ABSTRACT
STAT2 is a central component of the ISGF3 transcriptional complex downstream of type I interferon (IFN-I) signaling. The significance of in vivo IFN-I/STAT1 signals in cDCs is well-established in the generation of antitumor cytotoxic T cell (CTL) responses. However, the role of STAT2 has remained elusive. Here, we report a clinical correlation between cDC markers and STAT2 associated with better survival in human metastatic melanoma. In a murine tumor transplantation model, targeted Stat2 deletion in CD11c+cDCs enhanced tumor growth unaffected by IFNβ therapy. Furthermore, STAT2 was essential for both, the activation of CD8a+cDCs and CD11b+cDCs and antigen cross-presentation in vivo for the generation of robust T cell killing response. In contrast, STAT2 in CD11c+cDCs was dispensable for stimulating an antigen-specific humoral response, which was impaired in global Stat2 deficient mice. Thus, our studies indicate that STAT2 in cDCs is critical in host IFN-I signals by sculpting CTL responses against tumors.
KEYWORDS: STAT2, dendritic cells, T cell, interferon, tumor
Introduction
Type I interferons (IFN-I) play a central role in tumor immunity.1 They inhibit tumor growth by orchestrating immunomodulatory effects that bridge the innate and adaptive immune response.2–4 Major functions of IFN-I include induction of cell-intrinsic antiviral/antitumor responses and activation of adaptive antigen-specific B and T cell responses, modulated by IFN-stimulated genes (ISGs).5 Activation of the IFN-I signaling pathway requires the assembly of the canonical transcriptional complex ISGF3 consisting of STAT1, STAT2 and IRF9. The prevalent view is that Janus kinases JAK1 and TYK2 tyrosine phosphorylate STAT1 and STAT2. STAT1/STAT2 heterodimers form and bind IRF9 to translocate to the nucleus and drive ISG expression.6 IFN-I can also induce the formation of STAT1 homodimers as well as non-canonical heterodimers of STAT1/IRF9 and unphosphorylated STAT2/IRF9. Most recently, assembly of the ISGF3 complex was revisited. Under homeostatic conditions, preformed STAT2/IRF9 heterodimers were found to sustain basal ISG expression in a ligand-independent manner that rapidly switched to ISGF3 in the nucleus following IFN-I stimulation.7
Conventional dendritic cells (cDCs) are central regulators of tumor immunity.8 Two independent studies identified host IFN-I signaling in cDCs as essential for cross-presentation of tumor antigens to CD8 + T cells by employing mice with CD11c+cDCs lacking either IFN-I receptor or STAT1.9,10 We previously reported that STAT2 suppressed tumor growth and was required for bone marrow-derived DCs generated in the presence of GM-CSF, to become activated and cross-present tumor antigen to CD8 + T cells.11,12 Because some IFN-I responses are STAT1-independent,13,14 the role of STAT2 in cDCs in vivo has remained unclear.
cDCs are key players in humoral immunity for directly stimulating follicular T helper cells, which promote the classic antigen-specific antibody response; they provide help to B cells and support their survival.15,16 IFN-I are essential in humoral immunity as IFN-I receptor-deficient mice do not produce antigen-specific antibodies after immunization.17 While these results emphasize the importance of IFN-I-in antibody production, it is unknown if STAT2 signaling in general, and in cDCs specifically, is needed to stimulate a humoral response.
Here, we present novel evidence that intrinsic STAT2 signaling in cDCs is essential for the generation of an effective CTL response but nonessential in stimulating an antibody response. Importantly, human metastatic melanoma tumors display a STAT2 transcriptional signature in cDCs associated with better survival. Altogether, our study establishes STAT2 as a key molecule in DC-mediated antitumor immunity.
Materials and methods
Patient sample information and bioinformatic analysis
Publicly available TCGA-SKCM datasets were used for gene expression correlations and overall patient survival analysis.18 Characteristics of the melanoma patient cohort are presented in supplementary table 1. There are 470 samples represented, of which two patients are counted twice as they have both, a primary and a metastatic sample. Principal component analysis (PCA) was performed on gene expression values (measured in standardized counts-per-million (CPM)) using the R prcomp function. The set of STAT2-dependent ISGs used in the analysis was compiled from multiple publications19–27 and only those genes found expressed in DCs (VAV1, LAPTM5, WDFY4, HCLS1, CD86, SPI1, CD72, IL2RB, SLA2, IFI30, CXCL9, CD40, CD74 and CCL4) were used for PCA analysis. Correlations between principal component (PC1) and BATF3/IRF8 gene expression were calculated and significance was assessed using the R cor.test function. To perform survival analysis based on levels of the first PC1, we first noted that time from SKCM diagnosis to death or last follow-up was not the correct survival time to use as it gives rise to anomalous results (i.e. survival time for primary SKCM appears to be significantly shorter than for metastatic SKCM). To address this problem, we used the observed survival interval (OBS) defined in Xiong et al.,28 which replaces time of diagnosis with time of TCGA specimen sampling. Kaplan-Meier survival analysis using OBS was performed based on values of PC1 (low or high, based on median split). To assess potential confounders, we also fit Cox proportional hazards models including age and tumor stage, in addition to PC1. We limited attention to PC1 as it accounted for 73.9% of total variation in the samples.
