Abstract
The installation of HVAC systems in building is meant to enhance indoor air quality as well as increase comfort to occupants. However, HVAC systems have also become a vehicle of contamination of indoor air with potentially pathogenic microorganisms. DNA was extracted from ten HVAC filter dust samples collected from two buildings and subjected to high throughput sequencing analysis to determine the bacterial community structure. Further, the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) software was used to predict the potential functional capabilities of the bacterial communities. Sequencing analysis led to the identification of five major bacterial phyla, including Proteobacteria, Cyanobacteria, Actinobacteria, Firmicutes and Bacteroidetes. At genus level, Mycobacterium, Bacillus, Cupriavidus, Hyphomicrobium and Mesorhizobium were the most dominant. With the exception of the later two bacterial genera, the first three are potential pathogens whose presence in HVAC systems poses a significant public health risk, especially among immunocompromised individuals. Nine pathways associated with antibiotics resistance and bacterial pathogenicity were identified, including polymyxin resistance and peptidoglycan biosynthesis pathways. Further, investigation of the relationship between the detected bacterial meta-communities and predicted potential virulence factors (antibiotic resistance and pathogenic genes) led to the detection of 350 positive associations among 43 core bacteria, 2 pathogenic genes (sitA and uidA) and 14 resistance genes. Overall, the heterogeneous nature of microorganisms found in HVAC systems observed in this study shows that HVAC systems are the origin of airborne infections in indoor environments, and must be periodically cleaned and disinfected to avoid the build-up of pathogens, and the subsequent exposure of human occupants of these pathogens.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10661-020-08823-z.
Keywords: HVAC systems, Bioaerosols, Indoor air, Public health, Sequencing analysis, PICRUSt2
Introduction
Lifestyle changes are resulting in most people spending 30 to 90% of their time inside buildings (Adams et al. 2016; Andualem et al. 2019; Stryjakowska-Sekulska et al. 2007). The quality of indoor air in private and public buildings has, therefore, become an essential determinant of people’s health and well-being (Dumala and Dudzinska 2013; Nunes et al. 2005). Deterioration of indoor air quality has been attributed mostly to the presence of aeroallergens and bio-aerosols, including bacteria, fungi and viruses (Brągoszewska et al. 2018; Fekadu and Getachewu 2015; Macher et al. 1991). The installation of heating, ventilation and air conditioning (HVAC) systems in offices and other buildings is therefore designed to enhance comfort by maintaining ideal levels of humidity (typically between 40 and 60%) and temperature (Eleyowo and Amusa 2019; Ito et al. 2018), as well as improving the quality of indoor air by filtering out bio-aerosols, dust, allergens and reducing odours.
In addition, due to the design of some modern buildings, parts of which may lack direct natural ventilation, HVAC systems not only increase comfort but also enable maximum space utilization as well as improve indoor air quality by removing up to 80% of bioaerosols. But, the indoor air of many such buildings has been reported to be polluted with a myriad of dynamic microbial populations, at times with as much as 106 bacteria/m3 of air (Hewitt et al. 2012). Various research findings have shown that HVAC systems provide favourable breeding grounds for some biological agents like bacteria and fungi leading to subsequent but unintentional pollution of indoor air (Levetin et al. 2001; Schmidt et al. 2012; Wu et al. 2016). Of particular, public health concern is the potential growth or presence of pathogenic bacteria and mycotoxigenic fungi in the dark and consistently moist conditions presented by the cooling coils and drip pans of HVAC systems (Mendell 2004), and their subsequent discharge into ambient air on initial startup (Prussin and Marr 2015; Schmidt et al. 2012). Furthermore, emerging pathogens such as noroviruses and coronaviruses with potential for airborne transmission over short distances could easily be transmitted from person to person in overcrowded buildings equipped with HVAC system (Carducci et al. 2020).
The combination of prolonged exposure to HVAC systems and the potential of airborne bioaerosols to cause disease has been attributed to an increase in building-related diseases, including allergic rhinitis, asthma, hypersensitivity pneumonitis, acute toxicosis and cancer (Eleyowo and Amusa 2019; Levetin et al. 2001), most of which are a result of respiratory exposure to fungal biomass and mycotoxins (Bernstein et al. 2010; Yamamoto et al. 2015). Unfortunately, these ailments are seldom traced back to HVAC exposures (Mendell 2004), nor are they linked, by any measure of accuracy, to certain microbial species which make up part of the microbiome of those HVAC systems. Determining the microbiomes of HVAC systems is, therefore, important for indoor risk exposure assessment (Grisoli et al. 2019), as well as risk management activities related to occupational health. This particularly applies to public buildings whose bioaerosol compositions are influenced by the influx of microbes brought in by workers and visitors alike (Hewitt et al. 2012; Hospodsky et al. 2012), making for a highly dynamic microbiome (Takaoka and Norbäck 2020).
The primary goal of this research, therefore, was to determine the bacterial diversity of HVAC systems using next generation sequencing (NGS) platform in two university buildings. The first building houses offices and meeting rooms, and hence characterisation of bacterial communities in terms of diversity and functional genes in its HVAC system was meant to highlight the risk posed to its occupants. The second building is a laboratory, where students do their research, handling and processing plant, animal and microbiological samples. We therefore hypothesised that there would be a large variation on the microbiome of HVAC systems of the two building due to their different uses, over and above the dictates of the greater outdoor environment. Such insight can help inform how frequent HVAC systems should be cleaned and disinfected to reduce human exposure to microorganisms that cause adverse health effects.
Methods and materials
Sample collection
Two buildings within the University of South Africa, Florida Science Campus located in the Roodepoort Area, Johannesburg, South Africa (S 26° 9.501, E 27° 54.113) were chosen for the study. Both buildings, situated approximately 100 m from each other, are fitted with HVAC systems. The first building (Calabash Building, hereafter referred to as the office building) is a university office complex housing the College of Agriculture Environmental Sciences (CAES), which consist mainly of staff rooms, meeting halls, board rooms and toilets. The second building (Eureka Building, hereafter referred to as the laboratory building) houses different research laboratories including botany, chemistry, anatomy and microbiology. Before embarking on this study, permission and ethical clearance (2017/CAES/110) was obtained from the University Ethics Committee. To characterize the indoor microbial community, ten samples of HVAC filter dust were collected from two buildings. For the office building, six samples were collected randomly from offices, board room and meeting hall. While from the laboratory building, 4 samples were collected from each floor. Prior to the sample collection, HVAC filters were cleaned from selected system and placed in the air handling units for 30 days. Samples were collected using sterile swabs and immediately transferred into isotonic phosphate buffer solution and transferred immediately to the laboratory. All filter samples were stored at – 20 °C until further analysis.
DNA extraction, library preparation and Illumina Miseq high throughput sequencing
Environmental DNA (eDNA) was extracted from 5 ml of each collected samples using Faecal/Soil Total DNA™ extraction kit (Zymo Research Corporation, CA, USA) according to the manufacturer’s instructions. The resultant DNA concentration and quality were checked at 260 nm wavelength and absorbance ratios of 260/280 nm on a NanoDrop Spectrophotometer (NanoDrop Technology, USA), before preserving the DNA at – 20 °C until further processing.
