Abstract
Background
Mesenchymal stromal cells (MSCs) have been introduced as promising cell source for regenerative medicine. Besides their multilineage differentiation capacity, MSCs release a wide spectrum of bioactive factors. This secretome holds immunomodulatory and regenerative capacities. In intervertebral disc (IVD) cells, application of MSC secretome has been shown to decrease the apoptosis rate, induce proliferation, and promote production of extracellular matrix (ECM). For clinical translation of secretome-based treatment, characterization of the secretome composition is needed to better understand the induced biological processes and identify potentially effective secretomes.
Methods
This study aimed to investigate the proteome released by bone marrow-derived MSCs following exposure to a healthy, traumatic, or degenerative human IVD environment by mass spectroscopy and quantitative immunoassay analyses. Exposure of MSCs to the proinflammatory stimulus interleukin 1β (IL-1β) was used as control.
Results
Compared to MSC baseline secretome, there were 224 significantly up- or downregulated proteins following healthy, 179 following traumatic, 223 following degenerative IVD, and 160 proteins following IL-1β stimulus. Stimulation of MSCs with IVD conditioned media induced a more complex MSC secretome, involving more biological processes, compared to stimulation with IL-1β. The MSC response to stimulation with IVD conditioned medium was dependent on their pathological status.
Conclusions
The MSC secretome seemed to match the primary need of the IVD: homeostasis maintenance in the case of healthy IVDs, versus immunomodulation, adjustment of ECM synthesis and degradation disbalance, and ECM (re) organization in the case of traumatic and degenerative IVDs. These findings highlight the importance of cell preconditioning in the development of tailored secretome therapies.
Graphical abstract
The secretome of human bone marrow-derived mesenchymal stromal cells (MSCs) stimulated with intervertebral disc (IVD) conditioned medium was analyzed by proteomic profiling. Depending on the pathological state of the IVD, the MSC secretome protein composition indicated immunomodulatory or anabolic activity of the secretome. These findings may have implications for tailored secretome therapy for the IVD and other tissues.

Keywords: Mesenchymal stromal cells, Secretome, Intervertebral disc, Conditioned medium, Intervertebral disc degeneration
Background
Mesenchymal stromal cells (MSCs) have been the subject of extended research in the field of regenerative medicine. Defined as fibroblast-like non-hematopoietic cells by the International Society for Stem Cell Research (ISSCR), this pluripotent cell population holds differentiation potential towards various lineages [1]. In the field of musculoskeletal regeneration, MSCs have gained interest for their ability to differentiate towards chondrogenic and osteogenic phenotypes [2]. In the intervertebral disc (IVD), injection of naïve MSCs into degenerated IVDs has been associated with a regenerative effect, both in preclinical and clinical trials [3, 4]. However, the exact underlying mechanism remains unknown. On the one hand, host IVD cells can induce a discogenic phenotype in implanted MSCs, as evidenced by upregulated gene expression and synthesis of extracellular matrix (ECM) molecules. On the other hand, MSCs can influence IVD cells by secreting bioactive factors which cause a shift from a degenerative towards a healthier disc cell phenotype [5–7]. Nonetheless, in a degenerative IVD, MSCs face a challenging hypoxic and acidic milieu with limited nutrient supply [8, 9]. In vivo studies indicated that MSC survival under such conditions is limited [10, 11]. It is therefore hypothesized that the main regenerative effect of MSCs would be mediated by paracrine stimulation rather than by engrafting, differentiation, and de novo ECM production [12].
Paracrine stimulation is mediated by substances secreted by the MSCs as a response to the perceived environment. The secreted bioactive substances can be termed secretome. Released factors include soluble proteins, free nucleic acids, lipids, and extracellular vesicles which can be further subdivided into apoptotic bodies, micro-particles, and exosomes [13]. The secretome from MSCs is believed to hold great potential for tissue repair and regenerative medicine [14–17]. Application of naïve MSC secretome in animal models has shown to significantly improve the pathology of various diseases such as graft versus host disease, autoimmune, and inflammatory diseases [18]. Clinical application of naïve MSC secretome has already been investigated in small patient groups suffering from alveolar bone atrophy, alopecia, or skin damage following ablative fractional carbon dioxide laser resurfacing; in all patient groups, application of MSC secretome led to improved recovery, with no reported adverse effects [19–21].
The secretome composition is influenced by the environment which the MSCs are exposed to [17, 18, 22]. For instance, hypoxic preconditioning was associated with increased production of growth factors, including vascular endothelial growth factor (VEGF), fibroblast growth factor 2 (FGF-2), hepatocyte growth factor (HGF), and insulin-like growth factor 1 (IGF-1) [23]. Exposure to an inflammatory stimulus such as interleukin 1-beta (IL-1β), tumor necrosis factor-alpha (TNF-α), or interferon-gamma (IFN-γ) was shown to initiate the production of immune-modulatory factors. These include granulocyte colony-stimulating factor (G-CSF) [24], factor H which inhibits complement activation [25], and galectin-9 which suppresses T-cell proliferation [26], among others. Interestingly, culturing of MSCs in three-dimensional (3D) arrangement was also associated with an induced secretion of different potentially therapeutic factors compared to two-dimensional (2D) culture, including G-CSF, VEGF, IL-1 receptor antagonist (IL-1Ra), or FGF-1 [27–29]. Preconditioning of adipose-derived stem cells (ASC) with lipopolysaccharide resulted in the production of a secretome that was superior in hepatic regeneration compared to the secretome of unstimulated ASC [30]. Also, preconditioning of ASC with TNF-α potentiated the exosome efficacy for bone regeneration [31]. Furthermore, in vivo mouse models indicated accelerated skin wound healing following application of secretome from MSCs primed by hypoxia compared to MSC secretome obtained under normoxic conditions [32].
Concerning the IVD, so far only the application of unprimed MSC secretome (mainly in form of extracellular vesicles) has been investigated as a potential cell-free treatment strategy. The studies described the beneficial effect of secretome application on IVD cells, including prevention of cell death, decrease in apoptosis rate, enhanced cell proliferation, and ECM production [33]. The composition of the secretome, however, remains unknown. Aiming for clinical translation of secretome-based treatments, characterization of the secretome composition is needed to better understand its biological effect.
In the present study, we analyzed the protein composition within the secretome of MSCs exposed to healthy, traumatic, and degenerative human IVD conditioned medium. Exposure to a supraphysiological concentration of IL-1β was further used as a pro-inflammatory priming control. We hypothesized that distinct differences existed between the protein profiles of secretomes from MSCs primed with different IVD conditioned media or IL-1β as a single pro-inflammatory stimulus. Proteomic profiling by mass spectroscopy (LC-MS/MS) and quantitative immunoassays were used to identify proteins within the MSC secretome. Gene set enrichment analysis (GSEA) allowed us to identify enriched biological processes in MSCs following the different priming strategies (Fig. 1).
Fig. 1.
Experimental setup. a Mesenchymal stromal cells (MSCs; N = 12) were isolated from vertebral bone marrow aspirates obtained with written consent from patients undergoing spine surgery. b Intervertebral disc (IVD) tissue from patients suffering from spinal trauma (referred to as traumatic), from patients with disc degeneration (referred to as degenerative), and non-degenerated IVDs from organ donors (referred to as healthy) were obtained with written patient and/or familial consent. Tissue was incubated in basal medium for 48 h to collect released factors (referred to as IVD conditioned medium (CM)). Basal medium supplemented with IL-1β (10 ng/mL) was prepared as proinflammatory control. c MSCs were seeded in 6-well plates. After overnight attachment and 6 h of starvation, MSCs were stimulated with healthy CM (N = 4, pooled), traumatic CM (N = 4, pooled), degenerative CM (N = 4, pooled), IL-1β, and basal medium (baseline control), respectively. After 24 h of stimulation, stimulants were removed, and fresh basal medium was added to collect the MSC secretome during the following 24 h. MSC secretome was analyzed by LC-MS/MS and immunoassay. MSCs were analyzed by CellTiter-Blue, lactate dehydrogenase (LDH) assay, DNA quantification. BM = basal medium (low glucose-DMEM, 1% L-Ascorbic acid 2-phosphate, 1% Glutamax)
Methods
MSC isolation and expansion
MSCs were isolated by Ficoll® gradient centrifugation and adherence to tissue culture plastic from vertebral bone marrow aspirates obtained with written consent from patients undergoing spine surgery. Standardized methods were applied for cell isolation as previously described [34, 35]. MSCs from 12 different donors were used for this study (Suppl. Fig. 1A). Cells were expanded in growth medium composed of alpha minimal essential medium (α-MEM, Gibco) supplemented with 10% fetal bovine serum (FBS+, Sera Plus, Pan Biotech), 1% penicillin-streptomycin (P/S, 100x, Gibco), and 5 ng/mL FGF-2 (Fitzgerald Industries) according to standardized procedures [36, 37]. Passage 3 MSCs were used in this study.
