Abstract
Nucleocytoplasmic transport of signaling modulators is essential for regulating cellular responses to extracellular stimulation and stress, as well as pathogen infection. Exportin 1 (XPO1), also known as chromosomal maintenance 1 (CRM1), mediates nuclear export of proteins, rRNAs, snRNAs, and some mRNAs. In this study, we have identified an essential role of XPO1 in regulating Kaposi’s sarcoma-associated herpesvirus (KSHV) lytic replication during primary infection of primary human umbilical vein endothelial cells. Treatment with an XPO1 inhibitor KPT-8602 and short hairpin RNA (shRNA)-mediated knockdown of XPO1 reduced KSHV lytic replication but had no effect on KSHV entry and trafficking. XPO1 inhibition induced retention of autophagy adaptor protein p62 (SQSTM1) in the nucleus, which enhanced activation of TBK1 and IRF3. As a result, nuclear accumulation of p62 increased expression of innate immune-related genes including IRF7, ISG15, IFIT1, IFIT2, and IFIT3, leading to a reduction of KSHV lytic replication. These results illustrate a novel mechanism by which XPO1 mediates innate immune response and KSHV replication, and identify XPO1 as a potential therapeutic target and KPT-8602 as a promising therapeutic agent for KSHV infection.
Subject terms: Mechanisms of disease, Infection
Introduction
Kaposi’s sarcoma-associated herpesvirus (KSHV) is a gammaherpesvirus etiologically associated with Kaposi’s sarcoma and several other lymphoproliferative diseases1–3. KSHV has both latent and lytic replication cycles regulated by intracellular and extracellular signals4. In lytic replication-permissive cells such as primary human umbilical vein endothelial (HUVEC), activation of ERK, p38, and JNK pathways promotes expression of viral lytic genes, and KSHV undergoes robust lytic replication before entering into latency5–8. However, activation of NF-κB pathway promotes the expression of latent genes and KSHV enters into latency in nonpermissive cells such as primary human dermal microvascular endothelial cells and human foreskin fibroblasts9–12.
The exchange of signaling molecules between nuclear and cytoplasmic compartments is essential for cellular functions. This process is mediated by a nuclear pore complex (NPC) inserted across the nuclear envelope (NE). Unlike smaller molecules, which diffuse through NPCs, the shuttling of proteins larger than 40 kDa is regulated by karyopherins (Kaps), which interact with NPC components nucleoporins (Nups) to cross NE13.
A Kap consists of a cargo-binding domain, an NPC-binding domain, and an N-terminal Ras-related nuclear protein (Ran) binding domain14. Ran is a small GTPase, switching between a cytoplasmic RanGDP form and a nuclear RanGTP form by nucleotide exchange and GTP hydrolysis. Kaps include both importins and exportins. For import, after importins transport cargo molecules through NPCs into the nucleus, RanGTP stimulates dissociation of the importin-cargo complex to release cargos15. For export, exportin-cargo complex formation is induced by RanGTP. After transportation into the cytoplasm, hydrolysis of GTP to GDP results in a switch from RanGTP to RanGDP to release cargos16.
There are seven members of exportin, including XPO1 (CRM1), XPO2 (CSE1L), XPO3 (XPOt), XPO4, XPO5, XPO6, and XPO713. XPO1, XPO2, XPO4, XPO6, and XPO7 primarily mediate the export of proteins, whereas XPO3 and XPO5 are involved in the export of tRNAs and precursor microRNAs, respectively. XPO1 is the major exportin that mediates protein nuclear export, which is important for the replication of several viruses including retroviruses, orthomyxoviruses, paramyxoviruses, flaviviruses, coronaviruses, rhabdoviruses, and herpesviruses17,18. Hence, XPO1 is a potential target for antiviral therapy.
XPO1 transports proteins containing leucine-rich nuclear export signals (NESs)19,20. A novel class of selective inhibitor of nuclear export (SINE) compounds that bind to XPO1 NES groove was identified21–23. Unlike leptomycin B (LMB), SINE compounds form a reversible covalent bond with XPO1 cysteine 528, making it less cytotoxic than LMB to cells. KPT-8602 (Eltanexor) is a second-generation SINE with reduced penetration across blood–brain barrier and an enhanced tolerability profile24.
Here, we have identified a critical role of XPO1 in KSHV lytic replication during primary infection. We show that XPO1 inhibition with KPT-8602 and by shRNA knockdown significantly reduces infectious virions by inhibiting viral gene expression. XPO1 inhibition induces p62 nuclear retention, which blocks KSHV lytic replication by activating TBK1 and IRF3 and enhancing the expression of innate immune-related genes. These results identify XPO1 as an essential protein for KSHV lytic replication by regulating p62 nuclear shuttling and controlling antiviral immune responses.
Materials and methods
Cells and drug
HUVEC (ATCC) was cultured in VascuLife VEGF complete medium (Lifeline). KSHV-infected iSLK (iSLK-BAC16) cells were from Dr. Jae Jung and cultured as previously described25. KPT-8602 (Selleck) was dissolved in dimethyl sulfoxide (DMSO).
Virus preparation and titration
KSHV was prepared from iSLK-BAC16 cells as previously described25. KSHV in VascuLife VEGF complete medium without Heparin was titrated by infecting HUVEC as previously described5,26.
Virus infection
HUVEC were infected with KSHV at a multiplicity of infection of two IUs per cell5,26. To determine KPT-8602’s effect, HUVEC were pretreated with KPT-8602 or vehicle control DMSO for 1 h. Cells were then infected with KSHV with KPT-8602 or vehicle. At 4 h post infection (hpi), the medium was replaced with fresh medium. Cells were fixed at 6 hpi for examining virus entry and trafficking, at 24 hpi for examining viral infectivity, and at 48 hpi for examining viral transcripts and proteins. To examine infectious virions, supernatants were collected at 96 hpi and titrated5,26.
Immunofluorescence assay
Immunofluorescence assay was performed as previously described5 using antibodies to ORF655, LANA (Abcam), p62 (Cell Signaling), phosphorylated IRF3 (pIRF3, Novus Biologicals), and pTBK1 (Cell Signaling). Signal was detected with an Alexa488-, Alexa555-, or Alexa647-conjugated secondary antibody (Life Technologies). Images were acquired using an Olympus FV2000 Confocal Microscope with CellSens Software. Magenta of Alexa647 was shown as green pseudocolor for better presentation.
Western blotting
Western blotting was performed as previously described27 using primary antibodies against LANA (Abcam), K-bZIP (Santa Cruz), β-actin (Santa Cruz), XPO1 (Sigma), p62 (Cell Signaling), pTBK1 (Cell Signaling), TBK1 (Novus Biologicals), pIRF3 (Novus), IRF3 (Novus), STING (Novus), and LC3B (Sigma). ORF65 and RTA antibodies were previously described5,28.
RNA extraction and RT-qPCR
Expression of viral genes was analyzed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) as previously described8. qPCR primers were listed in Supplementary Table 1. Three biological repeats were performed for each treatment.
DNA extraction
DNA extraction was performed by using QIAamp DNA Mini Kit (Qiagen).
shRNA knockdown
Lentiviral production and knockdown with shRNAs were performed as previously described29. Supernatants were collected 72 h after transfection. HUVEC were infected with lentiviruses by spinning infection at 500 × g in 10 µg/ml Polybrene (Sigma). The effect of shRNA knockdown was examined at 48 hpi.
Statistical analysis
Results are expressed as means and standard deviations. Statistical analyses were performed using Prism 5.01 (GraphPad Software). Significance was determined by two-way ANOVA followed by Tukey’s multiple comparisons test.
Ethics statement
All authors had access to all data and have reviewed and approved the final paper.
