprise, also ABCE1 (Fig 1E). The classes representing 43S PIC and 48S IC were refined to a resolution of 5.3 and 6.2 Å, respectively, allowing us to fit molecular models of existing structures as rigid bodies (Fig 1D and E, Appendix Fig S3, Appendix Table S1).
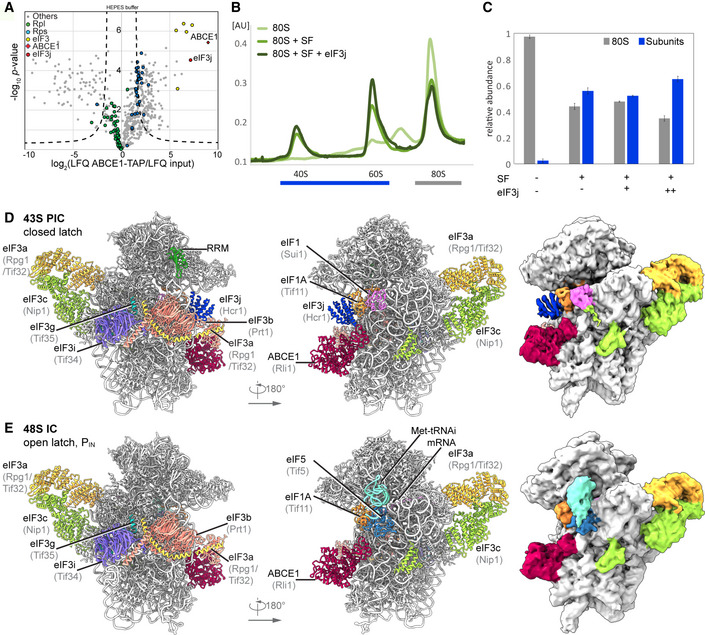
In the human sample, we also found 40S subunits associated with initiation factors, similar to the yeast sample. After classification, four major stable eIF3‐containing classes could be obtained (Fig 2A). The 40S in State I resembled the state of an empty 40S subunit with a closed latch (Heuer et al, 2017; Passmore et al, 2007), and only the core eIF3 subunits and weakly bound eIF1 were found. State II had a similar conformation, and we found extra densities in the ISS for eIF1, eIF3j, and ABCE1. State III additionally contained eIF1A and the ternary eIF2‐GTP‐tRNAi complex (TC) in the open POUT conformation (Llacer et al, 2015), whereas State IV was similar to State III but lacked ABCE1. Notably, in contrast to the yeast sample, we did not find any 48S classes containing mRNA. Thus, our human sample mainly represented 43S post‐splitting or pre‐initiation complexes prior to mRNA recruitment.
Figure 2. Cryo‐EM structures of human 43S PICs in different assembly states.
- Overview of four selected compositional states present in the human 43S PIC data set.
- Composite map of the complete human 43S PIC after focused and multi‐body refinements on individual sub‐complexes, filtered at local resolution.
- Composite model of the complete human 43S PIC, as represented by state III.
Independent focused classification and multi‐body refinements focusing on individual sub‐complexes (Fig EV2 and Appendix Fig S2) enabled us to obtain molecular resolution for large parts of the human 43S sub‐complexes. Therefore, we were able to build models for the octameric eIF3 PCI‐MPN core at the backside of the 40S, parts of the YLC at the mRNA entry site and most factors located in the ISS, including ABCE1, eIF3j, eIF1 (including the N‐terminal tail), eIF1A, the full eIF2 TC, and the eIF3c N‐terminal domain, thus resulting in a near‐complete molecular model of the human 43S particle bound to ABCE1 (Fig 2B and C, Appendix Table S2).
Figure EV2. Resolution and model fitting of the human 43S PIC.
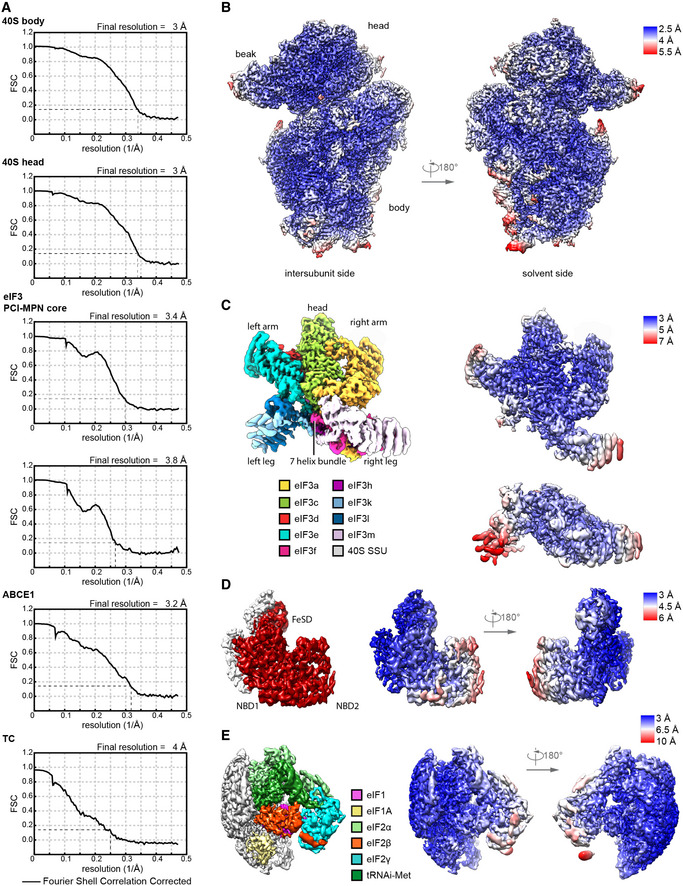
- Gold standard Fourier Shell Correlation (FSC) curve for individual bodies in multi‐body refinements (40S head including eIF3d, 40S body including eIF3c‐NTD and eIF1A, two bodies of the eIF3 PCI‐MPN core) and focused refinements using soft binary masks (ABCE1 and the TC).
- Composite map of 40S head and body after multi‐body refinement colored and low‐pass filtered according to local resolution.
- Composite map of the eIF3 MPN‐PCI core (left) after multi‐body refinement with two bodies (right), filtered according to local resolution. Structural hallmarks are color coded in the composite map as shown in Fig 5A, and the two bodies are colored according to local resolution.
- Focused refined map of the masked region containing ABCE1, filtered according to local resolution, and colored for ABCE1 (left). Two views are shown colored according to local resolution (right).
- Same as in (D) for eIF1, eIF1A, and the TC.
Conformation of ABCE1‐bound 40S‐initiation complexes
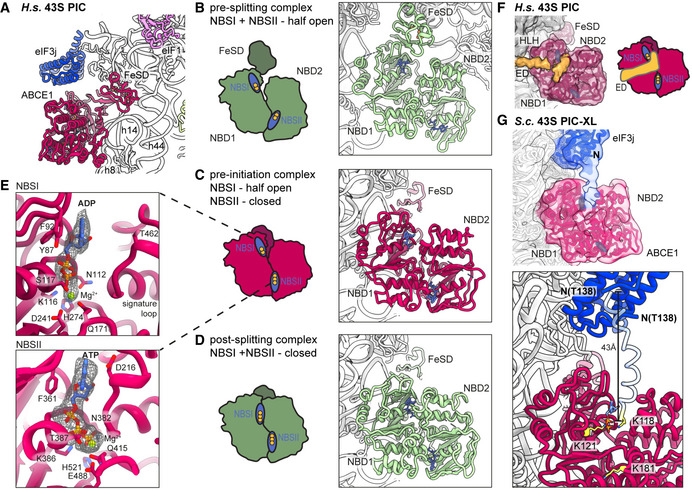
Strikingly, we observed ABCE1 associated with 40S subunits during all stages of 43S PIC assembly in humans and even with 48S IC complexes in the yeast sample. In all complexes, the FeSD of ABCE1 was in the extended conformation packed against h44, and the ATPase body occupied the universal translation factor binding site on the 40S, which is highly similar to previous observations of non‐native complexes (h8‐h14 junction; h5‐h15 junction) (Heuer et al, 2017; Mancera‐Martinez et al, 2017; Nürenberg‐Goloub et al, 2020) (Fig 3A). Here, the 40S subunit is engaged in a very similar way as in the archaeal 30S‐ABCE1 structure (Nürenberg‐Goloub et al, 2020) via the ABCE1‐specific helix‐loop‐helix (HLH) domain and the open conformation with respect to the composite hinge regions (h1 and h2). Surprisingly, however, in all structures we observed the ATPase in a novel state that has not yet been described for ABC‐type ATPases (Figs 3, EV3 and 3, EV3 and 3, EV3): Compared to the closed conformation as observed in in vitro reconstituted 30S and 40S PSCs (Heuer et al, 2017; Nürenberg‐Goloub et al, 2020), we found that only NBSII is closed whereas NBSI adopts a half‐open conformation comparable to the one observed in several 80S pre‐splitting complexes (Fig 3B) (Becker et al, 2012; Brown et al, 2015; Preis et al, 2014). When analyzing our best‐resolved human map, which was obtained after focused classification on ABCE1, we unambiguously identified an Mg2+‐ATP (Fig 3E) occluded in NBSII, similar to the archaeal 30S‐ABCE1 structure with Mg2+‐AMP‐PNP (Nürenberg‐Goloub et al, 2020). In the human structure, residues of the typical conserved motifs of ABC‐type ATPases are involved: Lys386 of the Walker A, Gly220 of the NBD1‐Signature loop, and His521 of H‐loop contact the ‐phosphate, and the Mg2+ ion is coordinated by Thr387 (Walker A) and Gln415 (Q‐loop). In contrast, for NBSI we observed Mg2+‐ADP bound exactly as observed in the crystal structures of open archaeal ABCE1 (Barthelme et al, 2011; Karcher et al 2008): Y87 of the A‐loop stacks on the adenine base, F92 on the ribose, the Walker A‐loop (Asn112‐Ser117) binds the ‐ and ‐phosphates, and the Mg2+ ion is coordinated by the ‐phosphate, Ser117, Gln171 (Q‐loop) and Asp241, Glu242 (Walker B). Importantly, the signature loop of NBD2 (Leu463‐Glu467), which occludes ATP in the catalytically active closed state, is moved by 3.5 Å away from NBD1. In conclusion, our data suggest that—in contrast to the nucleotide‐occluded state observed in vitro—in native SSU‐ABCE1 complexes, ATP hydrolysis in NBSI has already occurred, whereas NBSII is still inhibited.
Figure 3. Conformation of ABCE1 in native 40S initiation complexes.
-
AOverall position of ABCE1 in 40S initiation complexes, here representatively shown for the human State II with eIF3j.
-
B–DSchematic representation and structure of semi‐open ABCE1 as in 80S‐pre‐splitting complexes (Brown et al, 2015, PDB 3JAH) (B), hybrid semi‐open/closed ABCE1 as in native 40S initiation complexes (C) and fully closed ABCE1 as in in vitro reconstituted post‐splitting complexes (Nürenberg‐Goloub et al, 2020, PDB 6TMF) (D). Nucleotide‐binding sites colored in blue and bound nucleotide indicated with yellow circles (one circle per phosphate group).
-
EZoom into NBSI and NBSII showing bound Mg2+‐ADP (in NBSI) and Mg2+‐ATP (in NBSII) fit in density as obtained after focused classification on ABCE1 and refinement.
-
FView focusing on the NBDs and the unassigned extra density (ED) reaching from the 40S via the HLH into NBSI. The ABCE1 map was low‐pass filtered at 6 Å. Schematic representation highlighting the position of the ED with respect to the NBSs.
-
GUpper panel: Position of eIF3j and ABCE1 in the crosslinked yeast 43S‐PIC (43S‐PIC‐XL) sample. View focusing on the ABCE1‐eIF3j interaction (same view as (F)): An extra density attributing to the eIF3j N‐terminal region is connecting the eIF3j 6‐helix bundle with NBSI of ABCE1. The map was low‐pass filtered at 8 Å. Lower panel: N‐terminally extended model of eIF3j (transparent blue) highlighting the position of K118, which was found crosslinked to K121 and K181 of ABCE1 (atoms colored in yellow).
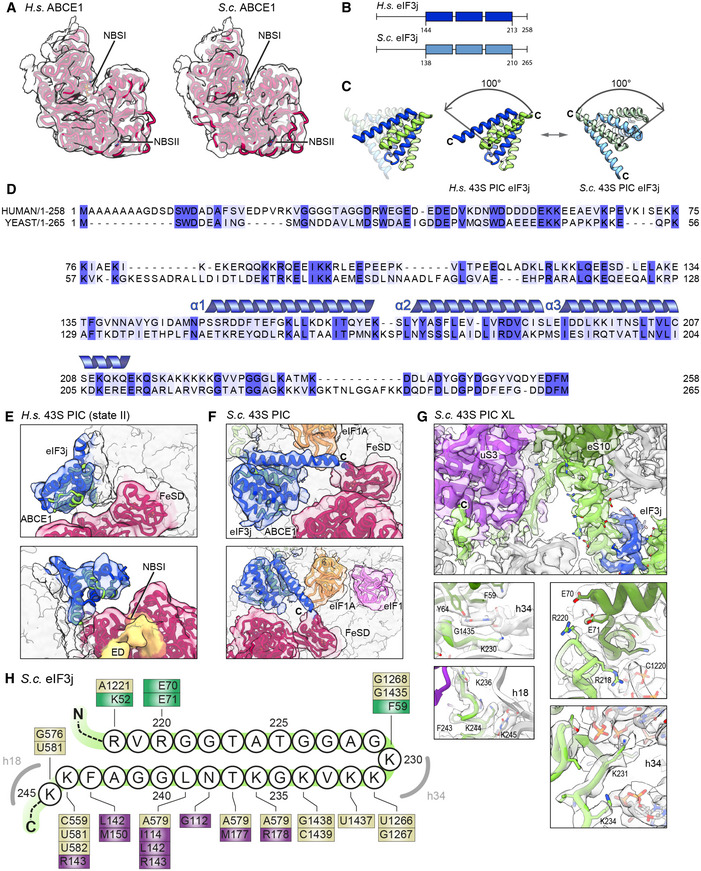
Figure EV3. Density fits of ABCE1 and alignment, model and ribosome binding of human and yeast eIF3j.
-
AModel for ABCE1 fit into low‐pass‐filtered density to demonstrate the hybrid semi‐open/closed conformation of ABCE1 in native yeast and human 43S PICs.
-
BSchemes of H.s. and S.c. eIF3j indicating the length of N‐ and C‐termini; the three helices present in the crystal structure of human eIF3j (PDB 3BPJ) are indicated.
-
CView highlighting the rearrangement (100‐degree rotation) of eIF3 in the human (eIF1A‐lacking) and yeast (eIF1A‐containing) 43S PICs.
-
DAlignment between H. s. and S. c. eIF3j shows 24.6% identity and 53.1% similarity for the full‐length protein. Dark blue boxes indicate conservation, light blue boxes indicate similarity. For the sequence (three α‐helices) present in the human X‐ray structure (PDB 3BPJ from residues 144‐213 in protomer 1 and 144‐216 in protomer 2), identity/similarity is 32.4%/66.2%, corroborating the reliability of the yeast homology model.
-
E, FFits of the human eIF3j crystal structure and the yeast homology model into the corresponding density.
-
GInteractions of the eIF3j C‐terminus (protomer 2) with the 40S head and body shown with models fit into the cryo‐EM map of the crosslinked yeast 43S‐PIC.
-
HSchematic representation summarizing interactions of the eIF3j C‐terminus with the 40S. Colored boxes indicate residues interacting with eIF3j, with brown representing 18S rRNA, purple representing uS3, and green representing eS10.
