Abstract
The novel betacoronavirus, SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), has spread across the globe at an unprecedented rate since its first emergence in Wuhan City, China in December 2019. Scientific communities around the world have been rigorously working to develop a potent vaccine to combat COVID-19 (coronavirus disease 2019), employing conventional and novel vaccine strategies. Gene-based vaccine platforms based on viral vectors, DNA, and RNA, have shown promising results encompassing both humoral and cell-mediated immune responses in previous studies, supporting their implementation for COVID-19 vaccine development. In fact, the U.S. Food and Drug Administration (FDA) recently authorized the emergency use of two RNA-based COVID-19 vaccines. We review current gene-based vaccine candidates proceeding through clinical trials, including their antigenic targets, delivery vehicles, and route of administration. Important features of previous gene-based vaccine developments against other infectious diseases are discussed in guiding the design and development of effective vaccines against COVID-19 and future derivatives.
Keywords: Coronavirus, COVID-19, SARS-CoV-2, Vaccines, DNA vaccines, RNA vaccines, Replicative viral vaccines, Non-replicative viral vaccines
Abbreviations: a.a, amino acid; AAV, adeno-associated virus; ACE2, angiotensin converting enzyme 2; ADE, antibody dependent enhancement; BGH, bovine growth hormone; ChAd, chimpanzee adenovirus; COVID-19, coronavirus disease 2019; CMV, cytomegalovirus; DPP4, dipeptidyl peptidase 4; E, envelope; EP, electroporation; GBV, gene-based vaccine; GP, glycoprotein; GC, guanine-cytosine; HA, hemagglutinin; HAI, hemagglutinin inhibition; IFN, interferon; IIV, inactivated influenza virus vaccine; IL, interleukin; LNP, lipid nanoparticle; Lunar, lipid-enabled and unlocked nucleomonomer agent modified RNA; M, membrane; MIV, monovalent inactivated virus; MERS-CoV, Middle East respiratory syndrome coronavirus; MV, measles virus; MVA, modified vaccinia virus Ankara; N, nucleocapsid; OGN, oligodeoxynucleotide; ORF, open reading frame; polyA, polyadenylation; RSV, respiratory syncytial virus; saRNA, self-amplifying RNA; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome 2; SLE, systemic lupus erythematosus; S, spike; RBD, receptor binding domain; Th, T helper; TLR, toll-like receptor; TNF, tumor necrosis factor; tPA, tissue plasminogen activator; VSV, vesicular stomatitis virus; WNV, West Nile virus
Graphical abstract
1. Introduction
The 2019 novel coronavirus outbreak (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) coronavirus, was declared a global pandemic on March 11, 2020 by the World Health Organization (WHO) and has since imparted significant morbidity and fatalities as governing bodies scramble to mitigate looming economic fallout. As of December 2020, this highly transmissible virus has already infected over 65 million worldwide, resulting in more than 1.5 million deaths [1]. Born out of mutation, SARS-CoV-2 will continue to drift as a response to evolutionary pressures, and as the outbreak continues, it will spread and kill indiscriminately. The need for protective solutions and, in particular, a safe and effective vaccine is of highest priority and paramount urgency.
As a member of the Coronavirdae family, SARS-CoV-2 is classified as a betacoronavirus which is characterized by having a single-stranded, positive-sense, enveloped RNA genome varying from 26.5 to 31.7 kb in size [2]. The virus is one of six human transmissible coronaviruses discovered to date [3]. COVID-19 presents similar features and symptoms to previous outbreaks of related betacoronaviruses: severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), which emerged in 2002 and 2012, respectively [4,5]. Although these viruses share considerable sequence similarity, SARS-CoV-2 has thus far demonstrated higher infection rates, longer incubation periods, and increased levels of asymptomatic transmission [5].
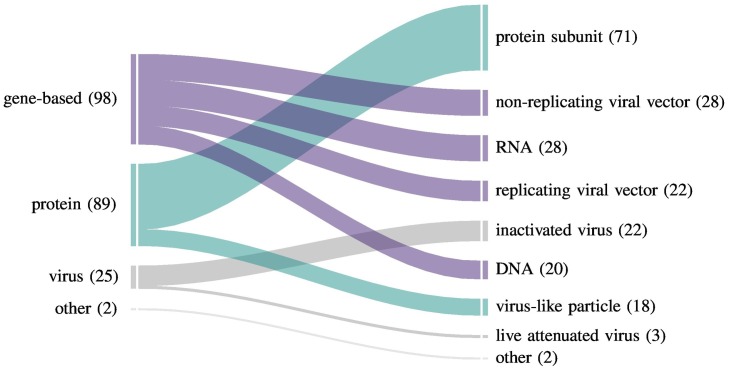
As of December 2020, WHO reports 215 COVID-19 vaccine candidates in development [6]. Of these, 52 are undergoing clinical evaluation and 163 are still in preclinical development. These candidates encompass a diverse selection of vaccine platforms: protein-based (subunit and virus-like particle), virus-based (live attenuated and inactivated), and novel gene delivery strategies such as nucleic acid (DNA and RNA), and viral vector (replicative and non-replicative) (Fig. 1 ). Conventional strategies, such as inactivated viral, live attenuated viral, and protein subunit vaccines have demonstrated successful outcomes in the past, but tend to confer complications with safety, limited cross-protection and immunogenicity [7]. Novel vaccine platforms include virus-like particles, viral vectors, and nucleic acid vaccines [8]. Gene-based vaccines (GBVs), including both viral vectors and nucleic acid vaccines, genetically encode an antigen to be delivered. They depend on the successful expression of delivered gene cassettes to peptide antigen(s), which must then be presented to immune cells to stimulate an immune response. Viral vector vaccines have previously been approved by health authorities [9]. The first two RNA-based vaccines against COVID-19 were recently approved by the U.S. FDA for emergency use [10,11,12]. This renders DNA-based vaccines as the only completely untested therapeutic vaccine approach for human application [13]. Heightened interest in the development of GBVs is attributed to some significant advantages over conventional vaccine platforms: greater safety and stability, potent cell-mediated protective immunity, specificity, ease of manipulation, low production costs, and simpler and more rapid development [9,14,15,16,17].
Fig. 1.
Overview of current COVID-19 vaccine development efforts. For each vaccine platform, the number of corresponding vaccine candidates are identified in parentheses. While protein subunit vaccines are the single most common vaccine candidate, gene-based vaccine development efforts overall outnumber all other platforms. Vaccine candidate numbers are based on the WHO Draft landscape of COVID-19 Candidate Vaccines [6].
In this review, we examine the development of COVID-19 GBV candidates under clinical evaluation. We highlight important facets of COVID-19 GBV design that should be considered including antigen selection, vaccine platform and route of administration. We examine the immunogenicity and safety profiles of earlier GBVs to see how this knowledge has been applied to the rapid development of current COVID-19 GBV candidates. Finally, we address potential safety challenges regarding COVID-19 vaccination and briefly comment on possible solutions.
2. COVID-19 vaccine targets
Predicting potent immunogenic targets is an essential step in the development of an effective and safe COVID-19 vaccine. Previous research on protective immune responses toward SARS-CoV and MERS-CoV can serve to guide COVID-19 development efforts, due to the similarity between these strains and overlapping etiology [18]. This section will outline the general process and importance of optimal antigen selection for vaccine development, in addition to identifying specific SARS-CoV-2 immunogenic targets. Sequence alignments of these targets will be compared to related coronavirus strains to estimate conservation levels.
2.1. Optimal antigen selection
GBVs encode specific SARS-CoV-2 antigenic epitopes and/or proteins rather than the entire viral genome [19]. As such, antigen selection is essential in the application of this strategy as it determines the strength, type, and cross-reactivity of the immune response [20]. For a vaccine to stimulate antibody (Ab) responses, the target antigen must contain B cell receptor epitopes; similarly, cell-mediated responses require sequences that allow T-cell epitope formation [21].
Modern methods, including genome-based reverse vaccinology, have proven to be efficient for the selection of optimal antigenic targets [22]. This technique involves analyzing a pathogen genome sequence to identify relevant protein sequences that can be screened for protective immunity [23]. The sequence information can then be compared between pathogenic strains to identify conservation levels, enabling the development of highly specialized vaccines specific to one strain, or cross-reactive vaccines conferring protection against multiple derivatives [24]. The sequence regions that are conserved and have not changed significantly over time are generally targeted by broadly neutralizing antibodies (NAbs), which have the ability to neutralize infecting viruses [25]. Therefore, using highly conserved regions could promote the development of a universal vaccine to protect against wild type or mutated betacoronavirus strains [25,26].
2.2. COVID-19 immunogenic targets
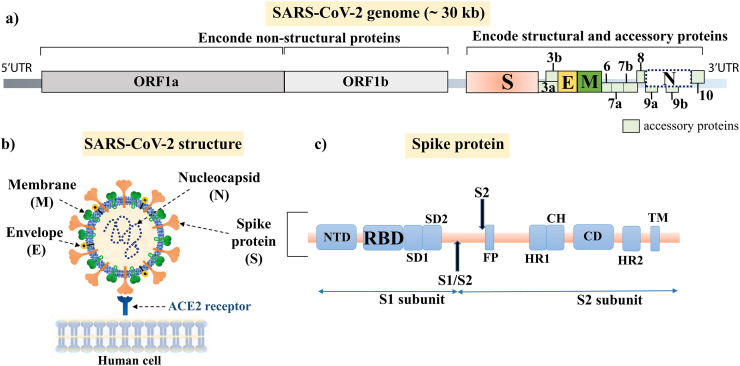
The genome of SARS-CoV-2 varies from 29.8 to 29.9 kb and contains 14 open reading frames (ORFs) encoding 27 proteins [27,28]. The major OFR1a and ORF1b encode non-structural proteins that form the replicase-transcriptase complex [29]. The four essential structural proteins encoded are: the spike (S), membrane (M), envelope (E) and nucleocapsid (N) proteins (Fig. 2a). The S protein facilitates viral uptake into host cells by binding to the host receptor, angiotensin-converting enzyme 2 (ACE2), found in the lower respiratory tract of humans (Fig. 2b) [30,31]. Specifically, it is the receptor binding domain (RBD), contained within the S1 subunit of the S protein that mediates cell attachment, while the S2 subunit regulates fusion of viral and host cell membranes (Fig. 2c) [32]. Interestingly, the RBD of SARS-CoV-2 has been shown to bind to the ACE2 target receptor with considerably higher affinity than that seen by SARS-CoV [32,33]. The M protein regulates the shape of the viral envelope and interacts with other structural proteins [30,31,34,35]. The small E protein is involved in viral maturation, assembly, and budding [34]. The N protein binds to the SARS-CoV-2 genome and is predominantly involved in genome-related processes [30,31,35].
Fig. 2.
SARS-CoV-2 genome and schematic representation of the structural proteins. a) The open reading frames (ORFs) that encode for the replicase polyprotein (ORF1a and 1b), structural and accessory proteins are indicated in boxes. b) Schematic representation of the four major structural proteins Spike, Envelope, Membrane and Nucleocapsid produced to assemble progeny virions. c) The S1 and S2 subunits of the Spike protein are indicated in blue boxes. The Receptor Binding Domain (RBD) which directly binds to the angiotensin converting enzyme 2 (ACE2) receptor of the host cell is highlighted.
The dynamic nature of the coronavirus perfusion form of the S protein exposes the RBD, making it an accessible and optimal target for NAbs [36,37]. In past studies, the immunogenicity of the S protein was demonstrated by the induction of NAbs and protective T-cell responses in animals and humans, in addition to protection against SARS-CoV challenge in mice [38,39,40,41,42]. Similarly, infection by MERS-CoV was found to confer increased neutralizing activities against the RBD of the S protein as opposed to other epitopes in humans and non-human primates [43]. Moreover, significant levels of Abs against the RBD were detected in recovered SARS-CoV patients [44]. Compared to the S protein, the N protein is highly conserved and less tolerant of mutation, but similarly confers strong immunogenicity as evidenced by induction of Abs, particularly immunoglobulin G (IgG) against the N protein of SARS-CoV [45,46,47,48]. The E protein is also immunogenic as demonstrated by induced CD4+ and CD8+ T-cell immune responses within peripheral blood mononuclear cells from SARS-recovered patients, following stimulation by E peptides [49]. The M protein has been shown to be an important stimulator of virus-specific humoral and cellular immune responses, and can induce high levels of NAbs [50,51]. In one study employing DNA vaccines encoding gene fragments of the SARS-CoV M gene, heightened levels of cellular immunity in mice were reported [52]. Although studies have found increased levels of immune responses elicited by the N, M and E proteins, the expression of these proteins from SARS-CoV in the absence of the S protein cannot confer adequate protection against viral challenge, as demonstrated in hamsters [41]. However, slightly elevated immune responses were observed when the S protein was coexpressed with M or E.
Overall, the S protein is a suitable antigenic target for GBVs as it has previously demonstrated potent and protective immunity in human and animal models. Moreover, considering its predominant role in facilitating viral uptake into host cells, vaccines stimulating the generation of NAbs against the S protein should be highly effective [53].
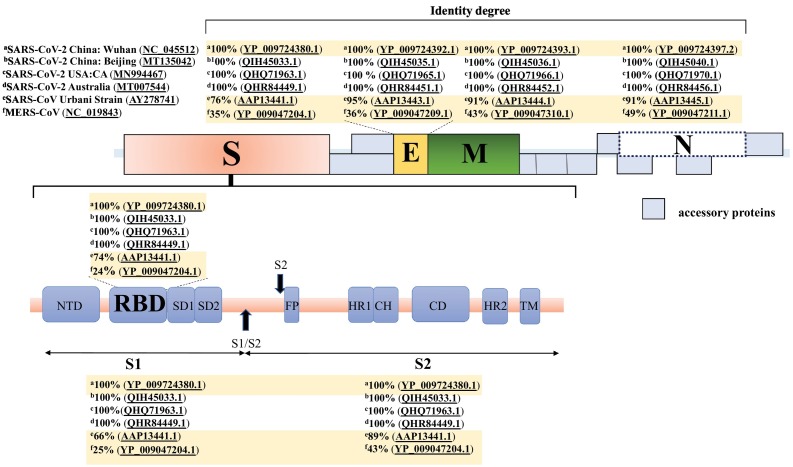
2.3. Amino acid sequence alignment of structural proteins from Coronavirus
We have aligned the amino acid (a.a.) sequences of the structural proteins S, M, N, and E from SARS-CoV-2 strains isolated from different regions of the world and related betacoronaviruses (Fig. 3 ). Firstly, 100% identity between strains isolated from Wuhan, Beijing, California, and Australia, indicate that any of these sequences could be used as the basis of vaccine design. The SARS-CoV-2 S protein a.a. sequence exhibited higher similarity and conservation levels with SARS-CoV (76%) compared to MERS-CoV (35%). The S protein a.a. sequence similarity between SARS-CoV-2 and SARS-CoV is lower than that of the E, M, and N proteins. However, SARS-CoV S protein NAbs demonstrated cross-reactivity evidenced by successfully preventing SARS-CoV-2 S-mediated entry into host cells [54], again suggesting that the S protein is an ideal vaccine target.
Fig. 3.
Protein identity comparison between SARS-CoV-2 strains from different countries and related betacoronavirus strains. The SARS-CoV-2 isolates demonstrate 100% identity across all regions, whereas the SARS-CoV and MERS-CoV strains show some variance. Amino acid sequences for all alignments were obtained from NCBI GenBank. Percent identities were obtained using BLASTp and are listed above or below their corresponding regions: spike (S), membrane (M), nucleocapsid (N), envelope (E), receptor binding domain (RBD), S1 and S2.
In addition, the RBD, S1 and S2 a.a. sequences from the S protein were aligned. A lower sequence similarity between the S1 and RBD regions of SARS-CoV-2, SARS-CoV and MERS-CoV were found, suggesting these regions and their corresponding Abs are subtype-specific. Moreover, it has been demonstrated that the S1 domain and RBD revealed low cross-reactivity between SARS-CoV-2 and other human coronaviruses [55]. In contrast, the S2 domain Abs are more cross-reactive [55], which corresponds with the increased sequence similarity between the different betacoronavirus strains (Fig. 3). In general, if generating a strain-specific COVID-19 GBVs, the S1 and RBD domains should be utilized, whereas the S2 domain is more suitable for universal coronavirus vaccine development.
3. Novel gene-based vaccine platforms
Upon selection of a SARS-CoV-2 antigenic target, its nucleic acid sequence, encoded as DNA or RNA, is used as a potential GBV. GBVs encompass DNA, RNA, and viral vector platforms, which each confer specific advantages and disadvantages. Here, we briefly identify their associated benefits and limitations, intracellular uptake, and storage stability.
3.1. Viral vector vaccines
A viral vector vaccine will consist of a recombinant virus, often attenuated, engineered to encode an antigen sequence for delivery into host cells for the endogenous production of high levels of that antigen. As a result, high levels of humoral and cellular immune responses are induced [56]. Viral vector vaccines confer high gene transduction capabilities due to the natural ability of viruses to infect host cells [56,57]. These vaccines do not require adjuvants due to the presence of viral components that stimulate the innate immune system 57]. Viral vector vaccines can either be replicating or non-replicating [9]. After infection of host cells, the replicating viral vector generates infectious progeny permitting the antigenic sequence to be amplified and produced in each replication cycle [58]. Common replicative viral vectors include measles virus (MV), adenovirus (Ad), and vesicular stomatitis virus (VSV) [59]. Although this platform confers high levels of immunogenicity, potential reversion to virulence remains a safety concern [60]. In comparison, non-replicative viral vectors are unable to produce infectious progeny, since all genetic material for replication is removed, ensuring a preferable safety profile [61]. However, due to replication deficiency, higher doses are needed to confer immunity, which can result in undesirable immune reactions to the vector itself. The most common non-replicative viral vectors include Ads (eg. human adenovirus serotype 5 (Ad5) and serotype 26 (Ad26), adeno-associated virus (AAV), alphavirus and modified vaccinia virus Ankara (MVA) [61]. Among them, Ad viral vectors have been extensively studied for their application as gene delivery vectors. They are non-enveloped, linear, double-stranded DNA viruses with a transgene packaging capacity up to 8 kbps. Most human cells can be easily infected with Ad vectors since they express cell surface Ad receptors (involved in host cell attachment) and integrin entry receptors (for endocytosis and viral entry). So far, they are the most common viral vectors used in clinical trials for vaccine, gene therapy and oncology applications [62,63]. Several non-human Ads, such as chimpanzee (e.g. ChAd3, ChAd6, ChAd63), are also undergoing testing in human and animal models [59,64,65].
Storage conditions for viral vector vaccines are highly dependent on the particular characteristics of each recombinant virus, which will determine the vaccine's stability, and consequently, its shelf-life [66]. Some viruses including Ads, are more sensitive to thermal degradation, causing a loss of infectivity [66,67]. Exposure to freeze-thaw cycles and free radical oxidation are other factors that can promote viral instability and degradation [68,69]. Thus, determining optimal storage conditions for viral-based vaccine distribution is crucial to maintaining pharmaceutical activity in the long-term [70,71,72].
Generally, licensed viral vector vaccines are stored at approximately 2–8 °C or at low freezing temperatures (−80 to −55 °C) with a shelf-life of over a year [66]. Promisingly, liquid formulations containing sucrose, polysorbate-80, and free radical oxidation inhibitors have been applied to an Ad vector vaccine which demonstrated minimal loss of Ad virus infectivity and sustained stability for 24 months at 4 °C [68]. A dehydration strategy known as lyophilization may enhance viral vector vaccine stability and eliminate the requirement of storage at freezing temperatures [66,72]. Protectants such as polyethylene glycol (PEG) and dextran applied during lyophilization can protect the virus from damage and help maintain viral activity [72,73]. Lyophilization in sucrose has demonstrated minimal loss of viral titer over a 1 year period when stored at 4 °C [74].
3.2. Nucleic acid vaccines
Nucleic acid vaccines represent a completely novel approach to imparting protective immunity, but DNA and RNA vectors are emerging as highly promising strategies. As non-viral vectors, these vaccines are thought to confer greater safety than their viral counterparts [60]. Although considered high-risk untested technologies [9,12], nucleic acid-based vaccines can have shorter development cycles, enabling quick deployment during pandemics.
The use of recombinant DNA vaccines requires successful transfer of the DNA vector into cell nuclei, transcription into messenger RNA (mRNA), and finally translation into the antigen of interest [75]. “Naked” or purified plasmid DNA is a highly attractive vehicle for antigen presentation as it is very simple to manipulate and inexpensive to generate [76]. A plasmid DNA vector typically consists of fundamental genetic components including a transcriptional promoter, RNA processing elements (polyadenylation (poly A) tail), and the gene encoding the antigen [77]. Plasmid DNA is an attractive biopharmaceutical as it can be amplified in large quantities in inexpensive prokaryotic hosts, although it must be purified [78,79]. The predominant challenge of utilizing DNA vaccines is that they generally impart low immunogenic responses in humans and larger animals compared to small animal systems [80,81].
An RNA vaccine already consists of an mRNA molecule which encodes the selected antigen, removing the need for transcription [82]. Upon delivery into a human cell, the immunogen sequence only requires translation to produce the antigen of interest. An RNA vaccine includes the mRNA transcript encoding the gene of interest surrounded by 5′ and 3′ untranslated regions and a polyA tail [83]. Some RNA vaccines can be self-amplifying (saRNA); the RNA molecule can direct its own replication and translation within the host upon delivery. As a result, more immunogen can be expressed [84].
However, there are concerns regarding the instability of delivering naked RNA, in addition to the size of the delivered molecules [83,85]. The instability of mRNA is prominently due to the ubiquitous presence of ribonucleases that actively degrade RNA [85,86]. The addition of a 5′ cap (7-methylguanosine cap) and 3′ polyA tail are essential in maintaining the stability and translation of mRNA within the cytosol [82,87]. Additionally, to protect from degradation, polymer and lipid formulations have been used [88]. Lipid nanoparticles (LNPs) are composed of ionizable cationic lipids, phospholipids, cholesterol and PEG that enable the assembly and formation of a stable lipid bilayer around the mRNA molecule [86]. Previous studies have shown sufficient protection of mRNA by effectively encapsulating it [89,90,91].
Lipo- or polyplexed DNA and RNA uptake is thought to occur primarily by endocytosis [92]. Uptake through other mechanisms such as pinocytosis and phagocytosis are more restricted [93]. LNPs enter cells through one of many endocytic pathways after interaction between the cell surface and the particle. Addition of targeting ligands to nanoparticles may therefore improve uptake through receptor-mediated endocytosis. Endocytosed DNA nanoparticles have been confirmed within the endosomal compartment through electron microscopy [94,92] and confocal microscopy [92]. Additionally siRNA delivered via LNPs have also been confirmed within the endosomal compartment through confocal microscopy [95,96]. Escape from the endosomal compartment into the cytosol is of paramount importance for subsequent gene expression [97]. Many nanoparticles have exhibited endosomal escape [98], although the mechanisms by which they do so remain unknown. A common hypothesis is the proton sponge effect [99], whereby the polymer buffers the low pH within an endosome. This leads to excessive pumping of ions into the endosome to compensate. Eventually, the endosome lyses from buildup of osmotic pressure, releasing its captive nanoparticles into the cytosol [99].
Methods to enhance vaccine uptake and expression have been more thoroughly investigated for DNA vaccines compared to RNA, as DNA needs to bypass two cellular membranes to arrive at the nucleus, while RNA only needs to bypass one to enter the cytoplasm [9]. DNA transfection is more effective in actively dividing cells, where the nuclear membrane has broken down, compared to quiescent cells [100]. The inclusion of a nuclear localization signal [101] can facilitate active transport across the nuclear membrane, enabling gene expression in quiescent cells.
As opposed to protein pharmaceuticals, tertiary structure is not generally important for DNA or RNA function, simplifying their storage requirements. When storing nucleic acids, the key is maintenance of the molecule's chemical integrity [69,71,102,103].
DNA itself appears to be robustly stable. Over a seven year period, supercoiled plasmid DNA was found to be stably maintained and demonstrated no difference to freshly prepared DNA when stored in 0.9% sodium chloride at −20 °C [104]. While short-term storage is possible in a refrigerator (2–8 °C) or room temperature (20–30 °C), DNA will eventually deteriorate within months (2–8 °C) or days (≥20 °C) [102,105,106]. Low temperature and pH are vital to preservation of DNA integrity in the long-term. In contrast, RNA products are very sensitive to temperature and should always be kept at extremely cold temperatures (−70 °C) during storage and distribution. The presence of ribonucleases can destroy the RNA vaccine product; therefore synthesis, purification, and storage of the product must be done in absence of enzymes to elongate its shelf-life [71]. Inexpensive excipients like PEG, trehalose, and sucrose have shown promising results in protecting RNA vaccines and increasing their stability and half-life from hours to days at ambient temperatures [69,71].
Additionally, DNA and RNA vaccines are likely formulated with cationic lipids or polymers and, as such, will have different storage conditions depending on the polymer. In general, lipoplexed DNA formulations appear to be stable at 4 °C for up to 3 months [107], which is still insufficient for widespread vaccine distribution. However, certain lipoplexed nucleic acid formulations have shown better stability, such as up to a year when frozen [108]. Lyophilization can also be employed for long-term stability [103] and disaccharide excipients may improve stabilization of lipoplexed nucleic acids [109,110]. Lyophilized polyplexed RNA particles were even stable at 40 °C with sugar excipients [110]. Clearly, choice of excipient and the formulation of the cationic carrier will greatly affect stability.
