Abstract
Introduction
In December 2019, the first COVID-19 case, caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) was reported in Wuhan, China. The SARS-CoV-2 rapidly disseminated throughout the world via community spread, acquiring pandemic status with significant fatality.
Observations
Rapid SARS-CoV-2 diagnosis was soon perceived critical for arresting community spread and effective therapy development. Human SARS-CoV-2 infection can be diagnosed either by nucleic acid identification or specific antibody detection. Contrary to nucleic acid identification confirmed active SARS-CoV-2 infection; antibody detection confirms a past infection, even in asymptomatic subjects. SARS-CoV-2 specific antibodies augment the ability to effectively counter the virus. A crucial hurdle limiting the steadfast implementation of antibody detection is the time required for threshold B lymphocyte population generation. This process is dependent on precise antigen recognition and MHC class I molecules presentation.
Conclusions
Thus, nucleic acid and antibody dependent tests complement each other in identifying human SARS-CoV-2 infection and shaping up subsequent immunological responses. This article discusses the complimentary association of nucleic acid identification (corresponding to an active infection) and antibody testing (the yester CoV-2 infection vulnerability) as the diagnostic and screening measures of SARS-CoV-2 infection.
Highlights
Nucleic acid (RNA) identification and specific antibody detection against SARS-CoV-2 are the noted diagnostic mechanisms for screening human SARS-CoV-2 infection.
While nucleic acid identification screens prevailing SARS-CoV-2 infection, detection of SARS-CoV-2 specific antibodies signifies a past infection, even in asymptomatic subjects.
Antibodies against SARS-CoV-2 provide a potential therapeutic option via transfer from antibody rich plasma of a recovered subject to an infected individual.
Nucleic acid identification may not absolutely confirm the infection because of frequent SARS-CoV-2 genome mutations and possible technical errors, while specific antibody detection also needs at least (8–14) days for detectable screening of B-cell generated antibodies.
Nucleic acid and antibody tests are complementary to each other as an early stage diagnostic assay for SARS-CoV-2 infection and possible therapy (antibodies).
Sufferers with a high clinical suspicion but negative RT-PCR screening could be examined via combined imaging and repeated swab test.
Keywords: Severe acute respiratory syndrome Coronavirus 2: real-time reverse transcriptase polymerase chain reaction: antibody, specific high sensitivity enzymatic reporter unlocking technique: Rapid Diagnostic Test, Enzyme Linked Immunosorbent Assay, neutralization assay, chemiluminescent immunoassay
Introduction
On 31 December 2019, the World Health Organization (WHO) was notified about a cluster of pneumonia cases in Wuhan, China. Based on recognized pathogens, the disease was initially named as Novel Coronavirus 2019 (2019-nCoV)1. On 11 February, 2019, WHO officially coined the terminology as Coronavirus Disease 2019 (COVID-19). Subsequently the International Committee of Viral Taxonomy proposed the name on the basis of causative agent as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)2.
Electron microscopic analysis of SARS-CoV-2 identified spikes protruding from the envelope (periphery) that collectively resembled into a crown (or corona in Latin). The serotype and genomic characteristics indicated coronaviruses of the Order: Nidovirales, Family: Coronaviridiae, Subfamily: Coronavirinae and Genera: Betacoronavirus3,4.
Recent phylogenetic analysis of full-length genome sequences from infected subjects revealed each SARS-CoV-2 particle as (60–160) nm in length, enveloped, with an unsegmented, single-stranded sense RNA. Coronaviruses have some of the largest RNA genomes (26–32) Kb, of all viruses5. At least 10 open reading frames have been identified and characterized in COVID-19. The two primary ones, ORF1a and ORF1b, are translated from the full-length genomic RNA (29,903 nt) that also serves as an mRNA. The ORF1a produces polypeptide1a (pp1a, (440–500 kDa) that is cleaved into 11 NSPs (non-structural proteins). The ORF1b, on the other hand, produces a large polypeptide (pp1ab, (740–810 kDa) which is cleaved into 15 NSPs. In addition to genomic RNA, nine major sub-genomic RNAs are produced6. These serve as non-canonical ORFs and have been linked to SARS-CoV-2 pathogenicity (ORFs’ 3a, E, M, 6, 7a, 7b, N, S, 10). It is noteworthy that these ORFs’ further produce the N-terminal truncated and frameshift ORFs, making the transcriptome architecture unusually complex. The single stranded genomic RNA of coronavirus has a cap like structure at the 5′-UTR and a poly(A)-tail at the 3′ UTR. These features allow the virus to assume a structure similar to mRNA of host cells5. Wang et al. further reported that ORFs of SARS-CoV-2 have an extremely low CG dinucleotide count. Thus, the secondary structure formed by SARS-CoV-2 genomic RNA is less stable than many other coronaviruses. This makes SARS-CoV-2 more efficient in reproduction than other coronaviruses, as less energy is required to disrupt the stem-loop structure of its genomic RNA. Genome sequence homology data indicated approximately 88% SARS-CoV-2 similarity to bat-SL-CoVZC45 and bat-SL-CoVZXC21, collected in 2018 in Zhoushan, Eastern China7. Further analysis divulged ∼79 and 50% sequence homology with Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV), respectively7. Since identification, considerable genetic diversity and rapid mutagenesis of this novel coronavirus have been observed8,9. Different SARS-CoV-2 mutated versions have been identified throughout the world.
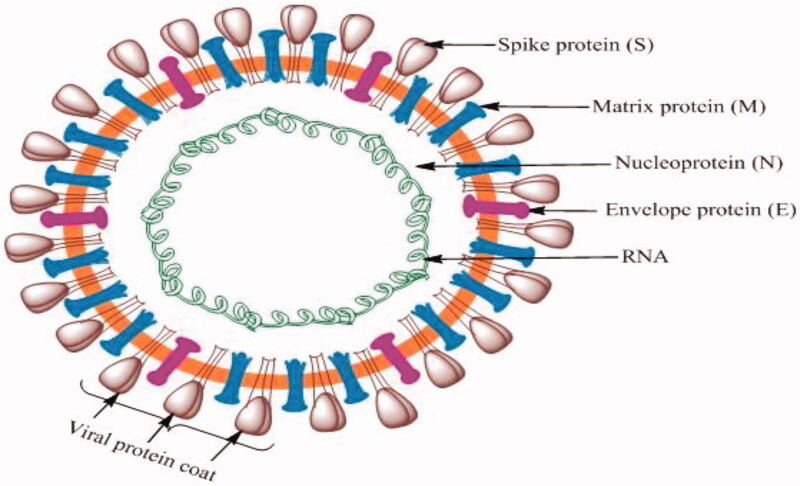
The coronavirus genome remains the largest of all RNA viruses, comprising multiple ORFs, followed by the Nucleocapsid (N), Spike (S), Envelope (E) and Matrix (M) proteins (Figure 1)10. The S protein is divergent with less than 75% nucleotide sequence similarity to previously identified SARS-associated coronaviruses11. The N, E and M structural proteins are more conserved and are essential for virus survival. These proteins encase the RNA and are essential for budding, envelope formation and pathogenesis12–14. The M protein binds the nucleocapsid, facilitating viral assembly and generation of new virus particles. The E protein is implicated in morphogenesis, liberation, and pathogenesis while the S protein develops the homotrimeric spikes which recognize receptor(s) through which invasion to a potential host is mediated12,15,16. A notable aspect herein pertains to distinctive M protein prevalence in elongated and compact forms. While the former contributes to rigidity via acting on clustered spikes and a rather unconventional narrow membrane curvature domain, the latter regulates the flexibility and S protein interactions. The M protein regulates the virion size via interaction with S, N proteins and genomic RNA, facilitating their involvement in virus assembly12.
