ABSTRACT
Placenta-specific 1 (PLAC1) is expressed primarily in placental trophoblasts but not in normal tissues and is a targetable candidate for cancer immunotherapy because it is a cancer testis antigen known to be up-regulated in various tumors. Although peptide epitopes capable of stimulating CD8 T cells have been previously described, there have been no reports of PLAC1 CD4 helper T lymphocyte (HTL) epitopes and the expression of this antigen in head and neck squamous cell carcinoma (HNSCC). Here, we show that PLAC1 is highly expressed in 74.5% of oropharyngeal and 51.9% of oral cavity tumors from HNSCC patients and in several HNSCC established cell lines. We also identified an HTL peptide epitope (PLAC131-50) capable of eliciting effective antigen-specific and tumor-reactive T cell responses. Notably, this peptide behaves as a promiscuous epitope capable of stimulating T cells in the context of more than one human leukocyte antigen (HLA)-DR allele and induces PLAC1-specific CD4 T cells that kill PLAC1-positive HNSCC cell lines in an HLA-DR-restricted manner. Furthermore, T-cells reactive to PLAC131-50 peptide were detected in the peripheral blood of HNSCC patients. These findings suggest that PLAC1 represents a potential target antigen for HTL based immunotherapy in HNSCC.
KEYWORDS: Placenta-specific 1, PLAC1, head and neck squamous cell carcinoma, CD4 T cells, helper T lymphocytes, epitope, peptide vaccine, immunotherapy
Introduction
Head and neck tumors, including cancers of the lip, oral cavity, nasal and sinonasal cavity, pharynx, larynx and salivary glands, are very frequent worldwide, with an estimated 890,000 new cases and 450,000 deaths in 2018.1,2 The most common histological type of head and neck cancer is squamous cell carcinoma (head and neck squamous cell carcinoma or HNSCC). Improvements in combination therapy with surgery, chemotherapy, radiotherapy and molecular targeting drugs (cetuximab) have reduced the mortality associated with HNSCC. However, more than 65% of HNSCC patients develop recurrent and/or metastatic disease.2,3 Recently, programmed death-1 immune-checkpoint inhibitors were approved as a treatment for platinum chemotherapy-treated patients with recurrent or metastatic HNSCC and have shown durable responses and survival improvements, albeit in a small number of patients.2 Thus, there is a medical need to improve immunotherapies for HNSCC.
Cancer vaccines capable of eliciting tumor-specific T-cell responses, such as the use of tumor antigen-derived synthetic peptides, are a potential therapeutic approach that is being explored.4–6 While peptide vaccines might be regarded as insufficient to elicit robust anti-tumor responses because of disappointing results in past clinical trials, recent research progress has shown that the weak efficacy of peptide vaccines might be overcome by optimization of peptide conformation, routes of administration and the use of effective immune adjuvants.5–7 Moreover, combination of peptide vaccines with immune-checkpoint inhibitors has demonstrated significant anti-tumor effects in preclinical studies.8,9 Thus, the time has arrived to reconsider the clinical effects of peptide vaccines as well as their advantages, such as ease of synthesis and cost-effectiveness compared to other immunotherapeutic agents. In regard to the components of tumor-specific T cells induced by peptide vaccines, both CD8 cytotoxic T lymphocytes (CTLs) and CD4 helper T lymphocytes (HTLs) play important roles in generating effective anti-tumor responses. The differentiation, growth and cytotoxic activity of CTLs require the support of HTLs.10–12 In addition, several investigators, including ourselves, have reported that tumor-reactive HTLs can exhibit direct tumor cell cytotoxicity via perforin and granzyme B.13–17 Interestingly, the recent report by Hashimoto et al.18 revealed that a marked increase of CD4 T cells having cytotoxic features is observed in supercentenarians, people who have reached 110 years of age, compared to younger people, suggesting that cytotoxic CD4 T cells might be related to protection against the development of tumors and the achievement of exceptional longevity. The latest study reported by Oh et al.19 also revealed that cytotoxic CD4 T cells were clonally expanded in bladder cancer and killed autologous tumor cells in a human leukocyte antigen (HLA) class II-dependent manner. Furthermore, it is known that HTLs play a critical role in the activation of innate cells, such as macrophages and NK cells, to contribute to anti-tumor responses.20 Therefore, the identification of helper epitope peptides derived from tumor antigens capable of inducing tumor-reactive HTLs could be a significant improvement for developing effective immunotherapies for HNSCC.
It would be desirable that a selected tumor antigen exhibits high immunogenicity and is essential for tumor development, as well as being expressed at high levels in tumor cells and is absent (or expressed at extremely low levels) in normal tissues.4,5,21 Cancer testis antigens (CTAs) are defined as proteins expressed in various types of cancer cells, but not in normal tissues, except for testis and/or placenta, and are ideal antigen targets for developing peptide vaccines.5,21,22 Placenta-specific 1 (PLAC1) is an X-linked gene product that plays a role in placental development.23 PLAC1 is expressed primarily in trophoblast lineage cells during pregnancy and its expression is tightly restricted in normal tissues.21,24 Recent studies have shown that PLAC1 is also expressed in various cancers, such as breast,25–27 lung,25,28,29 gastrointestinal,30–36 gynecological,37–40 and prostate,41 and can modulate tumor progression. Thus, PLAC1 is regarded as a CTA that may have potential to induce anti-tumor immunity with minimal risk of autoimmunity as a target of peptide vaccines. Some researchers have identified HLA-A2-restricted CTL epitopes of PLAC1 and evaluated the responses against tumor cells.33,35,42 However, to the best of our knowledge, there are no studies examining the capacity of CD4 HTL to recognize PLAC1 and whether this antigen is expressed in HNSCC.
Here, we report that PLAC1 is clearly expressed in HNSCC using patient tissues and cultured cell lines. In addition, we have identified a peptide epitope capable of inducing HTL responses. These HTLs released effector cytokines and exerted cytotoxicity against PLAC1-expressing HNSCC cell lines in an HLA-DR-restricted manner. Furthermore, we revealed that precursor T cells responding to PLAC1 helper epitope peptide were present in HNSCC patients. These results suggest that PLAC1 has potential as a target antigen for immunotherapy in HNSCC patients.
Materials and methods
Tissue samples
Tissue samples of oropharyngeal and oral cavity squamous cell carcinoma were respectively obtained at the pretreatment period from 59 and 52 Japanese patients who were diagnosed and treated at Asahikawa Medical University (Asahikawa, Japan) between 2004 and 2019. Clinical characteristics of the patients are summarized in Supplemental Tables S1 and S2. Staging of tumors was classified according to the 8th edition of the International Union Against Cancer TNM staging system. Patients were considered current or former smokers if they had smoked at least 100 cigarettes in their lifetime, and current or former drinkers if they had consumed alcoholic beverages at least once a week for 1 year or more during their lifetime, as previously described by other investigators.43 Patients considered former smokers and drinkers had quit for at least 1 year prior to presentation. Informed consent was obtained by the opt-out method on the Asahikawa Medical University website. Tissue studies and use of clinical data were conducted with the approval of the Institutional Review Board at Asahikawa Medical University.
Immunohistochemistry
Immunohistochemical (IHC) analysis of the expression of PLAC1 and HLA-DR was carried out on 4-μm-thick formalin-fixed, paraffin-embedded (FFPE) tissues from HNSCC patients. Anti-PLAC1 (G-1, 1:200, Santa Cruz Biotechnology, Santa Cruz, CA) and anti-HLA-DR (TAL.1B5, 1:100, DAKO, Glostrup, Denmark) mouse monoclonal antibodies (mAbs) were used as the primary antibodies (Abs). FFPE specimens were stained in a VENTANA Benchmark GX (Roche Diagnostics, Rotkreuz, Switzerland) using Cell Conditioning 1 or 2 buffer (Roche Diagnostics) as antigen retrieval solutions and a VENTANA ultraView Universal DAB Detection Kit (Roche Diagnostics). Representative images were acquired using a BZ-X710 microscope (Keyence, Tokyo, Japan). Staining intensity scores for PLAC1 and HLA-DR in tumor cells were graded as follows: 0, no staining; 1, weak; 2, moderate; 3, strong. Quantity scores for PLAC1 and HLA-DR consisted of the percentage of positively stained tumor cells and were graded as 0, <5%; 1, 5–25%; 2, 26–50%; 3, >50% and 0, <10%; 1, 10–25%; 2, 26–50%; 3, >50%, respectively. The IHC score was calculated by the sum of the staining intensity score and the quantity score in reference to previous reports.16,41,44 IHC scores ≥4 were defined as high expression and <4 as low expression. Two pathologists independently scored all stained sections.