4. Gene-based vaccine administration
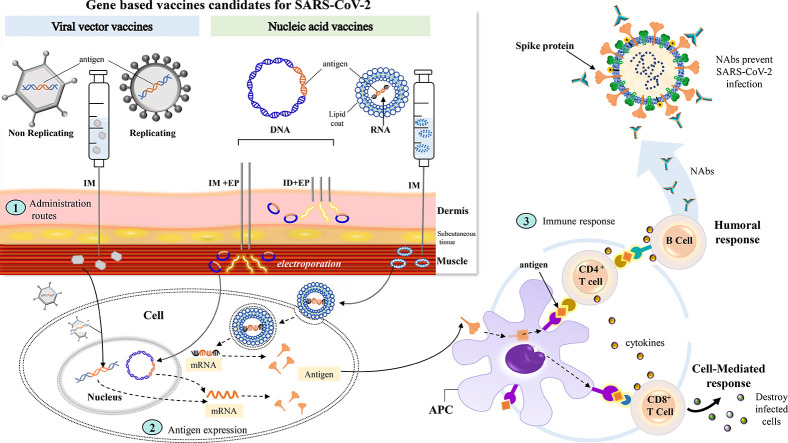
Methods and routes of administration can profoundly impact the aforementioned GBV platforms and are conventionally focused on parenteral and mucosal routes. Most commonly, parenteral routes include intramuscular (IM), intradermal (ID) and subcutaneous (SC); while common mucosal routes include oral and nasal, both of which are non-invasive [111,112]. Furthermore, administration using novel delivery tools have been investigated to increase the transfer, expression, and immunity of GBVs. Here, we examine different routes of administration and delivery tools from previous GBV developments and assess their applicability for COVID-19 vaccine development.
4.1. Route of administration
4.1.1. Mucosal vaccination
Since the vast majority of pathogens, including SARS-CoV-2, invade the host through mucosal sites [30,111], using this route of administration may be an effective way to control infection and prevent disease [113]. Both humoral and cellular immune responses are induced here, both systemically and locally. The immune response is characterized primarily by cytotoxic T lymphocyte and immunoglobulin A (IgA) responses [114]. Present across many mucosal sites, IgA may aid in the prevention of pathogenic entry into the body [115]. Varied levels of dendritic cells (DCs), which are professional antigen presenting cells (APCs), also inhabit mucosal sites, thereby stimulating antigen presentation to T-cells and modulating the immune response [116].
The expression of GBVs at mucosal sites requires penetration across the mucous and epithelial layers to access the underlying immune cells. Hence, vaccines carrying genetic material often require complexing with suitable delivery vehicles, such as micro- or nanoparticles [117,118]. Oral delivery of chitosan-encapsulated DNA vaccines appears effective, since chitosan confers a high affinity for DNA, contains mucoadhesive properties, and is able to reside within the intestinal mucosa for extended periods of time [119,120]. Ad vaccines administered via the oral route have demonstrated increased CD8+ T-cell and humoral responses, although CD4+ T-cell responses are generally not stimulated [121,122,123,124]. While oral administration of GBVs seems promising, studies are sparse and thorough investigation is still required.
Intranasal (IN) delivery is a suitable choice for respiratory-related diseases, although defense mechanisms of the nasal cavity presents a challenge for vaccine entry [125]. IN delivery of a polyethylenimine formulated DNA vaccine encoding hemagglutinin (HA) induced high levels of HA-specific Ig, but was absent when immunized with unformulated DNA [126]. IN naked, LNP-encapsulated, and polymer-based mRNA vaccination previously demonstrated enhanced humoral and cell-mediated immune responses [127,128,129]. LNP and polymer-formulated vaccines have been shown to access the nasal associated lymphoid tissues, an integral location for the induction of nasal immune responses [128,130,131]. Due to close proximity to the brain, there have been concerns that IN-delivered recombinant viral vector-based vaccines could spread to the central nervous system (CNS) via olfactory tissue, imparting neurological side-effects [132,133]. Gene transfer of recombinant Ads within the olfactory bulb has been observed, but in the absence of transfer levels within the CNS, spread into the CNS may not be a major safety concern for all viral vector vaccines [133,134]. Studies have shown that delivery of viral vector vaccines through the IN route has been associated with enhanced immunogenicity and high levels of safety [135,136,137,138].
4.1.2. Parenteral vaccination
Certain parenteral routes of administration are preferred for GBV delivery. Similar to mucosal sites, varied levels of DCs also inhabit parenteral sites, promoting varied immune response levels [112]. Administration through the ID route is highly effective as the skin contains enhanced levels of APCs that can serve as targets for the transfection of DNA and RNA vaccines [139,140,141]. An LNP-formulated nucleoside modified mRNA influenza H10 vaccine induced comparable protective titers in rhesus macaques when administered by the ID and IM route, although the ID route enhanced the speed of this response [142]. An enhanced immune response was found after three ID doses of a Lassa virus DNA vaccine when compared to IM in guinea pigs [143]. Additionally, a Phase I trial for a seasonal influenza DNA vaccine demonstrated that ID administration was more favourably immunogenic, but was also associated with greater risk of adverse events (AEs) [144]. In a clinical trial assessing the CUTHIVAC 001 HIV-1 DNA vaccine, the transcutaneous (TC) route was compared to the ID route. HIV-1 negative participants were dosed with DNA via IM injection supplemented with either ID injection or TC application. Distinct cytokine profiles were observed between each route. IM followed by ID (IM + ID) administration was found to be the least immunogenic, while IM + TC induced greater CD4+ and CD8+ T-cell responses, including higher levels of tumor necrosis factor (TNF)-α, interleukin (IL)-17a, and CD154 [145]. The TC route may improve the breadth of contact to the rich immune population in the skin compared to ID injection. TC administration of vaccines has been linked to greater cytotoxic T lymphocyte responses, although it remains limited by its invasiveness, laborious technique, and small deliverable volumes [146,147].
Studies employing viral vector vaccines have demonstrated similar immune responses between IM and ID routes [148], with heightened ID-mediated T-cell responses [149]. Although the dermis may be more enriched in APCs compared to the muscle [150,151], muscle tissue is able to efficiently recruit immune cells due to its extensive network of blood vessels and the onset of temporal local inflammation upon vaccine administration [9,88,152,153]. Additionally, local adverse reactions may be more severe with ID administration compared to IM, as demonstrated in studies employing DNA and Ad5-based vaccines [112,144,154]. This may partially explain why most viral vectors generate a more favourable response via the IM route.
The SC region is located within the adipose tissue layer beneath the dermis, where injection of GBVs primarily results in transfection of fibroblasts and keratinocytes [155]. Although the SC region harbours fewer APCs compared to the dermis [150,156,157], the loose adipose tissue contained here enables increased injection volumes [88,158], making it a beneficial route. Considering the instability of many mRNA vaccines, the SC route of administration may not be a suitable choice. The SC area contains a low level of blood vessels, limiting the absorption rate of vaccines and drugs, which may increase the chances of mRNA degradation prior to cell uptake [88,159,160].
Considering SARS-CoV-2 is transmitted via respiratory droplets and its site of infection is within the lower respiratory tract, mucosal administration may better target the virus, specifically through the IN route [30,161]. IN vaccination can induce mucosal responses such as Ab production within nasal- and bronchus-associated lymphoid tissue of the lower respiratory tract, that could be more effective against SARS-CoV-2 infection [111,162,163]. Overall, both mucosal and parenteral administration of GBVs have shown promising results. So far, many of the current COVID-19 vaccine candidates in ongoing trials are being administered parenterally, with just a few candidates using the mucosal route.
4.2. Novel delivery tools
Compared to viral vectors, in general, exogenous DNA and RNA are not readily taken up by cells as efficiently. Numerous physical delivery methods have been developed to enhance cellular uptake [164]. Physical gene transfection systems are delivery platforms that transport genetic material through mechanical processes, such as biojector and electroporation (EP) devices [165]. Biojector devices use CO2 pressure to deliver therapeutics via ID, IM and SC administration, which forces the vaccine into the skin, eliminating the requirement of a needle [166]. As it is needle-free, it confers several benefits over traditional needle injection, namely reduction in: side effects, needlestick injuries, and needle cross-contamination [112]. Two Zika virus DNA vaccines were recently investigated in a Phase I clinical trial demonstrating enhanced T-cell responses after needle-free administration compared to needle and syringe application [167]. Delivery of an mRNA vaccine against rabies using a biojector exhibited increased Ab responses compared to needle injection [168]. The enhanced vaccine efficacy via jet injection could be attributed to a wider distribution of the vaccine, consequently inducing better uptake by APCs [168,169].
Currently, IM or ID injection followed by EP are commonly used to deliver DNA vaccines in clinical trials [170]. EP involves the application of electrical pulses, generating pores in skin cells to enhance cell uptake of genetic material [171]. IM EP was first conducted by Aihara and Miyazaki in 1998 [172], and is able to permeabilize muscle cells to enhance the penetration and distribution of DNA vaccines [173,174,175]. Several reports have shown improved antigen expression and enhanced antigen-specific immune responses by in vivo EP [176,177]. The HIV DNA vaccine, ADVAX (Aaron Diamond AIDS Research Center, New York), demonstrated enhanced immunogenicity when delivered via EP [178], compared to IM injection [179]. Despite these advantages, EP has an added risk of cell death attributed to the application of high voltages [165].
Novel delivery strategies, like EP, are still under active investigation for RNA vaccines. The utility of EP for delivery of saRNA vaccines versus plasmid DNA vaccines was previously explored and noted comparable expression and immune responses in mice [180]. However, EP did not improve delivery of a conventional non-replicating RNA vaccine in an earlier study [14], likely limiting its utility to replicating RNA vectors.
In general, if DNA or RNA COVID-19 vaccines are designed to be administered parenterally, tools such as bioinjectors and EP may improve outcomes significantly, especially for DNA vaccines. These strategies are already part of many current DNA COVID-19 vaccine protocols, increasing the potential for enhanced expression and delivery of these vaccine candidates.
5. Immunity and safety of previous gene-based vaccines
GBVs remain novel pharmaceutical technologies. Knowledge gleaned from previous efforts evaluating GBV immunity and safety against other infectious diseases will direct the development of a vaccine against COVID-19. Table 1, Table 2, Table 3 summarize these clinical studies, discussed in detail below.
Table 1.
Immunity and safety of previous viral vector vaccines.
| Developer | Disease/vaccine | Vector/antigen | Delivery and dosage | Number of participants (age range) | Latest phase trial and description | Outcomes | Reference |
|---|---|---|---|---|---|---|---|
| Non-replicative viral vector | |||||||
| CanSino Biologics |
Ebola *AD5-ZEBOV |
Ad5 Antigen: GP of the 2014 Ebola strain |
IM 1.6 × 1011 vp, 8.0 × 1010 vp |
500 (18–50) |
Phase II (PACTR201509001259869) Two-dose levels. |
Safety: No vaccine-related SAEs. Immunogenicity: High humoral immune responses of GP-specific Abs peaked on day 28 and decreased about 85% 6 months after injection. T-cell immune responses were not measured. 8.0 × 1010 vp was the optimal dose. |
[184] |
| Gamaleya Research Institute |
Ebola *GamEvac- Combi |
Ad5 and VSV Antigen: GP of the Ebola virus |
IM 2.5 × 107 PFU |
84 (18–55) |
Phase I/II (No. 0373100043215000055) Heterologous prime-boost: VSV on day 1 and Ad5 on day 21. |
Safety: No vaccine-related SAEs. Immunogenicity: Seroconversion rate of 100%. NAbs, CD4+ and CD8+ T-cell responses detected in participants. |
[188] |
|
University of Oxford |
Influenza ChAdOx1 NP + M1/ MVA-NP + M1 |
ChAdOx1 and MVA Antigens: conserved influenza NP and M protein |
IM 5 × 108 vp, 5 × 109 vp, 2.5 × 1010 vp, 5 × 1010 vp (ChAdOx1)/ 1.5 × 108 PFUs (MVA) |
15 (18–50) |
Phase I (NCT01623518) Dose escalation. Heterologous prime-boost on day 7 or 14. |
Safety: At the highest dose two volunteers experienced severe local and systemic reactions. Immunogenicity: High levels of antigen specific T-cells and NAbs were found with the 2.5 × 1010 vp dose. The heterologous regimen increased the immune response. |
[202] |
| IM 2.5 × 1010 vp (ChAdOx1)/1.5 × 108 PFUs (MVA-NP + M1) |
49 (18–46) 24 (≥50) |
Phase I (NCT01818362) Heterologous two-dose schedule at an interval of 8 or 52 weeks. |
Safety: No vaccine-related SAEs. Immunogenicity: Better T-cell durability with the MVA/ChAdOx1 regimen. In young and older adults, an increase in antigen-specific IFN-γ+ and T-cell responses were observed. |
[203] | |||
| University of Oxford |
Middle East Respiratory Syndrome ChAdOx1 MERS |
ChAdOx1 Antigen: S surface GP |
IM 5 × 109 vp, 2.5 × 1010 vp, 5 × 1010 vp |
24 (18–50) |
Phase I (NC03399578) Dose escalation. |
Safety: The high dose group demonstrated an increased proportion of moderate and severe AEs. Immunogenicity: T-cell response peaked on day 14 and Abs on day 28. NAbs were present in 44% of the high dose group 18 days after vaccination. |
[204] |
| Johnson & Johnson |
Ebola **AD26.ZEBOV/ MVA-BN-Filo |
Ad26 and MV Antigens: Ebola virus GP and NP |
IM 5 × 1010 vp, 1 × 108 vp |
87 from UK, 72 from Africa, (18–50) |
Phase I (NCT02313077)(NCT02376400) Heterologous two-dose. Ad26.ZEBOV followed by MVA-BN-Filo or the reverse regimen, with intervals of 28 or 56 days. |
Safety: No vaccine-related SAEs. Immunogenicity: The two-dose regimen induced the highest elevation of Ebola GP-specific Abs at either 28 or 56 days. Phase I, II and III trials were completed (results not published). Another phase I, II and III trial is ongoing in multiple countries. |
[[197], [198], [199]] |
| Replicative viral vector | |||||||
| Themis Bioscience |
ChikungunyaFever MV-CHIK |
Measles virus Antigens: CHIK structural proteins |
IM 5 × 104 TCID (low dose), 5 × 105 (high dose) TCID |
263 (18–55) |
Phase II (NCT02861586) Prime–boost immunization schedule (1 and 6 months). |
Safety: SAEs related to arthritis were all unsolicited. Immunogenicity: Induction of higher concentrations of NAbs against CHIK with the 5 × 105 dose. The second vaccination induced high titers of NAbs. |
[209] |
This table only lists clinical trials discussed in the text. Ab: antibody; Ad5: adenovirus; AE: adverse event; ChAdOx1:chimpanzee adenovirus-vectored vaccine; GP: Glycoprotein; CHIK: chikungunya virus; IFN-y: interferon gamma; IM: intramuscular; M1: matrix protein 1; MV: Measles virus; MVA-BN-Filo: Modified Vaccinia Ankara-Bavarian Nordic; NAb: neutralizing antibody; NP: nucleoprotein; PFU: plaque forming unit; SAEs: serious adverse events; S: spike; TCID: tissue culture infectious dose; VSV: vesicular stomatitis virus; vp: viral particles; *Licensed in country of origin. ** Approved for medical use in the European Union.
Table 2.
Immunity and safety of previous DNA-based vaccines.
| Developer | Disease/vaccine | Vector/antigen | Delivery and dosage | Number of participants (age range) | Latest phase trial and description | Outcomes | Reference |
|---|---|---|---|---|---|---|---|
| Inovio Pharmaceuticals |
Middle East Respiratory Syndrome INO-4700 |
Plasmid Antigen: MERS-CoV S protein |
IM + EP with CELLECTRA 5P) 0.67 mg, 2 mg, 6 mg |
75 (18–50) |
Phase I (NCT02670187) Three-dose schedule at 0, 4 and 12 weeks. |
Safety: No vaccine-related SAEs. 27/75 patients reported infection and 6 were vaccine-related. Immunogenicity: Dose-independent immune responses in over 85% of participants after 2 doses. NAbs in 43% of participants and T-cell responses in 76%. |
[220] |
| Inovio Pharmaceuticals |
Ebola INO-4201 and INO-4202 |
Plasmid (SynCon DNA) Antigen: Ebola GP |
IM/ID + EP with CELLECTRA®. 2–4 mg |
240 (18–50) |
Phase I (NCT02464670) Three-dose schedule at 0, 4 and 12 weeks with or without a plasmid encoded human IL-12. |
Safety: No vaccine-related SAEs. Immunogenicity: ID delivery correlates with higher Ab titers, EBOV GP-specific T-cells producing TNF-α. Overall, T-cell responses ranged from 53.3%–93.3% across groups. |
[224] |
| NIAID |
Severe Acute Respiratory Syndrome VRC SARS |
Plasmid Antigen: SARS-CoV S protein |
IM (Biojector® 2000) 4 mg |
10 (21–49) |
Phase I (NCT0009946) Three-dose schedule on day 9, 28 and 56. |
Safety: No vaccine-related SAEs. Immunogenicity: NAbs and SARS-CoV-specific CD4+ T-cells detected in all participants. |
[39] |
| NIAID |
West Nile Virus VRC- WNVD |
Plasmid Antigens: prM and E proteins of the West Nile virus GP |
IM (Biojector® 2000) 4 mg |
30 (18–65) |
Phase I (NCT0030417) Three-dose schedule on day 0, 28 and 56. |
Safety: No vaccine-related SAEs. Immunogenicity: Induction of NAbs peaked at week 12. T-cell responses against WNV-E and WNV-M |
[227] |
| NIAID |
Influenza A, B VRC- FLUDNA063- 00-VP |
Plasmid 3 constructs. Antigens: influenza HA sequences |
IM/ID (Biojector® 2000) 1 mg or 4 mg DNA prime + IIV3 booster |
131 (18–70) 316 (18–70) 75 (6–17) |
Phase I (NCT01676402) (NCT01609998) (NCT01498718) DNA prime on day 0 + IIV3 booster at different schedules (14, 18, 36 or 44 weeks). |
Safety: No vaccine-related SAEs. Mild to moderate AEs. Immunogenicity: Generally high pre-existing anti-HA immunity. Higher anti-HA B titers in adults after DNA priming versus IIV3. Similar or superior anti-HA Ab titers to IIV3 control in juveniles. |
[144,235,234] |
| NIAID |
Influenza A (H1N1) VRC- FLUDNA057- 00-VP |
Plasmid Antigen: HA of A/California/04/2009(H1N1) |
IM (Biojector® 2000) 4 mg |
20 (24–70) |
Phase I (NCT00973895) Three dose schedule on day 0, 28 and 56. MIV boost 3–17 weeks after final DNA dose. |
Safety: No vaccine related SAEs. Four grade 1 AEs (migraine & erosion). Immunogenicity: Increased NAb responses after MIV booster but did not influence CD4+ T cell responses. |
[236] |
| Imperial College London |
Human Immunodeficiency Virus GTU®- MultiHIV B |
Plasmid Antigens: fusion of Rev, Nef, Tat, p17, p24 and CTL regions of the HAN2 HIV-1 B clade |
IM + EP via Trigid Ichor device IM + TC IM + ID IM/EP: 4 mg ID/TC: 0.4 mg |
30 (18–45) |
Phase I (NCT02075983) Three groups each vaccinated at 0, 4 and 12 weeks. |
Safety: No vaccine-related SAEs. Immunogenicity: IFN-ɣ responses greatest after IM + EP (9/10 participants) against Nef and Gag. CD4+ & CD8+ T-cell responses tended to be greater after IM + TC and IM + EP. Higher levels of TNF-α, IL-17a and CD154 observed after IM + TC. |
[145] |
This table only lists clinical trials discussed in the text. Ab: antibody; AE: adverse event; EP: electroporation; E: envelope; GP: glycoprotein; HA: hemagglutinin; IFN-y: interferon gamma; IL-12: interleukin-12; IM: Intramuscular; ID: Intradermal; IIV3: trivalent inactivated influenza virus; MIV: Monovalent inactivated virus; mg: milligram; NAbs: neutralizing antibodies; NIAID: National Institute of Allergy and Infectious Diseases; prM: protein premembrane; S: spike; SAE: serious adverse event; TC: transcutaneous; TNF-a: tumor necrosis factor-ɑ.
Table 3.
Immunity and safety of previous RNA-based vaccines.
| Developer |
Disease/ vaccine |
Vector/antigen | Delivery and dosage | Number of participants (age range) | Latest phase trial and description | Outcomes | Reference |
|---|---|---|---|---|---|---|---|
| CureVac AG |
Rabies virus CV7201 RNActive® |
mRNA Antigen: Rabies virus GP |
IM, ID 80–640 μg needle syringe, needle-free device |
101 (18–40) |
Phase I (NCT02241135) Dose escalation. Booster dose after 1 year. Different route of administration. |
Safety: One SAE that was possibly vaccine-related. Immunogenicity: Induction of NAb responses when applied needle-free. No cell-mediated immune responses detected. |
[168] |
| Moderna Therapeutics | Zika virus mRNA-1325 |
mRNA Antigens: variant Zika virus structural genes |
IM 10 μg, 25 μg, 100 μg |
90 (18–49) |
Phase I (NCT03014089) Escalating dose levels. Two-dose schedule (28 days apart). |
Studies completed, but not published. Next-generation vaccine mRNA-189 (NCT04064905) will replace the mRNA-1325 vaccine, which will not be further developed. | – |
| Moderna Therapeutics | Influenza virus VAL-506440 (mRNA-1440) |
LNP-mRNA Antigen: HA-GP from the H10N8 influenza strain |
IM 25 μg, 50 μg, 75 μg, 100 μg, 400 μg ID 25 μg, 50 μg |
201 (18–64) |
Phase I (NCT03076385) Dose escalation. Two-dose schedule on day 1 and 21. |
Safety: Dose-dependent AEs. Two participants experienced grade 3 solicited AEs. Immunogenicity: Induced robust humoral immune responses and high seroconversion rates. |
[248] |
| Moderna Therapeutics |
Influenza virus VAL-339851 (mRNA-1851) |
LNP-mRNA Antigen: HA-GP from the H7N9 influenza strain |
IM 10 μg, 25 μg, 50 μg |
156 (18–49) |
Phase I (NCT03345043) Dose escalation. Two-dose schedule on day 1 and 22. Booster dose at 6 months (25 and 50 μg). |
Safety: Three participants in the high dose group demonstrated severe injection site pain after the second vaccination. Immunogenicity: Induced humoral immune responses and high seroconversion rates. Persistent HAI. |
[248] |
| Moderna Therapeutics |
Chikungunya mRNA-1944 |
LNP-mRNA Antigen: IgG Ab with activity against CHIK |
IV 0.1 mg/kg, 0.3 mg/kg, 0.6 mg/kg, 1 mg/kg |
22 (18–50) |
Phase I (NCT03829384) Dose escalation. |
Ongoing | – |
| Moderna Therapeutics | Chikungunya VAL-181388 (mRNA-1388) |
mRNA Antigens: chikungunya structural C and E proteins |
IM 25 μg, 50 μg, 100 μg |
60 (18–49) |
Phase I (NCT03325075) Dose escalation. Two-dose schedule on day 0 and 28. |
Safety: Well-tolerated at all dose levels. Immunogenicity: NAb titers increased significantly. A boost after the second vaccination was observed. |
[247] |
This table only lists clinical trials discussed in the text. Ab: antibody; AE: adverse event; CHIK: chikungunya virus; GP: glycoprotein; HAI: hemagglutinin inhibition; ID, Intradermal; IM, Intramuscular; LNP: lipid nanoparticle; μg: microgram; mg: milligram; NAb: neutralizing antibodies; SAE: serious adverse event; IV: intravenous.
5.1. Previous viral vector vaccine immunity and safety
5.1.1. Non-replicative viral vector vaccines
The most frequently used Ad vector is Ad5 since it is able to induce potent CD8+ T-cell and Ab responses [181,182]. After the Ebola epidemic in 2014, the Ad5-EBOV vaccine (CanSino Biologics, China) based on the Ad5 vector expressing the Ebola envelope glycoprotein (GP), was developed (Table 1) [57,183]. The results of the Phase I and II trials proved Ad5-EBOV to be safe and highly immunogenic [183,184], considering a single dose induced the production of GP-specific Abs in 14 days in 85% of the participants, regardless of pre-existing immunity against the Ad5 vector. The vaccine conferred substantial protection, at least in the short term, raising the need for prime-boost immunization [184]. Phase III trials were not conducted as the Ebola epidemic ended in 2016. In 2017, China's food and drug authority approved the Ad5-EBOV vaccine [185].
Pre-existing immunity against the Ad5 vector produces NAbs, T-cells and type I IFN activated natural killer cells that inactivate the viral vector, reducing the efficacy of Ad5-based vaccines [56,186]. In order to circumvent this problem, heterologous prime-boost regimens are a potential solution [187].
