ABSTRACT
Chemokine-like factor (CKLF)-like MARVEL transmembrane domain containing 6 (CMTM6) modulates degradation of a number of proteins, including programmed death ligand-1 (PD-L1) by protecting it from ubiquitin-mediated degradation. In this role, it could modulate the effectiveness of immunotherapy. Here, for the first time, we characterize CMTM6 expression in melanoma and evaluate its association with response to immune checkpoint inhibitors (ICI). We evaluated the expression of CMTM6, PD-L1 and other immune-related proteins in 60 pretreatment biopsies from metastatic melanoma patients who received immunotherapy, in a tissue microarray (TMA) using quantitative immunofluorescence (QIF). Expression of mRNA from control patients obtained from The Cancer Genome Atlas (TCGA) database was also compared. CMTM6 expression was positively correlated with PD-L1, CD3, CD20, and CD68 markers, at protein (Pearson’s r = 0.53–0.81, all P < .0001) and mRNA (Spearman’s r = 0.15–0.44, all P < .002, except for CD68 where P = .26) levels. CMTM6 protein was associated with longer survival after immunotherapy when measured in the stromal (P = .007) and all the immune compartments tested (T cells, B cells, and macrophages). Multivariable analyses also revealed significant CMTM6 survival associations when measured in stromal (Hazard Ratio (HR) = 0.12, P = .001) and CD68-positive (HR = 0.30, P = .043) compartments. Additionally, PD-L1 but not CMTM6 showed prognostic value in control patients. Finally, high CMTM6 and PD-L1 co-expression in the stromal compartment was significantly associated with longer survival in treated patients (P = .028). Consequently, CMTM6 expression shows potential as a predictive factor for ICI treatments.
KEYWORDS: CMTM6, PD-L1, melanoma, immune cells, quantitative immunofluorescence
Introduction
Immune checkpoint inhibitors (ICIs), such as anti-programmed cell death 1 (PD-1) and cytotoxic T-lymphocyte associated protein 4 (CTLA-4) monoclonal antibodies, have had a tremendous impact on treatment of metastatic melanoma, reaching up to 60% response in combined therapy.1 However, there are still patients that do not benefit.1–5 The US Food and Drug Administration (FDA) has approved programmed death ligand-1 (PD-L1) assays to measure PD-L1 expression by immunohistochemistry for specific drugs and cancers such non-small cell lung cancer, bladder cancer, and head and neck squamous cell carcinoma, among others.6 However, patients with absence or low expression of PD-L1 have also shown clinical benefit to ICIs.5 Identifying biomarkers that can select patients that will benefit from immunotherapy in a more accurate way is necessary.
Chemokine-like factor (CKLF)-like MARVEL transmembrane domain containing family member 6 (CMTM6) was identified in 20037 and is widely expressed at the plasma membrane of several cell types, but only recently it has been identified as a PD-L1 protein stabilizer in two independent large-scale genetic screenings, which regulates T cell function.8,9 CMTM6 inhibits ubiquitination of PD-L1 and subsequent degradation via lysosomes, promoting stabilization of PD-L1 in the membrane. In addition, downregulation of CMTM6 gives rise to PD-L1 reduction, constitutively and interferon gamma (IFN-ɣ)-dependent, in cancer cells, dendritic cells, and xenografts derived from patients with melanoma, with no effect on PD-L1 mRNA levels.9 Furthermore, CMTM6, alone or in combination with PD-L1, has shown prognostic10–12 and predictive13,14 value in a variety of cancers. Despite all these findings, there have been few efforts to determine the predictive value of CMTM6, alone or colocalizing with PD-L1 for melanoma immunotherapy.
Given the potential value as a predictive marker, we decided to determine whether CMTM6 expression, alone and co-expressed with PD-L1, is associated with response to ICI therapy in metastatic melanoma. We explored CMTM6 and PD-L1 protein levels by quantitative immunofluorescence (QIF) in a cohort of ICI-treated patients with metastatic melanoma with known clinical outcomes. Additionally, we assessed the prognostic value of CMTM6 expression and immune-related genes utilizing a TCGA RNA-seq data set of control melanoma patients and performing QIF on tissue from a historic non-immunotherapy-treated melanoma cohort from Yale.
Materials and methods
Patient cohort and TMA construction
Tissue specimens were prepared in a tissue microarray (TMA) format as previously described.15 Pretreatment samples from 60 metastatic melanoma patients treated with immune checkpoint blockers (pembrolizumab, nivolumab, or ipilimumab plus nivolumab) from 2011 to 2017 were collected from Yale Pathology archives (cohort ITx1). The patients from the combined-treatment group received both drugs simultaneously in similar doses and cycles among the patients. The clinicopathological characteristics of the patients obtained from clinical records and pathology reports are included in Table 1, where stage refers to the moment of diagnosis. Response Evaluation Criteria in Solid Tumors (RECIST) 1.1 were used to classify best overall response as complete response (CR), partial response (PR), stable disease (SD), or progressive disease (PD), and clinical records were reviewed to determine progression-free survival (PFS). Tumor tissue was obtained from formalin-fixed, paraffin-embedded (FFPE) specimens, and 0.6 mm cores were arrayed in a recipient block in TMA format. Three additional cores from placenta were inserted in the TMA. Two independent blocks of this TMA were analyzed and averaged. If the number of cases in which target proteins were quantified differs from the total number of cases included in the TMA (n = 60), the reasons are: loss of core during TMA slide production or exclusion of cases after visual inspection for stain quality control. Additionally, a historic cohort of 131 untreated melanoma patients was used as the control group. The clinicopathological characteristics of the patients for both cohorts obtained from clinical records and pathology reports are included in Table 1. A Melanoma Index TMA was used for antibody validation. It consists of tumor tissue from 30 untreated melanoma patients from a historic cohort and control cores including placenta, tonsil, and melanoma cell lines (Yugen8, MEL624 WT, MEL624 B7-H1, MEL1335). The total of cores was 40 and two-fold redundancy was placed in the TMA. Tissues were collected with written-informed or waiver consent from patients under the approved Yale Human Investigation Committee protocol #9505008219 and conducted in accordance with the Declaration of Helsinki of 1975.
Table 1.
