Summary
Deciphering how neuronal diversity is established and maintained requires a detailed knowledge of neuronal gene expression throughout development. In contrast to mammalian brains1,2, the large neuronal diversity of the Drosophila optic lobes3 and its connectome4–6 are almost completely characterized. However, a molecular characterization of this diversity, particularly during development, has been lacking. We present novel insights into brain development through a nearly exhaustive description of the transcriptomic diversity of the optic lobes. We acquired the transcriptome of 275,000 single-cells at adult and five pupal stages, and developed a machine learning framework to assign them to almost 200 cell-types at all timepoints. We discovered two large neuronal populations that wrap neuropils during development but die just before adulthood, as well as neuronal subtypes that partition dorsal and ventral visual circuits by differential Wnt signaling throughout development. Moreover, we showed that neurons of the same type but produced days apart synchronize their transcriptomes shortly after being produced. We also resolved during synaptogenesis neuronal subtypes that converge to indistinguishable transcriptomic profiles in adults while greatly differing in morphology and connectivity. Our datasets almost completely account for the known neuronal diversity of the optic lobes and serve as a paradigm to understand brain development across species.
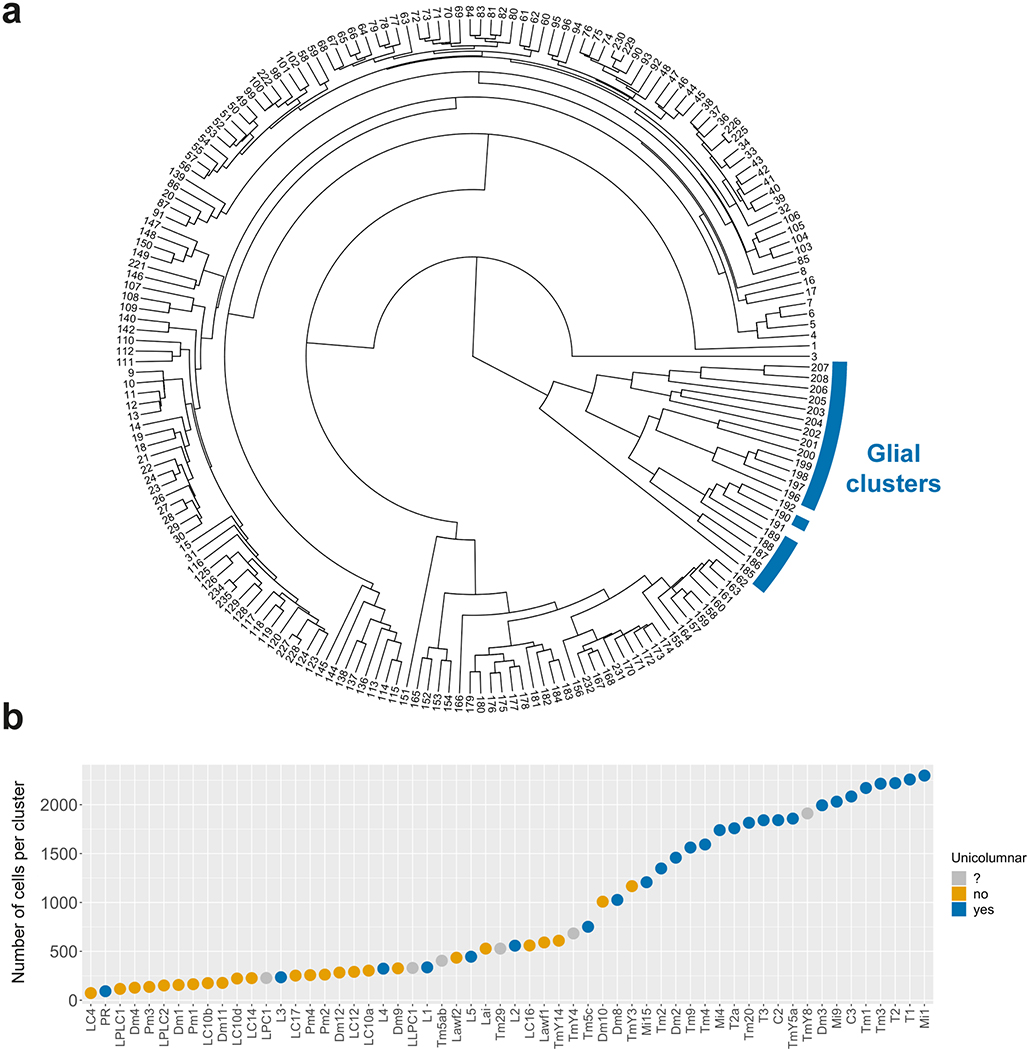
The optic lobes constitute two thirds of the fly brain with approximately 60,000 neurons per lobe and around 200 morphologically distinct neuronal types3,7–12. The visual information detected by the photoreceptors is integrated in its four neuropils: lamina, medulla, lobula and lobula plate (Fig.1a), which are each divided into ~750 columns, corresponding to the ~750 ommatidia in the compound eye. This highly parallel processing depends on neurons that are either present at a 1:1 ratio with the number of columns (unicolumnar neurons), or are present at a lower ratio but contact multiple columns to cover the entire visual map (multicolumnar neurons)3,7,13. This modular structure in which the same cell types are present multiple times renders the system particularly amenable to single-cell sequencing studies that rely heavily on clustering of numerous cells of the same type.
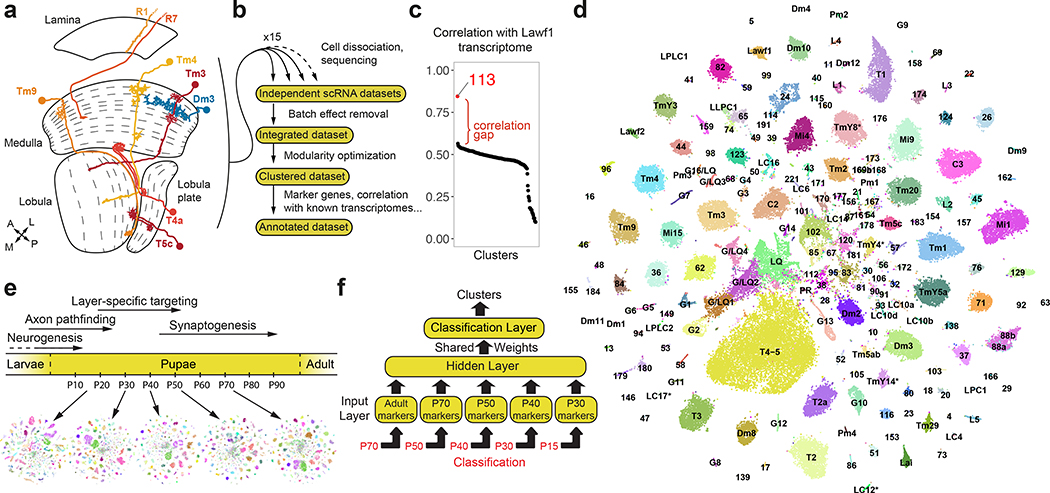
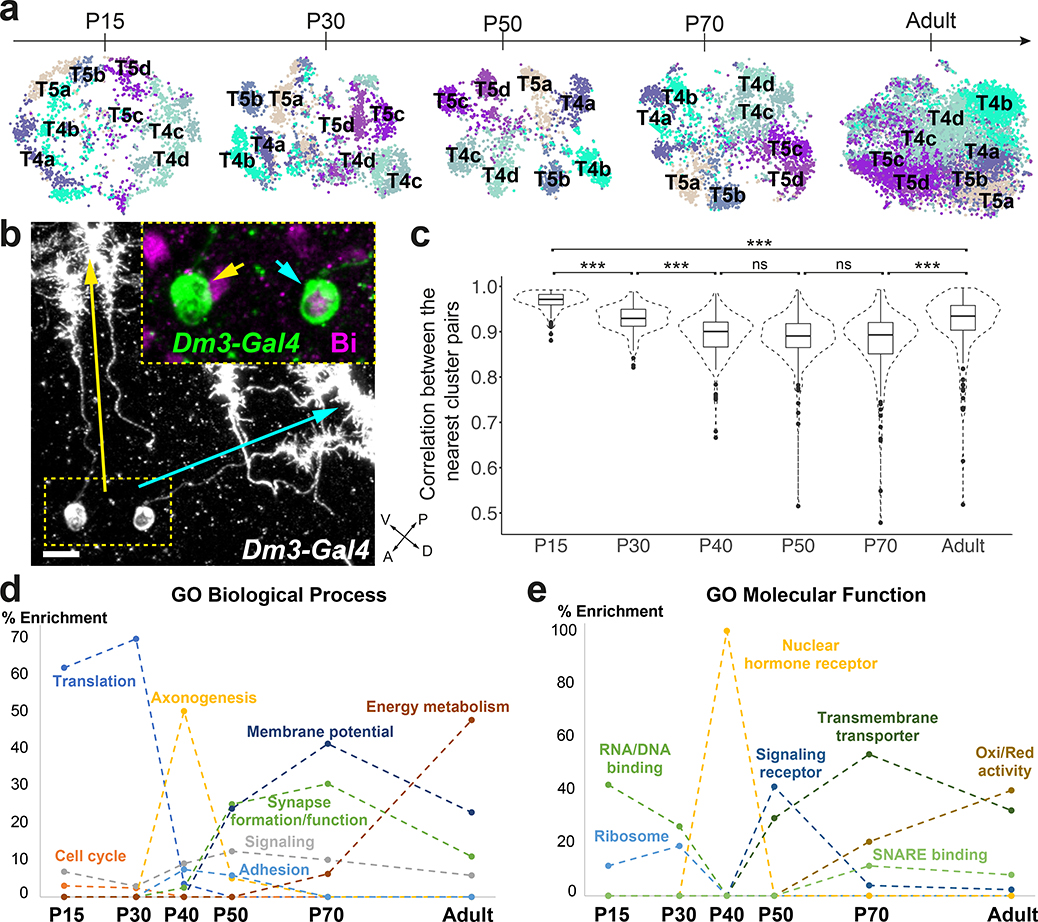
Figure 1: High resolution transcriptomic atlases of the optic lobe across development.
a, Optic lobe cross-section3, with drawings of unicolumnar (orange shades) and multicolumnar (blue) neurons. Dashed lines: boundaries between layers. A: anterior, L: lateral, M: medial, P: posterior. b, Approach followed to produce the adult dataset. c, Pearson correlation between the average gene expression of the adult dataset clusters (x-axis) and the transcriptome of isolated Lawf1 neurons (Methods). d, tSNE visualization of the final adult dataset, using 120 principal components calculated on the log-normalized integrated gene expression. The 61 identified neuronal clusters are labeled by their standard abbreviation, G1–16: glial clusters, LQ: low-quality cells, G/LQ1–4: glial clusters with some features of low-quality cells, *: clusters with less confident annotations (Suppl. Table 1). e, Approximate time frames of different steps of optic lobe development, and tSNE visualizations of the pupal datasets. Colors match to the adult dataset as classified by the neural network. f, Multi-task neural network classifier used at each stage to sequentially match developing cells to the adult clusters, as detailed in Methods.
Most optic lobe neurons are produced continuously through larval and early pupal development from two crescent-shaped neuroepithelia called the Outer and Inner Proliferation Centers (OPC and IPC). Immediately after birth, neurons project their processes to build the columns and protolayers of all neuropils, which is completed by 30% of pupal development (P30)14. Synapse formation commences around P45 and continues until eclosion15. These processes are genetically hardwired16 and appear to be strictly determined by the initial specification of neuronal identity as both the morphology of specific cell types and the identity of their synaptic partners are largely invariant4. Although we have a good understanding of the molecular mechanisms that specify neuronal identity17, it remains unclear how morphological and functional diversity is established and maintained throughout development and how brain wiring is achieved. Characterizing both the common and cell type-specific genetic programs implemented by neurons at different stages of their development is thus essential.
Developmental atlases of the optic lobe
The single-cell transcriptomic atlases of the adult Drosophila optic lobe previously generated by us18 and others19, or bulk sequencing of isolated cell types18,20, did not fully account for its cellular diversity. To produce an exhaustive catalog of neurons in the adult optic lobe (Fig.1a–b), we obtained 109,743 single-cell transcriptomes (Methods) using the Chromium system (10x Genomics). We used the Seurat v3 integration pipeline21 to remove batch effects between libraries (Extended Data Fig.1a–b, Methods). The single-cell transcriptomes were clustered21,22 using parameters that optimized the biological significance of the resulting clusters. We then tested the statistical significance of the clusters and merged improperly separated clusters that did not display biologically significant differences in gene expression, resulting in 193 final clusters (Extended Data Fig.1c–f, Suppl. Table 1, Methods).
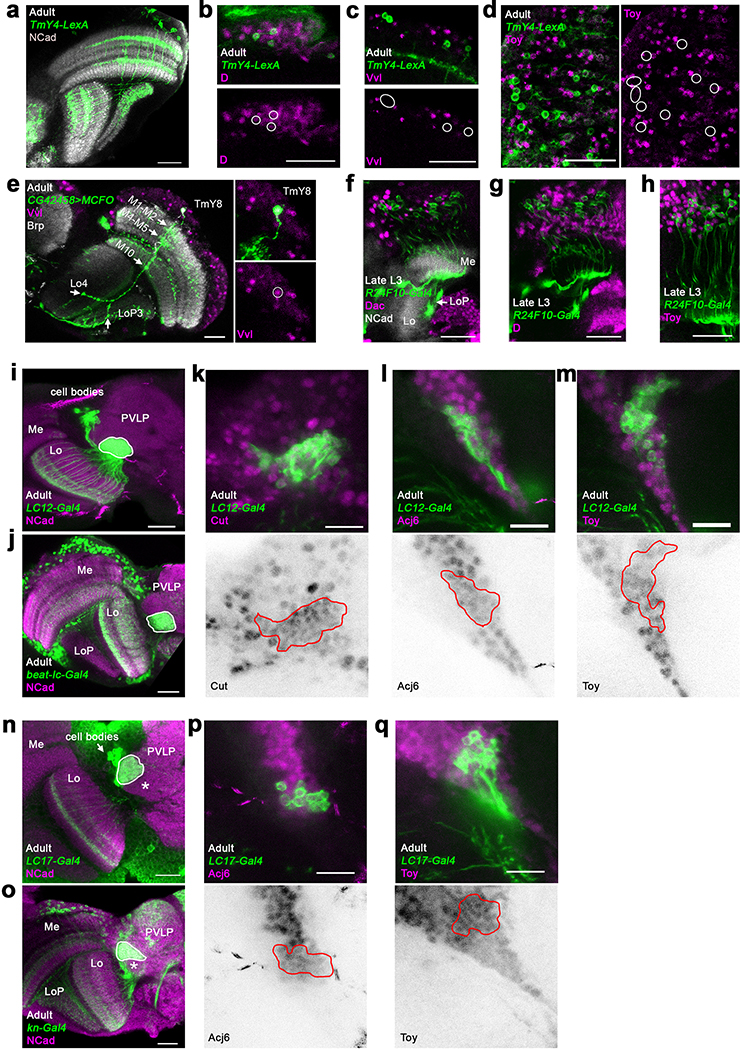
We identified 172 clusters as optic lobe neurons and 19 clusters as glia that were sharply separated within the hierarchical tree of the clusters (Extended Data Fig.5a), and one cluster of low-quality transcriptomes (Extended Data Fig.2, Suppl. Table 1, Methods). We annotated neuronal clusters by calculating the Pearson correlations between the average gene expression of each cluster and 52 published bulk-transcriptomes obtained from purified optic lobe neuronal types18,20 and 2 additional (Pm2 and T4) sequenced for this study (Methods). A clear match between a cell-type and a cluster should result in an obvious correlation gap between the best and second-best matches (Fig.1c). We could thereby identify the corresponding clusters for 53 of these 54 neurons (Extended Data Fig.3, Methods), with only one exception (LPi3–4). In addition, we identified the clusters corresponding to LC12, LC14, LC17, Pm1, T2a, TmY4, TmY8 and TmY14 neurons using the binarized expression (Methods) of combinations of protein markers identified by antibody or reporter line stainings (Extended Data Fig.4, Suppl. Table 1). In summary, we were able to identify 61 of our 172 neuronal clusters (Fig.1d). The relative size of the identified clusters was consistent with the known abundance of these cell types (Extended Data Fig.5b). The very high resolution of this atlas is highlighted by the fact that we found distinct cluster matches even for rare neuronal types, such as Dm4 and Dm1 that are represented by only ~40 cells in each optic lobe7. Thus, our dataset is likely to contain independent clusters for almost all neuron types present at more than 30 cells per optic lobe. This criterion is satisfied by all unicolumnar and most multicolumnar neuron types7, i.e. >95% of the cells in our dataset.
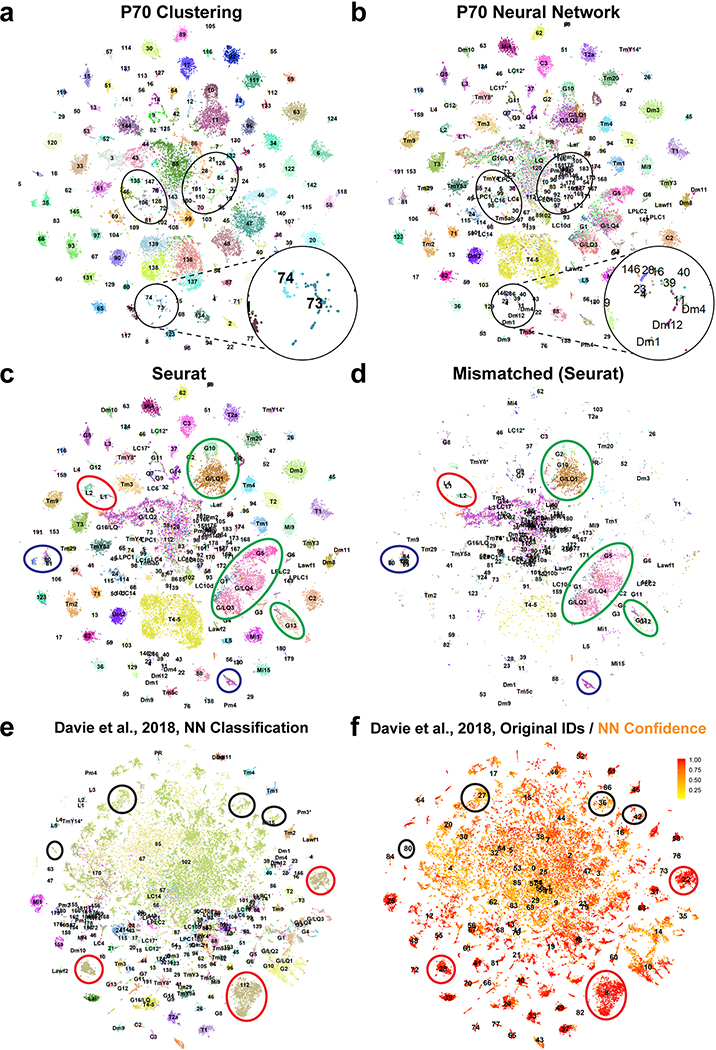
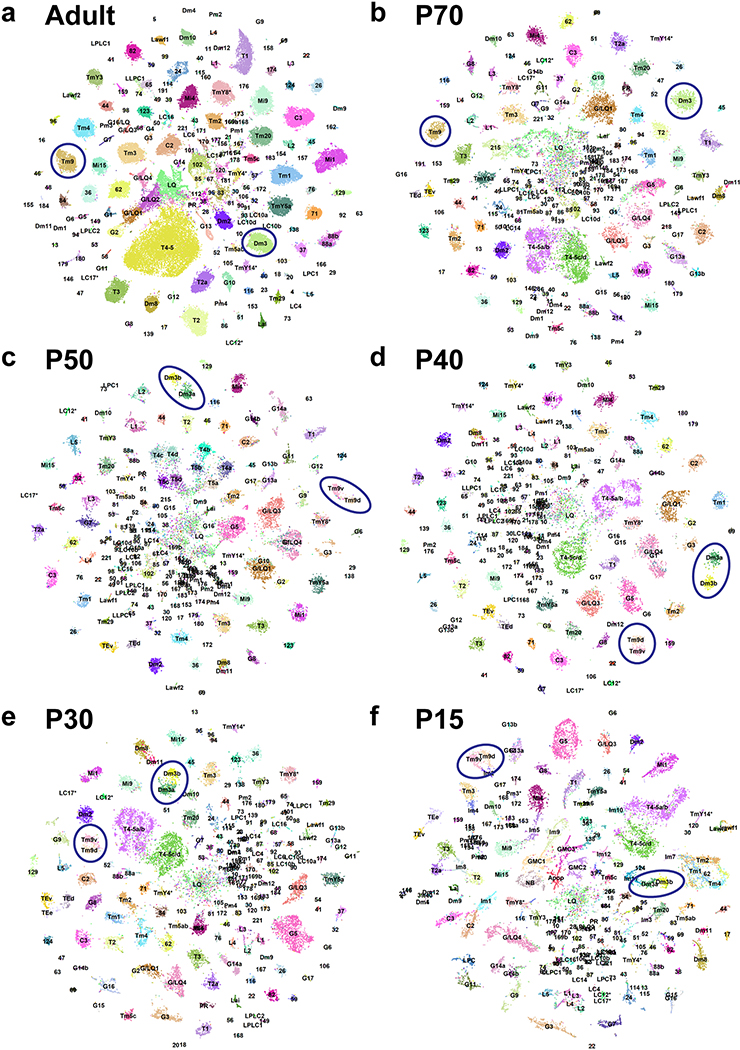
We next extended our single-cell atlas to developing neurons, sequencing optic lobe cells at five pupal stages (~30,000 cells each, Methods), covering all stages of neuronal differentiation (Fig.1e). Because developing neurons most often lack marker genes or available bulk transcriptomes, we adopted a supervised approach to annotate these datasets (Fig.1f). We first trained a two-layer neural network (NN) on our labeled adult dataset to classify P70 cells into corresponding adult clusters (Methods). This allowed us to sequence a lower number of cells at each pupal stage while retaining the high resolution of the adult dataset. Our method was able to resolve less abundant cell types that were grouped together by unsupervised clustering at P70 (Extended Data Fig.6a–b). It was more robust in distinguishing closely related cell types than Seurat v3 label transfer (Methods, Extended Data Fig.6c–d). However, supervised classifications are inherently unfit to identify new cell types. To overcome this drawback and assess the accuracy of NN classifications, we compared them to an unsupervised clustering of the P70 dataset and performed manual adjustments where appropriate (Methods).