Generation of conditional Stat2KO mouse
All animal studies were conducted with the approval of the Animal Care and Use Committee at Temple University. Wild type (WT) and Stat2KO mice on the B6 genetic background11 were bred in our animal facility under a pathogen-free environment. Stat2 floxed (Stat2fl/fl) mice were generated in the C57BL/6 J genetic background. Specific Stat2 deletion in DC (Stat2Δ-DC) was achieved by crossing B6-CD11c-Cre-GFP mice (Jackson Labs, Bar Harbor, ME) with Stat2fl/fl mice. Different set of primers (supplementary Table 2) were used to distinguish wild type from Stat2fl/fl allele and Stat2Δ-DC.
Tumor cell lines and primary bone marrow-derived cDCs
Murine B16-F1 melanoma cell line was maintained in DMEM medium (Mediatech, Inc; Herndon, VA) supplemented with 5% heat-inactivated FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 100 U/mL penicillin and 100 µg/mL streptomycin (Invitrogen Corp, CA) at 37°C and 5% CO2. Murine EL-4 lymphoma cell line was maintained in DMEM medium with 10% heat-inactivated horse serum, glutamine and sodium pyruvate. Mouse primary DC cultures were generated from bone marrow precursors as previously described.12 All experiments were performed with mycoplasma-free cells.
Antibodies and cytokines
Anti-STAT1 antibody (Cat# sc-346) was purchased from Santa Cruz Biotechnologies (Santa Cruz, CA). Anti-STAT2 antibody (Cat# 07–140) and anti-OVA antibody (Clone OVA-14) were obtained from Millipore-Sigma (St. Louis, MO). Anti-Actin-HRP antibody (Cat# HRP-60008) was obtained from Proteintech (Rosemont, IL) and HRP-conjugated anti-mouse (Cat#7076S) and anti-rabbit secondary antibodies (Cat#7074S) were purchased from Cell Signaling (Danvers, MA). For flow cytometry, anti-CD16/CD32 (clone 2.4G2) was purchased from BioLegend (San Diego, CA), anti-B220-PE (clone RA3-6B2), anti-CD11b-PE-Cy7 (cloneM1/70), anti-CD3-FITC (clone 17A2), anti-CD4-PE (clone RM4-5), anti-CD8a-APC (clone 53–6.7) and anti-CD40 (clone HM40-3) were obtained from BD Biosciences (San Jose, CA). Anti-CD11c-APC (clone N418) and anti-CD86 clone GL1 were purchased from eBioscience, Inc. (San Diego, CA). Recombinant murine IFN-β was provided by Biogen-Idec.
Tumor transplantation
One million B16-F1 tumor cells or 3 × 105 EL4 cells resuspended in 200 µL of endotoxin-free saline solution were injected subcutaneously (s.c.) on the dorsal flank of 6–8 weeks old C57BL/6 mice. Three days later, mice received IFN-β (2x104U) intratumorally and around the site of tumor injection twice weekly in 100 µL of PBS. Tumor measurements were started on day 7 using a digital caliper. Tumor volume was determined with the formula: V = a2b, where a is the shorter diameter and b is the longer diameter of the tumor. Study was terminated when tumors reached a size of 20 mm in diameter. No mice died during the study.
Western blot analysis
Cells and tissues were disrupted in lysis buffer as described previously.29 Protein extracts were resolved on precast SurePAGE 4–12% gradient gels (Genscript, Piscataway, NJ) and transferred onto PVDF membranes. Membranes were blocked with Blocker Casein TBS and incubated with the corresponding primary and HRP-conjugated secondary antibodies in TBS + 3% BSA. Signal was developed using enhanced chemiluminescence reagent (BioRad, Hercules, CA) and images captured with BioRad ChemiDoc imaging system. Actin was used as internal loading control.
Immunization with chicken egg ovalbumin
Mice were immunized by intramuscular injection with 50 μg of chicken egg ovalbumin (OVA) in Complete Freund’s Adjuvant (CFA, Sigma), followed by two booster injections of 100 μg of OVA and 50 μg of OVA in Incomplete Freund’s Adjuvant (IFA, Sigma) as 2nd and 3rd immunization, respectively, given two weeks apart. Blood was collected prior to initial immunization with OVA antigen in CFA, 2 weeks after the initial immunization before the first booster injection (2nd immunization), and 2 weeks after that, before the third injection. Tail bleeds were obtained, and blood was allowed to coagulate at room temperature. Serum was collected after centrifugation and stored at −80°C for antibody measurements.
Measurement of OVA antibodies by ELISA
Ninety-six-well plates were coated with 100 μl of OVA (30 μg/ml) resuspended in borate buffer saline (BBS) and incubated overnight at 4°C. Plates were then blocked with 3% BSA in BBS for 2 h at 37°C. Serum samples diluted 1:500 in BBS containing 1% Tween 80 (BBT) or a commercial mouse anti-OVA antibody standard were added to plate and incubated at 4°C overnight. Following incubation with secondary antibody (Alkaline Phosphatase-conjugated anti-mouse IgG diluted at 1:5000) signal was developed using 1 mg/ml of p-nitrophenyl phosphate (PNPP, Sigma). Optical density (OD) was measured at 405 nm with wavelength correction at 650 nm.