Clinicopathologic characteristics of the melanoma cohort treated with immune checkpoint inhibitors and non-immunotherapy-treated melanoma cohort
| Yale ITx melanoma cohort (YTMA376 – n = 60) |
Yale non-immunotherapy-treated cohort (YTMA76 – n = 131) |
|
|---|---|---|
| Characteristics | n (%) | n (%) |
| Sex | ||
| Female | 26 (43) | 73 (56) |
| Male | 34 (57) | 50 (38) |
| Unknown | 8 (6) | |
| Age | ||
| <60 | 24 (40) | |
| ≥60 | 36 (60) | |
| Mean±SD (range) | 61 ± 16.8 (6–86) | |
| Stage | ||
| I | 10 (17) | 63 (48) |
| II | 11 (18) | 38 (29) |
| III | 22 (37) | 18 (14) |
| IV | 12 (20) | |
| Unknown | 4 (8) | 12 (9) |
| RECIST groups | ||
| CR/PR | 26 (43) | |
| SD | 18 (30) | |
| PD | 16 (27) | |
| Treatment | ||
| Nivolumab | 11 (18) | |
| Pembrolizumab | 24 (40) | |
| Ipilimumab plus Nivolumab |
25 (42) | |
| Prior immune checkpoint blockade | ||
| Yes | 16 (27) | |
| No | 44 (73) | |
| Mutation status | ||
| BRAF | 19 (32) | |
| NRAS | 8 (13) | |
| CKIT | 1 (3) | |
| None detected | 31 (52) | |
|
Location of primary tumor |
||
| Extremity | 57 (44) | |
| Trunk | 66 (50) | |
| Unknown | 8 (6) | |
|
Follow-up in months DSOS – Median (IQT) |
76.9 (64) |
Multiplexed immunofluorescence staining protocol
CMTM6 (RCT6, Absea) antibody has been previously validated by our lab and others.9,13 Briefly, TMA sections were deparaffinized, then subjected to antigen retrieval with 1 mM EDTA (pH 8) at 97°C for 20 min in a Lab Vision PT Module (Thermo Scientific, Waltham, MA, USA). Endogenous peroxidases were blocked with 2.5% hydrogen peroxide in methanol for 30 min, followed by additional 30 min of incubation with 0.3% bovine serum albumin with 0.05% Tween-20 blocking solution. Subsequently, we performed a sequential multiplexed immunofluorescence staining combining CMTM6 (1 µg/ml) with different primary antibodies: panel 1 – PD-L1 (clone SP142, 0.1 µg/ml, Abcam), CD68 (clone PG-M1, 1:200, Agilent); panel 2 – CD3 (clone SP7, 1:100, Novus Biologicals), CD20 (clone L26, 1:150, Agilent). Isotype-specific horseradish peroxidase (HRP)-conjugated secondary antibodies were incubated with the TMAs at room temperature for 1 h before tyramide-based labeling for 10 min followed by 1 mM benzoic hydrazide with 0.15% hydrogen peroxide for 7 min twice to quench HRP activity. For panel 1, the secondary antibodies were anti-mouse IgG1 (1:100, Thermo Fisher Scientific), anti-rabbit EnVision reagent (Agilent), and anti-mouse IgG3 (1:1000, Abcam), and the substrates were TSA Plus cyanine (Cy)5 tyramide, TSA Plus Cy3 tyramide, and biotin tyramide, respectively, all from PerkinElmer. Sections were then treated with streptavidin–Alexa Fluor 750 conjugate (1:100, Invitrogen). For panel 2, the secondary antibodies were anti-mouse IgG1 (1:100, Thermo Fisher Scientific), anti-rabbit EnVision reagent (Agilent), and anti-mouse IgG2a (1:200, Abcam), and the substrates were TSA Plus Cy5 tyramide, biotin tyramide (1:50, PerkinElmer), and TSA Plus Cy3 tyramide, respectively, all from PerkinElmer. Sections were then treated with streptavidin–Alexa Fluor 750 conjugate (Invitrogen). Finally, both panels used mouse anti-S100 (clone 15E2E2, 1:100, BioGenex) and HMB45 (1:100, BioGenex) and then goat anti-mouse Alexa Fluor 488 antibody (1:100, Thermo Fisher Scientific) for 1 h to identify melanoma cells, as counterstaining 4ʹ,6-diamino-2-phenylindole (DAPI) to visualize nuclei and ProLong Gold Antifade (Invitrogen) was used for mounting. Control slides from index melanoma TMA were included in each staining experiment to verify reproducibility.
Fluorescence signal quantification and cutpoint selection
We used the Automated Quantitative Analysis (AQUA™) method (Navigate BioPharma) to quantify the fluorescent signal of CMTM6, PD-L1, macrophages and TILs as previously described.16 AQUA method allows to create molecular compartments based on fluorescent signal of specific cell markers (e.g., CD3+ for T cells) and quantify the expression of a given target in the corresponding compartment. For example, after defining cells positive for S100 and HMB45, a tumor mask will be generated and the intensity of our target, CMTM6, will be measured. CMTM6 was measured in five compartments: tumor compartment, created by binarizing the S100 and HMB45 signal; stromal compartment, created by excluding the tumor mask (a dilated tumor compartment) from a dilated DAPI mask representing the total tissue; and three immune compartments such as CD68-positive macrophage compartment, CD3-positive T cell compartment, and CD20-positive B cell compartment. QIF score was calculated by dividing the target pixel intensity by the area of the compartment of interest and then normalized to the exposure time and bit depth at which the images were captured. Cases with staining artifacts or presence of less than 3% compartment area were excluded after visual inspection.
We performed staining and target measurement in two independent TMA blocks, with each block containing one nonadjacent tumor core per patient; the average target QIF scores were calculated for each case. Merged images displayed in Figures 1–3 were combined using ImageJ (RRID:SCR_001935). Different cutpoints, specified in the text, were used to split tumors into high and low expression for each marker. X-tile software17 (Yale University, New Haven, CT, USA) was used to determine an optimal cutpoint.
Figure 1.
