Abstract
Background: SARS-CoV-2 is the causal agent of the current coronavirus disease 2019 (COVID-19) pandemic. They are enveloped, positive-sense, single-stranded RNA viruses of the Coronaviridae family. Proteases of SARS-CoV-2 are necessary for viral replication, structural assembly, and pathogenicity. The approximately 33.8 kDa M pro protease of SARS-CoV-2 is a non-human homologue and is highly conserved among several coronaviruses, indicating that M pro could be a potential drug target for Coronaviruses.
Methods: Herein, we performed computational ligand screening of four pharmacophores (OEW, remdesivir, hydroxychloroquine and N3) that are presumed to have positive effects against SARS-CoV-2 M pro protease (6LU7), and also screened 50,000 natural compounds from the ZINC Database dataset against this protease target.
Results: We found 40 pharmacophore-like structures of natural compounds from diverse chemical classes that exhibited better affinity of docking as compared to the known ligands. The 11 best selected ligands, namely ZINC1845382, ZINC1875405, ZINC2092396, ZINC2104424, ZINC44018332, ZINC2101723, ZINC2094526, ZINC2094304, ZINC2104482, ZINC3984030, and ZINC1531664, are mainly classified as beta-carboline, alkaloids, and polyflavonoids, and all displayed interactions with dyad CYS145 and HIS41 from the protease pocket in a similar way as other known ligands.
Conclusions: Our results suggest that these 11 molecules could be effective against SARS-CoV-2 protease and may be subsequently tested in vitro and in vivo to develop novel drugs against this virus.
Keywords: SARS-CoV-2, protease, virtual screening, pharmacophore, inhibitors, natural compounds
Introduction
Coronaviruses (CoVs) are enveloped, positive-sense, single-stranded RNA viruses of the Coronaviridae family 1. Based on their antigenic properties, they were classified into three main groups 2: i) alpha-CoVs, responsible for gastrointestinal disorders; ii) beta-CoVs, which include: (a) Bat coronavirus (BCoV), (b) human severe acute respiratory syndrome (SARS) virus, (c) Middle Eastern respiratory syndrome (MERS) virus; and iii) gamma-CoVs, which mainly infect avian species. The most well-known of these coronaviruses is the SARS-CoV, responsible for causing an outbreak in 2002–2003 3 and MERS-CoV, causing severe respiratory symptoms, which was identified in 2012 4.
In December 2019, a series of unusual pneumonia cases caused by a novel coronavirus, recently renamed as SARS-CoV-2, was identified in Wuhan, China 5, 6. The disease caused by SARS-CoV-2 is now called COVID-19, and displays vast pathophysiological aspects, which include symptoms, such as fever and coughing, and severe acute respiratory failure 7. Since the infection crossed geographical barriers, the World Health Organization (WHO) declared a pandemic situation in March 2020, reaching a worldwide mortality rate of approximately 3% 6.
The SARS-CoV-2 ORF1ab code for polyprotein 1ab (pp1ab), where the main protease M pro is found, which is similar to a key enzyme in the processing of the picornavirus family polyprotein. The protease M pro, digests more than 11 conserved sites starting from its autolytic cleavage in pp1ab, and is a protein with extreme functional importance in the viral cycle 8. Due to its great importance in the coronavirus cycle, the M pro sequence of SARS-CoV-2 shows more than 90% similarity with the enzymes of other coronaviruses 9 and shares 96% identity with SARS-CoV. Although M pro is conserved among SARS-CoVs, it has a loop that can make it difficult for an inhibitor to access the catalytic pocket, and mutations in this loop can generate drug resistance 10. Thus, even though M pro is one of the most conserved SARS-CoV group proteins, point mutational aspects can lead to a possible drug resistance, so that a wide range of inhibitor options is necessary for the treatment of COVID-19.
ORF1ab is characteristic of members of the Coronaviridae family 11 and is equivalent to two-thirds of the SARS-CoV-2 virus genome 12. Each of these ORFs encodes a polyprotein (pp), which, when cleaved by proteases contained in the sequence, will generate 11 proteins (pp1a) and 5 proteins (pp1ab), respectively. The functions associated with these proteins are related to the virus replication processes and the modulation of the immune response in the host, among other essential functions for the development of the pathogen within the host cell 6.
Virus resistance to drugs can lead to the emergence of new epidemics, such as influenza A virus (IAV). In this case, two drug classes have been related: M2 channel inhibitors (amantadine and rimantadine) and neuraminidase inhibitors (NAIs; oseltamivir, zanamivir, peramivir, and Laninamivir). Both drug classes act by inhibiting proteins that are located in the viral envelope, and this region is in greater contact with the external environment and is prone to suffer from greater evolutionary pressure and, consequently, mutations 13. Drug resistance can occur when rapid viral replication is not repressed completely 14. In contrast, virus proteases play a crucial role during virus replication and, therefore, they are a good target for drug discovery 15.
During viral replication, proteases are necessary for the assembly of the viral structure, and there have already been suggested to have relationships with the mechanism of infection and pathogenicity of SARS-CoV-2 5, 16. Proteases are enzymes found in all cellular organisms and viruses and are classified according to their catalytic nature. Proteases are divided into four groups: serine, cysteine, aspartyl and metalloproteases. Different types of proteases can perform the same activity through different catalytic mechanisms 15, and a protease commonly has a binding site and a catalytic site that are very close in the protein structure 15. Furthermore, proteases are present in several types of viruses and are widely found in human viruses 17.
In coronaviruses, pp1 is essential for the replication of the virus, as it encodes the protease M pro, which is also called the "main protease" 18, 19. M pro is classified as a chymotrypsin-like cysteine protease (3CLpro), EC: 3.4.19.12, 9, 18, and the M pro protease of SARS-CoV-2, which has a mass of approximately 33.8 kDa 19, is characterized by a self-cleavage protein 20, 21. It consists of a homodimer subdivided into two protomers (A and B) that have three distinct domains 22. The first and second domains have antiparallel β-sheets while the third domain contains five α-helices forming a globular group, which is connected in parallel with the domain-II through a loop region 19. The M pro of SARS-CoV-2 has a catalytic cleft, consisting of a Cys-His dyad in the place of the protease substrate interaction, which is situated between domains -I and -II 19. It also has non-canonic specificity to the substrate in the C-terminal portion. Furthermore, there is no homologue of M pro in the human genome 19, 23, and it is highly conserved amongst coronaviruses 24. Therefore, M pro is a potential target for studying inhibitors.
Antiviral therapy considers three main approaches for the control and avoidance of viral infections: (a) vaccination, (b) stimulation of host resistance mechanisms, and (c) antiviral chemotherapy. Antivirals are drugs that inhibit certain virus-specific events, such as binding to host cells, which is how SARS-Cov-2 binds to ACE2 and TMPRSS2 25, and MERS binds to the DPP415 receptor 26. Antiviral chemotherapy can involve interfering with any or all of these viral replication steps. Most antiviral drugs are primarily targeted to the synthesis of nucleic acids in viruses. As viral replication and host cell processes are closely linked, one of the main problems of viral therapy would be to find a drug capable of being selectively toxic only for the virus. Antivirals are frequently more effective in prevention than in the treatment itself, and are ineffective in eliminating latent or non-replicating viruses 27. In addition, when selecting an antiviral drug, viral resistance must also be considered since it is one of the main causes of therapeutic failure.
