Abstract
RYR1 encodes the type 1 ryanodine receptor, an intracellular calcium release channel (RyR1) on the skeletal muscle sarcoplasmic reticulum (SR). Pathogenic RYR1 variations can destabilize RyR1 leading to calcium leak causing oxidative overload and myopathy. However, the effect of RyR1 leak has not been established in individuals with RYR1-related myopathies (RYR1-RM), a broad spectrum of rare neuromuscular disorders. We sought to determine whether RYR1-RM affected individuals exhibit pathologic, leaky RyR1 and whether variant location in the channel structure can predict pathogenicity. Skeletal muscle biopsies were obtained from 17 individuals with RYR1-RM. Mutant RyR1 from these individuals exhibited pathologic SR calcium leak and increased activity of calcium-activated proteases. The increased calcium leak and protease activity were normalized by ex-vivo treatment with S107, a RyR stabilizing Rycal molecule. Using the cryo-EM structure of RyR1 and a new dataset of >2200 suspected RYR1-RM affected individuals we developed a method for assigning pathogenicity probabilities to RYR1 variants based on 3D co-localization of known pathogenic variants. This study provides the rationale for a clinical trial testing Rycals in RYR1-RM affected individuals and introduces a predictive tool for investigating the pathogenicity of RYR1 variants of uncertain significance.
Introduction
RYR1-related myopathies (RYR1-RM) comprise a spectrum of rare neuromuscular disorders for which there is no approved treatment [25]. Causative variants in RYR1 (19q13.2) lead to defects in the skeletal muscle ryanodine receptor (RyR1) protein and result in RYR1-RM that affects at least 1:90,000 individuals in the United States [1]. RyR1 is an intracellular Ca2+ release channel in the sarcoplasmic reticulum (SR), that is required for excitation-contraction coupling (ECC) [30,23,43,2,45]. Calstabin1 binds to RyR1 and stabilizes the closed state of the channel [7]. Defects in RyR1 that decrease the binding affinity of calstabin1, including PKA phosphorylation, oxidation, and Cys-nitrosylation, result in leaky channels [2,13,11,21,33,40,41]. RyR1-mediated SR Ca2+ leak causes muscle dysfunction via impaired ECC, activation of Ca2+ dependent caspases, and mitochondrial Ca2+ overload resulting in downstream production of pathologic reactive oxygen species (ROS). The oxidative environment also creates a feed-forward mechanism that promotes further RyR1 dysfunction {Andersson, 2011 #43.
Rycals are a class of compounds derived from a 1,4-benzothiazepine backbone that prevent intracellular Ca2+overload in disease states by stabilizing leaky ryanodine receptors. These improve cardiac muscle function in mice with heart failure by stabilizing the binding between calstabin2 and RyR2 {Lehnart, 2006 #164}[44,42]. Rycals improve skeletal muscle function by stabilizing calstabin1 binding to RyR1 and reducing RyR1 Ca2+ leak [43]. Subsequent studies demonstrated that treating leaky RyR1 with Rycals improves muscle function in Duchenne muscular dystrophy [5,14], diaphragmatic weakness during mechanical ventilation [31], and limb-girdle muscular dystrophy [3]. Elucidating whether RYR1-RM affected individuals exhibit calstabin1 depletion from RyR1 and Ca2+ leak is important for predicting whether such individuals may respond to Rycal therapy.
RyR1 is a homotetramer comprised of four identical 565,000 dalton protomers. Inherited mutations in RYR1 cause skeletal muscle myopathies of varying clinical phenotypes, including proximal versus and/or muscle weakness, hypotonia, contractures, scoliosis, respiratory insufficiency, and ophthalmoplegia. The relationship between particular pathologic variants and clinical phenotype, including rhabdomyolysis, intermittent periodic paralysis, [10,27,32] and risk for malignant hyperthermia (MH), a potentially fatal hypermetabolic response to pharmacological triggers, is not well understood. We compiled an RYR1 dataset from thousands of individuals with reported RYR1 variants and clinical suspicion for RYR1-RM.
The rarity of RYR1-RM makes it difficult to assign pathogenicity to variants of unknown significance (VUS) and provide individuals with a definitive diagnosis, prognosis and therapeutic strategy. We therefore also sought to develop an algorithm, based on a new RYR1 dataset and the 3D structure of RyR1, for predicting the pathogenicity of RYR1 VUS.
Results
Twenty-eight RYR1 variants were identified in 17 affected individuals whose muscle tissue was analyzed in this study. Of these variants, 26 were distinct (p.Arg4861His and p.Ser4028Leu were present twice, Table 1 and Supplementary Figure 1). Variant subtypes were as follows: missense substitutions (n= 22), deletions (n= 1), duplications (n=1), and nonsense variations (n= 2). Bioinformatic analyses of missense substitutions included in this study and 10 benign variants (>0.3% incidence in gnomAD database) are presented in Supplementary Table 1. The variants were also mapped to the RyR1 channel complex (Table 1, Supplementary Figure 1).
Table 1: Genetic data of individuals included in this study.
RYR1 sequencing methods. Mode of inheritance (MOI) determined through pedigree data. For cases without parental genetic testing, a plausible mode of inheritance was established through careful evaluation of clinical manifestations. ClinVar clinical significance as reported at https://www.ncbi.nlm.nih.gov/clinvar. Structure-based variant pathogenicity assignment provided in arbitrary units (AU) and as the probability of pathogenicity as described.
| Case | Sex | MOI | RYR1 sequencing | Nucleotide change | Amino acid change | Variant classification (based on ClinVar) | Structure-based variant pathogenicity assignment | Protein Location | |
|---|---|---|---|---|---|---|---|---|---|
| Score (AU) | Pathogenicity probability | ||||||||
| 1 | M | Recessive | Complete | c.4999C>T | p.Arg1667Cys | Conflicting | 0.000 | 0.682 | Jsol |
| c.5140_5142del | p.Leu1714del | VUS | - | - | Jsol | ||||
| c.14126C>T | p.Thr4709Met | Pathogenic | 0.001 | 0.686 | pVSD (S2S3) | ||||
| 2 | M | Dominant | Complete | c.12553G>A | p.Ala4185Thr | Conflicting | 0.001 | 0.685 | TaF |
| 3 | F | Dominant | Targeted | c.7354C>T | p.Arg2452Trp | Pathogenic | 0.189 | 0.973 | BSol |
| 4 | F | De novo | Complete | c.14693T>C | p.Ile4898Thr | VUS | 0.280 | 0.993 | Pore |
| 5 | F | Dominant | Targeted | c.14818G>A | p.Ala4940Thr | Pathogenic | 0.159 | 0.958 | Pore (S6c) |
| 6 | F | Dominant | Targeted | c.14582G>A | p.Arg4861His | Pathogenic | 0.195 | 0.975 | Pore |
| 7 | M | Recessive | Complete | c.6721C>T | p.Arg2241* | Pathogenic | - | - | Bsol |
| c.325C>T | p.Arg109Trp | Pathogenic | 0.081 | 0.878 | NTD-A | ||||
| c.2122G>A | p.Asp708Asn | Conflicting | 0.000 | 0.682 | SPRY1 | ||||
| c.1453A>G | p.Met485Val | Conflicting | 0.000 | 0.684 | Nsol | ||||
| 8 | F | Dominant a | Complete | c.14582G>A | p.Arg4861His | Pathogenic | 0.195 | 0.975 | Pore |
| c.13331_13351dup | p. Gly4444–50dup | VUS | - | - | Unresolved | ||||
| 9 | M | Dominant | Targeted | c.12083C>T | p.Ser4028Leu | VUS | 0.091 | 0.893 | Csol |
| 10 | F | Dominant | Targeted | c.14558C>T | p.Thr4853Ile | Pathogenic | 0.179 | 0.969 | Pore |
| 11 | F | Dominant | Exome | c.14731G>A | p.Glu4911Lys | VUS | 0.096 | 0.900 | Pore |
| 12 | M | Recessive | Exome | c.1589G>A | p.Arg530His | VUS | 0.161 | 0.959 | Nsol |
| c.3127C>T | p.Arg1043Cys | VUS | 0.001 | 0.685 | RY1&2 | ||||
| c.7007G>A | p.Arg2336His | Pathogenic | 0.155 | 0.956 | Bsol | ||||
| 13 | M | Dominant | Targeted | c.14681C>A | p.Ala4894Asp | VUS | 0.269 | 0.992 | Pore |
| 14 | M | Recessive | Complete | c.1993del | p.Val665* | Likely pathogenic | - | - | SPRY1 |
| c.4816C>A | p.Arg1606Ser | VUS | 0.001 | 0.686 | SPRY3 | ||||
| 15 | M | De novo | Exome | c.12083C>T | p.Ser4028Leu | VUS | 0.091 | 0.893 | Csol |
| 16 | M | Dominant | Complete | c.14209C>T | p.Arg4737Trp | Pathogenic | 0.174 | 0.966 | pVSD (S2S3) |
| 17 | M | Recessive | Targeted | c.10097G>A | p.Arg3366His | VUS | 0.109 | 0.916 | Bsol |
| c.11798A>G | p.Tyr3933Cys | VUS | 0.002 | 0.689 | Csol | ||||
| c.14645C>T | p.Thr4882Met | Conflicting | 0.018 | 0.736 | Pore | ||||
Abbreviations: M, male; F, female; MOI, mode of inheritance; VUS, variant of uncertain significance; del, deletion
premature stop codon; Jsol, junctional solenoid; pVSD, pseudo-voltage-sensing domain; S2S3, Helical-bundle domain between S2 and S3; TaF, Thumb and Forefingers domain; Bsol, bridging solenoid; Pore, channel pore domain; S6c, Cytoplasmic extension of S6; NTD-A, N-terminal domain A; SPRY1, SP1a / Ryanodine receptor domain 1; Nsol, N-terminal solenoid; Csol, core solenoid; RY1&2, RYR Repeats 1 and 2; SPRY3, SP1a / Ryanodine receptor domain.