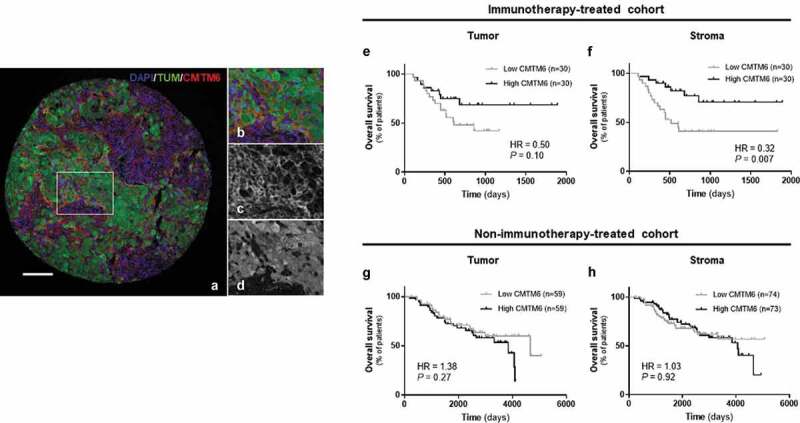
Distribution of CMTM6 expression and its indicative value in human melanoma tissue. A-D) Representative case with tumor and immune cells positive for CMTM6, being a) an overlapped image of the whole core with DAPI (nuclei) in blue, TUM (tumor cells = HMB45+ and S100+) in green, and CMTM6 in red (scale bar = 100 µm); b-d) region from (a) image showing the three channels overlapped (b), only CMTM6 (c), and only TUM (d). Overall survival based on CMTM6 expression in the tumor (e) and stromal (f) compartments in patients treated with immunotherapy, and overall survival for the non-immunotherapy-treated cohort based on CMTM6 expressed in tumor (g) or in stroma (h)
Figure 2.
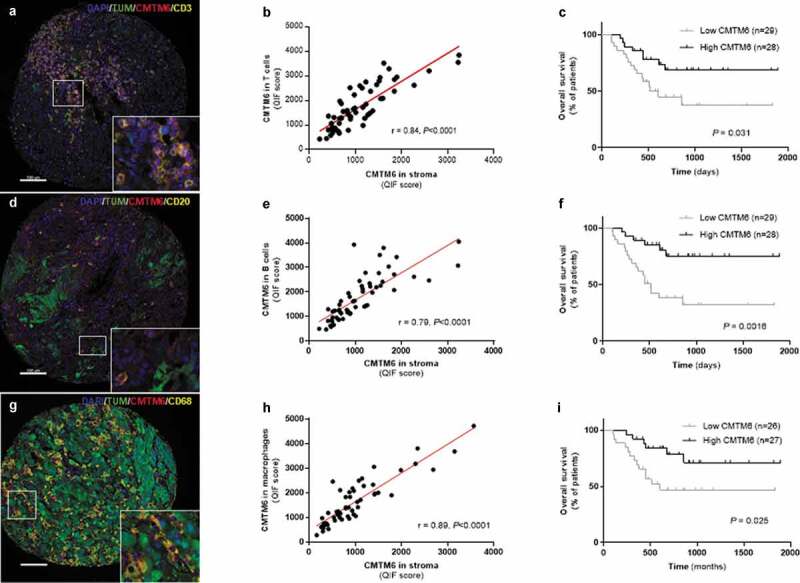
CMTM6 in the immune populations from immunotherapy-treated patients with melanoma. Representative images showing expression of CMTM6 in T cells (a), B cells (d), and macrophages (g) by colocalization of CMTM6 and CD3, CD20, and CD68 markers, respectively. Correlation between CMTM6 QIF scores in the stromal compartment versus CMTM6 expression in T cells (b), in B cells (e), and in macrophages (h). Overall survival based on CMTM6 expression in T cells (c), in B cells (f), and in macrophages (i) in patients treated with immunotherapy. r = Pearson’s correlation coefficient; DAPI, 4ʹ6-diamino-2-phenylindole; TUM (tumor cells = HMB45+ and S100+) (scale bar = 100 µm)
Figure 3.
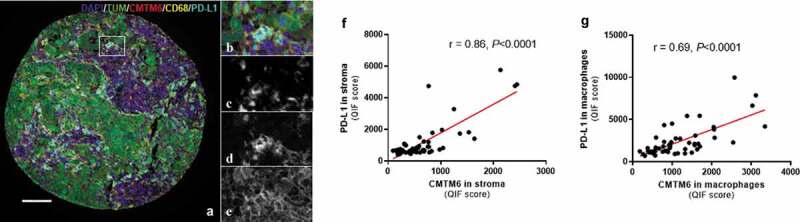
Expression of CMTM6 and PD-L1 in melanoma. Representative image of melanoma tissue expressing CD68 (c), PD-L1 (d), and CMTM6 (e) individually and overlapping (a, b) (scale bar = 100 µm). Correlation between CMTM6 and PD-L1 expression in stromal (f) and macrophage (g) compartments. r = Pearson’s correlation coefficient; DAPI, 4ʹ6-diamino-2-phenylindole; TUM (tumor cells = HMB45+ and S100+)
Gene expression data analysis
RNA-Seq expression data for human melanoma were downloaded from the TCGA data set through www.cbioportal.org. In this study, RNA-Seq by Expectation-Maximization (RSEM) expression values of relevant genes were applied. Correlation plots were generated by GraphPadTM Prism® v7.0 for Windows. After assessing normality of the data set by D’Agostino & Pearson and Shapiro-Wilk tests, Pearson’s or Spearman’s correlation coefficients were calculated whether the data set was normally distributed or not respectively, including statistical significance.
Statistical analysis
Based on D’Agostino & Pearson and Shapiro-Wilk tests results for normality, Pearson’s or Spearman’s correlation coefficients (r) were used to analyze the linear association between two continuous variables and used the non-parametric t-test Mann-Whitney to compare groups. Overall survival (OS) and PFS curves were constructed using the Kaplan-Meier analysis and statistical significance was determined using the log-rank test. Unadjusted univariable analysis was conducted. Multivariable Cox proportional hazards models included age, sex, mutation status, stage, specimen category, treatment, and prior immune checkpoint blockade as covariates.18–20 For statistical analysis, the average AQUA scores from two independent cores of each case was used. All statistical tests were two-sided, and P-values below 0.05 were considered statistically significant. All statistical analyses were performed using GraphPadTM Prism® v7.0 software, and JMP Pro software (version Pro 13, SAS Institute Inc, Cary, NC).
