Abstract
Background
Treatment strategies inhibiting BRAF in combination with EGFR have been developed in patients with BRAFV600E mutant metastatic colorectal cancer, but intrinsic and secondary resistance remains a challenge. We aimed to investigate which genetic alterations cause intrinsic non-response and/or acquired resistance in these patients receiving therapies consisting of a backbone of BRAF and EGFR inhibition.
Methods
This was a cohort study on genetic alterations in patients with BRAFV600E mutant advanced colorectal cancer treated with inhibitors of the MAPK pathway. We examined tumour tissue for genetic alterations at baseline, during treatment and at progression.
Results
In total, 37 patients were included in this cohort. Genetic alterations in EGFR and in PIK3CA are associated with non-response. A greater fraction of non-responders (75%) versus responders (46%) had at least one genetic alteration in other genes than TP53, APC or BRAF. Secondary resistance mutations (n = 16 patients) were observed most frequently in the PI3K pathway (n = 6) and in receptor tyrosine kinases (n = 4), leading to increased upstream signalling.
Conclusions
Genetic alterations in the PI3K and upstream receptor tyrosine kinases were mostly associated with intrinsic and acquired resistance. By understanding these alterations, simultaneous or alternating treatments with targeted inhibitors might improve response duration.
Subject terms: Colon cancer, Tumour biomarkers
Background
Colorectal cancer (CRC) is one of the leading causes of mortality in the world and was responsible for almost 900,000 deaths worldwide in 2018.1 At initial diagnosis, metastasised disease is found in 25% of the patients and 50% of all patients will develop metastases during their disease course.2 Approximately 8–15% of metastatic CRC harbour a BRAFV600E mutation, which results in failure of standard chemotherapy and a dismal prognosis.3 The BRAF gene encodes a serine/threonine protein kinase, which is part of the signal transduction pathway RAS-RAF-MEK-ERK, also known as the mitogen-activated protein kinase (MAPK) pathway. Activating BRAF mutations are leading to signalling via this pathway by phosphorylation of the downstream MEK 1/2 proteins. MEK 1/2 subsequently phosphorylates the ERK1/2 kinases, resulting in gene transcription that drives cell proliferation and survival.4 The BRAFV600E mutation is the most common mutation among different tumour types and is caused by the substitution of valine to glutamic acid within codon 600.5 BRAFV600E mutations were initially reported by Davies et al. in 2002. They discovered that these mutations in melanoma led to an overactive MAPK pathway and could therefore be an interesting drug target.6 During the past decades, researchers intensively studied the BRAFV600E mutation to understand its role in tumour development and to explore possible treatment strategies for BRAFV600E mutated carcinoma, including CRC. Initially, the BRAF inhibitor vemurafenib had been investigated with observed responses in only 5% of the patients with BRAFV600E mutated metastatic CRC.7 This lack of response was found to be the result of feedback reactivation of EGFR after BRAF inhibition, thereby limiting the response to BRAF inhibitors.8,9 To optimise response rates, BRAF inhibitors have been combined with EGFR inhibitors and other targeted agents in doublet and triplet regimens.10–13 So far the combination of the BRAF inhibitor encorafenib and the EGFR inhibitor cetuximab with or without the MEK inhibitor binimetinib showed the best outcome with an overall response rate of 23% (doublet) and 29% (triplet) with manageable toxicity. Progression free survival was 4.2 months for the doublet and 4.3 months for the triplet regimen.11 The combination of encorafenib and cetuximab was recently approved by the Food and Drug Administration (FDA).14 Although response rates are improved and acquired resistance delayed, progression free survival remains short and resistance is still a major challenge.15
Since not all BRAFV600E mutant tumours are responsive to MAPK inhibitors and resistance patterns differ among preclinical studies, it is likely that BRAFV600E mutated tumours are highly heterogeneous.16 Moreover, two gene expression subtypes were earlier identified in a clustered analysis of 218 biopsies from BRAFV600E mutant tumours. BRAFV600E mutant subtype 1 (BM1) harboured KRAS/mTOR/AKT/4EBP1 activation with high levels of immune infiltration and epithelial-mesenchymal transition (EMT) and BRAFV600E mutant subtype 2 (BM2) was mainly dysregulated in cell-cycle checkpoints.17 A higher sensitivity for BRAF, MEK and EGFR inhibition with dabrafenib, trametinib and panitumumab was found in BM1.18 These results suggest that BRAFV600E mutated CRC is indeed a heterogeneous disease with different molecular patterns, responses to and targets for therapy.