RyR1 Channel Activity and Properties
Biochemistry and electrophysiology
We assessed calstabin1 binding to RyR1 and RyR1 function in skeletal muscle from the RYR1-RM affected individuals in this study. Co-immunoprecipitation with immunoblotting demonstrated increased channel oxidation and reduced RyR1-calstabin1 binding to mutant channels relative to control (Figure 1a–b). Reduced RyR1-calstabin1 binding was consistently observed in all patient samples (n=17) regardless of where the RYR1 variants were located and ranged from 10–25% of control (Figure 1b). Case 2 with the p.Ala4185Thr variant and a less certain clinical diagnosis of RYR1-RM exhibited channels with only modestly reduced calstabin1 binding, consistent with our computational modeling that assigned the variant a low probability of pathogenicity score (Table 1). We previously showed that RyR1 oxidation causes a reduction in calstabin1 binding to RyR1 and promotes channel leak [13,11,41,3,26,20,8,46,35,36] (Figure 1a,b). Treating RYR1-RM skeletal muscle ex-vivo with S107 improved calstabin1 binding to RyR1 in all samples (Figure 1). In addition, calpain (a Ca2+-activated protease that can cause muscle damage) activity was increased in the presence of leaky RyR1 channels (Figure 1c).
Figure 1. Reduced calstabin1 binding to RYR1-RM channels restored by ex-vivo treatment with Rycal.
(a) RyR1 was immunoprecipitated using a RyR1-specific antibody from 0.1 mg of skeletal muscle lysates obtained from RYR1-RM affected individuals included in this study in the absence and presence of 1.0 μM S107. Co-immunoprecipitated calstabin1 is assessed using RyR antibody and calstabin specific antibodies. Oxidation of RyR1 is determined by first derivatizing the carbonyl groups in the protein side chains in the immunoprecipitate with dinitrophenylhydrazone (DNP, Abcam, 178020) and then detecting the DNP signal associated with RyR using an anti-DNP antibody. See Table 1 for RYR1 variants. (b) Quantification of immunoblot data. Control Calstabin/RyR1 set to 4/1. * p<0.01 compared to control, ** p<0.01 S107 treated vs. baseline lysates. (c). Calpain activity of muscle lysates determined using a commercial kit (Abcam). The calpain activity assay protocol is based on the detection of cleavage of calpain substrate and the relative change in fluorescence signal/μg lysate after 1 h at 37°C is plotted (control arbitrarily set to 1.0).
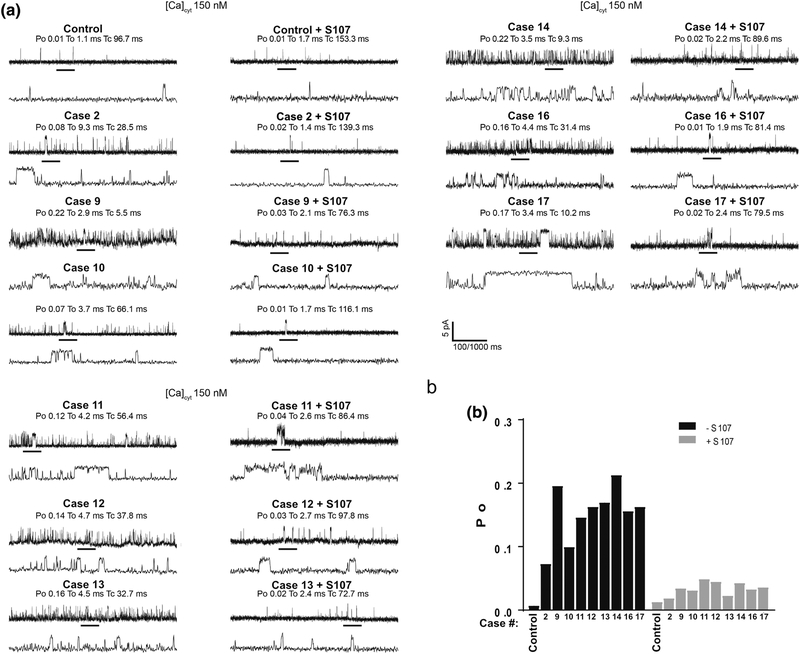
Single-channel recordings of RyR1 in planar lipid bilayers (Figure 2a,b) revealed increased Po at basal non-activating cis (cytosolic) Ca2+ levels (~150 nM), consistent with leaky channel behavior [30]. Case 2 with the p.Ala4185Thr variant exhibited increased leaky channel behavior but to a lesser degree compared to the other variants. Ca2+ leak was reversed by treating the patients’ muscle lysates with S107 prior to incorporating them into the bilayer (Figure 2a,b). SR Ca2+ leak was also assessed in microsomes isolated from RYR1-RM skeletal muscle as previously described [40,46,26]. SR microsomal Ca2+ leak was increased in skeletal muscle from RYR1-RM affected individuals compared to control (Figure 3 a,b). This leak was reversed by treating the patients’ muscle lysates with S107 (Figure 3 a,b).
Figure 2. Increased calcium leak in single RYR1-RM channels reconstituted in planar lipid bilayer.
SR microsomes containing single RyR1 channels isolated from individuals with RYR1-RM fused with planar lipid bilayer. Channel opening events are recorded as an upward deflection. Area on upper graph is expanded as the lower graph. Po = opening probability, To = time open, Tc = time closed. (a) Single-channel recordings of RyR1 from muscle lysates that were either treated or untreated with S107 (1.0 μM) from control and RYR1-RM affected individuals. Recordings were performed at 150 nM Ca2+. (b), Bar graph summarizing single-channel Po. N=3 per group. Limited availability of skeletal muscle precluded analyses in Cases 3–8 and 15.
Figure 3. Ca2+ leak from RYR1-RM SR microsomes.
(a) SR Ca2+ leak measured in microsomes (5μg/mL) from RYR1-RM muscle lysates. The Ca2+ leak was compared for control, RYR1-RM, and RYR1-RM treated with 1 uM S107 as indicated. Control data is shown in the first panel in blue and is represented by dotted line (....) in remaining traces for comparison. Inserts above traces show magnification of first 20 s after addition of thapsigargin. Bar graphs represent the quantification of the increase in Fluo-4/s over the first 5 seconds after addition of thapsigargin. (b) Quantification of Ca2+ leak experiments. N=3 in each group. Mutant channels at baseline exhibit continuous leak which does not occur in control channels or in mutant channels treated with S107. Limited availability of skeletal muscle precluded analyses in Cases 3–8 and 15.
Taken together, our data show that all 17 individuals in the present study have leaky skeletal muscle RyR1 channels in their lower extremities (no other muscle groups were examined due to the invasive nature of muscle biopsies). Case 2, (p.Ala4185Thr) had a less certain clinical phenotype characterized by post-exercise myalgias and occasional pigmenturia. Analysis of this individual’s skeletal muscle tissue revealed higher calstabin1 binding and less SR Ca2+ leak compared to other RYR1-RM affected individuals. Nonetheless, ex vivo Rycal treatment of skeletal muscle from Case 2 was still effective at stabilizing the closed state of RyR1 and reducing SR Ca2+ leak.