Results
CMTM6 protein expressed in stroma was associated with immunotherapy benefit in melanoma but not with prognosis
After finding the optimal concentration of anti-CMTM6 (RCT6) antibody for melanoma tissue, previously validated in lung tissue,13 we evaluated CMTM6 expression on pre-treatment samples from patients with melanoma who received immunotherapy. We detected CMTM6 in both tumor and stromal cells with a membranous or cytoplasmic localization (Figure 1(a-d)), as observed in NSCLC.13 Assessing CMTM6 expression quantitatively, the levels were significantly higher in CR/PR group than in SD or PD groups (Supplementary Fig. S1A and S1B). Although there were not significant differences when the patients were grouped based on durable clinical benefit (Supplementary Fig. S1F and S1G), CMTM6 expression was significantly higher in responders than in non-responders according to objective response rate (ORR) classification (Supplementary Fig. S1K and S1L), in both stromal and tumor compartments (P = .0028 and P = .015, respectively). Moreover, splitting the population at the median value showed that high levels of CMTM6 protein in the stromal, which includes mainly fibroblasts and immune cells, but not in the tumor compartment was significantly associated with a longer OS after PD-1 axis blockade (Figure 1(e,f), Table 2). These results were confirmed by multivariable analyses (Supplementary Table S1). Next, we measured CMTM6 expression in a historical cohort of melanoma patients that did not receive immunotherapy to assess its prognostic value. There were no statistically significant differences between high and low CMTM6 protein level in the control cohort, either in tumor nor stroma, in terms of survival (Figure 1(g,h), Table 2, Supplementary Table S1). Taking together, these results support CMTM6 as an indicative biomarker for immunotherapy in melanoma, a term that can be used instead of predictive, since interaction tests cannot be performed without an untreated control arm.
Table 2.
Summary of CMTM6 expression as a predictive marker for PFS and OS measured in tumor, stromal, T cell, B cell, and macrophage compartments using log-rank (Mantel-Cox) test for the immunotherapy-treated and the non-immunotherapy-treated melanoma cohorts. PFS = Progression-free survival; OS = Overall survival; DFS = Disease-free survival; HR = Hazard ratio; 95% CI = 95% Confidence interval; H = high; L = low (cut point = median). In bold text, statistically significant p values
| Tumor | Stroma | T lymphocytes | B lymphocytes | Macrophages | ||||||||||||
|
Variable (H/L) |
HR (95% CI) |
P value |
HR (95% CI) |
P value |
HR (95% CI) |
P value |
HR (95% CI) |
P value |
HR (95% CI) |
P value |
||||||
| Immunotherapy-treated cohort | PFS | 0.79 (0.42–1.47) |
0.45 | 0.69 (0.37–1.31) |
0.24 | 0.69 (0.36–1.32) |
0.25 | 0.69 (0.36–1.31) |
0.25 | 0.81 (0.40–1.62) |
0.54 | |||||
| OS | 0.50 (0.23–1.12) |
0.10 | 0.32 (0.14–0.72) |
0.007 | 0.41 (0.18–0.91) |
0.031 | 0.25 (0.11–0.57) |
0.0016 | 0.35 (0.14–0.87) |
0.025 | ||||||
|
Non-immunotherapy- treated cohort |
DFS | 1.03 (0.53–2.00) |
0.93 | 1.01 (0.56–1.82) |
0.98 | |||||||||||
| OS | 1.38 (0.77–2.47) |
0.27 | 1.03 (0.61–1.74) |
0.92 | ||||||||||||
Immune cells expressed CMTM6 and its level indicated response to immunotherapy in melanoma patients
The tumor microenvironment is composed of many different cell types, each of which plays some role either tumorigenesis or the immune response to control tumor progression. Many of those cells, such as lymphocytes (T cells, B cells, NK cells), macrophages, dendritic cells, and fibroblasts among others21 can be defined by expression of specific proteins. Here, we modified a previously published multiplexed TIL immunofluorescence panel22 (Figure 2(a,d)) to determine the association between CMTM6 and some stromal cells in melanoma. There was a high correlation between stromal CMTM6 expression and CMTM6 expressed in CD3+ T lymphocytes (r = 0.84, P < .0001) and in CD20+ B lymphocytes (r = 0.79, P < .0001) (Figure 2(b,e)). Furthermore, patients who responded to immunotherapy had significantly higher levels of CMTM6 protein in CD3+ cells and CD20+ cells compared with non-responders (Supplementary Figure 2(c,d,m,n)). PFS for CMTM6 levels expressed in both lymphocyte subpopulations were comparable between high and low protein groups (Table 2). However, OS was significantly longer in treated patients with high levels of CMTM6 expression in TILs (Figure 2(c,f); Table 2), especially in CD20 compartment (B cells) (P = .0016; HR = 0.25; 95% CI = 0.11–0.57). In unadjusted univariable Cox proportional hazard analysis, CMTM6 protein expressed by B cells was significantly associated with longer OS but not with PFS (Supplementary Table S1). However, it did not remain statistically significant by multivariable analysis (Supplementary Table S1). In Cox regression analyses, CMTM6 in CD3 compartment was not associated with either PFS nor OS (Supplementary Table S1). As expected, high CD3 [(PFS: P = .042; HR = 0.042; 95% CI = 0.27–1.76) (OS: P = .0004; HR = 0.20; 95% CI = 0.09–0.45)] but not CD20 [(PFS: P = .47; HR = 0.79; 95% CI = 0.41–1.51) (OS: P = .14; HR = 0.55; 95% CI = 0.25–1.22)] was associated with PFS and OS when measured in the stromal compartment, as previously described for this cohort.23,24 Taken together, we found CMTM6 protein was expressed in lymphocytes, and high levels of CMTM6 protein in these immune cells were associated with longer OS.
Additionally, we examined the relationship between CMTM6 and lymphocyte infiltration in TCGA RNA-seq data from 443 control patients with melanoma who did not receive immunotherapy. There was a statistically significant correlation between CMTM6 and CD3E (r = 0.15, P = .0019) and CD20 (= MS4A1) (r = 0.23, P < .0001) gene expression, corresponding to T and B lymphocytes, respectively (Supplementary Fig. S2A and S2B). This correlation was higher at protein level measured by QIF in the ITx-1 cohort (Supplementary Fig. S2F and S2G). When we analyzed the OS from the TCGA data set available for 434 patients using median as a cutpoint, CMTM6 was not prognostic (Supplementary Fig. S3A), whereas high levels of CD3E mRNA were correlated with a longer OS (P < .0001; HR = 0.58; 95% CI = 0.45–0.76) (Supplementary Fig. S3F). Similar results were observed for CD20 gene expression (P < .0001; HR = 0.56; 95% CI = 0.43–0.73) (Supplementary Fig. S3G). Additionally, we confirmed Ki67 (MKi67) as a poor prognostic marker in melanoma by analyzing its expression on the TCGA cohort as a control of our analysis (Supplementary Fig. S3H), as previously described.25 In summary, CMTM6 mRNA correlated with lymphocyte markers, such as CD3E and MS4A1, in RNA-seq data from TCGA for melanoma. Moreover, CMTM6 was not prognostic, supporting what we found at protein level in our historical control cohort survival (Figure 1(g,h), Table 2, Supplementary Table S1).
