Abstract
Of all microorganisms in the human body, the largest and most complex population resides in the gastrointestinal (GI) tract. The gut microbiota continuously adapts to the host environment and serves multiple critical functions for their hosts, including regulating host immunity, procuring energy from food, and preventing the colonization of pathogens. Mounting evidence has suggested gut microbial imbalance (dysbiosis) as a core pathophysiology in the development of GI motility and metabolic disorders, such as irritable bowel syndrome and diabetes. Current research has focused on discovering associations between these disorders and gut microbial dysbiosis; however, whether these associations are a consequence or cause is still mostly unexplored. State-of-the-art studies have investigated how gut microbes communicate with our body systems through microbiota-derived metabolites and how they are able to modulate host physiology. There is now mounting evidence that alterations in the composition of small intestinal microbes have an association with GI dysmotility and metabolic disorders. Although treatment options for gut microbial dysbiosis are currently limited, antibiotics, fecal microbiota transplantation, probiotics, and dietary interventions are currently the best options. However, treatment with broad-spectrum antibiotics has been viewed with skepticism due to the risk of developing antibiotic resistant bacteria. Studies are warranted to elucidate the cellular and molecular pathways underlying gut microbiota-host crosstalk and for the development of a powerful platform for future therapeutic approaches. Here, we review recent literature on gut microbial alterations and/or interactions involved in the pathophysiology of GI dysmotility and metabolic disorders.
Keywords: Diabetes mellitus, Fecal microbiota transplantation, Gastrointestinal microbiome, Irritable bowel syndrome, Metabolic diseases
Introduction
Trillions of microorganisms live inside every human being.1,2 Not surprisingly, these microbial inhabitants outnumber all of the human cells in the entire body by approximately one order of magnitude (1014 vs 1013, respectively).3 To better grasp the role that gut microbes play in health and disease, scientists around the globe are investigating gut microbiota and their metabolites that play a fundamental role in modulating metabolic, developmental, and physiological aspects.1,4 Many human diseases originate from distorted gut microbiota composition (dysbiosis) leading to dysregulation of physiological and metabolic processes.3-5 Gut microbial dysbiosis has been implicated in functional gastrointestinal (GI) disorders including irritable bowel syndrome (IBS), functional dyspepsia (FD), and inflammatory bowel diseases (Crohn’s disease and ulcerative colitis).6-9
In 2019 there were approximately 463 million people suffering from type 2 diabetes (T2D) and 4.2 million deaths due to T2D-related complications.10 Currently, T2D is projected to affect 700 million people worldwide by 2045, which could become a greater burden to the medical community already facing grim statistics.10 Approximately 50% of diabetic patients also suffer from GI motility disorders including but not limited to diarrhea, fecal incontinence, constipation, dyspepsia, and gastroparesis.11-13 GI motility disorders are also extremely common, with approximately 40% of adults suffering from functional bowel disorders worldwide.14 There is a great need to understand the molecular mechanisms that occurs in conditions associated with dysbiosis and how these altered pathways contribute to the development of GI motility disorders and T2D. There are currently many studies focused on creating a deeper understanding of microbiota-related mechanisms of disease pathogenesis with hope that it will lead to the development of effective, preventative, and therapeutic interventions.15-19 Here, we summarize the impact that the gut microbiome and its metabolites have on both GI motility disorders and metabolic diseases. Although, extensive review of current literature identifies a lack of knowledge, the question persists as to whether the disruption of gut microbial communities is a consequence or cause of chronic GI and metabolic diseases.
Function, Composition, and the Dysbiosis of the Gut Microbiota
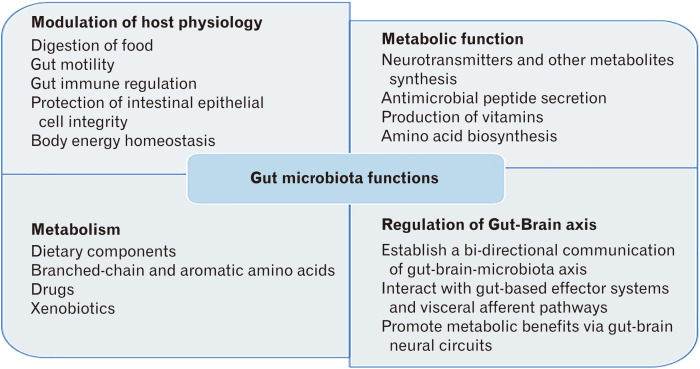
The GI tract is home to vast microbial communities including bacteria, fungi, archaea, and viruses.1,20 The microbe population is more sparse in the upper gut (stomach, duodenum, and jejunum) with approximately 103 bacteria per mL of aspirate, where there are approximately 107-1012 bacteria per mL of aspirate in the lower gut (ileum and proximal colon).21 Gut microbiota perform many diverse functions, such as aiding in the digestion of food, production of essential vitamins, synthesis of metabolites, prevention of pathogenic bacteria colonization, gut-immune regulation, drug metabolism, detoxification, and maintenance of GI physiological homeostasis (Fig. 1).20,22,23 Hence, maintaining a healthy proportion of beneficial microbes, also called eubiosis, is essential for human health.
Figure 1.
Functions of gut microbiota.
Gut microbial imbalance, known as dysbiosis, can include an increase in the proportion of small bowel bacteria, alteration in the relative proportion of benevolent microbes to pathogenic ones, as well as the translocation of colonic bacteria.4,24 At a fundamental level, there are many contributing factors for the progression of a diseased states including microbe-microbe interactions, microbial metabolites, host immune response, host physiology, diet, and the host environment.5,25,26
Gut microbiota composition and relative abundance changes throughout the varying microenvironments of the GI tract27 and over 50 bacterial phyla have been identified in the human GI tract so far.28 In the healthy host, Bacteroidetes and Firmicutes are the most predominantly found phyla in the gut, while Proteobacteria, Verrucomicrobia, Actinobacteria, Fusobacteria, and Cyanobacteria are found in much smaller proportions.29 Generally, the various segments of the GI tract are colonized by different microbial communities; Gram-positive bacteria are prevalent in the small intestine, while Gram-negative bacteria are predominate in the large intestine.21 Approximately 95% of bacteria presiding in the colon are strict anaerobes, which is determined by the available nutrients.30 Small intestinal bacterial overgrowth (SIBO) is largely determined by intrinsic and extrinsic factors.31 The most notable intrinsic factors preventing this overgrowth of bacteria are gastric acid and bile acid (BA) secretion, peristaltic movement, normal gut defense mechanisms, the production of mucin, gut antibacterial peptides, and prevention of bacterial retrograde translocation from the lower gut to the upper gut via the ileocecal valve.32,33 Extrinsic factors include nutrient intake and diet, bacterial and viral infection, medications altering motility (prokinetics), and drugs modulating the gut microbiota, for instance, pre and probiotics, proton pump inhibitors, H2 blockers, and antibiotics.34-36 If any one of the extrinsic factors cause imbalance to off-set the many protective mechanisms set in place, commensal and pathogenic gut microbiota may colonize disproportionately and lead to dysbiosis. One example of dysbiosis is the development of SIBO, diagnosed by overall bacterial overgrowth equal to or greater than 105 CFU per mL of upper gut (eg, jejunal) aspirate culture.37 However, overgrowth of bacteria equal to or greater than 103 CFU/mL of upper gut aspirate has also been included in the diagnosis of SIBO recently.38 Based on jejunal aspirate cultures, one study showed that Escherichia coli, Streptococcus species (spp.), Pseudomonas aeruginosa, Staphylococcus spp., Acinetobacter baumannii, Acinetobacter lwoffii, Enterococcus faecium, Klebsiella pneumoniae, and Enterococcus faecalis were predominantly found in patients with SIBO who suffered altered GI motility.39
Gut Microbiota Affect Host Physiology
The gut microbiota provide its host with essential health benefits primarily by maintaining healthy gut homeostasis.4,5 Currently, scientists are investigating what makes a healthy gut microbiome, and exploring the molecular mechanisms and signaling pathways that allow for crosstalk between gut microorganisms and the host. Many studies have shown that pathogens have the ability to impair the epithelial barrier function.40-42 In contrast, commensal gut microbiota have been found to act as gatekeepers to protect epithelial cell integrity from penetration and disease caused by pathogens.43 Other beneficial effects of commensal gut microbiota include micronutrient production, such as vitamin K and folate.44 Colonic bacteria ferment unabsorbed carbohydrates to short-chain fatty acids (SCFAs), which can be subsequently absorbed through the colonic mucosa and used as an additional energy source.45 Most importantly, commensal gut microbiota are essential to prevent colonization and translocation with pathogenic bacteria at the intestinal epithelial barrier.46
SIBO has been found to lead to several complications in affected hosts,28,47 including destruction of microvilli and heightened epithelial inflammatory response, which often results in impaired absorption.48,49 The bacteria responsible for the harmful effects of SIBO are often aerobes; however, in a healthy gut the small intestine primarily houses facultative anaerobes.50 This microbial shift in patients with SIBO leads to the malabsorption of fat and a deficiency in the fat-soluble vitamins D, E, A, and K.51 Common symptoms of SIBO are abdominal discomfort, gas, distension, and bloating and are likely caused by the dysregulated fermentation of carbohydrates and bacterial colonization in the small intestine, which produce methane, carbon dioxide, and hydrogen.52,53
Gut Microbial Alterations in Gastrointestinal Dysmotility and Metabolic Disorders
Gut Microbial Alterations and Gastrointestinal Dysmotility