DNA samples were subjected to high throughput sequencing (HTS) by Illumina MiSeq technology, according to Illumina 16S metagenomic Library Preparation and sequencing on the V3 chemistry (2 × 300 bp) recommendations (Illumina Inc., San Diego, CA, USA), with slight modifications. Briefly, the extracted DNA was first amplified to cover the whole variable region of bacterial 16S rDNA using universal 27F and 1492R primers under the following PCR conditions: initial denaturation at 95 °C for 5 min, 32 cycles consisting of 1 min at 95 °C, 1 min at 55 °C and 1 min at 72 °C, and a final extension of 7 min at 72 °C followed by cooling at 4 °C. This was followed by a second nested PCR to cover the bacterial 16S rRNA V1–V3 hypervariable region was performed using 27F and 518R primer pairs, fused with MiSeq adapters and heterogeneity spacers compatible with Illumina indexes for multiplex sequencing, as recommended by Illumina (https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf). Cleaning of the resultant PCR products, index library preparation, pooling and sequencing on Illumina Miseq 250® to generate paired 300-bp high-quality reads of the V1-V3 region were performed according to standard protocol (Illumina Inc., San Diego, CA, USA).
Processing of HTS datasets, diversity and statistical analyses
The demultiplexed paired-end sequencing files were first screened for quality using FastQC v0.11.5 (Babraham Institute, United Kingdom). Further quality-check was performed to eliminate sequencing reads with Phred Q score lower than 20 using Trimmomatic v0.38 (Bolger et al. 2014). The obtained reads were then subjected to processing using the Quantitative Insights into Microbial Ecology (QIIME) v2020.2 (Bolyen et al. 2019). DADA2 (Callahan et al. 2016) was used for further sequence denoising and clustering into amplicon sequence variants (ASVs). The obtained ASVs were futher binned into operational taxonomic units at 97% sequence similarity using VSEARCH (Rognes et al. 2016) based on open-reference OTU picking strategy against the SILVA v132 (Quast et al. 2013) 16S rRNA reference database. The representative sequences were assigned taxonomy using the default parameters of the QIIME VSEARCH consensus taxonomy classifier against the SILVA v132 reference database.
Prior to diversity analyses, the OTU table was rarefied to an even sequencing depth of 1388 reads. Also, low variance sequences were filtered by inter-quantile range while minimum OTU count was set to 4 to reduce the presence of sequences considered as low-level contaminants. Alpha diversity indices, Chao1 and Shannon-H, were calculated using phyloseq (McMurdie and Holmes 2013) while β-diversity was determined using Bray-Curtis dissimilarity matrices and principal coordinates analysis on Ampvis2 (Andersen et al. 2018) packages on R (v3.5.2) (R Core Team 2019). Pairwise variations in α-diversity was tested for significance (p ≤ 0.05) using two-sample Welch’s t test while PERMANOVA analysis of beta-diversity was carried out using Calinski-Harabasz index (pseudo-F) as the test statistics, to determine the significance of the sample clustering pattern. The OTUs at different taxonomic levels were used to generate stacked bar charts and heatmap using ggplot2 (Wickham 2016) and heatmap.2 packages (Warnes et al. 2015) in R version 3.5.2 (R Core Team 2019), respectively, to visualize the variations and distributions of present bacterial communities. Significant differences in the metacommunity of the two buildings were determined by considering significant (q ≤ 0.05) false discovery rate (FDR) scores using Aldex2 package (Fernandes et al. 2014) in R statistical software.
Functional prediction and co-network analysis
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) software package (Douglas et al. 2019) was used to predict and understand the potential functional capabilities and difference between the two built environment HVAC system bacterial communities. Towards this, the Nearest-Sequenced Taxon Index (NSTI) value was used to validate the reliability of predicted gene families and metabolic pathways as NSTI values ≥ 2 are considered noisy and unreliable. PICRUSt2 predicts gene families from the ASVs based on their nearest known taxonomic neighbour as interpolated from available fully sequenced genomes. In this case, the ASVs were first aligned to HMMER (Eddy 2011). The aligned sequences were subsequently placed into a reference tree using EPA-NG (Barbera et al. 2019) and gappa (Czech et al. 2019). Normalization of multiple 16S rRNA gene copies and prediction of gene families was achieved using Castor—a hidden state prediction tool (Louca and Doebeli 2018). The predicted gene families were subsequently collapsed into MetaCyc pathways using MinPath (Ye and Doak 2009) in order to have a simple overview of the HVAC community metagenome. Detected pathways were subjected to differential abundance analysis using Aldex2 package (Fernandes et al. 2014) in R statistical software. Pathways with significant p values (≥ 0.05), Benjamini-Hochberg’s FDR score ≤ 0.05 and effect size ≥ 0.6 were considered differentially abundant. The heatmap of the predicted relative abundances of genes related to different functions was generated using heatmap.2 package in R (v3.5.2) (R Core Team 2019). Bacterial interaction networks on the predicted functional activities and taxa involved were generated using CoNet (Faust and Raes 2016), based on significant correlations among taxa determined by Spearman rank correlation, Bray–Curtis dissimilarity and Kullback–Leibler dissimilarity. The resultant p values were adjusted and merged using Benjamini–Hochberg multiple test correction (Benjamini and Hochberg 1995) and Brown’s method as previously described by Selvarajan et al. (2019). Co-occurrence network models generated were visualized and customised using Cytoscape v3.7.2 (Shannon et al. 2003), to show the differential statistics such as the number of nodes (core OTUs), edges (associations), clustering coefficient, shortest paths, path length, density and heterogeneity between the two sampling sites.
Sequence accession numbers
The raw high throughput sequencing data was deposited into the NCBI Sequence Read Archive database (PRJNA630159).
Results
Diversity of bacterial communities on the HVAC systems
A total of 2,137,515 raw 16S rRNA sequences from the two buildings investigated were processed and quality checked using QIIME. Sequence processing resulted in the assignment of 109,328 high-quality bacterial sequences into 416 OTUs. Normalization of counts to an even sampling depth of 1388 (348 total OTUs) was sufficient to explain differences in diversity (Figure S1). A summary of OTUs and counts for all the samples can be found in Table S1. Merging of OTUs and counts (mean counts at genus level) from samples obtained from both the office building (OB) and laboratory building (LB) revealed that OTUs shared between the two buildings made up 6.4% of the total (Figure S2). One hundred and thirty-one OTUs were found to be unique to LB, 16 were unique to OB while 10 OTUs were present in both metacommunities.
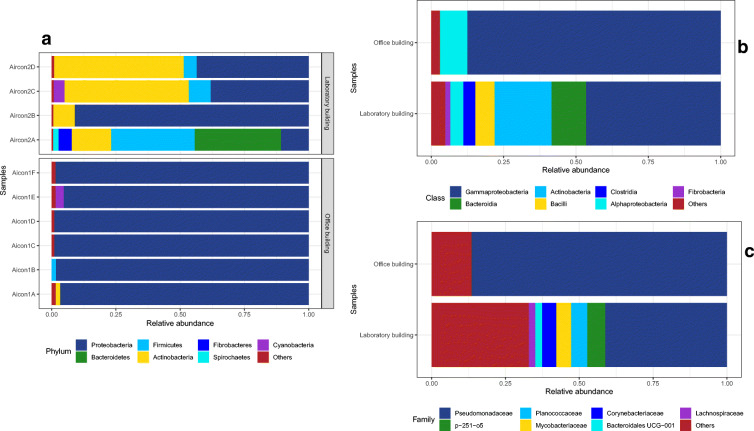
High-throughput sequencing analysis showed the bacterial communities in the OB were distributed among five major bacterial phyla. Of these, the phylum Proteobacteria represented 97.77% of the total sequences recovered followed by Cyanobacteria (0.93%), Actinobacteria (0.68%), Firmicutes (0.56%) and Bacteroidetes (0.06%). In contrast, more bacterial phyla were identified in the LB samples, the major phyla being Proteobacteria accounting for 45.83% of the recovered sequences followed by Actinobacteria (30.59%), Firmicutes (11.67%), Bacteroidetes (8.62%), Fibrobacteres (1.29%), Cyanobacteria (1.12%), Spirochaetes (0.52%), Epsilonbacteraeota (0.21%), Kiritimatiellaeota (0.11%), Verrucomicrobia (0.02%) and the phylum Tenericutes at 0.01% of the sequences, respectively. Detailed metataxonomic profiling of bacterial communities (phylum, class and family) in the HVAC systems of both the OB and LB is presented in Fig. 1.
