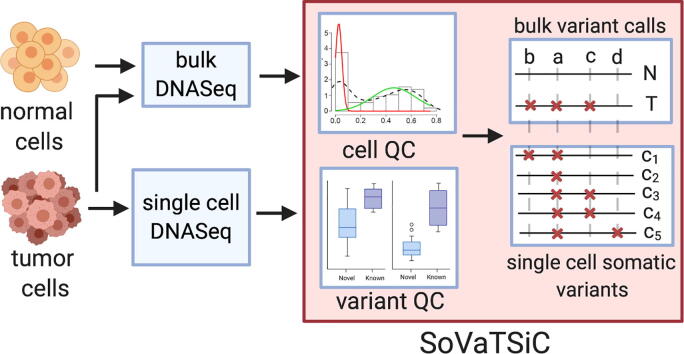
Graphical abstract
Abbreviations: ADO, Allelic dropout; GMM, Gaussian Mixture Model; FP, False positives; TP, True positives; WGA, Whole genome amplification; SNV, Single nucleotide variation; CNV, Copy number variation
Keywords: Single cell genomics, Whole genome amplification, Single cell somatic variant caller, Protocol aware bioinformatics
Abstract
Single cell genomics offers an unprecedented resolution to interrogate genetic heterogeneity in a patient’s tumour at the intercellular level. However, the DNA yield per cell is insufficient for today’s sequencing library preparation protocols. This necessitates DNA amplification which is a key source of experimental noise. We provide an evaluation of two protocols using micro-fluidics based amplification for whole exome sequencing, which is an experimental scenario commonly used in single cell genomics. The results highlight their respective biases and relative strengths in identification of single nucleotide variations. Towards this end, we introduce a workflow SoVaTSiC, which allows for quality evaluation and somatic variant identification of single cell data. As proof of concept, the framework was applied to study a lung adenocarcinoma tumour. The analysis provides insights into tumour phylogeny by identifying key mutational events in lung adenocarcinoma evolution. The consequence of this inference is supported by the histology of the tumour and demonstrates usefulness of the approach.
1. Background
Intra-tumour heterogeneity (ITH) poses a key challenge for clinical management of cancers as diagnosis and treatment are usually guided by a single snapshot representing the totality of the underlying disease. This single genomic snapshot, based on needle biopsy or resected tumour mass, does not reflect the complete genetic and phenotypic diversity, hence may miss cancer drivers and resistance mechanisms. As a result, personalized therapeutic intervention may fail without considering these unrepresented clones, leaving the door open for the tumour to grow unimpeded using its clonal diversity as a resource for treatment escape. In the long run, it would lead to patient relapse and potential metastases of these tumour cells to distant sites. Over the years, more focused efforts have been undertaken to better understand ITH by sampling multiple sectors within the same tumour [1], [2], [3], [4], [5], or by sequencing single homogenized samples at a greater depth[6], [7]. These efforts have provided key insights to understanding the evolutionary trajectory of these cancers. However, the mutations thus detected still only represent an average signal of cells within a tumour population. As a result, it hinders the identification of rare tumour cell populations and fails to define co-occurrence or mutual exclusivity of mutations amongst the clones. In order to overcome the aforementioned problems, single cell DNA sequencing has been recognized as a promising technique that can provide a better resolution to evaluate ITH.
Single cell DNA sequencing’s greater resolution comes at a technical cost. Exome-wide approaches with the ability to identify single nucleotide variants require whole genome amplification (WGA). Sequence and locus specific biases in WGA result in non-uniform distribution of reads across the genome and allelic dropouts, while base errors in early amplification cycles introduce false variants. There are three commonly used WGA strategies namely, Degenerate Oligonucleotide Primed PCR (DOP-PCR), Multiple Displacement Amplification (MDA), and Multiple Annealing and Looping Based Amplification Cycle (MALBAC). They differ in their chemistry, type of enzyme used, and protocol, which leads to differences in performance. A few studies have previously compared their performance[8], [9], [10], [11]. These comparisons have either been conducted on single cells that were isolated manually (in a tube, hereon abbreviated as tube based), or are based on nonhuman cells with different genome complexity (de Bourcy, De Vlaminck et al. 2014). Manual processing is labour intensive, time consuming and could be a source of operator specific variability. As a result, the microfluidics platforms that could automate this process are rapidly increasing in popularity and usage. The performance of the amplification protocols vary as the methods are translated from tube-based to microfluidics platforms or from whole genome to targeted exome sequencing. Previously, other studies have compared the performance of different WGA methods for exome sequencing[12]. However, the analysis was restricted to tube-based amplification. Recently, Marie and colleagues have presented an injection-moulded valveless microfluidic device for single cell isolation and DNA release[13]. Using this device followed by MDA and genome sequencing the authors demonstrate absence of contamination and high genome coverage. In the present study, we first evaluated the performance of two WGA protocols performed on a microfluidics device followed by sequencing the amplified products using exome sequencing. For this comparison, the fidelity of the two kits was compared based on their performance for single nucleotide variant calling.
Finally, as a clinical application, we performed exome sequencing using the best WGA protocol in the earlier survey on 200 single cells from a lung cancer patient. These cells were derived from two distant tumor sectors as well as a far normal tissue. One of the technical difficulties of a single cell cancer genomics experiment is to determine the somatic mutations accurately. To date, two single cell specific variant callers, MONOVAR[14]and SCcaller[15]were introduced for single cell genomics. However, these tools primarily aim to identify variants in general and are not specifically catered towards delineating somatic variants from germline ones. As such, we provide SoVaTSiC (Somatic Variant Tool for Single Cell), a workflow that allows quality evaluation of single cells and somatic variant identification. Practical guidelines were provided to fine tune the tool to the sample of study. These tweaks aim to counteract the biases specific to each dataset and leverage on the availability of less error prone bulk sequencing data which reflects the genetic landscape in broad strokes.
By applying SoVaTSiC to the lung cancer patient sample, the variants detected from the lung single cells show high accuracy and strong concordance with bulk samples. The unsupervised phylogenetic inference shows fidelity to the sector-of-origin and identifies key events in the patient specific tumor evolution. In addition, SoVaTSiC framework compares favorably to the state-of-the-art single cell specialized variant caller MONOVAR in sensitivity versus specificity tradeoff. In particular, the somatic inference steps provide substantial improvement in pruning MONOVAR results as well, thus highlighting their generalizability, and broad application. These results are consistent with the performance of the two methods on an independent dataset as well [16], where SoVaTSiC identifies cancer relevant mutations missed in previous analyses.
2. Results
2.1. Evaluation of the impact of amplification biases on single nucleotide variant calling in exome sequencing
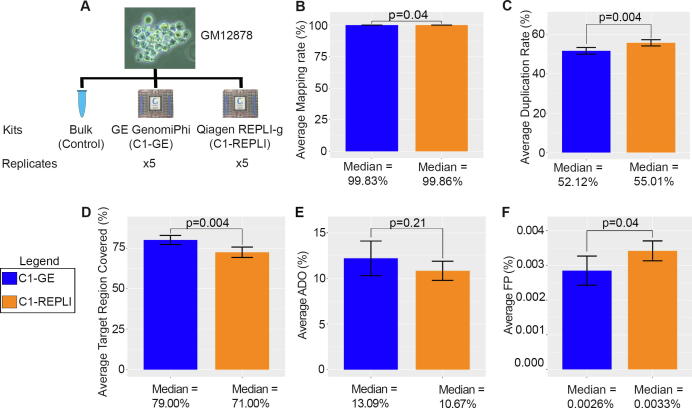
A single cell genomics experiment usually requires the sequencing of a large number of single cells. As a result, manual tube-based methods which require large volumes of chemicals and enzymes, are laborious and time-consuming may not be practical for such experiments. Droplet based single cell sequencing methods on the other hand are tailored for the analysis of thousands of cells in particular for single cell RNA-seq. As single cell whole genome sequencing at a depth that allows single nucleotide variant calling is still prohibitively expensive when hundreds of cells need to be sequenced single cell exome sequencing might currently be considered the most comprehensive nucleotide-resolution single cell sequencing approach that is realistic at medium scale. We first compared the two most common whole genome amplification concepts Multiple Displacement Amplification (MDA) and Multiple Annealing and Looping Based Amplification Cycle (MALBAC) for their fidelity using single cell shallow whole genome sequencing. For this analysis, we selected the lymphoblastoid cell line GM12878 as it has been intensely characterized by deep, whole-genome sequencing as part of a three-generation pedigree where a range of variant callers and haplotype transmission information were used to create a phased “Platinum” genome[17]. We found a higher genome coverage and a lower error rate per read for the MDA-based protocols (Supplementary Fig. 1 and Supplementary Material). In the interest of scalability, we transferred the MDA-based approaches to the microfluidics-based device, the C1 Autoprep System (Fluidigm), which allows the automatic processing of 96 cells at a time and noted comparable characteristics for the MDA approaches used in manual tube-based and the microfluidics protocols (Supplementary Fig. 1). We then focused on the microfluidics platform to compare two WGA chemistries, the C1 canonical illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare Life Sciences, hereafter C1-GE) and the commonly used REPLI-g single cell kit (Qiagen, hereafter C1-REPLI) in more detail (Fig. 1A). For this experiment, a total of five cells of the lymphoblastoid cell line GM12878 were used for the two amplification methods, and a pool of GM12878 cells was used as the bulk control.
