RcsB is a two-component response regulator in the Rcs phosphorelay system, and it plays versatile regulatory functions in Enterobacteriaceae. However, information on the function of the RcsB protein in bacteria, especially in S. marcescens, remains limited. In this work, we illustrated experimentally that the RcsB protein was involved in diverse cellular processes in S. marcescens, including prodigiosin synthesis, cell motility, capsular polysaccharide production, biofilm formation, and acid resistance. Additionally, the regulatory mechanism of the RcsB protein in these cellular processes was investigated. In conclusion, this work indicated that RcsB could be a regulator for prodigiosin synthesis and provides insight into the function of the RcsB protein in S. marcescens.
KEYWORDS: regulator RcsB, prodigiosin synthesis, cellular processes, Serratia marcescens
ABSTRACT
Prodigiosin (PG), a red linear tripyrrole pigment normally secreted by Serratia marcescens, has received attention for its reported immunosuppressive, antimicrobial, and anticancer properties. Although several genes have been shown to be important for prodigiosin synthesis, information on the regulatory mechanisms behind this cellular process remains limited. In this work, we identified that the transcriptional regulator RcsB encoding gene BVG90_13250 (rcsB) negatively controlled prodigiosin biosynthesis in S. marcescens. Disruption of rcsB conferred a remarkably increased production of prodigiosin. This phenotype corresponded to negative control of transcription of the prodigiosin-associated pig operon by RcsB, probably by binding to the promoter region of the prodigiosin synthesis positive regulator FlhDC. Moreover, using transcriptomics and further experiments, we revealed that RcsB also controlled some other important cellular processes, including swimming and swarming motilities, capsular polysaccharide production, biofilm formation, and acid resistance (AR), in S. marcescens. Collectively, this work proposes that RcsB is a prodigiosin synthesis repressor in S. marcescens and provides insight into the regulatory mechanism of RcsB in cell motility, capsular polysaccharide production, and acid resistance in S. marcescens.
IMPORTANCE RcsB is a two-component response regulator in the Rcs phosphorelay system, and it plays versatile regulatory functions in Enterobacteriaceae. However, information on the function of the RcsB protein in bacteria, especially in S. marcescens, remains limited. In this work, we illustrated experimentally that the RcsB protein was involved in diverse cellular processes in S. marcescens, including prodigiosin synthesis, cell motility, capsular polysaccharide production, biofilm formation, and acid resistance. Additionally, the regulatory mechanism of the RcsB protein in these cellular processes was investigated. In conclusion, this work indicated that RcsB could be a regulator for prodigiosin synthesis and provides insight into the function of the RcsB protein in S. marcescens.
INTRODUCTION
The Rcs phosphorelay system is one of the most complex two-component signal transduction systems in bacteria (1–3). At the core of this system are three multidomain proteins, including the hybrid sensor kinase RcsC, the phosphotransfer protein RcsD, and the response regulator RcsB. In addition, the outer membrane lipoprotein RcsF and the inner membrane protein IgaA (also known as GumB) are auxiliary proteins for receiving extracellular signals (4). Previous studies have reported that the response regulator RcsB controls expression of hundreds of bacterial genes associated with capsular polysaccharide biosynthesis (5, 6), flagellar biogenesis (7, 8), antibiotic resistance (9, 10), biofilm formation (11, 12), and virulence (13, 14) in Enterobacteriaceae. However, our understanding of the role of the regulator RcsB in bacteria, especially in Serratia marcescens is still limited.
S. marcescens, a Gram-negative rod-shaped bacterium of the Enterobacteriaceae family, is found in a wide range of environments, including soil, water, plants, insects, and foods, and is used for production of many high-value products, including prodigiosin (15), althiomycin (16), serratamolide (17), acetoin (18), and 2,3-butanediol (19). Prodigiosin (PG), a red linear tripyrrole pigment, has attracted attention due to its antimalarial, antibacterial, antifungal, antiprotozoal, and immunosuppressant activities (15). Studies on the metabolic regulation network of prodigiosin biosynthesis in S. marcescens have found that the enzymes related to prodigiosin production in this bacterium could be divided into groups, namely, enzymes involved in the biosynthetic pathway of prodigiosin synthesis and enzymes encoding transcriptional regulators. The prodigiosin biosynthesis pathway in S. marcescens consists of the genes pigA to pigN, a total of 14 genes, which are transcribed as a polycistronic mRNA from a promoter upstream of pigA. The genes pigB, pigD, and pigE encode proteins sufficient for the biosynthesis of 2-methyl-3-n-amyl-pyrrole (MAP), while the genes pigA, pigF, pigG, pigH, pigI, pigJ, pigK, pigL, pigM, and pigN are involved in the production of 4-methoxy-2,2′-bipyrrole-5-carbaldehyde (MBC). Finally, MAP and MBC condense to form prodigiosin via the terminal condensing enzyme PigC (15). The synthesis of prodigiosin in S. marcescens is also directly or indirectly controlled by transcriptional regulators, including the negative regulators MetR (20), SpnR (21), CopA (22), CRP (23), HexS (24), RssB (25), RcsB (26), and SmaR (27) and the positive regulators EepR (28), PigP (29), GumB (30), FlhDC (31), and RbsR (32). Although these nearly 30 genes have been reported to be involved in prodigiosin production in S. marcescens, knowledge of the regulatory mechanism behind prodigiosin biosynthesis in S. marcescens is still limited.
In this study, S. marcescens JNB5-1, a prodigiosin-producing strain isolated from soil samples (20), was used to investigate the regulatory mechanism behind prodigiosin synthesis in S. marcescens. By constructing a Tn5G insertion mutant library, the transcription regulator RcsB was identified to negatively regulate prodigiosin production in S. marcescens. Moreover, our study investigated the regulatory role of the RcsB protein in swarming and swimming motilities, capsular polysaccharide biosynthesis, biofilm formation, and acid resistance (AR) in S. marcescens. Our data showed an important role for the highly conserved RcsB protein in influencing prodigiosin production and investigated the mechanism by which RcsB regulates prodigiosin biosynthesis.
RESULTS
Identification of a regulator, RcsB, that negatively controls prodigiosin synthesis.
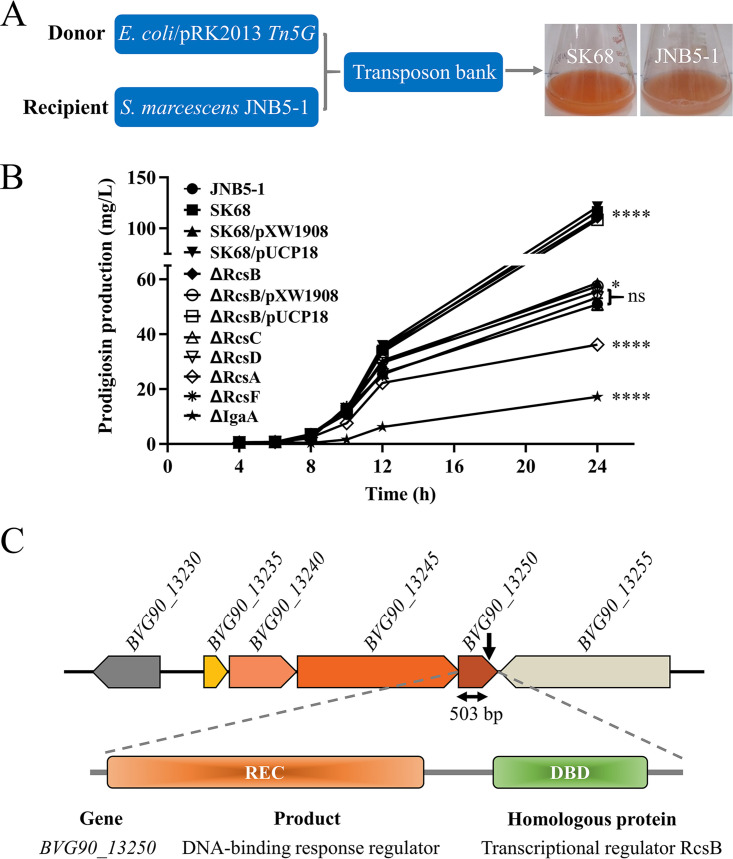
To identify genes that regulate prodigiosin production in S. marcescens, a Tn5G transposon insertion library was constructed using Escherichia coli/pRK2013 Tn5G as the donor strain and S. marcescens JNB5-1 as the recipient strain, and a prodigiosin-hyperproducing mutant, SK68, was isolated (Fig. 1A; see also Fig. S1 in the supplemental material). Shake flask fermentation analysis showed that the SK68 mutant could synthesize 116.48 mg/liter of prodigiosin after 24 h of fermentation, which was 2.28 times that synthesized by wild-type strain JNB5-1 (51.01 mg/liter) (P < 0.001; Fig. 1B).
FIG 1.
Regulator RcsB represses prodigiosin biosynthesis in S. marcescens. (A) A prodigiosin-hyperproducing mutant, SK68, was identified by Tn5G transposon insertion mutagenesis. (B) Shake flask fermentation to determine the ability of JNB5-1, SK68, SK68/pXW1908, SK68/pUCP18, ΔRcsB, ΔRcsB/pXW1908, ΔRcsB/pUCP18, ΔRcsA, ΔRcsC, ΔRcsD, ΔRcsF, and ΔIgaA strains to synthesize prodigiosin. JNB5-1 is a wild-type S. marcescens strain, SK68 is an rcsB-disrupted mutant, ΔRcsB is an rcsB deletion mutant, ΔRcsA is an rcsA deletion mutant, ΔRcsC is an rcsC deletion mutant, ΔRcsD is an rcsD deletion mutant, ΔRcsF is an rcsF deletion mutant, ΔIgaA is an igaA deletion mutant, SK68/pXW1908 and ΔRcsB/pXW1908 are rcsB complementary strains with plasmid pXW1908, and SK68/pUCP18 and ΔRcsB/pUCP18 are recombinant strains with empty vector pUCP18. (B) The experiment was performed independently three times. Error bars indicate standard deviations. One-way analysis of variance (ANOVA) was used to examine the mean differences between the data groups. ****, P < 0.001; *, P < 0.05; ns, no significant difference. (C) Genetic loci identified in mutant SK68. (Upper) Genetic map of the disrupted gene BVG90_13250 and its surrounding genes. The transposon insertion site is indicated by the black downward-pointing arrow. (Middle) Domain organization of the BVG90_13250 protein. (Lower) BVG90_13250 is highly homologous to the RcsB protein.
With inverse PCR, the insertion site of the Tn5G transposon in strain SK68 was identified between bp 503 and 504 in the coding region of the BVG90_13250 gene, which encodes a DNA binding response regulator. BVG90_13250 shares 100% identity with a predicted transcriptional regulator, RcsB, of a sequenced S. marcescens strain, Db11 (GenBank accession number NZ_HG326223) (Fig. 1C). Furthermore, the BVG90_13250 protein, containing a signal receiver domain (REC) at its N terminus and a DNA binding domain (DBD) at its C terminus, is 94.91%, 94.44%, 94.44%, 89.04%, and 92.13% identical to proven RcsB proteins of Escherichia coli K-12 (CQR81717.1) (33), Klebsiella pneumoniae ATCC 43816 (CEL83440.1) (5), Salmonella enterica subsp. enterica serovar Typhimurium strain ST4/74 (ADX18030.1) (34), Yersinia pestis KIM6+ (AKB89006.1) (35), and Yersinia ruckeri QMA0440 (ARZ00430.1) (36), respectively (see Fig. S2 in the supplemental material). Due to the high similarity to previously studied RcsB proteins from other Enterobacteriaceae species, we therefore referred to the BVG90_13250 open reading frame as RcsB. In the complementation experiment, the intact rcsB gene and the empty vector pUCP18 were separately introduced into the SK68 mutant, and a complementary strain, S. marcescens SK68/pXW1908, had significantly decreased prodigiosin production compared to that of mutant SK68 and recombinant strain SK68/pUCP18. This result demonstrated that RcsB was associated with the regulation of prodigiosin synthesis (Fig. 1B). To further confirm the function of the rcsB gene, a mutant strain, ΔRcsB, with the rcsB gene completely deleted was generated, and its ability to synthesize prodigiosin was analyzed. Results showed that prodigiosin production was significantly increased in the rcsB deletion mutant ΔRcsB compared to that in strains JNB5-1 and ΔRcsB/pXW1908 and was similar to that of rcsB-disrupted mutant SK68 (Fig. 1B). Together, these results suggested that RcsB functions as a prodigiosin synthesis repressor in strain JNB5-1.
