ABSTRACT
A better understanding of genetic influences on early white matter development could significantly advance our understanding of neurological and psychiatric conditions characterized by altered integrity of axonal pathways. We conducted a genome-wide association study (GWAS) of diffusion tensor imaging (DTI) phenotypes in 471 neonates. We used a hierarchical functional principal regression model (HFPRM) to perform joint analysis of 44 fiber bundles. HFPRM revealed a latent measure of white matter microstructure that explained approximately 50% of variation in our tractography-based measures and accounted for a large proportion of heritable variation in each individual bundle. An intronic SNP in PSMF1 on chromosome 20 exceeded the conventional GWAS threshold of 5 x 10−8 (p = 4.61 x 10−8). Additional loci nearing genome-wide significance were located near genes with known roles in axon growth and guidance, fasciculation, and myelination.
Keywords: diffusion tensor imaging, genome-wide association study, infant, magnetic resonance imaging
Introduction
Within the human brain, structurally segregated and functionally specialized regions communicate with each-other via a dense network of axonal pathways (Hagmann et al. 2008). Damage to these pathways, in the form of white matter lesions, has long been recognized to produce functional deficits or disconnection syndromes (Wernicke 1874; Dejerine 1892; Liepmann and Maas 1907; Aralasmak et al. 2006). More recently, diffusion tensor imaging (DTI) has been used to reconstruct the trajectories of fiber systems in three dimensional space and to assess the microstructural integrity of axonal pathways using measures such as fractional anisotropy (FA). Individual differences in FA have been linked to information processing speed and intelligence (Penke et al. 2010; Tamnes et al. 2010; Penke et al. 2012; Ritchie et al. 2015) and to the presence of psychiatric conditions including schizophrenia (Yao et al. 2013; Tamnes and Agartz 2016), autism (Ameis and Catani 2015), and attention deficit hyperactivity disorder (ADHD) (Chen et al. 2016a). Altered integrity of axonal pathways is also evident in many different neurological disorders including dyslexia (Norton et al. 2015), Angelman Syndrome (Peters et al. 2011), Rhett syndrome (Mahmood et al. 2010), and, unsurprisingly, various congenital disorders of axon guidance (Engle 2010; Poretti et al. 2013). Consequently, a better understanding of the factors influencing individual variation in white matter microstructure could have important implication for understanding variation in intelligence as well as the etiology of many psychiatric and neurological disorders.
The prenatal and early postnatal period represents the foundational period in the establishment of human brain connectivity (Stiles and Jernigan 2010). First, each growing axon must travel to its correct synaptic target while ignoring many inappropriate neuronal partners. This process relies on a variety of guidance molecules, both attractive and repellant, as well as a properly functioning growth cone (Kandel 2013). Extracellular matrix components may also play a role in axonal elongation and serve as guidance cues that distinguish afferent from efferent pathways (Pearlman and Sheppard 1996). During this phase, axons may adhere to (fasciculate) with other axons taking the same route, forming white matter bundles. This process relies on a variety of cell and substrate adhesion molecules such as NCAM1 (Van Vactor 1998) and is associated with increases in FA (Qiu et al. 2015). Several fiber bundles, including the corpus callosum (CC), fornix, and hemispheric stalk (containing all the projection fibers of the developing internal capsule) are present as early as 12 weeks gestational age (Vasung et al. 2010) and can be detected via DTI by 19 weeks of age (Huang et al. 2006). Between 20 and 23 weeks gestation, thalamocortical fibers reach the subplate zone (Kostovic and Goldman-Rakic 1983; Krmpotic-Nemanic et al. 1983; Kostovic and Rakic 1984, 1990), where they ‘wait’ for a prolonged period before growing into the cortical plate during gestational weeks 24 through 32 (Kostovic and Goldman-Rakic 1983). Callosal afferents and long corticocortical pathways reach the subplate somewhat later and grow into the cortical plate between 33 and 35 week gestation (Kostovic and Jovanov-Milosevic 2006). By birth, major commissural bundles (genu, body and splenium of the CC), projection bundles (corticofugal and corticothalamic tracts), limbic bundles (fornix and cingulate) and associative bundles (uncinate, arcuate, superior and inferior longitudinal fascicles) can be detected and tracked with DTI (Dubois et al. 2014). The next phase in white matter development, which begins prenatally and continues into the postnatal period, involves the proliferation and maturation of glial cells (Back et al. 2001; Sigaard et al. 2016). It has been argued that this process is relatively isotropic and hence should not change FA (Dubois et al. 2008). However, a study in perinatal rabbits suggests that developmental expansion of immature oligodendrocytes is associated with increasing FA (Drobyshevsky et al. 2005). Finally, the postnatal period witnesses both extensive axonal pruning and myelination (the ensheathment of axons in myelin protein by oligodendrocytes) (Yakovlev and Lecours 1967; Kinney et al. 1988; Lamantia and Rakic 1990; Galea and Darian-Smith 1995). Maturation of white matter during this phase is accompanied by increasing FA (Qiu et al. 2015).
All these processes involve the precise spatiotemporal regulation of gene expression. It is thus reasonable to suppose that genetic variation may influence individual differences in white matter development. Indeed, genetic mutations can produce a range of axonal guidance disorders including corpus callosum agenesis or dysgenesis, L1 syndrome, Kallmann syndrome, and Joubert syndrome (JS) (Engle 2010; Nugent et al. 2012). In addition, twin studies indicate significant genetic influences on whole brain FA (h2 between 0.24 and 0.88) and average FA within individual white matter tracts (h2 between 0 and 1.00) in infancy, childhood, and adulthood (Pfefferbaum et al. 2001; Brouwer et al. 2010; Chiang et al. 2011; Brouwer et al. 2012; Kanchibhotla et al. 2014; Kochunov et al. 2014; Budisavljevic et al. 2015; Kochunov et al. 2015; Lee et al. 2015; Budisavljevic et al. 2016; Vuoksimaa et al. 2017).
To further delineate how genetic variation impacts white matter microstructure in early infancy, we conducted a genome-wide association study (GWAS) of DTI phenotypes in a unique cohort of infants who received high-resolution magnetic resonance imaging (MRI) scans of the brain around 5 weeks ofage.
Materials and Methods
Subjects
Mothers were recruited from outpatient obstetrics and gynecology clinics at University of North Carolina hospitals and represent a subset of 1330 infants enrolled in the Early Brain Development Study (EBDS) based at the University of North Carolina at Chapel Hill (Gilmore et al. 2007; Knickmeyer et al. 2008; Gilmore et al. 2010; Knickmeyer et al. 2017). We acquired DTI scans from 857 children around five weeks of age with 663 passing neuroimaging quality control (77% success). We acquired buccal samples from 852 infants with 756 passing genotyping quality control (89% success rate). In total, 471 infants (259 male, 212 female) between 35.0 and 57.3 days of age had both high quality genetic data and high quality DTI images, encompassing 259 singletons or unpaired twins, 11 sibling pairs, and 95 twin pairs (51 same-sex DZ pairs, 12 opposite-sex DZ pairs, and 32 MZ pairs). See Table 1 for additional demographic information. See Figure 1 for a flow chart of dataloss.
Table 1.
Descriptive statistics for demographic and medical history variables
| Continuous Variables | Mean | SD | Min | Max | |
|---|---|---|---|---|---|
| Age at MRI (days post conception) | 293.4 | 16.6 | 245 | 401 | |
| Gestational Age at Birth (weeks) | 37 | 3 | 27 | 42 | |
| Birthweight (grams) | 2751.7 | 731.4 | 790 | 4820 | |
| 5 Minute Apgar Score | 8.6 | 0.8 | 3 | 10 | |
| Maternal Age (years) | 29.4 | 5.8 | 16 | 47 | |
| Paternal Age (years) | 32.0 | 6.7 | 18 | 64 | |
| Maternal Education (years) | 15.1 | 3.2 | 8 | 25 | |
| Paternal Education (years) | 15.0 | 3.3 | 2 | 26 | |
| Categorical Variables | No. | Percent | |||
| Gestation Number | Twin | 262 | 56 | ||
| Gender | Male | 259 | 55 | ||
| NICU Stay > 24 Hours | Yes | 92 | 20 | ||
| Caesarian Section | Yes | 253 | 54 | ||
| Maternal Ethnicity | White Black Asian Native American |
349 114 6 2 |
74 24 1 >1 |
||
| Paternal Ethnicity | White Black Asian Native American Missing |
327 121 12 6 5 |
69 26 3 1 1 |
||
| Maternal Psychiatric1 History | Yes | 176 | 37 | ||
| Paternal Psychiatric1 History | Yes | 58 | 12 | ||
| Income2 | High Middle Low Missing |
134 134 178 25 |
29 29 38 5 |
||
| Maternal Smoking | Yes | 56 | 12 | ||
1Maternal psychiatric history and paternal psychiatric history were treated as binary variables with a positive if mothers reported a diagnosis in any of the following DSM-V categories, or if maternity and pediatric medical records indicated such a diagnosis: schizophrenia spectrum and other psychotic disorders, bipolar and related disorders, depressive disorders, anxiety disorders, obsessive–compulsive and related disorders, attention-deficit hyperactivity disorders, Tourette’s syndrome, or autism-spectrum disorders. 2low income: at or below 200% of federal poverty level (FPL), middle income: between 200 and 400% of FPL, high income: above 400% ofFPL.
Figure 1 .
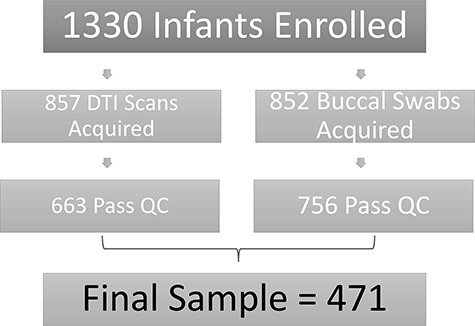
Flow chart showing data loss across the course of the study.