Fig. 1.
Taxonomic profiling of bacterial communities at the a phylum level, b class level and c family level in the HVAC systems of the office building (OB) and laboratory building (LB)
At class level, eight bacterial classes were identified in samples from the OB, whereas eighteen bacterial classes were identified in samples from the LB. Comparatively, sequences representing the class Gammaproteobacteria were the most abundant in all samples, constituting 89.05% and 36.79% of analysed sequences from OB and the LB samples, respectively. Other major bacterial classes identified in the OB included Alphaproteobacteria (8.72%), Oxyphotobacteria (0.94%) and Actinobacteria (0.68%). In the LB, the second most abundant bacterial class was Actinobacteria (30.53%), Alphaproteobacteria (9.04%), Bacteroidia (8.62%), Bacilli (7.53%), Clostridia (3.26%), Fibrobacteria (1.29%) and Oxyphotobacteria (1.11%) (Fig. 1b).
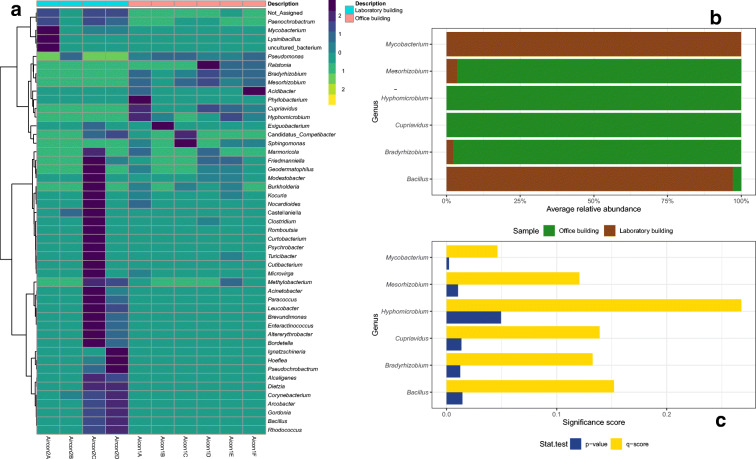
Other bacterial classes recovered in samples from the LB had an abundance of between 0.01 and 0.62%. These included the classes Erysipelotrichia > Spirochaetia > Negativicutes > Campylobacteria > Kiritimatiellae > Coriobacteriia > Verrucomicrobiae > MVP-15 > Melainabacteria > Mollicutes, listed according to sequence abundance. At family level, bacteria belonging to the family Pseudomonadaceae were particularly dominant in both samples, taking up 87.33% of recovered sequences from the OB, and 31.70% of sequences from the LB. Sequences belonging to the genus Pseudomonas were the most abundant in both samples, making up 87.32% of sequences from sample OB and 28% of sequences from sample LB. And, as was the trend with the phylum, class and family level distribution, more bacterial genera were recovered in sample LB than were recovered in sample OB (Fig. 2). Mycobacterium, Bacillus, Cupriavidus and Mesorhizobium were among bacteria with significant p values (≤ 0.05) on comparison of the metacommunity in the two buildings (Fig. 2b); however, only Mycobacterium passed the FDR cutoff (q ≤ 0.05) for differentially abundant species (Fig. 2c). Analysis of bacterial genera with at least 45% sample prevalence and 20% relative abundance (core microbiome) revealed Pseudomonas, Corynebacterium, Paenochrobactrum, Mycobacterium, Dietzia, Rhodococcus, Bacillus and Alcaligenes were the core microbiome in LB while OB comprised of three core bacterial genera (Pseudomonas, Bradyrhizobium and Mesorhizhobium).
Fig. 2.
Distribution of bacterial genera detected (a) and differentially abundant bacteria (b) and their corresponding p values and FDR q-scores (c) in the office and laboratory building
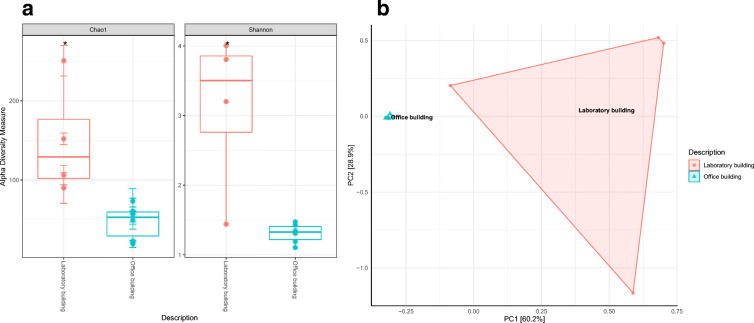
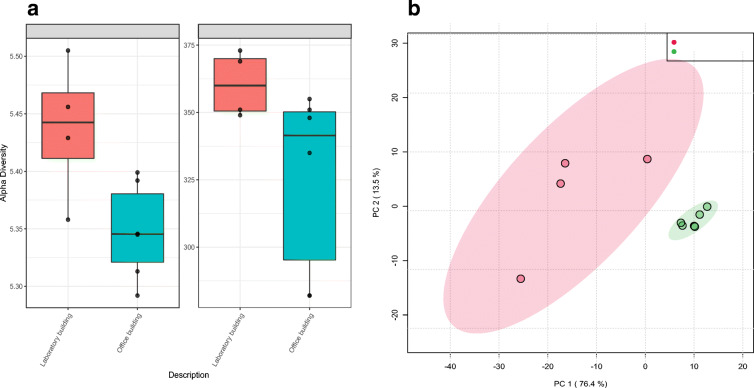
Sequences recovered from the two samples (OB and LB) were then subjected to alpha-diversity analysis to determine if indeed there were more bacterial communities in sample LB as compared to sample OB. Results indicated that sample LB harboured significantly higher bacterial diversity and richness (p ≤ 0.05) compared to sample OB. The two diversity indices (Shannon-H and Chao1) are presented in Fig. 3a. The alpha-diversity-based bacterial composition results were confirmed using principal coordinates analysis (PCoA) (Fig. 3b) which showed that bacterial communities recovered from all floors of the OB clustered together within the ordination space while samples drawn from the different floors of the LB had significantly different bacterial composition. PERMANOVA analysis further revealed that the observed clustering was indeed statistically significant (Ppermanova = 0.005) (Table S2).
Fig. 3.
a Statistical determinations of differences in bacterial richness (Chao1) and diversity (Shannon D) between the OB and LB. (ANOVA [p ≤ 0.05 (*); p ≤ 0.01 (**); p ≤ 0.001 (***)]). b Principal coordinates analysis (PCoA) determination of sample similarity
Predicted metabolic functions in HVACs bacterial communities
Predictive functional analysis using PICRUSt2 revealed the presence of 5414 KEGG orthologs (KOs) in the office building (OB) and 6148 KOs in the laboratory buildings (LB). Identified MetaCyc pathways included 358 and 384 pathways in the OB LB samples, respectively. The nearest-sequenced taxon index (NSTI) values for the predicted enzymes ranged from 0.05 to 0.33 (Table S3), suggesting a good prediction accuracy.
