Abstract
Purpose:
Known clinical and genetic markers have limitations in predicting disease course and outcome in juvenile myelomonocytic leukemia (JMML). DNA methylation (DNAme) patterns in JMML have correlated with outcome across multiple studies, suggesting it as a biomarker to improve patient stratification. However, standardized approaches to classify JMML based on DNAme patterns are lacking. We therefore sought to define an international consensus for DNAme subgroups in JMML and develop classification methods for clinical implementation.
Experimental Design:
Published DNAme data from 255 JMML patients were used to develop and internally validate a classifier model. Accuracy across platforms (EPIC-arrays and MethylSeq) was tested using a technical validation cohort (32 patients). The suitability of both methods for single-patient classification was demonstrated using an independent cohort (47 patients).
Results:
Analysis of pooled, published data established three DNAme subgroups as a de facto standard. Unfavorable prognostic parameters (PTPN11 mutation, elevated HbF and older age) were significantly enriched in the high methylation (HM) subgroup. A classifier was then developed that predicted subgroups with 98% accuracy across different technological platforms. Applying the classifier to an independent validation cohort confirmed an association of HM with secondary mutations, high relapse incidence and inferior overall survival while the low methylation subgroup was associated with a favorable disease course. Multivariable analysis established DNAme subgroups as the only significant factor predicting overall survival.
Conclusions:
The present study provides an international consensus definition for DNAme subgroups in JMML. We developed and validated methods which will facilitate the design of risk-stratified clinical trials in JMML.
Introduction
Juvenile myelomonocytic leukemia (JMML) is a myeloproliferative/myelodysplastic neoplasm (1) with an incidence of 1.3 per million children and a median age at diagnosis of 2 years (2). Older age, elevated fetal hemoglobin (HbF) levels, and thrombocytopenia at diagnosis are established clinical risk factors correlating with poor clinical outcome (3).
More than 90% of JMML patients harbor canonical mutations in PTPN11, KRAS, NRAS, CBL or NF1 that constitutively activate RAS signaling. Somatic PTPN11 mutations are detected in approximately 35 – 40% of patients, somatic KRAS and somatic NRAS mutations are found in about 15% each, while alterations of the CBL or NF1 locus are observed in about 15% and 10% of patients, respectively (1,4). Karyotypic abnormalities can be observed in 35% of patients, with monosomy 7 as the most frequent event occurring in 25% of children with JMML (5). Whole exome- or targeted-sequencing studies identified recurrent secondary mutations in a number of JMML patients, commonly affecting JAK3, SETBP1, and SH2B3 (6–10). While older patients with PTPN11-, NF1- or NRAS-driven disease have a high relapse incidence following allogeneic hematopoietic stem cell transplantation (HSCT), spontaneous disease regression is observed in some younger patients with NRAS and CBL mutations (11,12). Interestingly, infants with Noonan syndrome caused by germline PTPN11 mutations can experience a myeloproliferative disorder that can be indistinguishable from JMML but generally has a self-limiting course.
In an attempt to resolve this striking clinical and biological heterogeneity, gene expression profiling identified JMML patients with an acute myeloid leukemia (AML)-like expression signature that was associated with poor survival (13). However, the precise mechanisms regulating disease specific gene expression patterns in JMML are still obscure. A link between aberrant RAS signaling and epigenetic remodeling has been suggested in the literature (14). Consistent with this, DNA hypermethylation of candidate gene promoters was described in JMML, including CDKN2B, RASSF1A, CREBBP, RASA4, the β-globin promoter and others (15–21). The DNA methylation status of four candidate genes (BMP4, CALCA, CDKN2B, and RARB) was integrated into a prognostic model, with DNA methylation emerging as the strongest independent prognostic predictor in a multivariable analysis (22). More recently, three independent studies described DNA methylation subgroups in JMML based on genome-wide DNA methylation analysis (23–25). Study groups from Europe and USA identified three JMML subgroups with distinct clinical features whereas a Japanese group suggested a binary classification into hyper- or hypomethylated disease. In all three studies, the hypermethylation subgroup was associated with PTPN11 mutations and poor overall survival. Together, these studies suggested DNA methylation as a biomarker which might help to improve patient stratification in JMML. However, standardized approaches to identify DNA methylation JMML subclasses are lacking.
In the present study, we sought to define an international consensus for DNA methylation subgroups in JMML and to systematically describe the biological and clinical features associated with each subgroup. Furthermore, we developed and validated classification methods which are suitable for clinical implementation and will allow risk stratification in clinical trials.
Materials and Methods
Collection of data and patient samples
Illumina Infinium Human Methylation 450k Bead Chip raw data from 292 patients (203 male; 89 female) with JMML or Noonan syndrome and myeloproliferative disease (NS/MPD) patients were collected from three recent publications and clinical annotations were obtained from the respective study groups (Supplementary File 1, Supplementary Figure 1A) (23–25). Healthy references were obtained from GEO (accession number GSE36054). To ensure reproducibility on different technology platforms, 32 patients from this cohort were re-analyzed using targeted bisulfite sequencing (MethylSeq) and 31 out of these 32 patients were re-analyzed by Infinium Human MethylationEPIC Bead Chip (EPIC) arrays (“technical validation cohort”, Supplementary File 1). An independent validation cohort of 47 patients from the participating study centers was collected for classifier validation. Written informed consent from parents or legal guardians of all patients was obtained according to the Declaration of Helsinki. Patients’ material storage and collection was approved by institutional ethics committees.
Processing of DNA methylation array data
DNA methylation data was analyzed using the “RnBeads” Bioconductor package (26). Background correction (“methylumi.noob”)(27) and beta-mixture quantile normalization (BMIQ) were applied. Unreliable probes (Greedycut algorithm with detection p-value <0.01), cross-reactive probes and probes mapping to sex chromosomes were removed (Supplementary Figure 1A). Samples with outlier intensities in 450k/EPIC array control probes were removed from the dataset as described in the RnBeads vignette (26)(Supplementary Methods Figure 1–3). Hierarchical clustering based on SNP distances revealed study group-specific clusters that likely reflected ethnic differences (Supplementary Figure 1B). In line with this, initial exploratory analysis using principal component analysis (PCA) separated EWOG-MDS from USA and Japanese patients in the first principal component (PC; Supplementary Figure 1C). We therefore applied 1) SNPfiltering using dbSNP version 150 and 2) Combat batch correction using the “sva” R package (Supplementary Figure 1D) (28). The final RnBeads dataset was generated from the binary exponential of the batch corrected value. Finally, all CpG dinucleotides (CpGs) with variable DNA methylation across normal hematopoietic differentiation were excluded to account for potential differences in cell type composition of the samples (24). The pre-processing of EPIC array data was analogous to 450k data as described above. For the technical validation cohort, overlapping probes between 450k and EPIC arrays were determined. Patient-wise Pearson correlation was calculated across all probes and plotted as a correlation heatmap using the pheatmap R package (29).
Consensus clustering of DNA methylation array data
The 5000 most variable CpGs (mvCpGs), defined as the probes with the highest standard deviation between all samples, were identified and used for hierarchical consensus clustering using the ConsensusClusterPlus R package (30) with Ward’s linkage (ward.D2), Manhattan distance and 500 bootstrap iterations. Within each DNA methylation consensus cluster, patients were ordered according to similarity by hierarchical clustering. Results were plotted using the pheatmap R package (29).
Copy number variation analysis
Copy number variation was inferred from 450k or EPIC array data using the Conumee Bioconductor R package (31). Focal amplifications and deletions were determined by “Genomic Identification of Significant Targets in Cancer” (GISTIC2) (32). Data from healthy adult blood (EGAS00001002511; 450k array) and cord blood (GSE103189; EPIC array) were used as reference.
