Abstract
Background
Panax ginseng, as one of the most widely used herbal medicines worldwide, has been studied comprehensively in terms of the chemical components and pharmacology. The proteins from ginseng are also of great importance for both nutrition value and the mechanism of secondary metabolites. However, the proteomic studies are less reported in the absence of the genome information. With the completion of ginseng genome sequencing, the proteome profiling has become available for the functional study of ginseng protein components.
Methods
We optimized the protein extraction process systematically by using SDS-PAGE and one-dimensional liquid chromatography mass spectrometry. The extracted proteins were then analyzed by two-dimensional chromatography separation and cutting-edge mass spectrometry technique.
Results
A total of 2,732 and 3,608 proteins were identified from ginseng root and cauline leaf, respectively, which was the largest data set reported so far. Only around 50% protein overlapped between the cauline leaf and root tissue parts because of the function assignment for plant growing. Further gene ontology and KEGG pathway revealed the distinguish difference between ginseng root and leaf, which accounts for the photosynthesis and metabolic process. With in-deep analysis of functional proteins related to ginsenoside synthesis, we interestingly found the cytochrome P450 and UDP-glycosyltransferase expression extensively in cauline leaf but not in the root, indicating that the post glucoside synthesis of ginsenosides might be carried out when growing and then transported to the root at withering.
Conclusion
The systematically proteome analysis of Panax ginseng will provide us comprehensive understanding of ginsenoside synthesis and guidance for artificial cultivation.
Keywords: Cytochromes P450, Mass spectrometry, Panax ginseng, Proteomics, UDP-glycosyltransferase
1. Introduction
Ginseng is one of the oldest and most important medicinal plants in the Orient. It is a perennial herb of the family Araliaceae, which has the reputation of "king of hundred herbs." Ginseng has remarkable and powerful medicinal effects, which can relieve fatigue, regulate blood pressure and endocrine, as well as protect the heart. It can also be used to restore vitality, regulate the body and prolong life for the weak patients [[1], [2], [3], [4], [5]]. Ginseng usually takes 4–6 yrs to mature, and the requirements for the soil, temperature, precipitation, and illumination of the growing environment are high, which further reflected their preciousness and rarity. In recent years, unreasonable large-scale excavation and planting had caused serious damage to ginseng resources and environmental resources. Therefore, scientific cultivation of ginseng and research on its active ingredients brook no delay. The root is the officinal essence and the most important part of ginseng. After extensive analysis, ginsenosides were considered to be the main pharmacological ingredients of ginseng to reflect its biological activity [6,7].
In recent years, more and more research groups have put the focus of ginseng research on the level of proteomics, and the related research methods and the establishment of protein databases have been gradually improved. The proteomes of different varieties of ginseng have been analyzed comparatively, the identification of stress-responsive proteins and the researches on proteins related to ginsenoside biosynthesis have been also carried out [[8], [9], [10]]. Although there are numbers of reports on the protein component of ginseng, the systematical study of ginseng proteome has been limited because of the uncompleted genome resources [11]. Recently, the Chen and Yang teams have released the genome sequences individually, which greatly push the understanding of the biosynthetic pathway of ginsenosides [12,13]. The completion of whole genome sequencing provided the possibility for proteomic analysis. Protein is the direct executor of life activities and the ultimate embodiment of complexity. In-depth studies on physiological and metabolic pathways of ginseng are inseparable from protein-level analysis. Proteomics can be used to scientifically isolate and identify the proteins in different parts of ginseng tissues, so as to study the role of abundant proteins of ginseng in metabolic activities according to their different functions, explore the markers and related metabolic pathways involved in ginsenoside biosynthesis at the protein level, to provide new thought for ginseng cultivation and ginsenoside biosynthesis.
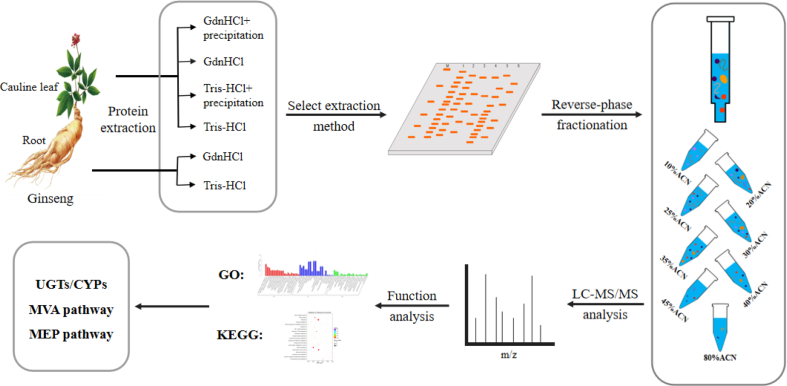
In this study, proteomic methods were used to identify and analyze the proteins in ginseng cauline leaf and root as shown in the workflow (Fig. 1). Firstly, the extraction methods of protein for different tissue part were optimized by different extraction buffers to increase the extraction efficiency and to eliminate the non-protein contaminants [14]. Secondly, protein samples were digested and fractionated by high-pH reverse chromatography to reduce the complexity [[15], [16], [17], [18]]. Thirdly, high-resolution mass spectrometry (MS) and latest database were used for protein identification. Finally, the biological functions and metabolic pathways of proteins in cauline leaf and root were classified and compared. We have preliminarily selected UDP-glycosyltransferase (UGT), cytochromes P450 (CYP), and related proteins involved in ginsenoside biosynthesis to better understand the related biological functions of cauline leaf and root in the process of ginsenoside biosynthesis.