NK cells have been shown to have a role in killing tumor cells by themselves and in supporting antitumor T cell activity through IFNɣ secretion.26 Moreover, they have been used in clinical trials as infusions leading to engineered NK cells with improved activity against tumor.26 There is a wide variety of surface markers that NK cells express depending on their state of maturation and activation. CD56 is expressed by NK cells along their maturation and it’s commonly used as general NK marker.27 We decided to explore the expression of NK-related genes from TCGA RNA-seq data of control melanoma patients. Supplementary Fig. S4 summarizes the correlation between CMTM6 and multiple NK-associated genes related to maturation and activation status. Overall, CMTM6 demonstrated a high statistically significant correlation with the majority of NK genes assessed, supporting the hypothesis of a relationship of CMTM6 and NK cells in melanoma. NKp44 was not correlated with CMTM6, most likely due to the low level of expression of this gene (n = 59). Interestingly, CD56 showed very low correlation or absent with CMTM6 and the rest of the NK genes, which could be explained by CD56 being expressed not only in NK cells but also in other immune cells and even tumors cells, as we observed in our melanoma samples (data not shown). Taken together, this indicates that CMTM6 could have a function in NK cells in melanoma.
Finally, we determined whether CMTM6 was associated with macrophages by multiplexing CMTM6 and CD68 antibodies using quantitative immunofluorescence in the immunotherapy-treated cohort (Figure 2(g)). As observed for TILs, there was a high correlation between CMTM6 protein measured in the stromal compartment and the CD68 compartment (r = 0.89, P < .0001, Figure 2(h)). Additionally, positive correlation was also seen between CD68 and CMTM6 total mRNA in the TCGA control cohort (Supplementary Fig. S2C) and CD68 and CMTM6 total protein in the immunotherapy-treated cohort (Supplementary Fig. S2H). When survival analysis was performed, high levels of CMTM6 protein expressed in macrophages were associated with longer survival in the immunotherapy cohort (Figure 2(i), Table 2), even upon multivariable analysis (Table S1). Similarly, CD68 and PD-L1 total levels of mRNA (Supplementary Fig. S2E) and protein (Supplementary Fig. S2J) were highly correlated. Altogether, patients with high levels of CMTM6 protein in macrophages showed a longer survival compared to the low expressers in the immunotherapy cohort.
Co-expression of PD-L1 and CMTM6 proteins did not improve CMTM6 protein alone performance as a predictive biomarker
The CMTM6 mechanism of action is not completely understood, but one of its functions is as a stabilizer of PD-L1 in the cell by protecting it from degradation.9 Since CMTM6 has been described to colocalize with PD-L1 in human lung cancer tissue,13 we assessed the association of these two molecules in tumor cells and macrophages from melanoma patients that received immunotherapy. Visually, CMTM6 and PD-L1 colocalization was mostly observed in macrophages (Figure 3(a-e)). There was a high correlation between CMTM6 and PD-L1 protein levels in stroma (Figure 3(f)) and in macrophages (Figure 3(g)), together with total mRNA level (r = 0.37, P < .0001; Supplementary Fig. S2D) and total protein level (r = 0.74, P < .0001; Supplementary Fig. S2I). PD-L1 alone was not associated with better outcome regardless the cutpoint used: median, optimal cutpoint or top tertile (Figure 4(a-c), Supplementary Table S2). Next, we assessed outcomes in four subgroups based on high or low CMTM6 and PD-L1 co-expression levels (Supplementary Fig. S5). Median PFS was comparable between the four phenotypes (Supplementary Fig. S5B, S5C, and S5D, Supplementary Table S1).13 However, OS was significantly longer in patients with high co-expression of CMTM6 and PD-L1 in the stroma compared to low levels of both and the rest of three phenotypes (Figure 4(d-f), Supplementary Fig. S5E, S5F, and S5G, Supplementary Table S1). No marker co-expression phenotype was significantly associated with survival by multivariable Cox proportional hazard analysis after adjustment for age, sex, mutation status, stage, treatment, specimen category, prior immune checkpoint blockade (Supplementary Table S1).
Figure 4.
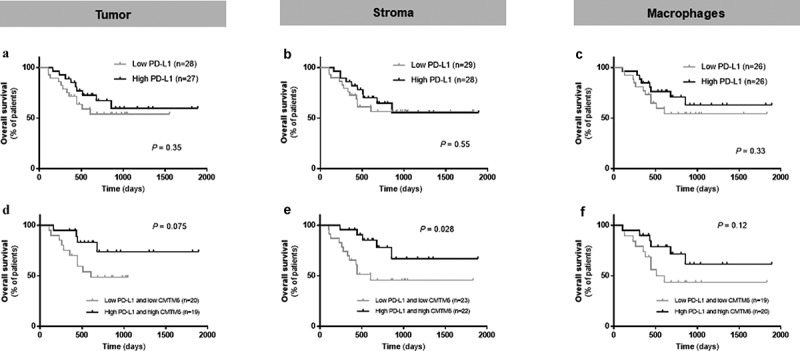
Predictive performance of CMTM6 and PD-L1 in immunotherapy-treated patients with melanoma. Overall survival corresponding to PD-L1 expression alone in the tumor compartment (a), the stromal compartment (b), and in macrophages (c) in patients who received immune checkpoint inhibitors. Overall survival in patients with high levels of both CMTM6 and PD-L1 compared with co-expression of low levels of CMTM6 and PD-L1 in the tumor compartment (d), the stromal compartment (e), and in macrophages (f)
PD-L1 protein colocalized with CMTM6 protein in macrophages at higher proportion than in tumor cells
To confirm the colocalization of CMTM6 and PD-L1, we used the recently published13 formula for AQUA analysis that we created for similar analysis in NSCLC. This formula allows calculation of the percentage of pixels per unit area where the pixels were above the threshold for both CMTM6 and PD-L1 and then divided that value by the number of pixels within the compartment of interest (tumor, stromal, or CD68). Macrophages showed a significantly higher colocalization of PD-L1 and CMTM6 than either tumor or stromal compartments (Supplementary Fig. S5A).