Extreme gradient boosting model
The “XGBoost” R package was used for a tree-based gradient boosting model (33). The patient cohort was split into a training (n = 229) and a testing cohort (n = 55), with random splits for each DNA methylation class to keep the class distribution balanced. CpGs which overlapped between 450k and EPIC methylation arrays were determined, and CpG sites with differentiation-dependent variation were removed. A multiclass classification model was trained for the prediction of DNA methylation subgroups using the 5000 mvCpGs (softprob objective and 100 iterations).
Hyper-parameter tuning was performed using a grid-search algorithm with 5-fold cross-validation (nrounds = 65, eta = 0.05, max_depth = 4, gamma = 1, colsample_bytree = 0.4, min_child_weight = 3, subsample = 0.75). The model accuracy was determined by predicting DNA methylation classes in the testing cohort. The feature importance for the tuned XGBoost model was calculated using the caret-wrapper function to determine the relative feature importance of the model CpGs (34). Out of 5000 model CpGs, 124 showed a feature importance of greater than zero (Supplementary File 2). A refined JMML DNA methylation classifier was trained based on these 124 CpGs and the hyper-parameters from the grid-search algorithm. The model accuracy was tested by comparing the DNA methylation class predictions of the testing cohort with the meta-analysis DNA methylation group assignments. Furthermore, 31/32 patients from the technical validation cohort were re-analyzed on EPIC methylation arrays. The model prediction accuracy was then tested with this cohort to validate the model accuracy across different technology platforms. The robustness of the refined 124-CpG classifier was tested by repeated re-training using only a subset of CpG sites. This approach enabled us to determine the model accuracy for DNA methylation class prediction of the training and testing cohorts for models based on 5 to 120 CpGs (each subset randomly selected 100x).
DNA panel sequencing
DNA samples (32 from the technical cohort and 47 from the independent validation cohort) were sequenced using a custom amplicon panel (Paragon Genomics, Hayward, CA, USA), targeting 26 genes that are recurrently mutated in JMML (Supplementary Methods Table 1). Ten nanograms of genomic DNA were used for each library and were processed according to the manufacturer’s protocol.
Library quality was assessed using the Bioanalyzer High Sensitivity DNA Analysis kit (Agilent, Santa Clara, CA, USA) and quantified using the Qubit HS assay (Thermo Fisher Scientific, Waltham, MA, USA). Dual-index libraries were sequenced on an Illumina MiSeq instrument using the 150 bp PE mode. Mean on-target sequencing depth was 1519x (median = 1297x). A minimum mutant allele fraction (MAF) of 0.03 was required for reporting. Patient samples where the MAF of the putative driver mutation was <0.25 were removed from the analysis to ensure only clonal samples were being analyzed.
Human MethylationEPIC Bead Chip sample processing
Genomic DNA (100–250ng) was subjected to the Genomics and Proteomics Core Facility at the German Cancer Research Center to perform genome-wide DNA methylation analysis using the Infinium Human MethylationEPIC Bead Chip platform (Illumina, San Diego, CA, USA).
Targeted MethylSeq library preparation and sequencing
3000 CpG loci were submitted for custom-capture assay design, of which 2992 CpGs were among the top ranked 5000 CpGs discriminating the JMML DNA methylation subgroups identified in the meta-analysis. Genomic DNA (300ng) was bisulfite converted using the TrueMethyl oxBS Module according to protocol (Tecan, Switzerland). Converted ssDNA was quantified using the Qubit ssDNA assay (Thermo Fisher Scientific, Waltham, MA, USA) and 100ng used as input for the custom-made Targeted MethylSeq assay (Tecan, Switzerland). Final libraries were quality checked using the Bioanalyzer High Sensitivity DNA Analysis kit (Agilent, Santa Clara, CA, USA) and quantified using the Qubit HS assay (Thermo Fisher Scientific, Waltham, MA, USA) before pooling for next generation sequencing. Single-index library pools were sequenced on the Illumina HiSeq 4000 with paired-end 150bp mode.
Hierarchical clustering-based sample classification using MethylSeq data
Sequence reads were trimmed using Cutadapt (35) to remove adapters, and CpG methylation status was called using Bismark (36). Only those 2992 CpG sites were considered for analysis and required to have a median 50x read coverage, resulting in removal of 150 CpG sites. From the remaining 2842 CpG sites, 1386 CpGs showed standard deviations greater than 0.25 across the 32 samples, which was the final data set used to analyze the technical validation cohort (Supplementary File 3). The beta-values of the 1386 CpG sites of the 32 patients from the technical validation cohort were hierarchically clustered in both directions (samples and probes) utilizing an unsupervised approach using the hclust function and Ward’s method. The samples were classified into one of three classes based on minimum distance to the centroid. The MethylSeq data from the independent validation cohort data were filtered accordingly and hierarchical clustering was performed using the list of 1386 CpG sites determined based on the analysis of the technical validation cohort. Methylation subgroup classification into one of three classes was based on the minimum distance to the centroid.
Statistical analysis
Clinical characteristics and mutation status were collected for all patients from the participating study groups. Pearson’s chi-square test was used to test categorical variables for independence. One-way analysis of variance (ANOVA) was utilized to test for differences among continuous variables. Fisher’s exact test was used to test for enrichment of canonical JMML driver mutations for each DNA methylation subgroup. The primary endpoint overall survival (OS) was defined as the time between diagnosis and death or last follow-up. The Kaplan-Meier method was used to estimate survival rates and the two-sided log-rank test was employed to evaluate the equality of the survivorship functions in different subgroups. For univariable and multivariable analysis, the relationships between overall survival and the variables PTPN11 (mutation or not), gender, age (≥1 year or not), mutations (>1 mutation or not), HbF (normal vs elevated), platelet count (≥50 or not), XGBoost subgroup (LM, IM or HM), and MethylSeq subgroup (LM, IM or HM) were examined. First, we tested all variables individually and then included significant variables in a multivariable model. P-values ≤ 0.01 were considered statistically significant, except in the multivariable analysis, where a cutoff of 0.05 was used.
Data Sharing Statement
The data used in this study are available for download from the European Genome-phenome archive (https://ega-archive.org/) under the accession numbers EGAS00001004682. All other relevant data are available from the authors upon request.
Results
Patients’ characteristics of the meta-analysis cohort
DNA methylome data of 292 patients with JMML (n = 263) and Noonan syndrome-associated myeloproliferative disorder (NS/MPD; n = 29) were combined from three previously published studies for an integrative analysis (23–25). Data of 284 patients (143 EWOG-MDS; 104 Japan; 37 USA) passed our quality control criteria (Supplementary Figure 1A). Of these, 255 patients (90%) were diagnosed with JMML and 29 (10%) with NS/MPD (Supplementary Table 1). NS/MPD patients were significantly younger than JMML patients with a median age at diagnosis of 0.1 years (range: 0.0 – 0.3, p = 8.2 × 10–7); all were diagnosed before the age of 2 years (Supplementary Table 1).
For JMML patients (pts), the tissue source for the DNA methylome analysis was either bone marrow (BM; 69%, n = 175 pts) or peripheral blood (PB; 31%, n = 80 pts; Supplementary Table 2). Median age at JMML diagnosis was 1.4 years (40% ≥ 2 years). 69% of patients were male and 31% were female. HbF levels at diagnosis were available for 210 (82%) patients and elevated for age in 145/210 (69%) patients (37). Median leukocyte and platelet counts at diagnosis were 29.9 x 109 /L [range: 2.9 – 563.0] and 65.5 × 109 /L [range: 5.0 – 730.0], respectively. Canonical JMML driver mutations were detected in 90% of JMML patients (38% PTPN11; 16% NRAS; 14% KRAS; 14% CBL; 9% NF1; 8% quintuple-negative; 1% NA). Monosomy 7 was detected in 14% and was the only parameter tested that showed differential distribution between the study groups (p = 2.4 × 10–3): monosomy 7 was present in 22% of EWOG-MDS patients, but was only found in 9% and 3% of the Japanese and USA patients, respectively (Supplementary Table 2).