Fig. 1.
Workflow illustration for ginseng protein extraction and analysis. UGTs, UDP-glycosyltransferase; CYPs, cytochromes P450; MVA, mevalonate; MEP, methyl-d-erythritol-4-phosphorylation; LC-MS/MS, liquid chromatography–tandem mass spectrometry.
2. Materials and methods
2.1. Protein extraction
The 5-yr-old ginseng samples were from ginseng culture base (Tonghua, Jilin Province, China) of Jilin Zixin Pharmaceutical Industrial Co., Ltd. The root and cauline leaf from 30 strains of ginseng were pooled together for the following experiments. Three biological replicates were used for each extracted protein condition. We designed and compared different extraction methods for ginseng cauline leaf and root samples as described in Table S1. The extract was treated with a sonicator at room temperature for 60 min, stirred at 4°C for 15 h, and centrifuged to remove the precipitation. For methanol--chloroform precipitation, the crude protein extract was sequentially mixed with four volumes of methanol, four volumes of chloroform, and three volumes of ddH2O. The suspension was centrifuged at 16,000g for 3 min. The precipitate was washed with four volumes of methanol and then reconstituted with distilled water to obtain a total protein extract. For acetone precipitation, the crude ginseng protein extract was added to the precooled acetone at a volume ratio of 1:5, left at −20°C overnight, and centrifuged at 5,000r/min for 30 min. After discarding the supernatant and evaporating the remaining acetone, it was reconstituted with distilled water to obtain a total protein extract. Each protein sample was loaded at the equivalent amounts and separated on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel for comparison.
2.2. Protein digestion and fractionation
In solution, digestion was performed as following: 1 mg of above ginseng protein samples were added, 200 μL denaturant (100mM NH4HCO3, 8M urea) and 10 μL DTT (1 mg/mL) shaking 2 h at 37°C to disrupt the disulfide bonds. Then, 1.4 mg iodoacetamide (IAA) were added in samples for 40 min at room temperature in the absence of light for alkylation, followed by addition of seven volumes of 100mM NH4HCO3 to dilute the denaturant. Then 20 μg of trypsin was added to at 1:50 (w/w) enzyme-to-protein ratio to digest the protein samples at 37°C overnight. The digested peptides were desalted using Sep-Pak C18 column and dried by speed vacuum for analysis.
For two-dimensional separation, the digested samples were loaded into the equilibrated Sep-Pak C18 column, and eight fractions were subsequently eluted with 200 μL 0.05% triethanolamine (TEA) buffer solution containing 10%, 20%, 25%, 30%, 35%, 40%, 45%, and 80% acetonitrile (ACN), respectively. Each fractionated sample was dried completely using speed vacuum and stored at −80°C until analysis.
2.3. LC-MS/MS analysis
The dried peptides were resolubilized in 0.1% (v/v) formic acid with 2% (v/v) acetonitrile and injected into nano-liquid chromatography system (UltiMate 3000 RSLCnano System; Thermo Scientific, Waltham, MA, US). Samples were separated on a 45-cm in-house packed column containing C18 resin (2.2 μm) by a constant flow rate of 0.35 μL/min with column oven temperature of 60oC. The mobile phase buffer consisted of 0.1% formic acid in ultrapure water (buffer A) with an eluting buffer of 0.1% formic acid in 80% (v/v) acetonitrile (buffer B). The separating gradient ramped linearly from 1% buffer B to 34% buffer B for 63 min, then 34% to 47% for 10 min, followed by 47% to 90% for 5 min, and finally 90% buffer B for 12 min. The UltiMate 3000 RSLCnano was coupled online with a hybrid higher solution Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Fisher Scientific). Full scan mass spectra were acquired in the Orbitrap cell in the mass range 300–1,500 m/z (positive polarity, with the resolution of 240,000, automatic gain control (AGC) = 4 × 105), followed by MS/MS of the 10 most intense ions.
2.4. Data processing
The MS data were searched against ginseng database [13] (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA385956/) using the Proteome Discoverer 2.2 software (Thermo Fisher Scientific). Enzyme was set with trypsin with three missed cleavages. Precursor mass tolerance was set at 10 ppm, and fragment mass tolerance was set at 0.6 Da. Search criteria included a oxidation (+15.9949 Da) of methionine residues in dynamic modifications, carbamidomethylation of cysteine (+57.0214 Da) in static modifications, and acetylation (+42.011 Da) at N-terminus of proteins in dynamic modifications (protein terminus). The false discovery rate was set at 0.01.