IVD conditioned medium
Human IVD tissues from patients with traumatic injury (“traumatic” sample) and from patients diagnosed with IVD degeneration (“degenerative” sample) were obtained with written consent from patients undergoing spine surgery. Non-degenerated (“healthy” sample) IVD tissues were obtained from organ donors after donor and familial consent by the McGill Scoliosis & Spinal Research Group via a collaboration with Transplant Quebec and approval by the McGill University’s Institutional Review Board (IRB# A04-M53-08B). Human IVDs from organ donors, degenerative and traumatic patients were used to produce IVD conditioned medium (CM) as previously described (Suppl. Fig. 1B) [38]. Briefly, the tissue was weighed and washed in red cell lysis buffer for 5 min. Tissue was then washed three times in phosphate buffered saline (PBS) supplemented with 1% P/S. Cartilaginous endplates were removed and IVD tissue was cut into pieces (approximately 4 × 4 × 4 mm). Basal medium, low glucose (1 g/L) Dulbecco’s modified Eagle’s medium (lg-DMEM, Gibco) supplemented with L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate (50 μg/mL, Sigma-Aldrich), 1% Glutamax (Gibco), and 0.1% Primocin™ (InvivoGen), was added to the tissue (3.5 mL/g tissue) in a 50-mL centrifugation tube and incubated for 48 h. Then, CM was filtered through a 100 μm pore size cell strainer (Falcon™) and aliquoted into 2 mL low binding protein tubes. Aliquots were stored at − 80 °C until use.
MSC stimulation and secretome collection
MSCs were plated in 6-well plates at a density of 10,000/cm2 and cultured in growth medium for 14 h. Following cell attachment, cells were washed 3 times with 1 mL PBS and subsequently starved for 6 h in 1 mL basal medium. Basal medium was removed and 1 mL of pooled IVD CMs (N = 4 for each degenerative, traumatic, or healthy CM) was added for MSC stimulation. As a pro-inflammatory positive control, cells were stimulated with 1 mL of basal medium containing 10 ng/mL IL-1β (PeproTech). As baseline control, cells were incubated with 1 mL of basal medium only. After 24 h, medium was removed, and cells were washed 3 times with 1 mL of lg-DMEM. To generate the secretome, 1 mL of basal medium without Primocin™ was added to each well. After 24 h, MSC secretome was collected in low binding protein tubes and stored at − 80 °C; MSCs were analyzed microscopically, for metabolic activity, lactate dehydrogenase (LDH) activity, and DNA content.
Metabolic activity
The resazurin-based CellTiter-Blue™ Cell Viability Assay (Promega) was used to measure the MSCs metabolic activity. Prior to the assay, MSCs were washed with 1 mL PBS. Subsequently, 1 mL of basal medium (without Primocin™) and 200 μL of CellTiter-Blue™ were added. Fluorescence intensity was measured after 3 h incubation using a VICTOR™ multilabel plate reader (Perkin Elmer). Values were normalized to the baseline treatment condition for each MSC donor.
Lactate dehydrogenase measurement
MSC viability was assessed using the LDH based cytotoxicity detection KitPlus™ (Roche) after secretome collection according to the manufacturer’s instructions. As a cytotoxic positive control, cells were treated with 1% Triton X-100 (Sigma-Aldrich) in basal medium without Primocin™. For the negative control, cells were left untreated with basal medium without Primocin™. Absorbance was measured at 490 nm using the VICTOR™ multilabel plate reader. For each MSC donor, cytotoxicity was calculated by dividing the difference of the sample and the negative control by the difference of the positive control and the negative control. This resulted in the negative control having 0% and the positive control 100% cytotoxicity.
DNA quantification
MSCs were digested with 500 μL of proteinase K (0.5 mg/mL, Roche) at 56 °C for 16 h. DNA quantification was performed with Qubit® 4 Fluorometer (Invitrogen) using the QubitR dsDNA HS assay kit according to the manufacturer’s instructions. DNA content after secretome collection was normalized to the DNA amount of the attached cells 14 h after seeding.
Cell morphology
For each condition and donor, microscopic images of one well of the 6-well plate were taken immediately before secretome collection using a × 5 magnification (Axiovert 40 CFL, Zeiss).
Sample processing for LC-MS/MS analysis
MSC secretomes from all 12 donors for all treatment conditions (healthy, degenerative, traumatic, baseline, and IL-1β) were analyzed. The samples were collected and measured in two batches of 48 samples (traumatic, degenerative, IL-1β, baseline) and 24 samples (healthy and baseline). In both batches, baseline samples were included to account for differences between batches. For each sample, the protein concentration was measured using the Qubit® Protein Assay Kit (Life Technologies, Switzerland). The samples were then prepared by using a commercial iST Phonix Kit (PreOmics, Germany). Mass spectrometry analysis was performed on a Q-Exactive HF-X mass spectrometer (Thermo Scientific) equipped with a Digital PicoView source (New Objective) and coupled to an M-Class UPLC (Waters). Solvent composition at the two channels was 0.1% formic acid for channel A and 0.1% formic acid, 99.9% acetonitrile for channel B. For each sample, 4 μL of peptides were loaded on a commercial MZ Symmetry C18 Trap Column (100 Å, 5 μm, 180 μm × 20 mm, Waters) followed by nanoEase MZ C18 HSS T3 Column (100 Å, 1.8 μm, 75 μm × 250 mm, Waters). The peptides were eluted at a flow rate of 300 nL/min by a gradient from 8 to 27% B in 82 min, 35% B in 5 min, and 80% B in 1 min. Samples were acquired in a randomized order. Only precursors with intensity above 110,000 were selected for MS/MS.
Quantification of chemokines and cytokines
As a complementary analysis technique, multiplex immunoassay was used to quantify cytokines and chemokines present in pooled IVD CM and released by MSCs following stimulation by IVD CM. All samples were analyzed with the proinflammatory panel 1, cytokine panel 1, and chemokine panel 1 assay kits (Meso Scale Discovery (MSD), Rockville, MD, USA) following the manufacturer’s protocol. Briefly, standards and samples (50 μL) were added to each well in technical duplicates. Plates were sealed and incubated under shaking for 2 h at room temperature (RT). Then, plates were washed, 25 μL of detection antibody solution was added to each well, and plates were incubated under shaking for 2 h at RT. After the final washing, 150 μL of MSD read buffer T (× 2) was added to each well and the plates were read using the MESO QuickPlex SQ 120 instrument (MSD). The concentration of each analyte in MSC secretomes was normalized to the DNA content of the respective sample. Finally, all values were normalized to the baseline control sample for each donor, and fold changes were calculated.
Bioinformatics and statistical analyses
We performed MS1 intensity-based label-free quantification to estimate protein fold changes among conditions. The acquired raw MS data were processed by MaxQuant (version 1.6.2.3), followed by protein identification using the integrated Andromeda search engine. Spectra were searched against a Uniprot human reference proteome (taxonomy 9906, canonical version from 2016-12-09), concatenated to its reversed decoyed fasta database and common protein contaminants. A peptide was considered quantified if it was observed in more than 50% of samples within one of the conditions. In addition, two quantified peptides per protein were required; proteins with only one quantified peptide were excluded.
Normalization was applied to remove systematic differences in protein abundance due to different amounts of sample loaded on the column. The z-score of the log2 transformed intensities was computed and updated by the average standard deviation of the log2 transformed intensities in all samples to preserve the original scale of the measurement. After normalization, all samples showed a similar distribution. As a next step, to obtain protein fold change among conditions, peptide intensities were modeled using a mixed linear model [39], to account for the repeated measures design. Log2 fold changes were computed based on the model parameters using the R package lmerTest [39]. The baseline samples were used as reference condition in all comparisons.
For the analysis R version 3.5.1 (2018-07-02) with the platform x86_64-w64-mingw32/x64 (64 bit) was used. Gene set enrichment analysis (GSEA) was performed, using the estimated protein fold changes to rank the proteins, with WebGestalt (http://webgestalt.org). The Gene Ontology (GO) classification for biological processes (GOBP) was used. The number of permutations was set to 1000. The IL-1β group was used to set a threshold for the “minimum number of genes for a category” in the conducted GSEA. The same threshold of a minimum of 10 genes for a category was used for GSEA of all groups. In addition to GSEA the protein list was filtered for elements with a log2 fold change ≥ 1.5 and a significance level of 0.05 [40]. Proteins were divided into the six categories (1) ECM, (2) anabolic, (3) catabolic, (4) growth factors, (5) immune system, and (6) other, with information retrieved from UniProt. Interaction network analysis of upregulated proteins was performed using STRING (http://string-db.org) [41]. Active interactions included text mining, experiments, and databases. Line thickness was set to indicate confidence.
For the DNA content, metabolic activity, LDH measurement, and immunoassay, statistical analysis of all data was performed using GraphPad Prism version 6.00. Normality was tested with D’Agostino-Pearson omnibus normality test, Shapiro-Wilk normality test, and Kolmogorov-Smirnov test with Dallal-Lilliefor P value. For parametric data, one-way ANOVA was performed. For non-parametric data, Kruskal-Wallis test was performed; P < 0.05 was regarded as significant.
Results
Viability of MSCs following priming with intervertebral disc conditioned medium
To investigate whether MSC viability was affected by the IVD CM or a proinflammatory environment, a cytotoxicity assay was performed (LDH), the DNA content and the metabolic activity were analyzed, and the cell morphology assessed. Measurements were performed at the end of the experiment following collection of MSC secretome. Stimulation of MSCs with healthy and degenerative IVD CM induced a significant increase in DNA content (1.78 ± 0.57 and 1.30 ± 0.27-fold, respectively) compared to the baseline condition (0.87 ± 0.24-fold) (P < 0.01), indicating enhanced cell proliferation. The traumatic stimulation induced a slight, although not significant, increase in DNA content (P = 0.241; 1.17 ± 0.26-fold). The DNA content following proinflammatory stimulus with IL-1β was similar to the baseline control (0.96 ± 0.30-fold, all normalized to day 0 after cell attachment) (Suppl. Fig. 2A).