The main classes of antiviral drugs used in clinical therapy to treat systemic viral infections include: a) synthetic nucleosides (e.g. acyclovir, famciclovir, ganciclovir, valacyclovir, and valganciclovir; b) pyrophosphate analogs (e.g. foscarnet); c) drugs for syncytial virus and influenza A (e.g. amantadine and rimantadine hydrochloride and ribavirin); d) nucleoside reverse transcriptase inhibitors (NRTI; e.g. abacavir, didanosine, emtricitabine, stavudine, lamivudine, zidovudine, tenofovir in combination with NRTI); e) non-nucleoside reverse transcriptase inhibitors (NNRTI; e.g. delavirdine, efavirenz, nevirapine); and f) protease inhibitors (e.g. amprenavir, atazanavir, darunavir, fosamprenavir, lopinavir and ritonavir, nelfinavir mesylate, saquinavir mesylate, ritonavir, indinavir sulfate and tipranavir) 28, 29
Computational studies of inhibitors that may reduce viral replication is a fast way for proposing drug candidates that can contribute to a reduction in severity and spread of the disease. Moreover, the use of antiviral compounds can assist in the prophylaxis of SARS-CoV-2 and reduce its spread 30. Therefore, screening for potential viral protease inhibitors may assist in the selection of new drugs with antiviral potential for SARS-CoV-2.
Methods
Ligand screening
For this study, we employed both ligand-based virtual screening (LBVS) and receptor-based virtual screening (RBVS) approaches, considering 50,000 structures of natural compounds from the ZINC Database, which has more than 900 million structures deposited, and includes millions of drug-like compounds that can be obtained for in vitro and in vivo tests 31. The ZINC molecules that were downloaded were those restricted to absorption, distribution, metabolism, excretion and toxicity characteristics (ADMET) for drug likeness: no more than 5 hydrogen bond donors, no more than 10 hydrogen bond acceptors, molecular weight between 160 and 500 Daltons and logP between -0.4 and 5.6 32, 33. For LBVS, we defined four known drugs divided in the following groups: 1) peptide-like crystallographic ligands (N3 and OEW); and 2) repurposed drugs (remdesivir (nucleoside) and hydroxychloroquine) for chemical comparison with our database.
Crystallographic ligand structures were obtained from their corresponding PDB files 6LU7 (N3) and 6Y7M (OEW). Additionally, these structures were used for re-docking validations. In the LBVS process, we used a simple run with vROCS (OpenEye) 34 for generating queries with the pharmacophoric map with the stereochemical characteristics for each known ligand. Another option for pharmacophore generation and searching is the free software PharmaGist 35. Afterwards, we submitted each ligand query for searching similar pharmacophore-like molecules using the Tanimoto Combo algorithm 36, 37 with a cutoff of 1.0, which returned the best 1,000 hits for each round. This procedure was repeated three times for each query, and, subsequently, redundant structures were discarded, generating, in the end, a total of 4,000 similar molecules for the docking experiment.
Docking studies
Considering PDB validation indices as crystallographic resolution, Ramachandran outliers, clash score, and release date, we selected the structure 6LU7 for RBVS, which is complexed with the peptide-like inhibitor N3 19. Furthermore, 6LU7 and 6Y7M 38 were used for re-docking validations with its corresponding crystallographic inhibitors.
The best LBVS hits were submitted to molecular docking calculations with 6LU7 structure using Autodock 4.2 virtual screening protocol 39. Ligand structures were prepared for virtual screening using Raccoon plugin 40 for Autodock Tools 40 according to the standard protocol 37, as well as the 6LU7 structure. The gridbox was defined on the active site region, considering the amino acids THR 190, GLU 166, GLN 189, GLY 143, HIS 163, HIS 164, CYS 145, PHE 140, and with accordance with previous studies with the crystallographic structure of the SARS-CoV-2 main protease 19, 38, 41. Each docking run was performed three times using the following specifications: flexible docking and Lamarckian Genetic Algorithm with 2,500,000 generations. Afterwards, the 10 best docking hits were selected using the Autodock Tools script summarize_results4.py, which can classify the best hits according to their lowest energy clustering conformations and root mean square deviation (RMSD) values. The results were organized according to the ligand pharmacophore relationship with the known structures in Table 2. Docking and re-docking results were evaluated at each docking position inside the 6LU7 active site using Pymol 2.1 42 and UCSF Chimera 1.14 43 in order to confirm molecule interactions with the amino acids within the protease active site. Furthermore, 2D interaction maps were generated by Discovery Studio 2019 44. Another option for 2D map generation is the LigPlot+ software 45.
Results
LBVS
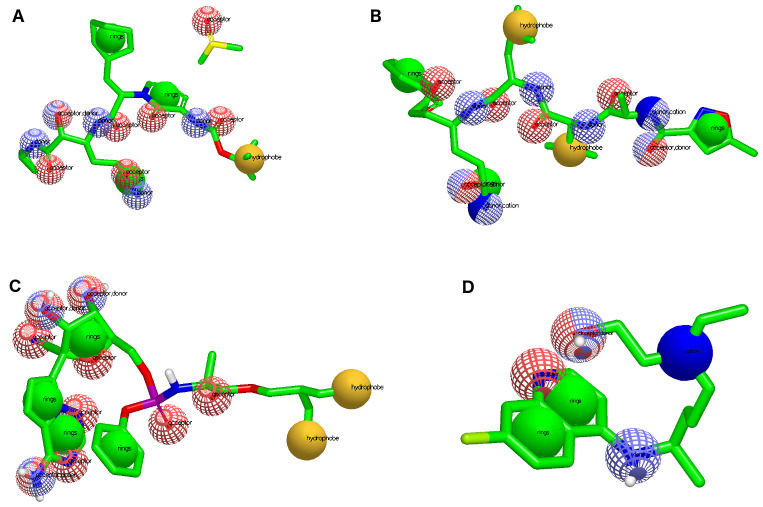
Different pharmacophoric characteristics were generated for each known ligand ( Figure 1), which allowed us to find molecules included in different chemical classifications and amplify the number of possible drug candidates. Table 1 shows the pharmacophoric characteristics for each known 6LU7 inhibitor, which permitted us to find natural ligands with pharmacophore-like regions. Additionally, we used the ADMET characteristics for molecular weight and LogP that are important for molecular druggability.
Figure 1. Pharmacophore representation for each known drug used for virtual screening.
( A) OEW, ( B) N3, ( C) remdesivir and ( D) hydroxychloroquine. In red spheres: hydrogen acceptors; blue spheres: hydrogen donors; yellow spheres: hydrophobic; and green spheres: aromatic.
Table 1. Pharmacophoric characteristics for each known inhibitor used for screening natural ligands from the ZINC Database.
| Inhibitor | Hb.A. | Hb.D. | Aromatic | Hydrophobic | M.W. | LogP |
|---|---|---|---|---|---|---|
| OEW | 7 | 5 | 3 | 1 | 663.8 | -0.71 |
| N3 | 6 | 4 | 3 | 2 | 680.8 | 2.32 |
| Remdesivir | 9 | 1 | 4 | 2 | 602.6 | 1.9 |
| Hydroxychloroquine | 2 | 2 | 2 | 0 | 335.9 | 3.6 |
Hb.A. = hydrogen acceptor; Hb.D. = hydrogen donor; M.W. = molecular weight.
Ligand based virtual screening and docking calculations of ZINC database compounds revealed the 40 best pharmacophore-like ligands that belong to different chemical classes, namely beta-carboline alkaloids, indole alkaloids, lupin alkaloids, harmala alkaloids, polyflavonoids, anthracenes, angular pyranocoumarins, and flavonoid-3-O-glycosides. Table 2 shows the detailed results on the average affinity energies, ZINC identification, and chemical classification of each selected ligand.