3. Results and discussion
3.1. Comparative analysis of the ginseng root and cauline leaf proteome
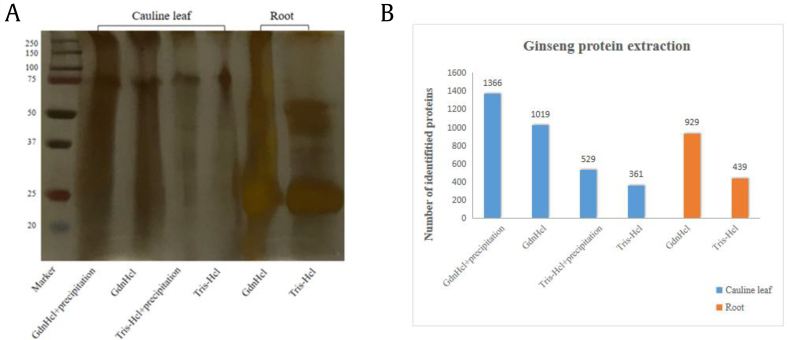
Several extraction approaches were compared to get the systematical analysis of different tissue proteome components. Optimizing the preparation of ginseng samples is critical to reducing impurities, increasing detection sensitivity, and expanding ginseng proteome coverage. For example, cell wall rupture effects and cell membrane lysate selection directly affect the efficiency of plant protein extraction, and the method of removing interfering substances directly affects the extent of protein purification. GdnHCl lysis buffer is reported to be an effective reagent for decomposing cell walls and extracting proteins from cells, and provides optimal yield for protein identification, because GdnHCl can eliminate interference from nucleic acids, phospholipids, light and pigments, and secondary metabolites, allowing good separation of proteins from interfering substances, thereby increasing the number of identified proteins [14]. To explore the efficiency of ginseng root and cauline leaf lysis strategies, we tested two commonly used lysis buffers GdnHCl and tris--HCl as shown in Table S1. Compared with root, cauline leaf protein detection is greatly disturbed by factors such as chlorophyll and secondary metabolites. Therefore, methanol–chloroform and acetone precipitation steps were further performed to purify the protein components. SDS-PAGE analysis on the protein extracts showed that the extraction efficiency of GdnHCl is significantly higher than that of tris--HCl (Fig. 2A). And for the cauline leaf, the combination of GdnHCl with methanol--chloroform precipitation got the most comprehensive extraction efficiency.
Fig. 2.
Proteomic analysis results of ginseng cauline leaf and root protein samples prepared using different extraction methods. (A) SDS-PAGE separation results. (B) Number of proteins identified by MS.
Each extract was then analyzed by nano--liquid chromatography tandem MS for protein identification. The results were consistent with the SDS-PAGE results as shown in Fig. 2B. For cauline leaf sample, 1,366 and 1,019 proteins were identified with GdnHCl extraction followed by methanol--chloroform precipitation, whereas only 529 and 361 proteins were identified from tris--HCl protocol with or without precipitation, respectively. For the extraction of ginseng root protein, 929 proteins were identified from GdnHCl protocol, which is over twice of that of tris–HCl lysis method (439 proteins). Although the cauline leaf has much more small molecules such as chlorophyll, the total protein identification is significantly higher than root, which is the main ingredient for herbal medicines. The SDS-PAGE results also showed very clearly that the cauline leaf proteins existed in the whole range of molecular weight, whereas the root proteins dominated around the 25 kDa. Therefore, we summarized the proteomic results of different region of ginseng to find out the highest abundant protein expressions.
The top 20 high-abundance proteins in MS identification results were listed in (Tables S2 and S3). To our surprise that, five of the top 20 proteins from root are ribonuclease-like storage proteins, which exactly corresponded to the 25 kDa band for SDS-PAGE analysis. It was reported that the ginseng root 28 kDa protein was composed of four abundant proteins with molecular weights of 28, 26, 21, and 20 KDa, also known as ginseng major proteins. Ginseng major proteins are also ribonuclease-like storage proteins whose main biological function is to store nutrients for the main root [19]. The root of ginseng stores energy to meet the needs of the fast-growing season (1–3 yrs), and the slow growth period (>5 yrs) of secondary metabolite synthesis and stress resistance because of harsh environments consumes a lot of energy. The rest of the high-abundance proteins in ginseng root are mainly involved in energy metabolism and stress response. In the related process of energy metabolism, mainly included glycolysis (enolase, fructose-diphosphate aldolase 6, glyceraldehyde-3-phosphate dehydrogenase), carbohydrate chemical reaction (β-amylase, α-1,4-glucan phosphorylase L-1 isozyme, 1,4-α-glucan branching enzyme), amino acid metabolism (adenosine homocysteine, 5-methyltetrahydropropionylglutamate-homocysteine methyltransferase), proton transport (ATP synthase subunit β), and so on. The heat shock homologous 70 kDa protein is a cytoprotective protein and these types of protein mainly act on the stress response of ginseng [20].
For the high-abundance proteins in cauline leaf, identification results showed that four of the top 20 proteins are ATP synthase subunits, which generated an electrochemical gradient through the transport of protons across a membrane that powers ATP synthesis. Another four proteins are subunits of glyceraldehyde-3-phosphate dehydrogenase, which mainly participates in the chemical reactions and pathways involving glucose with the concomitant removal or addition of electrons to or from substance [21]. Three of them are ribulose bisphosphate carboxylase family, which is the product controlled by both nuclear and chloroplast genes with main function for carbon dioxide fixation [22]. Others are catalase and heat shock cognate 70 kDa protein for stress response [23] and ribonuclease-like storage protein to store nutrients. Most high-abundance proteins play an important role in energy metabolism-related processes. To systematically analyze the protein functions, we further performed two-dimensional chromatography separation for comprehensive identification of the protein components.