Improvement in understanding this heterogeneity, resistance and moderate response rates of MAPK inhibitors in BRAFV600E mutated CRC is pivotal to optimise treatment outcomes. We here provide an overview of intrinsic and acquired mutations before and during treatment with targeted agents in patients with BRAFV600E mutant CRC. We present a cohort study of BRAFV600E mutant CRC patients treated with combinations of MAPK inhibitors in the Netherlands Cancer Institute-Antoni van Leeuwenhoek hospital (NKI-AVL). The research questions we aim to address are; (1) Which biomarkers are associated with non-responders? (2) Which secondary mutations causing acquired resistance are developed during targeted treatment of the MAPK pathway? (3) Can we take advantage of this secondary mutations for optimisation of subsequent treatment?
Methods
Cohort study
This retrospective cohort study was conducted in the NKI-AVL in patients with BRAFV600E mutated metastatic CRC between January 2012 and December 2019. All patients gave permission for the use of their residual material and data for research purposes. All patients included in the analyses were treated with targeted therapies, including the following combinations; encorafenib/cetuximab, encorafenib/cetuximab/alpelisib or encorafenib/cetuximab/ binimetinib. Encorafenib is an orally administered small molecule that inhibits BRAF.19 Cetuximab is an intravenously administered monoclonal antibody that binds specifically to the extracellular domain of EGFR. Binimetinib is an orally available small molecule and inhibits MEK 1/2.20 Alpelisib is an orally administered small molecule that inhibits PI3-kinase.10 The drugs were administered in the following doses: encorafenib 100–450 mg once daily continuously, cetuximab in an initial dose of 400 mg/m2 and thereafter 250 mg/m2 weekly, binimetinib 45 mg twice daily continuously and alpelisib 100–300 mg once daily continuously. We reviewed electronical medical records for information on demographics, anti-tumour response according to RECIST v 1.1. and mutational status. Results from histopathological reports of tumour biopsies before, during treatment or at progressive disease were collected, if available. On treatment tumour biopsies were collected after at least two weeks of treatment, which means that all drugs were on steady state concentrations. Results from different sequencing methods were included (Supplementary Table S1). If paired biopsies were available for patients, we carefully reviewed the sequencing methods used for overlap in genes present in the panels of the different sequencing methods. All results presented are restricted to genetic alterations of known clinical significance. This means that variants of unknown clinical significance (VUS) were excluded from this analysis.
Statistical methods
Descriptive statistics, including median along with percentages and frequencies for categorical variables were tabulated and presented in this paper. Responses were defined according to RECIST version 1.1. criteria.21 Time to progression was defined as the time from start of targeted treatment to date of first mentioning of progressive disease.
Results
Overall baseline characteristics
A total of 53 patients with BRAFV600E mutated metastatic CRC were screened for mutational status at baseline, on treatment and at progressive disease. The genetic alterations, including the variant of the mutation (e.g. KRASG12V), anti-tumour response and sequencing method per patient are summarised in Supplementary Table S2 for the total set of 53 patients. The current analysis is performed in 37 patients with at least one other detected genetic alteration at one-time point, besides the known BRAFV600E mutation.
The majority of patients was pre-treated with no more than two lines of anti-cancer therapy for advanced disease, including one patient with one line of immunotherapy, before the start of double or triple combined targeted agents. Seventeen patients were treated with encorafenib and cetuximab, seven patients with the combination of encorafenib, cetuximab and binimetinib and 13 patients with encorafenib, cetuximab and alpelisib. More than half of the tumours were microsatellite stable (Table 1).
