Abstract
Blood-derived microRNAs (miRNAs/miRs) are ideal clinical biomarkers, as they can be relatively non-invasively extracted and are stable across a range of storage conditions. However, the concentration and profile of miRNAs differ between specific patient groups and starting media, which must be a key consideration before embarking upon uses for clinical applications. The optimum blood-derived starting media for biomarker discovery involving pregnant women with an uncomplicated pregnancy has not been determined. Paired serum and plasma samples were collected from 10 pregnant women with uncomplicated low-risk pregnancies at three time points: i) During the second trimester of pregnancy; ii) during the third trimester; and iii) 6 weeks post-partum. Sample miRNA content was assessed using an Agilent Bioanalyzer Small RNA chip and reverse transcription-quantitative (RT-q)PCR using four constitutively expressed miRNAs: hsa-miR-222-3p, hsa-miR-23a, hsa-miR-30e-5p and hsa-miR-451a. Quality control spike-ins measured RNA extraction (UniSp2) and cDNA extraction (cel-miR-39-3p) efficiency. MiRNA concentration and percentage were significantly higher in the serum vs. plasma samples based on data obtained from the Bioanalyzer; however, RT-qPCR failed to replicate these differences in the majority of comparisons using the ΔCq values of the four constitutively expressed miRNAs. Using the standard deviations of the ΔCq values, the consistency of serum and plasma in terms of miRNA expression levels were equivalent. Thus, clinicians and researchers should take into consideration that different miRNA quantification methods can yield contrasting results with regards to the starting media utilized. Based on the equivalent performance of serum and plasma assessed using RT-qPCR, which is less likely to be influenced by the coagulation process or degraded long RNAs, both starting media assessed in the present study are equally suitable for ongoing biomarker discovery studies involving healthy pregnant women at any gestational time point or immediately postpartum.
Keywords: microRNA, blood serum, blood plasma, pregnancy, women, homo sapiens
Introduction
Over the past 10-15 years, scientific interest in microRNAs (miRNA/miRs) and their roles as modern, discriminatory biomarkers has grown exponentially (1-3). Now, this small (18-24 nucleotide) non-coding molecule encapsulates one of the most thriving, sought after areas of research. MiRNAs have been implicated in several disease processes, particularly in oncology, with the potential to facilitate diagnosis and predict disease development earlier than is currently possible (4-7). Furthermore, miRNAs have been suggested to confer overall prognosis, survival, drug sensitivity, treatment response and aid monitoring for disease progression and relapse (1,8-14). A growing body of literature is additionally emerging on the role of specific placental-derived miRNAs in pregnancy, particularly those complicated by processes such as gestational diabetes (15,16), gestational hypertension (17,18), pre-eclampsia (19-21) or congenital abnormalities (22,23).
miRNAs in biofluids, in particular serum and plasma derived from peripheral blood, are an attractive option for clinical biomarker development, as they are readily available, relatively non-invasive to obtain and widely processed in standard laboratory settings (24-29). Compared with other potential biomarkers, miRNAs are excellent candidates, due to their stable expression both in vivo across a range of tissues and biofluids (26,30-32), and ex vivo during differing storage conditions, being able to withstand ~48 h at room temperature or on ice (26,30), longer-term storage at -80˚C (33,34) and their ability to undergo multiple (between 4-8) freeze-thaw cycles (26,30,31). Furthermore, due to the wide range of downstream miRNA targets, with each mature miRNA being able to target ~200 mRNAs to exert their effects (9); their potential therapeutic capacity is simply vast (35).
In human pregnancy, there are three primary chromosomal miRNA clusters located on chromosome 19 (C19MC, miR-371-3) and chromosome 14 (C14MC) which are highly and widely expressed in placental tissue and as circulating markers, with the expression profile of these clusters varying during trimesters and gestational disease processes (21,36). MiRNAs are released from the trophoblastic layer into the maternal circulation in various forms, including microvesicle-enveloped, in apoptotic bodies, exosomally or as protein bound miRNAs (37-39), in order to avoid digestion by circulating RNAase enzymes (30,40-42). Several commercially available miRNA isolation kits using serum and plasma capture the whole biofluid, including the exosomal component; the latter only representing a subset of the miRNAs found in this media (43-45).
Although the discovery and validation of miRNA biomarkers remains at an experimental, pre-clinical stage, there is a growing body of literature concerning the role of miRNAs in specific pregnancy-associated conditions. However, there is little consensus regarding the optimal starting media or justification of the selection between serum or plasma (15,16,18,46). Most studies fail to acknowledge the potential differences in miRNA concentrations and profiles that may emerge when using serum vs. plasma as the starting biofluid (25-27,33,47-49), potentially inhibiting their successful applications in a clinical setting (26,27,47,48). Furthermore, several studies refer to serum or plasma interchangeably (25,41), whereas in fact they are quite different: Serum refers to the cell-free blood component obtained following the activation of platelets and factors within the coagulation cascade, whereas plasma refers to the cell-free blood component obtained prior to the coagulation process, hence collection tubes contain anticoagulants (EDTA or sodium citrate for downstream miRNA analysis) which inhibit the coagulation cascade (50). Heparinised tubes cannot be used for miRNA analysis as heparin inhibits downstream enzymatic reactions within the cDNA synthesis and reverse transcription (RT) steps of quantitative (q)PCR (33,45,49). The coagulation process itself can significantly alter a sample's miRNA profile and is a source of variability (for example, coagulation time and temperature) (27,33,47,49,50), which cannot be reliably controlled when using serum samples, leading several researchers to preferentially use plasma. Additionally, the coagulation process causes cell lysis and haemolysis, particularly from erythrocytes but also platelets, releasing RNA and miRNA into the serum, which affects the profile obtained (27,47,48). It is essential to acknowledge that haemolysis can also occur in both serum and plasma samples at the time of venepuncture, hence the need for standardised techniques to minimise this occurrence, including the use of larger gauge needles and well-trained phlebotomists (51). For these reasons, haemolysis monitoring is a part of the standard quality control steps of sample preparation, either involving spectrophotometric or specific miRNA analysis (33,52-54). Equally, the platelet content of plasma is an important consideration, given that their rich miRNA content may bias the determined outcome (24,27,47-49), highlighting the importance of two centrifugation steps during sample preparation to generate platelet-poor as opposed to platelet-rich plasma (7,49,55). One potential issue faced by miRNA researchers using pre-collected samples is that the majority of archived blood samples have been collected as serum.
