In 1973, Dr. Ralph Steinman first described a new cell type with a tree-like shape in the immune system and named it a “dendritic cell” (DC). Following this discovery, numerous studies confirmed the key role of DCs as antigen-presenting cells (APCs) in controlling immune responses, and then, Dr. Steinman won the Nobel Prize in Physiology or Medicine in 2011.1 When we have greater understanding of DC biology, more DC knowledge will be applied in medicine. DC-based therapies have been developed to treat various diseases, and DC vaccines for cancer therapy are the most prominent. In contrast to enhancing immunity, another type of DC vaccine is used to ameliorate autoimmune diseases and is called a “tolerogenic DC” vaccine.2 These discoveries suggest a “ying-yang” regulation of DCs in immune homeostasis. One theory is that DC precursors can differentiate into specialized subsets to initiate various immune responses. However, it is also possible that one DC subset may exert different functions based on external factors.
The ontogeny of DC is complicated and theories continue to be updated. In general, three major DC populations have been identified (Fig. 1). Conventional dendritic cells (cDCs) comprise two subsets, type 1 (cDC1) and type 2 (cDC2). Plasmacytoid DCs (pDCs) are specialized for type I interferon (IFN) secretion. Every subset expresses a specific profile of cell surface markers and transcription factors (TFs). In the classification of mouse cDCs, cDC1s are identified by their surface expression of XCR1, CD8α, CLEC9A, and DEC205, and they are developmentally dependent on IRF8, BATF3, Id2, and Nfil3. The cDC1s cross-present antigens and prime cytotoxic CD8+ T cell responses to intracellular pathogens. On the other hand, cDC2s, defined mainly by CD11b and CD172a (SIRPα) expression, constitute heterogeneous cell types with differential surface expression of ESAM, CD301b, CLEC12A, CXCR5, or CXCR3. cDC2s differentially require IRF4, Klf4, Zeb2, IRF2, RelB, Ikaros, or Notch and preferentially activate CD4+ helper T cell (Th) responses.3 However, under inflammatory conditions, classical circulating Ly6ChiCD11b+CD172a+ monocytes are recruited to the sites of injury and predominantly contribute to the development of DCs, which play major roles in infection control.4 These inflammatory DCs have been recognized as a special subset but are known by different names, such as monocyte-derived cells (MCs), TNF-α/iNOS-producing DCs (Tip-DCs), or monocyte-derived DCs (moDCs). All DCs initiate and modulate various immune responses.
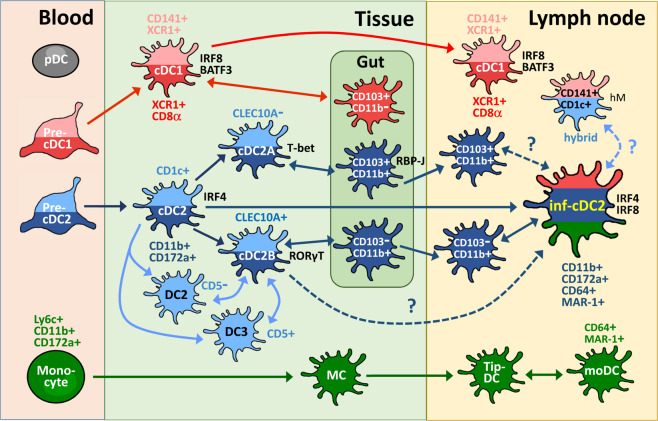
Fig. 1.
