ABSTRACT
Convergence of virulence and antibiotic-resistance has been reported in Klebsiella pneumoniae, but not in Klebsiella variicola. We, hereby, report the detection and genomic characterization of hypervirulent and hypermucoviscous K. pneumoniae and K.variicola recovered in Chile from health-care associated infections, which displayed resistance to broad-spectrum cephalosporins. One hundred forty-six K. pneumoniae complex isolates were screened by hypermucoviscosity by the “string test.” Two hypermucoid isolates, one hypermucoviscous K. pneumoniae (hmKp) and one K. variicola (hmKv), were further investigated by whole-genome sequencing. In vivo virulence was analyzed by the Galleria mellonella killing assay. In silico analysis of hmKp UCO-494 and hmKv UCO-495 revealed the presence of multiple antibiotic-resistance genes, such as blaCTX-M-1, blaDHA-1 and blaLEN-25 among others clinically relevant resistance determinants, including mutations in a two-component regulatory system related to colistin resistance. These genetic features confer a multidrug-resistant (MDR) phenotype in both strains. Moreover, virulome in silico analysis confirmed the presence of the aerobactin gene iutA, in addition to yersiniabactin and/or colicin V encoding genes, which are normally associated to high virulence in humans. Furthermore, both isolates were able to kill G. mellonella and displayed higher virulence in comparison with the control strain. In summary, the convergence of virulence and the MDR-phenotype in K. pneumoniae complex members is reported for the first time in Chile, denoting a clinical problem that deserves special attention and continuous surveillance in South America.
KEYWORDS: Klebsiella pneumoniae complex, virulence, hypermucoviscous, ESBL, multidrug-resistance
Introduction
Klebsiella pneumoniae complex includes K. pneumoniae sensu stricto, K. quasipneumoniae subsp. quasipneumoniae, K. quasipneumoniae subsp. similipneumoniae, K. variicola subsp. variicola, K. variicola subsp. tropica, K. quasivariicola, and K. africana, respectively [1]. Among members of this complex, K. pneumoniae and K. variicola have been widely recognized as important opportunistic human pathogens commonly involved in hospital-acquired infections (HAIs) [2,3]. The clinical importance of these species has been associated with multidrug-resistance, mediated by the expression of extended-spectrum β-lactamases (ESBLs) and carbapenemases [4,5], and more recently with colistin resistance [6,7]. Lately, convergence of virulence and antibiotic-resistance has been reported in K. pneumoniae [8]. In this regard, hypervirulent K. pneumoniae (hvKp) isolates have been defined under the following criteria: i) occurrence of the hypermucoviscous (hmKp) phenotype, as determined by a positive “string test”; ii) presence of the rmpA gene, which regulates the capsule biosynthesis; and iii) presence of the aerobactin genes iucA/iutA [9,10]. Similarly to K. pneumoniae, K. variicola can also display the hypermucoviscous (hmKv) and/or hypervirulent (hvKv) phenotypes [1]. Currently, hvKp isolates have been reported mainly in Asia, Europe and North America, and more recently in South America [9], where sporadic reports have been restricted to Argentina and Brazil [10–12]. Hence, the aim of our study was to detect and characterize hypervirulent and hypermucoviscous ESBL-producing K. pneumoniae and K. variicola isolates recovered from Chilean hospitals.
Materials and methods
K. pneumoniae complex isolates and antibiotic susceptibility testing
One hundred forty-six non-repetitive K. pneumoniae complex isolates collected between 2011 and 2018 in Chile, were investigated. All isolates were recovered from nosocomial infections and were initially identified by each hospital laboratory as third-generation cephalosporin-resistant K. pneumoniae. Species identification was confirmed by conventional PCR according to previously described [13]. Antibiotic susceptibility testing to imipenem, ertapenem, meropenem, ceftriaxone, cefpodoxime, cefotaxime, ceftazidime, amoxicillin/clavulanic acid, amikacin, tobramycin, kanamycin, gentamicin, ciprofloxacin, levofloxacin and tetracycline was performed by the Kirby-Bauer method. ESBL-production and colistin susceptibility were determined by the combined disc test and the broth microdilution method, respectively [14].
Phenotypic identification of hypermucoviscous isolates
The hypermucoviscous phenotype was determined by the “string test” [15]. In brief, when a bacteriological loop was able to generate a viscous filament ≥5 mm in length by stretching bacterial colonies growth at 37ºC by 18–24 h on a blood agar plate, the isolate was considered as positive, thus defined as hypermucoviscous. Two isolates resulted positive for the “string test,” therefore, subsequent experiments included both strains.
Whole-genome sequencing (WGS) and in silico analyses of hypermucoviscous isolates
Total DNA of both hypermucoviscous isolates was extracted for whole-genome sequencing (WGS) using the Wizard® Genomic DNA Purification kit (Promega, USA) following the manufacturer’s protocol. Sequencing was performed by the Illumina MiSeq platform (2 × 250 bp paired end reads) with libraries prepared by the NexteraXT kit (Illumina), with a coverage of 30x.