Fig. 1.
Comparison metrics for GM12878 single cells amplified by microfluidics based WGA methods for exome sequencing. (A) Summary of experimental setup for comparison of WGA methods for whole exome sequencing. Two microfluidics methods were used for this experiment and a total of five single cells evaluated for each method. (B-F) Barplots compare the mapping rate, duplication rate, target region coverage, allelic drop out (ADO) and per base false positive (FP) rate respectively. C1-GE outperforms C1-REPLI to varying degrees across majority of the core statistics related to SNV calling (panels B, C, D, and F).
Both methods show similar performance in terms of duplication and mapping rates with C1-GE doing slightly better on the former while C1-REPLI does marginally better on the latter (Fig. 1B, C). Moreover, when using only confidently mapped reads (mapping quality ≥ 20), cells amplified by the C1-GE method have a higher percentage of exome regions covered (Fig. 1D). The allelic drop out (ADO) rate and false positive (FP) rate were calculated by comparing SNVs detected in single cells against the gold standard GM12878 variants detected in the platinum genome project[17]. Based on this analysis, the C1-GE method has a slightly higher ADO rate (Fig. 1E). However, the difference is not significant (p-value = 0.21). On the other hand, the C1-REPLI method has a significantly higher false positive rate compared to C1-GE (p-value = 0.04) (Fig. 1F). Marie and colleagues defined p as the probability of an allele being detected as a measure of estimating allelic dropout performance [13]. Based on this measure, C1-GE significantly outperforms C1-REPLI (p -value = 0.0018) despite no significant difference in ADO (Supplementary Fig. 2B). This is due to a higher true positive variant detectability of the former protocol which is ignored in the traditional ADO definition. The uniformity of coverage of each method is depicted as Lorenz curves of the samples (Supplementary Fig. 2A). It is further quantified by Gini coefficients (Supplementary Fig. 2C) and Evenness score metrices (Supplementary Fig. 2D). C1-GE significantly outperforms C1-REPLI by both these measures (p-value = 0.0023 for Gini coefficients and p-value = 0.0082 for Evenness score). In summary, C1-GE outperforms C1-REPLI to varying degrees across majority of the core statistics related to SNV calling. Therefore, the C1-GE method is used for the following experiments.
2.2. Performance of single cell somatic variant calling in a lung cancer patient
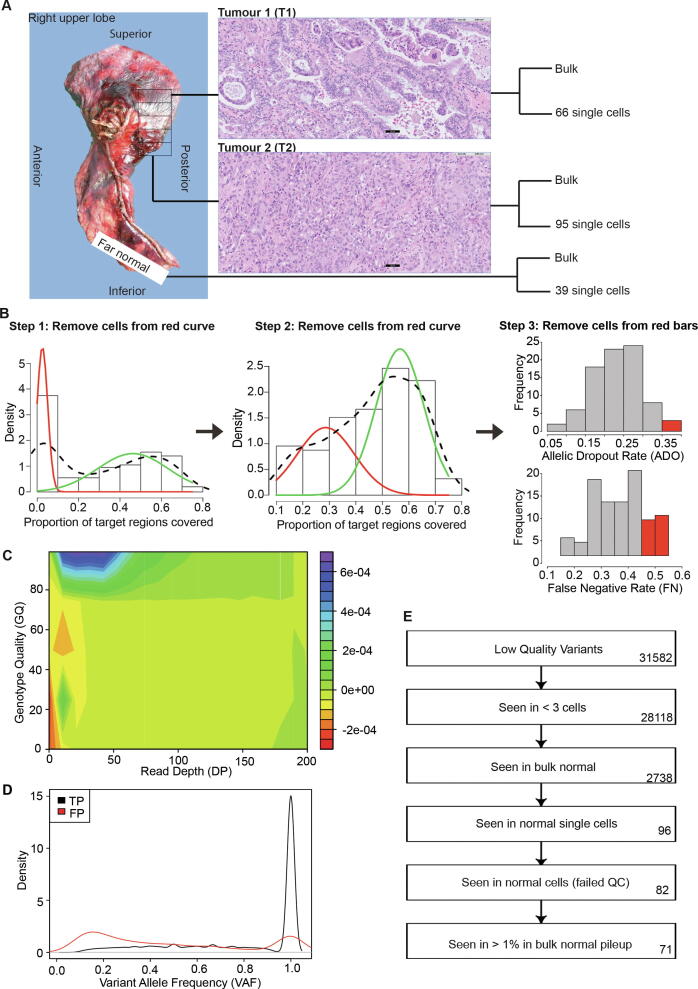
As a proof of concept for a single cell study of clinical application, the experimental and analytical framework was applied to study the intra-tumour heterogeneity in a treatment naive lung adenocarcinoma patient. Two distant sectors from the tumour (hereon abbreviated as T1 and T2) along with far normal tissue were interrogated using exome sequencing in both bulk and single cell analysis (Fig. 2A). The two sectors showed different histological characteristics. T1 showed a mixture of acinar, papillary and micropapillary histology while T2 showed a solid subtype (Fig. 2A). A total of 135 and 225 somatic mutations were detected in the two bulk sectors of which 42 were shared. The patient had been clinically screened for EGFR mutations and EGFR exon 19 deletion delE746_A750 was observed. EGFR activating mutations are known drivers of lung cancer that can be therapeutically targeted by tyrosine kinase inhibitors [18]. EGFR exon 19 in frame deletions, which result in constitutive activation of EGFR signalling, are the most abundant driver mutations in lung cancer patients [19]. Moreover, EGFR driver mutations are of particularly high frequency amongst lung cancer patients of Asian ethnicity [20], [21], [22]. We found the EGFR exon 19 deletion delE746_A750 in both sectors with high mutation allele frequency in the bulk analysis (42.5% and 35.5% in T1 and T2, respectively) indicating high likelihood of it being an early cancer development event and hence observed throughout the tumour. No other genes known to be frequently mutated in lung cancer such as TP53, KRAS, STK11, KEAP, or PIK3CA were found to be mutated in either sector.
Fig. 2.
Overview of lung cancer single cell experiment. (A) Location and histology of the tumour sectors is shown. Two different sectors and far normal tissue were evaluated in bulk and single cell sequencing. The number of single cells selected from each sector is indicated. Bar in microscopic image, 50 µm. (B) Description of quality control steps for single cells. Two iterations of Gaussian Mixture Model (GMM) were used to cluster the single cells based on exonic coverage. The low coverage clusters were removed from further analysis (cells with target coverage < 10%, and cells with coverage between 10% to < 42%). In addition, cells were removed based on the allelic dropout rate (ADO) and false negative (FN) rate. (C) Contour plot used to determine the threshold for filtering of low quality genotypes. Red indicates region enriched for false positive variants, whereas blue indicates region enriched for true positive variants. (D)The density plot shows the variant allele frequency distribution for true positive and false positive variants. (E) The flow chart shows the sequence of serial filters applied to remove germline variants. Numbers of variants that remained after each step are indicated on the right. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Exome sequencing was carried out on 66 single cells from T1, 95 single cells from T2 and 39 single cells from the far normal tissue following the C1-GE protocol (Supplementary Table 1). Patient samples tend to exhibit more experimental variability than the more docile established cell lines. This experimental noise stems from quality of cells obtained from primary tissue and the harsh environment they endure during and post-surgery. Each single cell library was thus first interrogated following a stringent quality control criterion based on coverage distribution, allelic dropout and false negative variant rates (see Fig. 2B, Methods for details). Of note, the latter two statistics are based on the true positive germline variant set derived from the bulk samples. 18 single cells from T1, 21 single cells from T2 and 27 single cells from the far normal passed this evaluation and were retained for further analysis. Key quality control measures of the retained cells estimating the coverage evenness and ADO are shown in Supplementary Fig. 3.
A key challenge in identifying somatic mutations in bulk tumour samples is the variability of mutant allele frequency due to the presence of normal cells and clonal heterogeneity. In particular it is difficult to discriminate between true somatic mutations and experimental noise at the lower end of the allele frequency spectrum. Single cell sequencing obviates this challenge as all variants (germline and somatic) are present at a germline-like allelic frequency albeit with a noisy distribution and potential focal amplifications. To identify somatic mutations in single cell data, a two-step procedure was adopted. In the first step, true positive variants (both SNPs and INDELs) were curated for each cell (Supplementary Figure 4). In the second step, this dataset was pruned of putative germline calls.
In the first step, to distinguish true variants observed in single cell data from false positives, once again the bulk samples were relied upon to define true positive and true negative sites (see Methods for details). It needs to be emphasized that the variant calls in the single cells are not restricted to the bulk observations. Rather these sets are used to define the discriminating characteristics to differentiate the good variants calls from the bad. An exhaustive search of the variant quality parameters computed by GATK revealed depth of coverage (DP), genotype quality (GQ) and variant allele frequency (VAF) to be the most predictive factors (Fig. 2C, D, Supplementary Figure 5). The exact cut-off for these parameters is experiment dependent impacted by multiple factors including DNA quality, amplification kit, and sequencing coverage (see Methods for details).