We then classified the earlier pupal datasets sequentially backwards throughout development (i.e. from P70 to P50, P50 to P40…). We used a multi-task extension of our NN (Fig.1f) in order to prevent over-fitting the classifiers, due to the smaller sample sizes of the pupal datasets (Methods). At each stage, the NN classifications were also manually assessed and adjusted before proceeding to the next step. Interestingly, several adult clusters corresponded to two or more unsupervised clusters at earlier timepoints (Suppl. Table 1, Methods), which we discuss below. Using this iterative “classify-adjust-retrain” approach, we successfully assigned correspondences between pupal cells and each of our adult clusters (Extended Data Fig.7). The final trained models are provided in Appendix 1 and can be used to annotate any scRNA-seq dataset containing optic lobe cells. As a benchmark, we tested our classifier on an atlas of the entire adult brain19. The optic lobe cells were classified with high confidence and at a much higher resolution than in the original study, while the central brain neurons were clearly set aside as low confidence predictions (Methods, Extended Data Fig.6e–f).
Together, these data represent the first single-cell atlas of a complex central nervous structure at near complete saturation and throughout development. Coupled with the detailed knowledge of optic lobe connectivity through EM reconstruction4–6 and the Drosophila genetic toolkit, this represents a unique resource to investigate general mechanisms underlying the function and development of neuronal circuits. In order to facilitate the exploration of our datasets, we provide summary tables of average gene expression, binarized gene expression, and differentially expressed genes for all our clusters at all stages in addition to the annotated objects with raw data (GEO: GSE142787). In addition, we provide a list of transcription factor (TF) markers that are maintained at all stages, whose combinations uniquely identify each neuronal cluster (Suppl. Table 2).
Discovery of Transient Extrinsic neurons
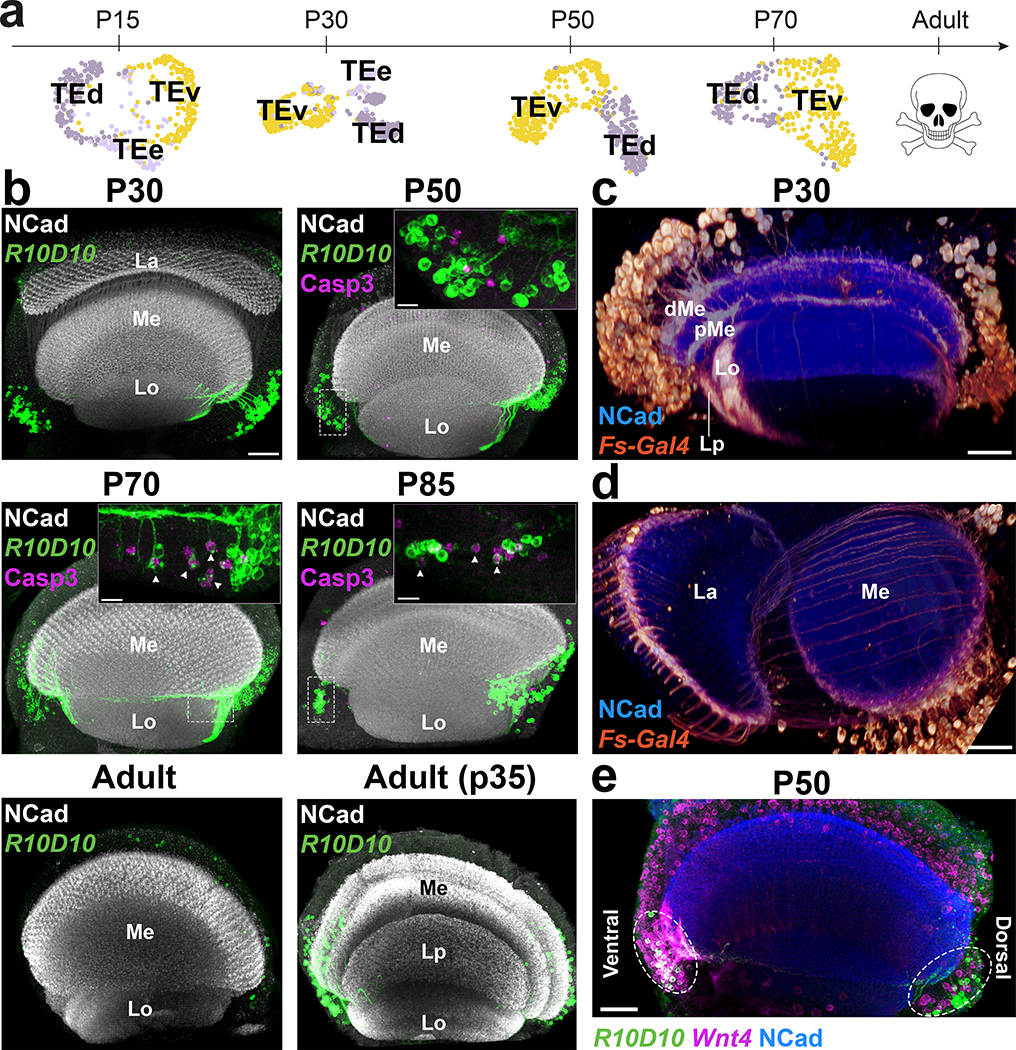
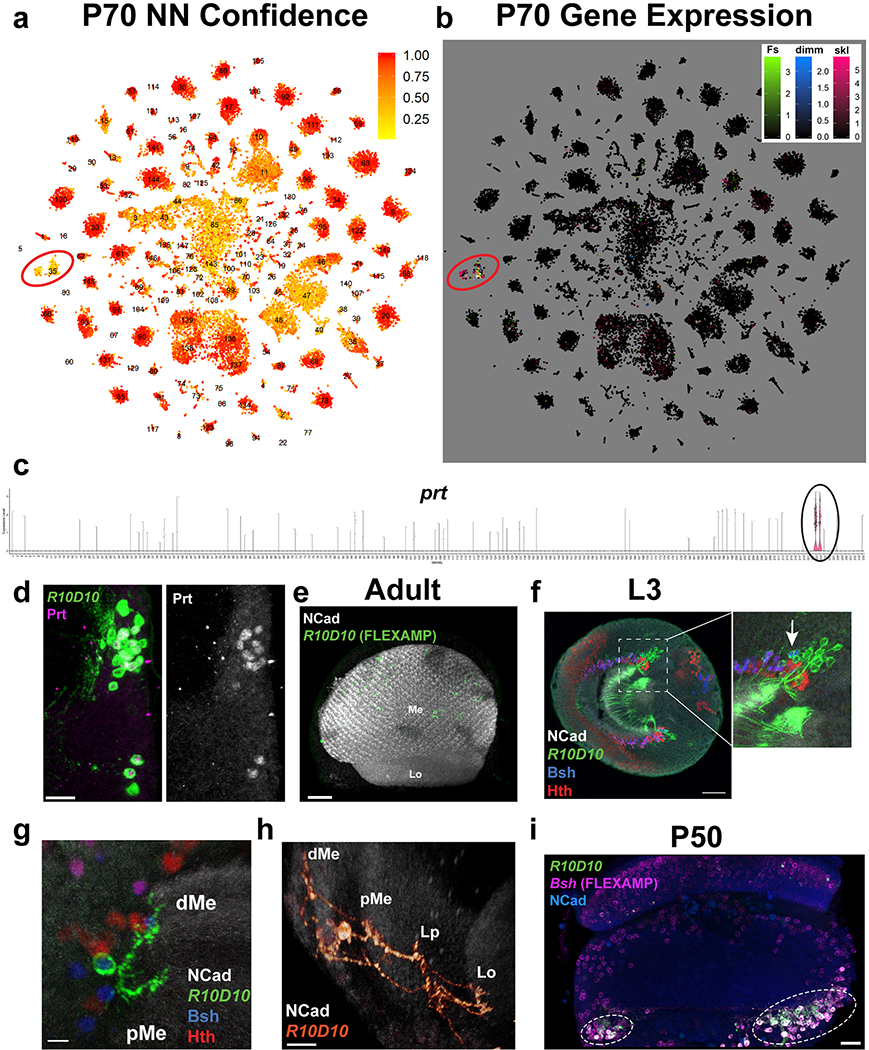
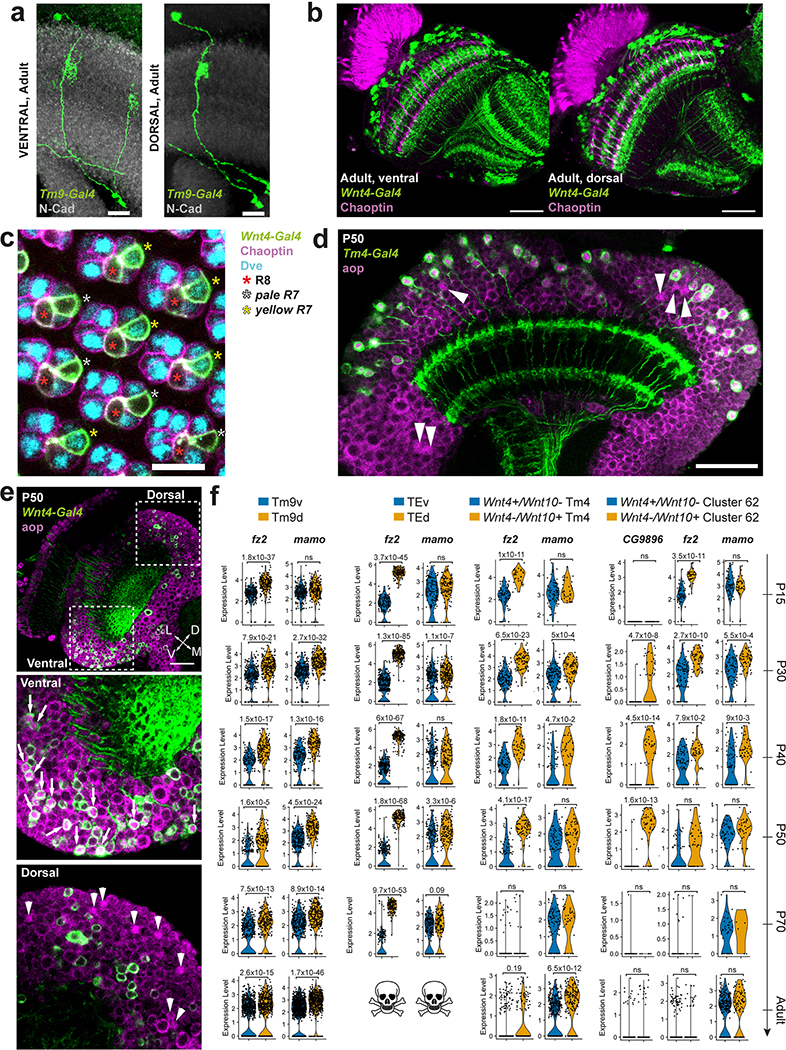
We identified two large neuronal clusters in the pupal datasets that received very low confidence scores from the NN classifier (indicating they may not exist in the adult dataset). These two clusters strongly expressed the pro-apoptotic gene sickle at P70 (Extended Data Fig.8a–b), suggesting that the corresponding cells die during late pupal stages. They specifically expressed throughout development genes for the secreted protein Follistatin (Fs), the pro-secretory TF dimmed (dimm)23 and the vesicular transporter portabella (prt) (Extended Data Fig.8b–c). R10D10(dimm)-Gal4 and Fs-Gal4 were expressed in pupae in two large sets of cells (Fig.2b–d) that were also labeled by anti-Prt antibody (Extended Data Fig.8d). Their projections wrapped the dorsal and ventral edges of all optic lobe neuropils, except the lamina: each neuron possessed multiple branches that contacted (but did not enter) the neuropils at different points (Extended Data Fig.8g–h). These neurons could not be found in adults and cleaved Dcp1 staining, a readout of Caspase-3 activity, confirmed that they were gradually cleared through apoptosis during late pupal stages (Fig.2b). Activation of a FLEXAMP memory cassette24 at L3 stage using R10D10-Gal4 (Extended Data Fig.8e) confirmed that our failure to observe these neurons in adult brains was not due to downregulation of reporter expression. Furthermore, expression of the anti-apoptotic protein p3525 caused the perdurance of these neurons in adult brains (Fig.2b). We thus named these cells transient extrinsic (TE) neurons.
Figure 2: Transient Extrinsic neurons demarcate the optic lobe neuropils and undergo apoptosis during late pupal development.
a, UMAP visualization of TE neurons, using 10 principal components calculated on the log-normalized integrated gene expression, across development. b, R10D10-Gal4 expression pattern (max projection) at the indicated stages (n=10 brains per stage), anterior view (with overexpression of anti-apoptotic protein p35 only in the last panel). Insets show z-restricted sections from the regions marked by dashed white rectangles. Staining of anti-NCad marks neuropils and anti-cleaved Dcp1 reports Caspase-3 (Casp3) activity, marking apoptotic cells. La = Lamina, Lo = Lobula, Me = Medulla, Lp = Lobula plate. c-d, Fs-Gal4 expression pattern (3D reconstruction) in P30 optic lobes (n=4 brains) with anti-NCad staining. d/pMe = distal/proximal Medulla. e, R10D10-LexA and Wnt4-Gal4 co-expression (white) pattern in a P50 optic lobe (max projection), with anti-NCad staining (n=8 brains). Dashed ellipses: TE neurons. Scale bars 7 μm (insets) or 30 μm (others).
Analysis of the genes differentially expressed between the two TE clusters revealed that only one of the clusters expressed Wnt4 while the other expressed Wnt10 (Fig.5c). Indeed, Wnt4-Gal4 was only expressed in the ventral cluster of TE neurons (Fig.2e) that we named TEv, while the Wnt10+ cluster was named TEd. Although we never observed TE neurons contacting the lamina, Fs-Gal4 was also expressed at P30 in photoreceptors from one row of ommatidia at the edge of the retina that connect to the edges of the lamina (Fig.2d) and are also apoptotically removed during development26. This suggests that Follistatin, an inhibitor of Activin signaling that has been implicated in regulating neuron growth27,28, is secreted at the edges of all neuropils by neurons that die before adulthood.
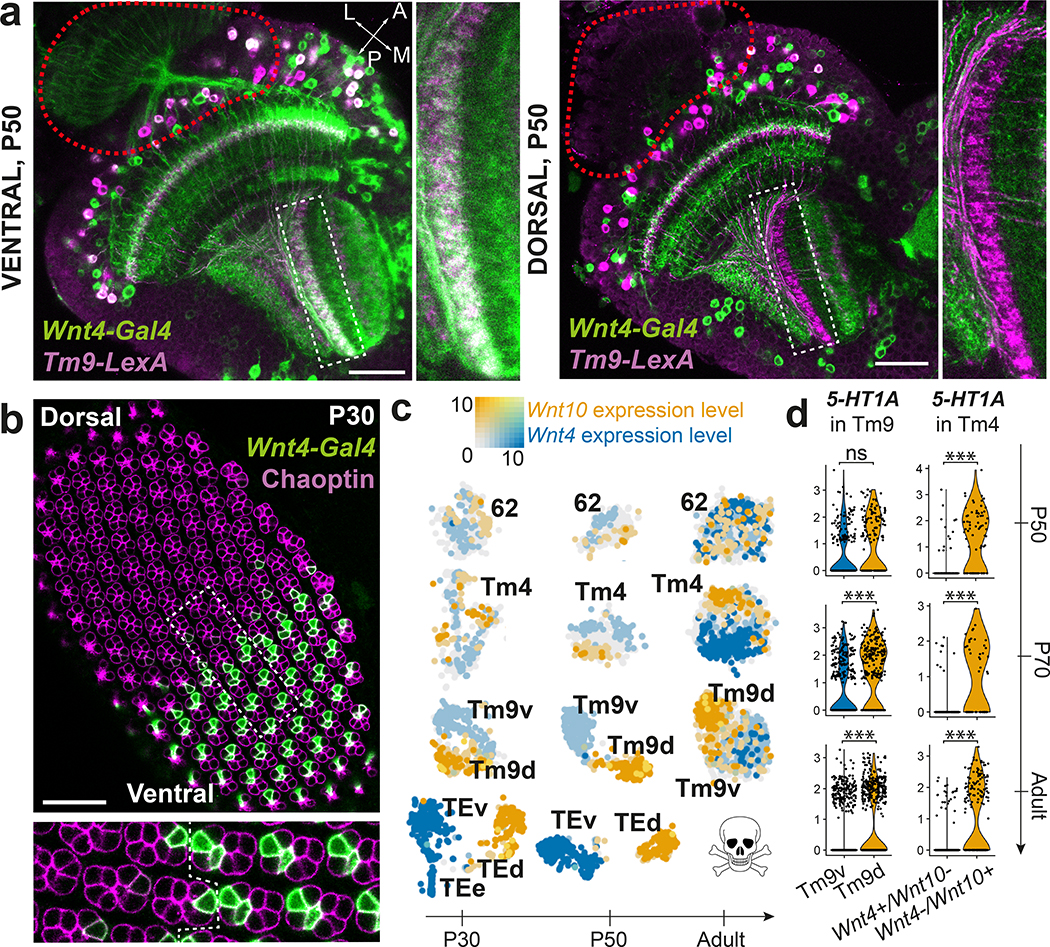
Figure 5: Dorsal and ventral visual circuits are partitioned by differential Wnt signaling.
a, Pattern of Wnt4-Gal4 and Tm9-LexA co-expression (white) at P50 in the ventral and dorsal part of the same optic lobe (n=8 brains). Red dashed line: location of photoreceptors. b, Wnt4-Gal4 expression pattern with anti-Chaoptin staining to mark photoreceptors in a P30 retina (n=6 eye discs). Dashed rectangle: inset. Dashed line within inset: equator of the retina. c, tSNE of the indicated clusters throughout development, with Wnt4 and Wnt10 log-normalized non-integrated expression levels. d, 5-HT1A differential expression between either Wnt4+/Wnt10- and Wnt4-/Wnt10+ Tm4 cells, or Tm9v and Tm9d cells (Methods). ***: adjusted p-value < 0.001 (P70 Tm9: 6×10−9, Adult Tm9: 3×10−18, P50 Tm4: 1×10−9, P70 Tm4: 2×10−7, Adult Tm4: 1×10−34), ns: not significant, two-sided Wilcoxon Rank Sum test. Scale bars = 30 μm.