Figure 1.
Representative structural make-up of SARS-CoV-2, depicting distinct surface receptor proteins interacting with invaded cells.
Interestingly, removal of group-specific SARS-CoV ORFs, either alone or in combinations, does not substantially affect the replication efficiency or RNA generation in cell culture. The maximum decline in the viral growth has been reported corresponding to the ORF3a removal. Moreover, the S protein in the SARS-CoV genome has no active contribution towards programming the rough endoplasmic reticulum (rough ER)/Golgi retention signal. It has also been reported that deletion of ORF3a hardly plays any role in targeting S protein localization within the rER/Golgi apparatus. So, it could be generalized here that ORF3a deletion in SARS-CoV-2 results in virus death not by intervening any of the s-protein functions and may be independently linked with native cellular activities of rough ER and Golgi apparatus17.
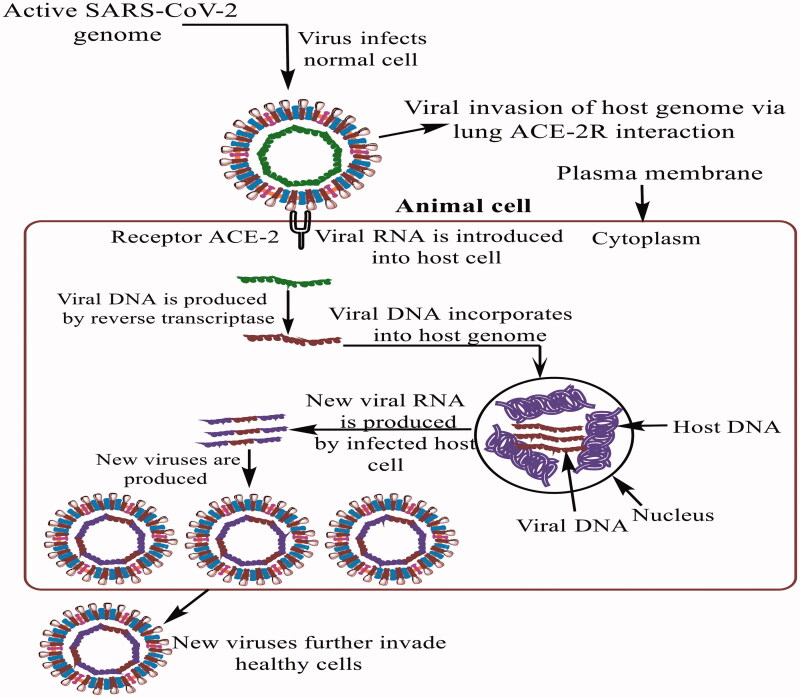
Structural analysis suggests that SARS-CoV-2 is able to bind the angiotensin converting enzyme 2 receptor (ACE2-R) on human cells (Figure 2). In this context, Zhang et al. found a high ACE2-R expression on type II (AT-II) alveolar epithelial cells, esophageal epithelium and stratified and absorptive intestinal (ileum and colon) epithelial cells. Inspection using Bioinformatics tools suggested digestive tract as the SARS-CoV-infection sanctuary18. Zhou et al. conducted infectivity studies by incubating SARS-CoV-2 with HeLa cells expressing ACE2-R. Analysis revealed an involvement of S glycoprotein’s receptor binding domain (RBD) in the SARS-CoV-2 binding with the ACE2-R11,16. Lu et al. also demonstrated that S glycoprotein driven ACE2-R binding and membrane fusion determined host tropism and transmission, in SARS-CoV-2 pathogenesis7. Studies on human infections identified Lys31 and Lys353 as critical ACE residues involved in SARS-CoV-2 binding via hydrophobic salt-bridge4. An intriguing aspect is the selective pressure driven mutational susceptibility of residues 479 and 487 in the C-terminal domain (CTD) of S proteins, for interacting with Lys31 and Lys353 amino acid residues of human ACE2-R19. Rigorous computational and modeling studies demonstrate K479N and S487T as natural viral mutations, modulating Lys31 and Lys353 structures and enhancing human ACE2-R binding with viral S1-CTD. Geography specific enhanced COVID-19 sufferers are being examined for enhanced viral S1-CTD-human ACE2-R binding, caused by the modifications in native Lys31 and Lys353confirmations. Thereby, exercising caution towards induced mutations in residues 479 and 487 in the CTD of S-protein emerges a guarding strategy towards enhanced infection spread. This monitoring could improve the one to one ACE-2 and CoVID S-protein interactions.
Figure 2.
Description of CoV2 invasion of a healthy organism (here, host). After uncoating, its RNA, the virus synthesizes its own proteins, followed by invasion of multiple native immunological activities.
It is further noted that RNA viruses are notorious for high mutation rates which can occur a million times faster than that of their hosts. This feature is a primary reason why we need an influenza vaccine every year. The mutagenic capability of a virus depends upon several factors, including the fidelity of viral enzymes that replicate nucleic acids. In the case of SARS-CoV-2, the fidelity of the RNA dependent RNA polymerase (RdRp) is critical. Mutation rates drive viral evolution and genome variability, thereby enabling the virus to escape host immunity and develop drug resistance20. Pachetti et al. reported that the viral genomes present different point mutations, distinguishable within different geographic areas. Three recurrent mutations were identified in Europe (in positions 3036, 14,408 and 23,403) and three different mutations were identified in North America (in positions 17,746, 17,857 and 18,060). These mutations are yet to be detected in Asia. However, the number and occurrence, as well as the median value of virus point mutations identified in Asia, is ever increasing.