De novo assembly was carried out by using the SPAdes software, version 3.9 (https://cge.cbs.dtu.dk/services/SPAdes/) with default values. Later, the assembled genomes were used to screen for genes for antibiotic-resistance, plasmids and virulence using the ResFinder v3.2, PlasmidFinder v2.1 and Virulence Finder v2.0 tools available at the Center for Genomic Epidemiology server (https://cge.cbs.dtu.dk/services/). Resistome (antibiotics, heavy metals, and disinfectants) was further predicted by the comprehensive antibiotic resistance database (CARD) (https://card.mcmaster.ca/), and ABRicate v0.9.8 (https://github.com/tseemann/abricate) using the BacMet2 database (http://bacmet.biomedicine.gu.se), respectively, considering a ≥ 90% similarity criteria. Genome annotation was accomplished using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) web-service (http://www.ncbi.nlm.nih.gov/genome/annotation_prok). Sequence types (STs) were determined for K. pneumoniae and K. variicola through the bioinformatic tools available at https://cge.cbs.dtu.dk/services/MLST/andhttp://mlstkv.insp.mx, respectively. Capsular serotypes (K-locus) and phylogenetic analysis of the ybt locus were predicted by Kleborate (https://github.com/katholt/Kleborate). Mutations in chromosomal genes mgrB, phoPQ, and pmrAB were analyzed with local BLAST+ DB using K. pneumoniae MGH78578 or K. variicola DSM 15,968 (accession number NC_009648.1 and NZ_CP010523.2) genomes as colistin-susceptible references. In order to predict the functional effect of amino acid substitutions, we used the PROVEAN web server (http://provean.jcvi.org/index.php).
We studied mutations in wzc, rcsAB, and lon genes in UCO-494 utilizing the K. pneumoniae (accession numbers LT174540 and JCMB01, respectively) genome as reference [16]. For all mutation, bioinformatic analysis was performed using the UGENE 1.32.0 Software.
Both UCO-494 and UCO-495 genomes have been deposited at DDBJ/ENA/GenBank under the accession numbers VSSY00000000.1 and VSSZ00000000.1, respectively.
Serum bactericidal assay and virulence behavior in the Galleria mellonella infection model
Serum bactericidal activity was analyzed according to previously described [17], with minor modifications. Briefly, 250 μL of a bacterial inoculum of 5 × 106 CFU/ml were mixed with 750 μL of fresh human serum. Then, viable bacterial cell count was performed in tryptone soy agar (TSA) plates. A K. pneumoniae isolate that was previously characterized as hypervirulent in our laboratory was used as positive control, while serum inactivated at 56°C for 30 min was utilized as blank. All experiments were performed in triplicate. A bacterial survival of <1% after 3 h of incubation with serum was considered as susceptible. On the other hand, survival percentages of 1–90% or >90% were considered as intermediate and resistant, respectively [18]. Additionally, in order to compare the levels of virulence of hmKp UCO-494 and hmKv UCO-495, the G. mellonella infection model was utilized [19]. K. quasipneumoniae subsp. similipneumoniae ATCC700,603 and hvKp k1/ST23 UCO-448 [10] were used as negative and positive hypervirulent controls, respectively. Larvae survival was analyzed during 96 h, and Kaplan-Meier killing curves of G. mellonella were generated using the log rank test with p < 0.05. Each assay was performed in triplicate.
Capsular-polysaccharide (CPS) quantification and estimation of capsular size
Total capsular-polysaccharide (CPS) of hmKp UCO-494 and hmKv UCO-495 was estimated according to the phenol-sulfuric acid method, after extraction using zwittergent 3–14 [20], and incubated in tryptone soy broth (TSB) at 37ºC for 18 h with agitation. The estimation of capsular size was carried out by transmission electron microscopy (TEM) of a bacterial inoculum incubated at 37ºC for 24 h [21]. Prior to microscopy, the samples were centrifuged at 3,000 rpm for 5 min and washed once with PBS buffer.
Biofilm assay
Biofilms-quantification was performed as previously described [22]. In brief, a colony from each strain was grown overnight in TSB at 37°C. From this culture, 10 μL of a bacterial suspension was used to inoculate 96-well polystyrene plates containing 90 μL of TSB, and these plates were incubated at 37°C for 24 h. Subsequently, the medium was removed from the plates and each wells was washed three times with water. Immediately, the samples were stained with 125 μL of 0.1% crystal violet for 15 min. Excess dye was removed by rinse 4 times in water, and dried during 10 min at 65°C. Afterward, 125 μL of acetic acid solution (30% v/v) were added and then incubated for 15 min at room temperature. Then, 125 μL of the solubilized crystal violet were transferred to a new 96-well polystyrene plates and color intensity was determined at a 550 nm using a spectrophotometer. K. pneumoniae ATCC 700603 strain was used as positive control, and acetic acid solution (30% v/v) was used as a negative control. Biofilm-formation abilities were defined as follows: i) absorbance values between 0.084 and 0.168 (2x – 4x blank absorbance) were considered as low biofilm-forming strains; ii) values ranging between 0.168 and 0.252 (4x – 6x blank absorbance) were considered as medium biofilm-forming strains, whereas iii) strains displayed absorbance values higher than 0.252 (>6x blank absorbance) were classified as high biofilm formers [23].
Results
Two hypermucoviscous isolates exhibiting a positive string test were identified as K. pneumoniae (UCO-494) and K. variicola (UCO-495) (Table 1). UCO-494 and UCO-495, belonging to the ST1161 and ST173 lineages, respectively, were isolated from blood and catheter cultures of ICU patients, admitted at two different hospitals located in southern Chile (Table 1). Both isolates were resistant to aminoglycosides and broad-spectrum cephalosporins. UCO-494 was additionally resistant to ertapenem, levofloxacin and ciprofloxacin, remaining susceptible to imipenem, meropenem, and tetracycline. Additionally, colistin-resistance in UCO-494 and UCO-495 was associated with MIC values of 8 and 16 μg/mL, respectively (Table 1). Resistome analysis revealed the presence of the ESBLs and cephalosporinases encoding genes blaCTX-M-1, blaSHV-187 and blaDHA-1 in K. pneumoniae UCO-494 and blaSHV-12 and the blaLEN-25 genes in K. variicola UCO-495 (Table 1). Moreover, ertapenem resistance in K. pneumoniae UCO-494 was associated with a deletion in the ompK35 gene, leading to porin deficiency, and also linked to the presence of the ompK37 gene, which has been associated with reduced permeability to carbapenems [24,25]. Additionally, K. pneumoniae UCO-494 harbored the aac(6ʹ)-Ib; aac(6ʹ)-Ib-cr, aadA1 and aadA2 and K. variicola UCO-495, the aph(3”)-Ia, aph(6)-Id and aph(3”)-Ib aminoglycosides resistance genes (Table 1).