3.2. In-depth analysis of the ginseng proteome
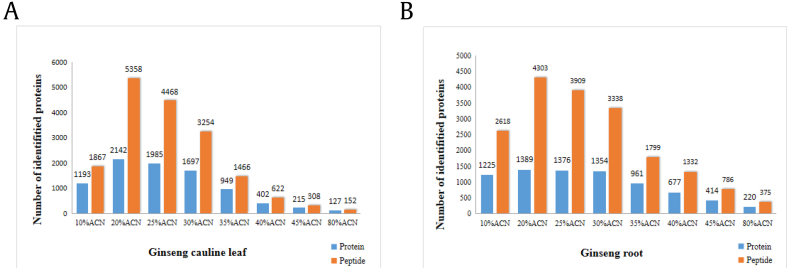
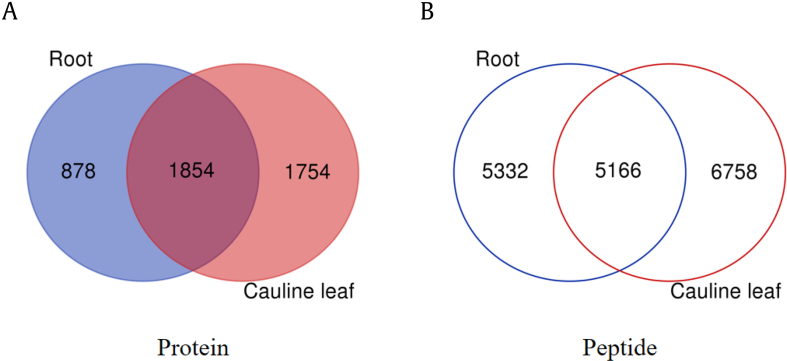
Currently, there are several approaches to improve the protein identification, such as prefraction on protein stage using SDS-PAGE or peptide level separation by different type of chromatography [15]. Recently, high-pH reversed-phase based fractionation method was explored as the first dimensional separation because of the high orthogonality with normally used low pH liquid chromatography, which is more comparable with the ionization process of MS [[16], [17], [18]]. The protein samples of root and cauline leaf were fractionated by high-pH C18 cartridge into eight fractions, and then each fraction was submitted for MS analysis. The number of protein and peptide identified in each fraction was shown in Fig. 3. It was found that the elution trend of protein in the samples of cauline leaf and root were basically consistent: most proteins were identified with the gradient from 20% to 30% ACN. After combination of all the proteins identified from eight fractions, totally 12, 552 peptides corresponding to 3,608 proteins and 11, 290 peptides corresponding to 2,732 proteins were identified in the cauline leaf and root protein samples, respectively. However, only around half peptides (5,166) overlapped between ginseng root and cauline leaf as shown in the Venn diagram of Fig. 4, which indicated significant difference between the tissue parts of ginseng. To explore the function difference between root and cauline leaf, gene ontology (GO) annotation and KEGG metabolic pathway analyses were carried out on the identified proteins.
Fig. 3.
The numbers of protein and peptide identified from each fraction by two-dimensional LC-MS/MS proteomic analysis of (A) cauline leaf and (B) root of ginseng samples.
Fig. 4.
The Venn diagram of (A) protein and (B) peptide identified from ginseng cauline leaf and root.
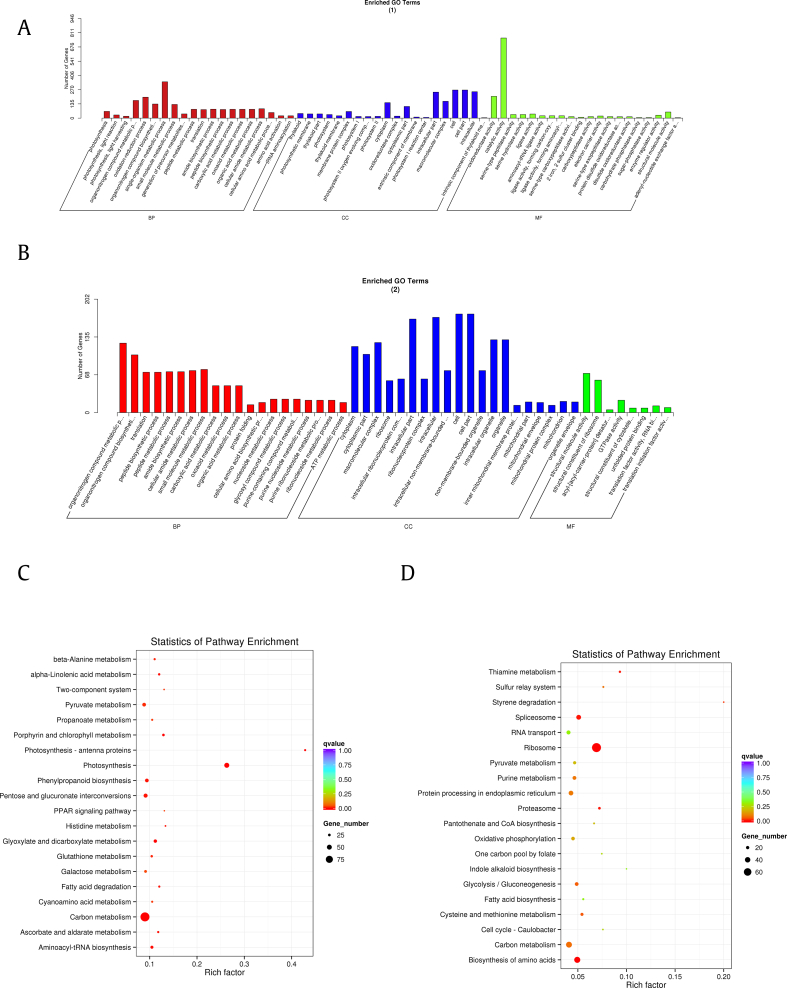
The KEGG enrichment and GO analysis of biological process (BP), cellular components, and molecular function were shown in Fig. 5. In the function analysis results of overlapped proteins between root and cauline leaf, we found that the GO results were mainly covered 20 BPs, which were basically involved in the biosynthesis and metabolism of organic compounds such as nucleosides, peptides, proteins, amides, and oxo acids, in which the highest protein abundance was the single-organism metabolic process, followed by organonitrogen compound and small molecule metabolic process. The molecular functions of common proteins mainly included structural molecule activity, cofactor and RNA binding, and as an enzyme to catalyze various enzymatic reactions such as oxidation--reduction reaction. These proteins are mainly present as cells, intracellular and cytoplasm parts, and include many macromolecular complexes (Fig. S1A). KEGG results showed that the pathway with the most common protein participation was ribosome and carbon metabolism, followed by biosynthesis and metabolism of amino acids, which is consistent with GO analysis results (Fig. S1B). These consensus proteins maintain the basic biological functions and metabolic activities of ginseng, provide the necessary substances for each tissue and organ to allow the functionalized reactions to proceed smoothly.