Table 2. The 40 best molecule hits of COVID-19 main protease inhibitor candidates from a dataset of 50,000 natural compounds from the ZINC Database.
| Known Drug | Ligand | Energ.
Binding (Kcal/Mol) |
Classification | RMSD
Å |
Pred.
IC50 (uM) |
Exp. IC50
(uM) |
|---|---|---|---|---|---|---|
| OEW | OEW * | -8.86 | Peptide-like | 1.97 | 0.320 | 0.670 |
| ZINC1845382 | -10.2 | β-carboline Alkaloid | ||||
| ZINC1875405 ** | -10.1 | β-carboline Alkaloid | ||||
| ZINC2092396 | -9.8 | β-carboline Alkaloid | ||||
| ZINC1900463 | -9.8 | β-carboline Alkaloid | ||||
| ZINC2149492 | -9.8 | β-carboline Alkaloid | ||||
| ZINC2112405 | -9.7 | β-carboline Alkaloid | ||||
| ZINC2095426 | -9.7 | β-carboline Alkaloid | ||||
| ZINC2094306 | -9.6 | β-carboline Alkaloid | ||||
| ZINC2144677 | -9.6 | Anthracene | ||||
| ZINC1095868 | -9.5 | Harmala Alkaloids | ||||
| N3 * | -9,77 | Peptide-like | 1.94 | 0.07 | 4.67 | |
| N3 | ZINC2104482 | -10.1 | β-carboline Alkaloid | |||
| ZINC3984030 | -9.9 | Polyflavonoid | ||||
| ZINC1531664 | -9.8 | Polyflavonoid | ||||
| ZINC2152199 | -9.8 | β-carboline Alkaloid | ||||
| ZINC4096847 | -9.6 | Flavonoid-3-O-glycoside | ||||
| ZINC3947428 | -9.6 | Flavonoid-3-O-glycoside | ||||
| ZINC2092587 | -9.6 | β-carboline Alkaloid | ||||
| ZINC2115924 | -9.5 | β-carboline Alkaloid | ||||
| ZINC2110081 | -9.5 | Lupin Alkaloid | ||||
| ZINC1898165 | -9.5 | Benzofuran | ||||
| HCQ | -7.90 | 4-aminoquinoline | ||||
| Hydroxychloroquine (HCQ) | ZINC2101723 | -10.2 | β-carboline Alkaloid | |||
| ZINC2094526 | -9.8 | β-carboline Alkaloid | ||||
| ZINC2094304 | -9.6 | β-carboline Alkaloid | ||||
| ZINC2091604 | -9.4 | β-carboline Alkaloid | ||||
| ZINC2113496 | -9.4 | β-carboline Alkaloid | ||||
| ZINC1460216 | -9.3 | Angular Pyranocoumarin | ||||
| ZINC2123008 | -9.2 | β-carboline Alkaloid | ||||
| ZINC682759 | -9.2 | Harmala Alkaloids | ||||
| ZINC2105243 | -9.2 | β-carboline Alkaloid | ||||
| ZINC2111696 | -9.1 | β-carboline Alkaloid | ||||
| REMD | -8.28 | Nucleoside | ||||
| Remdesivir (REMD) | ZINC2104424 | -10.6 | β-carboline Alkaloid | |||
| ZINC1875405 ** | -10.1 | β-carboline Alkaloid | ||||
| ZINC44018332 | -10.0 | Polyflavonoid | ||||
| ZINC2148932 | -9.9 | β-carboline Alkaloid | ||||
| ZINC2156531 | -9.9 | Indoles Alkaloid | ||||
| ZINC3197535 | -9.9 | Polyflavonoid | ||||
| ZINC2102620 | -9.9 | Indoles Alkaloid | ||||
| ZINC2123402 | -9.9 | β-carboline Alkaloid | ||||
| ZINC2149488 | -9.9 | β-carboline Alkaloid | ||||
| ZINC1531664 | -9.9 | Polyflavonoid |
*Re-docked crystallographic structures.
**Repeated ligand between OEW and Remdesivir pharmacophores.
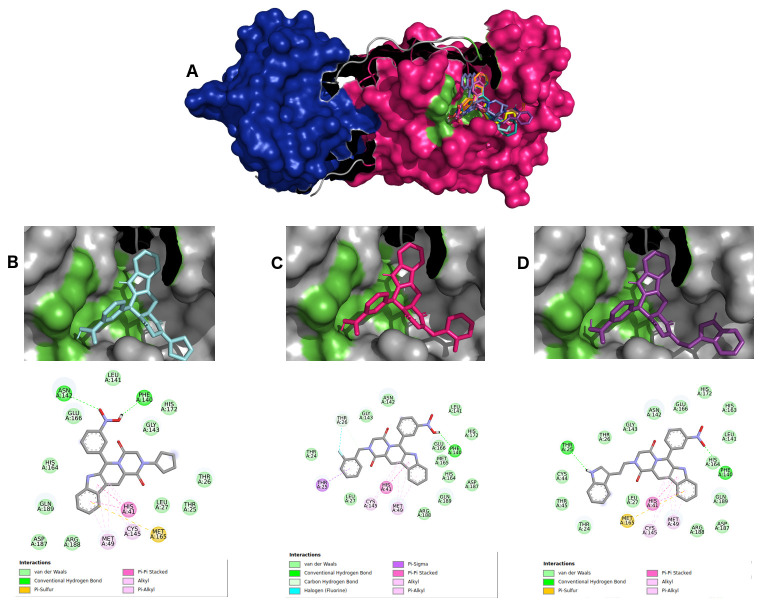
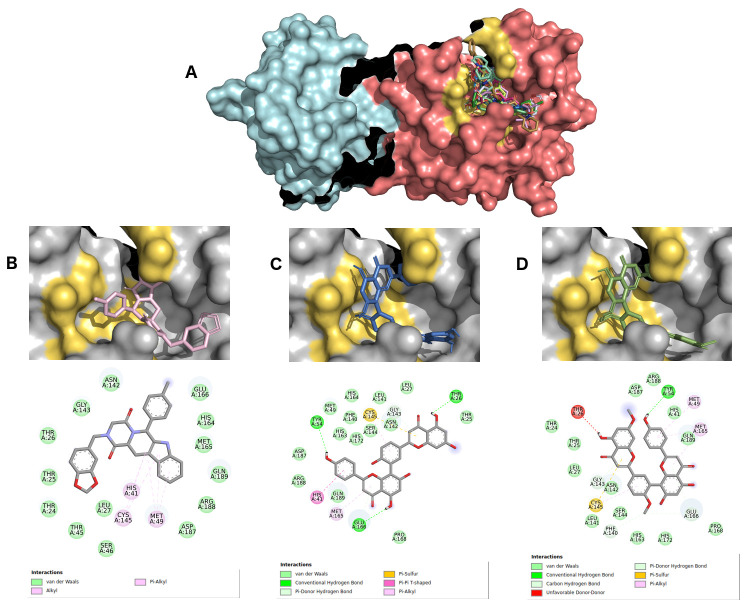
For selecting the best pharmacophore-like drug-candidates, we considered evaluating lower affinity energy values, as well as interactions with residues of the active site within the target. As can be seen in Figure 2A, all pharmacophore-like OEW ligand molecules formed a complex with the active pocket of 6LU7. The three best OEW ligands (ZINC1845382, ZINC1875405, ZINC2092396) are shown in complex with COVID-19 protease in Figure 2B–D with the detailed 2D interaction map. In this case, these top three hits are included in the beta-carboline alkaloid class.
Figure 2.