A third TE cluster, which we called TEe (early), could only be found at P15 and P30. This cluster was connected to both TE clusters on UMAP visualization (Fig.2a) and was bsh+/hth-. In L3 optic lobes, we could observe a few Bsh+/Hth- cells labeled by R10D10-Gal4 at the tips of the OPC (Extended Data Fig.8f), suggesting that TE cells are produced there (the only other Bsh+ neurons are Hth+ Mi129). Bsh was also expressed in a subset of TE neurons at P30 (Extended Data Fig.8g). FLEXAMP memory cassette driven by bsh-Gal4 labeled all TE neurons (Extended Data Fig.8i), confirming that TEe cluster contains younger bsh+ TE neurons that lose bsh expression as they mature. TE neurons expressing bsh were present in decreasing numbers until P50, but not at P70. We therefore propose that TE neurons are produced continuously from a relatively small number of progenitors at the OPC tips.
The transient nature of TE neurons, their superficial innervations and enrichment in secretory markers are reminiscent of mammalian Cajal-Retzius cells30 that are essential for neuronal migration and other developmental processes. Further investigations will determine whether TE cells serve comparable functions in fly brain development (Supplementary Discussion).
Synchronization of serially born neurons
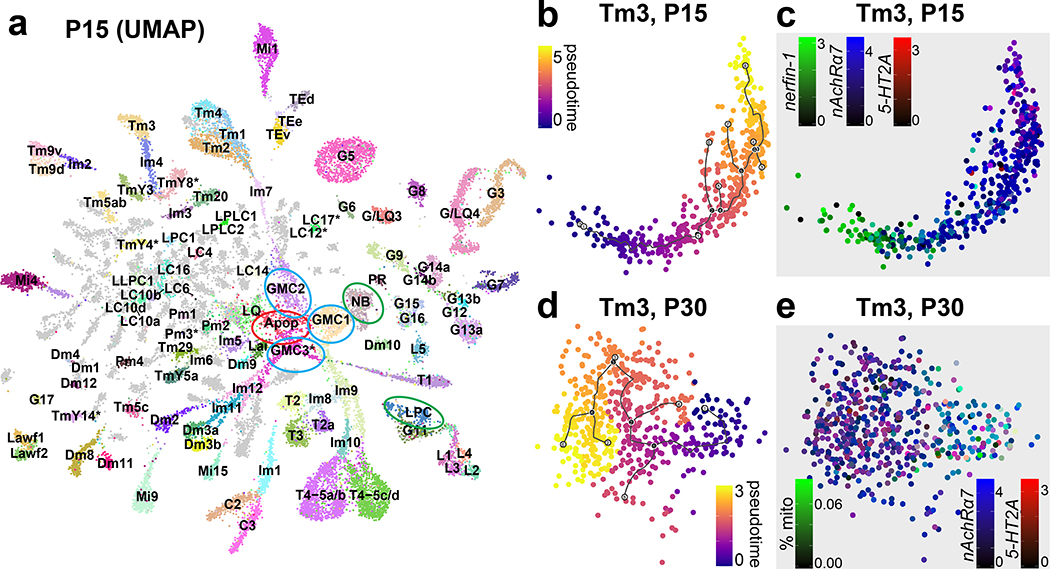
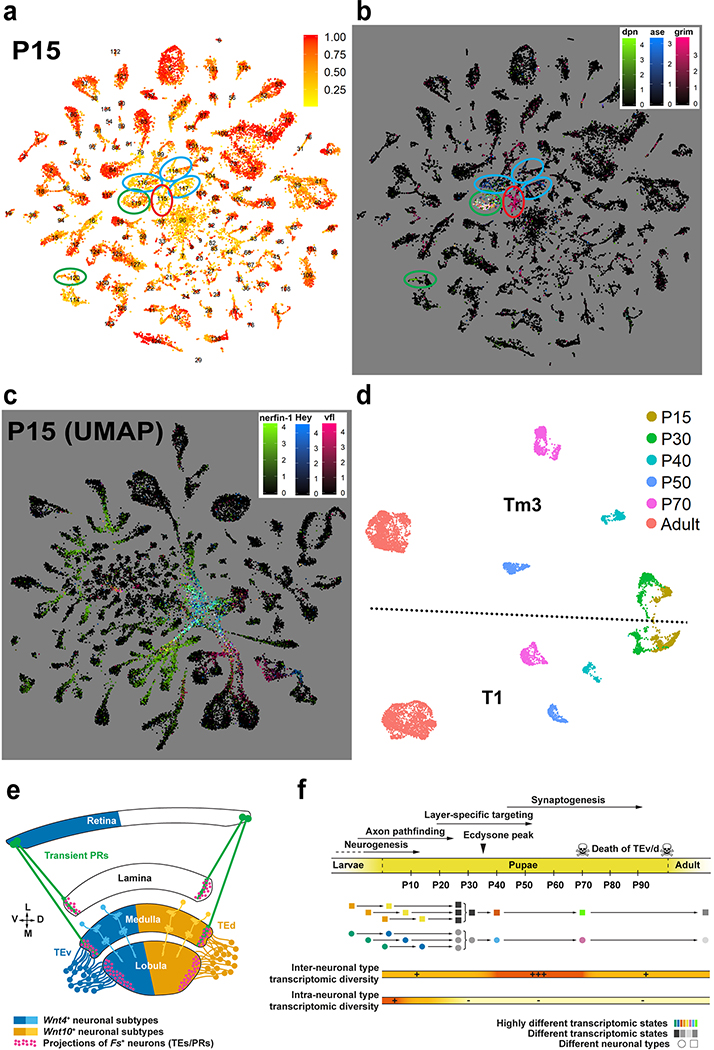
At larval stages, the IPC and medial OPC are progressively converted into neural stem cells (neuroblasts; NBs) that divide asymmetrically multiple times, each time self-renewing and producing a Ganglion Mother Cell (GMC), which in turn divides once to generate two neurons or glia31. Similar to photoreceptors that are produced sequentially from the posterior to anterior of the eye imaginal disc, optic lobe neurons are also produced sequentially, with the first born neurons connecting to the most posterior photoreceptors while later born neurons connect to more anterior photoreceptors32,33,. This is also true in the lateral OPC, where Lamina Precursor Cells (LPC) are progressively converted into lamina monopolar neurons34. As a result, neurons of the same types are produced days apart during development and the optic lobes contain neurons at various stages of their differentiation path. Accordingly, P15 neuronal clusters displayed ‘tails’ of differentiating cells (Fig.3a, Extended Data Fig.7f) that expressed several genes known to be transiently expressed by newborn neurons, including Hey35, nerfin-136, and zelda (vfl)37 (Extended Data Fig.9c). Moreover, whereas we could assign all P30-P70 cells directly to adult clusters (Extended Data Fig.7a–e) except for TE neurons and a few glia-like cells that were not investigated further (Methods), we found six clusters with low confidence scores from the NN at P15 (Extended Data Fig.9a). These clusters were identified by known markers (Extended Data Fig.9b, Suppl. Table 1) as NBs, GMCs (3 clusters), LPCs and undifferentiated apoptotic neurons (Fig.3a). On UMAP visualization, most P15 neuronal clusters converged towards the GMC and NB clusters while lamina monopolar cells L1-L4 converged specifically towards the LPC cluster (Fig.3a). This suggests that the cluster tails are intermediate states between progenitors and differentiated neurons. Since these convergent tails containing very young neurons might often be mixtures of different cell types, we separated them from the mature neurons wherever possible (Methods) and assigned them an “immature” (Im) designation (Fig.3a, Suppl. Table 1).
Figure 3: Transcriptomic synchronization of optic lobe neurons.
a, UMAP visualization of the P15 dataset (parameters as in Fig.1d). Ellipses: undifferentiated neurons undergoing apoptosis (red), stem cells (green), intermediate precursors (blue). b-e, UMAP visualization of the Tm3 cluster at P15 and P30. Cells are colored by their position on the pseudotime trajectory inferred by Monocle 3 (Methods) and shown here as black lines (b, d); or by their log-normalized non-integrated expression level of nerfin-1 (c), nAChRα7, 5-HT2A (c, e), and by the ratio of total mitochondrial transcripts they have (e). Scale bars = 30 μm.
Remarkably, these tails were not present at any of the later stages, when the clusters appeared homogenous despite containing neurons that were born more than 40 hours apart. To further investigate this phenomenon, we generated a UMAP plot containing only T1 and Tm3 neurons, combined from all stages with no batch correction (Extended Data Fig.9d). Due to continuous production of the neurons of the same type in the optic lobe, a Tm3/T1 cell born at P0 in the P30 dataset would be the same age as a Tm3/T1 cell born at P10 in the P40 dataset. Thus, if neurons were to maintain age-dependent differences, cells of the same type would form a continuous trajectory across timepoints; however, they do not. Indeed, we observed this only between P15 and P30, whereas all other timepoints remained robustly separated. To rule out batch effects between different stages, we also directly assessed the homogeneity of the Tm3 cluster. Using Monocle 338, we generated trajectories for Tm3 neurons at both P15 and P30 (Fig.3b,d) and calculated marker genes that varied with pseudotime (Methods). Analysis of these markers clearly indicated that cells were indeed ordered by age at P15. For instance, nerfin-136 was only expressed in the younger P15 cells (Fig.3c) (and in almost no P30 cells), whereas expression of nicotinic Acetylcholine Receptor α7 peaked later and serotonin receptor 5-HT2A could only be observed in the most mature cells at P15 (Fig.3c). On the other hand, all apparent sources of variation within the Tm3 cluster at P30 could be attributed to transcriptome quality, as measured by the percentage of mitochondrial UMIs observed (Fig.3e). Therefore, the youngest (10–15 hours old) and the oldest (>3 days old) Tm3 cells were no longer distinguishable at P30.
These data suggest that age information in neurons is lost within 15 hours or less after their birth, as they converge to a common transcriptomic state with older neurons of the same type. This generalizes observations previously made in photoreceptors39,40 (Supplementary Discussion), and implies that all brain wiring steps beyond initial neuropil targeting (i.e. axon pathfinding, Fig.1e) are executed synchronously in the optic lobe.
Increased diversity at synaptogenesis
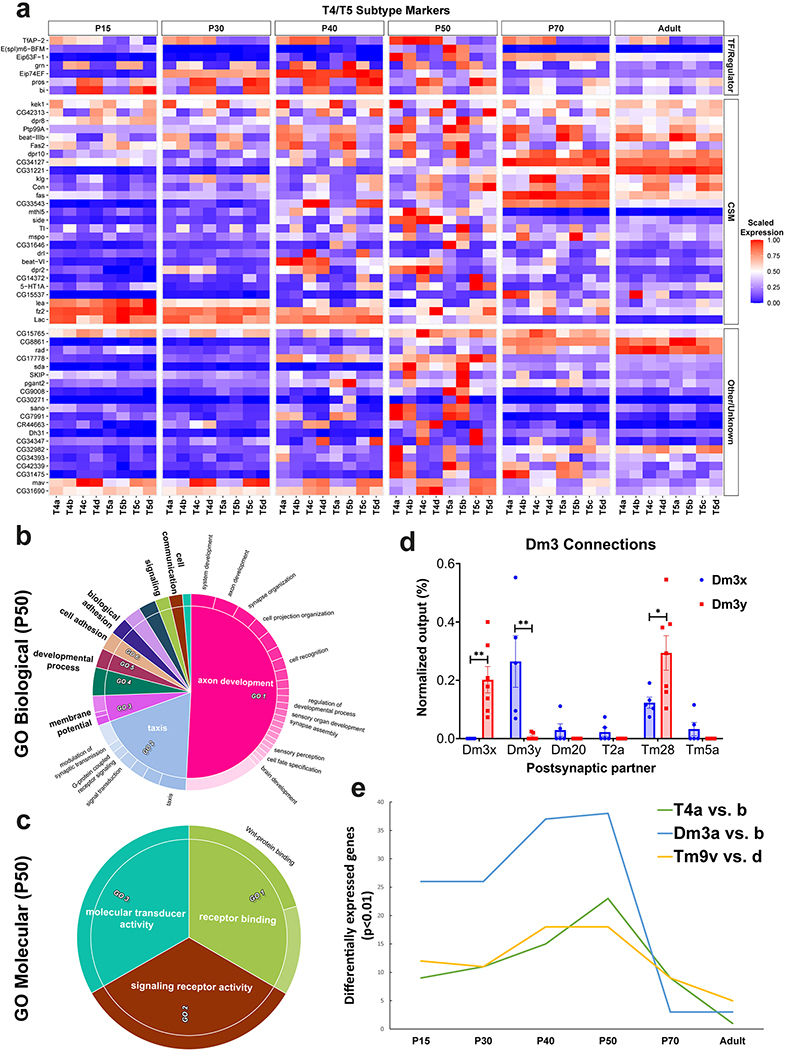
Both T4 and T5 neurons have 4 subtypes (a/b/c/d) that each processes motion in one of four cardinal directions41. Their polarized dendritic arbors mirror their direction selectivity6,42. Unsupervised clustering reliably distinguished these 8 subtypes only at P50. However, supervised annotation and subclustering of T4–5 cells (Fig. 4a, Methods) showed that a/b subtypes could be separated from c/d subtypes at all stages. Many of the P50 subcluster markers turned off or lost their specificity in adult brains (Extended Data Fig.10a), explaining why T4–5 subtypes were transcriptionally indistinguishable in adult brains (Fig.1d). GO analysis of these markers revealed exclusive enrichment for cell surface receptor terms involved in cell adhesion and axon/dendrite development (Extended Data Fig.10b–c).
Figure 4: Increased transcriptomic diversity during synaptogenesis.
a, tSNE visualizations of T4 and T5 neurons, using 20 principal components calculated on the log-normalized integrated gene expression, across development. b, R25F07(Dm3)-Gal4 sparse expression at P50 with anti-Bi (Omb) staining (n=10 brains). Yellow and cyan arrows/arrowheads indicate dendrites/cells bodies of a posterior-ventrally and a posterior-dorsally oriented Dm3, respectively. Scale bar = 5 μm. c, Plot of the highest Pearson correlation between each neuronal cluster (n=175) and all other neuronal clusters, at each stage, using all genes belonging to the top10 cluster markers of at least one stage. ***: adjusted p-value<0.001 (P15–30: 7X10−11, P30–40: 4X10−10, P70-Adult: 1X10−11, P15-Adult: 3X10−13), n.s: non-significant, one-way ANOVA with Tukey Honest Significant Differences. Boxplots display the first, second and third quartiles. Whiskers extend from the box to the highest or lowest values in the 1.5 inter-quartile range, and outlying datapoints are represented by a dot. d-e, Summary of GO enrichment analysis performed on the stage markers (Extended Data Fig.11a).
In addition, Dm3 and Tm9 cells were split in two subgroups only at P50 and earlier stages (Extended Data Fig.7). Subgroups of Tm9 have not been previously described (see last section), but Dm3 cells have two known sub-populations with orthogonal dendritic orientations7. Immunostainings against Bifid (Bi or Omb), which was differentially expressed between the Dm3 subclusters, showed that the dendrites of the Bi+ Dm3b cells were always oriented posterior-dorsally while those of Bi- Dm3a cells were oriented posterior-ventrally (n>100, Fig.4b). As Dm3 subtypes differentially expressed several cell surface molecules (CSM) during synapse formation, we asked whether they also differed in their connectivity. Analysis of the medulla connectome4 revealed that the two Dm3 subtypes connect differentially to several postsynaptic partners (Extended Data Fig.10d, Methods). Most notably, Dm3-Dm3 synapses were found almost exclusively between the different subtypes. Interestingly, Bi is also necessary and sufficient to specify the identity of T4-T5c/d subtypes that have dendrites with orthogonal directionality to those of T4-T5a/b neurons43. Thus, bi may specify subtypes with orthogonal dendritic orientations in neurons that have completely different origins and properties.
Attempting to project Dm3 or Tm9 subdivisions onto adult clusters by training binary classifiers on the P50 cells (Methods) proved very unreliable with out-of-bag errors (OOBE, Methods) >20%. Consistently, the number of differentially expressed genes between subgroups peaked around P50 and dropped sharply thereafter (Extended Data Fig.10e). Even though T4/T5, Dm3 and Tm9 may represent extreme cases, this increased transcriptional diversity during synapse formation is indeed a general phenomenon: Pearson correlations between the average gene expression profiles of clusters that are most similar to each other were significantly lower between P40-P70, across all neuronal clusters (Fig.4c). These results generalize the previous findings that olfactory projection neurons VA1d and DC3 are transcriptionally distinct during development but merge into a single cluster in adults44. They also call into question the common practice of cell-type identification based solely on adult transcriptomes and advocate for developmentally based approaches.
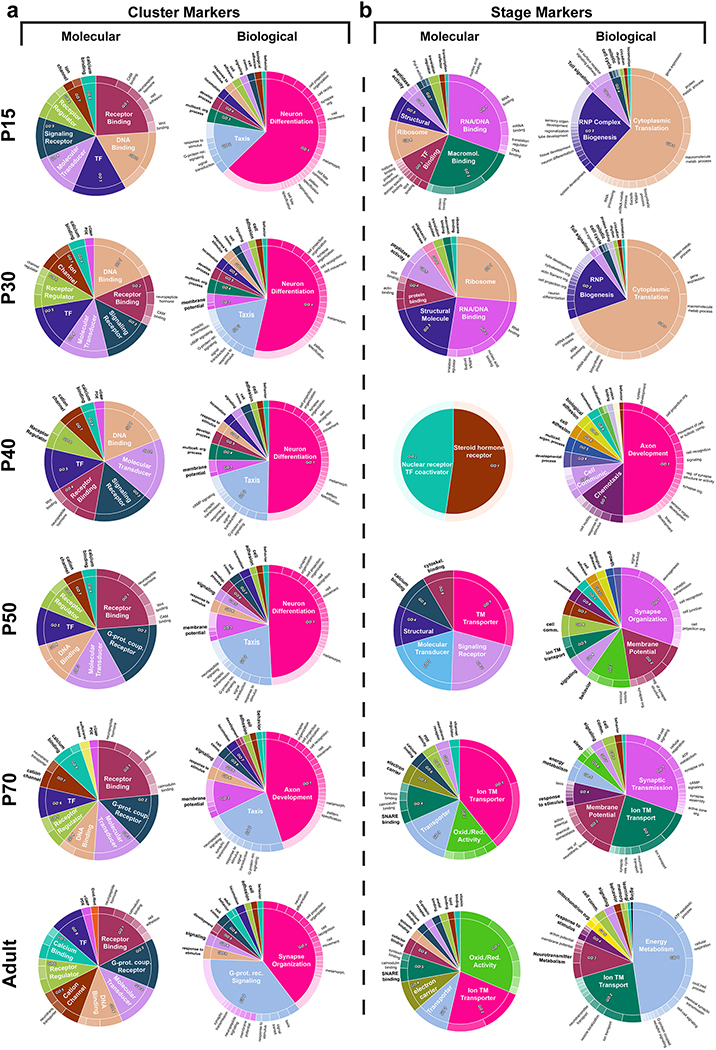
To investigate why neuronal types are more easily distinguishable during development, we performed GO analysis of the neuronal cluster markers at each stage. This consistently revealed at all stages overwhelming enrichment for receptor binding/activity terms related to axon/dendrite development and synapse formation, followed by TF and ion channel terms (Extended Data Fig.11a). If different neurons are distinguished largely by CSMs at all ages, what is the origin of the increased diversity we observe at mid-pupal stages? We performed GO analysis on ‘stage markers’, i.e. genes that were upregulated in neurons at a particular time point as compared to all other stages (Methods, Extended Data Fig.11b). CSM terms involved in synaptogenesis and membrane potential were particularly upregulated around P50–70 (Fig.4d–e), explaining the increased diversity at these stages. In contrast, early pupal markers were dominated by protein synthesis and adult markers by energy metabolism terms. This suggests that, as a general principle, the upregulation of CSMs necessary to enable synaptic specificity around P50 (Fig.1e) causes a peak of transcriptional diversity. This diversity is not maintained later, especially between subtypes that perform highly related functions and only differ based on their connectivity. Moreover, P40 was enriched in molecular terms related to nuclear hormone receptor, driven by ecdysone-responsive TFs, suggesting that global activation of a hormonal switch triggers the upregulation of cell-type specific CSMs at the onset of synaptogenesis (Supplementary discussion).