Table 1.
Baseline characteristics of patients included in the analyses.
| Patients n = 37 | |
|---|---|
| Sex, n (%) | |
| Female | 24 (65%) |
| Male | 13 (35%) |
| Age, median (range), years | 59 (38–74) |
| Number of prior treatment lines, n (%) | |
| 1 | 14 (38%) |
| 2 | 17 (46%) |
| ≥3 | 6 (16%) |
| Treatment arm, n (%) | |
| Encorafenib + cetuximab | 17 (46%) |
| Encorafenib + cetuximab + binimetinib | 7 (19%) |
| Encorafenib + cetuximab + alpelisib | 13 (35%) |
| Micro satellite stability, n (%) | |
| MSI | 1 (3%) |
| MSS | 20 (54%) |
| Unknown | 16 (43%) |
| Time points molecular analysis, n (%) | |
| Single: BL | 16 (43%) |
| Single: OT | 2 (5%) |
| Paired: BL and OT | 5 (14%) |
| Paired: BL and PD | 6 (16%) |
| Paired: OT and PD | 4 (11%) |
| Paired: BL, OT and PD | 4 (11%) |
n number, MSI micro satellite instable, MSS micro satellite stable, BL baseline, OT on treatment, PD progressive disease.
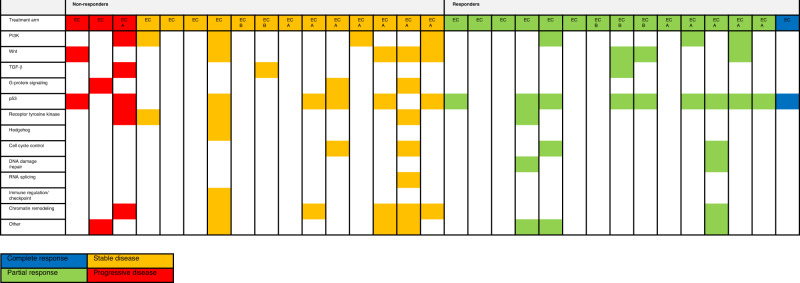
Best response on treatment and time to progression were collected for the total patient population and per treatment arm to correlate genetic alterations and anti-tumour activity (Table 2). The overall response rate was 46%, including one patient with a complete response (CR) and 16 patients with a partial response (PR). The median time to progression (TTP) was 9 months (range 1–26 months). Although significant correlations between mutational status and clinical outcomes were not found in this small sample size, some interesting trends were observed as described below. The genetic alterations per patient and anti-tumour response, including best response (BR) and TTP, is schematically shown in Fig. 1.
Table 2.
Anti-tumour activity.
| Encorafenib + cetuximab n = 17 | Encorafenib + cetuximab + binimetinib n = 7 | Encorafenib + cetuximab + alpelisib n = 13 | Total n = 37 | |
|---|---|---|---|---|
| Best response, n (%) | ||||
| CR | 1 (6%) | 0 (0%) | 0 (0%) | 1 (3%) |
| PR | 8 (473%) | 3 (43%) | 5 (38%) | 16 (43%) |
| SD | 6 (35%) | 3 (43%) | 7 (54%) | 16 (43%) |
| PD | 2 (12%) | 1 (14%) | 1 (8%) | 4 (10%) |
| TTP, median (range), months | 8 (1–26) | 14 (3–26) | 7 (2–16) | 9 (1–26) |
CR complete response, PR partial response, SD stable disease, PD progressive disease, TTP time to progression, n number of patients.
Fig. 1. Anti-tumour activity, including best response and time to progression, and mutational characterisation per patient and per treatment arm.
Genetic alterations are categorised per pathway; all included alterations are part of the pathway or directly influencing the pathway. TTP time to progression, BL baseline, OT on treatment, PD progressive disease.
Paired biopsies were available for 19 patients, of which 16 double paired biopsies and three triple paired biopsies. However, three patients were excluded from the analyses of paired biopsies, because of the lack of overlap in genes tested in the different sequencing methods per time point. In summary, baseline assessment was conducted in 16 non-responding and 15 responding patients and paired analyses comparing different time points in 16 patients (see Fig. S1 for the CONSORT diagram).