Consensus clustering confirms three DNA methylation subgroups in JMML
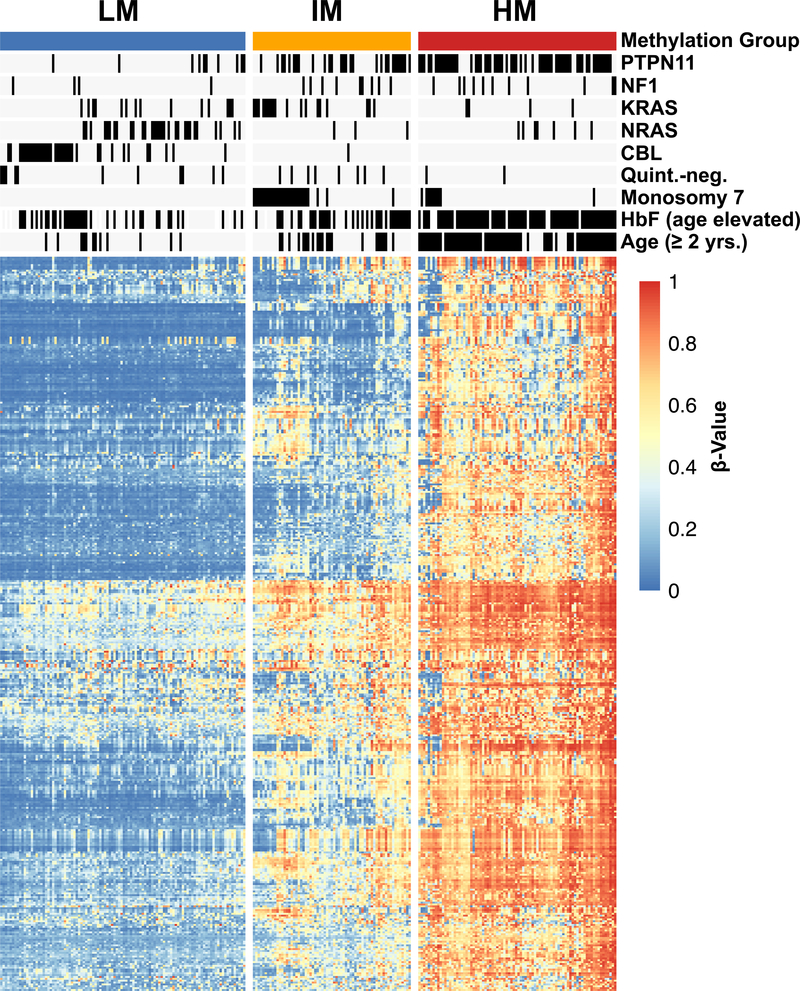
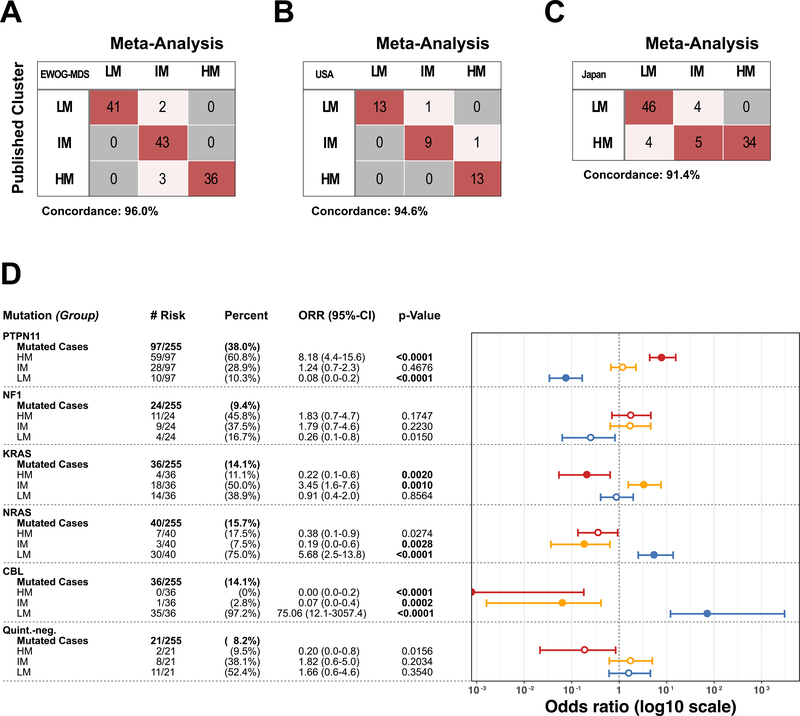
Unsupervised hierarchical consensus clustering using the 5000 most variable CpG sites identified three distinct DNA methylation subgroups, which were designated as low DNA methylation (LM), intermediate DNA methylation (IM) and high DNA methylation (HM) according to mean beta-values (Figure 1; Supplementary Figure 2A–J). When comparing our consensus approach to the assignments in the original study groups’ publications, we identified the same methylation subgroup assignment for 96% of EWOG-MDS (120/125), 95% of US (35/37) and 91% of Japanese (85/93) JMML patients (Figure 2A–C)(23–25). A global concordance of 96% was calculated when combining IM and HM subgroup as hypermethylated JMML, which allowed a comparison with the two methylation subgroups identified in the Japanese study (Supplementary Table 3)(25). All NS/MPD patients clustered in the LM subgroup and formed a sub-cluster that almost exclusively consisted of NS/MPD patients. Of note, the DNA methylomes of NS/MPD and LM patients were almost indistinguishable from those of healthy controls (Supplementary Figure 3). There was no significant association between sample source (bone marrow or peripheral blood) and DNA methylation subgroups (p = 0.9012, Chi-squared test; Supplementary Table 3 & Supplementary Figure 4).
Figure 1 |. DNA methylation patterns identify three biologically distinct JMML subgroups.
The heatmap displays three DNA methylation subgroups among patients with JMML (n=255; NS/MPD pts excluded). The patients (columns) were clustered by unsupervised consensus clustering (k=3) using the 5000 most variable CpGs. Known clinical and biological features are annotated for each patient. The heatmap shows beta-values of 395 representative CpG sites (rows).
(LM, low methylation; IM, intermediate methylation; HM, high methylation; HbF, fetal hemoglobin)
Figure 2 |. Enrichment of JMML driver mutations in DNA methylation subgroups.
(A-C) Confusion table comparing the published methylation subgroups to the meta-analysis consensus clustering. EWOG-MDS patients (A) and US patients (B) were classified in three methylation subgroups in the original publication whereas the Japanese study (C) separated patients in two methylation subgroups. (D) Absolute and relative numbers of patients with a specific JMML driver mutation and their distribution across the DNA methylation subgroups are depicted. Odd ratios, confidence intervals and p-values were calculated using Fisher’s exact test.