In the second step, a series of filters were employed to remove sites with either insufficient support across tumour cells, or some evidence of the presence of the variant in normal (bulk or single cell) libraries. The efficacy of these serial filters is demonstrated in Fig. 2E, where a candidate set of 31,582 variants were pruned down to a highly confident set of 71 somatic variants. To evaluate the performance of these calls, 28 of these loci were interrogated in deep targeted sequencing. With meagre to none amount of individual single cell DNA left, we relied on the more abundant bulk DNA from both tumour sectors as well as normal for this experimental validation. 24 (out of 28) loci were validated for both presence and absence in individual sectors (validation rate 86%) (Supplementary Table 2). 22 (out of 24, 92%) variants previously observed in both, bulk and single cell, were validated. 2 (out of 4, 50%) variants which had previously been missed in the bulk calls were now also confirmed. These validation numbers are likely a conservative estimate as the single-cell specific variants are observed in small number of cells and may not be present (or at least are underrepresented) in the cells pooled together for bulk DNA.
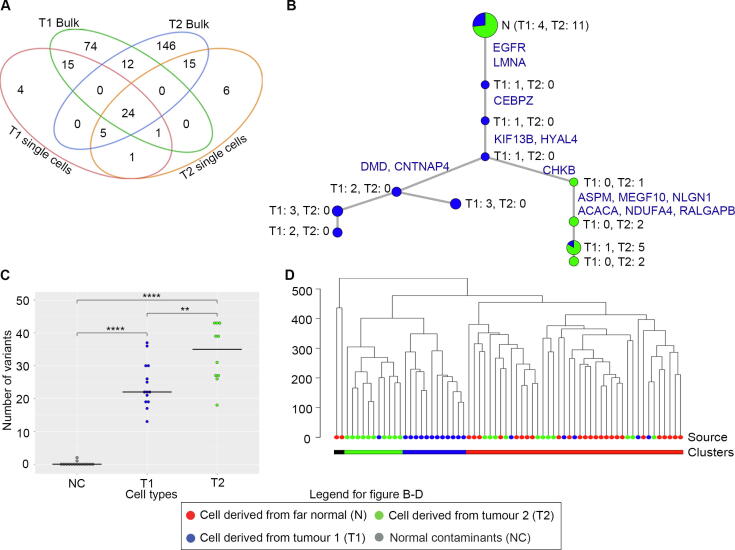
To better understand the detectability of variants in single cell data, all bulk single nucleotide somatic call sites were re-examined (Supplementary Figure 6). 31.7% of T1, and 21.8% of T2 loci of these have been recapitulated in the single cell data as well (Fig. 3A). In a large fraction of the sites called in bulk (26.2% in T1 and 58.9% in T2), no variant was detected in single cells even prior to Bioinformatics filtering (Supplementary Figure 6). This suggests that these variant alleles are not represented in the sequenced libraries. These missing variants tend to have lower variant allele frequencies suggesting their relative rarity in the tumour (Supplementary Figure 7). The intra-tumor heterogeneity of this lung tumor was particularly high since only 42 somatic mutations (13% of all somatic mutations) were shared in bulk exome sequencing between the two sectors T1 (135 somatic mutations) and T2 (225 somatic mutations). Genetic heterogeneity within the tumours, whereby none of the cells are expected to have all these mutations, is one contributing factor towards their absence. Even more so, since the SoVaTSiC protocol requires somatic variants to be detected in at least three cells. Allelic dropout/false negative calls (on average 0.22 and 0.34, respectively, Supplementary Table 1A) in single cells further contribute to the relatively low number of on average 14 somatic mutation calls per cell (median 17, Supplementary Table 7).
Fig. 3.
Lung cancer single cell analysis. (A) Venn diagram shows shared somatic point mutations between bulk tumour sectors and single cells. (B) Phylogenetic tree depicts the relationship between single cells derived from the two tumor sectors. The root of the tree (at the top, indicated by N) consists of putative normal contaminant cells without EGFR exon 19 delE746_A750 mutation. Progressing down the tree EGFR and LMNA mutations are acquired as early truncal events present in the remaining single cells below. A split in evolutionary trajectory is observed with a second (on the right) clone acquiring a high number of mutations. The size of each node is proportional to the number of cells it represents, with the color representing their source (blue from T1 and green from T2). These numbers are indicated next to each node as well. (C) Dot plot showing the number of mutations observed in cells from different sectors. The Y axis indicates the number of mutations. Blue indicate cells that were derived from T1, while green indicate cells derived from T2. Grey colour shows cells that were likely to be normal contaminants. ** indicates pvalue ≤ 0.01, *** for pvalue ≤ 0.001, and **** for pvalue ≤ 0.0001. (D) Clustering of single cells using copy number profiles. The node colors indicate the source of the cell: Red from far normal, blue from T1, and green from T2. Colour codes at the bottom represent the clustering categories: black indicates outliers, green indicates T2 cluster, blue indicates T1 cluster, while red indicates normal cluster which also includes the normal contaminant cells. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.3. Inferring tumour phylogeny from single cell data
Single cell sequencing provides an opportunity to infer their phylogeny and thereby the clonal evolution of a tumour. Phylogenetic clustering of the single cells based on somatic variants revealed 15 cells (4 T1 and 11 T2) that cluster near the root of the phylogenetic tree indicating high likelihood of them being normal contaminant cells from the patient (Fig. 3B). Lung cancers tend to have high normal contamination with the current observation of 15 / 39 (or 38.5%) normal contamination being on the lower end of the spectrum and not far off from the histological estimation of 70% tumour purity for this patient (TCGA lung cancer tumour purity estimates are ~ 40% on average[23]. The normal inference is further supported by the absence of EGFR exon 19 delE746_A750 mutation in these cells. This is a truncal event observed in all (except two) of the remaining single cells across both tumour sectors. In these two cells, based on the somatic mutation profile, this mutation is inferred and suspected missing due to low coverage and allelic dropout, respectively.
Amongst the tumour cells, a split in evolutionary trajectory is observed with most of the T1 cells arising from an earlier clone with lower mutation burden (Fig. 3C). T2 single cells on the other hand are representative of a secondary clone which harbours the truncal events but has a higher genomic instability which has led it to acquire a higher number of somatic mutations (p value = 0.007). This phenomenon is consistent with the observation in the bulk sample as well. Looking across the T2 specific events, a nonsense mutation in ASPM is potentially a contributing factor to this instability. ASPM plays an important role in mitotic cell division via the sonic hedgehog pathway and has been implicated in the growth of several cancers [24], [25], [26], [27], [28], [29]. Furthermore, deletion of ASPM has been shown to impair tumour growth and increase DNA damage in medulloblastoma [30]. To our knowledge, loss of function of ASPM has not been associated with increased DNA damage in lung cancer patients and this link remains to be validated. A predicted damaging missense mutation in MEGF10 is also observed which is unique to T2. MEGF10 expression has been linked to metastatic potential due to its role in cell adhesion and motility in other cancer types [31], [32]. The complete list of sector specific as well as shared somatic mutations can be found in Supplementary Tables 3-9.
Beyond SNVs and INDELs, the lung cancer single cell exome sequencing data was evaluated for variability in coverage reflective of copy number changes. Single cell clustering based on normalized coverage[33]using all normal and tumour cells nearly perfectly recapitulates the earlier somatic variant based results (Fig. 3D). The exception being a single cell from T1 harbouring EGFR and LMNA mutations but having a normal copy number profile. This cell is an intermediary between normal and other cancer cells in Fig. 3B as well. The tumour-derived cells which had grouped at the root of the somatic SNV based phylogenetic tree, again cluster with the normal single cells in the coverage based analysis. Thus, bringing further credence to our earlier hypothesis of them being normal contaminant cells. The tumour cell group is further split into two distinct subgroups representative of the two tumour sectors with one lone single cell from T1 breaking the perfect harmony by clustering across groups in both Fig. 3B and 3D, indicating a minor presence of the more unstable clone in sector T1 as well. Next, we compared the profiles of individual cells with the somatic copy number changes inferred in the two bulk sectors. Copy number changes inferred from bulk samples were observed at a low resolution in single cells as well (Supplementary Figure 8). The individual cells showed general concordance with the sector of origin in both shared as well as sector specific changes (Supplementary Figure 9) indicating that the single cell DNA sequencing pipeline allows consistent retrieval of both, point mutations and copy number alterations.
2.4. Comparison of somatic variant identification with Monovar
More recently, two single cell specific variant callers Monovar and SCcaller have been introduced. The latter identifies allelic drop out regions from neighboring heterozygous variants in long stretches (±10 kbp), and hence would be at a severe disadvantage in the targeted exome sequence data. Hence, it was not included in this evaluation. For the comparison with Monovar, two-pronged evaluation criteria were adopted based on the concordance with bulk (generally expected in true positives), and consistency with phylogeny (deviation indicative of false calls). Monovar run with default parameters identifies 1524 somatic variants of which only 92 (6.0%) were observed in the bulk data. In comparison, our pipeline showed 84.5% concordance. In addition, Monovar identified 70 putative somatic variants in the cells derived from far normal tissue (Supplementary Figure 10B). Although a very small number of these might be true events, the somatic inference in normal tissue in general is indicative of false positives. This overcalling of variants (213, 14.0%) is also observed in tumor derived cells inferred to be normal contaminants (Supplementary Figure 10B). To evaluate the generalizability of our recommended somatic inference pipeline, SoVaTSiC filters were applied to Monovar calls. The cleaned-up results now show high concordance with the bulk inferred somatic variants as well as our results (Supplementary Figure 10A). In total, 1459 variants were pruned out, with a large percentage (14.6%) initially called in normal contaminant cells as well.