As an additional difference to previous structures, we observed a rod‐like extra density (ED) after low‐pass filtering in all native 43S PIC structures, protruding from h17 of the 40S body via the HLH motif into the cleft between NBD1 and NBD2 of ABCE1 (Fig 3F). However, local resolution in both human and yeast samples was too low to identify this factor. To stabilize this assembly, we generated a chemically crosslinked yeast initiation complex sample derived from a strain harboring TAP‐tagged eIF3c (Nip1) and performed a cryo‐EM analysis focused on the ABCE1 and the adjacent eIF3j (Appendix Figs S1B and S4). Indeed, in this reconstruction, we clearly observed an extra density protruding from eIF3j into the composite NBSs of ABCE1. At a resolution of 3.0 Å, we built the model for yeast eIF3j (Fig 3G and 4, Appendix Fig S4) based on the human eIF3j dimer (unpublished; PDB 3BPJ; lacking 137 residues at the N‐terminus and 28 residues at the C‐terminus). In brief, this dimer folds into a stable entangled 6‐helix bundle that is arranged such that the N‐termini are in close vicinity. Yet, the C‐termini face into opposite directions, whereby the C‐terminal tail of one protomer reaches into the mRNA entry channel (see below). On this basis, we could assign the extra density in ABCE1 as a part of the eIF3j N‐terminus. This assignment was further confirmed by protein crosslinking coupled with mass spectrometry (XL‐MS) using a lysine‐specific BS2G crosslinker (Appendix Fig S5, Appendix Table S3). Two crosslinks between the Lys118 of eIF3j with Lys121 and Lys181 of ABCE1, both located near the ATP‐binding site of NBD1, were identified (Fig 3G). In this position, the eIF3j N‐terminus may easily modulate the ATPase activity of ABCE1 by restricting further movements of the HLH or the two NBDs with respect to each other. Interestingly, the position of the eIF3j‐NTD on ABCE1 is similar to the one observed in a recent structure of archaeal ABCE1 co‐crystallized with an 18‐mer fragment from the C‐terminus of the archaeal 50S stalk protein aP1 (Imai et al, 2018). This suggests that ABCE1 possesses a multivalent interaction patch in this region, which would allow for regulation of its ATPase activity. The observed stabilization of ABCE1 in the half‐open conformation with one ADP still bound in NBS1 may indicate an inhibition of ADP release, which would explain its rather stable association with the 40S subunit.
Figure 4. Two conformations of eIF3j in human and yeast 43S PICs.
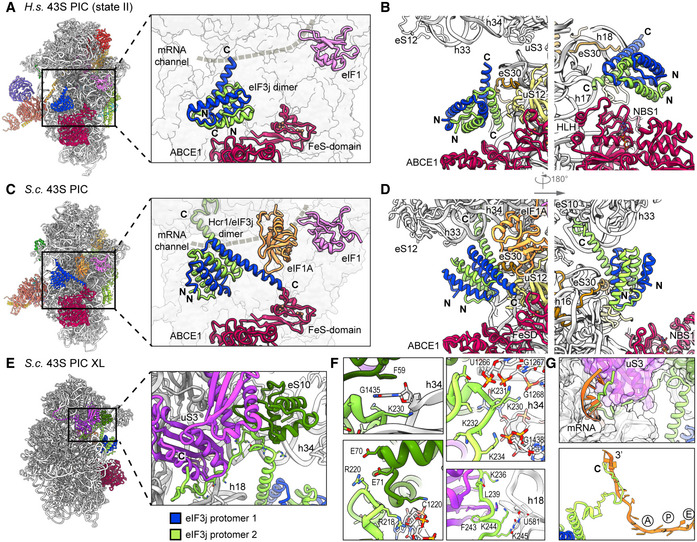
- Overview and zoomed view on the model of human 43S PIC II (lacking eIF1A), focusing on the two protomers of the dimeric eIF3j 6‐helix bundle in the ISS. eIF3j is in close vicinity to NBD1 of ABCE1 but only forms contacts to the 40S. The mRNA channel is indicated by a dashed gray line.
- Two different views showing the interaction of the two Homo sapiens (H.s.) eIF3j protomers with the 40S.
- Same views as in (A) on the model of the yeast 43S PIC. Here, eIF3j (Hcr1) is turned approximately 100 degrees around a pivot formed by the C‐terminal helices contacting eS30 and uS12. Protomer 1 thereby contacts eIF1A and the FeSD of ABCE1 and protomer 2 contacts h33.
- Two different views showing the interaction of the two S.c. eIF3j protomers with the 40S and ABCE1.
- Overview and zoomed view highlighting the position of the eIF3j C‐terminus in the yeast 43S‐PIC‐XL structure.
- Zoomed views focusing on interactions of the eIF3j C‐terminus with the 40S. The loop following the third helix of eIF3j protomer 2 is in a pocket formed by the 40S h33, h34, and eS10. Lys230 of eIF3j C‐terminus (protomer 2) and Phe59 of eS10 are sandwiching the flipped‐out G1435 base of h34 (upper left); Lys231 and Lys234 interact with h33 (U1266 and G1267) and h34 (G1438) (upper right); salt bridges between Arg220 and Glu70‐Glu71 of eS10 further stabilize the loop (lower left). Following the loop, the eIF3j C‐terminus bridges the 40S body and head in the latch and contacts are formed with h18 (via Lys236) and via hydrophobic interactions with uS3 (lower right). See Appendix Text 1 for more molecular details.
- Position of the ultimate eIF3j C‐terminus in the mRNA entry tunnel (upper panel) and steric clash with mRNA as positioned in an 80S ribosome stalled during translation (PDB 5MC6); for clarity, in the lower panel only eIF3j and mRNA are shown, A/P/E, respectively indicate the positions of aminoacyl, peptidyl, and exit site in the 80S ribosome.
Conformation of eIF3j in human and yeast 40S‐initiation complexes
As described above, we found yeast and human 43S PIC sub‐populations concomitantly bound to ABCE1 and eIF3j. The eIF3j subunit was positioned on the intersubunit side, roughly resembling the location previously described in low‐resolution maps (Aylett et al, 2015) (Fig 4). The main difference between the maps was the absence (human) or presence (yeast) of eIF1A. However, apart from a small rotation around the neck (approx. 3°), we did not observe significant conformational changes in the 40S when comparing the two structures.
In the low‐pass‐filtered human State II, which lacks eIF1A, we identified the eIF3j 6‐helix bundle located above the ABCE1 ATPase body and in close vicinity to NBD1 (Fig 4A and B), but no direct contacts were formed with ABCE1. On the 40S, eIF3j contacted the N‐terminal tail of eS30 (protomer 1) and the C‐terminus of uS12 (protomer 2). The C‐terminal helix of protomer 2 further projects toward the three‐way junction formed by h32, h33, and h34 at the 40S head, whereas in protomer 1 it points toward h17 and the HLH of ABCE1 (Figs 4B and EV3E). In this position, the N‐termini of eIF3j are located above the ABCE1 ATPase body close to the NBD1‐NBD2 cleft.
In the yeast 43S PIC, in which eIF1A was present, we found eIF3j in a similar position, but different conformation compared to the human structure (Fig 4C and D). Here, the 6‐helix bundle is stably anchored between the 40S beak at rRNA h33 on one side and the 40S body near the ABCE1 FeSD and eIF1A on the other side. The two sides of the anchor are formed again by the C‐terminal helices of eIF3j: protomer 2 contacts eS30 at a similar site as in the human structure but now the entire helix bundle was rotated by approximately 100 degrees (Fig EV3B, C, and D). Consequently, the tip of the protomer 2 C‐terminal helix now pointed toward the 40S head, whereas the C‐terminal helix of protomer 1 projected toward the ABCE1‐FeSD, thereby passing along eIF1A (Figs 4D and EV3F). Molecular details of the eIF3j‐40S interaction were derived from the high‐resolution structure of the crosslinked 43S‐PIC (Fig 4E). In brief, the 6‐helix bundle accommodates between the 40S body and head via interactions of both protomers. The body is contacted by the first and third helix of protomer 2 (to the h17‐h18 junction and eS30) mainly by basic residues. The third helix projects toward the beak to contact the phosphate backbone of h33 (G1264). Following this helix, the ultimate eIF3j C‐terminus forms a loop inside a pocket formed by h33, h34, and eS10 and from there runs along h18 and uS3, parallel to the latch, to position the ultimate C‐terminal tail inside the mRNA entry channel (Figs 4F, EV3G, and H; for a detailed description of molecular contacts see Appendix Text 1). In this position, eIF3j directly overlaps with the mRNA path and would possibly interfere with mRNA loading during 48S‐IC formation (Fig 4G).
Taken together, our structural data explain how eIF3j could exert its functions during key steps of translation initiation in conjunction with eIF1A.
Molecular architecture of the PCI‐MPN core and its interactions with 40S
State I of the human sample represented a stable class with mainly eIF3 and weak density for eIF1 bound to the 40S SSU. This appears plausible when considering that eIF3 activity during termination and ribosome recycling has been proposed (Beznoskova et al 2013; Pisarev et al, 2007; Valasek et al, 2017), which further indicates that eIF3 can already bind the 40S before eIF1A comes into play. The lack of ABCE1 in this complex may be a result of fast dissociation after splitting or of an alternative splitting mechanism. In any case, after accommodation of eIF1 and eIF1A, the eIF2 TC binds to the 43S to induce the POUT conformation (State III‐IV). Here, the improved resolution allowed us to describe the interaction network of these factors at unprecedented molecular detail.
The PCI‐MPN core is located at the backside of the 40S as observed before (des Georges et al, 2015; Hashem et al 2013; Srivastava et al 1992), and high resolution of the core was obtained by multi‐body refinement of State I and State II particles. The structure assembles into β‐sheets with the shape of an arc formed by PCI domains of eIF3 subunits a, c, e, l, k, and m. The arc wraps around a seven‐helix bundle formed by the C‐terminal helices of subunits c, e, f, h, k, and l (Figs 5A and EV4A), resulting in the typical five‐lobed structure (left and right arm, left and right leg and head), which was visualized at a local resolution of 3.4 Å (left arm, head and right arm) and 3.8 Å (left and right leg) (Fig EV2C). This allowed for an almost complete molecular interpretation (Fig EV4A, Appendix Table S4), thus refining previous low‐resolution models (des Georges et al, 2015; Eliseev et al, 2018; Erzberger et al, 2014), for example, by correcting the register of helices and extending molecular models (Appendix Fig S6).
Figure 5. Molecular interactions of the human PCI‐MPN core of eIF3 in the 43S PIC.
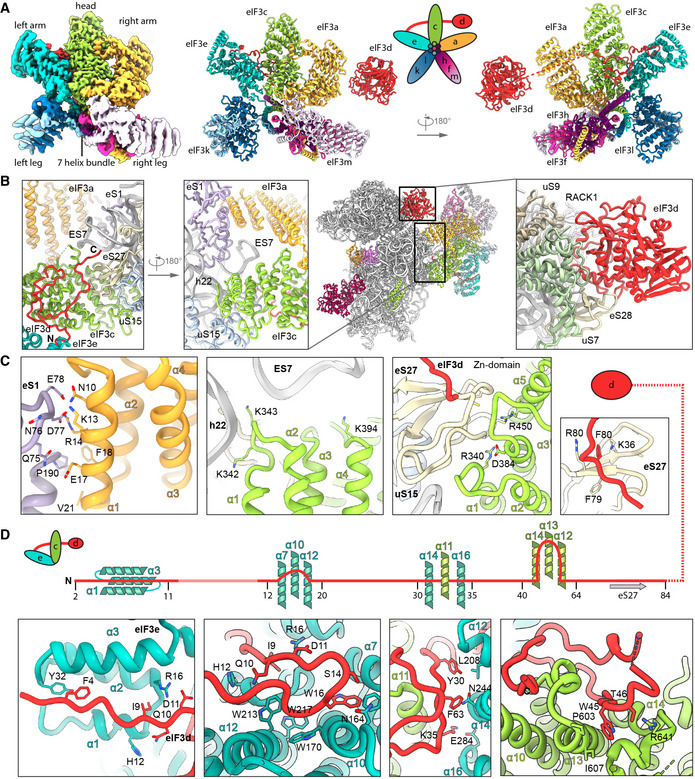
-
AIsolated map and molecular model of the eIF3 PCI‐MPN core color coded as in Fig 2. Structural hallmarks are indicated, and a scheme shows the composition of the lobes.
-
B, CInteractions of eIF3a, eIF3c, and eIF3d with the ribosome: (B) shows an overview of the structure and zoomed views highlighting the interactions of eIF3a, eIF3c, the eIF3d N‐terminal tail and the eIF3d cap‐binding domain with the 40S, (C) shows molecular details of eIF3a interacting with eS1; eIF3c interacting with rRNA h22 and eIF3c and the N‐terminal tail of eIF3d with the Zn‐knuckle domain of eS27.
-
DInteractions of the eIF3d N‐terminal tail with the PCI‐MPN core.
Figure EV4. Molecular interactions of eIF3d and the PCI‐MPN core in the 43S PIC shown with density.
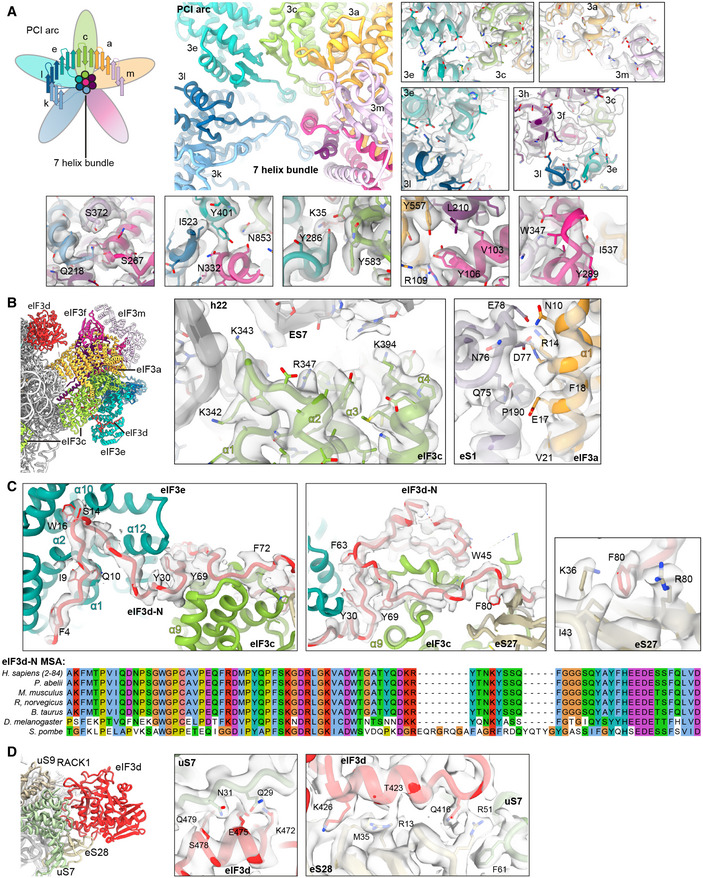
- PCI arc of the PCI‐MPN core; zoomed views highlighting fits of the eIF3 PCI arc into the refined density and interactions between the subunits in the PCI‐MPN core.
- eIF3a and eIF3c interactions with the 40S.