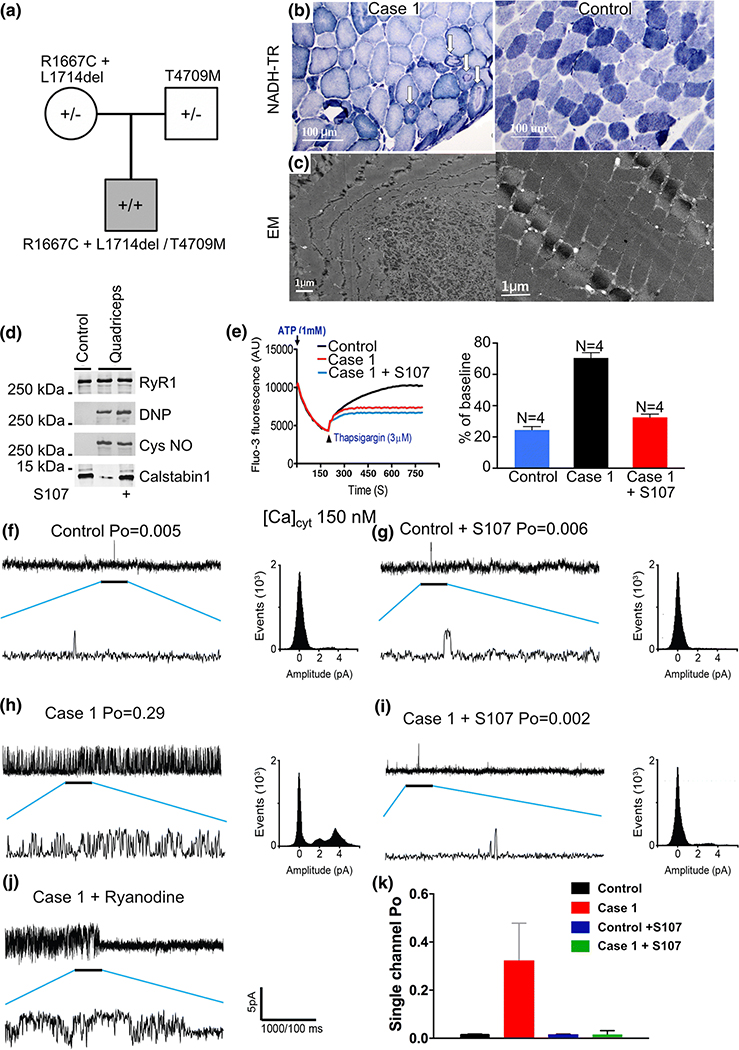
Case 1: Comprehensive analysis of RyR1 channel function
Case 1 is an individual who inherited two variants from his clinically asymptomatic mother, and one from his clinically asymptomatic father (Figure 4a). The father did, however, report profound statin intolerance (muscle pain). Case 1 exhibited myopathic histopathology with muscle fiber diameter variability and cores (Figure 4b). Electron microscopy revealed a reduced number of mitochondria in affected myofibers (Figure 4c). RyR1 isolated from this individual’s skeletal muscle exhibited increased oxidation, nitrosylation, and depletion of calstabin1 from the channel complex, assessed by co-immunoprecipitation and immunoblotting as described [2] (Figure 4d). RyR1-calstabin1 binding was restored by ex-vivo treatment of the individual’s muscle using the RyR stabilizing Rycal drug, S107 (Figure 4d) as previously reported [2,26,5,22,6]. Case 1’s muscle exhibited increased SR Ca2+ leak in a microsomal Ca2+ leak assay, which was reduced by ex-vivo treatment with S107 (Figure 4e). Similarly, single-channel recordings of RyR1 reconstituted into planar lipid bilayers [7] showed increased channel open probability (Po) in the presence of non-activating resting cytosolic [Ca2+]cyt compared to WT RyR1 channels (Figure 4f–h). The increased Po was normalized by ex-vivo treatment of the individual’s muscle with S107 (Figure 4i). Treating the isolated channels with ryanodine resulted in the channels being locked in a half-open sub-conductance state, diagnostic for RyR channels (Figure 4j).
Figure 4. Genotype-structure-function analysis of Case 1.
(a) Inheritance pattern of RYR1 variations. (b) NADH-TR stain and (c) electron microscopic of quadriceps muscle biopsy from Case 1 alongside control showing cores (arrows) of quadriceps muscle exhibiting a discrete area of myofibrillar derangement and paucity of mitochondria in affected myofibers. (d) RyR1 immunoprecipitated from muscle biopsies from control and RYR1-RM-affected muscle (quadriceps) comparing oxidation, nitrosylation, and calstabin1/RyR1 association (with and without Rycal S107 treatment). (e) Ca2+ leak assay in the presence and absence of Rycal S107 with bar graph, *P<0.05. (f) Single-channel data from normal control human quadriceps muscles in the absence and (g) presence of Rycal S107. (h) Single-channel data from Case 1’s quadriceps muscle in the absence (I) and presence of Rycal S107. There are multiple partial openings or subconductance states evident in the channels recorded from Case 1 in the absence of Rycal S107 which are seen in calstabin1 depleted channels that exhibit defective closing (leak). These subconductance states are not observed in the Rycal S107 treated muscle consistent with repair of channel leak. Scale bar represents current amplitude and time scale for compressed and expanded tracings. Amplitude histograms are shown for each experiment. (j) Channel from Case 1 treated with ryanodine which locks the channel in a sub-conductance state confirming the identity of the channel. (k) Bar graph shows quantification of single-channel Po data, N=5 per group, Tracings show samples of representative channel behavior in the bilayer. All analyses were performed on a minimum of two minutes of channel recording.
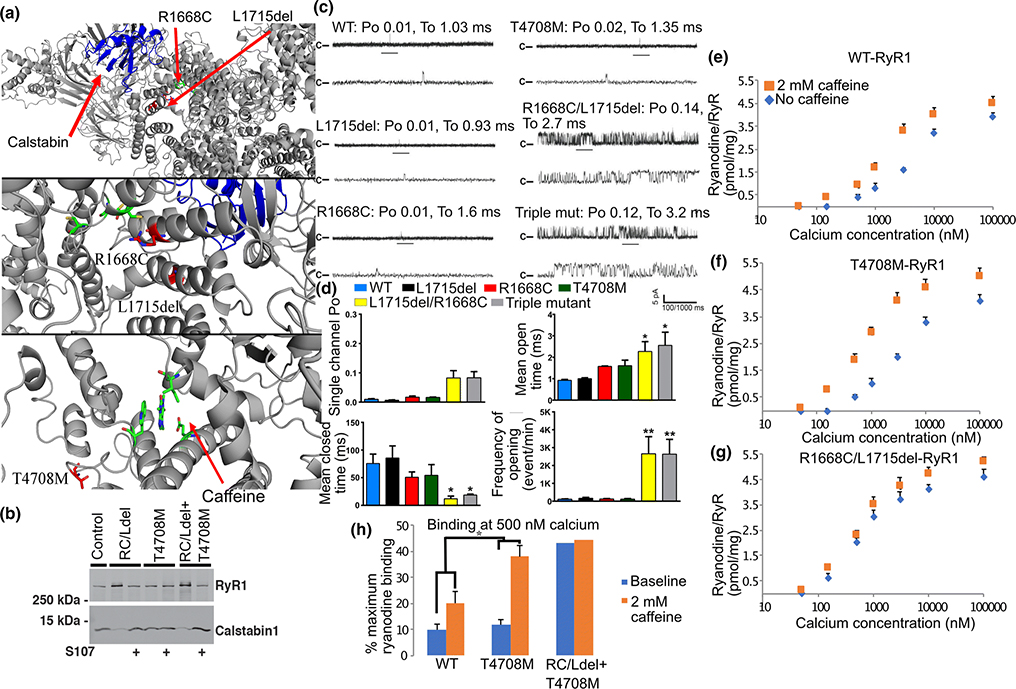
Having demonstrated structural (calstabin1 loss) and functional (leaky channel behavior) abnormalities exhibited by the mutant RyR1 in Case 1, and the positive response to the Rycal S107, we sought to determine which if any of the three RYR1 sequence variants cause the observed channel defects and could explain the individual’s symptoms of muscle weakness. Recombinant rabbit RyR1 channels were generated that expressed either the maternal genotype (p.Arg1667Cys + p.Leu1714del) or paternal genotype (p.Thr4709Met), or each variant individually. Based on the cryo-EM structure of the channel[47], p.Arg1667Cys and p.Leu1714del are near the calstabin1 binding site, and the p.Thr4709Met is near the caffeine and ATP binding sites (Figure 5a). Recombinant RyR1 channels with the triple mutants (p.Arg1667Cys + p.Leu1714del + p.Thr4709Met) and maternal mutants (p.Arg1667Cys + p.Leu1714del) exhibited decreased binding of calstabin1 to RyR1, but not the paternal mutant (p.Thr4709Met) (Figure 5b). When reconstituted in planar lipid bilayers the p.Arg1667Cys + p.Leu1714del allele resulted in increased channel Po (Figure 5c–d), consistent with leaky channels, whereas the p.Thr4709Met channels exhibited normal low Po under non-activating conditions. However, caffeine-treated channels expressing p.Thr4709Met had significantly increased channel activity compared to p.Arg1667Cys + p.Leu1714del channels, as assessed by radiolabeled ryanodine binding to the channel (Figure 5e–h) and a leftward shift in caffeine-dependent activation which is an indicator of increased sensitivity to Ca2+-dependent activation. Together, the channel structural and functional data indicate that the maternal genotype (p.Arg1667Cys + p.Leu1714del) is likely the cause of reduced calstabin 1 binding to the channel (see Fig. 5b) and leaky channel function (Fig. 5c–h). However, the paternal genotype (p.Thr4709Met) also results in altered channel function: increased sensitivity to the channel activator caffeine. Both findings are consistent with the high-resolution RyR1 structural location of the variants as the maternal variants are near the calstabin1 binding site and the paternal variant is near the caffeine binding site.