Fig. 5.
GO annotation of differentially expressed proteins in (A) cauline leaf and (B) root. KEGG enrichment of differentially expressed proteins in (C) cauline leaf and (D) root. GO, gene ontology.
Based on the GO function annotation and KEGG metabolic pathway analysis of specific proteins in cauline leaf and root (Fig. 5), we found that these specific proteins in different parts were involved in some of the same basic BPs to maintain the normal state of cells and tissues, such as biosynthesis and metabolism of organonitrogen compounds, metabolic processes of small molecules, organic acids, peptides and cellular amino acids and the biosynthetic process of peptides and amides. In addition, we found that the proteins unique to cauline leaf mainly specifically possess the BPs related to photosynthesis, such as the photoreaction phase and the light capture process. Quite a few cellular components in the annotation also belong to photosynthesis, like thylakoid and light reaction center. For example, the ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) was present in the chloroplast matrix, which is not only an important carboxylase in CO2 fixation, but also an oxygenase that catalyzes the reaction with O2 in photorespiration [22]. In terms of biological function, these proteins were mainly used as an enzyme to catalyze various redox reactions to maintain normal metabolism of the cauline leaf part and normal photosynthesis (Fig. 5A). The specific proteins in the root mainly involved in various nucleoside and ATP metabolic processes, and acted on the modification processes such as protein folding, mainly existed as the structural molecule in cells, and the cell components were also dominated by cell, intracellular, organelle, and cytoplasm part (Fig. 5B).
For the KEGG enrichment analysis of ginseng cauline leaf (Fig. 5C), we found that the most involved pathway was carbon metabolism, next were pentose and glucuronate interconversions and pyruvate metabolism, followed by the biosynthesis of aminoacyl−tRNA and phenylpropanoid, metabolism of galactose, glutathione, glyoxylate, and dicarboxylate, etc. Moreover, the pathways to photosynthesis and metabolism of chlorophyll got the highest enrichment as expected. Therefore, we supposed that the most important metabolic pathway of specific proteins in cauline leaf was the metabolism of various organic substances and the transformation and fixation of energy around photosynthesis. In the KEGG pathway analysis of root specific proteins (Fig. 5D), the most abundant of protein participation is ribosome, which mainly store nutrients and corresponded to the high abundant ribonuclease-like storage protein in root. The second one is carbon metabolism, followed by spliceosome, biosynthesis of amino acids and protein processing in endoplasmic reticulum, which are indispensable metabolic processes in the biosynthesis of peptides and proteins. In addition, root proteins also act on pathways such as proteasome, oxidative phosphorylation, glycolysis/gluconeogenesis, RNA transport, thiamine metabolism, and biosynthesis of some active substances, etc. These KEGG enrichment results reflected some of the major metabolic processes in the different parts of ginseng. Overall, the specific proteins in cauline leaf mainly acted on photosynthesis and related energy conversion, whereas the root had more kinds of proteins, had functions on the biosynthesis and modification of biomacromolecules, and provided energy for these processes in addition. These results were consistent with the functional analysis of high-abundance proteins in one-dimensional analysis.
3.3. Analysis of related proteins and pathways of ginsenoside biosynthesis
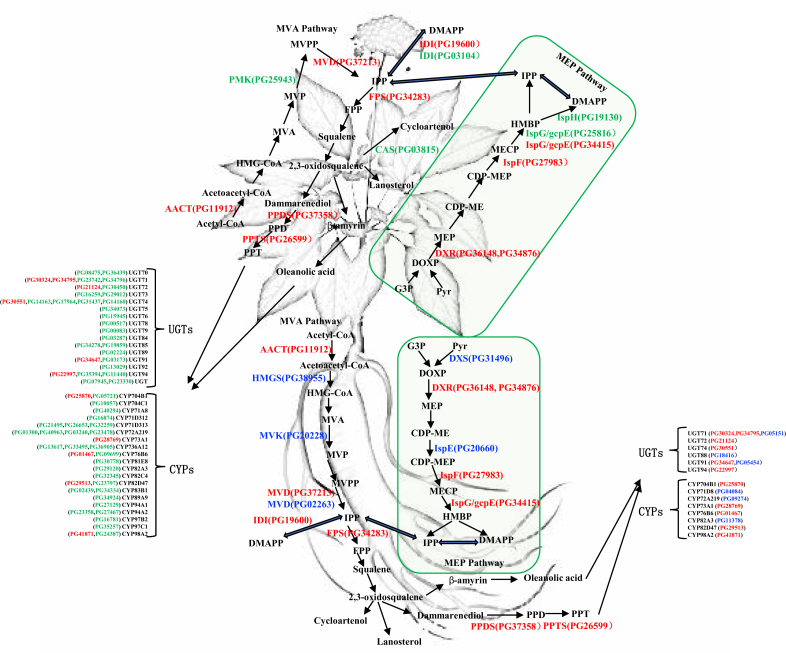
Ginsenosides are considered as the main bioactive components for Panax ginseng and the quality is highly related with the types and content of different ginsenosides. Therefore, we further screened proteins related to ginsenoside biosynthesis, including the well-known mevalonate (MVA) and 2C-methyl-d-erythritol-4-phosphorylation (MEP) pathway proteins, cytochromes P450 and UGT enzymes. Cytochrome P450 is a complex family of proteins that have powerful functions. As terminal oxygenases, they can insert an oxygen atom into hydrophobic molecules to change their hydrophilicity and activity. CYPs participate in a variety of secondary metabolic reactions in plants and play an important biological role [24]. During the biosynthesis of ginsenosides, they mainly carried out