Metabolic activity of MSCs following stimulation with all the IVD CM was increased compared to baseline control (Suppl. Fig. 2B). No significant changes were observed between different sources of IVD CM (P > 0.05) (healthy/traumatic/degenerative). LDH measurement in the MSC secretome showed no cytotoxicity in any culture condition (Suppl. Fig. 2C). Morphological analyses of MSCs supported findings from the metabolic and cytotoxicity assays; stimulation with healthy, traumatic, and degenerative IVD CM maintained a flattened cell morphology (Suppl. Fig. 2D).
Proteins identified in MSC secretome and identification of enriched biological processes (GSEA)
Secretomes of MSCs treated with healthy, traumatic, degenerative IVD CM, or IL-1β were compared to the secretome of MSCs incubated in basal medium (baseline control). Compared to the baseline control, there were 224 significantly up- or downregulated proteins in MSC secretomes following healthy (Suppl. Table 1), 179 following traumatic (Suppl. Table 2), 223 following degenerative IVD CM (Suppl. Table 3), and 160 proteins following IL-1β stimulus (Suppl. Table 4) (all comparisons as fold changes relative to the baseline control).
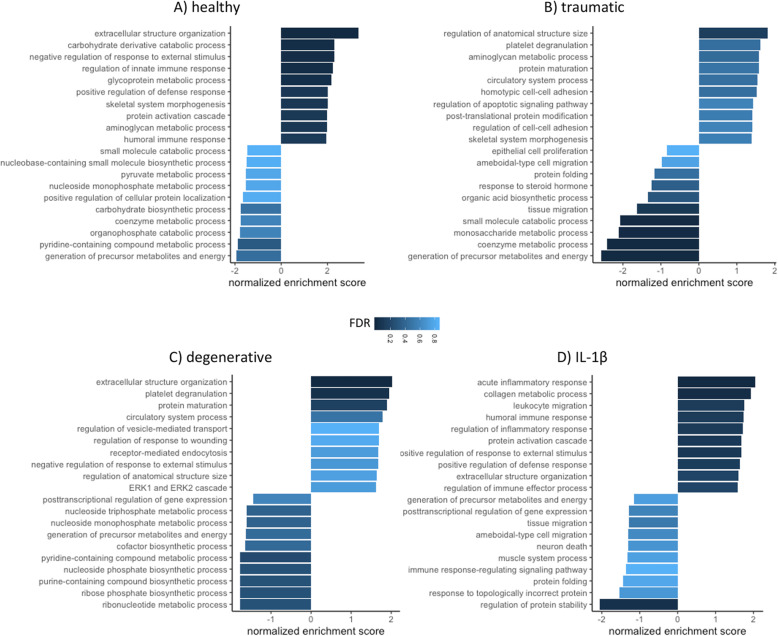
Enriched biological processes (GO terms) were identified based on the GO classification system (GOBP). To allow for a comparison of the different experimental groups (healthy, traumatic, degenerative, IL-1β), all data were normalized to the respective MSC donor baseline control prior analysis. Within the significantly upregulated processes (P < 0.05), the identified GSEA terms with a false discovery rate (FDR) < 0.05 are listed in Fig. 2. MSC secretome following stimulation with healthy IVD CM showed an upregulation of extracellular structure organization (normalized enrichment score (NES) = 3.34, FDR = 0), carbohydrate derivative catabolic process (NES = 2.29, FDR = 0.007), negative regulation of response to external stimulus (NES = 2.28, FDR = 0.004), regulation of innate immune response (NES = 2.23, FDR = 0.006), glycoprotein metabolic process (NES = 2.1, FDR = 0.001), positive regulation of defense response (NES = 2.01, FDR = 0.029), skeletal system morphogenesis (NES = 2.01, FDR = 0.025), protein activation cascade (NES = 1.98, FDR = 0.028), aminoglycan metabolic process (NES = 1.98, FDR = 0.026), and humoral immune response (NES = 1.93, FDR = 0.03). Following stimulation with traumatic IVD CM, downregulation was observed for: small molecule catabolic process (NES = − 2.08, FDR = 0.006), monosaccharide metabolic process (NES = − 2.11, FDR = 0.009), coenzyme metabolic process (NES = − 2.41, FDR = 0.001), and generation of precursor metabolites and energy (NES = − 2.57, FDR = 0); while regulation of anatomical structure size was upregulated. Analyzing the secretome following stimulation with degenerative IVD CM induced upregulation of extracellular structure organization (NES = 2.03, FDR = 0.035), and platelet degranulation (NES = 1.95, FDR = 0.047). The proinflammatory exposure to IL-1β resulted in an upregulation of acute inflammatory response (NES = 2.05, FDR = 0.003) and collagen metabolic process (NES = 1.94, FDR = 0.019), while a significant downregulation for the regulation of protein stability (NES = − 2.05, FDR = 0.044) was observed. Interestingly, only two significantly upregulated biological processes were observed among multiple groups, namely “extracellular structure organization” (healthy, degenerative, and IL-1β), and “aminoglycan metabolic process” (healthy and traumatic). Proteins involved in the top five significantly upregulated and downregulated biological processes are displayed within a chord diagram in Fig. 3a–d.
Fig. 2.
MSC secretome gene set enrichment analysis (GSEA). GSEA from LC-MS/MS data of released proteins following a healthy, b traumatic, c degenerative, and d IL-1β stimulus relative to baseline control. Figures display top 10 upregulated and downregulated gene ontology terms (functional database: biological processes). All reported GO terms have a P < 0.01. The false discovery rate (FDR) is color-coded with darker color indicating a low FDR
Fig. 3.
Gene set enrichment analysis (GSEA). Chord diagrams of top 5 upregulated and downregulated gene ontology (GO) terms (functional database: biological processes) found in a MSC “healthy” secretome gene set enrichment analysis; b MSC “traumatic” secretome gene set enrichment analysis; c MSC “degenerative” secretome gene set enrichment analysis; d MSC “IL-1β” secretome gene set enrichment analysis (LC-MS/MS data). All reported GO terms (right hemisphere) were significantly up- or downregulated (P < 0.01). Gene-box color (left hemisphere) indicates detection level as log2 fold change relative to the baseline control: upregulated detection = red, downregulated = blue. Strings indicate involvement of each gene in all respective GO-terms
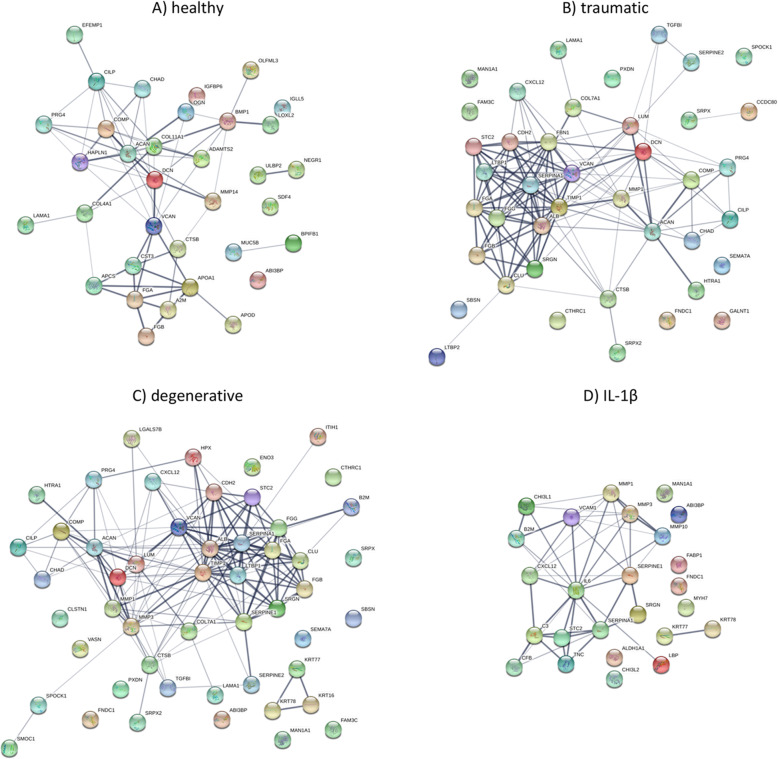
Proteins with a log2 fold-change > 1.5 (relative to the baseline) were further compared among the 4 experimental groups (Table 1). The most pronounced overlap in secreted proteins was found between the MSC secretomes following traumatic and degenerative stimulation (38 proteins). These two groups shared 11 (traumatic) and 13 (degenerative) secreted proteins with the secretome of MSCs stimulated by healthy CM (Fig. 4a). The highest overlap with the proinflammatory control was found in the secretome of MSCs stimulated with degenerative CM (14 proteins) followed by traumatic (8 proteins) and healthy (2 proteins) CM stimulation (Fig. 4a). Proteins were further categorized into anabolic, catabolic, ECM, growth factor, immune system, or other proteins (Fig. 4b). Of note, the percentage of secreted factors associated with immune response was almost doubled following exposure of MSCs to a healthy IVD CM environment (16%) compared to a traumatic or degenerative IVD CM stimulus (average: 9%) and was highest after MSC stimulation with IL-1β (41%). Network analysis of STRING revealed possible interactions among the upregulated proteins of healthy (Fig. 5a), traumatic (Fig. 5b), degenerative (Fig. 5c) CM, and IL-1β-induced MSC secretomes (Fig. 5d).