Figure 5. Characterization of defective RyR1 channel function Case 1.
(a) Localization of maternal variants (RYR1-R1668C, L1715del – numbering based on rabbit RyR1) from Case 1 to near the calstabin1 (yellow) binding site in RyR1 (top and middle panel). Localization of paternal variant (RYR1-T4708M) near the caffeine binding site (green). (b) Recombinant RYR1 variants (RC/Ldel: RYR1-R1668C, L1715del; T4708M: RYR1-T4708M and the combination of all three mutations: RC/Ldel on one cDNA and T4708M on another) immunoprecipitated from transfected HEK cell lysates and immunoblotted for calstabin in the presence or absence of Rycal S107. The RC/Ldel but not the T4708M result in decreased binding of calstabin1 to RyR1 that is restored by Rycal S107. (c) Single-channel current recordings from WT and mutant recombinant RyR1 at 150 nM [Ca2+]cis. (d) Bar graph shows quantification of data. N=6 per group, (* P<0.05). (e) 3H-ryanodine binding to recombinant RyR1 lysates expressing: (e) WT, (f) paternal, or (g) maternal variants, in the presence or absence of 2 mM caffeine and at the indicated [Ca2+]. (h) Bar graph shows quantification of 3H-ryanodine binding data at 500 nM [Ca2+]. *, P<0.05.
RYR1 dataset
A dataset of 2308 individuals (546 unique RYR1 variants) with presumed RYR1-RM was compiled consisting of genetic and clinical information (Supplementary Table 2). Analyses of this new RYR1 dataset revealed that the majority of reported variants localized to three distinct regions, consistent with known MH/CCD hotspot regions (Figure 6a). This is in contrast to variants reported in the gnomAD database [15], which distribute evenly across the channel (Figure 6a). Overall, only 179 of 3646 RYR1 variants in the gnomAD database have allele frequencies >0.01%. This is consistent with RYR1 having limited functional variation and being one of the genes most intolerant to sequence variation in the human genome [residual variation intolerance score: −8.29 (0.01%)] [34].
Figure 6. RYR1 dataset and localization of variants to the 3D RyR1 structure.
(A) Summary of variants in gnomAD database and RYR1 dataset with amino acid position on the X-axis. Channel structural domains highlighted based on references [9,47]. Each report of a variant in the RYR1 dataset is represented by a grey dot. Each variant reported in gnomAD is represented by a black dot. Inverted black triangles represent gnomAD variants with a frequency >0.01%. Inverted grey triangles represent variants included in this study. (B) Top - A single RyR1 protomer with locations of calstabin1 (yellow), the SPRY (cyan), Bsol (green), and pore (orange) domains highlighted. Bottom – Close-up of SPRY domain (cyan) with variants (red) present in the RYR1 dataset and their co-localization with calstabin1 (yellow). (C) Overview of the homotetrameric RyR1 channel. Variants in the Bsol domain are highlighted in red.
With the exception of Ala4185Thr, minor allele frequencies of all variants from the 17 individuals in this study were below 0.01% in gnomAD. When possible, family histories were obtained and pedigrees generated for individuals in this study (Supplementary Figure 1).
The distribution of variants across the channel highlights the limitation of adjudicating pathogenicity of variants based on localization in the primary structure of RyR1. Variants in the SPla and the RyR/SPRY1 domains localize to the calstabin1 binding site on the channel (Figure 6b). Variants in the Bridging-Solenoid (BSol) domain localize to the interprotomer region (Figure 6c). Variants that occur near the C-terminal domain (CTD) localize to both the pore region and the interprotomer region (Figure 6b) [47,9].
The 26 distinct variants from individuals in this study were mapped onto the high-resolution cryo-EM structure of RyR1 [47,9] (Supplementary Figure 2). Eight were in the RyR1 pore domain (Table 1). Sixteen of the variants fell within the three previously reported MH/CCD hotspots 1 (n= 5), 2 (n= 3) [24], and 3 (n= 8) (Figure 6a). Structure-function analyses of ten of these individuals have been reported elsewhere [38].
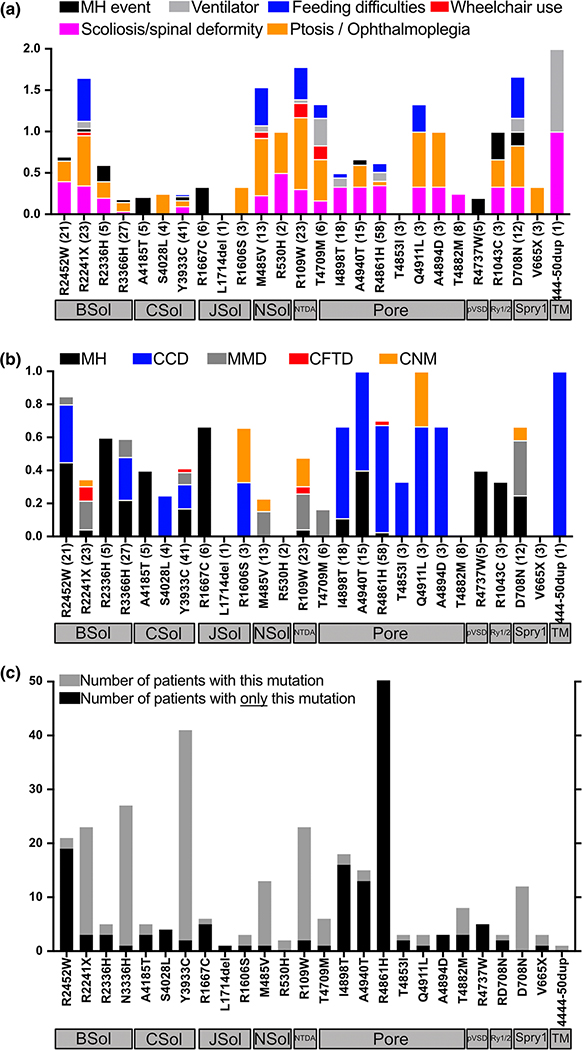
Clinical manifestations and histopathology associated with the 26 RYR1 variants included in this study across the RYR1 dataset are provided in Figures 7a and 7b. Variants associated with a history of MH events (Figure 7a) and variants with a positive in vitro contracture test diagnostic of MH susceptibility (Figure 7b) localized to the BSol, CSol, JSol, and SPRY domains, and pore region. The most frequent histopathological diagnosis was CCD, followed by MmD and CNM. Variants associated with CCD histopathology were more commonly found in the pore region (Figure 7b).