Image acquisition and analysis
Diffusion tensor images were acquired using a single shot echo planar spin echo sequence following two protocols. Under the first protocol (N = 260), 5 repetitions of 7 diffusion weighted images (in total 35) were generated: 1 without diffusion gradient (b = 0) and 6 with b = 1000 s/mm2 in unique directional diffusion gradients (TR = 5200 ms, TE = 73 ms, slice thickness = 2 mm, in-plane resolution = 2 mm x 2 mm). Under the second protocol (N = 211), a total of 49 images were acquired, 7 without diffusion gradients (b = 0) and 42 with b = 1000 s/mm2 in unique directional diffusion gradients (TR = 7680 ms, TE = 82 ms, slice thickness = 2 mm, in-plane resolution = 2 mm × 2 mm). 436 children were scanned on a 3 T Siemens Allegra head-only scanner (260 with protocol 1 and 176 with protocol 2). 35 children were scanned on a 3 T Siemens TIM Trio scanner (all with protocol 2).
Automated quality control was performed with DTIPrep (Oguz et al. 2014). This includes controlling for correct image dimensions and gradient directions, detecting slice-wise intensity change and excessive motion artifacts, and correcting for motion and eddy current effects. Diffusion images with large motion artifacts and missing or corrupted sections were excluded from analysis. Additional expert-guided QC was performed with 3DSlicer (Verde et al. 2013). Next, automatic brain masking was performed with manual corrections if necessary. See Figure 2 for an example DTI image after brain masking and a slice of the neonate DTI atlas.
Figure 2 .
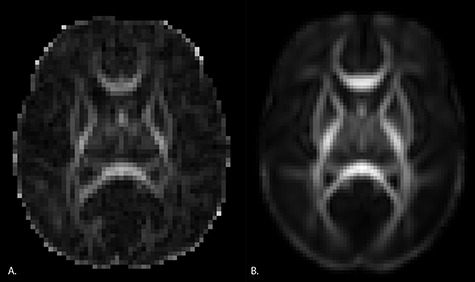
Example slices from an individual subject image and the neonate DTI atlas. Panel A. Sample axial slice from an individual subject. B. Axial slice from the neonate DTI atlas.
Functional fiber profile analysis was performed using a neonate specific adaptation of the UNC-Utah NA-MIC DTI pipeline (Verde et al. 2014). This includes (1) mapping of resulting images into a neonate DTI atlas space, (2) mapping atlas fibers into individual subject space, and (3) extracting FA profiles using DTIAtlasFiberAnalyzer (Goodlett et al. 2009). The neonate DTI atlas consists of the unbiased symmetric diffeomorphic transformed average of 144 neonatal DTI images upon which 47 fiber tracts of interest are defined. See Jha et al. (2016) for additional details. Individual FA profiles along each tract were correlated with atlas profiles to determine how well the fibers mapped into atlas space. Three tracts (the temporoparietal segment of the left arcuate, the segment of the right cingulum that originates in the posterior cingulate gyrus and terminates in the hippocampus, and the right corticofugal tract that connects to premotor cortex) were excluded from further analysis as more than 25% of the FA fiber profiles had correlation values below 0.7.
Genome-wide genotyping.
Genotyping of single nucleotide polymorphisms (SNPs) was carried out using Affymetrix Axiom Genome-Wide LAT and Exome arrays. We excluded samples with low DishQC as described at https://biobank.ctsu.ox.ac.uk/crystal/docs/axiom_geno_analguide.pdf (<0.82 for LAT array and < 0.79 for Exome array), low call rates (<95%), outliers for homozygosity, sex or zygosity from genotypes inconsistent with reported phenotypes, excessive relatedness (subjects with ≥6 related subjects, where relatedness is defined as proportion of identity by descent (IBD) > 0.125), and unexpected relatedness (IBD > 0.4 for subjects that were not known siblings). We also evaluated the sample for ancestry outliers, but none were detected. We removed individual SNPs that deviated from Hardy–Weinberg equilibrium (PHWE < 1x10−8), had low call rate (<95%), high Mendelian error rate (>0.1, based on 5 parent–child trios), high deviation of allele frequency compared to European American and African American subsets from the 1000 Genomes (1000G) Project (Abecasis et al. 2010; Abecasis et al. 2012) (either P < 1x10−5 and frequency difference > 0.07, or frequency difference > 0.15), and that did not match 1000G EUR/AFR founders. Population stratification was assessed using PCA (Price et al. 2006). Individuals of European ancestry constituted 59% of the sample. Remaining subjects were primarily of African ancestry. Imputation was performed with MACH-Admix (Liu et al. 2013) using 1000G reference panel (phase1_release_v3.20101123) (Abecasis et al. 2010; Abecasis et al. 2012). To evaluate the quality of imputed SNPs, we computed mean R2 for varying MAF categories and R2 cutoffs from 0.3 to 0.8 as described in Wright et al. (2014) and Xia et al. (2017). For each MAF category, we selected an R2 cutoff which produced an average R2 > 0.8 for the category. SNPs below this cutoff were excluded as were SNPs with minor allele frequency < 0.01.
Statistical Methods
Image analysis produced FA measures along 44 fiber tracts. To reduce the heterogeneity in variance scales over the whole brain (Jolliffe and Cadima 2016), FA measures in each tract were rescaled by dividing by the total standard deviation. To reduce the dimension of the outcomes, functional principal component analysis (Hall et al. 2006) was applied to each fiber tract and the first five functional principal components (fPCs) were extracted as these accounted for more than 70% of total variation. The first factor was used as the outcome in subsequent twin-based heritability analyses and GWAS (Zhang et al. 2017).
We first adopted the ACE model to calculate the heritability for individual tracts and the first tractography factor. For each tract curve, the heritabilities at all sampled points were calculated and an average value was reported. We also calculated the proportion of genetic variance after subtracting the common factor to the total variance for individual tracts as a comparison.
In the genome-wide association analysis, to control for population stratification, the first three genotypic principal components were included as covariates as was scanner type and DTI protocol. Additional covariates were selected via adaptive lasso from a comprehensive set of demographic and clinical variables (Ahn et al. 2012). These included gestational age at birth and age at MRI. To account for subject correlation within twin pairs, an ACE model was applied as described in Xia et al. (2017). Likelihood ratio statistics were calculated to test the significance of genetic associations. Variants with p-values less than 5 x 10−8 were considered as genome-wide significant. We also report genetic markers above the conventional suggestive threshold, i.e., SNPs with p-values less than 5 x 10−6. The phenotypic variance explained by each variant was calculated as h2 = β’2var(X)/(β’2var(X) + σa2+ σc2+ σe2), where β’ is the SNP effect size adjusted for the overestimation caused by winner’s curse (Ghosh et al. 2008) through an approximate conditional likelihood approach; var(X) is the variance of the genotype, and σa2, σc2 and σe2 are the variance explained by shared genetic effect, shared environment effect and random effect respectively. Linkage disequilibrium (LD)-pruning was then performed (using a 50 kbp moving window at a step of 5 kbp with r2 threshold of 0.5) to extract one representative variant at each locus. LD-independent SNPs with p-values less than 5 x 10−6 were annotated using GTEx (Consortium et al. 2017), HaploReg (Ward and Kellis 2012), and RegulomeDB (Boyle et al. 2012). The nearest protein coding gene for each SNP (± 500 kb) was annotated using GeneCards® (http://www.genecards.org/), Online Mendelian Inheritance in Man (http://www.omim.org), the NHGRI-EBI GWAS catalog (https://www.ebi.ac.uk/gwas/), and Mouse Genome Informatics (http://www.informatics.jax.org).
To examine potential confounding by population stratification, we calculated fixation index (FST) for all LD-independent variants passing the suggestive threshold. In addition, we calculated the frequencies of the effect alleles for different populations in the neonate samples using both self-reported ethnicity and genetic ancestry, and crosschecked our data with allele frequencies from the 1000 Genomes Project (1000G) (phase I, version 3) (Auton et al. 2015).
Sensitivity analyses were performed to examine the robustness of the GWAS findings. Specifically, we calculated the effect of the top SNPs in the following subsamples: individuals with European ancestry (based on PCA, N = 277), individuals with non-European ancestry (based on PCA, N = 194), female subjects (N = 212), male subjects (N = 259), individuals born with gestational age later than 32 weeks (N = 432), a subsample excluding children who spent time in neonatal intensive care (N = 345), a subsample excluding subjects with confirmed diagnosis of maternal schizophrenia or bipolar disorder (N = 443), and a subsample of unrelated individuals (N = 365). We also performed a sensitivity analysis with reduced covariates by only including age at MRI, scanner type, DTI direction, and the first three genetic principal components in the model. To further check the validity of our findings, we also performed a permutation test for the most significant SNP in our GWAS analysis. The empirical p-value is reported.
Enrichment tests of prenatally expressed genes in brain tissues were conducted using two genome-wide gene expression datasets, the microarray-based BrainCloud dorsolateral prefrontal cortex transcriptome (Colantuoni et al. 2011) and the RNA sequencing-based Brainspan transcriptome of 15 brain regions (Kang et al. 2011). Specifically, each LD-pruned SNP was mapped to its nearest gene within a 2Mbp distance. A log2 fold change equal to or greater than 0.5 in prenatal tissues compared to postnatal tissues was considered as elevated prenatal expression. The proportions of genes with elevated prenatal expression were calculated among all genes and among genes corresponding to variants passing a given p-value threshold. Then a two-sample z-test was performed to evaluate if genes selected by GWAS showed enrichment for prenatal expression. Various p-value cut-offs were adopted to flexibly incorporate signals at multiple levels. For all LD-independent variants with p-values smaller than 5 x 10−6, we also examined development gene expression patterns for the nearest protein-coding gene (± 500kbp) using Brainspan and BrainCloud data, and evaluated tissue-specific gene and protein expression using data from FANTOM (Lizio et al. 2015) and the human proteome map (Kim et al. 2014).
Gene-set-enrichment analysis (GSEA) was conducted using MAGMA (de Leeuw et al. 2015). Gene-level p-values were first calculated based on GWAS results and pathway analysis was then performed. We focused on candidate pathways involved in white matter development as represented in the Gene Ontology database of Biological Processes (Ashburner et al. 2000; The Gene Ontology Consortium 2017). This included axon ensheathment in the central nervous system, axonal fasciculation, oligodendrocyte development, negative and positive regulation of oligodendrocyte differentiation, regulation of oligodendrocyte differentiation, oligodendrocyte differentiation, negative and positive regulation of axon extension, axon extension, myelin assembly, positive regulation of myelination, regulation of myelination, negative regulation of axon guidance, regulation of axon guidance, negative and positive regulation of axonogenesis, central nervous system axonogenesis, regulation of axonogenesis, central nervous system projection neuron axonogenesis, and central nervous system development. We used version v4.0 for pathway and ontology data (accessed on 1/30/2015).