Table 1.
Proteins identified in secretomes of mesenchymal stromal cells stimulated with healthy, traumatic, or degenerative intervertebral disc conditioned medium or IL1-β, with a log2 fold-change > 1.5 (relative to the baseline control stimulation)
| gene_ID | protein_ID | Medium stimulus |
|---|---|---|
| CHAD | O15335 | Healthy, Traumatic, Degenerative |
| CILP | O75339 | Healthy, Traumatic, Degenerative |
| FGA | P02671 | Healthy, Traumatic, Degenerative |
| FGB | P02675 | Healthy, Traumatic, Degenerative |
| DCN | P07585 | Healthy, Traumatic, Degenerative |
| CTSB | P07858 | Healthy, Traumatic, Degenerative |
| VCAN | P13611 | Healthy, Traumatic, Degenerative |
| ACAN | P16112 | Healthy, Traumatic, Degenerative |
| LAMA1 | P25391 | Healthy, Traumatic, Degenerative |
| COMP | P49747 | Healthy, Traumatic, Degenerative |
| PRG4 | Q92954 | Healthy, Traumatic, Degenerative |
| IGHA1 | P01876 | Healthy, Degenerative, IL-1β |
| ABI3BP | Q7Z7G0 | Healthy, Degenerative, IL-1β |
| STC2 | O76061 | Traumatic, Degenerative, IL-1β |
| SERPINA1 | P01009 | Traumatic, Degenerative, IL-1β |
| MMP1 | P03956 | Traumatic, Degenerative, IL-1β |
| SRGN | P10124 | Traumatic, Degenerative, IL-1β |
| MAN1A1 | P33908 | Traumatic, Degenerative, IL-1β |
| CXCL12 | P48061 | Traumatic, Degenerative, L-1β |
| B2M | P61769 | Traumatic, Degenerative, IL-1β |
| FNDC1 | Q4ZHG4 | Traumatic, Degenerative, IL-1β |
| SRPX2 | O60687 | Traumatic, Degenerative |
| SEMA7A | O75326 | Traumatic, Degenerative |
| TIMP1 | P01033 | Traumatic, Degenerative |
| FGG | P02679 | Traumatic, Degenerative |
| ALB | P02768 | Traumatic, Degenerative |
| SERPINE2 | P07093 | Traumatic, Degenerative |
| CLU | P10909 | Traumatic, Degenerative |
| CDH2 | P19022 | Traumatic, Degenerative |
| LUM | P51884 | Traumatic, Degenerative |
| SRPX | P78539 | Traumatic, Degenerative |
| COL7A1 | Q02388 | Traumatic, Degenerative |
| SPOCK1 | Q08629 | Traumatic, Degenerative |
| LTBP1 | Q14766 | Traumatic, Degenerative |
| TGFBI | Q15582 | Traumatic, Degenerative |
| SBSN | Q6UWP8 | Traumatic, Degenerative |
| FAM3C | Q92520 | Traumatic, Degenerative |
| PXDN | Q92626 | Traumatic, Degenerative |
| HTRA1 | Q92743 | Traumatic, Degenerative |
| CTHRC1 | Q96CG8 | Traumatic, Degenerative |
| HAPLN1 | P10915 | Healthy |
| BPIFB1 | Q8TDL5 | Healthy |
| APOA1 | P02647 | Healthy |
| COL4A1 | P02462 | Healthy |
| COL11A1 | P12107 | Healthy |
| MUC5B | Q9HC84 | Healthy |
| IGKC | P01834 | Healthy |
| APOD | P05090 | Healthy |
| IGHG1 | P01857 | Healthy |
| APCS | P02743 | Healthy |
| IGLL5 | B9A064 | Healthy |
| ADAMTS2 | O95450 | Healthy |
| ULBP2 | Q9BZM5 | Healthy |
| OGN | P20774 | Healthy |
| EFEMP1 | Q12805 | Healthy |
| IGFBP6 | P24592 | Healthy |
| OLFML3 | Q9NRN5 | Healthy |
| A2M | P01023 | Healthy |
| LOXL2 | Q9Y4K0 | Healthy |
| NEGR1 | Q7Z3B1 | Healthy |
| SDF4 | Q9BRK5 | Healthy |
| BMP1 | P13497 | Healthy |
| CST3 | P01034 | Healthy |
| MMP14 | P50281 | Healthy |
| FBN1 | P35555 | Traumatic |
| GALNT1 | Q10472 | Traumatic |
| CCDC80 | Q76M96 | Traumatic |
| LTBP2 | Q14767 | Traumatic |
| HPX | P02790 | Degenerative |
| ITIH1 | P19827 | Degenerative |
| LGALS7; LGALS7B | P47929 | Degenerative |
| CLSTN1 | O94985 | Degenerative |
| SMOC1 | Q9H4F8 | Degenerative |
| ENO3 | P13929 | Degenerative |
| VASN | Q6EMK4 | Degenerative |
| KRT16 | P08779 | Degenerative |
| CFB | P00751 | IL1-β |
| IL6 | P05231 | IL1-β |
| C3 | P01024 | IL1-β |
| MMP10 | P09238 | IL1-β |
| MYH7 | P12883 | IL1-β |
| ALDH1A1 | P00352 | IL1-β |
| LBP | P18428 | IL1-β |
| FABP1 | P07148 | IL1-β |
| CHI3L1 | P36222 | IL1-β |
| VCAM1 | P19320 | IL1-β |
| CHI3L2 | Q15782 | IL1-β |
| TNC | P24821 | IL1-β |
Fig. 4.
Comparison of secretome composition. a Venn-diagram representing the overlap of the detected proteins among all four experimental groups. b Pie charts representing MSC secretome compositions, categorized into anabolic, catabolic, ECM, growth factor, immune system, and other proteins. Data includes the detected proteins (LC-MS/MS data) upregulated > 1.5 log2 fold relative to the baseline control
Fig. 5.
Interaction network of upregulated proteins in MSC secretome. Data includes detected proteins (LC-MS/MS data) upregulated > 1.5 log2 fold relative to baseline control. Upregulated proteins of a healthy, b traumatic, c degenerative, and d IL-1β stimulated MSC secretomes have been searched for possible interaction with the network analysis of String. Active interactions include text mining, experiments, and databases. Line thickness indicates confidence. Proteins are labeled with the corresponding gene name
Quantitative measurement of protein concentration by multiplex immunoassay
Levels of different cytokines and chemokines were measured in pooled IVD CM (Suppl. Table 5), and in secretomes of MSCs following culture in basal medium (baseline control) and following exposure to IVD CMs (healthy, traumatic, degenerative) and IL-1β (N = 9/ group). In MSC secretomes, IL-1α and IL-17A were either undetected or extremely low in all samples and could not be analyzed (Fig. 6). Interestingly, most of the proinflammatory and/or immunomodulatory cytokines were undetected in the secretomes of MSCs incubated with healthy IVD CM, including TNF-β, IL-5, IL-16, IL-12, IL-23p40, TNF-α, IL-4, IL-1β, IL-13, IL-12p70, IFN-γ, IL-8, and IL-10. In contrast, the concentration of several chemokines/cytokines such as GM-CSF, TNF-α, IL-12p70, IFN-γ (all P < 0.05), monocyte chemoattractant protein 1 (MCP-1), macrophage inflammatory protein 1 beta (MIP-1β), VEGF, IL-13, and IL-6 (all P < 0.01) was significantly higher in the secretomes of IL-1β primed MSCs than those of baseline control. There was a significant difference between traumatic IVD CM group and baseline control regarding the fold changes of IL-13, IL-8, IL-6 (P < 0.01), VEGF, MCP-1, and MIP-1β (P < 0.05). Moreover, the concentration of IL-13, IL-8, IL-6 (P < 0.01), TNF-α, IL-1β, VEGF, MCP-1, and MIP-1β (fold changes P < 0.05) was significantly higher in secretomes of MSCs exposed to degenerative IVD CM than the baseline control group. In both traumatic and degenerative IVD CM groups, a similar trend was observed for production of cytokines, including IL-13, IL-8, and IL-6 which were considerably higher than the healthy group (P < 0.01). On the other hand, the secretion of MIP-1β, a chemoattractant for inflammatory cells, was considerably higher in MSC secretome following stimulation by healthy IVD CM in comparison to other treatment groups (P < 0.05).
Fig. 6.
Immunoassay data of MSC secretomes. Cytokines and chemokines quantified in two immunoassay panels. Data includes quantified proteins as log2 fold-change relative to baseline control. Darker color indicates a higher upregulation. 0 = not detected
Discussion
The present study aimed to investigate the response of MSCs following exposure to a healthy, traumatic, or degenerative human IVD environment. Our primary finding was that stimulation with IVD CM induced a richer and more complex MSC secretome, involving more biological processes, compared to stimulation with a single cytokine (IL-1β). Secondly, the MSCs response to stimulation with IVD CM was different based on their status (i.e., healthy versus traumatic/degenerated). Thirdly, the MSC secretome seemed to match the primary need of the IVD tissue: homeostasis maintenance in the case of healthy IVDs, immunomodulation, adjustment of the matrix synthesis/degradation disbalance, and (re) organization of ECM in the case of traumatic and degenerative IVDs.