Biomarkers that predict non-response
The baseline samples of 16 patients with stable (n = 13) or progressive disease (n = 3) were analysed with the sequencing panels indicated in Table S1 to understand the genetic alterations that predict non-response. These 16 patients were compared to the baseline samples of 15 patients with response, including 14 patients with a PR and one patient with a CR. See Fig. 2 for the specific mutations in responding versus non-responding patients. No mutations other than BRAFV600E were found in eight patients, four non-responders and four responders. Six mutations were observed in the phosphoinositide-3-kinase (PI3K) pathway among four different non-responders and four mutations were observed among four different responders. Interestingly, PIK3CA mutations were found in non-responding patients and PTEN mutations in responding patients. Genetic alterations in or directly influencing the WNT pathway were detected in the tumour of five non-responding patients and three responding patients. The WNT pathway mutations in the three responding patients only included the commonly mutated APC gene. Given the fact that APC (and also TP53) mutation is frequent in colon cancer (seen in 35–70% of patients), it is unlikely that APC itself is a biomarker of response to BRAF inhibitor combinations. This means that especially relevant mutations in the WNT pathway or directly influencing the WNT pathway were observed in non-responding patients in the context of intrinsic resistance. Genetic alterations in genes related to chromatin remodelling were found in six non-responding patients and in only one responding patient. Moreover, in three patients mutations were observed in the epidermal growth factor receptor (EGFR) or another component of this receptor family (ERBB2 or ERBB4).
Fig. 2. Genetic alteration per pathway in tumours of non-responding patients versus responding patients.
Genetic alterations are categorised per pathway; all included alterations are part of the pathway or directly influencing the pathway. EC encorafenib and cetuximab, ECB encorafenib, cetuximab and binimetinib, ECA encorafenib, cetuximab and alpelisib.
Interestingly, if not counting TP53 and APC mutations, 73% of the tumours of non-responding patients were mutated in other genes than BRAF or TP53 and for responding patients this was only 46%. Furthermore, two pathways were mutated in more than one non-responding patients and not in responding patients, including mutations involved in RNA splicing (n = 2) and G-protein signalling (n = 3). No KRAS mutations were found at baseline in these 31 patients, which is not surprising since BRAF and KRAS mutations are considered mutually exclusive.22 In addition, no difference in outcome could be identified between microsatellite stable and instable tumours.
To summarise, genetic alterations in the following genes seem to predict non-response; alterations in or directly influencing the WNT pathway, alterations directly influencing chromatin remodelling, alterations in EGFR, components of EGFR or other receptor tyrosine kinases or alterations in PIK3CA. These mutations possibly bypass the MAPK pathway in BRAFV600E mutant CRC. A greater part of non-responding patients had at least one genetic alteration in other genes than TP53, APC or BRAF. Finally, genetic alterations in genes involved in G-protein signalling, immune regulation, RNA splicing and the Hedgehog pathway were only detected in the tumours of non-responding patients, but the significance of this remains uncertain.
Development of secondary mutations causing resistance
Since acquired resistance remains a major problem in the treatment with MAPK inhibitors, we explored the development of secondary mutations causing resistance by looking into available paired biopsies of 16 patients. From these 16 patients, ten were treated with encorafenib and cetuximab, four with encorafenib, cetuximab and binimetinib and two with the combination of encorafenib, cetuximab and alpelisib. Only mutations were included which were at least tested in molecular assessments at two time points. Several interesting trends were observed during the development of secondary resistance mutations (Fig. 3). One out of 16 patients did not develop a de novo mutation in a gene tested before and after treatment. The other 15 patients developed genetic alterations in different pathways. Four mutations were observed in receptor tyrosine kinases or their ligands (FGF, ERBB), which could lead to resistance by activation of upstream signalling. Six out of 15 patients (40%) developed mutations in the PI3K pathway, of which one patient was treated with the triple combination including alpelisib. This patient simultaneously developed genetic alterations in the MAPK and WNT pathways. We cannot exclude that the biopsy contained two clones that had acquired independent resistance mutations.