The optimum starting media is also crucially dependent upon the patient population under study, for example whether the individuals are healthy or diseased; pregnant or non-pregnant. Different starting media may be more appropriate for specific patient types, which explains the contradiction between existing studies suggesting that either serum (studies involving healthy males, females and pregnant women) (25,27) or plasma (research involving patients with primary liver cancer or benign liver disease) (25,47) are superior.
To the best of our knowledge, there is only one comparable study examining the difference in miRNA concentrations and profiles between serum and plasma derived samples taken from pregnant women (25). However, this previous study analysed only three paired serum and plasma samples at one time point in pregnancy (second trimester) and generated conflicting outcomes dependent on whether the proportion or absolute number of detected miRNAs was measured. This is a key starting point which must be established before miRNA biomarkers can be reliably pinpointed as hallmarks of gestational disease states. Therefore, the aim of the present study was to determine the optimum starting media at three key time points both intra and post-partum, involving women with an uncomplicated, healthy pregnancy.
Patients and methods
Sample collection and RNA isolation
Blood samples were obtained from 10 pregnant women (median age 29.5 years, range 22-34 years) with uncomplicated, low-risk pregnancies at the following time points: i) during the second trimester of pregnancy (18-24 weeks) n=2; ii) during weeks 36-40 of the third trimester n=3; and iii) 6 weeks post-partum, n=5. At each time point, blood was collected from each patient as follows: 13.5 ml plasma in 3x4.5 ml sodium citrate vacutainer tubes (NHS Supply Chain) and 5 ml serum in a 5 ml SST vacutainer tube (NHS Supply Chain). Standard venepuncture procedures were followed, according to the National Cancer Institute Early Detection Research Network, which involved using a 21-gauge needle to minimise haemolysis (51,56). All low-risk, healthy pregnant women referred to the antenatal clinic at the Jessop Wing, Sheffield Teaching Hospitals NHS Foundation Trust were eligible for inclusion in this study. Pregnant women were excluded from recruitment if: i) The pregnancy was dated >20 weeks gestation; ii) there was no foetal heartbeat detected on ultrasound imaging, or iii) there were known foetal anomalies. All samples were collected in the antenatal clinic of the Jessop Wing Hospital between January and September 2017.
Following collection, samples were kept upright and stored on ice (maintained at ~4˚C) to inhibit miRNA degradation by circulating RNases within the whole blood, and were processed within 4-6 h. Samples were centrifuged at 1,900 x g for 10 min at 4˚C and the recovered supernatant was aliquoted and immediately stored at -80˚C. Following gently thawing at room temperature, plasma samples underwent a second centrifugation step (16,000 x g for 10 min at 4˚C) to generate platelet deficient plasma. Total RNA (including miRNA) was extracted from serum and plasma samples using a prototype version of the Maxwell® Rapid Sample Concentrator (RSC) miRNA Tissue or Plasma Serum kit (cat. no. AS1460; Promega Corporation) (57) with minor modifications to the standard protocol. Specifically, lyophilised DNase I was resuspended with 275 µl nuclease free water, mixing gently through inversion. A total of 5 µl Blue Dye was added to the reconstituted DNase I as a visual indicator, swirling gently to mix. Aliquots were made and stored at 4˚C for a few weeks, or at -20˚C for longer storage. Subsequently, 2.5 µl UniSp2, UniSp4 and UniSp6 (reconstituted according to manufacturer's instructions; Qiagen RNA spike in kit for RT; cat. no. 339390; Qiagen, Inc.) (58) was added to 200 µl binding buffer and mixed thoroughly. A total of 500 µl pre-processed plasma was transferred to a 1.5 ml Eppendorf and 60 µl Proteinase K (Promega Corporation) was added. This was then combined with the binding buffer and spike-in mix, vortexing for 10 sec. This sample lysate was then incubated at 37˚C for 15 min. During this time, the Maxwell® RSC cartridges were prepared by removing their seals and loading them into the RSC deck tray. An RSC plunger was added to well 8 of each cartridge, 500 µl elution tubes were loaded into the deck and 60 µl nuclease-free water was added to each tube. A total of 10 µl reconstituted DNAse I was added to well 4 (yellow) of the cartridges, before the total volume of incubated sample lysate was transferred into well 1 of the cartridges. The Maxwell® RSC Instrument (cat. no. AS4500: Promega Corporation) (59) instrument and the ‘RSC miRNA Tissue' method (60) was used to initiate the automated purification run. Following processing, the eluate was stored at -80˚C. Extracted RNA was assessed for quality and quantity using an Agilent Small RNA Chip and 2100 Bioanalyzer (Agilent Technologies Deutschland GmbH), measuring miRNA percentage and concentration in pg/µl.
Ethics approval
Ethical approval for the present study (approval no. 16/NE/0292) was obtained from the United Kingdom North East Newcastle and North Tyneside 1 NHS Research Ethics Committee on 30/08/2016 and The Health Research Authority on 27/09/2016, with non-substantial amendments approved on 18/10/2018 and 01/05/2019. Written informed consent was obtained from all patients.
miRNA quantification
cDNA synthesis was performed on the 10 paired serum and plasma samples (2 replicates per sample; n=20 plasma cDNA, n=20 serum cDNA) using the qScript® microRNA cDNA Synthesis kit (Quantabio) according to the manufacturer's protocol, with one minor modification during the initial PolyA tailing reaction, specifically the addition of 1 µl cel-miR-30-3p and UniSp6 spike-in mix (Qiagen, Inc.) reconstituted as per manufacturer's protocol. A total of 6 µl extracted RNA was used in each 10 µl PolyA tailing reaction. Reverse transcription-quantitative (RT-q)RT-qPCR with a total reaction volume of 25 µl was performed as follows: 0.5 µl PerfeCTa Universal Primer within the cDNA synthesis kit (Quantabio), 12.5 µl ExiLENT SYBR® Green MasterMix (Exiqon; Qiagen, Inc.), 2 µl cDNA, 9.5 µl nuclease-free water and 0.5 µl of each of the 8 miRNA primers. Two quality control primers were used to assess the efficiency of RNA extraction (miRCURY LNA UniSp2 PCR assay) and cDNA synthesis (miRCURY LNA cel-miR-39-3p) (Qiagen, Inc.), whereas 4 miRNAs were chosen for the comparison between these starting media; hsa-miR-222-3p, hsa-miR-23a, hsa-miR-30e-5p and hsa-miR-451a (Integrated DNA Technologies, Inc.) (Table I). These four miRNAs were selected as they are known to be consistently and stably detected in biofluids such as serum and plasma (25,33,47); facilitating an assessment of their relative quantity. hsa-let-7i-3p and hsa-miR-148-3p were chosen for normalisation. A 3-step cycling qPCR protocol: 95˚C for 2 min; followed by 40 cycles of 95˚C for 5 sec, 60˚C for 15 sec and 70˚C for 15 sec was used with a Bio-Rad CFX96, Real time C1000 Touch Thermal Cycler (Bio-Rad Laboratories, Inc.). RT-qPCR involved no-template (NTC) and no-Reverse Transcriptase controls (NRTC).