We also used functional analysis of diffusion tensor tract statistics (FADTTS) (Zhu et al. 2011) to test for associations between our top loci and FA within and along the individual fiber tracts. FADTTS provides a global test statistic for each tract as well as local test statistics along each fiber tract. Test statistics and local p-values were merged onto the corresponding fiber locations for visualization.
Finally, to explore the effect of rare genetic copy number variations (CNVs), we ran a genome-wide burden analysis. We applied PennCNV and Partek Genomics Suite to perform CNV calling from signal intensity files of the Affymetrix Axiom Genotype array (Szatkiewicz et al. 2013). CNV detected in both PennCNV and Partek were considered as candidate calls. CNVs with probe coverage < 15, length < 100 kb or PennCNV confidence score < 100 were removed from the analysis. To further identify rare CNVs, we excluded calls that have more than 50% overlap with common CNVs (frequency > 1% in EUR population) or within regions of known rearrangement (chr14:105573485–107 197 774 and chr22:22473003–23 188 329). The ACE model was then applied to test the association between CNV burden (number of CNV-affected genes per subject) and the DTI tractography factor, with the same set of covariates as in the GWAS. We also identified subjects with rare CNVs that overlapped with known neuropsychiatric CNVs and examined their DTI factors.
Results
The first tractography factor is highly heritable (heritability equals 0.547, p = 5.10 x 10−7) and explains approximately 47.74% of phenotypic variation across all tracts (Fig. 3, Panel A.). The first tractography factor also explains a large portion of variance within the majority of tracts (Fig. 3, Panel B.). Tracts in which the common factor explains less than 20% of the variance include the right inferior longitudinal fasciculus, right corticofugal motor tracts, and the segment of the corpus callosum connecting the left and right motor cortex. The first tractography factor also accounts for a large proportion of heritable variation in each tract as can be seen in Figure 4.
Figure 3 .
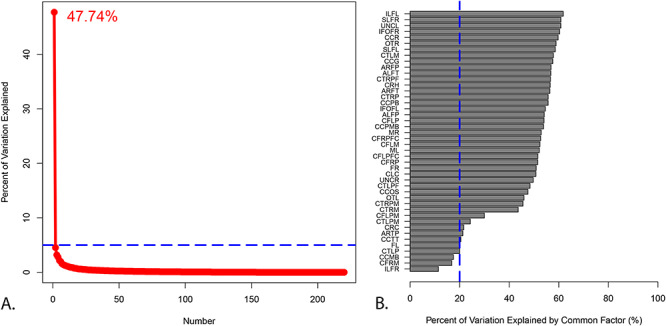
Factor analysis of diffusion tensor imaging data shows that the first tractography factor explains a substantial amount of variation across and within tracts. Panel A. Scree plot of the quantitative tractography factor analysis with the percent of variation explained by the first factor. Panel B. Percent of variation explained by the first factor in each individual tract. An explanation of these abbreviations can be found in Supplementary Table 6.
Figure 4 .
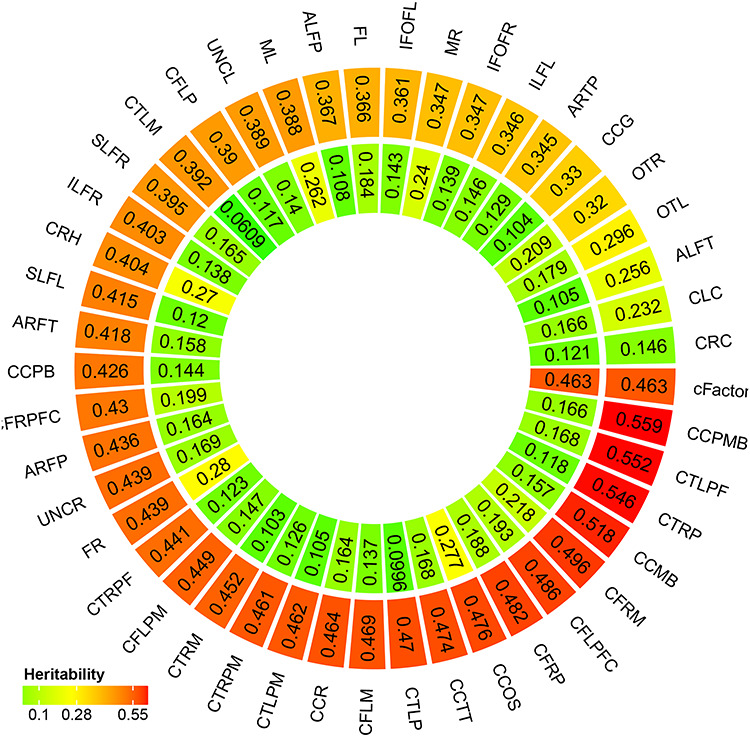
A latent measure of white matter microstructure accounts for a substantial proportion of heritable variation in individual tracts. Outer circle represents average heritability of each white matter tract. Inner circle represents proportion of genetic variation of each tract after subtracting the first tractography factor to the total variation. An explanation of these abbreviations can be found in Supplementary Table 6.
Table 2 displays all LD-independent SNPs with p-values less than 5 x 10−6. Manhattan plots and quantile-quantile plots are displayed in Figure 5. Lambda is 1.039 and Lambda-1000 is 1.083. Locus zoom plots are displayed in Supplementary Figure 1 with four histone marker tracts: H3K27me3, H3K4me3, H3K36me3 and H3K27ac. H3K27me3 indicates genes that may be repressed by Polycomb group proteins (Kharchenko et al. 2011). H3K4me3 indicates promoter and transcription start site (TSS)-proximal regions (Kharchenko et al. 2011). H3K36me3 enrichment is primarily found over exonic regions of transcribed genes (Kharchenko et al. 2011), and H3K27ac indicates enhancers (Pradeepa 2017).
Table 2.
LD-independent SNPs with p-values smaller than 5 x 10−6
| Best SNP | Chr | A11 | A2 | MAF | Effect size | p-value | Variance explained (%) | Function Class | Closest protein coding gene (± 500 kb) |
|---|---|---|---|---|---|---|---|---|---|
| rs6077860+ | 20 | A | C | 0.21 | 1.27 | 4.61E-08 | 2.13 | Intronic | PSMF1 |
| rs1446965+ | 1 | A | G | 0.11 | -1.73 | 7.60E-08 | 1.95 | Downstream | APCS |
| rs72830077+ | 5 | G | A | 0.02 | 4.01 | 3.32E-07 | 1.17 | Intronic | TENM2 |
| rs6777575 | 3 | G | T | 0.34 | 1.29 | 5.21E-07 | 0.92 | Intronic | MAP3K13 |
| rs151087896 | 11 | T | TTTC | 0.23 | 1.57 | 1.08E-06 | 0.33 | Intergenic | B3GAT1 |
| rs78070351 | 20 | G | A | 0.08 | -1.72 | 1.78E-06 | 0.05 | Intronic | UQCC1 |
| rs28627209 | 18 | G | A | 0.10 | -1.82 | 2.26E-06 | 0.03 | Intronic | NFATC1 |
| rs2216360 | 3 | C | T | 0.19 | 1.26 | 3.32E-06 | 0.02 | Intronic | MECOM |
| rs17004715+ | 21 | G | A | 0.04 | 2.69 | 3.34E-06 | 0.02 | Intronic; Splice region variant | ITGB2 |
| rs7366960 | 1 | A | T | 0.42 | 1.19 | 3.84E-06 | 0.02 | Downstream | SLC27A3 |
| rs114172604 | 6 | A | G | 0.03 | 2.64 | 3.94E-06 | 0.02 | In LOC105377975 (lncRNA) | MAN1A1 |
| rs11876680 | 18 | C | A | 0.08 | -1.71 | 4.39E-06 | 0.02 | Intronic | SETBP1 |
| rs79045984+ | 15 | G | A | 0.02 | -3.68 | 4.61E-06 | 0.02 | Intronic | SCAPER |
| rs2002371 | 1 | T | C | 0.27 | -1.07 | 4.62E-06 | 0.01 | Intronic | CNIH3 |
| rs16978169 | 18 | C | T | 0.08 | -1.70 | 4.74E-06 | 0.02 | Intronic | SETBP1 |
Note: We define LD-independent SNPs as those with low LD (r2 < 0.1) to a more significantly associated SNP within a 500 kb window. +directly genotyped (all others are imputed); 1A1 is referred to as the effect allele.
Figure 5 .
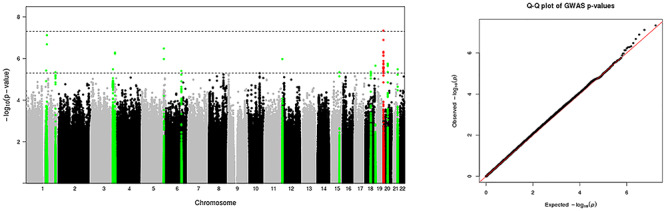
Common genetic variation associated with neonatal white matter microstructure. Green dots represent loci within +/− 500 kb of loci exceeding the “suggestive” association threshold (p-values less than 5 x 10–6). Red dots represent loci within +/− 500 kb of loci exceeding the conventional GWAS association threshold (p-values less than 5 x 10–8).
An intronic SNP in PSMF1 on chromosome 20 exceeded the conventional GWAS threshold of 5 x 10−8. Additional loci nearing genome-wide significance were located within 500 kb of genes with known roles in neurite outgrowth, fasciculation, and axonal pathfinding (Supplementary Table 1). Many loci were located in regulatory regions as indicated by specific histone modifications (See Supplementary Table 2 and Supplementary Fig. 1). Most of the SNPs in Table 2 alter regulatory motifs, though only one is considered likely to disrupt binding by RegulomeDB: the intronic SNP in NFATC1. An intronic SNP in TENM2 achieved a score in RegulomeDB of 3a (TF binding + any motif + DNase peak) and is notable in that the altered motif is for NFATc1 (Nuclear factor of activated T-cells, cytoplasmic 1), the protein product of NFATC1. NFATc1 targets are involved in axon growth, synaptic plasticity, neuronal survival, and myelination (Kipanyula et al. 2016; Vihma et al. 2016). Several SNPs were either known eQTLs or in high LD with known eQTLs, though most of these were not in brain. Exceptions include rs79045984, which is an eQTL for RCN2 in the putamen, and rs78070351, which is an eQTL for CEP250 in the cerebellum (Supplementary Table 2). LD-independent SNPs with p-values less than 5 x 10−6 account for 0.01%–2.13% of the phenotypic variance in the tract common factor.