Dorsoventral division of visual circuits
Differential gene expression analysis between the two Tm9 subgroups at P50 indicated that one of them exclusively expressed Wnt4 while the other expressed Wnt10, similarly to TEv/d neurons. Since Wnt4-Gal4 expression overlapped only with ventral Tm9s (Fig.5a), we named these subtypes Tm9v (Wnt4+) and Tm9d (Wnt10+). Sparse labeling of individual Tm9 neurons did not reveal any obvious morphological differences between these subtypes (Extended Data Fig.12a). However, differential expression of several cell adhesion molecules at P50 raise the possibility that they differ in their connectivity. Even though these subtypes were statistically mostly indistinguishable in P70 and adult datasets, Wnt4 and Wnt10 expression were found in separate parts of the same cluster on the tSNE (Fig.5c). We also noticed that Wnt4-Gal4 expression was restricted to the ventral photoreceptors R7/R8 in pupae, but not in adult (Fig.5a–b, Extended Data Fig.12b–c). Therefore, both the receptive visual field and its downstream circuitry are partitioned by differential Wnt signaling during development.
Flying insects are exposed to very different stimuli in their ventral vs. dorsal visual fields (ground vs. sky) that may need to be processed differently: Dorsoventral asymmetries could be a fundamental adaptation to flight since they have been described in the retinas of flies, butterflies, dragonflies and honeybees (reviewed in45). Our findings expand these differences to the downstream circuitry. In addition to the cell-autonomous differences during development that could enable these cells to connect with different synaptic partners, secreted Wnt ligands could differentially affect development and function of other neurons in ventral and dorsal parts of the brain (Supplementary Discussion).
Finally, we tested whether the differential expression of Wnt4 vs. Wnt10 observed in TE, Tm9 and R7/R8 neurons applied to other seemingly homogeneous neuronal types (Methods). We found two other neurons with dorsoventral asymmetries: Tm4 neurons and the unidentified cluster 62 could be separated into Wnt4+ and Wnt10+ populations, with only ventral Tm4 neurons overlapping with Wnt4-Gal4 expression (Extended Data Fig.12d–e). For both Tm4 and cluster 62, Wnt4+ and Wnt10+ cells remained separated on the tSNE (Fig.5c) but were highly similar: artificially separating them yielded an average OOBE across all stages around 25%. Despite this similarity, Wnt4+ and Wnt10+ neurons differentially expressed several genes throughout development (Extended Data Fig.12f). Interestingly, both Tm4d and Tm9d specifically expressed the serotonin receptor 5-HT1A in adult (Fig.5d), raising the possibility that neuromodulatory signals are processed differently in ventral vs. dorsal visual circuits.
Conclusions
We present here the first scRNA-seq dataset reaching near complete saturation of any complex nervous system throughout its development. Coupled with the available optic lobe connectomes4,6, this will provide an important resource for functional studies of adult neurons as well as for the identification of new mechanisms involved in circuit formation. Our analyses revealed two intriguing populations of pupae-specific neurons that share many characteristics with the mammalian Cajal-Retzius cells and could be involved in neuropil development (Extended Data Fig.9e). We made several observations with important implications on how neural circuits are built (Extended Data Fig.9f): We described the convergence of neuronal transcriptomes of the same type and generalized previous observations of increased transcriptomic diversity in neurons during development to the entire optic lobe circuit. We showed that this is due to a transient upregulation of cell-type specific CSMs involved in synapse formation, which explains how neurons with indistinguishable transcriptomes in adult brains could nevertheless serve different functions due to their developmental history. Lastly, we demonstrated that ventral and dorsal visual circuits are subjected to differential Wnt signaling, providing potential mechanisms for differential processing of ground vs. sky inputs, in extension of the asymmetries described in the retinas of several flying invertebrates.
Methods
Genetics
All sequencing experiments in this study were performed with female D. melanogaster Canton-S maintained at 18–25°C (unless indicated otherwise) and dissected within 3 days of eclosion (adult dissections) or selected at P0 (white pupae) and maintained at 25°C until the required stage (pupal dissections). Flies for the other experiments were maintained in the same way but were not selected for gender. Origin of all individual stocks is detailed in Supplementary Table 3.
Following final genotypes were used for the imaging experiments: Dm3 sparse labeling: hsFLP22/+; CyO/+; R25F07-Gal4/UAS-FSF-CD4-tdGFP. TE neuron labeling: 10xUAS-myr:GFP/+; R10D10-Gal4/+ or Fs-Gal4/+; UAS-CD4:tdGFP. Suppression of apoptosis in TE neurons: UAS-P35/+; UAS-myr-GFP/+; R10D10-Gal4/+. TE neuron FLEXAMP: UAS-Flp; Gal80ts/Cyo; Act>y+>LexA, LexAop-myr:GFP/R10D10-Gal4. Bsh FLEXAMP: UAS-Flp; Gal80ts/Cyo; Act>y+>LexA, LexAop-myr:GFP/Bsh-Gal4 TE neuron sparse labeling: hsFLP2:PEST/+; UAS-FSF-CD4:tdGFP/+; R10D10-Gal4/+. Tm9 sparse labelling: hsFLP2:PEST; UAS-FSF-CD4:tdGFP/R72F01-p65AD; R82F10-Gal4DBD/TM6B or MKRS. Wnt4-Gal4 and Tm9-LexA co-expression experiments: 10xUAS-IVSmCD8:RFP,13xLexAop2-mCD8:GFP; R82F10-LexA/Wnt4-Gal4; +/TM2. Tm4-Gal4 and aop co-expression:;10xUAS-myr:GFP; R35H01-Gal4. PR stainings: Wnt4-Gal4, UAS-myr:GFP/Cyo; Tm2/Tm6B. TmY4 labeling: Ac76E-LexA, P3-RFP/CyO; LexOpCD2-GFP/TM2. TmY8 sparse labeling: pBPhsFlp2::PEST;; HA_V5_FLAG/CG42458-Gal4. TmY14 labeling: 10xUAS-myr:GFP; R24F10-Gal4. LC12 labeling:; 10xUAS-myr:GFP/R35D04-p65.AD; R55F01-GAL4.DBD/+. LC17 labeling;; 10xUAS-myr:GFP/+; R92G12-Gal4/+ or; 10xUAS-myr:GFP/R21D03-p65.AD; R65C12-GAL4.DBD/+. Beat-1c expression:; beat 1c MI01467-T2A Gal4/Cyo; 20xUAS 6xGFP/TM6b. Kn expression:; 10xUAS-myr:GFP/ Mi-Trojan-GAL4.2-kn[MI15480-TG4.2];.
Single-cell RNA sequencing
Sample Preparation
Fly brains were dissected in ice-cold Schneider’s Insect Medium (SIM, Gibco) and incubated at 25°C for 1.5 hours (adult brains) or 30 minutes (pupal brains) in a dissociation solution of SIM with 2mg/mL Collagenase (Sigma) and 2mg/mL Dispase (Sigma). Towards the end of incubation, we placed all appropriate reagents (as indicated in the Chromium™ Single Cell 3’ Reagent Kits v2 User Guide – Rev D) to equilibrate at room temperature. We then washed once the whole brains with ice-cold SIM, separated the optic lobes, washed them 3 times with ice-cold Dulbecco’s Phosphate-Buffered Saline (DPBS, without calcium & magnesium, Corning/Fisher) + 0.04% Bovine Serum Albumin (BSA, Sigma), and transferred them into a low-bind tube on ice (150 μL ice-cold DPBS + 0.04% BSA for 12 optic lobes). We dissociated the optic lobes by vigorously pipetting up and down the content of the tube 2×50 times, resting the cells on ice for 1 minute in between. We then observed the cell suspension under a standard dissecting microscope and pipetted the suspension up to 50 times more, until no large chunks could be seen. Limiting the number of pipetting repetitions increased the number of cells recovered and limited the amount of ambient RNA released from damaged cells into the media. We then passed the cell suspension through a 20 μm strainer (pluriSelect). In order to process the cells as quickly as possible, one experimenter then estimated the concentration of an aliquot of the cell suspension with 1/2000 Hoeschst, using an epifluorescent microscope and a 0.02 mm deep cytometer, while the second experimenter proceeded with the preparation of the microfluidic chip.
All adult brains were dissected between 9–12am. Two adult experiments also included male flies; these cells were removed from the dataset in downstream analysis. Incubating the entire brains in the dissociation solution and removing the optic lobes afterwards improved the quality of the sequenced transcriptomes (number of genes recovered, proportion of UMIs from mitochondrial genes) compared to direct dissection of the optic lobes. We chose to neither centrifuge the dissociated cells and wash the pellets nor isolate cells from debris by FACS. While this could have reduced the amount of ambient RNA sequenced, by further lengthening the protocol these additional steps might have affected gene expression, increased batch effects, or created sampling biases affecting the frequency of sequenced cell types due to differential size or fragility.
Library preparation, sequencing and processing
Droplet-based purification, amplification and barcoding of single-cell transcriptomes were performed using Chromium™ Single Cell 3’ Reagent Kit v2 (10X Genomics) as described in the manufacturer’s manual (Rev D), with a target recovery of 7,000 cells per experiment. We prepared 15 libraries from adult brains, 5 libraries each from P15, P30 and P70 brains and 4 libraries each from P40 and P50 brains. The libraries were subjected to paired-end sequencing (26×8×98) with Illumina NextSeq 550 (Genomics Core at NYU CGSB) or NovaSeq 6000 (Genome Technology Center at NYU Langone Health) to on average 50,000 reads per cell sequenced (i.e. 350,000,000 reads for a 7,000 cells experiment).
We mapped the sequenced libraries to the D. melanogaster genome assembly BDGP6.88 using CellRanger 2.1.0 for the adult stage and 3.0.1 for the pupal stages. We kept all cell barcodes with 1,000 to 20,000 UMIs and less than 10% of UMIs corresponding to mitochondrial genes. Note that the incorporation of EmptyDrops algorithm in CellRanger v3 excluded further cells with >1000 UMIs predicted to contain only ambient RNA for pupal datasets. In addition, for the two experiments that included male cells at the adult stage, we removed all cell barcodes with at least one UMI of any of the genes of the male-specific lethal (MSL) complex (msl-1, msl-2, msl-3, mof, mle, roX1, roX2). These single-cell transcriptomes are provided in Adult_male.rds (GSE142787). Lastly, we kept only the genes that were expressed in at least 3 cells across all experiments of a given stage for further analysis. After processing, the adult dataset comprised 109,743 cells passing quality filters, with a median of 1805 UMIs and 903 genes per cell. The pupal dataset comprised 31,036 cells passing quality control filters at P15, 35,758 cells at P30, 24,084 cells at P40, 31,340 cells at P50 and 43,740 cells at P70.
RNA sequencing of FACS-sorted cell types
We crossed the Gal4 lines specific for individual cell types (Pm2-Gal4 (Bl stock no. 50240), T4-Gal4 (VT stock no. 37588)) as well as pan-neuronal marker line (Elav-Gal4) and pan-glial marker line (Repo-Gal4) to UAS-Red Stinger to fluorescently label nuclei of individual cell types, sort and sequence them as previously described18. Briefly, we first dissected adult optic lobes, then dissociated tissue while maintaining cell viability and finally, sorted cells by FACS (Facs Aria III) based on their differences in fluorescence intensity and cell size. RNA was extracted using the Arcturus PicoPure RNA Isolation Kit (Applied Biosystems) and the Smart-Seq v4 Ultra Low Input RNA Kit (Clontech) was used to generate full-length double stranded cDNA with 300 to 500 pg of total RNA input. The quality of RNA and cDNA was assessed by Bioanalyzer using RNA 6000 Pico and High sensitivity DNA assay (Agilent) respectively. Libraries were prepared using Nextera XT DNA Library Prep Kit (Illumina) and run on the Illumina HiSeq 2500 (CGSB at NYUAD). Three barcoded libraries were pooled per sequencing lane and paired-end 100 bp reads were generated. Sequences were mapped to the Drosophila melanogaster genome (BDGP6.81) using TopHat2 (v2.1.0). We obtained three biological replicates for each library.
Antibody generation
Polyclonal antibodies against Bsh, Hth, Dichaete and Vvl were generated by Genscript (https://www.genscript.com/).
The epitope used to immunize the rabbits was the full length Bsh protein:
MAMLNEASLS PADAHAHANA TTPTHSKAAA MASATTMLTT KTPFSIEHIL FQNLNSASNN NNSSDTNGIA ANTNNYAPKS SRNAVKSARS AFAHDNNPHK HPSQHSHPPQ SHPPASASAS ATATARSNQA ASGYAGEDYG KSMHSTPRSN HHSRHGTSHY NGDQISQQLG SGAAQHPPVP TTQPQPPPPP PLNGGSGASN GVLYPNAPYT DHGFLQMTLG YLSPSSGTYK SVDPYFLSQA SLFGGAPFFG APGCVPELAL GLGMGVNALR HCRRRKARTV FSDPQLSGLE KRFEGQRYLS TPERVELATA LGLSETQVKT WFQNRRMKHK KQLRRRDNAN EPVDFSRSEP GKQPGEATSS SGDSKHGKLN PGSVGGTPTQ PTSEQQLQMC LMQQGYSTDD YSDLEADSGD EDNSSDVDIV GDAKLYQLT
The epitope used to immunize the guinea pigs were the following amino acids of the Hth-PC isoform:
HGYHSGAGGHGTPSHVSPVGNHLMGAIPEVHKRDKDAIYEHPLFPLLALIFEKCELATCTPREPGVQGGDVCSSESFNEDIAMFSKQIRSQKPYYTADPEVDSLMVQAIQVLRFHLLELEKVHELCDNFCHRYISCLKGKMPIDLVIDERDTTKPPELGSANGEGRSNADSTSHTDGASTPDVRP
The epitope used to immunize the guinea pigs were the following amino acids of Vvl:
EVSVKGALEQHFHKQPKPSAQEITSLADSLQLEKEVVRVWFCNRRQKEKRMTPPNTLGGDMMDGMPPGHMHHGGYHPHHDMHGSPMGTHSHSHSPPMLSPQNMQSSAVAAHQLAAH
The epitope used to immunize the guinea pigs were the following amino acids of Dicheate:
SLATSPGQEGHIKRPMNAFMVWSRLQRRQIAKDNPKMHNSEISKRLGAEWKLLAESEKRPFIDEAKRLRALHMKEHPDYKYRPRRKPKNPLTAGPQGGLQ
Polyclonal antibody against Toy was generated by Thermo Scientific, using an epitope with the following amino acids to immunize rabbits:
MRTQRRSADTVDGSGRTSTANNPSGTTASSSVATSNNSTPGIVNSAINVAERTSSALVSN SLPEASNGPTVLGGEANTTHTSSESPPLQPAAPRLPLNSGFNTMYSSIPQPIATMAENYN SSLGSMTPSCLQQRDAYPYMFHDPLSLGSPYVSAHHRNTACNPSAAHQQPPQHGVYTNSS PMPSSNTGVISAGVSVPVQISTQNVSDLTGSNYWPRLQ
Immunohistochemistry
Brains were dissected in ice-cold SIM, fixed in 3.7% formaldehyde (in PBS) at room temperature for 30–50 minutes, washed in PBST (PBS + 0.3% Triton X-100) and incubated for an hour in blocking solution (PBST + 5% horse serum), which we also used to dilute all primary and secondary antibody solutions. They were incubated in primary antibodies for 1 or 2 days at 4°C, washed three times in PBST for 10 minutes and then incubated in secondary antibodies at 4°C overnight followed by washing in PBST again 3×10min. They were then mounted in Slowfade and imaged with a Leica SP8 confocal microscope using a 20x (NA=0.75) or a 63x (NA=1.3) glycerol objective. Images were processed with ImageJ or Imaris.
The following primary antibodies were used: Polyclonal rabbit anti-RFP (1:500), Polyclonal rabbit anti-cleaved Dcp-1 Antibody (1:100). sheep anti-GFP (1:200), chicken anti-GFP (1:100), Rabbit anti-PRT (1/100), rabbit anti-GFP (1:250), mouse anti-aop (1:100), rat anti-NCad (1:20), rabbit anti-Bi (1:400), mouse anti-Chaoptin (1:50), rabbit anti-Bsh (1:1800), guinea pig anti-Hth (1:100), rabbit anti-Dve (1:250), mouse anti-Cut (1:20), mouse anti-Acj6 (1:20), rabbit anti-Toy (1:250), rat anti-FLAG (1:200), guinea pig anti-Dichaete (D) (1:50), guinea pig anti-Vvl (1:100), mouse anti-Dac (1:20), mouse anti-Brp (nc82) (1:30).
The following secondary antibodies (all donkey, used at 1:400) were used: anti-sheep Alexa 488, anti-chicken Alexa 488, anti-rabbit Alexa 488, anti-rabbit Cy3, anti-mouse Cy3, anti-rat Alexa 555, anti-rat Alexa 647, anti-guinea pig Alexa 488, anti-guinea pig Alexa 647, anti-guinea pig Cy3, anti-rat Alexa 405, anti-rabbit Alexa 647, anti-mouse Alexa 647.
Origin of all antibodies used is detailed in Supplementary Table 3.
Data Analysis and Statistics
Integration of scRNA-Seq libraries
We used the procedure implemented in Seurat v321 to remove batch effects from our sequenced libraries. Briefly, it uses Canonical Correlation Analysis to project the libraries in a low dimensional space where their correlation is maximized. This keeps the variation shared between the libraries (biological variation) and removes the variation specific to each library (technical variation). The cells from different libraries that are mutual nearest neighbors in this shared low dimensional space are then used as anchors to calculate a matrix of ‘integrated’ gene expression, using the 2,000 most variable genes of the dataset, whose expression is corrected for batch-effect and used for downstream analysis.