Table 1.
Strain characteristic, MLST, capsular locus type, resistome and virulome of UCO-494 and UCO-495
| Phenotype | UCO-494 hmKp |
UCO-495 hmKv |
|---|---|---|
| Origin | Blood | Catheter |
| year | 2012 | 2012 |
| String test | + | + |
| ST | 1161 | 173 |
| K-locus* | KL19 | KL25 |
| ybt | ybt14 ICEKp5 | - |
| ybST | 327–1LV | - |
| O-locus | O1v2 | - |
| ESBL combined disc test | + | + |
| WGS data | ||
| Contig number | 462 | 226 |
| Genome size (bp) | 6,400,426 | 5,982,509 |
| GC% | 56,4% | 56,1% |
| CDS; pseudogenes; tRNA | 6668;203;95 | 5968;169;79 |
| Resistance profile | ERT, CIP, LEV AMK, KAN, GEN, TOB, AMP, CTX, CAZ, AMC | TET, STX, W, CPD, CRO, AMK, KAN, GEN, TOB, AMP, CTX, CAZ, FEP, AMC |
| MIC colistin | 8 μg/mL | 16 μg/mL |
| Resistome | ||
| Antibiotic resistance genes | sul1; sul2; arr-2; dfrA12; aadA1; aadA2; aac(6`)-Ib; aac(6`)-Ib-cr; oqxA/B; qnrB19; qnrB4; blaCTX-M-1; blaDHA-1; blaOXA-10; blaOXA-9; blaSHV-187; gyrA83L; gyrA87Y; parC80I | blaLEN-25; blaSHV-12; blaTEM-1B; oqxA; oqxB; aph(3”)-Ia; aph(6)-Id; aph(3”)-Ib; tet(D) |
| colistin mutation gen | PmrB: Gly256Arg (G766C) | PmrA: Thr146Ala (A436G); PmrB: Ser170Ala (G508T); PhoQ: Asp152Glu (T456G) |
| Heavy-metal resistance genes | arsenic (arsCDBAH); magnesium/cobalt/nickel/manganese (corA); glyphosate (phnMLKJI); quaternary ammonium (emrD – qacE∆1) | arsenic (arsBCRD), cobalt/manganese (corC), cobalt/magnesium (mgtA), magnesium/cobalt/nickel/manganese (corA): tellurium resistance gen (terW and terZCD) |
| Virulome | ||
| Virulence genes | Enterobactin (entB; entF; ycfH; entD), urea(ureA), alantoin (allS), aerobactin (iutA), fimbria type 1 (fimABCDFEGH), fimbria type 3 (mrkABCDF), yersiniabactin (irp1; irp2; fyuA; ybtAES), colicin V (cvpA; cvaA), biofilm (treC; sugE), ECP (ecpABCDE) | Urea (ureA), alantoin (allS), aerobactin (iutA), fimbria type 1 (fimABCDFEGH), fimbria type 3 (mrkABCDF), colicin V (cvpA; cvaA), biofilm (treC; sugE), ECP (ecpABCDE), KFU (kfuABC) |
| Plasmids | ColRNAI; IncA/C2; IncFIB (3); IncFII | IncFIB; IncFII; IncHI2; IncHI2A |
hmKP, hypermucoviscous Klebsiella pneumoniae; hvKP, hypervirulent Klebsiella pneumoniae; *Capsular polisacharide concentration in OD650nm 2.0. Significative difference with p-value equal to 0.0001 in t test. ERT, ertapenem; CIP, ciprofloxacin; LEV, levofloxacin; AMK, amikacin; KAN, kanamycin; GEN, gentamicin; TOB, tobramycin; AMP, ampicillin; CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; ; CPD, cefpodoxime; CRO, ceftriaxone; AMC, amoxicillin-clavulanic acid; W, trimethoprim; TET, tetracycline; SXT, sulphamethoxazole-trimethoprim .
Importantly, both isolates were resistant to colistin (Table 1). From WGS data, we predicted in K. pneumoniae UCO-494 (colistinMIC 8 ug/mL) a Gly256Arg (G766C) amino acid substitution in PmrB, while in K. variicola UCO-495 (colistinMIC 16 ug/mL) we predicted a Ser170Ala (G508T) amino acid substitution in PmrB, Thr146Ala (A436G) in PmrA and Asp152Glu (T456G) in PhoQ. All amino acid substitutions were neutral by PROVEAN.
Fluoroquinolone resistance in K. pneumoniae UCO-494 strain was mediated by aac(6ʹ)-Ib-cr, oqxA, oqxB, qnrB19 and qnrB4 genes and gyrA (83 L, 87Y) and parC (80I) mutations. Moreover, K. pneumoniae UCO-494 strain harbored the ColRNAI, IncA/C2, IncFIB and IncFII plasmids. On the other hand, K. variicola UCO-495 was susceptible to fluoroquinolones and additionally carried IncF-like plasmids (Table 1).
Furthermore, in K. variicola UCO-495, we found diverse metal-resistance systems, such as the arsenic (arsBCRD), cobalt/manganese (corC), cobalt/magnesium (mgtA), magnesium/cobalt/nickel/manganese (corA) and tellurium resistance genes terW and terZCD. Moreover, K. pneumoniae UCO-494 contained the arsenic (arsCDBAH) and magnesium/cobalt/nickel/manganese (corA) systems. Likewise, were identified the presence of resistance genes to glyphosate (phnMLKJI) and quaternary ammonium compounds (emrD – qacE∆1) in K. pneumoniae UCO-494.