The Ad5-vectored heterologous Ebola vaccine, GamEvac-Combi (Gamaleya Research Institute, Russia), was licensed in Russia in 2015 for emergency use. The vaccine regimen during the Phase I/II trials consisted of a priming immunization with a VSV vaccine followed by a boosting immunization with Ad5 21 days later (Table 1) [188]. The design was supported by multiple animal models and clinical trials showing that heterologous prime-boost immunization generally elicits superior immunogenicity in comparison to repeated doses of the same vaccine [112]. The results indicated that the vaccine has a good safety profile and elicited Ab responses in 100% of participants. A significant increase in anti–GP Ab titers was observed 28 days after the prime vaccination, and even higher titers on day 42. CD4+ and CD8+ T-cell responses were found in 82.8% and 58.6% of participants, respectively. A stronger Ab response was found with the heterologous VSV-prime Ad5-boost immunization strategy. The authors suggest that heterologous vaccination may compensate for the negative implications of pre-existing immunity to Ad5 [188]. Indeed, a large proportion of adults worldwide have already acquired such immunity, particularly among African populations [189].
Although the Ad vector confers a better safety profile compared with replicative viral vectors, there are some concerns about its use in clinical trials [56]. For example, an unexpected effect of pre-existing NAbs to the Ad vector was observed in a Phase II trial, where a vaccine candidate against human immunodeficiency virus (HIV) 1 showed increased HIV-1 acquisition risk in individuals with higher titers of pre-existing Ad5-NAbs [190]. In addition, predominant sequestration of Ad vaccines within the liver has been shown to induce hepatotoxic-associated adverse effects [191,192].
Avoiding pre-existing anti-Ad5 immunity can be addressed by the selection of rarer serotypes of Ad, such as Ad26, which has low prevalence in humans and induces enhanced memory conversion and more functional CD8+ T-cell responses compared to Ad5 [193,194,195]. Clinical trial results have shown that an Ad26 vector-based HIV vaccine was immunogenic and well-tolerated in healthy adults [195,196]. The AdVac® platform (Janssen, Belgium), serving as the template vector for the Ad26CoV2-S COVID-19 vaccine, was used previously to develop the Ad26.ZEBOV vaccine against Ebola (Table 1). During Phase I trials, the vaccine was tested alone or in a heterologous prime-boost regimen with the MVA vector vaccine (MVA-BN-Filo) demonstrating a favourable safety profile. A detectable Ebola GP-specific IgG response was observed 28 days after primary immunization with Ad26.ZEBOV in more than 90% of participants. Boosting by MVA-BN-Filo resulted in sustained elevation of specific IgG detectable 21 days post boost. EBOV-specific IgG responses were maintained at day 360 [197,198,199]. The two-dose regimen was approved for medical use in the European Union in July 2020.
ChAdOx1 is a replication-defective E1/E3 chimpanzee Ad-vector (University of Oxford, UK) developed to circumvent the issue of pre-existing anti-human Ad immunity [200]. Ads derived from chimpanzees have a low seroprevalence rate in most human populations [201]. ChAdOx1 is a promising vector that could be used to deliver vaccine antigens where strong cellular immune responses are required for protection. The first clinical use of the ChAdOx1 vector was in combination with the MVA poxvirus vector to develop a vaccine against influenza (Table 1). To date, two Phase I trials have been performed, both demonstrating good safety and T-cell immunogenicity profiles. In the first trial, different doses of the ChAdOx1 NP + M1 vaccine were evaluated. Encouraging increases in antigen-specific T-cell responses were observed in groups receiving up to a 2.5 × 1010 virus particle dose [202]. The same dose was used in the second Phase I trial where the heterologous MVA/ChAdOx1 regimen was highly immunogenic. In young adults, antigen-specific IFN-γ+ and T-cell responses increased and were maintained for up to 18 months after the first MVA or ChAdOx1 vaccination. In comparison, in older adults, the increase was observed 8 weeks after the boost with MVA and it was maintained up to 6 months after the first vaccination [203]. ChADOx1 has also been used to develop a MERS vaccine, which was tested in three doses in a Phase I trial (Table 1) with a significant increase in T-cell and IgG responses to the MERS-CoV S antigen observed with each dosing format. A single dose was able to elicit NAbs against live MERS-CoV in 44% of participants that received the high dose, although the highest dose elicited increased severity of reactogenicity. In the majority of participants, both humoral and cellular responses against MERS-CoV were induced and maintained for at least 1 year [204].
The data collected from the clinical studies mentioned here were relevant to support and accelerate SARS-CoV-2 vaccine development based on non-replicative viral vectors. Since most of these platforms are based on Ads, it is important to consider the pre-existing immunity problem in different populations. The use of non-human Ad vectors, heterologous prime-boost regimens and mucosal delivery of Ad vector vaccines are suitable alternatives to minimize this issue.
5.1.2. Replicative viral vector vaccines
To date, several replicative viral vector vaccines have been licensed for human use, including DENGVAXIA® (Dengue), IMVAMUNE® (smallpox), IMOJEV® (Japanese encephalitis) and ERVEBO® (rVSV-ZEBOV) for Ebola [9,57,205], supporting the use of this platform for SARS-CoV-2 vaccine development.
MV-based vaccines have been employed in a large preclinical track record and recently for the development of a vaccine against SARS-CoV-2 [206]. MV is able to induce potent CD4+ T-cell responses, unlike CD8+ T-cell dominated responses toward Ad vectors. Since MV can infect macrophages and DCs, its use as a vaccine could promote the delivery of target antigens directly to APCs [9]. The first evaluation in humans of the MV vaccine platform was done with the MV-CHIK vaccine candidate (Themis Bioscience, Austria) expressing structural genes of CHIK virus (Table 1). The Phase I and II clinical trials demonstrated strong safety and immunogenicity profiles with a homologous prime-boost regimen [207,208,209], where induction of NAbs were found in 50–93% of participants after the first immunization, and seroconversion rates were approximately 86–100% after the boost immunization at 1 or 6 months. Importantly, the immunogenicity of the vaccine did not appear to be affected by pre-existing anti-MV immunity [209]. In addition, recombinant vaccine candidates, MV-ZIKV as well as MV-HIV-1, have successfully completed Phase I clinical trials with the latter progressing to Phase II trials.
Although replicative viral vector vaccines generate strong immune responses, unfortunately there is a possibility for changes of the vector to occur, inducing vaccine virulence which can then cause infection [60,210]. This was demonstrated with the rVSV-ZEBOV Ebola vaccine (Merck Sharp & Dohme, U.S.), where replication in peripheral tissues promoted the onset of arthritis, rashes, dermatitis and vasculitis [211]. Additionally, viral replication in humans may enhance the negative implications of genomic integration of viral DNA into the host genome [59,212]. Although this is a predominant concern, MV replication is restricted to the cytoplasm, mitigating the risk of insertional mutagenesis [212,213,214]. In general, studies have shown that replicative viral vector vaccines successfully induce potent immune responses, and the technology already exists for large-scale production and storage.
5.2. Previous DNA vaccine immunity and safety
Plasmid-mediated Ab production was first reported in mice in 1992 [215]. A year later, plasmid-mediated cytotoxic T lymphocyte and Ab responses were also observed [216]. Immunization with naked plasmid DNA has since demonstrated both humoral and cell-mediated immunity in preclinical studies. However, DNA vaccines have not performed as well in humans. An early Phase I trial for a plasmid DNA vaccine against HIV showed promising cellular immunity in primates and humoral immunity in humans [217,218], but these results were not replicated. Later clinical trials would use larger doses, which may partially account for this inconsistency.
Developments against coronaviruses have been promising (Table 2). A DNA vaccine encoding the SARS-CoV S protein induced CD4+ and CD8+ T-cell and anti-S Ab responses in mice [42]. Another S protein plasmid was immunogenic in a Phase I trial: after 3 vaccinations, NAbs and CD4+ T-cell responses were detected across all participants, although CD8+ T-cell responses were present in only 20% of participants [39]. A DNA vaccine encoding a consensus MERS S protein similarly induced potent humoral and cell-mediated immune responses in mice, macaques, and camels. Encouragingly, MERS-immunized macaques were then protected against viral challenge within 6 weeks of vaccination [219]. More recently, a MERS DNA vaccine, INO-4700 (Inovio Pharmaceuticals, U.S.), demonstrated appreciable levels of MERS binding Abs in 94% of participants which persisted over a year in 77% of them, in a Phase I trial. CD8+ and CD4+ T-cell responses were similarly enduring in a majority of participants. Unfortunately, while NAbs were detected in 43% of participants by week 14, durability proved to be an issue as this dropped drastically (3%) after a year [220].
DNA vaccines have been examined against many pathogens without significant adverse effects including malaria [221,222], Ebola [223,224], dengue virus [225,226], West Nile virus (WNV) [227], and Zika virus [228,167]. Plasmid DNA vaccines against HIV-1 have similarly demonstrated high safety and appreciable dose-dependent CD4+ and CD8+ T-cell responses in human trials, although humoral immunity appeared to be weak in most cases [179,178,229,230,231,232,233]. In a WNV DNA vaccine Phase I trial for VRC-WNVD (National Institute of Allergy and Infectious Diseases [NIAID], U.S.), younger (18–50) and older (51–65) cohorts were contrasted. No vaccine-related serious adverse events (SAEs) were reported, indicating high safety even in the older cohort. NAbs were observed in over 96% of participants, while CD4+ T-cell responses were more prevalent than CD8+ [227]. Similarly, robust NAb titers and high frequency of CD4+ and CD8+ responses in a Phase I trial of an Ebola DNA vaccine were reported [224].
Seasonal influenza represents a favourable target for rapid vaccine development. VRC-FLUDNA064-00-VP (NIAID, U.S.), a trivalent DNA vaccine against seasonal influenza (Table 2), was investigated in prime-boost regimens across three trials in adults and children [144,234,235]. After priming with DNA, individuals were boosted with Fluzone, a trivalent inactivated influenza virus vaccine (IIV3). In one trial comparing younger (ages 18–50) and older (ages 51+) cohorts, participants were primed with either the plasmid vaccine or IIV3, or concurrent DNA/IIV3, followed by an IIV3 booster. Unfortunately, neither the DNA prime nor the concurrent DNA/IIV3 prime could improve upon a poor immune response in the older population [144]. The same DNA vaccine was assessed in a juvenile (ages 6–17) cohort. Here, participants were primed with an IM dose of either 1 mg of the DNA vaccine, 4 mg of the DNA vaccine, or IIV3, all followed by an IIV3 booster. In this cohort, priming with 4 mg of the DNA vaccine performed as well or better than with IIV3, demonstrating the potential for DNA-based vaccine use in an epidemic [234]. This was further demonstrated in a Phase I trial of GTU®-MultiHIV B (Imperial College London, UK), a DNA vaccine for the 2009 pandemic influenza A H1N1 virus (Table 2). Participants were first dosed with the DNA vaccine, then with the pandemic monovalent inactivated virus (MIV) vaccine upon availability. While few exhibited NAb responses after initial DNA immunization (6/20 subjects), the majority did so after the monovalent inactivated booster (5/18 subjects). Cell-mediated immunity, however, was superior as CD4+ T-cell responses were induced by DNA immunization alone and did not improve even after MIV boosting [236].
While generally well-tolerated, DNA vaccines may still carry other risks. Risk of insertional mutagenesis, while possible, appears low as no significant DNA vaccine integration into genomic DNA has yet been shown [237,238,239]. For industrial production of DNA vaccines, the FDA guidelines recommend that plasmid DNA be ≥80% supercoiled to prevent insertional mutagenesis [240].
The biggest risks related to DNA vaccines remain relating to genotoxicity and auto-immunity. Arguably the most important instance of anti-DNA immunity is as a hallmark of systemic lupus erythematosus (SLE), an autoimmune disease [241]. SLE is characterized by antinuclear Abs, most notably against double-stranded DNA and single-stranded DNA. Such anti-DNA Abs appear to bind specifically to antigenic DNA sequences. Interestingly, DNA isolated from SLE anti-DNA Ab/DNA complexes were found to be more highly guanine-cytosine (GC)-enriched (55% versus 38%), which more closely resembles bacterial GC content [242]. But in a comparison between SLE and non-SLE sera, SLE anti-DNA Abs bound indiscriminately to both eukaryotic and prokaryotic DNA, while non-SLE sera bound specifically to prokaryotic DNA [243]. Historically, administration of plasmid DNA to healthy animals has not generated appreciable levels of anti-DNA Abs [244] nor led to the onset of SLE [245]. However, as most plasmid DNA vectors are amplified in bacterial hosts, they will contain significant GC content in prokaryotic elements of the vector. Therefore, COVID-19 plasmid DNA vaccination should proceed with caution in individuals with, or at risk of, autoimmune disease.
Overall, plasmid DNA-based vaccines appear to be very safe, although not always immunogenic, where adequate immune response appears to require larger and more doses. Humoral immunity has not been consistent across human trials, although cell-mediated immunity appears more common. The safety profile of DNA vaccines in younger and older populations is extremely promising [144,227,234]. Importantly, DNA vaccines can be developed faster than their inactivated equivalents [236], as evidenced with current COVID-19 DNA vaccine developments. Even if a DNA vaccine does not induce sufficient immunity alone, it may be useful in prime-boost regimens as development of inactivated virus vaccines catch up.
5.3. Previous RNA vaccine immunity and safety
RNA-based vaccines, similar to DNA-based vaccines, can stimulate the synthesis of the immunogen by inducing both humoral and cellular immune responses [83]. The first mRNA vaccine to show efficacy in humans was mRNA-1944 (Moderna Therapeutics, U.S.) (Table 3), encapsulated with their proprietary LNP technology against the Chikungunya virus. Two mRNAs comprised mRNA-1944, encoding the sequences for the heavy and light chains of an anti-Chikungunya IgG respectively. This vaccine was delivered intravenously (IV) at four doses (0.1, 0.3, 0.6 and 1 mg/kg) and demonstrated linear dose-dependent production Abs in the human body, where 100% of participants demonstrated NAb activity against the Chikungunya virus. While the two lower doses did not induce vaccine-related SAEs, the higher doses induced infusion-related grade 3 AEs, indicating a sensitive balance between safety and efficacy [246]. More recently, Moderna's mRNA-1388 vaccine, also against the Chikungunya virus, was found to be well-tolerated and capable of eliciting dose-dependent increases in NAb titers. 100% seroconversion and a substantial boost in titers after the second vaccination was observed [247].
Moderna has also developed mRNA influenza vaccine candidates that have undergone Phase I trials evaluating two vaccines: one encoding the HA GP from H10N8 and the other encoding the GP from H7N9 (Table 3). While participants did not present significant HA-specific cell-mediated responses, they did show robust humoral immune responses and high seroconversion rates [248]. Previous H7N9 virus-like particle and subunit candidates required adjuvants to reach high seroconversion and seroprotective rates [249], alluding to a self-adjuvanting effect of the mRNA vaccines [248]. Additionally, the H7N9 mRNA vaccine demonstrated potential development of memory B cells, evidenced by persistent hemagglutinin inhibition (HAI) assay titers 6 months after vaccine administration [248]. A Phase I trial of Moderna's mRNA-1325 vaccine, against Zika virus has been completed, although results have not yet been published to date.
Boostable levels of functional Abs were observed in a Phase I clinical trial administering three doses of the rabies vaccine, CV7201 (CureVac, Germany), when delivered by needle-free administration, as aforementioned (Table 3). In some participants, NAb protective titers were above the expected protective threshold. Unfortunately, one SAE was noted after a 640 μg IM dose, but successfully resolved. T-cell immune responses were generally low, but these results were likely not representative due to blood samples that were derived two weeks from the last vaccination [168].
RNA vaccines have been generally safe in humans, although unintended adverse reactions are the biggest fear for RNA vaccines [84,250]. Clinical trials have demonstrated local and systemic moderate and severe AEs for some mRNA vaccines [168,248,251,252]. This could be due to the fact that pattern recognition receptors (PRRs) are able to detect single- and double-stranded RNA molecules and subsequently induce type 1 IFN responses resulting in unwanted innate immune responses and inflammation [253]. It has also been noted that the stimulation of potent type 1 IFN responses can promote the onset of autoimmunity [86,254,255]. The previously mentioned CV7201 vaccine reported one event of autoimmune thyroiditis a year after the last dose [164]. Furthermore, concerns regarding mRNA vaccination have been identified [81,86], due to the fact that extracellular RNA has been shown to alter endothelial cells and damage blood vessels, consequently promoting significant health issues including oedema, coagulation and pathological thrombus formation [256,257]. Although RNA-based vaccines carry additional safety concerns compared to their DNA counterparts, mRNA does not need to be transported into the nucleus for transcription, dramatically improving transfection efficiency and eliminating the possibility of oncogenic events arising from random integration into host DNA [258].
While RNA vaccines overall have demonstrated strong humoral immunity and high seroconversion rates, their inconsistent cell-mediated immunity responses merit concern. Due to strong induction of potent type 1 IFN responses [253], COVID-19 RNA vaccine candidates require thorough safety testing to ensure protection against the onset of unexpected side effects. Similarly with DNA, auto-immunity may be a concern [84,86]. Considering these caveats, close attention must be given to their formulation and more data is required to ensure their safety and efficacy in humans.
6. COVID-19 gene-based vaccines
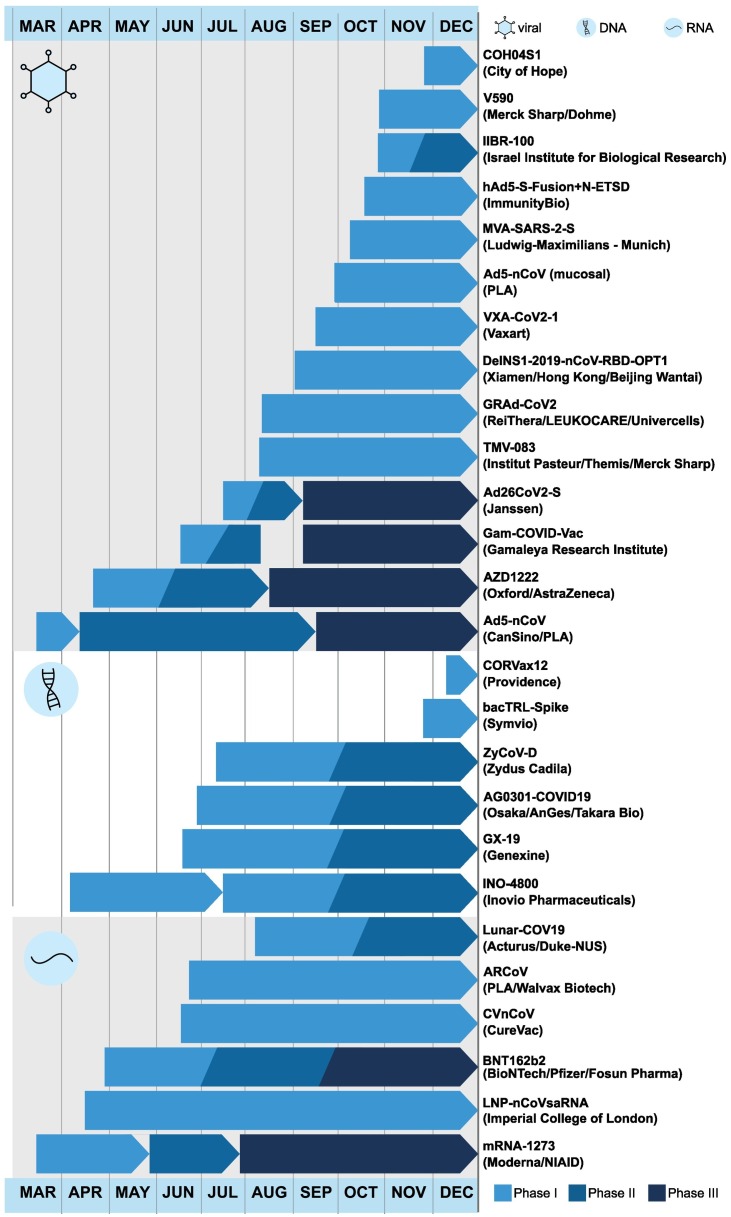
Foundational knowledge from previous GBV developments have enabled astonishingly rapid production of GBVs to combat COVID-19 in 2020. Moreover, previous studies on vaccines developed during the MERS-CoV and SARS-CoV pandemics have demonstrated the key roles of humoral and cellular immune responses to induce protective responses within a short period of time [259,260]. Overall, GBV platform development has progressed more rapidly than many other vaccine types, and now make up some of the most promising COVID-19 vaccine candidates (Fig. 4 ).
Fig. 4.
Timeline of clinical trials undertaken by GBV candidates. Vaccines are grouped based on platform: viral, DNA, then RNA. Based on early data, many vaccine candidates have begun subsequent phases before the end of earlier trials. Several vaccine candidates have concurrent clinical trials; for these, their earliest and most recent ongoing trials are depicted.
Here, we summarize current COVID-19 GBV candidates that have entered clinical trials as of December 2020 (Fig. 4), as listed by the WHO [6]. At this time, 26 of the 52 clinical COVID-19 candidates are GBVs: 10 are non-replicative viral-vectors (Table 4), 4 are replicative viral vectors (Table 4), 6 are DNA-based (Table 5), and 6 are RNA-based (Table 6). A large proportion of these vaccines encode the SARS-CoV-2 S protein, or a derivative thereof, and are being delivered intramuscularly.
Table 4.
Viral vector COVID-19 vaccine candidates in ongoing clinical trials.