( A) SARS-CoV-2 main protease complexed with the 10 best hits for OEW pharmacophore molecules. Protomer A is represented in marine blue surface and protomer B in dark pink surface. ZINC1845382 in cyan ( B), ZINC1875405 in dark pink ( C) and ZINC2092396 in purple ( D) inside 6LU7 binding site and their 2D interaction maps with pocket amino acids.
RBVS
The intermolecular interactions carried out by ligand ZINC1845382 exhibited a hydrogen bond with the residue of the active PHE140 protease site. The catalytic residues CYS145 and HIS41 represented interactions of the type π, π-π stacked and π-alkyl with the entire beta-carboline group, which was composed of three hydrophobic rings. The remaining residues were of the π-sigma type, hydrogen-carbon acceptors, and halogen acceptors from residues THR25, THR26, as well as other residues from the active site GLU166, GLN189, GLY143, HYS164, respectively.
Ligand ZINC1875405 represented two hydrogen bonding interactions with residues THR25 and PHE140. Additionally, four more polar interactions of the type π-π stacked, π-aquil, aquil and π-sulfur with residues HIS41, MET49, CYS145 and MET165, respectively, were formed. The other interactions were of hydrophobic van der Waals type.
Ligand ZINC2092396 interacted by hydrogen interaction with the residue PHE140, π and π-alkyl with CYS140, π-π stacked and π-alkyl HIS41, and van der Waals with GLN189, GLY143, HIS164, GLU166. Other interactions occurred with hydrogen bonds by the ligand nitrobenzene group with the ASN142 residue and a π-sulfur interaction of the beta-carboline group with MET165 residue.
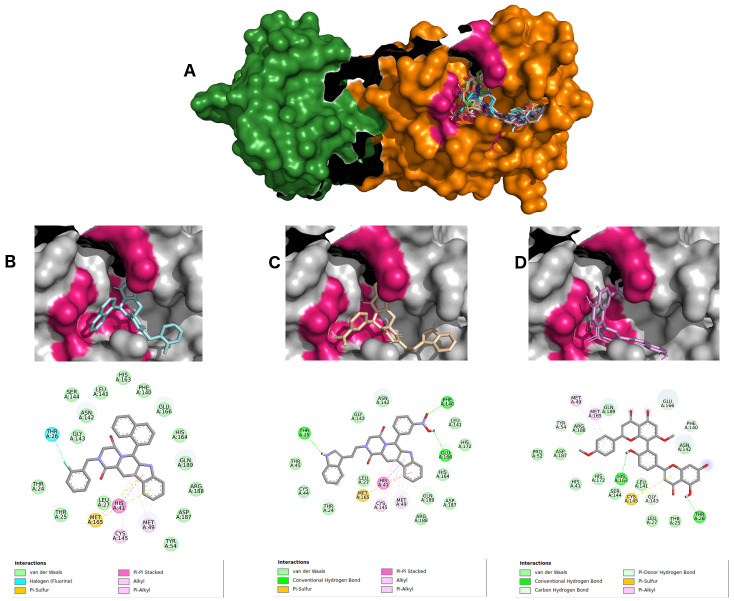
The remdesivir pharmacophore-like search returned two beta-carboline alkaloids (ZINC2104424 and ZINC1875405), as well as one polyflavonoid (ZINC44018332), which interacted with the COVID-19 main protease active pocket showing affinity energies below -10.0 kcal/Mol. Figure 3A–D shows the details of all ligand interactions, as well as the top three molecules interaction maps.
Figure 3.
( A) SARS-CoV-2 main protease complexed with 10 best hits for remdesivir pharmacophore molecules. Protomer A is represented in green surface, and protomer B in orange surface. ZINC2104424 in cyan ( B), ZINC1875405 in wheat ( C) and ZINC44018332 in violet ( D) inside 6LU7 binding site and their 2D interaction maps with pocket amino acids.
Ligand ZINC2104424 also occupied the region of the active site ( Figure 3B), showing polar interactions π, π-alkyl, π-π stacked and π-sulfur types from beta-carboline with HIS41, MET49, CYS145, and MET165 amino acids. Moreover, an interaction of THR26 halogen with the ligand fluorobenzene group also occurred. Other hydrophobic interactions were van der Waals, mostly with residues of the active site: PHE140, GLY143, HIS163, HIS164, GLU166 and GLN189.
Ligand ZINC1875405 ( Figure 3C) displayed three hydrogen interactions with the indole group, and two oxygen interactions from a nitrobenzene of THR25, PHE140 AND GLN166, respectively. Several van der Waals-type hydrophobic interactions were found with GLY143, HIS164 and GLN189 amino acids. Furthermore, four polar interactions (π, π-alkyl, π-π stacked and π-sulfur) with residues HIS41, MET49, CYS 145 and MET165, respectively, were also retrieved.
Ligand ZINC2092396 ( Figure 3D) exhibited two hydrogen interactions with HIS163 and THR26 by its hydroxyl from the flavonoid nucleus, as well as four more π-donor hydrogen bonding interactions with residues TYR54, PHE140, GLY143 and GLU166. Besides, three π-alkyl and π-sulfur interactions made with MET49, CYS145 and MET165 were also retrieved. Other hydrophobic interactions were of van der Waals type.
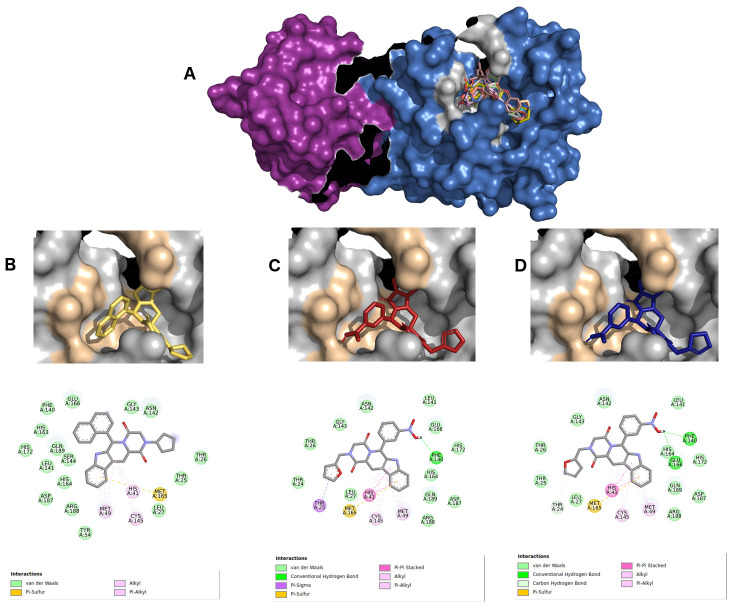
Figure 4 shows the interactions between 6LU7 active sites and the three best hits from derived molecules of hydroxychloroquine pharmacophore (ZINC2101723, ZINC2094526, ZINC2094304). These complexes displayed affinity energies varying from -10.2 kcal/Mol to -9.6 kcal/Mol, and all the ligands were classified as beta-carboline alkaloid derivatives.
Figure 4.
( A) Best hits for hydroxychloroquine pharmacophore. Protomer A is represented using a violet surface, and protomer B in marine blue surface. ZINC2101723 in yellow ( B), ZINC2094526 in red ( C), and ZINC2094304 in dark blue ( D) inside 6LU7 binding site and their 2D interaction maps with pocket amino acids.