In K. pneumoniae UCO-494, phylogenetic analysis of the ybt locus revealed 14 lineages (ybt locus sequence type YbST 327–1LV) with ICEKp5 element, were K-locus KL19 and O-locus O1v2, were also identified. On the other hand, we designated a new ST to MLST K. variicola, which corresponded to ST173 (allelic profile leuS10; pgi 9; pgk 6; phoE 1; pyrG 11; rpoB 1; fusA 2), whereas K. pneumoniae UCO-494 belonged to ST1161 (Table 1).
Virulome analysis of hvKv UCO-495 revealed the presence of the ferric uptake system kfuABC, which has been associated to hypervirulent Klebsiella strains [15]. Both isolates contained the aerobactin gene iutA, mannose-sensitive type 1 fimbriae (fimABCD operon), the mannose-resistant Klebsiella-like (type III) fimbriae cluster (mrkABCDFHIJ), and the E. coli common pilus operon (ecpABCDE) and biofilm related (treC, sugE) genes, which are associated with mucoviscosity and CPS production [26]. Only hmKp UCO-494 carried additionally the enterobactin (entB, entF, and ycfH), yersiniabactin siderophore cluster ybtAEPQSTUX and the siderophore genes irp1 and irp2, which are considered as genetic markers for high-pathogenicity island [27] (Table 1). It is important to highlight that in both strains the presence of rmpA/A2 was not identified.
Interestingly, hmKv UCO-495 was resistant to the bactericidal activity of human serum, while hmKp UCO-494 was susceptible, with 1% survival after 1 h interaction (Figure 1). Curiously, K. pneumoniae UCO-494 produced more CPS (155.44 ± 3.68 μg/mL) than K. variicola UCO-495 (30.26 ± 0.11 μg/mL). Likewise, UCO-494 displayed a capsular thickness of 0.124 ± 0.017 µm, whereas capsule thickness of UCO-495 was 0.097 ± 0.019 µm (Figure 2). Interestingly, in UCO-494 we predicted a F573S (T1718C) and R608T (G1823C) amino acid substitutions in wcz (deleterious by PROVEAN). Moreover, S35N (G104N) amino acid substitution in crsA in addition to E142Q (G424C) and R517C (T843C) in lon gen was identified. All of these genes were related with hypercapsule production [16].
Figure 1.
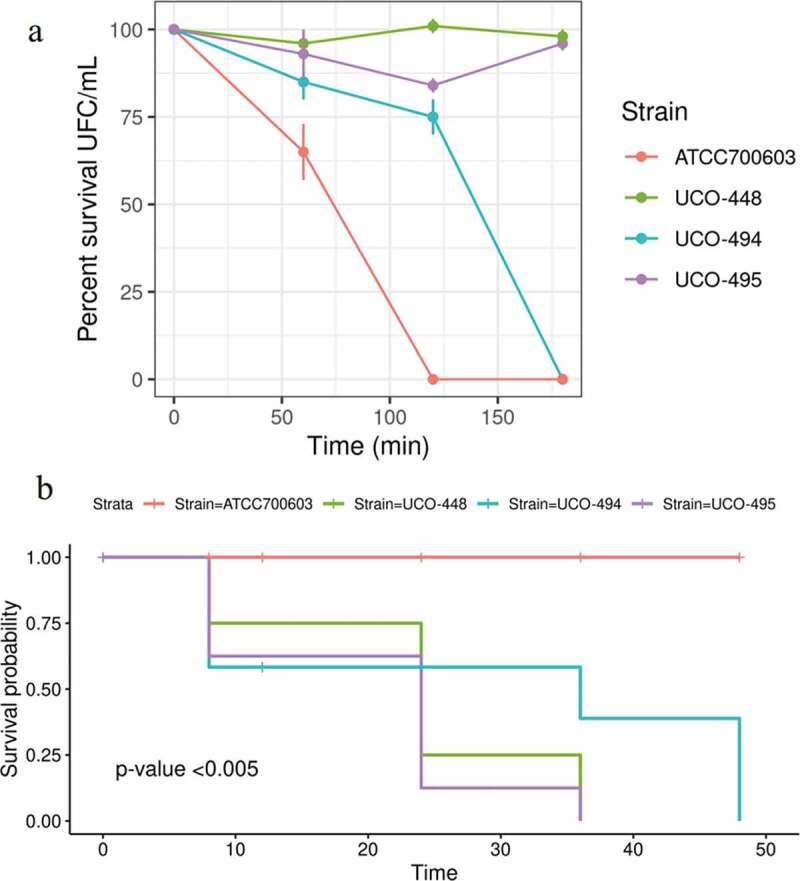
A) Serum bactericidal activity. K.quasipneumoniae subsp. similipneumoniae ATCC 700603 as negative control; K. pneumoniae hypervirulent UC-448 as positive control. b) K. pneumoniae UCO-494 and K. variicola UCO-495; Kaplan-Meier killing curves of G. mellonella larvae; ATCC 700603 as negative control; K. pneumoniae hypervirulent UC-448 as positive control; The assay was made with blank, inoculated the larvae with NaCl 0.9%. Data no showed
Figure 2.