Results
XPO1 inhibitor KPT-8602 decreases KSHV titer
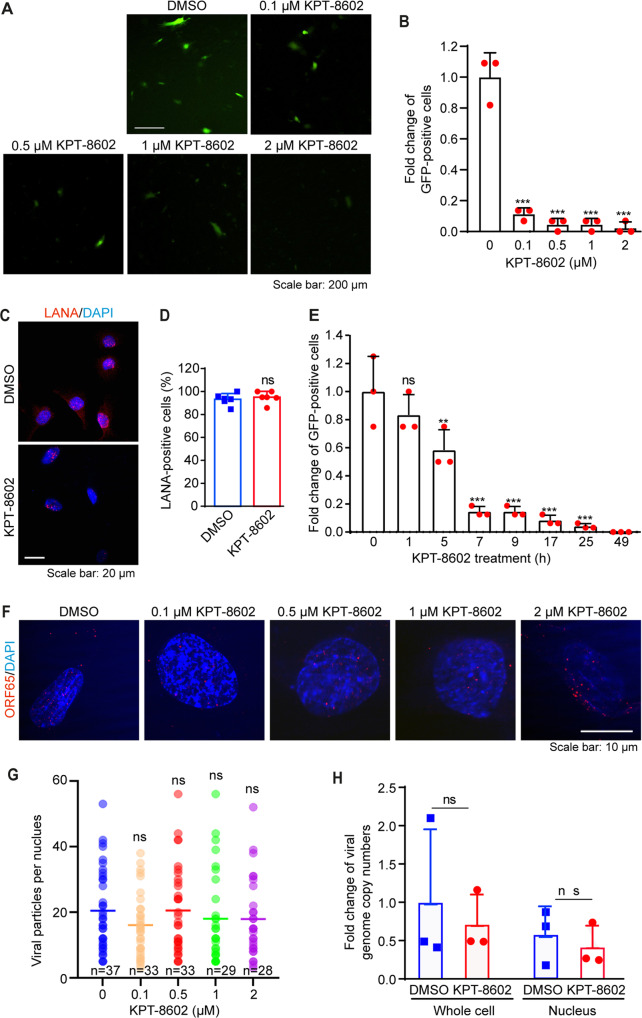
We used KPT-8602 to investigate XPO1’s role in KSHV primary infection in HUVEC as these cells support productive infection5. HUVEC treated with KPT-8602 for 1 h were infected with KSHV. At 48 hpi, cells were replaced with fresh medium without KPT-8602. At 96 hpi, supernatants were collected and titrated. KPT-8602 reduced KSHV titer at a dose-dependent fashion (Fig. 1A, B). KPT-8602 reduced titers by 84% at 0.1 µM and by 98% at 2 µM. At 0.1 µM, KPT-8602 had <30% cytotoxic effect on cells before 48 h, and >40% of the cells remained alive at 72 and 96 h (Supplementary Fig. 1). At 2 µM, >45% of the cells remained viable at any points examined. Thus, KPT-8602’s inhibitory effect on the production of KSHV virions was unlikely due to its cell cytotoxicity.
Fig. 1. XPO1 inhibitor KPT-8602 decreases the production of KSHV infectious virions but has no effect on KSHV infectivity, and virus entry and trafficking during primary infection.
A, B HUVEC were pretreated with the indicated doses of KPT-8602 for 1 h and then infected with KSHV in the presence of KPT-8602. At 4 hpi, cells were extensively washed and replaced with a new medium containing the indicated concentrations of KPT-8602. At 48 hpi, cells were extensively washed and replaced with a new medium without KPT-8602. Supernatants were collected at 96 hpi and used to titrate infectious virions by infecting fresh HUVEC. At 48 hpi, representative images were taken (A) to quantify the number of GFP-positive cells (B). C HUVEC pretreated with 0.5 µM KPT-8602 for 1 h were infected with KSHV in the presence of KPT-8602. At 48 hpi, cells were fixed and stained for LANA (red). The expression level and staining pattern of LANA are shown. Nuclei were stained with DAPI. Images were taken with confocal microscopy (magnification ×600). D Quantification of LANA-positive cells based on five images taken as described in (C). E HUVEC were pretreated with 0.5 µM KPT-8602 for 1 h and then infected with KSHV in the presence of KPT-8602 for the indicated times. Cells were extensively washed and replaced with a new medium without KPT-8602. At 50 hpi, cells were replaced with the new medium. Supernatants were collected at 96 hpi and used to titrate infectious virions by infecting fresh HUVEC. At 48 hpi, GFP-positive cells were quantified. F HUVEC pretreated with 0.5 µM KPT-8602 for 1 h were infected with KSHV in the presence of KPT-8602 for 6 h, fixed, and stained for nuclei with DAPI (blue) and KSHV particles with an antibody against ORF65 (red). Images were taken with a confocal microscopy (magnification ×1000). G The total number of KSHV particles successfully docked at the perinuclear region was determined and analyzed based on images taken as described in (F). H HUVEC pretreated with 0.5 µM KPT-8602 for 1 h were infected with KSHV in the presence of KPT-8602. Cells were harvested at 6 hpi. Cellular DNA and viral DNA were extracted from whole cells or the nuclear fraction for the detection of the viral genome by qPCR using LANA-specific primers. *, **, and *** indicate P values of <0.05, < 0.01, and <0.001, respectively; NS not significant.
KPT-8602 inhibits KSHV lytic replication at post-entry stage(s)
To identify step(s) of KSHV infection regulated by XPO1, we examined whether XPO1 inhibition might affect infectivity. We used 0.5 µM KPT-8602 based on the cytotoxicity results (Supplementary Fig. 1). Over 70% of cells survived at 24 and 48 h, and >40% of cells survived at 72 and 96 h. HUVEC pretreated with DMSO or 0.5 µM KPT-8602 for 1 h were infected with KSHV. At 4 hpi, cells were replaced with fresh medium without the inhibitor. At 48 hpi, cells were stained for LANA protein. Detection of LANA protein indicated successful viral infection. KPT-8602 treatment did not affect the percentage of LANA-positive cells (Fig. 1C, D), LANA staining pattern and intensity, and the number of LANA punctate dots per cell (Fig. 1C). Hence, KPT-8602 neither affected KSHV infectivity nor LANA protein expression. To confirm that KPT-8602 was unlikely to affect KSHV entry and trafficking, we treated HUVEC with 0.5 µM KPT-8602 for 1 h and then infected them with KSHV. Cells were replaced with fresh medium without the inhibitor at the indicated time points. Supernatants were titrated for infectious virions at 96 hpi. KPT-8602 treatment for 1 h had no significant effect on KSHV titer (Fig. 1E). However, KPT-8602 treatment for >5 h significantly reduced KSHV titer in a time-dependent fashion (Fig. 1E). Particularly, KPT-8602 treatment for >7 h reduced KSHV titer by 84%. These results confirmed that KPT-8602 was unlikely to affect the early steps of KSHV infection.
We then investigated whether KPT-8602 affected the trafficking of KSHV particles to the nucleus. HUVEC pretreated with DMSO or KPT-8602 for 1 h were infected with KSHV for 6 h and stained for ORF65 protein to directly visualize viral capsids (Fig. 1F)30. KPT-8602 treatment did not affect the numbers of viral particles reaching the nucleus (Fig. 1F, G). We further examined KSHV genomes injected into the nucleus by qPCR. HUVEC pretreated with DMSO or 0.5 µM KPT-8602 for 1 h were infected with KSHV with KPT-8602. At 6 hpi, the nuclear and cytoplasmic separation was performed, and both cellular and viral DNA were extracted and quantified by qPCR. XPO1 inhibition did not affect the numbers of viral genomes in cells and in the nucleus (Fig. 1H). These results confirmed that KPT-8602 had no effect on virus entry and trafficking. Thus, KPT-8602 likely reduced KSHV virion production at a post entry stage.