The beta-carboline group of the ligand ZINC2101723 ( Figure 4B) formed four π-alkyl, alkyl and π-sulfur type interactions with HIS41, MET49, CYS145 and MET165 residues, as well as other hydrophobic interactions from its naphthalene and beta-carboline groups with the active site amino acids PHE140, GLY143, HIS163 E164, GLU166 and GLN189. Ligand ZINC2094526 ( Figure 4C) displayed a hydrogen bond interaction with PHE140 by its nitrobenzene group. Five polar interactions (π-sigma, π-aquil, π-π stacked and π-sulfur) were observed with residues THR25, HIS41, MET49, CYS145 and MET165. For ligand ZINC2094304 ( Figure 4D ), two hydrogen bonds with residues PHE140 and GLU166 by its nitrobenzene group were formed. In addition, this ligand formed four polar interactions (π-π stacked, π-alkyl, alkyl and π-sulfur) with residues HIS41, MET49, CYS145 and MET165, respectively. Other van der Waals type interactions could also be identified.
The N3 pharmacophore revealed one beta-carboline alkaloid (ZINC2101723) and two polyflavonoids (ZINC2094526 and ZINC2094304). This group displayed affinity energies ranging from -9.8 kcal/Mol to -10.1 kcal/Mol. In Figure 5, the best complex interactions with the protease, as well as their positions inside the binding pocket are depicted.
Figure 5. Virtual screening results for the N3 pharmacophore.
( A) COVID-19 main protease is represented in cyan (protomer A) and dark salmon (protomer B). The best complexes are formed by the alkaloid ZINC2101723 in pink ( B) and two polyflavonoids ZINC2094526 in marine blue ( C) and ZINC2094304 in lemon green ( D), and their 2D interaction maps with pocket amino acids are shown below each complex.
Ligand ZINC2104482 ( Figure 5B) formed a large number of hydrophobic interactions (14 van der Waals interactions), surrounding the active site amino acid, such as GLY143, HIS164, GLU166 and GLN189. Furthermore, this ligand formed three π-alkyl and alkyl bonds with HIS41, MET49, CYS145 residues. Ligand ZINC3984030 ( Figure 5C) exhibited three hydrogen bonds with THR26, TYR54 and GLU166 residues by OH groups of flavonoid nuclei. A π-donor hydrogen bond interaction of the GLY143 residue was also observed. Moreover, three polar interactions (π-π stacked, π-alkyl and π-sulfur) were identified with HIS41, CYS145 and MET165. The rest of the interactions were van der Waals type. Ligand ZINC1531664 ( Figure 5D) showed a hydrogen bond by its OH group TYR54. Additionally, four π-donor hydrogen bond and hydrogen carbon bond interactions with residues PHE140, GLY143, GLU166 also occurred. Two polar interactions of the type π-alkyl and π-sulfur were observed with MET49, CY145, and MET165, and the other hydrophobic interactions were of van der Waals type.
Re-docking validation experiments
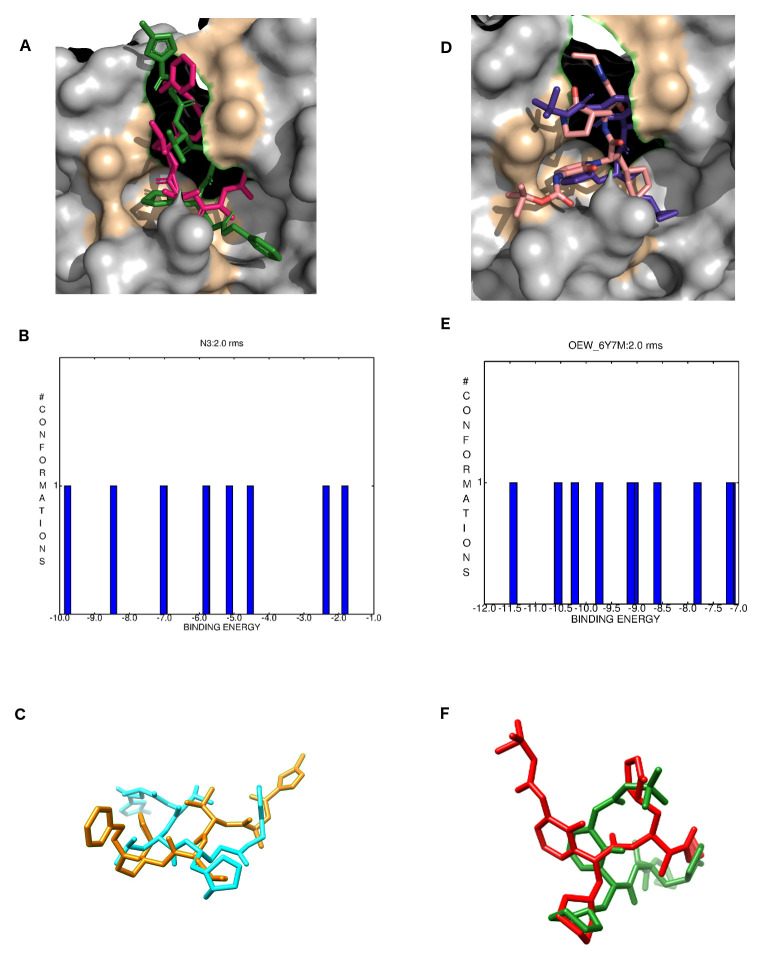
Crystallographic ligands N3 and OWE were re-docked with their respective M pro structures 6LU7 and 6Y7M. As can be seen in Figure 6A, both N3 and OEW molecules bound into similar positions in comparison to their original crystallographic forms. The Figure 6B depicts the best clustering conformations graph for N3 with a free energy of binding ranging from -1.83 kcal/Mol to -9.7 kcal/Mol. Figure 6C shows the superposition between the crystalized and re-docked N3 structure. Even though N3 is peptide-like with 13 routable bonds, it presented an RMSD of 1.94 Angstroms for its best conformation ( Table 2). OEW re-docking is shown in Figure 6D in the same way as for N3 where both the crystallized and docked structures bound into the same pocket. Conformational population of OEW clustering results returned a free energy of binding ranging from -7.0 kcal/Mol to -11.5 kcal/Mol but, on the other hand, the structure with binding energy of -8.86 kcal/Mol exhibited the smallest RMSD value ( Figure 6E). Additionally, OEW presented an RMSD of 1.97 Å in comparison to its crystallized form ( Figure 6F).
Figure 6. Re-docking validations for N3 and OEW.
( A) crystallographic (hot pink) and re-docked (green) N3 inhibitor of the 6LU7 SARS-CoV-2 M pro inside its binding pocket; ( B) N3 docking best clustering conformations; ( C) aligned N3 crystallized (yellow) and re-docked (cyan). ( D) crystallographic (pink) and re-docked (purple) OEW inhibitor of the 6LU7 SARS-CoV-2 M pro inside its binding pocket; ( E) OEW docking best clustering conformations; ( F) aligned OEW crystallized (green) and re-docked (red).
Discussion
Docking results revealed 39 pharmacophore-like natural ligands, which can be used as drug candidates for inhibiting SARS-CoV-2 main protease activity. Furthermore, we ranked the three best candidates for each known ligand pharmacophore as the best potential drug molecules (and totaling 12 molecules) for in vitro and in vivo assays purposes, but not excluding the other 28 molecules. For these cases, ligands are included in two most expressive chemical classes: β-carboline alkaloids and polyflavonoids. Additionally, all ligands exhibiting better affinity energies than the known drugs was used as references for construction of pharmacophoric characteristics: OEW 6, remdesivir 46, hydroxychloroquine 47, and N3 19.