Peristaltic movements are of paramount importance for food to properly travel through the gut.54 Peristalsis is generated by a coordination of both contraction and relaxation of the circular and longitudinal smooth muscles of the muscularis externa55 and are regulated by the enteric nervous system (ENS), GI smooth muscle cells, pacemaker cells called interstitial cells of Cajal, enterochromaffin (EC) cells, as well as other factors.56-60 In addition to host-specific genetic predispositions, diet and microbiota are critical regulators of GI physiology.16,17,61 Furthermore, altered microbiota composition of the lumen and mucus layer covering the epithelium often accompany GI disorders.7 As the complexities of the gut microbiota are being increasingly understood, it has revealed that microbe-host interaction, including immune and metabolic responses, are extremely important pathological factors of GI motility disorders. Previous research has shown significant changes in the gut microbiota of patients with IBS when compared to that of healthy individuals, which may contribute significantly to altered bowel habits caused by impaired colonic transit.62,63 However, it is still inconclusive whether the gut microbial signature is different between IBS patients and healthy controls. Also, more robust future studies are warranted to confirm whether this is an association and/or causation. A microbiome “signature” for diseases like IBS has been proposed and based upon previous literature would generally include reduced overall microbial diversity as well as an abundance of methanogenic or Clostridium spp., which are more commonly associated with increasing severity of IBS symptoms.64 Clostridium spp. have been shown to adversely affect normal GI activity due to their role in serotonin (5-hydroxytryptamine [5-HT]) synthesis, although more research is needed to confirm the causative relationship.6 Immune dysregulation, intestinal barrier dysfunction, and altered gut microbial signaling have been at the forefront of understanding the microbiome-related pathogenesis of GI disease.7,15,65
Further evidence of the relationship between microbial dysbiosis and GI motility is demonstrated by the association between SIBO and GI dysfunction. Proper GI motility allows for a constant flow of luminal material through the GI tract, which prevents bacterial overgrowth in the small intestine.66 However, patients presenting with GI dysmotility have a stagnant flow of luminal material, contributing to the development of SIBO.67 For example, patients with malabsorption syndromes have delayed upper gut motility and later also developed SIBO.67 Additionally, SIBO predisposes dysfunctional defense mechanisms of the gut.68 Gut defense mechanisms that prevent SIBO are mediated via secretory IgA, gastric acid, duodenal bile, and defensins.69 Defensins are host antimicrobial peptides, which contribute to the innate immunity of the gut and as one type of microbicidal agent, the adequate concentration of defensin plays a vital role in inhibiting pathogenic organism colonization and maintaining commensal bacteria.70,71 For instance, SIBO-induced GI dysfunction is a result of several mechanisms including disproportionate immune activation and inflammation, inadequate GI motility, intestinal epithelial barrier dysfunction, dysregulated BA deconjugation and serotonergic modulation.20,72,73 Individuals with SIBO often contain bacteria that is commonly found in the colon, including the Gram-negative, carbohydrate-fermenting, facultative aerobes and anaerobes such as E. coli, K. pneumonia, Enterococcus spp., and Proteus mirabilis.31,52,74-76 Not surprisingly, in patients with SIBO versus those without, the small intestinal luminal contents had different metabolomic profiles,77 and in another study patients with malabsorption syndrome had higher quantities of total BAs, lactate, acetate, and formate compared to controls.78 A similar study in subjects with SIBO found that patients were also unable to properly absorb these substances.35 Additionally, patients with malabsorption syndrome were found to have a positive correlation with the quantity of acetate and the degree of symptom severity of SIBO.35 In the same study, unconjugated BAs positively correlated with the degree of steatorrhea, or malabsorption of fat in the intestine.35 This indicates that bacteria commonly found in the small intestine of SIBO patients contributes to the excess production of acetate and deconjugated BA, leading to malabsorption of fat. The inability of intestinal epithelial cells to absorb SCFAs leads to further damage in small intestinal epithelial cells, inducing barrier dysfunction, ileal brake, leading to stasis and eventually bacterial colonization.24
Gut Microbial Alterations and Metabolic Disorders
There have been many studies demonstrating that gut microbiota contributes to the regulation of metabolic homeostasis and that microbial dysbiosis can lead to metabolic dysregulation and diseases; mechanisms include but are not limited to changes to gut barrier function and metabolic inflammation.5,79,80 Furthermore, there is surmounting evidence of the importance of the microbiota in regulating body weight and in the development of T2D. For example, fecal microbiota transplantation (FMT) with healthy gut bacteria improves insulin sensitivity and weight loss in mice and human subjects.81 The gut microbiota are able to produce and absorb host metabolism signaling molecules by regulating available energy extracted from indigestible carbohydrates to the host.82,83 While the primary phyla of the healthy gut remain relatively stable, colonization can be modified by diet, including prebiotics, and with probiotics and antibiotics, which have an effect on the production of microbial metabolites.84 Still, future studies are warranted to confirm the occurrence of a gut microbial shift caused by the administration of prebiotics and probiotics. Antibiotics have the ability to decimate microbial populations and have been associated with the development of metabolic disease, especially with early life exposure.80 Conversely, there has been recent evidence that the use of antibiotics may help regulate metabolic function by improving peripheral insulin sensitivity in obese patients.84 A considerable amount of experimental data has been produced backing the role of microbiota in metabolic regulation and in the genesis of obesity.85-87 Therefore, it should not be unexpected that several studies have found SIBO as a comorbidity in obese and diabetic patients alike.88 Further, among individuals with T2D, the presence of SIBO has been associated with delayed gut transit, indicating an association between gut microbial dysbiosis, GI dysmotility, and metabolic disorder.89 However, present knowledge is lacking robust and highly-controlled human studies examining the effects of microbial mechanisms on host metabolism. As discussed, these studies provide evidence that changes in the gut microbiota may provide an auspicious platform to treat diabetes-related metabolic disorders.
Microbial Signaling Uncovers Pathogenesis of Gastrointestinal Dysmotility and Metabolic Disease
Microbial signaling via metabolites or structural components of bacteria are transmitted across the intestinal epithelium to communicate with distant organs.26 Once these signals are transmitted, they are able to affect organs through subsequent signaling via nerves or hormones.90 Metabolic signaling from the gut microbiota have the potential to significantly affect the host, influencing health status.
Immune Signals
Immune signaling begins with the recognition of microbe-associated molecular patterns, which can include structural components such as lipopolysaccharide, flagellin, and peptidoglycan by pattern-recognition receptors. While there are many different types of pattern-recognition receptors, Toll-like receptors, retinoic acid-inducible gene-I-like receptors, and nucleotide-binding oligomerization domain-like receptors located on epithelial and immune cells are commonly used for host-microbe immune interactions.91 In addition, the aryl hydrocarbon receptor (AHR), which is a transcription factor important for coordinating cellular responses to external stimuli, is stimulated by the Lactobacilli tryptophan ligand, indole-3-aldehyde.92 Remarkably, it has been shown that in the dysbiosis-associated conditions, the microbiota fail to generate AHR ligands contributing to the pathogenesis of GI and metabolic disorders,93,94 as a recent study showed that AHR functions as a biosensor that connect the environment of the intestinal lumen to programming of the ENS via intestinal motility.17 The ENS regulates most aspects of gut physiology through intrinsic neural networks, which innervate throughout the GI system and is commonly called the second brain.57 Distinct neuronal transcriptomes have been identified in various delineations of the GI tract as well as microbiota communities in mice. These transcriptome data led to the discovery that AHR has a defined role in regulating microbe-associated intestinal peristalsis in the surveillance pathway of the ENS.17 Murine studies have also found that AHR deficiency enhances insulin sensitivity and reduces peroxisome proliferator-activated receptor-α, a key metabolic protein.95 Thus, modulating AHR signaling independently or though gut microbiota modulation could help to treat conditions commonly associated with impaired gut motility and metabolic diseases. Taken together, the studies reviewed demonstrate that gut microbial dysbiosis alters host immune signals, which is a central pathogenic mechanism for GI dysmotility and metabolic disorders.
Short-chain Fatty Acids
Butyrate, propionate, and acetate are the 3 most commonly studied SCFAs and are the major fermentation metabolites generated from gut microbial degradation of dietary fiber and help to provide up to 10% of the total energy required by the host.96 Butyrate, propionate, and acetate are also important multifunctional signals produced by the gut microbiota and can bind to the G-protein-coupled receptors (GPR43 and GPR41), also known as free fatty acid receptor 2 and 3 (FFAR2 and FFAR3), respectively. SCFAs binding to FFAR3 induces expression of the hormone peptide YY in enteroendocrine L-cells, which has been shown to normalize gut motility allowing for an increase in available energy harvested from food in mice.97 Binding of SCFAs to FFAR2 and FFAR3 in the epithelial cells of the small intestine and colon activates secretion of glucagon-like peptide-1 by L-cells, substantially impacting overall pancreatic function and insulin release, and hormonal effects regulating appetite.98,99 Independently, SCFAs perform a wide range of metabolic functions; propionate and butyrate are able to stimulate expression of intestinal gluconeogenic enzymes, and propionate is able to independently act as a precursor for intestinal gluconeogenesis.100 All 3 major SCFAs are able to activate FFAR2 in mouse white adipose tissue, suppressing insulin signaling and therefore decreasing fat accumulation and further stimulating energy expenditure in hepatocytes and myocytes.19,101
A study by Reigstad et al102 demonstrated that gut microbiota derived metabolites in human and mouse trigger tryptophan hydroxylase 1 (Tph1) gene expression and 5-HT production in the colon through stimulation of EC cells via SCFAs. In this study, to explore the association between intestinal microbes, gut contractility, and serotonergic gene expression, germ-free (GF) or humanized (HM; ex-GF mice colonized with human gut microbiota) mice were used. Findings showed that the microbiota from conventionally raised mice and HM mice had caused a significant increase in colonic mRNAs of TPH1 compared to the GF mice.102 These studies demonstrate that GI and metabolic homeostasis rely on SCFA production by the gut microbiota, which play a central role in regulating GI motility and metabolic functions.