The relationship of inflammaory cDC2s (inf-cDC2s) to other cDC2 subsets during DC development. The developmental pathways of DC populations are briefly illustrated. Both mouse (dark red) and human (light red) cDC1s and their major markers and transcription factors have been identified. The cDC2s are found in mice (dark blue) and humans (light blue), and the derivation (single arrow) or correlation (double arrow) is evidenced (solid line) or has been proposed (dashed line) for these cDC2 subsets. During inflammation, circulating mouse monocytes (green) are recruited and are transformed into inflammatory DCs (MC, monocyte-derived cell; Tip-DC, TNF-α/iNOS-producing DC; moDC, monocyte-derived DC). In particular, gut cDCs comprise distinct subsets and have unique phenotypes but are also classified into the cDC1 or cDC2 lineage. We focus on the cDC2-relevant subsets. Both cDC2A and cDC2B cells are characterized in mice and humans. Mouse inf-cDC2s are derived from the cDC2 precursor but share phenotypes with cDC1s and moDCs. However, the relationships of inf-cDC2s with cDC2B and gut CD103+CD11b+ cells are not known. In addition, further study is needed to determine whether CD141+CD1c+ DCs (in humanized mice, hM) are the counterparts of inf-cDC2s in humans
Single-cell technologies have been developed for extensively studying cell development, and they include multicolor flow cytometry, cytometry by time of flight, and single-cell RNA sequencing (scRNA-seq).5 Recently, the Lambrecht group at Ghent University reported in Immunity that a dominant DC population has great potential to migrate to draining lymph nodes (LNs) and prime CD4+ and CD8+ T cell responses to viral infection. This group called these cells inflammatory cDC2s (inf-cDC2s) because they express CD172a and CD11b; however, inf-cDC2s also express the Fc receptor CD64 and IRF8, similar to moDCs and cDC1s, respectively. In addition, this group found that Toll-like receptor (TLR) ligands and type 1 IFN induce the maturation of inf-cDC2s in an IRF8-dependent manner. Using other inflammatory disease models, the group also detected the accumulation of inf-cDC2s in other LNs, suggesting that inf-cDC2 represents a general APC in immune responses. Thus, inf-cDC2 likely becomes a new cDC2 subset during inflammation.6
Interestingly, the Rudensky group at the Memorial Sloan Kettering Cancer Center described two new cDC2 subtypes in Cell before finding inf-cDC2s by using single-cell “bulk” RNA-seq and specific gene reporter analyses.7 cDC2A and cDC2B cells mature through different developmental pathways regulated by the TFs T-bet and RORγt, respectively. cDC2A cells have a regulatory anti-inflammatory function, but cDC2B cells possess proinflammatory potential, suggesting a high degree of functional specialization in these two subsets. An immediate question based on the relationship of inf-cDC2s to cDC2A and/or cDC2B cells is raised. From a functional perspective, the immunogenic inf-cDC2s seem to be derived from the proinflammatory cDC2B cells, which produce more TNF-α and IL-6 than cDC2A cells do upon TLR activation. However, the expression of surface markers and transcriptional mediators, especially T-bet, in the cDC2B cells needs to be further examined in inf-cDC2s. It can certainly not be excluded that inf-cDC2s represent a distinct subset.
Do inf-cDC2s exist in humans? In human DC development, human cDCs are stratified into CD141+ cDC1s with the coexpression of XCR1, CLEC9A, BTLA, and NECL2, and IRF8 and BATF3 are hallmark TFs. Human CD1c+ cDC2s, on the other hand, are characterized by CD11c, SIRPα, and CLEC10A expression, and they rely on IRF4 during development. Recently, monocyte-like CD1c+CD5− cDC2s (DC2 cells), which induce the Th1 response, and CD1c+CD5+ cDC2s (DC3 cells), which stimulate Th2, Th17 and Treg differentiation, were further characterized.8 To address the counterpart cell type in humans, the Rudensky group demonstrated that CD1c+CLEC10A+CLEC4Alow cDC2s (including DC2 and DC3 cells) and CD1clowCLEC10A−CLEC4Ahigh cDC2s may resemble mouse cDC2B and cDC2A cells, respectively.9 Other evidence shows that cDC2A cells are also found in melanoma patients.7 Although the Lambrecht group did not identify inf-cDC2s in humans, the upregulation of CD1c was observed in human CD141+ DCs after immunization with adenovirus-based vaccines based on humanized mice with human hematopoietic stem cells.10 This “hybrid” CD141+CD1c+ DC likely provides a clue on the potential role of human inf-cDC2s; however, it is first necessary to characterize inf-cDC2s in humans with more studies, such as an exploration of IRF8 dependency.11
Another important functional marker that the Lambrecht group did not discuss in their study is CD103. Inf-cDC2s lack CD103 expression, which raises a question about inf-cDC2s in the gut immune system. Intestinal cDCs also comprise cDC1 and cDC2 subsets, which can be identified by the expression of XCR1 and CD172a, respectively. However, CD103 and CD11b are particularly used as markers for intestinal cDC subsets. In the mouse gut, cDC1s are usually classified as CD103+CD11b– cells, whereas cDC2s contain both CD103–CD11b+ and CD103+CD11b+ cell populations. The development of CD103+CD11b+ cDC2s requires the TF RBP-J, so they are related to cDC2A cells.9 In addition, the T-bet expression in intestinal cDC2A cells is dependent on the microbiota.7 Therefore, the functions of tolerogenic CD103+CD11b+ cDC2s and proinflammatory CD103–CD11b+ cDC2s are consistent with the definitions of cDC2A and cDC2B cells, respectively. In a steady state, both CD103+ cDC1s and CD103+ cDC2s play major roles in promoting Treg production and maintaining oral tolerance by expressing retinoic acid convertase Aldh1a2.12 When cDC2 subsets migrate into mesenteric LNs (MLNs), CD103–CD11b+ cDC2s induce a more potent Th response than is induced by CD103+CD11b+ cDC2s, indicating that CD103–CD11b+ cDC2s are analogs of inf-cDC2s. However, Flores-Langarica et al. reported that CD103+CD11b+ cDC2s are essential for the induction of primary T- and B-cell responses in the mucosa due to the accumulation of these cells in MLNs in a TLR5-dependent manner after immunization with soluble flagellin.13 We do not know how close the CD103+CD11b+ cDC2s are to the inf-cDC2s in the MLNs because CD103+CD11b+ cDC2s do not play a central role in the induction of these responses in the spleen. The expression patterns of other TFs and surface markers should be further examined in these gut cDC subsets.