Table I.
Description of the 8 miRNA primers used for reverse transcription-quantitative PCR.
| miRNA primer | Function | Sequence |
|---|---|---|
| UniSp2 | RNA extraction efficiency | Unavailable |
| cel-miR-39-3p | cDNA synthesis efficiency | 5'-UCACCGGGUGUAAAUCAGCUUG-3' |
| hsa-miR-451a | Stably expressed miRNA of interest and detection of haemolysis | 5'-AAACCGUUACCAUUACUGAGUU-3' |
| hsa-miR-23a | Stably expressed miRNA of interest and detection of haemolysis | 5'-AUCACAUUGCCAGGGAUUUCC-3' |
| hsa-let-7i-3p | Stably expressed miRNA used for normalisation | 5'-CUGCGCAAGCUACUGCCUUGCU-3' |
| hsa-miR-148-3p | Stably expressed miRNA used for normalisation | 5'-UCAGUGCAUCACAGAACUUUGU-3' |
| hsa-miR-222-3p | Stably expressed miRNA of interest | 5'-AGCUACAUCUGGCUACUGGGU-3' |
| hsa-miR-30e-5p | Stably expressed miRNA of interest | 5'-UGUAAACAUCCUUGACUGGAAG-3' |
Statistical analysis Outliers
For each miRNA analysed, the mean Cq value and variance within each replicate was calculated. RT-qPCR efficiency was calculated to be 2.0 and outliers (n=18) were removed accordingly (Table II).
Table II.
Method of identifying RT-qPCR data outliersa.
| Mean Cq of replicates | Maximal acceptable variance in Cq replicates |
|---|---|
| 25 | 0.5 |
| 26 | 0.5 |
| 27 | 0.5 |
| 28 | 0.5 |
| 29 | 0.5 |
| 30 | 0.5 |
| 31 | 0.5 |
| 32 | 0.7 |
| 33 | 0.9 |
| 34 | 1.3 |
| 35 | 1.9 |
aFor each miRNA replicate, the mean Cq value and variance was calculated and outliers removed accordingly. RT-qPCR efficiency was calculated to be 2.0. RT-qPCR, reverse transcription-quantitative PCR.
Quality control
RNA extraction efficiency, monitored using UniSp2, was acceptable with Cq values showing consistent values (Δ<3 Cq) across the dataset (61). Haemolysis was monitored using ΔCq=mean Cqhsa-miR-23a-mean Cqhsa-miR-451a, with a ΔCq >5 indicating possible haemolysis and a ΔCq >7 conferring a high risk of haemolysis affecting the data obtained (26,33,61). Samples with a ΔCq >7 were excluded prior to further analysis. cDNA efficiency was acceptable with consistent values of cel-miR-39-3p (Δ<2 Cq) across the dataset (61).
Relative expression
The relative expression (ΔCq) of each miRNA replicate was normalised against the geomean of two normalising miRNA (hsa-let-7i-3p and hsa-miR-148-3p), both of which are stably and consistently expressed in serum and plasma samples (33). Standard deviations and confidence intervals were calculated accordingly (62).
Analysis
Data were assessed for normality (Shapiro-Wilk test) prior to statistical testing. If the Gaussian distribution was satisfied, a paired t-test was performed, otherwise a Wilcoxon matched-pairs signed rank test was used. Statistics were performed in GraphPad Prism version 8 (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
Patient characteristics
Table III summarises the characteristics of the 10 patients (median age 29.5 years range 22-34 years) that provided paired serum and plasma samples taken at different time points during their uncomplicated low-risk pregnancy.
Table III.
Summary characteristics of recruited patientsa.
| Study time point | n | Average gestational age, weeks + days | Range, weeks + days |
|---|---|---|---|
| Second trimester | 2 | 18+4 | 17+5-19+1 |
| Third trimester | 3 | 38+1 | 37+5-39+2 |
| 6 weeks post-partum | 5 | 6.3 weeks post delivery | 5-7 weeks |
an=10.
Bioanalyzer miRNA content
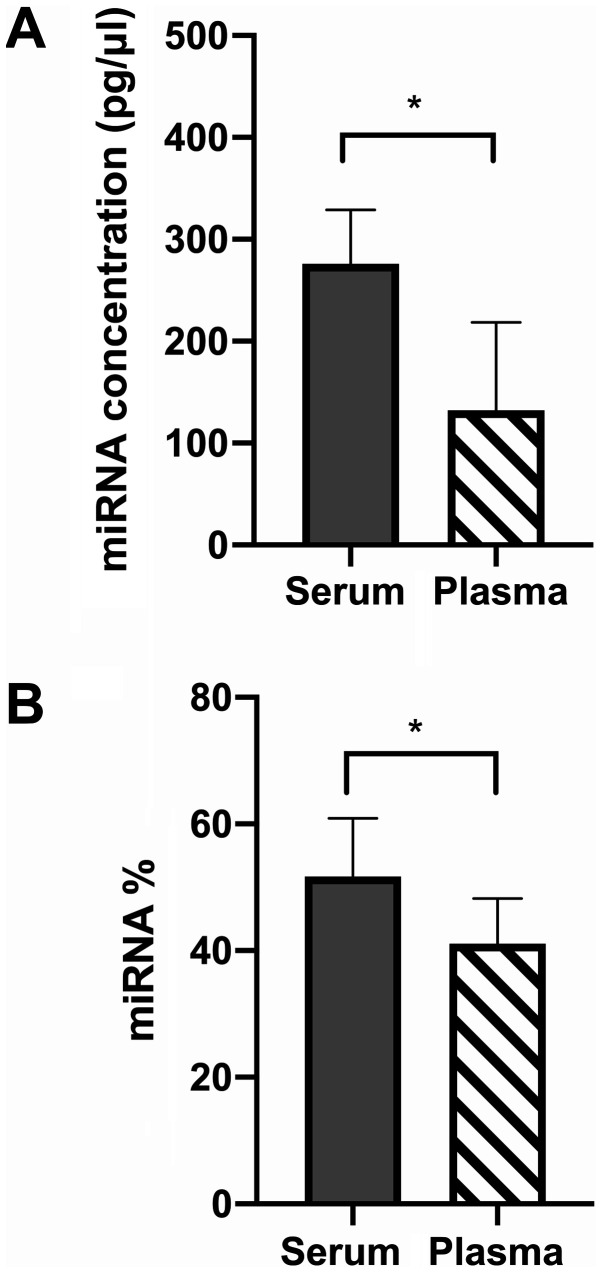
Both the average concentration and percentage of miRNA was higher within serum compared to plasma samples (Fig. 1).