Consensus DNA methylation subgroups correlate with disease biology
The association of consensus DNA methylation subgroups with the canonical JMML subgroups and clinical hematologic parameters was investigated for all JMML patients (Supplementary Table 3). Patients with somatic PTPN11 mutations were predominantly assigned to the HM subgroup (61%) and comprised more than 70% of HM patients (p = 3.2 × 10–13; Figure 2D, Supplementary Table 3). All HM patients had elevated HbF (100%) and were older at the time of diagnosis (median age: HM, 3.0 years; IM, 1.4 years; LM, 0.7 years; p < 2 × 10–16, AOV-test; Supplementary Table 3). The IM subgroup was mainly characterized by the presence of PTPN11 or KRAS mutations (42% and 27% of all IM patients). Of all monosomy 7 cases, 75% (27/36 pts) were found to cluster with the IM subgroup, whereas the remaining 25% of monosomy 7 cases clustered with the HM subgroup (p = 1.5 × 10–7, Chi-squared test). In contrast, almost all patients with CBL mutations (35/36 pts, 97%; p = 4.3 × 10–15 Chi-squared test) and the vast majority of patients with NRAS mutations (30/40 pts, 75%; p = 1.2 × 10–7, Chi-squared test) clustered in the LM subgroup (Figure 1 & 2D, Supplementary Table 3). The quintuple-negative JMML patients comprised a small proportion of all JMML patients (21/255 pts., 8%) and almost exclusively clustered with the LM (52%) or with the IM subgroup (38%; Figure 2D & Supplementary Table 3).
Genotype-specific enrichment analyses identified further associations between JMML driver mutations, clinical characteristics and DNA methylation subgroups. For example, patients who had a PTPN11 mutation and additional clinical high-risk factors (i.e. elevated HbF, thrombocytopenia, higher age) were more likely assigned to the HM subgroup, whereas patients with CBL mutations were most commonly assigned to the LM subgroup, independent of the presence or absence of additional clinical risk factors (Supplementary Figure 5A). In this retrospective analysis, all NF1 patients who presented with thrombocytopenia were assigned to the HM subgroup and none of the NF1 patients older than 2 years at the time of JMML diagnosis clustered with the LM subgroup (Supplementary Figure 5A). Monosomy 7 co-occurred with PTPN11 (16/36; 44%), KRAS (14/36; 39%) or NF1 (2/36; 6%) mutations but not with NRAS or CBL mutations. Patients with co-occurrence of monosomy 7 and a KRAS mutation were always assigned to the IM subgroup, while in the presence of PTPN11 or NF1 mutations patients with monosomy 7 were assigned either to the IM or HM subgroups. Of note, monosomy 7 was absent in the LM subgroup (Supplementary Table 3). In the group of KRAS-mutant patients, a skewed distribution of molecular features became evident across patients from different study groups: while 9/14 (64%) Japanese patients with KRAS mutations were assigned to the LM subgroup (p = 8.0 × 10–2, Fisher’s exact test), 13/19 (68%) EWOG-MDS patients with KRAS mutations were categorized into the IM subgroup (p = 5.5 × 10–3, Fisher’s exact test). This difference was paralleled by a more frequent co-occurrence of KRAS mutations with monosomy 7 in EWOG-MDS compared to Japanese patients (EWOG-MDS n = 12; Japan n = 2; p = 6.0 x 10–3, Fisher’s exact test; Supplementary Figure 5B).
Consensus DNA methylation subgroups predict clinical outcome and identify patients at high risk
We next analyzed the impact of the consensus DNA methylation subgroups on clinical outcome. Due to differences in treatment regimens and follow-up periods, clinical outcome was analyzed separately for each study group. Kaplan-Meier curves for overall-survival demonstrated that HM JMML had inferior 5 year overall survival in all study groups (EWOG-MDS: 62% [95% CI: 45 – 79]; Japan: 46% [95% CI: 29 – 62]; USA: 42% [95% CI: 22 – 79]), especially when compared to the LM subgroup (5-yr OS EWOG-MDS: 87% [95% CI: 76 – 98]; Japan: 74% [95% CI: 59 – 85]; USA: 100%; Supplementary Figure 6A,B&C). Long-term survival without hematopoietic stem cell transplantation (5-yr TFS) was only observed in patients assigned to the LM subgroup (EWOG-MDS: 24% [95% CI: 10 – 38]; Japan: 39% [95% CI: 24 – 53]; USA: 54% [95% CI: 33 – 89]; Supplementary Figure 6D,E&F).
Development and validation of a machine learning model for prospective JMML DNA methylation subgroup classification in the single-patient setting
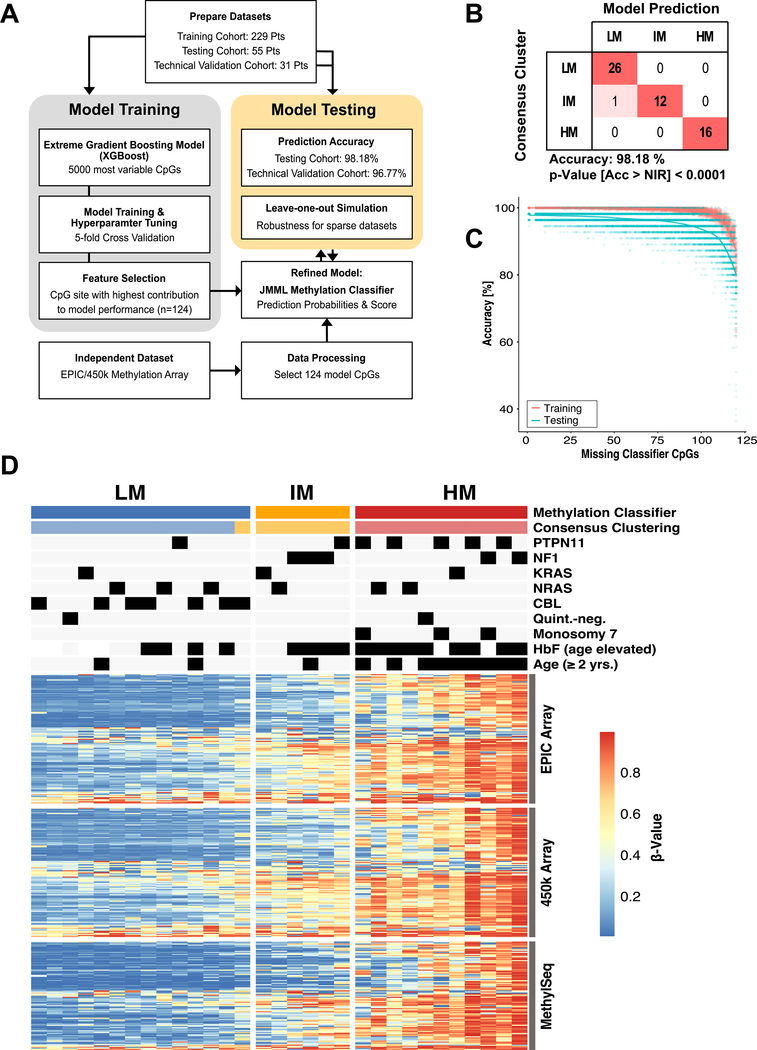
To prospectively classify single JMML patients into consensus DNA methylation subgroups as defined by this meta-analysis, we developed and validated a machine learning model. The combined JMML patient cohort was split into a training (n = 229) and a testing cohort (n = 55). A multiclass classification gradient boosting tree model (XGBoost) was developed based on the 5000 mvCpGs (Figure 3A, Supplementary Figure 1). To determine the model with the highest prediction accuracy, hyperparameter tuning was performed using a 5-fold cross-validation approach with subsequent feature selection based on variable importance. The refined JMML DNA methylation classifier was trained based on 124 CpG sites and validated by classifying the testing cohort (Figure 3B; Supplementary File 2). The model predicted consensus DNA methylation subgroups in the testing cohort with an accuracy of 98% [95%CI: 90.3%−99.9%]; that is, only one out of 55 patients was classified differently than by consensus clustering (Figure 3B).