The groups of OEW and hydroxychloroquine pharmacophores presented their three most promising ligands classified as β-carboline alkaloids. This class of molecules is reported by different authors with antiviral activities. According to Gonzalez et al. 43, β-carboline Alkaloids are widely distributed in nature, and its derivatives exhibited activity against Herpes Simplex Viruses by blocking virus replication. Additionally, Gonzalez et al. 43 demonstrated the action of these alkaloids in dengue virus RNA replication. Furthermore, several other studies suggest alkaloid activity against viral proteases 48– 50. Similarly, remdesivir pharmacophore revealed two β-carboline alkaloids (ZINC2104424 and ZINC1875405). In addition, we detected a polyflavonoid (ZINC44018332) as one probable active molecule from a different class against SARS-CoV-2 main protease, and several authors have already described flavonoid activity as viral protease inhibitors 51– 53, as well as antiviral molecules acting in different target classes 52, 54– 56. N3 pharmacophore displayed two flavonoids as the best molecules and just one β-carboline alkaloid. These results indicate that both classes of molecules could be explored for in vitro and in vivo tests to evaluate their potential antiviral activities for not only SARS-CoV-2 but also for other viruses of medical interest.
Other classes of molecules were found in our screening for protease activity that were previously described in antiviral studies: anthracenes 57, angular pyranocoumarin 58, 59, and flavonoid-3-O-glycoside 60. Interaction maps of these complexes are available as Extended data 61.
All the known ligands (OEW, remdesivir, hydroxychloroquine and N3), which were used for validating our computational screening, exhibited worse affinity energies in docking calculations (ranging from -7.8 kcal/Mol to -5.2 kcal/Mol) than the screened natural compounds (ranging from -10.6 kcal/Mol to -9.1 kcal/Mol). Moreover, all the 40 selected ligands docked inside M pro active site, as previously described in several antiviral studies, and interacted in the region of connection between domains I and II with amino acids HIS41 and CYS145 18, 19, 22, 38, 41, 62.
Novel M pro ketoamide inhibitors were recently proposed, including the OEW ligand (ligand 13b) that was used in our study, and the authors detected a reduction in RNA replication in human cells infected with SARS-CoV-2, and also described binding interactions with its main protease. Besides, the same study indicated a ketoamide as a probable drug candidate against this virus 38.
In a recent study, authors have proposed the peptidomimetic molecule N3 as a drug candidate against COVID-19, and described its binding interactions with the crystallographic structures of SARS-CoV-2 and other viral proteases. Their study reported that N3 can bind in all the active pockets from the main proteases of HCoV-NL63, SARS-CoV, and MERS-CoV 63.
Other molecules have also been tested as antivirals for effectiveness in inhibiting SARS-CoV-2 replication in cell culture. Two drugs exhibited a promising inhibitory effect: remdesivir, an experimental drug developed for the treatment of Ebola virus infection 41, 64, and hydroxychloroquine, a drug known for its effectiveness in the treatment of malaria and autoimmune diseases 41. Remdesivir is an adenosine triphosphate analogue initially described in the literature in 2016 as a potential treatment for Ebola 65, and this drug has been indeed considered as a potential treatment for SARS-CoV2, 64. Notably, remdesivir has demonstrated antiviral activity in the treatment of MERS and SARS 66 in animal models, both of which are caused by coronaviruses 67. Pharmacophore models are widely used in medicinal chemistry with the aim of amplifying the number of drug candidates, and according to this definition, they are represented by a 3D arrangement of abstract features instead of chemical groups 68. Remdesivir is a nucleotide analogue with capacity to inhibit RNA polymerase ( Table 1): this molecule displayed almost the same pharmacophoric features ( Figure 1) as for N3 and OEW, and, besides, both of them have already tested experimentally. Additionally, as can be seen in Table 2, the best hit is ZINC1875405, which was found in both OEW and remdesivir pharmacophore searching, and this could be explained by their similar characteristics. Hydroxychloroquine is an aminoquinoline-like chloroquine 69. It is a drug commonly prescribed for the treatment of uncomplicated malaria, rheumatoid arthritis, chronic discoid lupus erythematosus, and systemic lupus erythematosus 70. Chloroquine and hydroxychloroquine have been investigated for the treatment of SARS-CoV-2 71, and they have been reported to have direct antiviral effects, such as inhibition of flaviviruses, retroviruses (like HIV), and many coronaviruses. Additionally, hydroxychloroquine is capable of inhibiting the zika virus NS2B-NS3 protease, and exhibited good viral replication blocking in infected JEG3 cells in concentration of 80 µM of hydroxychloroquine 72. Furthermore, the use of chloroquine and its analogues can be corroborated by a recent study showing that, with EC 50 of 1.13 µmol/L and selectivity index (SI) greater than 88, chloroquine can effectively inhibit SARS-CoV-2 at the cellular level 73. Its effectiveness in the human body for SARS-CoV-2 infection; however, has not yet been clinically proven. Another in silico study with chloroquine detected its interactions with viral NSP-3B type protease 74.
The co-crystallized molecules N3 and OEW are both peptide analogs, which presented RMSD values of 1.94 Å and 1.97 Å in re-docking experiments, respectively. Generally, docking validations protocols use co-crystallized ligands, to test the accuracy of the program to predict the correct ligand docking poses in comparison to known conformations, and its RMSD varies 1.5 or 2 Å depending on ligand size for being considering acceptable 75. The number of studies using protein-peptide docking has been increasing rapidly, followed by the number of applied drug design programs and models. On the other hand, the use of RMSD validations with experimental structures is not always the best criterion of docking success, once it can be influenced by resolution quality, as well as the number of peptide residues 76. Thus, we can consider that N3 and OEW docking validations are in acceptable RMSD ranges.
Conclusions
In our study, we compared the pharmacophores of four well-tested human coronavirus (including SARS-Cov-2) main protease drug candidates to 50,000 structures of natural compounds from the ZINC Database. The three best molecules selected for each pharmacophore class are mainly classified as β-carboline alkaloids, and polyflavonoids. The best ligand-SARS-CoV-2 complexes exhibited better affinity energies in comparison to drug molecules used in this study. Furthermore, all the screened molecules bonded between domains -I and -II and formed interactions with the catalytic residues HIS41 and CYS145 in similar positions as previously described from other authors in viral protease inhibitor studies. Altogether, we propose these compounds as possible SARS-CoV-2 protease inhibitors, which can be used for subsequent in vitro and in vivo tests for finding novel drug candidates.
Data availability
Source data
Structures of natural compounds were downloaded from the ZINC Database.
Crystal structures of COVID-19 main protease were downloaded from the Protein Data Bank, accession numbers 6LU7 (in complex with N3) and 6Y7M (with OEW).
Extended data
Harvard Dataverse: Replication Data for: Computational screening for potential drug candidates against SARS-CoV-2 main protease. https://doi.org/10.7910/DVN/GYFXA0 61.
This project contains the following extended data:
2D interaction maps of all OEW pharmacophore-like ligands (PNG).
2D interaction maps of all Remdesivir pharmacophore-like ligands (PNG).
2D interaction maps of all Hydroxychloroquine pharmacophore-like ligands (PNG).
2D interaction maps of all N3 pharmacophore-like ligands (PNG).
Extended data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
Acknowledgements
We would like to thank The OpenEye Science for the OpenEye Software license, which made possible the ligand based virtual screening experiments.
This research was developed with the help of the Núcleo de Biologia Computacional e Gestão de Informações Biotecnológicas- NBCGIB ", with resources from FINEP / MCT, CNPQ and FAPESB and from Universdade Estadual de Santa Cruz – UESC, represented by Dr. Eduardo Almeida Costa. Additionally, the authors would like to acknowledge the SENAI CIMATEC Center for Industrial Innovation, with support from BG Brazil and the Brazilian Authority for Oil, Gas and Biofuels (ANP), for the provision and operation of computational facilities and the commitment to invest in Research & Development.