Figure 1.
Bioanalyzer data. (A) miRNA concentration as the median + 95% confidence interval of the serum vs. plasma samples measured using the Agilent Bioanalyzer Small RNA chip. n=20. Wilcoxon matched pairs signed rank test, P=0.0039. A Shapiro-Wilk test of normality was passed for plasma (W=0.95, P=0.65) but not for serum (W=0.65, P=0.0002). (B) MiRNA percentage as the mean + standard deviation of serum vs. plasma samples measured using the Agilent Bioanalyzer Small RNA chip. n=20. A two-tailed paired t-test was used to compare data (t=4.97, P=0.0008, df=9). Shapiro-Wilk test of normality was passed for both plasma and serum (plasma W=0.97, P=0.84; serum W=0.95, P=0.67). *P<0.05. miRNA, microRNA.
qPCR quality controls Control samples
RNA extraction efficiency was deemed efficient with a ΔCq of UniSp2=2.88 across the dataset. As expected, a Cq result was not obtained for the NTC or NRTC samples, indicating that neither the starting serum or plasma samples, or the qPCR reagents were contaminated.
Haemolysis detection
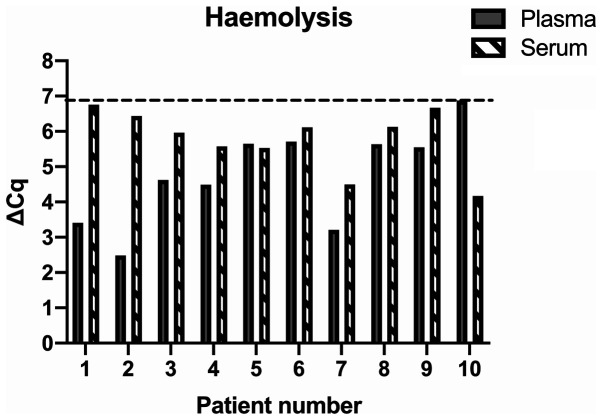
None of the included samples displayed significant haemolysis (ΔCq=>7 Fig. 2) and were all retained for downstream analysis.
Figure 2.
Haemolysis analysis for the included samples, where ΔCq=mean Cqhsa-miR-23a-mean Cqhsa-miR-451a. ΔCq >5 indicates possible haemolysis and ΔCq >7-8 confers a high risk of haemolysis.
Serum vs. plasma
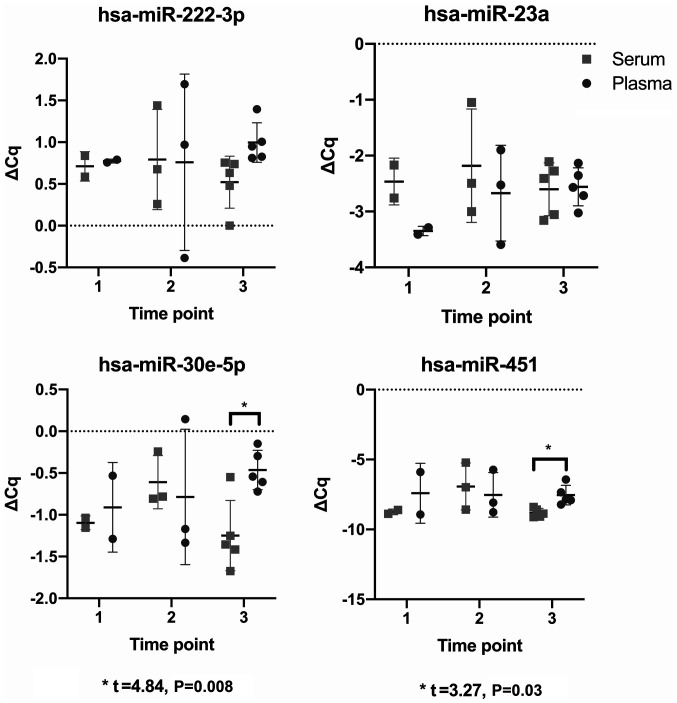
Using ΔCq values and paired t-tests, comparisons were made between paired serum and plasma samples taken at each study time point to determine whether the miRNA profile differed between media. Only two comparisons were significant; specifically, the comparison between the paired serum and plasma samples at time point three, studying hsa-miR-30e-5p (t=4.84, P=0.008) and hsa-miR-451a (t=3.27, P=0.03) (Fig. 3).
Figure 3.
Comparison between the ΔCq of paired serum and plasma samples, as the mean + standard deviation, across the three study time points, analysing four stably expressed miRNAs; hsa-miR-222-3p, hsa-miR-23a, hsa-miR-30e-5p and hsa-miR-451a. *P<0.05. miRNA/miR, microRNA; 1, second trimester of pregnancy (18-24 weeks); 2, weeks 36-40 of the third trimester; 3, 6 weeks post-partum.
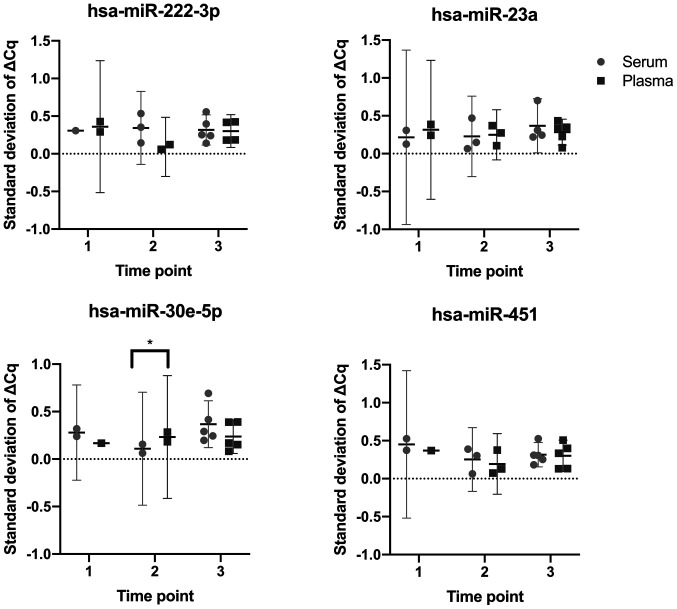
To determine whether serum or plasma samples yielded more consistent RT-qPCR results, the standard deviations of the ΔCq values for paired serum and plasma samples taken at each study time point were compared, analyzing the four miRNAs of interest. Only one comparison was found to be significant, specifically the comparison between serum and plasma samples at time point two, using hsa-miR-30e-5p. All other comparisons revealed no significant differences (P>0.05; Fig. 4).