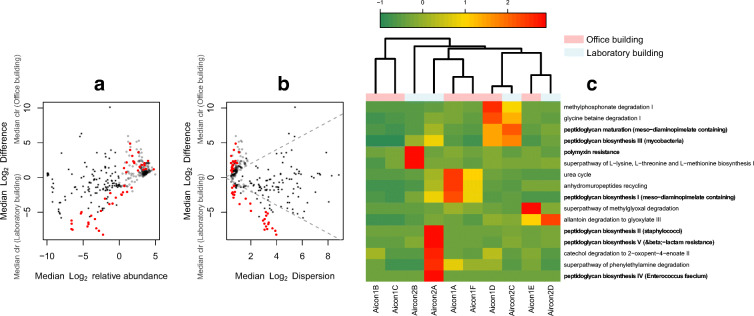
It was observed that of the 122 pathways with significant p values (Fig. 4 a and b), just 22 differentially abundant pathways had a Benjamini-Hochberg FDR < 0.05. Furthermore, only 9 of these 22 pathways passed the absolute effect size cutoff of ≥ 0.6. The nine pathways and other pathways of interest associated with antibiotics resistance and bacterial pathogenicity are presented in Fig. 4c. Notable enzyme pathways included the polymyxin resistance pathway which was detected in all HVAC systems of the OB on a scale of 1 to 2, while it was present in only one of the four HVAC systems of the LB. Pathways for peptidoglycan biosynthesis IV and V (beta lactam resistance) were also detected in all HVAC systems. However, the peptidoglycan biosynthesis IV pathway was particularly pronounced in the HVAC system 2A and 2C of the LB. Alpha-diversity analysis of enzyme distribution (KOs) between the OB and LB samples showed that there were significant differences (p ≤ 0.05) for the Shannon-H diversity index (Fig. 5). Differences in richness as measured using Chao1 were, however, not significant.
Fig. 4.
Relationship between pathway abundance and observed differences in the metacommunities of the two buildings (a). Effect plot displaying the relationship between dispersion and the observed differences in pathways within each building environment (b). Clustering of differentially enriched pathways (q ≤ 0.05; absolute effect size ≥ 0.6) and pathways associated with both resistance and pathogenicity. *Resistance/pathogenesis associated pathways are highlighted in bold face. The red coloured points in (a, b) are statistically significant (q ≤ 0.05) pathways in the two buildings; the grey coloured points are abundant but not significant while the black points are rare pathways
Fig. 5.
a Diversity based on the predicted functional profile of the bacterial communities in the two building-types investigated. (ANOVA [p ≤ 0.05 (*); p ≤ 0.01 (**); p ≤ 0.001 (***)]). b PCoA of predicted functional profiles based on Bray-Curtis dissimilarities grouped by building type
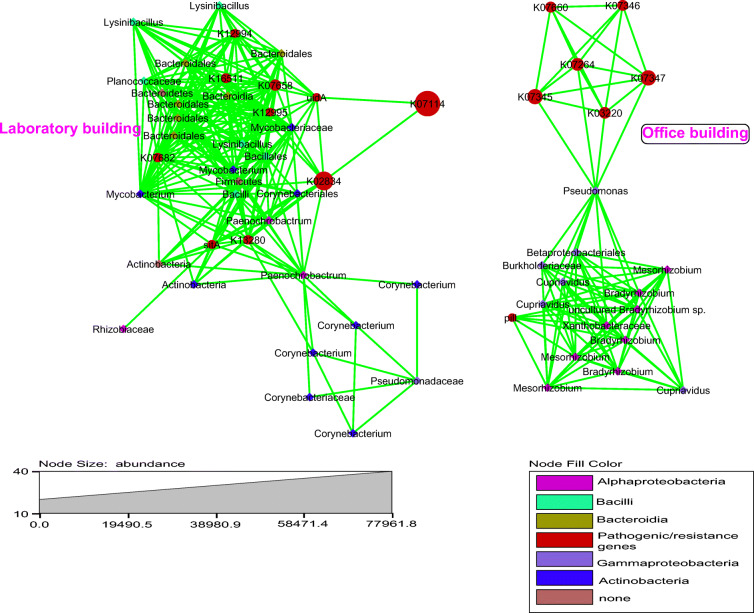
Investigation of the relationship between the detected bacterial meta-communities and PICRUSt predicted resistance and pathogenic genes led to the detection of 350 positive associations among 43 core bacteria, 3 pathogenic genes (sitA, pilL and uidA) and 14 resistance genes (Fig. 6). In this scenario too, as in all others investigated so far, the interactions formed clusters based on building-type (LB and/or OB). All the resistance genes with positive associations in the OB (6 resistance genes) only had Pseudomonas as first neighbour while the pathogenic pilL (type IV pili sensor histidine kinase and response regulator; KO2487) gene had Bradyrhizobium, Mesorhizobium and Xanthobacteraceae as immediate neighbours. In the LB, there were more associations (10 interactions) between both pathogenic and resistance genes and the core bacterial meta-community. For instance, the pathogenic gene sitA (manganese/iron transport system substrate-binding protein; K11604) was significantly correlated with Mycobacterium, and Corynebacterium. In contrast, pathogenic gene uidA (beta-glucuronidase; K01195) was considerably associated with Lysinibacillus and Paenochrobactrum in the LB meta-community.
Fig. 6.
Bacterial positive interactions and association with both pathogenic and resistance genes in the LB and OB HVAC samples—adapter protein, mecA1_2 (K16511); OmpR family alkaline phosphatase synthesis response regulator, PhoP (K07658, K07660); lipopolysaccharide biosynthesis proteins, wbdB, wbpY, mtfB (K12994) and wbdC, wbpZ, mtfC (K12995); NarL family sensor histidine kinase, DevS (K07682); manganese/iron transport system substrate-binding protein, sitA (K11604); uidA, beta-glucuronidase(K01195); type-IV pili sensor histidine kinase and response regulator, pilL (KO2487); signal peptidase I, sipW (K13280); ribosome-binding factor A, rbfA (K02834); Ca-activated chloride channel homolog, yfbK (K07114); fimbrial chaperone protein, fimC (K07346) and fimA (K07345); 4-amino-4-deoxy-l-arabinose transferase, arnT or pmrK (K07264); and type III secretion protein D, yscD, sctD, ssaD (K03220)
Discussion
The heating, ventilating and air-conditioning (HVAC) systems fitted in buildings tend to accumulate biocontaminants both from the outside environmental air as well as built environment indoor aerosols (Malebo and Shale 2013; Yamamoto et al. 2015). Consequently, indoor bioaerosol profiles are mostly shaped by three factors: the outdoor air; human occupancy; and building use (Adams et al. 2016; Hewitt et al. 2012). While HVAC systems may harbour a wide array of biocontaminants including insects, mites, nematodes and pollen, the primary biocontaminants of these systems include mostly fungi and bacteria, that are important due to their perceived public health implications. Bacterial community profiling from the HVAV systems of the two buildings in this study show the overwhelming presence of bacteria belonging to the phylum Proteobacteria. This observation corroborates the findings of Hoisington et al. (2016) who also reported this phylum as constituting over half of sequences obtained from the analysis of HVAC dust samples in retail stores.