Tryptamine
Similar to 5-HT, tryptamine is a monoamine metabolite produced from tryptophan by gut bacteria, particularly Ruminococcus gnavus and Clostridium sporogenes, and is found abundantly in human and rodent stool samples.103 In a study reported by Bhattarai et al,16 investigators determined that the role of tryptamine in the GI tract is facilitated by the 5-HT4 receptor that is only found in colonic epithelium. Tryptamine produced by both GF and HM mice were shown to increase movement across the colonic epithelium as well as fluid secretions in colonoids, validating the importance of tryptamine for proper intestinal secretion. Additionally, improved GI motility was seen in GF mice that were colonized with tryptamine-producing engineered Bacteroides thetaiotaomicron microbes.16 This study demonstrates that bacterial metabolites are able to control different facets of host physiology and could be used for localized treatments for GI disorders associated with constipation.
The aforementioned studies provide evidence that metabolites and byproducts essential to gut microbiome signaling fill a specific niche to maintain proper GI function and metabolic regulation. Further, these findings give insight into gut microbiota and host crosstalk alluding to future potential therapeutic options for GI dysmotility and metabolic disorders.
The Gut-Brain Axis and Gut Microbiota: Gastrointestinal Dysmotility and Metabolic Syndrome
The endocrine and nervous systems are able to conduct and coordinate with absolute synergy in each organ system in the body in order to maintain homeostasis.104 The processes between the brain and the gut are bi-directional; as the brain modulates gut physiology, the gut is also able to influence brain function.105 This bi-directional interaction has been demonstrated through the improvement in patients with hepatic encephalopathy after gut-microbiota directed antibiotic treatment.106 Moreover, animal and human studies have demonstrated how the gut microbiota is able to affect brain function. For example, the study by De Palma et al107 demonstrated that GF mice have altered hippocampal brain-derived neurotrophic factors (BDNF), dysregulated hypothalamic pituitary stress responses, impaired neurotransmission, diminished tryptophan availability and dysregulated metabolism, further demonstrating a connection between the gut microbiota and the gut-brain axis.
Bi-directional gut-brain interactions serve as important modulators of GI functionality influencing motility, gastric secretions, blood flow, immune activity, and visceral sensations.108 Brain-to-gut bi-directional signaling can also affect the GI tract through indirect signaling between the gut microbiota and the host. Microbial organisms residing in the GI tract can cause increased intestinal epithelial permeability and modulate the mucosal immune response, leading to changes in host physiology.109 Some evidences support that communication between the gut microbiota and intestinal epithelial cells occurs through luminal release from neurons, Paneth cells, and EC cells.110 The perception of gut stimuli and modulation of various gut functions are conducted through the emotional motor system. The emotional motor system is described as complimentary parallel outflow systems, which includes the sympathetic and parasympathetic branches of the autonomic system, and endogenous pain-modulation systems.108 A bi-directional gut-brain microbiota axis is established through the gut microbiota interactions with gut based effector systems and visceral afferent pathways.108 Also, current research demonstrates that the host 5-HT is synthesized in the gut via microbial-derived metabolites; these findings add further weight to the concept of a gut microbiota-gut-brain axis and its influence on GI homeostasis.111
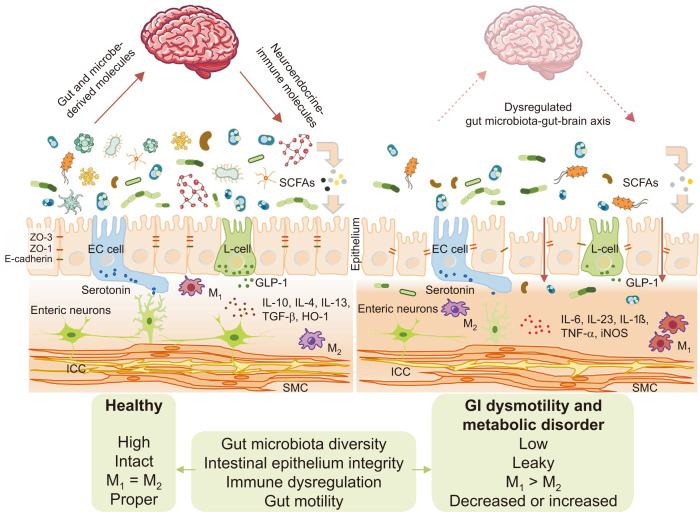
Connecting the dots between the gut microbiota, brain, and metabolic control of insulin secretion, Perry and colleagues19 explored a pathway involving the “rest-and-digest” and “feed-and-breed” processes. In this study, rats fed a high-fat diet had greater production and turnover of the SCFA, acetate, compared to normal control diet (NCD) rats. They found that by exposing stomachs of NCD rats with acetate, glucose-stimulated insulin secretion (GSIS) was increased, indicating a relationship between microbiota-derived acetate and insulin secretion.19 The vagus nerve largely controls parasympathetic activity through motor inputs that are sent to various organs and is able to control heart rate, regulate GI movement, aid in the digestion of food, and enhance insulin secretion. Perry et al19 demonstrated this by using parasympathetic blockers, atropine or methylatropine, or surgically severing portions of the vagus nerve connecting the gut, to prevent acetate from increasing GSIS. This indicates that the beneficial effects of acetate-inducing GSIS is controlled through the vagus nerve and parasympathetic nervous system. To elucidate the role of microbiota acetate turnover in this study, Perry et al19 performed FMT from NCD or high-fat diet donor rats into recipients on the opposite diet. Results showed that acetate levels and GSIS levels from the donor groups were transferred to recipient groups, implying that changes in the microbiota regulate acetate turnover, and therefore GSIS. These powerful findings confirmed the gut microbiota-gut-brain axis influences metabolic homeostasis (Fig. 2).
Figure 2.
Intestinal barrier dysfunction as a core pathophysiology of gastrointestinal (GI) dysmotility and metabolic disorder. SCFAs, short-chain fatty acids; ZO, zonula occludens; E-cadherin, epithelial cadherin; EC, enterochromaffin; GLP-1, glucagon-like peptide-1; IL, interleukin; M1, classically activated macrophages; M2, alternatively activated macrophages; TGF-β, transforming growth factor beta; HO-1, heme oxygenase-1; TNF-α, tumor necrosis factor alpha; iNOS, inducible nitric oxide synthase; ICC, interstitial cell of Cajal; SMC, smooth muscle cell; L-cell, enteroendocrine cell.
Strong pre-clinical data highlights that the gut microbiota is important for bi-directional interactions of the gut-brain axis in health and in disease. Therefore, gut microbial dysbiosis has pathophysiological effects on gut-brain bi-directional interactions leading to GI dysmotility and metabolic disorders. An emerging area that should be further explored is how the gut-brain axis is affected by the gut microbiota and their derived metabolites leading to metabolic benefits.
Gut Microbiota Mediated Immune Dysregulation in Gastrointestinal Dysmotility and Metabolic Disease
The most important function of the GI tract is to take in food particles, digest these to smaller molecules, absorb nutrients, and excrete the undigested byproducts. The GI tract also harnesses the benefits of commensal microbiota, which play a part in regulating the host metabolism and directing proper immunity.112 Like proper heart function, the functions of the GI tract, such as gut motility, are necessary for life. Current literature suggests proper gut motility is dependent on the interacting forces between the microbiome and ENS with the help of immune cells, like the tissue-resident muscularis macrophages (MMs).113 MMs are in close proximity to the myenteric plexus of GI smooth muscle.114 One study explored the functional role of MMs under normal physiological conditions and homeostatic crosstalk between ENS neurons and MMs.114 Muller et al115 showed that MMs are a unvarying subset of CX3CR1+ CD11cloMHCIIhi cells, dependent on the secretion of colony stimulatory factor 1 receptor (CSF-1R), a cytokine receptor. Intraperitoneal injection with a low-dose of anti-CSF-1R antibody specifically depleted 80% of MMs, leaving stromal cells and other GI resident macrophage populations unharmed. By depleting this specific subset of macrophages, gastric emptying was accelerated and colonic emptying was reduced in mice. Further, in vitro experiments demonstrated the homeostatic role of MMs in regulating peristalsis showing signs of deregulated and hyperreactive contractions of the muscularis externa. In addition to the distinctive role of MMs to establish the molecular mechanisms contributing to these effects, researchers analyzed the nonimmune MM transcripts and found novel expression of bone morphogenetic protein 2 (BMP2) that is known to stimulate the BMP receptors I and II (BMPRI and BMPRII). Neuronal and smooth muscle development relies on BMP receptors indicating BMP2 may be a candidate for MM-mediated control of peristalsis. It is well established that the intestinal microbiota possess the ability to instruct mucosal immune cells and therefore influence the composition of the gut microbiota.112 Studies have also shown that GF mice and mice treated with antibiotics experience GI dysmotility.115 However, how is it possible for the intestinal microbiota to influence MM function, given the distance between the intestinal lumen and MMs? To answer this question, Muller et al115 treated mice with antibiotics and found decreased expression of BMP2, CSF-1, and decreased numbers of MMs, alluding to reliance on the microbiome. Notably, treated mice also developed delayed GI transit. These findings advance our understanding of communication that occurs during inflammation between the nervous system and immune system and shows the significance of bi-directional neuroimmune communication for maintaining proper human body functions.116 An important component of this study was also in the identification of BMP2 as an additional neurotrophic factor produced by macrophages, in addition to BDNF.117 From this we can deduce that the intestinal microbiota is an essential component in the neuroimmune crosstalk and MMs act as intermediaries between the ENS and the gut microbiota (Fig. 2).