Figure 4.
Consistency comparison: Comparing the ΔCq standard deviations of paired serum and plasma samples across the three study time points, analysing four stably expressed miRNAs; hsa-miR-222-3p, hsa-miR-23a, hsa-miR-30e-5p and hsa-miR-451a. *P<0.05. miRNA/miR, microRNA; 1, second trimester of pregnancy (18-24 weeks); 2, weeks 36-40 of the third trimester; 3, 6 weeks post-partum.
Discussion
Selection of the most appropriate starting test sample for the identification of clinically relevant biomarkers is essential, and is dependent upon first establishing the underlying performance of different starting substrates (26,45,47,48). In the present study. within a cohort of patients with uncomplicated, low-risk, healthy pregnancies, across the three study time points, the Bioanalyzer Small RNA chip data revealed the miRNA content of serum samples to be higher than that of plasma. Dissimilar to the Bioanalyzer findings, the majority of paired comparisons using RT-qPCR showed no significant differences between the respective serum and plasma samples at any of the time points assessed. Only 2 comparisons out of 12 were significant, specifically between paired serum and plasma samples taken at time point three, using hsa-miR-30e-5p and hsa-miR-451a. Similarly, when comparing the consistency of serum vs. plasma samples, only 1 of the 12 comparisons was found to be statistically significant, involving time point two and hsa-miR-30e-5p.
The only truly comparable previous manuscript involving a healthy pregnant population both agrees and disagrees with our findings. This previous study reported a higher proportion of miRNA in plasma compared to serum, although the absolute number of miRNAs detected and occurrence of abundant miRNA was higher in the serum (25) This previous study included 32 samples (20 serum, 12 plasma), yet only three paired serum and plasma samples from the same patient were obtained (25), reducing the generalisability and reliability of the results obtained, given the known intra and inter-individual biological variability in the expression of certain miRNAs (63). Furthermore, the present study investigated three time points (and as such, determined whether the optimal starting media changed throughout pregnancy), as opposed to just one time point (second trimester) in the previous study (25).
Compared with the previous literature involving non-pregnant populations, the absence of a significant difference between serum and plasma samples using RT qPCR is in agreement with some studies (27,41,47,49,50), but in disagreement with others (7,25,33,48). However, direct comparisons cannot truly be made, given the known differences in miRNA expression profiles between different patient cohorts in terms of sex and disease states, with the above studies including lung, breast and colorectal cancer patients (7,41), healthy male or female subjects (7,27,41,48-50). It is evident that when using only the existing literature, it is extremely challenging for researchers to deduce which starting media they should preferentially use to answer their research question.
Previous studies have suggested that serum samples collected and extracted in optimal conditions may display less variation in the miRNA data than plasma samples (33); however the results of the present study did not replicate this, showing no significant differences between the standard deviations of normalised serum and plasma data based on RT-qPCR.
Regarding the results of the present study, the discrepancy between quantification techniques is potentially explained by the uncontrolled and variable release of miRNAs from erythrocytes and platelets during the coagulation process within serum samples, which may have contributed to the higher miRNA levels observed with the Small RNA chip data (27,33,45,47,48,50). This is corroborated by the higher mean haemolysis levels for serum samples compared with plasma, with the ΔCq (hsa-miR-23a and hsa-miR-451a) being 5.78 and 4.76, respectively. The chips used within the Agilent Bioanalyzer each comprise a network of capillary channels that separate the sample by means of gel electrophoresis. The principle is based upon the fact that small fragments migrate faster than larger ones, using fluorescent dye molecules that intercalate with the RNA strands. These hybrid molecules are detected by their fluorescence and translated into gel-like images (bands) and electropherograms (peaks). Small RNA chips measure RNA of 6-150 nucleotides in size, and quantify the concentration and percentage of miRNA within a sample based on the number of molecules falling within the 4-39 nucleotide region (64); however this pre-defined region for detection may additionally contain small interfering RNA (20-25 nucleotides in length) or degraded longer RNA molecules including small nuclear RNA (~100 nucleotides), primitive (pri-miRNA, several hundred nucleotides) or precursor miRNA (~70 nucleotides) (65), which could falsely elevate the quantification of miRNA. Dissimilarly, RT-qPCR using specific primers would only bind and amplify mature miRNA molecules with a sequence that precisely matches the primer assay, meaning that even isomers of the miRNA may not be detected (66). This is particularly problematic when profiling miRNAs, given there are >3,000 known miRNA variants, most of which differ from the official sequence as denoted on miRBASE, from which the majority of RT-qPCR primer assays are designed (67,68). Furthermore, existing studies have shown key differences between the varying methods of quantifying RNA and DNA libraries, with electrophoresis-based quantification techniques (including the Agilent Bioanalyzer) producing higher concentration estimates (69) or highly variable quantification results (70) compared with RT-qPCR or the Qubit fluorometer. Overall, RT-qPCR is suggested to generate more reliable results, notwithstanding the increased number of replicates performed using this technique and the high between-run variability that was noted with the Bioanalyzer in the present study.
Limitations of the present study include the relatively small sample size of 10 pregnant patients; however this is in keeping with the sample sizes of existing literature (27,48-50) and is strengthened by the presence of paired samples taken at exactly the same time points, and being stored and processed using the same conditions. Despite this, a larger patient cohort would have been preferable, particularly considering time point i), which involved only two patients, potentially limiting the generalisability of the data obtained and capacity to detect a true difference. Although a significant difference was not observed between the paired serum and plasma samples, it would have been preferable to match the baseline characteristics of the patients more closely, in terms of age, body mass index and the exact gestation at which the samples were taken, yet previously published literature have similarly not corrected for these factors (25,27,47). There is also no consensus on the optimal normalisation strategy when working with miRNA RT-qPCR data, leading to further variability and difficulty when comparing with existing studies (24,71,72). The approach taken here, to normalise to two stably expressed miRNAs (hsa-let-7i-3p and hsa-miR-148-3p) across the serum and plasma samples, is a recognised approach that is preferable to the use of synthetic spike-ins, such as cel-miR-39-3p. Such synthetic spike-ins are useful to identify differences in RNA extraction efficiency, but not to normalise the endogenous miRNA content of the sample (24,58,73). The use of two normalising miRNAs was further acceptable for the present study given that all RNA extractions were performed within the same batch. However, when this is not the case, and with greater resources, normalising to 3-5 miRNAs that are stably expressed within the starting media, or better still, normalising to the global mean of >90 miRNAs is preferred (33,45,71).