Funding Statement
This work was supported by the National Council for Scientific and Technological Development (CNPq) of Brazil: Luiz Alcantara receives research grant: 304387/2017-7; Aristóteles Góes-Neto receives research: grant 310764/2016-5; Vasco Azevedo receives research grant: 305093/2015-0.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 1 approved
References
- 1. Cui J, Li F, Shi ZL: Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schoeman D, Fielding BC: Coronavirus envelope protein: current knowledge. Virol J. 2019;16(1):69. 10.1186/s12985-019-1182-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peiris JS, Guan Y, Yuen KY: Severe acute respiratory syndrome. Nat Med. 2004;10(12 Suppl):S88–97. 10.1038/nm1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zaki AM, Van Boheemen S, Bestebroer TM, et al. : Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–20. 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- 5. Benvenuto D, Giovanetti M, Ciccozzi A, et al. : The 2019-new coronavirus epidemic: Evidence for virus evolution. J Med Virol. 2020;92(4):455–459. 10.1002/jmv.25688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu F, Zhao S, Yu B, et al. : A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng M, Gao Y, Wang G, et al. : Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17(5):533–535. 10.1038/s41423-020-0402-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jin Z, Du X, Xu Y, et al. : Structure of M pro from COVID-19 virus and discovery of its inhibitors. Nature. 2020. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- 9. Bzowka M, Mitusinska K, Raczynska A, et al. : Molecular Dynamics Simulations Indicate the COVID-19 Mpro Is Not a Viable Target for Small-Molecule Inhibitors Design. bioRxiv. 2020. 10.1101/2020.02.27.968008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bzówka M, Mitusińska K, Raczyńska A, et al. : Structural and Evolutionary Analysis Indicate That the SARS-CoV-2 Mpro Is a Challenging Target for Small-Molecule Inhibitor Design. Int J Mol Sci. 2020;21(9): pii: E3099. 10.3390/ijms21093099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng VC, Lau SK, Woo PC, et al. : Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20(4):660–94. 10.1128/CMR.00023-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo YR, Cao QD, Hong ZS, et al. : The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7(1):11. 10.1186/s40779-020-00240-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hussain M, Galvin HD, Haw TY, et al. : Drug resistance in influenza A virus: the epidemiology and management. Infect Drug Resist. 2017;10:121–134. 10.2147/IDR.S105473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McKeegan KS, Borges-Walmsley MI, Walmsley AR: Microbial and viral drug resistance mechanisms. Trends Microbiol. 2002;10(10 Suppl):S8–14. 10.1016/s0966-842x(02)02429-0 [DOI] [PubMed] [Google Scholar]
- 15. Sharma A, Gupta SP: Fundamentals of viruses and their proteases.In: Viral Proteases and Their Inhibitors. 2017;1–24. 10.1016/B978-0-12-809712-0.00001-0 [DOI] [Google Scholar]
- 16. Zhang T, Wu Q, Zhang Z: Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr Biol. 2020;30(7):1346–1351.e2. 10.1016/j.cub.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patel S: A critical review on serine protease: Key immune manipulator and pathology mediator. Allergol Immunopathol (Madr). 2017;45(6):579–591. 10.1016/j.aller.2016.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anand K, Palm GJ, Mesters JR, et al. : Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21(13):3213–24. 10.1093/emboj/cdf327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jin Z, Du X, Xu Y, et al. : Structure of M pro from COVID-19 virus and discovery of its inhibitors. bioRxiv. 2020. 10.1101/2020.02.26.964882 [DOI] [Google Scholar]
- 20. Meng T, Cao H, Zhang H, et al. : The insert sequence in SARS-CoV-2 enhances spike protein cleavage by TMPRSS. bioRxiv. 2020. 10.1101/2020.02.08.926006 [DOI] [Google Scholar]
- 21. Kang S, Yang M, Hong Z, et al. : Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. bioRxiv. 2020. 10.1101/2020.03.06.977876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang H, Yang M, Ding Y, et al. : The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci U S A. 2003;100(23):13190–5. 10.1073/pnas.1835675100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang P, Gao Q, Wang T, et al. : Evaluation of recombinant nucleocapsid and spike proteins for serological diagnosis of novel coronavirus disease 2019 (COVID-19). medRxiv. 2020. 10.1101/2020.03.17.20036954 [DOI] [Google Scholar]
- 24. Xu X, Chen P, Wang J, et al. : Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457–60. 10.1007/s11427-020-1637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoffmann M, Kleine-Weber H, Schroeder S, et al. : SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280.e8. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zumla A, Chan JF, Azhar EI, et al. : Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15(5):327–47. 10.1038/nrd.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crumpacker CS: Use of Antiviral Drugs to Prevent Herpesvirus Transmission. N Engl J Med. 2004;350(1):67–8. 10.1056/NEJMe038189 [DOI] [PubMed] [Google Scholar]
- 28. Ter Heine R, Mulder JW, Van Gorp EC, et al. : Intracellular and plasma steady-state pharmacokinetics of raltegravir, darunavir, etravirine and ritonavir in heavily pre-treated HIV-infected patients. Br J Clin Pharmacol. 2010;69(5):475–83. 10.1111/j.1365-2125.2010.03634.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Balayan T, Horvath H, Rutherford GW: Ritonavir-Boosted Darunavir Plus Two Nucleoside Reverse Transcriptase Inhibitors versus Other Regimens for Initial Antiretroviral Therapy for People with HIV Infection: A Systematic Review. AIDS Res Treat. 2017;2017: 2345617. 10.1155/2017/2345617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mitjà O, Clotet B: Use of antiviral drugs to reduce COVID-19 transmission. Lancet Glob Health. 2020;8(5):e639–e640. 10.1016/S2214-109X(20)30114-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Irwin JJ, Sterling T, Mysinger MM, et al. : ZINC: A free tool to discover chemistry for biology. J Chem Inf Model. 2012;52(7):1757–68. 10.1021/ci3001277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ghose AK, Viswanadhan VN, Wendoloski JJ: A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J Comb Chem. 1999;1(1):55–68. 10.1021/cc9800071 [DOI] [PubMed] [Google Scholar]
- 33. Lipinski CA, Lombardo F, Dominy BW, et al. : Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1–3):3–26. 10.1016/s0169-409x(00)00129-0 [DOI] [PubMed] [Google Scholar]
- 34. Hawkins PCD, Skillman AG, Nicholls A: Comparison of shape-matching and docking as virtual screening tools. J Med Chem. 2007;50(1):74–82. 10.1021/jm0603365 [DOI] [PubMed] [Google Scholar]
- 35. Schneidman-Duhovny D, Dror O, Inbar Y, et al. : PharmaGist: a webserver for ligand-based pharmacophore detection. Nucleic Acids Res. 2008;36(Web Server issue):W223–8. 10.1093/nar/gkn187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rácz A, Bajusz D, Héberger K: Life beyond the Tanimoto coefficient: Similarity measures for interaction fingerprints. J Cheminform. 2018;10(1):48. 10.1186/s13321-018-0302-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bajusz D, Rácz A, Héberger K: Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J Cheminform. 2015;7:20. 10.1186/s13321-015-0069-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang L, Lin D, Sun X, et al. : Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Morris GM, Dallakyan S: AutoDock — AutoDock. 2013. Reference Source [Google Scholar]