Cell lines
Four human HNSCC cell lines, HSC3 (tongue SCC, HLA-DR15), HSC4 (tongue SCC, HLA-DR1/4, 53), Sa-3 (gingival SCC, HLA-DR9/10, 53) and SAS (tongue SCC, HLA-DR9/15, 53); two human lung carcinoma cell lines, Lu65 (large cell carcinoma, HLA-DR4/15, 53) and EBC1 (lung SCC, HLA-DR1); and two human T-cell leukemia cell lines, Jurkat (HLA-DR negative) and Molt-4 were used in this study. HSC3, HSC4, Sa-3, Lu65 and EBC1 were supplied by the RIKEN BioResource Center (Tsukuba, Ibaraki, Japan). SAS, Jurkat and Molt-4 were purchased from the American Type Culture Collection (ATCC; Manassas, VA). Mouse fibroblast cell lines (L-cells) expressing individual human HLA-DR molecules (DR4, DR8, DR15 and DR53) were kindly provided by Dr. T. Sasazuki (Kyushu University, Fukuoka, Japan) and Dr. Robert W. Karr (Karr Pharma, St. Louis, MO). All cell lines were maintained in tissue culture as recommended by the supplier.
Western blotting
Tumor cell lines (2 × 106 cells) were washed in phosphate buffered saline (PBS). Cell lysates were extracted using a Total Protein Extraction Kit for Animal Cultured Cells and Tissues (Invent Biotechnologies, Inc., Plymouth, MN), subjected to electrophoresis in a 4–12% NuPAGE Bis-Tris SDS-PAGE gel (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA) and transferred to an Immunobilon-P membrane (Merck Millipore, Burlington, MA), followed by blocking of the membrane using PBS with 0.01% Tween 20 and 5% nonfat dry milk at room temperature. After 1 h, the membrane was incubated with anti-PLAC1 rabbit polyclonal Ab (Abgent, San Diego, CA) diluted 1:1000 in blocking buffer at 4°C overnight or anti-β-actin mouse mAb (C4, Santa Cruz Biotechnology) diluted 1:3000 in blocking buffer as the internal control for 2 h at room temperature. After washing, the membrane was incubated with horseradish peroxidase-labeled sheep anti-rabbit or anti-mouse IgG and visualized using an Amersham ECL Prime Western Blotting Detection System (GE Healthcare Life Sciences, Logan, UT).
Flow cytometry
HLA-DR expression on the surface of tumor cell lines was examined by flow cytometry using fluorescein isothiocyanate (FITC)-conjugated anti-HLA-DR mAb (G46-6, BD Pharmingen, San Diego, CA) as previously described.45 FITC-conjugated mouse IgG2a Ab (MOPC-173, BioLegend, San Diego, CA) was utilized as an isotype control. All tumor cell lines were incubated with or without 500 IU/ml interferon gamma (IFN-γ) for 48 h before analysis. The fluorescence of samples was measured using a BD Accuri C6 flow cytometer and the data were analyzed using the supplied software (BD Biosciences, San Jose, CA).
Synthetic peptides
The amino acid sequence of PLAC1 potentially binding common HLA-DR molecules, including DRB1*0101, DRB1*0401, DRB1*0701, DRB1*1101 and DRB1*1501, was predicted using two computer-based algorithms, SYFPEITHI (http://www.syfpeithi.de/)46 and Immune Epitope Database Analysis Resource (IEDB, https://www.iedb.org/).47 PLAC131-50 (SIDWFMVTVHPFMLNNDVCV) was selected as the potential epitope candidate in reference to sequences that showed high scores in both databases and was synthesized by GeneScript (Tokyo, Japan). Pan DR Epitope (PADRE) peptide (aK-Cha-VAAWTLKAAa, a = D-alanine; and Cha = I-cyclohex-ylalanine) was used as a positive control for activating CD4 T cells.48
In vitro generation of PLAC1-specific CD4 helper T cells with synthetic peptide
The procedure for the generation of peptide-specific CD4 helper T cells from peripheral blood mononuclear cells (PBMCs) of healthy individuals has been described in detail previously.49 Briefly, monocytes and CD4 T cells were purified from PBMCs using MACS microbeads for CD14 and CD4, respectively (Miltenyi Biotec, Cologne, Germany). Dendritic cells (DCs) were produced from the monocytes in the presence of granulocyte macrophage colony-stimulating factor (GM-CSF) (50 ng/ml) and interleukin (IL)-4 (1000 IU/ml) for 7 days. PLAC131-50 peptide-pulsed DCs (3 μg/ml for 3 h at room temperature) were co-cultured with autologous CD4 T cells in 96-well flat-bottomed culture plates. One week later, the CD4 T cells were restimulated in individual microcultures with PLAC131-50 peptide-pulsed (3 μg/ml) γ-irradiated autologous PBMCs, and 2 days later, recombinant human IL-2 (10 IU/ml) was added. After the two cycles of peptide stimulation, antigen reactivity of CD4 T cells to PLAC1 peptide was assessed by measuring cytokine production levels using ELISA kits for GM-CSF, IFN-γ (both BD Pharmingen) and granzyme B (MABTECH, Stockholm, Sweden). The absorption of the supernatants was measured at 450 nm using a Glomax Discover Microplate Reader (Promega, Madison, WI). AIM-V medium (Invitrogen, Carlsbad, CA) supplemented with 3% human male AB serum (Innovative Research, Novi, MI) was used as complete culture medium for all experiments. All blood materials were acquired after obtaining signed informed consent.
Analysis of PLAC1-specific responses with established CD4 T-cell lines
Established CD4 helper T cells (1–1.5 × 105) were mixed with antigen-presenting cells (APCs), which consisted of irradiated autologous PBMCs (1–1.5 × 105), HLA-DR-expressing L-cells (3 × 104) or tumor cell lines (3 × 104), in the presence or absence of various concentrations of PLAC131-50 peptide in 96-well culture plates. Tumor cell lines were exposed to IFN-γ (500 IU/ml) for 48 h to increase HLA-DR expression, and IFN-γ was then eliminated before the assay. To verify antigen specificity and HLA-DR restriction, the blockade of antigen presentation was evaluated by adding anti-HLA-DR mAb L243, which was prepared from the supernatants of hybridoma HB-55 (ATCC) or anti-HLA-A, B and C mAb W6/32 (ATCC) at 10 μg/ml through 48 h incubation. Supernatants were collected and evaluated for the production of GM-CSF, IFN-γ and granzyme B using commercial ELISA kits as mentioned above.
Cytotoxicity assay
Cytotoxic activity of PLAC1-specific CD4 T cells was evaluated by flow cytometry using a BD Accuri C6 flow cytometer and software (BD Biosciences). HSC3, HSC4, Lu65 (matched HLA-DR with PLAC131-50-specific CD4 T cells), and EBC1 (unmatched HLA-DR) were labeled using a CellTraceTM CFSE Cell Proliferation Kit (Invitrogen; Thermo Fisher Scientific, Inc.) after pretreatment with IFN-γ (500 IU/ml) for 48 h. After the labeled tumor cell lines were cultured with PLAC131-50-specific CD4 T cells for 6 h, they were collected and stained using 7-ADD viability staining solution (BioLegend) to detect dead cells. Cytotoxicity of PLAC131-50-specific CD4 T cells was measured at various effector/target cell (E:T) ratios (0:1, 10:1, 20:1, or 40:1).
Measurement of PLAC1 peptide-specific responses in HNSCC patients
PBMCs were collected from 12 HNSCC patients and cultured at 2–3 × 105/well with PLAC131-50 or PADRE peptide (10 μg/ml) in 96-well culture plates as described previously.50 One week later, the cultures were restimulated with peptide-pulsed (10 μg/ml) irradiated autologous PBMCs (5 × 104/well). After one week of restimulation, the amount of IFN-γ in the supernatants was assayed by ELISA. All HNSCC patients signed informed consent forms. The Institutional Review Board of Asahikawa Medical University approved this study.
Statistical analysis
An unpaired Student’s t-test was used to compare CD4 T-cell responses between two different conditions. P values <.05 were considered statistically significant. GraphPad Prism 7 (GraphPad Software, San Diego, CA) was used for analyses.