| Developer | Vaccine | Vector/antigen | Delivery and dosage | Number of participants (age range) | Phase trial and description | Outcomes | Reference |
|---|---|---|---|---|---|---|---|
| Non-replicating Viral Vector | |||||||
|
University of Oxford/ AstraZeneca |
AZD1222 |
ChAdOx1 Antigen: S protein (YP_009724390.1) |
IM 5 × 1010 vp |
1077 (18–55) |
Phase I/II (NCT04324606)(15281137) COV001 (UK). Single-dose or prime-boost 28 days apart. MenACWY (vaccine control). Included an efficacy cohort. |
Safety: No vaccine-related SAEs. Immunogenicity: Anti-S IgG responses peaked by day 28 post-prime. Induction of T-cell responses on day 14. Increase of NAbs after a booster dose. *Other Phase I/II trials are ongoing in the UK and Japan. |
[264] |
| IM 2.2 × 1010 vp (LD), 3.5–6.5 × 1010 vp (SD) |
160 (18–55) 160 (56–69) 240 (≥70) |
Phase II/ III (NCT04400838)(15281137) COV002 (UK). Age-escalation. Two doses (LD and SD). One or two-dose regimen 28 days apart. MenACWY (vaccine control). Efficacy results were not included. |
Safety: 13 SAEs that were not vaccine-related. Reduced reactogenicity in older adults (≥56). Immunogenicity: T-cell response peaked on day 14 after the first dose (regardless of age and vaccine dose). Similar Anti-S IgG responses 28 days after the boost in all age groups (regardless of vaccine dose). Nab responses in >99% of participants by day 14 after the boost vaccination. The boosted groups presented higher levels of NAbs. *Other Phase II/III trials are ongoing in the Uk and India. |
[265] | |||
| IM 2.2 × 1010 vp (LD), ~ 5 × 1010 vp (SD) |
23,848 (≥18) |
Phase I/II, II/III, III (NCT04324606)(NCT04400838)(NCT04444674) (ISRCTN89951424) Two doses (LD and SD). One or two-dose regimen ≥28 days apart. Pool interim analysis of 4 trials. Safety: COV001, COV002, COV003 (Brazil), COV005 (South Africa). Efficacy (n = 11,636): COV002 and COV003. COV002 doses: LD/SD or SD/SD). COV003 doses: SD/SD. In COV003, MenACWY and saline were used as a control. |
Safety: A possible neurological event that was vaccine-related. Efficacy: Vaccine efficacy was 62.1% in the SD/SD cohort (n: 8895) and 90.0% in the LD/SD cohort (n: 2741). Overall vaccine efficacy in both groups was 70.4%. Efficacy against asymptomatic infection was 58.9% and 3.8% in the LD/SD and SD/SD cohorts, respectively. *Other Phase III trials are ongoing in the U.S., Argentina, Chile, Colombia and Peru (n:40000). Suspended in Russia due to the occurrence of SUSAR in a phase II/III study. Waiting for the Russian MOH approval to continue. Pediatric studies have not started. |
[267] | |||
|
CanSino Biologics/ Beijing Institute of Biotechnology |
Ad5-nCoV |
Ad5 Antigen: S protein (YP_009724390) |
IM 5 × 1010 vp, 1 × 1011 vp, 1.5 × 1011 vp |
108 (18–60) |
Phase I: (NCT04313127) Dose escalation. |
Safety: Evidence of dose-dependent reactogenicity. 9% of participants in the high dose group presented fever. Immunogenicity: Induction of specific humoral responses peaked on day 28 and T-cell responses detected on day 14. *Two other Phase I trials are ongoing in China. |
[269] |
| IM 5 × 1010 vp (LD), 1 × 1011 vp (HD) |
508 (≥18) |
Phase II (NCT04341389) Dose escalation randomized trial. |
Safety: The 5 × 1010 vp dose showed a better safety profile. Reduced reactogenicity in older adults (≥55). Immunogenicity: Specific T-cell responses in 90% (HD) and 88% (LD) of participants. Seroconversion of NAbs in 59% (HD) and 47% (LD) of participants. RBD-specific Ab seroconversion in 96% (HD) and 97% (LD) of participants by day 28. *Two other Phase II trials are ongoing in China. |
[270] | |||
| IM 5 × 1010 vp |
40,000 (≥18)500 (18–85) |
Phase III (NCT04526990) (NCT04540419) |
Ongoing in multiple countries. | – | |||
| Gamaleya Research Institute | Gam- COVID-Vac (frozen) and Gam- COVID-Vac Lyo (lyophilized) |
Ad26, Ad5 Antigen: S protein |
IM 1011 vp (per dose for both rAds) |
76 (18–60) |
Phase I/II (NCT04436471) (NCT04437875) Phase I: single-dose (rAd26-S or rAd5-S). Phase II: heterologous prime-boost: rAd26-S on day 1 and rAd5-S on day 21. |
Safety: No vaccine-related SAEs. Immunogenicity: RBD-specific IgG responses on day 28 and 42 and NAbs on day 42 were found in all participants with both formulations. Antigen-specific CD4+ and CD8+ T-cells and increased IFN-γ secretion in 100% of participants were detected on day 28. *Another Phase II trial is ongoing in Russia. |
[272] |
| IM 0.5 ml/dose (each vaccine) |
40,000 (≥18) |
Phase III (NCT04530396) Heterologous prime-boost: rAd26-S on day 1 and rAd5-S on day 21. Second interim analysis to review the efficacy in 18,794 participants. |
Safety: As of November 24th, no vaccine-related SAEs were reported. Efficacy: 91.4% efficacy rate 7 days after the second dose. Over 95% efficacy 21 days after the second dose (preliminary data). The study is still ongoing in Russia. *Another Phase III trial is ongoing in India, Belarus and Venezuela. |
[274] | |||
| IM |
1045 (18–55 and ≥ 65) |
Phase I/IIa (NCT04436276) Single-dose or two-dose schedule at different intervals. |
Ongoing in U.S. and Belgium. *Another Phase I trial is ongoing in Japan. |
– |
|||
| Janssen Pharmaceutical Companies | Ad26Cov2-S |
Ad26 Antigen: S protein (MN996528.1) |
IM |
1210 (12-17, 18- ≥65) |
Phase II (NCT04535453) Three- dose levels. Two-dose schedules 28, 56, 84 days or 1 year after the first vaccination. |
Ongoing in Canada, Germany, Netherlands and Spain. |
– |
| IM 5 × 1010 vp |
6000 (≥18) |
Phase III (NCT04505722) A single-dose vaccine. |
Ongoing in multiple countries. *Another Phase III trial is ongoing. |
– |
|||
| ReiThera/ LEUKOCARE/ Univercells | GRAd-CoV2 |
Simian Ad (GRAd) Antigen: S protein |
IM 5 × 1010 vp, 1× 1011 vp, 2 × 1011 vp |
90 (18–85) |
Phase I (NCT04528641) Dose escalation. |
Ongoing | – |
| Institute of Biotechnology, Academy of Military Medical Sciences, PLA of China | Ad5-nCoV |
Ad5 Antigen: n on-specified |
IM/mucosal 5 × 1010 vp (IM) 2 × 1010 vp (mucosal), 1 × 1010 vp (mucosal) |
149 (18 and ≥ 60) |
Phase I (NCT04552366) Two doses. Different administration routes and schedules. |
Ongoing | – |
| Vaxart | VXA-CoV2-1 |
rAd5 Antigen: n on-specified |
Oral 1 × 1010 I.U., 1 × 1110 I.U. |
35 (18–54) |
Phase I (NCT04563702) Single-dose or two-dose (day 29). dsRNA adjuvant. |
Ongoing | – |
| ImmunityBio, Inc./ NantKwest Inc. | hAd5 S+ N |
Ad5 (E1/E2b/E3 deletions). Antigens: S + N- (ETSD) proteins |
SC 5 × 1010 vp, 1 × 1011 vp |
35 (18–33) |
Phase I (NCT04591717) |
Ongoing | – |
| Ludwig-Maximilians-University of Munich | MVA-SARS-2-S |
MVA Antigen: S protein |
IM 1 × 107 I.U., 1 × 108 I.U. |
30 (18–55) |
Phase I (NCT04569383) Two-dose schedule 28 days apart. |
Ongoing | – |
| City of Hope Medical Center | COH04S1 |
MVA Antigens: S and N protein |
IM 1 × 107, 1 × 108, 2.5 × 108 PFU/dose |
129 (18–55) |
Phase I (NCT04639466) Dose escalation. Three different dose levels. |
Ongoing | – |
| Replicating Viral Vector | |||||||
| Institut Pasteur/ Themis/ University of Pittsburgh CVR/ Merck Sharp & Dohme |
TMV-083 |
MV Antigen: non-specified |
IM | 90 (18–55) |
Phase I (NCT04497298) Dose escalation. |
Ongoing | – |
| Beijing Wantai Biological Pharmacy/ Xiamen University | DelNS1- 2019-nCoV- RBD-OPT1 |
Influenza Antigen: non-specified |
IN | 48 (≥18) | Phase I (ChiCTR2000037782) | Ongoing *A Phase II trial is ongoing. |
– |
| Israel Institute for Biological Research | IIBR-100 |
VSV Antigen: S protein |
IM 1 × 105, 1 × 106, 1 × 107 PFU/ml |
1040 (18–85) |
Phase I/II (NCT04608305) Dose escalation (phase I). Single-dose or two-dose schedule 28 days apart. |
Ongoing | – |
| Merck Sharp & Dohme/ IAVI | V590-001 |
VSV Antigen: S protein |
IM 5 × 105, 2.4 × 106, 1.15 × 107, 5.55 × 107 PFU/ml |
252 (≥18) |
Phase I (NCT04569786) A single-dose vaccine. |
Ongoing | – |
Ad: adenovirus; ChAdOx1:chimpanzee adenovirus-vectored vaccine; ETSD: enhanced T-cell stimulation domain; hAd: human adenovirus; HD: high dose; IFN-y: interferon gamma; Ig: immunoglobulin; IM: intramuscular; IN: intranasal; IU: International Unit; LD: low dose; MOH: Ministry of Health; MV: measles virus; MVA: modified vaccinia ankara; n: Number of participants; N: nucleoprotein; NAbs: neutralizing antibodies; PFU: plaque-forming unit; rAd: recombinant adenovirus; RBD: receptor binding domain; SAE: serious adverse event; S: spike; SC: subcutaneous; SD: standard dose; SUSAR: Suspected Unexpected Serious Adverse Reaction; vp: viral particles; VSV: vesicular stomatitis virus
Table 5.
DNA COVID-19 vaccine candidates in ongoing clinical trials.
| Developer | Vaccine |
Vector/ antigen |
Delivery and dosage | Number of participants (age range) | Phase trial and description | Outcomes | Reference |
|---|---|---|---|---|---|---|---|
|
Inovio Pharmaceuticals/ International Vaccine Institute |
INO-4800 |
PGX0001 expression vector Antigen: S protein |
IM (EP) 0.5 mg, 1 mg, 2 mg |
120 (≥18) |
Phase I (NCT04336410) Two-dose schedule on day 0 and 28. |
Ongoing | – |
| ID (EP) 1 mg, 2 mg |
160 (19–64) |
Phase I/IIa (NCT04447781) Two-dose schedule on day 0 and 28. |
Ongoing | – | |||
| ID (EP) 1 mg, 2 mg |
6578 (≥18) |
Phase II/III (NCT04642638) Two-dose schedule on day 0 and 28. |
Ongoing | – | |||
|
Osaka University/ AnGes/ Takara Bio |
AG0301-COVID19 |
Plasmid DNA Antigen: non-specified |
IM 1 mg, 2 mg |
30 (20–65) |
Phase I/II (NCT04463472) Two-dose schedule on day 0 and 14. |
Ongoing | – |
| IM 2 mg |
30 (20–65) |
Phase I/II (NCT04527081) Two-dose schedule at 2 or 4-week intervals. Three-dose schedule at 2-week intervals. |
Ongoing | – | |||
| Cadila Healthcare Limited | ZyCoV-D |
Plasmid DNA Antigen: non-specified |
ID | non-specified |
Phase I/II (CTRI/2020/07/026352) |
Ongoing | – |
| Genexine Consortium | GX-19 |
Plasmid DNA Antigen: S protein |
IM (EP or PharmaJet® Needle-Free) | 210 (18–50) |
Phase I/IIa (NCT04445389) Dose escalation. Two-dose schedule on day 0 and 28. |
Ongoing | – |
| Symvivo | bacTLR-Spike |
B. longum delivering plasmid DNA Antigen: S protein |
Oral 1, 3, 10 × 109 cfu B.longum |
12 (≥18) |
Phase I (NCT04334980) Single-dose. |
Ongoing | – |
| Providence Health and Services | CORVax12 |
Plasmid DNA Antigen: S protein |
ID | 36 (≥18) |
Phase I (NCT04627675) Vaccine administered with or without boost (IL-12p70 plasmid) 4 weeks apart. |
Ongoing | – |
B: bifidobacterium; cfu: colony forming units; EP: electroporation; ID: intradermal; IL: interleukin; IM: intramuscular; mg: milligram; S: spike.
Table 6.
RNA COVID-19 vaccine candidates in ongoing clinical trials.
| Developer | Vaccine |
Vector/ antigen |
Delivery and dosage |
Number of participants (age range) |
Phase trial and description | Outcomes | Reference |
|---|---|---|---|---|---|---|---|
|
Moderna/ NIAID |
mRNA-1273 |
LNP-encapsulated mRNA Antigen: S protein |
IM 25 μg, 100 μg, 250 μg |
45 (18–55) ++40 (56–70 or ≥ 71) |
Phase I (NCT04283461) Dose escalation. Two-dose schedule 28 days apart. ++Explanation of the trial in older adults (only doses of 25 or 100 μg were administered). |
Safety: Reactogenicity after the second vaccination. Immunogenicity: NAb responses were observed in all participants. The 100 μg dose showed high NAbs and CD4+ T-cell responses. |
[293,295] |
| IM 50 μg, 100 μg |
600 (≥18) |
Phase IIa (NCT04405076) Two-dose levels. |
Ongoing | – | |||
| IM 100 μg |
30,000 (≥18) |
Phase III (NCT04470427) Two-dose schedule on day 1 and 29. |
Ongoing | – | |||
| IM 10 μg, 20 μg, 30 μg, 100 μg |
195 (18–85) |
Phase I (NCT04368728) Dose escalation. Single-dose or two-dose schedule 21 days apart. |
Safety: No vaccine-related SAEs. Dose-dependent local and systemic AEs were mild to moderate. Immunogenicity: Dose-dependent NAb responses. *Another Phase II/III trial under the same identifier number (NCT436872) is ongoing in the U.S., Argentina, Brazil, Germany, South Africa and Turkey (n: 43998.) *Another Phase I trial is ongoing in China |
[301] |
|||
|
BioNTech/ Fosun Pharma/ Pfizer |
BNT162a1, b1, b2, c2. |
LNP-encapsulated mRNA vaccine Antigens: two encode the S protein and two encode the optimized RBD. |
IM 10 μg, 30 μg, 100 μg |
45 (18–55) |
Phase I/II, (NCT04368728) Dose escalation. Two-dose schedule on day 1 and 21. |
Safety: Increased reactogenicity with the 100 μg dose. One report of severe pain for the 100 μg dose. Immunogenicity: Increased RBD-binding IgG and NAb titers after a second dose. |
[300] |
| IM 1 μg, 10 μg, 30 μg, 50 μg |
456 (18–85) |
Phase I/II (NCT04380701) Two-part dose escalation. Single-dose or prime/boost immunization. |
Safety: No vaccine-related SAEs. Dose-dependent local and systemic AEs were generally mild to moderate. Immunogenicity: Increased RBD-binding IgG, NAb titers, CD4+ and CD8+ T-cell responses. *Other Phase I/II trials are ongoing in Japan (n:160) and Germany (n:444) |
[302] | |||
| IM 30 μg of BNT162b2 (0.3 ml/ per dose) or saline placebo |
43,548 (≥16) |
Phase II/III (NCT04368728) Two-dose schedule 21 days apart. |
Safety: Very low SAEs which were similar between vaccine and placebo groups. General mild to moderate AEs observed. Efficacy: Demonstrated 95% effectiveness in the prevention of COVID-19. 90–100% vaccine efficacy demonstrated across different subgroups. |
[303] | |||
|
CureVac |
CVnCoV |
LNP-encapsulated mRNA vaccine Antigen: S protein |
IM 2 μg, 4 μg, 8 μg |
168 (18–60) |
Phase I (NCT04449276) Dose escalation. Two-dose schedule on day 0 and 28. |
Ongoing | – |
| IM 6 μg, 12 μg |
660 ((≥18) |
Phase IIa (NCT04515147) Single-dose or two-dose schedule on day 1 and 29. |
Ongoing | – | |||
| Acturus/ Duke-NUS | ARCT-021 (Lunar-COV19) |
Replicating RNA Antigen: undisclosed epitopes |
IM | 92 (21–80) |
Phase I/II (NCT04480957) Phase I: escalating dose levels as a single injection. Phase 2: two-dose levels. |
Ongoing | – |
| Imperial College London | LNP-nCoVsaRNA |
LNP-sa RNA Antigen: S protein |
IM 0.1 μg, 0.3 μg, 1 μg |
(18–75) |
Phase I(ISRCTN17072692) Dose escalation. The participants in the expanded safety evaluation will receive the 1 μg dose. |
Ongoing | – |
| People's Liberation Army (PLA) Academy of Military Sciences/ Walvax Biotech. | ARCoV |
Non-specified RNA Antigen: RBD of S protein. |
Non-specified | (18–59 and ≥ 60) |
Phase I(ChiCTR2000034112) Dose escalation. |
Ongoing *Another phase I trial is ongoing in China. |
– |
AE: adverse events; Ig: immunoglobulin; IM: intramuscular; LNP: lipid nanoparticle; n: number of participants; NAbs: neutralizing antibodies; NIAID: National Institute of Allergy and Infectious Diseases; RBD: receptor binding domain; sa: self-amplifying; SAE: serious adverse event; S: spike; μg: microgram.
6.1. Viral vector vaccines
The AZD1222 vaccine candidate (University of Oxford, AstraZeneca, UK) (Table 4) is based on the ChAdOx1 vector encoding the S protein along with a tissue plasminogen activator (tPA) human leader sequence at the 5′ end, flanked by the cytomegalovirus (CMV) promoter and a bovine growth hormone (BGH) poly A signal sequence [261]. Leader tPA sequences have been shown to enhance immunogenicity and increase expression levels of recombinant proteins [262,263]. The AZD1222 vaccine reduced SARS-CoV-2 viral load in the lungs and was protective against pneumonia in a rhesus macaque challenge model, although it did not reduce viral loads in the upper respiratory tract [261]. The results of the Phase I/II trial performed in young adults, confirmed an acceptable safety profile for the vaccine. Mild to moderate local and systemic reactions such as pain, fever, chills, and aches were reported. As has been demonstrated with other Ad vectors, S-specific effector T-cell responses peaked on day 14 and were continued up to day 56. Anti-S IgG responses rose by day 28 after just a single dose. After the second dose, all participants had neutralizing activity, although a boost in cellular responses was not observed [264]. The dose employed in this study was based on the previous MERS clinical trials [204]. The Phase II/III trial involved both young and old adults. While its AEs profile was similar to the previous Phase I study, the vaccine was better tolerated in older adults compared with younger adults. After a booster dose, both anti-S SARS-CoV-2 IgG responses and NAbs titers were similar in young and older adults [265]. The efficacy report was published in an interim analysis with pooled data from the UK and Brazil studies. The vaccine had an efficacy of 90% against symptomatic COVID-19 disease in the group that received a low dose followed by a standard dose (Table 4). Vaccine efficacy in older age groups is still to be determined. Three cases of transverse myelitis briefly paused ongoing studies, but only one case was found to be possibly linked to AZD1222. After analysis, the vaccine was determined to have an acceptable safety profile. AZD1222 can be stored between 2 and 8 °C, which can simplify global distribution as ultra-cold temperatures are not required for transport [266,267].
The non-replicative Adn5-CoV vaccine candidate (CanSino Biologics, Beijing Institute of Biotechnology, China) containing an optimized full-length S sequence of SARS-CoV-2 along with the tpA leader sequence, is currently undergoing Phase III clinical trials (Table 4). This vector was chosen from previous experience with the Ebola vaccine. In preclinical studies, the vaccine was protective against SAR-CoV-2 infection in ferrets when administered via the IM or mucosal route [268]. The Phase I clinical trial of Ad5-nCoV, administered IM at three doses demonstrated specific humoral and T-cell responses against SARS-CoV-2. Although the high dose vaccine was more immunogenic, it also induced higher reactogenicity [269]. Therefore, the Phase II trial was conducted using the two lower doses. A single injection of Ad5-nCoV induced specific immune responses to the S protein at day 28, with no significant differences between the two dosage groups. A lower immune response was detected in older people. Among the participants that had low pre-existing anti-Ad5 NAbs, the levels of RBD-specific Abs were about two-times higher than the participants with high anti-Ad5 NAbs. Based on these results, the investigators expect the candidate Ad5-nCoV to confer superior immunogenicity in people with lower pre-existing anti-Ad5 immunity [270]. Additional Phase I (ChiCTR2000030906) and II (ChiCTR2000031781) trials are ongoing to evaluate a single dose of the vaccine. Phase I and II trials are ongoing to evaluate a booster dose 6 months after prime (NCT04568811) and the two-dose regimen (NCT04566770). Phase III trials are also currently ongoing, although the Chinese government has already approved Ad5-nCoV to be used by their armed forces [271].
Gam-COVID-Vac (Sputnik V), a heterologous vaccine candidate (Gamaleya Research Institute, Russia), consists of recombinant human rAd26 and rAd5 viral vectors both encoding the S protein in lyophilized (can be stored between 2 and 8 °C) or frozen form (Table 4). The Phase I/II studies compared single doses of either rAd26 or rAd5-S vaccines, or in a small number of participants, a single inoculum of rAd26-S then followed by a rAd5-S booster. The results revealed that the heterologous vaccine is safe, highly immunogenic and induced strong humoral and cellular immune responses in 100% of the volunteers. RBD-specific IgGs were detected in 88.9% of participants who received the rAd26-S vaccine, in 84.2% of participants immunized with rAd5-S at day 14, and in 100% of the participants from day 21. The single rAd26-S administration elicited the production of NAbs in only 61.1% of the participants, although boosting with rAd5-S increased the RBD-specific IgG titers. The prime-boost regimen induced the production of SARS-CoV-2 NAbs in 100% of participants with titers comparable to patients who had recovered from COVID-19. In comparison with the Adn5-CoV vaccine, the titers of RBD-specific IgGs were not affected by the presence of pre-existing immunity against rAd26 and rAd5. Headache, hyperthermia, pain at the injection site and asthenia were the most common systemic and local reactions noted, which are not unusual following recombinant viral vector vaccine administrations [272]. Although Phase III trials to evaluate the efficacy have not started yet, the Ministry of Health of the Russian Federation approved the registration of the Gam-COVID-Vac vaccine in August 2020 [271] under emergency use authorization [273]. According to the second interim analysis released by Gamaleya Institute (November 24), the Sputnik V vaccine showed an efficacy of 91.4% 7 days after the second dose in 18,794 volunteers from Russia. 1.2 billion doses of the vaccine have been requested from more than 50 countries [274].
Based on AdVac® viral vector technology (Janssen, Belgium), the Ad26 vector was used to develop the Ad26CoV2-S vaccine, encoding a prefusion stabilized S immunogen, which contains the wildtype leader sequence, the full-length S sequence with mutation of the furin cleavage site and two proline stabilizing mutations. Pre-clinical data showed that a single dose of the Ad26COVS1 vaccine in non-human primates elicited a strong immune response as demonstrated by NAbs that provide protection in the lower respiratory tract [275]. Phase I, II and III trials are ongoing in multiple countries (Table 4).
The GRAd-CoV2 vaccine candidate (ReiThera, Italy) is based on a novel replication-incompetent simian Ad strain, encoding the S protein of SARS-CoV-2, and is undergoing a Phase I trial (Table 4). Based on Vaxart's oral Ad platform (Vaxart, U.S.), the Ad5 vector was used to develop the VXA-CoV2-1 vaccine candidate, expressing one or more SARS-CoV-2 antigens. Pre-clinical studies demonstrated that the vaccine could induce protective mucosal immunity, as defined by significant IgA Ab responses found in the lungs of immunized mice. Antigen-specific CD4+ and CD8+ T-cells were also induced at both low and high doses, while humoral IgG responses were found to be dose-dependent. The candidate expressing full length wild-type S protein produced higher NAbs titers [276]. The Ad5-nCoV vaccine (Institute of Biotechnology, Academy of Military Medical Science, China) is another candidate based on the Ad5 vector that is undergoing a Phase I trial to compare IM and mucosal vaccination. According to previous reports, oral and nasal administration of Ad5 are less affected by pre-existing immunity to Ad and do not impart significant anti-vector immunity compared with IM administration [277,278,279]. As such, both VXA-CoV2-1 and Ad5-nCoV candidates are likely to be more suitable for repeated dosing [276].
The bivalent hAd5 S + N vaccine (ImmunityBio, Inc./ NantKwest Inc., US) based on the Ad5 vector, expresses both the SARS-CoV-2 S and N protein. The preliminary reports showed the vaccine is able to elicit NAbs and T-cell immunity in mice [280]. The vaccine antigens were recognized by antisera and T cells from previous SARS-CoV-2 infected patients. The presence of N protein was vital for the induction of CD4+ and CD8+ T cell memory recall [281]. The COH04S1 vaccine (City of Hope Medical Center, U.S.) based on the MVA vector, is the second viral vector vaccine currently in clinical trials that expresses SARS-CoV-2 S and N proteins. In pre-clinical studies conducted in mice, the vaccine elicited a robust humoral and cellular immune response to the S and N antigens [282]. Based on previous experience with the MVA-MERS-S vaccine [283], the MVA vector was used to develop the MVA-SARS-2-S vaccine (Ludwig Maximilian University of Munich, Germany) expressing the S protein. COH04S1 and MVA-SARS-2-S are the non-replicative viral vector vaccines currently in clinical trials that are utilizing the MVA vector instead of the Ad vector.
From their previous experiences with human MV technology, the Institut Pasteur, in collaboration with Themis, has developed a vaccine against SARS-CoV-2 (TMV-083) and is in the process of recruiting for their Phase I clinical trial. The first IN SARS-CoV-2 vaccine, DelNS1-2019-nCoV-RBD-OPT1 (Xiamen University, China), is based on a replicative influenza viral vector (Table 4 ). The Phase I clinical trial is expected to assess the safety of the vaccine and the potential effect of pre-existing Abs against the influenza viral vector. The VSV viral vector was used to develop the vaccines candidates IIBR-100 (Israel Institute for Biological Research) and V590-001 (Merck Sharp & Dohme/ IAVI) both expressing the S protein. A preliminary report conducted in hamsters suggested that the IIBR-100 vaccine was safe, effective and protective against SARS-CoV-2 infection [284].
6.2. DNA vaccines
The DNA vaccine candidate, INO-4800 (Inovio Pharmaceuticals, U.S.), encodes the SARS-CoV-2 S protein (Table 5 ). The vector, pGX001, encodes a consensus S sequence with an N-terminal immunoglobulin E (IgE) leader sequence under the control of the CMV immediate-early promoter [37]. Preclinical results in mice and guinea pigs exhibited both cellular and humoral immune responses. In a Phase I trial, two doses of INO-4800 were delivered through the ID route, supplemented by EP via Inovio Pharmaceutical's CELLECTRA®2000, which has shown some efficacy in previous studies in humans and non-human primates [285,286]. An “overall immunological response” based on Ab and T-cell responses in 34 out of 36 participants in their Phase I trial was reported. Only 10 reported AEs were observed (all grade 1 severity) and no SAEs were reported [287]. A Phase I/II trial began July 2020 to further assess the safety, tolerability, and immunogenicity of INO-4800. Previously, Inovio Pharmaceuticals reported their SynCon EBOV DNA vaccines to be stable for 1 month at 37 °C, over 1 year at room temperature (25 °C) and over 3 years at 4 °C [224]. INO-4800 has the same storage conditions [37], which is encouraging for easier vaccine dissemination.
Other COVID-19 DNA vaccine groups have also begun clinical trials (Table 5). A Phase I/IIa trial for GX-19 (Genexine, South Korea) began recruitment in June 2020. AG0301-COVID19, a DNA vaccine developed by the collaborative efforts of AnGes, Takaro Bio, and Osaka University (Japan), began recruitment for a Phase I/II trial in July 2020 to evaluate its safety and immunogenicity. AG0301-COVID19 will be delivered along with an adjuvant as a means to stimulate a greater immune response [288]. A Phase I/II trial to assess the safety and immunogenicity of three doses of ZyCoV-D (Cadila Healthcare Limited, India) is currently undergoing. In late November 2020, a Phase I trial began recruitment to evaluate a novel plasmid DNA vaccine, bacTRL-Spike. Based on Symvio Corporation's bacTRL platform, this oral vaccine encodes the S protein in plasmid DNA actively replicating in live Bifidobacterium longum, a known probiotic gut bacterium. Another Phase I trial began December 2020 to evaluate CORVax12, a plasmid DNA vaccine encoding the S protein, from the Providence Cancer Institute at Providence St. Joseph Health. In this trial, the efficacy of electroporated CORVax12 alone or supplemented with a second plasmid encoding IL-12 will be examined.