To ensure that these observations were not specific to our data, both Monovar and SoVaTSiC were applied to a published muscle-invasive bladder transitional carcinoma dataset[16]) . SoVaTSiC identifies a total of 151 somatic mutations (98 SNVs and 53 INDELs) amongst the single cells (Supplementary Figure 11A-C. To compare against Monovar, the analysis was restricted to SNVs. Our results exhibit high concordance with the bulk (70 out of 98; 71.4%). On the other hand, Monovar with default parameters identified 12,173 somatic mutations of which only 155 (1.27%) were observed in the bulk sample. Applying our filters to the set of somatic variations detected by Monovar, a large majority of the variants were removed, giving a final set of 117 somatic SNVs, of which 70 (59.8%) were shared with the bulk. Phylogenetic clustering of the single cells using the 151 variants detected using SoVaTSiC revealed a single branch from the normal, suggesting that the tumour cells were all derived from a single parental cell (Supplementary Figure 11D). No normal contaminant cells were bioinformatically inferred. This is as per expectation as the cells in this study have been carefully manually picked. Based on the phylogenetic inference, three different tumour sub-populations were observed, whereby the parent tumour clone gave rise to two different sub-clones which contain somatic mutations that are private to each clone. This observed phylogenetic tree (Supplementary Figure 11D) is consistent with the original publications[16], [34]. The advantage of our recommended filters in improving Monovar results was observed in this dataset as well (Supplementary Figure 11E). SoVaTSiC recapitulated 4/7 (57.1%) key events reported in the original study with less noise (39 versus 198 mutations per cell). A closer inspection revealed the remaining 3 somatic events in the original study are pruned out by SoVaTSiC because of evidence in the normal. It also recapitulates FGFR3 nonsynonymous mutation missed in the original publications but reported previously by Monovar. FGFR3 is a key cancer driver for bladder cancers with the activating S249C mutation observed in the patient as the most frequently mutated event [35], [36]. In addition, SoVaTSiC identified four mutations of disease relevance and/or potential therapeutic implication missed in the previous studies (Supplementary Table 10-13). Two of these are missense mutations in LSAMP and HELB which evade Monovar’s arbitrary cutoffs. While the other two are frameshift INDELs in AXL and FOXM1 genes, which are ignored by Monovar as it only identifies single base substitutions. LSAMP is a candidate tumor suppressor implicated in multiple cancer types [37]. HELB is part of cellular response to DNA damage. It is recruited to DNA damage sites and acts as an inhibitor for DNA end resection[38]. AXL is a receptor tyrosine kinase. Its inhibition has therapeutic consequence as it may enhance tumor response to cytotoxic agents[39]. While FOXM1 is a transcription factor which activates several cell cycle genes, it is a proto-oncogene with overexpression leading to cell proliferation while its loss leads to chromosomal instability[40]. In particular for muscle-invasive bladder cancer, its over expression is predictive of poor prognosis[41].
Beyond the somatic filters, our approach has the following added advantages: 1) It utilizes GATK’s more sophisticated haplotype driven engine which makes it more robust to local misalignment in comparison to Monovar’s pileup based approach. In addition, GATK parameters are fine tuned to single cell scenario. For example, it takes advantage of the commonality of underlying haplotype as all single cell libraries are from the same patient (see Methods for details). 2) SoVaTSiC’s data driven cell level QC and reliance on GATK’s data driven variant recalibration, both aim to account for the variability in noise across single cell experiments. In comparison, MONOVAR’s data agnostic default parameter settings do not cater to this variability. As an example, it does not account for sequencing depth of the libraries. Furthermore, it removes variants at low coverage but fails to demark outliers with high sequencing coverage (indicative of potential mismapped reads and hence prone to false variants). 3) SoVaTSiC aims to identify both SNPs and the more challenging INDELs. MONOVAR focuses only on the former and hence ignores EGFR delE746_A750, a key cancer driver event in the lung cancer patient. These factors in conjunction explain the added advantage of SoVaTSiC over MONOVAR.
3. Discussion
Single cell genomics is an area of active development. Various protocols have been introduced for the genomic DNA amplification to generate sufficient DNA material for downstream NGS library preparation. Each of these comes with its own strengths and weaknesses. In a head to head comparison between two MDA based protocols, GE outperforms REPLI in most statistics relevant to SNV calling. This comparison was conducted in a microfluidics system focusing on the exonic regions. These settings are of direct relevance to the current trend in single cell genomics of growing scalability demands coupled with affordability constraints. Recently, a few droplet-based high throughput single cell genome sequencing devices and protocols have been developed. Among commercial solutions, the Chromium Controller (10x Genomics) was designed and used for shallow whole genome sequencing and copy number analysis of mammalian cells while the Tapestri Platform (Mission Bio) performs targeted amplicon-based genome sequencing but not exome sequencing. Using a micro-capillary array (MiCA)-based centrifugal droplet generation technique with whole-genome amplification and target enrichment, Fu and colleagues report ADO of 20% [42]comparable to the average ADO of 22% we observe in our setting (Supplementary Table 1A). An increased focus on clonality in patient tumours dictates increase in scalability (in terms of number of cells per patient). The aim being to increase the likelihood of representing all therapeutically relevant clones in the single cell profile. Often enough during cancer progression, in particular in response to targeted therapy, the tumour composition shifts across the existing clones. Therefore, it is vital for any patient tailored therapy to not only be aware of, but also actively target, major as well as minor clones. However, costs have to be considered and rates of viable cells of clinical specimen that lead to high quality data have to be improved.
There is dearth of single cell somatic variant calling tools. The two current options available for this analysis[14], [15] are both tailored to germline calls without significant focus on differentiating the somatic mutations from germline variants. To address this challenge, we introduce a workflow for quality control and subsequent somatic variant inference. There is significant variability both within and amongst the single cell libraries. We guide the user to gain maximal benefit from the experiment at hand and make recommendations to fine tune the analysis to the experimental noise observed. In a direct comparison, our single cell somatic variant caller outperforms MONOVAR in both in-house and published data. Further, the somatic inference step when applied to MONOVAR show dramatic improvement. Thus, highlighting the generalisability of the method.
In the application case of a lung cancer patient, our analysis indicates that single cell whole exome sequencing is able to identify the clonal nature of this tumour based on either CNV or SNV profile. There is a good concordance in inferred phylogeny between the two approaches. In this specific case, the clonality generally coincides with sector specificity of the tumour cells. This allows us to directly compare the two clones based on sector specific information. The differences observed in single cell genomic profile concur with the histology of the two sectors. T1 constitutes a mixed histology whereas T2 comprises of primarily solid poorly differentiated carcinoma. Our hypothesis based on the phylogenetic inference is that EGFR exon 19 deletion (delE746_A750) is truncal and hence likely the cancer initiating event. This indel would have been missed by the current state-of-the-art informatics pipelines due to their base substitution only focus. The tumour cells during treatment naive evolution acquired the nonsense mutation in ASPM. This loss of function (along with other events) likely impaired the mitotic function of these cells leading to the mutator phenotype observed within this clone.
Cumulatively, this study provides guidelines on the trade-offs of various practical decisions faced in usage of single cell genomics in cancer patients with a meaningful application to understand the cancer evolution in a lung cancer patient. We provide SoVaTSiC a unified framework for single cell genomic analysis for cancer datasets. The workflow is provided open access at https://github.com/JoannaTan/SoVaTSiC. The current pipeline and the results presented are based on GATK3. With improvements in genomic variant calling methodologies, we expect further improvements in SoVaTSiC performance based on upgrade of the underlying GATK version.
4. Methods
4.1. GM12878 cell line preparation prior to WGA
GM12878 cells were cultured in complete media (RPMI (Gibco) supplemented with 20% FBS (Standard, Gibco), 1% 10,000U/ml Penicillin-Streptomycin antibiotics (Gibco) and 1% 200 nM L-glutamine (Gibco). The cells were spun down at 110 g for 5 min and washed once with phosphate buffered saline without calcium and magnesium before harvesting. Washed cells were passed through the pre-wet 40 µm cell strainers (BD Falcon, San Jose, CA, USA) for single cells, and counted using the Moxi™ Z Mini Automated Cell Counter (ORFLO Technologies).
For the whole exome sequencing experiment, a total of 10 single cells were isolated and a pool of GM12878 cells was used as the unamplified bulk control.
4.2. Clinical sample, pathological characteristics and cell preparation
The tissue samples were taken from a resected stage IIA lung adenocarcinoma of a 68 years old female non-smoking lung cancer patient of the National Cancer Centre Singapore. The adenocarcinoma consisted of predominantly acinar histology (55%) with minor papillary (20%), micropapillary (10%) and solid (15%) components. The tumour was tested negative for translocations of ALK, ROS1, and RET and amplification of MET and positive for an activating EGFR exon 19 deletion. Of the resected specimen, two tumour sectors and a morphologically normal lung sector more than 5 cm away from the tumour margin were obtained. The patient received no treatment before surgery and had given written informed consent to participate in this study. The biological samples were collected following the protocols approved by the Institutional Review Board (IRB).