Metabolic disorders, as in obesity and T2D, are associated with low-grade chronic inflammation in adipose tissue, liver, skeletal muscle, pancreas, brain, and intestines.118 Elevated levels of the circulating pro-inflammatory cytokines TNF-α, IL-1β, and IL-6 were found in high fat diet induced obese mice and humans, and was found to lead to insulin resistance and T2D.119 In obesity, there can be a disproportionate amount of visceral adipose tissue caused by the accumulated pro-inflammatory immune cells and reduced anti-inflammatory cells, which leads to chronic, low-grade inflammation. Visceral adipose tissue, which is hormonally active, can be considered as a major driver of insulin resistance in obese animal models and humans.118,120,121 Other organs also develop low-grade inflammation, which also contributes to insulin resistance.118,122 Billions of microbes are constantly interacting with epithelial mucosa in the gut, which keeps the intestinal immune system incessantly active. Intestinal barrier dysfunction is widely accepted as one of the first events in GI dysmotility and metabolic disorders and leads to immune cell infiltration and low-grade inflammation of the gut mucosa.118 Mainly, intestinal barrier dysfunction caused by microbial dysbiosis plays a critical component in the development of immune dysregulation leading to GI and metabolic disease.123 Recently, in a study published in Science by Thaiss et al,124 T2D and obesity mouse models showed that hyperglycemia led to intestinal barrier dysfunction through transcriptional restructuring of glucose transporter 2-dependent intestinal epithelial cells and altered tight junction and adherence protein integrity. As a consequence of hyperglycemia-mediated epithelial barrier disruption, a systemic influx of microbial products can enhance allocation of intestinal microbiome products leading to enteric infection. However, barrier function is restored and the microbiota is contained upon treatment and management of hyperglycemia, intestinal epithelial-specific glucose transporter 2 deletion, or by inhibition of glucose metabolism.124 Glycemic control, indicated by glycated hemoglobin levels, was shown to correlate with the systemic influx of intestinal microbiome products. Collectively, these results provide a mechanistic connection of the hyperglycemic condition and intestinal barrier dysfunction with the systemic inflammatory and potentially infectious consequences of obesity and T2D. Most importantly, this study shows that metabolic syndrome revolves around the gut, supports the hypothesis that imbalances in metabolic processes may begin in the gut, and highlights intestinal barrier dysfunction, mediated by gut microbial dysbiosis, as a core pathophysiology of GI dysmotility and metabolic disease (Fig. 2).
Gut Microbial Dysbiosis Connects Gastrointestinal Dysmotility With Metabolic Disorders
GI dysmotility is commonly diagnosed in patients with metabolic disorders.11,13 Accumulating evidence has shown that gut microbial dysbiosis and the resulting intestinal barrier dysfunction link these 2 conditions. For example, a recent study showed how duodenal microbial dysbiosis is linked with enteropathy and intestinal barrier dysfunction in the environmental enteric dysfunction (EED) condition.125 EED is a subclinical syndrome characterized by intestinal villous blunting, reduced absorptive capacity, and increased intestinal inflammation.125,126 Furthermore, there have been several metabolic consequences, such as malnutrition and stunting, in patients with EED. However, findings from this study raise additional questions: is environmental enteropathy caused by a form of bacterial overgrowth in the small intestine? If so, how does SIBO alter GI motility? In response to these questions, we reviewed an article published on tropical sprue (TS), a type of malabsorption syndrome that has clinical and histological features similar to EED.67 This study demonstrated that there was increased intestinal bacterial colonization in patients with TS. Furthermore, there is a vicious cycle of SIBO and small intestinal stasis due to the ileal brake induced by unabsorbed fat passing through the ileum.127 SIBO also deconjugates BAs, which further caused fat malabsorption and may even change gut motility. Malabsorption of fat induces ileal brake by liberating gut hormones such as peptide YY and neurotensin.67 Therefore, orocecal transit time was found to be longer in patients with TS than in controls.
Further evidence supporting intestinal barrier dysfunction as a pathogenic mechanism for metabolic disorders is illuminated by 2 recent studies. One study demonstrated that hyperglycemia drives intestinal barrier dysfunction and increases the risk for enteric infection.124 In contrast, a second study showed loss of the gut barrier integrity triggers activation of islet-reactive T cells and autoimmune diabetes.128 While it is currently unclear whether intestinal barrier dysfunction or metabolic disorder comes first, it is clear that there is a direct connection between these 2 conditions. Interestingly, research over the past few decades has indicated that intestinal barrier dysfunction is a major pathophysiological mechanism for the development of GI dysmotility and functional bowel disorders.129 Further studies provided evidence for this connection by demonstrating that the development of IBS follows acute infective gastroenteritis, a condition known as post-infectious IBS.130 Taken together, the aforementioned studies indicated that intestinal barrier dysfunction is associated with gut microbial dysbiosis and is likely a key pathophysiological mechanism that links metabolic disorders with GI dysmotility.
Furthermore, we reviewed some proof of concept studies that provide a direct connection between metabolic and GI motility disorders through microbial dysbiosis. Landmark studies have demonstrated that altering the gut microbiota during stages of critical developmental has lasting metabolic consequences.131,132 A large retrospective cohort study reported that prescription for antibiotics within the first 2 years of life is associated with the development of early childhood obesity.133 Also, antibiotic-induced depletion of the gut microbiota has been shown to induce changes in 5-HT biosynthesis and to delay GI motility.134 Moreover, studies have demonstrated that gut microbial alterations, especially alterations leading to increased abundance of methanogens, leads to the development of slow transit constipation.135 These state-of-the-art studies clearly indicate that intestinal barrier dysfunction, brought about by gut microbial dysbiosis, is a central pathogenic mechanism for metabolic as well as GI motility disorders.
Treatment Options for Gut Microbial Dysbiosis
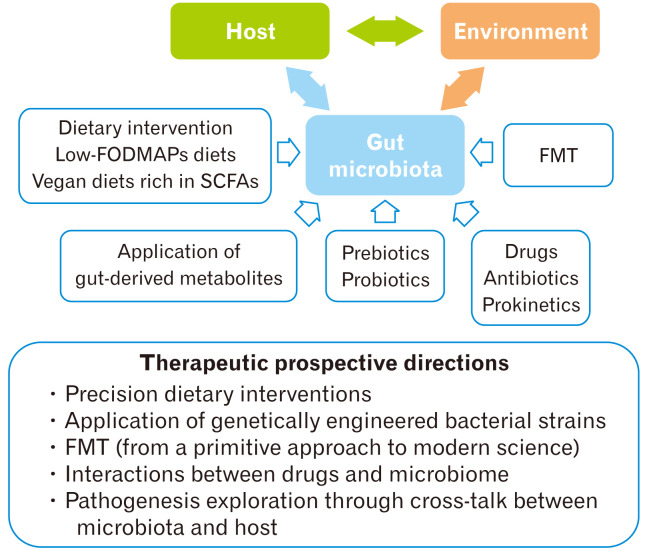
Current management options for gut microbial dysbiosis include antibiotics, FMT, as well as diet and probiotic interventions (Fig. 3).
Figure 3.
Therapeutic modulations of gut microbiota. FODMAPs, fermentable oligosaccharides, disaccharides, monosaccharides, and polyols; SCFAs, short-chain fatty acids; FMT, fecal microbiota transplantation.
Antibiotics
Minimally absorbed antibiotics neomycin and rifaximin led to improved colonic motility as evidenced in clinical trials in patients with IBS.136,137 In addition, several studies have shown that exposure to a combination of antibiotics for approximately 4-8 weeks in obesity mouse models or high-fat diet fed mice substantially improves metabolic parameters including: increased glucose tolerance, reduction in fat mass, and lowered hepatic steatosis in hepatic and systemic inflammation; these changes are also associated with changes in gut microbiota composition, gut barrier dysfunction, and metabolic endotoxemia.4,138 However, we desperately need well-designed human clinical trials to test if the efficacy of these intervention techniques are applicable to humans and may lead to manipulation of the gut microbiota contributing to hampered metabolism and the manifestation of metabolic disorders. The most studied treatment for patients with SIBO is rifaximin, a non-systemic antibiotic.139 A systematic review and meta-analysis (26 studies) of rifaximin reported that SIBO was improved or resolved in 70.8% of patients.140 However, systemic antibiotics, such as norfloxacin, have also been reported to also eradicate SIBO.52 A meta-analysis (10 prospective clinical studies) of non-systemic antibiotics found more normal hydrogen-breath tests in patients with SIBO that had been treated with an antibiotic compared to patients that received a placebo (51.1% vs 9.8%, respectively).141
Fecal Microbiota Transplantation
FMT is also thought to be beneficial in treating microbial dysbiosis as it could restore “healthy” microbes in diseased patients.142 However, whether FMT as a treatment for IBS is a panacea or placebo, is still debatable. A double-blind randomized controlled trial including 165 patients with IBS showed that after FMT, IBS symptom severity significantly improved when compared to a placebo control group, although there were no outstanding changes in the degree of overall dysbiosis.143 In a recent metanalysis, data pooled from 5 different randomized controlled trials found no significant improvements in IBS symptoms of patients who received FMT versus placebo.144 However, larger and more rigorous trials are needed; studies included in this meta-analysis were small and included potential for a high-risk of bias.145 Interestingly, FMT eliminated SIBO in 71% of patients with chronic intestinal pseudo-obstruction.146 Another study demonstrated the effect of a lean donor (allogenic) versus own (autologous) FMT to recipients with metabolic syndromes.147 Changes in blood plasma metabolites, such as gamma-aminobutyric acid, indicate metabolic responses responsible for the observed improved insulin sensitivity in the allogenic FMT group. However, this improvement is dependent on the fact that the patient had decreased fecal microbial diversity at the start of the study. Further, changes in intestinal microbiota composition is associated with the beneficial effects on glucose metabolism seen in patients in the allogenic group and may be predicted from fecal microbiota composition before treatment.147
Probiotics
Probiotics, live microorganisms, are believed to have favorable effects on the gut microbiota.148 However, there have only been a few clinical studies examining this option, and they lack consistency. Recently, a meta-analysis found that improved clearance of SIBO was associated with probiotic use.149 Probiotics have also been shown to confer health benefits in patients with IBS although the mechanism responsible for improved symptomology has yet to be elucidated.150 Another study found that probiotics from fermented camel milk significantly restored blood glucose and lipid levels back to healthy levels in the db/db T2D mouse model.151 In addition, researchers found that insulin secretion was improved in probiotic treated diabetic mice due to upregulation of GPR43/41, which improved glucose-triggered glucagon-like peptide-1 secretion.151 Taken together, the studies presented show promise for probiotic treatment in GI motility disorders as well as metabolic disease.