FST values larger than 0.05, indicating potential confounding by ancestry, were observed for rs78070351, rs7366960 and rs28627209. While FST values were below 0.05 for other top hits, we note that the minor allele frequency for rs79045984 was very low in individuals of European ancestry, and the minor allele frequency for rs72830077 was very low in individuals of non-European ancestry (Supplementary Table 3). We were unable to run relevant sensitivity analyses in those groups.
Effect sizes and significance in primary analyses were highly similar to effect sizes in sensitivity analyses (Supplementary Table 4) with a few exceptions. In individuals of European ancestry, rs17004715 was not associated with the DTI tract factor and rs79045984 showed no association in individuals who did not receive neonatal intensivecare.
Permutation testing yielded an empirical p-value of 7.40 x 10−8 for rs6077860’s association with the tract factor. The empirical P-value confirms that the asymptotic P-values are accurately estimated.
When we consider the spatiotemporal expression pattern of the nearest protein-coding genes for LD-independent SNPs with p-values less than 5 x 10−6, B3GAT1, TENM2, SCAPER, and PSMF1 all show strong protein expression in fetal brain tissue compared to non-brain tissue. SCAPER, SETBP1, B3GAT1, and MAP3K13 show strong gene expression in fetal brain tissue compared to non-brain tissue. Four genes, PSMF1, SCAPER, SLC27A3, and SETBP1 show elevated prenatal expression across all brain regions examined. UQCC1, MAN1A1, and NFATC1 show a strong increase in expression from prenatal to postnatal life. CNIH3, MAP3K13, B3GAT1, ITGB2, MECOM, and APCS show elevated prenatal expression in a subset of brain regions (Fig. 6). Tests for enrichment of prenatally expressed genes suggest significantly elevated expression at multiple thresholds in cerebellar cortex, but not in other brain regions (Fig. 7).
Figure 6 .
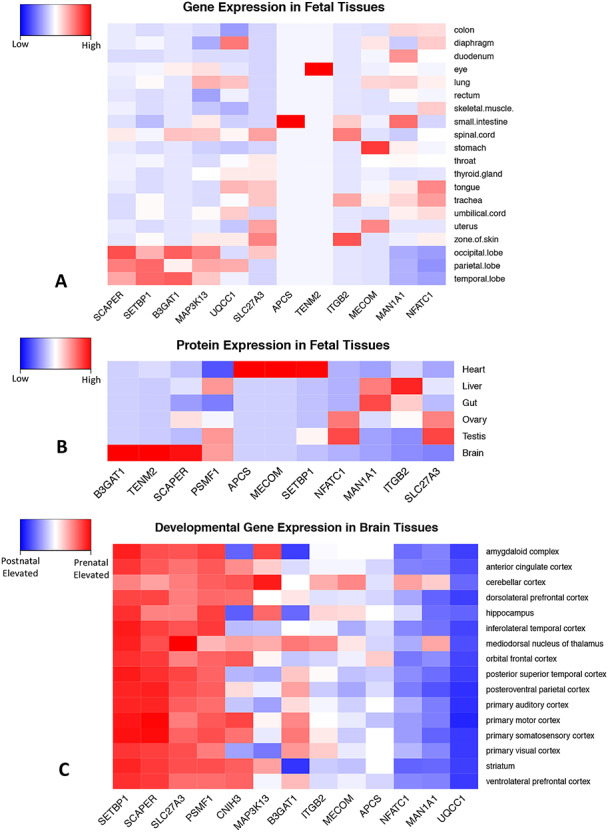
Spatiotemporal expression patterns for genes in close proximity to LD-independent SNPs with p-values smaller than 5 x 10−6. Figures include the closest protein coding gene (±500 kb) for each SNP. Panel A. Gene expression data in fetal tissues from FANTOM (Lizio M et al. 2015). Blue is low; red is high. Panel B. Protein expression data in fetal tissues from human proteome map (Kim MS et al. 2014). Blue is low; red is high. Panel C. Developmental gene expression patterns in human brain. Red represents elevated prenatal expression, blue indicates elevated postnatal expression. Developmental gene expression data is from Brainspan (Kang HJ et al. 2011).
Figure 7 .
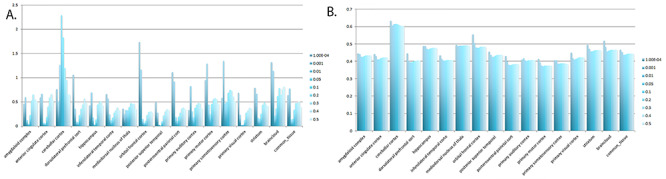
Enrichment of prenatally expressed genes is restricted to the cerebellum. After LD-pruning, each single-nucleotide polymorphism (SNP) was assigned to the nearest gene within 2 megabases. Elevated prenatal expression was defined as a fetal/postnatal log2 fold-change threshold of 0.5. Analyses were performed using both the RNA sequencing-based Brainspan transcriptome of 15 different brain regions and microarray-based ‘BrainCloud’ dorsolateral prefrontal cortex (DLPFC) transcriptome. To calculate values for “common_tissue”, we estimated the effect size of elevated expression in all brain tissues included in Brainspan while adjusting for tissue type. The estimated effect size is then used to do the sign test. Panel A. −log10 P-values. Panel B. percentage of genes with elevated prenatal expression.
In GSEA, the gene set related to central nervous system neuron axonogenesis showed a nominally significant association with the tract factor (p-value =0.0402), but no gene sets achieved significance after multiple comparison adjustments (Supplementary Table 5).
FADTTS analysis revealed that most of the variants listed in Table 2 have small global p-values in multiple tracts (Fig. 8). However, some specific tracts, including the right and left optic tract, the left and right medial lemniscus, and the superior portions of the left and right cingulum showed relatively weak associations with these variants. In addition, some variants appear to exert stronger effects in only a subset of tracts. In particular, the intronic SNP in CNIH3 shows stronger associations with various components of the arcuate fasciculus compared to other fiber tracts.
Figure 8 .
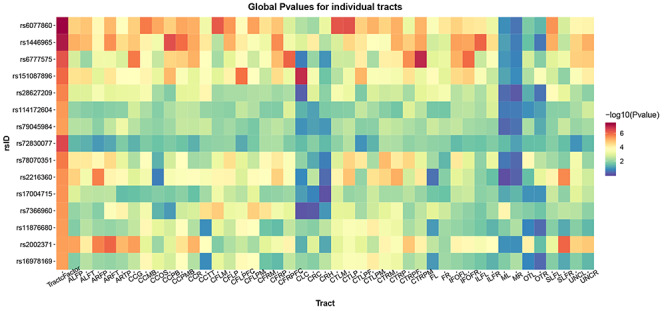
LD-independent SNPs with p-values smaller than 5 x 10−6 show significant associations with FA in multiple fiber tracts. We used functional analysis of diffusion tensor tract statistics (FADTTS) (Zhu H et al. 2011) to test for associations between our top hits and FA within individual fiber tracts. Most variants have small global p-values across multiple tracts. Associations with the tract factor are often stronger than associations with individual tracts, revealing the power of this analysis approach. Some tracts, including the right and left optic tract (OTL and OTR), the left and right medial lemniscus (ML and MR), and the superior portions of the left and right cingulum (CLC and CRC) show relatively weak associations with these variants. Some variants appear to exert stronger effects in only a subset of tracts. In particular, rs2002371 shows stronger associations with various components of the arcuate fasciculus compared to other fiber tracts. An explanation of these abbreviations can be found in Supplementary Table 6.
Examination of local p-values along each individual fiber tract showed that in most cases significance is observed along the majority of the arc length and the effect size retains the same directionality. However, there did appear to be a general pattern such that the central portions of the corpus callosum showed weak or nonexistent associations compared to the lateral portions (Fig. 9, Panels A. and C.). In addition, within the various corticofugal and corticothalamic tracts, there was a general pattern in which stronger associations were observed in the superior sections and weak or nonexistent asociations were observed in the inferior sections (Fig. 9, Panels B. andC.).
Figure 9 .
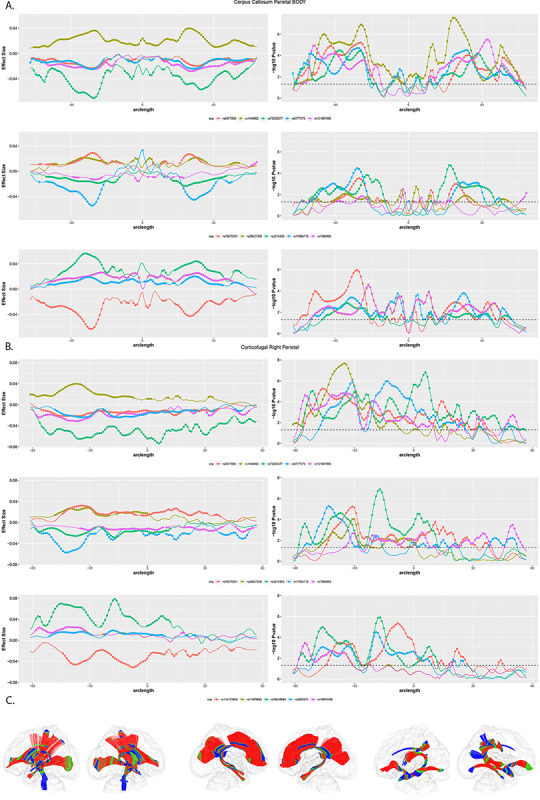
LD-independent SNPs with p-values smaller than 5 x 10−6 show significant associations with FA along multiple fiber tracts. Panel A. Local test statistics along the segment of the corpus callosum that connects left and right parietal cortex. Left side depicts effect size (Beta); right side depicts –log10 p-values. Circles represent significant associations. Note the drop in effect size and significance in the central portion of the tract. Panel B. Local test statistics along the corticofugal tract originating in right parietal cortex. Left side depicts effect size (Beta); right side depicts –log10 p-values. Circles represent significant associations. Note the drop in effect size and significance in the inferior portions of this tract. Panel C. 3D representations of major fiber tracts within a glass brain: projection tracts (left) commissural (middle), and association (right) tracts. Color represents –log10 p values for rs6077860, the intronic SNP in PSMF1. Red indicates low (more significant) p-values, blue represents high p-values.