Results
Evaluation of the expression levels of PLAC1 and HLA-DR in HNSCC tissues
To examine whether PLAC1 is expressed in HNSCC, we initially performed IHC analysis using tissue samples from 59 patients with oropharyngeal squamous cell carcinoma (OPSCC). The clinical characteristics of patients are summarized in Supplemental Table S1. PLAC1 was mainly localized in the cytoplasm of tumor cells, and the expression level was scored at four levels (0, no staining; 1, weak; 2, moderate; 3, strong) based on the staining intensity (Figure 1A). We simultaneously evaluated the percentage of PLAC1-positive tumor cells and classified them into four groups (0, <5%; 1, 5–25%; 2, 26–50%; 3, >50%) to determine the quantity score. As shown in Figure 1B, all patients were distributed into seven categories by the IHC score (IHC score 0, 1, 2, 3, 4, 5, or 6), which was calculated by the sum of the staining intensity and quantity scores. The number of patients in each IHC score was 6 (10.2%) in IHC score 0, 2 (3.4%) in score 1, 3 (5.1%) in score 2, 4 (6.8%) in score 3, 20 (33.8%) in score 4, 18 (30.5%) in score 5, and 6 (10.2%) in score 6. Similarly, we assessed the IHC score for HLA-DR by defining the staining intensity score (Figure 1A) and quantity score (0, <10%; 1, 10–25%; 2, 26–50%; 3, >50%). As shown in Figure 1C, the number of patients in each IHC score of HLA-DR was 28 (47.5%) in IHC score 0, 6 (10.2%) in score 1, 5 (8.4%) in score 2, 3 (5.1%) in score 3, 5 (8.4%) in score 4, 3 (5.1%) in score 5, and 9 (15.3%) in score 6. An IHC score ≥4 was considered as high expression and <4 as low expression; PLAC1 and HLA-DR were highly expressed in 74.5% (44/59) and 28.8% (17/59), respectively (Figure 1B, C and D). Furthermore, 12 of 59 cases (20.3%) showed high expression for both PLAC1 and HLA-DR (Figure 1D). In addition, we examined the expression of PLAC1 and HLA-DR in tumor tissues from 52 patients with oral cavity squamous cell carcinoma (OCSCC, summarized in Supplemental Table S2) in the same way (Supplemental Figure S1). Of the OCSCC patients, 51.9% (27/52) and 34.6% (18/52) highly expressed PLAC1 and HLA-DR, respectively (Supplemental Figure S1B, C and D). High expression for both PLAC1 and HLA-DR was found in 9 of 52 cases (17.3%, Supplemental Figure S1D). In both OPSCC and OCSCC samples, there was no correlation between PLAC1 and HLA-DR IHC scores (Supplemental Figure S2). We also examined the relationship between the expression of PLAC1 and clinical features of OPSCC and OCSCC patients, including tobacco, alcohol, human papillomavirus (HPV) status and tumor stage; however, no significant correlations were found (Supplemental Tables S3 and S4). Several studies51,52 have suggested that HNSCC exhibits molecular heterogeneity and has been classified into four distinct molecular subtypes (basal, mesenchymal, atypical and classical) by clustering analysis of gene expression of tumor tissues. Therefore, we compared the expression of PLAC1 and HLA-DR between the four molecular subtypes using the gene expression data of GSE39366 in the Gene Expression Omnibus (GEO) database,52 which contains 138 HNSCC samples and includes PLAC1, HLA-DRA and HLA-DRB5 gene expression. As shown in Supplemental Figure S3, there was no significant difference in the expression of these genes between the four molecular subtypes, except for the expression of HLA-DRB5 between mesenchymal and classical type (2-fold difference, p < .05), indicating that PLAC1 and HLA-DR expression in HNSCC may be remotely related to molecular subtypes. Overall, these results suggest an HTL-based vaccine could be developed for use in some OPSCC and OCSCC patients that co-express PLAC1 and HLA-DR if peptide epitopes were to be identified.
Figure 1.
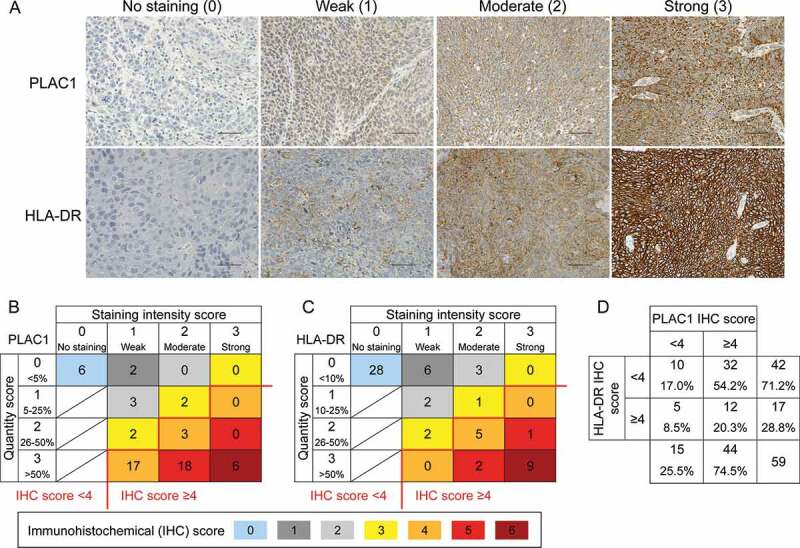
Expression of PLAC1 and HLA-DR in OPSCC specimens. (A) Representative immunohistochemical (IHC) images of PLAC1 and HLA-DR. Expression levels of tumor cells were classified into no, weak, moderate and strong staining by IHC staining intensity. Scale bar = 50 μm. (B) Distribution of IHC scores for PLAC1. The IHC score was calculated by the sum of the staining intensity score (0, no staining; 1, weak; 2, moderate; 3, strong) and the quantity score (the percentage of positively stained tumor cells: 0, <5%; 1, 5–25%; 2, 26–50%; 3, >50%). (C) Distribution of the IHC score of HLA-DR. The IHC score was calculated in the same way as described in B, although quantity was scored as 0, <10%; 1, 10–25%; 2, 26–50%; 3, >50%. In B and C, the IHC score could range from 0 to 6. An IHC score ≥4 was defined as high expression, and the others were considered as low expression. (D) Distribution of high and low expression of PLAC1 and HLA-DR based on IHC scores
Expression of PLAC1 and HLA-DR in HNSCC cell lines
We next assessed the expression of PLAC1 protein in cultured tumor cell lines, including HNSCC cells (HSC3, HSC4, SAS and Sa-3), using western blot analysis (Figure 2A). PLAC1 was expressed in all HNSCC tumor cell lines, but not in the Molt-4 T cell leukemia line or PBMCs from a healthy individual. The expression of HLA-DR on the surface of PLAC1-positive tumor cell lines, with or without IFN-γ pretreatment, was investigated using flow cytometric analysis (Figure 2B). HSC3, HSC4, SAS, Sa-3 and EBC1 cells had no detectable levels of HLA-DR; however, pretreatment with IFN-γ up-regulated HLA-DR expression. On the other hand, Lu65 cells constitutively expressed high levels of HLA-DR. Jurkat T cell leukemia cells did not express HLA-DR, regardless of pretreatment with IFN-γ. We further examined the expression of PLAC1 in HPV-positive HNSCC cell lines (SCC090, SCC152 and UM-SCC-47) because HSC3, HSC4, SAS and Sa-3 cells are negative for HPV. Supplemental Figure S4A shows that PLAC1 was expressed in all three HPV-positive HNSCC cell lines, whereas these cells did not express HLA-DR with or without IFN-γ treatment (Supplemental Figure S4B). These results indicate that HNSCC cell lines might express PLAC1 regardless of HPV status.
Figure 2.