Dietary Intervention
“You and your microbiome are what you eat.”152,153 Our gut microbiome is largely influenced by our diet because it modulates the richness of specific colonizers and their individual and collective functions. Microbial community changes facilitated by diet could have a detrimental effect for host health due to the essential role that the microbiome plays in regulating host physiology.154 A superlative option for a low-risk treatment intended to modulate the microbiome would be to change the patients’ diet. Therefore, utilizing diet and diet-based studies to change microbial communities could present novel therapeutic strategies for conditions in which the gut microbiota and its associated metabolic products have been shown to harm the host or be key in disease pathogenesis. An exciting question arises of whether the presence of the individual’s specific microbiome fingerprint can actually influence dietary preferences of the host, and therefore influencing positive feedback loops. In a study in which patients with obesity were assigned several different control diets, researchers found that the fecal microbiota profiles associated more by individual than by diet.155 On the contrary, in response to the dietary changes, marked changes were found in the relative abundance of dominant microbial phylotypes.155 Wu et al156 reported a correlation between long-term dietary habits and 2 enterotypes that were defined in 96 adults. High fiber intake conferred a “Prevotella-type” community and high protein intake was associated with a “Bacteroides-type” community. This implies that dietary patterns may influence the enterotypes found in the host. Another interesting study found that populations of Italian and African children had significant differences in fecal microbiota, which can be explained due to different dietary habits.157 In these samples, Italian children had higher intakes of starch and proteins compared to African populations and showed a higher proportion of Bacteroides spp. and Firmicutes, suggesting that both short-term and long-term dietary shifts can lead to compositional changes of the gut microbiota.
Dietary intervention has the potential to be a powerful tool in helping patients overcome microbial dysbiosis.86 In patients suffering from conditions as a result of gut dysbiosis, translocated colonic bacteria residing in the small bowel ferment carbohydrates causing excessive gas, abdominal pain, and bloating. Therefore, with the temporary restriction of dietary components, such as diets low in fermentable oligosaccharides, disaccharides, monosaccharides, and polyols (FODMAP), hence the FODMAP, has been suggested to improve symptoms of IBS and of dysbiosis.158 Low FODMAP diets have been associated with reduced absolute abundance of bacteria, which may have significant beneficial effects in the treatment of SIBO or other dysbiosis conditions.159 Moreover, a diet high in complex carbohydrates preferentially encourages the growth of less pathogenic bacteria than a diet rich in fats or protein.152 Primarily vegetarian diets abundant in fiber lead to increased production of SCFAs, which inhibit potentially invasive bacteria from colonizing the gut.160 While there is accumulating evidence supporting the role of diet on gut microbial composition, more research is needed to accurately deduce the effect of different diets on the gut microbiota.
Conclusions and Further Directions
Constant communication between the gut microbiota derived metabolites and human body systems regulates physiological aspects of health and disease. Current literature demonstrates that metabolic syndrome is highly influenced by the gut and supports the hypothesis that metabolic disorders may begin there. Metabolic disease is a multi-factorial condition that makes it difficult to unravel a causative effect of the microbiome on the pathogenesis of this condition. Further, gut microbiota mediated immune dysregulation and intestinal barrier dysfunction emerges as a core pathophysiology of GI dysmotility and metabolic disease. However, it should be noted that these disorders are not mutually inclusive; patients may have GI dysmotility without the co-occurrence of metabolic disorders and vice versa.
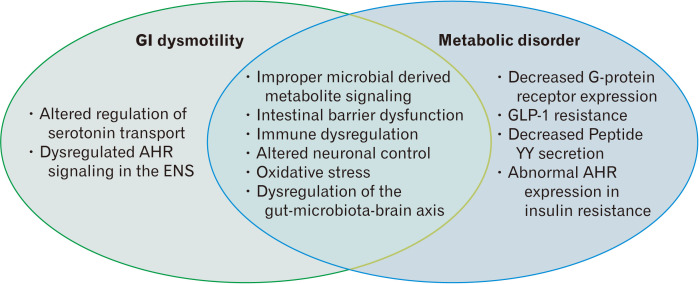
Evidence has emerged demonstrating a potential position of the gut microbiome in GI dysmotility and metabolic disorders (Fig. 4). A major challenge in studying the gut microbiota is in translating and applying data into physiologically relevant mechanisms. One way to go about facing this challenge is to isolate specific bacterial strains, or analyze how they are affected by specific macronutrients commonly found in humans, and use the information obtained to elucidate biomarkers that may be used to find better treatments for GI dysmotility and metabolic diseases. These biomarkers may also allow for the identification of mechanisms in which the microbial metabolites lead to or prevent the development of disease states. Also, due to obscure small intestinal microbiome research, more attention is needed on the pathogenesis of SIBO in GI dysmotility and metabolic diseases. Using next generation sequencing techniques to explore small intestinal microbiomes, together with better sampling of the small bowel aspirate, may allow us to prevent cases of antibiotic resistance and help better understand the microbial pathogenesis of SIBO.
Figure 4.
Pathogenesis in gastrointestinal (GI) dysmotility and metabolic disorder. AHR, aryl hydrocarbon receptor; ENS, enteric nervous system; GLP-1, glucagon-like peptide-1.
The collaborative work between microbiologists, gastroenterologists, endocrinologists, and epidemiologists along with improvements in the analysis of microbial markers, microbial metabolites, and molecular signals will lead to thrilling discoveries in the future. The closer we can get to the microbial pathogenesis of the gut microbial alterations and exploring crosstalk with the host, the more effectively we can treat and manage GI dysmotility and related metabolic manifestations. The statement from Hippocrates that “all disease begins in the gut” is becoming progressively more accepted with increasing knowledge of the gut microbiota-gut-brain axis and its influence on human health and disease when altered.
Footnotes
Financial support: None.
Conflicts of interest: None.
Author contributions: Rajan Singh, Hannah Zogg, Lai Wei, and Allison Bartlett reviewed the literature and drafted the original manuscript and figures; and Seungil Ro, Uday C Ghoshal, and Singh Rajender edited the manuscript and provided important intellectual directives.
References
- 1.Brody H. The gut microbiome. Nature. 2020;577:S5. doi: 10.1038/d41586-020-00194-2. [DOI] [PubMed] [Google Scholar]
- 2.Cani PD. Human gut microbiome: hopes, threats and promises. Gut. 2018;67:1716–1725. doi: 10.1136/gutjnl-2018-316723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prakash S, Rodes L, Coussa-Charley M, Tomaro-Duchesneau C. Gut microbiota: next frontier in understanding human health and development of biotherapeutics. Biologics. 2011;5:71–86. doi: 10.2147/BTT.S19099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cani PD. Gut microbiota: changes in gut microbes and host metabolism: squaring the circle? Nat Rev Gastroenterol Hepatol. 2016;13:563–564. doi: 10.1038/nrgastro.2016.135. [DOI] [PubMed] [Google Scholar]
- 5.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 6.Pittayanon R, Lau JT, Yuan Y, et al. Gut microbiota in patients with irritable bowel syndrome-a systematic review. Gastroenterology. 2019;157:97–108. doi: 10.1053/j.gastro.2019.03.049. [DOI] [PubMed] [Google Scholar]
- 7.Barbara G, Feinle-Bisset C, Ghoshal UC, et al. The intestinal microenvironment and functional gastrointestinal disorders. Gastroenterology. 2016;150:1305–1318.:e8. doi: 10.1053/j.gastro.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 8.Ghoshal UC. Marshall and Warren Lecture 2019: aparadigm shift in pathophysiological basis of irritable bowel syndrome and its implication on treatment. J Gastroenterol Hepatol. 2020;35:712–721. doi: 10.1111/jgh.15032. [DOI] [PubMed] [Google Scholar]
- 9.Pittayanon R, Lau JT, Leontiadis GI, et al. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: a systematic review. Gastroenterology. 2020;158:930–946.:e1. doi: 10.1053/j.gastro.2019.11.294. [DOI] [PubMed] [Google Scholar]
- 10.Cho NH, Shaw JE, Karuranga S, et al. IDF Diabetes Atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res Clin Pract. 2018;138:271–281. doi: 10.1016/j.diabres.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 11.Choung RS, Locke GR, 3rd, Schleck CD, Zinsmeister AR, Melton LJ, 3rd, Talley NJ. Risk of gastroparesis in subjects with type 1 and 2 diabetes in the general population. Am J Gastroenterol. 2012;107:82–88. doi: 10.1038/ajg.2011.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halland M, Bharucha AE. Relationship between control of glycemia and gastric emptying disturbances in diabetes mellitus. Clin Gastroenterol Hepatol. 2016;14:929–936. doi: 10.1016/j.cgh.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camilleri M, Chedid V, Ford AC, et al. Gastroparesis. Nat Rev Dis Primers. 2018;4:41. doi: 10.1038/s41572-018-0038-z. [DOI] [PubMed] [Google Scholar]
- 14.Sperber AD, Bangdiwala SI, Drossman DA, et al. Worldwide prevalence and burden of functional gastrointestinal disorders, results of Rome Foundation Global Study. Gastroenterology Published Online First. 2020 Apr 12;:doi: 10.1053/j.gastro.2020.04.014. doi: 10.1053/j.gastro.2020.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Öhman L, Törnblom H, Simrén M. Crosstalk at the mucosal border: importance of the gut microenvironment in IBS. Nat Rev Gastroenterol Hepatol. 2015;12:36–49. doi: 10.1038/nrgastro.2014.200. [DOI] [PubMed] [Google Scholar]