Using default parameters in Seurat 3.0.0.9000, we first normalized (NormalizeData) independently each sequenced library of a given stage and extracted their 2000 most variable features (FindVariableFeatures). For the adult stage, we then used the functions FindIntegrationAnchors, IntegrateData, ScaleData and RunPCA with default parameters, except for the dimensionality for which we tried the values 100, 150 and 200. To compare the results, we assessed how much the integrated dataset conserves the structure its individual libraries had before integration using the Seurat function LocalStruct with default parameters. This function counts, for each cell of a given library, how many of its 100 closest neighbors in this library are similar before and after integration. This is done and averaged for all libraries. The results obtained were 74%, 81% and 75%, respectively. We therefore chose a dimensionality of 150 for the adult dataset. This procedure conserved the local structure of each dataset, as the cells shared on average 81% of their closest neighbors before and after integration, and efficiently removed the batch effects, as our final adult clusters contained a uniform distribution of cells from each original library (Extended Data Fig.1b). Lastly, we used a dimensionality of 100 for all pupal datasets due to their smaller sizes. The values obtained with the LocalStruct function were P70 = 81% / P50 = 76% / P40 = 83% / P30 = 82% / P15 = 88%.
Clustering of the datasets
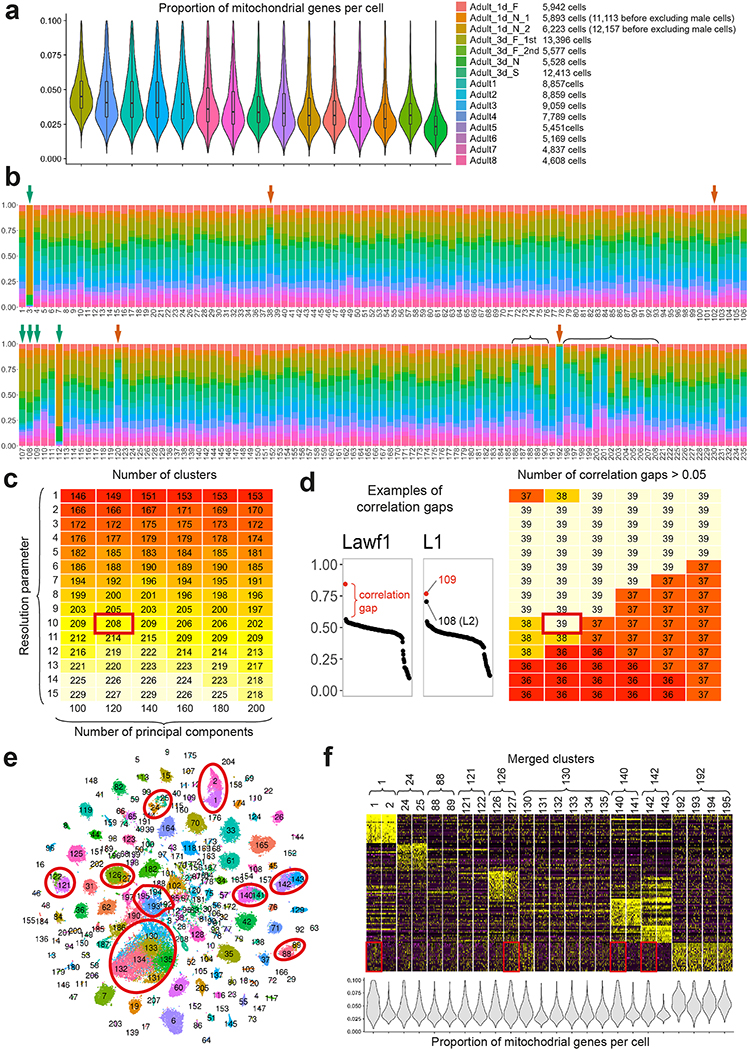
We used Seurat 3.0.0.9000 to cluster our integrated single-cell transcriptomes to identify groups of cells with similar gene expression patterns. For the adult dataset we used the functions FindNeighbors and FindClusters with default parameters except for the number of principal components (PCs), which are used to reduce the dataset dimensionality and calculate the distance between all pairs of cells, and the resolution, which is used to compensate for the tendency of modularity optimization algorithms to merge small clusters. Small resolution values favor big clusters, and high values favor small clusters but tend to artificially split large clusters (which can be corrected for by subsequent analysis). We therefore performed a grid search of 90 different combinations of parameters aimed to optimize the biological significance of the resulting clusters.
We varied the PC number between 100 and 200 (with a step size of 20), and the resolution between 1 and 15 (step size of 1), which yielded between 146 and 229 clusters (Extended Data Fig.1c). We assessed the biological significance of the clusters obtained with each pair of clustering parameters by using 54 log-normalized bulk-transcriptomes obtained from purified optic lobe neuronal types: 52 published before18,20 and 2 additional (Pm2 and T4) sequenced for this study. For each purified neuronal-type transcriptome, we calculated the Pearson correlation with the average log-normalized non-integrated gene expression of each cluster, using the list of the most variable genes found during the clustering step. We then ranked the clusters by decreasing value of Pearson correlation. If a cluster has a much higher correlation value than the others, the cells from this cluster are very likely to belong to this neuronal type. If several clusters have a much higher correlation value than the others, they might correspond to similar cell types or to an abundant cell type artificially separated in several clusters (i.e. “overclustering”). Thus, we counted for each pair of clustering parameters how many of the 54 isolated neuronal-types match to 1 to 5 clusters (i.e. we tested for the 6 best-correlated clusters for each neuronal type whether the difference in correlation value between a cluster and the subsequent one was higher than 0.05). The results are presented in Extended Data Figure 1d. Increasing the number of PCs decreased this estimate of the biological significance, probably because too many non-relevant principal components dilute the important information, as does increasing too much the resolution parameter. We chose the pair of clustering parameters (120 PCs and 10 resolution) that maximized both our estimation of biological significance and the number of clusters obtained, which resulted in 208 clusters.
For clustering the pupal datasets, we also used 120 PCs but a lower resolution of 6 due to their smaller size. We obtained 147 clusters from the P70 dataset, 137 clusters in P50, 135 clusters in P40, 145 clusters in P30 and 134 clusters in P15. We did not perform additional parameter tuning for either integration or clustering of these datasets, nor did we comprehensively assess the quality of these unsupervised clusters (as we do below for adult), because the pupal datasets were primarily annotated in a supervised manner using log-normalized non-integrated expression matrices.
Statistical significance of the clusters
Because keeping small clusters separate using a modularity optimization algorithm results in artificially splitting large clusters46, before annotation of the adult dataset we merged all pairs of clusters that were improperly separated (Extended Data Fig.1e–f). To do so, we first built a tree grouping the clusters based on the similarity of their average log-normalized non-integrated gene expression, using the Seurat v2 function BuildClusterTree. For each node, we then trained a random-forest model using the Seurat v2 function AssessNodes to predict to which branch of the node a given cell should be assigned. The accuracy of the model is measured by the out of bag error (OOBE), which is similar to the percent of cells misclassified by the model. If a node arbitrarily separates the cells, the random-forest model will not be able to “learn” this split and the OOBE will be high. Thus, we merged all pairs of clusters connected to a node for which the random-forest model prediction differed from the actual clustering with an OOBE > 5%, and if the two clusters did not have clearly differentially expressed genes. To do so, we calculated their cluster markers and decided whether to merge the clusters on a case by case basis based on the number of genes differentially expressed, the p-values, and the identity of these genes. Importantly, most of the merged clusters either matched to the same cell type during the annotation step (see below) and/or were clearly containing a different proportion a low-quality transcriptomes (i.e. the merged clusters differed not by gene expression but by their content of cells with high levels of mitochondrial genes, or low number of UMI/genes expressed). Lastly, although this is unlikely, we cannot entirely discard the possibility that merged clusters correspond to real neuronal subtypes. They are thus still accessible to study in our adult dataset since the metadata field ‘Clustering’ contains the initial unsupervised clusters identities.
Similarly, we used the Seurat v2 function BuildClusterTree on the T4/T5 subtypes (Extended Data Fig.10), and the function AssessNodes to evaluate the statistical significance of the separation between several pairs of cluster throughout this study (Dm3a/b, Tm9v/d, Tm4v/d, cluster 62 Wnt4+/Wnt10- and Wnt4-/Wnt10+ cells) and during the process of annotating the pupal stages (see below). Lastly, we used the Seurat 3.1.0 function BuildClusterTree to produce Extended Data Fig.5a.
Differential gene expression analysis
All differential gene expression analysis of this study was done using the FindAllMarkers function in Seurat 3.0.0.9000 or 3.1.0 with default parameters (two-sided Wilcox rank sum test) to find positive markers, on log-normalized non-integrated gene expression. We then selected the 5, 10 or 20 highest most differentially expressed genes based of their log fold-change (called top5–20 cluster markers), in either all clusters (when not indicated), a subset of them (when indicated) or two subpopulations of cells from the same cluster (when indicated).
Cluster_markers.xlsx (GSE142787) contains the differentially expressed genes for each stage, calculated as explained above using all clusters. It also includes a ‘curated’ version of these genes, in which we kept only the differentially expressed genes with an adjusted p-value below 0.001 and that are markers of 10 clusters or less. These should be the most specific genes, and therefore the best candidates to produce Gal-4 lines targeting specific cell-types.
Binarization of gene expression (Mixture modelling)
To identify clusters based on their expression of marker genes, we did not use transcript levels from the transcriptome as they are not directly indicative of protein expression. Indeed, the transcript level corresponding to protein expression can differ up to 50 fold from one gene to another20. In a given cluster, low UMI levels for a gene can indicate an expression level that is either sufficient or insufficient to produce a functional amount of protein, but also contamination of the cluster by another cell type or presence of this gene in the ambient RNA. We therefore inferred binarized expression matrices for all clusters using a mixture modeling approach20.
Briefly, to decide whether the observed expression level is meaningful, mixture modelling20 assumes that genes exist in two states, either ON or OFF, and compares multiple cell types to model the probability that a given expression level corresponds to the ON state. Mixture modelling discriminates between unimodal genes, that exist only in one state across cell types, and bimodal genes, that exist in both states. Importantly, ON or OFF state of unimodal genes is decided by comparison with the distribution of transcript abundance for ON and OFF states in confidently bimodal genes.
We performed mixture modelling for all our datasets using the code made available in20. As it was developed for replicated bulk-RNA sequencing data, we adapted our scRNA-seq data by separating all of our clusters in two replicates, each of them containing the averaged non-normalized non-integrated gene expression of half the libraries of a given dataset. Moreover, as cluster 192 contains ambient RNA, its expression levels are not representative of genuine cell types and we removed it from each dataset before mixture modelling. Similarly, we removed NBs, GMCs, and clusters of immature neurons in the P15 dataset as they could be mixtures of multiple cell types.
Despite its great usefulness, mixture modelling presents limitations. If a gene is expressed in more than 2 states (for instance low, medium and high expression), the intermediate expression levels could be wrongfully assigned to ON or OFF states. In case of doubts, we therefore advise to plot the expression levels across all clusters and assess them manually. Moreover, for genes not expressed in the optic lobe, a few cells in our dataset might still have a UMI of this gene by chance, and mixture modelling will consider the gene ON in the clusters containing these cells. If this affects a sufficient number of genes in a dataset, it will lower the average expression levels corresponding to the ON state across bimodal genes, and therefore artificially increase the number of unimodal genes considered to be ON (unimodal genes represent 6.4% of the genes in our adult dataset). This could be avoided by removing genes with very low expression levels in all clusters, but we decided against it to avoid using an arbitrary threshold. Thus, we added to our tables two columns, containing the frequency of cells with at least one UMI for a given gene for the clusters where the gene is ON (column ‘freq_cell_ON’) and for the ones where it is OFF (‘freq_cell_OFF’). It should be noted that some bimodal genes have an expression of 0 in all our clusters, and have therefore a value of NA (non-applicable) in the column ‘freq_cell_ON’. Therefore, for bimodal genes, the higher the ‘freq_cell_ON’ value is, the more confident we can be in the inferred ON and OFF states.
Mixture_modeling.xlsx contains mixture modelling for all genes at all stages, and Log_normalized_average_expression.xlsx log-normalized non-integrated average expression of all genes at all stages, for comparison (GSE142787).
Annotation of the adult dataset
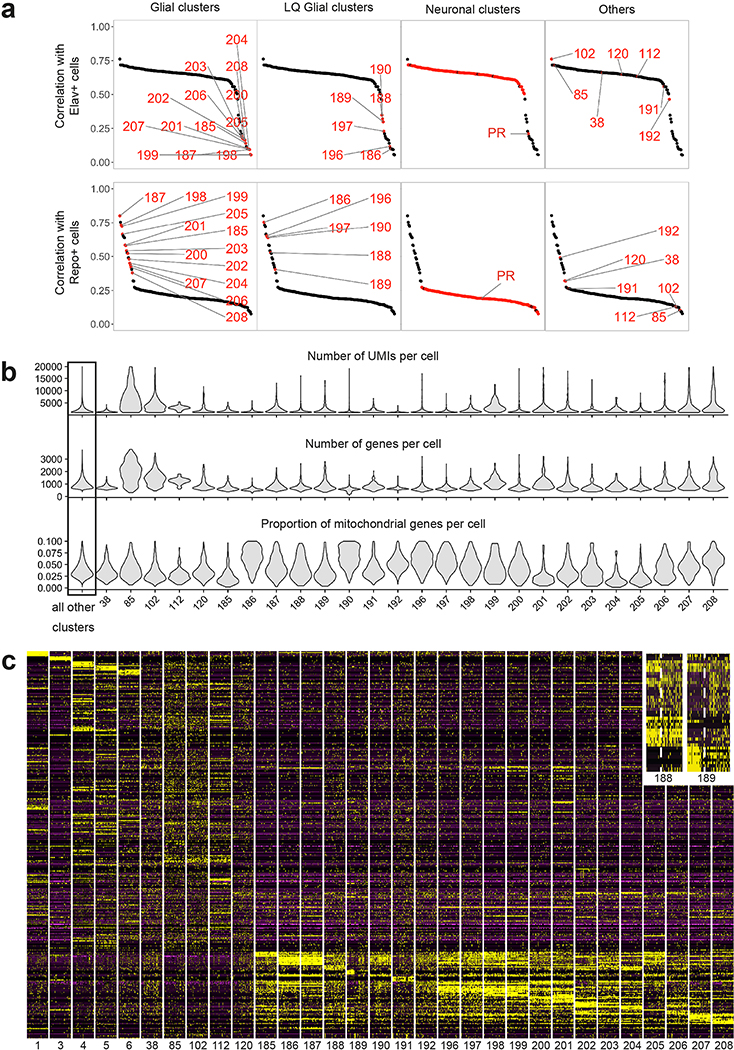
In order to assign the adult clusters to specific cell types, we first aimed at distinguishing neuronal and glial clusters by calculating Pearson correlations between the average gene expression profile of each cluster and transcriptomes sequenced from FACS sorted populations of neurons (elav-Gal4) or glia (repo-Gal4). In addition, we also identified clusters containing low quality transcriptomes using features tending to be higher (proportions of UMI from mitochondrial genes) or lower (number of UMIs or genes per cell) in low quality cells 47,48. Identification of neuronal clusters, glial clusters and low quality transcriptomes is detailed Supplementary Table 1, Extended Data Figs. 2–3. Cluster 192 is made mainly of low quality transcriptomes, which seem to originate largely from “ambient RNA” released by glial cells broken during brain dissociation (Suppl. Table 1). Probably due to this similarity between low quality and glial transcriptomes, 6 of the 19 glial clusters but only 4 of the 172 neuronal clusters (38, 85, 102, 120) were also identified to likely comprise a significant amount of low-quality transcriptomes (Extended Data Fig.2, Suppl. Table 1).
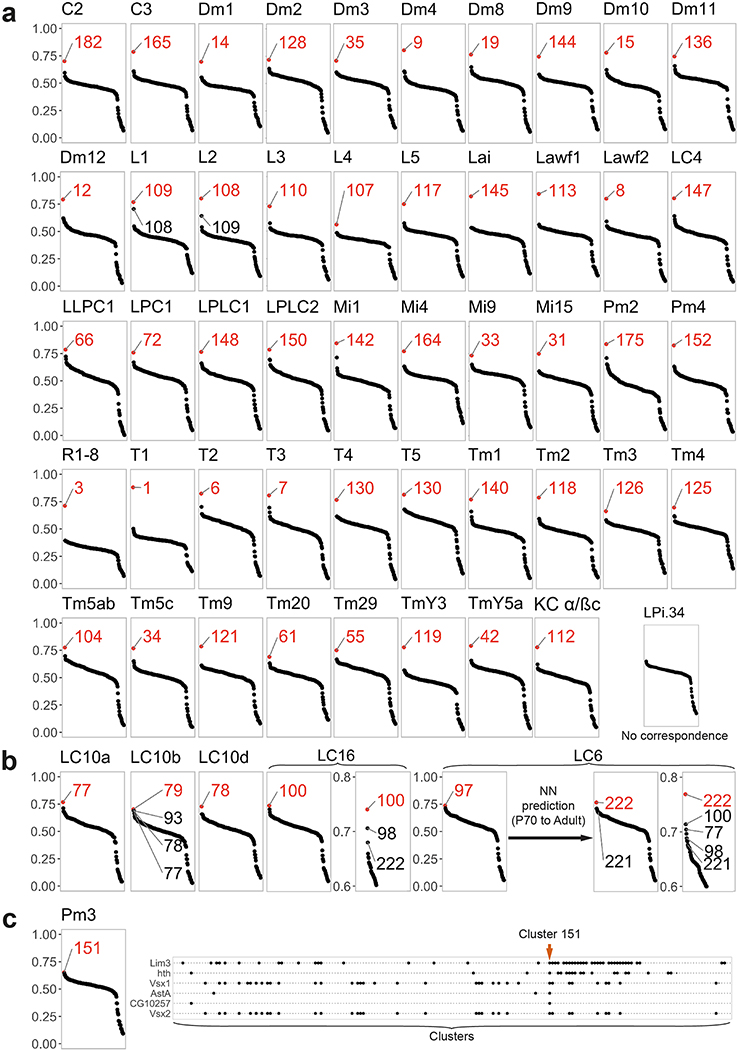
To further annotate neuronal transcriptomes, the Pearson correlations between the average log-normalized non-integrated gene expression of our clusters and log-normalized transcriptomes of isolated neuronal populations was first done using the 610 top10 cluster markers of the adult dataset (i.e. the genes ranked among the top 10 most differentially expressed genes for at least one of our clusters when compared to all other clusters). All neuronal types except Pm3, LC6, LC10b, LC16, and LPi3–4 displayed a clear match to a single cluster as evidenced by an obvious correlation gap (of at least 0.05) between the best and second-best cluster matches (Fig.1c, Extended Data Fig.3). T4 and T5 cells both matched to the largest cluster, a case discussed in detail in main text. The Pm3 transcriptome correlated best, but weakly, with cluster 151 which we verified to be the correct Pm3 cluster based on its expression of previously described markers 13,19 (Extended Data Fig.3). The LC10b transcriptome correlated best with cluster 79, but also well though more weakly with clusters 93, 78 (corresponding to LC10d) and 77 (LC10a). Thus, we assigned LC10b identity to cluster 79, cluster 93 likely being LC10c (for which we did not have a bulk transcriptome). LC6 matched best with cluster 97, which we split into clusters 221 and 222 (see the section “Backprojection of the developmental splits”). Cluster 222 then matched best with LC6, while the next best ranking clusters where other LC cells. LC16 matched best with cluster 100, but with a correlation gap of only 0.03. As LC transcriptomes seem very similar (as illustrated by the examples above), we believe this explains the relatively small correlation gap for LC16: the third best ranking clusters was cluster 222, which we identified as LC6. The only neurons we could not find a match for were LPi3–4 neurons. The resulting annotation was used to annotate the pupal cells. After this, and after backprojection of developmental splits (such as the separation of cluster 97 into 221 and 222, as described below), our final adult dataset contained 587 top10 cluster markers, which we used for the figures of this paper (including Extended Data Figs. 2 and 3). The identification of clusters without comparison to a reference transcriptome was primarily done using mixture modelling of known markers (Extended Data Fig.4), and is detailed in length for each cluster in Supplementary Table 1, Extended Data Fig. 4.