When exposed to a healthy IVD CM, MSCs response reflected the maintenance of homeostasis. This was expected, as in the absence of injury or degeneration, there is no need for tissue repair or regeneration. Successful IVD homeostasis involves a well-regulated, low dose of immunomodulatory cytokines and chemokines [42]. GSEA of healthy IVD-induced MSC secretome revealed enriched “regulation of innate immune response” and “humoral immune response.” Interestingly, the proportion of secreted proteins associated with the immune system was also higher following stimulation by healthy IVD CM compared to the traumatic and degenerative stimulation. For example, upregulation of Amyloid P was observed exclusively after the healthy stimulus (Table 1). Amyloid P inhibits the differentiation of fibrocytes, promotes the formation of immuno-regulatory macrophages, and inhibits neutrophil adhesion to ECM [43]. Immunoassay data indicated a significantly increased release of MIPs following exposure to a healthy environment. MIP-1β represents a ligand of C-C chemokine receptor type 5 (CCR5), which has been identified on IVD cells and may be involved in cell migration [44]. MIP-1α has been associated with inhibition of cell proliferation [45]. Release of MIP-1α might therefore represent an autoregulation of cell proliferation, as the intact healthy environment does not require an enriched cell population [46, 47]. Concerning ECM turnover, we observed an upregulated release of alpha 2 macroglobulin (A2M) (Table 1). A2M represents the major inhibitor of endoproteinases and its release might indicate that MSCs aim to stabilize the healthy IVD ECM in its physiological composition and structure.
The IVD depends on an intact ECM to maintain its primary mechanical function. Damage following traumatic injury represents a serious threat to the integrity of IVD ECM. Exposure of MSCs to traumatic IVD CM enriched the process “regulation of anatomical structure size.” MSCs secreted several proteins solely following traumatic stimulus, potentially participating in damage repair. Latent transforming growth factor-β1 binding protein 2 (LTBP-2, Table 1) has been reported to stimulate the expression of TGF-β1 in fibroblasts [48]. In the IVD, TGF-β1 has been associated with alleviation of the inflammatory response, inhibition of ECM degradation, inhibition of cell death, increased ECM synthesis, and promotion of cell proliferation [49]. Similarly, we observed an upregulation of cellular communication network factor 2/connective tissue growth factor (CCN2/CTGF) mainly following traumatic stimulus (Suppl. Table 2). CCN2/CTGF, also secreted by notochordal cells [50], can induce matrix production in nucleus pulposus (NP) cells [51]. After traumatic IVD CM stimulation, there were also upregulated levels of angiopoietin 1 (Suppl. Table 2), which represents the ligand of angiopoietin 1 receptor (Tie2) and is crucial for the survival of NP cells [52]. The overall MSCs response to traumatic IVD CM thus points towards cell growth and ECM repair.
In contrast to the immediate threat of ECM damage in traumatic IVDs, degeneration is a process evolving over decades, eventually resulting in a highly disorganized, fragmented ECM. As IVD degeneration is often associated with ingrowth of nerves and blood vessels, neural and vascular cells may have contributed to the composition of the degenerative IVD CM. Exposure of MSCs to a degenerative IVD environment did not enhance any immunomodulatory biological processes. However, secretion of certain proteins holding immunomodulatory functions such as keratin 16 (KRT16) and galectin-7 (LGALS7) was solely induced following degenerative stimulus (Table 1) [53, 54]. Concerning tissue regeneration, secreted modular calcium-binding protein 1 (SMOC-1, Table 1) was upregulated, which has been associated with TGF-β1 mediated fibrosis [55]. Interestingly, while the upregulated biological processes did differ between the four experimental groups, we did not observe any relevant differences in the downregulated processes (Fig. 2). Nevertheless, downregulation or absence of certain proteins might also be of importance for secretome mediated influence on biological processes.
Among all groups, the highest overlap in released proteins was observed following stimulation by traumatic and degenerative IVD CM. Both traumatic and degenerative stimuli induced secretion of matrix metalloproteinase (MMP)1, MMP2, MMP10, and MMP13 (Suppl. Table 2/3). MMPs represent matrix-degrading enzymes which can facilitate migration and recruitment of inflammatory cells or progenitor cells. Furthermore, MSCs are known to secrete MMPs to be able to migrate through the ECM [56]. Tissue inhibitors of metalloproteinases (TIMPs) are important MMP antagonists. They control MMP activities which results in a balanced matrix degradation. Interestingly, TIMP1 was upregulated following traumatic and degenerative stimulus (Table 1). This suggests that MSCs try to stabilize a dynamic balance of ECM turnover during reorganization following trauma or during regeneration.
MSCs also released interleukins (IL-1, IL-6, IL-8, IL-13), serglycin (SRGN), and complement component 1r following traumatic and degenerative IVD CM stimulus (Suppl. Table 2/3). IL-1β triggers the innate inflammation while IL-8 induces chemotaxis in neutrophils and MSCs, causing them to migrate towards the site of damage [57–59]. In contrast, IL-13 inhibits the production of proinflammatory cytokines and chemokines [60]. SRGN represents a binding partner for various chemokines and functions as a modulator of the immune system [61]. The immunoassay also revealed a significantly higher concentration of IL-6 following traumatic and degenerative compared to a healthy stimulus. IL-6 represents a key player of the primary inflammatory response where it functions as an important activator of pro- and anti-inflammatory cells [62]. Thus, the secreted proteins may have a role in modulating the inflammatory state in acute and chronic IVD pathologies.
STRING analysis revealed denser networks following traumatic and degenerative IVD CM stimulation, when compared to the healthy stimulus. Interestingly, the common center of both networks was alpha-1-antitrypsin, or SERPINA1, a broad-spectrum protease inhibitor that was associated with the regulation of inflammatory responses [63]. For the IVD, we can only speculate about the role of SERPINA1. Potential targets include its inhibition of proteinase 3 (PRTN3), MMP9, and a disintegrin and metalloprotease domain 17 (ADAM17), among others [64]. High levels of PRTN3 have been associated with calcifications in herniated IVD tissue [65]. Upregulated expression of MMP9 has been reported in cells from degenerative compared to traumatic IVDs [66]. Similarly, an increased concentration in MMP9 has been correlated with the degree of degeneration and with pain sensation in herniated IVD [66, 67]. Inhibition of ADAM17 has been associated with an upregulated TGFβ signaling [68]. In our data, degenerative IVD CM induced a higher release of SERPINA1 compared to traumatic stimulus, while healthy CM did not provoke SERPINA1 secretion. This might indicate that MSCs participate in the restoration of protease inhibitors known to deplete during IVD degeneration [69].
To validate the MSCs response, we also performed a simplistic stimulation with IL-1β. This resulted in enriched biological processes like “acute inflammatory response”, “regulation of inflammatory response,” and “leukocyte migration.” Also, IL-1β stimulus led to the secretion of various proinflammatory factors including elements of the complement system, TNF-α, IL-1β, and IL-6 (Suppl. Table 4). Elevated levels of TNF-α and IL-6 have previously been observed in MSCs following exposure to IL-1β [70]. Quantitative immunoassay revealed significantly higher concentrations of MCP-1 following IL-1β stimulus compared to stimulation by healthy IVD CM. MCP-1 contributes to the activation and recruitment of monocytes and has been detected in normoxic and hypoxic cultured MSCs [71].
The present study was performed using a 2D monolayer cell culture model. 3D culture systems require embedding MSCs within a carrier material (e.g., hydrogel) which might bind some of the released proteins and therefore alter the profile of proteins secreted by the cell-hydrogel construct. In addition, LC-MS/MS is very sensitive, and proteins released from the carrier material could potentially mask proteins released as part of the MSC secretome. However, it has been reported that MSC secretome composition may change following MSC culture within a 3D environment. For example, concentrations of hepatocyte growth factor (HGF) and intercellular adhesion molecule 1 (ICAM-1) were increased in 3D cultures compared to 2D cultures [29]. Furthermore, 3D culture of umbilical cord tissue MSCs has been associated with an enhanced expression of anti-inflammatory cytokines (IL-10 and LIF) and trophic factors involved in mechanisms leading to tissue regeneration compared to the secretome following 2D culture [72]. Further study should investigate the response of MSCs cultured within a 3D system to the different IVD environments. Besides, we investigated the composition of the MSC secretome at 24–48 h after stimulation. While this time frame is of relevance for short-term MSC preconditioning for secretome generation, it only partially reflects the situation occurring after therapeutic MSC injection into the IVD. In an in vivo or ex-vivo environment, it is likely that the MSC secretome may change over time and depending on the surrounding conditions.
As MSCs have been shown to have diverse properties based on age and gender, we analyzed whether the secretome compositions were different between young versus old and male versus female donors [73]. Interestingly, we neither observed any significant differences in secretome composition among the different donor age groups nor between female or male donors. This might indicate that the stimulus applied to the MSCs was more prominent to generate a certain type of secretome than the MSC source.
Limitations
Here we analyzed the molecular factors and the biological processes within the MSC secretome based on protein identification. Besides soluble proteins, secretome contains free nucleic acids, lipids, and extracellular vesicles. Our results therefore only reflect a portion of the MSC secretome. Secondly, IVD CM contains high amounts of ECM proteins, which potentially mask proteins present at low concentrations during LC-MS/MS. We therefore replaced the CM following 24 h stimulation to collect the proteins secreted by MSCs. It is likely that MSCs already secreted proteins during the first 24 h of stimulation by IVD CM which were not detected with this experimental setup. Thirdly, we did not perform any co-culture of MSCs and IVD cells, as the proteins released by both cell types could not have been differentiated in the analysis process. However, the use of cell free IVD conditioned medium omits the possible changes in secretome composition evolving from cell-cell or cell-ECM contact. Finally, we were not able to assign the degree of degeneration to IVDs from all donors since magnetic resonance imaging (MRI) scans were only available from donors with degenerative IVDs.