Representative transmission electronic microscopy images of exopolysaccharide capsular UCO-494_a (K. pneumoniae UCO-494); UCO-495_a (K. variicola UCO-495) and ATCC700603_a (K.quasipneumoniae negative control) without washes; UCO494_b (K. pneumoniae UCO-494); UCO-495_b (K. variicola UCO-495) and ATCC700603_b (negative control) after washes. We estimated of capsular size in 0.124 ± 0.017; 0.097 ± 0.019 µm and 0.091 ± 0.012 µm for UCO-494_b; UCO-495_b and ATCC700603_b, respectively
On the other hand, K. variicola UCO-495 killed >75% G. mellonella larvae at 24 h post-infection, while K. pneumoniae UCO-494 killed 50% of the larvae at 24 h post-infection. Moreover, 100% mortality was observed at 36 and 48 h, respectively (Figure 1). Finally, hmKp UCO-494 displayed a low biofilm-forming ability, since it showed an OD550 nm 0.130 ± 0.003, whereas hmKv UCO-495 was classified as medium biofilm-producer since it displayed an OD550 nm value of 0.246 ± 0.021 [23].
Discussion
Traditionally, K. variicola has been considered as susceptible to most antibiotic classes, but this description has change over time, due to an increase in the MDR-K. variicola reports [1]. In South America, there is a single report in Colombia describing a KPC-2-producing K. variicola strain, which was resistant to all β-lactams [5].
Worryingly, it is the emergence of hypervirulent-MDR phenotype, especially in K. variicola isolated. In this regard, Farzana R et al. describe a fatal MDR-hvKv outbreak in neonates in Bangladesh. The isolates contained a blaCTM-M-15 and blaNDM-1 genes, among others, in addition to several virulence genes like siderophore (kfuABC) and Enterobactin (entABCDEFHIJ) associated with hypervirulent phenotype [6]. On the other hand, Lu et al. described the first hvKv isolated from blood from a patient with cholangitis in China, which was resistant to colistin (MIC = 8 ug/mL) [28]. These are concordant with our study since we identified an MDR K. variicola that was resistant to colistin. In the case of K. pneumoniae, colistin-resistant hvKp isolates has been reported previously. Specifically, Lu et al. reported five colistin-resistant hmKp strains recovered from blood samples in China [29]. Similar to our findings, these isolates were colistin-resistant and carbapenems-susceptible. Moreover, Huang et al. characterized diverse colistin-resistant hmKp isolates that were also resistant to carbapenems, since they produced the KPC-2 carbapenemase [30].
Our findings described the convergent hypervirulent phenotype and colistin-resistance in K. pneumoniae and K. variicola MDR strains. In this sense, the mutations in genes involved in colistin-resistance might be mediating this phenotype. As described previously, point mutations or deletions in pmrA or pmrB genes result in the addition of phosphoethanolamine to the lipid A [31]. Moreover, it has been demonstrated in vivo the role of PmrAB system, in which it has been associated to intra-macrophage survival and virulence in K. pneumoniae [32]. In case of hvKp UCO-494, we identified a point mutation in pmrB, similarly to the description of Lagerbäck et al., where a NDM-1-producing K. pneumoniae isolate presented an amino acid substitution in G256R in the pmrB gen [33], which was related with colistin-resistance K. pneumoniae [34]. Furthermore, it is important to highlight that the mechanism of colistin-resistance in hvKv UCO-495 was mediated by chromosomal mutations in the two-component system PhoPQ, especially in the D150G substitution in PhoP. Even though mutations in these systems are associated to colistin-resistance [30], general data of molecular mechanisms of colistin-resistance in K. variicola are scarce; therefore, our results describe a non-classical pmrAB and phoQ mutations in this species [7]. In this regard, we determined that these mutations are neutral according to in silico models, in consequence, in vivo studies should be performed in order to determine if they have an impact on colistin-resistance [16,35].
WGS analyses reflect a widely diverse resistome. In this sense, the blaLEN-25 gene was detected in the K. variicola UCO-495 genome, which corresponds to an intrinsic-chromosomal β-lactamase. Furthermore, we found that hvKv UCO-495 strain was resistant to cephalosporins, which might be mediated by blaSHV-12, while hvKp UCO-494 resistance was mediated by blaCTX-M-1. In this case, there are some reports of convergence of hypervirulent phenotype and ESBL genes in K. pneumoniae. For instance, hypervirulent and ESBL-producing have been linked to several ESBLs genes, such as blaCTX-M-14, blaCTX-M-18, blaCTX-M-3 and blaSHV-12 [36,37,38].
In case of heavy-metal resistance genes, we found in hvKv UCO-495, the tellurium resistance genes terW and terZCD, which are related to the plasmid pKV8917 [39] in hvKp and hvKv strain [1,40]. These genes were not detected in hvKp UCO-494. Relevantly, we identified the presence of the quaternary-ammonium resistance gene emrD in K. pneumoniae UCO-494. As note, these compounds have been heavily used during the SARS-CoV-2 pandemic as disinfectants, which could have an important ecological impact on selecting MDR-bacterial isolates due to selective pressure [41].
Furthermore, Moura et al. identified a K. pneumoniae serotype K19 isolate in Brazil [10]. In this study, the authors determined that this serotype has a similar killing ability compared to hypervirulent K1-isolates [10]. Moreover, the Brazilian isolate produced the ESBL CTX-M-15, which belongs to the same group of the ESBL detected in hvKp UCO-494 isolate (CTX-M-1) [42]. These findings suggest that this serotype could be endemic to South America, where could being disseminated through the region. In addition, molecular epidemiology determined by MLST revealed that hvKp UCO-494 belonged to the ST1161, which is apparently endemic to Chile since it has been detected previously in the country [43]. In the case of hmKv UCO-495, it was designated as ST173, which corresponds to a new ST that could be endemic to this geographical area. In consequence, further epidemiological studies are needed, in order to understand their prevalence and epidemiology in South America.