KPT-8602 inhibits expression of KSHV lytic genes
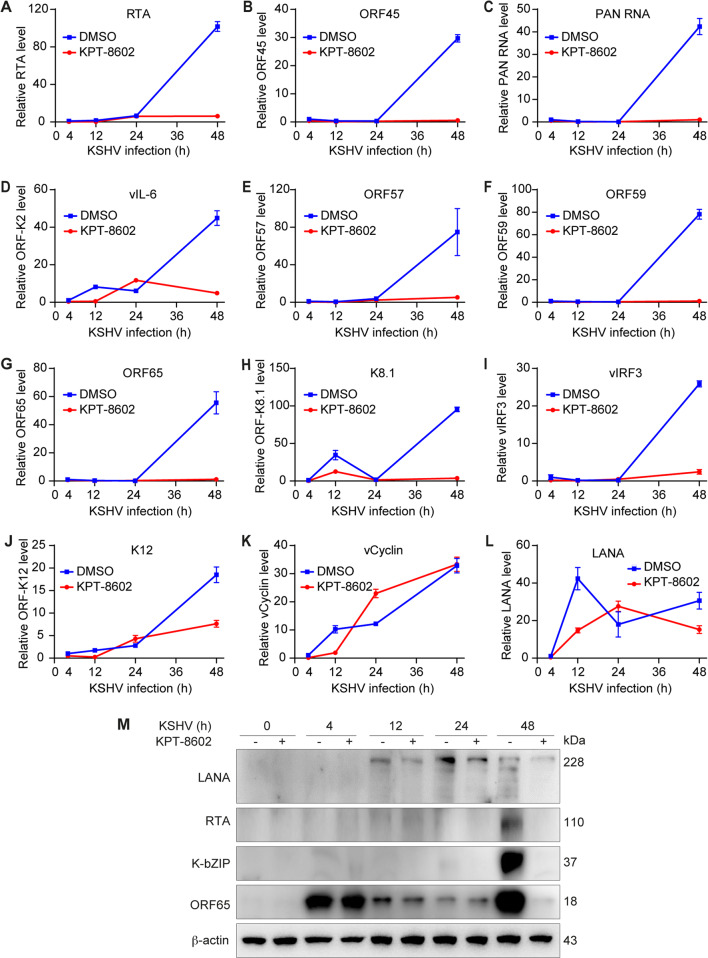
KSHV lytic genes include immediate-early (IE), early (E), and late (L) genes, whereas latent genes mainly consist of LANA, vFLIP, vCyclin, ORF-K12, and a cluster of microRNAs4. During KSHV primary infection in HUVEC, latent transcripts expressed first, followed by lytic transcripts in the order of IE, E, and L transcripts, which peaked at around 54 hpi8. We monitored the kinetics of KSHV lytic genes by RT-qPCR. KPT-8602 significantly reduced mRNA levels of lytic genes including RTA (ORF50), ORF45, vIL-6 (ORF-K2), MTA (ORF57), ORF59, ORF65, and ORF-K8.1 (Fig. 2A–H). For KSHV latent genes, KPT-8602 treatment also inhibited the expression of vIRF3 and ORF-K12 (Fig. 2I, J) but had no significant effect on vCyclin and LANA (Fig. 2K, L). The Western-blotting analysis confirmed that KPT8602 treatment reduced the expression of lytic proteins RTA, K-bZIP (ORF-K8), and ORF65 but only had a marginal effect on latent protein LANA (Fig. 2M), which was consistent with LANA’s immunostaining results (Fig. 1C, D). These results demonstrated that XPO1 had an essential role in the expression of KSHV lytic genes.
Fig. 2. XPO1 inhibitor KPT-8602 inhibits the expression of KSHV genes except for LANA and vCyclin genes during primary infection.
A–L HUVEC were pretreated with 0.5 µM KPT-8602 for 1 h and then infected with KSHV in the presence of KPT-8602 for 4, 12, 24, 36, and 48 h. Cells were collected for RNA extraction and examined for the expression of KSHV transcripts by RT-qPCR. M HUVEC were pretreated with 0.5 µM KPT-8602 for 1 h and then infected with KSHV in the presence of KPT-8602. The cell lysate was collected at 4, 12, 24, and 48 hpi and analyzed for the expression of viral proteins by Western blotting. β-actin served as the loading control.
XPO1 knockdown inhibits KSHV lytic replication
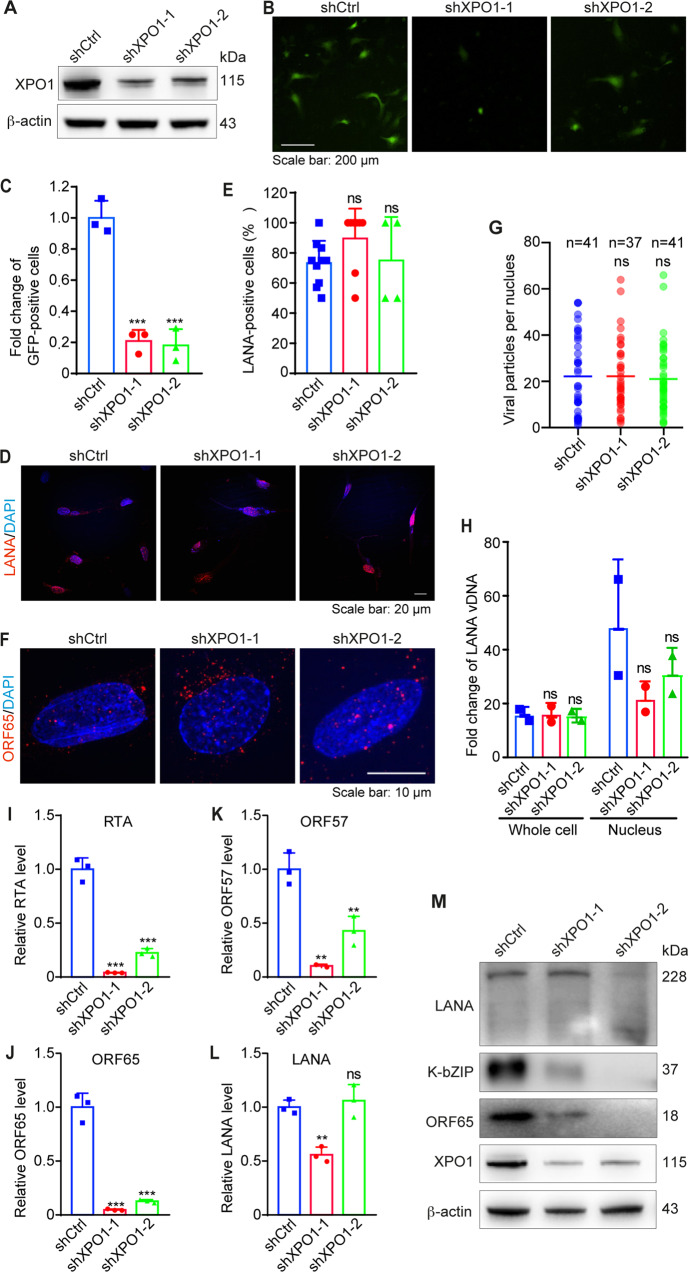
To confirm KPT-8602’s inhibitory effect on KSHV replication, we performed shRNA knockdown of XPO1. XPO1 protein was reduced by >80% in HUVEC transduced with lentiviruses expressing XPO1 shRNAs (Fig. 3A). XPO1 knockdown significantly reduced infectious virions by 80% compared to the scrambled control (Fig. 3B, C), confirming KPT-8602’s effect and XPO1’s critical role in KSHV replication.
Fig. 3. XPO1 knockdown blocks the KSHV lytic replication program but has no effect on KSHV infectivity, and virus entry and trafficking during primary infection.