It was interesting to note that after the phylum Proteobacteria, the phyla Firmicutes and Actinobacteria were the other dominant bacterial phyla to be recovered from the OB samples, which is consistent with the findings of Hewitt et al. (2012), who characterized office space bacterial abundance and diversity in three metropolitan areas of the USA. In addition to the phyla that were identified in the OB samples, other major bacterial phyla including Bacteroidetes, Cyanobacteria, Fibrobacteres, Spirochaetes, Epsilonbacteraeota, Kiritimatiellaeota, Verrucomicrobia and Tenericutes were also identified in the LB samples. These salient differences observed between OB and LB samples may point to the contribution of human occupancy and building use on the microbiome of the indoor environment. Congruent with our findings, Gaüzère et al. (2014) also identify the combination of the phyla Actinobacteria, Proteobacteria, Firmicutes and Bacteroidetes as a common indoor signature in office space, paediatric hospital and museum indoor environments. In other findings, Oberauner et al. (2013) and Mora et al. (2016) report that the hospital microbiome is dominated by a small number of prevalent bacterial taxa, mostly comprising of human-related commensals or pathogens belonging to phylum Acidobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Nitrospira and, most abundantly, Proteobacteria. Interestingly, bacterial diversity varied among different hospital areas, with the halls, living rooms, patient rooms and restrooms exhibiting more diverse bacterial compositions than that of the ICU (Oberauner et al. 2013).
At the genus level, bacteria of the genera Mycobacterium, Bacillus, Bradyrhizobium, Mesorhizobium, Cupriavidus and Hyphomicrobium were the most predominant in the HVAC systems of both buildings (Fig. 2). Mycobacteria are opportunistic pathogens whose source for human infection is the environment (Falkinham 2013; Nishiuchi et al. 2017). The presence of tuberculosis or nontuberculosis mycobacteria in HVAC systems raises greater concerns for public health than any other bacterium so far identified in this study because dead mycobacteria, just like live organisms, can influence inflammatory pathways involved in the pathogenesis of such diseases like asthma, type 1 diabetes mellitus, psoriasis and sarcoidosis among others by means of chemical products derived from dead cells (Hruska and Kaevska 2012). Besides this study, HVAC systems have previously been shown to be a potential source of aerosolized mycobacteria droplets (Choi and Choi 2017). Other known sources include natural waters, showerheads and water taps, hot tubs and spas, hydrotherapy pools and humidifiers (Falkinham 2013).
On the other hand, Bacillus are Gram-positive, rod-shaped, spore-forming aerobic bacteria that are widely distributed in nature, and are well-recognized causes of gastrointestinal as well as rare, non-gastrointestinal infections, usually in immunocompromised patients (Ohsaki et al. 2007). Inhalation of Bacillus spores from HVAC systems has been implicated in pneumonia and other respiratory tract infections (Anas et al. 2016), especially among individuals with pulmonary disease such as asthma and emphysema (Menetrez et al. 2006). Besides this study, the presence of Bacillus sp. in HVAC systems has previously been documented (Schmidt et al. 2012). The rhizobacterial genera Bradyrhizobium and Mesorhizobium that were part of the major bacterial genera present on the HVAC systems of both building are known primary contributors to biological nitrogen fixation (Fernández et al. 2019; Karaś et al. 2015; Ormeño-Orrillo and Martínez-Romero 2019), mainly associated with soil environments. While no previous record of their presence in HVAC systems could be found, it is possible that their real source in our HVAC systems could be the outdoor environmental air via soil dust contamination more than human occupancy or activities done in the buildings. Bacteria of the genus Cupriavidus are Gram-negative β-proteobacteria that are usually found from environmental and human clinical sources (Zhang et al. 2017). Bacteria of this genus have been identified as a risk factor in persons with cystic fibrosis where they are implicated in, together with other Gram-negative bacterial species, causing chronic airway infection (Kalka-Moll et al. 2009; Karafin et al. 2010). Finally, bacteria of the genus Hyphomicrobium are known facultative methylotrophs which are ubiquitous in water and soil (Fesefeldt and Gliesche 1997), and are mostly known for their denitrification capacities (Martineau et al. 2015). Interestingly, indoor biowall-associated Hyphomicrobium sp. have been successfully used to bioremediate volatile organic compounds (VOCs), removing as much as 96% of isopropanol from indoor air (Brown et al. 2018). While the present study did not specifically determine the presence or concentrations of VOCs in the HVAC systems, it can be hypothesised that the build-up of VOCs in the HVAC systems encouraged their growth. Overall, the heterogeneous nature of microorganisms found in HVAC systems as observed in this study and other studies shows that HVAC systems are the origin of airborne infections in indoor environments, and must be periodically cleaned and disinfected to avoid the build-up of pathogens, and the subsequent exposure of human occupants of these pathogens.
Secondly, as per our hypothesis, statistical analysis of sequence data from the two buildings proved that their HVAC systems harboured significantly different bacterial communities, with the LB having greater bacterial species diversity and richness as estimated by the Shannon-H and Chao1 indices (Figs. 3a and 5a), as well as the principal component analysis (PCoA) (Figs. 3b and 5b). In support of the above hypothesis, reduced microbial diversities coupled with enrichment of specific groups in indoor settings, as was observed in the OB HVAC systems, have been linked to the development of respiratory conditions including asthma and other allergies (Leung and Lee 2016), whereas higher diversities and richness tend to result in equalization between the good and the bad microbes, which ultimately promotes health. With the two building complexes located approximately 100 m apart, the net effect of soil conditions, land use, climate and primary productivity (collectively: the environment) on the microbiological quality of outdoor air around the two buildings is likely the same, as previously reported in other similar settings (Barberán et al. 2015; Leung and Lee 2016). This, therefore, implies that observed compositional differences in the indoor bacterial metastucture of the two buildings, both in terms of diversity and richness, was determined more by building use than environmental influences, especially when taking the human occupancy factor to be constant. Such an outcome had been predicted based on the hypothesis that the handling and processing of microbiological and other biological samples in the LB would contribute to a more dynamic and diverse indoor microbial populations in the LB than in the OB. For example, some inexperienced researchers in the microbiology laboratory have often been observed to remove their culture plates from incubators and open them on work benches, a practice that causes spores to escape and float in the indoor air, ultimately finding their way onto many surfaces in the laboratory, including HVAC systems. It is, therefore, expected that the diversity of sporogenic bacteria such as members of the phylum Firmicutes would even be much greater in the LB than in the OB since they produce spores as their main mode of reproduction and dispersal. As per this expectation, the relative abundance of sporogenic bacterial groups that include members of the genus Bacillus and Clostridium was ~ 20-fold higher (11.56% vs. 0.56%) in LB compared to OB samples. Conversely, the dominant presence of bacteria belonging to the phylum Proteobacteria, class Gammaproteobacteria, family Pseudomonadaceae and genus Pseudomonas in the HVAC systems of both buildings points to the effect of environmental air quality on the composition of microbial assemblages on the HVAC systems, as has been reported elsewhere (Stephens 2016).