The Rcs pathway is composed of the proteins RcsA, RcsC, RcsD, RcsF, and IgaA, and it is involved in regulating the phosphorylation and dephosphorylation of response regulator RcsB, thereby regulating the expression level of target genes (4). To study the contributions of other Rcs components to prodigiosin production in S. marcescens, we measured prodigiosin synthesis in several rcs mutants. Results showed that RcsC, RcsD, and RcsF conferred no significant effect on prodigiosin synthesis, while RcsA and IgaA positively regulated prodigiosin production in strain JNB5-1 (P < 0.001) (Fig. 1B). The result that IgaA positively regulates prodigiosin synthesis in S. marcescens was consistent with a previous report (30), and how RcsA regulates prodigiosin synthesis will be described elsewhere. Collectively, these results suggested that RcsB regulation of prodigiosin synthesis in S. marcescens probably does not require RcsACDF.
RcsB negatively regulates prodigiosin-associated pig operon transcription.
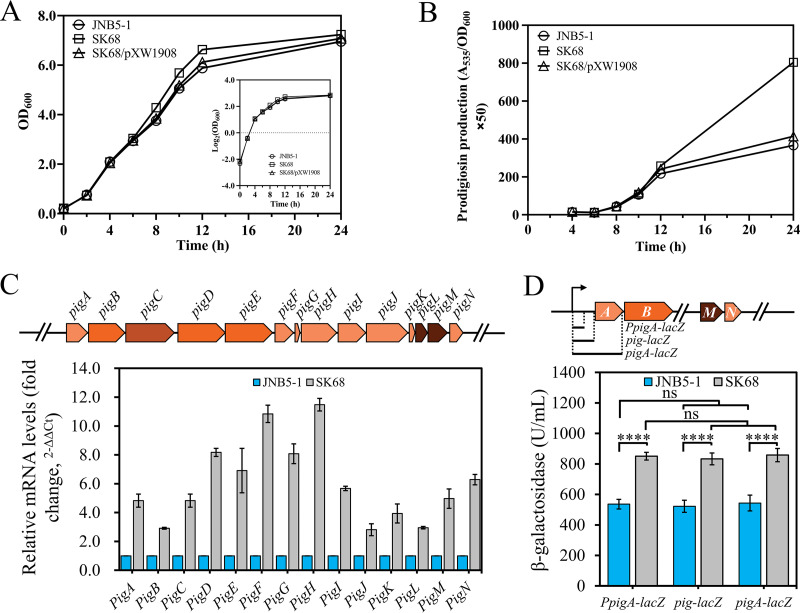
To analyze the cause of the higher production of prodigiosin in rcsB-disrupted mutant SK68, the cell growth of strains JNB5-1, SK68, and SK68/pXW1908 was determined. Results showed that there was no significant difference in cell growth between these three strains (Fig. 2A). Further analysis of single-cell synthesis of prodigiosin in these three strains found that the rcsB-disrupted mutant SK68 had significantly enhanced prodigiosin production compared to that of strains JNB5-1 and SK68/pXW1908 (Fig. 2B). These results indicated that the hyperproduction of prodigiosin in strain SK68 was possibly due to the higher expression levels of the prodigiosin-related pig gene cluster instead of to increased biomass.
FIG 2.
RcsB is a negative regulator for pig gene cluster transcription. (A) Growth curves of JNB5-1, SK68, and SK68/pXW1908 strains in LB medium. (B) Analysis of prodigiosin levels in JNB5-1, SK68, and SK68/pXW1908 strains. (C) Reverse transcription-quantitative PCR (RT-qPCR) analysis of the pig gene cluster expression level in the SK68 and JNB5-1 strains at an optical density at 600 nm (OD600) of 6.0. (Upper) Structure of the pig gene cluster; these 14 genes were transcribed as a polycistronic mRNA from a promoter upstream of the pigA gene. (Lower) Relative expression levels of the pig gene cluster in the strain SK68 compared with those in the JNB5-1 strain. (D) Analysis of β-galactosidase activity of strains SK68 and JNB5-1 harboring the PpigA-lacZ, pig-lacZ, or pigA-lacZ reporter fusion at an OD600 of 5.0. PpigA-lacZ indicates that the promoter of the pig operon was cloned upstream of the lacZ gene. pig-lacZ indicates that the upstream noncoding region, including the operator and promoter of the pig operon, was cloned upstream of the lacZ gene. pigA-lacZ indicates that the upstream noncoding region of the pig operon and coding sequences, except the termination codon of the pigA gene, was cloned upstream of the lacZ gene. (Upper) Schematic of PpigA-lacZ, pig-lacZ and pigA-lacZ. A, B, M and N indicate genes pigA, pigB, pigM, and pigN, respectively. (Lower) β-Galactosidase activity in strains SK68 and JNB5-1 harboring the PpigA-lacZ, pig-lacZ, or pigA-lacZ reporter fusion. (A to D) The experiment was performed independently three times. Error bars indicate standard deviations. Student’s t test was used to examine the mean differences between the data groups. ****, P < 0.001.
The synthesis of prodigiosin in S. marcescens is controlled by the pigA, pigB, pigC, pigD, pigE, pigF, pigG, pigH, pigI, pigJ, pigK, pigL, pigM, and pigN genes, which are transcribed as a polycistronic mRNA from a promoter upstream of pigA (Fig. 2C). To investigate how RcsB inhibits prodigiosin synthesis in JNB5-1, the expression levels of these 14 genes in JNB5-1 and rcsB-disrupted mutant SK68 were determined by real-time quantitative PCR (RT-qPCR) at the stationary phase (optical density at 600 nm [OD600] of 6.0), at which prodigiosin began to be produced in large quantities (Fig. 1B). As shown in Fig. 2C, compared to those in the wild-type strain, the expression levels of the pigABCDEFGHIJKLMN genes were upregulated by 2.91-fold to 11.48-fold in mutant SK68. In addition, a transcriptional reporter gene fusion of the pig operon was constructed by cloning the promoter region of the pig operon upstream of the lacZ gene, and β-galactosidase activity in strains JNB5-1 and SK68 was determined. Results showed that the level of transcription of the pig operon was significantly higher in the rcsB-disrupted mutant SK68 than in its parent strain, JNB5-1 (P < 0.001) (Fig. 2D). To further ascertain whether there is an indirect posttranscriptional regulation of the pig operon expression by RcsB, a pig-lacZ fusion reporter plasmid was constructed by cloning the upstream noncoding region, including the operator and the promoter of the pig operon upstream of the lacZ gene (Fig. 2D). Also, a pigA-lacZ fusion reporter plasmid was constructed by cloning the upstream noncoding region of the pig operon and coding sequences, except for the termination codon of the pigA gene upstream of the lacZ gene (Fig. 2D), and β-galactosidase activity in strains JNB5-1 and SK68 was determined. Results showed that the presence of the operator region of the pig operon or the coding sequences of the pigA gene had no significant effect on the expression level of the lacZ gene in both strains JNB5-1 and SK68 (Fig. 2D). These results suggested that the pig operon was not regulated by RcsB at the posttranscriptional level. Together, these results indicated that RcsB probably inhibited prodigiosin production by repressing the expression of the prodigiosin-associated pig operon at the transcriptional level.
Transcriptome analysis of the rcsB mutant strain.
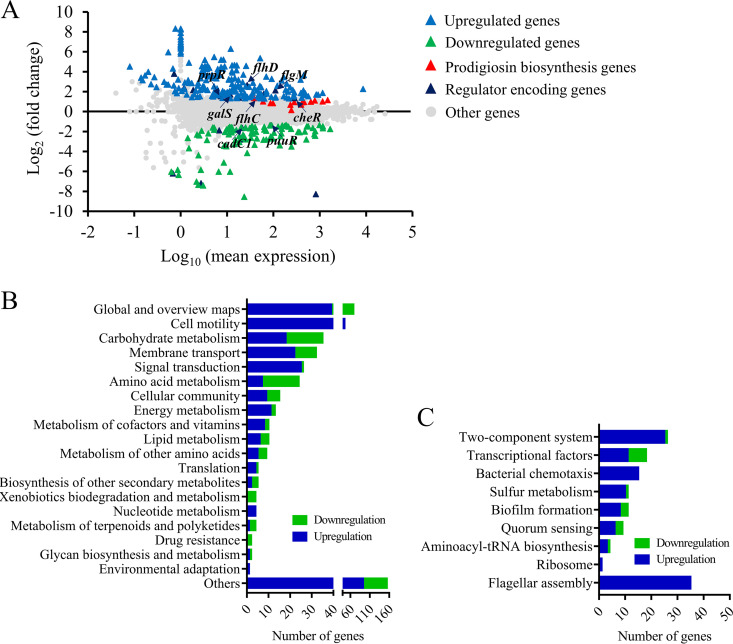
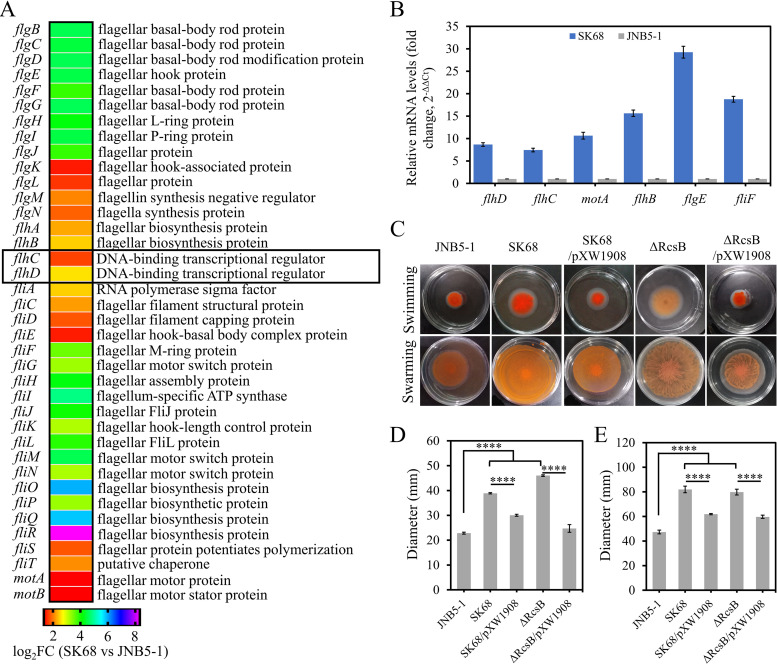
It has been confirmed that the RcsB regulator plays an important regulatory role in various bacterial metabolic pathways in Enterobacteriaceae (1, 4); however, our understanding of the role of RcsB in S. marcescens is still limited. Hence, to investigate the roles of RcsB in the cellular processes in S. marcescens, transcriptome sequencing (RNAseq) analysis was performed to evaluate gene expression differences between the rcsB-disrupted mutant SK68 and parent strain JNB5-1. Comparative transcriptomics data showed that the expression levels of 210 genes, including those of regulator-encoding genes flhC, flhD, flgM, cheR, gutM, prpR, and galS, were significantly upregulated and 106 genes, including regulator-encoding genes puuR and cadC1, were significantly downregulated in the rcsB-disrupted mutant SK68 (|log2[fold change {FC}]| ≥ 1, P ≤ 0.05) (Fig. 3A; see also Tables S4, S5, and S6). Based on the annotation of KEGG_B_class, these 316 significantly upregulated or downregulated genes were classified into 20 major cellular processes, such as cell motility, carbohydrate metabolism, and signal transduction (Fig. 3B). Moreover, these 316 genes could further be grouped using the Pathway Enrichment Analysis tool Omicshare into several major metabolic pathways, including flagellar assembly, two-component systems, and transcriptional factors, based on their annotations described in the pathway classification (Fig. 3C). Collectively, transcriptomics data suggested that RcsB possibly functions as a regulator controlling various cellular processes, including prodigiosin synthesis in S. marcescens.
FIG 3.
Transcriptome analysis of strains JNB5-1 and rcsB-disrupted mutant SK68. (A) Genome-wide analysis of gene expression differences between the rcsB-disrupted mutant SK68 and the wild-type strain JNB5-1 at an OD600 of 2.0. The x axis represents the logarithmic transformation value of gene expression levels in strain JNB5-1. The y axis represents log2-transformed values of gene expression fold changes between strains SK68 and JNB5-1. Light blue and green triangles represent the upregulated and downregulated genes, respectively. Dark blue represents the regulator-encoding genes. (B) Genes significantly differentially expressed by strains SK68 and JNB5-1 were classified into different cellular processes according to KEGG_B_class. (C) Expression profiles of the genes belonging to the indicated metabolic pathways. (B and C) Genes with significantly elevated and reduced expression levels in strain SK68 compared to strain JNB5-1 are shown in blue and green, respectively.