High PD-L1 mRNA levels are associated with longer survival in non-immunotherapy-treated patients with melanoma
Finally, we assessed whether the co-expression of CMTM6 and PD-L1 (CD274) had a prognostic significance at mRNA level on the TCGA control cohort. We used the median as a cutpoint for each marker individually and then combined them. When they were analyzed individually, PD-L1 but not CMTM6 was linked to longer survival (Supplementary Fig. S3A and S3B). Patients expressing high levels of PD-L1 mRNA showed a significantly higher median OS compared with the rest (low PD-L1 and low CMTM6: mOS = 50 months; low PD-L1 and high CMTM6: mOS = 62 months), either simultaneously expressing high (mOS = 111 months) or low (mOS = 117 months) levels of CMTM6 mRNA (P < .0001) (Supplementary Fig. S3C). Moreover, when we combined grouped patients based on high or low PD-L1 gene expression regardless CMTM6 mRNA level, the differences between median OS were statistically significant (high PD-L1 and low/high CMTM6: mOS = 111 months; low PD-L1 and low/high CMTM6: mOS = 58 months; P < .0001) (Supplementary Fig. S3D). This result indicates that PD-L1 is driving the prognostic value in the combination of CMTM6 and PD-L1 mRNA markers and it does not improve on survival stratification when PD-L1 mRNA is analyzed individually (117 months versus 58 months; P < .0001).
Discussion
In melanoma, immunotherapy based on immune checkpoint blockade has emerged as a highly effective treatment. However, the mechanisms of action of the checkpoints are not completely understood. As a result, the reasons why only a subset of patients respond to the therapy remain unclear.28 Recently, CMTM6 has been identified as a key positive regulator of PD-L1 expression8,9 and was related to prognosis and response to immunotherapy in a variety of tumor types.10,11,14 Here, we found that CMTM6 protein was broadly expressed in melanoma tissue, in both tumor and stromal compartments, and was associated with response to ICI therapy when measured in the stromal compartment. In multivariable analysis, CMTM6 was also found to be an independent predictor of response to immunotherapy. Previous work done by our lab on NSCLC showed a similar trend,13 and others have shown similar results by IHC assays also in NSCLC.14 The most probable explanation is that CMTM6 function will depend on the cell/tissue where it is expressed and, therefore, its role could be tumor-specific. To assess the prognostic value of CMTM6, we evaluated its expression in a historical cohort from our institution of melanoma patients who did not receive immunotherapy. We did not find an association between CMTM6 expression and survival at protein level. This result was supported by TCGA RNA-seq data from melanoma patients who did not receive immunotherapy. Next, we were able to measure CMTM6 protein expression in three immune cell populations: T lymphocytes, B lymphocytes, and macrophages. Additionally, protein expression of CMTM6 and markers for the above cell types (CD3, CD20, and CD68, respectively) were highly correlated and was associated with good outcome. CMTM6 might be stabilizing PD-L1 expression in these three immune populations, since macrophages8,9,13 and lymphocytes29 express PD-L1. This suggests an intricate relationship of suppressive and costimulatory function between different immune populations and/or tumor cells, without excluding the possibility of additional roles for CMTM6 in lymphocytes. Equally important, CMTM6 was correlated with NK-related genes at mRNA in a control melanoma cohort. Taken together, these results point out a broad role for CMTM6 in immune modulation, as previously suggested.12
Colocalization of CMTM6 and PD-L1 in tumor cells, stroma and CD68-positive macrophages seen here confirms previously published data.8,9,13 Both markers were correlated at both protein level in our ICI-treated cohort and at mRNA level from TCGA RNA-seq data of control patients. However, the association between both markers was statistically significant only when measured in the stromal compartment. Interestingly, the phenotypes with similar levels of CMTM6 and PD-L1, either high or low, represented the highest number of patients, although there were also some cases without this relationship, indicating that CMTM6 is not the only mediator of PD-L1 expression and is likely to have multiple functions beyond its interactions with PD-L1. CMTM6 expression has been shown to be independent of IFN-g pathway activation,8,10,30 which is a well-characterized regulator of PD-L1 expression.31 Recently, CMTM6 has been shown to be involved in lipid uptake during atherogenesis32,33 and regulation of activated and exhausted T cells in the TME and cancer stem cells.10 In addition, there have been described many regulators of PD-L1 expression such as protein stabilizers CMTM49 and COP9 signalosome 5,34 and PD-L1 motifs related to posttranslational modification regulation.35
The relationship between CMTM6 expression and outcome remains controversial. We did not find prognostic significance using TCGA RNA-seq data from control melanoma patients, supporting similar results obtained at protein level in NSCLC.13 Nevertheless, CMTM6 expression has been associated with poor prognosis for glioma,12 head and neck squamous cell carcinoma,10 and pancreatic cancer.11 On the other hand, high levels of CMTM6 have been linked to good prognosis in triple-negative breast cancer.11 These results together with the fact that CMTM6 is widely expressed in many cell types and multiple cancers11 suggest that CMTM6 might have variable roles depending on the cell type and tumor in which it is expressed. Interestingly, PD-L1 expression was associated with a good prognosis in the TCGA control melanoma cohort, which supports previously published data for melanoma36 and cervical cancer37. On the other hand, PD-L1 has shown negative prognostic value in other cancers.38–40
This study has a number of limitations. Firstly, the immunotherapy-treated cohort we evaluated by QIF is a retrospective collection with mixed therapies, not a clinical trial. In clinical trials, PD-L1 is a controversial predictive biomarker for immunotherapy in melanoma.41–43 In our study, we did not observe a significant association between PD-L1 expression and survival for any of the compartments in which it was measured even testing a range of cutpoints. Previously, our group found that PD-L1 expression in CD68-positive cells was a predictive marker for PFS and OS for the same cohort.24 This discrepancy could be explained by use of different methodologies (QIF versus NanoString GeoMx Digital spatial profiling, respectively). Further studies using larger independent cohorts will be needed to clarify the role of PD-L1 as a predictive marker in metastatic melanoma. Another limitation of the study is the use of median as a cutpoint. This cut-point was not optimized since we do not have a second independent validation set of ICI-treated patients. Lastly, TMAs were used to evaluate predictive value of biomarkers instead of whole tissue sections. In order to partially take into account tumor heterogeneity, experiments included two cores from separate regions of the same tumor for each patient. But this still represents a limitation since clinical biomarkers are always assessed on whole tissue slides.