the catalysis of the hydroxylation process [25]. Glycosyltransferase family can catalyze the glycosylation of receptor molecules, thereby altering their biological activity, solubility, and stability in vivo, thus play an important role in cell biological regulation and metabolite synthesis and storage [26]. UGTs mainly use UDP-glucose and UDP-glucuronic acid as donors to glycosylate receptor molecules and participate in important glycosylation steps in the biosynthesis of ginsenosides [27,28]. These complex biomolecular modification reactions enable the generation of diverse and complex ginsenoside molecules via MVA and MEP pathways. As listed in Tables S4 and S5, a total of 32 proteins related to ginsenoside synthesis were identified in ginseng root, including six enzymes in the MVA pathway, six enzymes in the MEP pathway and one FPS, one PPDS, one PPTS, eight CYPs, and nine UGTs in the downstream pathway. A total of 78 proteins related to ginsenoside synthesis were identified in the ginseng cauline leaf, including five enzymes in the MVA pathway, six enzymes in the MEP pathway and one FPS, one PPDS, one PPTS, 33 CYPs, and 31 UGTs in the downstream pathway. AACT (PG11912), MVD (PG37213), and IDI (PG19600) were co-identified in root and cauline leaf in the MVA pathway. HMGS (PG38955), MVK (PG20228), and MVD (PG02263) were only identified from root. PMK (PG25943) and IDI (PG03104) were only identified in cauline leaf. DXR (PG36148, PG34876), IspF (PG27983), and IspG/gcpE (PG34415) were co-identified in root and cauline leaf in the MEP pathway. DXS (PG31496) and IspE (PG20660) were only identified in the root. IspG/gcpE (PG25816) and IspH (PG19130) were only identified in cauline leaf. FPS (PG34283), PPDS (PG37358), and PPTS (PG26599) in the downstream pathway were co-identified from root and cauline leaf.
The aglycone skeleton of ginsenoside is classified into a dammarane type and an oleanane type. The triterpene skeleton is converted into various ginsenosides by hydroxylation of CYP and glycosylation of uridine diphosphate-dependent glycosyltransferase (UGT). A total of 33 CYPs were identified in the ginseng cauline leaf, whereas only eight CYPs were identified from the root. A total of five CYPs were co-identified, including CYP704B1 (PG25870), CYP73A1 (PG28769), CYP76B6 (PG01467), CYP82D47 (PG29513), and CYP98A2 (PG41871). There are three CYPs unique to ginseng root, including CYP71D8 (PG04084), CYP72A219 (PG09274), and CYP82A3 (PG11378). There are 28 unique CYPs in the cauline leaf, including CYP704B1 (PG05721), CYP704C1 (PG10057), CYP71A8 (PG40294), CYP71D312 (PG16874), CYP71D313 (PG21495, PG26653, PG32259), CYP72A219 (PG01300, PG40963, PG03240, PG23478), CYP736A12 (PG13617, PG33495, PG36905), CYP76B6 (PG09699), CYP81E8 (PG30778), CYP82A3 (PG29120), CYP82C4 (PG32345), CYP82D47 (PG23797), CYP83B1 (PG02439, PG34334), CYP89A9 (PG34924), CYP94A1 (PG27129), CYP94A2 (PG23358, PG27467), CYP97B2 (PG16781), CYP97C1 (PG35257), and CYP98A2 (PG24387). A total of 31 UGTs were identified in the ginseng cauline leaf, whereas only nine UGTs were identified from the root. A total of six UGTs were co-identified, including UGT71 (PG30324, PG34795), UGT72 (PG21124), UGT74 (PG30551), UGT91 (PG34647), and UGT94 (PG22997). There are three UGTs unique to ginseng root, including UGT71 (PG05151), UGT88 (PG18416), and UGT91 (PG05454). There are 25 unique UGT proteins in the cauline leaf, including UGT70 (PG08475, PG36439), UGT71 (PG23742, PG34796), UGT72 (PG30450), UGT73 (PG16259, PG29012), UGT74 (PG14163, PG17964, PG31437, PG14160), UGT75 (PG34073), UGT76 (PG15945), UGT78 (PG00517), UGT79 (PG00083), UGT84 (PG03287), UGT85 (PG34278, PG19859), UGT89 (PG02224), UGT91 (PG03173), UGT92 (PG13029), UGT94 (PG35394, PG11440), UGT (PG07945), and UGT (PG23330). Fig. 6 showed the spatial distribution of proteins related to ginsenoside biosynthesis pathway according to the location of the identified proteins [27,[29], [30], [31], [32]]. We hypothesized that the synthesis of ginsenosides are carried out in the root and cauline leaf simultaneously, whereas the post glucoside adduction of ginsenosides might be more conducted when growing in the cauline leaf and then transported to the root at withering every year. Our hypothesis is also consistent to the findings of ginsenoside distribution profile on the tissue part of ginseng. We believe that the realization of the protein expression localization will greatly help us on the understanding of the biosynthesis pathway of the ginsenosides.