The tumour tissues were transferred from the hospital to laboratory in cold washing buffer (DMEM/F12 (Gibco), 5% FBS (Standard, Gibco) supplemented with 1% 10,000U/ml penicillin–streptomycin (Gibco)). The tumour was washed thrice with cold washing buffer and chopped into smaller pieces with a sterile scalpel blade (Aesculap) followed by an incubation at 37 °C in 10 ml collagenase + dispase concoction (1 mg/ml) with shaking for 2 to 3 h. The suspensions were repeatedly washed with washing buffer for 5 times, and passed through the pre-wet 40 µm cell strainers (BD Falcon, San Jose, CA, USA) for single cells. The cells were counted using the Moxi™ Z Mini Automated Cell Counter (ORFLO Technologies).
4.3. WGA of single cell genomic DNA for whole exome sequencing
The GE Healthcare illustra GenomiPhi V2 DNA Amplification Kit and the Qiagen REPLI-g single cell kit were used on both tube and microfluidics platform. For the Qiagen REPLI-g single cell kit, the protocol recommended 8 h of incubation time.
4.4. Single cell WGA run on Fluidigm C1 Auto-prep platform
The medium-sized (10 to 17 µm) C1 chip (Fluidigm) was primed with C1 Harvest Reagent, Preloading Reagent, Blocking Reagent and C1 DNA Seq Cell Wash Buffer (Fluidigm) for 10 min before it was loaded with the dissociated single cells. The DTT Mix was prepared by the addition of DTT, Sample and Reaction Buffers (GE Healthcare). The Lysis Mix contained C1 DNA Seq Lysis Buffer and DTT (Fluidigm), while the Reaction-Enzyme Mix consisted of C1 DNA Seq Reaction Mix (Fluidigm), DTT Mix and Enzyme Mix (GE Healthcare). The Lysis Mix, Reaction-Enzyme Mix and C1 DNA Seq Stop Buffer were loaded on the C1 chip followed by the on-chip whole genome amplification experiment. The amplified DNA was harvested from the C1 chip and transferred into 96-well PCR plate. The DNA was quantified using PicoGreen dsDNA quantification assay (Thermo Fisher) on the Infinite 200Pro plate reader (Tecan).
4.5. Whole exome sequencing using Illumina Nextera Rapid Capture kit
The hybridization and library preparation for whole exome sequencing was carried out following the instructions provided by the Nextera Rapid Capture Enrichment kit (Illumina) with some modifications. A total of 10 ng input DNA were aliquoted into PCR plate followed by a 10-minute tagmentation at 58 °C. The dual indices were added into the tagmented DNA under the PCR amplification with the following thermal cycler’s setting: 72 °C for 3 min, 98 °C for 30 sec, 10 cycles of 98 °C for 10 sec, 60 °C for 30 sec, 72 °C for 30 sec, 72 °C for 5 min. The barcoded libraries were purified using magnetic Sample Purification Beads (SPB) and pooled together. Two rounds of hybridization with the Coding Exome Oligos (CEX) were carried out at 58 °C for 2 h. The enriched library was purified with Sample Purification Beads (SPB) and amplified with the supplied PCR Primer Cocktail (PPC) and Nextera Enrichment Amplification Mix (NEM) under the following thermal cycler’s setting: 98 °C for 30 sec, 10 cycles of 98 °C for 10 sec, 60 °C for 30 sec, 72 °C for 30 sec, 72 °C for 5 min. This final amplified library was purified by SPB and quantified using the KAPA Library Quantification Kit (KAPA Biosystems) on the LightCycler® 480 platform (Roche). The libraries were sequenced on Illumina Hiseq 2500 with paired-end read of 101 bp.
4.6. Read processing and mapping
4.6.1. Alignment of paired-end GM12878 exome sequencing reads
The sequenced reads were aligned to the Human reference genome hg19 using BWA MEM version 0.7.10-r789 [43] with default parameters. The mapped reads were sorted and duplicated reads were marked using Picard tool version 1.129 [44]. Lastly, Indel realignment and base recalibration were conducted using GATK Version 3.1–1[45]with default parameters to obtain the final BAM files for analysis. Due to differences in the sequencing depth, Picard tool version 1.129 was used to randomly down-sample the aligned reads. The resulting down-sampled BAM files were further used to compare the performance of different WGA kits for single nucleotide variants (SNVs) detection.
4.6.2. Alignment of paired-end lung cancer exome sequencing reads
BWA MEM version 0.7.5a-r405 [43] with default parameters was used to align sequencing reads to the Human reference genome hg19. Picard tool version 1.129 was used to sort and mark duplicated reads. GATK version 3.5 [45]with default parameters was used to perform indel realignment and base recalibration. For the lung cancer single cell data, an additional joint indel recalibration was performed using candidate sites obtain from all lung single cells.
4.6.3. Variant detection on GM12878 exome sequencing data
Variants from GM12878 single cells amplified by both C1-GE and C1-REPLI were detected using GATK haplotypeCaller version 3.1.1 using the following parameters: mapping quality (MQ) ≥ 40, base quality (BQ) ≥ 20). Joint genotyping was conducted for cells amplified by each kit separately so as to produce a single VCF file per kit. SNV sites were filtered using the following hard filters recommended by GATK: quality by depth (QD) < 2.0 or fisher strand (FS) greater than 60.0 or root mean square of the mapping quality (MQ) < 40.0 or mapping quality rank sum test (MQRankSum) < -12.5 or read position rank sum test (ReadPosRankSum) < -8.0). For INDEL sites, the following hard filters recommended by GATK were used to remove low quality sites: QD < 2.0 or FS greater than 200.0 or ReadPosRankSum < -20.0.
In order to compare the performance of C1-GE and C1-REPLI kit, all the following statistics were calculated using sites which were covered by at least 5 reads in each cell. ADO was calculated using the following formula Eq. (1):
| (1) |
Heterozygous sites detected in the platinum genome was obtained by downloading the gold standard GM12878 variants detected from the platinum genome project. The platinum genome version 8.0.1 dataset was downloaded from the platinum genome website[17].
Marie et al defined p as the probability of observing an allele as Eq. (2)
| (2) |
Where a is the number of times both alleles are detected, and b is the number of times only one of the two alleles was detected in the sample [13]. This was calculated for each sample using the heterozygous sites detected in the platinum genome.
FP rate was calculated using the following formula Eq. (3):
| (3) |
True negative sites were defined as sites within the exonic target regions whereby no variant was detected in the platinum genome project.
Evenness score E Eq. (4) was defined as the fraction of coverage that is correctly distributed[46].
| (4) |
Where Cave is the average coverage, NTP is the targeted position, and Pi is the number of targeted positions with at least coverage i.
Gini coefficients were calculated as Eq. (5)
| (5) |
Where A is the area under the Lorenz curve.
4.7. Variant detection in bulk lung cancer exome sequencing data
Germline variants (SNVs and INDELs) in the bulk tumour sectors and adjacent far normal tissue were detected using GATK haplotypeCaller v3.5 followed by hard filtering recommended by GATK best practices for both SNVs and INDELs. Germline SNVs were further filtered by removing variants which have DP < 8 or GQ < 30. For germline INDELs, we removed variants which have DP < 5 or GQ < 20.
Putative somatic SNVs were called by comparing bulk tumour samples with the adjacent normal tissue using Mutect with default parameters[47]. Somatic INDELS were detected by comparing bulk tumour samples with the adjacent tissue using Strelka[48]. Both somatic SNVs and indels with variant allele frequency<0.05 were removed to prevent spurious detection. The remaining variants were annotated via ANNOVAR [49].
Copy number variations (CNVs) were detected using EXCAVATOR2[50]after removing duplicated reads, secondary alignments and unmapped reads. The CNVs detected were annotated using PennCNV[51].
4.8. Quality control of lung single cells after sequencing
Exome sequencing was carried out on 66 single cells from tumour sector 1 (T1), 95 single cells from tumour sector 2 (T2) and 39 single cells from the far normal following the C1-GE protocol. Percentage of target regions covered (≥5 reads) was used as the first quality control criteria. The cells exhibited a bimodal distribution of coverage (Fig. 3B). Gaussian Mixture Model (GMM) was used twice to separate the cells into three groups: cells with coverage < 10%, 10% to < 42%, and ≥ 42% respectively. Cells belonging to the two lower coverage clusters were not considered in further analysis. The remaining cells were further filtered for allelic drop out (ADO) and false negative (FN) rates. For this the consensus heterozygous germline variant calls in the three bulk samples were used as the true variant set (see Supplementary Methods for details). In total 18 single cells from T1, 21 single cells from T2 and 27 single cells from the far normal passed the cell level filtering. These cells had at least 42% of the target regions covered (≥5 reads), ADO rate ≤ 0.35 and FN rate ≤ 0.45.
4.9. Variant detection in lung cancer single cell exome sequencing data
Variants in qualified tumour and normal single cells were detected via GATK haplotypeCaller v3.5 using the parameter (mapping quality (MQ) ≥ 40, base quality (BQ) ≥ 20). This was followed by joint genotyping and variant recalibration. All the tumour and normal cells were processed together to produce a single VCF file containing all potential variant sites. GATK variant recalibrator was used to filter the output at 99.9% sensitivity level. Recalibration training databases used include dbSNP build 138, Omni 2.5 M, 1000 genome phase 1 SNPs, Hapmap version 3.3, and Mills and 1000 genome gold standard INDELs. For SNVs, annotations used for recalibration training include variant quality score by read depth (QD), strand bias (FS), mapping quality rank sum score (MQRankSum), read position rank sum score (ReadPosRankSum), and mapping quality (MQ). For INDELS, the annotations used for recalibration training include variant quality score by read depth (QD), strand bias (FS), mapping quality rank sum score (MQRankSum), and read position rank sum score (ReadPosRankSum).