As explained in main text we could manually split Tm9, Tm4 and cluster 62 in a Wnt4+ and a Wnt10+ subpopulations at the adult stage. However, these subtypes do not appear as separate clusters. This is explained by the fact that for a given neuronal type, Wnt4+ and Wnt10+ cells are extremely similar: only a few genes (Wnt4, Wnt10, mamo, frizzled, and a few others out of thousands) show a statistically significant differential expression, which is not enough for unsupervised algorithm to differentiate the cells. However, since some of those genes are biologically significant and clearly differentially expressed (Fig.5, Extended Data Fig.12), the division into Wnt4+ and Wnt10+ subtypes is genuine. Similarly, it is likely that other subtypes too similar to appear as different unsupervised clusters could be discovered in the dataset by supervised methods based on prior knowledge.
An artificial neural network model to classify single cell transcriptomes
We sought to annotate the pupal datasets using our new, expanded adult atlas as a reference. Using supervised learning algorithms trained on one stage to classify another (even assuming identical cellular composition between stages) poses important challenges (generally referred to as domain adaptation) due to developmental changes in gene expression. Any such approach needs to be robust against these changes while still maintaining high accuracy to distinguish cell-types, in addition to being able to handle the sparseness inherent to single-cell datasets. One alternative is to try to minimize these domain differences beforehand, as adopted by Seurat v321, which uses an anchor based approach to find similarity between the datasets to “align” reference and query datasets using Canonical Correlation Analysis before performing a nearest-neighbor based label transfer. Another alternative, as we attempt here, is to train a sufficiently “underfit” model that is versatile enough to maintain its accuracy even if the query dataset structure is slightly different than the reference.
We explored different supervised learning algorithms by attempting to classify our previously published Drop-Seq dataset using simulated single-cells from bulk transcriptomes18. This task represented a significant domain adaptation problem in a dataset that is much sparser than the ones used in this study. As such, most algorithms including support vector machines, random forests and neural networks performed poorly (data not shown). However, we saw dramatic improvements upon incorporating dropout regularization49 in a 2-layer neural network (NN), which randomly removes a subset of nodes at each iteration of training. In particular, using dropout on the input layer in addition to the hidden layer, albeit against convention, appeared to increase the robustness of the network against natural “drop-out” events of single-cell datasets. In contrast, more complicated network architectures (e.g. additional hidden layers) only resulted in overfitting without significant improvements on the validation set accuracy. This suggests that classification of transcriptomic data is an inherently simple task which does not benefit from higher level embeddings of deep neural networks.
Using the Python library Keras, we therefore trained a NN with a single hidden layer to classify our adult dataset, using a randomly selected 90% sample of the dataset while the remaining 10% served as a validation set. As input features, we used log-normalized and mean-centered (non-integrated) expression values of 610 top10 cluster markers. As these marker genes were calculated on the initial adult dataset, before adjustments such as the separation of Tm9 cluster into Tm9v/d, these genes differ very slightly from the 587 top10 cluster markers of our final adult dataset. Further expansion of the feature space (e.g. top 30 markers, 1119 genes) did not result in significant improvements on the validation set accuracy. We used 200 rectified linear units (ReLU) in the hidden layer and a softmax classification layer corresponding to 193 adult clusters. L2 regularization (lambda=0.002) was applied at hidden and softmax layers and dropout regularization was applied at input (p=0.2) and hidden (p=0.5) layers. As the clusters had vast imbalances in the number of cells they contained (Extended Data Fig.5b), we computed “class weights” using the scikit-learn function compute_class_weight to balance the learning rates for clusters of different sizes. We trained the network for 30 epochs using stochastic gradient descent with Nesterov momentum (0.9) with early stopping based on validation loss (categorical cross-entropy) and kept the model with highest validation accuracy saved by the ModelCheckpoint callback. The initial learning rate of 0.05 was halved every 3 epochs as long as the training continued. We finally selected a model that achieved 92.9% accuracy on the training set and 92.3% accuracy on the validation set.
It has traditionally been difficult to calculate confidence measures on NN model classifications, which limited their usage in bioinformatics. However, recent work50 suggested that using dropout (which is normally used only during training) also during classification provides a Bayesian approximation of a probabilistic Gaussian process. We therefore modified this approach to turn on dropout at the classification stage, and calculated 500 independent predictions of classification for each cell in the target dataset using different random subnetworks. We calculated the classification “confidence” as the ratio of these predictions that agreed with the prediction generated by the regular Keras .predict() function which uses the entire trained network.
Supervised annotation of the P70 dataset
We used this NN model trained on the adult dataset to classify the P70 dataset, the pupal timepoint closest to adult. P70 cells were classified in general with high confidence (Extended Data Fig.8a), with the exception of those that corresponded to LQ clusters, some glia and several cases of biologically meaningful exceptions discussed below. Nevertheless, we did not have a direct way of assessing the accuracy of NN predictions without a preexisting gold standard. We therefore opted to comprehensively compare these results to the results of unsupervised clustering. For each “class” (the annotation predicted by the NN, corresponding to the 193 adult clusters), we calculated the percentage of cells from that class that were placed in the top 5 unsupervised clusters containing the most cells of that class. Similarly, for each unsupervised cluster we calculated the percentage of cells belonging to the 5 most numerous classes. Strikingly, in the case of large clusters (i.e. unicolumnar neurons), NN predictions and unsupervised clustering results overlapped near perfectly (Extended Data Fig.6a–b). The remaining clusters were handled in a case-by-case basis and manual adjustments to the NN predictions were applied when necessary using the following guidelines (as detailed in Suppl. Table 1):
In many cases, unsupervised clustering could not resolve multiple classes of highly similar cell types that occur at lower frequencies. This behavior is expected given the lower cell number in the pupal datasets. Our NN model trained on the high-resolution adult dataset could accurately classify and resolve cell types within these clusters (Extended Data Fig.6a–b, ellipses).
If a significant portion (>20%) of a class was placed in a secondary unsupervised cluster, we visually assessed the differential marker gene expression between subgroups using heatmaps. If the division was deemed genuine based on biologically relevant differences, we placed each group into new classes (leaving the original cluster number empty in the dataset). At P70, this was the case for class 188 (G4) which was divided into classes 212 and 213, 208 divided into 209 and 210, and 97 (LC6) divided into 221 and 222. T4–5 neurons existed in 4 different clusters, which were genuine divisions, but as these cells displayed fluctuating resolution across stages, we kept it as a single group (cluster 130) at that point.
If a significant portion of a class was instead distributed across multiple other clusters or was in one of the LQ clusters, we often “cleaned” these classes by keeping only the cells that were in the primary unsupervised cluster. For instance, the photoreceptor cluster in adult (class 3) was not pure, resulting in many LQ cells to be predicted as class 3 in P70, but we only kept the ones that were also in the unsupervised cluster 49, which represented real PRs, and discarded the rest from the dataset. This was performed more aggressively in the case of glia that were often classified with lower confidence, likely due to lower quality of these transcriptomes (Extended Data Fig.2); in almost all cases we only kept the cells that were also in the same primary unsupervised cluster and discarded the rest.
Non-glial clusters that received consistently lower confidence predictions (Extended Data Figs. 8a,9a ellipses) represented cell types with no adult correspondence (as verified experimentally or by known markers) and were assigned new identities. At P70, this was the case for Cluster 35 (TE cells).
After we completed all manual adjustments, we assessed the statistical significance of our divisions using the Seurat v2 function AssessNodes as described for the adult dataset above. We verified that all nodes had less than 15% error (OOBE), further validating the NN predictions. Finally, we calculated the P70 top10 cluster markers (642 genes, 445 of them overlapping with the 610 adult top10 markers) to serve as input features for the next iteration of NN training.
Benchmarking of the NN classifier
Comparison with Seurat label transfer
We aimed to assess how our NN model performs in comparison to established classification methods in the field for single-cell transcriptomes. Seurat v3 performs label transfer by identifying pairwise single-cell correspondences across two datasets (i.e. anchors) which are then used in a weighted vote classifier to predict cell identities; and it has been benchmarked to achieve higher accuracy than other recently developed methods21. We thus used FindTransferAnchors and TransferData functions (with default parameters of v3.1) to classify our P70 dataset by using the adult dataset as reference. We tried using both integrated and non-integrated expression matrices; for dimensional reduction we used either 150 CCs or 120 PCs (the values we optimized for integration and clustering above). The highest overlap (79%) with raw NN predictions (Extended Data Fig.6b, before manual adjustments described above) occurred with the PCA-project method using the integrate gene expression (Extended Data Fig.6c). As these were also the recommended defaults, we further analyzed these results.
Analysis of mismatched classifications between the two methods (Extended Data Fig.6d) revealed that the vast majority of disagreements happened within low-quality cells (center of the tSNEs), glia (green circles) or the clusters with no adult correspondence (blue circles), which were predicted with low confidence by both methods. Within the neuronal populations, Seurat was able to restore some lower quality cells (which were largely classified as LQ by the NN) back to their correct classes (based on their unsupervised clustering); however, it also appeared less robust in differentiating between relatively similar cell types. This was most obvious in the case of lamina neurons L1/L2 (red ellipse) because this division was captured by unsupervised clustering of the P70 dataset. 90% of L2 cells and 88% of L1 cells predicted by the NN were in these correct clusters. Seurat, however, misclassified 45% of the L1 cells (mostly as L2s) despite the fact that these were correctly placed in the L1 unsupervised cluster and correctly classified as L1s by the NN. This was observed, to a lesser extent, in the misclassification of some Dm11 cells as Dm8s as well. Interestingly, Seurat avoided these mistakes when using CCA projections instead of PCA, but this method in turn classified thousands of additional neurons as class 190 (glia/LQ) which again were classified correctly by the NN based on their unsupervised cluster assignments. Overall, these observations suggest that the NN classifier is more robust in resolving highly related cell types in our datasets.
Performance in other datasets
Next, we assessed the utility of our NN model in datasets beyond this study by classifying a scRNA-Seq dataset of 56,902 cells published by another group19. This dataset was generated from whole adult brains and thus included central brain tissue in addition to the optic lobes. Extended Data Figures 6e–f show that our NN model was able to handle this well: optic lobe neurons were classified with high confidence and at a much higher resolution (see below) than the clustering provided by the original study. In contrast, the clusters that were identified by the authors as central-brain neurons (circles) were clearly set aside as low-confidence predictions. The only obvious exception to this was Kenyon cells, which were classified with high confidence as class 112, further suggesting that our adult dataset might have included contaminating Kenyon cells.
Our classifications showed that 8 of the 21 clusters annotated as optic lobe neurons in this study in fact contained multiple cell types, which could now be resolved by our NN. Some of these (T2/T3, C2/C3, L4/L5) were already acknowledged by the authors; but we also observed, for instance, Pm1/Pm2 neurons constituted only 11% of the cluster that was annotated as such. Clusters annotated as TmY14 (11) and Tm9 (18) were also very heterogenous and in fact did not contain these cell types (Our TmY14 and Tm9 clusters mapped to their unannotated clusters 54 and 63, respectively). In addition, 3 optic lobe clusters in that study were homogenous but were misannotated: Tm5ab, Mt1 and Dm8/Tm5c clusters were in fact Tm3, Tm4 and Lai, respectively. Dm8 instead mapped to their unannotated cluster 52, where it had co-clustered with Dm11 and Dm12; Tm5c was in fact part of their cluster 11 mentioned above. Note that several of these annotations were made through mapping from our previous Drop-seq atlas18 and thus likely represent propagation of errors made in that much lower-resolution dataset. Further, we could see that out of the 46 unannotated clusters within their dataset, 19 of them contained at least some optic lobe neurons. Out of these, only 7 were homogenous (>80% of their cluster mapping to a single one of our clusters): T2a, Tm9, Dm9, C2 (the cluster identified as C2/C3 by the authors was mostly just C3) and our unidentified cell types 4, 24 and 26. The remaining 12 were mixtures of different neurons. These observations highlight the dramatic improvements in coverage and resolution our new atlas provides thanks to the increased sequencing depth and cell number.
Multitask extension of the NN classifier
After we completed the annotation of the P70 dataset, we aimed to extend our framework to the earlier pupal stages and annotate all pupal datasets by going backwards through development. While this would have been possible by re-training the NN with the annotated P70 dataset and classifying P50 (and so on) as described above, such a piecewise approach introduces important drawbacks. All supervised learning algorithms (but in particular neural networks) require large training datasets to be most effective. As our training sets became smaller (e.g. only 24k at P40), we would have faced increased overfitting that could reduce the effectiveness of our approach. This reduction in training set sizes would have been especially detrimental for accurately classifying low-frequency cell types. Even at P70 (the largest of the pupal datasets), 100 classes had fewer than 100 cells, and 44 classes had fewer than 50 cells. On the other hand, retraining the network with the aggregated data from all previous stages (e.g. Adult+P70 for classifying P50) would have ignored the higher relevance of the most closely placed dataset, i.e. P50 cells are expected to be much more similar to P70 cells than to adult cells.
We therefore designed a multi-task NN architecture that was able to use all available labeled data for training while still preserving the highest relevance of the immediately following stage (Fig.1f). We achieved this by creating separate models for all annotated stages (i.e. P70 and adult for P50 classification), with identical structures except for the input features (i.e. top 10 markers), which were chosen separately for each stage. The models were trained epoch-by-epoch in an alternating fashion, whereby the weights between ReLU and Softmax layers were copied from one model to another after each epoch (i.e. adult to P70, then P70 to P50, etc.). This setup effectively creates a larger, combined network with multiple input arms but with the same output; whereby the weights between the distinct input layers and the hidden layer are separate and learned independently for each stage but the weights between the hidden and classification layers are learned cooperatively. It started with two input arms (adult and P70) at the first step. After training, the P50 dataset was classified using the P70 model, and thus, with P70 markers as the input features, ensuring that the most proximal dataset was the one used directly but the adult dataset still contributed to the training of this model through information leakage at the second layer. After NN classification, the predictions were comprehensively compared to the unsupervised clustering results as described above for P70. All required manual adjustments were made before calculation of the cluster markers to serve as features to the newest input arm of the NN for the classification of the next stage. Supplementary Table 1 includes notes for each identity where such adjustments were applied. A new model combining P50, P70 and adult annotations was then computed and used to annotate the P40 dataset (and so on) until the P15 dataset. For P15 annotations, the NN model performed poorly on discriminating Lawf1 vs. Lawf2 neurons and also between Tm1/Tm2/Tm4/Cluster 62. We thus trained dedicated multitask networks at this stage, trained only on these cell types, to improve the classification power.
The multitask model provided slight improvements on classifier accuracy in addition to the expected decrease in overfitting. For instance, the multitask model trained on adult, P70, P50 and P40 datasets had achieved 95.9% accuracy on the P40 validation set, compared to the 95.2% accuracy of a model trained on the P40 dataset alone. Benefits are likely greater for classifying across stages due to lowered overfitting but were not possible to quantify without a developmental gold standard.
Backprojection of the developmental splits
In several cases (clusters 88, 97 (LC6), 169, Dm3, Tm9, T4–5, TE), we were able to find multiple clusters for an identity class in the developmental datasets, even though they were present as a single cluster in adults. These were assigned to new, distinct classes after verification and were thus automatically considered in the next iteration of the neural network. After we completed all annotations, in order to preserve ID consistency for downstream analyses, we aimed to transfer neuronal subdivisions to the older datasets where they were not captured by clustering but might still exist. We achieved this by training binary classifiers on these divisions: we isolated only that cell type at the stage where the split was first observed, calculated markers between the subgroups, and trained a dedicated NN (in the same way as the general classifier except with only 60 hidden units and 2 output classes) to classify the subdivision using the top 30 markers as input features. We then isolated the same cell type in the next older stage and classified its cells with this NN. If multiple steps were necessary to reach the adult dataset, we employed a multitask scheme as described above for the general classifier. This was done for classes 97 (LC6), 220 (TE cells, only from P50 to P70), 35 (Dm3), 121 (Tm9), 88 and 169. Note that even though Dm3 and Tm9 NNs were able to divide the corresponding clusters at P70 and adult into subtypes, these divisions were highly unreliable (OOBE P70/Adult: 27%/32% for Dm3 and 23%/22% for Tm9). We also calculated the number of differentially expressed genes (with an adjusted p-value of <0.01) between these subtypes at all stages (Extended Data Fig.10e), note that all groups were subsetted to 150 cells in this analysis for consistency. The rest of the divisions could be reliably carried back to the adult stage. We did not perform this for glial subdivisions.
In addition, we subclustered TE neurons at all pupal stages using 20 PCs and 0.5 resolution to search for any additional subtypes. In all stages, we identified a small number of cells that did not express several markers of TE neurons (Wnt4, Wnt10, Fs, dimm) but expressed Imp, which is not expressed in the rest of the TE cluster. These likely represented rare peptidergic neurons clustering with TE neurons due to their unique properties. Thus, we filtered out the subcluster containing these cells at each stage before generating the UMAP plots of Fig. 2a.
T4-T5 neurons (class 130), that each consist of 4 subtypes, also presented as a single cluster in adults and were subdivided in several unsupervised clusters for all developmental datasets, albeit inconsistently. As detailed in the next section, we attempted to subcluster these at all stages, but all 8 subtypes could only be reliably resolved at P50 while the a/b vs. c/d division was possible at all stages. We therefore trained a binary classifier on this division only and projected it to all stages (classes 234 and 235). The identities in the Seurat objects provided only includes this subdivision; however, we created a metadata field in the P50 object that also contains the full subclustering (classes 261 to 268).
Final datasets and classifier
After we finalized annotations at all stages, we trained a final multitask NN with newly calculated markers of all stages as features of each of 6 input arms. We provide this final model in the Appendix 1 along with all required files and instructions that could be used to classify datasets generated at any stage. Importantly, it is now possible to annotate new datasets using these models without a need to handle the original datasets. In addition, the cells discarded in the annotation process are still present in the Seurat objects we provide (GSE142787). These contain a metadata field ‘FinalIdents’ with the final annotations, in which the discarded cells have the identity ‘0’, a second metadata field ‘Clustering’ with the unsupervised clusters identities, and a third metadata field ‘NNPreds’ with the uncurated NN predictions.
Subclustering of T4-T5 neurons
Even though the subtype divisions of T4 and T5 neurons are well characterized both functionally and developmentally42, the unsupervised divisions within this cluster in the adult dataset were largely based on technical variation (Extended Data Fig.1f) and were merged. At P50, however, all 8 subtypes could be separately observed already on the tSNE (Fig. 4a, Extended Data Fig.7c). We thus isolated them at this stage and performed subclustering (resolution=1) using 8 hand-picked principle components whose top 10 feature loadings (positive or negative) included genes that were known markers of their subtypes51 (klg, bi, lea, Con, TfAP-2, grn, dpr10). We were able to obtain 8 clusters that corresponded to the known subtypes (see Results) and calculated the marker genes between them (Extended Data Fig.10a). As it was also recently reported by others51, we could observe that all T4-T5 subtypes can be defined by three distinct axes of division: T4 vs. T5 (TfAP-2), a/b vs. c/d (bi) and c/b vs. a/d (grn). We then attempted to subcluster all the other stages in a semi-supervised manner using independent component analysis (ICA). At each stage, we identified three ICs defined by the above axes and clustered T4-T5 neurons based exclusively on those. Details on the quality of these divisions are described below, stage by stage, but at no stage other than P50 we could reliably separate all 8 subtypes. P15 and Adult datasets were particularly difficult, in both cases the IC that defines b/c vs. a/d division (grn) was ranked particularly low (16th and 17th, respectively), resulting in very large errors over this axis. We then built cluster trees for all stages and report below the OOBE values calculated for some terminal nodes. Note that 50% OOBE would imply a completely random division and we usually require less than 5% (adult) or 10% (pupae, due to lower size datasets) OOBE to consider a division reliable, and consider any division with >20% OOBE as essentially unusable.