Fig. 6.
Spatial distribution of proteins identified related to ginsenoside biosynthesis pathway. The proteins are marked in red for co-identified, green for cauline leaf, and blue for ginseng root only.
4. Conclusion
In this report, we explored different methods to extract proteins from ginseng cauline leaf and root. State-of-art nano–LC-MS/MS was used to analyze the extracted proteins. The biological functions of the proteins expressed in different parts of ginseng were compared, and the enzymes involved in ginsenoside biosynthesis and modification were fished out for ginsenoside biosynthesis pathway analysis. Spatial distribution of CYPs, UGTs, and the proteins for MVA and MEP pathways was described to better understand the related processes of ginsenoside biosynthesis. Interestingly, a significant higher number of CYPs and UGTs were identified from ginseng cauline leaf than ginseng root, which indicated that this important glycosylation step in ginsenoside biosynthesis might be more conducted in cauline leaf. We believe that the in-depth proteomic analysis will help us on the knowledge of ginsenoside synthesis and improvement of ginseng cultivation.
Conflicts of interest
All authors have no conflicts of interest to declare.
Acknowledgments
Financial supports from National Science Foundation (21675061) and Jilin Provincial Science & Technology Department (20170101151JC) are gratefully acknowledged.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2020.01.009.
Appendix A. Supplementary data
A complete list of the protein and peptides identified in this article can be found on line. The raw data for mass spectrometry analysis can be downloaded via ftp://massive.ucsd.edu/MSV000084331/.
The following is the Supplementary data to this article:
References
- 1.Colzani M., Altomare A., Caliendo M., Aldini G., Righetti P.G., Fasoli E. The secrets of Oriental panacea: Panax ginseng. J Proteomics. 2016;130:150–159. doi: 10.1016/j.jprot.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 2.Kim S.W., Gupta R., Min C.W., Lee S.H., Cheon Y.E., Meng Q.F., Jang J.W., Hong C.E., Lee J.Y., Jo I.H. Label-free quantitative proteomic analysis of Panax ginseng leaves upon exposure to heat stress. J Ginseng Res. 2019;43:143–153. doi: 10.1016/j.jgr.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helms S. Cancer prevention and therapeutics: Panax ginseng. Altern Med Rev. 2004;9:259–274. [PubMed] [Google Scholar]
- 4.Kiefer D., Pantuso T. Panax ginseng. Am Fam Physician. 2003;68:1539–1542. [PubMed] [Google Scholar]
- 5.Kim J.H., Yi Y.S., Kim M.Y., Cho J.Y. Role of ginsenosides, the main active components of Panax ginseng, in inflammatory responses and diseases. J Ginseng Res. 2017;41:435–443. doi: 10.1016/j.jgr.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S.K., Park J.H. Trends in ginseng research in 2010. J Ginseng Res. 2011;35:389–398. doi: 10.5142/jgr.2011.35.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin B.K., Kwon S.W., Park J.H. Chemical diversity of ginseng saponins from Panax ginseng. J Ginseng Res. 2015;39:287–298. doi: 10.1016/j.jgr.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S.W., Lee S.H., Min C.W., Jo I.H., Bang K.H., Hyun D.-Y., Agrawal G.K., Rakwal R., Zargar S.M., Gupta R. Ginseng (Panax sp.)proteomics: an update. Applied Biol Chem. 2017;60:311–320. [Google Scholar]
- 9.Ma R., Sun L., Chen X., Jiang R., Sun H., Zhao D. Proteomic changes in different growth periods of ginseng roots. Plant Physiol Biochem. 2013;67:20–32. doi: 10.1016/j.plaphy.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 10.Nam M.H., Kim S.I., Liu J.R., Yang D.C., Lim Y.P., Kwon K.H., Yoo J.S., Park Y.M. Proteomic analysisof Korean ginseng (Panax ginseng C.A. Meyer) J Chromatogr B Analyt Technol Biomed Life Sci. 2005;815:147–155. doi: 10.1016/j.jchromb.2004.10.063. [DOI] [PubMed] [Google Scholar]
- 11.Jayakodi M., Choi B.S., Lee S.C., Kim N.H., Park J.Y., Jang W., Lakshmanan M., Mohan S.V.G., Lee D.Y., Yang T.J. Ginseng GenomeDatabase: an open-access platform for genomics of Panax ginseng. BMC PlantBiol. 2018;18:62. doi: 10.1186/s12870-018-1282-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim N.H., Jayakodi M., Lee S.C., Choi B.S., Jang W., Lee J., Kim H.H., Waminal N.E., Lakshmanan M., van Nguyen B. Genome and evolutionof the shade-requiring medicinal herb Panax ginseng. Plant Biotechnol J. 2018;16:1904–1917. doi: 10.1111/pbi.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J., Chu Y., Liao B., Xiao S., Yin Q., Bai R., Su H., Dong L., Li X., Qian j Panax ginseng genome examinationfor ginsenoside biosynthesis. Gigascience. 2017;6:1–15. doi: 10.1093/gigascience/gix093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu C.C., Zhu Y., Arrington J.V., Paez J.S., Wang P., Zhu P., Chen I.H., Zhu J.K., Tao W.A. Universal plantphosphoproteomics workflow and its application to tomato signaling inresponse to cold stress. Mol Cell Proteomics. 