CNV burden score was not significantly associated with the DTI factor (p = 0.8216). One individual carried a rare CNV affecting NDE1 that had 79.4% percent overlap with a known neuropsychiatric CNV (16p13.11 microdeletions). A scatter plot of the DTI factor suggested that this individual's phenotype was within the normal range among all subjects (Supplementary Fig. 2).
Discussion
The prenatal and early postnatal period witnesses several key events in human white matter development including axonogenesis, fasciculation, proliferation and maturation of glial cells, and the advent of myelination and axon elimination. Genetic mutations can disrupt these processes, resulting in a variety of neurological disorders. However, the role of common genetic variation in shaping white matter development remains poorly understood. In order to address this gap, we carried out the first GWAS of infant white matter microstructure, assessed via DTI. One of the challenges in carrying out a genome-wide association study of DTI phenotypes is the high-dimensionality of the data, which includes multiple fiber bundles with heterogeneous geometric structures and variation patterns. We addressed this challenge by applying a hierarchical functional principal regression model (Zhang et al. 2017) in order to create a tractography-based factor that captured shared variation in FA across 44 white matter bundles. This latent measure of white matter microstructure explained a substantial amount of variance across and within individual tracts. This may seem surprising as fiber tracts are structurally, functionally, and developmentally diverse. However, our results are similar to previous work in infants using tract average FA and more limited sets of fibers (Lee et al. 2017; Telford et al. 2017). We hypothesize that this latent factor is the end result of white matter maturational processes that occurred in utero including axon formation, axon guidance, and fasciculation. However, it is also possible that the latent factor captures maturational processes occurring around one month of age including proliferation and maturation of glial cells, axonal pruning, and myelination.
Notably, this latent measure accounts for a large proportion of heritable variation in each individual bundle. This suggests that genetic influences on white matter microstructure, measured during early infancy, primarily operate on a global scale, rather than being tract-specific. Subsequent analyses primarily focused on identifying genes and molecular pathways contributing to variation shared across tracts, rather than features that might be unique to specific tracts.
An intronic SNP in the gene PSMF1 exceeded the conventional GWAS threshold of 5 x 10−8 for the tractography-based factor. Intriguingly, PSMF1 encodes a protein that inhibits activation of the 26S proteasome, a multicatalytic proteinase complex that may play a role in developmental axonal pruning and synaptic plasticity (Hegde and Upadhya 2011). Specifically, the 26S proteasome degrades R subunits of cAMP-dependent protein kinase (Hegde et al. 1993), transcriptional repressors including CREB1b (Upadhya et al. 2004), and transcriptional activators including C/EBP (Yamamoto et al. 1999), as well as the transcriptional corepressor SnoN which promotes axonal growth (Konishi et al. 2004). Perturbations in the ubiquitin-proteasome pathway may play a causal role in Angelman syndrome (Hegde and Upadhya 2011), a neurological disorder associated with abnormalities in white matter microstructure (Peters et al. 2011).
Additional loci nearing genome-wide significance were located in or near genes with known roles in axon growth and guidance, fasciculation, and myelination including B3GAT1, TENM2, NFATC1, and MAP3K13. B3GAT1 is a key enzyme for the biosynthesis of the carbohydrate epitope HNK-1, which is involved in neurodevelopment and synaptic plasticity (Kizuka and Oka 2012), including fasciculation and axonal pathfinding (Becker et al. 2001). HNK-1 is almost entirely absent in the brains of B3GAT1 knockout mice, which show impaired synaptic plasticity and spatial learning (Yamamoto et al. 2002). TENM2 encodes a teneurin, one of a family of large cell surface glycoproteins that regulate the establishment of proper connectivity within the nervous system (Tucker et al. 2007; Young and Leamey 2009). Studies of knockout mice have shown that TENM2 is required for the establishment of ipsilateral projections in the mouse visual system (Young et al. 2013). NFATC1 is a transcription factor with potentially important roles in the developing nervous system (Kipanyula et al. 2016; Vihma et al. 2016). Targeted disruption of NFATc1 results in embryonic lethality (de la Pompa et al. 1998). However, deletion of other NFAT family members indicates that NFAT signaling is involved in axon growth (Graef et al. 2003). In addition, inhibition of Nfat activity disrupts oligodendrocyte differentiation in both rodents and human iPSC-derived oligodendrocytes (Weider et al. 2018). MAP3K13 is a member of serine/threonine protein kinase family that acts as a positive regulator of axon growth in mammalian neural cultures (Chen et al. 2016b).
GSEA suggested that the latent measure of white matter structure evaluated in this study might be related to central nervous system neuron axonogenesis, which showed a nominally significant association with the tract factor. However, we note that no gene sets achieved significance after multiple comparison adjustments. We also note that FA does not index a single biological process. As described in the introduction, FA is affected by diverse processes including fiber organization, proliferation of immature oligodendrocytes, and myelination (Qiu et al. 2015).
Integration of our data with gene expression data from FANTOM (Lizio et al. 2015), protein expression data from the human proteome map (Kim et al. 2014), and developmental gene expression data from Brainspan (Kang et al. 2011) and BrainCloud (Colantuoni et al. 2011) also highlighted potential genes of interest. B3GAT1, TENM2, SCAPER, and PSMF1 all show strong protein expression in fetal brain tissue compared to non-brain tissue. SCAPER, SETBP1, B3GAT1, and MAP3K13 show strong gene expression in fetal brain tissue compared to non-brain tissue. Four genes, PSMF1, SCAPER, SLC27A3, and SETBP1 show elevated gene expression during the prenatal period across all brain regions examined. Many other genes showed elevated prenatal expression in a subset of regions including B3GAT1 and MAP3K13. In contrast to our earlier GWAS of neonatal global brain tissue volumes (Xia et al. 2017), we did not observe significant prenatal expression enrichment for the tractography factor, except within the cerebellum. However, it should be noted that both BrainCloud (Colantuoni et al. 2011) and Brainspan (Kang et al. 2011) contain very little data for the second half of human gestation. Consequently, they may be better suited to the study of early neurodevelopmental processes such as neuron production and migration. It is unlikely they would capture gene expression changes associated with the formation and maturation of cerebral pathways, which primarily occurs during gestational weeks 20–45 (Kostovic and Jovanov-Milosevic 2006).
Although our primary focus in this project was on identifying genes and molecular pathways contributing to shared variation across tracts, we also performed secondary analyses investigating relationships between our top loci and FA within and along individual white matter tracts. In most cases, the loci we examined showed significant associations with FA in multiple fiber tracts. However, some specific tracts, including the right and left optic tract, the left and right medial lemniscus, and the superior portions of the left and right cingulum showed relatively weak associations with all the variants we examined. In addition, we found that some variants exert strong effects in only a subset of tracts. In particular, an intronic SNP in CNIH3 appeared to be particularly important for development of various components of the arcuate fasciculus, a complex fiber bundle involved in language processing (Phillips et al. 2011). Studies in rodents indicate that CNIH3 regulates the trafficking and gating properties of AMPA- selective glutamate receptors (AMPARs) (Herring et al. 2013; Shanks et al. 2014). This is relevant to the current study as signaling through AMPA receptors on oligodendrocyte precursors promotes oligodendrocyte survival, leading to more extensive myelination (Kougioumtzidou et al. 2017).
When we examined local effects within each fiber tract, we found that loci associated with the global tract factor also showed significant associations with FA along vast swathes of the individual fiber tracts. We also noted that central portions of the corpus callosum and inferior portions of the corticofugal and corticothalamic tracts showed weak or nonexistent associations compared to other tract regions. Post-mortem and neuroimaging studies suggest that myelination occurs in inferior and central regions before superior and peripheral regions (Yakovlev and Lecours 1967; Lebel and Deoni 2018). Consequently, we hypothesize that genes involved in myelination may be primary contributors to individual variation in central portions of the corpus callosum and inferior portions of the corticthalamci and corticofugal tracts at this age, while the global tract factor captures the effects of genes involved in axonogenesis and fasciculation, which are detectable in later maturing regions.
Finally, we did not observe a significant effect of CNV burden on our measures of white matter microstructure. We note that rare genic CNVs are increased in ASD (Pinto et al. 2010) and schizophrenia (Stone et al. 2008), and that both conditions are associated with altered white matter microstructure (Ameis and Catani 2015; Tamnes and Agartz 2016). In addition, altered white matter microstucture has been reported in individuals with specific neurocognitive CNVs including 22q11.2 deletion syndrome (Villalon-Reina et al. 2013) and 16p11.2 deletions (Berman et al. 2015). It is possible that CNV burden alters postnatal developmental processes, such as myelination, which are not fully captured by the neonate tract factor. It is also possible that different CNVs may influence distinct neuroimaging phenotypes, so no consistent effect emerges for overall CNV burden. This hypothesis is supported by a study by Stefansson et al. (2014) in which different neuropsychiatric CNVs affected different cognitive domains. One individual in the current study carried a rare CNV which overlapped with a known neurocognitive CNV, specifically a deletion in 16p13.11 impacting the NDE1 gene. Deletions in this area are associated with intellectual disability (Ullmann et al. 2007; Hannes et al. 2009; Loureiro et al. 2017), epilepsy (de Kovel et al. 2010; Heinzen et al. 2010), and possibly schizophrenia (Need et al. 2009). However, we note that the 16p13.11 deletion may not, in itself, be sufficient to cause a phenotype, acting istead as a susceptibility locus for neurocognitive disease. This individual had a tract factor value in the typical range as a neonate, but this does not rule out the possibility that his or her white matter development might deviate postnatally, or that other neuropsychiatric CNVs might influence neonatal white matter microstructure.