RcsB regulates the prodigiosin synthesis indirectly.
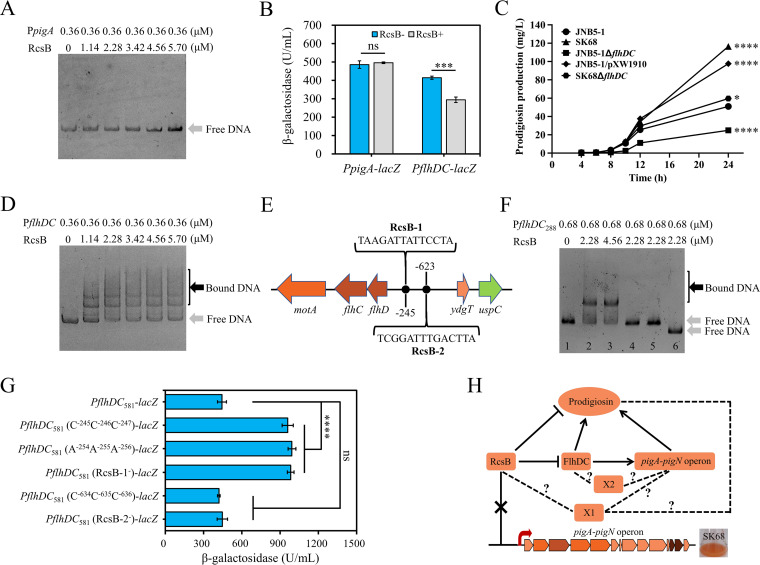
To explore whether RcsB directly controls the expression of the pig gene cluster, hence inhibiting prodigiosin synthesis, the rcsB gene was cloned with an N-terminal His tag in the pET28a plasmid, and the purified His-tagged RcsB was incubated with the 5′ noncoding region of the pigA gene (located between nucleotide positions −1 and −406 relative to the translational start site of the pigA gene) and subjected to electrophoretic mobility shift assay (EMSA). As shown in Fig. 4A, the result showed that the RcsB protein did not bind to the promoter region of the pig gene cluster. Furthermore, with a transcriptional lacZ reporter fusion, the expression level of the pig gene cluster was investigated in strains DH5α/pXW1908 (containing RcsB from strain JNB5-1 [RcsBSm+]) and DH5α/pUCP18 (lacking RcsB from strain JNB5-1 [RcsBSm−]) under the condition that strain DH5α has a functional RcsB protein that is very likely to be functionally redundant. Results showed that RcsB conferred no significant effect on pig gene cluster expression levels in E. coli (Fig. 4B). Taken together, these results suggested a possible indirect regulatory role of RcsB in prodigiosin synthesis genes.
FIG 4.
RcsB regulates the prodigiosin synthesis genes indirectly. (A) Electrophoretic mobility shift assay (EMSA) for RcsB protein binding to the promoter region of the pig operon. (B) Overexpression of RcsB protein has no significant effect on the expression of the pig gene cluster, but it inhibits the expression of the flhDC genes in E. coli. The transcription fusions PpigA-lacZ and PflhDC-lacZ were used in the E. coli strains DH5α/pXW1908 (rcsB+) and DH5α/pUCP18 (rcsB negative). The plasmid pXW1908 carries the intact rcsB gene. The samples were collected at an OD600 of 4.0. (C) Prodigiosin production analysis of strains JNB5-1, SK68, JNB5-1ΔflhDC, JNB5-1/pXW1910, and SK68ΔflhDC. JNB5-1 is a wild-type S. marcescens strain, SK68 is an rcsB-disrupted mutant, JNB5-1ΔflhDC is an flhDC deletion mutant, JNB5-1/pXW1910 is an rcsB-overexpressing mutant, and SK68ΔflhDC is an rcsB flhDC double mutant. (D) EMSA for RcsB protein binding to the promoter region of the flhDC genes. (E) Putative RcsB binding sites in the promoter region of flhDC genes in S. marcescens. The positions of the binding sites are numbered relative to the translational start site of the flhD gene. (F) EMSA results revealed that RcsB could specifically bind to the binding site RcsB-1 in vitro. PflhDC288, a 288-bp DNA fragment from nucleotide positions −86 to −373 relative to the translational start site of flhD gene. Lane 1, PflhDC288 without protein RcsB. Lane 2, PflhDC288 with 2.28 μM RcsB. Lane 3, PflhDC288 with 4.56 μM RcsB. Lane 4, PflhDC288 carrying mutations in positions TAA with protein RcsB. Lane 5, PflhDC288 carrying mutations in positions TCC with protein RcsB. Lane 6, PflhDC288 with complete deletion of RcsB-1. (G) In vivo analysis of the S. marcescens flhDC RcsB binding site by mutagenesis. The transcription fusions PflhDC581-lacZ, PflhDC581 (C−245C−246C−247)-lacZ, PflhDC581 (A−254A−255A−256)-lacZ, PflhDC581 (RcsB-1−)-lacZ, PflhDC581 (C−634C−635C−636)-lacZ, and PflhDC581 (RcsB-2−)-lacZ were used in the S. marcescens strain JNB5-1. PflhDC581, DNA fragment containing the binding sites RcsB-1 and RcsB-2; PflhDC581 (C−245C−246C−247), PflhDC581 carrying mutations in positions TAA; and PflhDC581 (A−254A−255A−256), PflhDC581 carrying mutations in positions TCC; PflhDC581 (RcsB-1−), PflhDC581 with complete deletion of RcsB-1; PflhDC581 (C−634C−635C−636), PflhDC581 carrying mutations in positions TTA; and PflhDC581 (RcsB-2−), PflhDC581 with complete deletion of RcsB-2. (H) The regulatory network of RcsB controls prodigiosin synthesis in S. marcescens. The pigA to pigN operon includes genes pigABCDEFGHIJKLMN, a total of 14 genes. RcsB is an inhibitor for prodigiosin synthesis, and FlhDC is an activator for prodigiosin synthesis. X1 indicates an unknown prodigiosin synthesis regulator controlled by RcsB. X2 indicates an unknown regulator which may be regulated by FlhDC. Arrows indicate positive regulation, and T-shaped lines indicate negative regulation. Dotted lines indicate additional pathways that influence RcsB or FlhDC control of prodigiosin production. (B and C) Experiments were performed in biological triplicates. Error bars indicate the standard deviations. (B) Student's t test was used for statistical analysis. (C) One-way ANOVA was used to examine the mean differences between the data groups. ****, P < 0.001; *, P < 0.05.
FlhDC has been shown to activate prodigiosin synthesis in Serratia sp. ATCC 39006 (31). However, the role of FlhDC in the regulation of prodigiosin biosynthesis in S. marcescens has not yet been tested. Transcriptome data showed that the expression levels of the flhC and flhD genes were upregulated by 3.42-fold and 6.29-fold, respectively, in the rcsB-disrupted mutant (Table S4). This result suggested that RcsB may regulate the expression of the pig gene cluster by directly binding to the promoter region of the flhDC genes, hence affecting prodigiosin synthesis in strain JNB5-1. To confirm this hypothesis, an flhDC deletion mutant, JNB5-1 ΔflhDC, and an flhDC-overexpressing mutant, JNB5-1/pXW1910, were constructed, and the ability of strains JNB5-1, JNB5-1 ΔflhDC, and JNB5-1/pXW1910 to synthesize prodigiosin was analyzed. As shown in Fig. 4C, in strain JNB5-1, FlhDC was also an activator in prodigiosin synthesis. Furthermore, through analysis by EMSA of the interaction between RcsB and the 5′ noncoding region of flhDC genes, located between nucleotide positions −1 and −983 relative to the translational start site of the flhD gene, it was found that RcsB could directly bind to the promoter region of the flhDC genes (Fig. 4D). This result was consistent with the result that RcsBSm (RcsB from strain JNB5-1) significantly reduced the expression of PSmflhDC-lacZ in E. coli (Fig. 4B). The direct binding of RcsB to the flhDC promoter has been also demonstrated previously in S. marcescens by Di Venanzio et al. (37). These results indicated that the regulation of RcsB in prodigiosin synthesis in S. marcescens probably depended on FlhDC.
A binding site for RcsB protein defined as “RcsB box,” with a consensus sequence of TaAGaatatTCctA (uppercase letters indicate that the degree of conservation of these bases in different RcsB binding sites was higher than 70%, while lowercase letters show that the degree of conservation of these bases was higher than 50% but lower than 70%), has been found in different species (1, 38). To further characterize the target motif of the RcsB protein within the flhDC promoter region, the FIMO tool of MEMEsuite (39) was used for the prediction of binding motifs in the promoter region of flhDC genes at locations between positions −1 and −983 relative to the translational start site of the flhD gene. As shown in Fig. 4E, two potential RcsB binding sites, RcsB-1 (located between positions −245 to −258 relative to the translational start site of the flhD gene) and RcsB-2 (located between positions −623 to −636 relative to the translational start site of the flhD gene), were identified in the 5′ noncoding region of the flhD gene. Accordingly, the reconstituted 288-bp DNA fragment PflhDC288, spanning nucleotide positions −86 to −373 and containing the binding site RcsB-1, and the reconstituted 134-bp DNA fragment PflhDC134, spanning nucleotide positions −532 to −666 and containing the binding site RcsB-2, were retarded by RcsB (Fig. 4F; see also Fig. S3 in the supplemental material). The retardation was eliminated in fragments PflhDC288 (C−245C−246C−247), PflhDC288 (A−254A−255A−256), and PflhDC288 (RcsB-1−), carrying mutations in three conserved positions or with complete deletion of the sequences of RcsB-1 (Fig. 4F), and in fragment PflhDC134 (C−634C−635C−636), carrying mutations in three conserved positions of binding site RcsB-2 (Fig. S3). These results demonstrated that the proposed sequences are essential for the in vitro binding of the RcsB protein to the promoter of flhDC genes. Moreover, to analyze in vivo the S. marcescens flhDC RcsB binding site, transcriptional reporter gene fusion of the flhDC operon was constructed by cloning the DNA fragment PflhDC581 (DNA fragment containing the binding sites RcsB-1 and RcsB-2 spanning nucleotide positions −86 to −666), PflhDC581 (C−245C−246C−247), PflhDC581 (A−254A−255A−256), PflhDC581 (RcsB-1−), PflhDC581 (C−634C−635C−636), and PflhDC581 (RcsB-2−) upstream of lacZ, and β-galactosidase activity in strain JNB5-1 was determined. Results showed that transcription of the flhDC operon was significantly higher expression levels for the DNA fragments when RcsB-1 was mutated, whereas the mutation of RcsB-2 had no significant effect on flhDC operon expression levels (Fig. 4G). Together, these results demonstrated the importance of the identified binding site RcsB-1 for the activation of flhDC expression and revealed that binding site RcsB-2 was probably not involved in the regulation of flhDC expression.
Also, hundreds of genes had altered expression in the rcsB-disrupted mutant (Fig. 3), suggesting that the increase in the prodigiosin synthesis in the mutant SK68 may partially be dependent on FlhDC. To determine whether the effect of RcsB on prodigiosin synthesis is entirely dependent on FlhDC, an rcsB flhDC double mutant, SK68 ΔflhDC, was constructed, and the prodigiosin synthesis of strains JNB5-1, SK68, and SK68 ΔflhDC was analyzed. Results showed that prodigiosin production by strain SK68 ΔflhDC was slightly higher than that in JNB5-1 (P < 0.05) (Fig. 4C) and significantly lower than that of the mutant SK68 (P < 0.001) (Fig. 4C). These results indicated that the regulation by RcsB of prodigiosin synthesis was largely or completely dependent on FlhDC. Furthermore, to reveal the molecular regulatory mechanism of FlhDC in the expression of the prodigiosin biosynthetic operon, RT-qPCR analysis, EMSA, and β-galactosidase activity analysis in different strains were used. Results showed that FlhDC positively regulates prodigiosin operon expression in strain JNB5-1. However, an indirect regulation between the RcsB protein and the prodigiosin synthesis genes probably existed in strain JNB5-1 (see Fig. S4 in the supplemental material). Collectively, our study supports our model that RcsB plays an inhibitory role on prodigiosin synthesis mainly through negative regulation of the expression of the pig gene cluster, probably by directly binding to the promoter region of the prodigiosin activator genes flhDC, although additional pathways independent of FlhDC may exist that control prodigiosin synthesis by RcsB (Fig. 4H). Also, an unknown regulator may be present in strain JNB5-1, mediating the regulation between the FlhDC regulator and the prodigiosin synthesis pathways (Fig. 4H).