Figure 3 |. Development of a DNA methylation classifier for JMML.
(A) The meta-analysis cohort was randomly split into a training (n = 229) and a testing cohort (n=55). A machine learning classifier was built based on the 5000 mvCpGs, which were covered by 450k and EPIC methylation arrays and trained on the training cohort. The fractional contribution of each CpG to the overall model performance was calculated and the CpGs with the highest gain were selected (n=124). This refined JMML methylation classifier was trained and the model performance evaluated on the testing cohort. Additionally, 31 patients were re-analyzed on EPIC arrays to test the model accuracy across different technical assays (“technical validation cohort”). (B) Confusion table for prediction of the testing cohort using the refined JMML methylation classifier (Acc, accuracy; NIR, no-information rate). (C) The JMML methylation classifier was re-trained with subsets of the 124 model CpGs, by repeatedly leaving out between 5 and 120 CpG sites. The model accuracy of these sparse models for predicting the training or testing cohort was determined. (D) Heatmap showing the DNA methylation beta-values for the 124 model CpG sites (rows) on 450k and EPIC methylation arrays for patients of the technical validation cohort (n=31; columns). Additionally, 104/124 CpGs assessed by the MethylSeq assay are shown. The predictions by the methylation classifier model are compared to the consensus clustering.
To test the power of the model for sparse datasets and the stability of the underlying JMML DNA methylation signature, a “leave-one-out” scenario was simulated (Figure 3C). Repeated model training and accuracy assessments using increasing numbers of CpG sites (between 4 and 119 out of the 124 model CpG sites) based on the training and testing cohorts was used for this simulation. We found that training the model with as few as 24/124 CpG sites still resulted in a prediction accuracy of about 85%. We confirmed the model performance across different technological platforms by re-analyzing 1/292 patients from the combined cohort (referred to as “technical validation cohort”) on the EPIC array platform (one sample did not meet quality control cutoffs). The genome wide DNA methylation measurements were highly correlated between the 450k and the EPIC array platforms (Supplementary Figure 7A). Accordingly, the classifier model predicted the consensus DNA methylation classes from EPIC array data with an accuracy of 97% in the technical validation cohort (Supplementary Figure 7B).
Targeted MethylSeq recapitulates consensus DNA methylation subgroup assignments
We also developed a targeted DNA methylation assay based on bisulfite sequencing (targeted MethylSeq) that could be incorporated into routine clinical workflows used for mutation testing. This assay encompassed 3000 CpG sites, 2992 of which overlapped with the 5000 most-variable CpGs used for consensus clustering in the present study (Supplementary File 3). The 32 patients of the technical validation cohort (for MethylSeq one additional patient was available) were analyzed using this assay (Supplementary File 4). After quality control filtering, 1386 CpGs with a standard deviation of greater than 0.25 between the samples were used for hierarchical clustering. Again, this resulted in three distinct DNA methylation subgroups that corresponded to the consensus assignments for all 32 samples analyzed (Supplementary Figure 7C). Furthermore, 104 CpG sites of the 3000 probes overlapped with the 124 CpG sites from the JMML methylation classifier, and their DNA methylation patterns were similar to the EPIC array data and the 450k array combined cohort analysis (Figure 3D).
Validation of the JMML DNA methylation classifier in an independent patient cohort
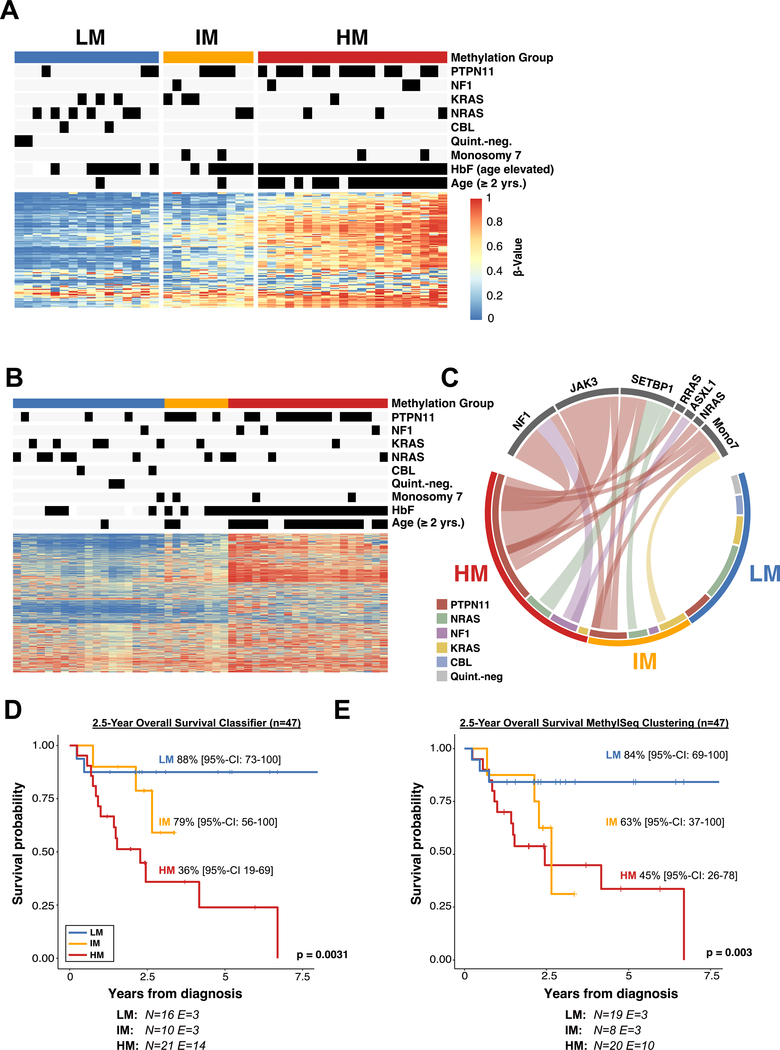
An independent patient cohort consisting of 47 patients (9 EWOG-MDS, 18 Japan and 20 USA) without prior DNA methylation analysis was collected in order to validate the DNA methylation classification methods. All patients from this independent validation cohort were tested for known JMML mutations using targeted deep-sequencing. The DNA methylation analysis was performed using both the EPIC array and the targeted MethylSeq platforms. In this cohort, 46 patients were diagnosed with JMML and 1 patient was diagnosed with NS/MPD. The median age at diagnosis was 1.7 years [range: 0.0 – 6.4]. HbF was elevated for age in 77.8% of patients. Median leukocyte and platelet counts were 30.2 x 109 /L [range: 5.0 – 166.4] and 67 × 109 /L [range: 11.0– 490.0], respectively (Supplementary Table 4). The median overall survival was 6.7 years. Canonical JMML driver mutations were detected in 96% of JMML patients (44% PTPN11, 15% KRAS, 24% NRAS, 4% CBL, and 9% NF1). Accordingly, 4% of patients were annotated as “quintuple-negative”. DNA methylation subgroups were determined by the 124 CpG XGBoost classifier in a single-patient setting (Figure 4A, Supplementary Table 4) and by minimum distance to the centroid based on the 1386 CpGs sites that were previously used to cluster the technical validation cohort (Figure 4B; Supplementary File 2&3). Based on the 124 CpG XGBoost classifier, 21 patients were classified as HM, 10 as IM, and 16 as LM JMML (Figure 4A). PTPN11 mutations, higher age, and elevated HbF levels were significantly enriched in HM JMML patients (Supplementary Table 4). When classifying the independent patient cohort based on the MethylSeq data, 20 patients were classified as HM, 8 as IM and 19 as LM. Importantly, the concordance of HM prediction was 95% (Figure 4B, Supplementary Figure 8A). Of note, variant-allele frequency measurements of canonical JMML driver mutations suggested that DNA methylation subgroups are not confounded by differences in tumor cell purity (Supplementary Figure 8B&C). Previous studies have suggested an enrichment of secondary mutations predominately in HM JMML (23,24). In the validation cohort 71% of HM patients and 50% of IM patients but none of the LM patients showed recurrent secondary mutations (Figure 4C). Interestingly, in JMML patients with PTPN11 mutations, most secondary mutations were detected in JAK3 and NF1 (79%; 11/14 patients with secondary mutation) whereas the most common secondary mutation in NRAS-mutant JMML patients was SETBP1 (100%; 3/3 patients with secondary mutation). In summary, we found a strong enrichment of secondary genetic hits in the HM subgroup.