Fig. 3. Pie charts of acquired mutations during treatment with MAPK inhibitors, per treatment arm.
a–d Show pie charts with the specific mutations. e Shows a pie chart with the involved pathways in which mutations were observed. E encorafenib, C cetuximab, B binimetinib, A alpelisib, N number of patients.
Heterogeneity
Besides interpatient variability in secondary mutations, heterogeneity was also observed within individual patients. For one patient treated with the triple combination of encorafenib, cetuximab and binimetinib, tumour tissue from three different liver metastases was available on treatment. All metastases harboured BRAFV600E, APCR1450* and PIK3CAE545K mutations, but only two of the lesions harboured a PTEN mutation and one of these lesions a KRASG12V mutation. Interestingly, the PTEN mutations were not identical, including a PTENR173C and PTENR233* mutation, marking the heterogeneity of the disease. When correlating these genetic alterations with radiological assessment, the lesion with the PTENR233* and KRASG12V mutation was progressive while the other two liver metastasis were in regression. The best response for this patient was stable disease with a TTP of 22 months. This patient demonstrates the heterogeneous character of the disease and heterogeneous response to treatment of different metastatic lesions.
In summary, the development of acquired resistance is common with intra- and interpatient heterogeneity and secondary mutations are observed on different levels in the MAPK pathway and interconnected pathways.
Taken all results together, genetic alterations in EGFR and in PIK3CA are associated with non-response. A greater fraction of non-responders (75%) versus responders (46%) had at least one genetic alteration in other genes than TP53, APC or BRAF. Secondary resistance mutations (n = 16 patients) were observed most frequently in the PI3K pathway (n = 6) and in receptor tyrosine kinases (n = 4), leading to increased upstream signalling.
Discussion
In this retrospective cohort study, patterns of intrinsic and acquired resistance in patients with BRAFV600E mutated metastatic CRC treated with double or triple combinations of inhibitors of the MAPK and PI3K pathways were observed. The majority of mutated pathways at baseline in our cohort study were similar to findings described in the literature. A total of six prior published clinical trials report molecular results of tumour tissue and circulating free DNA (cfDNA) in patients with BRAFV600E mutated metastatic CRC receiving therapies consisting of a backbone of BRAF and EGFR inhibition or BRAF inhibition monotherapy.7,10,13,18,23,24 No difference in outcome was described between microsatellite stable and instable tumours in this cohort nor in literature.7,12,13 The genetic alterations identified for acquired resistance arose in genes directly or indirectly activating signalling via the MAPK pathway or cross-linked pathways. In our cohort study, genetic alterations in or directly influencing the WNT pathway, directly influencing chromatin remodelling, in the PI3K pathway, and upstream in EGFR or other receptor tyrosine kinases seem to predict for non-response as these mutations probably are considered driver mutations in BRAFV600E mutant CRC. The earlier investigation of 15 molecularly analysed BRAFV600E CRC tumour samples of patients treated with the BRAF inhibitor dabrafenib and MEK inhibitor trametinib harboured alterations in the WNT and p53 pathways without a clear correlation with treatment outcome. Five out of 15 tumours had mutations in PIK3CA of which 60% of patients had a PR or CR. However, no clear correlation was reported for PTEN loss or EGFR expression and progression free survival.13 Remarkably, in our study cohort and in cfDNA profiling by Hong et al. it was observed that resistance might be caused by mutations in the PI3K or genes influencing the WNT pathway or at the level of EGFR signalling, resulting in signalling through these pathways.23 Activation of the WNT pathway or upstream receptor tyrosine kinases plays a pivotal role in epithelial-to-mesenchymal transition (EMT), often associated with a CSM4 subtype, in colorectal cancers leading to an adverse prognostic phenotype causing resistance to anti-cancer therapies. Another pathway involved in the development of the EMT subtype is TGF-β, which was probably activated in a minority of patients due to mutations in the SMAD gene.25 Surprisingly, Middleton et al. has shown that the majority of CSM4 subtype BRAFV600E mutant CRC represents a BM1 signature with a better response to combined treatment with dabrafenib, trametinib and panitumumab.18 Unfortunately, it was not feasible to actually determine molecular subtypes BM1 and BM2 in our patient cohort due to restrictions in sequencing data. It would nevertheless be very interesting to define those subtypes in future research on BRAF/MEK and EGFR inhibition to confirm if screening for BM1 or BM2 at baseline could be used as predictive biomarker for sensitivity to targeted treatments in BRAFV600E mutant CRC.