Figure 7. Analysis of RYR1 variants examined in the present study:
(A) Variants from individuals included in this study are on the X-axis. Number of individuals in the RYR1 dataset with that variant in parentheses. Variants are ordered based on their structural domain localization on the channel as depicted by the horizontal grey boxes under the variants. Y-axis represents the percentage of individuals with a specific symptom. For example, amongst 12 entries of D708N variant (located in the SPRY1 domain) in the RYR1 dataset: 35% have scoliosis; 40% have ptosis; 40% have feeding difficulties, 8% are wheelchair-bound, and 8% had an MH event. (B) Y-axis represents the percentage of individuals with a specific histopathologic finding (and/or laboratory diagnosis of MH who may or may not have had a clinical event). For example, for the 12 entries of D708N variant in RYR1 dataset, 25% have a laboratory diagnosis of MH, 40% have MmD, 8% of entries have CNM. (C) Y-axis grey bar is the total number of times each RYR1 variant appears in the RYR1 dataset. Y-axis black bar is the number of times a variant appears in the RYR1 dataset without additional variants.
Six of the 26 RYR1 variants included in the present study occurred more frequently in a dominant inheritance pattern in the RYR1 dataset whereas the remaining 20 variants occurred more frequently in a recessive inheritance pattern (Figure 7c). For example, p.Tyr3933Cys occurs 42 times but only twice as a unique variant without other variants. In contrast, p.Arg2452Trp frequently occurs on its own, without additional variants. Eight of the 26 RYR1 variants in the current study were present in >15 individuals in the RYR1 dataset (Figure 7c).
Structure-based variant pathogenicity assignment
The cryo-EM structure of RyR1 was used to develop a model for predicting the pathogenicity of RYR1 VUS based on their spatial proximity to known pathogenic variants. A subgroup of 165 variants from the RYR1 dataset (“training set”) was assigned pathogenic (Supplementary Table 3) status based on meeting specific criteria. These variants were localized to the channel structure. A score was generated for each VUS based on its proximity to these pathogenic variants (Supplementary Figure 3a). The ability for a particular score to predict the pathogenicity of a test set of known pathogenic and benign variants was determined.
ROC curves generated from this model were compared to ROC curves generated from bioinformatics-based-models: Polyphen-2 and SIFT (Supplementary Figure 3b and 3c). The ROC from the structure-based model for predicting pathogenicity was superior to the random model (P<0.05) and comparable to SIFT (P=0.2) and Polyphen (P=0.2).
Discussion
In this study, biochemical and functional analyses of mutant RyR1 channels from 17 RYR1-RM affected individuals consistently demonstrated reduced RyR1-calstabin1 binding, increased RyR1 oxidation, increased SR Ca2+ leak (determined by single-channel recordings and microsomal Ca2+ leak assays), and increased activity of Ca2+-activated proteases. These findings are consistent with reports of RyR1-mediated SR Ca2+ leak in murine models of muscular dystrophy (DMD and LGMD) [5,3], cancer-associated muscle weakness [41], and age-dependent loss of muscle function [13,33,2]. It is plausible that RyR1-mediated Ca2+ leak contributes to the clinical manifestations of RYR1-RM by reducing SR Ca2+ stores and causing damage to other cellular components (e.g. excessive mitochondrial Ca2+ uptake and lipid peroxidation) and exacerbating SR Ca2+ leak due to post-translational modification of RyR1 [2,13]. Although tissue yield from needle muscle biopsies is limited, it would be beneficial to further investigate the mechanism(s) underlying RYR1-RM skeletal muscle tissue by examining mitochondrial bioenergetics and ROS.
We explored the utility of therapeutic targeting of the SR Ca2+ leak observed in RYR1-RM patient skeletal muscle using a Rycal that stabilizes the RyR1 channel closed state. Rycals are a novel class of small molecules that stabilize the closed state of the RyR1 channel complex in vivo by restoring calstabin1 binding. In the present study ex vivo treatment of RYR1-RM skeletal muscle with a Rycal reduced RyR1-mediated SR Ca2+ leak. However, it remains to be determined whether Rycal treatment can improve muscle function and exercise capacity. An ongoing clinical trial in RYR1-RM affected individuals with the Rycal S48168/ARM210 (NCT04141670) is an important step toward addressing these questions.
Although our results show that RyR1-mediated SR Ca2+ leak is a common feature of RYR1-RM, alternative consequences of pathogenic RYR1 variations have been proposed including EC-uncoupling. The p.Ile4898Thr variant, identified in Case 2, is localized in the RyR1 Ca2+ conductance pathway [21,24,4]. Decreased Ca2+ permeation, EC-uncoupling, and an elevated unfolded protein response have been reported in heterozygous I4895T mice [4,24]. However, consistent with the findings of the present study, intracellular Ca2+ leak has been reported in HEK-293 cells expressing the p.Ile4898Thr variant and in myotubes isolated from individuals with p.Ile4898Thr [37,12]. Reported differences in the effects of RyR1 mutations on channel function (e.g. leak vs non-conductance) have raised the question of whether Rycal treatment (i.e. stabilization of the RyR1 closed state) could benefit individuals with variations linked to EC-uncoupling/decreased Ca2+ permeation. One possibility is that the stress of muscle dysfunction in the setting of one non-functioning RyR1 encoded by a mutant allele and one normal RyR1 encoded by a wild type allele could cause an oxidative state resulting in oxidation of the normal RyR1 and SR Ca2+ leak. Further clinical development of Rycal is expected to provide clarity in this regard.
Mice with the MH mutation Y524S exhibit increased SR Ca2+ release in response to simvastatin [19] and RyR1 destabilization has been reported in intact muscle fibers derived from patients treated with statins [28]. The underlying mechanism for statin-induced myopathy has been proposed to involve RyR1-calstabin1 disassociation, mitochondrial Ca2+ overload, and increased ROS/RNS-dependent Ca2+ sparks [28,39]. Results from our analysis of Case 1 further support this mechanism and also demonstrate that the combined expression of a variation linked to statin intolerance and increased RyR1 sensitivity (p.Thr4709Met) and variations shown to cause RyR1-mediated SR Ca2+ leak (p.Arg1667Cys + p.Leu1714del), can result in a severe phenotype characterized by profound and progressive muscle weakness. In case 1 the implication is that the mother has a low level of SR Ca2+ leak due to her two mutations and is asymptomatic. The father has RyR1 channels that are more sensitive to activation, but is also asymptomatic, except for his statin sensitivity. The combination of the two mutant alleles, however, results in the severe phenotype present in the son (progressive muscle weakness).
Using the high-resolution cryo-EM structure of RyR1 solved in our laboratory, we developed a method for predicting pathogenicity based on the co-localization of VUS with established pathogenic variants. This model is comparable to currently available bioinformatics-based pathogenicity prediction tools (Supplementary figure 3) in terms of predicting the pathogenicity of a test set of benign and pathogenic variants. Our structure-based prediction model will evolve as data regarding the pathogenicity of additional variants are obtained and programmed into the model.
All variants attributed to a dominant mode of inheritance in this study had a high probability of pathogenicity scores (Table 1), whereas those attributed to a recessive mode of inheritance had a lower probability of pathogenicity scores. This suggests that variants with a lower probability of pathogenicity can have a synergistic effect and cause disease (i.e. recessive inheritance). As noted above one example of this is Case 1, in which the mother’s variants alone (RYR1-R1668C and L1715del) exhibited a low probability of pathogenicity (Table 1), and each alone had little effect on channel function (Figure 5). However, the combination of variants resulted in decreased calstabin binding and leaky channels (Figure 5), suggesting a synergistic effect and when combined with the father’s mutation resulted in a severe phenotype.
In summary, skeletal muscle samples from 17 RYR1-RM affected individuals exhibited RyR1 channel oxidation, calstabin1 depletion from RyR1, and SR Ca2+ leak. Ex vivo treatment with Rycal restored RyR1-calstabin1 binding and reduced SR Ca2+ leak in all samples. This provides a rationale for a clinical trial testing Rycals in RYR1-RM affected individuals, which is currently underway. Our RYR1 dataset, which was used to develop a method for assigning pathogenicity probabilities to RYR1 VUS, has been made publicly available in this paper to support RYR1-RM translational research. Our goal is to translate this dataset into a fully functional online database that can grow over time and serve as a resource to investigators, clinicians, and the patient community.
Materials and Methods
Participants
Seventeen adults with genetic, clinical, and, if available, histologic evidence of RYR1-RM were included in this study (Table 1). RYR1-RM diagnostic confirmation was based on genetic testing, skeletal muscle histopathology from a previous biopsy, when available, and clinical assessment. Skeletal muscle biopsies, used in this study, were obtained from ten individuals who were enrolled in a clinical trial at the NIH Clinical Center, Bethesda MD, USA (NCT02362425). Written informed consent was provided and approved by an Institutional Review Board prior to study enrollment. Skeletal muscle tissue samples were also obtained from seven RYR1-RM affected individuals seen at McMaster University and Columbia University Irving Medical Center per clinical protocols. Data were shared with the research teams per approval of Hamilton Integrated Research Ethics Board and the CUIMC-IRB, respectively.