- Left, Middle: Interactions of the eIF3d N‐terminal tail with the PCI‐MPN core: Interactions of the ultimate eIF3d N‐terminus (Phe4‐Pro18) with eIF3e are established via Phe4 (to Tyr32 in loop between PCI helices α2 and α3), Gln10 (to His12 in α1), Ile9 and Asp11 (to Arg16 in α2), Ser14 (to Asn164 α9 and Phe132 in α7), Trp16 (to Gly171 in α10), and Gly17 (to Trp170 in α10). eIF3d residues 25‐36 are bridging eIF3e and eIF3c. Residues involved are Tyr30 (to Leu208 in eIF3e PCI helix α12), Phe33 (to the peptide backbone of Leu590 in eIF3c α11), and Lys35 (to Gln283 in eIF3e α16). eIF3c‐specific interactions are established by Leu39 (to Gln595), Trp45 (Pro603, Ile607, and Glu666), and Thr46 (to Arg641) (see Table S3). For clarity, only density for eIF3d is shown (gray transparent surface). Right: Detailed view of the interaction of F80 (eIF3d) with K36 and R80 (eS27). Density is shown for both proteins (gray transparent surface). Below: MSA of the conserved N‐terminal region of eIF3d is shown. Coloring according to default Clustal X color scheme (blue: hydrophobic, magenta: negative charge, red: positive charge, green: polar, orange: glycine, yellow: proline, pink: cysteine, cyan: aromatic).
- Interactions of the eIF3d C‐terminal domain with the 40S head and zoomed views of uS7 and eS28 interaction sites shown with density.
The main anchor of the eIF3 PCI‐MPN core to the 40S is provided by the eIF3a and eIF3c subunits, which form the “head” and the “right arm” of the PCI‐MPN core, respectively. eIF3a contacts eS1 via its N‐terminal PCI helix H1 and the loop between H1 and H2. Here, Arg14 forms salt bridges to Glu78 and Asp77 of eS1 (Fig 5B and C, see Appendix Table S4 for an inventory of observed molecular interactions). A second contact site was established between Glu17, Phe18, and Val21 of eIF3a and the eS1 Pro190 as well as adjacent residues. The loop H1‐H2 of eIF3c (residues 340‐345) interacts with rRNA h22 (G929, C930) and multiple sites at the Zn‐knuckle domain of eS27 (Figs 5C and EV4B). Furthermore, the ‐sheet insert between PCI helices 4 and 5 (residues 417‐441) of eIF3c forms interactions with uS15, and basic residues in the PCI loops of both eIF3a and eIF3c are positioned to interact with the flexible tip of rRNA ES7 (Fig 5B).
An additional anchor of the eIF3 PCI‐MPN to the 40S is provided by the N‐terminus of eIF3d (from A2 to D84) (Figs 5C and D, and EV4C). Interestingly, we found that it meanders along the PCI helices 1 to 3, 7, 9, 10, and 12 of eIF3e (left arm) and bridges eIF3e with eIF3c (head) by interacting with PCI helices 12, 14, and 16 (eIF3e) and PCI helix 11 (eIF3c). Another specific contact between eIF3c and eIF3e is formed by stacking of Y286 (eIF3e) to Y583 (eIF3c). Moreover, eIF3d also interacts with PCI helices 10, 13, and 14 of eIF3c by forming a large loop, which is anchored by the conserved Trp45 (interactions to Pro603, Ile607, and Glu666 of eIF3c). The interaction to eS27 is established via its Zn knuckle, where Phe80 of eIF3d is sandwiched between the side chains R80 and K36 of eS27.
Taken together, the PCI‐MPN core of eIF3 establishes a multi‐modal molecular interaction pattern with the 40S involving the eIF3a, c, and d subunits, which display an unexpected degree of inter‐connectivity.
Structure and location of the peripheral subunits
The peripheral subunits, which consist of the YLC, the eIF3c‐NTD, and in humans the eIF3d cap‐binding protein domain, are connected to the PCI‐MPN scaffold via flexible linkers. While eIF3a connects via its CTD to the YLC module located close to the mRNA entry site, the N‐terminus of eIF3c protrudes from the mRNA exit toward the ISS, where it interacts directly with eIF1. While the N‐terminus of eIF3d as an integral part of the PCI‐MPN core is anchored to the 40S body, the cap‐binding protein domain of eIF3d is located on the 40S head close to the mRNA exit site as observed before (Eliseev et al, 2018). Here, it contacts the 40S SSU via its highly conserved helix α10 (Lee et al 2016) that packs upon eS28 via Gln416, Thr423, and Lys426 and reaches into the interface between eS28 and uS7, where Gln416 stacks on Arg51 (eS28), which in turn stacks on Phe61 (uS7). The eIF3d helix α12 lies on top of uS7 and forms contacts via Lys472, Glu475, Ser478, and Gln479. Notably, since eIF3d is bridging the 40S head with the eIF3 PCI‐MPN core anchored to the 40S body, it could serve to relay conformational rearrangements of the 40S head—as occurring during the assembly of 43S and 48S complexes—to the PCI‐MPN core or, vice versa, allow the eIF3 complex to directly control the conformational state of the 40S head (Figs 5D and EV4C and D).
For eIF3c, only a part of its NTD could be located on the ISS of the 40S so far, where it forms a helix bundle (Llacer et al, 2015). We found a particularly stable arrangement of the eIF3c NTD in classes containing the eIF2 TC and, after multi‐body refinement, local resolution of 3 to 4 Å (Figs EV2B, EV5A and B) allowed us to determine the register of the four eIF3c‐NTD helices (Val47 to Y149) (Fig 6). A stretch preceding the first helix (47‐51) contacts h24 and h27 via R47 to the backbone phosphate of C1039 and the 2’‐OH of A1181. The peptide bond of Val49 of eIF3c stacks on base C1180, which is also contacted by the first helix (52‐74) of the bundle. Here, the two charged residues K55 and R56 interact with the backbone of rRNA G1179 and C1180. Backbone–phosphate interactions were also formed by the second helix (76‐92) to rRNA h11 (A364) and h27 (U1178), by the fourth helix (136‐143) to rRNA h11 via K136 (to U367), and finally by the peptide bond of Thr140 stacking upon the U367 base, as well as Gln143 hydrogen bonding to U367. Additional but less rigid contacts were established by the K‐rich loop between helix 3 and helix 4 of eIF3c (Figs 6E and F, and EV5A and B, Appendix Table S4).
Figure EV5. Model fitting and sequence alignment of the eIF3c‐NTD.
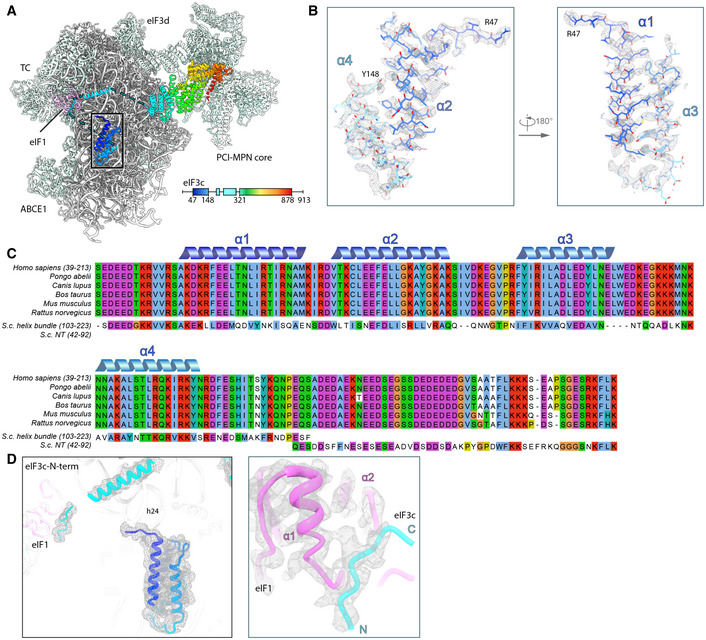
- Overview of the TC‐containing human 43S PIC as shown in Fig 6 and scheme indicating the parts of eIF3c modeled. Black box indicates section of eIF3c highlighted in (B).
- Zoomed views highlighting fits of the eIF3c‐NTD 4‐helix bundle into the refined density (gray transparent mesh). The N‐ and C‐terminal residues are marked.
- MSA of the conserved N‐terminal region of eIF3c in mammals (Ser39‐Lys213 in H.s.), aligned with segments of the NTD from S.c.. Coloring according to default Clustal X color scheme (blue: hydrophobic, magenta: negative charge, red: positive charge, green: polar, orange: glycine, yellow: proline, pink: cysteine, cyan: aromatic). The 4‐helix bundle shows 31.1/67.2% sequence identity/similarity, and the eIF1‐interacting stretch present in the N‐terminus of S.c. eIF3c (Gln42‐Lys92) shows 32.0/56.0% sequence identity/similarity with a mammalia‐specific insert C‐terminal of the conserved 4‐helix bundle.
- eIF3c‐NTD in the human 43S PIC fitted into the cryo‐EM map and zoomed view showing the fit of the eIF1‐interacting stretch of eIF3c into the cryo‐EM map.
Figure 6. Arrangement of the eIF3c‐NTD in human and yeast 43S PICs.
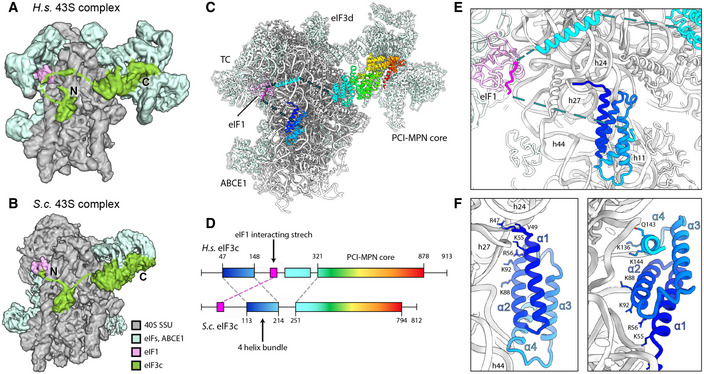
- Cryo‐EM map obtained after focused sorting of the human 43S PIC on the TC: when low‐pass filtered at 6 Å, it shows the density of almost complete eIF3c‐NTD in the ISS.
- Cryo‐EM map of the yeast 43S PIC low‐pass filtered at 6 Å.
- Model for human eIF3c in the TC‐containing 43S colored in rainbow (C) and scheme of the alignment between human and yeast eIF3c sequences, colored accordingly (D). The eIF1‐interacting stretch present in the N‐terminus of S.c. eIF3c shows 32.0/56.0% sequence identity/similarity with an insert C‐terminal of the conserved 4‐helix bundle conserved in mammals.
- Zoomed view highlighting the position of the eIF3c NTD and eIF1 in the 40S ISS.
- Molecular model for the 4‐helix bundle interacting with 40S rRNA and r‐proteins.
- Molecular model for the 4‐helix bundle interacting with 40S rRNA and r‐proteins.
Notably, when low‐pass filtered, a rod‐like extra density for the eIF3c N‐terminus became apparent, bridging the 4‐helix bundle with eIF1 near rRNA h23 and h24. This density was neither present in our nor in other (Llacer et al, 2015) yeast 43S/48S reconstructions, where the four‐helix bundle was directly connected to the eIF3c core moiety, and a site N‐terminal of this region interacted with eIF1 (Fig 6A and B). Sequence alignments of the yeast and human eIF3c N‐termini revealed an insertion on the C‐terminal side of the conserved four‐helix bundle in humans (Figs 6C and D, EV5C). This insertion from residue 165 to 213 displays 32.0% sequence identity and 56.0% sequence similarity with a stretch at the N‐terminus of yeast (42‐92), which was previously shown to be involved in the interaction of eIF3c with eIF1 by NMR studies (Obayashi et al, 2017). Here, chemical shift perturbation after eIF1 binding is observed for Glu51, Ala67, and a stretch between Lys68 and Lys77. Moreover, in our human complex one stretch of well‐resolved density for the eIF3c‐NTD was present at the eIF1 loop between helix 1 and helix 2 (Asp53‐Lys58) as well as Ile100 and Gly101 of 2 (Fig EV5D). This observation is highly consistent with the NMR study, in which the same interacting region on eIF1 is identified for the eIF3c‐NTD of yeast. Together, these observations lead us to the conclusion that the density observed near eIF1 in the human structure corresponds to this insertion C‐terminal of the helix bundle, fulfilling an analogous role to the previously characterized N‐terminal stretch of eIF3c in yeast.
From local classification, we also obtained one class with strong density for the YLC module including the eIF3a‐linker that connects it to the PCI‐MPN core (Appendix Fig S7). In brief, the YLC module contains two ‐propellers: the 7‐bladed WD40 repeat of eIF3i and the 9‐bladed WD40 repeat near the C‐terminus of eIF3b. The two propellers are held together by the C‐terminal helical domain of eIF3b, which is formed by 3 ‐helices: the most C‐terminal one binds to eIF3i, while the two preceding ‐helices are bracketing the eIF3a C‐terminus against the eIF3b ‐propeller (des Georges et al, 2015; Herrmannova et al 2012). N‐terminal of its ‐propeller, eIF3b contains a noncanonical RNA recognition motif (RRM) (ElAntak et al 2007) that can form further interactions with the eIF3a‐CTD (Dong et al 2013; Khoshnevis et al 2014; Valasek et al 2002; Valasek et al 2001) as well as the N‐terminus of eIF3j (Elantak et al, 2010; Valasek et al, 2001).
For the CTD of eIF3a, we could build a long ‐helix (residues 602‐743) into the elongated rod‐like density protruding from the PCI‐MPN core to contact uS2 and eS21 (Appendix Fig S7A). This helix extends further toward the YLC where it forms a hinge‐like structure and then connects to the stretch of the eIF3a helix that is bound to the eIF3b ‐propeller. It thereby contacts the tip of the otherwise flexible rRNA expansion segment ES6C, which in turn contacts the loop between the first two helices of the eIF3b helical domain. In this arrangement, the eIF3b WD40 is rigidly confined between rRNA h16 and uS4 on one side, and ES6C on the other side, and is thus well resolved in the proximity of the 40S (Appendix Fig S7B, Appendix Table S4). The eIF3i‐eIF3g complex and the eIF3b‐RRM, however, remained rather flexible as observed before (Erzberger et al, 2014). Nonetheless, we observed a stabilization of the eIF3b‐RRM in ABCE1‐ and eIF3j‐containing classes, possibly due to an interaction of the eIF3b‐RRM with the eIF3j N‐terminus (Elantak et al, 2010; Valasek et al, 2001).
In yeast, the positioning of the YLC module at the mRNA exit was the same, because here it was also held in place by ES6C (Appendix Fig S7C). However, in the majority of particles in the yeast dataset (approximately 85%), we could observe a conformational change in the eIF3i‐eIF3g module relative to the ES6 anchor point. Especially in the eIF3j‐containing 43S class, the eIF3i‐eIF3g entity rotates by approximately 120 degrees away from the mRNA entry toward ES6C and ES6B. The loop preceding the eIF3i‐contacting helix of eIF3b (Thr697‐Asp701) appears to serve as a hinge for this rotation (Appendix Fig S7D).