To determine the potential health effects of the bacterial assemblages inhabiting the HVAC systems of the two buildings, the recovered sequences were subjected to predictive functional analysis using PICRUSt2 pipeline. Both pathogenic and antibiotic resistance pathways were identified in both buildings (Figs. 4, 5 and 6), notable among them being pathways for polymyxin resistance and peptidoglycan biosynthesis IV and V (beta lactam resistance). Polymyxin is an antibiotic of last resort in the treatment of Gram-negative bacterial infections where its main mode of action is to break up bacterial cell membranes (Li et al. 2019; Srinivas and Rivard 2017). Identification of this pathway was linked to the dominance of microbial communities belonging to the phylum Proteobacteria, which houses all major Gram-negative bacterial pathogens. Their predominance in indoor air raises concerns about their effect on health and, particularly, the allergenic potential of their cell-wall fragments and endotoxins (Gaüzère et al. 2014). Among the pathogenic genes identified, the most prominent were the sitA, and uidA genes. The sitA gene is most prominent among bacterial pathogens which utilize it to mobilize iron and manganese inside eukaryotic cells and is therefore associated with bacterial virulence (Runyen-Janecky et al. 2003; Tivendale et al. 2009). In the present study, this gene was found associated with Mycobacterium and Corynebacterium. Corynebacterium sp. have been linked to cases of community-acquired pneumonia (CAP), especially in immunocompromised hosts (Yang et al. 2018). Likewise, mycobacteria are renowned etiological agents of disease as has already been discussed. The uidA gene, on the hand, is a known molecular marker gene used to identify pathogenic Escherichia coli (Feng and Lampel 1994; Godambe et al. 2017).
Finally, the threat posed by indoor airborne pathogens on public health cannot be overlooked. Estimates are that as much as 5% of patients who visit hospitals for treatment develop nosocomial (hospital-acquired) infections while in a hospital or other health care facility (Kishor and Lokesh 2018). Further, numerous studies have also shown that occupants of office buildings with HVAC systems consistently report more symptoms related to mucous membrane irritation, breathing difficulties, irritated skin and constitutional/neurological symptoms such as headache and fatigue in their buildings than do occupants of buildings with natural ventilation (Eleyowo and Amusa 2019; Mendell 2004; Prussin and Marr 2015; Yamamoto et al. 2015). Such symptoms could be linked to the production of allergens, endotoxins and bacterial volatile organic compounds (VOCs) by bacterial contaminants in HVAC systems. Additionally, HVAC systems occasionally spew musty odours which often can be traced to bacterial and fungal proliferations in the systems. Furthermore, direct inhalation of aerosolised bacteria such as Pseudomonas aeruginosa and mycobacteria may cause opportunistic infections.
Conclusion
Evaluation of the community structure and functional capabilities of bacteria inhabiting HVAC systems in buildings is important in providing insight into the exposure of human occupants to pathogens and associated health risks. This study ranks among a few that have explored the diversity of microbial communities in HVAC systems both in terms of their taxonomy and metabolic functions in relationship to type and use of a building, the microbial quality of the outdoor air, as well as human occupancy. We conclude that HVAC systems are veritable sources of potentially harmful bioaerosols, including pathogenic and antibiotic resistant bacteria. Immunocompromised persons may be at the most risk from contracting airborne infections in buildings ventilated by HVAC systems. Further, we conclude that the microbial metastructure of a building is significantly shaped by three factors, namely type and use of the building, the microbial quality of the outdoor air, as well as human occupancy. However, due to the complex interplay between architectural, meteorological and anthropogenic factors on built environment microbial assemblages, further investigations taking into account the contribution of occupants’ microbiomes (including bacterial, fungal and viral communities and their metatranscriptomic and proteomic signatures); indoor vis-à-vis outdoor environmental factors such as temperature, humidity and aerosol CO2, VOCs, PM2.5 and PM10 levels; and associated building use scenarios will be important to shed light on the community dynamics of indoor environments. Nevertheless, this study has provided insights into the microbial life in public building spaces and recommends periodic disinfection of HVAC systems to stem the build-up of pathogenic microbes which could be detrimental to the health of human occupants.
Supplementary information
Rarefaction plot to determine sufficiency of sample size for diversity analyses (PDF 48 kb)
Distribution of OTUs in the laboratory and office buildings using merged (mean) OTUs and counts from the individual samples. *Shared OTU analysis was done at the genus taxonomic level using the rarefied OTU table. (PDF 11 kb)
(DOCX 13.9 kb)
(DOCX 13.5 kb)
(DOCX 12.9 kb)
Acknowledgements
Authors wish to thank the University of South Africa (UNISA) estate office for granting permission, and helping to access the HVAC system during sample collection. Also, we thank staff members at the Calabash and Eureka buildings at UNISA for allowing us to take samples in their offices and laboratories respectively, and encouraging us to proceed with this project. Finally, the authors wish to thank the Centre for High Performance Computing (Pretoria, South Africa) for helping with computational analysis of our data.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adams RI, Bhangar S, Dannemiller KC, Eisen JA, Fierer N, Gilbert JA, Green JL, Marr LC, Miller SL, Siegel JA, Stephens B, Waring MS, Bibby K. Ten questions concerning the microbiomes of buildings. Building and Environment. 2016;109:224–234. doi: 10.1016/j.buildenv.2016.09.001. [DOI] [Google Scholar]
- Anas G, Aligbe DS, Suleiman G, Warodi FA. Studies on microorganisms associated with air-conditioned environments. IOSR Journal of Environmental Science. 2016;10:16–18. doi: 10.9790/2402-1007011618. [DOI] [Google Scholar]
- Andersen, K.S., Kirkegaard, R.H., Albertsen, M., 2018. ampvis2: an R package to analyse and visualise 16S rRNA amplicon data. bioRxiv. http://dx.plos.org/10.1371/journal.pone.0132783
- Andualem Z, Gizaw Z, Bogale L, Dagne H. Indoor bacterial load and its correlation to physical indoor air quality parameters in public primary schools. Multidisciplinary Respiratory Medicine. 2019;14:1–7. doi: 10.1186/s40248-018-0167-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera P, Kozlov AM, Czech L, Morel B, Darriba D, Flouri T, Stamatakis A. EPA-ng: Massively Parallel Evolutionary Placement of Genetic Sequences. Systematic Biology. 2019;68:365–369. doi: 10.1093/sysbio/syy054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberán, A., Dunn, R. R., Reich, B. J., Pacifici, K., Laber, E. B., Menninger, H. L., Morton, J. M., Henley, J. B., Leff, J. W., Miller, S. L., & Fierer, N. (2015). The ecology of microscopic life in household dust. Proceedings of the Royal Society B: Biological Sciences, 282. 10.1098/rspb.2015.1139. [DOI] [PMC free article] [PubMed]
- Benjamini, Y., Hochberg, Y., 1995. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B. 10.2307/2346101