Conclusions
Our results indicate that MSCs adapt their profile of secreted proteins depending on their environment. This allows them to take different roles tailored to the state of disease. In healthy IVDs, MSC-derived trophic factors may stabilize tissue homeostasis and act as immunomodulators. Following a traumatic injury, MSC secretome may support the recruitment of additional cells, modulate inflammation, cell survival, and secrete ECM proteins. A degenerative IVD milieu may induce factors initiating remodeling processes and synthesis of ECM proteins. However, characterization of the proteins released by MSCs represents only one part of the interaction between MSCs and the IVD milieu. The response of the resident IVD cells to the MSC secretome is currently under investigation and will provide important knowledge to identify the therapeutic secretome for specific IVD states of injury or degeneration.
Supplementary Information
Additional file 1: Supplementary Figure 1. Details of cell and tissue samples used for the experiments. (A) MSCs from twelve different donors were used. All MSCs were derived from vertebral bone marrow aspirates. Only donors younger than 50 years (age at isolation) were selected, representing four different age groups (average age 17, 26.33, 37.66 and 48.66 years). Gender was equally balanced (6 male; 6 female) and symmetrically distributed among age groups. (B) IVD conditioned medium donor overview. For MSC stimulation, IVD conditioned medium from different donors within one condition was pooled (n = 4/group).
Additional file 2: Supplementary Figure 2. Effect of IVD conditioned medium treatment on DNA content, metabolic activity and lactate dehydrogenase (LDH) release of MSCs. (A) DNA content of MSCs in 6-well plate normalized to timepoint zero after 14 h of cell attachment. *p < 0.05, ***p < 0.001 (Kruskal-Wallis test). (B) Metabolic activity was measured with CellTiter-Blue. Data was standardized to the treatment condition baseline within every MSC donor. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; One-way ANOVA. (C) LDH was measured in the MSC secretome to detect cytotoxic reactions. No significant differences were found (Kruskal-Wallis-test). (D-H) Images were taken just before secretome collection. Scale bar = 500 μm.
Additional file 3 : Supplementary Table 1. MSC secretome following healthy CM stimulation. Supplementary Table 2. MSC secretome following traumatic CM stimulation. Supplementary Table 3. MSC secretome following degenerative CM stimulation. Supplementary Table 4. MSC secretome following IL-1β stimulation.
Additional file 4 : Supplementary Table 5 Concentrations of cytokines and chemokines in pooled conditioned media from healthy, traumatic and degenerative intervertebral disc, measured by immunoassay technique (mean+/-sd of technical replicates; pg/mL).
Acknowledgements
The authors would like to acknowledge AO Foundation and AO Spine International for funding, and Dr. Paolo Nanni and Dr. Witold Wolski from the Functional Genomics Center Zurich for support acquisition and statistical analysis of the mass spectroscopy data. We thank Dr. Zhen Li, AO Research Institute Davos, for technical advice, and Dr. Katja Bärenfaller, Swiss Institute of Allergy and Asthma Research, Davos, for consulting in proteomics data analysis.
Abbreviations
- A2M
Alpha 2 macroglobulin
- ADAM
A disintegrin and metalloprotease domain
- a-MEM
Alpha minimal essential medium
- ASC
Adipose-derived stem cells
- CCN2
Cellular communication network factor 2
- CCR5
C-C chemokine receptor type 5
- CM
Conditioned medium
- CTGF
Connective tissue growth factor
- ECM
Extracellular matrix
- FBS
Fetal bovine serum
- FDR
False discovery rate
- FGF
Fibroblast growth factor
- G-CSF
Granulocyte colony-stimulating factor
- GO
Gene ontology
- GOBP
Gene ontology classification for biological processes
- GSEA
Gene set enrichment analysis
- HGF
Hepatocyte growth factor
- IGF-1
Insulin-like growth factor 1
- IL
Interleukin
- IL1-Ra
Interleukin 1 receptor antagonist
- INF
Interferon
- ISSCR
International society for stem cell research
- IVD
Intervertebral disc
- KRT16
Keratin 16
- LC-MS/MS
Liquid chromatography mass spectroscopy/ mass spectroscopy
- LDH
Lactate dehydrogenase
- LGALS7
Galectin 7
- Lg-DMEM
Low glucose Dulbecco’s modified Eagle’s medium
- LTBP-2
Latent transforming growth factor-β1 binding protein 2
- MCP
Monocyte chemoattractant protein
- MIP
Macrophage inflammatory protein
- MMP
Matrix metalloproteinase
- MRI
Magnetic resonance imaging
- MSC
Mesenchymal stromal cells
- NES
Normalized enrichment score
- NP
Nucleus pulposus
- P/S
Penicillin-Streptomycin
- PBS
Phosphate buffered saline
- PRTN3
Proteinase 3
- RT
Room temperature
- SMOC-1
Secreted modular calcium-binding protein 1
- SRGN
Serglycin
- TIMP
Tissue inhibitor of metalloproteinase
- TNF
Tumor necrosis factor
- VEGF
Vascular endothelial growth factor
Authors’ contributions
SW: Conception and design of the work, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of the manuscript. AK: Collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of the manuscript. CW: Conception and design of the work, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of the manuscript. KWK: Administrative support, data analysis and interpretation, manuscript revision, final approval of the manuscript. SH: Provision of study material, manuscript revision, final approval of the manuscript. CF: Data analysis and interpretation, manuscript revision, final approval of the manuscript. LMB: Provision of study material, manuscript revision, final approval of the manuscript. LH: Provision of study material, data analysis and interpretation, manuscript revision, final approval of the manuscript. RGR: Financial support; administrative support; final approval of the manuscript. MA: Conception and design of the work, data analysis and interpretation, manuscript revision, final approval of the manuscript. MP: Conception and design of the work, data analysis and interpretation, manuscript writing, final approval of the manuscript. SG: Conception and design of the work, data analysis and interpretation, administrative support, supervision, manuscript writing, final approval of the manuscript.
Funding
This study was funded by the AO Foundation and AOSpine International.
Availability of data and materials
Proteomics data are reported in the supplementary tables of the manuscript. All original data are available from the authors on request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD021281 [74].
Ethics approval and consent to participate
Vertebral bone marrow aspirates were obtained with written consent from patients undergoing spine surgery. IVD tissues from patients with traumatic injury and from patients diagnosed with IVD degeneration were obtained with written consent from patients undergoing spine surgery. Non-degenerated IVD tissues were obtained from organ donors after donor and familial consent by the McGill Scoliosis & Spinal Research Group via a collaboration with Transplant Quebec and approval by the McGill University’s Institutional Review Board (IRB# A04-M53-08B).
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13287-020-02062-2.