6.3. RNA vaccines
Lunar-COV19 (Acturus Therapeutics, U.S.; Duke-NUS, Singapore) is delivered by a lipid-enabled and unlocked nucleomoner agent modified RNA lipid mediated delivery system (Lunar) (Table 6 ), which has previously evidenced successful and efficient mRNA delivery in animal models [289,290]. Preclinical results demonstrated that a single prime vaccination in mice induced high NAb titers, which continually increased up to day 60, in addition to enhancement of CD8+ and CD4+ T-cell responses. The intermediate and high doses effectively protected SARS-CoV-2 challenged human ACE2 transgenic mice [291].
A Phase I trial for the vaccine candidate, LNP-nCoVsaRNA (Imperial College London, UK), which encodes a prefusion stabilized SARS-CoV-2 S protein, is currently ongoing (Table 6). Preclinical results were promising as the vaccine demonstrated effective viral neutralization which was directly related to the induction of SARS-CoV-2 specific IgG production. High levels of cell-mediated responses were also observed [292]. Recruitment for Phase I clinical trials for the CVnCoV (CureVac, Germany) and ARCoV (People's Liberation Army Academy of Military Sciences, China; Walvax Biotech, China) is currently undergoing (Table 6).
The RNA vaccine candidate, mRNA-1273 (Moderna Therapeutics, NIAID, U.S.) (Table 6), encodes the S protein which contains the S1-S2 cleavage site, essential for S-driven viral entry into lung cells [293,294,295]. The S protein is perfusion stabilized via proline substitutions, which has been shown to increase recombinant expression of viral fusion GPs [296,297]. The vaccine is encapsulated within LNPs [293], which as mentioned earlier, provides adequate protection of the mRNA molecule [89]. Vaccination in non-human primates demonstrated that a two-dose vaccination schedule with mRNA-1273 elicited robust NAb and T-cell responses and was protective against SARS-CoV-2 infection in the upper and lower airways [294]. Results of the Phase I trial conducted in adults (18–55) and older adults (≥ 56) showed that mRNA-1273 was safe and well tolerated. High NAb responses were elicited in a dose-dependent fashion in the absence of SAEs after the first vaccination. The importance of the second vaccination was indicated as the first vaccination only induced low levels of pseudovirus neutralization activity. The 100 μg dose elicited higher binding and NAb titers in all participants, supporting the use of this dose during the Phase III trial [293,295]. Interim data from the ongoing Phase III study involving 30,000 participants, indicated that mRNA-1273 is 94.5% effective in preventing COVID-19. From 95 confirmed infections, 90 were in the placebo group and only 8 were in the vaccinated group [298]. 11 confirmed severe cases were all in the placebo group. These results are extremely promising, especially for such novel untested technology.
Several RNA vaccine candidates have been developed by BioNTech, Germany in collaboration with Fosun Pharma, China and Pfizer, U.S.: BNT162a1, b1, b2, and c2. Two encode the RBD of the SARS-CoV-2 S protein, and two encode the larger S protein (Table 6). Altered forms of conventional RNA-based vaccine platforms, including saRNA and modified mRNA, may improve efficacy. Specifically, saRNA vaccines confer higher expression levels of the antigenic target [84], while modified mRNA incorporating pseudouridines can suppress adaptive immune response stimulation, circumventing RNA degradation and increasing efficacy [299]. BNT162b1, is a modified mRNA vaccine encoding the SARS-CoV-2 S RBD. In Phase I/II trials, the vaccine increased RBD-specific IgG and SARS-CoV-2 NAb levels after two doses, although participants did develop dose-dependent mild to moderate local and systemic AEs [300]. BNT162b2 encodes the full S protein stabilized in the prefusion conformation. In a Phase I trial comparing the two, both BNT162b1 and BNT162b2 were able to elicit dose-dependent NAb responses, but BNT162b2 produced fewer and less severe AEs [301]. In addition to Ab responses, a Phase I/II trial administering BNT162b1 demonstrated enhanced cell-mediated responses, evidenced by increased CD4+ and CD8+ T-cell expansion which augmented IFN-γ production [302]. The safe and effective stimulation of both cell-mediated and humoral immune responses confirmed in the aforementioned trials greatly supported the progression into Phase III trials. Interim data from Phase III trials indicated that BNT162b2 was approximately 95% effective in preventing COVID-19 [10,303]. From 170 confirmed infections, 162 were in the placebo group and 8 in the vaccinated group [10]. 9 out of 10 confirmed severe cases were in the placebo group [10,303].
Early December, the mRNA vaccine candidate, BNT162b2, was approved in the UK, making it the first RNA-based vaccine to ever be approved [10]. Shortly after, the U.S. FDA approved BNT162b2 for emergency use authorization [11]. Just a week after, mRNA-1237 was also approved by the U.S. FDA for emergency use authorization [12].
The impact of formulation design on stability is best exemplified in a comparison of the two currently leading RNA-based COVID-19 vaccine candidates: mRNA-1273 and BNT162b1. BNT162b1 must be kept frozen to prevent degradation and requires ultra-cold storage (−70 °C +/− 10 °C) and cold-chain transport. After thawing, BNT162b1 may be stored at 2–8 °C for up to 5 days [302,304]. In contrast, mRNA-1273 has been reported to remain stable at −20 °C for up to six months, at 2–8 °C for 30 days or at room temperature for up to 12 hours [305].
7. Adjuvant administration
Adjuvants are substances that can be employed along with vaccines to ensure immune recognition and enhance the immune response [81,306]. Among the ongoing trials for COVID-19 viral vector vaccines, the use of adjuvants has not been mentioned. Perhaps their use is not necessary, since viral vectors have demonstrated sufficient levels of immunogenicity without adjuvants by mimicking natural infection and activating innate immune responses [56]. Presumably, immune cells can recognize conserved pathogen associated molecular patterns (PAMP) of the viral DNA via toll-like receptors (TLRs), triggering antiviral immunity. Additionally, the viral capsid can be targeted by the adaptive immune response [307,308,309].
For COVID-19 nucleic acid based vaccines, the use of adjuvants has not been explicitly stated. RNA vaccines have demonstrated strong humoral immunity without the use of adjuvants, and can induce immune signals via recognition from PRRs [249,253]. Adjuvancy of LNPs has been suggested, and since many of the COVID-19 RNA vaccines are LNP-encapsulated, including mRNA-1273 and BNT162b2, this nanoparticle could confer additional adjuvant effects [307,310]. In contrast to RNA vaccines, adjuvants are particularly important for DNA vaccines as these vaccines in the past generally stimulate insufficient immune responses in large animals and human studies [81].
Aluminum salt (alum) is one of the most common chemical formulated adjuvants that has been used for conventional vaccine platforms, but it has also been employed in DNA vaccine studies [81]. Unfortunately, it does not appear that alum is effective when administered with DNA vaccines in larger species and humans [81,311]. Another well-known adjuvant, unmethylated CpG deoxynucleotides (ODNs), are rarely found within eukaryotic DNA, but are frequently present in prokaryotic DNA [312,313]. Therefore, plasmid DNA may have an inherent adjuvant effect considering most are produced in bacteria and will contain CpG motifs within prokaryotic elements of the plasmid vector [313]. The CpG-mediated immune response appears to be characterized by production of IL-6, IL-12, IL-18, TNF-ɑ, and IFN (interferon)-γ [314,315,316].
Some adjuvants have been shown to directly alter T-helper (Th) cell responses to maintain balance between Th1 and Th2 responses [306]. Both responses play an important role to protect against pathogens, but they are also involved in immunopathological reactions [317]. Th1 cells are primarily involved in the production of proinflammatory cytokines, whereas Th2 cells produce cytokines which enhance activation of B cells and induce isotype switching [318]. Some respiratory diseases, including SARS-CoV, have been associated with Th2 immunopathology [319], which will be discussed in the next section, suggesting the potential importance of employing adjuvants.
Delta inulin has been previously used for more conventional vaccines, but can balance Th1 and Th2 responses, thus minimizing the risk of Th2 immunopathology [320,321]. In a preclinical study, delta inulin helped alleviate lung immunopathology previously observed in mice after being challenged with SARS-CoV, while additionally enhancing NAb titers [321]. CpG ODNs also prevent the induction of excessive Th2 responses by the induction of IL-10, which supports Th1/Th2 balance [322].
Adjuvants that are specifically designed for IN vaccines may also be suitable for COVID-19 vaccine development, considering IN vaccines can directly target the respiratory tract [111,323]. Protollin, an adjuvant for IN vaccines has shown promising results for respiratory diseases, inducing both systemic and mucosal immune responses [324,325,326]. Stimulation of the immune response by Protollin occurs via TLR2 and TLR4 signaling, enhancing the production of cytokines and IFNs [323].
To combat SARS-CoV-2, the employment of adjuvants to enhance NAbs and antigen-specific T-cell responses is likely required, similar to immune responses required to combat influenza and SARS [327,328,329,330]. Therefore, the choice of a suitable adjuvant for GBVs, particularly DNA vaccines, can boost and stimulate these immune responses appropriately. Furthermore, the implementation of adjuvants may allow for lower vaccine dose administration, thereby enabling the immunization of a higher proportion of the population [323].
8. Immune enhancement of gene-based vaccines
Thus far, COVID-19 GBV candidates have demonstrated acceptable safety profiles in clinical trials. However, a very pressing safety concern in vaccine development has been immune enhancement. For over 60 years, immune backfiring has been observed for viruses such as SARS [319], dengue [331], RSV [332], and HIV [333]. In these cases, vaccination could predispose individuals toward exacerbated disease by priming an immune response that promotes infection or stimulates undesirable cell-mediated immunopathology. It is fundamental to consider these issues in order to polarize responses toward the most appropriate branch of cell-mediated immunity after immunization with COVID-19 GBVs.
8.1. Antibody dependent enhancement
Ab-dependent enhancement (ADE) has been implicated in the exacerbation of what is generally a mild viral manifestation of flu-like symptoms in the majority of SARS-CoV-2 infections. ADE has been demonstrated in previous accounts of dengue virus: individuals recovered from infection by a dengue serotype, while immune to that serotype, were up to 80-fold more likely to experience severe or even lethal symptoms upon secondary infection by a different dengue serotype [334]. While the mechanism is not completely understood, low level titers of non-NAbs produced following primary infection can bind and facilitate secondary infecting virus uptake by macrophages and DCs. Non-NAbs generated against the virus may bind to their viral targets and to receptors on DCs and macrophages, enhancing viral entry into these cells and facilitating replication [335]. In addition, Abs generated in response to original SARS-CoV infection may target counterpart epitopes on SARS-CoV-2 with lower affinity and/or consistency, enhancing the risk of ADE. Some believe that this may explain why some infected individuals with initially mild symptoms may progress to life-threatening respiratory illness.
The influence of ADE on SARS infections is slightly controversial. Although MERS-CoV and SARS-CoV are structurally and genetically similar, MERS-imparted fatality and pathogenicity is more than 3-fold greater than that of SARS (and likely SARS-CoV-2) [336], which can be partly attributed to its receptor, dipeptidyl peptidase 4 (DPP4). DPP4 is found on most host cells, including monocyte derived macrophages and DCs, conferring broad tissue tropism [121,337]. Perhaps the most important distinction between MERS-CoV and SARS-CoV is that MERS-CoV demonstrates productive infection in both these cell types, while for SARS-CoV, and presumably SARS-CoV-2, infection of these cells is abortive. Therefore, MERS-CoV may be more suitable to leverage ADE to facilitate infection than SARS-CoV, which may further explain the difference in mortality between MERS-CoV and the SARS viruses. Similar to MERS, dengue virus is also able to replicate within monocyte-derived macrophages and DCs [338], advocating the role of ADE in the pathogenesis of these viruses.
More recently it has been demonstrated that Abs from individuals infected by SARS-CoV could also block ACE2-mediated infection by SARS-CoV-2 into human lung cells in the absence of ADE [339]. In addition, rhesus macaques infected by SARS-CoV-2 were immune to subsequent reinfection by the virus [340]. Although, these results may suggest a lower likelihood that ADE contributes to the exacerbation of SARS-CoV, correlating SARS-CoV-2 severity with previous exposure to SARS-CoV will be most informative. While a recent study indicated that memory T-cells persist against the N protein in individuals recovered from SARS-CoV [341], this evidence could potentially be just as contributive to immune enhancement as it is to cross-reactive protection. More substantiating data as to whether SARS-CoV recovered individuals are susceptible to ADE is needed.
Vaccine designs that prevent the propagation and persistence of non-NAbs are preferential in mitigating the possibility of ADE against SARS-CoV-2. Strategies that target only critical segments of infection and minimize non-neutralizing epitopes, such as the RBD or highly conserved fusion sequences of the S2 domain, represent preferential alternatives to more conventional whole virus vaccine approaches.
8.2. Th2 immunopathology
Many virologists believe that the more pressing matter around vaccine enhancement is the cell-mediated dysfunction phenomenon of Th2 immunopathology. Upon infection, some viruses activate dysfunctional Th cell responses that impart powerful inflammatory signals and trigger the Ab-mediated complement cascade that can contribute to severe respiratory distress. These types of responses are initiated by dysregulated cytokines – a phenomenon that can be programmed quickly and results in lethal inflammation from influx of granulocytes and production of inappropriate Abs [319]. Viruses known to trigger such responses were first seen in RSVs, but also include SARS/MERS-CoVs, HIV, feline infectious peritonitis virus, and even influenza. More specifically to respiratory symptoms, Th2 immunopathology results in the recruitment of neutrophils and eosinophils to the lung tissue, promoting local inflammation [319]. Although the exact mechanism contributing to Th2 immunopathology is not fully understood, clues do exist to guide vaccine development.
Th2 immunopathology was first observed in the 1960s in preparation of a vaccine against RSV. Upon immunization many children developed severe illness, two of whom died from the vaccination [342]. Similar granulocyte penetration and inflammation have also been observed in RSV animal models [343]. Initial vaccines produced for SARS-CoV demonstrated inflammatory responses in the lungs, similar to those seen following immunization with the RSV vaccine. Tseng et al. [319] tested conventional SARS-CoV vaccine approaches of whole killed virus, subunit S protein and attenuated in mice. In all three approaches, lung inflammation, particularly induced by Th2 immunopathogenic responses and eosinophil and neutrophil accumulation in the lungs was generated. The induction of the memory CD8+ T-cell response is vital to inhibit the generation of a memory Th2 responses that impart pulmonary eosinophilia [344]. However, previous studies in RSV have demonstrated that the production of Th1-associated cytokines by memory CD8+ T-cells and/or memory Th1 cells can also lead to enhanced disease [345]. Human eosinophils express several PRRs including TLR1-5, TLR7 and TLR9 that upon receptor activation stimulate release of cytokines, oxidative burst factors, and other inflammation mediators [346]. Antagonists of TLR7 have been shown to relax airways and reduce eosinophilic inflammation, suggesting that activation of TLR7 in response to viral RNA recognition may contribute to Th2 immunopathology [347]. Furthermore, adjuvants that stimulate the Th2 pathway, appear to protect against Th2 immunopathology [319], as indicated earlier in the review, suggesting that prevention of Th2 immunopathology may not be as simple as polarization of the immune response toward the Th1 pathway.
MERS-CoV similarly induces a cytokine storm upon infection, which may be attributed to its ability to replicate within T-cells and DCs [348,349]. Again, the DPP4 target of MERS-CoV is implicated considering it is involved in T-cell activation and signal transduction and has been shown to induce activation of apoptotic pathways [349] that likely contributes to a dysfunctional cytokine profile. DDP4-transduced T-cell deficient mice demonstrated persistence of MERS-CoV in lungs following infection compared to wildtype, suggesting that T-cells are involved in pathogenesis of the virus [350].
The use of nucleic acids to encode limited and neutralizing sequences further stimulates protective and cross-reactive T-cell responses that with appropriate adjuvants polarize Th responses and cytokine profiles toward Th1 cytotoxic T-cell immunity and accompanying Ab responses, as opposed to Th2 responses that may impart unwanted Th2-mediated allergic inflammation.
T-cell memory is critical in generating cross-reactive protection against COVID-19. Cross-reactive memory T-cells generated in 18 individuals recovered from SARS-CoV and even other cold-causing betacoronaviruses are in fact protective against SARS-CoV-2 infection and may be the main reason for the wide differences in manifestation and pathogenicity seen between infected individuals [341]. Recent preclinical investigations of a universal influenza DNA vaccine demonstrate powerful cross-reactive protection against influenza serotypes in ferrets in the absence of remarkable Ab responses [351]. Thus, GBVs that can mimic endogenous viral protein production, minimize non-neutralizing epitopes, and adequately balance Th1 and Th2 responses and corresponding memory may enhance efficacy, durability and cross-reactiveness of vaccines, while further preventing vaccine enhancement.
9. Conclusions
The rapid worldwide dissemination of COVID-19 has emphasized the imperative need to develop an efficacious vaccine to prevent infection, disease, or transmission. Collaborative efforts around the world have contributed to unprecedented rapid vaccine development. GBVs encompass a significant proportion of COVID-19 vaccine candidates. Its quick development was feasible due to key aspects of previous GBVs studies including antigen selection, route of administration, delivery methods and dosage, among others.
Previous GBV developments certainly support the high potential of these platforms. However, they exert different immunological pros and cons. Viral vector vaccines are able to confer high levels of immunogenicity, although pre-existing vector immunity can limit their effectiveness. While DNA vaccines have demonstrated adequate immunogenicity in animals, they have yet to replicate this satisfactorily in humans. RNA vaccines may confer desirable immunological properties and significant benefits over DNA. Issues due to the unstable nature of RNA have been overcome by employing suitable formulations and storage strategies to protect from degradation.
Clinical study results for the COVID-19 GBVs outlined in this paper demonstrate a good immunization profile and for some, very high efficacy. However, further trials to analyze the efficacy and safety in high-risk groups are needed. So far, two COVID-19 viral vector vaccines were approved in their countries of origin. The Ad5-nCoV was approved by the Chinese government for use in their armed forces, in addition to Gam-COVID-Vac by the Ministry of Health of Russia for emergency use authorization. Furthermore, the University of Oxford and AstraZeneca have submitted data to regulators around the world for the approval of the AZD1222 viral vector vaccine. None of the COVID-19 DNA vaccines have published their Phase I clinical trial results or have been approved. BNT16b2 (BioNTech/Fosun Pharma/Pfizer) is the first ever RNA vaccine authorized by the U.S. FDA for the prevention of an infectious disease. A week after, the mRNA-1237 vaccine (Moderna Therapeutics/NIAID) was also authorized by the U.S FDA for emergency use. This could promote a new era for such novel technology.
The safety of vaccines is also of paramount importance and cannot be compromised for higher efficacy. A follow-up of the participants should continue. During this time, the induction of ADE and lung immunopathology must be assessed, especially considering these unfavorable events have occurred in previous coronaviruses and other respiratory-related disease models.
Finally, there are additional challenges that the available COVID-19 vaccines need to address including durability of protection, viral transmission prevention, efficacy within specific subgroups of the population, and public acceptance. In 2021, it is highly probable that COVID-19 vaccines will be globally accessible, as a result of extensive investment and diverse input from scientific communities around the world.
Ad: adenovirus; ChAdOx1:chimpanzee adenovirus-vectored vaccine; ETSD: enhanced T-cell stimulation domain; hAd: human adenovirus; HD: high dose; IFN-y: interferon gamma; Ig: immunoglobulin; IM: intramuscular; IN: intranasal; IU: International Unit; LD: low dose; MOH: Ministry of Health; MV: measles virus; MVA: modified vaccinia ankara; n: Number of participants; N: nucleoprotein; NAbs: neutralizing antibodies; PFU: plaque-forming unit; rAd: recombinant adenovirus; RBD: receptor binding domain; SAE: serious adverse event; S: spike; SC: subcutaneous; SD: standard dose; SUSAR: Suspected Unexpected Serious Adverse Reaction; vp: viral particles; VSV: vesicular stomatitis virus.
Acknowledgments
Acknowledgements
This work was supported in part by the National Sciences and Engineering Research Council of Canada [grant number 391457], Mitacs Canada, CONACYT Mexico, and Taibah University, Saudi Arabia.
Declaration of Interests
None.