- Bernstein RS, Sorenson WG, Garabrant D, Reaux C, Treitman RD. Exposures to respirable, airborne penicillium from a contaminated ventilation system: clinical, environmental and epidemiological aspects. American Industrial Hygiene Association Journal. 2010;44:161–169. doi: 10.1080/15298668391404581. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brągoszewska E, Biedroń I, Kozielska B, Pastuszka JS. Microbiological indoor air quality in an office building in Gliwice, Poland: analysis of the case study. Air Quality, Atmosphere & Health. 2018;11:729–740. doi: 10.1007/s11869-018-0579-z. [DOI] [Google Scholar]
- Brown, E., Enguillado, G., McDermott, R., Palumbo, N., Smith, J., Stanley, M., Sulzbach, M., Taylor, J., 2018. Bioremediation of volatile organic compounds in indoor spaces using a novel biowall design: A feasibility study. University of Maryland. 10.1007/978-981-10-7485-1_15
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carducci A, Federigi I, Verani M. Covid-19 airborne transmission and its prevention: Waiting for evidence or applying the precautionary principle? Atmosphere (Basel). 2020;11:1–21. doi: 10.3390/atmos11070710. [DOI] [Google Scholar]
- Choi SG, Choi MS. Isolation of Nontuberculous Mycobacteria (NTM) from Air Conditioner Dust. The Korean Journal of Clinical Laboratory Science. 2017;49:435–438. doi: 10.15324/kjcls.2017.49.4.435. [DOI] [Google Scholar]
- Czech, L., Barbera, P., Stamatakis, A., 2019. Genesis and Gappa: Library and Toolkit for Working with Phylogenetic (Placement) Data. bioRxiv 647958. 10.1101/647958
- Douglas, G.M., Maffei, V.J., Zaneveld, J., Yurgel, S.N., Brown, J.R., Taylor, C.M., Huttenhower, C., Langille, M.G.I., 2019. PICRUSt2: An improved and extensible approach for metagenome inference. bioRxiv 672295. 10.1101/672295
- Dumala S, Dudzinska M. Microbiological indoor air quality in Polish schools. MIddle Pomeranian. SCientific Society of the Environment Protection. 2013;15:231–244. [Google Scholar]
- Eddy SR. Accelerated Profile HMM Searches. PLoS Computational Biology. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleyowo OO, Amusa OD. Evaluation of air condition use and its health effects. Recent Adv. Biol. Med. 2019;5:1. doi: 10.18639/rabm.2019.858317. [DOI] [Google Scholar]
- Falkinham JO. Reducing human exposure to mycobacterium avium. Annals of the American Thoracic Society. 2013;10:378–382. doi: 10.1513/AnnalsATS.201301-013FR. [DOI] [PubMed] [Google Scholar]
- Faust K, Raes J. CoNet app: inference of biological association networks using Cytoscape. F1000Research. 2016;5:1519. doi: 10.12688/f1000research.9050.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekadu S, Getachewu B. Microbiological assessment of indoor air of teaching hospital wards: a case of Jimma University specialized hospital. Ethiopian Journal of Health Sciences. 2015;25:117–122. doi: 10.4314/ejhs.v25i2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng P, Lampel KA. Genetic analysis of uidA expression in enterohaemorrhagic Escherichia coli serotype O157:H7. Microbiology. 1994;140:2101–2107. doi: 10.1099/13500872-140-8-2101. [DOI] [PubMed] [Google Scholar]
- Fernandes AD, Reid JNS, Macklaim JM, McMurrough TA, Edgell DR, Gloor GB. Unifying the analysis of high-throughput sequencing datasets: Characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome. 2014;2:1–13. doi: 10.1186/2049-2618-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández, N., Cabrera, J.J., Varadarajan, A.R., Lutz, S., Ledermann, R., Roschitzki, B., Eberl, L., Bedmar, E.J., Fischer, H.M., Pessi, G., Ahrens, C.H., Mesa, S., 2019. An integrated systems approach unveils new aspects of microoxia-mediated regulation in bradyrhizobium diazoefficiens. Front. Microbiol. 10. 10.3389/fmicb.2019.00924 [DOI] [PMC free article] [PubMed]
- Fesefeldt A, Gliesche CG. Identification of Hyphomicrobium spp. using PCR-amplified fragments of the mxaF gene as a molecular marker. Systematic and Applied Microbiology. 1997;20:387–396. doi: 10.1016/S0723-2020(97)80007-0. [DOI] [Google Scholar]
- Gaüzère C, Godon J-J, Blanquart H, Ferreira S, Moularat S, Robine E, Moletta-Denat M. ‘Core species’ in three sources of indoor air belonging to the human micro-environment to the exclusion of outdoor air. Science of the Total Environment. 2014;485:508–517. doi: 10.1016/j.scitotenv.2014.03.117. [DOI] [PubMed] [Google Scholar]
- Godambe LP, Bandekar J, Shashidhar R. Species specific PCR based detection of Escherichia coli from Indian foods. 3 Biotech. 2017;7:1–5. doi: 10.1007/s13205-017-0784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisoli, P., Albertoni, M., & Rodolfi, M. (2019). Application of airborne microorganism indexes in offices, gyms, and libraries. Applied Sciences, 9. 10.3390/app9061101.
- Hewitt KM, Gerba CP, Maxwell SL, Kelley ST. Office space bacterial abundance and diversity in three metropolitan areas. PLoS One. 2012;7:3–9. doi: 10.1371/journal.pone.0037849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoisington A, Maestre JP, Kinney KA, Siegel JA. Characterizing the bacterial communities in retail stores in the United States. Indoor Air. 2016;26:857–868. doi: 10.1111/ina.12273. [DOI] [PubMed] [Google Scholar]
- Hospodsky, D., Qian, J., Nazaroff, W. W., Yamamoto, N., Bibby, K., Rismani-Yazdi, H., & Peccia, J. (2012). Human occupancy as a source of indoor airborne bacteria. PLoS One, 7. 10.1371/journal.pone.0034867. [DOI] [PMC free article] [PubMed]
- Hruska K, Kaevska M. Mycobacteria in water, soil, plants and air: A review. Vet. Med. (Praha) 2012;57:623–679. doi: 10.17221/6558-VETMED. [DOI] [Google Scholar]
- Ito K, Lane K, Olson C. Equitable access to air conditioning: A city health department’s perspective on preventing heat-related deaths. Epidemiology. 2018;29:749–752. doi: 10.1097/EDE.0000000000000912. [DOI] [PubMed] [Google Scholar]
- Kalka-Moll WM, LiPuma JJ, Accurso FJ, Plum G, Van Koningsbruggen S, Vandamme P. Airway infection with a novel Cupriavidus species in persons with cystic fibrosis. Journal of Clinical Microbiology. 2009;47:3026–3028. doi: 10.1128/JCM.00846-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafin M, Romagnoli M, Fink DL, Howard T, Rau R, Milstone AM, Carroll KC. Fatal infection caused by Cupriavidus gilardii in a child with aplastic anemia. Journal of Clinical Microbiology. 2010;48:1005–1007. doi: 10.1128/JCM.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaś MA, Turska-Szewczuk A, Trapska D, Urbanik-Sypniewska T. Growth and survival of Mesorhizobium loti inside Acanthamoeba enhanced its ability to develop more nodules on lotus corniculatus. Microbial Ecology. 2015;70:566–575. doi: 10.1007/s00248-015-0587-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishor S, Lokesh K. Comparative study of microbiological air quality of private and government hospitals in Mysuru City. International Journal of Engineering, Science and Technology. 2018;7:357–365. [Google Scholar]
- Leung MHY, Lee PKH. The roles of the outdoors and occupants in contributing to a potential pan-microbiome of the built environment: A review. Microbiome. 2016;4:1–15. doi: 10.1186/s40168-016-0165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levetin E, Shaughnessy R, Rogers CA, Scheir R. Effectiveness of germicidal UV radiation for reducing fungal contamination within air-handling units. Applied and Environmental Microbiology. 2001;67:3712–3715. doi: 10.1128/AEM.67.8.3712-3715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Cao Y, Yi L, Liu JH, Yang Q. Emergent polymyxin resistance: End of an era? Open Forum Infect. Dis. 2019;6:1–10. doi: 10.1093/ofid/ofz368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louca S, Doebeli M. Efficient comparative phylogenetics on large trees. Bioinformatics. 2018;34:1053–1055. doi: 10.1093/bioinformatics/btx701. [DOI] [PubMed] [Google Scholar]