In vivo CTL killing assay
Mice were immunized with OVA antigen using the protocol described above. One week after the third injection of OVA, in vivo CTL killing assay was performed as described.12 Percent specific lysis was calculated as 1-[rnaive/rimmunized]x100, where r = % CFSElo cells/% CFSEhi cells.
In vivo stimulation of DCs
Mice were injected intravenously with TLR7 ligand R848 (30 μg; InvivoGen) and euthanized 24 h later. Spleens were digested in collagenase type IV (Worthington, Lakewood, NJ) and DNAse from bovine pancreas type II (Sigma) for 45 min at 37°C in IMDM media and filtered through a 100 μm filter. Splenocytes were cleared of red blood cells using ammonium-chloride-potassium and immunostained for flow cytometry analysis.
Flow cytometry
Single-cell splenocytes were resuspended in cold PBS containing 1% BSA and incubated with rat anti-mouse CD16/CD32 mAb for 10 min on ice to block FcγR. Cells were then stained for 30 min on ice with antibodies against surface cell markers (B220, CD11c, CD11b, CD3, CD4, and/or CD8a clone) and activation markers (CD86 and CD40). Cells were analyzed on a FACSCanto cytometer (BD Biosciences). Data analysis was performed using FlowJo software.
Statistical analysis of animal studies
Prism software (GraphPad, San Diego, CA) was used for statistical analysis. Unpaired Student t- test was used for the comparison between two groups. One-way ANOVA or two-way ANOVA analysis followed by Dunnett’s test were applied for multiple comparisons. Values of p < .05 were considered statistically significant. A power calculation was set at the 5% significance with 80% power.
Results and discussion
Human melanoma tumors display a link between cDCs and STAT2-dependent gene transcripts
Evidence of IFN-I signaling in T cells infiltrating human melanoma tumors highlights the importance of host IFN-I in the generation of robust CD8+ cytolytic T cell (CTL) response.30 Therefore, we evaluated human melanoma samples for the presence of an IFN-I/STAT2 dependent signature in cDCs. Analysis of TCGA-SKCM datasets representing patients with primary and metastatic melanoma (Suppl. Table 1) based on STAT2 mRNA expression showed an association between high STAT2 and overall increased survival, seen only in metastatic patients (Figure 1a). Next, overall increased survival of patients with metastatic disease correlated with high PC1 of the expression values of 14 STAT2-dependent ISGs (Figure 1b). No statistical differences were found in overall survival of patients with primary melanoma based on PC1 expression (low PC1, n = 70 and high PC1, n = 31; p = .07). Interrogation of the same metastatic patient samples revealed that cDC markers BATF3 and IRF8 strongly correlated with PC1 (Figure 1c,d), indicating a significant role for cDCs in intra-tumor STAT2-dependent responses. PC1 and STAT2 remained significantly associated with survival in metastatic samples after adjusting for age and stage. These data provide further support to our previous finding in which we showed a requirement for STAT2 in cDC function and tumor growth control when studied in the context of global STAT2 deficiency.11
Figure 1.
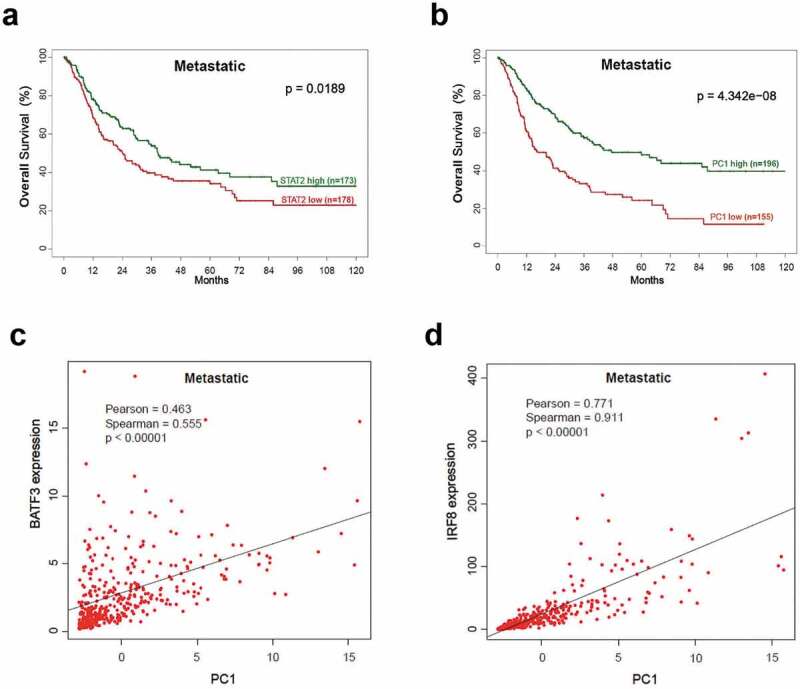