RYR1 Variant Identification
For all individuals, RYR1 sequencing was conducted using whole blood at laboratories certified by the Clinical Laboratory Improvement Amendments (CLIA) or the equivalent. RYR1 variants detailed in genetic test reports were evaluated using Alamut Visual software (version 2.9.0, Interactive Biosoftware, Rouen, France) and by consultation with a neurogenetics expert. Genetic screening methods included whole exome sequencing, complete RYR1 sequencing, partial RYR1 sequencing and targeted sequencing of familial RYR1 variants (targeted RYR1 sequencing).
Calcium leak assay
RyR1 functional studies were performed using methods well-established in the Marks laboratory [7,40,29]. Skeletal muscle SR microsomes were prepared by homogenizing muscle samples on ice using a Teflon-glass homogenizer (50 times) with 2 volumes of: 20 mmol/L (mM) Tris-maleate (pH 7.4), 1 mM EDTA, 1 mM DL-Dithiothreitol (DTT) and protease inhibitors (Roche). Homogenate was then centrifuged at 4,000 g for 15 min at 4°C and the following supernatant was centrifuged at 50,000 g for 45 min at 4°C. Pellets were resuspended in lysis buffer containing 300 mM sucrose. Microsomes (5 μg/mL) were diluted into a buffer (pH 7.2) containing 8 mM K-phosphocreatine, and 2 units/ml of creatine kinase, mixed with 3 μM Fluo-4 and added to multiple wells of a 96-well plate. Ca2+ loading of the microsomes was initiated by adding 1 mM ATP. After Ca2+ uptake (200 s), 3 μM thapsigargin was added to inhibit the Ca2+ reuptake by SERCA. SR Ca2+ leak was measured by the increase in intensity of the Fluo-4 signal (measured in a Tecan fluorescence plate reader). The Ca2+ leak was quantified as the increase in Fluo-4 signal after addition of thapsigargin.
RyR1 single-channel activity measurements
For single-channel studies SR microsomes containing RyR1 were fused to planar lipid bilayers formed by painting a lipid mixture of phosphatidylethanolamine and phosphatidylcholine (Avanti Polar Lipids) in a 3:1 ratio in decane; across a 200 μm hole in polysulfonate cups (Warner Instruments) separating 2 chambers. The trans chamber (1.0 ml), representing the intra- SR (luminal) compartment, was connected to the head stage input of a bilayer voltage clamp amplifier. The cis chamber (1.0 ml), representing the cytoplasmic compartment, was held at virtual ground. Solutions used for channel analysis were: cis solution 1 mM EGTA, 250 mM Hepes, 125 mM Tris, 50 mM KCl, 0.5 mM CaCl2, pH 7.35 and trans solution 53 mM Ca(OH)2, 50 mM KCl, 250 mM Hepes, pH 7.35. SR vesicles were added to the cis chamber. Making the cis chamber hyperosmotic by the addition of 400–500 mM KCl induced fusion of microsomes with the lipid bilayer. After the appearance of potassium and chloride channels, the cis chamber was perfused with the cis solution. Single-channel currents were recorded at 0 mV using a Bilayer Clamp BC-525D (Warner Instruments), filtered at 1 kHz using a Low-Pass Bessel Filter 8 Pole (Warner Instruments), and digitized at 4 kHz. All experiments were performed at room temperature (23°C). Open probability (Po) was determined using a minimum of 2 min of continuous recording. The recordings were analyzed using Clampfit 10.1 (Molecular Devices) and Prism (ver.7.0, GraphPad).
Heterologous expression of recombinant RyR1 channels
Mutant constructs RYR1-T4708M and RYR1-R1668C/L1715del were generated by introducing the relevant mutations into fragments of RyR1 using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent), using primers accggctggtgctcaac(atg)ccgtctttc (to introduce the T4708M mutation), tcgcacacgctg(tgc)ctctaccgcg (for R1668C mutation), and ggctactacgacc()tcatcagcatccacctg (for L1715del) – relevant codons are in parentheses and changed nucleotides in bold type; the reverse primers for each construct were reverse complementary to the listed ones. The relevant fragments were subsequently subcloned into full-length RyR1 construct in pcDNA3.1 vector, confirmed by sequencing, and expressed in 293T/17 cells using Lipofectamine 2000 (Thermo Fisher Scientific).
Co-immunoprecipitation and immunoblotting
Skeletal muscle biopsies (n= 17) were isotonically lysed in 0.25 mL of lysis buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 20 mM NaF, and protease inhibitors). Muscle lysates (100 mg) were treated with buffer or 10 mM Rycal for 1h at 4°C. RyR1 was immunoprecipitated from muscle lysates using an affinity-purified rabbit polyclonal antibody specific for RyR1 (RyR1–1327) [5] in 0.5 mL of a modified RIPA buffer (50 mM Tris-HCl pH 7.2, 0.9% NaCl, 5.0 mM NaF, 1% Triton-X100, and protease inhibitors) overnight at 4°C. The immunoprecipitants were size-fractionated on SDS-PAGE gels (6% for RyR, 15% for calstabin) and transferred onto nitrocellulose membranes for 2 hrs at 200 mA. Immunoblots were developed using a different anti-RyR antibody (5029, 1:5000 dilution) [17], or anti-Calstabin antibody (Santa Cruz, 1:2500) [42].
RyR1 oxidation
To determine RyR1 channel oxidation, RyR1 protein was immunoprecipitated and the carbonyl groups in side chains were derivatized to 2,4-dinitrophenylhydrazone (DNP) by reaction with 2,4-dinitrophenylhydrazine. The DNP signal associated with RyR1 was determined using an anti-DNP antibody, according to the manufacturer’s instructions (Millipore, Billerica, MA). All immunoblots were developed using the Odyssey system (LI-COR Biosciences, Lincoln, NE), with IR-labeled anti-mouse and anti-rabbit IgG (1:10000 dilution) secondary antibodies.
RyR1 Dataset: VUS
Data from >2200 RYR1-RM affected individuals were assembled in a dataset that includes genetic, structural, biophysical and clinical information. Data were obtained by reviewing 546 publications (Supplementary Table 4) identified via PubMed using the terms “ryr1 myopathy” and “malignant hyperthermia ryr1” (excluding review articles) and from neuromuscular disease clinics that care for RYR1-RM affected individuals located in Canada, Netherlands, United States, and the United Kingdom. The RYR1 dataset is available as Supplementary Table 2.
A subset of variants from the RYR1 dataset were adjudicated as pathogenic based on meeting any of the following criteria: 1) variants adjudicated as pathogenic by the European Malignant Hyperthermia Group (EMHG) (based on clinical symptoms, in vitro/ex vivo functional studies, and low prevalence in healthy populations)[16]; 2) variants associated with central cores on pathology (a hallmark of RYR1-RM); 3) variants that segregate with disease or myopathy within a family; 4) variants in RyR1 that are homologous to known CPVT-causative mutations in RYR2 [18] [catecholaminergic polymorphic ventricular tachycardia (CPVT) is an arrhythmogenic condition caused by mutations in the cardiac ryanodine receptor (RyR2). Due to homology between RyR1 and RyR2, the homologous variants on RyR1 will presumably also affect RyR1 function).
Using Chimera software (UCSF), a 10 Å sphere was centered on the alpha carbon of each pathogenic variant from the training set in the RyR1 cryo-EM structure. The value at the alpha carbon was 1 and decremented based on a Gaussian function which reached 0 at a distance of 10 Å. Every amino acid on RyR1 was assigned a value from 0–1 based on its localization within a sphere or group of spheres. Variants within overlapping spheres were assigned additive values (Supplementary Figure 3a).
A “test set” of pathogenic variants (adjudicated based on high prevalence in the RYR1 dataset) and benign variants (adjudicated based on a gnomAD prevalence of ≥ 0.1% in healthy populations - EMHG criteria [16]) were used to generate a receiver operating characteristic (ROC) ROC. The ROC Logistic regression was used to convert these arbitrary mutation density units into probabilities of pathogenicity (Supplementary Figure 3d).
Supplementary Material
Acknowledgments:
Funding: This work was supported by an RYR-1 Foundation Research Grant to AK and the National Institutes of Health, National Institute of Nursing Research, and Division of Intramural Research. The authors acknowledge the National Disease Research Interchange (NDRI) for supplying control skeletal muscle tissue. This work was also supported by grants from the NIH to ARM (T32HL120826, R01HL145473, R01DK118240, R01HL142903, R01HL061503, R01HL140934, R01AR070194, R25NS076445). This was also supported by a grant from the NIH UL1TR001873 to OC and AK.