References
- 1.WHO Coronavirus Disease (COVID-19) Dashboard 2020. https://covid19.who.int
- 2.Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K.-Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin M.D., Shukla S., Chung Y.H., Beiss V., Chan S.K., Ortega-Rivera O.A., et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020;15:646–655. doi: 10.1038/s41565-020-0737-y. [DOI] [PubMed] [Google Scholar]
- 5.Petersen E., Koopmans M., Go U., Hamer D.H., Petrosillo N., Castelli F., et al. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020;20:e238–e244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Draft Landscape of COVID-19 Candidate Vaccines 2020. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines
- 7.García-Sastre A., Mena I. Novel vaccine strategies against emerging viruses. Curr. Opin. Virol. 2013;3:210–216. doi: 10.1016/j.coviro.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallis J., Shenton D.P., Carlisle R.C. Novel approaches for the design, delivery and administration of vaccine technologies. Clin. Exp. Immunol. 2019;196:189–204. doi: 10.1111/cei.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rauch S., Jasny E., Schmidt K.E., Petsch B. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahase E. Covid-19: UK approves Pfizer and BioNTech vaccine with rollout due to start next week. BMJ. 2020;371 doi: 10.1136/bmj.m4714. [DOI] [PubMed] [Google Scholar]
- 11.O. of the Commissioner . FDA; 2020. Pfizer-BioNTech COVID-19 Vaccine.https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/pfizer-biontech-covid-19-vaccine [Google Scholar]
- 12.Moderna Announces FDA Authorization of Moderna COVID-19 Vaccine in the U.S. Moderna, Inc.; 2019. https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-fda-authorization-moderna-covid-19-vaccine-us [Google Scholar]
- 13.Lim M., Badruddoza A.Z.M., Firdous J., Azad M., Mannan A., Al-Hilal T.A., et al. Engineered nanodelivery systems to improve DNA vaccine technologies. Pharmaceutics. 2020;12 doi: 10.3390/pharmaceutics12010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson D.X., Ljungberg K., Kakoulidou M., Liljeström P. Intradermal electroporation of naked replicon RNA elicits strong immune responses. PLoS One. 2012;7 doi: 10.1371/journal.pone.0029732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodríguez-Gascón A., del Pozo-Rodríguez A., Solinís M.Á. Development of nucleic acid vaccines: use of self-amplifying RNA in lipid nanoparticles. Int. J. Nanomed. 2014;9:1833–1843. doi: 10.2147/IJN.S39810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geall A.J., Mandl C.W., Ulmer J.B. RNA: the new revolution in nucleic acid vaccines. Semin. Immunol. 2013;25:152–159. doi: 10.1016/j.smim.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Liu M.A. A comparison of plasmid DNA and mRNA as vaccine technologies. Vaccines. 2019;7 doi: 10.3390/vaccines7020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet Lond. Engl. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong E., Wong M.U., Huffman A., He Y. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. BioRxiv. 2020 doi: 10.1101/2020.03.20.000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flower D.R., Macdonald I.K., Ramakrishnan K., Davies M.N., Doytchinova I.A. Computer aided selection of candidate vaccine antigens. Immun. Res. 2010;6 doi: 10.1186/1745-7580-6-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rueckert C., Guzmán C.A. Vaccines: from empirical development to rational design. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liljeroos L., Malito E., Ferlenghi I., Bottomley M.J. Structural and computational biology in the design of immunogenic vaccine antigens. J. Immunol. Res. 2015;2015 doi: 10.1155/2015/156241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delany I., Rappuoli R., Seib K.L. Vaccines, reverse vaccinology, and bacterial pathogenesis. Cold Spring Harb. Perspect. Med. 2013;3 doi: 10.1101/cshperspect.a012476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seib K.L., Zhao X., Rappuoli R. Developing vaccines in the era of genomics: a decade of reverse vaccinology. Clin. Microbiol. Infect. 2012;18:109–116. doi: 10.1111/j.1469-0691.2012.03939.x. [DOI] [PubMed] [Google Scholar]
- 25.Nabel G.J. Designing tomorrow's vaccines. N. Engl. J. Med. 2013;368:551–560. doi: 10.1056/NEJMra1204186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vangelista L., Secchi M. Prepare for the future: dissecting the spike to seek broadly neutralizing antibodies and universal vaccine for pandemic coronaviruses. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19 doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., et al. Drug targets for corona virus: a systematic review. Indian J. Pharmacol. 2020;52:56–65. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Mil. Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ortiz-Prado E., Simbaña-Rivera K., Gómez- Barreno L., Rubio-Neira M., Guaman L.P., Kyriakidis N.C., et al. Clinical, molecular, and epidemiological characterization of the SARS-CoV-2 virus and the Coronavirus Disease 2019 (COVID-19), a comprehensive literature review. Diagn. Microbiol. Infect. Dis. 2020;98 doi: 10.1016/j.diagmicrobio.2020.115094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F., Kream R.M., Stefano G.B. An evidence based perspective on mRNA-SARS-CoV-2 vaccine development. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020;26:e924700-1–e924700-8. doi: 10.12659/MSM.924700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., et al. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV — a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin J.E., Louder M.K., Holman L.A., Gordon I.J., Enama M.E., Larkin B.D., et al. A SARS DNA vaccine induces neutralizing antibody and cellular immune responses in healthy adults in a phase I clinical trial. Vaccine. 2008;26:6338–6343. doi: 10.1016/j.vaccine.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang J., Cao Y., Du J., Bu X., Ma R., Wu C. Priming with SARS CoV S DNA and boosting with SARS CoV S epitopes specific for CD4+ and CD8+ T cells promote cellular immune responses. Vaccine. 2007;25:6981–6991. doi: 10.1016/j.vaccine.2007.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchholz U.J., Bukreyev A., Yang L., Lamirande E.W., Murphy B.R., Subbarao K., Collins P.L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. 2004;101:9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z.-Y., Kong W.-P., Huang Y., Roberts A., Murphy B.R., Subbarao K., et al. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L., Shi W., Chappell J.D., Joyce M.G., Zhang Y., Kanekiyo M., Becker M.M., et al. Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on the Middle East Respiratory Syndrome Coronavirus spike glycoprotein to avoid neutralization escape. J. Virol. 2018;92 doi: 10.1128/JVI.02002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao Z., Liu L., Du L., Zhang C., Jiang S., Li T., He Y. Potent and persistent antibody responses against the receptor-binding domain of SARS-CoV spike protein in recovered patients. Virol. J. 2010;7 doi: 10.1186/1743-422X-7-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dutta N.K., Mazumdar K., Gordy J.T. The Nucleocapsid protein of SARS–CoV-2: a target for vaccine development. J. Virol. 2020;94 doi: 10.1128/JVI.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Y., Liu M., Zhao W., Zhang J., Zhang X., Wang K., et al. Isolation of virus from a SARS patient and genome-wide analysis of genetic mutations related to pathogenesis and epidemiology from 47 SARS-CoV isolates. Virus Genes. 2005;30:93–102. doi: 10.1007/s11262-004-4586-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cong Y., Ulasli M., Schepers H., Mauthe M., V'kovski P., Kriegenburg F., et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 2020;94 doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee H.K., Lee B.H., Dutta N.K., Seok S.H., Baek M.W., Lee H.Y., et al. Detection of antibodies against SARS-coronavirus using recombinant truncated nucleocapsid proteins by ELISA. J. Microbiol. Biotechnol. 2008;18:1717–1721. [PubMed] [Google Scholar]
- 49.Peng H., Yang L., Li J., Lu Z., Wang L., Koup R.A., Bailer R.T., et al. Human memory T cell responses to SARS-CoV E protein. Microbes Infect. 2006;8:2424–2431. doi: 10.1016/j.micinf.2006.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J., Sun Y., Qi J., Chu F., Wu H., Gao F., et al. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J. Infect. Dis. 2010;202:1171–1180. doi: 10.1086/656315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pang H., Liu Y., Han X., Xu Y., Jiang F., Wu D., et al. Protective humoral responses to severe acute respiratory syndrome-associated coronavirus: implications for the design of an effective protein-based vaccine. J. Gen. Virol. 2004;85:3109–3113. doi: 10.1099/vir.0.80111-0. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z., Yuan Z., Matsumoto M., Hengge U.R., Chang Y.-F. Immune responses with DNA vaccines encoded different gene fragments of severe acute respiratory syndrome coronavirus in BALB/c mice. Biochem. Biophys. Res. Commun. 2005;327:130–135. doi: 10.1016/j.bbrc.2004.11.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salvatori G., Luberto L., Maffei M., Aurisicchio L., Roscilli G., Palombo F., et al. SARS-CoV-2 SPIKE PROTEIN: an optimal immunological target for vaccines. J. Transl. Med. 2020;18:222. doi: 10.1186/s12967-020-02392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. Biochemistry. 2020;181:281–291.e6. doi: 10.1101/2020.02.19.956581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan S., Nakajima R., Jain A., de Assis R.R., Jasinskas A., Obiero J.M., et al. Analysis of serologic cross-reactivity between common human coronaviruses and SARS-CoV-2 using coronavirus antigen microarray. BioRxiv. 2020 doi: 10.1101/2020.03.24.006544. [DOI] [Google Scholar]
- 56.Ura T., Okuda K., Shimada M. Developments in viral vector-based vaccines. Vaccines. 2014;2:624–641. doi: 10.3390/vaccines2030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sebastian S., Lambe T. Clinical advances in viral-vectored influenza vaccines. Vaccines. 2018;6 doi: 10.3390/vaccines6020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Riel D., de Wit E. Next-generation vaccine platforms for COVID-19. Nat. Mater. 2020;19:810–812. doi: 10.1038/s41563-020-0746-0. [DOI] [PubMed] [Google Scholar]
- 59.Robert-Guroff M. Replicating and non-replicating viral vectors for vaccine development. Curr. Opin. Biotechnol. 2007;18:546–556. doi: 10.1016/j.copbio.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi Y., Chang J. Viral vectors for vaccine applications. Clin. Exp. Vaccine Res. 2013;2:97–105. doi: 10.7774/cevr.2013.2.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinschewer D.D. Virally vectored vaccine delivery: medical needs, mechanisms, advantages and challenges. Swiss Med. Wkly. 2017;147 doi: 10.4414/smw.2017.14465. [DOI] [PubMed] [Google Scholar]
- 62.Lee C.S., Bishop E.S., Zhang R., Yu X., Farina E.M., Yan S., et al. Adenovirus-mediated gene delivery: potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 2017;4:43–63. doi: 10.1016/j.gendis.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stasiak A.C., Stehle T. Human adenovirus binding to host cell receptors: a structural view. Med. Microbiol. Immunol. (Berl.) 2020;209:325–333. doi: 10.1007/s00430-019-00645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Colloca S., Folgori A., Ammendola V., Capone S., Cirillo A., Siani L., et al. Generation and screening of a large collection of novel simian Adenovirus allows the identification of vaccine vectors inducing potent cellular immunity in humans. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wold W.S.M., Toth K. Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr. Gene Ther. 2013;13:421–433. doi: 10.2174/1566523213666131125095046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crommelin D.J.A., Volkin D.B., Hoogendoorn K.H., Lubiniecki A.S., Jiskoot W. The science is there: key considerations for stabilizing viral vector-based Covid-19 vaccines. J. Pharm. Sci. 2020;3549:30744–30749. doi: 10.1016/j.xphs.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rexroad J., Wiethoff C.M., Green A.P., Kierstead T.D., Scott M.O., Middaugh C.R. Structural stability of adenovirus type 5. J. Pharm. Sci. 2003;92:665–678. doi: 10.1002/jps.10340. [DOI] [PubMed] [Google Scholar]
- 68.Evans R.K., Nawrocki D.K., Isopi L.A., Williams D.M., Casimiro D.R., Chin S., et al. Development of stable liquid formulations for adenovirus-based vaccines. J. Pharm. Sci. 2004;93:2458–2475. doi: 10.1002/jps.20157. [DOI] [PubMed] [Google Scholar]
- 69.Pelliccia M., Andreozzi P., Paulose J., D'Alicarnasso M., Cagno V., Donalisio M., et al. Additives for vaccine storage to improve thermal stability of adenoviruses from hours to months. Nat. Commun. 2016;7 doi: 10.1038/ncomms13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bell K.N., Hogue C.J., Manning C., Kendal A.P. Risk factors for improper vaccine storage and handling in private provider offices. Pediatrics. 2001;107 doi: 10.1542/peds.107.6.e100. [DOI] [PubMed] [Google Scholar]
- 71.Jones K.L., Drane D., Gowans E.J. Long-term storage of DNA-free RNA for use in vaccine studies. BioTechniques. 2007;43:675–681. doi: 10.2144/000112593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen Y., Liao Q., Chen T., Zhang Y., Yuan W., Xu J., Zhang X. Freeze-drying formulations increased the adenovirus and poxvirus vaccine storage times and antigen stabilities. Virol. Sin. 2020:1–8. doi: 10.1007/s12250-020-00250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kraan H., van Herpen P., Kersten G., Amorij J.-P. Development of thermostable lyophilized inactivated polio vaccine. Pharm. Res. 2014;31:2618–2629. doi: 10.1007/s11095-014-1359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Croyle M.A., Cheng X., Wilson J.M. Development of formulations that enhance physical stability of viral vectors for gene therapy. Gene Ther. 2001;8:1281–1290. doi: 10.1038/sj.gt.3301527. [DOI] [PubMed] [Google Scholar]
- 75.Leitner W.W., Ying H., Restifo N.P. DNA and RNA-based vaccines: principles, progress and prospects. Vaccine. 1999;18:765–777. doi: 10.1016/s0264-410x(99)00271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams J.A. Vector design for improved DNA vaccine efficacy, safety and production. Vaccines. 2013;1:225–249. doi: 10.3390/vaccines1030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vogel F.R., Sarver N. Nucleic acid vaccines. Clin. Microbiol. Rev. 1995;8:406–410. doi: 10.1128/cmr.8.3.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ferreira G.N.M., Monteiro G.A., Prazeres D.M.F., Cabral J.M.S., Ferreira G.N.M., Monteiro G.A., et al. Downstream processing of plasmid DNA for gene therapy and DNA vaccine applications. Trends Biotechnol. 2000;18:380–388. doi: 10.1016/S0167-7799(00)01475-X. [DOI] [PubMed] [Google Scholar]
- 79.Prazeres D.M.F., Ferreira G.N.M., Monteiro G.A., Cooney C.L., Cabral J.M.S., Prazeres D.M.F., et al. Large-scale production of pharmaceutical-grade plasmid DNA for gene therapy: problems and bottlenecks. Trends Biotechnol. 1999;17:169–174. doi: 10.1016/S0167-7799(98)01291-8. [DOI] [PubMed] [Google Scholar]
- 80.Suschak J.J., Williams J.A., Schmaljohn C.S. Advancements in DNA vaccine vectors, non-mechanical delivery methods, and molecular adjuvants to increase immunogenicity. Hum. Vaccin. Immunother. 2017;13:2837–2848. doi: 10.1080/21645515.2017.1330236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grunwald T., Ulbert S. Improvement of DNA vaccination by adjuvants and sophisticated delivery devices: vaccine-platforms for the battle against infectious diseases. Clin. Exp. Vaccine Res. 2015;4(1):10. doi: 10.7774/cevr.2015.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schlake T., Thess A., Fotin-Mleczek M., Kallen K.-J. Developing mRNA-vaccine technologies. RNA Biol. 2012;9:1319–1330. doi: 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang C., Maruggi G., Shan H., Li J. Advances in mRNA vaccines for infectious diseases. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wadhwa A., Aljabbari A., Lokras A., Foged C., Thakur A. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics. 2020;12 doi: 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu S., Yang K., Li R., Zhang L. mRNA vaccine era—mechanisms, drug platform and clinical prospection. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21186582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gallie D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 88.Zeng C., Zhang C., Walker P.G., Dong Y. Springer; Berlin, Heidelberg: 2020. Formulation and Delivery Technologies for mRNA Vaccines; pp. 1–40. (n.d.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Midoux P., Pichon C. Lipid-based mRNA vaccine delivery systems. Expert Rev. Vaccin. 2015;14:221–234. doi: 10.1586/14760584.2015.986104. [DOI] [PubMed] [Google Scholar]
- 90.Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., et al. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. U. S. A. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Espeseth A.S., Cejas P.J., Citron M.P., Wang D., DiStefano D.J., Callahan C., et al. Modified mRNA/lipid nanoparticle-based vaccines expressing respiratory syncytial virus F protein variants are immunogenic and protective in rodent models of RSV infection. NPJ Vaccin. 2020;5 doi: 10.1038/s41541-020-0163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zabner J., Fasbender A.J., Moninger T., Poellinger K.A., Welsh M.J. Cellular and molecular barriers to gene transfer by a cationic lipid. J. Biol. Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- 93.Matsui H., Johnson L.G., Randell S.H., Boucher R.C. Loss of binding and entry of liposome-DNA complexes decreases transfection efficiency in differentiated airway epithelial cells. J. Biol. Chem. 1997;272:1117–1126. doi: 10.1074/jbc.272.2.1117. [DOI] [PubMed] [Google Scholar]
- 94.Zhou X., Huang L. DNA transfection mediated by cationic liposomes containing lipopolylysine: characterization and mechanism of action. Biochim. Biophys. Acta. 1994;1189:195–203. doi: 10.1016/0005-2736(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 95.Sahay G., Querbes W., Alabi C., Eltoukhy A., Sarkar S., Zurenko C., et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wittrup A., Ai A., Liu X., Hamar P., Trifonova R., Charisse K., et al. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 2015;33:870–876. doi: 10.1038/nbt.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shete H.K., Prabhu R.H., Patravale V.B. Endosomal escape: a bottleneck in intracellular delivery. J. Nanosci. Nanotechnol. 2014;14:460–474. doi: 10.1166/jnn.2014.9082. [DOI] [PubMed] [Google Scholar]
- 98.Cupic K.I., Rennick J.J., Johnston A.P., Such G.K. Controlling endosomal escape using nanoparticle composition: current progress and future perspectives. Nanomedicine. 2018;14:215–223. doi: 10.2217/nnm-2018-0326. [DOI] [PubMed] [Google Scholar]
- 99.Behr J.-P. The proton sponge: a trick to enter cells the viruses did not exploit. CHIMIA Int. J. Chem. 1997;51:34–36. [Google Scholar]
- 100.Lam A.P., Dean D.A. Progress and prospects: nuclear import of nonviral vectors. Gene Ther. 2010;17:439–447. doi: 10.1038/gt.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ludtke J.J., Zhang G., Sebestyén M.G., Wolff J.A. A nuclear localization signal can enhance both the nuclear transport and expression of 1 kb DNA. J. Cell Sci. 1999;112(Pt 12):2033–2041. doi: 10.1242/jcs.112.12.2033. [DOI] [PubMed] [Google Scholar]
- 102.Middaugh C.R., Evans R.K., Montgomery D.L., Casimiro D.R. Analysis of plasmid DNA from a pharmaceutical perspective. J. Pharm. Sci. 1998;87:130–146. doi: 10.1021/js970367a. [DOI] [PubMed] [Google Scholar]
- 103.Anchordoquy T.J., Molina M.C. Preservation of DNA. Cell Preserv. Technol. 2007;5:180–188. doi: 10.1089/cpt.2007.0511. [DOI] [Google Scholar]
- 104.Walther W., Schmeer M., Kobelt D., Baier R., Harder A., Walhorn V., et al. A seven-year storage report of good manufacturing practice–grade naked plasmid DNA: stability, topology, and in vitro/in vivo functional analysis. Hum. Gene Ther. Clin. Dev. 2013;24:147–153. doi: 10.1089/humc.2013.067. [DOI] [PubMed] [Google Scholar]
- 105.Murakami M. Evaluation of DNA plasmid storage conditions, open. Biotechnol. J. 2013;7 https://benthamopen.com/ABSTRACT/TOBIOTJ-7-10 [Google Scholar]
- 106.Evans R.K., Xu Z., Bohannon K.E., Wang B., Bruner M.W., Volkin D.B. Evaluation of degradation pathways for plasmid DNA in pharmaceutical formulations via accelerated stability studies. J. Pharm. Sci. 2000;89:76–87. doi: 10.1002/(SICI)1520-6017(200001)89:1<76::AID-JPS8>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 107.Anchordoquy T.J., Koe G.S. Physical stability of nonviral plasmid‐based therapeutics. J. Pharm. Sci. 2000;89:289–296. doi: 10.1002/(SICI)1520-6017(200003)89:3<289::AID-JPS1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 108.Zelphati O., Nguyen C., Ferrari M., Felgner J., Tsai Y., Felgner P.L. Stable and monodisperse lipoplex formulations for gene delivery. Gene Ther. 1998;5:1272–1282. doi: 10.1038/sj.gt.3300707. [DOI] [PubMed] [Google Scholar]
- 109.Anchordoquy T.J., Carpenter J.F., Kroll D.J. Maintenance of transfection rates and physical characterization of lipid/DNA complexes after freeze-drying and rehydration. Arch. Biochem. Biophys. 1997;348:199–206. doi: 10.1006/abbi.1997.0385. [DOI] [PubMed] [Google Scholar]
- 110.Kasper J.C., Troiber C., Küchler S., Wagner E., Friess W. Formulation development of lyophilized, long-term stable siRNA/oligoaminoamide polyplexes. Eur. J. Pharm. Biopharm. 2013;85:294–305. doi: 10.1016/j.ejpb.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 111.Miquel‐Clopés A., Bentley E.G., Stewart J.P., Carding S.R. Mucosal vaccines and technology. Clin. Exp. Immunol. 2019;196:205–214. doi: 10.1111/cei.13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang L., Wang W., Wang S. Effect of vaccine administration modality on immunogenicity and efficacy. Expert Rev. Vaccin. 2015;14:1509–1523. doi: 10.1586/14760584.2015.1081067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.De Magistris M.T. Mucosal delivery of vaccine antigens and its advantages in pediatrics. Adv. Drug Deliv. Rev. 2006;58:52–67. doi: 10.1016/j.addr.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 114.Hellfritzsch M., Scherließ R. Mucosal vaccination via the respiratory tract. Pharmaceutics. 2019;11 doi: 10.3390/pharmaceutics11080375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li Y., Jin L., Chen T. The effects of secretory IgA in the mucosal immune system. Biomed. Res. Int. 2020;2020:1–6. doi: 10.1155/2020/2032057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chang S.-Y., Ko H.-J., Kweon M.-N. Mucosal dendritic cells shape mucosal immunity. Exp. Mol. Med. 2014;46:e84. doi: 10.1038/emm.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Farris E., Brown D.M., Ramer-Tait A.E., Pannier A.K. Micro- and nanoparticulates for DNA vaccine delivery. Exp. Biol. Med. 2016;241:919–929. doi: 10.1177/1535370216643771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hobson P., Barnfield C., Barnes A., Klavinskis L.S. Mucosal immunization with DNA vaccines. Methods San Diego Calif. 2003;31:217–224. doi: 10.1016/s1046-2023(03)00139-7. [DOI] [PubMed] [Google Scholar]
- 119.Dudhani A.R., Kosaraju S.L. Bioadhesive chitosan nanoparticles: preparation and characterization. Carbohydr. Polym. 2010;81:243–251. doi: 10.1016/j.carbpol.2010.02.026. [DOI] [Google Scholar]
- 120.Sadeghi A.M.M., Dorkoosh F.A., Avadi M.R., Weinhold M., Bayat A., Delie F., et al. Permeation enhancer effect of chitosan and chitosan derivatives: comparison of formulations as soluble polymers and nanoparticulate systems on insulin absorption in Caco-2 cells. Eur. J. Pharm. Biopharm. 2008;70:270–278. doi: 10.1016/j.ejpb.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 121.Tatsis N., Ertl H.C.J. Adenoviruses as vaccine vectors. Mol. Ther. J. Am. Soc. Gene Ther. 2004;10:616–629. doi: 10.1016/j.ymthe.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xiang Z.Q., Yang Y., Wilson J.M., Ertl H.C. A replication-defective human adenovirus recombinant serves as a highly efficacious vaccine carrier. Virology. 1996;219:220–227. doi: 10.1006/viro.1996.0239. [DOI] [PubMed] [Google Scholar]
- 123.Sharpe S., Fooks A., Lee J., Hayes K., Clegg C., Cranage M. Single oral immunization with replication deficient recombinant adenovirus elicits long-lived transgene-specific cellular and humoral immune responses. Virology. 2002;293:210–216. doi: 10.1006/viro.2001.1281. [DOI] [PubMed] [Google Scholar]
- 124.Vela Ramirez J.E., Sharpe L.A., Peppas N.A. Current state and challenges in developing oral vaccines. Adv. Drug Deliv. Rev. 2017;114:116–131. doi: 10.1016/j.addr.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xu Y., Yuen P.-W., Lam J.K.-W. Intranasal DNA vaccine for protection against respiratory infectious diseases: the delivery perspectives. Pharmaceutics. 2014;6:378–415. doi: 10.3390/pharmaceutics6030378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Torrieri-Dramard L., Lambrecht B., Ferreira H.L., Van den Berg T., Klatzmann D., Bellier B. Intranasal DNA vaccination induces potent mucosal and systemic immune responses and cross-protective immunity against influenza viruses. Mol. Ther. 2011;19:602–611. doi: 10.1038/mt.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhuang X., Qi Y., Wang M., Yu N., Nan F., Zhang H., et al. mRNA vaccines encoding the HA protein of influenza a H1N1 virus delivered by cationic lipid nanoparticles induce protective immune responses in mice. Vaccines. 2020;8:123. doi: 10.3390/vaccines8010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li M., Zhao M., Fu Y., Li Y., Gong T., Zhang Z., Sun X. Enhanced intranasal delivery of mRNA vaccine by overcoming the nasal epithelial barrier via intra- and paracellular pathways. J. Control. Release. 2016;228:9–19. doi: 10.1016/j.jconrel.2016.02.043. [DOI] [PubMed] [Google Scholar]
- 129.Lorenzi J.C., Trombone A.P., Rocha C.D., Almeida L.P., Lousada R.L., Malardo T., et al. Intranasal vaccination with messenger RNA as a new approach in gene therapy: use against tuberculosis. BMC Biotechnol. 2010;10:77. doi: 10.1186/1472-6750-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reichmuth A.M., Oberli M.A., Jaklenec A., Langer R., Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lobaina Mato Y. Nasal route for vaccine and drug delivery: features and current opportunities. Int. J. Pharm. 2019;572 doi: 10.1016/j.ijpharm.2019.118813. [DOI] [PubMed] [Google Scholar]