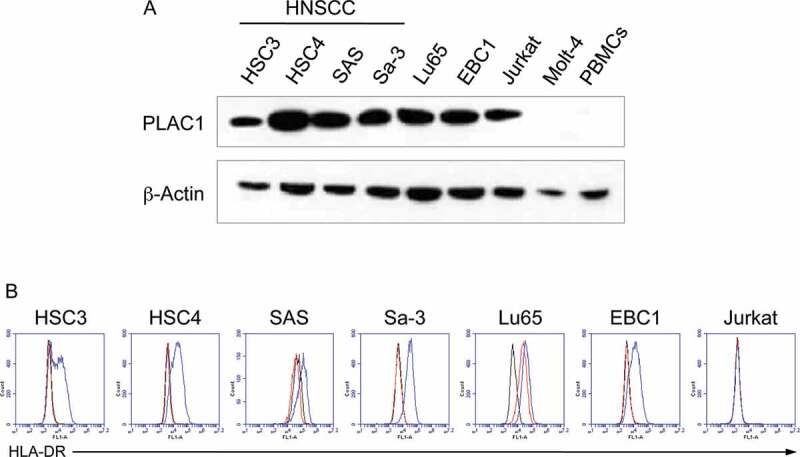
Expression of PLAC1 and HLA-DR in HNSCC cell lines. (A) Western blotting analysis of PLAC1 expression in tumor cell lines. β-Actin was used to confirm the amount of loaded protein. (B) Cell surface expression of HLA-DR on tumor cell lines as determined by flow cytometry. Cell lines were incubated with or without IFN-γ (500 IU/ml) for 48 h. The black, red and blue lines show isotype control staining, HLA-DR staining of IFN-γ-untreated cells and that of IFN-γ-treated cells, respectively
In vitro generation of PLAC1-specific CD4 helper T cells
In view of the above findings, we investigated whether PLAC1 peptide-specific CD4 HTLs could be generated in vitro from human PBMCs. To this end, a PLAC131-50 peptide (SIDWFMVTVHPFMLNNDVCV) was selected according to the two computer-based algorithms that predict the capacity of peptide sequences to bind to HLA-DR molecules. Analysis of genetic mutations in the Head and Neck Squamous Cell Carcinoma (The Cancer Genome Atlas (TCGA), PanCancer Atlas) dataset revealed that only 0.6% of HNSCC patients have mutations in PLAC1, and mutation sites were not detected in the PLAC131-50 peptide (Supplemental Figure S5). Interestingly, we observed that this peptide contained two HLA-A2-restricted CTL epitopes, PLAC131-39 (SIDWFMVTV) and PLAC141-50 (FMLNNDVCV).33,35,42 CD4 T cells isolated from PBMCs of healthy volunteers were stimulated with PLAC131-50 peptide-pulsed autologous DCs as described in the Materials and Methods. Three PLAC131-50-specific CD4 T-cell lines, T4, Y6 and H9, were induced from three healthy donors, T (HLA-DR8/15), Y (HLA-DR4/15, 53) and H (HLA-DR4/9, 53), respectively (Figure 3). A peptide titration curve was first performed to estimate the induced responses the CD4 T cells to the PLAC131-50 peptide. All three CD4 T-cell lines produced GM-CSF (Figure 3A) and IFN-γ (Supplemental Figure S6A) with increasing concentrations of PLAC131-50 peptide. To confirm the presentation of PLAC131-50 peptide through HLA-DR, PLAC131-50-specific CD4 T cells were incubated with peptide-pulsed autologous PBMCs in the presence of an anti-HLA-DR mAb (L243) that specifically reacts with HLA-DR and does not cross-react with other HLA class II molecules such as HLA-DP and DQ. Anti-HLA class I mAb (W6/32) was used as a negative control. As shown in Figure 3B and Supplemental Figure S6B, the response of the three CD4 T-cell lines to PLAC131-50 peptide was significantly blocked by L243 mAb but not by W6/32 mAb, indicating that the recognition of peptide by PLAC131-50-specific CD4 T cells was restricted by HLA-DR. To determine which HLA-DR alleles are responsible for antigen presentation, the reactivity of PLAC131-50-specific CD4 T cells was evaluated using several L-cells as APCs expressing individual HLA-DR molecules. L-cells expressing HLA-DR15 presented PLAC131-50 peptide to two T-cell lines, T4 and Y6 (Figure 3C and Supplemental Figure S6C), whereas L-cells expressing HLA-DR53 stimulated the H9 CD4 T cells (Figure 3C). These results suggest that the PLAC131-50 peptide has the capacity to bind and elicit HTL responses restricted by more than one HLA-DR allele and behaves as a typical promiscuous epitope.
Figure 3.
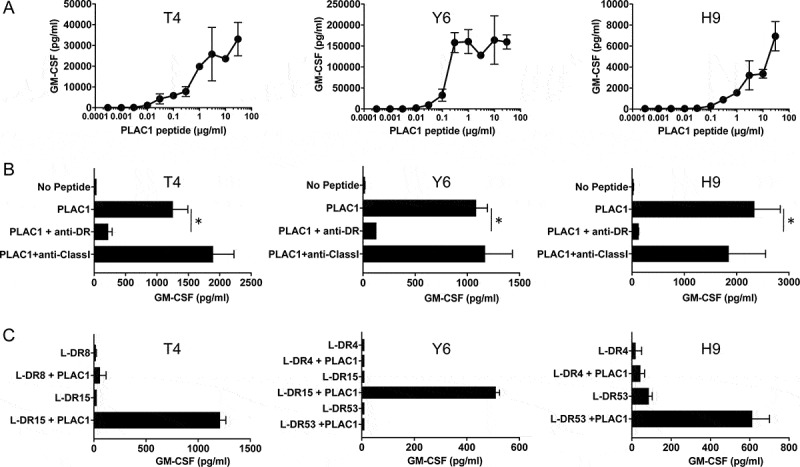
Characteristics of induced PLAC1-specific CD4 helper T cells. (A) Estimation of the peptide dose-responses of PLAC131-50-specific CD4 T cells (T4, Y6 and H9). These T cells were co-cultured with autologous PBMCs pulsed with various concentrations of PLAC131-50 peptide for 48 h. Supernatants were harvested and the secretion of GM-CSF was measured by ELISA. Points, mean of duplicate measurements; error bars, SD. (B) Determination of the HLA restriction of PLAC131-50-specific CD4 T cells. The response of T cells co-cultured with autologous PBMCs pulsed with PLAC131-50 peptide (3 μg/ml) was evaluated in the presence of anti-HLA-DR mAb L243 or anti-HLA class I mAb W6/32 (negative control). (C) Detailed determination of which HLA-DR molecules are responsible for PLAC131-50 peptide presentation. The response of CD4 T cells to PLAC131-50 peptide was measured using peptide-pulsed (3 μg/ml) L-cells transfected with individual HLA-DR alleles as APCs. In B and C, supernatants were harvested after 48 h and GM-CSF secretion was measured by ELISA. Columns, mean of triplicate measurements; error bars, SD. Statistical significance was assessed using an unpaired Student’s t-test (*p< .05)
Direct tumor recognition by and cytokine expression status of PLAC1-specific CD4 helper T cells
Next, we assessed whether the HTL epitope represented by PLAC131-50 peptide could be produced by PLAC1-expressing tumor cell lines through natural antigen processing and presentation in the major histocompatibility complex (MHC) class II pathway. Thus, we evaluated the direct reactivity of PLAC131-50-specific CD4 T cells against PLAC1-positive tumor cell lines that were treated with IFN-γ to enhance HLA-DR expression. As shown in Figure 4A and Supplemental Figure S6D, the HLA-DR15-restricted T-cell lines (T4 and Y6) recognized HLA-DR15-positive Lu65 and HSC3 cells, and this recognition was significantly inhibited by L243 mAb. Similarly, the HLA-DR53-restricted T-cell line (H9) responded to Lu65, HSC4 and Sa-3 cells positive for HLA-DR53, but hardly responded to these tumor cells in the presence of L243 mAb (Figure 4A). All three T-cell lines did not react to HLA-DR-unmatched EBC1 cells and HLA-DR-negative Jurkat cells, although both EBC1 and Jurkat cells sufficiently expressed PLAC1 protein (Figure 2A). These results suggest that PLAC131-50 peptide includes a naturally processed HTL epitope presented by HLA-DR in tumor cells and could efficiently stimulate CD4 helper T cells that respond to PLAC1-expressing tumor cells. We examined the production of granzyme B from PLAC131-50-specific CD4 T cells using the same experimental design as employed in Figure 4A. T4, Y6 and H9 cells released granzyme B in response to HLA-DR-matched tumor cell lines, but not HLA-DR-unmatched or -negative tumor cell lines, and the granzyme B release was significantly inhibited by L243 mAb (Figure 4B), indicating that PLAC131-50-specific CD4 T cells are not only able to recognize PLAC1-positive tumor cells but may also show cytotoxic potential against them. Additionally, we performed intracellular staining using flow cytometry to examine the cytokine expression status of PLAC131-50-specific CD4 T cells for peptide stimulation. As shown in Supplemental Figure S7, some PLAC131-50-specific CD4 T cells co-expressed two or all three of GM-CSF, IFN-γ and granzyme B, simultaneously.
Figure 4.