It is further reported that the RdRp mutation, located at position 14,408, which is present in European viral genomes starting from February 20th, 2020, is associated with a higher number of point mutations compared to viral genomes from Asia. Given that RdRp works in complex machinery that includes critical proofreading activities, it is tempting to speculate that this mutation has contributed to impairing its proofreading capability. On average, the coronavirus accumulates about two mutations per month in its genome. Coronaviruses have genetic proofreading mechanisms21,22, and SARS-CoV-2 sequence diversity is very low23. Still, natural selection can act upon rare but favorable mutations resulting in antigenic drift and the gradual accumulation of mutations. The complex interplay between immunological resistance and the fitness landscape enables antibody resistance to develop across populations. The most widely studied mutation of SARC-CoV-2 is that of the spike gene. The mutation at position 23,403 has drawn a rigorous attention, in part because it changed the virus’ spike, the protein on its surface that attaches to human cells. The mutation changed the amino acid at position 614 of the spike from an aspartic acid (D) to a glycine (G), thus, G614. It has been reported that a SARS-CoV-2 variant carrying the Spike protein amino acid change D614G has become the most prevalent form in the global pandemic. This mutation has substantially increased the COVID-19 infectivity24. Continuing surveillance of Spike mutations is, hence important for decoding the mechanistic understanding of the virus infection mechanism that could aid the developing vaccines and other relevant immunological interventions.
Molecular diagnosis of SARS-CoV-2 infection
Upon exhibiting symptoms consistent with SARS-CoV-2 infection, the screened positive subjects are promptly recommended for a chest x-ray and/or CT scan. Chest images of initial infection stages demonstrate interstitial changes with multiple small plaques, particularly along the lung periphery. Subsequently, involvement of the middle and outer lung regions (single or multiple lobe(s) become evident along with multiple infiltrating shadows or ground glass opacities25,26. Subjects suspected for COVID-19, should be screened through highly sensitive and specific SARS-CoV-2 diagnostic tests. Using viral cultures to establish the diagnosis is not practical, due to a minimal three days (72 h) requirement for manifesting SARS-CoV-2 characteristic cytopathic changes in selected cell lines. Moreover, SARS-CoV-2 isolation requires biosafety level 3 facilities, often unavailable at diagnostic laboratories11. Owing to these limitations, real-time reverse transcriptase-PCR (rRT-PCR) detection is currently the favored diagnostic approach. It is characterized by high specificity, reproducible quantitative assessment and procedural simplicity. Importantly, rRT-PCR is substantially more sensitive than conventional RT-PCR, with a lower template threshold, that is critical for early diagnosis27,28. The rRT-PCR technique is currently the predominant method for SARS-CoV-2 detection29,30. Of note, results from rRT-PCR using gene specific primers can be influenced by varied viral RNA sequences. False-negative results are likely when mutations prevail in the primer annealing sequences of the SARS-CoV-2 genome7–9. Moreover, rRT-PCR tests may provide false negative results corresponding to insufficient viral load. Owing to these bottlenecks, other molecular techniques such as reverse transcription loop-mediated isothermal amplification (RT-LAMP)31,32 and SARS-CoV-2 antibody dependent assays33 are in persuasion for confirming present or past SARS-CoV-2 infection. More recently, the clustered regularly interspaced short palindromic repeats (CRISPR)-based specific high sensitivity enzymatic reporter unlocking (SHERLOCK) methodology enables transferable, multiplexed and ultrasensitive RNA or DNA recognition from SARS-CoV-2 infected clinical samples.
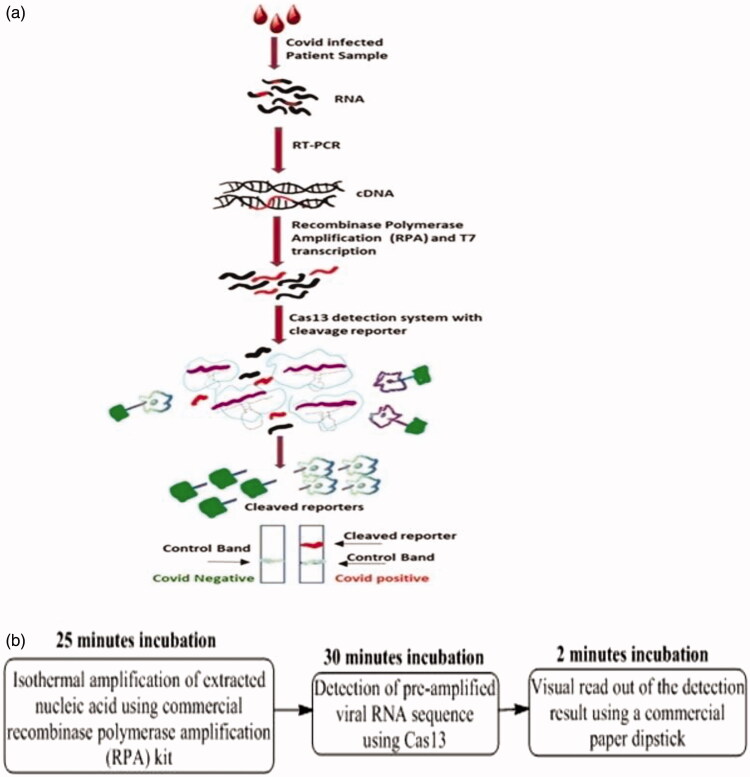
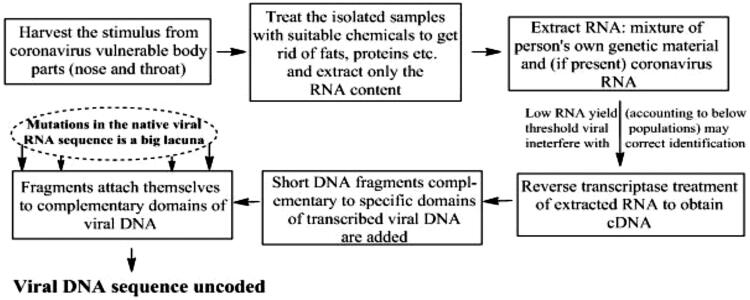
SHERLOCK assays use recombinase polymerase amplification (RPA) of DNA or RNA followed by Cas13- or Cas12-facilitatedrecognition via colorimetric read-outs or fluorescence. They provide results in slightly more than an hour (typically 75 min) (Figure 3)34. The mechanistic distinction of SHERLOCK technique is separately discussed in the subsequent section. The rRT-PCR technique is considered the “gold standard” of viral detection owing to its rapid, highly sensitive and genome specific attributes. The rRT-PCR based viral RNA detection is adequately sensitive for detecting early SARS-CoV-2 infection35. Screening requirements for rRT-PCR comprise primer design, synthesis, and availability in the diagnostic laboratories. Sample collection, RNA isolation, cDNA synthesis, cDNA to dDNA conversions, assay optimization and finally dDNA amplification are the chronological steps for accurate infection analysis (Figure 4)36,37.
Figure 3.