A total of three out of 16 patients in this cohort developed KRAS mutations on disease progression. Focal amplification of KRAS was earlier reported in one post-progression biopsy of a patient treated with RAF/MEK inhibition and in cfDNA samples of patients treated with vemurafenib and panitumumab.12,26 In addition, KRAS or NRAS clones or subclonal RAS mutations were detected in respectively 48% and 21% of patients on the time of disease progression.24 KRAS mutations activate CRAF leading to sustained phosphorylation of ERK and resistance, despite combined BRAF and EGFR or MEK inhibition.26 The simultaneous presence of KRAS and BRAF mutations also implies disease heterogeneity, since KRAS and BRAF mutations are mutually exclusive during primary tumour development.22 Clones sensitive and resistant to treatment might be present at the same time, which must be taken into account by switching to the next line of anti-tumour therapy. Adding an inhibitor to the current treatment regimen after development of resistance is recommendable to enhance treatment duration, if toxicity of the new drug combination is expected to be manageable. In the case of disease heterogeneity and expected severe or unpredictable toxicity of a combination, it might be better to start an alternating treatment regimen. This approach is currently investigated in resistant BRAFV600E mutated melanoma. In a proof of concept study, patients are treated with the histone deacetylase inhibitor vorinostat for 14 days upon resistance and thereafter BRAF and/or MEK inhibitors are reintroduced.27 This cohort contains unique data on paired molecular analyses in tumour tissue for patients with BRAFV600E mutant metastatic CRC treated with encorafenib and cetuximab with or without binimetinib or alpelisib. Since this treatment combination was recently approved by the FDA and the European Medicines Agency (EMA), the data are considered highly relevant for clinical practice. Despite these unique data, our study has some limitations. No statistically significant differences between non-responders and responders could be found due to the combination of the small sample size, difference in sequencing methods and the lack of molecular analyses on three different time points for all patients. Since molecular analysis on the different time points was not always performed in tumour tissue of the same lesion, it might be that mutations detected were not necessarily newly developed. Due to tumour heterogeneity, these mutations could already have been present at start of treatment. To strengthen our data, we decided to perform the analyses on paired biopsies in 16 patients to improve robustness of the results. Despite the presence of uncertainties, the trends in our and earlier published data provide insight into the mechanisms of resistance in this specific patient group and might generate opportunities for future studies.
In conclusion, our findings show that genetic alterations causing intrinsic and acquired resistance in this patient cohort were observed before and upon treatment with BRAFV600E targeted therapies. The genetic alterations revealed for intrinsic and acquired mutations arose in genes directly or indirectly activating signalling via the MAPK pathway or cross-linked pathways. Intrinsic and acquired resistance mechanisms are heterogeneous with a high intra- and interpatient variability. Based on these results, we suggest comprehensive molecular screening of BRAFV600E mutant metastatic CRC before start of first-line treatment in the palliative setting. Furthermore, it might be considered to closely monitor genetic alterations and accordingly switching therapy to a combined simultaneous or alternating treatment with a backbone of BRAF and EGFR inhibition combined with an inhibitor of the genetic alterations to optimise duration of treatment.
Monitoring of genetic alterations and switching therapies accordingly of course, could not be considered part of standard therapy but should be the scope of future studies. The transcriptional context, identification of responding and non-responding subtypes such as BM1 and BM2, real-time monitoring of tumour DNA and the effect of accordingly changing treatment strategies on response should be part of that.