After variant recalibration, variants within 10 bp of each other, tri-allelic sites, and singletons were removed to reduce the variant false positives rate (Supplementary Fig. 3). For SNVs, genotypes with read depth (DP) < 5, genotype quality (GQ) < 30, and variant allele frequency (VAF) < 0.15 were removed. Variant genotypes which failed the GQ filter were re-examined by comparing the difference in phred likelihood score (PL) between the homozygous reference genotype and the maximum of heterozygous genotype and homozygous alt genotype. If the difference is greater than 30, the genotype will be retained. For INDELS, genotypes with DP < 5, GQ < 40, and VAF < 0.2 were removed. Variant sites which have variants detected in at least 3 single cells were retained. The thresholds used for filtering of genotypes were determined by using variant calls in the three bulk samples (see Supplementary Methods for details).
Putative somatic variants were filtered (excluded) based on the following criteria within the sequencing data of the respective lung cancer patient: (I) variants were seen in<3 cells, (II) variants were detected in germline bulk normal tissue, (III) alternate allele was observed in more than one percent of total reads in germline bulk normal tissue pileup data, (IV) variants were detected in normal single cells (this criterion requires sites to be homozygous reference at the position of the putative somatic variant for all normal cells and have at least 3 normal cells covered), (V) variants were seen in normal single cells which failed QC. The flowchart showing the filters can be found in Fig. 2E. The final somatic variants were annotated via ANNOVAR.
To aid the understanding of tumour evolution, OncoNEM [34] was used to infer the relationship between the single cells.
4.10. Detection of copy number profiles from lung cancer single cell exome sequencing data
Sequencing reads within exonic target regions were counted and GC normalization was performed using Excavator2 EXCAVATORDataPrepare.pl script. To identify regions of copy number changes, the method by Patel et al. to detect copy number variations from single cell RNA-seq[33]was adopted. The exonic target regions were sorted based on their chromosomal location and a moving average of 2001 exonic target regions was used to estimate the copy number in each cell. 2001 was chosen based on the trade-off between higher resolution at lower numbers versus the better denoising at higher numbers. The following formula Eq. (6) was used to estimate the copy number for each region per chromosome in each cell:
| (6) |
Where i is the estimated average copy number change at target region i and j is the exonic region adjacent to i.
For each cell, a z-score is obtained per region using the following formula Eq. (7):
| (7) |
Euclidean distance between each cell using the z-score, followed by hierarchical clustering using R.
4.11. Validation of variants detected in lung single cells and bulk sequencing
28 sites were randomly selected for validation. 1 µg of genomic DNA was obtained by extracting DNA from a pool of dissociated cells from each sector. PCR reaction were conducted using the 28 primer sets and an agarose gel was used to validate the product size. The PCR products were then purified using MiniElute PCR Purification Kit (Qiagen) and PCR products were pooled for library preparation based on concentration measured by Agilent DNA 1000 kit (Agilent). Sequencing libraries were prepared using NEBnext DNA library Prep Master Mix Set for Illumina (NEB). Lastly, a final QC was done using KAPA (KAPA Biosystems) prior to sequencing. The libraries were sequenced using Illumina Miseq with paired-end reads of 151 bp.
The sequencing reads were aligned to the hg19 reference genome using BWA-MEM v0.7.10 with default parameters. The sequencing reads were sorted and duplicates were marked using Picard. INDEL realignment and base recalibration were done using GATK v3.5. SNVs were detected using Mutect and GATK HaplotypeCaller, whereas INDELs were detected using GATK HaplotypeCaller.
5. Comparison with Monovar
Monovar was run with default parameters using sequencing reads with mapping quality (MQ) ≥ 40[14] and base quality (BQ) ≥ 20. Filters recommended in the publication were then applied[14]. Thereby, variants within 10 bp of each other, tri-allelic sites, and singletons were removed to reduce the variant false positives rate. Putative somatic variants were filtered based on the following criteria: (I) variants were seen in<3 cells, (II) variants were detected in germline bulk normal tissue, (III) alternate allele was observed in more than one percent of total reads in germline bulk normal tissue pileup data, (IV) variants were detected in normal single cells (require sites to be homozygous reference for all normal cells and have at least 3 normal cells covered), (V) variants were seen in normal single cells which failed QC.
6. Declarations
6.1. Ethics approval and consent to participate
Written informed consent was obtained from the participating patient. The study was approved by the relevant Institutional Review Board (Singhealth Centralised IRB, Singapore; CIRB ref: 2007/444/B).
7. Availability of data and material
SoVaTSiC is implemented in Perl and R[52] and it is freely available and can be downloaded from https://github.com/JoannaTan/SoVaTSiC.
The GM12878 dataset generated and analysed during the current study are available in the European Nucleotide Archive (ENA) under the accession number of PRJEB22052. The lung adenocarcinoma dataset generated and analysed during the current study are available in the European Genome-phenome Archive (EGA) under the accession number of EGAS00001002972. The bladder cancer dataset analysed during the current study was downloaded from the NCBI short reads archive (SRA) under the accession number of SRA051489[16]. Additional file 2 is an excel file containing variants detected from both, bulk and single cells from lung adenocarcinoma dataset and bladder cancer dataset.
Funding
This work was supported by the Translational and Clinical Research Program “Non-Small Cell Lung Cancer: Targeting Cancer Stem Cell and Drug Resistance” by the National Medical Research Council, Singapore (NMRC/TCR/007-NCC/2013) and Strategic Positioning Fund (SPF2012/003) by Biomedical Research Council (BMRC), Singapore. J.H.J.T. was supported by National University of Singapore Research Scholarship. S.L.K. and A.J. were each supported by a Career Development Award (14302FG096 and 15302FG146 respectively) from Joint Council Office (JCO), Agency for Science, Technology and Research (A*STAR), Singapore.
Author contributions
S.L.K. and A.M.H. conceived the study. S.L.K. conducted or supervised all experiments with contributions from J.A.Y.T., H.M.P., F.Y., Y.Y.S., and .E K.H.L. J.H.J.T. analysed the data supervised by A.J. with contributions from A.M.T.P., and D.S.W.T. contributed to sample collection and interpretation of histological and clinical data. J.H.J.T and A.J. wrote the first draft with contributions from A.M.H. and S.L.K and approved by all co-authors. A.J. and A.M.H. directed the study.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank Elise T Courtois for help with the clinical samples, Burton K.H. Chia for help with first pass analysis, and Heng X. Ting for help with the figures. The authors would like to thank Siva Vijayakumar and Andreas Wilm for their comments on the manuscript. The authors would also like to thank Wang Lan Ying, Thum Chooi Mei, Maria Rosita Ibarola Negapatan for assistance in coordinating patient’s consent and sample’s collection.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.12.021.
Contributor Information
Asif Javed, Email: javed@hku.hk.