(a) Schematic representation of SHERLOCK technique, superseding PCR via isothermal working and recombinase polymerase assisted amplification, and (b) less than one hour screening cumulative incubation time.
Figure 4.
Procedural details of rRT-PCR technique to ascertain the presence of virus attack. The method is characterized by amplifying the estimated viral genetic content in a potential host.
The availability of the complete SARS-CoV-2 genome early in the outbreak provided a pathway for designing unambiguous primers and COVID-19 specific laboratory procedures7,38,39 On 23rd January 2020 Corman et al. published the sequence of a primers set and probes specific for SARS viral genomes40. The investigators observed three distinctive SARS-CoV-2 genome loci with conserved sequences identical with other coronaviruses. These sequences include the RNA-dependent RNA polymerase (RdRP) gene located in the ORF1ab region and the E and N protein genes. The detection thresholds of RdRP and E genes were 3.6 and 3.9 copies per reaction, i.e. a 95% finding probability. In contrast, the N genes were less sensitive, with 8.3 copies per reaction. Corman et al. proposed a two-target system assay, wherein one primer detects several coronaviruses including SARS-CoV, SARS-CoV-2 while the second detects only SARS-CoV-2. In the published assay, probe 1 was a “pan Sarbeco-Probe” capable of SARS-CoV-2, SARS-CoV and bat SARS-related coronavirus detection while probe 2 (“RdRp-P2” assay) was SARS-CoV-2 specific. It is worth noting yet again that these assays were designed and validated using synthetic nucleic acids in the absence of SARS-CoV-2 isolates or patient specimens41. The RdRp assays were screened in more than 30 European laboratories40. Owing to a high variability of S-protein RBD in the coronavirus genome, relatively few studies have listed S gene as an rRT-PCR target11,39.
After designing the primers and probes, the subsequent step required optimizing assay conditions (e.g. reagent usability, incubation time and temperatures), followed by control PCR validation. The RT-PCR can be performed as a single or two-step process. In the one-step configuration, reverse transcription and PCR amplification are conducted in a single reaction. This approach has significant advantages by providing speedy and reproducible outcomes for high-throughput analysis. The limitation pertains to reverse transcription and amplification optimization owing to their simultaneous conduct, generating a lower target amplicon. In contrast, the two-step model involves sequential reverse transcription and amplification, each in separate tubes42. This format is more sensitive but indeed requires additional time and multiple parameter validations42,43. Importantly, the control must be carefully chosen for minimizing experimental errors.
Samples from suspected COVID-19 subjects may be collected from nasopharyngeal or oropharyngeal swabs, expectorated sputum, endotracheal aspirates, bronchoalveolar lavages (BAL), blood or feces39. For diagnosis, sputum is the desired laboratory sample, followed by nasal swabs. Throat swabs are generally not recommended for diagnosis44. Since BAL collection needs an expert bronchoscope operator, BAL samples are not practical for routine laboratory diagnosis or disease monitoring. Samples should be obtained by Dacron or polyester flocked swabs and arrive at the laboratory immediately after collection.
In the United States, the Center for Disease Control and Prevention (CDC) recommends the one-step real time RT-PCR (rRT-PCR) assay, providing quantitative estimate of viral SARS-CoV-2 loads45. To perform the assay, viral RNA is extracted and added to a master mix (nuclease-free water + forward and reverse primers), a fluorophore-quenching probe and a reaction mix (reverse transcriptase + polymerase + Mg + nucleotides + buffer and additives)46. The master mix and extracted RNA are placed in a PCR thermocycler and the incubation temperatures are set. The CDC recommends rRT-PCR specific cycling conditions that cleave the fluorophore-quencher probe to generate a fluorescent response45. The fluorescence is detected by thermocycler while amplification is monitored in real time. The probe used by Guan et al. was Black Hole Quencher-1 (BHQ1) and fluorescein amidite (FAM). This reaction elapses nearly 45 min and can be run in 96-well plates, with each well carrying a different sample. For SARS-CoV-2, the CDC provides nCoVPC as a positive control sequence45. Several SARS-CoV-2 RT-PCR primers and probes from research laboratories and organizations across the world have been listed in a recent publication47. Most rRT-PCR assays are rapid, and are completed within (45-90) minutes.
RT-PCR, like any other diagnostic approach, can yield false positive and false negative results. The outcome is dependent on several factors including the concentration of template transcripts. Although it is at present the state-of-the-art diagnostics with high sensitivity, the complexity of the genomic and sub genomic COVID RNA with several non-canonical ORFs makes the process extremely complex. So, an accurate interpretation is indeed a challenge.
Feasible diagnostic assays for SARS-COV-2 infection
The SHERLOCK technique
Developed in 2017, the SHERLOCK technique aims at Cas13a facilitated highly sensitive rapid nucleic acid detection with single base specificity, for exclusive binding and cleavage of COVID-19 single stranded RNA48–51. Figure 3(a) depicts the SHERLOCK procedure, wherein RNA is isolated from viral population residing in the nasopharyngeal swabs of the patients followed by c-DNA preparation from RPA. The in vitro transcription is performed with T7 RNA polymerase after which the newly prepared samples are exposed to the Cas13 detection system. Viral transcripts if present activate the cleavage reporter molecule, thereby aiding the cleaved reporter detection using dipstick technique within one hour. The term SHERLOCK signifies a moveable platform, which in combination with isothermal Cas13, assisted pre-amplification besides RNA or DNA screening52.
Amongst the manifold subsequent advancements of SHERLOCK assay, the most significant involves amalgamation with Heating Unextracted Diagnostic Samples to Obliterate Nuclease (HUDSON). The method involves simultaneous thermal and chemical reduction assisted ribonuclease inactivation within body fluids. The inactivation is followed by viral envelope disruption mediated viral particle lysis and concomitant nucleic acid release. In 2018, Gootenberg et al. further advanced the methodology with the introduction of channel multiplexing, quantitative measurement (to as low as 2 aM), more than three-fold enhanced signal sensitivity accomplished via lateral flow readout52. On the basis of novel coronavirus RNA sequences, the researchers designed two guide RNAs, one recognizing the S gene while the other identifying the Orf1ab gene. To maximize the detection accuracy, new coronavirus specific sequences were screened. This approach also minimized the interference from other respiratory viral genomes. If the RNA being examined corresponds to a newly screened coronavirus sample, the guide RNA can distinguish it using concomitant Cas13a activation following which the Cas13a cleaves subsequently encountered RNA molecules. Thus, by confirming the RNA molecule digestion, the presence of new coronavirus can be ascertained in the samples. Using this approach, Kellner et al. continually screened the SARS-CoV-2 sequences within (20–200) aM concentrations at an extent of (10–100) copies per μl). The test can be read using a dipstick in <1 h without any need for elaborate instrumentation34.