RcsB negatively regulates swimming and swarming motility.
Based on the transcriptomics data, the importance of RcsB for other selected phenotypes was investigated. Swimming and swarming are two modes of motility found in S. marcescens, and multiple cellular systems have been found to contribute to these two motilities, especially the presence of functional flagella (40). To investigate the influence of RcsB on cell motility and flagellar biosynthesis in S. marcescens, the transcriptomic data of rcsB mutant SK68 and the wild-type strain JNB5-1 was analyzed. Results showed that the flagellar synthesis-related genes flgBCDEFGHIJKLMN, flhABCD, fliACDEFGHIJKLMNOPQRST, and motAB were significantly upregulated in the SK68 strain (log2FC > 1 and P value < 0.05) (Fig. 5A and Table S5). These results correlated with our findings that RcsB directly bound to the promoter region of the flagellar master regulator FlhDC (Fig. 3D). Furthermore, the differentially expressed genes (DEGs) flhD, flhC, motA, flhB, flgE, and fliF were selected, and the expression patterns of these genes in strains SK68 and JNB5-1 were validated by RT-qPCR. As shown in Fig. 5B, the results revealed that the expression of all six of these genes was significantly increased in the rcsB-disrupted mutant, indicating that RcsB was probably involved in flagellar synthesis and cell motility in S. marcescens.
FIG 5.
RcsB negatively controls flagellar gene expression and cell motility in S. marcescens. (A) Transcriptome data showed that the expression level of genes related to flagellum synthesis was significantly increased in the RcsB-disrupted mutant SK68. (B) RT-qPCR analysis of the relative expression levels of the target fliF, fliL, flgE, flgB, flhB, and motA genes in the rcsB-disrupted mutant SK68. (C) Swimming motility and swarming motility tests for JNB5-1, SK68, SK68/pXW1908, ΔRcsB, and ΔRcsB/pXW1908 strains. (Upper) Swimming motility assay. (Lower) Swarming motility assay. (D) Colony diameter of strains JNB5-1, SK68, SK68/pXW1908, ΔRcsB, and ΔRcsB/pXW1908 in swimming assay. (E) Colony diameter of strains JNB5-1, SK68, SK68/pXW1908, ΔRcsB, and ΔRcsB/pXW1908 in swarming assay. (B, D, and E) Experiments were performed in biological triplicates. Error bars indicate the standard deviations. One-way ANOVA was used to examine the mean differences between the data groups. ****, P < 0.001.
When testing the swimming motility of JNB5-1, SK68, ΔRcsB, SK68/pXW1908, and ΔRcsB/pXW1908 strains with 0.3% semisolid agarose plates, it was found that the swimming zone formed by the rcsB-disrupted mutant SK68 and the rcsB deletion mutant ΔRcsB had significantly increased (P < 0.001) (Fig. 5C, upper, and Fig. 5D). For the swarming test, the absence of RcsB in strains SK68 and ΔRcsB also significantly increased the swarming motility of the wild-type strain JNB5-1 (P < 0.001) (Fig. 5C, lower, and Fig. 5E). In the complementation assay, the intact rcsB gene decreased cell motility significantly in the mutant SK68 and ΔRcsB (Fig. 5C to E). Collectively, these data suggested that RcsB negatively regulates cell motility in S. marcescens by controlling flagellar biosynthesis.
RcsB is required for capsular polysaccharide biosynthesis and biofilm formation by S. marcescens.
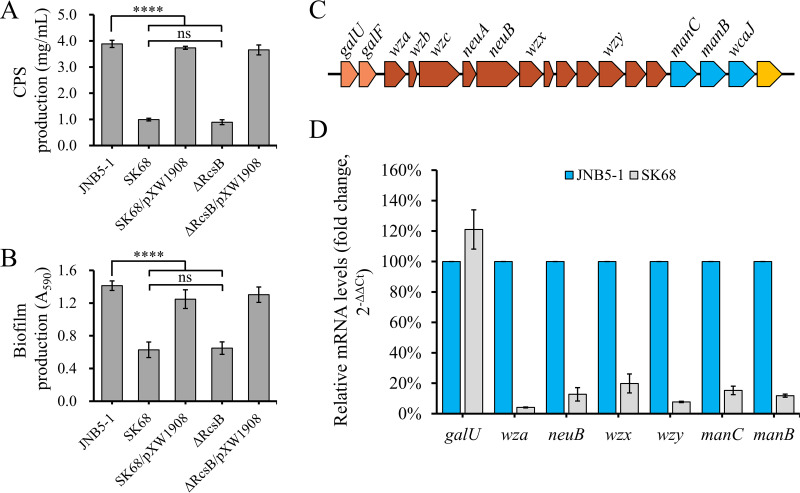
The regulator RcsB was first identified for its role in regulation of capsular polysaccharide biosynthesis in E. coli (41). To confirm whether RcsB is also necessary for capsular polysaccharide synthesis in S. marcescens, the ability of strains JNB5-1, SK68, ΔRcsB, SK68/pXW1908, and ΔRcsB/pXW1908 to synthesize extracellular polysaccharides (EPS), including capsular polysaccharide, was investigated. Results showed that the rcsB mutants SK68 (0.99 mg/ml) and ΔRcsB (0.89 mg/ml) produced significantly (P < 0.001) less extracellular polysaccharides than wild-type strain JNB5-1 (3.88 mg/ml) and complementary strains SK68/pXW1908 (3.73 mg/ml) and ΔRcsB/pXW1908 (3.65 mg/ml) (Fig. 6A). These results suggested that RcsB was required for extracellular polysaccharide synthesis in S. marcescens. At the same time, the expression levels of the bacterial capsular polysaccharide synthesis-related genes galU, wza, neuB, wzx, wzy, manC, and manB in strains JNB5-1 and SK68 were evaluated by RT-qPCR (Fig. 6C and D). Results showed that the expression levels of wza, neuB, wzx, wzy, manC, and manB genes decreased by 24.43-, 7.87-, 5.04-, 12.98-, 6.55-, and 8.48-fold, respectively, in the rcsB-disrupted mutant SK68 (Fig. 6D). Taken together, these results suggested that RcsB positively regulated capsular polysaccharide biosynthesis in S. marcescens through regulating the expression of capsular polysaccharide biosynthesis locus genes.
FIG 6.
RcsB contributes to capsular polysaccharide production (CPS) and biofilm formation by S. marcescens. (A) RcsB positively controls CPS production in S. marcescens. (B) RcsB positively regulates biofilm formation by S. marcescens. (C) Gene organization of the putative capsular polysaccharide biosynthesis gene clusters in S. marcescens. Arrows without gene designations represent genes that encode hypothetical proteins. Genes belonging to the same operon are represented by the same color, as indicated. (D) RT-qPCR analysis of the difference in expression of capsular polysaccharide biosynthesis-related genes galU, wza, neuB, wzx, wzy, manC, and manB between strain SK68 and its parent strain JNB5-1. (A, B, and D) Experiments were performed in biological triplicates. Error bars indicate the standard deviations. One-way ANOVA was used to examine the mean differences between the data groups. ****, P < 0.001.
Extracellular polysaccharide is one of the three major components of biofilms. As RcsB could suppress EPS formation, it may have an impact on biofilm formation. After 48 h of incubation in 96-well microtiter plates, biofilm production analysis showed that the rcsB mutants SK68 and ΔRcsB synthesized more biofilm than did their parent strain, JNB5-1, or complementary strains SK68/pXW1908 and ΔRcsB/pXW1908 (P < 0.001) (Fig. 6B), suggesting that RcsB was also required for S. marcescens biofilm biogenesis.
RcsB contributes to acid resistance in S. marcescens.
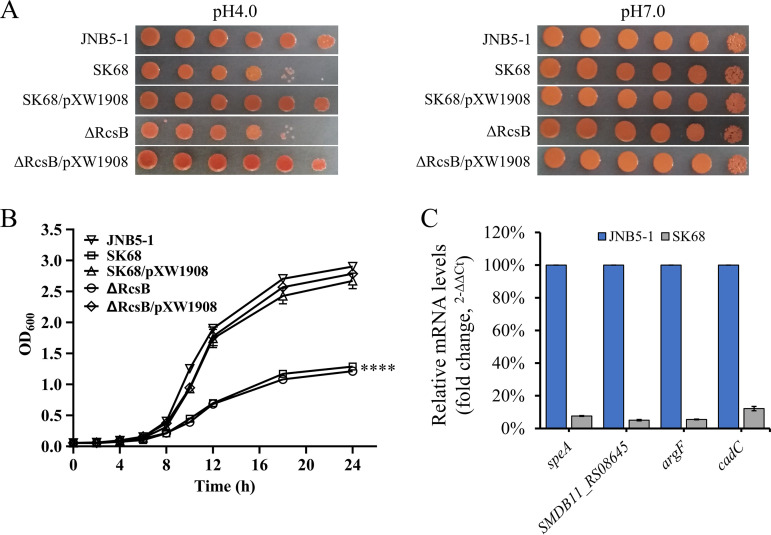
Adapting to acid resistance is a common feature for almost all living bacteria. To determine whether RcsB is required for acid resistance of S. marcescens, the strains JNB5-1, SK68, ΔRcsB, SK68/pXW1908, and ΔRcsB/pXW1908 were spotted and grown on LB medium at pH 4.0 and 7.0. It was observed that the cells lacking the rcsB gene (strains SK68 and ΔRcsB) showed significantly reduced growth at pH 4.0, whereas no significant difference in cell growth was observed between these five strains at pH 7.0 (Fig. 7A). Furthermore, the growth curves of these five strains at pH 4.0 were compared. Consistent with the results of the spot assay shown in Fig. 7A, growth of the rcsB mutants SK68 and ΔRcsB was much slower than that of the parent strain JNB5-1 (P < 0.001) (Fig. 7B). In the complementation experiment, introduction of the rcsB gene into the SK68 and ΔRcsB strains restored their cell growth at pH 4.0 (Fig. 7A and B). These data strongly suggested that RcsB plays a vital role in the acid resistance of S. marcescens.
FIG 7.
RcsB positively regulates acid resistance in S. marcescens. (A) Spot assays show that RcsB is required for S. marcescens to grow under low-pH conditions (pH 4.0). (B) Growth curves of the JNB5-1, SK68, SK68/pXW1908, ΔRcsB, and ΔRcsB/pXW1908 strains at pH 4.0. (C) Changes in acid resistance-related gene expression in the rcsB-disrupted mutant SK68 compared with that in the parent strain JNB5-1 at pH 4.0. The speA, SMDB11_RS08645, argF, and cadC genes encode arginine decarboxylase, agmatine deiminase, ornithine transcarbamylase, and transcriptional activator CadC, respectively. (B and C) Experiments were performed in biological triplicates. Error bars indicate the standard deviations. One-way ANOVA was used to examine the mean differences between the data groups. ****, P < 0.001.
Bacteria have developed five acid resistance (AR) systems, AR1 to AR5, to enable their survival in acidic environments (42). To reveal the mechanisms by which RcsB regulates acid resistance in S. marcescens, RT-qPCR analyses were conducted to determine the expression levels of the arginine decarboxylase-encoding gene speA, agmatine deiminase-encoding gene SMDB11_RS08645, ornithine transcarbamylase-encoding gene argF, and transcriptional activator CadC-encoding gene cadC in strains JNB5-1 and SK68. It was found that the expression levels of the speA, SMDB11_RS08645, argF, and cadC genes were decreased by 13.23-, 19.88-, 18.29-, and 8.24-fold, respectively, in the SK68 strain (Fig. 7C). Collectively, these results indicated that RcsB potentially affected acid resistance by regulating the expression of the speA, SMDB11_RS08645, argF, and cadC genes in S. marcescens.