Correlation between cDC markers and STAT2-dependent gene transcripts in human metastatic melanoma. Transcriptional signatures of human primary (n = 103) and metastatic melanoma (n = 367) samples were obtained from TCGA-skin cutaneous melanoma (SKCM) datasets. (a-b) Kaplan-Meier plots show significantly longer survival in samples with high values of STAT2 or high values of PC1 (first principal component based on expression of 14 STAT2-dependent ISGs). High and low values were based on median split. (c-d) Relationships between PC1 and IRF8/BATF3 expression. Gene expression levels are measured in counts per million
Stat2 ablation in CD11c+DCs facilitates tumor growth that is not suppressed by exogenous IFN-I
To determine intrinsic STAT2 signaling in cDCs, we generated mice with Stat2 deletion in CD11c+DCs, therein referred as Stat2Δ-DC (Suppl. Figure 1). We analyzed the three main DC subsets: CD11b+cDCs (B220− CD11c+ CD11b+, cDCs), plasmacytoid DCs (B220+ CD11cInt CD11b−, pDCs), and CD8a+cDCs (CD3− CD8a+ CD11c+ B220−), the latter being important for cross-priming CD8+ T cells. Total cellularity of the spleen and frequency of DC subsets in Stat2Δ-DC and Stat2KO mice were the same as in WT mice, indicating that STAT2 played no intrinsic role in DC development (Suppl. Figure 2a-D). We also showed that tumor growth was accelerated in Stat2Δ-DC mice as in Stat2KO mice (Figure 2a) and IFNβ treatment failed to restrict tumor growth (Figure 2b). Both findings illustrate a pivotal role for STAT2 in cDCs to activate the antitumor effects of IFN-I. Our data establish a salient feature of STAT2 that was inferred but not addressed in previous studies using mice lacking IFN-I receptor or Stat1.9,10 Moreover, this observation highlights cDCs as orchestrators of the anti-tumor immune responses.
Figure 2.
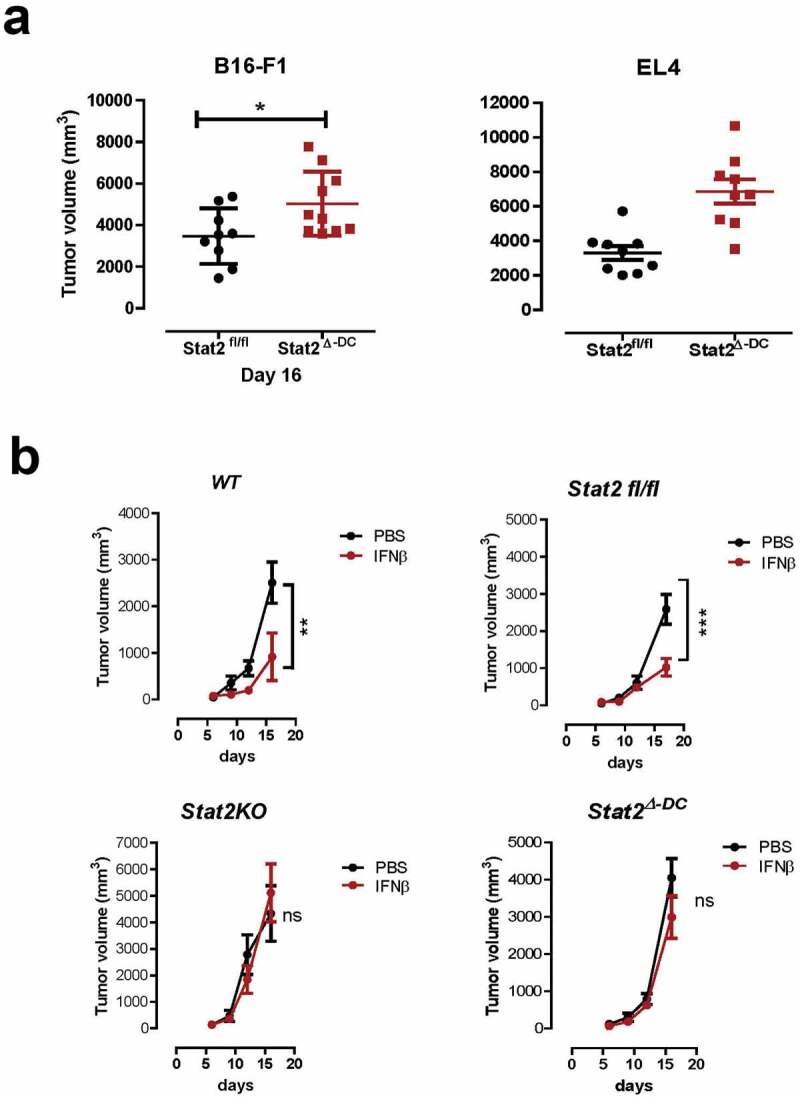
Intrinsic STAT2 signaling in cDCs is critical for tumor suppression and type I IFN antitumor response. (a) Tumor growth in Stat2fl/fl, and Stat2Δ-DC mice that were transplanted with B16-F1 (Stat2fl/fl; n = 9 and Stat2Δ-DC; n = 10) or EL4 tumor cells (n = 9 for each genotype). (b) Mice of the indicated genotypes (n = 8–9) were injected subcutaneously with B16-F1 cells. Three days later mice received injections of PBS or IFN-β around the site of tumor implantation twice weekly. Values are shown as mean tumor volume determined over 20 days. *, p < .05; **, p < .01; and ***, p < .001. ns, not statistically significant. The results are from two independent experiments that were combined with at least 4 mice per group
STAT2 signaling is required for DC activation in vivo
We determined whether Stat2 deletion in cDCs impaired their activation in vivo by TLR7 ligand R848. TLR7 stimulation enables antigen cross-presentation by DC subsets by causing a vigorous production of IFN-I and IL12,31,32 which are essential for the cross-priming of CD8+ T cells.33,34 The spleens of mice injected with R848 or PBS vehicle were analyzed after 24 h for changes in surface expression of activation markers CD86 and CD40 in CD11+cDCs (CD8a+cDCs and CD11b+cDCs) and B220+ CD11cint pDCs, gated as shown in Figure 3a,c. All three DC subsets in