A HUVEC were infected with lentiviruses expressing shRNAs targeting XPO1 (shXPO1-1 and shXPO1-2) or control shRNA (shCtrl). Cells were selected with puromycin (1 µg/ml) for 48 h and analyzed by Western-blotting to monitor the knockdown efficiency. β-actin was used as the loading control. B, C HUVEC with XPO1 knockdown were infected with KSHV for 96 h. Supernatants were collected and used to titrate infectious virions by infecting fresh HUVEC. At 48 hpi, representative images were taken (B) and used for quantifying the numbers of GFP-positive cells (C). D HUVEC with XPO1 knockdown were infected with KSHV for 48 h. Cells were fixed and stained for LANA (red). The expression level and pattern of LANA are shown. Nuclei were stained with DAPI. Images were taken with a confocal microscopy (magnification ×600). E Quantification of LANA-positive cells based on images taken as described in (D). F HUVEC with knockdown of XPO1 were infected with KSHV for 6 h, fixed, and stained for nuclei with DAPI (blue) and KSHV particles with an antibody to ORF65 (red). Images were taken with a confocal microscopy (magnification ×1000). G The total number of KSHV particles successfully docked at the perinuclear region of each nucleus was quantified and analyzed based on images taken as described in (F). H HUVEC with XPO1 knockdown infected with KSHV for 6 h were collected and analyzed for viral genome copy numbers by qPCR using LANA primers. I–L HUVEC with XPO1 knockdown were infected with KSHV and then collected for RNA extraction at 48 hpi. The expression levels of KSHV transcripts were then examined by RT-qPCR. M HUVEC with XPO1 knockdown were infected with KSHV and collected at 48 hpi. The expression of viral proteins was then analyzed by Western blotting. β-actin was used as a loading control. *, **, and *** indicate P values of < 0.05, < 0.01, and < 0.001, respectively; NS not significant.
To confirm the step of KSHV replication regulated by XPO1, we determined the effect of XPO1 knockdown on KSHV infectivity. HUVEC with XPO1 knockdown were infected with KSHV and stained for LANA protein at 48 hpi. XPO1 knockdown did not change the percentage of LANA-positive cells (Fig. 3D, E), and LANA staining pattern and intensity (Fig. 3D), indicating that XPO1 neither regulated KSHV infectivity nor LANA protein expression.
We further examined the effect of XPO1 knockdown on KSHV entry and trafficking. HUVEC with XPO1 knockdown were infected with KSHV for 6 h and stained for ORF65 protein (Fig. 3F). There was no difference in virus particles docked at the perinuclear region between cells transduced with XPO1 shRNAs or scrambled control (Fig. 3G). XPO1 knockdown also did not affect the internalization of the viral genome into cells and the nucleus (Fig. 3H). These results confirmed that XPO1 did not regulate virus entry and trafficking.
Finally, we investigated the effect of XPO1 knockdown on the expression of KSHV genes. HUVEC with XPO1 knockdown were infected with KSHV for 48 h. XPO1 knockdown significantly reduced mRNA levels of lytic genes RTA, ORF57, and ORF65 (Fig. 3I–K) but only had a minor effect on latent gene LANA (Fig. 3L). XPO1 knockdown also significantly decreased lytic proteins K-bZIP and ORF65 but not latent protein LANA (Fig. 3M). These results confirmed XPO1’s essential role in KSHV lytic replication during primary infection.
KPT-8602 retains p62 in the nucleus and enhances IRF3 and TBK1 activation
We have previously shown that p62 protein is retained in the nucleus in AGS and HUH7 cells following KPT-8602 treatment31. KPT-8602 treatment upregulated p62 protein level and retained p62 in nucleus of uninfected HUVEC (Supplementary Fig. 2). KSHV infection only had a marginal effect on the p62 level.
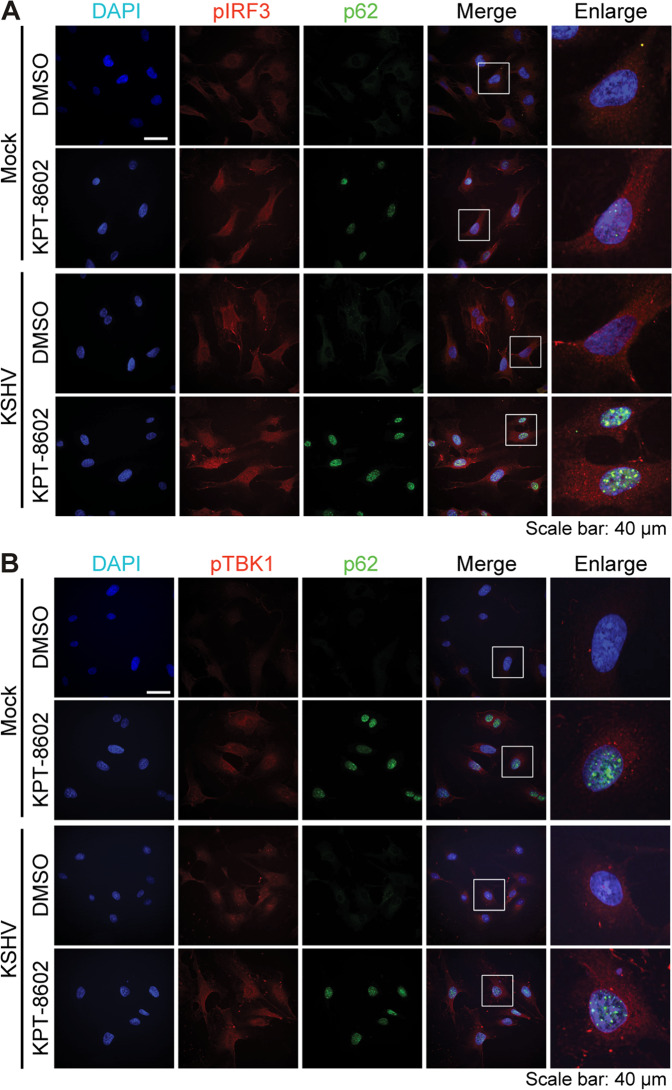
It was reported that cytosolic p62 could attenuate the cGAS-STING pathway and RIG-mediated type I interferon (IFN) signaling32,33. We investigated the activation of TBK-1 and IRF3 upon KPT-8602 treatment. HUVEC were pretreated with 0.5 µM of KPT-8602 or DMSO for 1 h, and then mock-infected or infected with KSHV. At 12 hpi, KSHV infection slightly activated both TBK-1 and IRF3 (Fig. 4A, B) as previously reported34,35. While infection by a DNA virus can activate an innate immune response, KSHV encodes numerous genes that antagonize this response34,36–40. KPT-8602 treatment alone was sufficient to increase levels of both pIRF3 and pTBK1 (Fig. 4A, B). Hence, KPT-8602-induced p62 nuclear accumulation and stabilization might lead to enhanced activation of TBK1 and IRF3.
Fig. 4. Treatment with XPO1 inhibitor KPT-8602 induces p62 nuclear retention and enhances IRF3 and TBK1 activation.
A, B HUVEC were pretreated with KPT-8602 at 0.5 µM or vehicle control DMSO for 1 h and then infected with KSHV for 12 h in the presence of KPT-8602 or DMSO. Cells were fixed and co-stained for pIRF3 (A) or pTBK1 (B) together with p62. Nuclei were stained with DAPI. Images were taken with confocal microscopy (magnification ×600). Magenta p62 was pseudo-colored to green for better observation.