DISCUSSION
RcsB, a well-studied response regulator of the Rcs phosphorelay system, is at the center of a complex network of regulatory inputs and outputs (4). As a global regulator, RcsB has been identified as being related to a series of core cellular processes in different microorganisms, such as the control of capsular polysaccharide production (43), carbapenem resistance (9), biofilm formation (44), cell motility (45), acid resistance (46), and heat resistance (47) in E. coli and the regulation of flagellar synthesis and virulence in Y. ruckeri (7). It mediates capsular polysaccharide production (6) and acid resistance (48) in Salmonella Typhimurium; inhibits cell motility (49) and cell elongation and morphology (50) in Proteus mirabilis; modulates biofilm formation and cell motility in Pantoea alhagi (8); is required for capsular polysaccharide production (5), acid resistance (51), and hypermucoviscosity (5) in K. pneumoniae; and is responsible for the type III secretion system in Yersinia pseudotuberculosis (14). However, our understanding of the function of the RcsB protein in bacteria, especially in S. marcescens, remains limited. In S. marcescens, RcsB was identified to play roles in controlling the biosynthesis of the pore-forming toxin ShlA (37) and of outer membrane vesicles (52). In this work, we confirmed that RcsB controls many unknown cellular processes in wild-type S. marcescens, including negative regulation of prodigiosin production (Fig. 1B) and of swimming and swarming motilities (Fig. 5C to E), as well as positive regulation of capsular polysaccharide production (Fig. 6A), biofilm formation (Fig. 6C), and acid resistance (Fig. 7A and B).
A number of transcription regulators have been identified as playing roles in prodigiosin synthesis in S. marcescens, such as the prodigiosin synthesis positive regulators EepR (28), PigP (29), GumB (30), FlhDC (31), and RbsR (32) and the prodigiosin synthesis negative regulators MetR (20), CopA (22), CRP (23), HexS (24), RssB (25), SmaR (27), RcsB (26), and SpnR (21). Among them, although regulator RcsB has been reported to be involved in prodigiosin production in S. marcescens CMS4441 (a gumB rcsB double mutant) (26), our understanding of the significance of this regulator in a wild-type background and the regulatory mechanism behind prodigiosin biosynthesis in S. marcescens is still limited. In this work, RcsB was identified as a negative regulator of prodigiosin production in the wild-type strain S. marcescens JNB5-1 (Fig. 1B). The molecular mechanism for negative regulation of prodigiosin production by RcsB was investigated, and results showed that the hyperproduction of prodigiosin by the rcsB mutant could probably be associated with RcsB repression of transcription of the pig operon by directly binding to the promoter region of the flhDC genes (Fig. 2C and D and Fig. 4). This direct binding of RcsB to the flhDC promoter has also been demonstrated in S. marcescens (37), Pectobacterium carotovorum (53), S. enterica serovar Typhimurium (54), and E. coli (55). Also, it remains possible that RcsB could interfere with binding of FlhDC to the pigA promoter and hence influence prodigiosin production in strain JNB5-1. More experiments are needed to further confirm this possibility. Interestingly, through the transcriptome data of strains JNB5-1 and SK68 at an OD600 of 2.0 (Table S6), it can be noted that the changes in pigABCDEFGHIJKLMN gene expression elicited by mutation of rcsB are not particularly dramatic; there was only an 111.98% to 255.76% increase in transcripts of these 14 genes. This is similar to what has been reported for regulators PigP (56), RssB (25), SpnR (21), and HexS (24), which affect prodigiosin, where minor changes in pig operon transcript levels have been shown to confer major phenotypic differences. Additionally, the result that the rcsC, rcsD, and rcsF mutants had no significant effect on prodigiosin production in strain JNB5-1 (Fig. 1B) suggested that RcsB-dependent prodigiosin regulation is probably independent of the classical Rcs phosphorelay cascade in S. marcescens.
Multiple cellular systems have been found to contribute to cell motility in bacteria, especially the presence of functional flagella (40). Using transcriptomics, RT-qPCR analysis, and EMSAs, we found that the RcsB protein directly bound to the promoter region of the flagellar master regulator FlhDC encoding genes flhDC (Fig. 4D) and repressed transcription of a variety of known flagellar synthesis-related genes in S. marcescens (Fig. 5A and B). Moreover, in this work, data showed that RcsB negatively regulated swarming and swimming motilities in S. marcescens (Fig. 5C to E). Collectively, this work showed that RcsB plays an important role in cell motility in S. marcescens, probably by controlling the transcription of flagellar synthesis-related genes.
The RcsB protein was first identified and named for its role in the regulation of capsular polysaccharide synthesis in E. coli (41), and it is increasingly evident that the RcsB proteins are required for normal capsular polysaccharide production in other bacteria, such as K. pneumoniae (5), Erwinia amylovora (57), S. enterica (58), and Pantoea stewartii (57). In this work, we found that RcsB also positively regulated capsular polysaccharide synthesis in S. marcescens (Fig. 6A). The expression levels of the wza, neuB, wzx, wzy, manC, and manB genes were decreased in the rcsB mutant, suggesting that the decreased capsular polysaccharide in mutant SK68 was probably a consequence of reduced expression of the biosynthetic genes (Fig. 6D). Importantly, the RT-qPCR results also indicated that the regulatory mechanism of RcsB in capsular polysaccharide synthesis by strain S. marcescens may differ from that of E. coli (41) and K. pneumoniae (5) strains. In E. coli and K. pneumoniae, the galF gene plays an important role in RcsB-dependent capsular polysaccharide synthesis. However, in S. marcescens, the galF gene probably has no effect on RcsB-dependent capsular polysaccharide synthesis (Fig. 6D).
Acid stress is an environmental stress that has important effects on microbial physiology and plays an important role in the process of microbial fermentation. To survive in acidic environments, microorganisms have evolved several distinct strategies (42). Also, a variety of transcription regulators have been reported to play roles in acid resistance in different microorganisms, such as transcriptional regulator GadR of Lactobacillus brevis (59), transcriptional regulator PsrB of E. coli (60), transcriptional regulator YthA of Lactococcus lactis (61), and transcriptional regulator RcsB of K. pneumoniae (51), S. Typhimurium (48), and E. coli (46, 62). However, our understanding of the molecular mechanism behind acid resistance in S. marcescens remains limited. In this work, we found that RcsB positively regulated acid resistance in S. marcescens (Fig. 7A and B). RT-qPCR analysis showed that RcsB affected the acid resistance in S. marcescens, likely by direct or indirect regulation of the expression of the arginine decarboxylase-encoding gene speA, agmatine deiminase-encoding gene SMDB11_RS08645, ornithine transcarbamylase-encoding gene argF, and transcriptional activator CadC-encoding gene cadC (Fig. 7C), suggesting that the production of alkali probably plays an important role in acid resistance in S. marcescens.
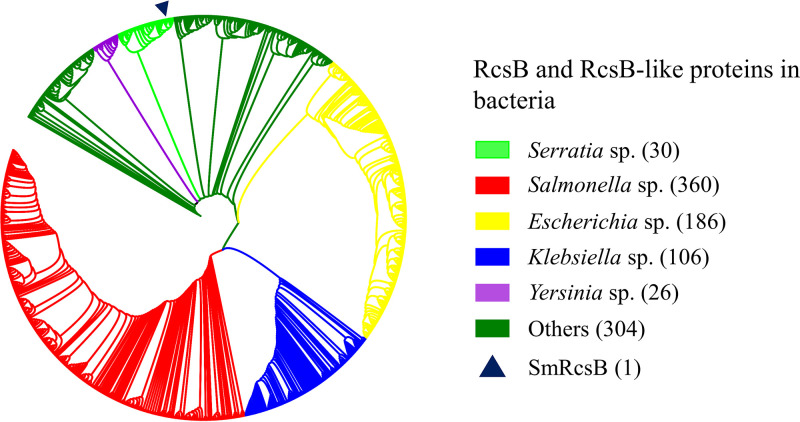
As confirmed in our study, RcsB controls various cellular processes, including prodigiosin synthesis, cell motility, acid resistance, biofilm formation, and exopolysaccharide production in S. marcescens JNB5-1. Transcriptome data showed that RcsB was involved in the regulation of 20 cellular processes in S. marcescens. Hence, further research on RcsB and RcsB-like proteins is necessary if they are widely conserved and distributed in other bacteria. Therefore, a search for homologs of RcsBSm (RcsB protein in S. marcescens JNB5-1) with an E value lower than 4E−139 was done. The top 1,000 proteins, showing similarities with RcsBSm ranging from 91.20% to 100%, were found in Serratia spp., Salmonella spp. (mostly S. enterica), Escherichia spp. (mostly E. coli), Klebsiella spp. (mostly K. pneumoniae), Yersinia spp., and other bacteria, including Cronobacter malonaticus, Ewingella americana, Edwardsiella tarda, etc. (Fig. 8). The fact that the RcsB-like proteins are highly homologous was consistent with the result that RcsB or RcsB-like proteins are functionally conserved, and it suggests that RcsB or RcsB-like proteins in other bacteria probably control cellular processes similar to those in S. marcescens. However, further studies need to be carried out to reveal the molecular mechanism by which RcsB regulates these phenotypes in different species, as in S. marcescens, the molecular mechanism of RcsB regulating the synthesis of capsular polysaccharide is not completely consistent with that in E. coli (41) and K. pneumoniae (5) strains.
FIG 8.
Phylogenetic tree of the top 1,000 homologs of RcsB with E values lower than 3E−134 showing that RcsB and RcsB-like proteins are widely distributed among bacteria. Colored rectangles indicate different species. Dark blue triangle indicates the RcsB protein studied in this study. “Others” indicates species not belonging to the indicated species.
In summary, this work describes a new regulator which regulates prodigiosin synthesis, cell motility, biofilm formation, capsular polysaccharide synthesis, and acid resistance in S. marcescens. Additionally, this work revealed the possible regulatory mechanism of the RcsB protein in these cellular processes. Further research is needed to understand the roles played by RcsB-like proteins in other bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. marcescens SK68 is a prodigiosin-hyperproducing strain isolated from a Tn5G transposon insertion mutant library of strain JNB5-1. E. coli strains were grown in LB medium at 37°C, and S. marcescens strains were grown in LB medium at 30°C. Where needed, the appropriate antibiotics were added at defined concentrations, as follows. Spectinomycin at 25 μg/ml, streptomycin at 25 μg/ml, kanamycin at 50 μg/ml, ampicillin at 50 μg/ml, or gentamicin at 10 μg/ml was used for the cultivation of E. coli strains. Spectinomycin at 50 μg/ml, streptomycin at 50 μg/ml, ampicillin at 150 μg/ml, apramycin at 50 μg/ml, or gentamicin at 50 μg/ml was used for the cultivation of S. marcescens strains. Bacterial strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source |
|---|---|---|
| E. coli strains | ||
| DH5α | hsdR recA lacZYAF80 lacZΔM15 | BRL |
| BL21(DE3) | F− dcm ompT hsdSB(rB− mB−) gal λ(DE3) | Laboratory collection |
| S17-1 | F− recA hsdR RP4-2 (Tc::Mu) (Km::Tn7) lysogenized with λpir phage | Laboratory collection |
| DH5α/pUCP18/pDN19lacΩ-PpigA | E. coli DH5α containing plasmids pUCP18 and pDN19lacΩ-PpigA | This study |
| DH5α/pUCP18/pDN19lacΩ-PflhDC | E. coli DH5α containing plasmids pUCP18 and pDN19lacΩ-PflhDC | This study |
| DH5α/pXW1908/pDN19lacΩ-PpigA | E. coli DH5α containing plasmids pXW1908 and pDN19lacΩ-PpigA | This study |
| DH5α/pXW1908/pDN19lacΩ-PflhDC | E. coli DH5α containing plasmids pXW1908 and pDN19lacΩ-PflhDC | This study |
| DH5α/pXW2005/pDN19lacΩ-PpigA | E. coli DH5α containing plasmids pXW2005 and pDN19lacΩ-PpigA | This study |
| S. marcescens strains | ||
| JNB5-1 | S. marcescens wild-type strain | 20 |
| SK68 | rcsB::Gmr mutant of JNB5-1, prodigiosin-hyperproducing mutant | This study |
| SK68/pXW1908 | Mutant SK68 containing plasmid pXW1908 | This study |
| SK68/pUCP18 | Mutant SK68 containing empty vector pUCP18 | This study |
| ΔRcsB | rcsB deletion mutant of S. marcescens JNB5-1 | This study |
| ΔRcsB/pXW1908 | Mutant ΔRcsB containing plasmid pXW1908 | This study |
| ΔRcsB/pUCP18 | Mutant ΔRcsB containing empty vector pUCP18 | This study |
| ΔRcsA | rcsA deletion mutant of S. marcescens JNB5-1 | This study |
| ΔRcsC | rcsC deletion mutant of S. marcescens JNB5-1 | This study |
| ΔRcsD | rcsD deletion mutant of S. marcescens JNB5-1 | This study |
| ΔRcsF | rcsF deletion mutant of S. marcescens JNB5-1 | This study |
| ΔIgaA | igaA deletion mutant of S. marcescens JNB5-1 | This study |
| JNB5-1ΔflhDC | flhDC deletion mutant of S. marcescens JNB5-1 | This study |
| SK68ΔflhDC | rcsB flhDC double mutant of S. marcescens JNB5-1 | This study |
| JNB5-1/pDN19lacΩ-PpigA | S. marcescens JNB5-1 containing plasmid pDN19lacΩ-PpigA | This study |
| SK68/pDN19lacΩ-PpigA | S. marcescens SK68 containing plasmid pDN19lacΩ-PpigA | This study |
| JNB5-1ΔflhDC/pDN19lacΩ-PpigA | S. marcescens JNB5-1ΔflhDC containing plasmid pDN19lacΩ-PpigA | This study |
| Plasmids | ||
| pRK2013Tn5G | Tn5G-carrying plasmid, Kmr Gmr | 67 |
| pMD18T | Cloning vector, 2,692 bp, Ampr, lacZ | TaKaRa |
| pET28a | E. coli expression vector, Kmr | Laboratory collection |
| pXW1907 | rcsB gene cloned in pET28a for expression of RcsB protein in E. coli BL21(DE3) | This study |
| pUCP18 | Broad-host-range shuttle vector, Ampr | 68 |
| pXW1908 | rcsB gene driven by Plac promoter cloned into pUCP18, Ampr | This study |
| pXW2005 | flhDC genes driven by Plac promoter cloned into pUCP18, Ampr | This study |
| pDN19lacΩ | Promoterless lacZ fusion vector, Spr Smr Tcr | 69 |
| pDN19lacΩ-ppigA | pig operon promoter cloned in pDN19lacΩ, Spr Smr Tcr | This study |
| pDN19lacΩ-PflhDC | flhDC genes promoter cloned in pDN19lacΩ, Spr Smr Tcr | This study |
| pUTKm | Tn5-based delivery plasmid with Kmr Ampr | 56 |
Gm, gentamycin; Km, kanamycin; Amp, ampicillin; Sp, spectinomycin; Sm, streptomycin; Tc, tetracycline.