Apart from the YLC, we observed an additional density near the mRNA entry at the tip of h16 in all of our 43S structures, which was previously assigned to the RRM of eIF4B (Eliseev et al, 2018) (Appendix Fig S8). This density is especially prominent in subclasses of the human dataset lacking the TC, in which we could unambiguously identify the typical RRM fold at a local resolution around 4 Å (Appendix Figs S8C and D). Notably, besides eIF4B, the largely flexible eIF3g subunit is a potential candidate for this density because it also contains an RRM, which shares very high structural and sequence similarity (50.0%) to eIF4B (Appendix Fig S8D and E), and it was crosslinked to the nearby proteins uS10 and uS3 (Cuchalova et al 2010). Unfortunately, at the current resolution we cannot unambiguously distinguish these two RRMs in our maps and it is possible that both compete for the same binding site. Next to this domain, we observed density reaching from the RRM into the mRNA channel in all human early 43S PIC structures with a closed latch (Appendix Fig S8A and B). Close to the RRM, this density forms a loop that shows multiple contacts to uS3 before winding along uS3 toward the mRNA channel. Within the channel, one side chain can clearly be identified as a tryptophan facing toward uS3 (contacting Lys148 and Met150) and further interacting with uS3 Leu142 and Val115. The stretch also contacts 18S rRNA G626, A628, and U630 of h18 as well as C1698 of h28, C1331, and A1489 of h34 (all in the A site). Thereby, this peptide stretch blocks the entire mRNA channel down to the P site where it contacts the flipped‐out base C1701 at the tip of h44. Unfortunately, local resolution in this region is insufficient to provide further molecular detail and clearly identify this entity, yet considerable candidates may be further missing parts of eIF3g, eIF4B, the C‐terminus of eIF3j as observed in yeast maps, the CTD of eIF3a, or the ribosome hibernation factor SERBP1 (Stm1 in yeast) (Anger et al 2013; Ben‐Shem et al 2011; Brown et al 2018). In any case, it is apparent that accommodation of mRNA in the 48S IC complex would require its relocation, which may allow for allosteric communication between the different eIFs.
Conformation of the ternary complex
After analyzing the eIF3 complex, we also gained molecular information on the human eIF2 TC by focused classification. The TC as well as eIF1 and eIF1A were observed on the intersubunit side in a similar overall position and conformation as described before for other ICs in POUT conformation at low resolution (PDB 6GSM, PDB 3JAQ (Llacer et al, 2015)) (Appendix Fig S2). Briefly, eIF2 consists of three subunits, The eIF2 subunit shares structural homology to EF‐Tu‐like translational GTPases (e.g., Schmitt et al, 2002) and consists of a G‐domain (domain I), including the regulatory switch loops (swI and swII), followed by two ‐barrel domains. eIF2 consists of an N‐terminal OB‐fold domain, a central helical domain, and a C‐terminal α‐β domain. The eIF2 subunit has an unstructured N‐terminal domain, followed by a central helix‐turn‐helix (HTH) domain and C‐terminal zinc binding domain (ZBD). In solution, tRNAi was shown to be bound to the TC in a distinct way different to canonical tRNA‐bound EF‐Tu/eEF1A by employing additional composite interactions with both eIF2 and eIF2 (Schmitt et al 2012). The eIF2 subunit, however, has never been sufficiently resolved to elucidate its molecular contribution to tRNAi binding and 43S PIC formation.
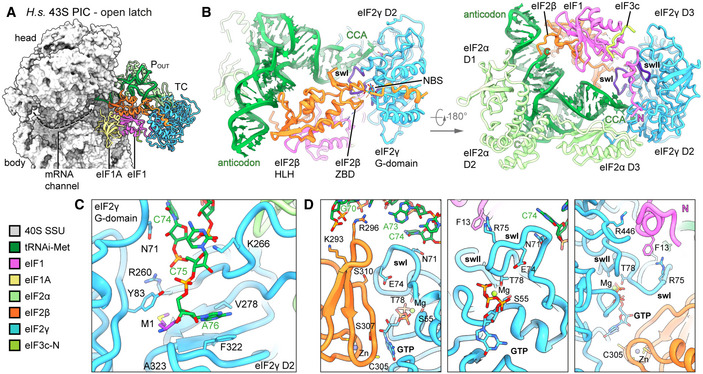
In our structure, we found the tRNAi embraced by all three eIF2 subunits (Fig 7A and B). Similar to the 5 Å resolution crystal structure (3V11 Schmitt et al 2012), the methionylated CCA‐end is sandwiched between the GTPase domain and domain II of eIF2. The terminal adenine base A76 is accommodated in a pocket formed by the ‐sheets of the eIF2 domain II including Val278, Phe322, Gly340, and Arg260 (Fig 7C, Appendix Fig S9C). The 2’‐OH group of the ribose moiety interacts with the carbonyl group of Ala323 and the methionyl side chain stacks on Tyr83 of eIF2 G‐domain. The CCA‐end is further stabilized by contacts including a cation‐ stack of Lys266 on tRNAi C75 and Asn71 of the eIF2 swI loop with tRNAi C74. Moreover, Arg296 of the eIF2 ZBD intercalates into the major groove of the acceptor‐stem helix (G70; supported by Lys293 contacting the phosphate backbone of U69) (Fig 7D, Appendix Fig S9C). eIF2 contacts the T‐ and D‐loops mainly via its central helical domain whereas the N‐terminal OB‐fold domain intercalates between anticodon stem and uS7 in the E site on the head of the 40S. The central eIF2 HTH domain contacts the anticodon from the A site and thereby forms multiple contacts to eIF1, also involving residues of the newly built C‐terminus (I314‐R329), which stretches below the tRNAi anticodon stem toward the E site and contacts C1057 of rRNA h24 (via N327).
Figure 7. Conformation of the TC in the complete human 43S PIC.
- Overview highlighting the positions of TC, eIF1, and eIF1A in the complete human 43S PIC.
- Interactions of eIF2 subunits and domains and eIF1 with methionylated tRNAi; switch loops (sw) of eIF2γ are labeled and colored in purple; nucleotide‐binding site (NBS) with Mg‐GTP bound; the de novo built N‐terminal tail of eIF1; and the C‐terminus of eIF2α and eIF2β are labeled with N and C, respectively.
- Molecular interactions of the methioninylated CCA‐end of tRNAi and eIF2γ.
- Molecular interactions within the nucleotide‐binding pocket and conformation of sw loops stabilized by the eIF1 N‐terminal tail, the eIF2β ZBD, and tRNAi.
Notably, in the GTP binding pocket of eIF2 we clearly identified a Mg2+‐GTP (Fig 7D). Ser55 of the conserved P‐loop and Thr78 of swI coordinate the Mg2+‐ion, whereas Asp134 and Pro135 of swII likely contact the ‐phosphate. Compared to the crystal structure of the archaeal TC (Schmitt et al, 2012), th, 2012 so that this citation matches the Reference List. Please confirm that this is correct."‐‐>e guanine base is rotated by 90° and accommodated in a pocket between Asn190 and Ala226 of eIF2 and Cys305 of the eIF2 ZBD, which is tightly packed upon the nucleotide‐binding pocket.
Interestingly, both switch loops were embedded in a tight interaction network involving interactions with tRNAi, eIF2, and the eIF1 N‐terminal tail, which we built de novo. The N‐terminal tail of eIF1 protrudes from the 5‐stranded ‐sheet and binds to Arg446 of eIF2 domain III, where it forms a loop and projects toward Arg75 of eIF2 swI, forming a cation‐ stack with Phe13 (Fig 7D, Appendix Fig S9C and D). Furthermore, the conformation of the swI loop was stabilized by the tRNAi via Asn71 (see above) and an interaction between conserved Ser310 of the ZBD of eIF2 with Glu74.
In close vicinity to the guanosine binding pocket, we find eIF2 Ser307, the equivalent of yeast eIF2 Ser264. In yeast, a Ser264Tyr mutation causes the Sui‐ (suppressor of initiation codon) phenotype, leading to increased utilization of UUG start codons (Huang et al 1997). This mutation was shown to increase GTP hydrolysis rates and stabilize the closed PIN conformation of the 43S PIC (Martin‐Marcos et al 2014). In the observed position, the tyrosine mutation of Ser307 could easily interfere with the bound nucleotide, for example, by stacking on the guanine base, and thus alter the geometry of the nucleotide‐binding pocket.
Taken together, we found the TC in a stable state within the 43S PIC, in an open conformation in the absence of mRNA. An intricate interaction framework is established by the 40S and eIF1 to accommodate the GTP‐bound eIF2‐tRNAi in a rigid position. The switch loops are kept in a rigid conformation stabilized by tRNAi, eIF2 and the eIF1 N‐terminal tail, and the GTPase pocket of eIF2 is closed by eIF2. This may prevent premature release of the bound nucleotide and, at the same time, may restrict access for eIF5‐NTD to avoid premature GAP activity.
Following TC assembly on 43S PIC and opening of the latch, mRNA can be threaded into the mRNA binding site, followed by scanning for the first AUG codon by the 48S particle. While we do not find scanning intermediates in either yeast or human datasets, in our yeast native 40S population we find one state containing eIF1A, tRNAi in the PIN state, and the eIF5‐NTD instead of eIF1 (yeast 43S PIC). Apart from weaker density for eIF2, this state is similar to one observed before (Llacer et al, 2018), where it was interpreted as a late state after start‐codon recognition. However, to our surprise we still find ABCE1 in this complex. This suggests that ABCE1 may play further roles even in later stages of initiation, or that its dissociation is not required at this stage.
Discussion
While the role of highly conserved ABCE1 during ribosome recycling has been studied in mechanistic details (Becker et al, 2012; Nurenberg‐Goloub et al, 2018; Nürenberg‐Goloub et al, 2020), its role after 60S dissociation remained largely elusive. However, when first characterized biochemically, ABCE1 was found associated with 43S/48S pre‐initiation complexes in yeast, humans, and Drosophila (Andersen & Leevers, 2007; Chen et al, 2006; Dong et al, 2004). Since then, it is a long‐standing question what the function of ABCE1 in these complexes is. Our extensive single particle analysis of native small subunits from yeast and human cells captured a variety of states throughout the assembly of the 43S PIC prior to mRNA loading, in which ABCE1 can stay associated with the 40S. Surprisingly, in yeast we even find ABCE1‐48S complexes beyond the stage of mRNA engagement and start‐codon recognition as indicated by the presence of the eIF5‐NTD (Fig 8).
Figure 8. Role of ABCE1 in eukaryotic translation initiation.
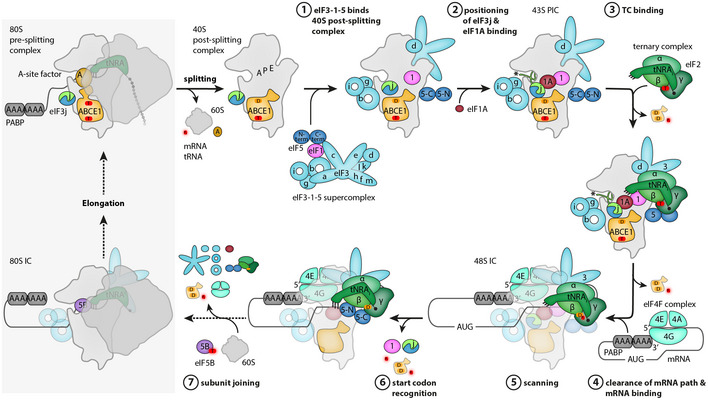
Schematic representation of eukaryotic translation initiation. ABCE1 is shown in orange, nucleotide states are symbolized with “T” for ATP bound and “D” for ADP bound; the two protomers of eIF3j are shown in blue and green, the eIF3j C‐terminus of protomer 2 in mRNA entry channel is marked with an asterisk. For details, see main text (discussion).
We further observe that in all ABCE1‐containing 43S structures its NBDs are in an unusual hybrid conformation, where NBS2 is closed and NBS1 is semi‐open. This is contrary to previous in vitro studies showing SSU‐associated ABCE1 in the ATP‐occluded fully closed state. Notably, the two NBSs in ABCE1 were shown to be highly asymmetric and NBSII has a low ATP‐turnover rate compared to NBSI (Gouridis et al, 2019; Nurenberg‐Goloub et al, 2018). Consistent with this behavior, we find Mg2+‐ATP still bound in the closed NBSII, whereas Mg2+‐ADP is present in NBSI. This is in agreement with the most recent model for the ABCE1 ATPase cycle, in which closure of the NBSII was discussed to be the decisive step for disassembly of 80S pre‐splitting complexes, a process that is then triggered by subsequent closure and ATP hydrolysis in NBS1. Subsequently, re‐opening of NBSI would be expected on the small subunit. But if ATP hydrolysis is prevented either by usage of a non‐hydrolyzable ATP analog or by hydrolysis‐deficient Walker B mutants, ABCE1 can be trapped in the fully closed state on the small subunit under facilitated splitting conditions (Heuer et al, 2017; Kiosze‐Becker et al, 2016; Nürenberg‐Goloub et al, 2020). In native ABCE1‐associated complexes, however, NBSI is already in a more open conformation and additionally obstructed by a part of the eIF3j N‐terminal domain, which intercalates between the two NBDs close to NBSI. Thus, eIF3j may keep NBSI from closing (after putative binding of another ATP), or alternatively, prevents further opening into a state as observed in free ABCE1. This brings up the question of why ATP hydrolysis in NBSII, which would lead to dissociation from the 40S SSU, is inhibited. We find NBSII in a very similar conformation as in the fully closed archaeal structure (Nürenberg‐Goloub et al, 2020), and the structure reveals no clues to explain why ATP hydrolysis is slowed down. Thus, we speculate that a further and likely only small‐scale allosteric signal into NBSII may be necessary for its activation. This may occur after dissociation of the eIF3j N‐terminus upon further opening of NBSI and be accompanied by changes in the ABCE1‐specific HLH and hinge regions.
The observation that ABCE1 dissociation can apparently be actively prevented points toward a direct role in 43S PIC and even 48S IC assembly, most likely in concert with eIF3j. We could corroborate the finding that eIF3j assists in ABCE1‐dependent splitting by in vitro dissociation assays, and furthermore, we established that eIF3j remains bound to the 40S together with ABCE1 after the splitting cycle. A high‐resolution structure of a crosslinked yeast 43S‐PIC revealed that dimeric eIF3j is highly stabilized in the presence of ABCE1, positioning the ultimate C‐terminus of one protomer in the mRNA channel near the entry site. This position explains, how eIF3j could exert its roles as an antagonist of mRNA binding, for example by recycling of mRNA from the 40S subunit (Pisarev et al 2007; Pisarev et al, 2010), or during initiation by preventing premature mRNA recruitment (Fraser et al 2007). Notably, its position close to eIF1A and thus near the A site may also explain its suggested role in regulating start‐site selection (Elantak et al, 2010). Moreover, the comparison of yeast with the human structures of early 43S PICs suggests that eIF3j and ABCE1 may be beneficial for binding of eIF1A. In the yeast conformation, eIF3j appears like a molecular ruler reading out the exact distance between the post‐splitting‐specific FeSD conformation of ABCE1 and the 40S head and beak conformation as adopted after eIF1A binding. Thus, it is tempting to speculate that the observed conformational change in eIF3j may play a role in priming the 40S for eIF1A binding and/or stabilizing the early closed‐latch conformation of the 43S PIC when eIF1A is bound. Notably, eIF1A is the only factor that was not found to be pre‐assembled in a 40S‐free multi‐factor complex (MFC) consisting of eIF1, eIF2‐tRNAi‐GTP, eIF3, and eIF5 in yeast (Asano et al 2000; Zeman et al, 2019), plants, and mammals (Sokabe et al 2012). While eIF1A is capable of binding 40S SSU independently and adopting a similar conformation as within the context of initiation (Yu et al 2009), it is possible that after binding of the MFC eIF3j binding between the 40S head and body in concert with rigidifying the latch structure may be constructive for its productive integration into the 43S complex.