Axel M. Hillmer, Email: ahillmer@uni-koeln.de.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Gerlinger M., Rowan A.J., Horswell S., Larkin J., Endesfelder D., Gronroos E. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Bruin E.C., McGranahan N., Mitter R., Salm M., Wedge D.C., Yates L. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346(6206):251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J., Fujimoto J., Zhang J., Wedge D.C., Song X., Zhang J. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yates L.R., Gerstung M., Knappskog S., Desmedt C., Gundem G., Van Loo P. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med. 2015;21(7):751–759. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahul Nahar, W. Z., Tong Zhang, Angela Takano, Alexis J Khng, Yin Yeng Lee, Xingliang Liu, Chong Hee Lim, Tina P T Koh, Zaw Win Aung, Tony Kiat Hon Lim, Lavanya Veeravalli, Ju Yuan, Audrey S M Teo, Cheryl X Chan, Huay Mei Poh, Ivan M L Chua, Audrey Ann Liew, Dawn Ping Xi Lau, Xue Lin Kwang, Chee Keong Toh, Wan-Teck Lim, Bing Lim, Wai Leong Tam, Eng-Huat Tan, Axel M Hillmer, Daniel S W Tan (2018). “Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing.” Nature Communications. [DOI] [PMC free article] [PubMed]
- 6.Campbell P.J., Pleasance E.D., Stephens P.J., Dicks E., Rance R., Goodhead I. Proceedings of the National Academy of Sciences of the United States of America. 2008. pp. 13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding L., Ley T.J., Larson D.E., Miller C.A., Koboldt D.C., Welch J.S. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Bourcy C.F., De Vlaminck I., Kanbar J.N., Wang J., Gawad C., Quake S.R. A quantitative comparison of single-cell whole genome amplification methods. PLoS ONE. 2014;9(8) doi: 10.1371/journal.pone.0105585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ning, L., G. Wang, Z. Li, W. Hu, Q. Hou, Y. Tong, M. Zhang, L. Qin, X. Chen, H. Man, P. Liu and J. He (2014). “Quantitative comparison of single-cell sequencing methods using hippocampal neurons.” bioRxiv. [DOI] [PMC free article] [PubMed]
- 10.Hou Y., Wu K., Shi X., Li F., Song L., Wu H. Comparison of variations detection between whole-genome amplification methods used in single-cell resequencing. GigaScience. 2015;4(1):1–16. doi: 10.1186/s13742-015-0068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L., Ma F., Chapman A., Lu S., Xie X.S. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genomics Hum Genet. 2015;16:79–102. doi: 10.1146/annurev-genom-090413-025352. [DOI] [PubMed] [Google Scholar]
- 12.Erik Borgström, M. P., Jeff E Mold, Jonas Frisen, Joakim Lundeberg (2017). “Comparison of whole genome amplification techniques for human single cell exome sequencing.” PLoS ONE. [DOI] [PMC free article] [PubMed]
- 13.Marie R., Podenphant M., Koprowska K., Baerlocher L., Vulders R.C.M., Wilding J. Sequencing of human genomes extracted from single cancer cells isolated in a valveless microfluidic device. Lab Chip. 2018;18(13):1891–1902. doi: 10.1039/c8lc00169c. [DOI] [PubMed] [Google Scholar]
- 14.Zafar H., Wang Y., Nakhleh L., Navin N., Chen K. Monovar: single nucleotide variant detection in single cells. Nat Methods. 2016;13(6):505–507. doi: 10.1038/nmeth.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong X., Zhang L., Milholland B., Lee M., Maslov A.Y., Wang T. Accurate identification of single-nucleotide variants in whole-genome-amplified single cells. Nat Methods. 2017;14(5):491–493. doi: 10.1038/nmeth.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Xu X., Song L., Hou Y., Li Z., Tsang S. Single-cell sequencing analysis characterizes common and cell-lineage-specific mutations in a muscle-invasive bladder cancer. GigaScience. 2012;1(1):12. doi: 10.1186/2047-217X-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eberle M., Fritzilas E., Krusche P., Kallberg M., Moore B., Bekritsky M. A reference dataset of 5.4 million phased human variants validated by genetic inheritance from sequencing a three-generation 17-member pedigree. Genome Res. 2017;27:157–164. doi: 10.1101/gr.210500.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maemondo M., Inoue A., Kobayashi K., Sugawara S., Oizumi S., Isobe H. Gefitinib or chemotherapy for non–small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;2010(362):2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 19.Costa D.B. Kinase inhibitor-responsive genotypes in EGFR mutated lung adenocarcinomas: moving past common point mutations or indels into uncommon kinase domain duplications and rearrangements. Translational Lung Cancer Research. 2016;5(3):331–337. doi: 10.21037/tlcr.2016.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Midha A., Dearden S., McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII) American Journal of Cancer Research. 2015;5(9):2892–2911. [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue A., Yoshida K., Morita S., Imamura F., Seto T., Okamoto I. Characteristics and overall survival of EGFR mutation-positive non-small cell lung cancer treated with EGFR tyrosine kinase inhibitors: a retrospective analysis for 1660 Japanese patients. Jpn J Clin Oncol. 2016;46(5):462–467. doi: 10.1093/jjco/hyw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jianbin Chen, H. Y., Audrey Su Min Teo, Lidyana Bte Amer, Faranak Ghazi Sherbaf, Chu Quan Tan, Jacob Josiah Santiago Alvarez, Bingxin Lu, Jia Qi Lim, Angela Takano, Rahul Nahar, Yin Yeng Lee, Cheryl Zi Jin Phua, Khi Pin Chua, Lisda Suteja, Pauline Jieqi Chen, Mei Mei Chang, Tina Puay Theng Koh, Boon-Hean Ong, Devanand Anantham, Anne Ann Ling Hsu, Apoorva Gogna, Chow Wei Too, Zaw Win Aung, Yi Fei Lee, Lanying Wang, Tony Kiat Hon Lim, Andreas Wilm, Poh Sum Choi, Poh Yong Ng, Chee Keong Toh, Wan-Teck Lim, Siming Ma, Bing Lim, Jin Liu, Wai Leong Tam, Anders Jacobsen Skanderup, Joe Poh Sheng Yeong, Eng-Huat Tan, Caretha L Creasy, Daniel Shao Weng Tan, Axel M Hillmer, Weiwei Zhai (2020). “Genomic landscape of lung adenocarcinoma in East Asians.” Nature Genetics. [DOI] [PubMed]
- 23.The Cancer Genome Atlas Research, N., E. A. Collisson, J. D. Campbell, A. N. Brooks, A. H. Berger, W. Lee, J. Chmielecki, D. G. Beer, L. Cope, C. J. Creighton, L. Danilova, L. Ding, G. Getz, P. S. Hammerman, D. Neil Hayes, B. Hernandez, J. G. Herman, J. V. Heymach, I. Jurisica, R. Kucherlapati, D. Kwiatkowski, M. Ladanyi, G. Robertson, N. Schultz, R. Shen, R. Sinha, C. Sougnez, M.-S. Tsao, W. D. Travis, J. N. Weinstein, D. A. Wigle, M. D. Wilkerson, A. Chu, A. D. Cherniack, A. Hadjipanayis, M. Rosenberg, D. J. Weisenberger, P. W. Laird, A. Radenbaugh, S. Ma, J. M. Stuart, L. Averett Byers, S. B. Baylin, R. Govindan, M. Meyerson, M. Rosenberg, S. B. Gabriel, K. Cibulskis, C. Sougnez, J. Kim, C. Stewart, L. Lichtenstein, E. S. Lander, M. S. Lawrence, G. Getz, C. Kandoth, R. Fulton, L. L. Fulton, M. D. McLellan, R. K. Wilson, K. Ye, C. C. Fronick, C. A. Maher, C. A. Miller, M. C. Wendl, C. Cabanski, L. Ding, E. Mardis, R. Govindan, C. J. Creighton, D. Wheeler, M. Balasundaram, Y. S. N. Butterfield, R. Carlsen, A. Chu, E. Chuah, N. Dhalla, R. Guin, C. Hirst, D. Lee, H. I. Li, M. Mayo, R. A. Moore, A. J. Mungall, J. E. Schein, P. Sipahimalani, A. Tam, R. Varhol, A. Gordon Robertson, N. Wye, N. Thiessen, R. A. Holt, S. J. M. Jones, M. A. Marra, J. D. Campbell, A. N. Brooks, J. Chmielecki, M. Imielinski, R. C. Onofrio, E. Hodis, T. Zack, C. Sougnez, E. Helman, C. Sekhar Pedamallu, J. Mesirov, A. D. Cherniack, G. Saksena, S. E. Schumacher, S. L. Carter, B. Hernandez, L. Garraway, R. Beroukhim, S. B. Gabriel, G. Getz, M. Meyerson, A. Hadjipanayis, S. Lee, H. S. Mahadeshwar, A. Pantazi, A. Protopopov, X. Ren, S. Seth, X. Song, J. Tang, L. Yang, J. Zhang, P.-C. Chen, M. Parfenov, A. Wei Xu, N. Santoso, L. Chin, P. J. Park, R. Kucherlapati, K. A. Hoadley, J. Todd Auman, S. Meng, Y. Shi, E. Buda, S. Waring, U. Veluvolu, D. Tan, P. A. Mieczkowski, C. D. Jones, J. V. Simons, M. G. Soloway, T. Bodenheimer, S. R. Jefferys, J. Roach, A. P. Hoyle, J. Wu, S. Balu, D. Singh, J. F. Prins, J. S. Marron, J. S. Parker, D. Neil Hayes, C. M. Perou, J. Liu, L. Cope, L. Danilova, D. J. Weisenberger, D. T. Maglinte, P. H. Lai, M. S. Bootwalla, D. J. Van Den Berg, T. Triche Jr, S. B. Baylin, P. W. Laird, M. Rosenberg, L. Chin, J. Zhang, J. Cho, D. DiCara, D. Heiman, P. Lin, W. Mallard, D. Voet, H. Zhang, L. Zou, M. S. Noble, M. S. Lawrence, G. Saksena, N. Gehlenborg, H. Thorvaldsdottir, J. Mesirov, M.