In conclusion, we have shown that common genetic variation influences white matter microstructure measured in early infancy using a GWAS design. Strengths of this study include 1) focus on a key period in human brain development, 2) use of a large and richly characterized cohort of human infants, 3) using a novel approach to address the high-dimensionality of DTI data, and 4) integration of genomics data with transcriptomic data to enhance our fundamental understanding of how genetic factors influence white matter development. The study also had certain limitations. First, the current analysis is cross-sectional and does not capture genetic influences on change in white matter microstructure over time. Second, although we used one of the largest infant neuroimaging cohorts ever assembled, our analysis is only powered to capture large genetic effects, not moderate or small effects. Finally, the current study was not sufficiently powered to run separate whole genome association analyses for 44 different white matter tracts. Instead, we used a hierarchical functional principal regression model (HFPRM) to identify a latent measure of white matter microstructure which explains a substantial amount of variation across and within tracts. In other words, the current analysis primarily focused on genetic influences that were shared across tracts, rather than features that might be unique to specific tracts. We note that there is both systematic and individual variation in the cytoarchitecture of the human cortex (Rajkowska and goldman-Rakic 1995; Economo et al. 2008) and that this variability may have important consequences for white matter microstructure in corticocortical and corticosubcortical connections. For example, neuron density in prefrontal cortex gray matter is strongly related to the density of myelinated axons in the underlying white matter (Zikopoulos et al. 2018). Future studies in larger samples should address how common genetic variation contributes to variation which is tract-specific.
This is the first study of its kind and independent replication is critical. Nevertheless, the current results suggest this approach will be fruitful. They also add to a growing body of research implicating the ubiquitin-proteasome system in white matter development. Ultimately, a better understanding of genetic influences on early white matter development could significantly advance our understanding of disorders of axon guidance as well as other neurological and psychiatric conditions characterized by altered white matter microstructure, and open up new treatment possibilities centered on normalizing adverse developmental trajectories.
Supplementary Material
Contributor Information
J Zhang, Department of Biostatistics, Harvard T.H. Chan School of Public Health, Boston, MA, USA.
K Xia, Department of Psychiatry, University of North Carolina, Chapel Hill, NC, USA.
M Ahn, Department of Mathematics and Statistics, University of Nevada, Reno, NV, USA.
S C Jha, Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA, USA.
R Blanchett, Genetics and Genome Sciences Program, Michigan State University, East Lansing, MI, USA.
J J Crowley, Department of Psychiatry, University of North Carolina, Chapel Hill, NC, USA; Department of Genetics, University of North Carolina, Chapel Hill, NC, USA; Department of Clinical Neuroscience, Karolinska Institute, Stockholm, Sweden.
J P Szatkiewicz, Department of Psychiatry, University of North Carolina, Chapel Hill, NC, USA; Department of Genetics, University of North Carolina, Chapel Hill, NC, USA.
F Zou, Department of Biostatistics, University of North Carolina, Chapel Hill, NC, USA.
H Zhu, Department of Biostatistics, University of North Carolina, Chapel Hill, NC, USA.
M Styner, Department of Psychiatry, University of North Carolina, Chapel Hill, NC, USA.
J H Gilmore, Department of Psychiatry, University of North Carolina, Chapel Hill, NC, USA.
R C Knickmeyer, Department of Psychiatry, University of North Carolina, Chapel Hill, NC, USA; Department of Pediatrics and Human Development, Michigan State University, East Lansing, MI, USA; Institute for Quantitative Health Sciences and Engineering, Michigan State University, East Lansing, MI, USA.
Notes
We thank the participating families that made this project possible as well as the staff of the UNC MRI Research Center, the UNC Neuro Image Research and Analysis Laboratories, the UNC Early Brain Development Program, and the UNC Biospecimen Processing Core (BSP), including Joseph Blocher, Dianne Evans, and Patricia Basta. We thank Patrick Sullivan for additional scientific guidance. Conflict of Interest: None declared.
Funding
This work was supported by the National Institutes of Health (MH064065, MH070890, and HD053000 to Dr Gilmore, MH083045 to Dr Knickmeyer, UL1RR025747 and MH086633 to Dr Zhu, HD003110 and EB005149 to Dr Styner, GM103650 to Dr Ahn, and NS007431 to Dr Jha) and the National Science Foundation (SES-1357666 and DMS-1407655 to Dr Zhu). The current study also makes use of data generated by the DECIPHER Consortium. A full list of centers who contributed to the generation of the data is available from https://decipher.sanger.ac.uk/ and via email from decipher@sanger.ac.uk. Funding for DECIPHER was provided by the Wellcome Trust.
Genomic Data Availability
The Biomedical IRB of the University of North Carolina at Chapel Hill has approved the sharing of genomic data on a subset of participants through the NIH GWAS repository (accession # phs001122.v1.p1).
References
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. 2010. A map of human genome variation from population-scale sequencing. Nature. 467:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature. 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn M, Zhang HH, Lu W. 2012. Moment-based method for random effects selection in linear mixed models. Stat Sin. 22:1539–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameis SH, Catani M. 2015. Altered white matter connectivity as a neural substrate for social impairment in autism Spectrum disorder. Cortex. 62:158–181. [DOI] [PubMed] [Google Scholar]
- Aralasmak A, Ulmer JL, Kocak M, Salvan CV, Hillis AE, Yousem DM. 2006. Association, commissural, and projection pathways and their functional deficit reported in literature. J Comput Assist Tomogr. 30:695–715. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JTet al. . 2000. Gene ontology: tool for the unification of biology. Nat Genet. 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. 2015. A global reference for human genetic variation. Nature. 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back SA, Luo NL, Borenstein NS, Levine JM, Volpe JJ, Kinney HC. 2001. Late oligodendrocyte progenitors coincide with the developmental window of vulnerability for human perinatal white matter injury. J Neurosci. 21:1302–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Becker CG, Schachner M, Bernhardt RR. 2001. Antibody to the HNK-1 glycoepitope affects fasciculation and axonal pathfinding in the developing posterior lateral line nerve of embryonic zebrafish. Mech Dev. 109:37–49. [DOI] [PubMed] [Google Scholar]
- Berman JI, Chudnovskaya D, Blaskey L, Kuschner E, Mukherjee P, Buckner R, Nagarajan S, Chung WK, Spiro JE, Sherr EHet al. . 2015. Abnormal auditory and language pathways in children with 16p11.2 deletion. Neuroimage Clin. 9:50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng Set al. . 2012. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer RM, Mandl RC, Peper JS, Baal GC, Kahn RS, Boomsma DI, Hulshoff Pol HE. 2010. Heritability of DTI and MTR in nine-year-old children. Neuroimage. 53:1085–1092. [DOI] [PubMed] [Google Scholar]
- Brouwer RM, Mandl RC, Schnack HG, Soelen IL, Baal GC, Peper JS, Kahn RS, Boomsma DI, Hulshoff Pol HE. 2012. White matter development in early puberty: a longitudinal volumetric and diffusion tensor imaging twin study. PLoS ONE. 7:e32316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budisavljevic S, Dell'Acqua F, Rijsdijk FV, Kane F, Picchioni M, McGuire P, Toulopoulou T, Georgiades A, Kalidindi S, Kravariti Eet al. . 2015. Age-related differences and heritability of the Perisylvian language networks. J Neurosci. 35:12625–12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budisavljevic S, Kawadler JM, Dell'Acqua F, Rijsdijk FV, Kane F, Picchioni M, McGuire P, Toulopoulou T, Georgiades A, Kalidindi Set al. . 2016. Heritability of the limbic networks. Soc Cogn Affect Neurosci. 11:746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Hu X, Ouyang L, He N, Liao Y, Liu Q, Zhou M, Wu M, Huang X, Gong Q. 2016a. A systematic review and meta-analysis of tract-based spatial statistics studies regarding attention-deficit/hyperactivity disorder. Neurosci Biobehav Rev. 68:838–847. [DOI] [PubMed] [Google Scholar]
- Chen M, Geoffroy CG, Wong HN, Tress O, Nguyen MT, Holzman LB, Jin Y, Zheng B. 2016b. Leucine zipper-bearing kinase promotes axon growth in mammalian central nervous system neurons. Sci Rep. 6:31482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MC, McMahon KL, Zubicaray GI, Martin NG, Hickie I, Toga AW, Wright MJ, Thompson PM. 2011. Genetics of white matter development: a DTI study of 705 twins and their siblings aged 12 to 29. Neuroimage. 54:2308–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DRet al. . 2011. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 478:519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTeX Consortium, Battle A, Brown CD, Engelhardt BE, Montgomery SB. 2017. Genetic effects on gene expression across human tissues. Nature. 550:204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovel CGF, Trucks H, Helbig I, Mefford HC, Baker C, Leu C, Kluck C, Muhle H, Spiczak S, Ostertag Pet al. . 2010. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 133:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pompa JL, Timmerman LA, Takimoto H, Yoshida H, Elia AJ, Samper E, Potter J, Wakeham A, Marengere L, Langille BLet al. . 1998. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature. 392:182–186. [DOI] [PubMed] [Google Scholar]
- Leeuw CA, Mooij JM, Heskes T, Posthuma D. 2015. MAGMA: generalized gene-set analysis of GWAS data. Plos Comput Biol. 11:e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejerine JJ. 1892. Contribution a l'etude anatomo-pathologique et clinique des differentes varietes de cecite verbale. Mem. Soc Biol. 4:61–90. [Google Scholar]
- Drobyshevsky A, Song SK, Gamkrelidze G, Wyrwicz AM, Derrick M, Meng F, Li LM, Ji XH, Trommer B, Beardsley DJet al. . 2005. Developmental changes in diffusion anisotropy coincide with immature oligodendrocyte progression and maturation of compound action potential. J Neurosci. 25:5988–5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J, Dehaene-Lambertz G, Kulikova S, Poupon C, Huppi PS, Hertz-Pannier L. 2014. The early development of brain white matter: a review of imaging studies in fetuses, newborns and infants. Neuroscience. 276:48–71. [DOI] [PubMed] [Google Scholar]