In the case of siderophore production, it has been demonstrated that yersiniabactin, salmochelin, and aerobactin are the most predominant in K. pneumoniae and K. variicola [44]. Specifically, the aerobactin system has four biosynthetic enzymes, iucABCD, and an outer membrane transporter, iutA [44]. Interestingly, epidemiological studies have shown a significant relationship between iucABCD-iutA with the hmKp phenotype; therefore, aerobactin is considered a substantive virulence factor in hvKp isolates [45]. However, the occurrence of multiple siderophore systems in hvKp strains suggests that siderophore systems in addition to Iuc-system play important roles in the pathogenesis of these microorganisms during either colonization or invasive processes [46].
Although all Klebsiella pneumoniae complex species could form mucoid colonies, it is well recognized the existence of two well-defined phenotypes. The classical (cKp/cKv) and hypermucoviscous (hmKp/Kv) phenotypes, both differentiated by their ability of forming a viscous and adhesive mucous string in solid media. Because of this, it is important to elucidate the mechanisms of CPS-production in hypermucoviscous K. pneumoniae strains that lack the rmpA/rmpA2 genes and do not belong to the predominant K1 or K2 serotypes [47]. In this sense, Ernst et al. studied the impact of single-nucleotide polymorphisms of the wzc gene in the capsule biosynthesis, which could confer a hypercapsule production phenotype, enhancing virulence [16]; and additionally, contribute to the resistance to polycationic peptides, such as colistin [48]. On the other hand, diverse mechanisms are related with hypercapsule production, such as mutation in wzc, rcsAB and lon protease genes [49]. Our results showed a mutation in all of this gen in hvKp UCO-494. In this sense, some authors suggest that a single amino acid substitution in wzc, rcsA or lon protease genes could increase capsule production [16], and this mechanism could be related to the hypermucoviscous phenotype in K. pneumoniae UCO-494; however, this phenomenon has not been studied in K. variicola.
In the case of virulence, the irp1 (polyketide synthetase) and irp2 (iron acquisition yersiniabactin synthesis enzyme) encode for iron-repressible high molecular weight proteins that are involved in yersiniabactin production [4]. This siderophore system was first described for Yersinia species; however, they could be also present in other Enterobacterales [50]. It is believed that its dissemination occurred via horizontal gene transfer events since the responsible genes have been identified within pathogenicity islands, such as ICEKp, which is frequently identified in K. pneumoniae [2]. The mannose-sensitive type 1 fimbriae are common in K. pneumoniae. These fimbriae are encoded by fim-like genes, in which the major components are fimA and fimH that confer its ability to adhere to human mucosal or epithelial surfaces [51]. Furthermore, other important adhesin in K. pneumoniae is the mannose-resistant Klebsiella-like (type III) codified in the fimbriae cluster mrkABCDFHIJ [52]. This is considered as a virulence factor and contributor to mucous adherence, tissue colonization, and biofilm [53]. In our case, only UCO-494 irp1 and irp2 genes.
Importantly, biofilm-formation ability of hmKp contributes to hypervirulence, since hypervirulent strains generate more biofilms in comparison with less virulent isolates [54]. Specifically, biofilms provide protection against environmental conditions, such as desiccation, and also protect bacteria from the immune system action [46]. Accordingly, diverse studies associate biofilm phenotype to capsule, and/or fimbriae; however, it has been also demonstrated that the lack of capsule enhances biofilm-formation in K. pneumoniae [46]. Our results revealed that K. pneumoniae UCO-494 presented a low biofilm-formation ability, and at the same time displayed a lower G. mellonella killing ability in comparison to K. variicola UCO-495. Moreover, hvKp UCO-494 was susceptible to the serum activity, in contrary to hvKv UCO-495 that was resistant. However, hvKp UCO-494 produced more CPS in comparison with hvKv UCO-495, which is concordant with the bacterial-size capsule, in which hvKp UCO-494 has a thicker capsule than hvKv UCO-495. These discordant results suggest that more research is needed in order to establish the specific role of biofilm-formation and virulence in Klebsiella species. In this regard, some studies have demonstrated no significant differences in biofilm-formation ability between invasive (more virulent) and noninvasive (less virulent) K. pneumoniae isolates [55]. In another study, K. pneumoniae mutant strains with decreased biofilm production ability did not show any difference in their ability to survive serum activity, which reaffirms the need for further studies in this regard.
In conclusion, we identified the convergence of hypermucoviscous phenotype and MDR K. pneumoniae and K. variicola isolates in Chile. It is important to consider the relevance of these phenotypes since they are not normally screened by a routine laboratory. Moreover, our results demonstrate the relevance of K. variicola as pathogen, due to its antibiotic-resistance and virulence features. Moreover, our results suggest that the hypermucoviscous/hypervirulent phenotype of K. pneumoniae-complex isolates is the results of multiple mechanisms, including siderophores and biofilm-production, which have not been well elucidated yet. Our results remark the need for more detailed research of the mechanisms and epidemiology of hypervirulent strains, in order to elucidate the role of high-risk K. pneumoniae-complex lineages.
Acknowledgments
The authors want to thank the microbiologists of the Hospital Dr. Hernán Henríquez Aravena, Temuco; Hospital Dr. Eduardo Schultz Schroeder, Puerto Montt; Hospital Clínico UC, Santiago; Hospital Clínico San Borja Arriarán, Santiago, Hospital Padre Hurtado, Santiago and Hospital Dr. Leonardo Guzmán Cortes, Antofagasta.