Concluding our cryo‐EM analysis of native initiation complexes, we can deduce a putative order of events during 43S PIC and 48S IC assembly by formation of several structural hallmarks (Fig 8). 80S ribosomes are recycled by ABCE1 after canonical or noncanonical termination. eIF3j may assist the recycling by actively aiding ABCE1 during splitting or by destabilizing mRNA while inserting with its C‐terminus into the mRNA channel. As a first step during initiation, the MFC binds to the recycled 40S as indicated by the highly populated eIF3‐eIF1 bound classes. While the PCI‐MPN core is stably anchored at the solvent side of the 40S, the eIF3c‐NTD locates into the ISS via the 4‐helix bundle, positioning eIF1 in the process. The YLC module is guided to the mRNA entry by stable positioning of the eIF3b ‐propeller between h16 and of rRNA expansion segment ES6c. Here, the eIF3i‐eIF3g complex can adopt variable positions that may be important for the role of eIF3g‐eIF3i during scanning (Cuchalova et al, 2010). Concomitantly, the RRM of either eIF3g or eIF4B accommodates on the mRNA entry, and in the human 43S complexes, the mRNA entry channel is blocked by a yet unidentified density. After eIF1A accommodation, the TC can be stably integrated to form the complete mRNA‐free POUT state 43S. This opens up the latch and leads to clearance of the mRNA path, since in POUT complexes no density in the mRNA path is visible.
With respect to a fully accommodated TC, our structure reveals for the first time a network of interactions between the tRNAi and all subunits of eIF2 as well as eIF1 at molecular resolution. The eIF2 switch loops are highly confined, and the GTPase pocket is closed by the ZBD of eIF2, thus restricting the access for the eIF5‐NTD to exert its GAP activity. Notably, GTP hydrolysis in eIF may already occur during scanning. This would require that the eIF5 N‐terminal tail could reach into the eIF2 GTPase pocket and, thus, result in a rearrangement of the eIF2 ZBD. A structure of a scanning 48S, however, is still lacking. Yet, large structural rearrangements have been observed after start‐codon recognition, during which the 48S IC adopts the closed PIN state. Here, the entire TC rearranges, and especially, eIF2 alters its location on the 40S head and relative to eIF1 and eIF1A. It is likely that this conformational switch could already partially occur during scanning and that this would also affect the position of the eIF2 ZBD, which was too flexible to be resolved in all previous cryo‐EM structures (Llacer et al, 2015; Llacer et al 2018; Simonetti et al 2016; Eliseev et al, 2018). After eIF5‐dependent GTP hydrolysis, release of inorganic phosphate (Pi) would still be inhibited until start‐codon recognition. During or after this process, the eIF5 NTD replaces the gatekeeper eIF1 and leads to a further stabilization and compaction of the PIN state, which may be a prerequisite for the following step of eIF5B‐mediated subunit joining (Llacer et al, 2018).
Our analysis shows that ABCE1 can still be associated with initiating 40S. Yet, which role might ABCE1 play during formation of the full 43S and—as observed in yeast—even in context of the eIF5‐accommodated partial 48S? Currently, ABCE1 is assumed to act as an anti‐association factor, ensuring that premature 60S interaction is prevented after termination and ribosome splitting. However, in this function it would likely become redundant after the formation of the 43S PIC, failing to explain its presence in later stages of initiation. Another possibility is that its observed interplay with eIF3j as early as during the splitting reaction supports the timely recruitment of the remaining eIFs to the vacant 40S. Furthermore, we speculate that the inhibiting peptide close to NBSI would need to be ejected to facilitate ATP‐hydrolysis in NBSII. Here, it is possible that dynamics of the rather flexible YLC module could play a role. In fact, this module is able to relocate into the ISS to occupy the position of ABCE1 (Llacer et al, 2015). With this steric competition in mind, it would be plausible that it contributes to ABCE1 dissociation, although it is not entirely clear at which stage this relocation happens. In addition, eIF3j, which is still present at least as fuzzy density in the fully assembled 43S, may also contribute in coordinating such events, for example, via its known interaction with eIF1A and the eIF3b‐RRM (Elantak et al, 2010). Finally, since ABCE1 is even present on 48S IC complexes after start‐codon recognition, events during subunit joining may be the final trigger for ABCE1 dissociation. In this context, the P proteins of the 60S subunit may not only play a role during ribosome splitting as suggested before (Imai et al 2018), but also for ABCE1 removal after initiation. Yet to reveal exact timing of these events and the mechanistic interplay of these factors, future work will be needed.
Materials and Methods
Yeast strains
Saccharomyces cerevisiae ribosomes for biochemical assays were purified from a wild‐type BY4741 strain, which was grown on YPD medium.
Samples for LC‐MS/MS analyses were purified from a BY4741 (MATa, ura3∆0, his3∆1, leu2∆0, met15∆0), Rli1‐TAP:HIS3MX6 strain (Ghaemmaghami et al 2003).
For the preparation of native yeast 40S initiation complexes, a BY4741 strain containing genomic TAP‐tagged SKI3 and a plasmid overexpressing SKA1 (pCM190) (Zhang et al 2019) were used; the crosslinked yeast 43S pre‐initiation complex was derived from a S.c. W303 strain (MATa, ade2∆1, trp1∆1, can1∆100, leo2∆3,112, his3∆11, ura3, GAL) expressing genomically TAP‐tagged Nip1 (eiF3c).
ABCE1‐TAP polysome profile and sucrose density gradient fractionation
Yeast (Saccharomyces cerevisiae; S.c.) cells from the BY4741 strain expressing C‐terminally TAP‐tagged ABCE1 (Rli1) were grown in 200 ml YPD to an OD600 of 0.8. The cells were then treated with 50 µg ml–1 cycloheximide on ice for 5 min. and collected by centrifugation. The cells were lysed in lysis buffer (20 mM Tris–HCl, pH 7.4, 50 mM KCl, 10 mM MgCl2, 50 µg ml–1 cycloheximide, and EDTA‐free protease inhibitors (Roche)) by vortexing them with glass beads (12 cycles of 30 sec. vortex/30 sec. on ice). The lysate was cleared by centrifugation for 10 min. at 16,000 g, 4 °C and stored at −80 °C. Ten A260 units were loaded on a 10–50% sucrose gradient and centrifuged at 187,813 g for 2.75 h at 4 °C in a SW41Ti rotor (Beckman Coulter). The fractions of the gradient were collected, and proteins were precipitated with trichloroacetic acid and separated on a 10% acrylamide gel. The proteins were detected with antibodies after Western blotting: ABCE1‐TAP with peroxidase anti‐peroxidase (PAP) complex (Sigma‐Aldrich) at 1:2,000, and Nog1 with a rabbit anti‐Nog1 antibody at 1:5,000 dilution.
ABCE1‐TAP tandem affinity purifications
Cells expressing C‐terminally TAP‐tagged ABCE1 (Rli1) were cultivated in rich medium (YPD) until OD600 of 2, and cultures were centrifuged at 4 °C, rinsed in cold water, and frozen at −80 °C. Cells were thawed on ice, resuspended in lysis buffer (50 mM Tris–HCl pH 8.0, 100 mM NaCl, 10 mM MgCl2, complete EDTA‐free protease inhibitor mix or: 20 mM HEPES/KOAc pH 7.4, 100 mM KOAc, 10 mM MgCl2, complete EDTA‐free protease inhibitor mix), and lysed with glass beads using a Magnalyser. The lysates were clarified by centrifugation at 16,000 g for 10 min. at 4 °C. Supernatants were collected, and triton (0.5% final) or NP‐40 (0.1% final) was added to the lysate. Binding to magnetic beads coupled with IgG was performed on a wheel at 4 °C overnight. Beads were collected on a magnet, flow‐through was discarded, and beads were washed in lysis buffer. Elution was performed by resuspension in 2% SDS, 1× Tris‐EDTA buffer and incubation at 65 °C for 10 min. Eluted beads were discarded on a magnet, and eluate was purified on HiPPR Detergent Removal Resin (Thermo Scientific, 88305). Purified proteins were eluted in PBS. The rest of the eluates was precipitated by the methanol/chloroform technique (Wessel & Flugge, 1984) and analyzed by mass spectrometry.
To control the quality of the affinity purification, a sample of eluates (3%) was separated on acrylamide NuPAGE Novex 4–12% Bis‐Tris gels (Life Technologies) and analyzed by silver staining.
Mass spectrometry: data acquisition and analysis
After reduction and alkylation, protein samples were treated with endoprotease Lys‐C (Wako) and trypsin (Trypsin Gold Mass Spec Grade; Promega). Peptide samples were desalted by OMIX C18 pipette tips (Agilent Technologies) and then analyzed by LC‐MS/MS on an LTQ‐Orbitrap velos instrument (Thermo Fisher Scientific) connected online to an EASY‐nLC system (Thermo Fisher Scientific). Raw mass spectrometry (MS) data from the LTQ‐Orbitrap were analyzed using MaxQuant software (Cox & Mann, 2008) version 1.6.10.43, which uses Andromeda search engine (Cox et al 2011). Bioinformatic analysis of the MaxQuant/Andromeda workflow output and the analysis of the abundances of the identified proteins were performed with the Perseus module (Tyanova et al 2016) version 1.6.10.43. Only protein identifications based on a minimum of two peptides were selected for further quantitative studies. After data processing, label‐free quantification (LFQ) values from the “proteinGroups.txt” output file of MaxQuant were further analyzed. To distinguish specifically enriched proteins from the background, protein abundances were compared between sample and control groups using Student’s t‐test statistic, and results were visualized as volcano plots (Hubner & Mann, 2011).
Preparation of puromycin‐treated 80S ribosomes from yeast
S.c. BY4741 wild‐type cells were grown in YP medium with 2% glucose to an OD600 of 2.5, then harvested by spinning at 4,400 g for 10 min. Cells were washed first with water, then 1% KCl, then resuspended in 30 ml lysis buffer (20 mM HEPES/KOH pH 7.4, 100 mM KOAc, 7.5 mM Mg(OAc)2, 1 mM DTT, 0.5 mM PMSF, complete EDTA‐free protease inhibitor mix). Lysis was performed using a Microfluidics M‐110L microfluidizer at 15k psi.
The lysate was cleared by centrifugation first at 26,892 g for 15 min., then at 140,531 g for 30 min. 15 ml of cleared lysate was loaded on a layered sucrose cushion consisting of 4 ml 2 M sucrose and 4 ml 1.5 M sucrose (buffer: 20 mM HEPES/KOH pH 7.4, 500 mM KOAc, 5 mM Mg(OAc)2, 1 mM DTT, 0.5 mM PMSF) and centrifuged at 246,468 g for 21 h and 15 min.
The pellet containing ribosomal components was resuspended in water and mixed with 2× puromycin buffer (40 mM HEPES pH 7.5, 1 M KOAc, 25 mM Mg(OAc)2, 2 mM puromycin, 2 mM DTT, 1 U/ml SUPERase‐In RNase Inhibitor (Invitrogen)). The mixture was incubated for 30 min at room temperature and then loaded on 10‐40% sucrose density gradients (20 mM HEPES/KOH pH 7.4, 500 mM KOAc, 5 mM Mg(OAc)2, 1 mM DTT, 0.5 mM PMSF). Gradients were centrifuged at 20,755 g in an SW 32 Ti rotor (Beckman Coulter) for 20 h. 80S ribosomal fractions were identified using a Biocomp Gradient station ip and a Triax Flow cell and were manually collected. Fractions were then pelleted in a TLA110 rotor at 417,200 g for 45 min and resuspended in storage buffer (20 mM HEPES/KOH pH 7.5, 100 mM KOAc, 5 mM Mg(OAc)2, 1 mM DTT). Aliquots were frozen in liquid nitrogen and stored at ‐80 °C until use.
Protein expression and purification
eIF3j (Hcr1) purification
Escherichia coli (E. coli) BL21(DE3) cells were transformed with the pTYB2 plasmid containing full‐length HCR1 and selected on LB plates containing ampicillin. Cells from a pre‐culture were inoculated into 1.5 l of LB medium with ampicillin, and cell growth was monitored at 37 °C. At an OD600 of 0.6, the cultures were transferred to an ice‐water bath and incubated for 20 min. 0.1 mM IPTG was added to induce protein expression, and cells were incubated for 15 h at 16 °C while shaking. Cells were harvested by centrifugation at 3,500 g for 10 min and washed with 1% KCl, then resuspended in lysis buffer (20 mM HEPES pH 7.5, 500 mM NaCl). Cells were then pelleted again at 2,600 g, frozen in liquid nitrogen, and stored at ‐80 °C until further use.
Frozen cell pellets were thawed, resuspended in lysis buffer, and lysed using a Microfluidics M‐110L microfluidizer at 15k psi. The lysate was cleared by centrifugation at 20,000 g for 30 min. Clear lysate fraction was added to 1.5 ml magnetic chitin beads (NEB E8036S) equilibrated in lysis buffer. Binding was performed for 1.5 h at 4 °C on a wheel. Beads were harvested on a magnet and washed once using 5 ml lysis buffer, twice using washing buffer (20 mM HEPES pH 7.4, 1 M NaCl, 1 mM EDTA) and once again using lysis buffer. The protein was then eluted from the beads using 5 ml elution buffer (20 mM HEPES pH 7.4, 500 mM KCl, 50 mM DTT) by incubating on a wheel at 4 °C overnight. A second elution step was performed using the same buffer for one hour after removal of the first elution fraction. Both elution volumes were combined and concentrated using an Amicon Ultra 10k MWCO concentrator. Aliquots of pure eIF3j were flash‐frozen in liquid nitrogen and stored at ‐80°C.
ABCE1 (Rli1) purification
ABCE1 (Rli1) was overexpressed in S. cerivisiae strain WCGα using the pYes2‐ABCE1‐His6 plasmid (kindly provided by R. Green, Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine) (Shoemaker & Green, 2011). Cells were grown in YP medium lacking uracil and containing 2% galactose, 1% raffinose at 30 °C to mid‐log phase and were harvested at a final OD600 of 1.0 by centrifugation at 3,500 g for 10 min. Cells were washed once with 1% KCl, pelleted again, and resuspended in lysis buffer (75 mM HEPES pH 8.0, 300 mM NaCl, 5mM beta‐mercaptoethanol (β‐ME), 1% Tween, 20 mM imidazole, 2 mM MgCl2, 10% glycerol). Excess buffer was removed by centrifugation at 2,600 g, and the cells were frozen in liquid nitrogen. Frozen cells were ground using a Spex SamplePrep Freezer Mill and the powder stored at ‐80 °C until further use. The cell powder was thawed and resuspended in lysis buffer. Cell debris was removed by centrifugation at 47,807.6 g for 30 min and filtered using a 1.6‐ m membrane.
ABCE1 was purified first by metal affinity chromatography. Cleared lysate was applied to a HisTrap HP column (GE 5 ml column). The column was washed with 15 column volumes (CV) wash buffer (50 mM HEPES pH 8.0, 500 mM NaCl, 5 mM β‐ME, 20 mM imidazole, 2 mM MgCl2, 10% glycerol), and the protein was eluted with 4 CV over a gradient from 20 mM to 300 mM imidazole. Fractions containing ABCE1 were combined and dialyzed against Buffer A (20 mM HEPES pH 7.6, 100 mM KCl, 5 mM β‐ME, 0.1 mM EDTA, 10% glycerol, 0.1 mM PMSF) overnight. The sample was diluted to 50 ml and loaded onto a cation exchange column (HiTrap SP 5 ml, GE). The column was washed with 6 CV Buffer A, and ABCE1 was eluted over gradient from 100 mM to 1 M KCl over 8 CV. ABCE1‐containing fractions were concentrated using Amicon® 50k MWCO concentrator before loading onto a gel filtration column (Superdex200) for size‐exclusion chromatography. The fractions containing ABCE1 were concentrated, and aliquots of pure ABCE1 in 20 mM HEPES pH 7.5, 200 mM KCl, 1.5 mM MgCl2, 2 mM β‐ME, and 5% glycerol were flash‐frozen and stored at ‐80 °C.
eIF6 purification
E. coli BL21 (DE3) cells were transformed with a p7XC3GH plasmid expressing eIF6 fused to 3C protease cleavage site, GFP, and 10‐His. Cells were grown on LB medium to mid‐log phase (OD600 = 0.7‐0.8) at 37 °C and induced with 1 mM IPTG at 16 °C for 20 h. Cells were harvested by centrifugation at 4,400 g and 4 °C for 8 min, washed with PBS, and resuspended in lysis buffer (20 mM Tris–HCl pH 8.0, 300 mM NaCl, 2 mM β‐ME) with 10% glycerol. Resuspended cells were flash‐frozen in liquid nitrogen and stored at ‐80 °C until further use. For purification, frozen cells were thawed and resuspended in lysis buffer without glycerol. Lysis was performed using a Microfluidics M‐100L microfluidizer at 15k psi. Crude lysate was cleared by centrifugation at 30,596 g for 20 min. TALON metal affinity resin was equilibrated in lysis buffer and added to the cleared lysate, then incubated at 4 °C for 40 min on a wheel. After collection of the flow‐through, the column was washed using lysis buffer with 10 mM imidazole. Elution was performed by incubating the resin with lysis buffer with 10 mM imidazole and 0.25 mg ml–1 3C protease for 30 min at 4 °C on a wheel. The elution fraction was concentrated using an Amicon 10k MWCO concentrator and loaded onto a Superdex200 column for size‐exclusion chromatography using storage buffer (50 mM HEPES/KOH pH 7.5, 500 mM KCl, 2 mM MgCl2, 2 mM β‐ME). The purified protein in storage buffer was flash‐frozen in liquid nitrogen and stored at ‐80 °C.