- Dubois J, Dehaene-Lambertz G, Perrin M, Mangin JF, Cointepas Y, Duchesnay E, Le Bihan D, Hertz-Pannier L. 2008. Asynchrony of the early maturation of white matter bundles in healthy infants: quantitative landmarks revealed noninvasively by diffusion tensor imaging. Hum Brain Mapp. 29:14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economo C, Koskinas GN, Triarhou LC. 2008. Atlas of cytoarchitectonics of the adult human cerebral cortex. Basil. New York: Karger. [Google Scholar]
- Engle EC. 2010. Human genetic disorders of axon guidance. Csh Perspect Biol. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galea MP, Darian-Smith I. 1995. Postnatal maturation of the direct corticospinal projections in the macaque monkey. Cereb Cortex. 5:518–540. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Zou F, Wright FA. 2008. Estimating odds ratios in genome scans: an approximate conditional likelihood approach (vol 82, pg 1064, 2008). Am J Hum Genet. 82:1224–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore JH, Lin W, Prastawa MW, Looney CB, Vetsa YS, Knickmeyer RC, Evans DD, Smith JK, Hamer RM, Lieberman JAet al. . 2007. Regional gray matter growth, sexual dimorphism, and cerebral asymmetry in the neonatal brain. J Neurosci. 27:1255–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore JH, Schmitt JE, Knickmeyer RC, Smith JK, Lin W, Styner M, Gerig G, Neale MC. 2010. Genetic and environmental contributions to neonatal brain structure: a twin study. Hum Brain Mapp. 31:1174–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodlett CB, Fletcher PT, Gilmore JH, Gerig G. 2009. Group analysis of DTI fiber tract statistics with application to neurodevelopment. Neuroimage. 45:S133–S142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef IA, Wang F, Charron F, Chen L, Neilson J, Tessier-Lavigne M, Crabtree GR. 2003. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell. 113:657–670. [DOI] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen V, Sporns O. 2008. Mapping the structural core of human cerebral cortex. Plos Biol. 6:1479–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall P, Muller HG, Wang JL. 2006. Properties of principal component methods for functional and longitudinal data analysis. Ann Stat. 34:1493–1517. [Google Scholar]
- Hannes FD, Sharp AJ, Mefford HC, Ravel T, Ruivenkamp CA, Breuning MH, Fryns JP, Devriendt K, Van Buggenhout G, Vogels Aet al. . 2009. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. J Med Genet. 46:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN, Goldberg AL, Schwartz JH. 1993. Regulatory subunits of cAMP-dependent protein kinases are degraded after conjugation to ubiquitin: a molecular mechanism underlying long-term synaptic plasticity. Proc Natl Acad Sci U S A. 90:7436–7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN, Upadhya SC. 2011. Role of ubiquitin-proteasome-mediated proteolysis in nervous system disease. Biochim Biophys Acta. 1809:128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen EL, Radtke RA, Urban TJ, Cavalleri GL, Depondt C, Need AC, Walley NM, Nicoletti P, Ge D, Catarino CBet al. . 2010. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am J Hum Genet. 86:707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring BE, Shi Y, Suh YH, Zheng CY, Blankenship SM, Roche KW, Nicoll RA. 2013. Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron. 77:1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhang J, Wakana S, Zhang W, Ren T, Richards LJ, Yarowsky P, Donohue P, Graham E, Zijl PCet al. . 2006. White and gray matter development in human fetal, newborn and pediatric brains. Neuroimage. 33:27–38. [DOI] [PubMed] [Google Scholar]
- Jha SC, Meltzer-Brody S, Steiner RJ, Cornea E, Woolson S, Ahn M, Verde AR, Hamer RM, Zhu H, Styner Met al. . 2016. Antenatal depression, treatment with selective serotonin reuptake inhibitors, and neonatal brain structure: a propensity-matched cohort study. Psychiatry Res. 253:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe IT, Cadima J. 2016. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci. 374:20150202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchibhotla SC, Mather KA, Thalamuthu A, Zhuang L, Schofield PR, Kwok JB, Ames D, Wright MJ, Trollor JN, Wen Wet al. . 2014. Genetics of microstructure of the corpus callosum in older adults. PLoS ONE. 9:e113181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. 2013. Principles of neural science. New York: McGraw-Hill. [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak Get al. . 2011. Spatio-temporal transcriptome of the human brain. Nature. 478:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu Tet al. . 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 471:480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain Set al. . 2014. A draft map of the human proteome. Nature. 509:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney HC, Brody BA, Kloman AS, Gilles FH. 1988. Sequence of central nervous system myelination in human infancy. II. Patterns of myelination in autopsied infants. J Neuropathol Exp Neurol. 47:217–234. [DOI] [PubMed] [Google Scholar]
- Kipanyula MJ, Kimaro WH, Seke Etet PF. 2016. The emerging roles of the Calcineurin-nuclear factor of activated T-lymphocytes pathway in nervous system functions and diseases. J Aging Res. 2016:5081021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizuka Y, Oka S. 2012. Regulated expression and neural functions of human natural killer-1 (HNK-1) carbohydrate. Cell Mol Life Sci. 69:4135–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knickmeyer R, Gouttard S, Kang C, Evans D, Wilber K, Smith JK, Hamer RM, Lin W, Gerig G, Gilmore JH. 2008. A structural MRI study of human brain development from birth to 2 years. J Neurosci. 28:12176–12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knickmeyer RC, Xia K, Lu ZH, Ahn M, Jha SC, Zou F, Zhu HT, Styner M, Gilmore JH. 2017. Impact of demographic and obstetric factors on infant brain volumes: a population neuroscience study. Cereb Cortex. 27:5616–5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Jahanshad N, Marcus D, Winkler A, Sprooten E, Nichols TE, Wright SN, Hong LE, Patel B, Behrens Tet al. . 2015. Heritability of fractional anisotropy in human white matter: a comparison of human Connectome project and ENIGMA-DTI data. Neuroimage. 111:300–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Jahanshad N, Sprooten E, Nichols TE, Mandl RC, Almasy L, Booth T, Brouwer RM, Curran JE, Zubicaray GIet al. . 2014. Multi-site study of additive genetic effects on fractional anisotropy of cerebral white matter: comparing meta and megaanalytical approaches for data pooling. Neuroimage. 95:136–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y, Stegmuller J, Matsuda T, Bonni S, Bonni A. 2004. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 303:1026–1030. [DOI] [PubMed] [Google Scholar]
- Kostovic I, Goldman-Rakic PS. 1983. Transient cholinesterase staining in the mediodorsal nucleus of the thalamus and its connections in the developing human and monkey brain. J Comp Neurol. 219:431–447. [DOI] [PubMed] [Google Scholar]
- Kostovic I, Jovanov-Milosevic N. 2006. The development of cerebral connections during the first 20-45 weeks' gestation. Semin Fetal Neonatal Med. 11:415–422. [DOI] [PubMed] [Google Scholar]
- Kostovic I, Rakic P. 1984. Development of prestriate visual projections in the monkey and human fetal cerebrum revealed by transient cholinesterase staining. J Neurosci. 4:25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostovic, Rakic P. 1990. Developmental history of the transient subplate zone in the visual and somatosensory cortex of the macaque monkey and human brain. J Comp Neurol. 297:441–470. [DOI] [PubMed] [Google Scholar]
- Kougioumtzidou E, Shimizu T, Hamilton NB, Tohyama K, Sprengel R, Monyer H, Attwell D, Richardson WD. 2017. Signalling through AMPA receptors on oligodendrocyte precursors promotes myelination by enhancing oligodendrocyte survival. Elife. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krmpotic-Nemanic J, Kostovic I, Kelovic Z, Nemanic D, Mrzljak L. 1983. Development of the human fetal auditory cortex: growth of afferent fibres. Acta Anat (Basel). 116:69–73. [PubMed] [Google Scholar]
- Lamantia AS, Rakic P. 1990. Axon overproduction and elimination in the Corpus-callosum of the developing rhesus-monkey. J Neurosci. 10:2156–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel C, Deoni S. 2018. The development of brain white matter microstructure. Neuroimage. 182:207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Steiner RJ, Luo S, Neale MC, Styner M, Zhu H, Gilmore JH. 2015. Quantitative tract-based white matter heritability in twin neonates. Neuroimage. 111:123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Steiner RJ, Yu Y, Short SJ, Neale MC, Styner MA, Zhu H, Gilmore JH. 2017. Common and heritable components of white matter microstructure predict cognitive function at 1 and 2 y. Proc Natl Acad Sci U S A. 114:148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepmann H, Maas O. 1907. Fall von linksseitiger Agraphie und Apraxie bei rechtsseitiger Lahmung. J Psychol Neurol. 10:214–227. [Google Scholar]
- Liu EY, Li M, Wang W, Li Y. 2013. MaCH-admix: genotype imputation for admixed populations. Genet Epidemiol. 37:25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, Abugessaisa I, Fukuda S, Hori F, Ishikawa-Kato Set al. . 2015. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 16:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro S, Almeida J, Cafe C, Conceicao I, Mouga S, Beleza A, Oliveira B, Sa J, Carreira I, Saraiva Jet al. . 2017. Copy number variations in chromosome 16p13.11-the neurodevelopmental clinical spectrum. Current Pediatric Research. 21. [Google Scholar]
- Mahmood A, Bibat G, Zhan AL, Izbudak I, Farage L, Horska A, Mori S, Naidu S. 2010. White matter impairment in Rett syndrome: diffusion tensor imaging study with clinical correlations. AJNR Am J Neuroradiol. 31:295–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Ge DL, Weale ME, Maia J, Feng S, Heinzen EL, Shianna KV, Yoon W, Kasperaviciute D, Gennarelli Met al. . 2009. A genome-wide investigation of SNPs and CNVs in schizophrenia. Plos Genet. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton ES, Beach SD, Gabrieli JD. 2015. Neurobiology of dyslexia. Curr Opin Neurobiol. 30:73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent AA, Kolpak AL, Engle EC. 2012. Human disorders of axon guidance. Curr Opin Neurobiol. 22:837–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguz I, Farzinfar M, Matsui J, Budin F, Liu Z, Gerig G, Johnson HJ, Styner M. 2014. DTIPrep: quality control of diffusion-weighted images. Front Neuroinform. 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlman AL, Sheppard AM. 1996. Extracellular matrix in early cortical development. Prog Brain Res. 108:117–134. [PubMed] [Google Scholar]