Antibody dependent SARS-CoV-2 detection
After entering the body, SARS-CoV-2 is endocytosed and processed for antigen presentation. Although the role of ACE2 receptor has been implicated in viral entry53, uncertainty looms regarding the very first responding antigen presenting cell. Furthermore, the process by which the virus is routed to the lysosomes via early and late endosomes formation and subsequent autophagic modulation remains uncertain. Characteristically, the viral genome must be recognized by the single stranded RNA specific intracellular endosomal TLR7 (Toll Like Receptor 7) of the innate immune system, triggering a downstream signal cascade. This is followed by the recognition of specific viral antigens and MHC I presentation to the naive B lymphocytes. Concomitantly, the B lymphocytes are activated, proliferated and differentiated into memory and plasma cells. The most critical hallmark is clonal selection and the viral antigen implicit B-cell expansion, requiring at least (8–14) days depending on viral epitope antigenicity and the kinetics of variable region binding with the newly synthesized surface bound IgM molecules. Upon epitope binding, the B cells undergo class switching to release soluble IgG molecules into the circulation. At this juncture, a small B cell population becomes dormant and is preserved as memory cells, for future response. Plasma cells subsequently release specific antibodies (Abs) into the circulation, which gradually enter other body fluids. In general, the time from infection to the Abs formation and their release into the circulation is (8–14) days, varying viz-a-viz infected host implicit immune response. IgM is the first antibody generated in the early weeks, followed by IgG in the middle and late infection stages. There is a continuous evolution of the Abs due to somatic hypermutation, progressively increasing the specificities besides ensuring a steady titer rise. SomeSARS-CoV-2 infected individuals may even never exhibit symptoms but though, have a detectable antibody response. While it is likely that Abs may provide immunity to future infections, there is not yet sufficient data to state this conclusively. Additionally, Abs also have variable half-lives. Although Abs are generally stable in the blood for months or in some cases even years, the half-life of the SARS-CoV-2 specific Abs is not known. Since Abs comprise a formidable aspect of the body’s immune response to exposure and do not indicate the virus itself, mere Ab detection is unable to diagnose an active infection but indeed can screen a yester infection. Recently, several interesting SARS-CoV-2 Ab detection approaches, including Rapid Diagnostic Tests (RDT), Enzyme Linked Immunosorbent Assays (ELISA), chemiluminescent and neutralization assays have been proposed29. Table 1 summarizes basic features of these tests. The approvals have been granted earlier this year, with some being permitted under the “Emergency Use Authorization (EUA)” clause.
Table 1.
Molecular approaches for diagnosing viral infections, the target molecule, underlying detection mechanism, amplified product, screened viral infection, commercialization platforms and salient operational aspects are listed.
| Diagnostic test/assay with conduct time | Information revealed/implicit requirements and merits | Major limitations | Manufacturer’s specification (United States) |
|---|---|---|---|
| rRT-PCR/completes in 2 hours | Detection of viral RNA in the serum, rapid and confirmatory screening pf live infection, mandates mRNA to dDNA conversion and finally dDNA amplification | May be false negative with low viral load or in early stages of infection | Lab Corp., Abbott Molecular Inc. |
| SHERLOCK/completes in one hour | Detects active infection, Exploits CRISPR (bacterial immune system) to screen viral nuc | Begins with RNA/cDNA harvested from patient’s sample | Sherlock Biosciences |
| Rapid Diagnostic Test (RDT)/completes within half an hour | Rapid detection of host Abs generated on previous viral exposure, overcomes costly PCR conditioning by isothermal working | Non-quantitative assay, error prone in terms of specificity, may not be exclusive for SARS-CoV2 | Luminex Molecular Diagnostics, Inc. |
| Enzyme Linked Immunosorbent Assay (ELISA)/completes within (2–5) hours | Quantitative/qualitative estimation of specific Ab titer, higher specificity conferred via enzyme linkages | No assurance about specific Ag-Ab interaction | Thermo Fisher Scientific, Inc. |
| Neutralization/completes in (3–5) days | Ascertains neutralizing Abs in patient serum, instrumental in preventive enhancement of viral population | Long duration and an escapism of antibodies that are not against viral replication | Bioreference, Quest Diagnostics Infectious Diseases Inc. |
| Chemiluminescent immunoassay/completes in (1–2) hours | High specificity and reagent stability, low cost method with less reagent consumption, reduced incubation time | Restricted Ag detection, high operative costs, may be inaccessible for catering the need of increasing CoVID2 sufferers | Cellex, Inc. |
CRISPR, Clustered regularly interspaced short palindromic repeats; SARS-CoV2, severe acute respiratory syndrome coronavirus 2; SHERLOCK, specific high-sensitivity enzymatic reporter unlocking; Ab, antibody; Abs, antibodies; Ag, antigen; Ag-Ab, antigen-antibody.
Rapid diagnostic test (RDT)
This is typically a qualitative lateral flow assay capable of being performed in small, portable, point of care (POC) systems. Samples are obtained via finger prick, salivary or nasal swab. Similar to pregnancy tests, RDTs are monitored via colored lines to indicate positive or negative response. For COVID-19, this assay allows rapid detection of host IgG and IgM Abs. Though several commercial kits are available, only a few are FDA approved. Some of these detect either IgM or IgG while a few kits are also equipped with dual detection. If possible, it is preferable to have baseline (prior to infection) IgG and IgM titers.
Enzyme-linked immunosorbent assay (ELISA)
This assay allows qualitative as well as quantitative assessment of antibody titer and can examine whole blood, plasma or serum samples. It is a sandwich antibody detection assay in which a plate is coated with a relevant viral protein, such as S protein of SARS-COV-2. The samples are then incubated in the plates to allow the Abs binding with the coat protein. Bound antibody-protein complex is subsequently detected using another set of Abs generating fluorescence sensitive readout of the binding complex. For COVID-19, this assay detects IgG and IgM, within (2-5) hour time span.
Neutralization assay
This assay determines the efficacy of patient Abs to prevent viral infection in a laboratory setting and can be performed even when the infection has been cleared. Input samples comprise whole blood, serum or plasma. This assay is conducted in cell culture under conditions allowing SARS-CoV-2 growth (like Vero E6 cells). Concerns with this assay include the 3 to 5 days requirement besides detecting the non-ARS-CoV-2 Abs. When virus and healthy cells are grown with serially diluted patient Abs, the antibody titer can be computed for inhibiting viral replication. Neutralizing antibodies may be used to treat a SARS-CoV-2 infected person. The immunoglobulin heavy chain gene (IGHV) mutation status correlates with the clinical outcome of patients presenting with certain diseases54. A treatment option for SARS-COV-2 patients in past 6 months has been convalescent plasma therapy. Convalescent plasma is a ready source of antibody and may help patients recover. The low somatic hypermutation of BCR genes should provide a greater opportunity for success of the convalescent plasma therapy, as it can be more widely used in patients with different degrees (mild to severe) of infection. There are reports of the presence of antibodies to IGHV3 in patients recovering from SARS-COV-2, inferring that antibodies with low somatic mutation of IGHV are sufficient to neutralize viral antigens and may possibly be successfully used as therapy55.