Adult
All terminal nodes corresponding to T4 vs. T5, a vs. b or c vs. d divisions had errors larger than 20%. a/b vs. c/d division had an OOBE of 18%. Note that we could achieve a much lower error (7%) on this division when performed in a supervised manner using the P50 dataset as reference (see previous section). Therefore, in the objects we provide we only include the a/b vs. c/d division made by the neural network.
P70
a/b vs. c/d division was very reliable. However, a vs. b and c vs. d divisions had OOBE of 13% and 18% respectively. Terminal divisions T4c vs. T5c and T4b vs. T5b had OOBEs of 24% and 16%.
P50
All subtypes could be reliably resolved with all nodes having less than 10% OOBE.
P40
Most subtypes could be reliably resolved except for T4c vs. T4d and T5c vs. T5d, with OOBE 13% and 29%, respectively.
P30
a/b vs. c/d division remained very reliable with variable performance in other splits. T4 vs. T5 divisions all had OOBE >10%, largest being T4c vs. T4c at 19%.
P15
a/b vs. c/d division was reliable with OOBE of 5%. a/d vs. c/b and T4 vs. T5 divisions had OOBEs of 10% and 20% respectively.
Stable transcription factor markers in neuronal clusters
Our supervised approach allowed us to assign fully consistent identities to all of our cells at all stages. To determine neuron-specific markers (to eliminate pan-neuronal genes), we isolated 175 neuronal clusters (including the backprojected developmental splits described above, excluding the photoreceptors) and re-calculated the clusters markers at each stage.
We filtered the marker lists for each stage using a list of 629 genes annotated as transcription factors in FlyBase. Next, we determined for each cluster which TFs were consistently significant markers between stages. Since some markers for smaller clusters may be randomly dropped out due to lower sample sizes in pupal stages and at P15 some clusters have not fully acquired their identity, we allowed some flexibility: We took the TFs that were markers for each cluster in the adult dataset and at least four out of the five pupal datasets. The resulting combinations of 113 TFs (Suppl. Table 2) were unique to each cluster, with only the exception of Tm9v/d subtypes. The median number of TFs in each cluster is 8, the minimum per cluster is 2 and maximum is 13 TFs for a cluster.
Trajectory analysis
We used Monocle 338 to determine the sources of age-dependent, continuous heterogeneity (if any) within the Tm3 cluster at P15 and P30. After creating CellDataSet objects with standard preprocessing, we copied UMAP loadings directly from Seurat as we found Seurat integration was better in reducing biologically irrelevant variations than the batch correction methods implemented in Monocle. We then ordered the cells in pseudotime using learn_graph (minimal branch length=10) and order_cells functions, calculated the genes that varied across pseudotime with graph_test function and filtered those by q value (< 0.05). Finally, we clustered these marker genes based on their temporal expression patterns using find_gene_modules function (resolution=0.01).
GO enrichment analysis of the markers
In order to determine the genes that were differentially expressed across different stages, we subsetted all objects (from P15 to adult) to contain only neuronal clusters. These were merged into a single object which was re-scaled and identities for all cells were set as their stage of origin. This object was subsetted to include 15,000 cells from each 6 stages and stage markers were calculated with FindAllMarkers function. The number of significant markers for each stage ranged from 193 in P40 to 746 in adult. We also calculated independently at each stage the cluster markers between neuronal clusters only. We included only the top 20 markers (by logFC) for each cluster in order to have a similar number of genes per stage as the stage markers. 332 of these markers genes (about half the total at each stage) were shared between all pupal and adult stages, and as a result, GO terms largely overlapped as well (Extended Data Fig.11a).
We then performed GO enrichment analysis on both the stage markers and the cluster markers, and calculated enrichment for both Biological Process and Molecular Function terms using The Gene Ontology Resource (http://geneontology.org/). We filtered the terms to include only those with fold enrichment greater than 2. We used REVIGO (http://revigo.irb.hr/) to remove redundant terms and group the related ones52 with a similarity index of 0.5. The TreeMap tables were then exported from REVIGO and inputted to the Python package CirGO53 to create the summary graphs of Extended Data Fig.11. Briefly, CirGO determines the relative size of each slice based on the absolute value of log10(p-value) of each term. To produce the Figure 4 plots, for each stage, the percent enrichment of each summary term as determined by REVIGO (inner rings in Extended Data Fig.11) were manually combined into 8 super-terms for Biological Process and 7 super-terms for Molecular Function, and plotted accordingly.
Connectome Analysis
Processed data for the 7-column medulla connectome4 with cartesian coordinates of all synaptic sites assigned to the cells traced in the original study was downloaded from https://github.com/connectome-neuprint/neuPrint/blob/master/fib25_neo4j_inputs.zip (Retrieved from April 2019 commit). Due to very elongated nature of Dm3 arbors, 2D projection and plotting of these coordinates for each Dm3 cell readily revealed its directionality. We thus plotted the synaptic coordinates of all Dm3 neurons together, each individual cell colored differently, to identify the two subtypes with orthogonal orientations. Cells with parallelly oriented arbors were placed into the same class, called either Dm3x or Dm3y, as we could not assign anatomical directions without examining the EM images.
We then calculated the values of normalized inputs/outputs to/from particular cell types for each Dm3 and compared their averages between the two subtypes, excluding the cells with less than 20 total input or outputs in each case. Normalization was performed separately for inputs and outputs. In each case the number of synapses to/from a specific neuronal identity (pooled total of all cells belonging to that type) was divided by the total number of input/output synapses of that Dm3 neuron. As we did not see clear differences in the average normalized inputs between subtypes (besides the synapses between Dm3s), Figure 4d only plots the average normalized outputs to the postsynaptic cell types where we identified differences.
Extended Data
Extended Data Figure 1: Batch effect removal and biological significance of the adult clusters.
a. The proportions of UMIs from mitochondrial genes per cell (n = number of cells in each library, indicated on the right) and the total number of cells passing filters in each of the 15 libraries comprising the adult dataset. Names indicated correspond to the names in the Seurat object provided (Adult.rds, GSE142787). Boxplots display the first, second and third quartiles. Whiskers extend from the box to the highest or lowest values in the 1.5 inter-quartile range, and outlying datapoints are represented by a dot. b, Origin of the cells in the final adult clusters, colored as in (a). Green arrows: clusters whose unique library distribution can be explained by variable contamination from surrounding tissues (cluster 3 is photoreceptors, 112 is likely Kenyon Cells from the central brain) or the number of lamina neuropils dissociated (clusters 107, 108, 109 are lamina neurons). Red arrows: clusters likely enriched in low quality transcriptomes, as they are enriched in cells from libraries with high number of mitochondrial genes (38, 120, 192) or high number of cells sequenced (102, likely corresponding to multiplets). Brackets: Glial clusters, some of them enriched in libraries with high number of mitochondrial genes as ambient RNA is more similar to RNA from glial vs. neuronal cells (Extended Data Fig.2). c, Number of clusters obtained with different pairs of clustering parameters. Red rectangle: pair of parameters used. d, Left: Legend as in Fig.1C. Right: Number of isolated neuronal type transcriptomes matching to 1–5 of our adult clusters, for each pair of parameters in (c), which we used as a measure of the biological relevance of our clusters. Matching was defined by the presence of a correlation gap above 0.05 (Methods). We took into account any correlation gap between the 6 best correlated clusters, since similar cell types or overclustering can affect the size of the first correlation gap as illustrated on the left graphs. Red rectangle: pair of parameters used. e, tSNE visualization of the adult optic lobe single-cell transcriptomes, using 120 principal components calculated on the log-normalized integrated gene expression. Cell colors indicate the cluster they belonged to before we merged artificially split clusters (red circles, Methods). f, Heatmap showing scaled log-normalized non-integrated expression of the top20 cluster markers between the merged clusters. Merged clusters had almost indistinguishable gene expression patterns, but often differed by their proportions of UMI from mitochondrial genes per cell or the expression levels of the genes highlighted in red, which are enriched in the “ambient RNA cluster” 192 (see also Extended Data Fig.3).
Extended Data Figure 2: Identification of neuronal and glial clusters.
a, Pearson correlation between the average log-normalized non-integrated expression of the top10 cluster markers of the adult dataset clusters (x-axis) and the transcriptome of isolated Repo+ (glial marker) or Elav+ (neuronal marker) populations. LQ = clusters containing a proportion of cells with features of lower quality transcriptomes. b, Violin plots of features tending to be higher (proportions of UMI from mitochondrial genes) or lower (number of UMIs or genes per cell) in low quality cells 47,48. c, Heatmap showing the scaled log-normalized non-integrated expression of the top5 cluster markers of the adult dataset. The first 5 neuronal adult clusters (1 to 6, cluster 1 and 2 having been merged) are plotted for reference as they clearly have specific gene expression patterns. Clusters 38, 85, 102 and 120 present much less defined gene expression patterns and likely contain low quality neuronal transcriptomes (see also Extended Data Fig.1). Clusters 188 and 189 could be further separated in two groups with different gene expression patterns, as illustrated by the dashed line in the insert. Cluster 191 expresses several markers found in no other clusters and likely correspond to neither glia nor optic-lobe neuron. Cluster 192 expresses mainly low levels of glia-specific genes, without specific markers. It likely corresponds to ambient RNA, which would be enriched in RNA from burst glial cells.
Extended Data Figure 3: Identification of the adult neuronal clusters.
a, Pearson correlation between the average log-normalized non-integrated expression of the top10 cluster markers of the adult dataset clusters (x-axis) and the transcriptome of isolated neurons18,20. We represented Dm3, Tm9, T4 and T5 before their split into Dm3a/b, Tm9v/d, T4/T5ab and T4/T5cd. When two transcriptomes were published for a given neuronal type, the one presenting the highest correlation gap is displayed in this figure. R1–8: average gene expression of all photoreceptors20. KC: Kenyon Cells, cluster 112 therefore likely corresponds to contamination from the central brain. b, Legend as in (a). We indicated several matching clusters to highlight the high similarity between LC cells transcriptomes, which explains the lower correlation gaps observed for these neurons. c, Left: Legend as in (a). Right: mixture modelling of Pm3 markers19 (y axis). Clusters are spread on the x-axis, with the probability of expression of the markers figured by the size of the black dots.
Extended Data Figure 4: Marker gene expressions in TmY4, TmY8, TmY14, LC12 and LC17 neurons.
a, Expression pattern of TmY4-LexA (green) in the adult optic lobe (n=7 brains) with anti-NCad immunostaining (white). The LexA line shows weak or no expression in the most anterior medulla region. b-d, Expression pattern of TmY4-LexA (green) in the adult optic lobe with anti-Dichaete (D) (b), anti-Vvl (c) and anti-Toy (d) immunostainings (magenta), n=15 neurons for each. TmY4 cell bodies express Dichaete but not Vvl or Toy. e, Sparse labeling of cell types (anti-FLAG, green) expressing CG42548 in the adult optic lobe using MCFO and immunostained with anti-Brp (white) and anti-Vvl (magenta). A TmY8 neuron (n=4 neurons) is labeled (arborized layers shown by arrows) and expresses Vvl. Adult flies were heat-shocked for 5 minutes and dissected after 6 days. f-h, Expression pattern of R24F10-Gal4 (green) in late L3 optic lobes with anti-NCad (white, f), anti-Dac (magenta, f), anti-Dichaete (magenta, g) and anti-Toy (magenta, h) immunostainings, n=15 neurons for each. R24F10-Gal4 is expressed in TmY14 neurons during larval stage (unpublished data). TmY14 neurons express Toy (h) but not Dac or D (f-g). i, Expression pattern of LC12-Gal4 (green) in the adult optic lobe (n=7 brains) with anti-NCad immunostaining (magenta) showing processes in lobula layers 2–4, and a projection to an optic glomerulus in the posterior ventrolateral protocerebrum (PVLP) of the central brain (white outline). The cell bodies of LC12 are located in the lateral cell body rind, the region separating the optic lobe and central brain. j, Expression pattern of beat-Ic (green) in the adult optic lobe (n=6 brains) with anti-NCad immunostaining (magenta) showing a strong expression profile in the optic glomerulus corresponding to LC12 in the PVLP (white outline). k-m. Expression pattern of LC12-Gal4 (green) in the adult optic lobes with anti-Cut (k, magenta-top panel, grey-bottom panel), anti-Acj6 (l, magenta-top panel, grey-bottom panel), and anti-Toy (m, magenta-top panel, grey-bottom panel) immunostainings, n=77 neurons. All LC12 cells bodies are cut+, Acj6+ and toy+. Red line delineates LC12 cell body location. n, Expression pattern of LC17-Gal4 (green) in the adult optic lobe (n=5 brains) with anti-NCad immunostaining (magenta) showing processes in lobula layers 2–5. LC17 project to the PVLP optic glomerulus (white outline) adjacent to that of LC12 (asterisk, see also i). o, Expression pattern of Kn-Gal4 (green) in the adult optic lobe (n=3 brains) with anti-NCad immunostaining (magenta) showing a strong expression profile in the optic glomerulus corresponding to LC17 in the PVLP (white outline). Note its absence from LC12 neurons (asterisk). p-q, Expression pattern of LC17-Gal4 (green) in the adult optic lobes with anti-Acj6 (p, magenta-top panel, grey-bottom panel) and anti-Toy (q, magenta-top panel, grey-bottom panel) immunostainings, n=62 neurons. All LC17 cells bodies are Acj6+ and toy+. Red line delineates LC17 cell body location (p-q). Me, medulla; Lo, lobula; LoP, lobula plate. Scale bars correspond to 25 μm in a-i, j, n, o and to 15 μm in k-m and p-q.
Extended Data Figure 5: Overview of the final adult clusters.
a, Hierarchical clustering tree of our adult clusters, based on the average log-normalized non-integrated expression of the 2,000 most variable genes of the dataset (the genes used to define the unsupervised clusters, Methods). Indicated in blue are the glial clusters. b, Number of cells in our identified neuronal clusters, excluding the T4/T5 cluster that contains 10,780 cells, with unicolumnar neurons in blue and multicolumnar neurons in orange. Importantly, photoreceptors (PR) and lamina neurons L1-L5 clusters contain fewer cells, as these neuronal types were not included in equal proportions in all libraries. As the number of cells per optic lobe for a given neuronal type is rarely formally counted, unicolumnar vs. multicolumnar character is based both on general knowledge and the following references7,8,12,54.
Extended Data Figure 6: Benchmarking of the neural network (NN) classifier.
a-b, tSNE visualization of the P70 optic lobe single-cell transcriptomes, using 120 principal components calculated on the log-normalized integrated gene expression. Cells colors indicate the clusters they belonged to according to unsupervised clustering (a), or the adult clusters they were classified as by the neural network (b, same as in Fig.1e). Black circles indicate high granularity regions, where less frequent cell types were grouped together by unsupervised clustering but could be resolved accurately by the neural network (b). c, Same as in (a-b) but cells are named and colored by the adult cluster they were classified as by Seurat label transfer (Methods). d, tSNE visualizations (same as c) including only the cells that were assigned inconsistent identities by Seurat and the neural network. Highest rates of inconsistencies were observed in the center (LQ cells), in L1 and L2 clusters (red ellipses), in most glia clusters (green ellipses), the TE neurons and a glia-like cluster (identity 214, Suppl. Table 1) with no adult correspondence (blue ellipses). e-f, tSNE visualizations of 56,902 cells sequenced from whole fly brains19, using 120 principal components calculated on the log-normalized gene expression. e, Cells are named and colored by the clusters they were classified as by our neural network. f, Cells are named by the cluster identities from the original study and colored by the confidence score they received from our neural network. Black circles mark the following central brain clusters (from left to right): Poxn, OPN, clock neurons and dopaminergic neurons, that all received low scores from the neural network. Kenyon cells (red circles) were assigned with high confidence as our adult dataset was contaminated by them (cluster 112).
Extended Data Figure 7: High resolution transcriptomic atlases of the optic lobe across development.
tSNE visualizations of all optic lobe single-cell transcriptomes acquired for this study, using 120 principal components calculated on the log-normalized integrated gene expression. The cells are named and colored consistently at all stages by the neural network classifications with manual adjustments as detailed in Methods. Blue ellipses: Dm3 and Tm9 neuronal subtypes, which could only be resolved at P50 and earlier.
Extended Data Figure 8: Transient Extrinsic neurons.
a-b, tSNE visualization of the P70 optic lobe single-cell transcriptomes, using 120 principal components calculated on the log-normalized integrated gene expression. Cells are named by the unsupervised cluster they were assigned to and colored by (a) the confidence score they received from the neural network (NN) or by (b) the log-normalized non-integrated expression of Fs (green), dimm (blue), and skl (red), which are co-expressed in TE neurons (red ellipses). c, Violin plot of log-normalized non-integrated prt expression in all clusters at P50. TE neuron clusters are indicated by circle. d, R10D10-Gal4 co-expression with anti-Prt staining in a P50 optic lobe (n=15 neurons). Scale bar: 10 μm. e, FLEXAMP memory cassette labeling of R10D10-Gal4 in an adult optic lobe (n=28 brains) with anti-NCad staining. Scale bar: 30 μm. f, R10D10-Gal4 expression pattern in L3 optic lobe (n=15 brains), with anti-NCad, anti-Bsh and anti-Hth staining. Arrow: Bsh+, Hth- neurons labeled by R10D10-Gal4. Scale bar: 30 μm. g-h, R10D10-Gal4 sparse expression at P30 (n=40 neurons), with anti-NCad, anti-Bsh and anti-Hth staining. Scale bars = 5 μm (g) and 15 μm (h). d/pMe: distal/proximal Medulla, Lo: Lobula, Lp: Lobula plate. I, Co-labeling of R10D10-LexA expression and bsh-Gal4 FLEXAMP memory cassette with anti-nCad staining in a P50 optic lobe (n=13 brains). Dashed ellipses: TE neurons. Scale bar: 20 μm.
Extended Data Figure 9: Early differentiation and transcriptomic synchronization of optic lobe neurons and summary of the main findings.
a-b, tSNE visualization of the P15 optic lobe single-cell transcriptomes, using 120 principal components calculated on the log-normalized integrated gene expression. Cells are named by the unsupervised cluster they were assigned to and colored by (a) the confidence score they received from the neural network or by (b) the log-normalized non-integrated expression of dpn (green), ase (blue), and grim (red). Circles match to those of Figure 3a. c, UMAP visualization of the P15 optic lobe single-cell transcriptomes, using 120 principal components calculated on the log-normalized integrated gene expression. Cells are colored by the log-normalized non-integrated expression of nerfin-1 (green), Hey (blue), and vfl (red) d, UMAP visualization of Tm3 and T1 cells (above and below the dashed line, respectively) from all stages sequenced in this study, using 25 principal components calculated on the log-normalized non-integrated gene expression. Cells are colored by their developmental stage. e, Ventral and dorsal Transient Extrinsic (TE) neurons as well as transient photoreceptors (PRs) line the edges of all optic lobe neuropils and express Follistatin (Fs). Moreover, TE and at least 3 other neuronal types express Wnt4 in the ventral medulla/lobula but express Wnt10 in the dorsal part of these neuropils. f, The transcriptome of neurons from the same neuronal type but produced days apart converge towards a similar transcriptomic state, which they reach by P30. Moreover, the inter-neuronal type transcriptomic diversity is highest during P40-P70.