- 16.Bhattarai Y, Williams BB, Battaglioli EJ, et al. Gut microbiota-produced tryptamine activates an epithelial g-protein-coupled receptor to increase colonic secretion. Cell Host Microbe. 2018;23:775–785.:e5. doi: 10.1016/j.chom.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obata Y, Castaño Á, Boeing S, et al. Neuronal programming by microbiota regulates intestinal physiology. Nature. 2020;578:284–289. doi: 10.1038/s41586-020-1975-8. [DOI] [PubMed] [Google Scholar]
- 18.Kimura I, Miyamoto J, Ohue-Kitano R, et al. Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science. 2020;367:eaaw8429. doi: 10.1126/science.aaw8429. [DOI] [PubMed] [Google Scholar]
- 19.Perry RJ, Peng L, Barry NA, et al. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature. 2016;534:213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cani PD. Gut microbiota - at the intersection of everything? Nat Rev Gastroenterol Hepatol. 2017;14:321–322. doi: 10.1038/nrgastro.2017.54. [DOI] [PubMed] [Google Scholar]
- 21.Ghoshal UC, Ghoshal U. Small intestinal bacterial overgrowth and other intestinal disorders. Gastroenterol Clin North Am. 2017;46:103–120. doi: 10.1016/j.gtc.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Eisenstein M. The hunt for a healthy microbiome. Nature. 2020;577:S6–S8. doi: 10.1038/d41586-020-00193-3. [DOI] [PubMed] [Google Scholar]
- 23.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 24.Ghoshal UC, Srivastava D. Irritable bowel syndrome and small intestinal bacterial overgrowth: meaningful association or unnecessary hype. World J Gastroenterol. 2014;20:2482–2491. doi: 10.3748/wjg.v20.i10.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Curr Opin Gastroenterol. 2015;31:69–75. doi: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroeder BO, Bäckhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22:1079–1089. doi: 10.1038/nm.4185. [DOI] [PubMed] [Google Scholar]
- 27.Shin A, Preidis GA, Shulman R, Kashyap PC. The gut microbiome in adult and pediatric functional gastrointestinal disorders. Clin Gastroenterol Hepatol. 2019;17:256–274. doi: 10.1016/j.cgh.2018.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dukowicz AC, Lacy BE, Levine GM. Small intestinal bacterial overgrowth: a comprehensive review. Gastroenterol Hepatol (NY) 2007;3:112–122. [PMC free article] [PubMed] [Google Scholar]
- 29.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagier JC, Million M, Hugon P, Armougom F, Raoult D. Human gut microbiota: repertoire and variations. Front Cell Infect Microbiol. 2012;2:136. doi: 10.3389/fcimb.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghoshal UC, Shukla R, Ghoshal U. Small intestinal bacterial overgrowth and irritable bowel syndrome: a bridge between functional organic dichotomy. Gut Liver. 2017;11:196–208. doi: 10.5009/gnl16126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gabrielli M, D'Angelo G, Di Rienzo T, Scarpellini E, Ojetti V. Diagnosis of small intestinal bacterial overgrowth in the clinical practice. Eur Rev Med Pharmacol Sci. 2013;17(suppl 2):30–35. [PubMed] [Google Scholar]
- 33.Riordan SM, McIver CJ, Wakefield D, Duncombe VM, Thomas MC, Bolin TD. Small intestinal mucosal immunity and morphometry in luminal overgrowth of indigenous gut flora. Neurogastroenterol Motil. 2001;96:494–500. doi: 10.1111/j.1572-0241.2001.03533.x. [DOI] [PubMed] [Google Scholar]
- 34.Erdogan A, Rao SS, Gulley D, Jacobs C, Lee YY, Badger C. Small intestinal bacterial overgrowth: duodenal aspiration vs glucose breath test. Neurogastroenterol Motil. 2015;27:481–489. doi: 10.1111/nmo.12516. [DOI] [PubMed] [Google Scholar]
- 35.Peralta S, Cottone C, Doveri T, Almasio PL, Craxi A. Small intestine bacterial overgrowth and irritable bowel syndrome-related symptoms: experience with rifaximin. World J Gastroenterol. 2009;15:2628–2631. doi: 10.3748/wjg.15.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziegler TR, Cole CR. Small bowel bacterial overgrowth in adults: a potential contributor to intestinal failure. Curr Gastroenterol Rep. 2007;9:463–467. doi: 10.1007/s11894-007-0060-x. [DOI] [PubMed] [Google Scholar]
- 37.Ghoshal UC, Goel A, Quigley EMM. Gut microbiota abnormalities, small intestinal bacterial overgrowth, and non-alcoholic fatty liver disease: an emerging paradigm. Indian J Gastroenterol. 2020;39:9–21. doi: 10.1007/s12664-020-01027-w. [DOI] [PubMed] [Google Scholar]
- 38.Ghoshal UC, Baba CS, Ghoshal U, et al. Low-grade small intestinal bacterial overgrowth is common in patients with non-alcoholic steatohepatitis on quantitative jejunal aspirate culture. Indian J Gastroenterol. 2017;36:390–399. doi: 10.1007/s12664-017-0797-6. [DOI] [PubMed] [Google Scholar]
- 39.Ghoshal UC, Srivastava D, Ghoshal U, Misra A. Breath tests in the diagnosis of small intestinal bacterial overgrowth in patients with irritable bowel syndrome in comparison with quantitative upper gut aspirate culture. Eur J Gastroenterol Hepatol. 2014;26:753–760. doi: 10.1097/MEG.0000000000000122. [DOI] [PubMed] [Google Scholar]
- 40.Odenwald MA, Turner JR. The intestinal epithelial barrier: a therapeutic target? Nat Rev Gastroenterol Hepatol. 2017;14:9–21. doi: 10.1038/nrgastro.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vancamelbeke M, Vermeire S. The intestinal barrier: a fundamental role in health and disease. Expert Rev Gastroenterol Hepatol. 2017;11:821–834. doi: 10.1080/17474124.2017.1343143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luissint AC, Parkos CA, Nusrat A. Inflammation and the intestinal barrier: leukocyte-epithelial cell interactions, cell junction remodeling, and mucosal repair. Gastroenterology. 2016;151:616–632. doi: 10.1053/j.gastro.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Köhler H, McCormick BA, Walker WA. Bacterial-enterocyte crosstalk: cellular mechanisms in health and disease. J Pediatr Gastroenterol Nutr. 2003;36:175–185. doi: 10.1097/00005176-200302000-00005. [DOI] [PubMed] [Google Scholar]
- 44.Jørgensen JR, Fitch MD, Mortensen PB, Fleming SE. In vivoabsorption of medium-chain fatty acids by the rat colon exceeds that of short-chain fatty acids. Gastroenterology. 2001;120:1152–1161. doi: 10.1053/gast.2001.23259. [DOI] [PubMed] [Google Scholar]
- 45.Shindo K, Machida M, Koide K, Fukumura M, Yamazaki R. Deconjugation ability of bacteria isolated from the jejunal fluid of patients with progressive systemic sclerosis and its gastric pH. Hepatogastroenterology. 1998;45:1643–1650. [PubMed] [Google Scholar]
- 46.Gorbach SL. Probiotics and gastrointestinal health. Am J Gastroenterol. 2000;95(1 suppl):S2–S4. doi: 10.1016/S0002-9270(99)00806-0. [DOI] [PubMed] [Google Scholar]
- 47.Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaufman SS, Loseke CA, Lupo JV, et al. Influence of bacterial overgrowth and intestinal inflammation on duration of parenteral nutrition in children with short bowel syndrome. J Pediatr. 1997;131:356–361. doi: 10.1016/S0022-3476(97)80058-3. [DOI] [PubMed] [Google Scholar]
- 49.Ament ME, Shimoda SS, Saunders DR, Rubin CE. Pathogenesis of steatorrhea in three cases of small intestinal stasis syndrome. Gastroenterology. 1972;63:728–747. doi: 10.1016/S0016-5085(19)33206-8. [DOI] [PubMed] [Google Scholar]
- 50.Riepe SP, Goldstein J, Alpers DH. Effect of secreted Bacteroides proteases on human intestinal brush border hydrolases. J Clin Invest. 1980;66:314–322. doi: 10.1172/JCI109859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giannella RA, Rout WR, Toskes PP. Jejunal brush border injury and impaired sugar and amino acid uptake in the blind loop syndrome. Gastroenterology. 1974;67:965–974. doi: 10.1016/S0016-5085(19)32751-9. [DOI] [PubMed] [Google Scholar]
- 52.Sachdev AH, Pimentel M. Gastrointestinal bacterial overgrowth: pathogenesis and clinical significance. Ther Adv Chronic Dis. 2013;4:223–231. doi: 10.1177/2040622313496126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dibaise JK, Young RJ, Vanderhoof JA. Enteric microbial flora, bacterial overgrowth, and short-bowel syndrome. Clin Gastroenterol Hepatol. 2006;4:11–20. doi: 10.1016/j.cgh.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 54.Sanders KM, Koh SD, Ro S, Ward SM. Regulation of gastrointestinal motility--insights from smooth muscle biology. Nat Rev Gastroenterol Hepatol. 2012;9:633–645. doi: 10.1038/nrgastro.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanders KM. Spontaneous electrical activity and rhythmicity in gastrointestinal smooth muscles. Adv Exp Med Biol. 2019;1124:3–46. doi: 10.1007/978-981-13-5895-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanders KM, Kito Y, Hwang SJ, Ward SM. Regulation of gastrointestinal smooth muscle function by interstitial cells. Physiology (Bethesda) 2016;31:316–326. doi: 10.1152/physiol.00006.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spencer NJ, Hu H. Enteric nervous system: sensory transduction, neural circuits and gastrointestinal motility. Nat Rev Gastroenterol Hepatol. 2020;17:338–351. doi: 10.1038/s41575-020-0271-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spencer NJ, Nicholas SJ, Robinson L, et al. Mechanisms underlying distension-evoked peristalsis in guinea pig distal colon: is there a role for enterochromaffin cells? Am J Physiol Gastrointest Liver Physiol. 2011;301:G519–G527. doi: 10.1152/ajpgi.00101.2011. [DOI] [PubMed] [Google Scholar]