The process of antibody production and subsequent antigen neutralization varies between individuals and is guided by the immune health of the patients. The data arising over the past several months indicates that many individuals suffer a second infection suggesting either the neutralizing antibodies are short lived or are incapable of providing a strong neutralizing response to viral antigens. Thus, in the case of COVID-19, a positive antibody test does not guarantee immunity. It has not yet been established that the antibodies produced are in fact neutralizing antibodies. It is possible that an antibody may bind to a virus that is not required for the virus to infect cells. To be neutralizing, an antibody must prevent the virus from infecting cells. In general, as the infection progresses the antigen binding specificity of antibodies are enhanced by somatic hypermutation and appropriate clonal selection. Such reports are yet to come COVID 19 infections and, in fact, the presence of low somatic hypermutation of B cell receptor genes has been reported.
Intracellular antigens including viral antigens are primarily presented by MHC (major histocompatibility complex/human leucocyte antigens) class 1 molecules. The different sub groups of HLA molecules (HLA A; HLA B; HLA C) present in different individuals have different antigen presenting abilities and can thus trigger stronger or weaker humoral responses. Clones producing deleterious antibodies are usually not selected (clonal selection) and are programmed to initiate apoptosis. Deleterious antibodies should not be present in immunologically strong individuals.
Arvin et al. have discussed the potential for antibody dependent enhancement of infection and inflammation (ADE) in context to SARS-CoV-2 infections56. It is yet to be determined whether antibodies predispose, trigger, or assist the progression of viral infections. Most studies of other viruses elucidate that ADE is related to inflammation that can be manifested by low affinity or cross-reactive antibodies with limited or no neutralizing activity. Rather the endemic nature of coronavirus infections suggests that infection in the presence of low antibody levels is common, providing a possible opportunity for ADE of disease-although these illnesses are mild, and infer that cross-protection may be transient. It is of noteworthy that neither low neutralizing-antibody titers nor heterologous virus challenge are associated with enhanced disease in human SARS-COV-2 studies.
The molecular mechanism that triggers the cytokine storm around (10–14) days post infection resulting in multi-organ damages has not yet been elucidated, and its correlation with the neutralizing antibodies has not been established. It is certainly possible that CD8+ T cells mediated immunity is the culprit that triggers cytokine production after (10–14) days of infection.
Chemiluminescent immunoassay
This procedure is a quantitative, laboratory-based assay employing whole blood, plasma or serum samples. Typically, subject samples are mixed with an identified viral protein, buffer reagents and enzyme-labeled Abs, allowing a luminescent read-out. Sample Abs react with viral protein to form an antigen-antibody (Ag–Ab) complex. Subsequently secondary enzyme-labeled Abs are added which bind to previously formed Ag-Ab complexes in a sandwich regime, detected via optically sensitive chemical reaction. The light emitted from each sample is used to quantify the Abs in screened sample(s). This test can detect IgG, IgM and IgA Abs.
Table 2 comprises the top 30 FDA approved companies for commercialized availability COVID-19 diagnostic kits, with most of the approvals have been granted under the “Emergency Use Authorization (EUA)” clause. The information is collected from US FDA official website (on 4 October, 2020) and most of the companies are US based. Although the time required for assay procedure varies, but most of the methods can generate results within one-two days. Each kit has its implicit advantages and drawbacks and efforts to overcome the concurrent limitations are in continual persuasion.
Table 2.
Top 30 FDA approved nucleic acid and antibody based SARS-CoV-2 commercialized diagnostic kits, with their manufacturer’s specifications.
| Sr. No. | Company/Organization | Date of Approval | Product Nomenclature | Company’s HQs |
|---|---|---|---|---|
| 01 | Tempus Labs, Inc. | 01-10-2020 | iC SARS-CoV-2 Test | Chicago |
| 02 | Aeon Global Health | 30-09-2020 | Aeon Global Health SARS-CoV-2 Assay | Gainesville (GA) |
| 03 | Alimetrix, Inc. | 30-09-2020 | Alimetrix SARS-CoV-2 RT-PCR Assay | Huntsville, Alabama |
| 04 | Centogone US, LLC | 29-09-2020 | CentoSure SARS-CoV-2 RT-PCR Assay | Massachusetts |
| 05 | Akron Children’s Hospital | 29-09-2020 | Akron Children’s Hospital SARS-CoV-2 Assay | Akron, Ohio |
| 06 | National Jewish Health | 29-09-2020 | SARS-CoV-2 Mass Array Test | Denver, Colorado |
| 07 | Genetrack Biolabs, Inc. | 25-09-2020 | Genetrack SARS-CoV-2 Molecular Assay | Seattle, WA |
| 08 | Cepheid Inc. | 24-09-2020 | Xpert Xpress SARS-CoV-2/Flu/RSV | Sunnyvale, California |
| 09 | Clear Labs, Inc. | 23-09-2020 | Clear Dx SARS-CoV-2 Test | San Carlos, California |
| 10 | Quadrant Biosciences Inc. | 22-9-2020 | Clarifi COVID-19 Test Kit | Syracuse, New York |
| 11 | Vela Operations Singapore Pte. Ltd. | 22-9-2020 | ViroKey SARS-CoV-2 RT-PCR Test v2.0 | New Jersey |
| 12 | KimForest Enterprise Co., Ltd. | 21-9-2020 | KimForest SARS-CoV-2 Detection Kit | Xizhi Dist., New Taipei City 221 |
| 13 | GK Pharmaceuticals Contract Manufacturing Operations | 18-09-2020 | GK ACCU-RIGHT SARS-CoV-2 RT-PCR KIT | Manatí, Puerto Rico |
| 14 | Visby Medical, Inc. | 16-9-2020 | Visby Medical COVID-19 | San Jose, California |
| 15 | Roche Molecular Systems, Inc. | 14-09-2020 | cobas SARS-CoV-2 & Influenza A/B Nucleic Acid Test for use on the cobas Liat System | Pleasanton, California |
| 16 | Beijing Wantai Biological Pharmacy Enterprise Co., Ltd. | 09-09-2020 | Wantai SARS-CoV-2 RT-PCR Kit | Beijing, China |
| 17 | Billion To One, Inc. | 04-09-2020 | qSanger-COVID-19 Assay | Menlo Park, California, |
| 18 | Roche Molecular Systems, Inc. | 03-09-2020 | cobas SARS-CoV-2 & Influenza A/B | Pleasanton, California |
| 19 | Bioeksen R&D Technologies Ltd. | 02-09-2020 | Bio-Speedy Direct RT-qPCR SARS-CoV-2 | KOSGEB-İT, Istanbul, Turkey |
| 20 | Detectachem Inc. | 01-09-2020 | MobileDetect Bio BCC19 (MD-Bio BCC19) Test Kit | Stafford, Texas |
| 21 | OPTOLANE Technologies, Inc. | 01-09-2020 | Kaira 2019-nCoV Detection Kit | Seongnam city, Gyeonggi-do, Korea |
| 22 | T2 Biosystems, Inc. | 31-08-2020 | T2SARS-CoV-2 Panel | Lexington, Massachusetts. |
| 23 | Mammoth Biosciences, Inc. | 31-08-2020 | SARS-CoV-2 DETECTR Reagent Kit | South San Francisco, California |
| 24 | Color Genomics, Inc. | 31-08-2020 | Biomeme SARS-CoV-2 Real-Time RT-PCR Test | Burlingame, California |
| 25 | MiraDx | 31-08-2020 | MiraDx SARS-CoV-2 RT-PCR assay |
Los Angeles, California |
| 26 | BayCare Laboratories, LLC | 31-08-2020 | BayCare SARS-CoV-2 RT PCR Assay | Florida |
| 27 | Cuur Diagnostics | 26-08-2020 | Cuur Diagnostics SARS-CoV-2 Molecular Assay | Las Vegas, NV, United States |
| 28 | Fluidigm Corporation | 25-08-2020 | Advanta Dx SARS-CoV-2 RT-PCR Assay | South San Francisco, California |
| 29 | Guardant Health, Inc. | 21-08-2020 | Guardant-19 | Redwood City, California |
| 30 | DxTerity Diagnostics, Inc. | 21-08-2020 | DxTerity SARS-CoV-2 RT-PCR Test | Rancho Dominguez, California |
Conduct time, precision of estimation and robustness with respect to patient status and operational procedures are the major performance determining criterion. In all, there are 176 companies having been granted approval (till October 4, 2020).