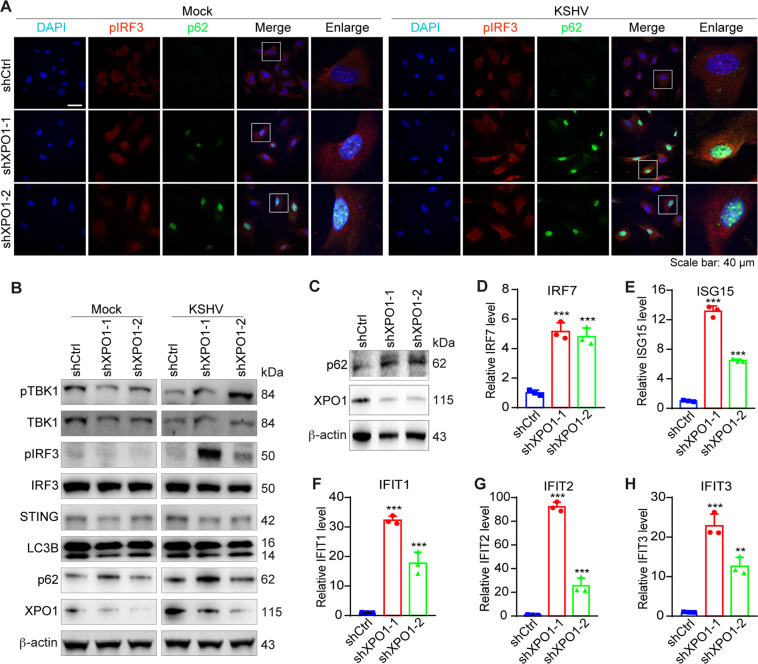
XPO1 knockdown upregulates p62 expression and enhances type I IFN signaling
Next, we confirmed XPO1’s role in the innate immune response by performing XPO1 knockdown. Similar to KPT-8602 treatment, XPO1 knockdown resulted in p62 nuclear accumulation (Supplementary Fig. 3), increased pIRF3 level, and enhanced KSHV activation of pIRF3 and pTBK1 (Fig. 5A, B). However, XPO1 knockdown did not affect STING protein level. There was no obvious change of LC3B cleavage or p62 level following XPO1 knockdown (Fig. 5B), indicating that XPO1 knockdown did not affect autophagy flux and STING was unlikely degraded by p62-mediated autophagy. These results indicated that the enhanced TBK1 and IRF3 activation by XPO1 inhibition were unlikely due to p62’s cytosolic function, rather it might be the result of p62 nuclear accumulation.
Fig. 5. XPO1 knockdown induces p62 nuclear accumulation, enhances IRF3 and TBK1 activation, and upregulates the expression of immune-related genes.
A HUVEC with XPO1 knockdown were infected with KSHV for 12 h, fixed and co-stained for pIRF3 and p62. Nuclei were stained with DAPI. Images were taken with a confocal microscopy (magnification ×600). Magenta p62 was presented as green pseudocolor for better observation. B HUVEC with XPO1 knockdown were infected with KSHV. Cell lysate was collected at 12 hpi and analyzed for levels of pTBK1, TBK1, pIRF3, IRF3, STING, LC3B, p62, and XPO1 by Western-blotting. β-actin was used as a loading control. C HUVEC with XPO1 knockdown were infected with KSHV for 48 h. Cell lysates were collected and analyzed for the expression of p62 by Western-blotting. β-actin was used as a loading control. D–H HUVEC with XPO1 knockdown were infected with KSHV for 48 h. Cells were collected for RNA extraction and examined for the expression of innate immune-related genes IRF7 (D), ISG15 (E), IFIT1 (F), IFIT2 (G), and IFIT3 (H) by RT-qPCR. *, **, and *** indicate P values of < 0.05, < 0.01, and < 0.001, respectively; NS not significant.
To determine whether the increased TBK1 and IRF3 activation led to enhanced innate immune responses following XPO1 knockdown, we examined innate immune-related genes. IFIT1, IFIT2, and IFIT3, which play important roles in KSHV lytic replication41. Compared to control cells, XPO1 knockdown significantly increased mRNA levels of IRF7, ISG15, IFIT1, IFIT, and IFIT3 at 48 hpi in KSHV-infected HUVEC (Fig. 5C–H). These results indicated that XPO1 inhibition indeed enhanced innate immune responses.
p62 knockdown restores KSHV lytic replication program but has no effect on virus entry, trafficking, and infectivity
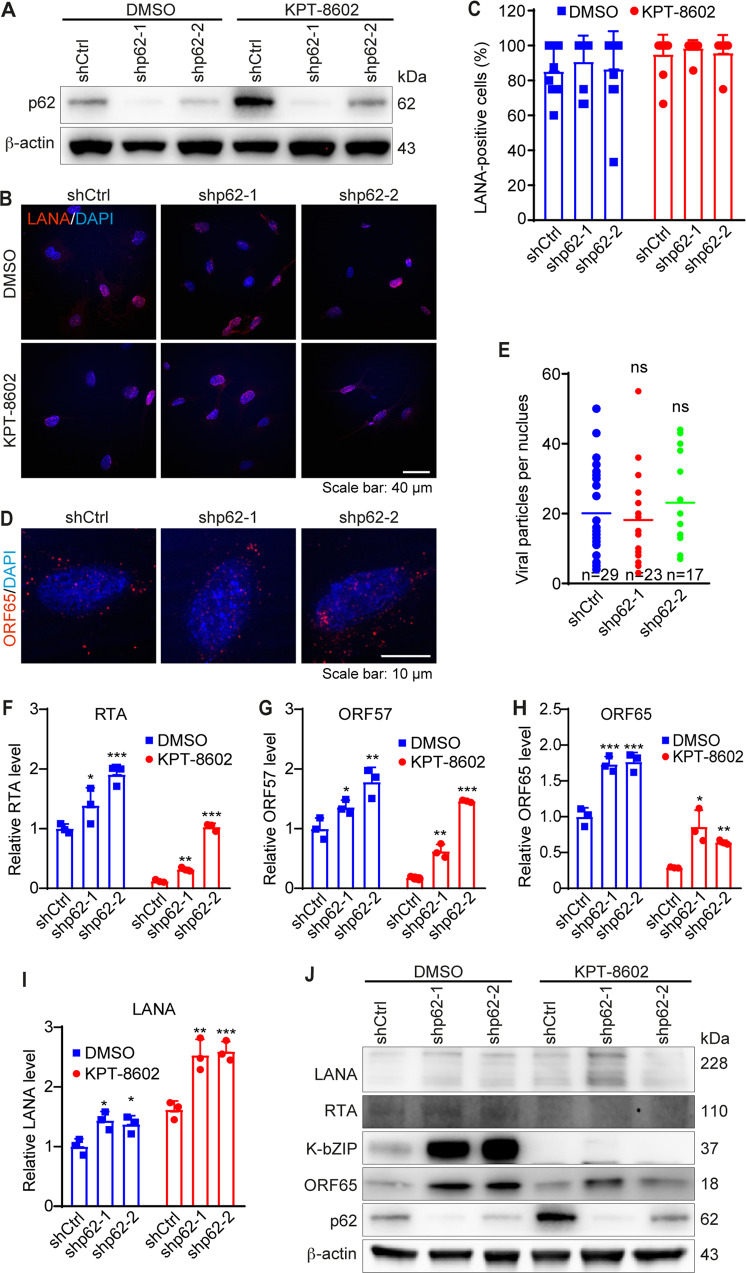
Our results so far showed that XPO1 inhibition reduced KSHV lytic replication by inhibiting the expression of viral lytic genes, which was correlated with p62 nuclear accumulation and enhanced innate immune responses. We further examined p62’s role in KSHV primary infection. p62 knockdown had no effect on KSHV infectivity as there was no obvious change in LANA-positive cells with or without KPT-8602 treatment (Fig. 6A–C). p62 knockdown also had no effect on LANA staining pattern and intensity (Fig. 6B), and the number of KSHV particles docked at the perinuclear region (Fig. 6D, E). These results indicated that p62 had no effect on KSHV entry, trafficking, and infectivity. However, p62 knockdown significantly increased mRNA levels of lytic genes RTA, Kb-ZIP, and ORF65 in the presence of KPT-8602, and rescued the inhibitory effect of KPT-8602 on these viral genes (Fig. 6F, H), indicating that p62 regulated expression of KSHV lytic genes. Interestingly, latent gene LANA was also increased, which persisted in the presence of KPT-8602 despite it had no noticeable effect on LANA mRNA (Fig. 6I), suggesting possible different mechanisms regulating KSHV genes following p62 knockdown and KPT-8602 inhibition. p62 knockdown also significantly increased levels of KSHV RTA, Kb-ZIP, and ORF65 proteins (Fig. 6J). Because of KPT-8602’s strong inhibitory effect on lytic proteins, the enhanced effect of p62 knockdown was not obvious for RTA and Kb-ZIP but was still visible for ORF65 protein (Fig. 6J). Consistent with the mRNA results, p62 knockdown also slightly increased LANA protein level without KPT-8602 treatment albeit it was less consistent with KPT-8602 treatment (Fig. 6J). These results indicated that increased p62 nuclear accumulation might at least partially mediate the inhibitory effect of XPO1 inhibition on KSHV lytic genes.