Supplementary information
Acknowledgements
We thank all patients for participation, the Clinical Research Unit of the NKI-AVL and the pathology department for conduct of the study.
Author contributions
The methods and analyses were developed and performed by S.H. with help of M.B. S.H. was in charge of final analyses of all the available data with help of R.B., F.O. and M.B. All authors reviewed and approved the paper.
Ethics approval and consent to participate
For these patients, no specific informed consent has been asked as this hospital has a general informed consent procedure in which patients give permission for the use of their residual material and data for research purposes. The general informed consent procedure has been approved by the ethics committee of the Antoni van Leeuwenhoek hospital, METC-AVL. The principles of the Declaration of Helsinki have been followed in this retrospective cohort study.
Data availability
The authors are committed to share sequencing data and patient characteristics in an anonymised manner according to applicable privacy regulations and laws. All data requests are reviewed and approved by the Institutional Review Board of the NKI-AVL on the basis of scientific relevance.
Competing interests
The authors declare no competing interests.
Funding information
No funding or other financial support was received for this paper.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sanne C. F. A. Huijberts, Email: s.huijberts@nki.nl
Frans L. Opdam, Email: f.opdam@nki.nl
Supplementary information
Supplementary information is available for this paper at 10.1038/s41416-020-01147-2.
References
- 1.Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019;14:89–103. doi: 10.5114/pg.2018.81072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016;27:1386–1422. doi: 10.1093/annonc/mdw235. [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E, Huijberts S, Grothey A, Yaeger R, Cuyle P, Elez E, et al. Binimetinib, Encorafenib, and Cetuximab Triplet therapy for patients with BRAF V600E-mutant metastatic colorectal cancer: safety lead-in results from the Phase III BEACON Colorectal Cancer Study. J. Clin. Oncol. 2019;37:1460–1469. doi: 10.1200/JCO.18.02459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caputo F, Santini C, Bardasi C, Cerma K, Casadei-Gardini A, Spallanzani A, et al. BRAF-mutated colorectal cancer: clinical and molecular insights. Int J. Mol. Sci. 2019;20:5369. doi: 10.3390/ijms20215369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahrami A, Hesari A, Khazaei M, Hassanian S, Ferns G, Avan A, et al. The therapeutic potential of targeting the BRAF mutation in patients with colorectal cancer. J. Cell Physiol. 2018;233:2162–2169. doi: 10.1002/jcp.25952. [DOI] [PubMed] [Google Scholar]
- 6.Davies H, Bignell G, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 7.Kopetz S, Desai J, Chan E, Hecht J, O’Dywer P, Maru D, et al. Phase II Pilot Study of Vemurafenib in patients with metastatic BRAF-mutated colorectal cancer. J. Clin. Oncol. 2015;33:4032–4038. doi: 10.1200/JCO.2015.63.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 9.Corcoran R, Ebi H, Turke A, Coffee E, Nishino M, Cogdill A, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Disco. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Geel R, Tabernero J, Elez E, Bendell J, Spreafico A, Schuler M, et al. A Phase Ib dose-escalation study of encorafenib and cetuximab with or without alpelisib in metastatic BRAF-mutant colorectal cancer. Cancer Discov. 2017;7:610–619. doi: 10.1158/2159-8290.CD-16-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-mutated colorectal cancer. N. Engl. J. Med. 2019;381:1632–1643. doi: 10.1056/NEJMoa1908075. [DOI] [PubMed] [Google Scholar]
- 12.Yaeger R, Cercek A, O’Reilly E, Reidy D, Kemeny N, Wolinsky T, et al. Pilot trial of combined BRAF and EGFR inhibition in BRAF-mutant metastatic colorectal cancer patients. Clin. Cancer Res. 2015;21:1313–1320. doi: 10.1158/1078-0432.CCR-14-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corcoran R, Atreya C, Falchook G, Kwak E, Ryan D, Bendell J, et al. Combined BRAF and MEK inhibition with dabrafenib and trametinib in BRAF V600-mutant colorectal cancer. J. Clin. Oncol. 2015;33:4023–4031. doi: 10.1200/JCO.2015.63.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Food and Drug Administration. FDA approves encorafenib in combination with cetuximab for metastatic colorectal cancer with a BRAF V600E mutation. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-encorafenib-combination-cetuximab-metastatic-colorectal-cancer-braf-v600e-mutation (2020).