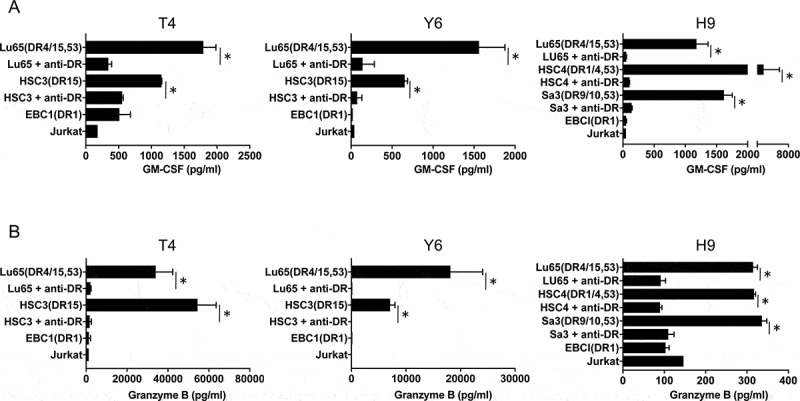
Direct recognition of PLAC1-expressing tumor cells by PLAC1-specific CD4 helper T cells. To evaluate the capacity to recognize naturally processed PLAC1 antigen in tumor cells, PLAC131-50-specific CD4 T cells (DR15-restricted T-cell lines, T4 and Y6; DR53-restricted T-cell line, H9) were co-cultured with HLA-DR15-positive (Lu65 and HSC3), HLA-DR53-positive (Lu65, HSC4 and Sa-3), HLA-DR-unmatched (EBC1) and HLA-DR-negative (Jurkat) tumor cells expressing PLAC1 with or without anti-HLA-DR mAb L243. Tumor cells were treated with IFN-γ (500 IU/ml) for 48 h before co-culturing with T cells to enhance HLA-DR expression. EBC1 and Jurkat were used as negative controls. Supernatants were harvested and subjected to ELISA for GM-CSF (A) and granzyme B (B) release after 48 h co-culture. Columns, mean of triplicate measurements; error bars, SD. Statistical significance was assessed using an unpaired Student’s t-test (*p< .05)
Cytotoxic activity of PLAC1-specific CD4 helper T cells
To confirm whether PLAC131-50-specific CD4 T cells could produce a direct anti-tumor effect by killing cancer cells, we assessed the cytotoxic activity of Y6 and H9 cells against PLAC1-positive tumor cell lines expressing HLA-DR following pretreatment with IFN-γ. The HLA-DR15-restricted T-cell line Y6 showed cytotoxicity against the HLA-DR-matched tumor cell lines Lu65 and HSC3, but not against the HLA-DR-unmatched tumor cell line EBC1 (Figure 5A). Similarly, the HLA-DR53-expressing tumor cell lines Lu65 and HSC4 were killed by the HLA-DR53-restricted T-cell line H9, whereas EBC1 cells not expressing HLA-DR53 were not (Figure 5B). These results demonstrate that the CD4 T-cell lines can directly exert HLA-DR-restricted cytotoxicity, which requires the presentation of the specific HLA-DR-peptide complexes.
Figure 5.
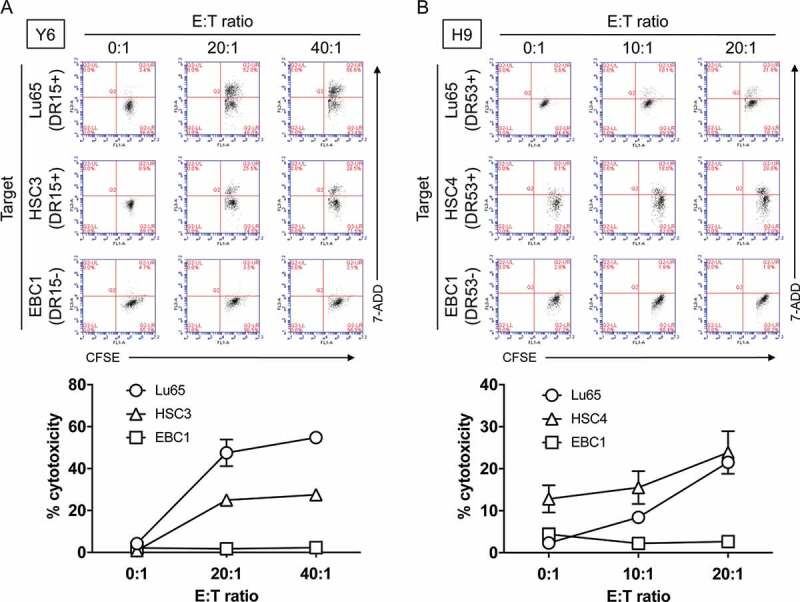
Evaluation of cytotoxic activity of PLAC1-specific CD4 helper T cells against HLA-DR-matched tumor cells expressing PLAC1. (A) HLA-DR15-restricted CD4 T cells (Y6) were co-cultured with HLA-DR-matched tumor cell lines (Lu65 and HSC3) expressing PLAC1. The HLA-DR-unmatched tumor cell line EBC1 was used as a negative control. Tumor cells were treated with IFN-γ (500 IU/ml) for 48 h before co-culturing with T cells to enhance HLA-DR expression, followed by CFSE labeling. After 6 h co-culture, the cells were collected to evaluate the percentage of CFSE-labeled dead cells by flow cytometry with 7-AAD. Effector:Target ratio (E:T ratio) was 0:1, 20:1 and 40:1. (B) Similarly, HLA-DR53-restricted CD4 T cells (H9) were co-cultured with HLA-DR-matched tumor cell lines (Lu65 and HSC4) expressing PLAC1 for 6 h. The HLA-DR-unmatched tumor cell line EBC1 was used as a negative control. The pretreatment of tumor cells and flow cytometric analysis were performed in the same way as described in A. E:T ratio was 0:1, 10:1 and 20:1. In A and B, upper panels indicate representative scatter plots of flow cytometric analysis. Lower panels indicate the average cytotoxicity of PLAC131-50-specific CD4 T cells against each tumor cell. Points, mean of triplicate measurements; error bars, SD
Recognition of PLAC1 peptide by PBMCs from HNSCC patients
Lastly, we evaluated the presence of T-cell responses to PLAC131-50 peptide in HNSCC patients because it is important to determine whether PLAC1-specific precursor T cells exist in HNSCC patients and can be activated by PLAC131-50 peptide stimulation in clinical applications. Since we were able to obtain only small blood volumes from HNSCC patients and were unable to establish and analyze long-term T-cell lines, the T-cell responses to PLAC131-50 peptide were measured by short-term cultures using peptide-stimulated PBMCs from the 12 HNSCC patients (as described in the Materials and Methods). The main clinical characteristics of the 12 HNSCC patients are summarized in the table in Figure 6. Sufficient T-cell responses to PLAC131-50 and PADRE (positive control) peptides compared with no peptide stimulation were observed in 7 of 12 HNSCC patients (#4, #5, #7, #9, #10, #11 and #12) and were significantly inhibited by L243 mAb (Figure 6), suggesting that PLAC131-50 peptide could elicit the responses of precursor T cells existing in HNSCC patients. We also examined the relationship between T-cell responses to PLAC131-50 peptide and the clinicopathological features of the 12 HNSCC patients (Supplemental Table S5). T-cell responses significantly correlated with lymph node metastasis (p= .0455). In addition, the correlation between T-cell responses and high expression of PLAC1 in tumor cells showed a trend toward statistical significance (p= .0606).
Figure 6.
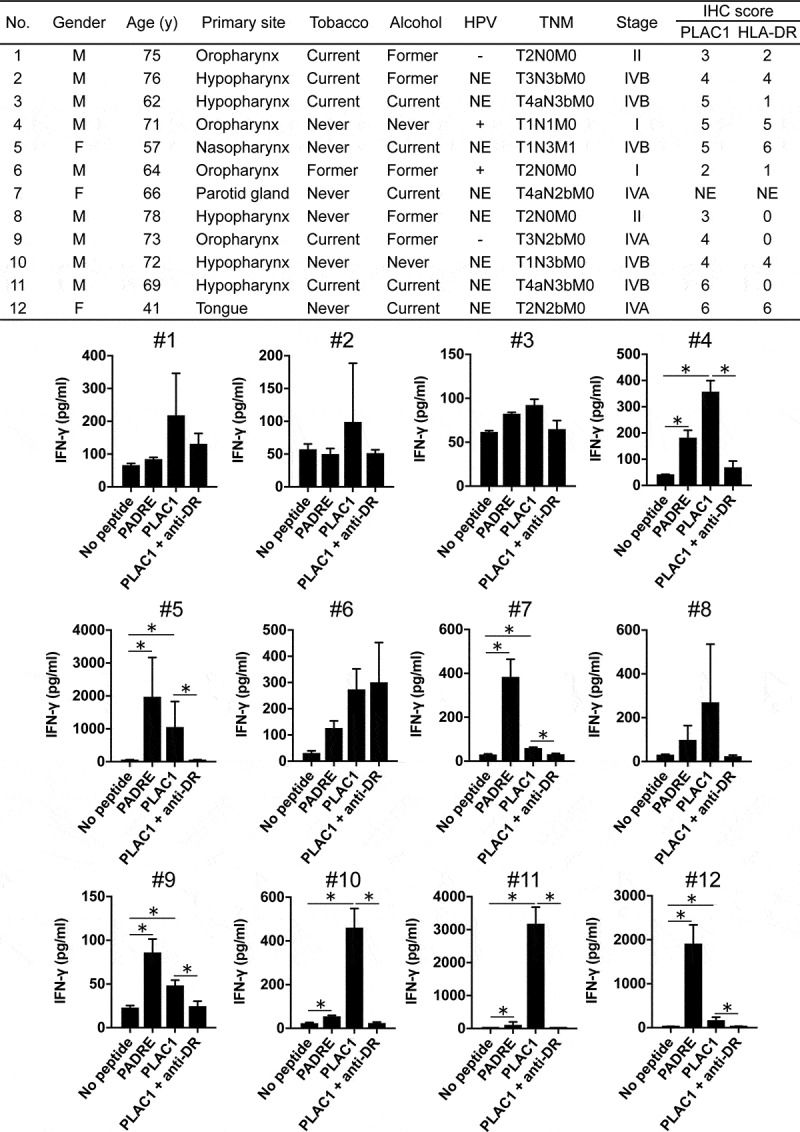
Evaluation of T-cell responses to PLAC1 peptide in HNSCC patients. The characteristics of the 12 HNSCC patients are summarized in the upper table (NE stands for not examined). PBMCs derived from HNSCC patients were cultured in the presence of PLAC131-50 or PADRE peptide (10 μg/ml) for the first 7 days and restimulated with peptide-pulsed (10 μg/ml) irradiated autologous PBMCs for the next 7 days. PADRE peptide was used as a positive control for activating CD4 T cells. Anti-HLA-DR mAb L243 was used to block the peptide presentation by HLA-DR. Supernatants were harvested and analyzed by ELISA for IFN-γ release. Columns, mean of triplicate measurements; error bars, SD. Statistical significance was assessed using an unpaired Student’s t-test (*p< .05)
Discussion
To the best of our knowledge, this is the first report of the identification of a PLAC1-derived HTL epitope. PLAC131-50-specific CD4 T cells recognized not only peptide-pulsed autologous APCs, but more importantly PLAC1-positive tumor cell lines in an HLA-DR15 or HLA-DR53-restricted manner, indicating that this promiscuous epitope is naturally processed in tumor cells. HLA-DR15 is a relatively common HLA-DRB1 molecule, having a population frequency of 20–30% in various racial groups.53 On the other hand, HLA-DR53 is an allele co-expressed by individuals with HLA-DR4, DR7 and DR9 alleles and has a high population frequency (~50%).50 Thus, this peptide has the potential to cover a large population of patients. Additionally, one of the notable results of this study is that PLAC131-50-specific CD4 T cells exhibited direct cytotoxicity against PLAC1-positive, HLA-DR-matched tumor cell lines. In regard to the cytotoxic activity by HTLs, granzyme B is one of the important mediators to directly kill target cells.19,54 Therefore, the cytotoxicity of PLAC131-50-specific CD4 T cells might depend on the secretion of granzyme B, although other factors such as the expression levels of HLA-DR on cancer cells and the production of other cytotoxic cytokines might also have cooperative roles. Furthermore, some PLAC131-50-specific CD4 T cells co-expressed 2 or more cytokines simultaneously in response to peptide stimulation (Supplemental Figure S7), suggesting that PLAC131-50-specific CD4 T cells may include multifunctional CD4 T cells with both cytotoxic and helper activity. Another interesting observation is that PLAC131-50 peptide contains two previously described HLA-A2-restricted CTL epitopes, PLAC131-39 and PLAC141-50.33,35,42 Our results, and the previous data reported by other investigators, raise the possibility that the administration of PLAC131-50 peptide as a vaccine for PLAC1-positive cancers could induce both PLAC1-reactive CTLs and HTLs in patients having HLA-A2 and one of the HLA-DR alleles capable of presenting this peptide.