Funding Statement
This study was supported by Universidad de Concepción, Grant [VRID N° 218.074.061-1.0], the National Agency for Research and Development (ANID)/Scholarship Program/DOCTORADO NACIONAL/2016 21160336 (for FML), the Merck Investigator Studies Program (MISP) USA. NLD-128156, the National Fund for Scientific and Technological Development (FONDECYT) of Chile (FONDECYT-Iniciación, grant number 11190602 to AOC) and by the ANID Millennium Science Initiative/Millennium Initiative for Collaborative Research on Bacterial Resistance, MICROB-R, NCN17_081.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Rodríguez-Medina N, Barrios-Camacho H, Duran-Bedolla J, et al. Klebsiella variicola: an emerging pathogen in humans. Emerg Microbes Infect. 2019;8(1):973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lam MMC, Wick RR, Wyres KL, et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genomics. 2018;4. DOI: 10.1099/mgen.0.000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Piepenbrock E, Higgins PG, Wille J, et al. Klebsiella variicola causing nosocomial transmission among neonates: an emerging pathogen? J Med Microbiol. 2020. DOI: 10.1099/jmm.0.001143. [DOI] [PubMed] [Google Scholar]
- [4].Wyres KL, Lam MMC, Holt KE.. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18:344. [DOI] [PubMed] [Google Scholar]
- [5].Barrios-Camacho H, Aguilar-Vera A, Beltran-Rojel M, et al. Molecular epidemiology of Klebsiella variicola obtained from different sources. Sci Rep. 2019;9:10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Farzana R, Jones LS, Rahman MA, et al. Outbreak of hypervirulent multidrug-resistant Klebsiella variicola causing high mortality in neonates in Bangladesh. Clin Infect Dis. 2019;68:1225–1227. [DOI] [PubMed] [Google Scholar]
- [7].Lu Y, Feng Y, McNally A, et al. Occurrence of colistin-resistant hypervirulent Klebsiella variicola. J Antimicrob Chemother. 2018;73:3001–3004. [DOI] [PubMed] [Google Scholar]
- [8].Araújo BF, Ferreira ML, de Campos PA, et al. Hypervirulence and biofilm production in KPC-2-producing Klebsiella pneumoniae CG258 isolated in Brazil. J Med Microbiol. 2018;67:523–528. [DOI] [PubMed] [Google Scholar]
- [9].Guerra JM, de Fernandes NCCA, Dos Santos ALM, et al. Detection of hypermucoviscous Klebsiella pneumoniae sequence type 86 capsular type K2 in South America as an unexpected cause of a fatal outbreak in captivity marmosets. bioRxiv. 2020. DOI: 10.1101/2020.02.02.930685. [DOI] [Google Scholar]
- [10].Moura Q, Esposito F, Fernandes MR, et al. Genome sequence analysis of a hypermucoviscous/hypervirulent and MDR CTX-M-15/K19/ST29 Klebsiella pneumoniae isolated from human infection. Pathog Dis. 2017;75. DOI: 10.1093/femspd/ftx121. [DOI] [PubMed] [Google Scholar]
- [11].Cejas D, Canigia LF, Cruz GR, et al. First isolate of KPC-2-producing Klebsiella pneumonaie sequence type 23 from the Americas. J Clin Microbiol. 2014;52:3483–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Coutinho RL, Visconde MF, Descio FJ, et al. Community-acquired invasive liver abscess syndrome caused by a K1 serotype Klebsiella pneumoniae isolate in Brazil: A case report of hypervirulent ST23. Mem Inst Oswaldo Cruz. 2014;109:973–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Garza-Ramos U, Silva-Sánchez J, Martínez-Romero E, et al. Development of a Multiplex-PCR probe system for the proper identification of Klebsiella variicola. BMC Microbiol. 2015;15:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Institute Clinical & Laboratory Standards . Performance standars for antimicrobial susceptibility testing: twenty-fourth informational supplement CLSI document M100-S24. Wayne: Clinical and Laboratory Standars Institute; 2018. [Google Scholar]
- [15].Catalán-Nájera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence. 2017;8::1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ernst CM, Braxton JR, Rodriguez-Osorio CA, et al. Adaptive evolution of virulence and persistence in carbapenem-resistant Klebsiella pneumoniae. Nat Med. 2020;26:705–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ullman U, Fischer A, Podschun R. Expression of putative virulence factors by clinical isolates of Klebsiella planticola. J Med Microbiol. 2000;49:115–119. [DOI] [PubMed] [Google Scholar]
- [18].Benge GR. Bactericidal activity of human serum against strains of Klebsiella from different sources. J Med Microbiol. 1988;27:11–15. [DOI] [PubMed] [Google Scholar]
- [19].Insua JL, Llobet E, Moranta D, et al. Modeling Klebsiella pneumoniae pathogenesis by infection of the Wax Moth Galleria mellonella. Infect Immun. 2013;81:3552–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cheng HY, Chen YS, Wu CY, et al. RmpA Regulation of Capsular Polysaccharide Biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol. 2010;192:3144–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schembri MA, Blom J, Krogfelt KA, et al. Capsule and fimbria interaction in Klebsiella pneumoniae. Infect Immun. 2005;73:4626–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].O’Toole GA. Microtiter dish Biofilm formation assay. J Vis Exp. 2010. DOI: 10.3791/2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cusumano JA, Caffrey AR, Daffinee KE, et al. Weak biofilm formation among carbapenem-resistant Klebsiella pneumoniae. Diagn Microbiol Infect Dis. 2019;95:114877. [DOI] [PubMed] [Google Scholar]