- 15.Obaid N, Bedard K, Huang W. Strategies for overcoming resistance in tumours harboring BRAF mutations. Int J. Mol. Sci. 2017;18:585. doi: 10.3390/ijms18030585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oddo D, Sennott E, Barault L, Valtorta E, Arena S, Cassingena A, et al. Molecular landscape of acquired resistance to targeted therapy combinations in BRAF-mutant colorectal cancer. Cancer Res. 2016;76:4504–4515. doi: 10.1158/0008-5472.CAN-16-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barras D, Missiaglia E, Wirapati P, Sieber O, Jorissen R, Love C, et al. BRAF V600E mutant colorectal cancer subtypes based on gene expression. Clin. Cancer Res. 2017;23:104–115. doi: 10.1158/1078-0432.CCR-16-0140. [DOI] [PubMed] [Google Scholar]
- 18.Middleton G, Yang Y, Campbell C, Andre T, Atreya C, Schellens J, et al. BRAF-mutant transcriptional subtypes predict outcome of combined BRAF, MEK, and EGFR blockade with dabrafenib, trametinib, and panitumumab in patients with colorectal cancer. Clin. Cancer Res. 2020;26:2466–2476. doi: 10.1158/1078-0432.CCR-19-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Center for Biotechnology Information. PubChem Database. Encorafenib, CID=50922675. https://pubchem.ncbi.nlm.nih.gov/compound/Encorafenib (2020).
- 20.Huijberts S, van Geel R, Bernards R, Beijnen J, Steeghs N. Encorafenib, binimetinib and cetuximab combined therapy for patients with BRAFV600E mutant metastatic colorectal cancer. Future Oncol. 2020;16:161–173. doi: 10.2217/fon-2019-0748. [DOI] [PubMed] [Google Scholar]
- 21.Eisenhauer E, Therasse P, Bogaerts J, Schwartz L, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur. J. Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 22.Cisowski J, Sayin V, Liu M, Karlsson C, Bergo M. Oncogene-induced senescence underlies the mutual exclusive nature of oncogenic KRAS and BRAF. Oncogene. 2016;35:1328–1333. doi: 10.1038/onc.2015.186. [DOI] [PubMed] [Google Scholar]
- 23.Hong D, Morris V, El Osta B, Sorokin A, Janku F, Fu S, et al. Phase IB Study of Vemurafenib in combination with Irinotecan and Cetuximab in patients with metastatic colorectal cancer with BRAFV600E mutation. Cancer Discov. 2016;6:1352–1365. doi: 10.1158/2159-8290.CD-16-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corcoran R, Andre T, Atreya C, Schellens J, Yoshino T, Bendell J, et al. Combined BRAF, EGFR, and MEK inhibition in patients with BRAF(V600E)-mutant colorectal cancer. Cancer Discov. 2018;8:428–443. doi: 10.1158/2159-8290.CD-17-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahronian L, Sennot E, Van Allen E, Wagle N, Kwak E, Faris J, et al. Clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discov. 2015;5:358–367. doi: 10.1158/2159-8290.CD-14-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huijberts S, Wang L, de Oliveira R, Rosing H, Nuijen B, Beijnen J, et al. Vorinostat in patients with resistant BRAF(V600E) mutated advanced melanoma: a proof of concept study. Future Oncol. 2020;16:619–629. doi: 10.2217/fon-2020-0023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors are committed to share sequencing data and patient characteristics in an anonymised manner according to applicable privacy regulations and laws. All data requests are reviewed and approved by the Institutional Review Board of the NKI-AVL on the basis of scientific relevance.