Fig. 6. p62 knockdown reverses the inhibitory effect of XPO1 inhibitor KPT-8602 on KSHV lytic replication but has no effect on virus infectivity, and entry and trafficking during primary infection.
A HUVEC were infected with lentiviruses expressing shRNAs targeting p62 (shp62-1 and shp62-2) or a control shRNA (shCtrl). Cells were selected with puromycin (1 µg/ml) for 48 h and analyzed by Western-blotting to monitor the knockdown efficiencies. β-actin served as the loading control. B HUVEC with p62 knockdown were pretreated with 0.5 µM KPT-8602. Cells infected with KSHV for 48 h in the presence of KPT-8602 (0.5 µM) were fixed and stained for LANA (red). The expression level and staining pattern of LANA protein are shown. Nuclei were stained with DAPI. Images were taken with a confocal microscopy (magnification ×600). C Quantification of LANA-positive cells using images taken as described in (B). D HUVEC with p62 knockdown were pretreated with 0.5 µM KPT-8602 for 1 h, and then infected with KSHV in the presence of KPT-8602 (0.5 µM) for 6 h. Cells were fixed and stained for nuclei with DAPI (blue) and KSHV particles with an antibody to ORF65 (red). Images were taken with a confocal microscopy (magnification ×1000). E The total number of KSHV particles successfully docked at the perinuclear region of each nucleus was quantified and analyzed based on images taken as described in (D). F–I HUVEC with p62 knockdown was pretreated with 0.5 µM KPT-8602 for 1 h and then infected with KSHV in the presence of KPT-8602 (0.5 µM). Cells were collected for RNA extraction at 48 hpi and examined for the expression of KSHV genes including RTA (F), ORF57 (G), ORF65 (H), and LANA (I) by RT-qPCR. J HUVEC with p62 knockdown were pretreated with 0.5 µM KPT-8602 for 1 h and then infected with KSHV for 48 h in the presence of KPT-8602 (0.5 µM). Cell lysate was collected and analyzed for the expression of p62 and KSHV proteins by Western-blotting. β-actin served as the loading control. *, **, and *** indicate P values of < 0.05, < 0.01, and < 0.001, respectively; NS not significant.
p62 nuclear accumulation induces an innate immune response to block KSHV lytic replication
To investigate p62’s role in innate immune response, we infected HUVEC with KSHV following p62 knockdown. At 48 hpi, p62 knockdown was confirmed (Fig. 7A). p62 knockdown significantly reduced levels of IRF7, ISG15, IFIT1, IFIT2, and IFIT3 genes (Fig. 7B–F), suggesting that p62 was required for maximal activation of the innate immune response during KSHV primary infection.
Fig. 7. Knockdown of p62 reduces the expression of innate immune-related genes during KSHV primary infection.
A–F HUVEC with p62 knockdown were infected with KSHV for 48 h. Cells were collected for RNA extraction and examined for the expression of p62 (A), IRF7 (B), ISG15 (C), IFIT1 (D), IFIT2 (E), and IFIT3 (F) by RT-qPCR. *, **, and *** indicate P values of < 0.05, < 0.01, and < 0.001, respectively; NS not significant.
To explore the role of p62 nuclear accumulation in innate immune response upon XPO1 inhibition, HUVEC with p62 knockdown were pretreated with KPT-8602 at 0.5 µM for 1 h and infected with KSHV for 6 or 12 h. Both pIRF3 and pTBK1 levels were reduced in HUVEC with p62 knockdown compared to cells transduced with the scrambled control (Fig. 8A, B). We observed increased levels of pTBK1, pIRF3, and p62 after KPT-8602 treatment in KSHV-infected HUVEC transduced with the control shRNA. However, p62 knockdown reversed levels of pTBK1 and pIRF3 (Fig. 8C). LC3B cleavage was not increased in HUVEC treated with KPT-8602 (Fig. 8C). These results indicated that p62 nuclear accumulation caused by XPO1 inhibition enhanced TBK1 and IRF3 activation. Finally, RNA was extracted and qRT-PCR was performed at 48 hpi to detect p62 and immune-related genes. Treatment of 0.5 µM KPT-8602 increased mRNA levels of p62, IRF7, ISG15, IFIT1, IFIT2, and IFIT3 (Fig. 7D–I). However, p62 knockdown significantly reversed levels of immune-related genes induced by XPO1 inhibition (Fig. 7D–I). Taken together, these results indicated that XPO1 inhibition caused p62 nuclear accumulation, which enhanced type I IFN signaling and blocked KSHV replication.
Fig. 8. p62 nuclear retention mediates induction of innate immune responses and inhibits KSHV lytic replication during primary infection.
A, B HUVEC with p62 knockdown were pretreated with KPT-8602 at 0.5 µM for 1 h and then infected with KSHV for 6 or 12 h in the presence of KPT-8602. Cells were fixed and co-stained for pIRF3 (A) or pTBK1 (B) together with p62. Nuclei were stained with DAPI. Images were taken with a confocal microscopy (magnification ×600). Magenta p62 was presented as green pseudocolor for better observation. C HUVEC with p62 knockdown were pretreated with KPT-8602 at 0.5 µM or control vehicle DMSO for 1 h, and then infected with KSHV for 6 or 12 h in the presence of KPT-8602 or DMSO. Cells were collected and analyzed for levels of pTBK1, TBK1, pIRF3, IRF3, and LC3B by Western-blotting. β-actin served as the loading control. D–I HUVEC with p62 knockdown was pretreated with KPT-8602 at 0.5 µM or DMSO for 1 h and infected with KSHV in the presence of KPT-8602 or DMSO. Cells were collected for RNA extraction at 48 hpi and examined for the expression of p62 (D), IRF7 (E), ISG15 (F), IFIT1 (G), IFIT2 (H), and IFIT3 (I) by RT-qPCR. *, **, and *** indicate P values of < 0.05, < 0.01, and < 0.001, respectively; NS not significant.
Discussion
Viruses often hijack cell machinery to facilitate infection. XPO1 mediates the replication of numerous viruses by regulating the nuclear export of viral and cellular proteins17,18. KSHV LANA2 and ORF45 harbor NESs and are regulated by XPO142,43. Nuclear shuttling of KSHV proteins could implicate important functions of importins and exportins in KSHV infection. Nevertheless, XPO1’s role in KSHV primary infection remains unknown.
We show that XPO1 inhibition by KPT-8602 or shRNA-mediated knockdown reduces KSHV replication during primary infection without affecting virus entry and trafficking. Numerous XPO1 inhibitors are available including LMB, ratjadone A, thymoquinone, plumbagin, and piperlongumine18,44–48. Recently, a novel class of SINE compounds has been identified21–23. Unlike LMB, which permanently binds to XPO1 NES groove, SINE compounds such as KPT-330 (selinexor), KPT-335 (verdinexor), KPT-185, and KPT-350 form a reversible covalent bond with the cysteine 528 residue, and hence are less cytotoxic than LMB21,22,49. SINE compounds reduce replication of influenza virus, Venezuelan equine encephalitis virus, and respiratory syncytial virus22,49,50. As the second generation of SINE, KPT-8602 manifests improved in vivo efficacy and tolerability in hematologic malignancies compared to KPT-33024,51,52. We demonstrate KPT-8602 as a promising therapeutic agent against KSHV replication. Furthermore, our previous study has identified KPT-8602 as an effective agent of KSHV-transformed cells31.