In summary, we demonstrated that CMTM6 is expressed in both tumor and immune cells in melanoma and is a predictive marker of OS. Additionally, CMTM6 is highly correlated with PD-L1 at both protein and mRNA level.
Supplementary Material
Acknowledgments
The authors thank Lori A. Charette and the staff of Yale Pathology Tissue Service for expert histology services.
Funding Statement
This work was funded by grants from Navigate Biopharma and the NIH/NCI including the Yale SPORE in Skin Cancer P50 CA121974 (PIs: M. Bosenberg and H. Kluger) and the Yale SPORE in Lung Cancer P50 CA196530 (PI: Roy Herbst) and the Yale Cancer Center Support Grant, P30CA016359 (PI: Charles Fuchs)
Author contributions
SMM and DLR designed the study, planned the experiments, interpreted the data, and drafted the manuscript. JZ and SMM developed the methodology. SMM performed the experiments and analyzed the data. PFW and HMK acquired and managed patients. All the authors critically reviewed the manuscript.
Disclosure of potential conflicts of interest
JZ has received consulting honoraria from Guardant Health. PFW currently works at Verily Life Sciences, Alphabet Inc. (CA, USA). HMK reports receiving commercial research grants from Merck, Bristol-Myers Squibb, and Apexigen, and is a consultant/advisory board member for Nektar, Biodesix, Genentech, Merck, Celldex, Pfizer, Iovance, Elevate Bio, Array Biopharma and Immunocore. DLR declares that in the last two years he has served as a consultant to Astra Zeneca, Agendia, Amgen, Bethyl Labs, Biocept, BMS, Cell Signaling Technology, ClearSight, Daiichi Sankyo, InVicro/Konica-Minolta, Navigate BioPharma, Merck, OptraScan, Perkin Elmer, and Ultivue. No potential conflicts of interest were disclosed by SMM.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D, Linette GP, Meyer N, Giguere JK, Agarwala SS, et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med. 2015;372:2006–10. doi: 10.1056/NEJMoa1414428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. New England J Medicine. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber JS, D’Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B, Hoeller C, Khushalani NI, Miller WH, Lao CD, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncology. 2015;16:375–384. doi: 10.1016/S1470-2045(15)70076-8. [DOI] [PubMed] [Google Scholar]
- 4.Larkin J, Hodi FS, Wolchok JD, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P.. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma REPLY. New England J Medicine. 2015;373:1270–1271. doi: 10.1056/NEJMoa1504030. [DOI] [PubMed] [Google Scholar]
- 5.Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob JJ, Cowey CL, Lao CD, Wagstaff J, Schadendorf D, Ferrucci PF, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. New England J Medicine. 2017;377:1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diggs LP, Hsueh EC.. Utility of PD-L1 immunohistochemistry assays for predicting PD-1/PD-L1 inhibitor response. Biomark Res. 2017;5:12. doi: 10.1186/s40364-017-0093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han W, Ding P, Xu M, Wang L, Rui M, Shi S, Liu Y, Zheng Y, Chen Y, Yang T, et al. Identification of eight genes encoding chemokine-like factor superfamily members 1-8 (CKLFSF1-8) by in silico cloning and experimental validation. Genomics. 2003;81:609–617. doi: 10.1016/S0888-7543(03)00095-8. [DOI] [PubMed] [Google Scholar]
- 8.Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, Lam EYN, Henderson MA, Bell CC, Stolzenburg S, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101–105. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, Logtenberg MEW, Slagter M, Rozeman EA, Hofland I, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:106–110. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Yang QC, Li YC, Yang LL, Liu JF, Li H, Xiaou Y, Bu LL, Zhang WF, Sun ZJ. Targeting CMTM6 suppresses stem cell-like properties and enhances antitumor immunity in head and neck squamous cell carcinoma. Cancer Immunol Res. 2019;8(2):179-191. [DOI] [PubMed] [Google Scholar]
- 11.Mamessier E, Birnbaum DJ, Finetti P, Birnbaum D, Bertucci F. CMTM6 stabilizes PD-L1 expression and refines its prognostic value in tumors. Ann Transl Med. 2018;6:54. doi: 10.21037/atm.2017.11.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan X, Zhang C, Zhao J, Sun G, Song Q, Jia W. CMTM6 overexpression is associated with molecular and clinical characteristics of malignancy and predicts poor prognosis in gliomas. EBioMedicine. 2018;35:233–243. doi: 10.1016/j.ebiom.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zugazagoitia J, Liu Y, Toki M, McGuire J, Ahmed FS, Henick BS, Gupta R, Gettinger SN, Herbst RS, Schalper KA, et al. Quantitative assessment of CMTM6 in the tumor microenvironment and association with response to PD-1 pathway blockade in advanced-stage non-small cell lung cancer. J Thorac Oncol. 2019;14:2084–2096. doi: 10.1016/j.jtho.2019.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh YW, Han JH, Haam S, Jung J, Lee HW. Increased CMTM6 can predict the clinical response to PD-1 inhibitors in non-small cell lung cancer patients. Oncoimmunology. 2019;8:e1629261. doi: 10.1080/2162402X.2019.1629261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giltnane JM, Rimm DL. Technology insight: identification of biomarkers with tissue microarray technology. Nat Clin Pract Oncol. 2004;1:104–111. doi: 10.1038/ncponc0046. [DOI] [PubMed] [Google Scholar]
- 16.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 17.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 18.Eton O, Legha SS, Moon TE, Buzaid AC, Papadopoulos NE, Plager C, Burgess AM, Bedikian AY, Ring S, Dong Q, et al. Prognostic factors for survival of patients treated systemically for disseminated melanoma. J Clin Oncol. 1998;16:1103–1111. doi: 10.1200/JCO.1998.16.3.1103. [DOI] [PubMed] [Google Scholar]