Figure 4 |. Validation of the DNA methylation classifier in an independent patient cohort.
(A) Heatmap showing the DNA methylation beta-values of the 124 model CpG sites for an independent validation cohort (n = 47). Methylation subgroups were assigned by classifier predictions. (B) Determination of the DNA methylation subgroups using targeted amplicon-bisulfite sequencing (MethylSeq). Patients were clustered using hierarchical clustering with Ward’s method. (C) Circos plot displaying the association between known driver mutations and secondary mutations in the three DNA methylation subgroups as determined by the 124 CpG machine learning classifier. (D+E) Kaplan-Meier curves showing the overall survival of JMML patients stratified by DNA methylation subgroups. DNA methylation subgroups were assigned based on the JMML methylation classifier (D) or MethylSeq clustering (E). Probabilities and confidence intervals are indicated for each DNA methylation subgroup. The number of individuals at risk (N) and the number of events (E) is depicted at the bottom. Statistical significance was calculated using log-rank test.
The clinical relevance of the 124 CpG XGBoost DNA methylation classifier was highlighted by significant differences in prognosis (log-rank test: p-value = 0.003). Overall survival (OS) at 2.5 years was significantly lower for HM JMML patients as compared to IM and LM JMML patients (2.5-year OS HM: 36% [95% CI: 19 – 69], IM: 79% [95% CI: 56 – 100], LM: 88% [95% CI: 73 – 100]; Figure 4D). Similarly, DNA methylation subgroups identified by the targeted MethylSeq assay were also associated with significant differences in prognosis (log-rank test: p-value = 0.003; 2.5-year OS HM: 45% [95% CI: 26 – 78], IM: 63% [95% CI: 37 – 100], LM: 84% [95% CI: 69 – 100]; Figure 4E).
Methylation status is independently predictive of outcome.
In univariable analyses (Table 1), the characteristics that reached significance at the 0.05 level for OS, were age at diagnosis of >12 months (HR = 2.75, 95% CI = 1.08–7.01, p = 0.03), platelets of ≤50 (HR = 0.35, CI = 0.14–0.89, p = 0.026), and DNA methylation group (p = 0.0026 for XGBoost and 0.0015 for MethylSeq). When a multi-variable model was applied using age, platelets and DNA methylation group, the DNA methylation grouping retained statistical significance for OS (p = 0.046 for XGBoost and 0.039 for MethylSeq; Table 1) whereas neither age (p = 0.51 for XGBoost and 0.82 for MethylSeq) nor platelets (p = 0.13 for XGBoost and 0.16 for MethylSeq) were significant.
Table 1 |. Univariable and multivariable analysis for overall survival in the biological validation cohort.
An univariable analysis including age at diagnosis, platelet count at diagnosis, XGBoost Cluster, MethylSeq Cluster, somatic PTPN11 mutations, sex, number of somatic mutations and HbF status was conducted for predictors of overall survival (OS). P-values lower than 0.05 were considered as statistically significant. A multivariable analysis including the significant features was calculated thereafter including either the XGBoost or MethylSeq methylation cluster assignments. (N = Number of pts; HR = Hazard ratio; CI = Confidence interval).
| Univariable Analysis | OS from date of diagnosis | |||
|---|---|---|---|---|
| N | HR | 95% CI | p | |
| Age at diagnosis (months) | ||||
| ≤12 months | 27 | 1 | ||
| >12 months | 20 | 2.75 | 1.08–7.01 | 0.03 |
| Platelet count at diagnosis ×109 | ||||
| ≤50 | 17 | 1 | ||
| >50 | 30 | 0.35 | 0.14–0.89 | 0.026 |
| XGBoost Cluster | ||||
| Low | 16 | 1 | ||
| Intermediate | 10 | 2.96 | 0.48–18.22 | 0.0026 |
| High | 21 | 8.07 | 1.81–36.01 | |
| MethylSeq Cluster | ||||
| Low | 19 | 1 | ||
| Intermediate | 8 | 6.76 | 1.20–38.01 | 0.0015 |
| High | 20 | 8.93 | 1.99–40.0 | |
| Somatic PTPN11 mutation | ||||
| No | 26 | 1 | ||
| Yes | 21 | 1.79 | 0.72–44.45 | 0.21 |
| Sex | ||||
| Male | 34 | 1 | ||
| Female | 13 | 0.84 | 0.28–2.57 | 0.76 |
| Somatic Mutations at Diagnosis | ||||
| <=1 | 29 | 1 | ||
| >1 | 18 | 2.33 | 0.93–5.79 | 0.068 |
| HbF at diagnosis | ||||
| Not elevated for age | 10 | 1 | ||
| Elevated for age | 35 | 1.36 | 0.39–4.73 | 0.61 |
| Multivariable Analysis (XGBoost) | ||||
| Age at diagnosis (months) | ||||
| ≤12 months | 27 | 1 | ||
| >12 months | 20 | 0.61 | 0.15–2.47 | 0.51 |
| Platelet count at diagnosis ×109 | ||||
| ≤50 | 17 | 1 | ||
| >50 | 30 | 0.49 | 0.19–1.25 | 0.13 |
| XGBoost Cluster | ||||
| Low | 16 | 1 | ||
| Intermediate | 10 | 3.09 | 0.49–19.65 | 0.046 |
| High | 21 | 10.82 | 1.56–74.84 | |
| Multivariable Analysis (MethylSeq) | ||||
| Age at diagnosis (months) | ||||
| ≤12 months | 27 | 1 | ||
| >12 months | 20 | 0.85 | 0.20–3.53 | 0.82 |
| Platelet count at diagnosis ×109 | ||||
| ≤50 | 17 | 1 | ||
| >50 | 30 | 0.52 | 0.20–1.31 | 0.16 |
| MethylSeq Cluster | ||||
| Low | 19 | 1 | ||
| Intermediate | 8 | 6.17 | 1.06–35.94 | 0.039 |
| High | 20 | 8.92 | 1.25–63.48 | |
DISCUSSION
The diagnosis of JMML has so far been based on clinical and genetic criteria. The unifying feature is pathologic activation of the RAS signaling pathway but the clinical course of JMML is highly heterogeneous. Patients with identical mutations may either experience spontaneous remission of the disease or relapse after allogeneic hematopoietic stem cell transplantation, indicating that genotype does not always dictate phenotype in this disease (38,39). Outcomes can be partly predicted using a combination of clinical and genetic parameters (3,6). However, well-established cut-offs for those parameters or even prognostic scores are missing. In addition, parameters such as fetal hemoglobin and thrombocytopenia may not be informative in cases where supportive treatment has been initiated before referral to a specialist center. Hence, robust, objective and reproducible molecular assays are needed to advance the diagnosis, risk-stratification and treatment of JMML. The potential of DNA methylation to sub-classify tumors such as glioma, acute myeloid leukemia, acute lymphoblastic leukemia, chronic lymphocytic leukemia (40,41) and Waldenström macroglobulinemia (42) has been demonstrated in several publications (43–46). In recent years, a number of publications have highlighted the correlation of DNA methylation with disease biology and prognosis in JMML (18,19,22–25).