When considering utilizing PLAC1 epitope peptides identified by us and other investigators as therapeutic vaccines in the clinic, novel immunization strategies capable of inducing rapid and vast peptide-specific T-cell responses will be required. For example, combined administration of synthetic peptides with strong immune adjuvants and costimulatory agonists could be useful to efficiently activate DCs, resulting in appropriate stimulation and expansion of antigen-reactive T cells.5,6 We have developed two vaccine strategies consisting of synthetic epitope peptides mixed with Toll-like receptor 3 (TLR3) agonist, polyinosinic–polycytidylic acid (poly-IC), and anti-CD40 Ab (called TriVax) or with poly-IC alone (called BiVax) capable of eliciting vast CTL responses in mouse cancer models.5–9 Furthermore, TriVax with the TLR7 agonist gardiquimod, instead of poly-IC, generated vast anti-tumor HTL responses by adding an anti-OX40 agonist Ab.55 Importantly, effective T-cell responses could be induced by injecting these vaccines systemically (intravenously or intramuscularly) with two sequential immunizations (prime and boost protocol),5–7,9 indicating that for practical use of peptide vaccines, systemic immunizations may be more effective than subcutaneous local administration. Thus, clinical application of PLAC1-derived HTL and CTL epitope peptides in patients with PLAC1-positive malignancies, including HNSCC, may become more likely in combination with optimal immune modulating agents and selecting optimal routes and protocols of administration based on knowledge gained in TriVax, BiVax and similar vaccine strategies.
The analysis of tumor antigen-specific T-cell receptors (TCRs) has provided significant benefits for the development of cancer immunotherapies. Adoptive immunotherapy using TCR-transduced T cells against CTAs mediates objective tumor regression. In regard to CD8 T cell-derived TCRs specific for PLAC1, Li et al.56 recently reported that HLA-A2-restricted and PLAC1-specific TCR-engineered CTLs could be generated by transfection of TCR genes isolated from a PLAC128-36 peptide-specific CTL clone into purified CD8 T cells. These cells exhibited anti-tumor effects against HLA-A2 and PLAC1-positive cancer cells in both in vitro cultures and in vivo xenograft mice, suggesting that PLAC1 represents a viable potential target for adoptive transfer of TCR-gene-transduced T cells. Up to now, few adoptive immunotherapies using MHC class II-restricted HTLs have been tested clinically, but have clearly shown long-term tumor regression.57,58 In addition, a recent clinical study demonstrated the safety and efficacy of an adoptive CD4 T-cell therapy using an MHC class II-restricted TCR that recognized melanoma-associated antigen-A3.59 Because tumor cells often lose the processing and presentation components of MHC class I to escape from the attack of CTLs,60 the use of genetically modified CD4 T cells in adoptive immunotherapy against MHC class II-expressing tumors, as well as the induction and expansion of endogenous CD4 T cells by a cancer vaccine using helper epitope peptides, may be an effective alternative.20,59 Therefore, future detailed TCR analysis of PLAC1-specific HTLs induced in this study may be useful for the development of adoptive immunotherapy using TCR-engineered HTLs against PLAC1 in patients with MHC class II-expressing HNSCC.
An interesting recent finding regarding the role of PLAC1 in cancer immunity is that PLAC1 might regulate adaptive immunity. Yuan et al.61 found, using a mouse mammary tumor model, that the knockdown of PLAC1 suppressed tumor growth in wild-type mice but not in SCID mice. In the gene expression profile of PLAC1 knockdown tumor cells, the reduction of chemokine (C-X-C motif) ligand 1 (CXCL1), which is the ligand of chemokine (C-X-C motif) receptor 2 (CXCR2), was detected. Furthermore, treatment with a CXCR2 antagonist impaired tumor growth in wild-type mice and reduced regulatory T cells and myeloid-derived suppressor cells in the tumor tissue. Their results suggest that PLAC1 might induce immune tolerance of the tumor microenvironment by modulating the CXCL1/CXCR2 chemokine pathway. Additionally, it was reported that several HNSCC cell lines, including PLAC1-positive HSC4 and SAS cells, secret CXCL1 in cell culture supernatants.62 Taken together with the results from these two studies, PLAC1 might modulate the expression of CXCL1 in human HNSCC cell lines. Thus, it may be possible to restore a tolerogenic state to the immunogenic tumor microenvironment in various PLAC1-positive cancers by developing drugs targeting PLAC1 itself or chemotactic mediators induced by PLAC1 expression. However, much remains to be elucidated concerning the detailed function of PLAC1 associated with the modulation of the tumor microenvironment in HNSCC.
In conclusion, PLAC1 was highly expressed in OPSCC and OCSCC patients (74.5% and 51.9%, respectively). We identified the first helper epitope from PLAC1 that could generate antigen-specific HTL responses and contains previously reported HLA-A2-restricted CTL epitopes. PLAC1-specific CD4 T cells induced by the epitope peptide clearly exhibited cytotoxic activity against PLAC1-positive tumor cell lines, including HNSCC cells, in an HLA-DR-restricted manner. In addition, the precursor of PLAC1-reactive HTLs was detected in HNSCC patients. The present findings indicate that the helper epitope peptide derived from PLAC1 could be viable as a therapeutic vaccine to effectively stimulate antigen-specific HTLs against HNSCC as well as other PLAC1-positive cancers. We further believe that PLAC1 has high potential as a target for various immunotherapy approaches.
Supplementary Material
Acknowledgments
The authors thank Ms. Rie Matsumoto (Department of Pathology, Asahikawa Medical University) and Ms. Keiko Nishikura (Department of Dermatology, Asahikawa Medical University) for their technical assistance.
Funding Statement
This work was supported by the Japan Society for the Promotion of Science [18K09310];Japan Society for the Promotion of Science [19K07452];Japan Society for the Promotion of Science [18H02948];Japan Society for the Promotion of Science [19K18755].