2018;17:2068–2080. doi: 10.1074/mcp.TIR118.000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang F., Shen Y., Camp D.G., 2nd, Smith R.D. High-pH reversed-phase chromatography with fraction concatenation for 2D proteomic analysis. Expert Rev Proteomics. 2012;9:129–134. doi: 10.1586/epr.12.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., Yang F., Gritsenko M.A., Wang Y., Clauss T., Liu T., Shen Y., Monroe M.E., Lopez-Ferrer D., Reno T. Reversed-phasechromatography with multiple fraction concatenation strategy for proteomeprofiling of human MCF10A cells. Proteomics. 2011;11:2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song C., Ye M., Han G., Jiang X., Wang F., Yu Z., Chen R., Zou H. Reversed-phase liquidchromatography approach with high orthogonality for multidimensionalseparation of phosphopeptides. Anal Chem. 2010;82:53–56. doi: 10.1021/ac9023044. [DOI] [PubMed] [Google Scholar]
- 18.Han D., Moon S., Kim Y., Kim J., Jin J., Kim Y. In-depth proteomic analysis of mouse microglia using a combination of FASP and StageTip-based, high pH, reversed-phase fractionation. Proteomics. 2013;13:2984–2988. doi: 10.1002/pmic.201300091. [DOI] [PubMed] [Google Scholar]
- 19.Kim S.I., Kweon S.M., Kim E.A., Kim J.Y., Kim S., Yoo J.S., Park Y.M. Characterization ofRNase-like major storage protein from the ginseng root by proteomicapproach. J Plant Physiol. 2004;161:837–945. doi: 10.1016/j.jplph.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Duck N.B., Folk W.R. Hsp70 heat shock protein cognate is expressed and stored in developing tomato pollen. Plant Mol Biol. 1994;26:1031–1039. doi: 10.1007/BF00040686. [DOI] [PubMed] [Google Scholar]
- 21.Gani Z., Boradia VM Raghu Ram J, Suryavanshi PM, Patil P, Kumar S, SinghR, RajeM, RajeCI.Purification and characterization of glyceraldehyde-3-phosphate-dehydrogenase(GAPDH) from pea seeds. Protein Expr Purif. 2016;127:22–27. doi: 10.1016/j.pep.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 22.Andersson I., Backlund A. Structure and function of rubisco. Plant Physiol Biochem. 2008;46:275–291. doi: 10.1016/j.plaphy.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Yuan H.M., Liu W.C., Lu Y.T. CATALASE2 coordinates SA-mediated repression of both auxin accumulation and JA biosynthesis in plant defenses. Cell Host Microbe. 2017;21:143–155. doi: 10.1016/j.chom.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Siminszky B., Corbin F.T., Ward E.R., Fleischmann T.J., Dewey R.E. Expression of a soybean cytochrome P450 monooxygenase cDNA in yeast and tobacco enhances the metabolism of phenylurea herbicides. Proc Natl Acad Sci U S A. 1999;96:1750–1755. doi: 10.1073/pnas.96.4.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Li X., Lin Y., Wang Y., Wang K., Sun C., Lu T., Zhang M. Structural variation, functional differentiation, and activity correlation of the cytochrome P450gene superfamily revealed in ginseng. Plant Genome. 2018:11. doi: 10.3835/plantgenome2017.11.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y., Walker S. Remarkable structural similarities between diverse glycosyltransferases. Chem Biol. 2002;9:1287–1296. doi: 10.1016/s1074-5521(02)00295-8. [DOI] [PubMed] [Google Scholar]
- 27.Kang K.B., Jayakodi M., Lee Y.S., Nguyen V.B., Park H.S., Koo H.J., Choi I.Y., Kim D.H., Chung Y.J., Ryu B. Identificationof candidate UDP-glycosyltransferases involved in protopanaxadiol-typeginsenoside biosynthesis in Panax ginseng. Sci Rep. 2018;8:11744. doi: 10.1038/s41598-018-30262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo S.L., Dang L.Z., Zhang K.Q., Liang L.M., Li G.H. Cloning and heterologous expression of UDP-glycosyltransferase genes from Bacillus subtilis and its application in the glycosylation of ginsenoside Rh1. Lett Appl Microbiol. 2015;60:72–78. doi: 10.1111/lam.12339. [DOI] [PubMed] [Google Scholar]
- 29.Lu J., Li J., Wang S., Yao L., Liang W., Wang J., Gao W. Advances in ginsenosidebiosynthesis and metabolic regulation. Biotechnol Appl Biochem. 2018;65:514–522. doi: 10.1002/bab.1649. [DOI] [PubMed] [Google Scholar]
- 30.Zhao S., Wang L., Liu L., Liang Y., Sun Y., Wu J. Both the mevalonate and the non-mevalonate pathways are involved in ginsenoside biosynthesis. Plant Cell Rep. 2014;33:393–400. doi: 10.1007/s00299-013-1538-7. [DOI] [PubMed] [Google Scholar]
- 31.Kim Y.J., Zhang D., Yang D.C. Biosynthesis and biotechnological production of ginsenosides. Biotechnol Adv. 2015;33:717–735. doi: 10.1016/j.biotechadv.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Kim Y.J., Jeon J.N., Jang M.G., Oh J.Y., Kwon W.S., Jung S.K., Yang D.C. Ginsenoside profilesand related gene expression during foliation in Panax ginseng Meyer. J Ginseng Res. 2014;38:66–72. doi: 10.1016/j.jgr.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.