Blackspot on the testing system between RT-PCR and antibody mediated detection of SARS-CoV-2 infection
COVID-19 detection assays were developed under tremendous pressure and hence blackspots in various detection systems are highly probable. While it could take several years to systematically revisit all of the current flaws and develop better assays to circumvent them, several blackspots have been identified and are actively being addressed by investigators and pharmaceutical companies. As one example, it was observed that even after the virus is cleared by the patients’ immune system, and the affected individual is no longer contagious, the RT-PCR test continues to be positive for several weeks owing to the presence of circulating viral RNA transcripts, which takes longer time to clear. The Center for Disease Control (CDC) recently established a diagnostic test to distinguish between individuals carrying the intact virus (contagious) and those having the circulating RNA transcripts (not contagious). This RT-PCR-based approach identifies a unique target on circulating viral transcripts. Use of this assay (SMART Assay) helps individuals return to work earlier and perhaps, reduce the socio-economic burden.
It is widely appreciated that antibody tests are not recommended for detection of infection. These tests help in knowing whether the individual being tested was previously infected even in asymptomatic subjects. Assays to determine serum antibody concentration can be used to support clinical assessment of an infected patient late in the course of their illnesses. Moreover, if a patient is suspected to have a post-infectious syndrome caused by SARS-CoV-2 infection (e.g. Multisystem Inflammatory Syndrome in Children; MIS-C), serologic assays may be used. Serum IgM and IgG production takes anywhere from ten to twenty-one days after initial infection to develop, depending on the immune health of the patient. Hence it is helpful in determining the onset of infection, or disease progression at early time points. The test establishes prior infections of either symptomatic or asymptomatic patients. Scientists have yet to determine the properties of the COVID-19 specific immunoglobulins in terms of half-life and antigen binding/neutralizing characteristics. Thus, the presence of antibody does not assure protection against re-infection. This limitation compounded with the rapid mutating nature of RNA viruses, is perhaps the greatest challenge in vaccine production and is considered as a serious blackspot of the assay.
Conclusions
As of 13 October 2020 more than thirty eight million SARS-CoV-2 infections have been detected worldwide, with well above one million deaths. The United States is disappointedly, the worst hit with over eight million infections and more than two lakh deaths. The position of India is no better, with greater than seven million infections and over one lakh deaths. Indeed, COVID-19 socio-economic impacts have been far reaching, and the research endeavors need to be robust. Intensive research is necessary to identify effective methods of prevention and treatment.
To prevent spread within communities, early virus detection, both for symptomatic and asymptomatic (∼ one-third of the patients) cases is essential. While SARS-CoV-2 specific Abs in body fluids pinpoint the previous infection, screening the genetic material (SARS-CoV-2 RNA) remains the best approach for prompt detection. An important distinction and rather necessity for rRT-PCR is the presence of viral burdens adequate for PCR initiation. Another challenge in PCR screening is the frequent SARS-CoV-2 mutations. Thus, while a PCR approach indeed assists clinical diagnosis and evaluation, it may not absolutely confirm the presence of an infection. Subjects with a high clinical suspicion but negative RT-PCR screening should be examined via repeated swab test. Antibody tests are of substantial epidemiological significance. Abs also portray a likelihood therapeutic option via administration of COVID-19 Abs rich plasma of a recovered subject to an infected individual. However, antibody tests cannot screen an active SARS-CoV-2 infection, owing to the minimal requirement of one to two weeks for detectable antibody titer. Furthermore, immuno-compromised subjects may even take longer to develop Abs. Thus, rRT-PCR and antibody tests are complementary to each other as early stage diagnosis and comprehensive epidemiology assessment of SARS-CoV-2 infection and the consequent therapeutic response.
Acknowledgements
Authors wish to oblige the uninterrupted internet facilities accorded hereby from Central University of Gujarat, (Gandhinagar), India and University of Utah, Salt Lake City, Utah, USA.
Transparency
Declaration of funding
The study was not funded.
Declaration of financial/other relationships
The authors declare no financial or other relationships that may cause any conflict of interest.
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Author contributions
All authors unanimously accept the responsibility for description and discussion covered in this manuscript and approval of its final version.