Identification of the Tn5G inserted gene in mutant SK68.
The Tn5G transposon was used to mutate S. marcescens JNB5-1 to isolate the prodigiosin-hyperproducing mutant SK68 as described previously (20). The Tn5G insertion site of the SK68 strain was identified by the inverse PCR method as described previously (63). Briefly, genomic DNA of strain SK68 was digested by the restriction nuclease TaqI, self-ligated, and amplified using the primers OTn1 and OTn2 (Table 2). The amplified DNA fragment was then cloned into the pMD18T vector for sequencing, and the sequences obtained were compared with the NCBI GenBank database to identify the insertion sites. The target gene identified was amplified using the primers RcsB-F1 and RcsB-R1, listed in Table 2, and cloned into the pUCP18 plasmid to obtain recombinant plasmid pXW1908. The recombinant plasmid was introduced into strains SK68 and ΔRcsB for complementation experiments.
TABLE 2.
Primers used in this study
| Function | Primer | Primer sequence (5′–3′)a |
|---|---|---|
| Identification of Tn5G in mutant SK68 | OTn1 | GATCCTGGAAAACGGGAAAG |
| OTn2 | CCATCTCATCAGAGGGTAGT | |
| Amplification of metR gene for complement expt | RcsB-F1 | CGCGAATTCATGAATAACCTGAACGTAATTATTGCTGATGACC |
| RcsB-R1 | CGCGGATCCTTAGTCTTTGTCCAACGGCGTCATG | |
| Overexpression of metR in plasmid pET28a | RcsB-F2 | CGCGGATCCATGAATAACCTGAACGTAATTATTGCTGATGACC |
| RcsB-R2 | CGCAAGCTTTTAGTCTTTGTCCAACGGCGTCATG | |
| Amplification of the pig operon promoter region | PigA-F1 | CGCGGATCCGTGTTATTTATACACAAAAATTTACATATAATAAAAATCTTAATTTGATC |
| PigA-R1 | CGCGAATTCTTTTTCCTCCGGAATGCTCCTGC | |
| Amplification of the pig operon promoter and operator region | PigA-F2 | CGCGGATCCGACGAACTCCGCCATTGGGTTG |
| PigA-R2 | CGCGAATTCTTTTTCCTCCGGAATGCTCCTGC | |
| Amplification of the pig operon promoter region, operator region, and coding sequences of pigA | PigA-F3 | CGCGGATCCTGCGCCTCCCCGCAGACC |
| PigA-R3 | CGCGAATTCTTTTTCCTCCGGAATGCTCCTGC | |
| Amplification of the flhDC gene promoter region | FlhDC-F | CGCGGATCCATTCCCCATCCCGACAGACTATG |
| FlhDC-R | CGCGAATTCGCTGCTTGCGTTGAAAAACCG |
Underlining indicates the added restriction enzyme sites.
Construction of rcsA, rcsB, rcsC, rcsD, rcsF, igaA, and flhDC mutants.
The rcsA, rcsB, rcsC, rcsD, rcsF, igaA, and flhDC genes of strain JNB5-1 or SK68 were inactivated by using a gene replacement method as described previously (20). In brief, the upstream and downstream fragments of target genes and aacC3 resistance gene DNA fragments were amplified. After integration of the aacC3 gene into the middle of the upstream and downstream fragments of the target gene by overlap extension PCR, the PCR product was cloned into a pUTKm vector. The resulting plasmid was transformed into E. coli S17-1 and then introduced into the JNB5-1 or SK68 strain by conjugation to knock out the genes mentioned above. These deletions removed M1 to S208 of the total 209 amino acids in RcsA, M1 to D216 of the total 216 amino acids in RcsB, M1 to Q944 of the total 953 amino acids in RcsC, M1 to R890 of the total 900 amino acids in RcsD, M1 to K134 of the total 134 amino acids in RcsF, M1 to P709 of the total 711 amino acids in IgaA, M1 to A192 of the total 193 amino acids in FlhC, and M1 to A116 of the total 116 amino acids in FlhD. Primers used for gene deletion are listed in Table S3 in the supplemental material.
Growth curve assays.
The growth curves of strains JNB5-1, SK68, and SK68/pXW1908 were determined as described previously (20). Exponential-phase cells (OD600 of 0.8) of the indicated strains were transferred into fresh LB medium at 3% inoculum and continuously incubated at 30°C. Optical densities of cultures were measured at a wavelength of 600 nm at time intervals of 0, 2, 4, 6, 8, 10, 12, and 24 h. The growth curves of these three strains were plotted as the values of OD600 versus the incubation time.
Real-time quantitative PCR assay.
The exponential-phase cells (OD600 of 0.8) of strains JNB5-1 and SK68 were grown in LB medium (pH 7.0) at 200 rpm and 30°C for 4 h to reach an OD600 of 2.0 before collection to analyze the expression levels of cell motility-related genes and capsular polysaccharide synthesis-related genes. The cultures of strains JNB5-1, SK68, and JNB5-1 ΔflhDC were collected after 12 h of shake flask fermentation with an OD600 of 6.0 to analyze the expression levels of prodigiosin synthesis-related genes. To assess the expression levels of the acid resistance-related genes, the exponential-phase cells (OD600 of 0.8) of strains JNB5-1 and SK68 were inoculated into fresh LB medium (pH 4.0) and incubated at 30°C and 200 rpm to reach an OD600 of 1.0. Then, the collected cells were subjected to total RNA extraction using an RNAprep Pure kit (Tiangen). After treating the total RNA with DNase I, cDNA was synthesized using the HiScript II Q RT SuperMix kit (Vazyme). Finally, the cDNA was diluted to 200 ng/μl and subjected to RT-qPCR analysis using a ChamQ Universal SYBR qPCR mastermix kit (Vazyme) with the primers listed in Table S2 in the supplemental material.
Construction of transcription reporters.
The promoter regions of the pig gene cluster and the flhDC genes were amplified by the primers listed in Table 2. Then, the 228-bp-long PCR products upstream of the pig operon and the 983-bp-long PCR products upstream of the flhDC genes were cloned into the lacZ-containing pDN19lacΩ vector to obtain the recombinant plasmids pDN19lacΩ-PpigA and pDN19lacΩ-PflhDC, respectively. The resulting recombinant plasmids pDN19lacΩ-PpigA and pDN19lacΩ-PflhDC were transformed into the indicated S. marcescens or E. coli strains by electroporation (Table 1). The expression levels of the pigABCDEFGHIJKLMN genes and flhDC genes were determined by measuring β-galactosidase activities of the transformants. To ascertain whether there is an indirect posttranscriptional regulation by RcsB of the pig operon expression, a pig-lacZ fusion report plasmid was constructed by cloning the upstream noncoding region of the pig operon upstream of the lacZ gene with the primer pair PigA-F2/PigA-R2, as listed in Table 2. Also, a pigA-lacZ fusion report plasmid was constructed by cloning the upstream noncoding region of the pig operon and coding sequences, except for the termination codon of the pigA gene upstream of the lacZ gene, with the primer pair PigA-F3/PigA-R3, as listed in Table 2. For analysis of the S. marcescens flhDC RcsB binding site in vivo, a transcriptional reporter gene fusion of the flhDC operon was constructed by cloning the DNA fragments PflhDC581 (DNA fragment containing the binding sites RcsB-1 and RcsB-2, spanning nucleotide positions −86 to −666 relative to the translational start site of the flhD gene), PflhDC581 (C−245C−246C−247) (carrying mutations in positions TAA), PflhDC581 (A−254A−255A−256) (carrying mutations in positions TCC), PflhDC581 (RcsB-1−) (with a complete deletion of RcsB-1), PflhDC581 (C−634C−635C−636) (carrying mutations in positions TTA), and PflhDC581 (RcsB-2−) (with a complete deletion of RcsB-2) upstream of lacZ, respectively, with the primers listed in Table S1 in the supplemental material, and β-galactosidase activity in the strain JNB5-1 was determined.
Transcriptomic analysis by RNAseq.
To reveal the roles of RcsB in the cellular processes of S. marcescens, total RNA was isolated from the samples of JNB5-1 and SK68 cells of three biological replicates, and transcriptome sequencing (RNAseq) analysis was performed to evaluate gene expression differences between strains JNB5-1 and SK68 as previously described with slight modification (64). Briefly, S. marcescens JNB5-1 and SK68 cells were grown in LB medium at 200 rpm and 30°C for 4 h to an OD600 of 2.0 before collection. One milliliter of the collected cells was treated with the RNAprep Pure kit (Tiangen) to extract total bacterial RNA and delivered in dry ice to Genewiz (South Plainfield, NJ) for RNAseq analysis. The rRNA in the total bacterial RNA was removed, and the obtained mRNA was used as the template for cDNA synthesis. The cDNA library was sequenced using the Illumina HiSeq platform. The genome of S. marcescens WW4 (GenBank accession number NC_020211.1) was used as a reference for annotation. The genes that had significantly different levels of expression by SK68 and JNB5-1 were determined using the standards of a P value of ≤0.05 and a fold change |log2FC| of ≥1. The Pathway Enrichment Analysis tool Omicshare was used to classify genes at the level of KEGG_B_class and pathway (https://www.omicshare.com/tools/Home/Soft/pathwaygsea).
Electrophoretic mobility shift assay.
The EMSA method was performed as described previously (65). Briefly, the rcsB gene was amplified with the primers RcsB-F2 and RcsB-R2, listed in Table 2, and the PCR product was cloned into the pET28a expression vector for overexpression of a His-tagged RcsB protein. To determine whether RcsB directly binds to the pig operon or flhDC genes, the promoters of the pig operon and flhDC genes were amplified with the primer pairs PigA-F2/PigA-R2 and FlhDC-F/FlhDC-R, respectively, listed in Table 2. For the analysis of RcsB cis elements in the promoter region of the flhDC genes, DNA fragments PflhDC288 and PflhDC134 containing the predicted RcsB binding sites RcsB-1 and RcsB-2 were amplified by the primer pairs FlhDC288-F/FlhDC288-R and FlhDC134-F/FlhDC134-R, respectively, listed in Table S1 in the supplemental material. DNA fragments PflhDC288 (C−245C−246C−247), PflhDC288 (A−254A−255A−256), and PflhDC288 (RcsB-1−), carrying mutations in three conserved positions or with complete deletion of the sequences of RcsB-1 (Fig. 4F), and DNA fragment PflhDC134 (C−634C−635C−636), carrying mutations in three conserved positions of binding site RcsB-2, were amplified by overlap extension PCR with the primers listed in Table S1. Then, the purified DNA fragments were incubated with serial dilutions of the RcsB proteins and kept at room temperature in 2× binding buffer (40 mM Tris-HCl, 4 mM MgCl2, 100 mM NaCl, 10% glycerol, 2 mM dithiothreitol [DTT], 0.2 mg/ml bovine serum albumin [BSA], and 1 mM EDTA) for 30 min. Electrophoresis was performed on a 5% native PAGE gel, and to visualize the DNA bands, the gels were stained with ethidium bromide.