- 132.Illum L. Nasal drug delivery—possibilities, problems and solutions. J. Control. ReleaseRelease Soc. 2003;87:187–198. doi: 10.1016/s0168-3659(02)00363-2. [DOI] [PubMed] [Google Scholar]
- 133.Damjanovic D., Zhang X., Mu J., Fe Medina M., Xing Z. Organ distribution of transgene expression following intranasal mucosal delivery of recombinant replication-defective adenovirus gene transfer vector. Genet. Vaccin. Ther. 2008;6:5. doi: 10.1186/1479-0556-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lemiale F., Kong W., Akyürek L.M., Ling X., Huang Y., Chakrabarti B.K., et al. Enhanced mucosal immunoglobulin a response of intranasal adenoviral vector human immunodeficiency virus vaccine and localization in the central nervous system. J. Virol. 2003;77:10078–10087. doi: 10.1128/JVI.77.18.10078-10087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang J., Thorson L., Stokes R.W., Santosuosso M., Huygen K., Zganiacz A., et al. Single mucosal, but not parenteral, immunization with recombinant adenoviral-based vaccine provides potent protection from pulmonary tuberculosis. J. Immunol. Baltim. Md. 2004;1950(173):6357–6365. doi: 10.4049/jimmunol.173.10.6357. [DOI] [PubMed] [Google Scholar]
- 136.Xing Z., Lichty B.D. Use of recombinant virus-vectored tuberculosis vaccines for respiratory mucosal immunization. Tuberc. Edinb. Scotl. 2006;86:211–217. doi: 10.1016/j.tube.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 137.Dietrich J., Andersen C., Rappuoli R., Doherty T.M., Jensen C.G., Andersen P. Mucosal administration of Ag85B-ESAT-6 protects against infection with Mycobacterium tuberculosis and boosts prior bacillus Calmette-Guerin immunity. J. Immunol. Baltim. Md. 2006;1950(177):6353–6360. doi: 10.4049/jimmunol.177.9.6353. [DOI] [PubMed] [Google Scholar]
- 138.Van Kampen K.R., Shi Z., Gao P., Zhang J., Foster K.W., Chen D.-T., et al. Safety and immunogenicity of adenovirus-vectored nasal and epicutaneous influenza vaccines in humans. Vaccine. 2005;23:1029–1036. doi: 10.1016/j.vaccine.2004.07.043. [DOI] [PubMed] [Google Scholar]
- 139.Bolhassani A., Yazdi S.R. DNA immunization as an efficient strategy for vaccination. Avicenna J. Med. Biotechnol. 2009;1:71–88. [PMC free article] [PubMed] [Google Scholar]
- 140.Kashem S.W., Haniffa M., Kaplan D.H. Antigen-presenting cells in the skin. Annu. Rev. Immunol. 2017;35:469–499. doi: 10.1146/annurev-immunol-051116-052215. [DOI] [PubMed] [Google Scholar]
- 141.Schwingshackl P., Obermoser G., Nguyen V.A., Fritsch P., Sepp N., Romani N. Distribution and maturation of skin dendritic cell subsets in two forms of cutaneous T-cell lymphoma: mycosis fungoides and Sézary syndrome. Acta Derm. Venereol. 2012;92:269–275. doi: 10.2340/00015555-1220. [DOI] [PubMed] [Google Scholar]
- 142.Lindgren G., Ols S., Liang F., Thompson E.A., Lin A., Hellgren F., et al. Induction of robust B cell responses after influenza mRNA vaccination is accompanied by circulating Hemagglutinin-specific ICOS+ PD-1+ CXCR3+ T follicular helper cells. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cashman K.A., Wilkinson E.R., Shaia C.I., Facemire P.R., Bell T.M., Bearss J.J., et al. A DNA vaccine delivered by dermal electroporation fully protects cynomolgus macaques against Lassa fever. Hum. Vaccin. Immunother. 2017;13:2902–2911. doi: 10.1080/21645515.2017.1356500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Carter C., Houser K.V., Yamshchikov G.V., Bellamy A.R., May J., Enama M.E., et al. Safety and immunogenicity of investigational seasonal influenza hemagglutinin DNA vaccine followed by trivalent inactivated vaccine administered intradermally or intramuscularly in healthy adults: An open-label randomized phase 1 clinical trial. PLoS One. 2019;14 doi: 10.1371/journal.pone.0222178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Haidari G., Cope A., Miller A., Venables S., Yan C., Ridgers H., et al. Combined skin and muscle vaccination differentially impact the quality of effector T cell functions: the CUTHIVAC-001 randomized trial. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-13331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vogt A., Mahé B., Costagliola D., Bonduelle O., Hadam S., Schaefer G., et al. Transcutaneous anti-influenza vaccination promotes both CD4 and CD8 T cell immune responses in humans. J. Immunol. 2008;180:1482–1489. doi: 10.4049/jimmunol.180.3.1482. [DOI] [PubMed] [Google Scholar]
- 147.Combadière B., Vogt A., Mahé B., Costagliola D., Hadam S., Bonduelle O., et al. Preferential amplification of CD8 effector-T cells after transcutaneous application of an inactivated influenza vaccine: a randomized phase I trial. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Meyer J., Harris S.A., Satti I., Poulton I.D., Poyntz H.C., Tanner R., et al. Comparing the safety and immunogenicity of a candidate TB vaccine MVA85A administered by intramuscular and intradermal delivery. Vaccine. 2013;31:1026–1033. doi: 10.1016/j.vaccine.2012.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Abadie V., Bonduelle O., Duffy D., Parizot C., Verrier B., Combadière B. Original encounter with antigen determines antigen-presenting cell imprinting of the quality of the immune response in mice. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lambert P.H., Laurent P.E. Intradermal vaccine delivery: will new delivery systems transform vaccine administration? Vaccine. 2008;26:3197–3208. doi: 10.1016/j.vaccine.2008.03.095. [DOI] [PubMed] [Google Scholar]
- 151.Ols S., Yang L., Thompson E.A., Pushparaj P., Tran K., Liang F., et al. Route of vaccine administration alters antigen trafficking but not innate or adaptive immunity. Cell Rep. 2020;30:3964–3971.e7. doi: 10.1016/j.celrep.2020.02.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liang F., Lindgren G., Lin A., Thompson E.A., Ols S., Röhss J., et al. Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in Rhesus Macaques. Mol. Ther. J. Am. Soc. Gene Ther. 2017;25:2635–2647. doi: 10.1016/j.ymthe.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liang F., Loré K. Local innate immune responses in the vaccine adjuvant-injected muscle. Clin. Transl. Immunol. 2016;5 doi: 10.1038/cti.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Enama M.E., Ledgerwood J.E., Novik L., Nason M.C., Gordon I.J., Holman L., et al. Phase I randomized clinical trial of VRC DNA and rAd5 HIV-1 vaccine delivery by intramuscular (IM), subcutaneous (SC) and intradermal (ID) administration (VRC 011) PLoS One. 2014:9. doi: 10.1371/journal.pone.0091366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hobernik D., Bros M. DNA vaccines—how far from clinical use? Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19113605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ibrahim M.M. Subcutaneous and visceral adipose tissue: structural and functional differences. Obes. Rev. 2010;11:11–18. doi: 10.1111/j.1467-789X.2009.00623.x. [DOI] [PubMed] [Google Scholar]
- 157.Bonnotte B., Gough M., Phan V., Ahmed A., Chong H., Martin F., et al. Intradermal injection, as opposed to subcutaneous injection, enhances immunogenicity and suppresses tumorigenicity of tumor cells. Cancer Res. 2003;63:2145–2149. [PubMed] [Google Scholar]
- 158.Kündig T.M., Johansen P., Foged C., Rades T., Perrie Y., Hook S. Subunit Vaccine Deliv. Springer Verlag; New York: 2015. Parenteral vaccine administration: tried and true; pp. 261–286. [DOI] [Google Scholar]
- 159.Kim H., Park H., Lee S.J. Effective method for drug injection into subcutaneous tissue. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-10110-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gradel A.K.J., Porsgaard T., Lykkesfeldt J., Seested T., Gram-Nielsen S., Kristensen N.R., et al. Factors affecting the absorption of subcutaneously administered insulin: effect on variability. J. Diabetes Res. 2018;2018:1–17. doi: 10.1155/2018/1205121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Morawska L., Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Partidos Intranasal vaccines: forthcoming challenges. Pharm. Sci. Technol. Today. 2000;3:273–281. doi: 10.1016/s1461-5347(00)00281-9. [DOI] [PubMed] [Google Scholar]
- 163.Jeyanathan M., Afkhami S., Smaill F., Miller M.S., Lichty B.D., Xing Z. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020;20:615–632. doi: 10.1038/s41577-020-00434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mellott A.J., Forrest M.L., Detamore M.S. Physical non-viral gene delivery methods for tissue engineering. Ann. Biomed. Eng. 2013;41:446–468. doi: 10.1007/s10439-012-0678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gulce-Iz S., Saglam-Metiner P. Immune Response Activation and Immunomodulation. 2019. Current state of the art in DNA vaccine delivery and molecular adjuvants: Bcl-xL Anti-apoptotic protein as a molecular adjuvant. [DOI] [Google Scholar]
- 166.Jorritsma S.H.T., Gowans E.J., Grubor-Bauk B., Wijesundara D.K. Delivery methods to increase cellular uptake and immunogenicity of DNA vaccines. Vaccine. 2016;34:5488–5494. doi: 10.1016/j.vaccine.2016.09.062. [DOI] [PubMed] [Google Scholar]
- 167.Gaudinski M.R., Houser K.V., Morabito K.M., Hu Z., Yamshchikov G., Rothwell R.S., et al. Safety, tolerability, and immunogenicity of two Zika virus DNA vaccine candidates in healthy adults: randomised, open-label, phase 1 clinical trials. Lancet Lond. Engl. 2018;391:552–562. doi: 10.1016/S0140-6736(17)33105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Alberer M., Gnad-Vogt U., Hong H.S., Mehr K.T., Backert L., Finak G., Gottardo R., Bica M.A., Garofano A., Koch S.D., Fotin-Mleczek M., Hoerr I., Clemens R., von Sonnenburg F. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet Lond. Engl. 2017;390:1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]
- 169.Williams J., Fox-Leyva L., Christensen C., Fisher D., Schlicting E., Snowball M., et al. Hepatitis A vaccine administration: comparison between jet-injector and needle injection. Vaccine. 2000;18:1939–1943. doi: 10.1016/s0264-410x(99)00446-6. [DOI] [PubMed] [Google Scholar]
- 170.Sardesai N.Y., Weiner D.B. Electroporation delivery of DNA vaccines: prospects for success. Curr. Opin. Immunol. 2011;23:421–429. doi: 10.1016/j.coi.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Saroja C., Lakshmi P., Bhaskaran S. Recent trends in vaccine delivery systems: a review. Int. J. Pharm. Investig. 2011;1:64–74. doi: 10.4103/2230-973X.82384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Aihara H., Miyazaki J. Gene transfer into muscle by electroporation in vivo. Nat. Biotechnol. 1998;16:867–870. doi: 10.1038/nbt0998-867. [DOI] [PubMed] [Google Scholar]
- 173.Rizzuto G., Cappelletti M., Maione D., Savino R., Lazzaro D., Costa P., et al. Efficient and regulated erythropoietin production by naked DNA injection and muscle electroporation. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6417–6422. doi: 10.1073/pnas.96.11.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dupuis M., Denis-Mize K., Woo C., Goldbeck C., Selby M.J., Chen M., et al. Distribution of DNA vaccines determines their immunogenicity after intramuscular injection in mice. J. Immunol. 2000;165:2850–2858. doi: 10.4049/jimmunol.165.5.2850. [DOI] [PubMed] [Google Scholar]
- 175.Sokołowska E., Błachnio-Zabielska A.U. A critical review of electroporation as a plasmid delivery system in mouse skeletal muscle. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20112776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yan J., Harris K., Khan A.S., Draghia-Akli R., Sewell D., Weiner D.B. Cellular immunity induced by a novel HPV-18 DNA vaccine encoding an E6/E7 fusion consensus protein in mice and rhesus macaques. Vaccine. 2008;26:5210–5215. doi: 10.1016/j.vaccine.2008.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yan J., Reichenbach D.K., Corbitt N., Hokey D.A., Ramanathan M.P., McKinney K.A., et al. Induction of antitumor immunity in vivo following delivery of a novel HPV-16 DNA vaccine encoding an E6/E7 fusion antigen. Vaccine. 2009;27:431–440. doi: 10.1016/j.vaccine.2008.10.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Vasan S., Hurley A., Schlesinger S.J., Hannaman D., Gardiner D.F., Dugin D.P., et al. In vivo electroporation enhances the immunogenicity of an HIV-1 DNA vaccine candidate in healthy volunteers. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Vasan S., Schlesinger S.J., Huang Y., Hurley A., Lombardo A., Chen Z., et al. Phase 1 safety and immunogenicity evaluation of ADVAX, a multigenic, DNA-based clade C/B' HIV-1 candidate vaccine. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cu Y., Broderick K.E., Banerjee K., Hickman J., Otten G., Barnett S., et al. Enhanced delivery and potency of self-amplifying mRNA vaccines by electroporation in situ. Vaccines. 2013;1:367–383. doi: 10.3390/vaccines1030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Catanzaro A.T., Koup R.A., Roederer M., Bailer R.T., Enama M.E., Moodie Z., et al. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 candidate vaccine delivered by a replication-defective recombinant adenovirus vector. J. Infect. Dis. 2006;194:1638–1649. doi: 10.1086/509258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Humphreys I.R., Sebastian S. Novel viral vectors in infectious diseases. Immunology. 2018;153:1–9. doi: 10.1111/imm.12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhu F.-C., Hou L.-H., Li J.-X., Wu S.-P., Liu P., Zhang G.-R., et al. Safety and immunogenicity of a novel recombinant adenovirus type-5 vector-based Ebola vaccine in healthy adults in China: preliminary report of a randomised, double-blind, placebo-controlled, phase 1 trial. Lancet Lond. Engl. 2015;385:2272–2279. doi: 10.1016/S0140-6736(15)60553-0. [DOI] [PubMed] [Google Scholar]
- 184.Zhu F.-C., Wurie A.H., Hou L.-H., Liang Q., Li Y.-H., Russell J.B.W., et al. Safety and immunogenicity of a recombinant adenovirus type-5 vector-based Ebola vaccine in healthy adults in Sierra Leone: a single-centre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Lond. Engl. 2017;389:621–628. doi: 10.1016/S0140-6736(16)32617-4. [DOI] [PubMed] [Google Scholar]
- 185.Matz K.M., Marzi A., Feldmann H. Ebola vaccine trials: progress in vaccine safety and immunogenicity. Expert Rev. Vaccin. 2019;18:1229–1242. doi: 10.1080/14760584.2019.1698952. [DOI] [PubMed] [Google Scholar]
- 186.Fausther-Bovendo H., Kobinger G.P. Pre-existing immunity against Ad vectors. Hum. Vaccin. Immunother. 2014;10:2875–2884. doi: 10.4161/hv.29594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lemckert A.A.C., Sumida S.M., Holterman L., Vogels R., Truitt D.M., Lynch D.M., et al. Immunogenicity of heterologous prime-boost regimens involving recombinant adenovirus serotype 11 (Ad11) and Ad35 vaccine vectors in the presence of anti-Ad5 immunity. J. Virol. 2005;79:9694–9701. doi: 10.1128/JVI.79.15.9694-9701.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dolzhikova I.V., Zubkova O.V., Tukhvatulin A.I., Dzharullaeva A.S., Tukhvatulina N.M., Shcheblyakov D.V., et al. Safety and immunogenicity of GamEvac-Combi, a heterologous VSV- and Ad5-vectored Ebola vaccine: an open phase I/II trial in healthy adults in Russia. Hum. Vaccin. Immunother. 2017;13:613–620. doi: 10.1080/21645515.2016.1238535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Barouch D.H., Kik S.V., Weverling G.J., Dilan R., King S.L., Maxfield L.F., et al. International seroepidemiology of adenovirus serotypes 5, 26, 35, and 48 in pediatric and adult populations. Vaccine. 2011;29:5203–5209. doi: 10.1016/j.vaccine.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Buchbinder S.P., Mehrotra D.V., Duerr A., Fitzgerald D.W., Mogg R., Li D., et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet Lond. Engl. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yang Y., Nunes F.A., Berencsi K., Furth E.E., Gönczöl E., Wilson J.M. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc. Natl. Acad. Sci. U. S. A. 1994;91:4407. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lozier J.N., Csako G., Mondoro T.H., Krizek D.M., Metzger M.E., Costello R., et al. Toxicity of a first-generation adenoviral vector in rhesus macaques. Hum. Gene Ther. 2002;13:113–124. doi: 10.1089/10430340152712665. [DOI] [PubMed] [Google Scholar]
- 193.Penaloza-MacMaster P., Provine N.M., Ra J., Borducchi E.N., McNally A., Simmons N.L., et al. Alternative serotype adenovirus vaccine vectors elicit memory T cells with enhanced anamnestic capacity compared to Ad5 vectors. J. Virol. 2013;87:1373–1384. doi: 10.1128/JVI.02058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tan W.G., Jin H.-T., West E.E., Penaloza-MacMaster P., Wieland A., Zilliox M.J., et al. Comparative analysis of simian immunodeficiency virus gag-specific effector and memory CD8+ T cells induced by different adenovirus vectors. J. Virol. 2013;87:1359–1372. doi: 10.1128/JVI.02055-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Baden L.R., Walsh S.R., Seaman M.S., Tucker R.P., Krause K.H., Patel A., et al. First-in-human evaluation of the safety and immunogenicity of a recombinant adenovirus serotype 26 HIV-1 Env vaccine (IPCAVD 001) J. Infect. Dis. 2013;207:240–247. doi: 10.1093/infdis/jis670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Baden L.R., Liu J., Li H., Johnson J.A., Walsh S.R., Kleinjan J.A., et al. Induction of HIV-1-specific mucosal immune responses following intramuscular recombinant adenovirus serotype 26 HIV-1 vaccination of humans. J. Infect. Dis. 2015;211:518–528. doi: 10.1093/infdis/jiu485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Milligan I.D., Gibani M.M., Sewell R., Clutterbuck E.A., Campbell D., Plested E., et al. Safety and immunogenicity of novel adenovirus type 26– and modified Vaccinia Ankara–vectored Ebola vaccines: a randomized clinical trial. JAMA. 2016;315:1610–1623. doi: 10.1001/jama.2016.4218. [DOI] [PubMed] [Google Scholar]
- 198.Winslow R.L., Milligan I.D., Voysey M., Luhn K., Shukarev G., Douoguih M., et al. Immune responses to novel adenovirus type 26 and modified Vaccinia Virus Ankara–vectored Ebola vaccines at 1 year. JAMA. 2017;317:1075–1077. doi: 10.1001/jama.2016.20644. [DOI] [PubMed] [Google Scholar]
- 199.Anywaine Z., Whitworth H., Kaleebu P., Praygod G., Shukarev G., Manno D., et al. Safety and immunogenicity of a 2-dose heterologous vaccination regimen with Ad26.ZEBOV and MVA-BN-filo Ebola vaccines: 12-month data from a phase 1 randomized clinical trial in Uganda and Tanzania. J. Infect. Dis. 2019;220:46–56. doi: 10.1093/infdis/jiz070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Dicks M.D.J., Spencer A.J., Edwards N.J., Wadell G., Bojang K., Gilbert S.C., et al. A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Guo J., Mondal M., Zhou D. Development of novel vaccine vectors: chimpanzee adenoviral vectors. Hum. Vaccin. Immunother. 2018;14:1679–1685. doi: 10.1080/21645515.2017.1419108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Antrobus R.D., Coughlan L., Berthoud T.K., Dicks M.D., Hill A.V., Lambe T., et al. Clinical assessment of a novel recombinant simian adenovirus ChAdOx1 as a vectored vaccine expressing conserved Influenza A antigens. Mol. Ther. J. Am. Soc. Gene Ther. 2014;22:668–674. doi: 10.1038/mt.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Coughlan L., Sridhar S., Payne R., Edmans M., Milicic A., Venkatraman N., et al. Heterologous two-dose vaccination with simian adenovirus and poxvirus vectors elicits long-lasting cellular immunity to influenza virus a in healthy adults. EBioMedicine. 2018;29:146–154. doi: 10.1016/j.ebiom.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Folegatti P.M., Bittaye M., Flaxman A., Lopez F.R., Bellamy D., Kupke A., et al. Safety and immunogenicity of a candidate Middle East respiratory syndrome coronavirus viral-vectored vaccine: a dose-escalation, open-label, non-randomised, uncontrolled, phase 1 trial. Lancet Infect. Dis. 2020;20:816–826. doi: 10.1016/S1473-3099(20)30160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Thomas S.J., Yoon I.-K. A review of Dengvaxia®: development to deployment. Hum. Vaccin. Immunother. 2019;15:2295–2314. doi: 10.1080/21645515.2019.1658503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Frantz P.N., Teeravechyan S., Tangy F. Measles-derived vaccines to prevent emerging viral diseases. Microbes Infect. 2018;20:493–500. doi: 10.1016/j.micinf.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ramsauer K., Schwameis M., Firbas C., Müllner M., Putnak R.J., Thomas S.J., et al. Immunogenicity, safety, and tolerability of a recombinant measles-virus-based chikungunya vaccine: a randomised, double-blind, placebo-controlled, active-comparator, first-in-man trial. Lancet Infect. Dis. 2015;15:519–527. doi: 10.1016/S1473-3099(15)70043-5. [DOI] [PubMed] [Google Scholar]
- 208.Gerke C., Frantz P.N., Ramsauer K., Tangy F. Measles-vectored vaccine approaches against viral infections: a focus on Chikungunya. Expert Rev. Vaccin. 2019;18:393–403. doi: 10.1080/14760584.2019.1562908. [DOI] [PubMed] [Google Scholar]
- 209.Reisinger E.C., Tschismarov R., Beubler E., Wiedermann U., Firbas C., et al. Immunogenicity, safety, and tolerability of the measles-vectored chikungunya virus vaccine MV-CHIK: a double-blind, randomised, placebo-controlled and active-controlled phase 2 trial. Lancet Lond. Engl. 2019;392:2718–2727. doi: 10.1016/S0140-6736(18)32488-7. [DOI] [PubMed] [Google Scholar]
- 210.Barry M. Single-cycle adenovirus vectors in the current vaccine landscape. Expert Rev. Vaccin. 2018;17:163–173. doi: 10.1080/14760584.2018.1419067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ledgerwood J.E. Use of low dose rVSV-ZEBOV: safety issues in a Swiss cohort. Lancet Infect. Dis. 2015;15:1117–1119. doi: 10.1016/S1473-3099(15)00222-4. [DOI] [PubMed] [Google Scholar]
- 212.Larocca C., Schlom J. Viral vector-based therapeutic cancer vaccines. Cancer J. Sudbury Mass. 2011;17:359–371. doi: 10.1097/PPO.0b013e3182325e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.David R.M., Doherty A.T. Viral vectors: the road to reducing genotoxicity. Toxicol. Sci. 2017;155:315–325. doi: 10.1093/toxsci/kfw220. [DOI] [PubMed] [Google Scholar]
- 214.Baldo A., Galanis E., Tangy F., Herman P. Biosafety considerations for attenuated measles virus vectors used in virotherapy and vaccination. Hum. Vaccin. Immunother. 2016;12:1102–1116. doi: 10.1080/21645515.2015.1122146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tang D.C., DeVit M., Johnston S.A. Genetic immunization is a simple method for eliciting an immune response. Nature. 1992;356:152–154. doi: 10.1038/356152a0. [DOI] [PubMed] [Google Scholar]
- 216.Ulmer J.B., Donnelly J.J., Parker S.E., Rhodes G.H., Felgner P.L., Dwarki V.J., et al. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993;259:1745–1749. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- 217.Boyer J.D., Chattergoon M., Shah A., Ginsberg R., MacGregor R.R., Weiner D.B. HIV-1 DNA based vaccine induces a CD8 mediated cross-clade CTL response. Dev. Biol. Stand. 1998;95:147–153. [PubMed] [Google Scholar]
- 218.Ugen K.E., Nyland S.B., Boyer J.D., Vidal C., Lera L., Rasheid S., et al. DNA vaccination with HIV-1 expressing constructs elicits immune responses in humans. Vaccine. 1998;16:1818–1821. doi: 10.1016/S0264-410X(98)00180-7. [DOI] [PubMed] [Google Scholar]
- 219.Muthumani K., Falzarano D., Reuschel E.L., Tingey C., Flingai S., Villarreal D.O., et al. A synthetic consensus anti–spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aac7462. 301ra132-301ra132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Modjarrad K., Roberts C.C., Mills K.T., Castellano A.R., Paolino K., Muthumani K., et al. Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect. Dis. 2019;19:1013–1022. doi: 10.1016/S1473-3099(19)30266-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Le T.P., Coonan K.M., Hedstrom R.C., Charoenvit Y., Sedegah M., Epstein J.E., et al. Safety, tolerability and humoral immune responses after intramuscular administration of a malaria DNA vaccine to healthy adult volunteers. Vaccine. 2000;18:1893–1901. doi: 10.1016/s0264-410x(99)00407-7. [DOI] [PubMed] [Google Scholar]
- 222.Sedegah M., Hedstrom R., Hobart P., Hoffman S.L. Protection against malaria by immunization with plasmid DNA encoding circumsporozoite protein. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9866–9870. doi: 10.1073/pnas.91.21.9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Martin J.E., Sullivan N.J., Enama M.E., Gordon I.J., Roederer M., Koup R.A., et al. A DNA vaccine for Ebola virus is safe and immunogenic in a phase I clinical trial. Clin. Vaccine Immunol. 2006;13:1267–1277. doi: 10.1128/CVI.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Tebas P., Kraynyak K.A., Patel A., Maslow J.N., Morrow M.P., Sylvester A.J., et al. Intradermal SynCon® Ebola GP DNA vaccine is temperature stable and safely demonstrates cellular and humoral immunogenicity advantages in healthy volunteers. J. Infect. Dis. 2019;220:400–410. doi: 10.1093/infdis/jiz132. [DOI] [PubMed] [Google Scholar]
- 225.Beckett C.G., Tjaden J., Burgess T., Danko J.R., Tamminga C., Simmons M., et al. Evaluation of a prototype dengue-1 DNA vaccine in a phase 1 clinical trial. Vaccine. 2011;29:960–968. doi: 10.1016/j.vaccine.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 226.Danko J.R., Kochel T., Teneza-Mora N., Luke T.C., Raviprakash K., Sun P., et al. Safety and immunogenicity of a tetravalent dengue DNA vaccine administered with a cationic lipid-based adjuvant in a phase 1 clinical trial. Am. J. Trop. Med. Hyg. 2018;98:849–856. doi: 10.4269/ajtmh.17-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ledgerwood J.E., Pierson T.C., Hubka S.A., Desai N., Rucker S., Gordon I.J., et al. A West Nile virus DNA vaccine utilizing a modified promoter induces neutralizing antibody in younger and older healthy adults in a phase I clinical trial. J. Infect. Dis. 2011;203:1396–1404. doi: 10.1093/infdis/jir054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Tebas P., Roberts C.C., Muthumani K., Reuschel E.L., Kudchodkar S.B., Zaidi F.I., et al. Safety and immunogenicity of an anti–Zika virus DNA vaccine — preliminary report. N. Engl. J. Med. 2017;0 doi: 10.1056/NEJMoa1708120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Krohn K., Stanescu I., Blazevic V., Vesikari T., Ranki A., Ustav M. A DNA HIV-1 vaccine based on a fusion gene expressing non-structural and structural genes of consensus sequence of the A-C subtypes and the ancestor sequence of the F-H subtypes. Preclinical and clinical studies. Microb. Infect. 2005;7:1405–1413. doi: 10.1016/j.micinf.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 230.Nilsson C., Heijdeman B., Godoy-Ramirez K., Tecleab T., Scarlatti G., Brave A., et al. HIV-DNA given with or without intradermal electroporation is safe and highly immunogenic in healthy Swedish HIV-1 DNA/MVA Vaccinees: a phase I randomized trial. PLoS One. 2015;10 doi: 10.1371/journal.pone.0131748. e0131748–e0131748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Palma P., Romiti M.L., Li Pira G., Montesano C., Mora N., Aquilani A., et al. PEDVAC trial: preliminary data from the first therapeutic DNA vaccination in HIV-infected children. Vaccine. 2011 doi: 10.1016/j.vaccine.2010.12.058. https://agris.fao.org/agris-search/search.do?recordID=US201500142120 [DOI] [PubMed] [Google Scholar]