References
- 1.Chen N, Zhou M, Dong X, et al. . Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses . The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2 (Consensus Statement). Nat. Microbiol. 2020;5:536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss SR, Leibowitz JL.. Coronavirus pathogenesis. Adv Virus Res. 2011;81:85–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3(1):237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Mao JM, Wang GD, et al. . Human SARS-CoV-2 has evolved to reduce CG dinucleotide in its open reading frames. Sci Rep. 2020;10(1):12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim D, Lee JY, Yang JS, et al. . The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Lu R, Zhao X, Li J, et al. . Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Phan T, Genetic diversity and evolution of SARS-CoV-2, Infect. Genet Evol. 2020; 81:104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen Z, Xiao Y, Kang L, et al. . Genomic diversity of SARs-CoV-2 in coronavirus disease 2019 patients. Clin Infect Dis. 2020;ciaa203. DOI: 10.1093/cid/ciaa203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou P, Yang XL, Wang XG, et al. . A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim YX, Ng YL, Tam JP, et al. . Human coronaviruses: a review of virus-host interactions, diseases. Diseases. 2016;4(4):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neuman BW, Kiss G, Kunding AH, et al. . A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoeman D, Fielding BC.. Coronavirus envelope protein: current knowledge. Virol J. 2019;16(1):10–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92(4):418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wrapp D, Wang N, Corbett KS, et al. . Cryo-EM Structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yount B, Roberts RS, Sims AC, et al. . Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J Virol. 2005;79(23):14909–14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Kang Z, Gong H, et al. . The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. bioRxiv 2020. DOI: 10.1101/2020.01.30.927806 [DOI] [Google Scholar]
- 18.Wu K, Chen L, Peng G, et al. A virus-binding hot spot on human angiotensin-converting enzyme 2 is critical for binding of two different coronaviruses. J Virol. 2011;85(11):5331–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen K, Yang Y, Wang T, Global Pediatric Pulmonology Alliance, et al. Diagnosis, treatment, and prevention of 2019 novel coronavirus infection in children: experts' consensus statement. World J Pediatr. 2020;16(3):223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pachetti M, Marini B, Benedetti F, et al. . Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18:179. DOI: 10.1186/s12967-020-02344-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sevajol M, Subissi L, Decroly E, et al. . Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014;194:90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith EC, Blanc H, Vignuzzi M, et al. . Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLOS Pathog. 2013;10(7):e1004342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fauver JR, Petrone ME, Hodcroft EB, et al. . Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell. 2020;181(5):990–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korber B, Fischer WM, Gnanakaran S, Sheffield COVID-19 Genomics Group, et al.. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan Y, Guan H, Zhou S, et al. . Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur Radiol. 2020;30(6):3306–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noh JY, Yoon SW, Kim DJ, et al. . Simultaneous detection of severe acute respiratory syndrome, middle east respiratory syndrome, and related bat coronaviruses by real-time reverse transcription PCR. Arch Virol. 2017;162(6):1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wan Z, Zhang Y, He Z, et al. . A melting curve-based multiplex RT-qPCR assay for simultaneous detection of four human coronaviruses. IJMS. 2016;17(11):1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu XY, Whitaker B, Sakthivel SKK, et al. . Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J Clin Microbiol. 2014;52(1):67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corman VM, Eckerle I, Bleicker T, et al. . Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17:20285. [DOI] [PubMed] [Google Scholar]
- 30.Corman VM, Landt O, Kaiser M, et al. . Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhadra S, Jiang YS, Kumar MR, et al. . Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV). PLOS One. 2015;10(4):e0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan JFW, Choi GKY, Tsang AKL, et al. . Development and evaluation of novel real-time reverse transcription-PCR Assays with locked nucleic acid probes targeting leader sequences of human-pathogenic Coronaviruses. J Clin Microbiol. 2015;53(8):2722–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long QX, Deng HJ, Chen J, et al. . Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. medRXiv. 2020. DOI: 10.1101/2020.03.18.20038018 [DOI] [Google Scholar]
- 34.Kellner MJ, Koob JG, Gootenberg JS, et al. . Author Correction: SHERLOCK: nucleic acid detection with CRISPR nucleases. Erratum in. Nat Protoc. 2020;15(3):1311. [DOI] [PubMed] [Google Scholar]
- 35.Zou L, Ruan F, Huang M, et al. . SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freeman WM, Walker SJ, Vrana KE.. Quantitative RT-PCR: pitfalls and potential. Bio Techniques. 1999;26:124–125. [DOI] [PubMed] [Google Scholar]
- 37.Kageyama T, Kojima S, Shinohara M, et al. . Broadly reactive and highly sensitive assay for norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol. 2003;41(4):1548–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.(a) World Health Organization (WHO) . Coronavirus. Geneva: WHO; 2020. (b) To KKW, Tsang OTY, Leung WS, et al. . Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reusken CBEM, Broberg EK, Haagmans B, et al. . Laboratory readiness and response for novel coronavirus (2019-nCoV) in expert laboratories in 30 EU/EEA countries. Eurosurveill. 2020;25:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong ML, Medrano JF.. Real-time PCR for mRNA quantitation. Bio Techniques. 2005;39:75–85. [DOI] [PubMed] [Google Scholar]
- 42.Bustin S. A–Z of Quantitative PCR. San Diego, CA: International University Line; 2004. [Google Scholar]
- 43.Yang Y, Peng F, Wang R, et al. . The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun. 2020;109:102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Centers for Disease Control and Prevention . Research use only real-time RT-PCR protocol for identification of 2019-nCoV; 2020. [cited 2020 Mar 6]. Available from: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-detection-instructions.html.
- 45.Real-time RT-PCR Diagnostic Panel; Division of Viral Diseases . CDC 2019-Novel Coronavirus (2019-nCoV). Atlanta (GA): U.S. Centers for Disease Control and Prevention; 2020. [Google Scholar]
- 46.Udugama B, Kadhiresan P, Kozlowski HN, et al. . Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14(4):3822–3835. [DOI] [PubMed] [Google Scholar]
- 47.Wright AV, Nunez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing Nature's Toolbox for Genome Engineering. Cell. 2016;164(1–2):29–44. [DOI] [PubMed] [Google Scholar]
- 48.Gootenberg JS, Abudayyeh OO, Lee JW, et al. . Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freije CA, Myhrvold C, Boehm CK, et al. . Programmable inhibition and detection of RNA viruses using Cas13. Mol Cell. 2019;76(5):826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.(a) Gronowski AM. Who or what is Sherlock? J Intl Fed Clin Chem Lab Med. 2018;29:201–204. (b) Myhrvold C, Freije CA, Gootenberg JS, et al. . Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360(6387): 444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang F, Abudayyeh OO, Gootenberg JS. A protocol for detection of COVID-19 using CRISPR diagnostics. v.20200321
- 52.Gootenberg JS, Abudayyeh OO, Kellner MJ, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia HP, Look DC, Shi L, et al. . ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol. 2005;79(23):14614–14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rozovski U, Keating MJ, Estrov Z.. Why is the immunoglobulin heavy chain gene mutation status a prognostic indicator in chronic lymphocytic leukemia? Acta Haematol. 2018;140(1):51–54. [DOI] [PubMed] [Google Scholar]
- 55.Zhang F, Gan R, Zhen Z, et al. . Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct Target Ther. 2020;5(1):156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arvin AM, Fink K, Schmid MA, et al. . A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584(7821):353–363. [DOI] [PubMed] [Google Scholar]