Future work should involve replicating the experiments using a larger patient population, and using a more stringent cut-off of ≤5 for the ΔCq for the haemolysis markers on RT-qPCR, which may reduce the potential to falsely elevate the miRNA content of serum samples, yet most clinical studies would accept ΔCq <7 (26,33).
It is clear that the choice of starting media for ongoing experiments is crucially dependent on three main factors: i) sample type; ii) the patient cohort under investigation (healthy vs. disease; pregnant vs. non-pregnant); and iii) the miRNA quantification technique. As seen in the present study, different miRNA quantification techniques can produce conflicting information concerning the optimal starting media, and scientists should be astutely aware of this when designing studies, particularly those with a clinical end point. In the healthy pregnant population using the Agilent Bioanalyzer, the present study showed there to be a higher miRNA concentration and percentage in serum samples. However, this was not replicated in the RT-qPCR results, which, due to the detection of specific miRNAs, is suggested to be more reliable and less liable to the incorrect inclusion of fragmented long RNA molecules or miRNAs released during the coagulation process. Furthermore, RT-qPCR involves a higher number of replicates, and this increases the reliability of the results, producing consistent data unlike the high between run variability that was observed in the present study using the Bioanalyzer. In view of this, it is concluded that neither serum nor plasma are superior starting media during the intra or post-partum period in the healthy pregnant population, and as such, either would be suitable for studies and downstream analyses investigating the miRNA profiles or use of clinical biomarkers in this population.
Acknowledgements
The authors would like to thank Dr Sarah Waite, Senior Research Technician in the Department of Oncology and Metabolism (University of Sheffield) for technical assistance with the laboratory work performed in the present study.
Funding
The present study was supported by funding from Weston Park Cancer Charity, Sheffield, U.K (grant nos. CA154 and CA184).
Availability of data and materials
The datasets used and/or analysed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
VLP conceived and designed the study, collected the data, managed the project, analysed the data and wrote the manuscript. EG and BM assisted in designing the study, in deciding the methodology and analysed the data. AP and PRH assisted in designing the methodology used, supervised the study and reviewed and edited the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Ethical approval was obtained from the North East Newcastle and North Tyneside 1 NHS Research Ethics Committee, UK (approval no. 16/NE/0292). No patient identifiable information is presented within the manuscript and thus patient consent was not required.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mishra PJ. MicroRNAs as promising biomarkers in cancer diagnostics. Biomark Res. 2014;2(19) doi: 10.1186/2050-7771-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nair VS, Pritchard CC, Tewari M, Ioannidis JP. Design and analysis for studying micrornas in human disease: A primer on-omic technologies. Am J Epidemiol. 2014;180:140–152. doi: 10.1093/aje/kwu135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Wang L, Qu J, Guan NN, Li JQ. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics. 2018;34:4256–4265. doi: 10.1093/bioinformatics/bty503. [DOI] [PubMed] [Google Scholar]
- 5.Papadaki C, Stratigos M, Markakis G, Spiliotaki M, Mastrostamatis G, Nikolaou C, Mavroudis D, Agelaki S. Circulating microRNAs in the early prediction of disease recurrence in primary breast cancer. Breast Cancer Res. 2018;20(72) doi: 10.1186/s13058-018-1001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rapado-Gonzalez O, Alvarez-Castro A, Lopez-Lopez R, Iglesias-Canle J, Suarez-Cunqueiro MM, Muinelo-Romay L. Circulating microRNAs as promising biomarkers in colorectal cancer. Cancers (Basel) 2019;11(898) doi: 10.3390/cancers11070898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Schooneveld E, Wouters MC, Van der Auwera I, Peeters DJ, Wildiers H, Van Dam PA, Vergote I, Vermeulen PB, Dirix LY, Van Laere SJ. Expression profiling of cancerous and normal breast tissues identifies microRNAs that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res. 2012;14(R34) doi: 10.1186/bcr3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan W, Liu B, Qu S, Liang G, Luo W, Gong C. MicroRNAs and cancer: Key paradigms in molecular therapy. Oncol Lett. 2018;15:2735–2742. doi: 10.3892/ol.2017.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 10.Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, function and role in cancer. Curr Genomics. 2010;11:537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Peng R, Wang J, Qin Z, Xue L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin Epigenetics. 2018;10(59) doi: 10.1186/s13148-018-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghasabi M, Mansoori B, Mohammadi A, Duijf PH, Shomali N, Shirafkan N, Mokhtarzadeh A, Baradaran B. MicroRNAs in cancer drug resistance: Basic evidence and clinical applications. J Cell Physiol. 2019;234:2152–2168. doi: 10.1002/jcp.26810. [DOI] [PubMed] [Google Scholar]
- 13.Gomes BC, Rueff J, Rodrigues AS. MicroRNAs and cancer drug resistance. Methods Mol Biol. 2016;1395:137–162. doi: 10.1007/978-1-4939-3347-1_9. [DOI] [PubMed] [Google Scholar]
- 14.Si W, Shen J, Zheng H, Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics. 2019;11(25) doi: 10.1186/s13148-018-0587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao C, Dong J, Jiang T, Shi Z, Yu B, Zhu Y, Chen D, Xu J, Huo R, Dai J, et al. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS One. 2011;6(e23925) doi: 10.1371/journal.pone.0023925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y, Tian F, Li H, Zhou Y, Lu J, Ge Q. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int J Gynaecol Obstet. 2015;130:49–53. doi: 10.1016/j.ijgo.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Hromadnikova I, Kotlabova K, Ivankova K, Vedmetskaya Y, Krofta L. Profiling of cardiovascular and cerebrovascular disease associated microRNA expression in umbilical cord blood in gestational hypertension, preeclampsia and fetal growth restriction. Int J Cardiol. 2017;249:402–409. doi: 10.1016/j.ijcard.2017.07.045. [DOI] [PubMed] [Google Scholar]