Swimming and swarming motility assays.
For swimming and swarming assay, strains JNB5-1, SK68, SK68/pXW1908, ΔRcsB, and ΔRcsB/pXW1908 were grown to an OD600 of 0.6, and 1 μl of the culture was spotted onto 0.3% and 0.5% semisolid LB medium, respectively. After incubation at 30°C for 18 h and 10 h, respectively, the diameters of the swarming zones and swimming zones were measured.
Analysis of extracellular polysaccharide production.
Extracellular polysaccharide, which included capsular polysaccharide produced by strains JNB5-1, SK68, SK68/pXW1908, ΔRcsB, and ΔRcsB/pXW1908, was measured as described previously (20, 30). Briefly, overnight cultures (18 h) of the indicated strains were centrifuged at 12,000 rpm for 15 min to harvest the bacterial cells. Then, the pelleted cells were resuspended with 30 ml phosphate-buffered saline (PBS) solution and 6 ml 1% Zwittergent 3-14 citric acid solution (100 mM; pH 2.0) and incubated at 50°C for 20 min. After centrifugation at 12,000 rpm for 30 min, the supernatant cells were transferred to four 50-ml centrifugal bottles, and four volumes of cold ethanol (−20°C) were added to each sample before incubation at −20°C overnight. After overnight precipitation, the samples were centrifuged at 14,000 rpm and 4°C for 45 min, and the supernatants were discarded. The exopolysaccharides in the sediment of each sample were air dried in a chemical fume hood, and the exopolysaccharides were weighed.
Acid resistance assay.
For the acid resistance assay, bacterial cultures of JNB5-1, SK68, SK68/pXW1908, ΔRcsB, and ΔRcsB/pXW1908 were grown to an OD600 of 0.6. An aliquot (1 μl) of the serially diluted (10-fold) culture was then spotted onto LB plates at pH 4.0 and incubated at 30°C for 24 h. For growth curve assays of strains JNB5-1, SK68, SK68/pXW1908, ΔRcsB, and ΔRcsB/pXW1908 under acidic conditions (pH 4.0), exponential-phase cells (OD600 of 0.6) were inoculated into fresh LB medium at pH 4.0. Samples were taken at time intervals of 0, 2, 4, 6, 8, 10, 12, and 24 h, and the A600 values were recorded.
Biofilm analysis.
Biofilm amounts for strains JNB5-1, SK68, SK68/pXW1908, ΔRcsB, and ΔRcsB/pXW1908 were measured by the crystal violet staining method as described previously (66). In brief, 20 μl exponential-phase culture (OD600 of 0.6) was transferred into a 96-well microtiter plate (Corning, Corning, NY) containing 200 μl 3-fold-diluted LB medium. After incubation at 37°C for 48 h without shaking, the cultures in the wells were gently removed and washed three times with distilled water. Biofilm in the walls was stained with 0.1% crystal violet solution for 15 min, and 95% ethanol was used to extract the biofilm in the crystal violet. Optical density at a wavelength of 595 nm was measured using a BioTek Epoch 2 microplate to determine the biofilm content.
Phylogenetic tree construction for RcsB-like proteins.
Position-Specific Iterated BLAST (PSI-BLAST) in the NCBI database was used to search the non-redundant protein sequence (nr) database and analyze the distribution of RcsB homologues in microorganisms. The top 1,000 sequences homologous to the RcsB protein studied in this work were selected, and the distance tree was generated by using BLAST pairwise alignments in NCBI. Online iTOL software (http://itol.embl.de/) was used to modify the phylogenetic tree.
Statistical analysis.
Each experiment in this study was performed independently three times, and data are expressed as means and standard deviations (SDs). Student’s t test or one-way analysis of variance (ANOVA) was used for comparing statistical differences between the groups of experiment data.
Data availability.
The RNA-seq data were submitted to the GEO database of NCBI under accession number GSE149400. Sequences of the rcsB gene from strain JNB5-1 were deposited in GenBank under accession number MT385774.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the National Natural Science Foundation of China (grants 31870066 and 21778024), the “10 Thousand Plan” national high-level talents special support plan (grant 2018RA2218), the national first-class discipline program of Light Industry Technology and Engineering (grant LITE2018-06), the Program of Introducing Talents of Discipline to Universities (grant 111-2-06), the Program of the Key Laboratory of Industrial Biotechnology, Ministry of Education, China (grant KLIB-KF201705), and the Postgraduate Research and Practice Innovation Program of Jiangsu Province (grant KYCX17_1419).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol 59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 2.Wolfe AJ. 2010. Physiologically relevant small phosphodonors link metabolism to signal transduction. Curr Opin Microbiol 13:204–209. doi: 10.1016/j.mib.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke DJ. 2010. The Rcs phosphorelay: more than just a two-component pathway. Future Microbiol 5:1173–1184. doi: 10.2217/fmb.10.83. [DOI] [PubMed] [Google Scholar]
- 4.Wall E, Majdalani N, Gottesman S. 2018. The complex Rcs regulatory cascade. Annu Rev Microbiol 72:111–139. doi: 10.1146/annurev-micro-090817-062640. [DOI] [PubMed] [Google Scholar]
- 5.Walker KA, Miner TA, Palacios M, Trzilova D, Frederick DR, Broberg CA, Sepulveda VE, Quinn JD, Miller VL. 2019. A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. mBio 10:e00089-19. doi: 10.1128/mBio.00089-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pando JM, Karlinsey JE, Lara JC, Libby SJ, Fang FC. 2017. The Rcs-regulated colanic acid capsule maintains membrane potential in Salmonella enterica serovar Typhimurium. mBio 8:e00808-17. doi: 10.1128/mBio.00808-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jozwick AKS, LaPatra SE, Graf J, Welch TJ. 2019. Flagellar regulation mediated by the Rcs pathway is required for virulence in the fish pathogen Yersinia ruckeri. Fish Shellfish Immunol 91:306–314. doi: 10.1016/j.fsi.2019.05.036. [DOI] [PubMed] [Google Scholar]
- 8.Li S, Liang H, Wei Z, Bai H, Li M, Li Q, Qu M, Shen X, Wang Y, Zhang L. 2019. An osmoregulatory mechanism operating through OmpR and LrhA controls the motile-sessile switch in the plant growth-promoting bacterium Pantoea alhagi. Appl Environ Microbiol 85:e00077-19. doi: 10.1128/AEM.00077-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Logre E, Denamur E, Mammeri H. 2020. Contribution to carbapenem resistance and fitness cost of DcuS/DcuR, RcsC/RcsB, and YehU/YehT two-component systems in CTX-M-15-producing Escherichia coli. Microb Drug Resist 26:349–352. doi: 10.1089/mdr.2019.0027. [DOI] [PubMed] [Google Scholar]
- 10.Laubacher ME, Ades SE. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J Bacteriol 190:2065–2074. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Latasa C, Garcia B, Echeverz M, Toledo-Arana A, Valle J, Campoy S, Garcia-del Portillo F, Solano C, Lasa I. 2012. Salmonella biofilm development depends on the phosphorylation status of RcsB. J Bacteriol 194:3708–3722. doi: 10.1128/JB.00361-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casino P, Miguel-Romero L, Huesa J, Garcia P, Garcia-Del Portillo F, Marina A. 2018. Conformational dynamism for DNA interaction in the Salmonella RcsB response regulator. Nucleic Acids Res 46:456–472. doi: 10.1093/nar/gkx1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howery KE, Clemmer KM, Rather PN. 2016. The Rcs regulon in Proteus mirabilis: implications for motility, biofilm formation, and virulence. Curr Genet 62:775–789. doi: 10.1007/s00294-016-0579-1. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Hu Y, Francis MS, Chen S. 2015. RcsB positively regulates the Yersinia Ysc-Yop type III secretion system by activating expression of the master transcriptional regulator LcrF. Environ Microbiol 17:1219–1233. doi: 10.1111/1462-2920.12556. [DOI] [PubMed] [Google Scholar]
- 15.Williamson NR, Fineran PC, Leeper FJ, Salmond GP. 2006. The biosynthesis and regulation of bacterial prodiginines. Nat Rev Microbiol 4:887–899. doi: 10.1038/nrmicro1531. [DOI] [PubMed] [Google Scholar]
- 16.Gerc AJ, Stanley-Wall NR, Coulthurst SJ. 2014. Role of the phosphopantetheinyltransferase enzyme, PswP, in the biosynthesis of antimicrobial secondary metabolites by Serratia marcescens Db10. Microbiology (Reading) 160:1609–1617. doi: 10.1099/mic.0.078576-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webber MA, Shanks RMQ, Stella NA, Lahr RM, Wang S, Veverka TI, Kowalski RP, Liu X. 2012. Serratamolide is a hemolytic factor produced by Serratia marcescens. PLoS One 7:e36398. doi: 10.1371/journal.pone.0036398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao S, Guo W, Shi L, Yu Y, Zhang C, Yang H. 2014. Characterization of acetoin production in a budC gene disrupted mutant of Serratia marcescens G12. J Ind Microbiol Biotechnol 41:1267–1274. doi: 10.1007/s10295-014-1464-x. [DOI] [PubMed] [Google Scholar]
- 19.Rao B, Zhang LY, Sun J, Su G, Wei D, Chu J, Zhu J, Shen Y. 2012. Characterization and regulation of the 2,3-butanediol pathway in Serratia marcescens. Appl Microbiol Biotechnol 93:2147–2159. doi: 10.1007/s00253-011-3608-5. [DOI] [PubMed] [Google Scholar]
- 20.Pan X, Sun C, Tang M, You J, Osire T, Zhao Y, Xu M, Zhang X, Shao M, Yang S, Yang T, Rao Z. 2020. LysR-Type transcriptional regulator MetR controls prodigiosin production, methionine biosynthesis, cell motility, H2O2 tolerance, heat tolerance, and exopolysaccharide synthesis in Serratia marcescens. Appl Environ Microbiol 86:e02241-19. doi: 10.1128/AEM.02241-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horng YT, Deng SC, Daykin M, Soo PC, Wei JR, Luh KT, Ho SW, Swift S, Lai HC, Williams P. 2002. The LuxR family protein SpnR functions as a negative regulator of N-acylhomoserine lactone-dependent quorum sensing in Serratia marcescens. Mol Microbiol 45:1655–1671. doi: 10.1046/j.1365-2958.2002.03117.x. [DOI] [PubMed] [Google Scholar]
- 22.Williamson NR, Simonsen HT, Harris AK, Leeper FJ, Salmond GP. 2006. Disruption of the copper efflux pump (CopA) of Serratia marcescens ATCC 274 pleiotropically affects copper sensitivity and production of the tripyrrole secondary metabolite, prodigiosin. J Ind Microbiol Biotechnol 33:151–158. doi: 10.1007/s10295-005-0040-9. [DOI] [PubMed] [Google Scholar]
- 23.Stella NA, Shanks RM. 2014. Cyclic-AMP inhibition of fimbriae and prodigiosin production by Serratia marcescens is strain-dependent. Arch Microbiol 196:323–330. doi: 10.1007/s00203-014-0970-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stella NA, Fender JE, Lahr RM, Kalivoda EJ, Shanks RM. 2012. The LysR transcription factor, HexS, is required for glucose inhibition of prodigiosin production by Serratia marcescens. Adv Microbiol 2:10.4236/aim.2012.24065. doi: 10.4236/aim.2012.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horng YT, Chang KC, Liu YN, Lai HC, Soo PC. 2010. The RssB/RssA two-component system regulates biosynthesis of the tripyrrole antibiotic, prodigiosin, in Serratia marcescens. Int J Med Microbiol 300:304–312. doi: 10.1016/j.ijmm.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Brothers KM, Callaghan JD, Stella NA, Bachinsky JM, AlHigaylan M, Lehner KL, Franks JM, Lathrop KL, Collins E, Schmitt DM, Horzempa J, Shanks RMQ. 2019. Blowing epithelial cell bubbles with GumB: ShlA-family pore-forming toxins induce blebbing and rapid cellular death in corneal epithelial cells. PLoS Pathog 15:e1007825. doi: 10.1371/journal.ppat.1007825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coulthurst SJ, Williamson NR, Harris AK, Spring DR, Salmond GP. 2006. Metabolic and regulatory engineering of Serratia marcescens: mimicking phage-mediated horizontal acquisition of antibiotic biosynthesis and quorum-sensing capacities. Microbiology (Reading) 152:1899–1911. doi: 10.1099/mic.0.28803-0. [DOI] [PubMed] [Google Scholar]