References
- 1.Ullah I, Subbarao RB, Rho GJ. Human mesenchymal stem cells - current trends and future prospective. Biosci Rep. 2015;35(2):e00191. [DOI] [PMC free article] [PubMed]
- 2.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 3.Sakai D, Schol J. Cell therapy for intervertebral disc repair: clinical perspective. J Orthop Translat. 2017;9:8–18. doi: 10.1016/j.jot.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Migliorini F, Rath B, Tingart M, Baroncini A, Quack V, Eschweiler J. Autogenic mesenchymal stem cells for intervertebral disc regeneration. Int Orthop. 2019;43(4):1027–1036. doi: 10.1007/s00264-018-4218-y. [DOI] [PubMed] [Google Scholar]
- 5.Strassburg S, Richardson SM, Freemont AJ, Hoyland JA. Co-culture induces mesenchymal stem cell differentiation and modulation of the degenerate human nucleus pulposus cell phenotype. Regen Med. 2010;5(5):701–711. doi: 10.2217/rme.10.59. [DOI] [PubMed] [Google Scholar]
- 6.Le Maitre CL, Baird P, Freemont AJ, Hoyland JA. An in vitro study investigating the survival and phenotype of mesenchymal stem cells following injection into nucleus pulposus tissue. Arthritis Res Ther. 2009;11(1):R20. doi: 10.1186/ar2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strassburg S, Hodson NW, Hill PI, Richardson SM, Hoyland JA. Bi-directional exchange of membrane components occurs during co-culture of mesenchymal stem cells and nucleus pulposus cells. PLoS One. 2012;7(3):e33739. doi: 10.1371/journal.pone.0033739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wuertz K, Godburn K, Neidlinger-Wilke C, Urban J, Iatridis JC. Behavior of mesenchymal stem cells in the chemical microenvironment of the intervertebral disc. Spine (Phila Pa 1976) 2008;33(17):1843–1849. doi: 10.1097/BRS.0b013e31817b8f53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang YC, Leung VY, Lu WW, Luk KD. The effects of microenvironment in mesenchymal stem cell-based regeneration of intervertebral disc. Spine J. 2013;13(3):352–362. doi: 10.1016/j.spinee.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Loibl M, Wuertz-Kozak K, Vadala G, Lang S, Fairbank J, Urban JP. Controversies in regenerative medicine: should intervertebral disc degeneration be treated with mesenchymal stem cells? JOR Spine. 2019;2(1):e1043. doi: 10.1002/jsp2.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hang D, Li F, Che W, Wu X, Wan Y, Wang J, et al. One-stage positron emission tomography and magnetic resonance imaging to assess mesenchymal stem cell survival in a canine model of intervertebral disc degeneration. Stem Cells Dev. 2017;26(18):1334–1343. doi: 10.1089/scd.2017.0103. [DOI] [PubMed] [Google Scholar]
- 12.Maguire G. Stem cell therapy without the cells. Commun Integr Biol. 2013;6(6):e26631. doi: 10.4161/cib.26631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beer L, Mildner M, Ankersmit HJ. Cell secretome based drug substances in regenerative medicine: when regulatory affairs meet basic science. Ann Transl Med. 2017;5(7):170. doi: 10.21037/atm.2017.03.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baglio SR, Pegtel DM, Baldini N. Mesenchymal stem cell secreted vesicles provide novel opportunities in (stem) cell-free therapy. Front Physiol. 2012;3:359. doi: 10.3389/fphys.2012.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Tredget EE, Wu PY, Wu Y. Paracrine factors of mesenchymal stem cells recruit macrophages and endothelial lineage cells and enhance wound healing. PLoS One. 2008;3(4):e1886. doi: 10.1371/journal.pone.0001886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.L PK, Kandoi S, Misra R, S V, K R, Verma RS. The mesenchymal stem cell secretome: a new paradigm towards cell-free therapeutic mode in regenerative medicine. Cytokine Growth Factor Rev 2019;46:1–9. [DOI] [PubMed]
- 17.Madrigal M, Rao KS, Riordan NH. A review of therapeutic effects of mesenchymal stem cell secretions and induction of secretory modification by different culture methods. J Transl Med. 2014;12:260. doi: 10.1186/s12967-014-0260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vizoso FJ, Eiro N, Cid S, Schneider J, Perez-Fernandez R. Mesenchymal stem cell secretome: toward cell-free therapeutic strategies in regenerative medicine. Int J Mol Sci. 2017;18(9):1852. [DOI] [PMC free article] [PubMed]
- 19.Zhou BR, Xu Y, Guo SL, Xu Y, Wang Y, Zhu F, et al. The effect of conditioned media of adipose-derived stem cells on wound healing after ablative fractional carbon dioxide laser resurfacing. Biomed Res Int. 2013;2013:519126. doi: 10.1155/2013/519126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katagiri W, Osugi M, Kawai T, Hibi H. First-in-human study and clinical case reports of the alveolar bone regeneration with the secretome from human mesenchymal stem cells. Head Face Med. 2016;12:5. doi: 10.1186/s13005-016-0101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukuoka H, Suga H. Hair regeneration treatment using adipose-derived stem cell conditioned medium: follow-up with Trichograms. Eplasty. 2015;15:e10. [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira JR, Teixeira GQ, Santos SG, Barbosa MA, Almeida-Porada G, Gonçalves RM. Mesenchymal stromal cell secretome: influencing therapeutic potential by cellular pre-conditioning. Front Immunol. 2018;9:2837. doi: 10.3389/fimmu.2018.02837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crisostomo PR, Wang Y, Markel TA, Wang M, Lahm T, Meldrum DR. Human mesenchymal stem cells stimulated by TNF-alpha, LPS, or hypoxia produce growth factors by an NF kappa B- but not JNK-dependent mechanism. Am J Physiol Cell Physiol. 2008;294(3):C675–C682. doi: 10.1152/ajpcell.00437.2007. [DOI] [PubMed] [Google Scholar]
- 24.Redondo-Castro E, Cunningham C, Miller J, Martuscelli L, Aoulad-Ali S, Rothwell NJ, et al. Interleukin-1 primes human mesenchymal stem cells towards an anti-inflammatory and pro-trophic phenotype in vitro. Stem Cell Res Ther. 2017;8(1):79. doi: 10.1186/s13287-017-0531-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu Z, Li Q, Bu H, Lin F. Mesenchymal stem cells inhibit complement activation by secreting factor H. Stem Cells Dev. 2010;19(11):1803–1809. doi: 10.1089/scd.2009.0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gieseke F, Kruchen A, Tzaribachev N, Bentzien F, Dominici M, Müller I. Proinflammatory stimuli induce galectin-9 in human mesenchymal stromal cells to suppress T-cell proliferation. Eur J Immunol. 2013;43(10):2741–2749. doi: 10.1002/eji.201343335. [DOI] [PubMed] [Google Scholar]
- 27.Redondo-Castro E, Cunningham CJ, Miller J, Brown H, Allan SM, Pinteaux E. Changes in the secretome of tri-dimensional spheroid-cultured human mesenchymal stem cells in vitro by interleukin-1 priming. Stem Cell Res Ther. 2018;9(1):11. doi: 10.1186/s13287-017-0753-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ylöstalo JH, Bartosh TJ, Coble K, Prockop DJ. Human mesenchymal stem/stromal cells cultured as spheroids are self-activated to produce prostaglandin E2 that directs stimulated macrophages into an anti-inflammatory phenotype. Stem Cells. 2012;30(10):2283–2296. doi: 10.1002/stem.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter K, Lee HJ, Na KS, Fernandes-Cunha GM, Blanco IJ, Djalilian A, et al. Characterizing the impact of 2D and 3D culture conditions on the therapeutic effects of human mesenchymal stem cell secretome on corneal wound healing in vitro and ex vivo. Acta Biomater. 2019;99:247–257. doi: 10.1016/j.actbio.2019.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SC, Jeong HJ, Lee SK, Kim SJ. Lipopolysaccharide preconditioning of adipose-derived stem cells improves liver-regenerating activity of the secretome. Stem Cell Res Ther. 2015;6(1):75. doi: 10.1186/s13287-015-0072-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Z, Chen Y, Dunstan C, Roohani-Esfahani S, Zreiqat H. Priming adipose stem cells with tumor necrosis factor-alpha preconditioning potentiates their exosome efficacy for bone regeneration. Tissue Eng A. 2017;23(21–22):1212–1220. doi: 10.1089/ten.tea.2016.0548. [DOI] [PubMed] [Google Scholar]
- 32.Jun EK, Zhang Q, Yoon BS, Moon JH, Lee G, Park G, et al. Hypoxic conditioned medium from human amniotic fluid-derived mesenchymal stem cells accelerates skin wound healing through TGF-β/SMAD2 and PI3K/Akt pathways. Int J Mol Sci. 2014;15(1):605–628. doi: 10.3390/ijms15010605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piazza N, Dehghani M, Gaborski TR, Wuertz-Kozak K. Therapeutic potential of extracellular vesicles in degenerative diseases of the intervertebral disc. Front Bioeng Biotechnol. 2020;8(311). [DOI] [PMC free article] [PubMed]
- 34.Bara JJ, Herrmann M, Menzel U, Benneker L, Alini M, Stoddart MJ. Three-dimensional culture and characterization of mononuclear cells from human bone marrow. Cytotherapy. 2015;17(4):458–472. doi: 10.1016/j.jcyt.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 35.Herrmann M, Hildebrand M, Menzel U, Fahy N, Alini M, Lang S, et al. Phenotypic characterization of bone marrow mononuclear cells and derived stromal cell populations from human iliac crest, vertebral body and femoral head. Int J Mol Sci. 2019;20(14):3454. [DOI] [PMC free article] [PubMed]
- 36.Gardner OFWA, M.; Stoddart, M. Mesenchymal stem cells derived from human bone marrow. In: Doran PM, editor. Cartilage tissue engineering: methods and protocols. New York: Springer; 2015. pp. 41–52. [DOI] [PubMed] [Google Scholar]
- 37.Della Bella E, Menzel U, Basoli V, Tourbier C, Alini M, Stoddart MJ. Differential regulation of circRNA, miRNA, and piRNA during early osteogenic and chondrogenic differentiation of human mesenchymal stromal cells. Cells. 2020;9(2):398. [DOI] [PMC free article] [PubMed]
- 38.Jim B, Steffen T, Moir J, Roughley P, Haglund L. Development of an intact intervertebral disc organ culture system in which degeneration can be induced as a prelude to studying repair potential. Eur Spine J. 2011;20(8):1244–1254. doi: 10.1007/s00586-011-1721-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuznetsova AB, P.B.; Christensen, R.H.B. lmerTest package: tests in linear mixed effects models. J Stat Software. 2017;82(13):26.