Combining published data from three JMML study groups, we analyzed the genetic and DNA methylation landscape in the largest JMML patient cohort studied to date.
The present meta-analysis sheds some light on controversial topics that have been discussed in the JMML research community. It confirmed the presence of at least three JMML DNA methylation subgroups with an obvious underrepresentation of Japanese patients in the IM subgroup. This might explain why a previous Japanese study identified two, instead of three, JMML DNA methylation subgroups (25). Similarly, we found that most KRAS-mutant EWOG-MDS patients clustered in the IM subgroup, while most KRAS-mutant Japanese patients clustered in the LM subgroup. Biological differences between the regional patient groups were further supported by the observation that monosomy 7 is rare in patients from Japan as compared to EWOG-MDS (monosomy 7 Japan: 9%; monosomy 7 EWOG-MDS: 22%). This difference cannot be explained by technical differences as the copy number variation status was uniformly inferred from the 450k or EPIC array data for all patients. In conclusion, we observed differences in the genetic and epigenetic landscape of KRAS patients between Europe (USA) and Japan which might, at least in part, explain previously discussed differences in the clinical course of these patients (12,47).
Our re-analysis established three DNA methylation subgroups as a de facto standard, which will allow the uniform classification of JMML into biologically and clinically meaningful risk groups. Furthermore, we developed and validated tools that allow for systematic and reproducible classification of JMML in the clinical setting and demonstrated that DNA methylation classification in JMML is stable across orthogonal technology platforms. Analysis of an independent patient cohort validated these classification tools. Using multivariable analysis, we confirmed the prognostic relevance of DNA methylation subgroups and provide further evidence suggesting that DNA methylation subgroup is the strongest independent prognostic factor in JMML.
The present meta-analysis provides a molecular rationale to define high-risk patients for whom allogeneic hematopoietic stem cell transplantation is not curative and who are therefore candidates for clinical trials testing innovative treatment options. On the other hand, there likely exists a subset of patients, characterized by an LM phenotype, for whom a watch-and-wait strategy plus supportive care might be the appropriate intervention.
In summary, the present meta-analysis of 255 JMML patients provides a consensus definition for DNA methylation subgroups in JMML. We have developed a DNA methylation classifier and validated its performance in an independent patient cohort. This classifier allows the prospective identification of DNA methylation subgroups for newly diagnosed patients based on the consensus definition described here. This work will support patient stratification and development of risk-adapted treatment strategies in the context of clinical trials and will improve the comparison of results obtained with different treatment strategies across study groups.
Supplementary Material
Statement of translational relevance.
Known clinical and genetic markers have limitations in predicting disease course and treatment outcome in juvenile myelomonocytic leukemia (JMML). Aberrant DNA methylation patterns correlate with clinical outcome in JMML across multiple studies, suggesting it as a biomarker to improve patient stratification. However, standardized approaches to identify DNA methylation subclasses are lacking. In the present study, we defined an international consensus for DNA methylation subgroups in JMML, and developed and validated classification methods that are suitable in the single-patient setting. Our DNA methylation classifier is ready for clinical implementation and will enable the design of risk-stratified clinical trials in JMML.
Acknowledgements
We first want to thank the patients and their families for participating in research studies that allowed for this work to be accomplished. We thank the Microarray unit of the Genomics and Proteomics Core Facility, German Cancer Research Center (DKFZ), for providing the Illumina Infinium MethylationEPIC arrays and related services. We also want to thank all members of the Division of Cancer Epigenomics (DKFZ) and the Division of Translational Medical Oncology (DKFZ & NCT) as well as Dr. Manuel Wiesenfarth (Division of Biostatistics, DKFZ) for helpful discussions related to this study. We want to thank Oliver Mücke for his excellent technical support and Jahan Parsa of Tecan for his assistance with pipeline development. We also acknowledge the Hilda Biobank at the University of Freiburg for specimen processing. This work was supported by the German José Carreras Leukemia Foundation (DJCLS) grant DJCLS R 15/01 (C.P., D.B.L. and C.F.); the German Research Foundation (DFG) grant CRC992-C05 (C.F.); the German Federal Ministry of Education and Research (BMBF) grant “MyPred” (C.F. and C.N.); the National Institutes of Health, National Cancer Institute grant 1U54CA196519 (M.L.L., E.S.); National Institutes of Health, National Heart, Lung, and Blood Institute grant K08HL135434 (E.S.); the Pediatric Cancer Research Foundation (E.S.); the V Foundation (E.S.); the UCSF Catalyst Program (E.S.); the California Cancer League (E.S); Dueling for Lincoln (E.S. and M.L.L); Lemonade 4 Leukemia (E.S. and M.L.L); the Frank A. Campini Foundation (E.S. and M.L.L.); the Leukemia and Lymphoma Society grant R6511-19 (M.L.L.); the Center for Advanced Technologies at UCSF and the National Cancer Institute Cancer Center Support grant 5P30CA082103.
Financial Support:
This work was supported by the German José Carreras Leukemia Foundation (DJCLS) grant DJCLS R 15/01 (C.P., D.B.L. and C.F.); the German Research Foundation (DFG) grant CRC992-C05 (C.F.); the German Federal Ministry of Education and Research (BMBF) grant “MyPred” (C.F. and C.N.); the National Institutes of Health, National Cancer Institute grant 1U54CA196519 (M.L.L., E.S.); National Institutes of Health, National Heart, Lung, and Blood Institute grant K08HL135434 (E.S.); the Pediatric Cancer Research Foundation (E.S.); the V Foundation (E.S.); the UCSF Catalyst Program (E.S.); the California Cancer League (E.S); Dueling for Lincoln (E.S. and M.L.L); Lemonade 4 Leukemia (E.S. and M.L.L); the Frank A. Campini Foundation (E.S. and M.L.L.); the Leukemia and Lymphoma Society grant R6511–19 (M.L.L.); the Center for Advanced Technologies at UCSF and the National Cancer Institute Cancer Center Support grant 5P30CA082103.
Footnotes
Conflict of interest statement:
C.M.N. received honoraria from BMS & Novartis. All other authors declare no competing interests.