Footnotes
Conflict of interest statement: Columbia University and ARM own stock in ARMGO, Inc. a company developing compounds targeting RyR and have patents on Rycals.
Data and materials availability: Antibodies and Rycals used in this study can be purchased from companies (as indicated in the methods section) not affiliated with any of the authors (none of the authors receive any income from selling any of the reagents used in this study), some of the reagents can be obtained from the Marks laboratory using an MTA with Columbia University.
References and Notes:
- 1.Amburgey K, McNamara N, Bennett LR, McCormick ME, Acsadi G, Dowling JJ (2011) Prevalence of congenital myopathies in a representative pediatric united states population. Ann Neurol 70:662–665. doi: 10.1002/ana.22510 [DOI] [PubMed] [Google Scholar]
- 2.Andersson DC, Betzenhauser MJ, Reiken S, Meli AC, Umanskaya A, Xie W, Shiomi T, Zalk R, Lacampagne A, Marks AR (2011) Ryanodine Receptor Oxidation Causes Intracellular Calcium Leak and Muscle Weakness in Aging. Cell metabolism 14:196–207. doi: 10.1016/j.cmet.2011.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson DC, Meli AC, Reiken S, Betzenhauser MJ, Umanskaya A, Shiomi T, D’Armiento J, Marks AR (2012) Leaky ryanodine receptors in beta-sarcoglycan deficient mice: a potential common defect in muscular dystrophy. Skelet Muscle 2:9. doi: 10.1186/2044-5040-2-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avila G, O’Brien JJ, Dirksen RT (2001) Excitation--contraction uncoupling by a human central core disease mutation in the ryanodine receptor. Proc Natl Acad Sci U S A 98:4215–4220. doi: 10.1073/pnas.071048198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellinger AM, Reiken S, Carlson C, Mongillo M, Liu X, Rothman L, Matecki S, Lacampagne A, Marks AR (2009) Hypernitrosylated ryanodine receptor calcium release channels are leaky in dystrophic muscle. Nat Med 15:325–330. doi: 10.1038/nm.1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellinger AM, Reiken S, Dura M, Murphy PW, Deng SX, Landry DW, Nieman D, Lehnart SE, Samaru M, LaCampagne A, Marks AR (2008) Remodeling of ryanodine receptor complex causes “leaky” channels: a molecular mechanism for decreased exercise capacity. Proc Natl Acad Sci U S A 105:2198–2202. doi: 10.1073/pnas.0711074105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brillantes AB, Ondrias K, Scott A, Kobrinsky E, Ondriasova E, Moschella MC, Jayaraman T, Landers M, Ehrlich BE, Marks AR (1994) Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. Cell 77:513–523 [DOI] [PubMed] [Google Scholar]
- 8.Bussiere R, Lacampagne A, Reiken S, Liu X, Scheuerman V, Zalk R, Martin C, Checler F, Marks AR, Chami M (2017) Amyloid beta production is regulated by beta2-adrenergic signaling-mediated post-translational modifications of the ryanodine receptor. J Biol Chem 292:10153–10168. doi: 10.1074/jbc.M116.743070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.des Georges A, Clarke OB, Zalk R, Yuan Q, Condon KJ, Grassucci RA, Hendrickson WA, Marks AR, Frank J (2016) Structural Basis for Gating and Activation of RyR1. Cell 167:145–157 e117. doi: 10.1016/j.cell.2016.08.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dlamini N, Voermans NC, Lillis S, Stewart K, Kamsteeg EJ, Drost G, Quinlivan R, Snoeck M, Norwood F, Radunovic A, Straub V, Roberts M, Vrancken AF, van der Pol WL, de Coo RI, Manzur AY, Yau S, Abbs S, King A, Lammens M, Hopkins PM, Mohammed S, Treves S, Muntoni F, Wraige E, Davis MR, van Engelen B, Jungbluth H (2013) Mutations in RYR1 are a common cause of exertional myalgia and rhabdomyolysis. Neuromuscul Disord 23:540–548. doi: 10.1016/j.nmd.2013.03.008 [DOI] [PubMed] [Google Scholar]
- 11.Dowling JJ, Arbogast S, Hur J, Nelson DD, McEvoy A, Waugh T, Marty I, Lunardi J, Brooks SV, Kuwada JY, Ferreiro A (2012) Oxidative stress and successful antioxidant treatment in models of RYR1-related myopathy. Brain 135:1115–1127. doi: 10.1093/brain/aws036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ducreux S, Zorzato F, Muller C, Sewry C, Muntoni F, Quinlivan R, Restagno G, Girard T, Treves S (2004) Effect of ryanodine receptor mutations on interleukin-6 release and intracellular calcium homeostasis in human myotubes from malignant hyperthermia-susceptible individuals and patients affected by central core disease. J Biol Chem 279:43838–43846. doi: 10.1074/jbc.M403612200 [DOI] [PubMed] [Google Scholar]
- 13.Durham WJ, Aracena-Parks P, Long C, Rossi AE, Goonasekera SA, Boncompagni S, Galvan DL, Gilman CP, Baker MR, Shirokova N, Protasi F, Dirksen R, Hamilton SL (2008) RyR1 S-nitrosylation underlies environmental heat stroke and sudden death in Y522S RyR1 knockin mice. Cell 133:53–65. doi: 10.1016/j.cell.2008.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fauconnier J, Meli AC, Thireau J, Roberge S, Shan J, Sassi Y, Reiken SR, Rauzier JM, Marchand A, Chauvier D, Cassan C, Crozier C, Bideaux P, Lompre AM, Jacotot E, Marks AR, Lacampagne A (2011) Ryanodine receptor leak mediated by caspase-8 activation leads to left ventricular injury after myocardial ischemia-reperfusion. Proc Natl Acad Sci U S A 108:13258–13263. doi: 10.1073/pnas.1100286108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. [Accessed March 6, 2018]; https://gnomad.broadinstitute.org/
- 16. [Accessed 07/03/2019]; https://www.emhg.org/diagnostic-mutations. https://www.emhg.org/diagnostic-mutations.