Disclosure of Potential Conflicts of Interest
Esteban Celis is a consultant for Oncovir, Inc. The rest of the authors do not have any conflicts of interest to declare.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A.. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–12. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chow LQM. Head and neck cancer. N Engl J Med. 2020;382(1):60–72. doi: 10.1056/NEJMra1715715. [DOI] [PubMed] [Google Scholar]
- 3.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371(9625):1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirayama M, Nishimura Y. The present status and future prospects of peptide-based cancer vaccines. Int Immunol. 2016;28(7):319–328. doi: 10.1093/intimm/dxw027. [DOI] [PubMed] [Google Scholar]
- 5.Kumai T, Kobayashi H, Harabuchi Y, Celis E. Peptide vaccines in cancer-old concept revisited. Curr Opin Immunol. 2017. 45:1–7. doi: 10.1016/j.coi.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumai T, Fan A, Harabuchi Y, Celis E. Cancer immunotherapy: moving forward with peptide T cell vaccines. Curr Opin Immunol. 2017. 47:57–63. doi: 10.1016/j.coi.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sultan H, Kumai T, Nagato T, Wu J, Salazar AM, Celis E. The route of administration dictates the immunogenicity of peptide-based cancer vaccines in mice. Cancer Immunol Immunother. 2019;68(3):455–466. doi: 10.1007/s00262-018-02294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho HI, Lee YR, Celis E. Interferon gamma limits the effectiveness of melanoma peptide vaccines. Blood. 2011;117(1):135–144. doi: 10.1182/blood-2010-08-298117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho HI, Barrios K, Lee YR, Linowski AK, Celis E. BiVax: a peptide/poly-IC subunit vaccine that mimics an acute infection elicits vast and effective anti-tumor CD8 T-cell responses. Cancer Immunol Immunother. 2013;62(4):787–799. doi: 10.1007/s00262-012-1382-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marzo AL, Kinnear BF, Lake RA, Frelinger JJ, Collins EJ, Robinson BW, Scott B. Tumor-specific CD4 + T cells have a major “Post-licensing” role in CTL mediated anti-tumor immunity. J Immunol. 2000;165(11):6047–6055. doi: 10.4049/jimmunol.165.11.6047. [DOI] [PubMed] [Google Scholar]
- 11.Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421(6925):852–856. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- 12.Perez SA, von Hofe E, Kallinteris NL, Gritzapis AD, Peoples GE, Papamichail M, Baxevanis CN. A new era in anticancer peptide vaccines. Cancer. 2010;116(9):2071–2080. doi: 10.1002/cncr.24988. [DOI] [PubMed] [Google Scholar]
- 13.Quezada SA, Simpson TR, Peggs KS, Merghoub T, Vider J, Fan X, Blasberg R, Yagita H, Muranski P, Antony PA, et al. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med. 2010. 207(3):637–650. doi: 10.1084/jem.20091918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishibashi K, Kumai T, Ohkuri T, Kosaka A, Nagato T, Hirata Y, Ohara K, Oikawa K, Aoki N, Akiyama N, et al. Epigenetic modification augments the immunogenicity of human leukocyte antigen G serving as a tumor antigen for T cell-based immunotherapy. Oncoimmunology. 2016. 5(6):e1169356. doi: 10.1080/2162402X.2016.1169356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohara K, Ohkuri T, Kumai T, Nagato T, Nozaki Y, Ishibashi K, Kosaka A, Nagata M, Harabuchi S, Ohara M, et al. Targeting phosphorylated p53 to elicit tumor-reactive T helper responses against head and neck squamous cell carcinoma. Oncoimmunology. 2018. 7(9):e1466771. doi: 10.1080/2162402X.2018.1466771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirata-Nozaki Y, Ohkuri T, Ohara K, Kumai T, Nagata M, Harabuchi S, Kosaka A, Nagato T, Ishibashi K, Oikawa K, et al. PD-L1-specific helper T-cells exhibit effective antitumor responses: new strategy of cancer immunotherapy targeting PD-L1 in head and neck squamous cell carcinoma. J Transl Med. 2019. 17(1):207. doi: 10.1186/s12967-019-1957-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohara M, Ohara K, Kumai T, Ohkuri T, Nagato T, Hirata-Nozaki Y, Kosaka A, Nagata M, Hayashi R, Harabuchi S, et al. Phosphorylated vimentin as an immunotherapeutic target against metastatic colorectal cancer. Cancer Immunol Immunother. 2020. 69(6):989–999. doi: 10.1007/s00262-020-02524-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashimoto K, Kouno T, Ikawa T, Hayatsu N, Miyajima Y, Yabukami H, Terooatea T, Sasaki T, Suzuki T, Valentine M, et al. Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians. Proc Natl Acad Sci U S A. 2019. 116(48):24242–24251. doi: 10.1073/pnas.1907883116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh DY, Kwek SS, Raju SS, Li T, McCarthy E, Chow E, Aran D, Ilano A, Pai CS, Rancan C, et al. Intratumoral CD4(+) T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell. 2020. 181(7):1612–1625 e1613. doi: 10.1016/j.cell.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inderberg EM, Walchli S. Long-term surviving cancer patients as a source of therapeutic TCR. Cancer Immunol Immunother. 2020;69(5):859–865. doi: 10.1007/s00262-019-02468-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahmoudian J, Ghods R, Nazari M, Jeddi-Tehrani M, Ghahremani MH, Ghaffari-Tabrizi-Wizsy N, Ostad SN, Zarnani AH. PLAC1: biology and potential application in cancer immunotherapy. Cancer Immunol Immunother. 2019;68(7):1039–1058. doi: 10.1007/s00262-019-02350-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5(8):615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 23.Cocchia M, Huber R, Pantano S, Chen EY, Ma P, Forabosco A, Ko MS, Schlessinger D. PLAC1, an Xq26 gene with placenta-specific expression. Genomics. 2000;68(3):305–312. doi: 10.1006/geno.2000.6302. [DOI] [PubMed] [Google Scholar]
- 24.Fant M, Farina A, Nagaraja R, Schlessinger D. PLAC1 (Placenta-specific 1): a novel, X-linked gene with roles in reproductive and cancer biology. Prenat Diagn. 2010;30(6):497–502. doi: 10.1002/pd.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koslowski M, Sahin U, Mitnacht-Kraus R, Seitz G, Huber C, Tureci O. A placenta-specific gene ectopically activated in many human cancers is essentially involved in malignant cell processes. Cancer Res. 2007;67(19):9528–9534. doi: 10.1158/0008-5472.CAN-07-1350. [DOI] [PubMed] [Google Scholar]
- 26.Yuan H, Chen V, Boisvert M, Isaacs C, Glazer RI. PLAC1 as a serum biomarker for breast cancer. PLoS One. 2018;13(2):e0192106. doi: 10.1371/journal.pone.0192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Chu J, Li J, Feng W, Yang F, Wang Y, Zhang Y, Sun C, Yang M, Vasilatos SN, et al. Cancer/testis antigen-Plac1 promotes invasion and metastasis of breast cancer through Furin/NICD/PTEN signaling pathway. Mol Oncol. 2018. 12(8):1233–1248. doi: 10.1002/1878-0261.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva WA Jr., Gnjatic S, Ritter E, Chua R, Cohen T, Hsu M, Jungbluth AA, Altorki NK, Chen YT, Old LJ, et al. 2007. PLAC1, a trophoblast-specific cell surface protein, is expressed in a range of human tumors and elicits spontaneous antibody responses. Cancer Immun. 7:18. [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L, Zha TQ, He X, Chen L, Zhu Q, Wu WB, Nie FQ, Wang Q, Zang CS, Zhang ML, et al. Placenta-specific protein 1 promotes cell proliferation and invasion in non-small cell lung cancer. Oncol Rep. 2018. 39(1):53–60. doi: 10.3892/or.2017.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong XY, Peng JR, Ye YJ, Chen HS, Zhang LJ, Pang XW, Li Y, Zhang Y, Wang S, Fant ME, et al. Plac1 is a tumor-specific antigen capable of eliciting spontaneous antibody responses in human cancer patients. Int J Cancer. 2008. 122(9):2038–2043. doi: 10.1002/ijc.23341. [DOI] [PubMed] [Google Scholar]
- 31.Wu Y, Lin X, Di X, Chen Y, Zhao H, Wang X. Oncogenic function of Plac1 on the proliferation and metastasis in hepatocellular carcinoma cells. Oncol Rep. 2017;37(1):465–473. doi: 10.3892/or.2016.5272. [DOI] [PubMed] [Google Scholar]
- 32.Liu F, Shen D, Kang X, Zhang C, Song Q. New tumour antigen PLAC1/CP1, a potentially useful prognostic marker and immunotherapy target for gastric adenocarcinoma. J Clin Pathol. 2015;68(11):913–916. doi: 10.1136/jclinpath-2015-202978. [DOI] [PubMed] [Google Scholar]