- [24].Doménech-Sánchez A, Martínez-Martínez L, Hernández-Allés S, et al. Role of Klebsiella pneumoniae OmpK35 porin in antimicrobial resistance. Antimicrob Agents Chemother. 2003;47:3332–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Poulou A, Voulgari E, Vrioni G, et al. Outbreak caused by an ertapenem-resistant, CTX-M-15-producing Klebsiella pneumoniae sequence type 101 clone carrying an OmpK36 porin variant. J Clin Microbiol. 2013;51:3176–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu M-C, Lin T-L, Hsieh P-F, et al. Isolation of genes involved in biofilm formation of a Klebsiella pneumoniae strain causing pyogenic liver abscess. PLoS One. 2011;6:e23500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Amaretti A, Righini L, Candeliere F, et al. Antibiotic resistance, virulence factors, phenotyping, and genotyping of non-escherichia coli enterobacterales from the gut microbiota of healthy subjects. Int J Mol Sci. 2020;21:1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lu Y, Feng Y, McNally A, et al. The occurence of colistin-resistant hypervirulent Klebsiella pneumoniae in China. Front Microbiol. 2018;9. DOI: 10.3389/FMICB.2018.02568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Huang Y-HY-W, Chou S-H, S-W L, et al. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother. 2018;73:2039–2046. [DOI] [PubMed] [Google Scholar]
- [30].Minh-Duy P, Nhu NTK, Achard MES, et al. Modifications in the pmrB gene are the primary mechanism for the development of chromosomally encoded resistance to polymyxins in uropathogenic Escherichia coli. J Antimicrob Chemother. 2017;72:2729–2736. [DOI] [PubMed] [Google Scholar]
- [31].Cheng HY, Chen YF, Peng HL. Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J Biomed Sci. 2010;17. DOI: 10.1186/1423-0127-17-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lagerbäck P, Khine WWT, Giske CG. Evaluation of antibacterial activities of colistin, rifampicin and meropenem combinations against NDM-1-producing Klebsiella pneumoniae in 24 h in vitro time–kill experiments. J Antimicrob Chemother. 2016;71:2321–2325. [DOI] [PubMed] [Google Scholar]
- [33].Huang J, Li C, Song J, et al. Regulating polymyxin resistance in Gram-negative bacteria: roles of two-component systems PhoPQ and PmrAB. Future Microbiol. 2020;15:445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Poirel L, Jayol A, Polymyxins: NP. Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30:557–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Surgers L, Boyd A, Girard PM, et al. ESBL-producing strain of hypervirulent Klebsiella pneumoniae K2, France. Emerg Infect Dis. 2016;22:1687–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lin Z, Zheng J, Bai B, et al. Characteristics of hypervirulent Klebsiella pneumoniae: does low expression of rmpA contribute to the absence of hypervirulence? Front Microbiol. 2020;11:436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Passet V, Brisse S. Association of tellurite resistance with hypervirulent clonal groups of Klebsiella pneumoniae. J Clin Microbiol. 2015;53:1380–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rodrigues C, Passet V, Rakotondrasoa A, et al. Description of Klebsiella africanensis sp. nov., Klebsiella variicola subsp. tropicalensis subsp. nov. and Klebsiella variicola subsp. variicola subsp. nov. Res Microbiol. 2019;170:165–170. [DOI] [PubMed] [Google Scholar]
- [39].Rodríguez-Medina N, Barrios-Camacho H, Duran-Bedolla J, Garza-Ramos U. Klebsiella variicola : an emerging pathogen in humans. Emerg Microbes Infect. 2019;8:973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rezazadeh M, Baghchesaraei H, Peymani A. Plasmid-mediated quinolone-resistance (qnr) genes in clinical isolates of Escherichia coli collected from several hospitals of Qazvin and Zanjan Provinces, Iran. Osong Public Heal Res Perspect. 2016;7:307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Barría-Loaiza C, Pincheira A, Quezada M, et al. Molecular typing and genetic environment of the blaKPC gene in Chilean isolates of Klebsiella pneumoniae. J Glob Antimicrob Resist. 2016;4:28–34. [DOI] [PubMed] [Google Scholar]
- [42].Harada S, Doi Y. Hypervirulent Klebsiella pneumoniae: a call for consensus definition and international collaboration. J Clin Microbiol. 2018;56:e00959–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80:629–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Choby JE, Howard‐Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae – clinical and molecular perspectives. J Intern Med. 2020;287:283–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cubero M, Grau I, Tubau F, et al. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007-2013). Clin Microbiol Infect. 2016;22:154–160. [DOI] [PubMed] [Google Scholar]
- [46].Aghapour Z, Gholizadeh P, Ganbarov K, et al. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect Drug Resist. 2019;12:965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gottesman S, Stout V. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol Microbiol. 1991;5:1599–1606. [DOI] [PubMed] [Google Scholar]
- [48].Carniel E. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 2001;3:561–569. [DOI] [PubMed] [Google Scholar]
- [49].Murphy CN, Mortensen MS, Krogfelt KA, et al. Role of Klebsiella pneumoniae type 1 and type 3 fimbriae in colonizing silicone tubes implanted into the bladders of mice as a model of catheter-associated urinary tract infections. Infect Immun. 2013;81:3009–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Roe CC, Vazquez AJ, Esposito EP, et al. Diversity, virulence, and antimicrobial resistance in isolates from the newly emerging Klebsiella pneumoniae ST101 lineage. Front Microbiol. 2019;10:542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].El Fertas-Aissani R, Messai Y, Alouache S, et al. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol Biol. 2013;61:209–216. [DOI] [PubMed] [Google Scholar]
- [52].Kong Q, Beanan JM, Olson R, et al. Biofilm formed by a hypervirulent (hypermucoviscous) variant of Klebsiella pneumoniae does not enhance serum resistance or survival in an in vivo abscess model. Virulence. 2012;3:309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Soto E, Dennis MM, Beierschmitt A, et al. Biofilm formation of hypermucoviscous and non-hypermucoviscous Klebsiella pneumoniae recovered from clinically affected African green monkey (Chlorocebus aethiops sabaeus). Microb Pathog. 2017;107:198–201. [DOI] [PubMed] [Google Scholar]