Dom34 and Hbs1 were purified as described before (Lee et al 2007).
Splitting assays
In vitro splitting assays
Ribosome splitting assays were carried out to test the influence of eIF3j (Hcr1) on the canonical splitting reaction mediated by Dom34, Hbs1, and ABCE1 in yeast. For each reaction, 5 pmol of yeast 80S ribosomes (see above) was mixed with fivefold molar excess of splitting factors Dom34, Hbs1, and ABCE1 as well as the anti‐association factor eIF6 under physiological buffer conditions (20 mM HEPES/KOH pH 7.5, 100 mM KOAc, 4 mM Mg(OAc)2, 5 mM β‐ME, 1 mM ATP, 1 mM GTP). Varying amounts of eIF3j were added to the reactions, ranging from twofold to twentyfold molar excess over the 80S ribosomes.
The samples were incubated on ice for 30 min and then loaded on 10‐50% sucrose density gradients (20 mM HEPES/KOH pH 7.5, 100 mM KOAc, 5 mM Mg(OAc)2, 1 mM DTT, 10‐50% (w/v) sucrose). Gradients were spun in an SW 40 Ti rotor (Beckman Coulter) at 202,048 g for 4 h and fractionated at a BioComp Gradient Station ip using a Triax Flow Cell for UV measurement.
Ribosomal peak fractions were collected manually, and from each fraction, proteins were precipitated using 0.015% sodium deoxycholate and 7.2% trichloroacetic acid at 4 °C.
Proteins were separated on a 15% SDS‐PAGE gel and visualized using SimplyBlue staining reagent.
“Facilitated” splitting assays
“Facilitated” splitting assays were performed to test the association of yeast ABCE1 and eIF3j to ribosomal particles under non‐physiological high‐salt conditions and in the presence of ATP or the non‐hydrolyzable ATP analog AMP‐PNP. To induce splitting, purified 80S ribosomes were mixed with tenfold molar excess of ABCE1 in splitting facilitating buffer (20 mM HEPES/KOH pH 7.4, 500 mM KCl, 1.5 mM MgCl2, 1 mM DTT). Depending on the experiment, 0.5 mM AMP‐PNP or ATP and 10‐fold molar excess of eIF3j were added. For the experiments described here, approx. 50 pmol ribosomes in a total reaction volume of 250 were used. The samples were incubated for 20 min at 25°C and then cooled down to 4 °C on ice and loaded on 10‐50% sucrose density gradients (20 mM HEPES/KOH pH 7.5, 100 mM KOAc, 5 mM Mg(OAc)2, 1 mM DTT, 10‐50% (w/v) sucrose). All following procedures were carried out as described above for splitting assays.
Cryo‐EM sample preparation
Preparation of native yeast 40S complexes
A BY4741 strain containing genomic TAP‐tagged SKI3 and a plasmid overexpressing SKA1 (pCM190) (Zhang et al 2019) was used for generation of the cryo‐EM sample.
Yeast cells were grown in synthetic medium lacking uracil (SL‐Ura) with 2% glucose at 30 °C to an OD600 of 3.0, whereupon the cultures were chilled in ice water. The cells were harvested by centrifugation at 4,422 g for 10 min in a Sorvall SLC‐6000 rotor, washed with water, and resuspended in lysis buffer (20 mM HEPES/KOH pH 7.4, 100 mM KOAc, 5 mM Mg(OAc)2, 1 mM DTT, 0.5 mM PMSF, complete EDTA‐free protease inhibitor mix). Cells were frozen in liquid nitrogen and ground using a Spex SamplePrep Freezer/Mill.
Frozen cell powder was resuspended in lysis buffer (1:3 w/v), and the lysate was cleared by centrifugation in an SS‐34 rotor (Thermo Scientific) at 26,891.8 g for 15 min.
Approximately 150 A260 absorption units were loaded on a 10‐50% sucrose density gradient (buffer composition identical to lysis buffer). Gradients were spun in an SW40 Ti rotor (Beckman Coulter) at 202,048 g for 3 h, and the 40S peak was harvested manually using a Triax Flow Cell.
Total A260 of the collected 40S fraction from yeast lysate was measured, and the buffer was exchanged to cryo‐EM grid buffer (20 mM HEPES/KOH pH 7.4, 100 mM KOAc, 5 mM Mg(OAc)2, 1 mM DTT, 0.5 mM PMSF, complete EDTA‐free protease inhibitor mix, 0.05% Nikkol) by three successive rounds of concentration and dilution by a factor of approx. 1:5 using an Amicon Ultra Centrifugal Filter (MWCO 100k) (total dilution factor approx. 1:125). The sample was then concentrated again. The A260 was measured as A260/ ml = 6.3.
Freshly prepared sample was diluted to approx. 1.25 A260 / ml, and 3.5 l was applied to 2 nm pre‐coated Quantifoil R3/3 holey carbon support grids and vitrified in liquid ethane using a Vitrobot mark IV (FEI Company, Netherlands). (wait time 45 s, blotting time 2 s).
Preparation of crosslinked yeast 43S pre‐initiation complexes
S.c. W303 cells expressing genomically TAP‐tagged Nip1 (eIF3c) were grown in YP medium with 2% glucose at 30 °C to an OD600 of 2.0 and harvested by centrifugation at 4,422 g for 10 min in a Sorvall SLC‐6000 rotor, washed with water, and resuspended in lysis buffer (50 mM HEPES/KOH pH 7.4, 100 mM KOAc, 1.5 mM Mg(OAc)2, 1 mM DTT, 0.5 mM PMSF, complete EDTA‐free protease inhibitor mix). Cells were frozen in liquid nitrogen and ground using a Spex SamplePrep Freezer/Mill.
Frozen powder was resuspended in lysis buffer with 0.15% (v/v) NP‐40, and the lysate was cleared by centrifugation first at 20,000 g for 30 min in an SS‐34 rotor and then at 26,891.8 g for 15 min in the same rotor.
The cleared lysate was applied to IgG Sepharose (GE17‐0969‐01) equilibrated in lysis buffer and incubated for 1 h at 4 °C on a rotating wheel. After binding, the supernatant was removed by centrifugation at 1,383 g for 3 min and the sepharose was transferred to a Bio‐Rad Micro Bio‐Spin Chromatography Column. The sepharose was sequentially washed with 10 column volumes each of lysis buffer containing 0.15% NP‐40 and elution buffer (50 mM HEPES/KOH pH 7.4, 100 mM KOAc, 1.5 mM Mg(OAc)2, 1 mM DTT).
Elution was performed by incubating the sepharose with elution buffer and approx. 2.4 U/µl AcTEV (Invitrogen) for 90 min at 20 °C. The eluted sample was harvested by spinning the columns at 380 g for 2 min.
The eluted sample was transferred onto a 10‐50% sucrose density gradient (50 mM HEPES/KOH pH 7.4, 100 mM KOAc, 5 mM Mg(OAc)2, 1 mM DTT). Gradients were spun in an SW40 Ti rotor (Beckman Coulter) at 202,048 g for 3 h, and the 40S peak was harvested manually using a Triax Flow Cell for UV absorption measurement.
The 40S fraction was concentrated and the buffer exchanged to elution buffer using an Amicon Ultra Centrifugal Filter (MWCO 100k). The sample was crosslinked by shaking with 0.5 mM BS3 at 10 °C, 1,200 rpm for 10 min and then further incubated at 4 °C for approx. 10 min. The reaction was quenched by addition of 40 mM Tris–HCl. To the sample, 0.05% β‐OG was added and cryogenic freezing was performed.
Preparation of native human 40S complexes
Human 40S initiation complexes were found as byproducts in an affinity purification using internally tagged RIOK1 and mutant RIOK1‐D324A as bait. In brief, HEK Flp‐In 293 T‐Rex (Invitrogen) was grown in a 10‐cm cell‐culture dish to approximately 70% confluency and transfected with 0.5 µg of a pcDNA5/FRT/TO vector containing RIOK1 or RIOK1‐D324A and 4.5 µg pOG44 (Invitrogen), using 20 µg polyethylenimine (PEI). Cells were selected using 150 µg ml‐1 hygromycin B (Thermo Scientific) and maintained in DMEM (Thermo Scientific) containing 10% fetal calf serum, 100 µg ml‐1 hygromycin B, 10 µg ml‐1 blasticidin and 1× penicillin/streptomycin and GlutaMAX (Thermo Scientific). Stable cell lines were subsequently grown in multiple 15‐cm cell‐culture dishes, protein expression induced with 1.6 µg ml‐1 tetracycline and harvested in 0.025% trypsin/EDTA (Thermo Scientific) after 24 h. Cells were washed one in 1x phosphate‐buffered saline (PBS) and subsequently pelleted at 1,600 g at 4 °C. Cells were then resuspended in lysis buffer (20 mM HEPES pH 7.6, 150 mM potassium acetate, 5 mM MgCl2, 1 mM DTT, 0.5 mM NaF, 0.1 mM Na3VO4, 1× protease inhibitor (Sigma‐Aldrich), 0.5% NP‐40 substitute) and incubated for 30 min in an over‐head rotator at 4 °C, before centrifugation at 4,000 g for 15 min at 4 °C. The cleared lysate was then added to 100 µl of anti‐Flag affinity beads (Sigma‐Aldrich) and rotated for 2 h at 4 °C. Beads were harvested and 4 times washed with 1 ml wash buffer (20 mM HEPES pH 7.6, 150 mM potassium acetate, 5 mM MgCl2, 1 mM DTT, 0.5 mM NaF, 0.1 mM Na3VO4, 1× protease inhibitor (Sigma‐Aldrich)), before bound complexes were eluted 6 times with 100 µl of 20 mM HEPES pH 7.6, 150 mM potassium acetate, 5 mM MgCl2, 1 mM DTT, 0.05% Nikkol, and 0.2 mg ml‐1 3x Flag peptide (Sigma‐Aldrich). All eluate fractions were combined and concentrated on 300 kDa molecular mass cut‐off filters (Sartorius).
3.5 µl of the concentrated sample was applied to glow discharged copper grids with holey carbon support and a 2 nm continuous carbon layer (R3/3, Quantifoil). Grids were blotted in a Vitrobot Mark IV (FEI Company) for 2 s after incubation for 45 s at 4°C and frozen in liquid ethane.
Cryo‐EM data collection and processing
Data collection and processing of the yeast 40S complex sample
Cryo‐EM data were collected on a Titan Krios TEM, using a Falcon II DED at 300 kV, with an electron dose of approx. 2.5 e‐/Å2 per frame for 10 frames (defocus range of 1.1 to 2.3 µm). The magnified pixel size was 1.084 Å/pixel.
Micrograph stacks collected at the TEM were summed and corrected using MotionCor2 (Zheng et al 2017). Micrograph quality was assessed individually, and CTF parameters were estimated using GCTF (Zhang, 2016). Particle picking was performed using Gautomatch (http://www.mrc‐lmb.cam.ac.uk/kzhang/), and all further processing was performed using RELION 3.0 (Scheres, 2012; Zivanov et al 2018).
Data collection and processing of the crosslinked yeast 43S PIC sample
For the crosslinked yeast sample, 5126 micrograph movies were collected at a Titan Krios at 300 kV, at a nominal pixel size of 1.059 Å, and a defocus range from 0.5 to 2.5 µm. Movies were recorded on a K2 Summit direct electron detector using low‐dose conditions with 40 frames at approximately 1.12 e‐/Å2. each. All frames were gain corrected and subsequently aligned and summed using MotionCor2 (Zheng et al, 2017), and CTF parameters were determined using CTFFIND (Rohou & Grigorieff, 2015) and Gctf (Zhang, 2016). Particles were picked using Gautomatch (http://www.mrc‐lmb.cam.ac.uk/kzhang/), and particle images were extracted in RELION 3.1 (Zivanov et al, 2018). 2D classification was performed using a previously generated cryo‐EM map of an idle 40S subunit as reference, and all particles with recognizable features of the small subunit were selected and subjected to an initial 3D refinement using the same reference, followed by 3D classification. All classes with density corresponding to ABCE1 were selected, grouped, and classified again using an ellipsoid mask around ABCE1. Particles with ABCE1 in the semi‐open conformation as observed in the native yeast sample were selected, refined, and subjected to one round of CTF refinement and Bayesian polishing. Thereupon, these particles were further sub‐classified using an ellipsoid mask around eIF3j, and all particles containing eIF3j were selected as the final class containing both semi‐open ABCE1 and eIF3j. This class represented 4.8% of the total dataset. Particles in this class were subjected to one more round of CTF refinement and Bayesian polishing, before 3D refinement and post‐processing as well as local resolution estimation (all within RELION 3.1) yielded the final reconstructions at 3.0 Å average resolution, as shown in (Appendix Fig S1).
Data collection and processing of the human 40S complex sample
Data collection was performed on a Titan Krios at 300 kV, where 7,365 and 4,499 movies were collected for RIOK1‐D324A and RIOK1, respectively, at a nominal pixel size of 1.059 Å and at a defocus range from 0.5 to 2.5 µm. Movies were recorded on a K2 Summit direct electron detector using low‐dose conditions with 48 frames at approximately 1 e‐/Å2. All frames were gain corrected and subsequently aligned and summed using MotionCor2 (Zheng et al, 2017), and CTF parameters were determined using CTFFIND (Rohou & Grigorieff, 2015) and Gctf (Zhang, 2016). Particles were then picked using Gautomatch (http://www.mrc‐lmb.cam.ac.uk/kzhang/). Particle images were extracted in RELION 3.0 (Zivanov et al, 2018) and subjected to reference‐free 2D classification. Good particles were selected, 3D refined, and classified. Besides the expected pre‐40S classes (unpublished), one class containing the initiation complex was obtained in both datasets, comprising approximately 2% (RIOK1‐D324A data set) and 8.7% (RIOK1 data set) of the total particle number. The two datasets were subsequently subjected to Bayesian polishing and CTF refinement, combined and further classified extensively as shown in Appendix Fig S2. Final reconstructions were then B‐factor sharpened with RELION and the local resolution estimated. Where indicated (Appendix Fig S2), local or multi‐body refinement was performed.
Model building and refinement
For rigid body fits and figures, Chimera version 1.13.1 (Pettersen et al 2004) and ChimeraX version 0.91 (Goddard et al 2018) were used. Homology models were created using SWISS‐MODEL Repository (Bienert et al 2017; Waterhouse et al 2018).