WT, Stat2fl/fl and CD11c-Cre mice showed upregulated CD86 and CD40 by R848 (Figure 3).12,34 In contrast, the same three DC subsets in Stat2KO mice showed impaired upregulation of both costimulatory markers, indicating that global STAT2 was required for in vivo DC responses to TLR7. The same defect was observed in Stat2Δ-DC mice but was restricted to CD11b+ and CD8a+cDC subsets with the CD8a+ cDC subset having the most severe impairment (Figure 3d and E vs. Figure 3g,h), which is closely aligned with Stat2KO mice. Although Stat2Δ-DC pDCs showed less upregulation, it did not reach statistical significance (figure 3f,i), possibly because CD11c+Cre is expressed less in pDCs.35 Hence, our results corroborate our previous study with Stat2KO mice12 and show conclusively that intrinsic STAT2 in cDCs is critical for their responses to systemic TLR activation mediated by autocrine/paracrine IFN-I.
Figure 3.
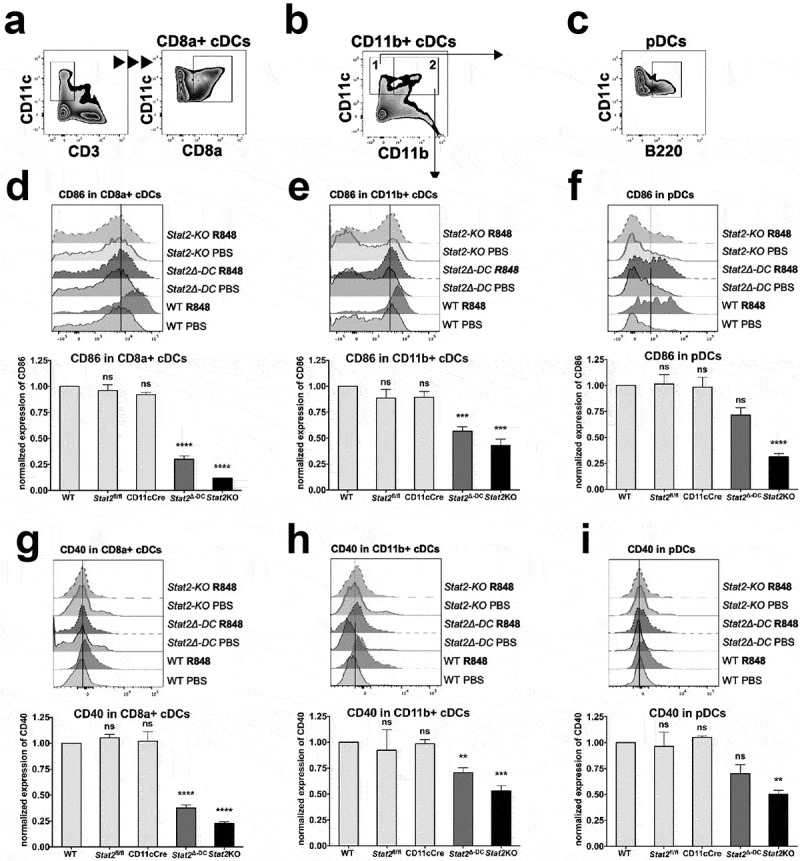
STAT2 is critical for DC subsets to respond to TLR stimulation in vivo. WT, Stat2KO, Stat2fl/fl, CD11c-Cre+ and Stat2Δ-DC mice were injected i.v. with PBS or TLR7 ligand R848. DC subsets in splenocytes were analyzed 24 h later by flow cytometry for surface lineage and activation markers. Normalized expression of CD40 and CD86 to WT mice set at 1, is shown for (d, g) CD8a+ cDCs, (e, h) CD11b+cDCs and (f, i) pDCs, gated as shown in (a, b, c). A representative histogram for CD86 and CD40 expression is shown for all 3 DC subsets. n = 3–4 mice per group. Error bars indicate mean ± SEM. **, p < .01; ***, p < .001; and ****, p < .0001, ns, not significant
CTL killing response is dependent on intrinsic STAT2 signaling in cDCs in vivo
To test whether STAT2 signaling in cDCs was needed for generating CD8 + T cell responses, we analyzed CTL killing in vivo of OVA-specific CFSE labeled target cells by flow cytometry. We immunized mice with OVA plus adjuvant and seven days after the last immunization, we performed CTL killing in vivo assay by injecting mice intravenously with targets consisting of a mix of unpulsed and OVA-pulsed syngeneic splenocytes, recognizable by differential CFSE staining. After 24 h, we found WT mice had effective specific target lysis of 75–80% when compared to naïve unimmunized mice (Figure 4b). In stark contrast, Stat2Δ-DC and Stat2KO mice displayed attenuated CTL responses with specific killing of 22.5% and 14.5% on average, respectively. Thus, our findings indicate that STAT2 deficiency in CD11c+cDCs impedes development of an effective CTL response in vivo. These observations together with our previous studies,11,12 in which DCs generated from bone marrow of Stat2KO mice were suboptimal in inducing antigen-specific CTL killing, strongly support the essential role of STAT2 signaling in CD11c+cDCs in inducing adaptive anti-tumor immunity. Intriguingly, individuals with homozygous STAT2 deficiency have been identified in the world who presented with severe and recurrent viral infections in childhood and exhibited defective IFN-I signaling.36 An outstanding question is whether STAT2 deficiency heightens the risk of these patients for developing cancer later in life that, at this moment, is unknown.