- Penke L, Maniega SM, Bastin ME, Hernandez MC, Murray C, Royle NA, Starr JM, Wardlaw JM, Deary IJ. 2012. Brain-wide white matter tract integrity is associated with information processing speed and general intelligence. Mol Psychiatry. 17:955. [DOI] [PubMed] [Google Scholar]
- Penke L, Munoz Maniega S, Murray C, Gow AJ, Hernandez MC, Clayden JD, Starr JM, Wardlaw JM, Bastin ME, Deary IJ. 2010. A general factor of brain white matter integrity predicts information processing speed in healthy older people. J Neurosci. 30:7569–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters SU, Kaufmann WE, Bacino CA, Anderson AW, Adapa P, Chu Z, Yallampalli R, Traipe E, Hunter JV, Wilde EA. 2011. Alterations in white matter pathways in Angelman syndrome. Dev Med Child Neurol. 53:361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan EV, Carmelli D. 2001. Genetic regulation of regional microstructure of the corpus callosum in late life. Neuroreport. 12:1677–1681. [DOI] [PubMed] [Google Scholar]
- Phillips OR, Clark KA, Woods RP, Subotnik KL, Asarnow RF, Nuechterlein KH, Toga AW, Narr KL. 2011. Topographical relationships between arcuate fasciculus connectivity and cortical thickness. Hum Brain Mapp. 32:1788–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BSet al. . 2010. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 466:368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poretti A, Meoded A, Rossi A, Raybaud C, Huisman TA. 2013. Diffusion tensor imaging and fiber tractography in brain malformations. Pediatr Radiol. 43:28–54. [DOI] [PubMed] [Google Scholar]
- Pradeepa MM. 2017. Causal role of histone acetylations in enhancer function. Transcription. 8:40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 38:904–909. [DOI] [PubMed] [Google Scholar]
- Qiu AQ, Mori S, Miller MI. 2015. Diffusion tensor imaging for understanding brain development in early life. Annual Review of Psychology. 66(66):853–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowska G, goldman-Rakic PS. 1995. Cytoarchitectonic definition of prefrontal areas in the normal human cortex: II. Variability in locations of areas 9 and 46 and relationship to the Talairach coordinate system. Cereb Cortex. 5:323–337. [DOI] [PubMed] [Google Scholar]
- Ritchie SJ, Bastin ME, Tucker-Drob EM, Maniega SM, Engelhardt LE, Cox SR, Royle NA, Gow AJ, Corley J, Pattie Aet al. . 2015. Coupled changes in brain white matter microstructure and fluid intelligence in later life. J Neurosci. 35:8672–8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks NF, Cais O, Maruo T, Savas JN, Zaika EI, Azumaya CM, Yates JR 3rd, Greger I, Nakagawa T. 2014. Molecular dissection of the interaction between the AMPA receptor and cornichon homolog-3. J Neurosci. 34:12104–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigaard RK, Kjaer M, Pakkenberg B. 2016. Development of the cell population in the brain white matter of Young children. Cereb Cortex. 26:89–95. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir B, Morgen K, Arnarsdottir S, Bjornsdottir G, Walters GB, Jonsdottir GA, Doyle OMet al. . 2014. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 505:361–366. [DOI] [PubMed] [Google Scholar]
- Stiles J, Jernigan TL. 2010. The basics of brain development. Neuropsychol Rev. 20:327–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JL, O'Donovan MC, Gurling H, Kirov GK, Blackwood DHR, Corvin A, Craddock NJ, Gill M, Hultman CM, Lichtenstein Pet al. . 2008. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 455:237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatkiewicz JP, Neale BM, O'Dushlaine C, Fromer M, Goldstein JI, Moran JL, Chambert K, Kahler A, Magnusson PK, Hultman CMet al. . 2013. Detecting large copy number variants using exome genotyping arrays in a large Swedish schizophrenia sample. Mol Psychiatry. 18:1178–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamnes CK, Agartz I. 2016. White matter microstructure in early-onset schizophrenia: a systematic review of diffusion tensor imaging studies. J Am Acad Child Adolesc Psychiatry. 55:269–279. [DOI] [PubMed] [Google Scholar]
- Tamnes CK, Ostby Y, Walhovd KB, Westlye LT, Due-Tonnessen P, Fjell AM. 2010. Intellectual abilities and white matter microstructure in development: a diffusion tensor imaging study. Hum Brain Mapp. 31:1609–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford EJ, Cox SR, Fletcher-Watson S, Anblagan D, Sparrow S, Pataky R, Quigley A, Semple SI, Bastin ME, Boardman JP. 2017. A latent measure explains substantial variance in white matter microstructure across the newborn human brain. Brain Struct Funct. 222:4023–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium . 2017. Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP, Kenzelmann D, Trzebiatowska A, Chiquet-Ehrismann R. 2007. Teneurins: transmembrane proteins with fundamental roles in development. Int J Biochem Cell Biol. 39:292–297. [DOI] [PubMed] [Google Scholar]
- Ullmann R, Turner G, Kirchhoff M, Chen W, Tonge B, Rosenberg C, Field M, Vianna-Morgante AM, Christie L, Krepischi-Santos ACet al. . 2007. Array CGH identifies reciprocal 16p13.1 duplications and deletions that predispose to autism and/or mental retardation. Hum Mutat. 28:674–682. [DOI] [PubMed] [Google Scholar]
- Upadhya SC, Smith TK, Hegde AN. 2004. Ubiquitin-proteasome-mediated CREB repressor degradation during induction of long-term facilitation. J Neurochem. 91:210–219. [DOI] [PubMed] [Google Scholar]
- Van Vactor D. 1998. Adhesion and signaling in axonal fasciculation. Curr Opin Neurobiol. 8:80–86. [DOI] [PubMed] [Google Scholar]
- Vasung L, Huang H, Jovanov-Milosevic N, Pletikos M, Mori S, Kostovic I. 2010. Development of axonal pathways in the human fetal fronto-limbic brain: histochemical characterization and diffusion tensor imaging. J Anat. 217:400–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde AR, Berger JB, Gupta A, Farzinfar M, Kaiser A, Chanon VW, Boettiger C, Goodlett C, Shi Y, Zhu Het al. . 2013. UNC-Utah NA-MIC DTI framework: atlas based Fiber tract analysis with application to a study of nicotine smoking addiction. Proc SPIE Int Soc Opt Eng. 8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde AR, Budin F, Berger JB, Gupta A, Farzinfar M, Kaiser A, Ahn M, Johnson H, Matsui J, Hazlett HCet al. . 2014. UNC-Utah NA-MIC framework for DTI fiber tract analysis. Front Neuroinform. 7:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihma H, Luhakooder M, Pruunsild P, Timmusk T. 2016. Regulation of different human NFAT isoforms by neuronal activity. J Neurochem. 137:394–408. [DOI] [PubMed] [Google Scholar]
- Villalon-Reina J, Jahanshad N, Beaton E, Toga AW, Thompson PM, Simon TJ. 2013. White matter microstructural abnormalities in girls with chromosome 22q11.2 deletion syndrome, fragile X or Turner syndrome as evidenced by diffusion tensor imaging. Neuroimage. 81:441–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuoksimaa E, Panizzon MS, Hagler DJ Jr, Hatton SN, Fennema-Notestine C, Rinker D, Eyler LT, Franz CE, Lyons MJ, Neale MCet al. . 2017. Heritability of white matter microstructure in late middle age: a twin study of tract-based fractional anisotropy and absolute diffusivity indices. Hum Brain Mapp. 38:2026–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, Kellis M. 2012. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40:D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weider M, Starost LJ, Groll K, Kuspert M, Sock E, Wedel M, Frob F, Schmitt C, Baroti T, Hartwig ACet al. . 2018. Nfat/calcineurin signaling promotes oligodendrocyte differentiation and myelination by transcription factor network tuning. Nat Commun. 9:899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernicke C. 1874. Der aphasische Symptomencomplex. Eine psychologische Studie auf anatomischer Basis. Breslau: Cohn. [Google Scholar]
- Wright FA, Sullivan PF, Brooks AI, Zou F, Sun W, Xia K, Madar V, Jansen R, Chung W, Zhou YHet al. . 2014. Heritability and genomics of gene expression in peripheral blood. Nat Genet. 46:430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K, Zhang J, Ahn M, Jha S, Crowley JJ, Szatkiewicz J, Li T, Zou F, Zhu H, Hibar Det al. . 2017. Genome-wide association analysis identifies common variants influencing infant brain volumes. Transl Psychiatry. 7:e1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev P, Lecours A. 1967. The myelogenetic cycles of regional maturation of the brain. In: Minkowski A, editor. Regional development of the brain in early life. Oxford: Blackwell, pp. 3–70. [Google Scholar]
- Yamamoto N, Hegde AN, Chain DG, Schwartz JH. 1999. Activation and degradation of the transcription factor C/EBP during long-term facilitation in Aplysia. J Neurochem. 73:2415–2423. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Oka S, Inoue M, Shimuta M, Manabe T, Takahashi H, Miyamoto M, Asano M, Sakagami J, Sudo Ket al. . 2002. Mice deficient in nervous system-specific carbohydrate epitope HNK-1 exhibit impaired synaptic plasticity and spatial learning. J Biol Chem. 277:27227–27231. [DOI] [PubMed] [Google Scholar]
- Yao L, Lui S, Liao Y, Du MY, Hu N, Thomas JA, Gong QY. 2013. White matter deficits in first episode schizophrenia: an activation likelihood estimation meta-analysis. Prog Neuropsychopharmacol Biol Psychiatry. 45:100–106. [DOI] [PubMed] [Google Scholar]
- Young TR, Bourke M, Zhou X, Oohashi T, Sawatari A, Fassler R, Leamey CA. 2013. Ten-m2 is required for the generation of binocular visual circuits. J Neurosci. 33:12490–12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TR, Leamey CA. 2009. Teneurins: important regulators of neural circuitry. Int J Biochem Cell Biol. 41:990–993. [DOI] [PubMed] [Google Scholar]
- Zhang J, Huang C, Ibrahim JG, Jha S, Knickmeyer RC, Gilmore JH, Styner M, Zhu H. 2017. HFPRM: hierarchical functional principal regression model for diffusion tensor image bundle statistics. Inf Process Med Imaging. 10265:478–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Kong L, Li R, Styner M, Gerig G, Lin W, Gilmore JH. 2011. FADTTS: functional analysis of diffusion tensor tract statistics. Neuroimage. 56:1412–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikopoulos B, Garcia-Cabezas MA, Barbas H. 2018. Parallel trends in cortical gray and white matter architecture and connections in primates allow fine study of pathways in humans and reveal network disruptions in autism. Plos Biol. 16:e2004559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