Yeast 43S PIC and 48S IC model
The atomic models PDB 5NDG (Prokhorova et al 2017), 6FYY, 6FYX (Llacer et al, 2018), and 6TB3 (Buschauer et al 2020) containing the models for S.c. 40S rRNA, r‐proteins, and eIFs were fitted as rigid bodies into the cryo‐EM maps of the S.c. 43S PIC and 48S IC. For the 43S PIC, the 40S rRNA and ribosomal proteins were fitted from PDB 5NDG and eIFs were fitted from PDB 6FYY. For the 48S IC, the 40S rRNA and ribosomal proteins were fitted from PDB 6TB3 and eIFs were fitted from PDB 6FYX. For ABCE1, the hybrid semi‐open/closed model derived from the human 43S PIC (see below) was fitted into the density. For Hcr1, a homology model was created based on the structure of the human eIF3j dimer (PDB 3BPJ). The C‐terminus of protomer 1 was extended by 3 amino acids, and the C‐terminus of protomer 2 was extended by 19 amino acids based on a comparison with the data from the yeast 43S PIC‐XL sample with the density observed for the native sample.
Models for the “mRNA entry position” of the YLC were obtained by fitting the crystal structure of eIF3i/g (PDB 4U1E, Erzberger et al, 2014) to the observed density as a rigid body and matching it to the structure of eIF3b CTD from PDB 6FYY; to obtain the “ES6 position”, the eIF3i‐eIF3g moiety bound to the C‐terminal helix of eIF3b was rotated by 120 degrees around the Thr697‐Asp701 hinge in the CTD of eIF3b as a rigid body.
Yeast 43S PIC‐XL model
The atomic model 6TB3 (Buschauer et al, 2020) containing the models for S.c. 40S rRNA and r‐proteins was split into 40S head and body and fitted as rigid bodies into the best‐resolved cryo‐EM map of the complex using ChimeraX version 1.0 (Goddard et al 2018). The homology models for eIF3j and ABCE1 previously generated for the native 43S complex were also fitted, and the entire model was adjusted in WinCoot 0.8.9.2 (Emsley & Cowtan, 2004). The C‐terminus of eIF3j protomer 2 was built de novo. A focused refined cryo‐EM map for the NBD2 of ABCE1 was consulted to improve accuracy in the model for this domain during initial model building. The model was real space refined using Phenix 1.18 (Afonine et al 2018; Liebschner et al 2019).
Human 43S PIC
To obtain the atomic model, the best‐resolved maps as obtained after local focused refinement or multi‐body refinement (Fig EV2, Appendix Fig S2) were used to build the different parts of the H.s. 43S PIC. The 40S subunit was fit into maps of 40S body and 40S head obtained from multi‐body refinement III (Appendix Fig S2) starting with the 40S model (PDB 6G5H, Ameismeier et al 2018). After rigid body fitting, side chains of ribosomal proteins and rRNA were adjusted using Coot (version 0.8.9.2) (Emsley & Cowtan, 2004). Further, the 60S ribosomal protein eL41 was added to the model using PDB 6EK0 (chain h, Natchiar et al 2017). For eIF1A, the homology model based on PDB 3J81 (Hussain et al 2014) was fitted and adjusted using the 40S body map. The N‐terminal helix bundle of eIF3c (47‐149) was built de novo into the same map.
The homology model of the crystal structure of the C‐terminal part of eIF3d (162‐527; PDB 5K4B, Lee et al, 2016) was fitted into the map for the 40S head obtained from multi‐body refinement III (Appendix Fig S2). The atomic model was only modified in the regions interacting with the 40S head. Similarly, the model for eIF3b (PDB 5K1H, Simonetti et al, 2016) was only adjusted in blades 5 and 6, which contact the 40S body. Here, the best‐resolved cryo‐EM map, obtained by focused classification on the YLC, could be used (Appendix Fig S2). Also, the homology model of eIF3i (PDB 5K0Y, Simonetti et al, 2016) and an ‐helix corresponding to the C‐terminal part of eIF3a were fitted into this map.
The eIF3‐PCI‐MPN core (including eIF3a, c, e, f, h, k, l, m) was modeled into the two maps of multi‐body refinement II (Appendix Fig S2) using the human homology model based on PDB 5A5T (des Georges et al, 2015) as starting model. eIF3d‐N (2‐84) was built de novo into the density.
For eIF3j, the unpublished crystal structure of the human eiF3j dimer (PDB 3BPJ) was fitted as rigid body into the density of 43S PIC state II.
Classification of the entire 43S dataset focusing on ABCE1 followed by focused refinement yielded a well‐resolved map, which could be used for model building. A homology model based on the closed‐state yeast ABCE1 bound to the 40S (PDB 5LL6, Heuer et al, 2017) was used as starting model. ATP and ADP were added to the NBSs.
One class obtained by focused classification on the YLC represents a very stable 43S complex in POUT state and yielded a well‐resolved map of the TC after focused refinement. The models of tRNAi (PDB 6FEC, Eliseev et al, 2018), eIF2 and eIF2 (PDB 6O85, Kenner et al 2019) and the homology models of eIF2 and eIF1 (based on PDB 6GSM) were fitted into the map and adjusted using Coot. Further, a stretch of 8 amino acids was modeled into the density adjacent to eIF1, which corresponds to eIF3c. For the unassigned RRM on top of 18S rRNA h16, we generated a poly‐alanine model.
All models were real space refined using Phenix (version 1.17 and 1.18) (Afonine et al, 2018; Liebschner et al, 2019). In order to model the interfaces between the different parts of the structure, maps before and after multi‐body refinement were used. Furthermore, neighboring parts were included in the real space refinement using focused cryo‐EM maps. The final composite model was subjected to a final refinement using the overall cryo‐EM map of state II and state III (Appendix Fig S2, Appendix Table S2). In regions with local resolution lower than 4 Å, side chains were not modeled.
Chemical crosslinking and mass spectrometry
For the crosslinking analysis followed by mass spectrometry of the ABCE1 43S pre‐initiation complex, the sample was prepared as described above (see section Preparation of crosslinked yeast 43S pre‐initiation complexes). After the sucrose density gradient, the buffer was exchanged to 50 mM HEPES/KOH pH 7.4, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, and crosslinking was performed using an equimolar mixture of isotopically labeled BS2G (H6/D6) (bis(sulfosuccinimidyl) 2,2,4,4‐glutarate, Creative molecules) for 30 min at 1,200 rpm and 10°C. The reaction was quenched with 100 mM ammonium bicarbonate for 10 min. Digestion and peptide clean‐up were performed using the EasyPep Mini MS Sample Prep Kit (Thermo Scientific), according to the manufacturer’s protocol. Crosslinks were further enriched on a Superdex Peptide PC 3.2/30 column (300 × 3.2 mm), and the fractions were analyzed by liquid chromatography coupled to tandem mass spectrometry with an LTQ‐Orbitrap Elite (Thermo Scientific) instrument (Herzog et al 2012). Identification of the crosslinked peptides was done using xQuest (Walzthoeni et al 2012). The results were filtered with an MS1 tolerance window of −4 to 4 ppm and score ≥ 22 followed by manual validation.
Sequence alignments
In order to quantify the conservation of protein sequences between human and yeast proteins of interest, pairwise alignments were conducted using the T‐Coffee implementation at https://toolkit.tuebingen.mpg.de (Notredame et al 2000; Zimmermann et al 2018) and visualized using JalView (Waterhouse et al 2009). Multiple sequence alignments of the conserved elements of the eIF3c N‐terminus were created using MAFFT (Katoh et al 2019).
Author contributions
HK, T M‐K, TB, and RB designed the study. MA prepared the sample for the human and TM‐K for the yeast native 40S complexes. TM‐K and HK prepared components for in vitro splitting assays and TM‐K performed splitting assays. MA, TM‐K, and OB collected and MA and TM‐K processed the cryo‐EM data. HK, JC, and TM‐K built and refined the model. HK, TM‐K, TB, and RB analyzed and interpreted the structures. ED, AN, and MF‐R performed ABCE1‐TAP purification and quantitative label‐free MS. MP and FH performed crosslinking mass spectrometry. TB, HK, TM‐K, and RB wrote the manuscript with contributions from all authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Figure 1
Acknowledgements
The authors thank H. Sieber, J. Musial, C. Ungewickell, and S. Rieder for technical assistance, L. Kater and K. Best for support with the pre‐processing pipeline of cryo‐EM data, R. Buschauer for assistance in model building, L. Valášek and A. Jacquier for critical reading of the manuscript, J. Wells for support during the setup of splitting assays and J. Zeman for support with the XL‐MS experiments. This work was supported by German Research Council (BE1814/15‐1 and TRR174), the Center for Integrated Protein Science Munich (CiPS‐M), the ANR‐17‐CE11‐0049‐01 and the ANR‐17‐CE12‐0024‐02 grants, the Pasteur Institute and the Centre National de la Recherche Scientifique. H.K., M.A., and M.P. are supported by a DFG fellowship through the Graduate School of Quantitative Bioscience Munich (QBM). Open Access funding enabled and organized by ProjektDEAL.
The EMBO Journal (2021) 40: e105179.
[Correction added on 5 February 2021 after first online publication: Publishing license has been changed]
Contributor Information
Thomas Becker, Email: becker@genzentrum.lmu.de.
Roland Beckmann, Email: beckmann@genzentrum.lmu.de.
Data availability
Cryo‐EM density map of the yeast 43S PIC: Electron Microscopy Data Bank EMD‐11160 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐11160).
Atomic model of the yeast 43S PIC: Protein Data Bank 6ZCE (http://www.rcsb.org/structure/6ZCE).
Cryo‐EM density map of the yeast 48S IC: Electron Microscopy Data Bank EMD#x02010;11439 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐11439).
Atomic model of the yeast 48S IC: Protein Data Bank 6ZU9 (http://www.rcsb.org/structure/6ZU9).
Cryo‐EM density map of the yeast 43S PIC‐XL: Electron Microscopy Data Bank EMD‐11608 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐11608).
Atomic model of the yeast 43S PIC: Protein Data Bank 7A1G (http://www.rcsb.org/structure/7A1G).
Cryo‐EM density map of the human 43S PIC—state II: Electron Microscopy Data Bank EMD‐11458 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐11458).
Atomic model of the human 43S PIC—state II: Protein Data Bank 6ZVJ (http://www.rcsb.org/structure/6ZVJ).
Cryo‐EM density map of the human 43S PIC—state III: Electron Microscopy Data Bank EMD‐11602 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐11602).
Atomic model of the human 43S PIC—state III: Protein Data Bank 7A09 (http://www.rcsb.org/structure/7A09).
Mass spectrometry proteomics data of the yeast 43S PIC: ProteomeXchange Consortium PXD020849 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD020849).
The cryo‐EM density maps of the yeast 43S PIC, the yeast 48S IC, the yeast 43S PIC‐XL, the human 43S‐PIC state II, and the human 43S‐PIC state III have been deposited in the Electron Microscopy Data Bank under accession numbers EMD‐11160, EMD‐11429, EMD‐11608, EMD‐11458, and EMD‐11602, respectively (https://www.emdataresource.org/). Atomic coordinates for the atomic models have been deposited in the Protein Data Bank under accession number PDB ID 6ZCE, 6ZU9, 7A1G, 6ZVJ, and 7A09, respectively. (https://www.wwpdb.org/) (see Appendix Table S1 and S2). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez‐Riverol et al, 2019) partner repository with the dataset identifier PXD020849. Correspondence and requests for materials should be addressed to T.B. (becker@genzentrum.lmu.de) or R.B. (beckmann@genzentrum.lmu.de).