Figure 4.
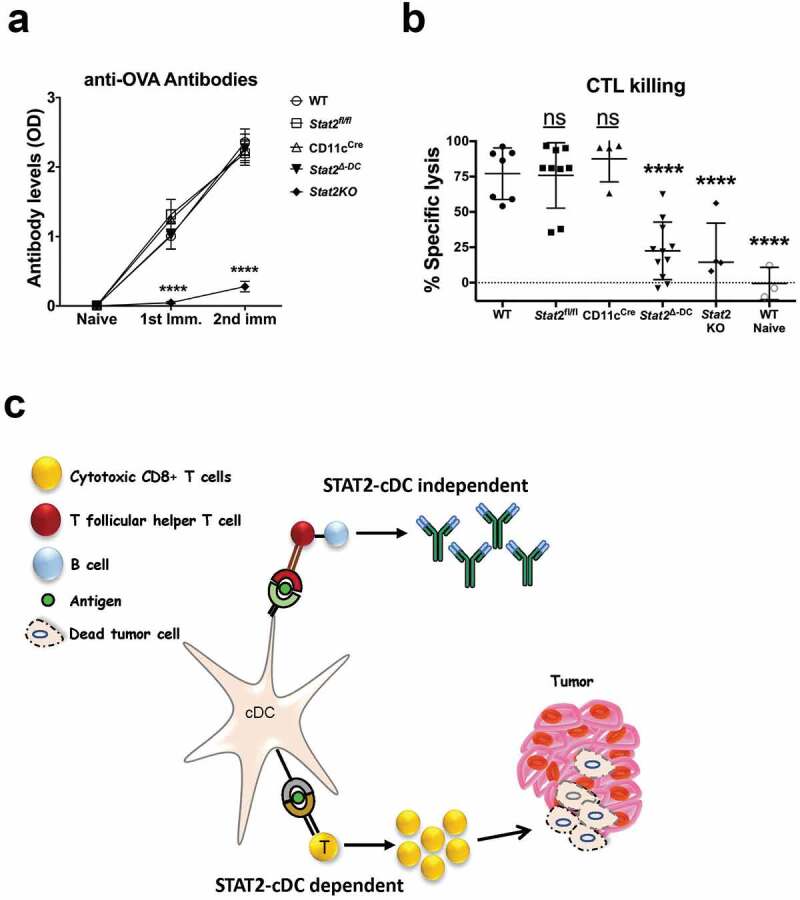
Intrinsic STAT2 signaling in cDCs is dispensable for antibody production but essential for CTL killing responses in vivo. Mice of different genotypes were immunized with OVA protein in CFA followed by one injection of OVA in IFA two weeks later. (a) Serum collection from individual mice was done prior to immunization, two weeks after the initial immunization, and two weeks after the second immunization. Anti-OVA antibody levels were determined by ELISA and data are shown as mean optical density (OD). WT, n = 14; Stat2fl/fl, n = 9; CD11c-Cre, n = 10; and Stat2Δ-DC, n = 8; Stat2KO, n = 7 for the two time points and n = 4 for the last time point. The mice tested were divided in two independent experiments. (b) Two weeks after the second immunization, mice received a third immunization for OVA. CTL killing activity was measured 7 days later by injecting mice i.v. with an equal mixture of splenocytes labeled with high or low CFSE and pulsed with or without OVA peptide. Spleens were analyzed 24 h later by flow cytometry and the percentage of specific lysis was calculated. Error bars show mean ± SD. Mice were tested divided in two independent experiments; each dot represents one individual mouse. ****, p < .0001. ns, not significant. (c) Proposed model of STAT2 role in cDC in tumor control
STAT2 signaling in cDCs is dispensable for the generation of antigen-specific antibodies
To further our understanding of the role of STAT2 signaling in cDC competence to stimulate humoral immunity, we assessed the production of antibodies against OVA, a well-characterized T cell-dependent antigen.37 Stat2Δ-DC mice produced similar levels of serum anti-OVA IgG as WT, CD11c-Cre+ and Stat2fl/fl mice following the first and second immunizations (Figure 4a). In contrast, Stat2KO mice displayed impaired production of anti-OVA IgG. These results indicate that STAT2 function is critical in the development of humoral responses regardless of the presence or absence of STAT2 signals in CD11c+cDCs.