-D. Nazaire, J. Robinson, G. Getz, W. Lee, B. Arman Aksoy, G. Ciriello, B. S. Taylor, G. Dresdner, J. Gao, B. Gross, V. E. Seshan, M. Ladanyi, B. Reva, R. Sinha, S. Onur Sumer, N. Weinhold, N. Schultz, R. Shen, C. Sander, S. Ng, S. Ma, J. Zhu, A. Radenbaugh, J. M. Stuart, C. C. Benz, C. Yau, D. Haussler, P. T. Spellman, M. D. Wilkerson, J. S. Parker, K. A. Hoadley, P. K. Kimes, D. Neil Hayes, C. M. Perou, B. M. Broom, J. Wang, Y. Lu, P. Kwok Shing Ng, L. Diao, L. Averett Byers, W. Liu, J. V. Heymach, C. I. Amos, J. N. Weinstein, R. Akbani, G. B. Mills, E. Curley, J. Paulauskis, K. Lau, S. Morris, T. Shelton, D. Mallery, J. Gardner, R. Penny, C. Saller, K. Tarvin, W. G. Richards, R. Cerfolio, A. Bryant, D. P. Raymond, N. A. Pennell, C. Farver, C. Czerwinski, L. Huelsenbeck-Dill, M. Iacocca, N. Petrelli, B. Rabeno, J. Brown, T. Bauer, O. Dolzhanskiy, O. Potapova, D. Rotin, O. Voronina, E. Nemirovich-Danchenko, K. V. Fedosenko, A. Gal, M. Behera, S. S. Ramalingam, G. Sica, D. Flieder, J. Boyd, J. Weaver, B. Kohl, D. Huy Quoc Thinh, G. Sandusky, H. Juhl, E. Duhig, P. Illei, E. Gabrielson, J. Shin, B. Lee, K. Rodgers, D. Trusty, M. V. Brock, C. Williamson, E. Burks, K. Rieger-Christ, A. Holway, T. Sullivan, D. A. Wigle, M. K. Asiedu, F. Kosari, W. D. Travis, N. Rekhtman, M. Zakowski, V. W. Rusch, P. Zippile, J. Suh, H. Pass, C. Goparaju, Y. Owusu-Sarpong, J. M. S. Bartlett, S. Kodeeswaran, J. Parfitt, H. Sekhon, M. Albert, J. Eckman, J. B. Myers, R. Cheney, C. Morrison, C. Gaudioso, J. A. Borgia, P. Bonomi, M. Pool, M. J. Liptay, F. Moiseenko, I. Zaytseva, H. Dienemann, M. Meister, P. A. Schnabel, T. R. Muley, M. Peifer, C. Gomez-Fernandez, L. Herbert, S. Egea, M. Huang, L. B. Thorne, L. Boice, A. Hill Salazar, W. K. Funkhouser, W. Kimryn Rathmell, R. Dhir, S. A. Yousem, S. Dacic, F. Schneider, J. M. Siegfried, R. Hajek, M. A. Watson, S. McDonald, B. Meyers, B. Clarke, I. A. Yang, K. M. Fong, L. Hunter, M. Windsor, R. V. Bowman, S. Peters, I. Letovanec, K. Z. Khan, M. A. Jensen, E. E. Snyder, D. Srinivasan, A. B. Kahn, J. Baboud, D. A. Pot, K. R. Mills Shaw, M. Sheth, T. Davidsen, J. A. Demchok, L. Yang, Z. Wang, R. Tarnuzzer, J. Claude Zenklusen, B. A. Ozenberger, H. J. Sofia, W. D. Travis, R. Cheney, B. Clarke, S. Dacic, E. Duhig, W. K. Funkhouser, P. Illei, C. Farver, N. Rekhtman, G. Sica, J. Suh and M.-S. Tsao (2014). “Comprehensive molecular profiling of lung adenocarcinoma.” Nature 511: 543-550 [DOI] [PMC free article] [PubMed]
- 24.Horvath S., Zhang B., Carlson M., Lu K., Zhu S., Felciano R. Proceedings of the National Academy of Sciences of the United States of America. 2006. pp. 17402–17407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin S.-Y., Pan H.-W., Liu S.-H., Jeng Y.-M., Hu F.-C., Peng S.-Y. ASPM is a novel marker for vascular invasion, early recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res. 2008;14(15):4814–4820. doi: 10.1158/1078-0432.CCR-07-5262. [DOI] [PubMed] [Google Scholar]
- 26.Bikeye S.N.N., Colin C., Marie Y., Vampouille R., Ravassard P., Rousseau A. ASPM-associated stem cell proliferation is involved in malignant progression of gliomas and constitutes an attractive therapeutic target. Cancer Cell International. 2010;10:1. doi: 10.1186/1475-2867-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brüning-Richardson A., Bond J., Alsiary R., Richardson J., Cairns D., McCormack L. ASPM and microcephalin expression in epithelial ovarian cancer correlates with tumour grade and survival. Br J Cancer. 2011;104(10):1602–1610. doi: 10.1038/bjc.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vulcani-Freitas T.M., Saba-Silva N., Cappellano A., Cavalheiro S., Marie S.K., Oba-Shinjo S.M. ASPM gene expression in medulloblastoma. Child's Nervous System. 2011;27(1):71–74. doi: 10.1007/s00381-010-1252-5. [DOI] [PubMed] [Google Scholar]
- 29.Wang W.Y., Hsu C.C., Wang T.Y., Li C.R., Hou Y.C., Chu J.M. A gene expression signature of epithelial tubulogenesis and a role for ASPM in pancreatic tumor progression. Gastroenterology. 2013;145(5):1110–1120. doi: 10.1053/j.gastro.2013.07.040. [DOI] [PubMed] [Google Scholar]
- 30.Williams S.E., Garcia I., Crowther A.J., Li S., Stewart A., Liu H. Aspm sustains postnatal cerebellar neurogenesis and medulloblastoma growth in mice. Development. 2015;142(22):3921–3932. doi: 10.1242/dev.124271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Yang Y., Chen L., Zhang J. Expression analysis of genes and pathways associated with liver metastases of the uveal melanoma. BMC Med Genet. 2014;15(1):29. doi: 10.1186/1471-2350-15-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koehne A.L., Sayles L.C., Breese M.R., Vaka D., Sweet-Cordero A. Abstract A35: Characterization of the genomic landscape of osteosarcoma metastasis. Cancer Res. 2016;76(5 Supplement):A35. [Google Scholar]
- 33.Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross E.M., Markowetz F. OncoNEM: inferring tumor evolution from single-cell sequencing data. Genome Biol. 2016;17:69. doi: 10.1186/s13059-016-0929-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cappellen D., De Oliveira C., Ricol D., de Medina S., Bourdin J., Sastre-Garau X. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23(1):18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 36.Junker K., van Oers J.M., Zwarthoff E.C., Kania I., Schubert J., Hartmann A. Fibroblast growth factor receptor 3 mutations in bladder tumors correlate with low frequency of chromosome alterations. Neoplasia. 2008;10(1):1–7. doi: 10.1593/neo.07178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kresse S.H., Ohnstad H.O., Paulsen E.B., Bjerkehagen B., Szuhai K., Serra M. LSAMP, a novel candidate tumor suppressor gene in human osteosarcomas, identified by array comparative genomic hybridization. Genes Chromosomes Cancer. 2009;48(8):679–693. doi: 10.1002/gcc.20675. [DOI] [PubMed] [Google Scholar]
- 38.Gu J., Xia X., Yan P., Liu H., Podust V.N., Reynolds A.B. Cell cycle-dependent regulation of a human DNA helicase that localizes in DNA damage foci. Mol Biol Cell. 2004;15(7):3320–3332. doi: 10.1091/mbc.E04-03-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma A., Warner S.L., Vankayalapati H., Bearss D.J., Sharma S. Targeting Axl and Mer kinases in cancer. Mol Cancer Ther. 2011;10(10):1763–1773. doi: 10.1158/1535-7163.MCT-11-0116. [DOI] [PubMed] [Google Scholar]
- 40.Laoukili J., Kooistra M.R., Brás A., Kauw J., Kerkhoven R.M., Morrison A. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7(2):126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 41.Rinaldetti S., Wirtz R.M., Worst T.S., Eckstein M., Weiss C.A., Breyer J. FOXM1 predicts overall and disease specific survival in muscle-invasive urothelial carcinoma and presents a differential expression between bladder cancer subtypes. Oncotarget. 2017;8(29):47595–47606. doi: 10.18632/oncotarget.17394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu Y.S., Zhang F.L., Zhang X.N., Yin J.L., Du M.J., Jiang M.C. High-throughput single-cell whole-genome amplification through centrifugal emulsification and eMDA. Communications Biology. 2019;2 doi: 10.1038/s42003-019-0401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li, H. (2013). “Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM.” arXiv:1303.3997.
- 44.Broad Institute. “Picard Tools Version 1.129.” 2014 - 2018, from http://broadinstitute.github.io/picard.
- 45.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., Banks E., Garimella K.V., Altshuler D., Gabriel S., DePristo M.A. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Current Protocols in Bioinformatics. 2013;43(1) doi: 10.1002/0471250953.bi1110s43. 11.10.11-11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mokry M., Feitsma H., Nijman I.J., de Bruijn E., van der Zaag P.J., Guryev V. Accurate SNP and mutation detection by targeted custom microarray-based genomic enrichment of short-fragment sequencing libraries. Nucleic Acids Res. 2010;38(10) doi: 10.1093/nar/gkq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saunders C.T., Wong W.S., Swamy S., Becq J., Murray L.J., Cheetham R.K. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28(14):1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 49.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16) doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D'Aurizio R., Pippucci T., Tattini L., Giusti B., Pellegrini M., Magi A. Enhanced copy number variants detection from whole-exome sequencing data using EXCAVATOR2. Nucleic Acids Res. 2016;44(20) doi: 10.1093/nar/gkw695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F.A. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17(11):1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Team, R. C. (2015). R: A language and environment for statistical computing [Internet]. Vienna, Austria: R Foundation for Statistical Computing; 2014.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SoVaTSiC is implemented in Perl and R[52] and it is freely available and can be downloaded from https://github.com/JoannaTan/SoVaTSiC.
The GM12878 dataset generated and analysed during the current study are available in the European Nucleotide Archive (ENA) under the accession number of PRJEB22052. The lung adenocarcinoma dataset generated and analysed during the current study are available in the European Genome-phenome Archive (EGA) under the accession number of EGAS00001002972. The bladder cancer dataset analysed during the current study was downloaded from the NCBI short reads archive (SRA) under the accession number of SRA051489[16]. Additional file 2 is an excel file containing variants detected from both, bulk and single cells from lung adenocarcinoma dataset and bladder cancer dataset.