- 18.Hromadnikova I, Dvorakova L, Kotlabova K, Krofta L. The Prediction of gestational hypertension, preeclampsia and fetal growth restriction via the first trimester screening of plasma exosomal C19MC microRNAs. Int J Mol Sci. 2019;20(2972) doi: 10.3390/ijms20122972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheikh AM, Small HY, Currie G, Delles C. Systematic review of Micro-RNA expression in Pre-eclampsia identifies a number of common pathways associated with the disease. PLoS One. 2016;11(e0160808) doi: 10.1371/journal.pone.0160808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skalis G, Katsi V, Miliou A, Georgiopoulos G, Papazachou O, Vamvakou G, Nihoyannopoulos P, Tousoulis D, Makris T. MicroRNAs in preeclampsia. Microrna. 2019;8:28–35. doi: 10.2174/2211536607666180813123303. [DOI] [PubMed] [Google Scholar]
- 21.Cai M, Kolluru GK, Ahmed A. Small molecule, big prospects: MicroRNA in pregnancy and its complications. J Pregnancy. 2017;2017(6972732) doi: 10.1155/2017/6972732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Z, Han S, Hu P, Zhu C, Wang X, Qian L, Guo X. Potential role of maternal serum microRNAs as a biomarker for fetal congenital heart defects. Med Hypotheses. 2011;76:424–426. doi: 10.1016/j.mehy.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Duenas A, Exposito A, Aranega A, Franco D. The role of non-coding RNA in congenital heart diseases. J Cardiovasc Dev Dis. 2019;6(15) doi: 10.3390/jcdd6020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tay JW, James I, Hughes QW, Tiao JY, Baker RI. Identification of reference miRNAs in plasma useful for the study of oestrogen-responsive miRNAs associated with acquired Protein S deficiency in pregnancy. BMC Res Notes. 2017;10(312) doi: 10.1186/s13104-017-2636-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge Q, Shen Y, Tian F, Lu J, Bai Y, Lu Z. Profiling circulating microRNAs in maternal serum and plasma. Mol Med Rep. 2015;12:3323–3330. doi: 10.3892/mmr.2015.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, Dahlsveen IK. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013;59 (Suppl):S1–S6. doi: 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 27.Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. Comparing the MicroRNA spectrum between serum and plasma. PLoS One. 2012;7(e41561) doi: 10.1371/journal.pone.0041561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He S, Zeng S, Zhou ZW, He ZX, Zhou SF. Hsa-microRNA-181a is a regulator of a number of cancer genes and a biomarker for endometrial carcinoma in patients: A bioinformatic and clinical study and the therapeutic implication. Drug Des Devel Ther. 2015;9:1103–1175. doi: 10.2147/DDDT.S73551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable Blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3(e3148) doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kontomanolis EN, Kalagasidou S, Fasoulakis Z. MicroRNAs as potential serum biomarkers for early detection of ectopic pregnancy. Cureus. 2018;10(e2344) doi: 10.7759/cureus.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Exiqon: Biofluids Guideline: Analysing microRNAs in liquid biopsies. https://www.gene-quantification.de/exiqon-biofluids-guidelines-2016.pdf. [Google Scholar]
- 34.Rounge TB, Lauritzen M, Langseth H, Enerly E, Lyle R, Gislefoss RE. microRNA biomarker discovery and high-throughput DNA sequencing are possible using long-term archived serum samples. Cancer Epidemiol Biomarkers Prev. 2015;24:1381–1387. doi: 10.1158/1055-9965.EPI-15-0289. [DOI] [PubMed] [Google Scholar]
- 35.Kreth S, Hubner M, Hinske LC. MicroRNAs as clinical biomarkers and therapeutic tools in perioperative medicine. Anesth Analg. 2018;126:670–681. doi: 10.1213/ANE.0000000000002444. [DOI] [PubMed] [Google Scholar]
- 36.Morales-Prieto DM, Ospina-Prieto S, Chaiwangyen W, Schoenleben M, Markert UR. Pregnancy-associated miRNA-clusters. J Reprod Immunol. 2013;97:51–61. doi: 10.1016/j.jri.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Bidarimath M, Khalaj K, Wessels JM, Tayade C. MicroRNAs, immune cells and pregnancy. Cell Mol Immunol. 2014;11:538–547. doi: 10.1038/cmi.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo SS, Ishibashi O, Ishikawa G, Ishikawa T, Katayama A, Mishima T, Takizawa T, Shigihara T, Goto T, Izumi A, et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81:717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- 39.Mouillet JF, Ouyang Y, Coyne CB, Sadovsky Y. MicroRNAs in placental health and disease. Am J Obstet Gynecol. 2015;213 (4 Suppl):S163–S172. doi: 10.1016/j.ajog.2015.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno-Moya JM, Vilella F, Simon C. MicroRNA: Key gene expression regulators. Fertil Steril. 2014;101:1516–1523. doi: 10.1016/j.fertnstert.2013.10.042. [DOI] [PubMed] [Google Scholar]
- 41.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 42.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Qiagen: miRNeasy Serum/Plasma-Handbook for miRNeasy Serum/Plasma Kit. Journal. [Google Scholar]
- 44. Promega: Maxwell RSC miRNA Plasma and Serum Kit: Instructions for use of product AS1680. https://www.promega.co.uk/products/nucleic-acid-extraction/rna/maxwell-rscmirna-tissue-plasma-serum-kit/?catNum=AS1460#protocols. [Google Scholar]
- 45. Exiqon: Profiling of microRNA in blood (serum/plasma). Guidelines for the miRCURY LNA Universal RT microRNA PCR system. https://www.gene-quantification.de/microRNA-serum-plasma-guidelines-exiqon.pdf. [Google Scholar]
- 46.Zou P, Luo L, Zhao C, Chen Z, Dong R, Li N, Wang Y, Wang J, Wang T, Chen M, et al. The serum microRNA profile of intrahepatic cholestasis of pregnancy: Identification of novel noninvasive biomarkers. Cell Physiol Biochem. 2018;51:1480–1488. doi: 10.1159/000495595. [DOI] [PubMed] [Google Scholar]