- 19.Manola J, Atkins M, Ibrahim J, Kirkwood J. Prognostic factors in metastatic melanoma: a pooled analysis of Eastern cooperative oncology group trials. J Clin Oncol. 2000;18:3782–3793. doi: 10.1200/JCO.2000.18.22.3782. [DOI] [PubMed] [Google Scholar]
- 20.Joosse A, Collette S, Suciu S, Nijsten T, Patel PM, Keilholz U, Eggermont AMM, Coebergh JWW, de Vries E. Sex is an independent prognostic indicator for survival and relapse/progression-free survival in metastasized stage III to IV melanoma: a pooled analysis of five European organisation for research and treatment of cancer randomized controlled trials. J Clin Oncol. 2013;31:2337–2346. doi: 10.1200/JCO.2012.44.5031. [DOI] [PubMed] [Google Scholar]
- 21.Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B, McGraw T, Mittal V. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer. 2019;19:9–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown JR, Wimberly H, Lannin DR, Nixon C, Rimm DL, Bossuyt V. Multiplexed quantitative analysis of CD3, CD8, and CD20 predicts response to neoadjuvant chemotherapy in breast cancer. Clin Cancer Res. 2014;20:5995–6005. doi: 10.1158/1078-0432.CCR-14-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong PF, Wei W, Smithy JW, Acs B, Toki MI, Blenman KRM, Zelterman D, Kluger HM, Rimm DL. Multiplex quantitative analysis of tumor-infiltrating lymphocytes and immunotherapy outcome in metastatic melanoma. Clin Cancer Res. 2019;25:2442–2449. doi: 10.1158/1078-0432.CCR-18-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toki MI, Merritt CR, Wong PF, Smithy JW, Kluger HM, Syrigos KN, Ong GT, Warren SE, Beechem JM, Rimm DL, et al. High-plex predictive marker discovery for melanoma immunotherapy-treated patients using digital spatial profiling. Clin Cancer Res. 2019;25:5503–5512. doi: 10.1158/1078-0432.CCR-19-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vereecken P, Laporte M, Heenen M. Significance of cell kinetic parameters in the prognosis of malignant melanoma: a review. J Cutan Pathol. 2007;34:139–145. doi: 10.1111/j.1600-0560.2006.00588.x. [DOI] [PubMed] [Google Scholar]
- 26.Shimasaki N, Jain A, Campana D. NK cells for cancer immunotherapy. Nat Rev Drug Discov. 2020;19(3):200-218. [DOI] [PubMed] [Google Scholar]
- 27.Van Acker HH, Capsomidis A, Smits EL, Van Tendeloo VF. CD56 in the immune system: more than a marker for cytotoxicity? Front Immunol. 2017;8:892. doi: 10.3389/fimmu.2017.00892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamazaki T, Akiba H, Iwai H, Matsuda H, Aoki M, Tanno Y, Shin T, Tsuchiya H, Pardoll DM, Okumura K, et al. Expression of programmed death 1 ligands by murine T cells and APC. J Immunol. 2002;169:5538–5545. doi: 10.4049/jimmunol.169.10.5538. [DOI] [PubMed] [Google Scholar]
- 30.Brockmann M, Blomen VA, Nieuwenhuis J, Stickel E, Raaben M, Bleijerveld OB, Altelaar AFM, Jae LT, Brummelkamp TR. Genetic wiring maps of single-cell protein states reveal an off-switch for GPCR signalling. Nature. 2017;546:307–311. doi: 10.1038/nature22376. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X, et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19(6):1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, et al.Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domschke G, Linden F, Pawig L, Hafner A, Akhavanpoor M, Reymann J, Doesch AO, Erbel C, Weber C, Katus HA, et al. Systematic RNA-interference in primary human monocyte-derived macrophages: A high-throughput platform to study foam cell formation. Sci Rep. 2018;8(1):10516. doi: 10.1038/s41598-018-28790-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu Y, Chang -S-S, Lin W-C, Hsu J-M, Hsu Y-H, et al. Deubiquitination and stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30:925–939. doi: 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gato-Canas M, Zuazo M, Arasanz H, Ibanez-Vea M, Lorenzo L, Fernandez-Hinojal G, Vera R, Smerdou C, Martisova E, Arozarena I, et al. PDL1 signals through conserved sequence motifs to overcome interferon-mediated cytotoxicity. Cell Rep. 2017;20(8):1818–1829. doi: 10.1016/j.celrep.2017.07.075. [DOI] [PubMed] [Google Scholar]
- 36.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, Chen S, Klein AP, Pardoll DM, Topalian SL, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4(127):127ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karim R, Jordanova ES, Piersma SJ, Kenter GG, Chen L, Boer JM, Melief CJM, van der Burg SH. Tumor-expressed B7-H1 and B7-DC in relation to PD-1+ T-cell infiltration and survival of patients with cervical carcinoma. Clin Cancer Res. 2009;15:6341–6347. doi: 10.1158/1078-0432.CCR-09-1652. [DOI] [PubMed] [Google Scholar]
- 38.Fakih M, Ouyang C, Wang C, Tu TY, Gozo MC, Cho M, Sy M, Longmate JA, Lee PP. Immune overdrive signature in colorectal tumor subset predicts poor clinical outcome. J Clin Invest. 2019;129:4464–4476. doi: 10.1172/JCI127046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohigashi Y, Sho M, Yamada Y, Tsurui Y, Hamada K, Ikeda N, Mizuno T, Yoriki R, Kashizuka H, Yane K, et al. Clinical significance of programmed death-1 ligand-1 and programmed death-1 ligand-2 expression in human esophageal cancer. Clin Cancer Res. 2005;11:2947–2953. doi: 10.1158/1078-0432.CCR-04-1469. [DOI] [PubMed] [Google Scholar]
- 40.Thompson RH, Kuntz SM, Leibovich BC, Dong H, Lohse CM, Webster WS, Sengupta S, Frank I, Parker AS, Zincke H, et al. Tumor B7-H1 is associated with poor prognosis in renal cell carcinoma patients with long-term follow-up. Cancer Res. 2006;66(7):3381–3385. doi: 10.1158/0008-5472.CAN-05-4303. [DOI] [PubMed] [Google Scholar]
- 41.Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372(4):320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- 42.Daud AI, Wolchok JD, Robert C, Hwu WJ, Weber JS, Ribas A, Hodi FS, Joshua AM, Kefford R, Hersey P, et al. Programmed death-ligand 1 expression and response to the anti-programmed death 1 antibody pembrolizumab in melanoma. J Clin Oncol. 2016;34:4102–4109. doi: 10.1200/JCO.2016.67.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