In summary, the Lambrecht group identified a new cDC2 subset, inf-cDC2, that shares characteristics with cDC1 and moDC to boost immunity during inflammation in mice. Although the study of DC development has been facilitated by new techniques, it is still limited in aspect of analyses of human DC subsets in vivo. Recently, engrafted engineered mesenchymal stromal cells were used to create a unique platform recapitulating the full spectrum of cDC subsets in vivo.14 This technique may be helpful for exploring human DC development. Generally, a commonly held supposition suggests that the heterogeneity of cDC2s is likely caused by specific transcriptional profiles in response to different tissue environments, in terms of either tolerance or immunity. Hence, the critical features to understand these distinct subsets of cDC2s include the environmental factors involved in cDC2 differentiation, the cDC2 developmental pathways driven by these factors, the distinct TF profiles of cDC2s in different pathways, the relationship of lineage dependency or an alternative activation state among the various cDC2 subsets, and T-cell differentiation programs induced by distinct cDC2s.9 In the future, we believe that researchers will continue identifying new DC subsets, which will surely facilitate the design of new strategies for DC-based immunotherapy.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Jenn-Yeu Shin, Chen-Yu Wang
References
- 1.Steinman RM. Annu. Rev. Immunol. 2012;30:1–22. doi: 10.1146/annurev-immunol-100311-102839. [DOI] [PubMed] [Google Scholar]
- 2.Fucikova J, Palova-Jelinkova L, Bartunkova J, Spisek R. Front. Immunol. 2019;10:2393. doi: 10.3389/fimmu.2019.02393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macri C, Pang ES, Patton T, O’Keeffe M. Semin. Cell Dev. Biol. 2018;84:11–21. doi: 10.1016/j.semcdb.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Bieber K, Autenrieth SE. Mol. Immunol. 2020;121:111–117. doi: 10.1016/j.molimm.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Dress RJ, Liu Z, Ginhoux F. Mol. Immunol. 2020;122:186–192. doi: 10.1016/j.molimm.2020.04.020. [DOI] [PubMed] [Google Scholar]
- 6.Bosteels C, et al. Immunity. 2020;52:1039–1056.e9. doi: 10.1016/j.immuni.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown CC, et al. Cell. 2019;179:846–863.e24. doi: 10.1016/j.cell.2019.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amon L, Lehmann CHK, Heger L, Heidkamp GF, Dudziak D. Mol. Immunol. 2020;120:122–129. doi: 10.1016/j.molimm.2020.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Bosteels C, Scott CL. Mol. Immunol. 2020;121:38–46. doi: 10.1016/j.molimm.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coelho-Dos-Reis JGA, et al. J. Infect. Dis. 2020;221:201–213. doi: 10.1093/infdis/jiz432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tussiwand R, Rodrigues PF. Immunity. 2020;52:892–894. doi: 10.1016/j.immuni.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Sun T, Nguyen A, Gommerman JL. J. Immunol. 2020;204:1075–1083. doi: 10.4049/jimmunol.1900710. [DOI] [PubMed] [Google Scholar]
- 13.Flores-Langarica A, et al. Front. Immunol. 2018;9:2409. doi: 10.3389/fimmu.2018.02409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anselmi G, et al. Nat. Commun. 2020;11:2054. doi: 10.1038/s41467-020-15937-y. [DOI] [PMC free article] [PubMed] [Google Scholar]