- 47.Foye C, Yan IK, David W, Shukla N, Habboush Y, Chase L, Ryland K, Kesari V, Patel T. Comparison of miRNA quantitation by Nanostring in serum and plasma samples. PLoS One. 2017;12(e0189165) doi: 10.1371/journal.pone.0189165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Max KEA, Bertram K, Akat KM, Bogardus KA, Li J, Morozov P, Ben-Dov IZ, Li X, Weiss ZR, Azizian A, et al. Human plasma and serum extracellular small RNA reference profiles and their clinical utility. Proc Natl Acad Sci USA. 2018;115:E5334–E5343. doi: 10.1073/pnas.1714397115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glinge C, Clauss S, Boddum K, Jabbari R, Jabbari J, Risgaard B, Tomsits P, Hildebrand B, Kääb S, Wakili R, et al. Stability of circulating Blood-based MicroRNAs-Pre-analytic methodological considerations. PLoS One. 2017;12(e0167969) doi: 10.1371/journal.pone.0167969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dufourd T, Robil N, Mallet D, Carcenac C, Boulet S, Brishoual S, Rabois E, Houeto JL, de la Grange P, Carnicella S. Plasma or serum? A qualitative study on rodents and humans using high-throughput microRNA sequencing for circulating biomarkers. Biol Methods Protoc. 2019;4(bpz006) doi: 10.1093/biomethods/bpz006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. The Early Detection Research Network (EDRN): Standard Operating Procedure (SOP) for collection of EDTA plasma. https://edrn.nci.nih.gov/resources/standard-operating-procedures/standard-operating-procedures/plasma-sop.pdf. [Google Scholar]
- 52. Qiagen: miRCURY LNA miRNA PCR-Exosomes, serum/plasma and other biofluid samples handbook. https://www.qiagen.com/fi/resources/resourcedetail?id=7ab5f614-f5d6-4bdc-b22b-246ec3601588&lang=en. [Google Scholar]
- 53.Shah JS, Soon PS, Marsh DJ. Comparison of methodologies to detect low levels of hemolysis in serum for accurate assessment of serum microRNAs. PLoS One. 2016;11(e0153200) doi: 10.1371/journal.pone.0153200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kirschner MB, Edelman JB, Kao SCH, Vallely MP, van Zandwijk N, Reid G. The impact of hemolysis on Cell-free microRNA biomarkers. Front Genet. 2013;4(94) doi: 10.3389/fgene.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.GmbH T. TechNote TN-01: The impact of sample type (serum and EDTA-plasma) and platelet contamination on osteomiR detection. [Google Scholar]
- 56. The Early Detection Research Network (EDRN): Standard Operating Procedure (SOP) For collection of serum. https://edrn.nci.nih.gov/resources/standard-operating-procedures/standard-operating-procedures/serum-sop.pdf. [Google Scholar]
- 57. Promega: Maxwell® RSC miRNA Plasma and Serum Kit: Instructions for use of product AS1680. https://www.promega.co.uk/products/nucleic-acid-extraction/rna/maxwell-rscmirna-tissue-plasma-serum-kit/?catNum=AS1460#protocols. [Google Scholar]
- 58. Qiagen: RNA Spike-In Kit, For RT handbook. https://www.qiagen.com/us/products/discovery-and-translational-research/pcr-qpcr-dpcr/qpcr-assays-and-instruments/mirna-qpcr-assay-and-panels/rna-spike-in-kit-for-rt/?clear=true#resources. [Google Scholar]
- 59. Promega: Maxwell® RSC Instrument, AS4500. https://www.promega.co.uk/products/lab-automation/maxwell-instruments/maxwell-rsc-instrument/?catNum=AS4500. [Google Scholar]
- 60. Promega: Maxwell® RSC miRNA Tissue Kit: Instructions for use of product. https://www.promega.co.uk/products/nucleic-acid-extraction/rna/maxwell-rscmirna-tissue-plasma-serum-kit/?catNum=AS1460#protocols. [Google Scholar]
- 61. Exiqon: miRCURY microRNA QC PCR Panel Instruction Manual 203887-203892. http://www.exiqon.com/ls/Documents/Scientific/QC-PCR-Panel-Manual.pdf. [Google Scholar]
- 62.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 63.Yoon H, Belmonte KC, Kasten T, Bateman R, Kim J. Intra- and Inter-individual Variability of microRNA levels in human cerebrospinal fluid: Critical implications for biomarker discovery. Sci Rep. 2017;7(12720) doi: 10.1038/s41598-017-13031-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Agilent: Bioanalyzer Small RNA Analysis. https://www.agilent.com/en/product/automated-electrophoresis/bioanalyzer-systems/bioanalyzer-rna-kits-reagents/bioanalyzer-small-rna-analysis-228257#support. [Google Scholar]
- 65.Masotti A, Caputo V, Prudente S, Bottazzo GF. Analysis of small RNAs with the Agilent 2100 Bioanalyzer. Application note. Agilent Technologies, 2006. https://www.agilent.com/cs/library/applications/5989-5215EN.pdf. [Google Scholar]
- 66.Magee R, Telonis AG, Cherlin T, Rigoutsos I, Londin E. Assessment of isomiR discrimination using commercial qPCR methods. Noncoding RNA Vol. 2017;3(18) doi: 10.3390/ncrna3020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuchenbauer F, Morin RD, Argiropoulos B, Petriv OI, Griffith M, Heuser M, Yung E, Piper J, Delaney A, Prabhu AL, et al. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res. 2008;18:1787–1797. doi: 10.1101/gr.077578.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. University of Manchester: miRBase: the microRNA database. http://www.mirbase.org/. [Google Scholar]
- 69.Hussing C, Kampmann ML, Mogensen HS, Børsting C, Morling N. Quantification of massively parallel sequencing libraries-a comparative study of eight methods. Sci Rep. 2018;8(1110) doi: 10.1038/s41598-018-19574-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garcia-Elias A, Alloza L, Puigdecanet E, Nonell L, Tajes M, Curado J, Enjuanes C, Díaz O, Bruguera J, Martí-Almor J, et al. Defining quantification methods and optimizing protocols for microarray hybridization of circulating microRNAs. Sci Rep. 2017;8(1110) doi: 10.1038/s41598-017-08134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Ronde MWJ, Ruijter JM, Lanfear D, Bayes-Genis A, Kok MGM, Creemers EE, Pinto YM, Pinto-Sietsma SJ. Practical data handling pipeline improves performance of qPCR-based circulating miRNA measurements. RNA. 2017;23:811–821. doi: 10.1261/rna.059063.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hardikar AA, Farr RJ, Joglekar MV. Circulating microRNAs: Understanding the limits for quantitative measurement by real-time PCR. J Am Heart Assoc. 2014;3(e000792) doi: 10.1161/JAHA.113.000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the present study are available from the corresponding author on reasonable request.