Based on our findings, we propose a model wherein intrinsic STAT2 signaling in cDCs is essential for promoting their maturation and capacity to cross-present tumor antigen to CTLs to generate a strong anti-tumor immune response (Figure 4c). We conclude that STAT2 mediates the antitumor effects of IFN-I via cDC activity while STAT2 signaling in cDCs is redundant in humoral responses by a STAT2-dependent process involving other cell types. Overall, our study underscores the prominent role of STAT2 in IFN-I-DC-CTL cell axis to control tumor growth.
Supplementary Material
Funding Statement
This work was supported by Temple University Bridge Funds and partial support by R03 CA215929 to AMG and R21AI119947 to SG. Bioinformatics support was by P30 CA006927.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Data availability statement
The data that supports the findings of this study are available in the main document and supplementary material of this article. TGCA-SKCM datasets are publicly available at https://portal.gdc.cancer.gov/and GDAC Firehose (https://gdac.broadinstitute.org/).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G.. Type I interferons in anticancer immunity. Nat Rev Immunol. 2015;15:405–8. doi: 10.1038/nri3845. [DOI] [PubMed] [Google Scholar]
- 2.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy DE, Darnell JE. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 4.Yarilina A, Park-Min KH, Antoniv T, Hu X, Ivashkiv LB. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol. 2008;9:378–387. doi: 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- 5.MacMicking JD. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat Rev Immunol. 2012;12:367–382. doi: 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stark GR, Darnell JE. The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Platanitis E, Demiroz D, Schneller A, Fischer K, Capelle C, Hartl M, Gossenreiter T, Müller M, Novatchkova M, Decker T. A molecular switch from STAT2-IRF9 to ISGF3 underlies interferon-induced gene transcription. Nat Commun. 2019;10:2921. doi: 10.1038/s41467-019-10970-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner A, Ruffell B. Dendritic cells and cancer immunity. Trends Immunol. 2016;37:855–865. doi: 10.1016/j.it.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, Lee H, Arthur CD, White JM, Kalinke U, et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med. 2011;208:1989–2003. doi: 10.1084/jem.20101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuertes MB, Kacha AK, Kline J, Woo SR, Kranz DM, Murphy KM, Gajewski TF. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8α+ dendritic cells. J Exp Med. 2011;208:2005–2016. doi: 10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue C, Xu J, Tan Estioko MD, Kotredes KP, Lopez-Otalora Y, Hilliard BA, Baker DP, Gallucci S, Gamero AM. Host STAT2/type I interferon axis controls tumor growth. Int J Cancer. 2015;136:117–126. doi: 10.1002/ijc.29004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Lee MH, Chakhtoura M, Green BL, Kotredes KP, Chain RW, Sriram U, Gamero AM, Gallucci S. STAT2 is required for TLR-induced murine dendritic cell activation and cross-presentation. J Immunol. 2016;197:326–336. doi: 10.4049/jimmunol.1500152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majoros A, Platanitis E, Kernbauer-Hölzl E, Rosebrock F, Müller M, Decker T. Canonical and non-canonical aspects of JAK-STAT signaling: lessons from interferons for cytokine responses. Front Immunol. 2017;8:29. doi: 10.3389/fimmu.2017.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueno H, Schmitt N, Palucka AK, Banchereau J. Dendritic cells and humoral immunity in humans. Immunol Cell Biol. 2010;88:376–380. doi: 10.1038/icb.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wykes M, MacPherson G. Dendritic cell-B-cell interaction: dendritic cells provide B cells with CD40-independent proliferation signals and CD40-dependent survival signals. Immunology. 2000;100:1–3. doi: 10.1046/j.1365-2567.2000.00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Bon A, Schiavoni G, D’Agostino G, Gresser I, Belardelli F, Tough DF. Type I interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity. 2001;14:461–470. doi: 10.1016/S1074-7613(01)00126-1. [DOI] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas N . Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blaszczyk K, Olejnik A, Nowicka H, Ozgyin L, Chen YL, Chmielewski S, Kostyrko K, Wesoly J, Balint BL, Lee CK, et al. STAT2/IRF9 directs a prolonged ISGF3-like transcriptional response and antiviral activity in the absence of STAT1. Biochem J. 2015;466:511–524. doi: 10.1042/BJ20140644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perry ST, Buck MD, Lada SM, Schindler C, Shresta S. STAT2 mediates innate immunity to dengue virus in the absence of STAT1 via the type I interferon receptor. PLoS Pathog. 