- 17.Jayaraman T, Brillantes AM, Timerman AP, Fleischer S, Erdjument-Bromage H, Tempst P, Marks AR (1992) FK506 binding protein associated with the calcium release channel (ryanodine receptor). J Biol Chem 267:9474–9477 [PubMed] [Google Scholar]
- 18.Kapplinger JD, Pundi KN, Larson NB, Callis TE, Tester DJ, Bikker H, Wilde AAM, Ackerman MJ (2018) Yield of the RYR2 Genetic Test in Suspected Catecholaminergic Polymorphic Ventricular Tachycardia and Implications for Test Interpretation. Circ Genom Precis Med 11:e001424. doi: 10.1161/CIRCGEN.116.001424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knoblauch M, Dagnino-Acosta A, Hamilton SL (2013) Mice with RyR1 mutation (Y524S) undergo hypermetabolic response to simvastatin. Skelet Muscle 3:22. doi: 10.1186/2044-5040-3-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lacampagne A, Liu X, Reiken S, Bussiere R, Meli AC, Lauritzen I, Teich AF, Zalk R, Saint N, Arancio O, Bauer C, Duprat F, Briggs CA, Chakroborty S, Stutzmann GE, Shelanski ML, Checler F, Chami M, Marks AR (2017) Post-translational remodeling of ryanodine receptor induces calcium leak leading to Alzheimer’s disease-like pathologies and cognitive deficits. Acta Neuropathol 134:749–767. doi: 10.1007/s00401-017-1733-7 [DOI] [PubMed] [Google Scholar]
- 21.Lee CS, Hanna AD, Wang H, Dagnino-Acosta A, Joshi AD, Knoblauch M, Xia Y, Georgiou DK, Xu J, Long C, Amano H, Reynolds C, Dong K, Martin JC, Lagor WR, Rodney GG, Sahin E, Sewry C, Hamilton SL (2017) A chemical chaperone improves muscle function in mice with a RyR1 mutation. Nat Commun 8:14659. doi: 10.1038/ncomms14659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehnart SE, Mongillo M, Bellinger A, Lindegger N, Chen BX, Hsueh W, Reiken S, Wronska A, Drew LJ, Ward CW, Lederer WJ, Kass RS, Morley G, Marks AR (2008) Leaky Ca2+ release channel/ryanodine receptor 2 causes seizures and sudden cardiac death in mice. J Clin Invest 118:2230–2245. doi: 10.1172/JCI35346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehnart SE, Wehrens XH, Reiken S, Warrier S, Belevych AE, Harvey RD, Richter W, Jin SL, Conti M, Marks AR (2005) Phosphodiesterase 4D deficiency in the ryanodine-receptor complex promotes heart failure and arrhythmias. Cell 123:25–35. doi: 10.1016/j.cell.2005.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesh RE, Marks AR, Somlyo AV, Fleischer S, Somlyo AP (1993) Anti-ryanodine receptor antibody binding sites in vascular and endocardial endothelium. Circ Res 72:481–488 [DOI] [PubMed] [Google Scholar]
- 25.Levin TR, Corley DA, Jensen CD, Marks AR, Zhao WK, Zebrowski AM, Quinn VP, Browne LW, Taylor WR, Ahlquist DA, Lidgard GP, Berger BM (2017) Genetic Biomarker Prevalence Is Similar in Fecal Immunochemical Test Positive and Negative Colorectal Cancer Tissue. Dig Dis Sci 62:678–688. doi: 10.1007/s10620-016-4433-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X, Betzenhauser MJ, Reiken S, Meli AC, Xie W, Chen BX, Arancio O, Marks AR (2012) Role of leaky neuronal ryanodine receptors in stress-induced cognitive dysfunction. Cell 150:1055–1067. doi: 10.1016/j.cell.2012.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loseth S, Voermans NC, Torbergsen T, Lillis S, Jonsrud C, Lindal S, Kamsteeg EJ, Lammens M, Broman M, Dekomien G, Maddison P, Muntoni F, Sewry C, Radunovic A, de Visser M, Straub V, van Engelen B, Jungbluth H (2013) A novel late-onset axial myopathy associated with mutations in the skeletal muscle ryanodine receptor (RYR1) gene. J Neurol 260:1504–1510. doi: 10.1007/s00415-012-6817-7 [DOI] [PubMed] [Google Scholar]
- 28.Lotteau S, Ivarsson N, Yang Z, Restagno D, Colyer J, Hopkins P, Weightman A, Himori K, Yamada T, Bruton J, Steele D, Westerblad H, Calaghan S (2019) A Mechanism for Statin-Induced Susceptibility to Myopathy. JACC: Basic to Translational Science 4:509–523. doi: 10.1016/j.jacbts.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marks AR (2002) Ryanodine receptors, FKBP12, and heart failure. Front Biosci 7:d970–977 [DOI] [PubMed] [Google Scholar]
- 30.Marx SO, Reiken S, Hisamatsu Y, Jayaraman T, Burkhoff D, Rosemblit N, Marks AR (2000) PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell 101:365–376 [DOI] [PubMed] [Google Scholar]
- 31.Matecki S, Dridi H, Jung B, Saint N, Reiken SR, Scheuermann V, Mrozek S, Santulli G, Umanskaya A, Petrof BJ, Jaber S, Marks AR, Lacampagne A (2016) Leaky ryanodine receptors contribute to diaphragmatic weakness during mechanical ventilation. Proc Natl Acad Sci U S A 113:9069–9074. doi: 10.1073/pnas.1609707113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthews E, Neuwirth C, Jaffer F, Scalco RS, Fialho D, Parton M, Raja Rayan D, Suetterlin K, Sud R, Spiegel R, Mein R, Houlden H, Schaefer A, Healy E, Palace J, Quinlivan R, Treves S, Holton JL, Jungbluth H, Hanna MG (2018) Atypical periodic paralysis and myalgia: A novel RYR1 phenotype. Neurology 90:e412–e418. doi: 10.1212/wnl.0000000000004894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michelucci A, De Marco A, Guarnier FA, Protasi F, Boncompagni S (2017) Antioxidant Treatment Reduces Formation of Structural Cores and Improves Muscle Function in RYR1(Y522S/WT) Mice. Oxid Med Cell Longev 2017:6792694. doi: 10.1155/2017/6792694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB (2013) Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet 9:e1003709. doi: 10.1371/journal.pgen.1003709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santulli G, Xie W, Reiken SR, Marks AR (2015) Mitochondrial calcium overload is a key determinant in heart failure. Proc Natl Acad Sci U S A 112:11389–11394. doi: 10.1073/pnas.1513047112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shan J, Betzenhauser MJ, Kushnir A, Reiken S, Meli AC, Wronska A, Dura M, Chen BX, Marks AR (2010) Role of chronic ryanodine receptor phosphorylation in heart failure and beta-adrenergic receptor blockade in mice. J Clin Invest 120:4375–4387. doi: 10.1172/JCI37649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tilgen N, Zorzato F, Halliger-Keller B, Muntoni F, Sewry C, Palmucci LM, Schneider C, Hauser E, Lehmann-Horn F, Müller CR, Treves S (2001) Identification of four novel mutations in the C-terminal membrane spanning domain of the ryanodine receptor 1: association with central core disease and alteration of calcium homeostasis. Human Molecular Genetics 10:2879–2887. doi: 10.1093/hmg/10.25.2879 [DOI] [PubMed] [Google Scholar]
- 38.Todd JJ, Sagar V, Lawal TA, Allen C, Razaqyar MS, Shelton MS, Chrismer IC, Zhang X, Cosgrove MM, Kuo A, Vasavada R, Jain MS, Waite M, Rajapakse D, Witherspoon JW, Wistow G, Meilleur KG (2018) Correlation of phenotype with genotype and protein structure in RYR1-related disorders. J Neurol 265:2506–2524. doi: 10.1007/s00415-018-9033-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner RM, Pirmohamed M (2019) Statin-Related Myotoxicity: A Comprehensive Review of Pharmacokinetic, Pharmacogenomic and Muscle Components. J Clin Med 9. doi: 10.3390/jcm9010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Umanskaya A, Santulli G, Xie W, Andersson DC, Reiken SR, Marks AR (2014) Genetically enhancing mitochondrial antioxidant activity improves muscle function in aging. Proc Natl Acad Sci U S A 111:15250–15255. doi: 10.1073/pnas.1412754111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waning DL, Mohammad KS, Reiken S, Xie W, Andersson DC, John S, Chiechi A, Wright LE, Umanskaya A, Niewolna M, Trivedi T, Charkhzarrin S, Khatiwada P, Wronska A, Haynes A, Benassi MS, Witzmann FA, Zhen G, Wang X, Cao X, Roodman GD, Marks AR, Guise TA (2015) Excess TGF-beta mediates muscle weakness associated with bone metastases in mice. Nat Med 21:1262–1271. doi: 10.1038/nm.3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wehrens XH, Lehnart SE, Huang F, Vest JA, Reiken SR, Mohler PJ, Sun J, Guatimosim S, Song LS, Rosemblit N, D’Armiento JM, Napolitano C, Memmi M, Priori SG, Lederer WJ, Marks AR (2003) FKBP12.6 deficiency and defective calcium release channel (ryanodine receptor) function linked to exercise-induced sudden cardiac death. Cell 113:829–840 [DOI] [PubMed] [Google Scholar]
- 43.Wehrens XH, Lehnart SE, Reiken S, van der Nagel R, Morales R, Sun J, Cheng Z, Deng SX, de Windt LJ, Landry DW, Marks AR (2005) Enhancing calstabin binding to ryanodine receptors improves cardiac and skeletal muscle function in heart failure. Proc Natl Acad Sci U S A 102:9607–9612. doi: 10.1073/pnas.0500353102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wehrens XH, Lehnart SE, Reiken SR, Deng SX, Vest JA, Cervantes D, Coromilas J, Landry DW, Marks AR (2004) Protection from cardiac arrhythmia through ryanodine receptor-stabilizing protein calstabin2. Science 304:292–296. doi: 10.1126/science.1094301 [DOI] [PubMed] [Google Scholar]
- 45.Witherspoon JW, Meilleur KG (2016) Review of RyR1 pathway and associated pathomechanisms. Acta Neuropathol Commun 4:121. doi: 10.1186/s40478-016-0392-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie W, Santulli G, Reiken SR, Yuan Q, Osborne BW, Chen BX, Marks AR (2015) Mitochondrial oxidative stress promotes atrial fibrillation. Sci Rep 5:11427. doi: 10.1038/srep11427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zalk R, Clarke OB, des Georges A, Grassucci RA, Reiken S, Mancia F, Hendrickson WA, Frank J, Marks AR (2015) Structure of a mammalian ryanodine receptor. Nature 517:44–49. doi: 10.1038/nature13950 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.