- 59.Keating DJ, Spencer NJ. What is the role of endogenous gut serotonin in the control of gastrointestinal motility? Pharmacol Res. 2019;140:50–55. doi: 10.1016/j.phrs.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 60.Grover M, Farrugia G, Stanghellini V. Gastroparesis: a turning point in understanding and treatment. Gut. 2019;68:2238–2250. doi: 10.1136/gutjnl-2019-318712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dickson I. Microbiota modulate ENS maturation. Nat Rev Gastroenterol Hepatol. 2018;15:454–455. doi: 10.1038/s41575-018-0040-7. [DOI] [PubMed] [Google Scholar]
- 62.Shukla R, Ghoshal U, Dhole TN, Goshal UC. Fecal microbiota in patients with irritable bowel syndrome compared with healthy controls using real-time polymerase chain reaction: an evidence of dysbiosis. Dig Dis Sci. 2015;60:2953–2962. doi: 10.1007/s10620-015-3607-y. [DOI] [PubMed] [Google Scholar]
- 63.Malinen E, Rinttilä T, Kajander K, et al. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–382. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 64.Tap J, Derrien M, Törnblom H, et al. Identification of an intestinal microbiota signature associated with severity of irritable bowel syndrome. Gastroenterology. 2017;152:111–123.:e8. doi: 10.1053/j.gastro.2016.09.049. [DOI] [PubMed] [Google Scholar]
- 65.Salem AE, Singh R, Ayoub YK, Khairy AM, Mullin GE. The gut microbiome and irritable bowel syndrome: state of art review. Arab J Gastroenterol. 2018;19:136–141. doi: 10.1016/j.ajg.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 66.Ghoshal UC, Ghoshal U, Das K, Misra A. Utility of hydrogen breath tests in diagnosis of small intestinal bacterial overgrowth in malabsorption syndrome and its relationship with oro-cecal transit time. Indian J Gastroenterol. 2006;25:6–10. [PubMed] [Google Scholar]
- 67.Ghoshal UC, Ghoshal U, Ayyagari A, et al. Tropical sprue is associated with contamination of small bowel with aerobic bacteria and reversible prolongation of orocecal transit time. J Gastroenterol Hepatol. 2003;18:540–547. doi: 10.1046/j.1440-1746.2003.03006.x. [DOI] [PubMed] [Google Scholar]
- 68.Nakamura K, Sakuragi N, Takakuwa A, Ayabe T. Paneth cell alpha-defensins and enteric microbiota in health and disease. Biosci Microbiota Food Health. 2016;35:57–67. doi: 10.12938/bmfh.2015-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simeonova D, Ivanovska M, Murdjeva M, et al. Recognizing the leaky gut as a trans-diagnostic target for neuroimmune disorders using clinical chemistry and molecular immunology assays. Curr Top Med Chem. 2018;18:1641–1655. doi: 10.2174/1568026618666181115100610. [DOI] [PubMed] [Google Scholar]
- 70.Ghosh D, Porter E, Shen B, et al. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol. 2002;3:583–590. doi: 10.1038/ni797. [DOI] [PubMed] [Google Scholar]
- 71.Salzman NH, Ghosh D, Huttner KM, Peterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 72.Ghoshal UC, Shukla R, Ghoshal U, et al. The gut microbiota and irritable bowel syndrome: friend or foe? Int J Inflam. 2012;2012:151085. doi: 10.1155/2012/151085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pimentel M, Kong Y, Park S. IBS subjects with methane on lactulose breath test have lower postprandial serotonin levels than subjects with hydrogen. Dig Dis Sci. 2004;49:84–47. doi: 10.1023/B:DDAS.0000011607.24171.c0. [DOI] [PubMed] [Google Scholar]
- 74.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 75.Bouhnik Y, Alain S, Attar A, et al. Bacterial populations contaminating the upper gut in patients with small intestinal bacterial overgrowth syndrome. Am J Gastroenterol. 1999;94:1327–1331. doi: 10.1111/j.1572-0241.1999.01016.x. [DOI] [PubMed] [Google Scholar]
- 76.Sachdev AH, Pimentel M. Antibiotics for irritable bowel syndrome: rationale and current evidence. Curr Gastroenterol Rep. 2012;14:439–445. doi: 10.1007/s11894-012-0284-2. [DOI] [PubMed] [Google Scholar]
- 77.Posserud I, Stotzer PO, Björnsson ES, Abrahamsson H, Simrén M. Small intestinal bacterial overgrowth in patients with irritable bowel syndrome. Gut. 2007;56:802–808. doi: 10.1136/gut.2006.108712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bala L, Ghoshal UC, Ghoshal U, et al. Malabsorption syndrome with and without small intestinal bacterial overgrowth: a study on upper-gut aspirate using 1H NMR spectroscopy. Magn Reson Med. 2006;56:738–744. doi: 10.1002/mrm.21041. [DOI] [PubMed] [Google Scholar]
- 79.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. 2018;24:392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 81.Kootte RS, Levin E, Salojärvi J, et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 2017;26:611–619.:e6. doi: 10.1016/j.cmet.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 82.Muscogiuri G, Balercia G, Barrea L, et al. Gut: a key player in the pathogenesis of type 2 diabetes? Crit Rev Food Sci Nutr. 2018;58:1294–1309. doi: 10.1080/10408398.2016.1252712. [DOI] [PubMed] [Google Scholar]
- 83.Bouter KE, van Raalte DH, Groen AK, Nieuwdorp M. Role of the gut microbiome in the pathogenesis of obesity and obesity-related metabolic dysfunction. Gastroenterology. 2017;152:1671–1678. doi: 10.1053/j.gastro.2016.12.048. [DOI] [PubMed] [Google Scholar]
- 84.Johnson AM, Olefsky JM. The origins and drivers of insulin resistance. Cell. 2013;152:673–684. doi: 10.1016/j.cell.2013.01.041. [DOI] [PubMed] [Google Scholar]
- 85.Roland BC, Lee D, Miller LS, et al. Obesity increases the risk of small intestinal bacterial overgrowth (SIBO) Neurogastroenterol Motil. 2018;30 doi: 10.1111/nmo.13199. [DOI] [PubMed] [Google Scholar]
- 86.Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 88.Adamska A, Nowak M, Piłaciński S, et al. Small intestinal bacterial overgrowth in adult patients with type 1 diabetes: its prevalence and relationship with metabolic control and the presence of chronic complications of the disease. Pol Arch Med Wewn. 2016;126:628–634. doi: 10.20452/pamw.3501. [DOI] [PubMed] [Google Scholar]
- 89.Rana SV, Malik A, Bhadada SK, Sachdeva N, Morya RK, Sharma G. Malabsorption, orocecal transit time and small intestinal bacterial overgrowth in type 2 diabetic patients: a connection. Indian J Clin Biochem. 2017;32:84–89. doi: 10.1007/s12291-016-0569-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 91.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 92.Zelante T, Iannitti RG, Cunha C, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 93.Xu CX, Wang C, Zhang ZM, et al. Aryl hydrocarbon receptor deficiency protects mice from diet-induced adiposity and metabolic disorders through increased energy expenditure. Int J Obes (Lond) 2015;39:1300–1309. doi: 10.1038/ijo.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bock KW. Aryl hydrocarbon receptor (AHR) functions in NAD(+) metabolism, myelopoiesis and obesity. Biochem Pharmacol. 2019;163:128–132. doi: 10.1016/j.bcp.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 95.Wang C, Xu CX, Krager SL, Bottum KM, Liao DF, Tischkau SA. Aryl hydrocarbon receptor deficiency enhances insulin sensitivity and reduces PPAR-alpha pathway activity in mice. Environ Health Perspect. 2011;119:1739–1744. doi: 10.1289/ehp.1103593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- 97.Samuel BS, Shaito A, Motoike T, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tolhurst G, Heffron H, Lam YS, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61:364–371. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fava S. Glucagon-like peptide 1 and the cardiovascular system. Curr Diabetes Rev. 2014;10:302–310. doi: 10.2174/1573399810666141030125830. [DOI] [PubMed] [Google Scholar]
- 100.De Vadder F, Kovatcheva-Datchary P, Goncalves D, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 101.Kimura I, Ozawa K, Inoue D, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reigstad CS, Salmonson CE, Rainey JF, 3rd, et al. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015;29:1395–1403. doi: 10.1096/fj.14-259598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Williams BB, Van Benschoten AH, Cimermancic P, et al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe. 2014;16:495–503. doi: 10.1016/j.chom.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ghoshal UC. Gut microbiota-brain axis modulation by a healthier microbiological microenvironment : facts and fictions. J Neurogastroenterol Motil. 2018;24:4–6. doi: 10.5056/jnm17150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Quigley EMM. Microbiota-brain-gut axis and neurodegenerative diseases. Curr Neurol Neurosci Rep. 2017;17:94. doi: 10.1007/s11910-017-0802-6. [DOI] [PubMed] [Google Scholar]
- 106.Garcovich M, Zocco MA, Roccarina D, Ponziani FR, Gasbarrini A. Prevention and treatment of hepatic encephalopathy: focusing on gut microbiota. World J Gastroenterol. 2012;18:6693–700. doi: 10.3748/wjg.v18.i46.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.De Palma G, Collins SM, Bercik P. The microbiota-gut-brain axis in functional gastrointestinal disorders. Gut Microbes. 2014;5:419–429. doi: 10.4161/gmic.29417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rhee SH, Pothoulakis C, Mayer EA. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat Rev Gastroenterol Hepatol. 2009;6:306–314. doi: 10.1038/nrgastro.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sharkey KA, Beck PL, McKay DM. Neuroimmunophysiology of the gut: advances and emerging concepts focusing on the epithelium. Nat Rev Gastroenterol Hepatol. 2018;15:765–784. doi: 10.1038/s41575-018-0051-4. [DOI] [PubMed] [Google Scholar]