- 33.Liu FF, Dong XY, Pang XW, Xing Q, Wang HC, Zhang HG, Li Y, Yin YH, Fant M, Ye YJ, et al. The specific immune response to tumor antigen CP1 and its correlation with improved survival in colon cancer patients. Gastroenterology. 2008. 134(4):998–1006. doi: 10.1053/j.gastro.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 34.Shantha Kumara HM, Grieco MJ, Caballero OL, Su T, Ahmed A, Ritter E, Gnjatic S, Cekic V, Old LJ, Simpson AJ, et al. 2012. MAGE-A3 is highly expressed in a subset of colorectal cancer patients. Cancer Immun. 12:16. [PMC free article] [PubMed] [Google Scholar]
- 35.Liu F, Zhang H, Shen D, Wang S, Ye Y, Chen H, Pang X, Song Q, He P. Identification of two new HLA-A*0201-restricted cytotoxic T lymphocyte epitopes from colorectal carcinoma-associated antigen PLAC1/CP1. J Gastroenterol. 2014;49(3):419–426. doi: 10.1007/s00535-013-0811-4. [DOI] [PubMed] [Google Scholar]
- 36.Yin Y, Zhu X, Huang S, Zheng J, Zhang M, Kong W, Chen Q, Zhang Y, Chen X, Lin K, et al. Expression and clinical significance of placenta-specific 1 in pancreatic ductal adenocarcinoma. Tumour Biol. 2017. 39(6):1010428317699131. doi: 10.1177/1010428317699131. [DOI] [PubMed] [Google Scholar]
- 37.Tchabo NE, Mhawech-Fauceglia P, Caballero OL, Villella J, Beck AF, Miliotto AJ, Liao J, Andrews C, Lele S, Old LJ, et al. 2009. Expression and serum immunoreactivity of developmentally restricted differentiation antigens in epithelial ovarian cancer. Cancer Immun. 9:6. [PMC free article] [PubMed] [Google Scholar]
- 38.Devor EJ, Gonzalez-Bosquet J, Warrier A, Reyes HD, Ibik NV, Schickling BM, Newtson A, Goodheart MJ, Leslie KK. p53 mutation status is a primary determinant of placenta-specific protein 1 expression in serous ovarian cancers. Int J Oncol. 2017;50(5):1721–1728. doi: 10.3892/ijo.2017.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devor EJ, Leslie KK. The oncoplacental gene placenta-specific protein 1 is highly expressed in endometrial tumors and cell lines. Obstet Gynecol Int. 2013. 2013:807849. doi: 10.1155/2013/807849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Devor EJ, Reyes HD, Gonzalez-Bosquet J, Warrier A, Kenzie SA, Ibik NV, Miller MD, Schickling BM, Goodheart MJ, Thiel KW, et al. Placenta-specific protein 1 expression in human papillomavirus 16/18-positive cervical cancers is associated with tumor histology. Int J Gynecol Cancer. 2017. 27(4):784–790. doi: 10.1097/IGC.0000000000000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghods R, Ghahremani MH, Madjd Z, Asgari M, Abolhasani M, Tavasoli S, Mahmoudi AR, Darzi M, Pasalar P, Jeddi-Tehrani M, et al. High placenta-specific 1/low prostate-specific antigen expression pattern in high-grade prostate adenocarcinoma. Cancer Immunol Immunother. 2014. 63(12):1319–1327. doi: 10.1007/s00262-014-1594-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W, Zhai M, Wu Z, Qi Y, Wu Y, Dai C, Sun M, Li L, Gao Y. Identification of a novel HLA-A2-restricted cytotoxic T lymphocyte epitope from cancer-testis antigen PLAC1 in breast cancer. Amino Acids. 2012;42(6):2257–2265. doi: 10.1007/s00726-011-0966-3. [DOI] [PubMed] [Google Scholar]
- 43.Dahlstrom KR, Little JA, Zafereo ME, Lung M, Wei Q, Sturgis EM. Squamous cell carcinoma of the head and neck in never smoker-never drinkers: a descriptive epidemiologic study. Head Neck. 2008;30(1):75–84. doi: 10.1002/hed.20664. [DOI] [PubMed] [Google Scholar]
- 44.Bi C, Liu M, Rong W, Wu F, Zhang Y, Lin S, Liu Y, Wu J, Wang L. High Beclin-1 and ARID1A expression corelates with poor survival and high recurrence in intrahepatic cholangiocarcinoma: a histopathological retrospective study. BMC Cancer. 2019;19(1):213. doi: 10.1186/s12885-019-5429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumai T, Nagato T, Kobayashi H, Komabayashi Y, Ueda S, Kishibe K, Ohkuri T, Takahara M, Celis E, Harabuchi Y. CCL17 and CCL22/CCR4 signaling is a strong candidate for novel targeted therapy against nasal natural killer/T-cell lymphoma. Cancer Immunol Immunother. 2015;64(6):697–705. doi: 10.1007/s00262-015-1675-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3–4):213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 47.Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, Peters B. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47(D1):D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexander J, Sidney J, Southwood S, Ruppert J, Oseroff C, Maewal A, Snoke K, Serra HM, Kubo RT, Sette A, et al. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity. 1994. 1(9):751–761. doi: 10.1016/s1074-7613(94)80017-0. [DOI] [PubMed] [Google Scholar]
- 49.Kobayashi H, Wood M, Song Y, Appella E, Celis E. Defining promiscuous MHC class II helper T-cell epitopes for the HER2/neu tumor antigen. Cancer Res. 2000;60:5228–5236. [PubMed] [Google Scholar]
- 50.Kobayashi H, Nagato T, Oikawa K, Sato K, Kimura S, Aoki N, Omiya R, Tateno M, Celis E. Recognition of prostate and breast tumor cells by helper T lymphocytes specific for a prostate and breast tumor-associated antigen, TARP. Clin Cancer Res. 2005;11(10):3869–3878. doi: 10.1158/1078-0432.CCR-04-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D, Butterfoss D, Xiang D, Zanation A, Yin X, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004. 5(5):489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 52.Walter V, Yin X, Wilkerson MD, Cabanski CR, Zhao N, Du Y, Ang MK, Hayward MC, Salazar AH, Hoadley KA, et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS One. 2013. 8(2):e56823. doi: 10.1371/journal.pone.0056823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Battiwalla M, Ellis K, Li P, Pavletic SZ, Akpek G, Hematti P, Klumpp TR, Maziarz RT, Savani BN, Aljurf MD, et al. HLA DR15 antigen status does not impact graft-versus-host disease or survival in HLA-matched sibling transplantation for hematologic malignancies. Biol Blood Marrow Transplant. 2012. 18(8):1302–1308. doi: 10.1016/j.bbmt.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marshall NB, Swain SL. Cytotoxic CD4 T cells in antiviral immunity. J Biomed Biotechnol. 2011. 2011:954602. doi: 10.1155/2011/954602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumai T, Lee S, Cho HI, Sultan H, Kobayashi H, Harabuchi Y, Celis E. Optimization of peptide vaccines to induce robust antitumor CD4 T-cell responses. Cancer Immunol Res. 2017;5(1):72–83. doi: 10.1158/2326-6066.CIR-16-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Q, Liu M, Wu M, Zhou X, Wang S, Hu Y, Wang Y, He Y, Zeng X, Chen J, et al. PLAC1-specific TCR-engineered T cells mediate antigen-specific antitumor effects in breast cancer. Oncol Lett. 2018. 15(4):5924–5932. doi: 10.3892/ol.2018.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hunder NN, Wallen H, Cao J, Hendricks DW, Reilly JZ, Rodmyre R, Jungbluth A, Gnjatic S, Thompson JA, Yee C. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N Engl J Med. 2008;358(25):2698–2703. doi: 10.1056/NEJMoa0800251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014. 344(6184):641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu YC, Parker LL, Lu T, Zheng Z, Toomey MA, White DE, Yao X, Li YF, Robbins PF, Feldman SA, et al. Treatment of patients with metastatic cancer using a major histocompatibility complex class II-restricted T-cell receptor targeting the cancer germline antigen MAGE-A3. J Clin Oncol. 2017. 35(29):3322–3329. doi: 10.1200/JCO.2017.74.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016. 375(9):819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan H, Wang X, Shi C, Jin L, Hu J, Zhang A, Li J, Vijayendra N, Doodala V, Weiss S, et al. Plac1 is a key regulator of the inflammatory response and immune tolerance in mammary tumorigenesis. Sci Rep. 2018. 8(1):5717. doi: 10.1038/s41598-018-24022-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolff HA, Rolke D, Rave-Frank M, Schirmer M, Eicheler W, Doerfler A, Hille A, Hess CF, Matthias C, Rodel RM, et al. Analysis of chemokine and chemokine receptor expression in squamous cell carcinoma of the head and neck (SCCHN) cell lines. Radiat Environ Biophys. 2011. 50(1):145–154. doi: 10.1007/s00411-010-0341-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