- 40. Forli S, Huey R, Pique ME, et al. : Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016;11(5):905–919. 10.1038/nprot.2016.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang M, Cao R, Zhang L, et al. : Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–71. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schrödinger L: The PyMol Molecular Graphics System, Versión 1.8.Thomas Holder.2015. Reference Source [Google Scholar]
- 43. Pettersen EF, Goddard TD, Huang CC, et al. : UCSF Chimera - A visualization system for exploratory research and analysis. J Compu Chem. 2004;25(13):1605–12. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 44. Discovery Studio: Dassault Systemes BIOVIA, Discovery Studio Modelling Environment, Release 4.5.Accelrys Softw Inc.2015. Reference Source [Google Scholar]
- 45. Laskowski RA, Swindells MB: LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778–2786. 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- 46. Martinez MA: Compounds with Therapeutic Potential against Novel Respiratory 2019 Coronavirus. Antimicrob Agents Chemother. 2020;64(5): pii: e00399–20. 10.1128/AAC.00399-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gonzalez MM, Cabrerizo FM, Baiker A, et al. : β-Carboline derivatives as novel antivirals for herpes simplex virus. Int J Antimicrob Agents. 2018;52(4):459–68. 10.1016/j.ijantimicag.2018.06.019 [DOI] [PubMed] [Google Scholar]
- 48. Ahmad I, Fatima I, Afza N, et al. : Urease and serine protease inhibitory alkaloids from Isatis tinctoria. J Enzyme Inhib Med Chem. 2008;23(6):918–21. 10.1080/14756360701743580 [DOI] [PubMed] [Google Scholar]
- 49. ul Qamar MT, Mumtaz A, Ashfaq UA, et al. : Potential of plant alkaloids as dengue ns3 protease inhibitors: Molecular docking and simulation approach. Bangladesh J Pharmacol. 2014;9(3):262–7. 10.3329/bjp.v9i3.18555 [DOI] [Google Scholar]
- 50. Powers CN, Setzer WN: An In-Silico Investigation of Phytochemicals as Antiviral Agents Against Dengue Fever. Comb Chem High Throughput Screen. 2016;19(7):516 536. 10.2174/1386207319666160506123715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qamar MT, Ashfaq UA, Tusleem K, et al. : In-silico identification and evaluation of plant flavonoids as dengue NS2B/NS3 protease inhibitors using molecular docking and simulation approach. Pak J Pharm Sci. 2017;30(6):2119–37. [PubMed] [Google Scholar]
- 52. Shimizu JF, Lima CS, Pereira CM, et al. : Flavonoids from Pterogyne nitens Inhibit Hepatitis C Virus Entry. Sci Rep. 2017;7(1):16127. 10.1038/s41598-017-16336-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hawas UW, Abou El-Kassem LT, Shaher F, et al. : In vitro inhibition of Hepatitis C virus protease and antioxidant by flavonoid glycosides from the Saudi costal plant Sarcocornia fruticosa. Nat Prod Res. 2019;33(23):3364–3371. 10.1080/14786419.2018.1477153 [DOI] [PubMed] [Google Scholar]
- 54. Kaul TN, Middleton E, Jr, Ogra PL: Antiviral effect of flavonoids on human viruses. J Med Virol. 1985;15(1):71–9. 10.1002/jmv.1890150110 [DOI] [PubMed] [Google Scholar]
- 55. González-Búrquez MD, González-Díaz FR, García-Tovar CG, et al. : Comparison between in Vitro Antiviral Effect of Mexican Propolis and Three Commercial Flavonoids against Canine Distemper Virus. Evidence-based Complement Altern Med. 2018;2018; 7092416. 10.1155/2018/7092416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dai W, Bi J, Li F, et al. : Antiviral Efficacy of Flavonoids against Enterovirus 71 Infection in vitro and in Newborn Mice. Viruses. 2019;11(7): pii: E625. 10.3390/v11070625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tomlinson SM, Watowich SJ: Anthracene-based inhibitors of dengue virus NS2B-NS3 protease. Antiviral Res. 2011;89(2):127–35. 10.1016/j.antiviral.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barnard DL, Xu Z, Stowell VD, et al. : Coumarins and pyranocoumarins, potential novel pharmacophores for inhibition of measles virus replication. Antivir Chem Chemother. 2002;13(1):39–59. 10.1177/095632020201300104 [DOI] [PubMed] [Google Scholar]
- 59. Hassan MZ, Osman H, Ali MA, et al. : Therapeutic potential of coumarins as antiviral agents. Eur J Med Chem. 2016;123:236–55. 10.1016/j.ejmech.2016.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Behbahani M, Sayedipour S, Pourazar A, et al. : In vitro anti-HIV-1 activities of kaempferol and kaempferol-7-O-glucoside isolated from Securigera securidaca. Res Pharm Sci. 2014;9(6):463–9. [PMC free article] [PubMed] [Google Scholar]
- 61. Andrade B: "Replication Data for: Computational screening for potential drug candidates against SARS-CoV-2 main protease". Harvard Dataverse, V1. 2020. 10.7910/DVN/GYFXA0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ren Z, Yan L, Zhang N, et al. : The newly emerged SARS-Like coronavirus HCoV-EMC also has an “Achilles’’ heel": Current effective inhibitor targeting a 3C-like protease”. Protein Cell. 2013;4(4):248–50. 10.1007/s13238-013-2841-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dai W, Zhang B, Su H, et al. : Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020; pii: eabb4489. 10.1126/science.abb4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. de Wit E, Feldmann F, Cronin J, et al. : Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc Natl Acad Sci U S A. 2020;117(12):6771–6776. 10.1073/pnas.1922083117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Warren TK, Jordan R, Lo MK, et al. : Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381–5. 10.1038/nature17180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gordon CJ, Tchesnokov EP, Feng JY, et al. : The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295(15):4773–4779. 10.1074/jbc.AC120.013056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sheahan TP, Sims AC, Graham RL, et al. : Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9(396): pii: eaal3653. 10.1126/scitranslmed.aal3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kaserer T, Beck KR, Akram M, et al. : Pharmacophore models and pharmacophore-based virtual screening: Concepts and applications exemplified on hydroxysteroid dehydrogenases. Molecules. 2015;20(12):22799–832. 10.3390/molecules201219880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Furst DE: Pharmacokinetics of hydroxychloroquine and chloroquine during treatment of rheumatic diseases. Lupus. 1996;5 Suppl 1:S11–5. 10.1177/0961203396005001041 [DOI] [PubMed] [Google Scholar]
- 70. Shippey EA, Wagler VD, Collamer AN: Hydroxychloroquine: An old drug with new relevance. Cleve Clin J Med. 2018;85(6):459–467. 10.3949/ccjm.85a.17034 [DOI] [PubMed] [Google Scholar]
- 71. Devaux CA, Rolain JM, Colson P, et al. : New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int J Antimicrob Agents. 2020;105938. 10.1016/j.ijantimicag.2020.105938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kumar A, Liang B, Aarthy M, et al. : Hydroxychloroquine Inhibits Zika Virus NS2B-NS3 Protease. ACS Omega. 2018;3(12):18132–41. 10.1021/acsomega.8b01002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang Q, Zhang Y, Wu L, et al. : Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020; pii: S0092-8674(20)30338-X. 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wu C, Liu Y, Yang Y, et al. : Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hevener KE, Zhao W, Ball DM, et al. : Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J Chem Inf Model. 2009;49(2):444–60. 10.1021/ci800293n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ciemny M, Kurcinski M, Kamel K, et al. : Protein-peptide docking: opportunities and challenges. Drug Discov Today. 2018;23(8):1530–7. 10.1016/j.drudis.2018.05.006 [DOI] [PubMed] [Google Scholar]