2011;7:e1001297. doi: 10.1371/journal.ppat.1001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Hofer MJ, Songkhunawej P, Jung SR, Hancock D, Denyer G, Campbell IL. Type I interferon-regulated gene expression and signaling in murine mixed glial cells lacking signal transducers and activators of transcription 1 or 2 or interferon regulatory factor 9. J Biol Chem. 2017;292:5845–5859. doi: 10.1074/jbc.M116.756510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Testoni B, Vollenkle C, Guerrieri F, Gerbal-Chaloin S, Blandino G, Levrero M. Chromatin dynamics of gene activation and repression in response to interferon alpha (IFN(alpha)) reveal new roles for phosphorylated and unphosphorylated forms of the transcription factor STAT2. J Biol Chem. 2011;286:20217–20227. doi: 10.1074/jbc.M111.231068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rauch I, Rosebrock F, Hainzl E, Heider S, Majoros A, Wienerroither S, Strobl B, Stockinger S, Kenner L, Müller M, et al. Noncanonical effects of IRF9 in intestinal inflammation: more than type I and type III interferons. Mol Cell Biol. 2015;35:2332–2343. doi: 10.1128/MCB.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog PJ. INTERFEROME v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2012;41:D1040–D6. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gopal R, Lee B, McHugh KJ, Rich HE, Ramanan K, Mandalapu S, Clay ME, Seger PJ, Enelow RI, Manni ML, et al. STAT2 signaling regulates macrophage phenotype during influenza and bacterial super-infection. Front Immunol. 2018;9:2151. doi: 10.3389/fimmu.2018.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.T-N N-P, Lim M-S, Nguyen TAT, Lee Y-K, Jin C-J, Lee HJ, Hong CY, Ahn J-S, Yang D-H, Kim Y-K, et al. Type I and II interferons enhance dendritic cell maturation and migration capacity by regulating CD38 and CD74 that have synergistic effects with TLR agonists. Cell Mol Immunol. 2011;8:341–347. doi: 10.1038/cmi.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spurrell DR, Luckashenak NA, Minney DC, Chaplin A, Penninger JM, Liwski RS, Clements JL, West KA. Vav1 regulates the migration and adhesion of dendritic cells. J Immunol. 2009;183:310–318. doi: 10.4049/jimmunol.0802096. [DOI] [PubMed] [Google Scholar]
- 28.Xiong J, Bing Z, Guo S. Observed survival interval: a supplement to TCGA pan-cancer clinical data resource. Cancers (Basel). 2019;11:280. doi: 10.3390/cancers11030280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yue C, Soboloff J, Gamero AM. Control of type I interferon-induced cell death by Orai1-mediated calcium entry in T cells. J Biol Chem. 2012;287:3207–3216. doi: 10.1074/jbc.M111.269068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harlin H, Meng Y, Peterson AC, Zha Y, Tretiakova M, Slingluff C, McKee M, Gajewski TF. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69:3077–3085. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doxsee CL, Riter TR, Reiter MJ, Gibson SJ, Vasilakos JP, Kedl RM. The immune response modifier and Toll-like receptor 7 agonist S-27609 selectively induces IL-12 and TNF-alpha production in CD11c+CD11b+CD8- dendritic cells. J Immunol. 2003;171:1156–1163. doi: 10.4049/jimmunol.171.3.1156. [DOI] [PubMed] [Google Scholar]
- 32.Saitoh SI, Abe F, Kanno A, Tanimura N, Mori Saitoh Y, Fukui R, Shibata T, Sato K, Ichinohe T, Hayashi M, et al. TLR7 mediated viral recognition results in focal type I interferon secretion by dendritic cells. Nat Commun. 2017;8:1592. doi: 10.1038/s41467-017-01687-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curtsinger JM, Lins DC, Mescher MF. Signal 3 determines tolerance versus full activation of naive CD8 T cells: dissociating proliferation and development of effector function. J Exp Med. 2003;197:1141–1151. doi: 10.1084/jem.20021910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh JZ, Kurche JS, Burchill MA, Kedl RM. TLR7 enables cross-presentation by multiple dendritic cell subsets through a type I IFN-dependent pathway. Blood. 2011;118:3028–3038. doi: 10.1182/blood-2011-04-348839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J Exp Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hambleton S, Goodbourn S, Young DF, Dickinson P, Mohamad SM, Valappil M, McGovern N, Cant AJ, Hackett SJ, Ghazal P, et al. STAT2 deficiency and susceptibility to viral illness in humans. Proc Natl Acad Sci U S A. 2013;110:3053–3058. doi: 10.1073/pnas.1220098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasmussen IB, Lunde E, Michaelsen TE, Bogen B, Sandlie I. The principle of delivery of T cell epitopes to antigen-presenting cells applied to peptides from influenza virus, ovalbumin, and hen egg lysozyme: implications for peptide vaccination. Proc Natl Acad Sci U S A. 2001;98:10296–10301. doi: 10.1073/pnas.181336898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that supports the findings of this study are available in the main document and supplementary material of this article. TGCA-SKCM datasets are publicly available at https://portal.gdc.cancer.gov/and GDAC Firehose (https://gdac.broadinstitute.org/).