- Macher JM, Fan-Yen HM, Flores M. A two-year study of microbiological indoor air quality in a new apartment. Archives of Environmental Health An International Journal. 1991;46:25–29. doi: 10.1080/00039896.1991.9937425. [DOI] [PubMed] [Google Scholar]
- Malebo N, Shale K. MALDI Biotyper Characterization of microorganisms colonizing heating ventilation air-conditioning systems at a South African hospital. Life Sci. J. 2013;10:413–417. [Google Scholar]
- Martineau C, Mauffrey F, Villemur R. Comparative analysis of denitrifying activities of Hyphomicrobium nitrativorans, Hyphomicrobium denitrificans, and Hyphomicrobium zavarzinii. Applied and Environmental Microbiology. 2015;81:5003–5014. doi: 10.1128/AEM.00848-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell MJ. Commentary: Air conditioning as a risk for increased use of health services. International Journal of Epidemiology. 2004;33:1123–1126. doi: 10.1093/ije/dyh264. [DOI] [PubMed] [Google Scholar]
- Menetrez MY, Foarde KK, Webber TD, Dean TR, Betancourt DA. Efficacy of UV irradiation on eight species of Bacillus. Journal of Environmental Engineering and Science. 2006;5:329–334. doi: 10.1139/S05-041. [DOI] [Google Scholar]
- Mora M, Mahnert A, Koskinen K, Pausan MR, Oberauner-Wappis L, Krause R, Perras AK, Gorkiewicz G, Berg G, Moissl-Eichinger C. Microorganisms in confined habitats: microbial monitoring and control of intensive care units, operating rooms, cleanrooms and the International Space Station. Frontiers in Microbiology. 2016;7:1573. doi: 10.3389/fmicb.2016.01573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiuchi Y, Iwamoto T, Maruyama F. Infection sources of a common non-tuberculous mycobacterial pathogen, Mycobacterium avium complex. Frontiers in Medicine. 2017;4:27. doi: 10.3389/fmed.2017.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes ZG, Martins AS, Altoe ALF, Nishikawa MM, Leite MO, Aguiar PF, Fracalanzza SEL. Indoor air microbiological evaluation of offices, hospitals, industries, and shopping centers. Mem. Inst. Oswaldo Cruz. 2005;100:351–357. doi: 10.1590/S0074-02762005000400003. [DOI] [PubMed] [Google Scholar]
- Oberauner L, Zachow C, Lackner S, Högenauer C, Smolle K-H, Berg G. The ignored diversity: complex bacterial communities in intensive care units revealed by 16S pyrosequencing. Scientific Reports. 2013;3:1413. doi: 10.1038/srep01413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y, Koyano S, Tachibana M, Shibukawa K, Kuroki M, Yoshida I, Ito Y. Undetected Bacillus pseudo-outbreak after renovation work in a teaching hospital. The Journal of Infection. 2007;54:617–622. doi: 10.1016/j.jinf.2006.10.049. [DOI] [PubMed] [Google Scholar]
- Ormeño-Orrillo E, Martínez-Romero E. A genomotaxonomy view of the bradyrhizobium genus. Frontiers in Microbiology. 2019;10:1–13. doi: 10.3389/fmicb.2019.01334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prussin AJ, Marr LC. Sources of airborne microorganisms in the built environment. Microbiome. 2015;3:1–10. doi: 10.1186/s40168-015-0144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Research. 2013;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A language and environment for statistical computing (Version 3.5. 2) Vienna: R Foundation for Statistical Computing; 2019. p. 2018. [Google Scholar]
- Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runyen-Janecky LJ, Reeves SA, Gonzales EG, Payne SM. Contribution of the Shigella flexneri sit, iuc, and feo iron acquisition systems to iron acquisition in vitro and in cultured cells. Infection and Immunity. 2003;71:1919–1928. doi: 10.1128/IAI.71.4.1919-1928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MG, Attaway HH, Terzieva S, Marshall A, Steed LL, Salzberg D, Hamoodi HA, Khan JA, Feigley CE, Michels HT. Characterization and control of the microbial community affiliated with copper or aluminum heat exchangers of HVAC systems. Current Microbiology. 2012;65:141–149. doi: 10.1007/s00284-012-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvarajan R, Sibanda T, Venkatachalam S, Ogola HJO, Christopher Obieze C, Msagati TA. Distribution, interaction and functional profiles of epiphytic bacterial communities from the rocky intertidal seaweeds, South Africa. Scientific Reports. 2019;9:1–13. doi: 10.1038/s41598-019-56269-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B., & Ideker, T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research. 10.1101/gr.1239303.metabolite. [DOI] [PMC free article] [PubMed]
- Srinivas P, Rivard K. Polymyxin resistance in Gram-negative pathogens. Current Infectious Disease Reports. 2017;19:7–9. doi: 10.1007/s11908-017-0596-3. [DOI] [PubMed] [Google Scholar]
- Stephens B. What have we learned about the microbiomes of indoor environments? Appl. Environ. Sci. 2016;1:e00083–e00016. doi: 10.1128/mSystems.00083-16.Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryjakowska-Sekulska M, Piotraszewska-Pajak A, Szyszka A, Nowicki M, Filipiak M. Microbiological qualtiy of indoor air in university rooms. Polish Journal of Environmental Studies. 2007;16:623–632. [Google Scholar]
- Takaoka, M., Norbäck, D., 2020. The indoor environment in schools, kindergartens and day care centres, in: Kishi, R., Norbäck, D., Araki, A. (Eds.), Indoor Environmental Quality and Health Risk toward Healthier Environment for All. Current Topics in Environmental Health and Preventive Medicine. Springer, Singapore, pp. 87–112.
- Tivendale KA, Allen JL, Browning GF. Plasmid-borne virulence-associated genes have a conserved organization in virulent strains of avian pathogenic Escherichia coli. Journal of Clinical Microbiology. 2009;47:2513–2519. doi: 10.1128/JCM.00391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes, G.R., Bolker, B., Bonebakker, L., Gentleman, R., Liaw, W.H.A., Lumley, T., Maechler, M., Magnusson, A., Moeller, S., Schwartz, M., 2015. gplots: Various R programming tools for plotting data.
- Wickham, H., 2016. ggplot2: elegant graphics for data analysis. Springer.
- Wu Y, Chen A, Luhung I, Gall ET, Cao Q, Chang VWC, Nazaroff WW. Bioaerosol deposition on an air-conditioning cooling coil. Atmospheric Environment. 2016;144:257–265. doi: 10.1016/j.atmosenv.2016.09.004. [DOI] [Google Scholar]
- Yamamoto N, Hospodsky D, Dannemiller KC, Nazaroff WW, Peccia J. Indoor emissions as a primary source of airborne allergenic fungal particles in classrooms. Environmental Science & Technology. 2015;49:5098–5106. doi: 10.1021/es506165z. [DOI] [PubMed] [Google Scholar]
- Yang K, Kruse RL, Lin WV, Musher DM. Corynebacteria as a cause of pulmonary infection: a case series and literature review. Pneumonia. 2018;10:1–8. doi: 10.1186/s41479-018-0054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Doak TG. A parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLoS Computational Biology. 2009;5:e1000465. doi: 10.1371/journal.pcbi.1000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Deng W, Wang S, Xu L, Yan L, Liao P. First case report of infection caused by Cupriavidus gilardii in a non-immunocompromised Chinese patient. IDCases. 2017;10:127–129. doi: 10.1016/j.idcr.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rarefaction plot to determine sufficiency of sample size for diversity analyses (PDF 48 kb)
Distribution of OTUs in the laboratory and office buildings using merged (mean) OTUs and counts from the individual samples. *Shared OTU analysis was done at the genus taxonomic level using the rarefied OTU table. (PDF 11 kb)
(DOCX 13.9 kb)
(DOCX 13.5 kb)
(DOCX 12.9 kb)