- 28.Shanks RM, Stella NA, Lahr RM, Aston MA, Brothers KM, Callaghan JD, Sigindere C, Liu X. 2017. Suppressor analysis of eepR mutant defects reveals coordinate regulation of secondary metabolites and serralysin biosynthesis by EepR and HexS. Microbiology (Reading) 163:280–288. doi: 10.1099/mic.0.000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shanks RM, Lahr RM, Stella NA, Arena KE, Brothers KM, Kwak DH, Liu X, Kalivoda EJ. 2013. A Serratia marcescens PigP homolog controls prodigiosin biosynthesis, swarming motility and hemolysis and is regulated by cAMP-CRP and HexS. PLoS One 8:e57634. doi: 10.1371/journal.pone.0057634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stella NA, Brothers KM, Callaghan JD, Passerini AM, Sigindere C, Hill PJ, Liu X, Wozniak DJ, Shanks RMQ. 2018. An IgaA/UmoB family protein from Serratia marcescens regulates motility, capsular polysaccharide biosynthesis, and secondary metabolite production. Appl Environ Microbiol 84:e02575-17. doi: 10.1128/AEM.02575-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hampton HG, McNeil MB, Paterson TJ, Ney B, Williamson NR, Easingwood RA, Bostina M, Salmond GP, Fineran PC. 2016. CRISPR-Cas gene-editing reveals RsmA and RsmC act through FlhDC to repress the SdhE flavinylation factor and control motility and prodigiosin production in Serratia. Microbiology (Reading) 162:1047–1058. doi: 10.1099/mic.0.000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee CM, Monson RE, Adams RM, Salmond GPC. 2017. The LacI-family transcription factor, RbsR, is a pleiotropic regulator of motility, virulence, siderophore and antibiotic production, gas vesicle morphogenesis and flotation in Serratia. Front Microbiol 8:1678. doi: 10.3389/fmicb.2017.01678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pruss BM. 2017. Involvement of two-component signaling on bacterial motility and biofilm development. J Bacteriol 199:e00259-17. doi: 10.1128/JB.00259-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson EJ, Limaye B, Inamdar H, Datta A, Manjari KS, Pullinger GD, Thomson NR, Joshi RR, Watson M, Stevens MP. 2011. Genome sequences of Salmonella enterica serovar Typhimurium, Choleraesuis, Dublin, and Gallinarum strains of well-defined virulence in food-producing animals. J Bacteriol 193:3162–3163. doi: 10.1128/JB.00394-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun YC, Guo XP, Hinnebusch BJ, Darby C. 2012. The Yersinia pestis Rcs phosphorelay inhibits biofilm formation by repressing transcription of the diguanylate cyclase gene hmsT. J Bacteriol 194:2020–2026. doi: 10.1128/JB.06243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barnes AC, Delamare-Deboutteville J, Gudkovs N, Brosnahan C, Morrison R, Carson J. 2016. Whole genome analysis of Yersinia ruckeri isolated over 27 years in Australia and New Zealand reveals geographical endemism over multiple lineages and recent evolution under host selection. Microb Genom 2:e000095. doi: 10.1099/mgen.0.000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Venanzio G, Stepanenko TM, Garcia Vescovi E. 2014. Serratia marcescens ShlA pore-forming toxin is responsible for early induction of autophagy in host cells and is transcriptionally regulated by RcsB. Infect Immun 82:3542–3554. doi: 10.1128/IAI.01682-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wehland M, Bernhard F. 2000. The RcsAB box: characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J Biol Chem 275:7013–7020. doi: 10.1074/jbc.275.10.7013. [DOI] [PubMed] [Google Scholar]
- 39.Wang K, Sybers D, Maklad HR, Lemmens L, Lewyllie C, Zhou X, Schult F, Brasen C, Siebers B, Valegard K, Lindas AC, Peeters E. 2019. A TetR-family transcription factor regulates fatty acid metabolism in the archaeal model organism Sulfolobus acidocaldarius. Nat Commun 10:1542. doi: 10.1038/s41467-019-09479-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura S, Minamino T. 2019. Flagella-driven motility of bacteria. Biomolecules 9:279. doi: 10.3390/biom9070279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brill JA, Quinlan-Walshe C, Gottesman S. 1988. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J Bacteriol 170:2599–2611. doi: 10.1128/jb.170.6.2599-2611.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Tang H, Lin Z, Xu P. 2015. Mechanisms of acid tolerance in bacteria and prospects in biotechnology and bioremediation. Biotechnol Adv 33:1484–1492. doi: 10.1016/j.biotechadv.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Gottesman S, Trisler P, Torres-Cabassa A. 1985. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J Bacteriol 162:1111–1119. doi: 10.1128/JB.162.3.1111-1119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szczesny M, Beloin C, Ghigo JM. 2018. Increased osmolarity in biofilm triggers RcsB-dependent lipid A palmitoylation in Escherichia coli. mBio 9:e01415-18. doi: 10.1128/mBio.01415-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pannen D, Fabisch M, Gausling L, Schnetz K. 2016. Interaction of the RcsB response regulator with auxiliary transcription regulators in Escherichia coli. J Biol Chem 291:2357–2370. doi: 10.1074/jbc.M115.696815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castanie-Cornet MP, Cam K, Bastiat B, Cros A, Bordes P, Gutierrez C. 2010. Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res 38:3546–3554. doi: 10.1093/nar/gkq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carter MQ, Parker CT, Louie JW, Huynh S, Fagerquist CK, Mandrell RE. 2012. RcsB contributes to the distinct stress fitness among Escherichia coli O157:H7 curli variants of the 1993 hamburger-associated outbreak strains. Appl Environ Microbiol 78:7706–7719. doi: 10.1128/AEM.02157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torrez Lamberti MF, Farizano JV, Lopez FE, Martinez Zamora MG, Pescaretti MM, Delgado MA. 2019. Cross-talk between the RcsCDB and RstAB systems to control STM1485 gene expression in Salmonella Typhimurium during acid-resistance response. Biochimie 160:46–54. doi: 10.1016/j.biochi.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Morgenstein RM, Rather PN. 2012. Role of the Umo proteins and the Rcs phosphorelay in the swarming motility of the wild type and an O-antigen (waaL) mutant of Proteus mirabilis. J Bacteriol 194:669–676. doi: 10.1128/JB.06047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Little K, Tipping MJ, Gibbs KA. 2018. Swarmer cell development of the bacterium Proteus mirabilis requires the conserved enterobacterial common antigen biosynthesis gene rffG. J Bacteriol 200:e00230-18. doi: 10.1128/JB.00230-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu CJ, Lin CT, Chiang JD, Lin CY, Tay YX, Fan LC, Peng KN, Lin CH, Peng HL. 2019. RcsB regulation of the YfdX-mediated acid stress response in Klebsiella pneumoniae CG43S3. PLoS One 14:e0212909. doi: 10.1371/journal.pone.0212909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McMahon KJ, Castelli ME, Garcia Vescovi E, Feldman MF. 2012. Biogenesis of outer membrane vesicles in Serratia marcescens is thermoregulated and can be induced by activation of the Rcs phosphorelay system. J Bacteriol 194:3241–3249. doi: 10.1128/JB.00016-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andresen L, Sala E, Koiv V, Mae A. 2010. A role for the Rcs phosphorelay in regulating expression of plant cell wall degrading enzymes in Pectobacterium carotovorum subsp. carotovorum. Microbiology (Reading) 156:1323–1334. doi: 10.1099/mic.0.033936-0. [DOI] [PubMed] [Google Scholar]
- 54.Wang Q, Zhao Y, McClelland M, Harshey RM. 2007. The RcsCDB signaling system and swarming motility in Salmonella enterica serovar typhimurium: dual regulation of flagellar and SPI-2 virulence genes. J Bacteriol 189:8447–8457. doi: 10.1128/JB.01198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Francez-Charlot A, Laugel B, Van Gemert A, Dubarry N, Wiorowski F, Castanie-Cornet MP, Gutierrez C, Cam K. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol Microbiol 49:823–832. doi: 10.1046/j.1365-2958.2003.03601.x. [DOI] [PubMed] [Google Scholar]
- 56.Herrero M, de Lorenzo V, Timmis KN. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. J Bacteriol 172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wehland M, Kiecker C, Coplin DL, Kelm O, Saenger W, Bernhard F. 1999. Identification of an RcsA/RcsB recognition motif in the promoters of exopolysaccharide biosynthetic operons from Erwinia amylovora and Pantoea stewartii subspecies stewartii. J Biol Chem 274:3300–3307. doi: 10.1074/jbc.274.6.3300. [DOI] [PubMed] [Google Scholar]
- 58.Cano DA, Dominguez-Bernal G, Tierrez A, Portillo FG, Casadesus J. 2002. Regulation of capsule synthesis and cell motility in Salmonella enterica by the essential gene igaA. Genetics 162:1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gong L, Ren C, Xu Y. 2019. Deciphering the crucial roles of transcriptional regulator GadR on gamma-aminobutyric acid production and acid resistance in Lactobacillus brevis. Microb Cell Fact 18:108. doi: 10.1186/s12934-019-1157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang J, Russell TW, Hocking DM, Bender JK, Srikhanta YN, Tauschek M, Robins-Browne RM. 2015. Control of acid resistance pathways of enterohemorrhagic Escherichia coli strain EDL933 by PsrB, a prophage-encoded AraC-like regulator. Infect Immun 83:346–353. doi: 10.1128/IAI.02758-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu H, Liu J, Miao S, Zhao Y, Zhu H, Qiao M, Saris PEJ, Qiao J. 2018. Contribution of YthA, a PspC family transcriptional regulator of Lactococcus lactis F44 acid tolerance and nisin yield: a transcriptomic approach. Appl Environ Microbiol 84:e02483-17. doi: 10.1128/AEM.02483-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson MD, Burton NA, Gutierrez B, Painter K, Lund PA. 2011. RcsB is required for inducible acid resistance in Escherichia coli and acts at gadE-dependent and -independent promoters. J Bacteriol 193:3653–3656. doi: 10.1128/JB.05040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, Mushegian A, Lory S, Jin S. 1996. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc Natl Acad Sci U S A 93:10434–10439. doi: 10.1073/pnas.93.19.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.You J, Sun L, Yang X, Pan X, Huang Z, Zhang X, Gong M, Fan Z, Li L, Cui X, Jing Z, Jin S, Rao Z, Wu W, Yang H. 2018. Regulatory protein SrpA controls phage infection and core cellular processes in Pseudomonas aeruginosa. Nat Commun 9:1846. doi: 10.1038/s41467-018-04232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang X, Zhang Z, Huang Z, Zhang X, Li D, Sun L, You J, Pan X, Yang H. 2019. A putative LysR-type transcriptional regulator inhibits biofilm synthesis in Pseudomonas aeruginosa. Biofouling 35:541–550. doi: 10.1080/08927014.2019.1627337. [DOI] [PubMed] [Google Scholar]
- 66.Djordjevic D, Wiedmann M, McLandsborough LA. 2002. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl Environ Microbiol 68:2950–2958. doi: 10.1128/aem.68.6.2950-2958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nunn DN, Lory S. 1992. Components of the protein-excretion apparatus of Pseudomonas aeruginosa are processed by the type IV prepilin peptidase. Proc Natl Acad Sci U S A 89:47–51. doi: 10.1073/pnas.89.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schweizer HP. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 69.Totten PA, Lory S. 1990. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J Bacteriol 172:7188–7199. doi: 10.1128/jb.172.12.7188-7199.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data were submitted to the GEO database of NCBI under accession number GSE149400. Sequences of the rcsB gene from strain JNB5-1 were deposited in GenBank under accession number MT385774.