- 110.Salzman NH, Underwood MA, Bevins CL. Paneth cells, defensins, and the commensal microbiota: a hypothesis on intimate interplay at the intestinal mucosa. Semin Immunol. 2007;19:70–83. doi: 10.1016/j.smim.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 111.Fung TC, Vuong HE, Luna CDG, et al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat Microbiol. 2019;4:2064–2073. doi: 10.1038/s41564-019-0540-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Robinette ML, Colonna M. GI motility: microbiota and macrophages join forces. Cell. 2014;158:239–240. doi: 10.1016/j.cell.2014.06.040. [DOI] [PubMed] [Google Scholar]
- 114.Mikkelsen HB. Interstitial cells of cajal, macrophages and mast cells in the gut musculature: morphology, distribution, spatial and possible functional interactions. J Cell Mol Med. 2010;14:818–832. doi: 10.1111/j.1582-4934.2010.01025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Muller PA, Koscsó B, Rajani GM, et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. 2014;158:300–313. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Olofsson PS, Rosas-Ballina M, Levine YA, Tracey K. Rethinking inflammation: neural circuits in the regulation of immunity. Immunol Rev. 2012;248:188–204. doi: 10.1111/j.1600-065X.2012.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Parkhurst CN, Yang G, Ninan I, et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell. 2013;155:1596–1609. doi: 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Winer DA, Luck H, Tsai S, Winer S. The intestinal immune system in obesity and insulin resistance. Cell Metab. 2016;23:413–426. doi: 10.1016/j.cmet.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 119.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 120.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117:175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brestoff JR, Kim BS, Saenz SA, et al. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature. 2015;519:242–246. doi: 10.1038/nature14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Garidou L, Pomié C, Klopp P, et al. The gut microbiota regulates intestinal CD4 T cells expressing RORγt and controls metabolic disease. Cell Metab. 2015;22:100–112. doi: 10.1016/j.cmet.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 123.Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 124.Thaiss CA, Levy M, Grosheva I, et al. Hyperglycemia drives intestinal barrier dysfunction and risk for enteric infection. Science. 2018;359:1376–1383. doi: 10.1126/science.aar3318. [DOI] [PubMed] [Google Scholar]
- 125.Chen RY, Kung VL, Das S, et al. Duodenal microbiota in stunted undernourished children with enteropathy. N Engl J Med. 2020;383:321–333. doi: 10.1056/NEJMoa1916004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gehrig JL, Venkatesh S, Chang HW, et al. Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science. 2019;365:eaau4732. doi: 10.1126/science.aau4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ghoshal UC, Srivastava D, Verma A, Ghoshal U. Tropical sprue in 2014: the new face of an old disease. Curr Gastroenterol Rep. 2014;16:391. doi: 10.1007/s11894-014-0391-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sorini C, Cosorich I, Lo Conte M, et al. Loss of gut barrier integrity triggers activation of islet-reactive T cells and autoimmune diabetes. Proc Natl Acad Sci USA. 2019;116:15140–15149. doi: 10.1073/pnas.1814558116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Camilleri M. Leaky gut: mechanisms, measurement and clinical implications in humans. Gut. 2019;68:1516–1526. doi: 10.1136/gutjnl-2019-318427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Barbara G, Grover M, Bercik P, et al. Rome foundation working team report on post-infection irritable bowel syndrome. Gastroenterology. 2019;156:46–58.:e7. doi: 10.1053/j.gastro.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jess T. Microbiota, antibiotics, and obesity. N Engl J Med. 2014;371:2526–2528. doi: 10.1056/NEJMcibr1409799. [DOI] [PubMed] [Google Scholar]
- 132.Cox LM, Yamanishi S, Sohn J, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Scott FI, Horton DB, Mamtani R, et al. Administration of antibiotics to children before age 2 years increases risk for childhood obesity. Gastroenterology. 2016;151:120–129.:e5. doi: 10.1053/j.gastro.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yano JM, Yu K, Donaldson GP, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shah A, Morrison M, Holtmann G. A novel treatment for patients with constipation: dawn of a new age for translational microbiomeresearch? Indian J Gastroenterol. 2018;37:388–391. doi: 10.1007/s12664-018-0912-3. [DOI] [PubMed] [Google Scholar]
- 136.Low K, Hwang L, Hua J, Zhu A, Morales W, Pimentel M. A combination of rifaximin and neomycin is most effective in treating irritable bowel syndrome patients with methane on lactulose breath test. J Clin Gastroenterol. 2010;44:547–550. doi: 10.1097/MCG.0b013e3181c64c90. [DOI] [PubMed] [Google Scholar]
- 137.Ghoshal UC, Srivastava D, Misra A. A randomized double-blind placebo-controlled trial showing rifaximin to improve constipation by reducing methane production and accelerating colon transit: apilot study. Indian J Gastroenterol. 2018;37:416–423. doi: 10.1007/s12664-018-0901-6. [DOI] [PubMed] [Google Scholar]
- 138.Reijnders D, Goossens GH, Hermes GD, et al. Effects of gut microbiota manipulation by antibiotics on host metabolism in obese humans: a randomized double-blind placebo-controlled trial. Cell Metab. 2016;24:63–74. doi: 10.1016/j.cmet.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 139.Rao SSC, Bhagatwala J. Small intestinal bacterial overgrowth: clinical features and therapeutic management. Clin Transl Gastroenterol. 2019;10:e00078. doi: 10.14309/ctg.0000000000000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gatta L, Scarpignato C. Systematic review with meta-analysis: rifaximin is effective and safe for the treatment of small intestine bacterial overgrowth. Aliment Pharmacol Ther. 2017;45:604–616. doi: 10.1111/apt.13928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shah SC, Day LW, Somsouk M, Sewell JL. Meta-analysis: antibiotic therapy for small intestinal bacterial overgrowth. Aliment Pharmacol Ther. 2013;38:925–934. doi: 10.1111/apt.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Khoruts A, Sadowsky MJ. Understanding the mechanisms of faecal microbiota transplantation. Nat Rev Gastroenterol Hepatol. 2016;13:508–516. doi: 10.1038/nrgastro.2016.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.El-Salhy M, Hatlebakk JG, Gilja OH, Kristoffersen AB, Hausken T. Efficacy of faecal microbiota transplantation for patients with irritable bowel syndrome in a randomised, double-blind, placebo-controlled study. Gut. 2020;69:859–867. doi: 10.1136/gutjnl-2019-319630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ianiro G, Eusebi LH, Black CJ, Gasbarrini A, Cammarota G Ford ACl, author. Systematic review with meta-analysis: efficacy of faecal microbiotatransplantation for the treatment of irritable bowel syndrome. Aliment Pharmacol Ther. 2019;50:240–248. doi: 10.1111/apt.15330. [DOI] [PubMed] [Google Scholar]
- 145.Xu D, Chen VL, Steiner CA, et al. Efficacy of fecal microbiota transplantation in irritable bowel syndrome: a systematic review and meta-analysis. Am J Gastroenterol. 2019;114:1043–1050. doi: 10.14309/ajg.0000000000000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Park JH. Clinical usefulness of fecal microbiota transplantation. J Neurogastroenterol Motil. 2017;23:149–150. doi: 10.5056/jnm17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vrieze A, Van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–916.:e7. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 148.Hemarajata P, Versalovic J. Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Therap Adv Gastroenterol. 2013;6:39–51. doi: 10.1177/1756283X12459294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhong C, Qu C, Wang B, Liang S, Zeng B. Probiotics for preventing and treating small intestinal bacterial overgrowth: a meta-analysis and systematic review of current evidence. J Clin Gastroenterol. 2017;51:300–311. doi: 10.1097/MCG.0000000000000814. [DOI] [PubMed] [Google Scholar]
- 150.Enck P, Aziz Q, Barbara G, et al. Irritable bowel syndrome. Nat Rev Dis Primers. 2016;2:16014. doi: 10.1038/nrdp.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang Y, Dilidaxi D, Wu Y, Sailike J, Sun X, Nabi XH. Composite probiotics alleviate type 2 diabetes by regulating intestinal microbiota and inducing GLP-1 secretion in db/db mice. Biomed Pharmacother. 2020;125:109914. doi: 10.1016/j.biopha.2020.109914. [DOI] [PubMed] [Google Scholar]
- 152.Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019;16:35–56. doi: 10.1038/s41575-018-0061-2. [DOI] [PubMed] [Google Scholar]
- 153.Pulipati P, Sarkar P, Jakkampudi A, et al. The Indian gut microbiota-is it unique? Indian J Gastroenterol. 2020;39:133–140. doi: 10.1007/s12664-020-01037-8. [DOI] [PubMed] [Google Scholar]
- 154.Delzenne NM, Bindels LB. Food for thought about manipulating gut bacteria. Nature. 2020;577:32–34. doi: 10.1038/d41586-019-03704-z. [DOI] [PubMed] [Google Scholar]
- 155.Walker AW, Ince J, Duncan SH, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mehtab W, Agarwal A, Singh N, Malhotra A, Makharia G. All that a physician should know about FODMAPs. Indian J Gastroenterol. 2019;38:378–390. doi: 10.1007/s12664-019-01002-0. [DOI] [PubMed] [Google Scholar]
- 159.Altobelli E, Del Negro V, Angeletti PM, Letella G. Low-FODMAP diet improves irritable bowel syndrome symptoms: a meta-analysis. Nutrients. 2017;9:940. doi: 10.3390/nu9090940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tomova A, Bukovsky I, Rembert E, et al. The effects of vegetarian and vegan diets on gut microbiota. Front Nutr. 2019;6:47. doi: 10.3389/fnut.2019.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]