References
- Algire MA, Maag D, Lorsch JR (2005) Pi release from eIF2, not GTP hydrolysis, is the step controlled by start‐site selection during eukaryotic translation initiation. Mol Cell 20: 251–262 [DOI] [PubMed] [Google Scholar]
- Acker MG, Shin BS, Dever TE, Lorsch JR (2006) Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J Biol Chem 281: 8469–8475 [DOI] [PubMed] [Google Scholar]
- Acker MG, Shin BS, Nanda JS, Saini AK, Dever TE, Lorsch JR (2009) Kinetic analysis of late steps of eukaryotic translation initiation. J Mol Biol 385: 491–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, Adams PD (2018) Real‐space refinement in PHENIX for cryo‐EM and crystallography. Acta Crystallogr D Struct Biol 74: 531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameismeier M, Cheng J, Berninghausen O, Beckmann R (2018) Visualizing late states of human 40S ribosomal subunit maturation. Nature 558: 249–253 [DOI] [PubMed] [Google Scholar]
- Andersen DS, Leevers SJ (2007) The essential Drosophila ATP‐binding cassette domain protein, pixie, binds the 40 S ribosome in an ATP‐dependent manner and is required for translation initiation. J Biol Chem 282: 14752–14760 [DOI] [PubMed] [Google Scholar]
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R (2013) Structures of the human and Drosophila 80S ribosome. Nature 497: 80–85 [DOI] [PubMed] [Google Scholar]
- Asano K, Clayton J, Shalev A, Hinnebusch AG (2000) A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev 14: 2534–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett CH, Boehringer D, Erzberger JP, Schaefer T, Ban N (2015) Structure of a yeast 40S‐eIF1‐eIF1A‐eIF3‐eIF3j initiation complex. Nat Struct Mol Biol 22: 269–271 [DOI] [PubMed] [Google Scholar]
- Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, Tampe R (2011) Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin‐ATPase ABCE1. Proc Natl Acad Sci USA 108: 3228–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D, Scheele U, Dinkelaker S, Janoschka A, Macmillan F, Albers SV, Driessen AJ, Stagni MS, Bill E, Meyer‐Klaucke W, et al (2007) Structural organization of essential iron‐sulfur clusters in the evolutionarily highly conserved ATP‐binding cassette protein ABCE1. J Biol Chem 282: 14598–14607 [DOI] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache J‐P, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al (2012) Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482: 501–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M (2011) The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334: 1524–1529 [DOI] [PubMed] [Google Scholar]
- Beznoskova P, Cuchalova L, Wagner S, Shoemaker CJ, Gunisova S, von der Haar T, Valasek LS (2013) Translation initiation factors eIF3 and HCR1 control translation termination and stop codon read‐through in yeast cells. PLoS Genet 9: e1003962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert S, Waterhouse A, de Beer TA, Tauriello G, Studer G, Bordoli L, Schwede T (2017) The SWISS‐MODEL Repository‐new features and functionality. Nucleic Acids Res 45: D313–D319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block KL, Vornlocher HP, Hershey JW (1998) Characterization of cDNAs encoding the p44 and p35 subunits of human translation initiation factor eIF3. J Biol Chem 273: 31901–31908 [DOI] [PubMed] [Google Scholar]
- Brown A, Baird MR, Yip MC, Murray J, Shao S (2018) Structures of translationally inactive mammalian ribosomes. Elife 7: e40486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V (2015) Structural basis for stop codon recognition in eukaryotes. Nature 524: 493–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate JH (2017) Human eIF3: from 'blobology' to biological insight. Philos Trans R Soc Lond B Biol Sci 372: 20160176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschauer R, Matsuo Y, Sugiyama T, Chen YH, Alhusaini N, Sweet T, Ikeuchi K, Cheng J, Matsuki Y, Nobuta R, et al (2020) The Ccr4‐Not complex monitors the translating ribosome for codon optimality. Science 368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZQ, Dong J, Ishimura A, Daar I, Hinnebusch AG, Dean M (2006) The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. J Biol Chem 281: 7452–7457 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10: 1794–1805 [DOI] [PubMed] [Google Scholar]
- Cuchalova L, Kouba T, Herrmannova A, Danyi I, Chiu WL, Valasek L (2010) The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol Cell Biol 30: 4671–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Ghosh R, Maitra U (2001) Eukaryotic translation initiation factor 5 functions as a GTPase‐activating protein. J Biol Chem 276: 6720–6726 [DOI] [PubMed] [Google Scholar]
- des Georges A, Dhote V, Kuhn L, Hellen CUT, Pestova TV, Frank J, Hashem Y(2015) Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 525: 491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG (2004) The essential ATP‐binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem 279: 42157–42168 [DOI] [PubMed] [Google Scholar]
- Dong Z, Qi J, Peng H, Liu J, Zhang JT (2013) Spectrin domain of eukaryotic initiation factor 3a is the docking site for formation of the a:b:i:g subcomplex. J Biol Chem 288: 27951–27959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElAntak L, Tzakos AG, Locker N, Lukavsky PJ (2007) Structure of eIF3b RNA recognition motif and its interaction with eIF3j: structural insights into the recruitment of eIF3b to the 40 S ribosomal subunit. J Biol Chem 282: 8165–8174 [DOI] [PubMed] [Google Scholar]
- Elantak L, Wagner S, Herrmannova A, Karaskova M, Rutkai E, Lukavsky PJ, Valasek L (2010) The indispensable N‐terminal half of eIF3j/HCR1 cooperates with its structurally conserved binding partner eIF3b/PRT1‐RRM and with eIF1A in stringent AUG selection. J Mol Biol 396: 1097–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliseev B, Yeramala L, Leitner A, Karuppasamy M, Raimondeau E, Huard K, Alkalaeva E, Aebersold R, Schaffitzel C (2018) Structure of a human cap‐dependent 48S translation pre‐initiation complex. Nucleic Acids Res 46: 2678–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Stengel F, Pellarin R, Zhang S, Schaefer T, Aylett CHS, Cimermancic P, Boehringer D, Sali A, Aebersold R, et al (2014) Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex. Cell 159: 1227–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, Berry KE, Hershey JW, Doudna JA (2007) eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell 26: 811–819 [DOI] [PubMed] [Google Scholar]
- Fraser CS, Lee JY, Mayeur GL, Bushell M, Doudna JA, Hershey JW (2004) The j‐subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40 S ribosomal subunits in vitro. J Biol Chem 279: 8946–8956 [DOI] [PubMed] [Google Scholar]
- Gerovac M, Tampe R (2019) Control of mRNA Translation by Versatile ATP‐Driven Machines. Trends Biochem Sci 44: 167–180 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE (2018) UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27: 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouridis G, Hetzert B, Kiosze‐Becker K, de Boer M, Heinemann H, Nurenberg‐Goloub E, Cordes T, Tampe R (2019) ABCE1 Controls Ribosome Recycling by an Asymmetric Dynamic Conformational Equilibrium. Cell Rep 28: 723–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Hellen CUT, Pestova TV, Frank J(2013) Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell 153: 1108–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmannova A, Daujotyte D, Yang JC, Cuchalova L, Gorrec F, Wagner S, Danyi I, Lukavsky PJ, Valasek LS (2012) Structural analysis of an eIF3 subcomplex reveals conserved interactions required for a stable and proper translation pre‐initiation complex assembly. Nucleic Acids Res 40: 2294–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, Walzthoeni T, Leitner A, Beck M, Hartl FU, Ban N, et al (2012) Structural probing of a protein phosphatase 2A network by chemical cross‐linking and mass spectrometry. Science 337: 1348–1352 [DOI] [PubMed] [Google Scholar]
- Heuer A, Gerovac M, Schmidt C, Trowitzsch S, Preis A, Kötter P, Berninghausen O, Becker T, Beckmann R, Tampé R (2017) Structure of the 40S–ABCE1 post‐splitting complex in ribosome recycling and translation initiation. Nature Structural & Molecular Biology 24: 453–460 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG (2006) eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci 31: 553–562 [DOI] [PubMed] [Google Scholar]
- Hopfner KP (2016) Invited review: Architectures and mechanisms of ATP binding cassette proteins. Biopolymers 105: 492–504 [DOI] [PubMed] [Google Scholar]
- Huang HK, Yoon H, Hannig EM, Donahue TF (1997) GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev 11: 2396–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner NC, Mann M (2011) Extracting gene function from protein‐protein interactions using Quantitative BAC InteraCtomics (QUBIC). Methods 53: 453–459 [DOI] [PubMed] [Google Scholar]
- Hussain T, Llacer JL, Fernandez IS, Munoz A, Martin‐Marcos P, Savva CG, Lorsch JR, Hinnebusch AG, Ramakrishnan V (2014) Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell 159: 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H, Abe T, Miyoshi T, Nishikawa SI, Ito K, Uchiumi T (2018) The ribosomal stalk protein is crucial for the action of the conserved ATPase ABCE1. Nucleic Acids Res 46: 7820–7830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher A, Schele A, Hopfner KP (2008) X‐ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi. J Biol Chem 283: 7962–7971 [DOI] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20: 1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenner LR, Anand AA, Nguyen HC, Myasnikov AG, Klose CJ, McGeever LA, Tsai JC, Miller‐Vedam LE, Walter P, Frost A (2019) eIF2B‐catalyzed nucleotide exchange and phosphoregulation by the integrated stress response. Science 364: 491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnevis S, Gunisova S, Vlckova V, Kouba T, Neumann P, Beznoskova P, Ficner R, Valasek LS (2014) Structural integrity of the PCI domain of eIF3a/TIF32 is required for mRNA recruitment to the 43S pre‐initiation complexes. Nucleic Acids Res 42: 4123–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiosze‐Becker K, Ori A, Gerovac M, Heuer A, Nürenberg‐Goloub E, Rashid UJ, Becker T, Beckmann R, Beck M, Tampé R (2016) Structure of the ribosome post‐recycling complex probed by chemical cross‐linking and mass spectrometry. Nature Communications 7: 13248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Kranzusch PJ, Doudna JA, Cate JH (2016) eIF3d is an mRNA cap‐binding protein that is required for specialized translation initiation. Nature 536: 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Kim YS, Kim KH, Heo I, Kim SK, Kim O, Kim HK, Yoon JY, Kim HS, Kim DJ, et al (2007) Structural and functional insights into Dom34, a key component of no‐go mRNA decay. Mol Cell 27: 938–950 [DOI] [PubMed] [Google Scholar]
- Lee JH, Pestova TV, Shin BS, Cao C, Choi SK, Dever TE (2002) Initiation factor eIF5B catalyzes second GTP‐dependent step in eukaryotic translation initiation. Proc Natl Acad Sci USA 99: 16689–16694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al (2019) Macromolecular structure determination using X‐rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75: 861–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llacer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, Hinnebusch AG, Ramakrishnan V (2015) Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell 59: 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llacer JL, Hussain T, Saini AK, Nanda JS, Kaur S, Gordiyenko Y, Kumar R, Hinnebusch AG, Lorsch JR, Ramakrishnan V (2018) Translational initiation factor eIF5 replaces eIF1 on the 40S ribosomal subunit to promote start‐codon recognition. Elife 7: e39273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera‐Martinez E, Brito Querido J, Valasek LS, Simonetti A, Hashem Y (2017) ABCE1: A special factor that orchestrates translation at the crossroad between recycling and initiation. RNA Biol 14: 1279–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Marcos P, Nanda JS, Luna RE, Zhang F, Saini AK, Cherkasova VA, Wagner G, Lorsch JR, Hinnebusch AG (2014) Enhanced eIF1 binding to the 40S ribosome impedes conformational rearrangements of the preinitiation complex and elevates initiation accuracy. RNA 20: 150–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad MP, Munzarova Pondelickova V, Zeman J, Gunisova S, Valasek LS (2017) In vivo evidence that eIF3 stays bound to ribosomes elongating and terminating on short upstream ORFs to promote reinitiation. Nucleic Acids Res 45: 2658–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I, Klaholz BP (2017) Visualization of chemical modifications in the human 80S ribosome structure. Nature 551: 472–477 [DOI] [PubMed] [Google Scholar]
- Nielsen KH, Valasek L, Sykes C, Jivotovskaya A, Hinnebusch AG (2006) Interaction of the RNP1 motif in PRT1 with HCR1 promotes 40S binding of eukaryotic initiation factor 3 in yeast. Mol Cell Biol 26: 2984–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J (2000) T‐Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217 [DOI] [PubMed] [Google Scholar]
- Nürenberg‐Goloub E, Heinemann H, Gerovac M, Tampé R (2018) Ribosome recycling is coordinated by processive events in two asymmetric ATP sites of ABCE1. Life Science Alliance 1: e201800095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürenberg‐Goloub E, Kratzat H, Heinemann H, Heuer A, Kötter P, Berninghausen O, Becker T, Tampé R, Beckmann R (2020) Molecular analysis of the ribosome recycling factor ABCE1 bound to the 30S post‐splitting complex. The EMBO Journal 39: e103788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurenberg‐Goloub E, Tampe R (2019) Ribosome recycling in mRNA translation, quality control, and homeostasis. Biol Chem 401: 47–61 [DOI] [PubMed] [Google Scholar]
- Obayashi E, Luna R, Nagata T, Martin‐Marcos P, Hiraishi H, Singh C, Erzberger J, Zhang F, Arthanari H, Morris J, et al (2017) Molecular Landscape of the Ribosome Pre‐initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5. Cell Reports 18: 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V (2007) The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50 [DOI] [PubMed] [Google Scholar]
- Paulin FE, Campbell LE, O'Brien K, Loughlin J, Proud CG (2001) Eukaryotic translation initiation factor 5 (eIF5) acts as a classical GTPase‐activator protein. Curr Biol 11: 55–59 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Perez‐Riverol Y, Csordas A, Bai J, Bernal‐Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47: D442–D450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335 [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV (2007) Recycling of eukaryotic posttermination ribosomal complexes. Cell 131: 286–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV (2010) The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell 37: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV (2011) Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J 30: 1804–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preis A, Heuer A, Barrio‐Garcia C, Hauser A, Eyler DE, Berninghausen O, Green R, Becker T, Beckmann R (2014) Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1‐eRF3 or eRF1‐ABCE1. Cell Rep 8: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhorova I, Altman RB, Djumagulov M, Shrestha JP, Urzhumtsev A, Ferguson A, Chang CT, Yusupov M, Blanchard SC, Yusupova G (2017) Aminoglycoside interactions and impacts on the eukaryotic ribosome. Proc Natl Acad Sci USA 114: E10899–E10908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N (2015) CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192: 216–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012) RELION: implementation of a Bayesian approach to cryo‐EM structure determination. J Struct Biol 180: 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al (2000) Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 102: 615–623 [DOI] [PubMed] [Google Scholar]
- Schmitt E, Blanquet S, Mechulam Y (2002) The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors. EMBO J 21: 1821–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E, Panvert M, Lazennec‐Schurdevin C, Coureux PD, Perez J, Thompson A, Mechulam Y (2012) Structure of the ternary initiation complex aIF2‐GDPNP‐methionylated initiator tRNA. Nat Struct Mol Biol 19: 450–454 [DOI] [PubMed] [Google Scholar]
- Sha Z, Brill LM, Cabrera R, Kleifeld O, Scheliga JS, Glickman MH, Chang EC, Wolf DA (2009) The eIF3 interactome reveals the translasome, a supercomplex linking protein synthesis and degradation machineries. Mol Cell 36: 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Brown A, Santhanam B, Hegde RS (2015) Structure and assembly pathway of the ribosome quality control complex. Mol Cell 57: 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R (2011) Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci USA 108: E1392–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti A, Brito Querido J, Myasnikov AG, Mancera‐Martinez E, Renaud A, Kuhn L, Hashem Y (2016) eIF3 Peripheral Subunits Rearrangement after mRNA Binding and Start‐Codon Recognition. Mol Cell 63: 206–217 [DOI] [PubMed] [Google Scholar]
- Sokabe M, Fraser CS, Hershey JW (2012) The human translation initiation multi‐factor complex promotes methionyl‐tRNAi binding to the 40S ribosomal subunit. Nucleic Acids Res 40: 905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Verschoor A, Frank J (1992) Eukaryotic initiation factor 3 does not prevent association through physical blockage of the ribosomal subunit‐subunit interface. J Mol Biol 226: 301–304 [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13: 731–740 [DOI] [PubMed] [Google Scholar]
- Valasek L, Hasek J, Trachsel H, Imre EM, Ruis H (1999) The Saccharomyces cerevisiae HCR1 gene encoding a homologue of the p35 subunit of human translation initiation factor 3 (eIF3) is a high copy suppressor of a temperature‐sensitive mutation in the Rpg1p subunit of yeast eIF3. J Biol Chem 274: 27567–27572 [DOI] [PubMed] [Google Scholar]
- Valasek L, Mathew AA, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG (2003) The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev 17: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Nielsen KH, Hinnebusch AG (2002) Direct eIF2‐eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J 21: 5886–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG (2004) Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol Cell Biol 24: 9437–9455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Phan L, Schoenfeld LW, Valaskova V, Hinnebusch AG (2001) Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J 20: 891–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek LS, Zeman J, Wagner S, Beznoskova P, Pavlikova Z, Mohammad MP, Hronova V, Herrmannova A, Hashem Y, Gunisova S (2017) Embraced by eIF3: structural and functional insights into the roles of eIF3 across the translation cycle. Nucleic Acids Res 45: 10948–10968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SE, Zhou F, Mitchell SF, Larson VS, Valasek L, Hinnebusch AG, Lorsch JR (2013) Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N‐terminal and internal repeat domains. RNA 19: 191–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzthoeni T, Claassen M, Leitner A, Herzog F, Bohn S, Forster F, Beck M, Aebersold R (2012) False discovery rate estimation for cross‐linked peptides identified by mass spectrometry. Nat Methods 9: 901–903 [DOI] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al (2018) SWISS‐MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46: W296–W303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, Flugge UI (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Singh CR, Marintchev A, Hall NS, Hannig EM, Wagner G, Asano K (2005) The eukaryotic initiation factor (eIF) 5 HEAT domain mediates multifactor assembly and scanning with distinct interfaces to eIF1, eIF2, eIF3, and eIF4G. Proc Natl Acad Sci USA 102: 16164–16169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Guydosh NR (2019) Hcr1/eIF3j Is a 60S Ribosomal Subunit Recycling Accessory Factor In Vivo. Cell Rep 28: 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Marintchev A, Kolupaeva VG, Unbehaun A, Veryasova T, Lai SC, Hong P, Wagner G, Hellen CU, Pestova TV (2009) Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res 37: 5167–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman J, Itoh Y, Kukacka Z, Rosulek M, Kavan D, Kouba T, Jansen ME, Mohammad MP, Novak P, Valasek LS (2019) Binding of eIF3 in complex with eIF5 and eIF1 to the 40S ribosomal subunit is accompanied by dramatic structural changes. Nucleic Acids Res 47: 8282–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang E, Khanna V, Dacheux E, Namane A, Doyen A, Gomard M, Turcotte B, Jacquier A, Fromont‐Racine M (2019) A specialised SKI complex assists the cytoplasmic RNA exosome in the absence of direct association with ribosomes. EMBO J 38: e100640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016) Gctf: Real‐time CTF determination and correction. J Struct Biol 193: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache J‐P, Verba KA, Cheng Y, Agard DA (2017) MotionCor2: anisotropic correction of beam‐induced motion for improved cryo‐electron microscopy. Nature methods 14: 331–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V (2018) A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol 430: 2237–2243 [DOI] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH (2018) New tools for automated high‐resolution cryo‐EM structure determination in RELION‐3. Elife 7: e42166 [DOI] [PMC free article] [PubMed] [Google Scholar]