REFERENCES
- 1.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- 2.Ries LAG, Smith MA, Gurney JG, Linet M, Tamra T, Young JL, et al. Cancer incidence and survival among children and adolescents: United States SEER Program 1975–1995. NCI, SEER Progr. 1999. [Google Scholar]
- 3.Locatelli F, Niemeyer CM. How I treat juvenile myelomonocytic leukemia. Blood. 2015;125:1083–90. [DOI] [PubMed] [Google Scholar]
- 4.Niemeyer CM, Flotho C. Juvenile myelomonocytic leukemia: who’s the driver at the wheel? Blood. 2019;133:1060–70. [DOI] [PubMed] [Google Scholar]
- 5.Niemeyer CM, Arico M, Basso G, Biondi A, Cantu Rajnoldi A, Creutzig U, et al. Chronic myelomonocytic leukemia in childhood: a retrospective analysis of 110 cases. European Working Group on Myelodysplastic Syndromes in Childhood (EWOG-MDS). Blood. 1997;89:3534–43. [PubMed] [Google Scholar]
- 6.Stieglitz E, Taylor-Weiner AN, Chang TY, Gelston LC, Wang Y-D, Mazor T, et al. The genomic landscape of juvenile myelomonocytic leukemia. Nat Genet. 2015;47:1326–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caye A, Strullu M, Guidez F, Cassinat B, Gazal S, Fenneteau O, et al. Juvenile myelomonocytic leukemia displays mutations in components of the RAS pathway and the PRC2 network. Nat Genet. 2015;47:1334–40. [DOI] [PubMed] [Google Scholar]
- 8.Pérez B, Kosmider O, Cassinat B, Renneville A, Lachenaud J, Kaltenbach S, et al. Genetic typing of CBL, ASXL1, RUNX1, TET2 and JAK2 in juvenile myelomonocytic leukaemia reveals a genetic profile distinct from chronic myelomonocytic leukaemia. Br J Haematol. 2010;151:460–8. [DOI] [PubMed] [Google Scholar]
- 9.Sakaguchi H, Okuno Y, Muramatsu H, Yoshida K, Shiraishi Y, Takahashi M, et al. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nat Genet. 2013;45:937–41. [DOI] [PubMed] [Google Scholar]
- 10.Stieglitz E, Troup CB, Gelston LC, Haliburton J, Chow ED, Yu KB, et al. Subclonal mutations in SETBP1 confer a poor prognosis in juvenile myelomonocytic leukemia. Blood. 2015;125:516–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niemeyer CM, Kang MW, Shin DH, Furlan I, Erlacher M, Bunin NJ, et al. Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat Genet. 2010;42:794–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuda K, Shimada A, Yoshida N, Ogawa A, Watanabe A, Yajima S, et al. Spontaneous improvement of hematologic abnormalities in patients having juvenile myelomonocytic leukemia with specific RAS mutations. Blood. 2007;109:5477–80. [DOI] [PubMed] [Google Scholar]
- 13.Bresolin S, Zecca M, Flotho C, Trentin L, Zangrando A, Sainati L, et al. Gene Expression–Based Classification As an Independent Predictor of Clinical Outcome in Juvenile Myelomonocytic Leukemia. J Clin Oncol. 2010;28:1919–27. [DOI] [PubMed] [Google Scholar]
- 14.MacLeod A, Rouleau J, Szyf M. Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem. 1995;270:11327–37. [DOI] [PubMed] [Google Scholar]
- 15.Hasegawa D, Manabe A, Kubota T, Kawasaki H, Hirose I, Ohtsuka Y, et al. Methylation status of the p15 and p16 genes in paediatric myelodysplastic syndrome and juvenile myelomonocytic leukaemia. Br J Haematol. 2005;128:805–12. [DOI] [PubMed] [Google Scholar]
- 16.Johan MF, Bowen DT, Frew ME, Goodeve AC, Reilly JT. Aberrant methylation of the negative regulators RASSFIA, SHP-1 and SOCS-1 in myelodysplastic syndromes and acute myeloid leukaemia. Br J Haematol. 2005;129:60–5. [DOI] [PubMed] [Google Scholar]
- 17.Liu YL, Castleberry RP, Emanuel PD. PTEN deficiency is a common defect in juvenile myelomonocytic leukemia. Leuk Res. 2009;33:671–7. [DOI] [PubMed] [Google Scholar]
- 18.Poetsch AR, Lipka DB, Witte T, Claus R, Nöllke P, Zucknick M, et al. RASA4 undergoes DNA hypermethylation in resistant juvenile myelomonocytic leukemia. Epigenetics. 2014;9:1252–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilhelm T, Lipka DB, Witte T, Wierzbinska JA, Fluhr S, Helf M, et al. Epigenetic silencing of AKAP12 in juvenile myelomonocytic leukemia. Epigenetics. 2016;11:110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fluhr S, Boerries M, Busch H, Symeonidi A, Witte T, Lipka DB, et al. CREBBP is a target of epigenetic, but not genetic, modification in juvenile myelomonocytic leukemia. Clin Epigenetics. 2016;8:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fluhr S, Krombholz CF, Meier A, Epting T, Mücke O, Plass C, et al. Epigenetic dysregulation of the erythropoietic transcription factor KLF1 and the β-like globin locus in juvenile myelomonocytic leukemia. Epigenetics. 2017;12:715–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olk-Batz C, Poetsch AR, Nöllke P, Claus R, Zucknick M, Sandrock I, et al. Aberrant DNA methylation characterizes juvenile myelomonocytic leukemia with poor outcome. Blood. 2011;117:4871–80. [DOI] [PubMed] [Google Scholar]
- 23.Stieglitz E, Mazor T, Olshen AB, Geng H, Gelston LC, Akutagawa J, et al. Genome-wide DNA methylation is predictive of outcome in juvenile myelomonocytic leukemia. Nat Commun. 2017;8:2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipka DB, Witte T, Toth R, Yang J, Wiesenfarth M, Nöllke P, et al. RAS-pathway mutation patterns define epigenetic subclasses in juvenile myelomonocytic leukemia. Nat Commun. 2017;8:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami N, Okuno Y, Yoshida K, Shiraishi Y, Nagae G, Suzuki K, et al. Integrated molecular profiling of juvenile myelomonocytic leukemia. Blood. 2018;131:1576–86. [DOI] [PubMed] [Google Scholar]
- 26.Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods. 2014;11:1138–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Triche TJ, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41:e90–e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolde R pheatmap: Pretty Heatmaps. CRAN R Package; 2015. [Google Scholar]
- 30.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hovestadt V, Zapatka M. conumee: Enhanced copy-number variation analysis using Illumina DNA methylation arrays. Bioconductor R Package; 2017. [Google Scholar]
- 32.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. arXiv. 2016; [Google Scholar]
- 34.Kuhn M Building Predictive Models in R Using the caret Package. J Stat Softw. 2008;28. [Google Scholar]
- 35.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10. [Google Scholar]
- 36.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huehns ER, Beaven G. Developmental changes in human haemoglobins In: Benson P, editor. Biochem Dev. London: William Heinemann Medical Books Ltd.; 1971. page 175–203. [Google Scholar]
- 38.Niemeyer CM, Strahm B, Dworzak M, De Moerloose B, Hasle H, Stary J, et al. JMML Revisited: Role Und Outcome of Hematopoietic Stem Cell Transplantation in Subtypes of Juvenile Myelomonocytic Leukemia (JMML). Blood. 2012;120:955–955. [Google Scholar]
- 39.Locatelli F, Algeri M, Merli P, Strocchio L. Novel approaches to diagnosis and treatment of Juvenile Myelomonocytic Leukemia. Expert Rev Hematol. Taylor & Francis; 2018;11:129–43. [DOI] [PubMed] [Google Scholar]
- 40.Oakes CC, Seifert M, Assenov Y, Gu L, Przekopowitz M, Ruppert AS, et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat Genet. 2016;48:253–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wierzbinska JA, Toth R, Ishaque N, Rippe K, Mallm J-P, Klett LC, et al. Methylome-based cell-of-origin modeling (Methyl-COOM) identifies aberrant expression of immune regulatory molecules in CLL. Genome Med. 2020;12:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roos-Weil D, Giacopelli B, Armand M, Della-Valle V, Ghamlouch H, Decaudin C, et al. Identification of 2 DNA methylation subtypes of Waldenström macroglobulinemia with plasma and memory B-cell features. Blood. 2020;136:585–95. [DOI] [PubMed] [Google Scholar]
- 43.Bolouri H, Farrar JE, Triche T, Ries RE, Lim EL, Alonzo TA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24:103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nordlund J, Bäcklin CL, Wahlberg P, Busche S, Berglund EC, Eloranta M-L, et al. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013;14:r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, et al. Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell. 2017;32:520–537.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flotho C, Kratz CP, Bergsträsser E, Hasle H, Starý J, Trebo M, et al. Genotype-phenotype correlation in cases of juvenile myelomonocytic leukemia with clonal RAS mutations. Blood. 2008;111:966–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.