Extended Data Figure 10: Increased transcriptomic diversity during synaptogenesis.
a, Log-normalized and scaled (to the max of 1 for each gene across all time points) expression of top10 (by logFC, calculated at P50) subcluster markers between T4 and T5 subtypes at all stages. TF: Transcription Factor, CSM: Cell Surface Molecule. b-c, GO enrichment analysis of all (269) differentially expressed genes between the T4-T5 subclusters at P50. All Biological Process (b) and Molecular Function (c) terms with greater than 2-fold enrichment were summarized by REVIGO to eliminate redundant terms and group related ones together. Inner rings in the CirGO plots indicate the summarized terms (names in bold). Some individual terms (outer ring) relevant to neuronal function and development are also labeled. Areas within the graphs are determined by the p-values of the terms. d, Average normalized number of presynaptic sites from Dm3 neurons with orthogonally oriented dendritic arbors (Since we could not assign the anatomical directions from the EM data, we plotted them as Dm3x and Dm3y) to indicated neuronal types (Methods). Only the outputs showing differences between the subtypes are plotted. Error bars denote SEM. p-value: 0.004 (Dm3x), 0.006 (Dm3y), 0.04 (Tm28), unpaired parametric two-tailed t-tests (n = 5 and 7 Dm3 cells per type). e, Number of genes differentially expressed between the indicated subtypes with a p-value < 0.01 (two-sided Wilcoxon Rank Sum), calculated after down-sampling the number of cells in each group to 150 for consistency.
Extended Data Figure 11: GO enrichment analysis of the cluster and stage markers.
GO terms were determined from a, the most differentially expressed genes between neuronal clusters (ranked top20 by logFC for at least one cluster) at all stages separately or b, all genes differentially expressed between stages (Methods) in all neurons (aggregated).. All terms with greater than 2-fold enrichment were summarized by REVIGO to eliminate redundant terms and group related ones together. Inner rings (labeled within the graphs or in bold) indicate the summarized terms. Some individual terms (outer ring, not bold), if non-redundant with the summary terms, are also labeled. Areas within the graphs are determined by the p-values of the terms.
Extended Data Figure 12. Dorsal and ventral visual circuits are partitioned by differential Wnt signaling.
a, Tm9-Gal4 sparse labeling in the ventral and dorsal part of the same adult optic lobe (n=4 neurons) with anti-NCad immunostaining (gray). b, Expression pattern of Wnt4-Gal4 (green) with anti-Chaoptin immunostaining (magenta) in the adult optic lobe (n=8 brains). c, Expression pattern of Wnt4-Gal4 (green) with anti-Chaoptin (magenta) and anti-Dve (cyan) immunostainings, in the ventral retina at P30 (n=6 eye discs). Chaoptin marks all photoreceptors, and Dve is expressed in the photoreceptors R1–6 and a subset of R7 (yellow R7), but not in R8 and the rest of R755. d, Expression pattern of Tm4-Gal4 (green) with anti-Aop immunostaining (magenta) at P50 (n=3 brains), showing that almost all Aop+ neurons are co-labelled by Tm4-Gal4. Arrow heads: Aop+ neurons not co-labelled by Tm4-Gal4. e, Wnt4-Gal4 expression pattern with anti-Aop staining (Tm4 marker, see d) at P50 (n=6 brains). Aop+ neurons co-express Wnt4-Gal4 in the ventral (white arrows) but not dorsal (white arrowheads) optic lobe. f, fz2, mamo or CG9896 differential expression between either Tm9v and Tm9d cells, TEv and TEd cells, Wnt4+/Wnt10- and Wnt4-/Wnt10+ Tm4 cells or Wnt4+/Wnt10- and Wnt4-/Wnt10+ cluster 62 cells (log-normalized non-integrated expression). Two-sided Wilcoxon Rank Sum test, p-values are indicated on the figure, ns: not significant. Scale bars = 10 μm (a, c) or 30 μm (b, d, e).
Supplementary Material
Acknowledgements
We would like to thank all members of the Desplan Lab, Shin-ya Takemura and Fred P. Davis for helpful discussions. We thank Liqun Luo, Chris Doe, Filipe Pinto Teixeira, Jan Slabbaert and Sergio Cordoba for critical reading of the manuscript. We further thank Iris Salecker and Robin Hiesinger for reagents. C.D.’s laboratory is supported by NYSTEM (DOH01-C32604GG), the National Eye Institute (R01 EY017916) and by ADHPG-CGSB1 to the CGSB of the NYU Abu Dhabi Research Institute. M.N.O. is a Leon Levy Neuroscience Fellow. F.S. and Y.C.C. are supported by New York University (MacCracken Fellowship). S.J. is supported by the Swedish Research Council (Vetenskapsrådet VR grant 2016- 06726). I.H. is supported by a Human Frontier Science Program postdoctoral fellowship (LT000757/2017). J.M is supported by the National Eye Institute (F32 F32EY028012). N.K. is supported by the National Eye Institute (K99 EY029356-01).
Footnotes
Declaration of Interests
Authors declare no conflicts of interest.
Data Availability
All raw and processed data referenced were uploaded to GEO. Accession number: GSE142789. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142789. Following publicly accessible datasets were also used: GSE103771, GSE103772 and GSE116969.
References
- 1.Lodato S & Arlotta P Generating Neuronal Diversity in the Mammalian Cerebral Cortex. Annual Review of Cell and Developmental Biology 31, 699–720, doi: 10.1146/annurev-cellbio-100814-125353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cembrowski MS & Spruston N Heterogeneity within classical cell types is the rule: lessons from hippocampal pyramidal neurons. Nature Reviews Neuroscience 20, 193–204, doi: 10.1038/s41583-019-0125-5 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Fischbach KF & Dittrich APM The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 258, 441–475, doi: 10.1007/BF00218858 (1989). [DOI] [Google Scholar]
- 4.Takemura S. y. et al. Synaptic circuits and their variations within different columns in the visual system of Drosophila. Proceedings of the National Academy of Sciences 112, 13711–13716, doi: 10.1073/pnas.1509820112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivera-Alba M et al. Wiring economy and volume exclusion determine neuronal placement in the Drosophila brain. Curr Biol 21, 2000–2005, doi: 10.1016/j.cub.2011.10.022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shinomiya K et al. Comparisons between the ON- and OFF-edge motion pathways in the. Elife 8, doi: 10.7554/eLife.40025 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nern A, Pfeiffer BD & Rubin GM Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proceedings of the National Academy of Sciences 112, E2967–E2976, doi: 10.1073/pnas.1506763112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otsuna H & Ito K Systematic analysis of the visual projection neurons of Drosophila melanogaster. I. Lobula-specific pathways. J Comp Neurol 497, 928–958, doi: 10.1002/cne.21015 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Panser K et al. Automatic Segmentation of Drosophila Neural Compartments Using GAL4 Expression Data Reveals Novel Visual Pathways. Curr Biol 26, 1943–1954, doi: 10.1016/j.cub.2016.05.052 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raghu SV & Borst A Candidate Glutamatergic Neurons in the Visual System of Drosophila. PLOS ONE 6, e19472, doi: 10.1371/journal.pone.0019472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varija Raghu S, Reiff DF & Borst A Neurons with cholinergic phenotype in the visual system of Drosophila. J Comp Neurol 519, 162–176, doi: 10.1002/cne.22512 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Wu M et al. Visual projection neurons in the Drosophila lobula link feature detection to distinct behavioral programs. Elife 5, doi: 10.7554/eLife.21022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erclik T et al. Integration of temporal and spatial patterning generates neural diversity. Nature 541, 365–370, doi: 10.1038/nature20794 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ngo KT, Andrade I & Hartenstein V Spatio-temporal pattern of neuronal differentiation in the Drosophila visual system: A user’s guide to the dynamic morphology of the developing optic lobe. Developmental Biology 428, 1–24, doi: 10.1016/j.ydbio.2017.05.008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Özel MN et al. Serial Synapse Formation through Filopodial Competition for Synaptic Seeding Factors. Developmental Cell 50, 447–461.e448, doi: 10.1016/j.devcel.2019.06.014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiesinger PR et al. Activity-Independent Prespecification of Synaptic Partners in the Visual Map of Drosophila. Current Biology 16, 1835–1843, doi: 10.1016/j.cub.2006.07.047 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holguera I & Desplan C Neuronal specification in space and time. Science 362, 176–180, doi: 10.1126/science.aas9435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konstantinides N et al. Phenotypic Convergence: Distinct Transcription Factors Regulate Common Terminal Features. Cell 174, 622–635 e613, doi: 10.1016/j.cell.2018.05.021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davie K et al. A Single-Cell Transcriptome Atlas of the Aging Drosophila Brain. Cell 174, 982–998 e920, doi: 10.1016/j.cell.2018.05.057 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis FP et al. A genetic, genomic, and computational resource for exploring neural circuit function. eLife 9, e50901, doi: 10.7554/eLife.50901 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stuart T et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821, doi: 10.1016/j.cell.2019.05.031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature biotechnology 36, 411–420, doi: 10.1038/nbt.4096 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hewes RS, Gu T, Brewster JA, Qu C & Zhao T Regulation of Secretory Protein Expression in Mature Cells by DIMM, a Basic Helix–Loop–Helix Neuroendocrine Differentiation Factor. The Journal of Neuroscience 26, 7860–7869, doi: 10.1523/jneurosci.1759-06.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertet C et al. Temporal Patterning of Neuroblasts Controls Notch-Mediated Cell Survival through Regulation of Hid or Reaper. Cell 158, 1173–1186, doi: 10.1016/j.cell.2014.07.045 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hay BA, Wolff T & Rubin GM Expression of baculovirus P35 prevents cell death in Drosophila. Development (Cambridge, England) 120, 2121–2129 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Lin HV, Rogulja A & Cadigan KM Wingless eliminates ommatidia from the edge of the developing eye through activation of apoptosis. Development (Cambridge, England) 131, 2409–2418, doi: 10.1242/dev.01104 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Pentek J, Parker L, Wu A & Arora K Follistatin preferentially antagonizes activin rather than BMP signaling in Drosophila. genesis 47, 261–273, doi: 10.1002/dvg.20486 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Ting CY et al. Photoreceptor-derived activin promotes dendritic termination and restricts the receptive fields of first-order interneurons in Drosophila. Neuron 81, 830–846, doi: 10.1016/j.neuron.2013.12.012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasegawa E et al. Concentric zones, cell migration and neuronal circuits in the Drosophila visual center. Development (Cambridge, England) 138, 983–993, doi: 10.1242/dev.058370 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Soriano E & Del Rio JA The cells of cajal-retzius: still a mystery one century after. Neuron 46, 389–394, doi: 10.1016/j.neuron.2005.04.019 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Yu F, Kuo CT & Jan YN Drosophila neuroblast asymmetric cell division: recent advances and implications for stem cell biology. Neuron 51, 13–20, doi: 10.1016/j.neuron.2006.06.016 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Hofbauer A & Campos-Ortega JA Proliferation pattern and early differentiation of the optic lobes in Drosophila melanogaster. Roux’s archives of developmental biology 198, 264–274, doi: 10.1007/bf00377393 (1990). [DOI] [PubMed] [Google Scholar]
- 33.Apitz H & Salecker I A region-specific neurogenesis mode requires migratory progenitors in the Drosophila visual system. Nat Neurosci 18, 46–55, doi: 10.1038/nn.3896 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selleck SB & Steller H The influence of retinal innervation on neurogenesis in the first optic ganglion of Drosophila. Neuron 6, 83–99, doi: 10.1016/0896-6273(91)90124-i (1991). [DOI] [PubMed] [Google Scholar]
- 35.Monastirioti M et al. Drosophila Hey is a target of Notch in asymmetric divisions during embryonic and larval neurogenesis. Development (Cambridge, England) 137, 191–201, doi: 10.1242/dev.043604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J et al. Prevention of medulla neuron dedifferentiation by Nerfin-1 requires inhibition of Notch activity. Development (Cambridge, England) 144, 1510–1517, doi: 10.1242/dev.141341 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Nien C-Y et al. Temporal Coordination of Gene Networks by Zelda in the Early Drosophila Embryo. PLOS Genetics 7, e1002339, doi: 10.1371/journal.pgen.1002339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao J et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502, doi: 10.1038/s41586-019-0969-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langen M et al. The Developmental Rules of Neural Superposition in Drosophila. Cell 162, 120–133, doi: 10.1016/j.cell.2015.05.055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Özel MN, Langen M, Hassan BA & Hiesinger PR Filopodial dynamics and growth cone stabilization in Drosophila visual circuit development. eLife 4, e10721, doi: 10.7554/eLife.10721 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maisak MS et al. A directional tuning map of Drosophila elementary motion detectors. Nature 500, 212–216, doi: 10.1038/nature12320 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Pinto-Teixeira F et al. Development of Concurrent Retinotopic Maps in the Fly Motion Detection Circuit. Cell 173, 485–498.e411, doi: 10.1016/j.cell.2018.02.053 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Apitz H & Salecker I Spatio-temporal relays control layer identity of direction-selective neuron subtypes in Drosophila. Nat Commun 9, 2295, doi: 10.1038/s41467-018-04592-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H et al. Classifying Drosophila Olfactory Projection Neuron Subtypes by Single-Cell RNA Sequencing. Cell 171, 1206–1220.e1222, doi: 10.1016/j.cell.2017.10.019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wernet MF, Perry MW & Desplan C The evolutionary diversity of insect retinal mosaics: common design principles and emerging molecular logic. Trends Genet 31, 316–328, doi: 10.1016/j.tig.2015.04.006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
References (online-only)
- 46.Lancichinetti A & Fortunato S Limits of modularity maximization in community detection. Phys Rev E Stat Nonlin Soft Matter Phys 84, 066122, doi: 10.1103/PhysRevE.84.066122 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Stoeckius M et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol 19, 224, doi: 10.1186/s13059-018-1603-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ilicic T et al. Classification of low quality cells from single-cell RNA-seq data. Genome Biol 17, 29, doi: 10.1186/s13059-016-0888-1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Srivastava N, Hinton G, Krizhevsky A, Sutskever I & Salakhutdinov R Dropout: A Simple Way to Prevent Neural Networks from Overfitting. Vol. 15 (2014). [Google Scholar]
- 50.Yarin G & Zoubin G 1050–1059 (PMLR, 2016). [Google Scholar]
- 51.Kurmangaliyev YZ, Yoo J, LoCascio SA & Zipursky L Modular transcriptional programs separately define axon and dendrite connectivity. eLife 8, e50822, doi: 10.7554/eLife.50822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Supek F, Bošnjak M, Škunca N & Šmuc T REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLOS ONE 6, e21800, doi: 10.1371/journal.pone.0021800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuznetsova I, Lugmayr A, Siira SJ, Rackham O & Filipovska A CirGO: an alternative circular way of visualising gene ontology terms. BMC Bioinformatics 20, 84, doi: 10.1186/s12859-019-2671-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karuppudurai T et al. A hard-wired glutamatergic circuit pools and relays UV signals to mediate spectral preference in Drosophila. Neuron 81, 603–615, doi: 10.1016/j.neuron.2013.12.010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnston RJ Jr. et al. Interlocked feedforward loops control cell-type-specific Rhodopsin expression in the Drosophila eye. Cell 145, 956–968, doi: 10.1016/j.cell.2011.05.003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brunet Avalos C, Maier GL, Bruggmann R & Sprecher SG Single cell transcriptome atlas of the Drosophila larval brain. eLife 8, e50354, doi: 10.7554/eLife.50354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Croset V, Treiber CD & Waddell S Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. eLife 7, e34550, doi: 10.7554/eLife.34550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Allen AM et al. A single-cell transcriptomic atlas of the adult Drosophila ventral nerve cord. eLife 9, e54074, doi: 10.7554/eLife.54074 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Konstantinides N, Degabriel S & Desplan C Neuro-evo-devo in the single cell sequencing era. Curr Opin Syst Biol 11, 32–40, doi: 10.1016/j.coisb.2018.08.001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jenett A et al. A GAL4-Driver Line Resource for Drosophila Neurobiology. Cell reports 2, 991–1001, doi: 10.1016/j.celrep.2012.09.011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Diao F et al. Plug-and-play genetic access to drosophila cell types using exchangeable exon cassettes. Cell reports 10, 1410–1421, doi: 10.1016/j.celrep.2015.01.059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hobert O & Kratsios P Neuronal identity control by terminal selectors in worms, flies, and chordates. Current Opinion in Neurobiology 56, 97–105, doi: 10.1016/j.conb.2018.12.006 (2019). [DOI] [PubMed] [Google Scholar]
- 63.La Manno G et al. RNA velocity of single cells. Nature 560, 494–498, doi: 10.1038/s41586-018-0414-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Togane Y et al. Spatio-temporal pattern of programmed cell death in the developing Drosophila optic lobe. Dev Growth Differ 54, 503–518, doi: 10.1111/j.1440-169X.2012.01340.x (2012). [DOI] [PubMed] [Google Scholar]
- 65.Dearborn R Jr. & Kunes S An axon scaffold induced by retinal axons directs glia to destinations in the Drosophila optic lobe. Development (Cambridge, England) 131, 2291–2303, doi: 10.1242/dev.01111 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Handler AM Ecdysteroid titers during pupal and adult development in Drosophila melanogaster. Developmental Biology 93, 73–82, doi: 10.1016/0012-1606(82)90240-8 (1982). [DOI] [PubMed] [Google Scholar]
- 67.Zhou Y et al. Broad Promotes Neuroepithelial Stem Cell Differentiation in the Drosophila Optic Lobe. Genetics 213, 941–951, doi: 10.1534/genetics.119.302421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hara Y et al. Ecdysone-dependent and ecdysone-independent programmed cell death in the developing optic lobe of Drosophila. Dev Biol 374, 127–141, doi: 10.1016/j.ydbio.2012.11.002 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Riddiford LM, Truman JW & Nern A Juvenile hormone reveals mosaic developmental programs in the metamorphosing optic lobe of Drosophila melanogaster. Biology Open 7, bio034025, doi: 10.1242/bio.034025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uyehara CM & McKay DJ Direct and widespread role for the nuclear receptor EcR in mediating the response to ecdysone in Drosophila. Proceedings of the National Academy of Sciences 116, 9893–9902, doi: 10.1073/pnas.1900343116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stoiber M, Celniker S, Cherbas L, Brown B & Cherbas P Diverse Hormone Response Networks in 41 Independent Drosophila Cell Lines. G3 (Bethesda) 6, 683–694, doi: 10.1534/g3.115.023366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takemura SY et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature 500, 175–181, doi: 10.1038/nature12450 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wernet Mathias F. et al. Genetic Dissection Reveals Two Separate Retinal Substrates for Polarization Vision in Drosophila. Current Biology 22, 12–20, doi: 10.1016/j.cub.2011.11.028 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wolf R, Gebhardt B, Gademann R & Heisenberg M Polarization sensitivity of course control inDrosophila melanogaster. Journal of comparative physiology 139, 177–191 (1980). [Google Scholar]
- 75.Sancer G et al. Modality-Specific Circuits for Skylight Orientation in the Fly Visual System. Current Biology 29, 2812–2825.e2814, doi: 10.1016/j.cub.2019.07.020 (2019). [DOI] [PubMed] [Google Scholar]
- 76.Mazzoni EO et al. Iroquois complex genes induce co-expression of rhodopsins in Drosophila. PLoS Biol 6, e97, doi: 10.1371/journal.pbio.0060097 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Courgeon M & Desplan C Coordination between stochastic and deterministic specification in the Drosophila visual system. Science, eaay6727, doi: 10.1126/science.aay6727 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sato M, Umetsu D, Murakami S, Yasugi T & Tabata T DWnt4 regulates the dorsoventral specificity of retinal projections in the Drosophila melanogaster visual system. Nat Neurosci 9, 67–75, doi: 10.1038/nn1604 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Dacks AM, Green DS, Root CM, Nighorn AJ & Wang JW Serotonin modulates olfactory processing in the antennal lobe of Drosophila. Journal of neurogenetics 23, 366–377, doi: 10.3109/01677060903085722 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.