- 232.Palma P., Romiti M.L., Montesano C., Santilli V., Mora N., Aquilani A., et al. Therapeutic DNA vaccination of vertically HIV-infected children: report of the first pediatric randomised trial (PEDVAC) PLoS One. 2013;8 doi: 10.1371/journal.pone.0079957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Vardas E., Stanescu I., Leinonen M., Ellefsen K., Pantaleo G., Valtavaara M., et al. Indicators of therapeutic effect in FIT-06, a phase II trial of a DNA vaccine, GTU(®)-multi-HIVB, in untreated HIV-1 infected subjects. Vaccine. 2012;30:4046–4054. doi: 10.1016/j.vaccine.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 234.Houser K.V., Yamshchikov G.V., Bellamy A.R., May J., Enama M.E., Sarwar U., et al. DNA vaccine priming for seasonal influenza vaccine in children and adolescents 6 to 17 years of age: a phase 1 randomized clinical trial. PloS One. 2018;13 doi: 10.1371/journal.pone.0206837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Ledgerwood J.E., Bellamy A.R., Belshe R., Bernstein D.I., Edupuganti S., Patel S.M., et al. DNA priming for seasonal influenza vaccine: a phase 1b double-blind randomized clinical trial. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Crank M.C., Gordon I.J., Yamshchikov G.V., Sitar S., Hu Z., Enama M.E., et al. Phase 1 study of pandemic H1 DNA vaccine in healthy adults. PLoS One. 2015;10 doi: 10.1371/journal.pone.0123969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Wang Z., Troilo P.J., Wang X., Griffiths T.G., Pacchione S.J., Barnum A.B., et al. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther. 2004;11:711–721. doi: 10.1038/sj.gt.3302213. [DOI] [PubMed] [Google Scholar]
- 238.Ledwith B.J., Manam S., Troilo P.J., Barnum A.B., Pauley C.J., Griffiths T.G., et al. Plasmid DNA vaccines: investigation of integration into host cellular DNA following intramuscular injection in mice. Intervirology. 2000;43:258–272. doi: 10.1159/000053993. [DOI] [PubMed] [Google Scholar]
- 239.Sheets R.L., Stein J., Manetz T.S., Duffy C., Nason M., Andrews C., et al. Biodistribution of DNA plasmid vaccines against HIV-1, Ebola, severe acute respiratory syndrome, or West Nile virus is similar, without integration, despite differing plasmid backbones or gene inserts. Toxicol. Sci. 2006;91:610–619. doi: 10.1093/toxsci/kfj169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Lee J., Arun Kumar S., Jhan Y.Y., Bishop C.J. Engineering DNA vaccines against infectious diseases. Acta Biomater. 2018;80:31–47. doi: 10.1016/j.actbio.2018.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Rahman A., Isenberg D.A. Systemic lupus erythematosus. N. Engl. J. Med. 2008;358:929–939. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- 242.Sano H., Morimoto C. Dna isolated from DNA/anti-DNA antibody immune complexes in systemic lupus erythematosus is rich in guanine-cytosine content. J. Immunol. 1982;128:1341–1345. [PubMed] [Google Scholar]
- 243.Karounos D.G., Grudier J.P., Pisetsky D.S. Spontaneous expression of antibodies to DNA of various species origin in sera of normal subjects and patients with systemic lupus erythematosus. J. Immunol. 1988;140:451–455. [PubMed] [Google Scholar]
- 244.Jiao S., Williams P., Berg R.K., Hodgeman B.A., Liu L., Repetto G., et al. Direct gene transfer into nonhuman primate myofibers in vivo. Hum. Gene Ther. 1992;3:21–33. doi: 10.1089/hum.1992.3.1-21. [DOI] [PubMed] [Google Scholar]
- 245.Madaio M.P., Hodder S., Schwartz R.S., Stollar B.D. Responsiveness of autoimmune and normal mice to nucleic acid antigens. J. Immunol. 1984;132:872–876. [PubMed] [Google Scholar]
- 246.Moderna Announces Positive Phase 1 Results for the First Systemic Messenger RNA Therapeutic Encoding a Secreted Protein (mRNA-1944) Moderna, Inc.; 2019. https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-positive-phase-1-results-first-systemic/ [Google Scholar]
- 247.Shaw C., Panther L., August A., Zaks T., Smolenov I., Bart S., et al. Safety and immunogenicity of a mRNA-based chikungunya vaccine in a phase 1 dose-ranging trial. Int. J. Infect. Dis. 2019;79:17. doi: 10.1016/j.ijid.2018.11.058. [DOI] [Google Scholar]
- 248.Feldman R.A., Fuhr R., Smolenov I., Mick Ribeiro A., Panther L., Watson M., et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37:3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- 249.Hu Z., Jiao X., Liu X. Antibody immunity induced by H7N9 avian influenza vaccines: evaluation criteria, affecting factors, and implications for rational vaccine design. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.01898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Iavarone C., O'hagan D.T., Yu D., Delahaye N.F., Ulmer J.B. Mechanism of action of mRNA-based vaccines. Expert Rev. Vaccin. 2017;16:871–881. doi: 10.1080/14760584.2017.1355245. [DOI] [PubMed] [Google Scholar]
- 251.Leal L., Guardo A.C., Morón-López S., Salgado M., Mothe B., Heirman C., et al. Phase I clinical trial of an intranodally administered mRNA-based therapeutic vaccine against HIV-1 infection. AIDS Lond. Engl. 2018;32:2533–2545. doi: 10.1097/QAD.0000000000002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tatematsu M., Funami K., Seya T., Matsumoto M. Extracellular RNA sensing by pattern recognition receptors. J. Innate Immun. 2018;10:398–406. doi: 10.1159/000494034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Theofilopoulos A.N., Baccala R., Beutler B., Kono D.H. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 255.Nestle F.O., Conrad C., Tun-Kyi A., Homey B., Gombert M., Boyman O., et al. Plasmacytoid predendritic cells initiate psoriasis through interferon-α production. J. Exp. Med. 2005;202:135–143. doi: 10.1084/jem.20050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Kannemeier C., Shibamiya A., Nakazawa F., Trusheim H., Ruppert C., Markart P., et al. Extracellular RNA constitutes a natural procoagulant cofactor in blood coagulation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6388–6393. doi: 10.1073/pnas.0608647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Fischer S., Gerriets T., Wessels C., Walberer M., Kostin S., Stolz E., et al. Extracellular RNA mediates endothelial-cell permeability via vascular endothelial growth factor. Blood. 2007;110:2457–2465. doi: 10.1182/blood-2006-08-040691. [DOI] [PubMed] [Google Scholar]
- 258.Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. Three decades of messenger RNA vaccine development. Nano Today. 2019;28 doi: 10.1016/j.nantod.2019.100766. [DOI] [Google Scholar]
- 259.Mubarak A., Alturaiki W., Hemida M.G. Middle East Respiratory Syndrome Coronavirus (MERS-CoV): infection, immunological response, and vaccine development. J. Immunol. Res. 2019;2019 doi: 10.1155/2019/6491738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Xu X., Gao X. Immunological responses against SARS-coronavirus infection in humans. Cell. Mol. Immunol. 2004;1:119–122. [PubMed] [Google Scholar]
- 261.van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Kou Y., Xu Y., Zhao Z., Liu J., Wu Y., You Q., et al. Tissue plasminogen activator (tPA) signal sequence enhances immunogenicity of MVA-based vaccine against tuberculosis. Immunol. Lett. 2017;190:51–57. doi: 10.1016/j.imlet.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 263.B O. Vector delivery-dependant effect of human tissue plasminogen activator signal peptide on vaccine induction of T cells. J. HIV AIDS. 2016;2 doi: 10.16966/2380-5536.130. [DOI] [Google Scholar]
- 264.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Ramasamy M.N., Minassian A.M., Ewer K.J., Flaxman A.L., Folegatti P.M., Owens D.R., et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet. 2020;0 doi: 10.1016/S0140-6736(20)32466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Knoll M.D., Wonodi C. Oxford–AstraZeneca COVID-19 vaccine efficacy. Lancet. 2020;0 doi: 10.1016/S0140-6736(20)32623-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2020;0 doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Wu S., Zhong G., Zhang J., Shuai L., Zhang Z., Wen Z., et al. A single dose of an adenovirus-vectored vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-17972-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Zhu F.-C., Li Y.-H., Guan X.-H., Hou L.-H., Wang W.-J., Li J.-X., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet Lond. Engl. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zhu F.-C., Guan X.-H., Li Y.-H., Huang J.-Y., Jiang T., Hou L.-H., et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Mahase E. Covid-19: where are we on immunity and vaccines? BMJ. 2020 doi: 10.1136/bmj.m3096. [DOI] [PubMed] [Google Scholar]
- 272.Logunov D.Y., Dolzhikova I.V., Zubkova O.V., Tukhvatullin A.I., Shcheblyakov D.V., Dzharullaeva A.S., et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet. 2020 doi: 10.1016/S0140-6736(20)31866-3. S0140673620318663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.The First Interim Data Analysis of the Sputnik V Vaccine Against COVID-19 Phase III Clinical Trials in the Russian Federation Demonstrated 92% Efficacy. 2020. http://sputnikvaccine.com/newsroom/pressreleases/the-first-interim-data-analysis-of-the-sputnik-v-vaccine-against-covid-19-phase-iii-clinical-trials/
- 274.Second Interim Analysis of Clinical Trial Data Showed a 91.4% Efficacy for the Sputnik V Vaccine on Day 28 After the First Dose; Vaccine Efficacy is Over 95% 42 Days After the First Dose. 2020. http://sputnikvaccine.com/newsroom/pressreleases/second-interim-analysis-of-clinical-trial-data-showed-a-91-4-efficacy-for-the-sputnik-v-vaccine-on-d/
- 275.Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Moore A.C., Dora E.G., Peinovich N., Tucker K.P., Lin K., Cortese M., et al. Pre-clinical studies of a recombinant adenoviral mucosal vaccine to prevent SARS-CoV-2 infection. BioRxiv. 2020 doi: 10.1101/2020.09.04.283853. 2020.09.04.283853. [DOI] [Google Scholar]
- 277.Richardson J.S., Pillet S., Bello A.J., Kobinger G.P. Airway delivery of an adenovirus-based Ebola virus vaccine bypasses existing immunity to homologous adenovirus in nonhuman primates. J. Virol. 2013;87:3668–3677. doi: 10.1128/JVI.02864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Liebowitz D., Lindbloom J.D., Brandl J.R., Garg S.J., Tucker S.N. High titre neutralising antibodies to influenza after oral tablet immunisation: a phase 1, randomised, placebo-controlled trial. Lancet Infect. Dis. 2015;15:1041–1048. doi: 10.1016/S1473-3099(15)00266-2. [DOI] [PubMed] [Google Scholar]
- 279.Kim L., Liebowitz D., Lin K., Kasparek K., Pasetti M.F., Garg S.J., et al. Safety and immunogenicity of an oral tablet norovirus vaccine, a phase I randomized, placebo-controlled trial. JCI Insight. 2018;3 doi: 10.1172/jci.insight.121077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Rice A., Verma M., Shin A., Zakin L., Sieling P., Tanaka S., et al. A next generation bivalent human Ad5 COVID-19 vaccine delivering both spike and nucleocapsid antigens elicits Th1 dominant CD4+, CD8+ T-cell and neutralizing antibody responses. BioRxiv. 2020 doi: 10.1101/2020.07.29.227595. 2020.07.29.227595. [DOI] [Google Scholar]
- 281.Sieling P., Zakin L., Shin A., Morimoto B., Adisetiyo H., Garban H., et al. Th1 dominant nucleocapsid and spike antigen-specific CD4+ and CD8+ memory T cell recall induced by hAd5 S-fusion + N-ETSD infection of autologous dendritic cells from patients previously infected with SARS-CoV-2. MedRxiv. 2020 doi: 10.1101/2020.11.04.20225417. 2020.11.04.20225417. [DOI] [Google Scholar]
- 282.Chiuppesi F., d'Alincourt Salazar M., Contreras H., Nguyen V.H., Martinez J., Park Y., et al. Development of a multi-antigenic SARS-CoV-2 vaccine candidate using a synthetic poxvirus platform. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-19819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Koch T., Dahlke C., Fathi A., Kupke A., Krähling V., Okba N.M.A., et al. Safety and immunogenicity of a modified vaccinia virus Ankara vector vaccine candidate for Middle East respiratory syndrome: an open-label, phase 1 trial. Lancet Infect. Dis. 2020;20:827–838. doi: 10.1016/S1473-3099(20)30248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Yahalom-Ronen Y., Tamir H., Melamed S., Politi B., Shifman O., Achdout H., et al. A single dose of recombinant VSV-ΔG-spike vaccine provides protection against SARS-CoV-2 challenge. BioRxiv. 2020 doi: 10.1101/2020.06.18.160655. 2020.06.18.160655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Amante D.H., Smith T.R.F., Mendoza J.M., Schultheis K., McCoy J.R., Khan A.S., et al. Skin transfection patterns and expression kinetics of electroporation-enhanced plasmid delivery using the CELLECTRA-3P, a portable next-generation dermal electroporation device. Hum. Gene Ther. Methods. 2015;26:134–146. doi: 10.1089/hgtb.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Diehl M.C., Lee J.C., Daniels S.E., Tebas P., Khan A.S., Giffear M., et al. Tolerability of intramuscular and intradermal delivery by CELLECTRA® adaptive constant current electroporation device in healthy volunteers. Hum. Vaccin. Immunother. 2013;9:2246–2252. doi: 10.4161/hv.24702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.INOVIO Announces Positive Interim Phase 1 Data For INO-4800 Vaccine for COVID-19. 2020. http://ir.inovio.com/news-releases/news-releases-details/2020/INOVIO-Announces-Positive-Interim-Phase-1-Data-For-INO-4800-Vaccine-for-COVID-19/default.aspx
- 288.Speiser D.E., Bachmann M.F. COVID-19: mechanisms of vaccination and immunity. Vaccines. 2020;8:404. doi: 10.3390/vaccines8030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Li B., Luo X., Deng B., Wang J., McComb D.W., Shi Y., et al. An orthogonal Array optimization of lipid-like nanoparticles for mRNA delivery in vivo. Nano Lett. 2015;15:8099–8107. doi: 10.1021/acs.nanolett.5b03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., et al. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.de Alwis R., Gan E.S., Chen S., Leong Y.S., Tan H.C., Zhang S.L., et al. A single dose of self-transcribing and replicating RNA based SARS-CoV-2 vaccine produces protective adaptive immunity in mice. BioRxiv. 2020 doi: 10.1101/2020.09.03.280446. 2020.09.03.280446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., et al. An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. 2020;0 doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Mahase E. Covid-19: moderna vaccine is nearly 95% effective, trial involving high risk and elderly people shows. BMJ. 2020;371 doi: 10.1136/bmj.m4471. [DOI] [Google Scholar]
- 299.Pardi N., Weissman D. In: RNA Vaccines Methods Protoc. Kramps T., Elbers K., editors. Springer; New York, NY: 2017. Nucleoside modified mRNA vaccines for infectious diseases; pp. 109–121. [DOI] [PubMed] [Google Scholar]
- 300.Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Phase 1/2 study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020:1–8. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 301.Walsh E.E., Frenck R.W., Falsey A.R., Kitchin N., Absalon J., Gurtman A., et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;0 doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., et al. COVID-19 vaccine BNT162b1 elicits human antibody and T H 1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 303.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.COVID-19 Vaccine U.S. Distribution Fact Sheet. Pfizer; 2020. https://www.pfizer.com/news/hot-topics/covid_19_vaccine_u_s_distribution_fact_sheet [Google Scholar]
- 305.Moderna Announces Longer Shelf Life for Its COVID-19 Vaccine Candidate at Refrigerated Temperatures. Moderna, Inc.; 2020. https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-longer-shelf-life-its-covid-19-vaccine/ [Google Scholar]
- 306.Coffman R.L., Sher A., Seder R.A. Vaccine adjuvants: putting innate immunity to work. Immunity. 2010;33:492–503. doi: 10.1016/j.immuni.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Chung Y.H., Beiss V., Fiering S.N., Steinmetz N.F. COVID-19 vaccine frontrunners and their nanotechnology design. ACS Nano. 2020;14:12522–12537. doi: 10.1021/acsnano.0c07197. [DOI] [PubMed] [Google Scholar]
- 308.Coughlan L. Factors which contribute to the immunogenicity of non-replicating adenoviral vectored vaccines. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Li L., Petrovsky N. Molecular mechanisms for enhanced DNA vaccine immunogenicity. Expert Rev. Vaccin. 2016;15:313–329. doi: 10.1586/14760584.2016.1124762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Perrie Y., Crofts F., Devitt A., Griffiths H.R., Kastner E., Nadella V. Designing liposomal adjuvants for the next generation of vaccines. Adv. Drug Deliv. Rev. 2016;99:85–96. doi: 10.1016/j.addr.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 311.Quirk E.K., Brown E.L., Leavitt R.Y., Mogg R., Mehrotra D.V., Evans R.K., et al. Safety profile of the Merck human immunodeficiency virus-1 clade B gag DNA plasmid vaccine with and without adjuvants. Open Forum Infect. Dis. 2014;1 doi: 10.1093/ofid/ofu016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Cardon L.R., Burge C., Clayton D.A., Karlin S. Pervasive CpG suppression in animal mitochondrial genomes. Proc. Natl. Acad. Sci. U. S. A. 1994;91:3799–3803. doi: 10.1073/pnas.91.9.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Zhang A., Jin H., Zhang F., Ma Z., Tu Y., Ren Z., et al. Effects of multiple copies of CpG on DNA vaccination. DNA Cell Biol. 2005;24:292–298. doi: 10.1089/dna.2005.24.292. [DOI] [PubMed] [Google Scholar]
- 314.Bohle B., Jahn‐Schmid B., Maurer D., Kraft D., Ebner C. Oligodeoxynucleotides containing CpG motifs induce IL-12, IL-18 and IFN-γ production in cells from allergic individuals and inhibit IgE synthesis in vitro. Eur. J. Immunol. 1999;29:2344–2353. doi: 10.1002/(SICI)1521-4141(199907)29:07<2344::AID-IMMU2344>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 315.Donnelly J.J., Ulmer J.B., Shiver J.W., Liu M.A. Dna vaccines. Annu. Rev. Immunol. 1997;15:617–648. doi: 10.1146/annurev.immunol.15.1.617. [DOI] [PubMed] [Google Scholar]
- 316.Krieg A.M. The role of CpG motifs in innate immunity. Curr. Opin. Immunol. 2000;12:35–43. doi: 10.1016/s0952-7915(99)00048-5. [DOI] [PubMed] [Google Scholar]
- 317.Romagnani S. TH1 and TH2 in human diseases. Clin. Immunol. Immunopathol. 1996;80:225–235. doi: 10.1006/clin.1996.0118. [DOI] [PubMed] [Google Scholar]
- 318.Van Eden W., Van Der Zee R., Van Kooten P., Berlo S.E., Cobelens P.M., Kavelaars A., et al. Balancing the immune system: Th1 and Th2. Ann. Rheum. Dis. 2002;61(Suppl. 2):ii25–ii28. doi: 10.1136/ard.61.suppl_2.ii25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Tseng C.-T., Sbrana E., Iwata-Yoshikawa N., Newman P.C., Garron T., Atmar R.L., et al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Honda-Okubo Y., Saade F., Petrovsky N. AdvaxTM, a polysaccharide adjuvant derived from delta inulin, provides improved influenza vaccine protection through broad-based enhancement of adaptive immune responses. Vaccine. 2012;30:5373–5381. doi: 10.1016/j.vaccine.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Honda-Okubo Y., Barnard D., Ong C.H., Peng B.-H., Tseng C.-T.K., Petrovsky N. Severe acute respiratory syndrome-associated coronavirus vaccines formulated with delta inulin adjuvants provide enhanced protection while ameliorating lung eosinophilic immunopathology. J. Virol. 2014;89:2995–3007. doi: 10.1128/JVI.02980-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Kitagaki K., Jain V.V., Businga T.R., Hussain I., Kline J.N. Immunomodulatory effects of CpG oligodeoxynucleotides on established Th2 responses. Clin. Diagn. Lab. Immunol. 2002;9:1260–1269. doi: 10.1128/CDLI.9.6.1260-1269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Gupta T., Gupta S.K. Potential adjuvants for the development of a SARS-CoV-2 vaccine based on experimental results from similar coronaviruses. Int. Immunopharmacol. 2020;86 doi: 10.1016/j.intimp.2020.106717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Jones T., Cyr S., Allard F., Bellerose N., Lowell G.H., Burt D.S. Protollin: a novel adjuvant for intranasal vaccines. Vaccine. 2004;22:3691–3697. doi: 10.1016/j.vaccine.2004.03.035. [DOI] [PubMed] [Google Scholar]
- 325.Hu M.C., Jones T., Kenney R.T., Barnard D.L., Burt D.S., Lowell G.H. Intranasal Protollin-formulated recombinant SARS S-protein elicits respiratory and serum neutralizing antibodies and protection in mice. Vaccine. 2007;25:6334–6340. doi: 10.1016/j.vaccine.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Chabot S., Brewer A., Lowell G., Plante M., Cyr S., Burt D.S., et al. A novel intranasal Protollin-based measles vaccine induces mucosal and systemic neutralizing antibody responses and cell-mediated immunity in mice. Vaccine. 2005;23:1374–1383. doi: 10.1016/j.vaccine.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 327.Holzer B., Morgan S.B., Matsuoka Y., Edmans M., Salguero F.J., Everett H., et al. Comparison of Heterosubtypic protection in ferrets and pigs induced by a single-cycle influenza vaccine. J. Immunol. 2018;1950(200):4068–4077. doi: 10.4049/jimmunol.1800142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Li C.K., Xu X. Host immune responses to SARS coronavirus in humans. Mol. Biol. SARS-Coronavirus. 2009:259–278. doi: 10.1007/978-3-642-03683-5_16. [DOI] [Google Scholar]
- 329.Lambert N.D., Ovsyannikova I.G., Pankratz V.S., Jacobson R.M., Poland G.A. Understanding the immune response to seasonal influenza vaccination in older adults: a systems biology approach. Expert Rev. Vaccin. 2012;11:985–994. doi: 10.1586/erv.12.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Zhou G., Zhao Q. Perspectives on therapeutic neutralizing antibodies against the Novel Coronavirus SARS-CoV-2. Int. J. Biol. Sci. 2020;16:1718–1723. doi: 10.7150/ijbs.45123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Guzman M.G., Alvarez M., Halstead S.B. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: an historical perspective and role of antibody-dependent enhancement of infection. Arch. Virol. 2013;158:1445–1459. doi: 10.1007/s00705-013-1645-3. [DOI] [PubMed] [Google Scholar]
- 332.Becker Y. Respiratory syncytial virus (RSV) evades the human adaptive immune system by skewing the Th1/Th2 cytokine balance toward increased levels of Th2 cytokines and IgE, markers of allergy—a review. Virus Genes. 2006;33:235–252. doi: 10.1007/s11262-006-0064-x. [DOI] [PubMed] [Google Scholar]
- 333.Tirado S.M.C., Yoon K.-J. 2004. Antibody-dependent Enhancement of Virus Infection and Disease.https://Home.Liebertpub.Com/Vim [DOI] [PubMed] [Google Scholar]
- 334.Halstead S.B. Immune enhancement of viral infection. Prog. Allergy. 1982;31:301–364. [PubMed] [Google Scholar]
- 335.Kulkarni R. Antibody-dependent enhancement of viral infections. Dyn. Immune Act. Viral Dis. 2019:9–41. doi: 10.1007/978-981-15-1045-8_2. [DOI] [Google Scholar]
- 336.Cheng V.C.C., Lau S.K.P., Woo P.C.Y., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Chen Y.-C., Wang S.-Y. Activation of terminally differentiated human monocytes/macrophages by dengue virus: productive infection, hierarchical production of innate cytokines and chemokines, and the synergistic effect of lipopolysaccharide. J. Virol. 2002;76:9877–9887. doi: 10.1128/JVI.76.19.9877-9887.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 340.Deng W., Bao L., Liu J., Xiao C., Liu J., Xue J., et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science. 2020;369:818–823. doi: 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 342.Kim H.W., Canchola J.G., Brandt C.D., Pyles G., Chanock R.M., Jensen K., et al. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 343.Waris M.E., Tsou C., Erdman D.D., Zaki S.R., Anderson L.J. Respiratory synctial virus infection in BALB/c mice previously immunized with formalin-inactivated virus induces enhanced pulmonary inflammatory response with a predominant Th2-like cytokine pattern. J. Virol. 1996;70:2852–2860. doi: 10.1128/jvi.70.5.2852-2860.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Stevens W.W., Sun J., Castillo J.P., Braciale T.J. Pulmonary eosinophilia is attenuated by early responding CD8+ memory T cells in a murine model of RSV vaccine-enhanced disease. Viral Immunol. 2009;22:243–251. doi: 10.1089/vim.2009.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Castilow E.M., Varga S.M. Overcoming T cell-mediated immunopathology to achieve safe RSV vaccination. Future Virol. 2008;3:445–454. doi: 10.2217/17460794.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Kvarnhammar A.M., Cardell L.O. Pattern-recognition receptors in human eosinophils. Immunology. 2012;136:11–20. doi: 10.1111/j.1365-2567.2012.03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Lebold K.M., Jacoby D.B., Drake M.G. Toll-like receptor 7-targeted therapy in respiratory Disease. Transfus. Med. Hemother. 2016;43:114–119. doi: 10.1159/000445324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Zhou J., Chu H., Chan J.F.-W., Yuen K.-Y. Middle East respiratory syndrome coronavirus infection: virus-host cell interactions and implications on pathogenesis. Virol. J. 2015;12 doi: 10.1186/s12985-015-0446-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Chu H., Zhou J., Wong B.H.-Y., Li C., Chan J.F.-W., Cheng Z.-S., et al. Middle East Respiratory Syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J. Infect. Dis. 2016;213:904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Zhao J., Li K., Wohlford-Lenane C., Agnihothram S.S., Fett C., Zhao J., et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. U. S. A. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Koday M.T., Leonard J.A., Munson P., Forero A., Koday M., Bratt D.L., et al. Multigenic DNA vaccine induces protective cross-reactive T cell responses against heterologous influenza virus in nonhuman primates. PLoS One. 2017;12 doi: 10.1371/journal.pone.0189780. [DOI] [PMC free article] [PubMed] [Google Scholar]