- 40.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 2012;8(2):e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D3d8. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Cao X. Cellular and molecular regulation of innate inflammatory responses. Cell Mol Immunol. 2016;13(6):711–721. doi: 10.1038/cmi.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pilling D, Gomer RH. The development of serum amyloid P as a possible therapeutic. Front Immunol. 2018;9:2328. doi: 10.3389/fimmu.2018.02328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu W, Liu D, Zheng J, Shi P, Chou PH, Oh C, et al. Annulus fibrosus cells express and utilize C-C chemokine receptor 5 (CCR5) for migration. Spine J. 2017;17(5):720–726. doi: 10.1016/j.spinee.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev. 2002;13(6):455–481. doi: 10.1016/s1359-6101(02)00045-x. [DOI] [PubMed] [Google Scholar]
- 46.Johnson ZI, Schoepflin ZR, Choi H, Shapiro IM, Risbud MV. Disc in flames: roles of TNF-α and IL-1β in intervertebral disc degeneration. Eur Cell Mater. 2015;30:104–116. doi: 10.22203/ecm.v030a08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wangler S, Peroglio M, Menzel U, Benneker LM, Haglund L, Sakai D, et al. Mesenchymal stem cell homing into intervertebral discs enhances the Tie2-positive progenitor cell population, prevents cell death, and induces a proliferative response. Spine (Phila Pa 1976) 2019;44(23):1613–1622. doi: 10.1097/BRS.0000000000003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sideek MA, Smith J, Menz C, Adams JRJ, Cowin AJ, Gibson MA. A central bioactive region of LTBP-2 stimulates the expression of TGF-β1 in fibroblasts via Akt and p38 signalling pathways. Int J Mol Sci. 2017;18(10):2114. [DOI] [PMC free article] [PubMed]
- 49.Chen S, Liu S, Ma K, Zhao L, Lin H, Shao Z. TGF-β signaling in intervertebral disc health and disease. Osteoarthr Cartil. 2019;27(8):1109–1117. doi: 10.1016/j.joca.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 50.Gantenbein B, Calandriello E, Wuertz-Kozak K, Benneker LM, Keel MJB, Chan SCW. Activation of intervertebral disc cells by co-culture with notochordal cells, conditioned medium and hypoxia. BMC Musculoskelet Disord. 2014;15:422. [DOI] [PMC free article] [PubMed]
- 51.Tran CM, Schoepflin ZR, Markova DZ, Kepler CK, Anderson DG, Shapiro IM, et al. CCN2 suppresses catabolic effects of interleukin-1beta through alpha5beta1 and alphaVbeta3 integrins in nucleus pulposus cells: implications in intervertebral disc degeneration. J Biol Chem. 2014;289(11):7374–7387. doi: 10.1074/jbc.M113.526111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakai D, Nakamura Y, Nakai T, Mishima T, Kato S, Grad S, et al. Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nat Commun. 2012;3:1264. doi: 10.1038/ncomms2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lessard JC, Pina-Paz S, Rotty JD, Hickerson RP, Kaspar RL, Balmain A, et al. Keratin 16 regulates innate immunity in response to epidermal barrier breach. Proc Natl Acad Sci U S A. 2013;110(48):19537–19542. doi: 10.1073/pnas.1309576110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rabinovich GA, Liu FT, Hirashima M, Anderson A. An emerging role for galectins in tuning the immune response: lessons from experimental models of inflammatory disease, autoimmunity and cancer. Scand J Immunol. 2007;66(2–3):143–158. doi: 10.1111/j.1365-3083.2007.01986.x. [DOI] [PubMed] [Google Scholar]
- 55.Gao Q, Mok HP, Zhuang J. Secreted modular calcium-binding proteins in pathophysiological processes and embryonic development. Chin Med J. 2019;132(20):2476–2484. doi: 10.1097/CM9.0000000000000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steingen C, Brenig F, Baumgartner L, Schmidt J, Schmidt A, Bloch W. Characterization of key mechanisms in transmigration and invasion of mesenchymal stem cells. J Mol Cell Cardiol. 2008;44(6):1072–1084. doi: 10.1016/j.yjmcc.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 57.Hammond ME, Lapointe GR, Feucht PH, Hilt S, Gallegos CA, Gordon CA, et al. IL-8 induces neutrophil chemotaxis predominantly via type I IL-8 receptors. J Immunol. 1995;155(3):1428–1433. [PubMed] [Google Scholar]
- 58.Dinarello CA. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev. 2018;281(1):8–27. doi: 10.1111/imr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ringe J, Strassburg S, Neumann K, Endres M, Notter M, Burmester GR, et al. Towards in situ tissue repair: human mesenchymal stem cells express chemokine receptors CXCR1, CXCR2 and CCR2, and migrate upon stimulation with CXCL8 but not CCL2. J Cell Biochem. 2007;101(1):135–146. doi: 10.1002/jcb.21172. [DOI] [PubMed] [Google Scholar]
- 60.Punnonen J, Carballido JM, Aversa G, de Vries JE. Interleukin 13 and its receptor. In: Delves PJ, editor. Encyclopedia of immunology. 2. Oxford: Elsevier; 1998. pp. 1489–1492. [Google Scholar]
- 61.Korpetinou A, Skandalis SS, Labropoulou VT, Smirlaki G, Noulas A, Karamanos NK, et al. Serglycin: at the crossroad of inflammation and malignancy. Front Oncol. 2014;3:327. doi: 10.3389/fonc.2013.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weiss ARR, Dahlke MH. Immunomodulation by mesenchymal stem cells (MSCs): mechanisms of action of living, apoptotic, and dead MSCs. Front Immunol. 2019;10:1191. doi: 10.3389/fimmu.2019.01191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Janciauskiene S, Welte T. Well-known and less well-known functions of alpha-1 antitrypsin. Its role in chronic obstructive pulmonary disease and other disease developments. Ann Am Thorac Soc. 2016;13(Suppl 4):S280–S288. doi: 10.1513/AnnalsATS.201507-468KV. [DOI] [PubMed] [Google Scholar]
- 64.Duranton J, Bieth JG. Inhibition of proteinase 3 by [alpha]1-antitrypsin in vitro predicts very fast inhibition in vivo. Am J Respir Cell Mol Biol. 2003;29(1):57–61. doi: 10.1165/rcmb.2002-0258OC. [DOI] [PubMed] [Google Scholar]
- 65.Shao J, Yu M, Jiang L, Wu F, Liu X. Sequencing and bioinformatics analysis of the differentially expressed genes in herniated discs with or without calcification. Int J Mol Med. 2017;39(1):81–90. doi: 10.3892/ijmm.2016.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zigouris A, Alexiou GA, Batistatou A, Voulgaris S, Kyritsis AP. The role of matrix metalloproteinase 9 in intervertebral disc degeneration. J Clin Neurosci. 2011;18(10):1424–1425. doi: 10.1016/j.jocn.2011.01.036. [DOI] [PubMed] [Google Scholar]
- 67.Basaran R, Senol M, Ozkanli S, Efendioglu M, Kaner T. Correlation of matrix metalloproteinase (MMP)-1, -2, -3, and -9 expressions with demographic and radiological features in primary lumbar intervertebral disc disease. J Clin Neurosci. 2017;41:46–49. doi: 10.1016/j.jocn.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 68.Malapeira J, Esselens C, Bech-Serra JJ, Canals F, Arribas J. ADAM17 (TACE) regulates TGFbeta signaling through the cleavage of vasorin. Oncogene. 2011;30(16):1912–1922. doi: 10.1038/onc.2010.565. [DOI] [PubMed] [Google Scholar]
- 69.Rajasekaran S, Tangavel C, K SS, Soundararajan DCR, Nayagam SM, Matchado MS, et al. Inflammaging determines health and disease in lumbar discs-evidence from differing proteomic signatures of healthy, aging, and degenerating discs. Spine J 2020;20(1):48–59. [DOI] [PubMed]
- 70.Carrero R, Cerrada I, Lledó E, Dopazo J, García-García F, Rubio MP, et al. IL1β induces mesenchymal stem cells migration and leucocyte chemotaxis through NF-κB. Stem Cell Rev Rep. 2012;8(3):905–916. doi: 10.1007/s12015-012-9364-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Linero I, Chaparro O. Paracrine effect of mesenchymal stem cells derived from human adipose tissue in bone regeneration. Plos One. 2014;9(9):e107001. doi: 10.1371/journal.pone.0107001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miranda JP, Camoes SP, Gaspar MM, Rodrigues JS, Carvalheiro M, Barcia RN, et al. The Secretome derived from 3D-cultured umbilical cord tissue MSCs counteracts manifestations typifying rheumatoid arthritis. Front Immunol. 2019;10:18. doi: 10.3389/fimmu.2019.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang YK. Aging of mesenchymal stem cells: implication in regenerative medicine. Regen Ther. 2018;9:120–122. doi: 10.1016/j.reth.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–DD50. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figure 1. Details of cell and tissue samples used for the experiments. (A) MSCs from twelve different donors were used. All MSCs were derived from vertebral bone marrow aspirates. Only donors younger than 50 years (age at isolation) were selected, representing four different age groups (average age 17, 26.33, 37.66 and 48.66 years). Gender was equally balanced (6 male; 6 female) and symmetrically distributed among age groups. (B) IVD conditioned medium donor overview. For MSC stimulation, IVD conditioned medium from different donors within one condition was pooled (n = 4/group).
Additional file 2: Supplementary Figure 2. Effect of IVD conditioned medium treatment on DNA content, metabolic activity and lactate dehydrogenase (LDH) release of MSCs. (A) DNA content of MSCs in 6-well plate normalized to timepoint zero after 14 h of cell attachment. *p < 0.05, ***p < 0.001 (Kruskal-Wallis test). (B) Metabolic activity was measured with CellTiter-Blue. Data was standardized to the treatment condition baseline within every MSC donor. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; One-way ANOVA. (C) LDH was measured in the MSC secretome to detect cytotoxic reactions. No significant differences were found (Kruskal-Wallis-test). (D-H) Images were taken just before secretome collection. Scale bar = 500 μm.
Additional file 3 : Supplementary Table 1. MSC secretome following healthy CM stimulation. Supplementary Table 2. MSC secretome following traumatic CM stimulation. Supplementary Table 3. MSC secretome following degenerative CM stimulation. Supplementary Table 4. MSC secretome following IL-1β stimulation.
Additional file 4 : Supplementary Table 5 Concentrations of cytokines and chemokines in pooled conditioned media from healthy, traumatic and degenerative intervertebral disc, measured by immunoassay technique (mean+/-sd of technical replicates; pg/mL).
Data Availability Statement
Proteomics data are reported in the supplementary tables of the manuscript. All original data are available from the authors on request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD021281 [74].