Our results show that XPO1 inhibition does not affect KSHV entry, trafficking, infectivity, and expression of latent gene LANA, rather it inhibits the expression of lytic genes. ORF45 is an IE gene-regulating lytic replication53. However, the negative effect of XPO1 inhibition on lytic replication is unlikely due to the inhibition of ORF45 nuclear shuttling or ORF45’s direct participation in lytic replication as an expression of RTA, which is required for the expression of other downstream lytic genes including ORF45, precedes that of ORF454. Instead, we observe that XPO1 inhibition leads to p62 nuclear accumulation, and enhanced innate immune response accompanying increased expression of innate immune-related genes including IFIT1, IFIT2, and IFIT3 known to inhibit KSHV lytic replication41. Nevertheless, since encapsidated ORF45 protein inhibits innate immune response38, it remains possible that XPO1 inhibition might affect encapsidated ORF45 protein function, leading to enhanced innate immune responses and decreased KSHV lytic replication.
The effect of KSHV replication on autophagy remains controversial. RTA overexpression induces autophagy in 293 T cells54. While chemical induction of KSHV lytic replication increases autophagy in PEL cells, it is blocked at a late stage of viral replication55. Interestingly, KSHV inhibits autophagy by activating mTORC1 during nutrient-deprived conditions, which is important for viral replication56. We did not observe any LC3II and p62 changes, indicating that there was no effect on autophagy initiation and flux during the KSHV primary infection of HUVEC. Different cell types and experimental systems likely contribute to these discrepancies.
Autophagy adaptor protein p62 is involved in multiple cellular processes, including autophagy, inflammatory responses, and redox homeostasis57–61. p62 dysfunction is associated with Parkinson’s disease, Alzheimer’s disease, and tumorigenesis62–65. Interestingly, p62 is categorized as a new group of innate immunity receptors66. p62 delivers specific cytosolic components and ubiquitinated targets to autophagic organelles for removal67,68. The role of cytosolic p62 has recently been expanded. p62-mediated autophagy selectively degrades RIG-I and STING, attenuating type I IFN signaling32,33. p62 harbors active nuclear import and export signals and shuttles continuously between the nucleus and cytoplasm69. Nevertheless, p62’s role within the nucleus remains unknown. Our results show that XPO1 inhibition enhances the activation of TBK1 and IRF3 and innate immune responses. While p62 nuclear retention following XPO1 inhibition might lead to reduced cytosolic p62 and enhanced innate immune response32,33, there is no change of STING. Hence, the enhanced innate immune responses following XPO1 inhibition are unlikely due to the compromised cytosolic function of p62, rather it might be due to the gain-of-function of nuclear p62, leading to an enhanced innate immune response and inhibition of KSHV replication. Hence, similar to p62, XPO1 appears to function to control activation of the innate immune response in normal conditions. This function is likely important for controlling hyperactivation of the innate immune system in conditions without pathogen invasion. Indeed, genetic mutations of both p62 and XPO1 are associated with various diseases including inflammatory diseases70–72. Hence, precaution should be taken when targeting p62 and XPO1 for therapy. It remains unclear how XPO1 inhibition and nuclear p62 enhance innate immune responses. Furthermore, p62 and XPO1 might be directly involved in the infection and replication of other viruses, which might warrant further investigations.
Supplementary information
Acknowledgements
We thank members of Dr. Gao’s laboratory for technical assistance and helpful discussions. This work was supported by grants from NIH (CA096512, CA124332, CA132637, CA213275, DE025465, and CA197153) to S.-J. Gao. This work was supported in part by award P30CA047904.
Author contributions
Wen Meng designed and performed experiments, analyzed and interpreted data, wrote drafts and revised the paper, and approved the final submitted paper. Shou-Jiang Gao secured funding for the project, planned and managed the project, designed experiments, analyzed and interpreted data, wrote and revised the paper, approved the final submitted paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by H.-U. Simon
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-03303-1).
References
- 1.Chang Y, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Soulier J, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. doi: 10.1182/blood.V86.4.1276.bloodjournal8641276. [DOI] [PubMed] [Google Scholar]
- 4.Ye F, Lei X, Gao SJ. Mechanisms of Kaposi’s sarcoma-associated herpesvirus latency and reactivation. Adv. Virol. 2011;2011:193860. doi: 10.1155/2011/193860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao SJ, Deng JH, Zhou FC. Productive lytic replication of a recombinant Kaposi’s sarcoma-associated herpesvirus in efficient primary infection of primary human endothelial cells. J. Virol. 2003;77:9738–9749. doi: 10.1128/JVI.77.18.9738-9749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan H, Xie J, Ye F, Gao SJ. Modulation of Kaposi’s sarcoma-associated herpesvirus infection and replication by MEK/ERK, JNK, and p38 multiple mitogen-activated protein kinase pathways during primary infection. J. Virol. 2006;80:5371–5382. doi: 10.1128/JVI.02299-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie J, Pan H, Yoo S, Gao SJ. Kaposi’s sarcoma-associated herpesvirus induction of AP-1 and interleukin 6 during primary infection mediated by multiple mitogen-activated protein kinase pathways. J. Virol. 2005;79:15027–15037. doi: 10.1128/JVI.79.24.15027-15037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoo SM, Zhou FC, Ye FC, Pan HY, Gao SJ. Early and sustained expression of latent and host modulating genes in coordinated transcriptional program of KSHV productive primary infection of human primary endothelial cells. Virology. 2005;343:47–64. doi: 10.1016/j.virol.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grossmann C, Ganem D. Effects of NFkappaB activation on KSHV latency and lytic reactivation are complex and context-dependent. Virology. 2008;375:94–102. doi: 10.1016/j.virol.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnan HH, et al. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi’s sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 2004;78:3601–3620. doi: 10.1128/JVI.78.7.3601-3620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadagopan S, et al. Kaposi’s sarcoma-associated herpesvirus induces sustained NF-kappaB activation during de novo infection of primary human dermal microvascular endothelial cells that is essential for viral gene expression. J. Virol. 2007;81:3949–3968. doi: 10.1128/JVI.02333-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye FC, et al. Kaposi’s sarcoma-associated herpesvirus latent gene vFLIP inhibits viral lytic replication through NF-kappaB-mediated suppression of the AP-1 pathway: a novel mechanism of virus control of latency. J. Virol. 2008;82:4235–4249. doi: 10.1128/JVI.02370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu. Rev. Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 14.Matsuura Y. Mechanistic insights from structural analyses of Ran-GTPase-driven nuclear export of proteins and RNAs. J. Mol. Biol. 2016;428:2025–2039. doi: 10.1016/j.jmb.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 15.Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 16.Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Sage V, Mouland AJ. Viral subversion of the nuclear pore complex. Viruses. 2013;5:2019–2042. doi: 10.3390/v5082019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathew C, Ghildyal R. CRM1 inhibitors for antiviral therapy. Front. Microbiol. 2017;8:1171. doi: 10.3389/fmicb.2017.01171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 20.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/S0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 21.London CA, et al. Preclinical evaluation of the novel, orally bioavailable Selective Inhibitor of Nuclear Export (SINE) KPT-335 in spontaneous canine cancer: results of a phase I study. PLoS ONE. 2014;9:e87585. doi: 10.1371/journal.pone.0087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perwitasari O, et al. Verdinexor, a novel selective inhibitor of nuclear export, reduces influenza a virus replication in vitro and in vivo. J. Virol. 2014;88:10228–10243. doi: 10.1128/JVI.01774-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranganathan P, et al. Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood. 2012;20:1765–1773. doi: 10.1182/blood-2012-04-423160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Etchin J, et al. KPT-8602, a second-generation inhibitor of XPO1-mediated nuclear export, is well tolerated and highly active against AML blasts and leukemia-initiating cells. Leukemia. 2017;31:143–150. doi: 10.1038/leu.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brulois KF, et al. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J. Virol. 2012;86:9708–9720. doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou FC, et al. Efficient infection by a recombinant Kaposi’s sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 2002;76:6185–6196. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao SJ, et al. Seroconversion to antibodies against Kaposi’s sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi’s sarcoma. N. Engl. J. Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Q, et al. Kaposi’s sarcoma-associated herpesvirus-encoded replication and transcription activator impairs innate immunity via ubiquitin-mediated degradation of myeloid differentiation factor 88. J. Virol. 2015;89:415–427. doi: 10.1128/JVI.02591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He M, et al. Cancer angiogenesis induced by Kaposi sarcoma-associated herpesvirus is mediated by EZH2. Cancer Res. 2012;72:3582–3592. doi: 10.1158/0008-5472.CAN-11-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greene W, Gao SJ. Actin dynamics regulate multiple endosomal steps during Kaposi’s sarcoma-associated herpesvirus entry and trafficking in endothelial cells. PLoS Pathog. 2009;5:e1000512. doi: 10.1371/journal.ppat.1000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gruffaz M, et al. CRISPR-Cas9 screening of Kaposi’s sarcoma-associated herpesvirus-transformed cells identifies XPO1 as a vulnerable target of cancer cells. mBio. 2019;10:e00866–00819. doi: 10.1128/mBio.00866-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prabakaran T, et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018;37:e97858. doi: 10.15252/embj.201797858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du Y, et al. LRRC25 inhibits type I IFN signaling by targeting ISG15-associated RIG-I for autophagic degradation. EMBO J. 2018;37:351–366. doi: 10.15252/embj.201796781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Z, et al. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc. Natl Acad. Sci. USA. 2015;112:E4306–E4315. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iqbal J, et al. Histone H2B-IFI16 recognition of nuclear herpesviral genome induces cytoplasmic interferon-beta responses. PLoS Pathog. 2016;12:e1005967. doi: 10.1371/journal.ppat.1005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefort S, Soucy-Faulkner A, Grandvaux N, Flamand L. Binding of Kaposi’s sarcoma-associated herpesvirus K-bZIP to interferon-responsive factor 3 elements modulates antiviral gene expression. J. Virol. 2007;81:10950–10960. doi: 10.1128/JVI.00183-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cloutier N, Flamand L. Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen inhibits interferon (IFN) beta expression by competing with IFN regulatory factor-3 for binding to IFNB promoter. J. Biol. Chem. 2010;285:7208–7221. doi: 10.1074/jbc.M109.018838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Q, et al. ORF45 of Kaposi’s sarcoma-associated herpesvirus inhibits phosphorylation of interferon regulatory factor 7 by IKKepsilon and TBK1 as an alternative substrate. J. Virol. 2012;86:10162–10172. doi: 10.1128/JVI.05224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobs SR, et al. Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor 1 interacts with a member of the interferon-stimulated gene 15 pathway. J. Virol. 2015;89:11572–11583. doi: 10.1128/JVI.01482-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang G, et al. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc. Natl Acad. Sci. USA. 2016;113:E1034–E1043. doi: 10.1073/pnas.1516812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li D, Swaminathan S. Human IFIT proteins inhibit lytic replication of KSHV: a new feed-forward loop in the innate immune system. PLoS Pathog. 2019;15:e1007609. doi: 10.1371/journal.ppat.1007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munoz-Fontela C, et al. Identification of a nuclear export signal in the KSHV latent protein LANA2 mediating its export from the nucleus. Exp. Cell Res. 2005;311:96–105. doi: 10.1016/j.yexcr.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 43.Li X, Zhu F. Identification of the nuclear export and adjacent nuclear localization signals for ORF45 of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2009;83:2531–2539. doi: 10.1128/JVI.02209-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamamoto T, Gunji S, Tsuji H, Beppu T. Leptomycins A and B, new antifungal antibiotics. I. Taxonomy of the producing strain and their fermentation, purification and characterization. J. Antibiot. 1983;36:639–645. doi: 10.7164/antibiotics.36.639. [DOI] [PubMed] [Google Scholar]
- 45.Fleta-Soriano E, et al. The myxobacterial metabolite ratjadone A inhibits HIV infection by blocking the Rev/CRM1-mediated nuclear export pathway. Microb. Cell Fact. 2014;13:17. doi: 10.1186/1475-2859-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forouzanfar F, Bazzaz BS, Hosseinzadeh H. Black cumin (Nigella sativa) and its constituent (thymoquinone): a review on antimicrobial effects. Iran. J. Basic Med. Sci. 2014;17:929–938. [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X, et al. CRM1 is a direct cellular target of the natural anti-cancer agent plumbagin. J. Pharm. Sci. 2014;124:486–493. doi: 10.1254/jphs.13240FP. [DOI] [PubMed] [Google Scholar]
- 48.Niu M, et al. Piperlongumine is a novel nuclear export inhibitor with potent anticancer activity. Chem. Biol. Interact. 2015;237:66–72. doi: 10.1016/j.cbi.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 49.Lundberg L, et al. Selective inhibitor of nuclear export (SINE) compounds alter New World alphavirus capsid localization and reduce viral replication in mammalian cells. PLoS Negl. Trop. Dis. 2016;10:e0005122. doi: 10.1371/journal.pntd.0005122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pickens JA, Tripp RA. Verdinexor targeting of CRM1 is a promising therapeutic approach against RSV and influenza viruses. Viruses. 2018;10:48. doi: 10.3390/v10010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hing ZA, et al. Next-generation XPO1 inhibitor shows improved efficacy and in vivo tolerability in hematological malignancies. Leukemia. 2016;30:2364–2372. doi: 10.1038/leu.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vercruysse T, et al. The second-generation exportin-1 inhibitor KPT-8602 demonstrates potent activity against acute lymphoblastic leukemia. Clin. Cancer Res. 2017;23:2528–2541. doi: 10.1158/1078-0432.CCR-16-1580. [DOI] [PubMed] [Google Scholar]
- 53.Zhu FX, Li X, Zhou F, Gao S-J, Yuan Y. Functional characterization of Kaposi’s sarcoma-associated herpesvirus ORF45 by bacterial artificial chromosome-based mutagenesis. J. Virol. 2006;80:12187–12196. doi: 10.1128/JVI.01275-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wen HJ, Yang Z, Zhou Y, Wood C. Enhancement of autophagy during lytic replication by the Kaposi’s sarcoma-associated herpesvirus replication and transcription activator. J. Virol. 2010;84:7448–7458. doi: 10.1128/JVI.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Granato M, et al. The activation of KSHV lytic cycle blocks autophagy in PEL cells. Autophagy. 2015;11:1978–1986. doi: 10.1080/15548627.2015.1091911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pringle ES, Robinson CA, McCormick C. Kaposi’s sarcoma-associated herpesvirus lytic replication interferes with mTORC1 regulation of autophagy and viral protein synthesis. J. Virol. 2019;93:e00854–00819. doi: 10.1128/JVI.00854-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on Huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 59.Lau A, et al. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol. Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong Z, et al. NF-kappaB restricts inflammasome activation via elimination of damaged mitochondria. Cell. 2016;164:896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carroll B, et al. Oxidation of SQSTM1/p62 mediates the link between redox state and protein homeostasis. Nat. Commun. 2018;9:256. doi: 10.1038/s41467-017-02746-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakaso K, et al. Transcriptional activation of p62/A170/ZIP during the formation of the aggregates: possible mechanisms and the role in Lewy body formation in Parkinson’s disease. Brain Res. 2004;1012:42–51. doi: 10.1016/j.brainres.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 63.Duran A, et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 64.Ramesh Babu J, et al. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J. Neurochem. 2008;106:107–120. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- 65.Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deretic V. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr. Opin. Immunol. 2012;24:21–31. doi: 10.1016/j.coi.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng YT, et al. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- 68.Ponpuak M, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–341. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pankiv S, et al. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J. Biol. Chem. 2010;285:5941–5953. doi: 10.1074/jbc.M109.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bartolome F, et al. Pathogenic p62/SQSTM1 mutations impair energy metabolism through limitation of mitochondrial substrates. Sci. Rep. 2017;7:1666. doi: 10.1038/s41598-017-01678-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cosin-Roger J, et al. Hypoxia ameliorates intestinal inflammation through NLRP3/mTOR downregulation and autophagy activation. Nat. Commun. 2017;8:98. doi: 10.1038/s41467-017-00213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.